Abstract
The differential soil microbial assimilation of common nitrogen (N) fertilizer compounds into the soil organic N pool is revealed using novel compound-specific amino acid (AA) 15N-stable isotope probing. The incorporation of fertilizer 15N into individual AAs reflected the known biochemistry of N assimilation—e.g. 15N-labelled ammonium (15NH4+) was assimilated most quickly and to the greatest extent into glutamate. A maximum of 12.9% of applied 15NH4+, or 11.7% of ‘retained’ 15NH4+ (remaining in the soil) was assimilated into the total hydrolysable AA pool in the Rowden Moor soil. Incorporation was lowest in the Rowden Moor 15N-labelled nitrate (15NO3−) treatment, at 1.7% of applied 15N or 1.6% of retained 15N. Incorporation in the 15NH4+ and 15NO3− treatments in the Winterbourne Abbas soil, and the 15N-urea treatment in both soils was between 4.4% and 6.5% of applied 15N or 5.2% and 6.4% of retained 15N. This represents a key step in greater comprehension of the microbially mediated transformations of fertilizer N to organic N and contributes to a more complete picture of soil N-cycling. The approach also mechanistically links theoretical/pure culture derived biochemical expectations and bulk level fertilizer immobilization studies, bridging these different scales of understanding.
Keywords: 15N stable isotope tracing (SIP), amino acids, nitrate, ammonium, urea, immobilization
Adapted from: Charteris AF. Overview and Future Work. In 15N Tracing of Microbial Assimilation, Partitioning and Transport of Fertilisers in Grassland Soils. Springer Theses, Springer Nature, Switzerland AG, 2019, 239.
Introduction
Nitrogen (N) fertilizers are essential to modern food production and 105 Tg N fertilizers were used in 2016 (FAO 2019). It is estimated, however, only 17% of N applied to crops ultimately supports human nutrition, with the remainder being lost to the environment during food production and processing (Leach et al. 2012, Fowler et al. 2013). This brings the low nutrient use efficiency of the human food-chain into critical focus. The interaction of applied fertilizer N with the soil N-cycle, and influence on soil organic N, represents an important determinant of the fate of fertilizer N, the N balance of soil and eventual efficiency of production systems. Major gaps exist regarding the biological processing of N fertilizers in soils, particularly the routes and proportions of conversion into soil organic N.
Processing of N fertilizers has traditionally been quantified using isotope pool dilution to determine rates of N mineralization, immobilization, and nitrification in soils. However, even in agricultural soils, N stored in organic forms dominates inorganic N (Dungait et al. 2012). This large and heterogeneous soil N pool still underpins soil N dynamics and the supply of N to microorganisms, plants, and loss pathways (in some cases providing 30%–50% of the inorganic N for crop uptake; Macdonald et al. 1997, Murphy et al. 2000, Dungait et al. 2012). In order to provide a new perspective on the biomolecular fate and partitioning of different common N fertilizer compounds into soil organic N, we herein describe the application of compound-specific amino acid (AA) 15N-stable isotope probing (SIP) to investigate N-cycling into the soil protein pool (Charteris et al. 2016). The approach combines compound-specific gas chromatography–combustion-isotope ratio mass spectrometry (GC–C-IRMS) and 15N-SIP in the meta-metabolome of the whole soil system (Knowles et al. 2010; or other complex media, e.g. river water; Mena-Rivera et al. 2022) and is essentially a targeted 15N fluxomics approach (Cascante and Marin 2008). The soil protein pool is the largest (20%–50% of total soil N), and arguably most important, identifiable class of soil organic N (Stevenson 1982). Microbially mediated N transformations through the AA glutamate (Glu; Santero et al. 2012) represent the gateway between the inorganic and organic soil N pools (Supplementary Fig. S1). The extent to which fertilizer N is incorporated into soil protein has implications for its temporal availability to plants and loss pathways [e.g. nitrate (NO3−) leaching, ammonia (NH3) volatilization, and nitrous oxide (N2O) emissions] and we can now reveal distinct differences between three different fertilizer N compounds in two different grassland soils.
Materials and methods
We explore whether differences exist in the processing of three different 10 atom % 15N-labelled fertilizer N compounds—potassium nitrate (K15NO3), ammonium chloride (15NH4Cl), and urea (CO(15NH2)2); henceforth referred to as the 15NO3− treatment; the 15NH4+ treatment, and the 15N-U treatment, respectively—in two different soils, identified by site—Rowden Moor (RM) and Winterbourne Abbas (WA)—using soil microcosms (Table 1).
Table 1.
Table summarizing the laboratory incubation experiments conducted.
| Soil | Substrate | Key | Labelling | Substrate applied 10−1 g soil | Mass N 10−1 g soil | Equivalenta fertilization rate kg−1 N ha−1 year−1 | Incubation periods |
|---|---|---|---|---|---|---|---|
| RM | K15NO3 | RM-15NO3− | 10 atom % 15N | 400 µg in 200 µl DDW | 55 µg | 100 | 1.5, 3, 6, and 12 hours and 1, 2, 4, 8, 16, and 32 days |
| 15NH4Cl | RM-15NH4+ | 10 atom % 15N | 400 µg in 200 µl DDW | 105 µg | 190 | 1.5, 3, 6, and 12 hours and 1, 2, 4, 8, 16, and 32 days | |
| CO(15NH2)2 | RM-15N-U | 10 atom % 15N | 400 µg in 200 µl DDW | 187 µg | 340 | 3 hours and 2, 16, and 32 days | |
| Negative control | RM-C | ― | 200 µl DDW | ― | ― | 0 hours and 32 days | |
| WA | K15NO3 | WA-15NO3− | 10 atom % 15N | 400 µg in 200 μl DDW | 55 µg | 100 | 1.5, 3, 6, and 12 hours and 1, 2, 4, 8, 16, and 32 daysb |
| 15NH4Cl | WA-15NH4+ | 10 atom % 15N | 400 µg in 200 μl DDW | 105 µg | 190 | 1.5, 3, 6, and 12 hours and 1, 2, 4, 8, 16, and 32 daysc | |
| CO(15NH2)2 | WA-15N-U | 10 atom % 15N | 400 µg in 200 μl DDW | 187 µg | 340 | 2 hours and 2, 16, and 32 days | |
| Negative control | WA-C | ― | 200 µl DDW | ― | ― | 0 hours and 32 days |
Equivalent fertilization rate calculated based on a 0.3-m soil depth and an average of five to six treatments between February and October. The rates are generally within the range recommended for grasslands for dairy grazing (140–340 kg N ha−1 year−1; Defra 2010).
