Abstract
Faithful transmission of genetic material is crucial for the survival of all organisms. In many eukaryotes, a feedback control mechanism called the spindle checkpoint ensures chromosome segregation fidelity by delaying cell cycle progression until all chromosomes achieve proper attachment to the mitotic spindle. Kinetochores are the macromolecular complexes that act as the interface between chromosomes and spindle microtubules. While most eukaryotes have canonical kinetochore proteins that are widely conserved, kinetoplastids such as Trypanosoma brucei have a seemingly unique set of kinetochore proteins including KKT1–25. It remains poorly understood how kinetoplastids regulate cell cycle progression or ensure chromosome segregation fidelity. Here, we report a crystal structure of the C-terminal domain of KKT14 from Apiculatamorpha spiralis and uncover that it is a pseudokinase. Its structure is most similar to the kinase domain of a spindle checkpoint protein Bub1. In addition, KKT14 has a putative ABBA motif that is present in Bub1 and its paralogue BubR1. We also find that the N-terminal part of KKT14 interacts with KKT15, whose WD40 repeat beta-propeller is phylogenetically closely related to a direct interactor of Bub1/BubR1 called Bub3. Our findings indicate that KKT14–KKT15 are divergent orthologues of Bub1/BubR1–Bub3, which promote accurate chromosome segregation in trypanosomes.
Keywords: kinetochore, chromosome segregation, spindle checkpoint, kinetoplastid, Trypanosoma brucei
1. Introduction
Accurate transmission of genetic material from mother to daughter cells is essential for the survival of all organisms. Segregation of replicated chromosomes in eukaryotes is achieved by a macromolecular protein complex called the kinetochore, which links centromeric DNA to microtubules [1,2]. Once all chromosomes have achieved proper attachments, the multi-subunit E3 ubiquitin ligase complex called the anaphase-promoting complex/cyclosome (APC/C) gets activated. This leads to the degradation of anaphase inhibitors securin and cyclin B, triggering sister chromatid separation and exit from mitosis [3,4].
The spindle checkpoint is a surveillance system that monitors defects in kinetochore–microtubule attachments and delays the onset of anaphase [5]. It works by inhibiting the activity of the APC/C that is in complex with its co-activator protein Cdc20. Spindle checkpoint components include Mps1, Bub1, BubR1 (Mad3), Bub3, Mad1 and Mad2 [6–8]. The mitotic checkpoint complex is a potent inhibitor of APC/CCdc20, which in humans consists of Mad2, Cdc20, BubR1 and Bub3 [9]. Unattached kinetochores recruit these checkpoint proteins to catalyse the formation of the mitotic checkpoint complex [10]. Kinetochore recruitment of spindle checkpoint proteins therefore needs to be under tight control. Bub3, a WD40 repeat domain protein, directs Bub1 and BubR1 (Mad3) to kinetochores by recognizing the Mps1-phosphorylated MELT motif of the outer kinetochore protein KNL1 [11–15]. Bub1 and BubR1 are paralogous proteins [16], which have a kinase and pseudokinase domain in their C-terminus, respectively, while Mad3, a Bub1 paralog in yeast, does not have a kinase domain. The kinase activity of Bub1 is largely dispensable for its spindle checkpoint function [17,18]. The pseudokinase domain in human BubR1 is thought to play a role in ensuring its protein stability [16]. Bub1, BubR1 and Mad3 carry the Gle2-binding sequence (GLEBS) motif that binds Bub3 and the ABBA motif that interacts with Cdc20 [19–22].
Despite its importance in ensuring accurate chromosome segregation, some organisms (e.g. yeasts, fruit flies and human HAP1 cells) do not require the spindle checkpoint for their proliferation or development under normal conditions [23–27]. It is thought that these organisms do not require a feedback-induced mitotic delay because all chromosomes can establish proper kinetochore–microtubule attachments before the APC/C gets fully activated. Furthermore, spindle checkpoint components are apparently absent in some organisms, including Trypanosoma brucei, which cannot halt cell cycle progression in response to spindle defects [28–32]. Trypanosoma brucei is an experimentally tractable parasite that belongs to kinetoplastids, a class of unicellular flagellated eukaryotes that are highly divergent from traditional model eukaryotes [33–35]. They include the parasitic order Trypanosomatida (e.g. T. brucei, Trypanosoma cruzi and Leishmania), free-living Bodonida (e.g. Bodo saltans), both belonging to the subclass Metakinetoplastina, and the subclass Prokinetoplastina (e.g. Apiculatamorpha spiralis, Papus ankaliazontas and Perkinsela) [36]. Very little is known about how kinetoplastids regulate cell cycle progression or ensure accurate chromosome segregation without a functional spindle checkpoint.
Like spindle checkpoint components, kinetochore proteins are widely conserved among eukaryotes [37,38]. However, unique kinetochore proteins called Kinetoplastid KineTochore 1–25 (KKT1–25) and KKT-Interacting Protein 1–12 (KKIP1–12) are present in T. brucei [39–43]. These proteins are conserved among kinetoplastids but do not have a significant sequence similarity to canonical kinetochore proteins, meaning that they are attractive drug targets against diseases caused by kinetoplastid parasites [44,45]. Understanding the structure and function of kinetoplastid kinetochore proteins also has the potential to shed light on fundamental requirements for the chromosome segregation machinery in eukaryotes.
In this study, we focus on KKT14 and KKT15, proteins of unknown functions that localize at the kinetochore from G2 until the end of anaphase in T. brucei [39]. Previous bioinformatics analysis using advanced hidden Markov model (HMM) searches (e.g. HMMER and HHpred) of T. brucei KKT14 failed to identify any obvious conserved domain. KKT15 has WD40 repeats that likely form a beta-propeller, a domain found in many different proteins, including Bub3, Cdc20 and mRNA export factor Rae1/Gle2 [46]. Here, we discover that KKT14 has a pseudokinase domain in its C-terminus, which is most similar to the kinase domain of Bub1. We also identify a putative ABBA motif in KKT14. The N-terminal part of KKT14 interacts with KKT15, which we suggest to be a Bub3 orthologue. These results reveal that kinetoplastids possess divergent Bub1/BubR1 and Bub3 proteins.
2. Results
2.1. Crystal structure of KKT14 C-terminal domain reveals similarity to Bub1
To gain insights into the function and evolutionary origin of KKT14, we aimed to obtain its high-resolution structural information. By screening four kinetoplastid species (T. brucei, T. cruzi, Paratrypanosoma confusum and Apiculatamorpha spiralis), we succeeded in determining a 2.2 Å resolution crystal structure for the C-terminal domain of the KKT14 protein from the prokinetoplastid A. spiralis (clone PhF-6) (figure 1a , table 1 and electronic supplementary material, table S1). Apiculatamorpha spiralis KKT14365−640 crystallized with two molecules in an asymmetric unit. Both molecules were essentially identical except for minor variations in flexible loops.
Figure 1.
Crystal structure of A. spiralis KKT14 reveals similarity to the Bub1 kinase domain. (a,b) Cartoon representation of A. spiralis KKT14365–640 (a) and human Bub1 kinase domain (PDB accession 6F7B [47]) (b). The fold nomenclature of the N-terminal extension and the kinase domain of Bub1 is based on [48]. (c) Structure-based pairwise alignment of A. spiralis KKT14 and human Bub1 kinase domain based on the DALI search output. Structurally equivalent residues are in uppercase, while structurally non-equivalent residues (e.g. in loops) are in lowercase. Secondary structures were assigned using DSSP [49].
Table 1.
Data collection and refinement statistics for Apiculatamorpha spiralis KKT14365−640.
| data collection | |
|---|---|
| beamline | Diamond Light Source I03 |
| wavelength (Å) | 0.9763 |
| space group (Z) | P 1 2 1 1 |
| unit cell | 49.76 Å 87.58 Å 71.88 Å 90◦ 93.61◦ 90◦ |
| resolution range (Å) | 49.66–1.95 (1.95–1.99) |
| unique reflections | 41 803 (2243) |
| completeness (%) | 93.3 (100.0) |
| multiplicity | 6.7 (6.7) |
| I/Iσ | 6.9 (0.7) |
| Rmeas | 0.188 (2.640) |
| CC1/2 | 1.0 (0.6) |
| Wilson B-factor (Å2) | 30.14 |
| refinement | |
| no. reflections | 28 797 (1146) |
| Rwork | 0.22 (0.36) |
| Rfree | 0.24 (0.40) |
| number of atoms | 4571 |
| protein | 4308 |
| solvent | 263 |
| RMS bonds (Å) | 0.009 |
| RMS angles (◦) | 1.23 |
| Ramachandran favoured (%) | 96.13 |
| Ramachandran allowed (%) | 3.87 |
| Ramachandran outliers (%) | 0.00 |
| average B-factor (Å2) | 39.00 |
Notes: Values in parentheses correspond to the highest resolution shell.
Interestingly, a structural homology search using the distance-matrix alignment (DALI) server [50] revealed similarity to a protein kinase fold with an N-lobe and C-lobe (figure 1a and electronic supplementary material, table S2). The N-lobe contains a five-stranded β-sheet, a helix termed the C-helix (αC), and loops that correspond to the catalytic loop and activation loop, while the C-lobe comprises a bundle of α-helices (figure 1a ). Although most protein kinases share a similar fold [51], we found that the most similar structure of A. spiralis KKT14365−640 in the PDB database was the Bub1 kinase domain (figure 1b ). Human Bub1 was the top hit in the DALI search with a Z-score of 15.5, while the next best hit was the MST3 kinase (Z-score 13.0) (electronic supplementary material, table S2). We obtained a similar result for an AlphaFold2-predicted structure of T. brucei KKT14358–685 (electronic supplementary material, figure S1a), showing a Z-score of 15.9 for human Bub1 and 12.9 for the next best hit, the PAK3 kinase (electronic supplementary material, table S2). Moreover, searches with Foldseek against the AlphaFold2-predicted structure database [52] produced congruent results (see §4). The higher structural similarity of KKT14 to the kinase domain of Bub1 rather than other kinases is owing to an N-terminal extension that is present in Bub1 and KKT14 (figure 1a–c ; electronic supplementary material, table S2) [47,48,53–55]. These results show that the C-terminal domain of KKT14 has a kinase fold with the most similar structure being the Bub1 kinase domain, raising a possibility that KKT14 is a Bub1/BubR1 orthologue.
