Abstract
Sequence-specific cytidine to uridine (C-to-U) and adenosine to inosine editing tools can alter RNA and DNA sequences and utilize a hydrolytic deamination mechanism requiring an active site zinc ion and a glutamate residue. In plant organelles, DYW-PG domain containing enzymes catalyze C-to-U edits through the canonical deamination mechanism. Proteins developed from consensus sequences of the related DYW-KP domain family catalyze what initially appeared to be uridine to cytidine (U-to-C) edits leading to this investigation into the U-to-C editing mechanism. The synthetic DYW-KP enzyme KP6 was found sufficient for C-to-U editing activity stimulated by the addition of carboxylic acids in vitro. Despite addition of putative amine/amide donors, U-to-C editing by KP6 could not be observed in vitro. C-to-U editing was found not to be concomitant with U-to-C editing, discounting a pyrimidine transaminase mechanism. RNAs containing base modifications were highly enriched in interphase fractions consistent with covalent crosslinks to KP6, KP2, and KP3 proteins. Mass spectrometry of purified KP2 and KP6 proteins revealed secondary peaks with mass shifts of 319 Da. A U-to-C crosslinking mechanism was projected to explain the link between crosslinking, RNA base changes, and the ∼319 Da mass. In this model, an enzymatic lysine attacks C4 of uridine to form a Schiff base RNA–protein conjugate. Sequenced RT-PCR products from the fern Ceratopteris richardii indicate U-to-C base edits do not preserve proteinaceous crosslinks in planta. Hydrolysis of a protonated Schiff base conjugate releasing cytidine is hypothesized to explain the completed pathway in plants.
Keywords: U-to-C RNA editing, C-to-U RNA editing, RNA crosslinking, metalloenzymes, Ceratopteris richardii
Eukaryotes have evolved to specifically modify DNA and RNA sequences for various purposes. The most common forms of editing are cytidine to uridine (C-to-U) and adenosine to inosine; these base changes can be catalyzed through a hydrolytic mechanism involving a zinc ion and an active site glutamate residue (1). Another form of editing present in trypanosomes inserts and deletes nucleotides creating functional mRNA sequences through an alternate mechanism (2). Additional base changes have been observed in cDNA sequences that might represent other forms of editing, but the biochemical mechanisms are unresolved.
Many organelle mRNAs in most land plants are edited C-to-U to properly encode proteins (3). Contemporary land plant organelle genomes are riddled with conserved T-to-C mutations that would lead to deleterious consequences without “repair” by C-to-U RNA editing factors. The RNA editing factors that specifically target cytidines are comprised of two main parts: (1) a series of pentatricopeptide repeat (PPR) domains often called the PPR tract which is integral for proper target recognition through sequence-specific RNA binding (4, 5, 6, 7) and (2) an enzymatic domain called the DYW domain capable of hydrolytic deamination of C-to-U (8, 9, 10, 11, 12, 13, 14).
In addition to C-to-U editing, many seedless plants have been discovered to edit the base of uridine to form cytidine (U-to-C) in the mRNA transcripts of organelles (15, 16, 17, 18). A variant of the DYW deaminase domain called the DYW-KP domain was found associated with U-to-C RNA editing in plants (15, 19) and was sufficient for U-to-C editing when expressed in exogenous systems (20). Since the overall reaction from U-to-C appears to be the reverse of the better characterized C-to-U editing, it often is called “reverse” editing.
Direct quantification of equilibrium values for DYW-catalyzed C-to-U editing have not been published. However, research using bacterial cytidine deaminase determined C-to-U deamination to be irreversible under physiological conditions with an equilibrium constant (Keq) = [uridine] [NH3]/[cytidine] [H2O] = 78 (21). When methylamine was substituted for ammonia as the initial nucleophile, the equilibrium was favored in the reverse direction [uridine] [CH3NH2]/[N4-methylcytidine] [H2O] with Keq = 0.46 (21). Applied to U-to-C editing, primary amines would be more favorable nitrogen donors than free ammonia, though the reaction pathway would have to be different than the reverse of cytidine deamination.
Potential biotechnological applications for a programmable site-specific RNA editor have driven the development of designer C-to-U editing enzymes (22). The PPR tract is comprised of serial PPRs, and each repeat contains two amino acid positions that are primarily responsible for specific binding to ribobases (4, 23). Arrangements of synthetic PPRs can yield a sequence-specific RNA-binding protein scaffold that can be tailored for specific tasks through the inclusion of C-terminal enzymatic domains (22). Though the C-to-U DYW editing platform was initially developed, synthetic U-to-C editing factors have also been devised (20). The DYW-KP domains of designer U-to-C editing factors KP2, KP3, and KP6 were constructed based on consensus sequences using different bioinformatic libraries and thus are chimeric (20). Unlike the obligate U-to-C editors KP2 and KP3, KP6 has both C-to-U and U-to-C RNA editing capabilities in mammalian cells while in bacteria only U-to-C activity was reported (20).
The DYW-KP domain has many similarities with the C-to-U RNA editing enzymes that have been differentiated by the name DYW-PG (20). The projected zinc coordinating residues and catalytic glutamate are conserved in DYW-PG and DYW-KP domains (20). The DYW-KP name comes from the presence of a conserved lysine and proline (KP) at the N terminus of the domain nearby the location of the often-conserved proline and glycine residues of the critical PG-box region in DYW-PG domains (24). Complicating existing nomenclatures, the DYW-KP does not share the conserved eponymic aspartate-tyrosine-tryptophan (DYW) motif at the C terminus. DYW-PG differs from DYW-KP by conservation of a serine residue upstream of the catalytic glutamate versus the respective presence of a hydrophobic, often alanine residue (20).
Investigations into the U-to-C apparatus and its biochemical mechanism have been hampered by the absence of a tractable plant model and a robust in vitro assay. Several ferns have been reported to have U-to-C activity (17, 25, 26) and Ceratopteris richardii has been developed as a fern model with a draft partial nuclear genome sequence (27). Based on the chloroplast genome annotation in GenBank: KM052729.1, C. richardii has 13 C-to-U and 27 U-to-C editing sites. In this manuscript, we use a nomenclature for editing sites that includes a genus and species abbreviation followed by the name of the gene edited, the nucleotide edited, its nucleotide position from the A of the initiation codon, and the nucleotide resulting from editing. Thus, the U-to-C editing site labeled Cr_ycf2 U3676C indicates C. richardii, ycf2, uridine-target, nucleotide position 3676, and resulting in cytidine.
