Abstract
Hyperactivation of the NLRP3 inflammasome has been implicated in the pathogenesis of numerous diseases. However, the precise molecular mechanisms that modulate the transcriptional regulation of NLRP3 remain largely unknown. In this study, we demonstrated that S-nitrosoglutathione reductase (GSNOR) deficiency in macrophages leads to significant increases in the Nlrp3 and Il-1β expression levels and interleukin-1β (IL-1β) secretion in response to NLRP3 inflammasome stimulation. Furthermore, in vivo experiments utilizing Gsnor−/− mice revealed increased disease severity in both lipopolysaccharide (LPS)-induced septic shock and dextran sodium sulfate (DSS)-induced colitis models. Additionally, we showed that both LPS-induced septic shock and DSS-induced colitis were ameliorated in Gsnor−/− Nlrp3−/− double-knockout (DKO) mice. Mechanistically, GSNOR deficiency increases the S-nitrosation of mitogen-activated protein kinase 14 (MAPK14) at the Cys211 residue and augments MAPK14 kinase activity, thereby promoting Nlrp3 and Il-1β transcription and stimulating NLRP3 inflammasome activity. Our findings suggested that GSNOR is a regulator of the NLRP3 inflammasome and that reducing the level of S-nitrosylated MAPK14 may constitute an effective strategy for alleviating diseases associated with NLRP3-mediated inflammation.
Keywords: GSNOR, S-nitrosation, NLRP3 inflammasome, MAPK14, Septic shock, Colitis
Subject terms: Inflammasome, Sepsis
Introduction
The NOD-like receptor protein 3 (NLRP3) inflammasome has been the subject of many studies in recent years and is by far the most extensively studied inflammasome [1–3]. The NLRP3 inflammasome complex is composed mainly of the sensor protein NLRP3, caspase-1, and the adaptor molecule apoptosis-associated speck-like protein containing a caspase recruitment domain (ASC) [1, 2]. Activation of the NLRP3 inflammasome involves two distinct signaling cascades known as priming and activation. The priming signal originates from the stimulation of pathogen recognition receptors (PRRs), which in turn activate the nuclear factor κB (NF-κB) and mitogen-activated protein kinase (MAPK) pathways, leading to the transcriptional upregulation of NLRP3 inflammasome components [4–7]. The second activation step, triggered by various danger signals such as potassium efflux, adenosine 5′-triphosphate (ATP), mitochondrial dysfunction and lysosomal rupture, results in the assembly of the NLRP3 inflammasome complex [4]. Upon activation, the NLRP3 inflammasome facilitates the processing of pro-caspase-1 into its mature form, thereby mediating the cleavage of pro-IL-1β to form mature IL-1β and the secretion of mature IL-1β [1, 6, 8]. Dysregulation of the NLRP3 inflammasome is associated with the progression of various inflammation-related diseases, including infections, neurodegenerative diseases, multiple sclerosis, colitis, atherosclerosis and gouty arthritis [9–11]. Therefore, maintaining a delicate balance between NLRP3 inflammasome activation and inhibition is crucial for preventing excessive inflammatory damage to the host.
The intracellular protein level of NLRP3 is critical for inflammasome assembly and constitutes a key rate-limiting element for inflammasome activation [12, 13]. The expression of NLRP3 is tightly controlled by a number of different regulators. Some directly inhibit NLRP3 protein expression, thus attenuating inflammasome activation. For instance, ubiquitin-specific peptidase 22 (USP22) suppresses NLRP3 inflammasome activation through the degradation of NLRP3 via ATG5-dependent autophagy [14]. Bile acids inhibit NLRP3 inflammasome activation via the TGR5-cAMP-PKA axis and PKA kinase activation, leading to the ubiquitination and degradation of NLRP3 [15]. However, other regulators have been shown to increase NLRP3 protein expression and inflammasome activation. For example, the E3 SUMO ligase tripartite motif-containing protein 28 (TRIM28) increases NLRP3 inflammasome activation by promoting SUMO1, SUMO2 and SUMO3 modification of NLRP3, thereby inhibiting NLRP3 ubiquitination and proteasomal degradation [12]. Most of the previously identified regulatory factors appear to act at the posttranscriptional and posttranslational levels, but the molecular mechanisms modulating NLRP3 inflammasome activation, especially at the priming step [5, 16], are still insufficiently understood.
S-nitrosoglutathione reductase (GSNOR) is a highly conserved denitrosylase that modulates S-nitrosation through the catabolism of S-nitrosoglutathione (GSNO) [17]. Deletion of GSNOR has been shown to significantly increase protein S-nitrosation, which in turn affects downstream signaling. Multiple studies, including our own [18–20], have highlighted the crucial role of GSNOR dysregulation in various diseases [21, 22]. Notably, dysregulation of S-nitrosation has been implicated in antiviral innate immunity [19], Parkinson’s disease [18], cardiovascular diseases [23, 24], cancer, and asthma [22].
In this study, we hypothesized that GSNOR plays a key role in modulating NLRP3 priming. We provided evidence showing that GSNOR affects the expression of NLRP3 inflammasome components and that GSNOR deficiency leads to significant increases in the expression levels of Nlrp3 and Il-1β, which is attributed to the S-nitrosation of the MAPK14 protein at the Cys211 residue and the consequent increase in its kinase activity. We also showed that Gsnor−/− mice exhibited heightened susceptibility to NLRP3-dependent inflammatory diseases and that this propensity was ameliorated in Gsnor−/− Nlrp3−/− double-knockout (DKO) mice and SB203580-pretreated mice. Our findings indicate that GSNOR is a critical regulator of the NLRP3 inflammasome via the modulation of MAPK14 S-nitrosylation.
Results
Increased Nlrp3 and Il-1β gene expression in GSNOR-deficient macrophages
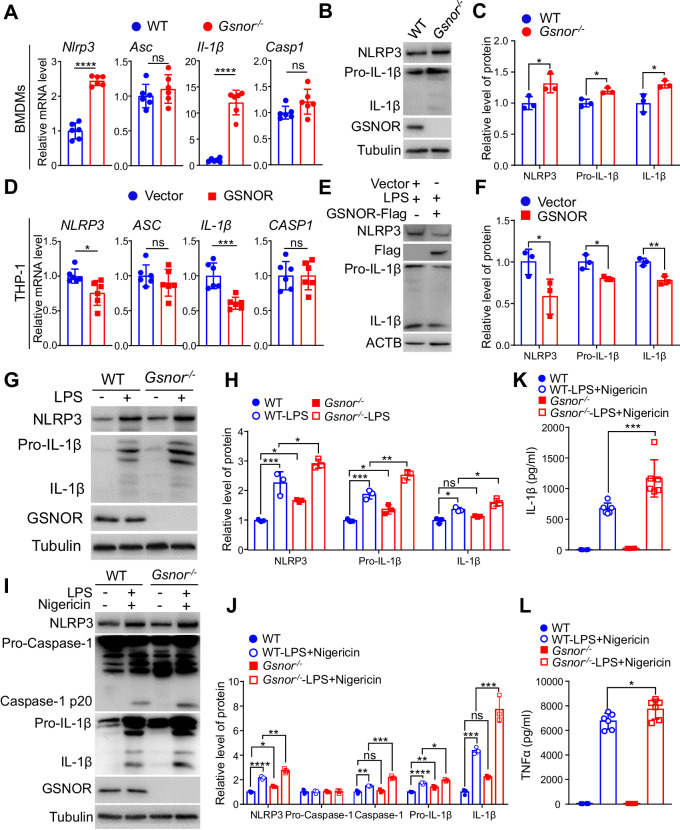
We used quantitative real-time PCR (qRT‒PCR) to measure the mRNA expression levels of Nlrp3, Il-1β, Caspase-1 and Asc in bone marrow-derived macrophages (BMDMs) isolated from Gsnor−/− mice and their wild-type (WT) littermates. The Nlrp3 and Il-1β mRNA expression levels were significantly elevated in Gsnor−/− BMDMs compared to WT BMDMs. In contrast, the mRNA levels of other inflammasome components, such as Asc and Caspase-1, were not significantly different between Gsnor−/− and WT BMDMs (Fig. 1A). The elevated protein levels of NLRP3 and IL-1β in Gsnor−/− BMDMs were confirmed by Western blotting (Fig. 1B-C). We further validated the effect of GSNOR on modulating the NLRP3 inflammasome using human THP-1 cells. Overexpression of GSNOR in THP-1 cells significantly reduced the NLRP3 and IL-1β mRNA levels compared with those in cells transfected with the empty vector (Fig. 1D). Consistently, GSNOR overexpression decreased the protein levels of NLRP3 and IL-1β in the presence of lipopolysaccharide (LPS) (Fig. 1E-F). As LPS has been reported to trigger the expression of Nlrp3 and pro-Il-1β [25], we treated BMDMs with LPS and found that LPS treatment further increased the protein levels of NLRP3 and pro-IL-1β in Gsnor−/− BMDMs compared to WT control BMDMs (Fig. 1G-H). These results indicate that GSNOR affects the expression of Nlrp3 and Il-1β.
Fig. 1.
