Summary
Iberdomide is a next-generation cereblon (CRBN)-modulating agent in the clinical development in multiple myeloma (MM). The analysis of biomarker samples from relapsed/refractory patients enrolled in CC-220-MM-001 (ClinicalTrials.gov: NCT02773030), a phase 1/2 study, shows that iberdomide treatment induces significant target substrate degradation in tumors, including in immunomodulatory agent (IMiD)-refractory patients or those with low CRBN levels. Additionally, some patients with CRBN genetic dysregulation who responded to iberdomide have a similar median progression-free survival (PFS) (10.9 months) and duration of response (DOR) (9.5 months) to those without CRBN dysregulation (11.2 month PFS, 9.4 month DOR). Iberdomide treatment promotes a cyclical pattern of immune stimulation without causing exhaustion, inducing a functional shift in T cells toward an activated/effector memory phenotype, including in triple-class refractory patients and those receiving IMiDs as a last line of therapy. This analysis demonstrates that iberdomide’s clinical mechanisms of action are driven by both its cell-autonomous effects overcoming CRBN dysregulation in MM cells, and potent immune stimulation that augments anti-tumor immunity.
Keywords: iberdomide, cereblon, myeloma, IMiD refractory, CC-220, immune stimulatory
Graphical abstract

Highlights
-
•
Iberdomide is pharmacodynamically active in late-line myeloma post-IMiDs
-
•
Iberdomide can overcome cereblon dysregulation associated with IMiD resistance
-
•
Iberdomide stimulates immune cell proliferation
-
•
Iberdomide shifts immunophenotype toward activated/effector memory T cells
Amatangelo et al. provide a biomarker analysis of myeloma patient samples from the phase 1/2 trial of iberdomide, a next-generation immunomodulatory drug. This work focuses on how iberdomide functions in tumor and immune cells and provides scientific insight for further clinical development.
Introduction
Iberdomide (CC-220) is a new cereblon (CRBN) E3 ligase modulator (CELMoD) that induces the ubiquitination and degradation of the ligase substrates Aiolos and Ikaros and is being investigated in patients with multiple myeloma (MM).1 Iberdomide binds CRBN with unique interactions and >20-fold higher affinity compared to the previous CRBN-targeting immunomodulatory agents (IMiDs) lenalidomide and pomalidomide.2 These interactions induce more efficient allosteric rearrangement of CRBN to a conformation required for Aiolos/Ikaros binding, promoting faster and deeper degradation of substrates and enhanced anti-proliferative and pro-apoptotic activity compared to IMiDs.3,4,5,6,7 Additionally, iberdomide has demonstrated more potent immunostimulatory effects and anti-myeloma activity in pre-clinical models of lenalidomide and pomalidomide resistance.3,4,5,6,7,8,9,10
Patients treated with anti-myeloma therapies including IMiDs ultimately become resistant and progress.11 Resistance to IMiDs due to reduced expression of CRBN or loss of CRBN E3 ligase activity (e.g., loss of COP9 signalosome activity) has been previously described.12,13,14,15,16,17,18 Immunohistochemical analysis of paired tumor samples demonstrated that 77% of patients had a significant reduction in CRBN protein upon the development of lenalidomide resistance vs. diagnosis.16 Recently, a systematic evaluation of relapsed and refractory MM (RRMM) patient tumor genomes characterized CRBN mutations, copy loss, and expression of a CRBN exon10 splice variant (CRBN-del-exon10), whose prevalence increased with repeated IMiD treatment and was associated with poor outcome.12 Similarly, copy loss of the chromosome 2q37 locus was also identified as a mechanism of IMiD resistance in RRMM due to the loss of two genes (COPS7b, COPS8) of the COP9 signalosome, whose function is essential for the maintenance of the CUL4-DDB1-CRBN E3 ubiquitin ligase.19 Together, these changes represent a functional loss of CRBN activity that we designate as a “CRBN dysregulation” that appears to be one of the major sources of therapy-related IMiD resistance. Additionally, molecular disease features, including gain/amplification of 1q [Amp1q], double hit events, high-risk (HR) del17p (cancer clonal fraction [CCF] ≥ 0.55), and HR t(4;14), may also indirectly contribute to therapeutic resistance, including for IMiDs.20,21,22,23
The transcription factors Aiolos and Ikaros are critical for MM cell proliferation and survival and play important roles in regulating immune cell development and homeostasis.24,25 Their targeted degradation results in broad immunomodulatory activities that are observed following lenalidomide and pomalidomide treatment, including increased activation of T and natural killer cells leading to enhanced cytotoxicity, increased secretion of interleukin-2 and interferon-gamma by T cells, and inhibition of pro-inflammatory cytokines (e.g., tumor necrosis factor-α, interleukin-6) by monocytes.10,26,27,28,29 Notably, treatment-induced immune changes are also associated with response and development of resistance to IMiDs. In a study of lenalidomide in patients with RRMM, failure to achieve a very good partial response (VGPR) or greater was associated with the decreased frequency of CD8+ T cells on treatment,30 and the clinical response to pomalidomide+dexamethasone was significantly associated with the increased activation of CD8+ T cells.26 Key translational questions for iberdomide are whether and how it functions in patients with MM and overcomes clinical IMiD resistance and to identify its molecular anti-tumor and immunostimulatory mechanisms driving clinical activity.
Here, we report the analysis of molecular and pharmacodynamic biomarker data from a subset of patients in an ongoing phase 1/2 study, CC-220-MM-001 (ClinicalTrials.gov: NCT02773030), evaluating iberdomide in RRMM. Our analysis focused on the baseline and on-treatment immunophenotypic characterization of peripheral blood (PB) immune cells and immunohistochemistry and genomic analyses from bone marrow (BM) tumor cells. These data demonstrate iberdomide’s broad pharmacodynamic activity in tumor and immune cells in lenalidomide-, pomalidomide-, and triple-class-refractory (defined as refractory to ≥1 IMiD, ≥1 proteasome inhibitor, and ≥1 anti-CD38 antibody) patients and provides rationale for its combination with other immune-based MM therapies. More specifically, we provide evidence of iberdomide’s ability to overcome IMiD resistance attributed to CRBN dysregulation, revealing its potency and suggesting a dual mechanism of clinical activity that involves direct effects on MM cells and immune stimulation.
Results
Pre-treatment immune and tumor profiles
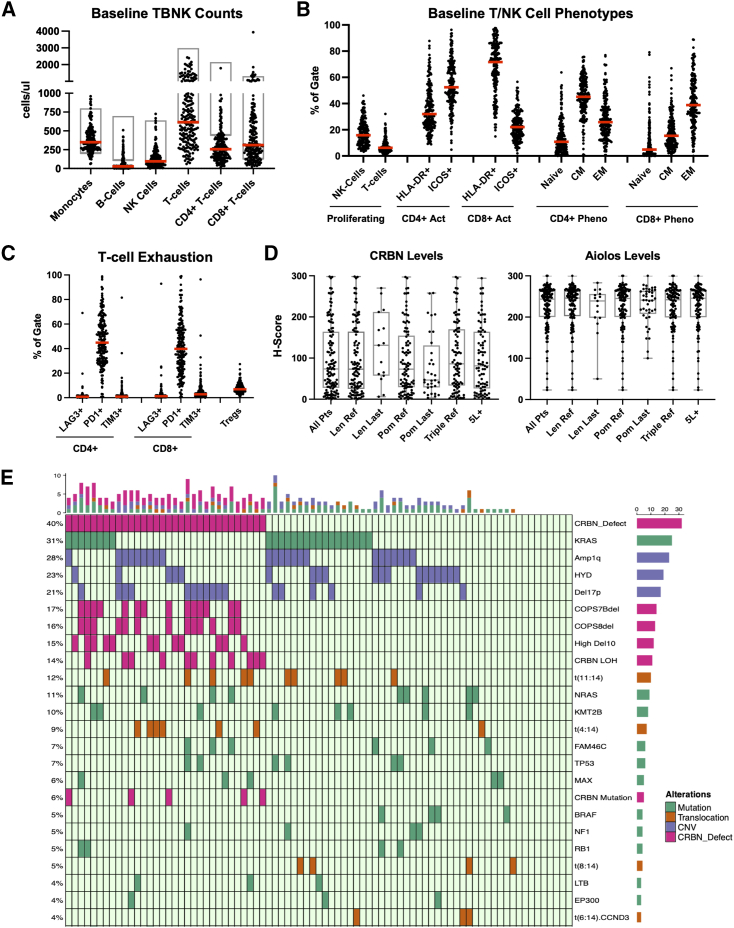
Patient demographics of the biomarker subsets and the intent-to-treat population previously published were comparable, including ∼80% of patients receiving prior stem cell transplant, ∼75% daratumumab exposed, and >95% refractory to an IMiD (Figure S1; Table S1).1 Analysis of RRMM patient pre-treatment baseline immunophenotypes showed that the median absolute counts of several immune cell types were below the lower limit of normal (LLN) reference range for healthy individuals per the reporting laboratory, including B cells (CD3−CD19+), NK cells (CD3−CD56+/CD16+), total T cells (CD3+), and CD4+ T cells (CD3+CD4+) (median: 27, 95, 611, and 256 cells/μL, respectively) (Figure 1A). The median number of CD4+ T cells was approximately half of the LLN (500 cells/μL), and median ratio of CD4+ to CD8+ T cells was ∼0.75. Phenotypic assessment of proliferating (Ki67+) NK cells, CD3+ T cells, activated T cells, and memory T cells showed that these patients also exhibited evidence of immune stimulation at enrollment, with a high relative proportion of T cells expressing the activation marker HLA-DR (71.7% CD8+ cells, 32% CD4+ cells) and exhibiting a memory phenotype with CD4+ T cells skewed toward a central memory (CM) phenotype (median: 45.1%) and CD8+ T cells skewed toward an effector memory (EM) phenotype (median: 38.5%) (Figure 1B). Conversely, a low proportion of pre-treatment T cells were observed to be in a proliferative state (median: 6.2% of CD3+ cells) despite evidence of activation, suggesting immune dysfunction. An analysis of regulatory T cells (Tregs) and T cell exhaustion markers, specifically Lag-3 (CD223), PD-1 (CD279), and Tim-3 (CD366), showed that a high percentage of both CD4+ and CD8+ cells expressed PD-1 (median: 43.9% and 39.8%, respectively) and that 6.9% of CD4+ T cells expressed a regulatory phenotype (Figure 1C). In contrast, an analysis of patients with newly diagnosed MM (NDMM) from the same study showed a significantly less immunodeficient profile at baseline (Figure S2; supplemental information). While patients with RRMM had similar CD8+ T cell profiles, they showed significantly less total CD4+ T cells and a lower proportion of naive CD4+ T cells (p < 0.001) and a significantly higher proportion of PD-1+ CD4+ T cells (p = 0.002) and CD4+ Tregs (p = 0.005) vs. NDMM subjects. These data illustrate that the patients with late-line RRMM enrolled in this study had appreciable immunosuppression, suggestive of exhaustion (low Ki-67/high PD-1), with particular dysfunction in the CD4+ helper T cell compartment when compared to healthy individuals or patients with NDMM.31,32
Figure 1.
