Abstract
Background & Aims
The functional maturation of the liver largely occurs after birth. In the early stages of life, the liver of a newborn encounters enormous high-fat metabolic stress caused by the consumption of breast milk. It is unclear how the maturing liver adapts to high lipid metabolism. Liver sinusoidal endothelial cells (LSECs) play a fundamental role in establishing liver vasculature and are decorated with many glycoproteins on their surface. The Slc35a1 gene encodes a cytidine-5'-monophosphate (CMP)-sialic acid transporter responsible for transporting CMP-sialic acids between the cytoplasm and the Golgi apparatus for protein sialylation. This study aimed to determine whether endothelial sialylation plays a role in hepatic vasculogenesis and functional maturation.
Methods
Endothelial-specific Slc35a1 knockout mice were generated. Liver tissues were collected for histologic analysis, lipidomic profiling, RNA sequencing, confocal immunofluorescence, and immunoblot analyses.
Results
Endothelial Slc35a1-deficient mice exhibited excessive neonatal hepatic lipid deposition, severe liver damage, and high mortality. Endothelial deletion of Slc35a1 led to sinusoidal capillarization and disrupted hepatic zonation. Mechanistically, vascular endothelial growth factor receptor 2 (VEGFR2) in LSECs was desialylated and VEGFR2 signaling was enhanced in Slc35a1-deficient mice. Inhibition of VEGFR2 signaling by SU5416 alleviated lipid deposition and restored hepatic vasculature in Slc35a1-deficient mice.
Conclusions
Our findings suggest that sialylation of LSECs is critical for maintaining hepatic vascular development and lipid homeostasis. Targeting VEGFR2 signaling may be a new strategy to prevent liver disorders associated with abnormal vasculature and lipid deposition.
Keywords: Slc35a1, Liver Sinusoidal Endothelial Cell, Lipid Deposition, Liver Injury, VEGFR2
Graphical abstract
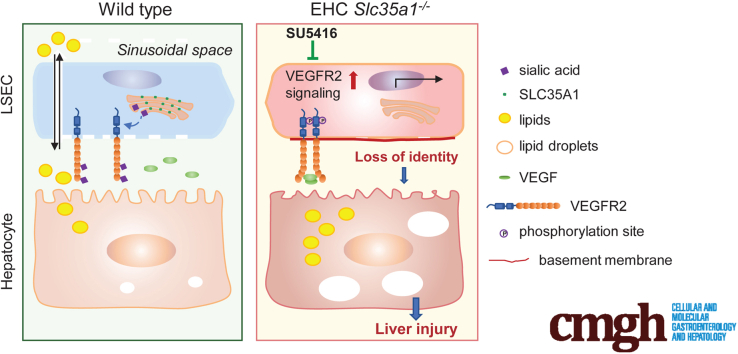
Summary.
Endothelial Slc35a1 deficiency aggravates neonatal hepatic lipid deposition, subsequently causing liver injury. Reduced sialylation of vascular endothelial growth factor receptor 2 enhances its signaling, which alters liver sinusoidal endothelial cell identity and hepatic zonation in endothelial Slc35a1-deficient mice. Vascular endothelial growth factor receptor 2 inhibition restores the hepatic vasculature and alleviates neonatal lipid deposition in Slc35a1-deficient mice.
The liver is a highly dynamic and pluripotent organ that functions differently from fetal to postnatal life. Before birth in mice, the fetal liver mainly serves as a hemopoietic organ during embryonic day (E)10.5–E16.5.1,2 The liver arises from the liver bud adjacent to septum transversum mesenchyme at approximately E8.5 in mice.3 Hepatoblasts in the ventral foregut endoderm start to express liver-specific proteins, such as albumin, α-fetoprotein, and hepatocyte nuclear factor 4a, and differentiate into hepatocytes at approximately E15.5.4 After that, the liver undergoes further functional cell proliferation to gain liver mass and gradually becomes a metabolic organ.5 Recently, single-cell RNA-sequencing (RNA-seq) has revealed that functional maturation of the liver largely occurs after birth.6,7
During liver development, the hepatic sinusoid is the first vascular system to form in the liver, which emerges at approximately E10 and is fully developed at the end of the embryonic period.2,3,8 This unique vasculature is crucial for establishing the structural and functional architecture of the liver and is essential for liver homeostasis under both physiological and pathologic conditions.9, 10, 11 Liver sinusoidal endothelial cells (LSECs), lining the sinusoids, are the most abundant nonparenchymal cells in the liver (∼15% of liver cells).12 LSECs play an instructive and supportive role for hepatogenesis.13, 14, 15 During the earliest stage of liver organogenesis, endothelial cells promote hepatoblast migration and outline the liver bud before differentiating into functional blood vessels.13,14 Differentiated LSECs are highly fenestrated and lack a basement membrane, and they act as gatekeepers, controlling bidirectional exchange between blood and hepatocytes.12,16 LSECs can be divided into periportal LSECs that have fewer and larger fenestrae, and pericentral LSECs that have more and smaller fenestrae.17 The structural differences of LSECs and changes in blood supply around birth confer functional heterogeneity to hepatic parenchymal cells.18 Genomic and metabolic profiling data have revealed that hepatocytes are spatially organized in a compartmentalized manner, termed liver zonation, and can be divided into periportal, intermediate, and pericentral metabolic zones.19,20 In recent years, LSEC-derived angiocrine signals have been shown to control liver maturation and regeneration of the adult liver after liver injury.21, 22, 23, 24 Several hepatotropic proteins released by LSECs, such as hepatocyte growth factor, R-spondin 3, bone morphogenetic protein 2, Wnt2, and Wnt9b, have been found to regulate liver zonation.25, 26, 27 Proper liver zonation is related closely to the establishment and maintenance of liver metabolic function, especially lipid metabolism.28,29 The strong link between hepatic zone disruption and dysregulation of lipid homeostasis has been demonstrated in several diseases such as nonalcoholic fatty liver disease (NAFLD), liver fibrosis, obesity, and liver cancer.30, 31, 32, 33 However, whether and how LSECs contribute to the establishment of lipid homeostasis remains largely unknown.
Lipid metabolism is one of the prominent metabolic functions of the liver. The underlying mechanisms breaking hepatic lipid homeostasis include decreased mitochondrial fatty acid β-oxidation, increased de novo lipogenesis, and enhanced uptake of fatty acids to the liver.34,35 The excessive accumulation of lipid metabolites causes lipotoxicity and eventually induces liver injury.36 Although the pathogenesis of fatty liver has received much attention over the past few decades, the primary study results are from adult animal models fed with a high-fat diet.37,38 There still are large gaps in understanding the regulation of the transition from fetal liver to mature liver after birth. During the neonatal period, the liver of a newborn encounters enormous challenges, including the cessation of the umbilical vein supply and high-fat metabolic stress caused by consumption of breast milk, which requires the liver to metabolize these lipids efficiently.39,40 It is unclear how the maturing liver at birth adapts to high lipid metabolism and whether the hepatic vasculature is involved in this process.
Protein glycosylation is a post-translational modification in which glycans are attached to proteins.41,42 Sialylation is one of the most important glycosylation processes, in which sialic acids are connected to terminal positions of the N- and O-linked glycans. The cytidine-5'-monophosphate (CMP)-sialic acid transporter, encoded by the Slc35a1 gene, transports CMP-sialic acids between cytoplasm and the Golgi apparatus for protein sialylation.43 Accumulating studies have demonstrated that sialylation of endothelial cells contributes to the modulation of endothelial functions including cell proliferation, survival, intercellular crosstalk, and angiogenesis.44, 45, 46, 47, 48 A large number of N-glycoproteins on the LSEC surface have been identified.49 However, the roles of sialylation in regulating LSECs and liver function have not been elucidated.
In this study, we aimed to explore the roles of endothelial sialylation in hepatic vasculature development and liver function. We generated endothelial/hematopoietic cell (EHC)-specific mice lacking Slc35a1 (EHC Slc35a1-/-). We found that EHC Slc35a1-/- mice showed exacerbated neonatal hepatic lipid deposition and developed severe liver injury. We further demonstrated that endothelial Slc35a1 deficiency led to loss of LSEC’s identity and disrupted hepatic zonation. Mechanistically, vascular endothelial growth factor receptor 2 (VEGFR2) in LSECs was desialylated, which enhanced VEGFR2 signaling in Slc35a1-deficient livers. Inhibition of VEGFR2 signaling with a VEGFR2 inhibitor SU5416 alleviated neonatal hepatic lipid deposition and restored liver vasculature in EHC Slc35a1-/- mice.
Results
EHC Slc35a1-/- Mice Die Postnatally and Show Progressive Liver Injury
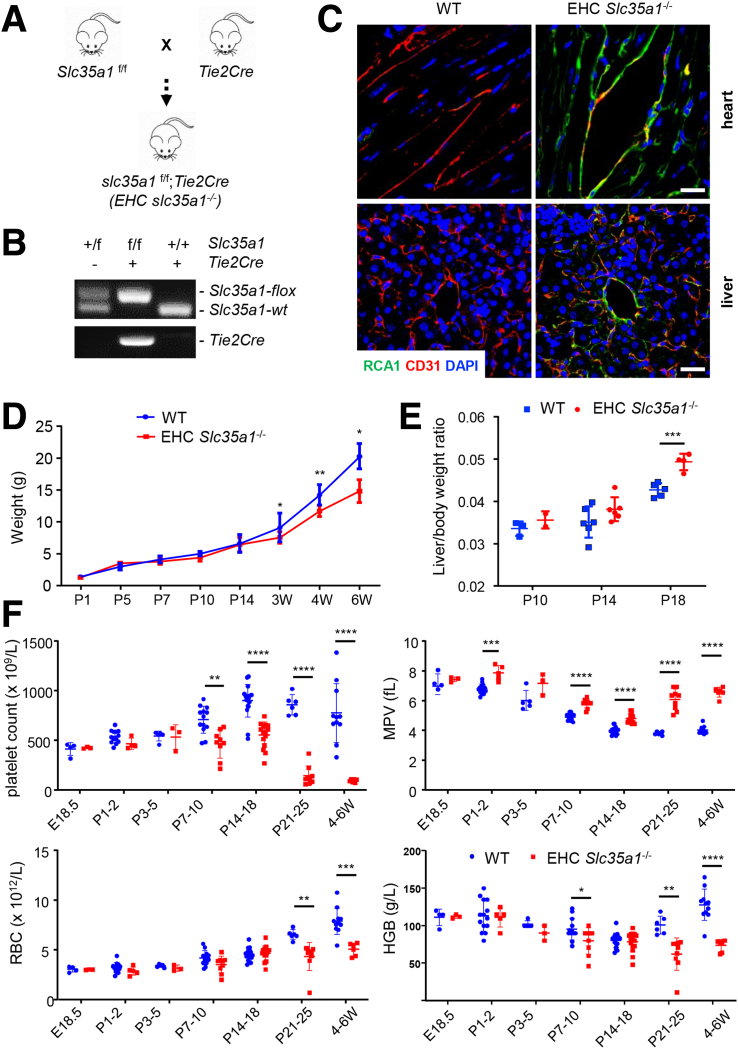
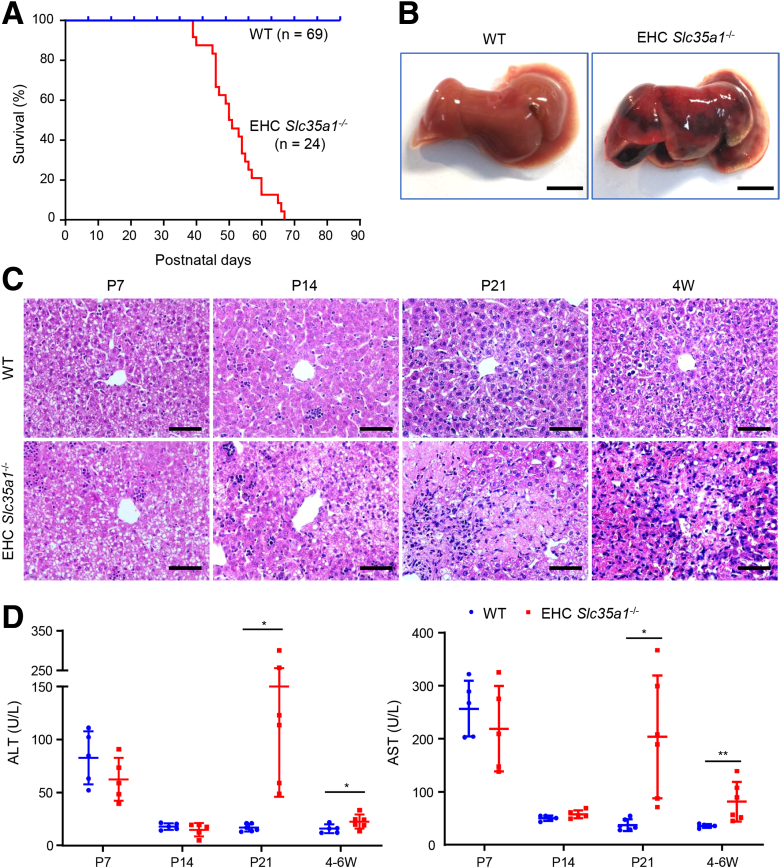
To investigate the role of endothelial sialylation in liver homeostasis, we generated EHC Slc35a1-/- mice (Figure 1A and B). Immunostaining with Ricinus communis agglutinin Ⅰ lectin, which specifically binds to terminal galactose exposed after loss of the capping sialic acids, revealed the loss of sialylation in both vascular and sinusoidal endothelial cells in the heart and liver tissues of EHC Slc35a1-/- mice, respectively (Figure 1C). EHC Slc35a1-/- mice appeared grossly normal before reaching 2 weeks of age and started to lose body weight from 3 weeks of age (Figure 1D). EHC Slc35a1-/- mice had high liver/body weight ratios after postnatal day (P)14, indicating hepatomegaly (Figure 1E). They also exhibited thrombocytopenia from P7 and anemia from P21 (Figure 1F). EHC Slc35a1-/- mice died as early as approximately 5 weeks of age, and all died before 10 weeks of age (Figure 2A). Severe liver damage was observed in EHC Slc35a1-/- mice before death (Figure 2B), which might be a direct cause of their death. Histologic analysis of liver sections revealed that the EHC Slc35a1-/- liver exhibited much greater steatosis at P7 and developed necrosis at P21 (Figure 2C) compared with wild-type (WT) mice. Furthermore, more blood cells and less-pronounced sinusoid spaces were found in P7 EHC Slc35a1-/- livers than in WT livers (Figure 2C), suggesting a disorganized vascular network in EHC Slc35a1-/- mice. EHC Slc35a1-/- mice exhibited severe steatosis at P14 and progressed to extensive necrosis at approximately P21 (Figure 2C). However, other organs of EHC Slc35a1-/- mice (heart, lung, and kidney) did not show significant histologic abnormalities. Biochemical analysis of the EHC Slc35a1-/- plasma revealed significantly increased levels of alanine aminotransferase and aspartate aminotransferase at P21 and 4 weeks of age (Figure 2D), which is consistent with the extensive hepatic necrosis that we observed at P21 (Figure 2C). These results indicate that endothelial Slc35a1 deficiency leads to a progressive hepatic steatosis and liver injury in mice.
Figure 1.
