Abstract
The proton pumping V-ATPase drives essential biological processes, such as acidification of intracellular organelles. Critically, the V-ATPase domains, V1 and VO, must assemble to produce a functional holoenzyme. V-ATPase dysfunction results in cancer, neurodegeneration, and diabetes, as well as systemic acidosis caused by reduced activity of proton-secreting kidney intercalated cells (ICs). However, little is known about the molecular regulation of V-ATPase in mammals. We identified a novel interactor of the mammalian V-ATPase, Drosophila melanogaster X chromosomal gene-like 1 (Dmxl1), aka Rabconnectin-3A. The yeast homologue of Dmxl1, Rav1p, is part of a complex that catalyzes the reversible assembly of the domains. We, therefore,hypothesized that Dmxl1 is a mammalian V-ATPase assembly factor. Here, we generated kidney IC-specific Dmxl1 knockout (KO) mice, which had high urine pH, like B1 V-ATPase KO mice, suggesting impaired V-ATPase function. Western blotting showed decreased B1 expression and B1 (V1) and a4 (VO) subunits were more intracellular and less colocalized in Dmxl1 KO ICs. In parallel, subcellular fractionation revealed less V1 associated B1 in the membrane fraction of KO cells relative to the cytosol. Furthermore, a proximity ligation assay performed using probes against B1 and a4 V-ATPase subunits also revealed decreased association. We propose that loss of Dmxl1 reduces V-ATPase holoenzyme assembly, thereby inhibiting proton pumping function. Dmxl1 may recruit the V1 domain to the membrane and facilitate assembly with the VO domain and in its absence V1 may be targeted for degradation. We conclude that Dmxl1 is a bona fide mammalian V-ATPase assembly factor.
Keywords: V-ATPase, Dmxl1/Rabconnectin-3A, acidification, proton pump, kidney collecting duct
Graphical Abstract
Graphical Abstract.

Introduction
The V-ATPase is a multisubunit, transmembrane proton pump whose primary job is to create an electrochemical gradient across eukaryotic cell membranes, which drives essential cell biological processes. The V-ATPase is composed of two domains, one containing transmembrane subunits (VO) and one comprised of soluble, cytosolic subunits (V1), that must assemble into a functional holoenzyme at the membrane for proton pumping activity to occur. This process is regulated by reversible assembly/disassembly of the two domains in response to a variety of physiological cues.1–6
Although the structure and function of the V-ATPase are highly conserved across eukaryotes, there are multiple isoforms and splice variants of most of the V-ATPase subunits that differ by species, and show tissue, cell, and subcellular specificity in their expression patterns.7,8 Typically, there is one ubiquitous isoform of each V-ATPase subunit expressed within the endomembrane system, which acidifies intracellular vesicles and organelles.8 On the other hand, alternate subunit isoforms are expressed at the plasma membrane of specialized proton secreting cells, which use membrane-localized V-ATPases to perform physiological activities that depend on extracellular acidification, such as maintenance of proper blood pH, bone remodeling, and male fertility.9–12 Here, we focus on the proton-secreting A-type intercalated cells (A-ICs), which are kidney collecting duct epithelial cells that express high levels of the B1 and a4 isoforms of the V-ATPase at their apical plasma membrane.11 The primary function of A-ICs is to maintain acid base balance in the body, which they accomplish by adjusting the amount of V-ATPase holoenzyme at the cell surface to increase or decrease proton secretion according to prevailing physiological conditions and systemic cues.11
Unlike the ubiquitous isoforms that are essential for life, dysfunction or mutations in the alternate isoforms are associated with serious, but non-fatal diseases such as distal renal tubular acidosis (dRTA).13 Distal renal tubular acidosis is caused by improper V-ATPase-dependent acid secretion into the urine, resulting in an abnormally and inappropriately low blood pH. Not surprisingly, A-IC dysfunction, primarily caused by V-ATPase mutations in the IC-specific subunits B1 and a4, results in dRTA.13 However, there are cases of genetic dRTA for which mutations in V-ATPase subunits have not been identified, suggesting that there could be other dRTA genes coding for proteins that interact with and regulate the V-ATPase. Despite this, very little is known about the protein–protein interactions that regulate V-ATPase function in the kidney, and indeed in other organs. To address this gap in knowledge, we performed a proteomic analysis to identify B1 V-ATPase associated proteins in mouse kidney lysates using coimmunoprecipitation and mass spectrometry. These studies revealed many previously unknown interactors of the V-ATPase, including Drosophila melanogaster X chromosomal gene-like 1 protein (Dmxl1), also known as Rabconnectin-3A (Rbcn3a), which surprisingly had higher interaction scores with the B1 isoform than some of the V-ATPase subunits themselves.14
Dmxl1 was first identified on the X chromosome of Drosophila,15 hence its name, but its function was first characterized in yeast, where its homologue, Rav1p, was identified as a central component of the RAVE complex (Regulator of H+-ATPase of Vacuolar and Endosomal membranes).16,17 The RAVE complex was discovered in response to glucose deprivation, which leads to a disassembly of the V-ATPase in yeast and is accompanied by a release of V1 from the membrane and binding of the RAVE complex to the cytosolic V1 subcomplexes. When glucose concentrations are restored, RAVE rapidly catalyzes the reassembly of the V-ATPase holoenzyme, thus re-establishing proton-pumping activity.17 Although reversible disassembly was first characterized in the tobacco hornworm18 and yeast,19 it has become evident that it is a conserved mechanism across species to regulate V-ATPase activity in response to diverse physiological cues. In mammals there are only a few examples of this phenomenon, such as in cultured mammalian cells where glucose starvation was shown to drive assembly of the V-ATPase,20,21 while amino acid starvation in human embryonic kidney cells led to assembly of the V-ATPase on lysosomes.22 Finally, a study in cultured neurons showed reversible V-ATPase assembly on synaptic vesicles.23 However, the molecular details of reversible V-ATPase disassembly are as yet poorly understood in mammals.
Humans, mice, zebrafish, and other higher eukaryotes have two Dmxl/Rbcn3a isoforms: Dmxl1 and Dmxl2, while in Drosophila and yeast there is only one.24 Dmxl1 and Dmxl2 show specificity in their expression patterns with Dmxl2 highly enriched in the brain, while Dmxl1 is widely expressed, but particularly highly expressed in the kidney, prostate, and thyroid.25 In mammals, Dmxl2 was originally co-immunoprecipitated from rat brain with a Rab3A GTPase-activating protein and Guanine nucleotide exchange factor,26 which are involved in synaptic vesicle release, which is why these proteins are also known as Rabconnectins. The Rabconnectin-3 complex is a heterodimer consisting of Dmxl1/Dmxl2 (Rbcn3a) and WDR7 (Rbcn3b), which is ubiquitously expressed.24 So far, pathologic mutations in the Rabconnectin-3 complexes are primarily in Dmxl2. Mutations in Dmxl2 have been connected to early infantile epileptic encephalopathy, non-syndromic deafness, delayed puberty, and decreased fertility.27–29 On the other hand, mutations in Dmxl1 are linked to the development of a variety of cancers30–34 and copy number variations in Dmxl1 have been associated with glaucoma.35 Importantly, these pathological phenotypes are reminiscent of what is seen in humans with V-ATPase mutations.
It has been shown that knockdown of Dmxl1 or WDR7 in cultured cells significantly impairs V-ATPase-dependent acidification of intracellular organelles, possibly by preventing V0/V1 assembly.14,36 Furthermore, in cultured kidney cells, Dmxl2 knockdown affected acidification to a lesser degree,14 and in zebrafish Dmxl2 is reported to promote assembly of V-ATPase holoenzymes on synaptic vesicles.37 On the other hand, there have been reports of conserved function of five mammalian orthologues of the ER-associated assembly factors that catalyze the assembly of the VO domain in yeast: CCDC115/Vma22p, TMEM199/Vma12p, VMA21/Vma21p, and ATP6AP1/ATP6AP2/Voa1. Mutations in these proteins lead to impaired assembly of the VO domain and patients display generalized protein glycosylation and autophagy defects and fatty-liver disease.38–44
Based on these data, using kidney intercalated cells (ICs) as a model system, we set out to determine whether Dmxl1 also functions as a mammalian assembly factor for the V-ATPase holoenzyme in vivo, like its homologue Rav1p in yeast. Thus, loss of Dmxl1 in A-ICs would disrupt recruitment of the cytosolic V1 to membrane-bound VO. This would prevent assembly of the functional holoenzyme and inhibit proton secretion into the urine, ultimately resulting in the development of dRTA. Since global knockout of Dmxl1 is embryonic lethal in mice, we utilized the Cre-Lox system to generate IC-specific Dmxl1 knockout mice to interrogate the role of Dmxl1 in regulating V-ATPase function in the mammalian kidney. We found that IC-specific Dmxl1 knockout mice developed incomplete dRTA, which presents as an inability to properly acidify the urine, similar to the phenotype of B1 V-ATPase knockout mice.45 We observed a decrease in the V1 domain B1 subunit expression, but not in other V-ATPase subunits or other acid base related proteins. Furthermore, using subcellular fractionation and a proximity ligation assay (PLA), we confirmed that loss of Dmxl1 prevents the assembly of the two domains of the V-ATPase, which explains the inability of these mice to properly acidify their urine in response to changes in blood pH. Thus, we conclude that Dmxl1 is a bona fide mammalian V-ATPase assembly factor in vivo.
Materials and Methods
Animals
All animal studies were approved by the Massachusetts General Hospital Subcommittee on Research Animal Care, in accordance with the NIH, Department of Agriculture, and Association for the Assessment and Accreditation of Laboratory Animal Care requirements. Mice were housed in the specific pathogen-free room at the Massachusetts General Hospital Animal Research Facility. Both male and female mice were used in the experiments. For all experiments, control mice were either littermates or age- and sex-matched mice from the same colony. The experimental unit (n) was always a single mouse.
C57BL/6 mice carrying the Dmxl1 floxed allele were custom-made by Biocytogen (Beijing, China), utilizing their proprietary CRISPR/Cas9-based Extreme Genome Editing (EGE™) technology. Briefly, sgRNAs and a targeting vector with LoxP sites flanking exons 3 and 4 of mouse Dmxl1 gene were designed, constructed, and co-injected together with Cas9 mRNA into C57BL/6 zygotes to generate F0 founder mice. The LoxP sites were inserted into the non-conserved regions of the big introns 2 and 4 so that they do not interfere with Dmxl1 RNA transcription and splicing in floxed mice. The choice of exons was based on the prediction that the Cre-mediated removal of the flanked exons 3 and 4 would lead to a Dmxl1 reading frameshift and result in a 75 amino acid (aa)-long (71 native aa plus 4 frameshift aa) truncated protein (for comparison Dmxl1 full-length protein is 3025 a.a. long) with the following sequence: MNLHQVLTGAVNPGDHCFAVGSVGEQRFTAYASGCDIVILGSNFERLQIIPGAKHGNIQVGCVDCSMQGKVIVF, which is non-functional and most likely short-lived. Both PCR and Southern blot analyses were performed to screen for founders and exclude random insertions. F0 mice were then intercrossed with C57BL/6 mice to generate F1 and F2 Dmxl1fl/+ heterozygotes. Two breeding pairs of F2 Dmxl1fl/+ heterozygotes were received from Biocytogen and bred in-house to generate Dmxl1fl/fl homozygotes and establish the Dmxl1 floxed mouse colony. For genotyping, genomic DNA was extracted from mouse tails using the KAPA Express Extract Kit (Kapa Biosystems, Wilmington, MA). Genotyping was performed by PCR using the KAPA Mouse Genotyping Kit (Kapa Biosystems) with the F1/R1 primer pair used to detect the insertion of 5′ LoxP site (F1:5′-AGGGTCTTAACTGTGTATCTGATGCTGA-3′, R1:5′-TCGTTGGAGTTCCTAATTTTCCCAACA-3′) and F2/R2 primer pair used to detect the insertion of 3′ LoxP site (F2:5′-ACTCCTACACCCTGGCCTTTCCTTA-3′, R2: 5′-CTGGGAGAGAGCCCATACTCTTGGA-3′).
Then, we attempted to generate global Dmxl1−/− knockout mice by intercrossing Dmxl1fl/fl and CMV-Cre mice that express Cre recombinase under the global CMV promoter facilitating deletion of floxed genes in all tissues, including germ cells.46 CMV-Cre mice are well characterized and widely used to generate global knockout mice.46 To generate global Dmxl1 knockout mice, Dmxl1fl/fl male mice were mated with females expressing Cre recombinase under the global cytomegalovirus (CMV) promoter (CMV-Cre mice: B6.C-Tg(CMV-cre)1Cgn/J; JAX strain #006054, The Jackson Laboratory, Bar Harbor, ME).46 In their offspring, the deletion of Dmxl1 LoxP-flanked exons 3-4 is expected to occur in all tissues, including germ cells. As expected, the Dmxl1+/−, CMV-Cre+ offspring were obtained, and males were crossed with C57BL/6 J females (JAX strain #000664, The Jackson Laboratory) to ensure germline transmission and removal of the CMV-Cre transgene. The next generation Dmxl1+/−, CMV-Cre− males and females were intercrossed to generate littermate Dmxl1+/+, Dmxl1+/−, and Dmxl1−/− mice. However, despite numerous attempts, we were unable to obtain Dmxl1−/− mice, suggesting that global Dmxl1−/− knockout mice are embryonic lethal. Dmxl1+/− heterozygous mice were born and grew normally, and overall were indistinguishable from their Dmxl1+/+ littermates. In these experiments, genotyping was performed by PCR using the F1/R1 primer pair to detect the “+” allele, the F1/R2 primer pair to detect the excised “-” allele, and general Cre-specific primers to detect the CMV-Cre transgene: Cre-F 5′-CATTACCGGTCGATGCAACGAG-3′ and Cre-R 5′-TGCCCCTGTTTCACTATCCAGG-3′.
