Abstract
Oncolytic adenoviruses (Ads) stand out as a promising strategy for the targeted infection and lysis of tumor cells, with well-established clinical utility across various malignancies. This study delves into the therapeutic potential of oncolytic Ads in the context of neurofibromatosis type 1 (NF1)-associated malignant peripheral nerve sheath tumors (MPNSTs). Specifically, we evaluate conditionally replicative adenoviruses (CRAds) driven by the cyclooxygenase 2 (COX2) promoter, as selective agents against MPNSTs, demonstrating their preferential targeting of MPNST cells compared with non-malignant Schwann cell control. COX2-driven CRAds, particularly those with modified fiber-knobs exhibit superior binding affinity toward MPNST cells and demonstrate efficient and preferential replication and lysis of MPNST cells, with minimal impact on non-malignant control cells. In vivo experiments involving intratumoral CRAd injections in immunocompromised mice with human MPNST xenografts significantly extend survival and reduce tumor growth rate compared with controls. Moreover, in immunocompetent mouse models with MPNST-like allografts, CRAd injections induce a robust infiltration of CD8+ T cells into the tumor microenvironment (TME), indicating the potential to promote a pro-inflammatory response. These findings underscore oncolytic Ads as promising, selective, and minimally toxic agents for MPNST therapy, warranting further exploration.
Keywords: MT: Regular Issue, neurofibromatosis type 1 (NF1), oncolytic adenovirus, malignant peripheral nerve sheath tumor (MPNST), conditionally replicative adenovirus (CRAd), cyclooxygenase 2 (COX2)
Graphical abstract

Oncolytic adenoviruses exhibit enhanced binding and cytocidal activity against MPNST cells compared with controls. Intratumoral administration of oncolytic viruses leads to a reduction in tumor growth rate and enhanced survival of tumor-bearing mice. In an immunocompetent mouse model, intratumoral injection of oncolytic adenoviruses induces T cell infiltration into the tumor microenvironment.
Introduction
Malignant peripheral nerve sheath tumors (MPNSTs) are aggressive and invasive soft tissue sarcomas that manifest either spontaneously or in association with neurofibromatosis type 1 (NF1).1,2,3 NF1, an autosomal dominant disorder, is linked to a germline loss-of-function mutation in the NF1 tumor suppressor gene, predisposing patients to the development of both benign and malignant tumors within the peripheral nervous system.2,3 A benign precursor lesion of Schwann cell origin, initiated by a homozygous loss of NF1, is poised for malignant transformation into MPNST upon acquiring additional pathogenic molecular alterations such as the loss of cell cycle regulator, cyclin dependent kinase inhibitor 2A (CDKN2A), and SUZ12, a subunit of polycomb repressive complex 2 (PRC2).4,5 NF1 patients face a lifetime risk of approximately 10%–13% for developing MPNSTs, making it the leading cause of death in this patient population.6
MPNSTs pose a profound therapeutic challenge due to their aggressive nature, the absence of targeted therapies, and a poor response to systemic treatments.3,7 Surgical removal currently stands as a sole potentially curative intervention.8 However, despite its curative intent, approximately 40%–60% of patients undergoing surgery will develop metastatic disease.9,10,11 Moreover, a notable 10% of all MPNST cases present as unresectable or metastatic at the time of diagnosis.9,12,13 In light of these challenges, there is a pressing need for innovative strategies to target MPNSTs and enhance overall treatment outcomes.
Oncolytic viruses constitute an emerging class of therapeutics with the capacity to induce tumor regression through multiple mechanisms.14 First, they achieve regression by directly lysing tumor cells, facilitated by tumor-selective replication and widespread dissemination within the tumor tissue.14,15,16 Second, they contribute to tumor regression by fostering a pro-inflammatory tumor microenvironment (TME), which counteracts the immune evasiveness of tumor cells through the release of tumor antigens.16,17,18 Considering the challenges posed by the invasiveness, location, or size of MPNSTs, rendering surgical resection often unattainable and resulting in a low rate of negative resection margins, the application of virotherapy emerges as a viable approach for MPNST treatment.19 Additionally, MPNSTs are frequently characterized by an "immune-cold" tumor microenvironment, rendering them resistant to many conventional immunomodulatory therapies.20,21,22,23 Leveraging oncolytic viruses as superior immunotherapeutic agents holds promise in potentiating an effective anti-tumor immune response, thus addressing the immunotherapy challenges inherent to MPNSTs.
Adenoviruses (Ads) stand at the forefront of extensively researched and commonly utilized therapeutic vectors for cancer treatment.24,25 Through targeted genetic modifications, Ads can be customized to be either replication-deficient, achieved by introducing deletions in essential virus genes such as E1A, or replication-competent.26,27,28 While replication-deficient Ads find applications in vaccine development and therapeutic gene delivery, replication-competent Ads, being naturally lytic, drive self-amplification and viral progeny spread, making them a natural choice for anti-cancer applications.29 A specialized subset of replication-competent Ads, conditionally replicative adenoviruses (CRAds), is engineered to enhance the safety and cancer selectivity of replication-competent Ad vectors while preserving their lytic and self-amplifying capacity.30 This results in a cancer-selective vector with a strong predilection for infecting and replicating within tumor cells, minimizing damage to surrounding normal tissue.
The cancer selectivity of Ad vectors can be attained by manipulating various stages of the viral life cycle, including transduction and replication steps.29 Transductional re-targeting leverages cell surface proteins overexpressed on cancer cells, improving the delivery of viral vectors. Simultaneously, the replication of therapeutic vectors can be regulated by controlling the production of essential viral proteins such as E1A.31
Transductional re-targeting entails modifying the adenoviral fiber-knob domain to bind to cell surface receptors overexpressed on cancer cells, skewing transduction and infectivity significantly toward neoplastic cells.32,33 For instance, the binding of wild-type (WT) Ad5 to target cells is mediated by the interaction of the viral fiber-knob domain with coxsackievirus and adenovirus receptor (CAR) ubiquitously expressed on many normal cells while absent or low expressed on many cancer cells.34,35,36 Therefore, fiber-knob modifications allowing for CAR-independent binding of Ad5-based vectors are needed to achieve efficient tumor cell transduction and minimize damage to normal cells.37 Efficient tumor cell transduction is crucial not only for initial virus internalization but also for subsequent intratumoral spread. Noteworthy modifications, such as incorporating the arginine-glycine-aspartic acid (RGD) tripeptide motif into the HI loop of the fiber-knob, results in increased binding capacity of Ads to cellular integrins, specifically αVβ3 and αVβ5—molecules frequently upregulated in various cancer types.37,38 Another fiber-knob modification, Ad5/3, is achieved through the substitution of Ad5 knob domain with that of Ad3. This alteration produces a chimeric Ad5/Ad3 fiber-knob adept at binding to desmoglein 2 (DSG-2), a component of the cell-cell adhesion structure highly expressed in numerous advanced malignancies.35,39
The control of cancer-selective viral replication can be achieved through various methods, with transcriptional control being a notable approach. This approach involves placing the viral E1A gene under the regulation of a human tumor/tissue-specific promoter (TSP), generating CRAds that predominantly replicate within cells where the TSP is active.26,40,41 TSPs are derived from gene promoters that are either overexpressed or aberrantly regulated in cancer cells compared with their normal counterparts, rendering them overactive in specific tumor cells while being less active or completely inactive in normal cells. Given that E1A is indispensable in the adenoviral life cycle, minimal or negligible TSP activity within normal cells hampers E1A production, thereby restricting viral replication primarily to tumor cells and minimizing the impact on surrounding non-neoplastic tissues.
This study utilizes the promoter of the cyclooxygenase-2 (COX2) gene as a TSP that drives selective replication of Ads within MPNST cells. COX2 is an inducible enzyme essential for the catalysis of arachidonic acid to prostaglandins, which are involved in various physiological processes, including inflammation and cell signaling.42,43 Under normal conditions, COX2 expression is typically maintained at low or negligible levels within most normal cells but can be transiently upregulated in response to specific pro-inflammatory stimuli.44 Furthermore, the prevalent constitutive overexpression of COX2 in various tumor types, including MPNST, renders it an appealing candidate as a TSP for orchestrating targeted viral replication in tumor cells.44,45,46
To date, oncolytic Ads have been evaluated in numerous human clinical trials targeting a diverse range of cancers.47 While the results from the trials are promising, the literature contains only a single report evaluating oncolytic adenovirus against MPNST in the context of a human clinical trial for patients with advanced sarcomas.48 A single patient with MPNST in the sarcoma trial who received oncolytic adenovirus (ONYX-015) achieved a partial response in their primary lesion and in two other uninjected lesions. This provides further evidence that oncolytic adenoviruses may potentiate systemic anti-MPNST immunity. In addition, due to their mild side effects, and minimal non-tumor cell cytotoxicity, oncolytic adenoviruses may be combined with standard care therapies.47
In this study, we confirm COX2 expression in MPNST models and explore the potential of COX2 promoter-driven CRAds as targeted agents against MPNST. Specifically, we focus on RGD fiber-modified vector leaving in-depth exploration of Ad5/3 chimeric fiber for future work. Our investigation included assessing the efficient binding of CRAds to MPNST, particularly those with modified fiber-knob domains. Comprehensive in vitro assays evaluated viral infectivity, progeny production, and spread in both MPNST and non-cancer Schwann cell controls. In vivo experiments, using mouse models of MPNST, examined the impact of intratumoral virus injections on tumor growth rates, mouse survival, and intratumoral immune infiltrate. The results presented herein demonstrate the effectiveness of COX2 promoter-driven CRAds against MPNST cells, showcasing robust replication and killing of tumor cells in vitro with an attenuated impact on non-malignant cells. Additionally, we observed a reduction in human tumor xenograft growth rates in vivo within immunocompromised mouse models. Finally, these CRAds also promote intratumoral CD8+ T cell infiltration in immunocompetent mouse models of MPNST. Collectively, these findings underscore the promising potential of COX2 promoter-driven CRAds against MPNST, warranting further investigation and optimization in vector development.
