Abstract
Background
Applications for artificial intelligence (AI) in ophthalmology are continually evolving. Fundoscopy is one of the oldest ocular imaging techniques but remains a mainstay in posterior segment imaging due to its prevalence, ease of use, and ongoing technological advancement. AI has been leveraged for fundoscopy to accomplish core tasks including segmentation, classification, and prediction.
Main body
In this article we provide a review of AI in fundoscopy applied to representative chorioretinal pathologies, including diabetic retinopathy and age-related macular degeneration, among others. We conclude with a discussion of future directions and current limitations.
Short conclusion
As AI evolves, it will become increasingly essential for the modern ophthalmologist to understand its applications and limitations to improve patient outcomes and continue to innovate.
Keywords: Artificial intelligence, Machine learning, Deep learning, Fundus, Fundoscopy, Choroid, Age-related macular degeneration, Diabetic retinopathy
Background
Artificial intelligence (AI) is reshaping ophthalmology, especially in fundus imaging, aiding in segmentation, classification, and prediction of chorioretinal diseases like diabetic retinopathy (DR) and age-related macular degeneration (AMD). Fundus imaging is the technique of creating a two-dimensional representation of the three-dimensional semi-transparent retinal tissues using reflected yield [1–4]. Digital technology has supplanted stereo film, used for image capture, allowing for faster processing times and better image editing and acquisition [5, 6]. There are various subcategories within fundus imaging, such as stereo fundus, widefield fundus (WF), ultra-widefield fundus (UWF), fundus autofluorescence (FAF), color fundus (CF), standard fundus, and angiographic applications [1].
Defining AI tasks in chorioretinal disease
AI is a powerful tool capable of learning a near universal set of tasks, but no single algorithm is universally successful. Algorithms are instead carefully designed to perform a specific task. AI in ophthalmology is most frequently used to automate three important tasks: image segmentation, classification, and prediction.
Segmentation
In the field of image processing, image segmentation refers to the partitioning of an image into multiple segments defined by a set of pixels. In medical imaging, segmentation plays an important role in identifying and highlighting regions of interest [7]. Classification and prediction models frequently use parameters and features obtained from analysis of regions of interest. As a result, segmentation is often performed as a first step in in the AI and image processing pipeline [8]. While this task could be performed using classic computer vision approaches such as tensor voting, AI-based learning techniques have shown great promise in improving accuracy and efficiency of segmentation.
In imaging of the chorioretinal space, segmentation plays a key role in delineating important structures, yielding biomarkers such as retinal thickness, intraretinal fluid volume, and choroidal thickness. In research, automated segmentation saves time and effort while improving accuracy. When dealing with large datasets, which are often required in DL tasks, automated segmentation is practically a necessity.
Classification
Classification is the task of assigning a category to a given input [9]. Classification is well-described problem not exclusive to AI. People classify objects regularly: for example, separating groceries into different food groups such as fruits, vegetables, breads, and meats. These groups are referred to as classes, categories, labels, or groups in AI. In ophthalmology, classifying a fundus image as polypoidal choroidal vasculopathy or age-related macular degeneration is a classification task [10]. Classification problems can be very specific, such as differentiating between stages of proliferative diabetic retinopathy or broad such as determining whether a patient needs referral or not [11]. These questions are of significant interest to clinicians as they can directly impact decision making. As a result, classification forms the backbone of AI use in medicine and is paving the way for automated diagnosis [12].
Prediction
Prediction is closely related to classification, but outputs a value or outcome rather than a category. Tasks that evaluate future outcomes, such as the chance of fluid recurrence or response to treatment, fall under the category of prediction. Models designed to solve prediction tasks may yield a continuous range of values: for example, the best-corrected visual acuity based on a set of clinical factors and treatment [13]. Even traditional classification models may be rethought as prediction models. Rather than discretely classifying AMD as intermediate or late stage, a prediction model may provide a continuous numeric value reflecting the severity of disease. Prediction models will serve as the foundation for automated assessment of disease progression and treatment outcomes.
AI in chorioretinal pathology through fundoscopy
Segmentation
Fundoscopy can visualize structures including retinal blood vessels, optic disc, optic cup, and macula [14]. Retinal blood vessel segmentation remains the key chorioretinal segmentation tasking using AI, along with the identification of chorioretinal lesions like microaneurysms, hemorrhages, and exudates [15]. However, there are relatively few articles regarding chorioretinal lesion segmentation using AI.
Segmentation of vessels
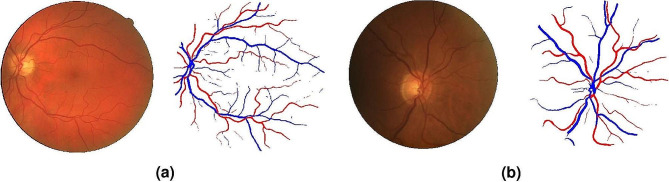
Retinal vessel segmentation is an important fundus task for the diagnosis and treatment of various ocular and cardiovascular pathologies [16] (Fig. 1).
Fig. 1.
Fundus vessel segmentation using a W-Net tested on a) DRIVE and b) LES-AV datasets. Reprinted with permission from Galdran et al. Galdran, A., Anjos, A., Dolz, J. et al. State-of-the-art retinal vessel segmentation with minimalistic models. Sci Rep 12, 6174 (2022) under Creative Commons Attribution 4.0 International License (https://creativecommons.org/licenses/by/4.0/legalcode)
Large datasets, such as DRIVE, STARE, and CHASE_DB1, have simplified vessel segmentation using machine learning (ML) and deep learning (DL) approaches, allowing for easy comparisons against other methods that have been similarly validated (Table 1) [17].
Table 1.
Commonly Used Public Fundus Datasets for AI Applications
| Dataset | Number of Images | Pathologies | Link |
|---|---|---|---|
| Digital Retinal Images for Vessel Extraction (DRIVE) | 40 | 7 | https://drive.grand-challenge.org/ |
| Structured Analysis of the Retina (STARE) | 397 | 13 | http://cecas.clemson.edu/~ahoover/stare/ |
| Child Hearth Health Study in England (CHASE_DB1) | 28 | Healthy | https://blogs.kingston.ac.uk/retinal/chasedb1/ |
| High-Resolution Fundus Image Database (HRF) | 45 | DR, glaucoma | https://www5.cs.fau.de/research/data/fundus-images/ |
| IOSTAR Retinal Vessel | 30 | N/A | http://www.retinacheck.org/download-iostar-retinal-vessel-segmentation-dataset |
| Standard Diabetic Retinopathy Database Calibration Level 0 (DIARETDB0) | 130 | DR | http://www.it.lut.fi/project/imageret/diaretdb0/ |
| Standard Diabetic Retinopathy Database Calibration Level 1 (DIARETDB1) | 89 | DR | http://www.it.lut.fi/project/imageret/diaretdb1/index.html |
| Automated Retinal Image Analysis (ARIA) | 143 | AMD, DR | http://www.damianjjfarnell.com/?page_id=276 |
| Age-Related Eye Disease Study 1/2 (AREDS1/2) | > 134,500 | AMD, cataract | https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000001.v3.p1 |
| Methods to Evaluate Segmentation and Indexing Techniques in the Field of Retinal Ophthalmology 1 (MESSIDOR1) | 1,200 | DR | https://www.adcis.net/en/third-party/messidor/ |
| Methods to Evaluate Segmentation and Indexing Techniques in the Field of Retinal Ophthalmology 2 (MESSIDOR2) | 1,748 | DR | https://www.adcis.net/en/third-party/messidor2/ |
| e-ophtha | 463 | DR | https://www.adcis.net/en/third-party/e-ophtha/ |
To our knowledge, Liskowski and Krawiec published the first example of AI segmentation of blood vessels from fundus images, achieving great success with a CNN validated of multiple datasets, including DRIVE, STARE, and CHASE_DB1 datasets [18]. This work paved the way for a significant rise in AI retinal vessel segmentation articles from 2017 to 2022.
The methods most used for retinal blood vessel segmentation include ML and DL techniques such as support vector machines (SVM), k-nearest neighbor, and U-net architecture [19–28]. Thin vessel problems are addressed by innovations such as SSANet and HHNet, which strike a balance between computing demands and accuracy [14, 29–35].