Deletion of Slc35a1 in endothelial/hematopoietic cells in mice. (A) Mice with specific deletion of Slc35a1 in murine EHC (Slc35a1-/-) were generated by breeding Slc35a1f/f mice with Tie2Cre transgenic mice. (B) Genotyping PCR confirms the successful generation of EHC Slc35a1-/- mice. The primers are listed in Table 1. (C) Representative immunofluorescence images showing loss of sialylation on vascular endothelial cells in heart and sinusoidal endothelial cells in liver of EHC Slc35a1-/- mice compared with WT mice. Frozen heart and liver sections from WT mice and EHC Slc35a1-/- mice at 4 weeks were analyzed using immunostaining of Ricinus communis agglutinin Ⅰ (RCAⅠ) (specific for nonreducing terminal β-galactose) and CD31. Scale bars: 20 μm. (D) Body weights of WT and EHC Slc35a1-/- mice over time with 4–11 mice per time point. (E) Liver-to–body weight ratios in EHC Slc35a1-/- mice were higher than those in WT mice at P18. Four to 6 mice per group. (F) Peripheral blood cell counts indicate that EHC Slc35a1-/- mice had thrombocytopenia and anemia. Each group had 3–11 mice. Data represent means ± SD, and ∗P < .05, ∗∗P < .01, ∗∗∗P < .001, and ∗∗∗∗P < .0001 indicate significant differences between WT and EHC Slc35a1-/- mice in D-F. DAPI, 4′,6-diamidino-2-phenylindole; HGB, hemoglobin; MPV, mean platelet volume; RBC, red blood cell; W, postnatal week.
Figure 2.
EHC Slc35a1-/-mice exhibit high mortality and severe liver injury. (A) Postnatal survival curves of WT (n = 69) and EHC Slc35a1-/- mice (n = 24). (B) Representative gross images showing severe liver injury in an EHC Slc35a1-/- mouse compared with its WT littermate at 6 weeks of age. Scale bar: 0.5 mm. (C) Representative microscopic images of H&E-stained mouse livers at indicated ages. Scale bars: 20 μm. (D) Plasma alanine aminotransferase (ALT) and aspartate aminotransferase (AST) levels in WT (n = 5) and EHC Slc35a1-/- mice (n = 6) at indicated ages. Data represent means ± SD. Mann–Whitney U test was performed between WT and EHC Slc35a1-/- mice. ∗P < .05 and ∗∗P < .01 indicate significant differences. W, postnatal week.
Deficient Endothelial Sialylation Exacerbates Neonatal Hepatic Lipid Deposition and Alters Hepatic Lipid Profile
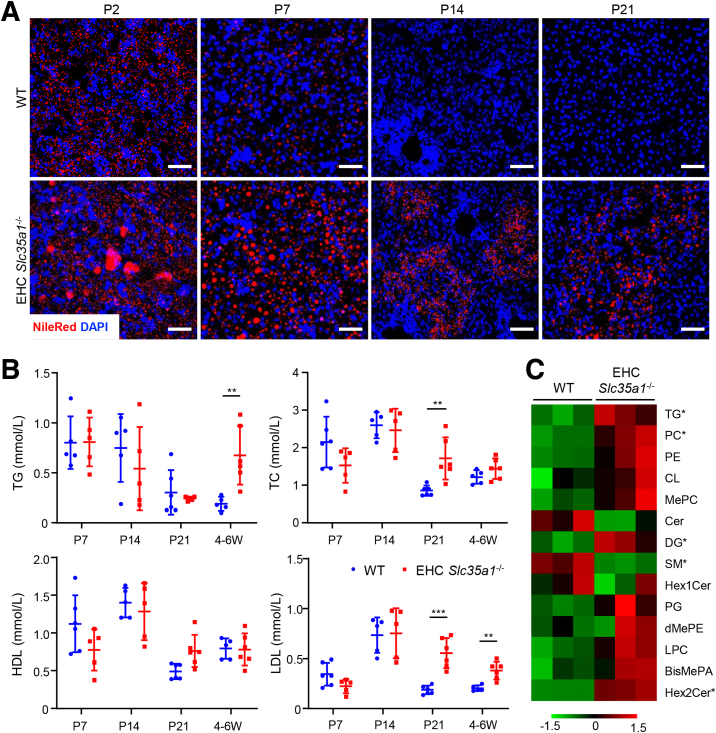
At P14, all EHC Slc35a1-/- mice showed hepatic steatosis. Thus, we further investigated the abnormal lipid deposition phenotype in the EHC Slc35a1-/- liver. Nile Red staining showed that hepatic lipid deposition before birth was faint and similar in both WT and EHC Slc35a1-/- mice (data not shown). Lipid droplets in EHC Slc35a1-/- livers were much more abundant and larger in size than those in WT livers at P2 and P7 (Figure 3A). Consistent with the histologic findings in Figure 2C, lipid deposition was completely resolved in WT livers, but persisted in EHC Slc35a1-/- livers from P14 (Figure 3A). Biochemical analysis further revealed that EHC Slc35a1-/- mice had higher levels of plasma total cholesterol and low-density lipoproteins than WT mice at P21. At 4 weeks of age, EHC Slc35a1-/- mice showed significantly increased levels of plasma total triglycerides (TGs) and low-density lipoproteins compared with WT mice (Figure 3B), suggesting a pronounced dyslipidemia. Lipidomic analysis of P14 livers demonstrated accumulation of neutral lipids in EHC Slc35a1-/- mice (Figure 3C). Among the 2445 lipid metabolites identified, 54 were found to be dysregulated significantly (38 up-regulated and 16 down-regulated) between WT and EHC Slc35a1-/- livers (Table 2). The fold-change of these metabolites is illustrated as a heatmap in Figure 3C. The 38 up-regulated lipids included 15 TGs, 9 dihexosylceramides, 6 phosphatidylethanolamines, 4 mono-hexosylceramides, 2 bis-methyl phosphatidic acids, 1 methyl phosphatidylcholine, and 1 phosphatidylcholine, and the 16 down-regulated lipids included 2 TGs, 1 phosphatidylethanolamine, 3 phosphatidylcholines, and 10 sphingomyelins (Table 2 and Figure 3C). These results indicate that endothelial deficiency of sialylation exacerbated neonatal hepatic lipid deposition characterized with a fatty liver–like lipid profile.
Figure 3.
Endothelial Slc35a1 deficiency exacerbates neonatal hepatic lipid droplet deposition and alters hepatic lipid metabolism. (A) Nile Red staining showed neonatal hepatic lipid deposition in WT and EHC Slc35a1-/- livers. Frozen liver sections from WT mice and EHC Slc35a1-/- mice at P2, P7, P14, and P21 were analyzed. Scale bars: 50 μm. (B) Biochemical analysis of plasma total triglyceride (TG), total cholesterol (TC), high-density lipoprotein (HDL), and low-density lipoprotein (LDL) levels in WT (n = 5 ∼ 6) and EHC Slc35a1-/- mice (n = 5 ∼ 6) at P7, P14, P21, and 4–6 weeks of age. Data represent means ± SD. ∗∗P < .01 and ∗∗∗P < .001 indicates significant differences between WT and EHC Slc35a1-/- mice by Mann–Whitney U test. (C) Lipidomic profile of livers from WT and EHC Slc35a1-/- mice at P14. ∗Significant difference of individual lipid species between WT and EHC Slc35a1-/- mice (P < .05). BisMePA, Bis-methyl phosphatidic acids; Cer, ceramide; CL, cardiolipin; DG, diglyceride; dMePE, dimethylphosphatidylethanolamine; Hex1Cer, mono-hexosylceramide; Hex2Cer, dihexosylceramide; LPC, lysophosphatidylcholine; MePC, methyl phosphatidylcholine; PC, phosphatidylcholine; PE, phosphatidylethanolamine; PG, phosphatidylglycerol; SM, sphingomyelins; W, postnatal week.
Table 2.
Significantly Dysregulated Lipid Metabolites Identified in EHC Slc35a1-/- Livers
| Lipid metabolite | VIP | Fold change, EHC Slc35a1-/- vs WT | P value |
|---|---|---|---|
| TG(12:0_14:1_18:2) | 4.7957 | 5.8037 | .0479 |
| TG(12:0_14:0_18:2) | 1.6409 | 3.0198 | .0124 |
| TG(16:0_12:0_16:1) | 3.9672 | 2.8233 | .0417 |
| TG(15:0_12:0_18:2) | 1.0161 | 2.2506 | .0318 |
| TG(12:0_18:2_18:2) | 1.4432 | 2.2056 | .0090 |
| TG(12:0_14:0_20:5) | 2.4420 | 2.0757 | .0491 |
| TG(12:0_18:2_20:4) | 1.4433 | 2.0315 | .0251 |
| TG(16:0_13:0_20:5) | 1.0011 | 1.9264 | .0338 |
| TG(18:0_10:0_18:2) | 2.9179 | 1.7843 | .0288 |
| TG(15:0_18:2_18:2) | 2.3143 | 1.7080 | .0182 |
| TG(18:1_22:4_22:5) | 3.3209 | 1.6466 | .0063 |
| TG(19:0_18:1_20:3) | 1.1493 | 1.6106 | .0160 |
| TG(15:0_18:2_18:3) | 1.1160 | 1.5922 | .0290 |
| TG(16:0_16:0_18:2) | 10.3136 | 1.3847 | .0440 |
| TG(18:1_18:2_20:2) | 6.6144 | 1.1699 | .0011 |
| TG(12:0e_10:4_20:5) | 1.0905 | 0.8203 | .0330 |
| TG(26:1_11:4_18:2) | 1.1657 | 0.4022 | .0332 |
| SM(d43:2) | 2.6716 | 0.7532 | .0467 |
| SM(d18:0_24:1) | 1.6608 | 0.7521 | .0324 |
| SM(d37:1) | 1.9055 | 0.7423 | .0003 |
| SM(d42:6) | 1.0775 | 0.6727 | .0204 |
| SM(d18:1_24:0) | 1.5733 | 0.6172 | .0015 |
| SM(d20:1_24:3) | 1.6078 | 0.5960 | .0008 |
| SM(t40:0) | 1.0871 | 0.5390 | .0038 |
| SM(d38:0) | 2.8404 | 0.4520 | .0151 |
| SM(d42:0) | 1.0482 | 0.3986 | .0089 |
| SM(d43:0) | 1.2753 | 0.2583 | .0132 |
| PE(16:0p_20:1) | 1.5378 | 1.4929 | .0486 |
| PE(18:1_22:1) | 1.3869 | 1.4507 | .0012 |
| PE(18:0p_18:2) | 1.4497 | 1.3651 | .0281 |
| PE(18:0e_20:4) | 1.3651 | 1.2447 | .0066 |
| PE(18:1p_22:4) | 1.6034 | 1.1963 | .0306 |
| PE(18:1e_20:4) | 1.2700 | 1.1596 | .0309 |
| PE(16:0_22:4) | 8.2155 | 0.7615 | .0205 |
| PC(32:4) | 1.5162 | 1.3590 | .0087 |
| PC(36:6) | 8.9242 | 0.8537 | .0110 |
| PC(15:0_22:6) | 6.5292 | 0.8459 | .0370 |
| PC(34:5) | 1.5918 | 0.7256 | .0199 |
| MePC(39:2) | 1.3809 | 1.4569 | .0303 |
| Hex2Cer(d18:1_22:1) | 1.0905 | 13451000.0000 | .0000 |
| Hex2Cer(d18:1_26:0) | 1.3906 | 30.8680 | .0035 |
| Hex2Cer(d18:0_16:0) | 1.3842 | 18.5060 | .0062 |
| Hex2Cer(d18:1_22:0) | 1.9497 | 6.6523 | .0007 |
| Hex2Cer(d18:1_24:0) | 3.4449 | 5.1272 | .0015 |
| Hex2Cer(d18:1_23:0) | 1.3381 | 3.7613 | .0014 |
| Hex2Cer(d18:0_22:0) | 1.0409 | 3.3269 | .0403 |
| Hex2Cer(d18:1_24:1) | 2.4272 | 3.2069 | .0001 |
| Hex2Cer(d18:1_16:0) | 1.0325 | 2.4814 | .0016 |
| Hex1Cer(t36:0) | 1.0696 | 1.3442 | .0475 |
| Hex1Cer(d18:1_22:0) | 3.4659 | 1.2570 | .0010 |
| Hex1Cer(d18:1_24:1) | 3.2160 | 1.1175 | .0416 |
| Hex1Cer(d18:1_24:2) | 1.3122 | 1.1070 | .0230 |
| BisMePA(18:3e_18:1) | 1.4532 | 1.3672 | .0279 |
| BisMePA(40:7e) | 1.6034 | 1.1963 | .0306 |
NOTE. Lipidomic analyses were performed on P14 livers of EHC Slc35a1-/- and WT mice by liquid chromatography–mass spectrometry with an ACQUITY UPLC BEH C18 column and a Thermo Q Exactive Focus mass spectrometer. Data-dependent acquisition tandem mass spectrometry experiments were performed with a high-energy, collision-induced, dissociation scan. Lipid metabolites with VIP > 1 and P < .05 were considered to be significantly different between EHC Slc35a1-/- and WT mice.
BisMePA, bis-methyl phosphatidic acids; Hex1Cer, mono-hexosylceramide; Hex2Cer, dihexosylceramide; MePC, methyl phosphatidylcholine; PC, phosphatidylcholine; PE, phosphatidylethanolamine; SM, sphingomyelins; TG, triglyceride; VIP, variable importance in the projection score.
Table 3.
Antibodies Used for Western Blot and Immunofluorescence
| Antibodies | Catalog no. | Vendor | Usage | RRID |
|---|---|---|---|---|
| Armenian hamster anti-CD31 | ab119341 | Abcam | 1:200 IF | AB_10900179 |
| Anti-Lyve-1 antibody | ab218535 | Abcam | 1:200 IF | AB_2927473 |
| Goat anti-mouse Lyve-1 | AF2125 | R&D | 1:50 IF | AB_2297188 |
| Antilaminin antibody | ab11575 | Abcam | 1:200 IF | AB_298179 |
| Anti-VEGF receptor 2 | 2479 | Cell Signaling Technology | 1:1000 for WB; 1:100 for IP | AB_2212507 |
| Anti-VEGF receptor 2 (phospho-Y1054 + Y1059) | ab5473 | Abcam | 1:250 IF | AB_304917 |
| Anti–phospho-VEGF receptor 2 (Tyr1175) | 2478 | Cell Signaling Technology | 1:1000 WB | AB_331377 |
| Anti–β-tubulin antibody | sab4200715 | Sigma | 1:2000 WB | AB_2827403 |
| Anti–cytochrome P450 2E1 antibody | ab28146 | Abcam | 1:500 IF | AB_2089985 |
| Anti-CD36 antibody | ab252923 | Abcam | 1:1000 WB | NA |
| Cy3 AffiniPure goat anti-Armenian hamster IgG (H+L) | 127-165-099 | Jackson Lab | 1:400 IF | AB_2338988 |
| Goat anti-rat IgG (H+L) Alexa Fluor 647) | A-21247 | Invitrogen | 1:500 IF | AB_141778 |
| Donkey anti-goat IgG H&L (Alexa Fluor 488) | ab150133 | Abcam | 1:200 IF | AB_2832252 |
| Donkey anti-rat IgG (H+L) (Alexa Fluor 647) | A-21244 | Invitrogen | 1:200 IF | AB_2535812 |
| Goat anti-rabbit IgG H&L (Alexa Fluor 488) | ab150077 | Abcam | 1:200 IF | AB_2630356 |
| RCA I-fluorescein | FL1081 | Vector Lab | 1:500 IF | AB_2336708 |
| Biotinylated maackia amurensis lectin II | B-1265 | Vector Lab | 1:200 IF | AB_2336569 |
| Biotinylated sambucus nigra lectin | B-1305-2 | Vector Lab | 1:200 IF | AB_2336718 |
| Horseradish peroxidase–conjugated streptavidin | 405210 | BioLegend | 1:2000 IF | NA |
| Anti–glutamine synthetase antibody | ab176562 | Abcam | 1:200 IF | AB_2868472 |
| Anti-mouse E-cadherin (clone DEMCA-1) (Alexa Fluor 594) | 147306 | BioLegend | 1:200 IF | AB_2563230 |
| Anti-CD34 antibody | ab81289 | Abcam | 1:50 IF | AB_1640331 |
DEMCA-1, clone number; H&L, heavy and light chains of immunoglobulin G; IF, immunofluorescence; IP, immunoprecipitation. NA, not available; RCAⅠ, Ricinus communis agglutinin Ⅰ; RRID, Research Resource Identifier; WB, Western blot.
