Summary
Background
Urine cytology is an important non-invasive examination for urothelial carcinoma (UC) diagnosis and follow-up. We aimed to explore whether artificial intelligence (AI) can enhance the sensitivity of urine cytology and help avoid unnecessary endoscopy.
Methods
In this multicentre diagnostic study, consecutive patients who underwent liquid-based urine cytology examinations at four hospitals in China were included for model development and validation. Patients who declined surgery and lacked associated histopathology results, those diagnosed with rare subtype tumours of the urinary tract, or had low-quality images were excluded from the study. All liquid-based cytology slides were scanned into whole-slide images (WSIs) at 40 × magnification and the WSI-labels were derived from the corresponding histopathology results. The Precision Urine Cytology AI Solution (PUCAS) was composed of three distinct stages (patch extraction, features extraction, and classification diagnosis) and was trained to identify important WSI features associated with UC diagnosis. The diagnostic sensitivity was mainly used to validate the performance of PUCAS in retrospective and prospective validation cohorts. This study is registered with the ChiCTR, ChiCTR2300073192.
Findings
Between January 1, 2018 and October 31, 2022, 2641 patients were retrospectively recruited in the training cohort, and 2335 in retrospective validation cohorts; 400 eligible patients were enrolled in the prospective validation cohort between July 7, 2023 and September 15, 2023. The sensitivity of PUCAS ranged from 0.922 (95% CI: 0.811–0.978) to 1.000 (0.782–1.000) in retrospective validation cohorts, and was 0.896 (0.837–0.939) in prospective validation cohort. The PUCAS model also exhibited a good performance in detecting malignancy within atypical urothelial cells cases, with a sensitivity of over 0.84. In the recurrence detection scenario, PUCAS could reduce 57.5% of endoscopy use with a negative predictive value of 96.4%.
Interpretation
PUCAS may help to improve the sensitivity of urine cytology, reduce misdiagnoses of UC, avoid unnecessary endoscopy, and reduce the clinical burden in resource-limited areas. The further validation in other countries is needed.
Funding
National Natural Science Foundation of China; Key Program of the National Natural Science Foundation of China; the National Science Foundation for Distinguished Young Scholars; the Science and Technology Planning Project of Guangdong Province; the National Key Research and Development Programme of China; Guangdong Provincial Clinical Research Centre for Urological Diseases.
Keywords: Artificial intelligence, Urine cytology, Urothelial carcinoma, Multicentre study, Prospective validation
Research in context.
Evidence before this study
A PubMed search was performed for relevant articles published in any language from the database inception to December 30 2023, using the following search string: “(“artificial intelligence” OR “deep learning” OR “machine learning”) AND (“urine” OR “voided urine” OR “cytology” OR “urine cytology”) AND (“urothelial carcinoma” OR “bladder carcinoma” OR “bladder cancer” OR “upper tract urothelial carcinoma”)”. We systematically reviewed the 44 search results and identified 14 relevant original articles. 13 studies primarily applied artificial intelligence (AI) to assist urine cytology for the diagnosis of urothelial carcinoma (UC), whereas one study focused on predicting recurrence. Among the 13 diagnostic studies, eight mainly used AI to automate the Paris System for Reporting Urinary Cytology at the whole slide image level. Five studies evaluated the diagnostic performance of AI models using UC histopathology results as the gold standard, with the goal of enhancing the sensitivity of urine cytology. However, the above studies had limited clinical impact due to small-sized samples with a narrow range of inclusion of UC subtypes, and three of them were single-centre studies. Furthermore, the performance of the AI model in the atypical urine cell (AUC) subgroup or different clinical scenarios was not evaluated. And prospective validation was also lacked.
Added value of this study
We developed and validated a model called the Precision Urine Cytology AI Solution (PUCAS) in a large, multi-centre observation cohort—which incorporated a wide range of UC types and more comprehensive clinical characteristics—using histopathology as the gold standard. Utilising a multistage framework and the weighted output from multiple models, PUCAS achieved favourable diagnostic performance in both retrospective and prospective validation cohorts. In patients with AUC, PUCAS presented satisfying sensitivity (>0.8). In upper-tract UC, pTa and low-grade, minimal (<1.5 cm), residual, and recurrent tumours subgroups (residual and recurrent tumours subgroups included diagnosis of papillary urothelial neoplasm of low malignant potential, low-grade, and high-grade UC), PUCAS achieved a substantial improvement in sensitivity compared to cytology (increased sensitivity by 24.2% to 49.0%) and fluorescence in situ hybridisation (FISH) (increased sensitivity by 12.6% to 33.5%). In the recurrence detection scenario, PUCAS could reduce the use of endoscopy by 57.5% with a negative predictive value of 96.4%.
Implications of all the available evidence
The use of PUCAS in clinical settings may help to improve the sensitivity of urine cytology, reduce misdiagnoses of UC, and avoid unnecessary endoscopy. PUCAS can serve as a supplementary tool or an alternative to FISH, as it offers higher sensitivity and can be evaluated using a cloud-based system. In clinical practice, its automatability also aids cytopathologists in minimising repetitive and time-consuming tasks.
Introduction
Urothelial carcinoma (UC) is one of the most common malignancies of the urinary system.1,2 Upper tract urothelial carcinoma (UTUC), comprising tumours in the pyelocalyceal cavities and ureter, constitutes 5%–10% of all UCs.3 UCs have a high tendency to recur, with significant variations in prognosis across diverse tumour grades. Early diagnosis and regular monitoring play pivotal roles in improving the prognosis of patients with UC.4 Presently, in clinical practice, the histopathological reports from endoscopy with biopsy is considered the gold standard for diagnosing UC,2,3 and regular endoscopic examinations are frequently required for detection of recurrence. Per recommendations, a high-risk tumour patient should undergo at least 15 endoscopic examinations over a 5-year period. However, endoscopy is invasive, costly, and often associated with discomfort. And in secondary resection scenario, it is still hard to detect residual tumour. Consequently, there is an urgent need to develop non-invasive, effective, and economical methods for the detection of UCs. The presently available non-invasive tools, including urine cytology, fluorescence in situ hybridisation (FISH), urine methylation assay, and Cxbladder, do not completely meet the clinical requirements.5,6
Urine cytology plays a crucial role in the clinical management of UC; it is convenient to use, non-invasive, cost-effective, and highly specific.2,3,7 The main limitations of urine cytology are as follows: potential inability to capture atypical or malignant cells and the challenge of reconciling between the cytological and histological findings8,9; low sensitivity, particularly for the detection of low-grade or early-stage UC5,10; examination of cytology slides under a microscope is a repetitive and time-consuming task for cytopathologists; and final diagnosis relies heavily on the cytopathologist’s experience, with poor inter-observer reproducibility.11,12 Although the Paris System for Reporting Urinary Cytology (TPS) is used to standardise diagnostic criteria,13,14 the reporting rate of atypical urothelial cells (AUC) remains high. The clinical implications of the AUC remain unclear, which causes significant confusion for urologists and introduces additional procedures for patients. Additional robust methods are needed to improve the diagnostic approach for urine cytology, improve diagnostic sensitivity, and identify patients who may not require endoscopy.