GSNOR deficiency increases Nlrp3 and Il-1β expression in mouse bone marrow-derived macrophages (BMDMs). A Relative mRNA levels of NLRP3 inflammasome components in BMDMs from Gsnor−/− mice and their wild-type (WT) littermates were measured by quantitative real-time PCR (n = 6 mice per group). B, C Protein levels of NLRP3 and IL-1β in BMDMs from Gsnor−/− mice and their WT littermates (B; n = 3 mice per group) and the related quantitative analysis (C). D Relative mRNA levels of NLRP3 inflammasome components in THP-1 cells with or without GSNOR overexpression (n = 6 biological replicates). Cells were transfected with the GSNOR expression vector (GSNOR) or empty vector (Vector) for 24 h before harvesting. E, F Protein levels of NLRP3 and IL-1β in THP-1 cells with or without GSNOR overexpression (E) and the related quantitative analysis (F). Cells were transfected with the GSNOR expression vector (GSNOR-Flag) or empty vector (Vector) for 36 h and then treated with lipopolysaccharide (LPS; 2 μg/mL) for 6 h before being harvested for immunoblotting (n = 3 biological replicates). The data shown in (D–F) are representative of three independent experiments with similar results. G, H Protein levels of NLRP3 and IL-1β in BMDMs from Gsnor−/− mice and their WT littermates (G) with or without LPS treatment, and the related quantitative analysis (H). Lysates of BMDMs treated with or without LPS (2 μg/mL) for 6 h were collected for immunoblot analysis (n = 3 mice per group). I, J Protein levels of NLRP3 inflammasome components in BMDMs from Gsnor−/− mice and their WT littermates stimulated with or without LPS and nigericin (I) and the related quantitative analysis (J). BMDMs were treated with LPS (500 ng/mL) for 6 h and were then treated with nigericin (15 μM) for 30 min before harvesting (n = 3 mice per group). K, L ELISA of IL-1β and TNF-α in the culture supernatants of BMDMs from Gsnor−/− mice and their WT littermates treated with or without LPS and nigericin (n = 6 mice per group). Treatment with LPS and nigericin significantly (P < 0.0001, not labeled in the bar graphs) increased the levels of IL-1β and TNF-α compared to those in the untreated BMDMs from both Gsnor−/− mice and their WT littermates. The data are presented as the means ± SDs. ns, not significant; *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. Two‐tailed unpaired Student’s t test was used for (A, C, D, F). One‐way ANOVA was used for (H, K, J, L)
Subsequently, we investigated whether GSNOR affects caspase-1 and IL-1β activity during NLRP3 inflammasome activation. We observed substantial increases in the protein levels of Caspase-1 p20 and IL-1β in LPS-primed Gsnor−/− BMDMs treated with nigericin compared with WT BMDMs under the same treatment conditions (Fig. 1I-J). Moreover, the secretion of IL-1β (Fig. 1K) and TNF-α (Fig. 1L) into the culture supernatant was significantly greater for LPS-primed Gsnor−/− BMDMs than for WT BMDMs after nigericin treatment. These results indicate that GSNOR activates an inhibitory signal to attenuate Nlrp3 and Il-1β expression in macrophages.
GSNOR deficiency aggravates DSS-induced colitis in mice
Previous studies have reported that the NLRP3 inflammasome plays an important role in dextran sulfate sodium (DSS)-induced colitis, and targeting the NLRP3 inflammasome showed obvious effects in a colitis mouse model [26–29]. To test whether GSNOR deficiency can aggravate DSS-induced colitis and NLRP3 activation in vivo, we treated Gsnor−/− mice and their WT littermates with 3% DSS in the drinking water for 6 days (Fig. 2A). Compared with WT mice, Gsnor−/− mice had higher mortality, greater body weight loss and higher disease activity index (DAI) scores (Fig. 2B–D), which are the crucial parameters indicating the severity of colitis [30]. In Gsnor−/− mice, we also observed colon shortening, which has previously been described as an indicator of the severity of colitis [31, 32] (Fig. 2E, F). Histological analysis revealed that GSNOR deficiency exacerbated the destruction of the intestinal epithelium and increased the infiltration of immune cells (Fig. 2G). Western blot analysis revealed that GSNOR deficiency increased NLRP3 inflammasome activation, as indicated by the increased levels of the NLRP3, Caspase-1 and IL-1β proteins in intestinal tissues from Gsnor−/− mice compared with those from WT mice (Fig. 2H, I). Similarly, an increase in IL-1β secretion in the colon organ culture supernatant was observed for Gsnor−/− mice compared to their WT littermates (Fig. 2J). Collectively, these results indicate that GSNOR deficiency increases NLRP3 inflammasome activation and exacerbates DSS-induced colitis in mice.
Fig. 2.
GSNOR deficiency exacerbates DSS-induced colitis in mice. A, B Schematic profile of the animal experiments (A) and survival curves of Gsnor−/− mice and their WT littermates after DSS treatment (n = 8 mice per group) (B). C–I Effects of DSS treatment for 6 consecutive days on Gsnor−/− mice and their WT littermates (n = 8 mice per group). The disease activity index (C) and body weight loss (D) were determined for the surviving animals on the indicated days. In the Gsnor−/− group, after DSS treatment, one mouse died on Day 5, and the other died on Day 6. No animals died in the WT group. All surviving animals were euthanized on Day 6 to evaluate morphological changes in the colon (E), measure the colon length (F), and evaluate pathological changes in colon tissue sections by hematoxylin and eosin staining (G). The protein levels of NLRP3, Caspase-1 (p20) and IL-1β in colon tissues from Gsnor−/− mice and their WT littermates (n = 3 per group) (H) were quantified (I). Scale bar, 100 μm. J ELISA of secreted IL-1β protein in colon organ culture supernatants. After 6 days of DSS treatment, surviving Gsnor−/− mice and their WT littermates (n = 4 mice per group) were euthanized for colon harvesting. The cleaned colonic tissues were cultured in RPMI 1640 medium supplemented with 1× penicillin/streptomycin for 24 h, and culture supernatants were collected for ELISA. The data are presented as the means ± SEMs for C and D and as the means ± SDs for (F, I, J). Statistical significance was determined by the log-rank Mantel‒Cox test in (B). Two-tailed unpaired Student’s t test was used for comparisons between the Gsnor−/− and WT groups on the indicated days (C, D) and between the Gsnor−/− and WT groups (F, I, J). ns, not significant; *P < 0.05; **P < 0.01
GSNOR deficiency increases susceptibility to LPS-induced septic shock in mice
We next tested the effect of GSNOR deficiency on the NLRP3 inflammasome using an LPS-induced septic shock mouse model (Fig. 3A) [33]. The results showed that Gsnor−/− mice were more susceptible to LPS and had higher mortality than their WT littermates (Fig. 3B). The protein expression of NLRP3 and IL-1β induced by LPS injection was significantly greater in the liver and spleen tissues of Gsnor−/− mice than in those of WT mice (Fig. 3C, D). We observed greater immune cell infiltration and liver tissue impairment in Gsnor−/− mice than in their WT littermates (Fig. 3E). Flow cytometry confirmed that the proportions of neutrophils and monocytes were significantly greater in the liver tissues of Gsnor−/− mice than in those of their WT littermates (Fig. 3F, G). In agreement with these observations, Gsnor−/− mice had a greater serum IL-1β protein level than did their WT littermates (Fig. 3H). These results indicate that GSNOR is a critical regulator of NLRP3 inflammasome activation in vivo.
Fig. 3.
GSNOR deficiency exacerbates LPS-induced septic shock in mice. A Schematic diagram illustrating the design of the LPS treatment experiment. Two cohorts of animals (Gsnor−/− mice and WT littermate mice) were intraperitoneally (i.p.) injected with LPS (20 mg/kg). One cohort was used for survival analysis, and the other cohort was used for tissue collection 6 h after LPS injection. B Survival curves of Gsnor−/− mice and their WT littermates after LPS treatment (n = 10 mice per group). C–E Changes in the levels of NLRP3 inflammasome components (C, D) and pathological changes (E) in Gsnor−/− mice and their WT littermates after LPS treatment (n = 3 mice per group). Protein levels of NLRP3, IL-1β and Pro-IL-1β in liver tissues and spleen tissues (C) and the related quantitative analysis (D), as determined by Western blotting. Pathological changes in liver tissue sections were evaluated by hematoxylin and eosin staining (E). F, G Percentages of liver-infiltrating cells (neutrophils, CD11b+Gr-1+; monocyte-macrophages, CD11b+Gr-1−) in Gsnor−/− mice and their WT littermates treated with LPS (n = 4 mice per group). Fresh liver tissues were used to isolate the infiltrating cells for flow cytometry. (H) ELISA of serum IL-1β in animals from (C, F) (n = 4–5 mice per group, depending on the availability of serum samples). Statistical significance was determined by the log-rank Mantel‒Cox test in (B). Two-tailed unpaired Student’s t test was used for (D, G, H). ns, not significant; *P < 0.05; **P < 0.01; ***P < 0.001
GSNOR deficiency increases NLRP3 inflammasome activation via MAPK signaling
To investigate the molecular mechanisms underlying GSNOR-mediated regulation of NLRP3 inflammasome activation, we performed RNA-seq analysis on BMDMs sorted from Gsnor−/− mice and their WT littermates after LPS treatment. Principal component analysis revealed a clear difference between the BMDM samples from the two groups (Fig. 4A). Comparison of the heatmaps showing the differentially expressed genes between Gsnor−/− and WT BMDMs revealed a clearly different signature in BMDMs from each group (Fig. 4B and Fig. S1). A greater number of upregulated genes and downregulated genes were detected in Gsnor−/− BMDMs than in the corresponding control cells; among these differentially expressed genes, Nlrp3, Il-1β, Caspase-1 and Mapk14 exhibited significantly increased expression in Gsnor−/− BMDMs (Fig. 4C, D and Fig. S1). KEGG enrichment analysis revealed that the activity of several signaling pathways, including the MAPK and NOD-like receptor signaling pathways, was upregulated in Gsnor−/− BMDMs compared with WT BMDMs (Fig. 4E). Indeed, we observed that in Gsnor−/− BMDMs, not only did the MAPK14 level increase, but this increase also affected the expression of genes related to NLRP3 priming, such as TLR4, TRAF6, MAP3K7, and IKKβ [5, 34]; in addition, the expression levels of molecules associated with NLRP3 activation, such as NEK7 [3], were increased compared to those in WT BMDMs (Fig. S1). These data suggest that GSNOR regulates inflammasome activity through multiple signaling pathways, including the MAPK signaling pathway.
