Abstract
DNA hypomethylating agents (HMAs) such as decitabine and 5-azacytidine have established roles in the treatment paradigms for myelodysplastic syndrome and acute myelogenous leukemia, where they are considered to exert their anticancer effects by restoring the expression of tumor suppressor genes. Due to their relatively favorable adverse effect profile and known ability to pass through the blood-brain barrier, applications in the treatment of glioblastoma (GBM) and other central nervous system malignancies are under active investigation. The present review examines the types of HMAs currently available, their known and less-understood antineoplastic mechanisms, and the evidence to date of their preclinical and clinical efficacy in glioblastoma and other solid malignancies. The present review discusses the potential synergies HMAs may have with established and emerging GBM treatments, including temozolomide, immune checkpoint inhibitors and cancer vaccines. Recent successes and setbacks in clinical trials for newly diagnosed and recurrent GBM are summarized in order to highlight opportunities for HMAs to improve therapeutic responses. Challenges for future clinical trials are also assessed.
Keywords: hypomethylating agent, decitabine, glioblastoma, temozolomide, immunotherapy
1. Introduction
Abnormal DNA methylation is a hallmark of a multitude of pathological processes, including neurodevelopmental imprinting disorders, atherosclerosis, autoimmune diseases, and cancer. Notably, abnormal DNA hypermethylation of tumor suppressor genes plays a critical role in the malignant transformation of myelodysplastic syndrome (MDS) (1) into acute myeloid leukemia (AML), with the possibility of this process being slowed or reversed using hypomethylating agents (HMAs). There is accumulating data that HMAs may have a role to play as well in the treatment of solid tumors, where they have primarily been explored in combination with other antineoplastic drugs, and may act through a variety of possible mechanisms, not just limited to the re-expression of tumor suppressor genes (2,3).
Glioblastoma, IDH-wildtype, (GBM) is most frequently diagnosed in adults in the seventh decade of life, and accounts for approximately 15% of all intracranial neoplasms and 50% of all primary malignant brain tumors (4). Despite advances in treatment over the past twenty years, median overall survival remains well under two years (5). The current paradigm for first-line treatment consists of maximal safe surgical resection when possible, followed by conformal external beam radiotherapy with concurrent and adjuvant temozolomide (TMZ), an alkylating chemotherapy agent that crosses the blood-brain barrier. Approximately 40% of GBMs exhibit elevated methylation levels at the methylguanine methyltransferase (MGMT) gene promoter, which generally predicts a favorable response to TMZ, although it is not the sole determining factor and some MGMT-unmethylated GBMs benefit from TMZ as well (6). Nonetheless, the lack of meaningful and lasting responses to TMZ in the majority of GBM patients emphasizes the critical need to identify and develop new treatment strategies (7).
This work explores the antineoplastic potential of current HMAs as well as established data of their preclinical and clinical effectiveness in GBM and other solid tumors. Although genome-wide hypermethylation, as seen in IDH-mutant gliomas, is not characteristic of GBM, a multitude of evidence points to the role HMAs might have in reversing focal genomic methylation aberrations that contribute to GBM treatment resistance. Additionally, we review possible synergies these drugs may have with current and emerging GBM therapies with a focus on temozolomide and immunotherapy. Challenges for future clinical trials are also assessed.
2. Mechanism of HMAs
Epigenetic alterations cause heritable changes in gene expression without changes in the DNA nucleotide sequence (8). Thus, an epigenetic mechanism can be thought of as a system for selectively using genetic information to turn ‘on’ and ‘off’ various functional genes in order to carry out key processes during normal embryonal development (9), including chromosome X inactivation (10), the maintenance of genomic stability (11), and transcriptional regulation (12). Since the early 1980s, DNA methylation has been recognized as one such epigenetic mechanism that plays a significant role in controlling cellular differentiation states (13,14).
DNA methylation is a tightly regulated gene silencing or activation process mediated through DNA methyltransferases (DNMTs). The DNMT family consists of 5 members, including DNMT1, DNMT2, DNMT3A, DNMT3B, and DNMT3L, of which DNMT1 is the best characterized (Table I). DNMT1, also referred to as maintenance DNMT, binds to newly synthesized DNA and acts to maintain the methylation pattern of the template DNA strand after replication. DNMT1 is also recruited to sites of DNA damage, including base mismatches and double-strand breaks, to prevent loss of DNA methylation and gene dysregulation after DNA repair (15). DNMT2 is primarily a tRNA methyltransferase, which acts to protect tRNA against fragmentation. DNMT3A, DNMT3B, and DNMT3L, also referred to as de novo DNMTs, can establish new methylation patterns during normal development and in response to environmental cues. These de novo DNMTs also form part of chromatin-remodeling complexes and help complete the process of establishing and maintaining cell-specific methylation arrangements (16,17).
Table I.
Summary of the function and role of DNMTs in human diseases.
| Gene | Function | Role in human disease |
|---|---|---|
| DNMT1 | De novo methylating activity; maintains DNA methylation patterns during embryo development. | Missense mutations linked to hereditary sensory and autonomic neuropathy type 1E; knockout in mouse models causes embryonic lethality. |
| DNMT2 | Also known as TRDMT1; mainly a tRNA methyltransferase; protects tRNA against fragmentation; restricts the activation of cryptic promoters. | Possible links to aberrant hematopoiesis; knockout mice are viable and fertile. |
| DNMT3A/B | De novo methylating activity; methylates previously unmethylated regions of the genome in a non-selective manner; role in transcription activation at enhancers. | DNMT3A mutations are common in AML; DNMT3A mutations linked to Tatton-Brown-Rahman syndrome; DNMT3B mutations linked to immunodeficiency, centromeric instability and facial anomalies (ICF syndrome). |
| DNMT3L | Stimulates methyltransferase activity of DNMT3A and DNMT3B; cofactor for retrotransposon methylation in male individuals; expressed during brain development and in the thymus in adulthood. | Reduced activity linked to AML. |
AML, acute myeloid leukemia; DNMT, DNA methyltransferase; ICF syndrome, immunodeficiency, centromere instability and facial anomalies syndrome; TRDMT1, tRNA aspartic acid methyltransferase 1; tRNA, transfer RNA.
DNMTs covalently transfer a methyl group to the C-5 position of cytosine residues within CpG dinucleotides (18). DNA regions with a high frequency of CpG sites are called CpG islands, which can range from 200 to 3,000 base pairs and are typically associated with gene promoters (19). In the normal mammalian cell, CpG islands are usually hypomethylated and have activating histone modifications, which allows for unhindered DNA accessibility and facilitated gene expression (20,21). DNA methylation can take place in the promoter of a gene, generally resulting in the repression of gene transcription, or in the gene body, where the usual result is promotion of gene transcription (14,22,23). Gene promoter methylation has several known effects. It may prevent RNA polymerase and transcription factors from binding to active regulatory sites. Alternatively, methylation can lure methyl-CpG binding domain proteins that recruit histone deacetylases, leading to the removal of gene-activating acetylation marks and chromatin condensation (24). Gene silencing from methylation-induced heterochromatization also results from the recruitment of polycomb repressor complexes (25) and nucleosome complexes (26-28).
On the other hand, methylation of CpG islands within the gene body may promote normal gene transcription through several interrelated mechanisms, including slowing the kinetics of RNA polymerase II for proper splice site recognition and inhibiting spurious transcription from ectopic promoters. Recent studies have shown that aberrant gene body methylation may have varying effects depending on cell type and differentiation state. The effects may also be gene specific. For example, aberrant gene body hypermethylation of the stem cell lineage marker brachyury has been associated with precancerous intestinal metaplasia, while global hypomethylation occurs when these same cells undergo neoplastic transformation into gastric adenocarcinoma (29). Contrastingly, hypomethylation can have antitumor effects when it occurs within the gene body of oncogenes, where a physiologic level of CpG methylation may act to promote the expression of oncogenic factors (30). Overall, the normal role of methylation and the consequences of aberrant methylation in gene bodies is not fully understood and is an active area of investigation.
3. Types of HMAs
HMAs are pharmacological agents that can inhibit methylation by trapping DNMTs, resulting in the expression of a previously hypermethylated silenced gene (31,32), and possibly also repression or modification of transcription at sites within the gene body. Developed beginning in the 1960s (33,34), HMAs currently in use include decitabine, azacytidine, guadecitabine and ASTX727(35), all of which have demonstrated effects on cell cycle control, DNA repair, cell signaling, apoptosis and metastasis (36). In general, these agents are cytosine analogs that exert their effects once they have integrated into newly synthesized DNA or RNA.
