Abstract
Human milk oligosaccharides (HMOs) are a diverse class of carbohydrates which support the health and development of infants. The vast health benefits of HMOs have made them a commercial target for microbial production; however, producing the approximately 200 structurally diverse HMOs at scale has proved difficult. Here we produce a diversity of HMOs by leveraging the robust carbohydrate anabolism of plants. This diversity includes high-value and complex HMOs, such as lacto-N-fucopentaose I. HMOs produced in transgenic plants provided strong bifidogenic properties, indicating their ability to serve as a prebiotic supplement with potential applications in adult and infant health. Technoeconomic analyses demonstrate that producing HMOs in plants provides a path to the large-scale production of specific HMOs at lower prices than microbial production platforms. Our work demonstrates the promise in leveraging plants for the low-cost and sustainable production of HMOs.
Subject terms: Molecular engineering in plants, Dietary carbohydrates, Mass spectrometry
Human milk oligosaccharides (HMOs) are essential carbohydrates for the early development of infants. In this study, the intrinsic carbohydrate anabolism of plants is leveraged to produce a variety of HMOs, highlighting the potential of this method for commercial applications.
Main
Human milk is a complete and comprehensive food evolved to nourish and protect infants. A key component to the distinct bioactive properties of human milk is the presence of a wide diversity of human milk oligosaccharides (HMOs) which are well documented in establishing the nascent gut microbiota of infants to prevent diseases and ensure healthy development1–4. While 75% of infants are supplemented with or exclusively fed infant formula in the first 6 months of life, current infant formulas are either devoid of HMOs or only contain one to two of the ~200 HMOs found in human milk, limiting the health outcomes of formula-fed infants3,5,6. In addition to their use for infant health, HMOs are being studied for their beneficial roles in adult health as a prebiotic to improve intestinal barrier function, lower gastrointestinal inflammation and treat irritable bowel diseases7–11; however, the study of HMO benefits in adults has been limited to a small subset of HMOs. Currently, commercial HMO production relies on microbial fermentation but, to date, microbial fermentation is only able to commercially produce two to five simple HMOs of the ~200 HMOs found in human milk at a scale suitable to supplement food products6,12,13. While five simple HMOs constitute a large portion of HMO mass in human milk, diverse HMOs with a range of linkages and degrees of polymerization enable the growth of beneficial gut microbes which have preferences for specific HMOs14,15. Thus, there is a need to develop biological platforms to produce a wider diversity of HMOs found in human milk, enabling the supplementation of food products for both infants and adults.
The combinatorial nature of glycosidic linkages, nucleotide sugar donors and oligosaccharide acceptor molecules enables the large diversity of HMOs found in human milk16. HMOs are composed of five distinct sugars—d-glucose (Glc), d-galactose (Gal), N-acetylglucosamine (GlcNAc), l-fucose (Fuc) and N-acetylneuraminic acid (Neu5Ac)—connected through various glycosidic linkages to generate a diverse range of molecular structures (Fig. 1a). HMO biosynthesis begins with the production of lactose which can be decorated with fucose or Neu5Ac to form a variety of trisaccharides and tetrasaccharides. Lactose can also be extended by glycosyltransferases through the addition of a GlcNAc-ß-1,3 or GlcNAc-ß-1,4 to form unbranched HMOs. Unbranched HMOs can be further extended by glycosyltransferases through the addition of a GlcNAc-ß-1,6 to produce branched HMOs (Fig. 1b). GlcNAc can undergo subsequent addition of Gal-ß-1,3 or Gal-ß-1,4 to form type I and type II HMOs, respectively (Fig. 1b). While HMOs can consist of all five monosaccharides, they are generally classified in three broad HMO groups on the basis of their composition: (1) neutral HMOs contain Glc, Gal and GlcNAc, (2) fucosylated HMOs contain a neutral core with one or more Fuc additions and (3) acidic HMOs contain a neutral core with one or more additions of Neu5Ac. Owing to the need for high amounts of nucleotide sugars and glycosylation potential in HMO biosynthesis, a suitable host must have robust sugar metabolism capable of managing the metabolic burden of HMO production.
Fig. 1. Production of all three HMO classes in planta.
a, HMOs are composed of Glc, Gal, Fuc, GlcNAc and/or Neu5Ac connected via Gal-ß-1,3/4, GlcNAc-ß-1,3/6, Fuc-α-1,2/3/4 or Neu5Ac-α-2,3/6 glycosidic linkages. b, HMOs can be divided into branched, unbranched, type I and/or type II HMOs. c, HMO biosynthetic pathways used in this study for HMO production in planta. d, Extracted ion chromatograms (EIC) showing the identification of 2′FL, 3′SL, 6′SL, LNFPI, LSTa, LSTc and LNT/LNnT in extracts of individual plant leaves using LC–MS/MS (Q Exactive, Thermo Fisher Scientific). Red, blue and purple colouring denote fucosylated, neutral and acidic HMOs, respectively. Some further peaks are present due to in-source fragmentation of larger oligosaccharides with no available standards.
Unlike many microbes used in commercial fermentation, plants have evolved to create a wide range of glycans which encompass a diversity of nucleotide sugars from photosynthetically fixed CO2. As masters of sugar anabolism, plants are able to create vast amounts of complex oligosaccharides and polysaccharides17. This has led to commercial operations for the production of prebiotic oligosaccharides from plant biomass, such as ß-glucan, xylooligosaccharides, inulin or soy oligosaccharides18–21. Many of these products can either be purified or directly consumed as a food, providing an easy means of ingestion. Additionally, plants can be grown in open fields, requiring minimal inputs, limiting the need for expensive substrates and axenic conditions22. The robust sugar metabolism of plants and ability to be grown at agricultural scales make plants an ideal platform for the large-scale production of HMOs.
Owing to the unique advantages of plants as a platform for carbohydrate production, we tested their ability to produce a range of HMOs using both transient and stable expression in Nicotiana benthamiana. Here, we report in planta production of neutral, fucosylated and acidic HMOs showcasing the intrinsic advantages of a plant-based production platform. Furthermore, we show that plant-produced HMOs provide selective growth of key bifidobacteria, indicating their potential prebiotic efficacy. Finally, we assess the economic viability of HMO production in planta compared to current microbial platforms.
Results
Production of all three HMO classes in planta
Production of HMOs requires the expression of glycosyltransferases capable of creating specific glycosidic linkages. While HMO biosynthesis in humans takes place in the Golgi by means of a largely unknown pathway23, various microbial enzymes are capable of generating HMOs in the cytosol. To produce HMOs in plants, we localized bacterial HMO biosynthetic enzymes to the cytosol for the production of neutral, fucosylated and acidic HMOs (Fig. 1c). We tested these pathways through transient expression in N. benthamiana. Transient expression permits relatively high throughput screening of biosynthetic pathways in planta by injecting strains of Agrobacterium tumefaciens into plant leaves, allowing the Agrobacterium to insert genes for HMO biosynthetic genes into the plant cell24. Leaves transiently expressing HMO biosynthetic pathways were subjected to liquid–liquid extraction, C18 solid-phase extraction (SPE) and porous graphitic carbon (PGC) SPE before characterization by mass spectrometry (MS) (Supplementary Fig. 1).
Neutral HMOs function as the core scaffolds of other more complex HMOs (fucosylated and acidic); thus, we first targeted the type I and type II neutral HMO core structures, lacto-N-tetraose (LNT) and lacto-N-neotetraose (LNnT). Expression of a neutral HMO biosynthetic pathway using two ß-1,4-galactosyltransferases (GalTPM1141 (ref. 25), Hp0826 (ref. 26)), one ß-1,3-galactosyltransferase (Cvß3GalT27) and one ß-1,3-N-acetylglucosaminyltransferease (NmLgtA28) resulted in the production of lactose and various neutral HMOs with degrees of polymerization ranging from three to seven. Notably, N. benthamiana transiently expressing this pathway produced the tetrasaccharides LNT (m/z 708.2559) and LNnT (m/z 708.2559), which represent principal type I and type II HMOs in human milk, respectively29 (Fig. 1d and Supplementary Table 1). Additionally, we identified the production of larger neutral oligosaccharides with varying degrees of polymerization using tandem mass spectrometry (MS/MS) fragmentation to determine the number of hexose and HexNAc sugars (Supplementary Table 1). This included several neutral isomers of pentasaccharides and heptasaccharides (Supplementary Table 1). Our findings demonstrate that plants have the ability to produce previously inaccessible oligosaccharides with various degrees of polymerization which may expand HMO functional bioactivity.
