Abstract
Tuberculosis (TB) remains a global health challenge with the emergence of drug-resistant Mycobacterium tuberculosis variants, necessitating innovative drug molecules. One potential target is the cell wall synthesis enzyme decaprenylphosphoryl-β-D-ribose 2′-epimerase (DprE1), crucial for virulence and survival. This study employed virtual screening of 111 Protein Data Bank (PDB) database molecules known for their inhibitory biological activity against DprE1 with known IC50 values. Six compounds, PubChem ID: 390820, 86287492, 155294899, 155522922, 162651615, and 162665075, exhibited promising attributes as drug candidates and validated against clinical trial inhibitors BTZ043, TBA-7371, PBTZ169, and OPC-167832. Concurrently, this research focused on DprE1 mutation effects using molecular dynamic simulations. Among the 10 mutations tested, C387N significantly influenced protein behavior, leading to structural alterations observed through root-mean-square deviation (RMSD), root-mean-square fluctuation (RMSF), radius of gyration (Rg), and solvent-accessible surface area (SASA) analysis. Ligand 2 (ID: 390820) emerged as a promising candidate through ligand-based pharmacophore analysis, displaying enhanced binding compared with reference inhibitors. Molecular dynamic simulations highlighted ligand 2’s interaction with the C387N mutation, reducing fluctuations, augmenting hydrogen bonding, and influencing solvent accessibility. These collective findings emphasize ligand 2’s efficacy, particularly against severe mutations, in enhancing protein-ligand complex stability. Integrated computational and pharmacophore methodologies offer valuable insights into drug candidates and their interactions within intricate protein environments. This research lays a strategic foundation for targeted interventions against drug-resistant TB, highlighting ligand 2’s potential for advanced drug development strategies.
Keywords: Mycobacterium tuberculosis, DprE1, virtual screening, 3D pharmacophore, resistance
Introduction
Tuberculosis (TB) remains a formidable global public health challenge due to its remarkable ability to persist within the human body in a clinically latent state. Mycobacterium tuberculosis (MTb), the causative agent of TB, ranks among the world’s most deadly diseases. In 2022, an estimated 10.6 million new TB cases were reported globally, and the number of cases of the disease has risen significantly by 4.5% compared with 2020, cementing TB status as a leading cause of mortality among infectious diseases. This toll accounted for 1.3 million deaths in individuals without HIV infection and an additional 187 000 deaths among those co-infected with HIV. The lingering presence of MTB often leads to latent infections that further compound the global burden.1,2
Notwithstanding considerable advances in health care infrastructure, including enhanced diagnostics, treatment regimens, and preventive measures, TB continues to cast a shadow over public health. The rise of multidrug-resistant (MDR) strains, combined with the absence of an effective vaccine and prolonged, potentially toxic treatment courses, has magnified the challenges in tackling this relentless ailment. Consequently, the development of innovative therapeutic agents has emerged as an urgent imperative.1 -5
Central to this pursuit is the targeting of decaprenylphosphoryl-β-D-ribose 2′-epimerase (DprE1) as a key therapeutic avenue against TB. Belonging to the vanillyl-alcohol oxidase family, DprE1 boasts a 2-domain structure encompassing a flavin adenine dinucleotide (FAD)-binding domain and a substrate-binding domain. Notably, DprE1 plays a pivotal role in the biosynthesis of MTb cell walls by orchestrating the essential epimerization process. Situated within the bacterium’s periplasm and absent in human physiology, DprE1 represents a promising target for the development of novel antitubercular agents. The distinct advantage of DprE1 inhibitors lies in their selectivity for MTB while exhibiting minimal adverse effects on human health. This feature renders DprE1 inhibitors particularly promising for addressing drug-resistant infections that necessitate prolonged therapeutic interventions.2,6,7
The intricate enzymatic conversion of decaprenylphosphoryl-β-D-ribose (DPR) into decaprenylphosphoryl-β-D-arabinose (DPA) defines the role of DPR 2′-oxidase. This intricate cascade involves a series of oxidation-reduction reactions, with the intermediate decaprenylphosphoryl-2-keto-β-D-erythro-pentofuranose (DPX) serving as both the product of DPR oxidation and the precursor to DPA. The coordinated interplay between DprE1 and DprE2 proteins dictates the functionality of this enzyme complex, where precise alignment of these polypeptides is imperative for effective epimerization. 2 ,8 -10
In light of these subtleties, the aim of this study to further in silico investigates the potential of DprE1 as a novel therapeutic target for the treatment of TB, thus identifying potential inhibitors with high affinity and stability superior to those of reference ligands by taking into account the mutational profile of this enzyme and taking advantage of computational techniques, virtual screening, molecular docking, and molecular dynamic (MD) simulations, aiming to provide information that could lead to innovative drug development strategies, addressing the global health threat posed by drug-resistant TB.
Material and Methods
Workflow
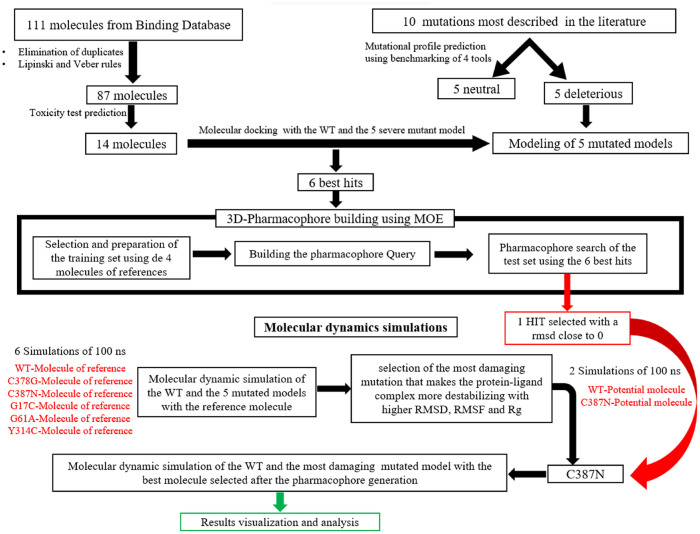
Figure 1 describes the workflow adopted in this study showing the approaches used.
Figure 1.
The workflow used in this study.
Database construction
The collection of DprE1 3D structure, mutations, and the generation of mutant models
Using the Protein Data Bank (PDB) database, the X-ray crystallographic three-dimensional (3D) structure of DprE1 was found completely crystallized with a resolution of 2.55 Å (PDB ID: 4P8T). 11
Ten known mutations were selected from literatures. These mutations were found in total: C387A, C387G, C387N, C387S, C387T, G17C, G61A, G248A, L368P, and Y314C.12 -16 The mutant model was generated using the UCSF Chimera program V1.1626 17 (University of California, San Francisco Chimera program) for each of them.
The collection and selection of potential and reference inhibitors
In total, 111 compounds were retrieved from the Binding database known for their inhibitory biological activity against DprE1 with known IC50 values. The selection of ligands was based on the rules of Lipinski and Veber. Then, the toxicity was checked using a comparative analysis of the Mcule toxicity checker, 18 the ProTox-II web server, 19 and the STOPTOX web server,20,21 including each toxic compound was eliminated. To compare the results obtained from these selected compounds, 4 reference inhibitors against DprE1 were selected from the literature: BTZ043, OPC_167832, TBA_7371, and PBTBZ169.22 -25
Prediction of mutational effects on protein function, stability, and flexibility
This step was carried out using the web servers Meta-SNP 26 It uses a benchmarking of 4 predictors, PhD-SNP (Single Nucleotide Polymorphism), SIFT (Sorting Intolerant From Tolerant), SNAP (Screening for Non-Acceptable Polymorphisms), and Meta-SNP. This web server predicts each single point protein function, determining whether it is deleterious or neutral.
For the study of the effect of mutations on protein stability and flexibility, the web server DynaMut was employed. It makes predictions on potential changes in protein stability (DDG: Delta Delta Gibbs) and flexibility (DDS: Difference in Distance Distribution). 27
Molecular docking
Using AutoDockTools-V.1.5.7, 28 the protein structure and the mutant models were prepared by adding polar hydrogens, Kollman United Atom charges, and eliminating water molecules. The file was then saved in “pdbqt” format. The ligands have been prepared and contain the atom types supported by the AutoDock tools by adding gasteiger charges, as well as additional records specifying rotating bonds, then saved in “pdbqt” format.
