Abstract
In natural niches, bacteria are forced to spend most of their lives under various environmental stresses, such as nutrient limitation, heavy metal pollution, heat and antibiotic stress. To cope with adverse environments, bacterial genome can during the life cycle, produce potential adaptive mutants. The genomic changes, especially mutations, in the genes that encode RNA polymerase and transcription factors, might lead to variations in the transcriptome. These variations enable bacteria to cope with environmental stresses through physiological adaptation in response to stress. This paper reviews the recent contributions of genomic and transcriptomic analyses in understanding the adaption mechanism of Escherichia coli to environmental stresses. Various genomic changes have been observed in E. coli strains in laboratory or under natural stresses, including starvation, heavy metals, acidic conditions, heat shock and antibiotics. The mutations include slight changes (one to several nucleotides), deletions, insertions, chromosomal rearrangements and variations in copy numbers. The transcriptome of E. coli largely changes due to genomic mutations. However, the transcriptional profiles vary due to variations in stress selections. Cellular adaptation to the selections is associated with transcriptional changes resulting from genomic mutations. Changes in genome and transcriptome are cooperative and jointly affect the adaptation of E. coli to different environments. This comprehensive review reveals that coordination of genome mutations and transcriptional variations needs to be explored further to provide a better understanding of the mechanisms of bacterial adaptation to stresses.
Keywords: E. coli, Environmental stress, Genome mutations, Transcriptome variation, Adaptation
Graphical Abstract
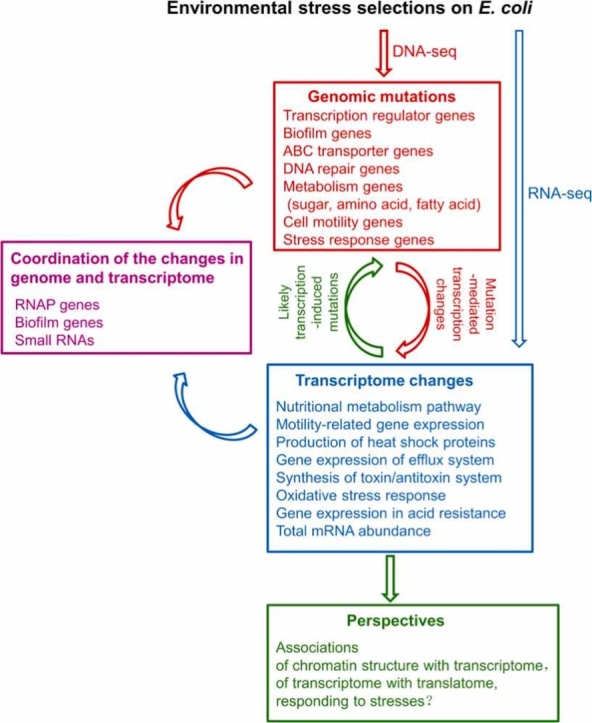
A summary of research fields for the adaptation of E. coli in response to environmental stress. The red dashed box indicates mutational mechanisms that cause genomic changes during environmental selection; the blue dashed box indicates transcriptomic changes during environmental selection; the purple dashed box represents coordination of the changes in genome and transcriptome; and the green dashed box indicates multi-omics cross-analysis during environmental selection.
1. Introduction
All organisms face adverse environmental challenges and stresses during their life cycle. To cope with these stresses, organisms have evolved cellular pathways of adaptations according to the stress [1]. These stress adaptation mechanisms usually include two common themes. First is the timely regulation of mutagenesis by cellular stress responses, which induces random mutations specifically the cells that are poorly adapted stressed. The second theme involves the possible restriction of random mutagenesis in genomic space, achieved via coupling of mutation-generating machinery to local events such as DNA repair or transcription. Furthermore, there are other important aspects that are still not well-understood. It is well known that either the alterations in the genome sequence of an organism or the processes resulting from the modifications are most likely responsible for the adaptation of organisms to environmental changes [2]. The genome mutations include base pair alternations (single nucleotide polymorphism, SNPs), insertions (INSs), deletions (DELs), duplications (DUPs) and inversions (INVs) of a fragment. The joint impact of these genome mutations largely shapes the dynamics of phenotype alterations in the organism. The different types of mutations have different impact on the fitness of populations to the stress selections. For example, non-synonymous single nucleotide polymorphisms (nsSNPs) are more likely to occur than synonymous SNPs to influence the protein functions.
Nevertheless, certain mutations enable the organism to adjust to harsh settings. Most changes in the DNA of an organism have detrimental impacts on its adaptability. Therefore, the mutation rate (i.e., number of genetic changes within a given unit of time) needs to be strictly regulated and it often serves as a quantitative indicator of the adaptability of organism [3]. However, Swings et al. demonstrated that genomic mutation rates can be unexpectedly flexible to changes in selective stresses. When cells experience higher stress, the mutation rates in the population rise, resulting in adaptation of population to stress [4]. Sprouffske et al. measured the mutation rate in E. coli under original environment as well as more than 90 novel chemical environments and showed the effects of mutation rate on the balance between favorable and detrimental impacts of mutations in terms of adaptation [5]. Putnins et al. reported that bacteria with high mutation rates may not respond to environmental stress more effectively regarding. However, wild-type bacteria exhibit a sharp rise in mutation rates in response to environmental stresses. Due to the adaptive gradient that exists and changes within each population, there is a higher chance of mutation, resulting in higher phenotypic diversity in the population [6].
Swift adaptation to fluctuating nutrient conditions is vital for survival of E. coli in diverse environments. The strict regulation of bacterial gene expression facilitates higher recycling of resources and energy, which further prevents the accumulation of harmful substances and maintains the adaptability of organisms [7]. Kim et al. suggested that the adaptability of E. coli might be linked to its capacity of transcriptional change, which may further influence the impact of mutations on the fitness of E. coli and its transcriptional dissimilarities [8]. A recent report indicated that the relative transcriptional levels remained constant in most experimental strains growing cultured under the growth condition of Long-Term Evolution Experiment (LTEE). In contrast, genes with altered expression levels showed similar modifications [9]. This suggests that the changes in transcriptional level may also serve as a mechanism for bacteria to adapt to environmental stress.
2. Indirect genomic mutations and direct adaptation in E. coli under stress selections
Genome plasticity accelerates the adaptive evolution of E. coli in diverse environments. The key driving force behind this evolution is genetic mutability. Genome mutations provide chances for natural selection during adaptive evolution.
E. coli is frequently exposed to starvation stresses in natural environment. Ratib et al. analyzed the genomic mutations in E. coli under nutrient stress and observed 679 mutations including 147 nsSNPs, 48 indels (1 to 16 bp), 376 novel insertion sequences (ISs), 14 DELs (100 bp to 64 kb) and 14 DUPs/amplifications (∼50 to ∼500 kb) (Table 1). Most mutant genes belonged to three functional categories: metabolic enzyme encoding genes, transcriptional regulator encoding genes, or the genes involved in metabolite transport. Mutant genes associated with central metabolism include included sdhB, acs and mhpF, encoding a subunit of succinate dehydrogenase, an acetyl-coenzyme A synthetase and an acetaldehyde dehydrogenase, respectively. Several transcriptional regulators mutant genes like glpR, paaX, lrp and crp participate in regulation of catabolism-associated genes. This implies that these transcriptional factors may affect the metabolism of organism. The transport of amino acids can also be severely affected by the mutations in putP, ybbP and sstT genes. Among these genes, putP gene product is responsible for the proline uptake, while ybbP encodes for a putative ABC transporter permease; and sstT is likely probably to a sodium ion-coupled serine/threonine symporter [10]. Mutations affecting transporter proteins may improve the fitness of organism by allowing more efficient material exchange between cells and the environment. Mutations in metabolism-related genes may enable the cells to use nutrients and/or expel metabolic waste more effectively to achieve greater fitness. These beneficial mutations are expected to provide a competitive edge to organisms by improving their efficiency to utilize nutrients, either directly or indirectly. Behringer et al. tracked the E. coli populations evolving to either 1-day or 10-day feast/famine cycles and detected 82 SNPs, 16 indels, 14 ISs and 5 DELs, with an average of about 43 mutations per clone [11] (Table 1). Most mutant genes were found to be related to fatty acid metabolism (fabF, plsX and putP), nutrient uptake (ompF, ompR, oppA, oppC and sstT) or acquisition of metals (fur and pitA). Modifying the cellular activities related to fatty acid biosynthesis and decreased metal acquisition can be advantageous in energy-restricted settings.
Table 1.
