Abstract
Given multifunction of copper (Cu) contributing to all stages of the physiology of wound healing, Cu-based compounds have great therapeutic potentials to accelerate the wound healing, but they must be limited to a very low concentration range to avoid detrimental accumulation. Additionally, the cellular mechanism of Cu-based compounds participating the healing process remains elusive. In this study, copper oxide nanoparticles (CuONPs) were synthesized to mimic the multiple natural enzymes and trapped into PEG-b-PCL polymersomes (PS) to construct cupric-polymeric nanoreactors (CuO@PS) via a direct hydration method, thus allowing to compartmentalize Cu-based catalytic reactions in an isolated space to improve the efficiency, selectivity, recyclability as well as biocompatibility. While nanoreactors trafficked to lysosomes following endocytosis, the released Cu-based compounds in lysosomal lumen drove a cytosolic Cu+ influx to mobilize Cu metabolism mostly via Atox1-ATP7a/b-Lox axis, thereby activating the phosphorylation of mitogen-activated protein kinase 1 and 2 (MEK1/2) to initiate downstream signaling events associated with cell proliferation, migration and angiogenesis. Moreover, to facilitate to lay on wounds, cupric-polymeric nanoreactors were finely dispersed into a thermosensitive Pluronic F127 hydrogel to form a composite hydrogel sheet that promoted the healing of chronic wounds in diabetic rat models. Hence, cupric-polymeric nanoreactors represented an attractive translational strategy to harness cellular Cu metabolism for chronic wounds healing.
Keywords: Cupric-polymeric nanoreactors, Copper metabolism, Atox1-ATP7a/b-Lox axis, MEK1/2 phosphorylation, Chronic wound healing
Graphical abstract
1. Introduction
Skin as the largest organ in the body provides the physical end border to protect internal tissues from extreme temperature, water loss, ultraviolet radiation, microbial and chemical insults, and injury. Chronic non-healing wound is one of the most common diabetes complications, and about 20 % of diabetic patients develop a chronic non-healing wound, suffering long-term pain, limited mobility, infection, and even amputation [1,2]. The standard of cares in clinic are mostly conservative treatments, such as tissue debridement, management of ischemia, infection control, and appropriate wound bed preparation [3,4]. Due to the poor managements by patients or in the community as opposed to under hospital settings, it is normally difficult to achieve the favorable clinical outcomes [4]. Therefore, it is urgent to develop more effective and convenient therapeutic modalities with high patient compliance, on the base of understanding the pathophysiology of non-healing wound occurring in diabetes.
Wound recovery is a complex process that can be divided into four successive and overlapped stages, i.e., hemostasis, inflammation, proliferation and remodeling [5,6]. However, many factors may impede or prolong the healing process, such as oxidative stress, inflammation and pathogenic infections [7]. Notably, the essential trace element copper (Cu) participates in all stages of the wound healing process [8,9]. Since Cu is a key cofactor to drive catalysis in microorganisms and mammalian, manipulation of Cu-dependent signaling pathways may offer benefits to accelerate wound healing [[10], [11], [12]]. Indeed, colloidal Cu particles as artificial nanozymes can mimic natural metalloenzymes, which not only promote angiogenesis and fibroblasts proliferation [13,14], but also exert the superior antioxidative and antimicrobial effects during healing [15,16]. Yet the underlying mechanism of Cu nanoparticles on wound repair remains to be elucidated.
Intracellular Cu concentration is restricted within a relatively low range and tightly controlled by multiple Cu transporters and chaperones to maintain the Cu homeostasis [17]. After taken up by copper transporter 1 (Ctr1) into cells, Cu is bound with various chaperones in cytosol, i.e., antioxidant-1 (Atox1), copper chaperone for superoxide dismutase (CCS), cytochrome c oxidases (Coxs), which further transferred Cu to the specific Cu-containing enzymes such as Cu, Zn-superoxide dismutase (SOD), or lysyl oxidase (Lox) via Cu transporter ATPase copper-transporting alpha or beta (ATP7a/b). Excessive amounts of Cu trigger oxidative stress and cell damages, which is also defined as a new type of cell death, cuproptosis [18]. Although low-concentrations of Cu nanoparticles may contribute to wound repair process, caution should be taken while applying them to skin wounds. Particularly, Cu nanoparticles directly contact with the wound surface without intact skin protection, therefore, excessive and prolonged exposure may induce skin irritation and allergies, and even systemic adverse effects. Moreover, instability of Cu-based nanoparticles, such as aggregation, dissociation or oxidation in aqueous solution may also hinder their medical use [[19], [20], [21]].
Heavy metal nanoparticles can be absorbed into polymeric matrix to reduce the toxicity and improve their stability [22]. Due to the vesicular structure, tunable membrane permeability and colloidal stability, polymersomes (PS) have become a versatile system to encapsulate diverse therapeutic compounds either in the aqueous core or in the hydrophobic membrane bilayer [23,24]. Moreover, polymersomes have been also employed as preferrable candidates to formulate nanoreactors via incorporating a variety of catalysts including natural enzymes or artificial nanozymes [[25], [26], [27], [28]] into a confined space to conduct catalytic reactions, thus avoiding the interferences from biological fluids, extracellular matrix, or cell/tissue interactions [29]. Particularly, some polymersomes constructed by biodegradable polymers, i.e., poly (caprolactone) (PCL), poly (lactic acid) (PLA) intrinsically possess semi-permeability for small molecules substrates including gases, ions or reactive oxygen species (ROS) [30]. Therefore, polymersomes can be potentially transformed into nanoreactors via the incorporation of copper-based catalysts for wound healing.
Hydrogel has been extensively investigated for wound dressing and also been exploited to deliver a diverse range of drugs, due to its high biocompatibility and biosafety [31]. Polymersomes-based drug delivery carriers can be combined with hydrogel not only to modulate the drug load and release by tuning the composition and/or ratio, but also to increase the adhesion of polymersomes with skin wounds [32]. Pluronics, as amphiphilic tri-block copolymers formed by poly (ethylene oxide)-poly (propylene oxide)-poly (ethylene oxide) (PEO-PPO-PEO), have been approved by the U.S. Food and Drug Administration (FDA) for pharmaceutical constituents, and are widely used in biomedical fields [33]. Amongst Pluronics, PF-127 has gained increasing attentions in wound dressing applications, due to its excellent properties such as thermo-sensitivity, injectability and biodegradability [34].
Herein, copper (II) oxide nanoparticles (CuONPs) with multi-enzyme activity were synthesized and enclosed into polymersomes in the concurrence with a self-assembly process of poly (ethylene glycol)-block-poly (ε-caprolactone) (PEG-b-PCL) via a direct hydration method, thus transforming them into cupric-polymeric nanoreactors. These newly generated nanoreactors can be further embedded into PF-127 hydrogel with an aim of wound management, which allowed not only to maintain the catalytic activity of CuONPs without toxic exposures, but also to manipulate Cu-based signaling pathways for promoting chronic wound healing.
2. Materials and methods
2.1. Materials
Poly (ethylene glycol)-block-poly (ε-caprolactone) (PEG-b-PCL, PEG Mw = 2000, PCL Mw = 4300) was synthesized based on our previous work [35]; Copper (II) acetate, Poly (ethylene glycol) 550 (PEG 550), hydrogen peroxide (H2O2, 30 %), Ferrous chloride (FeCl2), Rhodamine B were purchased form Macklin, China; Sephadex G-50 was bought from Shanghai Yuanye Bio-Technology, China; phosphotungstic acid hydrate, lipopolysaccharide (LPS), Pluronic F-127, fluorescein isothiocyanate (FITC), Bis(cyclohexanone)oxaldihydrazone (BCO) and Streptozotocin (STZ) were purchased from Sigma Aldrich, USA. O2.- scavenging assay kit (WST-8 method), cell counting kit-8 (CCK-8) were purchased from Abbkine Scientific, China. H2O2 scavenging assay kit, 2′,7′-Dichlorofluorescein diacetate (DCFH-DA) and BeyoClick™ EdU Cell Proliferation Kit were purchased from Beyotime Biotechnology, China. LysoTracker™ Red DND-99, LysoTracker™ Green DND-26 and 4′,6-Diamidino-2-phenylindole dihydrochloride (DAPI) were purchased from Thermo Fisher Scientific, USA. Long-chain carbocyanine dye DiD and Cu1+ probe Coppersensor-1 (CS-1) were from MedChemExpress, USA. Primary antibodies used in this study were Anti-TGF beta 1 Rabbit pAb (Proteintech, 21898-1-AP), Anti-alpha smooth muscle Actin Rabbit pAb (Servicebio, GB111364), Anti-VEGFA Rabbit pAb (Proteintech, 19003-1-AP), MEK1/2 (L38C12) Mouse mAb (Cell signaling technology, 4694S) and Phospho-MEK1/2 (Ser217/221) (41G9) Rabbit mAb (Cell signaling technology, 9154S).
2.2. Synthesis of copper (II) oxide nanoparticles
Copper (II) oxide nanoparticles (CuONPs) were synthesized using a rapid hydrolysis precipitation method [36]. Briefly, 0.1 mL of glacial acetic acid was added to 30 mL of 20 mM copper (II) acetate aqueous solution. The mix solution was heated to boiling under conditions of magnetic stirring. 2 mL of 1 M NaOH aqueous solution was quickly added into reaction, and then a large number of black precipitates were formed immediately. The precipitates were collected by centrifuge (12,000 rpm, 15 min) and washed three times with anhydrous ethanol to obtain the purified CuONPs. After drying, CuONPs were weighed and resuspended with ultrapure water to obtain a stock solution. The particle size and distribution were determined by using dynamic light scattering (DLS) and transmission electron microscopy (TEM), which were detailly described in supplementary materials.
2.3. Preparation of cupric-polymeric nanoreactors
The cupric-polymeric nanoreactors (CuO@PS) were prepared by a direct hydration method with some modifications [37]. 5 mg of PEG-b-PCL was dissolved into 50 μL of PEG 550 under stirring for 25 min at 60 °C. While the temperature adjusted to 50 °C, 50 μL of CuONPs (5 mg/mL in water) was added dropwise into polymers solution, and allowed to stir another 30 min. Subsequently, 100 μL and 350 μL of water were successively added into the mixture at every 30 min. After adding water, the obtained solution was purified through a Sephadex G-50 column. CuO@PS was collected from the first eluted fraction with light brown color, while the following fraction with dark brown color was CuONPs with smaller particle size. The methods to measure the loading efficiency and encapsulation efficiency of CuONPs in polymersomes were present in supplementary materials.
2.4. Free radicals scavenging assay
Scavenging abilities of CuONPs and CuO@PS to remove superoxide radicals (O2.-), hydrogen peroxide (H2O2), and hydroxyl radicals (·OH) were determined by different colorimetry assays following the kit instructions.
O2.- concentration was determined using a WST-8 method that 20 μL of each sample was added with 5 μL of xanthine, 5 μL of WST-8, and 75 μL of assay buffer, and mixed well to generate WST-8 formazan which quantitatively reflects the concentration of O2.-. The absorbance of WST-8 formazan in solution was measured with a microplate reader (Synergy H1, BioTek) at 450 nm every 10 s at 37 °C.
H2O2 scavenging ability of CuONPs or CuO@PS was evaluated by mixing 10 μL of the samples with 75 μL of assay buffer, 5 μL of chromogenic substrate, and 10 μL of 200 mM H2O2 for rapidly measuring absorbance at 520 nm every 10 s at 37 °C. The absorbance of benzoquinone monoamine derivative was positively correlated with the concentrations of H2O2 concentration.
Prior to assess the scavenging activity of CuONPs or CuO@PS for ·OH, 20 μL of 10 mM FeCl2 and 20 μL of 10 mM H2O2 were mixed well to freshly prepare ·OH. Then a mixture solution containing 10 μL of sample to be tested and 100 μL of rhodamine B (5 μM) were added and mixed well, and subjected to measure the absorbance of rhodamine B at 554 nm every 10 s at 37 °C. The absorbance of rhodamine B was inversely proportional to ·OH concentration, since rhodamine B can be easily removed by ·OH.
The changes in absorbance values of CuONPs or CuO@PS both at a CuONPs concentration of 1, 2.5, 5, 7.5, 10, 15 and 20 μg/mL were plotted over time, and analyzed using Graphpad (Version 8.0.2). For multiple cycle testing, CuONPs or CuO@PS were collected by centrifugation after each cycle and further subjected to a new reaction solution for another round.
2.5. In vitro scratch assay
While the cell confluency reaching around 90 %, NIH-3T3 cells were stimulated with 500 μM of H2O2 and 500 ng/mL of LPS for 12 h. Afterwards, the cell monolayer was scraped with a 200-μL pipette tip prior to addition of CuONPs or CuO@PS at the same CuONPs concentrations of 1, 2.5, 5 and 10 μg/mL. The scratch areas were monitored over time by optical imaging (EVOS M7000, Thermo Fisher Scientific). The scratch healing rate was calculated using Image J (Version 1.53).