Analysis of baseline immune and tumor characteristics of subjects enrolled in cohorts A, B, and D of the CC-220-MM-001 study
(A) Dot plot of absolute (ABS) cells per μL in peripheral blood of subjects on cycle 1 day 1 (C1D1) for monocytes (CD14+), B cells (CD3−CD19+), NK cells (CD3−CD56+/CD16+), T cells (CD3+), CD4+ T cells (CD3+CD4+), and CD8+ T cells (CD3+CD8+). Each dot represents one subject, the red line represents the median, and the gray box represents normal laboratory ranges for each assessment.
(B) Dot plot of proportion of cells from indicated lineage positive for phenotypic markers for subjects on C1D1, including proportions of NK and T cells proliferating (Ki-67+), CD4+ T cells and CD8+ T cells activated (HLA-DR+ or ICOS+), CD4+ T cells and CD8+ T cells in naive state (CD45RO−CCR7+), CD4+ T cells and CD8+ T cells in central memory (CM) state (CD45RO+CCR7+), and CD4+ T cells and CD8+ T cells in effector memory (EM) state (CD45RO+CCR7−). Each dot represents one subject, and the red line represents the median.
(C) Dot plot of proportion of cells from indicated lineage positive for exhaustion markers for subjects on C1D1, including proportion of CD4+ and CD8+ T cells expressing LAG-3 (CD223), PD-1 (CD279), and Tim-3 (CD366) and proportion of CD4+ Tregs (CD25+CD127−/loFoxP3+). Each dot represents one subject, and the red line represents the median.
(D) Box and whisker plots of CD138+ involved H-score for total CRBN expression and nuclear Aiolos expression in indicated patient subsets. Each dot represents a single patient value, the line represents the median, the top and bottom of the box represent the 25th and 75th percentiles, respectively, and whiskers represent the minimum and maximum. Data are shown for patients who had lenalidomide or pomalidomide (POM) in their last line (Len last, Pom last), who were refractory to Pom (Pom R), who were triple-class refractory (defined as refractory to ≥1 immunomodulatory agent, ≥1 proteasome inhibitor, and ≥1 anti-CD38 antibody), or who were fifth line or later (5L+).
(E) Oncoplot of most prevalent mutations (green), copy-number aberrations (blue) and translocations (orange), or CRBN dysregulation (CRBN mutation, CRBN loss of heterozygosity, high expression of CRBN-del-exon10, or 2q [COPS7b/COPS8] deletion) at baseline in patients who had whole-genome sequencing (WGS) and RNA sequencing (RNA-seq) data available. Prevalence is shown on left y axis and is limited to aberrations present in ≥3% of patients (note that there were no aberrations present at 3%, so the plot shows 4% and above). High Del10 = high expression of the CRBN-del-exon10 transcript.
Immunohistochemical staining of BM biopsies (n = 114) showed that despite being refractory to one or both IMiDs, CRBN was quantifiably detectable in CD138+ cells in almost all samples (median histology score [H-score] = 74) (Figure 1D). High levels of Aiolos were also detected pre-treatment (median H-score = 243.5) (Figure 1D). Notably, CRBN and Aiolos protein levels were not significantly different in patients who were refractory to IMiDs, received an IMiD in their last line of treatment, were triple-class refractory (defined as refractory to ≥1 IMiD, ≥1 proteasome inhibitor, and ≥1 anti-CD38 antibody), or had >5 prior lines of therapy. However, a trend toward lower CRBN levels was observed in patients who received pomalidomide in a last line of treatment (Figure 1D). These data show that the molecular targets of iberdomide are generally expressed in different RRMM patient subgroups, including those treated with prior CRBN-modulating therapies.
Whole-genome sequencing plus RNA sequencing was available from 81 patients to characterize the prevalence of translocations, hyperdiploidy, copy-number aberrations (CNAs), mutations, and CRBN dysregulation events (≥3% of patients) (Figure 1E). 40% of patients had CRBN dysregulation including 14 (17%) with loss of COPS7b, 13 (16%) with loss of COPS8, 12 (15%) with high CRBN-del-exon10 expression, 11 (14%) with CRBN loss of heterozygosity (LOH), and 5 (6%) with a CRBN mutation (some patients had multiple aberrations). 23% of the 81 patients had hyperdiploidy. Among the key translocation groups, t(11;14) (12%) and t(4;14) (9%) were the most common. KRAS (31%) and NRAS (11%) were the most frequent mutations.23,33,34 Amp1q (28%) and del17p (21%) were the most common CNAs, similar to our recent report.23 A larger cohort of patients with whole-genome data only are described in Figure S3. Thus, patients entering the iberdomide study presented with enrichment of HR and CRBN-related genomic aberrations, consistent with a late-line patient population.
Pharmacodynamic activity of iberdomide and iberdomide-dexamethasone combination
Immune pharmacodynamics by dose
We analyzed longitudinal changes in immune subpopulations in patients treated with iberdomide monotherapy or in combination with dexamethasone, combining adjacent dosing groups together as one group (previously shown to have overlapping drug exposures as described).1 Consistent with the function of Aiolos and Ikaros, which are required for B cell maturation, and their known pattern of degradation following treatment with IMiDs, a greater depletion in the absolute number of mature B cells in PB was observed in subjects treated with higher doses of iberdomide (Figures 2A and S4A).35,36,37 In subjects treated with iberdomide and dexamethasone, B cells decreased by ∼25% with a 0.3 mg dose vs. an ∼80% median reduction in subjects dosed at 0.75 mg. These changes saturated above 1.0 mg (trends demonstrated significant decreases [p < 0.05] from the 0.3–1.0 mg to the 1.6 mg dose level) with a median reduction of 95.8% at the higher dose (Figures 2A, S4A, and S5). At doses ≥1.3 mg, depletion in B cells was fully sustained after a week of drug holiday (black arrow, Figure 2A), suggesting that the continued biological effect of iberdomide in B cells carried though drug holidays at the highest doses.
Figure 2.
Iberdomide immune pharmacodynamic effects
Longitudinal analysis of iberdomide-induced changes in immune cell subsets.
(A) Line graph of median percentage of change with standard error in ABS B cell (CD3−CD19+) counts from patients treated with iberdomide+dexamethasone in cohorts B and D.
(B) Box and whisker plots showing proportion of proliferating NK cells (CD3−CD56/CD16+ %Ki67+) (left) and proportion of proliferating T cells (CD3+ %Ki-67+) (right) by enrollment dose group and cohort. Adjacent doses with overlapping exposure were combined. The middle line represents the median, the top and bottom of the box represent the 25th and 75th percentiles, respectively, and whiskers represent 95% confidence interval (CI).
(C) Line graph of median percentage of change with standard error in proportion of CD4+ T cells in naive phenotype (CD45RO−CCR7+) (left), in CM phenotype (CD45RO+CCR7+) (middle), and in EM phenotype (CD45RO+CCR7−) (right) by enrollment dose group counts from patients treated with iberdomide+dexamethasone in cohorts B and D. Adjacent doses with overlapping exposure were combined. Red arrows indicate the peak of iberdomide induction of activated/EM state by mid-cycle 2/4 after 2 weeks of dosing, and blue arrows indicate changes after a 1 week drug holiday.
(D) Box and whisker plots of percentage of change from C1D1 to C2D15 for patients dosed above 0.75 mg with iberdomide+dexamethasone in cohorts B and D in proportion of CD4+ (green) and CD8+ (blue) T cells positive (left) and median fluorescence intensity (right) for activation marker HLA-DR. Each dot represents a single patient value, the line represents the median, the top and bottom of the box represent the 25th and 75th percentiles, respectively, and whiskers represent 95% CI.
(E) Box and whisker plots of percentage of change from C1D1 to C2D15 for patients dosed above 0.75 mg with iberdomide+dexamethasone in cohorts B and D in proportion of cells from indicated phenotypic markers who were naive to POM (blue) or directly post-POM (orange) (patients with POM as a last line of therapy <3 months prior to enrollment). Each dot represents a single patient value, the line represents the median, the top and bottom of the box represent the 25th and 75th percentiles, respectively, and whiskers represent 95% CI.
(F) Granzyme B levels in bone marrow aspirate plasma at screening and C2D15.
Iberdomide treatment as monotherapy and in combination with dexamethasone also resulted in an increase in the relative percentage of proliferating (Ki67+) NK and T cells, demonstrating its immunostimulatory effects and an ability to overcome immune dysfunction observed in late-line patients as well as the immunosuppressive effects of dexamethasone (Figures 2B and S4B). Although the per-dose group size was small, the median increase in NK cell proliferation in cohort B suggested a point of inflection for maximal induction of NK cell proliferation in the range from 0.9 to 1.0 mg, with a possible hook effect (decreased magnitude of change) observed at the 1.6 mg dose (Figure S5). However, this effect was not confirmed in cohort D, where 85% of patients at 1.6 mg still exhibited NK cell stimulation in the dose-expansion cohort (Figure 2B). The median increases in proportion of proliferating T cells were nearly 2× greater than that observed in NK cells and appeared to plateau at doses >1.0 mg (without a hook effect); however, limited data and high variability likely contributed to a lack of statistical significance between doses (Figures S5 and 2B). Unlike the effect of iberdomide treatment on B cells, a cyclical pattern of T cell proliferation was observed, peaking at day 15 (mid-cycle) at each cycle and returning toward pre-treatment trough levels measured pre-dose on day 1 of the next cycle at the end of the 7 day drug holiday (Figure S4C).