The docking process of the 10 mutational models and the wild-type (WT) protein with the 18 ligands (14 best compounds after filtration + 4 references ligands) was performed using Autodock vina. 29 A grid box with a spacing of 1 Å was used for each macromolecular protein, and the center coordinates were fixed at X = 12.818 Å, Y = −26.038 Å, and Z = 2.694 Å, with a size of X = 26 Å, Y = 26 Å, Z = 26 Å.
The 3D visualization of the protein-ligand interaction results was done using PyMol version 2.5.2. 30
Three-dimensional pharmacophoric map generation and training set screening process
To generate the pharmacophoric map that identifies common features among the 4 reference inhibitor molecules, we used the Molecular Operating Environment (MOE) software. 31 This approach involves a pharmacophore query methodology, which includes searching the 3D distances between the features. The map was generated through energy optimization and flexible alignment, considering 200 possible conformations. The best model was then selected using the pharmacophore query editor tool.
To create the training set, the 4 reference molecules were used to generate e a pharmacophore map identifying the features responsible for inhibitory activity, and then, the test set was formed by selecting the top 6 molecules from the docking process. Subsequently, we screened these molecules against the pharmacophoric map to identify the best matches based on root-mean-square deviation (RMSD). This crucial step enabled us to pinpoint the compounds that shared common features with the map and showed promising inhibitory activity. Among these compounds, we selected the best one with the lowest RMSD value for further investigation using MDs.
Molecular dynamic simulation
To analyze the protein-ligand interaction energy for the WT protein and the 5 severe mutant models described as deleterious with the reference drug and the best compounds identified through the 3D pharmacophore screening, in total, we conducted 8 MD simulations (WT—reference ligand, C387G—reference ligand, Y314C—reference ligand, G61A—reference ligand, G17C—reference ligand, C387N—reference ligand, C387N—reference molecule, and C387N—potential molecule) each spanning 100-ns intervals. GROMACS v.2020.4 32 was employed for these simulations, allowing us to gain insights into the stability and behavior of the complexes.
This enabled us to examine the interaction energy between the protein and the selected compound during MD simulations, providing valuable information on their binding dynamics and stability.
The interactions between the protein and the solvent were described using the CHARMM27 force field, 33 with the TIP3P 34 water model employed for the water molecules. To solvate the protein, a cubic simulation box with an edge length of 1.0 nm was used. To achieve a neutral system, an equal number of positive and negative ions were added. The system underwent a minimization process using the steepest descent algorithm, followed by equilibration at 300 000 for 100 ps in an NVT (constant Number of particles, Volume, and Temperature) ensemble using V-rescale. Subsequently, production runs were performed in an NPT (constant Number of particles, Pressure, and Temperature) ensemble, with a time step of 100 ps, and further equilibration at 1 atm pressure using the Parrinello-Rahman algorithm. 35 During the simulation, the LINCS (Linear Constraint Solver) algorithm was applied for bond constraints, and a distance cutoff using Verlet was implemented. 32
Results
Data set generation
Table 1 presents the 14 selected potential inhibitors with their IC50, toxic/non-toxic reports, alongside the 4 commonly used inhibitors of the protein, which serve as test references.
Table 1.
The top inhibitors selected with the 4 reference molecules with their PubChem ID, molecular weight, IUPAC identifier, IC50, and their toxicity report.
| Ligand number | PubChem ID | Molecular weight (g/mol) | IUPAC | IC50 (μM) | Toxicity report using Stoptox | Toxicity class using ProToxII |
|---|---|---|---|---|---|---|
| Ligand 1 | 390818 | 377.3 | 3-[(4-methoxyphenyl)methylamino]-6-(trifluoromethyl)quinoxaline-2-carboxylic acid | 0.041 | Non-toxic | 4 |
| Ligand 2 | 390820 | 365.28 | 3-[(4-fluorophenyl)methylamino]-6-(trifluoromethyl)quinoxaline-2-carboxylic acid | 0.072 | Non-toxic | 5 |
| Ligand 3 | 67402849 | 492 | 7-chloro-2-ethyl-N-[[4-[4-(4-fluorophenyl)piperazin-1-yl]phenyl]methyl]imidazo[1,2-a]pyridine-3-carboxamide | 0.017 | Non-toxic | 4 |
| Ligand 4 | 86287487 | 415.3 | 6-(trifluoromethyl)-3-[[4-(trifluoromethyl)phenyl]methylamino]quinoxaline-2-carboxylic acid | 0.12 | Non-toxic | 4 |
| Ligand 5 | 86287489 | 391.3 | 3-[(4-ethoxyphenyl)methylamino]-6-(trifluoromethyl)quinoxaline-2-carboxylic acid | 0.088 | Non-toxic | 4 |
| Ligand 6 | 86287490 | 395.3 | 3-[(3-fluoro-4-methoxyphenyl)methylamino]-6-(trifluoromethyl)quinoxaline-2-carboxylic acid | 0.08 | Non-toxic | 4 |
| Ligand 7 | 86287491 | 381.7 | 3-[(4-chlorophenyl)methylamino]-6-(trifluoromethyl)quinoxaline-2-carboxylic acid | 0.05 | Non-toxic | 4 |
| Ligand 8 | 86287492 | 372.3 | 3-[(4-cyanophenyl)methylamino]-6-(trifluoromethyl)quinoxaline-2-carboxylic acid | 0.067 | Non-toxic | 4 |
| Ligand 9 | 155294899 | 436.5 | 2-[4-(cyclohexylmethyl)piperazin-1-yl]-4-oxo-6-(trifluoromethyl)-1,3-benzothiazine-8-carbonitrile | 1.7 | Non-toxic | 4 |
| Ligand 10 | 155522922 | 379.4 | N-[3-[(4-methylpyrimidin-2-yl)carbamoyl]thiophen-2-yl]-[1,2,4]triazolo[4,3-a]pyridine-7-carboxamide | 0.03 | Non-toxic | 4 |
| Ligand 11 | 162651562 | 464.5 | 3-[2-[4-(cyclohexylmethyl)piperazin-1-yl]-4-oxo-6-(trifluoromethyl)-1,3-benzothiazin-8-yl]propanenitrile | 13.2 | Non-toxic | 4 |
| Ligand 12 | 162651615 | 369.5 | 2-[4-(cyclohexylmethyl)piperazin-1-yl]-4-oxopyrido[3,2-e][1,3]thiazine-7-carbonitrile | 7.72 | Non-toxic | 4 |
| Ligand 13 | 162657633 | 482.6 | 3-[2-[4-(cyclohexylmethyl)piperazin-1-yl]-4-oxo-6-(trifluoromethyl)-1,3-benzothiazin-8-yl]propanamide | 16.7 | Non-toxic | 4 |
| Ligand 14 | 162665075 | 370.5 | 2-[4-(cyclohexylmethyl)piperazin-1-yl]-4-oxopyrimido[5,4-e][1,3]thiazine-7-carbonitrile | 20 | Non-toxic | 4 |
| Ligand 15 | BTZ043 (42609849) | 431.4 | 2-[(3S)-3-methyl-1,4-dioxa-8-azaspiro[4.5]decan-8-yl]-8-nitro-6-(trifluoromethyl)-1,3-benzothiazin-4-one | 0.002 | Toxic (+) | 3 |
| Ligand 16 | OPC_167832 (118904282) | 456.8 | 5-[[(3R,4R)-1-(4-chloro-2,6-difluorophenyl)-3,4-dihydroxypiperidin-4-yl]methoxy]-8-fluoro-3,4-dihydro-1H-quinolin-2-one | 0.26 | Toxic (+) | 2 |
| Ligand 17 | TBA_7371 (72792692) | 355.4 | N-(2-hydroxyethyl)-1-[(6-methoxy-5-methylpyrimidin-4-yl)methyl]-6-methylpyrrolo[3,2-b]pyridine-3-carboxamide | 0.01 | Non-toxic | 4 |
| Ligand 15 | PBTZ169 (57331386) | 456.5 | 2-[4-(cyclohexylmethyl)piperazin-1-yl]-8-nitro-6-(trifluoromethyl)-1,3-benzothiazin-4-one | 0.002 | Toxic (+) | 3 |
Prediction of the mutational effects
To evaluate the impact of mutations on protein-ligand binding, we collected 10 mutations from the literature. After benchmarking and assessing compatibility between predictors, the following mutations were identified as deleterious: C387G, C387N, G17C, G61A, and Y314C, whereas the mutations C387A, C387S, C387T, G248A, and L368P were classified as neutral (Table 2).
