Abstract
Ovarian cancer, ranking as the seventh most prevalent malignancy among women globally, faces significant challenges in diagnosis and therapeutic intervention. The difficulties in early detection are amplified by the limitations and inefficacies inherent in current screening methodologies, highlighting a pressing need for more efficacious diagnostic and treatment strategies. Phage display technology emerges as a pivotal innovation in this context, utilizing extensive phage-peptide libraries to identify ligands with specificity for cancer cell markers, thus enabling precision-targeted therapeutic strategies. This technology promises a paradigm shift in ovarian cancer management, concentrating on targeted drug delivery systems to improve treatment accuracy and efficacy while minimizing adverse effects. Through a meticulous review, this paper evaluates the revolutionary potential of phage display in enhancing ovarian cancer therapy, representing a significant advancement in combating this challenging disease. Phage display technology is heralded as an essential instrument for developing effective immunodiagnostic and therapeutic approaches in ovarian cancer, facilitating early detection, precision-targeted medication, and the implementation of customized treatment plans.
Keywords: ovarian cancer, phage display technology, targeted drug delivery, immunodiagnostics, personalized therapeutic strategies
1. Introduction
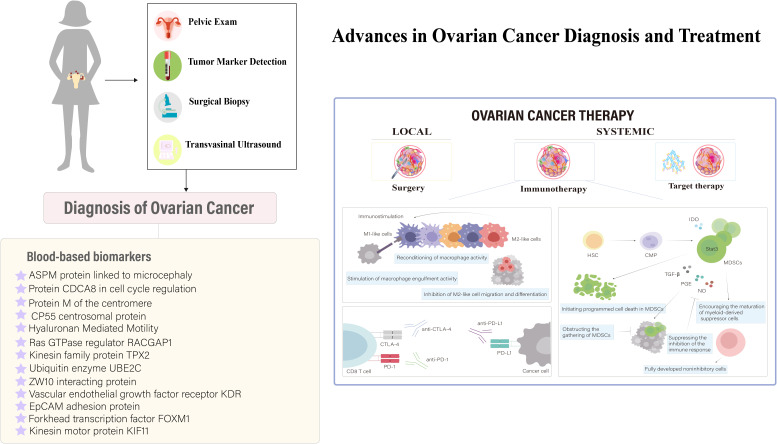
Ovarian cancer is identified as the seventh most common malignancy affecting women across the globe (1). Its significance is highlighted by GLOBOCAN estimates, which report that in 2020, approximately 314,000 women were diagnosed with ovarian cancer, leading to 207,000 fatalities. This positions ovarian cancer as the eighth leading cause of cancer-related incidence and mortality among women internationally (2). Forecasts for the year 2040, based on the assumption that incidence rates remain constant at the 2020 levels, predict around 428,000 new ovarian cancer cases and 307,000 fatalities annually. Achieving a 30% reduction in these figures by 2040 necessitates an annual decrease of 2% in the global incidence and mortality rates of this disease (3). The urgent need for the development of efficacious treatments for gynecological cancers, particularly ovarian cancer, which is known as the deadliest gynecological malignancy, continues to be a significant clinical challenge. This is largely attributed to the subtle, non-specific symptoms of ovarian cancer and the commonality of late-stage diagnoses (4). The complexity of ovarian cancer diagnosis and treatment has ignited intensive efforts to develop screening methodologies that enable early detection in asymptomatic individuals. Despite this, the effectiveness of current leading methods, including CA125 blood tests and transvaginal ultrasound (TVS), has not been conclusively shown to lower ovarian cancer mortality rates in the general population through randomized controlled trials (5). This urgency is further intensified by the challenges presented by advanced, recurrent, and drug-resistant forms of ovarian cancer, highlighting an acute need for novel therapeutic solutions (5). Consequently, this multifaceted dilemma has motivated the research community to push the boundaries of ovarian cancer research, seeking innovative solutions informed by a comprehensive understanding of the intricate dynamics of cancer progression (6). In this context, the advent of targeted diagnostic and therapeutic strategies emerges as a promising development, potentially heralding significant advancements in the management of ovarian cancer. Particularly in the domain of ovarian cancer therapeutics, there has been a notable increase in the development of targeted agents, often aimed at “shared antigens” found across a range of tumors (7). However, a detailed analysis suggests a complex scenario where the effectiveness of these agents may be limited in eliciting strong immune responses due to the ubiquitous nature of these “shared antigens” across various body tissues. This complexity introduces a risk of unintended autoimmune reactions and could contribute to the development of drug resistance (7). As research delves into the nuanced interaction between targeted therapies and the immune microenvironment, it becomes clear that customizing precision medicine strategies to minimize off-target effects and immune-related adverse events is crucial for improving the therapeutic outcomes of these interventions. This approach underscores the importance of advancing targeted treatments within a framework that carefully considers the broader implications on the immune system and patient health.
In the landscape of oncology research, phage display technology has recently stood out as a significant breakthrough, particularly in the realms of immunodiagnostics and therapeutic development for ovarian cancer. This method allows for the comprehensive screening of extensive phage-peptide libraries, facilitating the identification of ligands that bind with high specificity and affinity to cancerous cells, thus enabling precise targeting strategies (8). Such an approach promises to enhance the accuracy of drug targeting, reduce off-target effects, and minimize associated adverse events. Nonetheless, the path to fully leveraging this technology is fraught with obstacles. Phage-peptide display presents short peptides via bacteriophages, screening large libraries efficiently but with limited binding affinity and specificity. Nanobodies from camelids offer high specificity and stability on phages, though they face production challenges. Single-chain variable fragments (scFvs) provide smaller, tissue-penetrating antibody forms with potential stability issues. Fragment antigen-binding (Fab) displays, incorporating complete antibody domains, ensure high stability and affinity but may incur higher costs and reduced tissue penetration. The quest to discover novel and effective phage-peptides is a demanding endeavor that requires a detailed and laborious process. This journey involves repeated cycles of screening and sequencing against tumor cells or tissues, along with stringent validation of ligand binding affinities. Achieving success in this domain necessitates the synthesis of phage-peptides, which are integral to the development of precision-targeted cancer therapeutics (9). This review is dedicated to a comprehensive examination and integration of the existing literature on the application of phage display technology in the immunodiagnostics and treatment of ovarian cancer. Its goal is to delineate the significant progress made, highlight existing knowledge gaps, and outline prospective research trajectories within this area. By doing so, it seeks to furnish insights that are of critical relevance to both the scientific community and clinical practitioners.
2. Applications of phage display in cancer research
2.1. Phage display vectors: diversity and applications
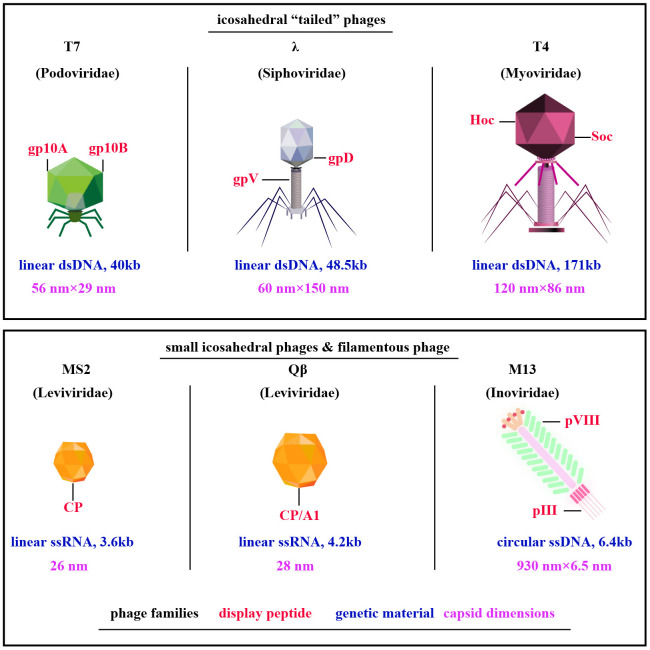
Exploration of phage display technologies has unveiled a plethora of vector systems, including but not limited to M13, T4, T7, λ, Qβ, and MS2 phages (10) ( Figure 1 ). The M13 phage, a filamentous non-lytic virus, excels in phage display due to its unique structure and compatibility with Escherichia coli, enabling the presentation of diverse peptides and proteins (12). T4 bacteriophage, lytic with a slender icosahedral head and complex tail, is essential for vaccine delivery, expressing coat proteins like gp23, gp24, gp20, Soc, and Hoc that aid DNA encapsulation and antigen presentation (13, 14). T7, a lytic Podoviridae member, efficiently displays large proteins thanks to its 60 nm head and 40kb DNA, with proteins like gp10A and gp10B enhancing secretion and replication (15–17). The Qβ phage, a minimalist Leviviridae member, features a simple 28nm diameter and a 4.2 kb ssRNA genome that encodes key proteins such as CP, A2, and A1, critical for infecting Escherichia coli and display technologies (18, 19). Similarly, the MS2 phage, also from the Leviviridae family, presents a lytic 26 nm icosahedral structure with a 3.6 kb ssRNA genome coding for proteins like CP, A, replicase, and lysis protein L, essential for assembly and display applications (18, 19). M13, T4, T7, λ, Qβ, and MS2 phages are tailored for distinct biotechnological uses ( Table 1 ). M13 facilitates phage display for peptide and protein engineering through non-lytic DNA packaging, supporting drug development and gene therapy. T4 combats antibiotic-resistant bacteria and aids molecular biology research. T7, known for its strong promoter, is essential for high-level protein expression and CRISPR applications. Lambda phage plays a critical role in molecular cloning and E. coli lysis, enhancing phage therapy. Qβ contributes to vaccine development and RNA research, while MS2 is vital in RNA studies and virology education due to its straightforward RNA genome. M13’s non-lytic replication enables the continuous production of large peptide or protein libraries.
Figure 1.
Diverse Phage Types Exploited in Phage Display. Among them, filamentous M13 phages are prominent, along with “tail” phages like T7, λ, and T4, and compact ssRNA icosahedral phages MS2 and Qβ. Notably, key capsid proteins for peptides display are highlighted in red, phage families in black, genetic material in blue, and capsid dimensions in purple [adapted from (11)].
