Abstract
Background
This study aimed to identify the risk factors associated with mortality among patients co-infected with human immunodeficiency virus (HIV) and Talaromyces marneffei (TM) in China, and develop a risk prediction model.
Methods
In this retrospective cohort analysis conducted from 2013 to 2024, comprehensive clinical data from 160 patients were analyzed using a logistic regression model to identify mortality predictors and construct a predictive model. An additional 36 patients constituted the validation cohort, which was specifically designed to evaluate the predictive value of the model. Model performance was assessed using the area under the curve (AUC).
Results
The overall mortality rate for hospitalized patients with HIV/TM co-infection was 17.35 %. The median age was 35.0 years, and 89.30 % were male. Additionally, 89.80 % of the patients reported fever and 87.76 % presented with lymphadenopathy. Key independent risk factors associated with mortality included age (odds ratio (OR): 1.103, 95 % confidence interval (CI) = 1.033–1.178, P = 0.003), procalcitonin (PCT) levels (OR: 1.270, 95 % CI = 1.052–1.534, P = 0.013), and urea to albumin ratio (UAR) (OR: 1.491, 95 % CI = 1.175–1.892, P < 0.001). Advanced age, elevated PCT levels, and increased UAR were identified as independent risk factors of mortality. Furthermore, the mortality prediction probability combining age, PCT, and UAR exhibited a high predictive value in patients with HIV/TM co-infection. Additionally, the AUC showed a good discrimination ability in the validation group (AUC, 0.898).
Conclusions
Advanced age, elevated PCT levels, and increased UAR significantly determine mortality in patients with HIV/TM co-infection. These findings underscore the potential of using laboratory parameters as predictive indicators of mortality, facilitating the early identification of HIV/TM co-infection cases in clinical practice.
Keywords: Talaromyces marneffei, HIV, Risk factors, Mortality
1. Introduction
Talaromyces marneffei (TM) is an opportunistic pathogenic fungus that poses a significant risk to individuals with compromised immune systems. Known as thallomycosis, this fungal infection is common in Southeast Asian countries and certain areas of southern China, including Guangxi, Guangdong, Fujian, and Yunnan [1,2]. TM is the leading cause of opportunistic infections and mortality among AIDS patients, with a persistently high mortality rate of up to 20 % despite standardized antifungal treatments [[3], [4], [5], [6]]. Consequently, there is a pressing need for healthcare professionals to have readily accessible tools to promptly identify patients at heightened risk of mortality following TM infection. Numerous studies have examined the risk factors for mortality in patients with HIV/TM co-infection. However, it is worth noting that these studies did not comprehensively investigate the associated factors, nor do they include a substantial sample size [3,4,6]. Considering these gaps, we conducted a retrospective cohort study based on hospital data to identify the risk factors and develop a predictive model for mortality in patients with HIV/TM co-infection. Our study includes a wide range of factors, including demographic features, clinical symptoms, and laboratory parameters. By analyzing these factors, our findings contribute to a deeper understanding of HIV/TM co-infection and assist in the early detection of symptoms that may indicate an increased risk of mortality.
2. Methods
This study was guided by the Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) guidelines. Following the established frameworks ensures a comprehensive and transparent presentation of our observational study and predictive model, promoting the reliability and interpretability of our findings.
2.1. Study design and patient selection
We conducted a retrospective cohort study of hospitalized patients with HIV/TM co-infection at Hangzhou Xixi Hospital, a designated HIV/AIDS care hospital in Zhejiang Province. All patients were confirmed to be HIV positive through enzyme-linked immunosorbent assay (ELISA) and Western blot testing and the diagnosis was in line with national HIV diagnostic criteria [7]. ELISA, a highly sensitive screening test, detects the presence of anti-HIV antibodies. As ELISA is a screening test, a confirmatory test is required to validate the results using Western blot, a confirmatory test used to evaluate positive or indeterminate ELISA results. The diagnostic criteria for TM infection include the isolation and cultivation of samples from blood, skin tissue, bone marrow, lymph nodes, and other bodily fluids. Mycelia cultivation was performed at 25 °C, while yeast-like structures were cultured at 37 °C. According to the national HIV diagnostic criteria, a combination of HIV antibody tests, HIV RNA tests, CD4 cell counts, and the patient's epidemiological history are used alongside symptoms to confirm HIV infection.
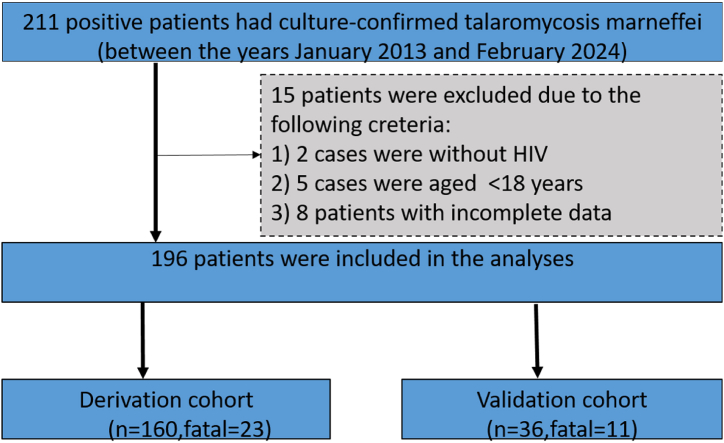
We analyzed the clinical and demographic data from 160 patients collected between January 2013 and December 2022, which included demographics, clinical manifestations, and laboratory test results at admission. For validation, we selected a total of 36 patients with HIV/TM co-infection as independent samples during their hospitalization from January 2023 to February 2024, with a median follow-up time of 374 days. Their prognoses were assessed using a predictive probability model for mortality. Out of 211 patients with culture-confirmed Talaromyces marneffei, 15 were excluded due to incomplete data, absence of HIV, or age under 18 years. Finally, patient characteristics and all required information were extracted from 196 medical records and included in the analyses (Fig. 1).