An analysis of T cell subsets revealed a shift from naive to an activated, EM T cell phenotype (in both CD4+ and CD8+) during iberdomide treatment with a dramatic decrease in naive T cells observed by mid-cycle 2 and a corresponding increase in EM T cells (Figures 2C and S4D–4F). Iberdomide treatment also resulted in an increased proportion of activated T cells and increased expression of HLA-DR on T cells by mid-cycle 2 (Figure 2D). This observed shift to an activated EM phenotype was greater at higher doses, increasing between 0.3 and 1.0 mg and plateauing above the 1.0 mg dose (trends demonstrated significant increases [p < 0.05] from 0.3 to 1.0 mg, but changes from 1.0 to 1.6 mg were not significantly different; Figures S4D–S4F and 5). Consistent with a cyclical pattern of T cell stimulation, during the dosing period (days 1–21 in each cycle), iberdomide promoted the activation and differentiation of T cells (peak of EM cells mid-cycle, Figure 2C, right, red arrows) and with drug holiday (days 22–28 in each cycle) the pool of T cells shifted to less differentiated CM cells (Figure 2C, middle, blue arrows). The timing of peaks and valleys of EM and activated phenotypes mirrored the opposite pattern of peaks and valleys of CM phenotypes (Figures 2C and S4D–S4F). Notably, these immunostimulatory effects of iberdomide were unaffected by prior treatment history, as potent NK and T cell proliferation and activation were observed in triple-class-refractory patients and in patients who progressed on pomalidomide or daratumumab <3 months prior to enrollment (Figures 2E and S6A–S6C). In a subset of patients treated with iberdomide (1.6 mg) and dexamethasone, a trend for increases in granzyme-B secretion was also observed in BM plasma, suggesting that iberdomide-induced immune changes shifted the BM microenvironment of the tumor to a more immune cytotoxic state (Figure 2F). A complete analysis of iberdomide-induced changes in BM immunophenotyping has been submitted as a separate manuscript (Van Oekelen et al.38 [this issue of Cell Reports Medicine]). Together, these data suggest broad and potent immune stimulation by iberdomide directly post-pomalidomide or -daratumumab treatment and in late-line patients with a likely dose-dependent ceiling of immune stimulation above the 1.0 mg dose. Remarkably, iberdomide did not push the pool of T cells into an unresponsive state, as the phenotypes appeared to reset through drug holidays and could be restimulated in the subsequent treatment cycles.
Figure 5.
Patient case studies: CRBN-dysregulated responders
(A–C) CRBN dysregulation patient case studies. Patient identification number and type of CRBN dysregulation are listed: (A) Pt. 109-1010, (B) Pt. 104-1004, and (C) Pt. 101-1026. The boxed region contains the best response, duration of response (DOR), and PFS and the CRBN immunohistochemistry (IHC) score at baseline. Protein expression (H-score) of Aiolos and/or Ikaros in the tumor at screening and on treatment from IHC (left), immune pharmacodynamic (PD) changes from baseline in B cells, CD4+ activated and EM cells from peripheral blood (middle), and immune PD changes in the bone marrow from CyTOF analysis are shown for each patient (right).
(D) Model depicting the anti-tumor (blue line) and immunomodulatory effects (red line) contributing to iberdomide mechanism of clinical activity. Myeloma cells with WT CRBN (purple), IMiD-resistant CRBN mutant/dysregulated CRBN cells (red), T cells (green), and NK cells (yellow).
Pharmacodynamic modulation of Aiolos and Ikaros
In a subset of patients (n = 10), we observed a decrease of Aiolos and Ikaros levels in PB T and B cell populations 3–6 h post-dose at the 1.6 mg dose of iberdomide (Figure S7A). In patient tumors, longitudinal analysis of CRBN levels from screening to end of treatment showed no significant changes (n = 9). While sample numbers were limited, only 1 patient showed a decrease in CRBN levels >50% (Figure 3A). To assess iberdomide’s pharmacodynamic activity on the CRBN substrates, the levels of Aiolos in tumor cells were analyzed mid-cycle 2 or 3 in 50 patients and compared to baseline levels. Aiolos levels significantly decreased following iberdomide (cohort A, 0.45–1.0 mg, p < 0.0001) or iberdomide+dexamethasone (0.3–1.6 mg, cohorts B [p < 0.0001] and D [p = 0.063]), with no clear dose-dependent trends (Figures 3B and S7B). Similar decreases in Ikaros levels were also observed (Figure S7C). Treatment with iberdomide+dexamethasone consistently reduced Aiolos levels irrespective of refractoriness to IMiDs, IMiD use in last line, or triple-class-refractory status and was similar in patients with highest vs. lowest quartile CRBN expression at baseline (Figures S7D and S7E). Together, these data demonstrate (1) continued expression of CRBN in patients with late-line MM and (2) iberdomide’s pharmacodynamic activity in patient’s tumor cells, including those who are pomalidomide- or triple-class refractory or received lenalidomide or pomalidomide in the immediate prior line of therapy. These results are especially noteworthy because a significant fraction of the patients harbor CRBN dysregulation (see the later section iberdomide response vs. genome biomarkers).
Figure 3.
Iberdomide tumor pharmacodynamic effects and response outcomes
Analysis of CRBN and Aiolos protein expression in patient tumor biopsies from screening, on treatment (C2D15 or C3D15 if C2D15 sample unavailable) and end of treatment scored for staining intensity to generate a histology score (H-score; see STAR Methods).
(A) Line graph of CRBN levels in longitudinal samples. Each dot represents one patient, with lines connecting samples from the same patient over time.
(B) Line graph of Aiolos expression in longitudinal samples. Each dot represents one patient, with lines connecting samples from the same patient over time. Cohort A (left) and cohort B (right) with doses color coded as indicated.
(C) Box and whisker plots of baseline CRBN levels in the tumor by response vs. non-response and by depth of response. Each dot represents a single patient value (colored by enrollment dose), the top and bottom of the box represent the 25th and 75th percentiles, respectively, and whiskers represent 95% CI.
(D) Line graph of Aiolos levels at screening and on treatment in responders and non-responders.
(E) Kaplan-Meier plot of PFS and relation to Aiolos degradation in cohorts B and D. See the main text for more details.
(F) Scatterplot showing cohort B and D patients’ percentage change in Aiolos, with censored data points shown as diamonds and patient response shown as plotting color. Trend is shown as the dotted black line. A log hazard ratio was determined by Wald test.
Correlative analysis of iberdomide activity with clinical response and PFS
Baseline CRBN protein levels trended lower, but were not significant, in non-responders vs. responders (p = 0.1211, Figure 3C). While higher CRBN expression appeared to correlate with the depth of response, many subjects treated with lower doses of iberdomide (<1.3 mg) had lower levels of CRBN protein (65% H-score < 50), and 47% of subjects whose best response was progressive disease were dosed with <1.3 mg, making it difficult to interpret association with response. Responders and non-responders at doses ≥1.3 mg did not show a significant difference in CRBN protein levels (Figure S8A). Baseline Aiolos levels did not correlate with clinical response (Figure S8B).
We analyzed the correlation of Aiolos degradation with progression-free survival (PFS) by levels of its degradation in patient tumors. Forty-four patients in the B and D cohorts with available data for the Aiolos level at screening and degradation at post-therapy were partitioned into two groups based on the median percentage change in Aiolos levels < or ≥ −45.98% from baseline. The curves (and 95% confidence intervals) were plotted, and differences in the two estimated Kaplan-Meier curves were found to be statistically significant (log-rank test, p = 0.01) (Figure 3E). The median PFS for the group with lesser Aiolos degradation (% change ≥ −45.98%) was estimated to be 83 days, and the median PFS for the group with greater Aiolos degradation (% change < −45.98%) was estimated to be 152 days. While an on-treatment decrease of Aiolos was consistently observed in all responders, it was also observed in a subset of non-responders. Thus, some patients showed a potent Aiolos decrease (>75%) but had shorter PFS (<200 days) (Figure 3F). In these patients, we observed a robust immune response induced by iberdomide treatment, with median increases in the proportions of proliferating and activated CD4+ T cells of 123.1% and 140.0%, respectively, a median decrease in naive CD4+ T cells of 91.4%, and a 113% increase in granzyme B (GZMB) levels in patient BM plasma (Figure S8C). We additionally stained a few samples from patients with short PFS for proteins downstream of Aiolos/Ikaros degradation, such as IRF4, and observed no downregulation in protein levels, suggesting that, as a previously described mechanism of IMiD resistance, their regulation had been disconnected from Aiolos/Ikaros degradation in these patients (Figure S8D).3 These data suggest that high levels of Aiolos degradation (over 45%) may be a potential pharmacodynamic marker for longer PFS, but it is only one of many factors contributing to clinical outcomes, especially in individual patients with lower levels of substrate degradation but longer PFS.
Supporting an immune-mediated mechanism of efficacy for iberdomide, the analysis of all patients with available immune pharmacodynamic data demonstrated that in responders, iberdomide induced significantly greater increases in proliferating CD4+ T cells (Ki-67+) (p < 0.001 for response; p = 0.002 for PFS) and CD8+ EM T cells (CD8+CD45RO+CCR7−) at cycle 2 day 15 (p = 0.016 for response; Tables 1 and S2D). However, these PD results were confounded, as the same baseline immune phenotypes were also associated with clinical response, including lower baseline levels of absolute CD4+ T cells and B cells observed in non-responders (p < 0.001 and p = 0.003, respectively; Tables 1 and S2A). An analysis of immune subpopulations showed the non-responders had higher proportions of proliferating T cells (Ki-67+), activated CD4+ T cells (HLA-DR+ and CD38+), and EM CD4+ T cells (p < 0.001, p = 0.017, and p = 0.009), suggesting that high levels of CD4+ T cell stimulation in the context of progressive disease may be a poor prognostic indicator (Tables 1 and S2B). Additionally, non-responders had a significantly higher proportion of Tregs in the CD4+ compartment vs. responders at baseline (p < 0.001; Tables 1 and S2C), suggesting that immune dysfunction in the CD4+ compartment may be an underlying feature contributing to non-response. Importantly, iberdomide was still immunostimulatory in non-responders, inducing T cell proliferation and activation, providing a rationale for its potential to enhance other immuno-oncology therapies (Figures S8E–S8G).
Table 1.
