Abstract
Esophageal squamous cell carcinoma (ESCC) is a prevalent and deadly malignancy of the digestive tract. Recent research has identified long non-coding RNAs (lncRNAs) as crucial regulators in the pathogenesis of ESCC. These lncRNAs, typically exceeding 200 nucleotides, modulate gene expression through various mechanisms, including the competing endogenous RNA (ceRNA) pathway and RNA-protein interactions. The current study reviews the multifaceted roles of lncRNAs in ESCC, highlighting their involvement in processes such as proliferation, migration, invasion, epithelial-mesenchymal transition, cell cycle progression, resistance to radiotherapy and chemotherapy, glycolysis, apoptosis, angiogenesis, autophagy, tumor growth, metastasis and the maintenance of cancer stem cells. Specific lncRNAs like HLA complex P5, LINC00963 and non-coding repressor of NFAT have been shown to enhance resistance to radio- and chemotherapy by modulating pathways such as AKT signaling and microRNA interaction, which promote cell survival and proliferation under therapeutic stress. Furthermore, lncRNAs like family with sequence similarity 83, member A antisense RNA 1, zinc finger NFX1-type containing 1 antisense RNA 1 and taurine upregulated gene 1 are implicated in enhancing invasive and proliferative capabilities of ESCC cells through the ceRNA mechanism, while interactions with RNA-binding proteins further influence cancer cell behavior. The comprehensive analysis underscores the potential of lncRNAs as biomarkers for prognosis and therapeutic targets in ESCC, suggesting avenues for future research focused on elucidating the detailed molecular mechanisms and clinical applications of lncRNAs in ESCC management.
Key words: epithelial-mesenchymal transition, esophageal squamous cell carcinoma, long non-coding RNAs, metastasis
1. Introduction
Long non-coding RNAs (lncRNAs) are transcribed by either RNA polymerase II (PolII) or PolIII, with a typical length exceeding 200 nucleotides, possessing a cap structure at the 5' end and being polyadenylated at the 3' end. It has been observed that the majority of lncRNAs cannot encode proteins (1). However, certain lncRNAs can produce short peptides that have a role in cellular physiology; for instance, HOXB cluster antisense RNA 3 can encode a conserved 53-amino acid peptide (2). Several mechanisms, including epigenetic modifications, competitive endogenous RNA (ceRNA) mechanisms (3), RNA-protein interactions, as well as the involvement of exosomes containing lncRNAs, contribute to the regulation of gene expression within cells.
Esophageal cancer (ESCA) primarily consists of two major subtypes, namely esophageal squamous cell carcinoma (ESCC) and esophageal adenocarcinoma. It has been established that lncRNAs play a crucial role in the progression of ESCC. Clinically, lncRNAs have been associated with various aspects of ESCC, including lymph node metastasis, tumor size (4), tumor node metastasis (TNM) stage (3), prognosis (5), overall survival (OS) rate (6), pathological grade of differentiation (7), survival duration (8), malignancy grade (9), distant metastasis (10), depth of invasion (11) and postoperative metastasis (12). In ESCC cell lines, lncRNAs have been implicated in processes such as cell proliferation (13), migration (14), invasion (15), epithelial-to-mesenchymal transition (16), cell cycle progression (17), radiosensitivity (18), glycolysis (19), chemotherapy resistance, apoptosis (20), angiogenesis (21), maintenance of stem cell characteristics (22), as well as autophagy-related events (23). In both cell-derived xenograft (CDX) models and patient-derived tumor xenografts (PDX), lncRNAs have been linked to tumor growth (24), lymph node metastasis (25), liver metastasis (16), lung metastasis (9), bone metastasis (13), chemotherapy resistance (26) and resistance to radiotherapy (27). The expanding knowledge of lncRNAs and their mechanisms has shed light on their diverse functions. This review aims to summarize recent studies on the involvement of lncRNAs in ESCC, providing valuable insight into the clinical diagnosis, treatment and prevention of ESCC.
2. Regulatory mechanisms of lncRNAs in ESCC
CeRNAs
In the ceRNA pathway, lncRNAs act as ceRNAs, sequestering micro (mi)RNAs and preventing them from binding to target mRNAs, thereby enhancing mRNA expression (28). The ceRNA mechanism has garnered significant attention in recent years for its role in regulating cellular protein expression.
RNA-protein interactions
Classic examples of ncRNA-protein complexes include ribosomal RNAs and transfer RNAs, which are essential for protein translation within cells. MiRNAs can bind to various proteins such as Dicer, transactivation response element RNA-binding protein, protein kinase activating protein, Gemin3 and Argonaute to form the RNA-induced protein-coating complex (RISC) that influences gene expression (29). Recent studies have found that ncRNAs, including lncRNAs, can modulate protein stability and activity by binding to proteins, thereby influencing mRNA expression, translational control and cellular localization.
3. LncRNAs upregulated in ESCC
In general, lncRNAs that are highly expressed in ESCC tend to promote tumorigenesis and the development of ESCC. However, abnormally upregulated lncRNAs may act as oncogenic factors in normal cells. Abnormally upregulated lncRNAs are often positively correlated with ESCC phenotypes such as proliferation, migration, invasion, chemo-resistance and radiotherapy resistance.
LncRNAs based on the ceRNA mechanism
Family member with sequence similarity 83 antisense RNA/miR-214/cell division cycle 25B (CDC25B) were shown to promote the proliferation, migration and invasion of KYSE30 and EC109 cells and to be associated with TNM staging and tumor differentiation grading (3). Zinc finger NFX1-type containing 1 antisense RNA 1 (ZF-AS1)/miR-124/signal transducer and activator of transcription 3 promoted the proliferation, migration and invasion of ECA109 cells and enhanced the growth of CDX (ECA109) tumors (13). Taurine upregulated 1/miR-498/CDC42 promoted the proliferation and invasion of TE-1 and KYSE30 cells (30). miR205HG/miR-214/SRY-box transcription factor 4 (SOX4) aided in cell cycle progression, proliferation, invasion and migration of KYSE30 and EC109 cells, with its function related to tumor size and lymphatic metastasis (4). Opa interacting protein 5-AS1/miR-1297 was able to promote the proliferation, migration and invasion of ECA109 cells and was linked to unfavorable clinical outcomes (14). Deleted in lymphocytic leukemia 1 (DLEU1)/miR-671-5p/high mobility group AT-hook 1 could promote the proliferation, migration and invasion of ECA109 cells (15).
LOC146880/miR-328-5p/Fascin actin-bundling protein 1 (FSCN1) has been shown to activate the MAPK pathway and promote the growth and liver metastasis of CDX (KYSE30) as well as the proliferation, invasion, migration and epithelial to mesenchymal transition (EMT) of KYSE30 and TE-1 cells (16). It was also shown to inhibit apoptosis in KYSE150 cells. LINC00680/miR-423-5p/p21 (also known as RAC1) activated kinase 6 promoted proliferation, colony formation, migration and invasion of KYSE140 and KYSE510 cells and enhanced the growth of CDX (KYSE510), correlating with tumor volume, stage and prognosis (31). LINC01711/miR-326/FSCN1 promoted TE-1 proliferation, migration, invasion and growth, enhanced CDX (TE-1) growth and inhibited apoptosis (24). High expression in hepatocellular carcinoma/miR-4458/PBX homeobox 3 enhanced EC109 cell viability and invasion (32). Gastric cancer associated transcript 3/miR-149/forkhead box (FOX)M1 promoted the proliferation, migration and invasion of KYSE150 and KYSE510 cells and increased CDX (KYSE510) growth (33). Hepatocyte nuclear factor 1 α-AS1/miRNA-298/transcription factor 4 could promote EMT and stem cell stemness of EC109 and KYSE70 cells (22).
Actin filament associated protein 1-AS1/miR-498/vascular endothelial growth factor (VEGF) A could promote the survival of ECA109 and KYSE30 cells and aid in their growth and migration (34). LINC00467/miR-485-5p/dolichyl-phosphate N-acetylglucosaminephosphotransferase 1 helped KYSE510 and TE-5 cells proliferate, promoted CDX (KYSE510) growth and prevented apoptosis of cancer cells (35). Epidermal growth factor receptor-AS1/miR-145/Rho associated coiled-coil containing protein kinase 1 promoted EC109 migration and invasion and was associated with OS (6). Small nucleolar RNA host gene 17 (SNHG17)/miR-338-3p/SOX4 promoted proliferation, invasion and EMT of ECA109 and TE-1, and enhanced CDX (ECA109) growth (36). Motor neuron and pancreas homeobox 1-AS1/miR-34a/Sirtuin 1 (SIRT1) promoted growth, migration, invasion and cell cycle progression in KYSE30 and KYSE150 cells, inhibited apoptosis and were linked to lymph node metastasis (37). THAP domain-containing protein 9 (THAP9)-AS1/miR-335-5p/Sphingomyelin synthase 2 aided the growth, migration, invasion, EMT and survival of KYSE150 and TE-10 cells (38).
MAF BZIP transcription factor G-AS1/miR-765/pancreatic and duodenal homeobox 1 have been shown to promote the proliferation, migration, invasion and aerobic glycolysis of EC109 cells (39). SNHG6/miR-101-3p/enhancer of zeste 2 polycomb repressive complex 2 subunit (EZH2) inhibited apoptosis in EC1 and KYSE30 cells (40). FAM83H-AS1/miR-10a-5p/Girdin increased proliferation, migration, invasion and EMT in KYSE150 and TE-1 and was linked to TNM staging, pathologic staging and lymph node metastasis (41). Cancer susceptibility candidate 15 (CASC15)/miR-33a-5p/prostaglandin-endoperoxide synthase 2 promoted the proliferation, migration and invasion of KYSE150 and KYSE450 cells (42). Juxtaposed with X-inactivation center/miR-516b-5p/VEGFA stimulated EC9706 and KYSE150 cell growth, angiogenesis, migration and invasion, as well as the growth of CDX (EC9706) (43). SNHG6/miR-186-5p/hypoxia inducible factor 1 subunit α (HIF1α) promoted the proliferation, migration and invasion of EC109 and KYSE150 cells (44). LINC00858/miR-425-5p/ABL proto-oncogene 2, non-receptor tyrosine kinase enhanced the proliferation, invasion, migration and EMT of ECA109 and EC9706 cells (45).
Differentiation antagonizing non-protein coding RNA/miR-33a-5p/zinc finger E-box binding homeobox 1 (ZEB1) could promote the proliferation and metastasis of EC1 and KYSE150 cells, which was shown to correlate with prognosis (46). Breast cancer anti-estrogen resistance 4 (BCAR4)/miR-139-3p/ELAV like RNA binding protein 1 suppressed the p53/p21 signaling pathway, facilitated EC109 and TE-1 cells growth and migration, and prevented their death (47). Chromosome 9 open reading frame 72/miR-661/histone deacetylase 11 promoted proliferation, migration and invasion of TE-1 and ECA109 cells through the nuclear factor κB (NF-κB) pathway, promoted CDX (TE-1) growth and inhibited apoptosis (48). miR31HG/miR-34a/MET proto-oncogene, receptor tyrosine kinase helped CDX (KYSE30) grow and stopped apoptosis (49). LINC01980/miR-190a-5p/myosin VA promoted proliferation, migration, invasion and EMT of KYSE150 and ECA109 cells and was shown to be associated with cancer prognosis (50). THAP9-AS1/miR-133b/SOX4 promoted the proliferation, migration and invasion of ECA109 and KYSE30, and CDX (ECA109) growth, inhibited apoptosis and were shown to be associated with tumor size, TNM staging, lymph node metastasis and cancer prognosis. SOX4 in turn bound to the THAP9-AS1 promoter and facilitated transcription, forming a positive feedback loop (51).
MCEI (ENST00000441932)/miR-6759-5p/Insulin like growth factor 2 (IGF2) has been shown to promote KYSE30 and EC109 cells' proliferation, migration, cisplatin resistance and invasion through the PI3K/AKT signaling pathway. It also made CDX (KYSE30 and EC109) resistant to cisplatin and inhibited apoptosis (52). Nicotinamide nucleotide transhydrogenase-AS1/miR-382-5p promoted the expression of NNT, the proliferation and migration of ECA109 and KYSE30 cells, as well as inhibiting cell apoptosis, and promoting the growth of CDX (ECA109) (53). Homo sapiens uncharacterized LOC100507002/miR-6785-5p/neurensin 2 promoted the proliferation, colony formation, migration and invasion of KYSE150, KYSE510 and KYSE140, and promoted the growth of CDX (KYSE510). It has been associated with gender, deep invasion, lymph node metastasis and poor prognosis in patients with ESCC (54). LncSUMO1P3/miR-486-5p/phosducin (PHD) or PHD finger protein 8 regulated CD151 [CD151 molecule (Raph blood group)] expression and promoted the proliferation, migration, invasion, and EMT of KYSE150 and KYSE450 (55). Prostate cancer associated transcript 5/miR-4295/PHD finger protein 20 upregulated the proliferation, migration and invasion of EC109 and KYSE450 cells (56). X-inactive specific transcript (XIST)/miR-34a/ZEB1 promoted the proliferation, migration, invasion and EMT of KYSE150 cells. It has been shown to promote the growth of CDX (KYSE150) and was associated with metastasis and reduced survival (57).
Zinc finger protein, FOG family member 2-AS1/miR-3612/TNF receptor-associated factor 4 could promote proliferation, migration and invasion, and inhibit apoptosis in EC9706 and TE-10 cells through NF-κB (58). LINC00662/miR-340-5p/homeobox B2 enhanced the proliferation, migration and invasion of KYSE510 cells, which was associated with cancer prognosis (59). Colon cancer associated transcript 2/miR-200b/IGF2 mRNA binding protein 2 (IGF2BP2) modulated thymidine kinase 1 (TK1) mRNA through m6A and IGF2BP2, leading to stabilized TK1 expression, enhanced ESC410 migration and spread, and promotion of CDX (ESC410) tumor formation (60). SET binding factor 2-AS1/miR-494/profilin 2 promoted proliferation, migration, invasion and EMT in ECA109 and KYSE150 cells (61). LINC00941/miR-877-3p/prostate transmembrane protein, androgen induced 1 facilitated proliferation, invasion, migration and EMT of KYSE510 and KYSE30 cells (62). LINC00152/miR-153-3p/FYN proto-oncogene, Src family tyrosine kinase supported the proliferation and colony formation of EC109 and KYSE150 cells, and CDX (EC109) growth, while inhibiting apoptosis, and showing a correlation with TNM staging, lymph node metastasis and prognosis (63).
Proteasome 20S subunit α 3-AS1/miR-101/EZH2 could stimulate the proliferation, invasion and migration of KYSE150 and KYSE450 cells, and has been associated with tumor size, distant metastasis and prognosis (10). LINC00963/miR-214-5p/RAB14, member RAS oncogene family enhanced the proliferation and invasion of KYSE150 and TE-1 cells, thereby promoting CDX (KYSE150) growth and showing an association with TNM staging, metastasis and prognosis (64). Gamma-butyrobetaine hydroxylase 1 (BBOX1)-AS1/miR-506-5p/eukaryotic translation initiation factor 5A boosted the proliferation and stem cell stemness of KYSE450 and EC109 cells by stabilizing patched 1 expression and activating the Hedgehog signaling pathway (65). Forkhead box D2-AS1/miR-204-3p stimulated the proliferation, migration and invasion of KYSE150 and EC109 cells, as well as promoting CDX (KYSE150) growth (66). Kinectin 1 (KTN1)-AS1/miR-885-5p/striatin 3 enhanced the proliferation, invasion and EMT of KYSE150 cells through the YAP1-dependent Hippo signaling pathway (67).