Besides a kinase/pseudokinase domain, Bub1 and BubR1 have various conserved domains and motifs [19,20,22,56]. We identify a putative ABBA motif (consensus: Fx[ILV][FHY]x[DE]) in KKT14, which is highly conserved among trypanosomatids (figure 2), as well as KEN boxes in some kinetoplastids (electronic supplementary material, figure S2). In contrast, other domains such as a TPR, CDI or a KARD domain were not found. The presence of an ABBA motif and a C-terminal Bub1-like kinase fold strongly supports the possibility that KKT14 is a divergent Bub1-like protein.
Figure 2.
![KKT14 has a putative ABBA motif. Schematic of the T. brucei KKT14 protein and a multiple sequence alignment showing a putative ABBA motif (consensus Fx[ILV][FHY]x[DE]](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/e4be/11286163/793b86a67655/rsob.240025.f002.jpg)
KKT14 has a putative ABBA motif. Schematic of the T. brucei KKT14 protein and a multiple sequence alignment showing a putative ABBA motif (consensus Fx[ILV][FHY]x[DE] based on [19]) conserved in trypanosomatid KKT14 proteins.
2.2. KKT14 C-terminal domain is an inactive pseudokinase
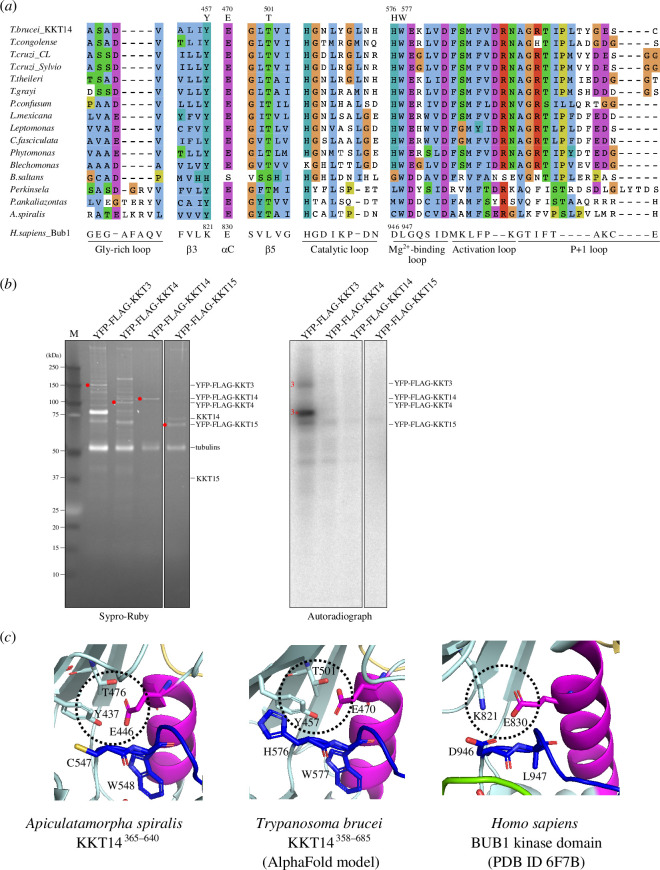
Despite the structural similarity, however, a structure-based sequence alignment of KKT14 with Bub1 shows a lack of conserved residues that play key roles in catalytically active protein kinases (figure 1c ). Most notably, none of KKT14 orthologues in kinetoplastids has a conserved β3 lysine (K821 in Bub1) (figure 3a ), whose mutation results in an inactive kinase [57,58]. In addition, KKT14 does not appear to have the Gly-rich motif (GxGxxG in most kinases, which is essential for stabilizing ATP phosphates during catalysis), and the HRD motif in the catalytic loop (usually His-Arg-Asp: HGD in Bub1) is HGN in T. brucei and HCA in A. spiralis (figure 3a ). Furthermore, the DFG motif (usually Asp-Phe-Gly: DLG in Bub1), which is required for Mg2+ coordination, is HWE in T. brucei and CWD in A. spiralis KKT14 (figure 3a ). Given that even a single amino acid change of key residues in these motifs results in inactive kinases [59], these findings strongly suggest that the C-terminal domain of KKT14 is a catalytically inactive pseudokinase. Consistent with this possibility, KKT14 purified from trypanosomes did not have any detectable auto-phosphorylation activity, while the KKT3 kinase did (figure 3b ).
Figure 3.
KKT14 lacks key residues that are present in active protein kinases. (a) Multiple sequence alignment of kinetoplastid KKT14 sequences highlighting the regions that correspond to the key parts of the Bub1 kinase domain. Note that T501 and W577 in T. brucei are conserved among kinetoplastids. (b) Lack of detectable auto-phosphorylation activity for KKT14. Indicated proteins were immunoprecipitated from trypanosomes using FLAG antibodies and eluted with FLAG peptides. The left panel shows a Sypro-Ruby stained SDS-PAGE gel (red circles indicate FLAG-tagged proteins), while the right panel shows phosphorylation detected by autoradiography. A degradation product of YFP-FLAG-KKT3 is indicated by 3*. (c) The C-helix has an ‘in’ conformation in the A. spiralis KKT14 crystal structure and AlphaFold2-predicted T. brucei KKT14 structure (electronic supplementary material, dataset S1). Note that E446 in the C-helix is in close proximity with Y437 of β3 and T476 of β5 in A. spiralis (E470, T501 and Y457 in T. brucei, respectively), while E830 in the C-helix forms a salt bridge with the conserved β3 lysine (K821) in Bub1.
Although KKT14 appears to be an inactive pseudokinase, features of active protein kinase conformations are found in its structure. In the active conformation of protein kinases, the DFG motif and the C-helix have an ‘in’ conformation, where the conserved phenylalanine or leucine of the DFG motif (L947 in Bub1) points out of the active site and the aspartic acid (D946 in Bub1) faces the ATP-binding site (figure 3c ) [57]. In addition, the β3 lysine (K821 in Bub1) forms a salt bridge with the glutamate (E830 in Bub1) in the C-helix (figure 3c ). In the crystal structure of A. spiralis KKT14, W548 (the phenylalanine equivalent of the DFG motif, which is CWD in A. spiralis KKT14), strictly conserved among kinetoplastids, faces away from the active site. Even though KKT14 in almost all kinetoplastids has the conserved glutamate in the C-helix (E446 in A. spiralis), the position of β3 lysine has tyrosine (Y437 in A. spiralis). Nonetheless, the C-helix sits close to β3, mediated by apparent interactions among Y437, E446 and T476 in β5. Similar interactions were observed in the AlphaFold2-predicted structure of T. brucei KKT14 (figure 3c ), and the threonine is strictly conserved among kinetoplastids (T501 in T. brucei) (figure 3a ). We also note that the positions of the N-terminal extension, C-helix, catalytic loop and Mg2+-binding loop are very similar in between the crystal structure of A. spiralis KKT14 and the AlphaFold2-predicted structure of T. brucei KKT14 (electronic supplementary material, figure S1), despite a limited similarity between the KKT14 sequences of the two species (20.1% identical, 31.5% similar). These findings suggest that the catalytically inactive pseudokinase domain of KKT14 takes an active-like conformation of a kinase fold.
2.3. KKT15 is a divergent Bub3 protein
Our previous sequence similarity searches were unsuccessful in finding homology between T. brucei KKT14 and Bub1/BubR1. Using HHpred [60] with a sensitive alignment of KKT14 proteins from an extended kinetoplastid dataset (electronic supplementary material, table S3), we were now able to observe a link to human Bub1, albeit with a non-significant E-value (electronic supplementary material, table S4). Owing to the low level of sequence similarities, conducting a phylogenetic analysis was not feasible. In contrast, searches with a KKT15 alignment retrieved many WD40 domain proteins with highly significant E-values and had Bub3 as its best hit (electronic supplementary material, table S4). The WD40 beta-propeller is a domain present in Bub3 and many other proteins [46]. To investigate to which WD40 group KKT15 might belong, we conducted a phylogenetic analysis using two multiple sequence alignments of WD40 proteins (see §4). In both cases, KKT15 orthologues cluster with Bub3 and Rae1, which are known to be closely related to one another [38]. One alignment yields all KKT15 orthologues as sisters to Bub3 and Rae1 (figure 4 and electronic supplementary material, figure S3a), whereas the other places only a particular subset, those of B. saltans, A. spiralis and P. ankaliazontas, next to Bub3 and Rae1 (electronic supplementary material, figure S3b), suggesting that trypanosomatid KKT15 sequences differ more from Bub3/Rae1 than those of bodonids and prokinetoplastids. Importantly, KKT15 proteins do not cluster within Bub3 in either tree, so we cannot unequivocally designate them as Bub3 orthologues. However, the fact that kinetoplastids have clear Rae1 orthologues (electronic supplementary material, figure S3a,b) [61], in combination with KKT15’s kinetochore localization and interaction with KKT14 (see below), analogous to Bub1/BubR1-Bub3, prompts us to propose that KKT15 is a Bub3 orthologue.
Figure 4.
Phylogeny of KKT15 and related WD40 repeat proteins. Subtree of the full phylogeny presented in electronic supplementary material, figure S3a. Note that in the alignment approach applied here, KKT15 proteins were prompted to form a single group (see §4).