Decitabine (5-aza-2-deoxycytidine) acts as a cytosine analogue, replacing cytosine in the CpG dinucleotide pair, which is the typical target of DNMTs. Unlike cytosine, decitabine possesses a nitrogen molecule instead of carbon at the fifth carbon position, preventing the transfer of a methyl group to this site. Decitabine also forms a covalent bond with the methyltransferase enzyme leading to its inactivation. Covalently trapped DNMTs are targeted for degradation by the proteasome, leading to a genome-wide decrease in CpG methylation levels. This may enable the re-expression of aberrantly repressed genes by preventing the re-methylation of CpG islands over the course of multiple cell cycles. It is critical to note that incorporation of decitabine into DNA requires the transition to S-phase of the cell cycle in the target cell (37,38); it has a limited effect on CpG methylation in non-proliferating cells, thus making it useful as an antineoplastic agent (39).
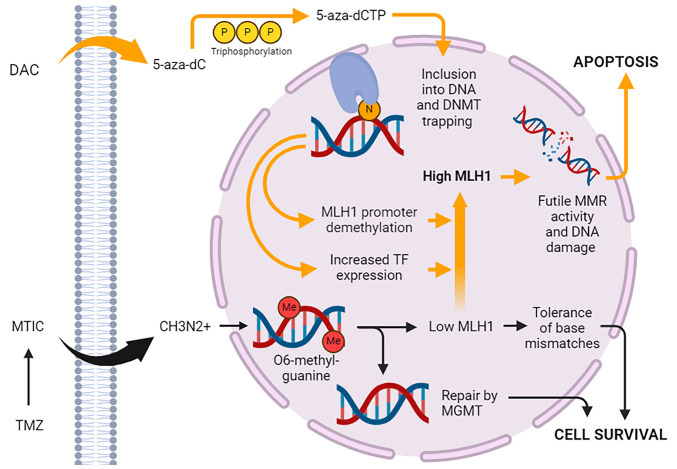
DAC reaches a maximum plasma concentration of about 65-77 ng/ml when given at standard intravenous dosing of 15 mg/m2 every 8 h in patients with AML and MDS (40). Cellular uptake of the drug is dependent on the nucleoside-specific transport mechanism. Rapid equilibration between the intra- and extracellular compartments results in a short alpha half-life of 5 min. In plasma, the drug is quickly inactivated by high levels of cytidine deaminase in the liver, spleen, intestinal epithelia, and blood, which accounts for its short plasma beta half-life of 15 to 25 min. Pharmacokinetic studies in rabbits and dogs show that DAC crosses the blood-brain barrier (41). Human pharmacokinetic studies have not been performed, but data from clinical trials of DAC in MDS indicate rates of neurological and psychiatric adverse reactions suggestive of CNS activity (42). Upon cellular entry, the prodrug form (5-AZA-CdR) undergoes phosphorylation by a series of kinases into its final active triphosphate form (5-AZA-CtR), which acts as a substrate for DNA polymerase (Fig. 1). 5-AZA-CtR is then incorporated into the cell DNA and asserts its effects. At lower dosages, the drug induces DNA hypomethylation and reactivation of genes leading to cell differentiation. Conversely, higher concentrations of the drug lead to a cytotoxic effect by blocking DNA synthesis.
Figure 1.
Proposed mechanism of action of DAC and TMZ. Once DAC enters the cell, it undergoes triphosphorylation, converting it to 5-aza-dCTP, which is incorporated into DNA during S-phase in place of cytosine. 5-aza-dCTP traps and inactivates DNMTs, causing exome-wide changes in gene expression mediated by promoter demethylation, gene body demethylation and changes in TF expression. MLH1 is upregulated, increasing levels of O6-methylguanine produced by TMZ and futile MMR activity. This ultimately results in enhanced cytotoxicity due to DNA double-strand break formation, cell cycle arrest and apoptosis. The effects of TMZ are exerted through its spontaneous decarboxylation to MTIC, which is unstable and degrades into the reactive CH3N2+. 5-aza-dC, 5-aza-2'-deoxycytidine; 5-aza-dCTP, 5-aza-2'-deoxycytidine-5'-triphosphate; CH3N2+, methyldiazonium ion; DAC, decitabine; DNMT, DNA methyltransferase; Me, methyl group; MGMT, methylguanine methyltransferase; MLH1, mutL homolog 1; MMR, mismatch repair; MTIC, 5-(3-methyltriazen-1-yl) imidazole-4-carboxamide; TF, transcription factor; TMZ, temozolomide.
The antileukemic effect of DAC was first demonstrated in 1968 in mouse models, which prompted further investigation into the drug's clinical potential in decades to follow. Early phase 1 clinical trials in the early 1990s determined a maximally tolerated dose of 2,250 mg/m2 with myelosuppression as the primary adverse effect. This was followed by several single-arm phase 2 trials that demonstrated good responses in AML, MDS, and chronic myelomonocytic leukemia (CMML) even at low dosing schedules of 20 mg/m2/day for 5 days (43). Finally, a North American phase 3 trial comparing DAC to supportive care in patients with intermediate- and high-risk MDS demonstrated a significant survival benefit at a dose of 45 mg/m2/day, leading to approval in the US of DAC for the treatment MDS in 2006(44). An oral form of DAC in combination with cedazuridine, a novel cytidine deaminase inhibitor that prevents drug inactivation in the digestive tract, was approved in the US in 2020 (ASTX727) (42).
Azacytidine (5-azacitidine) (AZA), in contrast to DAC, is a cytidine analogue that integrates preferentially into RNA after entering the cell via nucleoside transporters, although 10-20% is reduced by ribonucleotide reductase into DAC and incorporated into DNA (45,46). Once incorporated into RNA by RNA polymerase, AZA interferes with gene expression and protein synthesis by hampering RNA stability and correct folding (47), ultimately promoting apoptosis in tumor cells.
AZA absorption into tissues is fast and complete after IV or subcutaneous administration, with a peak concentration in 30 min and about 90% bioavailability (48). Unlike DAC, it does not cross the blood-brain barrier, limiting its potential for use in CNS cancers. It is metabolized in the liver and excretion is mostly through the kidneys with a half-life of 4 h (49).
In a 2004 randomized open-label, phase 3 multicenter trial conducted by the Cancer and Leukemia Group B, which included patients with all five MDS subtypes, treatment with AZA resulted in an overall response rate of 15.7%, compared to 0% response in the observation arm. Responses included partial or complete normalization of blood cells and bone marrow structure. On the basis of this study and two other smaller single-arm trials, AZA received approval in the US for all MDS subtypes (48).
Guadecitabine (SGI-110) is a second-generation HMA currently being investigated as an alternative to DAC and AZA in MDS and acute myeloid leukemia (50). Because the incorporation of DAC into DNA is S-phase dependent, its relatively short half-life of 20 min due to degradation by cytidine deaminase limits its ability to enter a large proportion of tumor cells after a single IV dose, necessitating three doses every 8 h. As a dinucleotide of DAC and deoxyguanosine linked by a 3'-5' phosphodiester bond, guadecitabine is resistant to cytidine deaminase (51). After subcutaneous administration, guadecitabine is cleaved into DAC in a slow, sustained fashion, resulting in a prolonged exposure period and better tolerated toxicity profile (52).
4. HMAs and tumor suppressors
Aberrant DNA methylation can provide survival benefits to cancer cells by silencing essential genes for anti-tumor activity, known as tumor suppressors. DNA demethylating agents therefore provide a possible means of reactivating silenced tumor suppressor genes and epigenetically reprogramming neoplastic cells to therapeutic advantage (53). In MDS, aberrant methylation patterns due to loss-of-function mutations in a number of epigenetic regulators, including DNMT3A and TET2, result in ineffective hematopoiesis and peripheral blood cytopenias by disrupting hematopoietic stem cell differentiation homeostasis (54,55). The sequestration of methyltransferases using HMAs may counteract the development of these aberrant patterns, as well as induce the re-expression of various tumor suppressor genes often silenced in MDS, including p15INK4B and p16INK4A (56), TP53(57) and DAPK1(58). Overall, this yields anti-proliferative and pro-apoptotic effects. Genes without CpG-island-containing promoters have also been shown to be upregulated by DAC in MDS and AML cells, emphasizing the role methylation-independent effects may have as well (59).
