Abstract
Mistranslation is the misincorporation of an amino acid into a polypeptide. Mistranslation has diverse effects on multicellular eukaryotes and is implicated in several human diseases. In Drosophila melanogaster, a serine transfer RNA (tRNA) that misincorporates serine at proline codons (P→S) affects male and female flies differently. The mechanisms behind this discrepancy are currently unknown. Here, we compare the transcriptional response of male and female flies to P→S mistranslation to identify genes and cellular processes that underlie sex-specific differences. Both males and females downregulate genes associated with various metabolic processes in response to P→S mistranslation. Males downregulate genes associated with extracellular matrix organization and response to negative stimuli such as wounding, whereas females downregulate aerobic respiration and ATP synthesis genes. Both sexes upregulate genes associated with gametogenesis, but females also upregulate cell cycle and DNA repair genes. These observed differences in the transcriptional response of male and female flies to P→S mistranslation have important implications for the sex-specific impact of mistranslation on disease and tRNA therapeutics.
Keywords: RNA sequencing, mistranslation, proteotoxicity, Drosophila melanogaster, translation, stress response, tRNA
Introduction
Accurate and efficient translation of mRNA into proteins is required for correct cell function and organism development. Errors during translation can decrease lifespan, induce neurodegeneration, and cause behavioral issues and developmental defects (Lee et al. 2006; Liu et al. 2014; Lu et al. 2014; Reverendo et al. 2014). Transfer RNAs (tRNAs) play a major role in determining the fidelity of translation, as do aminoacyl-tRNA synthetases (aaRSs) that aminoacylate tRNAs with their corresponding amino acid (reviewed in Pang et al. 2014). aaRSs recognize specific bases, base pairs, or motifs in their cognate tRNAs to ensure accurate aminoacylation (Hou and Schimmel 1988; Francklyn and Schimmel 1989; Normanly et al. 1992; Xue et al. 1993; Larkin et al. 2002). Since tRNA decoding potential is determined by the anticodon (the nucleotides at positions 34–36 of the tRNA that base pair with mRNA codons), the anticodon is an identity element for many tRNAs (Schulman and Pelka 1989; Jahn et al. 1991; Ruff et al. 1991; Tamura et al. 1992; Kholod et al. 1997; Giegé et al. 1998; Zamudio and José 2018; Giegé and Eriani 2023). However, some aaRSs do not use the anticodon to recognize their cognate tRNA. For example, tRNASer and tRNAAla are recognized through an elongated variable arm and a G3:U70 base pair, respectively (McClain and Foss 1988; Francklyn and Schimmel 1989; Achsel and Gross 1993). Because of this, anticodon mutations in tRNASer or tRNAAla genes cause the tRNA to decode noncognate mRNA codons and misincorporate serine or alanine in place of the amino acid normally specified by that codon. This error leads to mistranslation: the incorporation of an amino acid not specified by the standard genetic code. Mistranslation normally occurs at a rate of once per 103–106 codons (Joshi et al. 2019; Mordret et al. 2019), but tRNA variants or mutant aaRSs can dramatically increase mistranslation (Zimmerman et al. 2018; Berg, Zhu, et al. 2019; Zhang et al. 2021).
Humans have ∼66 tRNA variants per person, some of which cause mistranslation (Berg, Giguere, et al. 2019; Lant et al. 2021; Hasan et al. 2023; Davey-Young et al. 2024). Mistranslation induces aberrant phenotypes in a variety of organisms, including slow growth in yeast, deformities and decreased lifespan in flies, and cardiac abnormalities and neurodegeneration in mice (Liu et al. 2014; Lu et al. 2014; Berg, Zhu, et al. 2019; Berg, Isaacson, et al. 2021; Isaacson et al. 2022). Previous work in Saccharomyces cerevisiae demonstrated that mistranslation affects various biological processes, including translation, stress response, carbohydrate metabolism, and DNA replication (Paredes et al. 2012; Berg, Zhu, et al. 2021). The impact of mistranslation is likely more complex in multicellular organisms as codon usage and gene expression vary by tissue and developmental stage (Moriyama and Powell 1997; Dittmar et al. 2006; Vicario et al. 2008; Allen et al. 2022). Transient expression of mistranslating serine tRNA variants in zebrafish embryos upregulated stress response and DNA repair pathways (Reverendo et al. 2014), whereas transfection of human cells with mistranslating tRNAs upregulated protein folding and small-molecule catabolism genes (Hou et al. 2024). Some mistranslating tRNA variants reduce overall protein synthesis (Lant et al. 2021) and alter expression of other tRNAs (Hou et al. 2024). Not surprisingly, mistranslating tRNAs have been linked to disease (Goto et al. 1990; Shoffner et al. 1990; reviewed in Abbott et al. 2014; Lant et al. 2019).
Drosophila melanogaster is a popular model to study how defects in translational machinery affect developmental processes. Included in these are the Minute loci that code for the ribosomal proteins and when mutated extend development and reduce fertility (Bridges and Morgan 1923; Saeboe-Larssen and Lambertsson 1996; Marygold et al. 2007). Similarly, other aspects of translational control have been shown to impact fly development and behavior (e.g. Dorn et al. 1993; Lachance et al. 2002; Wilhelm and Smibert 2005; Fan et al. 2010). Flies have been used to study the physiological effects of mistranslation caused by mutant tRNA or aaRS genes (Laski et al. 1989; Garza et al. 1990; Lu et al. 2014). However, sex remains an understudied but important influence on organismal response to mistranslation, as male and female physiology differ dramatically due to different metabolic and reproductive requirements (reviewed in Millington and Rideout 2018). Supporting this idea, we previously found that a tRNASer variant, which causes proline-to-serine (P→S) mistranslation, increased morphological defects and impaired climbing performance in female fruit flies more than males (Isaacson et al. 2022). The mechanisms underlying this difference in male and female response to mistranslation are unknown. The goal of this work is to characterize the impact of P→S mistranslation on the transcriptome of male and female D. melanogaster to identify and compare genes and cellular processes that are disrupted in 1 or both sexes. Using a fly line containing a serine tRNA variant (tRNASerUGG, G26A) that induces P→S mistranslation, we found male mistranslating flies primarily downregulate metabolic, developmental, and extracellular matrix organization genes and upregulate genes associated with spermatogenesis. Female mistranslating flies downregulate genes associated with metabolism and ATP synthesis and upregulate genes associated with gametogenesis, cell cycle regulation, and DNA repair. As tRNA variants influence disease and are also being assessed as possible therapeutics (reviewed in Anastassiadis and Köhrer 2023 and Coller and Ignatova 2024; Hou et al. 2024), it is vital to understand differences in how males and females respond to mistranslating tRNA variants.
Methods
Fly stocks and husbandry
All fly stocks were maintained on standard Bloomington recipe food medium (Bloomington Drosophila Stock Center, Bloomington, IN, USA) under a 14:10 light:dark cycle at 24° and 70% relative humidity. The tRNA insertion lines used in this study were the same as those described in Isaacson et al. (2022). Two fly lines were used: a line containing the wild-type tRNASerUGA and a line containing the P→S mistranslating tRNASerUGG, G26A (Isaacson et al. 2022). The lines have the same genetic background and only differ in the type of tRNA transgene that was inserted. The genotype of both lines is as follows: w1118; P{CaryP}attP40[v+=tRNA]/CyO, P{w+mC=2xTb1-RFP}CyO; MKRS/TM6B, Tb1. Note that the lines used in this study are heterozygous for the inserted tRNA. The attP40 landing site was selected as it is relatively inert while allowing for strong expression of transgenes (Markstein et al. 2008).
