Abstract
Overexpression of the longevity gene Klotho prolongs lifespan, while its knockout shortens lifespan and impairs cognition via perturbation of myelination and synapse formation. However, comprehensive analysis of Klotho knockout effects on mammalian brain transcriptomics is lacking. Here, we report that Klotho knockout alters the levels of aging- and cognition related mRNAs, long non-coding RNAs, microRNAs and tRNA fragments. These include altered neuronal and glial regulators in murine models of aging and Alzheimer’s disease and in human Alzheimer’s disease post-mortem brains. We further demonstrate interaction of the knockout-elevated tRNA fragments with the spliceosome, possibly affecting RNA processing. Last, we present cell type-specific short RNA-seq datasets from FACS-sorted neurons and microglia of live human brain tissue demonstrating in-depth cell-type association of Klotho knockout-perturbed microRNAs. Together, our findings reveal multiple RNA transcripts in both neurons and glia from murine and human brain that are perturbed in Klotho deficiency and are aging- and neurodegeneration-related.
Subject terms: Molecular neuroscience, Small RNAs, Neural ageing, Microglia
Transcriptomic profiling shows that Klotho knockout perturbs brain short non-coding RNAs, such as microRNA and tRNA fragments, in both neurons and glia, that mimics the changes associated with Alzheimer’s disease and aging in humans and murine models.
Introduction
Age is a key risk factor for many neurodegenerative diseases1, and identifying transcripts distinguishing between healthy aging and age-associated cognitive decline remains a major research goal. The Klotho protein exhibits remarkable properties of lifespan regulation2, which has attracted significant interest in aging research for over two decades. Klotho-deficient mice show premature aging symptoms, including cognitive decline3, whereas Klotho overexpression improves amyloid-β clearance and cognition in one Alzheimer’s disease (AD) mouse model4, and long-term potentiation (LTP) and behavior in another5. Klotho excess also improved remyelination, decreased motor neuron death and lowered neuroinflammation in a mouse model of multiple sclerosis6,7. Furthermore, injected Klotho enhances cognition in aged rhesus macaques8, while elevated levels of functional Klotho protein in humans is associated with both longevity9 and lower AD risk10. On the contrary, human AD patients show lower Klotho levels in the cerebrospinal fluid (CSF) than age-matched controls11. Thus, the neuroprotective effects of Klotho are evolutionary conserved and functional in the mammalian brain.
While Klotho’s involvement is well recognized in multiple brain processes, including myelination and synaptic plasticity5,12, the impact of Klotho deficiency on both coding and non-coding RNA transcriptomes is incompletely understood. To address this issue, we compared short and long RNA-sequencing profiles of brain tissues from a Klotho knockout (KO) murine model to those of wild-type (WT) mice (n = 5 in each group), and identified changes in numerous RNA fragments. We sought correlations of the observed transcriptomic changes with reported aging and AD alterations, and pursued the cell type-specificity of the observed differences. Among short RNAs, we focused on microRNAs (miRs) and transfer RNA fragments (tRFs), due to their suggested involvement in brain aging and neurodegeneration13–15. We further established that the tRF upregulated by Klotho knockout interacts with proteins involved in major RNA processing pathways, such as splicing and mRNA decay, providing the first functional evidence for the involvement of tRFs in Klotho-associated cognitive decline. Finally, we developed a novel cell type-specific short RNA dataset from live human brain tissues and used it to demonstrate that microRNAs changed by Klotho KO represent both neuron- and microglia-specific responses.
Results
Klotho knock-out induces significant perturbations in mRNA and long non-coding RNA profiles
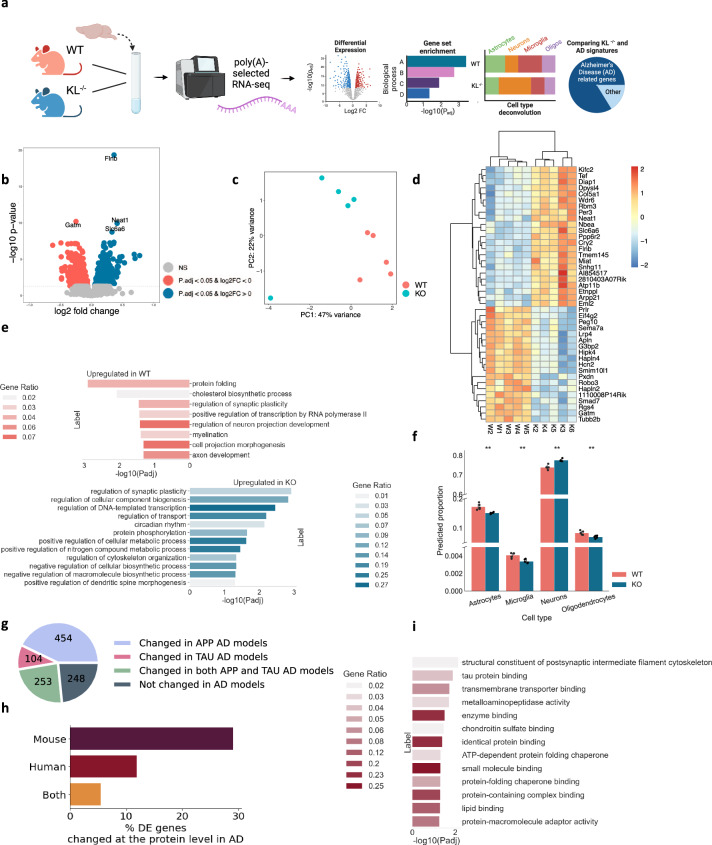
Klotho deficiency perturbed 10% (1,059 out of 10,334) of all of the expressed polyadenylated RNA transcripts in Klotho KO murine brains compared to WT (padj < 0.05; Fig. 1a,b, Supplementary Data 1). Klotho mRNA itself was predictably downregulated in the brains of KO mice (padj«0.001; Supplementary Fig. 1). Other differentially expressed (DE) transcripts showed smaller albeit significant changes, possibly representing the averaged expression differences between diverse cell types and brain regions in the analyzed samples. Briefly, Klotho deficiency led to impaired functioning of multiple cellular compartments, for example, by modulating filamin B (Flnb), which is involved in cytoskeleton formation of developing neurons16; Neat1, a biomarker of nuclear paraspeckles17; Slc6a6, involved in synaptic GABA uptake18; and Gatm, a mitochondrial enzyme participating in creatinine synthesis19. Together, those differences clearly distinguished Klotho-knockout from wild-type samples in principal component analysis (PCA) (Fig. 1c).
Fig. 1. Klotho KO induces numerous long RNA transcriptome changes.
a Graphical representation of the analysis shown in Fig. 1. Created with BioRender.com. b Volcano plot of polyadenylated RNAs DE in murine Klotho-knockout (n = 5) compared to wildtype (n = 5) brains; significantly upregulated transcripts in blue, significantly downregulated transcripts in pink; padj < =0.05. c PCA of polyadenylated RNA reads colored by genotype. d Heatmap showing normalized and centered counts of the most significantly altered transcripts (padj <= 5e-6; colormap - z-score of normalized count levels, centered to the mean). e GO annotation of biological processes enriched in up- and down-regulated transcripts (blue and pink, respectively) under Klotho-knockout. f AutoGeneS prediction of the cell type proportions in WT and KO profiles based on the scRNA-seq atlas21; **: pval < 0.01, ns: non-significant. g Pie chart proportions of DE genes in AD studies26. h Bar plot showing the percent of DE mRNAs encoding proteins that are altered in a mouse model AD study31, in a compilation of human AD proteomic studies32 and in both. i. GO annotation of molecular functions enriched in protein-coding transcripts whose associated proteins were altered in a compilation of human AD proteomic studies32. The error bands denote 95% confidence intervals.
Using the Gene Ontology (GO) resource20 we identified the biological pathways of the differentially expressed (DE) genes. Klotho KO reduced protein folding and phosphorylation processes, indicating disrupted protein metabolism and exacerbated protein aggregation (Fig. 1d,e). Myelination and neuron projection development were also decreased in Klotho KO, while genes involved in cytoskeleton organization, transport, and dendritic spine morphogenesis were elevated, reflecting general morphological changes in neuronal structure (Fig. 1d,e). Considering that the analyzed bulk RNA samples represent mixtures of multiple brain cell types, we further deconvoluted the bulk RNA profiles using a single-cell RNA-seq mouse brain atlas21. An AutoGeneS package22-based cellular heterogeneity search indicated that Klotho KO caused a signaling decline in all glial cell types (Fig. 1f).
Out of 10,334 expressed polyadenylated RNA transcripts, 70 belonged to the long non-coding RNA (lncRNA) family (Supplementary Data 1). The change in their levels caused by Klotho KO was sufficient to separate the samples by genotype using PCA analysis (Supplementary Fig. 2a). Furthermore, the 14 differentially expressed lncRNAs included Neat1, Miat and Meg3, all upregulated in Klotho KO and involved in neurogenesis, neuron apoptosis regulation and neural inflammation23–25 (padj « 0.05; Supplementary Fig. 2b).
The transcriptional perturbation caused by Klotho KO further mimicked changes seen in AD-mouse models. Meta-analysis of the AD-associated transcriptome26 (Materials and Methods) revealed that the DE transcripts identified in the Klotho KO profiles highly overlapped with the mouse model AD-altered transcripts (Fig. 1g). Briefly, 811 out of the 1059 DE genes in Klotho KO ( ~ 77%) were altered in at least one of the mouse models, with 454 modulated in models with mutated amyloid precursor protein (APP; 43%), 104 – in models with mutated microtubule-associated protein tau (MAPT; 10%), and 253 in both (24%). The overlapping genes included known AD markers which changed in more than 7 different studies26, including the lipid metabolism genes Abca1 and Apoe27; complement immune system members C4b, Ctsd, C1qb and C1qa28, and the astrocytic markers Gfap and Aqp429 (Supplementary Data 1). Thus, the transcriptional signature of murine Klotho KO shared basic features with the transcriptomic profiles associated with diverse mouse AD models.