To generate conditional kidney intercalated cell specific Dmxl1 knockout mice, we used our transgenic mice expressing Cre under the promoter of the B1 subunit isoform of the V-ATPase (B1-Cre). We intercrossed Dmxl1fl/fl mice, described above, and B1-Cre+ mice to produce Dmxl1fl/+, B1-Cre+ mice that were further crossed to generate littermate Dmxl1fl/fl, B1-Cre+ knockout (or kidney intercalated cell specific Dmxl1 conditional knockout) mice and Dmxl1fl/fl, B1-Cre− control mice and to establish the kidney intercalated cell specific Dmxl1 knockout mouse colony. The genotyping of mice in this colony was performed by PCR, using the F1/R1 5′ LoxP specific primers and the general Cre-specific primer pair Cre-F/Cre-R to detect the B1-Cre transgene.
Antibodies
Commercial primary antibodies used in this study were rabbit polyclonal anti-Dmxl1 at 1:100 for IF (cat # NBP1-90998, Novus Biologicals, Littleton, CO), rabbit polyclonal anti-Dmxl1 at 1:500 for WB (cat # 24413-1-AP, Proteintech, Rosemont, IL), rabbit polyclonal anti-a4 V-ATPase at 1:200 (IF and PLA) and 1:1000 (WB) (cat # 21570-1-AP, Proteintech, Rosemont, IL), mouse monoclonal anti-B1 V-ATPase at 1:200 (IF and PLA) (cat # TA502519, Origene, Rockville, MD), rabbit monoclonal anti-AE1 at 1:100 (IF) (cat # 20112S, Cell Signaling, Danvers, MA), mouse monoclonal anti-cortactin at 1:100 (IF) (cat # 05-180, EMD Millipore, Burlington, MA), rabbit monoclonal anti-CA2 at 1:1000 (WB) (cat # ab124687, Abcam, Waltham, MA), rabbit polyclonal anti-pendrin at 1:100 (IF) (cat # PA5-42060, ThermoFisher Scientific, Waltham, MA), and β-Actin-HRP at 1:1000 (WB) (cat # 5125S, Cell Signaling, Danvers, MA). The actin cytoskeleton was labeled with Rhodamine-phalloidin at 1:1000 (IF) (cat # R415, ThermoFisher Scientific, Waltham, MA) and nuclei with 4,6-diamidino-2-phenylindole (DAPI) (Vector Laboratories, Burlingame, CA). Importantly, the immunogenic sequence used to generate the Dmxl1 antibodies has a sequence homology of only 34% (Proteintech) and 42% (Novus Bio) with Dmxl2, which makes it highly unlikely that the anti-Dmxl1 antibody cross-reacts with Dmxl2.
Commercial secondary antibodies used in this study were for immunofluorescence donkey anti-rabbit Alexa 555 (cat # A32794, 1:600, Invitrogen, Waltham, MA), goat anti-mouse Alexa 555 (cat # A21424, 1:600, Invitrogen, Waltham, MA), goat anti-chicken Alexa 555 (cat # A32932, 1:600, Invitrogen, Waltham, MA), goat anti-rabbit Alexa 488 (cat # A32731, 1:600, Invitrogen, Waltham, MA), goat anti-mouse Alexa 488 (cat # A32723, 1:600, Invitrogen, Waltham, MA), goat anti-chicken Alexa 488 (cat # A32931, 1:600, Invitrogen, Waltham, MA), and for western blotting goat anti-rabbit-HRP (cat # ab97080, 1:5000, Abcam, Waltham, MA) and sheep anti-mouse-HRP (cat # NXA931, Cytiva, Marlborough, MA).
The rabbit polyclonal and chicken polyclonal anti-B1 V-ATPase (56-kDa subunit) antibodies (1:400, IF and 1:1000, WB) were raised against a 13-amino acid peptide (QGAQQDPASDTAL), corresponding to the COOH-terminal sequence of mouse V-ATPase. The chicken polyclonal anti-A V-ATPase (70-kDa subunit) antibody (1:1000, WB) was raised against a 10-amino acid peptide (MQNAFRSLED), corresponding to the C-terminal sequence of mouse V-ATPase. The chicken polyclonal anti-B2 V-ATPase (56-kDa subunit) antibody (1:1000, WB) was raised against a 10-amino acid peptide (EFYPRDSAKH), corresponding to the C-terminal sequence of mouse V-ATPase. These antibodies were produced, affinity purified, and characterized previously in our laboratory.14,47–49
RNA Isolation and End-Point Reverse Transcription PCR
To confirm the presence of the B1-Cre-excised Dmxl1 mRNA transcripts in the kidneys of Dmxl1fl/fl, B1-Cre+ mice, end-point reverse transcription PCR (RT-PCR) was carried out. Briefly, total kidney RNA was isolated by using QIAshredder and RNeasy purification kit (Qiagen, Germantown, MD). RNA concentration was measured with a NanoDrop 2000 Spectrophotometer (ThermoFisher Scientific, Waltham, MA). First-strand cDNA was then synthesized from 2 µg RNA using the High-Capacity RNA-to-cDNA Kit (Applied Biosystems, Waltham, MA) according to the manufacturer’s instructions. PCR reactions were performed using the Taq DNA Polymerase with Standard Taq Buffer (New England Biolabs, Ipswich, MA) on a T100 Thermal Cycler (Bio-Rad, Hercules, CA). The Dmxl1 exon 2-specific primer F3 5′-CAGATCATCCCAGGCGCTAA-3′ and Dmxl1 exon 5-specific primer R3 5′-CTGCAAACAGCTGGAACCAG-3′ were used to detect Dmxl1 excised transcripts alongside with non-excised wild-type transcripts.
Magnetic Activated Cell Sorting Isolation of ICs Using CD117/c-Kit Beads
Four- to five-mo-old control (Dmxl1fl/fl, B1-Cre-) and IC-specific Dmxl1 KO (Dmxl1fl/fl, B1-Cre+) mice were terminally anesthetized with inhalation of CO2. Kidneys were extracted, capsules removed, and placed in phosphate buffered saline (PBS) on ice. Kidneys were placed into 1 mL of digestion buffer (15 mg collagenase-I (cat # 17100-017, Gibco, Grand Island, NY), 15 mg collagenase-II (cat # C6885, Sigma Aldrich, St. Louis, MO), in 9.8 mL RPMI media (cat # 22400-089, Gibco, Grand Island, NY)) and thoroughly minced. Minced tissue was placed on a ThermoMixer C (Eppendorf, Enfield, CT) at 37°C and mixed for 10 s/min pipetting up and down 10 times every 10 min for 30-45 min until a single cell suspension was achieved. Homogenate was passed through a 40-µm mesh strainer (cat # 352 340, Corning, Corning, NY) into a 50 mL Falcon tube and rinsed through with 30 mL wash buffer (2% FBS, 2 mM EDTA, 1X PBS). Homogenates were centrifuged for 5 min at 500 × g, supernatant was poured off, and red blood cells were lysed using 2 mL ACK lysing buffer (cat # A10492-01, Gibco, Grand Island, NY). Reaction was stopped by adding 30 mL ice-cold PBS and homogenates were centrifuged for 5 min at 500 × g. Pellet was resuspended in 1000 µL wash buffer and passed through 35-µm filter tubes (cat # 352 235, Corning, Corning, NY). Cell suspension was centrifuged at 300 × g for 10 min and pellet was resuspended in 80 µL of magnetic activated cell sorting (MACS) buffer/107 cells (dilute MACS BSA Stock Solution (cat # 130-091-222, Miltenyi Biotec, Gaithersburg, MD) 1:20 in autoMACS Rinsing Solution (cat # 130-091-376, Miltenyi Biotec, Gaithersburg, MD)). A volume of 20 µL CD117/c-Kit magnetic microbeads (cat # 130-091-224, Miltenyi Biotec) per 107 cells were added, mixed well, and incubated for 15 min at 4°C with overhead rotation. A volume of 1 mL MACS buffer was added to the cell suspension and centrifuged at 300 × g for 10 min and the cell pellet was resuspended in 500 µL MACS buffer. The cell suspension was applied to a pre-washed MS column (cat # 130-042-201, Miltenyi Biotec, Gaithersburg, MD), placed in an OctoMACS separator (cat # 130-042-109, Miltenyi Biotec, Gaithersburg, MD), and the column washed with 3 × 500 µL MACS buffer. The column was removed from the magnetic field, and 1 mL of MACS buffer was added, and cells were flushed from the column with a plunger.
Blood and Urine Analysis
In control experiments Dmxl1fl/fl, B1-Cre+ (conditional knockout) and Dmxl1fl/fl, B1-Cre− (control) 4-5 mo old mice were maintained on a standard rodent diet (Prolab® Isopro® RMH 3000, LabDiet, St. Louis, MO). Both male and female mice were used, and they had free access to water and food. For the acid loading challenge, drinking water was substituted with 0.28 M NH4Cl/1% sucrose for 3 d. For the alkali loading challenge, drinking water was substituted with 0.28 M NaHCO3/1% sucrose for 3 d. For measurement of blood gases, pH, sodium, chloride, free calcium, and potassium, whole blood was collected from the submandibular vein of conscious mice directly into the BD microtainer blood collection tube with heparin (cat # 365 965, Becton Dickinson and Co, Franklin Lakes, NJ). It was then immediately tested using the SC80 ABL80 cassette (50-test, 30-d full panel with QC3 (no Glu), cat # 945-670, Radiometer America, Westlake, OH) and the SP80 ABL80 Solution Pack (cat # 944-174, Radiometer America) on a Radiometer ABL80Flex analyzer (Radiometer America). Blood bicarbonate concentration was calculated from the measured pH and pCO2 values using the Henderson–Hasselbalch equation. For spot urine pH measurement, mouse urine was collected directly onto Hydrion Urine and Saliva pH Paper 5.5-8.0 (Micro Essential Laboratory Inc., Brooklyn, NY) or ColorpHast pH 7.5-14.0 strips (MilliporeSigma, Burlington, MA), and pH was read immediately.
Whole Body Perfusion Fixation and Tissue Preparation
Four- to five-mo-old control (Dmxl1fl/fl, B1-Cre-) and IC-specific Dmxl1 KO (Dmxl1fl/fl, B1-Cre+) mice were anaesthetized by IP injection 50 mg/kg body wt Nembutal (NDC: 11695-4862-5, Covetrus, Dublin, OH). Animals were allowed to reach proper anesthetic depth over a period of 5 min, which was confirmed by lack of a response to a toe pinch. Mice were then perfused through the left ventricle of the heart for 1-2 min at 17 mL/min with 37°C PBS. For western blotting, the renal artery feeding the right kidney was clamped with a hemostat and the right kidney was excised, capsule removed, rinsed with PBS, and flash frozen in liquid nitrogen and stored at −80°C until ready for lysate preparation (see below). For immunofluorescence, the left kidney was perfusion fixed in situ with 37°C paraformaldehyde-lysine-periodate fixative (PLP; 4% paraformaldehyde, 75 mM lysine-HCl, 10 mM sodium periodate, and 0.15 M sucrose, in 37.5 mM sodium phosphate) for 5 min at 17 mL/min. The kidney was extracted, sectioned into five circumferential pieces, and fixed by immersion in PLP overnight at 4°C with rocking, then rinsed 5 × 1 h at RT in PBS, and stored at 4°C in PBS + 0.02% sodium azide until ready for cryosectioning. Tissues prepared, as described above, were cryoprotected in 30% sucrose in PBS overnight at 4°C, and then embedded in Tissue-Tek OCT compound on a specimen disk (Sakura Finetek USA, Torrance, CA), and frozen at −20°C. Tissues were sectioned at 5 µm on a Leica CM3050 S cryostat (Leica Microsystems, Bannockburn, IL), collected onto Superfrost Plus microscope slides (Thermo Fisher Scientific, Rockford, IL), and stored at 4°C until ready for use.
Lysate Preparation and Western Blotting
Lysis of a single transverse section from the central region of the kidney was performed in 1 mL/100 mg of tissue ice-cold Triton lysis buffer (25 mM Tris–HCL, 150 mM NaCl, 0.5 mL Triton X-100 in H2O, pH 7.4), containing cOmplete Protease Inhibitor cocktail (cat # 11 697 498 001, Roche Applied Science, Indianapolis, IN) by 40 up and down strokes with a Dounce homogenizer and clarified by centrifugation at 16 000 × g for 20 min at 4°C. Lysates were pre-cleared by passing them through a 0.2 µm syringe filter (cat # 4602, Pall Corporation, Port Washington, NY). Lysates were diluted in NuPage Sample Buffer (cat # NP0007, Thermo Fisher Scientific, Waltham, MA) + 100 mM DTT and incubated at 80°C for 15 min, followed by centrifugation for 7 min at 6000 × g at 4°C. An equal volume of each protein sample was run on NuPAGE 4-12% Bis–Tris gels (cat # NP0321, ThermoFisher Scientific, Waltham, MA) and analyzed via Western blotting.