RNA extraction, library preparation, and sequencing
Adult, virgin flies were aged 1–3 days and separated by sex. Ten flies were aspirated into a vial and flash frozen using liquid nitrogen. Males and females from the tRNASerUGA and tRNASerUGG, G26A (P→S) lines were collected and processed at the same time. Three replicates were collected in this manner for each genotype. RNA was extracted from fly tissue using the protocol outlined in Allen (2016), though volumes of all reagents were halved to account for using less tissue than the protocol specified. Following TRIzol extraction, RNA was measured in a NanoPhotometer P300 (Implen, Inc.) and concentration, 260/280 ratio, and 260/230 ratio were recorded to assess purity (Supplementary Table 1). To ensure RNA was free of genomic DNA, the remaining 25 µL of RNA was treated with dsDNAse (New England Biolabs Inc.) for 30 min at 37°. RNA was recovered through a second TRIzol extraction, and samples were assessed again using the NanoPhotometer to ensure the RNA remained pure. Up to 20 µg of RNA was loaded into RNA-stabilizing tubes, vacuum dried, and shipped to GeneWiz (South Plainfield, NJ, USA) for total RNA sequencing. If the total amount of RNA was less than 20 µg, then the entire sample was sequenced. Illumina HiSeq 2 × 150 bp RNA libraries with polyA selection were prepared from each sample. Number of raw reads obtained from each sample ranged from 12.7 million to 68.7 million.
RNA sequence data processing
Analysis of RNA sequencing data was performed using similar methods to those described in Berg, Zhu, et al. (2021). Short and/or low-quality reads were filtered out using a custom bioinformatics pipeline that utilized Trimmomatic v0.39 (Bolger et al. 2014) and FASTQC v0.11.9 (Andrews 2010) to produce filtered libraries containing 8.4–35.4 million reads per sample. Reads were aligned to the D. melanogaster reference genome (release r6.41_FB2021_04, downloaded from FlyBase.org; Öztürk-Çolak et al. 2024) using STAR v2.7.9a (Dobin et al. 2013). Read count data for each gene were summarized using featureCounts v2.0.0 (Liao et al. 2014). Only protein-coding genes were included in further analysis. List of protein-coding genes was based on the fly genome assembly BDGP6.46 (Celniker et al. 2002; Celniker and Rubin 2003). Parameters and commands used for this pipeline can be found in the extended methods section of Supplementary File 2.
Gene expression and Gene Ontology analysis
Statistical tests, principal component analysis (PCA), and RNA-seq data analyses were conducted using R Studio v1.2.5001. RNA sequencing sample normalization and differential gene expression analysis were performed using the DESeq2 R package (v1.26.0; Love et al. 2014), with a Benjamini–Hochberg false discovery rate (FDR) P-value cutoff < 0.05. To control for the batch effect identified by the PCA, we specified sample collection day as a covariate in the statistical model fit by ComBat-seq (Zhang et al. 2020). Analysis of differentially expressed genes was performed using WebGestalt's 2024 release (Liao et al. 2019). Lists of down- or upregulated genes were processed by ViSEAGO to produce Gene Ontology (GO) term heatmaps clustered by semantic similarity using Wang's method (Wang et al. 2007; Brionne et al. 2019; Gene Ontology Consortium et al. 2023). Significantly enriched GO terms were identified by ViSEAGO using the “weight01” algorithm and assessed with Fisher's exact test. Background gene lists composed of all genes with nonzero read counts for a given sample set (e.g. all female tRNASerUGA and tRNASerUGG, G26A samples) were provided to WebGestalt and ViSEAGO during enrichment analysis of that sample set as recommended by Timmons et al. (2015) and Wijesooriya et al. (2022). Figures were produced using RStudio and Inkscape v1.0.1.
Validation of RNA sequencing results using RT-qPCR
RNAs from 3 new replicates of 10 male or female virgin flies containing tRNASerUGA or tRNASerUGG, G26A (P→S) aged 1–3 days were extracted using the protocol described above. cDNA was synthesized from RNA using a Maxima H- First Strand cDNA Synthesis Kit (Thermo Scientific). Quantitative PCRs were performed on 3 independent replicates in duplicate using 10 ng/µL cDNA template, 500 ng/µL primers, and 1× PowerUp SYBR Green Master Mix for qPCR (Applied Biosystems) in a Bio-Rad CFX96 Touch Real-Time PCR Detection System (Bio-Rad Laboratories, Inc.). The Ct values of experimental genes were compared to the Ct values of αTub84B (FBgn0003884) for normalization and statistical analysis, which was performed by the Bio-Rad CFX Manager 3.0 software (Bio-Rad Laboratories, Inc.). A full list of qPCR primers can be found in Supplementary Table 2.
Clustering analysis
Clustering analysis was performed on the relative fold change of gene expression for tRNASerUGG, G26A (P→S) compared to control tRNASerUGA lines and the relative fold change of gene expression between treatment lines and controls within the microarray data described in Zhou et al. (2012). Normalized count data were obtained for all samples, and relative fold changes compared to controls were calculated for each gene within each treatment. Duplicate genes, genes with <10 normalized reads, or genes with a relative fold change > |5| were excluded from analysis as Z-score transformation is sensitive to outliers. Relative expression fold change values within each sample were Z-transformed and clustered using the ComplexHeatmap package in RStudio (Gu et al. 2016) using Ward's method (Ward 1963). Male and female data were clustered separately.
Results
Identifying mistranslation-induced differentially expressed genes
To analyze the transcriptomic response to serine mistranslation at proline codons in D. melanogaster, we sequenced polyA-enriched RNA from 1–3-day-old virgin adult male and female flies containing a single copy of either a wild-type tRNASerUGA or a tRNASerUGG, G26A variant that mistranslates proline to serine at a frequency of ∼0.6% per codon (Isaacson et al. 2022). The secondary G26A mutation was included in the mistranslating tRNASer variant as it disrupts a key modification in tRNASer species, reducing mistranslation to survivable levels based on work in yeast (Berg, Isaacson, et al. 2021; Boccaletto et al. 2022) and flies (Isaacson et al. 2022). tRNA insertion lines were maintained as heterozygotes because naturally occurring mistranslating tRNA variants are likely to arise as single alleles. PCA was performed on the male and female tRNASerUGA and tRNASerUGG, G26A (P→S) transcriptomic data (Supplementary Fig. 1). The first 2 principal components (PC1 and PC2) summarize ∼55% of the variance of both male and female data. The variation in PC1 captures the batch effect related to the day each sample was collected, as RNA from replicate 1 was harvested a day before replicates 2 and 3. Samples belonging to tRNASerUGA or tRNASerUGG, G26A (P→S) cluster together along the PC2 axis, indicating that the variance explained by PC2 likely represents differences due to the mistranslating tRNA (Supplementary Fig. 1a and b). We corrected the batch effect using ComBat-seq (Zhang et al. 2020), and the resulting PCA plots show that samples cluster well and PC1, which represents presence of mistranslation, explains 30–35% of the variance in the data (Supplementary Fig. 1c and d).