Notably, the observed links of altered RNA levels between Klotho KO and the AD-associated pathology are insufficient to suggest similarity between these phenotypes, since the levels of protein and their coding transcripts are not fully correlated30. Hence, we next evaluated the degree to which the mRNA changes induced by Klotho knockout are mirrored in the AD-associated proteome. Specifically, we calculated the proportion of the protein-coding transcripts, which were DE in Klotho KO and featured altered protein levels in a proteomic dataset from the mouse 5XFAD model31 and in the compilation of proteomic datasets from human samples32 (Methods). We found that 29% of the Klotho-DE transcripts encoded a protein that changed in a mouse 5XFAD model, 12% encoded a protein that changed in human AD proteomic studies, and 5.5% encoded a protein that changed in both (Fig. 1h; Supplementary Data 1). Notably, the DE mRNAs encoding for proteins with altered levels in human AD studies were statistically enriched in postsynaptic cytoskeleton organization and in tau protein binding, exposing the major common molecular cascades shared between the AD and the Klotho KO phenotypes (Fig. 1i).
Klotho knock-out perturbs brain microRNA (miR) signatures
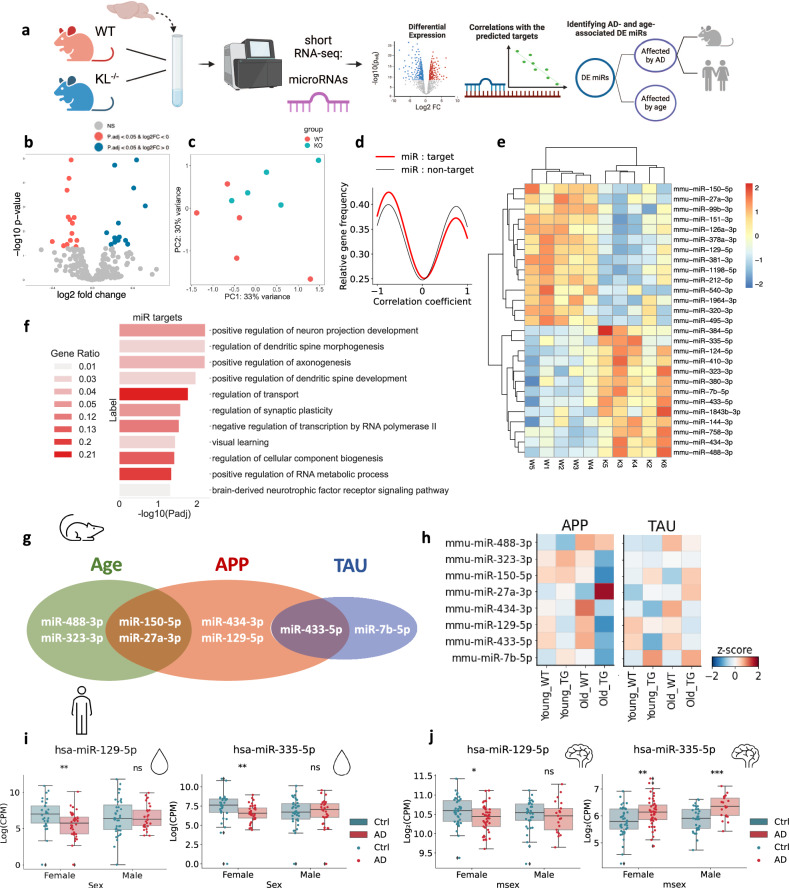
Apart from mRNAs, Klotho KO altered the levels of 27 miRs (Fig. 2a,b; Supplementary Data 2), whose signatures separated the samples on the PCA plane based on their genotype (Fig. 2c). Moreover, unsupervised clustering based on the profiles of the DE miRs reliably separated the samples to genotype-specific groups (Fig. 2d). Altered miRs include mmu-miR-212-5p, linked to longevity via regulating Sirt1 and cooperating with the AD-depleted acetylcholinesterase-targeting mmu-miR-132-5p33 (Fig. 2d). Identifying DE miRs targets using miRNet34 revealed the experimentally validated targets whose levels were repressed by the corresponding miRs in similar experimental systems35 (Materials and Methods; Supplementary Data 3). Further calculation of Pearson correlation coefficients between every pair of a DE miR and its DE-validated mRNA target enabled comparing the correlation coefficients between the miR – target pairs and miR – non-target pairs36. Intriguingly, miR-target profiles were more negatively correlated than miR- non-target pairs (Fig. 2e), demonstrating that some of the vast mRNA perturbations following Klotho KO might be explained by miR regulation. Using the Gene Ontology resource, we have further identified DE miR target genes involved in major neuronal pathways, such as axonogenesis, dendritic spine development, and regulation of synaptic plasticity (Fig. 2f). Further supporting active miR involvement in the Klotho-KO-activated downstream processes, we found Klotho KO -elevated levels of the microprocessor-associated protein DDX5, which is actively involved in miR processing37, and is targeted by four different downregulated miRs (Supplementary Data 3).
Fig. 2. Klotho depletion perturbs miR signatures, resembling AD and aging-associated changes.
a Graphical representation of the analysis shown in Fig. 2. Created with BioRender.com. b Volcano plot of miRs DE in Klotho-knockout (n = 5) compared to wildtype (n = 5) mice, significantly upregulated transcripts in blue, significantly downregulated transcripts in pink, padj < =0.05. c PCA based on miR counts and colored by genotype. d Gaussian kernel density estimates of Pearson correlation coefficients between klotho-affected miRs and their targets (red), and klotho-miRs and DE mRNAs which are not their targets (black). X axis: Pearson correlation coefficient. Y axis: relative frequency of miR-mRNA pairs with the corresponding coefficient. e Heatmap showing normalized and centered counts of DE miRs (colormap: z-score of the normalized counts, centered to the mean). f Gene Ontology annotation of miR targets from enriched biological processes. g Venn diagram of DE miRs that co-change in the murine hippocampus with age and/or in one of the AD pathology models38 (APP - APPswe/PS1L166P; TAU - THY-Tau22). h Scaled heatmap of miR alterations from (G) across age and genotype conditions of both APP and TAU mouse models38. i Log2(CPM) counts of hsa-miR-129-5p and hsa-miR-335-5p in CSF from AD patients and cognitively healthy controls, divided by sex41 (**: padj < 0.01). j Log2(CPM) counts of hsa-miR-129-5p and hsa-miR-335-5p in the nucleus accumbens of AD patients (cogdx=4), compared to cognitively healthy individuals (cogdx=1), divided by sex15 (**: padj < 0.01). The error bands denote 95% confidence intervals.
To determine out if Klotho DE miRs are also altered in aging or AD pathology, we analyzed short RNA-seq data from AD mouse models, including young and aged (4 and 10 months) wildtype and transgenic mice carrying either APP (APPswe/PS1L166P) or TAU (THY-Tau22) mutations38. For each strain, we compared miR profiles from wildtype and age-matched transgenic mice, and between ages within the same genotype (Supplementary Data 4). This identified Klotho-associated miRs whose levels were affected by age (e. g. miR-488-3p; miR-323-3p), by both age and APP pathology (miR-150-5p, miR-27a-3p), by APP pathology alone (miR-434-3p, miR-129-5p), by TAU pathology alone (miR-7b-5p), or by both APP and TAU pathologies (miR-433-5p; Fig. 2g,h), together reflecting altered regulation of cognition.
Next, we sought putative human links between DE miRs, cognition, and AD neuropathology. Intriguingly, two miRs that were DE in Klotho KO, miR-129-5p, which is associated with the Fragile X syndrome39 and the muscarinic receptors-targeting miR-335-5p40, were reduced in the CSF of female, but not male AD patients41 (Fig. 2i). Considering our recent findings of cognition-related female-specific AD declines in tRF regulators of cholinergic mRNAs26, this suggests links between Klotho-KO-induced transcriptomic perturbations and sex differences in dementia. Indeed, both miR-129-5p and miR-335-5p are also altered in the nucleus accumbens of female AD patients, compared to controls with no cognitive impairment15 (Fig. 2j; Supplementary Data 5), and with the same direction of change in human AD and murine Klotho KO brains. These down-regulated transcripts and processes hence emerge as co-related to the cognitive impairments shared by Klotho KO and AD.
Klotho knockout alters the levels of short tRNA fragments (tRFs)
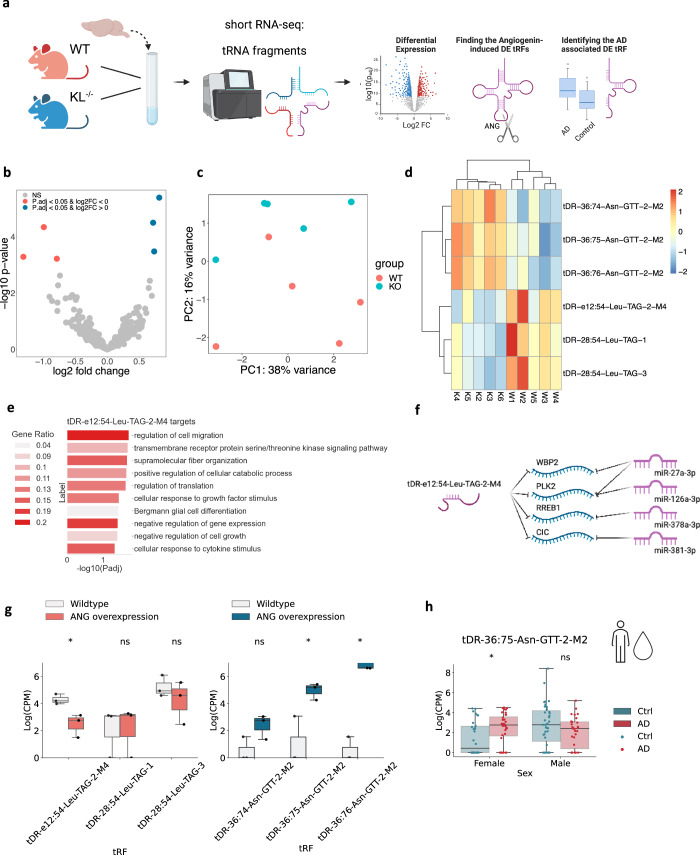
Apart from miRs, we also identified Klotho-associated expression changes in another short non-coding RNA family – tRFs11 (Fig. 3a). Six nuclear genome-originated tRFs were altered, three upregulated and three downregulated (Fig. 3b; Supplementary Data 6). Strikingly, all three upregulated tRFs were 3’ halves originated from Asparagine-GTT tRNA, representing matching 3’-end sequences with single nucleotide differences, whereas the three downregulated tRFs were Leucine-TAG tRNA-originated i-tRFs (Fig. 3b; Supplementary Data 6). These subtle differences allowed PCA to separate the samples (Fig. 3c), and unsupervised hierarchical clustering based on the DE tRFs recognized the knockout and wildtype genotypes as two distinct clusters (Fig. 3d).