Fig. 4.
Transcriptomic profiling of LPS-stimulated BMDMs from Gsnor−/− mice and their WT littermates. A Principal component analysis scatterplot based on the differentially expressed genes between LPS-treated WT and Gsnor−/− BMDMs (n = 4 mice per group). B, C Heatmap (B) and volcano plot (C) showing the differentially expressed genes between LPS-stimulated WT and Gsnor−/− BMDMs. A detailed list of the differentially expressed genes in (B) is shown in Supplementary Fig. 1. D Bar graphs showing the mRNA expression levels of the selected genes in A. E KEGG pathway enrichment analysis of the differentially expressed genes in A. The data are presented as the means ± SDs. *, P < 0.05; **, P < 0.01; ****, P < 0.0001; two-tailed unpaired Student’s t test
S-nitrosation of MAPK14 at Cys211 increases MAPK14 phosphorylation and NLRP3 inflammasome activation
To characterize the potential effect of the upregulated MAPK signaling pathway in BMDMs with GSNOR deficiency compared to those without, we examined the effects of GSNOR on MAPK14 kinase activity. We found that GSNOR deficiency increased NLRP3 protein expression and the level of phosphorylated MAPK14 in BMDMs (Fig. 5A, B), whereas GSNOR overexpression in human THP-1 cells decreased NLRP3 protein expression and the level of phosphorylated MAPK14 (Fig. 5C, D). We further determined whether GSNOR regulates the activity of the NLRP3 inflammasome by regulating the activity of MAPK14. We pretreated WT and Gsnor−/− BMDMs with SB203580, a specific small molecule inhibitor of MAPK14 [35], and found that pretreatment with SB203580 inhibited NLRP3 inflammasome activity induced by GSNOR deficiency (Fig. 5E, F). These results suggest that GSNOR negatively regulates MAPK14 kinase activity and NLRP3 inflammasome activity.
Fig. 5.
S-nitrosation of MAPK14 by GSNOR affects the phosphorylation of MAPK14 and NLRP3 inflammasome activity. A Gsnor deficiency increases MAPK14 phosphorylation and the NLRP3 protein level in BMDMs from Gsnor−/− mice and their WT littermates (n = 3 mice per group). B Quantification of the protein levels in (A). C, D GSNOR overexpression inhibits MAPK14 phosphorylation and NLRP3 protein expression in human THP-1 cells (C) and the related quantitative analysis (D). Cells were transfected with the GSNOR expression vector (GSNOR-Flag) or empty vector (Vector) for 36 h before being harvested for immunoblot analysis (n = 3 biological replicates). E, F Pretreatment with the MAPK14 inhibitor SB203580 inhibits NLRP3 inflammasome activity in BMDMs from Gsnor−/− mice and their WT littermates (E) and the related quantitative analysis of protein levels (F) (n = 3 mice per group). A significant difference between BMDMs treated with or without SB203580 in the presence of LPS and nigericin was found and is marked in (F). G, H Levels of S-nitrosated exogenous MAPK14 and exogenous MAPK14 (detected via the HA tag) in HEK293T cells after GSNO treatment. Cells were transfected with wild-type MAPK14 expression vector (MAPK14-HA), MAPK14 mutant expression vectors (p.C39A, MAPK14-C39A-HA; p.C119A, MAPK14-C119A-HA; p.C162A, MAPK14-C162A-HA; and p.C211A, MAPK14-C211A-HA), or empty vector (Vector) for 48 h. Cell lysates were incubated with GSNO (500 μM) for 30 min before the biotin switch assay (G) (n = 3 biological replicates). H Quantification of the protein levels in (G). I Evolutionary conservation of the Cys211 and Cys39 residues in four vertebrate species. J S-nitrosation of MAPK14 at the Cys211 residue increases MAPK14 kinase activity in HEK293T cells (n = 3 biological replicates). K Quantification of the protein levels in (J). L S-nitrosation of MAPK14 at the Cys211 residue increases NLRP3 inflammasome activity in NLRP3-reconstituted HEK293T cells (n = 3 biological replicates). M Quantification of the protein levels in (L). The data shown in (C, G, J, L) are representative of three independent experiments with similar results. The data are presented as the means ± SDs. ns, not significant; *P < 0.05; **P < 0.01; ****P < 0.0001. Two‐tailed unpaired Student’s t test was used for (B, D, F). One‐way ANOVA was used for (H, K, M)
Next, we examined how GSNOR regulates MAPK14 kinase activity. As GSNOR is a key metabolic enzyme for S-nitrosation homeostasis [22], we hypothesized that the inhibitory effect of GSNOR on MAPK14 kinase activity might be mediated by its effect on S-nitrosation. Coimmunoprecipitation revealed no direct interaction between GSNOR and MAPK14 or between GSNOR and MAPK14 p.C211A (Fig. S2). LPS treatment decreased the enzymatic activity of GSNOR in RAW264.7 cells (Fig. S3A). Similarly, LPS treatment increased the SNO-MAPK14 level in RAW264.7 cells overexpressing either GSNOR with the p.R115D mutation (GSNOR-115D mutation, which abolishes the enzymatic activity of GSNOR [36, 37]) or empty vector compared with cells overexpressing Flag-tagged GSNOR (Fig. S3B). These results suggested that the enzymatic activity of GSNOR is essential for regulating the S-nitrosation of MAPK14.
The MAPK14 protein has four cysteine residues (Fig. S4A, B), which are potential sites for S-nitrosation [38]. We created mutants in which each of these cysteine residues was individually mutated to alanine (Ala), which cannot be S-nitrosylated [39]. HEK293T cells were transfected with the indicated HA-tagged wild-type MAPK14 (HA-MAPK14-WT), mutant MAPK14 (p.C39A, p.C119A, p.C162A or p.C211A), or empty (vector-HA) expression vector for 48 h before harvesting. The cell lysates were incubated with GSNO for 30 min before the biotin switch assay. GSNO treatment significantly increased the S-nitrosation of HA-MAPK14-WT and all four cysteine mutants of MAPK14 (p.C39A, p.C119A, p.C162A and p.C211A). Specifically, the mutations at Cys211 and Cys39 significantly decreased the S-nitrosation of MAPK14, suggesting that these cysteine residues are major S-nitrosation sites of MAPK14 (Fig. 5G, H). The Cys211 and Cys39 residues are evolutionarily conserved in humans, chimpanzees, rats and mice (Fig. 5I).
Next, we explored whether S-nitrosation of MAPK14 affects its kinase activity. HEK293T cells were transfected with an expression vector for HA-MAPK14-WT or one of the four mutants (p.C39A, p.C119A, p.C162A and p.C211A). We found that overexpression of the MAPK14 p.C211A mutant decreased the level of phospho-MAPK14, suggesting the decreased kinase activity of this mutant (Fig. 5J, K). Moreover, in NLRP3-reconstituted HEK293T cells, overexpression of the MAPK14 p.C211A mutant decreased the abundance of the Caspase-1 p20 protein in the supernatant, suggesting that expression of this mutant decreased NLRP3 inflammasome activity (Fig. 5L, M). Notably, the NLRP3 expression level was unaffected when NLRP3-reconstituted HEK293T cells were cotransfected with GSNOR due to the overexpression of these proteins. These results indicate that GSNOR modulates NLRP3 inflammasome activity by regulating the S-nitrosation of MAPK14 at the Cys211 residue. Notably, overexpression of the MAPK14 p.C39A mutant slightly increased the level of phosphorylated MAPK14 and the activity of the NLRP3 inflammasome (Fig. 5J–M). We speculated that mutation of this residue may cause a change in the MAPK14 protein structure, which in turn might expose the Cys211 residue for additional S-nitrosation, ultimately promoting the activity of MAPK14 and the NLRP3 inflammasome. Future studies should be performed to confirm this possibility.
Knockout of NLRP3 ameliorated LPS-induced septic shock and DSS-induced colitis in Gsnor−/− mice
To confirm that the severity of inflammatory diseases caused by GSNOR deficiency is mediated by the NLRP3 inflammasome, we evaluated LPS-induced septic shock and DSS-induced colitis in Gsnor−/− Nlrp3−/− mice. Deletion of Nlrp3 in Gsnor−/− mice decreased the DAI score, mortality, and weight loss and attenuated DSS-induced colon shortening (Fig. 6A–E). Moreover, Gsnor−/− Nlrp3−/− DKO mice exhibited attenuated destruction of the intestinal epithelium and infiltration of immune cells (Fig. 6F). The Western blot results showed that NLRP3 deficiency could (albeit partially) inhibit the activation of the NLRP3 inflammasome caused by Gsnor deletion (Fig. 6G, H). Furthermore, NLRP3 deficiency reduced mortality in the LPS-induced septic shock model (Fig. 6I). Compared with Gsnor−/− mice, Gsnor−/− Nlrp3−/− (DKO) mice exhibited attenuated immune cell infiltration and liver tissue damage (Fig. 6J). Moreover, NLRP3 deficiency decreased NLRP3 inflammasome activation induced by Gsnor deficiency in liver and spleen tissues (Fig. S5A, B).
Fig. 6.