In a number of solid cancers, investigators have similarly demonstrated the ability of HMAs to re-express silenced tumor suppressor genes with favorable effects on tumor cell growth and gene expression profile. These include the Ras association domain family 1A gene (RASSF1A) in lung cancer (60), the DNA double-strand break repair gene BRCA1 in breast cancer (61), and the homeobox transcription factor HOXA10 in ovarian cancer (62). In GBM, several tumor suppressor genes have been found to be hypermethylated at their promoter regions and downregulated, including the microtubule associated tumor suppressor gene (MTUS1) (63), esophageal-cancer related gene (ECRG4) (64), epithelial membrane protein 3 (EMP3) (65), and SOCS1/3(66). In various GBM cell lines, the dampened expression of these genes was associated with increased cellular survival, invasion, proliferation and reduced apoptosis. DAC treatment was able to reverse hypermethylation and re-express these genes both at the mRNA and protein level.
HMAs can also silence pro-oncogenic pathways through demethylation of gene body CpGs. In colorectal carcinoma, DAC downregulated genes involved in the regulation of c-MYC signaling pathways, leading to a suppression of tumor growth; this effect reversed after withdrawal of treatment (67). In GBM, Sanaei and Kavoosi (66) demonstrated that DAC treatment significantly downregulated expression of the anti-apoptotic Bcl-2 protein, mirroring a key mechanism of TMZ-induced cytotoxicity (68) and suggesting DAC could have a cytotoxicity potentiating effect. DAC also inhibited JAK/STAT signaling, leading to reductions in cell proliferation and growth.
The expression of Promonin-1 or CD133, a known marker for GBM-initiating cancer stem cells (GSCs), correlates with increased WHO grade in gliomas and exhibits abnormal, but often variable, promoter methylation patterns even among different cell subpopulations within the same GBM (69). Through promoter demethylation, DAC treatment has been shown to upregulate CD133 promoter expression in multiple GBM cell lines (70), which could suggest a possible undesired pro-tumorigenic effect.
Although there is accumulating preclinical evidence that HMAs have the ability to alter the expression of tumor suppressors and oncogenes in a way that, on balance, could yield overall antitumor effects, further investigation will be required to translate these findings into the clinic. AZA has been tested clinically in recurrent IDH-mutant gliomas, but did not produce measurable clinical responses as a single agent (71), possibly due to lack of CNS penetration; to our knowledge, monotherapy with DAC or another HMA has not yet been tested clinically in GBM. There are several other challenges that could curtail therapeutic responses to HMAs, including relatively lower penetrance into solid tumors compared to hematologic malignancies (72), unpredictable off-target effects on other gene networks, and the intrinsic heterogeneity of GBM as opposed to the clonal nature of hematologic neoplasms. Demethylation alone may also be insufficient to reliably re-express a tumor suppressor gene if, for example, the required activating transcription factor is not expressed.
5. HMA-mediated chemosensitization
For the past 20 years, the alkylating agent temozolomide has remained the mainstay systemic agent used for GBM and other diffuse gliomas. In light of this, a large number of patients would potentially benefit from the identification of a subset of GBM patients where the unique gene expression-modifying properties of HMAs could act to potentiate the cytotoxic effects of this chemotherapy. The strategy of chemosensitization using epigenetic agents is an active line of investigation in a number of solid cancers, most notably ovarian cancer, where low-dose DAC was used successfully in a phase 2 clinical trial to overcome resistance to platinum-based chemotherapy. Methylation array profiling of patients in this study with progression-free survival (PFS) greater than 6 months compared to those less than 6 months suggested that demethylation of MLH1, RASSF1A, HOXA10, and HOXA11 were associated with longer PFS (73). Hypothesized mechanisms of this effect include the reactivation of genes involved in mitochondrial apoptosis, MAPK signaling, and membrane transporter pumps (74). Although not yet clinically tested in bladder cancer, experiments using urothelial carcinoma cells have also implicated HMA-induced reactivation of the tumor suppressor RASSF1A and consequent downstream activation of the Hippo pathway, which acts to slow cell proliferation, inhibit cancer stem cell maintenance, and augment sensitivity to cisplatin and doxorubicin (75).
Unfortunately, many systemically active chemotherapeutic agents, including cisplatin, are unable to cross the blood-brain barrier efficiently enough to penetrate into gliomas without dosing at levels that would be toxic to other end organs. Thus, the orally bioavailable and well-tolerated TMZ is strongly favored. The cytotoxic effects of TMZ are primarily mediated by the formation of O6-methylguanine adducts in DNA. The DNA repair enzyme MGMT acts to reverse these adducts, but if expressed at low levels or silenced, O6-methylguanine will mispair with thymine during DNA replication, triggering the cell's DNA mismatch repair (MMR) machinery, of which MLH1 is an essential player. MMR complexes excise the mispaired base, but because O6-methylguanine will continue to mispair with thymine, a futile loop of attempted mismatch repair is initiated, ultimately triggering DNA double-strand break formation, DNA damage checkpoints, and apoptosis (Fig. 1). MMR deficiency from epigenetic silencing or somatic mutation of component genes is closely linked with TMZ resistance, as is MGMT overexpression via hypomethylation of its promoter and/or methylation of its gene body (76). Thus, use of HMAs to reverse TMZ resistance in GBM has spurred significant interest.
The use of DAC to potentiate the effects of temozolomide via re-expression of MLH1 was first demonstrated by Plumb et al (77) in ovarian and colon cancer xenografts with MMR deficiency due to MLH1 promoter methylation. Subsequently, a phase 1/2 trial in non-resectable stage IIIB/C or IV melanoma was the first clinical demonstration that a 14-day regimen of low-dose DAC (0.15 mg/kg/day or 6 mg/m2/day daily for 5 days) could upregulate MMR genes and increase TMZ sensitivity (78). Complete responses were seen in 2 of 33 patients, with an overall clinical benefit rate of 61% and a median overall survival of 12.4 months, suggesting the safety and potential efficacy of the combination compared to historical controls. Pre- and post-treatment tumor tissue and peripheral blood mononuclear cells from 6 participating patients were analyzed for HMA-induced changes in MGMT and MMR genes, but none were clearly identified. However, overall gene expression changes were similar to those seen in MDS, AML, and sickle cell disease patients after treatment with DAC.
In GBM, using older methylation-specific PCR techniques, MLH1 promoter methylation has been detected in up to 15% of tumor tissue samples (79-81). With the advent of highly sensitive next-generation long-read methylation sequencing techniques, the actual rate is likely higher, presenting an opportunity to make meaningful improvements in TMZ response rates in the vast majority of GBM patients who will ultimately develop TMZ resistance. Work in our own laboratory using GBM stem cell cultures derived from fresh surgical specimens provides evidence that DAC increases MLH1 expression in a subset of GBMs regardless of MGMT methylation status, and that this effect mediates significant reductions in TMZ IC50. Interestingly, full DNA methylation sequencing of the MLH1 promoter region revealed an association between elevated baseline methylation and a lack of TMZ sensitization, while the absence of any baseline methylation at the promoter appeared necessary for sensitization. This suggests that DAC may increase MLH1 levels not through direct demethylation of the promoter but, rather, indirectly via induction of an upstream transcription factor (82) (Fig. 1). MLH1 promoter methylation levels may therefore serve as a clinically useful predictive biomarker for GBM patients who might respond well to DAC-based chemosensitization. Moving forward, biomarker-informed patient selection will be critical to the success of clinical trials testing this approach, since the high molecular heterogeneity of GBM tends to produce negative studies due to underpowering (i.e., beta error).
6. HMAs and immunotherapy
To date, cancer immunotherapy has seen the most success in the treatment of melanoma, lung adenocarcinoma, colorectal adenocarcinoma, and other malignancies with high mutational burden (i.e., ‘hot’ tumors). Such malignancies exhibit relatively high levels of tumor-specific neoantigen expression, which are potentially detectable by the immune system (83-85). GBM, however, is notoriously an immunologically ‘cold’ cancer because of its relatively low mutational burden and low level of neoantigen expression. Moreover, the immunosuppressive tumor microenvironment of GBM provides multiple pathways for tumor immune evasion, leading to low cytotoxic T-cell infiltration and generally poor responses to immunotherapy (86,87). Hence, much recent work in the field of GBM immunotherapy focuses on devising strategies to convert GBMs from ‘cold’ to ‘hot’ in order to enhance either adaptive or innate immune responses.