Over the past decade, artificial intelligence (AI) has advanced rapidly in the field of image diagnosis.15 AI has been reported to match or outperform human experts in numerous medical scenarios, owing to its strength in image texture analysis.16, 17, 18 Regarding cytopathology, AI has been applied for the identification of lesion cells in cervical and oesophageal cancers and has achieved promising performance.19,20 However, the use of AI-based detection of UC with urine cytology remains limited. Most similar studies have focused on automating TPS reports and simulating the diagnostic process of cytopathologists.21,22 In addition to TPS, five previous studies assessed the diagnostic performance of AI models using histopathology as the gold standard.8, 9, 10,23,24 However, the above AI models had limited clinical impact due to their restriction to single-centre,8,9,24 small-scale samples with a narrower range of UC types inclusion (i.e., didn’t include UTUC10,23,24 or papillary urothelial neoplasm of low malignant potential [PUNLMP]), and also the suboptimal sensitivity.9,10,23,24 Furthermore, the above studies did not evaluate the performance of the AI model in the AUC subgroup and in different clinical scenarios (i.e., recurrence detection). And prospective validation was also lacked.
In this study, we aimed to develop and validate an AI-based model called “Precision Urine Cytology AI Solution (PUCAS)” with multi-stage framework algorithms in a large, multi-centre observation cohort and incorporate a wide range of UC types and comprehensive clinical characteristics, using histopathology as the gold standard. Prospective validation and subgroup analyses were also conducted to explore its clinical applicability and robustness.
Methods
Study design and participants
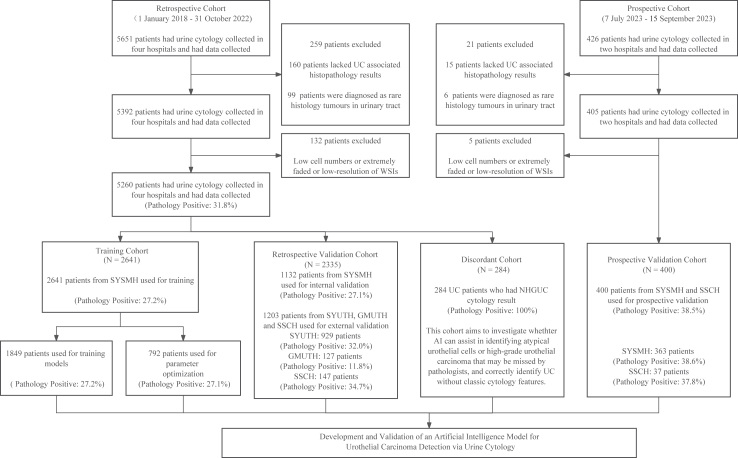
In this multicentre observational study, patients who underwent liquid-based voided urine cytology examinations in four hospitals in China were included for model development and validation (Fig. 1). The retrospective cohort was collected from January 1, 2018 to October 31, 2022, which included Sun Yat-sen Memorial Hospital of Sun Yat-Sen University (SYSMH, January 1, 2018 to October 31, 2022), the Third Affiliated Hospital of Sun Yat-Sen University (SYUTH, January 1, 2019 to October 31, 2022), the Third Affiliated Hospital of Guangzhou Medical University (GMUTH, March 1, 2021 to October 31, 2022), and Shen-Shan Central Hospital (SSCH, December 1, 2021 to October 31, 2022). The slides in the prospective cohort were collected from July 7, 2023 to September 15, 2023, which included SYSMH and SSCH. The following patients were excluded: those who declined UC-associated surgery and without histopathology reports despite radiology or urine cytology indicating the need for it; and those with rare subtypes of tumours of the urinary tract, such as squamous cell carcinoma and adenocarcinoma, because urinary cytology and TPS primarily detect urothelial carcinoma.13 The collection process of golden standard and AI prediction in prospective trial was designed to be independent and blinded to each other. The prospective observational study was registered with ChiCTR2300073192. And the detailed process of prospective validation trial is shown in Appendix (p 2).
Fig. 1.
Flowchart of the study. WSIs, whole-slide images; UC, urothelial carcinoma; SYSMH, Sun Yat-sen Memorial Hospital of Sun Yat-sen University; SYUTH, Third Affiliated Hospital of Sun Yat-sen University; GMUTH, Third Affiliated Hospital of Guangzhou Medical University; SSCH, Central Hospital of Shen Shan; NHGUC, negative for high-grade urothelial carcinoma; AI, artificial intelligence.
The requirements for informed consent for both retrospective and prospective studies were waived because of the observational design; however, each participant had provided written informed assent for the Collection and Application of Clinical Sample and Medical Data certified. This study was approved by the ethics committee of all participating hospitals.
Procedures
All liquid-based cytology slides, stained using the Papanicolaou method, were obtained from the pathology department’s archive and subsequently scanned into whole-slide images (WSIs) at 40 × magnification. The preparatory technique method for slides making is shown in Appendix (p 2). To enhance and validate the robustness of the PUCAS, five types of scanners were adopted, and the details are shown in Appendix (p 3). We excluded low-quality slides owing to low cell numbers (less than 20), extreme fading, and low-resolution WSIs caused by poor scanning quality.
The baseline characteristics of the study participants, including demographic information, clinical symptoms and scenarios, and histopathological results, were extracted from the medical record archives of the four participating hospitals. Race/ethnicity data were not collected. WSI-level labels were derived from histopathological reports. Positive cases were defined as ‘patients with confirmed UCs based on histopathological reports of endoscopic biopsy, transurethral resection of bladder tumour (TURBT), radical cystectomy, or radical nephroureterectomy’. Negative cases included patients with negative histopathological results or negative radiological or endoscopic findings. All negative cases were followed-up for a 3-month period to avoid misdiagnosis. For development of PUCAS, initially in retrospective cohort, positive cases that had negative for high-grade urothelial carcinoma (NHGUC) cytology were extracted separately. This group was defined as the ‘discordant cohort’, aiming to investigate whether PUCAS could assist in identifying suspicious cells missed by cytopathologists and correctly identify UC without classic cytology features. In the remaining cases of retrospective cohort, patients from the SYSMH were stratified in a 7:3 ratio by sex, slide staining year, and positive rate, and then divided into a ‘training cohort’ and an ‘internal validation cohort’. Similarly, patients from the training cohort were stratified into ‘model training’ and ‘parameter optimisation cohorts’ in a 7:3 ratio. Patients from SYUTH, GMUTH, and SSCH of retrospective cohort were assigned to three independent ‘external validation cohorts’. The whole prospective cohort was defined as ‘prospective validation cohort’.
