Abstract

The current investigation deployed Mendelian randomization (MR) to elucidate the causal relationship between circulating proteins and sepsis. A rigorous two-sample MR analysis evaluated the effect of plasma proteins on the sepsis susceptibility. To affirm the integrity of MR findings, a suite of supplementary analyses, including Bayesian colocalization, Steiger filtering, the assessment of protein-altering polymorphisms, and the correlation between expression quantitative trait loci and protein quantitative trait loci (pQTLs), was employed. The study further integrated the examination of protein–protein interactions and pathway enrichment, along with the identification of pharmacologically actionable targets, to advance our comprehension and outline potential sepsis therapies. Subsequent analyses leveraging cis-pQTLs within MR studies unveiled noteworthy relationships: 94 specific proteins exhibited significant links with sepsis-related 28 day mortality, while 96 distinct proteins correlated with survival outcomes in sepsis. Furthermore, incorporating both cis- and trans-pQTLs in MR investigations revealed more comprehensive findings, associating 201 unique proteins with sepsis-related 28 day mortality and 199 distinct proteins with survival outcomes in sepsis. Markedly, colocalization analyses confirmed that eight of these proteins exhibited prominent evidence for colocalization, emphasizing their potential criticality in sepsis pathophysiology. Further in silico analyses were conducted to delineate putative regulatory networks and to highlight prospective drug targets among these proteins. Employing the MR methodology has shed light on plasma proteins implicated in the etiopathogenesis of sepsis. This novel approach unveiled numerous biomarkers and targets, providing a scientific rationale for the development of new therapeutic strategies and prophylactic measures against sepsis.
Introduction
Sepsis constitutes a complex, life-threatening condition characterized by organ dysfunction and stems from a dysregulated host response to infection.1 Data from 2020 reveal approximately 48.9 million sepsis cases globally, causing 11 million fatalities which represent 19.7% of worldwide deaths.2 Recognizing its gravity, the World Health Organization has prioritized sepsis as a global health concern, urging international efforts to mitigate its prevalence and impact.3 Unfortunately, as the pathogenesis of sepsis is complicated and has not yet been fully elucidated, no drugs have successfully slowed its progression.4 Therefore, exploring the potential mechanism and drug targets for sepsis holds significant importance.
Proteins are quintessential molecules regulating various life processes, impacting both health and disease.5,6 The human plasma proteome comprises an intricate network of proteins, either secreted or shed into the bloodstream, facilitating essential functions and intertissue communication.7 Dysregulation of these proteins often signals disease, positioning them as crucial targets for pharmacological intervention.8 Given the ease of blood collection compared to other tissues, circulating proteins offer a promising resource for identifying disease-specific molecular signatures in large population cohorts.7
Advancements in proteomic methods now allow researchers to simultaneously quantify numerous circulating proteins in large-scale study populations.9 Genome-wide association studies (GWAS) assessing the levels of these proteins facilitate the identification of protein quantitative trait loci (pQTLs), enhancing our understanding of the genetic architecture governing protein expression.6 Integrating pQTL data with genetic associations related to various diseases yields robust mechanistic insights into the pathological roles of proteins. This integration is achieved by using a statistical method known as Mendelian randomization (MR). As a cornerstone technique in genetic epidemiology, MR leverages genetic variants as instrumental variables, refining causal inferences between exposures and outcomes while effectively controlling for confounding factors and addressing reverse causality.10 Utilizing pQTLs as such variables, MR facilitates the efficient exploration of causal relationships between circulating protein biomarkers and disease phenotypes, opening avenues for targeted therapeutic strategies and improving disease prognostication.
While prior investigations have explored the relationships between circulating proteins and sepsis incidence, they are often limited by their reliance on low-throughput protein assays and small sample sizes.11 Additionally, being observational, they inherently carry the risk of confounding variables and potential reverse causality, suggesting that associations with circulating proteins may not necessarily imply a direct causal relationship with sepsis. To deepen our understanding of the role of serum proteins in sepsis, more robust evidence is needed. In this context, our study employed an MR analytical framework to identify circulating proteins with a potential causal link to sepsis, aiming to reveal novel pharmacological targets. Thus, it provides crucial insights into sepsis prevention and therapeutic intervention.
Methods
Study Exposures
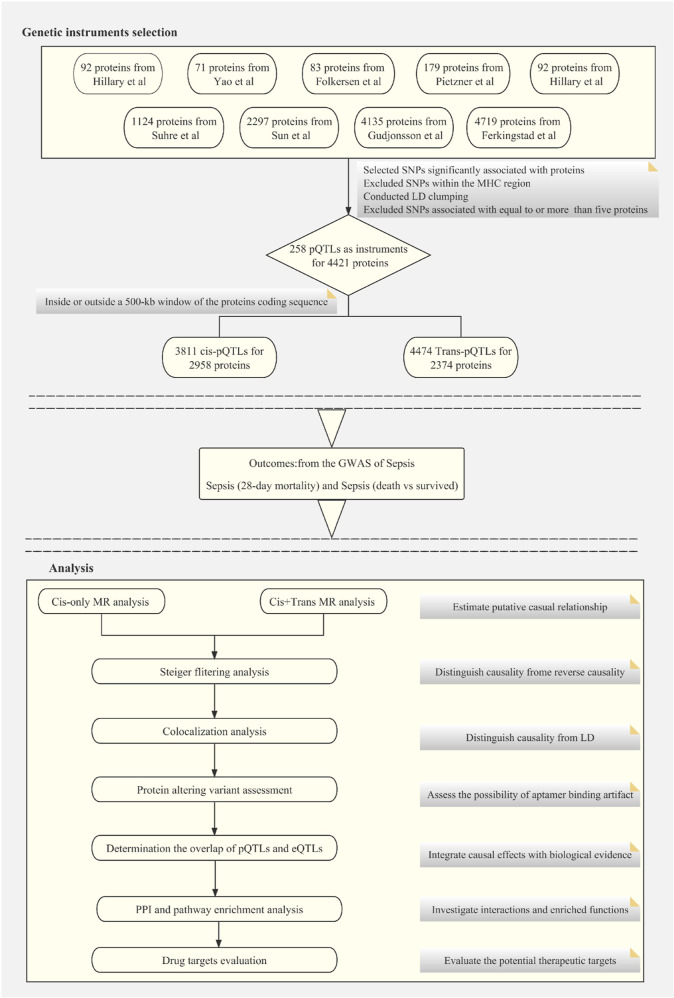
In this study, we focused on the levels of circulating proteins as predictor variables. These protein levels were hypothesized to be genetically determined. Genetic instruments for these proteins were curated using data from nine large-scale proteomic GWAS, each involving cohorts of over 500 individuals and analyzing over 50 proteins.6−8,12−17 The comprehensive details for these nine studies are available in Table S1.
