Abstract
Soluble di‐iron monooxygenases (SDIMOs) are multi‐component enzymes catalysing the oxidation of various substrates. These enzymes are characterized by high sequence and functional diversity that is still not well understood despite their key role in biotechnological processes including contaminant biodegradation. In this study, we analysed a mutant of Rhodoccocus aetherivorans BCP1 (BCP1‐2.10) characterized by a transposon insertion in the gene smoA encoding the alpha subunit of the plasmid‐located SDIMO SmoABCD. The mutant BCP1‐2.10 showed a reduced capacity to grow on propane, lost the ability to grow on butane, pentane and n‐hexane and was heavily impaired in the capacity to degrade chloroform and trichloroethane. The expression of the additional SDIMO prmABCD in BCP1‐2.10 probably allowed the mutant to partially grow on propane and to degrade it, to some extent, together with the other short‐chain n‐alkanes. The complementation of the mutant, conducted by introducing smoABCD in the genome as a single copy under a constitutive promoter or within a plasmid under a thiostreptone‐inducible promoter, allowed the recovery of the alkanotrophic phenotype as well as the capacity to degrade chlorinated n‐alkanes. The heterologous expression of smoABCD allowed a non‐alkanotrophic Rhodococcus strain to grow on pentane and n‐hexane when the gene cluster was introduced together with the downstream genes encoding alcohol and aldehyde dehydrogenases and a GroEL chaperon. BCP1 smoA gene was shown to belong to the group 6 SDIMOs, which is a rare group of monooxygenases mostly present in Mycobacterium genus and in a few Rhodococcus strains. SmoABCD originally evolved in Mycobacterium and was then acquired by Rhodococcus through horizontal gene transfer events. This work extends the knowledge of the biotechnologically relevant SDIMOs by providing functional and evolutionary insights into a group 6 SDIMO in Rhodococcus and demonstrating its key role in the metabolism of short‐chain alkanes and degradation of chlorinated n‐alkanes.
In this study, we analysed a mutant of Rhodoccocus aetherivorans BCP1 characterized by a transposon insertion in the gene operon smoABCD encoding a soluble di‐iron monooxygenase of the rare SDIMO group 6. The smo mutant showed a reduced capacity to grow on propane, lost the ability to grow on butane, pentane and n‐hexane and was heavily impaired in the capacity to degrade chloroform and trichloroethane. Phylogenetic analyses showed that SmoABCD originally evolved in Mycobacterium and was then acquired by Rhodococcus through horizontal gene transfer events.

INTRODUCTION
Alkanes are widely distributed in the environment as they are the most abundant hydrocarbons in crude oil (with an estimated abundance of 20%–50%). They are also naturally produced by living organisms (plants, algae and bacteria) as waste products, chemo‐attractants or for structural and defence purposes (Cappelletti, Fedi, & Zannoni, 2019). Short‐chain alkanes are important greenhouse gases, referring not only to methane but also to other gaseous alkanes (like ethane and propane) (Farhan Ul Haque et al., 2022). Due to their widespread presence in aquatic and soil ecosystems, bacterial oxidation of alkanes is a quite common process in natural environments and a major process in geochemical terms (van Beilen et al., 2003).
As alkanes are nonpolar and chemically inert, their utilization by microorganisms faces significant challenges concerning the energy required to activate them, their limited solubility, and their tendency to accumulate in cell membranes (Rojo, 2009). The study of alkanotrophic microbes (microbes that can utilize alkanes as growth substrate) and of the mechanisms allowing alkane degradation/metabolism is of great interest as the microbial degradation of n‐alkanes remains the main process for the remediation of oil‐contaminated areas (Brooijmans et al., 2009). Microbial growth on n‐alkanes can also induce co‐metabolic processes that allow the biodegradation of xenobiotics like halogenated hydrocarbons (Cappelletti et al., 2012, 2018; Holmes & Coleman, 2008).
In aerobic bacteria, the ability to utilize n‐alkanes as growth compounds is conferred by monooxygenases (MOs), a group of enzymes that catalyse the addition of one oxygen atom into organic compounds, activating the inert hydrocarbon molecule and forming an alcohol that is further oxidized and introduced in the central metabolism. Alkane hydroxylases involved in alkane metabolism typically belong to the alkane MO families AlkB, CYP153‐type cytochrome P450 and soluble di‐iron monooxygenases (SDIMOs). SDIMOs are multi‐component enzymes with wide applications in biodegradation and biocatalysis processes (Petkevičius et al., 2021). They are known to catalyse the initial oxidation of short‐chain alkanes, alkenes, halogenated alkanes, and aromatics (Coleman et al., 2006). Different SDIMOs are classified into different groups (1–6) based on gene organization, substrate specificity, and sequence similarity (Coleman et al., 2006, 2011; Holmes & Coleman, 2008; Leahy et al., 2003). While most of the knowledge on SDIMOs involved short‐chain n‐alkanes relies on studies of methane monooxygenases, SDIMOs involved in the metabolism of other short chain alkanes (gaseous and liquid n‐alkanes with carbon chain < C7) is less well understood. This catabolic activity is particularly abundant in the actinobacterial group CMNR including Rhodococcus together with the genera Corynebacterium, Nocardia and Mycobacterium.
Among alkanotrophic bacteria, members of the Rhodococcus genus are known to play an important role in the degradation of aliphatic hydrocarbons in natural environments and biotechnological settings like in enrichment bacterial mixed cultures exposed to alkanes as only carbon and energy source (Ciavarelli et al., 2012; Hamamura et al., 2006). Rhodococcus spp. strains are known to be able to transform a wide range of natural and xenobiotic compounds (Cappelletti et al., 2018; Ivshina et al., 2019; Larkin et al., 2005; Martínková et al., 2009; Orro et al., 2015; Presentato et al., 2018) in association with unique metabolic flexibility and high resistance/ tolerance to toxic compounds (Cappelletti et al., 2016; de Carvalho et al., 2014). The presence of large and complex genomes (up to 10.1 Mbp) together with catabolic gene redundancy is often considered the basis of the Rhodococcus catabolic versatility, functional robustness, adaptation to polluted and extreme environments and high‐performing environmental competition (Cappelletti, Zampolli, et al., 2019; Firrincieli et al., 2022). An increasing number of omics studies of microbial communities and alkanotrophic isolates from contaminated sites have extended the knowledge on the diversity of alkane MO in actinomycetes, mostly Mycobacterium and Rhodococcus (Coleman et al., 2006, 2011). On the other hand, the correlation between alkane MOs and xenobiotics co‐metabolism mostly involved only whole‐cell assays to determine biodegradation kinetic rates and enzymatic competition between the primary substrate (the alkane) and the co‐metabolized compound (e.g. halogenated hydrocarbons). Furthermore, only a few studies have applied molecular approaches to get insights into functional and regulatory aspects of the alkane MOs not only in Rhodococcus but also in other strains of CMNR group (Deng et al., 2018; Kotani et al., 2003; Martin et al., 2014; Sharp et al., 2007). This is mostly caused by the difficulties in genetic manipulation of these strains due to the presence of a robust mycolic acid‐containing cell wall, genomic instability, effective endogenous restriction systems that recognize unmethylated sites in exogenous DNA, the lack of universal molecular tools, high genome GC content (generally >60%) and low transformation and recombination efficiencies (Cappelletti, Zampolli, et al., 2019; Donini et al., 2021).
In this work, we analysed a mutant of Rhodoccocus aetherivorans BCP1 characterized by transposon insertion in the gene cluster smoABCD that encodes a monooxygenase belonging to group 6 SDIMOs. These are rare monooxygenases that have been scarcely studied, although they were previously suggested to be involved in co‐metabolic processes of recalcitrant pollutants (Cappelletti et al., 2015; Coleman et al., 2011; He et al., 2018). Here, we conducted growth and biodegradation assays and heterologous expression analyses to demonstrate for the first time the key role of a group 6 SDIMO in the metabolism of short‐chain n‐alkanes and degradation of chlorinated hydrocarbons. Phylogenetic and bioinformatic analyses also provide information on the evolutionary history of this SDIMO group that has been acquired by Rhodococcus from Mycobacterium through horizontal gene transfer (HGT) events.
EXPERIMENTAL PROCEDURES
Bacterial strains and culturing conditions
Rhodococcus aetherivorans BCP1 (DSM 44980) was initially isolated from a butane‐utilizing microbial consortium able to co‐metabolically degrade chloroform in batch slurry reactors (Frascari et al., 2006). R. erythropolis MTF was isolated from oil tailing ponds (Golby et al., 2014), while R. erythropolis SQ1 was a derivative strain of the type‐strain ATCC4277 with increased transformability (Dabbs et al., 1990). They were both found not to grow on short‐chain n‐alkanes in preliminary experiments. For the growth and degradation assays, Rhodococcus spp. strains and mutants were firstly pre‐cultured in 250 mL Erlenmeyer Baffled Flasks for 48 h at 30°C with shaking (150 rpm), containing 25 mL of LB medium [composed of (g L−1) NaCl, 10; Yeast Extract, 5; Tryptone, 10] supplied with the suitable antibiotic in the case of the mutants. The pre‐culture was then used to inoculate 50 mL of Mineral Salt Medium (MSM) supplied with the SL6 source of trace elements (as previously described by Presentato et al., 2018), at an initial OD600 of 0.05. Bottle microcosms (120 mL) containing 40 mL of MSM were incubated under shaking at 30°C and sealed with butyl rubber after the addition of alkanes at a final concentration of 0.1% (v/v) (Cappelletti et al., 2011). When necessary, antibiotics were added to select Rhodococcus transformants in the culture medium (thiostrepton at 10 μg/mL, tetracycline at 10 μg/mL and/or apramycin at 70 μg/mL). Bacterial growth was monitored by measuring optical density at 600 nm (OD600).
Escherichia coli DH5α was used as a host strain for the cloning of DNA fragments and the cells were cultivated in LB broth or on LB agar with ampicillin (50 μg/mL) when necessary. The bacterial strains and plasmids used in this work are listed in Table S1.