For model training, we obtained cell-level annotations from two expert cytopathologists, each with >15 years of experience in urine cytology diagnosis; they carefully reviewed and labelled urothelial cells into normal, atypical, or malignant categories following the established diagnostic criteria for TPS.13 Additionally, other cell types present in urine, such as glandular and squamous cells, were annotated. Uncertain cells identified by the cytopathologists and degenerated cells were annotated as ‘others’ (The representative examples are shown in Appendix p 18). Finally, 119,131 cells were annotated in the training cohort. For WSI-level cytology diagnosis in both training and validation cohorts, we applied a central review from a consensus to guarantee objectivity. The primary diagnosis was consensus-based and determined through the collaborative assessment of two expert cytopathologists. If intra-variability existed, another expert cytopathologist (>20 years of experience) was invited to give a final decision. High-grade urothelial carcinoma (HGUC) or suspected HGUC was defined as cytology-positive, whereas AUC and NHGUC were defined as cytology-negative for UC.
The architecture of PUCAS was composed of three distinct stages, as depicted in the Appendix (p 19). In the initial stage, high-resolution WSIs, consisting of 50,000 × 50,000 pixels, were divided into numerous non-overlapping patches using a sliding window approach. This preprocessing was performed to facilitate efficient training and validation of the models on the graphics processing unit. The crop size was set to 1024 × 1024 pixels, corresponding to a spatial resolution of 0.25 μm per pixel (40 × ), ensuring optimal compatibility with subsequent operations.
Subsequently, individual patches extracted from the WSIs were subjected to patch-level feature extraction. Three distinct feature extraction models (YOLOv7,25 EfficientNet,26 and ConvNeXt-B27) were integrated and trained to identify atypical or malignant cells and to extract crucial features. Upon completion of this stage, all features extracted from both the atypical cells and the patch as a whole were integrated to generate a comprehensive and informative patch result.
In the final stage, all patch results generated from the same WSI were merged to form WSI-level feature maps that served as the input for the diagnosis stage. This stage comprised three distinct models, namely, the attention bi-directional long short-term memory,28 Transformer,29 and Top-N Feature model30 (Appendix p 3), which independently predicted the confidence of each class. The combination of these diverse models afforded a more in-depth analysis of complex WSI-level feature maps and provided a comprehensive and robust diagnosis. The final confidence of each class was determined by averaging the confidence scores derived from each WSI-level feature extraction model with the appropriate weighting. More details about the network training process, including data balance, data augmentation methods, loss function setting, and associated training equipment, are provided in the Appendix (pp 3–5, 20–21).
We employed retrospective and prospective validation cohorts for performance validation of the PUCAS, and a discordant cohort to assess the model’s generalisability. Subgroup analyses were performed in retrospective validation cohort and discordant cohort separately. First, we evaluated the diagnostic efficacy of PUCAS across diverse clinical subgroups defined by factors such as centre, AUC cytology results, age, sex, haematuria, and smoking. Furthermore, we examined its effectiveness in various clinical scenarios including routine screening, secondary resection, and recurrence detection monitoring (including diagnosis of PUNLMP, low-grade, and high-grade UC in each scenario). Additionally, we tested the diagnostic sensitivity of different tumour subgroups, including UTUC, tumour grade, stage, number, and size to evaluate its robustness. In the discordant cohort, suspicious cells annotated by PUCAS were also reviewed by a centre review. At last, a multi-modal model was built with PUCAS and clinical factors to investigate whether PUCAS could be further improved. Logistic regression model was conducted. The method and flowchart of building multi-modal model are shown in Appendix (pp 5, 22).
Outcomes
Sensitivity of PUCAS was determined as the primary endpoint in this study. Other diagnostic performances of PUCAS, including area under the receiver operation characteristic curve (AUROC), specificity, accuracy, positive predictive value (PPV), and negative predictive value (NPV) were determined as the secondary endpoints. The performance of PUCAS was primarily compared with cytology and FISH in subgroup analyses.
Statistical analysis
All statistical analyses and data visualisation were conducted using R software (version 4.2.0) and Prism 9 (GraphPad Software). Receiver operating characteristic (ROC) curves were used to assess the discriminatory ability of the PUCAS prediction score (pROC package). Comparison of the two ROCs was based on the Delong test.31 The optimal positive cut-off value of the PUCAS was investigated using the ROC curve to achieve the highest Youden index in the internal validation cohort. The clinical utilities of the different subgroups were visualised using decision clinical analysis curves (rmda package). Performance indices, including sensitivity, specificity, accuracy, PPV, and NPV, with their 95% confidence intervals (CI), were calculated using the epiR package. The McNemar’s test was employed to assess the discrepancy in sensitivity, specificity, and NPV between various diagnostic methods. Net reclassification improvement (NRI) and integrated discrimination improvement (IDI) were used to assess the improved predictive ability of PUCAS compared to those of cytology and FISH. Logistic regression model was built based on rms package. All analyses were prespecified. Two-sided P values less than 0.050 were considered statistically significant.
Role of the funding source
The funders of the study had no role in the study design, data collection, data analysis, data interpretation, or writing of the report. All authors reviewed the manuscript, approved the submitted version, had full access to all the data reported in the study, and had final responsibility for the decision to submit for publication.
Results
Between January 1, 2018, and October 31, 2022, 5651 patients were retrospectively recruited in the retrospective cohort; 426 patients were enrolled in the prospective study between July 7, 2023 and September 15, 2023 (Fig. 1). 391 (6.9%) of retrospective cohort and 26 (6.1%) of prospective cohort were excluded according to the pre-defined exclusion criteria.
Consequently, 5260 urine cytology images in retrospective cohort were used to develop and validate the PUCAS. Initially, 284 images (5.4%) from patients with positive UC histopathology and NHGUC cytology results were chosen for the discordant cohort. The remaining images from SYSMH (3773 images, 71.7%) were stratified into training (2641 images, 50.2%) and internal validation cohorts (1132 images, 21.5%). The training cohort was then stratified into model training (1849 images, 35.2%) and parameter optimisation cohorts (792 images, 15.1%). The remaining images from SYUTH (929 images, 17.7%), GMUTH (127 images, 2.4%), and SSCH (147 images, 2.8%) were used as external validation cohorts (1203 images, 22.9%). 400 urine cytology images in prospective cohort (SYSMH [363 images, 90.8%] and SSCH [37 images, 9.3%]) were defined as prospective validation cohort. The clinical characteristics of the training, validation, and discordant cohorts are summarised in Table 1 and Appendix (pp 7–9). There were no missing data in all cohorts.
Table 1.