Results
MPNST cell lines have higher binding affinity for Ad vectors compared with non-cancer Schwann cell controls
To assess the binding capacity of Ad vectors to MPNST cells compared with non-cancer Schwann cell controls (immortalized human Schwann cells, iHSC1λ), we utilized replication-deficient Ad vectors (ΔE1A) to conduct a virus binding assay using quantitative PCR (qPCR). These vectors were equipped with one of the following fiber-knob domains: WT (Ad5), RGD-modified (RGD), or chimeric Ad5/Ad3 (Ad5/3) fiber-knob domain (Figure 1A). Our findings indicate that Ad vectors with the WT fiber-knob exhibit relatively low binding affinity across the board compared with those with modified fiber-knobs, which displayed preferential binding to MPNST cells compared with Schwann cells (Figure 1B).
Figure 1.
Adenovirus binding affinity to MPNST cell lines and viral receptor expression
(A) Schematic of viral genome for Ad vectors used in binding assay. (B) Each cell line was infected with Ad vectors equipped with either WT (Ad5), RGD fiber-modified (RGD), or chimeric Ad5/Ad3 fiber (Ad5/3) at 100 VP/cell. Binding was allowed to proceed for 2 h at 4°C, then was assessed by qPCR. (C) Flow cytometry analysis of CAR and integrin expression. MPNST cell lines were incubated with fluorescent antibodies against αVβ3 and αVβ5 integrins and coxsackie adenovirus receptor (CAR). The data are shown as a relative percentage of positive cells scored among at least 10,000 cells assessed. (D) Flow cytometry plots by cell line with respective isotype control for viral entry receptors in MPNST cell lines and iHSC1λ controls. Error bars represents ± standard deviation. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001, Student's t test.
The fluorescence-activated cell sorting analysis of cell populations revealed consistent expression of CAR and αVβ5 integrins across most cells within the population for all cell lines assessed, while αVβ3 integrins were predominantly present on Schwann cells but not on MPNST cells (Figures 1C, 1D, and S1). Despite observing a substantial increase in vector binding affinity when utilizing fiber-modified vectors, we did not discern a clear correlation between integrin expression and binding affinity. Notably, this enhanced binding affinity was evident for MPNST cells but not for Schwann cells, with fiber-modified Ads demonstrating a statistically significant preference for binding to MPNST cells (Figures 1B and 1C). The highest overall binding affinity was noted with the Ad5/3 chimeric fiber, followed closely by the RGD-modified fiber. Conversely, the lowest binding affinity was observed with the WT fiber, where, except for S462 cells, no statistically significant differences were detected between MPNST and Schwann cells (Figure 1B).
MPNST cells produce more viral proteins after infection with COX2-driven CRAds compared with Schwann cells
During a productive infection, viral proteins, such as E1A, hexon, and fiber accumulate in the cytosol of the infected cell, allowing for an indirect estimation of viral replication and progeny production through immunoblotting of virally infected lysates. Our analysis reveals that all human cell lines, including MPNSTs (S462-TY and STS26T), immortalized human Schwann cells (iHSC1λ and ipn02.3λ), primary human Schwann cells (pHSC), and the COX2-overexpressing lung carcinoma cell line, A549 (included as a positive control) express the COX2 gene, as confirmed by reverse transcriptase PCR (RT-PCR) (Figures 2A and S2). This indicates their theoretical capacity for driving COX2-CRAd replication. However, our data demonstrate that MPNST cells exhibit a higher production of viral proteins upon infection with RGD-COX2-CRAd when compared with Schwann cells (Figure 2B). This dose-dependent effect is particularly pronounced at MOIs of 10 and 50 VP/cell.
Figure 2.
COX2 expression and viral protein production in MPNST cell lines and Schwann cells
(A) Analysis of COX2 mRNA in MPNST cell lines (S462-TY and STS26T) and Schwann cells (ipn02.3 2λ, iHSC1λ and pHSC) via RT-PCR. A549 cell line represents a positive control. A total of 500 ng of RNA from each sample was used as template input. (B) Western blot analysis of viral protein production in infected cells. A total of 250,000 cells were infected with RGD-COX2-CRAd at MOI 0 VP/cell (negative control) 10 VP/cell, and 50 VP/cell), 48 h post transduction whole cell lysates were probed by immunoblotting for early (E1A) and late viral proteins (hexon and fiber). GRB2 was used as a loading control.
RGD-COX2-CRAds are significantly more selective toward MPNST cells compared with human Schwann cells
To assess the selectivity of RGD-COX2-CRAd toward MPNST cells compared with Schwann cells, we conducted cell viability assays 5 days post-viral infection at increasing MOIs using a standard MTS protocol. Our results reveal that MPNSTs are significantly more susceptible to RGD-CRAd-COX2-induced killing, particularly at lower MOIs, such as MOI of 50 and below (Figures 3A and S3). Remarkably, MPNSTs exhibit a susceptibility to RGD-COX2-CRAd killing comparable to that observed in the positive control A549 cells (Figures 3A–3C). Calculating the half maximal inhibitory concentration (IC50) demonstrated that MPNSTs, similar to A549 cells, require significantly fewer viral particles than Schwann cells to reach IC50 response (Figure 3B). The most prominent separation between the cell viability curves was observed at MOI of 50, where cancer cells were mostly cleared and non-cancer Schwann cell controls exhibited only a mild decrease in viability (Figures 3A, 3C, and 3D). Representative images of monolayers infected with RGD-CRAd-COX2 at 50 MOI revealed a much higher fraction of viable cells at day 5 post-infection in Schwann cells compared with cancer cells (Figure 3D). Notably, the degree of monolayer clearance in MPNST cells was similar to that observed in the positive control A549 cells (Figure 3D).
Figure 3.
Selectivity of RGD-COX2-CRAd toward MPNST cells
(A) Cell viability curves for MPNST cell lines (S462-TY and STS26T) compared with immortalized human Schwann cells (iHSC1λ and ipn02.3λ), and primary human Schwann cells (pHSC). Cell lines were infected with RGD-COX2-CRAd at various MOIs (0–200 PFU/cell, n = 6), and percent viable cells was determined 5 days post-infection utilizing MTS assay protocol. Error bars represent ± standard deviation. (B) The IC50 values for MPNSTs and Schwann cells were calculated based on cell viability curves. (C) Percent viable cells remaining within monolayer after 5 days of infection with RGD-CRAd-COX2 at 50 MOI, relative to corresponding uninfected controls (0 MOI) were quantified using Cytation5 software. p < 0.0001, Student's t test. (D) Representative images of cell monolayers stained with crystal violet 5 days post-infection with RGD-COX2-CRAds at 50 MOI and corresponding uninfected controls (0 MOI). The COX2-overexpressing lung cancer cell line, A549, was used as a positive control.
Cytotoxicity and viral spread analyses revealed more robust and selective viral replication and spread in MPNST cell lines compared with Schwann cells
To evaluate the selectivity of COX2-driven vectors toward MPNST cells and assess the production and spread of infectious viral progeny, we conducted crystal violet cell viability assays spanning a 35-day period (Figure 4A). This assay relies on the initial infection of a small subset of cells within the monolayer, which then serve as the primary producers of viral progeny. The spread of these progeny throughout the well leads to cell death and subsequent detachment from the monolayer, providing insights into viral replication rates, progeny production, and spread. However, it is important to note a technical limitation of the crystal violet assay, as it requires continuous cell culturing without medium change, which may be impractical for certain cell lines. S462-TY cells were chosen as a representative model for studying infection, progeny production, and spread of Ad vectors in MPNST, owing to their compatibility with long-term culturing. Cell viability was assessed following infection with four different Ad vectors, engineered with either WT fiber (Ad5-WT, COX2-CRAd) or RGD-modified fiber (RGD-WT, RGD-COX2-CRAd), and with either WT promoter (Ad5-WT, RGD-WT) or COX2 promoter (COX2-CRAd, RGD-COX2-CRAd) (Figure 4B). Vectors equipped with WT promoters served as non-selective controls.
Figure 4.
In vitro analysis of COX2-CRAd replication and viral progeny spread
(A) S462-TY, iHSC1λ (non-cancer control), and A549 (positive COX2-overexpression control) were used. The cells were infected at low MOI of 0.01, 0.1, and 1 VP/cell and cultured without medium change (to allow viral progeny production and spread) for up to 35 days or until cell monolayer was cleared. Cytotoxicity was assessed by affixing viable cell monolayers to the well, staining them with crystal violet dye, and quantifying images using FIJI software. Error bars represent ± standard deviation. The WT promoter-driven viruses (Ad5 and RGD-WT) were utilized as non-selective positive controls and exhibited robust viral spread and cell clearance rate. iHSC1λ cells served as non-cancer controls. Both COX2-CRAd and RGD-COX2-CRAd selectively killed COX-2-positive control cell line, A549 and MPNST cell line, S462-TY while leaving iHSC1λ mainly unharmed. RGD-COX2-CRAd showed a slightly better cytocidal effect compared with its WT fiber counterpart (COX2-CRAd). (B) Schematic representation of the four replication-competent Ad vectors. Vectors were engineered to have either WT or COX2 promoter and either WT or RGD fiber. Four replication-competent vectors were used. Vectors with WT promoter were used as non-selective controls. Vectors with WT fiber-knob domains (Ad5WT and COX2-CRAd) were employed for comparison with RGD fiber-modified vectors (RGD-WT and RGD-COX2-CRAd) to assess the degree of infectivity enhancement compared with WT fiber.