Epigenetic alterations and global hypermethylation contribute to an immunosuppressive landscape in GBM through a number of mechanisms. Downregulation of MHC class I in GBM cancer stem cells secondary to promoter methylation by EZH2, a methyltransferase, results in resistance to NK cell killing and subsequent innate immune escape (88). EZH2 has also been implicated in downregulating AP-2a, a transcription factor that blocks PD-L1 expression when bound to its promoter (89). HMAs, by suppressing AP-2a methylation, may have the ability to reduce levels of PD-L1 in glioma cells, thereby enhancing immune checkpoint blockade. Progressive methylation of genes can also impair inflammatory pathways in GBM, increasingly inhibiting the adaptive immune response with time. For example, methylation at promoter regions of the IL-7 gene and its receptor has been shown to be significantly more elevated in recurrent compared to newly diagnosed GBM (90).
These observations indicate that HMAs may have the potential to synergize with tumor immunotherapy via multiple, parallel mechanisms. Currently, clinical trials testing HMAs in combination with a variety of immunotherapies are ongoing in malignances such as AML and MDS. In the following sections, we discuss their applications in the enhancement of neoantigen expression, immune checkpoint blockade, cancer vaccines, and innate immune responses (91).
Neoantigen expression
Similar to their effects on tumor suppressor and oncogene expression, HMAs may directly modify neoantigen expression through their inhibition of CpG methylation at the promoters and bodies of neoantigen genes. Cancer-specific neoantigens, including those derived from TP53, KRAS, IDH1/2 and MLH1, may be clonal or highly subclonal throughout a tumor and therefore ideal targets for cancer immunotherapy. At the same time, undesired effects of the known oncogenic functions of these gene products must be weighed in any approach that attempts to increase their expression for heightened immune detection.
In one recent study, autologous neoepitopes generated from the mutational hotspot region of TP53, the most commonly mutated gene across all cancers, were found to be immunogenic in 39% of patients (92). The development of adoptive cell therapy using ex vivo expanded tumor-infiltrating T-cells targeted against TP53 mutations and other public neoepitopes is an active area of investigation (93), and HMAs may be a promising means of increasing the efficacy of this approach. In U87 and GBM patient-derived cell lines, Ma et al (94) demonstrated that DAC induces upregulation of genes encoding for both HLA-A2-restricted neoantigens and tumor-associated cancer testis antigens, leading to an enhanced CD8+ T-cell mediated toxicity response by healthy donor cells in a TCR:MHC class I-dependent manner. The authors also found that DAC generally increased the activation of preexisting cytotoxic T lymphocytes in GBM patients, improving the endogenous recognition of cancer cells.
Another novel approach that has been explored in GBM hijacks the ability of HMAs to upregulate certain oncogenes to instead enhance oncolytic virotherapy. Okemoto et al (95) used an engineered version of the neurotropic HSV1 virus that had been placed under the replicative control of a nestin promoter-enhancer sequence. Nestin is a glioma-specific intermediate filament known to be overexpressed in glioma cells and is used as a marker of the cancer stem cell compartment. The investigators identified several CpG islands within the nestin promoter that became hypermethylated after the virus entered glioma cells. AZA was able to reverse this hypermethylation, significantly improving viral replication both in vitro and in vivo in an orthotopic mouse xenograft model. These studies illustrate the innovative ways in which the wide-ranging effects of HMAs on tumor-associated gene expression can be channeled into anticancer treatment strategies.
Immune checkpoint inhibition
Immune checkpoint blockade (ICB) is an immunotherapeutic approach widely recognized for its potential to produce long-term and deep responses in a subset of cancer patients. The most potent example of checkpoint inhibition is the targeting of the programmed cell death protein 1 (PD-1)/programed cell death ligand 1 (PD-L1) axis and CTLA-4 to unleash a powerful T-cell response and eliminate cancer cells (96). Checkpoint inhibitors have shown promising results in melanoma, where PD-1 blockade with the IgG monoclonal antibody pembrolizumab has improved survival by inhibiting binding to the PD-L1 ligand expressed by neoplastic cells. The same effect has been observed in Hodgkin's lymphoma (97,98), bladder cancer (99), and renal cell carcinoma (100) among others. T-cell activation requires co-stimulatory molecules to induce signaling pathways that lead to chemokine production and proliferation. PD-1 is a co-inhibitory surface molecule present on T cells that helps regulate homeostasis during inflammatory states to prevent autoimmunity. Activation of PD-1 by its ligands leads to T-cell anergy, exhaustion, and apoptosis.
Tumor cells, particularly in glioblastoma, express high levels of the PD-L1 ligand leading to myriad immunosuppressive effects within the tumor microenvironment, many of which have proven challenging to overcome, even with ICB.
In this context, HMAs have garnered interest for their ability in preclinical studies to rejuvenate exhausted T-cells by reversing the acquisition of genomic methylation patterns that act to restrict T-cell expansion and diversification (101). Nie et al (98) demonstrated the clinical translation of this concept in a phase 2 study that examined the effects of adding low dose DAC (10 mg/d x5 days every 3 weeks) to camrelizumab, an anti-PD-1 monoclonal antibody, in patients with classic Hodgkin's lymphoma who were ICB-naïve, and another cohort of patients that had developed resistance to other ICBs. They found that combination therapy was tolerable and increased complete response rates significantly in both cohorts, indicating that DAC might reverse acquired and primary ICB resistance. This effect was associated with a broadened peripheral T-cell receptor repertoire, suggesting that increased tumor immunogenicity might be one responsible mechanism. In follow up in vitro experiments, the authors also found evidence that DAC prevents loss of JunD transcription factor expression in CD8+ T-cells, abrogating their tendency to develop an exhaustion phenotype during ICB therapy (102). Working with CD19-targeted chimeric antigen receptor (CAR) T-cells in a mouse model of non-Hodgkin's lymphoma, Wang et al (103) observed similar anti-exhaustion effects in vivo when low dose (10 nM) DAC was added to the CAR T-cell culture for 7 days.
Although checkpoint inhibition therapies have received FDA approval in the US for use in several solid cancers, including melanoma, hepatocellular carcinoma, and renal cell carcinoma (104,105), all randomized clinical trials for GBM to date have failed to demonstrate a survival benefit over standard chemoradiation. The CheckMate 143 phase 3 randomized trial (106) assessed the efficacy of the PD-1 inhibitor nivolumab compared to bevacizumab alone in patients with GBM at first recurrence, and showed no significant improvement in overall survival with nivolumab (mOS was about 10 months in both arms). More recently, in a phase 3 trial randomizing 560 patients (CheckMate 498), Omuro et al (107) compared combined nivolumab and radiotherapy (RT) to standard-of-care TMZ and RT in newly-diagnosed MGMT-unmethylated GBM, demonstrating significantly shorter overall survival in the nivolumab arm (13.4 vs. 14.9 months). Finally, in a companion phase 3 study that randomized 716 patients (CheckMate 548), the addition of nivolumab to standard radiotherapy plus TMZ in newly-diagnosed MGMT-methylated or indeterminate GBM patients did not improve overall or progression-free survival (108). Based on these negative results, it is becoming increasingly clear that ICB monotherapy as an addition to radiotherapy, with or without temozolomide, is unable to overcome T-cell exhaustion and the powerfully immunosuppressive tumor microenvironment of GBM. However, given the encouraging preclinical and clinical data emerging in hematologic malignancies, using HMAs to address the hurdle of ICB resistance seems to be a promising next tactic that merits exploration by investigators.
Cancer vaccines
Recent studies have shown that the use of personalized cancer vaccines to boost the host immune response are feasible even in tumors that are recognized as insensitive to immunotherapy, such as GBM (109,110) and pancreatic cancer (111). In these clinical trials, patients may receive vaccines containing peptides that match the amino acid sequences of their own tumor-specific antigens, known as ‘personalized neoantigen-targeting vaccines’, or a pre-determined ‘off-the-shelf’ panel of one or more public tumor-specific antigens (112).