Baseline characteristics of the training and validation cohorts.
| Retrospective cohort |
Prospective cohort |
|||||
|---|---|---|---|---|---|---|
| SYSMH training cohort (N = 2641) | SYSMH validation cohort (N = 1132) | SYUTH validation cohort (N = 929) | GMUTH validation cohort (N = 127) | SSCH validation cohort (N = 147) | Prospective validation cohort (N = 400) | |
| Age | 62.00 [53.00, 70.00] | 62.00 [54.00, 70.00] | 64.00 [54.00, 73.00] | 57.00 [44.00, 71.00] | 65.00 [58.00, 73.00] | 64.00 [56.00, 70.00] |
| Sex | ||||||
| Male | 1892 (71.6%) | 814 (71.9%) | 655 (70.5%) | 72 (56.7%) | 107 (72.8%) | 292 (73.0%) |
| Female | 749 (28.4%) | 318 (28.1%) | 274 (29.5%) | 55 (43.3%) | 40 (27.2%) | 108 (27.0%) |
| Haematuria | ||||||
| Yes | 936 (35.4%) | 393 (34.7%) | 578 (62.2%) | 31 (24.4%) | 83 (56.5%) | 166 (41.5%) |
| No | 1705 (64.6%) | 739 (65.3%) | 351 (37.8%) | 96 (75.6%) | 64 (43.5%) | 234 (58.5%) |
| Smoking | ||||||
| Yes | 446 (16.9%) | 193 (17.0%) | 193 (20.8%) | 16 (12.6%) | 53 (36.1%) | 67 (16.8%) |
| No | 2195 (83.1%) | 939 (83.0%) | 736 (79.2%) | 111 (87.4%) | 94 (63.9%) | 333 (83.2%) |
| Clinical scenario | ||||||
| Routine screening | 2065 (78.2%) | 894 (79.0%) | 851 (91.6%) | 101 (79.5%) | 122 (83.0%) | 275 (68.8%) |
| Recurrence detection | 370 (14.0%) | 151 (13.3%) | 53 (5.7%) | 18 (14.2%) | 18 (12.2%) | 114 (28.4%) |
| Secondary resection | 206 (7.8%) | 87 (7.7%) | 25 (2.7%) | 8 (6.3%) | 7 (4.8%) | 11 (2.8%) |
| Cytology results | ||||||
| NHGUC | 1551 (58.7%) | 693 (61.2%) | 445 (47.9%) | 92 (72.4%) | 77 (52.4%) | 239 (59.8%) |
| AUC | 601 (22.8%) | 217 (19.2%) | 304 (32.7%) | 20 (15.7%) | 37 (25.2%) | 65 (16.2%) |
| SHGUC | 101 (3.8%) | 50 (4.4%) | 76 (8.2%) | 9 (7.1%) | 5 (3.4%) | 22 (5.5%) |
| HGUC | 388 (14.7%) | 172 (15.2%) | 104 (11.2%) | 6 (4.7%) | 28 (19.0%) | 74 (18.5%) |
| Pathology positive | ||||||
| Positive | 718 (27.2%) | 307 (27.1%) | 297 (32.0%) | 15 (11.8%) | 51 (34.7%) | 154 (38.5%) |
| Negative | 1923 (72.8%) | 825 (72.9%) | 632 (68.0%) | 112 (88.2%) | 96 (65.3%) | 246 (61.5%) |
Data are n (%) or median (IQR). SYSMH, Sun Yat-sen Memorial Hospital of Sun Yat-sen University; SYUTH, The Third Affiliated Hospital of Sun Yat-sen University; GMUTH, The Third Affiliated Hospital of Guangzhou Medical University; SSCH, The Central Hospital of Shen-Shan; NHGUC, negative for high-grade urothelial carcinoma; AUC, atypical urothelial cells; SHGUC, suspicious for high-grade urothelial carcinoma; HGUC, positive for high-grade urothelial carcinoma.
The interactive system interface of PUCAS is shown in Appendix (p 23). The PUCAS was an integrated model and had a higher AUROC (0.970, 95% CI: 0.958–0.981) than any individual internal structural model within the internal validation cohort (Fig. 2A). Based on the optimal Youden index, 0.5 was defined as the cut-off value for binary classification. Across the three external validation cohorts, the AUROCs of the PUCAS ranged from 0.928 (95% CI: 0.874–0.982) to 0.963 (95% CI: 0.931–0.994) (Fig. 2B). In the prospective validation cohort, PUCAS also got a favourable result (AUROC: 0.917 [95% CI: 0.885–0.949], Fig. 2C). In retrospective and prospective validation cohorts, the sensitivities ranged from 0.896 (95% CI: 0.837–0.939) to 1.000 (95% CI: 0.782–1.000), while the NPV ranged from 0.932 (95% CI:0.892–0.961) to 1.000 (0.964–1.000) (Appendix p 10). Although PUCAS was mainly designed to improve sensitivity, it also showed satisfying specificity in retrospective (varying from 0.893 [95% CI: 0.820–0.943] to 0.942 [95% CI: 0.924–0.957]) and prospective validation cohorts (0.894 [95% CI: 0.849–0.930]) (the comparison of specificity with cytology and FISH is shown in Appendix p 11). In patients with AUC cytology result in retrospective (N = 578) and prospective validation cohort (N = 65), PUCAS showed superior AUROC and sensitivity results compared with FISH (Appendix p 24).
Fig. 2.
Performance of PUCAS in validation cohorts. (A) ROC curves of different deep learning models and integrated PUCAS in detecting UC in SYSMH internal validation cohort. (B) ROC curves of PUCAS in detecting UC were analysed separately for each centre and collectively for the entire external validation cohort. (C) ROC curves of PUCAS in detecting UC in prospective validation cohorts. (D) Corresponding sensitivities, specificities, NPVs, and AUROCs in different subgroups of retrospective validation cohorts, classified by age, sex, haematuria, smoking, and clinical scenarios. PUCAS, The Precision Urine Cytology AI Solution; ROC, receiver operating curve; UC, urothelial carcinoma; SYSMH, Sun Yat-sen Memorial Hospital of Sun Yat-sen University; SYUTH, The Third Affiliated Hospital of Sun Yat-sen University; GMUTH, The Third Affiliated Hospital of Guangzhou Medical University; SSCH, The Central Hospital of Shen-Shan; LSTM, long short-term memory; NPV, negative predictive value; AUROC, area under the receiver operating characteristic curve; FISH, fluorescence in situ hybridisation; CI, confidence interval.