Indirect Immunofluorescent Labeling and Confocal Image Capture
For each experiment, cryosections from at least 4 control and Dmxl1 IC-specific KO mice were incubated in parallel. Sections were rehydrated for 3 × 5 min in PBS. For immunofluorescence with V-ATPase antibodies sections were incubated with 1% (wt/vol) SDS for 4 min for antigen retrieval. For immunofluorescence with the Dmxl1 antibody, antigen retrieval was performed by heating slides for 10 min in the microwave in citrate buffer (9.5 mM citric acid, 0.5 mM Na-citrate, pH 6). After antigen retrieval, sections were washed for 3 × 5 min in PBS and incubated for 30 min in 1% (wt/vol) bovine serum albumin (BSA) in PBS. The sections were incubated for 90 min at RT, or overnight at 4°C, with the primary antibody diluted in 1% (wt/vol) BSA in PBS. After primary antibody incubation sections were washed 3 × 5 min with PBS and the secondary antibody was applied for 1 h at room temperature. Finally, the slides were rinsed again 3 × 5 min in PBS and mounted with SlowFade Diamond Antifade Mountant (ThermoFisher Scientific, Waltham, MA) with DAPI as a nuclear stain (Vector Laboratories, Burlingame, CA). Confocal images were acquired using an LSM 800 confocal laser scanning microscope (Carl Zeiss Microscopy, Thornwood, NY), controlled by ZEN 2 (blue edition) software (Carl Zeiss Microscopy). For comparative semiquantitative analysis (see below), confocal images of control and IC-specific Dmxl1 KO kidneys were acquired under identical conditions and imaging parameters. Images were postprocessed using Adobe Photoshop CS4 image-editing software (Adobe Systems, San Jose, CA).
Fluorescence Image Analysis
A-type ICs were identified by their localization in the inner medulla and expression of the B1 V-ATPase subunit. B-type ICs were identified by their localization in the cortex and apical expression of pendrin. Cells for which the nucleus or the apical membrane were not clearly visible due to the orientation of the cut were excluded from analysis. Images from four animals per genotype and sex were quantified for each parameter. Images were analyzed using ImageJ version 1.53a (NIH, Bethesda, MD) and ZenBlue software (Carl Zeiss Microscopy, White Plains, NY), and data were imported into Microsoft Excel version 16 (Microsoft, Redmond, WA), and GraphPad Prism version 10 (GraphPad Software, San Diego, CA) was used for further graphical and statistical analysis.
V-ATPase Apical Domain Fluorescence Intensity
Apical domain fluorescence intensity of the B1 and a4 subunits of the V-ATPase was quantified separately. Briefly, using the ImageJ “line tool,” a line of 0.5 µm width was drawn across each cell starting from, and perpendicular to, the apical membrane and adjacent to the nucleus. From this line, a “line intensity profile” was generated in ImageJ, which plots fluorescence intensity on the y-axis and distance from the apical membrane in microns on the x-axis. The mean fluorescence intensity of a circular ROI drawn adjacent to the apical membrane of the cell was subtracted from intensity values as background. The estimated area under the curve for each 0.05 µm rectangle was calculated using the equation:
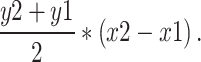 |
The sum of all the estimated areas under the curve was considered the total area under the curve. The apical domain was defined as the sum of the estimated areas under the curve from 0 to 2 µm. Apical domain fluorescence intensity was calculated as the percentage of apical domain fluorescence intensity relative to the total cell fluorescence intensity, which was defined as the total area under the curve. The percentage of apical domain fluorescence intensity for 10 cells was averaged per animal.
VO/V1 Domain Subunit Colocalization
A Pearson’s correlation coefficient for the B1 and a4 subunits of the V-ATPase was calculated using ZenBlue software. Briefly, a .czi file of an intercalated cell was opened in ZenBlue and “Colocalization” was selected from the toolbar. The 555 channel (B1 V-ATPase) was set as the horizontal axis and the 488 (a4 V-ATPase) channel was set as the vertical axis. The “Bezier” free drawing tool was used to outline the entire intercalated cell and thresholding was performed automatically using the “Costes” function. Pearson’s coefficient was recorded for the “# spline contour,” which is the value for the outlined cell. The Pearson’s coefficient for 20 cells was averaged per animal.
Corrected Total Cell Fluorescence
Corrected total cell fluorescence of Dmxl1 and the B1 V-ATPase subunit was measured on tissues from control and IC-specific KO mice double stained for Dmxl1 and B1. The “freehand selection” tool in ImageJ was used to draw an ROI around the entire cell of interest, and the area (A) and integrated density (ID) values were measured using the analyze > measure function. The mean intensity value of a circular ROI drawn away from the cell was subtracted as mean background fluorescence (MBF). The corrected total cell fluorescence (CTCF) for an individual cell was calculated using the equation:
 |
The CTCF of 20 cells was averaged per animal. Corrected total cell fluorescence values for Dmxl1 and B1 from each cell were used to generate an XY plot using Prism, and a simple linear regression analysis was performed in Prism to identify whether there was a correlation between Dmxl1 and B1 total cell fluorescence.
Cell Size Analysis
Cell size was measured on tissues from control and IC-specific KO mice stained for B1. The “freehand selection” tool in ImageJ was used to draw an ROI around the entire cell of interest, and the area (A) values were measured using the analyze > measure function. The outline of each cell was readily discernible using the intracellular fluorescence signal under all conditions examined, and no other marker was, therefore, necessary to establish the area of individual ICs for the purpose of this quantification. A cell size of 20 cells was averaged per animal. A histogram of the frequency distribution of cell sizes was generated and significant differences in the distribution were calculated using a Kolmogorov–Smirnov test.
Subcellular Fractionation Assay
Perfused kidneys from control and Dmxl1 IC-specific KO mice were homogenized in ice-cold homogenization buffer containing 50 mM Tris–HCL, 250 mM sucrose, 1 mM EDTA, and cOmplete Protease Inhibitor cocktail (cat # 11697498001, Roche Applied Science, Indianapolis, IN). Cell fractionation was performed using a sequential centrifugation procedure. Briefly, samples were centrifuged at 1000 × g for 5 min, the supernatant was collected and centrifuged again at 1000 × g for 5 min, the supernatant was extracted again and centrifuged at 6000 × g for 15 min, a fraction of this supernatant was saved for input and the rest was transferred into ultracentrifugation tubes and spun at 100 000 × g for 1 h. All centrifugation steps were performed at 4°C. After ultracentrifugation supernatants (cytosol) and pellets (membrane) were flash frozen on dry ice and stored at −80°C until ready for analysis by Western blotting.
Proximity Ligation Assay
In situ interactions were detected using the Duolink Proximity Ligation Assay (PLA) kit (cat # DUO92102, Sigma Aldrich, St. Louis, MO). The anti-rabbit plus probe was used to bind to the a4 subunit antibody, whereas the anti-mouse minus probe was used to bind to the B1 subunit antibody. The a4 and B1 subunit antibodies were used at 1:6000 dilution in 1% BSA o/n at 4°C prior to the PLA reaction. After the completion of the PLA reaction, sections were post-stained with primary antibodies against AE1 to identify A-type ICs or pendrin to identify B-type ICs. Total area of PLA signal per cell was quantified using ImageJ. A total of 25 cells were averaged per animal.
Transmission and Immunogold Electron Microscopy
Conventional transmission electron microscopy (TEM) was performed on 4-mo-old mouse kidney medullas (n = 2 mice per genotype) as described elsewhere.48 Briefly, for conventional TEM, following perfusion/immersion fixation with periodate–lysine–paraformaldehyde reagent, small pieces of Dmxl1fl/fl, B1-Cre− and Dmxl1fl/fl, B1-Cre+ kidney medullas were immersed in 2.0% glutaraldehyde in 0.1 M sodium cacodylate buffer, pH 7.4. They were postfixed for 1 h in 1% osmium tetroxide, dehydrated through a graded series of ethanol solutions up to 100%, and embedded in Eponate 12 (Ted Pella Inc., Redding, CA). Thin (∼70 nm) sections were collected onto copper slot grids and stained with uranyl acetate and Reynold’s lead citrate before examination. Sections were examined in a JEM-1011 transmission electron microscope (JEOL USA, Peabody, MA) at 80 kV, and images were collected using an AMT digital imaging system with proprietary image capture software (Advanced Microscopy Techniques, Danvers, MA).
Data and Statistical Analysis
Statistical significance was determined using a two-tailed unpaired t-test, a one-way analysis of variance (ANOVA), or a Kolmogorov–Smirnov test using Prism 9 software (GraphPad Software, San Diego, CA). A P-value ≤ .05 was considered statistically significant. Graphs were plotted with Prism 9 software. Experimental values are reported as means ± standard error of the mean (SEM).
Results
Generation of Dmxl1+/−, Dmxl1fl/fl, and Dmxl1fl/fl, B1-Cre Conditional Intercalated Cell Knockout Mice
First, we attempted to generate global Dmxl1−/− knockout mice by intercrossing Dmxl1fl/fl and CMV-Cre mice that express Cre recombinase under the global CMV promoter facilitating deletion of floxed genes in all tissues, including germ cells.46 However, while we were able to produce Dmxl1+/+ and Dmxl1+/− offspring as assessed by genotyping of mouse tails (Figure 1C), no mice with Dmxl1−/− genotype were obtained. This suggests that they do not survive beyond prenatal stages of development and complete knockout of the Dmxl1 gene resulted in an inability to survive and therefore it is an essential gene in mice and probably other mammals, in general. Our results are consistent with the previously reported data that Dmxl1 homozygous knockout mice were embryonic lethal, although the details were only published in an abstract from several years ago.50
Figure 1.
Generation of Dmxl1fl/fl, Dmxl1+/−, and Dmxl1fl/fl, B1-Cre intercalated cell knockout mice. (A) Schematic representation of Dmxl1 gene and its wild type, floxed and excised alleles. Dmxl1 gene is located at mouse chromosome 18: 49 965 737-50 098 540 forward strand (Genome Build GRCm39: CM001011.3) and contains 43 exons. For gene targeting, LoxP sites flanking exons 3 and 4 of mouse Dmxl1 gene were inserted into introns 2 and 4 using CRISPR/Cas9-based Extreme Genome Editing (EGE™) technology by Biocytogen (Beijing, China). Cre-mediated removal of exons 3 and 4 from the floxed allele produces the excised allele. Positions of the forward and reverse primer pairs used for mouse genotyping (F1/R1 and F2/R2) and end-point RT-PCR (F3/R3) are indicated. LoxP sites and primers are not drawn to scale. (B) The Dmxl1fl/+ genotype of four Dmxl1 colony founders (1-4) was confirmed by PCR analysis of tail genomic DNA. Presence of both the 5′LoxP and 3′LoxP sites was verified by PCR using the F1/R1 or F2/R2 primer pairs, respectively. The F1/R1 or F2/R2 primer pairs recognize both wild-type (378 and 492 bp) and floxed (722 and 807 bp) alleles. (C) Representative results of PCR genotyping of genomic DNA extracted from tails of Dmxl1+/+ and Dmxl1+/− mice. For each sample, two separate PCR reactions using either the F1/R1 or F1/R2 primer pairs were run. Primer pair F1/R1 is specific to non-excised alleles (wild-type and floxed) and amplifies a 378 bp DNA fragment in wild-type mice, while primer pair F1/R2 amplifies a 697 bp fragment within the excised allele. The inferred genotypes are shown above. (D) The excision of Dmxl1 transcripts in kidneys of Dmxl1fl/fl, B1-Cre+ intercalated cell conditional knockout mice was confirmed by end-point RT-PCR using F3 and R3 primers, specific for Dmxl1 exons 2 and 5, respectively. Total RNA was isolated from four Dmxl1fl/fl, B1-Cre− control and four Dmxl1fl/fl, B1-Cre+ mouse kidneys, reverse transcribed, and amplified by PCR. Primer pair F3/R3 amplified a 258 bp fragment within the wild-type Dmxl1 RNA transcript and a 107 bp fragment within the excised transcript. Both excised and non-excised Dmxl1 RNA are present in Dmxl1fl/fl, B1-Cre+ mice.
Since global knockout Dmxl1−/− mice were not viable, we focused on generation and characterization of Dmxl1 conditional knockout mice. Dmxl1fl/+ heterozygous mice, carrying one copy of the Dmxl1 floxed allele with LoxP sites flanking exons 3 and 4 were designed and produced by Biocytogen (Figure 1A and “Methods”). We received two breeding pairs of Dmxl1fl/+ heterozygotes (F2 generation) from Biocytogen and confirmed the presence of both 5′ and 3′ LoxP sites in these mice by PCR genotyping on genomic DNA from tails using primer pairs F1/R1 and F2/R2 (Figure 1B). The original F2 Dmxl1fl/+ heterozygotes were then intercrossed to generate Dmxl1fl/fl homozygous mice along with Dmxl1fl/+ and Dmxl1+/+ mice and establish the Dmxl1 floxed mouse colony. Dmxl1fl/fl mice were viable and fertile, without gross anatomical or physiological abnormalities, suggesting that the insertion of LoxP sites did not affect Dmxl1 gene function.
To study the role of Dmxl1 in kidney ICs, we generated intercalated cell-specific Dmxl1 knockout mice by intercrossing Dmxl1fl/fl and B1-Cre mice expressing Cre recombinase under the promoter of the B1 subunit isoform of the V-ATPase, which is specifically expressed in ICs in the kidney.51 Efficiency and specificity of Cre-mediated recombination using B1-Cre mice were evaluated previously by crossing them with reporter mice and were confirmed to be highly specific to ICs in the mouse kidney.51 We intercrossed Dmxl1fl/fl and B1-Cre mice to produce Dmxl1fl/+, B1-Cre+ mice that were further crossed to generate littermate Dmxl1fl/fl, B1-Cre+ knockout (or intercalated cell specific Dmxl1 conditional KO) mice and Dmxl1fl/fl, B1-Cre− control mice. The genotyping was performed by PCR, using F1/R1 5′ LoxP- and Cre-specific primers (see “Methods”). Then, Dmxl1 recombination was evaluated by end-point RT-PCR of RNA isolated from total kidney samples, using primer pair F3/R3. We were able to detect the presence of excised Dmxl1 RNA in Dmxl1fl/fl, B1-Cre+ mice (lower band, Figure 1D) by end-point PCR, as well as the wild-type, non-excised RNA that was still present in the kidneys of these mice (upper band, Figure 1D). However, we could not detect significant Dmxl1 knockdown at the RNA or protein level, using total kidney samples of Dmxl1fl/fl, B1-Cre+ compared to Dmxl1fl/fl, B1-Cre− mice by qPCR or Western blotting (Figure S1). Similar results were previously reported for the gene coding for the BK channel α subunit (BKα) that was conditionally knocked down using B1-Cre mediated excision.52 The inability to detect knockdown of BKα was explained by the fact that BKα expression is not restricted to ICs. Therefore, mild knockdown in ICs, which constitute only a very small proportion of all cells in the kidney, is masked by the unaltered gene expression in the other cell types.52 Similarly, while Dmxl1 expression in the kidney is highest in ICs, it is not restricted to them. Dmxl1 is widely expressed in all tubules of mouse kidney53 (https://esbl.nhlbi.nih.gov/MRECA/Nephron/), which may explain the fact that we were not able to detect its knockdown in Dmxl1fl/fl, B1-Cre+ mice using total kidney lysate preparations (Figure S1). However, in subsequent experiments, we were able to show that Dmxl1 expression was reduced in kidney ICs, as described in the next sections.