Association of baseline immunophenotypes and pharmacodynamic effects with response and PFS
| ORR p value | R LQ, MDN, UQ | NR LQ, MDN, UQ | PFS LogHR | PFS p value | |
|---|---|---|---|---|---|
| Baseline parent populations | |||||
| CD3+CD4+_ABS | 9.16E−05 | [191, 360, 454] | [135, 211, 314] | −0.00140 | 0.01086 |
| CD3+_ABS | 0.00094 | [436, 822, 1379] | [306, 528, 842] | −0.00039 | 0.06051 |
| CD3−CD19+_ABS | 0.00265 | [24, 42, 90] | [10, 24, 48] | −0.00177 | 0.10052 |
| CD3+CD8+_ABS | 0.01450 | [197, 422, 843] | [149, 298, 486] | −0.00035 | 0.12619 |
| Baseline subpopulations | |||||
| CD3+CD4+CD25+CD127−/loFoxP3+_% | 0.00020 | [4.2, 6.0, 7.8] | [5.5, 7.6, 10.9] | 0.08129 | 0.00019 |
| CD3+CD4+Ki67+_%(CD4+) | 0.00029 | [3.9, 5.1, 7.4] | [5.3, 7.7, 11.5] | 0.05868 | 5.98E−08 |
| CD3+Ki67+_%(CD3+) | 0.00038 | [3.3, 4.4, 6.4] | [4.3, 6.9, 10.5] | 0.06773 | 2.15E−08 |
| CD3+CD8+CD38+_(%CD8) | 0.00050 | [10.4, 49.8, 80.7] | [52.6, 78.4, 88.5] | 0.00880 | 0.00021 |
| CD3+CD8+Ki67+_%(CD8+) | 0.00112 | [2.3, 4.2, 6.2] | [3.6, 5.8, 10.1] | 0.06465 | 9.75E−09 |
| CD3+CD4+CD45RA-CD45RO+CCR7−_(%CD4) | 0.00870 | [12.4, 19.1, 34.1] | [20.1, 27.4, 37.2] | 0.00643 | 0.17376 |
| CD3−CD7+CD56+Ki67+_%(CD3−CD56+/CD16+) | 0.00955 | [3.7, 6.4, 10.8] | [5.2, 9.3, 16.15] | 0.01899 | 0.10075 |
| CD3+CD8+HLA-DR_APC_MFI | 0.01001 | [279.0, 780.0, 1,786.5] | [500.0, 1,514.0, 3,554.0] | 8.70E−05 | 0.02036 |
| CD3+CD4+HLA-DR+_(%CD4) | 0.01714 | [20.3, 26.0, 42.4] | [25.0, 34.6, 47.7] | 0.01132 | 0.00536 |
| CD3+CD4+CD38+_(%CD4) | 0.02026 | [10.6, 47.5, 66.4] | [36.7, 59.7, 69.4] | 0.00674 | 0.01461 |
| CD3+CD8+CD45RA−CD45RO+CCR7−_(%CD8) | 0.03261 | [21.6, 34.3, 50.0] | [30.9, 40.7, 55.2] | 0.00782 | 0.03735 |
| C2D15 pharmacodynamic effects on subpopulations | |||||
| CD3+CD4+Ki67+_%(CD4+) | 0.00251 | [84.8, 171.6, 300.0] | [31.3, 80.6, 151.4] | −0.00145 | 0.00189 |
| CD3+CD4+CD223+_(%CD4) | 0.00863 | [100.6, 274.6, 587.5] | [14.3, 108.3, 247.1] | −1.31E−05 | 0.86123 |
| CD3+CD8+CD45RA−CD45RO+CCR7−_(%CD8) | 0.01557 | [35.7, 70.4, 127.6] | [13.9, 39.16, 87.71] | −0.00051 | 0.52963 |
| CD3+CD8+_CD279_BV421_MFI | 0.02696 | [−129.3, −69.1, 2.3] | [−77.2, −19.0, 58.5] | 0.00030 | 0.09246 |
| CD3+CD8+HLA-DR+_(%CD8) | 0.05244 | [14.2, 29.2, 62.3] | [9.3, 17.6, 37.9] | −0.00387 | 0.12778 |
| CD3+CD4+ICOS+_(%CD4) | 0.10286 | [−0.6, 13.1, 45.2] | [−17.7, 5.1, 26.7] | −0.00534 | 0.00550 |
Statistical association of baseline (C1D1) and pharmacodynamic (percentage of change C2D15) immune measurements in subjects enrolled in cohorts B and D of the CC-220-MM-001 study. The table shows the p value for the indicated immune measurement association with overall response (OR) using Wilcoxon rank-sum test, lower quartile (LQ), median (MDN), and upper quartile (UQ) for responders (R) and non-responders (NR), progression-free survival (PFS) log(hazard ratio (HR)), and PFS using Wald’s test.
Iberdomide response vs. genome biomarkers
We next analyzed genomic subgroups in 82 patients where both genome and outcome data were available. Amp1q (≥4 copies, n = 21 [22%]) and P53 aberrations (del17p CCF ≥ 0.5519,21 or biallelic inactivation,20 n = 16 [19%]) were the most common HR subgroups (Figure S9; Table S3). While the numbers were small, examples of durable responses were observed in each of these HR groups (Amp1q: n = 21, 2 responders, median duration of response [mDOR] = 5.6 months; HR del17p/biallelic P53 inactivation: n = 17, 2 responders, mDOR = 7.2 months) (Figure S10). Other translocation subgroups and double hit/biallelic events were excluded from the response analysis due to small patient numbers (n < 10). Iberdomide was also active in patients addressable by targeted therapy, including those with any KRAS/NRAS/BRAF mutation (n = 37, objective response rate [ORR] 8/37 [22%], mDOR = 12.2 months) or t(11;14) (n = 13, ORR 3/13 [23%], mDOR = 8.3 months). The 5 most common genomic aberrations in responders and non-responders were the same and included CRBN dysregulation, KRAS mutation, Amp1q, hyperdiploidy, and HR del17p (Figure S11A). The median PFS across molecular subgroups was similar to the cohort as a whole (2.76–3.72 months) (Figure 4B; Table S3). These preliminary data show that iberdomide has activity across HR and targetable patient segments without an enrichment of any feature in responders or non-responders and highlights the opportunity for iberdomide combinations in these difficult-to-treat patients. Out of the six patient samples analyzed at progression, 1 non-responder patient had 2 de novo CRBN mutations detected (Figures S11B and S11C).
Figure 4.
Analysis of response to iberdomide in molecularly defined patient segments
(A) Patient-level best response data in IMiD-resistant patient segments with breakdown of type of CRBN dysregulation (copy loss, mutation, CRBN-del-exon10) and COPS deletions shown. For each bar, the total number of patients called as being positive for that segment are shown at the end of the bar (number at risk), followed by the total number of patients with available data (total subjects). The farthest set of numbers (farthest from bar) reports the percentage of patients with best response ≥ partial response (PR) and ≥ very good partial response (VGPR). The best response is color coded as shown below the bar graph.
(B) Kaplan-Meier curve for PFS in iberdomide-treated patients with CRBN dysregulation/2q loss responders (≥PR, green), CRBN dysregulation/2q loss non-responders (<PR, pink), wild-type (WT) CRBN/2q responders (blue), and WT CRBN/2q responders (purple). Summary table of number of responders (≥PR) vs. non-responders (<PR) in CRBN WT vs. dysregulated (CRBN locus or 2q abnormality) is shown.
Finally, we focused on the 34 CRBN-dysregulated subjects vs. 48 without any observable CRBN defects (designated as CRBN wild type) to understand the impact of CRBN dysregulation on iberdomide activity. The CRBN dysregulation subgroup included previously described IMiD-resistant patients based on CRBN genetic aberrations12 (CRBN-del-exon10, CRBN LOH, CRBN mutation) or loss of 2q (COPS7b/8).19 Responses to iberdomide+dexamethasone were observed in all CRBN-dysregulated subgroups at doses between 1.0 and 1.6 mg (6 responders, 28 non-responders) (Figures 4A and 4B; Tables S3 and S4). Thirteen (of 48) wild-type patients also responded, while 35/48 did not (Figures 4B; Table S5). Analysis of PFS and DOR in responders revealed similar outcomes for CRBN-dysregulated (mDOR 9.5 months, mPFS 10.9 months) vs. wild-type patients (mDOR 9.4 months, mPFS 11.2 months) (Figure 4B; Tables S3 and S4). Conversely, CRBN-dysregulated and wild-type non-responders had similarly short PFS (mPFS 1.9 vs. 2.3 months, respectively, Figure 4B). While the numbers are small, these observations suggest that in responding patients, iberdomide could overcome the adverse prognosis associated with CRBN dysregulation associated with IMiD-based therapies.12,19
Discussion
We explored the clinical mechanisms of action of iberdomide by analyzing patient biomarker samples collected from a large clinical study in MM, CC-220-MM-001. Baseline profiles of the late-line patients (median >5 lines of therapy, median >7 years from diagnosis) showed significant immunodeficiency, particularly in the CD4+ T cell compartment, and tumor genomes enriched for HR markers (∼30%) and CRBN dysregulation (∼40%), significantly higher than newly diagnosed or early-relapse patients.12,19,20,33 Analysis of immune and tumor biomarkers allowed us to interrogate iberdomide’s mechanisms of action, including understanding the basis of clinical response in CRBN-dysregulated, IMiD-resistant patients, a large emerging subset of RRMM.
Iberdomide showed pharmacodynamic activity in the immune compartment across a wide range of doses in patients regardless of prior therapies, including immediately post-pomalidomide and daratumumab, suggesting that it could be an immune-enhancing combination partner for immune-based therapies, including monoclonal antibodies, T cell engagers, and chimeric antigen receptor (CAR)-T cells. Supporting this further, recent pre-clinical evidence has shown the ability of iberdomide to enhance daratumumab and BCMA targeting T cell engagers and CAR-T cells.39,40,41,42 This activity appeared to saturate at a >1.0 mg dose and reverted toward baseline after drug holiday, highlighting the dynamic nature of iberdomide’s immune stimulation and providing evidence to design dosing and scheduling strategies to optimize immune stimulation for combination treatment. Furthermore, iberdomide induced these changes in patients who had baseline immunophenotypes suggestive of exhaustion and CD4+ T cell dysfunction, including a high proportion of PD1+, HLA-DR+, and effector T cell pre-treatment. Emerging evidence has demonstrated the essential role of CD4-mediated anti-tumor immune responses for the activity of T cell-redirecting therapies, including CAR-T cells.43,44,45,46,47,48 One possible implication of our data is that if these therapies are less effective in late-line RRMM vs. NDMM due to increased CD4+ T cell dysfunction, then their anti-tumor activity could be improved by adding iberdomide to enhance the immune stimulatory effect on CD4+ T cells. While the immunostimulatory activity of iberdomide is directionally consistent with what was observed during lenalidomide and pomalidomide treatment, it was achieved at ∼4-fold lower doses and ∼15-fold lower drug exposure with iberdomide in a later-line population.9,37,49 Taken together, these observations are consistent with a more potent pharmacological profile of iberdomide compared to IMiDs. These results support further investigation of iberdomide combinations, at a target dose >1.0 mg, with immune therapies whose suboptimal responses in patients could be driven in part by underlying T cell dysfunction.