Segmentation of lesions
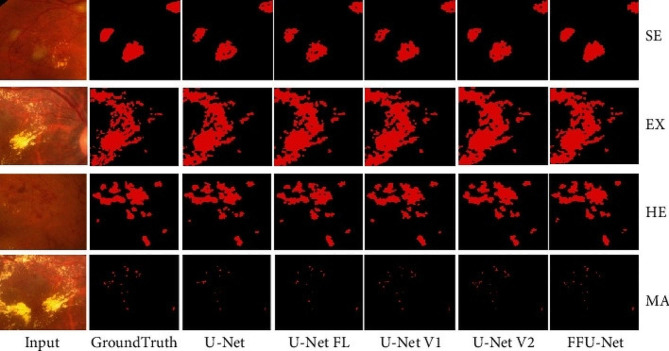
AI segmentation of lesions is an important, but less-investigated realm of chorioretinal fundus segmentation (Fig. 2). Four relevant studies were found (Table 2). Random forest, SVM, and CNN-based algorithms have been employed in research to segment a variety of findings, including geographic atrophy, drusen segmentation, macular edema, and retinal vein blockage [36–39].
Fig. 2.
Automated segmentation of soft exudates (SE), hard exudates (EX), hemorrhage (HE), and microaneurysms (MA) using multiple U-Net architectures. Reprinted with permission from Xu et al. Xu Y, Zhou Z, Li X, Zhang N, Zhang M, Wei P. FFU-Net: Feature Fusion U-Net for Lesion Segmentation of Diabetic Retinopathy. Biomed Res Int. 2021 Jan 2;2021:6644071 under Creative Commons Attribution License (https://creativecommons.org/licenses/by/4.0/legalcode)
Table 2.
Lesion Segmentation with Fundus
| Algorithm | Number of Articles | Results* | Groups |
|---|---|---|---|
| CNN | 2 | AUC: | Chen et al., Hassan et al. |
| Acc: 0.94 | |||
| Sens: | |||
| Spec: | |||
| ML/Multimodal** | 2 | AUC: | Feeny et al., Khalid et al. |
| Acc: 0.98 | |||
| Sens: 1.00 | |||
| Spec: 0.97 | |||
*Average values across articles **ML algorithm(s) or combined use of ML and DL AUC, area under curve. Acc, accuracy. Sens, sensitivity. Spec, specificity
Utility of fundus segmentation
Fundus segmentation remains a key area of research for lesion diagnosis and biomarkers, even with ongoing advancements in algorithms for other imaging modalities. Recent studies have shown new therapeutic uses. OTNet, a CNN method with an AUC of 0.806 that grades arteriosclerosis based on retinal vascular segmentation, was described by Bai et al. [40]. AI’s usefulness in surgical settings was demonstrated by Xu et al.‘s description of an enhanced few-shot learning framework for accurate retinal vascular localization during central serous chorioretinopathy (CSCR) laser surgery [41]. As segmentation technology develops, real-time data may facilitate clinicians in carrying out essential tasks.
Classification
AI classification of ocular pathologies from fundus images has focused on single and multiple pathologies. AMD and DR have received significant attention, with DL techniques prevailing.
Classification of a single pathology
Since the late 1990s, clinicians have been fascinated by automated classification of single diseases such as retinal vascular tortuosity and microaneurysms [42–44]. Commonly used datasets for validating classification algorithms include ARIA, AREDS1/2 for AMD, and MESSIDOR, DIARETDB0/1 for DR. STARE has been utilized for classification methods, as well as segmentation validation.
Many AI classifications of fundus images have been designed to diagnose AMD automatically (Table 3). Mookiah et al. published two of the first automatic AMD classification approaches from fundus images using a mixed methods approach, including decision tree, k-nearest neighbor, probabilistic neural network, and SVM [45, 46]. Their work achieved accuracies ranging from 90.19% to 97.78%, with later improvements achieving 100% accuracy via Locality Sensitive Discriminant Analysis.
Table 3.
AMD Classification with Fundus
| Algorithm | Number of Articles | Results* | Groups |
|---|---|---|---|
| CNN | 5 | AUC: 0.95 | Burlina et al., Chen et al. (a), Chen et al. (b), Keel et al., Matsuba et al. |
| Acc: 0.54 | |||
| Sens: 0.98 | |||
| Spec: 0.97 | |||
| ML/Multimodal** | 5 | AUC: 0.93 | Acharya et al., Govindaiah et al., Mookiah et al. (a), Mookiah et al. (b), Yoo et al. |
| Acc: 0.96 | |||
| Sens: 0.90 | |||
| Spec: 0.98 | |||
*Average values across articles **ML algorithm(s) or combined use of ML and DL AUC, area under curve. Acc, accuracy. Sens, sensitivity. Spec, specificity
CNNs like ResNet, Inception-ResNet-V2, and DeepSeeNet have achieved great accuracies in AMD classification, with AUCs above 0.970 [47–50]. Innovative techniques such as UWF-based CNN and multimodal frameworks improve AMD detection, resulting in impressive AUCs [51, 52].
DR has historically been a secondary emphasis of AI classification of a single pathology (Table 4). DL techniques, particularly, IDx-DR X2.1, result in FDA-approved AI diagnostic systems, while Gayathri et al. use ML approaches to obtain high precision and recall [53–58].
Table 4.
DR Classification with Fundus
| Algorithm | Number of Articles | Results* | Groups |
|---|---|---|---|
| CNN | 8 | AUC: 0.97 | Abràmoff et al. (a), Abràmoff et al. (b), Gargeya et al., Gulshan et al., Singh and Gorantla, Tang et al., Ting et al., Zhang et al. |
| Acc: 0.98 | |||
| Sens: 0.91 | |||
| Spec: 0.93 | |||
| ML/Multimodal** | 5 | AUC: 0.95 | Cao et al., Gayathri et al., Long et al., Yu et al., Zhang et al. |
| Acc: 0.95 | |||
| Sens: 0.90 | |||
| Spec: 0.96 |
*Average values across articles **ML algorithm(s) or combined use of ML and DL AUC, area under curve. Acc, accuracy. Sens, sensitivity. Spec, specificity
Cao et al. published the first paper focusing on AI detection of microaneurysms, critical for early DR diagnosis with an AUC of 0.985 [59]. Yu et al. and Tang et al. classified neovascularization with 95.23% and 99.48% accuracy, respectively [60, 61]. In DR, Sahlsten et al’s Inception-v3 model detected macular edema with an AUC of 0.987 [62]. Singh and Gorantla’s DMENet attained 96.12% accuracy in early macular edema identification [63].
In addition to AMD and DR, several other pathologies have been classified using AI on fundus photos (Table 5). For pathologic myopia (PM), Lu et al. and Rauf et al. developed CNN techniques that achieved similar AUCs of 0.979 and 0.9845, respectively [64, 65] Du et al. performed a similar feature-based classification for PM and achieved a high overall detection rate of 92.08%, demonstrating AI’s potential for identifying various PM lesions [66].
Table 5.
Classification of Non-AMD, Non-DR Pathologies with Fundus
| Algorithm | Number of Articles | Results* | Groups |
|---|---|---|---|
| CNN | 7 | AUC: 0.97 | Brown et al., Cai et al., Du et al., Lu et al., Lu et al., Rauf et al., Zhen et al. |
| Acc: 0.97 | |||
| Sens: 0.82 | |||
| Spec: 0.96 | |||
| ML/Multimodal** | 0 |
*Average values across articles **ML algorithm(s) or combined use of ML and DL AUC, area under curve. Acc, accuracy. Sens, sensitivity. Spec, specificity
Zhen et al. identified CSCR using Inception-V3, achieving AUC 0.934 [67]. Brown et al. used two CNNs, Inception-v1 and U-Net, for diagnose of plus disease in retinopathy of prematurity, with a 100% sensitivity and 94% specificity [68]. Cai et al. used Inception-v3 to automatically classify sea fan neovascularization in sickle cell hemoglobinopathy patients, attaining an AUC of 0.988, sensitivity of 97.4%, and specific of 97.0% [69].
Classification of multiple pathologies
Since 2017, multi-pathology classification using ML and DL has gained popularity, with the goal of rapidly detecting many diseases from a single image, which is critical for new patients. The first such study classified imagines into 10 retinal diseases using DL models like VGG-19, with VGG-19 transfer learning-random forest surpassing the others [70]. While a promising study on multiple disease classification, the authors acknowledged that their pilot study did not fully demonstrate the benefits of using DL for this task, owing to the limited sample size.
For the rest of this section, we will go over additional attempts at multiple disease classification utilizing a range of AI algorithms for many diseases (Table 6).
Table 6.