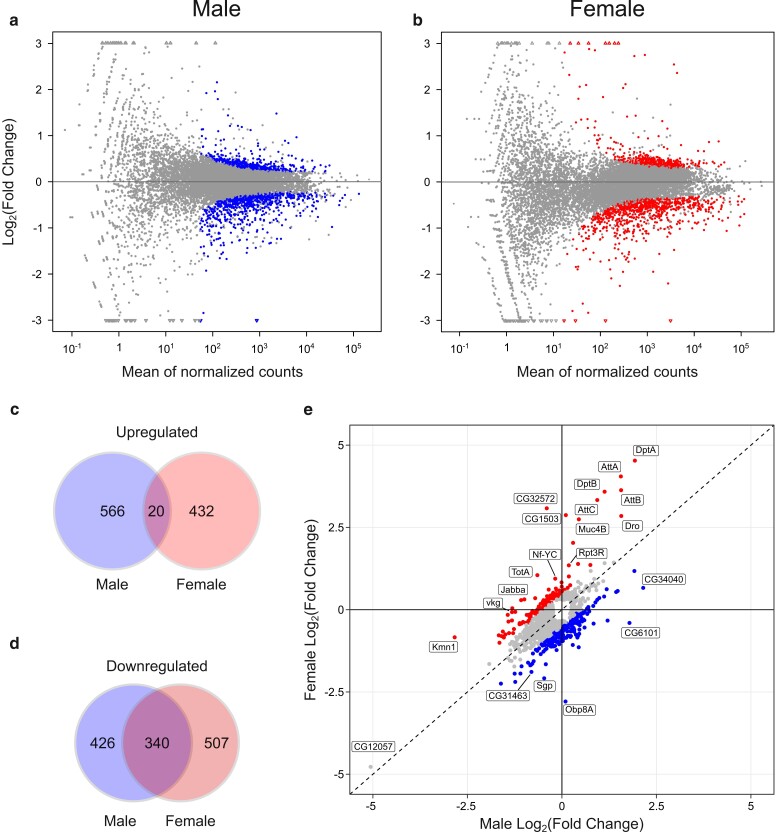
Differentially expressed genes between tRNASerUGA and tRNASerUGG, G26A (P→S) were identified using the R package DESeq2 (Love et al. 2014). Male and female samples were analyzed separately to determine the effects of tRNASerUGG, G26A (P→S) on each sex. We evaluated 13,202 genes with nonzero total read counts in male samples, whereas 12,893 genes were evaluated in female samples. MA plots constructed from male or female RNA sequencing data show that the majority of genes have a log2 fold change near zero, as expected (Fig. 1a and b). RNA sequencing revealed substantial sex-specific alterations to gene expression in response to mistranslation, as 426 genes were downregulated and 566 genes were upregulated uniquely in males, whereas 507 genes were downregulated and 432 genes upregulated uniquely in females (Wald test performed by DEseq2, Benjamini–Hochberg-adjusted P < 0.05, Fig. 1c and d). Only 20 genes were upregulated in both male and female flies containing tRNASerUGG, G26A (P→S) (Fig. 1c), whereas 340 genes were downregulated in both sexes in the mistranslating line (Fig. 1d). As shown in Fig. 1e, the relative expression of many of the differentially expressed genes differed substantially between the sexes. To identify genes that showed a significant interaction between sex and presence of tRNASerUGG, G26A (P→S), we analyzed the RNA sequencing data of both males and females simultaneously and found that transcriptional response to mistranslation of 251 genes significantly depended on fly sex (Supplementary File 1). These results show that P→S mistranslation disrupts expression of largely different sets of genes in males and females.
Fig. 1.
Differentially expressed genes in male or female flies containing tRNASerUGG, G26A (P→S). a) MA plot visualizing the relationship between transcript abundance and the difference in fold change of expression between male tRNASerUGG, G26A (P→S) and control tRNASerUGA samples. Blue points represent genes that are significantly differentially expressed between mistranslating tRNASerUGG, G26A (P→S) and control tRNASerUGA samples, whereas gray points represent genes where the expression change was not statistically significant. Triangular points at the edge of the y-axis indicate genes that have a fold change exceeding the limits of the y-axis. b) MA plot visualizing the relationship between transcript abundance and fold change of expression difference between female tRNASerUGG, G26A (P→S) and control tRNASerUGA samples. Red points represent genes that are significantly differentially expressed between mistranslating tRNASerUGG, G26A (P→S) and control tRNASerUGA samples. c) Venn diagram showing the number of significantly upregulated (FDR-adjusted P < 0.05) genes unique to tRNASerUGG, G26A (P→S) males, females, or genes upregulated in both sexes. d) Venn diagram showing the number of significantly downregulated (FDR-adjusted P < 0.05) genes unique to tRNASerUGG, G26A (P→S) males, females, or genes downregulated in both sexes. e) Scatterplot showing male vs female relative expression for all 1,705 genes that were identified as differentially expressed and not filtered out from analysis in either sex. Blue points represent genes that have higher relative expression in mistranslating males compared to females (log2 fold change difference > 0.5); red points represent genes with higher relative expression in mistranslating females compared to males. Genes that demonstrate sex-biased patterns of relative expression (log2 fold change difference > 1) in response to tRNASerUGG, G26A (P→S) are labeled. CG12057 is also labeled due to its strong downregulation in both sexes. The dashed line represents identical fold changes in expression for both males and females.
To provide further support for the transcriptomic data, we confirmed differential expression of 6 genes using RT-qPCR with RNA extracted from 3 independent replicates of both male and female flies. We analyzed 3 genes that were downregulated in both sexes (CG12057, CG11911, and fiz), 1 gene that was upregulated in both sexes (CG4650), 1 gene significantly upregulated in males (Pif1A), and 1 gene that was differentially expressed between males and females (CG1503). All genes showed the same pattern of expression in both qPCR and RNA sequencing analyses for both sexes except for CG11911, where the difference between flies containing tRNASerUGA or tRNASerUGG, G26A (P→S) was nonsignificant (Supplementary Fig. 2). This rate of nonconcordance matches the nonconcordance rate of 15–19% between RNA sequencing and RT-qPCR analysis observed by Everaert et al. (2017), who also found nonconcordance was more common for short 1-exon genes such as CG11911.
Proline-to-serine mistranslation causes sex-specific transcriptional responses
We analyzed the lists of differentially expressed genes using 2 different tools to identify cellular processes affected by the presence of tRNASerUGG, G26A (P→S). WebGestalt (Liao et al. 2019) was used to identify the 10 most enriched GO terms in the list of genes affected by tRNASerUGG, G26A (P→S). We also used ViSEAGO (Brionne et al. 2019) to construct a heatmap of enriched (GO) terms for males and females, allowing for visualization of sex differences in the fly response to P→S mistranslation. All enriched GO terms, their associated P-values, and the genes identified in our analysis that belong to those categories are reported in Supplementary File 1. The list of GO terms produced by WebGestalt showed similarities and differences between male and female responses to P→S mistranslation. Both males and females downregulated various metabolic processes (Fig. 2a), with females primarily downregulating aerobic respiration (e.g. ox, ND-23, ND-24, UQCR-6.4, Cyt-C1, and COX4) and males downregulating lipid and fatty acid metabolism (e.g. Lip4, Lsd-1, Hacl, FASN1, and CDase).
Fig. 2.
The top 10 significantly enriched GO terms in the list of genes a) downregulated or b) upregulated in male or female flies containing tRNASerUGG, G26A (P→S) compared to control tRNASerUGA flies. Higher enrichment ratios indicate that the set of genes associated with that GO term were more highly represented in our gene set. Note the differences in scale. Lists were produced using WebGestalt (Liao et al. 2019). A list of significantly enriched GO terms and associated statistics is found in Supplementary File 1.
There was limited overlap in the biological processes enriched in the upregulated genes shared between males and females, consistent with our observation that relatively few genes were upregulated in both sexes (Fig. 2b). Females upregulated genes associated with cell cycle regulation and cell division (e.g. CycA, CycB, Cdc16, APC7, and Mink) as well as genes involved in response to DNA replication (e.g. DNAlig1, PolA1, Prim1, RecQ4, and Fen1). Only 3 biological processes were significantly enriched in the list of upregulated genes in male tRNASerUGG, G26A (P→S) flies. This may arise because most upregulated genes (438 of 586) are uncharacterized (Supplementary File 1). The 3 enriched male terms all correspond to male gamete generation and development (e.g. fan, ProtA, Pif1A, and ntc).
We next used ViSEAGO to construct heatmaps of GO terms enriched in the set of genes down- or upregulated gene in tRNASerUGG, G26A (P→S). ViSEAGO clusters GO terms by semantic similarity, so GO terms corresponding to similar biological processes are near each other in the dendrogram (Brionne et al. 2019). Functional enrichment was determined using Fisher's exact test. Figure 3a further emphasizes the downregulation of genes involved in metabolic processes in response to P→S mistranslation, with different aspects of metabolism being affected in each sex (Fig. 3a). In agreement with the WebGestalt results, females downregulated genes associated with oxidative phosphorylation and ATP synthesis whereas males downregulated genes involved in fatty acid and carboxylic acid catabolism. In addition, both males and females downregulated genes involved in chemical or ion transport (e.g. nrv2, blw, rumpel, and snu). In contrast, biological processes such as response to negative stimuli like wounding (e.g. Atg2, PPO2, Hml, and Tg) and extracellular structure organization (Cad99C, LanA, LanB1, LanB2, Col4a1, and vkg) were downregulated only in males. Females uniquely downregulated genes associated with muscle function and development, such as myosin (Mhc, Mlc1, and Mlc2), troponin (up and wupA), and tropomyosin (Tm1 and Tm2) genes.