Table 2.
Prediction results of mutational effects on DrpE1 protein.
| C387A | C387G | C387N | C387S | C387T | G17C | G61A | G248A | L368P | Y314C | |
|---|---|---|---|---|---|---|---|---|---|---|
| PhD-SNP | Neutral | Disease | Disease | Disease | Disease | Disease | Disease | Neutral | Disease | Disease |
| SIFT | Neutral | Disease | Disease | Neutral | Neutral | Disease | Disease | Neutral | Neutral | Disease |
| SNAP | Neutral | Disease | Disease | Neutral | Neutral | Disease | Disease | Neutral | Neutral | Disease |
| Meta-SNP | Neutral | Disease | Disease | Neutral | Neutral | Disease | Disease | Neutral | Disease | Disease |
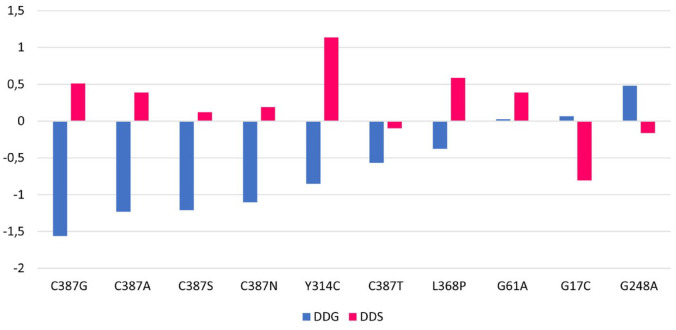
The evaluation of stability was measured by DDG (in kcal/mol), with positive values indicating stabilizing effects (DDG > 0) and negative values suggesting destabilizing effects (DDG < 0). The impact of the mutations on flexibility was assessed using DDS (in kcal/mol.K), where positive values indicate an increase in flexibility, and negative values indicate a decrease in stability (Figure 2).
Figure 2.
Prediction results of mutational effects on DprE1 stability and flexibility.
Docking and scoring
To identify small molecules with high affinity for both the WT DprE1 protein and the 10 mutated models, 111 compounds were extracted from the Binding DataBase. Among them, 87 compounds obeyed Lipinski and Veber rules and only 14 compounds were found to be non-toxic molecules. These 14 molecules, along with reference compounds, were subjected to molecular docking process with both the WT and the 10 mutated models of the protein.
The results (Table 3) indicate that the compounds with PubChem ID: 390820, 86287492, 155294899, 155522922, 162651615, and 162665075 exhibited significantly better affinity compared with the reference compounds.
Table 3.
Docking results of DprE1 wild-type protein and mutant-type models.
| Ligand number | PubChem ID | 4P8T | C387G | C387N | G17C | G61A | Y314C |
|---|---|---|---|---|---|---|---|
| Ligand 1 | 390818 | −9.1 | −9.3 | −9.5 | −9.2 | −9.2 | −9 |
| Ligand 2 | 390820 | −10.6 | −9.6 | −10.4 | −10.3 | −10.4 | −10.2 |
| Ligand 3 | 67402849 | −9.6 | −9.6 | −10 | −10 | −9.9 | −9.5 |
| Ligand 4 | 86287487 | −8.8 | −9.1 | −9.3 | −8.9 | −8.8 | −8.8 |
| Ligand 5 | 86287489 | −9.2 | −9.6 | −9.9 | −9.3 | −9.2 | −9 |
| Ligand 6 | 86287490 | −9.3 | −9.3 | −9.5 | −9.3 | −9.3 | −8.9 |
| Ligand 7 | 86287491 | −9.7 | −9.7 | −9.8 | −9.7 | −9.4 | −9.4 |
| Ligand 8 | 86287492 | −10.5 | −10.4 | −10.5 | −10.6 | −10.5 | −10.2 |
| Ligand 9 | 155294899 | −10.6 | −10.6 | −10.6 | −10.6 | −10.6 | −10.3 |
| Ligand 10 | 155522922 | −10.2 | −10 | −10.2 | −10.2 | −10.3 | −9.9 |
| Ligand 11 | 162651562 | −9.6 | −9.5 | −9.6 | −9.6 | −9.6 | −9.3 |
| Ligand 12 | 162651615 | −10.6 | −10.6 | −10.6 | −10.6 | −10.6 | −10.3 |
| Ligand 13 | 162657633 | −9.5 | −9.5 | −9.4 | −9.5 | −9.5 | −9.4 |
| Ligand 14 | 162665075 | −10.5 | −10.4 | −10.5 | −10.6 | −10.6 | −10.2 |
| Ligand 15 | BTZ043 (42609849) | −10.2 | −10 | −10.2 | −10.2 | −10.3 | −10 |
| Ligand 16 | OPC_167832 (118904282) | −10.1 | −10.2 | −10.1 | −10.1 | −10.8 | −10.4 |
| Ligand 17 | TBA_7371 (72792692) | −10.1 | −10.2 | −10.1 | −10.2 | −10.8 | −9.9 |
| Ligand 15 | PBTZ169 (57331386) | −8.2 | −8.5 | −8.1 | −8.2 | −8.2 | −8.3 |
Three-dimensional pharmacophore map generation
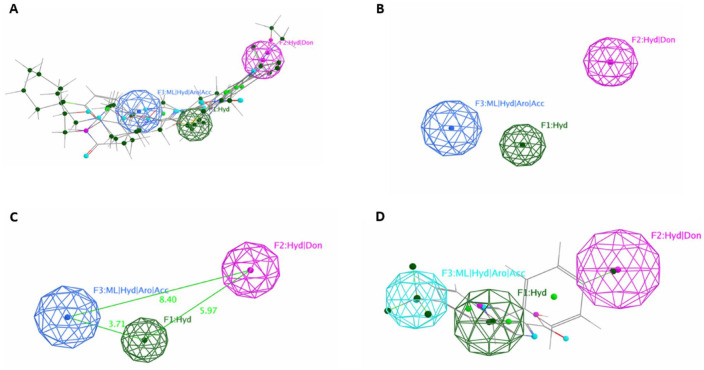
A pharmacophoric map was generated to identify common features involved in the inhibitory activity shared among the 4 known reference molecules, as depicted in Figure 3. The map revealed 3 features shared among the 4 aligned molecules: F1: hydrophobic, F2: hydrophobic/donor, and F3: metal ligator/hydrophobic/acceptor (Figure 3A and B), with distances of 5.97 Å between F1 and F2, 3.71 Å between F1 and F3, and 8.40 Å between F3 and F2 (Figure 3C).
Figure 3.
Pharmacophoric map generation: (A and B) The map generated with and without aligned ligand; (C) the distance between each feature in the map; and (D) the best ligand found with the pharmacophore query search with RMSD close to zero.
To select the most promising molecule of the 6 compounds identified after the molecular docking process, for further investigation using MD simulation, these 6 molecules were screened against the pharmacophoric map, and the results showed that ligand 2 (ID: 390820) had an RMSD close to 0 (RMSD = 0.06) compared with the other molecules. This ligand was deemed as the most suitable candidate, as it contained all the essential features identified in the map and exhibited significant affinity with the mutant models (Figure 3D).