Not all time-points analysed for AAs, only 3 and 6 hours and 2, 4, 16, and 32 days.
Not all time-points analysed for AAs, only 1.5, 3, and 12 hours and 2, 8, and 32 days.
Sites and soil sampling
Soil was sampled to a depth of 15 cm along a random W transect from plot six of RM experimental site at Rothamsted Research North Wyke, Devon, UK (50°46'42" N, 3°54'47" W) and from Little Broadheath field of Longlands Dairy Farm, near WA in Dorset, UK (50°42'46" N, 2°34'55" W). The RM soil is classified as a Stagni-vertic cambisol (FAO), a clayey noncalcareous Pelostagnogley of the Hallsworth series (British Classification), or a Typic haplaquept (USDA; Harrod and Hogan 2008). The Little Broadheath soil is a lime-rich clay loam of variable depth (0.3–0.8 m), underlain by chalk.
The RM site was a long-term grassland (>40 years) dominated by Lolium spp. interspersed with Cynosurus, Festuca, Agrostis, Holcus, and Dactylis spp. It had been grazed by cattle for around 25 years and had received ~200–250 kg N ha−1 year−1 as cattle slurry. The WA site, on the other hand, had been used for spring cropping before being converted to a grass ley (Lolium perenne and Trifolium repens) and used for dairying with a mobile milking parlour for 2 years prior to sampling. The ley was fertilized with 40 kg N ha−1 (previously as ammonium sulfate [(NH4)2SO4] and then as sulfur-coated urea [CH4N2O]) every 40 days from spring until the start of the ‘closed period’ on 15th September. which prohibits N fertilizer application on grasslands in nitrate vulnerable zones (Defra 2013). The samples of each soil were combined in equal weights and homogenized to produce a pooled soil sample for each site. Pooled samples were air-dried to allow sieving to <2 mm and then double distilled water (DDW) added to attain 50% water holding capacity (WHC).
Incubations
Each experimental unit consisted of 10 g soil at 50% WHC contained in a 10-cm high by 2-cm diameter glass tube. Maintenance of the soil at 50% WHC was selected to prevent leaching and the tubes were fitted with furnaced and pierced aluminium foil lids to minimize volatile and evaporative losses. All incubations were carried out in triplicate so there were three tubes for each time point of each treatment. Incubation treatments and periods are summarized in Table 1. Treatments were injected into the soil and distributed over the full core depth. Incubations were halted at the required time by immersion in liquid nitrogen (N2) and stored at −20°C prior to freeze-drying. Whole freeze-dried soil cores were finely ground and homogenized using a pestle and mortar and stored in sealed 28 ml vials at −20°C.
Extraction, isolation, and derivatization of hydrolysable AAs
Freeze-dried and ground incubation soil samples (100 mg) with an added internal standard of 100 µl norleucine in hydrochloric acid (400 µg ml−1 Nle in 0.1 M HCl) were hydrolyzed with 5 ml 6 M HCl at 100°C for 24 hours under an atmosphere of N2 (Fountoulakis and Lahm 1998, Roberts and Jones 2008). Acid hydrolysis extracts both free and proteinaceous AAs as well as catalyzing the breakdown of living microbial biomass (Roberts and Jones 2008). The relatively harsh conditions are necessary for the cleavage of peptide bonds between hydrophobic residues [e.g. isoleucine (Ile), leucine (Leu), and valine (Val)], but also result in the deamination of asparagine (Asn) to Asp and glutamine (Gln) to Glu and the complete destruction of cysteine (Cys) and tryptophan (Trp; Fountoulakis and Lahm 1998, Roberts and Jones 2008). The technique may also partially destroy serine (Ser; ca. 10% loss), threonine (Thr; ca. 5% loss), and tyrosine (Tyr; loss depends on level of trace impurities in hydrolysis agent; Fountoulakis and Lahm 1998) and has the potential to hydrolyse AA chains from nonproteinaceous sources, such as peptidoglycan, resulting in an overestimation of some AAs, mostly alanine (Ala), Glu, glycine (Gly), and lysine (Lys; Roberts and Jones 2008). The technique is, however, considered the most reliable method for determining the total protein content of soils (Roberts and Jones 2008) and as such, it is reasonable to equate total hydrolysable AA concentrations to the size of the soil protein pool. The hydrolysis is performed under N2 as the presence of O2 can induce the thermal breakdown of hydroxyl- and sulfur-containing AAs [e.g. Ser, Thr, Tyr, and methionine (Met); Roberts and Jones 2008]. Hydrolysates were collected by centrifugation, dried at 60°C under a stream of N2, and stored at −20°C under 1 ml 0.1 M HCl. Cation-exchange column chromatography with acidified Dowex 50WX8 200–400 mesh ion-exchange resin was used to isolate AAs from the hydrolysates (Metges and Petzke 1997). Finally, the hydrolysed soil AA mixtures were converted to their N-acetyl, O-isopropyl derivatives for analysis (Corr et al. 2007).