Fig. 3.
Heatmap of enriched GO terms from the differentially expressed genes in male or female flies containing tRNASerUGG, G26A (P→S). a) Heatmap of enriched GO terms in the list of downregulated genes in male and female flies containing tRNASerUGG, G26A (P→S). Each horizontal bar represents a GO term identified as significantly enriched in male and/or female data. GO terms were clustered by semantic similarity according to ViSEAGO using Wang's method (Wang et al. 2007; Brionne et al. 2019). Dendrogram clades of the same color represent semantically similar GO terms. Darker bars within the heatmap represent lower P-values as determined through Fisher's exact test. Notable groups of enriched processes are labeled in blue if enriched in males, red if enriched in females, or purple if enriched in both sexes. b) The same as a) but using the list of upregulated genes. A full list of enriched GO terms and their associated genes can be found in Supplementary File 1.
When examining the lists of genes upregulated in male or female flies containing tRNASerUGG, G26A (P→S), ViSEAGO did not identify any GO terms that were significantly enriched in both males and females though we note that gametogenesis and metabolic processes were affected in both sexes (Fig. 3b). Of the upregulated genes with identified function, only genes associated with spermatogenesis, protein localization to microtubules, the electron transport chain, and maltose metabolism were enriched in males. For females, in addition to genes associated with cell cycle regulation and DNA repair (discussed above), genes associated with protein and mRNA localization (e.g. Nup154, Elys, and Fmr1), development (e.g. glu, mor, and fz), and regulation of gene expression (e.g. bcd, Marf1, and pum) were upregulated. Genes involved in antibacterial immune response (e.g. DptA, Dro, AttA, and BomS5) were also upregulated in females but not males. These results emphasize that the cellular response to P→S mistranslation differs between male and female flies, and that the difference is particularly pronounced when comparing upregulated genes.
tRNA-induced P→S mistranslation clusters with heat shock and nutrient stress
Clustering analysis groups genes or treatments based on similarity and is useful to predict functions of uncharacterized genes or identify treatments that produce similar cellular effects (reviewed in Oyelade et al. 2016). To identify which environmental or physiological conditions resemble tRNA-induced P→S mistranslation in flies, we clustered the gene expression data from male and female flies containing tRNASerUGG, G26A with the microarray gene expression data from Zhou et al. (2012), containing the transcriptional response of male and female flies from the same genetic background exposed to 20 different nutritional, chemical, and physiological conditions (Fig. 4).
Fig. 4.
Clustering proline-to-serine mistranslation-induced transcriptome changes with transcriptome changes due to various other physiological or environmental conditions. a) Z-score normalized gene expression changes in tRNASerUGG, G26A (P→S) males relative to tRNASerUGA (wild-type) males clustered with normalized male gene expression changes from Zhou et al. (2012). Genes with fewer than 10 normalized reads or fold changes > |5| for any condition were excluded from analysis. Clustering was performed using the “ComplexHeatmap” R package using Ward's method (Ward 1963; Gu et al. 2016). The P→S mistranslation condition is highlighted in green. b) The same as a) but clustering female data.
As the transcriptomic data acquisition method differed between this study and Zhou et al. (2012), we used Z-transformed relative fold changes to compare these data sets. Clustering analysis of male data revealed that tRNA-induced P→S mistranslation induced a transcriptional response most resembling starvation (Fig. 4a). Mistranslating males also clustered with temperature or chemical stressors such as heat shock, chill coma, and ethanol exposure. In females, the transcriptional response of tRNA-induced P→S mistranslation most resembled flies reared on high yeast or high sugar and high yeast diets (Fig. 4b). Both male and female flies containing tRNASerUGG, G26A (P→S) clustered with treatments affecting nutrition, which aligns with our observations that various metabolic processes are affected by tRNASerUGG, G26A (Fig. 2a).
Discussion
Proline-to-serine mistranslation exerts sex-specific transcriptomic effects
In this study, we examined how D. melanogaster males and females alter their transcriptome when exposed to a mistranslating tRNASerUGG, G26A variant that causes P→S mistranslation. While some biological processes such as carboxylic acid metabolism, chemical transport, and germ cell production were affected in both sexes, we observed a disparity between male and female transcriptional responses to P→S mistranslation. This result is consistent with the different physiological and nutritional requirements of male and female flies. Female flies are larger, require a greater quantity and variety of nutrients, and store more triglycerides and glycogen than male flies (Bakker 1959; Wu et al. 2020, reviewed in Millington and Rideout 2018). These requirements are largely due to the increased cost of gamete production in females, which also affects virgin flies as they still devote resources to egg production and laying (Partridge et al. 1986; Wu et al. 2020). Disruptions to proteostasis, such as mistranslation, would exacerbate this discrepancy between males and females, as maintaining proteostasis requires a substantial proportion of all energy produced by the cell (Buttgereit and Brand 1995; Lahtvee et al. 2014). The relatively mild phenotypes previously observed in male flies containing tRNASerUGG, G26A (P→S) compared to females (Isaacson et al. 2022) may in part be due to having more cellular resources available to maintain homeostasis.
One notable group of sex-specific upregulated genes in females was associated with DNA repair and cell cycle regulation. Genes involved with DNA repair are often upregulated in response to cellular stress (Mendez et al. 2000; Pregi et al. 2017; Sottile and Nadin 2018; Clementi et al. 2020). Our observation that DNA repair and cell cycle genes are disrupted in mistranslating flies is consistent with the genetic instability observed by Kalapis et al. (2015) in response to mistranslation in yeast. Genetic interactions with mistranslation in yeast and transcriptional responses to mistranslation in human cells also identified the importance of genes involved in cell cycle and DNA damage response (Shcherbakov et al. 2019; Berg, Zhu, et al. 2021). Furthermore, mistranslation causes aneuploidy and aberrant nuclear division in yeast species and increases mutation rate in Escherichia coli (Al Mamun et al. 2002; Balashov and Humayun 2002; Kimata and Yanagida 2004; Silva et al. 2007). Mistranslation caused by tRNASerUGG, G26A may be exerting similar effects in female flies. Interestingly, female flies are less susceptible to sources of DNA damage such as oxidative stress or radiation and are better able to decompose reactive oxygen species than male flies (Parashar et al. 2008; Edman et al. 2009; Moskalev et al. 2011; Niveditha et al. 2017). The upregulation of DNA repair genes in mistranslating females may result from their observed increased resistance to stress and DNA damage relative to male flies (reviewed in Pomatto et al. 2018). Future studies should examine if flies containing tRNASerUGG, G26A (P→S) show similar genome instability as mistranslating yeast or E. coli.
Similarity to other transcriptomic studies of tRNA-induced mistranslation
Other studies have examined the transcriptomic effects of tRNA-induced mistranslation on organisms including yeast (Paredes et al. 2012; Berg, Zhu, et al. 2021), zebrafish (Reverendo et al. 2014), and human cells (HEK293; Hou et al. 2024), though none investigated how males and females differ in their response to mistranslation. Paredes et al. (2012) engineered a tRNASer variant that mistranslates leucine to serine in yeast and observed upregulation of stress response chaperone genes and downregulation of protein synthesis. When clustered with various environmental stresses, the mistranslating yeast transcriptome most resembled nutrient stresses such as nitrogen deprivation and amino acid starvation, which agrees with our results in flies. Zebrafish embryos transiently expressing mistranslating tRNASer variants similarly downregulate protein synthesis and upregulate stress response genes and genes associated with DNA damage and repair (Reverendo et al. 2014). Human cells transfected with mistranslating tRNAArg variants upregulate genes involved in protein folding and endoplasmic reticulum stress (Hou et al. 2024). Interestingly, some mistranslating tRNAArg variants have minimal effects on the transcriptome. While we did not observe significant downregulation of genes involved in protein synthesis in males or females containing tRNASerUGG, G26A (P→S), female flies containing tRNASerUGG, G26A upregulated genes involved in DNA damage and repair, which aligns with the previous studies. Overall, our data are consistent with previous work characterizing the transcriptomic effects of mistranslation in other organisms while uncovering novel sex-specific differences in these general responses.