Table 1.
The pros and cons of distinct phage display vectors.
| Phage Vector | Pros | Cons |
|---|---|---|
| M13 | High-level peptide display | Limited to filamentous phage biology |
| Non-lytic, continuous display | May require specialized host strains | |
| Useful for panning and affinity maturation | ||
| T4 | High payload capacity | Lytic cycle can complicate library maintenance |
| Robust, large capsid for display | Less common for peptide display | |
| Effective for vaccine development | ||
| T7 | Extremely efficient expression | Lytic nature requires careful handling |
| Rapid lytic cycle speeds up experiments | Smaller display capacity compared to T4 | |
| Suitable for high-throughput screening | ||
| λ (Lambda) | High DNA packaging efficiency | Complex lifecycle can be a challenge |
| Allows for large DNA insertions | Lytic and lysogenic cycles | |
| Well-studied genetics and molecular biology | ||
| Qβ | RNA phage, useful for RNA display | RNA genome stability issues |
| Can display peptides and proteins | Limited host range | |
| Useful for vaccine development | ||
| MS2 | Small RNA phage, simple genome | Limited display capacity |
| Useful for studying RNA-protein interactions | Stability and robustness issues | |
| Potential applications in targeting RNA |
2.2. Phage display for cancer immunodiagnostics and therapy
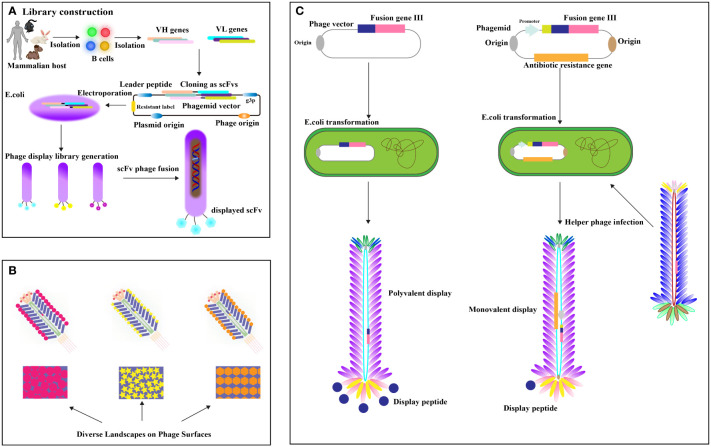
Phage display antibody (PDA) libraries created from immune donors are a direct method for isolating high-affinity antibodies against tumor-specific antigens, leveraging the antibody repertoire of cancer patients (20–22). This involves extracting mRNA from B lymphocytes, cloning it into phage coat protein genes like pIII, and expressing these in Escherichia coli to display antigen-binding domains (23, 24) ( Figure 2A ). Cancer patients frequently develop high-affinity antibodies due to tumors overexpressing or harboring mutated antigens (26). Utilizing immune phage display libraries from humans, researchers compile extensive antibody gene repertoires from cancer patients to isolate antibodies with specific binding capabilities (27). Studies include constructing scFv libraries from patient PBMCs, identifying high-affinity antibodies to EphA2 (28), and using cell panning to bind EGFR-affine fragments to esophageal cancer cells (29). Further developments involve producing nanobodies targeting HIF-1 and CD20/CD3 bispecific nanobodies from immunized camels, showing potential in cancer diagnostics and treatment (30–32). Camel-derived CD16a-specific antibodies conjugated with anti-CEA antibodies have effectively inhibited CEA-positive tumor growth (33), and nanobodies against EpCAM have significantly reduced MCF-4 cell proliferation (34). A VNAR antibody library from nurse sharks yielded antibodies binding to GPC3, HER2, and PD-1 (35), demonstrating phage display’s potential despite immunogenicity concerns (36). The phage display targets crucial cancer biomarkers and therapy targets like EGFR (37), its mutant EGFRvIII (38, 39). The Vascular Endothelial Growth Factor (VEGF), essential for angiogenesis, is targeted to inhibit tumor growth and metastasis (31), with phage display libraries from mice immunized against recombinant human VEGF (40). Landscape phage, defined as M13 phages with 5-mer phage peptides in all p8 protein copies, display thousands of ligands (41), providing a rich source of essential binding units (EBUs) for interacting with physiologically relevant proteins via short linear motifs (SLiMs) (42) ( Figure 2B ). Notable examples include Lei Han et al. (2018)’s high-affinity phage probe for prostate-specific antigen, applicable in ovarian cancer antigen detection (43), and V. Petrenko (2018)’s discussion on landscape phage’s role in nanobiotechnology, emphasizing their utility in cancer biomarker detection (41). The valency differences between phagemid and phage displays are significant ( Figure 2C ). Recombinant human antibody libraries, produced in bacterial or yeast hosts through recombinant DNA technology, mimic the human immune response, targeting diseases impartially using synthetic genes or unselected human B cells. Displayed on phages, yeast, or bacteria, antibodies are screened through biopanning against disease-specific targets and refined for high affinity and specificity. This approach generates antibodies effective against various diseases like cancer, autoimmune disorders, and infections, suitable when traditional immunization is impractical. Such technologies, which contribute to therapies like adalimumab, play a crucial role in advancing personalized medicine and improving treatment outcomes. Valency in phage and phagemid systems indicates the number of displayed peptides or antibodies. Phage systems like M13 often enable polyvalent displays, increasing binding strength through multivalent target interactions. In contrast, phagemid systems typically offer a monovalent display, with each particle showing a single antibody due to helper phage infection of plasmids with a phage origin of replication, allowing precise affinity measurements. Although phage systems are ideal for initial high-avidity screenings, phagemid systems are favored for their ease of genetic manipulation, accurate affinity assessment, and scalability, making them preferred for developing high-affinity therapeutic antibodies in advanced screenings.
Figure 2.
(A) Establishing the Phage Peptide Variant or Antibody Gene Library. Antibody libraries, fused with phage coat protein, are transformed into (E) coli. Enrichment of specific scFv-phage via target binding occurs, followed by removal of nonspecific or improperly folded scFvs through washing. Elution methods release specific scFv-phage fusions [adapted from (25)]. (B) Landscape Phage Concept: Illustrated is a distinct peptide “landscape” exhibited on the phage surface [adapted from (11)]. (C) Phage vector and selection: display valency is pivotal. Phage vectors carry recombinant genes for polyvalent display, while helper phages yield lower valence (typically pIII univalent) [adapted from (11)].
Challenges include antigen heterogeneity and immunogenicity from non-human sources like camels and sharks. Future efforts should refine library construction and enhance screening for greater specificity and reduced immune reactions. Additionally, integrating disease-agnostic recombinant human antibody libraries could widen their application across various cancers. Exploring differences in valency between phage and phagemid systems is crucial, as phage systems facilitate high-throughput screening and phagemids provide precise affinity measurements essential for therapy. Advancing these technologies will require integrating both systems to address these challenges and improve cancer treatment outcomes.
2.3. From in situ to human-specific targeting
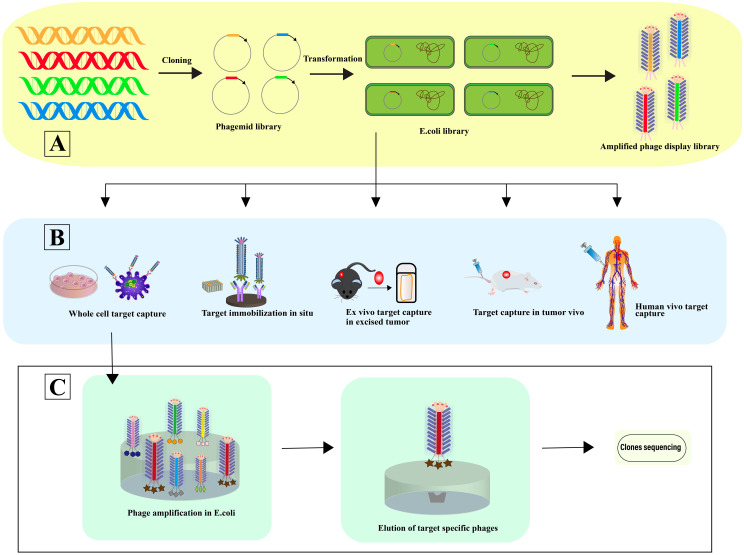
Phage selection involves a systematic four-step process: 1) Co-incubation with the target for selective binding; 2) Removal of non-specifically bound phages; 3) Elution of specifically bound phages; 4) Amplification of these phages for subsequent rounds of selection. This selection can occur in various contexts: In situ selection, a straightforward and commonly employed method, utilizes surfaces such as plates or beads for binding (44). In vitro cell selection is designed to identify phage-peptides specific to cell types, maintaining their biological activity, structural integrity, and receptor interactions, crucial for preserving their functional relevance (45). In vivo selection within animal models facilitates the isolation of phage-peptides that are specific to certain organs, providing insights under physiological conditions (46). Here, phage libraries are administered intravenously, allowing phages to circulate and bind organ-specifically. Phages are then harvested from targeted organs, with sequences identified post-homogenization (47). Ex vivo screening is particularly useful for identifying specific phage-peptides in rare cells within mixed populations, such as peripheral blood mononuclear cells (PBMCs) in hematologic malignancies (48). Utilizing human phage display libraries helps to minimize the variability in phage-peptide binding affinity that may arise due to interspecies differences (49), thereby enhancing the precision of cancer targeting in patient-specific therapies (50) ( Figure 3 ).
Figure 3.
Phage selection. (A) In this process, a vast array of library variants is incorporated into phagemids, facilitating the transformation of (E) coli and subsequent phage rescue. The result is the creation of extensive phage libraries, which support the iterative biopanning process for the selection and isolation of phages that bind specifically to target antibodies. (B) The diagram illustrates Various Strategies for Harvesting High-Affinity Peptides Through Phage Display Screening: In situ screening involves the application of targets onto plates. Ex vivo screening is designated for isolating rare cells amidst heterogeneity. Screening using samples from human patients helps minimize species mismatches. In vitro screening is tailored to identify peptides specific to adherent cells, while organ-specific peptides are derived through in vivo biopanning and selection within living organisms [adapted from (50)]. (C) This is followed by further screening and sequencing of the identified phages, advancing towards the pinpointing of optimal target-specific ligands.