Of the 19 responders with genomic data, the 13 wild-type and 6 CRBN-dysregulated patients did not share common genomic features, specific immune deficiency, or clinical history (Table S4). Pharmacodynamic changes in tumor and immune cells in CRBN-dysregulated patients for 3 representative cases illustrate the complex multifactorial determinants of iberdomide clinical efficacy (Figure 5). Patient 109-1010 (COPS7b/8 deletion) expressed high baseline CRBN (H-score = 212) but had limited Aiolos degradation in the tumor (18.5%). However, iberdomide induced robust immunostimulation in this patient with an ∼800% increase in proliferating CD4+ T cells, ∼600% increase in CD4+ EM T cells, and ∼350% increase in CD4+ activated T cells. In the BM microenvironment, iberdomide decreased levels of TIGIT+ NK and CD8+ T cells and increased levels of NKG2D+ NK cells and HLA-DR+ CD8+ T cells (comprehensive analysis of BM microenvironment changes in a larger cohort has been submitted [Van Oekelen et al.38]) (Figure 5A). Despite limited substrate degradation in the tumor, the patient achieved VGPR, 10.6 month DOR and 11.6 month PFS, suggestive of a significant immune-driven clinical outcome. Patient 104-1004 had a complex genotype that included multiple CRBN aberrations (CRBN-del-exon10, CRBN LOH, COPS7b/8 deletion) and an HR del17p/TP53 double hit. The tumor expressed low levels of CRBN (H-score = 19), but the degradation of Aiolos and Ikaros on treatment was 78.7% and 15%, respectively. Immune stimulation in PB was modest; however, on-treatment reduction in TIGIT+ NK and CD8+ T cells and increases in NKG2D+ and HLA-DR+ CD8+ T cells were observed in the BM (Figure 5B). This patient achieved partial response (PR), 15.7 month DOR, and 19.4 month PFS, providing evidence that iberdomide can overcome CRBN dysregulation and HR molecular features in the tumor.12 Finally, patient 101-1026 (CRBN-del-exon10) had high baseline tumor expression of CRBN (H-score = 187) and achieved 96.8% and 27% reductions of Aiolos and Ikaros and concomitant immunostimulation in PB (∼75% increase in activated CD4+ T cells; ∼50% increase in CD4+ EM T cells). While increased NKG2D+ cells in the BM were not observed on treatment, a decrease in TIGIT+ NK and T cells and an increase in HLADR+ CD8+ T cells were seen (Figure 5C). This patient achieved PR, 14.8 month DOR, and 19.4 month PFS (censoring time point), suggesting both tumor-intrinsic and immune-mediated mechanisms driving clinical outcome.
The data presented in this study reveal complex mechanisms that underlie iberdomide’s clinical activity exemplified by the induction of potent substrate degradation in tumors and the stimulation of NK and T cells. Notably, the comparable PFS of responders irrespective of CRBN status suggests possible mechanisms operating in the tumor and immune microenvironments of MM BM (Figure 5D). In the CRBN-dysregulated patients, the enhanced pharmacology of iberdomide compared to IMiDs may allow it to function efficiently with less or dysregulated CRBN associated with prior IMiD-based therapy. We hypothesize that once a response is achieved, it is sustained by continued immune-mediated cytotoxicity and/or direct anti-tumor activities, delaying progression. Thus, the initial achievement of a response may be a critical factor in iberdomide’s ability to contain the tumor from escaping therapy by expansion of resistant tumor clones. While the relative contribution of the tumor-intrinsic and -immune effects of iberdomide treatment to clinical response is difficult to tease out in this dataset, the sustained immune stimulation and pharmacodynamic shift to a more cytotoxic/effector T cell phenotype, particularly in patients with CRBN dysregulation in tumor cells, supports a concomitant cell-autonomous and immune-based mechanism acting in concert to drive the clinical response of iberdomide.
Limitations of the study
While the analyses presented in this work are comprehensive, the study has many limitations. One key limitation is that the small subsets of patient samples in genetic or immune biomarker data were insufficient to perform statistical analyses adequately. This precluded a fuller analysis of non-responders in the study, especially those with potent immune stimulation and degradation of CRBN substrates. Another limitation was the confounding effects of prognostic immune biomarkers (such as CD4 levels) for establishing correlation of treatment-induced immune pharmacodynamic changes in immune subsets with clinical outcome. Data from randomized controlled trials are needed to address these issues. Notably, iberdomide is being studied in an NDMM randomized study (ClinicalTrials.gov: NCT05827016) to compare it with lenalidomide and in an RRMM trial (ClinicalTrials.gov: NCT04975997) in combination with daratumumab and dexamethasone against daratumumab, bortezomib, and dexamethasone. These studies may answer how iberdomide’s activities described in this study perform compared to another CRBN targeting agent or as a combination partner with its immune-enhancing role.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Biological samples | ||
| Bone marrow specimens (FFPE) from myeloma patients | Multiple Clinical Study Sites | NCT02773030 |
| Bone marrow aspirates (BMA) from myeloma patients | Multiple Clinical Study Sites | NCT02773030 |
| Peripheral blood from myeloma patients | Multiple Clinical Study Sites | NCT02773030 |
| Critical commercial assays | ||
| CD138+ cell enrichment of BMA | Labcorp | NSSO6495_10936 |
| IHC analysis of FFPE samples | CellCarta | TE-IHC-374, TE-IHC-372, TE-IHC-3748, TE-IHC-346 |
| Dual Extraction of DNA/RNA from CD138+ BMA | Q2 Genomics | IS640 |
| WGS of DNA from CD138+ BMA | Q2 Genomics | SQ1803 |
| RNA-seq of RNA from CD138+ BMA | Q2 Genomics | SQ1102 |
| Flow cytometry of peripheral blood | Q2 Solutions | A167, A175, A326, A262 and A262 |
| Cytokine Analysis of BMA plasma | Ampersand Biosciences | HCD8MAG-17k |
| Deposited data | ||
| WGS and RNA-seq data | This paper | Study: EGAS50000000265 https://ega-archive.org/studies/EGAS50000000265 Dataset: EGAD50000000388 https://ega-archive.org/datasets/EGAD50000000388 |
| Software and algorithms | ||
| GraphPad Prism 10 | GraphPad | https://www.graphpad.com |
| Cytobank | Beckman Coulter, Inc | https://premium.cytobank.org/cytobank/login |
| BWA-mem | https://github.com/lh3/bwa | |
| GATK v4/MuTect2 | https://gatk.broadinstitute.org/hc/en-us/sections/360007226651-Best-Practices-Workflows | |
| ANNOVAR | https://annovar.openbioinformatics.org/en/latest/ | |
| MANTA | https://github.com/Illumina/manta/releases/tag/v1.6.0 | |
| Battenberg | Gooding et al. Ref. 12 | https://github.com/Wedge-lab/battenberg |
| STAR | Dobin et al. Ref. 50 | https://support.illumina.com/help/BS_App_RNASeq_Alignment_OLH_1000000006112/Content/Source/Informatics/STAR_RNAseq.htm |
| Salmon | Patro et al. Ref. 51 | https://salmon.readthedocs.io/en/latest/salmon.html |
| Python | www.python.org | |
| SciPy | www.scipy.org | |
| numpy | www.numpy.org | |
| statsmodels | www.statsmodels.org | |
| pandas | www.pandas.pydata.org | |
| lifelines | www.lifelines.readthedocs.io/en/latest/ | |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Michael Amatangelo (michael.amatangelo@bms.com).
Materials availability
This study did not generate new unique reagents.
Data and code availability
-
•
De-identified human/patient WGS and RNA-seq data have been deposited at EGA, and accession numbers are listed in the key resources table. They are available upon request.
-
•
The datasets used in this study are from an ongoing clinical trial of iberdomide in multiple myeloma that is sponsored by Bristol Myers Squibb (CC-220-MM-001 [NCT02773030]). The data from this study are not publicly available due to patient privacy concerns and because the clinical trial is currently ongoing. To request access to the data, please contact the lead contact, and also submit a request for access to ctt.group@bms.com. This is the mailbox for BMS' Clinical Trial Transparency (CTT) organization who manages the external distribution of data generated as part of a BMS study. Upon receiving a request, the CTT reviews the request and if approved, notifies EGA to release the data to the requestor. The lead contact can help connect you with a member of the CTT organization. Detailed questions on submitting a request can also be directed to ctt.group@bms.com. The requestor must describe the objectives of the research project for which the data will be used. Data access will be considered for research purposes and non-commercial use only. In order to ensure patient privacy, access to personally identifiable information or sensitive clinical information will not be provided, and requests for data access must rigorously adhere to the consent agreements established with study participants.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Experimental model and study participant details
Biomarker analyses were performed on RRMM patient samples collected on the open-label phase 1/2 study, CC-220-MM-001 (NCT02773030), with informed consent. This study is designed as a Phase 1b/2a study consisting of two parts: dose escalation (Part 1) and the expansion of the RP2D (Part 2). The study was conducted in accordance with the International Council for Harmonisation (ICH) guidelines on Good Clinical Practice (GCP) and the general principles outlined in the Declaration of Helsinki. The protocol was approved by each participating center’s institutional review board or ethics committee before initiation, and all patients provided written informed consent for participation in study and collection of biomarker samples. Details on the patient population, number of subjects and samples collected are provided in Table S1 and Figure S1. During the study the investigational drug iberdomide was administered orally on days 1–21 of each 28-day cycle. Samples were collected from Part 1 Cohorts A (0.3–1.0 mg dose escalation as monotherapy) and B (0.3–1.6 mg dose escalation in combination with dexamethasone) and Part 2 Cohort D (1.6 mg dose expansion in combination with dexamethasone). Supplementary analysis of transplant-ineligible NDMM patients at baseline was also performed on subjects enrolled in Cohort J1 and K (iberdomide with bortezomib and dexamethasone or daratumumab and dexamethasone, respectively).
Method details
Immunophenotyping
Peripheral blood samples were collected on study using sodium heparin and cytochex tubes for immunophenotyping pre-dose on C1D1, C2D1, C2D15, C4D1 and C4D15 per protocol. Samples were shipped same day as collection to be analyzed by flow cytometry at Q2 Solutions (Marietta, GA and Livingston, UK) as described. Flow cytometry panels included in the analysis were: A167 QTBNK, A175 TNK (KI67), A326 TACT TMEM (ICOS, HLA-DR, CD38), A262 T cell CHECKPOINT/EXHAUSTION and A262 T cell CHECKPOINT/EXHAUSTION. Data were analyzed for descriptive statistics using GraphPad PRISM software.
Granzyme B analysis
Plasma was isolated and frozen down from bone marrow aspirate at screening and cycle (C) 2 days (D) 15 or C3D15 (if unable to collect at C2D15) prior to CD138+ enrichment for patients enrolled in Cohort D. Samples were analyzed at Ampersand Biosciences (Lake Clear, NY) using Millipore (Burlington, MA) kit HCD8MAG-17k 17-plex cytokine assay. Data were analyzed for descriptive statistics using GraphPad PRISM software.
Immunohistochemistry analysis
Bone marrow samples (biopsies or aspirate clots) were collected at screening, cycle (C) 2 days (D) 15 or C3D15 (if unable to collect at C2D15), and treatment (EOT). Samples were decalcified in EDTA, formalin fixed/paraffin embedded and processed into 4mm thick slides. IHC analysis was performed at CellCarta (Antwerpan, Belgium) where samples were dual-stained for CD138 and either Aiolos, Ikaros, CRBN or IRF4. Nuclear proteins were digitally scored for intensity of nuclear staining (H-score = Histochemical Score = (% at 1+) x 1 + (% at 2+) x 2+ (% at 3+) x 3) [score = 300 maximum]) within CD138+ staining plasma cells, CRBN was scored for whole cell staining within CD138+ staining plasma cells. H-scores were provided by CellCarta and data were analyzed for descriptive statistics using GraphPad PRISM software.