Dmxl1 is Depleted in A-Intercalated Cells from Dmxl1fl/fl, B1-Cre+ Mice
Since we were unable to see a reduction in Dmxl1 in total kidney lysates from Dmxl1fl/fl, B1-Cre+ mice (Figure S1), likely due to incomplete knockout and expression of Dmxl1 in other cells in the kidney, we confirmed the reduction in Dmxl1 expression at the protein level in isolated ICs from Dmxl1fl/fl, B1-Cre+ mice. ICs were isolated from a single cell suspension prepared from total mouse kidney homogenate using MACS using c-kit/CD117 labeled beads, which is highly differentially expressed at the plasma membrane of ICs.54 We ran the input (before MACS sorting), flow through (cells not attached to magnetic beads), and elution (ICs isolated with c-kit beads) fractions on an sodium dodecyl-sulfate polyacrylamide gel electrophoresis (SDS-PAGE) gel and performed western blotting with antibodies against Dmxl1 and B1 V-ATPase (Figure 2A). This revealed a significant decrease in the amount of Dmxl1 in the elution fraction, which is made up of the c-kit labeled ICs, from the Cre+ mice relative to the Cre− mice, indicating that indeed there is a knockdown of Dmxl1 in the ICs of these mice. However, there is not a complete knockdown, as shown by the continued presence of a Dmxl1 band in the Cre+ elution fraction (Figure 2A and B). Importantly, levels of the IC-specific B1 subunit of the V-ATPase were greatly increased in the elution fractions indicating that we were indeed enriching ICs during our MACS preparation (Figure 2A and C).
Figure 2.
Dmxl1 fl/fl, B1-Cre+ mice have decreased expression of Dmxl1 in intercalated cells (ICs) isolated by magnetic activated cell sorting (MACS) from total kidney. (A) Western blot of lysates from Cre− and Cre+ mice prepared from a single cell suspension prepared from total kidney prior to MACS (input, Inp), from cells not bound to CD117/c-Kit beads (flow through, FT), and from ICs isolated by MACS using CD117/c-Kit microbeads (elution, Elu) probed with specific antibodies against Dmxl1 (top), B1 V-ATPase (middle), and β-Actin (bottom). (B) Quantification of Dmxl1 band intensity in Cre− and Cre+ mice normalized to β-Actin band intensity shows significantly reduced expression of Dmxl1 in ICs isolated from Cre+ mice. (C) Quantification of B1 V-ATPase band intensity in all mice normalized to β-Actin band intensity shows that B1-V-ATPase subunit, which is expressed mainly in ICs, was significantly enriched in eluted cells. Data were analyzed by t-test with values reported as means ± SEM with P-values ≤ .05 considered significant, with * denoting a P-value ≤ .05 and *** ≤ .0001.
Intercalated Cell Specific Knockdown of Dmxl1 in Mouse Kidney Results in Incomplete dRTA, Which is More Severe in Male Than in Female Mice
Both male and female Dmxl1fl/fl, B1-Cre+ mice were viable and fertile without obvious morphological or physiological abnormalities. In spite of an incomplete knockdown of Dmxl1 in ICs of Dmxl1fl/fl, B1-Cre+ mice, urine pH was significantly higher in Dmxl1fl/fl, B1-Cre+ than in Dmxl1fl/fl, B1-Cre− control mice even on a standard diet (pH 6.50 ± 0.09 vs. pH 5.96 ± 0.05 in males, Figure 3 and Table 1; and pH 6.28 ± 0.08 vs. pH 5.92 ± 0.05 in females; Figure S2 and Table S1). For both males and females on the standard diet, there was no significant difference between Dmxl1fl/fl, B1-Cre+ and Dmxl1fl/fl, B1-Cre− mice in venous blood pH, HCO3−, or pCO2 levels (Figure 3, Table 1, Figure S2, Table S1). In addition, on a standard diet, concentrations of Na+, Cl−, Ca2+, and K+ were not significantly different between genotypes in either male or female mice (Figure 4, Table 2, Figure S3, Table S2). Thus, Dmxl1fl/fl, B1-Cre+ mice did not develop acidemia on the standard diet, despite their elevated urinary pH. These results are similar to data previously reported for V-ATPase B1 subunit knockout mice.55
Figure 3.
Dmxl1 fl/fl, B1-Cre+ male mice demonstrate a reduced ability to maintain acid-base homeostasis after an ammonium chloride challenge. (A) Blood pH in Dmxl1fl/fl, B1-Cre+ male mice is not significantly different from Dmxl1fl/fl, B1-Cre− control males on a standard rodent diet. Both Dmxl1fl/fl, B1-Cre+ and Dmxl1fl/fl, B1-Cre− control male mice developed acidemia after acid loading with 0.28 M NH4Cl for 3 d. Note, that acidemia is more pronounced in Dmxl1fl/fl, B1-Cre+ males and their blood pH is significantly lower than in Dmxl1fl/fl, B1-Cre− controls. Blood pH did not change significantly after alkali loading with 0.28 M NaHCO3 for 3 d in either Dmxl1fl/fl, B1-Cre+ or Dmxl1fl/fl, B1-Cre− males. (B) Blood HCO3− levels were similar in Dmxl1fl/fl, B1-Cre+ and Dmxl1fl/fl, B1-Cre− males maintained on a standard diet. After acid loading, blood HCO3− levels were significantly reduced in mice of both genotypes to a similar degree. After alkali loading, blood HCO3− levels did not change significantly in either Dmxl1fl/fl, B1-Cre+ or Dmxl1fl/fl, B1-Cre− males. (C) Blood partial pressure of carbon dioxide (pCO2) was similar in Dmxl1fl/fl, B1-Cre+ and Dmxl1fl/fl, B1-Cre− males maintained on a standard diet. After acid loading, blood pCO2 levels were significantly reduced in Dmxl1fl/fl, B1-Cre−, but not in Dmxl1fl/fl, B1-Cre+ males, suggesting that the respiratory compensation to acidosis is weakened in Dmxl1fl/fl, B1-Cre+ in comparison with Dmxl1fl/fl, B1-Cre− males. After alkali loading, blood pCO2 levels did not change significantly in either Dmxl1fl/fl, B1-Cre+ or Dmxl1fl/fl, B1-Cre− males. (D) Urine pH was significantly higher in Dmxl1fl/fl, B1-Cre+ males compared to Dmxl1fl/fl, B1-Cre− controls on a standard diet. Acid loading resulted in a more acidic urine pH in Dmxl1fl/fl, B1-Cre+, but not in Dmxl1fl/fl, B1-Cre− control males, nevertheless urine pH remained significantly higher in Dmxl1fl/fl, B1-Cre+ males than in controls. Alkali loading resulted in a significantly higher alkaline urine pH in Dmxl1fl/fl, B1-Cre−, but not in Dmxl1fl/fl, B1-Cre+ males. Data were analyzed by t-test with values reported as means ± SEM with P-values ≤ .05 considered significant, with * denoting a P-value ≤ .05, ** ≤ .01, *** ≤ .001, and **** ≤ .0001. NS = non-significant. The actual P-values and other details can be found in Tables 1 and 2. The results comparing the same parameters in female mice are shown in Figure S2 and detailed in Tables S1 and S2.
Table 1.
Summary of NH4Cl—Loading of Male Mice
| Standard diet | Acid load | P (B1-Cre− standard vs. B1-Cre− acidified) | P (B1-Cre+ standard vs. B1-Cre+ acidified) | |||||
|---|---|---|---|---|---|---|---|---|
| Dmxl1 fl/fl, B1-Cre− | Dmxl1 fl/fl, B1-Cre+ | P (B1-Cre− vs. B1-Cre+) | Dmxl1 fl/fl, B1-Cre− | Dmxl1 fl/fl, B1-Cre+ | P (B1-Cre− vs. B1-Cre+) | |||
| Venous pH | 7.33 ± 0.01 (n = 13) | 7.32 ± 0.01 (n = 12) | 0.4705 | 7.26 ± 0.02 (n = 8) | 7.19 ± 0.02 (n = 8) | .0345 (*) | .0067 (**) | <.0001 (****) |
| pCO2, (mmHg) | 43.39 ± 1.15 (n = 12) | 42.41 ± 1.09 (n = 12) | 0.5404 | 39.28 ± 1.43 (n = 8) | 43.5 ± 1.03 (n = 8) | .0307 (*) | .0370 (*) | 0.4976 |
| HCO3−(mM) | 22.40 ± 0.70 (n = 13) | 21.11 ± 0.43 (n = 12) | 0.1344 | 16.94 ± 0.66 (n = 8) | 15.93 ± 0.85 (n = 8) | 0.3638 | <.0001 (****) | <.0001 (****) |
| Na+ (mM) | 149.83 ± 0.61 (n = 12) | 149.92 ± 0.4 (n = 12) | 0.9103 | 155.37 ± 0.98 (n = 8) | 161.00 ± 1.43 (n = 8) | .0058 (**) | <.0001 (****) | <.0001 (****) |
| K+ (mM) | 5.95 ± 0.21 (n = 13) | 5.84 ± 0.26 (n = 11) | 0.7406 | 5.76 ± 0.30 (n = 8) | 5.75 ± 0.16 (n = 8) | 0.9707 | 0.5905 | 0.7812 |
| Ca2+ ionized (mg/dL) | 5.11 ± 0.03 (n = 12) | 5.05 ± 0.05 (n = 12) | 0.3396 | 5.44 ± 0.05 (n = 8) | 5.72 ± 0.07 (n = 8) | .0039 (**) | <.0001 (****) | <.0001 (****) |
| Cl− (mM) | 110.25 ± 0.74 (n = 12) | 110.92 ± 1.13 (n = 12) | 0.6267 | 117.13 ± 1.33 (n = 8) | 123.00 ± 1.41 (n = 8) | .0090 (**) | .0001 (***) | <.0001 (****) |
| Urine pH | 5.96 ± 0.05 (n = 17) | 6.50 ± 0.09 (n = 17) | <.0001 (****) | 5.81 ± 0.01 (n = 8) | 5.95 ± 0.05 (n = 8) | .0124 (*) | 0.0574 | .0005 (***) |
Notes: Venous blood gases, blood pH, major electrolytes, and urine pH of Dmxl1fl/fl, B1-Cre+ conditional knockout and Dmxl1fl/fl, B1-Cre− control male mice on a standard rodent diet and after acid loading with 0.28 M NH4Cl for 3 d. All values are means ± SEM. * is for P-value ≤ .05, ** is for P-value ≤ .01, *** is for P-value ≤ .001, **** is for P-value ≤ .0001; data analyzed by t-test.
Figure 4.
Dmxl1 fl/fl, B1-Cre+ male mice demonstrate a reduced ability to maintain Na+, Cl−, and Ca2+ homeostasis when challenged with ammonium chloride, while K+ homeostasis is preserved. (A) Na+ concentration in Dmxl1fl/fl, B1-Cre+ male mice is not significantly different from Dmxl1fl/fl, B1-Cre− control males on a standard rodent diet. Both Dmxl1fl/fl, B1-Cre+ and Dmxl1fl/fl, B1-Cre− control male mice developed hypernatremia after acid loading with 0.28 M NH4Cl for 3 d, and it was more pronounced in Dmxl1fl/fl, B1-Cre+ males than in Dmxl1fl/fl, B1-Cre− controls. After alkali loading with 0.28 M NaHCO3 for 3 d, Na+ concentration was significantly increased in Dmxl1fl/fl, B1-Cre−, but not in Dmxl1fl/fl, B1-Cre+ males. (B) Cl− concentration in Dmxl1fl/fl, B1-Cre+ male mice is not significantly different from Dmxl1fl/fl, B1-Cre- control males on a standard rodent diet. Both Dmxl1fl/fl, B1-Cre+ and Dmxl1fl/fl, B1-Cre− control male mice developed hyperchloremia after acid loading with 0.28 M NH4Cl for 3 d, and it was more pronounced in Dmxl1fl/fl, B1-Cre+ males than in Dmxl1fl/fl, B1-Cre− controls. Cl− concentration did not change significantly after alkali loading with 0.28 M NaHCO3 for 3 d in either Dmxl1fl/fl, B1-Cre+ or Dmxl1fl/fl, B1-Cre− males. (C) Free Ca2+ concentration in Dmxl1fl/fl, B1-Cre+ male mice is not significantly different from Dmxl1fl/fl, B1-Cre− control males on a standard rodent diet. Both Dmxl1fl/fl, B1-Cre+ and Dmxl1fl/fl, B1-Cre− control male mice developed hypercalcemia after acid loading with 0.28 M NH4Cl for 3 d, and it was more pronounced in Dmxl1fl/fl, B1-Cre+ males than in Dmxl1fl/fl, B1-Cre− controls. Free Ca2+ concentration did not change significantly after alkali loading with 0.28 M NaHCO3 for 3 d in either Dmxl1fl/fl, B1-Cre+ or Dmxl1fl/fl, B1-Cre− males. (D) K+ concentration in Dmxl1fl/fl, B1-Cre+ male mice is not significantly different from Dmxl1fl/fl, B1-Cre− control males on a standard rodent diet. K+ concentration did not change significantly after acid loading with 0.28 M NH4Cl for 3 d or after alkali loading with 0.28 M NaHCO3 for 3 d in either Dmxl1fl/fl, B1-Cre+ or Dmxl1fl/fl, B1-Cre− males and there was no statistically significant difference between genotypes. Data were analyzed by t-test with values reported as means ± SEM with P-values ≤ .05 considered significant, with * denoting a P-value ≤ .05, ** ≤ .01, *** ≤ .001, and **** ≤ .0001. NS = non-significant. The actual P-values and other details can be found in Tables 1 and 2. The results comparing the same parameters in female mice are shown in Figure S3 and detailed in Tables S1 and S2.