Genomic changes of genes during varied environmental selections.
| Environments | Genes, types and features of mutations |
References | Strain IDs | ||
|---|---|---|---|---|---|
| Genes | Types | Features | |||
| Nutrient stress |
hfq, rpoS, paaX, lrp, sdhB, dtpA, glpR |
nsSNP, indel, IS, deletion, amplification | Parallelism | Ratib et al. (2021) | K-12 W3110 |
| Feast/famine cycle | crp, fusA, ompR, rpoS, rpoB | SNP, indel, IS, deletion | Parallelism | Behringer et al. (2022) | K-12 MG1655 |
| Heavy metal | araD, ulaD, galM, filI, emrB, nusA, araG, nepI | BPS, indel | Metal specificity, consistency | Ba et al. (2022) | K-12 MG1655 |
| Antibiotic environment | mrdAB, mreBCD, pykF, spoT, topA | \ | Parallelism, antagonistic pleiotropy | Lamrabet et al. (2019) | REL606 (B) |
| Ampicillin | ftsI, phoQ, dauA/prs, mgrB/yobH | nsSNP, sSNP, indel, intergenic, IS | Stability, parallelism | Cisneros-Mayoral et al. (2022) | K-12 MG1655 |
| Imipenem | dacC, mrdA, dacD, mrcB, ftsI, zwf, ttuC, lpxD, secF, envZ | sSNP, nsSNP, intergenic, insertion, deletion | \ | Geng et al. (2022) | Sx181-0-1, ATCC25922 |
| Adaptive laboratory evolution | pykF, zwf, spoT, mrdA, hns/tdk, rpoC, rpoB | SNP, indel, insertion, deletion | Consistency | Kavvas et al. (2022) | K-12 MG1655/W3110, BL21, C, W, Crooks |
| osmotic stress | dnaQ, mutS, mutL, mutH, uvrD, mutM, mutY, mutT | sSNP, nsSNP, insertion, indel | strong hypermutator phenotypes | Callens et al. (2023) | DH10B |
| Luria-Bertani broth | cadC, rseB, cytR, iscR, gcvA, sspA, arcA, proQ, rbsR, rplA, fimH, fimG, proQ, lsrK | nsSNP | Parallelism | Ho et al. (2021) | K-12 MG1655 |
Long-term exposure to heavy metals can induce genetic mutations in bacteria, causing diversification and adaptive changes. Ba et al. analyzed the genomic changes in E. coli populations after continuous exposure to low concentration of six heavy metals (i.e., As2O3, CdCl2, Cr2K2O7, CuSO4, NiCl2 and Pb (CH3CO2)2). Long-term multigenerational exposure to all six heavy metals induced mutations in the genome, resulting in 3680 mutations compared to136 mutations in the control lines. Mutations in Pb, Cu and Cr lines were 5-fold, 3.63-fold and 4.6-fold higher than the control lines. Base pair substitutions (BPSs) were the most common among all mutations after heavy metal exposure (83.35%), which was approximately 5-fold higher than that of the control group. BPSs incidence rate was comparatively higher in Pb-exposed lines and lower in the lines exposed to Cd, Cu or As. The average similarities of mutated genes were 54.4% and 38.8% for the same and different exposure patterns, respectively. This indicates that mutated genes can be diverse or convergent during the adaptation of organism to heavy metals. 14 mutations were observed to be specific to a single metal. Specifically, in Cu-exposed lines, mutations were observed in nusA, pagP, atpC, dpiA, tatE and dpiB. In Cd-exposed lines, mutations in hflB, araG, napB and araH were witnessed. In Pb-exposed lines, mutations in chaB and lolD were detected, while nepI gene was found to be mutated in Cr-exposed lines. These mutations were associated with biosorption, extracellular deposition and extracellular efflux pathways, which may all contribute to the metal tolerance of microbes [12]. Mutant microbial strains harbor different genes with a broad range of cellular functions, mainly metabolism. Surprisingly, certain functions have been specifically linked to a specific metal, like ABC transporter to Pb, nitrogen metabolism to Ni, organic acid metabolism to Cr, cell motility and movement to Cd, or folate biosynthesis to Cu. This suggests that microorganisms can evolve a variety of integrated adaptation mechanisms for different heavy metals, such as expelling the heavy metals out of cells, inhibiting contact with metals, or modifying cell membrane composition. These findings show that heavy metal exposure may lead to both common and unique genetic and functional adaptations in microorganisms [13] (Table 1).
One of the most important concerns to human health is the antibiotic resistance of microbes, which is a result of commercialization and unreasonable use of antibiotics. Therefore, understanding the mechanism of anti-bacterial drugs is crucial for clinical treatment and development of drugs for bacterial infections. Lamrabet et al. showed that the E. coli strains evolved in an antibiotic-free environment for more than 50000 generations exhibited higher susceptibility to most antibiotics than their ancestor [14] (Table 1). Similarly, Card et al. confirmed that mutants evolved in a fluctuating drug environment including Ampicillin (AMP), Ceftriaxone (CRO), Ciprofloxacin (CIP) and Tetracycline (TET), had lower adaptive antibiotic resistance compared to their ancestors [15]. Antibiotic resistance develop through mutations that change the gene regulation and expression, cell permeability and efflux and metabolism [16]. Several signature genes have been identified that contributed significantly to antibiotic treatment. For instance, ompR (TET-resistant lines) and ompF (CRO-resistant lines) genes encode a transcriptional regulator that controls the responses of microbe to osmotic and acid stress. Mutations in ompR leads to changes in the expression of OmpF major porin, which further affect the antibiotic resistance of organism. Similarly, alaT gene encodes a tRNA (CIP-resistant lines), while hns gene encodes an outer-membrane porin (CRO-resistant lines). The phoE gene encodes an outer membrane porin (AMP-resistant lines). Cisneros-Mayoral et al. reported higher resistance in microbes under mild selection pressure (MS, AMP, IC=50) as compared to strong selection pressure (SS, AMP, ∼ IC90), which was consistent with other studies [17], [18], [19]. This suggests that the greater selection stress leads to faster elimination of antibiotic resistance in the population as long as the antibiotic is obliterated Mutations in phoQ and ftsI genes during selection for MS were continuously maintained at all stages. Mutations in these two genes lead to modified β-lactam resistance in strains during drug exposures [20]. In natural habitats, highly resistant mutations are kept at low frequencies in a population through sub-lethal antibiotic dosages, which, in theory, only impose a mild selection pressure for resistance. This increases the rate of resistance development in the population during exposure to high doses of antibiotics again.
Among all mutations, nsSNPs are to understand drug-resistance mechanisms of microorganisms and implement the research on targeted therapy [21]. The nsSNPs mutations alter the amino acid sequence of a protein. During screening of nsSNPs associated with drug resistance and susceptibility. Geng et al. identified 60 nsSNPs in the strain with lower resistance levels and 156 nsSNPs in the highly resistant strain. It is clear that the higher number of point mutations is accompanied by the higher level of drug resistance. This indicates a positive correlation between the number of mutations and the degree of drug resistance in an organism. In support of these facts, 19 out of 60 nsSNPs, were predicted to be harmful. These nsSNPs were identified in drug-resistant strains by predicting whether the identified amino acid substitutions affect the protein function (i.e., sorting intolerant strains from tolerant). These mutated genes are associated with the production of peptidoglycan (dacC, mrdA, dacD, mrcB and ftsI), material and energy metabolism (zwf, ttuC and lpxD) and protein export (secF and envZ). 5 genes (dacC, mrdA, lpxD, mrcB and ftsI) were linked to the formation of cell walls. On the other hand, higher resistance to imipenem was attributed to mutations in mrdA, envZ or ftsI genes [22] (Table 1). These mutations enable bacteria to specifically resist the effects of some antibiotics, such as carbapenem, which mainly inhibits cell-wall synthesis, leading to bacterial death [23]. These findings may clarify the molecular mechanism of antibiotic resistance in E. coli.
Understanding the mechanisms of microbial adaptation would be helpful in combating the drug-resistant microbes and enabling the predictive genome design. The bacterial genomes may change rapidly in response to environmental stress (such as excess of nutrients) through gene-gain and -loss, leading to convergence in adaption. Kavvas et al. reported consistency in the evolution of E. coli strains, with different strains displaying mutations in the same specific genes (i.e., pykF, zwf, spoT, mrdA, hns/tdk, rpoC and rpoB) during environmental selection period. Some of these genes (spoT, rpoC, rpoB and mrdA) had SNPs modifications while rest of the genes were observed to have indel mutations. Mobile element INSs and larger deletions were observed in zwf gene [24] (Table 1). These genes were involved in transcription, ppGpp synthesis, NADPH dehydrogenase, cell wall synthesis, glycolysis, folate metabolism and pentose phosphate metabolism. The consistency in mutations suggests that these strains might converge genetically under similar evolutionary constraints. In conclusion, the mutations in E. coli are linked to systems-level changes in NADPH balance and expression of genes related to stress response. The systems-level adaptation patterns were consistent across various E. coli strains, which indicates that proteome reallocation and cofactor balance are the main constraints controlling the microbial adaptation.