Microscopic signs of oncolysis were evident in COX2-CRAd-infected S462-TY cells by day 4 post-infection (data not shown). By day 6, over 50% of S462-TY cells infected with COX2-driven vectors at MOI of 1 VP/cell were cleared, while the COX2-overexpressing cell line A549 showed no macroscopically visible cytopathic effects (Figures 4A and S4A–S4C). By day 12, all S462-TY cells infected with 1 VP/cell as well as most cells infected with 0.1 VP/cell were cleared, while A549 cells were cleared in all groups. S462-TY cells infected with 0.01 VP/cell (COX2-driven vectors) were mostly cleared by day 16.
Notably, non-malignant Schwann cells (iHSC1λ) exhibited minimal cytopathic effects post-infection with COX2-driven vectors, highlighting their relative resistance (Figures 4A and S4A). In contrast, WT promoter-driven vectors induced rapid and less selective killing of iHSC1λ cells, with most cells cleared by day 12 post-infection. All S462-TY cells were cleared by day 9 post-infection with WT promoter-driven vectors and by day 16 with COX2 promoter-driven vectors (Figures 4A and S4B). A549 cells were mostly cleared by day 8 post-infection with WT promoter-driven viruses and by day 12 with COX2 promoter-driven viruses (Figures 4A and S4C).
Virus-induced cytotoxicity for all other MPNST cell lines was assessed using the MTS assay, which demonstrated variable sensitivity among different MPNSTs to Ad vectors. However, all MPNST cells displayed greater sensitivity to Ad infection when compared with non-cancer controls (iHSC1λ, ipn02.3λ, and primary human Schwann cells pHSC) (Figure S3).
In vivo analysis of tumor growth kinetics showed a significant reduction in tumor growth rate and a survival benefit in mice
To assess the capacity of COX2 promoter-driven CRAds to reduce tumor growth rates in vivo and enhance the survival of tumor-bearing mice, we initiated subcutaneous tumor xenografts with a minimum volume of 200 mm3. Two distinct cell lines were employed to represent both PRC2-deficient status (S462-TY), prevalent in the majority of MPNST patients, as well as PRC2-proficient status (STS26T), offering valuable insights into the activity of Ad vectors across diverse disease-associated genotypes. Mice were randomly assigned to treatment or control arms and received three consecutive doses (3.3 × 1010 VP/dose) of the respective Ad vectors or vehicle control (PBS) via intratumoral injection on days 0, 2, and 4.
A notable reduction in tumor growth rates was evident in mice injected with oncolytic viruses compared with PBS. (Figures 5A, 5B, S5A, and S5B). The average reduction in tumor volumes, within 95% confidence intervals ranged between 437 and 938 mm3 for RGD-COX2-CRAd, between 405 and 934 mm3 for RGD-WT, and 293 and 822 mm3 for COX2-CRAd, when compared with PBS-treated tumor-bearing mice (Figure 5B) Tumor growth rates in mice treated with COX2-driven CRAds were comparable to those treated with RGD-WT (non-tumor-selective positive control), with no statistically significant differences observed among virus-treated groups (Figure 5B) (RGD-WT/COX2-CRAd p = 0.8675, RGD-WT/RGD-COX2-CRAd p = 0.9993, CRAD-COX2/RGD-COX2-CRAd p = 0.7879, Mixed Effects Model). However, S462-TY xenografts exhibited a non-significantly more robust response to virus treatment compared with STS26T xenografts for all vector types (p = 0.177748 Mixed Effects Model) (Figure S5B). Additional details on statistical analysis are available in Figures S5A and S5B.
Figure 5.
In vivo analysis of tumor growth kinetics and viral protein production
(A) NOD-Rag1nullIL2rgnull (NRG) mice were inoculated subcutaneously with MPNST cell lines (S462-TY or STS26T) at 2.0 × 106 tumor cells/mouse. Virus (3.3 × 1010 VP/dose) or PBS was intratumorally injected when nodules reached approximately 200 mm3. The injections were repeated two more times every other day for a total of three injections and cumulative viral dose of 1.0 × 1011 VP/animal. Tumor size is shown in mm3 starting on day 0 (first viral injection). S462-TY/PBS and S462-TY/RGD-COX2-CRAd n = 6; S462-TY/COX2-CRAd and S462-TY/RGD-WT n = 4; all STS26T treatment and control groups n = 5. (B) Tumors injected with any of the viral vectors demonstrated reduced tumor growth rates compared with those injected with PBS. The treatment effects on tumor volume were calculated using a linear mixed effects model where each group was compared with PBS. All treatments had a significant negative effect on tumor volume. An average reduction of tumor volumes, within 95% confidence intervals is between 437 and 938 mm3 (RGD-CRAd-COX2), between 405 and 934 mm3 (RGD-WT), and between 293 and 822 mm3 (CRAd-COX2). For more information on statistical analysis see Figure S5. (C) Mice bearing S462-TY xenografts injected with virus vectors survived significantly longer compared with those injected with PBS (p < 0.0045, Log rank test). There were no significant differences in survival between S462-TY-bearing virus injected groups. Median survival in days was as follows: PBS = 19; COX2-CRAd = 41; RGD-COX2-CRAd = 47; RGD-WT = 45. (D) STS26T xenograft-bearing mice treated with either RGD-COX2-CRAd or RGD-WT demonstrated a statistically significant increase in survival probability compared with PBS (p = 0.002, Log rank test). Median survival of STS26T xenograft-bearing mice was PBS = 16 days; RGD-COX2-CRAd = 21 days COX2-CRAd = 16 days; RGD-WT = 22 days. RGD-WT vector was used as a non-selective positive control. (E) Immunohistochemistry analysis of tumor xenografts resected at endpoint from mice injected with RGD-COX2-CRAd and PBS. All tumors injected with RGD-COX2-CRAd were positive for viral proteins (E1A, fiber, and hexon) and markers of apoptosis (cleaved caspase 3). Some areas within tissues appeared torn due to necrotic tumor tissue. All tumors injected with PBS were uniformly negative for all viral proteins. STS26T tumors injected with PBS demonstrated a positive signal for cleaved caspase 3 in some cases. Representative example (Figure S5A).
Survival analysis revealed a significant increase in the survival of S462-TY-bearing mice treated with any of the oncolytic Ad vectors compared with PBS (p < 0.0045, Log rank test), extending median survival by over 20 days in each treatment arm compared with PBS (Figures 5C and S6A–S6C). A more modest but statistically significant survival benefit was observed in STS26T-bearing mice treated with either RGD-COX2-CRAd or RGD-WT (p = 0.002, Log rank test), but not in those injected with COX2-CRAd vectors (Figure 5D). The median survival of STS26T-bearing mice injected with RGD-COX2-CRAd, and RGD-WT increased by 5 and 6 days, respectively, compared with PBS, while no statistically significant survival benefit was observed in the COX2-CRAd-treated arm (Figures 5D and S6D–S6F). Although the reduction in tumor growth rate and was less pronounced with STS26T xenografts compared with S462-TY (Figure 5A), the effect was not statistically significant, suggesting potential tumor lytic activity of Ad vectors across genotypically disparate MPNST cell lines.
To estimate in vivo intratumoral viral spread, and viral progeny production, we performed immunohistochemistry analyses on tumor xenografts resected from mice. All tumors treated with RGD-COX2-CRAd were positive for early viral protein E1A and late viral proteins, fiber, and hexon, suggesting intratumoral viral replication, and positive for markers of apoptosis (cleaved caspase 3), suggesting virus-induced cell death (Figure 5E). Tumors injected with PBS were negative for all viral proteins. STS26T tumors injected with PBS were sometimes positive for cleaved caspase 3 (Figure S6G). In virus-treated tumor tissues, areas of dead tissue were formed adjacent to tissue positive for viral proteins, cleaved caspase 3, or both (Figure 5E). Additionally, some tissue areas appeared uninfected and lacked positive staining for viral proteins, indicating a non-uniform viral spread throughout the tumor.
RGD-COX2-CRAd vectors in syngeneic models of MPNST enhance T cell infiltration into the tumor microenvironment
The exploration of oncolytic viruses in both clinical and pre-clinical studies has revealed that their efficacy is, in part, attributable to their capacity to stimulate anti-tumor immune response by drawing in CD8+ T cells that might be otherwise excluded from tumors.38,49 Consequently, our subsequent experiments aimed to assess virus-induced changes in the TME using an immunocompetent model. Investigating immune response to adenoviral infection and replication in immunocompetent mouse models is challenging due to the restricted replication of human adenovirus in murine cells, hindering viral progeny production and spread.50 Nevertheless, certain transformed murine cell lines have been reported to be susceptible to adenoviral infection and capable of supporting some aspects of the viral life cycle. However, they may require a higher infection dose and may not produce significant amounts, if any, of infectious viral progeny.51
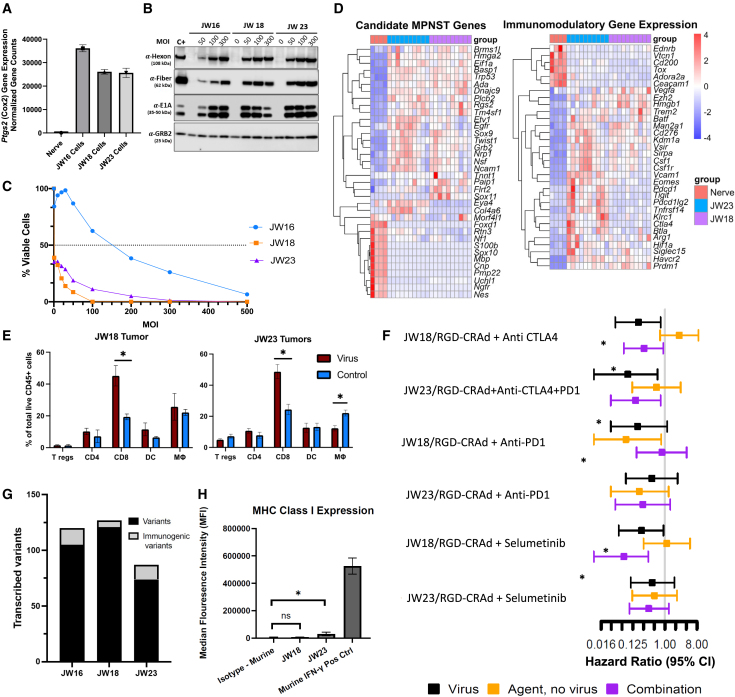
Utilizing murine MPNST-like cell lines (JW16, JW18, and JW2352) derived from C57BL/6J mice with Nf1+/− Trp53+/− in cis (NPCis model), we sought evidence indicating semi-permissiveness of JW cells to adenoviral infection and replication, including virus-induced cytopathic effects, cell killing, and viral protein production. RNA sequencing confirmed upregulation of the Cox2 gene in all JW cell lines compared with normal nerve tissue (Figure 6A), suggesting that the COX2-diven CRAds may be appropriate to drive E1A expression within those cells. Initial in vitro assessments using the MTS protocol revealed varying susceptibility among JW cell lines to virus-induced cell lysis with JW16 exhibiting increased resistance compared with JW18 and JW23 (Figure 6B). Immunoblotting for viral proteins post RGD-COX2-CRAd infection demonstrated robust viral protein production within murine cells, albeit at very high MOIs of 50 and above PFU/cell (Figure 6B). Based on lower IC50 values and enhanced viral protein production in JW18 and JW23 compared with JW16, these cell lines were selected for subsequent in vivo experiments (Figures 6B and 6C).
