Abstract
Cryptococcosis causes a high burden of disease worldwide. This systematic review summarizes the literature on Cryptococcus neoformans and C. gattii infections to inform the World Health Organization’s first Fungal Priority Pathogen List. PubMed and Web of Science were used to identify studies reporting on annual incidence, mortality, morbidity, antifungal resistance, preventability, and distribution/emergence in the past 10 years. Mortality rates due to C. neoformans were 41%–61%. Complications included acute renal impairment, raised intracranial pressure needing shunts, and blindness. There was moderate evidence of reduced susceptibility (MIC range 16–32 mg/l) of C. neoformans to fluconazole, itraconazole, ketoconazole, voriconazole, and amphotericin B. Cryptococcus gattii infections comprised 11%–33% of all cases of invasive cryptococcosis globally. The mortality rates were 10%–23% for central nervous system (CNS) and pulmonary infections, and ∼43% for bloodstream infections. Complications described included neurological sequelae (17%–27% in C. gattii infections) and immune reconstitution inflammatory syndrome. MICs were generally low for amphotericin B (MICs: 0.25–0.5 mg/l), 5-flucytosine (MIC range: 0.5–2 mg/l), itraconazole, posaconazole, and voriconazole (MIC range: 0.06–0.5 mg/l). There is a need for increased surveillance of disease phenotype and outcome, long-term disability, and drug susceptibility to inform robust estimates of disease burden.
Keywords: Cryptococcus neoformans, Cryptococcus gattii, cryptococcosis, cryptococcal meningitis, invasive fungal infection
Introduction
Invasive fungal infections pose a significant threat to global health. Although their burden is ill-defined, crude estimates suggest they cause over 1.6 million deaths annually.1 The absence of strong surveillance systems results in clinicians making decisions based on limited information about local epidemiology, antimicrobial resistance, and effective treatment strategies. In response to this growing threat, the World Health Organization (WHO) developed a Fungal Priority Pathogens List (FPPL). This list, published in 2022, was created through a comprehensive international consultation process, using a survey incorporating a discrete choice experiment. The individual fungal pathogens, including Cryptococcus neoformans and C. gattii, were ranked based on the results of systematic reviews, expert opinion, and data from the discrete choice experiments.
Cryptococcosis is a life-threatening invasive fungal infection, that poses a significant global health challenge. Historically, Cryptococcus was described as two species: C. neoformans (var. grubii and var. neoformans) and C. gattii. More recently, phylogenetic analyses have distinguished seven clades representing species (VNI-III and VGI-IV), and there are likely more, with varying virulence and regional distribution.2,3 For example, VGI is prevalent in Australia and Asia, VGII is particularly associated with the emergence in North America, VGIII is increasing among immunocompromised individuals in the United States, and VGIV is primarily found in Africa.3–5 Notably, the terminology of two cryptococcal ‘species complexes’ remains common in clinical practice as it is the most practicable for management purposes.
Cryptococcosis is best documented in people living with HIV/AIDS. However, it is increasingly recognized in other immunocompromised hosts, and occurs in people with various underlying conditions and even unrecognized risk factors.6–8 Members of the C. neoformans and C. gattii species complexes are the predominant causative agents,9 with species-specific differences in epidemiology: for example, C. neoformans species complex has traditionally been observed in HIV/AIDS patients, whilst C. gattii species complex infection has a propensity to occur in immunocompetent patients.10
Innate and adaptive responses work together to combat Cryptococcus spp., with CD4 + T-cells particularly important for an effective adaptive response.11,12 Symptomatic infection often indicates a compromised immune system, particularly in individuals with reduced CD4 + T-cell counts, such as people living with HIV.13–15 Latency and dormancy are also important aspects of cryptococcal pathogenesis. The fungus can remain dormant in the host due to both immune pressure and fungal factors,16–19 and in certain host environments, including granulomas, it can avoid immune detection.19 Reactivation of dormant cryptococci becomes a concern when the host's immune system becomes compromised, potentially leading to invasive disease.20 Improving our understanding of these and other factors is crucial for improving diagnostic, therapeutic, and preventive strategies.21
Cryptococcus neoformans and C. gattii species complexes are acquired via the respiratory tract, where they can cause local infection, although it is their tropism for the central nervous system (CNS) that is associated with the most serious manifestations of infection. Cryptococcal meningitis (CM) remains the most common cause of fungal meningitis worldwide with over 220 000 new cases and 180 000 deaths per annum.22 Consequently, CM is an infection of global relevance, with most deaths seen in sub-Saharan Africa and in South and Southeast Asia.23–25
Treatment options for invasive cryptococcosis are limited, and development of novel anti-cryptococcal agents has been slow in recent decades.26 Cryptococci are intrinsically resistant to echinocandins.27 Optimal induction treatment relies on amphotericin B and 5-flucytoscine despite their substantial toxicity and limited access associated with economic and logistical constraints. Prolonged treatment with azoles is required following induction therapy.28
In low- and middle-income countries (LMICs) where disease burden is highest, poor access to optimal therapeutics (i.e., 5-flucytosine and amphotericin B lipid formulations) increases the clinical challenges and contributes to the observed persistent poor clinical outcomes of cryptococcosis.29
This systematic review evaluates C. neoformans and C. gattii species complexe infections against a set of criteria, namely: mortality, hospitalization and disability, antifungal drug resistance, preventability, yearly incidence, global distribution, and emergence, based on data published between 2011 and 2021. The purpose is to determine knowledge gaps for both C. neoformans and C. gattii species complexes in the above areas to highlight research needs and to inform the WHO FPPL.
Materials and methods
Search strategies
We conducted a comprehensive search for studies published in English using the PubMed and Web of Science databases. These databases were chosen due to their extensive coverage of medical and scientific literature. The study was conducted according to PRISMA guidelines.30 All searches were limited to the last 10 years (from 1st January 2011 to 19th February 2021).
On PubMed, we used medical subject headings (MeSH) and/or keyword terms in the title/abstract for each pathogen and criterion.
For C. neoformans, the final search used (C. neoformans[Title] OR C. neoformans[Title]) combined; for C. gattii, the final search used (C. gattii [MeSH Terms]) combined, using AND term, with criteria terms including (mortality[MeSH Terms]) OR (morbidity[MeSH Terms]) OR (hospitalization[MeSH Terms]) OR (disability[All Fields]) OR (drug resistance, fungal[MeSH Terms]) OR (prevention and control[MeSH Subheading]) OR (disease transmission, infectious[MeSH Terms]) OR (diagnostic[Title/Abstract]) OR (antifungal agents[MeSH Terms]) OR (epidemiology[MeSH Terms]) OR (surveillance [Title/Abstract]).
On Web of Science, MeSH terms are not available and therefore topic search (TS), title (TI), or abstract (AB) search were used. The final search used [TI=(‘cryptococcus neoformans/cryptococcus gattii’) OR TI=(‘C. neoformans’) OR AB=(‘cryptococcus gattii’)], combined using AND term with criteria terms each as topic search, including (mortality) OR (case fatality) OR (morbidity) OR (hospitalization) OR (disability) OR (drug resistance) OR (prevention and control) OR (disease transmission) OR (diagnostic) OR (antifungal agents) OR (epidemiology) OR (surveillance). Symbol * allows a truncation search for variations of the term (e.g., hospitalization or hospitalization).
Study selection
We imported search results from each database into the online systematic review software, Covidence® (Veritas Health Innovation, Australia), and removed duplicates. The inclusion criteria were retrospective/prospective observational studies, randomized controlled trials, guidelines, epidemiology, surveillance reports, published within the last 10 years (2011–2021), reporting adults and paediatric data, including data on the fungal pathogen, and data on at least one criterion. Exclusion criteria were studies reporting on non-human data (e.g., animals, plants) or non-fungal data (e.g., bacteria), no data on relevant pathogens or criteria, case reports, conferences, abstracts, reviews, papers on drugs without marketing authorization, in vitro papers on resistance mechanisms, and papers published in non-English language. Identified articles underwent title and abstract screening based on the inclusion criteria. No reason was provided for exclusion during title and abstract screening. Two independent reviewers (AD and HYK) performed full text screening for the final eligible articles on Covidence®. A third reviewer resolved any discrepancies (JWA). Excluded articles were recorded with reasons when excluded during full text screening. If there were any additional articles identified from references of the included articles, these were added. The resulting articles were subject to the final analysis.
Data collection and synthesis
Data from the final included studies were extracted for relevant criteria (AD and HYK). The extracted data were checked by the second reviewer (JWA) (initially 10% check, then expanded to 20% and more if needed, depending on the type of extent of observed errors). The extracted data on the outcome criteria were qualitatively AND/OR quantitatively synthesized, depending on the amount and nature of the data.
Risk of bias assessment
We assessed risk of bias using the risk of bias tool for randomized trials version 2 (ROB 2) tool for randomized controlled trials.31 The risk of bias in non-randomized studies (RoBANS) tool was used to assess the non-randomized studies.32 For the overall risk, using ROB 2 tool, the studies were rated ‘low’, ‘high’, or ‘some’ concerns. Using the RoBANS tool, the studies were rated as ‘low’, ‘high’, or ‘unclear’ risk.
For the purposes of this review, we considered each criterion as an outcome of the study and assessed if any bias was expected based on the study design, data collection, and analysis methods for that outcome. Studies that were classified as having an unclear or high overall risk were still eligible for inclusion with cautious interpretation.
Results
Study selection
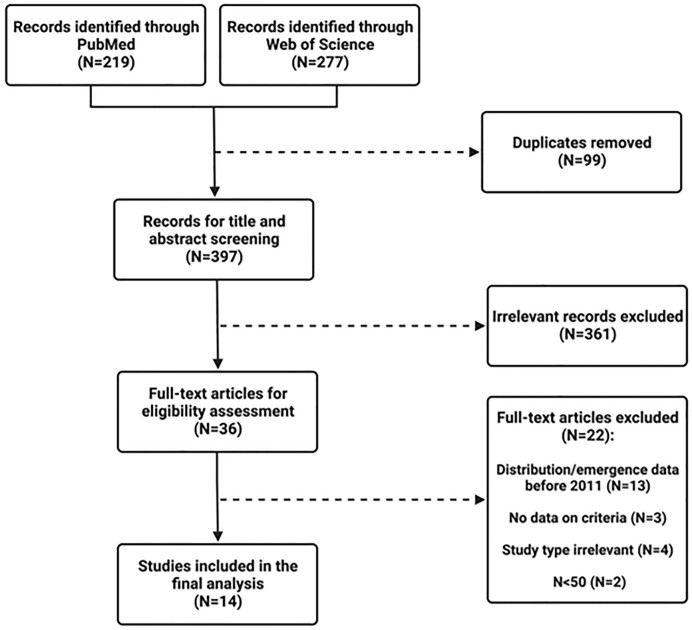
For C. neoformans, PubMed and Web of Science Core Collection databases searched between 1 January 2011 and 19 February 2021 yielded 287 and 388 articles, respectively (Fig. 1). For C. gattii, the search yielded 219 and 277 articles, respectively (Fig. 2). A total of 45 (C. neoformans) and 14 (C. gattii) articles were included in the final analysis.
Figure 1.
Flow diagram for selection of studies included in the systematic review for C. neoformans.
Figure 2.
Flow diagram for selection of studies included in the systematic review for C. gattii.
Risk of bias
For C. neoformans, the overall risk of bias for each study is presented in the Table 1A. Of the included studies, 22 studies were classified as low risk of bias in all domains assessed. Twenty-three studies were classified as unclear risk of bias, mostly due to the potential selection biases caused by unclear eligibility criteria or population groups, or unclear confirmation/consideration of confounding variables.
Table 1.
