Abstract

EIDD-1931 is the active form of molnupiravir, an orally effective drug approved by the United States Food and Drug Administration (USFDA) against COVID-19. Pharmacokinetic alteration can cause untoward drug interaction (drug–drug/disease–drug), but hardly any information is known about this recently approved drug. Therefore, we first investigated the impact of the arthritis state on the oral pharmacokinetics of EIDD-1931 using a widely accepted complete Freund’s adjuvant (CFA)-induced rat model of rheumatoid arthritis (RA) after ascertaining the disease occurrence by paw swelling measurement and X-ray examination. Comparative oral pharmacokinetic assessment of EIDD-1931 (normal state vs arthritis state) showed that overall plasma exposure was augmented (1.7-fold) with reduced clearance (0.54-fold), suggesting its likelihood of dose adjustment in arthritis conditions. In order to elucidate the effect of EIDD-1931 treatment at a therapeutic regime (normal state vs arthritis state) on USFDA-recommended panel of cytochrome P450 (CYP) enzymes (CYP1A2, CYP2B6, CYP2C8, CYP2C9, CYP2C19, CYP2D6, and CYP3A4) for drug interaction using the same disease model, we monitored protein and mRNA expressions (rat homologs) in liver tissue by western blotting (WB) and real time-polymerase chain reaction (RT-PCR), respectively. Results reveal that EIDD-1931 treatment could strongly influence CYP3A4 and CYP2C8 among experimental proteins/mRNAs. Although CYP2C8 regulation upon EIDD-1931 treatment resembles similar behavior under the arthritis state, results dictate a potentially reverse phenomenon for CYP3A4. Moreover, the lack of any CYP inhibitory effect by EIDD-1931 in human/rat liver microsomes (HLM/RLM) helps to ascertain EIDD-1931 treatment-mediated disease–drug interaction and the possibility of drug–drug interaction with disease-modifying antirheumatic drugs (DMARDs) upon coadministration. As elevated proinflammatory cytokine levels are prevalent in RA and nuclear factor-kappa B (NF-kB) and nuclear receptors control CYP expressions, further studies should focus on understanding the regulation of affected CYPs to subside unexpected drug interaction.
1. Introduction
Rheumatoid arthritis (RA) is a chronic inflammatory and autoimmune disorder characterized by severe joint inflammation.1 As per the World Health Organization (WHO) report, the prevalence of RA in different populations varies from less than 1% to almost 5%.2 Moreover, a recent analysis of Global Burden of Disease (GBD) 2019 data on rheumatic diseases showed that approximately 1.71 billion people worldwide have musculoskeletal conditions, where RA, a systemic musculoskeletal disease, is the significant contributor that affects around 18 million people globally with 2.4 million individuals having years lived with disability (YLD).3 Moreover, RA has the highest prevalence in India among the developing countries.4 During the disease progression of RA, various cytokines play a crucial role in exacerbating the inflammatory situation.5 It is established at in vitro/in vivo (preclinical/clinical) levels that inflammatory cytokines alter cytochrome P450 (CYP) expressions, which can lead to severe consequences.6,7 CYPs are responsible for the biotransformation of drugs. A combination of antiarthritic drugs like disease-modifying antirheumatic drugs (DMARDs)8 and nonsteroidal anti-inflammatory drugs (NSAIDs)9 is used simultaneously to curb the disease pathophysiology and signs/symptoms of RA, respectively. In addition, multiple drugs are used to tackle several comorbidities of RA patients.10 Under these circumstances, there are higher chances of a drug interaction that can either lead to therapeutic failure by lessening plasma exposure of the drug or precipitate severe dose-dependent adverse effects by enhancing plasma exposure of the drug.11 In this context, DMARDs, whether of synthetic or biological class, are vulnerable to causing severe side effects and are often needed for therapeutic drug monitoring, and subsequently, dose adjustment is required.11,12 Therefore, to get the desired therapeutic outcome, the CYP’s modulation in RA during the therapy of any new drug must be understood under the frame of drug interaction.
Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) infection has recently shaken the world due to the significant public health crisis from the coronavirus disease 2019 (COVID-19) pandemic.13 Molnupiravir (Figure 1) (EIDD-2801 or MK-4482) is the most effective oral medication for treating mild-to-moderate degrees of COVID-19 infection in nonhospitalized patients.14 Several countries have given emergency authorization for its use.15 EIDD-1931 (Figure 1) (β-d-N4-hydroxycytidine) is the active form of molnupiravir (prodrug) and has pronounced antiviral activity against a variety of deadly viruses like influenza A, influenza B, Ebola, respiratory syncytial, New World encephalitic alpha, chikungunya, etc.16−18 EIDD-1931 is a direct-acting antiviral agent against ribonucleic acid (RNA) viruses and has proven efficacy against remdesivir-resistant cases.19 However, hardly any information is known on the pharmacokinetic behavior of EIDD-1931 and its treatment-associated regulation of CYPs under any disease state. It could not only be beneficial for generating information regarding its dose adjustment in any particular disease state by avoiding untoward drug interaction (drug–drug/disease–drug) but also equally be beneficial to restrict the emergence of antimicrobial resistance (AMR) of such a crucial antiviral drug.20
Figure 1.
Chemical structure of molnupiravir and EIDD-1931.
In this pursuit, we aimed to investigate the pharmacokinetic behavior of EIDD-1931, followed by elucidating its impact on CYP’s regulation at protein and messenger ribonucleic acid (mRNA) levels under arthritis conditions (disease state) using a widely accepted rat model of arthritis. In parallel, we executed similar studies using healthy rats (normal state) to understand the possible influence of EIDD-1931 on its pharmacokinetics and particular CYPs.
2. Results
2.1. Affirmation of Arthritis Induction in the Rats
To investigate the pharmacokinetics and CYP expressions upon EIDD-1931 treatment in the arthritis state, the particular disease state was induced at first in rats by a widely used complete Freund’s adjuvant (CFA)-induced arthritis model.21 Two different studies were used to ascertain the occurrence of the arthritis state. Paw swelling is an external indicator of the severity of inflammation in arthritis.22 CFA treatment caused a significant increase in paw swelling by 2.2-fold (Figure 2A) compared to the left hind paw of the normal state animals. In comparison to the normal state (Figure 2B), radiological investigation of the inflamed left hind limb of CFA-treated animals showed bone proliferation and reduction in the joint space of metatarsals/phalanges affirming induction of arthritic condition (Figure 2C).21,22 Additionally, compared to the normal state (Figure 2D), the observed changes in the histopathological features of synovial tissue from the arthritic rats (Figure 2E) were desquamation of epithelial cells, synovial proliferation, and pannus formation, which authenticated the onset of the arthritic situation.21,22 We also observed similar alterations in the arthritic animals from CYP expression studies (Figure S1).
Figure 2.
Assessment of CFA-treated animals for induction of arthritis before pharmacokinetic studies of EIDD-1931. Changes in paw swelling behavior on 9th day (A); representative radiological images of left hind paw for the normal state (B) and the arthritis state (C); H&E-stained images of bone joints in the normal state (D) and the arthritis state (E), where arrow indicators dictate as follows: black—desquamation of epithelial cells, blue—synovial proliferation, and green—pannus formation. Data are represented as mean ± SEM (n = 5). Data were compared between the normal state and arthritis state. Statistical significance: p <*0.05/**0.01/***0.001.
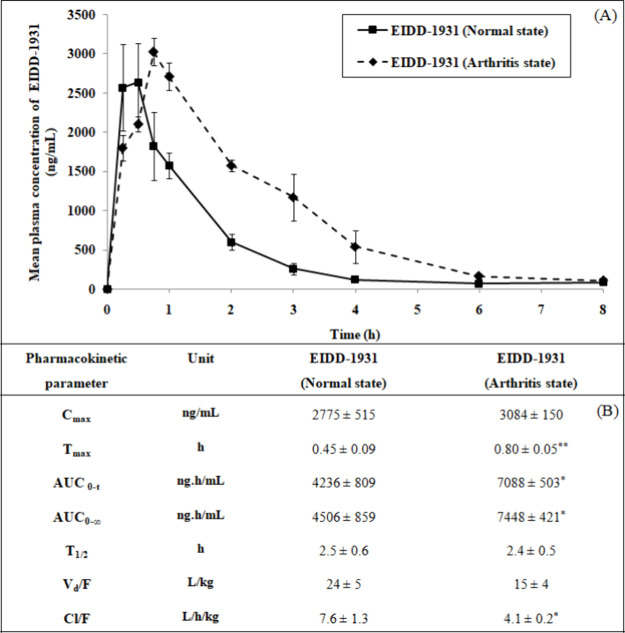
2.2. Alteration in the Pharmacokinetics of EIDD-1931 under the Arthritis State
The pharmacokinetic study of EIDD-1931 upon oral administration was executed in normal and arthritis states (Figure 3). Although the maximum plasma concentration (Cmax) of EIDD-1931 remained unaffected (1.1-fold) under the arthritis state compared to the normal state, the area under the curve (AUC) of EIDD-1931 was enhanced markedly (1.7-fold) along with a decline (0.54-fold) in the clearance after oral administration (Cl/F). In the arthritis state, the time to reach Cmax (Tmax) was notably increased (1.8-fold) compared to the normal state. There was a statistically insignificant alteration in the volume of distribution after oral administration (Vd/F) and elimination half-life (T1/2) of EIDD-1931 in the arthritis situation compared to normal conditions.
Figure 3.