Classification of Multiple Pathologies with Fundus
| Algorithm | Number of Articles | Results* | Groups |
|---|---|---|---|
| CNN | 10 | AUC: 0.97 | Cen et al., Choi et al., Chou et al., González-Gonzalo et al., Keel et al., Kim et al., Sahlsten et al., Son et al., Xu et al., Yu-Chuan Kang et al. |
| Acc: 0.724 | |||
| Sens: 0.89 | |||
| Spec: 0.94 | |||
| ML/Multimodal** | 6 | AUC: 0.89 | Antaki et al., Balasubramanian and Ananthamoorthy, Koh et al., Porwal et al., Standardization of Uveitis Nomenclature (SUN) Working Group, Tan et al. |
| Acc: 0.95 | |||
| Sens: 0.88 | |||
| Spec: 0.93 |
*Average values across articles **ML algorithm(s) or combined use of ML and DL AUC, area under curve. Acc, accuracy. Sens, sensitivity. Spec, specificity
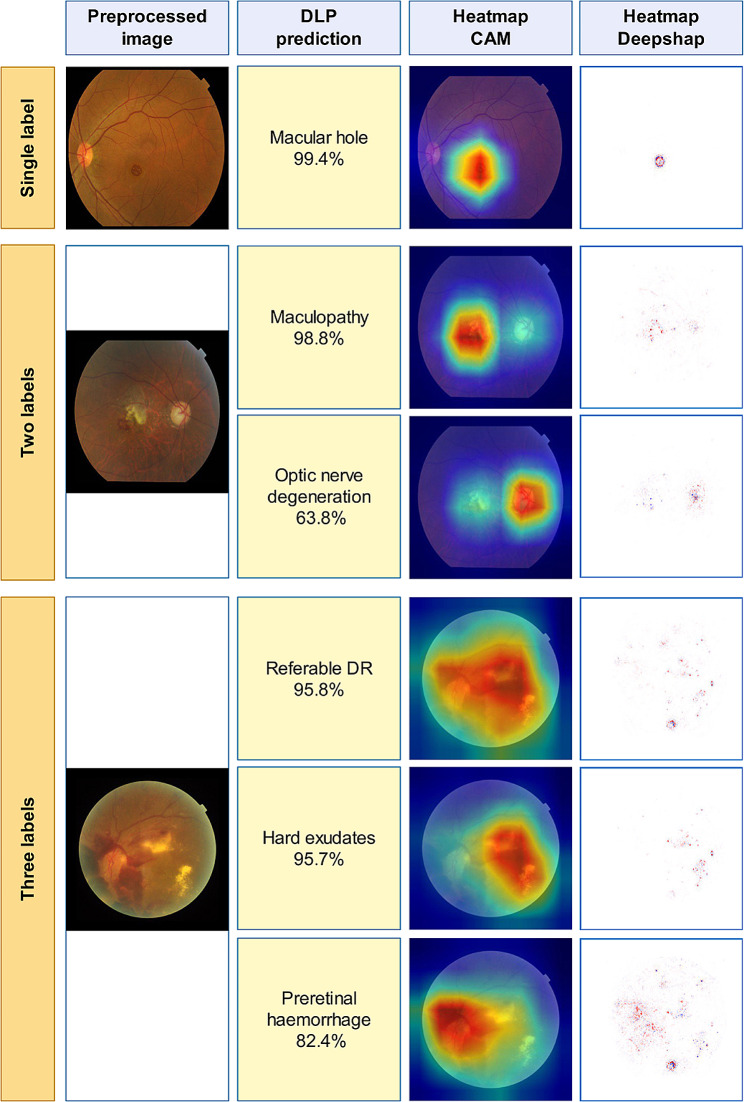
We found two algorithms for classification of AMD and DR from normal images. The first used multiple AI algorithms, including AdaBoost, c4.5, logistic regression, naive bayes, neural network, random forest, SVM [71]. The random forest classifier outperformed other methods with an AUC exceeding 0.995 and the authors contend that their approach is feasible even with a small image pool. González-Gonzalo et al. used RetCAD v.1.3.0 to attain AUC values of 95.1% for DR and 94.9% for AMD [72]. Studies also explored glaucoma classification alongside AMD-DR detection and focused on distinguishing between AMD and polypoidal choroidal vasculopathy (PCV), along with identifying less frequent pathologies and myopic conditions [73–77]. CNN-based approaches have been suggested for detecting significant findings in retinal images and classifying nine posterior segment pathologies [78–83] Cen et al. developed a CNN capable of detecting 39 common retinal diseases, showcasing AI’s potential in ophthalmologic practices [84] (Fig. 3).
Fig. 3.
Representative heatmap and feature detection for classification of multiple retinal pathologies. Reprinted with permission from Cen et al. Cen, LP., Ji, J., Lin, JW. et al. Automatic detection of 39 fundus diseases and conditions in retinal photographs using deep neural networks. Nat Commun 12, 4828 (2021) under Creative Commons Attribution License (https://creativecommons.org/licenses/by/4.0/legalcode)
Classification of stages in a pathology
AI staging of chorioretinal pathologies has focused on AMD, DR, and a few additional pathologies (Table 7). Burlina et al. pioneered AMD grading with DL models like AlexNet and OverFeat, achieving encouraging results but falling short of human grading [85–87].
Table 7.
Classification of Stages in a pathology with fundus
| Algorithm | Number of Articles | Results* | Groups |
|---|---|---|---|
| CNN | 8 | AUC: 0.96 | Alyoubi et al., Burlina et al. (a), Burlina et al. (b), Campbell et al., Heo et al., Peng et al., Shaban et al., Wan et al. |
| Acc: 0.82 | |||
| Sens: 0.88 | |||
| Spec: 0.95 | |||
| ML/Multimodal** | 4 | AUC: | Akbar et al., Grassman et al., Hosoda et al., Murugeswari and Sukanesh |
| Acc: 0.93 | |||
| Sens: | |||
| Spec: |
*Average values across articles **ML algorithm(s) or combined use of ML and DL AUC, area under curve. Acc, accuracy. Sens, sensitivity. Spec, specificity
Grassmann et al. used six CNNs (AlexNet, GoogLeNet, VGG, Inception-v3, ResNet, and Inception-ResNet-v2) to grade AMD with 63.3% accuracy [88]. DeepSeeNet outperformed physicians in identifying large drusen and pigmentary abnormalities in AMD, but was inferior in detecting late AMD (stage 5) [89]. VGG-16 identified wet vs. dry AMD with higher accuracy than first-year residents [90]. K-means cluster analysis identified pachychoroid features associated with improved visual acuity in AMD patients [91]. Despite their variable accuracy, AI algorithms demonstrated potential in a variety of classification systems. SVM achieved 98.33% accuracy in DR staging by analyzing fundus and OCT images [92]. CNNs achieved 88–89% accuracy in classifying DR severity, addressing issues like poor image quality and overfitting [93] (Fig. 4).
Fig. 4.
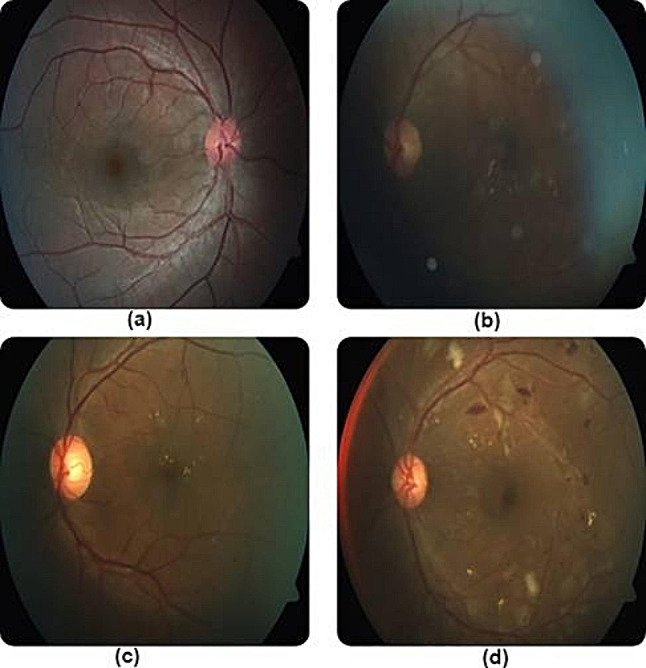
Misclassified images of DR in multiple stages due to poor lighting and contrast. Reprinted with permission from Shaban et al. Shaban M, Ogur Z, Mahmoud A, Switala A, Shalaby A, Abu Khalifeh H, Ghazal M, Fraiwan L, Giridharan G, Sandhu H, El-Baz AS. A convolutional neural network for the screening and staging of diabetic retinopathy. PLoS One. 2020 Jun 22;15 [6]:e0233514 under Creative Commons Attribution License (https://creativecommons.org/licenses/by/4.0/legalcode)
CNN512 and YOLOv3 achieved 89% accuracy in DR staging [11]. AI was also employed in assessing myopia risk, hypertensive retinopathy, and retinopathy of prematurity with high accuracies exceeding 95% in various modules and datasets, demonstrating AI’s potential in grading diverse eye pathologies [94–96].