Figure 6.
Investigation of CRAd in syngeneic models of MPNST
(A–C) Application of RGD-COX2-CRAd was investigated for murine MPNST-like syngeneic models. (A) RNA sequencing of JW cell lines shows upregulated Cox2 compared with normal murine sciatic nerve. (B) Western blot of RGD-COX2-CRAd-infected JW cells producing early (E1A) and late (hexon, fiber) viral proteins when infected at high MOIs. The human cell line was used as a positive control (C+) and was infected with RGD-COX2-CRAd at 3 MOI. (C) Both JW18 and JW23 are susceptible to cell killing by RGD-COX2-CRAd (IC50) < 1 as assessed by MTS assay 5 days post-infection, while JW16 is more resistant (six replicates performed per cell line per MOI). (D) Bulk RNA sequencing was performed on JW tumors implanted in C57BL/6 mice, and differential gene expression analysis performed against sciatic nerve of C57BL/6 mice, with all genes represented being significantly differentially expressed (p adjusted <0.05). The gene expression profile for JW cell lines is analogous to the gene expression profile in human MPNST, relative to normal nerve.53 Additionally, JW cell lines differentially express immunomodulatory genes, with JW23 showing the highest expression of both Pdcd1 (PD1) and Ctla4 (CTLA-4). (E) JW18 and JW23 flank allografts were established and subjected to virus or PBS injection to assess immune infiltration 10 days after implantation (six replicates per treatment group). In both JW18- and JW23-treated tumors, an increase in CD8+ T cell infiltration into the tumor microenvironment is observed (∗p < 0.05). (F) Six in vivo therapeutic trials were conducted (n = 6 mice per treatment group). Compared with intratumoral PBS injection (control), RGD-COX2-CRAd injections delayed tumor growth, although this was significant in only 50% of the trials (∗ represents hazard ratio of p < 0.05). CRAd was combined with immune checkpoint blockade or MEK inhibition with selumetinib, but no synergistic combination was identified possibly due to failure of MHCI presentation. (G) Whole exome and RNA sequencing identify nonsynonymous protein coding variants in these models, including variants predicted to bind to MHC class I H2-kb. (H) The baseline expression of MHC class 1 on the JW cell lines is low relative to IFN-γ stimulated cells.
Prior to in vivo therapeutic studies, we confirmed the fidelity of syngeneic models to human MPNST by analyzing tumor gene expression profiles. RNA sequencing of untreated JW18 and JW23 flank tumors (n = 10) relative to normal murine sciatic nerves (n = 4) from C57BL/6J mice revealed a gene expression pattern closely resembling that associated with human MPNST.54 Upregulation and downregulation of murine orthologs of canonical MPNST genes were observed. Genes including Tm4sf1, Rgs2, Etv1, Twist1, Ada, Basp1, Sox9, and Hmga2 were found to be upregulated, while genes like Sox10, Cnp, Ngfr, and Pmp22 showed downregulation in JW tumors. (Figure 6D). Additionally, we explored the expression of immunomodulatory genes within the syngeneic flank tumors, revealing the presence of immune checkpoints, including Ctla4 and Pdcd1, in both untreated JW tumors, with notably stronger expression observed in JW23 tumors (Figure 6D).55
After establishing susceptibility and MPNST-like nature of murine JW cell lines, we investigated the impact of RGD-COX2-CRAd treatment on intratumoral immune infiltrate. JW18 and JW23 flank tumors in syngeneic C57BL/6J mice were treated with four consecutive doses of RGD-COX2-CRAd (3.3 × 1010 VP/dose) on days 0, 2, 4, and 6. This dosing schedule aimed to sustain viral titers within the tumor microenvironment, given murine cells' relative resistance to human Ad vectors and lack of robust viral progeny production. Flow cytometry analysis of tumors harvested on day 10 revealed a robust increase CD8+ T cells within tumors treated with RGD-COX2-CRAd compared with those treated with the vehicle control (PBS). Notably, concurrent increase in T-regulatory cells or macrophages was not observed (Figure 6E).
Next, we selected clinically utilized checkpoint inhibitors given the expression of Pdcd1 and Ctla4 in these models with the rationale to prevent T cell exhaustion when used in combination with the virus which, theoretically can potentiate anti-tumor immune response and enhance survival. Additionally, we tested the virus in combination with selumetinib, which targets the activated mitogen-activated protein kinase (MAPK) pathway in Nf1-altered tumors. We found that 10 μM selumetinib significantly upregulates major histocompatibility class 1 (H2-Kb) in JW18 (p = 0.0014, two-sided t test) and JW23 (p = 0.0023, two-sided t test) as assessed by flow cytometry following 10 days of exposure in vitro (Figure S7). Therefore, the rationale for this combination was to enhance antigen presentation to facilitate a secondary anti-tumor immune response. We treated mice with virus alone, agent alone (selumetinib, anti- CTLA4, PD-L1, or PD1), combination of virus and agent, or PBS control. Across our in vivo experiments (Figures 6F and S8), we found that RGD-COX2-CRAd infection inconsistently led to statistically significant increase in survival time, with three out of six trials demonstrating a hazard ratio encompassing the 95% confidence interval of less than 1. Furthermore, we did not identify synergy utilizing anti-CTLA-4, anti-PD-1, or combination of anti-CTLA-4 and anti-PD-L1, nor combination with selumetinib.
To investigate whether these models contain predicted neoantigens, we performed whole exome and RNA sequencing on the cell lines to bioinformatically impute the number of transcribed immunogenic variants. Each cell line does express dozens of predicted neoantigens at the mRNA level (Figure 6G), including variants predicted to bind to the mouse H2 complex for antigen display (there are 6 and 13 predicted immunogenic variants in JW18 and JW23, respectively). However, flow cytometry reveals a low baseline expression of the class I major histocompatibility complex (MHC) relative to interferon-gamma (IFNγ)-stimulated cells, suggesting poor MHC class I presentation in this model. (Figure 6H).
Discussion
MPNSTs continue to present a significant therapeutic challenge, standing as the leading cause of NF1-related mortality and underscoring the urgent need for innovative therapeutic interventions.1,2,3,4 Their invasive growth, metastatic tendencies, and limited responsiveness to standard therapies lead to suboptimal outcomes. While surgery is a potentially curative intervention, it is not always achievable due to unresectable or metastatic disease.6,9,10,11 This highlights the pressing demand for novel treatments that are not only less cytotoxic and well-tolerated but also compatible with existing standard-of-care protocols. Oncolytic adenovirus vectors, as evaluated in numerous clinical trials across various malignancies, have demonstrated safety and tolerability with promising anti-tumor potential.56 In the context of MPNST, oncolytic viruses hold particular promise for meeting the specific needs of patients with refractory or unresectable tumors or those with positive post-surgical margins. Notably, oncolytic viruses operate through a dual mechanism, selectively eliminating tumor cells and reshaping the TME, thereby fostering an anti-tumor immune response. This underscores the critical role of advancing research and development in oncolytic virotherapy for MPNST.38,49
In this study, we evaluated the suitability of a COX2 promoter-driven oncolytic adenoviruses for selective targeting of MPNST cells and induction of intratumoral CD8+ T cell infiltration. The COX2 gene, previously identified as a potential therapeutic target for MPNST,45,46,57,58 exhibited robust activity in all human MPNST cell lines assessed during our investigation (Figures 2A and S2), prompting further exploration of COX2-driven oncolytic adenoviruses. Our data revealed that MPNST cell lines were significantly more susceptible to adenovirus infection compared with non-malignant Schwann cell controls (Figures 3 and S3). Specifically, Ads with modified fiber-knob domains displayed increased binding affinity to MPNST cells than Schwann cells (Figure 1B). Additionally, the COX2 promoter robustly and preferentially drove viral replication within MPNST cells, resulting in a strong cytolytic effect in vitro (Figures 3, 4, and S4) and a substantial anti-tumor effect in vivo (Figures 5A–5C). Treatment with RGD-COX2-CRAd significantly enhanced the survival of both tumor-bearing mouse models compared with controls (Figures 5C and 5D).