Results
Recombinant KP6 has C-to-U editing activity in vitro
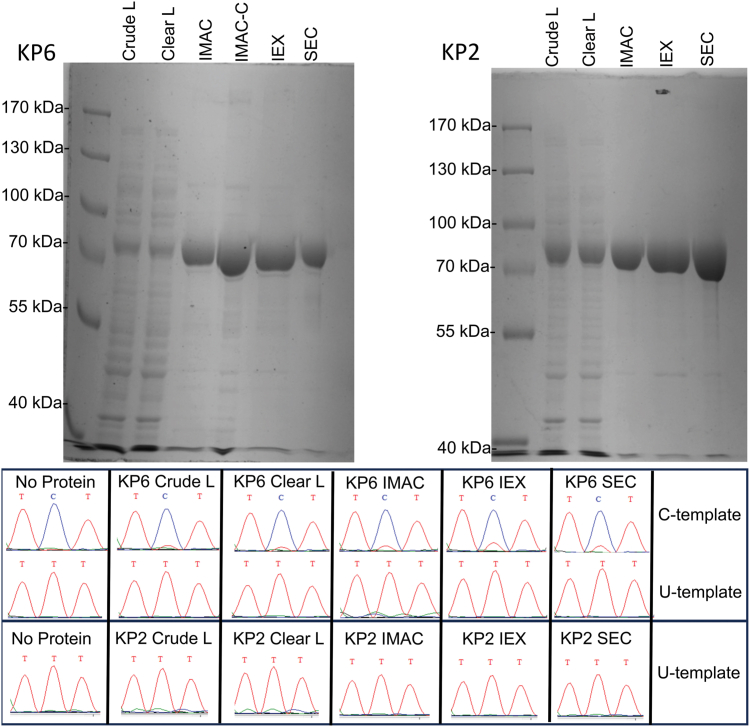
Original constructs for the expression of recombinant KP2 and KP6 were obtained though addgene (20). Plasmids were purified using the alkaline lysis method (28) and subcloned into Rosetta 2 (DE3) pLysS cells. Expression of DYW-KP proteins was induced using isopropyl β-d-1-thiogalactopyranoside (IPTG), and proteins were purified using immobilized metal-affinity chromatography (IMAC), ion-exchange chromatography (IEX), and size-exclusion chromatography (SEC). Mass estimates for KP6 and KP2 using mass spectrometry indicate a major species that matches theoretical predictions and a secondary minor species with an additional 319 Da mass (Fig. S1). Purified proteins were then assayed for C-to-U and U-to-C activity in vitro using the method previously described (9) and represented in a flowchart in Fig. S2. In this assay, recombinant proteins were mixed with RNA substrates that contain 149 upstream and 11 downstream nucleotides that include the pyrimidine editing target bordered by chloroplast sequences originating from Arabidopsis thaliana rpoA. Putative substrates containing either a C or a U at the editing site position were made by using the C or T plasmid vectors (20) as PCR templates to investigate C-to-U and U-to-C editing, respectively. RNAs were flanked with bacterial SK and KS sequences to allow specific amplification of RNAs added in vitro. After incubation with protein for 2.5 h at 28 °C, reactions were reverse transcribed, amplified using PCR, and then Sanger sequenced. Recombinant KP6 fractions from various steps in the same purification exhibited C-to-U editing activity, but no U-to-C activity could be observed (Fig. 1). Purified fractions of KP2 also had no observable U-to-C activity (Fig. 1).
Figure 1.
At top, an image of Coomassie-stained SDS-PAGE gels loaded with 10 μg of recombinant KP6 (left) and KP2 (right) purification fractions. Each marker lane is labeled with the reference masses in kDa to compare to expected masses for KP6 (83.8 kDa) and KP2 (83.4 kD). Crude L, ClearL, IMAC, IMAC-C, IEX, and SEC represent crude lysate, cleared lysate, immobilized metal-affinity chromatography, concentrated immobilized metal affinity chromatography, ion-exchange chromatography, and size-exclusion chromatography fractions, respectively. At bottom, C-to-U and U-to-C RNA editing was assayed using appropriate substrates (listed at right) using the following protein concentrations: KP6 crude lysate (3.6 μg/μl); KP6 cleared lysate (3.72 μg/μl); KP6 IMAC (1.1 μg/μl); KP6 IEX (0.4 μg/μl); KP6 SEC (0.1 μg/μl); KP2 crude lysate (9.0 μg/μl); KP2 cleared lysate (7.7 μg/μl); KP2 IMAC (7.5 μg/μl); KP2 IEX (0.5 μg/μl); KP2 SEC (1.2 μg/μl). Sequence traces from individual reactions are shown.
Absence of U-to-C editing activity observed with the addition of ammonia and putative amino/amine donors
Once purified fractions of DYW-KP proteins were available in the laboratory, C-to-U and U-to-C activity was studied in more detail. Increased levels of ammonia (NH3) up to 2 M can reverse the direction of catalysis by cytidine deaminase, an enzyme that uses a similar mechanism as proposed for the DYW domain (21), consistent with Le Chatelier's principle. Recombinant KP6 reactions in the presence of 2 M ammonium acetate were performed at pH 9.2 to favor the nonionized form but did not yield detectable U-to-C activity (Fig. S3). Since methylamine favors a reaction with uridine to produce N4-methylcytidine (21), a primary amine might better serve as an amine donor due to its increased basicity. Unfortunately, reactions with KP6 and the addition of 2 mM of putative amino/amine donors L-glutamine, L-asparagine, L-aspartic acid, L-glutamic acid, L-alanine, L-lysine, L-arginine, 6 amino hexanoic acid, and putrescine did not induce detectable U-to-C activity (Fig. S3).
C-to-U RNA editing of KP6 is stimulated by the addition of certain carboxylic acids
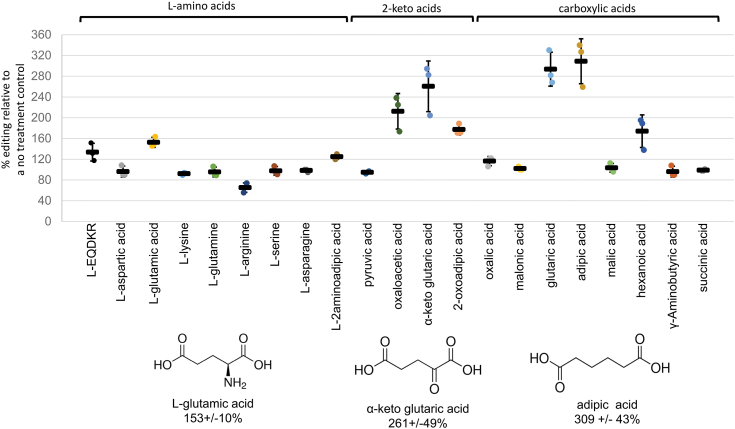
Due to the failure to observe U-to-C editing activity, focus shifted to the C-to-U editing activity by KP6 in vitro. We serendipitously discovered that a cocktail containing 2 mM of each of the putative amine donors L-glutamic acid, L-glutamine, L-aspartic acid, L-lysine, and L-arginine results in enhancement of activity (Fig. 2 and Table S1). The single addition of each amino acid in the cocktail indicated the enhancement was due to L-glutamic acid. Addition of L-2-aminoadipic acid with a terminal carboxylic acid group was also able to enhance editing. To test if alpha amino groups were involved in enhancement, 2-keto acids with terminal carboxylic acid groups were investigated for changes in C-to-U editing catalyzed by KP6. Addition of oxaloacetic acid and alpha-ketoglutaric acid enhanced editing to a greater extent than their alpha amino acid counterparts (Fig. 2). Finally, addition of carboxylic acids without 2-keto groups was studied. Glutarate and adipate were both found to stimulate C-to-U RNA editing by about 3-fold (Fig. 2). Carboxylic acids that stimulated C-to-U editing exhibited structural similarities including terminal carboxylic acids and a saturated 2 to 4 carbon backbone (Fig. S4). Reactions with KP2 in the presence of treatments shown to increase C-to-U editing by KP6 were unable to produce detectable C-to-U editing (Fig. S5).
Figure 2.