Prediction
In fundoscopy, prediction is used to predict future occurrences, such as the disease course, which is important for treatment planning and follow-up. The goal of five AI prediction experiments using fundus pictures was to forecast the progression of DR or AMD (Table 8). AI prediction in fundoscopy has demonstrated promise in improving ocular prognostics, despite being a relatively new subject.
Table 8.
Prediction with fundus
| Algorithm | Number of Articles | Results* | Groups |
|---|---|---|---|
| CNN | 3 | AUC: 0.84 | Arcadu et al., Hua et al., Peng et al. |
| Acc: 0.89 | |||
| Sens: 0.97 | |||
| Spec: 0.82 | |||
| ML/Multimodal** | 2 | AUC: | Bhuiyan et al., Govindaiah et al. |
| Acc: 0.81 | |||
| Sens: 0.83 | |||
| Spec: 0.80 |
*Average values across articles **ML algorithm(s) or combined use of ML and DL AUC, area under curve. Acc, accuracy. Sens, sensitivity. Spec, specificity
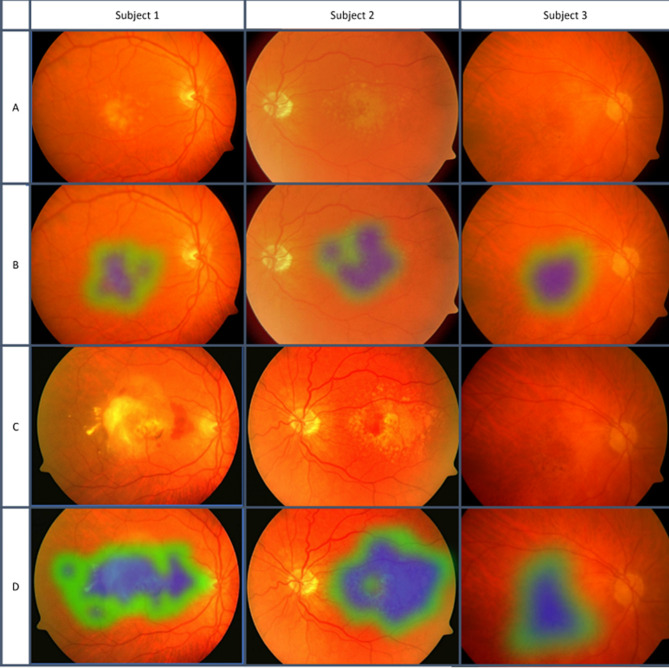
Arcadu et al. and Hua et al. employed DL to predict DR risk, with AUCs of 0.79 and 88.8%, respectively [97, 98]. The first example of AMD development was published in 2020 by Bhuiyan et al., who distinguished between early/none and intermediate/late AMD with 99.2% accuracy [99] (Fig. 5). Their two-year prediction model exhibited an overall accuracy of 86.36% for late-stage AMD progression. While this model performed well in predicting AMD in general, it was not as successful in differentiating between the wet and dry subtypes.
Fig. 5.
Classification of stages of AMD. Blue represents strong signs of AMD, while green represents weaker signs of AMD. Larger areas with more blue resulted in classification into a later stage. Reprinted with permission from Bhuiyan et al. Alauddin Bhuiyan, Tien Yin Wong, Daniel Shu Wei Ting, Arun Govindaiah, Eric H. Souied, R. Theodore Smith; Artificial Intelligence to Stratify Severity of Age-Related Macular Degeneration (AMD) and Predict Risk of Progression to Late AMD. Trans. Vis. Sci. Tech. 2020;9 [2]:25 under Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License (https://creativecommons.org/licenses/by-nc-nd/4.0/legalcode)
Peng et al. and Govindaiah et al. developed AI approaches for accurately predicting late AMD, highlighting AI’s potential to improve prognostics for eye disorders [100, 101].
Discussion
AI is transforming ophthalmology, particularly in forecasting disease progression like AMD and CSCR and treatment outcomes [102–106]. Despite challenges such as inconsistent therapy response, AI can enhance results and save costs. Enhancing OCT picture quality by denoising algorithms and image augmentation aids in diagnosis and therapy planning.
New developments in AI, such vision transformers (ViT), are broadening the range of uses for image processing. Convulsion layers have shown to be a particularly successful way for ViTs to incorporate image patches; in 2021, ViT-G beat previous models on the ImageNet dataset [107]. However, ViT networks pose difficulties for applications such as OCTs, as they demand large datasets and substantial processing resources [108]. Generative adversarial networks (GAN) are another type of deep learning technology that is becoming more and more popular. GAN is capable of image synthesis, superresolution, and picture-to-image translation [109, 110]. Deep convolutional GANs, or DCGANs, train the operators within CNNs, whereas conditional GANs (C-GANs) supply extra data to improve created data representations [111–113]. Zhang et al. highlighted the value of GANs in chorioretinal research by using them to remove retinal shadows and improve choroid area imaging [114].
AI in fundoscopy bridges access barriers to expensive imaging modalities because fundus images anticipate OCT biomarkers and offer angiographic images [115]. Although the creation of AI is made easier by automated machine learning (AutoML), high-quality datasets are still hard to come by. Federated learning and cooperative efforts may be able to solve this problem, increasing the application of AI in healthcare.
While many of AI’s applications in ophthalmology have been image-based to date, the rapid development and acceptance of large language models (LLMs), including, most famously, ChatGPT (OpenAI, San Francisco, USA), heralds an approaching era in which text-based generative algorithms are ubiquitous in clinical and research contexts [116]. LLMs offer the potential to guide clinical decision making for physicians, help patients self-triage and self-diagnose, generate novel research ideas for clinician-scientists, and assist in training the next generation of ophthalmologists, among other powerful benefits [117]. It should be noted that, like many AI applications across medicine and society, LLMs posit important ethical and implementation challenges alongside their potential to optimize clinical decision making and improve patient experiences. These include privacy concerns, especially for models trained using electronic medical record data, false/misleading responses, lack of accessible data to train a model specifically designed for ophthalmologic purposes, and other ethical concerns [118]. In other words, while LLMs will almost certainly find some role in clinical and investigative ophthalmology, and in fact have already seen preliminary studies explored, their use need be evaluated carefully to ensure continued quality and integrity in ophthalmic care [119].
Conclusions
Fundoscopy, despite its age and limitations, remains a valuable tool to image the posterior segment. While many AI applications in fundoscopy are still nascent, it is only a matter of time before these algorithms become commonplace in clinical and research settings. With innovation, acceptance, and understanding from modern day clinicians, our ability to treat and diagnose chorioretinal pathology is set to continue to improve in the golden age of AI.
Acknowledgements
Not applicable.
Author contributions
MD, AS, JO, JC: conceptualization. MD, AS, and JO prepared the first draft of the manuscript, while AY and JC contributed significantly to revision and finalization. All authors read and approved the final manuscript.
Funding
No funding available.
Data availability
Not applicable.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
JC: Allergan, Salutaris, Biogen, Erasca. All else: None.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Abràmoff MD, Garvin MK, Sonka M. Retinal imaging and image analysis. IEEE Rev Biomed Eng. 2010;3:169–208. doi: 10.1109/RBME.2010.2084567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Panwar N, Huang P, Lee J, Keane PA, Chuan TS, Richhariya A, et al. Fundus Photography in the 21st Century–A review of recent Technological advances and their implications for Worldwide Healthcare. Telemed J E Health. 2016;22(3):198–208. doi: 10.1089/tmj.2015.0068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.The Philadelphia photographer [Internet]. Philadelphia: Benerman & Wilson; 1864 [cited 2024 Mar 24]. 794 p. http://archive.org/details/philadelphiaphot18861phil.
- 4.Retinal Atlas. The - ClinicalKey [Internet]. [cited 2024 Mar 24]. https://www-clinicalkey-com.my.wvsom.edu:2443/#!/browse/book/3-s2.0-C20120022399.