2.6. Cellular reactive oxygen species detection
After stimulated with H2O2 (500 μM) and LPS (500 ng/mL) for 12 h, NIH-3T3 cells were replaced with fresh medium containing CuONPs or CuO@PS (at the same CuONPs concentration of 5 μg/mL) for 12 h incubation. Reactive oxygen species (ROS) from each group can be detected via a DCFH-DA assay. The cells were incubated with DCFH-DA (10 μM) for 30 min to generate fluorescent DCF in the presence of ROS, and further imaged with a confocal laser scanning microscope (CLSM, FV3000, Olympus). The fluorescent intensities of DCF were quantified by Image J (Version 1.53).
2.7. EdU cell proliferation assay
Upon exposed to H2O2 and LPS for 12 h, NIH-3T3 cells were further treated with CuONPs or CuO@PS at the same CuONPs concentration of 5 μg/mL for another 24 h. Then the proliferative cells were detected using an Alexa Fluor 488 EdU detection kit according to manufacturer's instructions. Briefly, NIH-3T3 cells were incubated with EdU (10 μM) for 2 h at 37 °C, and were further fixed to detect with Alexa Fluor® 488 azide. Cell nuclei were marked with DAPI prior to microscopic observations with a CLSM (FV3000, Olympus). EdU-positive cells from each group were analyzed using Image J (Version 1.53).
2.8. Cellular uptake test
After exposure to H2O2 and LPS for 12 h, NIH-3T3 cells were incubated with DiD-labeled CuO@PS containing 5 μg/mL of CuONPs for varying time points. Then the cells were stained with LysoTracker Red (500 nM) for 20 min at 37 °C, and immediately observed using a CLSM (FV3000, Olympus). The images were analyzed and quantified with Image J (Version 1.53).
2.9. Intracellular Cu (I) ion detection
NIH-3T3 cells, pre-stimulated with H2O2 and LPS for 12 h, were added with CuONPs or CuO@PS at the same CuONPs concentration of 5 μg/mL for 12 h incubation. After washing with cold 1 × PBS three times to remove the unbound particles, cells were incubated with Coppersensor-1 (5 μM) and LysoTracker green (500 nM) at predetermined time points for 20 min at 37 °C. Afterwards, the cells were immediately imaged using a CLSM (FV3000, Olympus). The images were analyzed with Image J (Version 1.53).
2.10. Preparation of a thermosensitive hydrogel sheet
Pluronic F-127 (PF-127) powder was slowly added into cold 1 × PBS (pH 7.4) and magnetically stirred for 12 h within ice-bath to obtain a homogeneous polymer solution containing 15 % (w/v) of PF-127, and stored at 4 °C. Then, CuONPs or CuO@PS solution both containing 250 μg of CuONPs were resuspended into 5 mL of PF-127 solution in an ice bath under stirring to obtain the composite hydrogel sheets CuOφGEL or CuOPSφGEL, respectively. The methods to characterize the sol-gel transition temperatures, rheological, structural and morphological properties as well as dissociation rates of various hydrogel sheets were detailly described in supplementary methods.
2.11. In vitro release profiling
CuO@PS containing 500 μg of CuONPs were suspended into 20 mL of 1 × PBS at pH 7.4 or pH 5.0, and placed in a shaker at 100 rpm at 37 °C. At predetermined intervals, 1 mL of sample was collected and were replenished with 1 mL of fresh buffer. Then the collected samples were centrifuged at 12,000 rpm for 15 min. Soluble Cu ions remained in the supernatant, while CuO@PS and CuONPs were precipitated. Then the obtained precipitates were resuspended into 1 × PBS and transferred to a Sephadex G-50 column to separate the intact CuO@PS and the released CuONPs. The amounts of different Cu compositions were quantified by a modified Bis(cyclohexanone)oxaldihydrazone (BCO) colorimetric method at a wavelength of 600 nm.
The release of Cu ions, CuONPs or CuO@PS from CuOPSφGEL were evaluated following the same procedure described above. Briefly, 1 mL of CuOPSφGEL (containing 50 μg/mL of CuONPs) was immersed into 19 mL of 1 × PBS. At predetermined time points, 1 mL of sample was collected and were replenished with 1 mL of fresh buffer. The collected release medium was firstly precipitated by high-speed centrifugation (12,000 rpm, 15 min) to obtain CuO@PS and CuONPs, while Cu ions remained in the supernatant ready to detect the concentration. After resuspending the precipitates, CuO@PS and CuONPs were further separated using a Sephadex G-50 column. The amounts of Cu ions, CuONPs or CuO@PS were also determined by a modified a modified Bis(cyclohexanone) oxaldihydrazone (BCO) colorimetric method at a wavelength of 600 nm.
2.12. In vitro anti-bacterial assay
Escherichia coli (E. coli, ATCC 25922) and Staphylococcus aureus (S. aureus, ATCC 25923) were used to evaluate the antimicrobial activity of GEL, Silvercel™ and CuOPSφGEL. First, 200 μL of bacterial suspensions (1 × 106 CFU/mL) of E. coli or S. aureus were applied on the surface of each hydrogel formulation. After incubated at 37 °C for 6 h, the surface of each sample was washed down thoroughly with 2 mL of 1 × PBS to obtain the bacterial suspensions, which were further inoculated on Luria-Bertani (LB) agar plates after 1 × 10−5 times of diluting. The bacterial suspension without incubation with hydrogels were used as a control. CFU number of each plate was recorded after 24 h of incubation and the antibacterial rates as compared with the control group were calculated.
2.13. In vivo evaluations
2.13.1. Experimental animals
Male SD rats (6–8 weeks old, 170–210 g) were purchased from the Laboratory Animal Center of Sun Yat-sen University (Guangzhou, China). Animal experiments were approved by the Institutional Animal Care and use Committee of Sun Yat-sen university (Approval Number: SYSU-IACUC-2022-000988).
2.13.2. Establish of diabetic wound model in rat
To establish the type I diabetic SD rat model, rats were fasted for 12 h and injected intraperitoneally with streptozotocin (STZ) for a single injection (60 mg/kg, dissolved in citrate buffer at pH 4.5). The blood glucose levels of the rats were monitored for 19 days using an electronic glucometer (model 580, Yuwell). While blood glucose level exceeding 16.7 mM for 7 consecutive days which were deem diabetic, the back of rat was shaved and sterilized with alcohol under isoflurane anesthesia, and then a full-thickness round wound with a diameter of 10 mm was created on the dorsal skin.
2.13.3. Evaluations of diabetic wound healing
For wound healing experiments, the animals were randomly divided into four groups (6 rats per group), and treated with 300 μL of PBS (Control), blank PF-127 hydrogel (GEL), PF-127 hydrogel embedded with CuONPs (CuOφGEL), PF-127 hydrogel loaded with CuO@PS (CuOPSφGEL), respectively. All hydrogel sheets based on CuONPs or CuO@PS contain 50 μg/mL of CuONPs. Various formulations were changed every 3 days, and the wounds were photographed at days 0, 3, 7, 11 and 14 post-surgeries. After 14 days of treatments, all rats were sacrificed to collect the tissues at the wound sites for further examinations, including hematoxylin and eosin (H&E) staining, Masson's trichrome staining, fluorescence staining with Coppersensor-1, immunocytochemistry/immunofluorescence staining for CD31, MEK1/2 and p-MEK1/2. The immunostaining method was detailly described in supplementary methods.
2.13.4. Infected wound healing assay
Ten microliter of S. aureus suspension (∼108 CFU/mL) was added to the wound sites in diabetic rats. One day after inoculation, the rats with visible pus in the wounds were selected for further treatments. The animals were randomly divided into 3 groups (6 rats per group) and treated with 300 μL of PBS, CuOPSφGEL (containing 50 μg/mL of CuONPs), or applied with a commercially available antimicrobial dressing (Silvercel™). All the treatments were changed every 3 days, and the photos of the wound site were taken at day 1, 4, 7, 11 and 15 post-inoculation. To evaluate the in vivo antibacterial effect of each group, secretion from the wound fluids were collected on day 4, 7 and 14, and spread on LB agar plates for CFUs analysis.
2.14. Statistical analysis
All data in this study were analyzed using one-way ANOVA followed by the t-test analysis and presented as mean ± standard deviation (SD). *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001 were considered statistically significant.
3. Results
3.1. Synthesis and characterization of cupric-polymeric nanoreactors
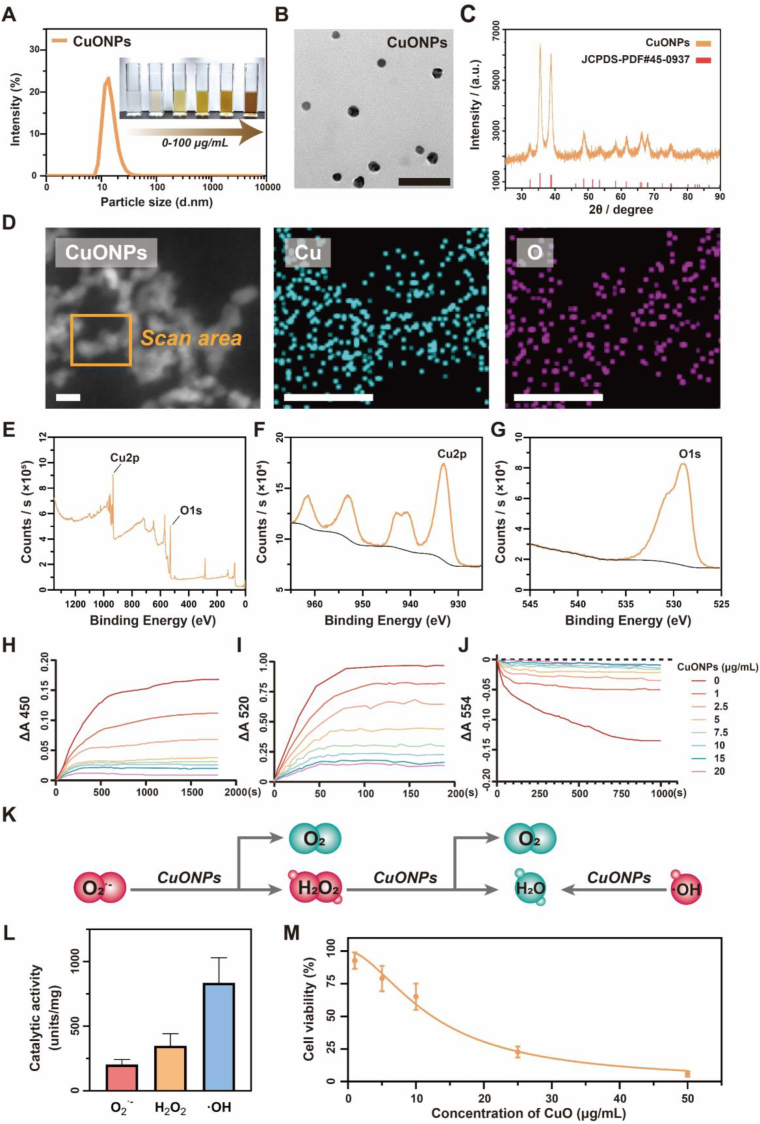
To mimic the catalytic activity of natural metalloenzymes against oxidized stress during wound healing process, copper oxide nanoparticles (CuONPs) were synthesized using a rapid precipitation method, achieving a considerable yield of 93.3 ± 3.8 % (Scheme S1) [36]. CuONPs showed a fine dispersibility in aqueous solution ranging from 1 to 100 μg/mL (Fig. 1A), probably because they displayed a narrow-distributed particle size lay in the range of 11.2 ± 1.5 nm (Fig. 1A–B, Table 1). To characterize the crystallite structure of CuONPs, the X-ray powder diffraction (XRD) analysis was applied to record the diffraction intensity in a 2θ range from 25° to 90° (Fig. 1C). It showed the crystal planes and the 2θ values of CuONPs highly matched to the standard (JCPDS card No. 45–0937). Additionally, high angle annular dark field scanning transmission electron microscope (HAADF-STEM) and energy dispersive spectrometry (EDS) mapping images further confirmed the formation of CuONPs which were composed of Cu and oxygen (O) (Fig. 1D). Consistently, the full X-ray photoelectron spectroscopy (XPS) spectrum also demonstrated that major components of the samples were Cu and O (Fig. 1E–G). Collectively, these results demonstrated CuONPs were successfully synthesized.
Fig. 1.
CuONPs displayed multienzymatic activities but associated with high toxicity. (A) Size distribution of CuONPs was measured by dynamic light scattering (DLS), and the inset digital photograph showing dispersity of CuONPs in aqueous solution at concentrations of 1, 5, 25, 50, 75 and 100 μg/mL. (B) Representative transmission electron microscopy (TEM) image of CuONPs. Scale bar, 50 nm. (C) XRD powder diffraction pattern of CuONPs. (D) HAADF-STEM and EDS mapping images for CuONPs. Cu (cyan), O (magenta). Scale bar, 20 nm. (E–G) Survey XPS spectrum of CuONPs (E), and the high-resolution XPS spectra of Cu 2p (F) and O 1s (G). (H–J) The absorbance changes of CuONPs incubated with WST-8 formazan at 450 nm (H), benzoquinone monoamine derivative at 520 nm (I), or unoxidized rhodamine B at 554 nm (J), which quantitatively reflects the amounts of O2•- H2O2, or •OH, respectively. (K) Schematic illustration of CuONPs exhibiting scavenging abilities to remove the main free radicals derived from oxygen. (L) The scavenging efficiency of CuONPs (5 μg/mL) for O2•-, H2O2 and •OH. (M) Viability of NIH-3T3 cells after incubated CuONPs at varying concentrations for 24 h. Three independent experiments were conducted for each result. All Data were presented as mean ± SD (n = 3 independent samples). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Table 1.