Instrumental analyses
Bulk soil percentage total N (% TN) and δ15N analyses were carried out by elemental analysis-isotope ratio mass spectrometry (EA-IRMS) at the Lancaster node of the Natural Environment Research Council Life Sciences Mass Spectrometry Facility (NERC LSMSF). AAs as their N-acetyl, O-isopropyl derivatives were quantified by comparison with the Nle internal standard using gas chromatography–flame ionization detection (GC–FID). The N-acetyl, O-isopropyl AAs were identified by their known elution order and by comparison with N-acetyl, O-isopropyl derivatized-AA standards. Data were acquired and analysed using Clarity chromatographic station for Windows by DataApex. The δ15N values of individual AAs as their N-acetyl, O-isopropyl derivatives were determined using GC–C-IRMS. Data were acquired and analysed using Isodat NT 3.0 (Thermo Electron Corporation). Bulk soil percentage total C (% TC) analyses were carried out on a Eurovector EA3000 elemental analyser.
Statistical information and calculations
AA plateau Δ15N values and % 15NR incorporations were determined by curve fitting with a simple exponential equation using Genstat® statistical software for biosciences (19th edition, VSNI):
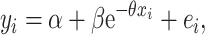 |
(1) |
where α is the plateau AA Δ15N value or % 15N incorporation, α + β is the AA Δ15N value or % 15N incorporation at t = 0 (which is 0 by definition for these parameters) and θ is the rate at which AA Δ15N values or % 15N incorporations increase. In addition, due to the temporal trend of Glx Δ15N values in the 15NH4+ and 15N-U treatments, these responses were also fitted with a critical exponential regression:
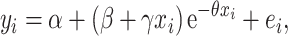 |
(2) |
where α is again the plateau AA Δ15N value or % 15N incorporation and α + β is again the AA Δ15N value or % 15N incorporation at t = 0 (again 0 by definition).The balance between γ (increase) and θ (decay) controls the height and positioning (x value) of the peak in the critical exponential function, where γ can be used to assess the rate of increase in AA Δ15N values or % 15N incorporated (larger γ = faster, although comparison between γ values becomes less clear where θ values differ).
Lack of error bar overlap between mean Δ15N values at t = 32 days was used as an indicator of significant statistical difference between final AA Δ15N values. This approach was used because formal statistical testing would confirm a significant statistical difference between means with separated error bars, and would, rather, only be useful to determine whether there were any statistically significant differences between means with some error bar overlap. This further level of inspection was not deemed to add sufficient value to the interpretation of this work as the complex statistical modelling required to rigorously determine the statistical difference between plateau Δ15N values (using constrained curve fitting) would not be proportionate for the additional information obtained. Simple t-tests or analysis of variance using final t = 32-day values would be based on very small datasets and would therefore only provide confirmation where errors bars are separated, which can already be observed.
The percentage of the applied 15N incorporated into each AA is as follows:
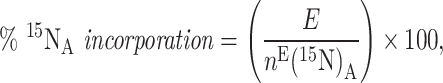 |
(3) |
where E is the 15N enrichment of the AA following application of a 15N-labelled substrate (taking into account the moles of N in the AA per gram of sample and the excess atom fraction of the AA after incubation, compared with the control). The percentage of retained 15N [based on nE(15 N)P/C, the excess moles of 15N present/retained per gram bulk sample at time, t] incorporated into each AA at time, t is as follows:
 |
(4) |
Finally, the percentage of applied/retained 15N incorporated into newly synthesized soil protein was determined by summing the results of Equations (3) or (4), respectively, for individual AAs.
Results and discussion
Ancillary data for the incubation experiments is given in Supplementary Note 1 and Supplementary Tables S1–S8. AA 15N-SIP exposes patterns in the biochemical assimilation pathways of applied 15N-labelled substrates via changes in the measured isotopic compositions (δ15N values) of each hydrolyzable AA over time (Charteris et al. 2016). AA δ15N values reflect the relative 15N content in the AA pool at that time, with any additional 15N (cf. t = 0 AA δ15N values, i.e. Δ15N values; Fig. 1A–F) being derived from the applied 15N-labelled substrate.
Figure 1.
Time–course plots of AA Δ15N values revealing 15N assimilation into individual AAs in the six treatments. (A) RM-15NO3− , (B) WA-15NO3− (error bars at t = 16 and 32 days are coloured to aid differentiation), (C) RM-15NH4+, (D) WA-15NH4+, (E) RM-15N-U, and (F) WA-15N-U. RM and WA refer to the two different soils from the two sites, RM and WA and the three amendments were potassium nitrate (K15NO3), ammonium chloride (15NH4Cl), and urea (CO(15NH2)2). Error bars are ± SE (n = 3). See Supplementary Fig. S3. For individual figures for each AA in each treatment for additional clarity.
Individual AAs demonstrated different levels and patterns of 15N incorporation in each treatment, but in both 15NO3− treatments, Δ15N values initially dipped before rising (Fig. 1A and B). All AAs exhibited a similar temporal pattern, but a range of responses (AA Δ15N values) was observed at all time points. In the 15NH4+ and 15N-U treatments (Fig. 1C–F), glutamate [abbreviated to ‘Glx’ since acid hydrolysis deaminates glutamine to glutamate, so the measured glutamate pool includes contributions from glutamic acid (Glu) and glutamine (Gln)] had a different trend from the two-phase rise of other AAs rising more quickly to an early peak (at ca. t = 2 days).