pQTLs selected through a multistep process were employed as potential instrumental variables. Initially, single-nucleotide polymorphisms (SNPs) with protein associations meeting significance levels in respective proteomic studies were identified (Table S1). Subsequently, to address complexities due to linkage disequilibrium (LD), we excluded SNPs within the human major histocompatibility complex region, defined as the locus spanning from 26 to 34 Mb on chromosome 6.
The third stage of SNP selection involved “LD clumping” to identify independent pQTLs, setting a threshold of r2 greater than 0.01. Additionally, a flank range was set to less than 5000 kb, both upstream and downstream. The final criterion involved excluding genetic instruments linked to five or more proteins to mitigate the potential confounding effects of pleiotropy on the result interpretability.
In this study, pQTLs were categorized based on their proximity to the gene encoding the protein of interest. cis-pQTLs were defined as those pQTLs located within a 500 kb interval flanking the protein-coding sequence, while trans-pQTLs were located beyond the 500 kb threshold relative to the coding gene sequence.
Study Outcomes
In our MR analysis, we selected two outcomes. The first was sepsis—28 day mortality, with data from 1896 European ancestry cases and 484 588 European ancestry controls.18 The second was sepsis—death versus survival, comprising 1896 European ancestry sepsis death cases and 9747 European ancestry sepsis survival cases.18 Detailed information about these outcomes can be found in Table S2.
MR Analysis
In the MR analysis, we hypothesized that genotype-predicted protein levels act as exposures, with sepsis—28 day mortality and sepsis—death vs survival as the respective outcomes. We utilized cis-pQTLs and a combination of cis- and trans-pQTLs to construct genetic instruments for MR analyses. To refine these genetic instruments, we excluded SNPs with a minor allele frequency below 0.01 and those that accounted for more variability in the outcome than in the exposure.
Instrument robustness was ensured using F-statistic applying the formula beta2/se2, where an F value > 10 indicated a strong genetic instrument. MR effects were estimated using the Wald ratio for single pQTL cases and the inverse variance weighted method for multiple pQTLs.
In cases of heterogeneity (detected via Cochran’s Q test) or horizontal pleiotropy (detected by the MR-Egger method), we applied the weighted median method or MR-Egger regression, respectively. The MR analyses were carried out using the “TwoSampleMR” R package (http://github.com/MRCIEU/TwoSampleMR).
Steiger Filtering Analysis
To evaluate reverse causality in MR associations exceeding the designated multiple-testing correction threshold, we applied the Steiger filtering method utilizing the “TwoSampleMR” package in R. The results of this analysis are expressed as categorical variables to improve interpretability: “true” signifies a directional effect from exposure to outcome with statistical significance at p-values <0.05; “false” indicates a reverse directional effect meeting the same significance criterion; and “uncertain” is attributed to findings where p-values ≥0.05.19
Colocalization Analysis
To identify plasma proteins with potential causal links to sepsis, we retained MR results with p-values <0.05. Results surpassing the multiple-testing threshold in the MR analysis underwent Bayesian colocalization analysis, evaluating the posterior probability that each genomic locus harbors a single variant influencing both protein levels and sepsis risk (as opposed to a coincidental sharing of the variant due to LD).19 The evaluation of colocalization was carried out using the “coloc” R package (http://cran.r-project.org/web/packages/coloc). This package calculates posterior probabilities for five hypotheses (0, 1, 2, 3, and 4), which determine whether a singular variant is jointly influencing two traits. Specifically, hypothesis 4, suggesting that a single genetic variant influences both traits,19 was considered supported if the posterior probability exceeded 0.5. Bayesian colocalization analysis assumes a single common causal SNP shared between the traits, a known limitation as, in practice, loci may have multiple causal SNPs.9
Protein-Altering Variant Annotation of cis-pQTLs
Affinity-based proteomics relies on stable binding epitopes. However, genetic polymorphisms altering protein structure, known as protein-altering variants (PAVs), can erroneously appear as cis-pQTLs due to altered aptamer interactions rather than genuine protein abundance variations. To differentiate potential artifacts from true binding discrepancies, cis-pQTLs supported by MR were examined for the presence of PAVs or linkage with PAVs (r2 ≥ 0.8) using the Ensembl Variant Effect Predictor (https://asia.ensembl.org/Tools/VEP). Variants were classified as PAVs if annotated as any of the following: coding sequence, frameshift, in-frame deletion or insertion, missense, splice acceptor, splice donor, splice region, start lost, stop gained, or stop lost.8
Evaluation of the Overlap between pQTLs and Expression Quantitative Trait Loci
Genetic variants exert a quantifiable effect on transcript and protein expression levels, with the alignment of expression quantitative trait loci (eQTLs) with pQTLs suggesting that genetic influences on protein expression may be mediated through mRNA transcription regulation. This underscores the biological relevance of the pQTL findings. To investigate the mechanisms through which pQTLs impact plasma protein levels, we conducted a genetic analysis examining the overlap between pQTLs and eQTLs. For pQTLs supported by MR, we scrutinized subject variants and their high-linkage proxies (r2 ≥ 0.8) for relevant eQTLs sharing the same allelic effects, using data from the GTEx Portal (V8, https://www.gtexportal.org). Furthermore, we assessed the concurrence of pQTLs with eQTLs from sepsis-associated tissues or cells, such as cartilage, synovium, fibroblasts, and macrophages, using eQTL data sourced from the Open Targets Genetics portal (https://genetics.opentargets.org/).
Protein–Protein Interaction and Functional Enrichment Analysis
Interactions among MR-selected proteins were identified by constructing protein–protein interaction (PPI) networks using STRING (V11.5, https://string-db.org/). Concurrently, “ClusterProfiler” in R (https://bioconductor.org/packages/release/bioc/html/clusterProfiler.html) was employed to conduct Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment, revealing pathways potentially associated with these proteins.