- 5.Yannuzzi LA, Ober MD, Slakter JS, Spaide RF, Fisher YL, Flower RW, et al. Ophthalmic fundus imaging: today and beyond. Am J Ophthalmol. 2004;137(3):511–24. doi: 10.1016/j.ajo.2003.12.035. [DOI] [PubMed] [Google Scholar]
- 6.Bernardes R, Serranho P, Lobo C. Digital ocular fundus imaging: a review. Ophthalmologica. 2011;226(4):161–81. doi: 10.1159/000329597. [DOI] [PubMed] [Google Scholar]
- 7.Pham DL, Xu C, Prince JL. Current methods in medical image segmentation. Annu Rev Biomed Eng. 2000;2:315–37. doi: 10.1146/annurev.bioeng.2.1.315. [DOI] [PubMed] [Google Scholar]
- 8.De Fauw J, Ledsam JR, Romera-Paredes B, Nikolov S, Tomasev N, Blackwell S, et al. Clinically applicable deep learning for diagnosis and referral in retinal disease. Nat Med. 2018;24(9):1342–50. doi: 10.1038/s41591-018-0107-6. [DOI] [PubMed] [Google Scholar]
- 9.Jena B, Saxena S, Nayak GK, Saba L, Sharma N, Suri JS. Artificial intelligence-based hybrid deep learning models for image classification: the first narrative review. Comput Biol Med. 2021;137:104803. doi: 10.1016/j.compbiomed.2021.104803. [DOI] [PubMed] [Google Scholar]
- 10.Chou YB, Hsu CH, Chen WS, Chen SJ, Hwang DK, Huang YM, et al. Deep learning and ensemble stacking technique for differentiating polypoidal choroidal vasculopathy from neovascular age-related macular degeneration. Sci Rep. 2021;11(1):7130. doi: 10.1038/s41598-021-86526-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alyoubi WL, Abulkhair MF, Shalash WM. Diabetic Retinopathy Fundus Image classification and lesions localization system using deep learning. Sens (Basel). 2021;21(11). [DOI] [PMC free article] [PubMed]
- 12.Schmidt-Erfurth U, Sadeghipour A, Gerendas BS, Waldstein SM, Bogunović H. Artificial intelligence in retina. Prog Retin Eye Res. 2018;67:1–29. doi: 10.1016/j.preteyeres.2018.07.004. [DOI] [PubMed] [Google Scholar]
- 13.Kawczynski MG, Bengtsson T, Dai J, Hopkins JJ, Gao SS, Willis JR. Development of Deep Learning models to predict best-corrected visual acuity from Optical Coherence Tomography. Translational Vis Sci Technol. 2020;9(2):51. doi: 10.1167/tvst.9.2.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guo S. Fundus image segmentation via hierarchical feature learning. Comput Biol Med. 2021;138:104928. doi: 10.1016/j.compbiomed.2021.104928. [DOI] [PubMed] [Google Scholar]
- 15.Wan C, Chen Y, Li H, Zheng B, Chen N, Yang W, et al. EAD-Net: a Novel Lesion Segmentation Method in Diabetic Retinopathy using neural networks. Dis Markers. 2021;2021:6482665. doi: 10.1155/2021/6482665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khanal A, Estrada R. Dynamic Deep Networks for Retinal Vessel Segmentation. Front Comput Sci [Internet]. 2020 Aug 26 [cited 2024 Mar 24];2. https://www.frontiersin.org/articles/10.3389/fcomp.2020.00035.
- 17.Khan SM, Liu X, Nath S, Korot E, Faes L, Wagner SK, et al. A global review of publicly available datasets for ophthalmological imaging: barriers to access, usability, and generalisability. Lancet Digit Health. 2021;3(1):e51–66. doi: 10.1016/S2589-7500(20)30240-5. [DOI] [PubMed] [Google Scholar]
- 18.Liskowski P, Krawiec K. Segmenting retinal blood vessels with deep neural networks. IEEE Trans Med Imaging. 2016;35(11):2369–80. doi: 10.1109/TMI.2016.2546227. [DOI] [PubMed] [Google Scholar]
- 19.Orlando JI, Prokofyeva E, Blaschko MB. A discriminatively trained fully connected conditional Random Field Model for blood vessel segmentation in Fundus images. IEEE Trans Biomed Eng. 2017;64(1):16–27. doi: 10.1109/TBME.2016.2535311. [DOI] [PubMed] [Google Scholar]
- 20.Rehman A, Harouni M, Karimi M, Saba T, Bahaj SA, Awan MJ. Microscopic retinal blood vessels detection and segmentation using support vector machine and K-nearest neighbors. Microsc Res Tech. 2022;85(5):1899–914. doi: 10.1002/jemt.24051. [DOI] [PubMed] [Google Scholar]
- 21.Yan Z, Yang X, Cheng KT. Joint segment-level and Pixel-wise losses for deep learning based Retinal Vessel Segmentation. IEEE Trans Biomed Eng. 2018;65(9):1912–23. doi: 10.1109/TBME.2018.2828137. [DOI] [PubMed] [Google Scholar]
- 22.Ding J, Zhang Z, Tang J, Guo F. A Multichannel Deep Neural Network for Retina Vessel Segmentation via a Fusion mechanism. Front Bioeng Biotechnol. 2021;9:697915. doi: 10.3389/fbioe.2021.697915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cheng YL, Ma MN, Zhang LJ, Jin CJ, Ma L, Zhou Y. Retinal blood vessel segmentation based on densely connected U-Net. Math Biosci Eng. 2020;17(4):3088–108. doi: 10.3934/mbe.2020175. [DOI] [PubMed] [Google Scholar]
- 24.Bhatia S, Alam S, Shuaib M, Hameed Alhameed M, Jeribi F, Alsuwailem RI. Retinal vessel extraction via assisted Multi-channel Feature Map and U-Net. Front Public Health. 2022;10:858327. doi: 10.3389/fpubh.2022.858327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li K, Qi X, Luo Y, Yao Z, Zhou X, Sun M. Accurate retinal vessel segmentation in Color Fundus images via fully attention-based networks. IEEE J Biomed Health Inf. 2021;25(6):2071–81. doi: 10.1109/JBHI.2020.3028180. [DOI] [PubMed] [Google Scholar]
- 26.Yuan Y, Zhang L, Wang L, Huang H. Multi-level attention network for retinal vessel segmentation. IEEE J Biomed Health Inf. 2022;26(1):312–23. doi: 10.1109/JBHI.2021.3089201. [DOI] [PubMed] [Google Scholar]
- 27.Yang T, Wu T, Li L, Zhu C. SUD-GAN: Deep Convolution Generative Adversarial Network combined with short connection and dense block for retinal vessel segmentation. J Digit Imaging. 2020;33(4):946–57. doi: 10.1007/s10278-020-00339-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hervella ÁS, Rouco J, Novo J, Penedo MG, Ortega M. Deep multi-instance heatmap regression for the detection of retinal vessel crossings and bifurcations in eye fundus images. Comput Methods Programs Biomed. 2020;186:105201. doi: 10.1016/j.cmpb.2019.105201. [DOI] [PubMed] [Google Scholar]
- 29.Guo S, Wang K, Kang H, Zhang Y, Gao Y, Li T. BTS-DSN: deeply supervised neural network with short connections for retinal vessel segmentation. Int J Med Inf. 2019;126:105–13. doi: 10.1016/j.ijmedinf.2019.03.015. [DOI] [PubMed] [Google Scholar]
- 30.Lin Z, Huang J, Chen Y, Zhang X, Zhao W, Li Y, et al. A high resolution representation network with multi-path scale for retinal vessel segmentation. Comput Methods Programs Biomed. 2021;208:106206. doi: 10.1016/j.cmpb.2021.106206. [DOI] [PubMed] [Google Scholar]
- 31.Noh KJ, Park SJ, Lee S. Scale-space approximated convolutional neural networks for retinal vessel segmentation. Comput Methods Programs Biomed. 2019;178:237–46. doi: 10.1016/j.cmpb.2019.06.030. [DOI] [PubMed] [Google Scholar]
- 32.Budak Ü, Cömert Z, Çıbuk M, Şengür A. DCCMED-Net: densely connected and concatenated multi encoder-decoder CNNs for retinal vessel extraction from fundus images. Med Hypotheses. 2020;134:109426. doi: 10.1016/j.mehy.2019.109426. [DOI] [PubMed] [Google Scholar]
- 33.Hua CH, Huynh-The T, Lee S. Retinal vessel segmentation using round-wise features aggregation on Bracket-shaped convolutional neural networks. Annu Int Conf IEEE Eng Med Biol Soc. 2019;2019:36–9. doi: 10.1109/EMBC.2019.8856552. [DOI] [PubMed] [Google Scholar]