Endothelial Slc35a1 Deficiency Causes Loss of LSEC Identity
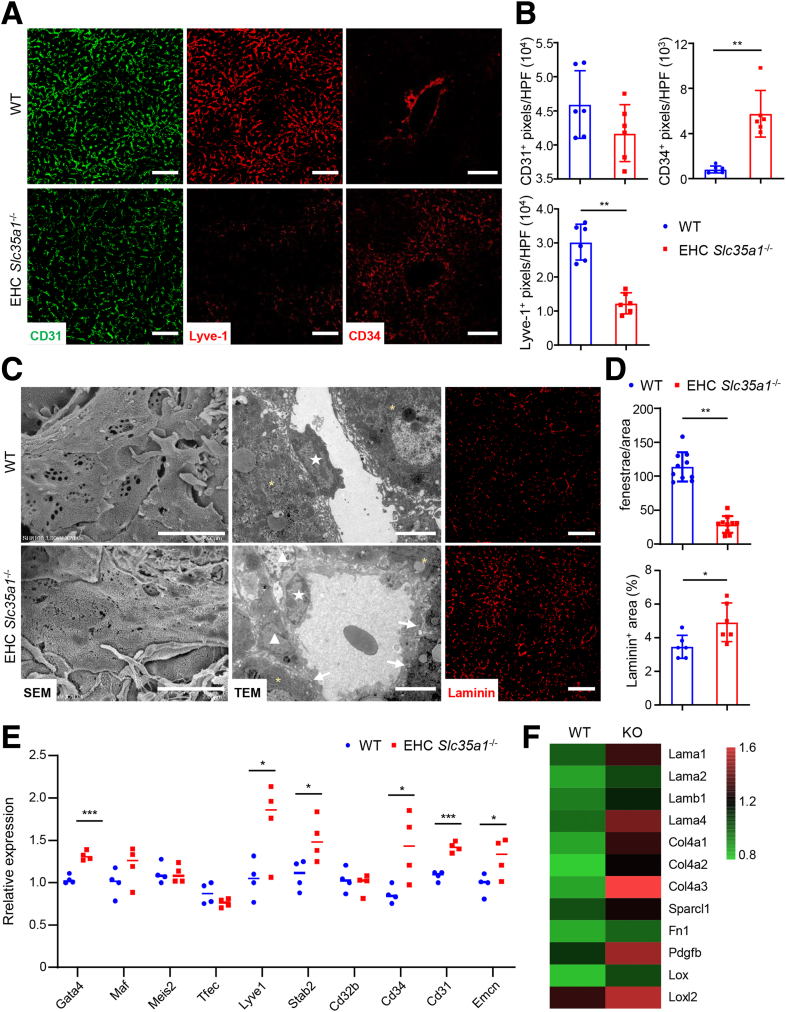
Because the hepatic vasculature is critical for maintaining liver lipid homeostasis,16,50, 51, 52, 53 we further investigated changes of hepatic vasculature in EHC Slc35a1-/- mice. Under normal conditions, differentiated LSECs strongly express their specific marker lymphatic vessel endothelial hyaluronan receptor-1 (Lyve-1) and do not express the continuous endothelial cell marker CD34.16,54 Loss of LSEC identity will lead to sinusoidal capillarization, a key feature of LSEC dysfunction in which LSECs lose their fenestrae and gain a basement membrane.55,56 In WT livers, Lyve-1 was strongly expressed in the sinusoids, whereas CD34 was observed mainly in the central vein and portal vein areas. In contrast, in EHC Slc35a1-/- livers, Lyve-1 expression was reduced markedly, whereas CD34 expression was enhanced dramatically in the sinusoids, indicating an increase in sinusoidal capillarization (Figure 4A and B). Additionally, CD31 immunostaining also revealed disordered sinusoidal vasculature (Figure 4A), and scanning electron micrographs demonstrated significant loss of fenestrae in EHC Slc35a1-/- livers (Figure 4C). Transmission electron micrographs and immunostaining showed a deposition of basement membrane laminin in EHC Slc35a1-/- mice (Figure 4C and D). Surprisingly, quantitative polymerase chain reaction (PCR) analysis revealed increased expression of transcriptional factor Gata4, as well as several common LSEC-specific genes (Lyve1, Stab2), and continuous endothelial cell genes (Cd34, Cd31, and Emcn) in EHC Slc35a1-/- livers (Figure 4E). Bulk RNA sequencing demonstrated that multiple laminin genes (Lama1, Lama2, Lamb1, and Lama4), collagen genes (Col4a1, Col4a2, and Col4a3), as well as profibrotic genes (Pdgfb, Lox, and Loxl2) were up-regulated in EHC Slc35a1-/- livers (Figure 4F), suggesting increased fibrosis and remodeling of the liver extracellular matrix. Overall, these results indicate that endothelial Slc35a1 deficiency causes loss of LSEC identity in EHC Slc35a1-/- mice, which is characterized by a phenotypic change toward continuous endothelial cells, such as disordered sinusoidal vasculature and loss of fenestrae, and deposition of basement membrane.
Figure 4.
Endothelial Slc35a1 deficiency causes loss of LSEC identity. (A) Immunostaining of CD31, Lyve-1, and CD34 on P7 liver cryosections. WT mice, n = 5; EHC Slc35a1-/- mice, n = 5. (B) The levels of CD31, Lyve-1, and CD34 in LSECs in panel A were compared quantitatively. Three images per mouse were chosen randomly for analysis. ∗∗P < .01 indicates significant differences between WT and EHC Slc35a1-/- mice by Mann–Whitney U test. (C) Representative scanning electron micrographs (SEM, left), transmission electron micrographs (TEM, middle), and laminin immunostaining images (right) of livers from WT and EHC Slc35a1-/- mice at P7. Asterisks in TEM images indicate hepatocytes. White arrows indicate capillary basement membrane. Pentagram indicates liver sinusoidal endothelial cells. Triangles indicate hepatic stellate cells. Scale bars in SEM and TEM images: 2 μm and 5 μm. Scale bars in immunostaining images: 50 μm. (D) The numbers of LSEC fenestrae and the level of laminin in panel C were compared quantitatively. Three images per mouse were chosen randomly for analysis. ∗P < .05 and ∗∗P < .01 indicate significant differences between WT and EHC Slc35a1-/- mice by Mann–Whitney U test. (E) Relative mRNA levels of LSEC-specific transcription factors and endothelial cell markers in livers from WT (n = 4) and EHC Slc35a1-/- mice (n = 4) at P7 were detected by real-time quantitative PCR. Glyceraldehyde-3-phosphate dehydrogenase served as an internal control. Bars indicate median values. ∗P < .05, ∗∗P < .01, and ∗∗∗P < .001 indicate significant differences between 2 groups by Mann–Whitney U test. (F) Bulk RNA sequencing showing the levels of multiple laminin genes, collagen genes, and profibrotic genes. HPF, high-power field; KO, knockout.
Endothelial Slc35a1 Deficiency Causes Disturbance of Hepatic Zonation
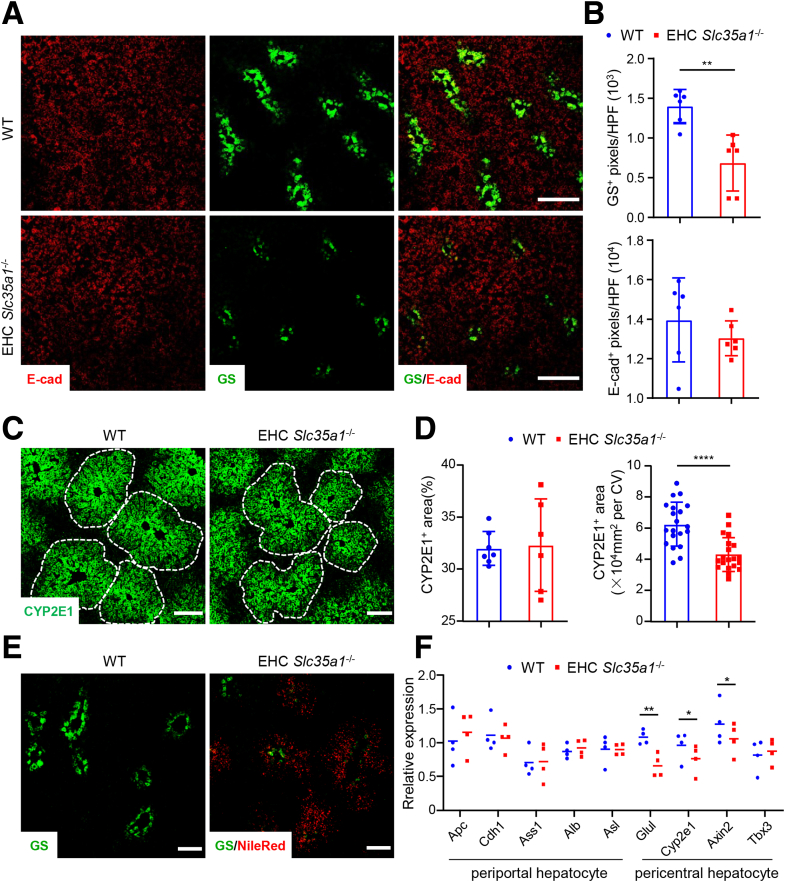
LSECs play a critical role in establishing and maintaining a proper spatial and metabolic architecture of the liver, known as hepatic zonation.20, 21, 22,57 To further investigate the role of endothelial Slc35a1 in hepatic zonation, we examined the expression of various cell markers specific to different hepatic zones in EHC Slc35a1-/- mice compared with WT mice. Immunostaining analysis revealed that E-cadherin, a marker for zone 1 periportal hepatocytes,23 was expressed in hepatocytes of P7 livers and did not exhibit significant differences between EHC Slc35a1-/- and WT mice (Figure 5A and B). However, the expression of glutaminase synthetase, a marker of zone 3 pericentral hepatocytes,23 was decreased markedly in EHC Slc35a1-/- livers (Figure 5A and B). Moreover, the expression of cytochrome P450 family 2 subfamily E member 1–positive hepatocytes (a marker for zone 2 hepatocytes23) in EHC Slc35a1-/- livers was lower than in WT livers (Figure 5C and D), indicating a reduction in the pericentral area. Further analysis using Nile Red staining revealed that lipid deposition in the livers of P10 EHC Slc35a1-/- mice was localized predominantly around glutaminase synthetase–positive hepatocytes (Figure 5E), suggesting a zonal pattern of lipid deposition. Quantitative PCR analysis of P7 liver tissues also revealed that the relative expression of several pericentral hepatocyte-specific genes, such as Glul, Cyp2e1, and Axin2, was significantly lower than that of WT livers (Figure 5F). Taken together, these results indicate that endothelial Slc35a1 deficiency disrupts hepatic zonation, leading to a decrease of pericentral hepatocytes.
Figure 5.
Endothelial Slc35a1 deficiency disrupts hepatic zonation. (A) Immunostaining of E-cadherin (E-cad, zone 1 periportal hepatocytes) and glutaminase synthetase (GS, zone 3 pericentral hepatocytes) on frozen liver sections from WT mice and EHC Slc35a1-/- mice at P7. Scale bars: 200 μm. (B) The levels of E-cad and GS in panel A were compared quantitatively. Three images per mouse were chosen randomly for analysis. ∗∗P < .01 indicates significant differences between WT and EHC Slc35a1-/- mice by Mann–Whitney U test. (C) Immunostaining of CYP2E1 (zone 2 pericentral hepatocytes) on frozen liver sections from WT mice and EHC Slc35a1-/- mice at P7. Scale bars: 100 μm. (D) CYP2E1+ area and CYP2E1+ area per central vein (CV) in panel C were compared quantitatively. Three images per mouse were chosen randomly for analysis. ∗∗∗∗P < .0001 indicates significant differences between WT and EHC Slc35a1-/- mice by Mann–Whitney U test. (E) Nile Red staining showed lipid deposition around GS-positive hepatocytes in EHC Slc35a1-/- livers compared with WT livers. Frozen liver sections from WT mice and EHC Slc35a1-/- mice at P10 were analyzed. Scale bars: 100 μm. (F) Relative mRNA levels of hepatocyte zonation–related genes in livers from WT (n = 4) and EHC Slc35a1-/- mice (n = 4) at P7 were detected by real-time quantitative PCR. Glyceraldehyde-3-phosphate dehydrogenase served as an internal control. Bars indicate median values. ∗P < .05 and ∗∗P < .01 indicate significant differences between 2 groups by Mann–Whitney U test. HPF, high-power field.