In further subgroup analyses of retrospective validation cohorts, we found that PUCAS had higher sensitivity in older groups (age ≥ 65 years, 0.953, 95% CI: 0.927–0.971; age < 65 years, 0.914, 95% CI: 0.874–0.945) and in women (0.949, 95% CI: 0.901–0.978; men: 0.934, 95% CI: 0.909–0.954). In patients with haematuria syndrome, PUCAS had a higher sensitivity (0.948, 95% CI: 0.925–0.966) than in those without haematuria (0.908, 95% CI: 0.857–0.946). For different clinical scenarios, the sensitivity of the PUCAS ranged from 0.934 (95% CI: 0.910–0.953) to 0.966 (95% CI: 0.822–0.999), and the NPV ranged from 0.964 (95% CI: 0.917–0.988) to 0.989 (95% CI: 0.943–1.000) (Fig. 2D, Appendix p 12). It showed that PUCAS showed varied sensitivity across different clinical factors but only age (P = 0.041) had statistical significance. For haematuria, the results showed borderline positive (P = 0.054). The heterogeneity of model performance could be explained by higher prevalence rate of UC in older and haematuria groups, which are also high-risk groups in clinic. The elevated prevalence rates in these subgroups provided an abundant supply of positive samples for model learning, which in turn yielded improved sensitivity in its diagnostic capabilities. This finding underscores the utility of PUCAS for UC diagnosis in older patients and those with haematuria. The decision clinical analysis curves of PUCAS in the retrospective and prospective validation cohorts are shown in the Appendix (pp 25–26).
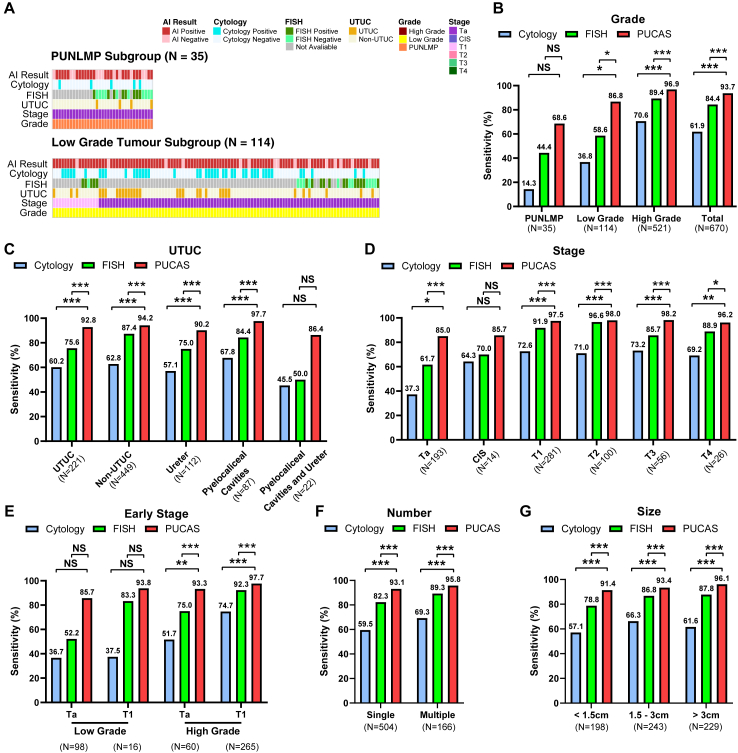
To compare the performances of PUCAS, urine cytology, and FISH, 670 patients with UC in the retrospective validation cohorts were included in the analysis. The landscape of clinical characteristics and the diagnostic status of the three methods for PUNLMP, and low-grade tumours are shown in Fig. 3A. In PUNLMP, PUCAS had a higher sensitivity (68.6%) than cytology (14.3%) and FISH (44.4%). Furthermore, in low-grade tumours, PUCAS had a significantly higher sensitivity (86.8%) than cytology (36.8%) and FISH (58.6%) (Fig. 3B). In UTUC, PUCAS achieved an improvement in sensitivity compared to cytology and FISH, regardless of the position (Fig. 3C). Regarding the cancer stage, PUCAS showed significantly higher sensitivity than cytology and FISH in the Ta and T1 stages (Fig. 3D). In pTa and low-grade (TaLG) UC, the sensitivity of PUCAS (85.7%) was 2-fold higher than that of cytology (36.7%) and 1.5-fold higher than that of FISH (52.2%) (Fig. 3E). The advantage of PUCAS was consistent in single-occurrence and minimal (<1.5 cm) tumours (Fig. 3F and G).
Fig. 3.
Sensitivity of PUCAS in the diagnosis of UC in comparison with urine cytology and FISH. (A) Distribution of PUCAS prediction results across patients with PUNLMP, and low-grade UC with associated tumour stages and grades, and cytology and FISH results in retrospective validation cohorts. (B–G) Sensitivity of PUCAS, urine cytology, and FISH in indicated grades (B), UTUC (C), stage (D), early-stage (E), number (F), and size (G) subgroups of retrospective validation cohorts. The comparison of sensitivity was assessed by McNemar test. ∗P < 0.050, ∗∗P < 0.010, ∗∗∗P < 0.0010. PUCAS, The Precision Urine Cytology AI Solution; UC, urothelial carcinoma; FISH, fluorescence in situ hybridisation; AI, artificial intelligence; UTUC, upper-tract urothelial carcinoma; PUNLMP, papillary urothelial neoplasm of low malignant potential; CIS, carcinoma in situ; NS, not significant.
To investigate the application of PUCAS in detecting residual tumours and monitoring the recurrence of UC, 127 (5.4%) patients in the secondary resection subgroup and 240 (10.3%) patients in the recurrence detection subgroup of the retrospective validation cohorts were included in the analysis (Fig. 4A). In secondary resection subgroup, PUCAS showed both higher sensitivity (96.6%) and NPV (98.9%) compared with cytology and FISH (Fig. 4B). PUCAS may serve as an accurate method to detect residual tumours and help select patients for re-TURBT. In the recurrence detection subgroup, we found that PUCAS had higher sensitivity (94.8%) and NPV (96.4%) than cytology and FISH (Fig. 4C). If the PUCAS-predicted positive status was referred for endoscopic examination, 57.5% of the endoscopies could be avoided, with an NPV of 96.4%. Furthermore, in recurrence detection, PUCAS showed improved sensitivity over cytology and FISH, especially in PUNLMP (71.4% vs. 14.3% vs. 50.0%, respectively), low-grade (100.0% vs. 57.1% vs. 40.0%, respectively), stage < T2 (93.8% vs. 60.5% vs. 73.9%, respectively), and minimal tumours (97.6% vs. 61.0% vs. 72.4%, respectively) (Fig. 4D–F). The NRIs and IDIs of PUCAS compared with the other two methods in retrospective and prospective validation cohorts are shown in the Appendix (p 13).
Fig. 4.