The risk of bias for each study of C. neoformans.
| Author | Publication year | Risk of bias (low, high, and unclear) | Reference |
|---|---|---|---|
| A | |||
| Andrade-Silva et al. | 2013 | Unclear | 33 |
| Andrade-Silva et al. | 2018 | Unclear | 34 |
| Ashton et al. | 2019 | Unclear | 35 |
| Bariao et al. | 2020 | Low | 36 |
| Beale et al. | 2015 | Low | 37 |
| Bertout et al. | 2012 | Unclear | 38 |
| Cao et al. | 2019 | Low | 39 |
| Chan et al. | 2014 | Low | 40 |
| Chen et al. | 2015 | Low | 41 |
| Chen et al. | 2018 | Low | 42 |
| Chowdhary et al. | 2011 | Unclear | 43 |
| Cogliati et al. | 2018 | Unclear | 44 |
| Córdoba et al. | 2016 | Low | 45 |
| de Oliveira et al. | 2017 | Low | 46 |
| Desnos-Ollivier et al. | 2015 | Unclear | 47 |
| Espinel-Ingroff et al. | 2012 | Unclear | 48 |
| Espinel-Ingroff et al. | 2012 | Unclear | 49 |
| Espinel-Ingroff et al. | 2015 | Unclear | 50 |
| Fan et al. | 2016 | Low | 51 |
| Gonzalez et al. | 2016 | Low | 52 |
| Govender et al. | 2011 | Unclear | 53 |
| Gutch et al. | 2015 | Unclear | 54 |
| Hagen et al. | 2016 | Unclear | 55 |
| Herkert et al. | 2018 | Unclear | 56 |
| Hurtado et al. | 2019 | Low | 57 |
| Kassi et al. | 2016 | Low | 58 |
| Lahiri et al. | 2020 | Unclear | 59 |
| Lin et al. | 2015 | Unclear | 60 |
| Mahabeer et al. | 2014 | Low | 61 |
| Mahabeer et al. | 2014 | Low | 62 |
| Martins et al. | 2011 | Low | 63 |
| Mdodo et al. | 2011 | Unclear | 64 |
| Miglia et al. | 2011 | Unclear | 65 |
| Naicker et al. | 2020 | Unclear | 66 |
| Nascimento et al. | 2017 | Low | 67 |
| Nishikawa et al. | 2019 | Low | 68 |
| Nyazika et al. | 2016 | Low | 69 |
| Pan et al. | 2012 | Unclear | 70 |
| Pfaller et al. | 2011 | Unclear | 71 |
| Prakash et al. | 2020 | Low | 72 |
| Rakotoarivelo et al. | 2020 | Unclear | 73 |
| Selb et al. | 2019 | Low | 74 |
| Smith et al. | 2015 | Low | 75 |
| Tewari et al. | 2012 | Unclear | 76 |
| Yoon et al. | 2020 | Low | 77 |
| B. The risk of bias for each study of C. gattii | |||
| Chen et al. | 2012 | Low | 78 |
| Chen et al. | 2013 | Low | 79 |
| Espinel-Ingroff et al. | 2012 | Unclear | 48 |
| Espinel-Ingroff et al. | 2012 | Unclear | 49 |
| Espinel-Ingroff et al. | 2015 | Unclear | 50 |
| Firacative et al. | 2016 | Unclear | 80 |
| Harris et al. | 2011 | Low | 81 |
| Hurtado et al. | 2019 | Unclear | 57 |
| Kassi et al. | 2016 | Unclear | 58 |
| Lahiri et al. | 2020 | Unclear | 59 |
| Lee et al. | 2019 | Unclear | 82 |
| Lockhart et al. | 2012 | Unclear | 83 |
| Phillips et al. | 2015 | Low | 84 |
| Smith et al. | 2014 | Low | 85 |
For C. gattii, the overall risk of bias for each study is presented in the Table 1B. Of the 14 studies, 5 studies were classified as low risk of bias in all domains assessed. Nine studies were classified as unclear risk of bias, mostly due to the selection biases caused by unclear eligibility criteria or population groups, or unclear confirmation/consideration of confounding variables.
Mortality rates
For C. neoformans, 13 studies reported on mortality (Table 2). The mortality rates due to C. neoformans were reported to be as high as 41%–61% for patients with HIV infection.37, 63, 64, 69, 73 Mortality rates specifically reported for HIV-negative patients were lower, ranging from 8% to 20%, but small patient numbers are noted (N = 12–44).63,40,77
Table 2.
The mortality rates due to C. neoformans and C. gattii infections.
| Author | Study period | Pathogen species | Country | Study design | Level of care | Population description | Patients (N=) | Mortality type, N/N, % |
|---|---|---|---|---|---|---|---|---|
| Desnos-Ollivier et al. 47 | 1997 to 2001 | Cryptococcus neoformans | France | Qualitative data and lab surveillance study (MC) | ND | Patients enrolled during the CryptoA/D study or the nationwide survey on cryptococcosis in France | 181 | % Patients who died within 90 days after diagnosis/total Serotype A: 21/82 (26%) Serotype D: 7/22 (32%) Serotype AD: 7/25 (28%) |
| Chan et al. 40 | 1999 to 2007 | Cryptococcus neoformans | Singapore | RCS (SC) | Tertiary | HIV with CD4 counts < 200 cells/mm3 | 62 | HIV+ 15/45 (33%) HIV- 1/12 (8%) OR (95% CI) 5.5 (0.65–46.69) P-value = 0.118 HIV+ Deaths at less or equal to 30 days = 9/46 (20%) HIV- Deaths at less or equal to 30 days = 1/12 (8%) OR (95% CI) 2.68 (0.30–127.77) P-value = 0.670 |
| Nascimento et al. 67 | 2000 to 2011 | Cryptococcus neoformans | Brazil | LSS (SC) | Tertiary | Patients with CM | 61 | 6/61 (10%) deaths (C. neoformans var. grubii) in AIDS patients |
| Govender et al. 53 | 2002 to 2008 | Cryptococcus neoformans | South Africa | PBS (MC) | Tertiary | Patients who had been diagnosed with the first episode of laboratory-confirmed cryptococcosis. Only 1033 out of 8439 met the selection criteria. |
1033 | Case fatality (non-30-day mortality) 2002–2003: 62/238 (26%) 2007–2008: 84/249 (36%) |
| Yoon et al. 77 | 2005 to 2017 | Cryptococcus neoformans | United States | RCS (SC) | Tertiary | All patients ≥ 18 years old with the diagnosis of cryptococcosis at Montefiore Medical Centre | 126 | 30-day mortality: HIV+ 4/68 (6%) HIV- 9/44 (20%) 1-year mortality HIV+ 7/55 (13%) HIV- 10/42 (24%) Cause of death due to cryptococcosis HIV + 3/7 (43%) HIV- 6/10 (60%) |
| Naicker et al. 66 | 2007 to 2008 and 2017 | Cryptococcus neoformans | South Africa | Prospective cohort study (MC) | ND | Patients with the first episode of culture-confirmed cryptococcal disease at 37 South African hospitals | 249 and 204 | In-hospital deaths 2007–2008: 84/249 (34%) 2017: 62/204 (30%) |
| Mdodo et al. 64 | 2008 to 2009 | Cryptococcus neoformans | Kenya | LSS (MC) | Tertiary | HIV-positive patients from Kenyatta National Hospital and Mbagathi District Hospital in Nairobi Kenya | 67 | In-hospital mortality 38/62 (61%) |
| Martins et al. 63 | 2008 to 2010 | Cryptococcus neoformans | Brazil | LSS (SC) | Tertiary | Patients diagnosed with mycological CM | 63 | Deaths occurred in 49% of the cases HIV+ 18/37 (49%) HIV- 13/26 (50%) The number is higher for patients infected by C. neoformans VNI genotype. Cryptococcus neoformans VNI predominated in HIV + patients |
| Smith et al. 75 | 2010 to 2014 | Cryptococcus neoformans | Uganda | LSS (MC) | Tertiary | HIV infected and was presenting with his or her first episode of CM. | 198 | Day 60 deaths: FLU susceptible: 29/58, 50% FLU dose-dependent: 11/27, 41% FLU resistant: 1/5, 20% AMB susceptible 41/89, 46% AMB resistant 0/1 |
| Nyazika et al. 69 | 2013 to 2014 | Cryptococcus neoformans | Zimbabwe | LSS (MC) | ND | HIV-infected adult inpatients from Parirenyatwa Group of Hospitals presenting signs and symptoms of meningitis. | 100 | Overall mortality rate 56% (30/54) AFLPI/VNI genotype 22/39 (56%) AFLPIA/VNB/VNII genotype 5/8 (63%) AFLPIB/VNII 3/7 genotype (43%) |
| Hurtado et al. 57 | 2013 to 2015 | Cryptococcus neoformans | Brazil | Autopsy study (MC) | Tertiary | 284 deceased patients; Cause of death assigned to a cryptococcal infection |
284 | Seventeen died from fatal cryptococcal infections. 7/17 patients (41%) died within the first 72 hours of admission. |
| Rakotoarivelo et al. 73 | 2014 to 2016 | Cryptococcus neoformans | Madagascar | CSS (MC) | Tertiary | Consecutive HIV-infected adults presenting with CD4 cell counts ≤ 200/µl | 129 | 90-day mortality: 30/129 (23%) 90-day mortality with CM: 8/14 (57%) 90-day mortality without CM: 22/115 (19%) |
| Beale et al. 37 | 2005 to 2010 | Cryptococcus neoformans | South Africa | RCS (MC) | ND | HIV infected individuals prior to the initiation of antifungal treatment | 230 | The overall mortality of 27% at 10 weeks, 41% at one year |
| Phillips et al. 84 | 1999 to 2007 | Cryptococcus gattii | British Columbia | RCS (MC) | ND | Patients with C. gattii infection, reported to BC Centre for Disease Control | 152 | Death due to C. gattii or where C. gattii contributed 11/47 (23.4%) (patients with CNS disease), 13/89 (14.6%) (with lung infection only) |
| Chen et al. 79,78 | 2000 to 2007 | Cryptococcus gattii | Australia | RCS (MC) | Tertiary | Adults with C. gattii infection | 86 | 11/85 (13%) (10 from C. gattii): within 4 months of diagnosis, 10/73 (13.6%) in CNS infection, 11% in CNS + lung infection, 17% in CNS infection only, (7/24) immunocompromised vs. (3/62) healthy hosts, 6/31 (19%) death at 12 months, patients with raised ICP |
| Smith et al. 85 | 2004 to 2011 | Cryptococcus gattii | United States Pacific Northwest (PNW) | RCS (MC) | ND | Patients with invasive C. gattii infection reported to CDC | 70 | 3-month mortality in patients 13/70 (19%) in all patients, 3/7 (43%) in bloodstream infections, 7/33 (21%) pulmonary infections, 3/30 (10%) CNS infections |
| Harris et al. 81 | 2004 to 2011 | Cryptococcus gattii | United States | RCS (MC) | Tertiary | Patients with C. gattii reported to the CDC, US | 76 | Died of or with infection 19/57 (33%) |
CSS = Cross sectional study; LSS = Lab surveillance study; MC = Multi-centre; ND = Not determined; PBS = Population-based surveillance; RSC = Retrospective cohort study; SC = Single centre
For C. gattii, four studies reported on mortality (Table 2). The mortality rate due to C. gattii-related bloodstream infection was 43% (N = 7) in the study by Smith et al.85 Other studies reported mortality rates of 10%–23.4% for CNS infections78, 79, 84, 85 and 14.6%–21% for pulmonary infections, acknowledging the relatively small cohorts.85,84
Antifungal susceptibilities
In total, 33 studies reported results of antifungal susceptibility testing on C. neoformans isolates (Table 3), and 6 studies for C. gattii (Table 4); methodologies included CLSI standard, EUCAST standard, Etest, Vitek 2 YST AST, and Sensititre YeastOne assays. Details of these studies are presented in the appendix (Tables A1 and A2).
Table 3.
Antifungal susceptibility of C. neoformans.