Following the success of generating type I and type II neutral HMOs, we examined the ability of plants to decorate neutral HMOs with fucose, as fucosylated HMOs are the most abundant class of HMO in human milk15. We transiently expressed an α-1,2-fucosyltransferase (Te2FT30) alongside the neutral HMO biosynthetic pathway to produce the most abundant fucosylated HMOs in human milk: 2′-fucosyllactose (2′FL) (m/z 489.1819) and lacto-N-fucopentaose I (LNFPI) (m/z 854.3136) (Fig. 1d and Supplementary Table 1). Additionally, several fucosylated hexasaccharide isomers were identified by m/z and MS/MS fragmentation (Supplementary Table 1). While the structure of each isomer could not be determined, each is composed of four hexoses, one HexNAc and one deoxyhexose, indicating that either LNFPI can be further decorated with hexose sugars or pentasaccharide neutral HMOs can be decorated with additional fucose.
While neutral and fucosylated HMOs represent most HMOs in human milk, acidic HMOs constitute the last main class found in mammalian milks which provide unique bioactivities as a result of the presence of N-acetylneuraminic acid31. Plants do not natively produce the donor molecule for production of acidic HMOs, CMP-Neu5Ac. To produce acidic HMOs, we simultaneously expressed the neutral HMO biosynthetic pathway, sialyltransferases and a mammalian pathway for the production of CMP-Neu5Ac (Supplementary Fig. 2)32. Expression of an α-2,6-sialyltransferase (St6; ref. 33) alongside the neutral HMO biosynthetic pathway produced the acidic trisaccharide, 6′-sialyllactose (6′SL) (m/z 634.2191), of type II acidic HMO, sialyllacto-N-neotetraose c (LSTc) (m/z 999.3505) (Fig. 1d and Supplementary Table 1). Expression of an α-2,3-sialyltransferase (PmST3; ref. 34) with the neutral HMO biosynthetic pathway produced a myriad of acidic HMOs, such as the acidic trisaccharide, 3′-sialyllactose (3′SL) (m/z 634.2187) and the acidic pentasaccharide, LSTd (m/z 999.3510) (Fig. 1d and Supplementary Table. 1). In addition to making several LST isomers in vivo, six isomers of acidic hexasaccharides were identified using m/z and MS/MS fragmentation. Each isomer was composed of four hexoses, one HexNAc and one Neu5Ac (Supplementary Table 1). Together, these results show the ability of plants to produce all three classes of HMOs combinatorially or simultaneously (Supplementary Fig. 3), including type I and type II structures, marking the greatest diversity of HMOs made in a single heterologous organism.
Optimized production of complex fucosylated HMOs
Microbial production platforms suffer from an inability to produce HMOs with higher degrees of polymerization at large scales, leaving many larger, more complex HMOs understudied. LNFPI is a fucosylated pentasaccharide that is the second most abundant fucosylated HMO. Despite its high abundance in breast milk, LNFPI has remained recalcitrant to fermentative production in microbes, limiting efforts to study its potential health benefits. Therefore, we sought to optimize the production of LNFPI in planta by overexpressing the requisite nucleotide sugar biosynthetic pathways. We transiently expressed the biosynthetic pathway for LNFPI (Fig. 2a) alongside pathways for the production of UDP-galactose, UDP-N-acetylglucosamine (UDP-GlcNAc) and GDP-fucose and quantified LNFPI production (Fig. 2b). Expression of the LNFPI pathway with the GDP-fucose pathway increased production of LNFPI by 32.9% (1,075.03 μg g−1 of dry weight) compared to the expression of the LNFPI pathway alone (808.91 μg g−1 of dry weight), indicating that GDP-fucose is limiting in N. benthamiana (Fig. 2c). Surprisingly, overexpression of the GDP-fucose pathway also resulted in the production of lactodifucotetraose (LDFT) (m/z 635.2394) and lacto-N-difuco-hexaose I (LNDFHI) (m/z 1,000.3720) (Supplementary Table 1) despite not expressing an α-1,3- or α-1,4-fucosyltransferase, indicating the presence of native plant fucosyltransferases capable of glycosylating HMOs. Overexpression of all other nucleotide sugar pathway combinations resulted in similar or lower levels of LNFPI production compared to expression of the LNFPI pathway alone.
Fig. 2. Manipulation of nucleotide sugar biosynthetic pathways modulates HMO profiles in planta.
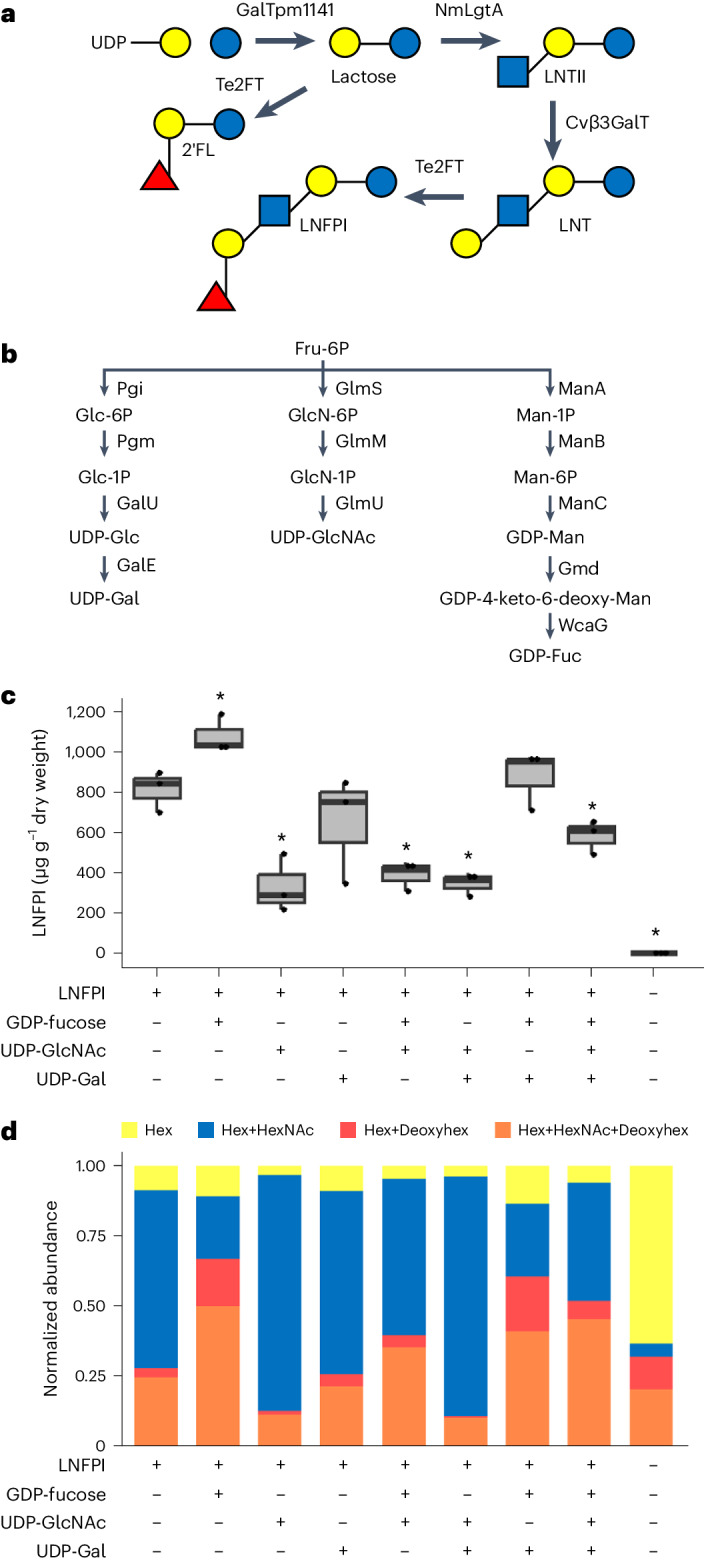
a, HMO biosynthetic pathway expressed for the production of LNFPI. b, Nucleotide sugar biosynthetic pathways expressed with LNFPI pathway. c, Quantification of LNFPI production through expression of LNFPI biosynthetic pathway alongside combinatorially expressed nucleotide sugar biosynthetic pathways using an internal calibration curve obtained with an Agilent 6530 Accurate-Mass Q-ToF MS. The middle bar represents the median. Upper and lower whiskers correspond to the largest and smallest values within 1.5 × the interquartile range, respectively. Upper and lower hinges represent the third and first quartiles, respectively. Statistical analysis was conducted using a heteroscedastic two-tailed Student’s t-test with the LNFPI pathway expressed alone used as the reference group. *P < 0.05. P values are: LNFPI + fucose, 0.030; LNFPI + GlcNAc, 0.012; LNFPI + fucose + GlcNAc, 0.01; LNFPI + GlcNAc + Gal, 0.01; LNFPI + fucose + GlcNAc + Gal, 0.043. A sample size of three leaves was used for each experiment. d, Effect of nucleotide sugar biosynthetic pathway overexpression on HMO profile produced using the LNFPI pathway. Values reflect normalized peak area. Hexose, HexNAc, deoxyhexose (Deoxyhex) determined using m/z and MS/MS fragmentation. We performed mass spectral analysis on an Agilent Q-ToF MS.