Future work and conclusions
These transcriptomic results provide intriguing avenues for future research. Drosophila melanogaster tissues have different codon usages and tRNA expression profiles and thus might be differently susceptible to tRNASer variants that cause P→S mistranslation (Dittmar et al. 2006; Allen et al. 2022). A focused transcriptomic approach centered on specific cell types, such as neurons or muscle, could reveal trends that are difficult to observe from whole fly transcriptomics. Testing other life stages could also reveal stage-specific transcriptomic responses to mistranslating tRNA variants. Different types of mistranslation exert unique cellular effects (Berg, Zhu, et al. 2021; Cozma et al. 2023; Davey-Young et al. 2024; Hou et al. 2024), so testing other amino acid substitutions will uncover which cellular responses are common to mistranslation and which are unique to specific substitutions.
The differentially expressed genes identified in this analysis can be targeted using available D. melanogaster knockout lines to determine which are necessary for the fly response to mistranslation. The uncharacterized gene CG12057 is worthy of further investigation as its expression was reduced >25-fold in both male and female tRNASerUGG, G26A (P→S) flies. CG12057 is primarily expressed in the midgut, and its expression is impacted by various stresses, including hypoxia, infection, and mitochondrial dysfunction (Carpenter et al. 2009; Fernández-Ayala et al. 2010; Mosqueira et al. 2010; Moskalev et al. 2015; Krause et al. 2022). Determining the function of CG12057 could provide insight into how flies cope with cellular stress. Further investigation into the cellular processes disrupted by P→S mistranslation may elucidate the genetic and physiological mechanisms behind sex-specific response to mistranslation and the striking phenotypes observed in mistranslating adult flies (Isaacson et al. 2022). Overall, this study demonstrates that sex strongly affects response to mistranslation and must be considered when studying mistranslation in sexually dimorphic organisms.
Supplementary Material
Acknowledgments
We would like to thank Dr. Patrick O’Donoghue, Dr. Robert Cumming, and Ecaterina Cozma for their feedback and guidance on the manuscript, as well as Dr. Gregory Gloor for his advice on data analysis and interpretation.
Contributor Information
Joshua R Isaacson, Department of Biology, Western University, London, Canada, N6A 5B7.
Matthew D Berg, Department of Genome Sciences, University of Washington, Seattle, WA 98195, USA.
William Yeung, Department of Biology, Western University, London, Canada, N6A 5B7.
Judit Villén, Department of Genome Sciences, University of Washington, Seattle, WA 98195, USA.
Christopher J Brandl, Department of Biochemistry, Western University, London, Canada, N6A 5B7.
Amanda J Moehring, Department of Biology, Western University, London, Canada, N6A 5B7.
Data availability
Fly lines are available upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and Supplementary material. A full list of all differentially expressed genes and all significantly enriched GO terms can be found in Supplementary File 1. Supplementary File 2 contains all supplementary figures, tables, and methods. Supplementary File 3 contains R code used to analyze RNA sequencing data and perform clustering and ViSEAGO analysis. All raw and processed data can be found at the NCBI GEO database using the accession number GSE256332.
Supplemental material available at G3 online.
Funding
This work was supported by the Natural Sciences and Engineering Research Council of Canada (NSERC) grants to CJB (RGPIN-2020-07046) and AJM (RGPIN-2020-06464); National Institutes of Health (NIH) grants to JV (R01AG056359, R56AG049494, and R35GM119536), and a University of Western Ontario Medical & Health Science Research Board seed grant to AJM. JRI and MDB were supported by an NSERC Postgraduate Scholarship (Doctoral) and NSERC Canada Graduate Scholarship (Doctoral), respectively.
Literature cited
- Abbott JA, Francklyn CS, Robey-Bond SM. 2014. Transfer RNA and human disease. Front Genet. 5:158. doi: 10.3389/fgene.2014.00158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achsel T, Gross HJ. 1993. Identity determinants of human tRNASer: sequence elements necessary for serylation and maturation of a tRNA with a long extra arm. EMBO J. 12(8):3333–3338. doi: 10.1002/j.1460-2075.1993.tb06003.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gene Ontology Consortium; Aleksander SA, Balhoff J, Carbon S, Cherry JM, Drabkin HJ, Ebert D, Feuermann M, Gaudet P, Harris NL, et al. 2023. The Gene Ontology knowledgebase in 2023. Genetics. 224(1):iyad031. doi: 10.1093/genetics/iyad031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen E. 2016. RNA extraction from Drosophila tissues using TRIzol reagent. protocols.io. doi: 10.17504/protocols.io.fgtbjwn. [DOI]
- Allen SR, Stewart RK, Rogers M, Ruiz IJ, Cohen E, Laederach A, Counter CM, Sawyer JK, Fox DT. 2022. Distinct responses to rare codons in select Drosophila tissues. Elife. 11:e76893. doi: 10.7554/eLife.76893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al Mamun AA, Marians KJ, Humayun MZ. 2002. DNA polymerase III from Escherichia coli cells expressing mutA mistranslator tRNA is error-prone. J Biol Chem. 277(48):46319–46327. doi: 10.1074/jbc.M206856200. [DOI] [PubMed] [Google Scholar]
- Anastassiadis T, Köhrer C. 2023. Ushering in the era of tRNA medicines. J Biol Chem. 299(10):105246. doi: 10.1016/j.jbc.2023.105246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews S. 2010. FASTQC: a quality control tool for high throughput sequence data [Online]. Available from http://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
- Bakker K. 1959. Feeding period, growth, and pupation in larvae of Drosophila melanogaster. Entomol Exp Appl. 2(3):171–186. doi: 10.1111/j.1570-7458.1959.tb00432.x. [DOI] [Google Scholar]
- Balashov S, Humayun MZ. 2002. Mistranslation induced by streptomycin provokes a RecABC/RuvABC-dependent mutator phenotype in Escherichia coli cells. J Mol Biol. 315(4):513–527. doi: 10.1006/jmbi.2001.5273. [DOI] [PubMed] [Google Scholar]
- Berg MD, Giguere DJ, Dron JS, Lant JT, Genereaux J, Liao C, Wang J, Robinson JF, Gloor GB, Hegele RA, et al. 2019. Targeted sequencing reveals expanded genetic diversity of human transfer RNAs. RNA Biol. 16(11):1574–1585. doi: 10.1080/15476286.2019.1646079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg MD, Isaacson JR, Cozma E, Genereaux J, Lajoie P, Villén J, Brandl CJ. 2021. Regulating expression of mistranslating tRNAs by readthrough RNA polymerase II transcription. ACS Synth Biol. 10(11):3177–3189. doi: 10.1021/acssynbio.1c00461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg MD, Zhu Y, Genereaux J, Ruiz BY, Rodriguez-Mias RA, Allan T, Bahcheli A, Villén J, Brandl CJ. 2019. Modulating mistranslation potential of tRNASer in Saccharomyces cerevisiae. Genetics. 213(3):849–863. doi: 10.1534/genetics.119.302525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg MD, Zhu Y, Ruiz BY, Loll-Krippleber R, Isaacson J, San Luis BJ, Genereaux J, Boone C, Villén J, Brown GW, et al. 2021. The amino acid substitution affects cellular response to mistranslation. G3 (Bethesda). 11(10):jkab218. doi: 10.1093/g3journal/jkab218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boccaletto P, Stefaniak F, Ray A, Cappannini A, Mukherjee S, Purta E, Kurkowska M, Shirvanizadeh N, Destefanis E, Groza P, et al. 2022. MODOMICS: a database of RNA modification pathways. 2021 update. Nucleic Acids Res. 50(D1):D231–D235. doi: 10.1093/nar/gkab1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridges CB, Morgan TH. 1923. The Third-Chromosome Group of Mutant Characters of Drosophila melanogaster. Washington: Carnegie Institution of Washington. [Google Scholar]