Mapping of the MR-Prioritized Proteins with Drug Targets
To explore the overlap between MR-identified proteins and potential drug targets, we aligned these proteins with Finan et al.’s druggable genome, which stratifies 4479 genes into three drug development tiers.20 Tier 1 comprises 1427 genes targeted by existing or trial-phase therapeutics, Tier 2 consists of 682 genes with bioactive ligands or significant similarity to current drug targets, Tier 3 encompasses 2370 genes involving secreted/extracellular proteins and additional druggable classes not covered in the first two tiers, with a further breakdown into Tier 3A for genes proximal to GWAS SNPs with extracellular protein coding; and Tier 3B for the rest. Our analysis assessed the MR-selected proteins against these tiers, their interaction with small molecules, and their biotherapeutic targeting potential, including monoclonal antibodies. Drug target classification was enhanced by utilizing the Therapeutic Target Database (TTD) (http://db.idrblab.net/ttd/), presenting 3578 drug targets across development stages—498 approved, 1342 in clinical trials, 185 under preclinical/patent review, and 1553 in research.21 The emphasis was placed on evaluating target classes, associated drugs, and diseases targeted by those drugs.
Results
Identifying Genetic Instruments for Plasma Proteins
We aggregated pQTLs from nine proteomic GWAS6−8,12−17 to create genetic instruments for our MR study, as summarized in Figure 1. Ultimately, we selected 8285 pQTLs linked to 4421 proteins (2518 unique) for subsequent MR analysis. These are listed in Table S3. These genetic instruments were classified into 3811 cis-pQTLs affecting 2958 proteins (1558 unique) and 4474 trans-pQTLs influencing 2374 proteins (1763 unique). Within the available data set, 911 proteins were impacted by both cis- and trans-pQTLs. In contrast, 2047 proteins were solely associated with cis-pQTLs and 1463 exclusively with trans-pQTLs.
Figure 1.
Flowchart of the MR-based analytical framework for evaluating the effect of plasma proteome on sepsis.
Estimating the Effects of Plasma Proteins on Sepsis by Using MR
Considering the higher likelihood of cis-pQTLs to exert specific biological effects compared to trans-pQTLs, we initially employed cis-pQTLs as genetic instruments in our MR analyses to systematically examine potential causal relationships between plasma protein levels and sepsis, as illustrated in Tables S4 and S5. In the MR analysis focusing on cis-pQTLs, 139 proteins (94 unique proteins) and 138 proteins (96 unique proteins) were significantly associated with sepsis outcomes concerning 28 day mortality and the dichotomy of death versus survival, respectively (p < 0.05) (Figure 2A,B and Tables S4 and S5). Consistent associations across both sepsis outcomes suggest a potential convergence of the underlying pathogenic mechanisms. Furthermore, sensitivity analyses of the cis-pQTL MR aligned with the primary findings, with no emergence or disappearance of significant protein–trait correlations, confirming the robustness of the results (Tables S6 and S7). Incorporating trans-pQTLs into our MR studies is expected to augment the reliability of inferred associations between proteins and phenotypic outcomes. Consequently, utilizing an exhaustive suite of instrumental variables that synergistically integrated both cis- and trans-pQTL insights, we discerned the significant associations of 258 proteins, encapsulating 201 unique proteins, and 254 proteins, embodying 199 unique proteins, with outcomes related to sepsis, specifically focusing on 28 day mortality and the critical distinction between death and survival (Figure 2C,D and Tables S8 and S9). This methodology revealed 278 protein-trait associations supported by MR evidence, most of which were not observed in analyses limited to cis-pQTLs, as detailed in Table S10. Additionally, sensitivity tests conducted on the MR analyses utilizing cis-pQTLs corroborated the initial results, showing no appearance or disappearance of significant protein–trait correlations, reinforcing the solidity of our findings (Tables S11 and S12).
Figure 2.
Volcano plot for the cis-only MR analyses, with circles representing the results for proteins on sepsis. (A) Volcano plot of cis-pQTL on sepsis (28 day mortality); (B) volcano plot of cis-pQTL on sepsis (death vs survived); (C) volcano plot of cis + trans-pQTL on sepsis (28 day mortality); and (D) volcano plot of cis + trans-pQTL-pQTL on sepsis (death vs survived).
Testing the Causal Direction by Steiger Filtering Analysis
To rigorously evaluate the direction of causality in predicted protein–phenotype associations and address potential confounding from reverse causation, we conducted a directionality assessment known as the Steiger filtering analysis. This analysis, implemented to determine the validity of the causal pathways identified by MR from proteins to sepsis, revealed that all associations, including those from sensitivity analyses, were accurately aligned, confirming that proteins serve as the causal factors for sepsis, as detailed in Table S13.
Colocalization of the pQTLs with Loci Associated with Sepsis Risk
To mitigate the confounding impact of LD on associations identified through MR, comprehensive colocalization analyses were undertaken. These analyses aimed to evaluate the probability that observed genetic correlations with both protein expression and phenotypic manifestations originated from a singular causal variant. Colocalization was specifically performed for proteins linked via a single instrumental variable where summary data from GWAS were available.
For the prioritized cis-MR (cis-MR) associations, colocalization analyses were executed on 139 associations linked to sepsis and 28 day mortality outcomes, revealing five significant colocalization signals, specifically denoted as apolipoprotein L3 (APOL3), nucleoside diphosphate kinase B (NDKB), and complement decay-accelerating factor (CD55), with CD55 being identified thrice. In addition, an examination of 138 associations pertinent to the dichotomous outcomes of sepsis, death versus survival, unveiled that seven exhibited a robust colocalization presence, precisely identified as low-density lipoprotein receptor-related protein 11 (LRP11), NDKB, PNP, 17-beta-hydroxysteroid dehydrogenase 14 (HSD17B14), amyloid beta precursor like protein 2 (APLP2), tumor necrosis factor-inducible gene 6 protein (TNFAIP6), and complement decay-accelerating factor (CD55). After excluding overlapping findings, we conclusively substantiated the presence of eight associations with compelling evidence of colocalization. These are documented in Table 1 and detailed in Table S14.