The overall profile of oligosaccharides produced was altered by overexpressing nucleotide sugar biosynthetic pathways (Fig. 2d). The number of hexose (Glc, Gal), HexNAc (GlcNAc) and deoxyhexose (Fuc) sugars in each oligosaccharide identified was determined through identification by means of m/z and MS/MS fragmentation and peak area was normalized using a LNFPI calibration curve. Overexpression of the UDP-GlcNAc and LNFPI pathways increased the relative amount of neutral oligosaccharides containing a hexose and HexNAc compared to expression of the LNFPI pathway alone (Fig. 2d). Overexpression of the GDP-fucose and LNFPI pathways resulted in a shift in the overall oligosaccharide composition, favouring the production of oligosaccharides containing at least one deoxyhexose, indicating an increase in the level of fucosylated oligosaccharides (Fig. 2d). These results demonstrate that tailoring the availability of nucleotide sugars enables control over the ratio of HMOs produced.
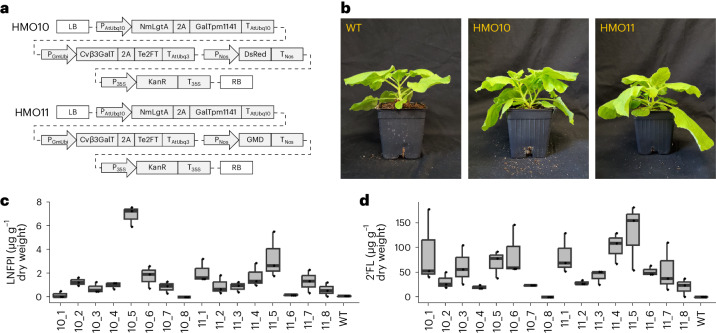
Because scaling HMO production in plants requires the growth of stably transformed plants, we developed transgenic lines of N. benthamiana expressing the LNFPI biosynthetic pathway. We generated two constructs for the constitutive production of 2′FL and LNFPI in transgenic N. benthamiana (Fig. 3a). HMO10 contains genes that encode four enzymes required to produce lactose, 2′FL, LNTII, LNT and LNFPI connected by means of 2A peptides35 to allow several coding sequences to be driven by a single constitutive promoter. To explore the effects of overexpressing portions of the GDP-fucose pathway, we also generated stable lines expressing HMO11, which contains a GDP-d-mannose-4,6-dehydratase (Gmd25) from the GDP-fucose pathway. Gmd transiently expressed alongside the neutral HMO pathway altered the HMO profile of plants in a similar way to expression of the full GDP-fucose pathway (Supplementary Fig. 4).
Fig. 3. Production of HMOs in stably transformed plants.
a, Constructs used in creation of stable lines containing biosynthetic enzymes for the production of LNFPI. b, Photos of 4-week-old transgenic N. benthamiana. c, Concentration of LNFPI produced in leaves of each stable line. d, Concentration of 2′FL produced in leaves of each stable line. For quantification, three leaves from each plant were analysed separately. Quantification of LNFPI and 2′FL obtained with a Thermo Fisher Scientific Q Exactive Mass spectrometer. LB, left border; 2A , 2A peptide; RB, right border; Px, promoter; Tx, terminator; WT, wild type. The middle bar represents the median. Upper and lower whiskers correspond to the largest and smallest values within 1.5 × the interquartile range, respectively. Upper and lower hinges represent the third and first quartiles, respectively. A sample size of three leaves was used for each experiment.
Transgenic T0 HMO-producing stable lines were assessed for fucosylated HMO yield. Most transgenic plants showed no drastic phenotypes compared to wild-type plants (Fig. 3b) and we conducted quantitative PCR with reverse transcription (RT–qPCR) analysis to confirm the expression transgenes (Supplementary Fig. 5). LNFPI and 2′FL were detected in leaves of transgenic N. benthamiana expressing both HMO10 and HMO11. Highest producing LNFPI lines accumulated an average concentration of 6.88 µg g−1 dry weight (Fig. 3c). Leaves from HMO11 no. 5 produced the highest concentration of 2′FL, reaching an average concentration of 130.35 µg g−1 dry weight (Fig. 3d). The low abundance of LNFPI in stable lines compared to transient expression could indicate that the ß-1,3-N-acetylglucosaminyltransferase and ß-1,3-galactosyltransferase suffer from altered activity due to the presence of 2A peptides or lower expression. Together, these results show the ability to produce and optimize a diversity of HMOs from photosynthetically fixed CO2, laying the foundation for future efforts to create high-HMO-yielding transgenic plants for commercial HMO production.
Purification and functional characterization of HMOs from plants
Mixtures of prebiotic sugars can have varying effects on the enrichment of beneficial gut microbes36. Therefore, we sought to assess the bifidogenic activity of extracts from HMO-producing plants; however, crude plant extracts can contain chemicals that interfere with bacterial growth assays, such as simple sugars (glucose, fructose and sucrose) and antimicrobial specialized metabolites. Therefore, we developed a method to extract and purify HMOs from N. benthamiana transiently expressing the biosynthetic pathways for LNFPI (Fig. 2a) and GDP-fucose (Fig. 2b) using an optimized extraction and purification process. Briefly, we performed a water extraction, yeast fermentation to remove simple sugars and a two-step resin adsorption with polyvinylpolypyrrolidone (PVPP) and C18 SPE. This resulted in an HMO-rich extract that contained negligible amounts of simple sugars and phenolic compounds (Supplementary Table 3). The HMO extract contained target fucosylated HMOs (Supplementary Fig. 6), including LNFPI, 2′FL and LNDFHI (Supplementary Table 4). The extract also contained a variety of additional oligosaccharides without assigned structures, which were composed of combinations of hexose, deoxyhexose and HexNAc sugars (Supplementary Fig. 6). These represent potentially non-natural oligosaccharide structures which could provide potential health benefits. Overall, these results show the ability to isolate HMOs from plants, improving their promise as an HMO production platform.
To assess the bifidogenic activity of plant-produced HMOs, we conducted growth assays to compare the effects of plant-produced HMOs to HMOs derived from human milk. We chose to assess the effects of plant-derived HMOs on Bifidobacterium longum subsp. infantis ATCC 15697 (BLI 15697) as it is a known HMO consumer37. We also included Bifidobacterium animalis subsp. lactis ATCC 27536 (BLAC 27536) as a negative control which does not consume HMOs38 but will grow on simple sugars that could be present in plant extracts or human milk. BLI 15697 grown in media containing plant-derived HMOs showed increases in optical density OD600 nm similar to that of BLI 15697 in HMO isolated from human milk, demonstrating that plant-produced HMOs possess the same selective bifidogenic activity as HMOs isolated from human milk (Fig. 4b). As expected, BLAC 27536 showed no growth in either plant-derived HMOs or HMO isolated from human milk, indicating that both extracts contained a minimal amount of simple sugars present (Fig. 4b). Together, these results demonstrate the ability of purified, plant-produced HMOs to mimic the bifidogenic activity of HMOs produced in humans.
Fig. 4. Optimized purification protocol developed for functional analysis of plant-produced HMOs.

a, Workflow of extraction, purification and characterization of HMOs from N. benthamiana leaves. b, Growth curves of HMO-consuming (BLI 15697) and control (BAL 27536) strains in media supplemented with HMO isolated from breast milk (HMO) or HMOs isolated from plants (pHMO). Error bars represent standard deviation.
Economic viability of plant-based HMO production
Plant-based HMO production can be commercially viable if it demonstrates cost-competitive or cost-advantage with current state-of-the-art production routes. To assess the economic viability of HMO production in a commercially relevant crop, we developed process models and compared the cost of HMO production in plants and microbes. To do this, we performed technoeconomic analysis (TEA) of the theoretical production of LNFPI. In the plant system, we adopted typical cellulosic biorefinery design using biomass from sorghum to coproduce HMOs along with biofuel because coproducing value-added bioproducts in biorefineries is a promising approach to maximize the use of biomass and hence improve the economics of biorefineries39,40. We assume that biomass sorghum can accumulate 0.31% dry weight of LNFPI in the entire biomass, as this was our highest yield following purification of LNFPI (Supplementary Table 4). We also developed process models and conducted TEA for the HMOs in Escherichia coli using the established processes and the highest reported yields of LNFPI41 from peer-reviewed papers at the time of conducting this TEA. The comparison between plant and microbial systems to produce the same product aims to provide indepth understanding of the cost–benefits of the systems.