Performance of PUCAS in detecting residual tumours and monitoring recurrence. (A) Distribution of PUCAS prediction results across patients in secondary resection and recurrence detection subgroups with associated pathology, tumour stages, grades, UTUC, and cytology and FISH results in retrospective validation cohorts. (B and C) Sensitivity and NPV of PUCAS in detecting UC in secondary resection (B) and recurrence detection (C) subgroups. (D–F) The sensitivity of PUCAS for the indicated grades (D), stages (E), and sizes (F) in recurrence detection subgroups of retrospective validation cohorts. The comparison of sensitivity and NPV was assessed by McNemar test. ∗P < 0.050, ∗∗P < 0.010, ∗∗∗P < 0.0010. PUCAS, the Precision Urine Cytology AI Solution; UC, urothelial carcinoma; AI, artificial intelligence; FISH, fluorescence in situ hybridisation; UTUC, upper-tract urothelial carcinoma; PUNLMP, papillary urothelial neoplasm of low malignant potential; CIS, carcinoma in situ; NS, not significant; NPV, negative predictive value.
In the discordant cohort, we found that PUCAS identified 105 (37.0%) UCs correctly. With the suspicious cells location function of the interactive system, PUCAS helped cytopathologists identify 1 HGUC (0.4%) and 15 AUCs (5.3%) missed before (Appendix p 14). The overall sensitivity of PUCAS (0.370, 95% CI: 0.313–0.429) in identifying UC in the discordant cohort was significantly higher than that of FISH (0.148, 95% CI: 0.100–0.207). This advantage was consistent in the subgroup analysis, especially in PUNLMP, low-grade, Ta, and TaLG UC (Appendix p 14). Furthermore, we found that PUCAS helped to detect seven UCs that were difficult to detect in the clinic, and these presented as NHGUC, FISH negative, computed tomography (CT) or magnetic resonance imaging (MRI) negative, and ultrasound negative (Appendix p 15). A hard-to-detect example is presented in the Appendix (p 27).
To interpret the PUCAS, we visualised the predictive results using heat maps based on cytology. Examples of HGUC and AUC and associations with their corresponding FISH and histopathology results are shown in the Appendix (pp 28–29). There was a high consistency between the AI-predicted areas and suspicious cells; most areas were located in the nucleus, cytoplasm, and some background area around suspicious cells.
The coefficients of multi-modal models are shown in Appendix (p 16). The ROC curves of multi-modal models are shown in Appendix (p 30). The results showed that the multi-modal model that included clinical factors had improved the AUROC than PUCAS alone in each cohort.
Discussion
Herein, we developed and validated an AI-based model for UC detection with urine cytology using a fully automated approach in a large cohort. Utilising a multi-stage framework and weighted output from multiple models, the PUCAS achieved good diagnostic performance in retrospective and prospective validation cohorts. In most tumour subgroup analyses, PUCAS showed higher sensitivity than cytology or FISH. Particularly notable were its performance in TaLG tumours, minimal tumours, UTUC, residual, and recurrent tumours.
The primary limitation of urine cytology is its low sensitivity, which often results in the misdiagnosis of low-grade, early-stage, and minimal tumours2; it ranges from 0.0% to 10.0% in PUNLMP and varies from 16.0% to 56.0% in low-grade tumours.5,32 These findings may be because PUNLMP has similar morphological characteristics as that of normal urothelial cells and it is difficult for cells of low-grade tumours and PUNLMP to exfoliate. Our PUCAS model enhanced the sensitivity of urine cytology for PUNLMP and low-grade and TaLG tumours in the validation and discordant cohorts. This can be explained by the fact that AI can identify subtle features by analysing extensive image data. The enhanced sensitivity of PUCAS is beneficial for early diagnosis and enables accurate screening of UC in both haematuria and non-haematuria cohorts.2 Moreover, PUCAS can serve as a warning signal when other diagnostic methods give negative results, particularly in cases of equivocal endoscopy results where false-negatives for carcinoma in situ are possible.33
PUCAS also showed improved sensitivity in detecting residual and recurrent tumours in secondary resection and recurrence detection scenarios. Secondary resection is recommended for T1 stage and incompletely resected tumours, because it can increase recurrence-free and progression-free survival.2,34 A systematic review reported a 56% risk of residual tumours and a 10% risk of upstaging to T2 after secondary resection of T1 bladder cancer.35 Herein, PUCAS could accurately detect residual tumours with a high sensitivity of 96.6%, and rule out blind secondary TURBT with an NPV of 98.9%. Given the high rate of recurrence and progression of UC, guidelines recommend repeat endoscopy every 3, 6, or 12 months for different risk tumours.2 However, this frequent surveillance schedule can cause significant discomfort and impose a substantial burden on patients. PUCAS, a preferable non-invasive tool, can accurately identify recurrent UC with a sensitivity of 94.8%; it can prevent endoscopy use by 57.5% with an NPV of 96.4%. Compared with the traditional diagnostic process, PUCAS can alleviate the burden on patients while minimising the risk of misdiagnosis.
According to guidelines, the diagnosis of UTUC relies mainly on radiology, cytological examination, and ureterorenoscopy biopsy.3 Due to anatomical constraints, voided urine cytology exhibits relatively limited diagnostic effectiveness compared with upper tract washing cytology.36 It serves as a supplementary test when lesions are not visible on radiology or ureterorenoscopy; however, ureterorenoscopy can be omitted when a clear lesion appears on a CT/MRI scan alongside a positive urine cytology, potentially allowing for direct radical surgery.7 Thus, the improved sensitivity of urine cytology aids in the accurate identification of UTUC and concurrently reduces the need for invasive examinations. Our results showed that PUCAS had a high sensitivity for UTUC (92.8%), which was significantly superior to cytology (60.2%) and FISH (75.6%), highlighting the potential for improved clinical decision-making.
The reported malignant rate in AUC ranges from 24% to 53%; AUC can also be caused by urolithiasis, hyperplasia, or intravesical therapy.13 This variability of malignancy risk makes AUC a dilemma for clinicians, resulting in a significant number of unnecessary endoscopic examinations that offer no additional benefits in haematuria investigation.37 TPS use and expert knowledge can reduce the reporting rate of AUC; however, in essence, it does not reduce the false-negative results for urine cytology. Thus, an accurate classification system for malignancy based on the AUC is clinically valuable. Our findings show that the sensitivity and AUROC of PUCAS exceeded those of FISH in retrospective and prospective validation cohorts. Based only on cytological images, PUCAS can aid in identifying high-risk patients in the AUC subgroup for further evaluation.
Notably, our study is the first to compare the AI model with FISH.8, 9, 10 Per guidelines, FISH is preferred over other diagnostic tests, such as uCyt1 or Cxbladder. Compared to cytology, FISH can detect genetic alterations, exhibit more straightforward biological interpretive features, and has higher sensitivity.5 Our results demonstrated that PUCAS had a higher sensitivity than FISH in terms of the AUC subgroup and tumour subgroups, implying that PUCAS can serve as a supplementary tool or an effective alternative to FISH, as it offers higher sensitivity and can be evaluated using a cloud-based system.