XIST/miR-129-5p/cyclin D1 have been shown to aid in the cell cycle progression, proliferation, migration and invasion of EC9706 and ECA109 cells, while preventing CDX (ECA109) growth by inhibiting apoptosis (68). LINC00473/miR-497-5p/protein kinase AMP-activated catalytic subunit α 1 promoted the proliferation, migration, EMT and growth of CDX (KYSE30) in KYSE30 and TE-1 cells (69). Nuclear paraspeckle assembly transcript 1/miR-590-3p/MDM2 proto-oncogene (MDM2) enhanced the proliferation, migration, invasion and angiogenesis of ECA109 and TE-13 cells, as well as promoting the growth of CDX (ECA109) (70). Lnc-ABCA12-3/miR-200b-3p/fibronectin 1 facilitated the migration, invasion and proliferation of KYSE510 and ECA109 cells and was linked to lymph node metastasis and prognosis (71). ESCC-related lncRNAs transcript 1 (ESCCAL-1)/miR-590/LDL receptor related protein 6 promoted the growth, invasion and migration of KYSE150 and KYSE450 cells (72). RAR related orphan receptor (ROR)/miR-145/FSCN1 enhanced EC109 migration and invasion and was shown to be associated with patient survival (73).
LOC440173/miR-30d-5p/histone deacetylase 9 could promote the proliferation, migration, invasion, as well as EMT in KYSE150 cells, as well as growth of CDX (KYSE150), and was shown to be associated with histological grading, tumor invasion, lymph node metastasis and TNM stage (74). Similarly, LINC00514/miR-378a-5p/pphingosine kinase 1 has been shown to promote proliferation and invasion in KYSE150 and KYSE30 cells, upregulate adipogenesis-related proteins, and to be associated with lymph node metastasis and prognosis (75). LINC00473/miR-374a-5p/spindlin 1 could promote radioresistance in TE-1 and EC9706 cells and has been linked to TNM staging (76). Meanwhile, LINC00473/miR-497-5p/CDC25A facilitated survival and radioresistance in KYSE30 and TE-5 cells, and was associated with T-stage, lymph node metastasis staging and tumor differentiation grading (77). NCK adaptor protein 1-AS1/miR-133b/glutamyl aminopeptidase enhanced proliferation, invasion and migration in EC109 and EC9706 cells, inhibited cell apoptosis and promoted CDX (EC109) growth (78).
Lipase H 4/miR-216b/IGF2BP2 promoted growth and cell cycle progression in KYSE150 and KYSE510 cells, as well as CDX (KYSE510) growth, and inhibited apoptosis, correlating with tumor size and prognosis (79). Testis-specific transcript, Y-linked 15/miRNA-337-3p/Janus kinase 2 (JAK2) promoted the proliferation, migration and invasion of TE-3 cells (80). LINC01232/miRNA-654-3p/Heparin binding growth factor (HDGF) enhanced the proliferation, migration and invasion of ECA109 and KYSE150 cells, while inhibiting apoptosis (81). LINC00152/miR-107/RAB10, member RAS oncogene family boosted migration and invasion of TE-1 and KYSE30 cells (82).
ROR/miR-204-5p/MDM2 heightened cell activity and proliferation of EC109 cells, whilst inhibiting cell apoptosis by promoting p53 ubiquitination (83). BCAR4/miRNA-181c-5p/LIM and SH3 protein 1 aided the invasion and movement of EC9706 and TE-1 cells, and CDX growth (EC9706), while preventing apoptosis (84). BBOX1-AS1/miR-513a-3p/Solute carrier family 7 member 11 was shown to promote proliferation, invasion and migration of TE-1 and KYSE150 cells, CDX (TE-1), inhibit apoptosis and ferroptosis, and to be associated with patient prognosis (85). Lastly, protein tyrosine phosphatase receptor type G-AS1/miR-599/pyruvate dehydrogenase kinase 1 (PDK1) boosted the proliferation, migration, glycolysis and stem cell stemness in TE-8 and KYSE150 cells (86).
LncRNAs regulated by RNA-protein interactions
Actin gamma 1 pseudogene interacted with 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3, enhancing its stability and preventing ubiquitination by APC/C. This interaction has been shown to promote cell cycle progression by activating glycolysis in KYSE150 and KYSE30 cells (19). Solute carrier family 25 member 21 (SLC25A21)-AS1 was reported to bind to nucleophosmin 1 (NPM1), facilitating c-Myc-mediated downstream gene transcription, leading to increased growth, migration and cisplatin resistance in KYSE30 and KYSE450 cells, as well as being associated with tumor grade and patient OS (7). LINC01554 was demonstrated to bind to GTPase activating protein (SH3 domain) binding protein 2 (G3BP2), protecting it from ubiquitin-mediated degradation. This interaction stabilized HDGF expression by binding to HDGF mRNA, promoting invasion and metastasis in KYSE410 and KYSE510 cells, as well as facilitating CDX metastasis (87). Similarly, LINC00239 bound to and inhibited c-Myc promoter-binding protein-1 expression, resulting in the activation of c-Myc transcription and promoting proliferation, metastasis, invasion and EMT in KYSE170 and KYSE150 cells, with implications for cancer prognosis (88). SNHG16 recruited and bound to eukaryotic translation initiation factor 4A3, controlling RhoU mRNA stability and promoting growth and spread in ECA109, KYSE30, KYSE140 and KYSE410 cells, as well as influencing tumor differentiation and staging (89). Binder of MAP3K1 and KLF4-AS1 bound to G3BP2, inhibiting the degradation of c-Myc mRNA and promoting proliferation, migration and invasion of KYSE450 and KYSE510 cells, with implications for malignant phenotype and c-Myc expression (90).
DiGeorge syndrome critical region gene 5 (DGCR5) bound to serine and arginine rich splicing factor 1, stabilizing it and regulating selective splicing. This interaction promoted myeloid cell leukemia 1 (Mcl-1) splicing and facilitated Mcl-1L isoform switching, promoting proliferation, migration and invasion in TE-1 and KYSE170 cells, as well as inhibiting apoptosis (91). DLEU1 bound to dynein light chain LC8-type 1, inhibiting its ubiquitination and degradation by ring finger protein 114, thereby inhibiting apoptosis in KYSE410 and EC109 cells, promoting cisplatin/cis-diamminedichloroplatinum (CDDP) resistance and conferring resistance in CDX (KYSE410) cells (92). GK intronic transcript 1 bound to mitogen-activated protein kinase 1, preventing its interaction with dual specificity phosphatase 6 and activating the ERK/MAPK signaling pathway, promoting proliferation, invasion and migration in TE-1 and TE-10 cells, as well as being associated with clinical staging and prognosis (23). SNHG17 bound to Jun proto-oncogene, AP-1 transcription factor subunit (c-Jun), facilitating c-Myc transcription and promoting proliferation, migration, invasion and EMT in ECA109 and KYSE150 cells, as well as inhibiting apoptosis and correlating with patient survival (93). Lastly, ESCCAL-1 prevented galectin 1 degradation through ubiquitination, promoting growth and cell cycle progression in EC9706, KYSE150 and KYSE450 cells (94).
Activation of LBX2-AS1 by ZEB1 enhanced ZEB1 and ZEB2 transcription through interaction with heterogeneous nuclear ribonucleoprotein C, leading to increased migration and EMT in KYSE150 and EC109 cells (95). LINC00337 bound to E2F transcription factor 4 to upregulate TPX2 microtubule nucleation factor expression, facilitating autophagy and cisplatin resistance in ECA109 cells, as well as cisplatin resistance in CDX (ECA109) (96). In the case of LINC01305, interaction with IGF2BP 2 and 3 stabilized mRNA coding 5-hydroxytryptamine receptor 3A, promoting migration and proliferation in KYSE510 and KYSE140 cells (97). On the other hand, LINC01296 interacted with EZH2 to repress KLF transcription factor 2 expression, fueling proliferation, clone formation, migration and invasion in EC106 and EC9706 cells, as well as contributing to lymph node metastasis, TNM staging and OS (98). SOX2 transcribed neurensin 2-AS1, which was bound to phosphoglycerate kinase 1 to inhibit its ubiquitination, thereby enhancing proliferation, migration and invasion, as well as correlating with survival in KYSE170 cells (99).
Transmembrane protein 44-AS1 was reported to bind to IGF2BP2 to stabilize glutathione peroxidase 4 mRNA, thereby suppressing ferroptosis and promoting proliferation, invasion and metastasis of ECA109, KYSE30 and KYSE150 cells (100). Protein disulfide isomerase family A member 3 pseudogene 1 (PDIA3P1) interacted with octamer-binding transcription factor 4 to prevent ubiquitination and degradation, thereby driving proliferation, invasion, metastasis and stem cell stemness in TE-1, ECA109, KYSE30 and KYSE150 cells. It was also found to inhibit cancer cell apoptosis and function as a transcription factor for PDIA3P1 (101). G077640 interacted with H2A.X variant histone to inhibit the degradation of HIF1α, leading to increased expression of glucose transporter type 4, hexokinase 2 and PDK1, reprogramming intracellular glycolysis metabolism, and promoting proliferation and migration in TE-11 and KYSE150 cells, ultimately supporting the growth of CDX (KYSE150) (102). Similarly, HOXC cluster-AS1 was reported to bind to IGF2BP2, thereby stabilizing SIRT1 expression, promoting proliferation and migration in ECA109 and KYSE30 cells, and supporting the growth of CDX (KYSE30) (103). Moreover, LINC00941 interacting with interleukin enhancer binding factor 2 and Y-box binding protein 1 boosted SOX2 expression, proliferation, stem cell differentiation, migration and invasion in TE-1 and KYSE170 cells, as well as promoting the growth of CDX (TE-1) (104).
Epigenetic regulation
Various lncRNAs are upregulated in ESCC, which modulates gene expression through epigenetic mechanisms. Double homeobox A pseudogene 10, expressed in KYSE30 and KYSE180 cells, recruited EZH2 to the cyclin-dependent kinase (CDK) inhibitor 1A (P21) promoter, leading to the silencing of P21 expression. This silencing in turn promoted cell proliferation, metastasis and cell cycle progression, as well as inhibiting apoptosis (8). FOXP4-AS1 bound to the FOXP4 promoter by enriching mixed lineage leukemia protein (MLL)2 and trimethylation of lysine 4 on histone H3 (H3K4me3), which promoted the expression of FOXP4. FOXP4 then acted as a transcription factor for β-catenin, promoting the proliferation, invasion and migration of YES-2 and KYSE150 cells. This lncRNA was also associated with lymph node metastasis and TNM staging (105). Thymopoietin (TMPO)-AS1 bound to FUS RNA binding protein (FUS) and recruited p300 to the TMPO promoter, resulting in histone h3 lysine 27 acetylation (H3K27ac) and activation of TMPO transcription. This lncRNA promoted proliferation, metastasis and the growth of CDX (KYSE150 and TE-11) cell lines, and correlated with patients' survival (106). LINC01419 bound to the glutathione S-transferase pi 1 (GSTP1) promoter region, leading to increased methylation of the GSTP1 promoter and decreased expression of GSTP1 by recruiting DNA methyltransferases. This promoted proliferation and 5-FU resistance, while inhibiting apoptosis in EC9706 and KYSE70 cells (107). H3K27ac activated the expression of Lymphoid enhancer binding factor 1-AS1, which interacted with programmed cell death 5, resulting in tumor protein p53 degradation. This promoted the proliferation of ECA109 cells and the growth of CDX (EC109), and was associated with OS (108).
Functional role of antisense lncRNAs
Briefly, in terms of antisense lncRNAs, SLC25A21-AS1 increased the stability of SLC25A21 mRNA by affecting tryptophan catabolism (7). Proliferating cell nuclear antigen (PCNA)-AS1 promoted the proliferation, invasion and migration of Eca109 and TE-1 cells by antagonizing the expression of miR-2467-3p, which in turn promoted PCNA expression (109).
M6A modulates the regulatory function of lncRNAs
N6-methyladenosine (M6A) is a common internal modification found in mRNAs, and recent research indicates that M6A also plays a role in modifying lncRNAs, influencing their stability either positively or negatively (110). For instance, M6A methylation of LINC00022 was suppressed by FTO α-ketoglutarate dependent dioxygenase (FTO), inhibiting its degradation by YTH N6-methyladenosine RNA binding protein F2. This interaction led to the binding of LINC00022 to p21, promoting p21 ubiquitination and degradation, ultimately facilitating the progression and proliferation of KYSE150 and TE-1 cells (17). In addition, CASC8 interacted with alkB homolog 5, RNA demethylase (m6A RNA demethylase) to stabilize its expression. This binding inhibited the ubiquitination and degradation of heterogeneous nuclear ribonucleoprotein L, activated the Bcl2/caspase3 pathway, reduced sensitivity to cisplatin in YES-2, KYSE450, KYSE30 and KYSE180, and promoted the growth of CDX (KYSE30) along with correlating with prognosis (111). Furthermore, CASC15 diminished single-minded 2 stability through demethylation mediation, thereby promoting the proliferation of ECA109 and KYSE450, hindering apoptosis and correlating with prognosis (112). LncRNA HLA complex P5 interacted with YTH N6-methyladenosine RNA binding protein F1 (YTHDF1) to enhance the m6A modification of HK2 mRNA by YTHDF1, leading to increased HK2 stability, which in turn promoted the proliferation, invasion and Warburg effect of EC109 and TE10 cells. This interaction also boosted the growth of CDX (EC109) and correlated with tumor volume, TNM stage, lymph node metastasis and survival time (113).
Resistance to radio- and chemotherapy
A study revealed that lncRNA HCP5/miR-216a-3p/PDK1 enhanced radiotherapy resistance in KYSE30 and TE-1 cells by activating the AKT signaling pathway (114). Similarly, LINC00963/miR-10a/spindle and kinetochore associated complex subunit 1 was found to increase cisplatin resistance in TE-1 and TE-1/DDP cells, thereby promoting CDX growth (115). Furthermore, after cisplatin treatment, lncRNA non-coding repressor of NFAT was upregulated, contributing to cisplatin resistance by downregulating miR-31 in KYSE510 (116). Another study showed that Linc01614/miR-4775 enhanced proliferation and cisplatin resistance in EC9706 and KYSE30 cells, while inhibiting cell apoptosis and promoting CDX growth in EC9706 (117). In addition, lncRNA HCP5 inhibited UTP3 small subunit processome component ubiquitination and degradation, leading to the recruitment of c-MYC to activate vesicle associated membrane protein 3 expression and inhibited caspase-dependent apoptosis, ultimately resulting in cisplatin resistance in CDX (KYSE150) (118). Researchers also found that FTO, in association with m6A modification, stabilized long intergenic noncoding RNA for kinase activation (LINK-A) expression, promoted CDK1-mediated phosphorylation of phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit α (MCM), relieved MCM-mediated HIF-1α transcriptional repression, and promoted glycolysis and cisplatin resistance in the KYSE450 and KYSE150 cell lines. This mechanism also facilitated cisplatin resistance in PDX and CDX (KYSE450 and KYSE150) (119). Furthermore, lncRNA SOX2 overlapping transcript (SOX2OT) was discovered to enhance the expression of stem cell-related genes, sphere formation ability and docetaxel resistance in KYSE30 and YM1 cells (120).