Genomics and RNA-seq analysis
FASTQ files were aligned to the human genome assembly hg38 using BWA-mem. Base recalibration of alignments was performed using GATK v4. SNVs and small insertions/deletions (indels) were called using MuTect2 (v4) and annotated with ANNOVAR. Matched normal samples were available. Kinship coefficient testing was performed to confirm that tumor and normal samples were taken from the same individual.52 Cytogenetic assessments from WGS were made by MANTA and used to identify translocation DNA breakpoint location. CNA calling was performed using Battenberg as previously described with loss of heterozygosity (LOH) called for cancer clonal fraction (CCF) ≥7%.12 RNASeq reads were mapped and gene expression was quantitated using STAR and Salmon.50,51 High-risk features and patient segments were called from genomics analysis using previously described approaches.12,19,20,21,23 Briefly, this includes high-risk and genomic aberrations called from WGS (CNAs, SVs, SNVs, biallelic events, and DNA-based CRBN dysregulation segments [2q37 loss, CRBN LOH, CRBN mutation]. Calls for high expression of the CRBN-del-exon10 transcript requires available RNASeq data as well as tumor purity data. Only a subset of patients with sufficient bone marrow aspirates had cytogenetic risk assessed by fluorescence in situ hybridization (FISH), which has been described previously.1 We focused our initial analysis on three categories of patients defined by molecular genomic features. The main text focuses on patients who had a complete dataset with WGS, RNASeq, and clinical outcomes data (n = 81), while a larger WGS-only plot for the patients with only WGS (n = 109) are provided in Figure S2.
Mass cytometry analysis
Cells were stained with a novel mass cytometry (CyTOF) antibody panel as outlined in Tables S6 and S7. All antibodies were either purchased pre-conjugated from Fluidigm or conjugated at the Human Immune Monitoring Center, Icahn School of Medicine at Mount Sinai, using commercial X8 polymer conjugation kits purchased from Fluidigm. All conjugated antibodies were tittered and validated on healthy donor PBMCs. For longitudinal monitoring of phenotypic changes, cells from selected timepoints were thawed, counted, and assessed for viability utilizing the Nexcelom Cellaca Automated Cell Counter (Nexcelom Bioscience, Lawrence, MA, USA) along with acridine orange/propidium iodine staining (Nexcelom Bioscience, Lawrence, MA, USA). For sample timepoint batching, live-cell CyTOF barcoding was performed using anti-B2M antibodies conjugated to cadmium isotopes (Cd 111, 112, 114 and 116). Rhodium-103 viability and Human TruStain FcX staining (BioLegend) was performed simultaneously at room temperature for 30 min. After cell washing in flow cytometry buffer (1x PBS+0.2%BSA+0.05%NaN3), patient timepoints were combined and cells were stained with surface antibodies. Surface-stained cells were then fixed with 1.6% formaldehyde. In a second step, each patient was barcoded with the CyTOF Cell-ID 20-Plex Palladium Barcoding Kit (Fluidigm) (using Pd 102, 104, 105, 106, 108 and 110 isotopes). Barcoded cells were fixed and permeabilized with Fix-Perm buffer (BD Biosciences) and stained with the remaining intracellular antibodies from the panel. Stained samples were finally fixed in freshly diluted 4% paraformaldehyde containing 125nM intercalator-Ir (Fluidgm) and 300nM OsO4 (ACROS Organics) and stored at −80°C in FBS+10%DMSO. Samples for mass cytometry acquisition were washed with cell staining buffer (Fluidigm) and re-suspended in CAS buffer containing EQ normalization beads (Fluidigm) and acquired on a CyTOF2 instrument (Fluidigm). Post-acquisition, the data was normalized using bead-based normalization algorithm in the CyTOF software (Fluidigm). Normalized and de-barcoded data was uploaded to Cytobank for final analysis as detailed below.
Data was visualized and manual gating was performed using Cytobank. All samples with less than 2,000 BMMC (i.e., CD45+CD66b-singlets) after initial quality control were excluded. Clustering was based on the FlowSOM algorithm using all extracellular protein markers from the panel on a 10x10 grid size with K = 20 clusters. In a first step, the clusters corresponding to CD45−cells, CD66b+ cells and plasma cells (i.e., clusters with expression of CD138) were removed for quality control and for consistency with the hierarchical gating approach. The remaining cells were then re-clustered using the FlowSOM algorithm using all extracellular protein markers from the panel on a 14x14 grid size with K = 40 clusters to create sufficient granularity and capture relevant phenotypic clusters. These clusters were visualized using heatmaps and t-distributed stochastic neighbor embedding (TSNE) dimension reduction and annotated based on canonical protein expression.
Quantification and statistical analysis
Descriptive statistical analysis and statistical hypothesis testing was performed using GraphPad Prism and Python (pandas, numpy, scipy, statsmodels and lifelines). Pharmacodynamic changes were calculated as a difference between baseline (pre-treatment) and the on-treatment timepoint indicated using t-test or Mann-Whitney-Wilcoxon test for significant difference.53 Statistical analysis for significant association with response was performed using Wilcoxon rank-sum test. Tables S2A–S2D identify statistically significant changes in immunophenotypic cell counts and related assays between patient groups partitioned based on retrospective (refractory status) variables, number of lines of therapy (5 lines or greater) clinical response to iberdomide using the Kruskal-Wallis test (equivalent to the Mann-Whitney Wilcoxon test for this analysis between 2 groups). We additionally performed Cox Proportional Hazards modeling for univariate linear association between PFS and different immuno-assays in Tables S2A–S2D. The log(hazard ratio) and its 95% confidence interval, and Wald’s test p value testing for the log(hazard ratio) to be zero are reported.
Correlative analyses of iberdomide dosage versus immunophenotypic changes were performed as percent change from baseline at Cycle 2 Day 15 counts were used in cohorts B and D (subjects treated with iberdomide + dexamethasone), except for counts for proliferating NK-cells which was limited to the B cohort to assess possible hook effect observed in this cohort. Due to the nature of iberdomide dosage in the trial (patients were dosed at one of 10 fixed dose levels), neither value or rank based correlations were deemed suitable for identifying association due to the large number of ties in values and ranks for dosage. Adjacent doses were grouped together based on overlapping exposures as previously published1 and were virtually assigned to 6 dosage groups, each group coalescing no more than two adjacent dosage levels: 0.30–0.45 mg, 0.60–0.75 mg, 0.90–1.0 mg, 1.1 mg, 1.2–1.3 mg, and 1.6 mg. Successive dosage groups were not tested for inferiority or superiority of their medians, due to possibility of overlapping immunophenotypic values in successive dosage groups. Instead, for testing increasing or decreasing association with dosage, dosage groups that were one or more dosage group apart were tested for superiority or inferiority of their medians depending on the hypothesis being tested, using the non-parametric Mann-Whitney U test. Based on the proposed immune mechanism of action of iberdomide,8,10 absolute counts for B cells and naive T cell percentages were tested for decrease in cell counts with dosage and percentages of effector memory and proliferating T-cells were tested for increase in cell counts with dosage. For testing for an inflection point, the 6 dosage groups were further coalesced into three supergroups: <0.90 mg, 0.90–1.1 mg, and >1.1 mg, and the groups were tested for opposing trends with respect to the dosage groups on either side using non-parametric Mann – Whitney U test of the medians. Proportion of proliferating NK-cells was tested for a potential inflection point (maxima) in the range (0.9–1.1mg) (a potential hook effect) by creating three buckets of dosage <0.9 mg, 0.9–1.1 mg, and >1.1 mg. Since only three dosage groups were used to test for the inflection point, successive dosage groups were tested for differences in medians for both the original dosage groups and the supergroups.
Since we did not perform trend testing for all assays and were limited to a deeper analysis of a smaller subset of biologically relevant cell types, multiple testing correction was not performed. Even then, our testing strategy carries a multiplicity burden due to testing multiple groups against each other for each assay. For some of the assays (absolute B-cell counts, naive T cell percentage, effector memory T cell percentage), multiple tests show statistical significance with p < 0.05, lending credence to the statistical association even in the context of the multiple testing burden.
For Correlation and survival analyses with respect to Aiolos degradation, change in Aiolos (measured as percent change post-therapy with respect to screening) was analyzed with respect to 44 patients in the B and D cohorts for whom Aiolos data was available both at screening and post-therapy. 3 of these patients had right censored PFS data. Spearman’s rank correlation was performed between PFS survival times and change in Aiolos on all uncensored data points. However, for additional clarity on how Aiolos levels impact PFS survival times, patients were partitioned into two groups of 22: those with Aiolos change percentage < median (−45.98%), and those with Aiolo change percentage ≥ median (−45.98%). Kaplan-Meier curves and median PFS times based on the curves were estimated on these two groups separately, and the log rank test was used to assess whether the difference in the two Kaplan-Meier curves were statistically significant. Spearman’s rank correlation was also performed between PFS survival times and change in Aiolos on all uncensored data points. However, for additional clarity on how Aiolos levels impact PFS survival times, a Cox Proportional Hazard regression model (Cox PH model) was fit on all data points, in a univariate fashion assuming a linear relationship between Aiolos percent change and PFS (the simplest model testing association). The log hazard ratio (and its 95% confidence interval), and p value for the Wald test, which tests whether the coefficient of a variable in the Cox PH model is equal to zero (showing that the variable is not a significant predictor of the outcome) is reported. The survival curves for different levels of Aiolos degradation are calculated and shown.
Additional resources
Additional details on the clinical study can be found at: https://clinicaltrials.gov/study/NCT02773030.
Acknowledgments
The authors thank Marissa Hirst, Hillary Mosso, and Joseph Ramirez from Rancho Biosciences for data curation, cleaning, and summary of patient demographics for this analysis. Thanks to Michael Sturniolo, Sarah Gooding, and Karthik Ramasamy for helpful comments. Thanks to Sheida Hayati for assistance with genomic analysis and to Chad Bjorklund for assisting in the biomarker collection strategy for this study. Thanks to Jim Pratt and Md Shamsuzzaman for comments on the statistical analysis plan. Thanks to Preeti Trisal for assistance with graphical analysis. We also thank the Myeloma Disease team members from Celgene who worked together to bring the CELMoDs into clinical development. Special thanks to Jennifer Hopwood, Brendan Delaney, Benjamin Winograd, and Amit Agarwal for their insight. We also thank Mark Alles and Rupert Vessey for supporting the Myeloma Genome Project.