3. Coordination between higher mutation rate and fitness of organism
Genotypes with a higher mutation rate are called hypermutators. These genotypes can confer an advantage by providing a greater pool of beneficial mutations that are crucial for microbial adaptation. Callens et al. measured the emergence of hypermutators in experimental E. coli populations under to varying degrees of osmotic or antibiotic stress. The study reported that under osmotic stress, a higher emergence rate of hypermutators was observed as compared to the population in the antibiotic stress group or control group. Remarkably, osmotic stress adaption was mediated by 43 loci, while antibiotic stress was mediated by 23 loci. Numerous loci were observed to be part of the genetic basis of microbial adaptation to osmotic stress. These loci included the genes that are known to be involved in responses to osmotic stress, such as proP, rcsF and wcaJ. The proP gene encodes an H-symporter that senses osmotic shifts and responds by importing osmolytes. On the other hand, rcsF and wcaJ genes are involved in the synthesis of the colonic acid capsule, which provides protection against the osmotic stress. Similarly, several genes known to produce resistance to aminoglycosides were also observed, including fhuA, fusA and energy metabolism related genes. The fhuA gene encodes the ribosomal elongation factor G, while fusA encodes an outer membrane ferrochrome transporter. Similarly, genes involved in energy metabolism (i.e., cyoA, nuoE, nuoG and nuoM) were also discovered at the loci identified as part of the genetic basis of adaptation to antibiotic stress (Table 1). These results provide a better understanding of the impact of mutation rates on the evolution of bacteria during adaptation and demonstrate the variations in hypermutators depending on the type of stress [25].
The batch cultured, E. coli strain frequently adapts to the settings and shows better fitness than its ancestors. Ho et al. showed stronger fixation of mutations (MMR−) in the hypermutant populations than that in WT populations in three different transfer dilutions (1/10, 1/104 and 1/107) [26]. Interestingly, evolved population showed parallelism in terms of fixed mutations. Mutations with high parallelism are ideal to understand the mechanisms of microbial adaption to complicated environments. Ho et al. found the genes that were overrepresented for fixed mutations using a sum of G-scores [27]. This helped in identifying the mutations that are most likely to be the drivers of adaptation. Gene Ontology (GO) analysis of the genes overrepresented for fixed nonsynonymous mutations revealed that significantly enriched GO were frequently associated with the regulation of transcription and biofilm production. Particularly, the ability to form biofilms is crucial for E. coli to survive in the harsh environments. Several genes exhibiting an abundance of non-synonymous mutations have become permanently fixed. These genes facilitate the production of type I fimbriae, including fimH, fimG, fimB, fimE, proQ and lsrK. The production of type I fimbriae is an essential factor that enables E. coli to form biofilms. Interestingly, several transcription regulators genes (arcA, cadC, cytR, rbsR, rseB and sspA) were found to be enriched in fixed nonsynonymous mutations. The genes related to transcription regulators (such as arcB, cadC, rpoS and nlpD) also enriched in structured variations. Mutations in these genes may facilitate rapid transition of microbes from stationary to exponential phase by reprogramming the transcriptomic. Loss-of-function mutation in these genes are beneficial in the laboratory, setting with constant supply of fresh media and continuous mild stress.
Under fluctuating settings, mutator alleles can become more frequent, either reaching fixation or resulting in mutation rate polymorphism [28]. Ramiro et al. followed the long-term evolution of E. coli in the mouse gut and observed mutation rate polymorphism (1000-fold higher than the wild-type). Sequencing of clones between day 136 and 190 post colonization revealed that the mutations in DNA polymerase III subunits sustained hypermutator fitness and boosted the mutation rate by about 1000 times. These included several mutations in subunits of DNA polymerase III: the proofreading (ε) subunit (DnaQ), the catalytic (α) subunit (DnaE) and the θ subunit (HolE). According to a previous study, nonsynonymous mutations in dnaQ can increase the mutation rate by up to > 5000-fold [29]. Mutations in dnaE and to some extent in holE linked to dnaQ mutation can lead to antimutator effects [30]. It was hypothesized that a mutation in dnaQ first raised the mutation rate, which was later reduced by subsequent mutations in dnaE, thus leading to the observed mutation rate polymorphism. As adaptive mutations target metabolism and stress resistance, 12 targets mutated in the mutator clones, were identified. Among these targets, ptsP, frlR and dgoR were found to be mutated in all mutators (include ancestor). The frlR and dgoR genes are negative regulators, which are respectively involved in the metabolism of fructoselysine and galactonate metabolism. On the other hand, ptsP encodes phosphoenolpyruvate-protein phosphotransferase involved in the gut's reaction to pressures, such as pH and salt, as well as nitrogen availability [31], [32]. It is worth noting that ISs, indels or nonsense mutations are frequently involved in the alterations that targeting dgoR and frlR. These mutations may inhibit the negative regulators, causing the regulated operons to overexpress. These studies suggest that genomic mutations in E. coli are not accidental and random, but are indirect, changes made in order to adapt. Adaptation, on the other hand, is a direct response to pressure selection.
4. Transcriptomic changes under environmental selections
Bacteria constantly face the changing nutrient conditions. Adaptation to these changes is particularly crucial during periods of carbon starvation. Morin et al. documented the transcriptomic changes in E. coli cells grown in media with fluctuating carbon (Exponential (P1); Glucose exhaustion/Transition (P2); Acetate consumption (P3); Starvation (P4)) [33]. The study reported the down-regulated expression of 1542 genes between P2 and P1, response to glucose depletion. The up-regulated genes (367) were mainly associated with fermentation, which relieves the starvation stress. Down-regulated expression of 855 genes was observed between P3 and P2, response to acetate depletion, while 15 up-regulated genes were associated with acetate metabolism. 178 genes were found to be down-regulated between P4 and P3. 107 genes associated with stress responses and motility regulation were up-regulated. The down-regulated genes at each stage of carbon fluctuations were mainly enriched in metabolic and transport pathways (Table 2). These observations suggest that E. coli cells up-regulate the metabolism of acetate after acetate ingestion and subsequent fasting. Furthermore, they subsequently turn off most metabolic pathways and nutrition uptake during a stress response. The primary post-transcriptional mechanism regulating the mRNA concentration is mRNA stabilization. A steady decline in mRNA concentrations was observed throughout the starving culture with glucose and acetate. Any variations in mRNA stability, transcription activity, or both may be the cause of variations in mRNA concentration. While most up-regulated gene expressions were under transcriptional control despite mRNA stability, some up-regulated gene expressions were subject to degradational regulation [33]. This suggests that the response of E. coli to environmental changes may involve a complex regulatory system including both mRNA stabilization and degradation. Murashko et al. found that fluoride can affect the gene expression post-transcriptionally by stabilizing the structured RNAs, which may impede the machinery responsible for RNA breakdown [34].
Table 2.
Transcriptomic changes of genes during varied environmental selections.
| Environments | Groups | DEGs and their involved metabolic pathways or biological functions* |
References | Strain IDs | |||
|---|---|---|---|---|---|---|---|
| Up-regulation | Down-regulation | ||||||
| Carbon starvation | P1-P2 | 367 | Fermentation and response to starvation | 1542 | Glucose transport | Morin et al. (2020) | K-12 MG1655 |
| P2-P3 | 15 | Acetate metabolism | 855 | Sugar and amino acid transport; Cell motility | |||
| P3-P4 | 107 | Stress response; Motility regulation | 178 | Amino acid/sugar and acid transport; Catabolism | |||
| pathway; Cell motility | |||||||
| Heat stress | Early | 20 (e.g., clpB, dnaKJ, groSL, grpE, hslVU, hspQ, htpG, ibpA) | Response to heat | 18 (e.g., purB, purD, purE, purL, purT) | Purine biosynthesis | Kim et al. (2020) | BL21 |
| Middle | 12 | Not detected | 8 | Not detected | |||
| Late | 47 (e.g., entCEB, entD, fes-entF) | Enterobactin metabolic process | 19 | Not detected | |||
| Exposure to Cr (VI) | MDR strain | 194 | 320 | Zeng et al. (2023) | LM13, ATCC25922 | ||
| arnA, mdtG, mdtL | Antibiotic resistance | mdtE, mdtF, mdfA | Antibiotic resistance | ||||
| rsmB, rlmJ, rsmJ | DNA and RNA methyltransferase | rlmI, rlmG, rlmF | DNA and RNA methyltransferase | ||||
| cybB, cysC, cysH, cysN | Oxidative stress response | ||||||
| Susceptible strain | 304 | 461 | |||||
| arnA, bcr, mdtE, mdtG, mdtO | Antibiotic resistance genes | rlmN, rlmI, rlmG, rlmF | DNA and RNA methyltransferase | ||||
| ssuB, ssuC | Sulfur transport | sodB, katG | Oxidative stress response | ||||
| cybB, sodA, cysC, cysH, cysJ, cysM | Oxidative stress response | ||||||
| Bovine digestive content | Rumen | 59 | 8 (e.g., argI, arnA, arnD/pmrJ, arnT) | Drug export | Segura et al. (2021) | EDL933 | |
| emrD, glpD, nemA | Drug export | ||||||
| ychH, yhcN | Multiple stress response | ||||||
| Small intestine | 59 | 8 (e.g., argI, arnA, arnC, arnT, arnD/pmrJ) | Drug export | ||||
| emrD, glpD, glpT, tnaB | Drug export | ||||||
| adiA, cadB, caiF | Acid resistance response | ||||||
| clpB, dnaK | Temperature-change response | ||||||
| Rectum | 32 (e.g., ahpC, ahpF, grxA, katG, sodB, trxC, yaaA, yhaK) | Oxidative stress response | 27 | ||||
| argI, arnA, arnC, arnT, arnD/pmrJ | Drug export | ||||||
| gadA, gadE/yhiE | Acid resistance response | ||||||
| slp, yhiM, yhiO | Multiple stress response | ||||||
| Guanylhydr-azone treatment | Pathogenic strain | 383 | 366 | Wang et al. (2022) | RM8082, ATCC25922 | ||
| marA, gadX | Multidrug efflux system | eptA, arnC | Multidrug efflux system | ||||
| cma, cba, clpF, eaeH | Virulence genes | rpl/rpm, rps | Ribosome assembly pathway | ||||
| flgB, flgC, motA, motB | Flagellar assembly pathway | ||||||
| Nonpathogenic strain | 787 | 1169 | |||||
| mdtM, gadX, mdtG, evgA, mdtH | Multidrug efflux system | eptA, yojI, msbA, arnC, mdfA, emrB, emrA, bacA | Multidrug efflux system | ||||
| rpl/rpm, rps | Ribosome assembly pathway | ||||||
| clbR, clbD, clbE, clbA, clbF, clbC, clbI, clbB, clbG, clbQ | Virulence genes | flgB, flgC | Flagellar assembly pathway | ||||
| murA, mrcA | peptidoglycan biosynthesis | ||||||
*Genes showing the expression levels of ≥ 2- or ≤ 0.5-fold were considered DEGs.