DNA extraction and manipulation
Total DNA from Rhodococcus aetherivorans BCP1 and the transposon mutants was extracted from cells grown in 50 mL liquid LB cultures incubated at 30°C for 48 h (up to OD600 of 1–1.2). After cell harvesting, BCP1 WT and mutant cells were lysed using the protocol reported by Cappelletti et al. (2011), while standard molecular techniques were used for DNA manipulation and cloning (Sambrook et al., 1989). Shortly, for molecular cloning 1.5–3 μg of DNA were digested with appropriate restriction endonucleases (1 U) (Roche) for 3–4 h at 37°C. In multi‐enzyme digestion, compatible reaction buffers were used. Ligation reactions were performed overnight in a final volume of 15 μL using the T4 ligase (Roche) based on the manufacturer instructions except for the use of a temperature gradient ranging from 37°C to 4°C. Recombinant plasmids were introduced into E. coli DH5α using chemical transformation standard protocol (Sambrook et al., 1989). PCR and plasmid purifications were carried out using the Plasmid Mini Kit and PCR purification kit (Qiagen) according to the manufacturer's instructions. QIAquick Gel Extraction Kit (Qiagen) was used for the recovery and purification of DNA fragments excised from agarose gel.
Generation of a transposon mutant library of Rhodococcus aetherivorans BCP1
Plasmid pTNR‐TA (Sallam et al., 2006) was used to construct the random transposon mutant library of Rhodococcus aetherivorans BCP1. pTNR‐TA is a transposable vector deriving from the non‐replicating transposon tool pTNR that carries the insertion sequence IS1415, a member of IS21 family initially identified in R. erythropolis. pTNR‐TA has a thiostrepton resistance gene (thio R ) as transposable‐marker gene and was introduced into Rhodococcus aetherivorans BCP1 cells through electroporation. Transformants were selected on the LB agar plates supplied with thiostrepton 5 μg/mL after 5 days of incubation at 30°C. The transformants were then re‐streaked on LB agar plates with thiostrepton 5 μg/mL for transposon insertion confirmation before being tested for their capacity to grow on C6 (0.1% v/v), C16 (0.1% v/v) or glucose (0.5% w/v) that were directly supplied on solid MSM plates as sole carbon and energy sources. Mutants that could not grow on glucose as the sole carbon source were identified as mutants that had transposon insertion in genes required for the growth on a minimal medium supplied with a single carbon and energy source and were omitted from subsequent analyses.
Electroporation of Rhodococcus aetherivorans BCP1
To generate electrocompetent cells, Rhodococcus spp. strains cultures (BCP1, BCP1‐2.10 and MTF) were grown up to an OD600 of 0.6–0.7 in LB medium supplied with glycine 3.5% w/v, sucrose 1.8% w/v, isoniazid 150 μg/mL under shaking (150 rpm) at 30°C (around 18 h). The culture was then supplied with 3 μg of ampicillin and incubated for an additional 1.5 h at 30°C under shaking (150 rpm) before being centrifuged for 10 min at 6000 rpm at 4°C. The cell pellet was then washed once with 25 mL of ice‐cold EPB1 (20 mM Hepes pH 7.2, 5% glycerol) and twice with 10 mL of ice‐cold EPB2 (5 mM Hepes pH 7.2, 15% glycerol) before the final centrifuge at 6000 rpm for 10 min at 4°C. After discarding the supernatant, cells were suspended in 1–2 mL EPB2 and then aliquots of 200 μL were transferred into cold 1.5 mL tubes to be mixed with 1 μg of foreign DNA. After 5 min of incubation on ice, cell preparations were transferred in cold 0.2 cm‐cuvettes (Biorad) for electroporation. The electroporation was performed with Eporator (Eppendorf) using the setting 2.5 kV, 25 μF, and 400 Ω. Pulsed cells were immediately supplied with 1 mL of LB and transferred into a new tube. Cells were regenerated for 5–6 h with shaking at 150 rpm at 30°C, before being plated onto LB agar plates containing the appropriate antibiotic.
Southern blotting
To confirm the single insertion of the transposon in the genomes of the mutant library, representative mutants were subjected to DNA extraction following the procedure previously described (Cappelletti et al., 2011). 5 μg of DNA was then subjected to enzymatic restriction with the enzyme PstI and ran on electrophoresis gel. The chromosomal DNAs from the agarose gels were then transferred to nylon membrane filters and Southern hybridization was performed according to the previously described protocol (Sambrook et al., 1989). The probe was made through PCR amplification using pTNR‐TA as template together with the primers TioR‐For (5′‐GGATCCGCCAGAGAGCGACGAC‐3′) and TioR‐Rev (5′‐CGCCTTCGAGGAGTGCCCG‐3′) to amplify part of the thiostrepton resistance gene that is comprised within the transposable region of the integrative vector. After PCR purification through Qiagen PCR cleaning kit, the DNA fragment was labelled with the DIG High Prime DNA labelling kit (Roche). Prehybridization and hybridization were carried out with DIG Easy Hyb (Roche) at 37°C overnight in a rotary oven. Washes were performed first in a 2× SSC (0.3 M NaCl plus 0.03 M sodium citrate)‐equivalent buffer for 5 min at room temperature, then in a 0.5x SSC‐equivalent buffer for 15 min at 65°C. The digoxigenin‐labelled probe was detected using CSPD (the alkaline phosphatase substrate) (Roche) as a chemiluminescent substrate according to instructions provided by the manufacturer.
Inverse PCR (iPCR)
To identify the genes interrupted by the transposon insertion, total DNA was extracted and 1–2 μg samples were digested with BglII or PstI, purified with phenol/chloroform method and then EtOH precipitated. Digested DNA was then pooled in 15 μL milliQ H2O and then self‐ligated in a 30 μL reaction mixture at room temperature for 2 h. iPCR amplification was carried out using 2 μL of ligation mixture as the template, the TioseqTn1 (5′‐CAGTCATGGTCGTCCTACCG‐3′) and TioseqTn2 (5′‐CGAGGTATGTAGGCGGTGCT‐3′) as the primers and the BIOTAQ™ DNA polymerase (Bioline) using the manufacturer protocol. The PCR program was the following: initial denaturation at 95°C for 3 min; followed by 30 cycles of denaturation at 95°C for 50 s, annealing at 59°C for 50 s and extension at 72°C for 3 min, with a final extension at 72°C for 15 min. The PCR products were sequenced with the primer TioseqTn1.
Plasmid construction for mutant complementation and heterologous expression
To carry out the complementation assay of the BCP1‐2.10 mutant strain, the mutant cells were transformed with two different constructs (visually displayed in Figure S1): (i) the self‐replicating pTipQT1‐smoABCD based on the shuttle vector pTipQT1 (Nakashima & Tamura, 2004a) and (ii) the transposable non‐replicating vector pTNR‐AA/pNitsmoABCD based on a modified version of the transposable tool pTNR‐TA (Sallam et al., 2007) (Table S1). To construct the plasmid pTipQT1‐smoABCD, the gene cluster smoABCD was PCR amplified from the BCP1 genome using the primers smoA‐NdeI‐For (5′‐TCACATATGACTACATCGGTCACAACTCAGCA‐3′) and smoD‐HindIII‐Rev (5′‐TGCAAGCTTTCACGCTGGGAGCTGGGCGGTTT‐3′) and the Taq polymerase Ex Taq (Takara) following manufacturer's instructions. After PCR clean‐up and restriction digestion with NdeI and HindIII, the smoABCD gene cluster was cloned in pTipQT1 within the compatible enzyme restriction sites NcoI‐HindIII. The pTipQT1 shuttle vector (Nakashima & Tamura, 2004a) contains a thiostrepton‐inducible promoter (PtipA), as well as the thiostrepton (thio R ) and tetracycline (tet R ) resistance genes for the selection in Rhodococcus and the ampicillin (amp R ) resistance cassette for the selection in E. coli. To construct the plasmid pTNR‐AA/pNitsmoABCD, the thior cassette in pTN‐TA was replaced with the apramycin (apra R ) resistance gene. The apra R gene was first PCR amplified from pIJ8600 (Takano et al., 1995) using the primers Apra‐BamHI‐For (5′‐CTCAGGATCCTCTGACGCTCAGTGGAAC‐3′) and Apra‐HindIII‐Rev (5′‐TCACAAGCTTACGTCGCGGTGAGTTCAG‐3′) and then cloned in pTNR‐TA within the restriction enzyme sites BamHI and HindIII (replacing thio R ). The gene cluster smoABCD amplified with the primers smoA‐NdeI‐For and smoD‐HindIII‐Rev was inserted within the restriction enzyme sites NcoI‐ HindIII in the plasmid pNitQT1 (Nakashima & Tamura, 2004b) before being amplified with the primers pNit‐StuI‐For (5′‐ACTCATATGTCACGCTGGGAGCTGGGCGGTT‐3′) and smoD‐HindIII‐Rev to obtain the pNit‐smoABCD fragment corresponding to the smoABCD operon under the control of the constitutive Pnit promoter. The PCR product was then inserted into pTNR‐AA within the StuI‐HindIII restriction sites, to finally obtain pTNR‐AA/pNitsmoABCD, whose genomic insertional event inserts pNitsmoABCD together with apra R .
For complementation experiments, R. erythropolis MTF was transformed using pTipQT1‐smoABCD and an additional construct including the entire gene cluster smoABCD/aldDH/alcDH/groEL (smoABCD together with the alcohol DH, aldehyde DH and GroEL chaperonin) (Table S1). To generate the construct for heterologous expression, the gene cluster smoABCD/aldDH/alcDH/groEL was amplified from the BCP1 genome by using the primers smoA‐NdeI‐For (5′‐TCACATATGACTACATCGGTCACAACTCAGCA‐3′) and groEL‐SpeI‐Rev (5′‐ATCACTAGTATCAGAGGGGAGGCATTTTCGA‐3′) using the Taq polymerase Ex Taq (Takara) following manufacturer's instructions. After PCR purification and enzymatic restriction digestion, the smoABCD/aldDH/alcDH/groEL gene cluster was cloned in the vector pTipQT1 within the enzymatic restriction sites NdeI‐SpeI to construct the vector pTipQT1‐smoABCD/aldDH/alcDH/groEL.