TEA results indicate that producing LNFPI from the plant system is economically favourable compared to the microbial system (Supplementary Fig. 7). In the plant system, the minimum selling prices (MSPs) of LNFPI are US$4.9 kg−1 when selling ethanol at the cellulosic ethanol selling price and US$18.4 kg−1 when ethanol is sold at the target fuel price, respectively (Fig. 5). However, microbial-based LNFPI results in MSP of US$45.0 kg−1 with downstream recovery and purification being the largest cost contributor, followed by the cost of glucose (Supplementary Fig. 7b). The high cost obtained in the microbial system is largely due to its extremely low highest reported yields (0.48%) and recovery rate after bioconversion (62%). These results indicate that when microbial hosts are unable to produce HMOs at a comparable yield, plants may be a cost-effective bioplatform for high-value products. In addition to relative cost advantages of plant systems, using biomass as the feedstock to coproduce high-value compounds and biofuel offers considerable environmental benefits because biomass can absorb CO2 from the atmosphere during its growth42. Although current HMO yields in stable lines of the model plant N. benthamiana (Fig. 3) are below the yields in transiently expressing tissue, high yields could be achieved by optimizing the HMO constructs used for the production of stable lines, finding an optimal crop species for HMO production and optimizing growing conditions.
Fig. 5. Plant-based platform improves the economics of producing the HMO, LNFPI.
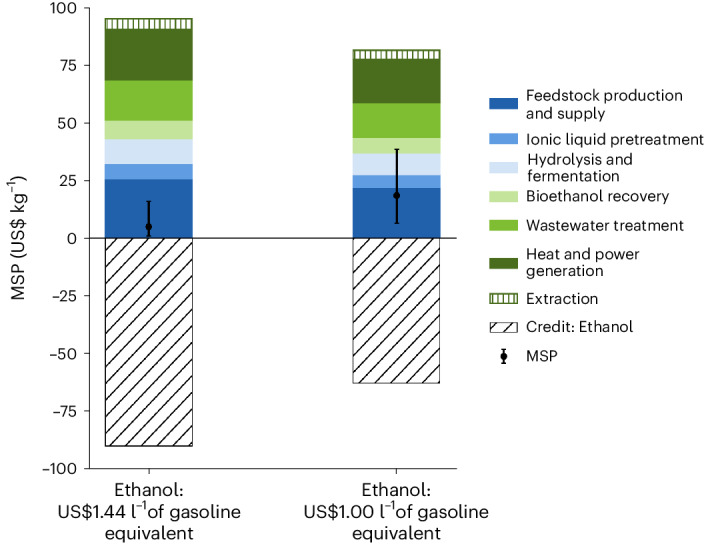
Estimated MSP of LNFPI produced using biomass sorghum as a model production platform in two bioethanol price scenarios. Error bands represent final values calculated with ±20% of input parameters.
Discussion
Human milk oligosaccharides are major contributors to the bioactive properties of human milk1,2. While the unique bioactivities of few HMOs have been investigated3,8–11, over 100 HMOs remain to be studied owing to lack of access to material, representing a wealth of potential bioactive molecules. Here, we report the production of all three classes of HMOs in planta. Notably, expression of HMO biosynthetic pathways in planta created a variety of complex HMOs, including oligosaccharides that at present are not produced in microbial platforms, which indicates plants could serve as a platform for the production of a range of HMOs which are not currently producible in microbial hosts. Furthermore, we optimized the production of both specific HMOs and HMO classes by overexpressing nucleotide sugar biosynthetic pathways. The diversity of HMOs produced from inserting a relatively small number of genes shows the ability of plants to generate complex sugars. Transient expression serves as a viable platform for testing HMO biosynthetic genes and small-scale production of HMOs for functional validation. Optimized l, laboratory-scale purification methods enabled the isolation of oligosaccharides, including HMOs, from plant tissue, showing the potential of plants as an industrial source of HMOs. HMOs purified from plants provided selective bifidogenic activity, indicating that they would serve as a potent prebiotic in vivo. Future pathway engineering could yield biosynthetic pathways for the production of all HMOs present in human milk, including complex, branched HMOs. This would enable the study and potential consumption of bioactive HMOs that are currently unavailable.
Despite the field of plant synthetic biology being in its nascent stages, plants have the potential to produce compounds of interest at lower costs than microbial platforms because of their intrinsic and unique metabolic capabilities39. Additionally, plants are capable of using atmospheric carbon dioxide during their growth cycle to produce target compounds and biofuels, improving the sustainability of target compound production. We demonstrate the ability of stably transformed plants to produce two HMOs that are abundant in breast milk of most mothers, 2′FL and LNFPI. While the feasibility of purifying HMOs from plants at industrial scales still needs to be validated, our TEA results indicate that HMO production in commercially relevant crops has the potential to be a more cost-effective platform than microbial production for complex HMOs.
As the demand for HMOs increases because of the growing infant formula and adult prebiotic markets, plants may emerge as a cost-competitive and sustainable platform for the production of diverse HMOs at agricultural scales. Furthermore, the diversity of plant-produced HMOs will provide researchers with access to HMOs that were previously inaccessible. Since the structure of an HMO determines its bioactivity, this could lead to the discovery of HMOs that treat various gastrointestinal illnesses. Additionally, production of HMOs in plants could permit direct consumption as food by directly ingesting the plant or products made from the plant. Such a product may serve as a consumable source of prebiotic HMOs for humans or be added to forage crops for animal consumption. Overall, the production of HMOs in planta provides the opportunity to simultaneously improve the scale of HMO production and expand the diversity of HMOs available to improve the gastrointestinal health of infants and adults.
Methods
Plant growth
N. benthamiana was grown from seed in 3.5 inch square pots in a controlled environment facility. Plants were grown with a 12 h/12 h day/night cycle at ~700 µmol of photons per m2 s−1. Daytime growth chamber temperatures were kept at 26 °C. Night-time growth chamber temperatures were kept at 25 °C. Relative humidity in the growth chamber was kept between 60% and 75%.
Plasmid construction and transient expression
For transient expression, the native sequences for candidate glycosyltransferases were PCR amplified and cloned into the binary vector, PMS057 (ref. 43), using Golden Gate assembly44, Gibson assembly45 or restriction-ligation (see Supplementary Table 5 for sequences). XL1-blue E. coli cells were transformed with the assembled plasmids via heat shock46. Transformed cells were selected by plating cells on Lysogeny broth (LB) agar plates containing 50 µg ml−1 of kanamycin. Plasmid assembly was confirmed by means of miniprep and Sanger sequencing (Azenta). A. tumefaciens str. GV3101 was transformed using sequence-verified plasmids by electroporation47. Transformed colonies were selected using LB agar plates containing 50 µg ml−1 of kanamycin, 50 µg ml−1 of rifampicin and 10 µg ml−1 of gentamicin. A. tumefaciens str. GV3101 harbouring individual candidate glycosyltransferases were grown in LB overnight to OD600 nm (VWR, V-1200) of 0.8–1.2. The cultures were centrifuged at 4,000g for 10 min and the supernatant was decanted. Cell pellets were resuspended in infiltration media (10 mM MES, 10 mM MgCl2, 500 µM acetosyringone, pH 5.6) and incubated at room temperature for 1 h with gentle rocking (Thermolyne, VariMix). A. tumefaciens strains harbouring each glycosyltransferase were mixed in equal amounts alongside a strain harbouring the p19 silencing suppressor48 to reach a final OD600 nm of 0.5. A. tumefaciens mixtures were injected into the abaxial side of a leaf on a 4-week-old N. benthamiana using a needleless syringe. Each experiment was performed with three biological replicates.
For the production of stable lines, HMO10 and HMO11 constructs were generated through a multipart Golden Gate assembly containing subcloned transcriptional units. Assembled plasmids were transformed and sequence verified as described above. N. benthamiana was transformed using A. tumefaciens str. EHA105 harbouring HMO10 or HMO11 by the UC Davis Plant Transformation Facility.
Quantitative PCR with reverse transcription
Total messenger RNA was extracted using E.Z.N.A. plant RNA kit (Omega Bio-tek) following manufacturer’s directions using the RB lysis buffer variation and on-column DNase digestion; complementary DNA synthesis was achieved with SSIV Vilo IV kit using random hexamers (Thermo Fisher Scientific). Quantitative PCR was performed using a CFX96 Real-Time thermocycler (Bio-Rad) programmed for detection of SYBR intercalating dye with the following temperature programming: 95 °C for 3 min, then 95 °C for 30 s, 60 °C for 45 s, repeated 34 times, then a gradual increase from 65 °C to 95 °C at 0.5 °C per minute to generate melt curves. Sso-Advanced Universal SYBR Green Supermix (Bio-Rad) was used for qPCR amplification. A previously validated primer set was used to amplify EF1α for internal normalization, primers for target genes were designed with Benchling’s qPCR primer design wizard and synthesized by IDT. One target gene from each transcriptional unit was chosen for both constructs (Supplementary Table 6). Melt curves for the product of all primer sets were unimodal and steep, suggesting only a single product was formed for each primer set. No reverse-transcriptase controls showed no amplification within the dynamic range of samples, confirming the efficacy of DNAse treatment and no template controls instituted at the beginning of RNA extraction with no plant matter and kept in parallel with real samples throughout all molecular steps did not amplify, confirming lack of contamination with extraneous DNA. Normalized relative expression was calculated using the ∆∆Cq method and normalized by setting the average level of amplification in the wild-type samples as 1.