Fig. 3. Klotho knockout alters tRF levels sex-dependently.
a Graphical representation of the analysis shown in Fig. 3. Created with BioRender.com. b Volcano plot of DE tRFs in murine Klotho-knockout (n = 5) compared to wildtype (n = 5) brains; significantly upregulated transcripts in blue, significantly downregulated transcripts in pink; padj < =0.05. c PCA of tRF counts colored by genotype. d Heatmap showing normalized and centered counts of altered tRFs (colormap: z-score of the normalized counts, centered to the mean). e Gene Ontology annotation of biological processes enriched in tDR-36:75-Asn-GTT-2-M2 targets, and negatively correlated with this tRF in Klotho KO data. f Schematically illustrated common targets of tDR-36:75-Asn-GTT-2-M2 and miRs. g Log2(CPM) counts of altered tRFs in HEK293 cells under angiogenin overexpression compared to wildtype; left panel, tRFs decreased, right panel tRFs increased under Klotho KO;43.*: padj < 0.1. h Log (CPM) counts of tDR-36:75-Asn-GTT-2-M2 in CSF of AD patients and cognitively healthy controls, divided by sex41,*: padj < 0.05. The error bands denote 95% confidence intervals.
Of all the DE tRFs, only tDR-e12:54-Leu-TAG-2-M4, downregulated in Klotho KO, was short enough to predict miR-like activity14. The tRFTar tool42 identified 81 predicted mRNA targets that were negatively correlated with tDR-e12:54-Leu-TAG-2-M4 (Supplementary Data 7). Notably, these tRF targets were enriched in biological processes distinct from those enriched by the Klotho-altered miR targets, and included cell migration, intracellular signaling, and translation regulation pathways (Fig. 3e). Moreover, tDR-e12:54-Leu-TAG-2-M4 shared 4 targets with downregulated miRs (Fig. 3f), possibly reflecting cooperation of these short non-coding RNA families in knockout-induced transcriptional changes.
The transcriptomic profiles further revealed slightly upregulated mRNA levels of the nuclease angiogenin, which is linked to stress-related tRF production (Supplementary Fig. 3b)43. To determine whether tRFs elevated by Klotho knockout were stress-induced products of angiogenin43,44, we sought parallel tRF changes in relevant model systems. Notably, all Klotho KO upregulated tRFs were found in angiogenin-overexpressing HEK293 cells, and one of the downregulated tRFs, tDR-e12:54-Leu-TAG-2-M4, decreases in U2OS cells with angiogenin overexpression (Fig. 3g) and increases in angiogenin knockout43 (Supplementary Fig. 3a). Thus, the tRF signature following Klotho knockout might represent a stress-induced and angiogenin-dependent response.
Comparing the identified tRF changes under Klotho knockout to those of AD brains, identified tDR-36:75-Asn-GTT-2-M2 as both upregulated in Klotho knockout and in the CSF of female AD patients compared to cognitively unimpaired controls41 (Fig. 3h). Mimicking the sex-specific miR changes and decline of tRFs in the nucleus accumbens from AD female brains15, tDR-36:75-Asn-GTT-2-M2 levels remained unchanged in male patient brains. In summary, both Klotho knockout-induced brain miR and tRF changes correspond to their parallel changes in AD pathology.
tDR-36:74-Asn-GTT-2-M2 interacts with proteins involved in RNA splicing and degradation
We further aimed to establish a functional role of the tRFs upregulated in Klotho KO, which are of particular interest for several reasons. First, these tRFs only differ in 1-2 nucleotides at the 3’ end, essentially representing the same sequence, which constitutes a highly specific response and may suggest a functional impact for the shared sequence. Second, these tRFs belong to the family of stress-related tRNA halves14 and might be involved in angiogenin-induced stress response, caused by Klotho KO (Fig. 3g; Supplementary Fig. 3a). Finally, tDR-36:75-Asn-GTT-2-M2 is elevated in the CSF of female AD patients (Fig. 3h), providing additional evidence for functional role in neurodegeneration and cognitive decline.
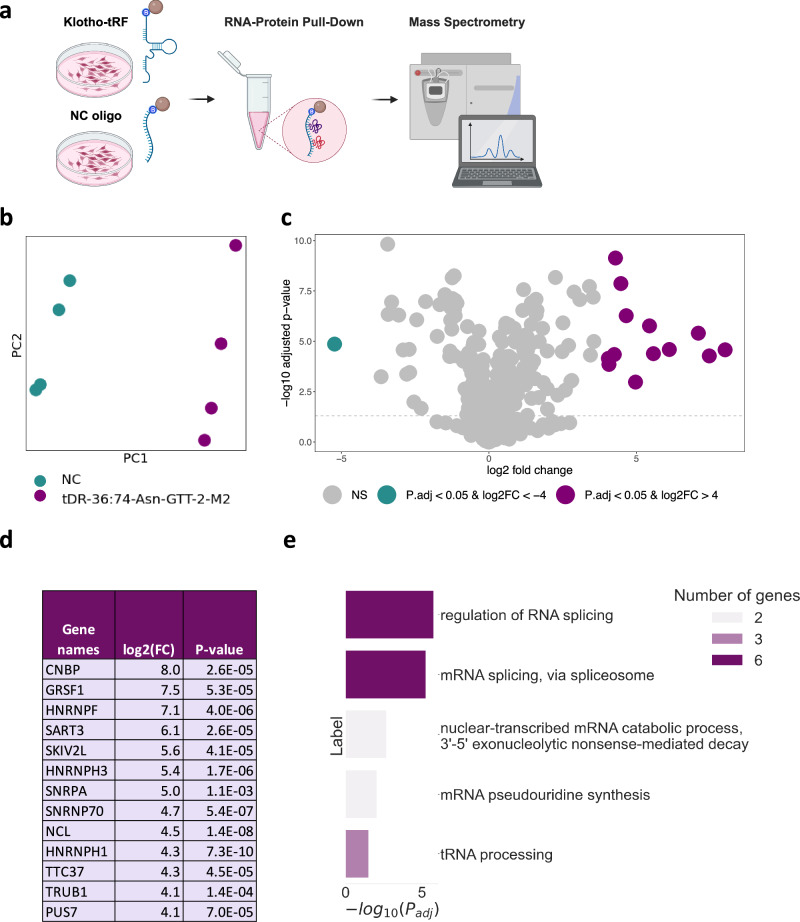
Over the past few years, tRNA halves are continuously reported to modulate crucial intracellular pathways, such as translation and apoptosis, by direct interaction with proteins14,45,46. Therefore, we sought proteins interacting with the tRFs of interest. To this aim, we established a pull-down assay in which we incubated a lysate of SH-SY5Y human neuroblastoma cells with a biotinylated sequence of the shortest tRF upregulated in Klotho KO, tDR-36:74-Asn-GTT-2-M2, representing the shared sequence between the three upregulated fragments. Importantly, this tRF is conserved between the human and mouse genomes, which enabled us to perform the experiment using a human cell line. The proteins bound to the sequence were then isolated using streptavidin-coated magnetic beads and identified by mass spectrometry (MS) (Fig. 4a). Close to 100 proteins were found to be significantly enriched in the tDR-36:74-Asn-GTT-2-M2 samples, compared to the samples treated with a negative control (NC) oligo, allowing for a clear separation on the PCA plane (Fig. 4b; Supplementary Data 8). We further focused our analysis on the 13 proteins which were most enriched in tDR-36:74-Asn-GTT-2-M2 compared to NC samples, using a fold change (FC) cutoff of log2(FC)≥4, to minimize analysis of proteins not markedly changed between the tRF and the NC (Fig. 4c,d; Materials and Methods).
Fig. 4. Pull-down assay revealing that tDR-36:74-Asn-GTT-2-M2 binds multiple proteins involved in splicing, mRNA decay and pseudouridylation.
a Graphical representation of the pull-down experiment: SH-SY5Y cells were plated, harvested after four days, and incubated with magnetic streptavidin beads to which biotinylated tDR-36:74-Asn-GTT-2-M2 (Klotho-tRF) or biotinylated negative control (NC) RNA-oligonucleotides were bound. After wash steps the beads were submitted to mass spectrometry and the proteins bound to each oligonucleotide were identified. Created with BioRender.com. b PCA showing separation of the proteins bound to the tDR-36:74-Asn-GTT-2-M2 (purple) vs. those bound to the NC oligo (green), 4 samples per group. c Volcano plot showing the differentially expressed proteins in biotinylated tDR-36:74-Asn-GTT-2-M2 (n = 4) vs biotinylated NC pulldown samples (n = 4). Proteins significantly upregulated with log2FC > =4 are colored in purple, proteins sinificantly downregulated with log2FC > =-4 are colored in green. The statistical analysis in was performed using the Perseus package. d List of the 13 proteins enriched by log2FC > =4 (16-fold) in tDR-36:74-Asn-GTT-2-M2 vs. NC samples. e Gene Ontology annotation of the biological processes enriched within the differentially expressed proteins in panel D.
Intriguingly, these 13 proteins represent a highly interconnected molecular network associated with RNA processing. The most enriched biological process among these proteins is regulation of RNA splicing (Fig. 4e). GRSF1, HNRNPF, HNRNPH3, SNRNP70, NCL and HNRNPH1 are annotated as parts of the spliceosome complex by the GO resource20, along with SART3 and SNRPA that are reported to be involved in RNA splicing as well47,48. Thus, tDR-36:74-Asn-GTT-2-M2 might be a potential regulator of alternative splicing, which represents a hallmark of aging and neurodegeneration49–51.