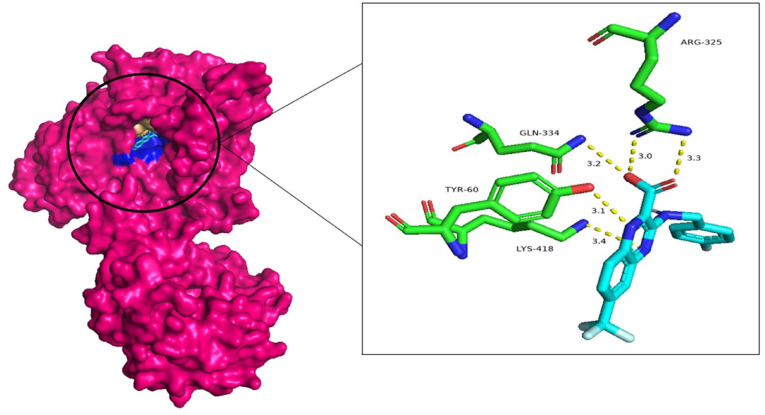
Figure 4 shows the 3D interaction of this ligand (ID: 390820) with the DprE1 protein.
Figure 4.
Three-dimensional visualization of the interaction between the potential ligand (ID: 390820) and the DprE1 protein, with the residues in green and the ligand in light blue.
MD simulations results
To comprehensively investigate the binding affinity of ligands, for both reference ligand and potential ligand, in complex with the WT and the deleterious mutant models of the protein, a total of 8 MD simulations were conducted, each spanning 100 ns.
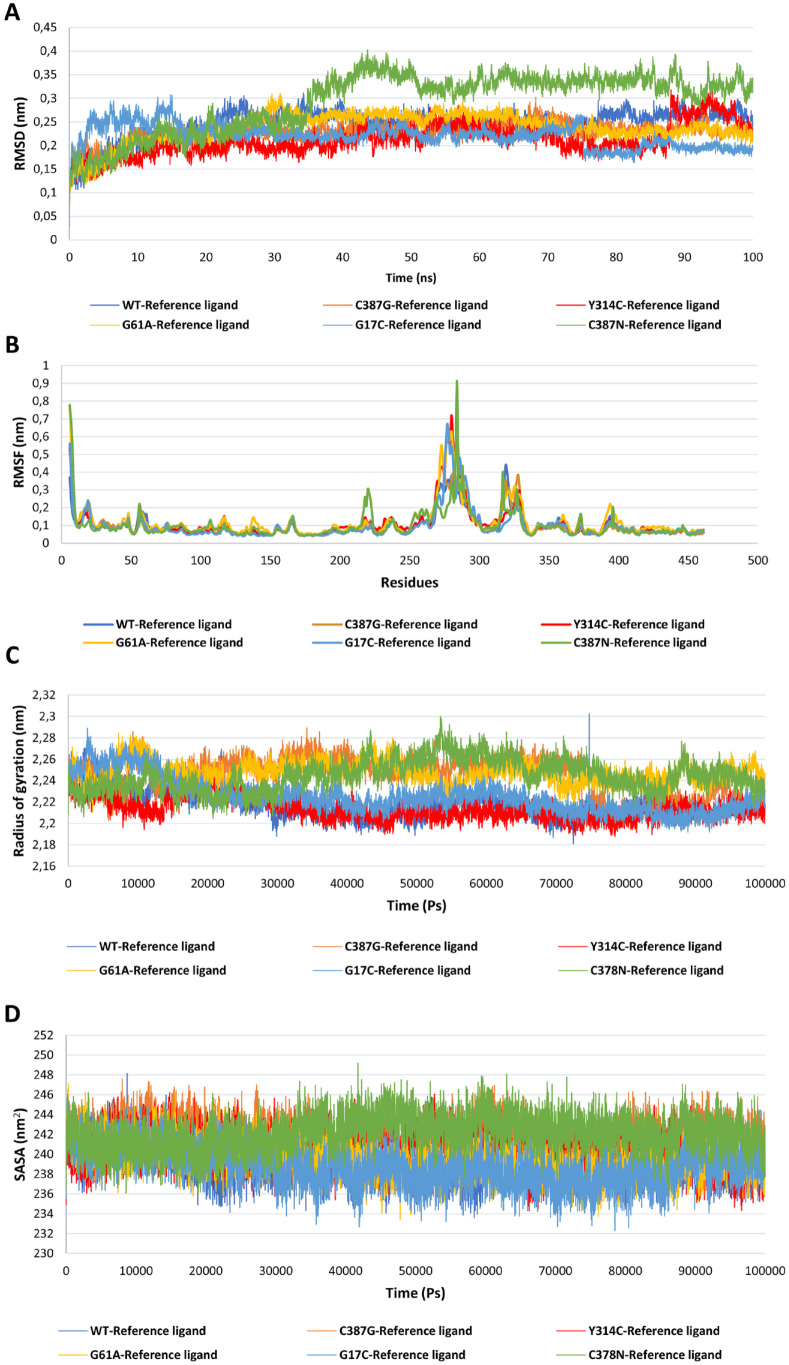
To assess the impact of the 5 identified deleterious mutations, 6 simulations were performed using the WT and the 5 mutant models, each complexed with the reference ligand: WT—reference ligand, C387G—reference ligand, C387N—reference ligand, G17C—reference ligand, G61A—reference ligand, and Y314C—reference ligand (Figure 5). Tables 4 and 5 present the mean and standard deviation for each complex. The result revealed that the mutation C387N exerts a remarkable influence on the protein-ligand complex (C387N—reference ligand) compared with the WT and the other mutant models, this was evident in the analyses of key structural parameters, such as RMSD with a high value (0.29 ± 0.07) compared with the other complexes (Figure 5A), while by measuring the fluctuations of the residues, root-mean-square fluctuation (RMSF) shows a higher value of 0.16 ± 0.08 compared with other complexes (Figure 5B). The radius of gyration (Rg) indicates more flexible shape for the C378N—reference ligand with a value of 2.40 ± 0.014 compared with other complexes which show that the shape remains more or less compact even with the existence of the mutation (Figure 5C), and finally, the solvent-accessible surface area (SASA) (Figure 5D) that revealed a greater exposed surface area for the complex C378N—reference ligand (245.15 ± 1.71). To summarize this part of 6 simulations, all parameters RMSD, RMSF, Rg, and SASA showed significantly higher values for the reference C387N—ligand complex compared with the reference WT—ligand and the other mutant model complexes (reference C378G—ligand, reference G17C—ligand, reference G61A—ligand, and reference Y314C—ligand). These increased values indicate greater structural fluctuations and flexibility in the reference C387N—ligand complex, potentially affecting ligand binding and stability in the binding site.
Figure 5.
Protein-ligand molecular dynamic analysis for the DprE1 protein, the WT version with reference ligand complex and the 5 mutant deleterious models with reference ligand complexes: WT—reference ligand (light blue), C387G—reference ligand (orange), C387N—reference ligand (green), G17C—reference ligand (dark blue), G61A—reference ligand (yellow), and Y314C—reference ligand (red): (A) RMSD analysis calculated for 100 ns; (B) RMSF of each complex as a function of residue number; (C) radius of gyration of protein-ligand complexes calculated during simulations expressed in pico-second; and (D) the SASA analysis calculated for the complexes for 100 000 ps.
Table 4.
The mean and standard deviation of the calculated parameters RMSD, RMSF, Rg, and SASA of the WT—reference ligand, C387G—reference ligand, Y314C—reference ligand, G61A—reference ligand, G17C—reference ligand, and C387N—reference ligand.
| WT—reference ligand | C387G—reference ligand | Y314C—reference ligand | G61A—reference ligand | G17C—reference ligand | C387N—reference ligand | |
|---|---|---|---|---|---|---|
| RMSD (nm) | 0.24 ± 0.03 | 0.23 ± 0.02 | 0.21 ± 0.03 | 0.24 ± 0.03 | 0.22 ± 0.02 | 0.29 ± 0.07 |
| RMSF (nm) | 0.09 ± 0.06 | 0.10 ± 0.08 | 0.10 ± 0.08 | 0.12 ± 0.09 | 0.09 ± 0.08 | 0.16 ± 0.08 |
| Rg (nm) | 2.22 ± 0.01 | 2.24 ± 0.01 | 2.21 ± 0.01 | 2.24 ± 0.009 | 2.22 ± 0.01 | 2.40 ± 0.014 |
| SASA (nm2) | 239.35 ± 0.1.76 | 241.64 ± 1.64 | 240.14 ± 1.73 | 239.65 ± 1.71 | 238.81 ± 1.88 | 245.15 ± 1.71 |
Table 5.