VEGFR2 Signaling Is Enhanced in EHC Slc35a1-/- Mice
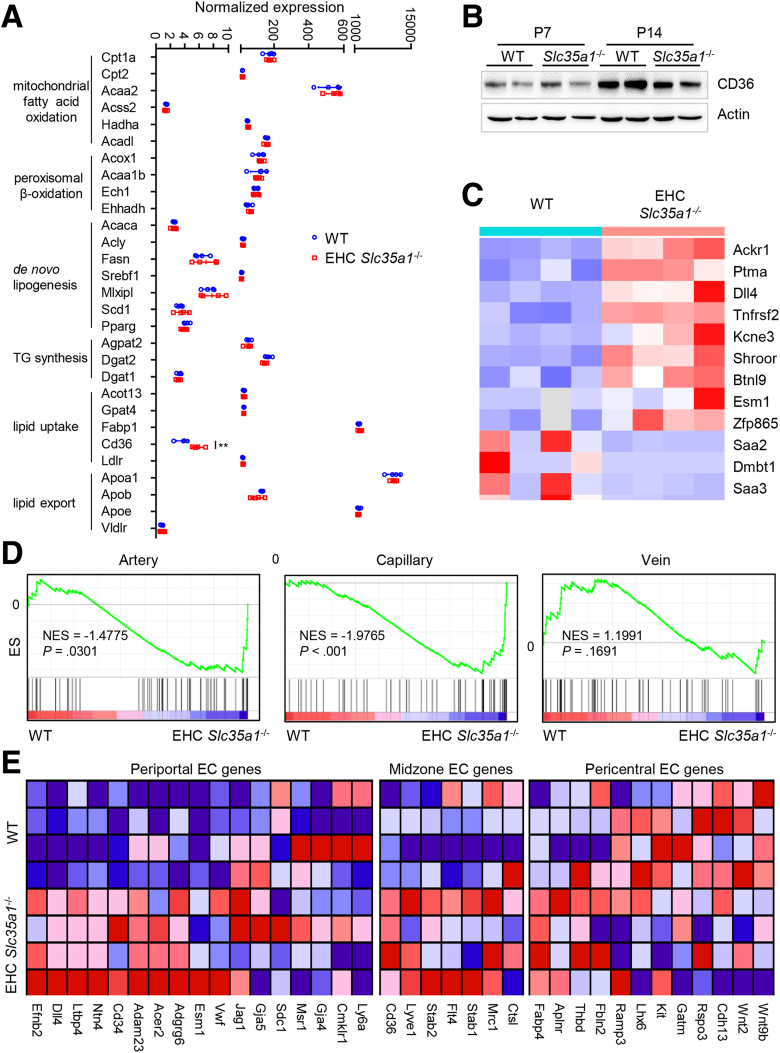
Because defective sinusoidal vasculature was observed in EHC Slc35a1-/- mice (Figure 4A), we next performed RNA sequencing to investigate the changes of gene profiles in the EHC Slc35a1-/- mice. Interestingly, the expression of most genes involved in lipid metabolism was unchanged, except CD36, which was higher in the mutant mice (Figure 6A). However, the protein level of liver CD36 was similar in EHC Slc35a1-/- and WT mice (Figure 6B). In the top 12 differentially expressed genes, several previously reported LSEC-specific genes, including Esm1, Dll4, and Ackr1, were up-regulated significantly in EHC Slc35a1-/- mice (Figure 6C). Gene set enrichment analysis (GSEA) showed that capillary genes were up-regulated significantly, whereas arterial venous genes were unchanged (Figure 6D). Further analysis showed that most periportal and midzone LSEC genes were up-regulated, whereas certain pericentral LSEC genes such as Wnt2, Wnt9b, and others were down-regulated (Figure 6E).
Figure 6.
Endothelial Slc35a1 deficiency preferentially disrupts LSEC gene signature. (A) The expression of lipid metabolism-related genes in P7 livers were analyzed with RNA-seq. WT mice, n = 4; EHC Slc35a1-/- mice, n = 4. Data represent means ± SD. ∗∗P < .01 indicates a significant difference between WT and EHC Slc35a1-/- mice. (B) Western blot of CD36 in P7 and P14 livers of WT and EHC Slc35a1-/- mice (n = 2 per group). Actin was used as a loading control. (C) Heatmap of the top 12 differentially expressed genes between WT and EHC Slc35a1-/- mice. Several previously reported LSEC-specific genes, including Ackr1, Dll4, and Esm1, were up-regulated significantly in EHC Slc35a1-/- mice (all P < .001). (D) GSEA analysis of artery, capillary, and vein-associated vascular landscape genes using RNA-seq data of P7 livers of WT (n = 4) and EHC Slc35a1-/- mice (n = 4). Normalized enrichment scores (NESs) and P values are shown. (E) Heatmaps of zonatic LSEC-specific genes in WT and EHC Slc35a1-/- livers at P7. EC, endothelial cell.
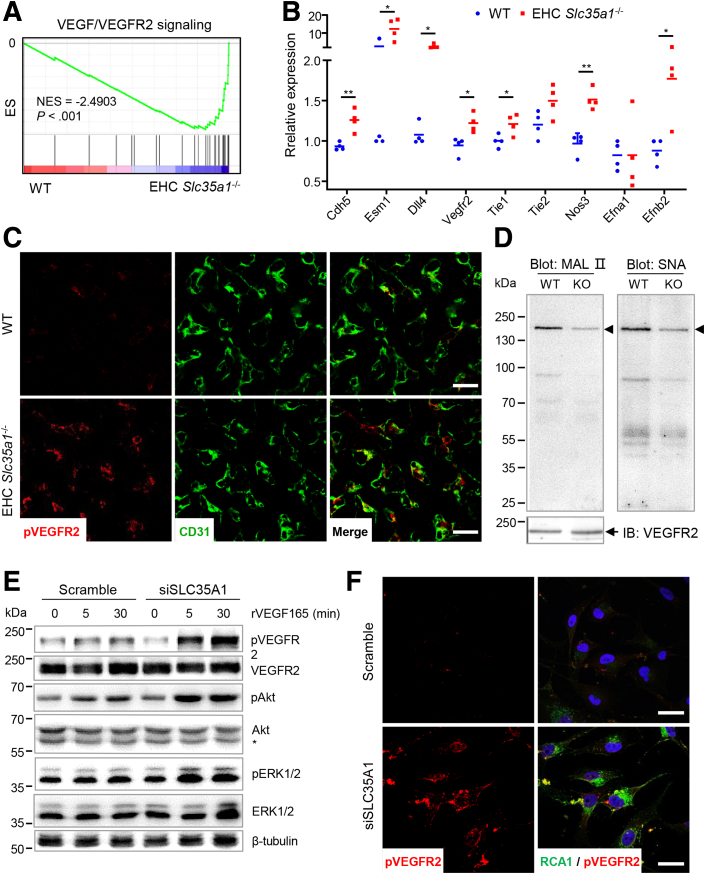
Because VEGFR2 signaling plays a key role in hepatic vascular development,58 we further found that most VEGFR2 signaling-related genes were up-regulated in EHC Slc35a1-/- livers (Figure 7A), which was validated by real-time quantitative PCR (Figure 7B). Immunostaining results showed increased phosphorylation of VEGFR2 in the liver of EHC Slc35a1-/- mice (Figure 7C), indicating enhanced VEGFR2 signaling. We further performed immunoprecipitation with anti-VEGFR2 antibody and blotting with maackia amurensis lectin II and sambucus nigra lectin that specifically bind to α2,3- and α2,6-sialic acid, respectively. The results showed that VEGFR2 in EHC Slc35a1-/- livers were desialylated dramatically (Figure 7D). To further verify the effect of Slc35a1 deficiency on VEGFR2 signaling, we also performed RNA interference to knockdown Slc35a1 expression in human umbilical vein endothelial cells (HUVECs). The results confirmed that Slc35a1 silencing resulted in enhanced VEGFR2 phosphorylation and activation of its downstream signaling (Figure 7E and F). Combined with previous reports that loss of VEGFR2 sialylation favors its activation in endothelial cells,59 our findings suggest that endothelial Slc35a1 deficiency led to VEGFR2 desialylation, thereby enhancing VEGFR2 signaling in LSECs.
Figure 7.
VEGFR2 signaling is enhanced in EHC Slc35a1-/-mice. (A) GSEA analysis of VEGF/VEGFR2 signaling-related genes using RNA-seq data of P7 livers of WT and EHC Slc35a1-/- mice. (B) Relative mRNA levels of VEGFR2 signaling-targeted genes in livers from WT (n = 4) and EHC Slc35a1-/- mice (n = 4) at P7 were detected by real-time quantitative PCR. Glyceraldehyde-3-phosphate dehydrogenase served as an internal control. ∗P < .05 and ∗∗P < .01 indicate significant differences between 2 groups by Mann–Whitney U test. (C) Immunostaining of phosphorylated VEGFR2 and CD31. P7 liver cryosections were collected from WT mice (n = 3) and EHC Slc35a1-/- mice (n = 3) 30 minutes after intraperitoneal injection of 500 ng rVEGF165 protein. Scale bars: 20 μm. (D) Immunoprecipitation of VEGFR2 and lectin blotting to detect desialylation of VEGFR2 in WT (left lane) and EHC Slc35a1-/- mice (right lane). VEGFR2 protein in P7 liver tissue lysates was immunoprecipitated with anti-VEGFR2 antibody and blotted with maackia amurensis lectin II (MAL II), and sambucus nigra lectin (SNA), respectively. (E) Western blot analysis of VEGFR2 activation in HUVECs transfected with Scramble or SLC35A1 siRNAs. After 24 hours of siRNA transfection, HUVECs were starved for 18 hours and stimulated with 25 ng/mL rVEGF165 protein for 5 and 30 minutes, respectively. ∗Nonspecific band in anti-Akt blotting. β-tubulin, loading control. (F) Immunostaining of phosphorylated VEGFR2 and RCAⅠ in HUVECs transfected with Scramble and SLC35A1 (siSLC35A1) siRNAs. After 24 hours of siRNA transfection, HUVECs were starved for 18 hours and stimulated with 25 ng/mL rVEGF165 protein for 5 minutes. RCAⅠ specifically binds to terminal galactose exposed after loss of the capping sialic acids. Representative figures from 3 independent expriments are shown. Scale bar: 25 μm. IB, immunoblot ; KO, knockout; pAkt, Akt phosphorylation at Ser473; pERK1/2, ERK1/2 phosphorylation at Thr202 and Tyr204; pVEGFR2, VEGFR2 phosphorylation at Tyr1054 and Tyr1059; RCAⅠ, Ricinus communis agglutinin Ⅰ lectin.
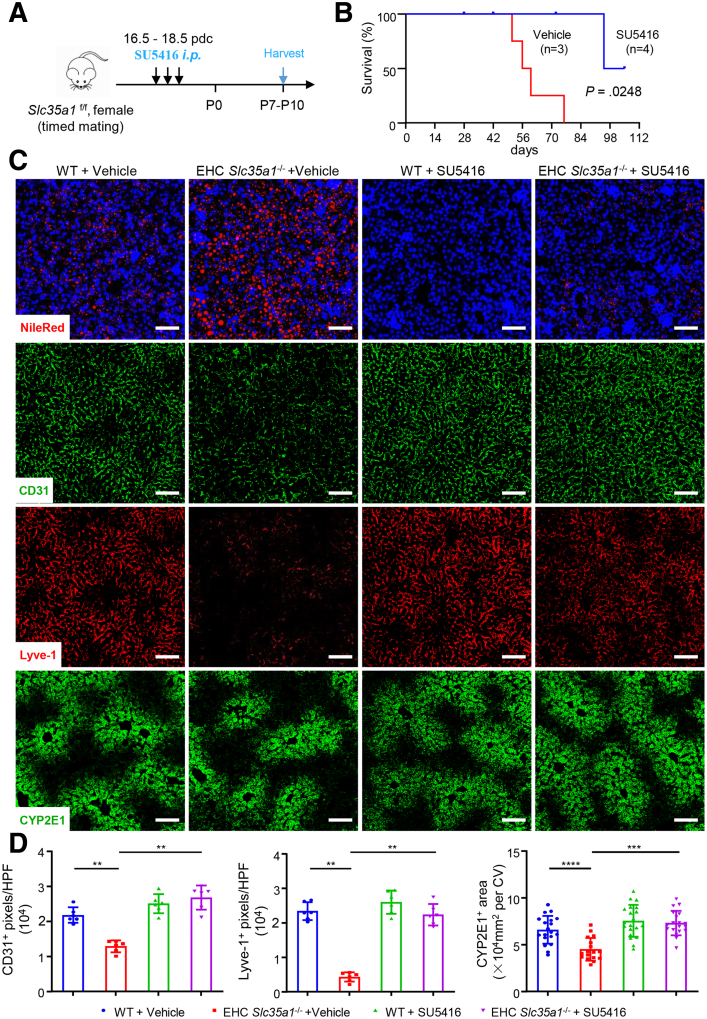
Inhibition of VEGFR2 Kinase Activity Attenuates Neonatal Hepatic Lipid Deposition and Restores Liver Homeostasis in EHC Slc35a1-/- Mice
To further validate the role of VEGFR2 signaling in regulating liver function in vivo, we used kinase inhibitor SU541660, 61, 62 to inhibit VEGFR2 signaling (Figure 8A). Because the hepatic vasculature predominantly is established before birth,2,3,8 and it is necessary to rule out the influence of milk sucking after birth, a daily dose of 25 mg/kg SU5416 was administered intraperitoneally to pregnant dams for 3 days at the late embryonic stage (approximately 16.5-18.5 days post coitum) (Figure 8A). The results showed that SU5416-treated EHC Slc35a1-/- offspring survived longer than vehicle-treated pups (Figure 8B). As shown in Figure 8C, SU5416 treatment significantly attenuated neonatal lipid deposition in P7 EHC Slc35a1-/- pups. Immunostaining revealed that SU5416 treatment partially restored the disrupted sinusoidal vasculature, as indicated by increased expression of Lyve-1 and CD31, and recovered hepatocyte zonation, as indicated by the increased number of cytochrome P450 family 2 subfamily E member 1–positive hepatocytes in P7 EHC Slc35a1-/- pups (Figure 8C and D). These results suggest that the VEGFR2 kinase inhibitor SU5416 can rescue the phenotypes of EHC Slc35a1-/- mice and restore their liver function.
Figure 8.
Inhibition of VEGFR2 signaling alleviates neonatal hepatic lipid deposition and restores liver homeostasis in EHC Slc35a1-/-mice. (A) Schematic of the experimental design. Pregnant Slc35a1f/f mice or WT mice as control at 16.5 days post coitum (dpc) were injected intraperitoneally with vehicle or VEGFR2 inhibitor (SU5416, 25 μg/g of body weight) daily for 3 days, and livers of the Slc35a1-/- pups and WT pups were harvested at P7–P10. (B) Survival curve showed the survival rate of EHC Slc35a1-/- mice from vehicle-treated (n = 3) and SU5416-treated dams (n = 4). (C) Nile Red staining and immunostaining of CD31, Lyve-1, and CYP2E1 in liver cryosections obtained from EHC Slc35a1-/- or WT pups born of pretreated Slc35a1f/f or WT mice, respectively. Scale bars: 100 μm. (D) CD31, Lyve-1 expression, and CYP2E1+ area in panel C were compared quantitatively. Three images per mouse were chosen randomly for analysis. ∗∗P < .01, ∗∗∗P < .001, and ∗∗∗∗P < .0001 indicate significant differences. CV, central vein; HPF, high-power field.
Discussion
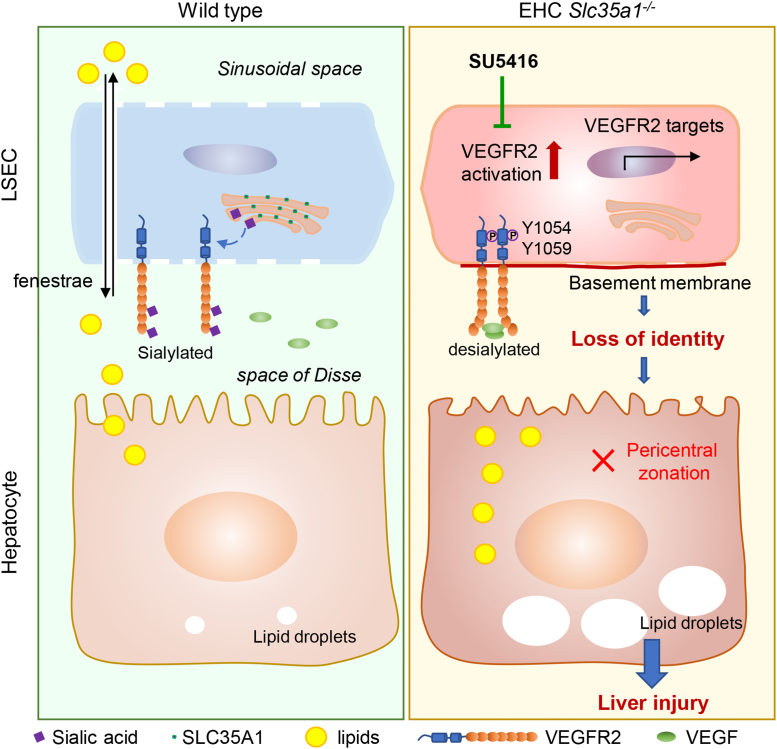
The primary findings of our study were that endothelial Slc35a1 deficiency leads to VEGFR2 desialylation on LSECs, which enhances VEGFR2 signaling and consequently causes loss of LSEC identity and disruption of hepatic zonation. These changes further exacerbate neonatal lipid deposition and lead to fatty liver and eventually liver injury. VEGFR2 inhibition at late embryonic stage by its kinase inhibitor SU5416 restores hepatic vasculature and liver lipid homeostasis in EHC Slc35a1-/- mice (Figure 9).