The heat shock response is a major defense mechanism used by bacteria to cope with sudden increase in temperature [35]. Kim et al. studied the changes in mRNA in the E. coli cells at different time points after rising the temperature to 42 ℃. The number of differentially expressed genes (DEGs) decreased with the increase in heat exposure time at 42 ℃. 455 DEGs were observed after 2 min, while only 289 DEGs were observed after 40 h at 42 ℃. This suggests that the transcriptomic changes are highly dynamic during continuous incubation of cells at high temperatures. The different time points were divided into three stages early, middle and late. The up-regulation of early-responsive genes in response to a temperature up-shift was characterized by rapid and instantaneous changes in the expression levels. These genes encode crucial chaperones (i.e., dnaKJ, grpE, ibpA and groESL) and proteases (i.e., htpG, hspQ, clpB and hslUV) that play vital roles in protein folding and degradation within cytoplasm. When heat stress persists, gene expression levels can be slowly stabilized. Thus, genes that are overexpressed in late stage might be involved in conferring heat tolerance to E. coli. Expressions of kps gene cluster associated with the biosynthesis of capsular polysaccharides and the genes (entCEB, entD and fes-entF) linked to enterobactin biosynthesis increased only at the late stage (Table 2). Enterobactin is an iron carrier that exhibits cytoprotective activity against heat-induced oxidative stress. High expression of capsular polysaccharide biosynthetic gene cluster may facilitate the cell growth in E. coli during prolonged exposure to high temperatures [36]. Lambros et al. demonstrated that fluctuation in temperatures (15–43 °C) resulted in a restorative response to optimal conditions (37 °C). Mutations in heat shock (e.g., lon and groEL-ES), cold shock (e.g., fabF and groEL-ES) and regulatory genes (e.g., rho: ATP-dependent DNA-RNA helicase) may be especially important for maintaining or restoring internal cell physiology of the strains at 37 °C. Therefore, even in the face of a more difficult adaptive challenge, the strains show a tendency to restore the original phenotype that existed in the ideal environment [37].
Hexavalent chromium (Cr (VI)) pollution can hinder the microbial growth, inducing alterations in the composition and function of microbial communities across different environments [38]. Under Cr (VI) stress, Zeng et al. observed up-regulated expression of 194 genes and down-regulation of 320 genes in the multidrug-resistant (MDR) E. coli strain. In contrast, 304 up-regulated and 461 down-regulated genes were observed in the susceptible E. coli strain. After being exposed to Cr (VI) stress, the MDR strain exhibited a significant enrichment of DEGs across 339 GO terms. The up-regulated genes were primarily responses to external stresses while the down-regulated genes were associated with material and energy metabolism. In comparison to MDR strain, the susceptible strain showed a higher number of down-regulated genes enriched in GO related to ribosomal and translational function. In the MDR strain, the up-regulated genes were enriched in GO associated with external stress responses, which were similar to those observed in the susceptible strain. Furthermore, significant up-regulation of the efflux pump genes (bcr, mdtG and mdtL) and the polymyxin resistance gene (arnA) were also observed in the MDR strain. Expressions of the mdtG, mdtF, mdtE and arnA genes in the MDR strain were significantly higher than their expression levels in the susceptible strain (Table 2). Thus, the increase in the expression of efflux pump genes may aid the MDR strain in preventing Cr (VI) from entering the cell [39].
Segura et al. highlighted the adaptive survival of enterohemorrhagic E. coli (EHEC) O157:H7 in the gastrointestinal tract of cattle. Transcriptomes of strains incubated in the sterile digestive fluid of cattle that did not receive any antibiotic injection for one year before slaughtered were examined and compared to the transcriptome of the strain grown in artificial media. The protective efflux systems necessary to protect E. coli from exposure to other metal ions were activated, alongside the genes responsible for detecting and responding to oxidative stress. Ferrous iron utilization is necessary for E. coli to survive in the anaerobic gut environment. During incubation of EHEC in bovine digestive fluid, many systems involved in iron uptake or utilization were activated or repressed. For instance, chu, efe, ent and fep gene clusters were down-regulated, while feo genes were over-expressed. During EHEC transit, ferrous iron intake was significantly influenced by the Feo transport mechanism. Du et. al found that among the most up-regulated DEGs, many DEGs were associated with iron metabolism (exbD, feoA, ftnA and feoB) under acid stress [40]. Up-regulation of genes involved in acid resistance, specifically arginine- and lysine-dependent acid resistance, in EHEC cells grown in the intestinal digestive fluid, may be attributed to the adaptation of these cells to the high concentrations of organic acids in the gut content. In addition, transcriptional up-regulation of genes encoding for three toxin/antitoxin systems (GhoT/GhoS, HicA/HicB and YhaV/PrlF) was induced, which helped EHEC to survive under unfavorable conditions in the rumen. In the rectum contents, genes associated with chemotaxis and motility (fliGMN, motAB and malE) were activated. Furthermore, genes related to adhesion systems and almost all LEE genes were observed to be up-regulated, which suggests that the rectum may be the major attachment site of the EHEC EDL933 [41] (Table 2). outcomes demonstrated the methods of reducing acid stress associated with amino acid metabolism as well as energy synthesis and conversion.
Riboflavin is produced by commensal bacteria in the human colon. Infants' immune system mature more quickly after exposure to riboflavin produced by gut microbiota. Since riboflavin cannot be produced by mammals, the large intestineal microbiota provides the riboflavin required to maintain the riboflavin homeostasis in host. Liu et al. found that EHEC O157:H7 senses microbiota-produced riboflavin and increased its virulence in the gut. The transcriptome results showed that expressions of z5692 and z5684 were up-regulated 33.0-fold and 9.1-fold respectively in response to riboflavin. Z5692/z5684 shared 43% and 56% similarity to TorS/R at the amino acid level. These data indicate that z5692 and z5684 encode a putative TCS, which can be activated by riboflavin, leading to higher expression of LEE gene. Z5684 and Z5692 are hence called RbfS/R. Additionally, O157:H7 and O145:H28 (two of the nine common EHEC serotypes) have rbfS/R. The virulence of other EHEC serotypes can be boosted by introducing rbfS/R. Therefore, gaining rbfS/R is a significant step in the development of O157:H7. However, rbfS/R may eventually increase the pathogenicity of other EHEC serotypes [42].
One common type of foodborne pathogen that can cause mild diarrhea, bloody diarrhea or potentially fatal hemolytic uremic syndrome is Shiga toxin-producing E. coli (STEC). To control and manage STEC infections, commercially available disinfectants are being used to disinfect or sterilize surfaces of medical and food processing equipment. Wang et al. analyzed the global transcriptional changes in the nonpathogenic ATCC25922 and pathogenic RM8082 after guanylhydrazone treatment. Most of the genes involved in the ribosomal (rpl/rpm and rps) and flagellar assembly pathways (flgB and flgC) were found to be repressed after in both strains after exposure to a lethal dose of guanosinehydrazone. Guanosinehydrazone may inhibit the STEC growth by suppressing protein synthesis and cell motility. The down-regulated genes of strain ATCC25922 were enriched in four pathways: aminoacyl-tRNA biosynthesis (lysS), peptidoglycan biosynthesis (murA and mtgA), biosynthesis of iron-carrier-motif of non-ribosomal peptides (entA-F and entH) and drug metabolism (gst and katG). These pathways are crucial for drug metabolism, chemotaxis, biosynthesis of proteins, cell walls and iron carriers. In addition, 172 virulence genes in strain RM8082 and 224 genes in strain ATCC25922 were identified. All genes located in the pks gene island (clbA-S) were observed to be up-regulated except clbN, clbP and clbS [43] (Table 2). The disinfectant treatment enhanced the expression of virulence genes encoding colibactin, colistin and adhesin. These findings show how cells employ persistence and survival strategies.