Phage display challenges include inefficient step execution and slow clinical translation due to procedural inconsistencies and variable experiments, leading to unreliable peptide identification. Enhancements in phage selection could involve advanced molecular and computational tools to refine peptide specificity and functionality. High-throughput sequencing and machine learning could improve peptide insights, while better in vivo models might enhance translational potential. Utilizing human-specific phage libraries could diminish immunogenicity and improve outcomes, enhancing cancer targeting precision in personalized medicine. Adopting an integrated approach with human-specific libraries is crucial to advancing personalized therapies in phage selection.
3. Advancements and challenges in ovarian cancer immunotherapy
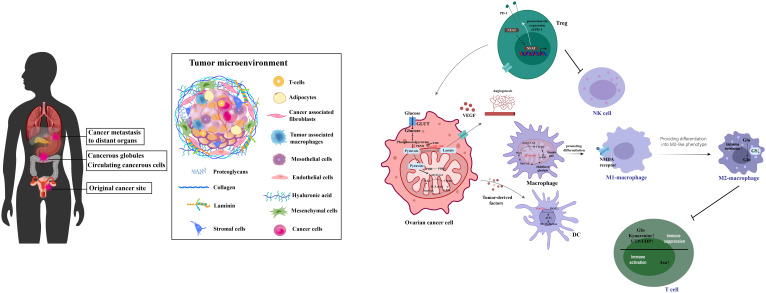
Ovarian cancer, a significant challenge within gynecologic oncology (51), is characterized by promising survival outcomes when identified at an early stage (52). Yet, the non-specific nature of its initial symptoms (53) frequently results in diagnoses during advanced stages (54). Survival rates for late-stage diagnoses fall under 20%, in stark contrast to the 90% survival rate associated with stage I detection (55, 56). Ovarian cancers mainly arise from the fallopian tube linings and metastasize to the abdominal cavity, where cells transition between epithelial and mesenchymal states to form complex secondary lesions with varied cell types and altered metabolism that supports rapid growth, as depicted in Figure 4 . Despite ongoing research endeavors, the complexities of ovarian cancer detection and management persist, underscoring the imperative need for novel diagnostic and therapeutic strategies (59).
Figure 4.
This illustration captures the process of ovarian cancer metastasis and the intricacies of its tumor microenvironment. It shows how ovarian cancer typically begins in the fallopian tube linings and spreads to the abdominal cavity. The diagram highlights the dynamic transitions between epithelial and mesenchymal states that are critical for the formation of secondary lesions. It further details the metabolic reprogramming within various cell types, including cancer and immune cells, contributing to a suppressive immune environment. This environment promotes tumor growth and progression by enhancing immunosuppression, facilitating robust cellular interactions, and enabling immune escape through a complex network of interconnected metabolites. [adapted from (57, 58)].
3.1. Surgical and chemoradiation therapies
Ovarian cancer treatment typically involves postoperative chemotherapy and surgery, especially important in early stages. Advanced-stage treatment generally consists of debulking surgery followed by adjuvant chemotherapy (60, 61). Patients unsuitable for optimal cytoreduction due to age or comorbidities might receive neoadjuvant chemotherapy and interval debulking surgery (62). The standard chemotherapy regimen often includes paclitaxel/carboplatin and pegylated liposomal doxorubicin/carboplatin (63), with olaparib recommended for remission patients with BRCA mutations post-initial platinum-based chemotherapy (± bevacizumab) (64). Despite initial responses, the high recurrence rate of ovarian cancer and platinum resistance complicate long-term management, with surgical complications and treatment variability demanding personalized care strategies to effectively address individual patient needs.
3.2. Utilization of targeted therapies and current challenges
In ovarian cancer treatment, targeted therapies like anti-angiogenic agents and PARP inhibitors offer significant hope (65). Bevacizumab is effective in reducing ascites and increasing progression-free survival initially (66), though its use is limited by high costs (67) and resistance (68). PARP inhibitors like Olaparib, Rucaparib, and Niraparib benefit ovarian cancer patients with BRCA1/2 mutations or homologous recombination deficiency (HRD) (69). Olaparib monotherapy yielded a 31.1% tumor response rate in platinum-resistant recurrent ovarian cancer (70), and Niraparib improved progression-free survival in late-stage patients (71). PARP inhibitors have been observed to enrich ovarian cancer stem cell populations marked by CD133 and CD117 in vitro and in vivo (72). ABBV-085 targeting LRRC15 has been effective in eliminating LRRC15-positive ovarian tumor cells in micrometastases and ascites (73). MFAP5 antibodies reduced fibrosis and enhanced chemosensitivity (74), while OMTX705 targeting FAPα with chemotherapy completely inhibited tumor growth and induced sustained regression in animal models (75). Research on the EZH2 inhibitor has focused on SCCOHT for ovarian cancer therapy (76), and the antibody-drug conjugate 3A5-MMAE targeting the MUC16-MSLN interaction significantly reduced ovarian cancer cell adhesion and invasion, demonstrating potent antitumor efficacy (77).
3.3. Applications and current challenges in immunotherapeutic approaches
Recent studies on ovarian cancer have shown that serum biomarkers and nanosensors are crucial for diagnosis. Conventional treatments like debulking surgery and chemotherapy modify the immune response, first by increasing immunosuppression for healing, then boosting immunostimulation. This understanding is key to refining immunotherapies to enhance efficacy ( Figure 5 ). In ovarian cancer treatment, immunotherapies like personalized vaccines using autologous dendritic cells and anti-PD-L1 antibodies are in clinical trials, showing potential to enhance adaptive immunity (80, 81). Genetically modified CAR-T cell therapy targeting tumor-associated markers like the folate receptor and mesothelin amplifies antitumor effects (82–84). Genetically engineered oncolytic viral therapies target ovarian tumors selectively, reducing non-tumor cell toxicity (85). Immunotherapy is increasingly integrated as a supplementary treatment, particularly against resistance to established therapies like bevacizumab and PARP inhibitors (86). Synergistic uses of αPD-1 and AMD3100 convert macrophages from M2 to M1, enhancing antitumor responses (87), while PI3K pathway inhibitors combined with bevacizumab (88), and VEGF antibodies with tumor vaccines enhance vaccine efficacy by modulating CD4+ CTL populations (89). Combining PARP inhibitors with PD-L1 inhibitors shows promise in recurrent ovarian cancer treatment by modulating the immune response (90), highlighting the disease’s heterogeneity and propensity for recurrence and metastasis (56).
Figure 5.
Innovations in Ovarian Cancer Immunodiagnostics and Therapeutic Strategies. This figure illustrates the critical role of serum biomarkers and nanosensors in diagnosing ovarian cancer, a condition typically managed through debulking surgery followed by systemic treatments like chemotherapy, targeted therapy, and immunotherapy. It highlights how conventional treatments modify the immune landscape, initially promoting immunosuppression for healing and subsequently enhancing immunostimulation to fight the disease. These dynamics underscore the importance of advancing immunotherapeutic strategies to maximize treatment effectiveness. [adapted from (78, 79)].
4. Phage display applications in ovarian cancer therapy and diagnosis
4.1. Specific targeting phage-peptide identification for ovarian cancer
Phage display emerges as a transformative tool in ovarian cancer research, offering the capability to identify ligands that precisely target cancer cells, potentially ushering in new paradigms in diagnosis and treatment by enhancing drug specificity and reducing side effects (91) ( Figure 6 ). Zhou et al. successfully leveraged phage display to pinpoint a unique phage-peptide, SWQIGGN, demonstrating its capacity to bind and impede the functions of ovarian cancer cells both in vitro and in vivo. This phage-peptide exhibited selective affinity for cancer cells, significantly curtailing their proliferation, migration, invasion, and adhesion. Moreover, it notably inhibited tumor growth and metastasis in animal models, setting the stage for further exploration into SWQIGGN’s clinical utility in ovarian cancer treatment (92). Wang et al. addressed the critical demand for phage-peptides with high specificity as targeting agents for the early detection of ovarian cancer. They proposed the use of phage display technology to identify phage-peptides uniquely associated with cancer cells. By integrating a novel microfluidic system, they streamlined the screening process, enhancing the efficiency and reducing the laborious nature of traditional methods. The identified phage-peptides displayed remarkable affinity and specificity to ovarian cancer cells, indicating its potential in targeted immunodiagnostics. This method’s increased efficiency, minimal sample requirements, and accelerated screening process hold promise for advancing ovarian cancer theranostics (93). Davidson et al. explored the application of phage display in biomaterials engineering, aiming to discover phage-peptides with high affinity for specific targets. These phage-peptides can be engineered to functionalize material surfaces, precisely guiding interactions within the biological milieu. By employing targeting phage-peptides, materials can be designed to selectively interact with proteins, cells, or tissues. Moreover, functional phage-peptides are capable of not just binding but also influencing target activities. The integration of targeting and functional phage-peptides paves the way for the development of dual-functional phage-peptides, enabling the bridging of distinct targets or the modulation of specific protein or cell functions (94), illustrating the versatile potential of phage display in advancing ovarian cancer research and treatment modalities.
Figure 6.
Phage Display Technology in Ovarian Cancer Treatment Customization. This diagram underscores the transformative role of phage display technology in ovarian cancer immunodiagnostics and therapy. It showcases how phage display facilitates the identification of specific ligands that target ovarian cancer cells, thus paving the way for innovative diagnostic and therapeutic approaches. This advancement enhances drug specificity and minimizes side effects, promising to revolutionize treatment modalities for ovarian cancer. [adapted from (57, 79)].