The constructs containing smoABCD and smoABCD/aldDH/alcDH/groEL were first obtained in E. coli DH5α and then the constructs and the empty vectors were transformed in Rhodococcus spp. cells by electroporation. Rhodococcus cells transformed with pTipQT plasmids were selected on LB agar plates supplied with tetracycline at a final concentration of 10 μg/mL.
Alkane and chlorinated alkane degradation assays
For the biodegradation assays, resting cell experiments were performed by growing first the bacterial cells on an inducer alkane and then testing the biodegradation activity in phosphate buffer. In particular, after the pre‐inoculum in LB and the cell washing with phosphate buffer (NaH2PO4 7.72 g/L, Na2HPO4 20.44 g/L, pH 7.2), an aliquot of cell suspension was used to inoculate 20 mL liquid MSM in 119 mL bottles that were then closed with rubber caps and sealed with metal rings, before being supplied with propane (the only alkane that allows the growth of the BCP1‐2.10 strain) as sole carbon and energy source. These cultures were then incubated for 168 h at 30°C, while shaking at 150 rpm. At the end of bacterial growth, cells were pelleted and washed once with 10 mL phosphate buffer. The biomass pellets were then resuspended in phosphate buffer to obtain an OD600 of 0.2–0.3. Before biomass addition, 12.3‐mL vials were pre‐filled with 1 mL of sterile phosphate buffer together with each substrate under analysis, then closed with Teflon caps (to limit the abiotic loss of chlorinated alkanes), sealed with metal rings and finally incubated for 1 h at 30°C for gas–liquid phase equilibrium. In each bottle, the following substrates were added separately using sterile syringes at the following concentration: propane 70 μM, butane 55 μM, pentane 12 μM, hexane 7 μM, chloroform (CF) 2 μM and 10 μM, 1,1,2‐trichloroethane (1,1,2‐TCA) 10 μM and 50 μM. After the first gas chromatography analysis (T0), the biomass aliquots (500 μL) were inoculated in the bottles, which were then incubated at 30°C and 150 rpm and monitored at different time points from the initial GC measurement (2.5, 5, 20 and 51 h). Biodegradation of n‐alkanes and chlorinated alkanes was assessed through gas chromatography by following the procedures described by Frascari et al. (2006). Specific biodegradation rates were calculated by considering the mass of compound degraded normalized over the amount of cellular biomass measured as mg of proteins. For protein quantification, 200 μL of the same cell suspension used to inoculate the 12.3 mL vials was analysed through Lowry assay using bovine serum albumin (BSA) as standard (Lowry et al., 1951).
Phylogenetic analysis to identify the SDIMO family of SmoABCD
Phylogenetic analysis of Mycobacteriaceae sDIMO was performed as follows. Non‐redundant protein accession of the Methane/Phenol/Toluene Hydroxylase (MPTh) protein family belonging to Actinobacteriota was retrieved from the NCBI Protein database at the following link: https://www.ncbi.nlm.nih.gov/protein/?term=%28%232%29+AND+%22actinobacteria%22%5Bporgn%3A__txid201174%5D+. The protein accessions (WP_*) were then used to retrieve all the unique SDIMO protein sequences that were associated to Actinobacteriota phylum from NCBI Identical Protein Group (IPG) database. The Actinobaceriota Assembly accessions obtained from the IPG database were finally filtered to keep only genomes and Methane/Phenol/Toluene Hydroxylase protein affiliated to the Mycobacteriaceae family according to the Genome Taxonomy Database v214. The identification and classification of the MPTh alpha subunits into group 1, group 2, group 3, group 3‐like, group 4, group 5 and group 6 were then carried out via phylogenetic analysis using as reference database the alpha‐subunit data set described in Zou et al. (2021). Briefly, the Zhou alpha‐subunit data set and Mycobacteriaceae MPTh were aligned between each other with MAFFT (Katoh et al., 2002) in auto mode (−‐auto) and the resulting alignment file was trimmed with TrimAl (Capella‐Gutiérrez et al., 2009) in automated mode (−automated1). The trimmed file was used to construct an unrooted maximum likelihood phylogenetic tree in FastTree (Price et al., 2009) with default parameters. The resulting phylogenetic tree was visually inspected to identify and classify the MPTh alpha subunits. Finally, a definitive maximum likelihood phylogenetic tree representing the Mycobacteriaceae MPTh alpha subunits was constructed in IQ‐TREE v.2.2.2.6 (Nguyen et al., 2015) using the Q.yeast+F + I + G4 substitution model.
Gene tree reconciliation analysis
GeneRax (Morel et al., 2020) was used to identify horizontal gene transfer events involving the alpha subunits of the group 6 SDIMOs between members of Mycobacteriaceae family. For this purpose, a phylogenetic tree representing the group 6 and group 3 alpha subunits (group 3 was included to generate a rooted binary tree) was reconciled against the GTDBtk‐based species tree of the Mycobcateriaceae family using the UndatedDTL probabilistic model for computing the reconciliation and the SPR tree search mode. The resulting transfer events from the Mycobacterium donor strain and within members of the Rhodococcus genus were finally visualized in iToL (https://itol.embl.de/).
RESULTS
Isolation and genetic characterization of a transposon insertion mutant of R. aetherivorans BCP1 defective in the growth on short‐chain n‐alkanes
A library of random insertional mutants of R. aetherivorans BCP1 was created and screened for the capacity to grow using n‐alkanes as the only carbon and energy source. A non‐auxotrophic mutant, named BCP1 2.10, was selected for the inability to grow on a solid minimal medium supplied with n‐hexane (C6). Conversely, this mutant retained the capacity to grow on n‐hexadecane (C16) and on other standard carbon sources (Figures S2A–C and S3). This initial screening suggested this mutant was deficient in the ability to grow on liquid short‐chain n‐alkanes. To characterize further BCP1 2.10, we extended the range of tested n‐alkanes by inoculating the mutant strain in liquid cultures with propane (C3), butane (C4), pentane (C5) or hexane (C6) as only carbon and energy source (Figure 1A–D). In these growth assays, in addition to the WT strain, the insertional mutant BCP1 2.13 was also utilized as a positive control as it was not compromised in the ability to grow on n‐alkanes and carried thio R gene, thus evaluating the possible influence of antibiotic resistance gene on the bacterial growth. As a result of the growth assays, BCP1 2.10 almost completely lost the capacity to grow on C4, C5 and C6 (Figure 1). On the other hand, 2.10 showed a certain capacity to grow on C3 although the growth performance was significantly lower than the controls (WT and 2.13 mutant strains). Mutant 2.10 maintained the capacity to grow on the alkanes' metabolic intermediates 1‐propanol, 1‐butanol and 1‐hexanol (Table S2), indicating that the mutation affected the first alkane oxidation reaction and not the subsequent oxidation steps leading to the conversion of the alcohol into aldehyde.
FIGURE 1.
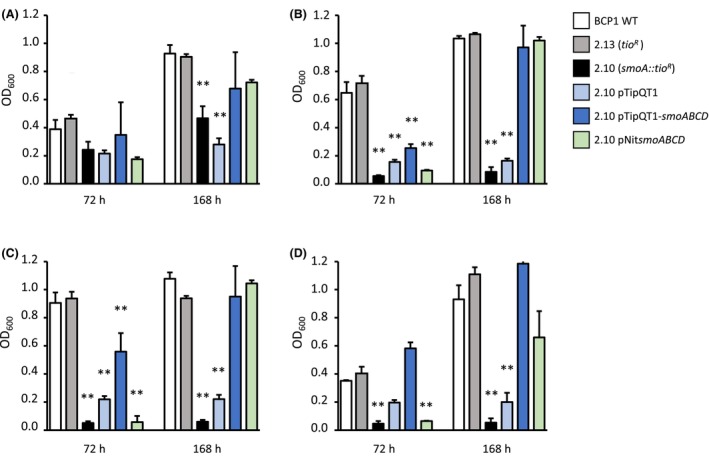
. Growth assays of R. aetherivorans BCP1 on short‐chain n‐alkanes. Growth was measured as OD600 values at two different time points (72 and 168 h) after the inoculation on minimal medium (MSM) supplied with C3 (A), C4 (B), C5 (C) or C6 (D) as only carbon and energy source. Results shown are means ± standard deviation with n = 3. Asterisks indicate statistically different groups respect BCP1 wild type (BCP1 WT) according to one‐way ANOVA analysis. *p < 0.5, **p < 0.01.
The single insertion of pTNR‐TA into the BCP1 genome was confirmed through the Southern blot analysis of several library mutants, including 2.10 and 2.13 (Figure S4). The genetic characterization of the mutant BCP1 2.10 was then performed by inverse PCR. As a result, the strain was found to be mutated at the level of the smoA gene (smoA::tio R ) that is included within the smoABCD operon encoding a soluble di‐iron monooxygenase (SDIMO) (Cappelletti et al., 2015). In particular, the transposon was found to interrupt the smoA gene at the level of the nucleotide 798 from the ATG start codon and two 9 bp direct repeats ‘GACAATTTC’ flanked the transposon insertion, as displayed in Figure 2A. As the transposon insertion did not shift the reading frame, no polar effects are expected on the expression of adjacent genes.
FIGURE 2.