The observed anti-tumor effect varied among different human MPNST cell lines, with PRC2-deficient cells (S462-TY) showing a more pronounced response compared with PRC2-proficient STS26T, although the difference was not statistically significant (Figures 5A and S5). Similarly, the in vitro cytolytic effect varied among MPNST cell lines but was consistently more robust compared with Schwann cells (Figures 3, 4, and S3). This suggests that COX2-driven CRAds may elicit a strong, albeit non-uniform anti-MPNST response, emphasizing the need for further vector development and predictive biomarker identification for MPNST. Nevertheless, treatment with RGD-COX2-CRAd significantly enhanced the survival of both tumor-bearing mouse models compared with controls (Figures 5C and 5D), suggesting that RGD modification is more effective across a range of MPNST models compared with WT fiber.
Previous studies have demonstrated the importance of anti-tumor immune response triggered by adenovirus-mediated cell lysis and subsequent release of tumor-associated antigens, promoting T cell activity.38 However, investigating this response is challenging due to lack of suitable immunocompetent models. Human adenoviruses do not replicate readily in murine cells, which hinders the pre-clinical oncolytic adenovirus development not only for MPNST, but also for many other malignancies.50 Additionally, as it relates to MPNST specifically, available mouse models do not fully capture all aspects of human disease. For instance, currently utilized mouse MPNST-like cells are derived from NPCis mice, carrying both Nf1 and Trp53 mutations in cis. This model differs from most human NF1-associated MPNSTs, which are usually characterized by WT P53, and having mutations in NF1, CDNK2A/B, and SUZ12 or EED.53 Nevertheless, our study aimed to evaluate aspects of CRAd-mediated anti-tumor immune response in an immunocompetent system. Here we utilized syngeneic mice to model adenovirus-induced changes in the TME and CD8+ T cell infiltration. Although we observed some anti-tumor activity in several in vivo tails, with positive effects on mouse survival, all tumors eventually overcame CRAd therapy, as expected due to the lack of virus replication with subsequent intratumoral spread. Nevertheless, we noted a substantial intratumoral infiltration of CD8+ T cells following CRAd injection, indicating the ability of the virus to remodel the TME into a more immune-permissive state. The low expression of MHC class I in these models may explain the lack of durable immune control over tumor growth, as processed tumor-associated antigens would not be effectively displayed on non-infected tumor cells (Figures 6G and 6H). Another limitation of the present model is its rapid in vivo growth kinetics, which, unlike usually slow-growing human MPNSTs, may restrict the murine immune system’s ability to process antigens and mount an adequate anti-tumor response (Figure S8). These characteristics of the murine model coupled with lack of efficient viral replication explain lack of durable response.
To address these shortcomings, we are developing a new murine model that progressively accumulates mutations in Nf1, Cdkn2A/B, and Suz12 genes, and more faithfully recapitulates human disease progression. Additionally, our lab is working on cytokine-expressing CRAds, such as those with IFN-γ transgene, to overcome low MPNST MHC class I expression. Moreover, CRAds with different tumor-specific promoters and different fiber-knob modifications, such as Ad5/3 chimeric fiber are under development for future studies. While Ad5/3 fiber demonstrated a robust binding affinity to MPNST cells (Figure 1B), future studies employing this modification are likely to be limited to immunodeficient models, as mice do not express the DSG-2 receptor required for Ad5/3 binding.35 Future investigations may be warranted in Syrian golden hamsters or porcine models of MPNST, which are fully permissive to adenoviral replication. However, the absence of syngeneic MPNST models in these species remains a challenge.
Conclusion
The data obtained in the present study underscore the potential of oncolytic Ads as a selective therapeutic agent for MPNST, which could be integrated into the current standard of care therapies or utilized in cases of unresectable, residual, or refractory tumors. Because virus-mediated tumor cell lysis and immune induction are both important for successful oncolytic virotherapy, we combined immunodeficient and immunoproficient mouse models to assess both aspects of oncolysis. The former effectively models tumor selectivity and tumor cell lysis, virus replication, and viral progeny spread in human tumor cells, while the latter can model some aspects of anti-tumor immune response. Taken together, these data offer insights into the suitability of oncolytic Ad vectors as a novel therapeutic option for the treatment of MPNST. Specifically, COX2-driven CRAds were shown to preferentially replicate and lyse MPNST cells while leaving non-cancer cells relatively unharmed. Simultaneously, COX2-CRAds can induce CD8+ T cell infiltrate into the TME, suggesting that anti-tumor immunity can be facilitated using CRAds. These results suggest that oncolytic Ads are a promising novel class of anti-MPNST therapeutics.
Materials and methods
Cell lines
Human MPNST cell lines STS26T, ST88–14, S462, and S462-TY were provided by Nancy Ratner (Cincinnati Children’s); 2-002 MP3, JHU 2–079, and JHU 2–103 were provided by Christine Pratilas (Johns Hopkins University); iHSC1λ was obtained from Peggy Wallace (University of Florida); primary human Schwann cells (pHSC) were purchased from ScienCell Research Laboratories. C57BL/6J mice on an Nf1+/−; Trp53+/− NPCis background develop MPNST-like syngeneic tumors (JW16, JW18, and JW23), and were kindly provided by Karlyne Reilly, PhD (Center for Cancer Research, National Cancer Institute, Frederick, MD). All other cell lines were purchased from the American Type Culture Collection (ATCC). All cell lines were maintained in Dulbecco’s Modified Eagle’s Medium (DMEM, Gibco, Life Technologies) with 10% fetal bovine serum (FBS, Gibco), 200 U/mL of Penicillin/Streptomycin (Gibco). Cells were incubated at 37°C with 5% CO2 under humidified conditions. All cells tested negative for mycoplasma (Lonza). Whenever possible, cell lines were authenticated by the University of Arizona Genetics Core.
Adenovirus binding analysis
To assess cell-virus binding, 250,000 cells/well were infected at an MOI of 100 viral particles (VP)/cell in 250 μL of DMEM. To allow viral binding but not internalization, infected cells were incubated at 4°C for 2 h. Cells were then washed with ice-cold PBS, and extracted DNA (QIAamp Blood Mini Kit, Qiagen) was analyzed by real-time PCR. To determine virus copy number using Ad5-Hexon-specific primers (Sense: 5′-CTTACCCCCAACGAGTTTGA-3′; Anti-sense 5′-GGAGTACATGCGGTCCTTGT-3′). For the standard curve, Ad5-Hexon template DNA with a known copy number of 108–102 VP/reaction was used. All RT-PCR reactions were performed using the LightCycler System (Roche). The thermal cycling conditions were as follows: Initial denaturation for 10 min at 95°C, followed by 40 cycles of 15 s at 95°C, and 1 min at 60°C.
In vitro analysis of cytocidal effect
Infectious viral progeny production, viral spread, and cytocidal capacity of adenoviruses were assessed via crystal violet staining. Briefly, cells were infected with adenovirus at either 1, 0.1, or 0.01 VP/cell. Two days later the infection medium was replaced with fresh DMEM containing 1% FBS; cells were then cultured for 6–35 days. Cell viability was determined via crystal violet staining on different days post-infection. Briefly, the cell monolayer was fixed using 10% buffered formalin (10 min), stained with 1% crystal violet in 70% ethanol (20 min), washed, air-dried, and imaged. Images were analyzed using FIJI ImageJ software.
Animal models
All in vivo procedures were approved by the Institutional Animal Care and Use Committee (IACUC) at the University of Minnesota. To establish MPNST xenografts, an equal number of male and female NOD-Rag1nullIL2rgnull (NRG) mice (The Jackson Laboratory; 6–7 weeks old) were inoculated with 2 × 106 human MPNST cells into flanks. Nodules reaching 200 mm3 were injected intratumorally with 1.0 × 1011 VP/tumor spread over three doses (3.30 × 1010 VP/dose every 48 h) of either adenoviruses or vehicle. Differences in tumor growth curves were assessed using a mixed effects model test where each group was compared with the PBS control group. Bonferroni correction was used to control family-wise error rate.
To establish syngeneic allografts, an equal number of male and female C57BL/6J mice (The Jackson Laboratory; 6–7 weeks old) were inoculated with 1 × 105 JW18 or JW23 cells into flanks. Nodules reaching 200 mm3 were injected intratumorally with 1.0 × 1011 VP/tumor or PBS control for four doses. In this immunocompetent model, CRAd therapy was tested in combination with immune checkpoint and targeted therapies, including anti-CTLA-4 (Bio X Cell, BE0164; 200 μg per dose intraperitoneally [i.p.] every 48 h for four doses), anti-PD-L1 (Bio X Cell, BP0101; 200 μg per dose i.p. every 48 h for four doses), anti-PD-1 (Bio X Cell, BP0146; 200 μg per dose i.p. every 48 h for four doses), and selumetinib (Selleck Chemicals, S1008; 100 μL of 100mM stock diluted in 815 μL of drug dilution buffer (30% PEG, 5% Tween80, 65% dH2O) and injected i.p. at 25 mg/kg daily until endpoint).
Tumors were measured five times/week and mice were euthanized when tumor reached 2,000 mm3 per IACUC requirements. Tumor volumes were calculated using a modified ellipsoid formula (Width2 ✕ Length)/2.59
Adenoviral vectors
Replication-proficient viruses used in the study were Ad5-WT, RGD-WT (positive controls), CRAd-COX2, and RGD-CRAd-COX2. All adenovirus vectors were designed and amplified in the laboratory of Dr. Masato Yamamoto (University of Minnesota).42
Cell viability assay
Cells were infected with RGD-CRAd at MOIs ranging from 1 to 500 plaque-forming units (PFU)/cell. Fresh medium was added after 2 h and cells were cultured for 5 days. The cell viability was assessed after 5 days using an MTS assay performed according to the manufacturer’s protocol (Corning).