Table 1. Outcomes of the Colocalization Analysis after MR Study with cis-pQTLsa.
| MR results |
||||||||
|---|---|---|---|---|---|---|---|---|
| PMID | Uniprot | protein names | outcomes | method | nSNP | OR(95% CI) | P value | PP.H4.abf |
| 34857953 | O95236 | APOL3 | sepsis (28 day mortality) | WR | 1 | 0.87(0.80–0.95) | 0.002046461 | 0.518702543 |
| 34857953 | P22392 | NDKB& | sepsis (28 day mortality) | WR | 1 | 0.41(0.26–0.65) | 0.000162427 | 0.922110503 |
| 35078996 | P08174 | complement decay-accelerating factor (CD55)# | sepsis (28 day mortality) | WR | 1 | 1.42(1.13–1.77) | 0.002465158 | 0.529747802 |
| 29875488 | P08174 | complement decay-accelerating factor (CD55)# | sepsis (28 day mortality) | WR | 1 | 1.25(1.08–1.46) | 0.003156166 | 0.507980692 |
| 28240269 | P08174 | complement decay-accelerating factor (CD55)# | sepsis (28 day mortality) | WR | 1 | 1.26(1.10–1.44) | 0.001082243 | 0.726193226 |
| 34857953 | Q86VZ4 | LRP11 | sepsis (death vs survived) | WR | 1 | 0.73(0.60–0.89) | 0.002113459 | 0.518673042 |
| 34857953 | P22392 | NDKB& | sepsis (death vs survived) | WR | 1 | 0.42(0.25–0.69) | 0.000781606 | 0.743190218 |
| 35078996 | P00491 | PNP | sepsis (death vs survived) | IVW | 2 | 1.52(1.19–1.95) | 0.000817079 | 0.824065552 |
| 35078996 | Q9BPX1 | 17-beta-hydroxysteroid dehydrogenase 14 (HSD17B14) | sepsis (death vs survived) | WR | 1 | 0.50(0.32–0.78) | 0.002569258 | 0.561121153 |
| 35078996 | Q06481 | APLP2 | sepsis (death vs survived) | WR | 1 | 1.48(1.13–1.93) | 0.00424199 | 0.512768759 |
| 28240269 | P98066 | TNFAIP6 | sepsis (death vs survived) | WR | 1 | 1.34(1.10–1.62) | 0.003008802 | 0.552163903 |
| 28240269 | P08174 | complement decay-accelerating factor (CD55)# | sepsis (death vs survived) | WR | 1 | 1.30(1.11–1.51) | 0.001140351 | 0.735431777 |
The symbols “#” and “&” indicate that the protein is concurrently present in two distinct outcome databases.
PAV Assessment for the cis-pQTLs
The variant in question or its proxy within the encoding gene may modify the amino acid composition, potentially altering the conformation of the protein. This transformation could influence the binding affinity with the aptamer, distorting measurements. To address this concern, we thoroughly assessed PAVs for pQTLs showing evidence in cis-MR analysis. Among 12 proteins linked to cis-pQTLs, 9 pQTLs were identified as PAVs or were in strong LD (r2 > 0.8) with variants influencing the associated gene. This finding indicates that such interactions could be misinterpreted as epitope-binding anomalies, a detail further elaborated on in Table S15.
Determining the Overlap of pQTLs and eQTLs
To ascertain whether the associations between specific pQTLs and plasma protein concentrations stem from transcriptional effects rather than alternate mechanisms, an analysis of pQTLs overlapping with eQTLs was carried out. In MR analyses restricted to cis-pQTLs, eight such cis-pQTLs were used as instruments for proteins, supported by MR findings. Within this subgroup, five cis-pQTLs exhibited a significant overlap with eQTLs in one or more tissue types, aligned in the same direction of influence, as indicated by the GTEx project. The remaining pQTLs possessed proxy markers coinciding with eQTLs in identical allelic directions (Table S16). We searched for two trans-pQTLs or their proxies, yet found no overlap with eQTLs (Table S16). Exploration of the Open Targets Genetics database for blood eQTLs revealed that only five pQTLs from the cis-MR study shared congruence with these eQTLs (Table S16).
Investigation of PPI and Enriched Pathways
To gain a more profound insight into the etiopathogenesis of sepsis, we conducted an extensive PPI analysis along with pathway elucidation, probing the functional interplay of MR-prioritized proteins. Although a medium–confidence interaction threshold with a score of 0.4 was established, this did not result in the formation of a robust interaction network among the eight proteins under consideration. GO pathway enrichment analysis did, however, unveil enriched biological pathways relevant to sepsis. Notably, MR-prioritized proteins are predominantly localized to specific cellular environments, including the ficolin-1-rich granule lumen, the extracellular region, extracellular exosomes, and the secretory granule lumen, which are instrumental in sepsis pathobiology. Additionally, KEGG pathway annotations hint at a putative involvement in the metabolism of nucleotides and purines, suggesting a nuanced layer of biological complexity that merits further exploration (Table S17).
Evaluating the Drug Targets of the MR-Prioritized Proteins
Recognizing that human proteins are primary therapeutic targets, we further evaluated those identified by MR to ascertain their existing drug target status or potential druggability. Initially, we aligned the MR-highlighted proteins against the druggable genome outlined by Finan et al. Among the 10 proteins assessed, 5 were classified as having druggable targets, comprising three at Tier 1, 1 at Tier 3A, and another at Tier 3B (Tables 2 and S18). Utilizing the Therapeutic Target Database, three proteins—thioredoxin domain-containing protein 12 (TXNDC12), PNP, and complement decay-accelerating factor (CD55)—have been pinpointed as critical targets within the sphere of current or potential therapeutic applications. Notably, PNP and CD55 are currently being evaluated in clinical trials, marking their significant potential for therapeutic interventions. Concurrently, TXNDC12 has garnered extensive documentation within the realm of scientific literature, highlighting its importance and affirming the crucial role these proteins play in the development and innovation of new therapeutic strategies. However, TNFAIP6 and 17-beta-hydroxysteroid dehydrogenase 14 (HSD17B14) are yet to be recognized as crucial targets within the landscape of contemporary or forthcoming therapeutic applications (Tables 2 and S18). Importantly, proteins associated with targeted drugs in accordance with the Therapeutic Target Database showed alignment with the druggable genes identified by Finan et al.