The mean and standard deviation of the calculated parameters RMSD, RMSF, Rg, SASA, and H-bond of WT—potential molecule, WT—reference molecule, C387N—potential molecule, and C387N—reference molecule complexes.
| WT—reference molecule | WT—potential molecule | C387N—reference molecule | C387N—potential molecule | |
|---|---|---|---|---|
| RMSD (nm) | 0.24 ± 0.03 | 0.20 ± 0.02 | 0.29 ± 0.07 | 0.21 ± 0.01 |
| RMSF (nm) | 0.09 ± 0.06 | 0.08 ± 0.05 | 0.16 ± 0.08 | 0.10 ± 0.05 |
| Rg (nm) | 2.22 ± 0.01 | 2.01 ± 0.01 | 2.40 ± 0.014 | 2.10 ± 0.01 |
| SASA (nm2) | 239.35 ± 0.1.76 | 238.26 ± 1.93 | 245.15 ± 1.71 | 239.71 ± 1.70 |
| H-bond | 0.47 ± 0.66 | 0.73 ± 1.01 | 0.52 ± 1.86 | 0.70 ± 1.01 |
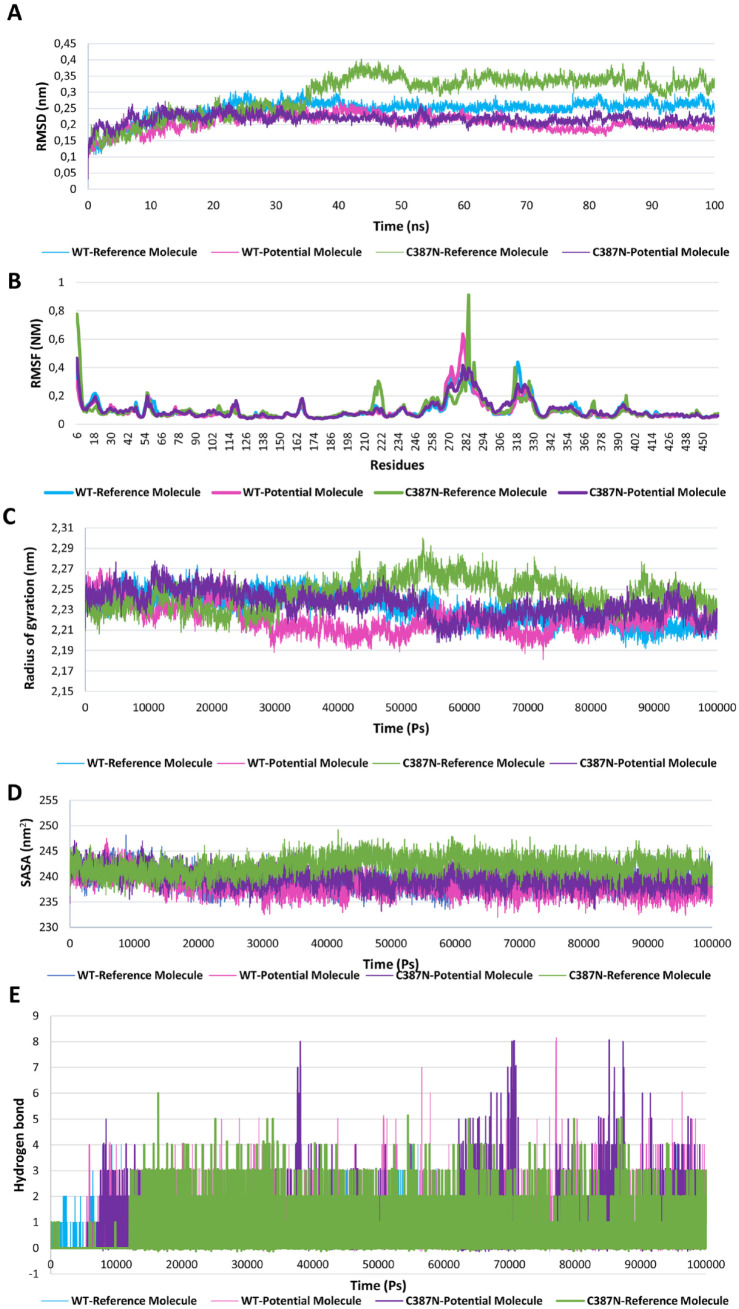
To evaluate the efficacy of the potential ligand, 2 additional simulations were performed: one between the WT—potential ligand, and another between the C387N—potential ligand (Figure 6). The results clearly demonstrated that the potential ligand positively impacted the complex’s stability on both the WT and mutant complexes, as evidenced by reduced RMSD values from 0.29 ± 0.07 to 0.21 ± 0.01 showing strong interactions with the mutant type, as revealed by the prominent hydrogen bonding and RMSD analysis (Figure 6A). Moreover, the RMSF analysis indicated that the potential ligand influenced the flexibility of specific residues in both the WT and mutant complexes (Figure 6B). Furthermore, the Rg analysis revealed that the potential ligand altered the compactness of both complexes with a value of 2.01 ± 0.0 for the WT—potential molecule and 2.10 ± 0.01 for the mutant type. In addition, the Rg for C387N—potential ligand is more reduced than that for C387N—reference ligand, making the complex more compact with the existence of the potential ligand (Figure 6C). Moreover, the SASA analysis provided valuable insights into the solvent accessibility in both C387N—potential ligand and WT—potential ligand complexes, with a value of 239.71 ± 1.70 and 238.26 ± 1.93, respectively (Figure 6D). Finally, The H-bond analysis uncovered crucial information regarding the stability of the ligand-protein complexes. The presence of the potential ligand resulted in an increased number of hydrogen bonds in both the WT (0.73 ± 1.01) and mutant-type complexes (0.70 ± 1.01), suggesting stronger and more favorable interactions compared with the WT—reference ligand (0.47 ± 0.66) (Figure 6E).
Figure 6.
Molecular dynamic analysis of the best ligand found in affinity ratio and screening of the pharmacophore model, complexed with the WT version of the protein and the most severe mutation found C387N: (A) RMSD analysis calculated for 100 ns in nm of the 4 complexes: WT—reference molecule (light blue), WT—potential molecule (pink), C387N—reference molecule (green), and C387N—potential molecule (purple); (B) RMSF of each complex as a function of the number of residues; (C) the radius of gyration in nm of protein-ligand complexes calculated during simulations expressed in pico-seconds; (D) SASA analysis calculated for the complexes for 100 000 ps; and (E) number of hydrogen bonds calculated for each complex.
As an overall summary of the results, the simulation results unveiled a marked effect of the C387N mutation on various key parameters. The RMSD, a measure of structural stability, demonstrated significant perturbations in the presence of the C387N mutation. Likewise, the RMSF, indicated the flexibility of specific residues such as Ala6, Leu 56, Gln220, Ile284, Leu317, and Asn373 with a value of 0.77, 0.22, 0.30, 0.94, 0.40, and 0.16 nm, respectively, exhibited notable variations compared with the WT complex. The Rg, which reflects the compactness of the protein structure, and the SASA, a measure of surface exposure, were also markedly altered due to the C387N mutation.
Discussion
Tuberculosis infection presents a considerable global health care challenge. Hence, it is crucial to create novel, potent therapeutic schemes against MTb, which must fight against the emergence of drug-resistant TB. decaprenylphosphoryl-β-D-ribose 2′-epimerase stands out as a promising drug target due to its location in the peri-plasmic region, wide-ranging interactions, and thorough biochemical and genetic understanding. This uniqueness makes it well-suited for developing small-molecule treatments. Presently, 4 DprE1 inhibitors are already in clinical trials, and the field is wide open with significant potential to develop new DprE1 inhibitors with higher specificity and improved pharmacokinetics This study aims to contribute to scaffold refinement and structure-based drug design, ultimately advancing novel anti-TB agents.