4.2. Phage-derived specific peptides for targeted drug delivery
Conjugating therapeutic agents to targeting phage-peptides or antibodies identified through phage display can directly target cancer cells, substantially reducing off-target effects and sparing healthy tissues. This technique enhances the potential for precise drug delivery, significantly advancing the pursuit of personalized medicine by allowing for the screening of novel targeting ligands tailored to individual cancer profiles. As innovations progress, phage display-mediated targeted drug delivery is set to transform cancer therapy, heralding a new era of efficient and customized treatments (95). Ma et al. unveiled a promising phage-peptide, WSGPGVWGASVK, identified via phage display for targeting human ovarian cancer, potentially revolutionizing cancer therapy. This phage-peptide ‘s in vivo application demonstrated selective tumor site accumulation post intravenous administration, minimizing normal organ distribution. Immunostaining revealed PC3–1 phage clone’s affinity for tumor cells and vasculature, ensuring comprehensive tumor binding. In vitro assessments further confirmed the phage-peptide ‘s tumor cell and angiogenic endothelial cell binding and internalization capabilities, with a dissociation constant (Kd) of 5.43 ± 0.4 μM, underscoring its therapeutic delivery potential (96). An innovative active targeting liposome system for ovarian cancer, developed using phage display, incorporates the ovarian-specific ligand WSGFPGVWGASVK (WSG) on thermosensitive phospholipids, forming WSG-modified thermosensitive liposomes (WSG-Lipo). These liposomes showcase enhanced drug release at 42°C, targeting SKOV-3 ovarian cancer cells and suppressing their activity in vitro. This strategy hints at the WSG-Lipo’s potential to refine thermosensitive liposome specificity, promising advances in combined chemotherapy and thermotherapy for ovarian cancer (97). Addressing the challenge of treating clear cell carcinoma, a notably aggressive and chemoresistant epithelial ovarian cancer subtype, researchers utilized phage display to isolate specific phage-peptides (OC-6 and OC-26) targeting this cancer form. Peptide-conjugated nanoparticles significantly improved tumor cell uptake and drug delivery internally, compared to non-targeted versions. Moreover, the targeted liposomal doxorubicin formulation achieved superior tumor suppression, indicating that phage display-derived subtype-specific therapies could offer more effective, targeted treatment options for varying epithelial ovarian cancer subtypes (98).
4.3. Imaging applications for ovarian cancer
Screening phage libraries has emerged as a pivotal method for identifying phage-peptides or ligands that target molecules uniquely expressed on ovarian cancer cells, facilitating early disease detection and precise diagnostics (99). Engineered phage-displayed peptides can be conjugated with imaging agents, such as fluorescent labels or radioactive markers, allowing for the direct visualization and mapping of ovarian cancer cells and lesions. Moreover, this technology is instrumental in creating innovative biosensors capable of detecting ovarian cancer biomarkers in bodily fluids, offering a minimally invasive, highly sensitive modality for ongoing disease surveillance. Wang et al. employed phage display to isolate NPMIRRQ, a phage-peptide that exhibits selective affinity towards HO-8910 ovarian cancer cells, demonstrating its utility as a diagnostic tool through specific ovarian cancer cell and tissue binding, potentially advancing the identification of reliable biomarkers for the disease’s early detection and management (100). Xing et al. discovered a phage-peptide, C7, via phage display, showcasing targeted binding to the folate receptor alpha (FRα), predominantly expressed in ovarian cancer cells. C7’s tumor-targeting capabilities were confirmed in in vivo studies, with structural modeling elucidating its interaction with FRα, underscoring C7’s therapeutic and diagnostic potential by leveraging FRα’s restricted tumor expression profile in ovarian cancer (101). Further, two phage clones, pJ18 and pJ24, along with their phage-peptides, J18 and J24, demonstrated enhanced affinity for SKOV-3 ovarian cancer cells relative to controls. AF680-labeled phage particles underwent biodistribution analysis, revealing tumor accumulation and retention, with optical imaging pinpointing SKOV-3 tumors post 2 and 4 hours. Particularly, pJ18 exhibited promising tumor-to-background contrast, suggesting its suitability for ovarian cancer detection and imaging (102). A dual-stage phage display technique was utilized to select phage-peptide J18 for its specificity to ovarian cancer cells. Radiolabeling J18 with (11) In and assessing its imaging prowess via SPECT in SKOV-3 xenografted mice, revealed efficient tumor localization with minimal nonspecific binding, indicating J18’s significant potential as a nuclear imaging agent for ovarian cancer diagnostics and treatment (103). Faintuch et al. investigated the NGR phage-peptide, identified through phage display, for radiolabeling with technetium-99m across different tumor models, noting similar uptake in ovarian and lung tumors. Biodistribution studies indicated renal clearance with higher ovarian tumor cell uptake, suggesting enhancements in tumor targeting and imaging by potentially combining NGR with RGD into a heterodimer (104). Babeker et al. reported on generating fully human antibodies against the MUC16 cancer biomarker via phage display. The derived antibody, M16Ab, was modified with p-SCN-Bn-DFO and labeled with 89Zr for immuno-PET imaging. Subsequent in vitro and in vivo assessments showcased specific, high-affinity MUC16 binding, especially in ovarian and pancreatic cancer models. Pharmacokinetic analysis revealed favorable attributes for the non-invasive imaging of these cancers (105), illustrating the broad and impactful applications of phage display in enhancing ovarian cancer diagnostics and targeted therapy.
4.4. Phage-based gene therapy for ovarian cancer
Yu et al. elucidated the utility of phage display in discovering anti-HE4 nanobodies, notably neutralizing the human epididymis protein 4 (HE4) and thereby attenuating the viability of epithelial ovarian cancer (EOC) cells while augmenting their responsiveness to cisplatin. This research underscores the autocrine pro-survival function of HE4 in EOC cells and positions the anti-HE4 nanobody, particularly 1G8, as an efficacious candidate to bolster cisplatin chemotherapy in EOC management (106). Huang et al. probed the fibroblast growth factor-2 (FGF2)’s contribution to ovarian cancer advancement and its therapeutic targeting viability. Elevated FGF2 levels in ovarian tumors were inversely related to patient survival, highlighting its prognostic significance. A heptapeptide-derived phage-peptide showcased inhibitory actions on FGF2-mediated proliferation, migration, and invasion in p53-null epithelial ovarian cancer cells, disrupting cell cycle progression, cyclin D1 upregulation, and MAPK/Akt pathway activation. Additionally, it showed promise in mitigating FGF2-driven doxorubicin resistance by downregulating antiapoptotic proteins and neutralizing FGF2’s antiapoptotic influence (107). Pu et al.’s work focused on devising targeted anti-metastatic therapies, pinpointing a phage clone pc3–1 that exhibited high affinity and specificity to SK-OV-3 cells via the phage-peptide WSGPGVWGASVK. This clone and its corresponding phage-peptide significantly hampered SK-OV-3 cell adhesion to the extracellular matrix and endothelial monolayers, also curtailing invasion (108). Chen et al. deployed a human phage display library to identify a potent anti-MSLN single-chain Fv antibody, facilitating the creation of a second-generation anti-MSLN CAR-T cell therapy. These CAR-T cells proved effective in vitro, eliminating ovarian tumor cells and suppressing MSLN-positive tumor growth, corroborated by elevated cytokine production. In vivo assays further confirmed their therapeutic potential against ovarian cancer xenografts (109). Qiao et al. aimed for enhanced affinity antibodies using phage display, employing 3D complex structural modeling of the HER2-antibody (MIL5) interface to guide the synthesis of a site-directed mutagenesis library. This approach yielded a higher affinity single-chain antibody (M5scFv_ph), maintaining the epitope specificity of its precursor and demonstrating comparable tumor-suppressive efficacy in ovarian carcinoma xenograft models (110). Bortot et al. explored M13 bacteriophage as a precision vector for photodynamic therapy (PDT) in ovarian cancer. By engineering M13 phage to present an EGFR-binding phage-peptide and coupling it with chlorin e6 (Ce6), they generated M13r-Ce6, which produced reactive oxygen species (ROS) upon irradiation, effectively eradicating EGFR-positive ovarian cancer cells. This modification ensured superior cellular uptake and mitochondrial localization, highlighting its potential to induce autophagy and augment PDT efficacy in ovarian cancer (111). Table 2 provides a comprehensive overview of ovarian cancer-targeting phage-peptides identified through phage display, underscoring a decade of significant advancements in targeted cancer therapy research.
Table 2.
Exploring Ovarian Cancer Targeting Peptides through Decade-long Phage Display Screening.
| peptide sequence | phage clone | Target | Year | Application | Reference |
|---|---|---|---|---|---|
| SVSVGMKPSPRP | Z3 | SKOV3 cells | 2011 | Diagnostics | (112) |
| GD3A10 | glycosaminoglycans | 2014 | Diagnostics | (113) | |
| OSTP | A2780 mice | 2015 | Diagnostics | (114) | |
| GD3A11 | Chondroitin sulfate (CS) | 2015 | Diagnostics | (115) | |
| NPMIRRQ | P2 | HO-8910 cells | 2016 | Diagnostics | (100) |
| CREB3 | Igs | 2019 | Diagnostics | (116) | |
| S36 | SKOV3 | 2021 | Diagnostics | (117) | |
| M16Ab | MUC16 | 2022 | Diagnostics | (105) | |
| WSGPGVWGASVK | pc3-1 | SKOV3 mice | 2013 | Therapy | (96) |
| OR2H1 scFv | OR2H1 | 2022 | Therapy | (118) | |
| A7 | HB-EGF | 2021 | Therapy | (119) | |
| OCSP-6 and OCSP-26 | OC-6&OC-26 | OC-3 cells | 2015 | Therapy | (98) |
| no. 7 and no. 29 | HB-EGF | 2019 | Therapy | (120) | |
| SWQIGGN | HO8910 cells | 2015 | Therapy | (92) | |
| P2 | FGF2 | 2019 | Therapy | (107) | |
| C7 | FRα | 2018 | Therapy | (101) | |
| 8G2/8G3 | HE4 | 2022 | Therapy | (106) | |
| anti-MSLN scFv | MSLN-His tag protein | 2023 | Therapy | (109) |
5. Future directions and challenges
5.1. Advances and applications in obtaining targeted phage-peptides
Utilizing a phenotypic library screening strategy enables the identification of ovarian cancer-specific surface markers in a non-biased fashion, capturing physiologically pertinent targets. The application of single-domain antibody libraries, notably VHH libraries, facilitates the unveiling of targets with diverse expression profiles. This method’s adaptability lends itself well to various detection and therapeutic modalities. Notably, Her2 and BCAM have been pinpointed as promising candidates for targeted ovarian cancer therapy. Her2, in particular, merits further investigation within specific ovarian cancer subsets. BCAM, distinguished by its pronounced expression in high-grade serous ovarian carcinoma (HGSOC) and minimal presence in normal tissue, emerges as a compelling target for precision therapy endeavors. Moreover, the integration of phage display-derived antibodies with chemotherapy agents, such as paclitaxel, suggests a pathway to enhancing treatment outcomes while mitigating additional toxicity. The principal benefits of employing phage display for targeted phage-peptide acquisition include its efficiency and cost-effectiveness (121).