Explanations for the temporal trends in soil AA δ15N values following 15NO3− and 15NH4+ treatments fit with the known biochemistry of N assimilation (Charteris et al. 2016, Supplementary Fig. S1). Ammonium is generally the preferred anabolic source of inorganic N for soil microorganisms as both NO3− uptake and reduction to NH4+ for incorporation into cell material require more energy (and thus C; Rice and Tiedje 1989, Recous et al. 1990, Magasanik 1993, Geisseler et al. 2010). The suggestion of toxically high NO3− concentrations inducing localized cell lysis (Charteris et al. 2016) may not adequately account for the early negative Δ15N values in the 15NO3− treatments. Much of the material released from lysed cells would require mineralization prior to assimilation into AAs, which would take time (Kuzyakov et al. 2000). Instead, KNO3 could have stimulated the release of some clay fixed NH4+ (by replacement of NH4+ with K+; Nieder et al. 2011). Fast assimilation of this apparently 15N-depleted NH4+ (cf. other N sources for AA biosynthesis, perhaps due to some isotope effect(s) associated with NH4+ fixation and subsequent release) resulted in the biosynthesis of transiently 15N-depleted AAs. Temporal trends in AA Δ15N values in the 15N-U treatments are similar to those of the 15NH4+ treatments and urea-15N was most likely hydrolyzed (Mobley et al. 1995) and assimilated as 15NH4+ [via reductive amination of α-ketoglutarate to L-Glu catalyzed by glutamate dehydrogenase (GDH) or via the glutamine synthetase-glutamate synthase (GS-GOGAT) pathway; Supplementary Fig. S1; Santero et al. 2012]. The relative contribution of Glu and Gln to the Glx pool could not be determined in this study and may have influenced which 15NH4+ assimilation pathway dominated in the different soils (Geisseler et al. 2009). The relative operation of these pathways is also affected by other factors (e.g. the C:N ratio of the amendment; Geisseler et al. 2009). The contribution of Glu and Gln to the Glx pool can be expected to have been the same at the start of each incubation in the same soil receiving the different treatments.
Since AA concentrations (and thus the balance of AA degradation/biosynthesis/turnover) did not change markedly during the incubation experiments (Supplementary Note 1; Supplementary Tables S3–S8), 15N may be expected to be distributed (after initial uptake) in proportion to the quantity of N in each AA pool. However, 15N can only be incorporated into actively cycling pools, so a large, but stable AA pool would incorporate less 15N than expected based on the amount of N in that AA pool. Deviations from a proportional distribution, therefore, resulted from activity differences between AA pools and from the different biochemical routing of 15N. These deviations are reflected in differing fitted (Equations 1 and 2) or ‘plateau’ AA Δ15N values (if 15N is distributed in proportion with AA concentration, AA Δ15N values would be approximately equal for all AAs in a given experiment; Supplementary Note 2; Supplementary Tables S9 and S10).
AA δ15N (and Δ15N) values indicate the proportion of N derived from the applied 15N but not the total flux of that 15N into in each hydrolyzable AA [or, therefore, the distribution of applied 15N or 15N still present in the soil (retained 15N) amongst the AAs]. AAs present in higher concentrations require larger amounts of 15N to raise the N isotopic composition of the whole pool. It is, therefore, useful to consider the excess moles of 15N in each AA and, to provide some context, in comparison with the excess moles 15N applied (Equation 3), or alternatively, the excess moles 15N retained in the soil at that time (Equation 4; Supplementary Fig. S2). Percentage applied 15N incorporations (% 15NA incorporation) are useful in providing an indication of the overall fate of applied 15N (affected by heterogenous treatment applications and any losses of 15N from the system, which would occur in a field). Percentage retained 15N incorporations (% 15NR incorporation) reflect the partitioning of 15N present (or retained) in the system at the time, but as these data are calculated based on bulk soil δ15N values, could be affected by volatile losses of lighter 14N raising values.
Temporal patterns in the % 15NR incorporation into each AA under each treatment (Fig. 2A–F) were similar to those of increasing AA Δ15N values (Fig. 1A–F) but were dependent on the quantity of AA N in each pool (Supplementary Tables S3–S8; to reflect the routing/partitioning of 15N) and smoothed by the availability of 15N in the bulk soil. As for AA plateau Δ15N values, AA plateau % 15NR incorporations were determined by fitting simple exponential regressions (as well as critical exponential regressions for Glx in the 15NH4+ and 15N-U treatments; Equations 1 and 2; Supplementary Tables S11 and S12). The largest plateau hydrolyzable AA % 15NR incorporations were found in Glx in five out of the six treatments, ranging from 2.65 ± 0.15% of retained 15N in RM-15NH4+ to 1.0 ± 0.21% in WA-15NO3− (Fig. 2A–F). Using an analogous experimental approach (kinetic flux profiling) on an Escherichia coli culture, Yuan et al. (2006) similarly found largest fluxes of 15N into Glu and Gln and surmized that Glu N was quickly transferred into other AAs (Reitzer 2003). The exception to this was the RM-15NO3− treatment, in which the highest % 15NR was observed in Ala (0.4% retained 15N). In general, and particularly in the 15NO3− treatments, AAs present at higher concentrations (Supplementary Tables S3–S8) demonstrated larger % 15NR incorporations (Fig. 2A–F), as might be expected to maintain the AA concentration profile of the soil, which did not vary. As highlighted by differences in AA Δ15N values, however, applied 15N was not homogeneously distributed across the AA pools due to differently responding subpools of AAs and/or the differential biochemical routing of 15N (Fig. 2A–F; Supplementary Note 3). That the plateau 15N levels (as depicted in the pie charts in Fig. 2C–F) for the 15NH4+ and 15N-U treatments are very similar, but those of the 15NO3− are different both from these four and one another, suggests that the two soils responded differently to nitrate, but similarly to the other two substrates.