Another highly relevant pathway, represented by PUS7 and TRUB1, is RNA pseudouridylation52,53 (Fig. 4e). G-rich pseudouridylated tRNA-fragments were shown to control translation, eventually determining the fate of the developing hematopoietic stem cells54 and predicting leukaemic progression55. Although this effect has been demonstrated for 5’ halves54,55, our current results indicate that 3’ halves, such as tDR-36:74-Asn-GTT-2-M2, might be modified as well, possibly exerting a parallel downstream function. Finally, SKIV2L and TTC37 represent components of the SKI complex, which is involved in 3’ to 5’ mRNA decay56.
Notably, PUS7, CNBP, GRSF1, and HNRNPF specifically bind to G-rich oligonucleotide sequences or G-quadruplexes54,55,57–59, with CNBP promoting their formation57. This is particularly relevant, since G-rich tRNA halves were demonstrated to form G-quadruplexes, thereby inhibiting translation and promoting neuroprotection via the formation of stress granules60. Guanine is indeed the most frequent base in the tDR-36:74-Asn-GTT-2-M2 sequence, consisting of 33% G, 28% C, 23% A, and 16% T (Materials and Methods). Therefore, we hypothesize that tDR-36:74-Asn-GTT-2-M2 and its analogous fragments, highly expressed under Klotho KO, have the potential to form G-quadruplexes. This secondary structure may facilitate their molecular roles, thus affecting splicing, mRNA decay or translation60.
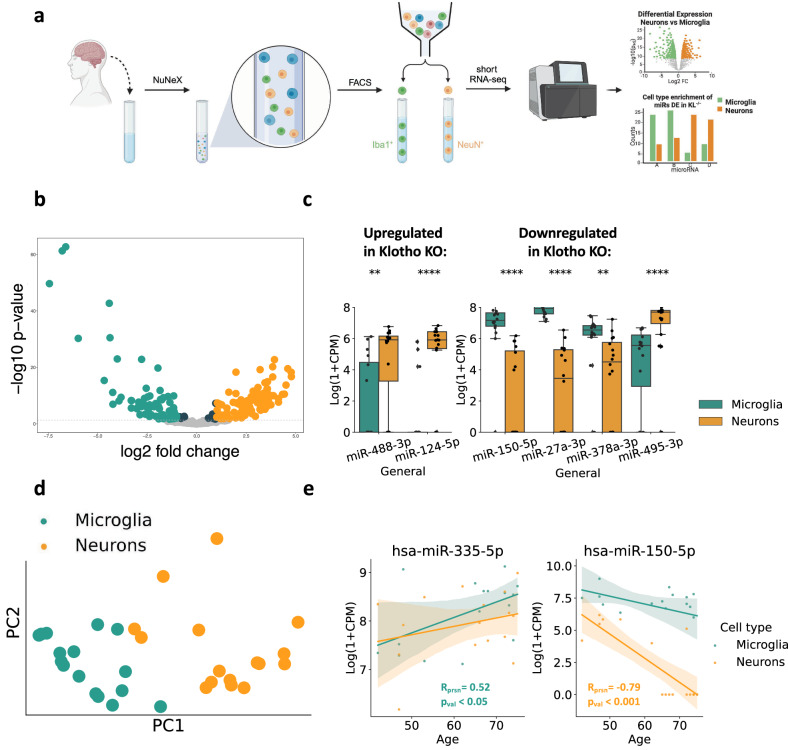
Klotho-miR profiles reveal neuronal and microglial signatures
Deconvoluting the bulk mRNA signal to cell type-specific signatures showed that Klotho KO led to reduced numbers of glial transcripts (Fig. 1f). However, deconvoluting the affected miRs, which are highly cell type-specific61, was challenging due to the lack of reference datasets with cell type resolution. Therefore, we developed novel neuron- and microglia-specific short RNA-seq datasets from live human brain samples resected during surgeries for the removal of non-infiltrative brain tumors. To assess the non-tumor origin of the cortical tissues, we excluded patients with infiltrative brain pathologies such as meningitis, glioma, or diffuse cerebral inflammation, and collected the tissues as far from the tumor as possible. We then validated that the acquired miR profiles differ from specimens of meningioma62, but rather resemble postmortem bulk miRNA profiles from various human brain regions63 (Supplementary Fig. 4a-c).
We collected 16 live human brain samples (6 females, 10 males; 42-75 years old) and recorded a wide range of patients’ additional medical parameters that could be relevant for future analyses (Supplementary Data 9). Following our inhouse NuNeX protocol64, the collected samples were exposed to a brief formalin fixation and separated into single nuclei suspensions, which retained some associated cytoplasm. Staining with neuron-specific (NeuN) and microglia-specific (Iba1) fluorescent markers followed by fluorescence-activated cell sorting (FACS; Supplementary Fig. 5a-e) yielded 32 neuron- and microglia-specific populations and their short RNA-seq profiles (Fig. 5a; Supplementary Datas 10, 11). Since Iba1 is also expressed by macrophages that infiltrate the brain65, we validated the microglial identity of the sorted Iba1+ populations using qPCR measurement of specific microglial RNA markers, such as P2y12, Sall1, and Tmem119 (Supplementary Fig. 6).
Fig. 5. Klotho KO affects miR levels in both neurons and microglia.
a Graphical representation of the pipeline used to produce bulk cell type-specific short transcriptome datasets of live brain cells. Using the NuNeX protocol64, samples extracted during neurosurgeries were broken down to single nuclei surrounded by thin cytoplasm layers. Nuclei stained with microglia- (Iba1) and neuron- (NeuN) specific fluorescent markers were FACS-sorted and short RNA-sequenced. Created with BioRender.com. b Volcano plot showing altered level miRs enriched in neurons (n = 16) in orange or microglia (n = 16) in green. c 6 miRS DE in Klotho KO were also enriched in one of the cell types. Left panel: miRs upregulated in Klotho KO; Right panel: miRs downregulated in Klotho KO, **: padj < =0.01; ****: padj < =0.0001. d PCA of the cell type-specific small transcripts (7 miRs DE in Klotho KO, colored by cell type). e Linear correlation of donor age with log2(CPM) count levels of hsa-miR-335-5p (significantly correlated in microglia) and hsa-miR-150-5p (significantly correlated in neurons), colored by cell type as in panel D. The error bands denote 95% confidence intervals.
Intriguingly, our novel short RNA-seq datasets revealed highly distinct miR profiles between live human brain neurons and microglia (Fig. 5b). Using the DESEQ2 package66, we found that 57% of the expressed miRs (173 of 303) were enriched in one of these cell types (Supplementary Data 10). Among them, 69 and 91 were upregulated in microglia or neurons with a log-fold change higher than 1, yielding highly valuable resources for deconvolution of bulk brain-derived short RNA profiles. Furthermore, the miRBase microRNA annotation database features 1830 mature human miRs and 1650 mature murine miRs of mouse origin, 493 of which are conserved and represent human-murine orthologs, which enables usage of our cell-type resolved human dataset for deconvoluting bulk data from murine tissues as well67.
Of the 27 DE miRs in murine Klotho KO, 7 were found in our newly produced cell type-specific human dataset, with 6 of those selectively enriched in one of those cell types (Fig. 5c; Supplementary Data 10). Parallel to the decline in glial mRNA transcripts under Klotho KO, 3 out of 4 miRs decreased in Klotho KO were enriched microglia from live human brain samples (Fig. 5c). In contrast, miRs upregulated in Klotho KO were enriched in human neurons from live human brain samples, mimicking the elevated neuron-specific mRNA transcripts in our mRNA-based deconvolution. Further, the cell type-dependent expression of the 7 DE miRs in Klotho KO sufficed to separate microglial and neuronal samples, showing a strong cell type-specific signal in the Klotho KO miR profiles (Fig. 5d) with shared cell type specificity of the DE miRs seen between the sexes (Supplementary Fig. 7a-c).
Notably, two DE miRs in Klotho KO showed cell type-dependent age correlation (Fig. 5e) The cholinergic miR-335-5p, which targets the muscarinic receptors and has been proposed to limit the cholinergic blockade of inflammation40,68, was upregulated with age in human brain microglia, murine Klotho KO and AD patients’ CSF and nucleus accumbens (Fig. 2i,j). In contrast, miR-150-5p, which targets the autism-associated Foxp169 transcript, declined with age in live human neurons, Klotho KO and by both age and APP pathology in AD mice models (Fig. 2g, h). Thus, our findings further strengthened the notion of miR-335-5p and miR-150-5p contributions to aging-related processes.
Aligning our neuron- and microglia-specific short RNA datasets to a tRF database (Materials and Methods) produced a novel resource of cell type-specific tRFs from live human brains. Unlike miRs, there were many more tRFs elevated in neurons than in microglia (Supplementary Fig. 8a). While none of the tRFs DE in Klotho KO were expressed in human neurons or microglia, other fragments originating from the same tRNAs exhibited a trend of elevation in neurons as compared to microglia (Supplementary Fig. 8b), possibly indicating that tRF signaling is generally more potent in neurons than in microglia. The neuron-specific tRF dataset is openly available (Supplementary Data 11).
Discussion
Klotho is an anti-aging gene with promising therapeutic potential70. To seek mechanisms by which Klotho exerts its CNS effects, we compared the RNA profiles of murine Klotho-deficient brains with those of healthy controls. This revealed a major Klotho knockout-induced transcriptional dysregulation affecting multiple cellular compartments and brain cell types. Importantly, many Klotho KO-altered mRNA transcripts and their protein products were also changed in brain aging and AD. Remarkably, short non-coding RNA transcripts, affected by Klotho KO, also exhibit parallel changes in aging and AD, reflecting an orchestrated transcriptional response shared by Klotho KO and aging-related cognitive decline.