Fig. 1.
Flowchart of the HIV/TM co-infected patient enrollment.
2.2. Sample size and data collection
To address potential sources of bias in this retrospective cohort study, several strategies were implemented. Selection bias was minimized by including all eligible patients with HIV/TM co-infection admitted to Hangzhou Xixi Hospital between January 2013 and February 2024. Information bias was reduced by using standardized data collection methods and verifying clinical and laboratory data through hospital records. The data was also preprocessed, some real data was deleted, and whether there were any abnormal values in the data was checked. Confounding was addressed through multivariable logistic regression analysis, which adjusted for potential confounders such as age, gender and comorbidities.
We adopt a temporal validation method, continuing to collect data during the process of developing the risk prediction model, and verifying the model with the newly collected data after the model development is completed. As the current study is a retrospective analysis, no sample size calculations were conducted. The primary outcome of this study is mortality among patients with HIV/TM co-infection. Potential confounders that might affect the study results include: age, gender, presence of comorbidities and other infections.
Patient data were meticulously reviewed item-by-item. To ensure accuracy, two investigators cross-checked all information against medical records rigorously. All study laboratories adhered to the standardization and certification programs to maintain their quality and reliability. We used a retrospective medical review for data collection, which encompasses the following three main categories. First, it involved capturing demographic features, such as age and gender. Secondly, it focused on documenting the clinical symptoms associated with the disease, including fever, cough, sputum, swollen lymph nodes, fatigue, and rashes. Lastly, it encompassed observing a range of laboratory parameters throughout the clinical course, such as platelet, C-reactive protein (CRP), albumin, urea, procalcitonin (PCT), CD4+T lymphocytes (CD4); CD8+T lymphocytes (CD8), HIVRNA, and others. Additionally, the study included multiple inflammatory indicators including the neutrophil to lymphocyte ratio (NLR), lymphocyte to monocyte ratio (LMR), platelet to lymphocyte ratio (PLR), PLR*neutrophil (SII), platelet*monocyte*NLR (AISI), C-reactive protein to lymphocyte ratio (CLR), and urea to albumin ratio (UAR).
2.3. Statistical analysis
Prior to data analysis, the predictor variables in the development and validation cohorts were inspected for missing values. Among the predictors, variables with a missing data rate exceeding 20 % were excluded, and for those with less than a missing data rate below 20 %, including CD4, CD8, HIV-RNA, albumin and platelet, mean imputation was employed. The study used a combination of direct participant contact and medical record tracking of participants over time. Loss to follow-up was minimal due to the retrospective nature of the data collection.
All data were analyzed using SPSS version 26.0 (IBM Corporation). To ensure accuracy, both investigators independently entered the data twice and compared the data later. Continuous variables are expressed as means ± S.D, if normally distributed, or as median (interquartile range), if not. Categorical variables are presented as numbers (percentages). Comparisons of prevalence between groups was conducted using the Pearson χ2 test, adjusted χ2 test, or Fisher' exact test, as appropriate. Furthermore, risk factors for mortality were analyzed using a binary logistic regression method. Odds ratios (OR) and 95 % confidence intervals (CIs) were also determined. The variables we selected may not fully capture the dependent variables, making bias inevitable. To minimize this bias, we implemented several measures, such as preprocessing the data, deleting certain data points, and checking for outliers. To exclude the effects of potential confounders, we use binary logistic regression analysis, the primary outcome was entered into the model as a dependent variable significant general and laboratory test results identified in the univariate analysis were included as independent variables in the forward stepwise multivariate analysis. Multivariable logistic regression was used to control for confounding by including potential confounders such as age, gender, and comorbidities in the model. Factors with a P-value < 0.01 were further selected and analyzed using the forward stepwise method “Forward LR” multivariate logistic regression analysis. Receiver operating characteristic (ROC) curves were used to evaluate the predictive value of age, PCT, and UAR, both separately and jointly, in patients with HIV/TM co-infection. The optimal threshold value for diagnosis was determined when the Youden index was at its maximum and diagnostic efficacy was evaluated using the area under the curve (AUC). Statistical significance was set at a two-sided P-value of < 0.05.
3. Results
3.1. Demographic data
The patient flow diagram is summarized in Fig. 1. A total of 196 patients diagnosed with HIV/TM co-infection were included. Most patients were male, and the median age of the entire cohort was 35.0 years. The average mortality rate during hospitalization from 2013 to 2024 is 17.35 % (34/196 cases). Characteristics of the patients in the development and validation cohorts are presented in Table 1. The demographic characteristics of the nonfatal group and fatal group in the validation cohort are presented in Table 2.
Table 1.
Characteristics of the patients in development and validation cohorts.