Characterization of CuONPs, PS and CuO@PS.
| Size (nm) | PDI | Zeta Potential (mV) | Loading Efficiency (%) | Encapsulation Efficiency (%) | |
|---|---|---|---|---|---|
| CuONPs | 11.2 ± 1.5 | 0.09 ± 0.01 | −15.6 ± 1.4 | / | / |
| PS | 123.3 ± 8.2 | 0.12 ± 0.02 | −8.8 ± 0.4 | / | / |
| CuO@PS | 129.1 ± 9.6 | 0.16 ± 0.02 | −9.4 ± 0.6 | 2.72 ± 0.34 | 40.45 ± 3.82 |
To evaluate the scavenging activity of CuONPs on three primary ROS, i.e., superoxide radical anion (O2•−), hydrogen peroxide (H2O2) and hydroxyl radical (•OH), different colorimetric methods were applied. The changes in absorbances of WST-8 formazan at 450 nm, benzoquinone monoamine derivative at 520 nm, and Rhodamine B at 554 nm after incubation with CuONPs were monitored to determine the scavenging activity of CuONPs on O2•−, H2O2, and •OH, respectively (Fig. 1H–J). CuONPs by increasing concentrations strongly reduced the changes in absorbance (ΔA) of each indicator, which were positively correlated with ROS contents, demonstrating an extensive scavenging activity of CuONPs for O2•−, H2O2 and extremely active •OH. However, there was no catalytic activities observe in copper (II) chloride (Fig. S1). Since O2•−, H2O2 and •OH are main toxic byproducts of various cellular O2-consuming redox processes [38], CuONPs displayed multi-enzymic activities to remove the most of ROS, thereby having the capability to recover the redox balance (Fig. 1K). Notably, CuONPs at the same CuONPs concentration (5 μg/mL) showed a superior catalytic efficiency to remove the highly toxic •OH in comparison with O2•− and H2O2 (Fig. 1L). In consistent with previous evidences concerning copper ions limited to a narrow therapeutic concentration range [39], CuONPs generated high toxicity in murine embryonic fibroblast cells NIH-3T3 while its concentrations exceeded above 10 μg/mL (Fig. 1M). Therefore, CuONPs-induced cytotoxicity appeared to impede its further medical applications.
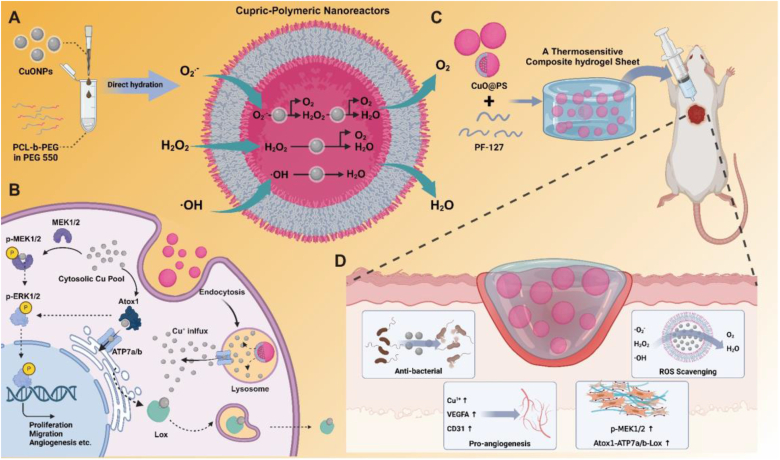
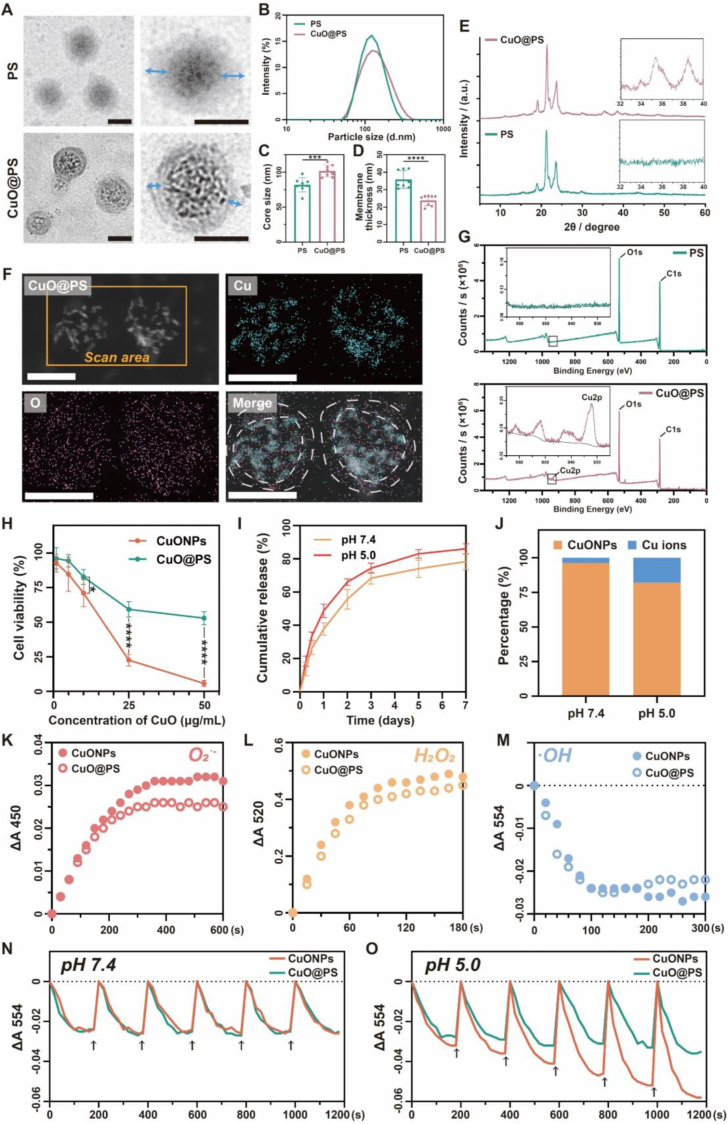
To address the overexposure risks induced by CuONPs, polymersomes fabricated by an amphiphilic block copolymer PEG-b-PCL, were employed to incorporate CuONPs to form cupric-polymeric nanoreactors (CuO@PS) via a modified direct hydration method [35,37], by which block polymers and payloads self-assembled into a polymersome in aqueous solution under benign conditions (50 °C, 2 h). In contrast to classical methods such as solvent-exchange or thin film hydration [40], the direct hydration method would allow CuONPs to directly load into the hydrophilic core of polymersomes without any organic solvents contaminated (Scheme 1A). The newly generated CuO@PS could be separated from unentrapped CuONPs by chromatography on a Sephadex G-50 column (Fig. S2), and Cu (II) concentrations in each fraction were determined based on a standard curve (Fig. S3). Through particle size measurement and transmission electron microscopy (TEM) observation, we found colloidal CuONPs were finely packed into the aqueous core of polymersomes, while achieving a loading efficiency (LE) of 2.72 ± 0.34 % and an encapsulation efficiency (EE) of 40.45 ± 3.82 % (Fig. 2A and Table 1). Moreover, CuONPs encapsulation seemed unlikely to disturb the core-shell structure, nanometer size, surface charge and integrality of polymersomes (Fig. 2A–B and Table 1), but enlarged the core diameter and reduced the membrane thickness (Fig. 2C–D). By XRD analysis, both CuO@PS and PS exhibited multiple strong peaks at 18°–25°, which were identified as characteristic peaks of the block copolymer PEG-b-PCL [41] (Fig. 2E). Moreover, CuO@PS also exhibited two peaks at similar positions between 32°–40° as CuONPs (Fig. 1, Fig. 2E), suggesting the presence of CuONPs in CuO@PS. Additionally, HAADF-STEM image and EDS elemental maps indicated Cu signals were mainly confined into an isolated space surrounding by a round shape of O signals probably attributed by CuO and PEG-b-PCL (Fig. 2F), which was in line with the core-shell morphological structure observed by TEM (Fig. 2A). Likewise, both PS and CuO@PS contained strong C and O signals in XPS spectra, however, only CuO@PS displayed a Cu 2p characteristic peak at almost the same binding energy as CuONPs (Fig. 1, Fig. 2G). Together, those data indicated CuONPs have been successfully encapsulated into PS, and been transformed to cupric-polymeric nanoreactors.
Scheme 1.
(A) Preparation of cupric-polymeric nanoreactors (CuO@PS) via a direct hydration method for in situ cascade catalytic reactions. (B) Cu+ released from lysosomal CuO@PS integrate into cytosolic Atox1-ATP7a/b-Lox axis, leading to direct and indirect activation of MEK1/2 signaling pathway. (C) Fabrication process of a thermosensitive hydrogel sheet CuOPSφGEL. (D) Multifunction of CuOPSφGEL contributing to the chronic wound healing.
Fig. 2.
Characterization of cupric-polymeric nanoreactors. (A) Representative TEM images of PEG-b-PCL polymersomes (PS) and CuONPs-encapsulated polymersomes (CuO@PS), and blue arrows indicating the thickness of polymeric membranes. Scale bar, 100 nm. (B) Size distribution of PS and CuO@PS. (C–D) Quantification of core diameter (C) and membrane thickness (D) from eight polymersomes randomly selected from TEM images. (E) XRD powder diffraction pattern of CuO@PS and PS in wide range. The inset figures indicated the high-resolution XRD patterns at 2θ range from 32° to 40°. (F) HAADF-STEM and EDS mapping images for CuO@PS. Cyan represents Cu signals, while magenta indicates O signals. White dash line shows the area of polymeric membrane in the merged image. Scale bar, 100 nm. (G) Survey XPS spectra of PS and CuO@PS. The inset figures show the high-resolution XPS spectrum of Cu 2p. (H) Cytotoxicity of NIH-3T3 cells after treated with different concentrations of CuONPs or CuO@PS for 24 h. At each concentration point, CuO@PS and unencapsulated CuONPs had equal weight concentrations of CuONPs. (I) In vitro copper release profile of CuO@PS at pH 7.4 or pH 5.0. (J) The amounts of CuONPs and Cu ions released from CuO@PS over 48 h at pH 7.4 or pH 5.0, respectively. (K–M) Time-dependent absorbance changes of WST-8 formazan at 450 nm (K), benzoquinone monoamine derivative at 520 nm (L), or unoxidized rhodamine B at 554 nm (M) in the presence of CuONPs or CuO@PS both at a CuONPs concentration of 5 μg/mL to quantify the unreacted O2•-, H2O2 and •OH. (N, O) Cycling performance of CuONPs or CuO@PS to remove •OH both at a CuONPs concentration of 5 μg/mL at pH 7.4 (N) or pH 5.0 (O). Black arrows indicate the time points to introduce •OH into reactions and each cycle lasts for 200 s. Three independent experiments were conducted for each result. Data represent mean ± SD. *p < 0.05, ***p < 0.001, ****p < 0.0001. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Notably, the cytotoxicity of CuONPs was reduced when the same amounts of CuONPs were encapsulated into polymersomes (Fig. 2H), probably because CuONPs with the smaller size and the greater surface area were more easily to cause severe cell damages in relative to CuO@PS [42]. Next, the cargo released from CuO@PS was profiled, illustrating CuO@PS did not show an initial burst release but exhibit a continuous slow release over 7 days (Fig. 2I). Moreover, the release rate of CuO@PS was moderately enhanced at acidic pH (pH 5.0) as compared to physiological pH (pH 7.4) (Fig. 2I). In release medium, most of copper element were detected in the form of colloid CuONPs, rather than free Cu ions (Fig. 2J). Significantly, the similar absorbance differences suggested CuO@PS still exerted the comparable abilities to scavenge free radicals including O2•−, H2O2 and •OH as CuONPs group (Fig. 2K–M). Additionally, there was no ROS scavenging ability observed in blank polymersomes (Fig. S1). Since most of CuONPs were shielded in the polymersome cavities from external environment, it could be inferred that CuO@PS enabled to conduct the tandem catalytic reactions in an isolated space with the permeable ROS across polymeric membrane. Therefore, it did not only preserve the activity and selectivity of CuONPs, but also enhanced their biocompatibility with cells. Strikingly, in contrast to CuONPs, CuO@PS persisted the comparable scavenging activity of ROS, e.g., upon repetitive cycles of •OH adding, either at pH 7.4 or at pH 5.0 (Fig. 2N–O).
