Summary
This review explores the hallmarks of cancer resistance, including drug efflux mediated by ATP-binding cassette (ABC) transporters, metabolic reprogramming characterized by the Warburg effect, and the dynamic interplay between cancer cells and mitochondria. The role of cancer stem cells (CSCs) in treatment resistance and the regulatory influence of non-coding RNAs, such as long non-coding RNAs (lncRNAs), microRNAs (miRNAs), and circular RNAs (circRNAs), are studied. The chapter emphasizes future directions, encompassing advancements in immunotherapy, strategies to counter adaptive resistance, integration of artificial intelligence for predictive modeling, and the identification of biomarkers for personalized treatment. The comprehensive exploration of these hallmarks provides a foundation for innovative therapeutic approaches, aiming to navigate the complex landscape of cancer resistance and enhance patient outcomes.
Subject areas: health sciences, microenvironment, biological sciences, molecular biology, epigenetics, cancer
Graphical abstract

Health sciences; Microenvironment; Biological sciences; Molecular biology; Epigenetics; Cancer.
Introduction
Despite significant advances in our understanding of cancer biology and the development of novel therapeutic approaches, the phenomenon of cancer resistance remains a persistent challenge. This resistance is characterized by the remarkable ability of cancer cells to evade the cytotoxic effects of treatments, thereby compromising the effectiveness of therapies and, ultimately, endangering the lives of countless individuals.1 In our pursuit of effective cancer management, it is imperative to unravel the intricate web of mechanisms underlying the hallmarks of cancer resistance.
Cancer resistance represents a multifaceted spectrum of mechanisms that enable malignant cells to withstand even the most potent therapeutic strategies. From the early days of chemotherapy to the emergence of targeted therapies and immunotherapies, cancer cells have displayed an exceptional capacity to adapt, evolve, and persist.1 This resistance can manifest in various forms, including inherent resistance, acquired resistance, and adaptive resistance, further complicating the landscape of cancer treatment.
Inherent resistance is often rooted in the genetic and epigenetic characteristics of cancer cells, predisposing certain tumors to be refractory to conventional treatments.2 Acquired resistance, on the other hand, emerges in response to the selective pressure exerted by therapeutic interventions, resulting in the emergence of treatment-resistant subpopulations within the tumor.3 Additionally, adaptive resistance occurs when cancer cells dynamically alter their behavior and signaling pathways in response to treatment, rendering once-potent therapies ineffective.4
The challenges posed by cancer resistance extend beyond the realm of chemotherapy and encompass the evolving field of targeted therapies and immunotherapies. Even therapies that initially elicit remarkable responses frequently encounter a formidable adversary in the form of resistance.5 The ongoing evolution of resistance mechanisms necessitates a holistic understanding of cancer biology that transcends reductionist views and embraces the complex, dynamic, and adaptable nature of cancer.
The significance of comprehending cancer resistance cannot be overstated. It holds the key to improving patient outcomes, enhancing the durability of treatment responses, and ultimately advancing the field of oncology.6 Without a profound understanding of resistance mechanisms, we are left with an incomplete picture of cancer’s resilience and a limited array of treatment options.7
By gaining insights into the hallmarks of cancer resistance, we can not only identify novel targets for therapeutic intervention but also develop strategies to mitigate or overcome resistance.8 This knowledge empowers clinicians to tailor treatment regimens to individual patients, ultimately ushering in the era of personalized medicine in oncology. Furthermore, it informs the development of combination therapies, which may prove more effective by targeting multiple facets of resistance simultaneously.9
In this review article, we will delve into the intricate web of resistance mechanisms that characterize cancer cells' tenacious survival tactics. We will explore the genetic and epigenetic alterations, the dynamic interactions within the tumor microenvironment (TME), the activation of adaptive signaling pathways, the evasion of apoptosis, and the various mechanisms of drug efflux. Additionally, we will discuss the roles of cancer stem cells, ncRNA regulation, and the heterogeneity that underpins resistance.
As we embark on this journey through the hallmarks of cancer resistance, our aim is to provide a comprehensive overview of the current state of knowledge, explore emerging research areas, and shed light on the strategies that hold promise for overcoming the formidable challenge of cancer resistance. The lessons learned and the insights gained from this exploration are essential in our ongoing battle against this relentless adversary, with the ultimate goal of improving the lives of patients with cancer worldwide.
Genetic and epigenetic mechanisms
Mutations and genetic variations
In this study, our focus centered on exploring eight fundamental hallmarks associated with cancer resistance. Genetic and epigenetic mechanisms occupy a central position in shaping the resilience of cancer cells, affording them the capacity to elude treatment strategies.10 Genetic mutations stand as the linchpin of cancer resistance, serving as pivotal drivers of cancer initiation and progression. Introducing genomic instability and diversity within tumor populations gives rise to a heterogeneous landscape.11 Genomic instability is a phenomenon that can cause DNA damage and activate DNA damage response (DDR) signaling pathways.12 It can be induced by various factors such as exposure to radiation, chemicals, and other environmental stressors.12 Genomic instability can lead to genetic alterations such as mutations, chromosomal rearrangements, and copy number variations, which can contribute to the development of cancer (Figure 1).13 In addition to its role in cancer initiation, genomic instability can also play a role in cancer resistance to therapy.13 For example, genomic instability can promote the generation of resistance mutations, which can reduce the efficacy of targeted therapies such as EGFR TKIs.13 Moreover, genomic instability can lead to the activation of bypass signaling pathways such as MET and AXL receptors, which can further support cancer progression.13 Single nucleotide variations, insertions, deletions, and changes in copy number make up this spectrum of genetic mutations. In many cases, these mutations affect genes involved in fundamental cellular functions, such as cell cycle regulation, DNA repair, apoptosis, and intracellular signaling pathways, contributing to therapeutic resistance.14
Figure 1.
Genomic instability in cancer biology and therapy resistance
The figure illustrates the pivotal role of genomic instability in cancer biology and therapy resistance. Genomic instability, triggered by factors such as radiation, chemicals, and environmental stressors, leads to DNA damage and subsequent activation of DNA damage response (DDR) signaling pathways. This genomic turmoil results in various genetic alterations, including mutations, chromosomal rearrangements, and copy number variations. These alterations contribute to the initiation and progression of cancer. Furthermore, genomic instability plays a crucial role in therapy resistance by promoting the generation of resistance mutations and activating bypass signaling pathways.
Among these genetic mutations, certain alterations, referred to as "driver mutations," confer a selective growth advantage to cancer cells, thereby propelling tumorigenesis. Driver mutations are genetic alterations in the DNA of cells that provide a growth advantage to those cells, allowing them to proliferate more rapidly or survive longer than normal cells. Of the utmost importance is their profound influence on the response to therapy.15 For example, driver mutations within the EGFR gene in non-small cell lung cancer or the BRAF V600E mutation in melanoma are well-recognized culprits associated with resistance to targeted therapies.16,17 Multiple mechanisms leading to acquired resistance to BRAF inhibitor therapy have been documented. These mechanisms encompass various alterations, such as the upregulation of tyrosine kinase activity, occurrence of NRAS mutations, amplification, or alternative splicing of mutant BRAF, and mutations in the MEK signaling pathway.18 Furthermore, beyond driver mutations, there exists a category of genes specifically implicated in mediating resistance.19 These genes often play pivotal roles in circumventing the effects of therapeutic targets or activating compensatory pathways.19 For instance, mutations within the ABL kinase domain are frequently the cause of the establishment of resistance to tyrosine kinase inhibitors in chronic myeloid leukemia. These mutations interfere with the drug’s ability to bind and be effective, presenting a significant obstacle to treatment.20 In addition to mutations, genetic variations, including polymorphisms, exert a substantial impact on the response to cancer therapies.21 These variations affect drug metabolism, transport, and target engagement, thereby leading to variations in drug efficacy and toxicity.22 Such as, genes that encode drug-metabolizing enzymes and transporters can significantly affect the body’s ability to absorb, metabolize, and eliminate cancer medications.22
Pharmacogenomics (the study of how drugs work in the body) explores the genetic influence on drug response, particularly in cancer treatment, identifying genetic markers to predict drug response and toxicity, ultimately improving treatment outcomes.23 Studies in this area provide insight into the causes of inter-individual differences in pharmacokinetics and drug metabolism.24 Variations in genes encoding drug-metabolizing enzymes, transporters, and drug targets all contribute to the diversity of responses to cancer therapies.25 Consequently, drug efficacy and toxicity may alter as a result of these differences effects on drug metabolism, transport, and target engagement. Gene polymorphisms, also known as genetic polymorphisms, refer to variations in the DNA sequence of a gene that occur naturally within a population. Gene polymorphisms encoding drug-metabolizing enzymes and transporters can impact the absorption, metabolism, and excretion of anticancer medications. Consequently, changed pharmacokinetics may significantly affect the toxicity and efficacy of drugs.25 Moreover, polymorphisms within genes encoding cytochrome P450 enzymes, the principal actors in drug metabolism, can significantly influence the pharmacokinetics of chemotherapeutic agents.26 These polymorphisms can cause abolished, quantitatively, or qualitatively altered, or enhanced drug metabolism. Ultrarapid metabolism is due to stable duplication, multiduplication, or amplification of active genes.27 The mechanism behind this is that cytochrome P450 enzymes are responsible for the metabolism of many drugs, including chemotherapeutic agents. Polymorphisms within these genes can lead to changes in the activity of these enzymes, which can affect the way drugs are metabolized and eliminated from the body. This can result in altered drug efficacy, toxicity, and side effects.26 CYP2D6 is an enzyme that plays a crucial role in the metabolism of tamoxifen, a drug used in the treatment of breast cancer. Genetic variations within CYP2D6 can lead to altered enzyme activity, which can impact the metabolism of tamoxifen and ultimately affect the treatment outcomes of patients with breast cancer.28 Tamoxifen relies on CYP2D6 to form active metabolites crucial for its effects. Genetic variations in CYP2D6 can diminish these metabolites, impacting drug efficacy. Concurrent use of CYP2D6-inhibiting drugs can further reduce effectiveness, especially in treating hot flashes.29 Genetic variations within CYP2D6, for example, hold the potential to affect the metabolism of tamoxifen in patients with breast cancer, thereby impacting the overall treatment outcomes.30 These variations can lead to differences in drug efficacy and toxicity, thereby impacting the overall treatment outcomes.31
Furthermore, genetic variations within genes encoding drug transporters, such as ABC transporters, may lead to alterations in drug efflux and intracellular drug concentrations.32 The proteins play a crucial role in regulating various physiological functions necessary for maintaining homeostasis in the body and responding to various pharmacological substrates by transporting both endogenous and exogenous substrates across biological membranes in body tissues.26 In consequence, genetic variations can result in multidrug resistance, primarily due to increased drug efflux by the ABC superfamily. Understanding these mechanisms is crucial for tailored treatment strategies. Precision medicine, utilizing genetic data, presents a hopeful approach to combat cancer resistance.
Epigenetic modifications in resistance
Epigenetic regulation represents a critical dimension of gene expression control, orchestrating intricate processes without modifying the underlying DNA sequence. It encompasses a diverse array of mechanisms, including DNA methylation, histone modifications, and the influence of non-coding RNAs, collectively shaping the complex landscape of cancer resistance.33
DNA methylation is a biochemical process in which methyl groups are added to the DNA molecule, typically at cytosine nucleotides, resulting in changes to gene expression without altering the underlying DNA sequence. DNA methylation involves adding methyl groups to cytosine residues, impacting gene expression in cancer cells' responsiveness to therapy. Promoter hypermethylation can silence tumor suppressor genes, aiding resistance. Histone modifications, including acetylation and methylation, regulate gene expression via chromatin structure changes, contributing to drug resistance in cancer cells.34
Non-coding RNAs are RNA molecules that are transcribed from DNA but do not encode proteins. Instead, they have diverse regulatory functions within the cell, including regulating gene expression, chromatin structure, and other cellular processes. Non-coding RNAs such as miRNAs and lncRNAs play pivotal roles in cancer resistance. MiRNAs target resistance-related genes, influencing cancer cell responses to therapy. Long non-coding RNAs act as oncogenes or tumor suppressors, impacting cancer cell sensitivity or resistance.35
Moreover, epigenetic modifications contribute to epithelial-mesenchymal transition (EMT), a process in which cancer cells become more invasive and drug-resistant. Changes in DNA methylation and histone modifications can serve as triggers for EMT, ultimately leading to increased resistance to various therapeutic strategies.36 One of the defining characteristics of cancer resistance is the adaptability of cancer cells to diverse microenvironments and therapeutic interventions. Epigenetic modifications play a pivotal role in this adaptability, facilitating swift changes in gene expression profiles. This epigenetic plasticity equips cancer cells with the capacity to develop resistance to multiple treatments over time, underscoring the formidable challenge in the effective management of cancer.33
Overall, epigenetic modifications are integral in shaping the development of cancer resistance. The intricate interplay between DNA methylation, histone modifications, and non-coding RNAs in driving resistance mechanisms is multifaceted and central to our understanding of cancer resistance. This epigenetic plasticity provides cancer cells with the ability to rapidly adapt to therapeutic interventions, rendering them formidable adversaries in the ongoing battle against cancer.
Tumor microenvironment
Role of immune cells
The TME represents a dynamic and intricate milieu residing within cancer tissues, comprising a myriad of components that profoundly influence cancer resistance. Immune cells in the TME play a pivotal role in shaping the equilibrium between tumor suppression and resistance to cancer therapies.37
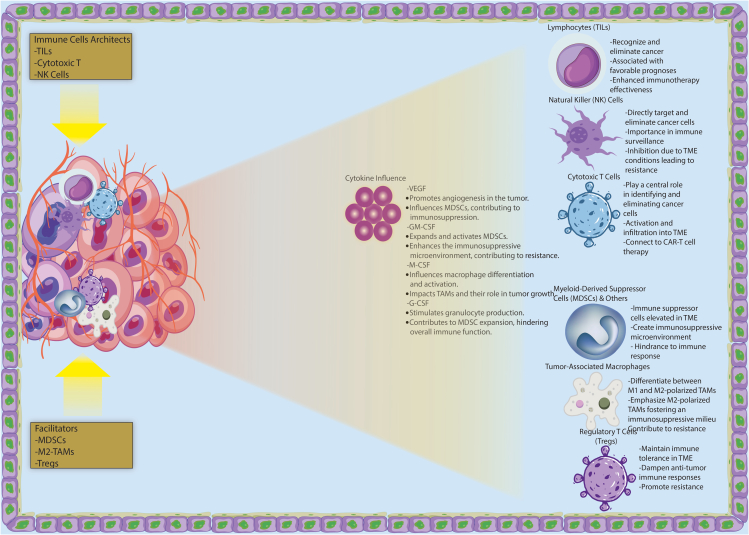
Within the TME, immune cells serve as both architects of restraint and facilitators of cancer growth (Figure 2). This array of immune cell populations includes those capable of mounting potent anti-tumor responses, as well as others with the capacity to exert immunosuppressive effects, ultimately aiding in tumor immune evasion and resistance to treatment.38 Tumor-infiltrating lymphocytes (TILs), primarily composed of T cells, are instrumental in recognizing and eliminating cancer cells.39 The presence of TILs within the TME is often associated with favorable prognoses in specific types of cancer.40 Their presence within the TME is frequently associated with favorable prognoses in specific cancer types and significantly enhances the effectiveness of immunotherapies.41
Figure 2.
Dynamics of the tumor microenvironment (TME) in cancer tissues
The figure illustrates the intricate dynamics of the tumor microenvironment (TME) within cancer tissues. The TME is divided into two main sections: immune cells serving as architects of restraint, such as Tumor-Infiltrating Lymphocytes (TILs), cytotoxic T cells, and Natural Killer (NK) cells, and immune cells facilitating cancer growth, including Myeloid-Derived Suppressor Cells (MDSCs), M2-polarized Tumor-Associated Macrophages (TAMs), and Regulatory T Cells (Tregs). TILs, primarily composed of T cells, are associated with favorable prognoses and enhanced immunotherapy effectiveness. NK cells, with their ability to directly target cancer cells, play a crucial role in immune surveillance, but their activity is inhibited in the hypoxic and acidic TME, leading to resistance. Cytotoxic T cells are central in identifying and eliminating cancer cells, connecting to CAR-T cell therapy. MDSCs, elevated in the TME, create an immunosuppressive microenvironment promoting resistance. M2-polarized TAMs foster an immunosuppressive milieu, contributing to resistance. Tregs maintain immune tolerance, dampening anti-tumor responses and promoting resistance. The figure also depicts an interaction network between immune cells and highlights the influence of cytokines such as VEGF, GM-CSF, M-CSF, and G-CSF on immune cell behavior in the TME.
Natural killer (NK) cells, as innate immune effectors, possess the unique ability to directly target and eliminate cancer cells.42 Their presence within the TME is of paramount importance for immune surveillance against cancer. However, the inhibition of NK cell activity within the TME can give rise to resistance.43 Therefore, tumor-derived factors and TME conditions suppress immune response, inhibiting NK cell activity, which fosters treatment resistance due to the immunosuppressive effects.43
In adaptive immune responses against cancer, cytotoxic T cells play a central role in identifying and eliminating cancer cells.44 Their activation and subsequent infiltration into the TME are indispensable for the orchestration of effective anti-tumor immunity.44 Immunotherapeutic modalities, such as CAR-T cell therapy, harness the potent cytotoxic abilities of these cells to combat resistance.45 The therapies have demonstrated impressive effectiveness in patients with chemotherapy-resistant blood cancers, although a substantial number still resist or relapse.45 This highlights the importance of further research into improving the effectiveness of these therapies.
Myeloid-derived suppressor cells (MDSCs), immune suppressor cells frequently elevated within the TME of various cancers, are adept at creating an immunosuppressive microenvironment that bolsters resistance to immunotherapies and cytotoxic agents.46 MDSCs are a significant hindrance to the immune response in the TME, due to their powerful immunosuppressive capabilities. They hinder the function of dendritic cells, NK cells, and T cells; promote the growth of regulatory T cells and tumor-associated macrophages; make it easier for the immune system to elude detection; and eventually aid in the spread and metastasis of tumors.47 It is well known that MDSCs play an important role in the development of tumor immunity. It has been demonstrated that cytokines released by tumor cells and activated immune cells in the TME promote MDSC recruitment, activation, expansion, and suppression in tumor progression. Based on their impact on MDSCs, these cytokines can be divided into two groups. MDSC expansion is mediated by vascular endothelial growth factor (VEGF), granulocyte-macrophage colony-stimulating factor (GM-CSF), macrophage colony stimulating factor (M-CSF), and granulocyte colony-stimulating factor (G-CSF).48 A study examines the limitations of immunotherapy for colorectal cancer, along with the mechanisms that contribute to drug resistance and other potential treatment options.49
Depending on how they are polarized, tumor-associated macrophages (TAMs) can either promote or inhibit tumor growth in the TME. M2-polarized TAMs, in particular, contribute significantly to resistance by fostering an immunosuppressive milieu. In addition to being anti-inflammatory immune cells, TAMs are usually associated with a poor prognosis and malignant disease progression.50 By fostering an immunosuppressive milieu, M2-polarized TAMs are particularly important for resistance. T cells, NK cells, and dendritic cells are responsible for mediated immune responses; M2-like TAMs assist in immune escape, generate regulatory T cells and tumor-associated macrophages, and ultimately result in tumor progression and metastasis. Therefore, M2-like TAMs are attractive targets for cancer immunotherapy.51
Regulatory T cells (Tregs) are a subset of T lymphocytes that play a crucial role in regulating the immune system. Tregs are involved in maintaining immune tolerance and preventing autoimmune diseases by suppressing excessive immune responses against self-antigens. They help prevent the immune system from attacking the body’s own tissues, thus maintaining immune homeostasis. Tregs in the TME maintain immune tolerance, dampen anti-tumor responses, and promote resistance to immunotherapies. Clinically, Tregs are targeted, alone or with other immunotherapies, to counter the immunosuppressive TME. Additionally, Tregs limit anti-tumor immune responses by interacting with peritumor cells, stroma, vasculature, and lymphatics.52 Tumor tissue with high levels of Tregs and circulating MDSCs exhibit poor responses to ICIs (Immune Checkpoint Inhibitors) treatment, suggesting these immunosuppressive cells may contribute both to ICIs resistance and can serve as indicators of treatment response.53
Immunotherapies have heralded a transformative era in cancer treatment, capitalizing on the immune system’s inherent capabilities to combat cancer.54 Strategies such as ICIs,55,56 CAR-T cell therapy,57 and oncolytic viruses,58 have been meticulously designed to target and modulate immune cells within the TME. An intricate understanding of the multifaceted interactions among immune cells in the TME is paramount to the success of these therapies.
Stroma and tumor-associated factors
The TME is a multifaceted and dynamic environment within cancer tissues, comprising various components, including stromal cells and tumor-associated factors. These elements play a central role in the development of cancer resistance, and comprehending their interactions is crucial for effective cancer treatment.59 Stromal cells, encompassing fibroblasts and endothelial cells, represent an integral part of the TME. Beyond providing structural support, they exert significant influence on cancer progression and resistance.60 Stromal cells exhibit a dual role, capable of either facilitating tumor growth or inhibiting cancer cells, contributing to the intricate nature of the TME.61 The dual interaction between stromal cells and immune cells within the TME can both have positive and negative effects on tumor development.62 For example, a recent study delves into the dual role of stromal cells within the TME, shedding light on their complex mechanisms in tumorigenesis, development, and treatment resistance.62 Understanding these interactions is essential for developing precise therapeutic approaches.60
Mesenchymal stem cells (MSCs) are multipotent cells capable of differentiating into various cell types, including bone, cartilage, and fat cells. Found in tissues such as bone marrow and adipose tissue, MSCs hold immense promise for regenerative medicine due to their ability to promote tissue repair, modulate immune responses, and reduce inflammation. Their capacity to migrate to sites of injury or inflammation further enhances their therapeutic potential. Easily isolated and expanded in culture, MSCs offer a versatile platform for developing innovative treatments targeting a range of conditions, from autoimmune disorders to tissue damage, making them a focal point in biomedical research and clinical applications. In cancer development, MSCs have been found to be associated with the growth of tumor cells as well as the angiogenesis and metastasis of cancer cells. Furthermore, recent research indicates that MSCs also play a role in therapeutic resistance.63 MSCs, obtained from various human tissues, are crucial in cancer therapy resistance due to their differentiation potential. Their interaction with tumor cells regulates behaviors such as proliferation and metastasis.64 MSCs secrete exosomes, influencing the tumor microenvironment and cancer resistance to various therapies. MSC-derived exosomes are implicated in multiple myeloma PI resistance, carrying specific upregulated markers.