| Variable | Development cohort(n = 160) | Validation cohort (n = 36) | Whole cohort (n = 196) | P-value |
|---|---|---|---|---|
| Demographics | ||||
| Male, n(%) | 148(92.5 ) | 27(75.0 ) | 175(89.3 ) | 0.006a |
| Age(years) | 35.0(28.9,47.0) | 35.5(31.0,45.0) | 35.0(29.0,46.0) | 0.649 |
| Symptoms, n(%) | ||||
| Fever | 146(91.30) | 30(83.33) | 176(89.80) | 0.266a |
| Lymphadenectasis | 143(89.38) | 29(80.56) | 172(87.76) | 0.239a |
| Cough | 122(76.25) | 27(75.00) | 149(76.02) | 0.874 |
| Sputum | 90(56.25) | 20(55.56) | 110(56.12) | 0.940 |
| Fatigue | 83(51.88) | 18(50.00) | 101(51.53) | 0.839 |
| Rash | 69(43.13) | 14(38.89) | 83(42.35) | 0.642 |
| Gastrointestinal symptoms | 57(35.63) | 12(33.33) | 69(35.20) | 0.795 |
| Candida albicans | 78(48.75) | 16(44.44) | 94(47.96) | 0.640 |
| Aspergillus | 65(40.63) | 13(36.11) | 78(39.80) | 0.617 |
| Syphilis | 40(25.00) | 8(22.22) | 48(24.49) | 0.726 |
| Cytomegalovirus | 40(25.00) | 9(25.00) | 49(25.00) | 1.000 |
| Weight loss | 52(32.50) | 12(33.33) | 64(332.65) | 0.923 |
| Serositis | 74(46.25) | 15(41.67) | 89(45.41) | 0.618 |
| Laboratory measures | ||||
| Platelet (109/L) | 118.00(70.00,202.50) | 93.00(56.50,160.75) | 114.50(69.00,193.75) | 0.157 |
| CRP(mg/L) | 58.00(20.23,91.75) | 50.10(29.48,92.25) | 56.85(21.00,91.75) | 0.817 |
| Albumin (g/L) | 25.80(22.20,31.00) | 26.35(22.98,33.48) | 26.00(22.65,31.35) | 0.525 |
| AST/ALT | 2.19(1.45,3.39) | 2.70(1.65,4.55) | 2.29(1.55,3.59) | 0.079 |
| Urea (mmol/L) | 4.30(3.20,6.25) | 5.10(3.10,13.00) | 4.40(3.20,6.85) | 0.115 |
| Procalcitonin (ng/mL) | 0.50(0.13,1.95) | 0.68(0.22,5.09) | 0.58(0.17,2.45) | 0.324 |
| WBC count(109/L) | 3.63(2.51,5.09) | 4.06(3.13,5.45) | 3.67(2.57,5.27) | 0.249 |
| Neutrophil count (109/L) | 2.96(2.00,4.35) | 3.49(2.61,4.93) | 3.08(2.07,4.42) | 0.145 |
| Lymphocyte count (109/L) | 0.34(0.19,0.50) | 0.26(0.14,0.47) | 0.33(0.19,0.50) | 0.198 |
| Monocyte count(109/L) | 0.18(0.10,0.31) | 0.12(0.07,0.27) | 0.17(0.10,0.31) | 0.056 |
| CD4 count (cells/uL) | 9.00(3.00,24.00) | 8.50(4.00,24.00) | 9.00(3.00,24.00) | 0.856 |
| CD8 count (cells/uL) | 194.00(122.00,324.25) | 154.00(84.25,322.50) | 185.50(101.75,321.75) | 0.173 |
| HIVRNA (copies/mL) | 67100.00(11100.00,309500.00) | 171000.00(12900.00,1000000.00) | 76200.00(11950.00,363750.00) | 0.066 |
| NLR | 9.73(5.64,16.24) | 12.03(6.69,26.03) | 9.83(5.71,17.28) | 0.088 |
| LMR | 1.84(1.27,2.98) | 2.57(1.57,3.55) | 2.00(1.27,3.00) | 0.073 |
| PLR | 371.52(216.54,596.11) | 377.43(168.45,669.00) | 371.52(216.54,599.04) | 0.935 |
| SII | 1039.03(509.81,2162.22) | 1481.94(459.08,2395.86) | 1068.45(503.05,2201.90) | 0.513 |
| AISI | 189.47(66.91,457.89) | 159.31(63.19,311.96) | 167.94(66.91,420.37) | 0.507 |
| CLR | 148.68(56.39,352.89) | 160.79(74.75,685.83) | 157.51(57.45,373.94) | 0.360 |
| UAR | 2.93(2.00,4.39) | 3.52(1.93,9.92) | 3.08(1.99,4.61) | 0.098 |
Abbreviations: CRP, C-reactive protein; WBC, white blood cell; CD4, CD4+T lymphocytes; CD8, CD8+T lymphocytes; NLR, neutrophil to lymphocyte ratio; LMR, lymphocyte to monocyte ratio; PLR, platelet to lymphocyte ratio; SII, PLRa neutrophil; AISI, plateleta monocyteaNLR; CLR, c-reactive protein to lymphocytes ratio; UAR, urea to albumin ratio.
The missing data for CD4, CD8, HIV-RNA, albumin and platelet were 3 %, 3 %, 5 %, 6 % and 2 %, respectively.
Continuous correction chi-square test.
Table 2.
Comparison of clinical parameters between nonfatal group and fatal group.
| Variable | Nonfatal group(n = 137) | Fatal group (n = 23) | All (n = 160) |
P-value |
|---|---|---|---|---|
| Demographics | ||||
| Male, n(%) | 125(91.2) | 23(100.0) | 148(92.5) | 0.295* |
| Age(years) | 34.0(28.0,44.0) | 46.0(32.0,54.0) | 35.0(28.9,47.0) | 0.007 |
| Symptoms, n(%) | ||||
| Fever | 127(92.70) | 19(82.60) | 146(91.30) | 0.236* |
| Lymphadenectasis | 123(89.78) | 20(86.96) | 143(89.38) | 0.967* |
| Cough | 108(78.83) | 14(60.87) | 122(76.25) | 0.051 |
| Sputum | 80(58.39) | 10(43.48) | 90(56.25) | 0.182 |
| Fatigue | 68(49.64) | 15(65.22) | 83(51.88) | 0.166 |
| Rash | 58(42.34) | 11(47.83) | 69(43.13) | 0.623 |
| Gastrointestinal symptoms | 48(35.04) | 9(39.13) | 57(35.63) | 0.704 |
| Candida albicans | 71(51.82) | 7(30.40) | 78(48.75) | 0.058 |
| Aspergillus | 54(39.42) | 11(47.83) | 65(40.63) | 0.447 |
| Syphilis | 38(27.74) | 2(8.70) | 40(25.00) | 0.051 |
| Cytomegalovirus | 32(23.36) | 8(34.78) | 40(25.00) | 0.242 |
| Weight loss | 40(29.20) | 12(52.17) | 52(32.50) | 0.029 |
| Serositis | 58(42.34) | 16(69.57) | 74(46.25) | 0.015 |
| Laboratory measures | ||||