5.2. Current limitations and obstacles
In phage display systems, phage-peptides are showcased by coupling them with the coat proteins of filamentous bacteriophages. Yet, the diminutive size of these phages imposes limitations on the magnitude of proteins that can be displayed (122). Short phage-peptides, due to their linear and compact nature, often demonstrate restrained affinity, which can hamper their efficacy in target binding. This limitation poses potential obstacles in leveraging these phage-peptides as therapeutic agents (50). Additionally, the complex lifecycle of bacteriophages introduces challenges in showcasing post-translationally modified proteins or phage-peptides, such as those undergoing phosphorylation and glycosylation, on the phage surface. These modifications are crucial for protein folding, interactions, and signal transduction, thereby limiting the widespread application of phage display in these essential functions (123). The effectiveness of a bacteriophage display library, characterized by the variety and representation of displayed proteins or phage-peptides, plays a critical role in its practical utility. Ongoing investigations in this field are poised to explore novel avenues in science and medicine, potentially fostering breakthroughs in ovarian cancer treatment and other areas. Phage display, a leading platform for antibody discovery, is robust and excels in quickly screening extensive libraries for high-affinity binders. However, it struggles with post-translational modifications and complex antibody structures. Integrating phage display with yeast and mammalian technologies helps overcome these limits. Yeast display uses eukaryotic systems for accurate protein folding and modifications, while mammalian display presents antibodies on mammalian cells, ensuring human-like modifications critical for therapeutic use. This integration enhances antibody properties like stability and pharmacokinetics, and facilitates the discovery of therapeutically effective antibodies. By merging these technologies, researchers can produce superior antibodies more suited for therapeutic use, speeding the development of advanced antibody therapies.
5.3. Enhancing the phage display platform in therapy
Phage display technology offers a versatile toolkit for a broad spectrum of applications, from material science to the development of therapeutic agents (124). This method boasts several advantages over alternative non-phage systems, such as facilitating the discovery of antibodies in nonphysiological settings, identifying pH-sensitive antibodies with distinct binding characteristics, and generating recombinant antibodies against potent toxins. Despite its successes, the technology faces hurdles, including the presentation of hydrophobic phage-peptides on the phage surface and the propensity for recombinant proteins to aggregate. Advancements in phage display techniques are sought to address these challenges, with the investigation of thermophilic bacteriophages presenting promising avenues (125). The awarding of the Nobel Prize in Chemistry in 2018 to phage display technology underscores its pivotal role and achievements in identifying high-affinity phage-peptides from complex libraries. Within the realm of ovarian cancer therapy, this review explores the advancements in phage display methodologies aimed at isolating cancer-specific ligands through varied screening processes. These developments highlight the technology’s potential to refine targeted therapeutic strategies in clinical settings.
6. Conclusion
Ovarian cancer represents a significant contributor to female mortality worldwide, yet the availability of specific and high-affinity agents for its early detection remains scarce. Through the identification of phage-peptides that bind selectively and strongly to ovarian cancer cells, phage display technology facilitates the swift isolation and recognition of these cells, enhancing the speed and efficiency of immunodiagnostics. This method also holds promise for utilizing phage-peptides as precision targeting agents in ovarian cancer treatment. Furthermore, phage display aids in uncovering novel oncogenic targets, such as BCAM, offering promising avenues for targeted interventions in high-grade serous ovarian cancers. This technology establishes a versatile foundation for the identification of cancer-specific targets and antibodies, contributing significantly to the advancement of targeted therapy strategies.
Author contributions
YL: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. X-ML: Methodology, Visualization, Software, Writing – original draft. K-DY: Conceptualization, Data curation, Investigation, Software, Writing – original draft. W-HT: Investigation, Software, Supervision, Writing – review & editing.
Funding Statement
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported by Scientific Research Program of Jilin Provincial Department of Education (No.JJKH20231194KJ); Research Project of Undergraduate Teaching Reform in Jilin University (No. 2021XZC087); Research Projects of Higher Education in Jilin Province (No. JGJX2021D53).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- 1. Ottevanger PB. Ovarian cancer stem cells more questions than answers. Semin Cancer Biol. (2017) 44:67–71. doi: 10.1016/j.semcancer.2017.04.009 [DOI] [PubMed] [Google Scholar]
- 2. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. (2021) 71:209–49. doi: 10.3322/caac.21660 [DOI] [PubMed] [Google Scholar]
- 3. Cabasag CJ, Fagan PJ, Ferlay J, Vignat J, Laversanne M, Liu L, et al. Ovarian cancer today and tomorrow: A global assessment by world region and Human Development Index using GLOBOCAN 2020. Int J Cancer. (2022) 151:1535–41. doi: 10.1002/ijc.34002 [DOI] [PubMed] [Google Scholar]
- 4. Zhang M, Cheng S, Jin Y, Zhao Y, Wang Y. Roles of CA125 in diagnosis, prediction, and oncogenesis of ovarian cancer Biochim biophys acta, rev cancer. Biochim Biophys Acta Rev Cancer. (2021) 1875(2):188503. doi: 10.1016/j.bbcan.2021.188503 [DOI] [PubMed] [Google Scholar]
- 5. Hurwitz LM, Pinsky PF, Trabert B. General population screening for ovarian cancer. Lancet. (2021) 397:2128–30. doi: 10.1016/s0140-6736(21)01061-8 [DOI] [PubMed] [Google Scholar]
- 6. Wang Q, Peng H, Qi X, Wu M, Zhao X. Targeted therapies in gynecological cancers: a comprehensive review of clinical evidence. Signal Transduct Target. (2020) 5:137. doi: 10.1038/s41392-020-0199-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Voelker R. Pursuing an effective ovarian cancer vaccine. JAMA. (2018) 320:858–60. doi: 10.1001/jama.2018.10822 [DOI] [PubMed] [Google Scholar]
- 8. Shen W, Shi P, Dong Q, Zhou X, Chen C, Sui X, et al. Discovery of a novel dual-targeting D-peptide to block CD24/Siglec-10 and PD-1/PD-L1 interaction and synergize with radiotherapy for cancer immunotherapy. J Immunother Cancer. (2023) 11(6):e007068. doi: 10.1136/jitc-2023-007068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Solomon PE, Bracken CJ, Carozza JA, Wang H, Young EP, Wellner A, et al. Discovery of VH domains that allosterically inhibit ENPP1. Nat Chem Biol. (2024) 20(1):30–41. doi: 10.1038/s41589-023-01368-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lemire S, Yehl KM, Lu TK. Phage-based applications in synthetic biology. Annu Rev Virol. (2018) 5:453–76. doi: 10.1146/annurev-virology-092917-043544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jaroszewicz W, Morcinek-Orłowska J, Pierzynowska K, Gaffke L, Węgrzyn G. Phage display and other peptide display technologies. FEMS Microbiol Rev. (2022) 46(2):fuab052. doi: 10.1093/femsre/fuab052 [DOI] [PubMed] [Google Scholar]
- 12. Chang C, Guo W, Yu X, Guo C, Zhou N, Guo X, et al. Engineered M13 phage as a novel therapeutic bionanomaterial for clinical applications: From tissue regeneration to cancer therapy. Materials Today Bio. (2023) 20:100612. doi: 10.1016/j.mtbio.2023.100612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Shivachandra SB, Li Q, Peachman KK, Matyas GR, Leppla SH, Alving CR, et al. Multicomponent anthrax toxin display and delivery using bacteriophage T4. Vaccine. (2007) 25:1225–35. doi: 10.1016/j.vaccine.2006.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tao P, Zhu J, Mahalingam M, Batra H, Rao VB. Bacteriophage T4 nanoparticles for vaccine delivery against infectious diseases. Adv Drug Delivery Rev. (2019) 145:57–72. doi: 10.1016/j.addr.2018.06.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Piggott AM, Karuso P. Identifying the cellular targets of natural products using T7 phage display. Nat Prod Rep. (2016) 33:626–36. doi: 10.1039/c5np00128e [DOI] [PubMed] [Google Scholar]
- 16. Yu T, Sun Z, Cao X, Pang Q, Deng H. Recent trends in T7 phage application in diagnosis and treatment of various diseases. Int Immunopharmacol. (2022) 110:109071. doi: 10.1016/j.intimp.2022.109071 [DOI] [PubMed] [Google Scholar]
- 17. Yue H, Li Y, Yang M, Mao C. T7 phage as an emerging nanobiomaterial with genetically tunable target specificity. Adv Sci (Weinh). (2022) 9:e2103645. doi: 10.1002/advs.202103645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hess KL, Jewell CM. Phage display as a tool for vaccine and immunotherapy development. Bioeng Transl Med. (2020) 5:e10142. doi: 10.1002/btm2.10142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jaroszewicz W, Morcinek-Orlowska J, Pierzynowska K, Gaffke L, Wegrzyn G. Phage display and other peptide display technologies [Review]. FEMS Microbiol Rev. (2022) 46(2):fuab052. doi: 10.1093/femsre/fuab052 [DOI] [PubMed] [Google Scholar]