Cancer-associated fibroblasts (CAFs) actively participate in the remodeling of the extracellular matrix and secrete growth factors and cytokines that enhance cancer cell survival and invasiveness.65 The presence of CAFs is often linked with a poor prognosis, as they substantially contribute to resistance against various anticancer therapies.66 Tumor cells interact with immune cells within the TME by secreting cytokines, growth factors, chemokines, exosomes, and other effector chemicals. By forming an immunosuppressive TME, cancer cells are able to evade immune monitoring.37 The presence of CAFs is often linked with a poor prognosis. They substantially contribute to resistance against various anticancer therapies.66 Colon cancer risk assessment may be aided by the identification of stroma-specific signatures. Based on the Consensus Molecular Subtype classification (CMS), the mesenchymal or CMS4 group of colon cancer is characterized by stromal invasion and remodeling of the extracellular matrix. Poor prognosis rates are associated with this group.67
Endothelial cells in the TME play a central role in angiogenesis that supply oxygen and nutrients to tumors.68 While angiogenesis is crucial for tumor survival, it may also contribute to treatment resistance by ensuring that cancer cells have access to vital resources.69 Tumor endothelial cells (TECs) not only induce angiogenesis in the TME but are important immune regulatory mediators. They play a vital role in regulating the migration of peripheral immune cells into the tumor compartment by acting as gatekeepers of cellular transmigration. In order to prepare, activate, and multiply T cells, TECs also serve as antigen-presenting cells.70
Tumor-associated factors encompass a wide array of molecules, including cytokines, growth factors, and chemokines. These factors, produced by both cancer cells and stromal components, establish a microenvironment that can promote drug resistance by influencing cancer cell behavior.66 CAFs can promote tumor growth, angiogenesis, invasion, metastasis, and even chemoresistance through multiple pathways.59 Various molecules, including cytokines, growth factors, chemokines, exosomes, and other effector molecules, are secreted by tumor cells within the TME to interact with immune cells. By interacting with the TME, cancer cells are able to evade immune surveillance as a result.71 TAMs are a key component of the tumor microenvironment that contributes to the immune escape of cancer cells as well as the failure of therapy. TAMs are associated with poor prognosis as well as drug resistance, including immunotherapy.72
Cytokines and growth factors, such as interleukin, tumor necrosis factor, epidermal growth factor (EGF), and transforming growth factor β (TGF-β), play essential roles in the TME.73 They can impact cancer cell proliferation, survival, and their responsiveness to treatment. These factors, produced by both cancer cells and stromal components, establish a microenvironment that can promote drug resistance by influencing cancer cell behavior.74 Understanding the roles of these cytokines and growth factors in the TME could provide valuable insights for developing more effective cancer treatments.75
Chemokines, responsible for the recruitment and regulation of immune cells, hold significant importance in the TME.76 Specific chemokines can attract immune cells to the tumor site, either by promoting an anti-tumor immune response or by creating an immunosuppressive environment,76 that favors resistance to therapeutic agents, including immunotherapy.71 By secreting chemokines, TAMs can contribute to tumor progression. They are involved in differentiation, polarization, and infiltration into tumors, as well as developing suppressive functions.76
The extracellular matrix (ECM) provides structural support to tissues and is frequently remodeled by cancer cells and stromal components in the TME.77 This impacts the behavior of cancer cells and may promote resistance. When the ECM density increases, a physical barrier can prevent therapeutic agents from penetrating.78 The increased density of ECM can create physical obstacles that hinder the penetration of therapeutic compounds. The ECM not only serves as a shelter for cancer and stroma cells but also stores growth factors and cytokines. In addition, ECM promotes tumor growth and metastasis by interacting with neighboring cells and triggering various cellular signaling pathways.79
A common feature of the TME is hypoxia, characterized by low oxygen levels. Hypoxia in the TME prompts metabolic changes in cancer cells, fostering resistance to chemotherapy. Adipocyte metabolism shifts under hypoxia, benefiting mammary epithelial and cancer cells. Hypoxia alters gene expression, contributing to drug resistance via a hypoxia-regulated gene network.80 Moreover, in a recent investigation, it was observed that hypoxia significantly influences cancer therapy through various mechanisms.81 These mechanisms include altering gene expression, activating oncogenes, deactivating suppressor genes, diminishing genomic stability, and facilitating clonal selection. Furthermore, hypoxia induces the secretion of cytokines and growth factors, thereby promoting angiogenesis and supporting tumor growth. Central to the hypoxic response is the pivotal role played by Hypoxia-Inducible Factor (HIF), which critically contributes to the development of hypoxia-induced resistance. HIF effectively regulates the expression of a diverse array of genes involved in angiogenesis, metabolism, cell survival, and apoptosis.81
Targeting hypoxia emerges as a promising strategy to surmount hypoxia-associated resistance in cancer treatment.82 One avenue of exploration involves the utilization of Hypoxia-Activated Prodrugs (HAPs). Specifically designed, these prodrugs selectively release cytotoxic agents in the hypoxic regions of tumors, effectively addressing resistance linked to hypoxia. Another approach focuses on targeting the Hypoxia-Inducible Factor (HIF) pathway, which holds a pivotal role in hypoxia-mediated resistance. Inhibitors designed to modulate HIF present a potential therapeutic strategy to disrupt the mechanisms contributing to resistance in cancer treatment.83
ICIs represent a distinct facet of therapeutic strategies. These monoclonal antibodies function by blocking inhibitory immune checkpoints, thereby enhancing the immune response against cancer cells. Recent studies indicate that hypoxia-targeted therapy has the potential to augment the efficacy of ICIs, offering a multifaceted approach to overcome resistance.82
Radiation therapy is also implicated in combating hypoxia-mediated resistance. By inducing hypoxia in tumors, radiation therapy promotes the release of tumor antigens, thereby stimulating the immune response against cancer cells. This multifaceted strategy addresses both the physical and immunological aspects of resistance.83 Additionally, gene therapy presents an innovative approach to intervene in hypoxia-mediated resistance. This involves the delivery of genes capable of regulating the expression of key genes involved in the mechanisms of resistance. By directly influencing the genetic underpinnings, gene therapy offers a targeted means to disrupt the pathways contributing to hypoxia-associated resistance in cancer treatment.83
Therapeutic strategies targeting TME have emerged as a promising avenue. Understanding the role of stromal cells and tumor-associated factors in cancer resistance has led to the development of therapies aimed at disrupting these processes. These strategies include targeted therapies that inhibit specific cytokines, growth factors, or chemokines, as well as approaches to normalize the ECM and combat hypoxia, all actively under investigation.84
Adaptive signaling pathways
Receptor tyrosine kinase signaling pathways
Cancer resistance is a multifaceted challenge, with adaptive signaling pathways at its core, enabling cancer cells to persist and proliferate despite therapeutic interventions. These pathways function as a symphony of resilience, allowing cancer cells to swiftly reconfigure their intracellular signaling networks in response to the stress induced by treatment. Among these pathways, receptor tyrosine kinase (RTK) signaling pathways play a pivotal role in the development of drug resistance.85 Cellular processes such as differentiation, growth, and survival are controlled by RTKs, which are receptors on the cell surface.86 However, in the context of cancer resistance, they assume a different role.87 Cancer cells often upregulate or abnormally activate RTKs, resulting in sustained pro-survival signaling and resistance to therapy. A profound understanding of the intricacies of these RTK signaling pathways is essential in the fight against resistance.86
One illustrative instance of RTK-driven resistance is the Epidermal Growth Factor Receptor (EGFR) pathway. EGFR is frequently overexpressed or mutated in various cancer types. These genetic alterations lead to uncontrolled downstream signaling, fostering cell survival and diminishing the effectiveness of therapies such as tyrosine kinase inhibitors (TKIs). While EGFR-targeted therapies have enhanced patient outcomes, the challenge of drug resistance remains substantial. For instance, the use of monoclonal antibodies and TKIs to target EGFR has dramatically improved disease prognoses. The majority of patients develop drug resistance after treatment.88
In breast cancer, the Human Epidermal Growth Factor Receptor 2 (HER2) pathway takes the spotlight. HER2 amplification or overexpression is linked with aggressive cancer behavior and resistance to targeted therapies.89 HER2 (Human Epidermal growth factor Receptor 2) plays an important role in several cellular processes important for cancer, including cell growth, differentiation, movement, and invasion. Around 25% of initial invasive breast tumors are characterized by HER2 overexpression or amplification. HER2 overexpression/amplification has been linked to more aggressive cancer behavior and poorer patient outcomes.90 Despite therapies such as trastuzumab significantly improving patient outcomes,91 the hurdle of drug resistance persists.92 Resistance can emerge through various mechanisms, such as the existence of clones with differing traits within the tumor. This diversity leads to the tumor’s ability to adapt in response to treatment exposure.93
Insulin-like Growth Factor 1 Receptor (IGF-1R) signaling represents another facet of cancer resistance, particularly in the context of resistance to endocrine therapy in breast cancer.94 In the last twenty years, the development of breast cancer and other solid tumors as well as several pathophysiological processes have been linked to the insulin-like growth factor-1 (IGF-1). The IGF-1 receptor is an important target for therapy because it is overexpressed and hyperphosphorylated in various subtypes of breast cancer, and it plays a vital role in the proliferation and spread of tumor cells. Therefore, blocking IGF-1R may conflict with other crucial treatment approaches such as mTOR inhibitors and anti-HER2 medications. Currently, a number of clinical trials are being carried out to ascertain how IGF-1R inhibition modifies target therapy resistance pathways.94 The landscape of resistance is made more complex by the interactions between the IGF-1R pathway and other signaling pathways. For instance, fibroblast migration requires IGF-1R expression and activation. Through β1 integrin and the scaffolding protein RACK1, this mechanism integrates signals from the extracellular matrix (ECM).95 αvβ3 integrin collaboration is necessary for IGF-1-stimulated cell migration and division in vascular smooth muscle cells.96 In prostate cancer cells, α5β1 integrin signaling interacts strongly with IGF-1R signaling, and it has been demonstrated that IGF-1R both binds to and stabilises β1 integrin.97
To effectively address these adaptive signaling pathways, combination therapies targeting multiple nodes within the network have emerged as a promising strategy.98 By simultaneously targeting multiple elements, the likelihood of resistance diminishes.1 For example, combining RTK inhibitors with downstream pathway inhibitors may enhance overall treatment effectiveness.99 It is crucial to acknowledge that cancer resistance does not exclusively arise from aberrations in the RTK signaling pathway. Multiple molecular pathways and mechanisms contribute to the development of resistance in cancer.84 It involves a multifaceted interplay of different mechanisms, including genetic mutations,1 epigenetic modifications,33 and the tumor microenvironment.84 For instance, mutations in downstream signaling components such as Ras or Raf can lead to the constitutive activation of the pathway, resulting in uncontrolled cell proliferation and resistance to apoptosis.86 In addition to genetic mutations, epigenetic modifications further modulate RTK signaling and contribute to cancer resistance. Epigenetic changes, such as DNA methylation or histone modifications, can impact the expression of genes within the RTK pathway. Aberrant epigenetic modifications may lead to the silencing of tumor suppressor genes or the activation of oncogenes, enhancing the survival and resistance capabilities of cancer cells.100 The intricate relationship between RTK signaling and these epigenetic modifications underscores the complexity of cancer resistance mechanisms.10 Adaptive signaling pathways interact with these mechanisms, creating a complex network of drug resistance that challenges treatment efforts. However, recent trials targeting RTKs signaling to overcome cancer resistance have shown promising results (Figure 3).
Figure 3.
Clinical trials targeting RTKs signaling to overcome cancer resistance
This figure illustrates about the clinical trials for targeting RTKs signaling to overcome cancer resistance.
PI3K/AKT/mTOR pathway
This section delves into the intricate domain of adaptive signaling pathways, with a specific emphasis on the PI3K/AKT/mTOR pathway, uncovering its pivotal role in the realm of cancer resistance.101 These adaptive signaling pathways can be likened to a symphony, orchestrating the strategies that cancer cells employ to survive and adapt rapidly to a diverse array of treatment approaches.102 Within this complex web of signaling pathways, the PI3K/AKT/mTOR pathway emerges as a central player in the resistance against therapeutic interventions.103 The PI3K/AKT/mTOR pathway, a well-established signaling cascade, regulates fundamental cellular processes including growth, proliferation, and survival under normal circumstances.101 However, in the context of cancer, deviations from the norm in this pathway often result in uncontrolled signaling, leading to resistance against therapeutic measures.101
Phosphoinositide 3-kinase (PI3K), a critical component of this pathway, is frequently mutated or hyperactivated in various cancer types. Such alterations lead to uncontrolled signaling, promoting cell survival and resistance to treatments.104 PI3K inhibitors have shown promise in preclinical studies.105 There are different types of inhibitors that can be classified into three categories: dual PI3K/mTOR inhibitors, pan-PI3K inhibitors, and isoform-specific inhibitors. Among these inhibitors, idelalisib is a PI3K delta-specific inhibitor that has been approved by the United States Food and Drug Administration (FDA) for cancer treatment and has been proven to be effective.106 While PI3K inhibitors have shown promise in preclinical studies, the development of drug resistance remains a formidable challenge.106
The serine/threonine kinase AKT, a downstream effector of PI3K, occupies a central role in this pathway. Activation of AKT promotes cell survival and proliferation. The overexpression or hyperactivation of AKT is commonly observed in cancer cells, contributing to drug resistance. Strategies to target AKT have emerged as potential approaches to counteract this resistance.107 Numerous AKT inhibitors have been produced since the first AKT inhibitor was discovered in 2000, and many of them are presently being assessed in early- or late-stage clinical trials. Of these, phase III clinical trials for cancer therapy are presently being conducted on two promising inhibitors, capivasertib and ipatasertib. These inhibitors make use of liquid biopsy and genomic or molecular profiling to achieve personalized cancer treatment.107
The macrophage target of rapamycin (mTOR) regulates protein synthesis and cell division.101 Dysregulation of mTOR signaling is a common occurrence in cancer and often correlates with resistance to both targeted therapies and conventional chemotherapy.108 These inhibitors participate in several signaling pathways to control apoptosis, autophagy, and cell proliferation.109 Newly discovered mTOR inhibitors have entered clinical trials in recent years, and drugs that combine with mTOR inhibitors have been shown to be highly effective.109 However, the development of drug resistance remains a formidable challenge.108 While mTOR inhibitors have demonstrated efficacy, their application is restricted by the emergence of resistance.
Given the intricate nature of adaptive signaling pathways and the propensity of cancer cells to evade single-agent therapies, combination treatments targeting multiple nodes within the PI3K/AKT/mTOR pathway have garnered considerable attention. Combining PI3K inhibitors, AKT inhibitors, and mTOR inhibitors can amplify the effectiveness of treatments and counteract resistance.110 However, it is imperative to acknowledge that cancer resistance is not solely confined to the PI3K/AKT/mTOR pathway. It is a multifaceted challenge involving genetic mutations, epigenetic modifications, and the intricacies of TME. For instance, mutations in the PIK3CA gene, which encodes the catalytic subunit of PI3K, lead to the sustained activation of the pathway, fostering cell survival and resistance to apoptosis. Likewise, mutations in PTEN, a negative regulator of the pathway, result in the hyperactivation of PI3K/AKT/mTOR signaling, contributing to resistance against targeted therapies.111 Epigenetic modifications also play a role in shaping PI3K/AKT/mTOR signaling and cancer resistance. Alterations such as DNA methylation or histone modifications influence the expression of genes within the pathway. Aberrant epigenetic changes may silence tumor suppressors or activate oncogenes in the PI3K/AKT/mTOR pathway, promoting cancer cell survival and resistance to treatment.112 Furthermore, the TME is a dynamic and influential participant in PI3K/AKT/mTOR-driven resistance. Inflammatory signals, growth factors, and hypoxia present in the TME can activate this pathway, creating a supportive environment that enhances cancer cell survival and resistance to therapies. The intricate crosstalk between cancer cells and the TME contributes to the sustained activation of PI3K/AKT/mTOR signaling.113 Adaptive signaling pathways interconnect with these aspects, giving rise to a complex network of resistance mechanisms.109
Wnt/β-catenin signaling
The Wnt/β-catenin pathway drives cancer resistance, orchestrating adaptive strategies for cells to endure and adapt to treatments. The Wnt/β-catenin pathway is a well-established signaling cascade that governs fundamental cellular processes. However, in the context of cancer, this pathway can be hijacked and exploited by cancer cells to promote their survival and resistance to treatment.114
This pathway contains secreted glycoproteins that initiate signaling by binding to cell surface receptors. Dysregulation of Wnt ligands and receptors is a common observation in various cancer types, leading to the persistent activation of the pathway. β-catenin stabilizes and moves to the nucleus in response to this activation, where it functions as a transcriptional co-activator.115 In the nucleus, β-catenin activates gene transcription by collaborating with the transcription factor T cell factor/lymphoid enhancing factor (TCF/LEF).116 These genes are involved in processes such as EMT, a phenomenon that enhances the invasive and drug-resistant properties of cancer cells.117
The Wnt/β-catenin pathway plays a pivotal role in the emergence of treatment resistance, encompassing targeted therapy and chemotherapy.118 Its activation is often linked to poor treatment outcomes and an elevated risk of disease recurrence.114 Tackling drug resistance driven by the Wnt/β-catenin pathway is a formidable challenge. Targeted therapies designed to disrupt this pathway have shown promise in early preclinical and clinical studies (Figure 4). However, the emergence of drug resistance to these targeted treatments poses a major obstacle.114 To effectively address this challenge, researchers are exploring combination therapies that concurrently target the Wnt/β-catenin pathway and other drug resistance mechanisms. Employing a multifaceted approach, researchers aim to enhance treatment effectiveness and prevent or overcome drug resistance.119 It is crucial to acknowledge that cancer resistance is not solely shaped by the Wnt/β-catenin pathway. For example, Genetic mutations within critical components of the Wnt/β-catenin pathway play a significant role in driving resistance. Mutations in the APC gene, a negative regulator of Wnt signaling, can result in the constitutive activation of the pathway, promoting uncontrolled cell growth and resistance to apoptosis. Similarly, mutations in β-catenin itself can lead to its stabilization and nuclear translocation, enhancing Wnt signaling and contributing to the development of cancer resistance.120 Epigenetic modifications also contribute to the modulation of the Wnt/β-catenin pathway. DNA methylation and histone modifications influence the expression of genes within this pathway. Aberrant epigenetic changes may silence negative regulators or activate components of the Wnt pathway, fostering cancer cell survival and resistance to therapeutic interventions.121 The TME plays a crucial role in influencing Wnt/β-catenin signaling and contributing to resistance. Inflammatory signals, growth factors, and interactions with stromal cells within the TME can activate or enhance Wnt signaling, creating a supportive microenvironment for cancer cells. The dynamic crosstalk between cancer cells and TME components, including immune cells and fibroblasts, establishes an environment that sustains Wnt/β-catenin activation, thereby promoting resistance to treatment.122 The interplay between different resistance mechanisms, including genetic mutations, epigenetic modifications, and the tumor microenvironment, adds layers of complexity to the drug resistance puzzle.9
Figure 4.
Clinical trial targeting the Wnt pathway to combat cancer resistance
This figure illustrates the clinical trial targeting the Wnt pathway to combat cancer resistance.
Monophosphate protein kinase pathway
The adenosine monophosphate protein kinase (MAPK) pathway is a ubiquitous signal transduction pathway that regulates all aspects of life and is frequently altered in disease, including cancer.123 The MAPK pathway plays a crucial role in modulating drug sensitivity and resistance in cancer.123 The extracellular signaling-regulated kinase (ERK) pathway is a main mechanism of resistance to RAF or MEK inhibitors.123 However, resistance mechanisms to combined RAF and MEK inhibition increasingly include the activation of alternative pathways that can drive cancer cell proliferation and survival.123 Recent studies have identified that the overexpression of antiapoptotic genes,124 stimulation of autophagy,125 AMPK activation, alterations in the tumor microenvironment,126 and changes in metabolic flux,127 also promote resistance and treatment failure. For example, overexpression of antiapoptotic genes such as Bcl-2 and Bcl-xL has been shown to promote resistance to chemotherapy and radiation therapy in cancer cells.124 Additionally, the stimulation of autophagy and activation of AMPK have been linked to resistance against these treatments in cancer cells.128 Alterations in the tumor microenvironment, including conditions such as hypoxia and acidosis, have also been implicated in promoting resistance to chemotherapy and radiation therapy.126 Furthermore, changes in metabolic flux, exemplified by the Warburg effect, have been identified as contributors to the development of resistance mechanisms in cancer cells.127
The MAPK pathway is a cascade of three kinases, where the most upstream kinase (MAPKKK) responds to various extra- and intracellular signals and activates the middle kinase (MAPKK) by direct phosphorylation. MAPKKs exclusively phosphorylate and activate a MAPK, which typically has many substrates that execute specific cell fate decisions adequate to the input signal.123
Janus kinase/signal transducer and activator of transcription pathway
The JAK/STAT (Janus kinase/signal transducer and activator of transcription) pathway, a critical signaling cascade, plays a pivotal role in regulating various physiological processes, including immune response, cell proliferation, and differentiation. In cancer, the JAK/STAT pathway’s dysregulation has emerged as a significant factor influencing tumor behavior. This pathway is initiated by extracellular signals, leading to the activation of Janus kinases (JAKs), phosphorylation of STAT (signal transducer and activator of transcription) proteins, and subsequent transcriptional regulation of target genes involved in crucial cellular processes.129
In cancer, the dysregulation of the JAK/STAT pathway contributes to both tumor promotion and suppression, making its role in cancer biology complex.130 Aberrant activation of the pathway is often observed in various cancers, resulting in the constitutive activation of downstream targets that promote cell survival, proliferation, and angiogenesis, fostering tumor growth. Genetic alterations, such as mutations or amplifications in JAK and STAT genes, can lead to persistent pathway activation, facilitating tumorigenesis.131
Conversely, the JAK/STAT pathway has tumor-suppressive functions, particularly in promoting an anti-tumor immune response. Activation of JAK/STAT signaling enhances the expression of cytokines and immune checkpoint molecules, contributing to the immune system’s ability to target and eliminate cancerous cells.132 Additionally, in certain contexts, JAK/STAT signaling induces the expression of pro-apoptotic genes, supporting the elimination of damaged or cancerous cells.133
In cancer, dysregulated signaling through the JAK/STAT pathway can significantly contribute to the development of resistance against chemotherapy.134 The JAK/STAT pathway plays a crucial role in mediating cellular responses to various external stimuli, including cytokines and growth factors.135 Dysregulation of this pathway can occur through multiple mechanisms, such as genetic mutations, abnormal activation of upstream signaling molecules, or alterations in the expression levels of pathway components.136
When the JAK/STAT pathway becomes aberrantly activated in cancer cells, it leads to the promotion of cell survival and the inhibition of apoptosis. This sustained activation can confer resistance to the effects of chemotherapeutic agents, which typically aim to induce cell death in cancer cells.137 The elevated levels of JAK/STAT signaling provide a survival advantage to cancer cells, enabling them to evade the cytotoxic effects of chemotherapy drugs and continue proliferating.
Furthermore, dysregulated JAK/STAT signaling impacts cancer cell survival and influences PD-L1 expression, enabling cancer cells to evade immune surveillance. This upregulation allows cancer cells to inhibit anti-tumor immune responses by engaging immune cells through PD-L1/PD-1 interaction, suppressing immune recognition and attack.138 This immune evasion strategy allows cancer cells to evade the immune system and develop resistance to immune-mediated destruction.
Efforts to harness the therapeutic potential of the JAK/STAT pathway in cancer treatment involve targeted therapies against JAK or STAT proteins. However, resistance to these targeted therapies can emerge through mechanisms such as secondary mutations or the activation of alternative survival pathways. Understanding the intricate involvement of the JAK/STAT pathway in cancer resistance is essential for developing more effective and tailored therapeutic strategies that can overcome or mitigate these resistance mechanisms.
Evasion of apoptosis
Bcl-2 family proteins
Among the hallmarks of cancer resistance, the evasion of apoptosis stands as a critical facet. In this chapter, we delve deeper into the pivotal role played by Bcl-2 family proteins in orchestrating resistance to apoptosis.139 Apoptosis serves as a fundamental process that upholds tissue homeostasis by removing damaged or surplus cells. In cancer, the evasion of apoptosis is a characteristic that empowers cancer cells to persist and proliferate uncontrollably, thwarting the effects of therapeutic interventions.
The family of Bcl-2 proteins is important in controlling apoptosis. It is made up of pro- and anti-apoptotic components that work in harmony to create a delicate equilibrium.139 Pro-apoptotic members, such as Bax and Bak, play a crucial role in apoptosis by permeabilizing the mitochondrial outer membranes. Without these proteins, apoptosis cannot take place.140 Anti-apoptotic members, such as Bcl-2 and Bcl-xL, counteract this process, inhibiting cell death.141 The past ten years have seen a significant amount of research on the BCL-2 protein family, which includes Bcl-2 and Bcl-xL, because of its vital function in controlling apoptosis, carcinogenesis, and cellular responses to anticancer therapy. This family’s members engage in either pro- or anti-apoptotic actions.139 Another study found that Bcl-2 and Bcl-xL are anti-apoptotic paralogues that decrease apoptosis brought on by a variety of stimuli and may be involved in the development of cancer.142
In cancer, the equilibrium of Bcl-2 family proteins is frequently disrupted, tilting the balance in favor of cell survival (Figure 5). This disruption can arise from various sources, including genetic mutations, gene amplifications, and aberrant protein expression.143 A classic example is chronic lymphocytic leukemia (CLL), where the common overexpression of Bcl-2 inhibits apoptosis, contributing to the survival and resistance of CLL cells.144 Targeting Bcl-2 with specific inhibitors, such as venetoclax, has shown promise in clinical trials across CLL and other malignancies, underscoring the significance of understanding and targeting Bcl-2 family proteins in cancer treatment.145 ABT-737, a promising candidate with structural similarities to Navitoclax, has demonstrated efficacy in preclinical studies. ABT-737 selectively binds to Bcl-2, Bcl-xL, and Bcl-w, disrupting their anti-apoptotic functions. The potential of ABT-737 lies in its ability to overcome resistance associated with single-agent Bcl-2 inhibition, presenting a compelling avenue for therapeutic intervention.146 Navitoclax, another well-known Bcl-2 inhibitor, targets both Bcl-2 and Bcl-xL. This dual inhibition strategy has shown promise in preclinical and clinical studies, offering a more comprehensive approach to tackling anti-apoptotic defenses. Understanding the advancements and challenges associated with Navitoclax is essential for contextualizing its role in current treatment paradigms.147 Furthermore, the pro-apoptotic member BIK deserves attention in this context. Studies have elucidated BIK’s ability to counteract the anti-apoptotic effects of Bcl-2, presenting a potential therapeutic strategy. Exploring the latest developments regarding BIK and its application in Bcl-2-targeted therapies adds depth to our understanding of the intricate regulatory network governing apoptosis.148 Furthermore, the intricate interplay between Bcl-2 family proteins and other apoptosis regulators, such as the p53 pathway, adds layers of complexity to the landscape of cancer resistance. The upregulation of anti-apoptotic Bcl-2 family members might result from the dysregulation of the p53 pathway, which hinders a cell’s capacity to initiate apoptosis in response to treatment-induced stress.139 For example, P53 directly affects the transcriptional activity of PUMA, a pro-apoptotic member of the BCL-2 family that is part of the BH3-only subgroup. When treated with radiation and chemotherapy, PUMA causes mitochondrial apoptosis.149 Furthermore, mitochondrial p53 can bind to and block the apoptotic antagonists BCL-2 and BCL-XL, just like the majority of BH3-only proteins.150
Figure 5.