Alternatively, antigen-presenting dendritic cells collected from patients through leukapheresis can be exposed ex vivo to tumor-specific antigens in the presence of immunostimulatory adjuvants, such as poly-ICLC or GM-CSF, which promote antigen delivery and dendritic cell maturation and activation (113). Once the dendritic cells are activated, they are reintroduced into the patient to stimulate the adaptive immune response (114). This was tested in newly-diagnosed GBM patients in a recent prospective externally controlled cohort trial where researchers added an autologous tumor lysate-loaded dendritic cell vaccine (DCVax-L) to standard-of-care chemoradiation. The trial was initially designed as a randomized phase 3 trial comparing vaccine to placebo, but due to a high rate of crossover diluting the control arm, could not be meaningfully analyzed as such. Compared to patients receiving only standard-of-care therapy in other GBM trials, who were matched by known prognostic factors, overall survival after initial surgery was modestly but significantly longer (19.3 vs. 16.5 months). The survival advantage was more pronounced when comparing the subset of patients who received the vaccine only after tumor progression to matched external control patients with recurrent GBM (13.2 vs. 7.8 months) (115).
As discussed above, HMAs enhance the expression of neoantigens by tumor cells for targeting by antigen-specific cytotoxic T-cells, presenting an opportunity for synergistic effects when combined with tumor vaccines. Although no trials using this strategy have yet been reported in GBM, a phase 1 study employed standard dose DAC to increase tumor-associated antigen expression in high-risk MDS patients receiving the CDX-1401 vaccine. CDX-1401 targets the cancer testis antigen NY-ESO-1, which is aberrantly expressed in a variety of solid and hematologic cancers. After the vaccine was administered every four weeks, alternating with cycles of DAC, the investigators observed increased NY-ESO-1 expression in myeloid cells and NY-ESO-1-specific CD4+ and CD8+ T-cell responses in a majority of the patients. In a separate preclinical study, DAC significantly increased the expression of NY-ESO-1 in cultured GBM cells and intracranial xenografts in mice after three 10 mg/kg doses. Adoptively transferred NY-ESO-1 TCR-transduced lymphocytes were then able to traffic from an injection site in the contralateral cerebral hemisphere to the xenograft, extending mouse survival significantly (116). Together, these studies provide a strong justification for testing the efficacy of CDX-1401 and other similar vaccines in combination with DAC in GBM.
Innate immune system
Although most studies to date have focused on harnessing and enhancing adaptive immune responses against malignant gliomas with HMAs, evidence also points to the potential for enhancement of the innate immune system. As a first-line defense of the immune system, natural killer (NK) cells are activated by stress-induced ligands and can distinguish ‘self’ from ‘missing-self’ by detecting the absence of the MHC class I molecule on cell membranes. Depending on the balance of activating and inhibitory signals from receptors at the cell surface, NK cells will be cytotoxic or tolerant.
Zhang et al (117) discovered that expression of NK cell-activating NKG2D ligands is suppressed in IDH-mutant gliomas due to their hypermethylated phenotype. DAC (1 µM) treatment of IDH-mutant glioma cells was able to restore NKG2D ligand expression, with the increase lasting up to 7 days after washout of DAC from the culture medium. In IDH-mutant glioma xenografts established in the flanks of athymic nude mice, DAC (10 mg/kg) increased NKG2D ligand expression, enabling NK cells to recognize glioma cells, triggering an NK cell-mediated anti-tumor response and slowing tumor growth (118). Although no such reversible deficit of NK cell-activating ligand expression has yet been demonstrated in GBM, which are IDH-wildtype, these findings raise the possibility that HMAs might be harnessed as a potent enhancer of immunotherapies based on the NK cell platform.
7. Conclusion
In conclusion, GBM is an aggressive tumor with poor prognosis, which currently lacks therapeutic options that can reliably extend survival beyond two years. Based on successes seen in hematologic and other solid malignancies, the use of HMAs in GBM holds promise as a versatile means of enhancing established treatment paradigms, but much work still needs to be done. With strong evidence supporting its clinical use in MDS and AML, DAC is perhaps the most well studied HMA that could also have epigenetic activity in the CNS with an acceptable toxicity profile. This review has examined how HMAs exert their effects by modifying the expression of tumor suppressor genes, oncogenes, tumor-associated antigens and neoantigens, and genes supporting T-cell diversification. A growing understanding of these mechanisms has led investigators to explore the synergies HMAs may have with alkylating chemotherapy, immune checkpoint blockade, cancer vaccines, and other forms of immunotherapy. Future research in GBM should focus on combining HMAs with temozolomide to overcome resistance mechanisms; exploring the utility HMAs might have in changing the immunosuppressive tumor microenvironment in GBM to a more favorable one for tumor-infiltrating lymphocytes; and improving the expression of GBM neoantigens for detection by the immune system. A more complete understanding of the varied mechanisms by which HMAs exert their effects will lead to the identification of biomarkers that will enable clinicians to select the patients most likely to benefit from epigenetic therapy, based on immune system and molecular tumor profiling. Personalized approaches that combine HMAs with rationally chosen, mechanistically complementary therapies offer hope for improving outcomes for GBM patients in the future.
Acknowledgements
Not applicable.
Funding Statement
Funding: Funding for this work was provided by the National Institute of Neurological Disorders and Stroke (Bethesda, Maryland; grant no. R03NS112572).
Availability of data and materials
Not applicable.
Authors' contributions
TJSH drafted the manuscript. JFI contributed to drafting the manuscript and critically edited the manuscript. RLY conceived the study and critically edited the manuscript. All authors read and approved the final manuscript. Data authentication is not applicable.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Di Croce L, Raker VA, Corsaro M, Fazi F, Fanelli M, Faretta M, Fuks F, Lo Coco F, Kouzarides T, Nervi C, et al. Methyltransferase recruitment and DNA hypermethylation of target promoters by an oncogenic transcription factor. Science. 2002;295:1079–1082. doi: 10.1126/science.1065173. [DOI] [PubMed] [Google Scholar]