| Platelet (109/L) | 126.00(85.00,210.50) | 47.00(23.00,84.00) | 118.00(70.00,202.50) | <0.001 |
| CRP(mg/L) | 49.00(17.45,84.94) | 123.00(62.47,159.27) | 58.00(20.23,91.75) | <0.001 |
| Albumin (g/L) | 27.00(23.60,31.90) | 20.85(16.37,25.43) | 25.80(22.20,31.00) | <0.001 |
| AST/ALT | 2.06(1.32,2.98) | 3.97(2.46,6.39) | 2.19(1.45,3.39) | <0.001 |
| Urea (mmol/L) | 4.00(3.00,5.35) | 9.20(7.40,13.90) | 4.30(3.20,6.25) | <0.001 |
| Procalcitonin (ng/mL) | 0.36(0.11,1.27) | 9.41(0.98,26.96) | 0.50(0.13,1.95) | <0.001 |
| WBC count(109/L) | 3.56(2.44,5.02) | 3.66(3.40,6.29) | 3.63(2.51,5.09) | 0.176 |
| Neutrophil count (109/L) | 2.94(1.90,4.33) | 3.19(2.40,4.68) | 2.96(2.00,4.35) | 0.244 |
| Lymphocyte count (109/L) | 0.34(0.20,0.50) | 0.32(0.18,0.56) | 0.34(0.19,0.50) | 0.848 |
| Monocyte count(109/L) | 0.18(0.11,0.31) | 0.13(0.06,0.30) | 0.18(0.10,0.31) | 0.134 |
| CD4 count (cells/uL) | 10.00(4.00,24.00) | 4.00(1.00,12.00) | 9.00(3.00,24.00) | 0.036 |
| CD8 count (cells/uL) | 195.00(125.00,322.00) | 104.00(34.00,258.00) | 194.00(122.00,324.25) | 0.023 |
| HIVRNA (copies/mL) | 36400.00(1400.00,277500.00) | 65500.00(143.50,194000.00) | 37700.00(1160.00,261000.00) | 0.712 |
| NLR | 9.51(5.55,16.15) | 10.94(6.33,19.73) | 9.73(5.64,16.24) | 0.228 |
| LMR | 1.70(1.26,2.66) | 2.75(1.50,4.50) | 1.84(1.27,2.98) | 0.031 |
| PLR | 381.25(235.58,644.72) | 209.09(75.34,408.33) | 371.52(216.54,596.11) | 0.001 |
| SII | 1048.65(571.27,2214.59) | 535.56(264.00,1508.95) | 1039.03(509.81,2162.22) | 0.015 |
| AISI | 200.51(78.86,467.63) | 74.78(16.47,196.16) | 189.47(66.91,457.89) | 0.004 |
| CLR | 138.24(39.59,293.93) | 310.53(181.82,600.00) | 148.68(56.39,352.89) | 0.003 |
| UAR | 2.76(1.92,4.00) | 7.61(3.66,14.77) | 2.93(2.00,4.39) | 0.000 |
3.2. Clinical characteristics
The fatal group exhibited a significantly higher median age than the non-fatal group (34.0 vs 46.0, P = 0.007). There was no statistically significant difference in sex between both groups (P = 0.295). The most prevalent symptoms were fever (91.30 %), lymphadenopathy (89.38 %), and coughing (76.25 %). Weight loss and serositis were more frequently observed in the fatal group and these differences were statistically significant (P = 0.029 and 0.015, respectively). However, other clinical symptoms did not show statistically significant differences. We conducted a comparative analysis of the four prevalent opportunistic infections—Candida albicans, Aspergillus, syphilis, and cytomegalovirus—no substantial differences were observed between the two groups (Table 2).
3.3. Laboratory features
We conducted a comprehensive analysis of 19 laboratory parameters in patients from both groups and compared their median levels. Among the fatal cases, CRP, AST/ALT, Urea, and PCT levels were significantly higher than those in the non-fatal cases (P < 0.001, <0.001, <0.001, and <0.001, respectively). Conversely, platelet count and albumin levels were significantly lower in fatal cases than in non-fatal cases (P < 0.001, <0.001, respectively). The detailed results are presented in Table 2.
3.4. Comparison of inflammatory indicators between the nonfatal and fatal groups
When evaluating hemogram-based inflammatory parameters, we observed significantly higher levels of LMR, CLR, and UAR in the fatal group than in the non-fatal group. Conversely, PLR, SII, and AISI were significantly lower in the fatal group than in the non-fatal group. A summary of the laboratory data of the study groups is presented in Table 2.
3.5. Risk factors for mortality in hospitalized patients
Using binary logistic regression analysis, significant general and laboratory test results identified in the univariate analysis were included as independent variables in the forward stepwise multivariate analysis. The results showed that age (OR: 1.103; 95 % CI: 1.033–1.178; P = 0.003), PCT (OR: 1.270; 95 % CI: 1.052–1.534; P = 0.013), and UAR (OR: 1.491; 95 % CI: 1.175–1.892; P = 0.001) were independent predictors of mortality in patients with HIV/TM co-infection. Among inflammatory parameters, the UAR exhibited the best mortality predicting performance. The predictive probability model for mortality in these patients was determined as follows: logit (P) = −9.035 + 0.098 × age + 0.239 × PCT + 0.400 × UAR. Detailed results are presented in Table 3. The observed absolute risks of mortality were 19.14 % for patients with increased age, 22.03 % for those with elevated PCT levels, and 25.86 % for individuals with elevated UAR measurements, respectively.
Table 3.