Several carboxylic acids enhance C-to-U RNA editing catalyzed by recombinant KP6. Relative C-to-U RNA editing was assayed in reactions with 2 mM additions of 20 different carboxylic acids. Percent editing was calculated from three reactions per treatment. Calculated values were then related to percent editing calculated from a no treatment reaction in the same batch using the same protein fraction (% editing treatment/% editing no treatment ×100). Ion-exchange and size-exclusion chromatography purification fractions of KP6 were used for this analysis from the initial protein purification shown in Figure 1. An X-Y scatterplot displays relative editing values calculated for each reaction with error bars representing 1 standard deviation from the mean for the triplicate reactions. Reactions were performed at pH 7.7, and all molecules shown would predominantly be in the ionized form. C-to-U, cytidine to uridine; U-to-C, uridine to cytidine.
The functional role of carboxylic acid enhancement of C-to-U editing was unclear. Amidotransferases are able to catalyze the release of ammonia as well as its transfer to a carboxylate group (29). If the DYW-KP was capable of catalyzing the transfer of the amine liberated from cytidine to a carboxylic acid, an amidated product might result. If this putative reaction was reversed, amides might serve as nitrogen donors for the U-to-C reaction. Addition of succinamide, adipamide, 5-amino 5 oxopentanoic acid, and succinamic acid were unable to induce measurable amounts of U-to-C editing in vitro (Fig. S3).
C-to-U RNA editing catalyzed by KP6 is not concomitant with U-to-C editing
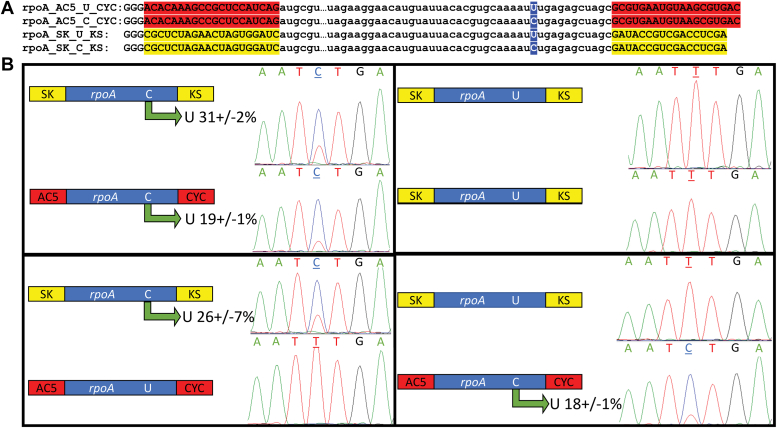
C-to-U and U-to-C editing activity was previously described for recombinant KP6 expressed in mammalian cells (20). If the enzyme has both activities, it might act as a transaminase capable of transferring an amine from cytidine to uracil and back depending on an equilibrium. To test this hypothesis, we generated C and U putative substrates flanked by SK/KS and AC5/CYC sequences used like “barcodes” to specifically amplify targeted substrates from mixtures (Fig. 3A). Two RNAs with rpoA C editing sites were added together in stochiometric equivalent amounts in the presence of KP6, and both were edited, though to different extents (top left, Fig. 3B). No U-to-C editing was observed by KP6 for two rpoA U putative substrates added in stoichiometric equivalent amounts (top right, Fig. 3B). When rpoA C substrates and RNAs containing rpoA U were mixed and added to reactions with KP6 in equivalent amounts, the C template was edited with no observed base changes on the U template (bottom, Fig. 3B). Ultimately, mixtures of RNAs and KP6 resulted in C-to-U editing of acceptable substrates without any observable U-to-C changes (Fig. 3). Thus, C-to-U editing by KP6 is not concomitant with U-to-C editing, and equilibria are determined by other reactants and products.
Figure 3.
C-to-U RNA editing activity catalyzed by KP6 is not concomitant with U-to-C RNA editing. A, the sequences of the RNA substrates are shown to represent differences between substrates. RNA substrates with C and U nucleotides (in blue) at the RNA editing position (underlined) were constructed with both terminal AC5/CYC (red) and SK/KS (yellow) sequences. B, reactions containing combinations of two putative substrates at equimolar concentrations are represented by cartoon boxes. The green arrow highlights C-to-U RNA editing with the mean value and 1 standard deviation from the mean shown for triplicate reactions. A representative Sanger sequencing trace is displayed next to each substrate cartoon to show the extent of conversion with the editing site underlined. Reactions were performed using size-exclusion chromatography purification fractions in the presence of 2 mM of adipate. C-to-U, cytidine to uridine; U-to-C, uridine to cytidine.
Base modification catalyzed by KP6, KP2, and KP3 in vivo involves protein crosslinking
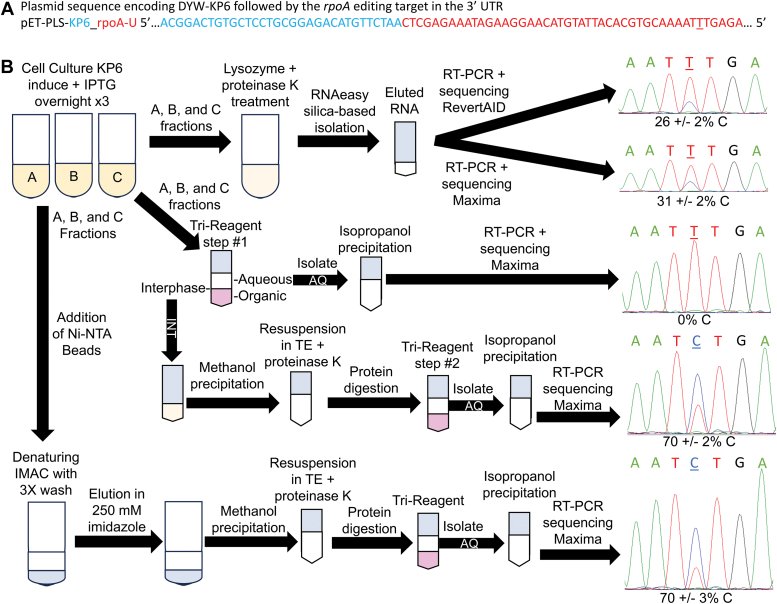
After obtaining the constructs for expression of KP6, we had difficulty recapitulating the U-to-C base editing using organic phase separation methods to isolate RNA from bacterial cell pellets. Several attempts always resulted in absence of observed U-to-C editing (Fig. 4). Typically, RNAs are isolated from the aqueous phase that represent free RNAs not tied to other macromolecules like proteins since noncovalent reactions are disrupted by the presence of the strong chaotrope guanidinium thiocyanate. A full recapitulation using the methods of the original publication (20) solved the initial problem, and U-to-C editing was observed, albeit at lower conversion extents (Fig. 4) than previously published (20). This led to investigation into the location of the missing U-to-C–edited RNAs. The sequencing of RT-PCR products from the interphase, the location of crosslinked RNA–protein conjugates reported in UV-crosslinking experiments (30), revealed changes in base-pairing consistent with U-to-C RNA editing (Fig. 4). Sanger sequencing traces of RT-PCR products from bacteria cultures expressing KP2 and KP3 also indicate base changes and crosslinking are correlated (Fig. S6). Thus, U-to-C editing and RNA crosslinking are concomitant by KP6, KP2, and KP3.
Figure 4.