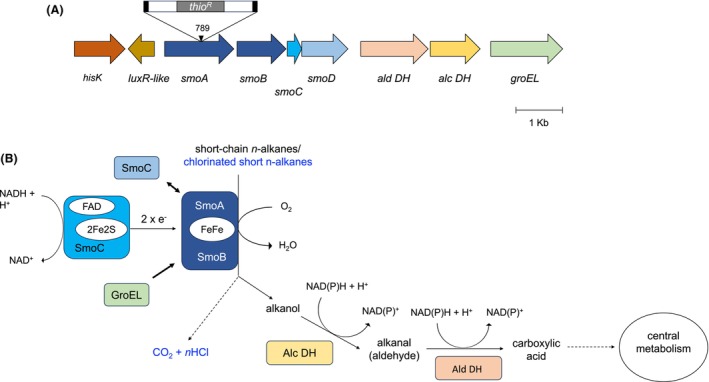
The soluble di‐iron monooxygenase (SDIMO) Smo from Rhodococcus aetherivorans BCP1. (A) Schematic organization of the gene cluster smoABCD encoding the soluble di‐iron monooxygenase (SDIMO) Smo and the flanking regions that are co‐localized on the megaplasmid pBMC2 in R. aetherivorans BCP1 genome. Gene are displayed as arrows. The single genes of the smoABCD operon code for smoA, the monooxygenase alpha subunit, smoB, the monooxygenase beta subunit, smoC the coupling protein, smoD, the reductase. The flanking genes code for ald DH, the aldehyde dehydrogenase (DH), alc DH the alcohol DH, groEL GroEL the chaperonin, hisK the histidine kinase, luxR‐like the cognate response regulator. The transposable element of pTNR‐TA that interrupt smoA gene in the mutant 2.10 is shown together with the 1070 nt‐long thiostrepton resistance gene (thio R ) and the insertion site (at the nucleotide 789 of smoA). The locus tags of the genes go from N505_0128845 (for hisK) to N505_0128885 (for groEL) B. (B) Cofactor composition of the Smo components and predicted enzymatic reactions catalysed by Smo, Alc DH and Ald DH. The metabolic pathway and reaction intermediates for the alkanes are typed in black, the reaction involved in the degradation of chlorinated alkanes are in blue. Dashed lines correspond to more reactions.
Complementation of the smoA mutant BCP1‐2.10 strain
The complementation of the smoA mutant 2.10 strain was carried out by transforming the mutant with two plasmid systems, separately, (i) the replicative plasmid pTipQT1 harbouring the smoABCD gene cluster under the control of the PtipA thiostrepton‐inducible promoter (BCP1 2.10 pTipQT1‐smoABCD), (ii) the non‐replicating plasmid pTNR‐AA/pNitsmoABCD harbouring the smoABCD operon under the constitutive promoter Pnit (Table S1). While the complementation with the first plasmid is expected to lead to the introduction of 40–60 copies of smoABCD inside the cell that are contemporary expressed at high level because under the inducible promoter (pTip) (Nakashima & Tamura, 2004a), the use of the non‐replicative plasmid is associated with the expression of only one single gene copy that is integrated in the genome under the control of a constitutive promoter (pNit) that has similar expression levels to pTip (Nakashima & Tamura, 2004b). Both the complemented strains were tested for the phenotype rescue by analysing the growth in liquid cultures in the presence of C3, C4, C5 and C6 as only carbon and energy sources. As a result, both 2.10 pTipQT1‐smoABCD and 2.10 pNitsmoABCD strains showed a complete recovery of the growth capacities after 168 h of incubation (Figure 1). In the case of the strain 2.10 pTipQT1‐smoABCD, the phenotype reacquisition was observed even after 72 h of growth, suggesting that the number of smoABCD copies and/or the control by the inducible promoter pTip, positively impacted the growth on n‐alkanes. The control strain harbouring the only plasmid (2.10 pTipQT1) could grow on alkanes slightly better than the mutant 2.10. A possible hypothesis about this behaviour regards the influence that the antibiotic resistance gene tet R (on pTipQT1) might have on global bacterial metabolism (Martínez & Rojo, 2011). All the complemented mutant strains showed the same growth when a standard carbon and energy source like glucose was added (Figure S3), demonstrating the specific role of the Smo protein on alkane metabolism.
Heterologous expression of smoABCD provides a Rhodococcus strain with the ability to grow on short‐chain n‐alkanes
To further investigate the role of SmoABCD in bacterial growth on short‐chain n‐alkanes, the heterologous expression of the monooxygenase gene cluster was performed using the two different Rhodococcus strains R. erytropolis MTF and R. erytropolis SQ1. These bacterial strains are unable to grow on short‐chain n‐alkanes (C3–C6) and were transformed via electroporation with the empty pTipQT1 vector, the pTipQT1 containing the only smo gene cluster (smoABCD), and the pTipQT1 containing the smoABCD together with the genes that in BCP1 genome are immediately downstream the cluster, which are the alcohol and aldehyde dehydrogenase genes and the GroEL chaperone gene. The results of the growth assays on liquid minimal salts medium supplemented with Thio and different short‐chain n‐alkanes, showed that the introduction of pTipQT1‐smoABCD did not allow either of the two strains to grow on short‐chain alkanes (data not shown). Conversely, R. erytropolis MTF (but not R. erytropolis SQ1) transformed with pTipQT1‐smoABCD/aldDH/alcDH/groEL acquired the ability to grow on C5 and C6 (Figure 3; Figure S5). This result demonstrated on one hand that the expression of the SDIMO Smo provides host strain with the ability to grow on some short‐chain n‐alkanes, and on the other that the genes involved in protein folding and in enzymatic reactions downstream of the initial alkane oxidation step are necessary for the alkane metabolism mediated by the SDIMO Smo.
FIGURE 3.
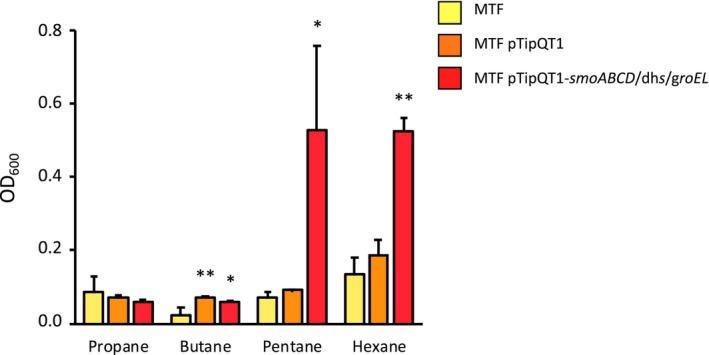
Heterologous expression of smoABCD in R. erythropolis MTF. Growth of MTF wild type (MTF) and MTF transformed with pTipQT1‐smoABCD/dhs/groEL (MTF pTipQT1‐smoABCD/dhs/groEL) or the empty vector (MTF pTipQT1) is shown as OD600 values measured after 72 h of culture in minimal medium (MSM) supplied with C3, C4, C5 and C6 (0.1% v/v) as only carbon and energy source. Results shown are means ± SD. Statistically significant differences (using one‐way ANOVA analysis) respect to the MTF wild type are shown with asterisks *p < 0.05, **p < 0.01.
Role of the Smo in the degradation of short‐chain alkanes and chlorinated aliphatic alkanes (ChlA) in R. aetherivorans BCP1
In order to define the role of monooxygenase Smo in the degradation of alkanes and in the co‐metabolism of chlorinated alkanes in BCP1, the mutant strain 2.10, the complemented strain 2.10 pNitSmoABCD and the WT strain were grown on propane and then exposed to short chain alkanes and chlorinated alkanes under resting cell conditions. Propane was used since it is the only short‐chain alkane that allowed the growth of the 2.10 mutant (although at a lower rate, Figure 1). This was needed to develop cell biomass of the 2.10 mutant with the same alkane‐induced enzymatic asset (needed for chlorinated hydrocarbon co‐metabolism) of the BCP1 WT strain except for a functional SDIMO SmoABCD. The cell growth of the 2.10 mutant could be attributed to the presence of the SDIMO PrmABCD that is transcriptionally induced at high levels by propane (Cappelletti et al., 2015). The activity of SmoABCD could be therefore provided by the difference observed between the WT strain (expressing both the SDIMOs SmoABCD and PrmABCD) and the 2.10‐BCP1 strain (expressing the only SDIMO PrmABCD) in terms of biodegradation performance. The biodegradation assay results are reported in Figure 4A,B which displays the specific degradation rates for all the tested compounds considering the concentration of the substrate degraded over time normalized over the biomass amount, to avoid interferences due to lower 2.10‐BCP1 biomass growth. As a result, the mutant strain 2.10 showed impaired capacity to degrade C3, C4, C5 and C6 as compared to the WT strain. Nevertheless, 2.10 did not completely lose the capacity to degrade any of these substrates, probably due to the presence of an intact Prm monooxygenase in the 2.10 mutant that can metabolize, to some extent, the tested substrates. The analysis of the complemented mutant strain, 2.10 pNitSmoABCD, showed that the expression of the Smo monooxygenase allowed recovery of the phenotype of growth on all the short alkanes even showing improved performance in the degradation of C3 (Figure 1). These results demonstrate the role of Smo monooxygenase in the degradation performance of BCP1 towards all the short alkanes under analysis.
FIGURE 4.
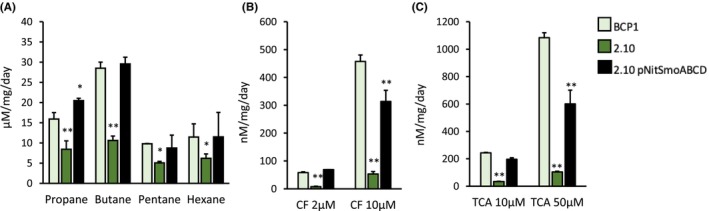
Role of the monooxygenase Smo in the degradation of short‐chain n‐alkanes and chlorinated alkanes in Rhodococcus aetherivorans BCP1. The capability of the wild‐type strain BCP1 (BCP1), the mutant 2.10 strain (2.10) and the complemented mutant strain (2.10 pNitsmoABCD) to degrade short chain alkanes (each supplied in MSM at 0.1% v/v) (A) and chlorinated n‐alkanes (chloroform, CF, and 1,1,2‐trichloroethane, TCA, each supplied at the concentrations 2 and 10 μM) (B, C) are shown in terms of specific degradation rates (i.e. concentration of the compound degraded per mg of cellular proteins per day). Asterisks indicate statistical significance (one‐way ANOVA analysis compared to BCP1). *p < 0.05, **p < 0.01.