Table 2. List of the MR-Prioritized Proteins That Were Drug Targets or to Be Druggable.
| Uniprot | protein names | HGNC names | drug ability tiera | target typeb |
|---|---|---|---|---|
| O95236 | apolipoprotein L3 | APOL3 | ||
| O95881 | thioredoxin domain-containing protein 12 | TXNDC12c | Tier 3B | literature-reported target |
| P00491 | purine nucleoside phosphorylase | PNP | Tier 1 | clinical trial target |
| P08174 | complement decay-accelerating factor | CD55 | Tier 1 | clinical trial target |
| P22392 | nucleoside diphosphate kinase B | NME2 | ||
| P98066 | tumor necrosis factor-inducible gene 6 protein | TNFAIP6 | Tier 3A | |
| Q06481 | amyloid beta precursor like protein 2 | APLP2 | ||
| Q15648 | mediator of RNA polymerase II transcription subunit 1 | MED1c | ||
| Q86VZ4 | low-density lipoprotein receptor-related protein 11 | LRP11 | ||
| Q9BPX1 | 17-beta-hydroxysteroid dehydrogenase 14 | HSD17B14 | Tier 1 |
Based on the druggable genes from Finan et al.
Based on the therapeutic target database.
Based on the colocalization results obtained from MR analysis of trans-pQTLs.
Discussion
Our study employed an integrative approach combining MR, colocalization, Steiger filtering analysis, PAV assessment, eQTL overlap determination, PPI analysis, pathway enrichment, and drug target evaluation to assess the causal effects of thousands of plasma proteins from nine large proteomics GWAS on sepsis. MR studies using cis-pQTLs revealed associations between 94 unique proteins and sepsis—28 day mortality. Additionally, the studies identified associations between 96 unique proteins and sepsis—death versus survival (p < 0.05). Advancing the complexity of the analysis framework to encompass both cis + trans-pQTLs has remarkably expanded the spectrum of identified proteins. Consequently, 201 unique proteins have been found to exhibit meaningful associations with 28 day mortality attributable to sepsis. Moreover, a comprehensive examination has revealed that 199 unique proteins possess statistically significant correlations with the bifurcated outcomes of death versus survival among sepsis patients. Colocalization analysis of these proteins revealed that eight exhibit robust evidence of colocalization, highlighting their potential role in sepsis pathophysiology. Subsequent in silico analyses were performed to confirm these findings, elucidate potential regulatory pathways, and identify drug targets among these specified proteins. Serum biomarkers of sepsis are a focal point in current clinical research trends; however, most of the studies are using low-throughput methods for detection. To the best of our knowledge, only a few studies have explored sepsis serum protein biomarkers using high-throughput methods.22 Thus, 12 serum proteins with varying abundance were successfully identified in individuals with sepsis compared to healthy controls.11 Nevertheless, this analysis was limited to 396 proteins detected in a moderate-sized cohort encompassing 59 patients with sepsis and 31 healthy individuals. While these proteins with differential abundance serve as biomarkers, their role as pathogenic agents in sepsis has not been definitively established. This limitation arises from the inherent constraints of observational studies, where causality cannot be reliably inferred from the observed correlations. In our investigative framework, we prioritized certain proteins when conducting MR analysis by employing cis-pQTLs.
Further exploration through colocalization analysis unveiled a set of eight proteins with pronounced colocalization, a subset of which has been previously established as contributory factors in the pathogenesis of sepsis. In particular, the TNFAIP6, which is integrated into the hyaluronan-binding protein cohort, serves as a key modulator within the intricate inflammatory protease matrix.23 Our systematic investigation delineated a pronounced positive correlation between TNFAIP6 expression and sepsis development. This discovery is consistent with and augments the body of evidence from other scholarly research, which has observed a regulatory upsurge of TNFAIP6 in individuals afflicted with sepsis. The congruent findings across multiple studies reinforce the pivotal role of TNFAIP6 as a biomolecular participant in the multifactorial landscape of sepsis etiology.
TNFAIP6 may modulate endopeptidase activity within apoptotic pathways, thereby potentially intensifying sepsis severity through its influence on molecular mechanisms. This indicates a crucial role for TNFAIP6 in sepsis progression, emphasizing the need for more research to explore its therapeutic implications and impact on sepsis severity reduction.24,25 Additionally, our investigation revealed a positive correlation between PNP and sepsis prognosis. Inherited anomalies of PNP, an enzyme pivotal for purine nucleoside cleavage, can lead to immunodeficiencies through the selective attrition of T lymphocytes. Subsequently, persistent infections escalating to sepsis coupled with neurological disturbances and heightened vulnerability to autoimmune pathologies may occur.26−28 However, the potential connection between the overexpression of PNP and the instigation of a hyperactive immune response culminating in sepsis remains less elucidated in comparison to the well-documented link with PNP deficiency. Sepsis is characteristically triggered by an overwhelming or dysregulated immune response to infection, orchestrated by the sophisticated interactions among a plethora of cytokines and immune cells, and is not solely contingent on the enzymatic functions of PNP. Although PNP plays a pivotal role in modulating metabolic pathways, its overexpression may perturb normal metabolic homeostasis. Presently, there is a lack of comprehensive evidence unequivocally associating PNP overexpression with the onset of an exaggerated immune response that leads to sepsis. Moreover, discrepancies have been noted between previous scholarly works and our observations, indicating a complex nexus that warrants further investigation to delineate the role of PNP overexpression in the pathophysiology of sepsis.
There are also inconsistencies between previous studies and our findings. Several studies have suggested that augmenting CD55 expression can ameliorate the severity of sepsis, implying a potential protective function for CD55.29,30 However, our research identified CD55 as a risk factor. This disparity may arise from the prevalence of animal experiments over clinical trials. CD55’s influence on the immune system represents a “double-edged sword” having conflicting roles across various diseases.31 A comprehensive understanding of CD55’s precise mechanism requires a thorough evaluation of all relevant factors. Consequently, further investigations into the correlation between CD55 and sepsis are imperative to ascertain its prospective therapeutic role in sepsis management.