Other mechanisms of action
Non-coding RNA activated by DNA damage (NORAD) has been shown to interact with pumilio RNA binding family member 1 on pri-miR-199a, leading to the inhibition of miR-199a-5p processing and maturation. This resulted in the attenuation of miR-199a-5p silencing of endonuclease/exonuclease/phosphatase family domain containing 1, ultimately enhancing radioresistance in KYSE150 and TE-1 cells (18). In addition, NORAD inhibited the formation of mature RISC by binding to Ago2, which inhibited metadherin silencing by miR-224-3p. This promoted β-catenin accumulation and contributed to CDDP resistance in KYSE30 and TE-1 cells, as well as cisplatin resistance in CDX (KYSE30 and TE-1) (26). LincIN has been shown to bind to interleukin enhancer binding factor 3 (NF90), enhancing NF90's binding to prim-miR-7 and further inhibiting miR-7. This resulted in increased homeobox B13 expression, promoting the growth, migration and invasion of TE-1 and ECA109 cells, which correlated with invasion, lymph node metastasis, TNM staging and prognosis (11). LINC02042 enhanced the viability, invasion and growth of KYSE30 and KYSE150 cells. This lncRNA stabilized c-Myc mRNA by doubling the Y-box binding protein 1 of the c-Myc mRNA 3'-UTR, leading to enhanced stability of c-Myc mRNA. C-Myc also enhanced LINC02042 transcription by binding the E-box proximal end to the transcription start site of the LINC02042 promoter (9). FOXP4-AS1 has been shown to bind to IGF2BP2, promoting stability of FOXP4 mRNA and inhibiting apoptosis, while promoting proliferation in KYSE150 and KYSE450 cells (121).
LINC01004 recruited Spi-1 proto-oncogene in tumor-associated macrophages (TAMs) to induce transcription of sialic acid binding Ig like lectin 9, leading to M2 polarization of TAMs and enhanced radio-resistance and immunosuppression in TE-1 and KYSE30 cells, and inhibiting radiation-induced ferroptosis (122). LINC00858 enhanced zinc finger protein 184 expression and upregulated FTO expression, ultimately enhancing MYC expression and promoting proliferation, migration, invasion and growth in TE-1 cells (123). N-acetyltransferase 10-mediated N4-acetylcytidine modification upregulated lncRNA CTC-490G23.2 expression, promoting CD44 pre-mRNA binding to polypyrimidine tract binding protein 1, inducing transformation of CD44 molecule (IN blood group) (CD44s) into CD44v and promoting invasion of KYSE150 and KYSE410 cells, as well as CDX (KYSE150) translocation (124). LINC02820 interacted with splicing factor 3b subunit 3 and synergistically mediated nuclear factor kappa-B signaling with tumor necrosis factor α (TNF-α)-like to promote migration and invasion of KYSE180, KYSE410, KYSE30 and EC109 cells (125). LncRNA prostate cancer associated transcript 6 activated the JAK/STAT signaling pathway, thereby promoting proliferation, migration, invasion and stem cell stemness in EC109 cells, which was shown to be associated with prognosis, clinical features and immune expansion (126).
4. LncRNAs downregulated in ESCC
Studies have shown that the number of genes downregulated in tumors is significantly less than that of genes that are upregulated. This pattern is also observed in lncRNA research in ESCC. During the literature review, genes with no expression difference were not included, suggesting more studies reporting differentially upregulated genes.
LncRNAs operating via the ceRNA mechanism
Various lncRNAs have been identified to exert regulatory effects through the ceRNA mechanism in ESCA. KLF transcription factor 3-AS1, acting through the miR-185-5p/KLF axis, demonstrated inhibitory effects on the clone formation, invasion and migration abilities of ECA109 cells, and the growth of corresponding xenografts (127). Similarly, prostate androgen-regulated transcript 1 (PART1), by sponging miR-18a-5p to modulate SOX6, suppressed the β-catenin/c-Myc signaling pathway, thereby impeding proliferation and invasion of ECA109 and EC9706 cells and correlating with clinical parameters such as TNM staging and lymph node metastasis (5). Metaxin 2-6, through the miR-574-5p/SMAD family member 4 axis, hindered proliferation and promoted apoptosis in EC1 and EC109 cells, with implications for tumor volume and prognosis (128). LET, interacting with miR-93-5p or miR-106b-5p to regulate suppressor of cytokine signaling 4, exhibited anti-proliferative effects in ECA109 and KYSE410 cells, as well as in corresponding xenografts (129). WD repeat and FYVE domain containing 3-AS2, via miR-2355-5p/suppressor of cytokine signaling 2, impeded the JAK2/STAT5 pathway, thereby suppressing proliferation, invasion and EMT in EC9706 and TE-1 cells, with implications for TNM staging, lymph node metastasis and prognosis (130). In addition, ribosomal protein L34 (RPL34)-AS1, through the miR-575/acetyl-CoA acyltransferase 2 (ACAA2) axis, demonstrated inhibitory effects on proliferation, clone formation, invasion and migration of EC109 cells, with an influence on OS (131).
Furthermore, LINC00261, by modulating the miR-545-3p/metallothionein 1M axis, attenuated proliferation and cisplatin resistance, and promoted apoptosis in TE-1 and ECA109 cells (132). Transmembrane protein 161B-AS1, through miR-23a-3p/HIF1α inhibitor, inhibited proliferation, invasion and glycolysis of ECA109 and KYSE30 cells, with implications for TNM staging, lymph node metastasis and prognosis (133). LncRNA transducer of ERBB2, 1-AS1, by sponging miR-103a-2-5p to modulate TOB1, inhibited the growth, migration and invasion of KYSE170 cells (134). Furthermore, small nucleolar RNA host gene 12 (SNHG12), through the miR-195-5p/BCL9 transcription coactivator axis, exhibited inhibitory effects on proliferation, colony formation, migration and invasion of EC109 cells, along with promoting apoptosis in corresponding xenografts (135). Zinc finger protein 667 (ZNF667)-AS1, via miRNA-1290/prune homolog 2 with BCH domain, attenuated malignancy and growth of ECA109 xenografts, correlating with patient prognosis (136).
In addition, the zinc finger protein 750 inhibited lncRNA DANCR expression to disrupt the miR-4707-3p/FOXC2 axis, thereby attenuating the pro-angiogenic capacity of KYSE150 and KYSE140 cells, as well as corresponding xenografts (137). Furthermore, GATA binding protein 2-AS1, through the miR-940/protein tyrosine phosphatase non-receptor type 12 axis, inhibited the proliferation, invasion, migration and EMT of KYSE170 cells and the growth of corresponding xenografts (138). Lastly, ZNF667-AS1, via miR-18b-5p/RAS p21 protein activator 1, inhibited the proliferation and invasion of ECA109 cells (139).
LncRNAs regulated by RNA-protein interactions
LINC00551 interacted with Heat shock protein 27 (HSP27), leading to decreased phosphorylation of HSP27 and inhibition of proliferation and invasion of KYSE150 and TE-1 cells. It has also been shown to impede the growth of CDX (TE-1) and to be associated with lymph node metastasis, advanced TNM staging, tumor size, progression-free survival and OS (140). Urothelial cancer associated 1 (UCA1) was reported to bind to heterogeneous nuclear ribonucleoprotein F and facilitate selective cleavage of fibroblast growth factor receptor 2 (FGFR2), which prevents the conversion of FGFR2 IIIb to FGFR2 IIIc. Activation of FGFR2 IIIc through the PI3K-AKT pathway promoted EMT in EC109 cells while inhibiting proliferation, migration and invasion. In addition, UCA1 suppressed the malignant transformation of Het-1A cells and precancerous lesions in the rat esophagus, thereby inhibiting tumor growth (141). LINC01088 was reported to bind to NPM1, targeting the NPM1-MDM2 proto-oncogene-p53 pathway. This was shown to reduce the expression of mut-p53, restore p53 transcriptional activity, inhibit migration and invasion in KYSE150 and EC9706 cells, and suppress CDX growth (KYSE150 and EC9706). Furthermore, LINC01088 was shown to be associated with vascular tumor embolization and post-operative OS (142).
Epigenetic regulation
Small nucleolar RNA host gene 5 (SNHG5) promoted metastasis associated 1 family member 2 ubiquitination, reversing EMT and inhibiting proliferation, migration and invasion of KYSE30 and KYSE510 cells. It also suppressed CDX growth (KYSE30) and lung metastasis, and was correlated with tumor progression, clinical outcome and survival (143). By inhibiting the interaction between LINC00886 and SIRT7, SNHG5, upregulated histone H3K18 acetylation in the promoter region of E74 like ETS transcription factor 3 (ELF3). This inhibited the binding of ELF3 to the promoter region of miR-144, leading to increased expression of miR-144-3p and suppression of ZEB1 and ZEB2 expression. SNHG5 also inhibited proliferation, migration, invasion and EMT in ECA109 and KYSE150 cells (144).
Diverse functions
Cancer Susceptibility 2 upregulated suppressor of cytokine signaling 1 (SOCS1) expression through the miR-155/SOCS1 ceRNA mechanism and bound to SOCS1 to prevent degradation. This inhibited the proliferation, migration, invasion and cisplatin tolerance of KYSE30 and KYSE150 cells (145). MiR-320 is contained within the novel lncRNA inhibiting proliferation and metastasis (NLIPMT) sequence, and both NLIPMT and miR-320 have been shown to suppress survivin expression, leading to reduced proliferation and migration in KYSE510 cells and promoting cell cycle arrest and apoptosis (20). The NF90 protein was shown to bind to and stabilize pri-miR-548k, increasing the expression of both pri-miR-548k and miR-548k. On the other hand, lncRNA-LET downregulated NF90 and miR-548k expression, creating a regulatory loop. NF90 enhanced HIF1α and VEGF expression, while miR-548k inhibited KLF transcription factor 10 and promoted EGFR expression. LncRNA-LET boosted p53 expression and hindered the proliferation and migration of KYSE30 cells (146).
5. Exosomal lncRNAs
Several lncRNAs have been identified in exosomes, including ZFAS1 (13), LINC01711 (24) and family with sequence similarity 225, member A (FAM225A) (21), which was shown to exert control over ESCC and other cells and tissues from a remote location. For instance, PART1 was found to promote gefitinib resistance in ESCC through exosomes (33). In addition, FAM225A/miR-206/neuropilin and tolloid like 2 and FOXP1 have been indicated to enhance the viability, migration, invasion and angiogenesis, and inhibit apoptosis in ECA109 and TE-1 cells. FOXP1 was also shown to promote the expression of FAM225A (21). Furthermore, the lncRNA Ras association domain family member 8-AS1 found in serum exosomes was found to promote the proliferation, migration and invasion of TE-1 and TE-13 cells, and to be associated with characteristics such as depth of invasion, lymph node metastasis, advanced tumor stage and poor prognosis (147). LINC02096, identified in plasma exosomes, was shown to bind to and stabilize MLL1 to enhance H3K4me3 levels in the promoter regions of programmed cell death 1 ligand 1 and indoleamine 2,3-dioxygenase 1, thereby promoting the expression of these genes in KYSE70 and KYSE520 cells. This lncRNA also inhibited the infiltration and activation of CD8+ T cells in a huPBMC-NOG-CDX/PDX model (148).
6. Effects of lncRNAs on ESCC progression
The expression of SNHG12 was elevated in CD133+ ESCC, while the SNHG12/miR-6835-3p/BMI1 proto-oncogene, polycomb ring finger axis promoted proliferation, migration and EMT in EC109 (CD133+) and KYSE410 (CD133+) cells. In addition, SNHG12 recruitment of IGF2BP2 enhanced the stability of β-catenin mRNA (149).
SOX2OT, containing the intron SOX2, was upregulated in ESCC and was shown to enhance the growth, proliferation and cisplatin resistance of KYSE150 and KYSE450 cells (150).
LINC00657 expression was increased in ESCC cells after radiotherapy, promoting the growth of cisplatin-resistant cells by mediating the miR-615-3p/JunB proto-oncogene, AP-1 transcription factor subunit ceRNA mechanism. This has been demonstrated to enhance migration, proliferation and radiotherapy resistance of KYSE cells (151). Peptides encoded by lncRNA lysine demethylase 4A-AS1 could increase reactive oxygen species levels, reduce the mitochondrial membrane potential and inhibit the viability and migration of KYSE150 and TE-1 cells (152). TAM secreted TNFα, which has been reported to induce the expression of lncRNA macrophage-associated long noncoding RNA in TE-9 and TE-15 cells. Furthermore, MALR was demonstrated to bind to the dsRBD1 domain of ILF3, which enhanced ILF3 stabilization and liquid-liquid phase separation. This led to the activation of the HIF1α signaling pathway, promoting aerobic glycolysis activity, angiogenesis and the growth of CDX (TE-9), and had an influence on patient prognosis (153). E2F transcription factor 1 activated the expression of LINC01224, which in turn mediated the Wnt/β-catenin signaling pathway through miR-6884-5p/dishevelled segment polarity protein 3. This promoted the proliferation, migration and invasion of TE-10 and KYSE30 cells (154). Upon stimulation with Wnt3a, β-catenin was shown to act as a cofactor of FUS to promote the generation of lncRNA DGCR5-S, which in turn promoted inflammation in PDX and CDX (KYSE140 and KYSE180) by inhibiting ZFP36 ring finger protein dephosphorylation (155). SOX2 directly activated the transcription of lncRNA KTN1-AS1, which could interact with RB binding protein 4, chromatin remodeling factor (RBBP4) to enhance the binding of RBBP4 and histone deacetylase 1. This promoted the proliferation, invasion and EMT of KYSE150 cells and enhanced the growth of CDX (KYSE150) cells (156).
7. Future perspectives
In recent years, studies on the function of lncRNAs in ESCC have predominantly focused on individual lncRNAs that are significantly upregulated in ESCC and their ceRNA mechanisms. References to online databases such as miRcode (http://www.mircode.org/) and starbase (starbase.sysu.edu.cn) have greatly aided in the functional study of lncRNAs. However, bioinformatics predictions based on available data can only offer preliminary references. Validation of the ceRNA mechanism primarily relies on the dual-luciferase reporter system, with auxiliary validation through real-time quantitative PCR (qPCR) and western blot (WB) analysis potentially required. Validation is typically performed on exogenous cells through in situ mutagenesis or knocking down theoretical binding sites, making the validation method relatively limited in scope.
Furthermore, functional studies focusing on RNA-protein binding regulation and epigenetic regulation have also been explored. Initial RNA pull-down assays and mass spectrometry (MS) analysis often yield numerous potentially interacting proteins, necessitating extensive time and effort for validation. The complexity of the validation process, despite available bioinformatics analysis and prediction databases, calls for modifications as research progresses. As mechanisms are being further explored, a substantial workload is required to support continued research.
One suggestion for future research is to mutate or knock out theoretical binding sites in situ and observe resulting phenotypic differences. In addition, investigating RNA-protein binding sites to analyze binding patterns and spatial conformation and construct molecular interaction models for functional studies could provide valuable insight.
While numerous studies focus on post-expression functional studies of lncRNAs in gene expression regulation, fewer studies have explored the regulation of lncRNA expression itself. The rationale behind cells expressing lncRNAs for gene regulation instead of miRNAs or direct regulation using cis- or trans-acting factors remains unclear. Another potential research avenue is exploring the use of lncRNAs as reinforcing elements to further up- or downregulate gene expression.