Logistic regression analysis of risk factors for death in patients with HIV and PM co-infection.
| Variable | B | Standard error | Waldχ2 | P-value | Crude OR | (95%CL) | Adjusted OR | (95%CL) |
|---|---|---|---|---|---|---|---|---|
| Age(years) | 0.098 | 0.034 | 8.571 | 0.003 | 1.055 | (1.018–1.095) | 1.103 | (1.033–1.178) |
| PCT (ng/mL) | 0.239 | 0.096 | 6.198 | 0.013 | 1.306 | (1.134–1.504) | 1.270 | (1.052–1.534) |
| UAR | 0.400 | 0.121 | 10.830 | 0.001 | 1.381 | (1.192–1.602) | 1.491 | (1.175–1.892) |
| Constant | −9.035 | 2.042 | 19.580 | 0.000 |
3.6. Predictive value of risk factors for mortality
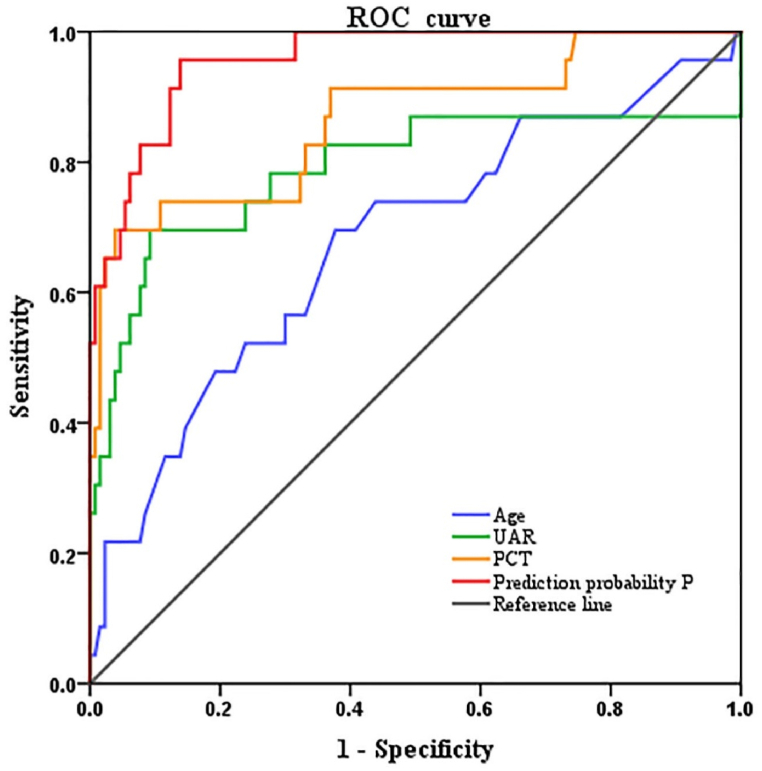
To evaluate the predictive values of age, PCT level, UAR, and mortality prediction probability for patient mortality, an ROC curve was constructed. The results indicated that UAR and PCT had moderate diagnostic values (0.7 < AUC< 0.9), whereas the mortality prediction probability had a high diagnostic value (AUC >0.9). Specifically, the predictive values were as follows: Age, AUC = 0.676, SE = 0.066, P = 0.007, 95 % CI: 0.548–0.805, cutoff value = 37.50, sensitivity = 0.696, specificity = 0.620; PCT, AUC = 0.865, SE = 0.048, P = 0.000, 95 % CI: 0.770–0.959, cutoff value = 6.19, sensitivity = 0.696, specificity = 0.962; UAR, AUC = 0.789, SE = 0.070, P = 0.000, 95 % CI: 0.652–0.926, cutoff value = 5.27, sensitivity = 0.696, specificity = 0.905. Furthermore, the mortality prediction probability combining age, PCT, and UAR exhibited a high predictive value for patients with HIV/TM co-infection, with an AUC of 0.958 (SE = 0.018, P = 0.000, 95 % CI: 0.920–0.992), sensitivity = 0.957, and specificity = 0.862 (Table 4/Fig. 2).
Table 4.
Predictive value of age, PCT, and UAR for mortality in patients with HIV and PM co-infection.
| Variable | AUC | Standard error | P-value | 95 %(CL) | Sensitivity | Specificity | Cutoff value | Youden index |
|---|---|---|---|---|---|---|---|---|
| Age | 0.676 | 0.066 | 0.007 | 0.548–0.805 | 0.696 | 0.620 | 37.50 | 0.316 |
| PCT | 0.865 | 0.048 | 0.000 | 0.770–0.959 | 0.696 | 0.962 | 6.19 | 0.657 |
| UAR | 0.789 | 0.070 | 0.000 | 0.652–0.926 | 0.696 | 0.905 | 5.27 | 0.601 |
| mortality prediction probability P | 0.958 | 0.018 | 0.000 | 0.923 | 0.992 |
Fig. 2.
Diagnostic value of different markers in predicting death.
3.7. Evaluation of the prediction model
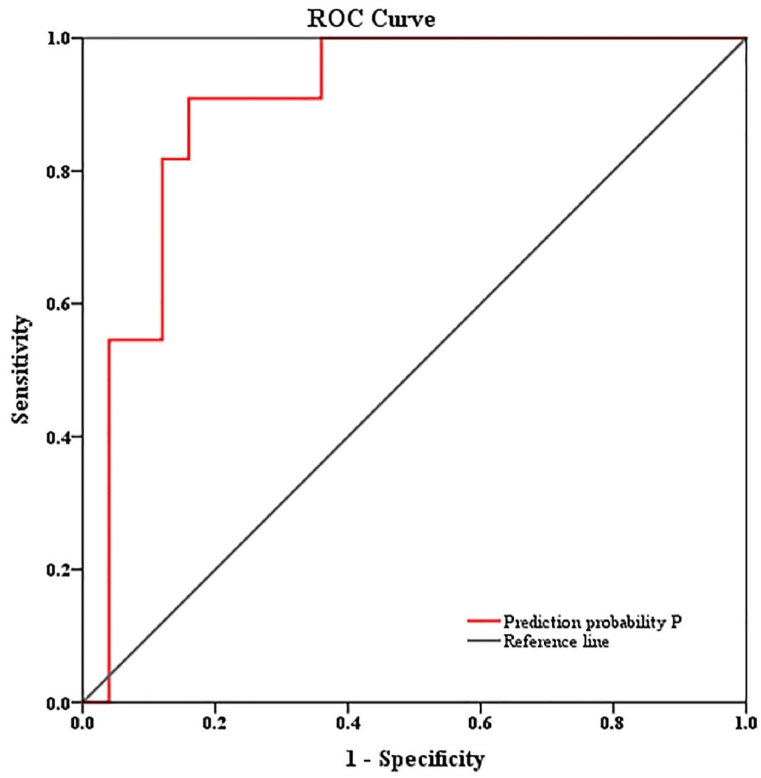
The validation group comprised 27 males and 9 females, with an average age of 35.50. Among them, 25 and 11 patients were in the non-fatal and fatal groups, respectively. The model calculated the P value for each patient, which was subsequently used to predict their respective prognoses. The AUC showed good discriminative ability (AUC = 0.898, SE = 0.054, P = 0.000, 95 % CI: 0.793–1.000), sensitivity = 0.909, and specificity = 0.840) (Fig. 3). The prognosis was predicted by the constructed model and compared with the actual prognosis. The model prediction had a sensitivity of 54.5 %, a specificity of 80 %, a positive predictive value (PPV): 54.5 %, a negative predictive value (NPV): 80 %, and an accuracy of 72.2 % (Table 5).