- Brionne A, Juanchich A, Hennequet-Antier C. 2019. ViSEAGO: a Bioconductor package for clustering biological functions using Gene Ontology and semantic similarity. BioData Min. 12(1):16. doi: 10.1186/s13040-019-0204-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buttgereit F, Brand MD. 1995. A hierarchy of ATP-consuming processes in mammalian cells. Biochem J. 312(1):163–167. doi: 10.1042/bj3120163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter J, Hutter S, Baines JF, Roller J, Saminadin-Peter SS, Parsch J, Jiggins FM. 2009. The transcriptional response of Drosophila melanogaster to infection with the sigma virus (Rhabdoviridae). PLoS One. 4(8):e6838. doi: 10.1371/journal.pone.0006838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celniker SE, Rubin GM. 2003. The Drosophila melanogaster genome. Annu Rev Genomics Hum Genet. 4(1):89–117. doi: 10.1146/annurev.genom.4.070802.110323. [DOI] [PubMed] [Google Scholar]
- Celniker SE, Wheeler DA, Kronmiller B, Carlson JW, Halpern A, Patel S, Adams M, Champe M, Dugan SP, Frise E, et al. 2002. Finishing a whole-genome shotgun: release 3 of the Drosophila melanogaster euchromatic genome sequence. Genome Biol. 3(12):research0079.1. doi: 10.1186/gb-2002-3-12-research0079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clementi E, Inglin L, Beebe E, Gsell C, Garajova Z, Markkanen E. 2020. Persistent DNA damage triggers activation of the integrated stress response to promote cell survival under nutrient restriction. BMC Biol. 18(1):36. doi: 10.1186/s12915-020-00771-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coller J, Ignatova Z. 2024. tRNA therapeutics for genetic diseases. Nat Rev Drug Discov. 23(2):108–125. doi: 10.1038/s41573-023-00829-9. [DOI] [PubMed] [Google Scholar]
- Cozma E, Rao M, Dusick M, Genereaux J, Rodriguez-Mias RA, Villén J, Brandl CJ, Berg MD. 2023. Anticodon sequence determines the impact of mistranslating tRNAAla variants. RNA Biol. 20(1):791–804. doi: 10.1080/15476286.2023.2257471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey-Young J, Hasan F, Tennakoon R, Rozik P, Moore H, Hall P, Cozma E, Genereaux J, Hoffman KS, Chan PP, et al. 2024. Mistranslating the genetic code with leucine in yeast and mammalian cells. RNA Biol. 21(1):1–23. doi: 10.1080/15476286.2024.2340297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittmar KA, Goodenbour JM, Pan T. 2006. Tissue-specific differences in human transfer RNA expression. PLoS Genet. 2(12):e221. doi: 10.1371/journal.pgen.0020221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorn R, Morawietz H, Reuters G, Saumweber H. 1993. Identification of an essential Drosophila gene that is homologous to the translation initiation factor eIF-4A of yeast and mouse. Mol Gen Genet MGG. 237–237(1–2):233–240. doi: 10.1007/BF00282805. [DOI] [PubMed] [Google Scholar]
- Edman U, Garcia AM, Busuttil RA, Sorensen D, Lundell M, Kapahi P, Vijg J. 2009. Lifespan extension by dietary restriction is not linked to protection against somatic DNA damage in Drosophila melanogaster. Aging Cell. 8(3):331–338. doi: 10.1111/j.1474-9726.2009.00480.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everaert C, Luypaert M, Maag JLV, Cheng QX, Dinger ME, Hellemans J, Mestdagh P. 2017. Benchmarking of RNA-sequencing analysis workflows using whole-transcriptome RT-qPCR expression data. Sci Rep. 7(1):1559. doi: 10.1038/s41598-017-01617-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y, Schlierf M, Gaspar AC, Dreux C, Kpebe A, Chaney L, Mathieu A, Hitte C, Grémy O, Sarot E, et al. 2010. Drosophila translational elongation factor-1γ is modified in response to DOA kinase activity and is essential for cellular viability. Genetics. 184(1):141–154. doi: 10.1534/genetics.109.109553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Ayala DJM, Chen S, Kemppainen E, O’Dell KMC, Jacobs HT. 2010. Gene expression in a Drosophila model of mitochondrial disease. PLoS One. 5(1):e8549. doi: 10.1371/journal.pone.0008549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francklyn C, Schimmel P. 1989. Aminoacylation of RNA minihelices with alanine. Nature. 337(6206):478–481. doi: 10.1038/337478a0. [DOI] [PubMed] [Google Scholar]
- Garza D, Medhora MM, Hartl DL. 1990. Drosophila nonsense suppressors: functional analysis in Saccharomyces cerevisiae, Drosophila tissue culture cells and Drosophila melanogaster. Genetics. 126(3):625–637. doi: 10.1093/genetics/126.3.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giegé R, Eriani G. 2023. The tRNA identity landscape for aminoacylation and beyond. Nucleic Acids Res. 51(4):1528–1570. doi: 10.1093/nar/gkad007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giegé R, Sissler M, Florentz C. 1998. Universal rules and idiosyncratic features in tRNA identity. Nucleic Acids Res. 26(22):5017–5035. doi: 10.1093/nar/26.22.5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto YI, Nonaka I, Horai S. 1990. A mutation in the tRNALeu(UUR) gene associated with the MELAS subgroup of mitochondrial encephalomyopathies. Nature. 348(6302):651–653. doi: 10.1038/348651a0. [DOI] [PubMed] [Google Scholar]
- Gu Z, Eils R, Schlesner M. 2016. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 32(18):2847–2849. doi: 10.1093/bioinformatics/btw313. [DOI] [PubMed] [Google Scholar]
- Hasan F, Lant JT, O’Donoghue P. 2023. Perseverance of protein homeostasis despite mistranslation of glycine codons with alanine. Philos Trans R Soc B Biol Sci. 378(1871):20220029. doi: 10.1098/rstb.2022.0029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou YM, Schimmel P. 1988. A simple structural feature is a major determinant of the identity of a transfer RNA. Nature. 333(6169):140–145. doi: 10.1038/333140a0. [DOI] [PubMed] [Google Scholar]
- Hou Y, Zhang W, McGilvray PT, Sobczyk M, Wang T, Weng SHS, Huff A, Huang S, Pena N, Katanski CD, et al. 2024. Engineered mischarged transfer RNAs for correcting pathogenic missense mutations. Mol Ther. 32(2):352–371. doi: 10.1016/j.ymthe.2023.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaacson JR, Berg MD, Charles B, Jagiello J, Villén J, Brandl CJ, Moehring AJ. 2022. A novel mistranslating tRNA model in Drosophila melanogaster has diverse, sexually dimorphic effects. G3 (Bethesda). 12:jkac035. doi: 10.1093/g3journal/jkac035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn M, Rogers MJ, Söll D. 1991. Anticodon and acceptor stem nucleotides in tRNAGln are major recognition elements for E. coli glutaminyl-tRNA synthetase. Nature. 352(6332):258–260. doi: 10.1038/352258a0. [DOI] [PubMed] [Google Scholar]
- Joshi K, Cao L, Farabaugh PJ. 2019. The problem of genetic code misreading during protein synthesis. Yeast. 36(1):35–42. doi: 10.1002/yea.3374. [DOI] [PubMed] [Google Scholar]