The analysis of the degradation of chlorinated alkanes showed that the absence of the Smo monooxygenase in mutant 2.10 strongly impaired the degradation rate of both chloroform (CF) and 1,1,2‐trichloroethane (TCA) (Figure 4B). The complementation of the mutant with a functional Smo monooxygenase allowed for a complete recovery of the degradation performance when the chlorinated alkanes were added at the lowest concentration under analysis. The complemented mutant showed an improvement but not a complete regain of the function when the chlorinated alkanes were tested at higher (10 μM) concentration. These results demonstrate a major role of Smo monooxygenase in the co‐metabolism of CF and TCA and a possible influence of the expression apparatus/regulatory mechanisms when high concentrations of the chlorinated alkanes are tested. Indeed, the smoABCD cluster is under the control of an inducible promoter in the WT strain, while the complemented strain has the Smo function under the control of the constitutive promoter pNit. The possible difference in Smo monooxygenase production in the two strains might explain the incapacity of the complemented strain to degrade the chlorinated alkanes with the same degradation rate as the WT one.
Phylogenetic analysis of the alpha subunit of the SDIMOs from R. aetherivorans BCP1
Initial orthologous analyses of the BCP1 SmoA protein carried out through BLAST search in the NCBI non‐redundant protein database, showed that this gene has significant similarity (>70%) only with alpha subunits of SDIMOs from strains of Rhodococcus and Mycobacterium genera of Mycobacteraceae family (Table S3). We therefore conducted a phylogenetic analysis including all the SDIMO alpha subunits carried by bacterial strains of the Mycobacteriaceae family strains present in the database to define the SDIMO family classification of BCP1 SmoA and to get insights into the evolutionary history of this biotechnologically important group of enzymes (Figures 5 and S6; Table S4). The phylogenetic tree showed the clustering of SDIMOs in 7 main groups, that is, group 1, group 2, group 2‐like, group 3‐like, group 4, group 5 and group 6. The clustering of the SDIMOs shows that the group 6 falls in a monophyletic clade together with the alpha subunits of SDIMOs group 3‐like and share a common ancestor with SDIMOs of group 1, group 2‐like and group 4. Conversely, SDIMOs of group 5 (including the BCP1 PrmA protein) belong to a separate clade that is composed of alpha subunits retrieved from different genera demonstrating a more widespread distribution among actinobacterial lineages despite the high sequence similarity (Figure S6).
FIGURE 5.
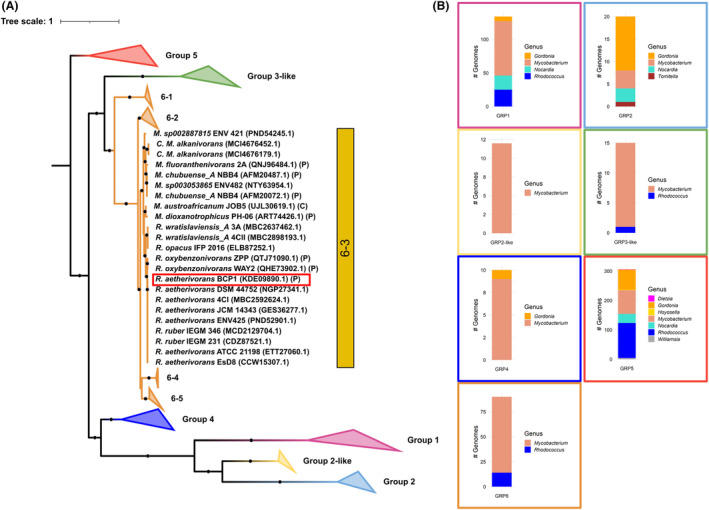
Maximum likelihood phylogenetic tree and distribution of the SDIMO groups (from 1 to 6) of Mycobacteriaceae family with an enlargement on the SDIMO group 6 sub‐clusters (from 6‐1 to 6‐5). (A) Phylogenetic distances in the phylogenetic tree were calculated using the Q.yeast+F + I + G4 substitution model. Ultrafast bootstrap support values >80 (1000 bootstrap replicates) are indicated as black dots. R. aetherivorans BCP1 SmoA is framed by a red box. Besides each strain name, NCBI protein accession is indicated between brackets together with genomic location, if known, corresponding to P, for plasmid or C, for chromosome. (B) The distribution of each SDIMO group among different Mycobacteriaceae genera is shown by framing each bar plot in boxes that are coloured in the same way as the phylogenetic tree.
The phylogenetic analysis clustered the BCP1 PrmA protein within the SDIMOs group 5, and the BCP1 SmoA protein within the SDIMO group 6 together with other smoA from several Mycobacterium spp. strains and a few Rhodococcus spp. strains (Figures 5 and S6). The SDIMOs group 6 was shown to be in turn sub‐grouped into 5 clusters (bootstrap support >80%) with the BCP1 SmoA included in the only sub‐cluster (i.e. sub‐group 6–3) comprising other alpha subunits from several Rhodococcus species (R. aetherivorans, R. ruber, R. oxybenzonivorans, R. wratislaviensis and R. opacus) together with the Mycobacterium species M. alkanivorans, M. dioxanotrophicus, M. sp002887815, M. chubuense, M. fluoroanthenivorans, M. sphagni, M. austroafricanum. All the other 4 sub‐clusters of the SDIMO group 6 included only Mycobacterium species (sub‐groups 6–1, 6–2, 6–4 and 6–5 in Figures 5 and S6; Table S4). Interestingly, the group 6 SDIMO alpha subunits SmoA detected in Mycobacterium complete genomes showed both chromosomal and plasmidic location, while in Rhodococcus, complete genomes smoA genes were exclusively located in plasmids (Figure 5; Table S4). On the other hand, all prmA genes (group 5 SDIMOs), including the one from BCP1, had chromosomal localization. Gene cluster analysis of the group 6 Rhodococcus‐Mycobacterium sub‐cluster 3 revealed complete conservation of the smoABCD coding genes, while the regulatory genes hisK and luxR‐like, and the accessory genes groEL, alc DH (encoding an NDMA‐dependent alcohol dehydrogenase) and ald DH (encoding an aldehyde dehydrogenase) were only partially conserved because missing in some of Mycobacterium strains carrying the sub‐cluster 6–3 smoA genes (Figure S7).
A gene tree/species tree reconciliation analysis was then performed to elucidate the phylogenetic history of the smoA genes in BCP1 and, more in general, in the Rhodococcus genus. From the analysis of the transfer events is clear that M. dioxanotrophicus is the donor bacterial species of the group 6 SDIMOs in the Rhodococcus genus with the recipient represented by the R. aetherivorans lineage. Later, R. aetherivorans was the donor species of the group 6 SDIMOs in strains of the R. opacus (IFP 2016), R. ruber (IEGM 231 and 346), R. oxybenzonivorans (WAY2 and ZPP), R. wratilasviensis_A (4CII and 3A) lineages (Figure 6). This reconciliation analysis together with the plasmidic localization of smoA in Rhodococcus strains suggests that group 6 SDIMO in Rhodococcus appeared for the first time in the R. aetherivorans lineage through a horizontal gene transfer involving M. dioxanotrophicus as the donor bacterial species.
FIGURE 6.
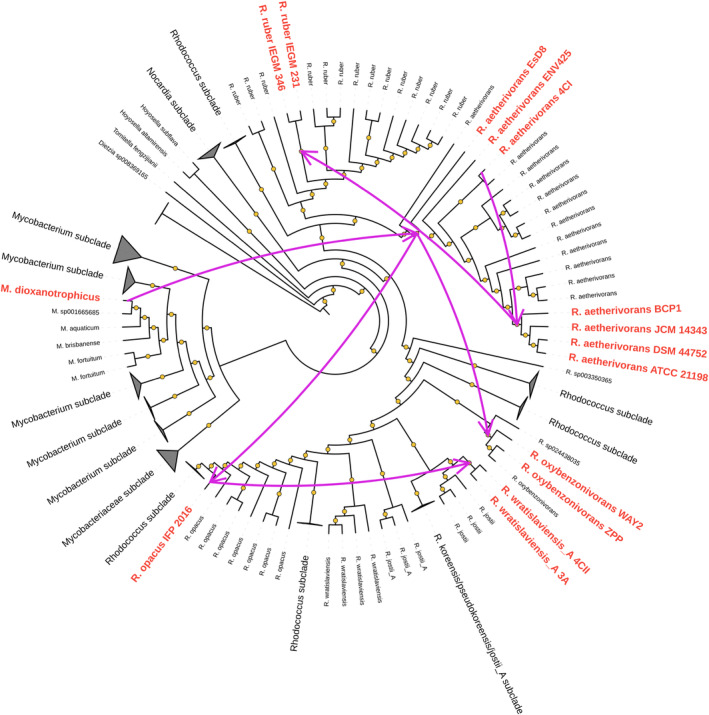
Predicted GeneRax horizontal gene transfer (HGT) events of group 6 SDIMOs between members of Rhodococcus and Mycobacterium (only these bacterial genera possess group 6 SDIMOs). HGT events occurring between Rhodococcus spp. strains are shown as pink arrows. Phylogenetic distances were calculated using the WAG substitution model. Ultrafast bootstrap support values >80 (1000 bootstrap replicates) are indicated as yellow dots.