Regarding MR-prioritized proteins not previously associated with sepsis, there is potential evidence indicating their involvement in sepsis pathogenesis. For example, APOL3, a protein essential in cholesterol and lipid transportation and closely associated with lipoproteins, is upregulated and positively correlated with the promotion of ferroptosis, an iron-regulated cell death.32−34 Ferroptosis plays a significant role in sepsis pathophysiology and progression.35,36 Consequently, it is hypothesized that APOL3 may act as a protective factor in sepsis by modulating the iron-mediated cell death pathway.
In analyzing the MR results, we implemented a series of analytical procedures. Employing Bayesian colocalization, we investigated the potential genetic confounding arising from LD between pQTLs and sepsis-associated SNPs. A minority of these associations exhibited robust indicators of colocalization, thereby bolstering the evidence for a causal relationship. Nevertheless, the underlying premise of Bayesian colocalization methods, assuming a singular association signal in each discrete region, may not align with practical complexities. This misalignment could lead to an underestimation of the prevalence and significance of colocalization phenomena.37 To explore potential reverse causality, we employed Steiger filtering analysis, revealing that all correlations identified in our MR investigation possessed direct causal connections, indicating that protein expressions were causally linked to sepsis-associated outcomes. Additionally, a PAV could induce modifications in protein conformation, influencing the affinity of aptamers and potentially causing artifactual measurement.6
Our analysis revealed that over half of the cis-MR-highlighted pQTLs were PAVs or in LD with PAVs, indicating the necessity for careful interpretation of these proteins’ causal links to phenotypic outcomes due to potential aptamer-related artifacts. While PPI and pathway enrichment did not show a strong protein network, GO analysis identified critical pathways associated with sepsis, and KEGG analysis linked to nucleotide and purine metabolism, underlining the significance and interpretability of our findings. The lack of approved sepsis therapies underscores the urgent need for drug development in this field. MR is increasingly recognized as a pivotal tool for the screening of novel drug targets.38 Consequently, we evaluated drug targets for proteins supported by MR evidence to aid in prioritizing drug discovery efforts and repurposing existing medications for the treatment of sepsis.
This investigation had several limitations. First, our analysis focused on the implications of circulating proteins, encompassing both deliberately and unintentionally released species. The blood levels of these proteins may vary significantly from their cellular or tissue concentrations, potentially leading to a lack of insights into the effects of cell- or tissue-specific protein abundances. Second, limited individual-level data in the sepsis GWAS hampered the ethnicity-based stratification of participants. Despite this, given the predominantly European descent cohort, the influence of ethnic variation on our results is considered minimal. Third, a coding variant in a cis-pQTL might alter measurable protein levels without affecting bioactivity or abundance, leading to potential misinterpretations. Consequently, PAV evaluations were conducted to detect such disparities among cis-pQTLs. Fourth, the Bayesian colocalization technique assessed the likelihood of LD influencing significant MR findings. Nonetheless, due to assumptions in Bayesian colocalization, there is a possibility of overemphasizing the impact of LD on our MR results. Last, reliance on publicly accessible data sets may limit innovativeness. However, thorough curation and examination of these data sets can still yield novel insights, enriching the current research landscape.
Conclusions
In summary, through two-sample MR analyses, we identified several circulating proteins causally linked to sepsis-associated traits. Our findings not only validate the MR outcomes but also suggest potential therapeutic interventions for sepsis depending on the employed analytical framework. Future research is crucial to understanding the pathogenic roles and foundational biological mechanisms of proteins causally implicated in sepsis.
Acknowledgments
We thank Bullet Edits Limited for the linguistic editing and proofreading of the manuscript. This study received funding and support from the Natural Science Foundation of Xinjiang Uygur Autonomous Region (no. 2023D01F26), the Wuxi Taihu Lake Talent Plan High Level Talent Training Project (no. HB2023036), the Scientific Research Project of Wuxi Municipal Health Commission (no. Q202221), the National Natural Science Foundation Incubation Plan of the Second Affiliated Hospital of Anhui Medical University (no. 2022GMFY09), the Clinical Research Cultivation Program of the Second Affiliated Hospital of Anhui Medical University (no. 2021LCYB12), and the Support Program for Elite Young Talents in Colleges and Universities of Anhui Province (no. gxyq2022006).
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.4c01934.
Source information on the pQTL; source information on the outcome; information of the instrumental variables; main MR analysis results of cis-pQTL on sepsis (death vs survived); main MR analysis results of cis-pQTL on sepsis (28 day mortality); sensitivity analysis results of cis-pQTL on sepsis (death vs survived); sensitivity analysis results of cis-pQTL on sepsis (28 day mortality); main MR analysis results of cis + trans-pQTL on sepsis (death vs survived); main MR analysis results of cis + trans-pQTL on sepsis (28 day mortality); main MR analysis results of trans-pQTL on sepsis; sensitivity analysis results of cis + trans-pQTL on sepsis (28 day mortality); sensitivity analysis results of cis + trans-pQTL on sepsis (death vs survived); Steiger filtering analysis of cis-pQTL on sepsis; colocalization of the pQTLs prioritized by the MR analyses with sepsis risk loci; PAV assessment for the pQTLs with MR evidence; overlapping of pQTLs prioritized by MR with eQTLs; GO/KEGG pathway enrichment analysis for proteins prioritized by cis-only MR; and drug targets of the MR-prioritized proteins (ZIP)