Fig. 3.
Evaluation of the prediction model.
Table 5.
Prediction results and actual situation of 36 cases.
| Model predicted results | The actual situation |
|
|---|---|---|
| fatal | nonfatal | |
| fatal | 6 | 5 |
| nonfatal | 5 | 20 |
| All | 11 | 25 |
4. Discussion
In this study, we examined the mortality and risk factors for death among hospitalized patients with HIV/TM co-infection in Hangzhou, Zhejiang Province, China. The findings revealed a mortality rate of 17.35 % [8]. Advanced age and elevated PCT and UAR levels have been identified as valuable predictors of mortality. Notably, this study made a novel discovery that UAR is a risk factor for mortality in patients with HIV/TM co-infection.
TM is a pathogenic fungus that poses a fatal risk, particularly to those with compromised immune function. It primarily invades the monocyte-macrophage system when the immune system is impaired [8], inducing inflammatory reactions in tissues and organs rich in monocytes, such as the lungs, liver, spleen, lymph nodes, skin, and bone marrow, by releasing cytokines, such as interleukin-8 (IL-8). Inflammation plays a crucial role in the prognosis of patients with HIV/TM and TM co-infection. The incidence of TM infection has been rapidly increasing owing to the increasing number of patients [9,10]. Research has shown that TM is associated with a poor prognosis in patients with HIV, and co-infection with TM further compromises the individual's immune function. TM has transformed from a rare human pathogen to one of the leading causes of AIDS-related deaths in epidemic areas [7,[11], [12], [13]]. This highlights the need for further investigation in China. Patients with HIV/TM co-infection are highly susceptible to other opportunistic infectious diseases because of the combined impact of HIV infection on the immune system and TM, affecting multiple organ systems throughout the body [14]. Our study identified Candida albicans, Aspergillus, syphilis, and cytomegalovirus as the primary opportunistic infections associated with this co-infection. Notably, our findings revealed a relatively low incidence of Candida albicans and syphilis infections in the fatal group. This can be attributed to the severe immune system impairment observed in this group, which hindered their ability to effectively combat opportunistic infections. Additionally, patients in the fatal group may have encountered additional complications throughout the disease progression, leading to a reduced incidence of opportunistic infections. However, further prospective studies are required to further validate and delve deeper into this relationship.
This study found no association between mortality and sex, but identified older age and elevated PCT levels as risk factors for mortality in patients with HIV/TM co-infection [6,15]. Globally, individuals living with HIV survive to an older age, and the predictive value of increasing age for prognosis should not be overlooked. PCT level is a reliable indicator of severe bacterial and fungal infections. Under normal physiological conditions, PCT levels in the body are either low or undetectable. However, in cases of infection and trauma, PCT levels significantly increase and remain elevated. Consequently, PCT levels can reflect the severity of a patient's condition and have gained attention as a predictor of disease mortality [16]. High PCT levels serve as predictors of sepsis-induced organ dysfunction, including kidney injury, and are negatively correlated with patient prognosis [17]. Our study found that PCT is a more accurate predictor of death in patients with HIV/TM co-infection than other inflammatory biomarkers such as CRP, WBC count, lymphocyte count, monocyte count, and neutrophil count. As individuals age, immune function naturally declines, which may have implications in HIV/TM co-infection. Furthermore, HIV/TM co-infection can contribute to chronic inflammation. In this scenario, the combination of aging and co-infection can further exacerbate chronic inflammation. This heightened state of inflammation can stimulate the increased production and release of PCT. Therefore, in older individuals with HIV/TM co-infection, the interplay between age-related immune decline and the inflammatory response associated with co-infection may contribute to elevated PCT levels.
The UAR, which represents the ratio of urea to albumin, demonstrates a higher correlation with prognosis than individual albumin or urea values. Studies have established a relationship between UAR and prognosis in various diseases, including cardiovascular diseases, pneumonia, and gastrointestinal diseases [[18], [19], [20], [21], [22]], and UAR levels can significantly affect adverse outcomes, including death [23,24]. A retrospective study including 1253 patients found that a UAR index greater than 6.25 can independently predict hospitalization mortality in elderly emergency patients [23]. In the present study, a higher UAR were associated with an increased risk of death in patients with HIV/TM co-infection. This may be attributed to elevated urea levels and hypoproteinemia. Urea is considered a predictive indicator of hemodynamic damage in critically ill patients, and high urea levels are often associated with mortality [25]. A study conducted in Vietnam suggested that urea is an independent risk factor for HIV/TM co-infection, highlighting the importance of considering kidney damage during simultaneous HIV/TM co-infection [26]. Moreover, albumin levels in the fatal group were significantly lower than those in the non-fatal group, indicating the potential significance of nutritional status in these patients. Hypoproteinemia is often indicative of progression to cachexia. The UAR indirectly reflects the urea and albumin levels in patients, and the results suggest that as the UAR increases, so does the risk of death in patients with HIV/TM co-infection.