- 20. Kumar R, Parray HA, Shrivastava T, Sinha S, Luthra K. Phage display antibody libraries: A robust approach for generation of recombinant human monoclonal antibodies. Int J Biol Macromol. (2019) 135:907–18. doi: 10.1016/j.ijbiomac.2019.06.006 [DOI] [PubMed] [Google Scholar]
- 21. Zhao H, Nie D, Hu Y, Chen Z, Hou Z, Li M, et al. Phage display-derived peptides and antibodies for bacterial infectious diseases therapy and diagnosis [Review]. Molecules. (2023) 28(6):2621. doi: 10.3390/molecules28062621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zheng M, Haeffner F, Gao J. N-Terminal cysteine mediated backbone-side chain cyclization for chemically enhanced phage display. Chem Sci. (2022) 13:8349–54. doi: 10.1039/d2sc03241d [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Guliy OI, Evstigneeva SS, Dykman LA. Recombinant antibodies by phage display for bioanalytical applications. Biosens Bioelectron. (2023) 222:114909. doi: 10.1016/j.bios.2022.114909 [DOI] [PubMed] [Google Scholar]
- 24. Wang XS, Chen PC, Hampton JT, Tharp JM, Reed CA, Das SK, et al. A genetically encoded, phage-displayed cyclic-peptide library. Angew Chem Int Ed Engl. (2019) 58:15904–9. doi: 10.1002/anie.201908713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Almagro JC, Escalona MP, Arrieta HI, Pérez-Tapia SM. Phage display libraries for antibody therapeutic discovery and development. Antibodies (Basel). (2019) 8:44. doi: 10.3390/antib8030044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ledsgaard L, Ljungars A, Rimbault C, Sørensen CV, Tulika T, Wade J, et al. Advances in antibody phage display technology. Drug Discovery Today. (2022) 27:2151–69. doi: 10.1016/j.drudis.2022.05.002 [DOI] [PubMed] [Google Scholar]
- 27. Dantas-Barbosa C, De Macedo Brigido M, Maranhao AQ. Antibody phage display libraries: contributions to oncology. Int J Mol Sci. (2012) 13:5420–40. doi: 10.3390/ijms13055420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yang Y, Nian S, Li L, Wen X, Liu Q, Zhang B, et al. Fully human recombinant antibodies against EphA2 from a multi-tumor patient immune library suitable for tumor-targeted therapy. Bioengineered. (2021) 12:10379–400. doi: 10.1080/21655979.2021.1996807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Takagi-Maeda S, Yajima S, Suzuki T, Usami K, Takahashi N, Niwa R, et al. Novel cancer-specific epidermal growth factor receptor antibody obtained from the serum of esophageal cancer patients with long-termsurvival. CancerSci. (2022) 113:2118–28. doi: 10.1111/cas.15350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Laustsen AH, Greiff V, Karatt-Vellatt A, Muyldermans S, Jenkins TP. Animal immunization, in vitro display technologies, and machine learning for antibody discovery. Trends Biotechnol. (2021) 39:1263–73. doi: 10.1016/j.tibtech.2021.03.003 [DOI] [PubMed] [Google Scholar]
- 31. Hu Y, Romão E, Vincke C, Brys L, Elkrim Y, Vandevenne M, et al. Intrabody targeting HIF-1α Mediates transcriptional downregulation of target genes related to solid tumors. Int J Mol Sci. (2021) 22(22):12335. doi: 10.3390/ijms222212335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Liu Y, Ao K, Bao F, Cheng Y, Hao Y, Zhang H, et al. Development of a bispecific nanobody targeting CD20 on B-cell lymphoma cells and CD3 on T cells. Vaccines (Basel). (2022) 10(8):1335. doi: 10.3390/vaccines10081335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhao Y, Li Y, Wu X, Li L, Liu J, Wang Y, et al. Identification of anti-CD16a single domain antibodies and their application in bispecific antibodies. Cancer Biol Ther. (2020) 21:72–80. doi: 10.1080/15384047.2019.1665953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Roshan R, Naderi S, Behdani M, Cohan RA, Ghaderi H, Shokrgozar MA, et al. Isolation and characterization of nanobodies against epithelial cell adhesion molecule as novel theranostic agents for cancer therapy. Mol Immunol. (2021) 129:70–7. doi: 10.1016/j.molimm.2020.10.021 [DOI] [PubMed] [Google Scholar]
- 35. Manoutcharian K, Gevorkian G. Shark VNAR phage display libraries: An alternative source for therapeutic and diagnostic recombinant antibody fragments. Fish Shellfish Immunol. (2023) 138:108808. doi: 10.1016/j.fsi.2023.108808 [DOI] [PubMed] [Google Scholar]
- 36. Liu H, Liu Y, Zhao Z, Li Y, Mustafa B, Chen Z, et al. Discovery of anti-PD-L1 human domain antibodies for cancer immunotherapy. Front Immunol. (2022) 13:838966. doi: 10.3389/fimmu.2022.838966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ahmadi M, Ahmadyousefi Y, Salimi Z, Mirzaei R, Najafi R, Amirheidari B, et al. Innovative diagnostic peptide-based technologies for cancer diagnosis: focus on EGFR-targeting peptides. Chemmedchem. (2023) 18:e202200506. doi: 10.1002/cmdc.202200506 [DOI] [PubMed] [Google Scholar]
- 38. Rahbarnia L, Farajnia S, Babaei H, Majidi J, Dariushnejad H, Hosseini MK. Isolation and characterization of a novel human scFv inhibiting EGFR vIII expressing cancers. Immunol Lett. (2016) 180:31–8. doi: 10.1016/j.imlet.2016.10.008 [DOI] [PubMed] [Google Scholar]
- 39. Xi X, Sun W, Su H, Zhang X, Sun F. Identification of a novel anti-EGFR nanobody by phage display and its distinct paratope and epitope via homology modeling and molecular docking. Mol Immunol. (2020) 128:165–74. doi: 10.1016/j.molimm.2020.10.014 [DOI] [PubMed] [Google Scholar]
- 40. Lai YD, Wu YY, Tsai YJ, Tsai YS, Lin YY, Lai SL, et al. Generation of potent anti-vascular endothelial growth factor neutralizing antibodies from mouse phage display library for cancer therapy. Int J Mol Sci. (2016) 17:214. doi: 10.3390/ijms17020214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Petrenko VA. Landscape phage: evolution from phage display to nanobiotechnology. Viruses. (2018) 10(6):311. doi: 10.3390/v10060311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gillespie JW, Yang L, De Plano LM, Stackhouse MA, Petrenko VA. Evolution of a landscape phage library in a mouse xenograft model of human breast cancer. Viruses. (2019) 11(11):988. doi: 10.3390/v11110988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Han L, Xia H, Yin L, Petrenko VA, Liu A. Selected landscape phage probe as selective recognition interface for sensitive total prostate-specific antigen immunosensor. Biosens Bioelectron. (2018) 106:1–6. doi: 10.1016/j.bios.2018.01.046 [DOI] [PubMed] [Google Scholar]
- 44. Gallo E, Kelil A, Bayliss PE, Jeganathan A, Egorova O, Ploder L, et al. In situ antibody phage display yields optimal inhibitors of integrin α11/β1. MAbs. (2020) 12:1717265. doi: 10.1080/19420862.2020.1717265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Asar MC, Franco A, Soendergaard M. Phage display selection, identification, and characterization of novel pancreatic cancer targeting peptides. Biomolecules. (2020) 10(5):714. doi: 10.3390/biom10050714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Pleiko K, Põšnograjeva K, Haugas M, Paiste P, Tobi A, Kurm K, et al. In vivo phage display: identification of organ-specific peptides using deep sequencing and differential profiling across tissues. Nucleic Acids Res. (2021) 49:e38. doi: 10.1093/nar/gkaa1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. André AS, Moutinho I, Dias JNR, Aires-da-Silva F. In vivo Phage Display: A promising selection strategy for the improvement of antibody targeting and drug delivery properties. Front Microbiol. (2022) 13:962124. doi: 10.3389/fmicb.2022.962124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sørensen MD, Kristensen P. Selection of antibodies against a single rare cell present in a heterogeneous population using phage display. Nat Protoc. (2011) 6:509–22. doi: 10.1038/nprot.2011.311 [DOI] [PubMed] [Google Scholar]
- 49. Krag DN, Shukla GS, Shen GP, Pero S, Ashikaga T, Weaver DL, et al. Selection of tumor-binding ligands in cancer patients with phage display libraries. Cancer Res. (2006) 66:7724–33. doi: 10.1158/0008-5472.Can-05-4441 [DOI] [PubMed] [Google Scholar]
- 50. Saw PE, Song EW. Phage display screening of therapeutic peptide for cancer targeting and therapy. Protein Cell. (2019) 10:787–807. doi: 10.1007/s13238-019-0639-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. An D, Banerjee S, Lee JM. Recent advancements of antiangiogenic combination therapies in ovarian cancer. Cancer Treat Rev. (2021) 98:102224. doi: 10.1016/j.ctrv.2021.102224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Davies M, Davey MG, Miller N. The potential of microRNAs as clinical biomarkers to aid ovarian cancer diagnosis and treatment. Genes. (2022) 13(11):2054. doi: 10.3390/genes13112054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Shaik B, Zafar T, Balasubramanian K, Gupta SP. An overview of ovarian cancer: molecular processes involved and development of target-based chemotherapeutics. Curr Top Med Chem. (2021) 21:329–46. doi: 10.2174/1568026620999201111155426 [DOI] [PubMed] [Google Scholar]