- 2.Turcan S, Rohle D, Goenka A, Walsh LA, Fang F, Yilmaz E, Campos C, Fabius AW, Lu C, Ward PS, et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012;483:479–483. doi: 10.1038/nature10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lu C, Ward PS, Kapoor GS, Rohle D, Turcan S, Abdel-Wahab O, Edwards CR, Khanin R, Figueroa ME, Melnick A, et al. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature. 2012;483:474–478. doi: 10.1038/nature10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Louis DN, Perry A, Wesseling P, Brat DJ, Cree IA, Figarella-Branger D, Hawkins C, Ng HK, Pfister SM, Reifenberger G, et al. The 2021 WHO classification of tumors of the central nervous system: A summary. Neuro Oncol. 2021;23:1231–1251. doi: 10.1093/neuonc/noab106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ostrom QT, Price M, Neff C, Cioffi G, Waite KA, Kruchko C, Barnholtz-Sloan JS. CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2016-2020. Neuro Oncol. 2023;25 (12 Suppl 2):iv1–iv99. doi: 10.1093/neuonc/noad149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alnahhas I, Alsawas M, Rayi A, Palmer JD, Raval R, Ong S, Giglio P, Murad MH, Puduvalli V. Characterizing benefit from temozolomide in MGMT promoter unmethylated and methylated glioblastoma: A systematic review and meta-analysis. Neurooncol Adv. 2020;2(vdaa082) doi: 10.1093/noajnl/vdaa082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Szklener K, Mazurek M, Wieteska M, Wacławska M, Bilski M, Mańdziuk S. New directions in the therapy of glioblastoma. Cancers (Basel) 2022;14(5377) doi: 10.3390/cancers14215377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Handy DE, Castro R, Loscalzo J. Epigenetic modifications: Basic mechanisms and role in cardiovascular disease. Circulation. 2011;123:2145–2156. doi: 10.1161/CIRCULATIONAHA.110.956839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ivanova E, Canovas S, Garcia-Martínez S, Romar R, Lopes JS, Rizos D, Sanchez-Calabuig MJ, Krueger F, Andrews S, Perez-Sanz F, et al. DNA methylation changes during preimplantation development reveal inter-species differences and reprogramming events at imprinted genes. Clin Epigenetics. 2020;12(64) doi: 10.1186/s13148-020-00857-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duncan CG, Grimm SA, Morgan DL, Bushel PR, Bennett BD. Roberts JD, Tyson FL, Merrick BA, Wade PA. Dosage compensation and DNA methylation landscape of the X chromosome in mouse liver. Sci Rep. 2018;8(10138) doi: 10.1038/s41598-018-28356-3. NISC Comparative Sequencing Program. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brabson JP, Leesang T, Mohammad S, Cimmino L. Epigenetic regulation of genomic stability by vitamin C. Front Genet. 2021;12(675780) doi: 10.3389/fgene.2021.675780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dhar GA, Saha S, Mitra P, Nag Chaudhuri R. DNA methylation and regulation of gene expression: Guardian of our health. Nucleus (Calcutta) 2021;64:259–270. doi: 10.1007/s13237-021-00367-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Compere SJ, Palmiter RD. DNA methylation controls the inducibility of the mouse metallothionein-I gene lymphoid cells. Cell. 1981;25:233–240. doi: 10.1016/0092-8674(81)90248-8. [DOI] [PubMed] [Google Scholar]
- 14.Moore LD, Le T, Fan G. DNA methylation and its basic function. Neuropsychopharmacology. 2013;38:23–38. doi: 10.1038/npp.2012.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mortusewicz O, Schermelleh L, Walter J, Cardoso MC, Leonhardt H. Recruitment of DNA methyltransferase I to DNA repair sites. Proc Natl Acad Sci USA. 2005;102:8905–8909. doi: 10.1073/pnas.0501034102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaneda M, Okano M, Hata K, Sado T, Tsujimoto N, Li E, Sasaki H. Essential role for de novo DNA methyltransferase Dnmt3a in paternal and maternal imprinting. Nature. 2004;429:900–903. doi: 10.1038/nature02633. [DOI] [PubMed] [Google Scholar]
- 17.Aapola U, Kawasaki K, Scott HS, Ollila J, Vihinen M, Heino M, Shintani A, Kawasaki K, Minoshima S, Krohn K, et al. Isolation and initial characterization of a novel zinc finger gene, DNMT3L, on 21q22.3, related to the cytosine-5-methyltransferase 3 gene family. Genomics. 2000;65:293–298. doi: 10.1006/geno.2000.6168. [DOI] [PubMed] [Google Scholar]
- 18.Jin B, Li Y, Robertson KD. DNA methylation: Superior or subordinate in the epigenetic hierarchy? Genes Cancer. 2011;2:607–617. doi: 10.1177/1947601910393957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rideout WM III, Coetzee GA, Olumi AF, Jones PA. 5-Methylcytosine as an endogenous mutagen in the human LDL receptor and p53 genes. Science. 1990;249:1288–1290. doi: 10.1126/science.1697983. [DOI] [PubMed] [Google Scholar]
- 20.Ramirez-Carrozzi VR, Braas D, Bhatt DM, Cheng CS, Hong C, Doty KR, Black JC, Hoffmann A, Carey M, Smale ST. A unifying model for the selective regulation of inducible transcription by CpG islands and nucleosome remodeling. Cell. 2009;138:114–128. doi: 10.1016/j.cell.2009.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brenet F, Moh M, Funk P, Feierstein E, Viale AJ, Socci ND, Scandura JM. DNA methylation of the first exon is tightly linked to transcriptional silencing. PLoS One. 2011;6(e14524) doi: 10.1371/journal.pone.0014524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hellman A, Chess A. Gene body-specific methylation on the active X chromosome. Science. 2007;315:1141–1143. doi: 10.1126/science.1136352. [DOI] [PubMed] [Google Scholar]
- 24.Bogdanović O, Veenstra GJ. DNA methylation and methyl-CpG binding proteins: Developmental requirements and function. Chromosoma. 2009;118:549–565. doi: 10.1007/s00412-009-0221-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Y, Zheng H, Wang Q, Zhou C, Wei L, Liu X, Zhang W, Zhang Y, Du Z, Wang X, Xie W. Genome-wide analyses reveal a role of polycomb in promoting hypomethylation of DNA methylation valleys. Genome Biol. 2018;19(18) doi: 10.1186/s13059-018-1390-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mohn F, Weber M, Rebhan M, Roloff TC, Richter J, Stadler MB, Bibel M, Schübeler D. Lineage-specific polycomb targets and de novo DNA methylation define restriction and potential of neuronal progenitors. Mol Cell. 2008;30:755–766. doi: 10.1016/j.molcel.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 27.Ghadiri Moghaddam F, Farajnia S, Karbalaei-Mahdi M, Monir L. Epigenetic insights in the diagnosis, prognosis, and treatment selection in CRC, an updated review. Mol Biol Rep. 2022;49:10013–10022. doi: 10.1007/s11033-022-07569-w. [DOI] [PubMed] [Google Scholar]
- 28.Collings CK, Anderson JN. Links between DNA methylation and nucleosome occupancy in the human genome. Epigenetics Chromatin. 2017;10(18) doi: 10.1186/s13072-017-0125-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang KK, Ramnarayanan K, Zhu F, Srivastava S, Xu C, Tan ALK, Lee M, Tay S, Das K, Xing M, et al. Genomic and epigenomic profiling of high-risk intestinal metaplasia reveals molecular determinants of progression to gastric cancer. Cancer Cell. 2018;33:137–150.e5. doi: 10.1016/j.ccell.2017.11.018. [DOI] [PubMed] [Google Scholar]
- 30.Wang Q, Xiong F, Wu G, Liu W, Chen J, Wang B, Chen Y. Gene body methylation in cancer: Molecular mechanisms and clinical applications. Clin Epigenetics. 2022;14(154) doi: 10.1186/s13148-022-01382-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Santini V, Ossenkoppele GJ. Hypomethylating agents in the treatment of acute myeloid leukemia: A guide to optimal use. Crit Rev Oncol Hematol. 2019;140:1–7. doi: 10.1016/j.critrevonc.2019.05.013. [DOI] [PubMed] [Google Scholar]
- 32.Holliday R, Ho T. DNA methylation and epigenetic inheritance. Methods. 2002;27:179–183. doi: 10.1016/s1046-2023(02)00072-5. [DOI] [PubMed] [Google Scholar]
- 33.Jabbour E, Issa JP, Garcia-Manero G, Kantarjian H. Evolution of decitabine development: Accomplishments, ongoing investigations, and future strategies. Cancer. 2008;112:2341–2351. doi: 10.1002/cncr.23463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sorm F, Veselý J. Effect of 5-aza-2'-deoxycytidine against leukemic and hemopoietic tissues in AKR mice. Neoplasma. 1968;15:339–343. [PubMed] [Google Scholar]
- 35.Xu K, Hansen E. Novel agents for myelodysplastic syndromes. J Oncol Pharm Pract. 2021;27:1982–1992. doi: 10.1177/10781552211037993. [DOI] [PubMed] [Google Scholar]
- 36.Kordella C, Lamprianidou E, Kotsianidis I. Mechanisms of action of hypomethylating agents: Endogenous retroelements at the epicenter. Front Oncol. 2021;11(650473) doi: 10.3389/fonc.2021.650473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Quintás-Cardama A, Santos FP, Garcia-Manero G. Therapy with azanucleosides for myelodysplastic syndromes. Nat Rev Clin Oncol. 2010;7:433–444. doi: 10.1038/nrclinonc.2010.87. [DOI] [PubMed] [Google Scholar]