Mean plasma concentration vs time profile (A) and pharmacokinetic parameters (B) of EIDD-1931 after oral administration in rats under the normal state and arthritis state. Data are presented as mean ± SEM (n = 5). Data were compared between the normal state and arthritis state. Statistical significance: *p <0.05.
2.3. Amendment in the Protein Expression of CYPs upon EIDD-1931 Treatment in the Arthritis State
Western blotting (WB) was performed using liver tissues of various study groups to determine the impact of EIDD-1931 treatment on the protein expression of experimental CYPs (Figure 4). Compared to the control group (normal state), the protein expression of the major CYPs in the control group (arthritis state) was downregulated in the following order: CYP3A4 ∼ CYP2C8 (0.3-fold) > CYP2C11 (0.6-fold) > CYP2C19 ∼ CYP1A2 (0.9-fold). However, upregulation was observed for the protein expression of CYP2B6 (1.1-fold) > CYP2D6 (1.01-fold) in the control group (arthritis state) compared to the control group (normal state). Upon EIDD-1931 treatment in the normal state compared to the control group (normal state), the protein expression for CYP enzymes was downregulated in the following order: CYP3A4 (0.4-fold) > CYP1A2 ∼ CYP2C8 (0.6-fold) > CYP2C11 (0.7-fold), while protein expression for CYP2B6 (1.5-fold) > CYP2C19 (1.3-fold) > CYP2D6 (1.1-fold) was upregulated. When EIDD-1931 was given to the arthritic animals, the downregulation in the protein expression for CYP enzymes was observed compared to the control group (arthritis state) in the following order: CYP2C8 (0.3-fold) > CYP2D6 (0.8-fold) > CYP2C11 ∼ CYP2C19 (0.9-fold). Conversely, CYP3A4 (2.3-fold) > CYP2B6 (1.6-fold) > CYP1A2 (1.5-fold) was upregulated in the same setup.
Figure 4.
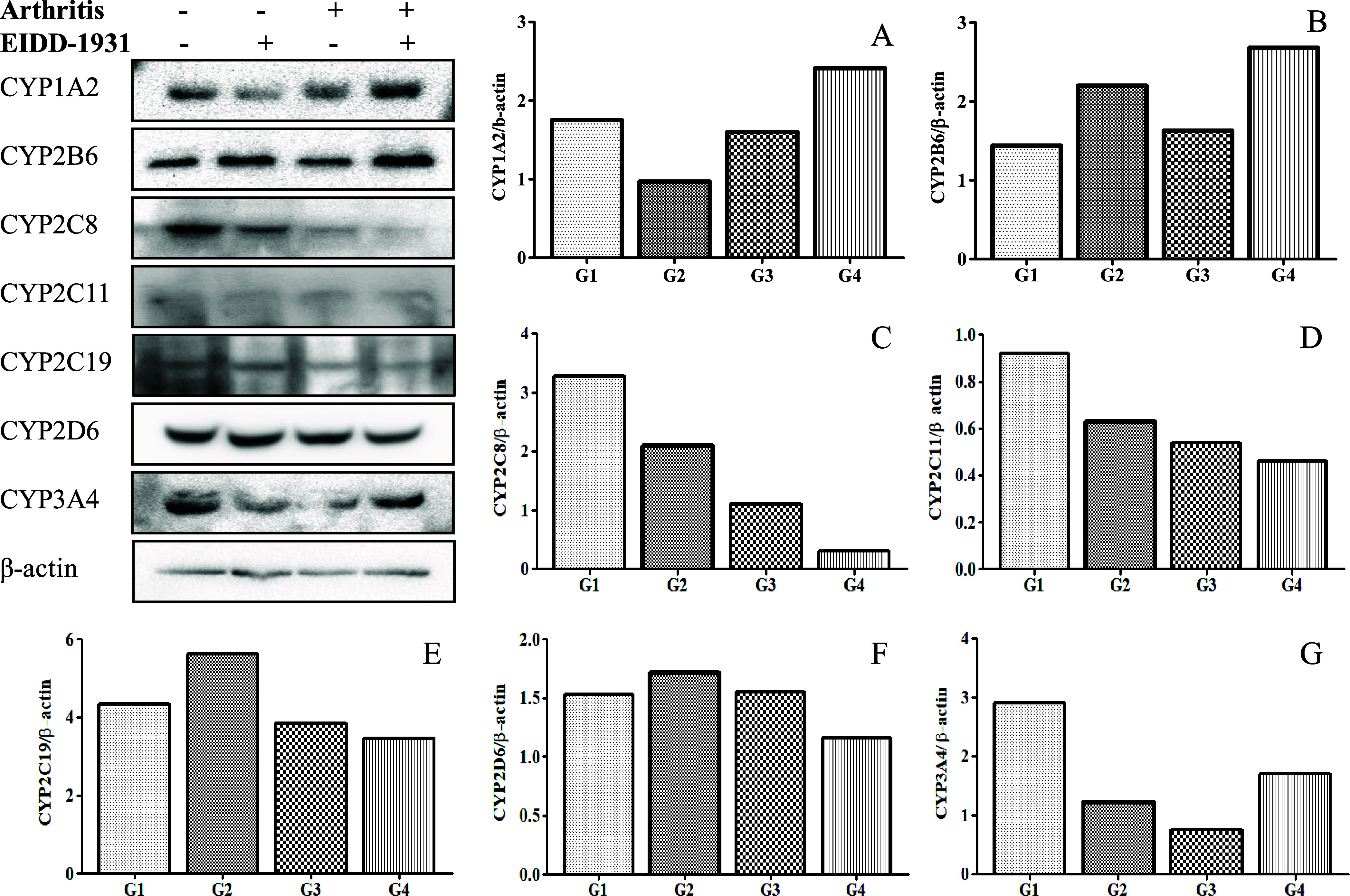
Western blot analysis with densitometry data for the protein expressions in the hepatic tissues of the animals in the different study groups using β-actin as a loading control: (A) CYP1A2, (B) CYP2B6, (C) CYP2C8, (D) CYP2C11, (E) CYP2C19, (F) CYP2D6, and (G) CYP3A4. G1 represents the control group (normal state), G2 represents the treated group (normal state), G3 represents the control group (arthritis state), and G4 represents the treated group (arthritis state).
2.4. Modulation in the mRNA Expression of CYPs upon EIDD-1931 Treatment in the Arthritis State
Reverse transcription real-time PCR was performed using the liver tissues of different study groups to assess the impact of EIDD-1931 treatment on the mRNA expression of various CYPs in the arthritis state (Figure 5). In the control group (arthritis state), the mRNA expression of the investigated CYPs was downregulated compared to the control group (normal state) in the following order: CYP2D1 ∼ CYP3A18 (below 0.1-fold) > CYP2C13 (0.1-fold) > CYP2C11 (0.2-fold) > CYP2B1 (0.3-fold) > CYP1A2 (0.4-fold) > CYP3A1 ∼ CYP2B2 (0.6-fold) > CYP3A9 (1.1-fold). Further, downregulation in the mRNA expression of the experimental CYPs was observed upon EIDD-1931 treatment in the treated group (normal state) compared to the control group (normal state) in the following order: CYP2D1 ∼ CYP3A18 ∼ CYP2C13 ∼ CYP1A2 ∼ CYP3A9 (below 0.1-fold) > CYP2B2 ∼ CYP2C11 (0.1-fold) > CYP3A1 ∼ CYP2B1 (0.2-fold). Upon EIDD-1931 treatment to arthritic animals compared to the control group (arthritis state), the mRNA expressions for CYPs were downregulated as follows: CYP2B1 ∼ CYP3A1 ∼ CYP2C11 (0.4-fold) > CYP3A9 (0.5-fold) > CYP2C13 ∼ CYP1A2 (0.6-fold) > CYP2B2 (0.8-fold) > CYP2D3 (0.9-fold). However, CYP3A18 (3.3-fold) and CYP2D1 (1.8-fold) were upregulated in the same setup.
Figure 5.
RT-PCR analysis for the mRNA expressions in the hepatic tissues of the animals in the different study groups using GAPDH as a loading control: (A) CYP1A2, (B) CYP2B1, (C) CYP2B2, (D) CYP2C11, (E) CYP2C13 (F) CYP2D1, (G) CYP3A1, (H) CYP3A18, and (I) CYP3A9. G1 represents the control group (normal state), G2 represents the treated group (normal state), G3 represents the control group (arthritis state), and G4 represents the treated group (arthritis state).
2.5. Lack of a Strong CYP Inhibitory Action of EIDD-1931
The CYP inhibitory action of EIDD-1931 was evaluated using the United States Food and Drug Administration (USFDA)-recommended CYP-specific marker reactions (in vitro) in human liver microsomes (HLM). The results obtained for standard inhibitors corroborate with the literature reports (Table 1).23,24 IC50 >10 μM is considered a weak or noninhibitor for any CYP isoforms. EIDD-1931 showed minimal inhibitory action on all experimental CYPs (IC50 >100 μM for all, except IC50: 74 μM for CYP1A2) (Table 1). Similarly EIDD-1931 showed an insignificant inhibitory effect on CYP3A4 and CYP2C8 using rat liver microsomes (RLM) (Figure S2).