Table 2.
Summary of NaHCO3—Loading of Male Mice
| Alkali load | P (B1-Cre− standard (from Table 1) vs. B1-Cre− alkalinized) | P (B1-Cre+ standard (from Table 1) vs. B1-Cre+ alkalinized) | |||
|---|---|---|---|---|---|
| Dmxl1 fl/fl, B1-Cre− | Dmxl1 fl/fl, B1-Cre+ | P (B1-Cre− vs. B1-Cre+) | |||
| Venous pH | 7.34 ± 0.04 (n = 4) | 7.35 ± 0.01 (n = 3) | 0.8963 | 0.8417 | 0.2799 |
| pCO2 (mmHg) | 44.98 ± 2.72 (n = 4) | 45.47 ± 1.41 (n = 3) | 0.8916 | 0.5375 | 0.2094 |
| HCO3− (mM) | 23.85 ± 2.03 (n = 4) | 24.10 ± 1.00 (n = 3) | 0.9254 | 0.3933 | .0092 (**) |
| Na+ (mM) | 153.25 ± 0.85 (n = 4) | 152.00 ± 1.53 (n = 3) | 0.4776 | .0117 (*) | 0.0706 |
| K+ (mM) | 6.6 ± 0.43 (n = 4) | 5.78 ± 0.19 (n = 3) | 0.1878 | 0.1655 | 0.9137 |
| Ca2+ ionized (mg/dL) | 5.04 ± 0.11 (n = 4) | 5.23 ± 0.04 (n = 3) | 0.2473 | 0.4414 | 0.1336 |
| Cl− (mM) | 109.00 ± 1.91 (n = 4) | 108.67 ± 0.88 (n = 3) | 0.8942 | 0.4649 | 0.3575 |
| Urine pH | 7.08 ± 0.32 (n = 5) | 6.74 ± 0.21 (n = 5) | 0.4026 | <.0001 (****) | 0.2396 |
Notes: Venous blood gases, blood pH, major electrolytes, and urine pH of Dmxl1fl/fl, B1-Cre+ conditional knockout and Dmxl1fl/fl, B1-Cre− control male mice on a standard rodent diet and after alkali loading with 0.28 M NaHCO3 for 3 d. All values are means ± SEM. * is for P-value ≤ .05, ** is for P-value ≤ .01, *** is for P-value ≤ .001, **** is for P-value ≤ .0001; data analyzed by t-test.
To study the ability of these mice to maintain systemic acid-base homeostasis during both an acid and alkali challenge, they were given either 0.28 M NH4Cl or 0.28 M NaHCO3 in their drinking water for 3 d. When Dmxl1fl/fl, B1-Cre+ and Dmxl1fl/fl, B1-Cre− mice were challenged with NH4Cl, urine pH was reduced in mice of both genotypes, as expected (Figure 3, Table 1, Figure S2, Table S1). However, urine pH stayed significantly higher in Dmxl1fl/fl, B1-Cre+ than in Dmxl1fl/fl, B1-Cre− control mice (pH 5.95 ± 0.05 vs. pH 5.81 ± 0.01 in males, Figure 3 and Table 1; and pH 5.85 ± 0.05 vs. pH 5.65 ± 0.06 vs. in females; Figure S2 and Table S1), suggesting an inability of Dmxl1fl/fl, B1-Cre+ conditional KO mice to maximally acidify their urine under acid challenged conditions, a hallmark of dRTA.13 In addition, after the NH4Cl challenge both Dmxl1fl/fl, B1-Cre+ and Dmxl1fl/fl, B1-Cre− male mice developed acidemia and hypobicarbonatemia and there was a significant difference between genotypes in blood pH (pH 7.19 ± 0.02 in Dmxl1fl/fl, B1-Cre+ males vs. pH 7.26 ± 0.02 in Dmxl1fl/fl, B1-Cre− males, Figure 3 and Table 1). After treatment with NH4Cl, Dmxl1fl/fl, B1-Cre− but not Dmxl1fl/fl, B1-Cre+ male mice had significantly lower pCO2 levels when compared to the untreated controls (39.28 ± 1.43 vs. 43.39 ± 1.15 mmHg), suggesting respiratory compensation for acidosis occurred only in the Cre− mice (Figure 3 and Table 1). Unlike in male mice, there was no significant difference in blood pH between acid-challenged Dmxl1fl/fl, B1-Cre+ and Dmxl1fl/fl, B1-Cre− female mice, although blood pH was slightly, but significantly, reduced only in Dmxl1fl/fl, B1-Cre− females in response to NH4Cl treatment (pH 7.33 ± 0.02 vs pH 7.25 ± 0.03) (Figure S2 and Table S1). Moreover, neither Dmxl1fl/fl, B1-Cre− nor Dmxl1fl/fl, B1-Cre+ female mice developed hypobicarbonatemia and there was no change in pCO2 levels in response to the acid challenge (Figure S2 and Table S1).
The NH4Cl-induced acidemia was accompanied by significant increase of Na+ and Cl− in the blood of male, and to a lesser degree, female mice of both genotypes (Figure 4, Table 1, Figure S3, Table S1). The increase in these electrolytes in mice in response to NH4Cl treatment is a known phenomenon, which we have observed previously.56 The difference between genotypes was significant only in male mice with Dmxl1fl/fl, B1-Cre+ males having a higher concentration of Na+ (161.00 ± 1.43 vs. 155.37 ± 0.98 mM, Figure 4 and Table 1) and Cl− (123.00 ± 1.41 vs. 117.13 ± 1.33 mM, Figure 4 and Table 1) than Dmxl1fl/fl, B1-Cre− males. After NH4Cl treatment, the ionized (free) Ca2+ was also significantly increased in the blood of males of both genotypes (5.44 ± 0.05 vs. 5.11 ± 0.03 mg/dL in Dmxl1fl/fl, B1-Cre− males; and 5.72 ± 0.07 vs. 5.05 ± 0.05 mg/dL in Dmxl1fl/fl, B1-Cre+ males; Figure 4 and Table 1), but not female mice (Figure S3 and Table S1). In addition, the Ca2+ concentration in the blood was significantly higher in acid-treated Dmxl1fl/fl, B1-Cre+ than in acid-treated Dmxl1fl/fl, B1-Cre− male mice (5.72 ± 0.07 vs. 5.44 ± 0.05 mg/dL, Figure 4 and Table 1). The observed hypercalcemia in males was probably a consequence of significant acidemia and calcium leakage from bones under lower-than-normal blood pH. Lastly, we did not detect a significant effect of NH4Cl treatment on the K+ concentration in blood of either male or female mice of either genotype (Figure 4 and Table 1, Figure S3 and Table S1). Finally, treatment with 0.28 M NaHCO3 for 3 d as an alkali loading challenge did not reveal any significant differences in the measured parameters between Dmxl1fl/fl, B1-Cre+ and Dmxl1fl/fl, B1-Cre− control mice in either males or females (Figures 3 and 4, Figure S2 and S3, Tables 2 and S2).
In summary, the NH4Cl challenge revealed a phenotype of incomplete dRTA in both male and female Dmxl1fl/fl, B1-Cre+ mice, although it was more severe in male than in female mice. In Dmxl1fl/fl, B1-Cre+ males, acidemia was also accompanied by significantly elevated levels of Na+, Cl−, and Ca2+ in blood relative to Dmxl1fl/fl, B1-Cre− males suggesting that they are less capable of maintaining their acid-base and electrolyte homeostasis.
Expression of the Intercalated Cell-Specific B1 Subunit of the V-ATPase is Decreased in Dmxl1fl/fl, B1-Cre+ Mice
Here, we focused on A-ICs, since dysfunction of these cells results in acidosis,13 which is what we observe in our Dmxl1 knockdown mice (Figures 3 and S2). To visualize expression of Dmxl1 protein in A-ICs, we performed indirect immunofluorescence using specific antibodies against Dmxl1 and the B1 V-ATPase. A-ICs were positively identified by their expression of B1 V-ATPase and their localization to the renal medulla. As shown in Figure 5A, Dmxl1 colocalizes with the B1 subunit of the V-ATPase at the apical pole of A-ICs in Dmxl1fl/fl, B1-Cre− control mice. This is perhaps not surprising given our previous findings that Dmxl1 interacts with the B1 subunit by coimmunoprecipitation from total mouse kidney lysates.14 Furthermore, immunostaining showed that indeed there was a specific depletion of Dmxl1 in B1-positive A-ICs from kidneys from Dmxl1fl/fl, B1-Cre+ mice relative to Cre− littermate controls, although some fluorescent signal is still apparent in the knockout ICs (Figure 5A). This remaining signal could be explained by incomplete recombination or excision of the LoxP sites flanking the Dmxl1 gene by Cre recombinase. This aligns with our PCR results showing incomplete recombination in the Dmxl1fl/fl, B1-Cre+ mice (Figure 1D), which would suggest incomplete penetrance of Cre function, as well as our MACS experiments, which showed an incomplete knockdown of Dmxl1 protein in ICs (Figure 2). This interpretation is further supported by our analysis of Dmxl1 and B1 fluorescence intensity by immunofluorescence. When quantifying average cellular fluorescence intensity by animal, we observe a slight, but not significant, trend toward decreased Dmxl1 expression in the A-ICs from Cre+ animals (Figure S4A). On the other hand, we do see a significant reduction in B1 fluorescence intensity in the A-ICs from Cre+ animals relative to controls, which we do not observe in B-ICs (Figure S4B and C). Importantly, however, when we performed a single cell analysis of A-ICs from the same images, the fluorescence intensity of Dmxl1 per cell showed a strong negative linear correlation with the fluorescence intensity of B1 (R2 = 0.72 for Cre− and 0.82 for Cre+). Thus, when there is decreased Dmxl1 intensity in a given cell, there is a corresponding and proportionate decrease in B1 intensity in the same cell in both control and Dmxl1 knockdown A-ICs (Figure 5B). Furthermore, whereas more than 80% of A-ICs from control animals had a B1 intensity of more than 1.0 × 105 relative fluorescence units (RFU), 60% of A-ICs from knockdown animals had B1 fluorescence below this threshold level (Figure 5B, shaded area). We did not observe a similar difference in the distribution of B1 fluorescence intensity by cell in pendrin positive B-ICs from control versus knockdown animals (Figure S4C). This strongly suggests that the reduction in B1 fluorescent intensity in A-ICs as seen by immunofluorescence is indeed due to decreased Dmxl1 expression and is variable from cell to cell.
Figure 5.
Dmxl1 fl/fl, B1-Cre+ male mice have decreased expression of Dmxl1, the IC-specific B1, and the ubiquitous A subunit of V-ATPase in A-type intercalated cells (A-ICs). (A) Immunofluorescence images taken from the outer medulla of kidneys from Cre− (top) and Cre+ (bottom) mice stained with specific antibodies against Dmxl1 (left) and the B1 V-ATPase (center). Both B1 and Dmxl1 staining are apically polarized in IC from Cre− mice, whereas they are distributed throughout the cytoplasm of IC from Cre+ mice. Some background fluorescence, including an apparent nucleolar pattern, is seen when using the Dmxl1 antibody, which is likely due to heat-mediated antigen retrieval in a low pH citrate buffer. B1 staining was used to identify A-ICs. Nuclei are labeled with DAPI (right). Tubule lumen indicated with an asterisk (*). Scale bars = 10 µm. (B) X–Y plot of corrected total cell fluorescence (CTCF) of Dmxl1 and B1 by cell with a linear regression analysis showing that B1 CTCF has a linear correlation with Dmxl1 CTCF. Shaded area represents a B1 intensity of less than 1.0 × 105 relative fluorescence units (RFU), which encompasses 60% of A-ICs from knockdown animals and only 20% of A-ICs from control animals. (C) Western blots of total kidney lysates prepared from Cre− and Cre+ male mice probed with specific antibodies against the B1 and A subunits of the V-ATPase and β-Actin. (D) Quantification of B1 and A V-ATPase band intensities normalized to β-Actin band intensity shows a significant reduction of B1 and A subunit expression in total kidney lysates from Cre+ male mice. Data were analyzed by t-test with values reported as means ± SEM with P-values ≤ .05 considered significant, with * denoting a P-value ≤ .05.
To confirm that knockdown of Dmxl1 in ICs affects the protein expression levels of the B1 V-ATPase, we performed SDS-PAGE electrophoresis followed by Western blotting of total kidney lysates. This revealed a significant decrease in B1 V-ATPase expression in both males (Figure 5C and D) and females (Figure S5B). Interestingly, we saw a significant decrease in the ubiquitous A subunit of the V1 domain in males, but not in females (Figure 5C-D and S5B), and not in the IC-specific VO subunit a4, other ubiquitous V-ATPase subunits, or carbonic anhydrase in either sex (Figure S5). This is intriguing since the B1 and A subunits are part of the cytosolic V1 domain, while a4 is part of the membrane bound VO domain. This suggests that the two domains of the V-ATPase are not affected equally by the loss of Dmxl1. However, the decrease in B1 expression alone could explain the inability of these mice to properly acidify their urine.