2.4. KKT14 interacts with KKT15
KKT14 and KKT15 localize at kinetochores from G2 to anaphase [39]. This localization pattern differs from the rest of transiently localized kinetoplastid kinetochore proteins that start to localize at kinetochores from the S phase, suggesting that KKT14 and KKT15 may directly interact with each other. This possibility is supported by our mass spectrometry analysis (figure 5a ). Although KKT14 does not appear to have a GLEBS motif present in the N-terminal region of Bub1/BubR1 that interacts with Bub3 [12,21,62], AlphaFold2 predicted an interaction between the N-terminal region of KKT14 and KKT15 (figure 5b,c ). The region of KKT14 predicted to interact with KKT15 is well conserved among trypanosomatids (residues 2–111 in T. brucei) (electronic supplementary material, figure S2). Interestingly, the confidence score (predicted local distance difference test: pLDDT) for this region improved when predicted as a KKT14–KKT15 complex, compared with KKT14 alone (figure 5d ), implying that this region of KKT14 is more likely to form a secondary structure (α-helices) in the complex. These results strongly support the idea that kinetoplastid kinetochore proteins KKT14 and KKT15 are divergent Bub1/BubR1 and Bub3 proteins, although they might have adopted a distinct interaction mode.
Figure 5.
KKT14 is predicted to interact with KKT15 directly. (a) A volcano plot showing relative enrichment and significance values between the immunoprecipitates of YFP-KKT14 and YFP-KKT22 (n = 4 each). KKT22 was used as a comparison, which mainly co-purifies with KKT23 and KKT3 [43]. See electronic supplementary material, table S5, for all proteins identified by mass spectrometry. (b) AlphaFold2 predictions of the KKT142–125–KKT15 complex in cartoon representation (electronic supplementary material, dataset S2). (c) The PAE plots for the rank 1 model for KKT14–KKT15, predicting interactions via the N-terminal part of KKT14. (d) pLDDT plots for KKT14–KKT15 (left) and KKT14 (right). AlphaFold2-predicted models are provided in the electronic supplementary material, datasets S3 and S4. PAE, predicted aligned error.
To better characterize KKT14, we ectopically expressed its fragments in trypanosomes. We found that KKT14N2–357 localized at kinetochores from G2 to anaphase, while KKT14C358–685 only had diffuse nuclear signals (figure 6a ). Immunoprecipitation of these fragments revealed that KKT14N co-purified with many kinetochore proteins, including KKT15 (figure 6b and electronic supplementary material, table S5). Furthermore, LacO/LacI-based tethering experiments show that KKT14N, not KKT14C, was able to recruit KKT15 to an ectopic locus in vivo (figure 6c ). These results suggest that the N-terminal region of KKT14 interacts with KKT15, as predicted by AlphaFold2 (figure 5b ).
Figure 6.

N-terminal region of KKT14 binds KKT15. (a) Ectopically expressed GFP-KKT14N2–357 localizes at kinetochores, while GFP-KKT14C358–685 does not. K and N stand for the kinetoplast (mitochondrial DNA) and nucleus, respectively. Cell cycle stages of individual cells were determined based on the number of K and N as described previously [63,64]. The GFP fusion proteins were expressed in trypanosomes using 10 ng ml−1 doxycycline for 1 day and fixed for microscopy. Cell lines: BAP2386, BAP2387. (b) Immunoprecipitation/mass spectrometry analysis shows that GFP-KKT14N co-purifies with many kinetochore proteins, while GFP-KKT14C does not. Immunoprecipitation was carried out using cells expressing the GFP fusion proteins using 10 ng ml−1 doxycycline for 1 day. See electronic supplementary material, table S5, for all proteins identified by mass spectrometry. (c) KKT14N2–357 is sufficient to recruit KKT15 in trypanosomes. Recruitment of tdTomato-KKT15 was observed in 100% or 0% of 1K1N (G1) cells that have GFP-KKT14N2–357-LacI or GFP-KKT14C358–685-LacI dots, respectively (n = 10 each). The GFP fusion proteins were expressed in trypanosomes using 10 ng ml−1 doxycycline for 1 day. Cell lines: BAP2655, BAP2656. Scale bars, 5 µm.
2.5. KKT14 and KKT15 are required for accurate chromosome segregation
We next performed an RNAi-mediated knockdown of KKT14 and KKT15 to assess their function for chromosome segregation. The RNAi construct for KKT14 is previously described [65], while that for KKT15 was established in this study (figure 7a–c). We found that kinetochore localization of KKT14 and KKT15 are mutually co-dependent (figure 7d–g ), further supporting the notion that they form a complex. Although KKT14 depletion caused severe growth defects (figure 7h ) [65], we failed to find obvious cell cycle profile changes at 8 or 16 h after induction of KKT14 RNAi, apart from a moderate increase in anaphase cells (figure 7i ). In contrast, we observed lagging kinetochores in almost all anaphase cells even at 8 h post-induction (figure 7j,k ). These results show that KKT14 is essential for accurate chromosome segregation and cell growth.
Figure 7.
KKT14 is essential for accurate chromosome segregation in trypanosomes. (a) Growth curve upon RNAi-mediated knockdown of KKT15 using an RNAi construct against its 3’ UTR. Data are presented as the mean ± s.d. of three replicates. RNAi was induced with 1 μg ml−1 doxycycline and cultures were diluted on day 2. Cell line: BAP2533. (b,c) Validation of KKT15 knockdown. RNAi was induced with 1 μg ml−1 doxycycline and cultures were diluted on day 2. K and N stand for the kinetoplast and nucleus, respectively. Cell line: BAP2535. At least 130 cells per condition were quantified. (d) Representative fluorescence micrographs showing the localization of YFP-KKT14 upon RNAi-mediated knockdown of KKT15 in G2/M (2K1N) and anaphase (2K2N) cells. RNAi was induced with 1 μg ml−1 doxycycline for 24 h. Cell line: BAP2533. (e) Quantification of 2K1N and 2K2N that have kinetochore-like dots of YFP-KKT14 upon RNAi-mediated depletion of KKT15. All graphs depict the means (bar) ± s.d. of three replicates (shown as dots). (f) Representative fluorescence micrographs showing the localization of YFP-KKT15 upon RNAi-mediated knockdown of KKT14 in 2K1N and 2K2N cells. RNAi was induced with 1 μg ml−1 doxycycline for 24 h. Cell line: BAP2534. (g) Quantification of 2K1N and 2K2N that have kinetochore-like dots of YFP-KKT15 upon RNAi-mediated depletion of KKT14. All graphs depict the means (bar) ± s.d. of three replicates (shown as dots). A minimum of 50 cells per replicate were quantified in each condition. (h) Growth curve upon RNAi-mediated knockdown of KKT14. Data are presented as the mean ± s.d. of three replicates. RNAi was induced with 1 μg ml−1 doxycycline and cultures were diluted at day 2. Cell line: BAP2534. (i) Cell cycle profile upon knockdown of KKT14. RNAi was induced with 1 μg ml−1 doxycycline and cells were fixed at 8 or 16 h. All graphs depict the means (bar) ± s.d. of three replicates. A minimum of 700 cells per replicate were quantified. Cell line: BAP680. (j) Representative fluorescence micrographs showing lagging kinetochores (marked by tdTomato-KKT2) in anaphase cells upon KKT14 knockdown at 8 and 16 h post-induction. RNAi was induced with 1 μg ml−1 doxycycline. Cell line: BAP680. (k) Quantification of lagging kinetochores in anaphase cells upon KKT14 knockdown. All graphs depict the means (bar) ± s.d. of three replicates (shown as dots). A minimum of 50 cells per replicate were quantified in each condition. *p < 0.05, **p ≤ 0.01, ***p ≤ 0.001 (two-sided, unpaired t‐test).
3. Discussion
Previous studies have shown that T. brucei has a Mad2-like protein, Cdc20 and components of the APC/C [66,67]. However, the Mad2-like protein localizes near basal bodies, not kinetochores, while Cdc20 lacks a well-conserved Mad2-interacting motif, suggesting that these proteins are unable to play a role in the canonical spindle checkpoint control [68,69]. Indeed, trypanosome cells cannot halt cell cycle progression in response to spindle defects [70]. By contrast, forced stabilization of cyclin B or treatment with proteasome inhibitors cause the nucleus to arrest in metaphase [70,71], raising a possibility that trypanosomes may possess an intrinsic mechanism that regulates the timing of nuclear division by controlling the APC/CCdc20 activity in a Mad2-independent manner.
In this study, we propose that the kinetoplastid kinetochore proteins KKT14 and KKT15 are divergent Bub1/BubR1 and Bub3 orthologues, respectively. The discovery of a kinase fold in KKT14 was surprising because our previous sequence-based approach or a previous study that comprehensively catalogued pseudokinases failed to identify KKT14 as a pseudokinase [59]. In other words, KKT14’s pseudokinase domain is highly divergent from other kinases or pseudokinases. It is therefore remarkable that KKT14 retains significant structural similarities to the kinase domain of Bub1, including its N-terminal extension. In human Bub1, this N-terminal extension acts as a ‘minicyclin’ by making extensive contacts with the N-lobe of the kinase domain and thereby promoting an active conformation [48]. Similarly, in both the crystal structure of A. spiralis KKT14 and the AlphaFold2-predicted structure of T. brucei KKT14, the N-terminal extension makes extensive contacts with the N-lobe, which may stabilize the pseudokinase structure. Although the function of the KKT14 pseudokinase domain remains unclear, it is striking that all known KKT14 orthologs in kinetoplastids conserved it.