Table 1. CYP Inhibitory Effect of EIDD-1931 in HLM.
| CYP isoform | EIDD-1931 (IC50) | standard inhibitor (IC50) |
|---|---|---|
| CYP1A2 | 74 μM | 0.39 μM (fluvoxamine) |
| CYP2B6 | >100 μM | 0.05 μM (ticlopidine) |
| CYP2C8 | >100 μM | 6.2 μM (quercetin) |
| CYP2C9 | >100 μM | 1.28 μM (sulfaphenazole) |
| CYP2C19 | >100 μM | 7.1 μM (tranylcypromine) |
| CYP2D6 | >100 μM | 0.02 μM (quinidine) |
| CYP3A4 | >100 μM | 0.02 μM (ketoconazole) |
3. Discussion
In the quest to explore the impact of EIDD-1931 treatment on alteration in the expression of CYP isoforms, we first studied the effect of the arthritis state on the pharmacokinetics of EIDD-1931 after single-dose oral administration to CFA-induced arthritic rats. We observed a substantially augmented plasma exposure of EIDD-1931 in the arthritis state compared to the normal state. A similar line of effect was also reported on the pharmacokinetics of verapamil (↑ AUC: 1.6-fold; ↓ Cl/F: 35%) under the arthritis state.25 Likewise, upadacitinib, an antiarthritic drug (Janus kinases/Signal transducer and activator of transcription proteins or JAK/STAT inhibitor), reduced the clearance behavior in human under the arthritic condition.26 Hence, dose adjustment is likely needed for EIDD-1931, and further studies should be conducted to assess the corresponding required dose. It is well-known that, in arthritis conditions, the level of inflammatory cytokines is elevated (Figure S3), which are reported to modulate the activity of drug metabolizing enzymes.27,28 The results of the present study displayed a marked decline in the clearance behavior of EIDD-1931, which suggests that it is possibly due to the impediment of the inflammation-linked CYP activities. Literature evidence also advocates that both protein and mRNA expressions of CYPs are lowered in the arthritis situation.28−30 Therefore, we looked into the effect of EIDD-1931 on all the CYP isoforms (recommended by the USFDA for CYP inhibition-mediated drug interaction) in the liver tissues using a preclinical model to investigate drug–drug interaction potential. Compared to the normal state, we observed a decreasing trend in the protein and mRNA expressions of CYPs in the arthritis state, which aligns with the available literature on specific CYP isoforms.29−31 However, results showed a varying degree of effect on the various CYPs, which corroborates with the reported effect under the arthritis state.29,32 It is also evidenced in the literature that the level of expression (protein/mRNA) for a particular CYP level could vary depending on the experimental time frame of sampling.29,31 Considering the effect on our experimental CYPs, results displayed the strongest influence (≥0.5-fold) of the arthritis state on CYP3A4 and CYP2C8 (protein expression), and CYP3A18, CYP2C11, CYP2C13, CYP2D1, CYP2B1, and CYP1A2 (mRNA expression). As mentioned above, a similar line of effect on the protein/mRNA expressions is also reported in the literature for the CFA-induced rat model of RA.29,30
Exploration of CYP expressions upon EIDD-1931 treatment to the arthritic rats was done to identify the influence on particular CYP isoforms (Figure 6). Results displayed the highest downregulating effect on the protein expression of CYP2C8 (CYP2C11/CYP2C19) compared to any other experimental CYP isoforms. The obtained results resemble the protein expression data under the arthritis state and are relatively lower than the expression levels observed under normal conditions after EIDD-1931 treatment. CYP2C11 is the common rat homolog of human CYP2C8, CYP2C9, and CYP2C19.33,34 In contrast, CYP2C13 has the maximum similarity (∼70%) with the human CYP2C8.34 Our experimental results displayed a reduction in the mRNA expression of CYP2C13 (CYP2C11), which is consistent with the observed decline in the protein expression of CYP2C8.29 This similarity signifies that the effect of EIDD-1931 treatment may occur at the pretranslational or transcriptional phases.35−37 Although limited data on CYP2C13 expression in RA exist in the literature, a similar impact on CYP2C13 was observed in the case of inflammation associated with diabetes.38 RA is associated with elevated levels of proinflammatory cytokines, like Interleukin-6 (IL-6) (Figure S3), which are reported to downregulate the CYP2C enzymes in rat and human hepatocytes.32,39 Alterations in inflammatory cytokines have been reported to regulate nuclear receptors, thereby modulating the CYPs in various inflammatory disorders, including RA.40 In the present study, we observed the downregulating and upregulating effects on constitutive androstane receptor (CAR) protein and mRNA expressions, respectively, after EIDD-1931 treatment in the arthritis state (Figures S3 and S4). Hydroxychloroquine is one of the effective DMARDs, but its treatment is associated with significant dose-dependent side effects like cardiac myopathy, retinal damage, corneal opacity, and many more.41,42 CYP2C8 plays a crucial role in its metabolism in the liver.43 Therefore, EIDD-1931 treatment-mediated decline in CYP2C8 expression in RA can have a pronounced upshot on plasma exposure and its treatment-linked toxic effects. Furthermore, the CYP2C family of enzymes metabolizes most of the NSAIDs that are prescribed to relieve signs/symptoms of inflammation in RA.44−46 Thus, EIDD-1931 treatment in RA can cause potential CYP2C-mediated drug interaction, and further studies are needed to assess its likelihood of precipitating untoward effects. In addition to the CYP2C, EIDD-1931 treatment in RA exhibited a minimal ramification in lowering the protein expression (<0.5-fold) of CYP2D6. The rat homolog of human CYP2D6 is CYP2D1, which has a similarity of >70% to the human gene.34 In the present study, we observed an upregulating effect in the mRNA expression of CYP2D1. However, mRNA expression of another rat homolog of human CYP2D6, i.e., CYP2D3, was downregulated similarly (Figure S4). The observed effect of EIDD-1931 treatment on the protein expression in the arthritis state is inversely correlated to the impact in normal conditions. The above changes imply the likelihood of involvement of multiple mechanisms at transcriptional and translational levels in regulating CYP2D6.29,32,47,48 CYP2D6 comprises around 5% of the total CYP enzymes and is responsible for the metabolism of 25% of all clinical drugs.7 In short, it is unlikely to cause drug interaction due to concomitant therapy with EIDD-1931 in RA.
Figure 6.
Pictorial representation of the impact of EIDD-1931 in the arthritis state toward drug interaction.
Considering the outcome of EIDD-1931 treatment to the arthritic rats on upregulation of CYP expressions, our experimental results depicted a considerable effect on the protein expression of CYP3A4. The rat homolog of human CYP3A4 is CYP3A1, having >75% similarity,33,34,49 for which mRNA expression notably declined. On the contrary, CYP3A18 is the predominant rat homolog of human CYP3A5,33 and the mRNA expression of CYP3A18 was enhanced. It helps to differentiate the effect of CYP3A4 from CYP3A5. There was a decrease in the mRNA expression of CYP3A9, another rat homolog of human CYP3A4, supporting the action on the CYP3A4. The results of protein and mRNA expressions of CYP3A4 dictate the possible involvement of multiple mechanisms at transcriptional or translational phases.29,32,47,50 Thus, we observed a decreased protein expression of CYP3A4 in arthritic rats (without EIDD-1931 treatment), but upon EIDD-1931 treatment, the protein expression behavior reversed. In this context, CYP3A4 comprises 60% of the total CYP enzymes present in the body and is responsible for the metabolism of more than 50% of the xenobiotics.51 Moreover, synthetic DMARDs from the class of JAK/STAT inhibitors (tofacitinib, baricitinib, and upadacitinib) and typical DMARDs like cyclosporine are mainly metabolized by CYP3A4 in the liver.52−54 Therefore, EIDD-1931 treatment-mediated augmentation in CYP3A4 expression in RA can have an appreciable consequence on plasma exposure to the subtherapeutic level, leading to treatment failure. In addition to the CYP3A4, EIDD-1931 treatment in RA exhibited a relatively low elevation (>1.5-fold) of the protein expression of CYP2B6 (CYP3A4 > CYP2B6). The observed effect of EIDD-1931 treatment on the protein expression in the arthritis state is similar to the outcome in the normal condition. The rat homolog of human CYP2B6 is CYP2B1, having >70% similarity.34 In the present study, we observed a downregulating effect on the mRNA expression of CYP2B1. The lowering of mRNA expression is also supported by another rat homolog of human CYP2B6, i.e., CYP2B2. The opposite trend in protein and mRNA expression is likely due to regulating the CYP2B enzyme family by multiple mechanisms at posttranscriptional or translational levels.29,32,47,48 CYP2B6 makes up 6–10% of all CYP enzymes and is responsible for metabolizing clinical drugs (<5%) and a few environmental chemicals.33,34 Therefore, precaution should be taken for concomitant treatment of EIDD-1931 with any other drug (CYP2B6 substrate) in RA. Furthermore, CYP1A2 constitutes around 15% of total hepatic CYP content and is responsible for the metabolism of around 10% of clinically used drugs.55 Considering the change in the protein (1.5-fold) and mRNA (0.6-fold) expression data, EIDD-1931 showed a modest impact on regulating CYP1A2 under arthritis conditions. Based on the literature evidence, posttranscriptional regulation possibly contributes to the CYP1A2 regulation.29 Hence, the occurrence of drug interaction is unlikely due to concomitant therapy with EIDD-1931 in RA.
Other than USFDA-recommended CYP isoforms, we also studied the effect of EIDD-1931 on CYP2E1 (Figures S3 and S4). CYP2E1 comprises about 5–16% of the total CYPs in the liver and is responsible for the metabolism of 3% of the clinically used drugs. CYP2E1 plays a role in the clearance of toxic chemicals from the body.56 It is activated during toxic manifestations compared to its little role in normal conditions. In the present study, we observed a slackening of protein and mRNA expressions in the control group (arthritis state) compared to the control group (normal state) (<0.5-fold), which corroborates with the reported findings.29,30 Treatment of EIDD-1931 did not cause a marked change (<0.5-fold) in the protein and mRNA expressions in the arthritis state. Based on literature facts, posttranscriptional regulation may exist in regulating CYP2E1.29 A negligible difference was observed between arthritis and normal conditions due to EIDD-1931 treatment. Under these circumstances, EIDD-1931 treatment is unexpected to influence CYP2E1, demonstrating the least propensity to cause drug interaction.