This study focused on the search for potential inhibitors for DprE1 as a new therapeutic target for the treatment of TB. Using virtual screening, a database of 111 ligands known for their inhibitory biological activity against DprE1 with known IC50 values was scrutinized. Among them, 88 obeyed to Lipinski and Veber rules, and only 14 passed toxicity tests. Subsequently, molecular docking via Autodock Vina was employed to identify compounds with favorable affinity for DprE1. Six compounds with good affinity for DprE1 protein, ranging from −10.6 to −10.2 kcal/mol, were selected: ligand 2 (ID: 390820), ligand 9 (ID: 155294899), and ligand 12 (ID: 162651615) with a docking score of −10.6 kcal/mol) with DprE1 protein, ligand 9 (ID: 155294899) and ligand 14 (ID: 162665075) with a score of −10.5 kcal/mol, and ligand 10 (ID: 155522922) with a score of −10.2 kcal/mol. These ligands exhibited superior affinities in comparison with reference inhibitors currently in clinical trials, namely BTZ-043, PBTZ-169, TBA-737120, and OPC167832. Interestingly, BTZ043 (ID: 42609849), a non-covalent DprE1 inhibitor in phase II clinical trials for pulmonary TB treatment, 36 displayed the most favorable affinity (approximately −10.2 kcal/mol) among the reference compounds (OPC_167832, PBTZ169, and TBA-737120). Thus, BTZ043 serves as the reference inhibitor for comparison.
To discern the most promising candidate ligand among the 6 identified compounds, characterized by their enhanced affinity, a ligand-based pharmacophore approach was employed by generating a pharmacophoric map based on the 4 reference ligands; the map shows 3 features shared which are as follows: F1: hydrophobic, F2: hydrophobic/donor, and F3: metal ligator/hydrophobic/acceptor, the hydrophobic and the donor/acceptor region play a crucial role in the binding. 37 To construct the training set, the 6 top-scoring compounds were used, and the analysis demonstrates that ligand 2 (ID: 390820) exhibited a remarkable RMSD score approaching zero.
This research explores novel inhibitors of the DprE1 protein to overcome the problem of resistance arising from mutations within this bacterium. To achieve this goal, computer prediction methods were employed to scrutinize the impact of 10 mutations, sourced from existing literature on the properties of the target protein. These mutations, namely C387G, C387N, G17C, G61A, Y314C, C387A, C387S, C387T, G248A, and L368P, were carefully examined to identify the deleterious ones. The outcomes demonstrate that when integrated into different domains, each mutation has distinct effects on stability, flexibility, and function. Through a consensus of predictor results, a comprehensive analysis unveiled that 5 mutations are deleterious. Interestingly, 7 mutations induce destabilization. In addition, 3 mutations reduce flexibility, while 7 increase it. Consequently, these mutations impact protein folding and interactions, be it with other proteins or ligands. This correlation is corroborated by multiple studies.38 -40 Intriguingly, several investigations have highlighted that diseases, including those linked to drug resistance, often involve mutations found in accessible regions. This suggests that analyzing such mutations could offer insights into the underlying disease mechanisms.41 -43
To further investigate and provide valuable insights into how these deleterious mutations disrupt the intricate interactions between the protein and the reference ligand, 6 MD simulations were conducted. These simulations aimed to provide a comprehensive understanding of how these mutations, C387G, C387N, G17C, G61A, and Y314C, influence the protein-ligand complex. Notably, among these mutations, C387N exhibited the most profound impact on the complex.
To assess the effectiveness of the ligand 2 (ID: 390820) on the most severe mutation, C387N, 2 additional MD simulations of 100 ns were conducted: one with C387N (referred to as C387N—potential molecule) and the other with the WT counterpart (referred to as WT—potential molecule). The results prominently indicated that the potential ligand exerted a positive influence on the protein-ligand complex. Notably, it led to a reduction in RMSD, indicative of improved stability, revealing that the potential ligand induced significant conformational changes in both complexes, leading to increased structural deviations compared with the reference ligand. These observations were further corroborated by comprehensive analyses of other parameters, such as RMSF, Rg, SASA, and hydrogen bond interactions, as illustrated in the graphs in Figure 6. The RMSF results clearly show that the binding of the potential ligand to the mutated protein reduces the fluctuations in the residues from Ala6 (0.77 nm), Leu 56 (0.22 nm), Gln220 (0.30 nm), Ile284 (0.94 nm), Leu317 (0.40 nm), and Asn373 (0.16 nm) to Ala6 (0.32 nm), Leu 56 (0.19 nm), Gln220 (0.09 nm), Ile284 (0.39 nm), Leu317 (0.23 nm), and Asn373 (0.08 nm). The results obtained from the MD simulations further highlight this enhanced interaction. Notably, the Rg curve displayed a significantly smaller fluctuation for the potential ligand when compared to the reference ligand. This observation underscores that the potential ligand forms a more stable and tightly bound complex with both the WT and mutated protein counterparts.
Furthermore, the results revealed a noteworthy increase in hydrogen bond interactions for the potential ligand in comparison with the reference ligand. Throughout the 100 ns MD simulations, the potential ligand consistently formed a maximum of 8 stable hydrogen bonds with the protein at various time intervals. This stands in contrast to the reference ligand, which reached a maximum of 6 hydrogen bonds. The sustained and elevated hydrogen bonding interactions observed with the potential ligand underscore its strong and consistent binding to the protein, suggesting a more robust and enduring association.
In addition, the analysis of SASA exhibited significant differences between the C387N—potential ligand and C387N—reference ligand simulations. The SASA curve for the C387N—potential ligand demonstrated a noticeable reduction compared with that of the C387N—reference ligand. This reduction indicates a more compact and stable conformation adopted by the protein-ligand complex when interacting with the potential ligand. The diminished SASA values imply a tighter association and less exposure to the solvent environment, highlighting the potential ligand’s ability to induce a favorable binding pocket conformation. This distinct behavior further supports the enhanced binding characteristics of the potential ligand, emphasizing its potential as a valuable therapeutic agent against the challenging C387N mutation.
Another element contributing to the lower performance of the reference ligand stems from its formation of a limited number of hydrogen bonds with both the WT and C378N proteins with higher SASA values suggest that the reference ligand is less deeply embedded within the complex and exhibits reduced protection from the solvent environment, ultimately resulting in increased instability of the complex. 44
In summary, these comprehensive analyses collectively highlight the profound impact exerted by the potential ligand on both the WT and mutant complexes, as indicated by detailed analyses of RMSD, RMSF, RG, SASA, and H-bond. The observed modifications in these pivotal structural parameters not only signify the potential ligand’s ability to induce conformational adjustments and improve flexibility within the binding site but also underline its capability to reinforce interactions. These outcomes highlight the potential ligand’s capacity to establish more favorable and enduring interactions within the protein’s binding pocket, thus holding promise for its application as a targeted therapeutic agent. Furthermore, it’s crucial to assess how well the potential ligand, especially in light of the severe C387N mutation, can enhance the stability and overall dynamics of the resulting protein-ligand complex. This strategic integration of ligand-based pharmacophore methods with dynamic simulations enriches our understanding of prospective therapeutic candidates and their interactions with the intricate protein landscape, ultimately paving a path toward drug development strategies.
The accumulated data strongly indicate that this potential ligand not only exhibits exceptional binding capabilities compared to the reference ligand but also displays a superior proficiency in stabilizing the mutant complex. The ligand (3-[(4-fluorophenyl)methylamino]-6-(trifluoromethyl)quinoxaline-2-carboxylic acid) emerges as a promising therapeutic intervention, possesses several characteristics that could contribute to its efficacy and affinity for a target protein with a IC50 of 0.072 µM.