Another advantage of the model is its interpretability and automatability. Regarding feature extraction, our framework employs a multi-level approach that captures features at both the patch and cell levels. This comprehensive extraction process encompasses not only relative information from the background contrast, such as size, colour, and brightness, but also fine-grained features of the cells, including hyperchromatic, coarse chromatin, and irregular chromatinic rims. Furthermore, our three-model design simulates the consensus process of voting opinions of multiple experts, thereby enhancing classification efficiency and accuracy. For automatability, cytopathologists only need to place the urine cytology sections into the slide scanner, and the scanner will complete the scan and upload the WSI images into a cloud-based system. PUCAS helps locate atypical or malignant cells based on WSI and generates a score for UC detection, which reduces repetitive and time-consuming tasks for cytopathologists.
This study has some limitations. First, the usage of PUCAS still relied on image scanning and digitization, which is still a time-consuming and cost barriers to real-world use. Our further goal is to integrate the model with microscope and realize real-time diagnosis. Second, the PUCAS was mainly built using cytological images alone, without considering other multi-modal data such as epidemiological factors, additional laboratory tests, and radiology. As a result, PUCAS cannot fully replicate the multi-modal diagnostic process in clinical settings, leading to increased misdiagnoses of PUNLMP and low-grade tumours. In future studies, we plan to build a multi-modal prediction model based on both urine cytology and other multi-modal data, particularly radiological data, to compensate for this limitation. Third, this study was mainly conducted in China and further validation of its generalisability to other countries and regions is necessary.
In conclusion, based on a large cohort that included different clinical scenarios, we developed an automatic AI model to diagnose UC with improved sensitivity. PUCAS can help reduce misdiagnoses, avoid unnecessary endoscopy, and reduce the burden on both cytopathologists and patients in resource-limited areas.
Contributors
TL, SW, RS, GH, YL and JH conceived and designed the study. RS, GH, HW, ZC, FJ, YW, CL, XL, BL, PQ, and YW did the data collection. RS, NO, HW, and YX screened patients and images. YL, and HW curated the pathological examinations. RS, CL, JF, XH and KL trained and developed the AI model. SW, RS, GH, YW, and NO did the data analysis, data interpretation, and wrote the original draft manuscript. TL, SW, and RS accessed and verified the data. TL provided study supervision. All authors reviewed the manuscript, approved the submitted version, had full access to all the data reported in the study, and had final responsibility for the decision to submit for publication.
Data sharing statement
See Appendix (p 17). To protect patient privacy, image datasets and other patient-related data are not publicly accessible, but all data are available upon reasonable request emailed to the corresponding author. To gain access, data requestors will need to sign a data-access agreement.
Declaration of interests
All authors declare no competing interests.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (grant numbers U21A20383 and 82003151), Key Program of the National Natural Science Foundation of China (82341018), the National Science Foundation for Distinguished Young Scholars (81825016), the Science and Technology Planning Project of Guangdong Province (2023B1212060013 and 2018B010109006), the National Key Research and Development Programme of China (2018YFA0902803), and the Guangdong Provincial Clinical Research Centre for Urological Diseases (2020B1111170006).
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.eclinm.2024.102566.
Appendix ASupplementary data
References
- 1.Siegel R.L., Miller K.D., Wagle N.S., Jemal A. Cancer statistics, 2023. CA Cancer J Clin. 2023;73(1):17–48. doi: 10.3322/caac.21763. [DOI] [PubMed] [Google Scholar]
- 2.Babjuk M., Burger M., Capoun O., et al. European association of urology guidelines on non-muscle-invasive bladder cancer (Ta, T1, and carcinoma in situ) Eur Urol. 2022;81(1):75–94. doi: 10.1016/j.eururo.2021.08.010. [DOI] [PubMed] [Google Scholar]
- 3.Rouprêt M., Seisen T., Birtle A.J., et al. European association of urology guidelines on upper urinary tract urothelial carcinoma: 2023 update. Eur Urol. 2023;84(1):49–64. doi: 10.1016/j.eururo.2023.03.013. [DOI] [PubMed] [Google Scholar]
- 4.Jubber I., Ong S., Bukavina L., et al. Epidemiology of bladder cancer in 2023: a systematic review of risk factors. Eur Urol. 2023;84(2):176–190. doi: 10.1016/j.eururo.2023.03.029. [DOI] [PubMed] [Google Scholar]
- 5.Chen X., Zhang J., Ruan W., et al. Urine DNA methylation assay enables early detection and recurrence monitoring for bladder cancer. J Clin Invest. 2020;130(12):6278–6289. doi: 10.1172/JCI139597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Konety B., Shore N., Kader A.K., et al. Evaluation of cxbladder and adjudication of atypical cytology and equivocal cystoscopy. Eur Urol. 2019;76(2):238–243. doi: 10.1016/j.eururo.2019.04.035. [DOI] [PubMed] [Google Scholar]
- 7.Shvero A., Hubosky S.G. Management of upper tract urothelial carcinoma. Curr Oncol Rep. 2022;24(5):611–619. doi: 10.1007/s11912-021-01179-8. [DOI] [PubMed] [Google Scholar]
- 8.Nojima S., Terayama K., Shimoura S., et al. A deep learning system to diagnose the malignant potential of urothelial carcinoma cells in cytology specimens. Cancer Cytopathol. 2021;129(12):984–995. doi: 10.1002/cncy.22443. [DOI] [PubMed] [Google Scholar]
- 9.Liu Y., Jin S., Shen Q., et al. A deep learning system to predict the histopathological results from urine cytopathological images. Front Oncol. 2022;12 doi: 10.3389/fonc.2022.901586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lebret T., Pignot G., Colombel M., et al. Artificial intelligence to improve cytology performances in bladder carcinoma detection: results of the VisioCyt test. BJU Int. 2022;129(3):356–363. doi: 10.1111/bju.15382. [DOI] [PubMed] [Google Scholar]
- 11.Lenis A.T., Lec P.M., Chamie K., Mshs M.D. Bladder cancer: a review. JAMA. 2020;324(19):1980–1991. doi: 10.1001/jama.2020.17598. [DOI] [PubMed] [Google Scholar]