COX2 expression analysis in MPNST cells
COX2 expression was analyzed by RNA sequencing and corroborated by RT-PCR. Total RNA, extracted from cells (High-Pure RNA kit; Roche) was used in a two-step RT-PCR. RT reaction was done using SuperScript VILO (Invitrogen) and 50 ng of cDNA. The primer sequences used to amplify complementary DNA (cDNA) transcript were as follows: COX2 sense: 5′-GGTCTGGTGCCTGGTCTGATGATG-3′; COX2 anti-sense: 5′-GTCCTTTCAAGGAGAATG-GTGC-3′; GAPDH sense: 5′-CAACTACATGGTTTACATGTTCCAA-3′; GAPDH anti-sense: 5′-GCCAGTGGACTCCACGACGT-3′. The transcripts were amplified using 50 nM of primers and the following cycling program: an initial step of 95°C for 105 s, then 35 cycles of denaturation at 95°C for 15 s, and annealing/extension at 60°C for 60 s. For GAPDH the number of cycles was reduced to 30 and annealing/extension was reduced to 30s.
Analysis of intratumoral viral spread via immunohistochemical analyses
Parafilm-imbedded sections of mouse tumor tissue were immunostained for antibodies specific to adenoviral goat-hexon (1:750; AB1056, Millipore), mouse-E1A (1:750; ab204123, Abcam), mouse-fiber (1:750; MA5-11222, Thermo), and rabbit-cleaved caspase 3 (1:200; Cell Signaling, 9661) following conventional procedures.
Assessment of virus entry receptors
Cells were plated and cultured for 24 h, non-enzymatically lifted off the plate (Sigma C5789), washed with PBS, and stained with antibodies for integrins ανβ3 (BS-1310R-Cy3, Bioss) or immunoglobulin (Ig)G isotype control (BS-025P); ανβ5 (565836-AF647, BD) or κ isotype control (565378-AF647, BD); and CAR (BS-2389R-A488, Bioss) or IgG Isotype control (BS-0295P-A488).
Immunoblotting
The cells were infected with RGD-COX2-CRAd at various MOIs (VP/cell). The samples were lysed 2 days post-infection and subjected to sodium dodecyl sulphate-Tris-glycine gel electrophoresis. The membranes were incubated with the following antibodies: E1A, (1:1,000, ab204123, Abcam), fiber (1:1,000, MA5-11222, Thermo), Hexon (1:5,000 b-12354), and GRB-2 (1:1,000; 610111, BD).
RNA sequencing
JW cell lines were grown in flanks of C57BL/6J mice until 1,000 mm3 and processed for RNA sequencing along with sciatic nerve control. RNA was extracted utilizing RNeasy Mini Kit (Qiagen, 74106) per manufacturer instructions. Library preparation and paired sequencing at 40 million read depth and 75 base pair read length was performed by the University of Minnesota Genomics Center. Quality control, read alignment with HISAT2, and read mapping with Stringtie were performed with support from the Minnesota Supercomputing Institute. Differential gene expression experiments were performed in R Statistical Software with the DESEQ2 package.55,56,57,58
Syngeneic model whole exome sequencing and prediction of transcribed variants
Genomic DNA of each JW cell line was extracted utilizing the QIAamp DNA Mini Kit (Qiagen, 51304) for whole exome sequencing. DNA was additionally extracted from the liver of a C57BL/6 mouse for reference. DNA fragmentation, library preparation, hybridization, PCR amplification, and sequencing was performed by Azenta Life Sciences. Sequence reads were trimmed to remove possible adapter sequences and nucleotides with poor quality using Trimmomatic v.0.38. The trimmed reads were aligned to the reference genomes using the Illumina Dragen Bio-IT Platform. Somatic variants were called using the Illumina Dragen Bio-IT Platform in somatic mode. Paired normal samples were used. The filtered VCF was then annotated with Ensembl Variant Effect Predictor (VEP) v95. Called variants were next cross referenced with the RNA-sequencing data to identify transcribed variants.
Next, we assessed whether cell line variants are potentially immunogenic. After variant calling and quality filtering, protein sequences were generated using SnpEff. Normal and variant sequences for 663 genes were analyzed for immunogenicity using NetMHCpan-4.1.60 The maximum immunogenic peptides were selected for each transcript and compared between variant and normal. A total of 48 genes contained variants that increased predicted immunogenicity by at least 0.1 eluted ligand rank percent. Variants unique to each cell line were selected for display as representing unique neoantigens rather than variants that may arise from genetic drift of the NPcis murine model.
Flow cytometry
Assessment of intra-tumor immune infiltrate was analyzed via flow cytometry on the CytoFLEX (Beckman Coulter). The following antibodies were utilized: BV785 anti-mouse F4/80 (BioLegend, 123141), BV711 anti-mouse Cd11c (BioLegend, 117349), AF488 anti-mouse Foxp3 (Invitrogen, 53-5773-82), PE-Cy7 anti-mouse Cd11b (BD Pharmingen, 552850), APC anti-mouse Cd45.2 (Cytek Biosciences, 20–0454), BUV396 anti-mouse Cd4 (BD Horizon, 563790), BV421 anti-mouse Cd3ε (BD Horizon, 562600), PE anti-mouse Cd8a (BD Pharmingen, 553033), eFluor780 fixable viability dye (eBioscience, 65-0865-14), and mouse FcR blocking reagent (Miltenyi, 65-0865-14). Flank tumors were treated with CRAd or PBS control as above and harvested on day 10. Fresh tumor samples in triplicate for each condition were isolated and dissociated into single-cell suspensions utilizing gentleMACS Octo Dissociator and mouse tumor dissociation kit (Miltenyi, 130-096-730) per manufacturer instructions; 1 × 109 cells per sample were aliquoted for analysis. Staining including fixation/permeabilization for the intranuclear antigen Foxp3 (eBioscience, 00-5521-00) was performed according to ThermoFisher Scientific protocol, with 1 μL of each antibody used per sample as appropriate (https://www.thermofisher.com/us/en/home/references/protocols/cell-and-tissue-analysis/protocols/staining-intracellular-antigens-flow-cytometry.html, Protocol B).
For analysis of major histocompatibility class 1 expression on JW cell lines, 2 μL of PE anti-MHC Class I H-2Kb (eBioscience, 12-5958-80) or PE mouse IgG2a kappa isotype control (eBioscience, 12-4724-82) was performed on 1 × 106 cells isolated with non-enzymatic dissociation (Sigma-Aldrich, C5789). For human cell lines, PE conjugated anti-HLA-ABC (eBioscience, 12-9983-42) and IgG2a kappa isotype control (eBioscience, 12-4724-81) were used. For MHC class 1 positive control upregulation, cells were stimulated with either murine (Abcam, ab9922) or human (R&D Systems, 285IF100) recombinant IFN-γ at 1 ng/mL for 24 h. Analysis of MHC class 1 expression on MPNST models was performed in triplicate on the CytoFLEX (Beckman Coulter) on the viable cell population.
Statistical analysis
Differences in viral binding, infectivity, and spread and cell immunophenotyping were analyzed using Student’s t test. The relative tumor volumes and tumor growth rates were analyzed using a linear mixed effects random intercept model using R Statistical Software (TidyVerse, and lmerTest). Differences in survival times were assessed with Kaplan-Meier and proportional hazard regression models using R Statistical Software (Survival, Survminer). The results were considered statistically significant at a p value of less than 0.05. All data are expressed as mean ± SD.
Data and code availability
Data supporting the findings of this study are available within the paper and its supplemental information. Additional data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus (Edgar et al., 200261) and are accessible through GEO Series accession number GSE246610 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE246610).
Acknowledgments
Funding for this work was provided by National Institutes of Health (CA151845, and CA154998 to D.L., CA276480 and CA228760 to M.Y.); American Cancer Society Research Professor Award (RP-17-216-06-COUN to D.L.); National Institutes of Health (CA099936 to R.T.G.); 2021 Children’s Cancer Research Fund Emerging Scientist Award (to R.T.G.).
Author contributions
Concept of study: J.A.N., M.Y., D.A.L. Experimental design: J.A.N., R.T.G., M.Y., K.L.J., D.A.L. Data acquisition: J.A.N., R.T.G., M.M.S., E.L.N., P.J.B. Data analysis: J.A.N., R.T.G., M.M.S., E.L.N., S.M.C., T.A.J. Draft of manuscript: J.A.N., R.T.G. Careful review of manuscript: D.A.L., M.Y., K.L.J., M.M.S., E.L.N.
Declaration of interests
D.A.L. is the co-founder and co-owner of NeoClone Biotechnologies, Inc., Discovery Genomics, Inc. (acquired by Immusoft, Inc.), B-MoGen Biotechnologies, Inc. (acquired by Bio-Techne corporation), and Luminary Therapeutics, Inc. D.A.L. holds equity in, is a Board of Directors member of, and serves as the Senior Scientific Advisor to Recombinetics, a genome-editing company, and Makana, a xenotransplantation company. D.A.L. consults for Styx Biotechnologies, Inc. and Genentech, Inc., which is funding some of his research. The business of all the companies above is unrelated to the contents of this manuscript.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omton.2024.200783.