Specific functional groups such as the presence of a fluorobenzylamine group and a trifluoromethyl group in the ligand’s structure may confer particular chemical properties, such as hydrophobic interactions or hydrogen bonding, which enhance its interaction with the target protein. The quinoxaline moiety may enable favorable interactions with the binding sites of the protein. The carboxylic group may allow the ligand to establish ionic interactions or hydrogen bonds with specific residues of the target protein. In addition, it is noteworthy that this quinoxaline core was the subject of a published study confirming our results. 12 These results lay the groundwork for extensive exploration and advancement of this ligand as a viable therapeutic contender.
Conclusions
In light of the escalating challenges posed by drug-resistant TB, this study’s multifaceted approach holds significant promise for the development of targeted therapeutic interventions. By focusing on DprE1 as a crucial target in MTb, the research has unveiled potential inhibitors with remarkable binding affinity and stability, particularly against the severe C387N mutation. The integration of computational techniques, encompassing virtual screening, molecular docking, and MD simulations, offers a strategic framework for designing advanced interventions against drug-resistant TB.
Supplemental Material
Supplemental material, sj-docx-1-bbi-10.1177_11779322241257039 for Targeting decaprenylphosphoryl-β-D-ribose 2′-epimerase for Innovative Drug Development Against Mycobacterium Tuberculosis Drug-Resistant Strains by Ghyzlane EL Haddoumi, Mariam Mansouri, Jouhaina Kourou, Lahcen Belyamani, Azeddine Ibrahimi and Ilham Kandoussi in Bioinformatics and Biology Insights
Supplemental material, sj-jpg-2-bbi-10.1177_11779322241257039 for Targeting decaprenylphosphoryl-β-D-ribose 2′-epimerase for Innovative Drug Development Against Mycobacterium Tuberculosis Drug-Resistant Strains by Ghyzlane EL Haddoumi, Mariam Mansouri, Jouhaina Kourou, Lahcen Belyamani, Azeddine Ibrahimi and Ilham Kandoussi in Bioinformatics and Biology Insights
Footnotes
Author Contributions: The conceptualization of the study was carried out by Ghyzlane EL Haddoumi, Azeddine Ibrahimi, and Ilham Kandoussi. Ghyzlane EL Haddoumi and Ilham Kandoussi were responsible for developing the research methodology. The software development was a collaborative effort between Ghyzlane EL Haddoumi, Mariam Mansouri, and Jouhaina Kourou. Ghyzlane EL Haddoumi and Ilham Kandoussi conducted the investigation, performed formal analysis, and curated the data. Ghyzlane EL Haddoumi was responsible for the original draft of the manuscript, while Ghyzlane EL Haddoumi, Mariam Mansouri, Azeddine Ibrahimi, and Ilham Kandoussi reviewed and edited the manuscript. Ghyzlane EL Haddoumi and Mariam Mansouri created the visualizations. Azeddine Ibrahimi and Ilham Kandoussi supervised the study, and Azeddine Ibrahimi and Lahcen Belyamani secured the funding.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was carried out under national funding from the Moroccan Ministry of Higher Education and Scientific Research (COVID-19 program) to A.I. This work was also supported by a grant from the Moroccan Institute of Cancer Research and the PPR-1 program to A.I. and Biocodex grant to A.I.
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
ORCID iD: Ilham Kandoussi  https://orcid.org/0000-0002-5894-7817
https://orcid.org/0000-0002-5894-7817
Supplemental Material: Supplemental material for this article is available online.
References
- 1. Hariguchi N, Chen X, Hayashi Y, et al. OPC-167832, a novel carbostyril derivative with potent antituberculosis activity as a DprE1 inhibitor. Antimicrob Agents Chemother. 2020;64:e02020-19. doi: 10.1128/AAC.02020-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gawad J, Bonde C. Decaprenyl-phosphoryl-ribose 2′-epimerase (DprE1): challenging target for antitubercular drug discovery. Chem Cent J. 2018;12:1-12. doi: 10.1186/s13065-018-0441-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Shirude PS, Shandil R, Sadler C, et al. Azaindoles: noncovalent DprE1 inhibitors from scaffold morphing efforts, kill Mycobacterium tuberculosis and are efficacious in vivo. J Med Chem. 2013;56:9701-9708. Accessed February 5, 2023. https://pubs.acs.org/doi/abs/10.1021/jm401382v [DOI] [PubMed] [Google Scholar]
- 4. Ma Z, Lienhardt C, McIlleron H, Nunn AJ, Wang X. Global tuberculosis drug development pipeline: the need and the reality. Lancet Lond Engl. 2010;375:2100-2109. doi: 10.1016/S0140-6736(10)60359-9 [DOI] [PubMed] [Google Scholar]
- 5. Koul A, Arnoult E, Lounis N, Guillemont J, Andries K. The challenge of new drug discovery for tuberculosis. Nature. 2011;469:483-490. doi: 10.1038/nature09657 [DOI] [PubMed] [Google Scholar]
- 6. Imran M, A S A, Thabet HK, Afroz Bakht M. Synthetic molecules as DprE1 inhibitors: a patent review. Expert Opin Ther Pat. 2021;31:759-772. doi: 10.1080/13543776.2021.1902990 [DOI] [PubMed] [Google Scholar]
- 7. Landge S, Mullick AB, Nagalapur K, et al. Discovery of benzothiazoles as antimycobacterial agents: synthesis, structure-activity relationships and binding studies with Mycobacterium tuberculosis decaprenylphosphoryl-β-D-ribose 2’-oxidase. Bioorg Med Chem. 2015;23:7694-7710. doi: 10.1016/j.bmc.2015.11.017 [DOI] [PubMed] [Google Scholar]
- 8. Lindenberger JJ, Veleti SK, Wilson BN, Sucheck SJ, Ronning DR. Crystal structures of Mycobacterium tuberculosis GlgE and complexes with non-covalent inhibitors. Sci Rep. 2015;5:12830. doi: 10.1038/srep12830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Manina G, Pasca MR, Buroni S, De Rossi E, Riccardi G. Decaprenylphosphoryl-β-D-ribose 2’-epimerase from Mycobacterium tuberculosis is a magic drug target. Curr Med Chem. 2010;17:3099-3108. doi: 10.2174/092986710791959693 [DOI] [PubMed] [Google Scholar]
- 10. Parchina A, Froeyen M, Margamuljana L, et al. Discovery of an acyclic nucleoside phosphonate that inhibits Mycobacterium tuberculosis ThyX based on the binding mode of a 5-alkynyl substrate analogue. ChemMedChem. 2013;8:1373-1383. Accessed February 6, 2023. https://pubmed.ncbi.nlm.nih.gov/23836539/ [DOI] [PubMed] [Google Scholar]
- 11. Burley SK, Berman HM, Kleywegt GJ, Markley JL, Nakamura H, Velankar S. Protein Data Bank (PDB): the single global macromolecular structure archive. In: Wlodawer A, Dauter Z, Jaskolski M, eds. Protein Crystallography: Methods and Protocols. Methods in Molecular Biology. Springer; 2017:627-641. doi: 10.1007/978-1-4939-7000-1_26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Neres J, Hartkoorn RC, Chiarelli LR, et al. 2-Carboxyquinoxalines Kill Mycobacterium tuberculosis through noncovalent inhibition of DprE1. ACS Chem Biol. 2015;10:705-714. doi: 10.1021/cb5007163 [DOI] [PubMed] [Google Scholar]
- 13. Piton J, Foo CS, Cole ST. Structural studies of Mycobacterium tuberculosis DprE1 interacting with its inhibitors. Drug Discov Today. 2017;22:526-533. doi: 10.1016/j.drudis.2016.09.014 [DOI] [PubMed] [Google Scholar]
- 14. Chen X, Li Y, Wang B, Lu Y. Identification of mutations associated with macozinone-resistant in Mycobacterium tuberculosis. Curr Microbiol. 2022;79:205. doi: 10.1007/s00284-022-02881-x [DOI] [PubMed] [Google Scholar]
- 15. Liu R, Lyu X, Batt SM, et al. Determinants of the inhibition of DprE1 and CYP2C9 by antitubercular thiophenes. Angew Chem Int Ed. 2017;56:13011-13015. doi: 10.1002/anie.201707324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Foo CS, Lechartier B, Kolly GS, et al. Characterization of DprE1-mediated benzothiazinone resistance in Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2016;60:6451-6459. doi: 10.1128/AAC.01523-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pettersen EF, Goddard TD, Huang CC, et al. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605-1612. doi: 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