KP6 activity attributed to U-to-C RNA editing is linked to RNA–protein crosslinking.A, the plasmid sequence is represented that once transcribed results in a U-target present in the 3′ UTR of the DYW-KP protein. B, a detailed flowchart shows three different experiments evaluating RNA editing from the same batch of three bacterial batch culture replicates A, B, and C. At top, the RNA purification from bacterial pellets proceeded using the techniques in the original report (20). Two different reverse transcriptase enzymes were compared (RevertAID RT versus Maxima RT). In the center, a diagram represents an experiment where RNA editing was calculated from RNAs isolated from the aqueous and interphase fractions of an organic phase separation. At bottom, a drawing represents isolation of RNAs from a denaturing immobilized metal affinity chromatography experiment in 6M guanidinium HCl. Mean percent RNA editing and one standard deviation from the mean were calculated from Sanger sequencing traces from three separate bacterial cultures. Representative images are displayed to the right of arrows with the editing site underlined. IMAC, immobilized metal affinity chromatography; IPTG, isopropyl β-d-1-thiogalactopyranoside; U-to-C, uridine to cytidine.
The RNA crosslinks could be with KP6 or potentially other proteins. Since KP6 has an internal His-tag, copurification of RNA targets and KP6 would indicate the interaction is direct. Fractions from the same bacterial cultures where U-to-C RNA editing was demonstrated were disrupted in a buffer containing 6M guanidinium hydrochloride, and KP6 was purified using denaturing IMAC. The protein was precipitated using methanol and digested using proteinase K. Released RNAs isolated from the aqueous phase exhibited changes in base pairing consistent with U-to-C RNA editing. Ratios from Sanger sequencing electrophoretograms of T versus C peaks at the editing site position were identical between denaturing IMAC and interphase RNA purifications (Fig. 4).
U-to-C–edited RNAs in C. richardii are not crosslinked to proteins
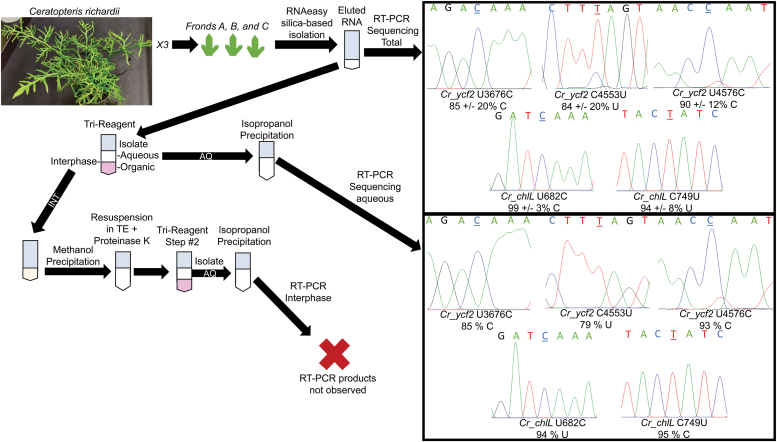
The crosslinking mechanism would seem to be problematic for a native editing mechanism since editing targets are largely in mRNA coding sequences and translation would likely be affected. RNAs were isolated from the model moss C richardii to investigate if the native edited RNA remains crosslinked to the native U-to-C enzymes. Both free and crosslinked RNAs should elute off the silica column from the Qiagen RNAeasy kit. An additional organic phase separation was used to collect free RNAs from the aqueous phase and RNAs in the interphase. Sequences of RT-PCR products from C. richardii for chloroplast genes ycf2 and chlL genes reveal RNAs from the aqueous phase are edited to the same extent as total RNA (Fig. 5). RT-PCR products from interphase RNAs were conspicuously absent (Fig. S7).
Figure 5.
U-to-C editing in the model fern C. richardii is not linked to crosslinking. At left, a detailed flowchart describes an experiment used to examine RNA editing in RNAs from different fronds and present in the aqueous versus the interphase fractions of an organic phase separation. At right, representative Sanger sequencing electrophoretograms are shown with the editing site underlined above editing site labels over mean percent editing values calculated from three fronds from different plants (at top). One standard deviation from the mean is shown. Since RNAs from fronds A, B, and C were combined to increase yield, mean editing values are shown (at bottom right) from only two sequencing reactions using a forward and a reverse primer with the editing sites underlined. U-to-C, uridine to cytidine.
Discussion
A proposed mechanism for U-to-C crosslinking performed by synthetic editing factors
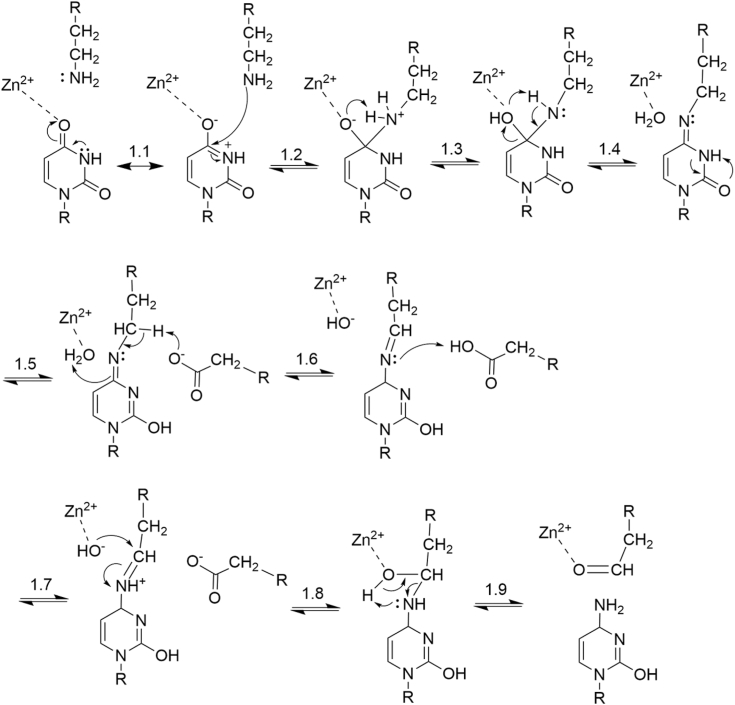
A mechanistic model was developed based on the following criteria: (1) it had to explain the observed crosslinks; (2) base-pairing changes needed to be represented; (3) the model needed to include a rationale for the additional ∼319 Da mass observed on KP2 and KP6 proteins; (4) the model needed to include a full mechanism resulting in a free cytidine; and (5) a scheme should use conserved features in the active site of DYW domains including zinc ions and a catalytic glutamate residue. We propose a U-to-C crosslinking mechanistic model that meets all these criteria (Fig. 6).
Figure 6.
A proposed mechanism for U-to-C crosslinking consistent with the observation that base differences in the Sanger sequencing traces are concomitant with RNA–protein crosslinking. The mechanism design was influenced by presence of local zinc ions and a catalytic glutamate in the DYW domain active site and the requirement for lysine as the only amino acid capable of both RNA–protein crosslinking and observed altered base pairing. Though the reaction mechanism does not appear to complete in bacteria, the full mechanism projects how hydrolysis of a protonated Schiff base can release the edited cytidine. Images were constructed using ChemDraw 23.0.1. U-to-C, uridine to cytidine.