Author contributions
The project was conceived and designed by M.A., E.F., and A.T. Funding for biomarker analyses was provided by Celgene, a Bristol Myers Squibb Company. Oversight and management of resources (data generation, collection, transfer, infrastructure, data processing) was done by M.A., E.F., N.S., M.W., M.O., P.M., S.L., N.W.C.J.v.d.D., T.P., S.P., O.V.O., and A.T. Analyses and interpretation were done by M.A., E.F., N.S., M.W., M.O., S.P., O.V.O., P.R., and A.T. Data visualization was performed by M.A., E.F., P.R., and N.S. Supervision and scientific direction were provided by A.T. The manuscript was written by M.A., E.F., and A.T. All authors provided critical review of the manuscript.
Declaration of interests
M.A., E.F., M.W., M.O., N.S., P.M., T.P., P.R., and A.T. report equity ownership in Bristol Myers Squib. M.A., E.F., N.S., M.W., M.O., P.M., T.P., and P.R. are current employees of Bristol Myers Squib. A.T. is a former employee of Bristol Myers Squibb. Funding for data generation, processing, and storage was provided by Bristol Myers Squib. S.L. reports consulting fees from AbbVie, Bristol Myers Squibb, GlaxoSmithKline, Janssen Pharmaceuticals, and Takeda; institution grants or contracts from Bristol Myers Squibb, Janssen, and Takeda; membership on an entity’s board of directors or advisory committees for TG Therapeutics; and stock ownership in TG Therapeutics. N.W.C.J.v.d.D. reports institution grants or contracts from Amgen, Cellectis, and Janssen Pharmaceuticals and membership on an entity’s board of directors or advisory committees for Adaptive Biotechnologies, Amgen, GlaxoSmithKline, Janssen Pharmaceuticals, Novartis, Roche Sanofi, and Takeda. S.P. reports consulting fees from Foundation Medicine and research funding from Bristol Myers Squibb (Celgene), Karyopharm, and Amgen. P.R. reports shares in Doloromics.
Published: May 21, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xcrm.2024.101571.
Contributor Information
Michael Amatangelo, Email: michael.amatangelo@bms.com.
Anjan Thakurta, Email: anjan.thakurta@ndorms.ox.ac.uk.
Supplemental information
References
- 1.Lonial S., Popat R., Hulin C., Jagannath S., Oriol A., Richardson P.G., Facon T., Weisel K., Larsen J.T., Minnema M.C., et al. Iberdomide plus dexamethasone in heavily pretreated late-line relapsed or refractory multiple myeloma (CC-220-MM-001): a multicentre, multicohort, open-label, phase 1/2 trial. Lancet. Haematol. 2022;9:e822–e832. doi: 10.1016/S2352-3026(22)00290-3. [DOI] [PubMed] [Google Scholar]
- 2.Watson E.R., Novick S., Matyskiela M.E., Chamberlain P.P., H de la Peña A., Zhu J., Tran E., Griffin P.R., Wertz I.E., Lander G.C. Molecular glue CELMoD compounds are regulators of cereblon conformation. Science. 2022;378:549–553. doi: 10.1126/science.add7574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bjorklund C.C., Lu L., Kang J., Hagner P.R., Havens C.G., Amatangelo M., Wang M., Ren Y., Couto S., Breider M., et al. Rate of CRL4CRBN substrate Ikaros and Aiolos degradation underlies differential activity of lenalidomide and pomalidomide in multiple myeloma cells by regulation of c-Myc and IRF4. Blood Cancer J. 2015;5:e354. doi: 10.1038/bcj.2015.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kronke J., Udeshi N.D., Narla A., Grauman P., Hurst S.N., McConkey M., Svinkina T., Heckl D., Comer E., Li X., et al. Lenalidomide causes selective degradation of IKZF1 and IKZF3 in multiple myeloma cells. Science. 2014;343:301–305. doi: 10.1126/science.1244851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lopez-Girona A., Mendy D., Ito T., Miller K., Gandhi A.K., Kang J., Karasawa S., Carmel G., Jackson P., Abbasian M., et al. Cereblon is a direct protein target for immunomodulatory and antiproliferative activities of lenalidomide and pomalidomide. Leukemia. 2012;26:2326–2335. doi: 10.1038/leu.2012.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lu G., Middleton R.E., Sun H., Naniong M., Ott C.J., Mitsiades C.S., Wong K.K., Bradner J.E., Kaelin W.G., Jr. The Myeloma Drug Lenalidomide Promotes the Cereblon-Dependent Destruction of Ikaros Proteins. Science. 2014;343:305–309. doi: 10.1126/science.1244917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Matyskiela M.E., Zhang W., Man H.-W., Muller G., Khambatta G., Baculi F., Hickman M., LeBrun L., Pagarigan B., Carmel G., et al. A Cereblon Modulator (CC-220) with Improved Degradation of Ikaros and Aiolos. J. Med. Chem. 2018;61:535–542. doi: 10.1021/acs.jmedchem.6b01921. [DOI] [PubMed] [Google Scholar]
- 8.Bjorklund C.C., Kang J., Amatangelo M., Polonskaia A., Katz M., Chiu H., Couto S., Wang M., Ren Y., Ortiz M., et al. Iberdomide (CC-220) is a potent cereblon E3 ligase modulator with antitumor and immunostimulatory activities in lenalidomide- and pomalidomide-resistant multiple myeloma cells with dysregulated CRBN. Leukemia. 2020;34:1197–1201. doi: 10.1038/s41375-019-0620-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thakurta A., Pierceall W.E., Amatangelo M.D., Flynt E., Agarwal A. Developing next generation immunomodulatory drugs and their combinations in multiple myeloma. Oncotarget. 2021;12:1555–1563. doi: 10.18632/oncotarget.27973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gandhi A.K., Kang J., Havens C.G., Conklin T., Ning Y., Wu L., Ito T., Ando H., Waldman M.F., Thakurta A., et al. Immunomodulatory agents lenalidomide and pomalidomide co-stimulate T cells by inducing degradation of T cell repressors Ikaros and Aiolos via modulation of the E3 ubiquitin ligase complex CRL4(CRBN.) Br. J. Haematol. 2014;164:811–821. doi: 10.1111/bjh.12708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yamamoto L., Amodio N., Gulla A., Anderson K.C. Harnessing the Immune System Against Multiple Myeloma: Challenges and Opportunities. Front. Oncol. 2020;10 doi: 10.3389/fonc.2020.606368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gooding S., Ansari-Pour N., Towfic F., Ortiz-Estevez M., Chamberlain P.P., Tsai K.-T., Flynt E., Hirst M., Rozelle D., Dhiman P., et al. Multiple Cereblon genetic changes associate with acquired resistance to Lenalidomide or Pomalidomide in Multiple Myeloma. Blood. 2020;137:232–237. doi: 10.1182/blood.2020007081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thakurta A., Gandhi A.K., Waldman M.F., Bjorklund C., Ning Y., Mendy D., Schafer P., Lopez-Girona A., Lentzsch S., Schey S.A., et al. Absence of mutations in cereblon (CRBN) and DNA damage-binding protein 1 (DDB1) genes and significance for IMiD therapy. Leukemia. 2014;28:1129–1131. doi: 10.1038/leu.2013.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kortüm K.M., Mai E.K., Hanafiah N.H., Shi C.X., Zhu Y.X., Bruins L., Barrio S., Jedlowski P., Merz M., Xu J., et al. Targeted sequencing of refractory myeloma reveals a high incidence of mutations in CRBN and Ras pathway genes. Blood. 2016;128:1226–1233. doi: 10.1182/blood-2016-02-698092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Egan J.B., Kortuem K.M., Kurdoglu A., Izatt T., Aldrich J., Reiman R., Phillips L., Baker A., Shi C.X., Schmidt J., et al. Extramedullary myeloma whole genome sequencing reveals novel mutations in Cereblon, proteasome subunit G2 and the glucocorticoid receptor in multi drug resistant disease. Br. J. Haematol. 2013;161:748–751. doi: 10.1111/bjh.12291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Franssen L.E., Nijhof I.S., Couto S., Levin M.D., Bos G.M.J., Broijl A., Klein S.K., Ren Y., Wang M., Koene H.R., et al. Cereblon loss and up-regulation of c-Myc are associated with lenalidomide resistance in multiple myeloma patients. Haematologica. 2018;103:e368–e371. doi: 10.3324/haematol.2017.186601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Neri P., Maity R., Keats J.J., Tagoug I., Simms J., Auclair D., Lonial S., Bahlis N.J. Cereblon Splicing of Exon 10 Mediates IMiDs Resistance in Multiple Myeloma: Clinical Validation in the CoMMpass Trial. Blood. 2016;128:120. [Google Scholar]
- 18.Skerget S., Benard B., Christofferson A., Legendre C., Aldrich J., Yesil J., Auclair D., Liang W., Lonial S., Keats J.J., et al. A Molecular Analysis of Cereblon-Related Immunomodulatory Drug Resistance in Commpass Multiple Myeloma Patients. Blood. 2017;130:1754. [Google Scholar]
- 19.Gooding S., Ansari-Pour N., Kazeroun M., Karagoz K., Polonskaia A., Salazar M., Fitzsimons E., Sirinukunwattana K., Chavda S., Ortiz Estevez M., et al. Loss Of COP9-Signalosome Genes At 2q37 Is Associated With IMiD Agent Resistance In Multiple Myeloma. Blood. 2022;140:1816–1821. doi: 10.1182/blood.2022015909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walker B.A., Mavrommatis K., Wardell C.P., Ashby T.C., Bauer M., Davies F., Rosenthal A., Wang H., Qu P., Hoering A., et al. A high-risk, Double-Hit, group of newly diagnosed myeloma identified by genomic analysis. Leukemia. 2019;33:159–170. doi: 10.1038/s41375-018-0196-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thakurta A., Ortiz M., Blecua P., Towfic F., Corre J., Serbina N.V., Flynt E., Yu Z., Yang Z., Palumbo A., et al. High subclonal fraction of 17p deletion is associated with poor prognosis in multiple myeloma. Blood. 2019;133:1217–1221. doi: 10.1182/blood-2018-10-880831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stong N., Ortiz Estevez M., Towfic F., Samur M., Agarwal A., Corre J., Flynt E., Munshi N., Avet-Loiseau H. Thakurta A.Location of the t(4;14) translocation breakpoint within the NSD2 gene identifies a subset of high-risk NDMM patients. Blood. 2022;141:1574–1583. doi: 10.1182/blood.2022016212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ansari-Pour N., Samur M., Flynt E., Gooding S., Towfic F., Stong N., Estevez M.O., Mavrommatis K., Walker B., Morgan G., et al. Whole-genome analysis identifies novel drivers and high-risk double-hit events in relapsed/refractory myeloma. Blood. 2023;141:620–633. doi: 10.1182/blood.2022017010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Georgopoulos K., Winandy S., Avitahl N. The role of the Ikaros gene in lymphocyte development and homeostasis Annual Review of Immunology. Annu. Rev. Immunol. 1997;15:155–176. doi: 10.1146/annurev.immunol.15.1.155. [DOI] [PubMed] [Google Scholar]