Gsnor−/− mice with and without NLRP3 knockout exhibit differences in disease severity in the LPS-induced septic shock and DSS-induced colitis models. A Survival curves of wild-type (WT), Gsnor−/−, Nlrp3−/− and Gsnor−/−Nlrp3−/− mice treated with DSS (n = 8 mice per group). The procedure is the same as that shown in Fig. 2A. B, C Disease activity index scores (B) and body weight loss (C) in WT, Gsnor−/−, Nlrp3−/− and Gsnor−/−Nlrp3−/− mice treated with DSS (n = 5–8 mice per group). D–F Assessment of morphological changes in the colon (D), colon length (E), and pathological changes in colon tissue sections by hematoxylin and eosin staining (F) in the animals described in (B, C). G, H Protein levels of NLRP3, Caspase-1 (p20) and IL-1β in colon tissues (G) and the related quantitative analysis (H) in the animals described in (B, C) (n = 3 mice per group). I Survival curves of LPS-treated WT, Gsnor−/−, Nlrp3−/− and Gsnor−/−Nlrp3−/− mice (n = 8 mice per group). The procedure is the same as that shown in Fig. 3A. J Hematoxylin and eosin staining of liver tissues from WT, Gsnor−/−, Nlrp3−/− and Gsnor−/−Nlrp3−/− mice treated with LPS (n = 3 mice per group). The data are presented as the means ± SEMs for (B, C) and the means ± SDs for (E, H). Statistical significance was determined by the log-rank Mantel‒Cox test in (A, I). Two-tailed unpaired Student’s t test was used to quantify the significance of differences between the Gsnor−/− and Gsnor−/−Nlrp3−/− groups and between the Gsnor−/− and Nlrp3−/− groups on the indicated days (B, C). One‐way ANOVA was used for (E, H). *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. Scale bar, 50 μm
To demonstrate the inhibitory function of GSNOR mediated through MAPK14, we tested whether pharmacological inhibition of MAPK14 can ameliorate LPS-induced septic shock. We intraperitoneally injected Gsnor−/− mice and their WT littermates with SB203580 for two days before LPS treatment. Pretreatment with SB203580 caused a significant reduction in mortality in the LPS-induced septic shock model (Fig. S6A, B). Western blot analysis confirmed that SB203580 pretreatment reduced the NLRP3 and Caspase-1 p20 protein levels in the liver tissues of Gsnor−/− mice and their WT littermates (Fig. S6C, D).
Taken together, the above in vivo results confirm the proposed mechanism through which GSNOR regulates the inflammatory response by regulating the activity of the NLRP3 inflammasome via MAPK14.
Sepsis patients have increased mRNA levels of NLRP3 inflammasome genes and MAPK14
We hypothesized that it might be possible to extend our observations in mice to patients with inflammatory-related diseases. If this were so, we anticipated a decreased level of GSNOR and increased levels of MAPK14, NLRP3 and IL-1β mRNA expression in patients with sepsis. We therefore examined the expression matrix associated with sepsis in a dataset publicly available under Gene Expression Omnibus (GEO) accession ID GSE100159 [40] to validate our speculation. Indeed, we found that the mRNA levels of MAPK14, NLRP3 and IL-1β were higher in patients with sepsis than in healthy subjects, whereas the mRNA level of GSNOR exhibited the opposite pattern of change (Fig. 7). These results were consistent with the above findings in mouse models and further supported the key role of GSNOR as an important player in inflammatory diseases, highlighting its potential as a therapeutic target.
Fig. 7.

Relative mRNA levels of GSNOR, MAPK14, NLRP3, and IL-1β in patients with sepsis. The expression dataset was retrieved from GEO (GSE100159) and reanalyzed. The data are presented on violin plots. ns, not significant; *P < 0.05; **P < 0.01; ****P < 0.0001; two-tailed unpaired Student’s t test
Discussion
The activation of the NLRP3 inflammasome is closely regulated at the transcriptional and posttranslational levels to avoid excessive inflammation in the host [1, 5, 6]. Thus, the identification and characterization of unknown factors that regulate the NLRP3 inflammasome may yield important insights into developing targeted therapeutics to combat NLRP3 inflammasome-associated diseases. In this study, we demonstrated that GSNOR, an evolutionarily conserved enzyme of the denitrosylating enzymatic system [17], modulates the S-nitrosation of MAPK14, which in turn affects Nlrp3 and Il-1β expression. We provided direct evidence that knockout of GSNOR increases the S-nitrosation of MAPK14 at Cys211, promotes the phosphorylation of MAPK14, and increases Nlrp3 and Il-1β expression. This kind of interaction between S-nitrosation and phosphorylation in the regulation of protein activity, as occurs for MAPK14, has also been shown previously for different proteins and in different cell types [41]. In addition, we performed in vivo assays based on Gsnor–/–, Nlrp3–/– and Gsnor–/–Nlrp3–/– DKO mouse models and pharmacological treatments to show that GSNOR deficiency results in susceptibility to LPS-induced septic shock and dextran sodium sulfate (DSS)-induced colitis in mice and that NLRP3 knockout or SB203580 pretreatment reverses the deleterious effects of GSNOR deficiency. All these results show that GSNOR is a modulator of the NLRP3 inflammasome and that this modulatory effect of GSNOR has potential utility in the treatment of NLRP3-related inflammatory diseases (Fig. 8). Notably, the roles of the NLRP3 inflammasome in the pathogenesis of DSS-mediated colitis in murine models are controversial. Several reports have shown that NLRP3 inflammasome activation has a protective effect on DSS-induced colitis [42–45]. The discrepancy in outcomes across independent studies might be caused by disparities in the gut microbiota composition, variability in colitis models, and differences in the methodologies used to induce intestinal inflammation [46].
Fig. 8.
A proposed mechanism by which GSNOR regulates NLRP3 inflammasome activity. Treatment with lipopolysaccharide (LPS) or dextran sodium sulfate (DSS) results in reactive nitrogen stress (RNS), which causes S-nitrosation (SNO) of the MAPK14 protein. S-nitrosation of MAPK14 at Cys211 affects its phosphorylation (P) and augments its downstream signaling to upregulate cytokine expression, as observed in NLRP3 inflammasome-related disorders. GSNOR inhibits NLRP3 inflammasome activity by regulating MAPK14 S-nitrosation
The NLRP3 inflammasome plays pivotal roles in the host defense against microbial pathogens and is tightly controlled [2, 5, 47, 48]. The protein levels of NLRP3 inflammasome components are the rate-limiting elements for inflammasome activation [4, 49]. Previous studies have reported diverse regulators of the NLRP3 protein level and NLRP3 inflammasome complex assembly, most of which act through a posttranslational mechanism. For example, SUMOylation of NLRP3 by the SUMO E3 ligase MAPL restrains NLRP3 inflammasome activation [50]. The PKA kinase induces the phosphorylation and subsequent ubiquitination of NLRP3, which serves as a critical brake of NLRP3 inflammasome activation [15]. In addition to SUMOylation and ubiquitination, NLRP3 also undergoes other posttranslational modifications, such as deacetylation, in which the cytosolic deacetylase Sirtuin 2 (SIRT2) deacetylates NLRP3 and suppresses its activity [51]. Our current study provides another line of evidence indicating that the expression of NLRP3 inflammasome components can be regulated via GSNOR deficiency-mediated S-nitrosation of components of the MAPK signaling pathway (Fig. 8), which is actively involved in the expression of NLRP3 inflammasome components [6, 52].
As a highly evolutionarily conserved enzyme of the denitrosylation system, GSNOR modulates S-nitrosation through the catabolism of S-nitrosoglutathione (GSNO) [17]. Previous studies have indicated that impaired GSNOR activity could influence the S-nitrosation levels of its target proteins, ultimately affecting the function of these proteins [21, 22, 53]. Our finding that GSNOR deficiency mediated the S-nitrosation of the MAPK14 protein in the MAPK signaling pathway, which is the most active pathway in inflammatory cells and can respond to external stress stimuli [54–56], constitutes an example of a posttranslational modification that increases MAPK14 activity. A previous study reported a similar pattern, i.e., loss of GSNOR increased MAPK signaling activity in Solanum lycopersicum L. [57]. However, whether GSNOR has a direct regulatory effect on the expression of NLRP3 inflammasome components remains to be determined, although the levels of genes encoding components of the MAPK signaling pathway were found to be increased by transcriptome analysis of Gsnor–/– and WT BMDMs.
The key role of GSNOR in modulating NLRP3 inflammasome activity was shown in the two inflammatory disease mouse models (LPS-induced septic shock model and DSS-induced colitis model) used in this study. In both models, the NLRP3 inflammasome has been implicated as a major contributor to the effects of the inducing agents [26, 28]. Hyperactivation of the NLRP3 inflammasome can also worsen DSS-induced intestinal inflammatory damage [27]. Here, we found that Gsnor−/− mice exhibited greater NLRP3 inflammasome activation and more severe colitis than did WT mice following DSS administration, and these deleterious effects were attenuated by NLRP3 knockout. LPS injection can induce NLRP3-dependent IL-1β production and septic shock in mice [58]. We found that Gsnor−/− mice injected with LPS had a higher mortality rate and more severe pathological changes than WT mice. This result was consistent with that of a previous study in which GSNOR knockout mice were found to be more likely to die after LPS treatment [59]. Consistent with our speculation regarding the GSNOR deficiency–MAPK14–NLRP3 activation axis, Nlrp3 knockout or SB203580 pretreatment significantly alleviated LPS-mediated lethality and inflammatory activation in Gsnor−/− mice.
There are several limitations of this study. First, we found that GSNOR regulated NLRP3 inflammasome activation via the S-nitrosation of MAPK14 at Cys211. It is appropriate to establish a MAPK14 p.C211A knock-in mouse model to further validate this observation. Second, GSNOR knockout might have a widespread effect on the S-nitrosation of proteins and affect many signaling pathways, as GSNOR deficiency has been reported to induce the S-nitrosation of numerous protein targets [60]. It is thus plausible that the S-nitrosation of other targets, in addition to MAPK14, as shown here, might regulate NLRP3 inflammasome activation via unknown pathways. Third, we did not investigate how the balance between S-nitrosation and denitrosation of MAPK14 is maintained in vivo under physiological and pathological conditions, which could be worthwhile to explore in future studies.
In brief, we identified GSNOR as a critical negative regulator of Nlrp3 and Il-1β. These findings suggest the potential for a novel therapeutic strategy targeting GSNOR-mediated signaling pathways along with other interventions to alleviate NLRP3-mediated inflammatory diseases in human patients. Furthermore, our study highlighted the importance of diverse regulatory mechanisms in the control of the NLRP3 inflammasome, expanding our understanding of the complex regulatory mechanisms governing this important immune pathway.