Inflammatory disorders like RA have been associated with elevated proinflammatory cytokines, which have been reported to downregulate nuclear receptors, like CAR, aryl hydrocarbon receptor (AhR), pregnane X receptor (PXR), liver X receptor (LXR), farnesoid X receptor (FXR), etc.30−32,57,58 We observed a substantial impact on CYP3A4 and CYP2C8 in the liver tissue of the present preclinical study (in vivo). Conversely, we observed a negligible effect of EIDD-1931 on CYP inhibition in HLM (Table 1) that helps to rule out any drug–drug interaction of EIDD-1931 with drugs that are substrate of particular CYP. A similar line of an insignificant inhibitory effect on CYP3A4 and CYP2C8 was also obtained in the RLM (Figure S2). These in vitro studies (HLM/RLM) help to clarify EIDD-1931 treatment-mediated disease–drug interaction. CAR is reported to regulate the gene expression of both CYP3A4 and CYP2C8 in primary human hepatocytes.31,59 Moreover, cross-talk between CAR and PXR is reported for the same CYPs using human hepatocytes.60,61 On the other hand, nuclear factor-kappa B (NF-κB) is activated by proinflammatory cytokines and is responsible for the transcriptional regulation of a wide array of inflammatory genes. According to the present findings, we also observed a mild impact to decrease protein expression of CAR (Figures S3 and S4) due to induction of arthritis and EIDD-1931 treatment. It is evident that cross-talk exists between various nuclear receptors and CYP expression.58,62 Hence, further investigations are needed to clarify the involvement of particular nuclear receptors in modulating the activity of specific CYP isoforms. Moreover, NF-kB can regulate CYPs directly/via nuclear receptors or posttranscriptional regulation.58 Hence, correlating the role of NF-kB in regulating nuclear receptors will also be advantageous.
4. Conclusions
To elucidate the impact of the arthritis state on the pharmacokinetics of EIDD-1931 and after that effect on CYP regulation, experiments were carried out using a widely used preclinical model of RA after ascertaining the disease state. Comparative assessment of oral pharmacokinetics of EIDD-1931 (normal state vs arthritis state) suggests dose adjustment of EIDD-1931 is likely to be required in arthritis conditions. To illustrate the effect of EIDD-1931 treatment at its therapeutic regime under arthritis situations on a USFDA-recommended panel of CYPs for drug interaction, results reveal that EIDD-1931 treatment could show a strong influence on CYP3A4 (upregulation) and CYP2C8 (downregulation) leading to precipitation of drug interaction during concomitant therapy with their respective substrates. Although the regulation of CYP2C8 due to EIDD-1931 treatment resembles the particular CYP behavior under the arthritis state, results dictate a reverse phenomenon in the case of CYP3A4. Lack of a CYP inhibitory effect by EIDD-1931 in HLM/RLM helps to ascertain the occurrence of EIDD-1931 treatment-mediated disease–drug interaction as well as the possibility of drug–drug interaction with DMARDs upon concomitant therapy. Further studies should be focused on bridging the role of NF-kB and nuclear receptors, in regulating these affected CYPs to subside untoward drug interaction, leading to precipitate dose-dependent side effects or emerging AMR.
5. Materials and Methods
5.1. Chemicals and Reagents
Heat-killed Mycobacterium tuberculosis (Cat No. 231141) was obtained from DIFCO Laboratories. Incomplete Freund’s adjuvant (Cat No. SLBM9341V), diazepam (purity ≥98%), phenacetin (purity ≥98%), fluvoxamine maleate (purity ≥97%), bupropion hydrochloride (purity ≥98%), (2S,3S)-hydroxybupropion hydrochloride (purity ≥98%), ticlopidine hydrochloride (purity ≥99%), amodiaquine dihydrochloride dihydrate (purity ≥97%), N-desethylamodiaquine dihydrochloride (purity ≥95%), quercetin hydrate (purity ≥95%), diclofenac sodium (purity ≥98%), 4′-hydroxy diclofenac (purity ≥98%), S-mephenytoin (purity ≥98%), 4-hydroxymephenytoin (purity ≥98%), tranylcypromine sulfate (purity ≥97%), dextromethorphan hydrobromide (purity ≥98%), quinidine anhydrous (purity ≥80%), testosterone (purity ≥98%), 6β-hydroxy testosterone (purity ≥97%), sulfaphenazole (purity ≥98%), dextrorphan tartrate (purity ≥97%), and nicotinamide adenine dinucleotide phosphate (NADPH) tetrasodium salt (purity ≥98%) were purchased from Sigma-Aldrich. Paracetamol (purity ≥98%) and ketoconazole (purity ≥98%) were purchased from Cayman Chemicals. HLM (Lot No# PL050C-E) and RLM (Lot No# RT062-D) were obtained from Gibco. Ketamine hydrochloride was obtained from Neon Laboratories. Methanol, acetonitrile, dimethyl sulfoxide of high-performance liquid chromatography-grade and NaCl, and ethylenediaminetetraacetic acid (EDTA) tetrasodium salt of analytical grade were procured from Merck. Acetonitrile and formic acid of mass spectrometry-grade was acquired from Thermo Fisher Scientific. MgCl2.7H2O was purchased from Rankem. Ultrapure/purified water for studies was obtained from the water purification system (make: Merck-Millipore, model: Direct-Q3).
5.2. Antibodies
CYP2E1 polyclonal antibody (pAb) (Cat No. AB1252) was purchased from Sigma-Aldrich. CYP2B6 pAb (Cat No. PA5-35032), CYP2C8 pAb (Cat No. PA5-101311), CYP2C11 pAb (Cat No. PA3-034), CYP2C19 pAb (Cat No. PA5-112395), CYP2D6 pAb (Cat No. PA5-75500), and CYP3A4 pAb (Cat No. PA5-14896) were purchased from Invitrogen. CAR monoclonal antibody (mAb) (Cat No. sc-373791) and IL-6 mAb (Cat No. sc-32296) were obtained from Santa Cruz Biotechnology. CYP1A2 mAb (Cat No. 14719S) and antirabbit IgG horseradish peroxidase (HRP)-linked pAb (Cat No. 7074-S) were obtained from Cell Signaling Technology.
5.3. Animal Care
The present experimentations were carried out using male Wistar rats (10–12 weeks of age) and bred and maintained in the pathogen-free environment at our institutional animal house facility. Animals were housed in an individually ventilated cage (IVC) system made of polypropylene using chipsi wood shaving bedding material under an environmentally controlled room at typical laboratory conditions, i.e., temperature (25 ± 2 °C), humidity (50 ± 20%), and circadian cycle (12 h light/12 h dark). Animals were fed a pellet diet and had free access to purified water.
5.4. Ethical Prerequisite
The study protocols were approved by our “Institutional Animal Ethics Committee” (IAEC) (IAEC approval no. 255/79/8/2021; 316/82/2/2023). Animal experiments were performed according to the guidelines issued by the “Committee for Control and Supervision of Experiments on Animals” (CCSEA), Government of India.
5.5. Test Article
EIDD-1931 was synthesized, purified, characterized in-house, and used as a test article.63 The purity of EIDD-1931 was >97% (Figure S5).
5.6. Induction of the Arthritis State
CFA was prepared as a suspension (5 mg/mL) by triturating heat-killed Mycobacterium tuberculosis in the incomplete Freund’s adjuvant. Arthritis was induced using healthy animals by injecting 50 μL of CFA into the subplantar region of the left hind paw.22 Within 2 weeks of dosing, induction of the arthritis state was confirmed, and then the animals were used in the subsequent experiments. The investigations were performed to ascertain the initiation of the arthritis state in animals as follows: (a) measurement of the left hind paw to assess paw swelling using a digital vernier caliper (make: Generic; model: LSHAZI03590); (b) radiological examination of the left hind paw was carried out using an X-ray instrument for veterinary use (make: Siemens; model: Heliophos-D) after anesthetizing the animals with ketamine hydrochloride (100 mg/kg) via the intraperitoneal route.22,64 In addition to that, the histopathological examination of the left hind limb was done at the end of the pharmacokinetic study using the hematoxylin and eosin (H&E) staining procedure followed by slides evaluation under a light microscope (make: Magnus; model: INVI).22 The studies mentioned above were also executed using the left hind paws of healthy animals in parallel for comparative assessment.
5.7. Effect of the Arthritis State on the Pharmacokinetics of EIDD-1931
5.7.1. Study Design
The pharmacokinetic study of EIDD-1931 was performed in animals under the normal state (healthy rats) and the disease state (arthritic rats) via the oral route. Considering earlier preclinical reports,17,65 the dose of EIDD-1931 was selected at 30 mg/kg and prepared freshly using normal saline (100%, v/v) as a vehicle at a dose volume of 10 mL/kg. Each study arm involved five animals. Study arms were as follows: group 1: normal state and group 2: arthritis state. After dosing of overnight (∼10 h) fasted animals with single-dose of EIDD-1931 through oral gavage, blood samples were collected from retro-orbital plexus at 0.25, 0.5, 0.75, 1, 2, 3, 4, 6, and 8 h into microcentrifuge tubes containing 5% aqueous EDTA solution (w/v) as an anticoagulant. Samples were centrifuged at 8000 rpm for 10 min to obtain plasma (50 μL) and stored in a deep freezer (−80 °C) until bioanalysis. At the end of the study, animals were sacrificed using carbon dioxide euthanasia followed by cervical dislocation to collect left hind limbs, which were fixed in 10% neutral buffered formalin solution before further processing for histopathological examination.