5. Coordination between the changes in genome and transcriptome: the molecular adaptation mechanism of E. coli to cope with environmental stresses
The coordination between genome mutations and transcriptome variations may reveal the mechanism of microbial adaptation to environmental stress on the molecular level. However, the interactions between the changes in genome and transcriptome have not been explored and discussed properly, specifically, RNAR σ subunit, cell membrane-associated genes and small RNAs (sRNAs).
Many stress selection experiments have shown that rapid adaptation of bacteria usually tends to occur through mutations in crucial housekeeping genes [44]. Such as the genes coding for RNAP. The σ subunit of RNAP (rpoD, rpoN, rpoS, rpoH, fliA, rpoE and fecI, encoding σ factors) controls the housekeeping and stress-responsive genes in E. coli by directing the core RNAP (α, β, β′and ω subunits (encoded by rpoA, rpoB, rpoC and rpoZ)) to their specific promoters [45]. Evolutionary rescue allows bacteria to genetically adapt and evade extinction in lethal environments [46]. Batarseh et al. sequenced the genomes of 8.8% (26 of 296) rescued E. coli populations, identified in a lethal environment of heat stress (43 °C) (Table 3). Four distinct mutations in rpoBC operon were identified in 6 out of 26 populations. All these point mutations caused nonsynonymous changes in the populations. Two of the rpoBC mutations (rpoB H447Land rpoC W1020G) were the only fixed mutations in at least one population, which suggested that these two mutations were sufficient to rescue a population. To elucidate the molecular mechanisms that leading evolutionary rescue, the gene expressions between REL1206 (rescue ancestor) and the rpoBC mutants were compared. In comparison to REL1206, the single mutations in rpoBC showed 220 to 250 up-regulated DEGs and 230 to 370 down-regulated DEGs. Previous research demonstrated that single mutations in RNAP could change the expression of around 1000 or more genes [47]. Therefore, the huge number of DEGs is not surprising. Among these DEGs, 40 up-regulated genes yielded significant observations. 24 of these 40 genes were involved in transport, ribosome assembly and amino acid and nucleotide pathways. This suggests that changes in gene expression support the translation process, perhaps to increase the efficiency of microbes at high temperatures [48]. In the future, proteomic analyses may provide deeper insights regarding the changes and evolutionary mechanisms induced by rescuing mutations.
Table 3.
Simultaneous changes in genome and transcriptome of genes during varied environmental selections.
| Environments | Genes | Genomic changes | Transcriptomic changes | References | Strain IDs | |
|---|---|---|---|---|---|---|
| Heat stress (43 ℃) | RNAP genes | rpoB, rpoC | nsSNP | \ | Batarseh et al. (2020) | REL1206 |
| Nutrient stress | rpoB, rpoC | nsSNP | \ | Avrani et al. (2017) | K-12 MG1655 | |
| rpoS | nsSNP, deletion | Down-regulation | Ratib et al. (2021) | K-12 W3110 | ||
| Ampicillin environment | rpoD | nsSNP | \ | Cisneros-Mayoral et al. (2022) | K-12 MG1655 | |
| Citrate-only environment | rpoA, rpoB, rpoC, rpoS | nsSNP | Down-regulation | Blount et al. (2020) | B | |
| Heat stress (42 ℃) | rpoD, rpoH | \ | Up-regulation | Kim et al. (2020) | BL21 | |
| Luria-Bertani broth | Biofilm-formation-associated genes | fimE, hns | Frameshift, IS, deletion | \ | Behringeret al. (2018) | K-12 MG1655 |
| flu | \ | Up-regulation | ||||
| Bovine digestive content | fimH, flhD, waaB, yqiG, ecpR, ecpD, | Deletion, non-synonymous substitutions, IS | \ | Yoshida et al. (2021) | K-12 MG1655 | |
| antibiotic stress | sRNAs | fnrS | \ | Down-regulation | kim et al. (2024) | K-12 MG1655, FS0402, BW25113 |
| tp2 | \ | Up-regulation | ||||
| microaerobic and anaerobic | CsrC, CsrB, RprA, GcvB | \ | Up-regulation | Liou et al. (2022) | K-12 MG1655 | |
| NaF treatment under anaerobic condition | RyhB | \ | Down-regulation | Murashko et al. (2023) | K-12 MG1655 | |
| Lysogeny Broth | mgrR | IS | \ | Wright et al. (2021) | E. fergusonii ATCC 35469 | |
| nutrient limitation | hfp | IS | \ | Maharjan et al. (2013) | BW2952, BW4001 | |
In a long-term stationary phase (LTSP) culture experiment under starvation, Avrani et al. studied the mutations in RNAP core enzyme. Almost all cells surviving to LTSP carried a mutation within the exact same gene complex. There were three distinct locations where 93.5% of the mutations were found, rpoB1272, rpoC334 and rpoC428 [49] (Table 3). The widespread presence of these mutations across the LTSP populations suggests that E. coli cells with those mutations possessed a significant advantage under LTSP conditions, exhibiting antagonistic pleiotropy. When resources were made available again, the antagonistic pleiotropy and the mutation accumulation prominently reduced the microbial growth under resource exhaustion. Past studies into LTSP have suggested that the microbial adaptation to LTSP often involves changes in the function of master regulator of stress response, RpoS. Therefore, absence of any mutations within the rpoS gene was somewhat surprising. One possibility could be that the role of RpoS in LTSP adaptation is mostly indirect. The E. coli populations undergo repeated replacement of parental genotypes with fitter variants deep in stationary phase. Nandy et al. isolated one such variant, which displayed a small colony phenotype and slow growth and was able to outcompete its ancestor over a narrow time window in stationary phase. One such variety was discovered by Nandy et al. It had a small colony phenotype, grew slowly, and was able to outcompete its ancestor during a limited time frame in the stationary phase. Despite never being exposed to the antibiotic before, the strain demonstrated resistance to beta-lactam medications. According to Nandy et al., the small colony and slow growth of the variant can be attributed to mutation in RpoC (A494V). The presence of the stationary-phase sigma factor σS determines whether this mutation can provide a growth advantage during stationary phase. The RpoC (A494V) mutation upregulates the σS regulon, thus providing a competitive advantage to its bearer during deep stationary phase [50]. This clarifies the key role of RNAP core enzyme in microbial adaptation to a range of situations.
Most bacteria exist in nature as a multicellular community known as biofilm, which has significant adaptive potential. Behringer et al. sequenced the genome of 100 wild type (MMR+) and repair-deficient (MMR−) E. coli populations after 1 year of evolution (800–3400 generations). The study reported major polymorphisms of note included insertion elements transposing into fimE (a repressor of the type 1 fimbriae) and upstream of hns (histone-like global regulator which regulates fimB and fimE and inhibits acrB (a subunit of the MDR family efflux pump AcrAB/TolC)). All these genes are known to regulate biofilm formation. Up-regulation of flu (an epigenetically regulated phase-change gene encoding a biofilm-promoting autotransporter) was observed in individual clones in which these mutations were located [51] (Table 3). According to Benyoussef et al., motile cells have a glaring drawback, demonstrating a prolonged latency in the biofilm-forming process relative to nonmotile cells. This indicates the function of cell motility in surface colonization and biofilm formation [52]. Yoshida et al. showed that the high frequency mutations in the fimH gene were present in the E. coli populations with improved biofilm formation capabilities. On abiotic surfaces, FimH have been demonstrated to be essential for biofilm production. The ∆f im populations revealed mutations in biofilm functions related genes, that were associated with chaperone-usher fimbrial surface structure (yqiG, ecpR and ecpD), flagellum (flgH and ecpR) or autotransporter adhesin (ycgV and flu/agn43). On the other hand, biofilm-promoting mutations in ecpD were only detected in the clones exhibiting IS insertion immediately after the inhibition of ecpR (the regulator of ecp operon) in E. coli K12 MG1655. This IS insertion mutations resulted in expression of ecp operon, inducing further mutations in ecpD and leading to increased biofilm formation in ∆f im population. This suggests that distinct alterations in surface structure and adhesins could result in increased biofilm formation capacity of E. coli in the absence of type 1 fimbriae [53]. Li et al. observed up-regulation of type 1 fimbrial-protein pilus (fimA and fimH) in an antibiotic-influenced environment [54]. These findings demonstrated that mutations in fimH are the main drivers of surface adhesion and biofilm formation capacity of E. coli during stress selection.