Together, our results demonstrated that cupric-polymeric nanoreactors CuO@PS wherein artificial metalloenzyme based on CuONPs were effectively incorporated into polymersomes, exhibited superoxide dismutase- and catalase-like activities, and more remarkable hydroxyl radical scavenging abilities to complete cascade catalytic reactions without producing toxic radical intermediates (Scheme 1A). Strikingly, CuO@PS as an intact nanoreactor enabled not only to selectively react with ROS in a separate space to circumvent the cytotoxicity of CuONPs, but also to achieve the recycling of CuONPs.
3.2. Cupric-polymeric nanoreactors exerted antioxidant, anti-inflammatory, proliferative, and angiogenic effects in vitro
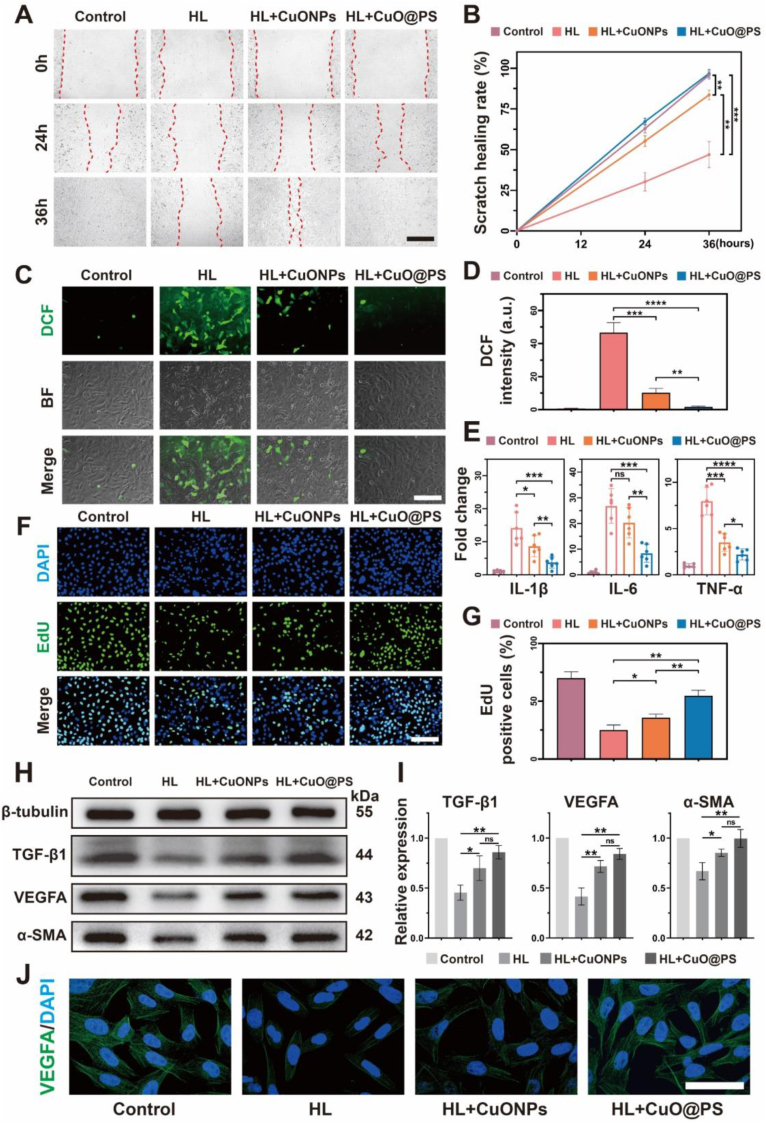
To select the optimal concentrations of CuONPs and CuO@PS in vitro, a scratch-wound assay was applied based on a fibroblast cell line NIH-3T3 following stimulation of 500 μM of H2O2 and 500 ng/mL of lipopolysaccharide (LPS) for 12 h. Either CuONPs or CuO@PS at the weight concentration of CuONPs (5 μg/mL) showed the most significant cell migration ability, therefore, 5 μg/mL was selected for further investigation (Fig. S4). Notably, CuO@PS remarkably heal the scratch area over 36 h, as compared to those H2O2/LPS (HL) stimulated cells without any treatment or treated with equal amounts of free CuONPs, and close to the extent of uninjured cells (Fig. 3A–B). Additionally, HL treatment may induce the ROS production, initiate the inflammatory response, and impair cell growth and proliferation in fibroblasts [43]. In consistent with the observations in free radical scavenging tests without cells, addition of CuONPs to HL injured cells moderately alleviated ROS productions, which could be detected using DCFH-DA assay (Fig. 3C–D). By contrast, CuO@PS strongly boosted the antioxidant properties of CuONPs, and the ROS levels were recovered to the unstimulated level (Fig. 3C–D). In addition, NIH-3T3 pre-treated with HL strongly increased the mRNA production of proinflammatory cytokines, i.e., IL-1β, IL-6 and TNF-α, however, CuO@PS and to a less extent CuONPs significantly attenuated the expression of those proinflammatory genes, showing the superior anti-inflammatory effects (Fig. 3E). Furthermore, the fibroblast proliferations in the presence of various formulations were determined utilizing EdU assay to assess the healing potential. As shown in Fig. 3F–G, EdU-positive 3T3 cells were significantly decreased upon exposure of HL for 12 h, however, the stimulated cells further processing with CuO@PS remarkably promoted cell proliferation as evidenced by the most EdU-positive cell percentages (approx. 55 %), although CuONPs were also able to mildly activate the proliferation. Except for the lack of redox activity (Fig. S1), in contrast, either free Cu2+ (63 μM, equal to the molar concentration of 5 μg/mL of CuONPs) or blank polymersomes (PS) showed no effects on cell migration and proliferation (Figs. S5–S6). Thereby, the results suggested that copper nanoparticles, rather than cupric ions or polymeric vesicles, exerted biological activities in fibroblasts.
Fig. 3.
Cupric-polymeric nanoreactors promoted cell migration and exerted antioxidant, anti-inflammatory, proliferative, and angiogenic effects in cells. NIH-3T3 cells were stimulated with 500 μM of H2O2 and 500 ng/mL of LPS (HL) for 12 h, and further treated with CuONPs or CuO@PS both containing 5 μg/mL of CuONPs followed by different measurements. (A) Cell migration of NIH-3T3 cells affected by different treatments was monitored within 36 h. Scale bar, 400 μm. (B) The scratch healing rates were calculated by the changes in the scratch areas from each group (A) over time. (C) Intracellular ROS levels in cells detected by DCFH-DA assay with a confocal laser scanning microscopy (CLSM). Scale bar, 200 μm. (D) Quantitative analysis of intracellular ROS based on DCF fluorescence intensities. (E) mRNA expressions of IL-1β, IL-6 and TNF-α after various treatments. (F) EdU cell proliferation assay on NIH-3T3 cells with various treatments as described above. Scale bar, 200 μm. (G) EdU positive cell percentages representing proliferative cell rate under different treatments. (H–I) Western blot and quantification analysis of TGF-β1, VEGFA and α-SMA expressions in NIH-3T3 cells. β-tubulin was used as a loading control. (J) Immunofluorescence staining of VEGFA in HUVEC cells after different treatments. Scale bar, 60 μm. All data were shown as means ± SD. Three independent experiments were conducted for each result. ns, not significant, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
Moreover, CuO@PS, significantly activated the expressions of growth factors in fibroblasts, including transforming growth factor-β (TGF-β) and vascular endothelial growth factor A (VEGFA), and further stimulated the production of alpha-smooth muscle actin (α-SMA) for accelerating the wound healing as compared to HL stimulated group (Fig. 3H–I). Correspondingly, we also found CuO@PS elevated VEGF expressions in an endothelial cell line HUVECs, validating the potent effects of CuO@PS in angiogenesis (Fig. 3J). However, the expressions of TGF-β, α-SMA and VEGFA induced by CuONPs were insignificantly lower than those in CuO@PS-treated cells under the same conditions. Thus, in comparison with CuONPs, cupric-polymeric nanoreactors CuO@PS promoted wound closure via in vitro scratch assay, and displayed remarkable antioxidant, anti-inflammatory, proliferative and angiogenic properties that might contribute to accelerate the healing process in vivo, however, it was unlikely to fully explain by the activation of TGF-β or VEGFA associated signaling pathways.
3.3. Cupric-polymeric nanoreactors integrated into copper metabolism to activate MEK1/2 phosphorylation
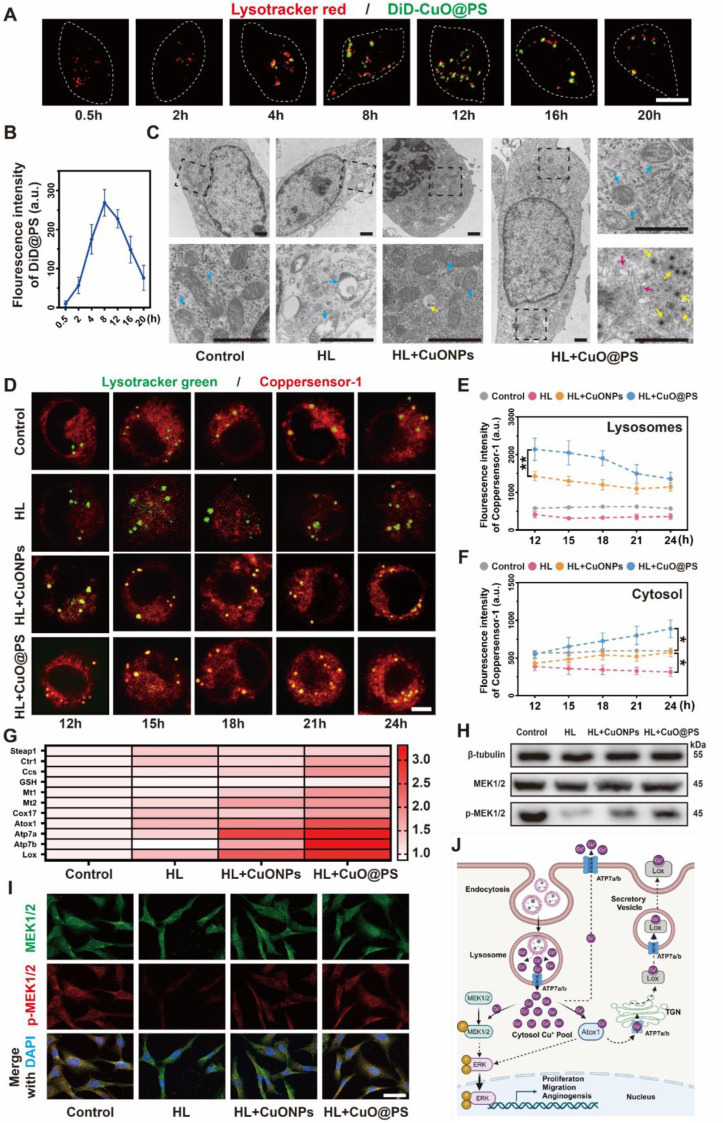
To elucidate the mechanisms underlying the therapeutic differences among cupric ions, CuONPs and CuO@PS, cellular uptake and copper distribution were further assessed. Firstly, cellular entry of cupric-polymeric nanoreactors in NIH-3T3 cells was tracked over time by fluorescent dye DiD labeling CuO@PS. Although CuONPs entered cells possibly through the processes of endocytosis or membrane diffusion [44], CuO@PS seemed to be internalized by cells via endocytosis, and continuously trafficked into the lysosomes marked by lysotracker probes, reaching an uptake peak at 12 h and followed by a gradual decline (Fig. 4A–B). Consistently, TEM analysis further confirmed CuO@PS extensively accumulated into the lysosomes (Fig. 4C, yellow arrows), that located close to the Golgi apparatus (Fig. 4C, magenta arrows), yet few CuONPs were detected in whole cells. Moreover, swelling cristae and vacuole-like mitochondria were present in NIH-3T3 cells that were damaged by H2O2/LPS (Fig. 4C, blue arrows), but this phenomenon was largely improved by the addition of CuONPs and CuO@PS.
Fig. 4.