5.7.2. Bioanalysis
Quantitation of EIDD-1931 in plasma was done after processing of samples using a plasma protein precipitation technique and analyzed by a matrix match calibration curve (25–5000 ng/mL) using our in-house developed and validated liquid chromatography-tandem mass spectrometry (LC-MS/MS) method (Figure S6).24,66
5.7.3. Pharmacokinetic Calculations
Mean plasma concentration data at corresponding time points were analyzed using the noncompartmental method by PK solution software (Summit Research Services, USA). The following pharmacokinetic parameters were calculated: T1/2, Cmax, Tmax, AUC for plasma concentration from time zero to last measurable concentration (AUC0–t) and infinity (AUC0–∞), Vd/F, and Cl/F.
5.8. Effect of EIDD-1931 Treatment on CYP Expressions in the Arthritis State
5.8.1. Study Design
The impact of EIDD-1931 treatment on the expression of various CYP isoforms in the arthritis state (arthritic rats) was evaluated in parallel to the normal state (healthy rats). Considering its therapeutic human dose (800 mg, twice daily, 5 days), the treatment regime in rats was selected as follows: (a) oral route, (b) dose of 80 mg/kg, (c) twice a day at around 8 am and 8 pm daily, and (d) consecutive treatment for 5 days. Dose formulation (vehicle) and dose volume were the same as mentioned above (Section 5.7.1). Each study arm involved three animals. Study arms were as follows: group 1: control (normal state), group 2: treated (normal state), group 3: control (arthritis state), and group 4: treated (arthritis state). Groups 2 and 4 were treated with EIDD-1931 at the above-mentioned treatment regime. After 24 h of the last treatment with EIDD-1931, animals were sacrificed to collect organ (liver), snap-frozen under liquid nitrogen, and stored in a deep freezer (−80 °C) for further protein and mRNA expression studies.
5.8.2. Western Blotting
WB was performed to check the protein expression of CYP1A2, CYP2B6, CYP2C8, CYP2C11, CYP2C19, CYP2D6, and CYP3A4 in the experimental liver tissues (equal proportion from each animal) of various study groups using the standard immunoblotting protocol.67,68 The present study was carried out using β-actin as a loading control, which is widely used by several researchers for WB analysis.69−71 The tissue homogenate was prepared using radioimmunoprecipitation assay (RIPA) buffer and protein content was determined using the Bradford method. The protein was separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE), transferred onto polyvinylidene fluoride (PVDF) membranes, blocked with skimmed milk (5% w/v), and probed with specific primary antibody (∼14 h at 4 °C). After washing with Tris-buffered saline and Tween 20 buffer, blots were incubated with respective HRP-conjugated secondary antibodies (2 h at room temperature). Protein bands were detected using an Immobilon Forte Western HRP substrate (Merck-Millipore), and images were taken in the Genaxy Chemidoc imaging system (make: Syngene; model; G-BOX, XT-4). Densitometry analysis was then done using ImageJ software.
5.8.3. Reverse Transcription Real-Time Polymerase Chain Reaction
The mRNA expression levels of CYP1A2, CYP2B1, CYP2B2, CYP2C11, CYP2C13, CYP2D1, CYP2D3, CYP3A1, CYP3A18, and CYP3A9 in the experimental liver tissues (equal proportion from each animal) were analyzed using the in-house standardized protocol.24,72 Briefly, the following steps were used: (a) isolation of total RNA from tissue using TRIzol reagent (Invitrogen) as per manufacturer’s protocol, (b) quantification of RNA by a NanoDrop spectrophotometer (make: Thermo Fisher Scientific; model: ND-2000), (c) RNA quality determination by using 2% (w/v) agarose gel electrophoresis, (d) performing mRNA quantitative polymerase chain reaction (qPCR) assays as described earlier,73 synthesis of cDNA was done by reverse transcribing 1 μg of DNase-treated (Ambion TURBO DNA-free kit, Life Technologies) total RNA using oligo dT primer and ImProm-II Reverse Transcription system, and (e) real time-polymerase chain reaction (RT-PCR) was carried out on a rotor-gene Q 2 plex HRM platform (Qiagen). Each PCR reaction (10 μL) contained a 2X Light Cycler 480 SYBR Green I Master mix, 1 μM of each primer (Table 2), and appropriately diluted cDNA. Thermal cycling conditions for the qPCR were as follows: preincubation at 95 °C for 5 min, followed by 40 cycles of 3-step amplification (95 °C for 10 s, 55–60 °C for 20 s, and 72 °C for 20 s) with data collection in the green channel at the annealing-extension step. The cycle threshold (Ct) of the amplification curve was used for the calculations. Ct values were normalized using glyceraldehyde 3-phosphate dehydrogenase (GAPDH). Gene expression analysis was performed in triplicate. The relative expression level was analyzed using the 2–ΔΔCT method.74
Table 2. List of Primers Used for the Estimation of mRNA Expression.
| gene | primer sequence | nucleotides |
|---|---|---|
| CYP1A2 | forward: GGAGATGCTCAACCTCG | 17 |
| reverse: TGCAGAAACAGCACAAAGTTA | 21 | |
| CYP2B1 | forward: ACATCATCTGCTCCATTGT | 19 |
| reverse: TGGGCACCAGGAAAGTA | 17 | |
| CYP2B2 | forward: CAACATCATCTGCTCCATTG | 20 |
| reverse: GGGCACCAGGAAAGTAT | 17 | |
| CYP2C11 | forward: CCACCTTTATCCTGGGC | 17 |
| reverse: AGTATTGCAGACCTGTAGC | 19 | |
| CYP2C13 | forward: GATGAACTATACACTTGAACACCT | 24 |
| reverse: TGGTCAATCTCTTCCTGGACTTTA | 24 | |
| CYP2D1 | forward: CCGAAATCTGACTGATGCC | 19 |
| reverse: CGGGTATAGAATCATGAGCAGTA | 23 | |
| CYP2D3 | forward: GATAACCTGGTGACTGAGC | 19 |
| reverse: GTCATTGACCACCAGACGTA | 20 | |
| CYP3A1 | forward: GAGATCACAGCCCAGTCA | 18 |
| reverse: TAGGTGGGAGGTGCCTTA | 18 | |
| CYP3A18 | forward: TCAAACCAGAAGAGCCG | 17 |
| reverse: GGTGCTTGTGGCATCATA | 18 | |
| CYP3A9 | forward: TGTGGAGATTGTGGCCC | 17 |
| reverse: TGGCATGTGCCTTATTGG | 18 | |
| CYP2E1 | forward: ACTGGACATCAACTGCG | 17 |
| reverse: CCCATATCTCAGAGTTGTGC | 20 | |
| CAR | forward: CACACACTTTGCAGATATCAAT | 22 |
| reverse: GTGTTGAGTGAGATATGCAAGA | 22 |
5.9. Effect of EIDD-1931 on Inhibition of CYP Isoforms
To evaluate any CYP inhibitory effect of EIDD-1931, we conducted in vitro studies on the USFDA-recommended CYP isoforms75 using our earlier reported protocols in HLM.23,24,76 The marker reactions were as follows: phenacetin O-deethylation for CYP1A2, bupropion hydroxylation for CYP2B6, amodiaquine N-deethylation for CYP2C8, diclofenac 4′-hydroxylation for CYP2C9, S-mephenytoin 4-hydroxylation for CYP2C19, dextromethorphan O-demethylation for CYP2D6, and testosterone 6β-hydroxylation for CYP3A4. Additionally, inhibitory potential for CYP3A4 and CYP2C8 was analyzed using RLM. The reaction mixture comprised phosphate buffer (100 mM, pH 7.4), MgCl2, probe substrate, HLM/RLM, test candidate, and NADPH (Table S1).23,24 Reactions were initiated by adding NADPH and incubating at 37 °C in a shaking water bath (make: New Brunswich Scientific; model: CLASSIC C76) at 120 rpm. After a specific incubation time frame (CYP-specific), the reaction was quenched by placing the sample tubes in the thermal block and adding ice-cold acetonitrile (100 μL). The samples were then vortex-mixed (2 min), centrifuged (14,000 rpm, 10 min), decanted into inner vials, and quantified the CYP-specific metabolites by LC–MS/MS (make: Thermo Fisher Scientific; model: TSQ Endura) using the matrix match calibration standards of individual metabolite. In parallel, CYP-specific standard inhibitors were used, and the reactions were performed in triplicate. The reaction without the standard/test inhibitor was considered as a control. The samples with the standard/test inhibitor were compared with the control to estimate % inhibition, and the half maximal inhibitory concentration (IC50) value was calculated using GraphPad Prism software.
5.10. Statistical Analysis
The unpaired Student’s t test was used for statistical analysis by GraphPad Prism software. The p-value of less than 0.001/0.01/0.05 was considered statistically significant.
Acknowledgments
M.B. and D.K. as well as D.M. are thankful to RGNF (UGC) and CSIR (New Delhi, India), respectively, in providing fellowship support. IIIM publication number: CSIR-IIIM/IPR/00606.