Figure 9.
Schematic diagram of the mechanism by which endothelial sialylation maintains hepatic vasculature and lipid homeostasis. In WT mice, VEGFR2 activation on LSECs is tightly regulated, with little VEGF binding to sialylated VEGFR2. In EHC Slc35a1-/- mice, endothelial Slc35a1 deficiency leads to VEGFR2 desialylation on LSECs, which enhances VEGFR2 signaling and consequently causes loss of LSEC identity and disrupts hepatic zonation. These changes further exacerbate neonatal lipid deposition and lead to fatty liver and eventually liver injury. VEGFR2 inhibition at the late embryonic stage by its kinase inhibitor SU5416 restores hepatic vasculature and liver lipid homeostasis in EHC Slc35a1-/- mice.
In this study, we found that endothelial Slc35a1-deficient mice exhibited fatty liver shortly after birth, with increased lipids in plasma and increased lipid deposition in the liver tissue, accompanied by progressive liver injury, leading to high postnatal mortality. This partially resembles endothelial and hematopoietic T-synthase–deficient mice (fatty liver, normal plasma lipids).63 EHC Slc35a1-/- mice exhibited defects primarily in the liver, unlike our previous megakaryocyte/platelet-specific Slc35a1 knockout mice presenting only with thrombocytopenia.64 This, along with other studies,65, 66, 67 highlights the organ-specificity of glycosylation defects and their varying disease phenotypes. In addition, differences also exist between EHC Slc35a1-/- mice and human SLC35A1–congenital disorders of glycosylation patients, who exhibit multi-organ dysfunction, including thrombocytopenia, opportunistic infections, psychomotor retardation, epilepsy, ataxia, microcephaly, and chorea, despite lacking reported liver issues.68,69 One possibility is that EHC Slc35a1-/- mice die before reaching adulthood to develop multi-organ abnormalities. This early lethality result also suggests that SLC35A1–congenital disorders of glycosylation patients with Slc35a1 mutations have sufficient residual CMP-sialic acid transporting activity to bypass the early mortality so that they exhibit abnormalities in multiple organs.
The metabolic function of the liver is not fully established during the neonatal stage of mice,6,7,70,71 suggesting that the immature liver at birth may not be able to meet the high demand of lipid metabolism after being fed with milk that contains high levels of fat. Our results showed that WT newborns had marked lipid deposition shortly after birth, which diminished rapidly after P7. In this study, transient lipid deposition was found in normal neonatal livers in mice. However, EHC Slc35a1-/- mice had more pronounced lipid deposition that failed to resolve. Both biochemical analysis and lipidomic profiling revealed fatty liver–like changes in EHC Slc35a1-/- mice. These results suggest that loss of sialylation disrupts the adaptation of neonatal immature liver to high lipid metabolic demands, leading to fatty liver disease. Therefore, EHC Slc35a1-/- mice may be a good model for studying the development of fatty liver disease.
The typical mechanisms for lipid deposition in fatty liver disease involve decreased mitochondrial fatty acid β-oxidation and increased de novo lipogenesis or enhanced uptake of fatty acids to the liver.34,35 Contrary to expectations, endothelial Slc35a1-deficient mice exhibited no significant changes in hepatocyte lipid metabolism genes except slightly up-regulated CD36. Although scavenger receptors and transporters54,72 also may be involved in compensatory lipid uptake enhancement, RNA-seq data revealed more profound endothelial-specific gene dysregulation, prompting investigation of LSEC function. Indeed, EHC Slc35a1-/- mice displayed loss of LSEC identity, including loss of fenestrae, basement membrane formation, and altered marker expression, suggesting disrupted hepatic vasculature.15,16 This aligns with reports linking LSEC fenestrae loss to impaired lipid uptake and hyperlipoproteinemia, phenotypes resembling EHC Slc35a1-/- mice.50, 51, 52, 53 Loss of LSEC fenestrae also might directly promote lipid accumulation in EHC Slc35a1-/- mice. This observation is consistent with findings in plasmalemma vesicle-associated protein-deficient mice, which also lack fenestrated LSECs and spontaneously develop steatosis alongside hyperlipoproteinemia.53 One possible explanation could be that diminished LSEC permeability restricts the outward passage of hepatocyte-derived very-low-density lipoprotein toward the sinusoids, thus exacerbating lipid accumulation, independent of reduced uptake from the blood.73 Notably, hypertriglyceridemia (P21) arose later than early onset lipid deposition (P2) in EHC Slc35a1-/- mice, implying a temporal separation. Before complete vasculature establishment, neonatal hepatocytes may bypass LSECs and directly acquire circulating lipids, exceeding processing capacity, causing transient lipid deposition that resolves with LSEC maturation in wild types. However, endothelial Slc35a1 deficiency disrupts this resolution, leading to persistent accumulation and liver injury. These findings suggest that LSECs, beyond limiting lipid influx, play a crucial role in early lipid clearance, highlighting their importance in maintaining hepatic lipid homeostasis and preventing fatty liver disease.
The intricate spatial organization of hepatocytes, known as liver zonation, plays a crucial role in efficient lipid metabolism.23,28,29,74 LSECs significantly contribute to establishing and maintaining this zonation through paracrine and angiocrine signaling.15,23,24,57,75 LSEC dysfunction often is implicated in altered zonation patterns observed in various liver diseases, including NAFLD and liver fibrosis.19,30 In this study, we found that liver zonation was disrupted in EHC Slc35a1-/- mice, evidenced by loss of pericentral hepatocytes and a reduction of expression of several pericentral hepatocyte-specific genes. Moreover, the predominant accumulation of lipids in the pericentral area suggests that pericentral hepatocytes primarily are affected in EHC Slc35a1-/- mice. This observation aligns with the known vulnerability of pericentral hepatocytes to damage.76 Our findings, therefore, suggest that LSEC dysfunction, by disrupting liver zonation, might contribute to impaired hepatic lipid metabolism in EHC Slc35a1-/- mice. However, the precise mechanisms by which LSECs orchestrate this complex zonation pattern remain to be investigated.
LSECs are highly heterogeneous, characterized by distinct transcriptional signatures governed by specific factors such as GATA-binding factor 4, Transcription factor Maf, and Homeobox protein Meis2.17,77,78 Despite loss of LSEC fenestrae in Slc35a1-deficient mice, quantitative PCR surprisingly revealed increased expression of LSEC-specific genes (Gata4, Lyve1, and Stab2) and continuous endothelial markers (Cd34, Cd31, and Emcn). This suggests that endothelial Slc35a1 deficiency does not directly inhibit LSEC specification but may influence specific LSEC subtypes, which is supported by expansion of periportal LSEC markers alongside reduced pericentral LSEC markers revealed by RNA-seq data. Potential explanations include compensatory expansion of LSECs and vascular endothelial cells after liver injury and Slc35a1 deletion, primarily affecting the distribution of LSEC subtypes, leading to impaired pericentral hepatocyte zonation. However, results from bulk RNA-seq and quantitative PCR using whole liver tissues may be biased owing to the low proportion of endothelial cells in the liver. Future studies using single-cell RNA sequencing after depletion of liver parenchymal cells following our published method79 will help address these issues.
VEGF–VEGFR2 signaling plays a pivotal role in LSEC biology and hepatic vascular development.53,78,80 Existing evidence suggests its involvement in maintaining LSEC identity, including fenestration formation and zonation.15,51,81 However, the specific contribution of VEGFR2 remains unclear. In this study, we found increased VEGFR2 phosphorylation and enhanced expression of several known VEGFR2-targeted genes, suggesting enhanced VEGFR2 signaling in EHC Slc35a1-/- mice. Furthermore, inhibition of VEGFR2 signaling with the VEGFR2 kinase inhibitor SU5418 effectively restored hepatic vasculature, attenuated neonatal liver lipid deposition, and promoted the survival of EHC Slc35a1-/- mice. Our findings suggest that hyperactivation of VEGFR2 signaling is the culprit in disrupting the hepatic vasculature and may be a therapeutic target for various liver diseases with compromised hepatic vasculature. This aligns with reports of VEGFR2 dysfunction in NAFLD and its potential reversal through VEGFR2 inhibition.82,83 However, discrepancies arise between our findings and the established role of VEGFR2 in LSEC identity. We propose several explanations. First, VEGFR2's role in LSECs might change with development, perhaps it may be less crucial postnatally, as suggested by down-regulation of VEGFR2 pathway genes in postnatal liver single-cell RNA sequencing data,84 and the lack of abnormalities in wild-type mice after late embryonic VEGFR2 inhibition in this study. VEGFR2 association with fenestrae proteins diminishes in adult LSECs compared with fetal stages.85 Second, VEGFR2 might differentially affect the development of different hepatic vascular beds, such as portal veins, hepatic arteries, and sinusoids. Consistent with this, VEGF plays distinct roles in different vessel types,85 and LSECs, with their unique venous characteristics, may require lower VEGFR2 activation compared with other ECs.86 Finally, although SU5416 remains a prominent VEGFR2 kinase inhibitor for both in vitro and in vivo vasculogenesis research,60,62,87 its potential off-target effects warrant further exploration through the application of diverse VEGFR2 inhibitors.88 Additionally, inducible endothelial VEGFR2 knockout mice could help differentiate its role at different developmental stages.
VEGFR2 acts as a key regulator of angiogenic signaling, and its activity is tightly regulated by its glycosylation.89 VEGFR2 bears both α2,3- and α2,6-sialic acid on its N-glycans.59,89,90 Chiodelli et al91 found that VEGFR2 sialylation plays an important role in modulating VEGF/VEGFR2 interaction and angiogenic activity. In this study, we found that VEGFR2 was desialylated significantly in EHC Slc35a1-/- livers, exhibiting loss of both α2,3- and α2,6-sialic acid. This is partially consistent with a previous report that α2,3-sialic acid was more affected in SLC35A1–congenital disorders of glycosylation patients.92 It has been reported that removal of sialic acid from N-linked glycans amplified ligand-dependent VEGFR2 activation and signaling on endothelial cells.59 In view of this, we speculated that desialylation of VEGFR2 led to enhanced VEGFR2 signaling in EHC Slc35a1-/- mice. In addition, N-glycosylation inhibitors have been reported to stimulate endothelial cell proliferation through c-Jun-N-terminal kinase activation and endoplasmic reticulum stress in vitro and to promote angiogenesis in vivo.93 The sialidase Neu1 was used to remove α2,6-sialic acid from platelet endothelial cell adhesion molecule, thereby disrupting endothelial tube formation.94 These results combined with our findings suggest that targeting glycosylation may be a novel strategy to modulate endothelial function.
Our study had some limitations. First, Tie2-Cre also mediates Slc35a1 deletion in hematopoietic lineages.95 Although our focus is on hepatic endothelium, potential contributions from affected hematopoietic cells cannot be definitively excluded. Future studies could use more relative endothelium-specific Cre drivers, such as Cdh5-Cre,96 to differentiate the role of endothelial Slc35a1 deficiency. Second, the precise impact of EHC Slc35a1 deficiency on specific LSEC and hepatocyte subtypes remains to be investigated. Future studies using isolated LSECs and single-cell RNA-seq would provide valuable insights into the diverse LSEC populations and their specific contributions to the observed phenotypes.
Taken together, we found that endothelial Slc35a1 deficiency aggravated neonatal lipid deposition and caused severe liver injury and high mortality. Endothelial Slc35a1 deficiency led to loss of identity of LSECs and disrupted hepatic zonation owing to desialylation of VEGFR2 enhanced VEGFR2 signaling. Inhibition of VEGFR2 signaling by its kinase inhibitor SU5416 restored hepatic vasculature and zonation, and alleviated postnatal lipid deposition in Slc35a1-deficient mice. Our findings underscore a fundamental role of sialylation for LSECs to support hepatic vascular development and maintain hepatic lipid homeostasis in the newborn. Targeting VEGFR2 signaling or its glycosylation may be a novel strategy to prevent liver disorders and to develop animal models for exploring the metabolic maturation of the liver.
Materials and Methods
Mouse Models
Mice with Slc35a1 gene flanked by locus of X-over P1 (loxP) sites (Slc35a1f/f) were generated as previously described.64 Slc35a1f/f mice then were crossed with Tie2-Cre transgenic mice (strain ID: T003764; GemPharmatech, Nanjing, China) (Figure 1A and B), which mediate deletion of floxed gene in both endothelial and hematopoietic cells, to generate mice lacking sialylation in endothelial and hematopoietic cells (EHC Slc35a1-/- mice). The genotypes of EHC Slc35a1-/- mice were confirmed by PCR with tail genomic DNA to amplify a 343-bp product for floxed Slc35a1 allele and a 411-bp product for the Tie2-Cre gene, respectively (Figure 1B). Mice were bred and maintained in the specific pathogen-free condition on a 12-hour light/dark cycle in the Laboratory Animal Experimental Center at Soochow University (Suzhou, China).
For SU5416 (VEGFR2 kinase inhibitor) treatment, timed mating was performed and pregnant Slc35a1f/f mice and WT littermate controls at 16.5 days post coitum were injected intraperitoneally with either vehicle or SU5416 (cat# S2845, 25 mg/kg/d; Selleck) for 3 days, and livers of EHC Slc35a1-/- and WT pups were harvested for further analyses.
For recombinant VEGF165 (rVEGF165) protein injection, WT and EHC Slc35a1-/- pups at P7 were injected intraperitoneally with either phosphate-buffered saline (PBS) or rVEGF165 (cat# 450-32, 500 ng/mouse; Peprotech) 30 minutes before liver harvest.
Ethics
All animal experiments were performed in accordance with Animal Research: Reporting of In Vivo Experiments guidelines. Mouse studies were approved by the Ethics Committee of Soochow University (no. 202009A303). All mice within a single test were raised in the same cage. Simple randomization was used for grouping. The investigators who performed the animal experiments were not blinded to the groups. To follow the Replacement, Reduction and Refinement principle, the number of mice in each group was limited according to our previous experience and a priori power analysis. The sample size was determined using G∗power software (v3.1.97; HHU, Düsseldorf, Germany) with a 0.05 α level, 0.8 power, and 0.2 SDs of effect size. All mice used in the study were age- and gender-matched unless indicated otherwise.