While the potential involvement of tRFs in major molecular pathways has been established, their role in neurodegenerative diseases and cognitive decline is incompletely understood. We provide the first experimental evidence of the functional involvement of tRFs in Klotho-related cognitive decline. Specifically, Klotho KO elevated the expression of tDR-36:74-Asn-GTT-2-M2, which we show to bind numerous spliceosome components and major parts of the SKI complex involved in mRNA decay. In addition, our results hint that this tRF may be subjected to pseudouridylation and thereby control translation. Further experimental validation is required to establish the functional impact of the interaction between tDR-36:74-Asn-GTT-2-M2 and the reported pathways, as well as the role of its pseudouridylation71.
Lastly, our study substantially contributes to the understanding of the cell type specificity of brain-derived short non-coding RNAs. The deconvolution analysis of Klotho KO-altered mRNA profiles using a single-cell RNA-seq atlas of the murine brain21,22 showed that Klotho knockout causes a slight decline in glial mRNA transcripts. By generating a novel cell type-resolved short RNA dataset from live human brain tissues, we managed to support this finding at the level of microRNA, revealing that Klotho KO led to reduced levels of microglia-enriched miRs while elevating the levels of neuronal miRs, thus providing a link to potential mechanisms of action explaining at least part of its aging-related roles. Our novel cell-type resolved dataset, exhibiting neuron- and microglia-specific expression profiles of miRs and tRFs, provides a key resource for aging researchers with a focus on Klotho-associated brain processes.
This study has several potential limitations. First, our discovery of overlapping molecular signatures between Klotho KO, human brain aging, and AD is correlative and requires experimental validation. Furthermore, our novel cell type-specific short RNA-seq dataset from live human brain tissues is prone to bias by the pathology effect. We have shown that the extracted profiles are closer to those of healthy postmortem brains than to meningioma samples (Supplementary Fig. 3a,b), similar to multiple previous studies on live human brain tissue, which suffered from the same limitation72–74. However, we cannot fully rule out the possibility that the extracted neuronal and microglial profiles are affected by the tumor. Finally, the NuNeX protocol yields mainly nuclear-enriched RNA profiles, introducing a bias into the deconvolution analysis and possibly omitting the signal from transcripts abundant in other cellular compartments such as synapses.
In summary, while the major phenotypic effect of Klotho deficiency on premature aging and dementia has already been established, our results contribute to the study of Klotho-associated aging processes by thoroughly describing the transcriptomic changes occurring in Klotho knock-out and providing the first notes referring to its impact on tRFs-affected brain proteins and their related pathways. Overall, the data presented here indicates that physiologically normal Klotho levels are necessary for white and gray matter development and maintenance throughout life. Since Klotho levels decrease with age, augmenting Klotho protein expression either by enhancing its endogenous expression or by delivering exogenous genetic material, could be beneficial in neurodegenerative and other age-related diseases. We believe that by exposing the transcriptional network perturbation caused by the deficiency of Klotho, our work will accelerate future drug discovery targeting aging-related processes.
Methods
RNA extraction from Klotho KO murine brains
Klotho knock-out (KL-/-) mice were generated as described previously75,76, by crossing KL+/- mice carrying an insert mutation at 5’ promoter region of the KL gene. WT and KL-/- female mice, n = 5 in each group, were sacrificed at 6 weeks of age by cervical dislocation and the whole brain removed and snap-frozen in liquid nitrogen before storage at -80°C. Brains were homogenized in QIAzol (Qiagen, 217004, kit component) using a Polytron PT 3000 (Kinematica) and stored at -80°C. RNA was extracted from a 700 µL aliquot of each homogenate using the miRNeasy Mini Kit (Qiagen, 217004) according to the manufacturer’s protocol. RNA concentration was determined (NanoDrop 2000, Thermo Scientific) and RIN was measured (Bioanalyzer 6000, Agilent), with all samples ranging from 8.3 to 8.9.
RNA sequencing and alignment of Klotho KO murine brain-derived RNA
All samples underwent small RNA-sequencing and poly(A)-selected long RNA-sequencing. For small RNA, libraries were constructed from 1000 ng total RNA (NEBNext Multiplex Small RNA library prep set for Illumina, New England Biolabs, NEB-E7560S), and the small RNA fraction sequenced on the NextSeq 500 System (Illumina) at the Center for Genomic Technologies Facility, the Hebrew University of Jerusalem. For poly(A)-selected RNA, libraries were constructed from 1000 ng total RNA (KAPA Stranded mRNA-Seq Kit, Kapa Biosystems, KR0960–v5.17), and the RNA sequenced as above. Small RNA was aligned to miRBase using miRexpress77 and to the tRNA transcriptome using the MINTmap78. Long RNA was aligned to the Mus Musculus reference genome (GRCm38) using STAR79. The nomenclature of tRFs was based on the recently published standardized ontology80.
Cell culture for tRF pull-down assay
The human-derived neuroblastoma line SH-SY5Y (ATCC, CRL-2266) was grown in standard conditions at 37°C and 5% CO2, in EMEM:F12 (Merck, M5650 and N4888, respectively). All media were supplemented with FCS (10% final concentration, Sartorius, 04-127), L-glutamine (2 mM final concentration, Sartorius, 03-020) and Penicillin-Streptomycin-Amphotericin (100 units/mL, 0.10 mg/mL, 0.25 µg/mL, final concentrations, respectively, Sartorius, 03-033). Cells were mycoplasma-free (Mycoblue® Mycoplasma Detection Kit, Vazyme, D101).
RNA-Protein pull-down assay
The Pierce™ Magnetic RNA-Protein Pull-Down Kit (Thermo Fisher Scientific, 20164) was used to identify proteins binding to tDR-36:74-Asn-GTT-2-M2. Cells were seeded in 100 ×15 mm plates (Thermo Fisher Scientific, 150350) at 10-12×106 cells/plate and harvested four days later in 500 µL lysis buffer/plate. Lysis buffer contained 25 mM Tris-HCl pH 7.4, 150 mM NaCl, 1% IGEPAL® CA-630, 1 mM EDTA pH 8.0, and 5% glycerol (identical to Pierce® IP Lysis Buffer, Thermo Fisher Scientific, 87787), and Protease Inhibitor Cocktail (1:100, Cell Signaling Technology, 5871). Plates were incubated on ice for 15 minutes, cells were scraped off, collected in Eppendorf test tubes, and incubated another 5 minutes on ice. After samples were clarified by high-speed centrifugation (10 minutes, 13,000 g, 4oC) and transferred to fresh test tubes, protein concentrations were determined using the Bradford assay (Sigma Aldrich, B6916). Standards and samples were prepared in diluted lysis buffer (1:1) to reduce IGEPAL® concentration to 0.5% which is compatible with the assay. Streptavidin magnetic beads (Dynabeads™ MyOne™ Streptavidin C1, Thermo Fisher Scientific, 65001), at 50 µL/sample, were pre-washed per kit instructions and 100 pmol/sample of 5’-biotinylated RNA oligonucleotide was bound to the beads per kit instructions (we did not use the 3’ desthiobiotinylation option available in the kit). tDR-36:74-Asn-GTT-2-M2 sequence was as follows: UAA CCG AAA GGU UGG UGG UUC GAG CCC ACC CAG GGA CGC. The control sequence (NC5, IDT) was as follows: GCG ACU AUA CGC GCA AUA UG. In both cases the penultimate base and the one preceding it were protected by 2’-O-methylation. Beads were washed again and 120 µg cell lysate was added to each sample, with glycerol and Tween® 20 at final concentrations of 15% and 0.1%, respectively, per the initial conditions suggested in the kit. After rotation for 1 hour at 4oC beads were washed once in kit Wash Buffer and another three times in the same buffer lacking detergent. 40% of the bead pellet from each sample (representing 48 µg protein/sample) was stored at -80°C until submission for mass spectrometry.
Mass spectrometry
On-bead digestion was performed as follows: packed beads were resuspended in buffer containing 8 M urea, 10 mM DTT, and 25 mM Tris-HCl pH 8.0, and incubated for 30 minutes. 55 mM iodoacetamide was then added and beads incubated for an additional 30 minutes. The urea was diluted in 8 volumes of 25 mM Tris-HCl pH 8.0 and 0.4 µg/sample trypsin added. Beads were incubated overnight at 37°C with gentle agitation, peptides were acidified in 0.38% formic acid, and desalted. MS was performed on a Q Exactive™ Plus mass spectrometer coupled on-line to a Dionex UltiMate™ 3000 system, with Xcalibur™ software for data acquisition (Thermo Fisher Scientific). Peptides were separated over an acetonitrile gradient (0 - 80%), flow rate 0.15-0.3 μl/min on a reverse phase 25-cm-long C18 column (75 μm ID, 2 μm, 100 Å, PepMap RSLC) for 120 minutes. Survey scans (380–2,000 m/z, target value 3E6 charges, maximum ion injection times 50 ms) were acquired and followed by higher energy collisional dissociation-based fragmentation. A resolution of 70,000 was used for survey scans and up to 15 dynamically chosen most abundant precursor ions with “peptide preferred” profile were fragmented (isolation window 1.8 m/z). The MS/MS scans were acquired at a resolution of 17,500 (target value 1E5 charges, maximum ion injection times 120 ms). Dynamic exclusion was 60 sec. To avoid carryover the column was washed with 80% acetonitrile and 0.1% formic acid for 25 min between samples. Data was processed using the MaxQuant computational platform. Peak lists were searched against the human reference proteome from UniProt (UP000005640) and allowed up to two mis-cleavages. Peptides with a length of at least seven amino acids were analyzed and the required FDR was set to 1% at the peptide and protein level. Relative protein quantification was determined using the label-free quantification (LFQ) algorithm. The Perseus statistical package with default software parameters was used for statistical computations. All analysis was performed at the Stein Family mass spectrometry center in the Silberman Institute of Life Sciences, The Hebrew University of Jerusalem.
Sampling of live human brain tissue
The study, as well as acquisition of cortical brain samples, was approved by the institutional review board (IRB protocol RMB-0713-19) and by the national ethical review board. All patients signed informed consent forms. The biopsies were extracted in operations for resection of non-neural pathologies, such as meningiomas and metastases. We excluded patients who were operated on for neural pathologies (e. g. gliomas). We recorded patient age, sex, the pathology for which the patient was operated on, and presence of edema in the biopsied brain specimen. We also recorded additional parameters including previous brain irradiation, past medical history, current medications, and monocyte counts and fractions. In the operating room, samples were collected from the cortex overlying the tumor. Sampled brain tissue was fixed in formaldehyde 4% (Sigma-Aldrich, HT501128) for 30 minutes, washed with phosphate buffered saline (PBS) and stored at 4°C in RNAlater® (Merck, R0901-100M) until homogenization.