- 18. Toxicity checker. Accessed June 11, 2022. https://mcule.com/apps/toxicity-checker/
- 19. Banerjee P, Eckert AO, Schrey AK, Preissner R. ProTox-II: a webserver for the prediction of toxicity of chemicals. Nucleic Acids Res. 2018;46:W257-W263. doi: 10.1093/nar/gky318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Borba JVB, Alves VM, Braga RC, et al. STopTox: an in silico alternative to animal testing for acute systemic and topical toxicity. Environ Health Perspect. 2022;130:27012. doi: 10.1289/EHP9341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Raschi E, Ceccarini L, De Ponti F, Recanatini M. HERG-related drug toxicity and models for predicting hERG liability and QT prolongation. Expert Opin Drug Metab Toxicol. 2009;5:1005-1021. doi: 10.1517/17425250903055070 [DOI] [PubMed] [Google Scholar]
- 22. Ramey ME, Kaya F, Bauman AA, et al. Drug distribution and efficacy of the DprE1 inhibitor BTZ-043 in the C3HeB/FeJ mouse tuberculosis model. Antimicrob Agents Chemother. 2023;67:e0059723. doi: 10.1128/aac.00597-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Reali F, Fochesato A, Kaddi C, et al. A minimal PBPK model to accelerate preclinical development of drugs against tuberculosis. Front Pharmacol. 2023;14:1272091. doi: 10.3389/fphar.2023.1272091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Li SY, Tyagi S, Soni H, et al. Bactericidal and sterilizing activity of novel regimens combining bedaquiline or TBAJ-587 with GSK2556286 and TBA-7371 in a mouse model of tuberculosis. Antimicrob Agents Chemother. 2024;68:e0156223. doi: 10.1128/aac.01562-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dube PS, Legoabe LJ, Jordaan A, Sigauke L, Warner DF, Beteck RM. Quinolone analogues of benzothiazinone: synthesis, antitubercular structure-activity relationship and ADME profiling. Eur J Med Chem. 2023;258:115539. doi: 10.1016/j.ejmech.2023.115539 [DOI] [PubMed] [Google Scholar]
- 26. Meta-SNP—meta-predictor of disease causing variants. Accessed February 20, 2023. https://snps.biofold.org/meta-snp/index.html
- 27. Rodrigues CH, Pires DE, Ascher DB. DynaMut: predicting the impact of mutations on protein conformation, flexibility and stability. Nucleic Acids Res. 2018;46:W350-W355. doi: 10.1093/nar/gky300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sanner MF. Python: a programming language for software integration and development. J Mol Graph Model. 1999;17:57-61. [PubMed] [Google Scholar]
- 29. Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31:455-461. doi: 10.1002/jcc.21334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yuan S, Chan HCS, Hu Z. Using PyMOL as a platform for computational drug design. Wires Comput Mol Sci. 2017;7:e1298. doi: 10.1002/wcms.1298 [DOI] [Google Scholar]
- 31. Molecular Operating Environment (MOE), 2022.02 . Chemical Computing Group ULC; 2023. [Google Scholar]
- 32. Berendsen HJC, van der Spoel D, van Drunen R. GROMACS: a message-passing parallel molecular dynamics implementation. Comput Phys Commun. 1995;91:43-56. doi: 10.1016/0010-4655(95)00042-E [DOI] [Google Scholar]
- 33. MacKerell AD, Jr, Bashford D, Bellott M, et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J Phys Chem B. 1998;102:3586-3616. Accessed January 31, 2023. https://pubs.acs.org/doi/abs/10.1021/jp973084f [DOI] [PubMed] [Google Scholar]
- 34. Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J Chem Phys. 2023;79:926-935. Accessed January 31, 2023. https://aip.scitation.org/doi/abs/10.1063/1.445869 [Google Scholar]
- 35. Parrinello M, Rahman A. Polymorphic transitions in single crystals: a new molecular dynamics method. J Appl Phys. 1981;52:7182-7190. Accessed January 31, 2023. https://aip.scitation.org/doi/abs/10.1063/1.328693 [Google Scholar]
- 36. Hoelscher M. A phase IIb, open-label, randomized controlled dose ranging multi-center trial to evaluate the safety, tolerability, pharmacokinetics and exposure-response relationship of different doses of BTZ-043 in combination with bedaquiline and delamanid in adult subjects with newly diagnosed, uncomplicated, drug-sensitive pulmonary tuberculosis. clinicaltrials.gov; 2023. Accessed August 16, 2023. https://clinicaltrials.gov/study/NCT05926466 [DOI] [PMC free article] [PubMed]
- 37. Haddoumi GE, Mansouri M, Bendani H, et al. Selective non-toxics inhibitors targeting DHFR for tuberculosis and cancer therapy: pharmacophore generation and molecular dynamics simulation. Bioinform Biol Insights. 2023;17:11779322231171778. doi: 10.1177/11779322231171778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ode H, Matsuyama S, Hata M, et al. Computational characterization of structural role of the non-active site mutation M36I of human immunodeficiency virus type 1 protease. J Mol Biol. 2007;370:598-607. doi: 10.1016/j.jmb.2007.04.081 [DOI] [PubMed] [Google Scholar]
- 39. Lorch M, Mason JM, Sessions RB, Clarke AR. Effects of mutations on the thermodynamics of a protein folding reaction: implications for the mechanism of formation of the intermediate and transition states. Biochemistry. 2000;39:3480-3485. doi: 10.1021/bi9923510 [DOI] [PubMed] [Google Scholar]
- 40. Reva B, Antipin Y, Sander C. Predicting the functional impact of protein mutations: application to cancer genomics. Nucleic Acids Res. 2011;39:e118. doi: 10.1093/nar/gkr407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Steward RE, MacArthur MW, Laskowski RA, Thornton JM. Molecular basis of inherited diseases: a structural perspective. Trends Genet. 2003;19:505-513. doi: 10.1016/S0168-9525(03)00195-1 [DOI] [PubMed] [Google Scholar]
- 42. George DC, Chakraborty C, Haneef SA, Nagasundaram N, Chen L, Zhu H. Evolution- and structure-based computational strategy reveals the impact of deleterious missense mutations on MODY 2 (maturity-onset diabetes of the young, type 2). Theranostics. 2014;4:366-385. doi: 10.7150/thno.7473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. El Haddoumi G, Mansouri M, Bendani H, et al. Facing antitubercular resistance: identification of potential direct inhibitors targeting InhA enzyme and generation of 3D-pharmacophore model by in silico approach. Adv Appl Bioinform Chem. 2023;16:49-59. doi: 10.2147/AABC.S394535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wróbel A, Baradyn M, Ratkiewicz A, Drozdowska D. Synthesis, biological activity, and molecular dynamics study of novel series of a trimethoprim analogs as multi-targeted compounds: dihydrofolate reductase (DHFR) inhibitors and DNA-binding agents. Int J Mol Sci. 2021;22:3685. doi: 10.3390/ijms22073685 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-docx-1-bbi-10.1177_11779322241257039 for Targeting decaprenylphosphoryl-β-D-ribose 2′-epimerase for Innovative Drug Development Against Mycobacterium Tuberculosis Drug-Resistant Strains by Ghyzlane EL Haddoumi, Mariam Mansouri, Jouhaina Kourou, Lahcen Belyamani, Azeddine Ibrahimi and Ilham Kandoussi in Bioinformatics and Biology Insights
Supplemental material, sj-jpg-2-bbi-10.1177_11779322241257039 for Targeting decaprenylphosphoryl-β-D-ribose 2′-epimerase for Innovative Drug Development Against Mycobacterium Tuberculosis Drug-Resistant Strains by Ghyzlane EL Haddoumi, Mariam Mansouri, Jouhaina Kourou, Lahcen Belyamani, Azeddine Ibrahimi and Ilham Kandoussi in Bioinformatics and Biology Insights