In the U-to-C crosslinking mechanistic model, initially resonance in step 1.1 allows for the nucleophilic attack of a lysine residue in step 1.2 from the DYW-KP protein on the C4 of a bound uracil. This results in a protein–RNA crosslink and resolves after step 1.4 in a Schiff base. Tautomerization of the C2 ketone forms a stabilizing quinoid intermediate after step 1.5. The catalytic glutamate can then facilitate a 1,3-proton shift/trans-imination reaction (31) allowing the formation of a protonated Schiff base with an iminium poised for hydrolysis by the zinc-bound hydroxide group (steps 1.5–1.7). Final steps 1.8 and 1.9 represent hydrolysis of the protonated Schiff base to release the new cytidine nucleotide of the RNA and a modified amino acid allysine group of the DYW-KP protein to explain native systems that do not retain the crosslinked C.
Since the actual final species is not known, there are several hypotheses that can explain the altered base pairing. The Schiff base resulting from step 1.4 is in the imino isomer conformation and would be predicted to either inhibit reverse transcriptase polymerization in the Z-imino isomer or base pair as a U with A in the E-imino isomer. Similar differences in base-pairing properties have been used to detect modified bases (32). Through tautomerization, the modified base potentially could form the amino isomer with a C2 ketone that could base pair with a G (Fig. S8). The amino form would not be directly susceptible to hydrolysis.
At present, it is unclear what explains the thermodynamic trap and the final stabilized crosslink species in bacteria. RNA hydrolysis during purification by ubiquitous RNases have likely left only a crosslinked CMP moiety on the protein (Fig. S8). The observed 319 Da mass present on KP2 and KP6 (Fig. S1) fits closely with the estimated mass shift of 322 Da for crosslinked intermediates from our model and does not overlap with common protein translational modifications.
Sequence traces showing complete U-to-C conversion (100% C) were not observed despite covalently crosslinked RNAs being uniquely able to survive 6M guanidinium hydrochloride conditions in the denaturing IMAC or the organic phase separation. This could represent an equilibrium between tautomer species or reduced specificity by reverse transcriptase for the modified base (Fig. 6). Additional mass spectrometry studies are required to discover the position of the crosslink.
Differences with the native mechanism
Unfortunately, mechanistic differences exist in the function of KP6 in bacteria and native U-to-C RNA editing enzymes in organelles. The completion of U-to-C editing by resolving the crosslink likely occurs through hydrolysis of the iminium ion of the protonated Schiff base conjugate resulting in an enzyme allysine, similar to the first steps in the reaction mechanism of lysyl oxidases (33). The resulting reactive aldehyde group could participate in additional crosslinking reactions, and it is unclear if the enzyme could be regenerated. U-to-C DYW-KP editing factors might be suicide enzymes that participate as reactants or an unknown amine carrier might regenerate the lysyl group.
The synthetic origins of KP2, KP3, and KP6 do not result from a single native enzyme but a consensus sequence and might be responsible for mechanistic differences compared with native enzymes in planta. In the absence of purifying selection, the consensus sequence selected might be incapable of completing catalysis.
Since all three proteins in this investigation seem to be stuck at a crosslinked intermediate, it is more likely the environment or expression system in bacteria may lead to an altered reaction mechanism. KP6 can act as both a U-to-C and C-to-U editing enzyme in mammalian cells, indicating a difference in catalysis based on environment (20). Also, the C-to-U reaction is well demonstrated in conditions in vitro (Figs. 1 and 2) and absent in bacteria (20).
Unlike the native editing systems that have editing targets in translated mRNAs, the transgenic editing targets in bacteria and mammalian cells are localized to untranslated 3′-UTR sequences. One hypothesis is that the ribosome might play a role in resolving the crosslinked species and explain why bacterial systems are unable to complete catalysis to the free cytidine. Further work is needed to explain the nature of the environmental effects on enzymatic activities and/or resolve a possible link to translation.
Use of KP6 as a crosslinking platform to study RNA–protein interactions or arrest expression
With an improved understanding of DYW-KP functions in bacteria, limitations and opportunities to the mechanism become clear. Targeting transcripts for U-to-C editing to repair pathogenic SNPs with DYW-KP-type enzymes would theoretically lead to issues due to crosslinking. On the other hand, this could also be exploited to arrest transcript expression. RNA–protein studies could be enhanced by the development of a site-specific crosslinking reagents like DYW-KP proteins. Researchers could have a new tool to investigate RNA–protein interactions and possibly track RNA conjugates with fluorescently tagged DYW-KP proteins.
C-to-U and U-to-C RNA editing by DYW-KP proteins and prevention of futile cycles
The C-to-U editing activity observed in vitro by KP6 is confounding. DYW-KP class enzymes are very similar to the C-to-U editing class and contain all essential features for the canonical zinc-dependent deamination reaction mechanism. Conspicuous differences present in the KP variant include (1) the presence of an alanine in place of serine at a position upstream of the active site glutamate and downstream of the zinc coordinating histidine; (2) a separate small 3 to 4 amino acid deletion in a region of sequence called the PG-box; and (3) conservation of the eponymous KP sequence (Fig. S9). It is likely the C-to-U editing mechanism of KP6 utilizes the canonical zinc-dependent deamination reaction.
If C-to-U editing and U-to-C editing activities were allowed to be concurrent, then this class of enzyme might be in danger of a futile cycle. C-to-U and U-to-C RNA editing were not found to be concomitant in vitro (Fig. 3). The U-to-C crosslinking mechanism potentially avoids futility by physically linking the modified base and enzyme. This would prevent activity by other DYW-KP enzymes on the product and prevents canonical C-to-U activity by the enzyme conjugate through occupation of the active site. Ferns and likely other U-to-C utilizing plants can release the crosslink, but further work is needed to determine how a native amino acid configuration, cofactor, coenzyme, or trans-acting factor could lead to release of the cytidine.
Evolution of C-to-U and U-to-C enzymes from the DYW domain family
The DYW domain family can be broken into two groups: the DYW-PG that catalyze C-to-U editing and the DYW-KP that catalyze U-to-C editing. There is extensive amino acid identity and similarity between the two groups (Fig. S9). The proposed U-to-C crosslinking mechanism is also functionally similar to canonical C-to-U deamination (Fig. S9). The zinc ion in both mechanisms plays a dual role in stabilizing intermediates and acting as a Lewis acid. The glutamate has essential tasks in both reactions shuttling protons. The elegant simplicity of using the zinc and glutamate ions in slightly different reaction pathways serves as a remarkable example of how enzymes can evolve to catalyze new reactions that seemingly overcome thermodynamic barriers.
Experimental procedures
Plasmids and expression of KP2 and KP6 for recombinant protein purification
KP2 (pET-PLS-KP2_rpoA-U), KP3 (pET-PLS-KP3_rpoA-U), KP6 (pET-PLS-KP6_rpoA-U), and KP6 with C-template (pET-PLS-KP6_rpoA-C) plasmids were obtained from addgene.org. Plasmids were isolated using alkaline lysis (28) and subcloned into Rosetta 2 (DE3) pLysS cells originally from Novagen. LB broth was inoculated to make 3 L cultures initially at an absorbance of A600 = 0.05 and allowed to grow at 37 °C until cultures reached A600 = 0.3. Cultures were transferred to an incubated shaker set to 18 °C and allowed to chill in the air for 30 min. Zinc acetate (0.4 mM final) and IPTG (1 mM final) were added, and cultures were incubated for 5 h at 18 °C. Cell pellets were harvested in 6× 250 ml bottles through centrifugation at 5000g for 15 min. Media were removed after pelleting, and bottles were stored in the −80 °C freezer.