Cupric-polymeric nanoreactors influenced intracellular copper metabolism after endocytosis. (A) Representative CLSM images of H2O2 and LPS (HL) stimulated NIH-3T3 cells incubating with CuO@PS labeled by DiD (green) for varying time points. At predetermined time points, the lysosomes were stained with lysotracker (red) and immediately for image acquisition. Scale bar, 10 μm. (B) Quantitative analysis of fluorescence intensities of fluorescently labeled CuO@PS over time. 50–100 individual cells were randomly selected from each sample. (C) Representative TEM images of the ultrastructure of NIH-3T3 cells followed by different treatments for 12 h (yellow arrows: Cu-based nanoparticles in lysosomes; blue arrows: mitochondria; magenta arrows: trans-Golgi network). Scale bar, 1 μm. (D) Representative fluorescent images of intracellular Cu+ distribution. Upon exposure to HL stimulation for 12 h, NIH-3T3 cells were treated with CuONPs or CuO@PS both containing CuONPs concentrations of 5 μg/mL for 12 h, and then the cells were washed and detected with Cu+ probe Coppersensor-1 (CS-1, red) at different time points. Lysosomes were stained with lysotracker (green). Scale bar, 10 μm. (E–F) Quantitative analysis of fluorescent intensities of Cu1+ in lysosomes and cytosol based on time-lapse images. For each sample, 50–100 individual cells were imaged and analyzed. (G) Heat map of the real-time quantitative PCR (qRT-PCR) analysis results of copper metabolism-related genes in NIH-3T3 cells after different treatments. (H) Western blot of MEK1/2 and phospho-MEK1/2 (p-MEK1/2) proteins in NIH-3T3 cells under different treatments. β-tubulin was used as a logading control. (I) Immunofluorescence staining of MEK1/2 and p-MEK1/2 in HUVEC cells after different treatments. Scale bar, 30 μm. (J) Schematic illustration of Cu+ derived from CuO@PS activating Atox1–ATP7a/b–Lox and MEK1/2 pathways involved in Cu metabolism. All experiments were performed in biological triplicates Results were presented as means ± SD. *p < 0.05. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Since copper ions primarily existed in a reduced state (Cu+) in the cell, a specific Cu+ probe coppersensor-1 was used to monitor Cu+ distribution after 12 h of CuONPs or CuO@PS incubation with NIH-3T3 cells. In healthy cells, Cu+ ions were mostly diffused throughout cytoplasm and merely detected in the lysosomes, however, the Cu+ levels were clearly decreased in the whole cells after H2O2/LPS treatment (Fig. 4D–F). Although supplementing of CuONPs moderately elevated the Cu+ levels in injured cells, CuO@PS seemed to significantly increase cellular Cu+ signals, especially in lysosomes, probably because polymeric coating facilitated the transport of inorganic nanoparticles into the lysosomes [45]. With time increasing, lysosomal Cu+ derived from CuO@PS were constantly exported into cytosol to provide an intracellular Cu pool (Fig. 4D–F). Conversely, the Cu+ efflux driven by CuONPs in lysosomes was moderately detected in cells, as similar to the level of normal cells.
Given Cu as a key modulator of cell signal transduction pathways, cellular Cu+ concentrations increased by CuO@PS, not by CuONPs, stimulated the gene expressions of Cu-dependent transporters, including Cu cell importer Ctr1 and exporter ATP7a/b, while cytosolic Cu chaperones such as CCS, Cox17, Atox1 were also elevated to bind the free Cu ions assuring the appropriate Cu level (Fig. 4G). Additionally, Cu cellular chelator metallothionein 1 and 2 (MT1/2), rather than another copper reservoir glutathione (GSH), were also enhanced by CuO@PS, which might explain CuO@PS did not induce the cytotoxicity by excessive copper. Since CuO@PS were directly endocytosed by cells, the expression of metalloreductases six-transmembrane epithelial antigen of the prostate 1 (Steap1), which was responsible for reducing divalent Cu2+ to monovalent Cu+ available for cellular uptake, was not affected. Remarkably, the gene expression levels of Atox1, ATP7a/b and Lox were the most significantly elevated. Previous study [46] demonstrated that Atox1 might promote cell migration via ATP7a/b-Lox axis, where Atox1 shuttled Cu via ATP7a/b to the trans-Golgi network (TGN) for the secretory of Cu-containing enzymes Lox. Lox plays important roles in extracellular matrix formation for healing process [47]. Additionally, cytosol Cu+ directly binds with mitogen-activated protein kinase (MEK) to initiate the cell proliferation and angiogenesis [48], therefore, phosphorylation levels of MEK1/2 were measured to evaluate the MEK1/2 activation. In compared with CuONPs, CuO@PS strongly phosphorylated MEK1/2 in LPS/H2O2 treated 3T3 cells and HUVEC cells (Fig. 4H–I), which presumably activated the downstream extracellular signal-regulated kinase (ERK) [49]. Therefore, CuO@PS localized in lysosomes might serve as a Cu+ depot to drive a copper influx into cytoplasm, leading to the transmission of growth signals to Atox1–ATP7a/b–Lox and MEK1/2 pathways (Fig. 4J).
3.4. Cupric-polymeric nanoreactors dispersed into a thermo-responsive hydrogel to form a composite hydrogel sheet
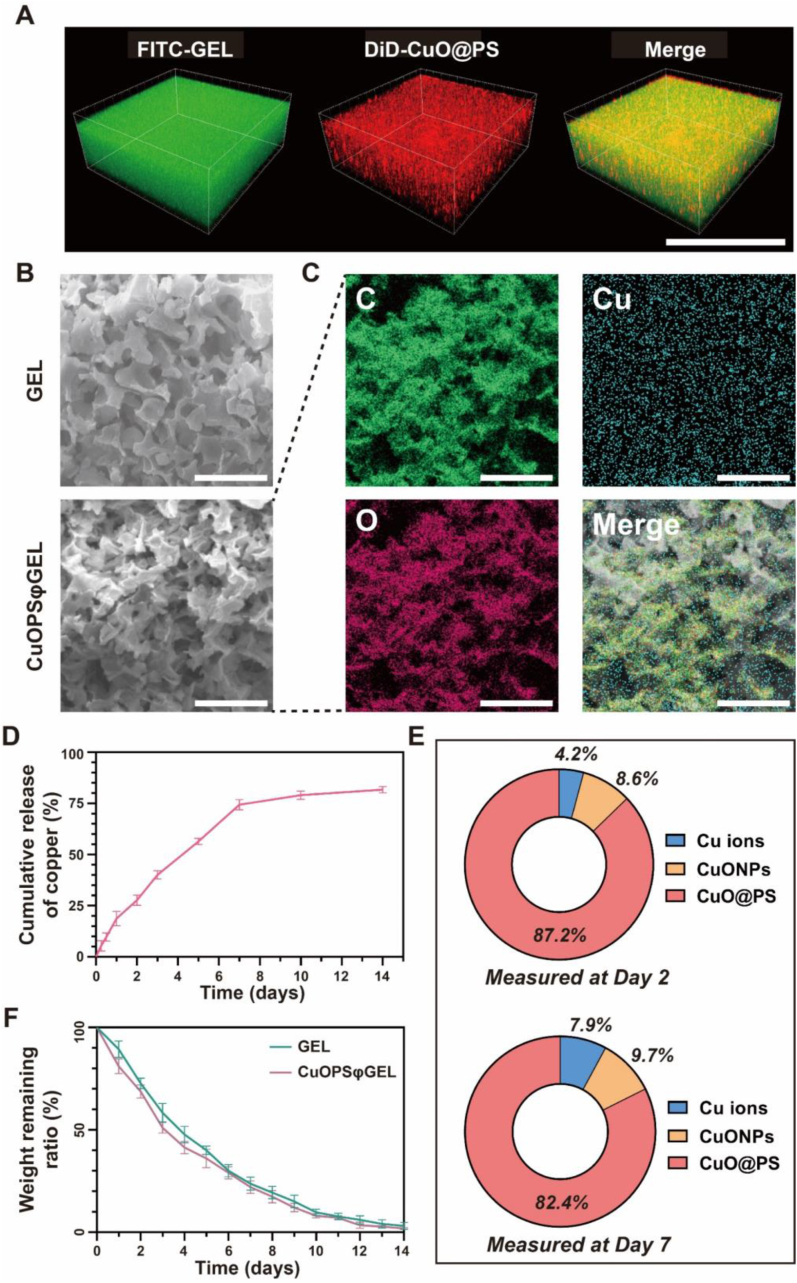
In order to precisely lay copper catalysts at the skin injury cite, CuONPs or CuO@PS can be loaded into 15 wt% of Pluronic F-127 (PF-127) hydrogel by a simple dispersion method to form the hydrogel sheets CuOPSφGEL or CuOφGEL both containing of 50 μg/mL CuONPs (Scheme 1C). Addition of CuO@PS did not affect the solution-to-gel transition property of PF-127, i.e., solution phase at 4 °C and gel phase at 37 °C (Fig. S7A). Moreover, CuOPSφGEL, similar as blank PF-127 (GEL), can be kept in syringes at 4 °C and injected into the wound site to form a fully-fitted hydrogel in 20 s (Fig. S7B). Surprisingly, gelation temperature (GT) of PF-127 hydrogel was visibly increase from 30 °C to 37 °C while mixing with CuO@PS, not with CuONPs (Fig. S8). Since the skin wound temperature maintains around 32–37 °C [50], CuOPSφGEL seemed to fit for topical administration. Moreover, rheological measurements including temperature sweep and frequency sweep, were conducted to estimate the thermo-responsive gelation behavior and mechanical properties of hydrogels. As shown in Fig. S9A, the storage modulus (G′) exhibited a significant rise with increasing temperature, while the loss modulus (G″) did not change with temperature, implying a clear temperature-dependent sol-gel transition property. Additionally, G′ exceeded G″ over the whole range of measured frequency, thus establishing the stability of all gel formulations (Fig. S9B). Additionally, Young's modulus and cytotoxicity were not markedly changed after dispersion of nanoparticles into PF-127 (Figs. S10 and S11). Moreover, we found hydrophobic dye DiD labeled-nanoreactors (red) remained homogeneous dispersion in the PF-127 gel labeled by hydrophilic dye fluorescein isothiocyanate (FITC, green) after 7 days storage at 37 °C (Fig. 5A). The hydrogel sheets exhibited a porous structure evaluated by scanning electron microscopy (SEM) (Fig. 5B). Then, the elemental composition of CuOPSφGEL was analyzed using EDS. Carbon (C) and Oxygen (O) signals were contributed by PF-127 and PEG-b-PCL, while Cu signals indicated cupric-polymeric nanoreactors, which further verified that cupric-polymeric nanoreactors homogeneously distributed throughout PF-127 hydrogel matrix (Fig. 5C). Correspondingly, the release of copper was more greatly impeded from CuOPSφGEL as compared to those from CuO@PS (Fig. 2, Fig. 5D). Additionally, most of CuONPs preserved in the polymersomes were released from CuOPSφGEL (Fig. 5E), probably because PF-127 hydrogel matrix can sustainably dissociate within 14 days (Fig. 5F). Thus, cupric-polymeric nanoreactor CuO@PS can be evenly dispersed into a thermosensitive PF-127 hydrogel to become a composite hydrogel sheet as a wound dressing, which might facilitate the topical application and sustainable release of CuO@PS for chronic wounds.
Fig. 5.
Characterization of a composite hydrogel sheet (CuOPSφGEL) for chronic wound healing. CuOPSφGEL was formed by CuO@PS dispersed into 15 wt% of Pluronic F-127 (PF-127) hydrogel with a final CuONPs concentration of 50 μg/mL. (A) Three-dimensional fluorescent imaging of CuOPSφGEL after 7 days of storage at 37 °C by using CLSM. FITC (green) was used to label hydrophilic gel networks, while DiD (red) was used to label CuO@PS. Scale bar, 100 μm. (B) Morphological characterization of GEL and CuOPSφGEL by scanning electron microscopy (SEM). Scale bar, 40 μm. (C) Elemental distribution analysis of carbon (C), oxygen (O) and copper (Cu) in CuOPSφGEL using EDS mapping. Scale bar, 40 μm. (D) In vitro release profile of CuOPSφGEL within 14 days. (E) Pie chart showing the percentages of the amounts of CuO@PS, CuONPs and Cu ions released from CuOPSφGEL at day 2 and day 7. (F) Weight loss of 1 mL of GEL or CuOPSφGEL samples after immersion in 19 mL PBS at 37 °C over time. Results were shown as means ± SD. Three independent experiments were conducted for each result. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.5. Cupric-polymeric nanoreactors accelerated chronic wound healing in diabetic rats
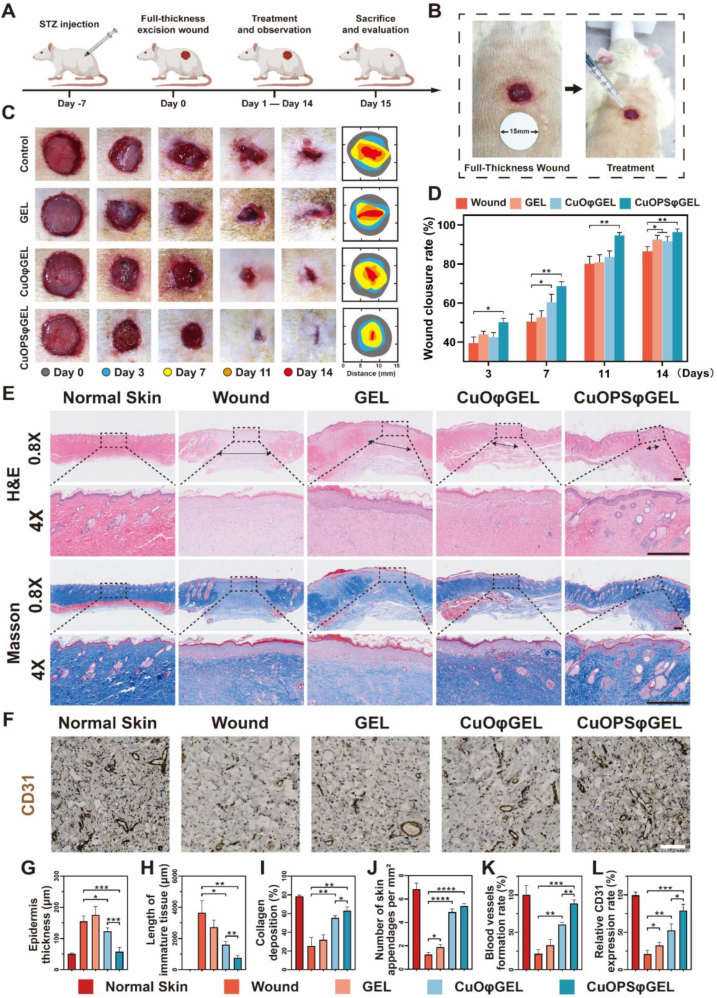
Streptozotocin (STZ)-induced type Ⅰ diabetic rats were employed to assess the pro-reparative effects of various formulations in vivo [51]. Rats were considered diabetic if their blood glucose levels exceeded 16.7 mM for 7 consecutive days after STZ injection (Fig. S12), then a full-thickness round excision wound with a diameter of 10 mm was established on the back of rats to mimic the diabetic foot ulcer [52] (Fig. 6A–B). All animals were randomly divided into four groups and further treated with 300 μL of PBS (control) and different hydrogel sheets. The administrated gels formed a transparent sheet with 3–4 mm thickness, and the gel dressings were changed to fresh every 3 days (Fig. 6A–B). The wound closure areas were monitored over time (Fig. 6C), demonstrating CuOPSφGEL strongly accelerated the early recovery process of wounds with the highest wound closure rate, as compared with other groups within 14 days of treatments (Fig. 6D). To be note, the regenerated skin by CuOPSφGEL treatment was smoother and flatter without scar hyperplasia by visual observation and touching than those in CuOφGEL group.