- 38.Hollenbach PW, Nguyen AN, Brady H, Williams M, Ning Y, Richard N, Krushel L, Aukerman SL, Heise C, MacBeth KJ. A comparison of azacitidine and decitabine activities in acute myeloid leukemia cell lines. PLoS One. 2010;5(e9001) doi: 10.1371/journal.pone.0009001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Seelan RS, Mukhopadhyay P, Pisano MM, Greene RM. Effects of 5-Aza-2'-deoxycytidine (decitabine) on gene expression. Drug Metab Rev. 2018;50:193–207. doi: 10.1080/03602532.2018.1437446. [DOI] [PubMed] [Google Scholar]
- 40.Cashen AF, Shah AK, Todt L, Fisher N, DiPersio J. Pharmacokinetics of decitabine administered as a 3-h infusion to patients with acute myeloid leukemia (AML) or myelodysplastic syndrome (MDS) Cancer Chemother Pharmacol. 2008;61:759–766. doi: 10.1007/s00280-007-0531-7. [DOI] [PubMed] [Google Scholar]
- 41.Chabot GG, Rivard GE, Momparler RL. Plasma and cerebrospinal fluid pharmacokinetics of 5-Aza-2'-deoxycytidine in rabbits and dogs. Cancer Res. 1983;43:592–597. [PubMed] [Google Scholar]
- 42.Kim N, Norsworthy KJ, Subramaniam S, Chen H, Manning ML, Kitabi E, Earp J, Ehrlich LA, Okusanya OO, Vallejo J, et al. FDA approval summary: Decitabine and cedazuridine tablets for myelodysplastic syndromes. Clin Cancer Res. 2022;28:3411–3416. doi: 10.1158/1078-0432.CCR-21-4498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kantarjian H, Oki Y, Garcia-Manero G, Huang X, O'Brien S, Cortes J, Faderl S, Bueso-Ramos C, Ravandi F, Estrov Z, et al. Results of a randomized study of 3 schedules of low-dose decitabine in higher-risk myelodysplastic syndrome and chronic myelomonocytic leukemia. Blood. 2007;109:52–57. doi: 10.1182/blood-2006-05-021162. [DOI] [PubMed] [Google Scholar]
- 44.Kantarjian H, Issa JP, Rosenfeld CS, Bennett JM, Albitar M, DiPersio J, Klimek V, Slack J, de Castro C, Ravandi F, et al. Decitabine improves patient outcomes in myelodysplastic syndromes: Results of a phase III randomized study. Cancer. 2006;106:1794–1803. doi: 10.1002/cncr.21792. [DOI] [PubMed] [Google Scholar]
- 45.Müller A, Florek M. 5-Azacytidine/azacitidine. Recent Results Cancer Res. 2010;184:159–170. doi: 10.1007/978-3-642-01222-8_11. [DOI] [PubMed] [Google Scholar]
- 46.Krawczyk J, Keane N, Freeman CL, Swords R, O'Dwyer M, Giles FJ. 5-Azacytidine for the treatment of myelodysplastic syndromes. Expert Opin Pharmacother. 2013;14:1255–1268. doi: 10.1517/14656566.2013.794222. [DOI] [PubMed] [Google Scholar]
- 47.Glover AB, Leyland-Jones BR, Chun HG, Davies B, Hoth DF. Azacitidine: 10 Years later. Cancer Treat Rep. 1987;71:737–746. [PubMed] [Google Scholar]
- 48.Kaminskas E, Farrell AT, Wang YC, Sridhara R, Pazdur R. FDA drug approval summary: Azacitidine (5-azacytidine, Vidaza) for injectable suspension. Oncologist. 2005;10:176–182. doi: 10.1634/theoncologist.10-3-176. [DOI] [PubMed] [Google Scholar]
- 49.Marcucci G, Silverman L, Eller M, Lintz L, Beach CL. Bioavailability of azacitidine subcutaneous versus intravenous in patients with the myelodysplastic syndromes. J Clin Pharmacol. 2005;45:597–602. doi: 10.1177/0091270004271947. [DOI] [PubMed] [Google Scholar]
- 50.Garcia-Manero G, Roboz G, Walsh K, Kantarjian H, Ritchie E, Kropf P, O'Connell C, Tibes R, Lunin S, Rosenblat T, et al. Guadecitabine (SGI-110) in patients with intermediate or high-risk myelodysplastic syndromes: phase 2 results from a multicentre, open-label, randomised, phase 1/2 trial. Lancet Haematol. 2019;6:e317–e327. doi: 10.1016/S2352-3026(19)30029-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chuang JC, Warner SL, Vollmer D, Vankayalapati H, Redkar S, Bearss DJ, Qiu X, Yoo CB, Jones PA. S110, a 5-Aza-2'-deoxycytidine-containing dinucleotide, is an effective DNA methylation inhibitor in vivo and can reduce tumor growth. Mol Cancer Ther. 2010;9:1443–1450. doi: 10.1158/1535-7163.MCT-09-1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Issa JJ, Roboz G, Rizzieri D, Jabbour E, Stock W, O'Connell C, Yee K, Tibes R, Griffiths EA, Walsh K, et al. Safety and tolerability of guadecitabine (SGI-110) in patients with myelodysplastic syndrome and acute myeloid leukaemia: A multicentre, randomised, dose-escalation phase 1 study. Lancet Oncol. 2015;16:1099–1110. doi: 10.1016/S1470-2045(15)00038-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ramakrishnan S, Hu Q, Krishnan N, Wang D, Smit E, Granger V, Rak M, Attwood K, Johnson C, Morrison C, et al. Decitabine, a DNA-demethylating agent, promotes differentiation via NOTCH1 signaling and alters immune-related pathways in muscle-invasive bladder cancer. Cell Death Dis. 2017;8(3217) doi: 10.1038/s41419-017-0024-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li M, Zhang D. DNA methyltransferase-1 in acute myeloid leukaemia: Beyond the maintenance of DNA methylation. Ann Med. 2022;54:2011–2023. doi: 10.1080/07853890.2022.2099578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pappalardi MB, Keenan K, Cockerill M, Kellner WA, Stowell A, Sherk C, Wong K, Pathuri S, Briand J, Steidel M, et al. Discovery of a first-in-class reversible DNMT1-selective inhibitor with improved tolerability and efficacy in acute myeloid leukemia. Nat Cancer. 2021;2:1002–1017. [PMC free article] [PubMed] [Google Scholar]
- 56.Quesnel B, Fenaux P. P15INK4b gene methylation and myelodysplastic syndromes. Leuk Lymphoma. 1999;35:437–443. doi: 10.1080/10428199909169608. [DOI] [PubMed] [Google Scholar]
- 57.Daver NG, Maiti A, Kadia TM, Vyas P, Majeti R, Wei AH, Garcia-Manero G, Craddock C, Sallman DA, Kantarjian HM. TP53-mutated myelodysplastic syndrome and acute myeloid leukemia: Biology, current therapy, and future directions. Cancer Discov. 2022;12:2516–2529. doi: 10.1158/2159-8290.CD-22-0332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Claus R, Hackanson B, Poetsch AR, Zucknick M, Sonnet M, Blagitko-Dorfs N, Hiller J, Wilop S, Brümmendorf TH, Galm O, et al. Quantitative analyses of DAPK1 methylation in AML and MDS. Int J Cancer. 2012;131:E138–E142. doi: 10.1002/ijc.26429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Flotho C, Claus R, Batz C, Schneider M, Sandrock I, Ihde S, Plass C, Niemeyer CM, Lübbert M. The DNA methyltransferase inhibitors azacitidine, decitabine and zebularine exert differential effects on cancer gene expression in acute myeloid leukemia cells. Leukemia. 2009;23:1019–1028. doi: 10.1038/leu.2008.397. [DOI] [PubMed] [Google Scholar]
- 60.Xie B, Peng F, He F, Cheng Y, Cheng J, Zhou Z, Mao W. DNA methylation influences the CTCF-modulated transcription of RASSF1A in lung cancer cells. Cell Biol Int. 2022;46:1900–1914. doi: 10.1002/cbin.11868. [DOI] [PubMed] [Google Scholar]
- 61.Tang Q, Cheng J, Cao X, Surowy H, Burwinkel B. Blood-based DNA methylation as biomarker for breast cancer: A systematic review. Clin Epigenetics. 2016;8(115) doi: 10.1186/s13148-016-0282-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cheng W, Jiang Y, Liu C, Shen O, Tang W, Wang X. Identification of aberrant promoter hypomethylation of HOXA10 in ovarian cancer. J Cancer Res Clin Oncol. 2010;136:1221–1227. doi: 10.1007/s00432-010-0772-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ranjan N, Pandey V, Panigrahi MK, Klumpp L, Naumann U, Babu PP. The tumor suppressor MTUS1/ATIP1 modulates tumor promotion in glioma: Association with epigenetics and DNA repair. Cancers (Basel) 2021;13(1245) doi: 10.3390/cancers13061245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Götze S, Feldhaus V, Traska T, Wolter M, Reifenberger G, Tannapfel A, Kuhnen C, Martin D, Müller O, Sievers S. ECRG4 is a candidate tumor suppressor gene frequently hypermethylated in colorectal carcinoma and glioma. BMC Cancer. 2009;9(447) doi: 10.1186/1471-2407-9-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Alaminos M, Dávalos V, Ropero S, Setién F, Paz MF, Herranz M, Fraga MF, Mora J, Cheung NK, Gerald WL, Esteller M. EMP3, a myelin-related gene located in the critical 19q13.3 region, is epigenetically silenced and exhibits features of a candidate tumor suppressor in glioma and neuroblastoma. Cancer Res. 2005;65:2565–2571. doi: 10.1158/0008-5472.CAN-04-4283. [DOI] [PubMed] [Google Scholar]