8. Conclusions
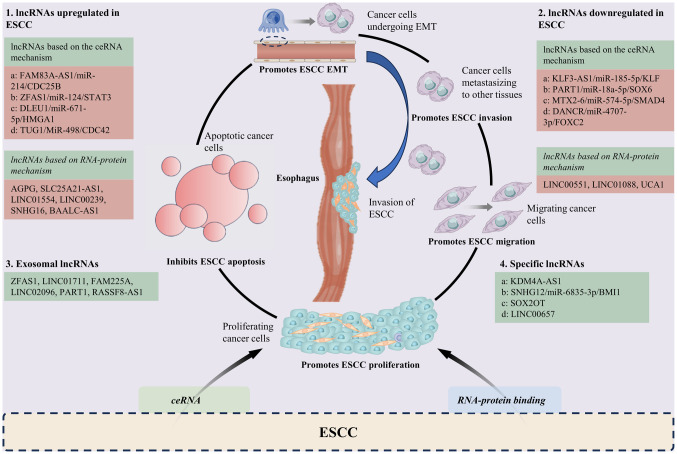
This review provides a brief overview of the lncRNAs and their mechanisms of action in ESCC in recent years (Tables I and II). Although research on these lncRNAs in ESCC is still in the early stages, there is potential for further exploration. The significance of lncRNA expression and their clinical roles in vivo are still evolving, with a focus on their regulatory functions in tumorigenesis, metastasis, progression and drug resistance in ESCC (Fig. 1). Through continued research, the current understanding of the potential roles of lncRNAs in the diagnosis and treatment of ESCC may be enhanced.
Table I.
List of lnRNAs upregulated in esophageal cell squamous carcinoma.
| LncRNA | Target | Oncological significance | Clinicopathological features influenced by lncRNA | (Refs.) |
|---|---|---|---|---|
| FAM83A-AS1 | miR-214/CDC25B | Proliferation, migration invasion | TNM staging, tumor differentiation grading | (1) |
| ZFAS1 | miR-124/STAT3 | Proliferation, migration, invasion | ND | (2) |
| TUG1 | miR-498/CDC42 | Proliferation, invasion | ND | (3) |
| miR205HG | miR-214/SOX4 | Cell cycle, proliferation, invasion, migration | Tumor size, lymphatic metastasis | (4) |
| OIP5-AS1 | miR-1297 | Proliferation, migration, invasion | ND | (5) |
| DLEU1 | miR-671-5p/HMGA1 | Proliferation, migration, invasion | ND | (6) |
| LOC146880 | miR-328-5p/FSCN1 | Proliferation, invasion, migration, EMT, liver metastasis | ND | (7) |
| LINC00680 | miR-423-5p/PAK6 | Proliferation, colony formation, migration, invasion | Tumor volume, tumor stage, prognosis | (8) |
| LINC01711 | miR-326/FSCN1 | Proliferation migration, invasion, growth | ND | (9) |
| HEIH | miR-4458/PBX3 | Viability, invasion | ND | (10) |
| GACAT3 | miR-149/FOXM1 | Proliferation, migration, invasion | ND | (11) |
| HNF1A-AS1 | miRNA-298/TCF4 | EMT, stem cell stemness | ND | (12) |
| AFAP1-AS1 | miR-498/VEGFA | Growth, migration | ND | (13) |
| LINC00467 | miR-485-5p/DPAGT1 | Proliferation | ND | (14) |
| EGFR-AS1 | miR-145/ROCK1 | Migration, invasion | Overall survival time | (15) |
| SNHG17 | miR-338-3p/SOX4 | Proliferation, invasion, EMT | ND | (16) |
| MNX1-AS1 | miR-34a/SIRTI | Grow, migration, invasion, cell cycle | ND | (17) |
| THAP9-AS1 | miR-335-5p/SGMS2 | Grow, migration, invasion, facilitation of EMT | Survival | (18) |
| MAFG-AS1 | miR-765/PDX1 | Proliferation, migration, invasion, aerobic glycolysis | ND | (19) |
| SNHG6 | miR-101-3p/EZH2 | Apoptosis | ND | (20) |
| FAM83H-AS1 | miR-10a-5p/Girdin | Proliferation, migration, invasion, EMT | TNM staging, pathologic staging, lymph node metastasis | (21) |
| CASC15 | miR-33a-5p/PTGS2 | Proliferation, migration, invasion | ND | (22) |
| JPX | miR-516b-5p/VEGFA | Growth, angiogenesis, migration, invasion | ND | (23) |
| SNHG6 | miR-186-5p/HIF1α | Proliferation, migration, invasion | ND | (24) |
| LINC00858 | miR-425-5p/ABL | Proliferation, invasion, migration, EMT | ND | (25) |
| DANCR | miR-33a-5p/ZEB1 | Proliferation, metastasis | Prognosis | (26) |
| BCAR4 | miR-139-3p/ELAVL1 | Growth, migration, evasion of apoptosis | ND | (27) |
| C9orF139 | miR-661/HDAC11 | Proliferation, migration, invasion | ND | (28) |
| miR31HG | miR-34a/c-Met | Growth, evasion of apoptosis | ND | (29) |
| LINC01980 | miR-190a-5p/MYO5A | Proliferation, migration, invasion, EMT | ND | (30) |
| THAP9-AS1 | miR-133b/SOX4 | Proliferation, migration, invasion | Tumor size, TNM staging, lymph node metastasis, cancer prognosis | (31) |
| MCEI | miR-6759-5p/IGF2 | Proliferation, migration, cisplatin resistance, invasion | ND | (32) |
| NNT-AS1 | miR-382-5p | Proliferation, migration | ND | (33) |
| LUESCC | miR-6785-5p/NRSN2 | Proliferation, colony formation, migration, invasion | Gender, deep invasion, lymph node metastasis, poor prognosis | (34) |
| LncSUMO1P3 | miR-486-5p/PHD or PHF8 | Proliferation, migration, invasion, EMT | ND | (35) |
| PCAT5 | miR-4295/PHF20 | Proliferation, migration, invasion | ND | (36) |
| XIST | miR-34a/ZEB1 | Proliferation, migration, invasion, EMT | Metastasis, reduced patients survival | (37) |
| ZFPM2-AS1 | miR-3612/TRAF4 | Proliferation, migration, invasion, inhibits apoptosis | ND | (38) |
| LINC00662 | miR-340-5p/HOXB2 | Proliferation, migration, invasion | Cancer prognosis | (39) |
| CCAT2 | miR-200b/IGF2BP2 | Migration | ND | (40) |
| BF2-AS1 | miR-494/PFN2 | Proliferation, migration, invasion, EMT | ND | (41) |
| LINC00941 | miR-877-3p/PMEPA1 | Proliferation, invasion, migration, EMT | ND | (42) |
| LINC00152 | miR-153-3p/FYN | Proliferation, colony formation | TNM staging, lymph node metastasis, prognosis | (43) |
| Psma3-as1 | miR-101/EZH2 | Proliferation, invasion, migration | Tumor size, distant metastasis, prognosis | (44) |
| LINC00963 | miR-214-5p/RAB14 | Proliferation, invasion | TNM staging, metastasis, prognosis | (45) |
| BBOX1-AS1 | miR-506-5p/EIF5A | Proliferation, stem cell stemness | ND | (46) |
| FOXD2-AS1 | miR-204-3p | Proliferation, migration, invasion | ND | (47) |
| KTN1-AS1 | miR-885-5p/STRN3 | Proliferation, invasion, EMT | ND | (48) |
| XIST | miR-129-5p/CCND1 | Cell cycle progression, proliferation, migration, invasion | ND | (49) |
| LINC00473 | miR-497-5p/PRKAA1 | Proliferation, migration, EMT | ND | (50) |
| NEAT1 | miR-590-3p/MDM2 | Proliferation, migration, invasion, angiogenesis | ND | (51) |
| abca12-3 | miR-200b-3p/FN1 | Migration, invasion, proliferation | Lymph node metastasis, prognosis | (52) |
| ESCCAL-1 | miR-590/LRP6 | Growth, invasion, migration | ND | (53) |
| ROR | miR-145/FSCN1 | Migration, invasion | Patient survival | (54) |
| LOC440173 | miR-30d-5p/HDAC9 | Proliferation, migration, invasion, EMT | Histological grading, tumor invasion, lymph node metastasis, TNM stage | (55) |
| LINC00514 | miR-378a-5p/SPHK1 | Proliferation, invasion | Lymph node metastasis, prognosis | (56) |
| LINC00473 | miR-374a-5p/SPIN1 | Radioresistance | TNM staging | (57) |
| LINC00473 | miR-497-5p/CDC25A | Survival, radioresistance | T-stage, lymph node metastasis staging, tumor differentiation grading | (58) |
| NCK1-AS1 | miR-133b/ENPEP | Proliferation, invasion, migration | ND | (59) |
| LIPH-4 | miR-216b/IGF2BP2 | Growth, cell cycle | Tumor size, prognosis | (60) |
| TTTY15 | miR-337-3p/JAK2 | Proliferation, migration, invasion | ND | (61) |
| LINC01232 | miR-654-3p/HDGF | Proliferation, migration, invasion | ND | (62) |
| LINC00152 | miR-107/Rab10 | Migration, invasion | ND | (63) |
| ROR | miR-204-5p/MDM2 | Activity, proliferation | ND | (64) |
| BCAR4 | miR-181c-5p/LASP1 | Migration, invasion | ND | (65) |
| BBOX1-AS1 | miR-513a-3p/SLC7A11 | Proliferation, invasion, migration | ND | (66) |
| PTPRG-AS1 | miR-599/PDK1 | Proliferation, migration, glycolysis, stem cell stemness | ND | (67) |
| AGPG | PFKFB3 | Cell cycle progression | ND | (68) |
| SLC25A21-AS1 | NPM1 | Tumor growth, migration, resistance to cisplatin | Tumor grade, overall patient survival | (69) |
| LINC01554 | G3BP2 | Migration, invasion | ND | (70) |
| LINC00239 | MBP-1 | Proliferation, metastasis, invasion, EMT | ND | (71) |
| SNHG16 | EIF4A3 | Tumor growth, migration | Tumor differentiation, T staging | (72) |
| BAALC-AS1 | G3BP2 | Proliferation, migration, invasion | ND | (73) |
| DGCR5 | SRSF1 | Proliferation, migration, invasion | ND | (74) |
| DLEU1 | DYNLL1 | Inhibition of apoptosis, promotion of DDP resistance | ND | (75) |
| GK-IT1 | USP6 | Proliferation, invasion, migration, autophagy | Clinical staging, prognosis | (76) |
| SNHG17 | c-Jun | Proliferation, migration, invasion, EMT | Survival | (77) |
| ESCCAL-1 | Gal-1 | Tumor growth, migration, cell cycle | ND | (78) |
| LBX2-AS1 | ZEB1 | Migration, EMT | ND | (79) |
| LINC00337 | E2F4 | Autophagy, cisplatin resistance | ND | (80) |
| LINC01305 | IGF2BP2 and IGF2BP3 | Migration, proliferation | ND | (81) |
| LINC01296 | EZH2 | Proliferation, clone formation, migration, invasion | Lymph node metastasis, TNM staging, overall survival | (82) |
| NRSN2-AS1 | PGK1 | Proliferation, migration, invasion | Survival | (83) |
| TMEM44-AS1 | IGF2BP2 | Ferroptosis, proliferation, invasion, metastasis | ND | (84) |
| PDIA3P1 | OCT4 | Proliferation, invasion, metastasis, stem cell stemness | ND | (85) |
| G077640 | H2AX | Glycolysis metabolism, promotion of proliferation, migration | ND | (86) |
| HOXC-AS1 | IGF2BP2 | Proliferation, migration | ND | (87) |
| LINC00941 | ILF2 and YBX1 | Proliferation, stem cell differentiation, migration, invasion | ND | (88) |
| DUXAP10 | EZH2 | Proliferation, metastasis, cell cycle progression, inhibition of apoptosis | ND | (89) |
| FOXP4-as1 | FOXP4 | Proliferation, invasion, migration | Lymph node metastasis, TNM staging | (90) |
| TMPO-as1 | FUS | Proliferation, metastasis | Survival | (91) |
| LINC01419 | GSTP1 | Proliferation, 5-FU resistance | ND | (92) |
| LEF1-AS1 | PDCD5 | Proliferation | Overall survival | (93) |
| SLC25A21-as1 | SLC25A21 | Tryptophan catabolism | ND | (69) |
| PCNA-as1 | miR-2467-3p/PCNA | Proliferation, invasion, migration | ND | (94) |
| LINC00022 | p21 | Cell cycle progression, proliferation | ND | (95) |
| CASC8 | hnRNPL | Cisplatin resistance | Prognosis | (96) |
| CASC15 | SIM2 | Proliferation | Prognosis | (97) |
| HCP5 | HK2 mRNA | Proliferation, invasion, Warburg | Tumor volume, TNM stage, lymph node metastasis, survival time | (98) |
| HCP5 | miR-216a-3p/PDK1 | Radiotherapy resistance | ND | (99) |
| LINC00963 | miR-10a/SKA1 | Cisplatin resistance | ND | (100) |
| NRON | miR-31 | Cisplatin resistance | ND | (101) |
| Linc01614 | miR-4775 | Cisplatin resistance | ND | (102) |
| HCP5 | VAMP3 | Cisplatin resistance | ND | (103) |
| LINK-A | CDK1 | Glycolysis, cisplatin resistance | ND | (104) |
| SOX2OT | Not known | Stem cell-related genes, sphere formation ability, docetaxel resistance | ND | (105) |
| NORAD | pri-miR-199a | Radioresistance | ND | (106) |
| NORAD | Ago2 | Cisplatin resistance | ND | (107) |
| LincIN | NF90 | Growth, migration, invasion | Invasion, lymph node metastasis, TNM staging, prognosis | (108) |
| LINC02042 | c-Myc mRNA | Viability, invasion, CDX growth, lung metastasis | ND | (109) |
| FOXP4-as1 | FOXP4 | Proliferation | ND | (110) |
| LINC01004 | SPI | Ferroptosis, radioresistance, immunosuppression | ND | (111) |
| LINC00858 | ZNF184 | Proliferation, migration, invasion | ND | (112) |
| CTC-490G23.2 | CD44 pre-mRNA and PTBP1 | Invasion | ND | (113) |
| LINC02820 | SF3B3 | Migration, invasion | ND | (114) |
| PCAT6 | Not known | Proliferation, migration, invasion, stem cell stemness | Prognosis, clinical features, immune expansion | (115) |
Please refer to Data S1 for the full definitions of gene/protein names. EMT, epithelial to mesenchymal transition; lncRNA, long non-coding RNA; miR, microRNA; ND, not determined.
Table II.