- Kalapis D, Bezerra AR, Farkas Z, Horvath P, Bódi Z, Daraba A, Szamecz B, Gut I, Bayes M, Santos MA, et al. 2015. Evolution of robustness to protein mistranslation by accelerated protein turnover. PLoS Biol. 13(11):e1002291. doi: 10.1371/journal.pbio.1002291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kholod NS, Pan'kova NV, Mayorov SG, Krutilina AI, Shlyapnikov MG, Kisselev LL, Ksenzenko VN. 1997. Transfer RNAPhe isoacceptors possess non-identical set of identity elements at high and low Mg2+ concentration. FEBS Lett. 411(1):123–127. doi: 10.1016/S0014-5793(97)00608-X. [DOI] [PubMed] [Google Scholar]
- Kimata Y, Yanagida M. 2004. Suppression of a mitotic mutant by tRNA-Ala anticodon mutations that produce a dominant defect in late mitosis. J Cell Sci. 117(11):2283–2293. doi: 10.1242/jcs.01078. [DOI] [PubMed] [Google Scholar]
- Krause SA, Overend G, Dow JAT, Leader DP. 2022. FlyAtlas 2 in 2022: enhancements to the Drosophila melanogaster expression atlas. Nucleic Acids Res. 50(D1):D1010–D1015. doi: 10.1093/nar/gkab971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachance PED, Miron M, Raught B, Sonenberg N, Lasko P. 2002. Phosphorylation of eukaryotic translation initiation factor 4E is critical for growth. Mol Cell Biol. 22(6):1656–1663. doi: 10.1128/MCB.22.6.1656-1663.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahtvee P-J, Seiman A, Arike L, Adamberg K, Vilu R. 2014. Protein turnover forms one of the highest maintenance costs in Lactococcus lactis. Microbiology. 160(7):1501–1512. doi: 10.1099/mic.0.078089-0. [DOI] [PubMed] [Google Scholar]
- Lant JT, Berg MD, Heinemann IU, Brandl CJ, O’Donoghue P. 2019. Pathways to disease from natural variations in human cytoplasmic tRNAs. J Biol Chem. 294(14):5294–5308. doi: 10.1074/jbc.REV118.002982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lant JT, Kiri R, Duennwald ML, O’Donoghue P. 2021. Formation and persistence of polyglutamine aggregates in mistranslating cells. Nucleic Acids Res. 49(20):11883–11899. doi: 10.1093/nar/gkab898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin DC, Williams AM, Martinis SA, Fox GE. 2002. Identification of essential domains for Escherichia coli tRNALeu aminoacylation and amino acid editing using minimalist RNA molecules. Nucleic Acids Res. 30(10):2103–2113. doi: 10.1093/nar/30.10.2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laski FA, Ganguly S, Sharp PA, RajBhandary UL, Rubin GM. 1989. Construction, stable transformation, and function of an amber suppressor tRNA gene in Drosophila melanogaster. Proc Natl Acad Sci U S A. 86(17):6696–6698. doi: 10.1073/pnas.86.17.6696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JW, Beebe K, Nangle LA, Jang J, Longo-Guess CM, Cook SA, Davisson MT, Sundberg JP, Schimmel P, Ackerman SL. 2006. Editing-defective tRNA synthetase causes protein misfolding and neurodegeneration. Nature. 443(7107):50–55. doi: 10.1038/nature05096. [DOI] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W. 2014. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 30(7):923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- Liao Y, Wang J, Jaehnig EJ, Shi Z, Zhang B. 2019. WebGestalt 2019: gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res. 47(W1):W199–W205. doi: 10.1093/nar/gkz401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Satz JS, Vo MN, Nangle LA, Schimmel P, Ackerman SL. 2014. Deficiencies in tRNA synthetase editing activity cause cardioproteinopathy. Proc Natl Acad Sci U S A. 111(49):17570–17575. doi: 10.1073/pnas.1420196111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Bergert M, Walther A, Suter B. 2014. Double-sieving-defective aminoacyl-tRNA synthetase causes protein mistranslation and affects cellular physiology and development. Nat Commun. 5(1):5650. doi: 10.1038/ncomms6650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markstein M, Pitsouli C, Villalta C, Celniker SE, Perrimon N. 2008. Exploiting position effects and the gypsy retrovirus insulator to engineer precisely expressed transgenes. Nat Genet. 40(4):476–483. doi: 10.1038/ng.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marygold SJ, Roote J, Reuter G, Lambertsson A, Ashburner M, Millburn GH, Harrison PM, Yu Z, Kenmochi N, Kaufman TC, et al. 2007. The ribosomal protein genes and Minute loci of Drosophila melanogaster. Genome Biol. 8(10):R216. doi: 10.1186/gb-2007-8-10-r216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClain WH, Foss K. 1988. Changing the identity of a tRNA by introducing a G-U wobble pair near the 3′ acceptor end. Science. 240(4853):793–796. doi: 10.1126/science.2452483. [DOI] [PubMed] [Google Scholar]
- Mendez F, Sandigursky M, Franklin WA, Kenny MK, Kureekattil R, Bases R. 2000. Heat-shock proteins associated with base excision repair enzymes in HeLa cells. Radiat Res. 153(2):186–195. doi: 10.1667/0033-7587(2000)153[0186:HSPAWB]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Millington JW, Rideout EJ. 2018. Sex differences in Drosophila development and physiology. Curr Opin Physiol. 6:46–56. doi: 10.1016/j.cophys.2018.04.002. [DOI] [Google Scholar]
- Mordret E, Dahan O, Asraf O, Rak R, Yehonadav A, Barnabas GD, Cox J, Geiger T, Lindner AB, Pilpel Y. 2019. Systematic detection of amino acid substitutions in proteomes reveals mechanistic basis of ribosome errors and selection for translation fidelity. Mol Cell. 75(3):427–441.e5. doi: 10.1016/j.molcel.2019.06.041. [DOI] [PubMed] [Google Scholar]
- Moriyama EN, Powell JR. 1997. Codon usage bias and tRNA abundance in Drosophila. J Mol Evol. 45(5):514–523. doi: 10.1007/PL00006256. [DOI] [PubMed] [Google Scholar]
- Moskalev AA, Plyusnina EN, Shaposhnikov MV. 2011. Radiation hormesis and radioadaptive response in Drosophila melanogaster flies with different genetic backgrounds: the role of cellular stress-resistance mechanisms. Biogerontology. 12(3):253–263. doi: 10.1007/s10522-011-9320-0. [DOI] [PubMed] [Google Scholar]
- Moskalev A, Zhikrivetskaya S, Krasnov G, Shaposhnikov M, Proshkina E, Borisoglebsky D, Danilov A, Peregudova D, Sharapova I, Dobrovolskaya E, et al. 2015. A comparison of the transcriptome of Drosophila melanogaster in response to entomopathogenic fungus, ionizing radiation, starvation and cold shock. BMC Genomics. 16(S13):S8. doi: 10.1186/1471-2164-16-Supplementary 13-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosqueira M, Willmann G, Ruohola-Baker H, Khurana TS. 2010. Chronic hypoxia impairs muscle function in the Drosophila model of Duchenne's muscular dystrophy (DMD). PLoS One. 5(10):e13450. doi: 10.1371/journal.pone.0013450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niveditha S, Deepashree S, Ramesh SR, Shivanandappa T. 2017. Sex differences in oxidative stress resistance in relation to longevity in Drosophila melanogaster. J Comp Physiol. B. 187(7):899–909. doi: 10.1007/s00360-017-1061-1. [DOI] [PubMed] [Google Scholar]
- Normanly J, Ollick T, Abelson J. 1992. Eight base changes are sufficient to convert a leucine-inserting tRNA into a serine-inserting tRNA. Proc Natl Acad Sci U S A. 89(12):5680–5684. doi: 10.1073/pnas.89.12.5680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyelade J, Isewon I, Oladipupo F, Aromolaran O, Uwoghiren E, Ameh F, Achas M, Adebiyi E. 2016. Clustering algorithms: their application to gene expression data. Bioinform Biol Insights. 10:237–253. doi: 10.4137/BBI.S38316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Öztürk-Çolak A, Marygold SJ, Antonazzo G, Attrill H, Goutte-Gattat D, Jenkins VK, Matthews BB, Millburn G, Dos Santos G, Tabone CJ, et al. 2024. FlyBase: updates to the Drosophila genes and genomes database. Genetics. 227(1):iyad211. doi: 10.1093/genetics/iyad211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang YLJ, Poruri K, Martinis SA. 2014. tRNA synthetase: tRNA aminoacylation and beyond. Wiley Interdiscip Rev RNA. 5(4):461–480. doi: 10.1002/wrna.1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parashar V, Frankel S, Lurie AG, Rogina B. 2008. The effects of age on radiation resistance and oxidative stress in adult Drosophila melanogaster. Radiat Res. 169(6):707–711. doi: 10.1667/RR1225.1. [DOI] [PubMed] [Google Scholar]