In contrast to KKT14, KKT15 has a readily discernible, common protein fold, namely a WD40 repeat beta-propeller. Our phylogenetic analysis places KKT15 close to Bub3. Taking into account also its interaction with the divergent Bub1/BubR1 protein KKT14, we propose that KKT15 is a Bub3 orthologue. Yet, its sequence is quite divergent from Bub3, particularly in trypanosomatids. In humans, Bub3 localizes at kinetochores by recognizing the phosphorylated KNL1 protein, which is not found in kinetoplastids. Furthermore, it remains unclear whether KKT15 binds a phosphorylated peptide because the regions of Bub3 that bind KNL1’s phosphorylated MELT motif [12] are not well conserved in KKT15 (electronic supplementary material, figures S4 and S5). It therefore remains unknown how the KKT14–KKT15 complex is recruited to kinetochores. Depletion of KKT2 disrupts KKT14 localization, suggesting that KKT14 and KKT15 are downstream of KKT2 [65]. Although our mass spectrometry data support a possibility that KKT14 and/or KKT15 directly interact with KKT2, AlphaFold2 fails to predict interactions between them. It will be important to identify direct interaction partners for KKT14 and KKT15 to reveal how these proteins function at kinetoplastid kinetochores.
In humans and C. elegans, some of the ABBA motifs in Bub1/BubR1 function by promoting kinetochore localization of Cdc20 and contribute to the strength of the checkpoint [19,72–75]. In trypanosomes, kinetochore localization of Cdc20 has not been reported [61] (and our unpublished data), and it remains unclear if, how, when, and where the ABBA motif of KKT14 may regulate the activity of Cdc20 and contribute to the cell cycle control. Addressing these questions will be key to understanding how KKT14 and KKT15 contribute to accurate chromosome segregation in trypanosomes that lack a canonical spindle checkpoint.
4. Material and methods
4.1. Trypanosomes and microscopy
All trypanosome cell lines used in this study were derived from T. brucei SmOxP927 procyclic form cells (TREU 927/4 expressing T7 RNA polymerase and the tetracycline repressor to allow inducible expression) [76] and are listed in electronic supplementary material, table S1. Cells were grown at 28°C in SDM-79 medium supplemented with 10% (v/v) heat-inactivated foetal calf serum, 7.5 µg ml−1 hemin [77], and appropriate drugs. Endogenous YFP tagging was performed using the pEnT5-Y vector [78] or a PCR-based method [79]. Endogenous 3FLAG-6HIS-YFP tagging was performed using pBA106 [70], while endogenous tdTomato tagging was performed using pBA892 [80]. LacO-LacI tethering experiments were performed as described previously using the LacO array inserted at the rDNA locus [80,81]. Inducible expression of GFP-NLS fusion and GFP-NLS-LacI fusion proteins was carried out using pBA310 [42] and pBA795 [80], respectively. Cell growth was monitored using a CASY cell counter (Roche). Expression of GFP fusion proteins and RNAi were induced with doxycycline at a final concentration of 10 ng ml−1 and 1 µg ml−1, respectively. All plasmids were linearized by NotI and transfected into trypanosomes by electroporation. Transfected cells were selected by the addition of 30 μg ml−1 G418 (Sigma), 50 μg ml−1 hygromycin (Sigma), 5 μg ml−1 phleomycin (Sigma) or 10 μg ml−1 blasticidin S (Insight Biotechnology).
4.2. Immunoprecipitation
For each experiment, 400 ml cultures of asynchronously growing cells (unless otherwise indicated) were grown to ~1 × 107 cells ml−1 and harvested. Ectopic expression of GFP-tagged KKT14N2–357 and KKT14C358–685 in T. brucei was induced with 10 ng ml−1 doxycycline for 24 h. YFP-KKT14 and YFP-KKT22 were expressed from the endogenous locus. Where indicated, 10 µM MG132 (to arrest cells prior to anaphase) or 2 µM 1NM-PP1 (to inhibit the AUK1 kinase activity) were added for 4 h prior to harvesting the cells. Note that there was no noticeable change in the amount of co-purifying proteins in these conditions, so we pooled all results for the volcano plot analysis (figure 5a and electronic supplementary material, table S5). Immunoprecipitation of GFP/YFP-tagged proteins was performed with anti-GFP antibodies (11814460001, Roche) using a method we previously described [80]. 3FLAG-6HIS-YFP-tagged proteins expressed from the endogenous locus were immunoprecipitated using anti-FLAG M2 antibodies (F3165, Sigma) [82] and eluted with 0.5 mg ml−1 3×FLAG peptide (F4799, Sigma) in BH0.15 (25 mM HEPES pH 8.0, 2 mM MgCl2, 0.1 mM EDTA pH 8.0, 0.5 mM EGTA pH 8.0, 1% NP-40, 150 mM KCl and 15% glycerol) supplemented with protease inhibitors (10 μg ml−1 leupeptin, 10 μg ml−1 pepstatin, 10 μg ml−1 E-64 and 0.2 mM PMSF) and phosphatase inhibitors (1 mM sodium pyrophosphate, 2 mM Na-β-glycerophosphate, 0.1 mM Na3VO4, 5 mM NaF and 100 nM microcystin-LR) with agitation for 25 min at room temperature. FLAG eluates were run on an SDS-PAGE gel, which was stained with Sypro-Ruby (Thermo Fisher).
4.3. In vitro kinase assay
To examine auto-phosphorylation activities in the immunoprecipitated 3FLAG-6HIS-YFP-KKT3/4/14/15 samples, 10 µl of FLAG eluates were mixed with 2.5 µl of 10× kinase buffer (500 mM Tris–HCl pH 7.4, 10 mM DTT, 250 mM β-glycerophosphate, 50 mM MgCl2, 50 μCi [32P] ATP and 100 μM ATP) in 25 µl volumes. The mixture was incubated at 30°C for 30 min, and the reaction was stopped by the addition of the LDS sample buffer (Thermo Fisher). The samples were run on an SDS-PAGE gel and stained with Coomassie Brilliant Blue R-250 (Bio-Rad) (not shown), which was subsequently dried and used for autoradiography using a phosphorimager screen. The signal was detected by an FLA 7000 scanner (GE Healthcare).
4.4. Mass spectrometry
Reduction of disulfide bridges in cysteine-containing proteins was performed with 10 mM DTT dissolved in 50 mM HEPES, pH 8.5 (56°C, 30 min). Reduced cysteines were alkylated with 20 mM 2-chloroacetamide dissolved in 50 mM HEPES, pH 8.5 (room temperature, in the dark, 30 min). Mass spectrometry samples were prepared using the SP3 protocol [83], and trypsin (Promega) was added in a 1:50 enzyme-to-protein ratio for overnight digestion at 37°C. Next day, peptide recovery was done by collecting supernatant on a magnet and combining with a second elution of beads with 50 mM HEPES, pH 8.5. For a further sample clean up, an OASIS HLB µElution Plate (Waters) was used. The samples were dissolved in 10 µl of reconstitution buffer (96:4 water:acetonitrile, 1% formic acid) and analysed by LC-MS/MS using QExactive (Thermo Fisher) in the proteomics core facility at EMBL Heidelberg (https://www.embl.org/groups/proteomics/). Peptides were identified by searching tandem mass spectrometry spectra against the T. brucei protein database with MaxQuant (v. 2.0.1) with carbamidomethyl cysteine set as a fixed modification and oxidization (Met), phosphorylation (Ser, Thr and/or Tyr) and acetylation (N-term and Lys) set as variable modifications. Up to two missed cleavages were allowed. The first peptide tolerance was set to 10 ppm (protein FDR 1%). Proteins identified with at least two peptides were considered significant and reported in electronic supplementary material, table S5. All raw mass spectrometry files and the custom database file used in this study have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository [84,85] with the dataset identifier PXD047806.
Differential enrichment analysis of YFP-KKT14 versus YFP-KKT22 was performed on iBAQ values using the DEP package in R [86] (electronic supplementary material, table S5). Reverse hits and contaminants were removed, and results were filtered for proteins that were identified in all replicates of at least one condition. The data were background corrected and normalized by variance stabilizing transformation (vsn). Missing values were imputed using the k-nearest neighbour approach (knn). Potential interactors were determined using t-tests, with threshold values set to lfc = 2 and alpha = 0.05. The volcano plot shown was constructed using the EnhancedVolcano package [87].
4.5. Expression and purification of Apiculatamorpha spiralis KKT14C
To make pBA2356 (6HIS-KKT14365–640 from Apiculatamorpha spiralis (clone PhF-6)), the DNA was amplified from BAG142 (a synthetic DNA that encodes A. spiralis KKT14, codon optimized for expression in E. coli) with primers BA3187/BA3188 and cloned into RSFDuet-1 using BamHI/EcoRI sites with the NEBuilder HiFi DNA Assembly kit (NEB) (electronic supplementary material, table S1). Escherichia coli BL21(DE3) cells were transformed with ~100 ng of plasmid DNA (pBA2356) and inoculated into 50 ml of 2×TY medium containing 50 μg ml−1 kanamycin and grown overnight at 37°C. The next morning, 6 l of 2×TY medium with 50 μg ml−1 of kanamycin was warmed at 37°C, and 5 ml of the overnight culture was inoculated into each litre. Cells were grown at 37°C with shaking (200 rpm) until the OD600 reached ~0.6. Protein expression was induced with 0.2 mM IPTG for 16 h at 20°C. Cells were spun down at 3400g at 4°C and resuspended in 200 ml of lysis buffer (50 mM sodium phosphate, pH 7.5, 500 mM NaCl and 10% glycerol) supplemented with protease inhibitors (20 μg ml−1 leupeptin, 20 μg ml−1 pepstatin, 20 μg ml−1 E-64 and 0.4 mM PMSF), benzonase nuclease (500 U per 1 l culture) and 0.5 mM TCEP. All subsequent steps were performed at 4°C. Bacterial cultures were mechanically disrupted using a French press (1 passage at 20 000 psi) and the soluble fraction was separated by centrifugation at 48 000g for 30 min. Supernatants were loaded on 5 ml of TALON beads (Takara Bio) pre-equilibrated with the lysis buffer. Next, the beads were washed with 300 ml of the lysis buffer with 0.5 mM TCEP, and proteins were eluted with 50 mM sodium phosphate pH 7.5, 500 mM NaCl, 10% glycerol, 250 mM imidazole and 0.5 mM TCEP. To cleave off the His-tag, samples were incubated with TEV protease in 1:50 (w/w) ratio overnight while being buffer-exchanged into 25 mM sodium phosphate, 250 mM NaCl, 5% glycerol, 5 mM imidazole and 0.5 mM TCEP by dialysis. To increase the sample purity and remove the His-tag, samples were re-loaded on TALON beads pre-equilibrated with the dialysis buffer and the flow-through was collected. Next, the sample was concentrated using 10-kD MW Amicon concentrator (Millipore), and loaded on Superdex 75 16/600 (GE Healthcare) columns to further purify and buffer exchange into 25 mM HEPES pH 7.5, 150 mM NaCl with 0.5 mM TCEP. Fractions containing the protein of interest were pooled, concentrated to 15.1 mg ml−1 using a 10-kD MW Amicon concentrator (Millipore), and flash-frozen in liquid nitrogen for −80°C storage.