(PDF)
Author Contributions
∥ J.M. and L.F. contributed equally to this work. J.M. contributed to formal analysis, software, visualization, and writing original draft. L.F. contributed to software, visualization, and validation. Z.L. contributed to data curation and resources. Z.L. contributed to methodology. Y.S. contributed to conceptualization, methodology, writing—review and editing, and funding acquisition. All authors have read and agreed to the published version of the manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Singer M.; Deutschman C. S.; Seymour C. W.; Shankar-Hari M.; Annane D.; Bauer M.; Bellomo R.; Bernard G. R.; Chiche J. D.; Coopersmith C. M.; et al. The Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3). JAMA 2016, 315 (8), 801–810. 10.1001/jama.2016.0287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudd K. E.; Johnson S. C.; Agesa K. M.; Shackelford K. A.; Tsoi D.; Kievlan D. R.; Colombara D. V.; Ikuta K. S.; Kissoon N.; Finfer S.; et al. Global, regional, and national sepsis incidence and mortality, 1990–2017: analysis for the Global Burden of Disease Study. Lancet 2020, 395 (10219), 200–211. 10.1016/S0140-6736(19)32989-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart K.; Daniels R.; Kissoon N.; Machado F. R.; Schachter R. D.; Finfer S. Recognizing Sepsis as a Global Health Priority - A WHO Resolution. N. Engl. J. Med. 2017, 377 (5), 414–417. 10.1056/NEJMp1707170. [DOI] [PubMed] [Google Scholar]
- Huang M.; Cai S.; Su J. The Pathogenesis of Sepsis and Potential Therapeutic Targets. Int. J. Mol. Sci. 2019, 20 (21), 5376. 10.3390/ijms20215376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emilsson V.; Gudmundsdottir V.; Gudjonsson A.; Jonmundsson T.; Jonsson B. G.; Karim M. A.; Ilkov M.; Staley J. R.; Gudmundsson E. F.; Launer L. J.; et al. Coding and regulatory variants are associated with serum protein levels and disease. Nat. Commun. 2022, 13 (1), 481. 10.1038/s41467-022-28081-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferkingstad E.; Sulem P.; Atlason B. A.; Sveinbjornsson G.; Magnusson M. I.; Styrmisdottir E. L.; Gunnarsdottir K.; Helgason A.; Oddsson A.; Halldorsson B. V.; et al. Large-scale integration of the plasma proteome with genetics and disease. Nat. Genet. 2021, 53 (12), 1712–1721. 10.1038/s41588-021-00978-w. [DOI] [PubMed] [Google Scholar]
- Gudjonsson A.; Gudmundsdottir V.; Axelsson G. T.; Gudmundsson E. F.; Jonsson B. G.; Launer L. J.; Lamb J. R.; Jennings L. L.; Aspelund T.; Emilsson V.; et al. A genome-wide association study of serum proteins reveals shared loci with common diseases. Nat. Commun. 2022, 13 (1), 480. 10.1038/s41467-021-27850-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun B. B.; Maranville J. C.; Peters J. E.; Stacey D.; Staley J. R.; Blackshaw J.; Burgess S.; Jiang T.; Paige E.; Surendran P.; et al. Genomic atlas of the human plasma proteome. Nature 2018, 558 (7708), 73–79. 10.1038/s41586-018-0175-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yazdanpanah N.; Yazdanpanah M.; Wang Y.; Forgetta V.; Pollak M.; Polychronakos C.; Richards J. B.; Manousaki D. Clinically Relevant Circulating Protein Biomarkers for Type 1 Diabetes: Evidence From a Two-Sample Mendelian Randomization Study. Diabetes Care 2022, 45 (1), 169–177. 10.2337/dc21-1049. [DOI] [PubMed] [Google Scholar]
- Sheehan N. A.; Didelez V.; Burton P. R.; Tobin M. D. Mendelian randomisation and causal inference in observational epidemiology. PLoS Med. 2008, 5 (8), e177 10.1371/journal.pmed.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao H.; Chen S.; Ding R. Evaluation of the Molecular Mechanisms of Sepsis Using Proteomics. Front. Immunol. 2021, 12, 733537. 10.3389/fimmu.2021.733537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suhre K.; Arnold M.; Bhagwat A. M.; Cotton R. J.; Engelke R.; Raffler J.; Sarwath H.; Thareja G.; Wahl A.; DeLisle R. K.; et al. Erratum: Connecting genetic risk to disease end points through the human blood plasma proteome. Nat. Commun. 2017, 8, 15345. 10.1038/ncomms15345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillary R. F.; McCartney D. L.; Harris S. E.; Stevenson A. J.; Seeboth A.; Zhang Q.; Liewald D. C.; Evans K. L.; Ritchie C. W.; Tucker-Drob E. M.; et al. Genome and epigenome wide studies of neurological protein biomarkers in the Lothian Birth Cohort 1936. Nat. Commun. 2019, 10 (1), 3160. 10.1038/s41467-019-11177-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietzner M.; Wheeler E.; Carrasco-Zanini J.; Raffler J.; Kerrison N. D.; Oerton E.; Auyeung V. P. W.; Luan J.; Finan C.; Casas J. P.; et al. Genetic architecture of host proteins involved in SARS-CoV-2 infection. Nat. Commun. 2020, 11 (1), 6397. 10.1038/s41467-020-19996-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao C.; Chen G.; Song C.; Keefe J.; Mendelson M.; Huan T.; Sun B. B.; Laser A.; Maranville J. C.; Wu H.; et al. Genome-wide mapping of plasma protein QTLs identifies putatively causal genes and pathways for cardiovascular disease. Nat. Commun. 2018, 9 (1), 3268. 10.1038/s41467-018-05512-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folkersen L.; Fauman E.; Sabater-Lleal M.; Strawbridge R. J.; Frånberg M.; Sennblad B.; Baldassarre D.; Veglia F.; Humphries S. E.; Rauramaa R.; et al. Mapping of 79 loci for 83 plasma protein biomarkers in cardiovascular disease. PLoS Genet. 2017, 13 (4), e1006706 10.1371/journal.pgen.1006706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilly A.; Park Y. C.; Png G.; Barysenka A.; Fischer I.; Bjørnland T.; Southam L.; Suveges D.; Neumeyer S.; Rayner N. W.; et al. Whole-genome sequencing analysis of the cardiometabolic proteome. Nat. Commun. 2020, 11 (1), 6336. 10.1038/s41467-020-20079-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton F. W.; Thomas M.; Arnold D.; Palmer T.; Moran E.; Mentzer A. J.; Maskell N.; Baillie K.; Summers C.; Hingorani A.; et al. Therapeutic potential of IL6R blockade for the treatment of sepsis and sepsis-related death: A Mendelian randomisation study. PLoS Med. 2023, 20 (1), e1004174 10.1371/journal.pmed.1004174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y. T.; Ou Y. N.; Wu B. S.; Yang Y. X.; Jiang Y.; Huang Y. Y.; Liu Y.; Tan L.; Dong Q.; Suckling J.; et al. Identifying causal genes for depression via integration of the proteome and transcriptome from brain and blood. Mol. Psychiatry 2022, 27 (6), 2849–2857. 10.1038/s41380-022-01507-9. [DOI] [PubMed] [Google Scholar]