Glossary
Abbreviations
- RA
rheumatoid arthritis
- WHO
World Health Organization
- GBD
global burden of disease
- YLD
years lived with disability
- CYP
cytochrome P450
- DMARDs
disease-modifying antirheumatic drugs
- NSAIDs
nonsteroidal anti-inflammatory drugs
- SARS-COV-2
severe acute respiratory syndrome coronavirus-2
- COVID-19
coronavirus disease 2019
- RNA
ribonucleic acid
- AMR
antimicrobial resistance
- mRNA
messenger ribonucleic acid
- CFA
complete Freund’s adjuvant
- Cmax
maximum plasma concentration
- AUC
area under the curve
- Cl/F
clearance after oral administration
- Tmax
time to reach Cmax
- Vd/F
volume of distribution after oral administration
- T1/2
elimination half-life
- WB
Western blotting
- USFDA
United States Food and Drug Administration
- HLM
human liver microsomes
- RLM
rat liver microsomes
- JAK/STAT
Janus kinases/signal transducer and activator of transcription proteins
- IL-6
interleukin-6
- CAR
constitutive androstane receptor
- AhR
aryl hydrocarbon receptor
- PXR
pregnane X receptor
- LXR
liver X receptor
- FXR
farnesoid X receptor
- NF-κB
nuclear factor-kappa B
- NADPH
nicotinamide adenine dinucleotide phosphate
- EDTA
ethylenediaminetetraacetic acid
- pAb
polyclonal antibody
- mAb
monoclonal antibody
- HRP
horseradish peroxidase
- IVC
individually ventilated cage
- IAEC
Institutional Animal Ethics Committee
- CCSEA
Committee for Control and Supervision of Experiments on Animals
- H&E
hematoxylin and eosin
- LC-MS/MS
liquid chromatography-tandem mass spectrometry
- RIPA
radioimmunoprecipitation assay
- SDS-PAGE
sodium dodecyl sulfate–polyacrylamide gel electrophoresis
- PVDF
polyvinylidene fluoride
- qPCR
quantitative polymerase chain reaction
- RT-PCR
real time-polymerase chain reaction
- cDNA
complementary DNA
- Ct
cycle threshold
- GAPDH
glyceraldehyde 3-phosphate dehydrogenase
- IC50
half maximal inhibitory concentration
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.3c09287.
Information on EIDD-1931 purity (HPLC), SRM chromatograms, arthritis confirmation data, WB and RT-PCR data, and CYP2C8 and CYP3A4 inhibition data and protocol (PDF)
Research support was received from Council of Scientific and Industrial Research (New Delhi, India) under research grant (MLP21006 and HCP41).
The authors declare no competing financial interest.
Supplementary Material
References
- Radu A.-F.; Bungau S. G. Management of rheumatoid arthritis: an overview. Cells 2021, 10 (11), 2857. 10.3390/cells10112857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO Scientific group Rheumatic diseases World Health Organization, 1992.
- WHO Musculoskeletal Health. World Health Organization; 2022. [Google Scholar]
- Finckh A.; Gilbert B.; Hodkinson B.; Bae S.-C.; Thomas R.; Deane K. D.; Alpizar-Rodriguez D.; Lauper K. Global epidemiology of rheumatoid arthritis. Nat. Rev. Rheumatol. 2022, 18 (10), 591–602. 10.1038/s41584-022-00827-y. [DOI] [PubMed] [Google Scholar]
- Kany S.; Vollrath J. T.; Relja B. Cytokines in inflammatory disease. International journal of molecular sciences 2019, 20 (23), 6008. 10.3390/ijms20236008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutant D. E.; Hall S. D. Disease–drug interactions in inflammatory states via effects on CYP-mediated drug clearance. Journal of Clinical Pharmacology 2018, 58 (7), 849–863. 10.1002/jcph.1093. [DOI] [PubMed] [Google Scholar]
- Stipp M. C.; Acco A. Involvement of cytochrome P450 enzymes in inflammation and cancer: a review. Cancer Chemotherapy and Pharmacology 2021, 87 (3), 295–309. 10.1007/s00280-020-04181-2. [DOI] [PubMed] [Google Scholar]
- Veeravalli V.; Dash R. P.; Thomas J. A.; Babu R. J.; Madgula L. M. V.; Srinivas N. R. Critical assessment of pharmacokinetic drug–drug interaction potential of tofacitinib, baricitinib and upadacitinib, the three approved janus kinase inhibitors for rheumatoid arthritis treatment. Drug safety 2020, 43, 711–725. 10.1007/s40264-020-00938-z. [DOI] [PubMed] [Google Scholar]
- Chowdhury S. R.; Gupta O. D.; Ghosh A. K.; Singha P. S.; Firdaus S. B.; Klarskov K. Genetic variations and epigenetic modulations in CYP genes: Implications in NSAID-treatment of arthritis patients. Nucleus 2021, 64, 1–12. 10.1007/s13237-021-00373-0. [DOI] [Google Scholar]
- Michaud K.; Wolfe F. Comorbidities in rheumatoid arthritis. Best practice & research Clinical rheumatology 2007, 21 (5), 885–906. 10.1016/j.berh.2007.06.002. [DOI] [PubMed] [Google Scholar]
- Pflugbeil S.; Böckl K.; Pongratz R.; Leitner M.; Graninger W.; Ortner A. Drug interactions in the treatment of rheumatoid arthritis and psoriatic arthritis. Rheumatology International 2020, 40, 511–521. 10.1007/s00296-020-04526-3. [DOI] [PubMed] [Google Scholar]
- Kumar D.; Trivedi N. Disease-drug and drug-drug interaction in COVID-19: Risk and assessment. Biomedicine & Pharmacotherapy 2021, 139, 111642 10.1016/j.biopha.2021.111642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciotti M.; Ciccozzi M.; Terrinoni A.; Jiang W.-C.; Wang C.-B.; Bernardini S. The COVID-19 pandemic. Critical reviews in clinical laboratory sciences 2020, 57 (6), 365–388. 10.1080/10408363.2020.1783198. [DOI] [PubMed] [Google Scholar]
- Bernal A. J.; Gomes da Silva M. M.; Musungaie D. B.; Kovalchuk E.; Gonzalez A.; Delos Reyes V.; Martín-Quirós A.; Caraco Y.; Williams-Diaz A.; Brown M. L.; Du J.; Pedley A.; Assaid C.; Strizki J.; Grobler J. A.; Shamsuddin H. H.; Tipping R.; Wan H.; Paschke A.; Butterton J. R.; Johnson M. G.; De Anda C. Molnupiravir for oral treatment of Covid-19 in nonhospitalized patients. N. Engl. J. Med. 2022, 386 (6), 509–520. 10.1056/NEJMoa2116044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imran M.; Kumar Arora M.; Asdaq S. M. B.; Khan S. A.; Alaqel S. I.; Alshammari M. K.; Alshehri M. M.; Alshrari A. S.; Ali A. M.; Al-Shammeri A. M.; Alhazmi B. D.; Ali Harshan A.; Alam T.; Abida Discovery, development, and patent trends on molnupiravir: a prospective oral treatment for COVID-19. Molecules 2021, 26 (19), 5795. 10.3390/molecules26195795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bluemling G. R.; Mao S.; Natchus M. G.; Painter W.; Mulangu S.; Lockwood M.; De La Rosa A.; Brasel T.; Comer J. E.; Freiberg A. N.; Kolykhalov A. A.; Painter G. R. The prophylactic and therapeutic efficacy of the broadly active antiviral ribonucleoside N4-Hydroxycytidine (EIDD-1931) in a mouse model of lethal Ebola virus infection. Antiviral Res. 2023, 209, 105453 10.1016/j.antiviral.2022.105453. [DOI] [PubMed] [Google Scholar]
- Painter G. R.; Bowen R. A.; Bluemling G. R.; DeBergh J.; Edpuganti V.; Gruddanti P. R.; Guthrie D. B.; Hager M.; Kuiper D. L.; Lockwood M. A.; Mitchell G.; Natchus M. G.; Sticher Z. M.; Kolykhalov A. A. The prophylactic and therapeutic activity of a broadly active ribonucleoside analog in a murine model of intranasal venezuelan equine encephalitis virus infection. Antiviral Res. 2019, 171, 104597 10.1016/j.antiviral.2019.104597. [DOI] [PubMed] [Google Scholar]
- Painter W. P.; Holman W.; Bush J. A.; Almazedi F.; Malik H.; Eraut N. C.; Morin M. J.; Szewczyk L. J.; Painter G. R. Human safety, tolerability, and pharmacokinetics of molnupiravir, a novel broad-spectrum oral antiviral agent with activity against SARS-CoV-2. Antimicrob. Agents Chemother. 2021, 65 (5), e02428–20. 10.1128/aac.02428-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheahan T. P.; Sims A. C.; Zhou S.; Graham R. L.; Pruijssers A. J.; Agostini M. L.; Leist S. R.; Schäfer A.; Dinnon K. H. III; Stevens L. J.; Chappell J. D.; Hughes T. M.; Saindane M.; Kolykhalov A. A.; Painter G.; Harcourt J.; Tamin A.; Thornbug N. J.; Swanstrom R.; Denison M. R.; Baric R. S. An orally bioavailable broad-spectrum antiviral inhibits SARS-CoV-2 in human airway epithelial cell cultures and multiple coronaviruses in mice. Sci. Transl. Med. 2020, 12 (541), eabb5883 10.1126/scitranslmed.abb5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattar C.; Edwards S.; Baraldi E.; Hood J. An overview of the global antimicrobial resistance research and development hub and the current landscape. Curr. Opin. Microbiol. 2020, 57, 56–61. 10.1016/j.mib.2020.06.009. [DOI] [PubMed] [Google Scholar]