4.6. Crystallization trials and structural determination
All crystals were obtained in sitting drop vapour diffusion experiments in 96-well plates, using drops of overall volume 200 nl, mixing protein and mother liquor in a 1:1 (v/v) ratio. Crystals of A. spiralis KKT14365−640 (15.1 mg ml−1) were grown at 4°C in MIDAS HT-96 B1 solution (Molecular Dimensions) containing 0.1 M sodium formate and 20% (w/v) SOKALAN CP 45. Crystals were briefly transferred into mother liquor prepared with the addition of 25% glycerol prior to flash-cooling by plunging into liquid nitrogen. Data collection and model-building X-ray diffraction data from A. spiralis KKT14365−640 were carried out at the I03 beamline at the Diamond Light Source (Harwell, UK). The structure was solved using the AlphaFold2-predicted structure of A. spiralis KKT14398−640 as a model with a molecular replacement software, PHASER [88], followed by initial model building with BUCCANEER [89]. The data were scaled to 2.2 Å based on I/Iσ parameters (I/Iσ value of 2.0 was used as a threshold). Further manual model building and refinement were completed iteratively using COOT [90] and PHENIX [91]. All images were made with PyMOL (v. 2.5.2, Schrödinger). Protein coordinates have been deposited in the RCSB protein data bank with the accession number 8QOH.
4.7. Bioinformatic analysis of KKT14 and KKT15
The protein sequences for KKT14 and KKT15 were retrieved from the TriTryp database [92] or published studies [93,94]. Searches for their homologous proteins were done using BLAST in the TriTryp database [92] or manual searches using hmmsearch (HMMER v. 3.0) on predicted proteomes using manually prepared hmm profiles [95]. Multiple sequence alignments were performed with MAFFT (L-INS-i method, v. 7) [96] and visualized with the clustalx colouring scheme in Jalview (v. 2.11) [97]. The pairwise sequence identity and similarity between A. spiralis KKT14365−640 and T. brucei KKT14358–685 were calculated using EMBOSS Needle [98]. Structures and interactions were predicted with AlphaFold2-Multimer-v. 2.3.1 [99,100] through ColabFold v. 1.5.3 using MMseqs2 with 24 recycles (UniRef + Environmental) [101]. The rank 1 model, predicted aligned error and pLDDT plots for each prediction are provided in the electronic supplemental material (dataset S1–S4). In all cases, similar results were obtained for five predictions. All structure figures were made using PyMOL v. 2.5.2 (Schrödinger, LLC). The following command was used to map the pLDDT score onto the AlphaFold2-predicted structure models: spectrum b, rainbow_rev, maximum = 100, minimum = 50. Foldseek searches were carried out using the Web server against AlphaFold2-predicted structure databases covering UniProt50, Swiss-Prot and Proteome (v. 4, Mode 3Di/AA) (https://search.foldseek.com/search) [52,102].
4.8. Phylogenetic analysis of KKT15 and related WD40 repeat-containing proteins
To conduct a phylogenetic analysis to determine to which WD40 repeat proteins KKT15 proteins are closely related, we selected the 30 best EggNOG KOG/COG hits resulting from online HHpred [60,103] of our best, most sensitive KKT15 multiple sequence alignment. The latter was obtained by iterative profile HMM searches combined with phylogenetic analysis among a local database of euglenozoa and some other eukaryotic protein sequences, as well as a local eukaryote-wide dataset [104] (electronic supplementary material, table S3). The trusted list of KKT15 orthologues across these euglenozoa was aligned using MAFFT (v. 7.505, option L-INS-i, alignment and profile HMM are included in electronic supplementary material, dataset S5) [96] and submitted to online HHpred to search in its COG_KOG_v. 1.0 database. Among these 30 best hits, there were only KOG families, and one duplicate, which we removed (electronic supplementary material, table S4). We collected the corresponding profile HMMs of these KOGs from EggNOG (v. 5.0) [105] and added our initial KKT15 HMM. We then executed hmmscan (hmmer.org) of these HMMs versus a subsampled (49 diverse eukaryotes) version of our local eukaryotic database, applying an E-value cut-off of 1 × 10–5. We assigned the retrieved eukaryotic proteins to their respective KOG (or KKT15), and appended the original euglenozoan KKT15 orthologues to that family as well. We subsequently, again for all KOGs/KKT15, used hmmsearch among the assigned hits, in order to be able to retrieve the 100 best hits per KOG/KKT15, and gathered of these 100 hits only the domain that was hit by the HMM (the putative WD40 repeat). For Rae1 (KOG0647), we also included the hit regions of some additional Discoba orthologue candidates, because these were not part of the 49 species selection but potentially informative in placing KKT15. Except for KKT15 and these additional Discoba sequences, we subjected the KOG sequence selections to CD-hit (v. 4.8.1, identity cut-off 70%) [106], in order to facilitate the phylogenetic analysis and interpretation. We then removed all sequences across all KOGs that were shorter than 50 amino acids (i.e. this was not applied to KKT15). Each KOG was separately aligned using MAFFT (L-INS-i).
We then employed two different strategies to combine all sequences into a single alignment, resulting in two different phylogenies (referred to as tree 1 and tree 2, found in electronic supplementary material, figure S3a,b, respectively). For tree 1, we similarly aligned KKT15, and subsequently used MAFFT’s –merge option to combine all individual alignments into a single alignment (parameters: --localpair --maxiterate 100 --merge). We also had identified five additional, potentially close KKT15 homologues of different species, which cannot be evidently classified as a particular WD40 repeat protein. We added them to the alignment using MAFFT option –add (parameters: --maxiterate 1000 --add). For tree 2, we used MAFFT –merge (parameters: --localpair --maxiterate 100 –merge) on just the KOG’s individual alignments, and subsequently used MAFFT –add (parameters: --maxiterate 1000 –add) to add KKT15 orthologues and the sequences of unknown identity to it. The alignments for tree 1 and tree 2 hence differ in the way the KKT15 sequences are aligned: either first among one another (tree 1), or only through adding the sequences to the already existing (merged) alignment of the KOGs. The first approach forces the KKT15 sequences to be monophyletic in the tree, while the second approach does not. We trimmed both alignments using trimAl [107] (v. 1.4.rev15, option -gappyout) and removed sequences with >85% gaps. We used the alignments to infer a maximum likelihood phylogeny with IQ-TREE (v. 2.0.3) [108], applying an evolutionary model selected by ModelFinder [109], also allowing for complex mixture (C-series) models to be selected. Branch support was estimated through ultrafast bootstraps (1000 replicates) [110]. The tree with the highest likelihood, either the maximum likelihood tree or the consensus tree, was selected for visualization in iTOL [111]. The phylogenies were initially rooted in a well-supported clade with a relatively long branch, which was not closely associated with Rae1, Bub3 or KKT15 (KOG2111). The phylogenies were annotated using, if available, the name of the human protein belonging to each KOG. If not available, the budding yeast protein name was used. The multiple sequence alignments and raw IQ-TREE output can be found in electronic supplementary material, dataset S5. The full, uncollapsed phylogenies can be inspected on iTOL: https://itol.embl.de/tree/62145194227274281702966749 (tree 1, associated with electronic supplementary material, figure S3a) and https://itol.embl.de/tree/62145194227421141702968325 (tree 2, associated with electronic supplementary material, figure S3b). Note that alongside KKT15, we performed a similar online HHpred search for a refined KKT14 multiple sequence alignment, the results of which are also reported in electronic supplementary material, table S4. The alignments used as input for these searches can be found in electronic supplementary material, dataset S6.
Acknowledgments
We thank Kyle Muir for comments on the manuscript. We thank the crystallography facility manager Edward Lowe, Micron Advanced Bioimaging Unit, and the proteomics core facility at EMBL, especially Mandy Rettel and Jennifer Schwarz, for their support. We also thank Miguel Navarro (Instituto de Parasitologıá y Biomedicina López-Neyra, Consejo Superior de Investigaciones Cientificas, Spain) for providing the pMig75 and pMig96 plasmids, and Sam Dean (Warwick Medical School, University of Warwick, UK) for pPOT plasmids. D.B. was supported by the Berrow Foundation. J.J.E.H. was supported by a Veni Fellowship from the Dutch Research Council (NWO, VI.Veni.212.099). E.C.T. was supported by a Veni Fellowship from the Dutch Research Council (NWO, VI.Veni.202.223). P.L. was supported by the Boehringer Ingelheim Fonds. B.A. was supported by a Wellcome Trust Senior Research Fellowship (grant 210622/Z/18/Z), a Wellcome Discovery Award (grant 227243/Z/23/Z), and a Centre Core Grant to the Wellcome Trust Centre for Cell Biology (203149).
Contributor Information
Daniel Ballmer, Email: daniel.ballmer@lincoln.ox.ac.uk.
William Carter, Email: william.carter2009@gmail.com.