Antibiotic-induced regulatory sRNAs promote the tolerant of bacteria to environmental variations. Kim et al. demonstrated that during exposure to sublethal dose of antibiotics, bacteria delayed oxygen consumption by switching from active aerobic respiration to anaerobic adaptation via sRNA (FnrS and Tp2) mediated sudden respiratory changes [55]. Liou et al. reported differential expression of several sRNAs in microaerobic and anaerobic environments. Abundance of 18 (out of 64) known sRNAs in E. coli, differed by more than 1.5 times between microaerobic and aerobic environments. Several sRNAs, including CsrB, CsrC, GcvB or RprA, were observed to be more prevalent in microaerobic environments, where CsrB and CsrC performed their regulatory duties by attaching to translational inhibitor CsrA and blocking the latter from interacting with translation initiation regions of several transcripts, including pgaABCD (Table 3). By competitively bidding CsrB and CsrC to CsrA, this operon got translated and activated, thus increasing the production of polysaccharide and promoting biofilm formation. Other genes (such as iraD and glgS) that were also elevated in microaerobic environments were probably translated through CsrB- and CsrC-competitive binding mechanisms. GlgS is widely recognized as an inhibitor of cell motility, while iraD gene encodes an anti-adapter protein that prevents the degradation of sigma stress factor RpoS mediated by RssB [56]. Additionally, Wright et al. suggested that having an alternative function enabled sRNA to retain in Escherichia fergusonii despite sustaining a large and potentially disruptive mutation. This finding is not surprising given the variety of roles that sRNAs play during RNA degradation, cell division and histone-like activities [57]. In many cases, RNA chaperones, such as Hfq and ProQ, facilitate complementary base pairing between sRNAs and target RNAs to control the corresponding gene expressions. This phenomenon is commonly observed during sRNA processes. Maharjan et al. did not find any null mutations in hfq, but observed promoter IS mutation which indicates that reduced Hfq protein amount may be advantageous at slower growth rates [58]. Several studies on bacterial regulatory mechanisms have concluded that sRNAs typically modify the target gene expression at post-transcriptional level [59]. Numerous sRNAs, in an analogous mechanism, are widespread in bacteria. Thus, sRNAs are critical to reveal the key changes in the gene expressions at transcriptional and post-transcriptional levels during stress adaptation.
6. Perspectives
Occurrence of SNPs in the genome has been considered a significant way of genetic variation. SVs include DELs, INSs, DUPs, INVs and translocations (TRAs) of at least 50 bp in size [60]. After subjecting the E. coli cells to long-term exposure to stress selection, Lato et al. discovered that more than 71% of the INVs led to 9.4 to 85.6 times higher numbers of DEGs than those in the control group [61]. Le et al. found that large chromosomal INVs affect the sensitivity of E. coli to sodium deoxycholate [62]. These results lend credibility to the idea that, in some circumstances, a big chromosomal INV can be advantageous to bacterial cells, as it increases the genetic variety available for selection throughout evolution. Delgado-Blas et al. witnessed higher multiplicity of resistance genes in E. coli in the wastewater as compared to that in river water. This suggests that the wastewater environment favors the selection of conservative chromosome structures. In contrast, genomic multiplicity of riverine E. coli showed greater variations in the chromosomal structure, including integration of mobile genetic elements and few plasmid types shared among different STs, which resulted in limited resistance genes [63]. Maddamsetti et al. also reported mutations in the genes associated with chromosome structure (such as fis, topA and other genes) in the LTEE-evolved strains [64]. These chromosomal structural variations may result in changes in gene transcription. El Houdaigui et al. has suggested that the structure of genome controls the global transcriptional regulation via DNA superhelix [65]. Furthermore, Shen et al. reported that chromatin can impact the transcription process by several distinct mechanisms, including occlusion of RNAP binding, roadblocking RNAP progression, constraining DNA topology, RNA-mediated interactions etc. [66].
Ahmed et al. showed that the distribution of DNA topoisomerases was guided by active transcription in vivo [67]. This result suggests that the transcriptional changes may provide a chance for genetic diversification. Huang et al. reported that histone-like proteins, H-Ns and IHF, in E. coli affected the higher-order DNA compaction while H-NS and IHF regulated the gene expression as transcription factors. Similarly, the transcription factor DnaA has also been shown to affect the compaction of nucleoid [68], [69] and RNA structures [70]. Le Berre et al. observed the link between gene expression and chromosomal spatial organization, which turned out to be essential for bacterial adaptation to changing environmental circumstances [71]. Further, Rashid et al. proposed that H-NS could act as a coupling sensor for transcriptional-hyperhelical dynamic. H-NS could also affect its binding properties to DNA by interacting with chaperone proteins (ProVWX and HlyCABD) and through post-translational modifications [72]. To fully understand the environmental adaptation of bacteria, it is important to establish an association between chromatin structure and transcriptional regulation.
By measuring the transcriptome and relative translation efficiency (RTE) of E. coli under distinct nutrient constraints, Zhang et al. found that the genes involved in ribosomal (rplA and rmf) and metabolic homeostasis (nagC, ascG and kdgR) exhibited a strong correlation between mRNA levels and RTEs, indicating coordination between transcription and translation [73]. Bacterial cells may employ a universal approach of synergistic regulation of transcription and translation to increase their ability to adapt to environmental variations [74]. During the process of translation elongation, ribosomes may experience deceleration or temporary halting caused by the secondary structure of mRNA and availability of tRNA. The fast-moving ribosomes would have the potential chance to encounter slow or paused ribosomes. Interestingly, Li et al. performed genome-wide ribosome profiling using a machine learning model to predict the role of SNPs in translation since moving ribosomes could collapse during translation upon an encounter with SNPs [75]. Thus, SNPs may modulate the occurrence of ribosome collisions by modifying the secondary structure of mRNA or the abundance of homologous tRNAs. These findings provide a clue to deepen and widen the investigations on bacterial adaptation to environmental selections. Exploring the direct associations between genome and transcriptome, transcriptome and translatome and genome and translatome would be helpful in understanding the molecular mechanism of environmental adaptation of bacteria, or other organisms during the selections.
CRediT authorship contribution statement
Jianlu Jiao: Conceptualization, Writing – review & editing, Writing – original draft, Formal analysis. Xiaoli Lv: Conceptualization, Writing – original draft. Chongjie Shen: Writing – original draft, Formal analysis. Morigen Morigen: Conceptualization, Funding acquisition, Supervision, Supervision.
Author’s contribution
JL, XL, CJ and M wrote and revised the manuscript. All authors contributed significantly, directly and intellectually to the work, provided substantial input and approved it for publication.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The study was supported by the National Natural Science Foundation of China (Grant no. 32060016 to M).