Fig. 6.
CuOPSφGEL promoted the chronic wound healing in diabetic rat models. (A) Scheme illustrating the establishments of excisional wounds on diabetic rats induced by streptozotocin (STZ), and the treatments of various hydrogel sheets for 14 days. (B) Photographs showing the full-thickness round excision wounds covered with CuOPSφGEL. (C) Representative photographs of the wounds following different treatments at varying days. (D) Wound closure rates from each group were measured at different days. (n = 6). (E) Representative 0.8 × and 4 × magnification of hematoxylin and eosin (H&E) and Masson's trichrome (Masson) staining of wound tissues after 14 days of different treatments. Black arrows in H&E images indicate the length of immature tissue. Scale bar, 500 μm. (F) Expressions of CD31 in wound tissues from different groups at day 14 detected by immunohistochemistry (IHC) staining. Scale bar, 100 μm. (G–I) Quantitative analysis of epidermis thickness (G), length of immature tissue (H), collagen deposition rate (I), number of skin appendages (J), blood vessels formation rate (K) and CD31 expressions (L) of each group based on H&E staining (E) and IHC staining (F). Areas of 1 mm2 randomly selected from each group (n = 6) were used for analysis. Results were presented as means ± SD. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
In the process of re-epithelialization, the thick immature epidermis will gradually transition to thin mature epidermis, therefore, epidermal thickness is an important indicator for wound healing [53]. In consistent with visual and touching evaluations, hematoxylin and eosin (H&E) pathological staining demonstrated that CuOPSφGEL significantly decreased the epidermis thickness, and promoted immature granulation tissue to transform into mature dermal tissue as compared with other groups, being the closest ones to the normal skin (Fig. 6E, G and H). Moreover, the wounds treated with CuOPSφGEL and CuOφGEL showed the remarkable collagen deposition evaluated by Masson's trichrome staining, and their collagen fibers were better arranged and denser, especially in the CuOPSφGEL group (Fig. 6E and I). In addition to the obvious enhancements on the density of skin appendages including sweat glands, sebaceous glands and hair follicles (Fig. 6E and J), the newly formed blood vessels labeled with endothelial markers CD31 were also strongly increased in CuOPSφGEL group (Fig. 6E, F, K and L). Additionally, CuOPSφGEL and CuOφGEL did not cause any observed harm to rats according to body weights and the H&E staining of different organs (Figs. S13A and S14).
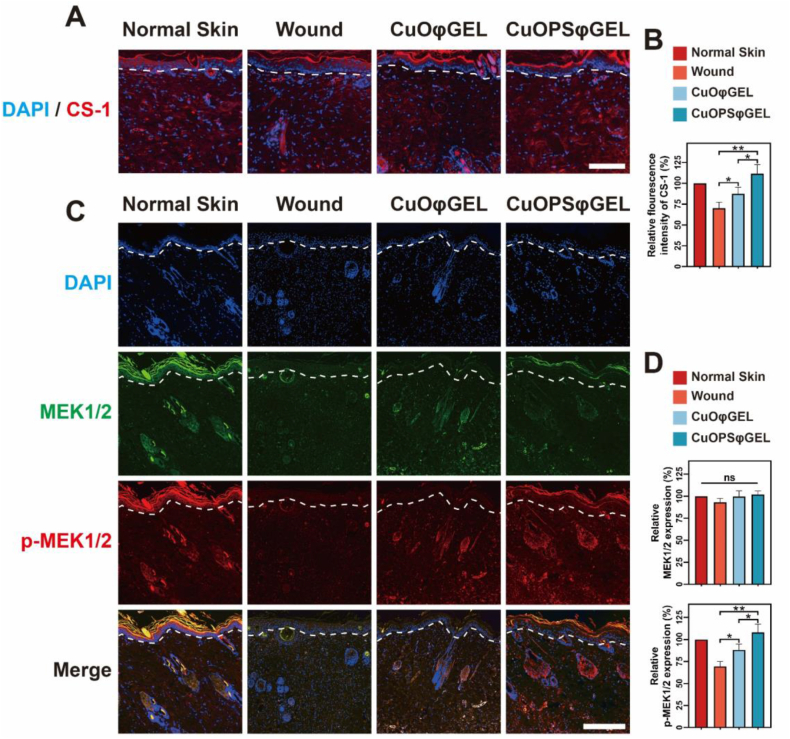
Moreover, after laid with various gel sheets for 24 h, the skins were excised to detect the Cu+ distribution by fluorescence staining. The strongest Cu+ signals were detected both in epidermis and dermis from the rats treated with CuOPSφGEL as compared to those rats treated with CuOφGEL or without treatments (Fig. 7A–B). Most significantly, we also found that MEK phosphorylation levels were remarkably enhanced in the wound area with abundant Cu+. These data might suggest accumulating Cu+ that were derived from CuOPSφGEL can positively impact on MEK activation to promote chronic wound healing in vivo (Fig. 7C–D). Therefore, CuOPSφGEL treatment accelerated would repair by enhancing re-epithelialization, collagen deposition, angiogenesis, which is driven by a Cu+ mild elevation to stimulate MEK phosphorylation.
Fig. 7.
Cu+ distribution and MEK1/2 expressions in wound tissues after various treatments. (A) Tissue distribution of Cu+ in wound tissues were probed by Coppersensor-1 (CS-1, red) after 24 h of treatments with different hydrogel sheets. White dashed lines indicate a boundary between epidermis and dermis. Scale bar, 200 μm. (B) Mean fluorescence intensities of CS-1 in the stained skins from each group (A) were determined by the analysis software (Image J). (C) Fluorescent staining of MEK1/2 (green) and p-MEK1/2 (red) in skin tissue sections of diabetic rats from each group. White dashed lines show a boundary between epidermis and dermis. Scale bar, 400 μm. (D) The relative expression levels of MEK1/2 (upper panel) and p-MEK1/2 (lower panel) were quantified based on fluorescent intensity analysis of (C). Cell nuclei were stained with DAPI (blue). Areas of 1 mm2 randomly selected from each group (n = 6) were used for quantitative analysis. Results were shown as means ± SD. ns, not significant, *p < 0.05, **p < 0.01. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.6. Cupric-polymeric nanoreactors promoted the healing of infected diabetic wounds
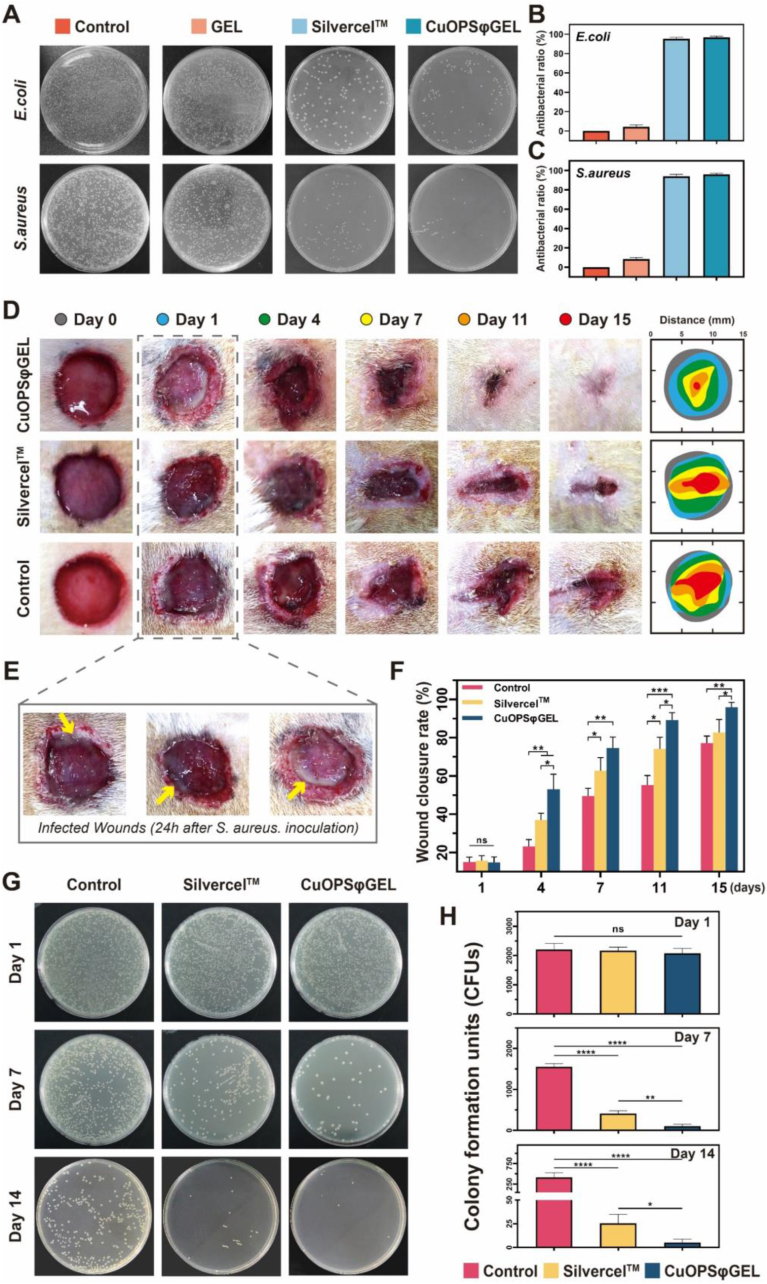
Since chronic open wounds are highly susceptible to bacterial infections, antibacterial experiments were further conducted to assess the clearance efficiency of CuOPSφGEL on Escherichia coli (E. coli) or Staphylococcus aureus (S. aureus) bacteria, which predominate in wound infections [54]. CuOPSφGEL containing CuONPs (50 μg/mL) strongly killed the bacterial approaching 100 % of inhibitions against both E. coli and S. aureus, as similar to those of the commercial alginate-silver ion (Ag+) antimicrobial wound dressing, Silvercel™ (Fig. 8A–C). Next, the infected wounds on diabetic rats were treated with PBS (control), Silvercel™ and CuOPSφGEL to assess the antibacterial efficiency in vivo, while a large amount of pus being present in the wounds after one day of infection (Fig. 8D and E, yellow arrows). While monitoring wound closure, the bacterial dispersal at varying times post inoculation was assessed as a number of colony-forming units (CFUs). With the extension of time, CuOPSφGEL group showed a significant higher antibacterial rate and body weight gain than those of Silvercel™ or PBS Control, as well as decreasing the bacterial load of wound fluid (Fig. 8D, F–H, S13B). Upon exposure to CuOPSφGEL for 14 days, significantly, there was barely few CFU detected in the wound exudates (Fig. 8G and H). Therefore, CuO@PS dispersed into a thermo-sensitive PF-127 hydrogel exhibited remarkable antibacterial capacity than commercial Ag-containing wound dressing.
Fig. 8.