Jolien J. E. van Hooff, Email: jolien.vanhooff@wur.nl.
Eelco C. Tromer, Email: e.c.tromer@rug.nl; ecet2@cam.ac.uk.
Midori Ishii, Email: midori.ishii@ed.ac.uk.
Patryk Ludzia, Email: patryk.ludzia@icloud.com.
Bungo Akiyoshi, Email: bungo.akiyoshi@ed.ac.uk.
Ethics
This work did not require ethical approval from a human subject or animal welfare committee.
Data accessibility
All raw mass spectrometry files and the custom database file used in this study have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository [84,85] with the dataset identifier PXD047806. Protein coordinates have been deposited in the RCSB protein data bank with the accession number 8QOH.
Supplementary material is available online [112].
Declaration of AI use
We have not used AI-assisted technologies in creating this article.
Authors’ contributions
D.B.: investigation, writing—review and editing; W.C.:investigation, writing—review and editing; J.J.E.H.: investigation, writing—review and editing; E.C.T.: writing—review and editing; M.I.: writing—review and editing; P.L.: writing—review and editing; B.A.: conceptualization, funding acquisition, investigation, project administration, resources, supervision, writing—original draft, writing—review and editing;
All authors gave final approval for publication and agreed to be held accountable for the work performed therein.
Conflict of interest declaration
We declare we have no competing interests.
Funding
This research was funded in whole, or in part, by the Wellcome Trust [210622/A/18/Z and 227243/Z/23/Z].
References
- 1. McAinsh AD, Marston AL. 2022. The four causes: the functional architecture of centromeres and kinetochores. Annu. Rev. Genet. 56 , 279–314. ( 10.1146/annurev-genet-072820-034559) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Musacchio A, Desai A. 2017. A molecular view of kinetochore assembly and function. Biology (Basel) 6 , 5. ( 10.3390/biology6010005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Alfieri C, Zhang S, Barford D. 2017. Visualizing the complex functions and mechanisms of the anaphase promoting complex/cyclosome (APC/C). Open Biol. 7 , 170204. ( 10.1098/rsob.170204) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pines J. 2011. Cubism and the cell cycle: the many faces of the APC/C. Nat. Rev. Mol. Cell Biol. 12 , 427–438. ( 10.1038/nrm3132) [DOI] [PubMed] [Google Scholar]
- 5. Murray AW. 2011. A brief history of error. Nat. Cell Biol. 13 , 1178–1182. ( 10.1038/ncb2348) [DOI] [PubMed] [Google Scholar]
- 6. Lara-Gonzalez P, Pines J, Desai A. 2021. Spindle assembly checkpoint activation and silencing at kinetochores. Semin. Cell Dev. Biol. 117 , 86–98. ( 10.1016/j.semcdb.2021.06.009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. London N, Biggins S. 2014. Signalling dynamics in the spindle checkpoint response. Nat. Rev. Mol. Cell Biol. 15 , 736–747. ( 10.1038/nrm3888) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. McAinsh AD, Kops GJPL. 2023. Principles and dynamics of spindle assembly checkpoint signalling. Nat. Rev. Mol. Cell Biol. 24 , 543–559. ( 10.1038/s41580-023-00593-z) [DOI] [PubMed] [Google Scholar]
- 9. Sudakin V, Chan GK, Yen TJ. 2001. Checkpoint inhibition of the APC/C in HeLa cells is mediated by a complex of BUBR1, BUB3, CDC20, and MAD2. J. Cell Biol. 154 , 925–936. ( 10.1083/jcb.200102093) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. De Antoni A, et al. 2005. The Mad1/Mad2 complex as a template for Mad2 activation in the spindle assembly checkpoint. Curr. Biol. 15 , 214–225. ( 10.1016/j.cub.2005.01.038) [DOI] [PubMed] [Google Scholar]
- 11. London N, Ceto S, Ranish JA, Biggins S. 2012. Phosphoregulation of Spc105 by Mps1 and PP1 regulates Bub1 localization to kinetochores. Curr. Biol. 22 , 900–906. ( 10.1016/j.cub.2012.03.052) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Primorac I, Weir JR, Chiroli E, Gross F, Hoffmann I, van Gerwen S, Ciliberto A, Musacchio A. 2013. Bub3 reads phosphorylated MELT repeats to promote spindle assembly checkpoint signaling. Elife 2 , e01030. ( 10.7554/eLife.01030) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Shepperd LA, Meadows JC, Sochaj AM, Lancaster TC, Zou J, Buttrick GJ, Rappsilber J, Hardwick KG, Millar JBA. 2012. Phosphodependent recruitment of Bub1 and Bub3 to Spc7/KNL1 by Mph1 kinase maintains the spindle checkpoint. Curr. Biol. 22 , 891–899. ( 10.1016/j.cub.2012.03.051) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Taylor SS, Ha E, McKeon F. 1998. The human homologue of Bub3 is required for kinetochore localization of Bub1 and a Mad3/Bub1-related protein kinase. J. Cell Biol. 142 , 1–11. ( 10.1083/jcb.142.1.1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yamagishi Y, Yang CH, Tanno Y, Watanabe Y. 2012. MPS1/Mph1 phosphorylates the kinetochore protein KNL1/Spc7 to recruit SAC components. Nat. Cell Biol. 14 , 746–752. ( 10.1038/ncb2515) [DOI] [PubMed] [Google Scholar]
- 16. Suijkerbuijk SJE, et al. 2012. The vertebrate mitotic checkpoint protein BUBR1 is an unusual pseudokinase. Dev. Cell 22 , 1321–1329. ( 10.1016/j.devcel.2012.03.009) [DOI] [PubMed] [Google Scholar]
- 17. Kawashima SA, Yamagishi Y, Honda T, Ishiguro K, Watanabe Y. 2010. Phosphorylation of H2A by Bub1 prevents chromosomal instability through localizing shugoshin. Science 327 , 172–177. ( 10.1126/science.1180189) [DOI] [PubMed] [Google Scholar]
- 18. Sharp-Baker H, Chen RH. 2001. Spindle checkpoint protein Bub1 is required for kinetochore localization of Mad1, Mad2, Bub3, and CENP-E, independently of its kinase activity. J. Cell Biol. 153 , 1239–1250. ( 10.1083/jcb.153.6.1239) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Di Fiore B, Davey NE, Hagting A, Izawa D, Mansfeld J, Gibson TJ, Pines J. 2015. The ABBA motif binds APC/C activators and is shared by APC/C substrates and regulators. Dev. Cell 32 , 358–372. ( 10.1016/j.devcel.2015.01.003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Diaz-Martinez LA, Tian W, Li B, Warrington R, Jia L, Brautigam CA, Luo X, Yu H. 2015. The Cdc20-binding Phe box of the spindle checkpoint protein BubR1 maintains the mitotic checkpoint complex during mitosis. J. Biol. Chem. 290 , 2431–2443. ( 10.1074/jbc.M114.616490) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Larsen NA, Al-Bassam J, Wei RR, Harrison SC. 2007. Structural analysis of Bub3 interactions in the mitotic spindle checkpoint. Proc. Natl Acad. Sci. USA 104 , 1201–1206. ( 10.1073/pnas.0610358104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tromer E, Bade D, Snel B, Kops GJPL. 2016. Phylogenomics-guided discovery of a novel conserved cassette of short linear motifs in BubR1 essential for the spindle checkpoint. R. Soc. Open Biol. 6 , 160315. ( 10.1098/rsob.160315) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Buffin E, Emre D, Karess RE. 2007. Flies without a spindle checkpoint. Nat. Cell Biol. 9 , 565–572. ( 10.1038/ncb1570) [DOI] [PubMed] [Google Scholar]
- 24. Burds AA, Lutum AS, Sorger PK. 2005. Generating chromosome instability through the simultaneous deletion of Mad2 and p53. Proc. Natl Acad. Sci. USA 102 , 11296–11301. ( 10.1073/pnas.0505053102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hoyt MA, Totis L, Roberts BT. 1991. S. cerevisiae genes required for cell cycle arrest in response to loss of microtubule function. Cell 66 , 507–517. ( 10.1016/0092-8674(81)90014-3) [DOI] [PubMed] [Google Scholar]
- 26. Li R, Murray AW. 1991. Feedback control of mitosis in budding yeast. Cell 66 , 519–531. ( 10.1016/0092-8674(81)90015-5) [DOI] [PubMed] [Google Scholar]
- 27. Raaijmakers JA, van Heesbeen R, Blomen VA, Janssen LME, van Diemen F, Brummelkamp TR, Medema RH. 2018. BUB1 Is essential for the viability of human cells in which the spindle assembly checkpoint is compromised. Cell Rep. 22 , 1424–1438. ( 10.1016/j.celrep.2018.01.034) [DOI] [PubMed] [Google Scholar]
- 28. Kops GJPL, Snel B, Tromer EC. 2020. Evolutionary dynamics of the spindle assembly checkpoint in eukaryotes. Curr. Biol. 30 , R589–R602, ( 10.1016/j.cub.2020.02.021) [DOI] [PubMed] [Google Scholar]
- 29. Markova K, Uzlikova M, Tumova P, Jirakova K, Hagen G, Kulda J, Nohynkova E. 2016. Absence of a conventional spindle mitotic checkpoint in the binucleated single-celled parasite Giardia intestinalis. Eur. J. Cell Biol. 95 , 355–367, ( 10.1016/j.ejcb.2016.07.003) [DOI] [PubMed] [Google Scholar]
- 30. Morrissette NS, Sibley LD. 2002. Disruption of microtubules uncouples budding and nuclear division in Toxoplasma gondii. J. Cell Sci. 115 , 1017–1025. ( 10.1242/jcs.115.5.1017) [DOI] [PubMed] [Google Scholar]
- 31. Ploubidou A, Robinson DR, Docherty RC, Ogbadoyi EO, Gull K. 1999. Evidence for novel cell cycle checkpoints in trypanosomes: kinetoplast segregation and cytokinesis in the absence of mitosis. J. Cell Sci. 112 (Pt 24), 4641–4650. ( 10.1242/jcs.112.24.4641) [DOI] [PubMed] [Google Scholar]
- 32. Vleugel M, Hoogendoorn E, Snel B, Kops GJPL. 2012. Evolution and function of the mitotic checkpoint. Dev. Cell 23 , 239–250. ( 10.1016/j.devcel.2012.06.013) [DOI] [PubMed] [Google Scholar]