Figure 2.
Time–course plots of AA % 15NR incorporations revealing 15N assimilation into individual AAs in the six treatments, alongside pie charts of the relative percentage of retained 15N in each AA pool this represented, based on the plateau partitioning of 15N in each total hydrolyzable AA pool (derived from simple exponential regressions of the % 15NR incorporated into AAs over time; Equation 1). (A) RM-15NO3− (error bars for Ala and Gly are coloured to aid differentiation), (B) WA-15NO3− (error bars for Glu, Asp, and Ala are coloured to aid differentiation), (C) RM-15NH4+, (D) WA-15NH4+, (E) RM-15N-U, and (F) WA-15N-U. Error bars are ± SE (n = 3). Adapted from Charteris (2019).
A summation of the results of Equations (3) and (4) for each hydrolyzable AA gives the % 15NA incorporation and % 15NR incorporation into the total hydrolyzable AA pool, respectively (Fig. 3). There were only minor differences between the % 15NA incorporation and % 15NR incorporation into the total hydrolyzable AA pool, which were due to bulk soil 15N contents (Supplementary Table S1). As before, plateau % 15N incorporations into the total hydrolyzable AA pool were determined by fitting simple exponential regressions (Equation 1; Supplementary Table S16). Differences between the three N sources and two soils are clear—the three substrates are assimilated to significantly different extents (15NH4+ > 15N-U > 15NO3−) in the RM soil, but not in the WA soil (based on error bar overlap).
Figure 3.
Percentage of 15N incorporated into the total hydrolyzable AA pool for all treatments, labelled with the plateau % 15NR incorporations determined by simple exponential regressions. (A) Percentage of applied 15N and (B) Percentage of the 15N still present in the soil or ‘retained’ at that time. Error bars are ± SE (n = 3), the error bars of the WA-15NO3− treatment are highlighted in red as the bar at t = 32 days is large and otherwise difficult to distinguish. Adapted from Charteris (2019).
Although the two soils in these experiments were sampled from cattle-grazed grasslands in southwest England, they had different management histories and contrasting compositions (non-calcareous versus calcareous), which affected the biotic and abiotic processing of applied N (Müller et al. 2011). The RM soil received only cattle slurry for the 25 years prior to soil sampling while the WA soil also received regular additions of ammonium sulfate or urea (since 2011 when it was converted from spring crops to grass ley). Manuring, and higher soil percentage total organic carbon (% TOC) and percentage total N (% TN) contents have been related to greater soil microbial biomass activity (RM > WA; t = 0% TOC 6.80% cf. 4.17% and % TN 0.63% cf. 0.45; Söderström et al. 1983, Černý et al. 2003, Edmeades 2003, Booth et al. 2005, Müller et al. 2011).
Substrate assimilation in the RM soil matched expectations based on N assimilation biochemistry and previous studies assessing fertilizer N immobilization with bulk measurements (e.g. Wickramasinghe et al. 1985, Jackson et al. 1989, Recous et al. 1990, Christie and Wasson 2001). NO3−-15N was not used extensively as an anabolic N source. Both NO3− uptake and incorporation into cell material (via reduction to NH4+) require more energy (and thus C) than NH4+ assimilation and NO3− uptake can be inhibited by only low concentrations of NH4+ (Rice and Tiedje 1989, Recous et al. 1990, Magasanik 1993, Geisseler et al. 2010). Urea-15N incorporation was slower and less extensive than 15NH4+ incorporation as urea must first be hydrolyzed. Urease is ubiquitous in soils, however, and urea hydrolysis can occur extra- or intracellularly (Mobley et al. 1995, Geisseler et al. 2010), at a lower metabolic cost than 15NO3− reduction.
The operation of a more active (or larger) soil microbial biomass in the RM soil is supported by the significantly higher (almost double) plateau level of incorporation of 15NH4+ (bioavailable N source) in this soil, compared with the WA soil. Alternatively, less of the applied 15NH4+ may have become unavailable (e.g. by organic matter adsorption or clay-fixation; Booth et al. 2005, Nieder et al. 2011) in the RM soil. Lower NH4+ availability in the WA soil, compared with the RM soil could also explain the significantly greater assimilation of the less favourable N source, NO3−(-15N), by the WA soil, (where not limited by C). In addition, the WA soil may have become better adapted to NO3−-anabolism due to historic inorganic fertilization (Inselsbacher et al. 2010, Bunch and Bernot 2012), which can result in soil NO3− accumulation from nitrification due to NH4+ assimilation saturation or out-competition under C-limitation (Robertson and Groffman 2007). Indeed, attunement to urea fertilization of this soil could also be responsible for the faster (initial and overall) assimilation of urea-15N compared with the RM soil through increased endogenous urease concentrations.
Further, differences in the active microbial community, such as relative bacterial and fungal ratios, arising from differing management, may also influence dynamics of uptake for differing N amendments. Other work at the RM site using amino sugar (AS) 15N-SIP allowed quantification of 15N assimilation in this smaller, but more specific soil organic N pool (Reay et al. 2019a, Joergensen 2018). Assimilation into bacterial AS pools reflected dynamics observed herein for AAs (Reay et al. 2019b), while fungal AS exhibited slower uptake, and a lower preference for NH4+ over NO3−, likely reflecting uptake of secondary N sources (Marzluf 1997, He et al. 2011). Hence the differing soil types, and management at the RM and WA sites herein likely resulted in differing microbial communities (Malik et al. 2018, Romdhane et al. 2022), and thus attunement to N amendments.