References
- 1.Rodriguez-Verdugo A., Tenaillon O., Gaut B.S. First-step mutations during adaptation restore the expression of hundreds of genes. Mol Biol Evol. 2016;33(1):25–39. doi: 10.1093/molbev/msv228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fitzgerald D.M., Rosenberg S.M. What is mutation? A chapter in the series: how microbes "jeopardize" the modern synthesis. PLoS Genet. 2019;15(4) doi: 10.1371/journal.pgen.1007995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang G., Campbell E.A., Minakhin L., Richter C., Severinov K., Darst S.A. The mutation rate as an evolving trait. Nat Rev Genet. 2023;24(1):3. doi: 10.1038/s41576-022-00547-9. [DOI] [PubMed] [Google Scholar]
- 4.Swings T., Van den Bergh B., Wuyts S., Oeyen E., Voordeckers K., Verstrepen K.J., et al. Adaptive tuning of mutation rates allows fast response to lethal stress in Escherichia coli. Elife. 2017;6 doi: 10.7554/eLife.22939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sprouffske K., Aguilar-Rodriguez J., Sniegowski P., Wagner A. High mutation rates limit evolutionary adaptation in Escherichia coli. PLoS Genet. 2018;14(4) doi: 10.1371/journal.pgen.1007324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Putnins M., Androulakis I.P. Self-selection of evolutionary strategies: adaptive versus non-adaptive forces. Heliyon. 2021;7(5) doi: 10.1016/j.heliyon.2021.e06997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bervoets I., Charlier D. Diversity, versatility and complexity of bacterial gene regulation mechanisms: opportunities and drawbacks for applications in synthetic biology. FEMS Microbiol Rev. 2019;43(3):304–339. doi: 10.1093/femsre/fuz001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim K., Kwon S.K., Kim P., Kim J.F. Transcriptional potential determines the adaptability of Escherichia coli strains with different fitness backgrounds. Microbiol Spectr. 2022;10(6) doi: 10.1128/spectrum.02528-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Favate J.S., Liang S., Cope A.L., Yadavalli S.S., Shah P. The landscape of transcriptional and translational changes over 22 years of bacterial adaptation. Elife. 2022;11 doi: 10.7554/eLife.81979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ratib N.R., Seidl F., Ehrenreich I.M., Finkel S.E. Evolution in long-term stationary-phase batch culture: emergence of divergent Escherichia coli lineages over 1,200 Days. mBio. 2021;12(1):e03337–e03420. doi: 10.1128/mBio.03337-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Behringer M.G., Ho W.C., Meraz J.C., Miller S.F., Boyer G.F., Stone C.J., et al. Complex ecotype dynamics evolve in response to fluctuating resources. mBio. 2022;13(3) doi: 10.1128/mbio.03467-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bruins M.R., Kapil S., Oehme F.W. Microbial resistance to metals in the environment. Ecotoxicol Environ Saf. 2000;45(3):198–207. doi: 10.1006/eesa.1999.1860. [DOI] [PubMed] [Google Scholar]
- 13.Ba Q., Zhou J., Li J., Cheng S., Zhang X., Wang H. Mutagenic characteristics of six heavy metals in Escherichia coli: the commonality and specificity. Environ Sci Technol. 2022;56(19):13867–13877. doi: 10.1021/acs.est.2c04785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lamrabet O., Martin M., Lenski R.E., Schneider D. Changes in intrinsic antibiotic susceptibility during a long-term evolution experiment with Escherichia coli. mBio. 2019;10(2) doi: 10.1128/mBio.00189-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Card K.J., LaBar T., Gomez J.B., Lenski R.E. Historical contingency in the evolution of antibiotic resistance after decades of relaxed selection. PLoS Biol. 2019;17(10) doi: 10.1371/journal.pbio.3000397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blair J.M., Webber M.A., Baylay A.J., Ogbolu D.O., Piddock L.J. Molecular mechanisms of antibiotic resistance. Nat Rev Microbiol. 2015;13(1):42–51. doi: 10.1038/nrmicro3380. [DOI] [PubMed] [Google Scholar]
- 17.Card K.J., Thomas M.D., Graves J.L., Jr., Barrick J.E., Lenski R.E. Genomic evolution of antibiotic resistance is contingent on genetic background following a long-term experiment with Escherichia coli. Proc Natl Acad Sci USA. 2021;118(5) doi: 10.1073/pnas.2016886118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barbosa C., Mahrt N., Bunk J., Grasser M., Rosenstiel P., Jansen G., et al. The genomic basis of rapid adaptation to antibiotic combination therapy in Pseudomonas aeruginosa. Mol Biol Evol. 2021;38(2):449–464. doi: 10.1093/molbev/msaa233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cisneros-Mayoral S., Grana-Miraglia L., Perez-Morales D., Pena-Miller R., Fuentes-Hernandez A. Evolutionary history and strength of selection determine the rate of antibiotic resistance adaptation. Mol Biol Evol. 2022;39(9) doi: 10.1093/molbev/msac185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Batra A., Roemhild R., Rousseau E., Franzenburg S., Niemann S., Schulenburg H. High potency of sequential therapy with only β-lactam antibiotics. eLife. 2021;10 doi: 10.7554/eLife.68876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Singh P., Jamal S., Ahmed F., Saqib N., Mehra S., Ali W., et al. Computational modeling and bioinformatic analyses of functional mutations in drug target genes in Mycobacterium tuberculosis. Comput Struct Biotechnol J. 2021;19:2423–2446. doi: 10.1016/j.csbj.2021.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Geng J., Liu H., Chen S., Long J., Jin Y., Yang H., et al. Comparative genomic analysis of Escherichia coli strains obtained from continuous imipenem stress evolution. FEMS Microbiol Lett. 2022;369(1) doi: 10.1093/femsle/fnac015. [DOI] [PubMed] [Google Scholar]
- 23.Barbarini D., Russello G., Brovarone F., Capatti C., Colla R., Perilli M., et al. Evaluation of carbapenem-resistant Enterobacteriaceae in an Italian setting: report from the trench. Infect Genet Evol. 2015;30:8–14. doi: 10.1016/j.meegid.2014.11.025. [DOI] [PubMed] [Google Scholar]
- 24.Kavvas E.S., Long C.P., Sastry A., Poudel S., Antoniewicz M.R., Ding Y., et al. Experimental evolution reveals unifying systems-level adaptations but diversity in driving genotypes. mSystems. 2022;7(6) doi: 10.1128/msystems.00165-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Callens M., Rose C.J., Finnegan M., Gatchitch F., Simon L., Hamet J., et al. Hypermutator emergence in experimental Escherichia coli populations is stress-type dependent. Evol Lett. 2023;7:252–261. doi: 10.1093/evlett/qrad019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ho W.C., Behringer M.G., Miller S.F., Gonzales J., Nguyen A., Allahwerdy M., et al. Evolutionary dynamics of asexual hypermutators adapting to a novel environment. Genome Biol Evol. 2021;13(12) doi: 10.1093/gbe/evab257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tenaillon O., Barrick J.E., Ribeck N., Deatherage D.E., Blanchard J.L., Dasgupta A., et al. Tempo and mode of genome evolution in a 50,000-generation experiment. Nature. 2016;536(7615):165–170. doi: 10.1038/nature18959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gordo I., Campos P.R. Evolution of clonal populations approaching a fitness peak. Biol Lett. 2013;9(1):20120239. doi: 10.1098/rsbl.2012.0239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fijalkowska I.J., Schaaper R.M. Mutants in the Exo I motif of Escherichia coli dnaQ: defective proofreading and inviability due to error catastrophe. Proc Natl Acad Sci USA. 1996;93:2856–2861. doi: 10.1073/pnas.93.7.2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Taft-Benz S.A., Schaaper R.M. The θ Subunit of Escherichia coli DNA Polymerase III: a role in stabilizing the ε proofreading subunit. J Bacteriol. 2004;186:2774–2780. doi: 10.1128/JB.186.9.2774-2780.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ramiro R.S., Durao P., Bank C., Gordo I. Low mutational load and high mutation rate variation in gut commensal bacteria. PLoS Biol. 2020;18(3) doi: 10.1371/journal.pbio.3000617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Winter S.E., Winter M.G., Xavier M.N., Thiennimitr P., Poon V., Keestra A.M., et al. Host-derived nitrate boosts growth of E. coli in the inflamed gut. Science. 2013;339(6120):708–711. doi: 10.1126/science.1232467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morin M., Enjalbert B., Ropers D., Girbal L., Cocaign-Bousquet M. Genomewide Stabilization of mRNA during a "Feast-to-Famine" Growth Transition in Escherichia coli. mSphere. 2020;5(3):e00276–e00320. doi: 10.1128/mSphere.00276-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murashko O.N., Yeh K.H., Yu C.A., Kaberdin V.R., Lin-Chao S. Sodium Fluoride Exposure Leads to ATP Depletion and Altered RNA Decay in Escherichia coli under Anaerobic Conditions. Microbiol Spectr. 2023;11(2) doi: 10.1128/spectrum.04158-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Richter K., Haslbeck M., Buchner J. The heat shock response: life on the verge of death. Mol Cell. 2010;40(2):253–266. doi: 10.1016/j.molcel.2010.10.006. [DOI] [PubMed] [Google Scholar]
- 36.Kim S., Kim Y., Suh D.H., Lee C.H., Yoo S.M., Lee S.Y., et al. Heat-responsive and time-resolved transcriptome and metabolome analyses of Escherichia coli uncover thermo-tolerant mechanisms. Sci Rep. 2020;10(1) doi: 10.1038/s41598-020-74606-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lambros M., Pechuan-Jorge X., Biro D., Ye K., Bergman A. Emerging adaptive strategies under temperature fluctuations in a laboratory evolution experiment of Escherichia coli. Front Microbiol. 2021;12 doi: 10.3389/fmicb.2021.724982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li D., Li G., Zhang D. Field-scale studies on the change of soil microbial community structure and functions after stabilization at a chromium-contaminated site. J Hazard Mater. 2021;415 doi: 10.1016/j.jhazmat.2021.125727. [DOI] [PubMed] [Google Scholar]