HMO extraction for identification of HMOs from individual leaves
N. benthamiana leaves transiently expressing HMO biosynthetic enzymes were harvested 5 days after infiltration. Three leaves of N. benthamiana stable lines transformed with HMO10 and HMO11 were harvested at 4 weeks old. Following harvest, vasculature was removed and leaves were frozen in liquid nitrogen and lyophilized (Labconco, Freezone 4.5) for 2 days. Lyophilized leaves were homogenized via a bead mill (Retsch, MM400) at 20 Hz for 10 min. Oligosaccharides were extracted from 20 mg of lyophilized leaf tissue by ethanol precipitation. To each sample, 1 ml of 80% ethanol was added before homogenization on a bead mill at 10 Hz for 1 min. Samples were then precipitated overnight at −20 °C and centrifuged at 10,000g for 15 min. The supernatant was transferred to a 2 ml screw-cap tube. The pellet was washed twice by adding 500 μl of 80% ethanol, homogenizing via bead mill for 1 min and centrifuging at 10,000g for 15 min. The supernatant and washes were combined and dried in a vacuum centrifuge (Genevac EZ-2, SP Scientific). Dried supernatants were reconstituted in 200 μl of water and subjected to both C18 and PGC SPE (Thermo Fisher Scientific) in 96-well plate format. C18 cartridges containing 25 mg of stationary phase were first conditioned by two additions of 250 μl of acetonitrile (ACN) followed by four additions of 250 μl of water. Samples were then loaded and eluted with two volumes of 200 μl of water. PGC cartridges containing 40 mg of stationary phase were conditioned by addition of 400 μl of water, 400 μl of 80% (v/v) ACN and water, followed by two volumes of 400 μl of water. The sample eluate from C18 SPE was then loaded, washed thrice with 500 μl of water and eluted using two volumes of 200 μl of 40% (v/v) ACN and water. The purified extracts were dried in a vacuum centrifuge and reconstituted in 100 μl of water before injecting 5 μl for liquid chromatography mass spectrometry (LC–MS) analysis.
For quantification of LNFPI in Figs. 2 and 3, LNFPI at known concentrations was added to the extraction solution. The extraction solution was then used on wild-type N. benthamiana. This was done to ensure accuracy of HMO quantification by accounting for HMO losses in the extraction processes and ion suppression that could occur due to the plant metabolites present.
LC–MS analysis of HMOs from individual leaves
For initial screening, chromatographic separation was carried out using a Thermo Scientific Vanquish UHPLC system equipped with a Waters BEH C18 Amide column (HILIC) (1.7 µm, 100 mm × 2.1 mm). A 10 min binary gradient was used based on ref. 49: 0.0–4.0 min, 25–35% A; 4.0–8.50 min, 35–65% A; 8.50–8.70 min, 25% A. Mobile phase A consisted of 3% ACN (v/v) in water with 0.1% formic acid and mobile phase B consisted of 95% ACN (v/v) in water with 0.1% formic acid.
For identification of HMOs produced, we performed LC–MS analysis using a Thermo Scientific Vanquish 3000 UPLC system connected to Thermo Scientific Q Exactive mass spectrometer. Chromatographic separation was carried out using a Hypercarb PGC column (5 µm, 150 mm × 1 mm, Thermo Scientific). A 40 min binary gradient using 3% ACN in water containing 0.1% formic acid (Solvent A) and 90% (v/v) ACN in water containing 0.1% formic acid was performed as follows: 100% A, 0–2.5 min; 100–84% A, 2.5–15 min; 84–42% A, 15–20 min; 42–0% A, 20–22 min; 0% A, 22–28 min; 0–100% A, 28–30 min; 100% A, 30–40 min.
For identification of HMOs, the Q Exactive mass spectrometer equipped with an electrospray ionization source was operated in positive ionization mode with the following parameters: scan range m/z 133.4–2,000; spray voltage 2.5 kV, capillary temperature 320 °C, aux gas heater temperature 325 °C, sheath gas flow rate 25, aux gas flow rate 8, sweep gas flow rate 3. MS/MS analysis was performed using stepped collision energies of 20, 30, 40 eV. MsDIAL was used for data analysis50.
For quantification of LNFPI and HMO profiling, mass spectral analysis was carried out on an Agilent 6530 Accurate-Mass Q-ToF MS operated in positive mode using data-dependent acquisition. The gas temperatures were held at 150 °C. The fragmentor, skimmer, octopole and capillary were operated at 70, 55, 750 and 1,800 V, respectively. The collision energy was based on the empirically derived linear formula (1.8 × (m/z/100) − 3.6). The reference mass used for calibration was m/z 922.009798. The Agilent MassHunter Qualitative software was used for data analysis. Oligosaccharides were identified using an inhouse library, their MS/MS spectra and comparison to either authenticated standards or a pool of HMOs of known composition.
Extraction and purification of HMOs from pooled leaves
Five grams of lyophilized and ground N. benthamiana leaves transiently expressing the LNFPI and GDP-fucose biosynthetic pathways was mixed with 150 ml of water and agitated for 15 min at room temperature in a stirring plate. The mixture was centrifuged at 4,000g for 5 min and the supernatant was separated. The extraction was repeated two more times, combining the supernatant each time. The final supernatant was filtered using a 0.22 µm Millipore Steritop vacuum filter. The extraction process was carried out in duplicate to ensure reproducibility.
Yeast fermentation was carried out to eliminate simple sugars (glucose, sucrose and fructose) from the extracts51. Briefly, autoclaved extracts were inoculated with 0.4 g l−1 of commercial active dry yeast Saccharomyces cerevisiae52 (UCD 522 Montrachet, Lallemand) at 30 °C, 150 rpm for 24 h (Excella E24 Incubator Shaker Series, New Brunswick Scientific). After 24 h, the samples were centrifuged at 4,000g for 5 min and filtered using a 0.22 µm Millipore Steritop vacuum filter to remove the yeast. Samples were concentrated using a vacuum concentrator (Genevac miVac Centrifugal Concentrator) at room temperature and frozen until their purification.
PVPP (Sigma-Aldrich) was used to bind phenolic compounds within the sample following a previous protocol52. Briefly, 3 g of PVPP was conditioned by mixing it with 100 ml of 12 M HCl at 100 °C for 30 min with constant stirring in a stirring plate. After cooling off, the slurry was centrifuged at 4,000g for 5 min and filtered using a 0.22 µm Millipore Steritop vacuum filter. Subsequently, the PVPP was washed with nanopure water until the flow-through reached pH 7. Activated PVPP was mixed with water to a final concentration of 20 mg of PVPP per ml.
PVPP suspension was added to the extracts at a concentration of 6 mg of PVPP per ml and agitated at room temperature for 15 min on a stirring plate. After the time had elapsed, the sample containing the plant extracts and the PVPP was centrifuged at 4,000g for 5 min and filtered using a 0.22 µm Millipore Steritop vacuum filter to separate the PVPP containing the bound phenolics. To eliminate residual phenolics from the extracts, more PVPP was added to the supernatant (6 mg of PVPP per ml) and the process was repeated. The filtrate containing the oligosaccharides was concentrated and frozen until further purification.
Two SPE columns of 60 ml were packed with 15 g of Bondesil-C18, 40 µm suspended in 20 ml of ACN. After the ACN was drained, a frit was added to the C18. Before loading the samples, the columns were conditioned with three volumes of ACN and three volumes of nanopore water. PVPP-treated extracts were loaded onto the conditioned C18 columns and the oligosaccharides were washed with 250 ml of nanopure water divided into four washes. To ensure the complete removal of interfering compounds, C18 SPE was repeated two more times. The purified HMO fractions were dried in a vacuum concentrator (Genevac miVac Centrifugal Concentrator) at room temperature and frozen.
Compositional analysis of plant material
Total carbohydrate content was assessed by the anthrone method with modifications53. In a 96-well microplate, 40 μl of purified and diluted extracts were combined with 100 μl of anthrone reagent (2 mg ml−1 (w/v) in cold 98% sulfuric acid) and mixed through pipette tip aspiration. The microplate was incubated for 3 min at 92 °C in a water bath followed by 5 min at a room temperature water bath and then 15 min in a 45 °C Thermolyne Benchtop muffle furnace (Thermo Fisher Scientific). The plate was cooled for 3 min at room temperature before measuring the absorbance with a SpectroMax M5 UV/Vis spectrophotometer (Molecular Devices) at 630 nm. Total carbohydrate quantification calculations were based on a glucose standard curve. Each plant extract was prepared in duplicate and each sample was further analysed in duplicate.
Total phenolic content of the extracts was determined according to the Folin–Ciocalteu spectrophotometric method as described by ref. 54.