List of lncRNAs downregulated in esophageal cell squamous carcinoma.
| LncRNA | Target | Oncological significance | Clinicopathological features influenced by lncRNA | (Refs.) |
|---|---|---|---|---|
| KLF3-AS1 | miR-185-5p/KLF | Clone formation ability, invasion, migration | ND | (116) |
| PART1 | miR-18a-5p/SOX6 | Proliferation, invasion | TNM staging, lymph node metastasis, prognosis | (117) |
| MTX2-6 | miR-574-5p/SMAD4 | Proliferation | ND | (118) |
| LET | miR-93-5p or miR-106b-5p/SOCS4 | Viability | ND | (119) |
| WDFY3-AS2 | miR-2355-5p/SOCS2 | Proliferation, invasion, EMT | TNM staging, lymph node metastasis, prognosis | (120) |
| RPL34-AS1 | miR-575/ACAA2 | Proliferation, clone formation, invasion, migration | Overall survival | (121) |
| LINC00261 | miR-545-3p/MT1M | Proliferation, cisplatin resistance | ND | (122) |
| TMEM161B-AS1 | miR-23a-3p/HIF1AN | Proliferation, invasion, glycolysis, reduces glucose consumption, lactate production | TNM staging, lymph node metastasis, prognosis | (123) |
| TOB1-AS1 | miR-103a-2-5p/TOB1 | Growth, migration, invasion | ND | (124) |
| SNHG12 | miR-195-5p/BCL9 | Proliferation, colony formation, migration, invasion | ND | (125) |
| ZNF667-AS1 | miR-1290/PRUNE2 | Growth | Prognosis | (126) |
| DANCR | miR-4707-3p/FOXC2 | Pro-angiogenic capacity | ND | (127) |
| GATA2-AS1 | miR-940/PTPN12 | Proliferation, invasion, migration, EMT | ND | (128) |
| ZNF667-AS1 | miR-18b-5p/RASA1 | Proliferation, invasion | ND | (129) |
| LINC00551 | HSP27 | Proliferation, invasion | Lymph node metastasis, advanced TNM stage, tumor size, progression survival, overall survival | (130) |
| UCA1 | hnRNP F | EMT, proliferation, migration, invasion | ND | (131) |
| LINC01088 | NPM1 | Growth, migration, invasion | Vascular tumor embolization, postoperative overall survival time | (132) |
| SNHG5 | MTA2 | EMT, proliferation, migration, invasion, lung metastasis | Tumor progression, clinical outcome, survival | (133) |
| LINC00886 | SIRT7 | Proliferation, migration, invasion, EMT | ND | (134) |
| CASC2 | miR-155/SOCS1 | Proliferation, migration, invasion, cisplatin resistance | ND | (135) |
| NLIPMT | Survivin | Proliferation, migration, promotion of cell cycle arrest, apoptosis | ND | (136) |
| lncRNA-LET | NF90 and miR-548k | Proliferation, migration | ND | (137) |
| FAM225A | miR-206/NETO2 and FOXP1 | Viability, migration, invasion, angiogenesis, inhibition of apoptosis | ND | (138) |
| RASSF8-AS1 | Not known | Proliferation, migration, invasion | Invasion, lymph node metastasis, advanced tumor stage, poor prognosis | (139) |
| LINC02096 | MLL1 | Infiltration, activation of CD8+ T cells | ND | (140) |
| SNHG12 | miR-6835-3p/BMI1 | Proliferation, migration, EMT | ND | (141) |
| SOX2OT | Not known | Growth, proliferation, cisplatin resistance | ND | (142) |
| LINC00657 | miR-615-3p/JunB | Migration, proliferation, radiotherapy resistance | ND | (143) |
| KDM4A-AS1 | Peptides | Viability, migration | ND | (144) |
| MALR | ILF3 | Aerobic glycolysis activity, angiogenesis, growth | Prognosis | (145) |
| LINC01224 | miR-6884-5p/DVL3 | Proliferation, migration, invasion | ND | (146) |
| DGCR5-S | TTP | Inflammation | ND | (147) |
| KTN1-AS1 | RBBP4 | Proliferation, invasion, EMT | ND | (148) |
Please refer to Data S1 for the full definitions of gene/protein names. EMT, epithelial to mesenchymal transition; lncRNA, long non-coding RNA; miR, microRNA; NF90, interleukin enhancer binding factor 3; ND, not determined.
Figure 1.
Dysregulation of lncRNAs involved in the development of ESCC. Refer to Data S1 for the full definitions of gene names. ESCC, esophageal squamous cell carcinoma; lncRNA, long non-coding RNA; EMT, epithelial to mesenchymal transition; ceRNA, competing endogenous RNA; miR, microRNA.
The role of lncRNAs in ESCC can be better understood by comparing them to studies of lncRNAs in other diseases. Utilizing databases such as The Cancer Genome Atlas and Gene Expression Omnibus, differentially expressed lncRNAs in ESCC can be identified for further transcriptome analysis through library construction. Mechanistic studies of lncRNAs typically involve bioinformatics analysis and experimental validation to verify predictive models. This includes analyzing expression levels and bioinformatics in clinical ESCC tissues, followed by molecular function verification using techniques such as MS, qPCR and WB. Functional validation of lncRNAs can be performed by manipulating their expression in ESCC cell lines and confirming these findings in animal models, such as the CDX model.
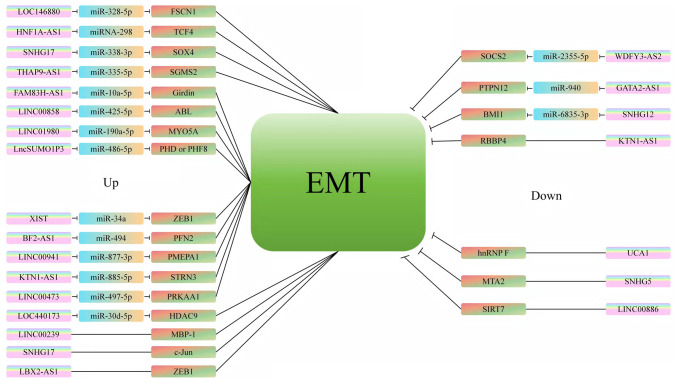
While studies on individual lncRNAs have focused on their roles in tumor proliferation, migration, EMT (Fig. 2) and drug resistance, the complexity of lncRNAs requires further exploration. Due to their long sequences, lncRNAs contain significantly more information than certain shorter biological factors, warranting more in-depth studies. In addition, the same lncRNAs can have different functional roles in various tissues or tumors, a fact often overlooked by researchers. Future analyses are needed to explore the functional and compartmental differences of lncRNAs across different diseases and tumors.
Figure 2.
Summary of lncRNAs associated with EMT. Refer to Data S1 for the full definitions of gene names. EMT, epithelial to mesenchymal transition; lncRNA, long non-coding RNA; miR, microRNA.
Supplementary Data
Acknowledgements
Not applicable.
Funding Statement
No funding was received.
Availability of data and materials
Not applicable.
Authors' contributions
QY, WW and LG prepared the manuscript and reviewed the literature. JY, DL, KYC reviewed and provided critical feedback on the manuscript. SD and JW revised and approved the manuscript for publication. All authors read and approved the final manuscript. Data authentication is not applicable.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Bridges MC, Daulagala AC, Kourtidis A. LNCcation: lncRNA localization and function. J Cell Biol. 2021;220:e202009045. doi: 10.1083/jcb.202009045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang JZ, Chen M, Chen D, Gao XC, Zhu S, Huang H, Hu M, Zhu H, Yan GR. A Peptide encoded by a putative lncRNA HOXB-AS3 suppresses colon cancer growth. Mol Cell. 2017;68:171–184.e6. doi: 10.1016/j.molcel.2017.09.015. [DOI] [PubMed] [Google Scholar]
- 3.Jia J, Li H, Chu J, Sheng J, Wang C, Jia Z, Meng W, Yin H, Wan J, He F. LncRNA FAM83A-AS1 promotes ESCC progression by regulating miR-214/CDC25B axis. J Cancer. 2021;12:1200–1211. doi: 10.7150/jca.54007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li H, Jia J, Yang L, Chu J, Sheng J, Wang C, Meng W, Jia Z, Yin H, Wan J, He F. LncRNA MIR205HG drives esophageal squamous cell carcinoma progression by regulating miR-214/SOX4 Axis. Onco Targets Ther. 2020;13:13097–13109. doi: 10.2147/OTT.S286627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhao Y, Zhang Q, Liu H, Wang N, Zhang X, Yang S. lncRNA PART1, manipulated by transcriptional factor FOXP2, suppresses proliferation and invasion in ESCC by regulating the miR-18a-5p/SOX6 signaling axis. Oncol Rep. 2021;45:1118–1132. doi: 10.3892/or.2021.7931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feng Z, Li X, Qiu M, Luo R, Lin J, Liu B. LncRNA EGFR-AS1 Upregulates ROCK1 by Sponging miR-145 to promote esophageal squamous cell carcinoma cell invasion and migration. Cancer Biother Radiopharm. 2020;35:66–71. doi: 10.1089/cbr.2019.2926. [DOI] [PubMed] [Google Scholar]
- 7.Liu Y, Li C, Fang L, Wang L, Liu H, Tian H, Zheng Y, Fan T, He J. Lipid metabolism-related lncRNA SLC25A21-AS1 promotes the progression of oesophageal squamous cell carcinoma by regulating the NPM1/c-Myc axis and SLC25A21 expression. Clin Transl Med. 2022;12:e944. doi: 10.1002/ctm2.944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Z, Ren B, Huang J, Yin R, Jiang F, Zhang Q. LncRNA DUXAP10 modulates cell proliferation in esophageal squamous cell carcinoma through epigenetically silencing p21. Cancer Biol Ther. 2018;19:998–1005. doi: 10.1080/15384047.2018.1470723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Du J, Zhang G, Qiu H, Yu H, Yuan W. A novel positive feedback loop of linc02042 and c-Myc mediated by YBX1 promotes tumorigenesis and metastasis in esophageal squamous cell carcinoma. Cancer Cell Int. 2020;20:75. doi: 10.1186/s12935-020-1154-x. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 10.Qiu BQ, Lin XH, Ye XD, Huang W, Pei X, Xiong D, Long X, Zhu SQ, Lu F, Lin K, et al. Long non-coding RNA PSMA3-AS1 promotes malignant phenotypes of esophageal cancer by modulating the miR-101/EZH2 axis as a ceRNA. Aging (Albany NY) 2020;12:1843–1856. doi: 10.18632/aging.102716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tan Z, Zhou P, Zhu Z, Wang Y, Guo Z, Shen M, Xiao Y, Shen W, Wu D. Upregulated long non-coding RNA LincIN promotes tumor progression via the regulation of nuclear factor 90/microRNA-7/HOXB13 in esophageal squamous cell carcinoma. Int J Mol Med. 2021;47:78. doi: 10.3892/ijmm.2021.4911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu J, Gao W. Long noncoding RNA PVT1 promotes tumour progression via the miR-128/ZEB1 axis and predicts poor prognosis in esophageal cancer. Clin Res Hepatol Gastroenterol. 2021;45:101701. doi: 10.1016/j.clinre.2021.101701. [DOI] [PubMed] [Google Scholar]
- 13.Li Z, Qin X, Bian W, Li Y, Shan B, Yao Z, Li S. Exosomal lncRNA ZFAS1 regulates esophageal squamous cell carcinoma cell proliferation, invasion, migration and apoptosis via microRNA-124/STAT3 axis. J Exp Clin Cancer Res. 2019;38:477. doi: 10.1186/s13046-019-1473-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang L, Ren X, Ma X, Yin L, Niu X, Xing S. LncRNA OIP5-AS1 promotes the development of esophageal squamous cell carcinoma by binding to miR-1297. Panminerva Med. 2022;64:589–590. doi: 10.23736/S0031-0808.19.03801-1. [DOI] [PubMed] [Google Scholar]
- 15.Lin P, Li Q, Lv X, Qu J, Wang D, Li A, Jiang G. HMGA1 promotes the development of esophageal squamous cell carcinoma by mediating miR-671-5p/lncRNA DLEU1. Panminerva Med. 2023;65:264–266. doi: 10.23736/S0031-0808.19.03843-6. [DOI] [PubMed] [Google Scholar]
- 16.Tang J, Xu H, Liu Q, Zheng J, Pan C, Li Z, Wen W, Wang J, Zhu Q, Wang Z, Chen L. LncRNA LOC146880 promotes esophageal squamous cell carcinoma progression via miR-328-5p/FSCN1/MAPK axis. Aging (Albany NY) 2021;13:14198–14218. doi: 10.18632/aging.203037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cui Y, Zhang C, Ma S, Li Z, Wang W, Li Y, Ma Y, Fang J, Wang Y, Cao W, Guan F. RNA m6A demethylase FTO-mediated epigenetic up-regulation of LINC00022 promotes tumorigenesis in esophageal squamous cell carcinoma. J Exp Clin Cancer Res. 2021;40:294. doi: 10.1186/s13046-021-02096-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun Y, Wang J, Ma Y, Li J, Sun X, Zhao X, Shi X, Hu Y, Qu F, Zhang X. Radiation induces NORAD expression to promote ESCC radiotherapy resistance via EEPD1/ATR/Chk1 signalling and by inhibiting pri-miR-199a1 processing and the exosomal transfer of miR-199a-5p. J Exp Clin Cancer Res. 2021;40:306. doi: 10.1186/s13046-021-02084-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu J, Liu ZX, Wu QN, Lu YX, Wong CW, Miao L, Wang Y, Wang Z, Jin Y, He MM, et al. Long noncoding RNA AGPG regulates PFKFB3-mediated tumor glycolytic reprogramming. Nat Commun. 2020;11:1507. doi: 10.1038/s41467-020-15112-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li D, Li D, Meng L, Liu J, Huang C, Sun H. LncRNA NLIPMT inhibits tumorigenesis in esophageal squamous-cell carcinomas by regulating miR-320/Survivin Axis. Cancer Manag Res. 2020;12:12603–12612. doi: 10.2147/CMAR.S253452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang C, Luo Y, Cao J, Wang X, Miao Z, Shao G. Exosomal lncRNA FAM225A accelerates esophageal squamous cell carcinoma progression and angiogenesis via sponging miR-206 to upregulate NETO2 and FOXP1 expression. Cancer Med. 2020;9:8600–8611. doi: 10.1002/cam4.3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Z, Huang YF, Yu L, Jiao Y. sh-HNF1A-AS1 reduces the epithelial-mesenchymal transition and stemness of esophageal cancer cells. Neoplasma. 2022;69:560–570. doi: 10.4149/neo_2022_211102N1555. [DOI] [PubMed] [Google Scholar]
- 23.Yang X, Zeng T, Liu Z, He W, Hu M, Tang T, Chen L, Xing L. Long noncoding RNA GK-IT1 promotes esophageal squamous cell carcinoma by regulating MAPK1 phosphorylation. Cancer Med. 2022;11:4555–4574. doi: 10.1002/cam4.4795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu ML, Liu TC, Dong FX, Meng LX, Ling AX, Liu S. Exosomal lncRNA LINC01711 facilitates metastasis of esophageal squamous cell carcinoma via the miR-326/FSCN1 axis. Aging (Albany NY) 2021;13:19776–19788. doi: 10.18632/aging.203389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Y, Zhang W, Liu W, Huang L, Wang Y, Li D, Wang G, Zhao Z, Chi X, Xue Y, et al. Long Noncoding RNA VESTAR regulates lymphangiogenesis and lymph node metastasis of esophageal squamous cell carcinoma by enhancing VEGFC mRNA Stability. Cancer Res. 2021;81:3187–3199. doi: 10.1158/0008-5472.CAN-20-1713. [DOI] [PubMed] [Google Scholar]