| Author | MIC methods | Fluconazole | Isavuconazole | Itraconazole | Ketoconazole | Posaconazole | Voriconazole | Amphotericin B | Flucytosine |
|---|---|---|---|---|---|---|---|---|---|
| Andrade-Silva et al. 33 | CLSI M27-A3 | MIC GM 9.7 MIC range 2–32 MIC50 8 MIC90 16 %R 0% 97.9% MIC ≤ ECV 16 |
NA | MIC GM 0.30 MIC range 0.06–2 MIC50 0.5 MIC90 1 %R 22% (21/95) 100% MIC ≤ ECV 1 |
MIC GM 0.16 MIC range 0.03–0.5 MIC50 0.12 MIC90 0.5 %R 0% 100% MIC ≤ ECV 0.5 |
NA | MIC GM 0.11 MIC range 0.06–0.25 MIC50 0.12 MIC90 0.25 %R 0% 97.9% ≤ ECV 1 |
MIC GM 0.69 MIC range 0.12–4 MIC50 1 MIC90 2 %R 11% (10/95) 97.9% ≤ ECV 2 |
NA |
| Bariao et al. 36 | CLSI M27-A3 | MIC GM 1.369 MIC range 0.25–16 MIC50 1 MIC90 4 |
NA | MIC GM 0.092 MIC range 0.031–0.25 MIC50 0.125 MIC90 0.125 |
NA | NA | MIC GM 0.089 MIC range 0.031–1 MIC50 0.062 MIC90 0.25 |
MIC GM 0.107 MIC range 0.031–1 MIC50 0.125 MIC90 0.25 |
MIC GM 1.079 MIC range 0.125–4 MIC50 1 MIC90 2 |
| Bertout et al. 38 | Sensititre YeastOne | MIC GM 3.22 MIC range 0.5–32 MIC50 2 MIC90 16 |
NA | MIC GM 0.05 MIC range < 0.008–0.12 MIC50 0.015 MIC90 0.12 |
MIC GM 0.08 MIC range < 0.008–0.25 MIC50 0.015 MIC90 0.12 |
MIC GM 0.10 MIC range 0.008–0.5 MIC50 0.06 MIC90 0.5 |
MIC GM 0.06 MIC range < 0.008–0.12 MIC50 0.015 MIC90 0.12 |
MIC GM 0.31 MIC range 0.06–1 MIC50 0.06 MIC90 0.5 |
MIC GM 2.76 MIC range 0.5–16 MIC50 2 MIC90 8 |
| Chen et al. 41 | CLSI M27-A3 | MIC range 2–64 30/89 (34%) fluconazole MIC ≥ 8 |
NA | NA | NA | NA | NA | NA | NA |
| Chen et al. 42 | ATBTM FUNGUS-3 kit | No isolates with MIC ≥ 16 MIC range 1–8 |
NA | No isolates with MIC ≥ 1 MIC range 0.125–0.5 |
NA | NA | No isolates with MIC ≥ 1 MIC range 0.06–0.25 |
No isolates with MIC ≥ 2 MIC range < 0.5–1 |
No isolates with MIC ≥ 32 MIC range < 4–4 |
| Chowdhary et al. 43 | CLSI M27-A3 | MIC GM 2.190 MIC range 0.5–8 MIC50 2 MIC90 4 |
NA | MIC GM 0.099 MIC range 0.031–0.250 MIC50 0.125 MIC90 0.250 |
NA | NA | MIC GM 0.053 MIC range 0.015–0.125 MIC50 0.062 MIC90 0.125 |
MIC GM 0.235 MIC range 0.031–1 MIC50 0.250 MIC90 0.5 |
MIC GM 1.450 MIC range 0.031–64 MIC50 2 MIC90 4 |
| Cogliati et al. 44 | Yeast nitrogen base (YNB) broth microdilution method | MIC GM 1.962 MIC range 0.12–16 MIC mode 4 98.6% MIC ≤ ECV 8 |
NA | MIC GM 0.096 MIC range 0.03–1 MIC mode 0.03 or 0.06 99.7% MIC ≤ ECV 0.5 |
NA | NA | MIC GM 0.049 MIC range 0.03–0.5 MIC mode 0.03 96.9% MIC ≤ ECV 0.12 |
NA | MIC GM 0.297 MIC range 0.12–64 MIC mode 0.25 MIC mean 0.297 97.8% MIC ≤ ECV 1 |
| Córdoba et al. 45 | EUCAST | MIC range 0.13–128 MIC50 8 MIC90 32 MIC mode 8 ECV95 32 (2.2% non-WT) ECV99 64 (0.7% non-WT) |
NA | MIC range 0.015–1 MIC50 0.03 MIC90 0.25 MIC mode 0.015 ECV95 0.5 (0.6% non-WT) ECV99 0.5 (0.6% non-WT) |
NA | MIC range 0.015–0.13 MIC50 0.015 MIC90 0.06 MIC mode 0.015 ECV95 0.06 (1.3% non-WT) ECV99 0.13 (0% non-WT) |
MIC range 0.015–2 MIC50 0.13 MIC90 0.25 MIC mode 0.13 ECV95 0.5 (1.9% non-WT) ECV99 1 (0.3% non-WT) |
MIC range 0.015–1 MIC50 0.25 MIC90 0.5 MIC mode 0.25 ECV95 0.5 (3.8% non-WT) ECV99 1 (0% non-WT) |
MIC range 0.13–128 MIC508 MIC90 16 MIC mode 4 ECV95 32 (2.4% non-WT) ECV99 128 (0% non-WT) |
| de Oliveira et al. 46 | EUCAST | NA | NA | NA | NA | NA | NA | MIC GM 0.4 MIC range 0.12–1 MIC mode 0.5 MIC50/90 0.5 |
NA |
| Espinel-Ingroff et al. 48 | CLSI M27-A3 | MIC range ≤ 0.12–≥ 64 MIC mode 4–8 98.3% MIC ≤ ECV 16 |
NA | MIC range ≤ 0.008–≥ 4 MIC mode 0.12 98.9% MIC ≤ ECV 0.5 |
NA | MIC range ≤ 0.008–≥ 2 MIC mode 0.12 94.3% MIC ≤ ECV 0.25 |
MIC range ≤ 0.008–≥ 4 MIC mode 0.06 96.5% MIC ≤ ECV 0.25 |
NA | NA |
| Espinel-Ingroff et al. 49 | CLSI M27-A3, Etest |
NA | NA | NA | NA | NA | NA | MIC range ≤ 0.03–4 MIC mode 0.25 ECV95 1 ECV99 2 |
MIC range 0.06– to ≥ 64 MIC mode 4 ECV95 = 16 ECV99 = 32 |
| Espinel-Ingroff et al. 50 | CLSI M27-A3 | NA | MIC range 0.008–0.5 MIC mode 0.03 ECV95 0.06–0.12 ECV97.5 0.12 |
NA | NA | NA | NA | NA | NA |
| Fan et al. 51 | Sensititre YeastOne | MIC GM 4.28 MIC range 0.5–64 MIC50 4 MIC90 8 WT 92.4% Non-WT 7.6% |
NA | MIC GM 0.057 MIC range 0.015–0.5 MIC50 0.06 MIC90 0.12 WT 99% Non-WT 1% |
NA | MIC GM 0.084 MIC range 0.008–0.5 MIC50 0.06 MIC90 0.25 WT 97.7% Non-WT 2.3% |
MIC GM 0.034 MIC range 0.008–0.5 MIC50 0.03 MIC90 0.12 WT 98.3% Non-WT 1.7% |
MIC GM 0.60 MIC range 0.25–1.0 MIC50 0.5 MIC90 1.0 WT 100% Non-WT 0% |
MIC GM 3.42 MIC range 0.06–16 MIC50 4 MIC90 8 WT 98.7% Non-WT 1.3% |
| Gonzalez et al. 52 | CLSI M27-A3 | MIC GM 1.335 MIC range 0.5–4 MIC50 2 MIC90 2 |
NA | NA | NA | NA | MIC GM 0.061 MIC range 0.03–0.125 MIC50 0.06 MIC90 0.125 |
MIC GM 0.343 MIC range 0.125–1 MIC50 0.25 MIC90 1 |
NA |
| Govender et al. 53 | CLSI M27-A3 | 2002–2003 MIC range 0.5–16 MIC50 1 MIC90 2 2007–2008 MIC range 0.25–8 MIC50 1 MIC90 2 |
NA | 2002–2003 MIC range 0.03–1 MIC50 0.12 MIC90 0.25 2007–2008 MIC range 0.015–0.5 MIC50 0.06 MIC90 0.12 |
NA | 2002–2003 MIC range 0.03–0.5 MIC50 0.12 MIC90 0.25 2007–2008 MIC range 0.03–1 MIC50 0.06 MIC90 0.12 |
2002–2003 MIC range 0.008–0.25 MIC50 0.015 MIC90 0.06 2007–2008 MIC range 0.008–0.25 MIC50 0.015 MIC90 0.03 |
2002–2003 MIC range 0.012–0.38 MIC50 0.094 MIC90 0.19 2007–2008 MIC range 0.008–0.94 MIC50 0.094 MIC90 0.19 |
2002–2003 MIC range 0.25–16 MIC50 1 MIC90 4 2007–2008 MIC range 0.05–8 MIC50 1 MIC90 2 |
| Tewari et al. 76 | Vitek 2, Etest, CLSI M27-A3 | CLSI MIC range 0.25–32 MIC90 16 Etest MIC range 1– >256 MIC90 16 Vitek 2 MIC range < 1–>64 MIC90 8 80%–90% of isolates MIC ≤ 8 |
NA | NA | NA | NA | NA | CLSI MIC range 0.06–5 MIC90 0.5 Etest MIC range 0.047–0.38 MIC90 0.25 Vitek 2 MIC range < 0.25–2 MIC90 1 98%–100% of isolates MIC < 1 |
NA |
| Gutch et al. 54 | CLSI M27-A | MIC range 0.063–64 MIC50 8 MIC90 32 Mean 6.93 MIC ≥ 64, 8.6% MIC 16–32, 31.1% MIC ≤ 8, 60.3% |
NA | MIC range 0.03–1 MIC50 0.125 MIC90 0.5 Mean 0.124 MIC ≥ 1, 5.2% MIC 0.25–0.5, 24.1% MIC ≤ 0.125, 70.7% |
MIC range 0.03–0.25 MIC500.064 MIC90 0.064 Mean 0.051 MIC ≥ 0.125, 6.9% MIC 0.0625, 55.2% MIC < 0.0625 37.9% |
NA | NA | NA | NA |
| Hagen et al. 55 | EUCAST | MIC GM 8.96 MIC range 0.5–> 32 MIC50 4 |
MIC GM 0.065 MIC range < 0.03–0.25 MIC50 0.06 |
NA | NA | NA | MIC GM 0.104 MIC range < 0.03–0.5 MIC50 0.06 |
MIC GM 0.180 MIC range < 0.03–1 MIC50 0.125 |
MIC GM 8.80 MIC range 1–> 32 MIC50 8 |
| Herkert et al. 56 | CLSI M27-A3 | MIC GM 0.516 MIC range 0.125–8 MIC50 0.5 MIC90 0.5 |
MIC GM 0.011 MIC range < 0.016–0.063 MIC50 < 0.016 MIC90 0.031 |
MIC GM 0.027 MIC range < 0.016–0.25 MIC50 0.031 MIC90 0.063 |
NA | MIC GM 0.027 MIC range < 0.016–0.125 MIC50 0.031 MIC90 0.063 |
MIC GM 0.021 MIC range < 0.016–0.125 MIC50 0.031 MIC90 0.031 |
MIC GM 0.098 MIC range < 0.016–0.125 MIC50 0.125 MIC90 0.125 |
MIC GM 2.42 MIC range 0.25–8 MIC50 2 MIC90 4 |
| Hurtado et al. 57 | Sensititre YeastOne | MIC range 4–16 | NA | MIC range 0.03–0.12 | NA | MIC range 0.06–0.25 | MIC range 0.06–0.25 | MIC range 0.5–1 | MIC range 1–16 |
| Kassi et al. 58 | CLSI M27-A3 | MIC range 0.125–8 | NA | NA | NA | NA | NA | MIC range 0.125–1 | MIC range 0.5–16 |
| Mahabeer et al. 61 | CLSI M27-A3, Etest, Vitek 2 | CLSI MIC range 0.25–4 MIC50 1 MIC90 2 Etest MIC range 0.25–4 MIC50 1 MIC90 2 Vitek 2 MIC range ≤ 1–16 MIC50 ≤ 1 MIC90 2 |
NA | NA | NA | NA | CLSI MIC range ≤ 0.002–0.064 MIC50 0.004 MIC90 0.016 Etest MIC range ≤ 0.002–0.064 MIC500.008 MIC90 0.016 |
CLSI MIC range ≤ 0.008–1 MIC50 0.125 MIC90 0.25 Etest MIC range ≤ 0.008–0.25 MIC50 0.06 MIC90 0.125 Vitek 2 MIC range ≤ 0.25–0.5 MIC50 ≤ 0.25 MIC90 0.5 |
CLSI MIC range ≤ 0.125–4 MIC50 1 MIC90 2 Etest MIC range ≤ 1–8 MIC50 ≤ 1 MIC90 2 |
| Mahabeer et al. 62 | CLSI M27-A3, Etest, Vitek-2 | CLSI MIC range 0.25–4 MIC50 1 MIC90 2 Etest MIC50 1 MIC90 2 MIC range 0.06–4 Vitek-2 MIC range ≤ 1–16 MIC50 ≤ 1 MIC90 2 |
NA | NA | NA | NA | CLSI MIC range ≤ 0.002–0.064 MIC50 0.004 MIC90 0.016 Etest MIC range ≤ 0.002–0.064 MIC50 0.008 MIC90 0.016 |
CLSI MIC range ≤ 0.008–1 MIC50 0.125 MIC90 0.25 Etest MIC range ≤ 0.008–0.25 MIC50 0.06 MIC90 0.125 Vitek-2 MIC range ≤ 0.25–0.5 MIC50 ≤ 0.25 MIC90 0.5 |
CLSI MIC range ≤ 0.125–4 MIC50 1 MIC90 2 Vitek-2 MIC range ≤ 1–8 MIC50 ≤ 1 MIC90 2 |
| Mdodo et al. 64 | CLSI M27-A3 | MIC range 0.25–16 MIC50 4 MIC90 8 |
NA | NA | NA | NA | MIC range 0.015–0.25 MIC50 0.06 MIC90 0.25 |
MIC range 0.5–1 MIC50 1 MIC90 1 |
MIC range 1–16 MIC50 2 MIC90 4 |
| Naicker et al. 66 | CLSI M27-A3 | 2007–2008 MIC GM: 2.08 MIC range 0.25–8 MIC50 1 MIC90 2 2017 MIC GM: 4.11 MIC range 0.5–64 MIC50 4 MIC90 8 |
NA | NA | NA | NA | NA | NA | NA |
| Nascimento et al. 67 | CLSI M27-A2, E–test |
CLSI MIC GM 0.30 MIC range 1–16 MIC50 0.25 MIC90 0.5 Etest MIC GM 0.20 MIC range 0.047–0.5 MIC50 0.19 MIC90 0.38 |
NA | CLSI MIC GM 0.13 MIC range 0.03–1.0 MIC50 0.06 MIC90 0.25 Etest MIC GM 0.44 MIC range 0.016–2.0 MIC50 0.38 MIC90 0.75 |
NA | NA | CLSI MIC GM 0.27 MIC range 0.03–0.5 MIC50 0.25 Broth MIC90 0.50 Etest MIC GM 0.14 MIC range 0.016–0.75 MIC50 0.094 MIC90 0.25 |
CLSI MIC GM 0.30 MIC range 0.13–0.5 MIC50 0.25 Broth MIC90 0.50 Etest MIC GM 0.20 MIC range 0.047–0.5 MIC50 0.19 MIC90 0.38 |
NA |
| Nishikawa et al. 68 | CLSI M27-A3, Etest, Vitek 2 |
CLSI MIC range 2–8 MIC50 4 MIC90 4 MIC mode 4 Etest MIC range 2–32 MIC50 4 MIC90 16 MIC mode 8 Vitek 2 MIC range ≤ 1–2 MIC50 1 MIC90 2 MIC mode 2 |