Simple sugars (glucose, sucrose and fructose) were quantified by high-performance anion exchange chromatography with pulsed amperometric detection on a Thermo Fisher Dionex ICS-5000+ HPAE-PAD system based on a method described by ref. 55 with modifications. Diluted extracts (1:100, v/v or 1:1,000 in nanopure water) were filtered through a 0.2 mm syringe filter (Agilent Captiva Econo Filter, PES, 13 mm, 0.2 μm) into 2 ml vials with septa. The samples (25 μl) were injected into a CarboPac PA200 guard column (3 × 50 mm) and a CarboPac PA200 analytical column (3 × 250 mm) and chromatographic separation was carried out with a 12 min gradient elution (from 0.6% to 25% B in 12 min), 0.5 ml min−1 flow rate. The solvent system consisted of A: 100% water; and B: 200 mM sodium hydroxide. Calibration curves (correlation coefficient ≥0.999) were prepared using glucose, sucrose and fructose standards.
Quantification of HMOs by QqQ LC–MS
Detection and quantitation of HMOs were performed using an Agilent 6470 triple quadrupole LC–MS system (QqQ LC–MS) equipped with an Advance Bio Glycan Map column (2.1 mm × 150 mm, 2.7 μm, Agilent). The mobile phase consisted of 10 mM ammonium acetate in 3% ACN, 97% water (v/v, pH 4.5; A) and 10 mM ammonium acetate in 95% ACN, 5% water (v/v, pH 4.5; B). The chromatographic separation was carried out at 35 °C with gradient elution at a flow rate of 0.3 ml min−1. The MS analysis was conducted in positive ion mode with source parameters as follows: the gas temperature was 150 °C at a flow rate of 10 l min−1; the nebulizer was 45 psi; the sheath gas temperature was 250 °C at a flow rate of 7 l min−1; capillary voltage was 2,200 V. See Supplementary Table 2 for gradient and multiple reaction monitoring transitions.
Characterization of HMOs by LC-QToF-MS
Oligosaccharides were purified by a two-step SPE using C18 (HyperSep C18–96, 50 mg bed weight; Thermo Fisher Scientific) and PGC (HyperSep Hypercarb-96, 25 mg bed weight; Thermo Fisher Scientific)56. The samples were filtered (Captiva Premium Syringe Filter PES membrane, 4 mm diameter, 0.2 µm pore size, LC/MS certified) into 200 µl vials.
Individual oligosaccharide compositions were analysed with an Agilent 6520 NanoChip LC-QToF mass spectrometer. Oligosaccharides separation was achieved with a microfluidic high-performance liquid chromatography PGC chip containing an enrichment (4 mm, 40 nl) and an analytical (75 μl × 43 mm) column as well as a nanoelectrospray tip, using a binary solvent gradient of solvent A (5 mM ammonium acetate in 3% ACN, 97% water (v/v)) and solvent B (5 mM ammonium acetate in 90% ACN, 10% water (v/v)) based on a previously optimized method55. The gradient was 0–16% B at 0–20 min, 16–44% B at 20–30 min, 44–100% B at 30–35 min, 100% B at 35–45 min and 100–0% B from 45 to 45.1 min, followed by a 15 min re-equilibration of 100% A57. The mass spectrometer was operated in positive ionization mode with a range of m/z 320–2,500 and an electrospray capillary voltage of 1,800–1,900 V. Reference masses of m/z 922.009 and 1,221.991 provided continuous internal calibration. All samples were analysed using MS/MS with tandem fragmented peaks selected by the automated precursor selection of the six ions with highest signal intensity with a medium isolation width. The Q-ToF MS had a ramped collision energy slope of 0.02 based on m/z values with an offset of −3.5 V. The acquisition rate of 1 spectrum per s was used for both MS and MS/MS. Each spectrum was manually examined and molecular masses were confirmed with Agilent MassHunter Qualitative Analysis B.07.00 software using the molecular feature extraction and a maximum tolerance of 20 ppm.
Bacterial strains and growth conditions
B. longum subsp. infantis ATCC 15697 and B. animalis subsp. lactis ATCC 27536 were cultured at 37 °C in a Coy vinyl anaerobic bubble with an atmosphere of 2.5% H2, ~5% CO2 and balance N2. Routine culturing was done with Difco MRS + 0.05% l-cysteine HCl (MRSC) and carbohydrate-specific culturing was done with modified MRS (mMRSC), which was prepared per litre as follows: 10 g of Bacto proteose peptone no. 3, 10 g of Bacto casitone, 5 g of Bacto yeast extract, 2 g of triammonium citrate, 5 g of sodium acetate trihydrate, 200 mg of magnesium sulfate hexahydrate, 34 mg of manganese sulfate monohydrate, 0.5 g of l-cysteine HCl and 1.063 g of Tween-80. Normally, 2 g of anhydrous dipotassium phosphate would also be added but it was precipitated by the plant HMO preparation.
Growth curves
One colony was used to inoculate 1 ml mMRSC + dipotassium phosphate + 1% lactose monohydrate and incubated for 24 h. The growth curve inoculum was cultured by diluting the 24 h culture 1:100 in mMRSC + dipotassium phosphate + 1% lactose, then incubating for 12–15 h. The inoculum was prepared by washing the cells twice with one volume of 1× PBS. Growth curve cultures (160 μl) were contained in flat-bottomed, optically clear, 96-well, lidded plates and they had a final inoculum and sugar concentration of 1% in mMRSC. Lactose was the growth control substrate, water was the no-growth control substrate and pooled HMO58 was the HMO-growth control substrate. Cultures were done in triplicate. Wells were overlaid with 40 μl of sterile mineral oil and incubated in a BMG SpectroStar Nano. The plate reader was set to read the OD600 nm of each well 30 times in a spiral pattern every half-hour with medium orbital shaking before each read. All media had uninoculated controls whose OD600 nm was subtracted from that of the inoculated medium.
Technoeconomic analysis
In this study, we used SuperPro Designer v.12 to develop technoeconomic models of HMOs which can be produced from both plant and microbial systems. To do this, we first developed process models and then applied discount cash flow analysis of the theoretical production of LNFPI in plants and microbes. The simplified process flow diagram can be found in Supplementary Fig. 8. In the plant system, we adopted integrated cellulosic biorefinery design to coproduce HMO and ethanol to maximize the use of plant biomass. Biomass sorghum was used as the representative plant because its characteristics, such as high yields and drought tolerance, are ideal for biofuel production. Previous studies demonstrated that using biomass sorghum as the representative plant to accumulate value-added bioproducts could improve the economic performance of an integrated biorefinery39,40. Since ethanol is coproduced in the biorefinery, two ethanol selling prices were considered: (1) baseline cellulosic biofuel selling price of US$1.44 l−1 of gasoline equivalent and (2) target fuel price of US$1.00 l−1 of gasoline equivalent.
Briefly, biomass sorghum with 0.31% dry weight HMOs accumulated in the plant biomass are harvested and transported to the biorefinery gate for preprocessing and short-term onsite storage. HMO extraction, separation and recovery is then conducted on the basis of our laboratory processes and as described in previous texts. Water is used to extract HMO from the biomass (room temperature for 6 h) since the industrial extraction process will last longer than the laboratory-scale process given the large quantity of biomass being processed. The extraction efficiency is assumed at 90% based on previous studies39. After extraction, the slurry is first cooled down to room temperature and is sent to centrifugation to remove water. This water is sent to the wastewater treatment unit located in the biorefinery for recycling and reusing. Afterwards, multistage ultrafiltration is used to recover HMO from the extraction stream. Before transporting to HMO onsite storage, another centrifuge is applied to ensure the recovery and purity of the final product. In this plant system, the extracted HMOs are considered as the main product with the purity of >95%. The remaining biomass from biomass sorghum is routed to ionic liquid pretreatment for biomass deconstruction. After ionic liquid pretreatment, enzymatic hydrolysis and ethanol fermentation are conducted to produce ethanol, followed by distillation and molecular sieve to remove excess water. Wastewater from the overall process is routed to the wastewater treatment sector to produce reusable process water and biogas, which can be combusted in the onsite turbogenerator, along with other solids from biomass, to generate heat and electricity that can satisfy the facility’s need. In the microbial system, HMOs were produced as the single product in the biorefinery. Unlike the plant system, pure sugar is used in the microbial system as the sole feedstock and no wastewater treatment sector and onsite combustion are designed for the microbial system. The downstream processing of microbial production is adopted from peer-reviewed publication41. After LNFPI is produced in the bioreactor under the bioconversion condition of 30 °C for 52 h, ultrafiltration is first applied to remove biomass and ion exchange chromatography to remove ions and other charged impurities. Multistage nanofiltration is further used to reduce the total volume and remove excess impurities. Gel filtration is used for the final purification of HMO from the microbial production process. The purity of the final product is >98% through this process.