NuNeX
Samples were homogenized with a Dounce tissue grinder (Sigma-Aldrich, D9063). The homogenate was passed through a 40 μm cell strainer (Corning, 352340) and pelleted by centrifugation at 900 g for 5 min. Cells were resuspended and stored in -80°C until FACS sorting.
Immunostaining and FACS
Before cell sorting, Fc block (Invitrogen,14-9161-73) was added according to manufacturer’s instructions at 20 μL/tube and incubated at 4°C for 20 min. For staining, anti-NeuN (1:500; Alexa Fluor®488-conjugated, Sigma, MAB377X) and anti-Iba1 (1:500; Abcam, ab178846) antibodies were added for 30 min incubation. Cells were then pelleted by centrifugation at 900 g for 5 min, resuspended in staining buffer, and stained with secondary antibody against anti-Iba1 (1:500; Alexa Fluor® 647- conjugated, Jackson ImmunoResearch, 711-605-152), followed by 30 min incubation. Re-pelleted cells were resuspended in staining buffer with DAPI (1:1000; Santa Cruz, sc-3598). All steps were performed on ice. Iba1- and NeuN-positive cells were sorted through a 85 μm nozzle with an approximate flow rate of 8,000 events/s. Sorted Iba1- and NeuN- positive cells were collected into tubes containing 500 μL staining buffer ( > 1,000 cells), then centrifuged at 900 g for 5 min and resuspended in 100 μL of protein kinase digestion (PKD) buffer. RNA was extracted using a specialized kit (RNEasy® FFPE Kit, Qiagen, 73504). Samples were collected with BD FACSAria III (BD Biosciences) and analyzed with FCS Express 7 Software (De Novo Software).
RNA sequencing and alignment of live human brain tissue-derived RNA
All samples underwent sequencing of small RNA. Libraries were constructed from 500 pg total RNA using D-Plex Small RNA-seq Kit for Illumina and Single Indexes for Illumina - Set B (Diagenode, C05030001). RIN after cell sorting was determined to be between 1.2 and 7.2 (Bioanalyzer 6000, Agilent) and the small RNA fraction was sequenced using the NextSeq 2000 System (Illumina) on the NextSeq 2000 System (Illumina) at the Center for Genomic Technologies Facility, the Hebrew University of Jerusalem. Small RNA was aligned to miRBase using miRexpress77 and to the tRNA transcriptome using the MINTmap78.
ROSMAP subjects
RNA-Sequencing (RNA-Seq) data was derived from participants in the Rush Alzheimer’s Disease Center’s religious order study (ROS) and Rush memory and aging project (MAP) cohorts63. Both studies were approved by an Institutional Review Board of Rush University Medical Center. All participants in ROSMAP enroll without known dementia and agree to annual clinical evaluation and brain donation at the time of death. Subjects are clinically classified for dementia, AD and Mild Cognitive Impairment (MCI) as previously reported81,82. Those without dementia or MCI were designated as no cognitive impairment (NCI)83. A summary diagnosis was made by a neurologist after death based on select clinical data blinded to all neuropathologic data. All cases received a neuropathological evaluation based upon Braak stage and CERAD which are combined to create NIA-Reagan pathologic criteria for AD84. Analysis shown in Fig. 2j includes 141 postmortem samples from the nucleus accumbens. The labels (AD and control) were assigned based on the summary diagnosis of AD and NCI. Persons with MCI were excluded. The summary metadata is provided in Supplementary Data 5.
Differential Expression (DE) analysis
DE analysis was performed in R using the DESEQ2 package66. Pre-filtered raw counts of long RNAs and miRs included fragments with median expression above 10 counts per million (CPM). The raw counts of tRFs were normalized using the DESEQ2 median of ratios method in which counts are divided by sample-specific size factors determined by median ratio of gene counts relative to geometric mean per gene66 and prefiltered to include fragments with median expression above 10 counts.
Gene set enrichment
Gene set enrichment to identify biological processes was performed with the PANTHER tool of Gene Ontology resource85. The list of all fragments expressed above (median CPM > = 0) was used as a reference gene list.
Cell type deconvolution
Cell type deconvolution was performed in python using the AUTOGENES package22 (Aliee et al, 2021), using a single-cell dataset from whole murine brain as a reference21.
Elucidating AD- related RNA transcripts
Identifying AD-associated transcripts among DE long RNA was performed using the meta-analysis resource26, combining lists of DE genes in 15 AD-APP mouse model studies and 12 AD-MAPT mouse model studies. The overlap of DE genes in Klotho KO with the DE genes in APP and MAPT models was calculated separately.
Elucidating the protein-coding RNA transcripts changed at the protein level in AD
In order to identify transcripts encoding proteins with altered levels in AD (Fig. 1h,i), we used a proteomic dataset from a mouse 5XFAD model31 and a meta-analysis of a series of human proteomic studies32. For mouse data, we took all the significantly changed proteins between 10-months-old 5XFAD mice and the age-matched wildtype mice (2961 proteins in total). For human data, we took all the proteins significantly altered in at least 5 bulk tissue proteomic studies (848 proteins in total).
miR and tRF target prediction
Predicted miR mRNA targets were identified using miRNet34. Our selection included targets validated in the Mus musculus host organism by at least one of the techniques below: 5’RACE, ChIP-seq, ELISA, luciferase assay, FACS, GFP reporter assay, HITS-CLIP, microarray, immunoblot, immunocytochemistry, immunohistochemistry, immunoprecipitaion, LacZ reporter assay, qPCR, Western blot, Northern blot, quantitative proteomic approach, sequencing.
Predicted targeted tRFs were identified using tRFTar42.
Elucidating AD related miRs
AD-associated miRs were identified using the analysis of the short RNA-seq data derived from AD mouse models38. MiRs, changed with age or AD mutations, were identified using the Wilcoxon signed-rank test, applied to log-transformed normalized counts. Fragments with padj < =0.05 after FDR correction were considered significant.
Statistics and reproducibility
To establish the transcriptomic effect of Klotho knockout, the RNA profiles of 5 mouse WT brains were compared to 5 mouse brains with Klotho knockout. Differential expression was performed separately for polyadenylated RNA, microRNA and tRNA fragments using a DESEQ2 package66. The Gene Ontology analyses were performed with the PANTHER software85,86 (https://geneontology.org). The deconvolution to cell types based on mRNA profiles was performed using the AUTOGENES package22, and the yielded cell type proportions were compared between WT and Klotho-KO brains using the nonparametric Mann-Whitney ranked sum test. The Mann-Whitney test was also used to detect differences between individual microRNA and tRF expression profiles in the external datasets such as in comparison between WT and AD mouse models, CSF and brain samples of human AD patients and controls, as well as between tRF levels in control HEK293 and U2OS cell cultures and parallel cultures with Angiogenin knockout/overexpression. The statistical analysis of the pull-down assay mass spectrometry output was performed using a PERSEUS software87. To generate neuron- and microglia-specific short RNA profiles from live human brain tissue, 16 samples with neurons were compared with 16 microglia samples. Differential expression analysis was performed using the DESEQ2 package66. The linear correlation of the patient age with the levels of has-miR-335-5p and has-miR-150-5p was established using a Pearson correlation coefficient.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Materials
Acknowledgements
The authors thank the study participants of the Religious Orders Study (ROS) and Rush Memory and Aging Project (MAP) and staff of the Rush Alzheimer’s Disease Center in Chicago. The authors also express their gratitude to Dr. Ola Karmi from the Research Infrastructures Center (Hebrew University of Jerusalem) for her guidance and assistance in FACS sorting, to Ms. Adi Tujerman from the Center for Genomic Technologies (Hebrew University of Jerusalem) for her assistance with RNA-seq experiments, to Dr. Hanan Schoffman from the Stein Family mass spectrometry center (Hebrew University of Jerusalem) for his assistance in Mass Spectrometry analysis, and Dr. Taylor Schmitz from the University of Western Ontario for inspiring discussions. This work was supported by the Israel Science Foundation (ISF; 1016/18; 3213/19 to H. Soreq), the National Institute of Health (NIH; Aging grant 5P01AG014449-21 to E. Mufson and H Soreq), Keter Holdings (to H. Soreq), Klogenix LLC (Boston, MA, USA), the Gatsby Charitable Foundation (to Y.L.), the K. Stein foundation (to H.S) and a joint grant by the Shaarei Zedek Medical center (to I. Paldor and H. Soreq). The ROS-MAP projects were supported by grants from the National Institute of Health (NIH; P30AG10161, P30AG72975, R01AG15819, R01AG17917, U01AG46152, U01AG61356). Serafima Dubnov is an awardee of PhD fellowships by Azrieli, Kaete-Klausner and TEVA foundations. The graphical abstracts in this article were created with BioRender.com.
Author contributions
Conceptualization: S.D., I.P., C.R.A., H.S. Data collection: S.D., N.Y., O.Y., D.A.B., S.S., E.M., H.S. Data curation: S.D., H.S. Formal analysis: S.D. Funding acquisition: D.G., I.P., C.R.A., H.S. Methodology: S.D., E.R.B., N.Y., O.Y., Y.T., M.K. Project administration: S.D., H.S. Supervision: D.G., C.R.A., H.S. Visualization: S.D. Writing – original draft: S.D., C.R.A., H.S. Writing – review and editing: S.D., E.R.B., H.S.
Peer review
Peer review information
Communications Biology thanks the anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editors: Alex Nord and Mengtan Xing. A peer review file is available.
Data availability
Raw aligned counts of polyadenylated RNA, microRNA and tRNA fragments from Klotho KO and WT mouse brains, as well as raw aligned counts of microRNA and tRNA fragments from neurons and microglia from live human brain tissue is available at 10.6084/m9.figshare.24100785. Differential expression results reported in the paper are provided in Supplementary Data.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s42003-024-06407-y.