NA | CLSI MIC range 0.06–0.5 MIC50 0.125 MIC90 0.125 MIC mode 0.125 Etest MIC range 0.032–1 MIC50 0.125 MIC90 0.75 MIC mode 0.125 |
NA | NA | CLSI MIC range 0.015–0.25 MIC50 0.06 MIC90 0.06 MIC mode 0.06 Etest MIC range 0.016–0.38 MIC50 0.047 MIC90 0.094 MIC mode 0.06 Vitek 2 MIC range ≤ 0.125 MIC50 ≤ 0.125 MIC90 ≤ 0.125 MIC mode ≤ 0.125 |
CLSI MIC range 0.5–2 MIC50 1 MIC90 1 MIC mode 1 Etest MIC range 0.012–0.25 MIC50 0.094 MIC90 0.125 MIC mode 0.094 or 0.125 Vitek 2 MIC range 1–2 MIC50 1 MIC90 1 MIC mode 1 |
CLSI MIC range 1–8 MIC50 2 MIC90 4 MIC mode 2, 4 Etest MIC range 0.125–>32 MIC50 4 MIC90 8 MIC mode 4 Vitek 2 MIC range ≤ 1–2 MIC50 ≤ 1 MIC90 2 MIC mode ≤ 1 |
| Pan et al. 70 | CLSI M27-A3 | MIC GM 2.294 MIC range 0.125–32 MIC50 2 MIC90 4 |
MIC GM 0.027 MIC range < 0.016–0.125 MIC50 0.031 MIC90 0.063 |
MIC GM 0.063 MIC range < 0.016–0.5 MIC50 0.063 MIC90 0.25 |
NA | MIC GM 0.061 MIC range < 0.016–0.5 MIC50 0.063 MIC90 0.125 |
MIC GM 0.049 MIC range < 0.016–0.5 MIC50 0.063 MIC90 0.125 |
MIC GM 0.251 MIC range 0.063–1 MIC50 0.25 MIC90 0.5 |
MIC GM 3.483 MIC range < 0.063–>64 MIC50 4 MIC90 8 |
| Pfaller et al. 71 | CLSI M27-A3 | MIC range 0.25–32 Mode 4 ECV 8 96.9% MIC ≤ ECV 8 |
NA | NA | NA | MIC range 0.03–0.5 Mode 0.12 ECV 0.25 96.5% MIC ≤ ECV 0.25 |
MIC range 0.008–0.5 Mode 0.06 ECV 0.12 95.1% MIC ≤ ECV 0.12 |
NA | NA |
| Prakash et al. 72 | CLSI M27-A3 | MIC GM 3.575 MIC range 0.06–64 MIC50 4 MIC90 8 |
MIC GM 0.03136 MIC range 0.016–0.25 MIC50 0.03 MIC90 0.063 |
MIC GM 0.517 MIC range 0.016–0.5 MIC50 0.06 MIC90 0.125 |
NA | MIC GM 0.06658 MIC range 0.016–0.5 MIC50 0.06 MIC90 0.125 |
MIC GM 0.051 MIC range 0.016–1 MIC50 0.06 MIC90 0.125 |
MIC GM 0.228 MIC range 0.03–4 MIC50 0.25 MIC90 0.5 |
MIC GM 4.660 MIC range 0.25–64 MIC50 4 MIC90 16 |
| Rakotoarivelo et al. 73 | Etest | MIC range 0.5–>256 MIC mode 12 ECV 32 |
NA | NA | NA | NA | MIC range 0.004–0.5 MIC mode 0.047 ECV 0.5 |
MIC range 0.032–0.5 MIC mode 0.250 ECV 1 |
MIC range 4–>32 MIC mode > 32 ECV 16 |
| Selb et al. 74 | CLSI M27-A3 | Serotype A MIC range 0.5–16 MIC50 1 MIC90 2 MIC mode 1 Serotype D MIC range 0.125–0.5 MIC50 0.5 MIC90 2 MIC mode 0.5 |
NA | NA | NA | Serotype A MIC range 0.03–0.5 MIC50 0.06 MIC90 0.125 MIC mode 0.06 Serotype D MIC range 0.03–0.125 MIC50 0.03 MIC90 0.06 MIC mode 0.03 |
Serotype A MIC range 0.03–0.125 MIC50 0.03 MIC90 0.03 MIC mode 0.03 Serotype D MIC range 0.03 MIC50 0.03 MIC90 0.03 MIC mode 0.03 |
Serotype A MIC range 0.125–0.5 MIC50 0.5 MIC90 0.5 MIC mode 0.5 Serotype D MIC range 0.25–0.5 MIC50 0.5 MIC90 0.5 MIC mode 0.5 |
Serotype A MIC range 0.25–>64 MIC50 1 MIC90 1 MIC mode 1 Serotype D MIC range 1–>64 MIC50 4 MIC90 8 MIC mode 2 |
| Smith et al. 75 | CLSI | MIC range 0.125–64 MIC mode 8 MIC50 8 MIC90 32 69% isolates MIC < 16 |
NA | NA | NA | NA | NA | MIC range 0.125–2 MIC mode 0.5 MIC50 0.5 MIC90 1 |
NA |
Data are reported as they appear in source documents. Susceptibility is expressed as mg/l unless indicated otherwise. ECV = epidemiological cutoff value, GM = geometric mean, MIC = minimum inhibitory concentration, NA = not available, MIC50 = MIC required to inhibit the growth of 50% of isolates, MIC90 = MIC required to inhibit the growth of 90% of isolates.
Table 4.
Antifungal susceptibility of C. gattii.
| Author | Year | MIC method | Fluconazole | Isavuconazole | Itraconazole | Posaconazole | Voriconazole | Amphotericin B | Flucytosine |
|---|---|---|---|---|---|---|---|---|---|
| Espinel-Ingroff et al. 48 | 2012 | CLSI M27A-3 | MIC mode Non-typed 4 VGI 4 VGII 8 VGIIa 4 VGIII 4 VGIV 8 |
NA | MIC mode Non-typed 0.12 VGI 0.25 VGII 0.12 VGIII 0.25 VGIV 0.5 |
MIC mode Non-typed 0.12 VGI 0.12 |
MIC mode Non-typed 0.06 VGI 0.12 VGII 0.12 VGIIa 0.12 |
NA | NA |
| Espinel-Ingroff et al. 49 | 2012 | CLSI M27A-3 | NA | NA | NA | NA | NA | MIC mode (range) Non-typed 0.5 (0.06–1) VGI 0.25 (0.03–1) VGII 0.5 (0.125–2) VGIIa 0.25 (0.06–1) |
MIC mode Non-typed 1 (0.25–8) VGI 2 (0.125–>64) VGII 2 (0.25–≥64) |
| Espinel-Ingroff et al. 50 | 2015 | CLSI | NA | MIC mode (range) 0.03 (0.008–0.5) | NA | NA | NA | NA | NA |
| Firacative et al. 80 | 2016 | Sensititre YeastOne | Clinical isolates GM MIC 5.384 |
NA | Clinical isolates GM MIC 0.0453 |
Clinical isolates GM MIC 0.06 987 | Clinical isolates GM MIC 0.04421 |
Clinical isolates GM MIC 0.2726 |
Clinical isolates GM MIC 1.927 |
| Lee et al. 82 | 2019 | Sensititre YeastOne | VGI GM MIC 1.46 MIC range 0.25–2 |
NA | VGI GM MIC 0.02 MIC range 0.015–0.06 |
VGI GM MIC 0.04 MIC range 0.008–0.12 |
VGI GM MIC 0.02 MIC range 0.008–0.06 |
VGI GM MIC 0.39 MIC range 0.12–1 |
VGI GM MIC 0.47 MIC range 0.25–2 |
| Lockhart et al. 83 | 2012 | CLSI | All isolates GM MIC 5.51 MIC range 0.5–32 VGI GM MIC 1.69 MIC range 0.5–8 VGII GM MIC 8.60 MIC range 1–32 VGIII GM MIC 3.48 MIC range 1–16 VGIV GM MIC 4.79 MIC range 0.5–32 |
NA | All isolates GM MIC 0.30 MIC range 0.03–2 VGI GM MIC 0.19 MIC range 0.03–1 VGII GM MIC 0.36 MIC range 0.06–2 VGIII GM MIC 0.28 MIC range 0.06–0.5 VGIV GM MIC 0.30 MIC range 0.06–2 |
All isolates GM MIC 0.31 MIC range 0.008–1 VGI GM MIC 0.20 MIC range 0.03–1 VGII GM MIC 0.33 MIC range 0.008–1 VGIII GM MIC 0.34 MIC range 0.12–1 VGIV GM MIC 0.34 MIC range 0.12–1 |
All isolates GM MIC 0.10 MIC range 0.008–1 VGI GM MIC 0.03 MIC range 0.008–0.25 VGII GM MIC 0.13 MIC range 0.015–1 VGIII GM MIC 0.07 MIC range 0.03–0.25 VGIV GM MIC 0.10 MIC range 0.03–0.5 |
NA | NA |
Data are reported as they appear in source documents. Susceptibility is expressed as mg/l unless indicated otherwise. ECV = epidemiological cutoff value, GM = geometric mean, MIC = minimum inhibitory concentration, NA = not available, MIC50 = MIC required to inhibit the growth of 50% of isolates, MIC90 = MIC required to inhibit the growth of 90% of isolates.
Cryptococcus neoformans susceptibility to antifungals
Before 2020, when EUCAST has provided a CBP for amphotericin B only, there were no interpretative clinical breakpoint (CBP) MICs for C. neoformans. It is also noteworthy that no causal relationship has been established between MIC and treatment failure.86 Consequently, interpretive criteria applied to antifungal MIC results for C. neoformans in the reviewed publications were highly variable both within and between publications. Examples of interpretive criteria included utilizing C. albicans CBPs, or breakpoints suggested with user manuals provided with testing kits, CLSI epidemiologic cutoff values (ECVs), and values selected from previous scientific publications.
Reported susceptibility of C. neoformans to fluconazole was variable, with two studies reporting no ‘resistance’ in their tested isolates33,42 and some others reporting higher ‘resistance’ rates of up to 30%.41,75 Fluconazole MIC90 values were variable between studies; however, were as high as 16 to 32 mg/l based on CLSI33,54, 75, 76 and EUCAST methods for MIC determination.45 Chen et al. observed significantly increasing numbers of isolates with fluconazole MIC ≥ 8 mg/l over the study period 2001–2012 (P < 0.001).41
Limited numbers of studies reported susceptibility to isavuconazole. Geometric mean MIC values from these studies ranged from 0.011 to 0.065 mg/l55, 56, 70, 72 and MIC90 values ranged from 0.031 to 0.063 mg/l.56, 70, 72 Reduced susceptibility to itraconazole (0.03–2 mg/l) was uncommon, ranging from 0% to 22%,42,54 with ≤ 1% non-wild type (non-WT) rates.45,48,51 ‘Resistance’ rates were lower for ketoconazole (0%–7%)33,54 and voriconazole (0%).33,42 For posaconazole and voriconazole, non-WT rates of 1.3%–5.7% were reported.45,48,51
For amphotericin B, Andrade-Silva et al. reported a resistance rate of 11% based on 95 isolates from HIV/AIDS patients in Brazil,33 in contrast to Tewari et al. reporting < 2% resistance rate in their Indian population (80% without HIV infection).76
Susceptibility to 5-flucytosine was only reported as non-WT rates of 1%–2%,45,51 and MIC90 values were highly variable between studies but were as high as 8–16 mg/l.45,70,72,51,38 Selb et al. observed a lower MIC90 of 1 mg/l for serotype A (genotype VNI) compared with MIC90 of 8 mg/l for serotype D (genotype VNIV).74
Cryptococcus gattii susceptibility to antifungals
For C. gattii, all studies reported MIC values without interpretive CBP MICs.
Studies by Espinel-Ingroff et al. and Lockhart et al. were conducted on large number of isolates (∼300) from multiple countries.48,49, 50, 83 Reported MICs for fluconazole were generally high (range: 0.5–32 mg/l), although variable, with isolates of molecular type VGII showing the highest modal or geometric mean MIC of > 8 mg/l compared with other molecular types (1.7–4.0 mg/l for VGI and VGIII).48,83 Modal MICs of itraconazole, posaconazole, and voriconazole for C. gattii ranged from 0.06 to 0.5 mg/l for both molecular-typed and non-typed isolates.48
For amphotericin B, modal or geometric mean MICs ranged from 0.25 to 0.5 mg/l for both typed and non-typed isolates.49,80,82 Susceptibility results for flucytosine were variable with modal or geometric mean MICs of 0.5–2 mg/l, and with higher values reported (> 64 mg/l) for molecular types VGI and VGII.49,80,82 No susceptibility data were available for echinocandins, but Cryptococcus species, like all basidiomycetes are intrinsically resistant to this class.