- 34.Boudegga H, Elloumi Y, Akil M, Hedi Bedoui M, Kachouri R, Abdallah AB. Fast and efficient retinal blood vessel segmentation method based on deep learning network. Comput Med Imaging Graph. 2021;90:101902. doi: 10.1016/j.compmedimag.2021.101902. [DOI] [PubMed] [Google Scholar]
- 35.Zhu C, Zou B, Zhao R, Cui J, Duan X, Chen Z, et al. Retinal vessel segmentation in colour fundus images using Extreme Learning Machine. Comput Med Imaging Graph. 2017;55:68–77. doi: 10.1016/j.compmedimag.2016.05.004. [DOI] [PubMed] [Google Scholar]
- 36.Feeny AK, Tadarati M, Freund DE, Bressler NM, Burlina P. Automated segmentation of geographic atrophy of the retinal epithelium via random forests in AREDS color fundus images. Comput Biol Med. 2015;65:124–36. doi: 10.1016/j.compbiomed.2015.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Khalid S, Akram MU, Hassan T, Jameel A, Khalil T. Automated segmentation and quantification of Drusen in Fundus and Optical Coherence Tomography images for detection of ARMD. J Digit Imaging. 2018;31(4):464–76. doi: 10.1007/s10278-017-0038-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hassan B, Hassan T, Li B, Ahmed R, Hassan O. Deep ensemble learning based objective grading of Macular Edema by extracting clinically significant findings from fused retinal imaging modalities. Sens (Basel). 2019;19(13). [DOI] [PMC free article] [PubMed]
- 39.Chen Q, Yu WH, Lin S, Liu BS, Wang Y, Wei QJ, et al. Artificial intelligence can assist with diagnosing retinal vein occlusion. Int J Ophthalmol. 2021;14(12):1895–902. doi: 10.18240/ijo.2021.12.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bai H, Gao L, Quan X, Zhang H, Gao S, Kang C, et al. OTNet: a CNN Method based on hierarchical attention maps for Grading arteriosclerosis of Fundus images with small samples. Interdiscip Sci. 2022;14(1):182–95. doi: 10.1007/s12539-021-00479-8. [DOI] [PubMed] [Google Scholar]
- 41.Xu J, Shen J, Wan C, Jiang Q, Yan Z, Yang W. A few-shot learning-based retinal vessel segmentation method for assisting in the Central Serous Chorioretinopathy laser surgery. Front Med (Lausanne) 2022;9:821565. doi: 10.3389/fmed.2022.821565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hart WE, Goldbaum M, Côté B, Kube P, Nelson MR. Automated measurement of retinal vascular tortuosity. Proc AMIA Annu Fall Symp. 1997;459–63. [PMC free article] [PubMed]
- 43.Hart WE, Goldbaum M, Côté B, Kube P, Nelson MR. Measurement and classification of retinal vascular tortuosity. Int J Med Inf. 1999;53(2–3):239–52. doi: 10.1016/S1386-5056(98)00163-4. [DOI] [PubMed] [Google Scholar]
- 44.Hipwell JH, Strachan F, Olson JA, McHardy KC, Sharp PF, Forrester JV. Automated detection of microaneurysms in digital red-free photographs: a diabetic retinopathy screening tool. Diabet Med. 2000;17(8):588–94. doi: 10.1046/j.1464-5491.2000.00338.x. [DOI] [PubMed] [Google Scholar]
- 45.Mookiah MRK, Acharya UR, Koh JEW, Chandran V, Chua CK, Tan JH, et al. Automated diagnosis of Age-related Macular Degeneration using greyscale features from digital fundus images. Comput Biol Med. 2014;53:55–64. doi: 10.1016/j.compbiomed.2014.07.015. [DOI] [PubMed] [Google Scholar]
- 46.Mookiah MRK, Acharya UR, Fujita H, Koh JEW, Tan JH, Noronha K, et al. Local configuration pattern features for age-related macular degeneration characterization and classification. Comput Biol Med. 2015;63:208–18. doi: 10.1016/j.compbiomed.2015.05.019. [DOI] [PubMed] [Google Scholar]
- 47.Burlina P, Joshi N, Pacheco KD, Freund DE, Kong J, Bressler NM. Utility of deep learning methods for referability classification of age-related Macular Degeneration. JAMA Ophthalmol. 2018;136(11):1305–7. doi: 10.1001/jamaophthalmol.2018.3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Govindaiah A, Smith RT, Bhuiyan A. A New and Improved Method for Automated Screening of Age-Related Macular Degeneration using ensemble deep neural networks. Annu Int Conf IEEE Eng Med Biol Soc. 2018;2018:702–5. doi: 10.1109/EMBC.2018.8512379. [DOI] [PubMed] [Google Scholar]
- 49.Chen Q, Peng Y, Keenan T, Dharssi S, Agro NE, Wong WT et al. A multi-task deep learning model for the classification of Age-related Macular Degeneration. AMIA Jt Summits Transl Sci Proc. 2019;2019:505–14. [PMC free article] [PubMed]
- 50.Keel S, Li Z, Scheetz J, Robman L, Phung J, Makeyeva G, et al. Development and validation of a deep-learning algorithm for the detection of neovascular age-related macular degeneration from colour fundus photographs. Clin Exp Ophthalmol. 2019;47(8):1009–18. doi: 10.1111/ceo.13575. [DOI] [PubMed] [Google Scholar]
- 51.Matsuba S, Tabuchi H, Ohsugi H, Enno H, Ishitobi N, Masumoto H, et al. Accuracy of ultra-wide-field fundus ophthalmoscopy-assisted deep learning, a machine-learning technology, for detecting age-related macular degeneration. Int Ophthalmol. 2019;39(6):1269–75. doi: 10.1007/s10792-018-0940-0. [DOI] [PubMed] [Google Scholar]
- 52.Chen Q, Keenan TDL, Allot A, Peng Y, Agrón E, Domalpally A, et al. Multimodal, Multitask, multiattention (M3) deep learning detection of reticular pseudodrusen: toward automated and accessible classification of age-related macular degeneration. J Am Med Inf Assoc. 2021;28(6):1135–48. doi: 10.1093/jamia/ocaa302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Abràmoff MD, Lou Y, Erginay A, Clarida W, Amelon R, Folk JC, et al. Improved Automated Detection of Diabetic Retinopathy on a publicly available dataset through integration of Deep Learning. Invest Ophthalmol Vis Sci. 2016;57(13):5200–6. doi: 10.1167/iovs.16-19964. [DOI] [PubMed] [Google Scholar]
- 54.Gayathri S, Gopi VP, Palanisamy P. Automated classification of diabetic retinopathy through reliable feature selection. Phys Eng Sci Med. 2020;43(3):927–45. doi: 10.1007/s13246-020-00890-3. [DOI] [PubMed] [Google Scholar]
- 55.Gargeya R, Leng T. Automated identification of Diabetic Retinopathy using deep learning. Ophthalmology. 2017;124(7):962–9. doi: 10.1016/j.ophtha.2017.02.008. [DOI] [PubMed] [Google Scholar]
- 56.Ting DSW, Cheung CYL, Lim G, Tan GSW, Quang ND, Gan A, et al. Development and validation of a Deep Learning System for Diabetic Retinopathy and Related Eye diseases using retinal images from multiethnic populations with diabetes. JAMA. 2017;318(22):2211–23. doi: 10.1001/jama.2017.18152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang X, Li F, Li D, Wei Q, Han X, Zhang B, et al. Automated detection of severe diabetic retinopathy using deep learning method. Graefes Arch Clin Exp Ophthalmol. 2022;260(3):849–56. doi: 10.1007/s00417-021-05402-x. [DOI] [PubMed] [Google Scholar]
- 58.Gulshan V, Peng L, Coram M, Stumpe MC, Wu D, Narayanaswamy A, et al. Development and validation of a deep learning algorithm for detection of Diabetic Retinopathy in Retinal Fundus photographs. JAMA. 2016;316(22):2402–10. doi: 10.1001/jama.2016.17216. [DOI] [PubMed] [Google Scholar]
- 59.Cao W, Czarnek N, Shan J, Li L. Microaneurysm Detection Using Principal Component Analysis and machine learning methods. IEEE Trans Nanobiosci. 2018;17(3):191–8. doi: 10.1109/TNB.2018.2840084. [DOI] [PubMed] [Google Scholar]