V-ATPase is Localized Intracellularly in A-Intercalated Cells Lacking Dmxl1
In A-ICs, the V-ATPase traffics between a pool of subapical vesicles and the apical plasma membrane in response to physiological cues, thereby adjusting the amount of V-ATPase holoenzyme at the cell surface, and thus regulating proton secretion into the urine. To examine whether loss of Dmxl1 affects the apical localization of the V-ATPase we used indirect immunofluorescence to examine the relative localization of the IC-specific B1 and a4 subunits of the V-ATPase in A-ICs from Dmxl1fl/fl, B1-Cre− and Dmxl1fl/fl, B1-Cre+ littermate mice (Figure 6). We quantified the percent apical localization of the V-ATPase subunits using a line intensity scan in FIJI. This revealed a significantly greater intracellular distribution with very little apical localization of both the B1 and a4 subunits of the V-ATPase in A-ICs from knockdown mice relative to control mice, which had a higher percentage of apically localized V-ATPase (Figure 6A and B). Of note, we did not observe a significant change in the localization of the B1 subunit of the V-ATPase in pendrin-positive B-ICs (Figure S6A). Furthermore, when we quantified colocalization of the B1 and a4 subunits in A-ICs using the Pearson’s coefficient of correlation we observed a significant decrease in colocalization of the B1 subunit, which is part of the V1 domain of the V-ATPase, and a4, which is part of the VO domain in Cre+ mice relative to Cre− mice (Figure 6C). These data show that Dmxl1 is likely required for the assembly of the VO and V1 domains of the V-ATPase, similar to what has been reported in yeast with the Dmxl1 homologue, Rav1p.17
Figure 6.
The B1 and a4 subunits of the V-ATPase are localized intracellularly and are less colocalized in A-type intercalated cells (A-ICs) from Dmxl1fl/fl, B1-Cre+ mice. (A) Immunofluorescence images taken from the outer medulla of kidneys from Cre− (top) and Cre+ (bottom) mice stained with specific antibodies against the V1 domain-associated B1 subunit of the V-ATPase (left) and the VO domain-associated a4 subunit of the V-ATPase (center). B1 and a4 are apically polarized in cells from Cre− mice but show a more diffuse intracellular distribution in A-ICs from Cre+ mice. V-ATPase staining was used to identify A-ICs. Nuclei are labeled with DAPI (right). Tubule lumen is indicated with an asterisk (*). Scale bars = 5 µm. (B) Quantification of % apical localization of B1 and a4 subunits relative to total cell localization shows significantly less apical polarization in Cre+ A-ICs. (C) Pearson’s coefficient of colocalization of the B1 and a4 subunits, as an estimate of domain assembly, shows significantly less colocalization in A-ICs from Cre+ mice. Data were analyzed by t-test with values reported as means ± SEM with P-values ≤ .05 considered significant, with * denoting a P-value ≤ .05, *** ≤ .0001, and **** ≤ .00001.
Loss of Dmxl1 Prevents Assembly of the V-ATPase Holoenzyme
To confirm that loss of Dmxl1 prevents assembly of the VO and V1 domains of the V-ATPase we used high speed centrifugation to prepare cytosolic and membrane fractions from whole kidney lysates from male Dmxl1fl/fl, B1-Cre− and Dmxl1fl/fl, B1-Cre+ mice (Figure 7A). We examined the distribution of the cytosolic B1 subunit (V1 domain), which must translocate to the membrane and assemble with the membrane-bound a4 subunit (VO domain). We observed a significant decrease in the amount of B1 subunit in the membrane fraction (mem) relative to the cytosolic fraction (cyto) from the Dmxl1fl/fl, B1-Cre+ kidney lysates indicating that less of the V1-associated B1 is assembling with the membrane bound a4 from the VO domain (Figure 7B). The presence of the membrane bound a4 subunit only in the membrane fraction confirms the purity of preparation (Figure 7A, bottom). Furthermore, we performed a proximity ligation assay (PLA) that detects interactions of two antibody-labeled proteins if they are within 40 nm of each other (Figure 7C). We labeled kidney sections with specific antibodies against the B1 (V1) and a4 (VO) subunits of the V-ATPase. Thus, a successful PLA reaction, which appears as a distinct punctum, should only be seen when the two domains are assembled. We quantified the area of PLA puncta per A-IC, which we identified by basolateral staining with an AE1 antibody (Figure 7C and D). As expected, we observed significantly more area of PLA puncta in the Cre− mice relative to the Cre+ mice. Taken together, these results demonstrate that Dmxl1 acts as an assembly factor for the two domains of the V-ATPase.
Figure 7.
The V1 and VO domains of the V-ATPase are less assembled in the kidneys of Dmxl1fl/fl, B1-Cre+ mice. (A) Western blots of subcellular fractionation experiments showing decreased membrane (mem) accumulation of the cytosolic V1-associated B1 subunit of the V-ATPase in Cre+ mice relative to Cre− mice (top). The membrane-bound VO-associated a4 subunit is only detected in the membrane fraction, as expected, confirming the purity of the preparation (bottom). (B) Quantification of the membrane: cytosol ratio of the cytosolic V1-associated B1 subunit of the V-ATPase. (C) Immunofluorescence images taken from the outer medulla of kidneys from Cre− (top) and Cre+ (bottom) mice after performing a proximity ligation assay (PLA, spots, center) using specific antibodies against the B1 and a4 V-ATPase and post-staining with a specific antibody against AE1 (left). Association of B1 with a4 is represented by the increased number of spots in A-type intercalated cells (A-ICs) from Cre− mice. Basolateral AE1 staining was used to identify A-ICs (indicated with arrowheads). Nuclei are labeled with DAPI (right). Scale bars = 10 µm. (D) Quantification of total area of PLA signal per A-IC shows a significant decrease of B1/a4 association in Cre+ mice. Values for 25 cells were averaged per animal. Data were analyzed by t-test with values reported as means ± SEM with P-values ≤ .05 considered significant, with * denoting a P-value ≤ .05, and ** ≤ .001.
Loss of Dmxl1 Leads to Changes in A-Intercalated Cell Size and Morphology
When analyzing the localization of the V-ATPase in Dmxl1 knockdown A-ICs by immunofluorescence we observed A-ICs that appeared much smaller than the average A-IC as well as, A-ICs that appeared much larger than the average A-ICs that are seen in control animals (Figure 8A). We quantified cell size by measuring cell area (µm2) using FIJI and compared the cumulative distribution of cell sizes using a Kolmogorov–Smirnov test (Figure 8B). This analysis revealed a significantly different distribution of A-IC size in the Dmxl1fl/fl, B1-Cre+ mice relative to the Dmxl1fl/fl, B1-Cre− mice. There were more A-ICs with a very small area (∼40 µm2, indicated with #), as well as more A-ICs with a very large area (≥100 µm2, indicated with arrowheads) in the Cre+ mice relative to the Cre− mice (Figure 8B). Of note, there were no A-ICs with an area ≥ 140 µm2 in the control animals, while 12% of the A-ICs quantified in the Dmxl1fl/fl, B1-/Cre+ mice were ≥ 140 µm2 in area. Surprisingly, even though we saw no difference in B1 fluorescence intensity or localization in B-ICs we saw a significantly smaller population of B-ICs in the knockdown animals, but no population of B-ICs that were bigger in knockdown animals, relative to control animals (Figure S6B).
Figure 8.
A-type intercalated cell (A-IC) size is affected in Dmxl1fl/fl, B1-Cre+ mice. (A) Immunofluorescence images taken from the outer medulla (top) and inner medulla (bottom) of kidneys from Cre− and Cre+ mice stained with specific antibodies against the B1 V-ATPase (left) and a4 V-ATPase (center). Nuclei are labeled with DAPI (right). Scale bars = 10 µm. (B) Histogram of A-IC size distribution in Cre− and Cre+ mice. Data were analyzed by Kolmogorov–Smirnov test with a P-value of ≤ .05 considered significant. Smaller cells in Cre+ animals (found principally in the inner medulla) are indicated with a number sign, while larger cells in Cre+ animals (found principally in the outer medulla) are indicated with arrowheads. There are reduced numbers of cells of “normal” size (60-80 µm2) in the Cre+ mice.
Intriguingly, the populations of ICs at either size extreme showed unusual morphological characteristics. The cells that were ∼40 µm2 in the Cre+ mice, found primarily in the inner medulla, had only a narrow band of perinuclear cytoplasm and were very rounded in appearance, much like a neutrophil, rather than adopting the more elongated, tapered shape typically seen with epithelial ICs (Figure 8A, bottom). Despite this, they appeared to be polarized as demonstrated by anion exchanger 1 (AE1) staining, which localizes specifically to the basolateral membrane of A-ICs under normal conditions (Figure S7A). On the other hand, the cells ≥ 140 µm2 often exhibited remarkable V-ATPase positive apical projections, protrusions, and ruffling (Figure 9 and Figure S7B), although there was a very small subpopulation of the ∼40 µm2 cells that also presented with apical protrusions (Figure S7C). Intriguingly, labeling the F-actin cytoskeleton with Rhodamine-phalloidin revealed that these apical projections were not F-actin based (Figure 9A). However, when we stained the same tissues with an antibody against cortactin, which has been reported to colocalize with the a3 subunit of the V-ATPase at the leading edge of metastatic breast cancer cells,57 we saw that the apical protrusions were cortactin-positive (Figure 9B). We did not observe these morphological anomalies in B-ICs. We confirmed that these morphological changes were occurring at the apical domain, by co-staining with an antibody against anion exchanger 1 (AE1), which specifically localizes to the basolateral membrane of A-ICs (Figure S7D). Furthermore, these unusual cells, also, frequently had large intracellular vacuoles (Figure 9B). These unusual apical morphologies and vacuolization were readily apparent by TEM, but the identity of, and mechanism by which these unusual structures form is at present unknown (Figure 10 and Figure S7E and F) and will be the subject of future studies.
Figure 9.
A-type intercalated cells (A-ICs) from Dmxl1fl/fl, B1-Cre+ kidneys have intracellular vacuoles and phalloidin-negative, cortactin-positive apical protrusions. Immunofluorescence images taken from the outer (top) and inner medulla (bottom) of kidneys from Cre+ mice labeled with specific antibodies against the B1 V-ATPase (left) and with (A) the actin cytoskeleton labeled with Rhodamine-phalloidin (center) or (B) stained with a specific antibody against cortactin (center). Apical protrusions, indicated with arrowheads, are phalloidin-negative (A), but cortactin-positive (B). Large, perinuclear vacuoles are often present (B). Nuclei are labeled with DAPI (right). Tubule is lumen indicated with an asterisk (*). Scale bars = 2 µm (A) and 5 µm (B).
Figure 10.
Transmission electron micrographs showing ultrastructural details of morphological differences between A-type intercalated cells (A-ICs) from Dmxl1fl/fl, B1-Cre− and Dmxl1fl/fl, B1-Cre+ medullary collecting ducts. Low magnification overview of representative collecting ducts from Cre− (A) and Cre+ (C) mice. The black boxes in (A) and (C) indicate the area magnified in (B) and (D), respectively. High magnification images showing morphological appearance of A-ICs from Cre− (B) and Cre+ (D) mice. Note that the Cre+ ICs (arrowhead in C) have large intracellular vacuoles and large, electron dense intracellular vesicles (D, inset). Scale bars = 8 µm (A, C) and 2 µm (B, D).
Discussion
To examine the role of the V-ATPase interacting protein, Dmxl1, in the biology of this important proton pumping enzyme, we developed a novel Dmxl1 floxed mouse model, which is critical to understanding the function of this protein in mammals. A global Dmxl1 knockout mouse model was previously reported in an abstract from the American Society of Human Genetics meeting in 200950 to examine the proposed role of Dmxl1 missense mutations in the pathogenesis of Prader-Willi-like syndrome. However, the authors reported that “homozygous knockout mice are embryonic lethal, while heterozygous mice were noted to have hyperactivity and deficits in social memory.”50 This has not been studied further, and these results were not published in a peer-reviewed journal. We, therefore, generated our own Dmxl1 global knockout mouse model and confirmed that these mice are not viable, as detailed in the Methods section above. To assess the role of Dmxl1 in acid-secreting, kidney ICs, we then generated a kidney IC knockout mouse line using a mouse line in which Cre recombinase is driven by the B1-V-ATPase subunit promoter, which is specific to ICs in the kidney. The floxed Dmxl1 mice that we generated also have the potential to be bred with mouse lines expressing Cre recombinases driven by diverse promoters, thus allowing future researchers to interrogate the role of Dmxl1 in specific cells, tissues, or organs, in vivo. Using these mice, our main finding is that Dmxl1 is a mammalian V-ATPase assembly factor in vivo, similar to its yeast homolog, Rav1.17 Although there are reports of assembly factors in mammals that regulate the assembly of the VO domain38–44 our new data now identify Dmxl1 as a mammalian factor that catalyzes the assembly of the two domains of the V-ATPase into a functional proton pumping holoenzyme, a role that had been previously suggested based on data from cell culture work using lysosomal acidification to monitor V-ATPase activity.14,36
While Dmxl1 global knockout mice were not viable as discussed above, Apt6v1b1 (the V-ATPase B1 catalytic subunit) global knockout mice are viable, and generally healthy, due to functional compensation by the B1-subunit homologue, B2, the second isoform of the V-ATPase B subunit, Atp6v1b2.58 In contrast, despite high similarity, the Dmxl1 homologue Dmxl2 is apparently unable to compensate for the loss of Dmxl1 in mice. Thus, the Dmxl1 gene is essential in mice and probably other mammals, while Apt6v1b1 is not. The Dmxl1 global knockout phenotype is, however, as severe as the knockout of ubiquitously expressed subunits of the V-ATPase, such as Atp6v0c, which is, also, embryonic lethal.59 Urine pH in Apt6v1b1 knockout mice reached pH 7.17 ± 0.08 and remained at pH 6.4 after an acid load with 0.28 M NH4Cl, while in Dmxl1 conditional knockout mice urine pH was pH 6.50 ± 0.09 (pH 5.95 ± 0.05 after acid load).55 This relatively mild phenotype of incomplete renal tubular acidosis in Dmxl1 conditional knockout mice can be explained by the incomplete excision of the Dmxl1 floxed exons by the B1-driven Cre recombinase, resulting in the incomplete Dmxl1 knockout in ICs. While the specificity of B1-Cre recombinase localization is very high, as was confirmed by us and others, its efficiency is apparently relatively low as observed in the current study and elsewhere.51,52 The future development and use of more efficient Cre-recombinases may result in a stronger phenotype in the Dmxl1 conditional knockout mice.