After developing technoeconomic models in SuperPro Designer, we performed mass and energy balance and then applied discounted cash flow analysis to quantify the MSP of HMOs (US$ kg−1). For both systems, we assume the biorefinery can operate 24 h per day and 330 days per year for 30 years. The unit price of biomass sorghum is assumed at US$95 per bone-dry tonne and we assume in the plant system the cellulosic biorefinery can intake 2,000 bone-dry tonnes of biomass sorghum per day. The unit price of ionic liquid is US$2 kg−1 with a range of US$1–5 kg−1. Total capital investment includes installed equipment cost, piping costs, engineering costs, warehouse, site development, construction fees, contingency costs, land costs, startup and working capital. Annual operating costs include raw materials costs, utility costs, labour costs and facility-dependent costs such as insurance. These parameters are kept constant as per the 2011 National Renewable Energy Laboratory report59.
Statistics and reproducibility
No statistical method was used to predetermine sample size as performing experiments in biological triplicate is standard of the field. No data were excluded from the analyses. The experiments were not randomized. The Investigators were not blinded to allocation during experiments and outcome assessment. All experiments looking at individual leaves were conducted in biological triplicate. For the laboratory-scale purifications, extractions were completed in duplicate and each duplicate was measured with two technical replicates. Microbial growth assays were conducted in biological triplicate. For statistical analysis of LNFPI optimization, a heteroscedastic two-tailed Student’s t-test with the LNFPI pathway expressed alone was used as the reference group in RStudio v.1.2.5033. GraphPad v.10 was used to plot the microbial growth assays.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Supplementary Figs. 1–8 and Tables 1–6.
Acknowledgements
C.R.B. acknowledges support from the National Institutes of Health NIGMS T32 Training Program and the US Department of Energy. C.R.B. and P.M.S. acknowledge support from the Joint BioEnergy Institute (http://www.jbei.org) supported by the US Department of Energy, Office of Science, Office of Biological and Environmental Research, through contract DE-AC02-05CH11231 between Lawrence Berkeley National Laboratory and the US Department of Energy. P.M.S. acknowledges support from grant no. R00AT009573 from the National Center for Complementary and Integrative Health at the National Institutes of Health. M.Y. acknowledges USDA NIFA HATCH NC02948. D.A.M. acknowledges support of the P. J. Shields, Chair in Dairy Food Science. D.B. acknowledges the Hatch project CA-D-FST-2744-H. We thank X. Chen for sharing plasmids with genes encoding several glycosyltransferases.
Author contributions
C.R.B. performed the construct design and agrobacterium infiltrations. C.R.B., G.C. and Y.C conducted the extractions and mass spectral analysis of single leaves. B.P. and Y.-P.H. conducted the laboratory-scale extractions and subsequent mass spectral analysis. C.M. conducted the microbial growth assays. M.Y. conducted the TEA. All authors contributed to writing the paper.
Peer review
Peer review information
Nature Food thanks Hao Du, Ted Johanson and Zhenhua Liu for their contribution to the peer review of this work.
Data availability
The main data supporting the findings of this study are available within the article and its Supplementary Information.
Competing interests
D.A.M., D.B. and C.B.L. are cofounders of Infinant Health, a company focused on probiotic-based manipulation of the infant gut microbiota. D.A.M. and C.B.L. are cofounders of One.Bio, a company advancing new bioactive glycans. None of these companies had any role in the conceptualization, design, analysis or preparation of this manuscript. The other authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s43016-024-00996-x.
References
- 1.Walsh C, Lane JA, van Sinderen D, Hickey RM. Human milk oligosaccharides: shaping the infant gut microbiota and supporting health. J. Funct. Foods. 2020;72:104074. doi: 10.1016/j.jff.2020.104074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kirmiz N, Robinson RC, Shah IM, Barile D, Mills DA. Milk glycans and their interaction with the infant-gut microbiota. Annu. Rev. Food Sci. Technol. 2018;9:429–450. doi: 10.1146/annurev-food-030216-030207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vandenplas Y, et al. Human milk oligosaccharides: 2′-fucosyllactose (2′-FL) and lacto-N-neotetraose (LNnT) in infant formula. Nutrients. 2018;10:1161. doi: 10.3390/nu10091161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wiciński M, Sawicka E, Gębalski J, Kubiak K, Malinowski B. Human milk oligosaccharides: health benefits, potential applications in infant formulas and pharmacology. Nutrients. 2020;12:266. doi: 10.3390/nu12010266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.2022 Breastfeeding Report Card (CDC, 2022); https://www.cdc.gov/breastfeeding/data/reportcard.htm
- 6.Urashima T, Horiuchi R, Sakanaka M, Katayama T, Fukuda K. Lactose or milk oligosaccharide: which is significant among mammals? Anim. Front. 2023;13:14–23. doi: 10.1093/af/vfad017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee S, et al. Human milk oligosaccharide 2′-fucosyllactose supplementation improves gut barrier function and signaling in the vagal afferent pathway in mice. Food Funct. 2021;12:8507–8521. doi: 10.1039/D1FO00658D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Šuligoj T, et al. Effects of human milk oligosaccharides on the adult gut microbiota and barrier function. Nutrients. 2020;12:2808. doi: 10.3390/nu12092808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elison E, et al. Oral supplementation of healthy adults with 2′-O-fucosyllactose and lacto-N-neotetraose is well tolerated and shifts the intestinal microbiota. Br. J. Nutr. 2016;116:1356–1368. doi: 10.1017/S0007114516003354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iribarren C, et al. Human milk oligosaccharide supplementation in irritable bowel syndrome patients: a parallel, randomized, double-blind, placebo-controlled study. Neurogastroenterol. Motil. 2020;32:e13920. doi: 10.1111/nmo.13920. [DOI] [PubMed] [Google Scholar]
- 11.Iribarren C, et al. The effects of human milk oligosaccharides on gut microbiota, metabolite profiles and host mucosal response in patients with irritable bowel syndrome. Nutrients. 2021;13:3836. doi: 10.3390/nu13113836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bych K, et al. Production of HMOs using microbial hosts—from cell engineering to large scale production. Curr. Opin. Biotechnol. 2019;56:130–137. doi: 10.1016/j.copbio.2018.11.003. [DOI] [PubMed] [Google Scholar]
- 13.Palur DSK, Pressley SR, Atsumi S. Microbial production of human milk oligosaccharides. Molecules. 2023;28:1491. doi: 10.3390/molecules28031491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shani G, et al. Fucosylated human milk oligosaccharide foraging within the species Bifidobacterium pseudocatenulatum is driven by glycosyl hydrolase content and specificity. Appl. Environ. Microbiol. 2022;88:e0170721. doi: 10.1128/AEM.01707-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Soyyılmaz B, et al. The mean of milk: a review of human milk oligosaccharide concentrations throughout lactation. Nutrients. 2021;13:2737. doi: 10.3390/nu13082737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen, X. in Advances in Carbohydrate Chemistry and Biochemistry (eds Baker, D. C. & Horton, D.) 113–190 (Academic Press, 2015).
- 17.Bar-Peled M, O’Neill MA. Plant nucleotide sugar formation, interconversion and salvage by sugar recycling. Annu. Rev. Plant Biol. 2011;62:127–155. doi: 10.1146/annurev-arplant-042110-103918. [DOI] [PubMed] [Google Scholar]
- 18.Zhu F, Du B, Xu B. A critical review on production and industrial applications of beta-glucans. Food Hydrocoll. 2016;52:275–288. doi: 10.1016/j.foodhyd.2015.07.003. [DOI] [Google Scholar]
- 19.Palaniappan A, Antony U, Emmambux MN. Current status of xylooligosaccharides: production, characterization, health benefits and food application. Trends Food Sci. Technol. 2021;111:506–519. doi: 10.1016/j.tifs.2021.02.047. [DOI] [Google Scholar]
- 20.Choct M, Dersjant-Li Y, McLeish J, Peisker M. Soy oligosaccharides and soluble non-starch polysaccharides: a review of digestion, nutritive and anti-nutritive effects in pigs and poultry. Asian-Australas. J. Anim. Sci. 2010;23:1386–1398. doi: 10.5713/ajas.2010.90222. [DOI] [Google Scholar]
- 21.Shoaib M, et al. Inulin: properties, health benefits and food applications. Carbohydr. Polym. 2016;147:444–454. doi: 10.1016/j.carbpol.2016.04.020. [DOI] [PubMed] [Google Scholar]
- 22.Barnum CR, Endelman BJ, Shih PM. Utilizing plant synthetic biology to improve human health and wellness. Front. Plant Sci. 2021;12:1824. doi: 10.3389/fpls.2021.691462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kellman BP, et al. Elucidating human milk oligosaccharide biosynthetic genes through network-based multi-omics integration. Nat. Commun. 2022;13:2455. doi: 10.1038/s41467-022-29867-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thompson MG, et al. Agrobacterium tumefaciens: a bacterium primed for synthetic biology. BioDesign Res. 2020;2020:8189219. doi: 10.34133/2020/8189219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parschat K, Schreiber S, Wartenberg D, Engels B, Jennewein S. High-titer de novo biosynthesis of the predominant human milk oligosaccharide 2′-fucosyllactose from sucrose in Escherichia coli. ACS Synth. Biol. 2020;9:2784–2796. doi: 10.1021/acssynbio.0c00304. [DOI] [PubMed] [Google Scholar]
- 26.Ooi K-E, Zhang X-W, Kuo C-Y, Liu Y-J, Yu C-C. Chemoenzymatic synthesis of asymmetrically branched human milk oligosaccharide lacto-N-hexaose. Front. Chem. 2022;10:905105. doi: 10.3389/fchem.2022.905105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McArthur JB, Yu H, Chen X. A bacterial β1–3-galactosyltransferase enables multigram-scale synthesis of human milk lacto-N-tetraose (LNT) and its fucosides. ACS Catal. 2019;9:10721–10726. doi: 10.1021/acscatal.9b03990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li Y, et al. Donor substrate promiscuity of bacterial β1–3-N-acetylglucosaminyltransferases and acceptor substrate flexibility of β1–4-galactosyltransferases. Bioorg. Med. Chem. 2016;24:1696–1705. doi: 10.1016/j.bmc.2016.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thurl S, Munzert M, Boehm G, Matthews C, Stahl B. Systematic review of the concentrations of oligosaccharides in human milk. Nutr. Rev. 2017;75:920–933. doi: 10.1093/nutrit/nux044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen, X., Zhao, C. & Yu, H. Te2ft enzyme for enzymatic synthesis of alpha1-2-fucosides. International patent application PCT/US2016/067601 (2017).