- 26.Jia Y, Tian C, Wang H, Yu F, Lv W, Duan Y, Cheng Z, Wang X, Wang Y, Liu T, et al. Long non-coding RNA NORAD/miR-224-3p/MTDH axis contributes to CDDP resistance of esophageal squamous cell carcinoma by promoting nuclear accumulation of β-catenin. Mol Cancer. 2021;20:162. doi: 10.1186/s12943-021-01455-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang P, Yang Z, Ye T, Shao F, Li J, Sun N, He J. lncTUG1/miR-144-3p affect the radiosensitivity of esophageal squamous cell carcinoma by competitively regulating c-MET. J Exp Clin Cancer Res. 2020;39:7. doi: 10.1186/s13046-019-1519-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang H, Bian C, Tu S, Yin F, Guo P, Zhang J, Song X, Liu Q, Chen C, Han Y. Integrated analysis of lncRNA-miRNA-mRNA ceRNA network in human aortic dissection. BMC Genomics. 2021;22:724. doi: 10.1186/s12864-021-08012-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu S, Li B, Liang Q, Liu A, Qu L, Yang J. Classification and function of RNA-protein interactions. Wiley Interdiscip Rev RNA. 2020;11:e1601. doi: 10.1002/wrna.1601. [DOI] [PubMed] [Google Scholar]
- 30.Wang Z, Liu J, Wang R, Wang Q, Liang R, Tang J. Long Non-Coding RNA Taurine Upregulated Gene 1 (TUG1) downregulation constrains cell proliferation and invasion through regulating cell division cycle 42 (CDC42) Expression Via MiR-498 in esophageal squamous cell carcinoma cells. Med Sci Monit. 2020;26:e919714. doi: 10.12659/MSM.919714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xue ST, Zheng B, Cao SQ, Ding JC, Hu GS, Liu W, Chen C. Long non-coding RNA LINC00680 functions as a ceRNA to promote esophageal squamous cell carcinoma progression through the miR-423-5p/PAK6 axis. Mol Cancer. 2022;21:69. doi: 10.1186/s12943-022-01539-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang D, You D, Pan Y, Liu P. Downregulation of lncRNA-HEIH curbs esophageal squamous cell carcinoma progression by modulating miR-4458/PBX3. Thorac Cancer. 2020;11:1963–1971. doi: 10.1111/1759-7714.13489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Su M, Tang J, Zhang B, Yang D, Wu Z, Wu J, Zhou Y, Liao Q, Wang H, Wang W, Xiao Y. LncRNA GACAT3 promotes esophageal squamous cell carcinoma progression through regulation of miR-149/FOXM1. Cancer Cell Int. 2021;21:478. doi: 10.1186/s12935-021-02192-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shen W, Yu L, Cong A, Yang S, Wang P, Han G, Gu B, Zhang W. Silencing lncRNA AFAP1-AS1 inhibits the progression of esophageal squamous cell carcinoma cells via regulating the miR-498/VEGFA Axis. Cancer Manag Res. 2020;12:6397–6409. doi: 10.2147/CMAR.S254302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu Z, Yang S, Chen X, Dong S, Zhou S, Xu S. LncRNA LINC00467 acted as an oncogene in esophageal squamous cell carcinoma by accelerating cell proliferation and preventing cell apoptosis via the miR-485-5p/DPAGT1 axis. J Gastroenterol Hepatol. 2021;36:721–730. doi: 10.1111/jgh.15201. [DOI] [PubMed] [Google Scholar]
- 36.Chen W, Wang L, Li X, Zhao C, Shi L, Zhao H, Huang C. LncRNA SNHG17 regulates cell proliferation and invasion by targeting miR-338-3p/SOX4 axis in esophageal squamous cell carcinoma. Cell Death Dis. 2021;12:806. doi: 10.1038/s41419-021-04093-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chu J, Li H, Xing Y, Jia J, Sheng J, Yang L, Sun K, Qu Y, Zhang Y, Yin H, et al. LncRNA MNX1-AS1 promotes progression of esophageal squamous cell carcinoma by regulating miR-34a/SIRT1 axis. Biomed Pharmacother. 2019;116:109029. doi: 10.1016/j.biopha.2019.109029. [DOI] [PubMed] [Google Scholar]
- 38.Pan Q, Li B, Zhang J, Du X, Gu D. LncRNA THAP9-AS1 accelerates cell growth of esophageal squamous cell carcinoma through sponging miR-335-5p to regulate SGMS2. Pathol Res Pract. 2021;224:153526. doi: 10.1016/j.prp.2021.153526. [DOI] [PubMed] [Google Scholar]
- 39.Qian CJ, Xu ZR, Chen LY, Wang YC, Yao J. LncRNA MAFG-AS1 accelerates cell migration, invasion and aerobic glycolysis of esophageal squamous cell carcinoma cells via miR-765/PDX1 Axis. Cancer Manag Res. 2020;12:6895–6908. doi: 10.2147/CMAR.S262075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang J, Yang X, Li R, Zhang R, Hu D, Zhang Y, Gao L. LncRNA SNHG6 Inhibits Apoptosis by Regulating EZH2 Expression via the Sponging of MiR-101-3p in Esophageal Squamous-Cell Carcinoma. Onco Targets Ther. 2020;13:11411–11420. doi: 10.2147/OTT.S275135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Feng B, Wang G, Liang X, Wu Z, Wang X, Dong Z, Guo Y, Shen S, Liang J, Guo W. LncRNA FAM83H-AS1 promotes oesophageal squamous cell carcinoma progression via miR-10a-5p/Girdin axis. J Cell Mol Med. 2020;24:8962–8976. doi: 10.1111/jcmm.15530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou WZ, Wang XW, Zhu J, Chen MZ, Jin H. LncRNA-CASC15 knockdown inhibits the progression of esophageal squamous cell carcinoma through targeting miR-33a-5p/PTGS2 axis. Histol Histopathol. 2023;38:223–232. doi: 10.14670/HH-18-517. [DOI] [PubMed] [Google Scholar]
- 43.He Y, Hua R, Yang Y, Li B, Guo X, Li Z. LncRNA JPX promotes esophageal squamous cell carcinoma progression by targeting miR-516b-5p/VEGFA Axis. Cancers (Basel) 2022;14:2713. doi: 10.3390/cancers14112713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Du F, Guo T, Cao C. Silencing of Long Noncoding RNA SNHG6 inhibits esophageal squamous cell carcinoma progression via miR-186-5p/HIF1α Axis. Dig Dis Sci. 2020;65:2844–2852. doi: 10.1007/s10620-019-06012-8. [DOI] [PubMed] [Google Scholar]
- 45.Li P, Ding H, Han S, Ding S, Yang Y. Long noncoding RNA LINC00858 aggravates the progression of esophageal squamous cell carcinoma via regulating the miR-425-5p/ABL2 axis. Heliyon. 2024;10:e27337. doi: 10.1016/j.heliyon.2024.e27337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang C, Wang L, Yang J, Fu Y, Li H, Xie L, Cui Y. MicroRNA-33a-5p suppresses esophageal squamous cell carcinoma progression via regulation of lncRNA DANCR and ZEB1. Eur J Pharmacol. 2019;861:172590. doi: 10.1016/j.ejphar.2019.172590. [DOI] [PubMed] [Google Scholar]
- 47.Yan S, Xu J, Liu B, Ma L, Feng H, Tan H, Fang C. Long non-coding RNA BCAR4 aggravated proliferation and migration in esophageal squamous cell carcinoma by negatively regulating p53/p21 signaling pathway. Bioengineered. 2021;12:682–696. doi: 10.1080/21655979.2021.1887645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang X, Shen Z, Tian M, Lin Y, Li L, Chai T, Zhang P, Kang M, Lin J. LncRNA C9orf139 can regulate the progression of esophageal squamous carcinoma by mediating the miR-661/HDAC11 axis. Transl Oncol. 2022;24:101487. doi: 10.1016/j.tranon.2022.101487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chu J, Jia J, Yang L, Qu Y, Yin H, Wan J, He F. LncRNA MIR31HG functions as a ceRNA to regulate c-Met function by sponging miR-34a in esophageal squamous cell carcinoma. Biomed Pharmacother. 2020;128:110313. doi: 10.1016/j.biopha.2020.110313. [DOI] [PubMed] [Google Scholar]
- 50.Liang X, Wu Z, Shen S, Niu Y, Guo Y, Liang J, Guo W. LINC01980 facilitates esophageal squamous cell carcinoma progression via regulation of miR-190a-5p/MYO5A pathway. Arch Biochem Biophys. 2020;686:108371. doi: 10.1016/j.abb.2020.108371. [DOI] [PubMed] [Google Scholar]
- 51.Cheng J, Ma H, Yan M, Xing W. THAP9-AS1/miR-133b/SOX4 positive feedback loop facilitates the progression of esophageal squamous cell carcinoma. Cell Death Dis. 2021;12:401. doi: 10.1038/s41419-021-03690-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu G, Guo W, Chen G, Li W, Cui Y, Qin J, Peng J. Lnc-MCEI mediated the chemosensitivity of esophageal squamous cell carcinoma via miR-6759-5p to competitively regulate IGF2. Int J Biol Sci. 2020;16:2938–2950. doi: 10.7150/ijbs.47051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pan X, Wang Q, Yu Y, Wu W, Chen L, Wang W, Li Z. Antisense lncRNA NNT-AS1 promoted esophageal squamous cell carcinoma progression by regulating its sense gene NNT expression. Cell Death Discov. 2022;8:424. doi: 10.1038/s41420-022-01216-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xue ST, Cao SQ, Ding JC, Li WJ, Hu GS, Zheng JC, Lin X, Chen C, Liu W, Zheng B. LncRNA LUESCC promotes esophageal squamous cell carcinoma by targeting the miR-6785-5p/NRSN2 axis. Cell Mol Life Sci. 2024;81:121. doi: 10.1007/s00018-024-05172-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang T, Yue G, Tian X, Xu Y, Li Z. LncSUMO1P3 exacerbates malignant behaviors of esophageal squamous cell carcinoma cells via miR-486-5p/PHF8/CD151. Heliyon. 2023;9:e19110. doi: 10.1016/j.heliyon.2023.e19110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu H, Yin H, Yang T, Lin J, Sun T. Long non-coding RNA PCAT5 regulates the progression of Esophageal Squamous Cell Carcinoma via miR-4295/PHF20. Heliyon. 2023;9:e22086. doi: 10.1016/j.heliyon.2023.e22086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Guo B, He M, Ma M, Tian Z, Jin J, Tian G. Long Non-coding RNA X-Inactive specific transcript promotes esophageal squamous cell carcinoma progression via the MicroRNA 34a/Zinc Finger E-box-Binding Homeobox 1 Pathway. Dig Dis Sci. 2024;69:1169–1181. doi: 10.1007/s10620-024-08269-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sun G, Wu C. ZFPM2-AS1 facilitates cell growth in esophageal squamous cell carcinoma via up-regulating TRAF4. Biosci Rep. 2020;40:BSR20194352. doi: 10.1042/BSR20194352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang Z, Liang X, Ren L, Zhang S, Li S, Wan T, Xu D, Lv S. LINC00662 promotes cell viability and metastasis in esophageal squamous cell carcinoma by sponging miR-340-5p and upregulating HOXB2. Thorac Cancer. 2020;11:2306–2315. doi: 10.1111/1759-7714.13551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wu X, Fan Y, Liu Y, Shen B, Lu H, Ma H. Long Non-Coding RNA CCAT2 promotes the development of esophageal squamous cell carcinoma by inhibiting miR-200b to Upregulate the IGF2BP2/TK1 Axis. Front Oncol. 2021;11:680642. doi: 10.3389/fonc.2021.680642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang Q, Pan X, You D. Overexpression of long non-coding RNA SBF2-AS1 promotes cell progression in esophageal squamous cell carcinoma (ESCC) by repressing miR-494 to up-regulate PFN2 expression. Biol Open. 2020 Mar 30; doi: 10.1242/bio.048793. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 62.Zhang Y, Zhu H, Sun N, Zhang X, Liang G, Zhu J, Xia L, Kou Y, Lu J. Linc00941 regulates esophageal squamous cell carcinoma via functioning as a competing endogenous RNA for miR-877-3p to modulate PMEPA1 expression. Aging (Albany NY) 2021;13:17830–17846. doi: 10.18632/aging.203286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liu D, Gao M, Wu K, Zhu D, Yang Y, Zhao S. LINC00152 facilitates tumorigenesis in esophageal squamous cell carcinoma via miR-153-3p/FYN axis. Biomed Pharmacother. 2019;112:108654. doi: 10.1016/j.biopha.2019.108654. [DOI] [PubMed] [Google Scholar]
- 64.Liu HF, Zhen Q, Fan YK. LINC00963 predicts poor prognosis and promotes esophageal cancer cells invasion via targeting miR-214-5p/RAB14 axis. Eur Rev Med Pharmacol Sci. 2020;24:164–173. doi: 10.26355/eurrev_202001_19907. [DOI] [PubMed] [Google Scholar]
- 65.Hu L, Cao H, Zheng L, Li R. BBOX1-AS1 activates hedgehog signaling pathway to facilitate the proliferation and stemness of esophageal squamous cell carcinoma cells via miR-506-5p/EIF5A/PTCH1 Axis. Curr Mol Pharmacol. 2023;16:894–904. doi: 10.2174/1874467216666230130132927. [DOI] [PubMed] [Google Scholar]
- 66.Luo D, Salai A, Lv H, Wang Y, Gao Y. FOXD2-AS1 acts an oncogene in esophageal squamous cell carcinoma through sponging miR-204-3p. Clin Transl Oncol. 2022;24:1954–1963. doi: 10.1007/s12094-022-02850-7. [DOI] [PubMed] [Google Scholar]
- 67.Chen L, Lu J, Li X, Wang X, Qiao R, Guo W, Ren Q. LncRNA KTN1-AS1 facilitates esophageal squamous cell carcinoma progression via miR-885-5p/STRN3 axis. Genes Genomics. 2024;46:241–252. doi: 10.1007/s13258-023-01451-0. [DOI] [PubMed] [Google Scholar]
- 68.Wang H, Li H, Yu Y, Jiang Q, Zhang R, Sun H, Xing W, Li Y. Long non-coding RNA XIST promotes the progression of esophageal squamous cell carcinoma through sponging miR-129-5p and upregulating CCND1 expression. Cell Cycle. 2021;20:39–53. doi: 10.1080/15384101.2020.1856497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.He Z. LINC00473/miR-497-5p regulates esophageal squamous cell carcinoma progression through targeting PRKAA1. Cancer Biother Radiopharm. 2019;34:650–659. doi: 10.1089/cbr.2019.2875. [DOI] [PubMed] [Google Scholar]
- 70.Luo J, Xie K, Gao X, Yao Y, Wang G, Shao C, Li X, Xu Y, Ren B, Hu L, Shen Y. Long Noncoding RNA nuclear paraspeckle assembly transcript 1 promotes progression and angiogenesis of esophageal squamous cell carcinoma through miR-590-3p/MDM2 Axis. Front Oncol. 2021;10:618930. doi: 10.3389/fonc.2020.618930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ma J, Xiao Y, Tian B, Chen S, Zhang B, Wu J, Wu Z, Li X, Tang J, Yang D, et al. Long noncoding RNA lnc-ABCA12-3 promotes cell migration, invasion, and proliferation by regulating fibronectin 1 in esophageal squamous cell carcinoma. J Cell Biochem. 2020;121:1374–1387. doi: 10.1002/jcb.29373. [DOI] [PubMed] [Google Scholar]