- Finan C.; Gaulton A.; Kruger F. A.; Lumbers R. T.; Shah T.; Engmann J.; Galver L.; Kelley R.; Karlsson A.; Santos R.; et al. The druggable genome and support for target identification and validation in drug development. Sci. Transl. Med. 2017, 9 (383), eaag1166 10.1126/scitranslmed.aag1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y.; Zhang Y.; Lian X.; Li F.; Wang C.; Zhu F.; Qiu Y.; Chen Y. Therapeutic target database update 2022: facilitating drug discovery with enriched comparative data of targeted agents. Nucleic Acids Res. 2022, 50 (D1), D1398–D1407. 10.1093/nar/gkab953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabri Faja N.; Calvo Lozano O.; Dey P.; Terborg R. A.; Estevez M. C.; Belushkin A.; Yesilköy F.; Duempelmann L.; Altug H.; Pruneri V.; et al. Early sepsis diagnosis via protein and miRNA biomarkers using a novel point-of-care photonic biosensor. Anal. Chim. Acta 2019, 1077, 232–242. 10.1016/j.aca.2019.05.038. [DOI] [PubMed] [Google Scholar]
- Guo F.; Yuan Y. Tumor Necrosis Factor Alpha-Induced Proteins in Malignant Tumors: Progress and Prospects. OncoTargets Ther. 2020, 13, 3303–3318. 10.2147/OTT.S241344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allantaz-Frager F.; Turrel-Davin F.; Venet F.; Monnin C.; De Saint Jean A.; Barbalat V.; Cerrato E.; Pachot A.; Lepape A.; Monneret G. Identification of biomarkers of response to IFNg during endotoxin tolerance: application to septic shock. PLoS One 2013, 8 (7), e68218 10.1371/journal.pone.0068218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H.; Li Y.; Li T.; Sun H.; Tan C.; Gao M.; Xing W.; Xiao X. Identification of Potential Transcriptional Biomarkers Differently Expressed in Both S. aureus- and E. coli-Induced Sepsis via Integrated Analysis. Biomed. Res. Int. 2019, 2019, 2487921. 10.1155/2019/2487921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bantia S.; Ananth S. L.; Parker C. D.; Horn L. L.; Upshaw R. Mechanism of inhibition of T-acute lymphoblastic leukemia cells by PNP inhibitor--BCX-1777. Int. Immunopharmacol. 2003, 3 (6), 879–887. 10.1016/S1567-5769(03)00076-6. [DOI] [PubMed] [Google Scholar]
- Markert M. L. Purine nucleoside phosphorylase deficiency. Immunodeficiency Rev. 1991, 3 (1), 45–81. [PubMed] [Google Scholar]
- Kütükçüler N.; Bölük E.; Tökmeci N.; et al. Recurrent infections, neurologic signs, low serum uric acid levels, and lymphopenia in childhood: Purine nucleoside phosphorylase deficiency, an emergency for infants. Turk. Pediatr. Ars. 2020, 55 (3), 320–327. 10.14744/TurkPediatriArs.2019.83788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh S. J.; Kim J. H.; Chung D. H. NOD2-mediated suppression of CD55 on neutrophils enhances C5a generation during polymicrobial sepsis. PLoS Pathog. 2013, 9 (5), e1003351 10.1371/journal.ppat.1003351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. H.; Oh S. J.; Ahn S.; Chung D. H. IFN-γ-producing NKT cells exacerbate sepsis by enhancing C5a generation via IL-10-mediated inhibition of CD55 expression on neutrophils. Eur. J. Immunol. 2014, 44 (7), 2025–2035. 10.1002/eji.201343937. [DOI] [PubMed] [Google Scholar]
- Dho S. H.; Lim J. C.; Kim L. K. Beyond the Role of CD55 as a Complement Component. Immune Network 2018, 18 (1), e11 10.4110/in.2018.18.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y.; Dai Y. APOL3-LDHA axis related immunity activation and cancer ferroptosis induction. Int. J. Biol. Sci. 2023, 19 (5), 1401–1402. 10.7150/ijbs.83342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv Y.; Tang W.; Xu Y.; Chang W.; Zhang Z.; Lin Q.; Ji M.; Feng Q.; He G.; Xu J. Apolipoprotein L3 enhances CD8+ T cell antitumor immunity of colorectal cancer by promoting LDHA-mediated ferroptosis. Int. J. Biol. Sci. 2023, 19 (4), 1284–1298. 10.7150/ijbs.74985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv Y.; Zheng P.; Mao Y.; Xu Y.; Chang W.; Lin Q.; Ji M.; Ye L.; Tang W.; Xu J. Intratumor APOL3 delineates a distinctive immunogenic ferroptosis subset with prognosis prediction in colorectal cancer. Cancer Sci. 2024, 115, 257–269. 10.1111/cas.16009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xl L.; Gy Z.; R G.; N C. Ferroptosis in sepsis: The mechanism, the role and the therapeutic potential. Front. Immunol. 2022, 13, 956361. 10.3389/fimmu.2022.956361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.; Tan S.; Wu Y.; Tan S. The Emerging Role of Ferroptosis in Sepsis. DNA Cell Biol. 2022, 41 (4), 368–380. 10.1089/dna.2021.1072. [DOI] [PubMed] [Google Scholar]
- Zheng J.; Haberland V.; Baird D.; Walker V.; Haycock P. C.; Hurle M. R.; Gutteridge A.; Erola P.; Liu Y.; Luo S.; et al. Phenome-wide Mendelian randomization mapping the influence of the plasma proteome on complex diseases. Nat. Genet. 2020, 52 (10), 1122–1131. 10.1038/s41588-020-0682-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folkersen L.; Gustafsson S.; Wang Q.; Hansen D. H.; Hedman Å. K.; Schork A.; Page K.; Zhernakova D. V.; Wu Y.; Peters J.; et al. Genomic and drug target evaluation of 90 cardiovascular proteins in 30,931 individuals. Nat. Metab. 2020, 2 (10), 1135–1148. 10.1038/s42255-020-00287-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.