Cell Culture and Transfection
Primary HUVECs (less than passage 7) were cultured in Endothelial Cell Medium-basal (cat# 1001-b; ScienCell) supplemented with 5% fetal bovine serum (Gibco), 1× Endothelial Cell Growth Supplement (cat# 1052; ScienCell), 1% streptomycin, and penicillin at 37°C in 5% CO2. For small interfering RNA (siRNA) nucleofection, 5 × 105 HUVECs were transfected with 150 nmol/L Scramble or SLC35A1 siRNAs (synthesized by Genepharma, Inc, Shanghai, China; siRNA sequences are provided in Table 1) by a Lonza 4D-Nucleofector (program: A-034). After 24 and 48 hours of transfection, cells were harvested to evaluate the messenger RNA (mRNA) and protein expression. For rVEGF165 (cat# 450-32; Peprotech) stimulation, HUVECs 24 hours after transfection with Scramble or SLC35A1 siRNAs were starved further for 18 hours in serum-free medium, and then stimulated with 25 ng/mL rVEGF165 protein for 5 and 30 minutes, respectively. Cells were harvested for further immunostaining or Western blot analysis.
Table 1.
Sequence of Primers Used for Genotyping and Real-Time Quantitative PCR and siRNA Sequences for Slc35a1 Gene Knockdown
| Genes | Forward primer, 5'-3' | Reverse primer, 5'-3' |
|---|---|---|
| Tie2Cre | GGGCAGTCTGGTACTTCCAAGCT | CTTGATTCACCAGATGCTGAGGTTA |
| Slc35a1 floxed allele | TCGCTTGCTTTCAGATCCGAGTT | ACAATTAAGTAGGATTATCGCC |
| Gapdh | GGTCTCCTCTGACTTCAACA | AGCCAAATTCGTTGTCATAC |
| PPARa | AGTTCGGGAACAAGACGTTG | CAGTGGGGAGAGAGGACAGA |
| cpt1a | CCTGGGCATGATTGCAAAG | ACGCCACTCACGATGTTCTTC |
| cpt2 | TCTTCCTGAACTGGCTGTCA | GTACCCACCATGCACTACCA |
| acc1 | GATGAACCATCTCCGTTGGC | GACCCAATTATGAATCGGGAGTG |
| acly | CAGCCAAGGCAATTTCAGAGC | CTCGACGTTTGATTAACTGGTCT |
| FASN | GGAGGTGGTGATAGCCGGTAT | TGGGTAATCCATAGAGCCCAG |
| SREBP1 | GATGTGCGAACTGGACACAG | CTGTCTCACCCCCAGCATAG |
| ChREBP | AGATGGAGAACCGACGTATCA | ACTGAGCGTGCTGACAAGTC |
| agpat2 | GAGCCTTCTACTTGGCCATCCA | TTGATTGTTCCTGAGGTGAAGAGC |
| dgat2 | AATAAAGGATCTGCCCTGTCACG | TTGAGCCAGGTGACAGAGAAGATG |
| atgl | GGATGGCGGCATTTCAGAC | CAAAGGGTTGGGTTGGTTCAG |
| hsl | ACGCTACACAAAGGCTGCTT | TCTCGTTGCGTTTGTAGTGC |
| lipa | TGTTCGTTTTCACCATTGGGA | CGCATGATTATCTCGGTCACA |
| acox1 | TAACTTCCTCACTCGAAGCCA | AGTTCCATGACCCATCTCTGTC |
| acaa1b | CAGGACGTGAAGCTAAAGCCT | CTCCGAAGTTATCCCCATAGGAA |
| fabp1 | ATGAACTTCTCCGGCAAGTACC | GGTCCTCGGGCAGACCTAT |
| abcg5 | AGGGCCTCACATCAACAGAG | GCTGACGCTGTAGGACACAT |
| abcg8 | CTGTGGAATGGGACTGTACTTC | GTTGGACTGACCACTGTAGGT |
| SREBF2 | GCAGCAACGGGACCATTCT | CCCCATGACTAAGTCCTTCAACT |
| hmgcr | AGCTTGCCCGAATTGTATGTG | TCTGTTGTGAACCATGTGACTTC |
| cyp7a1 | GGGATTGCTGTGGTAGTGAGC | GGTATGGAATCAACCCGTTGTC |
| baat | TGCTGGTGGATTGATGGAGT | CCGAGGACCTTAGGATGTCTC |
| wnt2 | CTCGGTGGAATCTGGCTCTG | CACATTGTCACACATCACCCT |
| slc35a1 | GCTCCGGCGAGAGAAAATGT | CACGGCAGTGGTTGAGAAGT |
| cdh5 | CACTGCTTTGGGAGCCTTC | GGGGCAGCGATTCATTTTTCT |
| esm1 | CTGGAGCGCCAAATATGCG | TGAGACTGTACGGTAGCAGGT |
| dll4 | TTCCAGGCAACCTTCTCCGA | ACTGCCGCTATTCTTGTCCC |
| vegfr2 | TTTGGCAAATACAACCCTTCAGA | GCAGAAGATACTGTCACCACC |
| tie1 | TTTTCTTGGCCTCTCATGTTGG | CGCACGATGCGATCATCCTT |
| tie2 | GAGTCAGCTTGCTCCTTTATGG | AGACACAAGAGGTAGGGAATTGA |
| nos3 | GGCTGGGTTTAGGGCTGTG | CTGAGGGTGTCGTAGGTGATG |
| efna1 | CCCGGAGAAGCTGTCTGAGA | ACATGGGCCTGGGGATTATGA |
| efnb2 | TTCTAGCACCGATGGCAACAG | CCCTGCGAATAAGGCCACT |
| Gata4 | CCCTACCCAGCCTACATGG | ACATATCGAGATTGGGGTGTCT |
| Maf | GGAGACCGACCGCATCATC | TCATCCAGTAGTAGTCTTCCAGG |
| Meis2 | GACCTCGTGATTGATGAGAGAG | GGTCGGCGAGATTTGTGGA |
| Tfec | GGTCTCACGGATGCTCCTTG | TCCAGCGCATATCAGGATCATTA |
| Cd34 | GGTAGCTCTCTGCCTGATGAG | TGGTAGGAACTGATGGGGATATT |
| Lyve1 | CAGCACACTAGCCTGGTGTTA | CGCCCATGATTCTGCATGTAGA |
| Cd31 | ACGCTGGTGCTCTATGCAAG | TCAGTTGCTGCCCATTCATCA |
| Stab2 | AGCTGCTGCCTTTAATCCTCA | ACTCCGTCTTGATGGTTAGAGTA |
| Cd32b | AGGGCCTCCATCTGGACTG | GTGGTTCTGGTAATCATGCTCTG |
| Emcn | AATACCAGGCATCGTGTCAGT | CCACTTCATGTTTTGGTGTTGTC |
| Apc | GGCGTGAAATCCGAGTCCTTC | CCACCTGCAATAACTCTGCAA |
| Cdh1 | CAGGTCTCCTCATGGCTTTGC | CTTCCGAAAAGAAGGCTGTCC |
| Ass1 | ACACCTCCTGCATCCTCGT | GCTCACATCCTCAATGAACACCT |
| Alb | GACGTGTGTTGCCGATGAGT | GTTTTCACGGAGGTTTGGAATG |
| Asl | CTATGACCGGCATCTGTGGAA | AGCAACCTTGTCCAACCCTTG |
| Cyp2f2 | GGACCCAAACCTCTCCCAATC | CCGTGAACACCGACCCATAC |
| Glul | TGAACAAAGGCATCAAGCAAATG | CAGTCCAGGGTACGGGTCTT |
| Cyp2e1 | CGTTGCCTTGCTTGTCTGGA | AAGAAAGGAATTGGGAAAGGTCC |
| Axin2 | ATGAGTAGCGCCGTGTTAGTG | GGGCATAGGTTTGGTGGACT |
| Cldn2 | CAACTGGTGGGCTACATCCTA | CCCTTGGAAAAGCCAACCG |
| Tbx3 | AGATCCGGTTATCCCTGGGAC | CAGCAGCCCCCACTAACTG |
| siSLC35A1 | GGUAGAUUCAAAGCAUCUUTT | AAGAUGCUUUGAAUCUACCTT |
| Scramble siRNA | UUCUCCGAACGUGUCACGUTT | ACGUGACACGUUCGGAGAATT |
Lipidomic Analysis by Liquid Chromatography–Mass Spectrometry
P14 livers were harvested and snap-frozen in liquid nitrogen and stored at -80°C before detection. The liquid chromatography–mass spectrometry–based lipidomic analysis was conducted by Suzhou BioNovoGene Co, Ltd. The lipids were isolated as previously described.97,98 Lipid separation was performed with an ACQUITY UPLC BEH C18 column (Water Co, Milford, MA) maintained at 50°C. After column equilibration, 2 μL each sample was loaded onto the column via autosampler, which was kept at 8°C. Gradient elution of analytes was performed with acetonitrile:water = 60:40 (with 0.1% formic acid and 10 mmol/L ammonium formate) (solvent C) and isopropanol:acetonitrile = 90:10 (with 0.1% formic acid and 10 mmol/L ammonium formate) (solvent D) at a flow rate of 0.25 mL/min. A linear gradient of solvent C (v/v) was used as follows: 0∼5 minutes, 70% - 57% C; 5 - 5.1 minutes, 57% - 50% C; 5.1 - 14 minutes, 50% - 30% C; 14 - 14.1 minutes, 30% C; 14.1 - 21 minutes, 30% - 1% C; 21 - 24 minutes, 1% C; 24 - 24.1 minutes, 1% - 70% C; and 24.1 - 28 minutes, 70% C. All solvents were liquid chromatography–mass spectrometry grade. Then, the analytes were loaded on a Thermo Q Exactive Focus mass spectrometer (ThermoFisher Scientific, Waltham, MA) with a spray voltage of 3.5 kV and -2.5 kV in positive and negative modes, respectively. Sheath gas and auxiliary gas were set at 30 and 10 arbitrary units, respectively. The capillary temperature was 325ºC. The Orbitrap analyzer scanned over a mass range of m/z 150–2000 for full scan at a mass resolution of 35,000. Data-dependent acquisition tandem mass spectrometry experiments were performed with a high-energy, collision-induced, dissociation scan. The normalized collision energy was 30 eV. Dynamic exclusion was implemented to remove some unnecessary information in tandem mass spectrometry spectra.
Gene Expression Profiling by Bulk RNA Sequencing
P7 livers were harvested and snap-frozen in liquid nitrogen. Total RNA was extracted using the MagBeads Total RNA Extraction Kit (cat# T02-096; Agilent Technologies, Santa Clara, CA) following the manufacturer’s instructions and checked for RNA integrity by an Agilent 2100 Bioanalyzer (Agilent Technologies). Total RNA was purified further with the RNAClean XP Kit (cat# A63987; Beckman Coulter) and RNase-Free DNase Set (cat# 79254; QIAGEN). Library construction and sequencing were performed by Shanghai Biotechnology Co (Shanghai, China) using an Illumina NovaSeq6000 sequencer with a PE150 sequencing mode. The raw reads obtained from sequencing were filtered using the Seqtk-1.3 (r106) toolkit to exclude unqualified reads such as low overall quality, containing sequencing primers, low end quality, contaminating ribosomal RNA, and so forth.99 The obtained clean reads were mapped to the mouse reference genome (version Genome Reference Consortium Mouse Build 38) using Hisat2 (version 2.0.4) with a spliced mapping algorithm.100
To make the gene expression levels comparable between different genes and samples, the reads were transformed into fragments per kilobase of transcript per million mapped reads (FPKM) for the normalization of gene expression.101 FPKM values were calculated by the following formula: FPKM = total exon fragments / (mapped reads [millions] × exon length [kB]). Differential gene expression analysis was performed using the Bioconductor R package edgeR (v4.0.1),102 the P value was obtained and adjusted through multiple testing by controlling the false discovery rate (FDR). Concurrently, the multiplicity of differential expression, represented by fold change, was computed based on the FPKM value. Genes with a fold change >2 and a q-value <0.05 were considered to be differentially expressed.
For data visualization, the pheatmap package (Raivo Kolde, version 1.0.12) and GraphPad Prism (GraphPad Software, version 8.3.0) were used to create heatmaps that visually represented the expression patterns of genes across samples. For transcriptional data analysis, GSEA software (UC San Diego and Broad Institute, version 4.3.2) was used to generate enrichment plots for predefined gene sets.103 The normalized enrichment score and FDR were calculated. The gene sets with FDR < 0.25, P < 0.05, and |normalized enrichment score| > 1 were considered to be enriched significantly. The predefined sets of genes used for GSEA enrichment in this study were obtained either from GSEA official websites (https://www.gsea-msigdb.org/gsea/msigdb/index.jsp) or meticulously curated through literature review.
Real-Time Quantitative PCR
Total RNA was extracted from cells and snap-frozen liver tissues using the TRIzol reagent (cat# 15596026; ThermoFisher Scientific) and reverse-transcribed into complementary DNA using the RevertAid first-strand complementary DNA synthesis kit (cat# k1622; ThermoFisher Scientific). Real-time quantitative PCR was performed using PowerUp SYBR Green Master Mix (cat# a25742; ThermoFisher Scientific) and Applied Biosystems 7500 real-time quantitative PCR system (Foster City, CA). Relative mRNA expression was normalized to Gapdh mRNA levels. The primer sequences are listed in Table 2.
Western Blot
Livers from WT and EHC Slc35a1-/- mice were collected and snap-frozen in liquid nitrogen. Liver tissue lysates were homogenized in RIPA lysis buffer containing protease inhibitor cocktail (cat# 11697498001; Roche) and phosphatase inhibitors (cat# P081; Beyotime). Total proteins were quantified using the BCA Protein Assay Kit (cat# P0010; Beyotime) and were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis and transferred to a nitrocellulose membrane (cat# 10600001; GE Healthcare) blocked with 5% bovine serum albumin (BSA) and incubated with primary antibodies overnight at 4°C and secondary antibodies for 1 hour at room temperature, and finally visualized by chemiluminescence detection using the Tanon 4600SF Imaging System (Tanon, Shanghai, China). All antibodies are listed in Table 3.
Immunoprecipitation and Lectin Blotting
Liver tissue lysates were obtained and quantified as described previously. For each immunoprecipitation reaction, 10 μL anti-VEGFR2 antibody (cat# 2479; Cell Signaling Technology) was added to 1 mg total protein lysate and the mixture was incubated for 1.5 hours with rotation at 4°C. A total of 20 μL protein A/G agarose beads (cat# sc-2003; Santa Cruz) were added and rotated overnight at 4°C. The mixture was centrifuged at 2500 rpm for 5 minutes to pellet the beads and washed for 5 times with RIPA buffer. The proteins immunoprecipitated on beads were denatured by boiling for 10 minutes and further resolved by sodium dodecyl sulfate–polyacrylamide gel electrophoresis and transferred to a nitrocellulose membrane. For lectin blotting, the membrane was blocked and blotted with 5 μg/mL biotinylated maackia amurensis lectin II (cat# B-1265; Vector Laboratories) or 10 μg/mL sambucus nigra lectin (cat# B-1305-2; Vector Laboratories) in blocking buffer (5% BSA in Tris-buffered saline with Tween 20) at 4°C overnight. After four 5-minute washes, the membrane was incubated in horseradish peroxidase–conjugated streptavidin (cat# 405210; BioLegend) and visualized by chemiluminescence detection using the Tanon 4600SF Imaging System.