- 33. Cavalier-Smith T. 2010. Kingdoms Protozoa and Chromista and the eozoan root of the eukaryotic tree. R. Soc. Biol. Lett. 6 , 342–345. ( 10.1098/rsbl.2009.0948) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Figueiredo LM, Cross GAM, Janzen CJ. 2009. Epigenetic regulation in African trypanosomes: a new kid on the block. Nat. Rev. Microbiol. 7 , 504–513. ( 10.1038/nrmicro2149) [DOI] [PubMed] [Google Scholar]
- 35. Wheeler RJ, Gull K, Sunter JD. 2019. Coordination of the cell cycle in trypanosomes. Annu. Rev. Microbiol. 73 , 133–154. ( 10.1146/annurev-micro-020518-115617) [DOI] [PubMed] [Google Scholar]
- 36. d’Avila-Levy CM, et al. 2015. Exploring the environmental diversity of kinetoplastid flagellates in the high-throughput DNA sequencing era. Mem. Inst. Oswaldo Cruz. 110 , 956–965. ( 10.1590/0074-02760150253) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. van Hooff JJ, Tromer E, van Wijk LM, Snel B, Kops GJ. 2017. Evolutionary dynamics of the kinetochore network in eukaryotes as revealed by comparative genomics. EMBO Rep. 18 , 1559–1571. ( 10.15252/embr.201744102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tromer EC, van Hooff JJE, Kops GJPL, Snel B. 2019. Mosaic origin of the eukaryotic kinetochore. Proc. Natl Acad. Sci. USA 116 , 12873–12882. ( 10.1073/pnas.1821945116) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Akiyoshi B, Gull K. 2014. Discovery of unconventional kinetochores in kinetoplastids. Cell 156 , 1247–1258. ( 10.1016/j.cell.2014.01.049) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Brusini L, D’Archivio S, McDonald J, Wickstead B. 2021. Trypanosome KKIP1 dynamically links the inner kinetochore to a kinetoplastid outer kinetochore complex. Front. Cell. Infect. Microbiol. 11 , 641174. ( 10.3389/fcimb.2021.641174) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. D’Archivio S, Wickstead B. 2017. Trypanosome outer kinetochore proteins suggest conservation of chromosome segregation machinery across eukaryotes. J. Cell Biol. 216 , 379–391. ( 10.1083/jcb.201608043) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Nerusheva OO, Akiyoshi B. 2016. Divergent polo box domains underpin the unique kinetoplastid kinetochore. R. Soc. Open Biol. 6 , 150206. ( 10.1098/rsob.150206) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Nerusheva OO, Ludzia P, Akiyoshi B. 2019. Identification of four unconventional kinetoplastid kinetochore proteins KKT22-25 in Trypanosoma brucei. R. Soc. Open Biol. 9 , 190236. ( 10.1098/rsob.190236) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Rao SPS, et al. 2019. Drug discovery for kinetoplastid diseases: future directions. ACS Infect. Dis. 5 , 152–157. ( 10.1021/acsinfecdis.8b00298) [DOI] [PubMed] [Google Scholar]
- 45. Saldivia M, et al. 2020. Targeting the trypanosome kinetochore with CLK1 protein kinase inhibitors. Nat. Microbiol. 5 , 1207–1216. ( 10.1038/s41564-020-0745-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Jain BP, Pandey S. 2018. WD40 Repeat proteins: signalling scaffold with diverse functions. Protein J. 37 , 391–406. ( 10.1007/s10930-018-9785-7) [DOI] [PubMed] [Google Scholar]
- 47. Siemeister G, et al. 2019. Inhibition of BUB1 kinase by BAY 1816032 sensitizes tumor cells toward taxanes, ATR, and PARP inhibitors in vitro and in vivo. Clin. Cancer Res. 25 , 1404–1414. ( 10.1158/1078-0432.CCR-18-0628) [DOI] [PubMed] [Google Scholar]
- 48. Kang J, Yang M, Li B, Qi W, Zhang C, Shokat KM, Tomchick DR, Machius M, Yu H. 2008. Structure and substrate recruitment of the human spindle checkpoint kinase Bub1. Mol. Cell 32 , 394–405. ( 10.1016/j.molcel.2008.09.017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kabsch W, Sander C. 1983. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 22 , 2577–2637. ( 10.1002/bip.360221211) [DOI] [PubMed] [Google Scholar]
- 50. Holm L, Laiho A, Törönen P, Salgado M. 2023. DALI shines a light on remote homologs: one hundred discoveries. Protein Sci. 32 , e4519. ( 10.1002/pro.4519) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Endicott JA, Noble MEM, Johnson LN. 2012. The structural basis for control of eukaryotic protein kinases. Annu. Rev. Biochem. 81 , 587–613. ( 10.1146/annurev-biochem-052410-090317) [DOI] [PubMed] [Google Scholar]
- 52. van Kempen M, Kim SS, Tumescheit C, Mirdita M, Lee J, Gilchrist CLM, Söding J, Steinegger M. 2024. Fast and accurate protein structure search with foldseek. Nat. Biotechnol. 42 , 243–246. ( 10.1038/s41587-023-01773-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Breit C, Bange T, Petrovic A, Weir JR, Müller F, Vogt D, Musacchio A. 2015. Role of intrinsic and extrinsic factors in the regulation of the mitotic checkpoint kinase Bub1. PLoS One 10 , e0144673. ( 10.1371/journal.pone.0144673) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Huang Y, et al. 2019. BubR1 phosphorylates CENP-E as a switch enabling the transition from lateral association to end-on capture of spindle microtubules. Cell Res. 29 , 562–578. ( 10.1038/s41422-019-0178-z) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lin Z, Jia L, Tomchick DR, Luo X, Yu H. 2014. substrate-specific activation of the mitotic kinase Bub1 through intramolecular autophosphorylation and kinetochore targeting. Structure 22 , 1616–1627. ( 10.1016/j.str.2014.08.020) [DOI] [PubMed] [Google Scholar]
- 56. Davey NE, Morgan DO. 2016. Building a regulatory network with short linear sequence motifs: lessons from the degrons of the anaphase-promoting complex. Mol. Cell 64 , 12–23. ( 10.1016/j.molcel.2016.09.006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Bayliss R, Fry A, Haq T, Yeoh S. 2012. On the molecular mechanisms of mitotic kinase activation. R. Soc. Open Biol. 2 , 120136. ( 10.1098/rsob.120136) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Tang Z, Shu H, Oncel D, Chen S, Yu H. 2004. Phosphorylation of Cdc20 by Bub1 provides a catalytic mechanism for APC/C inhibition by the spindle checkpoint. Mol. Cell 16 , 387–397. ( 10.1016/j.molcel.2004.09.031) [DOI] [PubMed] [Google Scholar]
- 59. Kwon A, Scott S, Taujale R, Yeung W, Kochut KJ, Eyers PA, Kannan N. 2019. Tracing the origin and evolution of pseudokinases across the tree of life. Sci. Signal. 12 , eaav3810. ( 10.1126/scisignal.aav3810) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zimmermann L, et al. 2018. A completely reimplemented MPI bioinformatics toolkit with a new HHpred server at its core. J. Mol. Biol. 430 , 2237–2243. ( 10.1016/j.jmb.2017.12.007) [DOI] [PubMed] [Google Scholar]
- 61. Billington K, et al. 2023. Genome-wide subcellular protein map for the flagellate parasite Trypanosoma brucei. Nat. Microbiol. 8 , 533–547. ( 10.1038/s41564-022-01295-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Larsen NA, Harrison SC. 2004. Crystal structure of the spindle assembly checkpoint protein Bub3. J. Mol. Biol. 344 , 885–892. ( 10.1016/j.jmb.2004.09.094) [DOI] [PubMed] [Google Scholar]
- 63. Siegel TN, Hekstra DR, Cross GAM. 2008. Analysis of the Trypanosoma brucei cell cycle by quantitative DAPI imaging. Mol. Biochem. Parasitol. 160 , 171–174. ( 10.1016/j.molbiopara.2008.04.004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Woodward R, Gull K. 1990. Timing of nuclear and kinetoplast DNA replication and early morphological events in the cell cycle of Trypanosoma brucei . J. Cell Sci. 95 , 49–57. ( 10.1242/jcs.95.1.49) [DOI] [PubMed] [Google Scholar]
- 65. Marcianò G, Ishii M, Nerusheva OO, Akiyoshi B. 2021. Kinetoplastid kinetochore proteins KKT2 and KKT3 have unique centromere localization domains. J. Cell Biol. 220 , e202101022. ( 10.1083/jcb.202101022) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Berriman M, et al. 2005. The genome of the African trypanosome Trypanosoma brucei. Science 309 , 416–422. ( 10.1126/science.1112642) [DOI] [PubMed] [Google Scholar]
- 67. Kumar P, Wang CC. 2005. Depletion of anaphase-promoting complex or cyclosome (APC/C) subunit homolog APC1 or CDC27 of Trypanosoma brucei arrests the procyclic form in metaphase but the bloodstream form in anaphase. J. Biol. Chem. 280 , 31783–31791. ( 10.1074/jbc.M504326200) [DOI] [PubMed] [Google Scholar]
- 68. Akiyoshi B. 2020. Analysis of a Mad2 homolog in Trypanosoma brucei provides possible hints on the origin of the spindle checkpoint. bioRxiv. ( 10.1101/2020.12.29.424754) [DOI]
- 69. Akiyoshi B, Gull K. 2013. Evolutionary cell biology of chromosome segregation: insights from trypanosomes. R. Soc. Open Biol. 3 , 130023. ( 10.1098/rsob.130023) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Hayashi H, Akiyoshi B. 2018. Degradation of cyclin B is critical for nuclear division in Trypanosoma brucei Biol. Open 7 , bio031609. ( 10.1242/bio.031609) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Mutomba MC, To WY, Hyun WC, Wang CC. 1997. Inhibition of proteasome activity blocks cell cycle progression at specific phase boundaries in African trypanosomes. Mol. Biochem. Parasitol. 90 , 491–504. ( 10.1016/S0166-6851(97)00197-7) [DOI] [PubMed] [Google Scholar]