Overall, a maximum of 12.9% of applied 15N (as 15NH4+), or 11.7% of ‘retained’ 15N was assimilated into the total hydrolyzable AA pool (in RM-15NH4+; Fig. 3; Supplementary Table S16). Incorporation was lowest in RM-15NO3−, at 1.7% of applied 15N, or 1.6% of retained 15N. These maximal plateau % 15N incorporations are unlikely to have been caused by 15N-substrate limitation during the incubations since 15N remained in the soil (based on bulk soil δ15N values) and other processes: are either considered poor competitors for NH4+ (e.g. nitrification); would not reduce 15N availability (e.g. denitrification or other gaseous losses, which were not observed to occur extensively, and would likely increase, rather than decrease, bulk soil δ15N values); or were not observed to occur (e.g. 15N loss via leaching). Maintenance of the soil at 50% WHC prevented leaching losses and made anaerobic microsites suitable for denitrification and dissimilatory nitrate reduction to ammonium (DNRA) less likely to develop (Tiedje et al. 1984, Sexstone et al. 1985). Rather, maximal 15N assimilations probably resulted from regulation of N uptake/assimilation as limitation by another essential nutrient (e.g. C or P) arose in the soil. Physical and chemical protection of soil organic C reduces microbial availability, resulting in C-limitation, which is consistent with lower NO3− assimilation observed in the WA soil, which had lower C content compared to the RM soil (Soong et al. 2019).
The application of our new 15N-AA SIP approach provides new insights into inorganic and organic N assimilation biochemistry by soil microbes. Critically, it provides vital mechanistic links between theoretical/pure culture derived biochemical expectations and bulk level fertilizer immobilization studies, bridging these different scales of understanding. Moreover, the work demonstrates that simple biochemical processes (N assimilation in this case) operating in physiologically relevant complex matrices are subject to additional biotic and abiotic environmental influences. This includes substrate supply by similarly influenced upstream processes and can overall result in quite different apparent process efficiencies in different settings (here, soils). Hence, the work constitutes a key step toward greater appreciation of the microbially mediated transformations of fertilizer N to organic N and contributes to a more complete picture of soil N-cycling in response to fertilizer N applications. Finally, the quantitative estimates regarding these transformations generated through time–course incubation experiments are vital parameters for the next generation of soil N-cycling models.
Supplementary Material
Acknowledgements
We thank J. Dungait and Rothamsted Research, which is supported by the UK Biotechnology and Biological Sciences Research Council (BBSRC), for providing soil from Rowden Moor, North Wyke.
Contributor Information
Alice F Charteris, Organic Geochemistry Unit, School of Chemistry, University of Bristol, Cantock’s Close, Bristol BS8 1TS, United Kingdom; Sustainable Agriculture Sciences, Rothamsted Research, North Wyke, Okehampton, Devon EX20 2SB, United Kingdom.
Timothy D J Knowles, Organic Geochemistry Unit, School of Chemistry, University of Bristol, Cantock’s Close, Bristol BS8 1TS, United Kingdom.
Andrew Mead, Computational and Analytical Sciences, Rothamsted Research, Harpenden, Hertfordshire AL5 2JQ, United Kingdom.
Michaela K Reay, Organic Geochemistry Unit, School of Chemistry, University of Bristol, Cantock’s Close, Bristol BS8 1TS, United Kingdom.
Katerina Michaelides, School of Geographical Sciences, University of Bristol, University Road, Bristol BS8 1SS, United Kingdom.
Richard P Evershed, Organic Geochemistry Unit, School of Chemistry, University of Bristol, Cantock’s Close, Bristol BS8 1TS, United Kingdom.
Author contributions
Alice F. Charteris (Conceptualization, Formal analysis, Investigation, Visualization, Writing – original draft, Writing – review & editing), Timothy D.J. Knowles (Conceptualization, Writing – review & editing), Andrew Mead (Formal analysis), Michaela K. Reay (Writing – review & editing), Katerina Michaelides (Conceptualization, Supervision, Writing – review & editing) and Richard P. Evershed (Conceptualization, Funding acquisition, Supervision, Writing – review & editing)
Conflict of interest
None.
Funding
This work was carried out during A. F. Charteris's UK Natural Environment Research Council (NERC) Open CASE PhD studentship between the University of Bristol and Wessex Water (NE/J017523/1). We thank NERC for partial funding of the mass spectrometry facilities at Bristol (contract number R8/H10/63; www.lsmsf.co.uk) and H. Grant of the UK NERC Life Sciences Mass Spectrometry Facility (Lancaster node) for stable isotopic characterization of reference standards and derivatizing agents and bulk soil N isotope analysis.
Data Availability
All relevant data are available in this article, its supplementary information files or at DOI (fnae041), which includes the full raw data for all figures.