Purification of recombinant KP2 and KP6
Cell pellets from 3 L cultures were suspended in 150 ml of lysis buffer (20 mM Tris pH 7.2 @ room temperature (RT), 250 mM NaCl, and 10% Glycerol). Crude cell suspensions were disrupted by sonication using 6× 20 s pulses. Lysates were cleared by centrifugation at 12,000g for 15 min. Cleared lysates were then passed over 5 ml of His Pure IMAC resin column (ThermoFisher) equilibrated with lysis buffer. The IMAC resin was washed using 200 ml of IMAC wash buffer (20 mM Tris pH 7.2@RT, 250 mM NaCl, 10% glycerol, and 20 mM imidazole) and then eluted using 20 ml of elution buffer (20 mM Tris pH 7.2 @ RT, 250 mM NaCl, 10% glycerol, and 250 mM imidazole). Elution fractions were concentrated with a 50,000 MCO Amicon Ultra-15 Centrifugal Filter Unit (Merck KGaA) and then dialyzed in low salt buffer (20 mM Tris pH 7.2 @ RT, 20 mM NaCl, and 10% glycerol) then passed through a HiTrap Q XL 5 ml column (Cytiva). Protein was eluted using a continuous gradient using a high salt buffer (20 mM Tris pH 7.2 @ RT, 1 M NaCl, and 10% glycerol) with a NGC Chromatography System (Bio-Rad Laboratories Inc). The protein was concentrated and injected into a 1 ml load loop leading into a Superdex 200 Increase 10/300 Gl column (GE Healthcare Bio-Sciences) using a single isocratic step using the lysis buffer. Fractions of 0.5 ml were collected, screened for the protein of interest using SDS-PAGE, pooled, aliquoted, and flash frozen with liquid nitrogen. Aliquots were stored in the −80 °C, with C-to-U activity for KP6 fractions retained after 6 months of storage.
Production of RNA substrates
DNA templates pET-PLS-KP6_rpoA-U and pET-PLS-KP6_rpoA-C were used to construct U and C putative RNA substrates. Primers rpoA_SK_F:CGCTCTAGAACTAGTGGATCATGCGTCAAAGATCATCTCC, rpoA_KS_R_U:TCGAGGTCGACGGTATCGCTAGCTCTCAAATTTTGCAC, rpoA_KS_R_C:TCGAGGTCGACGGTATCGCTAGCTCTCAGATTTTGCAC, rpoA_AC5_F:ACACAAAGCCGCTCCATCAGATGCGTCAAAGATCATCTCC, rpoA_CYC1_R_U:GTCACGCTTACATTCACGCGCTAGCTCTCAAATTTTGCAC, and rpoA_CYC1_R_C:GTCACGCTTACATTCACGCGCTAGCTCTCAGATTTTGCAC were used to create initial PCR products that lacked the T7 promoter. The T7 promoter sequences were then added in a second round of PCR using the appropriate primers, either T7SK: TAATACGACTCACTATAGGGGCGCTCTAGAACTAGTGGATC or T7AC5:TAATACGACTCACTATAGGGGACACAAAGCCGCTCCATCAG. PCR products were purified using the GeneJET PCR Purification Kit (Thermo Scientific). RNA was produced using the T7 TranscriptAid High sola Kit (ThermoFisher), DnaseI treated, and purified using the RNA Clean & Concentrator (Zymo Research).
RNA editing activity assay in vitro
Assay conditions for C-to-U and U-to-C were similar to previously published assays (9). For each 12.5 μl reaction, 6.25 μl of one of the following protein fractions was added: KP6 crude lysate (3.6 μg/μl); KP6 cleared lysate (3.72 μg/μl); KP6 IMAC (1.1 μg/μl); KP6 IMAC-C (12.9 μg/μl); KP6 IEX (0.4 μg/μl); KP6 SEC (0.1 μg/μl); KP2 crude lysate (9.0 μg/μl); KP2 cleared lysate (7.7 μg/μl); KP2 IMAC (7.5 μg/μl); KP2 IEX (0.5 μg/μl); and KP2 SEC (1.2 μg/μl). The protein was then added 1:1 to a 6.25 μl mixture comprised of 60 mM Hepes-KOH (pH 7.7), 6 mM magnesium acetate, 90 mm potassium acetate, 60 mM ammonium acetate, 4 mM ATP, 10 mM dithiothreitol, 2% polyethylene glycol 6000, 10% glycerol, 10 U of Rnase inhibitor (Fermentas), 2× proteinase inhibitor mixture (Complete EDTA-free; Boehringer Mannheim), and 20 fmol of mRNA substrate. Addition of experimental molecules was at a final concentration of 2 mM for the 12.5 μl reaction volume. Reaction sets always contained a reference untreated sample used to calculate relative editing per experiment. Reactions were incubated at 28 °C for 2.5 h and stopped through heating to 65 °C for 5 min.
Quantification of RNA editing activity
RNA was copied into cDNA using 10 μl reactions using Maxima Reverse Transcriptase (ThermoFisher) except in the stated cases where RevertAid Reverse Transcriptase was used (ThermoFisher). For the assay of editing in vitro, a sequence-specific primer, either KS: TCGAGGTCGACGGTATC or CYC: GTCACGCTTACATTCACGC, was used. For RNAs from bacterial cultures, the gene-specific primer MLH309: GTTAGCAGCCGGATCTCAG was used. For C. richardii editing, random hexamers were used in RT reactions.
Reaction products were then amplified in a PCR step using Dream Taq (ThermoFisher). For assay of editing in vitro primers, T7-SK and KS or T7AC5 and CYC were used. For KP6 culture editing, PCR included primers rpoA_SK_F and MLH309. For KP2 and KP3, rpoA_KP2_KP3_SK_F: CGCTCTAGAACTAGTGGATCCAGTTCAACCGAGATTATGTCAAG and MLH309 were used. For C. richardii editing analysis, the following primers were used: Ct_ycf2_Sub23_F: CGCTCTAGAACTAGTGGATCCCAATGCGCGGATTCTATTTG; Ct_ycf2_Sub23_R: TCGAGGTCGACGGTATCCTGACGTTGACGCCCG; Ct_chlL_Sub_F: CGCTCTAGAACTAGTGGATCCGTGAAACGAAAATAGCAGTTTAC; and Ct_chlL_Sub_R: TCGAGGTCGACGGTATCTAACTGTAAAACCAAGCGAAGG. Reactions were checked for product on a 1% agarose/0.25% Synergel gels stained with ethidium bromide and cleaned using the GeneJET PCR Purification Kit (Thermo Scientific). Cleaned PCR products were then sequenced by Retrogen, INC. Peaks from electrophoretograms were calculated using BioEdit, version 7.7.1.
Growth of cultures for U-to-C analysis in bacteria
Overnight cultures of Rosetta 2 (DE3) pLysS containing pET-PLS-KP6_rpoA-U were used to seed 3 ml cultures to an A600 of 0.05. Cells were grown at 37 °C until they reached an A600 0.5 After chilling in the air at 4 °C for 10 min, zinc acetate (0.4 mM final) and IPTG (0.4 mM final) were added. Cells were incubated with 200 rpm shaking overnight (18 h) at 16 °C. Fractions (0.8 ml) from each culture were either used for Rneasy Kit purification or centrifuged in a microfuge at 5000g, and media were removed from cell pellets. Cell pellets were frozen and stored in a −80 °C freezer.