Interplay of Bcl-2 family proteins and therapeutic intervention in cancer resistance
This illustration depicts the intricate interplay of Bcl-2 family proteins and the disrupted equilibrium in cancer cells leading to apoptosis resistance. The structure of Bcl-2 family proteins is delineated, showcasing pro-apoptotic members, alongside their anti-apoptotic counterparts. Disruptions in equilibrium, emanate from sources such as genetic mutations, gene amplifications, and aberrant protein expression, underscoring the multifaceted nature of cancer resistance. The targeted inhibitors, venetoclax, ABT-737, and Navitoclax, are elucidated with lines pointing to their respective positions. Venetoclax, ABT-737, and Navitoclax act through distinct mechanisms, inhibiting anti-apoptotic proteins or promoting pro-apoptotic activities, ultimately restoring apoptosis in cancer cells. This figure illustrates the critical role of Bcl-2 family proteins, the perturbations in their equilibrium, and the therapeutic interventions aimed at overcoming cancer resistance through targeted inhibition.
Overcoming drug resistance associated with Bcl-2 family proteins remains an active area of research. Combination therapies that concurrently target Bcl-2 alongside other resistance mechanisms, including kinases or immune checkpoints, are currently under investigation, holding promise for improved treatment efficacy.151 Therefore, a profound comprehension of the intricacies of these proteins and their interactions with other signaling pathways is imperative for devising effective therapeutic strategies. Whether by targeting Bcl-2 family proteins alone or in combination with other agents, this approach offers a promising avenue for combatting drug resistance and elevating outcomes for patients with cancer.
Inhibition of death receptors
Cancer resistance is a formidable challenge in the field of oncology, and among its numerous facets, evasion of the apoptosis is of paramount importance. This section investigates the mechanisms of cancer resistance, with a specific focus on the inhibition of death receptors, a strategy employed by cancer cells to evade programmed cell death.152 Apoptosis serves to regulate cell turnover and eliminate damaged or unwanted cells within an organism. In the context of cancer, the evasion of apoptosis is a key feature that empowers cancer cells to survive and proliferate uncontrollably despite therapeutic interventions.153 Death receptors play a critical role in initiating the extrinsic apoptosis pathway. These receptors, including Fas, TRAIL-R1, and TRAIL-R2, are activated by specific ligands, such as Fas ligand and TRAIL.154 Upon binding of these ligands, death receptors initiate a signaling cascade that ultimately culminates in cell death.
In the realm of cancer resistance, the inhibition of death receptors emerges as a pivotal mechanism that cancer cells employ to escape apoptotic cell death. Cancer cells often downregulate death receptors, leading to diminished sensitivity to death ligands. This reduced sensitivity can result from various mechanisms, including alterations in gene expression, post-translational modifications, and activation of survival signaling pathways.152 One way in which cancer cells survive is by silencing death receptors, which helps them avoid apoptosis through death receptor-induced pathways. This is achieved by mutating the death receptors, which still bind with ligands but are unable to participate in the apoptosis signaling pathway due to a lack of downstream processing.1 Furthermore, another study focuses on the two mechanisms by which cytotoxic T lymphocytes and Natural Killer cells can directly eliminate malignant or infected cells: death receptor-mediated cytotoxicity, which uses the death receptor ligands Fas ligand and TRAIL, or granule-mediated cytotoxicity, which uses perforin and granzyme B. However, malignant and infected cells can use both of these cell-mediated killing pathways to avoid cytolytic death.152
A classic illustration of death receptor inhibition in cancer resistance is the downregulation of Fas expression in various cancer types. The reduced expression of Fas compromises the cell’s ability to respond to Fas ligand-induced apoptosis, thereby contributing to the survival of cancer cells.155 Cancer cells can activate survival signaling pathways to prevent receptor-induced apoptosis in addition to downregulating death receptors. These pathways, which include the NF-κB and PI3K/AKT pathways, increase cell survival by preventing death receptors from starting the apoptotic cascade.156 Overcoming cancer resistance associated with death receptor inhibition is a formidable task. Researchers are actively exploring strategies to sensitize cancer cells to receptor-mediated apoptosis.157 This endeavor includes the development of agonistic antibodies or recombinant ligands capable of activating death receptors, thereby circumventing the inhibitory mechanisms employed by cancer cells.158
Drug efflux mechanisms
ATP-binding cassette transporters
Within the realm of drug resistance, the mechanisms of drug clearance, particularly those facilitated by ABC transporters, emerge as pivotal factors influencing disease recovery and cancer persistence. This chapter seeks to provide a deeper exploration of the intricate landscape of drug clearance mechanisms, spotlighting the pivotal role played by ABC transporters in mediating resistance to therapeutic interventions.159 ABC transporters, a diverse family of membrane proteins, play a fundamental role in cellular detoxification by actively expelling a variety of substrates, including chemotherapeutic agents.160 In the context of cancer resistance, these transporters act as gatekeepers, actively extruding drugs from cancer cells, thus diminishing the efficacy of therapeutic treatments.159 The upregulation of ABC transporter expression in cancer cells significantly contributes to their resistance against the cytotoxic effects of therapeutic agents.161 This upregulation can be attributed to genetic alterations, epigenetic modifications, and adaptive responses to prolonged drug exposure.
Prominent members of the ABC transporter family implicated in cancer resistance include ABCB1 (P-glycoprotein), ABCC1 (multidrug resistance-associated protein 1, MRP1), and ABCG2 (breast cancer resistance protein, BCRP), extensively studied in the context of drug resistance.162 P-glycoprotein encoded by the ABCB1 gene actively pumps out various chemotherapeutic agents, thus diminishing their intracellular concentrations and impeding their cytotoxic effects. Similarly, MRP1 and BCRP contribute to drug resistance by eliminating diverse drugs, consequently reducing their effectiveness against cancer cells.
Drug efflux inhibitors are pivotal components in the landscape of cancer treatment, addressing the challenge of MDR that often hampers the efficacy of chemotherapy.159 This resistance mechanism involves ABC transporters, including P-glycoprotein (P-gp), breast cancer resistance protein (BCRP), and multidrug resistance protein (MRP), which actively pump chemotherapeutic agents out of cancer cells.163 One approach to overcoming this resistance is the use of drug efflux inhibitors designed to counteract the activity of these transporters, thereby enhancing the intracellular concentration and effectiveness of chemotherapy drugs.
Several generations of drug efflux inhibitors have been developed, each with its unique characteristics and mechanisms of action. First-generation inhibitors, such as verapamil, cyclosporine A, and quinidine, have demonstrated efficacy in laboratory studies. However, their clinical application has been limited due to issues related to toxicity and drug interactions. These inhibitors, notably verapamil, primarily target P-gp (ABCB1) as an inhibitory agent.164
The second generation of drug efflux inhibitors includes more potent and selective compounds designed to overcome the limitations seen with their predecessors. Tariquidar, elacridar, and zosuquidar fall into this category and have shown promise in preclinical and clinical studies. Tariquidar, for instance, has been investigated in clinical trials to assess its potential in reversing MDR in various cancers, including lung, breast, and ovarian cancers.165
In recent advancements, Ko143 has emerged as a potent and selective inhibitor of ABCG2, which is associated with MDR. Ko143 offers a new avenue for addressing drug efflux mediated by ABCG2 and has shown effectiveness in preclinical studies. Its specific targeting of ABCG2 makes it a promising addition to the armamentarium of drug efflux inhibitors.166
Combining drug efflux inhibitors with standard chemotherapy regimens is a common strategy. This combination approach aims to sensitize cancer cells to the cytotoxic effects of anticancer drugs by preventing their rapid removal from the cells. Clinical trials exploring the efficacy and safety of these combination therapies are ongoing, providing valuable insights into the potential benefits of drug efflux inhibition in diverse cancer types.165
A nuanced understanding of the intricate regulatory mechanisms governing ABC transporter expression and activity is imperative for devising strategies to overcome drug efflux-mediated resistance.167 Inhibition of ABC transporters, whether through small molecule inhibitors or targeted approaches, holds promise as a means to enhance the efficacy of cancer treatments.159 Ongoing exploration of combination therapies that integrate ABC transporter inhibitors with conventional chemotherapeutic agents actively seeks to combat drug resistance mechanisms.159 Furthermore, advancements in nanotechnology, and drug delivery systems, aim to mitigate the impact of ABC transporters by improving the selective delivery of therapeutic agents to cancer cells.168 Moreover, a deep comprehension of the regulatory networks governing ABC transporter expression, coupled with the development of innovative strategies to mitigate their impact, is indispensable for advancing the field of cancer treatment.
Drug resistance protein families
Drug efflux mechanisms, orchestrated by specific protein families, serve as robust defenders exploited by cancer cells to expel therapeutic agents, thwarting their intended cytotoxic effects. A profound comprehension of the intricate network of these resistance protein families is imperative for deciphering the enigma of cancer resistance. Among the prominent families implicated in drug resistance, the ABC transporter family stands out. These membrane proteins actively extrude a diverse array of substrates, including chemotherapeutic agents, from cancer cells. The recurrent upregulation of ABC transporters is a prevalent theme in cancer resistance, significantly diminishing the efficacy of therapeutic interventions. Key players within this family, such as ABCB1, ABCC1, and ABCG2, underscore their pivotal roles in mediating resistance to various drugs.169
Another noteworthy contributor to drug efflux and resistance is the Solute Carrier transporter family. In contrast to ABC transporters that actively expel substrates, SLC transporters are responsible for importing nutrients and ions into cells. Alterations in the expression or function of specific SLC transporters can modulate the intracellular concentration of drugs, thereby influencing their therapeutic efficacy. Understanding the interplay between SLC transporters and drug resistance offers critical insights into the multifaceted nature of cancer resistance.170 Additionally, the Multidrug and Toxin Extrusion family of proteins significantly contributes to drug resistance by actively expelling drugs from cells.171 Their involvement in mediating multidrug resistance underscores the intricate mechanisms employed by cancer cells to evade therapeutic interventions. MATEs are adaptable proteins that carry out a variety of tasks and move different kinds of substrates. Their capacity to bind and efflux free aluminum ions from the environment in order to avoid aluminum toxicity is what makes them most well-known. Additionally, MATEs assist in iron solubilization and translocation to the xylem sap, and they are also involved in transporting secondary metabolites, detoxifying xenobiotics, and participating in biotic interactions and developmental pathways.172
The complexity of drug resistance protein families extends beyond the mentioned groups, encompassing the Resistance-Nodulation-Division family, the Major Facilitator Superfamily (MFS), and the Breast Cancer Resistance Protein (BCRP) family. Each family introduces layers of complexity to the drug resistance landscape, showcasing the adaptability of cancer cells to a spectrum of therapeutic agents. One of the biggest known membrane transporter families is the MFS. The regulation of the majority of currently available and developing molecular cancer treatment drugs' absorption, distribution, metabolism, and excretion (ADME) is one of the numerous vital roles that MFS transporters play.173 Several molecular cancer treatments are absorbed, distributed, metabolised, and excreted in large part by an efflux transporter called BCRP.174
Strategies designed to overcome drug efflux mechanisms and resistance involve the development of targeted therapies. Small molecule inhibitors, antibody-based approaches, and combination therapies, concurrently targeting multiple resistance protein families are actively explored to amplify treatment effectiveness. Innovative technologies, such as nanoparticle-based drug delivery systems, aim to alleviate the impact of drug efflux mechanisms by enhancing the selective delivery of therapeutic agents to cancer cells.175 Therefore, there is a need of specific focus on drug resistance protein families, providing a holistic perspective on the formidable challenge of cancer resistance. The intricate interplay between these families adds intricate layers to the adaptive strategies employed by cancer cells. Ongoing research and innovative therapeutic approaches targeting these protein families offer promising avenues to surmount the challenges posed by efflux-mediated drug resistance in cancer.
Metabolic reprogramming
Warburg effect in resistance
Metabolic reprogramming serves as a distinctive hallmark of cancer cells, enabling them to adapt to the demands of rapid proliferation and survival.176 Among the various metabolic alterations observed in cancer, the Warburg effect emerges as a prominent feature. Coined after Otto Warburg, who initially described it in the 1920s, this phenomenon entails cancer cells promoting glycolysis even in the presence of oxygen, a process conventionally associated with energy production under low oxygen conditions. The Warburg effect contributes to chemoresistance, and is thought to increase the survival and proliferation of cancer cells.177
In cancer resistance, the Warburg effect significantly contributes to the adaptive strategies employed by cancer cells. The Warburg effect, which describes how cancer cells metabolize glucose anaerobically instead of aerobically, even under normoxic conditions, is a contributing factor to chemoresistance. Research on the relationship between glycolysis and chemoresistance, as well as the molecular mechanisms underlying glycolysis-induced chemoresistance, is ongoing.178 Since glycolysis is the principal energy source for cancer cells, even under normoxia, pyruvate is primarily transformed into lactate instead of being absorbed into the tricarboxylic acid cycle.178 Hypoxia and acidosis in the tumor microenvironment are also induced by glycolysis, including PI3K and hypoxia-inducible factor-1 alpha (HIF-1A).178 One of the characteristics of cancer is that the cells need to re-programmed their energy metabolism in order to continue growing and proliferating endlessly.59 Cells use the glycolysis process to break down glucose in order to produce pyruvate and a negligible amount of ATP. Pyruvate can enter the tricarboxylic acid cycle to produce an abundance of energy when cells have enough oxygen. Nevertheless, tumor cells create lactate by activating lactate dehydrogenase (LDH) and blocking pyruvate metabolism in mitochondria, exhibiting high levels of glycolysis independent of oxygen levels.179 Tumor tissue has an acidic microenvironment due to anaerobic glycolysis producing lactate, which is the primary cause of extracellular acidification.180 Besides activating secreted lysosomal enzymes with an optimal pH in the acidic range, an acidic extracellular pH induces the expression of certain pro-metastatic genes by a different intracellular signaling pathway than hypoxia.180
Cancer cells often exhibit an increased reliance on glycolysis for energy production, even in the presence of oxygen. For example, some cancer cells upregulate glycolysis to meet their energy demands. This altered metabolism can lead to resistance against chemotherapeutic drugs that rely on specific cellular pathways.181 For instance, drugs such as cisplatin and doxorubicin target DNA and induce cell death. However, glycolytic cancer cells may evade their toxic effects by altering drug uptake or detoxification pathways.182
Glutamine is another essential nutrient for cancer cells. It fuels the tricarboxylic acid cycle and supports biosynthesis.183 For example, some cancer cells become addicted to glutamine metabolism. Inhibiting glutamine uptake or targeting enzymes involved in glutamine utilization can sensitize these cells to chemotherapy.184 Drugs such as DON (6-diazo-5-oxo-L-norleucine) interfere with glutamine metabolism and enhance drug efficacy.183
Moreover, altered mitochondrial function affects drug sensitivity. Dysfunctional mitochondria can impact cellular redox balance and alter drug activation. Such as, in melanoma, mitochondrial dysfunction contributes to resistance against BRAF inhibitors.185 These inhibitors target the MAPK pathway, but dysfunctional mitochondria reduce drug effectiveness.185
Also, Cancer cells adapt their metabolism in response to drug exposure.186 For example, lymphomas, which rely on oxidative metabolism, may switch to glycolysis upon exposure to certain drugs. This metabolic shift helps them survive and resist treatment.187
Another important aspect is the tumor microenvironment and metabolic crosstalk. The tumor microenvironment influences nutrient availability and metabolic pathways.188 For example, Hypoxic regions within tumors activate HIF-1α, promoting glycolysis.189 These hypoxic cells become resistant to therapies that target aerobic metabolism.190
Targeting the Warburg effect in chemoresistant cancer represents a promising frontier in the quest for more effective therapeutic interventions.191 Understanding the metabolic reprogramming characteristic of the Warburg effect provides a foundation for developing strategies aimed at exploiting the vulnerabilities associated with this phenomenon.191 One approach involves inhibiting key enzymes in the glycolytic pathway.192 Hexokinase, a pivotal enzyme initiating glucose metabolism, can be targeted with inhibitors such as 2-deoxyglucose (2-DG), a glucose analog that competes with glucose for hexokinase binding.192 Additionally, inhibitors of phosphofructokinase (PFK), another critical glycolytic enzyme, disrupt glycolytic progression, thereby sensitizing cancer cells to chemotherapy.191
Metabolic modulation strategies also hold promise in targeting the Warburg effect.193 Compounds inhibiting aerobic glycolysis, such as lonidamine, have demonstrated efficacy in disrupting glycolysis and enhancing the susceptibility of cancer cells to chemotherapy.194 Mitochondrial targeting agents seek to capitalize on the reduced reliance on mitochondrial oxidative phosphorylation in cancer cells exhibiting the Warburg effect.195
Inhibiting hypoxia-inducible factor (HIF), a transcription factor activated under hypoxic conditions associated with the Warburg effect, represents another avenue.196 Compounds such as digoxin and echinomycin have shown potential in inhibiting HIF and sensitizing cancer cells to chemotherapy.196 Combination therapies are gaining traction, where glycolytic inhibitors are paired with standard chemotherapy regimens to synergistically enhance cytotoxic effects.193 This approach aims to exploit both the vulnerabilities associated with the Warburg effect and the specific mechanisms targeted by chemotherapy.
Furthermore, the integration of immunotherapy with glycolytic inhibitors is being explored to potentiate the immune response against cancer cells.197 The altered metabolism in cancer cells can influence the tumor microenvironment, and targeting glycolysis may modulate immunosuppressive conditions, enhancing the effectiveness of immunotherapy.197 Personalized medicine approaches, involving metabolic profiling to identify specific metabolic dependencies, enable the tailoring of treatment strategies based on the unique metabolic characteristics of individual cancers.198
Adaptive therapeutic strategies are also being considered to address the challenge of acquired resistance. Continuous monitoring of metabolic changes and adaptation patterns in response to glycolytic inhibitors can guide adaptive treatment strategies, potentially overcoming resistance mechanisms that cancer cells may develop over time. In summary, the multifaceted approach to targeting the Warburg effect in chemoresistant cancer, encompassing enzymatic inhibition,192 metabolic modulation,197 combination therapies, immunotherapy integration, personalized medicine, and adaptive strategies, holds promise for advancing cancer treatment and improving patient outcomes.198
Comprehending the intricacies of the Warburg effect offers valuable insights into potential therapeutic strategies. Targeting key enzymes in the glycolytic pathway or exploiting metabolic vulnerabilities associated with this alteration represents a promising route for overcoming drug resistance. Integrating traditional cytotoxic agents with compounds disrupting altered metabolic pathways holds potential to enhance treatment efficacy and alleviate challenges posed by drug resistance.
Mitochondrial adaptations
Mitochondria, often referred to as the cellular powerhouse, play a pivotal role in cellular energy production, apoptosis regulation, and various metabolic processes. In cancer resistance, the intricate relationship between cancer cells and mitochondria has become a focal point of investigation.199 It is crucial to comprehend how cancer cells adapt mitochondrial functions to sustain resistance to therapeutic interventions. A significant facet of mitochondrial adaptation in cancer resistance involves the modulation of oxidative phosphorylation (OXPHOS).199 While normal cells predominantly rely on OXPHOS for energy production,200 cancer cells frequently undergo a metabolic shift toward increased glycolysis, known as the Warburg effect.201 This alteration is accompanied by changes in mitochondrial metabolism, as cancer cells strategically manipulate mitochondrial functions to support their aggressive phenotype.202
Mitochondrial dynamics,199 encompassing processes such as fusion and fission,202 also contribute to cancer resistance. Cancer cells may display heightened mitochondrial fission, resulting in the generation of fragmented mitochondria. The pro-fragmentation activity of MYC, for instance, has also been seen in mouse embryonic fibroblasts, where enhanced phosphorylation of DRP1 at S579, a post-translational change known to promote mitochondrial fragmentation, results from the overexpression of c-Myc. Mutants that are phosphomimetic at this site have shown this.202 These altered mitochondrial dynamics can influence cellular processes, including apoptosis and energy production,203 thereby contributing to drug resistance.199
Mitochondrial DNA (mtDNA) mutations have been observed in different types of cancer.204 It is also investigated whether somatic mitochondrial mutations are related to survival in breast cancer in another article.205 There are a variety of diseases associated with mitochondrial dysfunction. Mutations in mtDNA cause mitochondria to function abnormally.206 Another article discusses how mtDNA mutations and heteroplasmy shifting contribute to breast cancer development.205 Drug resistance in cancer is frequently caused by mitochondrial adaptation, and it has been shown that a number of these processes, including mitochondrial dynamics, may be targeted.199 Breast cancer progression is also discussed by another article regarding mitochondrial fission and fusion.207 The role of mitochondrial DNA variation in drug response has been discussed in a systematic review.208 Another article provides a summary of the most recent research on the molecular mechanisms underpinning mitochondrial stress adaptation and its complex relationship to cancer treatment resistance.199
Understanding mitochondrial mediated cancer resistance and targeting mitochondrial adaptation has emerged as a potential avenue to overcome cancer resistance (Figure 6). Ongoing research explores mitochondrial-targeted therapies, including compounds selectively inducing mitochondrial dysfunction in cancer cells. Additionally, combination therapies integrating mitochondria-targeting agents with conventional treatments aim to enhance overall treatment effectiveness.
Figure 6.
Mitochondrial biology and implications in cancer resistance
This figure discusses various aspects of mitochondrial biology and their implications in cancer resistance. Mitochondrial components such as the inner membrane, mtDNA, mitochondrial ribosomes, mitochondrial matrix, endoplasmic reticulum, and cristae play crucial roles in cancer cell adaptation and resistance to therapies. The inner membrane’s modulation of oxidative phosphorylation (OXPHOS) influences cancer cell responses to metabolic-targeted therapies and resistance against oxidative stress-induced cell death. mtDNA mutations impact tumor initiation and chemoresistance, contributing to cancer heterogeneity and altering treatment responses. Mitochondrial ribosomes are essential for protein synthesis crucial to energy metabolism and are linked to drug resistance in various cancers. The mitochondrial matrix is central to energy production, metabolism, redox homeostasis, and apoptotic pathways, shaping cancer cell resistance. The endoplasmic reticulum influences cellular functions, mitochondrial dynamics, apoptosis regulation, and stress resistance in cancer cells. Mitochondrial cristae, integral to maintaining mitochondrial membrane potential and influencing ATP synthesis and apoptosis regulation, may impact cancer cell resistance to apoptotic stimuli and certain therapies. This figure also outlines various strategies for targeting mitochondria in cancer cells to induce apoptosis, disrupt energy metabolism, and overcome resistance. These include mitochondrial uncouplers, electron transport chain inhibitors, ROS-inducing agents, modulation of the mitochondrial permeability transition pore, stimulation of mitophagy, targeting mitochondrial DNA replication, HSP90 inhibitors, peptide-based therapies, and photodynamic therapy. These strategies exploit specific vulnerabilities, offering potential therapeutic avenues, requiring further research to optimize and assess clinical applicability.