References
- Booth MS, Stark JM, Rastetter E. Controls on nitrogen cycling in terrestrial ecosystems: a synthetic analysis of literature data. Ecol Monogr. 2005;75:139–57. [Google Scholar]
- Bunch ND, Bernot MJ. Nitrate and ammonium uptake by natural stream sediment microbial communities in response to nutrient enrichment. Res Microbiol. 2012;163:137–41. [DOI] [PubMed] [Google Scholar]
- Cascante M, Marin S. Metabolomics and fluxomics approaches. Essays Biochem. 2008;45:67–81. 10.1042/bse0450067. [DOI] [PubMed] [Google Scholar]
- Černý J, Balík J, Pavlíková Det al. The influence of organic and mineral nitrogen fertilisers on microbial biomass nitrogen and extractable organic nitrogen in long-term experiments with maize. Plant Soil Environ. 2003;49:560–4. [Google Scholar]
- Charteris AF, Knowles TDJ, Michaelides Ket al. Compound-specific amino acid 15N stable isotope probing of nitrogen assimilation by the soil microbial biomass using gas chromatography/combustion/isotope ratio mass spectrometry. Rapid Commun Mass Spectrom. 2016;30:1846–56. 10.1002/rcm.7612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charteris AF. Biosynthetic routing, rates and extents of microbial fertiliser nitrogen assimilation in two grassland soils. In: 15N Tracing of Microbial Assimilation, Partitioning and Transport of Fertilisers in Grassland Soils. Springer Theses. Cham: Springer Nature Switzerland AG, 2019, 79–151. [Google Scholar]
- Christie P, Wasson EA. Short-term immobilization of ammonium and nitrate added to a grassland soil. Soil Biol Biochem. 2001;33:1277–8. [Google Scholar]
- Corr LT, Berstan R, Evershed RP. Optimisation of derivatisation procedures for the determination of δ13C values of amino acids by gas chromatography/combustion/isotope ratio mass spectrometry. Rapid Commun Mass Spectrom. 2007;21:3759–71. 10.1002/rcm.3252. [DOI] [PubMed] [Google Scholar]
- Defra . Fertiliser Manual (RB209). London: The Stationery Office, 2010. http://www.ahdb.org.uk/projects/CropNutrition.aspx (11 April 2020, date last accessed). [Google Scholar]
- Defra . Guidance on Complying with the Rules for Nitrate Vulnerable Zones in England for 2013 to 2016. London: The Stationery Office, 2013. http://www.gov.uk/nitrate-vulnerable-zones (13 January 2016, date last accessed). [Google Scholar]
- Dungait JAJ, Cardenas LM, Blackwell MSAet al. Advances in the understanding of nutrient dynamics and management in UK agriculture. Sci Total Environ. 2012;434:39–50. [DOI] [PubMed] [Google Scholar]
- Edmeades DC. The long-term effects of manures and fertilisers on soil productivity and quality: a review. Nutr Cycling Agroecosyst. 2003;66:165–80. [Google Scholar]
- FAO . World Fertilizer Trends and Outlook to 2022: summary report. 2019. http://www.fao.org/publications/card/en/c/CA6746EN (10 April 2020, date last accessed). [Google Scholar]
- Fountoulakis M, Lahm HW. Hydrolysis and amino acid composition analysis of proteins. J Chromatogr A. 1998;826:109–34. [DOI] [PubMed] [Google Scholar]
- Fowler D, Coyle M, Skiba Uet al. The global nitrogen cycle in the twenty-first century. Philos Trans R Soc B. 2013;368:20130164. 10.1098/rstb.2013.0164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisseler D, Doane TA, Horwath WR. Determining potential glutamine synthetase and glutamate dehydrogenase activity in soil. Soil Biol Biochem. 2009;41:1741–49. 10.1016/j.soilbio.2009.06.002. [DOI] [Google Scholar]
- Geisseler D, Horwath WR, Joergensen RGet al. Pathways of nitrogen utilization by soil microorganisms: a review. Soil Biol Biochem. 2010;42:2058–67. 10.1016/j.soilbio.2010.08.021. [DOI] [Google Scholar]
- Harrod TR, Hogan DV. The Soils of North Wyke and Rowden. Revised Edition of Original Report by Harrod TR, 1981. Bedford: Soil Survey of England and Wales (now the National Soil Resources Institute, Cranfield University, 2008. https://repository.rothamsted.ac.uk/item/96xqw/the-soils-of-north-wyke-and-rowden (14 July 2022, date last accessed). [Google Scholar]
- He H, Li XB, Zhang Wet al. Differentiating the dynamics of native and newly immobilized amino sugars in soil frequently amended with inorganic nitrogen and glucose. Eur J Soil Sci. 2011;62:144–51. 10.1111/j.1365-2389.2010.01324.x. [DOI] [Google Scholar]
- Inselsbacher E, Hinko-Najera Umana N, Stange FCet al. Short-term competition between crop plants and soil microbes for inorganic N fertilizer. Soil Biol Biochem. 2010;42:360–72. 10.1016/j.soilbio.2009.11.019. [DOI] [Google Scholar]
- Jackson LE, Schimel JP, Firestone MK. Short-term partitioning of ammonium and nitrate between plants and microbes in an annual grassland. Soil Biol Biochem. 1989;21:409–15. [Google Scholar]
- Joergensen RG. Amino sugars as specific indices for fungal and bacterial residues in soil. Biol Fert Soils. 2018;54:559–68. 10.1007/s00374-018-1288-3. [DOI] [Google Scholar]
- Knowles TDJ, Chadwick DR, Bol Ret al. Tracing the rate and extent of N and C flow from 13C,15N-glycine and glutamate into individual de novo synthesised soil amino acids. Org Geochem. 2010;41:1259–68. 10.1016/j.orggeochem.2010.09.003. [DOI] [Google Scholar]
- Kuzyakov Y, Friedel JK, Stahr K. Review of mechanisms and quantification of priming effects. Soil Biol Biochem. 2000;32:1485–98. [Google Scholar]
- Leach AM, Galloway JN, Bleeker Aet al. A nitrogen footprint model to help consumers understand their role in nitrogen losses to the environment. Environ Dev. 2012;1:40–66. 10.1016/j.envdev.2011.12.005. [DOI] [Google Scholar]