Annual incidence and global distribution
Annual global incidence rates for C. neoformans and C. gattii could not be assessed due to lack of denominator from all included studies. However, at a population level, there were estimated 220 000 cases of CM globally in 2014 (about 3 in 100 000 population).22 Chen et al. reported the annual incidence of C. gattii infections was 6 in 100 000 between 2000 and 2007 in Australia,78 but higher (nearly 10-fold) annual incidence rate was reported in Aboriginal Australians.78
Although its proportional contribution to total cases of cryptococcal disease varies by geographic region, it was evident that C. neoformans was globally distributed.87 The prevalence of C. neoformans among isolates causing CM was reported in three multi-centre studies from African countries73,58,65 and one single-centre study from India (Table 5).59 In Madagascar during 2014–2016, the proportion of cryptococcal infection caused by C. neoformans var. grubii (serotype A) in HIV-infected patients was 13.2%.73 A multi-centre lab surveillance study conducted in South Africa during 2005–2006 reported a high prevalence (82%) of C. neoformans serotype A (VNI) and a lower prevalence (0%–10%) of serotype A (VNB, VNII), serotype AD (VNIII), and serotype D (VNIV) among paediatric patients with cryptococcosis.65 Similarly, in Ivory Coast during 2012–2014, a study showed 86% of HIV-associated CM was caused by C. neoformans VNI genotype.58 In India, the majority of the CNS cryptococcosis patients were from Bangalore Urban, Karnataka, which is in the southern part of India; 80% of the clinical strains were C. neoformans VNI and 8.75% were C. neoformans VNII.59
Table 5.
The incidence and global distribution of C. neoformans and C. gattii.
| Study period | Pathogen | Country | Study design | Level of care | Population description | Patients (N=) | Incidence | References |
|---|---|---|---|---|---|---|---|---|
| 2005 to 2006 | Cryptococcus neoformans | South Africa | LSS (MC) | ND | Paediatric and adult patients with cryptococcosis during a 2-year period in South Africa | 199 | Paediatric cases Serotype A, VNI: 67/82 (82%) Serotype A, VNB: 8/82 (10%) Serotype A, VNII: 6/82 (7%) Serotype AD, VNIII: 1/82 (1%) Serotype D, VNIV: 0/82 |
Miglia et al. 65 |
| 2008 to 2017 | Cryptococcus gattii | Australia | RCS (MC) | ND | Patients with cryptococcal infections | ND | 13/55 (24%) C. gattii complex (majority VGI (11), VGII (2) uncommon) | Lee et al. 82 |
| 2012 to 2014 | Cryptococcus neoformans and C. gatii | Ivory Coast | LSS (MC) | Tertiary | Patients with HIV positive, and none of them received a systemic antifungal treatment | 61 | High prevalence of CM (86%) due to C. neoformans VNI among HIV-infected patients. The results show the prevalence (95%) of serotype A in Ivory Coast. 1/61 (C. gattii (VGII)) |
Kassi et al. 58 |
| 2013 to 2015 | Cryptococcus gattii | Mozambique and Brazil | Autopsy study (MC) | Tertiary | Deceased patients (for diagnostic autopsies) | 223 (Mozambique) and 61 (Brazil) | 5/15 (33%) C. gattii (VGI and VGIV molecular types) out of fatal cryptococcal infections | Hurtado et al. 57 |
| 2014 to 2016 | Cryptococcus neoformans | Madagascar | CSS (MC) | Tertiary | Consecutive HIV-infected adults presenting with CD4 cell counts ≤ 200/µl | 129 | The overall prevalence of cryptococcal infection was 13.2% (17/129, 95% CI7.9-20.3), and that of CM was 10.9% (14/129, 95% CI 6.1–17.5). | Rakotoarivelo et al. 73 |
| ND | Cryptococcus neoformans and C. gatii | India | Epidemiology study (SC) | Tertiary | CNS cryptococcosis patients attending the neurological and neurosurgical services of National Institute of Mental Health and Neurosciences. | 160 | 146/160 (91%) from Karnataka 14/160 (9%) from Tamil Nadu, Andhra Pradesh, West Bengal, Orissa, Bihar, and Pondicherry. 80% C. neoformans VNI, 8.75% VNII and 22.5% C . gattii (VGI), 8.75% C. gattii (VGIV). |
Lahiri et al. 59 |
| ND | Cryptococcus gattii | India | PCS (SC) | Tertiary | Patients with CNS cryptococcosis | 160 | 18/160 (11.25%) C. gattii (Of these, 14 (8.75%) were C. gattii genotype AFLP7/VGIV (serotype C), and 4 (2.5%) C. gattii AFLP4/VGI (serotype B). | Lahiri et al. 59 |
CSS = Cross sectional study; LSS = Lab surveillance study; MC = Multi-centre; ND = Not determined; PCS = Prospective cohort study; RSC = Retrospective cohort study; SC = Single centre
There was limited data available to assess the global distribution of C. gattii, four studies informed prevalence of C. gattii in patients with cryptococcal infections in different study locations, including Australia, India, Brazil, and Africa (Table 5). Overall, C. gattii accounted for 11%–33% of cryptococcal infections.82,59,57 In contrast, the earlier study conducted in Ivory Coast reported only one case of C. gattii infection in 61 HIV-positive patients with cryptococcal infections.58 Like C. neoformans, the distribution of C. gattii molecular types seems to vary across regions, although it was difficult to assess as few regions were represented. In Australia, genotype VGI caused the majority of the C. gattii cases,82 whereas in India, VGIV was the most commonly observed genotype.59
Inpatient care and the length of stay in hospital
The median hospital length of stay in patients with C. neoformans infection ranged from 18 to 39 days,40, 69, 73,39 with only Cao et al. 2019 reporting on HIV-negative patients (Table 6). Although Chan et al. reported a greater length of stay for HIV-negative patients with cryptococcosis (predominantly involving C. neoformans var. grubii VNI) compared with HIV positive cryptococcosis patients (31 days vs. 18.5 days), this difference was based on only 12 HIV-negative patients and was not statistically significant.40
Table 6.
The hospital length of stay due to C. neoformans and C. gattii infections.
| Study period | Pathogens | Study design | Country | Level of care | Population description | Patients (N=) | Length of stay | References |
|---|---|---|---|---|---|---|---|---|
| 1999 to 2007 | Cryptococcus neoformans | RCS (SC) | Singapore | Tertiary | HIV with CD4 counts < 200 cells/mm3 | 62 | HIV+ 18.5 days (13–33) (median IQR) HIV- 31 days (17.5–44.5) OR (95% CI) 0.99 days (0.97–1.01), P-value 0.192 |
Chan et al. 40 |
| 2000 to 2007 | Cryptococcus gattii | RCS (MC) | Australia | Tertiary | Adults with C. gattii infection | 86 | mean ICU stay: 9.1 days (range 1–29) (n = 18) | Chen et al. 79,78 |
| 2010 to 2016 | Cryptococcus neoformans | RCS (SC) | China | Tertiary | CM patients | 90 | Pulmonary nodule (PN) positive: 39 days (2–180) PN negative: 37 days (5–210) P-value 0.768 |
Cao et al. 39 |
| 2013 to 2014 | Cryptococcus neoformans | LSS (MC) | Zimbabwe | ND | HIV-infected adult inpatients from Parirenyatwa Group of Hospitals with signs and symptoms of meningitis. | 100 | 17.5 days of hospital stay IQR (10–22 days) |
Nyazika et al. 69 |
| 2014 to 2016 | Cryptococcus neoformans | CSS (MC) | Madagascar | Tertiary | Consecutive HIV-infected adults presenting with CD4cell counts ≤ 200/µl | 129 | Hospital stay, days, median, (IQR): 22 (11.0–35.0) | Rakotoarivelo et al. 73 |
CSS = Cross sectional study; LSS = Lab surveillance study; MC = Multi-centre; ND = Not determined; RSC = Retrospective cohort study; SC = Single centre
Only one study reported on the hospital length of stay in patients with C. gattii infection (Table 6). This nationwide retrospective study conducted in Australian hospitals described average intensive care unit (ICU) stay related to C. gattii infection in 18 adult patients as 9.1 days with a wide range of 1–29 days.78 It did not report overall hospital length of stay. Notably, 90% of patients in this study received amphotericin B for the first 14 days, which typically requires inpatient therapy.
Complications, sequelae, and disabilities
Both C. neoformans and C. gattii infections can lead to severe complications, sequelae, and disabilities (Table 7).
Table 7.
The complications, sequelae and disabilities caused by C. neoformans and C. gattii.
| Study period | Pathogens | Study design | Country | Level of care | Population description | Patients (N=) | Complications, sequelae, and disabilities | References |
|---|---|---|---|---|---|---|---|---|
| 1999 to 2007 | Cryptococcus neoformans | RCS (SC) | Singapore | Tertiary | HIV with CD4 counts < 200 cells/mm3 | 62 | Complications in 50 HIV+ patients: Raised ICP needing shunts (18%) of patients, blindness (12%), acute renal impairment (28%) |
Chan et al. 40 |
| 1999 to 2007 | Cryptococcus gattii | RCS (MC) | British Columbia | ND | Patients with C. gattii infection, reported to BC Centre for Disease Control | 152 | Persistent neurological symptoms at the end of 12-month follow-up in 8/47 (17%) of CNS patients: including gait or balance disturbance (n = 3), partial hearing loss (n = 2), cognitive impairment (n = 2), blindness (n = 1), and seizure disorder (n = 1) |
Phillips et al. 84 |
| 2000 to 2007 | Cryptococcus gattii | RCS (MC) | Australia | Tertiary | Adults with C. gattii infection | 86 | 20/73 (27%) with neurological sequelae at 12 months, including: visual impairment (n = 8), deafness (n = 3), limb weakness (n = 2), dysphasia (n = 2), IRIS (n = 8) after 6 weeks to 12 months |
Chen et al. 79,78 |
| 2010 to 2016 | Cryptococcus neoformans | RCS (SC) | China | Tertiary | CM patients | 90 | Unfavourable clinical outcome in pulmonary nodule (PN)-positive patients vs. PN-negative patients (72.5% vs. 48%, P = 0.019): Glasgow Outcome Scale score (on discharge) of 1 to 4, which indicates death, vegetative status, severe and moderate disability, was considered unfavourable clinical outcomes. [40/90 (44%) patients was PN-positive and 50/90 (56%) was PN-negative] |
Cao et al. 39 |
MC = Multi-centre; ND = Not determined; RSC = Retrospective cohort study; SC = Single centre
A 2017 review highlighted that neurosensorial impairment and disability are common sequelae 6 months to 1 year after diagnosis in C. neoformans infections. Symptoms mainly include residual headache, motor deficit, and vertigo.88 Other common complications may include anaemia, hypokalaemia, elevated aminotransferase levels, neutropenia, hypercreatinemia, and opportunistic infections.89
A study (n = 50) described complications from C. neoformans infection and treatment in HIV-positive individuals (mostly infected with C. neoformans var. grubii VNI genotype), including acute renal impairment, likely associated with antifungal therapies (28% of patients), raised intracranial pressure (ICP) needing shunts (18%), and blindness (12%).40 Cao et al. reported a higher rate of unfavourable clinical outcome (defined as death, vegetative status, or severe to moderate disability) in CM patients with pulmonary nodules compared with those without the pulmonary nodule involvement (72.5% vs. 48%, P = 0.019).39
Day et al. (2013) found that baseline fungal count and Glasgow Coma Scale (GCS) were independent predictors of 6-month survival for CM. Furthermore, the choice of therapy regimen affects the survival rate and complications. For instance, it was found that neutropenia was more frequent among patients receiving amphotericin B with fluconazole or flucytosine than patients receiving amphotericin B monotherapy. Also, fewer patients had severe anaemia and visual deficit when combined therapy of amphotericin B with fluconazole/flucytosine than amphotericin B therapy alone.89
Neurological sequelae at 12 months of treatment were reported in 17%–27% of patients with C. gattii infections, and included signs and symptoms of visual impairment, hearing loss, limb weakness or balance disturbance, and cognitive impairment.78,84
Immune reconstitution inflammatory syndrome (IRIS) was observed in 9.4% of patients with C. gattii infections from 6 weeks to as long as 12 months after the initiation of azole eradication therapy, and these patients presented with new or enlarging brain lesions.78
Preventability
Risk factors for C. neoformans infection were documented in two studies. HIV/AIDS, cell-mediated immunity-suppressive regimens without calcineurin inhibitors, and decompensated liver cirrhosis were risk factors for CM (adjusted OR of 181.4, 15.9, and 8.5, respectively) and cryptococcemia (adjusted OR of 216.3, 7.3, and 23.8, respectively).60 Autoimmune diseases (adjusted OR = 9.3) were an additional risk factor for cryptococcemia.60
HIV-infected patients and immunocompromised individuals are particularly vulnerable to cryptococcal infections and CM. Although not specific to C. neoformans, a retrospective review of routine cerebrospinal fluid laboratory records (N = 4702) between 2000 and 2014 in Botswana, South Africa, determined that antiretroviral therapy access alone did not lead to a significant decrease in the incident rate of HIV-associated CM.90 Furthermore, several systematic reviews have quantified the preventative effect of pre-emptive therapy on CM: Relative risk of 0.19 (P < 0.0001)91; incidence reduced from 21% to 5% in patients with CD4 < 100, relative risk 0.2392; and incidence reduced from 5% to 3% in patients with CD4 < 200, relative risk 0.6.93
A study by Harris et al. observed that patients with C. gattii outbreak strain infections had more pre-existing conditions compared with patients with non-outbreak strain infections (86% vs. 31%; P < 0.0001).81 The pre-existing conditions mainly involved immunosuppression or previous use of oral corticosteroids (during the year before infection) in 50% of patients and existing lung, renal, heart disease, or diabetes in 20%–30% of patients. It was also observed that patients with outbreak strain infections were older [median (range) of 56 (2–95) vs. 45 (18–56) years, P = 0.007].