Supplemental information
References
- 1.Le Guellec S., Decouvelaere A.V., Filleron T., Valo I., Charon-Barra C., Robin Y.M., Terrier P., Chevreau C., Coindre J.M. Malignant Peripheral Nerve Sheath Tumor Is a Challenging Diagnosis: A Systematic Pathology Review, Immunohistochemistry, and Molecular Analysis in 160 Patients from the French Sarcoma Group Database. Am. J. Surg. Pathol. 2016;40:896–908. doi: 10.1097/PAS.0000000000000655. [DOI] [PubMed] [Google Scholar]
- 2.Staedtke V., Bai R.Y., Blakeley J.O. Cancer of the Peripheral Nerve in Neurofibromatosis Type 1. Neurotherapeutics. 2017;14:298–306. doi: 10.1007/s13311-017-0518-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Korfhage J., Lombard D.B. Malignant Peripheral Nerve Sheath Tumors: From Epigenome to Bedside. Mol. Cancer Res. 2019;17:1417–1428. doi: 10.1158/1541-7786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Upadhyaya M., Kluwe L., Spurlock G., Monem B., Majounie E., Mantripragada K., Ruggieri M., Chuzhanova N., Evans D.G., Ferner R., et al. Germline and Somatic NF1 Gene Mutation Spectrum in NF1-Associated Malignant Peripheral Nerve Sheath Tumors (MPNSTs) Hum. Mutat. 2008;29:74–82. doi: 10.1002/humu.20601. [DOI] [PubMed] [Google Scholar]
- 5.Pemov A., Li H., Presley W., Wallace M.R., Miller D.T. Genetics of Human Malignant Peripheral Nerve Sheath Tumors. Neurooncol. Adv. 2020;2:i50–i61. doi: 10.1093/noajnl/vdz049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Evans D.G.R., Baser M.E., McGaughran J., Sharif S., Howard E., Moran A. Malignant Peripheral Nerve Sheath Tumours in Neurofibromatosis. J. Med. Genet. 2002;39:311–314. doi: 10.1136/jmg.39.5.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ferner R.E., Gutmann D.H. Neurofibromatosis Type 1 (NF1): Diagnosis and Management. Vol 115. Handb. Clin. Neurol. 2013;115:939–955. doi: 10.1016/B978-0-444-52902-2.00053-9. [DOI] [PubMed] [Google Scholar]
- 8.Somatilaka B.N., Sadek A., McKay R.M., Le L.Q. Malignant Peripheral Nerve Sheath Tumor: Models, Biology, and Translation. Oncogene. 2022;41:2405–2421. doi: 10.1038/s41388-022-02290-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wong W.W., Hirose T., Scheithauer B.W., Schild S.E., Gunderson L.L. Malignant Peripheral Nerve Sheath Tumor: Analysis of Treatment Outcome. Int. J. Radiat. Oncol. Biol. Phys. 1998;42:351–360. doi: 10.1016/S0360-3016(98)00223-5. [DOI] [PubMed] [Google Scholar]
- 10.Anghileri M., Miceli R., Fiore M., Mariani L., Ferrari A., Mussi C., Lozza L., Collini P., Olmi P., Casali P.G., et al. Malignant Peripheral Nerve Sheath Tumors: Prognostic Factors and Survival in a Series of Patients Treated at a Single Institution. Cancer. 2006;107:1065–1074. doi: 10.1002/cncr.22098. [DOI] [PubMed] [Google Scholar]
- 11.Zehou O., Fabre E., Zelek L., Sbidian E., Ortonne N., Banu E., Wolkenstein P., Valeyrie-Allanore L. Chemotherapy for the Treatment of Malignant Peripheral Nerve Sheath Tumors in Neurofibromatosis 1: A 10-year Institutional Review. Orphanet J. Rare Dis. 2013;8:127. doi: 10.1186/1750-1172-8-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carli M., Ferrari A., Mattke A., Zanetti I., Casanova M., Bisogno G., Cecchetto G., Alaggio R., De Sio L., Koscielniak E., et al. Pediatric Malignant Peripheral Nerve Sheath Tumor: The Italian and German Soft Tissue Sarcoma Cooperative Group. J. Clin. Oncol. 2005;23:8422–8430. doi: 10.1200/JCO.2005.01.4886. [DOI] [PubMed] [Google Scholar]
- 13.Valentin T., Le Cesne A., Ray-Coquard I., Italiano A., Decanter G., Bompas E., Isambert N., Thariat J., Linassier C., Bertucci F., et al. Management and Prognosis of Malignant Peripheral Nerve Sheath Tumors: The Experience of the French Sarcoma Group (GSF-GETO) Eur. J. Cancer. 2016;56:77–84. doi: 10.1016/j.ejca.2015.12.015. [DOI] [PubMed] [Google Scholar]
- 14.Cassady K.A., Haworth K.B., Jackson J., Markert J.M., Cripe T.P. To Infection and Beyond: The Multi-pronged Anti-cancer Mechanisms of Oncolytic Viruses. Viruses. 2016;8:43. doi: 10.3390/v8020043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moore A.E. Effect of Inoculation of the Viruses of Influenza A and Herpes Simplex on the Growth of Transplantable Tumors in Mice. Cancer. 1949;2:516–524. doi: 10.1002/1097-0142(194905). [DOI] [PubMed] [Google Scholar]
- 16.Russell S.J., Peng K.W., Bell J.C. Oncolytic Virotherapy. Nat. Biotechnol. 2012;30:658–670. doi: 10.1038/nbt.2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Raja J., Ludwig J.M., Gettinger S.N., Schalper K.A., Kim H.S. Oncolytic Virus Immunotherapy: Future Prospects for Oncology. J. Immunother. Cancer. 2018;6:140. doi: 10.1186/s40425-018-0458-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ribas A., Dummer R., Puzanov I., VanderWalde A., Andtbacka R.H.I., Michielin O., Olszanski A.J., Malvehy J., Cebon J., Fernandez E., et al. Oncolytic Virotherapy Promotes Intratumoral T Cell Infiltration and Improves Anti-PD-1 Immunotherapy. Cell. 2017;170:1109–1119.e10. doi: 10.1016/j.cell.2017.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu L.M.N., Lu Q.R. Therapeutic Targets for Malignant Peripheral Nerve Sheath Tumors. Future Neurol. 2019;14:FNL7. doi: 10.2217/fnl-2018-0026. [DOI] [Google Scholar]
- 20.Shurell E., Singh A.S., Crompton J.G., Jensen S., Li Y., Dry S., Nelson S., Chmielowski B., Bernthal N., Federman N., et al. Characterizing the Immune Microenvironment of Malignant Peripheral Nerve Sheath Tumor by PD-L1 Expression and Presence of CD8+ Tumor Infiltrating Lymphocytes. Oncotarget. 2016;7:64300–64308. doi: 10.18632/oncotarget.11734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee P.R., Cohen J.E., Fields R.D. Immune System Evasion by Peripheral Nerve Sheath Tumor. Neurosci. Lett. 2006;397:126–129. doi: 10.1016/j.neulet.2005.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weng W., Yu L., Li Z., Tan C., Lv J., Lao I.W., Hu W., Deng Z., Liu Z., Wang J., Xu M. The immune subtypes and landscape of sarcomas. BMC Immunol. 2022;23:46. doi: 10.1186/s12865-022-00522-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yan J., Chen Y., Patel A.J., Warda S., Lee C.J., Nixon B.G., Wong E.W., Miranda-Román M.A., Yang N., Wang Y., et al. Tumor-intrinsic PRC2 Inactivation Drives a Context-dependent Immune-desert Microenvironment and is Sensitized by Immunogenic Viruses. J. Clin. Invest. 2022;132:e153437. doi: 10.1172/JCI153437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ginn S.L., Amaya A.K., Alexander I.E., Edelstein M., Abedi M.R. Gene therapy clinical trials worldwide to 2017: An update. J. Gene Med. 2018;20:e3015. doi: 10.1002/jgm.3015. [DOI] [PubMed] [Google Scholar]
- 25.Mantwill K., Klein F.G., Wang D., Hindupur S.V., Ehrenfeld M., Holm P.S., Nawroth R. Concepts in Oncolytic Adenovirus Therapy. Int. J. Mol. Sci. 2021;22:10522. doi: 10.3390/ijms221910522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Savontaus M.J., Sauter B.V., Huang T.G., Woo S.L.C. Transcriptional Targeting Of Conditionally Replicating Adenovirus to Dividing Endothelial Cells. Gene Ther. 2002;9:972–979. doi: 10.1038/sj.gt.3301747. [DOI] [PubMed] [Google Scholar]
- 27.Wakayama M., Abei M., Kawashima R., Seo E., Fukuda K., Ugai H., Murata T., Tanaka N., Hyodo I., Hamada H., Yokoyama K.K. E1A, E1B Double-restricted Adenovirus with RGD-Fiber Modification Exhibits Enhanced Oncolysis for CAR-deficient Biliary Cancers. Clin. Cancer Res. 2007;13:3043–3050. doi: 10.1158/1078-0432.CCR-06-2103. [DOI] [PubMed] [Google Scholar]
- 28.Singh P.K., Doley J., Kumar G.R., Sahoo A.P., Tiwari A.K. Oncolytic Viruses & Their Specific Targeting to Tumour Cells. Indian J. Med. Res. 2012;136:571–584. [PMC free article] [PubMed] [Google Scholar]
- 29.Kirn D. Replication-selective Oncolytic Adenoviruses: Virotherapy Aimed at Genetic Targets in Cancer. Oncogene. 2000;19:6660–6669. doi: 10.1038/sj.onc.1204094. [DOI] [PubMed] [Google Scholar]
- 30.Baker A.T., Aguirre-Hernández C., Halldén G., Parker A.L. Designer Oncolytic Adenovirus: Coming of Age. Cancers. 2018;10:201. doi: 10.3390/cancers10060201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sharma A., Tandon M., Bangari D.S., Mittal S.K. Adenoviral Vector-based Strategies for Cancer Therapy. Curr. Drug Ther. 2009;4:117–138. doi: 10.2174/157488509788185123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krasnykh V., Belousova N., Korokhov N., Mikheeva G., Curiel D.T. Genetic Targeting of an Adenovirus Vector via Replacement of the Fiber Protein with the Phage T4 Fibritin. J. Virol. 2001;75:4176–4183. doi: 10.1128/JVI.75.9.4176-4183.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roelvink P.W., Mi Lee G., Einfeld D.A., Kovesdi I., Wickham T.J. Identification of a Conserved Receptor-Binding Site on the Fiber Proteins of CAR-Recognizing Adenoviridae. Science. 1999;286:1568–1571. doi: 10.1126/science.286.5444.1568. [DOI] [PubMed] [Google Scholar]