- 31.Hobbs M, Jahan M, Ghorashi SA, Wang B. Current perspective of sialylated milk oligosaccharides in mammalian milk: implications for brain and gut health of newborns. Foods. 2021;10:473. doi: 10.3390/foods10020473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Castilho A, et al. Construction of a functional CMP-sialic acid biosynthesis pathway in Arabidopsis. Plant Physiol. 2008;147:331–339. doi: 10.1104/pp.108.117572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Drouillard S, Mine T, Kajiwara H, Yamamoto T, Samain E. Efficient synthesis of 6′-sialyllactose, 6,6′-disialyllactose and 6′-KDO-lactose by metabolically engineered E. coli expressing a multifunctional sialyltransferase from the Photobacterium sp. JT-ISH-224. Carbohydr. Res. 2010;345:1394–1399. doi: 10.1016/j.carres.2010.02.018. [DOI] [PubMed] [Google Scholar]
- 34.Chen, X., Thon, V. & Yu, H. PmST3 enzyme for chemoenzymatic synthesis of alpha-2-3-sialosides. US patent application US14/356,376 (2017).
- 35.Ryan MD, Drew J. Foot-and-mouth disease virus 2 A oligopeptide mediated cleavage of an artificial polyprotein. EMBO J. 1994;13:928–933. doi: 10.1002/j.1460-2075.1994.tb06337.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ghoddusi HB, Grandison MA, Grandison AS, Tuohy KM. In vitro study on gas generation and prebiotic effects of some carbohydrates and their mixtures. Anaerobe. 2007;13:193–199. doi: 10.1016/j.anaerobe.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 37.Sela DA, et al. The genome sequence of Bifidobacterium longum subsp. infantis reveals adaptations for milk utilization within the infant microbiome. Proc. Natl Acad. Sci. USA. 2008;105:18964–18969. doi: 10.1073/pnas.0809584105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Underwood MA, et al. A comparison of two probiotic strains of bifidobacteria in premature infants. J. Pediatr. 2013;163:1585–1591. doi: 10.1016/j.jpeds.2013.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang M, et al. Accumulation of high-value bioproducts in planta can improve the economics of advanced biofuels. Proc. Natl Acad. Sci. USA. 2020;117:8639–8648. doi: 10.1073/pnas.2000053117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang M, et al. Comparing in planta accumulation with microbial routes to set targets for a cost-competitive bioeconomy. Proc. Natl Acad. Sci. USA. 2022;119:e2122309119. doi: 10.1073/pnas.2122309119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Derya SM, et al. Biotechnologically produced fucosylated oligosaccharides inhibit the binding of human noroviruses to their natural receptors. J. Biotechnol. 2020;318:31–38. doi: 10.1016/j.jbiotec.2020.05.001. [DOI] [PubMed] [Google Scholar]
- 42.Sandalow, D., Aines, R., Friedmann, J., McCormick, C. & Sanchez, D. Biomass Carbon Removal and Storage (BiRCS) Roadmap (US Department of Energy, 2021); https://www.osti.gov/servlets/purl/1763937/
- 43.Barnum CR, Endelman BJ, Ornelas IJ, Pignolet RM, Shih PM. Optimization of heterologous glucoraphanin production in planta. ACS Synth. Biol. 2022;11:1865–1873. doi: 10.1021/acssynbio.2c00030. [DOI] [PubMed] [Google Scholar]
- 44.Engler C, Gruetzner R, Kandzia R, Marillonnet S. Golden gate shuffling: a one-pot DNA shuffling method based on type IIs restriction enzymes. PLoS ONE. 2009;4:e5553. doi: 10.1371/journal.pone.0005553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gibson GR, et al. Expert consensus document: the International Scientific Association for Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of prebiotics. Nat. Rev. Gastroenterol. Hepatol. 2017;14:491–502. doi: 10.1038/nrgastro.2017.75. [DOI] [PubMed] [Google Scholar]
- 46.Froger, A. & Hall, J. E. Transformation of plasmid DNA into E. coli using the heat shock method. J. Vis. Exp.2007, 253 (2007). [DOI] [PMC free article] [PubMed]
- 47.Lin, J.-J. in Electroporation Protocols for Microorganisms (ed. Nickoloff, J. A.) 171–178 (Humana Press, 1995); 10.1385/0-89603-310-4:171
- 48.Lakatos L, Szittya G, Silhavy D, Burgyán J. Molecular mechanism of RNA silencing suppression mediated by p19 protein of tombusviruses. EMBO J. 2004;23:876–884. doi: 10.1038/sj.emboj.7600096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xu G, et al. Absolute quantitation of human milk oligosaccharides reveals phenotypic variations during lactation. J. Nutr. 2017;147:117–124. doi: 10.3945/jn.116.238279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tsugawa H, et al. MS-DIAL: data-independent MS/MS deconvolution for comprehensive metabolome analysis. Nat. Methods. 2015;12:523–526. doi: 10.1038/nmeth.3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.de Moura Bell JMLN, et al. An integrated bioprocess to recover bovine milk oligosaccharides from colostrum whey permeate. J. Food Eng. 2018;216:27–35. doi: 10.1016/j.jfoodeng.2017.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Magalhães PJ, et al. Isolation of phenolic compounds from hop extracts using polyvinylpolypyrrolidone: characterization by high-performance liquid chromatography–diode array detection–electrospray tandem mass spectrometry. J. Chromatogr. A. 2010;1217:3258–3268. doi: 10.1016/j.chroma.2009.10.068. [DOI] [PubMed] [Google Scholar]
- 53.Laurentin A, Edwards CA. A microtiter modification of the anthrone–sulfuric acid colorimetric assay for glucose-based carbohydrates. Anal. Biochem. 2003;315:143–145. doi: 10.1016/S0003-2697(02)00704-2. [DOI] [PubMed] [Google Scholar]
- 54.Singleton, V. L., Orthofer, R. & Lamuela-Raventós, R. M. Analysis of total phenols and other oxidation substrates and antioxidants by means of Folin–Ciocalteu reagent. Methods Enzymol.299, 152–178 (1999).
- 55.Huang Y-P, Paviani B, Fukagawa NK, Phillips KM, Barile D. Comprehensive oligosaccharide profiling of commercial almond milk, soy milk and soy flour. Food Chem. 2023;409:135267. doi: 10.1016/j.foodchem.2022.135267. [DOI] [PubMed] [Google Scholar]
- 56.Huang Y-P, Robinson RC, Barile D. Food glycomics: dealing with unexpected degradation of oligosaccharides during sample preparation and analysis. J. Food Drug Anal. 2022;30:62–76. doi: 10.38212/2224-6614.3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bhattacharya M, Salcedo J, Robinson RC, Henrick BM, Barile D. Peptidomic and glycomic profiling of commercial dairy products: identification, quantification and potential bioactivities. NPJ Sci. Food. 2019;3:4. doi: 10.1038/s41538-019-0037-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lee H, Garrido D, Mills DA, Barile D. Hydrolysis of milk gangliosides by infant-gut associated bifidobacteria determined by microfluidic chips and high-resolution mass spectrometry. Electrophoresis. 2014;35:1742–1750. doi: 10.1002/elps.201300653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Humbird, D. et al. Process Design and Economics for Biochemical Conversion of Lignocellulosic Biomass to Ethanol: Dilute-Acid Pretreatment and Enzymatic Hydrolysis of Corn Stover (US Department of Energy, 2011); https://www.osti.gov/servlets/purl/1013269/
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figs. 1–8 and Tables 1–6.
Data Availability Statement
The main data supporting the findings of this study are available within the article and its Supplementary Information.