Cancer stem cells
CSCs represent a distinct subset within tumors, demonstrating the capacity for self-renewal and the ability to generate diverse cell types within the tumor milieu. Identifying these cells proves challenging due to their heterogeneity and dynamic interactions with the tumor microenvironment. Established markers for CSC identification encompass surface proteins such as CD44, CD133, and ALDH1, supplemented by functional assays evaluating their tumorigenic potential in vivo. CD44 and ALDH1 are used as markers for the assessment of cancer risk in oral potentially malignant disorders (OPMDs) and lymph node metastasis in oral squamous cell carcinoma. CD44 is a membranous marker and ALDH1 is a cytoplasmic marker.209 A subpopulation of CD133+ cancer stem-like cells derived from SK-UT-1 cells has also been identified.210 CSCs share defining traits with normal stem cells, including self-renewal and differentiation capabilities.211 Their remarkable capacity to evade conventional therapies significantly contributes to treatment resistance. CSCs often exhibit heightened DNA repair mechanisms, reduced susceptibility to apoptosis, and a quiescent state shielding them from chemotherapy -induced cytotoxicity.212 Additionally, their plasticity enables seamless transitions between stem-like and differentiated states, facilitating adaptation to evolving microenvironmental cues.213
Resistance orchestrated by CSCs manifests as a complex interplay of intrinsic properties and interactions within the tumor microenvironment, creating a protective niche against therapeutic interventions. Active participation in driving resistance involves mechanisms such as upregulated drug efflux pumps, activation of pro-survival signaling pathways, and manipulation of immune responses.214 For example, one pivotal pathway associated with CSC behavior is the Wnt/β-Catenin pathway.4 Upon activation, this pathway prompts the nuclear translocation of β-Catenin, fostering the transcription of genes that drive cell proliferation and survival.215 CSCs exploit this pathway to perpetuate their self-renewal capabilities and maintain tumorigenic potential.216 The Notch signaling pathway is another key player in CSC dynamics, regulating cell fate determination and stem cell maintenance.217 Aberrant activation of Notch in CSCs enhances self-renewal capacities and confers resistance to apoptotic signals, contributing to tumor progression. Moreover, the Hedgehog pathway, involving components such as Sonic Hedgehog (SHH) and Gli transcription factors, is frequently dysregulated in CSCs.217 Activation of Hedgehog signaling promotes CSC self-renewal and is implicated in the initiation of various cancers and can play a role in mediating resistance to cancer therapy.218 The PI3K/Akt/mTOR pathway is a central node in cellular signaling, and its dysregulation is a common feature in CSCs.219 Activation of this pathway in CSCs supports cell survival, growth, and resistance to apoptosis, ultimately fostering their resilience to conventional cancer therapies.219 Nuclear Factor-kappa B (NF-κB) signaling is associated with inflammation and anti-apoptotic responses, and its activation in CSCs promotes their survival and self-renewal.220 This pathway contributes to the maintenance of CSC populations, enhancing their resistance to therapeutic interventions.220 The Signal Transducer and Activator of Transcription 3 (STAT3) pathway is frequently activated in CSCs, driving survival, self-renewal, and resistance to therapeutic interventions.221 Its dysregulation contributes to the aggressive behavior of CSCs in various cancers.221 Also, TGF-β signaling, while functioning as a tumor suppressor in early stages, can paradoxically support CSC characteristics in later stages of cancer. TGF-β activation contributes to the maintenance of CSCs and their ability to undergo EMT.4 The capability of CSCs to undergo EMT further enhances their migratory and invasive potential, contributing to metastasis and evading therapeutic targeting.222 The MAPK/ERK pathway is often dysregulated in CSCs, influencing cell proliferation, survival, and resistance to apoptosis. Its activation contributes to the aggressive behavior of CSC populations.223
Understanding the dynamic nature of CSCs in cancer resistance is pivotal for formulating effective therapeutic strategies. Targeting CSC-specific vulnerabilities, disrupting their supportive microenvironment, and implementing combination therapies addressing both bulk tumor cells and CSCs emerge as promising avenues.224 Advanced technologies such as single-cell sequencing and functional genomics continually unveil the intricacies of CSC biology, providing insights that may lead to innovative approaches for overcoming cancer resistance.225
Non-coding RNAs regulation
Long non-coding RNAs
Different ncRNAs, including lncRNAs, miRNAs, and circRNAs, play multifaceted roles in the resistance of cancer to drugs. This section explores the intricate landscape of cancer resistance, with a specific emphasis on the regulatory roles played by long noncoding RNAs (lncRNAs) and their significant impact on the development and sustenance of drug resistance mechanisms in cancer cells.226 For instance, It has been reported that lncRNAs can influence gene expression through chromatin remodeling, transcriptional regulation, and posttranscriptional processing.226 lncRNAs, once relegated to the sidelines of genetic research, have emerged as pivotal entities in the complex orchestration of gene regulation. Despite their lack of protein-coding potential, lncRNAs intricately coordinate various cellular processes, and their dysregulation is increasingly implicated in cancer initiation and progression.227 In the realm of cancer resistance, the regulatory influence of lncRNAs takes a critical role (Table 1). These transcripts, characterized by extensive nucleotide sequences, engage in diverse molecular mechanisms that govern gene expression,228 cellular pathways,229 and, notably, the adaptive responses of cancer cells in the face of therapeutic challenges.230 Table 1 provides information on how various types of lncRNAs can mediate resistance to cancer treatment.
Table 1.
Long non-coding RNAs (lncRNAs) in cancer associated drug resistance
| lncRNA | Target | Chemotherapy Drug | Correlation |
|---|---|---|---|
| MALAT1 | HIF-2α-MALAT1-miR-216b axis | 5-Fluorouracil (5-FU) | Chemoresistance in HCC was shown to be mediated by the HIF-2α/MALAT-1/miR-216b axis. |
| HOTAIR | ULK1 | Crizotinib | In NSCLC, the inhibition of HOTAIR expression mitigated resistance to crizotinib by dampening autophagy through the inhibition of the ULK1 pathway. |
| LINC01296 | ATG2B | Paclitaxel | reducing LINC01296 levels has a suppressive effect on non-small-cell lung cancer by interacting with miR-143-3p and ATG2B. |
| H19 | PI3K/Akt pathway | Doxorubicin | H19 overexpression has been linked to activation of the PI3K/Akt pathway, promoting resistance to Doxorubicin in various cancers. |
| ACTA2-AS1 | TSC2 | Cisplatin | long non-coding RNA ACTA2-AS1 counteracts cisplatin resistance in non-small cell lung cancer cells by suppressing autophagy through TSC2 inhibition. |
| ENST00000500843 | N/A | Paclitaxel | Reduced expression of ENST00000500843 contributes to chemoresistance to paclitaxel in lung adenocarcinoma. |
| UCA1 | mTOR pathway | Gemcitabine | Elevated expression of UCA1 is implicated in conferring resistance to oxaliplatin in hepatocellular carcinoma by suppressing miR-138-5p and activating the AKT/mTOR signaling pathway. |
| MA-linc1 | N/A | Paclitaxel | MA-linc1, a novel mitosis-associated long non-coding RNA (lncRNA), facilitates cell cycle progression and enhances cancer cell sensitivity to Paclitaxel. |
| XIST | Epigenetic regulation | Cisplatin | XIST plays a role in cisplatin resistance by modulating gene expression through epigenetic mechanisms in ovarian cancer. |
| DLX6-AS1 | N/A | Cisplatin | DLX6-AS1, activated by H3K4me1, plays a crucial role in promoting secondary cisplatin resistance in lung squamous cell carcinoma by modulating the miR-181a-5p/miR-382-5p/CELF1 axis. |
| GAS5 | Glucocorticoid receptor pathway | Dexamethasone | GAS5 acts as a tumor suppressor and enhances sensitivity to Dexamethasone in multiple myeloma. |
| NEAT1 | miR-770-5p/PARP1 Axis | Cisplatin | NEAT1 contributes to cisplatin resistance by miR-770-5p/PARP1 Axis in ovarian cancer. |
| TUG1 | Autophagy pathway | Cisplatin | TUG1 modulates autophagy and affects cisplatin resistance in colorectal cancer. |
| CCAT1 | MAPK signaling pathway | Oxaliplatin | CCAT1 promotes Wnt/β-catenin signaling and resistance to Oxaliplatin in HCC. |
| MEG3 | miR-141/PDCD4 axis | Oxaliplatin | Enhanced expression of MEG3 enhances the sensitivity of colorectal cancer cells to oxaliplatin by modulating the miR-141/PDCD4 axis. |
| ANRIL | N/A | Cisplatin | Suppression of the ANRIL hinders the emergence of multidrug resistance in gastric cancer cells. |
| HOTTIP | Chromatin remodeling | N/A | HOTTIP interacts with the WDR5/MLL complex, affecting chromatin remodeling and contributing to resistance in prostate cancer. |
| PVT1 | N/A | N/A | Exon 9 of PVT1 is upregulated in prostate cancer, promoting malignant transformation, and contributing to the development of castration resistance in prostate epithelial cells. |
| HULC | Protective autophagy | Oxaliplatin and 5-FU | The expression of HULC is induced by Oxaliplatin and 5-FU, leading to protective autophagy mediated by HULC, which contributes to resistance against chemotherapy. |
The regulatory roles of lncRNAs are diverse, encompassing functions as molecular sponges for microRNAs (miRNAs), For instance, miR-34a was found to inhibit cancer development by targeting NOTCH1 in glioblastoma,231 scaffolds for chromatin-modifying complexes, For example, the lncRNA HOTAIR binds Polycomb Repressive Complex 2 (PRC2) and the LSD1/CoREST/REST complex.232 In addition to regulating gene expression and chromatin conformation, they can also act as a decoy or scaffold for protein complexes,233 and modulators of crucial signaling pathways, lncRNAs are essential for multiple critical cellular processes, such as ubiquitination, phosphorylation, and glycolysis, which enable the movement of nuclear factor kappa B (NF-κB), type I interferons (IFN-I), and inflammatory factors to the antiviral action site, as well as the transport of these factors into the nucleus.229 A noteworthy aspect of lncRNA involvement in cancer resistance is their role in regulating the characteristics and behavior of CSCs, a subpopulation within tumors endowed with unique properties such as self-renewal, differentiation, and resistance to conventional therapies.234
Specific lncRNAs, including MALAT1, HOTAIR, and NEAT1, have been implicated in modulating these CSC features, thus influencing the overall landscape of tumor drug resistance.235 By interacting with miRNAs, lncRNAs act as molecular sponges,231 sequestering miRNAs and preventing them from inhibiting the translation of target mRNAs. This intricate regulatory network often involves key tumor-suppressive or oncogenic miRNAs, contributing to the fine-tuning of gene expression patterns associated with drug resistance mechanisms. Moreover, lncRNAs participate in chromatin remodeling, influencing the epigenetic landscape and, consequently, the transcriptional profile of cancer cells.10 Drug resistance is linked to the dysregulation of particular lncRNAs, which can affect essential physiological functions such as DNA repair, cell cycle regulation, and apoptosis. These lncRNAs play a part in coordinating drug resistance mechanisms due to genetic mutations, epigenetic modifications, and changes in their expression levels.
Understanding the complexity of lncRNA regulation and their specific roles in cancer resistance provides the basis for the development of targeted therapeutic strategies. Precision medicine approaches that take into account the unique lncRNA profile of tumors hold promise for disrupting drug resistance mechanisms and improving treatment outcomes. As research in this field advances, the potential to reveal new levels of complexity and identify clinically relevant targets will increase, providing hope for more effective strategies to address this formidable challenge.
Micro-RNAs
This section delves into the intricate realm of cancer resistance, focusing on the regulatory role of microRNAs (miRNAs) and their pivotal influence on the development and persistence of drug resistance mechanisms in cancer. MicroRNAs, small non-coding RNA molecules, have emerged as key players in complex gene regulatory networks. Despite their modest size, miRNAs orchestrate diverse cellular processes, and their dysregulation is increasingly recognized as a crucial factor in the initiation and progression of cancer.236 Importantly, miRNAs contribute significantly to the adaptive responses of cancer cells to therapeutic challenges.236
In the context of cancer resistance, miRNAs assume a central role in the regulatory symphony.237 These small molecules, often functioning as post-transcriptional regulators, modulate gene expression,236 cellular pathways, and the adaptive responses of cancer cells to therapeutic agents (Table 2). Table 2 elucidates how different classes of miRNAs play a role in conferring resistance to cancer treatment. Their regulatory roles are diverse, encompassing functions such as downregulating target mRNAs, influencing cellular pathways, and acting as mediators of crucial signaling cascades. In the intricate dance of cancer resistance, miRNAs play a pivotal role in modulating the characteristics and behavior of CSCs,238 a subpopulation within tumors endowed with unique properties such as self-renewal, differentiation, and resistance to conventional therapies. Specific miRNAs, including miR-34a,239 miR-21,240 and miR-155,241 are implicated in regulating CSC characteristics, thereby shaping the overall landscape of tumor drug resistance. By targeting key genes involved in drug response, cell survival, and DNA repair,242 miRNAs intricately mold the adaptive responses of cancer cells to therapeutic challenges.
Table 2.
Micro-RNAs (miRNAs) in cancer associated drug resistance
| miRNA | Target | Chemotherapy Drug | Correlation |
|---|---|---|---|
| miR-646 | CDK6 pathway | Cisplatin | hsa_circ_0081143 contributes to cisplatin resistance in gastric cancer by targeting the miR-646/CDK6 pathway. |
| miR-451 | YWHAZ | Paclitaxel | miR-451 plays a role in paclitaxel resistance by regulating YWHAZ in breast cancer. |
| miR-198 | PIK3R1 | Cisplatin | circAKT3 contributes to cisplatin resistance in gastric cancer by boosting PIK3R1 expression through miR-198 suppression. |
| miR-155 | N/A | Cisplatin | The delivery of miR-155 through exosomes contributes to the development of cisplatin resistance in oral cancer cells by promoting EMT. |
| miR-19b | mRNAs | 5-fluorouracil | miR-19b is upregulated in response to 5-fluorouracil (5-FU), and it influences the cell cycle by targeting specific mRNAs. |
| miR-208b | PDCD4 | Oxaliplatin | Exosomal miR-208b contributes to oxaliplatin resistance in colorectal cancer by promoting Treg expansion through targeting PDCD4. |
| miR-17-5p | Beclin1 | Paclitaxel | miR-17-5p downregulation is associated with paclitaxel resistance in lung cancer. It achieves this by directly altering Beclin1 expression, a crucial modulator of autophagy. |
| miR-873 | CDK3 | Tamoxifen | miR-873 acts as a tumor suppressor in ER-positive breast cancer by reducing ERα transcriptional activity and overcoming tamoxifen resistance through CDK3 targeting. |
| miR-449a | ADAM22 | Tamoxifen | miR-449a combats tamoxifen resistance in human breast cancer cells by targeting ADAM22, a key player in this process. |
| miR-34a-5p and miR-204-5p | ACSL4 | Docetaxel | LncRNA NEAT1 contributes to docetaxel resistance in prostate cancer by upregulating ACSL4 through miR-34a-5p and miR-204-5p interactions. |
| miR-375 | N/A | Paclitaxel | miR-375 exhibits increased expression in cervical cancer cells that have developed paclitaxel resistance. |
| miR-186 | CPEB2 | Methotrexate | TUG1 contributes to methotrexate resistance in colorectal cancer by modulating the miR-186/CPEB2 axis. |
| miR-200c and miR-141 | N/A | Oxaliplatin | Down-regulation of miR-200c and miR-141 contributes to selective resistance to oxaliplatin and EMT in colorectal cancer cells during repeated treatments with L-OHP. |
| miR-130b | Colony-stimulating factor 1 | Cisplatin, Paclitaxel | miR-130b is epigenetically silenced in ovarian cancer, leading to the development of multidrug resistance by targeting colony-stimulating factor 1. |
| let-7 | Hedgehog signaling | Erlotinib/Cisplatin | Blocking Hedgehog signaling enhances the sensitivity of NSCLC cells to conventional therapies by influencing the expression of miRNAs that regulate EMT. |
| miR-21 | PTEN/PI3K/Akt pathway | Paclitaxel | miR-21 overexpression associated with the suppression of PTEN, activating the PI3K/Akt pathway, and promoting resistance to Paclitaxel in various cancers. |
| miR-203 | ATM kinase | Oxaliplatin | miR-203 contributes to oxaliplatin resistance in colorectal cancer cells by negatively regulating ATM kinase. |
| miR-200b | FN1 | Doxorubicin | miR-200b modulates EMT in chemo-resistant breast cancer cells by directly targeting FN1. |
| miR-21 | PTEN | Trastuzumab | miR-21 overexpression contributes to trastuzumab resistance in HER2+ breast cancers. |
| miR-22 | PTEN | Tamoxifen | CD63+ cancer-associated fibroblasts (CAFs) promote tamoxifen resistance in breast cancer cells by secreting exosomes rich in miR-22. |
| miR-374a-5p | N/A | Oxaliplatin | miR-374a-5p, elevated in patients with gastric cancer, predicts therapy response. It induces drug resistance by targeting Neurod1. Exosome-delivered anti-miR-374a-5p restores sensitivity to oxaliplatin in gastric cancer cells. |
| MiR-99a and MiR-491 | CAPNS1 | Cisplatin | MiR-99a and MiR-491 play a crucial role in regulating cisplatin resistance in human gastric cancer cells. |
| miR-196a | CDKN1B and ING5 | Cisplatin | Exosomal miR-196a, originating from cancer-associated fibroblasts (CAFs), induces cisplatin resistance in head and neck cancer (HNC). |
| miR-29 | ERK 1/2 | Cisplatin | miR-29 downregulation is associated with cisplatin resistance in ovarian cancer cells. |
| MiR-148a | MSK1 | paclitaxel | MiR-148a plays a crucial role in reducing paclitaxel resistance in hormone-refractory, drug-resistant prostate cancer cells. |
| miR-100 and miR-125b | Wnt/β-catenin signaling pathway | Cetuximab | lncRNA MIR100HG, along with its embedded microRNAs miR-100 and miR-125b, contributes to cetuximab resistance by modulating the Wnt/β-catenin signaling pathway. |
| miR-192 | DNA damage response pathways | Doxorubicin | miR-192 downregulation linked to impaired DNA damage response, contributing to resistance to Doxorubicin in various cancers. |
| miR-19a | N/A | FOLFOX | Serum miR-19a serves as a potential molecular biomarker for predicting and monitoring resistance to first-line FOLFOX chemotherapy regimens in advanced patients with colorectal cancer. |
| miR-224 | N/A | Methotrexate | miR-224 is underexpressed in methotrexate-resistant human colon cancer cells. |
| miR-153 | FOXO3a | Platinum | miR-153 contributes to colorectal cancer progression by enhancing invasion and promoting chemotherapeutic resistance. |
| miR-200c | EMT pathway | Gemcitabine | miR-200c downregulation associated with EMT and resistance to Gemcitabine in pancreatic cancer. |
| miR-128 | ABCC5 and Bmi-1 | Cisplatin | miR-128 plays a role in promoting cisplatin resistance in ovarian cancer by reducing ABCC5 and Bmi-1 expression. |
| miR-193a-3p | LOXL4 | Pirarubicin, Paclitaxel, Adriamycin, Epirubicin Hydrochloride, Cisplatin | miR-193a-3p influences bladder cancer chemoresistance by directly targeting the LOXL4 gene and affecting the Oxidative Stress pathway. |
| miR-320c | SMARCC1 | Gemcitabine | miR-320c regulates gemcitabine resistance in pancreatic cancer by targeting SMARCC1, a core subunit of the chromatin remodeling complex. |
| miR-381 | MDR1 | Cisplatin | miR-381 combats cisplatin resistance in breast cancer by targeting MDR1, a key player in drug resistance. |
| miR-503 | Bcl-2 | Cisplatin | miR-503 modulates cisplatin resistance in non-small cell lung cancer cells by targeting Bcl-2. |
| miR-29b | N/A | Tamoxifen | miR-29b downregulation contributing in resistance to Tamoxifen in breast cancer. |
| miR-17-92 | TGF-β1 | Gemcitabine | The miR-17-92 cluster opposes the state of dormancy and resistance to chemotherapy in a specific subset of pancreatic cancer stem cells. |
| miR-619-5p | Wnt/β-catenin and autophagy pathway | Gemcitabine | LncRNA PVT1 contributes to gemcitabine resistance in pancreatic cancer by activating the Wnt/β-catenin and autophagy pathway through the modulation of the miR-619-5p/Pygo2 and miR-619-5p/ATG14 axes. |
| miR-634 | Ras-MAPK pathway | Cisplatin | miR-634 enhances drug sensitivity in resistant ovarian cancer cells by directly targeting the Ras-MAPK pathway. |
| miR-217 | N/A | Paclitaxel | chemotherapy-induced exosomal circBACH1 contributes to breast cancer resistance and stemness by interacting with miR-217 and upregulating the expression of G3BP2. |
| MiR-30a | AKT | Gemcitabine | MiR-30a modulates the responsiveness of cancer cells to chemotherapy by influencing the SNAI1/IRS1/AKT pathway. |
| miR-15b and miR-16 | BCL2 | Vincristine, Adriamycin, 5-fluorouracil, cisplatin, mitomycin C, etoposide | miR-15b and miR-16 regulate multidrug resistance in human gastric cancer cells by targeting BCL2. |
| miR-125a-3p | N/A | Gemcitabine | In PDAC, miR-125a-3p governs chemosensitivity by suppressing epithelial-mesenchymal transition through the inhibition of Fyn. |
| miR-153 | N/A | Gemcitabine | miR-153 augments the therapeutic efficacy of gemcitabine through the targeting of Snail in the context of pancreatic cancer. |
| miR-183 | N/A | Gemcitabine | A newly identified interplay between KLF4 and ZEB1 governs the resistance to gemcitabine in pancreatic ductal adenocarcinoma. |
| miR-200 | N/A | Gemcitabine | Elevation of miR-200 and let-7 expression induced by natural agents results in the reversal of epithelial-to-mesenchymal transition in pancreatic cancer cells resistant to gemcitabine. |
| MiR-130a and MiR-374a | PTEN | Cisplatin | MiR-130a and MiR-374a Serve as New Regulators of Cisplatin Resistance in A2780 Human Ovarian Cancer Cells. |
| miR-1252 | N/A | Paclitaxel | circCELSR1 (hsa_circ_0063809) plays a role in paclitaxel resistance of ovarian cancer cells by regulating FOXR2 expression via miR-1252. |
| miR-205 | N/A | Gemcitabine | Induction of epithelial-to-mesenchymal transition by macrophage migration inhibitory factor amplifies tumor aggressiveness and serves as a prognostic predictor in surgically resected pancreatic ductal adenocarcinoma. |
| miR-214 | PTEN | Cisplatin | miR-214 promotes cell survival and confers resistance to cisplatin by directly targeting PTEN. |
| miR-222 and miR-29a | N/A | Adriamycin and docetaxel | miR-222 and miR-29a play a role in fostering drug resistance in breast cancer cells. |
| miR-205 and miR-31 | BCL2L2 (encoding Bcl-w) and E2F6 | Docetaxel, Cisplatin | Reduced expression of miR-205 and miR-31 leads to resistance against apoptosis induced by chemotherapy in prostate cancer cells. |
| miR-497 | NF-κB1 | Gemcitabine | microRNA-497 averts gemcitabine resistance, migration, and invasion in pancreatic cancer stem cells by directly targeting nuclear factor kappa B-1. |
| MiR-487a | BCRP/ABCG2 | Mitoxantrone | MiR-487a reverses mitoxantrone (MX) resistance in breast cancer cells by targeting the breast cancer resistance protein (BCRP/ABCG2). |
| miR-506 | N/A | Gemcitabine | Reduced expression of NEAT1 heightens the sensitivity of gemcitabine-resistant pancreatic cancer cells to gemcitabine via the modulation of the miR-506-3p/ZEB2/EMT axis. |
| miR-106a and miR-591 | N/A | Paclitaxel | miR-106a and miR-591 play crucial roles in causing paclitaxel resistance in ovarian cancer cells. |
| miR-26a and miR-30b | CCNE2 | Trastuzumab | miR-26a and miR-30b are key players in HER2+ breast cancer trastuzumab resistance, with their target gene being CCNE2. |
| miR-509 | N/A | Gemcitabine | miR-509-5p and miR-1243 enhance gemcitabine sensitivity by suppressing epithelial-mesenchymal transition in pancreatic cancer. |
| miR-27a | N/A | Vincristine, Adriamycin, Cisplatin, 5-fludrouracil | Reducing miR-27a levels could potentially hinder the growth and enhance sensitivity to drugs in gastric cancer cells. |
| miR-1206 | N/A | Gemcitabine | Circular RNA circ_0092367 suppresses epithelial-mesenchymal transition (EMT) and mitigates gemcitabine resistance in pancreatic cancer through the regulation of the miR-1206/ESRP1 axis. |
| miR-509 | N/A | Gemcitabine | miR-509-5p and miR-1243 enhance gemcitabine sensitivity in pancreatic cancer by impeding epithelial-mesenchymal transition. |
Moreover, the crosstalk between miRNAs and other regulatory elements, such as lncRNAs and signaling pathways,243 adds another layer of complexity to the role of miRNAs in cancer resistance.236 Dysregulation of specific miRNAs is linked with the development of drug resistance, affecting crucial cellular processes such as apoptosis, cell cycle regulation, and DNA repair.244 Understanding the nuanced regulatory functions of miRNAs and their specific roles in cancer resistance provides a foundation for the development of targeted therapeutic strategies. Precision medicine approaches that consider the unique miRNA profile of tumors hold promise for disrupting drug resistance mechanisms and improving treatment outcomes. Therefore, decoding the regulatory functions of miRNAs, especially in the context of cancer resistance, unveils new therapeutic avenues. As research in this field progresses, the opportunity to explore new levels of complexity and identify clinically relevant targets will increase, providing hope for more effective strategies to address this formidable challenge.
Circular RNAs
Here we shed light on the multifaceted field of cancer resistance, with a primary focus on the regulatory role of circular RNAs (circRNAs) and their critical influence on the development and maintenance of resistance mechanisms in cancer. CircRNAs, once considered byproducts of splicing errors, have evolved into essential players in the complex coordination of gene regulation. Characterized by a unique circular structure, these covalently closed, single-stranded RNA molecules exhibit resistance to exonucleases, ensuring stability in the cellular environment.245 Despite their noncoding nature, circRNAs manifest complex regulatory functions, participating in diverse cellular processes, and their aberrant expression is increasingly recognized as a hallmark of cancer.246
In the context of cancer resistance, circRNAs assume a central role in the regulatory landscape (Table 3). These molecules participate in the regulation of gene expression at multiple levels,247 acting as sponges for miRNAs, interacting with RNA-binding proteins, and influencing various signaling pathways.248 Notably, circRNAs play a crucial role in shaping the adaptive responses of cancer cells to therapeutic challenges. Their diverse regulatory roles include functioning as competing endogenous RNAs (ceRNAs), sequestering miRNAs, and thereby relieving the repression of target mRNAs. Additionally, circRNAs interact with RNA-binding proteins, regulating their activity and influencing downstream signaling pathways involved in drug resistance.249 Table 3 outlines the ways in which distinct types of ceRNAs participate in the development of resistance to cancer treatments.