- 39.Zeng X., Cao Y., Wang L., Wang M., Wang Q., Yang Q. Viability and transcriptional responses of multidrug resistant E. coli to chromium stress. Environ Pollut. 2023;324 doi: 10.1016/j.envpol.2023.121346. [DOI] [PubMed] [Google Scholar]
- 40.Du B., Olson C.A., Sastry A.V., Fang X., Phaneuf P.V., Chen K., et al. Adaptive laboratory evolution of Escherichia coli under acid stress. Microbiol (Read) 2020;166(2):141–148. doi: 10.1099/mic.0.000867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Segura A., Bertin Y., Durand A., Benbakkar M., Forano E. Transcriptional analysis reveals specific niche factors and response to environmental stresses of enterohemorrhagic Escherichia coli O157:H7 in bovine digestive contents. BMC Microbiol. 2021;21(1):284. doi: 10.1186/s12866-021-02343-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu B., Liu Y., Yang B., Wang Q., Liu X., Qin J., et al. Escherichia coli O157:H7 senses microbiota-produced riboflavin to increase its virulence in the gut. Proc Natl Acad Sci USA. 2022;119(48) doi: 10.1073/pnas.2212436119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang Y., Hart-Cooper W.M., Rasooly R., Carter M.Q., Orts W.J., Gu Y., et al. Effect of an eco-friendly cuminaldehyde guanylhydrazone disinfectant on shiga toxin production and global transcription of Escherichia coli. Toxins (Basel) 2022;14(11):752. doi: 10.3390/toxins14110752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Avrani S., Katz S., Hershberg R. Adaptations accumulated under prolonged resource exhaustion are highly transient. mSphere. 2020;5(4) doi: 10.1128/mSphere.00388-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shimada T., Tanaka K., Ishihama A. The whole set of the constitutive promoters recognized by four minor sigma subunits of Escherichia coli RNA polymerase. PLoS One. 2017;12(6) doi: 10.1371/journal.pone.0179181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bell G. Evolutionary rescue. Annu Rev Ecol Evol Syst. 2017;48(1):605–627. [Google Scholar]
- 47.Conrad T.M., Frazier M., Joyce A.R., Cho B.K., Knight E.M., Lewis N.E., et al. RNA polymerase mutants found through adaptive evolution reprogram Escherichia coli for optimal growth in minimal media. Proc Natl Acad Sci USA. 2010;107(47):20500–20505. doi: 10.1073/pnas.0911253107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Batarseh T.N., Hug S.M., Batarseh S.N., Gaut B.S. Genetic mutations that drive evolutionary rescue to lethal temperature in Escherichia coli. Genome Biol Evol. 2020;12(11):2029–2044. doi: 10.1093/gbe/evaa174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Avrani S., Bolotin E., Katz S., Hershberg R. Rapid genetic adaptation during the first four months of survival under resource exhaustion. Mol Biol Evol. 2017;34(7):1758–1769. doi: 10.1093/molbev/msx118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nandy P., Chib S., Seshasayee A. A mutant RNA polymerase activates the general stress response, enabling Escherichia coli Adaptation to Late Prolonged Stationary Phase. mSphere. 2020;5(2) doi: 10.1128/mSphere.00092-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Behringer, M.G. , Choi, B.I. , Miller, S.F. , Doak, T.G. , Karty, J.A. , Guo, W. , et al. , Escherichia coli cultures maintain stable subpopulation structure during long-term evolution. Proc Natl Acad Sci U S A, 2018. 115(20): p. E4642-E4650. [DOI] [PMC free article] [PubMed]
- 52.Benyoussef W., Deforet M., Monmeyran A., Henry N. Flagellar Motility During E. coli Biofilm Formation Provides a Competitive Disadvantage Which Recedes in the Presence of Co-Colonizers. Front Cell Infect Microbiol. 2022;12 doi: 10.3389/fcimb.2022.896898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yoshida M., Thiriet-Rupert S., Mayer L., Beloin C., Ghigo J.M. Selection for nonspecific adhesion is a driver of FimH evolution increasing Escherichia coli biofilm capacity. Microlife. 2022;3 doi: 10.1093/femsml/uqac001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li D., Liang W., Hu Q., Ren J., Xue F., Liu Q., et al. The effect of a spontaneous induction prophage, phi458, on biofilm formation and virulence in avian pathogenic Escherichia coli. Front Microbiol. 2022;13:1049341. doi: 10.3389/fmicb.2022.1049341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kim D., Bhat A., Kim S.K., Lee S., Ryu C.M. Small RNA-modulated anaerobic respiration allows bacteria to survive under antibiotic stress conditions. Front Cell Infect Microbiol. 2024;14:1287557. doi: 10.3389/fcimb.2024.1287557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liou G.G., Chao Kaberdina A., Wang W.S., Kaberdin V.R., Lin-Chao S. Combined transcriptomic and proteomic profiling of E. coli under Microaerobic versus Aerobic conditions: the multifaceted roles of noncoding Small RNAs and oxygen-dependent sensing in global gene expression control. Int J Mol Sci. 2022;23(5) doi: 10.3390/ijms23052570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wright A.P., Dutcher H.A., Butler B., Nice T.J., Raghavan R. A small RNA is functional in Escherichia fergusonii despite containing a large insertion. Microbiol (Read) 2021;167(10) doi: 10.1099/mic.0.001099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Maharjan R., McKenzie C., Yeung A., Ferenci T. The basis of antagonistic pleiotropy in hfq mutations that have opposite effects on fitness at slow and fast growth rates. Hered (Edinb) 2013;110(1):10–18. doi: 10.1038/hdy.2012.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dutta T., Srivastava S. Small RNA-mediated regulation in bacteria: a growing palette of diverse mechanisms. Gene. 2018;656:60–72. doi: 10.1016/j.gene.2018.02.068. [DOI] [PubMed] [Google Scholar]
- 60.Abyzov A., Li S., Kim D.R., Mohiyuddin M., Stutz A.M., Parrish N.F., et al. Analysis of deletion breakpoints from 1,092 humans reveals details of mutation mechanisms. Nat Commun. 2015;6:7256. doi: 10.1038/ncomms8256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lato D.F., Zeng Q., Golding G.B. Genomic inversions in Escherichia coli alter gene expression and are associated with nucleoid protein binding sites. Genome. 2022;65(5):287–299. doi: 10.1139/gen-2021-0102. [DOI] [PubMed] [Google Scholar]
- 62.Le V.V.H., Leon-Quezada R.I., Biggs P.J., Rakonjac J. A large chromosomal inversion affects antimicrobial sensitivity of Escherichia coli to sodium deoxycholate. Microbiol (Read) 2022;168(8) doi: 10.1099/mic.0.001232. [DOI] [PubMed] [Google Scholar]
- 63.Delgado-Blas J.F., Ovejero C.M., David S., Montero N., Calero-Caceres W., Garcillan-Barcia M.P., et al. Population genomics and antimicrobial resistance dynamics of Escherichia coli in wastewater and river environments. Commun Biol. 2021;4(1):457. doi: 10.1038/s42003-021-01949-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Maddamsetti R., Grant N.A. Divergent evolution of mutation rates and biases in the long-term evolution experiment with Escherichia coli. Genome Biol Evol. 2020;12(9):1591–1603. doi: 10.1093/gbe/evaa178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.El Houdaigui B., Forquet R., Hindre T., Schneider D., Nasser W., Reverchon S., et al. Bacterial genome architecture shapes global transcriptional regulation by DNA supercoiling. Nucleic Acids Res. 2019;47(11):5648–5657. doi: 10.1093/nar/gkz300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shen B.A., Landick R. Transcription of bacterial chromatin. J Mol Biol. 2019;431(20):4040–4066. doi: 10.1016/j.jmb.2019.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ahmed W., Sala C., Hegde S.R., Jha R.K., Cole S.T., Nagaraja V. Transcription facilitated genome-wide recruitment of topoisomerase I and DNA gyrase. PLoS Genet. 2017;13(5) doi: 10.1371/journal.pgen.1006754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Huang T., Yuan H., Fan L., Morigen H-NS, IHF, and DnaA lead to changes in nucleoid organizations, replication initiation, and cell division. J Basic Microbiol. 2020;60(2):136–148. doi: 10.1002/jobm.201900497. [DOI] [PubMed] [Google Scholar]
- 69.Jianlu J., Morigen The QseB/QseC-mediated Quorum-Sensing In Response To Environmental Changes. Chin J Biochem Mol Biol. 2023;39(6):759–768. [Google Scholar]
- 70.Yao Y., Sun H., Gegeheng Wurihan, Skarstad Gezi, Fan K., Morigen L. A DnaA-dependent riboswitch for transcription attenuation of the his operon. mLife. 2023;2(2):126–140. doi: 10.1002/mlf2.12075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Le Berre D., Reverchon S., Muskhelishvili G., Nasser W. Relationship between the chromosome structural dynamics and gene expression-a chicken and egg dilemma? Microorganisms. 2022;10(5):846. doi: 10.3390/microorganisms10050846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rashid F.M., Dame R.T. Three-dimensional chromosome re-modelling: the integral mechanism of transcription regulation in bacteria. Mol Microbiol. 2023;120(1):60–70. doi: 10.1111/mmi.15062. [DOI] [PubMed] [Google Scholar]
- 73.Zhang D., Li S.H., King C.G., Wingreen N.S., Gitai Z., Li Z. Global and gene-specific translational regulation in Escherichia coli across different conditions. PLoS Comput Biol. 2022;18(10) doi: 10.1371/journal.pcbi.1010641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nguyen H.L., Duviau M.P., Laguerre S., Nouaille S., Cocaign-Bousquet M., Girbal L. Synergistic Regulation of Transcription and Translation in Escherichia coli Revealed by Codirectional Increases in mRNA Concentration and Translation Efficiency. Microbiol Spectr. 2022;10(1) doi: 10.1128/spectrum.02041-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Li Z., Chen L. Predicting functional consequences of SNPs on mRNA translation via machine learning. Nucleic Acids Res. 2023 doi: 10.1093/nar/gkad576. gkad576. [DOI] [PMC free article] [PubMed] [Google Scholar]