- 72.Guan H, Lv P, Han P, Zhou L, Liu J, Wu W, Yan M, Xing Q, Cao W. Long non-coding RNA ESCCAL-1/miR-590/LRP6 signaling pathway participates in the progression of esophageal squamous cell carcinoma. Cancer Med. 2022;12:445–458. doi: 10.1002/cam4.4915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shang M, Wang X, Zhang Y, Gao Z, Wang T, Liu R. LincRNA-ROR promotes metastasis and invasion of esophageal squamous cell carcinoma by regulating miR-145/FSCN1. Onco Targets Ther. 2018;11:639–649. doi: 10.2147/OTT.S157638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang G, Feng B, Niu Y, Wu J, Yang Y, Shen S, Guo Y, Liang J, Guo W, Dong Z. A novel long noncoding RNA, LOC440173, promotes the progression of esophageal squamous cell carcinoma by modulating the miR-30d-5p/HDAC9 axis and the epithelial-mesenchymal transition. Mol Carcinog. 2020;59:1392–1408. doi: 10.1002/mc.23264. [DOI] [PubMed] [Google Scholar]
- 75.Wang X, Liu H, Zhang Q, Zhang X, Qin Y, Zhu G, Dang J, Wang F, Yang X, Fan R. LINC00514 promotes lipogenesis and tumor progression in esophageal squamous cell carcinoma by sponging miR-378a-5p to enhance SPHK1 expression. Int J Oncol. 2021;59:86. doi: 10.3892/ijo.2021.5266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chen W, Zhang Y, Wang H, Pan T, Zhang Y, Li C. LINC00473/miR-374a-5p regulates esophageal squamous cell carcinoma via targeting SPIN1 to weaken the effect of radiotherapy. J Cell Biochem. 2019;120:14562–14572. doi: 10.1002/jcb.28717. [DOI] [PubMed] [Google Scholar]
- 77.Liu WH, Qiao HY, Xu J, Wang WQ, Wu YL, Wu X. LINC00473 contributes to the radioresistance of esophageal squamous cell carcinoma by regulating microRNA-497-5p and cell division cycle 25A. Int J Mol Med. 2020;46:571–582. doi: 10.3892/ijmm.2020.4616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.He XY, Wang XQ, Xiao QL, Liu D, Xu QR, Liu S. Long non-coding RNA NCK1-AS1 functions as a ceRNA to regulate cell viability and invasion in esophageal squamous cell carcinoma via microRNA-133b/ENPEP axis. Cell Cycle. 2023;22:596–609. doi: 10.1080/15384101.2022.2138416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Xiao Y, Tang J, Yang D, Zhang B, Wu J, Wu Z, Liao Q, Wang H, Wang W, Su M. Long noncoding RNA LIPH-4 promotes esophageal squamous cell carcinoma progression by regulating the miR-216b/IGF2BP2 axis. Biomark Res. 2022;10:60. doi: 10.1186/s40364-022-00408-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wang W, Yang J. Long noncoding RNA TTTY15 promotes growth and metastasis of esophageal squamous cell carcinoma by sponging microRNA-337-3p to upregulate the expression of JAK2. Anticancer Drugs. 2020;31:1038–1045. doi: 10.1097/CAD.0000000000000960. [DOI] [PubMed] [Google Scholar]
- 81.Zhao M, Cui H, Zhao B, Li M, Man H. Long intergenic non-coding RNA LINC01232 contributes to esophageal squamous cell carcinoma progression by sequestering microRNA-654-3p and consequently promoting hepatoma-derived growth factor expression. Int J Mol Med. 2020;46:2007–2018. doi: 10.3892/ijmm.2020.4750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhou Z, Huang F. Long Non-Coding RNA LINC00152 regulates cell proliferation, migration and invasion in esophageal squamous cell carcinoma via miR-107/Rab10 Axis. Onco Targets Ther. 2019;12:8553–8567. doi: 10.2147/OTT.S221515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gao H, Wang T, Zhang P, Shang M, Gao Z, Yang F, Liu R. Linc-ROR regulates apoptosis in esophageal squamous cell carcinoma via modulation of p53 ubiquitination by targeting miR-204-5p/MDM2. J Cell Physiol. 2020;235:2325–2335. doi: 10.1002/jcp.29139. [DOI] [PubMed] [Google Scholar]
- 84.Ke S, Fang M, Li R, Wang J, Lu J. Downregulation of long noncoding RNA breast cancer anti-estrogen resistance 4 inhibits cell proliferation, invasion, and migration in esophageal squamous cell carcinoma by regulating the microRNA-181c-5p/LIM and SH3 protein 1 axis. Bioengineered. 2022;13:12998–13010. doi: 10.1080/21655979.2022.2060720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pan C, Chen G, Zhao X, Xu X, Liu J. lncRNA BBOX1-AS1 silencing inhibits esophageal squamous cell cancer progression by promoting ferroptosis via miR-513a-3p/SLC7A11 axis. Eur J Pharmacol. 2022;934:175317. doi: 10.1016/j.ejphar.2022.175317. [DOI] [PubMed] [Google Scholar]
- 86.Wu K, Wang Z, Huang Y, Yao L, Kang N, Ge W, Zhang R, He W. LncRNA PTPRG-AS1 facilitates glycolysis and stemness properties of esophageal squamous cell carcinoma cells through miR-599/PDK1 axis. J Gastroenterol Hepatol. 2022;37:507–517. doi: 10.1111/jgh.15719. [DOI] [PubMed] [Google Scholar]
- 87.Zheng Y, Wu J, Deng R, Lin C, Huang Y, Yang X, Wang C, Yang M, He Y, Lu J, et al. G3BP2 regulated by the lncRNA LINC01554 facilitates esophageal squamous cell carcinoma metastasis through stabilizing HDGF transcript. Oncogene. 2022;41:515–526. doi: 10.1038/s41388-021-02073-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Liang X, Lu J, Wu Z, Guo Y, Shen S, Liang J, Dong Z, Guo W. LINC00239 Interacts with C-Myc Promoter-Binding Protein-1 (MBP-1) to Promote Expression of C-Myc in esophageal squamous cell carcinoma. Mol Cancer Res. 2021;19:1465–1475. doi: 10.1158/1541-7786.MCR-20-1025. [DOI] [PubMed] [Google Scholar]
- 89.Ren L, Fang X, Shrestha SM, Ji Q, Ye H, Liang Y, Liu Y, Feng Y, Dong J, Shi R. LncRNA SNHG16 promotes development of oesophageal squamous cell carcinoma by interacting with EIF4A3 and modulating RhoU mRNA stability. Cell Mol Biol Lett. 2022;27:89. doi: 10.1186/s11658-022-00386-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhang H, Wang Y, Zhang W, Wu Q, Fan J, Zhan Q. BAALC-AS1/G3BP2/c-Myc feedback loop promotes cell proliferation in esophageal squamous cell carcinoma. Cancer Commun (Lond) 2021;41:240–257. doi: 10.1002/cac2.12127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Duan Y, Jia Y, Wang J, Liu T, Cheng Z, Sang M, Lv W, Qin J, Liu L. Long noncoding RNA DGCR5 involves in tumorigenesis of esophageal squamous cell carcinoma via SRSF1-mediated alternative splicing of Mcl-1. Cell Death Dis. 2021;12:587. doi: 10.1038/s41419-021-03858-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Li Q, Zhang Z, Jiang H, Hou J, Chai Y, Nan H, Li F, Wang L. DLEU1 promotes cell survival by preventing DYNLL1 degradation in esophageal squamous cell carcinoma. J Transl Med. 2022;20:245. doi: 10.1186/s12967-022-03449-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Shen S, Liang J, Liang X, Wang G, Feng B, Guo W, Guo Y, Dong Z. SNHG17, as an EMT-related lncRNA, promotes the expression of c-Myc by binding to c-Jun in esophageal squamous cell carcinoma. Cancer Sci. 2022;113:319–333. doi: 10.1111/cas.15184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Cui Y, Yan M, Wu W, Lv P, Wang J, Huo Y, Lou Y, Ma X, Chang J, Guan F, Cao W. ESCCAL-1 promotes cell-cycle progression by interacting with and stabilizing galectin-1 in esophageal squamous cell carcinoma. NPJ Precis Oncol. 2022;6:12. doi: 10.1038/s41698-022-00255-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhang Y, Chen W, Pan T, Wang H, Zhang Y, Li C. LBX2-AS1 is activated by ZEB1 and promotes the development of esophageal squamous cell carcinoma by interacting with HNRNPC to enhance the stability of ZEB1 and ZEB2 mRNAs. Biochem Biophys Res Commun. 2019;511:566–572. doi: 10.1016/j.bbrc.2019.02.079. [DOI] [PubMed] [Google Scholar]
- 96.Yang C, Shen S, Zheng X, Ye K, Ge H, Sun Y, Lu Y. Long non-coding RNA LINC00337 induces autophagy and chemoresistance to cisplatin in esophageal squamous cell carcinoma cells via upregulation of TPX2 by recruiting E2F4. FASEB J. 2020;34:6055–6069. doi: 10.1096/fj.201900731RR. [DOI] [PubMed] [Google Scholar]
- 97.Huang GW, Chen QQ, Ma CC, Xie LH, Gu J. linc01305 promotes metastasis and proliferation of esophageal squamous cell carcinoma through interacting with IGF2BP2 and IGF2BP3 to stabilize HTR3A mRNA. Int J Biochem Cell Biol. 2021;136:106015. doi: 10.1016/j.biocel.2021.106015. [DOI] [PubMed] [Google Scholar]
- 98.Wang L, Meng D, Wang Y, Hu J. Long non-coding RNA LINC01296 promotes esophageal squamous cell carcinoma cell proliferation and invasion by epigenetic suppression of KLF2. Am J Cancer Res. 2018;8:2020–2029. [PMC free article] [PubMed] [Google Scholar]
- 99.Xu T, Yan Z, Lu J, Chen L, Li X, Li Y, Dong Z, Guo W. Long non-coding RNA NRSN2-AS1, transcribed by SOX2, promotes progression of esophageal squamous cell carcinoma by regulating the ubiquitin-degradation of PGK1. Clin Exp Metastasis. 2022;39:757–769. doi: 10.1007/s10585-022-10174-7. [DOI] [PubMed] [Google Scholar]
- 100.Yang R, Wan J, Ma L, Zhou F, Yang Z, Li Z, Zhang M, Ming L. TMEM44-AS1 promotes esophageal squamous cell carcinoma progression by regulating the IGF2BP2-GPX4 axis in modulating ferroptosis. Cell Death Discov. 2023;9:431. doi: 10.1038/s41420-023-01727-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Huang T, You Q, Huang D, Zhang Y, He Z, Shen X, Li F, Shen Q, Onyebuchi IC, Wu C, et al. A positive feedback between PDIA3P1 and OCT4 promotes the cancer stem cell properties of esophageal squamous cell carcinoma. Cell Commun Signal. 2024;22:60. doi: 10.1186/s12964-024-01475-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Huang X, Liu C, Li H, Dai T, Luo G, Zhang C, Li T, Lü M. Hypoxia-responsive lncRNA G077640 promotes ESCC tumorigenesis via the H2AX-HIF1α-glycolysis axis. Carcinogenesis. 2023;44:383–393. doi: 10.1093/carcin/bgad036. [DOI] [PubMed] [Google Scholar]
- 103.Yang Z, Wan J, Ma L, Li Z, Yang R, Yang H, Li J, Zhou F, Ming L. Long non-coding RNA HOXC-AS1 exerts its oncogenic effects in esophageal squamous cell carcinoma by interaction with IGF2BP2 to stabilize SIRT1 expression. J Clin Lab Anal. 2023;37:e24801. doi: 10.1002/jcla.24801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lu JT, Yan ZY, Xu TX, Zhao F, Liu L, Li F, Guo W. Reciprocal regulation of LINC00941 and SOX2 promotes progression of esophageal squamous cell carcinoma. Cell Death Dis. 2023;14:72. doi: 10.1038/s41419-023-05605-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Niu Y, Wang G, Li Y, Guo W, Guo Y, Dong Z. LncRNA FOXP4-AS1 promotes the progression of esophageal squamous cell carcinoma by interacting with MLL2/H3K4me3 to Upregulate FOXP4. Front Oncol. 2021;11:773864. doi: 10.3389/fonc.2021.773864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Luo XJ, He MM, Liu J, Zheng JB, Wu QN, Chen YX, Meng Q, Luo KJ, Chen DL, Xu RH, et al. LncRNA TMPO-AS1 promotes esophageal squamous cell carcinoma progression by forming biomolecular condensates with FUS and p300 to regulate TMPO transcription. Exp Mol Med. 2022;54:834–847. doi: 10.1038/s12276-022-00791-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Chen JL, Lin ZX, Qin YS, She YQ, Chen Y, Chen C, Qiu GD, Zheng JT, Chen ZL, Zhang SY. Overexpression of long noncoding RNA LINC01419 in esophageal squamous cell carcinoma and its relation to the sensitivity to 5-fluorouracil by mediating GSTP1 methylation. Ther Adv Med Oncol. 2019;11:1758835919838958. doi: 10.1177/1758835919838958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Xiao Z, Si M, Cao J, Chen X, Wang Z, Zhang T, Zhou Y, Li H, Dai J, Wang C, et al. Cancer-testis non-coding RNA LEF1-AS1 regulates the nuclear translocation of PDCD5 and suppresses its interaction with p53 signaling: A novel target for immunotherapy in esophageal squamous cell carcinoma. Carcinogenesis. 2023;44:263–276. doi: 10.1093/carcin/bgad006. [DOI] [PubMed] [Google Scholar]
- 109.Hu T, Niu Y, Fu J, Dong Z, He D, Liu J. Antisense lncRNA PCNA-AS1 promotes esophageal squamous cell carcinoma progression through the miR-2467-3p/PCNA axis. Open Med (Wars) 2022;17:1483–1494. doi: 10.1515/med-2022-0552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ban Y, Tan P, Cai J, Li J, Hu M, Zhou Y, Mei Y, Tan Y, Li X, Zeng Z, et al. LNCAROD is stabilized by m6A methylation and promotes cancer progression via forming a ternary complex with HSPA1A and YBX1 in head and neck squamous cell carcinoma. Mol Oncol. 2020;14:1282–1296. doi: 10.1002/1878-0261.12676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wu Q, Zhang H, Yang D, Min Q, Wang Y, Zhang W, Zhan Q. The m6A-induced lncRNA CASC8 promotes proliferation and chemoresistance via upregulation of hnRNPL in esophageal squamous cell carcinoma. Int J Biol Sci. 2022;18:4824–4836. doi: 10.7150/ijbs.71234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Qin B, Dong M, Wang Z, Wan J, Xie Y, Jiao Y, Yan D. Long non-coding RNA CASC15 facilitates esophageal squamous cell carcinoma tumorigenesis via decreasing SIM2 stability via FTO-mediated demethylation. Oncol Rep. 2021;45:1059–1071. doi: 10.3892/or.2020.7917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wang Y, Yu Z, Shi W, Shen J, Guan Y, Ni F. HLA complex P5 upregulation is correlated with poor prognosis and tumor progression in esophageal squamous cell carcinoma. Bioengineered. 2022;13:9301–9311. doi: 10.1080/21655979.2022.2051854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Guo Y, Wang L, Yang H, Ding N. Knockdown long non-coding RNA HCP5 enhances the radiosensitivity of esophageal carcinoma by modulating AKT signaling activation. Bioengineered. 2022;13:884–893. doi: 10.1080/21655979.2021.2014386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Hu D, Ma A, Lu H, Gao Z, Yu Y, Fan J, Liu S, Wang Y, Zhang M. LINC00963 Promotes Cisplatin Resistance in Esophageal Squamous Cell Carcinoma by Interacting with miR-10a to Upregulate SKA1 Expression. Appl Biochem Biotechnol. 2024 Mar 20; doi: 10.1007/s12010-024-04897-4. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 116.Liu B, Li X, Xie J, Feng Z, Lin N, Yu M. LncRNA NRON negatively regulates cisplatin-induced cell apoptosis via downregulating miR-31 in esophageal squamous cell carcinomas. In Vitro Cell Dev Biol Anim. 2022;58:37–43. doi: 10.1007/s11626-021-00638-7. [DOI] [PubMed] [Google Scholar]