- 12.Bakkar R., Mirocha J., Fan X., et al. Impact of the Paris system for reporting urine cytopathology on predictive values of the equivocal diagnostic categories and interobserver agreement. CytoJournal. 2019;16:21. doi: 10.4103/cytojournal.cytojournal_30_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wojcik E.M., Kurtycz D.F.I., Rosenthal D.L. We’ll always have Paris the Paris system for reporting urinary cytology 2022. J Am Soc Cytopathol. 2022;11(2):62–66. doi: 10.1016/j.jasc.2021.12.003. [DOI] [PubMed] [Google Scholar]
- 14.Barkan G.A., Wojcik E.M., Nayar R., et al. The Paris System for reporting urinary cytology: the quest to develop a standardized terminology. Acta Cytol. 2016;60(3):185–197. doi: 10.1159/000446270. [DOI] [PubMed] [Google Scholar]
- 15.Haug C.J., Drazen J.M. Artificial intelligence and machine learning in clinical medicine, 2023. N Engl J Med. 2023;388(13):1201–1208. doi: 10.1056/NEJMra2302038. [DOI] [PubMed] [Google Scholar]
- 16.Wu S., Hong G., Xu A., et al. Artificial intelligence-based model for lymph node metastases detection on whole slide images in bladder cancer: a retrospective, multicentre, diagnostic study. Lancet Oncol. 2023;24(4):360–370. doi: 10.1016/S1470-2045(23)00061-X. [DOI] [PubMed] [Google Scholar]
- 17.Lång K., Josefsson V., Larsson A.M., et al. Artificial intelligence-supported screen reading versus standard double reading in the Mammography Screening with Artificial Intelligence trial (MASAI): a clinical safety analysis of a randomised, controlled, non-inferiority, single-blinded, screening accuracy study. Lancet Oncol. 2023;24(8):936–944. doi: 10.1016/S1470-2045(23)00298-X. [DOI] [PubMed] [Google Scholar]
- 18.Yang Y., Cairang Y., Jiang T., et al. Ultrasound identification of hepatic echinococcosis using a deep convolutional neural network model in China: a retrospective, large-scale, multicentre, diagnostic accuracy study. Lancet Digital Health. 2023;5(8):e503–e514. doi: 10.1016/S2589-7500(23)00091-2. [DOI] [PubMed] [Google Scholar]
- 19.Cheng S., Liu S., Yu J., et al. Robust whole slide image analysis for cervical cancer screening using deep learning. Nat Commun. 2021;12(1):5639. doi: 10.1038/s41467-021-25296-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao Y., Xin L., Lin H., et al. Machine learning-based automated sponge cytology for screening of oesophageal squamous cell carcinoma and adenocarcinoma of the oesophagogastric junction: a nationwide, multicohort, prospective study. Lancet Gastroenterol Hepatol. 2023;8(5):432–445. doi: 10.1016/S2468-1253(23)00004-3. [DOI] [PubMed] [Google Scholar]
- 21.Kaneko M., Tsuji K., Masuda K., et al. Urine cell image recognition using a deep-learning model for an automated slide evaluation system. BJU Int. 2022;130(2):235–243. doi: 10.1111/bju.15518. [DOI] [PubMed] [Google Scholar]
- 22.Vaickus L.J., Suriawinata A.A., Wei J.W., Liu X. Automating the Paris System for urine cytopathology-A hybrid deep-learning and morphometric approach. Cancer Cytopathol. 2019;127(2):98–115. doi: 10.1002/cncy.22099. [DOI] [PubMed] [Google Scholar]
- 23.Lebret T., Paoletti X., Pignot G., et al. Artificial intelligence to improve cytology performance in urothelial carcinoma diagnosis: results from validation phase of the French, multicenter, prospective VISIOCYT1 trial. World J Urol. 2023;41(9):2381–2388. doi: 10.1007/s00345-023-04519-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tsuji K., Kaneko M., Harada Y., et al. A fully automated artificial intelligence system to assist pathologists’ diagnosis to predict histologically high-grade urothelial carcinoma from digitized urine cytology slides using deep learning. Eur Urol Oncol. 2023 doi: 10.1016/j.euo.2023.11.009. [DOI] [PubMed] [Google Scholar]
- 25.Wang C.-Y., Bochkovskiy A., Liao H.-Y.M. 2023. YOLOv7: trainable bag-of-freebies sets new state-of-the-art for real-time object detectors; pp. 7464–7475. (Proceedings of the IEEE/CVF conference on computer vision and pattern recognition; 2023). [Google Scholar]
- 26.Tan M.X., Le Q.V. Jmlr-Journal Machine Learning Research; Long Beach, CA: 2019. EfficientNet: rethinking model scaling for convolutional neural networks. (36th international conference on machine learning (ICML); 2019 Jun 09-15). [Google Scholar]
- 27.Liu Z., Mao H., Wu C.-Y., Feichtenhofer C., Darrell T., Xie S. 2022. A convnet for the 2020s; pp. 11976–11986. (Proceedings of the IEEE/CVF conference on computer vision and pattern recognition; 2022). [Google Scholar]
- 28.Wei S.J., Qu Q.Z., Zeng X.F., Liang J.D., Shi J., Zhang X.L. Self-attention Bi-LSTM networks for radar signal modulation recognition. IEEE Trans Microw Theory Tech. 2021;69(11):5160–5172. [Google Scholar]
- 29.Vaswani A., Shazeer N., Parmar N., et al. Neural Information Processing Systems (NIPS); Long Beach, CA: 2017. Attention is all you need. (31st annual conference on neural information processing systems (NIPS); 2017 dec 04-09). [Google Scholar]
- 30.Ilse M., Tomczak J., Welling M. PMLR; 2018. Attention-based deep multiple instance learning; pp. 2127–2136. (International conference on machine learning; 2018). [Google Scholar]
- 31.DeLong E.R., DeLong D.M., Clarke-Pearson D.L. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44(3):837–845. [PubMed] [Google Scholar]
- 32.Shefer H.K., Masarwe I., Bejar J., et al. Performance of CellDetect for detection of bladder cancer: comparison with urine cytology and UroVysion. Urol Oncol. 2023;41(6):296.e1–296.e8. doi: 10.1016/j.urolonc.2022.12.012. [DOI] [PubMed] [Google Scholar]
- 33.Wu S., Chen X., Pan J., et al. An artificial intelligence system for the detection of bladder cancer via cystoscopy: a multicenter diagnostic study. J Natl Cancer Inst. 2022;114(2):220–227. doi: 10.1093/jnci/djab179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gontero P., Sylvester R., Pisano F., et al. The impact of re-transurethral resection on clinical outcomes in a large multicentre cohort of patients with T1 high-grade/Grade 3 bladder cancer treated with bacille Calmette-Guérin. BJU Int. 2016;118(1):44–52. doi: 10.1111/bju.13354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Naselli A., Hurle R., Paparella S., et al. Role of restaging transurethral resection for T1 non-muscle invasive bladder cancer: a systematic review and meta-analysis. Eur Urol Focus. 2018;4(4):558–567. doi: 10.1016/j.euf.2016.12.011. [DOI] [PubMed] [Google Scholar]
- 36.Białek Ł., Bilski K., Dobruch J., et al. Non-invasive biomarkers in the diagnosis of upper urinary tract urothelial carcinoma-A systematic review. Cancers. 2022;14(6):1520. doi: 10.3390/cancers14061520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tan W.S., Sarpong R., Khetrapal P., et al. Does urinary cytology have a role in haematuria investigations? BJU Int. 2019;123(1):74–81. doi: 10.1111/bju.14459. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.