- 72. Kim T, Lara-Gonzalez P, Prevo B, Meitinger F, Cheerambathur DK, Oegema K, Desai A. 2017. Kinetochores accelerate or delay APC/C activation by directing Cdc20 to opposing fates. Genes Dev. 31 , 1089–1094. ( 10.1101/gad.302067.117) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Lara-Gonzalez P, Kim T, Oegema K, Corbett K, Desai A. 2021. A tripartite mechanism catalyzes Mad2-Cdc20 assembly at unattached kinetochores. Science 371 , 64–67. ( 10.1126/science.abc1424) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Piano V, Alex A, Stege P, Maffini S, Stoppiello GA, Huis in ’t Veld PJ, Vetter IR, Musacchio A. 2021. CDC20 assists its catalytic incorporation in the mitotic checkpoint complex. Science 371 , 67–71. ( 10.1126/science.abc1152) [DOI] [PubMed] [Google Scholar]
- 75. Vleugel M, Hoek TA, Tromer E, Sliedrecht T, Groenewold V, Omerzu M, Kops GJPL. 2015. Dissecting the roles of human BUB1 in the spindle assembly checkpoint. J. Cell Sci. 128 , 2975–2982. ( 10.1242/jcs.169821) [DOI] [PubMed] [Google Scholar]
- 76. Poon SK, Peacock L, Gibson W, Gull K, Kelly S. 2012. A modular and optimized single marker system for generating Trypanosoma brucei cell lines expressing T7 RNA polymerase and the tetracycline repressor. R. Soc. Open Biol. 2 , 110037. ( 10.1098/rsob.110037) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Brun R, Schönenberger M. 1979. Cultivation and in vitro cloning or procyclic culture forms of Trypanosoma brucei in a semi-defined medium. Acta Trop. 36 , 289–292. [PubMed] [Google Scholar]
- 78. Kelly S, et al. 2007. Functional genomics in Trypanosoma brucei: a collection of vectors for the expression of tagged proteins from endogenous and ectopic gene loci. Mol. Biochem. Parasitol. 154 , 103–109. ( 10.1016/j.molbiopara.2007.03.012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Dean S, Sunter J, Wheeler RJ, Hodkinson I, Gluenz E, Gull K. 2015. A toolkit enabling efficient, scalable and reproducible gene tagging in trypanosomatids. R. Soc. Open Biol. 5 , 140197. ( 10.1098/rsob.140197) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Ishii M, Akiyoshi B. 2020. Characterization of unconventional kinetochore kinases KKT10 and KKT19 in Trypanosoma brucei. J. Cell Sci. 133 , jcs240978. ( 10.1242/jcs.240978) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Landeira D, Navarro M. 2007. Nuclear repositioning of the VSG promoter during developmental silencing in Trypanosoma brucei. J. Cell Biol. 176 , 133–139. ( 10.1083/jcb.200607174) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Unnikrishnan A, Akiyoshi B, Biggins S, Tsukiyama T. 2012. An efficient purification system for native minichromosome from Saccharomyces cerevisiae. Methods Mol. Biol. 833 , 115–123. ( 10.1007/978-1-61779-477-3_8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Hughes CS, Moggridge S, Müller T, Sorensen PH, Morin GB, Krijgsveld J. 2019. Single-pot, solid-phase-enhanced sample preparation for proteomics experiments. Nat. Protoc. 14 , 68–85. ( 10.1038/s41596-018-0082-x) [DOI] [PubMed] [Google Scholar]
- 84. Deutsch EW, et al. 2023. The ProteomeXchange consortium at 10 years: 2023 update. Nucleic Acids Res. 51 , D1539–D1548. ( 10.1093/nar/gkac1040) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Perez-Riverol Y, et al. 2022. The PRIDE database resources in 2022: a hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res. 50 , D543–D552. ( 10.1093/nar/gkab1038) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Zhang X, Smits AH, van Tilburg GB, Ovaa H, Huber W, Vermeulen M. 2018. Proteome-wide identification of ubiquitin interactions using UbIA-MS. Nat. Protoc. 13 , 530–550. ( 10.1038/nprot.2017.147) [DOI] [PubMed] [Google Scholar]
- 87. Blighe K. 2023. Enhancedvolcano: publication-ready volcano plots with enhanced colouring and labeling. See https://github.com/kevinblighe/EnhancedVolcano.
- 88. McCoy AJ. 2017. Acknowledging errors: advanced molecular replacement with phaser. Methods Mol. Biol. 1607 , 421–453. ( 10.1007/978-1-4939-7000-1_18) [DOI] [PubMed] [Google Scholar]
- 89. Cowtan K. 2006. The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr. D 62 , 1002–1011. ( 10.1107/S0907444906022116) [DOI] [PubMed] [Google Scholar]
- 90. Emsley P, Lohkamp B, Scott WG, Cowtan K. 2010. Features and development of Coot. Acta Crystallogr. D 66 , 486–501. ( 10.1107/S0907444910007493) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Liebschner D, et al. 2019. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr. D 75 , 861–877. ( 10.1107/S2059798319011471) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Aslett M, et al. 2010. TriTrypDB: a functional genomic resource for the Trypanosomatidae. Nucleic Acids Res. 38 , D457–62. ( 10.1093/nar/gkp851) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Butenko A, et al. 2020. Evolution of metabolic capabilities and molecular features of diplonemids, kinetoplastids, and euglenids. BMC Biol. 18 , 23. ( 10.1186/s12915-020-0754-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Tikhonenkov DV, Gawryluk RMR, Mylnikov AP, Keeling PJ. 2021. First finding of free-living representatives of Prokinetoplastina and their nuclear and mitochondrial genomes. Sci. Rep. 11 , 2946. ( 10.1038/s41598-021-82369-z) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Eddy SR. 1998. Profile hidden Markov models. Bioinformatics 14 , 755–763. ( 10.1093/bioinformatics/14.9.755) [DOI] [PubMed] [Google Scholar]
- 96. Katoh K, Rozewicki J, Yamada KD. 2019. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 20 , 1160–1166. ( 10.1093/bib/bbx108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ. 2009. Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25 , 1189–1191. ( 10.1093/bioinformatics/btp033) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Madeira F, Pearce M, Tivey ARN, Basutkar P, Lee J, Edbali O, Madhusoodanan N, Kolesnikov A, Lopez R. 2022. Search and sequence analysis tools services from EMBL-EBI in 2022. Nucleic Acids Res. 50 , W276–W279. ( 10.1093/nar/gkac240) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Evans R, et al. 2022. Protein complex prediction with alphafold-multimer. bioRxiv. ( 10.1101/2021.10.04.463034) [DOI]
- 100. Jumper J, et al. 2021. Highly accurate protein structure prediction with AlphaFold. Nature 596 , 583–589. ( 10.1038/s41586-021-03819-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Mirdita M, Schütze K, Moriwaki Y, Heo L, Ovchinnikov S, Steinegger M. 2022. ColabFold: making protein folding accessible to all. Nat. Methods 19 , 679–682. ( 10.1038/s41592-022-01488-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Varadi M, et al. 2022. Alphafold protein structure database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 50 , D439–D444. ( 10.1093/nar/gkab1061) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Gabler F, Nam SZ, Till S, Mirdita M, Steinegger M, Söding J, Lupas AN, Alva V. 2020. Protein sequence analysis using the MPI bioinformatics toolkit. Curr. Protoc. Bioinform. 72 , e108. ( 10.1002/cpbi.108) [DOI] [PubMed] [Google Scholar]
- 104. de Potter B, Raas MWD, Seidl MF, Verrijzer CP, Snel B. 2023. Uncoupled evolution of the polycomb system and deep origin of non-canonical PRC1. Commun. Biol. 6 , 1144. ( 10.1038/s42003-023-05501-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Huerta-Cepas J, et al. 2019. eggNOG 5.0: a hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 47 , D309–D314. ( 10.1093/nar/gky1085) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Fu L, Niu B, Zhu Z, Wu S, Li W. 2012. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28 , 3150–3152. ( 10.1093/bioinformatics/bts565) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25 , 1972–1973. ( 10.1093/bioinformatics/btp348) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, Lanfear R. 2020. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 37 , 1530–1534. ( 10.1093/molbev/msaa015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods 14 , 587–589. ( 10.1038/nmeth.4285) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS. 2018. UFBoot2: improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 35 , 518–522. ( 10.1093/molbev/msx281) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Letunic I, Bork P. 2021. Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 49 , W293–W296. ( 10.1093/nar/gkab301) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Ballmer D, Carter W, van Hooff JJE, Tromer EC, Ishii M, Ludzia P, Akiyoshi B. 2024. Supplementary material from: Kinetoplastid kinetochore proteins KKT14–KKT15 are divergent Bub1/BubR1–Bub3 proteins. Figshare ( 10.6084/m9.figshare.c.7214471) [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All raw mass spectrometry files and the custom database file used in this study have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository [84,85] with the dataset identifier PXD047806. Protein coordinates have been deposited in the RCSB protein data bank with the accession number 8QOH.
Supplementary material is available online [112].