The area under the ROC curve for predicting death based on UAR alone was 0.789, indicating high diagnostic value. When age, PCT, and UAR were combined, their diagnostic values increased, with the area under the curve reaching 0.958; highlighting the importance of monitoring these indicators in patients with HIV/TM co-infection, particularly in high-risk patients, and emphasizing the need for prompt rescue measures when necessary. In the validation cohort, the AUC was 0.898 (SE = 0.054, P = 0.000, 95 % CI = 0.793–1.000), the sensitivity was 0.909, and the specificity was 0.840. The results validated the prediction model in an external cohort with good discriminant ability, indicating that the model could predict the mortality risk of patients with HIV/TM co-infection. The results of this study may aid in the early detection of mortality risk factors. For elderly patients with elevated PCT or UAR levels during the disease process, clinicians must pay more attention and provide timely treatment.
The research conducted on patients with HIV/TM co-infection in Hangzhou, Zhejiang Province, presents several limitations that should be acknowledged. Firstly, the study's retrospective observational design, spanning a period of 10 years, resulted in some variables were not included in the study. Secondly, as this is a single-center study conducted in non-epidemic areas of TM infection, the generalizability of the findings is limited. Therefore, these results should be validated through multicenter prospective studies to ensure broader applicability. Additionally, the study did not assess continuous changes in laboratory parameters over time, which could provide more dynamic insights into the disease progression. Despite the inherent limitations of retrospective studies, such as confounding and selection biases, this research offers valuable insights into the risk factors for mortality in patients with HIV/TM co-infection. However, further prospective studies are necessary to evaluate these and other prognostic factors comprehensively. The predictive model developed in this study requires further validation in diverse settings. The results should be interpreted with caution, considering the study's limitations. Future research should aim to confirm these findings and investigate additional factors influencing mortality in this patient population. Integrating the predictive model into a clinical decision support system could assist physicians in assessing patients' risk and developing personalized treatment plans. Continuous monitoring of the clinical outcomes of co-infected patients is crucial to evaluate the effectiveness of interventions. Moreover, additional research is needed to deepen our understanding of the pathological mechanisms and optimal treatments for HIV/TM co-infection.
5. Conclusion
In general, this retrospective cohort study identified key independent risk factors associated with mortality in patients with HIV/TM co-infection, including advanced age, elevated PCT levels, and increased UAR. The value of predictive models for the prognostic evaluation of HIV/TM co-infection using available data is increasingly recognized as a cost-effective approach to support clinical decision-making. However, there is a lack of well-developed studies with large sample sizes and high-performing predictive models in this area. Further large-scale, prospective studies across different contexts are recommended to strengthen the evidence base.
Funding
This research was funded by Hangzhou Biomedical and Health Industry Development Support Technology Special Project (2023WJC193). The funders had roles in the study design, data collection, analysis, decision to publish, or preparation of the manuscript.
Ethics approval and consent to participate
The study protocol was in accordance with the ethical standards of the institutional research committee and the ethics guidelines of the 1975 Declaration of Helsinki. Hangzhou Xixi Hospital's institutional ethics review committee approved the study (2023 Science Ethic No.65). Written informed consent was not required due to the retrospective nature of this study. All the data used in this study were anonymized.
Data availability statement
The data will be made available upon request by the corresponding author.
CRediT authorship contribution statement
Yan Zhang: Writing – original draft, Visualization, Project administration. Kailong Gu: Methodology, Investigation, Data curation. Wei Du: Validation, Software. Aifang Xu: Writing – review & editing, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Cao C., Liang L., Wang W., Luo H., Huang S., Liu D., et al. Common reservoirs for Penicillium marneffei infection in humans and rodents. China. Emerg. Infect. Dis. 2011;17(2):209–214. doi: 10.3201/eid1702.100718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Le T., Wolbers M., Chi N.H., Quang V.M., Chinh N.T., Lan N.P., et al. Epidemiology, seasonality, and predictors of outcome of AIDS-associated Penicillium marneffei infection in Ho Chi Minh City, Viet Nam. Clin. Infect. Dis. 2011;52(7):945–952. doi: 10.1093/cid/cir028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Son V.T., Khue P.M., Strobel M. Penicilliosis and AIDS in Haiphong, Vietnam: evolution and predictive factors of death. Med. Maladies Infect. 2014;44(11–12):495–501. doi: 10.1016/j.medmal.2014.09.008. [DOI] [PubMed] [Google Scholar]
- 4.Qin Y., Zhou Y., Lu Y., Chen H., Jiang Z., He K., et al. Multicentre derivation and validation of a prognostic scoring system for mortality assessment in HIV-infected patients with talaromycosis. Mycoses. 2021;64(2):203–211. doi: 10.1111/myc.13206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hien H.T.A., Thanh T.T., Thu N.T.M., Nguyen A., Thanh N.T., Lan N.P.H., et al. Development and evaluation of a real-time polymerase chain reaction assay for the rapid detection of Talaromyces marneffei MP1 gene in human plasma. Mycoses. 2016;59(12):773–780. doi: 10.1111/myc.12530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jiang J., Meng S., Huang S., Ruan Y., Lu X., Li J.Z., et al. Effects of Talaromyces marneffei infection on mortality of HIV/AIDS patients in southern China: a retrospective cohort study. Clin. Microbiol. Infect. 2019;25:233–241. doi: 10.1016/j.cmi.2018.04.018. [DOI] [PubMed] [Google Scholar]