Materials and methods
Antibodies and reagents
The following antibodies and chemicals were used in this study: rabbit polyclonal anti-GSNOR (Abcam; ab175406; 1:1000), rabbit monoclonal anti-NLRP3 (Cell Signaling Technology; D4D8T; 1:1000), rabbit polyclonal anti-IL-1β (Abcam; ab82558; 1:1000), mouse monoclonal anti-Caspase-1 (Adipogen; AG-20B-0042; 1:1000), rabbit polyclonal anti-ASC (Santa Cruz Biotechnology; sc-22514-R; 1:1000), mouse monoclonal anti-HA (Origene; TA180128; 1:1000), mouse monoclonal anti-Flag (Abmart; TT0003; 1:1000), rabbit monoclonal anti-p38α MAPK (Cell Signaling Technology; 9218; 1:1000), rabbit monoclonal anti-phospho-p38 MAPK (Cell Signaling Technology; 4511 T; 1:1000), mouse monoclonal anti-β-actin (Beijing Zhong Shan-Golden Bridge Biological Technology Co., Ltd.; TA-09; 1:10000), mouse monoclonal anti-tubulin (EnoGene; E1C601; 1:10000), BD Pharmingen™ FITC Rat Anti-CD11b (BD Biosciences; 561688; 1:200), BD Horizon™ BV605 Rat Anti-Mouse Ly-6G and Ly-6C (BD Biosciences; 563299; 1:200), peroxidase-conjugated anti-mouse (KPL; 474–1806; 1:10000), and peroxidase-conjugated anti-rabbit (KPL; 474–1516; 1:10000) antibodies. The following reagents were used in this study: LPS (Sigma; L2630), nigericin (Merck Millipore; 481990), DSS (Coolaber; CD4421), phorbol 12-myristate 13-acetate (PMA) (Sigma; P8139), SB203580 (Beyotime; S1863), methyl methanethiosulfonate (MMTS) (Sigma; 208795), biotin-HPDP (Glpbio; GC11037-100), Percoll (Solarbio; P8370), NADH (MedChemExpress; HY-F0001), GSNO (Santa Cruz; sc-200349A), SB203580 (Selleck; S1076), and ACK Lysis Buffer (Beyotime; C3702).
Cell preparation and stimulation
Human monocytes (THP-1 cell line), mouse fibroblasts (L929 cell line), monocytes/macrophages (RAW264.7 cell line) and HEK293T cells were obtained from the Kunming Cell Bank, Kunming Institute of Zoology, Chinese Academy of Sciences. THP-1, RAW264.7 and L929 cells were cultured in RPMI 1640 medium (Gibco). HEK293T cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Gibco-BRL, 11965–092). THP-1 cells were differentiated for 36 h with 50 nM phorbol 12-myristate 13-acetate (PMA) and subsequently replated. All culture media were supplemented with 10% fetal bovine serum (FBS; Gibco, 10099-141) and 1× penicillin/streptomycin (Gibco, 15140122) and maintained at 37 °C in a 5% CO2 atmosphere.
Mouse bone marrow-derived macrophages (BMDMs) were obtained via a modified protocol described previously [61]. Briefly, BMDMs were isolated from the femora and tibiae and cultured for 7 days in DMEM supplemented with 10% FBS, 20% culture supernatant from L929 mouse fibroblasts, and 1× penicillin/streptomycin (Gibco, 15140122). The culture medium was changed to fresh medium on Day 3 and Day 5. To stimulate NLRP3 inflammasome activation, BMDMs were primed with 500 ng/mL LPS (Sigma; L2630) for 6 h and were then stimulated with the NLRP3 inflammasome activator nigericin (15 µM; Merck Millipore, 481990) for 30 min, as previously reported [62, 63].
Reconstitution of the NLRP3 Inflammasome in HEK293T cells
NLRP3-reconstituted HEK293T cells were generated as previously described [64]. Briefly, HEK293T cells were seeded into 6-well plates at 5 × 105 cells per well in a complete cell culture medium. After 12 h, the cells were transfected with plasmids expressing Flag-Pro-caspase-1 (500 ng), Flag-ASC (150 ng) and Flag-NLRP3 (200 ng) using Lipofectamine 3000 (Invitrogen, L3000015). After 48 h, the cells were stimulated with 10 μM nigericin for 2 to 3 h, and pro-caspase-1 cleavage was analyzed in lysates and supernatants by Western blotting.
Plasmid construction and transfection
Expression vectors for GSNOR, MAPK14 and site-directed mutants of MAPK14 (p.C39A, p.C119A, p.C162A and p.C211A) were purchased from the Miaoling Plasmid Sharing Platform (MLPSP). A site-directed mutant of GSNOR (p.R115D) was constructed using the Easy Mutagenesis System (Beijing TransGen Biotech) according to the manufacturer’s protocols. All constructs were confirmed by DNA sequencing. Transient transfection of vectors was performed using Lipofectamine 3000 (Invitrogen, L3000015). Briefly, cells were seeded in either 6-well plates or 10 cm cell culture dishes. When the cells were 50–60% confluent, the growth medium was removed, and the cells were washed once with Opti-MEM (Gibco BRL, 31985–070) for transfection. The expression vectors or empty vector (2.5 μg/well or 10 μg/dish) were mixed with Opti-MEM (125 μL/well or 500 μL/dish) containing P3000™ reagent (5 μL/well or 20 μL/dish), and this mixture was then mixed with Lipofectamine™ 3000 (3.75 μL/well or 15 μL/dish) diluted in Opti-MEM (125 μL/well or 500 μL/dish). The final mixture was incubated at room temperature for 15 min and added to each well or dish together with additional Opti-MEM (750 μL/well or 3000 μL/dish). The medium was removed 6 h after transfection, and fresh growth medium (2 mL/well or 8 mL/dish) was added for growth until the cells were harvested 48 h after transfection.
Animal models of inflammation
Gsnor KO (Gsnor−/−) and Nlrp3 KO (Nlrp3−/−) mice on a C57BL/6J background were described in our previous studies [19, 20, 63]. All mice were housed under specific pathogen-free conditions within the Experimental Animal Center at the Kunming Institute of Zoology, Chinese Academy of Sciences. All animal experimental procedures and protocols were approved by the Institutional Review Board of the Kunming Institute of Zoology, Chinese Academy of Sciences (approval no: IACUC-PA-2022-03-020).
To induce and assess dextran sulfate sodium (DSS)-induced colitis, male C57BL/6 mice aged 8-10 weeks, as well as Nlrp3−/−, Gsnor−/−, and Nlrp3−/−Gsnor−/− DKO mice, were used. These mice were subjected to a six-day treatment regimen with 3% DSS (Coolaber; CD4421) dissolved in their drinking water followed by access to regular drinking water until the end of the experiment. Body weight was measured daily at consistent time points. Disease activity index (DAI) scores, ranging from 0 to 4, were assigned as follows: 0 for normal stool; 1 for soft stool; 2 for dark mucoid stool; 3 for liquid stool with slight bleeding; and 4 for gross bleeding [26]. Another cohort of surviving DSS-treated animals was euthanized on Day 6, after which the colon length was measured and the tissues were collected for hematoxylin and eosin (H&E) staining and Western blot analysis.
The lipopolysaccharide (LPS) challenge experiment was performed on male C57BL/6 mice aged 8 to 10 weeks. Briefly, the animals were intraperitoneally injected with LPS at a dose of 20 mg/kg body weight. After 6 h, the mice were euthanized, and serum, liver, and spleen samples were obtained for subsequent analysis. For the endotoxic shock model [12, 28], mice were intraperitoneally injected with LPS at a dose of 20 mg/kg body weight and monitored for lethality over a period of 3 days. For SB203580 pretreatment, mice were intraperitoneally injected with SB203580 at a dose of 25 mg/kg body weight for two consecutive days, and LPS was then injected intraperitoneally.
qRT‒PCR, Western blotting and immunoprecipitation
Total RNA was isolated from cells using TRIzol (Invitrogen, 15596-018). Single-strand cDNA was synthesized from 1 μg of total RNA using M-MLV Reverse Transcriptase (Promega, M170A) according to the manufacturer’s instructions. qRT‒PCR was performed to quantify the relative mRNA levels of Nlrp3, Caspase-1, Asc and Il-1β, with β-actin serving as the reference gene. qRT‒PCR was carried out in a total volume of 20 μL containing 2 μL of diluted template, 10 μL of SYBR Master Mix (Takara), and 0.2 μL of each primer (10 μM; Table S1) on a Bio-Rad real-time PCR detection system. The thermal cycling conditions included a denaturation step at 95 °C for 5 min, followed by 40 cycles at 95 °C for 10 s and 57 °C for 30 s.
Western blot analysis of the respective target proteins was performed using standard procedures as described in our previous studies [18, 19, 63]. Lysates of cultured mouse BMDMs, HEK293T cells, RAW264.7 cells and THP-1 cells were prepared, and protein concentrations were quantified by using a BCA protein assay kit (Beyotime Institute of Biotechnology, P0012). A total of 25 μg of protein was separated by 12% or 15% sodium dodecyl sulfate‒polyacrylamide gel electrophoresis (SDS‒PAGE), and the separated proteins were transferred to a polyvinylidene difluoride membrane (Bio-Rad, L1620177 Rev D). The membrane was soaked in 1x TBST with 5% BSA (for detection of phosphorylated proteins) or 5% (w/v) skim milk (for detection of other proteins) for 2 h at room temperature and then incubated with the indicated primary antibody overnight at 4 °C. Next, the membrane was washed with 1× TBST three times (for five minutes each) and incubated with the corresponding peroxidase-conjugated anti-mouse (lot number 474-1806) or anti-rabbit (lot number 474–1516) secondary antibody for 1 h at room temperature. After another round of three washes with 1× TBST, the immune complexes on the membrane were visualized using an ECL Western Blot Detection Kit (Millipore, WBKLS0500).