Discussion
Cryptococcosis is particularly common in HIV/AIDS patients. However, antiretroviral therapy (ART) access alone has not always decreased the incidence of HIV-associated CM significantly.90 This observation may be associated with late presentation and cumulative default from care by HIV/AIDS patients, suggesting that integrated interventions beyond simply providing ART are required to prevent cryptococcosis and CM.
Cryptococcosis can lead to prolonged hospitalization. The long length of stay in hospital may be partially attributed to treatment recommendations involving 14 days induction therapy with amphotericin B for most of the study period (although current WHO treatment recommendations for HIV-associated CM now favour shorter courses of amphotericin). Amphotericin B must be administered intravenously and, in most settings, is delivered as in-patient therapy. Although CM clearly causes significant morbidity and has a long-term impact on patients, the effect is poorly quantified, and future CM studies should continue to expand the evidence on short- and longer-term disability and quality of life.
There is clear evidence that cryptococcosis is associated with high mortality. Baddley et al. stated that the all-cause mortality rates were 18.8% at 3 months and 25.5% at 12 months.94 The rates described in this review are higher than those observed in clinical trials. For example, some studies have reported mortality rates for CM of around 20%.95–99 In trials, patients with significant co-morbidities or very advanced disease may be excluded, and interventions and investigations follow a strict protocol. These factors may contribute to the lower mortality.26,100 Furthermore, diagnoses such as toxoplasmosis, Pneumocystis jirovecii pneumonia, or other opportunistic infections may be more thoroughly screened for and managed in trial settings. This hypothesis is supported by Tenforde et al. (2020), who found that in sub-Saharan Africa, short-term mortality rate was 44% in observational studies and only 21% in randomized control trials.101 Regardless, the mortality rate is unacceptably high, and global research to improve outcomes is needed.
A detailed summary of antifungal susceptibility data is presented in this review. We observed rising MICs to azoles (e.g., itraconazole, ketoconazole, and voriconazole), including in vitro ‘resistance’ to fluconazole in up to 30%,41 with an increasing number of isolates with MIC ≥8 µg/ml between 2001 and 2012. However, the data are limited, and there is yet no clear association between MIC and clinical outcomes. Nonetheless, this observation calls for ongoing surveillance globally and investigation into the cause. Since Cryptococcus spp. are not transmitted from human to human, an environmental selection pressure for azole resistance could hypothetically be at play, as described for other fungal pathogens such as Aspergillus.102 Two studies reported that in patients with HIV/AIDS, 11% of the Cryptococcus strains showed non-WT MICs to amphotericin B. A much lower percentage (< 2%) of the Cryptococcus strains showed non-WT MICs to amphotericin B in HIV-negative patients.
Cryptococcus gattii susceptibility data varied with molecular type and, in general, showed higher MICs to fluconazole compared with other azoles, including isavuconazole, itraconazole, posaconazole, and voriconazole. MICs for amphotericin B (0.25–0.5 mg/l) and 5-flucytosine (0.5–2 mg/l) were low. Therefore, future studies should continue tracking antifungal susceptibility and resistance for C. gattii, and their correlation with clinical outcomes.
There have been significant developments in prevention of CM over the past decade. Strong evidence has emerged for the cost-effectiveness of screening for C. neoformans cryptococcal antigenaemia with point-of-care antigen tests and treating positive cases, especially in low-resource settings or high-prevalence areas with high number of HIV cases.103,104 However, there are no data on high-income countries, for C. gattii, or for patient groups outside of HIV/AIDS.
The systematic reviews of C. neoformans and C. gattii infections were characterized by sparse, frequently inconsistent data. For instance, there were few studies determining the incidence of infections in specific countries. However, it is known that C. neoformans is globally distributed, with some geographic variation between members of the species complex as the causative agent. For example, in Madagascar, 13.2% of HIV-infected patients had cryptococcal infection due to C. neoformans var. grubii (serotype A). Studies in South Africa, Ivory Coast, and India reported high prevalence of C. neoformans serotype A (VNI) (80%–86%) in adult and paediatric patients with cryptococcosis. C. gattii accounted for 11%–33% of cryptococcal infections overall in countries such as Australia, India, Brazil, and Africa.
Trends over the last 10 years for C. neoformans were difficult to assess due to incomplete data. However, the prevalence of C. neoformans serotype A VNI reported in two African countries and India was comparable and was consistently high (80%–86%) over the period of 2011–2020.58, 59, 65 Apart from that, there was also a lack of country-level or global surveillance studies reporting the emergence of C. gattii infections in the last 10 years. The studies reporting the prevalence of C. gattii did not provide adequate data to assess global trends. Although studies conducted in African countries (Ivory Coast and Mozambique, respectively) showed a greater prevalence of 33% in 2019 compared with 1.6% in 2016,58,57 these data are confounded by environmental and study population-related variables. Thus, it is not possible to make a conclusive statement about the trend in this region.
Our review has several limitations. In particular, we were unable to include non-English-language studies. We only included data from peer-reviewed and indexed publications and may therefore have missed valuable data.
Future perspectives
Future research on C. neoformans and C. gattii should focus on several key areas: (1) obtaining more robust clinical and microbiological data to support diagnosis and treatment; (2) developing new diagnostic tools and treatments; (3) understanding the genetic and molecular mechanisms of these pathogens; (4) understanding host-pathogen interactions and host’s immunological response to the infection; (5) understanding the epidemiology of these pathogens in different regions and populations to identify high-risk groups and develop targeted prevention and control strategies.
Stronger surveillance systems and epidemiology studies would better inform the disease burden and the global distribution of C. neoformans and C. gattii. These may allow more rigorous identification of at-risk populations, dispersion patterns, and preventative measures. Better understanding of clinical manifestations and susceptibility profiles for different molecular types is needed and could potentially inform individualized treatment options. Conducting trials in cryptococcosis is complex because disease is rare, and it is difficult to recruit sufficient patients into clinical trials to detect impacts on clinical outcome, especially in non-HIV populations. Several groups have investigated surrogate markers of treatment effect (such as early fungicidal activity)105,106 to allow smaller trials. Additional work in this area is needed.
Conclusion
Cryptococcus neoformans and C. gattii are important fungal pathogens. Both are globally distributed with significant incidence and mortality rates. Although rising MICs to antifungals have been reported, these are yet to show a clear impact on clinical outcomes. Careful ongoing systematic observations are warranted alongside detailed work to better define burden of infection in terms of both death and disability.
The knowledge gaps identified through this systematic review open avenues for future research studies to elucidate the genetic and molecular mechanisms underlying C. neoformans and C. gattii infections. Understanding host-pathogen interactions, the role of host immune responses, and the impact of specific molecular characteristics on disease outcomes can guide the development of targeted therapies and interventions. Furthermore, the observed disparities in global distribution and prevalence among different regions and populations emphasize the importance of region-specific surveillance and tailored public health strategies. By addressing these research gaps, the disease burden of cryptococcosis can be reduced, and the health outcomes of affected individuals across the globe can be improved.
Supplementary Material
Acknowledgements
This work, and the original report entitled ‘WHO Fungal Priority Pathogens List to Guide Research, Development, and Public Health Action’, was supported by funding kindly provided by the Governments of Austria and Germany (Ministry of Education and Science). We acknowledge all members of the WHO Advisory Group on the Fungal Priority Pathogens List (WHO AG FPPL), the commissioned technical group, and all external global partners, as well as Haileyesus Getahun (Director Global Coordination and Partnerships Department, WHO), for supporting this work. The authors alone are responsible for the views expressed in this article and they do not necessarily represent the decisions, policies, or views of the World Health Organization.
Contributor Information
Aiken Dao, Sydney Infectious Diseases Institute, The University of Sydney, Sydney, Australia; Westmead Institute for Medical Research, Westmead, Sydney, Australia; Westmead Clinical School, Westmead Hospital, Sydney, Australia.
Hannah Yejin Kim, Sydney Infectious Diseases Institute, The University of Sydney, Sydney, Australia; Sydney Pharmacy School, Faculty of Medicine and Health, The University of Sydney, Camperdown, Australia; Department of Pharmacy, Westmead Hospital, Westmead, Australia.
Katherine Garnham, Sydney Infectious Diseases Institute, The University of Sydney, Sydney, Australia; Sunshine Coast University Hospital, Birtinya, Qld 4575, Australia.
Sarah Kidd, National Mycology Reference Centre, Microbiology and Infectious Diseases, SA Pathology, Adelaide, Australia.
Hatim Sati, AMR Division, World Health Organization, Geneva, Switzerland.
John Perfect, Duke University Medical Center, Durham, NC, USA.
Tania C Sorrell, Sydney Infectious Diseases Institute, The University of Sydney, Sydney, Australia; Westmead Institute for Medical Research, Westmead, Sydney, Australia; Westmead Clinical School, Westmead Hospital, Sydney, Australia.
Thomas Harrison, Institute of Infection and Immunity, St George’s University London, London, UK; Medical Research Council Centre for Medical Mycology, University of Exeter, Exeter, UK.
Volker Rickerts, Robert Koch Institute, Berlin, Germany.
Valeria Gigante, AMR Division, World Health Organization, Geneva, Switzerland.
Ana Alastruey-Izquierdo, Mycology Reference Laboratory, National Centre for Microbiology, Instituto de Salud Carlos III, Madrid, Spain.
Jan-Willem Alffenaar, Sydney Infectious Diseases Institute, The University of Sydney, Sydney, Australia; Westmead Clinical School, Westmead Hospital, Sydney, Australia; Sydney Pharmacy School, Faculty of Medicine and Health, The University of Sydney, Camperdown, Australia.
C Orla Morrissey, Department of Infectious Diseases, Alfred Health, Melbourne, Australia; Monash University, Department of Infectious Diseases, Melbourne, Victoria, Australia.
Sharon C-A Chen, Centre for Infectious Diseases and Microbiology Laboratory Services, Institute of Clinical Pathology and Medical Research, New South Wales Health Pathology, Westmead Hospital, Westmead, Sydney, Australia.
Justin Beardsley, Sydney Infectious Diseases Institute, The University of Sydney, Sydney, Australia; Westmead Institute for Medical Research, Westmead, Sydney, Australia; Westmead Clinical School, Westmead Hospital, Sydney, Australia.
Author contributions
Aiken Dao (Data curation, Investigation, Project administration, Writing – original draft), Hannah Yejin Kim (Conceptualization, Investigation, Methodology, Project administration, Writing – original draft, Writing – review & editing), Katherine Garnham (Data curation, Formal analysis, Writing – original draft, Writing – review & editing), Sarah Kidd (Data curation, Validation, Writing – review & editing), Hatim Sati (Conceptualization, Data curation, Formal analysis, Funding acquisition, Methodology, Project administration, Writing – review & editing), John Perfect (Conceptualization, Formal analysis, Methodology, Writing – review & editing), Tania C. Sorrell (Conceptualization, Methodology, Writing – review & editing), Thomas Harrison (Conceptualization, Methodology, Writing – review & editing), Volker Rickerts (Conceptualization, Methodology, Writing – review & editing), Valeria Gigante (Data curation, Project administration, Writing – review & editing), Ana Alastruey-Izquierdo (Conceptualization, Formal analysis, Methodology, Project administration, Writing – review & editing), Jan-Willem Alffenaar (Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Project administration, Writing – review & editing), C. Orla Morrissey (Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Project administration, Writing – review & editing), Sharon C-A. Chen (Data curation, Formal analysis, Writing – review & editing), and Justin Beardsley (Conceptualization, Formal analysis, Funding acquisition, Methodology, Project administration, Visualization, Writing – original draft, Writing – review & editing)
Conflict of interest
AA-I has received personal fees for educational talks on behalf of Gilead and Pfizer.
None.