Organic phase separation of RNAs
Cell pellets were disrupted using 1 ml of RiboZol RNA Extraction Reagent (VWR) for 5 min at RT. Two hundred microliters of chloroform was added, and tubes were shaken. Phases were developed through centrifugation at 14,000g for 10 min. Aqueous phases (the top) were removed, and RNA isolated by 1:1 addition of isopropanol. Once the aqueous fraction was removed, the interphase was removed along with 120 μl of organic layer. Proteins were precipitated through the addition of 1.2 ml of methanol and incubated 10 min at −20 °C. Proteins were pelleted through centrifugation at 14,000g for 10 min. Cell pellets were dried and resuspended in 90 μl of TE followed by the addition of 10 μl (8 U) of Proteinase K (New England Biolabs). Protein was digested at 37 °C for 15 min. For the isolation of free RNA, 1 ml of RiboZol was added. Chloroform was added (200 μl), the aqueous phase isolated, and RNA precipitated using 1:1 isopropanol.
Purification of RNA using silica column-based techniques
Isolation of RNA from bacteria pellets using silica column techniques were followed from a recent publication (20). Cell RNA protect reagent (Qiagen) was added to cell culture, and cells were pelleted through centrifugation for 10 min at 5000g. Bacterial pellets were resuspended in 30 μl of lysozyme buffer (30 mM Tris-HCL, pH 8.0, 1 mM EDTA, and 15 mg/ml lysozyme) and 20 μl (16 U) of Proteinase K. Protease digestion reactions were incubated 10 min at RT. Purification then followed manufacturer instructions for the RNeasy Mini kit (Qiagen).
Denaturing immobilized metal chromatography
Cell pellets were resuspended in 500 ml of denaturing lysis buffer (6M guanidinium chloride, 20 mM Tris-HCl pH 7.3 @ RT, 300 mM NaCl, and 10 mM imidazole). His Pure Ni-NTA resin (ThermoFisher) was equilibrated in lysis buffer, and 150 μl of suspension was added per bacterial suspension. Microfuges containing cell and Ni-NTA mixtures were inverted several times and incubated 5 min at RT. Suspensions were centrifuged 5000g for 5 min and supernatant removed. Ni-NTA resin was washed three times with 1 ml denaturing lysis buffer. Bound proteins were eluted by addition of 120 μl of denaturing elution buffer (6M guanidinium chloride, 20 mM Tris-HCl pH 7.3 @ RT, 300 mM NaCl, and 250 mM imidazole). The supernatant was removed, and 1.2 ml of methanol was added to precipitate proteins during a 10 min precipitation at −20 °C. Methanol was removed after centrifugation at 14,000g for 10 min, and the pellets were dried. Protein pellets were resuspended in 90 μl TE buffer (10 mM Tris-HCl pH8.0, 1 mM EDTA), and 10 μl (8 U) of proteinase K was added. Protease reactions were incubated for 10 min at 37 °C. RNA was isolated from the aqueous fractions of organic phase separation using RiboZol.
C. richardii materials and growth conditions
C. richardii (RNWT1) spores were obtained from Carolina Biological Supply Company. Spores were germinated on C-Fern Medium. Gametophytes bred on plates and sporophytes were transferred to soil. Plants were maintained clonally for a year under four rows of 16 W 5000 K fluorescent lights in a 16 h day/8 h night cycle at 22 °C. Once older leaves reach senescence, new plantlets at frond bracts were transferred to new pots in soil. Plants were watered weekly with a general-purpose fertilizer.
QTOF mass spectrometry
Proteins were diluted to 1 mg/ml and dialyzed overnight against 20 mM ammonium acetate. Mass spectrometry and analysis was performed at the UCI Mass Spectrometry Facility using a Waters Acquity UPLC Xevo G2-XS instrument. Data were analyzed with Waters MassLynx v.4.1.
Data availability
Data are contained within the manuscript and supplementary information.
Supporting information
This article contains supporting information.
Conflict of interests
The authors declare no conflicts of interest with the contents of this article.
Acknowledgments
The authors convey special thanks to Jamil Momand and Robert Vellanoweth at CSULA for many discussions about the project. They thank undergraduate students Jesus Torres, Mark Terras, Gustavo Cervantes, and Dorsa Mohammadigerani. They also thank MS students Alfredo Salinas-Jimenez and Karen “Stephanie” Lemus for participation in discussions around the project. They wish to thank the UCI Mass Spectrometry Facility and Ben Katz for assistance with collection and analyses of protein mass spectrometry data. MS data were collected on a Waters Acquity UPLC Xevo G2-XS QTOF system [NIH supplemental funding support received by J. S. Nowick (NIGMS GM097562), Vy Y. Duong (NIH GM105938), and O. Cinquin (NIGMS GM102635)].
Author contributions
E. T. G., S. O. C., M. L. H., and M. S. investigation; E. T. G., S. O. C., M. L. H., and M. S. writing–review & editing. M. L. H. conceptualization; M. L. H. data curation; M. L. H. formal analysis; M. L. H. funding acquisition; M. L. H. methodology; M. L. H project administration; M. L. H resources; M. L. H. software; M. L. H. supervision; M. L. H. validation; M. L. H and M. S. visualization, M. L. H. writing–original draft.