Biochemical Assay
Plasma levels of alanine aminotransferase, aspartate aminotransferase, total cholesterol, triglyceride, high-density lipoprotein, and low-density lipoprotein were determined using commercially available kits (CAM-SU Genomic Resource Center of Soochow University, Suzhou, China).
Histology Analysis
Livers were fixed in 10% buffered formalin overnight at 4°C. After three 30-minute washes in PBS, the formalin-fixed samples were dehydrated gradually, paraffin-embedded, sectioned into 5-μm–thick slices, and stained with H&E following routine procedures. After H&E staining, the slices were examined and imaged under a microscope.
Immunostaining
Livers were fixed in 4% paraformaldehyde (PFA) overnight at 4°C. After three 30-minute washes in PBS, the PFA-fixed samples were dehydrated in 20% sucrose/PBS solution overnight at 4°C, embedded with optimum cutting temperature compound, and frozen at -80°C. The embedded frozen tissues were sectioned into 10- to 30-μm cryosections. The slices were washed in PBS and further fixed in 4% PFA for 15 minutes at room temperature. After three 10-minute washes in PBS, the slices were blocked in blocking buffer (3% donkey serum, 3% BSA, and 0.3% Triton X-100 in PBS) for 1 hour at room temperature. Immunofluorescence was performed using fluorescent antibodies (Table 3) and counterstained with 4′,6-diamidino-2-phenylindole Fluoromount-G (cat# 0100-20; SouthernBiotech). Images were taken with a confocal microscope (SP8; Leica, Wetzlar, Germany). The positive area of each target was quantified by ImageJ (1.53t; National Institutes of Health, Bethesda, MD). Six images per mouse were chosen randomly for analysis.
Nile Red Staining
Nile Red staining was conducted on PFA-fixed liver cryosections. Briefly, after three 10-minute washes in PBS, the sections were stained with 1 μg/mL Nile Red (cat# N3013; Sigma-Aldrich) in PBS for 30 minutes at room temperature and counterstained in 4′,6-diamidino-2-phenylindole Fluoromount-G. Images were taken with a confocal microscope (SP8; Leica).
Scanning Electron Microscopy and Transmission Electron Microscopy
P7 mice were killed and perfused with cold PBS and 4% PFA. Liver tissues (2∼3 mm2) were harvested, fixed with 2.5% glutaraldehyde, postfixed with osmium tetroxide, dehydrated with graded alcohols, dried with hexamethyldisilazane, sputter-coated with gold, and examined using a SU8100 field emission scanning electron microscope (HITACHI, Tokyo, Japan). The average number of fenestrae was determined from 10 random scanning electron micrographs of each group of 3 mice. The data are presented as the number of fenestrae per area. For transmission electron microscopy analysis, the samples were embedded in epoxy resin after dehydration with graded alcohols, and were cut into thin sections. Three samples from each group were examined with a transmission electron microscope (TECNAI 10; FEI, Tokyo, Japan) to examine changes in LSEC basement membranes and the integrity of the space of Disse.
Statistics
All data were analyzed using GraphPad software (v8.3.0; San Diego, CA) and presented as means ± SD, as noted in the figure legends. All data represent at least 3 independent experiments. The unpaired Student t test was used for comparison of differences between 2 different experimental settings (Mann–Whitney U test for nonparametric data). One-way analysis of variance was used for comparison of differences among multiple groups. A P value < .05 was considered significant.
All authors had access to the study data and reviewed and approved the final manuscript.
Acknowledgments
CRediT Authorship Contributions
Bin Zuo (Data curation: Equal; Formal analysis: Lead; Investigation: Equal; Methodology: Lead; Software: Lead; Validation: Equal; Writing – original draft: Lead; Writing – review & editing: Equal)
Fei Yang (Data curation: Supporting; Formal analysis: Supporting; Investigation: Equal; Methodology: Equal; Resources: Supporting; Validation: Equal)
Lulu Huang (Formal analysis: Supporting; Investigation: Supporting; Methodology: Supporting)
Jingjing Han (Formal analysis: Supporting; Investigation: Supporting; Resources: Equal)
Tianyi Li (Formal analysis: Supporting; Investigation: Supporting)
Zhenni Ma (Formal analysis: Supporting; Investigation: Supporting; Methodology: Supporting)
Lijuan Cao (Formal analysis: Supporting; Investigation: Supporting; Methodology: Supporting)
Yun Li (Formal analysis: Supporting; Investigation: Supporting; Resources: Supporting)
Xia Bai (Investigation: Supporting; Funding acquisition: Equal; Resources: Lead; Supervision: Supporting)
Miao Jiang (Formal analysis: Supporting; Supervision: Supporting)
Yang He (Conceptualization: Supporting; Funding acquisition: Equal; Supervision: Supporting; Writing – review & editing: Supporting)
Lijun Xia (Conceptualization: Lead; Investigation: Equal; Resources: Lead; Supervision: Equal; Writing – review & editing: Lead)
Footnotes
Conflicts of interest The authors disclose no conflicts.
Funding This work was supported by grants from the Priority Academic Program Development of Jiangsu Higher Education Institutions (X.B.) and National Natural Science Foundation of China (82070123) (Y.H.). The funders played no role in the study design, data collection, analysis and interpretation, and manuscript preparation.
Data Availability Statement Data are available from the corresponding author upon request.
Contributor Information
Yang He, Email: heyang1963@163.com.
Lijun Xia, Email: Lijun-xia@omrf.org.
References
- 1.Lewis K., Yoshimoto M., Takebe T. Fetal liver hematopoiesis: from development to delivery. Stem Cell Res Ther. 2021;12:139–145. doi: 10.1186/s13287-021-02189-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Soares-da-Silva F., Peixoto M., Cumano A., et al. Crosstalk between the hepatic and hematopoietic systems during embryonic development. Front Cell Dev Biol. 2020;8:612–622. doi: 10.3389/fcell.2020.00612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ober E.A., Lemaigre F.P. Development of the liver: insights into organ and tissue morphogenesis. J Hepatol. 2018;68:1049–1062. doi: 10.1016/j.jhep.2018.01.005. [DOI] [PubMed] [Google Scholar]
- 4.Yang L., Wang W.H., Qiu W.L., et al. A single-cell transcriptomic analysis reveals precise pathways and regulatory mechanisms underlying hepatoblast differentiation. Hepatology. 2017;66:1387–1401. doi: 10.1002/hep.29353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gruppuso P.A., Sanders J.A. Regulation of liver development: implications for liver biology across the lifespan. J Mol Endocrinol. 2016;56:R115–R125. doi: 10.1530/JME-15-0313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gong T., Zhang C., Ni X., et al. A time-resolved multi-omic atlas of the developing mouse liver. Genome Res. 2020;30:263–275. doi: 10.1101/gr.253328.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu J., Hao S., Shi Q., et al. Transcriptomic profile of the mouse postnatal liver development by single-nucleus RNA sequencing. Front Cell Dev Biol. 2022;10 doi: 10.3389/fcell.2022.833392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao R., Duncan S.A. Embryonic development of the liver. Hepatology. 2005;41:956–967. doi: 10.1002/hep.20691. [DOI] [PubMed] [Google Scholar]
- 9.Reichen J. The role of the sinusoidal endothelium in liver function. News Physiol Sci. 1999;14:117–121. doi: 10.1152/physiologyonline.1999.14.3.117. [DOI] [PubMed] [Google Scholar]
- 10.Wake K., Sato T. "The sinusoid" in the liver: lessons learned from the original definition by Charles Sedgwick Minot (1900) Anat Rec (Hoboken) 2015;298:2071–2080. doi: 10.1002/ar.23263. [DOI] [PubMed] [Google Scholar]
- 11.Gracia-Sancho J., Caparrós E., Fernández-Iglesias A., et al. Role of liver sinusoidal endothelial cells in liver diseases. Nat Rev Gastroenterol Hepatol. 2021;18:411–431. doi: 10.1038/s41575-020-00411-3. [DOI] [PubMed] [Google Scholar]
- 12.Colucci S., Altamura S., Marques O., et al. Liver sinusoidal endothelial cells suppress bone morphogenetic protein 2 production in response to TGFbeta pathway activation. Hepatology. 2021;74:2186–2200. doi: 10.1002/hep.31900. [DOI] [PubMed] [Google Scholar]
- 13.Han S., Tan C., Ding J., et al. Endothelial cells instruct liver specification of embryonic stem cell-derived endoderm through endothelial VEGFR2 signaling and endoderm epigenetic modifications. Stem Cell Res. 2018;30:163–170. doi: 10.1016/j.scr.2018.06.004. [DOI] [PubMed] [Google Scholar]
- 14.Han S., Dziedzic N., Gadue P., et al. An endothelial cell niche induces hepatic specification through dual repression of Wnt and Notch signaling. Stem Cells. 2011;29:217–228. doi: 10.1002/stem.576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Walter T.J., Cast A.E., Huppert K.A., et al. Epithelial VEGF signaling is required in the mouse liver for proper sinusoid endothelial cell identity and hepatocyte zonation in vivo. Am J Physiol Gastrointest Liver Physiol. 2014;306:G849–G862. doi: 10.1152/ajpgi.00426.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Poisson J., Lemoinne S., Boulanger C., et al. Liver sinusoidal endothelial cells: physiology and role in liver diseases. J Hepatol. 2017;66:212–227. doi: 10.1016/j.jhep.2016.07.009. [DOI] [PubMed] [Google Scholar]
- 17.Koch P.S., Lee K.H., Goerdt S., et al. Angiodiversity and organotypic functions of sinusoidal endothelial cells. Angiogenesis. 2021;24:289–310. doi: 10.1007/s10456-021-09780-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Collardeau-Frachon S., Scoazec J.Y. Vascular development and differentiation during human liver organogenesis. Anat Rec (Hoboken) 2008;291:614–627. doi: 10.1002/ar.20679. [DOI] [PubMed] [Google Scholar]
- 19.Cunningham R.P., Porat-Shliom N. Liver zonation - revisiting old questions with new technologies. Front Physiol. 2021;12 doi: 10.3389/fphys.2021.732929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ben-Moshe S., Itzkovitz S. Spatial heterogeneity in the mammalian liver. Nat Rev Gastroenterol Hepatol. 2019;16:395–410. doi: 10.1038/s41575-019-0134-x. [DOI] [PubMed] [Google Scholar]
- 21.Zhu S., Rao X., Qian Y., et al. Liver endothelial Heg regulates vascular/biliary network patterning and metabolic zonation via Wnt signaling. Cell Mol Gastroenterol Hepatol. 2022;13:1757–1783. doi: 10.1016/j.jcmgh.2022.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rocha A.S., Vidal V., Mertz M., et al. The angiocrine factor rspondin3 is a key determinant of liver zonation. Cell Rep. 2015;13:1757–1764. doi: 10.1016/j.celrep.2015.10.049. [DOI] [PubMed] [Google Scholar]
- 23.Ma R., Martinez-Ramirez A.S., Borders T.L., et al. Metabolic and non-metabolic liver zonation is established non-synchronously and requires sinusoidal Wnts. eLife. 2020;9 doi: 10.7554/eLife.46206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kostallari E., Shah V.H. Angiocrine signaling in the hepatic sinusoids in health and disease. Am J Physiol Gastrointest Liver Physiol. 2016;311:G246–G251. doi: 10.1152/ajpgi.00118.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang X.J., Olsavszky V., Yin Y., et al. Angiocrine hepatocyte growth factor signaling controls physiological organ and body size and dynamic hepatocyte proliferation to prevent liver damage during regeneration. Am J Pathol. 2020;190:358–371. doi: 10.1016/j.ajpath.2019.10.009. [DOI] [PubMed] [Google Scholar]
- 26.Apte U., Zeng G., Muller P., et al. Activation of Wnt/beta-catenin pathway during hepatocyte growth factor-induced hepatomegaly in mice. Hepatology. 2006;44:992–1002. doi: 10.1002/hep.21317. [DOI] [PubMed] [Google Scholar]
- 27.Duan J.L., Zhou Z.Y., Ruan B., et al. Notch-regulated c-Kit-positive liver sinusoidal endothelial cells contribute to liver zonation and regeneration. Cell Mol Gastroenterol Hepatol. 2022;13:1741–1756. doi: 10.1016/j.jcmgh.2022.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Behari J., Li H., Liu S., et al. beta-catenin links hepatic metabolic zonation with lipid metabolism and diet-induced obesity in mice. Am J Pathol. 2014;184:3284–3298. doi: 10.1016/j.ajpath.2014.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hijmans B.S., Grefhorst A., Oosterveer M.H., et al. Zonation of glucose and fatty acid metabolism in the liver: mechanism and metabolic consequences. Biochimie. 2014;96:121–129. doi: 10.1016/j.biochi.2013.06.007. [DOI] [PubMed] [Google Scholar]
- 30.Dobie R., Wilson-Kanamori J.R., Henderson B.E.P., et al. Single-cell transcriptomics uncovers zonation of function in the mesenchyme during liver fibrosis. Cell Rep. 2019;29:1832–1847 e8. doi: 10.1016/j.celrep.2019.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hall Z., Bond N.J., Ashmore T., et al. Lipid zonation and phospholipid remodeling in nonalcoholic fatty liver disease. Hepatology. 2017;65:1165–1180. doi: 10.1002/hep.28953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Behari J., Li H., Liu S., et al. β-catenin links hepatic metabolic zonation with lipid metabolism and diet-induced obesity in mice. Am J Pathol. 2014;184:1–10. doi: 10.1016/j.ajpath.2014.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sangineto M., Villani R., Cavallone F., et al. Lipid metabolism in development and progression of hepatocellular carcinoma. Cancers. 2020;12:1419. doi: 10.3390/cancers12061419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vancells Lujan P., Vinas Esmel E., Sacanella Meseguer E. Overview of non-alcoholic fatty liver disease (NAFLD) and the role of sugary food consumption and other dietary components in its development. Nutrients. 2021;13:1442. doi: 10.3390/nu13051442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gong Z., Tas E., Yakar S., et al. Hepatic lipid metabolism and non-alcoholic fatty liver disease in aging. Mol Cell Endocrinol. 2017;455:115–130. doi: 10.1016/j.mce.2016.12.022. [DOI] [PubMed] [Google Scholar]
- 36.Svegliati-Baroni G., Pierantonelli I., Torquato P., et al. Lipidomic biomarkers and mechanisms of lipotoxicity in non-alcoholic fatty liver disease. Free Radic Biol Med. 2019;144:293–309. doi: 10.1016/j.freeradbiomed.2019.05.029. [DOI] [PubMed] [Google Scholar]
- 37.Oligschlaeger Y., Shiri-Sverdlov R. NAFLD preclinical models: more than a handful, less of a concern? Biomedicines. 2020;8:28–42. doi: 10.3390/biomedicines8020028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jahn D., Kircher S., Hermanns H.M., et al. Animal models of NAFLD from a hepatologist's point of view. Biochim Biophys Acta Mol Basis Dis. 2019;1865:943–953. doi: 10.1016/j.bbadis.2018.06.023. [DOI] [PubMed] [Google Scholar]