For immunoprecipitation assays, the indicated antibodies were incubated with protein G-agarose beads (15920010; Life Technologies) for 2 h at room temperature to form complexes. Cells were lysed with RIPA lysis buffer on ice for 1 h prior to centrifugation at 12 000 × g for 10 min at 4 °C. Lysates were immunoprecipitated with the antibody–bead complexes at 4 °C overnight, and the precipitates were then washed with RIPA lysis buffer and resuspended in sample loading buffer for SDS‒PAGE as described above.
Biotin switch assay
S-nitrosylation was detected as previously described [18, 19, 65]. Briefly, cells were lysed with HENS buffer, and cysteine thiols were blocked by treatment with methyl methanethiosulfonate (MMTS) at room temperature for 30 min. Excess MMTS was removed through ice-cold acetone precipitation by centrifugation at 2000 × g for 10 min. The resulting precipitate was resuspended in HENS buffer containing 2.5% SDS, 0.4 mM biotin-maleimide, and 10 mM ascorbate and incubated at 37 °C for 1 h or at room temperature for 2 h. Excess biotin-maleimide was removed by ice-cold acetone precipitation. The resulting pellet was resuspended in HENS buffer supplemented with 200 mM DTT and incubated at 100 °C for 15 min. Subsequently, neutralization buffer (250 mM HEPES (pH 7.7), 100 mM NaCl, 0.1 mM EDTA, and 10 mM neocuproine) and streptavidin-agarose (50–100 μL/sample) were added to purify the biotinylated proteins. The mixture was incubated at room temperature for 2 h, and the eluate was analyzed using 12% SDS‒PAGE.
Histopathological staining
Liver and intestinal tissues from mice were fixed with 10% phosphate-buffered formalin, embedded in paraffin, sectioned, stained with hematoxylin and eosin (H&E) solution, and examined by microscopy (Leica, Germany) for histological analysis following a procedure described in our previous study [66].
Colon organ culture
Colon tissues were cultured as previously described [67, 68] with minor modifications. Briefly, colon tissues were flushed with ice-cold PBS containing 1× penicillin/streptomycin, opened longitudinally, and sliced into sections. Tissue segments weighing approximately 100 mg were placed in RPMI 1640 medium supplemented with 1× penicillin/streptomycin in a 24-well culture plate and incubated for 24 h at 37 °C with 5% CO2. The culture supernatants were collected and stored at −80 °C until analysis.
Enzyme-linked immunosorbent assay (ELISA)
Supernatants from cell culture, serum or colon tissue culture were assayed for mouse IL-1β (Elabscience; E-EL-M0037c) and mouse TNF-α (Elabscience; E-EL-M3063) according to the manufacturer’s instructions. Briefly, we added the test samples, together with serially diluted reference standard samples (each 25 µL), to each well of a 96-well plate and then incubated the plate at 37 °C for 90 min. After aspirating the solution in each well, we promptly added the biotinylated detection antibody working solution (50 µL) to each well, and the plate was then incubated for an additional 60 min at 37 °C. After three washes with washing buffer, we added horseradish peroxidase (HRP)-conjugated working solution (50 µL/well), incubated the plate at 37 °C for 30 min, and washed the plate five times before adding the substrate reagent (50 µL/well). After another 15 min incubation at 37 °C, stop solution (50 µL/well) was added, and the absorbance of each well was immediately measured at 450 nm on a fully automatic microplate reader (ELX808; BioTek Instruments, Inc.).
Determination of GSNOR enzymatic activity
GSNOR enzymatic activity was determined by monitoring GSNO-dependent NADH consumption at 340 nm as previously described [17]. Forty microliters of lysate (250 µg of total extracted protein) from LPS-treated and untreated RAW264.7 cells was added to 200 µL of reaction buffer (20 mM Tris-HCl, pH 8.0), and 30 µL of 2 mM NADH was then added at room temperature. The reaction was started by the addition of 30 µL of 4 mM GSNO, and 30 µL of water was used as a blank control. Then, the samples were monitored for a decrease in the absorbance at 340 nm in an Infinite M1000 Pro multimode microplate reader (30064852; Tecan) at 1–10 min intervals.
Flow cytometry
Mouse liver tissue was homogenized and passed through a 70 μm cell strainer (Corning). The resulting cell mixture was centrifuged at 200 × g for 5 min, after which the pellet was resuspended in ACK Lysis Buffer (Beyotime; C3702) and incubated at room temperature for 5 min to remove red blood cells. After another round of centrifugation at 200 × g for 5 min, the pellet was resuspended in 4 mL 30% percoll and laid onto 4 mL 80% percoll, followed by centrifugation at 900 × g for 30 min at minimal acc/dec speed. The interface layer was harvested and the isolated cells was resuspended in FACS buffer (PBS with 2% BSA), and antibody cocktails were added. After incubation at 4 °C for 30 min, the cells were washed three times with PBS (5 min each), resuspended in PBS supplemented with 1% PFA, and subjected to flow cytometric analysis using a flow cytometer (BD LSRFortessa) and FlowJo software.
RNA sequencing (RNA-seq) and data analysis
We performed RNA-seq analysis following a previously described procedure [69, 70]. Briefly, RNA was isolated from cells using TRIzol (Invitrogen, 15596-018), and quality control, library construction, and sequencing were performed by Biolinker Technology (Kunming) Co., Ltd. The raw sequencing reads were processed with Trimmomatic (version 0.39) software [71] to remove adapter sequences and low-quality reads, employing the parameters “LEADING:3 TRAILING:3 SLIDINGWINDOW:4:15 MINLEN:36”. After read filtering, the clean reads were aligned to the mouse reference genome GRCm39 (https://www.ensembl.org/Mus_musculus/Info/Index?db=core) using STAR software (version 2.7.3a) [72]. The aligned reads were then processed in binary alignment map (bam) format using the featureCounts function within the Subread software package [73]. With this function, the uniquely mapped reads corresponding to exons (-t exon -g gene id) were allocated and counted based on the GRCm39 annotation file (Mus_musculus.GRCm39.104.gtf). After combining the gene expression levels for each sample, genes with low expression levels (row means of gene read counts of less than 10) were filtered out. The rlogTransformation function in the R package DESeq2 [74] was used to normalize and scale the read counts. Principal component analysis and differential expression analysis were performed using the DESeq2 R package [74] based on the normalized counts. Differential expression was evaluated with DESeq2, and adjusted P values (P.adjust) were obtained using the Benjamini–Hochberg (BH) method. Genes with P.adjust < 0.05 were considered differentially expressed. Gene Ontology (GO) term enrichment and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses were performed by using the R package clusterProfiler (version 4.0) [75], with the differentially expressed genes identified by DESeq2 used as input. Data visualization was performed with the R packages ggplot2 (https://ggplot2.tidyverse.org/) and ComplexHeatmap [76].
Statistical analysis
All the statistical analyses were performed using GraphPad Prism v7.0. Student’s t test was used for statistical analysis of differences between two groups, one‐way ANOVA was used for statistical analysis of differences among multiple groups, and the log-rank (Mantel‒Cox) test was used for statistical analysis of survival data. The data are presented as the means ± SDs or means ± SEMs as appropriate for the analysis conditions. A P value < 0.05 was considered to indicate statistical significance.
Supplementary information
Acknowledgements
The authors thank Prof. Rongbin Zhou for sharing the Nlrp3−/− mice. We are grateful to Dr. Ian Logan for the helpful comments and language editing of this manuscript. This study was supported by Yunnan Fundamental Research Project (202305AH340006), the National Natural Science Foundation of China (32201018), the Basic Research Program and Key Project of Yunnan Province (202301AW070013 and 202003AD150009), and the Youth Innovation Promotion Association (2023403).
Author contributions
Yong-Gang Yao and Qianjin Liu conceived and designed the experiments. Qianjin Liu, Lijin Jiao, Zhiyu Ma, Jinsong Yu, Ling-Yan Su, Wei-Yin Zou and Lu-Xiu Yang performed the cellular and mouse experiments. Mao-Sen Ye performed the RNA-seq data analysis. Chang Chen provided mouse lines and provided constructive suggestions. Yong-Gang Yao and Qianjin Liu wrote the manuscript. All authors reviewed the content and approved the final version for publication.
Competing interests
The authors declare no competing interests.
Contributor Information
Qianjin Liu, Email: liuqianjin@mail.kiz.ac.cn.
Yong-Gang Yao, Email: yaoyg@mail.kiz.ac.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41423-024-01155-9.