- 60.Yu S, Xiao D, Kanagasingam Y. Machine learning based automatic neovascularization detection on Optic Disc Region. IEEE J Biomed Health Inf. 2018;22(3):886–94. doi: 10.1109/JBHI.2017.2710201. [DOI] [PubMed] [Google Scholar]
- 61.Tang MCS, Teoh SS, Ibrahim H, Embong Z. Neovascularization Detection and Localization in Fundus Images Using Deep Learning. Sensors [Internet]. 2021;21(16). https://www.mdpi.com/1424-8220/21/16/5327. [DOI] [PMC free article] [PubMed]
- 62.Sahlsten J, Jaskari J, Kivinen J, Turunen L, Jaanio E, Hietala K, et al. Deep Learning Fundus Image Analysis for Diabetic Retinopathy and Macular Edema Grading. Sci Rep. 2019;9(1):10750. doi: 10.1038/s41598-019-47181-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Singh RK, Gorantla R, DMENet Diabetic Macular Edema diagnosis using hierarchical ensemble of CNNs. PLoS ONE. 2020;15(2):e0220677. doi: 10.1371/journal.pone.0220677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lu L, Zhou E, Yu W, Chen B, Ren P, Lu Q, et al. Development of deep learning-based detecting systems for pathologic myopia using retinal fundus images. Commun Biology. 2021;4(1):1225. doi: 10.1038/s42003-021-02758-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rauf N, Gilani SO, Waris A. Automatic detection of pathological myopia using machine learning. Sci Rep. 2021;11(1):16570. doi: 10.1038/s41598-021-95205-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Du R, Xie S, Fang Y, Igarashi-Yokoi T, Moriyama M, Ogata S, et al. Deep Learning Approach for Automated Detection of Myopic Maculopathy and pathologic myopia in Fundus images. Ophthalmol Retina. 2021;5(12):1235–44. doi: 10.1016/j.oret.2021.02.006. [DOI] [PubMed] [Google Scholar]
- 67.Zhen Y, Chen H, Zhang X, Meng X, Zhang J, Pu J, ASSESSMENT OF CENTRAL SEROUS CHORIORETINOPATHY DEPICTED ON COLOR FUNDUS PHOTOGRAPHS USING DEEP LEARNING Retina. 2020;40(8):1558–64. doi: 10.1097/IAE.0000000000002621. [DOI] [PubMed] [Google Scholar]
- 68.Brown JM, Campbell JP, Beers A, Chang K, Ostmo S, Chan RVP, et al. Automated diagnosis of plus Disease in Retinopathy of Prematurity using deep convolutional neural networks. JAMA Ophthalmol. 2018;136(7):803–10. doi: 10.1001/jamaophthalmol.2018.1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cai S, Parker F, Urias MG, Goldberg MF, Hager GD, Scott AW. Deep learning detection of Sea Fan Neovascularization from Ultra-widefield Color Fundus photographs of patients with Sickle Cell Hemoglobinopathy. JAMA Ophthalmol. 2021;139(2):206–13. doi: 10.1001/jamaophthalmol.2020.5900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Choi JY, Yoo TK, Seo JG, Kwak J, Um TT, Rim TH. Multi-categorical deep learning neural network to classify retinal images: a pilot study employing small database. PLoS ONE. 2017;12(11):e0187336. doi: 10.1371/journal.pone.0187336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Porwal P, Pachade S, Kokare M, Giancardo L, Mériaudeau F. Retinal image analysis for disease screening through local tetra patterns. Comput Biol Med. 2018;102:200–10. doi: 10.1016/j.compbiomed.2018.09.028. [DOI] [PubMed] [Google Scholar]
- 72.González-Gonzalo C, Sánchez-Gutiérrez V, Hernández-Martínez P, Contreras I, Lechanteur YT, Domanian A, et al. Evaluation of a deep learning system for the joint automated detection of diabetic retinopathy and age-related macular degeneration. Acta Ophthalmol. 2020;98(4):368–77. doi: 10.1111/aos.14306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Koh JEW, Ng EYK, Bhandary SV, Hagiwara Y, Laude A, Acharya UR. Automated retinal health diagnosis using pyramid histogram of visual words and Fisher vector techniques. Comput Biol Med. 2018;92:204–9. doi: 10.1016/j.compbiomed.2017.11.019. [DOI] [PubMed] [Google Scholar]
- 74.Keel S, Wu J, Lee PY, Scheetz J, He M. Visualizing Deep Learning models for the detection of Referable Diabetic retinopathy and Glaucoma. JAMA Ophthalmol. 2019;137(3):288–92. doi: 10.1001/jamaophthalmol.2018.6035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Balasubramanian K, Ananthamoorthy NP. Analysis of hybrid statistical textural and intensity features to discriminate retinal abnormalities through classifiers. Proc Inst Mech Eng H. 2019;233(5):506–14. doi: 10.1177/0954411919835856. [DOI] [PubMed] [Google Scholar]
- 76.Antaki F, Coussa RG, Kahwati G, Hammamji K, Sebag M, Duval R. Accuracy of automated machine learning in classifying retinal pathologies from ultra-widefield pseudocolour fundus images. Br J Ophthalmol. 2023;107(1):90–5. doi: 10.1136/bjophthalmol-2021-319030. [DOI] [PubMed] [Google Scholar]
- 77.Tan TE, Anees A, Chen C, Li S, Xu X, Li Z, et al. Retinal photograph-based deep learning algorithms for myopia and a blockchain platform to facilitate artificial intelligence medical research: a retrospective multicohort study. Lancet Digit Health. 2021;3(5):e317–29. doi: 10.1016/S2589-7500(21)00055-8. [DOI] [PubMed] [Google Scholar]
- 78.Son J, Shin JY, Kim HD, Jung KH, Park KH, Park SJ. Development and Validation of Deep Learning models for Screening multiple abnormal findings in Retinal Fundus images. Ophthalmology. 2020;127(1):85–94. doi: 10.1016/j.ophtha.2019.05.029. [DOI] [PubMed] [Google Scholar]
- 79.Classification Criteria for Birdshot Chorioretinitis Am J Ophthalmol. 2021;228:65–71. doi: 10.1016/j.ajo.2021.03.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Classification Criteria for Multifocal Choroiditis With Panuveitis Am J Ophthalmol. 2021;228:152–8. doi: 10.1016/j.ajo.2021.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Classification Criteria for Punctate Inner Choroiditis Am J Ophthalmol. 2021;228:275–80. doi: 10.1016/j.ajo.2021.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Classification Criteria for Serpiginous Choroiditis Am J Ophthalmol. 2021;228:126–33. doi: 10.1016/j.ajo.2021.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Classification Criteria for Vogt-Koyanagi-Harada Disease Am J Ophthalmol. 2021;228:205–11. doi: 10.1016/j.ajo.2021.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cen LP, Ji J, Lin JW, Ju ST, Lin HJ, Li TP, et al. Automatic detection of 39 fundus diseases and conditions in retinal photographs using deep neural networks. Nat Commun. 2021;12(1):4828. doi: 10.1038/s41467-021-25138-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Burlina P, Pacheco KD, Joshi N, Freund DE, Bressler NM. Comparing humans and deep learning performance for grading AMD: a study in using universal deep features and transfer learning for automated AMD analysis. Comput Biol Med. 2017;82:80–6. doi: 10.1016/j.compbiomed.2017.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Burlina PM, Joshi N, Pekala M, Pacheco KD, Freund DE, Bressler NM. Automated grading of age-related Macular Degeneration from Color Fundus images using deep convolutional neural networks. JAMA Ophthalmol. 2017;135(11):1170–6. doi: 10.1001/jamaophthalmol.2017.3782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sermanet P, Eigen D, Zhang X, Mathieu M, Fergus R, LeCun Y, OverFeat. Integrated Recognition, Localization and Detection using Convolutional Networks [Internet]. arXiv; 2014 [cited 2024 Mar 24]. http://arxiv.org/abs/1312.6229.