CuOPSφGEL prevented the bacterial infections in diabetic chronic wounds. (A) In vitro antibacterial activities were estimated through which 1 mL of GEL or CuOPSφGEL containing 50 μg/mL of CuONPs were incubated with 200 μL of E. coli or S. aureus suspensions (106 CFU/mL) at 37 °C for 6 h, following by the CFU assay. One piece of Silvercel™ was used as positive control following the same procedure. (B–C) Antibacterial rates of various treatments were calculated by counting numbers of the bacterial colonies on agar plates as compared to the control plate incubated with 106 CFU/mL of E. coli or S. aureus suspensions from (A). (D) Representative photographs of the infected wounds upon exposure to various treatments over time. (E) Photographs showing the pus (yellow arrows) at wound sites after 24 h of S. aureus inoculation. (F) Wound closure rates were quantified based on the wound areas (D) over time. (G) Agar culture of wounds secretions (diluted 10−5 times with sterile PBS) from each rat at day 1, 7 and 14. (H) Quantitative analysis of CFUs on agars from (G). Data were presented as means ± SD. Three independent experiments were conducted for each result. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
4. Discussion
As far as 2600 BC, the Egyptians have discovered that Cu can sterilize wounds and water. Currently, a vast of biological activities of Cu-based compounds are increasingly being identified, i.e., antioxidant, anti-inflammatory, proliferative, antimicrobial, angiogenesis properties which are beneficial to chronic wound healing. In contrast to silver (Ag)-containing commercial wound dressings only useful to prevent or treat infections, Cu nanoparticles are cheap and easily prepared, and confer many benefits in each stage of wound healing. Due to high toxicity and low stability, e.g., oxidation or aggregation, however, the applications of Cu nanoparticle in medicine are still under debate. Cu nanoparticle can impair the cell membrane integrity or organelles functions by direct contact, or produce excessive ROS to indirectly damage biomacromolecules including lipids, proteins, or nucleic acids [[55], [56], [57], [58]]. Therefore, how to expand the therapeutic window of Cu nanoparticle but avoid its detrimental accumulation in the cells remains a major challenge. In this study, CuONP are efficiently encapsulated into the lumen of PEG-b-PCL polymersomes to form cupric-polymeric nanoreactors (CuO@PS), which ensures Cu-assisted catalytic reactions more benign and specific in a favorable space that reduces the toxic overexposure, leading to an extended therapeutic concentration range (Scheme 1). Additionally, CuO@PS can be recycled with persistent catalytic activity for multiple times. As results, CuO@PS strongly alleviated the cellular oxidative stress induced by LPS/H2O2, in turn promoting fibroblast cell proliferation and migration as compared to CuONPs, free cupric ions or blank polymersomes. After CuO@PS dispersed into a thermosensitive hydrogel based on PF-127 to obtain a composite hydrogel sheet, CuOPSφGEL exerted pro-healing and anti-bacterial effects in diabetic chronic wounds, in comparison with a commercial wound dressing Silvercel™ containing Ag nanoparticles (Scheme 1). Although many polymeric compositions [[59], [60], [61], [62], [63]] are used to stabilize CuONPs, and prevent them from oxidation and aggregations, our study showed that CuONPs simply distributed into PF127 hydrogel matrix (CuOφGEL) displayed less efficient in healing than CuOPSφGEL, probably because cupric-polymeric nanoreactors prevent the rapid release of free CuONPs, but enable an in-situ control of Cu-based catalytic reactions.
Strikingly, although CuO@PS increased the expressions of TGF-β, α-SMA or VEGFA in fibroblasts and endothelial cells, the mechanisms of CuO@PS behind pro-healing functions could not be simply attributed to the activations of TGF-β or VEGFA. In addition to being a nanoreactor for scavenging ROS in extracellular space, nanosized CuO@PS can enter cells via endocytosis and accumulate in the lysosomes. Acidic environments in lysosomes (pH 5.0) may not influence the catalytic activities of CuO@PS, but slightly accelerated the release of CuONPs release and the dissociation of copper ions as compared to those at pH 7.4, which drives a relatively flat gradient concentration of Cu+ from lysosomes into cytoplasm. We also found LPS/H2O2 stimulated cells underwent Cu deficiency, consistently, some researchers suspected that chronic wounds with vascular compromise failed to heal probably due to low levels of copper supply at the wound site [64]. Although this viewpoint remains controversial, supplements of Cu nanoparticles in our study, especially CuO@PS, appeared to provide the adequate cytosolic Cu+ to mobilize a complex Cu metabolic network including various Cu transporters and chaperones (Scheme 1). Most significantly, CuO@PS activated Atox1–ATP7a/b–Lox signaling axis, which may allow not only to export free Cu+ via secretory pathways mediated by trans-Golgi network (TGN) nearby lysosomes to maintain the Cu homeostasis, but also Atox1 served as transcription factors to promote cell proliferation and migration [65]. Although Atox1–ATP7a/b–Lox axis plays a critical role in tumorigenesis and progression [66], there were no abnormal wound healing or hyperplasia observed in the regenerated skins treated with CuOPSφGEL. Additionally, upregulation of Atox1 can potently phosphorylate and activate cytoplasmic proteins ERK [67,68], which enables to initiate a wide variety of cellular processes, including cell proliferation, migration, angiogenesis (Scheme 1) [69,70] Furthermore, Cu+ derived from CuO@PS may also directly interacts with MEK1/2 to promote the phosphorylation and activation of MEK1/2 and its downstream effector ERK. Taken together, intracellular CuO@PS deposited in lysosomes continuously and conservatively transmitted Cu signals to directly or indirectly activate MEK signaling pathway.
5. Conclusions
In conclusions, CuONPs can be embedded into polymersomes via a direct hydration method to form cupric-polymeric nanoreactors, which not only possess the multienzyme-like activities, but also create a nanoscale space isolated from the surrounding environment to ensure the selectivity, sustainability, cyclicity and safety of CuONPs-based catalytic reactions. Moreover, cupric-polymeric nanoreactors stimulated a complex Cu-dependent signaling pathway in cells, and exhibited antioxidant, proliferative, angiogenic and antibacterial activities to promote chronic diabetic wounds healing.
CRediT authorship contribution statement
Qi Tang: Writing – original draft, Methodology, Formal analysis. Yinqiu Tan: Writing – original draft, Methodology, Formal analysis. Shaolong Leng: Methodology, Formal analysis. Qi Liu: Writing – review & editing, Supervision, Funding acquisition. Linyu Zhu: Writing – review & editing, Supervision, Conceptualization. Cuifeng Wang: Writing – review & editing, Supervision, Funding acquisition, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported by National Natural Science Foundation of China (82073772), Guangdong Basic and Applied Basic Research Foundation (2019A1515010632, 2021B1515140044, 2023A1515010613), Science and Technology Planning Project of Guangdong Province of China (2021A0505030025), Jiangxi Provincial Natural Science Foundation (20212ACB206030) and The Science and Technology Foundation of Guangzhou (No. 202102080266), Medical Scientific Research Foundation of Guangdong Province (A2021196), Dongguan Science and Technology of Social Development Program (20221800905702), and shenzhen Science and Technology Project (JCYJ20210324122214040). The authors thank J. Sun (Instrumental Analysis and Research Center, Sun Yat-sen University) for assistance with TEM measurements.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.mtbio.2024.101087.
Contributor Information
Linyu Zhu, Email: zhuly35@mail.sysu.edu.cn.
Cuifeng Wang, Email: wangcf6@mail.sysu.edu.cn.
Appendix A. Supplementary data
The following is the Supplementary data to this article.
Data availability
No data was used for the research described in the article.
References
- 1.Armstrong D.G., Tan T.-W., Boulton A.J.M., Bus S.A. Diabetic foot ulcers: a review. JAMA. 2023;330:62–75. doi: 10.1001/jama.2023.10578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Falanga V., Isseroff R.R., Soulika A.M., Romanelli M., Margolis D., Kapp S., Granick M., Harding K. Chronic wounds. Nat. Rev. Dis. Prim. 2022;8:50. doi: 10.1038/s41572-022-00377-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bowers S., Franco E. Chronic wounds: Evaluation and management. Am. Fam. Physician. 2020;101:159–166. [PubMed] [Google Scholar]
- 4.Frykberg R.G., Banks J. Challenges in the treatment of chronic wounds. Adv. Wound Care. 2015;4:560–582. doi: 10.1089/wound.2015.0635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brem H., Tomic-Canic M. Cellular and molecular basis of wound healing in diabetes. J. Clin. Invest. 2007;117:1219–1222. doi: 10.1172/JCI32169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Studebaker S.A. Wound repair and regeneration. JAMA. 1994;272:1715. [Google Scholar]
- 7.Guo S., Dipietro L.A. Factors affecting wound healing. J. Dent. Res. 2010;89:219–229. doi: 10.1177/0022034509359125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sen C.K., Khanna S., Venojarvi M., Trikha P., Ellison E.C., Hunt T.K., Roy S. Copper-induced vascular endothelial growth factor expression and wound healing. Am. J. Physiol. Heart Circ. Physiol. 2002;282:H1821–H1827. doi: 10.1152/ajpheart.01015.2001. [DOI] [PubMed] [Google Scholar]
- 9.Borkow G. Using copper to improve the well-being of the skin, Curr. Chem. Biol. 2014;8:89–102. doi: 10.2174/2212796809666150227223857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tsang T., Davis C.I., Brady D.C. Copper biology. Curr. Biol. 2021;31:R421–R427. doi: 10.1016/j.cub.2021.03.054. [DOI] [PubMed] [Google Scholar]
- 11.Tapiero H., Townsend D.M., Tew K.D. Trace elements in human physiology and pathology. Copper, Biomed. Pharmacother. 2003;57:386–398. doi: 10.1016/s0753-3322(03)00012-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Salvo J., Sandoval C. Role of copper nanoparticles in wound healing for chronic wounds: literature review. Burns Trauma. 2022;10 doi: 10.1093/burnst/tkab047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sandoval C., Ríos G., Sepúlveda N., Salvo J., Souza-Mello V., Farías J. Effectiveness of copper nanoparticles in wound healing process using in vivo and in vitro studies: a Systematic review. Pharmaceutics. 2022;14:1838. doi: 10.3390/pharmaceutics14091838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alizadeh S., Seyedalipour B., Shafieyan S., Kheime A., Mohammadi P., Aghdami N. Copper nanoparticles promote rapid wound healing in acute full thickness defect via acceleration of skin cell migration, proliferation, and neovascularization. Biochem. Biophys. Res. Commun. 2019;517:684–690. doi: 10.1016/j.bbrc.2019.07.110. [DOI] [PubMed] [Google Scholar]
- 15.Ma X., Zhou S., Xu X., Du Q. Copper-containing nanoparticles: mechanism of antimicrobial effect and application in dentistry-a narrative review. Front. Surg. 2022;9 doi: 10.3389/fsurg.2022.905892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Din M.I., Arshad F., Hussain Z., Mukhtar M. Green Adeptness in the Synthesis and stabilization of copper nanoparticles: catalytic, antibacterial, cytotoxicity, and antioxidant activities. Nanoscale Res. Lett. 2017;12:638. doi: 10.1186/s11671-017-2399-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fukai T., Ushio-Fukai M., Kaplan J.H. Copper transporters and copper chaperones: roles in cardiovascular physiology and disease. Am. J. Physiol-Cell Ph. 2018;315:C186–C201. doi: 10.1152/ajpcell.00132.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen L., Min J., Wang F. Copper homeostasis and cuproptosis in health and disease. Signal Transduct. Targeted Ther. 2022;7:378. doi: 10.1038/s41392-022-01229-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Woźniak-Budych M.J., Maciejewska B., Przysiecka Ł., Wieczorek D., Staszak K., Jenczyk J., Jesionowski T., Jurga S. Comprehensive study of stability of copper oxide nanoparticles in complex biological media. J. Mol. Liq. 2020;319 [Google Scholar]
- 20.Conway J.R., Adeleye A.S., Gardea-Torresdey J., Keller A.A. Aggregation, dissolution, and transformation of copper nanoparticles in natural waters. Environ. Sci. Technol. 2015;49:2749–2756. doi: 10.1021/es504918q. [DOI] [PubMed] [Google Scholar]