Table 3.
circ_RNAs in cancer associated drug resistance
| Circ_RNA | Target | Chemotherapy Drug | Correlation |
|---|---|---|---|
| circ_0007385 | HMGB1 | Cisplatin | Circ_0007385 functions as a competitive endogenous RNA, exerting inhibitory effects on the malignant behaviors and cisplatin resistance of non-small cell lung cancer cells by sequestering miR-519d-3p. |
| circ 0008253 | TGF-β | Oxaliplatin | Exosomes derived from macrophages control oxaliplatin resistance in gastric cancer cells by encapsulating circ 0008253. |
| circ_0076305 | ABCC1 | Cisplatin | Circular RNA circ_0076305 enhances cisplatin (DDP) resistance in non-small cell lung cancer cells by modulating ABCC1 via miR-186-5p regulation. |
| circ-ZEB1 | N/A | Oxaliplatin and 5-fluorouracil (5-FU) | Circular RNA circ-ZEB1 modulates epithelial-mesenchymal transition and chemotherapy resistance in colorectal cancer by interacting with miR-200c-5p. |
| circRNA_103615 | ABCC1 | Cisplatin | Circular RNA circRNA_103615 plays a role in both the advancement of tumors and the development of cisplatin resistance in non-small cell lung cancer (NSCLC) by controlling ABCB1. |
| circ_0008928 | HK2 | Cisplatin | The biomarker circ_0008928 derived from serum exosomes modulates cisplatin sensitivity, tumor progression, and glycolysis metabolism through the miR-488/HK2 axis in cisplatin-resistant non-small cell lung carcinoma. |
| circ-RNF121 | SOX4 | Cisplatin | Circular RNA circ-RNF121 plays a role in the resistance of non-small cell lung cancer cells to cisplatin (DDP) by modulating the miR-646/SOX4 axis. |
| circ-PIP5K1A | N/A | Cisplatin | The mechanism involving the circ-PIP5K1A/miR-942-5p/NFIB axis in cisplatin resistance in ovarian cancer elucidated through imaging and molecular diagnostics. |
| hsa_circRNA_103809 | GOT1 | Cisplatin | A newly discovered circular RNA, hsa_circRNA_103809, orchestrates cisplatin resistance in non-small cell lung cancer (NSCLC) through the miR-377-3p/GOT1 pathway. |
| circ_0082182 | NFIB | Oxaliplatin | Circular RNA circ_0082182 enhances the expression of NFIB by sequestering miR-326, thereby promoting oxaliplatin resistance and the malignant progression of colorectal cancer cells. |
| circ-PRMT5 | REV3L | Cisplatin | Through the miR-4458/REV3L axis, circular RNA PRMT5 imparts cisplatin resistance in non-small-cell lung cancer. |
| circAKT3 | STAT3 | Cisplatin | In lung cancer cells, CircAKT3 suppresses glycolytic balance by controlling miR-516b-5p/STAT3, leading to the inhibition of cisplatin sensitivity. |
| circ-LDLRAD3 | SOX5 | Cisplatin | Reducing the expression of circ-LDLRAD3 diminishes cisplatin chemoresistance and hinders the progression of cisplatin-resistant gastric cancer by inhibiting SOX5 through the enrichment of miR-588. |
| circRNA-FOXO3 | Foxo3 | Cisplatin | Through the miR-543/Foxo3 axis, circular RNA FOXO3 promotes glycolysis and enhances cisplatin sensitivity in lung cancer cells. |
| hsa_circ_0014235 | CDK4 | Cisplatin | Exosomes transfer hsa_circ_0014235, fostering chemoresistance to DDP and exacerbating the progression of non-small cell lung cancer by orchestrating the miR-520a-5p/CDK4 pathway. |
| hsa_circ_0096157 | N/A | Cisplatin | Circular RNA hsa_circ_0096157 plays a role in cisplatin resistance by promoting proliferation, facilitating cell cycle progression, and suppressing apoptosis in non-small-cell lung carcinoma cells. |
| circRNA_100565 | ADAM28 | Cisplatin | Circular RNA circRNA_100565 contributes to cisplatin resistance in NSCLC cells by modulating proliferation, apoptosis, and autophagy through the miR-337-3p/ADAM28 axis. |
| circ_0076305 | ABCC1 | Cisplatin | Circular RNA circ_0076305 enhances resistance to cisplatin (DDP) in non-small cell lung cancer cells by modulating ABCC1 through miR-186-5p. |
| circ_PIP5K1A | ROCK1 | Cisplatin | Circular RNA Circ_PIP5K1A governs cisplatin resistance and malignant progression in non-small cell lung cancer cells and a xenograft murine model by relying on the miR-493-5p/ROCK1 axis. |
| hsa_circ_0085131 | ATG7 | Cisplatin | Circular RNA hsa_circ_0085131 participates in the cisplatin resistance of non-small-cell lung cancer cells by modulating autophagy. |
| circ-ABCB10 | AK4 | Cisplatin | Reducing the expression of circ-ABCB10 enhances the sensitivity of lung cancer cells to cisplatin through the miR-556-3p/AK4 axis. |
| circ-AnnexinA7 | Cyclin D1 | Cisplatin | Circular RNA-AnnexinA7 expedites cisplatin resistance in non-small cell lung cancer by manipulating microRNA-545-3p to mediate Cyclin D1. |
| Circ-ERBB2 | PTEN | 5-FU | Downregulation of Circ-ERBB2 renders colorectal cancer cells more susceptible to 5-FU by modulating the miR-181a-5p/PTEN/Akt pathway. |
| circ_0000079 | FXR1 | Cisplatin | Circ_0000079 acts as a decoy for the RNA-binding protein FXR1, disrupting the formation of the FXR1/PRCKI complex and diminishing their role in cell invasion and drug resistance in non-small cell lung cancer (NSCLC). |
| circ_0001821 | GRK5 | Paclitaxel | Suppression of Circ_0001821 through knockdown inhibits the growth, metastasis, and taxane resistance of non-small-cell lung cancer cells by modulating the miR-526b-5p/GRK5 axis. |
| circ_ZFR | KPNA4 | Paclitaxel | Circular RNA Circ_ZFR plays a role in both paclitaxel resistance and the progression of non-small cell lung cancer by elevating KPNA4 levels through the sequestration of miR-195-5p. |
| circANKRD17 | FOXR2 | Paclitaxel | Circular RNA circANKRD17 (also known as hsa_circ_0007883) induces paclitaxel resistance in ovarian cancer by interacting with FUS to stabilize FOXR2. |
| circ_0011292 | TRIM65 | Paclitaxel | Circular RNA Circ_0011292 boosts paclitaxel resistance in non-small cell lung cancer by modulating the miR-379-5p/TRIM65 axis. |
| circ_0010235 | E2F7 | Cisplatin | Circular RNA circ_0010235 imparts cisplatin resistance in lung cancer by elevating E2F7 levels through sequestering miR-379-5p. |
| hsa_circ_0002874 | MDM2/P53 pathway | Paclitaxel | The upregulation of hsa_circ_0002874 induces resistance in non-small cell lung cancer to paclitaxel by influencing the miR-1273f/MDM2/p53 pathway. |
| Circ_0001667 | N/A | Adriamycin | Circular RNA Circ_0001667 enhances resistance to Adriamycin and fosters malignant progression by targeting the miR-193a-5p/Rap2A molecular axis in breast cancer. |
| Circ_0078607 | N/A | Platinum | Circular RNA Circ_0078607 enhances sensitivity to platinum drugs through the miR-196b-5p/GAS7 axis in ovarian cancer. |
| hsa_circ_0030998 | N/A | Paclitaxel | A recently discovered circular RNA, hsa_circ_0030998, hinders tumorigenesis and Taxol resistance in lung cancer by acting as a sponge for miR-558. |
| circ_0002483 | GRB2, FOXO1, and FOXO3 | Taxol | Circular RNA hsa_circ_0002483 impedes the progression and improves Taxol sensitivity in non-small cell lung cancer by targeting miR-182-5p. |
| circ_0014130 | YAP1 | Docetaxel | Inhibiting circ_0014130 mitigated drug resistance and malignant behaviors in non-small cell lung cancer (NSCLC) cells with acquired docetaxel resistance by modulating the miR-545-3p-YAP1 axis. |
| circPOFUT1 | N/A | Cisplatin | Circular RNA circPOFUT1 amplifies malignant characteristics and chemoresistance associated with autophagy by capturing miR-488-3p to activate the PLAG1-ATG12 axis in gastric cancer. |
| circ_0003998 | CORO1C | Docetaxel | Circular RNA circ_0003998 governs the progression and docetaxel sensitivity of non-small cell lung cancer cells resistant to DTX through the miR-136-5p/CORO1C axis. |
| hsa_circ_0005909 | SOX4 | Adriamycin | Circular RNA hsa_circ_0005909 serves as a prognostic indicator for poor outcomes and facilitates the growth, metastasis, and drug resistance of non-small-cell lung cancer through the miRNA-338-3p/SOX4 pathway. |
| circRBM33 | N/A | Osimertinib | Circular RNA circRBM33 stimulates migration and invasion while mediating resistance to osimertinib in a non-small cell lung cancer cell line. |
| circRNA_0067717 | p53 | Paclitaxel | Circular RNA circRNA_0067717 induces resistance to paclitaxel in nasopharyngeal carcinoma by serving as a scaffold for the interaction between TRIM41 and p53. |
| circASK1 | N/A | Gefitinib | A recently identified protein encoded by circASK1 alleviates gefitinib resistance in lung adenocarcinoma by competitively activating ASK1-dependent apoptosis. |
| circSETD3 | ABCG2 | Gefitinib | Circular RNA circRNA_103615 plays a role in both the advancement of tumors and the development of cisplatin resistance in non-small cell lung cancer (NSCLC) through the regulation of ABCB1. |
| CircIFNGR2 | KRAS | Cetuximab | Circular RNA CircIFNGR2 promotes the proliferation and migration of colorectal cancer (CRC) and induces cetuximab resistance by indirectly targeting KRAS through sequestering MiR-30b. |
| circANKRD28 | N/A | Cisplatin | Circular RNA circANKRD28 suppresses cisplatin resistance in non-small-cell lung cancer by modulating the miR-221-3p/SOCS3 axis. |
| circSnx12 | N/A | Cisplatin | Circular RNA circSnx12 imparts cisplatin chemoresistance to ovarian cancer by suppressing ferroptosis through the miR-194-5p/SLC7A11 axis. |
| circRNA_102481 | ROR1 | EGFR-TKIs | Exosomal circRNA_102481, derived from tumors, contributes to resistance against EGFR-TKIs through the miR-30a-5p/ROR1 axis in non-small cell lung cancer. |
A crucial aspect of circRNA involvement in cancer resistance is their impact on the tumor microenvironment. CircRNAs contribute to the crosstalk between cancer cells and the surrounding stroma, establishing a supportive niche that promotes resistance.250 Their influence extends to the regulation of EMT and the promotion of metastatic potential, further complicating therapeutic interventions.251 Dysregulation of specific circRNAs are linked with the development of drug resistance, affecting key cellular processes such as apoptosis, DNA repair, and cell cycle regulation.252 Understanding the complex regulatory functions of circRNAs and their specific roles in cancer resistance delivers the basis for the development of targeted therapeutic strategies.
Precision medicine approaches that consider the unique circRNA profile of tumors hold promise for circumventing drug resistance mechanisms and improving treatment outcomes. As research in this field advances, the opportunity to explore new levels of complexity and identify clinically relevant targets will increase, offering hope for more effective strategies to address the formidable challenge of cancer resistance.
Therapeutic approaches
Targeted therapies
Various therapeutic approaches can be employed to address and overcome drug resistance in cancer. Such as, targeted, immune, combination and personalized therapeutic approaches (Figure 7). The complexity of cancer has led to the development of combination therapies as a strategic response. These therapies involve the use of distinct agents, each with unique mechanisms of action, with the aim of enhancing efficacy and circumventing resistance mechanisms.98 For instance, the combination of chemotherapy and targeted therapies can simultaneously tackle proliferating cancer cells and disrupt specific signaling pathways.98 Chemotherapy, which is designed to kill rapidly dividing cells, can be effective against proliferating cancer cells.253 On the other hand, targeted therapies are designed to interfere with specific signaling pathways that are often altered in cancer cells, thereby disrupting the growth and survival of these cells.254 The synergistic effects of these combination therapies can lead to more potent anti-cancer responses.98 Synergy in this context refers to the enhanced effect achieved when two or more drugs are used together, compared to the sum of their effects when used separately.253 This can result in a more effective suppression of cancer growth and limit the emergence of drug resistance.98 However, it is important to note that while combination therapies can be highly effective, they also pose challenges. These include the potential for increased toxicity, the complexity of managing multiple drugs, and the need for careful coordination of treatment schedules. Despite these challenges, combination therapies represent a promising approach in the ongoing battle against cancer.98
Figure 7.
Integrated therapeutic approaches in cancer treatment
The figure illustrates integrated therapeutic approaches in cancer treatment, highlighting key strategies. Targeted therapies involve the combination of chemotherapy and targeted agents to simultaneously address rapidly dividing cancer cells, utilizing chemotherapy, and disrupt specific signaling pathways, deploying targeted therapies. Immunotherapies focus on immune checkpoint inhibitors such as pembrolizumab and nivolumab, blocking signals for immune evasion, and CAR-T cell therapy for haematological malignancies. Combination therapies synergize to enhance anti-cancer responses and potent suppression of cancer growth. Personalized treatment involves molecular profiling to identify genetic alterations, tailoring treatment based on individual tumor profiles, and maximizing therapeutic efficacy through personalized regimens. This integrated approach harnesses the strengths of targeted therapies, immunotherapies, and personalized treatment, offering a comprehensive strategy against cancer.
Immunotherapies
Through the activation of the body’s immune system, immunotherapy has brought about a significant transformation in the treatment of cancer. ICIs, including pembrolizumab and nivolumab, help the body recognise and destroy cancer cells by blocking the signals that these cells use to elude the immune system.255 Adoptive cell therapy known as CAR-T cell therapy uses a patient’s own T cells that have been genetically modified to target and eradicate cancer cells. It has demonstrated impressive results in the treatment of haematological malignancies such as multiple myeloma (MM), non-Hodgkin lymphoma, and B-cell acute lymphocytic leukemia (B-ALL).256 These therapies provide a dynamic and adaptable strategy against cancer, with the potential for durable responses. However, the development of resistance to single-agent therapies remains a significant challenge. Tumor heterogeneity, dynamic adaptations, and various resistance mechanisms contribute to this issue.257 Tumor heterogeneity refers to the diversity of cancer cells within a single tumor, which can lead to different responses to treatment.258 Dynamic adaptations involve changes in cancer cells over time in response to treatment, leading to the development of resistance.259 Innovative solutions are being explored to overcome these challenges. For instance, understanding the tumor microenvironment, which includes various immune cells, the tumor histology, molecular subtype, clonal heterogeneity and evolution, can help improve the efficacy of immunotherapies.260 Furthermore, research is being done on synthetic lethality, a phenomenon in which the simultaneous occurrence of several genetic events causes cell death, as a possible method of overcoming medication resistance.261 Furthermore, research is ongoing to develop more effective targeted drugs and identify predictive factors for immunotherapeutic responses.260
Combination therapies
Indeed, the integration of targeted therapies, combination therapies, and immunotherapies is a promising approach in cancer treatment. This synergistic strategy leverages the strengths of each therapy, enhancing the chances of overcoming resistance.98 Targeted therapies aim to inhibit specific molecular vulnerabilities in cancer cells, thereby disrupting key pathways involved in cancer progression.262 Immunotherapies, on the other hand, work by stimulating the body’s immune system to recognize and destroy cancer cells. When these two approaches are combined, they can capitalize on the unleashed potential of the immune system while simultaneously targeting specific molecular vulnerabilities in cancer cells.263 The evolving landscape of precision medicine further propels the potential of combination therapies. Molecular profiling, which involves analyzing the genetic makeup of a tumor, enables the identification of specific genetic alterations.264 This information guides the selection of targeted therapies, ensuring that the chosen treatment is tailored to the unique genetic profile of the individual’s tumor.265 Personalized treatment regimens, which are designed based on an individual’s unique tumor profile, can maximize therapeutic efficacy. By tailoring the treatment to the specific characteristics of the tumor, healthcare providers can ensure that the therapy is as effective as possible.266 In conclusion, the combination of targeted therapies, immunotherapies, and personalized treatment regimens offers a promising approach in the fight against cancer. By leveraging the strengths of each therapy and tailoring the treatment to the individual’s unique tumor profile, this approach enhances the chances of overcoming resistance and maximizing therapeutic efficacy.98
Future directions
Cancer resistance remains a multifaceted challenge requiring ongoing exploration for effective solutions. The future trajectory of anticancer research is intricately linked to the advancements in precision medicine.267 This entails a more profound molecular profiling through genomics, transcriptomics, and proteomics, fostering a comprehensive understanding of individual tumor landscapes. The integration of diverse omics data is poised to contribute significantly to the development of personalized therapeutic strategies.265
Immunotherapy, a revolutionary approach in cancer treatment, is currently undergoing intensive efforts to refine its efficacy and applicability. The future objectives of immunotherapy research and development are centered around augmenting the specificity and durability of immune responses against cancer cells. This encompasses the pursuit of several key advancements, which include the creation of more precisely targeted ICIs, enhancements in adoptive cell therapies, and a comprehensive understanding of the intricate tumor microenvironment to fully unlock the immense potential of immunotherapy in combating cancer.268
To achieve these objectives, it is imperative to effectively address and overcome adaptive resistance, a crucial challenge encountered in immunotherapy. Accordingly, research initiatives will focus on unraveling the mechanisms by which cancer cells adapt and evolve in response to treatment stress over time. Scientists aim to decipher the intricate molecular processes underlying adaptive responses exhibited by cancer cells during immunotherapy. By gaining a comprehensive understanding of these adaptive mechanisms, researchers can develop interventions and therapeutic strategies to prevent or counteract such adaptations, potentially improving the outcomes and increasing the effectiveness of immunotherapeutic approaches in the fight against cancer.125
Cancer stem cells (CSCs), a subset of cells within tumors, have been recognized for their significant contribution to drug resistance and tumor relapse. Consequently, future research endeavors will place a paramount emphasis on unraveling the mysteries of CSCs and developing effective strategies to target and eliminate them.269 To achieve this goal, intensive efforts will be dedicated to identifying unique markers that can specifically distinguish CSCs from other tumor cells. Researchers aim to uncover molecular signatures or surface proteins that are exclusively expressed on CSCs, enabling their precise identification and isolation. By identifying these distinct CSC markers, scientists can improve early detection methods, potentially leading to more tailored and effective therapeutic interventions.269
Furthermore, comprehending the signaling pathways that sustain CSC characteristics will be a crucial aspect of future research. Scientists will delve into the intricate molecular mechanisms that allow CSCs to self-renew, differentiate, and resist therapeutic interventions. By deciphering these pathways, researchers can develop targeted treatments that disrupt the molecular processes supporting CSC survival and proliferation, thereby reducing their ability to drive tumor growth and therapy resistance.270
Building upon the understanding of CSC markers and signaling pathways, specific treatments will be developed to eradicate this resilient subpopulation. Novel therapeutic approaches will be designed to selectively target CSCs while sparing normal cells, aiming to eliminate the sources of drug resistance and minimize the risk of tumor relapse. These treatments may involve the use of targeted therapies, immunotherapies, or combination approaches tailored to exploit the vulnerabilities specific to CSCs.271
Ultimately, by intensifying research efforts in the field of CSCs, scientists aim to significantly improve cancer treatment outcomes by specifically eliminating the subpopulation responsible for therapy resistance and tumor recurrence. The comprehensive exploration of CSCs, including the identification of CSC markers, elucidation of signaling pathways, and development of specific treatments, holds the promise of advancing the field of cancer therapy and bringing us closer to effective and lasting solutions for patients with cancer.272
The integration of artificial intelligence (AI) and machine learning is emerging as a highly promising approach in the field of cancer research, particularly in the context of understanding and overcoming treatment resistance. AI algorithms, leveraging their exceptional ability to analyze vast datasets, offer the potential to uncover intricate patterns and relationships, predict treatment responses, and assist clinicians in making informed decisions regarding personalized treatment strategies for each individual patient. By processing and analyzing diverse data sources, including genomics, proteomics, and patient clinical profiles, AI algorithms can identify subtle molecular and clinical features that contribute to treatment resistance. This knowledge can then be utilized to develop predictive models that aid in anticipating how patients may respond to specific therapies, enabling treatment plans to be optimized based on each patient’s unique characteristics and expected outcomes.273
In parallel, significant attention is being given to enhancing the effectiveness of drug delivery systems through innovative technologies. Nanoparticle-based drug delivery is one such approach, where tiny particles are engineered to carry therapeutic agents and deliver them directly to tumor sites. This targeted drug delivery minimizes off-target effects, ensuring that healthy tissues are spared while maximizing drug concentration and efficacy within the tumor microenvironment.274
In addition to nanoparticle drug delivery, targeted drug encapsulation techniques are being developed to further optimize treatment efficacy. These methods involve encapsulating therapeutic agents within nanoparticles or other carriers that are specifically designed to release the drug at the tumor site, increasing local drug concentration and minimizing systemic side effects.275
Advancements in smart drug delivery systems are also gaining attention, seeking to disrupt the mechanisms of drug resistance. Such systems use responsive materials or stimuli-responsive nanoparticles that can release drugs in response to specific triggers present within the tumor microenvironment. These triggers may include changes in pH, temperature, or enzymatic activity, effectively bypassing resistance mechanisms and delivering therapeutics precisely where they are needed.276
Ultimately, the integration of AI and machine learning with innovative drug delivery technologies holds immense promise in revolutionizing cancer treatment. By harnessing the power of AI for predictive modeling and personalized therapy selection, coupled with advanced drug delivery systems that optimize treatment efficacy and disrupt resistance mechanisms, researchers and clinicians can develop more effective and precisely tailored strategies to combat cancer and improve patient outcomes.
In future research, the identification of novel biomarkers that can predict the development of drug resistance will play a pivotal role. These biomarkers encompass genetic, epigenetic, and proteomic factors that have the potential to serve as early indicators of the emergence of drug resistance.277 By identifying these biomarkers, researchers aim to enable timely adjustments and interventions in treatment plans, improving patient outcomes. To ensure effectiveness, future research will adopt a patient-centered approach that recognizes and accounts for the diversity of tumor genetic and molecular landscapes. It is understood that tumors can exhibit significant heterogeneity, and a one-size-fits-all approach may not be sufficient. By considering the unique characteristics of each patient’s tumor, researchers can design personalized therapeutic strategies that specifically target the underlying mechanisms of resistance.
To evaluate and validate these targeted treatment strategies, advanced models will be employed that closely mimic the clinical environment. Xenograft models, in which human tumor cells are implanted into immunodeficient animals, can provide valuable insights into tumor behavior and response to treatment. Similarly, organoids three-dimensional cell cultures derived from patient tumors allow for the study of tumor biology in a more physiologically relevant context. Additionally, patient-derived in vitro models, which involve growing tumor cells in a laboratory setting, provide key data on treatment response and enable the testing of various therapeutic approaches. The utilization of these advanced models in research will empower scientists to evaluate treatment strategies in settings that more accurately simulate the complexity of human tumors. By conducting experiments in clinically relevant environments, researchers can gain a comprehensive understanding of treatment efficacy, resistance mechanisms, and potential combinatorial approaches. This knowledge will lay the groundwork for the development of innovative treatments that can overcome drug resistance and improve patient outcomes.278
Conclusion
In conclusion, the hallmarks of cancer resistance present a multifaceted challenge requiring comprehensive strategies for effective therapeutic interventions. Key mechanisms, including drug efflux mediated by ABC transporters, metabolic reprogramming exemplified by the Warburg effect, and the intricate interplay between cancer cells and mitochondria, underscore the complexity of resistance. CSCs contribute significantly to treatment evasion, demanding targeted approaches. Non-coding RNAs, such as lncRNAs, miRNAs, and circRNAs, add additional layers of complexity to resistance. Future directions in research involve refining immunotherapy, combating adaptive resistance, leveraging artificial intelligence, and identifying new biomarkers. Patient-centered approaches and the integration of advanced technologies promise to translate insights into innovative, personalized therapeutic strategies to navigate the intricate landscape of cancer resistance and improve patient outcomes.
Acknowledgments
This work was supported by the National Natural Science Foundation of China of China (Grant No. 82170974) and the Central South University Research Program of Advanced Interdisciplinary Studies (Grant No. 2023QYJC038).
Author contributions
M.T.: conceptualization, original drafting, visualization, and writing - review & editing. J.-J.H.: review and editing and visualization. Jie Liang: review and editing and visualization. Y.-Q.H.: visualization and suggestions. C.-Y.H.: visualization and editing. Wen-Dong Wan: visualization and valuable suggestions. C.-H.J.: visualization. H.W.: visualization. N.L.: supervision, reviewed, and editing. All authors have reviewed and approved the final manuscript for publication.