In our previous proteomics studies, we identified Dmxl1 as a novel B1 V-ATPase interacting protein using co-immunoprecipitation and mass spectrometry. We also detected novel interactions of the V-ATPase with Dmxl2, the other mammalian Rbc3A isoform, and WDR7/Rabcn3b, the other subunit of the Rabconnectin-3 complex. Moreover, we demonstrated a functional role for Dmxl1 and WDR7 in regulating the acidification of lysosomes in cultured kidney cells, which is likely due to an impairment of holoenzyme assembly at the lysosomal membrane.14,36 In the case of Dmxl1 knockdown in kidney ICs, assembly of the V-ATPase holoenzyme is impaired at the apical plasma membrane, as shown by the increased intracellular localization of the V-ATPase and decreased colocalization of the two domains in our immunofluorescence and PLA studies. These results suggest that preventing assembly of the holoenzyme negatively impacts targeting of both subunits of the V-ATPase to the apical surface, which would explain why both B1 and a4 are more intracellular, although future studies would be necessary to confirm this. Overall, this would prevent protons from being extruded into the urine, thus explaining the inability of these mice to properly acidify their urine.
Although WDR7/Rbcn3b has been identified as another structural homolog of Rav1, and Sethi et al.60 and our lab14 have demonstrated that it is required for V-ATPase activity in mammalian cells, future studies will be needed to determine whether this is due to impaired V-ATPase assembly. Intriguingly, WDR72, the closest human homolog of WDR7, shows strong tissue-specific expression, with high levels in the kidney and bladder.61 At present, there is no indication that WDR72 interacts with the V-ATPase or the Rbcn3 complex, but mutations in WDR72 have been shown to lead to the development of dRTA in humans.62 This suggests a role in regulating the V-ATPase, potentially by acting as a functional homolog of Rbcn3b, but this remains to be determined. This and the fact that there are two Rbcn3a isoforms (Dmxl1 and Dmxl2) in humans and mice while there is only one isoform in some other eukaryotes, such as in Drosophila, raises the possibility that a variety of Rbcn3 complexes could be formed in humans with tissue, cell, or subcellular specificity in their function.
In this study, we analyzed both male and female mice and found that several of the physiological parameters that we measured were not statistically different in Dmxl1fl/fl, B1-Cre+ vs. Dmxl1fl/fl, B1-Cre− female mice in contrast to male mice. Of note, Cre− male mice had a significantly lower pCO2 after being acid challenged, while there was no apparent change in Cre+ male mice and this phenomenon was not seen in female mice. This could be due to an impaired ability of the Cre+ mice to use respiratory compensation to overcome their metabolic acidosis. It is possible that Dmxl1 regulates the assembly of the V-ATPase holoenzyme in the B1 V-ATPase expressing cells of the small airways of these animals where the V-ATPase is required for acidification of the airway surface liquid,63 which is required for normal respiratory function. This raises the interesting possibility that Dmxl1 regulates V-ATPase assembly in a variety of tissues that contain specialized V-ATPase rich cells, such as ICs in the kidney, clear cells in the epididymis, or ionocytes in the lung.64–66 Intriguingly, the physiological differences we observe between the male and female mice correlate with our cell biological observations. For example, the apical projections and the intracellular localization of V-ATPase subunits were more readily apparent and more dramatic in Cre+ male mice than in females, and in male mice (but not females), the ubiquitous A-subunit of the V-ATPase was also downregulated, in addition to the B1 subunit. However, it is unclear why Dmxl1fl/fl, B1-Cre+ female mice have a less severe phenotype than Dmxl1fl/fl, B1-Cre+ male mice in comparison with their corresponding B1-Cre− controls. But apparently upon acid challenge, both Dmxl1fl/fl, B1-Cre+ and Dmxl1fl/fl, B1-Cre− female mice are less susceptible to developing acidosis than male mice and can maintain their acid-base and electrolyte homeostasis better than males. We recently reported that upon dehydration, wild-type female mice maintain their water homeostasis better than wild-type male mice, but intriguingly, this advantage was lost in knockout Atp6v1b1−/− females, which behaved the same as knockout Atp6v1b1−/− males with respect to water homestasis.67 These results imply a complex relationship between water homeostasis and acid base balance and underscore the importance of studying kidney function in both male and female mice because there are clear sex-dependent differences in response to various physiological challenges.
Of note, in this study, we observed a specific depletion of the intercalated cell enriched V1-associated B1 subunit of the V-ATPase in both sexes, and of the A subunit in males, but not of any other intercalated cell-specific or ubiquitous V-ATPase subunits. We and others have reported that protein expression of V-ATPase subunits can be differentially regulated.58,68 Furthermore, in yeast the RAVE complex binds specifically to the V1 domain before catalyzing the recruitment to the membrane and assembly with the VO domain.17 In the absence of RAVE, or in our study Dmxl1, the V1 domain would not be bound and stabilized and likely would be targeted for degradation, which could explain why we see a consistent reduction only in V1-associated B1 expression. It is possible we are unable to see a reduction in most other V1-associated subunits because they are ubiquitously expressed, thus the change in expression in ICs would be masked by expression in other cell types in the kidney. However, we did see a significant decrease in the V1-associated A subunit in males, but not females, suggesting that indeed the expression of the subunits making up the V1 domain is more affected by loss of Dmxl1 than those in the VO domain.
We also observed obvious morphological changes in the Dmxl1 knockout A-intercalated cells, including many cells with a very small size and reduced cytoplasmic volume, as well as larger cells with unusual cortactin-positive apical protrusions. Many studies have reported the direct binding of the V-ATPase to filamentous (F) actin and found that loss of V-ATPase activity resulted in a reorganization of the actin cytoskeleton. Furthermore, cortactin has been associated with the V-ATPase positive leading-edge protrusions of metastatic breast cancer cells.57 Intriguingly, we did not observe F-actin in the apical protrusions in Dmxl1 knockout ICs, even though they were positive for cortactin and the V-ATPase, which can both bind to F-actin. How the absence of Dmxl1 leads to changes in cell morphology is a question to be addressed in future studies, perhaps using a more efficient Cre recombinase to allow a more severe knockout to be developed.
Finally, the phenotype resulting from loss of Dmxl1 in B-type ICs was less obvious relative to A-type ICs. Whereas, the A-ICs showed decreased V-ATPase fluorescence intensity, increased intracellular localization, decreased colocalization of subunits B1 and a4, and dramatically altered cell size and morphology, we observed a significant decrease only in cell size of B-ICs. Although, by RNA sequencing, Dmxl1 is expressed at similar levels in both A- and B-ICs, the B1 subunit of the V-ATPase is less expressed in B-ICs.54 Thus, it is likely that the B1 promoter is more active in A-ICs and would drive higher Cre expression leading to more robust excision of the floxed Dmxl1 gene and decreased protein expression relative to B-ICs. Moreover, Dmxl2 expression is much higher in B-ICs than A-ICs by RNA sequencing, which could play a partial compensatory role in the absence of Dmxl1. The lack of any detectable effect of Dmxl1 depletion on V-ATPase localization in B-ICs is reflected in the acid/base challenge studies in which we were unable to reveal any differences resulting from HCO3− loading between Dmxl1fl/fl, B1-Cre+ and Dmxl1fl/fl, B1-Cre− mice, in either males or females. B-ICs are responsible for distal HCO3− excretion in the kidney and appear to maintain this function in our mice, again possibly due to reduced Cre expression and Dmxl1 excision in B-ICs.
In conclusion, we generated a new floxed Dmxl1 mouse that successfully produced conditional kidney intercalated cell knockout mice when crossed with mice expressing Cre driven by the B1 V-ATPase subunit promoter in ICs. This allowed us to demonstrate that Dmxl1, the mammalian homolog of Rav1, acts as a V-ATPase holoenzyme assembly factor, the first to be functionally described in mammals in vivo. In the future, our novel mouse model can be used to generate conditional knockout mice in other organs, such as the brain. Both Dmxl1 and Dmxl2 are expressed in brain, and while Dmxl2 is known to play a role in brain function and mutations result in neurological diseases27,29 this remains to be determined for Dmxl1. Furthermore, our model could be harnessed to understand whether Rabconnectin complexes formed with Dmxl1 show tissue, cell, or compartment specific function, which could be mediated by binding of the complex to particular subunit isoforms of the V-ATPase, similar to the yeast RAVE complex.69 These insights could allow specific subpopulations of V-ATPases to be targeted, thus providing the groundwork for developing therapeutics against cancer and glaucoma, both of which have been linked to Dmxl1 dysfunction.
Supplementary Material
Acknowledgement
We are grateful to Dr. Susan Wall from Emory University for providing us with the B1-Cre mice for breeding, which were used to generate our knockout colony. A.F.E, M.M., and D.B. conceived and designed research; A.F.E., E.C.D., D.C., and M.M. performed experiments; A.F.E. and M.M. analyzed data; A.F.E., M.M., and D.B. interpreted results of experiments; A.F.E. and M.M. prepared figures; A.F.E. drafted manuscript; A.F.E., E.C.D., M.M., and D.B. edited and revised manuscript; A.F.E., E.C.D., D.C., M.M., and D.B. approved final version of manuscript.
Contributor Information
Amity F Eaton, Program in Membrane Biology and Division of Nephrology, Massachusetts General Hospital and Harvard Medical School, Boston, MA 02114, USA.
Elizabeth C Danielson, Program in Membrane Biology and Division of Nephrology, Massachusetts General Hospital and Harvard Medical School, Boston, MA 02114, USA.
Diane Capen, Program in Membrane Biology and Division of Nephrology, Massachusetts General Hospital and Harvard Medical School, Boston, MA 02114, USA.
Maria Merkulova, Program in Membrane Biology and Division of Nephrology, Massachusetts General Hospital and Harvard Medical School, Boston, MA 02114, USA.
Dennis Brown, Program in Membrane Biology and Division of Nephrology, Massachusetts General Hospital and Harvard Medical School, Boston, MA 02114, USA.
Funding
This work was supported by National Institutes of Health (NIH) Grants R01 DK121848 (D.B.) and T32 DK007540 (A.F.E). The Zeiss LSM 800 with Airyscan confocal was purchased using NIH Shared Instrumentation Grant OD021577 (D.B.). Additional support for the Program in Membrane Biology Microscopy Core comes from the Boston Area Diabetes and Endocrinology Research Center (DK135043) and the Massachusetts General Hospital (MGH) Center for the Study of Inflammatory Bowel Disease (DK043351).
Conflict of Interest
D.B. holds the position of Senior Consulting Editor for Function and was blinded from reviewing or making decisions for the manuscript.
Data Availability
All data underlying this article are available in the main text or the online supplementary materials.