DISCUSSION
In this work, we first demonstrate the role of a group 6 soluble di‐iron monooxygenase (SDIMO) in the metabolism and degradation of gaseous and short‐chain liquid n‐alkanes and chlorinated alkanes (chloroform and 1,1,2‐trichloroethane) by analysing a transposon mutant strain of Rhodococcus aetherivorans BCP1. Previously, apart from the methane monooxygenases (group 3 SDIMOs) and soluble butane monooxygenase of Thauera butanovora, the only studies that characterized mutants of SDIMOs associated with n‐alkane metabolism targeted (i) the SDIMO group 5 (prmABCD gene cluster) that was found to be fundamental for the growth on propane by Mycobacterium, Rhodococcus, and Gordonia strains (Kotani et al., 2003; Sharp et al., 2007), (ii) the SDIMO group 3‐like in a Rhodococcus strain that was reported to play a key role in the growth on ethane and propane (Zou et al., 2021). The R. aetherivorans BCP1 mutant (2.10 mutant) that we analysed in this study has a transposon insertion in the gene smoA coding for the large subunit of the hydroxylase component of the SDIMO SmoABCD (group 6 SDIMO). The mutant was found to have a reduced growth on propane, while it almost completely lost the capacity to grow on butane, pentane and n‐hexane. These results are in line with a previous study that reported the over‐expression of the smoABCD gene cluster in BCP1 cells exposed to diverse gaseous and liquid short‐chain n‐alkanes (Cappelletti et al., 2015).
In addition to the phenotypic analyses of the mutant strain, the heterologous expression of smoABCD conducted in a non‐alkanotrophic Rhodococcus host strain provided the capacity to grow on pentane and hexane when the gene cluster was introduced together with the flanking genes encoding an alcohol dehydrogenase (DH), an aldehyde DH and a GroEL chaperonin. This result demonstrates the possibility to transfer the metabolic function associated with this SDIMO in other Rhodococcus strains with biotechnological relevance to create recombinant strains for in situ or ex situ bioremediation applications. Furthermore, it demonstrates the key role of the genes flanking smoABCD. This is in line with the positional conservation that included not only smoABCD but also the downstream genes encoding an aldehyde DH, an alcohol DH and a GroEL chaperon (Figure S7). The two DHs are thought to catalyse the oxidation reactions that convert the alkanol (produced by Smo) into carboxylic acid which enters the central metabolism (Figure 2B). Conversely, the GroEL chaperonin might have a fundamental function in the correct folding of the hydroxylase subunit of SmoABCD (Furuya et al., 2003; McCarl et al., 2018). In line with these results, the same aldehyde DH and the alcohol DH were previously found to be over‐expressed in the proteome of BCP1 cells grown on butane and hexane (Cappelletti et al., 2015). Furthermore, the co‐expression of a chaperonin‐like gene was reported to be necessary for the activity of other SDIMOs expressed in host strains (Furuya et al., 2003; Kotani et al., 2003; Kurth et al., 2008; Stafford et al., 2003).
The recombinant Rhodococus strain carrying smoABCD/aldDH/alcDH/groEL gene cluster could grow using pentane and hexane, but it could not grow on the gaseous n‐alkanes butane and propane. Possible hypotheses concern the absence of additional genetic functions in the host strain that are needed for effective use of gaseous alkanes or their oxidation products as carbon sources (including enzymes involved in the metabolism of the alcohols and aldehydes produced by the first alkane oxidation). Furthermore, heterologous expression of Smo provided n‐alkane growth capacity to one of the two non‐alkanotrophic Rhodococcus strains that were tested in this study (to R. erythropolis MTF but not to R. erythropolis SQ1). Although genome‐based indications are not available for the two strains, it is known that different R. eryhtropolis strains can have wide catabolic and genetic diversity (de Carvalho & da Fonseca, 2005). Therefore, this result might be ascribed to the presence in MTF strain (but not in SQ1) of specific genetic functions possibly associated to alkane uptake, oxidation products metabolism and/or mechanisms supporting resistance to the toxicity of liquid short‐chain n‐alkanes (Cappelletti, Fedi, & Zannoni, 2019; de Carvalho & da Fonseca, 2004).
The mutation of the group 6 SDIMO Smo heavily impaired the biodegradation capacities of R. aetherivorans BCP1 not only towards short‐chain n‐alkanes but also towards the chlorinated hydrocarbons chloroform and trichloroethane. The biodegradation experiments were conducted using resting cells that were grown on propane before being tested in whole‐cell assays. This is probably the reason why the mutation of smoA did not completely erase biodegradation capacities towards the tested n‐alkanes. Indeed, BCP1 cells grown on propane are known to express the other SDIMO gene cluster prmABCD (Cappelletti et al., 2015). After being produced, the SDIMO Prm could be able to recognize and oxidize butane, pentane and n‐hexane although they are not the primary substrates/inducers thanks to a certain level of aspecificity that is known for SDIMO enzymes, allowing the co‐oxidation of structural analogues (Coleman et al., 2006; McDonald et al., 1997; Sluis et al., 2002). On the other hand, group 6 SDIMO seems to have a major role in the degradation of chloroform and trichloroethane as its mutation determined a 90% decrease in biodegradation rate.
Therefore, alkane and chlorinated hydrocarbon metabolism in BCP1 involve the activity of the two SDIMOs Smo and Prm which have partly overlapped alkane substrate specificity and inducer range but different degradation kinetics. A third alkane monooxygenase that belongs to the AlkB family also participates in alkane metabolism in BCP1 by targeting n‐alkanes from C6 to C32 (Cappelletti et al., 2011). This alkane range also partially overlaps with the one associated with smoABCD (C6, C7 and C8 induce both Smo and AlkB), suggesting a specificity of the different monooxygenases that is related to the alkane chain length. The coexistence of multiple alkane hydroxylase genes in Rhodoccocus could be the result of evolution processes aimed at differentiating and widening the catabolic processes of members of this genus in response to the presence of specific classes of hydrocarbons.
The phylogenetic and comparative analyses of the alpha subunit of the smoABCD and prmABCD gene clusters classified the two BCP1 SDIMOs into groups 6 and 5, respectively, which show distinct evolutionary relationships. This can be at least in part associated with their different substrate specificity. Indeed, group 6 SDIMOs shared the common ancestor with the group 3‐like SDIMOs that include two SDIMO gene clusters named smoXYB1C1Z from Mycobacterium chubuensis NB44 and Rhodococcus sp. ZPP (Zou et al., 2021), which has similar substrate specificity to BCP1 SmoABCD (including various gaseous alkanes and chlorinated hydrocarbons) (Coleman et al., 2012; Martin et al., 2014; Zou et al., 2021). On the other hand, group 5 has a phylogenetic history distinct from the other SDIMO groups probably in association with its high catalytic specificity towards propane. Furthermore, the presence of group 6 SDIMOs only in the genera Mycobacterium and Rhodococcus and, in some cases, the plasmidic location suggest a more recent acquisition of the group 6 as compared to group 5 SDIMO (that is distributed among different genera and have only chromosomal location) (Vial & Hommais, 2020). The group 6 SDIMO in Rhodococcus spp. strains was probably acquired through a horizontal gene transfer (HGT) event from the M. dioxanotrophicus lineage to the R. aetherivorans lineage. Rhodococcus spp. strains frequently carry megaplasmids that act as reservoirs of catabolic functions which greatly contribute to the metabolic diversity and versatility of strains of this genus (Larkin et al., 2005).
The genetic, functional and phylogenetic results from the present work can assist in future strategies of bioremediation by providing information on (i) the molecular targets to detect and quantify in microbial communities applied in co‐metabolic processes of chloroform and trichloroethane and (ii) the genetic functions to introduce in possible microbial cell factories designed to be applied in biodegradation processes (through bioaugmentation in situ or in bioreactors) of soils or waters contaminated by short‐chain n‐alkanes and chlorinated hydrocarbons.
CONCLUSIONS
This study demonstrates the role of the rare SDIMO SmoABCD in the growth and biodegradation of gaseous and liquid n‐alkanes and in the biodegradation of chlorinated hydrocarbons in Rhodococcus aetherivorans BCP1. The combination of this functional information together with previous genetic and transcriptional analyses indicates that SmoABCD is one of the different alkane hydroxylase systems which are present in R. aetherivorans BCP1 and have complementary and integrated roles in the metabolism of alkanes and degradation of chlorinated alkanes. Phylogenetic and comparative approaches indicate that SmoABCD belongs to the group 6 SDIMOs. The latter has been recently acquired by R. aetherivorans BCP1 through a horizontal gene transfer (HGT) conferring advantages in terms of alkane degradation kinetics and cellular growth rate. In general, this study provides fundamental information on the functions of a group 6 SDIMO, providing also novel insights into the biological and evolutionary significance of the alkane hydroxylation‐functional redundancy that is at the basis of the metabolic versatility of some biotechnologically relevant bacteria like Rhodococcus bacterial strains.
AUTHOR CONTRIBUTIONS
Eleonora Ferrari: Data curation; formal analysis; investigation; visualization; writing – original draft; writing – review and editing. Giulio Di Benedetto: Data curation; formal analysis; investigation; writing – original draft. Andrea Firrincieli: Software; writing – original draft; writing – review and editing. Alessandro Presentato: Investigation; methodology; writing – review and editing. Dario Frascari: Formal analysis; methodology; resources; writing – review and editing. Martina Cappelletti: Conceptualization; funding acquisition; resources; supervision; visualization; writing – original draft; writing – review and editing.
FUNDING INFORMATION
This research was supported by internal funding from the University of Bologna (RFO). Open access publishing fees were partly covered by a dedicated funding support provided by the Department of Pharmacy and Biotechnology (FaBit) University of Bologna.
CONFLICT OF INTEREST STATEMENT
The authors declare no conflict of interest.
Supporting information
Data S1.
Table S3.
Table S4.
ACKNOWLEDGMENTS
The authors thank Dr. Tomohiro Tamura (Bioproduction Research Institute, National Institute of Advanced Industrial Sciences and Technology) for providing pTipQT and pTNR‐TA vectors and Prof. Nicholas Coleman (The University of Sydney) for providing the strain Rhodococcus erythropolis SQ1.
Ferrari, E. , Di Benedetto, G. , Firrincieli, A. , Presentato, A. , Frascari, D. & Cappelletti, M. (2024) Unravelling the role of the group 6 soluble di‐iron monooxygenase (SDIMO) SmoABCD in alkane metabolism and chlorinated alkane degradation. Microbial Biotechnology, 17, e14453. Available from: 10.1111/1751-7915.14453
DATA AVAILABILITY STATEMENT
Raw data used to generate Figures 6 and S7 and the SDIMOs phylogenetic trees in Figures 5A,B and S6 are available under the https://doi.org/10.6084/m9.figshare.24972594.