References
- 1.Hou Y, et al. Ageing as a risk factor for neurodegenerative disease. Nat. Rev. Neurol. 2019;15:565–581. doi: 10.1038/s41582-019-0244-7. [DOI] [PubMed] [Google Scholar]
- 2.Kuro-o M. Klotho and aging. Biochim. Biophys. Acta BBA - Gen. Subj. 2009;1790:1049–1058. doi: 10.1016/j.bbagen.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nagai T, et al. Cognition impairment in the genetic model of aging klotho gene mutant mice: a role of oxidative stress. FASEB J. 2003;17:50–52. doi: 10.1096/fj.02-0448fje. [DOI] [PubMed] [Google Scholar]
- 4.Zhao, Y. et al. Klotho overexpression improves amyloid‐β clearance and cognition in the APP/PS1 mouse model of Alzheimer’s disease. Aging Cell19, e13239 (2020). [DOI] [PMC free article] [PubMed]
- 5.Dubal DB, et al. Life Extension Factor Klotho Enhances Cognition. Cell Rep. 2014;7:1065–1076. doi: 10.1016/j.celrep.2014.03.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zeldich E, Chen C-D, Avila R, Medicetty S, Abraham CR. The Anti-Aging Protein Klotho Enhances Remyelination Following Cuprizone-Induced Demyelination. J. Mol. Neurosci. 2015;57:185–196. doi: 10.1007/s12031-015-0598-2. [DOI] [PubMed] [Google Scholar]
- 7.Zeldich E, et al. Klotho Is Neuroprotective in the Superoxide Dismutase (SOD1G93A) Mouse Model of ALS. J. Mol. Neurosci. 2019;69:264–285. doi: 10.1007/s12031-019-01356-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Castner, S. A. et al. Longevity factor klotho enhances cognition in aged nonhuman primates. Nat. Aging3, 931–937 (2023). [DOI] [PMC free article] [PubMed]
- 9.Arking DE, et al. Association of human aging with a functional variant of klotho. Proc. Natl Acad. Sci. 2002;99:856–861. doi: 10.1073/pnas.022484299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Neitzel J, et al. KL-VS heterozygosity is associated with lower amyloid-dependent tau accumulation and memory impairment in Alzheimer’s disease. Nat. Commun. 2021;12:3825. doi: 10.1038/s41467-021-23755-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Semba RD, et al. Klotho in the cerebrospinal fluid of adults with and without Alzheimer’s disease. Neurosci. Lett. 2014;558:37–40. doi: 10.1016/j.neulet.2013.10.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen C-D, et al. The Antiaging Protein Klotho Enhances Oligodendrocyte Maturation and Myelination of the CNS. J. Neurosci. 2013;33:1927–1939. doi: 10.1523/JNEUROSCI.2080-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hébert SS, De Strooper B. Alterations of the microRNA network cause neurodegenerative disease. Trends Neurosci. 2009;32:199–206. doi: 10.1016/j.tins.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 14.Dubnov, S. & Soreq, H. Transfer RNA fragments, from structure to function. in RNA Structure and Function 1–19 (2023).
- 15.Shulman, D. et al. Sex‐specific declines in cholinergic‐targeting tRNA fragments in the nucleus accumbens in Alzheimer’s disease. Alzheimers Dement. 19, alz.13095 (2023). [DOI] [PMC free article] [PubMed]
- 16.Sheen VL. Filamin A and Filamin B are co-expressed within neurons during periods of neuronal migration and can physically interact. Hum. Mol. Genet. 2002;11:2845–2854. doi: 10.1093/hmg/11.23.2845. [DOI] [PubMed] [Google Scholar]
- 17.Hanan M, et al. A Parkinson’s disease Circ RNA s Resource reveals a link between circ SLC 8A1 and oxidative stress. EMBO Mol. Med. 2020;12:e11942. doi: 10.15252/emmm.201911942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tomi M, Tajima A, Tachikawa M, Hosoya K. Function of taurine transporter (Slc6a6/TauT) as a GABA transporting protein and its relevance to GABA transport in rat retinal capillary endothelial cells. Biochim. Biophys. Acta BBA - Biomembr. 2008;1778:2138–2142. doi: 10.1016/j.bbamem.2008.04.012. [DOI] [PubMed] [Google Scholar]
- 19.Baker SA, Gajera CR, Wawro AM, Corces MR, Montine TJ. GATM and GAMT synthesize creatine locally throughout the mammalian body and within oligodendrocytes of the brain. Brain Res. 2021;1770:147627. doi: 10.1016/j.brainres.2021.147627. [DOI] [PubMed] [Google Scholar]
- 20.Ashburner M, et al. Gene Ontology: tool for the unification of biology. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zeisel A, et al. Molecular Architecture of the Mouse Nervous System. Cell. 2018;174:999–1014.e22. doi: 10.1016/j.cell.2018.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aliee H, Theis FJ. AutoGeneS: Automatic gene selection using multi-objective optimization for RNA-seq deconvolution. Cell Syst. 2021;12:706–715.e4. doi: 10.1016/j.cels.2021.05.006. [DOI] [PubMed] [Google Scholar]
- 23.Meydan C, Üçeyler N, Soreq H. Non-coding RNA regulators of diabetic polyneuropathy. Neurosci. Lett. 2020;731:135058. doi: 10.1016/j.neulet.2020.135058. [DOI] [PubMed] [Google Scholar]
- 24.Zhang H, Tao J, Zhang S, Lv X. LncRNA MEG3 Reduces Hippocampal Neuron Apoptosis via the PI3K/AKT/mTOR Pathway in a Rat Model of Temporal Lobe Epilepsy. Neuropsychiatr. Dis. Treat. 2020;16:2519–2528. doi: 10.2147/NDT.S270614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ni X, et al. Knockdown lncRNA NEAT1 regulates the activation of microglia and reduces AKT signaling and neuronal apoptosis after cerebral ischemic reperfusion. Sci. Rep. 2020;10:19658. doi: 10.1038/s41598-020-71411-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wan Y-W, et al. Meta-Analysis of the Alzheimer’s Disease Human Brain Transcriptome and Functional Dissection in Mouse Models. Cell Rep. 2020;32:107908. doi: 10.1016/j.celrep.2020.107908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wahrle, S. E. et al. Overexpression of ABCA1 reduces amyloid deposition in the PDAPP mouse model of Alzheimer disease. J. Clin. Invest. 118, JCI33622 (2008). [DOI] [PMC free article] [PubMed]
- 28.McGeer PL, Akiyama H, Itagaki S, McGeer EG. Immune System Response in Alzheimer’sDisease. Can. J. Neurol. Sci. J. Can. Sci. Neurol. 1989;16:516–527. doi: 10.1017/S0317167100029863. [DOI] [PubMed] [Google Scholar]
- 29.Zeppenfeld DM, et al. Association of Perivascular Localization of Aquaporin-4 With Cognition and Alzheimer Disease in Aging Brains. JAMA Neurol. 2017;74:91. doi: 10.1001/jamaneurol.2016.4370. [DOI] [PubMed] [Google Scholar]
- 30.Liu Y, Beyer A, Aebersold R. On the Dependency of Cellular Protein Levels on mRNA Abundance. Cell. 2016;165:535–550. doi: 10.1016/j.cell.2016.03.014. [DOI] [PubMed] [Google Scholar]
- 31.Kim DK, et al. Deep proteome profiling of the hippocampus in the 5XFAD mouse model reveals biological process alterations and a novel biomarker of Alzheimer’s disease. Exp. Mol. Med. 2019;51:1–17. doi: 10.1038/s12276-019-0326-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Askenazi M, et al. Compilation of reported protein changes in the brain in Alzheimer’s disease. Nat. Commun. 2023;14:4466. doi: 10.1038/s41467-023-40208-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hadar A, et al. SIRT1, miR-132 and miR-212 link human longevity to Alzheimer’s. Dis. Sci. Rep. 2018;8:8465. doi: 10.1038/s41598-018-26547-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chang L, Zhou G, Soufan O, Xia J. miRNet 2.0: network-based visual analytics for miRNA functional analysis and systems biology. Nucleic Acids Res. 2020;48:W244–W251. doi: 10.1093/nar/gkaa467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Karagkouni D, et al. DIANA-TarBase v8: a decade-long collection of experimentally supported miRNA–gene interactions. Nucleic Acids Res. 2018;46:D239–D245. doi: 10.1093/nar/gkx1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Somel M, et al. MicroRNA, mRNA, and protein expression link development and aging in human and macaque brain. Genome Res. 2010;20:1207–1218. doi: 10.1101/gr.106849.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Davis BN, Hilyard AC, Lagna G, Hata A. SMAD proteins control DROSHA-mediated microRNA maturation. Nature. 2008;454:56–61. doi: 10.1038/nature07086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sierksma A, et al. Deregulation of neuronal miRNAs induced by amyloid-β or TAU pathology. Mol. Neurodegener. 2018;13:54. doi: 10.1186/s13024-018-0285-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zongaro S, et al. The 3’ UTR of FMR1 mRNA is a target of miR-101, miR-129-5p and miR-221: implications for the molecular pathology of FXTAS at the synapse. Hum. Mol. Genet. 2013;22:1971–1982. doi: 10.1093/hmg/ddt044. [DOI] [PubMed] [Google Scholar]
- 40.Madrer N, Soreq H. Cholino‐ncRNAs modulate sex‐specific‐ and age‐related acetylcholine signals. FEBS Lett. 2020;594:2185–2198. doi: 10.1002/1873-3468.13789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Burgos K, et al. Profiles of Extracellular miRNA in Cerebrospinal Fluid and Serum from Patients with Alzheimer’s and Parkinson’s Diseases Correlate with Disease Status and Features of Pathology. PLoS ONE. 2014;9:e94839. doi: 10.1371/journal.pone.0094839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou Y, Peng H, Cui Q, Zhou Y. tRFTar: Prediction of tRF-target gene interactions via systemic re-analysis of Argonaute CLIP-seq datasets. Methods. 2021;187:57–67. doi: 10.1016/j.ymeth.2020.10.006. [DOI] [PubMed] [Google Scholar]