References
- 1.Swanson KV, Deng M, Ting JP. The NLRP3 inflammasome: molecular activation and regulation to therapeutics. Nat Rev Immunol. 2019;19:477–89. doi: 10.1038/s41577-019-0165-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Strowig T, Henao-Mejia J, Elinav E, Flavell R. Inflammasomes in health and disease. Nature. 2012;481:278–86. doi: 10.1038/nature10759. [DOI] [PubMed] [Google Scholar]
- 3.Huang Y, Xu W, Zhou R. NLRP3 inflammasome activation and cell death. Cell Mol Immunol. 2021;18:2114–27. doi: 10.1038/s41423-021-00740-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fu J, Wu H. Structural mechanisms of NLRP3 inflammasome assembly and activation. Annu Rev Immunol. 2023;41:301–16. doi: 10.1146/annurev-immunol-081022-021207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paik S, Kim JK, Silwal P, Sasakawa C, Jo EK. An update on the regulatory mechanisms of NLRP3 inflammasome activation. Cell Mol Immunol. 2021;18:1141–60. doi: 10.1038/s41423-021-00670-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cornut M, Bourdonnay E, Henry T. Transcriptional regulation of inflammasomes. Int J Mol Sci. 2020;21:8087. doi: 10.3390/ijms21218087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Luo F, Li H, Ma W, Cao J, Chen Q, Lu F, et al. The BCL-2 inhibitor APG-2575 resets tumor-associated macrophages toward the M1 phenotype, promoting a favorable response to anti-PD-1 therapy via NLRP3 activation. Cell Mol Immunol. 2024;21:60–79. doi: 10.1038/s41423-023-01112-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu Q, Zhao B, Ye Y, Li Y, Zhang Y, Xiong X, et al. Relevant mediators involved in and therapies targeting the inflammatory response induced by activation of the NLRP3 inflammasome in ischemic stroke. J Neuroinflammation. 2021;18:123. doi: 10.1186/s12974-021-02137-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coll RC, Schroder K, Pelegrin P. NLRP3 and pyroptosis blockers for treating inflammatory diseases. Trends Pharm Sci. 2022;43:653–68. doi: 10.1016/j.tips.2022.04.003. [DOI] [PubMed] [Google Scholar]
- 10.Mangan MSJ, Olhava EJ, Roush WR, Seidel HM, Glick GD, Latz E. Targeting the NLRP3 inflammasome in inflammatory diseases. Nat Rev Drug Discov. 2018;17:588–606. doi: 10.1038/nrd.2018.97. [DOI] [PubMed] [Google Scholar]
- 11.Zeng J, Xie X, Feng XL, Xu L, Han JB, Yu D, et al. Specific inhibition of the NLRP3 inflammasome suppresses immune overactivation and alleviates COVID-19 like pathology in mice. EBioMedicine. 2022;75:103803. doi: 10.1016/j.ebiom.2021.103803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qin Y, Li Q, Liang W, Yan R, Tong L, Jia M, et al. TRIM28 SUMOylates and stabilizes NLRP3 to facilitate inflammasome activation. Nat Commun. 2021;12:4794. doi: 10.1038/s41467-021-25033-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haneklaus M, O’Neill LA, Coll RC. Modulatory mechanisms controlling the NLRP3 inflammasome in inflammation: recent developments. Curr Opin Immunol. 2013;25:40–5. doi: 10.1016/j.coi.2012.12.004. [DOI] [PubMed] [Google Scholar]
- 14.Di Q, Zhao X, Tang H, Li X, Xiao Y, Wu H, et al. USP22 suppresses the NLRP3 inflammasome by degrading NLRP3 via ATG5-dependent autophagy. Autophagy. 2023;19:873–85. doi: 10.1080/15548627.2022.2107314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guo C, Xie S, Chi Z, Zhang J, Liu Y, Zhang L, et al. Bile acids control inflammation and metabolic disorder through inhibition of NLRP3 inflammasome. Immunity. 2016;45:802–16. doi: 10.1016/j.immuni.2016.09.008. [DOI] [PubMed] [Google Scholar]
- 16.McKee CM, Coll RC. NLRP3 inflammasome priming: a riddle wrapped in a mystery inside an enigma. J Leukoc Biol. 2020;108:937–52. doi: 10.1002/JLB.3MR0720-513R. [DOI] [PubMed] [Google Scholar]
- 17.Liu L, Hausladen A, Zeng M, Que L, Heitman J, Stamler JS. A metabolic enzyme for S-nitrosothiol conserved from bacteria to humans. Nature. 2001;410:490–4. doi: 10.1038/35068596. [DOI] [PubMed] [Google Scholar]
- 18.Jiao L, Su LY, Liu Q, Luo R, Qiao X, Xie T, et al. GSNOR deficiency attenuates MPTP-induced neurotoxicity and autophagy by facilitating CDK5 S-nitrosation in a mouse model of Parkinson’s disease. Free Radic Biol Med. 2022;189:111–21. doi: 10.1016/j.freeradbiomed.2022.07.016. [DOI] [PubMed] [Google Scholar]
- 19.Liu Q, Gu T, Su LY, Jiao L, Qiao X, Xu M, et al. GSNOR facilitates antiviral innate immunity by restricting TBK1 cysteine S-nitrosation. Redox Biol. 2021;47:102172. doi: 10.1016/j.redox.2021.102172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Y, Wu K, Su W, Zhang DF, Wang P, Qiao X, et al. Increased GSNOR expression during aging impairs cognitive function and decreases S-Nitrosation of CaMKIIα. J Neurosci. 2017;37:9741–58. doi: 10.1523/JNEUROSCI.0681-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chatterji A, Banerjee D, Billiar TR, Sengupta R. Understanding the role of S-nitrosylation/nitrosative stress in inflammation and the role of cellular denitrosylases in inflammation modulation: implications in health and diseases. Free Radic Biol Med. 2021;172:604–21. doi: 10.1016/j.freeradbiomed.2021.07.015. [DOI] [PubMed] [Google Scholar]
- 22.Barnett SD, Buxton ILO. The role of S-nitrosoglutathione reductase (GSNOR) in human disease and therapy. Crit Rev Biochem Mol Biol. 2017;52:340–54. doi: 10.1080/10409238.2017.1304353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tang X, Pan L, Zhao S, Dai F, Chao M, Jiang H, et al. SNO-MLP (S-Nitrosylation of muscle LIM protein) facilitates myocardial hypertrophy through TLR3 (Toll-Like Receptor 3)-mediated RIP3 (Receptor-Interacting Protein Kinase 3) and NLRP3 (NOD-Like Receptor Pyrin Domain Containing 3) inflammasome activation. Circulation. 2020;141:984–1000. doi: 10.1161/CIRCULATIONAHA.119.042336. [DOI] [PubMed] [Google Scholar]
- 24.Tang X, Zhao S, Liu J, Liu X, Sha X, Huang C, et al. Mitochondrial GSNOR alleviates cardiac dysfunction via ANT1 denitrosylation. Circ Res. 2023;133:220–36. doi: 10.1161/CIRCRESAHA.123.322654. [DOI] [PubMed] [Google Scholar]
- 25.Patel MN, Carroll RG, Galvan-Pena S, Mills EL, Olden R, Triantafilou M, et al. Inflammasome priming in sterile inflammatory disease. Trends Mol Med. 2017;23:165–80. doi: 10.1016/j.molmed.2016.12.007. [DOI] [PubMed] [Google Scholar]
- 26.Chen Y, He H, Lin B, Chen Y, Deng X, Jiang W, et al. RRx-001 ameliorates inflammatory diseases by acting as a potent covalent NLRP3 inhibitor. Cell Mol Immunol. 2021;18:1425–36. doi: 10.1038/s41423-021-00683-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bauer C, Duewell P, Mayer C, Lehr HA, Fitzgerald KA, Dauer M, et al. Colitis induced in mice with dextran sulfate sodium (DSS) is mediated by the NLRP3 inflammasome. Gut. 2010;59:1192–9. doi: 10.1136/gut.2009.197822. [DOI] [PubMed] [Google Scholar]
- 28.Xu G, Fu S, Zhan X, Wang Z, Zhang P, Shi W, et al. Echinatin effectively protects against NLRP3 inflammasome-driven diseases by targeting HSP90. JCI Insight. 2021;6:e134601. doi: 10.1172/jci.insight.134601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin Y, Li Z, Wang Y, Tian T, Jia P, Ye Y, et al. CCDC50 suppresses NLRP3 inflammasome activity by mediating autophagic degradation of NLRP3. EMBO Rep. 2022;23:e54453. doi: 10.15252/embr.202154453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cooper HS, Murthy SN, Shah RS, Sedergran DJ. Clinicopathologic study of dextran sulfate sodium experimental murine colitis. Lab Invest. 1993;69:238–49. [PubMed] [Google Scholar]
- 31.Ma H, Hu T, Tao W, Tong J, Han Z, Herndler-Brandstetter D, et al. A lncRNA from an inflammatory bowel disease risk locus maintains intestinal host-commensal homeostasis. Cell Res. 2023;33:372–88. doi: 10.1038/s41422-023-00790-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chassaing B, Aitken JD, Malleshappa M, Vijay-Kumar M. Dextran sulfate sodium (DSS)-induced colitis in mice. Curr Protoc Immunol. 2014;104:15 25 1–15 25 14. doi: 10.1002/0471142735.im1525s104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sutterwala FS, Ogura Y, Szczepanik M, Lara-Tejero M, Lichtenberger GS, Grant EP, et al. Critical role for NALP3/CIAS1/Cryopyrin in innate and adaptive immunity through its regulation of caspase-1. Immunity. 2006;24:317–27. doi: 10.1016/j.immuni.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 34.Blevins HM, Xu Y, Biby S, Zhang S. The NLRP3 inflammasome pathway: a review of mechanisms and inhibitors for the treatment of inflammatory diseases. Front Aging Neurosci. 2022;14:879021. doi: 10.3389/fnagi.2022.879021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen Y, Wang CY, Zan GY, Yao SY, Deng YZ, Shu XL, et al. Upregulation of dynorphin/kappa opioid receptor system in the dorsal hippocampus contributes to morphine withdrawal-induced place aversion. Acta Pharm Sin. 2023;44:538–45. doi: 10.1038/s41401-022-00987-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Engeland K, Höög JO, Holmquist B, Estonius M, Jörnvall H, Vallee BL. Mutation of Arg-115 of human class III alcohol dehydrogenase: a binding site required for formaldehyde dehydrogenase activity and fatty acid activation. Proc Natl Acad Sci USA. 1993;90:2491–4. doi: 10.1073/pnas.90.6.2491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu K, Ren R, Su W, Wen B, Zhang Y, Yi F, et al. A novel suppressive effect of alcohol dehydrogenase 5 in neuronal differentiation. J Biol Chem. 2014;289:20193–9. doi: 10.1074/jbc.C114.561860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hess DT, Matsumoto A, Kim SO, Marshall HE, Stamler JS. Protein S-nitrosylation: purview and parameters. Nat Rev Mol Cell Biol. 2005;6:150–66. doi: 10.1038/nrm1569. [DOI] [PubMed] [Google Scholar]
- 39.Yang L, Calay ES, Fan J, Arduini A, Kunz RC, Gygi SP, et al. S-Nitrosylation links obesity-associated inflammation to endoplasmic reticulum dysfunction. Science. 2015;349:500–6. doi: 10.1126/science.aaa0079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Altman MC, Baldwin N, Whalen E, Al-Shaikhly T, Presnell S, Khaenam P et al. A transcriptome fingerprinting assay for clinical immune monitoring. bioRxiv. 2019:587295. 10.1101/587295.