Genome sequence and annotation of Rhodococcus aetherivorans BCP1 are available under the NCBI RefSeq assembly GCF_000470885.1.
REFERENCES
- Brooijmans, R.J. , Pastink, M.I. & Siezen, R.J. (2009) Hydrocarbon‐degrading bacteria: the oil‐spill clean‐up crew. Microbial Biotechnology, 2(6), 587–594. Available from: 10.1111/j.1751-7915.2009.00151.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capella‐Gutiérrez, S. , Silla‐Martínez, J.M. & Gabaldón, T. (2009) TrimAl: a tool for automated alignment trimming in large‐scale phylogenetic analyses. Bioinformatics, 25(15), 1972–1973. Available from: 10.1093/bioinformatics/btp348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cappelletti, M. , Fedi, S. , Frascari, D. , Ohtake, H. , Turner, R.J. & Zannoni, D. (2011) Analyses of both the alkB gene transcriptional start site and alkB promoter‐inducing properties of Rhodococcus sp. strain BCP1 grown on n‐alkanes. Applied and Environmental Microbiology, 77(5), 1619–1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cappelletti, M. , Fedi, S. , Zampolli, J. , Di Canito, A. , D'Ursi, P. , Orro, A. et al. (2016) Phenotype microarray analysis may unravel genetic determinants of the stress response by Rhodococcus aetherivorans BCP1 and Rhodococcus opacus R7. Research in Microbiology, 167(9–10), 766–773. Available from: 10.1016/j.resmic.2016.06.008 [DOI] [PubMed] [Google Scholar]
- Cappelletti, M. , Fedi, S. & Zannoni, D. (2019) Degradation of alkanes in Rhodococcus . In: Alvarez, H. (Ed.) Biology of Rhodococcus, Vol. 16. Cham, Switzerland: Springer International Publishing, pp. 137–171. Available from: 10.1007/978-3-030-11461-9_6 [DOI] [Google Scholar]
- Cappelletti, M. , Frascari, D. , Zannoni, D. & Fedi, S. (2012) Microbial degradation of chloroform. Applied Microbiology and Biotechnology, 96, 1395–1409. Available from: 10.1007/s00253-012-4494-1 [DOI] [PubMed] [Google Scholar]
- Cappelletti, M. , Pinelli, D. , Fedi, S. , Zannoni, D. & Frascari, D. (2018) Aerobic co‐metabolism of 1,1,2,2‐tetrachloroethane by Rhodococcus aetherivorans TPA grown on propane: kinetic study and bioreactor configuration analysis. Journal of Chemical Technology & Biotechnology, 93(1), 155–165. Available from: 10.1002/jctb.5335 [DOI] [Google Scholar]
- Cappelletti, M. , Presentato, A. , Milazzo, G. , Turner, R.J. , Fedi, S. , Frascari, D. et al. (2015) Growth of Rhodococcus sp. strain BCP1 on gaseous n‐alkanes: new metabolic insights and transcriptional analysis of two soluble di‐iron monooxygenase genes. Frontiers in Microbiology, 6, 393. Available from: 10.3389/fmicb.2015.00393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cappelletti, M. , Zampolli, J. , Di Gennaro, P. & Zannoni, D. (2019) Genomics of Rhodococcus . In: Alvarez, H. (Ed.) Biology of Rhodococcus, Vol. 16. Cham, Switzerland: Springer International Publishing, pp. 23–60. Available from: 10.1007/978-3-030-11461-9_2 [DOI] [Google Scholar]
- Ciavarelli, R. , Cappelletti, M. , Fedi, S. , Pinelli, D. & Frascari, D. (2012) Chloroform aerobic cometabolism by butane‐growing Rhodococcus aetherovorans BCP1 in continuous‐flow biofilm reactors. Bioprocess and Biosystems Engineering, 35(5), 667–681. Available from: 10.1007/s00449-011-0647-3 [DOI] [PubMed] [Google Scholar]
- Coleman, N. , Le, N. , Ly, M. , Ogawa, H.E. , McCarl, V. , Wilson, N.L. et al. (2012) Hydrocarbon monooxygenase in mycobacterium: recombinant expression of a member of the ammonia monooxygenase superfamily. ISME Ournal, 6, 171–182. Available from: 10.1038/ismej.2011.98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman, N.V. , Bui, N.B. & Holmes, A.J. (2006) Soluble di‐iron monooxygenase gene diversity in soils, sediments and ethene enrichments. Environmental Microbiology, 8, 1228–1239. Available from: 10.1111/j.1462-2920.2006.01015.x [DOI] [PubMed] [Google Scholar]
- Coleman, N.V. , Yau, S. , Wilson, N.L. , Nolan, L.M. , Migocki, M.D. , Ly, M.A. et al. (2011) Untangling the multiple monooxygenases of mycobacterium chubuense strain NBB4, a versatile hydrocarbon degrader. Environmental Microbiology Reports, 3, 297–307. Available from: 10.1111/j.1758-2229.2010.00225.x [DOI] [PubMed] [Google Scholar]
- Dabbs, E.R. , Gowan, B. & Andersen, S.J. (1990) Nocardioform arsenic resistance plasmids and construction of Rhodococcus cloning vectors. Plasmid, 23(3), 242–247. Available from: 10.1016/0147-619x(90)90056-i [DOI] [PubMed] [Google Scholar]
- de Carvalho, C.C.C.R. & da Fonseca, M.M.R. (2004) Solvent toxicity in organic‐aqueous systems analysed by multivariate analysis. Bioprocess and Biosystems Engineering, 26, 361–375. Available from: 10.1007/s00449-004-0381-1 [DOI] [PubMed] [Google Scholar]
- de Carvalho, C.C.C.R. & da Fonseca, M.M.R. (2005) The remarkable Rhodococcus erythropolis . Applied Microbiology and Biotechnology, 67, 715–726. Available from: 10.1007/s00253-005-1932-3 [DOI] [PubMed] [Google Scholar]
- de Carvalho, C.C.C.R. , Marques, M.P.C. , Hachicho, N. & Heipieper, H.J. (2014) Rapid adaptation of Rhodococcus erythropolis cells to salt stress by synthesizing polyunsaturated fatty acids. Applied Microbiology and Biotechnology, 98, 5599–5606. Available from: 10.1007/s00253-014-5549-2 [DOI] [PubMed] [Google Scholar]
- Deng, D. , Li, F. & Li, M. (2018) A novel propane monooxygenase initiating degradation of 1, 4‐dioxane by mycobacterium dioxanotrophicus PH‐06. Environmental Science & Technology Letters, 5(2), 86–91. Available from: 10.1021/acs.estlett.7b00504 [DOI] [Google Scholar]
- Donini, E. , Firrincieli, A. & Cappelletti, M. (2021) Systems biology and metabolic engineering of Rhodococcus for bioconversion and biosynthesis processes. Folia Microbiologica, 66, 701–713. Available from: 10.1007/s12223-021-00892-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farhan Ul Haque, M. , Hernández, M. , Crombie, A.T. & Murrell, C. (2022) Identification of active gaseous‐alkane degraders at natural gas seeps. ISME Journal, 16, 1705–1716. Available from: 10.1038/s41396-022-01211-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firrincieli, A. , Zannoni, D. , Donini, E. , Dostálová, H. , Rädisch, R. , Iommarini, L. et al. (2022) Transcriptomic analysis of the dual response of Rhodococcus aetherivorans BCP1 to inorganic arsenic oxyanions. Applied and Environmental Microbiology, 88(7), e02209–e02221. Available from: 10.1128/aem.02209-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frascari, D. , Pinelli, D. , Nocentini, M. , Fedi, S. , Pii, Y. & Zannoni, D. (2006) Chloroform degradation by butane‐grown cells of Rhodococcus aetherovorans BCP1. Applied Microbiology and Biotechnology, 73(2), 421–428. Available from: 10.1007/s00253-006-0433-3 [DOI] [PubMed] [Google Scholar]
- Furuya, T. , Hayashi, M. & Kino, K. (2003) Reconstitution of active mycobacterial binuclear iron monooxygenase complex in Escherichia coli . Applied and Environmental Microbiology, 79(19), 6033–6039. Available from: 10.1128/AEM.01856-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golby, S. , Ceri, H. , Marques, L.L.R. & Turner, R.J. (2014) Mixed‐species biofilms cultured from an oil sand tailings pond can biomineralize metals. Microbial Ecology, 68(1), 70–80. [DOI] [PubMed] [Google Scholar]
- Hamamura, N. , Olson, S.H. , Ward, D.M. & Inskeep, W.P. (2006) Microbial population dynamics associated with crude‐oil biodegradation in diverse soils. Applied and Environmental Microbiology, 72, 6316–6324. Available from: 10.1128/AEM.01015-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan, D. (1983) Studies on transformation of Escherichia coli with plasmids. Journal of Molecular Biology, 166, 557–580. [DOI] [PubMed] [Google Scholar]
- He, Y. , Mathieu, J. , da Silva, M.L.B. , Li, M. & Alvarez, P.J.J. (2018) 1,4‐Dioxane‐degrading consortia can be enriched from uncontaminated soils: prevalence of mycobacterium and soluble di‐iron monooxygenase genes. Microbial Biotechnology, 11(1), 189–198. Available from: 10.1111/1751-7915.12850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes, A.J. & Coleman, N.V. (2008) Evolutionary ecology and multidisciplinary approaches to prospecting for monooxygenases as biocatalysts. Antonie Van Leeuwenhoek, 94, 75–84. Available from: 10.1007/s10482-008-9227-1 [DOI] [PubMed] [Google Scholar]