- 88.Grassmann F, Mengelkamp J, Brandl C, Harsch S, Zimmermann ME, Linkohr B, et al. A Deep Learning Algorithm for Prediction of Age-Related Eye Disease Study Severity Scale for Age-Related Macular Degeneration from Color Fundus Photography. Ophthalmology. 2018;125(9):1410–20. doi: 10.1016/j.ophtha.2018.02.037. [DOI] [PubMed] [Google Scholar]
- 89.Peng Y, Dharssi S, Chen Q, Keenan TD, Agrón E, Wong WT, et al. DeepSeeNet: a Deep Learning Model for Automated classification of patient-based Age-related Macular Degeneration Severity from Color Fundus photographs. Ophthalmology. 2019;126(4):565–75. doi: 10.1016/j.ophtha.2018.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Heo TY, Kim KM, Min HK, Gu SM, Kim JH, Yun J et al. Development of a deep-learning-based Artificial Intelligence Tool for Differential diagnosis between dry and neovascular age-related Macular Degeneration. Diagnostics (Basel). 2020;10(5). [DOI] [PMC free article] [PubMed]
- 91.Hosoda Y, Miyake M, Yamashiro K, Ooto S, Takahashi A, Oishi A, et al. Deep phenotype unsupervised machine learning revealed the significance of pachychoroid features in etiology and visual prognosis of age-related macular degeneration. Sci Rep. 2020;10(1):18423. doi: 10.1038/s41598-020-75451-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Murugeswari S, Sukanesh R. Investigations of severity level measurements for diabetic macular oedema using machine learning algorithms. Ir J Med Sci. 2017;186(4):929–38. doi: 10.1007/s11845-017-1598-8. [DOI] [PubMed] [Google Scholar]
- 93.Shaban M, Ogur Z, Mahmoud A, Switala A, Shalaby A, Abu Khalifeh H, et al. A convolutional neural network for the screening and staging of diabetic retinopathy. PLoS ONE. 2020;15(6):e0233514. doi: 10.1371/journal.pone.0233514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wan C, Li H, Cao GF, Jiang Q, Yang WH. An Artificial Intelligent Risk classification method of high myopia based on Fundus images. J Clin Med. 2021;10:19. doi: 10.3390/jcm10194488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Akbar S, Akram MU, Sharif M, Tariq A, Yasin UU. Arteriovenous ratio and papilledema based hybrid decision support system for detection and grading of hypertensive retinopathy. Comput Methods Programs Biomed. 2018;154:123–41. doi: 10.1016/j.cmpb.2017.11.014. [DOI] [PubMed] [Google Scholar]
- 96.Campbell JP, Chiang MF, Chen JS, Moshfeghi DM, Nudleman E, Ruambivoonsuk P, et al. Artificial Intelligence for Retinopathy of Prematurity: validation of a vascular severity scale against International Expert diagnosis. Ophthalmology. 2022;129(7):e69–76. doi: 10.1016/j.ophtha.2022.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Arcadu F, Benmansour F, Maunz A, Willis J, Haskova Z, Prunotto M. Deep learning algorithm predicts diabetic retinopathy progression in individual patients. NPJ Digit Med. 2019;2:92. doi: 10.1038/s41746-019-0172-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hua CH, Huynh-The T, Kim K, Yu SY, Le-Tien T, Park GH, et al. Bimodal learning via trilogy of skip-connection deep networks for diabetic retinopathy risk progression identification. Int J Med Inf. 2019;132:103926. doi: 10.1016/j.ijmedinf.2019.07.005. [DOI] [PubMed] [Google Scholar]
- 99.Bhuiyan A, Wong TY, Ting DSW, Govindaiah A, Souied EH, Smith RT. Artificial Intelligence to stratify severity of age-related Macular Degeneration (AMD) and predict risk of progression to late AMD. Transl Vis Sci Technol. 2020;9(2):25. doi: 10.1167/tvst.9.2.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Peng Y, Keenan TD, Chen Q, Agrón E, Allot A, Wong WT, et al. Predicting risk of late age-related macular degeneration using deep learning. Npj Digit Med. 2020;3(1):111. doi: 10.1038/s41746-020-00317-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Govindaiah A, Baten A, Smith RT, Balasubramanian S, Bhuiyan A. Optimized prediction models from Fundus Imaging and Genetics for late age-related Macular Degeneration. J Pers Med. 2021;11(11). [DOI] [PMC free article] [PubMed]
- 102.Pfau M, Sahu S, Rupnow RA, Romond K, Millet D, Holz FG, et al. Probabilistic forecasting of Anti-VEGF treatment frequency in Neovascular Age-Related Macular Degeneration. Transl Vis Sci Technol. 2021;10(7):30. doi: 10.1167/tvst.10.7.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Arrigo A, Calamuneri A, Aragona E, Bordato A, Grazioli Moretti A, Amato A, et al. Structural OCT parameters Associated with Treatment Response and Macular Neovascularization Onset in Central Serous Chorioretinopathy. Ophthalmol Ther. 2021;10(2):289–98. doi: 10.1007/s40123-021-00336-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Bogunovic H, Waldstein SM, Schlegl T, Langs G, Sadeghipour A, Liu X, et al. Prediction of Anti-VEGF treatment requirements in Neovascular AMD using a machine learning Approach. Invest Ophthalmol Vis Sci. 2017;58(7):3240–8. doi: 10.1167/iovs.16-21053. [DOI] [PubMed] [Google Scholar]
- 105.Feng D, Chen X, Zhou Z, Liu H, Wang Y, Bai L, et al. A preliminary study of Predicting effectiveness of Anti-VEGF injection using OCT images based on deep learning. Annu Int Conf IEEE Eng Med Biol Soc. 2020;2020:5428–31. doi: 10.1109/EMBC44109.2020.9176743. [DOI] [PubMed] [Google Scholar]
- 106.Gallardo M, Munk MR, Kurmann T, De Zanet S, Mosinska A, Karagoz IK, et al. Machine learning can predict Anti-VEGF treatment demand in a treat-and-extend regimen for patients with Neovascular AMD, DME, and RVO Associated Macular Edema. Ophthalmol Retina. 2021;5(7):604–24. doi: 10.1016/j.oret.2021.05.002. [DOI] [PubMed] [Google Scholar]
- 107.Zhai X, Kolesnikov A, Houlsby N, Beyer L. Scaling Vision Transformers [Internet]. arXiv; 2022 [cited 2024 Mar 24]. http://arxiv.org/abs/2106.04560.
- 108.Khan S, Naseer M, Hayat M, Zamir SW, Khan FS, Shah M. Transformers in Vision: a Survey. ACM Comput Surv. 2022;54(10s):1–41. doi: 10.1145/3505244. [DOI] [Google Scholar]
- 109.Goodfellow IJ, Pouget-Abadie J, Mirza M, Xu B, Warde-Farley D, Ozair S et al. Generative Adversarial Networks [Internet]. arXiv; 2014 [cited 2024 Mar 24]. http://arxiv.org/abs/1406.2661.
- 110.Creswell A, White T, Dumoulin V, Arulkumaran K, Sengupta B, Bharath AA. Generative adversarial networks: an overview. IEEE Signal Process Mag. 2018;35(1):53–65. doi: 10.1109/MSP.2017.2765202. [DOI] [Google Scholar]
- 111.Wang K, Gou C, Duan Y, Yilun L, Zheng X, Wang FY. Generative Adversarial Networks: Introduction Outlook. 2017;4:588–98. [Google Scholar]
- 112.Radford A, Metz L, Chintala S. Unsupervised Representation Learning with Deep Convolutional Generative Adversarial Networks [Internet]. arXiv; 2016 [cited 2024 Mar 24]. http://arxiv.org/abs/1511.06434.
- 113.Mirza M, Osindero S. Conditional Generative Adversarial Nets [Internet]. arXiv; 2014 [cited 2024 Mar 24]. http://arxiv.org/abs/1411.1784.
- 114.Zhang H, Yang J, Zhou K, Li F, Hu Y, Zhao Y, et al. Automatic segmentation and visualization of Choroid in OCT with knowledge infused Deep Learning. IEEE J Biomed Health Inf. 2020;24(12):3408–20. doi: 10.1109/JBHI.2020.3023144. [DOI] [PubMed] [Google Scholar]
- 115.Alimanov A, Islam MB. Denoising Diffusion Probabilistic Model for Retinal Image Generation and Segmentation [Internet]. arXiv; 2023 [cited 2024 Mar 24]. http://arxiv.org/abs/2308.08339.
- 116.OpenAI, Achiam J, Adler S, Agarwal S, Ahmad L, Akkaya I et al. GPT-4 Technical Report [Internet]. arXiv; 2024 [cited 2024 Mar 24]. http://arxiv.org/abs/2303.08774.
- 117.Ting DSJ, Tan TF, Ting DSW. ChatGPT in ophthalmology: the dawn of a new era? Eye. 2024;38(1):4–7. doi: 10.1038/s41433-023-02619-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Betzler BK, Chen H, Cheng CY, Lee CS, Ning G, Song SJ, et al. Large language models and their impact in ophthalmology. Lancet Digit Health. 2023;5(12):e917–24. doi: 10.1016/S2589-7500(23)00201-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wei L, Mohammed ISK, Francomacaro S, Munir WM. Evaluating text-based generative artificial intelligence models for patient information regarding cataract surgery. J Cataract Refract Surg. 2024;50(1):95–6. doi: 10.1097/j.jcrs.0000000000001288. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.