- 43.Su Z, Kuscu C, Malik A, Shibata E, Dutta A. Angiogenin generates specific stress-induced tRNA halves and is not involved in tRF-3–mediated gene silencing. J. Biol. Chem. 2019;294:16930–16941. doi: 10.1074/jbc.RA119.009272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Emara MM, et al. Angiogenin-induced tRNA-derived Stress-induced RNAs Promote Stress-induced Stress Granule Assembly. J. Biol. Chem. 2010;285:10959–10968. doi: 10.1074/jbc.M109.077560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ivanov P, Emara MM, Villen J, Gygi SP, Anderson P. Angiogenin-Induced tRNA Fragments Inhibit Translation Initiation. Mol. Cell. 2011;43:613–623. doi: 10.1016/j.molcel.2011.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Saikia M, et al. Angiogenin-Cleaved tRNA Halves Interact with Cytochrome c, Protecting Cells from Apoptosis during Osmotic Stress. Mol. Cell. Biol. 2014;34:2450–2463. doi: 10.1128/MCB.00136-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Novotný I, et al. SART3-Dependent Accumulation of Incomplete Spliceosomal snRNPs in Cajal Bodies. Cell Rep. 2015;10:429–440. doi: 10.1016/j.celrep.2014.12.030. [DOI] [PubMed] [Google Scholar]
- 48.Mo Z, et al. Splicing factor SNRPA associated with microvascular invasion promotes hepatocellular carcinoma metastasis through activating NOTCH1 /Snail pathway and is mediated by circSEC62 / miR ‐625‐5p axis. Environ. Toxicol. 2023;38:1022–1037. doi: 10.1002/tox.23745. [DOI] [PubMed] [Google Scholar]
- 49.Deschênes M, Chabot B. The emerging role of alternative splicing in senescence and aging. Aging Cell. 2017;16:918–933. doi: 10.1111/acel.12646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nikom D, Zheng S. Alternative splicing in neurodegenerative disease and the promise of RNA therapies. Nat. Rev. Neurosci. 2023;24:457–473. doi: 10.1038/s41583-023-00717-6. [DOI] [PubMed] [Google Scholar]
- 51.Hsieh Y-C, et al. Tau-Mediated Disruption of the Spliceosome Triggers Cryptic RNA Splicing and Neurodegeneration in Alzheimer’s Disease. Cell Rep. 2019;29:301–316.e10. doi: 10.1016/j.celrep.2019.08.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhao Y, Rai J, Li H. Regulation of translation by ribosomal RNA pseudouridylation. Sci. Adv. 2023;9:eadg8190. doi: 10.1126/sciadv.adg8190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Borchardt EK, Martinez NM, Gilbert WV. Regulation and Function of RNA Pseudouridylation in Human Cells. Annu. Rev. Genet. 2020;54:309–336. doi: 10.1146/annurev-genet-112618-043830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Guzzi N, et al. Pseudouridylation of tRNA-Derived Fragments Steers Translational Control in Stem Cells. Cell. 2018;173:1204–1216.e26. doi: 10.1016/j.cell.2018.03.008. [DOI] [PubMed] [Google Scholar]
- 55.Guzzi N, et al. Pseudouridine-modified tRNA fragments repress aberrant protein synthesis and predict leukaemic progression in myelodysplastic syndrome. Nat. Cell Biol. 2022;24:299–306. doi: 10.1038/s41556-022-00852-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Maquat LE. Skiing Toward Nonstop mRNA Decay. Science. 2002;295:2221–2222. doi: 10.1126/science.1071285. [DOI] [PubMed] [Google Scholar]
- 57.Chen S, et al. Mechanistic studies for the role of cellular nucleic-acid-binding protein (CNBP) in regulation of c-myc transcription. Biochim. Biophys. Acta BBA - Gen. Subj. 2013;1830:4769–4777. doi: 10.1016/j.bbagen.2013.06.007. [DOI] [PubMed] [Google Scholar]
- 58.Noh JH, et al. GRSF1 suppresses cell senescence. Aging. 2018;10:1856–1866. doi: 10.18632/aging.101516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Huang H, Zhang J, Harvey SE, Hu X, Cheng C. RNA G-quadruplex secondary structure promotes alternative splicing via the RNA-binding protein hnRNPF. Genes Dev. 2017;31:2296–2309. doi: 10.1101/gad.305862.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ivanov P, et al. G-quadruplex structures contribute to the neuroprotective effects of angiogenin-induced tRNA fragments. Proc. Natl Acad. Sci. 2014;111:18201–18206. doi: 10.1073/pnas.1407361111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sood P, Krek A, Zavolan M, Macino G, Rajewsky N. Cell-type-specific signatures of microRNAs on target mRNA expression. Proc. Natl Acad. Sci. 2006;103:2746–2751. doi: 10.1073/pnas.0511045103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Harmancı AS, et al. Integrated genomic analyses of de novo pathways underlying atypical meningiomas. Nat. Commun. 2017;8:14433. doi: 10.1038/ncomms14433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bennett DA, et al. Religious Orders Study and Rush Memory and Aging Project. J. Alzheimers Dis. 2018;64:S161–S189. doi: 10.3233/JAD-179939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yayon N, et al. High‐throughput morphometric and transcriptomic profiling uncovers composition of naïve and sensory‐deprived cortical cholinergic VIP/CHAT neurons. EMBO J. 2023;42:e110565. doi: 10.15252/embj.2021110565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jurga AM, Paleczna M, Kuter KZ. Overview of General and Discriminating Markers of Differential Microglia Phenotypes. Front. Cell. Neurosci. 2020;14:198. doi: 10.3389/fncel.2020.00198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kozomara A, Birgaoanu M, Griffiths-Jones S. miRBase: from microRNA sequences to function. Nucleic Acids Res. 2019;47:D155–D162. doi: 10.1093/nar/gky1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Borovikova LV, et al. Vagus nerve stimulation attenuates the systemic inflammatory response to endotoxin. Nature. 2000;405:458–462. doi: 10.1038/35013070. [DOI] [PubMed] [Google Scholar]
- 69.Bacon C, et al. Brain-specific Foxp1 deletion impairs neuronal development and causes autistic-like behaviour. Mol. Psychiatry. 2015;20:632–639. doi: 10.1038/mp.2014.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Abraham CR, Li A. Aging-suppressor Klotho: Prospects in diagnostics and therapeutics. Ageing Res. Rev. 2022;82:101766. doi: 10.1016/j.arr.2022.101766. [DOI] [PubMed] [Google Scholar]
- 71.Begik O, et al. Quantitative profiling of pseudouridylation dynamics in native RNAs with nanopore sequencing. Nat. Biotechnol. 2021;39:1278–1291. doi: 10.1038/s41587-021-00915-6. [DOI] [PubMed] [Google Scholar]
- 72.Young AMH, et al. A map of transcriptional heterogeneity and regulatory variation in human microglia. Nat. Genet. 2021;53:861–868. doi: 10.1038/s41588-021-00875-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Braun PR, et al. Genome-wide DNA methylation comparison between live human brain and peripheral tissues within individuals. Transl. Psychiatry. 2019;9:47. doi: 10.1038/s41398-019-0376-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nishitani S, et al. Cross-tissue correlations of genome-wide DNA methylation in Japanese live human brain and blood, saliva, and buccal epithelial tissues. Transl. Psychiatry. 2023;13:72. doi: 10.1038/s41398-023-02370-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kuro-o M, et al. Mutation of the mouse klotho gene leads to a syndrome resembling ageing. Nature. 1997;390:45–51. doi: 10.1038/36285. [DOI] [PubMed] [Google Scholar]
- 76.Kurosu H, et al. Suppression of Aging in Mice by the Hormone Klotho. Science. 2005;309:1829–1833. doi: 10.1126/science.1112766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang W-C, et al. miRExpress: Analyzing high-throughput sequencing data for profiling microRNA expression. BMC Bioinforma. 2009;10:328. doi: 10.1186/1471-2105-10-328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Loher P, Telonis AG, Rigoutsos I. MINTmap: fast and exhaustive profiling of nuclear and mitochondrial tRNA fragments from short RNA-seq data. Sci. Rep. 2017;7:41184. doi: 10.1038/srep41184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Holmes AD, et al. A standardized ontology for naming tRNA-derived RNAs based on molecular origin. Nat. Methods. 2023;20:627–628. doi: 10.1038/s41592-023-01813-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bennett DA, et al. Decision Rules Guiding the Clinical Diagnosis of Alzheimer’s Disease in Two Community-Based Cohort Studies Compared to Standard Practice in a Clinic-Based Cohort Study. Neuroepidemiology. 2006;27:169–176. doi: 10.1159/000096129. [DOI] [PubMed] [Google Scholar]
- 82.Bennett DA, et al. Natural history of mild cognitive impairment in older persons. Neurology. 2002;59:198–205. doi: 10.1212/WNL.59.2.198. [DOI] [PubMed] [Google Scholar]
- 83.Bennett DA, et al. Neuropathology of older persons without cognitive impairment from two community-based studies. Neurology. 2006;66:1837–1844. doi: 10.1212/01.wnl.0000219668.47116.e6. [DOI] [PubMed] [Google Scholar]
- 84.Schneider JA, Arvanitakis Z, Leurgans SE, Bennett DA. The neuropathology of probable Alzheimer disease and mild cognitive impairment. Ann. Neurol. 2009;66:200–208. doi: 10.1002/ana.21706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Thomas PD, et al. PANTHER: A Library of Protein Families and Subfamilies Indexed by Function. Genome Res. 2003;13:2129–2141. doi: 10.1101/gr.772403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Thomas PD, et al. PANTHER: Making genome‐scale phylogenetics accessible to all. Protein Sci. 2022;31:8–22. doi: 10.1002/pro.4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tyanova S, et al. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods. 2016;13:731–740. doi: 10.1038/nmeth.3901. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Materials
Data Availability Statement
Raw aligned counts of polyadenylated RNA, microRNA and tRNA fragments from Klotho KO and WT mouse brains, as well as raw aligned counts of microRNA and tRNA fragments from neurons and microglia from live human brain tissue is available at 10.6084/m9.figshare.24100785. Differential expression results reported in the paper are provided in Supplementary Data.