Declaration of interests
There is no competing interest to declare.
References
- 1.Haider T., Pandey V., Banjare N., Gupta P.N., Soni V. Drug resistance in cancer: mechanisms and tackling strategies. Pharmacol. Rep. 2020;72:1125–1151. doi: 10.1007/s43440-020-00138-7. [DOI] [PubMed] [Google Scholar]
- 2.Keyvani-Ghamsari S., Khorsandi K., Rasul A., Zaman M.K. Current understanding of epigenetics mechanism as a novel target in reducing cancer stem cells resistance. Clin. Epigenetics. 2021;13:120. doi: 10.1186/s13148-021-01107-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alkhatib H., Rubinstein A.M., Vasudevan S., Flashner-Abramson E., Stefansky S., Chowdhury S.R., Oguche S., Peretz-Yablonsky T., Granit A., Granot Z., et al. Computational quantification and characterization of independently evolving cellular subpopulations within tumors is critical to inhibit anti-cancer therapy resistance. Genome Med. 2022;14:120. doi: 10.1186/s13073-022-01121-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zeng Z., Fu M., Hu Y., Wei Y., Wei X., Luo M. Regulation and signaling pathways in cancer stem cells: implications for targeted therapy for cancer. Mol. Cancer. 2023;22:172. doi: 10.1186/s12943-023-01877-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cao D., Song Q., Li J., Jiang Y., Wang Z., Lu S. Opportunities and challenges in targeted therapy and immunotherapy for pancreatic cancer. Expert Rev. Mol. Med. 2021;23:e21. doi: 10.1017/erm.2021.26. [DOI] [PubMed] [Google Scholar]
- 6.Pienta K.J. Perspectives in Oncology: a new article type for Medical Oncology. Med. Oncol. 2020;37:21. doi: 10.1007/s12032-020-01349-x. [DOI] [PubMed] [Google Scholar]
- 7.Evans Webb M., Murray E., Younger Z.W., Goodfellow H., Ross J. The Supportive Care Needs of Cancer Patients: a Systematic Review. J. Cancer Educ. 2021;36:899–908. doi: 10.1007/s13187-020-01941-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Forbes A.N., Xu D., Cohen S., Pancholi P., Khurana E. Discovery of novel therapeutic targets in cancer using patient-specific gene regulatory networks. bioRxiv. 2022 doi: 10.1101/2022.01.31.478503. Preprint at. [DOI] [Google Scholar]
- 9.Zhou Z., Edil B.H., Li M. Combination therapies for cancer: challenges and opportunities. BMC Med. 2023;21:171. doi: 10.1186/s12916-023-02852-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wajapeyee N., Gupta R. Epigenetic Alterations and Mechanisms That Drive Resistance to Targeted Cancer Therapies. Cancer Res. 2021;81:5589–5595. doi: 10.1158/0008-5472.CAN-21-1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Raynaud F., Mina M., Tavernari D., Ciriello G. Pan-cancer inference of intra-tumor heterogeneity reveals associations with different forms of genomic instability. PLoS Genet. 2018;14 doi: 10.1371/journal.pgen.1007669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu Y., Li H., Yang F., Yang D., Zhou B.-B.S. Cell plasticity and genomic instability in cancer evolution. Genome Instab. Dis. 2020;1:301–309. doi: 10.1007/s42764-020-00023-w. [DOI] [Google Scholar]
- 13.Liu B., Duenas D., Zheng L., Reckamp K., Shen B. Genomic instability as a major mechanism for acquired resistance to EGFR tyrosine kinase inhibitors in cancer. Protein Cell. 2022;13:82–89. doi: 10.1007/s13238-021-00855-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu Z., Sun R., Curtis C. A population genetics perspective on the determinants of intra-tumor heterogeneity. Biochim. Biophys. Acta. Rev. Cancer. 2017;1867:109–126. doi: 10.1016/j.bbcan.2017.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Greenbaum B.D., Hoyos D., Thomas P. Tumour driver mutations compromise between cancer growth and immune responses. Nature. 2022;37:21. doi: 10.1038/d41586-022-01138-8. [DOI] [PubMed] [Google Scholar]
- 16.Feng Y., Song F., Zhang P., Fan G., Zhang T., Zhao X., Ma C., Sun Y., Song X., Pu H., et al. Prediction of EGFR Mutation Status in Non–Small Cell Lung Cancer Based on Ensemble Learning. Front. Pharmacol. 2022;13 doi: 10.3389/fphar.2022.897597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tanda E.T., Vanni I., Boutros A., Andreotti V., Bruno W., Ghiorzo P., Spagnolo F. Current State of Target Treatment in BRAF Mutated Melanoma. Front. Mol. Biosci. 2020;7 doi: 10.3389/fmolb.2020.00154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arozarena I., Wellbrock C. Overcoming resistance to BRAF inhibitors. Ann. Transl. Med. 2017;5:387. doi: 10.21037/atm.2017.06.09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Y., Wang Z., Ajani J.A., Song S. Drug resistance and Cancer stem cells. Cell Commun. Signal. 2021;19:19. doi: 10.1186/s12964-020-00627-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Soverini S., Abruzzese E., Bocchia M., Bonifacio M., Galimberti S., Gozzini A., Iurlo A., Luciano L., Pregno P., Rosti G., et al. Next-generation sequencing for BCR-ABL1 kinase domain mutation testing in patients with chronic myeloid leukemia: a position paper. J. Hematol. Oncol. 2019;12:131. doi: 10.1186/s13045-019-0815-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zaal E.A., Berkers C.R. The Influence of Metabolism on Drug Response in Cancer. Front. Oncol. 2018;8 doi: 10.3389/fonc.2018.00500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bertholee D., Maring J.G., van Kuilenburg A.B.P. Genotypes Affecting the Pharmacokinetics of Anticancer Drugs. Clin. Pharmacokinet. 2017;56:317–337. doi: 10.1007/s40262-016-0450-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miteva-Marcheva N.N., Ivanov H.Y., Dimitrov D.K., Stoyanova V.K. Application of pharmacogenetics in oncology. Biomark. Res. 2020;8:32. doi: 10.1186/s40364-020-00213-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tracy T.S., Chaudhry A.S., Prasad B., Thummel K.E., Schuetz E.G., Zhong X.-b., Tien Y.-C., Jeong H., Pan X., Shireman L.M., et al. Interindividual Variability in Cytochrome P450–Mediated Drug Metabolism. Drug Metab. Dispos. 2016;44:343–351. doi: 10.1124/dmd.115.067900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li J., Bluth M.H. Pharmacogenomics of drug metabolizing enzymes and transporters: implications for cancer therapy. Pharmgenomics. Pers. Med. 2011;4:11–33. doi: 10.2147/PGPM.S18861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kulma I., Boonprasert K., Na-Bangchang K. Polymorphisms of genes encoding drug transporters or cytochrome P450 enzymes and association with clinical response in cancer patients: a systematic review. Cancer Chemother. Pharmacol. 2019;84:959–975. doi: 10.1007/s00280-019-03932-0. [DOI] [PubMed] [Google Scholar]
- 27.Ingelman-Sundberg M. Implications of polymorphic cytochrome p450-dependent drug metabolism for drug development. Drug Metab. Dispos. 2001;29:570–573. [PubMed] [Google Scholar]
- 28.Yu K.D., Shao Z.M. Genetic matters of CYP2D6 in breast cancer: copy number variations and nucleotide polymorphisms. Nat. Rev. Cancer. 2009;9:842. doi: 10.1038/nrc2683-c1. [DOI] [PubMed] [Google Scholar]
- 29.Jin Y., Desta Z., Stearns V., Ward B., Ho H., Lee K.-H., Skaar T., Storniolo A.M., Li L., Araba A., et al. CYP2D6 Genotype, Antidepressant Use, and Tamoxifen Metabolism During Adjuvant Breast Cancer Treatment. J. Natl. Cancer Inst. 2005;97:30–39. doi: 10.1093/jnci/dji005. [DOI] [PubMed] [Google Scholar]
- 30.Brooks J.D., Comen E.A., Reiner A.S., Orlow I., Leong S.F., Liang X., Mellemkjær L., Knight J.A., Lynch C.F., John E.M., et al. CYP2D6 phenotype, tamoxifen, and risk of contralateral breast cancer in the WECARE Study. Breast Cancer Res. 2018;20:149. doi: 10.1186/s13058-018-1083-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chan C.W.H., Law B.M.H., So W.K.W., Chow K.M., Waye M.M.Y. Pharmacogenomics of breast cancer: highlighting CYP2D6 and tamoxifen. J. Cancer Res. Clin. Oncol. 2020;146:1395–1404. doi: 10.1007/s00432-020-03206-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Juan-Carlos P.-D.M., Perla-Lidia P.-P., Stephanie-Talia M.-M., Mónica-Griselda A.-M., Luz-María T.-E. ABC transporter superfamily. An updated overview, relevance in cancer multidrug resistance and perspectives with personalized medicine. Mol. Biol. Rep. 2021;48:1883–1901. doi: 10.1007/s11033-021-06155-w. [DOI] [PubMed] [Google Scholar]
- 33.Lu Y., Chan Y.-T., Tan H.-Y., Li S., Wang N., Feng Y. Epigenetic regulation in human cancer: the potential role of epi-drug in cancer therapy. Mol. Cancer. 2020;19:79. doi: 10.1186/s12943-020-01197-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jin M.L., Jeong K.W. Histone modifications in drug-resistant cancers: From a cancer stem cell and immune evasion perspective. Exp. Mol. Med. 2023;55:1333–1347. doi: 10.1038/s12276-023-01014-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou X., Ao X., Jia Z., Li Y., Kuang S., Du C., Zhang J., Wang J., Liu Y. Non-coding RNA in cancer drug resistance: Underlying mechanisms and clinical applications. Front. Oncol. 2022;12 doi: 10.3389/fonc.2022.951864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dong B., Qiu Z., Wu Y. Tackle Epithelial-Mesenchymal Transition With Epigenetic Drugs in Cancer. Front. Pharmacol. 2020;11 doi: 10.3389/fphar.2020.596239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mao X., Xu J., Wang W., Liang C., Hua J., Liu J., Zhang B., Meng Q., Yu X., Shi S. Crosstalk between cancer-associated fibroblasts and immune cells in the tumor microenvironment: new findings and future perspectives. Mol. Cancer. 2021;20:131. doi: 10.1186/s12943-021-01428-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ayob A.Z., Ramasamy T.S. Cancer stem cells as key drivers of tumour progression. J. Biomed. Sci. 2018;25:20. doi: 10.1186/s12929-018-0426-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marofi F., Motavalli R., Safonov V.A., Thangavelu L., Yumashev A.V., Alexander M., Shomali N., Chartrand M.S., Pathak Y., Jarahian M., et al. CAR T cells in solid tumors: challenges and opportunities. Stem Cell Res. Ther. 2021;12:81. doi: 10.1186/s13287-020-02128-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maibach F., Sadozai H., Seyed Jafari S.M., Hunger R.E., Schenk M. Tumor-Infiltrating Lymphocytes and Their Prognostic Value in Cutaneous Melanoma. Front. Immunol. 2020;11 doi: 10.3389/fimmu.2020.02105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang S., Sun J., Chen K., Ma P., Lei Q., Xing S., Cao Z., Sun S., Yu Z., Liu Y., Li N. Perspectives of tumor-infiltrating lymphocyte treatment in solid tumors. BMC Med. 2021;19:140. doi: 10.1186/s12916-021-02006-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mitchison T.J. So many ways to naturally kill a cancer cell. BMC Biol. 2021;19:149. doi: 10.1186/s12915-021-01092-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Choi C., Finlay D.K. Optimising NK cell metabolism to increase the efficacy of cancer immunotherapy. Stem Cell Res. Ther. 2021;12:320. doi: 10.1186/s13287-021-02377-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Raskov H., Orhan A., Christensen J.P., Gögenur I. Cytotoxic CD8+ T cells in cancer and cancer immunotherapy. Br. J. Cancer. 2021;124:359–367. doi: 10.1038/s41416-020-01048-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lemoine J., Ruella M., Houot R. Born to survive: how cancer cells resist CAR T cell therapy. J. Hematol. Oncol. 2021;14:199. doi: 10.1186/s13045-021-01209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li Y., He H., Jihu R., Zhou J., Zeng R., Yan H. Novel Characterization of Myeloid-Derived Suppressor Cells in Tumor Microenvironment. Front. Cell Dev. Biol. 2021;9 doi: 10.3389/fcell.2021.698532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhao Y., Du J., Shen X. Targeting myeloid-derived suppressor cells in tumor immunotherapy: Current, future and beyond. Front. Immunol. 2023;14 doi: 10.3389/fimmu.2023.1157537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mi Y., Guo N., Luan J., Cheng J., Hu Z., Jiang P., Jin W., Gao X. The Emerging Role of Myeloid-Derived Suppressor Cells in the Glioma Immune Suppressive Microenvironment. Front. Immunol. 2020;11 doi: 10.3389/fimmu.2020.00737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shan J., Han D., Shen C., Lei Q., Zhang Y. Mechanism and strategies of immunotherapy resistance in colorectal cancer. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.1016646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gao J., Liang Y., Wang L. Shaping Polarization Of Tumor-Associated Macrophages In Cancer Immunotherapy. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.888713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee C., Jeong H., Bae Y., Shin K., Kang S., Kim H., Oh J., Bae H. Targeting of M2-like tumor-associated macrophages with a melittin-based pro-apoptotic peptide. J. Immunother. Cancer. 2019;7:147. doi: 10.1186/s40425-019-0610-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Scott E.N., Gocher A.M., Workman C.J., Vignali D.A.A. Regulatory T Cells: Barriers of Immune Infiltration Into the Tumor Microenvironment. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.702726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Saida Y., Watanabe S., Koyama S., Togashi Y., Kikuchi T. Editorial: Strategies to overcome tumor evasion and resistance to immunotherapies by targeting immune suppressor cells. Front. Oncol. 2023;13 doi: 10.3389/fonc.2023.1240926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ramos R.N., Amano M.T., Paes Leme A.F., Fox J.W., de Oliveira A.K. Editorial: Tumor microenvironment (TME) and tumor immune microenvironment (TIME): New perspectives for prognosis and therapy. Front. Cell Dev. Biol. 2022;10 doi: 10.3389/fcell.2022.971275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ciccolini J., Milano G. Immune check points in cancer treatment: current challenges and perspectives. Br. J. Cancer. 2023;129:1365–1366. doi: 10.1038/s41416-023-02478-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Marei H.E., Hasan A., Pozzoli G., Cenciarelli C. Cancer immunotherapy with immune checkpoint inhibitors (ICIs): potential, mechanisms of resistance, and strategies for reinvigorating T cell responsiveness when resistance is acquired. Cancer Cell Int. 2023;23:64. doi: 10.1186/s12935-023-02902-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lu J., Jiang G. The journey of CAR-T therapy in hematological malignancies. Mol. Cancer. 2022;21:194. doi: 10.1186/s12943-022-01663-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hemminki O., dos Santos J.M., Hemminki A. Oncolytic viruses for cancer immunotherapy. J. Hematol. Oncol. 2020;13:84. doi: 10.1186/s13045-020-00922-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Baghban R., Roshangar L., Jahanban-Esfahlan R., Seidi K., Ebrahimi-Kalan A., Jaymand M., Kolahian S., Javaheri T., Zare P. Tumor microenvironment complexity and therapeutic implications at a glance. Cell Commun. Signal. 2020;18:59. doi: 10.1186/s12964-020-0530-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mun J.-Y., Leem S.-H., Lee J.H., Kim H.S. Dual Relationship Between Stromal Cells and Immune Cells in the Tumor Microenvironment. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.864739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gilazieva Z., Ponomarev A., Rizvanov A., Solovyeva V. The Dual Role of Mesenchymal Stromal Cells and Their Extracellular Vesicles in Carcinogenesis. Biology. 2022;11 doi: 10.3390/biology11060813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhao Y., Shen M., Wu L., Yang H., Yao Y., Yang Q., Du J., Liu L., Li Y., Bai Y. Stromal cells in the tumor microenvironment: accomplices of tumor progression? Cell Death Dis. 2023;14:587. doi: 10.1038/s41419-023-06110-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lan T., Luo M., Wei X. Mesenchymal stem/stromal cells in cancer therapy. J. Hematol. Oncol. 2021;14:195. doi: 10.1186/s13045-021-01208-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lin Z., Wu Y., Xu Y., Li G., Li Z., Liu T. Mesenchymal stem cell-derived exosomes in cancer therapy resistance: recent advances and therapeutic potential. Mol. Cancer. 2022;21:179. doi: 10.1186/s12943-022-01650-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Loh J.J., Ma S. The Role of Cancer-Associated Fibroblast as a Dynamic Player in Mediating Cancer Stemness in the Tumor Microenvironment. Front. Cell Dev. Biol. 2021;9 doi: 10.3389/fcell.2021.727640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Feng B., Wu J., Shen B., Jiang F., Feng J. Cancer-associated fibroblasts and resistance to anticancer therapies: status, mechanisms, and countermeasures. Cancer Cell Int. 2022;22:166. doi: 10.1186/s12935-022-02599-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Herrera M., Berral-González A., López-Cade I., Galindo-Pumariño C., Bueno-Fortes S., Martín-Merino M., Carrato A., Ocaña A., De La Pinta C., López-Alfonso A., et al. Cancer-associated fibroblast-derived gene signatures determine prognosis in colon cancer patients. Mol. Cancer. 2021;20:73. doi: 10.1186/s12943-021-01367-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ebeling S., Kowalczyk A., Perez-Vazquez D., Mattiola I. Regulation of tumor angiogenesis by the crosstalk between innate immunity and endothelial cells. Front. Oncol. 2023;13 doi: 10.3389/fonc.2023.1171794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bani M., Decio A., Giavazzi R., Ghilardi C. Contribution of tumor endothelial cells to drug resistance: anti-angiogenic tyrosine kinase inhibitors act as p-glycoprotein antagonists. Angiogenesis. 2017;20:233–241. doi: 10.1007/s10456-017-9549-6. [DOI] [PubMed] [Google Scholar]
- 70.Nagl L., Horvath L., Pircher A., Wolf D. Tumor Endothelial Cells (TECs) as Potential Immune Directors of the Tumor Microenvironment – New Findings and Future Perspectives. Front. Cell Dev. Biol. 2020;8 doi: 10.3389/fcell.2020.00766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Khalaf K., Hana D., Chou J.T.-T., Singh C., Mackiewicz A., Kaczmarek M. Aspects of the Tumor Microenvironment Involved in Immune Resistance and Drug Resistance. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.656364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kumari N., Choi S.H. Tumor-associated macrophages in cancer: recent advancements in cancer nanoimmunotherapies. J. Exp. Clin. Cancer Res. 2022;41:68. doi: 10.1186/s13046-022-02272-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zalpoor H., Aziziyan F., Liaghat M., Bakhtiyari M., Akbari A., Nabi-Afjadi M., Forghaniesfidvajani R., Rezaei N. The roles of metabolic profiles and intracellular signaling pathways of tumor microenvironment cells in angiogenesis of solid tumors. Cell Commun. Signal. 2022;20:186. doi: 10.1186/s12964-022-00951-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Silva A.C., Lobo J.M.S. In: Current Applications of Pharmaceutical Biotechnology. Silva A.C., Moreira J.N., Lobo J.M.S., Almeida H., editors. Springer International Publishing; 2020. Cytokines and Growth Factors; pp. 87–113. [DOI] [Google Scholar]
- 75.Kusmartsev S., Gabrilovich D.I. Effect of tumor-derived cytokines and growth factors on differentiation and immune suppressive features of myeloid cells in cancer. Cancer Metastasis Rev. 2006;25:323–331. doi: 10.1007/s10555-006-9002-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Qin R., Ren W., Ya G., Wang B., He J., Ren S., Jiang L., Zhao S. Role of chemokines in the crosstalk between tumor and tumor-associated macrophages. Clin. Exp. Med. 2023;23:1359–1373. doi: 10.1007/s10238-022-00888-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Vishnoi M., Upadhyay R., Cho W.C. Editorial: The role of the extracellular matrix in tumor progression and therapeutic resistance. Front. Mol. Biosci. 2022;9 doi: 10.3389/fmolb.2022.994506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yuan Z., Li Y., Zhang S., Wang X., Dou H., Yu X., Zhang Z., Yang S., Xiao M. Extracellular matrix remodeling in tumor progression and immune escape: from mechanisms to treatments. Mol. Cancer. 2023;22:48. doi: 10.1186/s12943-023-01744-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jiang Y., Zhang H., Wang J., Liu Y., Luo T., Hua H. Targeting extracellular matrix stiffness and mechanotransducers to improve cancer therapy. J. Hematol. Oncol. 2022;15:34. doi: 10.1186/s13045-022-01252-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bao M.H.-R., Wong C.C.-L. Hypoxia, Metabolic Reprogramming, and Drug Resistance in Liver Cancer. Cells. 2021;10:1715. doi: 10.3390/cells10071715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.McDonald P.C., Chafe S.C., Dedhar S. Overcoming Hypoxia-Mediated Tumor Progression: Combinatorial Approaches Targeting pH Regulation, Angiogenesis and Immune Dysfunction. Front. Cell Dev. Biol. 2016;4 doi: 10.3389/fcell.2016.00027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jing X., Yang F., Shao C., Wei K., Xie M., Shen H., Shu Y. Role of hypoxia in cancer therapy by regulating the tumor microenvironment. Mol. Cancer. 2019;18:157. doi: 10.1186/s12943-019-1089-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ruan C., Su K., Zhao D., Lu A., Zhong C. Nanomaterials for Tumor Hypoxia Relief to Improve the Efficacy of ROS-Generated Cancer Therapy. Front. Chem. 2021;9 doi: 10.3389/fchem.2021.649158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tiwari A., Trivedi R., Lin S.-Y. Tumor microenvironment: barrier or opportunity towards effective cancer therapy. J. Biomed. Sci. 2022;29:83. doi: 10.1186/s12929-022-00866-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Butti R., Das S., Gunasekaran V.P., Yadav A.S., Kumar D., Kundu G.C. Receptor tyrosine kinases (RTKs) in breast cancer: signaling, therapeutic implications and challenges. Mol. Cancer. 2018;17:34. doi: 10.1186/s12943-018-0797-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sudhesh D.,S., Zainal Abidin S.A., Farghadani R., Othman I., Naidu R. Receptor Tyrosine Kinases and Their Signaling Pathways as Therapeutic Targets of Curcumin in Cancer. Front. Pharmacol. 2021;12:772510. doi: 10.3389/fphar.2021.772510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Du Z., Lovly C.M. Mechanisms of receptor tyrosine kinase activation in cancer. Mol. Cancer. 2018;17:58. doi: 10.1186/s12943-018-0782-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Engelman J.A., Jänne P.A. Mechanisms of acquired resistance to epidermal growth factor receptor tyrosine kinase inhibitors in non-small cell lung cancer. Clin. Cancer Res. 2008;14:2895–2899. doi: 10.1158/1078-0432.Ccr-07-2248. [DOI] [PubMed] [Google Scholar]
- 89.Singh H., Kang A., Bloudek L., Hsu L.-I., Corinna Palanca-Wessels M., Stecher M., Siadak M., Ng K. Systematic Literature Review and Meta-Analysis of HER2 Amplification, Overexpression, and Positivity in Colorectal Cancer. JNCI Cancer Spectr. 2024;8 doi: 10.1093/jncics/pkad082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wu X., Yang H., Yu X., Qin J.-J. Drug-resistant HER2-positive breast cancer: Molecular mechanisms and overcoming strategies. Front. Pharmacol. 2022;13 doi: 10.3389/fphar.2022.1012552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mouabbi J.A., Singareeka Raghavendra A., Bassett R.L., Jr., Hassan A., Tripathy D., Layman R.M. Survival Outcomes in Patients With Hormone Receptor–Positive Metastatic Breast Cancer With Low or No ERBB2 Expression Treated With Targeted Therapies Plus Endocrine Therapy. JAMA Netw. Open. 2023;6 doi: 10.1001/jamanetworkopen.2023.13017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bose R., Ma C.X. Breast cancer, HER2 mutations, and overcoming drug resistance. N. Engl. J. Med. 2021;385:1241–1243. doi: 10.1056/NEJMcibr2110552. [DOI] [PubMed] [Google Scholar]