- 54. Shimizu A, Sawada K, Kimura T. Pathophysiological role and potential therapeutic exploitation of exosomes in ovarian cancer. Cells. (2020) 9(4):814. doi: 10.3390/cells9040814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Menon U, Karpinskyj C, Gentry-Maharaj A. Ovarian cancer prevention and screening. Obstet Gynecol. (2018) 131:909–27. doi: 10.1097/aog.0000000000002580 [DOI] [PubMed] [Google Scholar]
- 56. Garcia J, Hurwitz HI, Sandler AB, Miles D, Coleman RL, Deurloo R, et al. Bevacizumab (Avastin®) in cancer treatment: A review of 15 years of clinical experience and future outlook. Cancer Treat Rev. (2020) 86:102017. doi: 10.1016/j.ctrv.2020.102017 [DOI] [PubMed] [Google Scholar]
- 57. Khetan R, Dharmayanti C, Gillam TA, Kübler E, Hoffmann MK, Ricciardelli C, et al. Using GPCRs as molecular beacons to target ovarian cancer with nanomedicines. Cancers (Basel). (2022) 14:2362. doi: 10.3390/cancers14102362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Lin Y, Zhou XT, Ni YH, Zhao X, Liang X. Metabolic reprogramming of the tumor immune microenvironment in ovarian cancer: A novel orientation for immunotherapy. Front Immunol. (2022) 13:1030831. doi: 10.3389/fimmu.2022.1030831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Barani M, Bilal M, Sabir F, Rahdar A, Kyzas GZ. Nanotechnology in ovarian cancer: Diagnosis and treatment. Life Sci. (2021) 266:118914. doi: 10.1016/j.lfs.2020.118914 [DOI] [PubMed] [Google Scholar]
- 60. Mao G, Xin D, Wang Q, Lai D. Sodium molybdate inhibits the growth of ovarian cancer cells via inducing both ferroptosis and apoptosis. Free Radic Biol Med. (2022) 182:79–92. doi: 10.1016/j.freeradbiomed.2022.02.023 [DOI] [PubMed] [Google Scholar]
- 61. Jorgensen K, Melamed A, Wu CF, Nitecki R, Pareja R, Fagotti A, et al. Minimally invasive interval debulking surgery for advanced ovarian cancer after neoadjuvant chemotherapy. Gynecol Oncol. (2023) 172:130–7. doi: 10.1016/j.ygyno.2023.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Armstrong DK, Alvarez RD, Bakkum-Gamez JN, Barroilhet L, Behbakht K, Berchuck A, et al. Ovarian cancer, version 2.2020, NCCN clinical practice guidelines in oncology. J Natl Compr Canc Netw. (2021) 19:191–226. doi: 10.6004/jnccn.2021.0007 [DOI] [PubMed] [Google Scholar]
- 63. Stewart C, Ralyea C, Lockwood S. Ovarian cancer: an integrated review. Semin Oncol Nurs. (2019) 35:151–6. doi: 10.1016/j.soncn.2019.02.001 [DOI] [PubMed] [Google Scholar]
- 64. O'Malley DM. New therapies for ovarian cancer. J Natl Compr Canc Netw. (2019) 17:619–21. doi: 10.6004/jnccn.2019.5018 [DOI] [PubMed] [Google Scholar]
- 65. Luvero D, Plotti F, Aloisia A, Montera R, Terranova C, Carlo De Cicco N, et al. Ovarian cancer relapse: From the latest scientific evidence to the best practice. Crit Rev Oncol Hematol. (2019) 140:28–38. doi: 10.1016/j.critrevonc.2019.05.014 [DOI] [PubMed] [Google Scholar]
- 66. Tewari KS, Burger RA, Enserro D, Norquist BM, Swisher EM, Brady MF, et al. Final overall survival of a randomized trial of bevacizumab for primary treatment of ovarian cancer. J Clin Oncol. (2019) 37:2317–28. doi: 10.1200/jco.19.01009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Lheureux S, Braunstein M, Oza AM. Epithelial ovarian cancer: Evolution of management in the era of precision medicine. CA Cancer J Clin. (2019) 69:280–304. doi: 10.3322/caac.21559 [DOI] [PubMed] [Google Scholar]
- 68. Ruiz M, Zhang N, Sood AK, An Z. Antibody therapeutics for epithelial ovarian cancer. Expert Opin Biol Ther. (2022) 22:1379–91. doi: 10.1080/14712598.2022.2141565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Kuroki L, Guntupalli SR. Treatment of epithelial ovarian cancer. BMJ. (2020) 371:m3773. doi: 10.1136/bmj.m3773 [DOI] [PubMed] [Google Scholar]
- 70. Guan LY, Lu Y. New developments in molecular targeted therapy of ovarian cancer. Discovery Med. (2018) 26:219–29. [PubMed] [Google Scholar]
- 71. González-Martín A, Pothuri B, Vergote I, DePont Christensen R, Graybill W, Mirza MR, et al. Niraparib in patients with newly diagnosed advanced ovarian cancer. N Engl J Med. (2019) 381:2391–402. doi: 10.1056/NEJMoa1910962 [DOI] [PubMed] [Google Scholar]
- 72. Bellio C, DiGloria C, Foster R, James K, Konstantinopoulos PA, Growdon WB, et al. PARP inhibition induces enrichment of DNA repair-proficient CD133 and CD117 positive ovarian cancer stem cells. Mol Cancer Res. (2019) 17:431–45. doi: 10.1158/1541-7786.Mcr-18-0594 [DOI] [PubMed] [Google Scholar]
- 73. Ray U, Jung DB, Jin L, Xiao Y, Dasari S, Sarkar Bhattacharya S, et al. Targeting LRRC15 inhibits metastatic dissemination of ovarian cancer. Cancer Res. (2022) 82:1038–54. doi: 10.1158/0008-5472.Can-21-0622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Yeung TL, Leung CS, Yip KP, Sheng J, Vien L, Bover LC, et al. Anticancer immunotherapy by MFAP5 blockade inhibits fibrosis and enhances chemosensitivity in ovarian and pancreatic cancer. Clin Cancer Res. (2019) 25:6417–28. doi: 10.1158/1078-0432.Ccr-19-0187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Fabre M, Ferrer C, Domínguez-Hormaetxe S, Bockorny B, Murias L, Seifert O, et al. OMTX705, a novel FAP-targeting ADC demonstrates activity in chemotherapy and pembrolizumab-resistant solid tumor models. Clin Cancer Res. (2020) 26:3420–30. doi: 10.1158/1078-0432.Ccr-19-2238 [DOI] [PubMed] [Google Scholar]
- 76. Jones BA, Varambally S, Arend RC. Histone methyltransferase EZH2: A therapeutic target for ovarian cancer. Mol Cancer Ther. (2018) 17:591–602. doi: 10.1158/1535-7163.Mct-17-0437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Zheng A, Wei Y, Zhao Y, Zhang T, Ma X. The role of cancer-associated mesothelial cells in the progression and therapy of ovarian cancer. Front Immunol. (2022) 13:1013506. doi: 10.3389/fimmu.2022.1013506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Gulia M, Nishal S, Maddiboyina B, Dutt R, Desu PK, Wadhwa R, et al. : Physiological Pathway, diagnosis and nanotechnology based treatment strategies for ovarian Cancer: A review. Med Omics Volume. (2023) 8:100020. doi: 10.1016/j.meomic.2023.100020 [DOI] [Google Scholar]
- 79. Rajtak A, Leśko MO, Żak K, Tarkowski R, Kotarski J, Okła K. Integration of local and systemic immunity in ovarian cancer: Implications for immunotherapy. Front Immunol. (2022) 13:1018256. doi: 10.3389/fimmu.2022.1018256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Tanyi JL, Bobisse S, Ophir E, Tuyaerts S, Roberti A, Genolet R, et al. Personalized cancer vaccine effectively mobilizes antitumor T cell immunity in ovarian cancer. Sci Transl Med. (2018) 10(436):eaao5931. doi: 10.1126/scitranslmed.aao5931 [DOI] [PubMed] [Google Scholar]
- 81. Odunsi K. Immunotherapy in ovarian cancer. Ann Oncol. (2017) 28:viii1–7. doi: 10.1093/annonc/mdx444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Schoutrop E, El-Serafi I, Poiret T, Zhao Y, Gultekin O, He R, et al. Mesothelin-specific CAR T cells target ovarian cancer. Cancer Res. (2021) 81:3022–35. doi: 10.1158/0008-5472.Can-20-2701 [DOI] [PubMed] [Google Scholar]
- 83. Liang Z, Dong J, Yang N, Li SD, Yang ZY, Huang R, et al. Tandem CAR-T cells targeting FOLR1 and MSLN enhance the antitumor effects in ovarian cancer. Int J Biol Sci. (2021) 17:4365–76. doi: 10.7150/ijbs.63181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Sánchez-Paulete AR, Mateus-Tique J, Mollaoglu G, Nielsen SR, Marks A, Lakshmi A, et al. Targeting macrophages with CAR T cells delays solid tumor progression and enhances antitumor immunity. Cancer Immunol Res. (2022) 10:1354–69. doi: 10.1158/2326-6066.Cir-21-1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Tian L, Xu B, Teng KY, Song M, Zhu Z, Chen Y, et al. Targeting fc receptor-mediated effects and the "Don't eat me" Signal with an oncolytic virus expressing an anti-CD47 antibody to treat metastatic ovarian cancer. Clin Cancer Res. (2022) 28:201–14. doi: 10.1158/1078-0432.Ccr-21-1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Hansen JM, Coleman RL, Sood AK. Targeting the tumour microenvironment in ovarian cancer. Eur J Cancer. (2016) 56:131–43. doi: 10.1016/j.ejca.2015.12.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Zeng Y, Li B, Liang Y, Reeves PM, Qu X, Ran C, et al. Dual blockade of CXCL12-CXCR4 and PD-1-PD-L1 pathways prolongs survival of ovarian tumor-bearing mice by prevention of immunosuppression in the tumor microenvironment. FASEB J. (2019) 33:6596–608. doi: 10.1096/fj.201802067RR [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Huang TT, Lampert EJ, Coots C, Lee JM. Targeting the PI3K pathway and DNA damage response as a therapeutic strategy in ovarian cancer. Cancer Treat Rev. (2020) 86:102021. doi: 10.1016/j.ctrv.2020.102021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Yang C, Xia BR, Zhang ZC, Zhang YJ, Lou G, Jin WL. Immunotherapy for ovarian cancer: adjuvant, combination, and neoadjuvant. Front Immunol. (2020) 11:577869. doi: 10.3389/fimmu.2020.577869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Lampert EJ, Zimmer A, Padget M, Cimino-Mathews A, Nair JR, Liu Y, et al. Combination of PARP inhibitor olaparib, and PD-L1 inhibitor durvalumab, in recurrent ovarian cancer: a proof-of-concept phase II study. Clin Cancer Res. (2020) 26:4268–79. doi: 10.1158/1078-0432.Ccr-20-0056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Ott C, Ainciburu M, Parent C, Michel S, Roesch C, Vourc'h P, et al. Development of monoclonal antibodies to human kallikrein-related peptidase 6 (KLK6) and their use in an immunofluorometric assay for free KLK6. Biol Chem. (2014) 395:1119–26. doi: 10.1515/hsz-2014-0133 [DOI] [PubMed] [Google Scholar]