References
- 1. Bongomin F, Gago S, Oladele RO, Denning DW. Global and multi-national prevalence of fungal diseases-estimate precision. J Fungi (Basel). 2017; 3(4): 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hagen F, Khayhan K, Theelen B et al. Recognition of seven species in the Cryptococcus gattii/Cryptococcus neoformans species complex. Fungal Genet Biol. 2015; 78: 16–48. [DOI] [PubMed] [Google Scholar]
- 3. Byrnes EJ III, Li W, Lewit Y et al. Emergence and pathogenicity of highly virulent Cryptococcus gattii genotypes in the Northwest United States. PLoS Pathog. 2010; 6(4): e1000850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chen S, Sorrell T, Nimmo G et al. Epidemiology and host- and variety-dependent characteristics of infection due to Cryptococcus neoformans in Australia and New Zealand. Australasian Cryptococcal Study Group. Clin Infect Dis. 2000; 31(2): 499–508. [DOI] [PubMed] [Google Scholar]
- 5. Kidd SE, Hagen F, Tscharke RL et al. A rare genotype of Cryptococcus gattii caused the cryptococcosis outbreak on Vancouver Island (British Columbia, Canada). Proc Natl Acad Sci USA. 2004; 101(49): 17258–17263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Firacative C, Trilles L, Meyer W. Recent advances in cryptococcus and cryptococcosis. Microorganisms. 2021; 10(1): 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ferreira-Paim K, Andrade-Silva L, Fonseca FM et al. MLST-based population genetic analysis in a global context reveals clonality amongst Cryptococcus neoformans var. grubii VNI isolates from HIV patients in southeastern Brazil. PLoS Negl Trop Dis. 2017; 11(1): e0005223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Henao-Martínez AF, Chastain DB, Franco-Paredes C. Treatment of cryptococcosis in non-HIV immunocompromised patients. Curr Opin Infect Dis. 2018; 31(4): 278–285. [DOI] [PubMed] [Google Scholar]
- 9. Chayakulkeeree M, Perfect JR. Cryptococcosis. Infect Dis Clin North Am. 2006; 20(3): 507–544. [DOI] [PubMed] [Google Scholar]
- 10. Kidd SE, Chen SC, Meyer W, Halliday CL. A new age in molecular diagnostics for invasive fungal disease: are we ready?. Front Microbiol. 2019; 10: 2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. McQuiston TJ, Williamson PR. Paradoxical roles of alveolar macrophages in the host response to Cryptococcus neoformans. J Infect Chemother. 2012; 18(1): 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Osterholzer JJ, Milam JE, Chen GH, Toews GB, Huffnagle GB, Olszewski MA. Role of dendritic cells and alveolar macrophages in regulating early host defense against pulmonary infection with Cryptococcus neoformans. Infect Immun. 2009; 77(9): 3749–3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jarvis JN, Harrison TS. HIV-associated cryptococcal meningitis. AIDS. 2007; 21(16): 2119–2129. [DOI] [PubMed] [Google Scholar]
- 14. Warkentien T, Crum-Cianflone NF. An update on Cryptococcus among HIV-infected patients. Int J STD AIDS. 2010; 21(10): 679–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rajasingham R, Govender NP, Jordan A et al. The global burden of HIV-associated cryptococcal infection in adults in 2020: a modelling analysis. Lancet Infect Dis. 2022; 22(12): 1748–1755. 10.1016/S1473-3099(22)00499-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. May RC, Stone NR, Wiesner DL, Bicanic T, Nielsen K. Cryptococcus: from environmental saprophyte to global pathogen. Nat Rev Micro. 2016; 14(2): 106–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Alanio A. Dormancy in Cryptococcus neoformans: 60 years of accumulating evidence. J Clin Invest. 2020; 130(7): 3353–3360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Alanio A, Vernel-Pauillac F, Sturny-Leclère A, Dromer F. Cryptococcus neoformans host adaptation: toward biological evidence of dormancy. mBio. 2015; 6(2): e02580–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Voelz K, May RC. Cryptococcal interactions with the host immune system. Euk Cell. 2010; 9(6): 835–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Perfect JR. Cryptococcus neoformans: the yeast that likes it hot. FEMS Yeast Res. 2006; 6(4): 463–468. [DOI] [PubMed] [Google Scholar]
- 21. Mukaremera L, Nielsen K. Adaptive immunity to Cryptococcus neoformans infections. J Fungi (Basel). 2017; 3(4): 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rajasingham R, Smith RM, Park BJ et al. Global burden of disease of HIV-associated cryptococcal meningitis: an updated analysis. Lancet Infect Dis. 2017; 17(8): 873–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fisher KM, Montrief T, Ramzy M, Koyfman A, Long B. Cryptococcal meningitis: a review for emergency clinicians. Intern Emerg Med. 2021; 16(4): 1031–1042. [DOI] [PubMed] [Google Scholar]
- 24. Oliveira LSS, Pinto LM, de Medeiros MAP et al. Comparison of Cryptococcus gattii/neoformans species complex to related genera (Papiliotrema and Naganishia) reveal variances in virulence associated factors and antifungal susceptibility. Front Cell Infect Microbiol. 2021; 11: 642658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nyazika TK, Robertson VJ, Nherera B, Mapondera PT, Meis JF, Hagen F. Comparison of biotyping methods as alternative identification tools to molecular typing of pathogenic Cryptococcus species in sub-Saharan Africa. Mycoses. 2016; 59( 3): 151–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Beardsley J, Wolbers M, Kibengo FM et al. Adjunctive dexamethasone in HIV-associated cryptococcal meningitis. N Engl J Med. 2016; 374(6): 542–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Iyer KR, Revie NM, Fu C, Robbins N, Cowen LE. Treatment strategies for cryptococcal infection: challenges, advances and future outlook. Nat Rev Micro. 2021; 19(7): 454–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chang CC, Hall V, Cooper C et al. Consensus guidelines for the diagnosis and management of cryptococcosis and rare yeast infections in the haematology/oncology setting, 2021. Intern Med J. 2021; 51: 118–142. [DOI] [PubMed] [Google Scholar]
- 29. Loyse A, Burry J, Cohn J et al. Leave no one behind: response to new evidence and guidelines for the management of cryptococcal meningitis in low-income and middle-income countries. Lancet Infect Dis. 2019; 19(4):e143–e147. [DOI] [PubMed] [Google Scholar]
- 30. Page MJ, McKenzie JE, Bossuyt PM et al. The PRISMA 2020 statement: an updated guideline for reporting systematic reviews. BMJ. 2021; 372: n71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sterne JAC, Savović J, Page MJ et al. RoB 2: a revised tool for assessing risk of bias in randomised trials. BMJ. 2019; 366: l4898. [DOI] [PubMed] [Google Scholar]
- 32. Kim SY, Park JE, Lee YJ et al. Testing a tool for assessing the risk of bias for nonrandomized studies showed moderate reliability and promising validity. J Clin Epidemiol. 2013; 66(44):408–414. [DOI] [PubMed] [Google Scholar]
- 33. Andrade-Silva L, Ferreira-Paim K, Mora DJ et al. Susceptibility profile of clinical and environmental isolates of Cryptococcus neoformans and Cryptococcus gattii in Uberaba, Minas Gerais, Brazil. Med Mycol. 2013; 51(6): 635–640. [DOI] [PubMed] [Google Scholar]
- 34. Andrade-Silva LE, Ferreira-Paim K, Ferreira TB et al. Genotypic analysis of clinical and environmental Cryptococcus neoformans isolates from Brazil reveals the presence of VNB isolates and a correlation with biological factors. PLoS One. 2018; 13(3): e0193237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ashton PM, Thanh LT, Trieu PH et al. Three phylogenetic groups have driven the recent population expansion of Cryptococcus neoformans. Nat Commun. 2019; 10(1): 2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bariao PHG, Tonani L, Cocio TA, Martinez R, Nascimento E, Kress MRV. Molecular typing, in vitro susceptibility and virulence of Cryptococcus neoformans/Cryptococcus gattii species complex clinical isolates from south-eastern Brazil. Mycoses. 2020; 63(12): 1341–1351. [DOI] [PubMed] [Google Scholar]
- 37. Beale MA, Sabiiti W, Robertson EJ et al. Genotypic Diversity Is Associated with Clinical Outcome and Phenotype in Cryptococcal Meningitis across Southern Africa. PLoS Negl Trop Dis. 2015; 9(6): e0003847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bertout S, Drakulovski P, Kouanfack C et al. Genotyping and antifungal susceptibility testing of Cryptococcus neoformans isolates from Cameroonian HIV-positive adult patients. Clin Microbiol Infect. 2013; 19(8): 763–769. [DOI] [PubMed] [Google Scholar]
- 39. Cao W, Jian C, Zhang H, Xu S. Comparison of clinical features and prognostic factors of cryptococcal meningitis caused by Cryptococcus neoformans in patients with and without pulmonary nodules. Mycopathologia. 2019; 184(1): 73–80. [DOI] [PubMed] [Google Scholar]
- 40. Chan M, Lye D, Win MK, Chow A, Barkham T. Clinical and microbiological characteristics of cryptococcosis in Singapore: predominance of Cryptococcus neoformans compared with Cryptococcus gattii. Int J Infect Dis. 2014; 26: 110–115. [DOI] [PubMed] [Google Scholar]
- 41. Chen YC, Chang TY, Liu JW et al. Increasing trend of fluconazole-non-susceptible Cryptococcus neoformans in patients with invasive cryptococcosis: a 12-year longitudinal study. BMC Infect Dis. 2015; 15: 277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chen YH, Yu F, Bian ZY et al. Multilocus sequence typing reveals both shared and unique genotypes of Cryptococcus neoformans in Jiangxi Province, China. Sci Rep. 2018; 8(1): 1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chowdhary A, Randhawa HS, Sundar G et al. In vitro antifungal susceptibility profiles and genotypes of 308 clinical and environmental isolates of Cryptococcus neoformans var. grubii and Cryptococcus gattii serotype B from north-western India. J Med Microbiol. 2011; 60(Pt 7): 961–967. [DOI] [PubMed] [Google Scholar]
- 44. Cogliati M, Prigitano A, Esposto MC et al. Epidemiological trends of cryptococcosis in Italy: molecular typing and susceptibility pattern of Cryptococcus neoformans isolates collected during a 20-year period. Med Mycol. 2018; 56(8): 963–971. [DOI] [PubMed] [Google Scholar]
- 45. Córdoba S, Isla MG, Szusz W, Vivot W, Altamirano R, Davel G. Susceptibility profile and epidemiological cut-off values of Cryptococcus neoformans species complex from Argentina. Mycoses. 2016; 59(6): 351–356. [DOI] [PubMed] [Google Scholar]
- 46. de Oliveira L, Cristina Silva Santos D, dos Anjos Martins M et al. Time-kill curves studies with amphotericin B against Cryptococcus neoformans/C. gattii species complex clinical isolates. Curr Fungal Infect Rep. 2017; 11: 158–162. [Google Scholar]
- 47. Desnos-Ollivier M, Patel S, Raoux-Barbot D, Heitman J, Dromer F. Cryptococcosis serotypes impact outcome and provide evidence of Cryptococcus neoformans speciation. mBio. 2015;6(3): e00311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Espinel-Ingroff A, Aller AI, Canton E et al. Cryptococcus neoformans-Cryptococcus gattii species complex: an international study of wild-type susceptibility endpoint distributions and epidemiological cutoff values for fluconazole, Itraconazole, posaconazole, and voriconazole. Antimicrob Agents Chemother. 2012; 56(11): 5898–5906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Espinel-Ingroff A, Chowdhary A, Cuenca-Estrella M et al. Cryptococcus neoformans-Cryptococcus gattii species complex: an international study of wild-type susceptibility endpoint distributions and epidemiological cutoff values for amphotericin B and flucytosine. Antimicrob Agents Chemother. 2012; 56(6): 3107–3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Espinel-Ingroff A, Chowdhary A, Gonzalez GM et al. Multicenter study of isavuconazole MIC distributions and epidemiological cutoff values for the Cryptococcus neoformans-Cryptococcus gattii species complex using the CLSI M27-A3 broth microdilution method. Antimicrob Agents Chemother. 2015; 59(1): 666–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Fan X, Xiao M, Chen S et al. Predominance of Cryptococcus neoformans var. grubii multilocus sequence type 5 and emergence of isolates with non-wild-type minimum inhibitory concentrations to fluconazole: a multi-centre study in China. Clin Microbiol Infect. 2016; 22(10): 887.e1. [DOI] [PubMed] [Google Scholar]
- 52. Gonzalez GM, Casillas-Vega N, Garza-Gonzalez E et al. Molecular typing of clinical isolates of Cryptococcus neoformans/Cryptococcus gattii species complex from Northeast Mexico. Folia Microbiol (Praha). 2016; 61(1): 51–56. [DOI] [PubMed] [Google Scholar]
- 53. Govender NP, Patel J, van Wyk M, Chiller TM, Lockhart SR. Trends in antifungal drug susceptibility of Cryptococcus neoformans isolates obtained through population-based surveillance in South Africa in 2002-2003 and 2007-2008. Antimicrob Agents Chemother. 2011; 55(6): 2606–2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Gutch RS, Nawange SR, Singh SM et al. Antifungal susceptibility of clinical and environmental Cryptococcus neoformans and Cryptococcus gattii isolates in Jabalpur, a city of Madhya Pradesh in Central India. Braz J Microbiol. 2015; 46(4): 1125–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Hagen F, Hare Jensen R, Meis JF, Arendrup MC. Molecular epidemiology and in vitro antifungal susceptibility testing of 108 clinical Cryptococcus neoformans sensu lato and Cryptococcus gattii sensu lato isolates from Denmark. Mycoses. 2016; 59(9): 576–584. [DOI] [PubMed] [Google Scholar]