Funding and additional information
The work was funded by the National Institutes of Health AREA15 GM144905 grant awarded to Michael L. Hayes. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Reviewed by members of the JBC Editorial Board. Edited by Sarah E. O'Connor
Supporting information
References
- 1.Fisher A.J., Beal P.A. Structural basis for eukaryotic mRNA modification. Curr. Opin. Struct. Biol. 2018;53:59–68. doi: 10.1016/j.sbi.2018.05.003. [DOI] [PubMed] [Google Scholar]
- 2.Aphasizheva I., Alfonzo J., Carnes J., Cestari I., Cruz-Reyes J., Göringer H.U., et al. Lexis and grammar of mitochondrial RNA processing in trypanosomes. Trends Parasitol. 2020;36:337–355. doi: 10.1016/j.pt.2020.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Knoop V. C-to-U and U-to-C: RNA editing in plant organelles and beyond. J. Exp. Bot. 2023;74:2273–2294. doi: 10.1093/jxb/erac488. [DOI] [PubMed] [Google Scholar]
- 4.Barkan A., Rojas M., Fujii S., Yap A., Chong Y.S., Bond C.S., et al. A combinatorial amino acid code for RNA recognition by pentatricopeptide repeat proteins. PLoS Genet. 2012;8 doi: 10.1371/journal.pgen.1002910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yan J., Yao Y., Hong S., Yang Y., Shen C., Zhang Q., et al. Delineation of pentatricopeptide repeat codes for target RNA prediction. Nucleic Acids Res. 2019;47:3728–3738. doi: 10.1093/nar/gkz075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yagi Y., Hayashi S., Kobayashi K., Hirayama T., Nakamura T. Elucidation of the RNA recognition code for pentatricopeptide repeat proteins involved in organelle RNA editing in plants. PLoS One. 2013;8 doi: 10.1371/journal.pone.0057286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Takenaka M., Zehrmann A., Brennicke A., Graichen K. Improved computational target site prediction for pentatricopeptide repeat RNA editing factors. PLoS One. 2013;8 doi: 10.1371/journal.pone.0065343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takenaka M., Takenaka S., Barthel T., Frink B., Haag S., Verbitskiy D., et al. DYW domain structures imply an unusual regulation principle in plant organellar RNA editing catalysis. Nat. Catal. 2021;4:510–522. doi: 10.1038/s41929-021-00633-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hayes M.L., Santibanez P.I. A plant pentatricopeptide repeat protein with a DYW-deaminase domain is sufficient for catalyzing C-to-U RNA editing in vitro. J. Biol. Chem. 2020;295:3497–3505. doi: 10.1074/jbc.RA119.011790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oldenkott B., Yang Y., Lesch E., Knoop V., Schallenberg-Rüdinger M. Plant-type pentatricopeptide repeat proteins with a DYW domain drive C-to-U RNA editing in Escherichia coli. Commun. Biol. 2019;2:85. doi: 10.1038/s42003-019-0328-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Salone V., Rüdinger M., Polsakiewicz M., Hoffmann B., Groth-Malonek M., Szurek B., et al. A hypothesis on the identification of the editing enzyme in plant organelles. FEBS Lett. 2007;581:4132–4138. doi: 10.1016/j.febslet.2007.07.075. [DOI] [PubMed] [Google Scholar]
- 12.Wagoner J.A., Sun T., Lin L., Hanson M.R. Cytidine deaminase motifs within the DYW domain of two pentatricopeptide repeat-containing proteins are required for site-specific chloroplast RNA editing. J. Biol. Chem. 2015;290:2957–2968. doi: 10.1074/jbc.M114.622084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boussardon C., Avon A., Kindgren P., Bond C.S., Challenor M., Lurin C., et al. The cytidine deaminase signature HxE(x)n CxxC of DYW1 binds zinc and is necessary for RNA editing of ndhD-1. New Phytol. 2014;203:1090–1095. doi: 10.1111/nph.12928. [DOI] [PubMed] [Google Scholar]
- 14.Hayes M.L., Dang K.N., Diaz M.F., Mulligan R.M. A conserved glutamate residue in the C-terminal deaminase domain of pentatricopeptide repeat proteins is required for RNA editing activity. J. Biol. Chem. 2015;290:10136–10142. doi: 10.1074/jbc.M114.631630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gerke P., Szövényi P., Neubauer A., Lenz H., Gutmann B., McDowell R., et al. Towards a plant model for enigmatic U-to-C RNA editing: the organelle genomes, transcriptomes, editomes and candidate RNA editing factors in the hornwort Anthoceros agrestis. New Phytol. 2020;225:1974–1992. doi: 10.1111/nph.16297. [DOI] [PubMed] [Google Scholar]
- 16.Grewe F., Herres S., Viehöver P., Polsakiewicz M., Weisshaar B., Knoop V. A unique transcriptome: 1782 positions of RNA editing alter 1406 codon identities in mitochondrial mRNAs of the lycophyte Isoetes engelmannii. Nucleic Acids Res. 2011;39:2890–2902. doi: 10.1093/nar/gkq1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Knie N., Grewe F., Fischer S., Knoop V. Reverse U-to-C editing exceeds C-to-U RNA editing in some ferns - a monilophyte-wide comparison of chloroplast and mitochondrial RNA editing suggests independent evolution of the two processes in both organelles. BMC Evol. Biol. 2016;16:134. doi: 10.1186/s12862-016-0707-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kugita M., Yamamoto Y., Fujikawa T., Matsumoto T., Yoshinaga K. RNA editing in hornwort chloroplasts makes more than half the genes functional. Nucleic Acids Res. 2003;31:2417–2423. doi: 10.1093/nar/gkg327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gutmann B., Royan S., Schallenberg-Rüdinger M., Lenz H., Castleden I.R., McDowell R., et al. The expansion and diversification of pentatricopeptide repeat RNA-editing factors in plants. Mol. Plant. 2020;13:215–230. doi: 10.1016/j.molp.2019.11.002. [DOI] [PubMed] [Google Scholar]
- 20.Ichinose M., Kawabata M., Akaiwa Y., Shimajiri Y., Nakamura I., Tamai T., et al. U-to-C RNA editing by synthetic PPR-DYW proteins in bacteria and human culture cells. Commun. Biol. 2022;5:968. doi: 10.1038/s42003-022-03927-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cohen R.M., Wolfenden R. The equilibrium of hydrolytic deamination of cytidine and N 4 -methylcytidine. J. Biol. Chem. 1971;246:7566–7568. [PubMed] [Google Scholar]
- 22.McDowell R., Small I., Bond C.S. Synthetic PPR proteins as tools for sequence-specific targeting of RNA. Methods. 2022;208:19–26. doi: 10.1016/j.ymeth.2022.10.003. [DOI] [PubMed] [Google Scholar]
- 23.Shen C., Zhang D., Guan Z., Liu Y., Yang Z., Yang Y., et al. Structural basis for specific single-stranded RNA recognition by designer pentatricopeptide repeat proteins. Nat. Commun. 2016;7 doi: 10.1038/ncomms11285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hayes M.L., Giang K., Berhane B., Mulligan R.M. Identification of two pentatricopeptide repeat genes required for RNA editing and zinc binding by C-terminal cytidine deaminase-like domains. J. Biol. Chem. 2013;288:36519–36529. doi: 10.1074/jbc.M113.485755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vangerow S., Teerkorn T., Knoop V. Phylogenetic information in the mitochondrial nad5 gene of pteridophytes: RNA editing and intron sequences. Plant Biol. 1999;1:235–243. [Google Scholar]
- 26.Guo W., Grewe F., Mower J.P. Variable frequency of plastid RNA editing among ferns and repeated loss of uridine-to-cytidine editing from vascular plants. PLoS One. 2015;10 doi: 10.1371/journal.pone.0117075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marchant D.B., Sessa E.B., Wolf P.G., Heo K., Barbazuk W.B., Soltis P.S., et al. The C-Fern (Ceratopteris richardii) genome: insights into plant genome evolution with the first partial homosporous fern genome assembly. Sci. Rep. 2019;9 doi: 10.1038/s41598-019-53968-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Birnboim H.C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7:1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Raushel F.M., Thoden J.B., Holden H.M. The amidotransferase family of enzymes: molecular machines for the production and delivery of ammonia. Biochemistry. 1999;38:7891–7899. doi: 10.1021/bi990871p. [DOI] [PubMed] [Google Scholar]
- 30.Queiroz R.M.L., Smith T., Villanueva E., Marti-Solano M., Monti M., Pizzinga M., et al. Comprehensive identification of RNA-protein interactions in any organism using orthogonal organic phase separation (OOPS) (vol 37, pg 169, 2019) Nat. Biotechnol. 2019;37:692. doi: 10.1038/s41587-018-0001-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schaufelberger F., Hu L., Ramstrom O. trans-Symmetric dynamic covalent systems: connected transamination and transimination reactions. Chemistry. 2015;21:9776–9783. doi: 10.1002/chem.201500520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Booth M.J., Raiber E.A., Balasubramanian S. Chemical methods for decoding cytosine modifications in DNA. Chem. Rev. 2015;115:2240–2254. doi: 10.1021/cr5002904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang H., Poe A., Pak L., Nandakumar K., Jandu S., Steppan J., et al. An in situ activity assay for lysyl oxidases. Commun. Biol. 2021;4:840. doi: 10.1038/s42003-021-02354-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are contained within the manuscript and supplementary information.