- 92. Zhou C, Kang J, Wang X, Wei W, Jiang W. Phage display screening identifies a novel peptide to suppress ovarian cancer cells in vitro and in vivo in mouse models. BMC Cancer. (2015) 15:889. doi: 10.1186/s12885-015-1891-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Wang CH, Weng CH, Che YJ, Wang K, Lee GB. Cancer cell-specific oligopeptides selected by an integrated microfluidic system from a phage display library for ovarian cancer diagnosis. Theranostics. (2015) 5:431–42doi: 10.7150/thno.10891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Davidson TA, McGoldrick SJ, Kohn DH. Phage display to augment biomaterial function. Int J Mol Sci. (2020) 21(17):5994. doi: 10.3390/ijms21175994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Newman MR, Benoit DSW. In vivo translation of peptide-targeted drug delivery systems discovered by phage display. Bioconjug Chem. (2018) 29:2161–9. doi: 10.1021/acs.bioconjchem.8b00285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Ma C, Yin G, Yan D, He X, Zhang L, Wei Y, et al. A novel peptide specifically targeting ovarian cancer identified by in vivo phage display. J Pept Sci. (2013) 19:730–6. doi: 10.1002/psc.2555 [DOI] [PubMed] [Google Scholar]
- 97. Wang X, Wang F, Li S, Yin G, Pu X. Preparation and in vitro evaluation of thermosensitive liposomes targeting ovarian cancer. Curr Drug Deliv. (2022) 19:940–8. doi: 10.2174/1567201819666220321110812 [DOI] [PubMed] [Google Scholar]
- 98. Shen YA, Liu CS, Chang YH, Chen PH, He CL, Wu HC, et al. Subtype-specific binding peptides enhance the therapeutic efficacy of nanomedicine in the treatment of ovarian cancer. Cancer Lett. (2015) 360:39–47. doi: 10.1016/j.canlet.2015.01.042 [DOI] [PubMed] [Google Scholar]
- 99. Deutscher SL. Phage display in molecular imaging and diagnosis of cancer. Chem Rev. (2010) 110:3196–211. doi: 10.1021/cr900317f [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Wang L, Hu Y, Li W, Wang F, Lu X, Han X, et al. Identification of a peptide specifically targeting ovarian cancer by the screening of a phage display peptide library. Oncol Lett. (2016) 11:4022–6. doi: 10.3892/ol.2016.4549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Xing L, Xu Y, Sun K, Wang H, Zhang F, Zhou Z, et al. Identification of a peptide for folate receptor alpha by phage display and its tumor targeting activity in ovary cancer xenograft. Sci Rep. (2018) 8:8426. doi: 10.1038/s41598-018-26683-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Asar M, Newton-Northup J, Deutscher S, Soendergaard M. Ovarian cancer targeting phage for in vivo near-infrared optical imaging. Diagnostics (Basel). (2019) 9. doi: 10.3390/diagnostics9040183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Soendergaard M, Newton-Northup JR, Deutscher SL. In vivo phage display selection of an ovarian cancer targeting peptide for SPECT/CT imaging. Am J Nucl Med Mol Imaging. (2014) 4:561–70. [PMC free article] [PubMed] [Google Scholar]
- 104. Faintuch BL, Oliveira EA, Targino RC, Moro AM. Radiolabeled NGR phage display peptide sequence for tumor targeting. Appl Radiat Isot. (2014) 86:41–5. doi: 10.1016/j.apradiso.2013.12.035 [DOI] [PubMed] [Google Scholar]
- 105. Babeker H, Ketchemen JP, Annan Sudarsan A, Andrahennadi S, Tikum AF, Nambisan AK, et al. Engineering of a fully human anti-MUC-16 antibody and evaluation as a PET imaging agent. Pharmaceutics. (2022) 14(12):2824. doi: 10.3390/pharmaceutics14122824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Yu J, Guo Y, Gu Y, Li F, Song H, Nian R, et al. Targeting and neutralizing human epididymis protein 4 by novel nanobodies to suppress ovarian cancer cells and attenuate cisplatin resistance. Int J Biol Macromol. (2022) 199:298–306. doi: 10.1016/j.ijbiomac.2022.01.015 [DOI] [PubMed] [Google Scholar]
- 107. Huang YS, Wang TX, Lin XM, Wang H, Li RZ, Zeng X, et al. Antitumor effect of a short peptide on p53-null SKOV3 ovarian cancer cells. Anticancer Drugs. (2019) 30:973–82. doi: 10.1097/cad.0000000000000830 [DOI] [PubMed] [Google Scholar]
- 108. Pu X, Ma C, Yin G, You F, Wei Y. Cell adhesion and invasion inhibitory effect of an ovarian cancer targeting peptide selected via phage display in vivo . Biochem Biophys Res Commun. (2014) 443:858–63. doi: 10.1016/j.bbrc.2013.12.058 [DOI] [PubMed] [Google Scholar]
- 109. Chen J, Hu J, Gu L, Ji F, Zhang F, Zhang M, et al. Anti-mesothelin CAR-T immunotherapy in patients with ovarian cancer. Cancer Immunol Immunother. (2023) 72:409–25doi: 10.1007/s00262-022-03238-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Qiao C, Lv M, Li X, Geng J, Li Y, Zhang J, et al. Affinity maturation of antiHER2 monoclonal antibody MIL5 using an epitope-specific synthetic phage library by computational design. J Biomol Struct Dyn. (2013) 31:511–21. doi: 10.1080/07391102.2012.706073 [DOI] [PubMed] [Google Scholar]
- 111. Bortot B, Apollonio M, Baj G, Andolfi L, Zupin L, Crovella S, et al. Advanced photodynamic therapy with an engineered M13 phage targeting EGFR: Mitochondrial localization and autophagy induction in ovarian cancer cell lines. Free Radic Biol Med. (2022) 179:242–51. doi: 10.1016/j.freeradbiomed.2021.11.019 [DOI] [PubMed] [Google Scholar]
- 112. Zhang L, Yin G, Yan D, Wei Y, Ma C, Huang Z, et al. In vitro screening of ovarian tumor specific peptides from a phage display peptide library. Biotechnol Lett. (2011) 33:1729–35. doi: 10.1007/s10529-011-0634-4 [DOI] [PubMed] [Google Scholar]
- 113. Vallen MJ, van Tilborg AA, Tesselaar MH, ten Dam GB, Bulten J, van Kuppevelt TH, et al. Novel single-chain antibody GD3A10 defines a chondroitin sulfate biomarker for ovarian cancer. biomark Med. (2014) 8:699–711. doi: 10.2217/bmm.14.6 [DOI] [PubMed] [Google Scholar]
- 114. Yang C, He X, Liu X, Tang Z, Liang X. OSTP as a novel peptide specifically targeting human ovarian cancer. Oncol Rep. (2015) 34:972–8. doi: 10.3892/or.2015.4066 [DOI] [PubMed] [Google Scholar]
- 115. van der Steen SC, van Tilborg AA, Vallen MJ, Bulten J, van Kuppevelt TH, Massuger LF. Prognostic significance of highly sulfated chondroitin sulfates in ovarian cancer defined by the single chain antibody GD3A11. Gynecol Oncol. (2016) 140:527–36. doi: 10.1016/j.ygyno.2015.12.024 [DOI] [PubMed] [Google Scholar]
- 116. Antony F, Deantonio C, Cotella D, Soluri MF, Tarasiuk O, Raspagliesi F, et al. High-throughput assessment of the antibody profile in ovarian cancer ascitic fluids. Oncoimmunology. (2019) 8:e1614856. doi: 10.1080/2162402x.2019.1614856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Zhao S, Li C, Gao Y, Qian L, Dong J, Zhai L, et al. Screening and identification of a specific binding peptide to ovarian cancer cells from a phage-displayed peptide library. Int J Pept Res Ther. (2021) 27:1741–9. doi: 10.1007/s10989-021-10206-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Martin AL, Anadon CM, Biswas S, Mine JA, Handley KF, Payne KK, et al. Olfactory receptor OR2H1 is an effective target for CAR T cells in human epithelial tumors. Mol Cancer Ther. (2022) 21:1184–94. doi: 10.1158/1535-7163.Mct-21-0872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Lü P, Qiu S, Pan Y, Shi S, Yu Q, Yu F, et al. Discovery of an heparin-binding epidermal growth factor domain antibody from a phage library and analysis of its inhibitory effects in SKOV3 cells. Cancer Biother Radiopharm. (2023) 38(8):572–9. doi: 10.1089/cbr.2021.0123 [DOI] [PubMed] [Google Scholar]
- 120. Shen Y, Ruan L, Lian C, Li R, Tu Z, Liu H. Discovery of HB-EGF binding peptides and their functional characterization in ovarian cancer cell lines. Cell Death Discovery. (2019) 5:82. doi: 10.1038/s41420-019-0163-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Wang J, Masehi-Lano JJ, Chung EJ. Peptide and antibody ligands for renal targeting: nanomedicine strategies for kidney disease. Biomaterials science. (2017) 5:1450–9. doi: 10.1039/c7bm00271h [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Mahdavi SZB, Oroojalian F, Eyvazi S, Hejazi M, Baradaran B, Pouladi N, et al. An overview on display systems (phage, bacterial, and yeast display) for production of anticancer antibodies; advantages and disadvantages. Int J Biol Macromol. (2022) 208:421–42. doi: 10.1016/j.ijbiomac.2022.03.113 [DOI] [PubMed] [Google Scholar]
- 123. Qi H, Ma M, Lai D, Tao SC. Phage display: an ideal platform for coupling protein to nucleic acid Acta biochim biophys sin (Shanghai). Acta Biochim Biophys Sin (Shanghai). (2021) 53(4):389–99. doi: 10.1093/abbs/gmab006 [DOI] [PubMed] [Google Scholar]
- 124. Smith GP. Filamentous fusion phage: novel expression vectors that display cloned antigens on the virion surface. Science. (1985) 228:1315–7. doi: 10.1126/science.4001944 [DOI] [PubMed] [Google Scholar]
- 125. Sobolewski I, Adamowicz K, Struck A, Zylicz-Stachula A, Skowron PMA. Method for isolation bacteriophage particles-free genomic DNA, exemplified by TP-84, infecting thermophilic geobacillus. Microorganisms. (2022) 10(9):1782. doi: 10.3390/microorganisms10091782 [DOI] [PMC free article] [PubMed] [Google Scholar]