- Ivshina, I.B. , Tyumina, E.A. , Kuzmina, M.V. & Vikhareva, E.V. (2019) Features of diclofenac biodegradation by Rhodococcus ruber IEGM 346. Scientific Reports, 9(1), 1–13. Available from: 10.1038/s41598-019-45732-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh, K. , Misawa, K. , Kuma, J. & Miyata, T. (2002) MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Research, 30(14), 3059–3066. Available from: 10.1093/nar/gkf436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotani, T. , Yamamoto, T. , Yurimoto, H. , Sakia, Y. & Kato, N. (2003) Propane monooxygenase and NAD+‐dependent secondary alcohol dehydrogenase in propane metabolism by Gordonia sp. strain TY‐5. Journal of Bacteriology, 185, 7120–7128. Available from: 10.1128/JB.185.24.7120-7128.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurth, E.G. , Doughty, D.M. , Bottomley, P.J. , Arp, D.J. & Sayavedra‐Soto, L.A. (2008) Involvement of BmoR and BmoG in n‐alkane metabolism in ‘Pseudomonas butanovora’. Microbiology, 154(1), 139–147. Available from: 10.1099/mic.0.2007/012724-0 [DOI] [PubMed] [Google Scholar]
- Larkin, M.J. , Kulakov, L.A. & Allen, C.C. (2005) Biodegradation and Rhodococcus ‐masters of catabolic versatility. Current Opinion in Biotechnology, 16(3), 282–290. Available from: 10.1016/j.copbio.2005.04.007 [DOI] [PubMed] [Google Scholar]
- Leahy, J.G. , Batchelor, P.J. & Morcomb, S.M. (2003) Evolution of the soluble diiron monooxygenases. FEMS Microbiology Reviews, 27, 449–479. Available from: 10.1016/S0168-6445(03)00023-8 [DOI] [PubMed] [Google Scholar]
- Lowry, O.H. , Rosebrough, N.J. , Farr, A.L. & Randall, R.J. (1951) Protein measurement with the Folin phenol reagent. Journal of Biological Chemistry, 193, 265–275. [PubMed] [Google Scholar]
- Martin, K.E. , Ozsvar, J. & Coleman, N.V. (2014) SmoXYB1C1Z of mycobacterium sp. strain NBB4: a soluble methane monooxygenase (sMMO)‐like enzyme, active on C2 to C4 alkanes and alkenes. Applied and Environmental Microbiology, 80(18), 5801–5806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez, J.L. & Rojo, F. (2011) Metabolic regulation of antibiotic resistance. FEMS Microbiology Reviews, 35, 768–789. Available from: 10.1111/j.1574-6976.2011.00282.x [DOI] [PubMed] [Google Scholar]
- Martínková, L. , Uhnáková, B. , Pátek, M. , Nešvera, J. & Křen, V. (2009) Biodegradation potential of the genus Rhodococcus . Environment International, 35(1), 162–177. Available from: 10.1016/j.envint.2008.07.018 [DOI] [PubMed] [Google Scholar]
- McCarl, V. , Somerville, M.V. , Ly, M.A. , Henry, R. , Liew, E.F. , Wilson, N.L. et al. (2018) Heterologous expression of mycobacterium alkene monooxygenases in gram‐positive and gram‐negative bacterial hosts. Applied and Environmental Microbiology, 84(15), e00397‐18. Available from: 10.1128/AEM.00397-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald, I.R. , Uchiyama, H. , Kambe, S. , Yagi, O. & Murrell, J.C. (1997) The soluble methane monooxygenase gene cluster of the trichloroethylene‐degrading methanotroph Methylocystis sp. strain M. Applied and Environmental Microbiology, 63, 1898–1904. Available from: 10.1128/aem.63.5.1898-1904.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel, B. , Kozlov, A.M. , Stamatakis, A. & Szöllősi, G.J. (2020) GeneRax: a tool for species‐tree‐aware maximum likelihood‐based gene family tree inference under gene duplication, transfer, and loss. Molecular Biology and Evolution, 37(9), 2763–2774. Available from: 10.1093/molbev/msaa141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima, N. & Tamura, T. (2004a) A novel system for expressing recombinant proteins over a wide temperature range from 4 to 35 C. Biotechnology and Bioengineering, 86(2), 136–148. Available from: 10.1002/bit.20024 [DOI] [PubMed] [Google Scholar]
- Nakashima, N. & Tamura, T. (2004b) Isolation and characterization of a rolling‐circle‐type plasmid from Rhodococcus erythropolis and application of the plasmid to multiple‐recombinant‐protein expression. Applied and Environmental Microbiology, 70(9), 5557–5568. Available from: 10.1128/AEM.70.9.5557-5568.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen, L.T. , Schmidt, H.A. , von Haeseler, A. & Minh, B.Q. (2015) IQ‐TREE: a fast and effective stochastic algorithm for estimating maximum‐likelihood phylogenies. Molecular Biology and Evolution, 32(1), 268–274. Available from: 10.1093/molbev/msu300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orro, A. , Cappelletti, M. , D'Ursi, P. , Milanesi, L. , Di Canito, A. , Zampolli, J. et al. (2015) Genome and phenotype microarray analyses of Rhodococcus sp. BCP1 and Rhodococcus opacus R7: genetic determinants and metabolic abilities with environmental relevance. PLoS One, 10(10), 1–41. Available from: 10.1371/journal.pone.0139467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petkevičius, V. , Vaitekūnas, J. , Gasparavičiūtė, R. , Tauraitė, D. & Meškys, R. (2021) An efficient and regioselective biocatalytic synthesis of aromatic N‐oxides by using a soluble di‐iron monooxygenase PmlABCDEF produced in the pseudomonas species. Microbial Biotechnology, 14(4), 1771–1783. Available from: 10.1111/1751-7915.13849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presentato, A. , Cappelletti, M. , Sansone, A. , Ferreri, C. , Piacenza, E. , Demeter, M.A. et al. (2018) Aerobic growth of Rhodococcus aetherivorans BCP1 using selected naphthenic acids as the sole carbon and energy sources. Frontiers in Microbiology, 9, 672. Available from: 10.3389/fmicb.2018.00672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price, M.N. , Dehal, P.S. & Arkin, A.P. (2009) FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Molecular Biology and Evolution, 26(7), 1641–1650. Available from: 10.1093/molbev/msp077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojo, F. (2009) Degradation of alkanes by bacteria. Environmental Microbiology, 11(10), 2477–2490. Available from: 10.1111/j.1462-2920.2009.01948.x [DOI] [PubMed] [Google Scholar]
- Sallam, K.I. , Mitani, Y. & Tamura, T. (2006) Construction of random transposition mutagenesis system in Rhodococcus erythropolis using IS1415. Journal of Biotechnology, 121, 13–22. [DOI] [PubMed] [Google Scholar]
- Sallam, K.I. , Tamura, N. & Tamura, T. (2007) A multipurpose transposon‐based vector system mediates protein expression in Rhodococcus erythropolis . Gene, 386(1–2), 173–182. Available from: 10.1016/j.gene.2006.09.006 [DOI] [PubMed] [Google Scholar]
- Sambrook, J. , Fritsch, E.F. & Maniatis, T. (1989) Molecular cloning: a laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Sharp, J.O. , Sales, C.M. , LeBlanc, J.C. , Liu, J. , Wood, T.K. , Eltis, L.D. et al. (2007) An inducible propane monooxygenase is responsible for N‐nitrosodimethylamine degradation by Rhodococcus sp. strain RHA1. Applied and Environmental Microbiology, 73, 6930–6938. Available from: 10.1128/AEM.01697-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sluis, M.K. , Sayavedra‐Soto, L.A. & Arp, D.J. (2002) Molecular analysis of the soluble butane monooxygenase from “pseudomonas butanovora”. Microbiology, 148, 3617–3629. Available from: 10.1099/00221287-148-11-3617 [DOI] [PubMed] [Google Scholar]
- Stafford, G.P. , Scanlan, J. , McDonald, I.R. & Murrell, J.C. (2003) rpoN, mmoR and mmoG, genes involved in regulating the expression of soluble methane monooxygenase in Methylosinus trichosporium OB3b. Microbiology, 149(7), 1771–1784. Available from: 10.1099/mic.0.26060-0 [DOI] [PubMed] [Google Scholar]
- Takano, E. , White, J. , Thompson, C.J. & Bibb, M.J. (1995) Construction of thiostrepton‐inducible, high‐copy‐number expression vectors for use in Streptomyces spp. Gene, 166, 133–137. [DOI] [PubMed] [Google Scholar]
- Van Beilen, J.B. , Li, Z. , Duetz, W.A. , Smits, T.H.M. & Witholt, B. (2003) Diversity of alkane hydroxylase systems in the environment. Oil & Gas Science and Technology, 58(4), 427–440. Available from: 10.2516/ogst:2003026 [DOI] [Google Scholar]
- Vial, L. & Hommais, F. (2020) Plasmid‐chromosome cross‐talks. Environmental Microbiology, 22(2), 540–556. Available from: 10.1111/1462-2920.14880 [DOI] [PubMed] [Google Scholar]
- Yanisch‐Perron, C. , Vieira, J. & Messing, J. (1985) Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene, 33(1), 103–119. Available from: 10.1016/0378-1119(85)90120-9 [DOI] [PubMed] [Google Scholar]
- Zou, B. , Huang, Y. , Zhang, P.P. , Ding, X.M. , Op den Camp, H.J.M. & Quan, Z.X. (2021) Horizontal gene transfer of genes encoding copper‐containing membrane‐bound monooxygenase (CuMMO) and soluble di‐iron monooxygenase (SDIMO) in ethane‐and propane‐oxidizing Rhodococcus bacteria. Applied and Environmental Microbiology, 87(14), e00227‐21. Available from: 10.1128/AEM.00227-21funding [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1.
Table S3.
Table S4.
Data Availability Statement
Raw data used to generate Figures 6 and S7 and the SDIMOs phylogenetic trees in Figures 5A,B and S6 are available under the https://doi.org/10.6084/m9.figshare.24972594.
Genome sequence and annotation of Rhodococcus aetherivorans BCP1 are available under the NCBI RefSeq assembly GCF_000470885.1.