References
- 1. Eaton AF, Merkulova M, Brown D. The H(+)-ATPase (V-ATPase): from proton pump to signaling complex in health and disease. Am J Physiol Cell Physiol. 2021;320(3):C392–C414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kane PM. Targeting reversible disassembly as a mechanism of controlling V-ATPase activity. Curr Protein Pept Sci. 2012;13(2):117–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Collins MP, Forgac M. Regulation and function of V-ATPases in physiology and disease. Biochim Biophys Acta Biomembr. 2020;1862(12):183341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Oot RA, Couoh-Cardel S, Sharma S, Stam NJ, Wilkens S. Breaking up and making up: the secret life of the vacuolar H(+)-ATPase. Protein Sci. 2017;26(5):896–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Forgac M. Vacuolar ATPases: rotary proton pumps in physiology and pathophysiology. Nat Rev Mol Cell Biol. 2007;8(11):917–929. [DOI] [PubMed] [Google Scholar]
- 6. Maxson ME, Grinstein S. The vacuolar-type H(+)-ATPase at a glance—more than a proton pump. J Cell Sci. 2014;127(Pt 23):4987–4993. [DOI] [PubMed] [Google Scholar]
- 7. Miranda KC, Karet FE, Brown D. An extended nomenclature for mammalian V-ATPase subunit genes and splice variants. PLoS One. 2010;5(3):e9531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sun-Wada GH, Wada Y. Vacuolar-type proton pump ATPases: roles of subunit isoforms in physiology and pathology. Histol Histopathol. 2010;25(12):1611–1620. [DOI] [PubMed] [Google Scholar]
- 9. Breton S, Brown D. Regulation of luminal acidification by the V-ATPase. Physiology (Bethesda). 2013;28(5):318–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Brown D, Smith PJ, Breton S. Role of V-ATPase-rich cells in acidification of the male reproductive tract. J Exp Biol. 1997;200(Pt 2):257–262. [DOI] [PubMed] [Google Scholar]
- 11. Brown D, Paunescu TG, Breton S, Marshansky V. Regulation of the V-ATPase in kidney epithelial cells: dual role in acid-base homeostasis and vesicle trafficking. J Exp Biol. 2009;212(Pt 11):1762–1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chu A, Zirngibl RA, Manolson MF. The V-ATPase a3 Subunit: structure, Function and therapeutic potential of an essential biomolecule in osteoclastic bone resorption. Int J Mol Sci. 2021;22(13):6934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wagner CA, Unwin R, Lopez-Garcia SC, Kleta R, Bockenhauer D, Walsh S. The pathophysiology of distal renal tubular acidosis. Nat Rev Nephrol. 2023;19(6):384–400. [DOI] [PubMed] [Google Scholar]
- 14. Merkulova M, Paunescu TG, Azroyan A, Marshansky V, Breton S, Brown D. Mapping the H(+) (V)-ATPase interactome: identification of proteins involved in trafficking, folding, assembly and phosphorylation. Sci Rep. 2015;5:14827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kraemer C, Weil B, Christmann M, Schmidt ER. The new gene DmX from Drosophila melanogaster encodes a novel WD-repeat protein. Gene. 1998;216(2):267–276. [DOI] [PubMed] [Google Scholar]
- 16. Seol JH, Shevchenko A, Shevchenko A, Deshaies RJ. Skp1 forms multiple protein complexes, including RAVE, a regulator of V-ATPase assembly. Nat Cell Biol. 2001;3(4):384–391. [DOI] [PubMed] [Google Scholar]
- 17. Smardon AM, Tarsio M, Kane PM. The RAVE complex is essential for stable assembly of the yeast V-ATPase. J Biol Chem. 2002;277(16):13831–13839. [DOI] [PubMed] [Google Scholar]
- 18. Sumner JP, Dow JA, Earley FG, Klein U, Jager D, Wieczorek H. Regulation of plasma membrane V-ATPase activity by dissociation of peripheral subunits. J Biol Chem. 1995;270(10):5649–5653. [DOI] [PubMed] [Google Scholar]
- 19. Kane PM. Disassembly and reassembly of the yeast vacuolar H(+)-ATPase in vivo. J Biol Chem. 1995;270(28):17025–17032. [PubMed] [Google Scholar]
- 20. McGuire CM, Forgac M. Glucose starvation increases V-ATPase assembly and activity in mammalian cells through AMP kinase and phosphatidylinositide 3-kinase/Akt signaling. J Biol Chem. 2018;293(23):9113–9123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sautin YY, Lu M, Gaugler A, Zhang L, Gluck SL. Phosphatidylinositol 3-kinase-mediated effects of glucose on vacuolar H+-ATPase assembly, translocation, and acidification of intracellular compartments in renal epithelial cells. Mol Cell Biol. 2005;25(2):575–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Stransky LA, Forgac M. Amino acid availability modulates vacuolar H+-ATPase assembly. J Biol Chem. 2015;290(45):27360–27369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bodzeta A, Kahms M, Klingauf J. The presynaptic V-ATPase reversibly disassembles and thereby modulates exocytosis but is not part of the fusion machinery. Cell Rep. 2017;20(6):1348–1359. [DOI] [PubMed] [Google Scholar]
- 24. Jaskolka MC, Winkley SR, Kane PM. RAVE and Rabconnectin-3 complexes as signal dependent regulators of organelle acidification. Front Cell Dev Biol. 2021;9:698190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fagerberg L, Hallstrom BM, Oksvold P et al. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol Cell Proteomics. 2014;13(2):397–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nagano F, Kawabe H, Nakanishi H et al. Rabconnectin-3, a novel protein that binds both GDP/GTP exchange protein and GTPase-activating protein for Rab3 small G protein family. J Biol Chem. 2002;277(12):9629–9632. [DOI] [PubMed] [Google Scholar]
- 27. Esposito A, Falace A, Wagner M et al. Biallelic DMXL2 mutations impair autophagy and cause Ohtahara syndrome with progressive course. Brain. 2019;142(12):3876–3891. [DOI] [PubMed] [Google Scholar]
- 28. Chen DY, Liu XF, Lin XJ et al. A dominant variant in DMXL2 is linked to nonsyndromic hearing loss. Genet Med. 2017;19(5):553–558. [DOI] [PubMed] [Google Scholar]
- 29. Tata B, Huijbregts L, Jacquier S et al. Haploinsufficiency of Dmxl2, encoding a synaptic protein, causes infertility associated with a loss of GnRH neurons in mouse. PLoS Biol. 2014;12(9):e1001952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Liu Y, Liu F, Hu X, He J, Jiang Y. Combining genetic mutation and expression profiles identifies novel prognostic biomarkers of lung adenocarcinoma. Clin Med Insights Oncol. 2020;14:1179554920966260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gorlova OY, Kimmel M, Tsavachidis S, Amos CI, Gorlov IP. Identification of lung cancer drivers by comparison of the observed and the expected numbers of missense and nonsense mutations in individual human genes. Oncotarget. 2022;13:756–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Li M, Zhao H, Zhang X et al. Inactivating mutations of the chromatin remodeling gene ARID2 in hepatocellular carcinoma. Nat Genet. 2011;43(9):828–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cleary SP, Jeck WR, Zhao X et al. Identification of driver genes in hepatocellular carcinoma by exome sequencing. Hepatology. 2013;58(5):1693–1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lan L, Xu B, Chen Q, Jiang J, Shen Y. Weighted correlation network analysis of triple-negative breast cancer progression: identifying specific modules and hub genes based on the GEO and TCGA database. Oncol Lett. 2019;18(2):1207–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Davis LK, Meyer KJ, Schindler EI et al. Copy number variations and primary open-angle glaucoma. Invest Ophthalmol Vis Sci. 2011;52(10):7122–7133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ratto E, Chowdhury SR, Siefert NS et al. Direct control of lysosomal catabolic activity by mTORC1 through regulation of V-ATPase assembly. Nat Commun. 2022;13(1):4848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Einhorn Z, Trapani JG, Liu Q, Nicolson T. Rabconnectin3alpha promotes stable activity of the H+ pump on synaptic vesicles in hair cells. J Neurosci. 2012;32(32):11144–11156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rujano MA, Panasyuk G et al. Mutations in the X-linked ATP6AP2 cause a glycosylation disorder with autophagic defects. J Exp Med. 2017;214(12):3707–3729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Larsen LE, van den Boogert MAW, WA R-O et al. Defective lipid droplet-lysosome interaction causes fatty liver disease as evidenced by human mutations in TMEM199 and CCDC115. Cell Mol Gastroenterol Hepatol. 2022;13(2):583–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Jansen JC, Timal S, van Scherpenzeel M et al. TMEM199 deficiency is a disorder of Golgi homeostasis characterized by elevated aminotransferases, alkaline phosphatase, and cholesterol and abnormal glycosylation. Am J Hum Genet. 2016;98(2):322–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Jansen JC, Cirak S, van Scherpenzeel M et al. CCDC115 deficiency causes a disorder of Golgi homeostasis with abnormal protein glycosylation. Am J Hum Genet. 2016;98(2):310–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Cannata Serio M, Graham LA, Ashikov A et al. Mutations in the V-ATPase assembly factor VMA21 cause a congenital disorder of glycosylation with autophagic liver disease. Hepatology. 2020;72(6):1968–1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Guida MC, Hermle T, Graham LA et al. ATP6AP2 functions as a V-ATPase assembly factor in the endoplasmic reticulum. Mol Biol Cell. 2018;29(18):2156–2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Jansen EJ, Timal S, Ryan M et al. ATP6AP1 deficiency causes an immunodeficiency with hepatopathy, cognitive impairment and abnormal protein glycosylation. Nat Commun. 2016;7:11600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Paunescu TG, Russo LM, Da Silva N et al. Compensatory membrane expression of the V-ATPase B2 subunit isoform in renal medullary intercalated cells of B1-deficient mice. Am J Physiol Renal Physiol. 2007;293(6):F1915–1926. [DOI] [PubMed] [Google Scholar]
- 46. Schwenk F, Baron U, Rajewsky K. A cre-transgenic mouse strain for the ubiquitous deletion of loxP-flanked gene segments including deletion in germ cells. Nucleic Acids Res. 1995;23(24):5080–5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Breton S, Wiederhold T, Marshansky V, Nsumu NN, Ramesh V, Brown D. The B1 subunit of the H+ATPase is a PDZ domain-binding protein. Colocalization with NHE-RF in renal B-intercalated cells. J Biol Chem. 2000;275(24):18219–18224. [DOI] [PubMed] [Google Scholar]
- 48. Paunescu TG, Ljubojevic M, Russo LM et al. cAMP stimulates apical V-ATPase accumulation, microvillar elongation, and proton extrusion in kidney collecting duct A-intercalated cells. Am J Physiol Renal Physiol. 2010;298(3):F643–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Da Silva N, Shum WW, El-Annan J et al. Relocalization of the V-ATPase B2 subunit to the apical membrane of epididymal clear cells of mice deficient in the B1 subunit. Am J Physiol Cell Physiol. 2007;293(1):C199–210. [DOI] [PubMed] [Google Scholar]
- 50. Gokhalel K, Kulkarni B, Chin E et al. Genetic and biochemical analysis of mutations in the DMXL1 gene causing a Prader-Willi like Syndrome. In: Unpublished: American Society of Human Genetics. 2009.
- 51. Miller RL, Lucero OM, Riemondy KA et al. The V-ATPase B1-subunit promoter drives expression of Cre recombinase in intercalated cells of the kidney. Kidney Int. 2009;75(4):435–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Carrisoza-Gaytan R, Ray EC, Flores D et al. Intercalated cell BKalpha subunit is required for flow-induced K+ secretion. JCI Insight. 2020;5(8):e130553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Chen L, Chou CL, Knepper MA. A comprehensive Map of mRNAs and their isoforms across All 14 renal tubule segments of mouse. J Am Soc Nephrol. 2021;32(4):897–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Chen L, Lee JW, Chou CL et al. Transcriptomes of major renal collecting duct cell types in mouse identified by single-cell RNA-seq. Proc Natl Acad Sci USA. 2017;114(46):E9989–E9998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Finberg KE, Wagner CA, Bailey MA et al. The B1-subunit of the H(+) ATPase is required for maximal urinary acidification. Proc Natl Acad Sci USA. 2005;102(38):13616–13621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Merkulova M, Paunescu TG, Nair AV et al. Targeted deletion of the Ncoa7 gene results in incomplete distal renal tubular acidosis in mice. Am J Physiol Renal Physiol. 2018;315(1):F173–F185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Cotter K, Liberman R, Sun-Wada G et al. The a3 isoform of subunit a of the vacuolar ATPase localizes to the plasma membrane of invasive breast tumor cells and is overexpressed in human breast cancer. Oncotarget. 2016;7(29):46142–46157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Vedovelli L, Rothermel JT, Finberg KE et al. Altered V-ATPase expression in renal intercalated cells isolated from B1 subunit-deficient mice by fluorescence-activated cell sorting. Am J Physiol Renal Physiol. 2013;304(5):F522–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Sun-Wada G, Murata Y, Yamamoto A, Kanazawa H, Wada Y, Futai M. Acidic endomembrane organelles are required for mouse postimplantation development. Dev Biol. 2000;228(2):315–325. [DOI] [PubMed] [Google Scholar]
- 60. Sethi N, Yan Y, Quek D, Schupbach T, Kang Y. Rabconnectin-3 is a functional regulator of mammalian Notch signaling. J Biol Chem. 2010;285(45):34757–34764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Husein D, Alamoudi A, Ohyama Y, Mochida H, Ritter B, Mochida Y. Identification of the C-terminal region in amelogenesis imperfecta causative protein WDR72 required for Golgi iocalization. Sci Rep. 2022;12(1):4640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Rungroj N, Nettuwakul C, Sawasdee N et al. Distal renal tubular acidosis caused by tryptophan-aspartate repeat domain 72 (WDR72) mutations. Clin Genet. 2018;94(5):409–418. [DOI] [PubMed] [Google Scholar]
- 63. Li X, Villacreses R, Thornell IM et al. V-Type ATPase mediates airway surface liquid acidification in pig small airway epithelial cells. Am J Respir Cell Mol Biol. 2021;65(2):146–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Brown D, Breton S. Mitochondria-rich, proton-secreting epithelial cells. J Exp Biol. 1996;199(Pt 11):2345–2358. [DOI] [PubMed] [Google Scholar]
- 65. Montoro DT, Haber AL, Biton M et al. A revised airway epithelial hierarchy includes CFTR-expressing ionocytes. Nature. 2018;560(7718):319–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Miller RL, Zhang P, Smith M et al. V-ATPase B1-subunit promoter drives expression of EGFP in intercalated cells of kidney, clear cells of epididymis and airway cells of lung in transgenic mice. Am J Physiol Cell Physiol. 2005;288(5):C1134–1144. [DOI] [PubMed] [Google Scholar]
- 67. Nair AV, Yanhong W, Paunescu TG, Bouley R, Brown D. Sex-dependent differences in water homeostasis in wild-type and V-ATPase B1-subunit deficient mice. PLoS One. 2019;14(8):e0219940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Vidarsson H, Westergren R, Heglind M, Blomqvist SR, Breton S, Enerback S. The forkhead transcription factor Foxi1 is a master regulator of vacuolar H-ATPase proton pump subunits in the inner ear, kidney and epididymis. PLoS One. 2009;4(2):e4471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Smardon AM, Diab HI, Tarsio M et al. The RAVE complex is an isoform-specific V-ATPase assembly factor in yeast. Mol Biol Cell. 2014;25(3):356–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data underlying this article are available in the main text or the online supplementary materials.