- 25.Heizmann B., Kastner P., Chan S. The Ikaros family in lymphocyte development. Curr. Opin. Immunol. 2018;51:14–23. doi: 10.1016/j.coi.2017.11.005. [DOI] [PubMed] [Google Scholar]
- 26.Sehgal K., Das R., Zhang L., Verma R., Deng Y., Kocoglu M., Vasquez J., Koduru S., Ren Y., Wang M., et al. Clinical and pharmacodynamic analysis of pomalidomide dosing strategies in myeloma: impact of immune activation and cereblon targets. Blood. 2015;125:4042–4051. doi: 10.1182/blood-2014-11-611426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Davies F.E., Raje N., Hideshima T., Lentzsch S., Young G., Tai Y.T., Lin B., Podar K., Gupta D., Chauhan D., et al. Thalidomide and immunomodulatory derivatives augment natural killer cell cytotoxicity in multiple myeloma. Blood. 2001;98:210–216. doi: 10.1182/blood.v98.1.210. [DOI] [PubMed] [Google Scholar]
- 28.Busch A., Zeh D., Janzen V., Mügge L.O., Wolf D., Fingerhut L., Hahn-Ast C., Maurer O., Brossart P., von Lilienfeld-Toal M. Treatment with lenalidomide induces immunoactivating and counter-regulatory immunosuppressive changes in myeloma patients. Clin. Exp. Immunol. 2014;177:439–453. doi: 10.1111/cei.12343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brioli A., Melchor L., Titley I., Vijayaraghavan G., Stephens C., Zeisig A., Pawlyn C., Cavo M., Morilla R., Davies F.E., Morgan G.J. The impact of long-term lenalidomide exposure on the cellular composition of bone marrow. Leuk. Lymphoma. 2014;55:2665–2668. doi: 10.3109/10428194.2014.900765. [DOI] [PubMed] [Google Scholar]
- 30.Lee S.-E., Lim J.-Y., Ryu D.-B., Kim T.W., Yoon J.H., Cho B.S., Eom K.S., Kim Y.J., Kim H.J., Lee S., et al. Circulating immune cell phenotype can predict the outcome of lenalidomide plus low-dose dexamethasone treatment in patients with refractory/relapsed multiple myeloma. Cancer Immunol. Immunother. 2016;65:983–994. doi: 10.1007/s00262-016-1861-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hong J.J., Amancha P.K., Rogers K., Ansari A.A., Villinger F. Re-Evaluation of PD-1 Expression by T Cells as a Marker for Immune Exhaustion during SIV Infection. PLoS One. 2013;8 doi: 10.1371/journal.pone.0060186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wherry E.J. T cell exhaustion. Nat. Immunol. 2011;12:492–499. doi: 10.1038/ni.2035. [DOI] [PubMed] [Google Scholar]
- 33.Walker B.A., Mavrommatis K., Wardell C.P., Ashby T.C., Bauer M., Davies F.E., Rosenthal A., Wang H., Qu P., Hoering A., et al. Identification of novel mutational drivers reveals oncogene dependencies in multiple myeloma. Blood. 2018;132:587–597. doi: 10.1182/blood-2018-03-840132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Walker B.A., Boyle E.M., Wardell C.P., Murison A., Begum D.B., Dahir N.M., Proszek P.Z., Johnson D.C., Kaiser M.F., Melchor L., et al. Mutational Spectrum, Copy Number Changes, and Outcome: Results of a Sequencing Study of Patients With Newly Diagnosed Myeloma. J. Clin. Oncol. 2015;33:3911–3920. doi: 10.1200/JCO.2014.59.1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kirstetter P., Thomas M., Dierich A., Kastner P., Chan S. Ikaros is critical for B cell differentiation and function. Eur. J. Immunol. 2002;32:720–730. doi: 10.1002/1521-4141(200203)32:3<720::AID-IMMU720>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 36.Schmitt C., Tonnelle C., Dalloul A., Chabannon C., Debré P., Rebollo A. Aiolos and Ikaros: Regulators of lymphocyte development, homeostasis and lymphoproliferation. Apoptosis. 2002;7(3):277–284. doi: 10.1023/a:1015372322419. [DOI] [PubMed] [Google Scholar]
- 37.Prabhala R., Pierceall W.E., Samur M., Potluri L.B., Hong K., Peluso T., Talluri S., Wang A., Katiki A., Vangala S.D., et al. Immunomodulation of NK, NKT and B/T cell subtypes in relapsed/refractory multiple myeloma patients treated with pomalidomide along with velcade and dexamethasone and its association with improved progression-free survival. Front. Oncol. 2023;13 doi: 10.3389/fonc.2023.1271807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Van Oekelen O., Amatangelo M., Guo M., Upadhyaya B., Cribbs A., Kelly G., Patel M., Kim-Schulze S., Flynt E., Lagana A., et al. Iberdomite increases innate and adaptive immune cell subsets in the bone marrow of relapsed/refractory multiple myeloma patients. Cell Rep Med. 2024;5 doi: 10.1016/j.xcrm.2024.101584. [DOI] [PubMed] [Google Scholar]
- 39.Paiva B., Gaffney B., Burnett K., Castiglioni P., Angelo M., Pierce D.W., Boss I.W. Synergistic Antitumor Activity of Alnuctamab (ALNUC; BMS-986349; CC-93269), a BCMA 2+1 T Cell Engager (TCE), and Celmod Agents in Multiple Myeloma (MM) Preclinical Models. Blood. 2022;140:7054–7055. [Google Scholar]
- 40.Aleman A., Kogan Zajdman A., Upadhyaya B., Van Oekelen O., Chen L., Leshchenko V., Jagannath S., Parekh S. P-175 Improving Anti-BCMA CAR-T functionality with novel immunomodulatory agent Iberdomide (CC220) in Multiple Myeloma. Clin. Lymphoma, Myeloma & Leukemia. 2023;23:S131–S132. [Google Scholar]
- 41.Soni N.B.A., Works M., Qin J., Balakrishnan A., Ports M. Iberdomide increased the potency of the anti-BCMA CAR T Cell product Orvacabtagene autoleucel (orva-cel) Mol. Ther. 2020;28:1–592. [Google Scholar]
- 42.Ma P., Sridharan V., Wollerman K., Dutta D., Singh R.K., Mukherjee S., Bjorklund C.C., Hagner P.R., Amatangelo M., Gandhi A.K. Iberdomide Enhances Dara Mediated Cytotoxicity through Upregulation of CDC Activity and Elevated NK Cell Mediated ADCC. Blood. 2023;142:3289. [Google Scholar]
- 43.Kravtsov D.S., Erbe A.K., Sondel P.M., Rakhmilevich A.L. Roles of CD4+ T cells as mediators of antitumor immunity. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.972021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang D., Aguilar B., Starr R., Alizadeh D., Brito A., Sarkissian A., Ostberg J.R., Forman S.J., Brown C.E. Glioblastoma-targeted CD4+ CAR T cells mediate superior antitumor activity. JCI Insight. 2018;3 doi: 10.1172/jci.insight.99048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Perez-Diez A., Joncker N.T., Choi K., Chan W.F.N., Anderson C.C., Lantz O., Matzinger P. CD4 cells can be more efficient at tumor rejection than CD8 cells. Blood. 2007;109:5346–5354. doi: 10.1182/blood-2006-10-051318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Adusumilli P.S., Cherkassky L., Villena-Vargas J., Colovos C., Servais E., Plotkin J., Jones D.R., Sadelain M. Regional delivery of mesothelin-targeted CAR T cell therapy generates potent and long-lasting CD4-dependent tumor immunity. Sci. Transl. Med. 2014;6:261ra151. doi: 10.1126/scitranslmed.3010162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sommermeyer D., Hudecek M., Kosasih P.L., Gogishvili T., Maloney D.G., Turtle C.J., Riddell S.R. Chimeric antigen receptor-modified T cells derived from defined CD8+ and CD4+ subsets confer superior antitumor reactivity in vivo. Leukemia. 2016;30:492–500. doi: 10.1038/leu.2015.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Turtle C.J., Hanafi L.-A., Berger C., Hudecek M., Pender B., Robinson E., Hawkins R., Chaney C., Cherian S., Chen X., et al. Immunotherapy of non-Hodgkin’s lymphoma with a defined ratio of CD8+ and CD4+ CD19-specific chimeric antigen receptor-modified T cells. Sci. Transl. Med. 2016;8:355ra116. doi: 10.1126/scitranslmed.aaf8621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pierceall W.E., Amatangelo M.D., Bahlis N.J., Siegel D.S., Rahman A., Van Oekelen O., Neri P., Young M., Chung W., Serbina N., et al. Immunomodulation in Pomalidomide, Dexamethasone, and Daratumumab-Treated Patients with Relapsed/Refractory Multiple Myeloma. Clin. Cancer Res. 2020;26:5895–5902. doi: 10.1158/1078-0432.CCR-20-1781. [DOI] [PubMed] [Google Scholar]
- 50.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Patro R., Duggal G., Love M.I., Irizarry R.A., Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods. 2017;14:417–419. doi: 10.1038/nmeth.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Manichaikul A., Mychaleckyj J.C., Rich S.S., Daly K., Sale M., Chen W.-M. Robust relationship inference in genome-wide association studies. Bioinformatics. 2010;26:2867–2873. doi: 10.1093/bioinformatics/btq559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kruskal W.H., Wallis W.A. Use of Ranks in One-Criterion Variance Analysis. J. Am. Stat. Assoc. 1952;47:583–621. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
De-identified human/patient WGS and RNA-seq data have been deposited at EGA, and accession numbers are listed in the key resources table. They are available upon request.
-
•
The datasets used in this study are from an ongoing clinical trial of iberdomide in multiple myeloma that is sponsored by Bristol Myers Squibb (CC-220-MM-001 [NCT02773030]). The data from this study are not publicly available due to patient privacy concerns and because the clinical trial is currently ongoing. To request access to the data, please contact the lead contact, and also submit a request for access to ctt.group@bms.com. This is the mailbox for BMS' Clinical Trial Transparency (CTT) organization who manages the external distribution of data generated as part of a BMS study. Upon receiving a request, the CTT reviews the request and if approved, notifies EGA to release the data to the requestor. The lead contact can help connect you with a member of the CTT organization. Detailed questions on submitting a request can also be directed to ctt.group@bms.com. The requestor must describe the objectives of the research project for which the data will be used. Data access will be considered for research purposes and non-commercial use only. In order to ensure patient privacy, access to personally identifiable information or sensitive clinical information will not be provided, and requests for data access must rigorously adhere to the consent agreements established with study participants.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.