- 39.Ganeshalingam M., Enstad S., Sen S., et al. Role of lipidomics in assessing the functional lipid composition in breast milk. Front Nutr. 2022;9 doi: 10.3389/fnut.2022.899401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ward Platt M., Deshpande S. Metabolic adaptation at birth. Semin Fetal Neonatal Med. 2005;10:341–350. doi: 10.1016/j.siny.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 41.Reily C., Stewart T.J., Renfrow M.B., et al. Glycosylation in health and disease. Nat Rev Nephrol. 2019;15:346–366. doi: 10.1038/s41581-019-0129-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li F., Ding J. Sialylation is involved in cell fate decision during development, reprogramming and cancer progression. Protein Cell. 2019;10:550–565. doi: 10.1007/s13238-018-0597-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nji E., Gulati A., Qureshi A.A., et al. Structural basis for the delivery of activated sialic acid into Golgi for sialyation. Nat Struct Mol Biol. 2019;26:415–423. doi: 10.1038/s41594-019-0225-y. [DOI] [PubMed] [Google Scholar]
- 44.Cheng W.K., Oon C.E. How glycosylation aids tumor angiogenesis: an updated review. Biomed Pharmacother. 2018;103:1246–1252. doi: 10.1016/j.biopha.2018.04.119. [DOI] [PubMed] [Google Scholar]
- 45.Park E.J., Grabińska K.A., Guan Z., et al. NgBR is essential for endothelial cell glycosylation and vascular development. EMBO Rep. 2016;17:167–177. doi: 10.15252/embr.201540789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.D'Addio M., Frey J., Otto V.I. The manifold roles of sialic acid for the biological functions of endothelial glycoproteins. Glycobiology. 2020;30:490–499. doi: 10.1093/glycob/cwaa008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kitazume S., Imamaki R., Ogawa K., et al. Alpha2,6-sialic acid on platelet endothelial cell adhesion molecule (PECAM) regulates its homophilic interactions and downstream antiapoptotic signaling. J Biol Chem. 2010;285:6515–6521. doi: 10.1074/jbc.M109.073106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chandler K.B., Costello C.E., Rahimi N. Glycosylation in the tumor microenvironment: implications for tumor angiogenesis and metastasis. Cells. 2019;8:544–554. doi: 10.3390/cells8060544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li J., Gao J., Jiang M., et al. Rat liver sinusoidal surface N-linked glycoproteomic analysis by affinity enrichment and mass spectrometric identification. Biochemistry (Mosc) 2015;80:260–275. doi: 10.1134/S0006297915030025. [DOI] [PubMed] [Google Scholar]
- 50.Clark S.A., Angus H.B., Cook H.B., et al. Defenestration of hepatic sinusoids as a cause of hyperlipoproteinaemia in alcoholics. Lancet. 1988;2:1225–1227. doi: 10.1016/s0140-6736(88)90813-6. [DOI] [PubMed] [Google Scholar]
- 51.Carpenter B., Lin Y., Stoll S., et al. VEGF is crucial for the hepatic vascular development required for lipoprotein uptake. Development. 2005;132:3293–3303. doi: 10.1242/dev.01902. [DOI] [PubMed] [Google Scholar]
- 52.Hagberg C.E., Falkevall A., Wang X., et al. Vascular endothelial growth factor B controls endothelial fatty acid uptake. Nature. 2010;464:917–921. doi: 10.1038/nature08945. [DOI] [PubMed] [Google Scholar]
- 53.Herrnberger L., Hennig R., Kremer W., et al. Formation of fenestrae in murine liver sinusoids depends on plasmalemma vesicle-associated protein and is required for lipoprotein passage. PLoS One. 2014;9 doi: 10.1371/journal.pone.0115005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pandey E., Nour A.S., Harris E.N. Prominent receptors of liver sinusoidal endothelial cells in liver homeostasis and disease. Front Physiol. 2020;11:873. doi: 10.3389/fphys.2020.00873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ruan B., Duan J.L., Xu H., et al. Capillarized liver sinusoidal endothelial cells undergo partial endothelial-mesenchymal transition to actively deposit sinusoidal ECM in liver fibrosis. Front Cell Dev Biol. 2021;9 doi: 10.3389/fcell.2021.671081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kus E., Kaczara P., Czyzynska-Cichon I., et al. LSEC fenestrae are preserved despite pro-inflammatory phenotype of liver sinusoidal endothelial cells in mice on high fat diet. Front Physiol. 2019;10:6. doi: 10.3389/fphys.2019.00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Paris J., Henderson N.C. Liver zonation, revisited. Hepatology. 2022;76:1219–1230. doi: 10.1002/hep.32408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang X., Bove A.M., Simone G., et al. Molecular bases of VEGFR-2-mediated physiological function and pathological role. Front Cell Dev Biol. 2020;8 doi: 10.3389/fcell.2020.599281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chandler K.B., Leon D.R., Kuang J., et al. N-glycosylation regulates ligand-dependent activation and signaling of vascular endothelial growth factor receptor 2 (VEGFR2) J Biol Chem. 2019;294:13117–13130. doi: 10.1074/jbc.RA119.008643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fong T.A., Shawver L.K., Sun L., et al. SU5416 is a potent and selective inhibitor of the vascular endothelial growth factor receptor (Flk-1/KDR) that inhibits tyrosine kinase catalysis, tumor vascularization, and growth of multiple tumor types. Cancer Res. 1999;59:99–106. [PubMed] [Google Scholar]
- 61.Honda M., Asai T., Umemoto T., et al. Suppression of choroidal neovascularization by intravitreal injection of liposomal SU5416. Arch Ophthalmol. 2011;129:317–321. doi: 10.1001/archophthalmol.2011.12. [DOI] [PubMed] [Google Scholar]
- 62.Edelbauer M., Datta D., Vos I.H., et al. Effect of vascular endothelial growth factor and its receptor KDR on the transendothelial migration and local trafficking of human T cells in vitro and in vivo. Blood. 2010;116:1980–1989. doi: 10.1182/blood-2009-11-252460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fu J., Gerhardt H., McDaniel J.M., et al. Endothelial cell O-glycan deficiency causes blood/lymphatic misconnections and consequent fatty liver disease in mice. J Clin Invest. 2008;118:3725–3737. doi: 10.1172/JCI36077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ma X., Li Y., Kondo Y., et al. Slc35a1 deficiency causes thrombocytopenia due to impaired megakaryocytopoiesis and excessive platelet clearance in the liver. Haematologica. 2021;106:759–769. doi: 10.3324/haematol.2019.225987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hennet T., Chui D., Paulson J.C., et al. Immune regulation by the ST6Gal sialyltransferase. Proc Natl Acad Sci U S A. 1998;95:4504–4509. doi: 10.1073/pnas.95.8.4504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hellbusch C.C., Sperandio M., Frommhold D., et al. Golgi GDP-fucose transporter-deficient mice mimic congenital disorder of glycosylation IIc/leukocyte adhesion deficiency II. J Biol Chem. 2007;282:10762–10772. doi: 10.1074/jbc.M700314200. [DOI] [PubMed] [Google Scholar]
- 67.Stanley P. What have we learned from glycosyltransferase knockouts in mice? J Mol Biol. 2016;428:3166–3182. doi: 10.1016/j.jmb.2016.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Martinez-Duncker I., Dupré T., Piller V., et al. Genetic complementation reveals a novel human congenital disorder of glycosylation of type II, due to inactivation of the Golgi CMP-sialic acid transporter. Blood. 2005;105:2671–2676. doi: 10.1182/blood-2004-09-3509. [DOI] [PubMed] [Google Scholar]
- 69.Mohamed M., Ashikov A., Guillard M., et al. Intellectual disability and bleeding diathesis due to deficient CMP-sialic acid transport. Neurology. 2013;81:681–687. doi: 10.1212/WNL.0b013e3182a08f53. [DOI] [PubMed] [Google Scholar]
- 70.Su X., Shi Y., Zou X., et al. Single-cell RNA-seq analysis reveals dynamic trajectories during mouse liver development. BMC Genomics. 2017;18:946. doi: 10.1186/s12864-017-4342-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cui J.Y., Renaud H.J., Klaassen C.D. Ontogeny of novel cytochrome P450 gene isoforms during postnatal liver maturation in mice. Drug Metab Dispos. 2012;40:1226–1237. doi: 10.1124/dmd.111.042697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang X., Sessa W.C., Fernandez-Hernando C. Endothelial transcytosis of lipoproteins in atherosclerosis. Front Cardiovasc Med. 2018;5:130. doi: 10.3389/fcvm.2018.00130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hammoutene A., Rautou P.E. Role of liver sinusoidal endothelial cells in non-alcoholic fatty liver disease. J Hepatol. 2019;70:1278–1291. doi: 10.1016/j.jhep.2019.02.012. [DOI] [PubMed] [Google Scholar]
- 74.Jungermann K., Katz N. Functional specialization of different hepatocyte populations. Physiol Rev. 1989;69:708–764. doi: 10.1152/physrev.1989.69.3.708. [DOI] [PubMed] [Google Scholar]
- 75.Leibing T., Geraud C., Augustin I., et al. Angiocrine Wnt signaling controls liver growth and metabolic maturation in mice. Hepatology. 2018;68:707–722. doi: 10.1002/hep.29613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Su T., Yang Y., Lai S., et al. Single-cell transcriptomics reveals zone-specific alterations of liver sinusoidal endothelial cells in cirrhosis. Cell Mol Gastroenterol Hepatol. 2021;11:1139–1161. doi: 10.1016/j.jcmgh.2020.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.de Haan W., Øie C., Benkheil M., et al. Unraveling the transcriptional determinants of liver sinusoidal endothelial cell specialization. Am J Physiol Gastrointest Liver Physiol. 2020;318:G803–G815. doi: 10.1152/ajpgi.00215.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nagy D., Maude H., Birdsey G.M., et al. RISING STARS: liver sinusoidal endothelial transcription factors in metabolic homeostasis and disease. J Mol Endocrinol. 2023;71 doi: 10.1530/JME-23-0026. [DOI] [PubMed] [Google Scholar]
- 79.Shi H., Gao L., Kirby N., et al. Clearance of VWF by hepatic macrophages is critical for the protective effect of ADAMTS13 in sickle cell anemia mice. Blood. 2024;143(13):1293–1309. doi: 10.1182/blood.2023021583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mousavi S.A., Skjeldal F., Fonhus M.S., et al. Receptor-mediated endocytosis of VEGF-A in rat liver sinusoidal endothelial cells. Biomed Res Int. 2019;2019 doi: 10.1155/2019/5496197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Funyu J., Mochida S., Inao M., et al. VEGF can act as vascular permeability factor in the hepatic sinusoids through upregulation of porosity of endothelial cells. Biochem Biophys Res Commun. 2001;280:481–485. doi: 10.1006/bbrc.2000.4148. [DOI] [PubMed] [Google Scholar]
- 82.Coulon S., Francque S., Colle I., et al. Evaluation of inflammatory and angiogenic factors in patients with non-alcoholic fatty liver disease. Cytokine. 2012;59:442–449. doi: 10.1016/j.cyto.2012.05.001. [DOI] [PubMed] [Google Scholar]
- 83.Coulon S., Legry V., Heindryckx F., et al. Role of vascular endothelial growth factor in the pathophysiology of nonalcoholic steatohepatitis in two rodent models. Hepatology. 2013;57:1793–1805. doi: 10.1002/hep.26219. [DOI] [PubMed] [Google Scholar]
- 84.Liang Y., Kaneko K., Xin B., et al. Temporal analyses of postnatal liver development and maturation by single-cell transcriptomics. Dev Cell. 2022;57:398–414.e5. doi: 10.1016/j.devcel.2022.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Auvinen K., Lokka E., Mokkala E., et al. Fenestral diaphragms and PLVAP associations in liver sinusoidal endothelial cells are developmentally regulated. Sci Rep. 2019;9 doi: 10.1038/s41598-019-52068-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gage B.K., Liu J.C., Innes B.T., et al. Generation of functional liver sinusoidal endothelial cells from human pluripotent stem-cell-derived venous angioblasts. Cell Stem Cell. 2020;27:254–269.e9. doi: 10.1016/j.stem.2020.06.007. [DOI] [PubMed] [Google Scholar]
- 87.Johnson B.M., Johnson A.M., Heim M., et al. Biomechanical stimulation promotes blood vessel growth despite VEGFR-2 inhibition. BMC Biol. 2023;21:290. doi: 10.1186/s12915-023-01792-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mezrich J.D., Nguyen L.P., Kennedy G., et al. SU5416, a VEGF receptor inhibitor and ligand of the AHR, represents a new alternative for immunomodulation. PLoS One. 2012;7 doi: 10.1371/journal.pone.0044547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Croci D.O., Cerliani J.P., Dalotto-Moreno T., et al. Glycosylation-dependent lectin-receptor interactions preserve angiogenesis in anti-VEGF refractory tumors. Cell. 2014;156:744–758. doi: 10.1016/j.cell.2014.01.043. [DOI] [PubMed] [Google Scholar]
- 90.Chandler K.B., Leon D.R., Meyer R.D., et al. Site-specific N-glycosylation of endothelial cell receptor tyrosine kinase VEGFR-2. J Proteome Res. 2017;16:677–688. doi: 10.1021/acs.jproteome.6b00738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chiodelli P., Rezzola S., Urbinati C., et al. Contribution of vascular endothelial growth factor receptor-2 sialylation to the process of angiogenesis. Oncogene. 2017;36:6531–6541. doi: 10.1038/onc.2017.243. [DOI] [PubMed] [Google Scholar]
- 92.Hipgrave Ederveen A.L., de Haan N., Baerenfaenger M., et al. Dissecting total plasma and protein-specific glycosylation profiles in congenital disorders of glycosylation. Int J Mol Sci. 2020;21:7635–7643. doi: 10.3390/ijms21207635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhong C., Li P., Argade S., et al. Inhibition of protein glycosylation is a novel pro-angiogenic strategy that acts via activation of stress pathways. Nat Commun. 2020;11:6330. doi: 10.1038/s41467-020-20108-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lee C., Liu A., Miranda-Ribera A., et al. NEU1 sialidase regulates the sialylation state of CD31 and disrupts CD31-driven capillary-like tube formation in human lung microvascular endothelia. J Biol Chem. 2014;289:9121–9135. doi: 10.1074/jbc.M114.555888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Tang Y., Harrington A., Yang X., et al. The contribution of the Tie2+ lineage to primitive and definitive hematopoietic cells. Genesis. 2010;48:563–567. doi: 10.1002/dvg.20654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Naranjo O., Osborne O., Torices S., et al. In vivo targeting of the neurovascular unit: challenges and advancements. Cell Mol Neurobiol. 2022;42:2131–2146. doi: 10.1007/s10571-021-01113-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Werner E.R., Keller M.A., Sailer S., et al. A novel assay for the introduction of the vinyl ether double bond into plasmalogens using pyrene-labeled substrates. J Lipid Res. 2018;59:901–909. doi: 10.1194/jlr.D080283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Narváez-Rivas M., Zhang Q. Comprehensive untargeted lipidomic analysis using core–shell C30 particle column and high field orbitrap mass spectrometer. J Chromatogr A. 2016;1440:123–134. doi: 10.1016/j.chroma.2016.02.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Shen W., Le S., Li Y., Hu F. SeqKit: A Cross-Platform and Ultrafast Toolkit for FASTA/Q File Manipulation. PloS one. 2016;11(10) doi: 10.1371/journal.pone.0163962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kim D., Langmead B., Salzberg S.L. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Mortazavi A., Williams B.A., McCue K., et al. Mapping and quantifying mammalian transcriptomes by RNA-seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 102.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Subramanian A., Tamayo P., Mootha V.K., et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]