- Paredes JA, Carreto L, Simões J, Bezerra AR, Gomes AC, Santamaria R, Kapushesky M, Moura GR, Santos MA. 2012. Low level genome mistranslations deregulate the transcriptome and translatome and generate proteotoxic stress in yeast. BMC Biol. 10(1):55. doi: 10.1186/1741-7007-10-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partridge L, Fowler K, Trevitt S, Sharp W. 1986. An examination of the effects of males on the survival and egg-production rates of female Drosophila melanogaster. J Insect Physiol. 32(11):925–929. doi: 10.1016/0022-1910(86)90140-X. [DOI] [Google Scholar]
- Pomatto LCD, Tower J, Davies KJA. 2018. Sexual dimorphism and aging differentially regulate adaptive homeostasis. J Gerontol Ser A. 73(2):141–149. doi: 10.1093/gerona/glx083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pregi N, Belluscio LM, Berardino BG, Castillo DS, Cánepa ET. 2017. Oxidative stress-induced CREB upregulation promotes DNA damage repair prior to neuronal cell death protection. Mol Cell Biochem. 425(1–2):9–24. doi: 10.1007/s11010-016-2858-z. [DOI] [PubMed] [Google Scholar]
- Reverendo M, Soares AR, Pereira PM, Carreto L, Ferreira V, Gatti E, Pierre P, Moura GR, Santos MA. 2014. tRNA mutations that affect decoding fidelity deregulate development and the proteostasis network in zebrafish. RNA Biol. 11(9):1199–1213. doi: 10.4161/rna.32199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruff M, Krishnaswamy S, Boeglin M, Poterszman A, Mitschler A, Podjarny A, Rees B, Thierry JC, Moras D. 1991. Class II aminoacyl transfer RNA synthetases: crystal structure of yeast aspartyl-tRNA synthetase complexed with tRNAAsp. Science. 252(5013):1682–1689. doi: 10.1126/science.2047877. [DOI] [PubMed] [Google Scholar]
- Saeboe-Larssen S, Lambertsson A. 1996. A novel Drosophila Minute locus encodes ribosomal protein S13. Genetics. 143(2):877–885. doi: 10.1093/genetics/143.2.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulman LH, Pelka H. 1989. The anticodon contains a major element of the identity of arginine transfer RNAs. Science. 246(4937):1595–1597. doi: 10.1126/science.2688091. [DOI] [PubMed] [Google Scholar]
- Shcherbakov D, Teo Y, Boukari H, Cortes-Sanchon A, Mantovani M, Osinnii I, Moore J, Juskeviciene R, Brilkova M, Duscha S, et al. 2019. Ribosomal mistranslation leads to silencing of the unfolded protein response and increased mitochondrial biogenesis. Commun Biol. 2(1):381. doi: 10.1038/s42003-019-0626-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoffner JM, Lott MT, Lezza AM, Seibel P, Ballinger SW, Wallace DC. 1990. Myoclonic epilepsy and ragged-red fiber disease (MERRF) is associated with a mitochondrial DNA tRNALys mutation. Cell. 61(6):931–937. doi: 10.1016/0092-8674(90)90059-N. [DOI] [PubMed] [Google Scholar]
- Silva RM, Paredes JA, Moura GR, Manadas B, Lima-Costa T, Rocha R, Miranda I, Gomes AC, Koerkamp MJ, Perrot M, et al. 2007. Critical roles for a genetic code alteration in the evolution of the genus Candida. EMBO J. 26(21):4555–4565. doi: 10.1038/sj.emboj.7601876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sottile ML, Nadin SB. 2018. Heat shock proteins and DNA repair mechanisms: an updated overview. Cell Stress Chaperones. 23(3):303–315. doi: 10.1007/s12192-017-0843-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Himeno H, Asahara H, Hasegawa T, Shimizu M. 1992. In vitro study of E. coli tRNAArg and tRNALys identity elements. Nucleic Acids Res. 20(9):2335–2339. doi: 10.1093/nar/20.9.2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmons JA, Szkop KJ, Gallagher IJ. 2015. Multiple sources of bias confound functional enrichment analysis of global -omics data. Genome Biol. 16(1):186. doi: 10.1186/s13059-015-0761-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicario S, Mason CE, White KP, Powell JR. 2008. Developmental stage and level of codon usage bias in Drosophila. Mol Biol Evol. 25(11):2269–2277. doi: 10.1093/molbev/msn189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JZ, Du Z, Payattakool R, Yu PS, Chen CF. 2007. A new method to measure the semantic similarity of GO terms. Bioinformatics. 23(10):1274–1281. doi: 10.1093/bioinformatics/btm087. [DOI] [PubMed] [Google Scholar]
- Ward JH. 1963. Hierarchical grouping to optimize an objective function. J Am Stat Assoc. 58(301):236–244. doi: 10.1080/01621459.1963.10500845. [DOI] [Google Scholar]
- Wijesooriya K, Jadaan SA, Perera KL, Kaur T, Ziemann M. 2022. Urgent need for consistent standards in functional enrichment analysis. PLoS Comput Biol. 18(3):e1009935. doi: 10.1371/journal.pcbi.1009935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm JE, Smibert CA. 2005. Mechanisms of translational regulation in Drosophila. Biol Cell. 97(4):235–252. doi: 10.1042/BC20040097. [DOI] [PubMed] [Google Scholar]
- Wu Q, Yu G, Cheng X, Gao Y, Fan X, Yang D, Xie M, Wang T, Piper MDW, Yang M. 2020. Sexual dimorphism in the nutritional requirement for adult lifespan in Drosophila melanogaster. Aging Cell. 19(3):e13120. doi: 10.1111/acel.13120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue H, Shens W, Giegeq R, Tze J, Wongii F. 1993. Identity elements of tRNATrp. Identification and evolutionary conservation. J Biol Chem. 268(13):9316–9322. doi: 10.1016/S0021-9258(18)98352-3. [DOI] [PubMed] [Google Scholar]
- Zamudio GS, José MV. 2018. Identity elements of tRNA as derived from information analysis. Orig Life Evol Biosph. 48(1):73–81. doi: 10.1007/s11084-017-9541-6. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Parmigiani G, Johnson WE. 2020. ComBat-seq: batch effect adjustment for RNA-seq count data. NAR Genomics Bioinforma. 2(3):lqaa078. doi: 10.1093/nargab/lqaa078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Wu J, Lyu Z, Ling J. 2021. Impact of alanyl-tRNA synthetase editing deficiency in yeast. Nucleic Acids Res. 49(17):9953–9964. doi: 10.1093/nar/gkab766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou S, Campbell TG, Stone EA, Mackay TFC, Anholt RRH. 2012. Phenotypic plasticity of the Drosophila transcriptome. PLoS Genet. 8(3):e1002593. doi: 10.1371/journal.pgen.1002593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman SM, Kon Y, Hauke AC, Ruiz BY, Fields S, Phizicky EM. 2018. Conditional accumulation of toxic tRNAs to cause amino acid misincorporation. Nucleic Acids Res. 46(15):7831–7843. doi: 10.1093/nar/gky623. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Fly lines are available upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and Supplementary material. A full list of all differentially expressed genes and all significantly enriched GO terms can be found in Supplementary File 1. Supplementary File 2 contains all supplementary figures, tables, and methods. Supplementary File 3 contains R code used to analyze RNA sequencing data and perform clustering and ViSEAGO analysis. All raw and processed data can be found at the NCBI GEO database using the accession number GSE256332.
Supplemental material available at G3 online.