- 117.Wei W, Wei X, Xie X. Linc01614 regulates the proliferation, apoptosis, and chemotherapy resistance in esophageal squamous cell carcinoma by targeting Mir-4775. Iran J Public Health. 2023;52:1170–1180. doi: 10.18502/ijph.v52i6.12959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Nan Y, Luo Q, Wu X, Chang W, Zhao P, Liu S, Liu Z. HCP5 prevents ubiquitination-mediated UTP3 degradation to inhibit apoptosis by activating c-Myc transcriptional activity. Mol Ther. 2023;31:552–568. doi: 10.1016/j.ymthe.2022.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Nan Y, Liu S, Luo Q, Wu X, Zhao P, Chang W, Zhang R, Li Y, Liu Z. m(6)A demethylase FTO stabilizes LINK-A to exert oncogenic roles via MCM3-mediated cell-cycle progression and HIF-1α activation. Cell Rep. 2023;42:113273. doi: 10.1016/j.celrep.2023.113273. [DOI] [PubMed] [Google Scholar]
- 120.Haghi B, Saghaeian Jazi M, Khosravi A, Jafari SM, Asadi J. SOX2OT lncRNA inhibition suppresses the stemness characteristics of esophageal tumorspheres. Noncoding RNA. 2022;8:80. doi: 10.3390/ncrna8060080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Li Y, Li T, Yang Y, Kang W, Dong S, Cheng S. YY1-induced upregulation of FOXP4-AS1 and FOXP4 promote the proliferation of esophageal squamous cell carcinoma cells. Cell Biol Int. 2020;44:1447–1457. doi: 10.1002/cbin.11338. [DOI] [PubMed] [Google Scholar]
- 122.Zhao F, Tian H, Wang Y, Zhang J, Liu F, Fu L. LINC01004-SPI1 axis-activated SIGLEC9 in tumor-associated macrophages induces radioresistance and the formation of immunosuppressive tumor microenvironment in esophageal squamous cell carcinoma. Cancer Immunol Immunother. 2023;72:1835–1851. doi: 10.1007/s00262-022-03364-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ke S, Wang J, Lu J, Fang M, Li R. Long intergenic non-protein coding RNA 00858 participates in the occurrence and development of esophageal squamous cell carcinoma through the activation of the FTO-m6A-MYC axis by recruiting ZNF184. Genomics. 2023;115:110593. doi: 10.1016/j.ygeno.2023.110593. [DOI] [PubMed] [Google Scholar]
- 124.Yu XM, Li SJ, Yao ZT, Xu JJ, Zheng CC, Liu ZC, Ding PB, Jiang ZL, Wei X, Zhao LP, et al. N4-acetylcytidine modification of lncRNA CTC-490G23.2 promotes cancer metastasis through interacting with PTBP1 to increase CD44 alternative splicing. Oncogene. 2023;42:1101–1116. doi: 10.1038/s41388-023-02628-3. [DOI] [PubMed] [Google Scholar]
- 125.Wang J, Huang TJ, Mei Y, Luo FF, Xie DH, Peng LX, Liu BQ, Fan ML, Zhang JB, Zheng ST, et al. Novel long noncoding RNA LINC02820 augments TNF signaling pathway to remodel cytoskeleton and potentiate metastasis in esophageal squamous cell carcinoma. Cancer Gene Ther. 2023;30:375–387. doi: 10.1038/s41417-022-00554-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Luo Q, Kuang Y, Tao X, Zhu X, Zhao X, Lu G, Zhang W, Sun E, Chen B. Pan-cancer analysis of PCAT6 and its effect on oesophageal squamous cell carcinoma cell proliferation and migration. Med Oncol. 2023;40:125. doi: 10.1007/s12032-023-01982-2. [DOI] [PubMed] [Google Scholar]
- 127.Liu JQ, Deng M, Xue NN, Li TX, Guo YX, Gao L, Zhao D, Fan RT. lncRNA KLF3-AS1 Suppresses Cell Migration and Invasion in ESCC by Impairing miR-185-5p-Targeted KLF3 Inhibition. Mol Ther Nucleic Acids. 2020;20:231–241. doi: 10.1016/j.omtn.2020.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Li J, Han X, Gu Y, Wu J, Song J, Shi Z, Chang H, Liu M, Zhang Y. LncRNA MTX2-6 suppresses cell proliferation by acting as ceRNA of miR-574-5p to Accumulate SMAD4 in esophageal squamous cell carcinoma. Front Cell Dev Biol. 2021;9:654746. doi: 10.3389/fcell.2021.654746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Su X, Xue C, Xie C, Si X, Xu J, Huang W, Huang Z, Lin J, Chen Z. lncRNA-LET regulates glycolysis and glutamine decomposition of esophageal squamous cell carcinoma through miR-93-5p/miR-106b-5p/SOCS4. Front Oncol. 2022;12:897751. doi: 10.3389/fonc.2022.897751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Zhang Q, Guan F, Fan T, Li S, Ma S, Zhang Y, Guo W, Liu H. LncRNA WDFY3-AS2 suppresses proliferation and invasion in oesophageal squamous cell carcinoma by regulating miR-2355-5p/SOCS2 axis. J Cell Mol Med. 2020;24:8206–8220. doi: 10.1111/jcmm.15488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Zhang H, Pan E, Zhang Y, Zhao C, Liu Q, Pu Y, Yin L. LncRNA RPL34-AS1 suppresses the proliferation, migration and invasion of esophageal squamous cell carcinoma via targeting miR-575/ACAA2 axis. BMC Cancer. 2022;22:1017. doi: 10.1186/s12885-022-10104-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Wang L, Wang X, Yan P, Liu Y, Jiang X. LINC00261 suppresses cisplatin resistance of esophageal squamous cell carcinoma through miR-545-3p/MT1M Axis. Front Cell Dev Biol. 2021;9:687788. doi: 10.3389/fcell.2021.687788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Shi Z, Li G, Li Z, Liu J, Tang Y. TMEM161B-AS1 suppresses proliferation, invasion and glycolysis by targeting miR-23a-3p/HIF1AN signal axis in oesophageal squamous cell carcinoma. J Cell Mol Med. 2021;25:6535–6549. doi: 10.1111/jcmm.16652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Dong Z, Zhang G, Lu J, Guo Y, Liang J, Shen S, Guo W. Methylation mediated downregulation of TOB1-AS1 and TOB1 correlates with malignant progression and poor prognosis of esophageal squamous cell carcinoma. Dig Dis Sci. 2023;68:1316–1331. doi: 10.1007/s10620-022-07664-9. [DOI] [PubMed] [Google Scholar]
- 135.Liang M, Pan Z, Yu F, Chen C. Long noncoding RNA SNHG12 suppresses esophageal squamous cell carcinoma progression through competing endogenous RNA networks. Clin Transl Oncol. 2020;22:1786–1795. doi: 10.1007/s12094-020-02317-7. [DOI] [PubMed] [Google Scholar]
- 136.Zheng YJ, Liang TS, Wang J, Zhao JY, Zhai SN, Yang DK, Wang LD. Long non-coding RNA ZNF667-AS1 retards the development of esophageal squamous cell carcinoma via modulation of microRNA-1290-mediated PRUNE2. Transl Oncol. 2022;21:101371. doi: 10.1016/j.tranon.2022.101371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Bi Y, Guo S, Xu X, Kong P, Cui H, Yan T, Ma Y, Cheng Y, Chen Y, Liu X, et al. Decreased ZNF750 promotes angiogenesis in a paracrine manner via activating DANCR/miR-4707-3p/FOXC2 axis in esophageal squamous cell carcinoma. Cell Death Dis. 2020;11:296. doi: 10.1038/s41419-020-2492-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Niu Y, Guo Y, Li Y, Shen S, Liang J, Guo W, Dong Z. LncRNA GATA2-AS1 suppresses esophageal squamous cell carcinoma progression via the mir-940/PTPN12 axis. Exp Cell Res. 2022;416:113130. doi: 10.1016/j.yexcr.2022.113130. [DOI] [PubMed] [Google Scholar]
- 139.Wei L, Gu W, Hu L, Wang K, Huang H, Shen Y. Regulation of IncRNA ZNF667-AS1 in proliferation and invasion of esophageal squamous cell carcinoma cells via mediating ceRNA Network. Crit Rev Eukaryot Gene Expr. 2022;32:57–68. doi: 10.1615/CritRevEukaryotGeneExpr.2022042267. [DOI] [PubMed] [Google Scholar]
- 140.Peng X, Zhou Y, Chen Y, Tang L, Wang G, Jiang H, Wang X, Tao Y, Zhuang W. Reduced LINC00551 expression promotes proliferation and invasion of esophageal squamous cancer by increase in HSP27 phosphorylation. J Cell Physiol. 2021;236:1418–1431. doi: 10.1002/jcp.29947. [DOI] [PubMed] [Google Scholar]
- 141.Wang X, Sun M, Gao Z, Yin L, Pu Y, Zhu Y, Wang X, Liu R. N-nitrosamines-mediated downregulation of LncRNA-UCA1 induces carcinogenesis of esophageal squamous by regulating the alternative splicing of FGFR2. Sci Total Environ. 2023;855:158918. doi: 10.1016/j.scitotenv.2022.158918. [DOI] [PubMed] [Google Scholar]
- 142.Liang F, Luo Q, Han H, Zhang J, Yang Y, Chen J. Long noncoding RNA LINC01088 inhibits esophageal squamous cell carcinoma progression by targeting the NPM1-HDM2-p53 axis. Acta Biochim Biophys Sin (Shanghai) 2023;55:367–381. doi: 10.3724/abbs.2023021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Wei S, Sun S, Zhou X, Zhang C, Li X, Dai S, Wang Y, Zhao L, Shan B. SNHG5 inhibits the progression of EMT through the ubiquitin-degradation of MTA2 in oesophageal cancer. Carcinogenesis. 2021;42:315–326. doi: 10.1093/carcin/bgaa110. [DOI] [PubMed] [Google Scholar]
- 144.Dong Z, Yang L, Lu J, Guo Y, Shen S, Liang J, Guo W. Downregulation of LINC00886 facilitates epithelial-mesenchymal transition through SIRT7/ELF3/miR-144 pathway in esophageal squamous cell carcinoma. Clin Exp Metastasis. 2022;39:661–677. doi: 10.1007/s10585-022-10171-w. [DOI] [PubMed] [Google Scholar]
- 145.Sun K, Zhang G. Long noncoding RNA CASC2 suppresses esophageal squamous cell carcinoma progression by increasing SOCS1 expression. Cell Biosci. 2019;9:90. doi: 10.1186/s13578-019-0353-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Lin J, Chen Z, Wu S, Huang W, Chen F, Huang Z. An NF90/long noncoding RNA-LET/miR-548k feedback amplification loop controls esophageal squamous cell carcinoma progression. J Cancer. 2019;10:5139–5152. doi: 10.7150/jca.30816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Xie K, Zheng C, Gu W, Jiang Z, Luo C, Luo J, Diao Y, Wang G, Cong Z, Yao X, et al. A RASSF8-AS1 based exosomal lncRNAs panel used for diagnostic and prognostic biomarkers for esophageal squamous cell carcinoma. Thorac Cancer. 2022;13:3341–3352. doi: 10.1111/1759-7714.14690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Liu J, Zhou WY, Luo XJ, Chen YX, Wong CW, Liu ZX, Bo Zheng J, Yu Mo H, Chen JQ, Li JJ, et al. Long noncoding RNA Regulating ImMune Escape regulates mixed lineage leukaemia protein-1-H3K4me3-mediated immune escape in oesophageal squamous cell carcinoma. Clin Transl Med. 2023;13:e1410. doi: 10.1002/ctm2.1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Wu D, He X, Wang W, Hu X, Wang K, Wang M. Long noncoding RNA SNHG12 induces proliferation, migration, epithelial-mesenchymal transition, and stemness of esophageal squamous cell carcinoma cells via post-transcriptional regulation of BMI1 and CTNNB1. Mol Oncol. 2020;14:2332–2351. doi: 10.1002/1878-0261.12683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Wu Y, Chen X, Liang Y, Li J, Zhang K, Dai L, Guan X, Wang K, Bai Y. Overexpression of long non-coding RNA SOX2OT promotes esophageal squamous cell carcinoma growth. Cancer Cell Int. 2018;18:76. doi: 10.1186/s12935-018-0570-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Sun Y, Wang J, Pan S, Yang T, Sun X, Wang Y, Shi X, Zhao X, Guo J, Zhang X. LINC00657 played oncogenic roles in esophageal squamous cell carcinoma by targeting miR-615-3p and JunB. Biomed Pharmacother. 2018;108:316–324. doi: 10.1016/j.biopha.2018.09.003. [DOI] [PubMed] [Google Scholar]
- 152.Zhou B, Wu Y, Cheng P, Wu C. Long noncoding RNAs with peptide-encoding potential identified in esophageal squamous cell carcinoma: KDM4A-AS1-encoded peptide weakens cancer cell viability and migratory capacity. Mol Oncol. 2023;17:1419–1436. doi: 10.1002/1878-0261.13424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Liu J, Liu ZX, Li JJ, Zeng ZL, Wang JH, Luo XJ, Wong CW, Zheng JB, Pu HY, Mo HY, et al. The Macrophage-Associated LncRNA MALR Facilitates ILF3 Liquid-Liquid Phase Separation to Promote HIF1α signaling in esophageal cancer. Cancer Res. 2023;83:1476–1489. doi: 10.1158/0008-5472.CAN-22-1922. [DOI] [PubMed] [Google Scholar]
- 154.Qiang G, Yu Q, Su K, Guo Y, Liu D, Liang C. E2F1-activated LINC01224 drives esophageal squamous cell carcinoma cell malignant behaviors via targeting miR-6884-5p/DVL3 axis and activating Wnt/β-catenin signaling pathway. Pathol Res Pract. 2022;235:153873. doi: 10.1016/j.prp.2022.153873. [DOI] [PubMed] [Google Scholar]
- 155.Li Y, Chen B, Jiang X, Li Y, Wang X, Huang S, Wu X, Xiao Y, Shi D, Huang X, et al. A Wnt-induced lncRNA-DGCR5 splicing switch drives tumor-promoting inflammation in esophageal squamous cell carcinoma. Cell Rep. 2023;42:112542. doi: 10.1016/j.celrep.2023.112542. [DOI] [PubMed] [Google Scholar]
- 156.Chen L, Lu J, Xu T, Yan Z, Guo Y, Dong Z, Guo W. KTN1-AS1, a SOX2-mediated lncRNA, activates epithelial-mesenchymal transition process in esophageal squamous cell carcinoma. Sci Rep. 2022;12:20186. doi: 10.1038/s41598-022-24743-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Not applicable.