- 7.Zhao W. Chinese guidelines for the diagnosis and treatment of HIV/AIDS (2021 edition) Infectious Diseases & Immunity. 2022;2(3):145–167. [Google Scholar]
- 8.Shi M., Lin J., Wei W., Qin Y., Meng S., Chen X., et al. Machine learning-based in-hospital mortality prediction of HIV/AIDS patients with Talaromyces marneffei infection in Guangxi, China. PLoS Neglected Trop. Dis. 2022;16(5) doi: 10.1371/journal.pntd.0010388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ning C., Lai J., Wei W., Zhou B., Huang J., Jiang J., et al. Accuracy of rapid diagnosis of Talaromyces marneffei: a systematic review and meta-analysis. PLoS One. 2018;13(4) doi: 10.1371/journal.pone.0195569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qin Y., Huang X., Chen H., Liu X., Li Y., Hou J., et al. Burden of Talaromyces marneffei infection in people living with HIV/AIDS in Asia during ART era: a systematic review and meta-analysis. BMC Infect. Dis. 2020;20(1):551. doi: 10.1186/s12879-020-05260-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Limper A.H., Adenis A., Le T., Harrison T.S. Fungal infections in HIV/AIDS. Lancet Infect. Dis. 2017;17(11):e334–e343. doi: 10.1016/S1473-3099(17)30303-1. [DOI] [PubMed] [Google Scholar]
- 12.AIDS-associated opportunistic infections research group of the National Science and Technology Major Project of China during the 13th Five-Year Plan period. Expert consensus on the diagnosis and treatment of Talaromycosis in AIDS patients in China. J. Southwest Univ. (Nat. Sci. Ed.) 2020;42(7):61–75. [Google Scholar]
- 13.Nor-Hayati S., Sahlawati M., Suresh-Kumar C., Lee K.C. A retrospective review on successful management of Penicillium marneffei infections in patients with advanced HIV in Hospital Sungai Buloh. Med. J. Malaysia. 2012;67(1):66–70. [PubMed] [Google Scholar]
- 14.Chen J., Zhang R., Shen Y., Liu L., Qi T., Wang Z., et al. Clinical characteristics and prognosis of Penicilliosis among human accepted article immunodeficiency virus-infected patients in eastern China. Am. J. Trop. Med. Hyg. 2017;96(6):1350–1354. doi: 10.4269/ajtmh.16-0521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wei H.Y., Liang W.J., Li B., Wei L.Y., Jiang A.Q., Chen W.D., et al. Clinical characteristics and risk factors of Talaromyces marneffei infection in human immunodeficiency virus-negative patients: a retrospective observational study. World J Emerg Med. 2021;12(4):281–286. doi: 10.5847/wjem.j.1920-8642.2021.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xin Q., Xie T., Chen R., Wang H., Zhang X., Wang S., et al. Construction and validation of an early warning model for predicting the acute kidney injury in elderly patients with sepsis. Aging Clin. Exp. Res. 2022;34(12):2993–3004. doi: 10.1007/s40520-022-02236-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen L., Wu X., Qin H., Zhu H. The PCT to albumin ratio predicts mortalityin patients with acute kidney injury caused by abdominal infection-evoked sepsis. Front. Nutr. 2021;8 doi: 10.3389/fnut.2021.584461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oh D.H., Kim Y.C., Kim E.J., Jung I.Y., Jeong S.J., Kim S.Y., et al. Multidrug-resistant Acinetobacter baumannii infection in lung transplant recipients: risk factors and prognosis. Inf. Disp. 2019;51(7):493–501. doi: 10.1080/23744235.2018.1556400. [DOI] [PubMed] [Google Scholar]
- 19.Ugajin M., Yamaki K., Iwamura N., Yagi T., Asano T. Blood urea nitrogen to serum albumin ratio independently predicts mortality and severity of community-acquired pneumonia. Int. J. Gen. Med. 2012;5:583–589. doi: 10.2147/IJGM.S33628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Viasus D., Garcia-Vidal C., Simonetti A., Manresa F., Dorca J., Gudiol F., et al. Prognostic value of serum albumin levels in hospitalized adults with community-acquired pneumonia. J. Infect. 2013;66(5):415–423. doi: 10.1016/j.jinf.2012.12.007. [DOI] [PubMed] [Google Scholar]
- 21.Zou X.L., Feng D.Y., Wu W.B., Yang H.L., Zhang T.T. Blood urea nitrogen to serum albumin ratio independently predicts 30-day mortality and severity in patients with Escherichia coli bacteraemia. Med. Clin. 2021;157(5):219–225. doi: 10.1016/j.medcli.2020.06.060. [DOI] [PubMed] [Google Scholar]
- 22.Zhao D., Chen S., Liu Y., Xu Z., Shen H., Zhang S., et al. Blood urea nitrogen-to-albumin ratio in predicting long-term mortality in patients following coronary artery bypass grafting: an analysis of the MIMIC-III database. Front. Surg. 2022;9 doi: 10.3389/fsurg.2022.801708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dundar Z.D., Kucukceran K., Ayranci M.K. Blood urea nitrogen to albumin ratio is a predictor of in-hospital mortality in older emergency department patients. Am. J. Emerg. Med. 2021;46:349–354. doi: 10.1016/j.ajem.2020.10.008. [DOI] [PubMed] [Google Scholar]
- 24.Ponti G., Maccaferri M., Ruini C., Tomasi A., Ozben T. Biomarkers associated with COVID-19 disease progression. Crit. Rev. Clin. Lab Sci. 2020;57(6):389–399. doi: 10.1080/10408363.2020.1770685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tjendra Y., Al Mana A.F., Espejo A.P., Akgun Y., Millan N.C., Gomez-Fernandez C., et al. Predicting disease severity and outcome in COVID-19 patients: a review of multiple biomarkers. Arch. Pathol. Lab Med. 2020;144(12):1465–1474. doi: 10.5858/arpa.2020-0471-SA. [DOI] [PubMed] [Google Scholar]
- 26.Larsson M., Nguyen L.H., Wertheim H.F., Dao T.T., Taylor W., Horby P., et al. Clinical characteristics and outcome of Penicillium marneffei infection among HIV-infected patients in northern Vietnam. AIDS Res. Ther. 2012;9(1):24. doi: 10.1186/1742-6405-9-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data will be made available upon request by the corresponding author.