- Snekhalatha U.; Anburajan M.; Venkatraman B.; Menaka M. Evaluation of complete Freund’s adjuvant-induced arthritis in a Wistar rat model. Z. Rheumatol 2013, 72 (4), 375–382. 10.1007/s00393-012-1083-8. [DOI] [PubMed] [Google Scholar]
- Dogra A.; Kour D.; Bhardwaj M.; Dhiman S.; Kumar A.; Vij B.; Kumar A.; Nandi U. Glabridin plays dual action to augment the efficacy and attenuate the hepatotoxicity of methotrexate in arthritic rats. ACS omega 2022, 7 (38), 34341–34351. 10.1021/acsomega.2c03948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuthakki V. K.; Choudhary S.; Reddy C. N.; Bhatt S.; Jamwal A.; Jotshi A.; Raghuvanshi R.; Sharma A.; Thakur S.; Jadhav H. R.; Bharate S. S.; Nandi U.; Kumar A.; Bharate S. B. Design, Synthesis, and Pharmacological Evaluation of Embelin–Aryl/alkyl Amine Hybrids as Orally Bioavailable Blood–Brain Barrier Permeable Multitargeted Agents with Therapeutic Potential in Alzheimer’s Disease: Discovery of SB-1448. ACS Chem. Neurosci. 2023, 14 (6), 1193–1219. 10.1021/acschemneuro.3c00030. [DOI] [PubMed] [Google Scholar]
- Manhas D.; Bhatt S.; Rai G.; Kumar V.; Bharti S.; Dhiman S.; Jain S. K.; Sharma D. K.; Ojha P. K.; Gandhi S. G.; Goswami A.; Nandi U. Rottlerin renders a selective and highly potent CYP2C8 inhibition to impede EET formation for implication in cancer therapy. Chem.-Biol. Interact. 2023, 380, 110524 10.1016/j.cbi.2023.110524. [DOI] [PubMed] [Google Scholar]
- Ling S.; Jamali F. The Effect of Infliximab on Hepatic Cytochrome P450 and Pharmacokinetics of Verapamil in Rats with Pre-Adjuvant Arthritis: A Drug–Disease and Drug–Drug Interaction. Basic & clinical pharmacology & toxicology 2009, 105 (1), 24–29. 10.1111/j.1742-7843.2009.00405.x. [DOI] [PubMed] [Google Scholar]
- Mohamed M.-E. F.; Klünder B.; Othman A. A. Clinical pharmacokinetics of upadacitinib: review of data relevant to the rheumatoid arthritis indication. Clinical Pharmacokinetics 2020, 59, 531–544. 10.1007/s40262-019-00855-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanada H.; Sekimoto M.; Kamoshita A.; Degawa M. Changes in expression of hepatic cytochrome P450 subfamily enzymes during development of adjuvant-induced arthritis in rats. Journal of toxicological sciences 2011, 36 (2), 181–190. 10.2131/jts.36.181. [DOI] [PubMed] [Google Scholar]
- Dunvald A. C. D.; Järvinen E.; Mortensen C.; Stage T. B. Clinical and molecular perspectives on inflammation-mediated regulation of drug metabolism and transport. Clinical Pharmacology & Therapeutics 2022, 112 (2), 277–290. 10.1002/cpt.2432. [DOI] [PubMed] [Google Scholar]
- Projean D.; Dautrey S.; Vu H. K.; Groblewski T.; Brazier J.-L.; Ducharme J. Selective downregulation of hepatic cytochrome P450 expression and activity in a rat model of inflammatory pain. Pharm. Res. 2005, 22 (1), 62–70. 10.1007/s11095-004-9010-6. [DOI] [PubMed] [Google Scholar]
- Kawase A.; Tateishi S.; Kazaoka A. Profiling of hepatic metabolizing enzymes and nuclear receptors in rats with adjuvant arthritis by targeted proteomics. Biopharmaceutics & drug disposition 2018, 39 (6), 308–314. 10.1002/bdd.2147. [DOI] [PubMed] [Google Scholar]
- Assenat E.; Gerbal-Chaloin S.; Larrey D.; Saric J.; Fabre J. M.; Maurel P.; Vilarem M. J.; Pascussi J. M. Interleukin 1β inhibits CAR-induced expression of hepatic genes involved in drug and bilirubin clearance. Hepatology 2004, 40 (4), 951–960. 10.1002/hep.20387. [DOI] [PubMed] [Google Scholar]
- Aitken A. E.; Morgan E. T. Gene-specific effects of inflammatory cytokines on cytochrome P450 2C, 2B6 and 3A4 mRNA levels in human hepatocytes. Drug Metab. Dispos. 2007, 35 (9), 1687–1693. 10.1124/dmd.107.015511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer H.; Schmidt F.; Marx-Stoelting P.; Pötz O.; Braeuning A. Cross-species analysis of hepatic cytochrome P450 and transport protein expression. Archives of toxicology 2021, 95, 117–133. 10.1007/s00204-020-02939-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souček P.; Gut I. Cytochromes P-450 in rats: structures, functions, properties and relevant human forms. Xenobiotica 1992, 22 (1), 83–103. 10.3109/00498259209053106. [DOI] [PubMed] [Google Scholar]
- Abd El Gayed E. M.; Rizk M. S.; Ramadan A. N.; Bayomy N. R. mRNA expression of the CUB and sushi multiple domains 1 (CSMD1) and its serum protein level as predictors for psychosis in the familial high-risk children and young adults. ACS omega 2021, 6 (37), 24128–24138. 10.1021/acsomega.1c03637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan T.; Pan L.; Liu Z.; Chen J.; Ge Z.; Lu L.; Zhang X.; Cui S.; Zhang C.; Liu W.; Zhuang S. Metabolic susceptibility of 2-chlorothioxanthone and its toxic effects on mRNA and protein expression and activities of human CYP1A2 and CYP3A4 enzymes. Environ. Sci. Technol. 2018, 52 (20), 11904–11912. 10.1021/acs.est.8b04643. [DOI] [PubMed] [Google Scholar]
- Sewer M. B.; Morgan E. T. Nitric oxide-independent suppression of P450 2C11 expression by interleukin-1β and endotoxin in primary rat hepatocytes. Biochemical pharmacology 1997, 54 (6), 729–737. 10.1016/S0006-2952(97)00226-8. [DOI] [PubMed] [Google Scholar]
- Cheng P.-Y.; Morgan E. T. Hepatic cytochrome P450 regulation in disease states. Current drug metabolism 2001, 2 (2), 165–183. 10.2174/1389200013338676. [DOI] [PubMed] [Google Scholar]
- Chen J.-Q.; Ström A.; Gustafsson J.-A.; Morgan E. T. Suppression of the constitutive expression of cytochrome P-450 2C11 by cytokines and interferons in primary cultures of rat hepatocytes: Comparison with induction of acute-phase genes and demonstration that CYP2C11 promoter sequences are involved in the suppressive response to interleukins 1 and 6. Mol. Pharmacol. 1995, 47 (5), 940–947. [PubMed] [Google Scholar]
- Kawase A.; Yoshida I.; Tsunokuni Y.; Iwaki M. Decreased PXR and CAR inhibit transporter and CYP mRNA Levels in the liver and intestine of mice with collagen-induced arthritis. Xenobiotica 2007, 37 (4), 366–374. 10.1080/00498250701230534. [DOI] [PubMed] [Google Scholar]
- Lenfant T.; Costedoat-Chalumeau N. Hydroxychloroquine dose: balancing toxicity and SLE flare risk. Nature Reviews Rheumatology 2023, 19 (1), 6–7. 10.1038/s41584-022-00868-3. [DOI] [PubMed] [Google Scholar]
- Stokkermans T. J.; Goyal A.; Bansal P.; Trichonas G., Chloroquine and hydroxychloroquine toxicity StatPearls StatPearls Publishing. 2019. [PubMed]
- Paludetto M.-N. I. A.; Kurkela M.; Kahma H.; Backman J. T.; Niemi M.; Filppula A. M. Hydroxychloroquine is metabolized by CYP2D6, CYP3A4, and CYP2C8, and inhibits CYP2D6, while its metabolites also inhibit CYP3A in vitro. Drug Metab. Dispos. 2023, 51, 293. 10.1124/dmd.122.001018. [DOI] [PubMed] [Google Scholar]
- Gan T. J. Diclofenac: an update on its mechanism of action and safety profile. Current medical research and opinion 2010, 26 (7), 1715–1731. 10.1185/03007995.2010.486301. [DOI] [PubMed] [Google Scholar]
- Vane J.; Botting R. The mechanism of action of aspirin. Thrombosis research 2003, 110 (5–6), 255–258. 10.1016/S0049-3848(03)00379-7. [DOI] [PubMed] [Google Scholar]
- Agúndez J. A.; García-Martín E.; Martínez C. Genetically based impairment in CYP2C8-and CYP2C9-dependent NSAID metabolism as a risk factor for gastrointestinal bleeding: is a combination of pharmacogenomics and metabolomics required to improve personalized medicine?. Expert opinion on drug metabolism & toxicology 2009, 5 (6), 607–620. 10.1517/17425250902970998. [DOI] [PubMed] [Google Scholar]
- Hakkola J.; Hu Y.; Ingelman-Sundberg M. Mechanisms of down-regulation of CYP2E1 expression by inflammatory cytokines in rat hepatoma cells. Journal of Pharmacology and Experimental Therapeutics 2003, 304 (3), 1048–1054. 10.1124/jpet.102.041582. [DOI] [PubMed] [Google Scholar]