- 21.Woźniak-Budych M.J., Staszak K., Staszak M. Copper and copper-based nanoparticles in medicine—perspectives and challenges. Molecules. 2023;28:6687. doi: 10.3390/molecules28186687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao G., Huang X., Tang Z., Huang Q., Niu F., Wang X. Polymer-based nanocomposites for heavy metal ions removal from aqueous solution: a review. Polym. Chem. 2018;9:3562–3582. [Google Scholar]
- 23.Iqbal S., Blenner M., Alexander-Bryant A., Larsen J. Polymersomes for therapeutic delivery of protein and nucleic acid macromolecules: from design to therapeutic applications. Biomacromolecules. 2020;21:1327–1350. doi: 10.1021/acs.biomac.9b01754. [DOI] [PubMed] [Google Scholar]
- 24.Discher D.E., Ahmed F. Polymersomes. Annu. Rev. Biomed. Eng. 2006;8:323–341. doi: 10.1146/annurev.bioeng.8.061505.095838. [DOI] [PubMed] [Google Scholar]
- 25.Li G., Huang Y., Zhao L., Yang B., Guo J., Hu J., Wang J., Wang H., Liu B., Zhang A., Sun F., Luo Q. Targeting and microenvironment-activated nanoreactor for diabetic chronic wound healing via multienzyme cascade reactions. ACS Appl. Mater. Interfaces. 2024;16(5):6315–6326. doi: 10.1021/acsami.3c12427. [DOI] [PubMed] [Google Scholar]
- 26.Che H., Cao S., van Hest J.C.M. Feedback-induced temporal control of “breathing” polymersomes to create self-adaptive nanoreactors. J. Am. Chem. Soc. 2018;140:5356–5359. doi: 10.1021/jacs.8b02387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oerlemans R.A.J.F., Shao J., Huisman S.G.A.M., Li Y., Abdelmohsen L.K.E.A., van Hest J.C.M. Compartmentalized intracellular click chemistry with biodegradable polymersomes. Macromol. Rapid Commun. 2023;44 doi: 10.1002/marc.202200904. [DOI] [PubMed] [Google Scholar]
- 28.Louzao I., van Hest J.C.M. Permeability effects on the efficiency of antioxidant nanoreactors. Biomacromolecules. 2013;14:2364–2372. doi: 10.1021/bm400493b. [DOI] [PubMed] [Google Scholar]
- 29.Miller A.J., Pearce A.K., Foster J.C., O'Reilly R.K. Probing and tuning the permeability of polymersomes. ACS Cent. Sci. 2021;7:30–38. doi: 10.1021/acscentsci.0c01196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhu Y., Cao S., Huo M., van Hest J.C.M., Che H. Recent advances in permeable polymersomes: fabrication, responsiveness, and applications. Chem. Sci. 2023;14:7411–7437. doi: 10.1039/d3sc01707a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liang Y., He J., Guo B. Functional hydrogels as wound dressing to enhance wound healing. ACS Nano. 2021;15:12687–12722. doi: 10.1021/acsnano.1c04206. [DOI] [PubMed] [Google Scholar]
- 32.Lee J.S., Zhou W., Meng F., Zhang D., Otto C., Feijen J. Thermosensitive hydrogel-containing polymersomes for controlled drug delivery. J. Contr. Release. 2010;146:400–408. doi: 10.1016/j.jconrel.2010.06.002. [DOI] [PubMed] [Google Scholar]
- 33.Akash M.S.H., Rehman K. Recent progress in biomedical applications of Pluronic (PF127): pharmaceutical perspectives. J. Contr. Release. 2015;209:120–138. doi: 10.1016/j.jconrel.2015.04.032. [DOI] [PubMed] [Google Scholar]
- 34.Li S., Yang C., Li J., Zhang C., Zhu L., Song Y., Guo Y., Wang R., Gan D., Shi J., Ma P., Gao F., Su H. Progress in pluronic F127 derivatives for application in wound healing and repair. Int. J. Nanomed. 2023;18:4485–4505. doi: 10.2147/IJN.S418534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu H., Cui W., Zong Z., Tan Y., Xu C., Cao J., Lai T., Tang Q., Wang Z., Sui X., Wang C. A facile method for anti-cancer drug encapsulation into polymersomes with a core-satellite structure. Drug Deliv. 2022;29:2414–2427. doi: 10.1080/10717544.2022.2103209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhu J., Li D., Chen H., Yang X., Lu L., Wang X. Highly dispersed CuO nanoparticles prepared by a novel quick-precipitation method. Mater. Lett. 2004;58:3324–3327. [Google Scholar]
- 37.Sui X., Kujala P., Janssen G.-J., de Jong E., Zuhorn I.S., van Hest J.C.M. Robust formation of biodegradable polymersomes by direct hydration. Polym. Chem. 2015;6:691–696. [Google Scholar]
- 38.Collin F. Chemical basis of reactive oxygen species reactivity and involvement in neurodegenerative diseases. Int. J. Mol. Sci. 2019;20:2407. doi: 10.3390/ijms20102407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bassett D.C., Madzovska I., Beckwith K.S., Melø T.B., Obradovic B., Sikorski P. Dissolution of copper mineral phases in biological fluids and the controlled release of copper ions from mineralized alginate hydrogels. Biomed. Mater. 2014;10 doi: 10.1088/1748-6041/10/1/015006. [DOI] [PubMed] [Google Scholar]
- 40.Lefley J., Waldron C., Becer C.R. Macromolecular design and preparation of polymersomes. Polym. Chem. 2020;11:7124–7136. [Google Scholar]
- 41.Yan X., Shi Y., Jiang Q., Ping G.-F., Deng Z. Design of amphiphilic PCL-PEG-PCL block copolymers as vehicles of Ginkgolide B and their brain-targeting studies. J. Biomater. Sci. Polym. Ed. 2017;28:1497–1510. doi: 10.1080/09205063.2017.1332470. [DOI] [PubMed] [Google Scholar]
- 42.Sukhanova A., Bozrova S., Sokolov P., Berestovoy M., Karaulov A., Nabiev I. Dependence of nanoparticle toxicity on their physical and chemical properties. Nanoscale Res. Lett. 2018;13:44. doi: 10.1186/s11671-018-2457-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Michels M., Córneo E., Rocha L.B.G., Dias R., Voytena A.P.L., Rossetto M., Ramlov F., Dal-Pizzol F., Jesus G.F.A. Paraprobiotics strains accelerate wound repair by stimulating re-epithelialization of NIH-3T3 cells, decreasing inflammatory response and oxidative stress. Arch. Microbiol. 2023;205:134. doi: 10.1007/s00203-023-03469-0. [DOI] [PubMed] [Google Scholar]
- 44.Ameh T., Sayes C.M. The potential exposure and hazards of copper nanoparticles: a review. Environ. Toxicol. Pharmacol. 2019;71 doi: 10.1016/j.etap.2019.103220. [DOI] [PubMed] [Google Scholar]
- 45.Kim B.Y.S., Jiang W., Oreopoulos J., Yip C.M., Rutka J.T., Chan W.C.W. Biodegradable quantum dot nanocomposites enable live cell labeling and imaging of cytoplasmic targets. Nano Lett. 2008;8:3887–3892. doi: 10.1021/nl802311t. [DOI] [PubMed] [Google Scholar]
- 46.Blockhuys S., Zhang X., Wittung-Stafshede P. Single-cell tracking demonstrates copper chaperone Atox1 to be required for breast cancer cell migration. Proc. Natl. Acad. Sci. USA. 2020;117:2014–2019. doi: 10.1073/pnas.1910722117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cai L., Xiong X., Kong X., Xie J. The role of the lysyl oxidases in tissue repair and remodeling: a concise review, tissue eng. Regen. Med. 2017;14:15–30. doi: 10.1007/s13770-016-0007-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grasso M., Bond G.J., Kim Y.-J., Boyd S., Matson Dzebo M., Valenzuela S., Tsang T., Schibrowsky N.A., Alwan K.B., Blackburn N.J., Burslem G.M., Wittung-Stafshede P., Winkler D.D., Marmorstein R., Brady D.C. The copper chaperone CCS facilitates copper binding to MEK1/2 to promote kinase activation. J. Biol. Chem. 2021;297 doi: 10.1016/j.jbc.2021.101314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang W., Liu H.T. MAPK signal pathways in the regulation of cell proliferation in mammalian cells. Cell Res. 2002;12:9–18. doi: 10.1038/sj.cr.7290105. [DOI] [PubMed] [Google Scholar]
- 50.Yang C., Yang C., Chen Y., Liu J., Liu Z., Chen H.-J. The trends in wound management: sensing, therapeutic treatment, and “theranostics,”. J. Sci-Adv Mater. Dev. 2023;8 [Google Scholar]
- 51.Furman B.L. Streptozotocin‐induced diabetic models in mice and rats. Curr. Protoc. 2021;1 doi: 10.1002/cpz1.78. [DOI] [PubMed] [Google Scholar]
- 52.Wang W., Bai D., Wu C., Li H., Xie X., Ji W., Gao J. A protocol for constructing a rat wound model of type 1 diabetes. J. Vis. Exp. 2023 doi: 10.3791/64914. [DOI] [PubMed] [Google Scholar]
- 53.Rousselle P., Braye F., Dayan G. Re-epithelialization of adult skin wounds: cellular mechanisms and therapeutic strategies. Adv. Drug Deliv. Rev. 2019;146:344–365. doi: 10.1016/j.addr.2018.06.019. [DOI] [PubMed] [Google Scholar]
- 54.Puca V., Marulli R.Z., Grande R., Vitale I., Niro A., Molinaro G., Prezioso S., Muraro R., Di Giovanni P. Microbial species isolated from infected wounds and antimicrobial resistance analysis: data emerging from a three-years retrospective study. Antibiotics. 2021;10:1162. doi: 10.3390/antibiotics10101162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kumari P., Panda P.K., Jha E., Kumari K., Nisha K., Mallick M.A., Verma S.K. Mechanistic insight to ROS and Apoptosis regulated cytotoxicity inferred by Green synthesized CuO nanoparticles from Calotropis gigantea to Embryonic Zebrafish. Sci. Rep. 2017;7 doi: 10.1038/s41598-017-16581-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Naz S., Gul A., Zia M. Toxicity of copper oxide nanoparticles: a review study. IET Nanobiotechnol. 2020;14:1–13. doi: 10.1049/iet-nbt.2019.0176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sajjad H., Sajjad A., Haya R.T., Khan M.M., Zia M. Copper oxide nanoparticles: in vitro and in vivo toxicity, mechanisms of action and factors influencing their toxicology. Comp. Biochem. Physiol. C. 2023;271 doi: 10.1016/j.cbpc.2023.109682. [DOI] [PubMed] [Google Scholar]
- 58.Pohanka M. Copper and copper nanoparticles toxicity and their impact on basic functions in the body, Bratisl. Med. J. 2019;120:397–409. doi: 10.4149/BLL_2019_065. [DOI] [PubMed] [Google Scholar]
- 59.Ou L., Wu Z., Hu X., Huang J., Yi Z., Gong Z., Li H., Ke P., Chang S., Koole L.H. A tissue adhesive F127 hydrogel delivers antioxidative copper-selenide nanoparticles for the treatment of dry eye disease. Acta Biomater. 2024;175:353–368. doi: 10.1016/j.actbio.2023.12.021. [DOI] [PubMed] [Google Scholar]
- 60.Essa A.M.M., Khallaf M.K. Antimicrobial potential of consolidation polymers loaded with biological copper nanoparticles. BMC Microbiol. 2016;16:144. doi: 10.1186/s12866-016-0766-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jessop I.A., Pérez Y.P., Jachura A., Nuñez H., Saldías C., Isaacs M., Tundidor-Camba A., Terraza C.A., Araya-Durán I., Camarada M.B., Cárcamo-Vega J.J. New hybrid copper nanoparticles/conjugated polyelectrolyte composite with antibacterial activity. Polymers. 2021;13:401. doi: 10.3390/polym13030401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gorginpour F., Zali-Boeini H., Rudbari H.A. A quinoxaline based porous organic polymer containing copper nanoparticles CuNPs@Q POP as a robust nanocatalyst toward C–N coupling reaction. RSC Adv. 2021;11:3655–3665. doi: 10.1039/d0ra10741g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Anyaogu K.C., Fedorov A.V., Neckers D.C. Synthesis, characterization, and antifouling potential of functionalized copper nanoparticles. Langmuir. 2008;24:4340–4346. doi: 10.1021/la800102f. [DOI] [PubMed] [Google Scholar]
- 64.Borkow G., Gabbay J., Zatcoff R.C. Could chronic wounds not heal due to too low local copper levels? Med. Hypotheses. 2008;70:610–613. doi: 10.1016/j.mehy.2007.06.006. [DOI] [PubMed] [Google Scholar]
- 65.Das A., Sudhahar V., Chen G.-F., Kim H.W., Youn S.-W., Finney L., Vogt S., Yang J., Kweon J., Surenkhuu B., Ushio-Fukai M., Fukai T. Endothelial antioxidant-1: a key mediator of copper-dependent wound healing in vivo. Sci. Rep. 2016;6 doi: 10.1038/srep33783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shanbhag V., Jasmer-McDonald K., Zhu S., Martin A.L., Gudekar N., Khan A., Ladomersky E., Singh K., Weisman G.A., Petris M.J. ATP7A delivers copper to the lysyl oxidase family of enzymes and promotes tumorigenesis and metastasis. Proc. Natl. Acad. Sci. USA. 2019;116:6836–6841. doi: 10.1073/pnas.1817473116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kim Y.-J., Bond G.J., Tsang T., Posimo J.M., Busino L., Brady D.C. Copper chaperone ATOX1 is required for MAPK signaling and growth in BRAF mutation-positive melanoma. Metallomics. 2019;11:1430–1440. doi: 10.1039/c9mt00042a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vitaliti A., De Luca A., Rossi L. Copper-dependent kinases and their role in cancer inception, progression and metastasis. Biomolecules. 2022;12:1520. doi: 10.3390/biom12101520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yaeger R., Corcoran R.B. Targeting alterations in the RAF–MEK pathway. Cancer Discov. 2019;9:329–341. doi: 10.1158/2159-8290.CD-18-1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Guo Y., Pan W., Liu S., Shen Z., Xu Y., Hu L. ERK/MAPK signalling pathway and tumorigenesis. Exp. Ther. Med. 2020:8454. doi: 10.3892/etm.2020.8454. S. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No data was used for the research described in the article.