- 34.Sato-Dahlman M., Yamamoto M. The development of Oncolytic Adenovirus Therapy in the Past and Future - For the Case of Pancreatic Cancer. Curr. Cancer Drug Targets. 2018;18:153. doi: 10.2174/1568009617666170222123925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koodie L., Robertson M.G., Chandrashekar M., Ruth G., Dunning M., Bianco R.W., Davydova J. Rodents Versus Pig Model for Assessing the Performance of Serotype Chimeric Ad5/3 Oncolytic Adenoviruses. Cancers. 2019;11:198. doi: 10.3390/cancers11020198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bergelson J.M., Cunningham J.A., Droguett G., Kurt-Jones E.A., Krithivas A., Hong J.S., Horwitz M.S., Crowell R.L., Finberg R.W. Isolation of a Common Receptor for Coxsackie B Viruses and Adenoviruses 2 and 5. Science. 1997;275:1320–1323. doi: 10.1126/science.275.5304.1320. [DOI] [PubMed] [Google Scholar]
- 37.Dmitriev I., Krasnykh V., Miller C.R., Wang M., Kashentseva E., Mikheeva G., Belousova N., Curiel D.T. An Adenovirus Vector with Genetically Modified Fibers Demonstrates Expanded Tropism via Utilization of a Coxsackievirus and Adenovirus Receptor-Independent Cell Entry Mechanism. J. Virol. 1998;72:9706–9713. doi: 10.1128/JVI.72.12.9706-9713.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lang F.F., Conrad C., Gomez-Manzano C., Yung W.K.A., Sawaya R., Weinberg J.S., Prabhu S.S., Rao G., Fuller G.N., Aldape K.D., et al. Phase I Study of DNX-2401 (Delta-24-RGD) Oncolytic Adenovirus: Replication and Immunotherapeutic Effects in Recurrent Malignant Glioma. J. Clin. Oncol. 2018;36:1419–1427. doi: 10.1200/JCO.2017.75.8219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang H., Li Z.Y., Liu Y., Persson J., Beyer I., Möller T., Koyuncu D., Drescher M.R., Strauss R., Zhang X.B., et al. Desmoglein 2 is a Receptor for Adenovirus Serotypes 3, 7, 11 and 14. Nat. Med. 2011;17:96–104. doi: 10.1038/nm.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Connolly J.B. Conditionally Replicating Viruses in Cancer Therapy. Gene Ther. 2003;10:712–715. doi: 10.1038/sj.gt.3301898. [DOI] [PubMed] [Google Scholar]
- 41.Gómez-Navarro J., Curiel D.T. Conditionally Replicative Adenoviral Vectors for Cancer Gene Therapy. Lancet Oncol. 2000;1:148–158. doi: 10.1016/s1470-2045(00)00030-9. [DOI] [PubMed] [Google Scholar]
- 42.Davydova J., Le L.P., Gavrikova T., Wang M., Krasnykh V., Yamamoto M. Infectivity-enhanced Cyclooxygenase-2-based Conditionally Replicative Adenoviruses for Esophageal Adenocarcinoma Treatment. Cancer Res. 2004;64:4319–4327. doi: 10.1158/0008-5472.CAN-04-0064. [DOI] [PubMed] [Google Scholar]
- 43.Simon L.S. Role and Regulation of Cyclooxygenase-2 During Inflammation. Am. J. Med. 1999;106:37S–42S. doi: 10.1016/S0002-9343(99)00115-1. [DOI] [PubMed] [Google Scholar]
- 44.Hashemi Goradel N., Najafi M., Salehi E., Farhood B., Mortezaee K. Cyclooxygenase-2 in Cancer: A Review. J. Cell. Physiol. 2019;234:5683–5699. doi: 10.1002/jcp.27411. [DOI] [PubMed] [Google Scholar]
- 45.Hakozaki M., Tajino T., Konno S., Kikuchi S., Yamada H., Yanagisawa M., Nishida J., Nagasawa H., Tsuchiya T., Ogose A., et al. Overexpression of Cyclooxygenase-2 In Malignant Peripheral Nerve Sheath Tumor and Selective Cyclooxygenase-2 Inhibitor-induced Apoptosis by Activating Caspases in Human Malignant Peripheral Nerve Sheath Tumor Cells. PLoS One. 2014;9 doi: 10.1371/journal.pone.0088035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Staedtke V., Gray-Bethke T., Riggins G.J., Bai R.Y. Preventative effect of mebendazole against malignancies in neurofibromatosis 1. Genes. 2020;11:762. doi: 10.3390/genes11070762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sato-Dahlman M., LaRocca C.J., Yanagiba C., Yamamoto M. Adenovirus and Immunotherapy: Advancing Cancer Treatment by Combination. Cancers. 2020;12:1295. doi: 10.3390/cancers12051295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Antoszczyk S., Rabkin S.D. Prospect and Progress of Oncolytic Viruses for Treating Peripheral Nerve Sheath Tumors. Expert Opin. Orphan Drugs. 2016;4:129–138. doi: 10.1517/21678707.2016.1128322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Andtbacka R.H.I., Collichio F., Harrington K.J., Middleton M.R., Downey G., Ӧhrling K., Kaufman H.L. Final analyses of OPTiM: a Randomized Phase III trial of Talimogene Laherparepvec Versus Granulocyte-macrophage Colony-Stimulating Factor in Unresectable Stage III–IV Melanoma. J. Immunother. Cancer. 2019;7:145. doi: 10.1186/s40425-019-0623-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Blair G.E., Dixon S.C., Griffiths S.A., Zajdel M.E. Restricted Replication of Human Adenovirus Type 5 in Mouse Cell Lines. Virus Res. 1989;14:339–346. doi: 10.1016/0168-1702(89)90026-9. [DOI] [PubMed] [Google Scholar]
- 51.Martínez-Vélez N., Garcia-Moure M., Marigil M., González-Huarriz M., Puigdelloses M., Gallego Pérez-Larraya J., Zalacaín M., Marrodán L., Varela-Guruceaga M., Laspidea V., et al. The oncolytic virus Delta-24-RGD elicits an antitumor effect in pediatric glioma and DIPG mouse models. Nat. Commun. 2019;10 doi: 10.1038/s41467-019-10043-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Inoue A., Janke L.J., Gudenas B.L., Jin H., Fan Y., Paré J., Clay M.R., Northcott P.A., Hirbe A.C., Cao X. A Genetic Mouse Model with Postnatal Nf1 and p53 Loss recapitulates the Histology and Transcriptome of Human Malignant Peripheral Nerve Sheath Tumor. Neurooncol. Adv. 2021;3:vdab129. doi: 10.1093/noajnl/vdab129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Miller S.J., Rangwala F., Williams J., Ackerman P., Kong S., Jegga A.G., Kaiser S., Aronow B.J., Frahm S., Kluwe L., et al. Large-Scale Molecular Comparison of Human Schwann Cells to Malignant Peripheral Nerve Sheath Tumor Cell Lines and Tissues. Cancer Res. 2006;66:2584–2591. doi: 10.1158/0008-5472.CAN-05-3330. [DOI] [PubMed] [Google Scholar]
- 54.Hirbe A.C., Dahiya S., Miller C.A., Li T., Fulton R.S., Zhang X., McDonald S., DeSchryver K., Duncavage E.J., Walrath J., et al. Whole Exome Sequencing Reveals the Order of Genetic Changes during Malignant Transformation and Metastasis in a Single Patient with NF1-plexiform Neurofibroma. Clin. Cancer Res. 2015;21:4201–4211. doi: 10.1158/1078-0432.CCR-14-3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Galvin R.T., Jena S., Maeser D., Gruener R., Huang R.S. Revealing Pan-histology Immunomodulatory Targets in Pediatric Central Nervous System Tumors. Cancers. 2023;15:5455. doi: 10.3390/cancers15225455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhao Y., Liu Z., Li L., Wu J., Zhang H., Zhang H., Lei T., Xu B. Oncolytic Adenovirus: Prospects for Cancer Immunotherapy. Front. Microbiol. 2021;12 doi: 10.3389/fmicb.2021.707290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hakozaki M., Hojo H., Sato M., Tajino T., Yamada H., Kikuchi S., Abe M. Establishment and Characterization of a Novel Human Malignant Peripheral Nerve Sheath Tumor Cell Line, FMS-1, that Overexpresses Epidermal Growth Factor Receptor and Cyclooxygenase-2. Virchows Arch. 2009;455:517–526. doi: 10.1007/s00428-009-0848-1. [DOI] [PubMed] [Google Scholar]
- 58.Deadwyler G.D., Dang I., Nelson J., Srikanth M., De Vries G.H., De Vries G. Prostaglandin E2 Metabolism is Activated In Schwann Cell Lines Derived From Human Nf1 Malignant Peripheral Nerve Sheath Tumors. Neuron Glia Biol. 2004;1:149–155. doi: 10.1017/S1740925X04000262. [DOI] [PubMed] [Google Scholar]
- 59.Tomayko M.M., Reynolds C.P. Determination of Subcutaneous Tumor Size in Athymic (Nude) Mice. Cancer Chemother. Pharmacol. 1989;24:148–154. doi: 10.1007/BF00300234. [DOI] [PubMed] [Google Scholar]
- 60.Reynisson B., Alvarez B., Paul S., Peters B., Nielsen M. NetMHCpan-4.1 and NetMHCIIpan-4.0: Improved Predictions of MHC Antigen Presentation by Concurrent Motif Deconvolution and Integration of MS MHC Eluted Ligand Data. Nucleic Acids Res. 2020;48:W449–W454. doi: 10.1093/nar/gkaa379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Eddgar R., Domrachev M., Lash A.E. Gene Expression Omnibus: NCBI gene expression and hybridization array data reposotory. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data supporting the findings of this study are available within the paper and its supplemental information. Additional data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus (Edgar et al., 200261) and are accessible through GEO Series accession number GSE246610 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE246610).