- Kojima S.; Green C. B. Circadian genomics reveal a role for post-transcriptional regulation in mammals. Biochemistry 2015, 54 (2), 124–133. 10.1021/bi500707c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J.; Liu Y.-T.; Xiao L.; Zhu L.; Wang Q.; Yan T. Anti-inflammatory effects of apigenin in lipopolysaccharide-induced inflammatory in acute lung injury by suppressing COX-2 and NF-kB pathway. Inflammation 2014, 37 (6), 2085–2090. 10.1007/s10753-014-9942-x. [DOI] [PubMed] [Google Scholar]
- White E. J.; Brewer G.; Wilson G. M. Post-transcriptional control of gene expression by AUF1: mechanisms, physiological targets, and regulation. Biochimica et Biophysica Acta (BBA)-Gene Regulatory Mechanisms 2013, 1829 (6–7), 680–688. 10.1016/j.bbagrm.2012.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denisov I. G.; Grinkova Y. V.; McLean M. A.; Camp T.; Sligar S. G. Midazolam as a probe for heterotropic drug-drug interactions mediated by CYP3A4. Biomolecules 2022, 12 (6), 853. 10.3390/biom12060853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowty M. E.; Lin J.; Ryder T. F.; Wang W.; Walker G. S.; Vaz A.; Chan G. L.; Krishnaswami S.; Prakash C. The pharmacokinetics, metabolism, and clearance mechanisms of tofacitinib, a janus kinase inhibitor, in humans. Drug Metab. Dispos. 2014, 42 (4), 759–773. 10.1124/dmd.113.054940. [DOI] [PubMed] [Google Scholar]
- Kuriya B.; Cohen M. D.; Keystone E. Baricitinib in rheumatoid arthritis: evidence-to-date and clinical potential. Therapeutic advances in musculoskeletal disease 2017, 9 (2), 37–44. 10.1177/1759720X16687481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gremese E.; Ferraccioli G. Benefit/risk of cyclosporine in rheumatoid arthritis. Clin. Exp. Rheumatol. 2004, 22, S101–S101. [PubMed] [Google Scholar]
- Tornio A.; Backman J. T. Cytochrome P450 in pharmacogenetics: an update. Adv. Pharmacol. 2018, 83, 3–32. 10.1016/bs.apha.2018.04.007. [DOI] [PubMed] [Google Scholar]
- Esteves F.; Rueff J.; Kranendonk M. The central role of cytochrome P450 in xenobiotic metabolism—a brief review on a fascinating enzyme family. Journal of xenobiotics 2021, 11 (3), 94–114. 10.3390/jox11030007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y.; Tang J.; Wu B.; Yang B.; Ou Q.; Lin J. Correlation between albumin to fibrinogen ratio, C-reactive protein to albumin ratio and Th17 cells in patients with rheumatoid arthritis. Clin. Chim. Acta 2020, 500, 149–154. 10.1016/j.cca.2019.10.009. [DOI] [PubMed] [Google Scholar]
- Zordoky B. N.; El-Kadi A. O. Role of NF-κB in the regulation of cytochrome P450 enzymes. Current drug metabolism 2009, 10 (2), 164–178. 10.2174/138920009787522151. [DOI] [PubMed] [Google Scholar]
- Klein M.; Thomas M.; Hofmann U.; Seehofer D.; Damm G.; Zanger U. M. A systematic comparison of the impact of inflammatory signaling on absorption, distribution, metabolism, and excretion gene expression and activity in primary human hepatocytes and HepaRG cells. Drug Metab. Dispos. 2015, 43 (2), 273–283. 10.1124/dmd.114.060962. [DOI] [PubMed] [Google Scholar]
- Pascussi J.-M.; Gerbal-Chaloin S.; Pichard-Garcia L.; Daujat M.; Fabre J.-M.; Maurel P.; Vilarem M.-J. Interleukin-6 negatively regulates the expression of pregnane X receptor and constitutively activated receptor in primary human hepatocytes. Biochemical and biophysical research communications 2000, 274 (3), 707–713. 10.1006/bbrc.2000.3219. [DOI] [PubMed] [Google Scholar]
- Ferguson S. S.; Chen Y.; LeCluyse E. L.; Negishi M.; Goldstein J. A. Human CYP2C8 is transcriptionally regulated by the nuclear receptors constitutive androstane receptor, pregnane X receptor, glucocorticoid receptor, and hepatic nuclear factor 4α. Molecular pharmacology 2005, 68 (3), 747–757. 10.1124/mol.105.013169. [DOI] [PubMed] [Google Scholar]
- De Bosscher K.; Vanden Berghe W.; Haegeman G. Cross-talk between nuclear receptors and nuclear factor κB. Oncogene 2006, 25 (51), 6868–6886. 10.1038/sj.onc.1209935. [DOI] [PubMed] [Google Scholar]
- Mukherjee D.; Ahmed Q. N.; Ahmed A.; Banday J. S.. Non infringing process for the synthesis of N4-hydroxycytidine and its derivative. IN2021110206782022.
- Mohammed S. M.; Taqa G. A.; Sulaiman M. S.. Effect of flavonoid luteolin on primary oral wound healing in rats, In AIP Conf. Proc., AIP Publishing LLC: 2022; p 020007. [Google Scholar]
- Toots M.; Yoon J. J.; Cox R. M.; Hart M.; Sticher Z. M.; Makhsous N.; Plesker R.; Barrena A. H.; Reddy P. G.; Mitchell D. G.; Shean R. C.; Bluemling G. R.; Kolykhalov A. A.; Greninger A. L.; Natchus M. G.; Painter G. R.; Plemper R. K. Characterization of orally efficacious influenza drug with high resistance barrier in ferrets and human airway epithelia. Sci. Transl. Med. 2019, 11 (515), eaax5866 10.1126/scitranslmed.aax5866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik S.; Mintoo M. J.; Reddy C. N.; Kumar R.; Kotwal P.; Bharate S. B.; Nandi U.; Mondhe D. M.; Shukla S. K. In vitro and in vivo anticancer potential and molecular targets of the new colchicine analog IIIM-067. Journal of Integrative Medicine 2023, 21 (1), 62–76. 10.1016/j.joim.2022.09.006. [DOI] [PubMed] [Google Scholar]
- Kotwal P.; Khajuria P.; Dhiman S.; Kour D.; Dhiman S. K.; Kumar A.; Nandi U. Molecular mechanism for the involvement of CYP2E1/NF-κB axis in bedaquiline-induced hepatotoxicity. Life Sciences 2023, 315, 121375 10.1016/j.lfs.2023.121375. [DOI] [PubMed] [Google Scholar]
- Bhatt S.; Sharma A.; Dogra A.; Sharma P.; Kumar A.; Kotwal P.; Bag S.; Misra P.; Singh G.; Kumar A.; Sangwan P. L.; Nandi U. Glabridin attenuates paracetamol-induced liver injury in mice via CYP2E1-mediated inhibition of oxidative stress. Drug Chem. Toxicol. 2022, 45 (5), 2352–2360. 10.1080/01480545.2021.1945004. [DOI] [PubMed] [Google Scholar]
- Guo P.; Wang Q.; Chen L.; Dingya K.; Wang B. Ultrasound-Responsive Micelle-Encapsulated Mesenchymal Stem Cell-Derived EVs for the Treatment of Lower Limb Microcirculation Disease. ACS Omega 2023, 4, 49406. 10.1021/acsomega.3c08133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keskin E.; Gezen-Ak D.; Dursun E. Amyloid β, α-Synuclein and Amyloid β-α-Synuclein Combination Exert Significant but Different Alterations in Inflammatory Response Profile in Differentiated Human SH-SY5Y Cells. ACS Omega 2023, 45519. 10.1021/acsomega.3c05585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annušová A.; Labudová M.; Truchan D.; Hegedűšová V.; Švajdlenková H.; Mičušík M.; Kotlár M.; Slušná L. B.; Hulman M.; Salehtash F.; Kálosi A.; Csáderová L.; Švastová E.; Šiffalovič P.; Jergel M.; Pastoreková S.; Majková E. Selective Tumor Hypoxia Targeting Using M75 Antibody Conjugated Photothermally Active MoO x Nanoparticles. ACS Omega 2023, 8 (47), 44497–44513. 10.1021/acsomega.3c01934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dogra A.; Gour A.; Bhatt S.; Sharma P.; Sharma A.; Kotwal P.; Wazir P.; Mishra P.; Singh G.; Nandi U. Effect of rutin on pharmacokinetic modulation of diclofenac in rats. Xenobiotica 2020, 50 (11), 1332–1340. 10.1080/00498254.2020.1773008. [DOI] [PubMed] [Google Scholar]
- Awasthi P.; Mahajan V.; Jamwal V. L.; Kapoor N.; Rasool S.; Bedi Y. S.; Gandhi S. G. Cloning and expression analysis of chalcone synthase gene from Coleus forskohlii. Journal of genetics 2016, 95, 647–657. 10.1007/s12041-016-0680-8. [DOI] [PubMed] [Google Scholar]
- Livak K. J.; Schmittgen T. D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2021, 25, 402–408. 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- USFDA Bioanalytical Method Validation Guidance for Industry; US Department of Health and Human Services, Food and Drug Administration, Center for Drug Evaluation and Research (CDER), Center for Veterinary Medicine (CVM). Biopharm 2018, 44. [Google Scholar]
- Bhatt S.; Dhiman S.; Kumar V.; Gour A.; Manhas D.; Sharma K.; Ojha P. K.; Nandi U. Assessment of the CYP1A2 Inhibition-Mediated Drug Interaction Potential for Pinocembrin Using In Silico, In Vitro, and In Vivo Approaches. ACS Omega 2022, 20321. 10.1021/acsomega.2c02315. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.