- Macdonald AJ, Poulton PR, Powlson DSet al. Effects of season, soil type and cropping on recoveries, residues and losses of 15N-labelled fertilizer applied to arable crops in spring. J Agri Sci. 1997;129:125–54. [Google Scholar]
- Magasanik B. The regulation of nitrogen utilization in enteric bacteria. J Cell Biochem. 1993;51:34–40. [DOI] [PubMed] [Google Scholar]
- Malik AA, Puissant J, Buckeridge KMet al. Land use driven change in soil pH affects microbial carbon cycling processes. Nat Commun. 2018;9:3591, 10.1038/s41467-018-05980-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzluf GA. Genetic regulation of nitrogen metabolism in the fungi. Microbiol Mol Biol Rev. 1997;61:17–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mena-Rivera L, Lloyd CEM, Reay MKet al. Tracing carbon and nitrogen microbial assimilation in suspended particles in freshwaters. Biogeochemistry. 2022;164:277–93. 10.1007/s10533-022-00915-x. [DOI] [Google Scholar]
- Metges CC, Petzke K-J. Measurement of 15N/14N isotopic combustion in individual plasma free amino acids of human adults at natural abundance by gas chromatography-combustion-isotope ratio mass spectrometry. Anal Biochem. 1997;247:158–64. [DOI] [PubMed] [Google Scholar]
- Mobley HLT, Island MD, Hausinger RP. Molecular biology of microbial ureases. Microbiol Rev. 1995;59:451–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller C, Laughlin RJ, Christie Pet al. Effects of repeated fertilizer and cattle slurry applications over 38 years on N dynamics in a temperate grassland soil. Soil Biol Biochem. 2011;43:1362–71. [Google Scholar]
- Murphy DV, Macdonald AJ, Stockdale EAet al. Soluble organic nitrogen in agricultural soils. Biol Fert Soils. 2000;30:374–87. [Google Scholar]
- Nieder R, Benbi DK, Scherer HW. Fixation and defixation of ammonium in soils: a review. Biol Fertil Soils. 2011;47:1–14. 10.1007/s00374-010-0506-4. [DOI] [Google Scholar]
- Reay MK, Knowles TDJ, Jones DLet al. Development of alditol acetate derivatives for the determination of 15N-enriched amino sugars by gas chromatography-combustion-isotope ratio mass spectrometry. Anal Chem. 2019a;91:3397–404. 10.1021/acs.analchem.8b04838. [DOI] [PubMed] [Google Scholar]
- Reay MK, Charteris AF, Jones DLet al. 15N-amino sugar stable isotope probing (15N-SIP) to trace the assimilation of fertiliser-N by soil bacterial and fungal communities. Soil Biol Biochem. 2019b;138:107599. 10.1016/j.soilbio.2019.107599. [DOI] [Google Scholar]
- Recous S, Mary B, Faurie G. Microbial immobilisation of ammonium and nitrate in cultivated soils. Soil Biol Biochem. 1990;22:913–22. [Google Scholar]
- Reitzer L. Nitrogen assimilation and global regulation in Escherichia coli. Annu Rev Microbiol. 2003;57:155–76. 10.1146/annurev.micro.57.030502.090820. [DOI] [PubMed] [Google Scholar]
- Rice CW, Tiedje JM. Regulation of nitrate assimilation by ammonium in soils and in isolated soil microorganisms. Soil Biol Biochem. 1989;21:597–602. [Google Scholar]
- Roberts P, Jones DL. Critical evaluation of methods for determining total protein in soil solution. Soil Biol Biochem. 2008;40:1485–95. 10.1016/j.soilbio.2008.01.001. [DOI] [Google Scholar]
- Robertson GP, Groffman PM. Nitrogen transformations. In: Paul EA (ed.), Soil Microbiology, Ecology, and Biochemistry. 3rd edn. Oxford: Academic Press Elsevier, 2007, 353–55. [Google Scholar]
- Romdhane S, Spor A, Banerjee Set al. Land-use intensification differentially affects bacterial, fungal and protist communities and decreases microbiome network complexity. Environ Microbiome. 2022;17:1–15. 10.1186/S40793-021-00396-9/FIGURES/5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santero E, Hervás A, Canosa Iet al. Glutamate dehydrogenases: enzymology, physiological role and biotechnological relevance. In: Canuto RA (ed.), Dehydrogenases. London: InTech, Published online 2012, 289–91. [Google Scholar]
- Sexstone AJ, Parkin TB, Tiedje JM. Temporal response of soil denitrification rates to rainfall and irrigation. Soil Sci Soc Am J. 1985;49:99–103. [Google Scholar]
- Söderström B, Bååth E, Lundgren B. Decrease in soil microbial activity and biomasses owing to nitrogen amendments. Can J Microbiol. 1983;29:1500–6. [Google Scholar]
- Soong JL, Fuchslueger L, Marañon-Jimenez Set al. Microbial carbon limitation: the need for integrating microorganisms into our understanding of ecosystem carbon cycling. Glob Change Biol. 2019;26:1953–61. 10.1111/gcb.14962. [DOI] [PubMed] [Google Scholar]
- Stevenson FJ. Organic forms of soil nitrogen. In: Stevenson FJ (ed.), Nitrogen in Agricultural Soils. Madison: American Society of Agronomy, 1982, 67–74. [Google Scholar]
- Tiedje JM, Sexstone AJ, Parkin TBet al. Anaerobic processes in soil. Plant Soil. 1984;76:197–212. [Google Scholar]
- Wickramasinghe KN, Rodgers GA, Jenkinson DS. Transformations of nitrogen fertilisers in soil. Soil Biol Biochem. 1985;17:625–30. [Google Scholar]
- Yuan J, Fowler WU, Kimball Eet al. Kinetic flux profiling of nitrogen assimilation in Escherichia coli. Nat Chem Biol. 2006;2:529–30. 10.1038/nchembio816. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All relevant data are available in this article, its supplementary information files or at DOI (fnae041), which includes the full raw data for all figures.