- 93.Ocaña A., Amir E., Pandiella A. HER2 heterogeneity and resistance to anti-HER2 antibody-drug conjugates. Breast Cancer Res. 2020;22:15. doi: 10.1186/s13058-020-1252-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ianza A., Sirico M., Bernocchi O., Generali D. Role of the IGF-1 Axis in Overcoming Resistance in Breast Cancer. Front. Cell Dev. Biol. 2021;9 doi: 10.3389/fcell.2021.641449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rieger L., O’Connor R. Controlled Signaling—Insulin-Like Growth Factor Receptor Endocytosis and Presence at Intracellular Compartments. Front. Endocrinol. 2020;11 doi: 10.3389/fendo.2020.620013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Takada Y., Takada Y.K., Fujita M. Crosstalk between insulin-like growth factor (IGF) receptor and integrins through direct integrin binding to IGF1. Cytokine Growth Factor Rev. 2017;34:67–72. doi: 10.1016/j.cytogfr.2017.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pang X., He X., Qiu Z., Zhang H., Xie R., Liu Z., Gu Y., Zhao N., Xiang Q., Cui Y. Targeting integrin pathways: mechanisms and advances in therapy. Signal Transduct. Target. Ther. 2023;8:1. doi: 10.1038/s41392-022-01259-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhu S., Zhang T., Zheng L., Liu H., Song W., Liu D., Li Z., Pan C.-x. Combination strategies to maximize the benefits of cancer immunotherapy. J. Hematol. Oncol. 2021;14:156. doi: 10.1186/s13045-021-01164-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Montor W.R., Salas A.R.O.S.E., Melo F.H.M.d. Receptor tyrosine kinases and downstream pathways as druggable targets for cancer treatment: the current arsenal of inhibitors. Mol. Cancer. 2018;17:55. doi: 10.1186/s12943-018-0792-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Du Y., Zhang P., Liu W., Tian J. Optical Imaging of Epigenetic Modifications in Cancer: A Systematic Review. Phenomics. 2022;2:88–101. doi: 10.1007/s43657-021-00041-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Peng Y., Wang Y., Zhou C., Mei W., Zeng C. PI3K/Akt/mTOR Pathway and Its Role in Cancer Therapeutics: Are We Making Headway? Front. Oncol. 2022;12:819128. doi: 10.3389/fonc.2022.819128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lee S., Rauch J., Kolch W. Targeting MAPK Signaling in Cancer: Mechanisms of Drug Resistance and Sensitivity. Int. J. Mol. Sci. 2020;21:1102. doi: 10.3390/ijms21031102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Burris H.A. Overcoming acquired resistance to anticancer therapy: focus on the PI3K/AKT/mTOR pathway. Cancer Chemother. Pharmacol. 2013;71:829–842. doi: 10.1007/s00280-012-2043-3. [DOI] [PubMed] [Google Scholar]
- 104.Zhang Z., Richmond A. The Role of PI3K Inhibition in the Treatment of Breast Cancer, Alone or Combined With Immune Checkpoint Inhibitors. Front. Mol. Biosci. 2021;8 doi: 10.3389/fmolb.2021.648663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Tufail M., Hu J.-J., Liang J., He C.-Y., Wan W.-D., Huang Y.-Q., Jiang C.-H., Wu H., Li N. Predictive, preventive, and personalized medicine in breast cancer: targeting the PI3K pathway. J. Transl. Med. 2024;22:15. doi: 10.1186/s12967-023-04841-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Yang J., Nie J., Ma X., Wei Y., Peng Y., Wei X. Targeting PI3K in cancer: mechanisms and advances in clinical trials. Mol. Cancer. 2019;18:26. doi: 10.1186/s12943-019-0954-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Hua H., Zhang H., Chen J., Wang J., Liu J., Jiang Y. Targeting Akt in cancer for precision therapy. J. Hematol. Oncol. 2021;14:128. doi: 10.1186/s13045-021-01137-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ali E.S., Mitra K., Akter S., Ramproshad S., Mondal B., Khan I.N., Islam M.T., Sharifi-Rad J., Calina D., Cho W.C. Recent advances and limitations of mTOR inhibitors in the treatment of cancer. Cancer Cell Int. 2022;22:284. doi: 10.1186/s12935-022-02706-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zou Z., Tao T., Li H., Zhu X. mTOR signaling pathway and mTOR inhibitors in cancer: progress and challenges. Cell Biosci. 2020;10:31. doi: 10.1186/s13578-020-00396-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sun K., Jin L., Karolová J., Vorwerk J., Hailfinger S., Opalka B., Zapukhlyak M., Lenz G., Khandanpour C. Combination Treatment Targeting mTOR and MAPK Pathways Has Synergistic Activity in Multiple Myeloma. Cancers. 2023;15:2373. doi: 10.3390/cancers15082373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Vanhaesebroeck B., Bilanges B., Madsen R.R., Dale K.L., Lau E., Vladimirou E. Perspective: Potential Impact and Therapeutic Implications of Oncogenic PI3K Activation on Chromosomal Instability. Biomolecules. 2019;9 doi: 10.3390/biom9080331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Glaviano A., Foo A.S.C., Lam H.Y., Yap K.C.H., Jacot W., Jones R.H., Eng H., Nair M.G., Makvandi P., Geoerger B., et al. PI3K/AKT/mTOR signaling transduction pathway and targeted therapies in cancer. Mol. Cancer. 2023;22:138. doi: 10.1186/s12943-023-01827-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Mardanshahi A., Gharibkandi N.A., Vaseghi S., Abedi S.M., Molavipordanjani S. The PI3K/AKT/mTOR signaling pathway inhibitors enhance radiosensitivity in cancer cell lines. Mol. Biol. Rep. 2021;48:1–14. doi: 10.1007/s11033-021-06607-3. [DOI] [PubMed] [Google Scholar]
- 114.Zhang Y., Wang X. Targeting the Wnt/β-catenin signaling pathway in cancer. J. Hematol. Oncol. 2020;13:165. doi: 10.1186/s13045-020-00990-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Chen C., Luo L., Xu C., Yang X., Liu T., Luo J., Shi W., Yang L., Zheng Y., Yang J. Tumor specificity of WNT ligands and receptors reveals universal squamous cell carcinoma oncogenes. BMC Cancer. 2022;22:790. doi: 10.1186/s12885-022-09898-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Robinson K.F., Narasipura S.D., Wallace J., Ritz E.M., Al-Harthi L. β-Catenin and TCFs/LEF signaling discordantly regulate IL-6 expression in astrocytes. Cell Commun. Signal. 2020;18:93. doi: 10.1186/s12964-020-00565-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.De Las Rivas J., Brozovic A., Izraely S., Casas-Pais A., Witz I.P., Figueroa A. Cancer drug resistance induced by EMT: novel therapeutic strategies. Arch. Toxicol. 2021;95:2279–2297. doi: 10.1007/s00204-021-03063-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Leung R.W.H., Lee T.K.W. Wnt/β-Catenin Signaling as a Driver of Stemness and Metabolic Reprogramming in Hepatocellular Carcinoma. Cancers. 2022;14:5468. doi: 10.3390/cancers14215468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.G Lindström H.J., Friedman R. The effects of combination treatments on drug resistance in chronic myeloid leukaemia: an evaluation of the tyrosine kinase inhibitors axitinib and asciminib. BMC Cancer. 2020;20:397. doi: 10.1186/s12885-020-06782-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kishore C., Zi X. Wnt Signaling and Therapeutic Resistance in Castration-Resistant Prostate Cancer. Curr. Pharmacol. Rep. 2023;9:261–274. doi: 10.1007/s40495-023-00333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Sharma A., Mir R., Galande S. Epigenetic Regulation of the Wnt/β-Catenin Signaling Pathway in Cancer. Front. Genet. 2021;12 doi: 10.3389/fgene.2021.681053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Patel S., Alam A., Pant R., Chattopadhyay S. Wnt Signaling and Its Significance Within the Tumor Microenvironment: Novel Therapeutic Insights. Front. Immunol. 2019;10 doi: 10.3389/fimmu.2019.02872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Lee S., Rauch J., Kolch W. Targeting MAPK Signaling in Cancer: Mechanisms of Drug Resistance and Sensitivity. Int. J. Mol. Sci. 2020;21 doi: 10.3390/ijms21031102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Usta S.Z., Uchihashi T., Kodama S., Kurioka K., Inubushi T., Shimooka T., Sugauchi A., Seki S., Tanaka S. Current Status and Molecular Mechanisms of Resistance to Immunotherapy in Oral Malignant Melanoma. Int. J. Mol. Sci. 2023;24 doi: 10.3390/ijms242417282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Bai R., Chen N., Li L., Du N., Bai L., Lv Z., Tian H., Cui J. Mechanisms of Cancer Resistance to Immunotherapy. Front. Oncol. 2020;10 doi: 10.3389/fonc.2020.01290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Tufail M. Unlocking the potential of the tumor microenvironment for cancer therapy. Pathol. Res. Pract. 2023;251 doi: 10.1016/j.prp.2023.154846. [DOI] [PubMed] [Google Scholar]
- 127.Liang L., Sun F., Wang H., Hu Z. Metabolomics, metabolic flux analysis and cancer pharmacology. Pharmacol. Ther. 2021;224 doi: 10.1016/j.pharmthera.2021.107827. [DOI] [PubMed] [Google Scholar]
- 128.Han F., Li C.-F., Cai Z., Zhang X., Jin G., Zhang W.-N., Xu C., Wang C.-Y., Morrow J., Zhang S., et al. The critical role of AMPK in driving Akt activation under stress, tumorigenesis and drug resistance. Nat. Commun. 2018;9:4728. doi: 10.1038/s41467-018-07188-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Valle-Mendiola A., Gutiérrez-Hoya A., Soto-Cruz I. JAK/STAT Signaling and Cervical Cancer: From the Cell Surface to the Nucleus. Genes. 2023;14:1141. doi: 10.3390/genes14061141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Liang D., Wang Q., Zhang W., Tang H., Song C., Yan Z., Liang Y., Wang H. JAK/STAT in leukemia: a clinical update. Mol. Cancer. 2024;23:25. doi: 10.1186/s12943-023-01929-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.2023. https://www.imt.ie/news/cancer-incidence-and-survival-rates-rise-in-children-and-young-people-01-09-2023/
- 132.Owen K.L., Brockwell N.K., Parker B.S. JAK-STAT Signaling: A Double-Edged Sword of Immune Regulation and Cancer Progression. Cancers. 2019;11 doi: 10.3390/cancers11122002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Xiong H., Zhang Z.G., Tian X.Q., Sun D.F., Liang Q.C., Zhang Y.J., Lu R., Chen Y.X., Fang J.Y. Inhibition of JAK1, 2/STAT3 signaling induces apoptosis, cell cycle arrest, and reduces tumor cell invasion in colorectal cancer cells. Neoplasia. 2008;10:287–297. doi: 10.1593/neo.07971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Stevens L.E., Peluffo G., Qiu X., Temko D., Fassl A., Li Z., Trinh A., Seehawer M., Jovanović B., Alečković M., et al. JAK-STAT Signaling in Inflammatory Breast Cancer Enables Chemotherapy-Resistant Cell States. Cancer Res. 2023;83:264–284. doi: 10.1158/0008-5472.Can-22-0423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Hu Q., Bian Q., Rong D., Wang L., Song J., Huang H.S., Zeng J., Mei J., Wang P.Y. JAK/STAT pathway: Extracellular signals, diseases, immunity, and therapeutic regimens. Front. Bioeng. Biotechnol. 2023;11 doi: 10.3389/fbioe.2023.1110765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Rah B., Rather R.A., Bhat G.R., Baba A.B., Mushtaq I., Farooq M., Yousuf T., Dar S.B., Parveen S., Hassan R., et al. JAK/STAT Signaling: Molecular Targets, Therapeutic Opportunities, and Limitations of Targeted Inhibitions in Solid Malignancies. Front. Pharmacol. 2022;13 doi: 10.3389/fphar.2022.821344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Mengie Ayele T., Tilahun Muche Z., Behaile Teklemariam A., Bogale Kassie A., Chekol Abebe E. Role of JAK2/STAT3 Signaling Pathway in the Tumorigenesis, Chemotherapy Resistance, and Treatment of Solid Tumors: A Systemic Review. J. Inflamm. Res. 2022;15:1349–1364. doi: 10.2147/jir.s353489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Zhu J., Li Y., Lv X. IL4I1 enhances PD-L1 expression through JAK/STAT signaling pathway in lung adenocarcinoma. Immunogenetics. 2023;75:17–25. doi: 10.1007/s00251-022-01275-4. [DOI] [PubMed] [Google Scholar]
- 139.Qian S., Wei Z., Yang W., Huang J., Yang Y., Wang J. The role of BCL-2 family proteins in regulating apoptosis and cancer therapy. Front. Oncol. 2022;12 doi: 10.3389/fonc.2022.985363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Yao Y., Gong J.-N., Segal D., Riffkin C.D., van Delft M.F., Roberts A.W., Huang D.C. The Role of BAX/BAK-Mediated Apoptosis for the Cytotoxic Action of Anti-Myeloma Agents. Blood. 2016;128:5706. doi: 10.1182/blood.V128.22.5706.5706. [DOI] [Google Scholar]
- 141.Roberts A.W. Therapeutic development and current uses of BCL-2 inhibition. Hematology. 2020;2020:1–9. doi: 10.1182/hematology.2020000154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Fiebig A.A., Zhu W., Hollerbach C., Leber B., Andrews D.W. Bcl-XL is qualitatively different from and ten times more effective than Bcl-2 when expressed in a breast cancer cell line. BMC Cancer. 2006;6:213. doi: 10.1186/1471-2407-6-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Kaloni D., Diepstraten S.T., Strasser A., Kelly G.L. BCL-2 protein family: attractive targets for cancer therapy. Apoptosis. 2023;28:20–38. doi: 10.1007/s10495-022-01780-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Liu L., Cheng X., Yang H., Lian S., Jiang Y., Liang J., Chen X., Mo S., Shi Y., Zhao S., et al. BCL-2 expression promotes immunosuppression in chronic lymphocytic leukemia by enhancing regulatory T cell differentiation and cytotoxic T cell exhaustion. Mol. Cancer. 2022;21:59. doi: 10.1186/s12943-022-01516-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Gao X., Zeng H., Zhao X., Wu H., Yan M., Li Y., Zhang G., Sun F. Efficacy and safety of venetoclax in patients with relapsed/refractory multiple myeloma: a meta-analysis. BMC Cancer. 2023;23:1058. doi: 10.1186/s12885-023-11553-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Klymenko T., Brandenburg M., Morrow C., Dive C., Makin G. The novel Bcl-2 inhibitor ABT-737 is more effective in hypoxia and is able to reverse hypoxia-induced drug resistance in neuroblastoma cells. Mol. Cancer Ther. 2011;10:2373–2383. doi: 10.1158/1535-7163.mct-11-0326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Ploumaki I., Triantafyllou E., Koumprentziotis I.-A., Karampinos K., Drougkas K., Karavolias I., Trontzas I., Kotteas E.A. Bcl-2 pathway inhibition in solid tumors: a review of clinical trials. Clin. Transl. Oncol. 2023;25:1554–1578. doi: 10.1007/s12094-022-03070-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Adams C.M., Clark-Garvey S., Porcu P., Eischen C.M. Targeting the Bcl-2 Family in B Cell Lymphoma. Front. Oncol. 2018;8 doi: 10.3389/fonc.2018.00636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Li M. The role of P53 up-regulated modulator of apoptosis (PUMA) in ovarian development, cardiovascular and neurodegenerative diseases. Apoptosis. 2021;26:235–247. doi: 10.1007/s10495-021-01667-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Hao Q., Chen J., Lu H., Zhou X. The ARTS of p53-dependent mitochondrial apoptosis. J. Mol. Cell Biol. 2023;14 doi: 10.1093/jmcb/mjac074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Xu Y., Ye H. Progress in understanding the mechanisms of resistance to BCL-2 inhibitors. Exp. Hematol. Oncol. 2022;11:31. doi: 10.1186/s40164-022-00283-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Tuomela K., Ambrose A.R., Davis D.M. Escaping Death: How Cancer Cells and Infected Cells Resist Cell-Mediated Cytotoxicity. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.867098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Kari S., Subramanian K., Altomonte I.A., Murugesan A., Yli-Harja O., Kandhavelu M. Programmed cell death detection methods: a systematic review and a categorical comparison. Apoptosis. 2022;27:482–508. doi: 10.1007/s10495-022-01735-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Guicciardi M.E., Gores G.J. Essentials of apoptosis: a guide for basic and clinical research. Springer; 2003. The death receptor family and the extrinsic pathway; pp. 67–84. [Google Scholar]
- 155.Xiao W., Ibrahim M.L., Redd P.S., Klement J.D., Lu C., Yang D., Savage N.M., Liu K. Loss of Fas expression and function is coupled with colon cancer resistance to immune checkpoint inhibitor immunotherapy. Mol. Cancer Res. 2019;17:420–430. doi: 10.1158/1541-7786.MCR-18-0455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Jiang N., Dai Q., Su X., Fu J., Feng X., Peng J. Role of PI3K/AKT pathway in cancer: the framework of malignant behavior. Mol. Biol. Rep. 2020;47:4587–4629. doi: 10.1007/s11033-020-05435-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Zaitseva O., Anany M., Wajant H., Lang I. Basic characterization of antibodies targeting receptors of the tumor necrosis factor receptor superfamily. Front. Immunol. 2023;14 doi: 10.3389/fimmu.2023.1115667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Lim J.L., Augustinus R., Plomp J.J., Roya-Kouchaki K., Vergoossen D.L.E., Fillié-Grijpma Y., Struijk J., Thomas R., Salvatori D., Steyaert C., et al. Development and characterization of agonistic antibodies targeting the Ig-like 1 domain of MuSK. Sci. Rep. 2023;13:7478. doi: 10.1038/s41598-023-32641-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Zheng Y., Ma L., Sun Q. Clinically-Relevant ABC Transporter for Anti-Cancer Drug Resistance. Front. Pharmacol. 2021;12:648407. doi: 10.3389/fphar.2021.648407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Guo B., Xu Z., Yan X., Buttino I., Li J., Zhou C., Qi P. Novel ABCB1 and ABCC Transporters Are Involved in the Detoxification of Benzo(α)pyrene in Thick Shell Mussel, Mytilus coruscus. Front. Mar. Sci. 2020;7 doi: 10.3389/fmars.2020.00119. [DOI] [Google Scholar]
- 161.Theile D., Wizgall P. Acquired ABC-transporter overexpression in cancer cells: transcriptional induction or Darwinian selection? Naunyn-Schmiedeberg’s Arch. Pharmacol. 2021;394:1621–1632. doi: 10.1007/s00210-021-02112-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Fletcher J.I., Williams R.T., Henderson M.J., Norris M.D., Haber M. ABC transporters as mediators of drug resistance and contributors to cancer cell biology. Drug Resist. Updat. 2016;26:1–9. doi: 10.1016/j.drup.2016.03.001. [DOI] [PubMed] [Google Scholar]
- 163.Xiao H., Zheng Y., Ma L., Tian L., Sun Q. Clinically-Relevant ABC Transporter for Anti-Cancer Drug Resistance. Front. Pharmacol. 2021;12 doi: 10.3389/fphar.2021.648407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Sharom F.J. ABC multidrug transporters: structure, function and role in chemoresistance. Pharmacogenomics. 2008;9:105–127. doi: 10.2217/14622416.9.1.105. [DOI] [PubMed] [Google Scholar]
- 165.Robey R.W., Pluchino K.M., Hall M.D., Fojo A.T., Bates S.E., Gottesman M.M. Revisiting the role of ABC transporters in multidrug-resistant cancer. Nat. Rev. Cancer. 2018;18:452–464. doi: 10.1038/s41568-018-0005-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Robey R.W., Polgar O., Deeken J., To K.W., Bates S.E. ABCG2: determining its relevance in clinical drug resistance. Cancer Metastasis Rev. 2007;26:39–57. doi: 10.1007/s10555-007-9042-6. [DOI] [PubMed] [Google Scholar]
- 167.He P., Gelissen I.C., Ammit A.J. Regulation of ATP binding cassette transporter A1 (ABCA1) expression: cholesterol-dependent and – independent signaling pathways with relevance to inflammatory lung disease. Respir. Res. 2020;21:250. doi: 10.1186/s12931-020-01515-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Siva V., Ravikumar C.K.R.P., Arasu P.T., Yadav N.N., Murugan A., Yadav H.S., Bahadur S.A., Balamurali S. In: Saravanan M., Barabadi H., editors. Vol. 1. Springer International Publishing; 2021. Cancer Nanotechnology for Drug Targeting and Delivery Approaches; pp. 53–91. (Cancer Nanotheranostics). [DOI] [Google Scholar]
- 169.Sodani K., Patel A., Kathawala R.J., Chen Z.-S. Multidrug resistance associated proteins in multidrug resistance. Chin. J. Cancer. 2012;31:58–72. doi: 10.5732/cjc.011.10329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Puris E., Fricker G., Gynther M. The Role of Solute Carrier Transporters in Efficient Anticancer Drug Delivery and Therapy. Pharmaceutics. 2023;15:364. doi: 10.3390/pharmaceutics15020364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Nimmy M.S., Kumar V., Suthanthiram B., Subbaraya U., Nagar R., Bharadwaj C., Jain P.K., Krishnamurthy P. A Systematic Phylogenomic Classification of the Multidrug and Toxic Compound Extrusion Transporter Gene Family in Plants. Front. Plant Sci. 2022;13 doi: 10.3389/fpls.2022.774885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Kar D., Pradhan A.A., Dutta A., Bhagavatula L., Datta S. In: Plant Metal and Metalloid Transporters. Kumar K., Srivastava S., editors. Springer Nature Singapore; 2022. The Multidrug and Toxic Compound Extrusion (MATE) Family in Plants and Their Significance in Metal Transport; pp. 151–177. [DOI] [Google Scholar]
- 173.dos Santos S.C., Teixeira M.C., Dias P.J., Sá-Correia I. MFS transporters required for multidrug/multixenobiotic (MD/MX) resistance in the model yeast: understanding their physiological function through post-genomic approaches. Front. Physiol. 2014;5 doi: 10.3389/fphys.2014.00180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Zhang H., Han X., Wang Z., Wang Z., Cui Y., Tian R., Zhu Y., Han B., Liu H., Zuo X., et al. Mitochondrial Breast Cancer Resistant Protein Sustains the Proliferation and Survival of Drug-Resistant Breast Cancer Cells by Regulating Intracellular Reactive Oxygen Species. Front. Cell Dev. Biol. 2021;9 doi: 10.3389/fcell.2021.719209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Zhang J., Wang S., Zhang D., He X., Wang X., Han H., Qin Y. Nanoparticle-based drug delivery systems to enhance cancer immunotherapy in solid tumors. Front. Immunol. 2023;14 doi: 10.3389/fimmu.2023.1230893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Zhu Y., Li X., Wang L., Hong X., Yang J. Metabolic reprogramming and crosstalk of cancer-related fibroblasts and immune cells in the tumor microenvironment. Front. Endocrinol. 2022;13 doi: 10.3389/fendo.2022.988295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Shimi G. Dietary approaches for controlling cancer by limiting the Warburg effect: a review. Nutr. Rev. 2023;1 doi: 10.1093/nutrit/nuad130. [DOI] [PubMed] [Google Scholar]
- 178.Liu C., Jin Y., Fan Z. The Mechanism of Warburg Effect-Induced Chemoresistance in Cancer. Front. Oncol. 2021;11 doi: 10.3389/fonc.2021.698023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Ganapathy-Kanniappan S., Geschwind J.-F.H. Tumor glycolysis as a target for cancer therapy: progress and prospects. Mol. Cancer. 2013;12:152. doi: 10.1186/1476-4598-12-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Kwon Y.-J., Seo E.-B., Jeong A.J., Lee S.-H., Noh K.H., Lee S., Cho C.-H., Lee C.-H., Shin H.M., Kim H.-R., et al. The acidic tumor microenvironment enhances PD-L1 expression via activation of STAT3 in MDA-MB-231 breast cancer cells. BMC Cancer. 2022;22:852. doi: 10.1186/s12885-022-09956-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Peng J., Cui Y., Xu S., Wu X., Huang Y., Zhou W., Wang S., Fu Z., Xie H. Altered glycolysis results in drug-resistant in clinical tumor therapy (Review) Oncol. Lett. 2021;21:369. doi: 10.3892/ol.2021.12630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Varghese E., Samuel S.M., Líšková A., Samec M., Kubatka P., Büsselberg D. Targeting Glucose Metabolism to Overcome Resistance to Anticancer Chemotherapy in Breast Cancer. Cancers. 2020;12:2252. doi: 10.3390/cancers12082252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Jin J., Byun J.-K., Choi Y.-K., Park K.-G. Targeting glutamine metabolism as a therapeutic strategy for cancer. Exp. Mol. Med. 2023;55:706–715. doi: 10.1038/s12276-023-00971-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Gong T., Zheng C., Ou X., Zheng J., Yu J., Chen S., Duan Y., Liu W. Glutamine metabolism in cancers: Targeting the oxidative homeostasis. Front. Oncol. 2022;12 doi: 10.3389/fonc.2022.994672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Golub K., Bai W., Zhang Z., Xiao H., Sun R., Shen J., Sun J. The mechanism and consequences of BRAF inhibitor resistance in melanoma. Genome Instab. Dis. 2023;4:266–274. doi: 10.1007/s42764-023-00105-5. [DOI] [Google Scholar]