- 41.Irie T, Sips PY, Kai S, Kida K, Ikeda K, Hirai S, et al. S-Nitrosylation of calcium-handling proteins in cardiac adrenergic signaling and hypertrophy. Circ Res. 2015;117:793–803. doi: 10.1161/CIRCRESAHA.115.307157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bauer C, Duewell P, Lehr HA, Endres S, Schnurr M. Protective and aggravating effects of Nlrp3 inflammasome activation in IBD models: influence of genetic and environmental factors. Dig Dis. 2012;30:82–90. doi: 10.1159/000341681. [DOI] [PubMed] [Google Scholar]
- 43.Zaki MH, Boyd KL, Vogel P, Kastan MB, Lamkanfi M, Kanneganti TD. The NLRP3 inflammasome protects against loss of epithelial integrity and mortality during experimental colitis. Immunity. 2010;32:379–91. doi: 10.1016/j.immuni.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Allen IC, TeKippe EM, Woodford RM, Uronis JM, Holl EK, Rogers AB, et al. The NLRP3 inflammasome functions as a negative regulator of tumorigenesis during colitis-associated cancer. J Exp Med. 2010;207:1045–56. doi: 10.1084/jem.20100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang X, Feng Z, Jiang Y, Li J, Xiang Q, Guo S, et al. VSIG4 mediates transcriptional inhibition of Nlrp3 and Il-1β in macrophages. Sci Adv. 2019;5:eaau7426. doi: 10.1126/sciadv.aau7426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Meng G, Kirschning CJ, Zhou R. Editorial: patho- and physiological roles of inflammasomes. Front Immunol. 2022;13:857929. doi: 10.3389/fimmu.2022.857929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hise AG, Tomalka J, Ganesan S, Patel K, Hall BA, Brown GD, et al. An essential role for the NLRP3 inflammasome in host defense against the human fungal pathogen Candida albicans. Cell Host Microbe. 2009;5:487–97. doi: 10.1016/j.chom.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gao Y, Yu S, Chen M, Wang X, Pan L, Wei B, et al. cFLIP(S) regulates alternative NLRP3 inflammasome activation in human monocytes. Cell Mol Immunol. 2023;20:1203–15. doi: 10.1038/s41423-023-01077-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huai W, Zhao R, Song H, Zhao J, Zhang L, Zhang L, et al. Aryl hydrocarbon receptor negatively regulates NLRP3 inflammasome activity by inhibiting NLRP3 transcription. Nat Commun. 2014;5:4738. doi: 10.1038/ncomms5738. [DOI] [PubMed] [Google Scholar]
- 50.Barry R, John SW, Liccardi G, Tenev T, Jaco I, Chen CH, et al. SUMO-mediated regulation of NLRP3 modulates inflammasome activity. Nat Commun. 2018;9:3001. doi: 10.1038/s41467-018-05321-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.He M, Chiang HH, Luo H, Zheng Z, Qiao Q, Wang L, et al. An acetylation switch of the NLRP3 inflammasome regulates aging-associated chronic inflammation and insulin resistance. Cell Metab. 2020;31:580–91. doi: 10.1016/j.cmet.2020.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen MY, Ye XJ, He XH, Ouyang DY. The signaling pathways regulating NLRP3 inflammasome activation. Inflammation. 2021;44:1229–45. doi: 10.1007/s10753-021-01439-6. [DOI] [PubMed] [Google Scholar]
- 53.Li J, Zhang Y, Zhang Y, Lu S, Miao Y, Yang J, et al. GSNOR modulates hyperhomocysteinemia-induced T cell activation and atherosclerosis by switching Akt S-nitrosylation to phosphorylation. Redox Biol. 2018;17:386–99. doi: 10.1016/j.redox.2018.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hale KK, Trollinger D, Rihanek M, Manthey CL. Differential expression and activation of p38 mitogen-activated protein kinase alpha, beta, gamma, and delta in inflammatory cell lineages. J Immunol. 1999;162:4246–52. doi: 10.4049/jimmunol.162.7.4246. [DOI] [PubMed] [Google Scholar]
- 55.Luo Q, Schnoder L, Hao W, Litzenburger K, Decker Y, Tomic I, et al. p38alpha-MAPK-deficient myeloid cells ameliorate symptoms and pathology of APP-transgenic Alzheimer’s disease mice. Aging Cell. 2022;21:e13679. doi: 10.1111/acel.13679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hotamisligil GS, Davis RJ. Cell signaling and stress responses. Cold Spring Harb Perspect Biol. 2016;8:a006072. doi: 10.1101/cshperspect.a006072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang X, Du H, Shi QH, Gong BA. Loss of GSNOR increases abiotic stress sensitivity via regulating MAPK-ethylene cascade signaling in Solanum lycopersicum L. Environ Exp Bot. 2022;199.
- 58.He Y, Franchi L, Nunez G. TLR agonists stimulate Nlrp3-dependent IL-1beta production independently of the purinergic P2X7 receptor in dendritic cells and in vivo. J Immunol. 2013;190:334–9. doi: 10.4049/jimmunol.1202737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu L, Yan Y, Zeng M, Zhang J, Hanes MA, Ahearn G, et al. Essential roles of S-nitrosothiols in vascular homeostasis and endotoxic shock. Cell. 2004;116:617–28. doi: 10.1016/S0092-8674(04)00131-X. [DOI] [PubMed] [Google Scholar]
- 60.Rizza S, Cardaci S, Montagna C, Di Giacomo G, De Zio D, Bordi M, et al. S-nitrosylation drives cell senescence and aging in mammals by controlling mitochondrial dynamics and mitophagy. Proc Natl Acad Sci USA. 2018;115:E3388–e97. doi: 10.1073/pnas.1722452115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.He H, Jiang H, Chen Y, Ye J, Wang A, Wang C, et al. Oridonin is a covalent NLRP3 inhibitor with strong anti-inflammasome activity. Nat Commun. 2018;9:2550. doi: 10.1038/s41467-018-04947-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Huang B, Qian Y, Xie S, Ye X, Chen H, Chen Z, et al. Ticagrelor inhibits the NLRP3 inflammasome to protect against inflammatory disease independent of the P2Y(12) signaling pathway. Cell Mol Immunol. 2021;18:1278–89. doi: 10.1038/s41423-020-0444-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liu Q, Su LY, Sun C, Jiao L, Miao Y, Xu M, et al. Melatonin alleviates morphine analgesic tolerance in mice by decreasing NLRP3 inflammasome activation. Redox Biol. 2020;34:101560. doi: 10.1016/j.redox.2020.101560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Guo C, Chi Z, Jiang D, Xu T, Yu W, Wang Z, et al. Cholesterol homeostatic regulator SCAP-SREBP2 integrates NLRP3 inflammasome activation and cholesterol biosynthetic signaling in macrophages. Immunity. 2018;49:842–56. doi: 10.1016/j.immuni.2018.08.021. [DOI] [PubMed] [Google Scholar]
- 65.Yi W, Zhang Y, Liu B, Zhou Y, Liao D, Qiao X, et al. Protein S-nitrosylation regulates proteostasis and viability of hematopoietic stem cell during regeneration. Cell Rep. 2021;34:108922. doi: 10.1016/j.celrep.2021.108922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yu D, Long Y, Xu L, Han JB, Xi J, Xu J, et al. Infectivity of SARS-CoV-2 and protection against reinfection in rats. Zool Res. 2022;43:945–48. doi: 10.24272/j.issn.2095-8137.2022.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang X, Cai J, Lin B, Ma M, Tao Y, Zhou Y, et al. GPR34-mediated sensing of lysophosphatidylserine released by apoptotic neutrophils activates type 3 innate lymphoid cells to mediate tissue repair. Immunity. 2021;54:1123–36. doi: 10.1016/j.immuni.2021.05.007. [DOI] [PubMed] [Google Scholar]
- 68.Allen IC, Wilson JE, Schneider M, Lich JD, Roberts RA, Arthur JC, et al. NLRP12 suppresses colon inflammation and tumorigenesis through the negative regulation of noncanonical NF-κB signaling. Immunity. 2012;36:742–54. doi: 10.1016/j.immuni.2012.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Luo R, Fan Y, Yang J, Ye M, Zhang DF, Guo K, et al. A novel missense variant in ACAA1 contributes to early-onset Alzheimer’s disease, impairs lysosomal function, and facilitates amyloid-β pathology and cognitive decline. Signal Transduct Target Ther. 2021;6:325. doi: 10.1038/s41392-021-00748-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ye MS, Zhang JY, Yu DD, Xu M, Xu L, Lv LB, et al. Comprehensive annotation of the Chinese tree shrew genome by large-scale RNA sequencing and long-read isoform sequencing. Zool Res. 2021;42:692–709. doi: 10.24272/j.issn.2095-8137.2021.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–20. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–30. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 74.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wu T, Hu E, Xu S, Chen M, Guo P, Dai Z, et al. clusterProfiler 4.0: a universal enrichment tool for interpreting omics data. Innovation (Camb) 2021;2:100141. doi: 10.1016/j.xinn.2021.100141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gu Z, Eils R, Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016;32:2847–9. doi: 10.1093/bioinformatics/btw313. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.