- 56. Herkert PF, Meis JF, Lucca de Oliveira Salvador G et al. Molecular characterization and antifungal susceptibility testing of Cryptococcus neoformans sensu stricto from southern Brazil. J Med Microbiol. 2018; 67(4): 560–569. [DOI] [PubMed] [Google Scholar]
- 57. Hurtado JC, Castillo P, Fernandes F et al. Mortality due to Cryptococcus neoformans and Cryptococcus gattii in low-income settings: an autopsy study. Sci Rep. 2019; 9(1): 97493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kassi FK, Drakulovski P, Bellet V et al. Molecular epidemiology reveals genetic diversity among 363 isolates of the Cryptococcus neoformans and Cryptococcus gattii species complex in 61 Ivorian HIV-positive patients. Mycoses. 2016; 59(12): 811–817. [DOI] [PubMed] [Google Scholar]
- 59. Lahiri S, Manjunath N, Bhat M et al. Clinical insights and epidemiology of central nervous system infection due to Cryptococcus neoformans/gattii species complexes: a prospective study from South India. Med Mycol. 2020; 58(5): 600–608. [DOI] [PubMed] [Google Scholar]
- 60. Lin YY, Shiau S, Fang CT. Risk factors for invasive Cryptococcus neoformans diseases: a case-control study. PLoS One. 2015; 10(3): e0119090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Mahabeer Y, Chang CC, Naidu D et al. Comparison of Etests and Vitek 2 ® to broth microdilution for the susceptibility testing of Cryptococcus neoformans. Diagn Microbiol Infect Dis. 2014; 80(4): 294–298. [DOI] [PubMed] [Google Scholar]
- 62. Mahabeer Y, Chang CC, Naidu D et al. Comparison of Etests and Vitek 2 (R) to broth microdilution for the susceptibility testing of Cryptococcus neoformans. Diagn Microbiol Infect Dis. 2014; 80(4): 294–298. [DOI] [PubMed] [Google Scholar]
- 63. Martins LMS, Wanke B, Lazera MD et al. Genotypes of Cryptococcus neoformans and Cryptococcus gattii as agents of endemic cryptococcosis in Teresina, Piaui (northeastern Brazil). Mem Inst Oswaldo Cruz. 2011; 106(6): 725–730. [DOI] [PubMed] [Google Scholar]
- 64. Mdodo R, Moser SA, Jaoko W et al. Antifungal susceptibilities of Cryptococcus neoformans cerebrospinal fluid isolates from AIDS patients in Kenya. Mycoses. 2011; 54(5): E438–E442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Miglia KJ, Govender NP, Rossouw J, Meiring S, Mitchell TG, Group for Enteric, Respiratory, and and Meningeal Disease Surveillance in South Africa . Analyses of pediatric isolates of Cryptococcus neoformans from South Africa. J Clin Microbiol. 2011; 49(1): 307–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Naicker SD, Mpembe RS, Maphanga TG et al. Decreasing fluconazole susceptibility of clinical South African Cryptococcus neoformans isolates over a decade. PLoS Negl Trop Dis. 2020; 14(3): e0008137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Nascimento E, Vitali LH, Kress M, Martinez R. Cryptococcus neoformans and C. gattii isolates from both HIV-infected and uninfected patients: antifungal susceptibility and outcome of cryptococcal disease. Rev Inst Med Trop Sao Paulo. 2017; 59: e49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Nishikawa MM, Almeida-Paes R, Brito-Santos F et al. Comparative antifungal susceptibility analyses of Cryptococcus neoformans VNI and Cryptococcus gattii VGII from the Brazilian Amazon Region by the Etest, Vitek 2, and the Clinical and Laboratory Standards Institute broth microdilution methods. Med Mycol. 2019; 57(7): 864–873. [DOI] [PubMed] [Google Scholar]
- 69. Nyazika TK, Hagen F, Machiridza T et al. Cryptococcus neoformans population diversity and clinical outcomes of HIV-associated cryptococcal meningitis patients in Zimbabwe. J Med Microbiol. 2016; 65(11): 1281–1288. [DOI] [PubMed] [Google Scholar]
- 70. Pan W, Khayhan K, Hagen F et al. Resistance of Asian Cryptococcus neoformans serotype A is confined to few microsatellite genotypes. PLoS One. 2012; 7(3): e32868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Pfaller MA, Castanheira M, Diekema DJ, Messer SA, Jones RN. Wild-type MIC distributions and epidemiologic cutoff values for fluconazole, posaconazole, and voriconazole when testing Cryptococcus neoformans as determined by the CLSI broth microdilution method. Diagn Microbiol Infect Dis. 2011; 71(3): 252–259. [DOI] [PubMed] [Google Scholar]
- 72. Prakash A, Sundar G, Sharma B, Hagen F, Meis JF, Chowdhary A. Genotypic diversity in clinical and environmental isolates of Cryptococcus neoformans from India using multilocus microsatellite and multilocus sequence typing. Mycoses. 2020; 63(3): 284–293. [DOI] [PubMed] [Google Scholar]
- 73. Rakotoarivelo RA, Raberahona M, Rasamoelina T et al. Epidemiological characteristics of cryptococcal meningoencephalitis associated with Cryptococcus neoformans var. grubii from HIV-infected patients in Madagascar: A cross-sectional study. PLoS NeglTrop Dis. 2020; 14(1): e0007984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Selb R, Fuchs V, Graf B et al. Molecular typing and in vitro resistance of Cryptococcus neoformans clinical isolates obtained in Germany between 2011 and 2017. Int J Med Microbiol. 2019; 309(6): 151336. [DOI] [PubMed] [Google Scholar]
- 75. Smith KD, Achan B, Hullsiek KH et al. Increased antifungal drug resistance in clinical isolates of Cryptococcus neoformans in Uganda. Antimicrob Agents Chemother. 2015; 59(12): 7197–7204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Tewari A, Behera B, Mathur P, Xess I. Comparative analysis of the Vitek 2 antifungal susceptibility system and E-test with the CLSI M27-A3 broth microdilution method for susceptibility testing of indian clinical isolates of Cryptococcus neoformans. Mycopathologia. 2012; 173(5-6): 427–433. [DOI] [PubMed] [Google Scholar]
- 77. Yoon HA, Felsen U, Wang T, Pirofski LA. Cryptococcus neoformans infection in Human Immunodeficiency Virus (HIV)-infected and HIV-uninfected patients at an inner-city tertiary care hospital in the Bronx. Med Mycol. 2020; 58(4): 434–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Chen SCA, Slavin MA, Heath CH et al. Clinical manifestations of Cryptococcus gattii infection: determinants of neurological sequelae and death. Clin Infect Dis. 2012; 55(6): 789–798. [DOI] [PubMed] [Google Scholar]
- 79. Chen SC, Korman TM, Slavin MA et al. Antifungal therapy and management of complications of cryptococcosis due to Cryptococcus gattii. Clin Infect Dis. 2013; 57(4): 543–551. [DOI] [PubMed] [Google Scholar]
- 80. Firacative C, Roe CC, Malik R et al. MLST and whole-genome-based population analysis of Cryptococcus gattii VGIII links clinical, veterinary and environmental strains, and reveals divergent serotype specific sub-populations and distant ancestors. PLoS NeglTrop Dis. 2016; 10(8): e0004861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Harris JR, Lockhart SR, Debess E et al. Cryptococcus gattii in the United States: clinical aspects of infection with an emerging pathogen. Clin Infect Dis. 2011; 53(12): 1188–1195. [DOI] [PubMed] [Google Scholar]
- 82. Lee GA, Arthur I, Merritt A, Leung M. Molecular types of Cryptococcus neoformans and Cryptococcus gattii in Western Australia and correlation with antifungal susceptibility. Med Mycol. 2019; 57(8): 1004–1010. [DOI] [PubMed] [Google Scholar]
- 83. Lockhart SR, Iqbal N, Bolden CB et al. Epidemiologic cutoff values for triazole drugs in Cryptococcus gattii: correlation of molecular type and in vitro susceptibility. Diagn Microbiol Infect Dis. 2012; 73(2): 144–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Phillips P, Galanis E, MacDougall L et al. Longitudinal clinical findings and outcome among patients with Cryptococcus gattii infection in British Columbia. Clin Infect Dis. 2015; 60(9): 1368–1376. [DOI] [PubMed] [Google Scholar]
- 85. Smith RM, Mba-Jonas A, Tourdjman M et al. Treatment and outcomes among patients with Cryptococcus gattii infections in the United States Pacific Northwest. PLoS One. 2014; 9(2):e88875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. O'Connor L, Van Anh D, Chau TTH et al. Antifungal susceptibility does not correlate with fungal clearance or survival in AIDS-associated cryptococcal meningitis. Clin Infect Dis. 2021; 73(7): e2338–e2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Cogliati M. Global molecular epidemiology of Cryptococcus neoformansand Cryptococcus gattii: an atlas of the molecular types. Scientifica (Cairo). 2013; 2013: 675213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Pasquier E, Kunda J, De Beaudrap P et al. Long-term mortality and disability in cryptococcal meningitis: a systematic literature review. Clin Infect Dis. 2018; 66(7): 1122–1132. [DOI] [PubMed] [Google Scholar]
- 89. Day JN, Chau TTH, Wolbers M et al. Combination antifungal therapy for cryptococcal meningitis. N Engl J Med. 2013; 368(14): 1291–1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Tenforde MW, Mokomane M, Leeme T et al. Advanced Human Immunodeficiency Virus disease in Botswana following successful antiretroviral therapy rollout: incidence of and temporal trends in cryptococcal meningitis. Clin Infect Dis. 2017; 65(5): 779–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Ssekitoleko R, Kamya MR, Reingold AL. Primary prophylaxis for cryptococcal meningitis and impact on mortality in HIV: a systematic review and meta-analysis. Future Virol. 2013; 8(9): 917–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Temfack E, Bigna JJ, Luma HN et al. Impact of routine cryptococcal antigen screening and targeted preemptive fluconazole therapy in antiretroviral-naive Human Immunodeficiency Virus-infected adults with CD4 cell counts <100/μl: a systematic review and meta-analysis. Clin Infect Dis. 2019; 68(4):688–698. [DOI] [PubMed] [Google Scholar]
- 93. Li Y, Huang X, Chen H et al. The prevalence of cryptococcal antigen (CrAg) and benefits of pre-emptive antifungal treatment among HIV-infected persons with CD4+ T-cell counts < 200 cells/μl: evidence based on a meta-analysis. BMC Infect Dis. 2020; 20(1): 410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Baddley JW, Chen SC, Huisingh C et al. MSG07: an international cohort study comparing epidemiology and outcomes of patients with Cryptococcus neoformans or Cryptococcus gattii infections. Clin Infect Dis. 2021; 73(7): 1133–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Hevey MA, Presti RM, OʼHalloran JA et al. Mortality after cryptococcal infection in the modern antiretroviral therapy era. J Acquir Immune Defic Syndr. 2019; 82(1):81–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Alves SE, Lazera MDS, Wanke B et al. Mortality by cryptococcosis in Brazil from 2000 to 2012: a descriptive epidemiological study. PLoS Negl Trop Dis. 2019; 13(7): e0007569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Lee Y-C, Wang J-T, Sun H-Y, Chen Y-C. Comparisons of clinical features and mortality of cryptococcal meningitis between patients with and without human immunodeficiency virus infection. J Microbiol Immunol Infect. 2011; 44(5):338–345. [DOI] [PubMed] [Google Scholar]
- 98. Pasquier E, Kunda J, De Beaudrap P et al. Long-term mortality and disability in cryptococcal meningitis: a systematic literature review. Clin Infect Dis. 2018; 66(7): 1122–1132. [DOI] [PubMed] [Google Scholar]
- 99. Pan D, Wong N, Toovey O, Hills G, Stephenson I. A multicenter, longitudinal cohort study of cryptococcosis in Human Immunodeficiency Virus-negative people in the United States. Clin Infect Dis. 2020; 71(11):3014–3015. [DOI] [PubMed] [Google Scholar]
- 100. Molloy SF, Kanyama C, Heyderman RS et al. Antifungal combinations for treatment of cryptococcal meningitis in Africa. N Engl J Med. 2018; 378( 11): 1004–1017. [DOI] [PubMed] [Google Scholar]
- 101. Tenforde MW, Gertz AM, Lawrence DS et al. Mortality from HIV-associated meningitis in sub-Saharan Africa: a systematic review and meta-analysis. J Int AIDS Soc. 2020; 23(1): e25416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Verweij PE, Lucas JA, Arendrup MC et al. The one health problem of azole resistance in Aspergillus fumigatus: current insights and future research agenda. Fung Biol Rev. 2020; 34(4):202–214. [Google Scholar]
- 103. Mfinanga S, Chanda D, Kivuyo SL et al. Cryptococcal meningitis screening and community-based early adherence support in people with advanced HIV infection starting antiretroviral therapy in Tanzania and Zambia: an open-label, randomised controlled trial. Lancet. 2015; 385( 9983): 2173–2182. [DOI] [PubMed] [Google Scholar]
- 104. WHO Guidelines Approved by the Guidelines Review Committee . Guidelines for the Diagnosis, Prevention and Management of Cryptococcal Disease in HIV-Infected Adults, Adolescents and Children: Supplement to the 2016 Consolidated Guidelines on the Use of Antiretroviral Drugs for Treating and Preventing HIV Infection. Geneva: World Health Organization, © World Health Organization 2018; 2018. [PubMed] [Google Scholar]
- 105. Pullen MF, Hullsiek KH, Rhein J et al. Cerebrospinal fluid early fungicidal activity as a surrogate endpoint for cryptococcal meningitis survival in clinical trials. Clin Infect Dis. 2020; 71(7): e45–e49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Tshepiso M, Aude S-L, Kwana L et al. 2023; Innovative quantitative PCR assays for the assessment of HIV-associated cryptococcal meningoencephalitis in Sub-Saharan Africa. medRxiv. 2023.08.24.23294467. 10.1101/2023.08.24.23294467. https://www.medrxiv.org/content/10.1101/2023.08.24.23294467v1. Date accessed May 26, 2024. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.