- 186.Vander Heiden M.G., Cantley L.C., Thompson C.B. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Tennant D.A., Durán R.V., Gottlieb E. Targeting metabolic transformation for cancer therapy. Nat. Rev. Cancer. 2010;10:267–277. doi: 10.1038/nrc2817. [DOI] [PubMed] [Google Scholar]
- 188.Dutta S., Sahoo S.K. In: Hypoxia in Cancer: Significance and Impact on Cancer Therapy. Mukherjee S., Kanwar J.R., editors. Springer Nature Singapore; 2023. Hypoxic Tumor Microenvironment: Driver for Cancer Progression; pp. 65–88. [DOI] [Google Scholar]
- 189.Sharma A., Sinha S., Shrivastava N. Therapeutic Targeting Hypoxia-Inducible Factor (HIF-1) in Cancer: Cutting Gordian Knot of Cancer Cell Metabolism. Front. Genet. 2022;13 doi: 10.3389/fgene.2022.849040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Su P., Yu L., Mao X., Sun P. Role of HIF-1α/ERRα in Enhancing Cancer Cell Metabolism and Promoting Resistance of Endometrial Cancer Cells to Pyroptosis. Front. Oncol. 2022;12 doi: 10.3389/fonc.2022.881252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Yoshida G.J. Metabolic reprogramming: the emerging concept and associated therapeutic strategies. J. Exp. Clin. Cancer Res. 2015;34:111. doi: 10.1186/s13046-015-0221-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Hellemann E., Walker J.L., Lesko M.A., Chandrashekarappa D.G., Schmidt M.C., O’Donnell A.F., Durrant J.D. Novel mutation in hexokinase 2 confers resistance to 2-deoxyglucose by altering protein dynamics. PLoS Comput. Biol. 2022;18 doi: 10.1371/journal.pcbi.1009929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Yoshida G.J. Metabolic reprogramming: the emerging concept and associated therapeutic strategies. J. Exp. Clin. Cancer Res. 2015;34:111. doi: 10.1186/s13046-015-0221-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Coothankandaswamy V., Cao S., Xu Y., Prasad P.D., Singh P.K., Reynolds C.P., Yang S., Ogura J., Ganapathy V., Bhutia Y.D. Amino acid transporter SLC6A14 is a novel and effective drug target for pancreatic cancer. Br. J. Pharmacol. 2016;173:3292–3306. doi: 10.1111/bph.13616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Vyas S., Zaganjor E., Haigis M.C. Mitochondria and cancer. Cell. 2016;166:555–566. doi: 10.1016/j.cell.2016.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Semenza G.L. Hypoxia-inducible factors in physiology and medicine. Cell. 2012;148:399–408. doi: 10.1016/j.cell.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Ganapathy V., Thangaraju M., Prasad P.D. Nutrient transporters in cancer: relevance to Warburg hypothesis and beyond. Pharmacol. Ther. 2009;121:29–40. doi: 10.1016/j.pharmthera.2008.09.005. [DOI] [PubMed] [Google Scholar]
- 198.Vander Heiden M.G., DeBerardinis R.J. Understanding the intersections between metabolism and cancer biology. Cell. 2017;168:657–669. doi: 10.1016/j.cell.2016.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Jin P., Jiang J., Zhou L., Huang Z., Nice E.C., Huang C., Fu L. Mitochondrial adaptation in cancer drug resistance: prevalence, mechanisms, and management. J. Hematol. Oncol. 2022;15:97. doi: 10.1186/s13045-022-01313-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Foo B.J.-A., Eu J.Q., Hirpara J.L., Pervaiz S. Interplay between Mitochondrial Metabolism and Cellular Redox State Dictates Cancer Cell Survival. Oxid. Med. Cell. Longev. 2021;2021 doi: 10.1155/2021/1341604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Lengauer F., Geisslinger F., Gabriel A., von Schwarzenberg K., Vollmar A.M., Bartel K. A metabolic shift toward glycolysis enables cancer cells to maintain survival upon concomitant glutamine deprivation and V-ATPase inhibition. Front. Nutr. 2023;10 doi: 10.3389/fnut.2023.1124678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Sessions D.T., Kashatus D.F. Mitochondrial dynamics in cancer stem cells. Cell. Mol. Life Sci. 2021;78:3803–3816. doi: 10.1007/s00018-021-03773-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.McInnes J. Insights on altered mitochondrial function and dynamics in the pathogenesis of neurodegeneration. Transl. Neurodegener. 2013;2:12. doi: 10.1186/2047-9158-2-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Cavalcante G.C., Ribeiro-Dos-Santos Â., de Araújo G.S. Mitochondria in tumour progression: a network of mtDNA variants in different types of cancer. BMC Genom. Data. 2022;23:16. doi: 10.1186/s12863-022-01032-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Pérez-Amado C.J., Tovar H., Gómez-Romero L., Beltrán-Anaya F.O., Bautista-Piña V., Dominguez-Reyes C., Villegas-Carlos F., Tenorio-Torres A., Alfaro-Ruíz L.A., Hidalgo-Miranda A., Jiménez-Morales S. Mitochondrial DNA Mutation Analysis in Breast Cancer: Shifting From Germline Heteroplasmy Toward Homoplasmy in Tumors. Front. Oncol. 2020;10:572954. doi: 10.3389/fonc.2020.572954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Fu L., Luo Y.-X., Liu Y., Liu H., Li H.-z., Yu Y. Potential of Mitochondrial Genome Editing for Human Fertility Health. Front. Genet. 2021;12 doi: 10.3389/fgene.2021.673951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Wang W., Scheffler K., Esbensen Y., Strand J.M., Stewart J.B., Bjørås M., Eide L. Addressing RNA integrity to determine the impact of mitochondrial DNA mutations on brain mitochondrial function with age. PLoS One. 2014;9 doi: 10.1371/journal.pone.0096940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Jones S.W., Ball A.L., Chadwick A.E., Alfirevic A. The Role of Mitochondrial DNA Variation in Drug Response: A Systematic Review. Front. Genet. 2021;12 doi: 10.3389/fgene.2021.698825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Dhumal S.N., Choudhari S.K., Patankar S., Ghule S.S., Jadhav Y.B., Masne S. Cancer Stem Cell Markers, CD44 and ALDH1, for Assessment of Cancer Risk in OPMDs and Lymph Node Metastasis in Oral Squamous Cell Carcinoma. Head Neck Pathol. 2022;16:453–465. doi: 10.1007/s12105-021-01384-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Gao J., Yang T., Wang X., Zhang Y., Wang J., Zhang B., Tang D., Liu Y., Gao T., Lin Q., et al. Identification and characterization of a subpopulation of CD133+ cancer stem-like cells derived from SK-UT-1 cells. Cancer Cell Int. 2021;21:157. doi: 10.1186/s12935-021-01817-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Lai X., Guo Y., Chen M., Wei Y., Yi W., Shi Y., Xiong L. Caveolin1: its roles in normal and cancer stem cells. J. Cancer Res. Clin. Oncol. 2021;147:3459–3475. doi: 10.1007/s00432-021-03793-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Lee S.H., Reed-Newman T., Anant S., Ramasamy T.S. Regulatory Role of Quiescence in the Biological Function of Cancer Stem Cells. Stem Cell Rev. Rep. 2020;16:1185–1207. doi: 10.1007/s12015-020-10031-8. [DOI] [PubMed] [Google Scholar]
- 213.Zheng X., Yu C., Xu M. Linking Tumor Microenvironment to Plasticity of Cancer Stem Cells: Mechanisms and Application in Cancer Therapy. Front. Oncol. 2021;11 doi: 10.3389/fonc.2021.678333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Phi L.T.H., Sari I.N., Yang Y.G., Lee S.H., Jun N., Kim K.S., Lee Y.K., Kwon H.Y. Cancer Stem Cells (CSCs) in Drug Resistance and their Therapeutic Implications in Cancer Treatment. Stem Cells Int. 2018;2018 doi: 10.1155/2018/5416923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Clevers H., Nusse R. Wnt/β-catenin signaling and disease. Cell. 2012;149:1192–1205. doi: 10.1016/j.cell.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 216.Vermeulen L., De Sousa E Melo F., Van Der Heijden M., Cameron K., De Jong J.H., Borovski T., Tuynman J.B., Todaro M., Merz C., Rodermond H., et al. Wnt activity defines colon cancer stem cells and is regulated by the microenvironment. Nat. Cell Biol. 2010;12:468–476. doi: 10.1038/ncb2048. [DOI] [PubMed] [Google Scholar]
- 217.Bocci F., Onuchic J.N., Jolly M.K. Understanding the principles of pattern formation driven by Notch signaling by integrating experiments and theoretical models. Front. Physiol. 2020;11:929. doi: 10.3389/fphys.2020.00929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Kovall R.A., Gebelein B., Sprinzak D., Kopan R. The canonical Notch signaling pathway: structural and biochemical insights into shape, sugar, and force. Dev. Cell. 2017;41:228–241. doi: 10.1016/j.devcel.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Glaviano A., Foo A.S.C., Lam H.Y., Yap K.C.H., Jacot W., Jones R.H., Eng H., Nair M.G., Makvandi P., Geoerger B., et al. PI3K/AKT/mTOR signaling transduction pathway and targeted therapies in cancer. Mol. Cancer. 2023;22:138. doi: 10.1186/s12943-023-01827-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Vazquez-Santillan K., Melendez-Zajgla J., Jimenez-Hernandez L., Martínez-Ruiz G., Maldonado V. NF-κB signaling in cancer stem cells: a promising therapeutic target? Cell. Oncol. 2015;38:327–339. doi: 10.1007/s13402-015-0236-6. [DOI] [PubMed] [Google Scholar]
- 221.Li Y.-R., Fang Y., Lyu Z., Zhu Y., Yang L. Exploring the dynamic interplay between cancer stem cells and the tumor microenvironment: implications for novel therapeutic strategies. J. Transl. Med. 2023;21:686. doi: 10.1186/s12967-023-04575-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Huang Y., Hong W., Wei X. The molecular mechanisms and therapeutic strategies of EMT in tumor progression and metastasis. J. Hematol. Oncol. 2022;15:129. doi: 10.1186/s13045-022-01347-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Moon H., Ro S.W. MAPK/ERK Signaling Pathway in Hepatocellular Carcinoma. Cancers. 2021;13 doi: 10.3390/cancers13123026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Clara J.A., Monge C., Yang Y., Takebe N. Targeting signalling pathways and the immune microenvironment of cancer stem cells — a clinical update. Nat. Rev. Clin. Oncol. 2020;17:204–232. doi: 10.1038/s41571-019-0293-2. [DOI] [PubMed] [Google Scholar]
- 225.Han Y., Wang D., Peng L., Huang T., He X., Wang J., Ou C. Single-cell sequencing: a promising approach for uncovering the mechanisms of tumor metastasis. J. Hematol. Oncol. 2022;15:59. doi: 10.1186/s13045-022-01280-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.He J., Zhu S., Liang X., Zhang Q., Luo X., Liu C., Song L. LncRNA as a multifunctional regulator in cancer multi-drug resistance. Mol. Biol. Rep. 2021;48:1–15. doi: 10.1007/s11033-021-06603-7. [DOI] [PubMed] [Google Scholar]
- 227.Hong Y., Zhang Y., Zhao H., Chen H., Yu Q.-Q., Cui H. The roles of lncRNA functions and regulatory mechanisms in the diagnosis and treatment of hepatocellular carcinoma. Front. Cell Dev. Biol. 2022;10 doi: 10.3389/fcell.2022.1051306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Fu X., Zhao B., Tian K., Wu Y., Suo L., Ba G., Ciren D., De J., Awang C., Gun S., Yang B. Integrated analysis of lncRNA and mRNA reveals novel insights into cashmere fineness in Tibetan cashmere goats. PeerJ. 2020;8 doi: 10.7717/peerj.10217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Liu J., Ji Q., Cheng F., Chen D., Geng T., Huang Y., Zhang J., He Y., Song T. The lncRNAs involved in regulating the RIG-I signaling pathway. Front. Cell. Infect. Microbiol. 2022;12 doi: 10.3389/fcimb.2022.1041682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Pan X., Li C., Feng J. The role of LncRNAs in tumor immunotherapy. Cancer Cell Int. 2023;23:30. doi: 10.1186/s12935-023-02872-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Rajakumar S., Jamespaulraj S., Shah Y., Kejamurthy P., Jaganathan M.K., Mahalingam G., Ramya Devi K.T. Long non-coding RNAs: an overview on miRNA sponging and its co-regulation in lung cancer. Mol. Biol. Rep. 2023;50:1727–1741. doi: 10.1007/s11033-022-07995-w. [DOI] [PubMed] [Google Scholar]
- 232.Tsai M.-C., Manor O., Wan Y., Mosammaparast N., Wang J.K., Lan F., Shi Y., Segal E., Chang H.Y. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Guh C.-Y., Hsieh Y.-H., Chu H.-P. Functions and properties of nuclear lncRNAs—from systematically mapping the interactomes of lncRNAs. J. Biomed. Sci. 2020;27:44. doi: 10.1186/s12929-020-00640-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Castro-Oropeza R., Melendez-Zajgla J., Maldonado V., Vazquez-Santillan K. The emerging role of lncRNAs in the regulation of cancer stem cells. Cell. Oncol. 2018;41:585–603. doi: 10.1007/s13402-018-0406-4. [DOI] [PubMed] [Google Scholar]
- 235.Zangouei A.S., Zangoue M., Taghehchian N., Zangooie A., Rahimi H.R., Saburi E., Alavi M.S., Moghbeli M. Cell cycle related long non-coding RNAs as the critical regulators of breast cancer progression and metastasis. Biol. Res. 2023;56 doi: 10.1186/s40659-022-00411-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Si W., Shen J., Zheng H., Fan W. The role and mechanisms of action of microRNAs in cancer drug resistance. Clin. Epigenetics. 2019;11:25. doi: 10.1186/s13148-018-0587-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Kousar K., Ahmad T., Abduh M.S., Kanwal B., Shah S.S., Naseer F., Anjum S. miRNAs in Regulation of Tumor Microenvironment, Chemotherapy Resistance, Immunotherapy Modulation and miRNA Therapeutics in Cancer. Int. J. Mol. Sci. 2022;23 doi: 10.3390/ijms232213822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Pedroza-Torres A., Romero-Córdoba S.L., Justo-Garrido M., Salido-Guadarrama I., Rodríguez-Bautista R., Montaño S., Muñiz-Mendoza R., Arriaga-Canon C., Fragoso-Ontiveros V., Álvarez-Gómez R.M., et al. MicroRNAs in Tumor Cell Metabolism: Roles and Therapeutic Opportunities. Front. Oncol. 2019;9:1404. doi: 10.3389/fonc.2019.01404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Li W.J., Wang Y., Liu R., Kasinski A.L., Shen H., Slack F.J., Tang D.G. MicroRNA-34a: Potent Tumor Suppressor, Cancer Stem Cell Inhibitor, and Potential Anticancer Therapeutic. Front. Cell Dev. Biol. 2021;9 doi: 10.3389/fcell.2021.640587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Zhao Y., Tao Z., Chen X. Identification of the miRNA-mRNA regulatory pathways and a miR-21-5p based nomogram model in clear cell renal cell carcinoma. PeerJ. 2020;8 doi: 10.7717/peerj.10292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Nascimento N.P.G., Gally T.B., Borges G.F., Campos L.C.G., Kaneto C.M. Systematic review of circulating MICRORNAS as biomarkers of cervical carcinogenesis. BMC Cancer. 2022;22:862. doi: 10.1186/s12885-022-09936-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Zhang H., Zhang M., Meng L., Guo M., Piao M., Huang Z., Yu H. Investigation of key miRNAs and their target genes involved in cell apoptosis during intervertebral disc degeneration development using bioinformatics methods. J. Neurosurg. Sci. 2022;66:125–132. doi: 10.23736/s0390-5616.20.04773-6. [DOI] [PubMed] [Google Scholar]
- 243.Bhattacharjee R., Prabhakar N., Kumar L., Bhattacharjee A., Kar S., Malik S., Kumar D., Ruokolainen J., Negi A., Jha N.K., Kesari K.K. Crosstalk between long noncoding RNA and microRNA in Cancer. Cell. Oncol. 2023;46:885–908. doi: 10.1007/s13402-023-00806-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Fu Z., Wang L., Li S., Chen F., Au-Yeung K.K.-W., Shi C. MicroRNA as an Important Target for Anticancer Drug Development. Front. Pharmacol. 2021;12 doi: 10.3389/fphar.2021.736323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Nisar S., Bhat A.A., Singh M., Karedath T., Rizwan A., Hashem S., Bagga P., Reddy R., Jamal F., Uddin S., et al. Insights Into the Role of CircRNAs: Biogenesis, Characterization, Functional, and Clinical Impact in Human Malignancies. Front. Cell Dev. Biol. 2021;9 doi: 10.3389/fcell.2021.617281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Wang C., Liu W.-R., Tan S., Zhou J.-K., Xu X., Ming Y., Cheng J., Li J., Zeng Z., Zuo Y., et al. Characterization of distinct circular RNA signatures in solid tumors. Mol. Cancer. 2022;21:63. doi: 10.1186/s12943-022-01546-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Wang S., Qian L., Cao T., Xu L., Jin Y., Hu H., Fu Q., Li Q., Wang Y., Wang J., et al. Advances in the Study of CircRNAs in Tumor Drug Resistance. Front. Oncol. 2022;12 doi: 10.3389/fonc.2022.868363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Cui C., Yang J., Li X., Liu D., Fu L., Wang X. Functions and mechanisms of circular RNAs in cancer radiotherapy and chemotherapy resistance. Mol. Cancer. 2020;19:58. doi: 10.1186/s12943-020-01180-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Wei L., Liu L., Bai M., Ning X., Sun S. CircRNAs: versatile players and new targets in organ fibrosis. Cell Commun. Signal. 2023;21:90. doi: 10.1186/s12964-023-01051-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Liu T., Long K., Zhu Z., Song Y., Chen C., Xu G., Ke X. Roles of circRNAs in regulating the tumor microenvironment. Med. Oncol. 2023;40:329. doi: 10.1007/s12032-023-02194-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Yu C.-Y., Kuo H.-C. The emerging roles and functions of circular RNAs and their generation. J. Biomed. Sci. 2019;26:29. doi: 10.1186/s12929-019-0523-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Xu T., Wang M., Jiang L., Ma L., Wan L., Chen Q., Wei C., Wang Z. CircRNAs in anticancer drug resistance: recent advances and future potential. Mol. Cancer. 2020;19:127. doi: 10.1186/s12943-020-01240-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Tang S., Li S., Tang B., Wang X., Xiao Y., Cheke R.A. Hormetic and synergistic effects of cancer treatments revealed by modelling combinations of radio - or chemotherapy with immunotherapy. BMC Cancer. 2023;23:1040. doi: 10.1186/s12885-023-11542-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Zhou Z., Li M. Targeted therapies for cancer. BMC Med. 2022;20:90. doi: 10.1186/s12916-022-02287-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Zhu A., Garcia J.A., Faltas B., Grivas P., Barata P., Shoag J.E. Immune Checkpoint Inhibitors and Long-term Survival of Patients With Metastatic Urothelial Cancer. JAMA Netw. Open. 2023;6:e237444. doi: 10.1001/jamanetworkopen.2023.7444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Pan K., Farrukh H., Chittepu V.C.S.R., Xu H., Pan C.-x., Zhu Z. CAR race to cancer immunotherapy: from CAR T, CAR NK to CAR macrophage therapy. J. Exp. Clin. Cancer Res. 2022;41:119. doi: 10.1186/s13046-022-02327-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Peterson C., Denlinger N., Yang Y. Recent Advances and Challenges in Cancer Immunotherapy. Cancers. 2022;14:3972. doi: 10.3390/cancers14163972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Wu P., Gao W., Su M., Nice E.C., Zhang W., Lin J., Xie N. Adaptive Mechanisms of Tumor Therapy Resistance Driven by Tumor Microenvironment. Front. Cell Dev. Biol. 2021;9 doi: 10.3389/fcell.2021.641469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Wu Y., Song Y., Wang R., Wang T. Molecular mechanisms of tumor resistance to radiotherapy. Mol. Cancer. 2023;22:96. doi: 10.1186/s12943-023-01801-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Wang S., Xie K., Liu T. Cancer Immunotherapies: From Efficacy to Resistance Mechanisms – Not Only Checkpoint Matters. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.690112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Topatana W., Juengpanich S., Li S., Cao J., Hu J., Lee J., Suliyanto K., Ma D., Zhang B., Chen M., Cai X. Advances in synthetic lethality for cancer therapy: cellular mechanism and clinical translation. J. Hematol. Oncol. 2020;13:118. doi: 10.1186/s13045-020-00956-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Cheng H.S., Lee J.X.T., Wahli W., Tan N.S. Exploiting vulnerabilities of cancer by targeting nuclear receptors of stromal cells in tumor microenvironment. Mol. Cancer. 2019;18:51. doi: 10.1186/s12943-019-0971-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Bergholz J.S., Wang Q., Kabraji S., Zhao J.J. Integrating Immunotherapy and Targeted Therapy in Cancer Treatment: Mechanistic Insights and Clinical Implications. Clin. Cancer Res. 2020;26:5557–5566. doi: 10.1158/1078-0432.CCR-19-2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Zhang Q., Fu Q., Bai X., Liang T. Molecular Profiling–Based Precision Medicine in Cancer: A Review of Current Evidence and Challenges. Front. Oncol. 2020;10 doi: 10.3389/fonc.2020.532403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Malone E.R., Oliva M., Sabatini P.J.B., Stockley T.L., Siu L.L. Molecular profiling for precision cancer therapies. Genome Med. 2020;12:8. doi: 10.1186/s13073-019-0703-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Gambardella V., Tarazona N., Cejalvo J.M., Lombardi P., Huerta M., Roselló S., Fleitas T., Roda D., Cervantes A. Personalized Medicine: Recent Progress in Cancer Therapy. Cancers. 2020;12:1009. doi: 10.3390/cancers12041009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Berger M.F., Van Allen E.M. Delivering on the promise of precision cancer medicine. Genome Med. 2016;8:110. doi: 10.1186/s13073-016-0373-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Yi M., Li T., Niu M., Mei Q., Zhao B., Chu Q., Dai Z., Wu K. Exploiting innate immunity for cancer immunotherapy. Mol. Cancer. 2023;22:187. doi: 10.1186/s12943-023-01885-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Rezayatmand H., Razmkhah M., Razeghian-Jahromi I. Drug resistance in cancer therapy: the Pandora's Box of cancer stem cells. Stem Cell Res. Ther. 2022;13:181. doi: 10.1186/s13287-022-02856-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Ju F., Atyah M.M., Horstmann N., Gul S., Vago R., Bruns C.J., Zhao Y., Dong Q.-Z., Ren N. Characteristics of the cancer stem cell niche and therapeutic strategies. Stem Cell Res. Ther. 2022;13:233. doi: 10.1186/s13287-022-02904-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Mai Y., Su J., Yang C., Xia C., Fu L. The strategies to cure cancer patients by eradicating cancer stem-like cells. Mol. Cancer. 2023;22:171. doi: 10.1186/s12943-023-01867-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Gao Q., Zhan Y., Sun L., Zhu W. Cancer Stem Cells and the Tumor Microenvironment in Tumor Drug Resistance. Stem Cell Rev. Rep. 2023;19:2141–2154. doi: 10.1007/s12015-023-10593-3. [DOI] [PubMed] [Google Scholar]
- 273.Alowais S.A., Alghamdi S.S., Alsuhebany N., Alqahtani T., Alshaya A.I., Almohareb S.N., Aldairem A., Alrashed M., Bin Saleh K., Badreldin H.A., et al. Revolutionizing healthcare: the role of artificial intelligence in clinical practice. BMC Med. Educ. 2023;23:689. doi: 10.1186/s12909-023-04698-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Hong L., Li W., Li Y., Yin S. Nanoparticle-based drug delivery systems targeting cancer cell surfaces. RSC Adv. 2023;13:21365–21382. doi: 10.1039/d3ra02969g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Manzari M.T., Shamay Y., Kiguchi H., Rosen N., Scaltriti M., Heller D.A. Targeted drug delivery strategies for precision medicines. Nat. Rev. Mater. 2021;6:351–370. doi: 10.1038/s41578-020-00269-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Bedoya D.A., Figueroa F.N., Macchione M.A., Strumia M.C. In: Advanced Biopolymeric Systems for Drug Delivery. Nayak A.K., Hasnain M.S., editors. Springer International Publishing; 2020. Stimuli-Responsive Polymeric Systems for Smart Drug Delivery; pp. 115–134. [DOI] [Google Scholar]
- 277.Karimi Kelaye S., Najafi F., Kazemi B., Foruzandeh Z., Seif F., Solali S., Alivand M.-R. The contributing factors of resistance or sensitivity to epigenetic drugs in the treatment of AML. Clin. Transl. Oncol. 2022;24:1250–1261. doi: 10.1007/s12094-022-02776-0. [DOI] [PubMed] [Google Scholar]
- 278.Kaur R., Bhardwaj A., Gupta S. Cancer treatment therapies: traditional to modern approaches to combat cancers. Mol. Biol. Rep. 2023;50:9663–9676. doi: 10.1007/s11033-023-08809-3. [DOI] [PubMed] [Google Scholar]