- 66.Sanaei M, Kavoosi F. The effect of 5-aza,2'-deoxyCytidine (5 AZA CdR or decitabine) on extrinsic, intrinsic, and JAK/STAT pathways in neuroblastoma and glioblastoma cells lines. Asian Pac J Cancer Prev. 2023;24:1841–1854. doi: 10.31557/APJCP.2023.24.6.1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yang X, Han H, De Carvalho DD, Lay FD, Jones PA, Liang G. Gene body methylation can alter gene expression and is a therapeutic target in cancer. Cancer Cell. 2014;26:577–590. doi: 10.1016/j.ccr.2014.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ochs K, Kaina B. Apoptosis induced by DNA damage O6-methylguanine is Bcl-2 and caspase-9/3 regulated and Fas/caspase-8 independent. Cancer Res. 2000;60:5815–5824. [PubMed] [Google Scholar]
- 69.Tabu K, Sasai K, Kimura T, Wang L, Aoyanagi E, Kohsaka S, Tanino M, Nishihara H, Tanaka S. Promoter hypomethylation regulates CD133 expression in human gliomas. Cell Res. 2008;18:1037–1046. doi: 10.1038/cr.2008.270. [DOI] [PubMed] [Google Scholar]
- 70.Yi JM, Tsai HC, Glöckner SC, Lin S, Ohm JE, Easwaran H, James CD, Costello JF, Riggins G, Eberhart CG, et al. Abnormal DNA methylation of CD133 in colorectal and glioblastoma tumors. Cancer Res. 2008;68:8094–8103. doi: 10.1158/0008-5472.CAN-07-6208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Federici L, Capelle L, Annereau M, Bielle F, Willekens C, Dehais C, Laigle-Donadey F, Hoang-Xuan K, Delattre JY, Idbaih A, et al. 5-Azacitidine in patients with IDH1/2-mutant recurrent glioma. Neuro Oncol. 2020;22:1226–1228. doi: 10.1093/neuonc/noaa074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sato T, Issa JJ, Kropf P. DNA Hypomethylating drugs in cancer therapy. Cold Spring Harb Perspect Med. 2017;7(a026948) doi: 10.1101/cshperspect.a026948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Matei D, Fang F, Shen C, Schilder J, Arnold A, Zeng Y, Berry WA, Huang T, Nephew KP. Epigenetic resensitization to platinum in ovarian cancer. Cancer Res. 2012;72:2197–2205. doi: 10.1158/0008-5472.CAN-11-3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Glaysher S, Gabriel FG, Johnson P, Polak M, Knight LA, Parker K, Poole M, Narayanan A, Cree IA. Molecular basis of chemosensitivity of platinum pre-treated ovarian cancer to chemotherapy. Br J Cancer. 2010;103:656–662. doi: 10.1038/sj.bjc.6605817. NHS Collaborative Research Programme for Predictive Oncology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hannon CE, Eisen MB. Intrinsic protein disorder is insufficient to drive subnuclear clustering in embryonic transcription factors. Elife. 2024;12(RP88221) doi: 10.7554/eLife.88221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Moen EL, Stark AL, Zhang W, Dolan ME, Godley LA. The role of gene body cytosine modifications in MGMT expression and sensitivity to temozolomide. Mol Cancer Ther. 2014;13:1334–1344. doi: 10.1158/1535-7163.MCT-13-0924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Plumb JA, Strathdee G, Sludden J, Kaye SB, Brown R. Reversal of drug resistance in human tumor xenografts by 2'-deoxy-5-azacytidine-induced demethylation of the hMLH1 gene promoter. Cancer Res. 2000;60:6039–6044. [PubMed] [Google Scholar]
- 78.Tawbi HA, Beumer JH, Tarhini AA, Moschos S, Buch SC, Egorin MJ, Lin Y, Christner S, Kirkwood JM. Safety and efficacy of decitabine in combination with temozolomide in metastatic melanoma: A phase I/II study and pharmacokinetic analysis. Ann Oncol. 2013;24:1112–1119. doi: 10.1093/annonc/mds591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Skiriutė D, Vaitkienė P, Ašmonienė V, Steponaitis G, Deltuva VP, Tamašauskas A. Promoter methylation of AREG, HOXA11, hMLH1, NDRG2, NPTX2 and Tes genes in glioblastoma. J Neurooncol. 2013;113:441–449. doi: 10.1007/s11060-013-1133-3. [DOI] [PubMed] [Google Scholar]
- 80.Rodríguez-Hernández I, Garcia JL, Santos-Briz A, Hernández-Laín A, González-Valero JM, Gómez-Moreta JA, Toldos-González O, Cruz JJ, Martin-Vallejo J, González-Sarmiento R. Integrated analysis of mismatch repair system in malignant astrocytomas. PLoS One. 2013;8(e76401) doi: 10.1371/journal.pone.0076401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Fukushima T, Katayama Y, Watanabe T, Yoshino A, Ogino A, Ohta T, Komine C. Promoter hypermethylation of mismatch repair gene hMLH1 predicts the clinical response of malignant astrocytomas to nitrosourea. Clin Cancer Res. 2005;11:1539–1544. doi: 10.1158/1078-0432.CCR-04-1625. [DOI] [PubMed] [Google Scholar]
- 82.Gallitto M, Cheng He R, Inocencio JF, Wang H, Zhang Y, Deikus G, Wasserman I, Strahl M, Smith M, Sebra R, Yong RL. Epigenetic preconditioning with decitabine sensitizes glioblastoma to temozolomide via induction of MLH1. J Neurooncol. 2020;147:557–566. doi: 10.1007/s11060-020-03461-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mehnert JM, Panda A, Zhong H, Hirshfield K, Damare S, Lane K, Sokol L, Stein MN, Rodriguez-Rodriquez L, Kaufman HL, et al. Immune activation and response to pembrolizumab in POLE-mutant endometrial cancer. J Clin Invest. 2016;126:2334–2340. doi: 10.1172/JCI84940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Panda A, Betigeri A, Subramanian K, Ross JS, Pavlick DC, Ali S, Markowski P, Silk A, Kaufman HL, Lattime E, et al. Identifying a clinically applicable mutational burden threshold as a potential biomarker of response to immune checkpoint therapy in solid tumors. JCO Precis Oncol. 2017;2017(PO.17.00146) doi: 10.1200/PO.17.00146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rizvi NA, Hellmann MD, Snyder A, Kvistborg P, Makarov V, Havel JJ, Lee W, Yuan J, Wong P, Ho TS, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348:124–128. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.DeCordova S, Shastri A, Tsolaki AG, Yasmin H, Klein L, Singh SK, Kishore U. Molecular heterogeneity and immunosuppressive microenvironment in glioblastoma. Front Immunol. 2020;11(1402) doi: 10.3389/fimmu.2020.01402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zaidi N, Jaffee EM. Immune cells track hard-to-target brain tumours. Nature. 2019;565:170–171. doi: 10.1038/d41586-018-07728-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhong J, Yang X, Chen J, He K, Gao X, Wu X, Zhang M, Zhou H, Xiao F, An L, et al. Circular EZH2-encoded EZH2-92aa mediates immune evasion in glioblastoma via inhibition of surface NKG2D ligands. Nat Commun. 2022;13(4795) doi: 10.1038/s41467-022-32311-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Long S, Huang G, Ouyang M, Xiao K, Zhou H, Hou A, Li Z, Zhong Z, Zhong D, Wang Q, et al. Epigenetically modified AP-2α by DNA methyltransferase facilitates glioma immune evasion by upregulating PD-L1 expression. Cell Death Dis. 2023;14(365) doi: 10.1038/s41419-023-05878-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tompa M, Kraboth Z, Galik B, Kajtar B, Gyenesei A, Kalman B. Epigenetic suppression of the IL-7 pathway in progressive glioblastoma. Biomedicines. 2022;10(2174) doi: 10.3390/biomedicines10092174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Héninger E, Krueger TE, Lang JM. Augmenting antitumor immune responses with epigenetic modifying agents. Front Immunol. 2015;6(29) doi: 10.3389/fimmu.2015.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Malekzadeh P, Pasetto A, Robbins PF, Parkhurst MR, Paria BC, Jia L, Gartner JJ, Hill V, Yu Z, Restifo NP, et al. Neoantigen screening identifies broad TP53 mutant immunogenicity in patients with epithelial cancers. J Clin Invest. 2019;129:1109–1114. doi: 10.1172/JCI123791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tran E, Robbins PF, Lu YC, Prickett TD, Gartner JJ, Jia L, Pasetto A, Zheng Z, Ray S, Groh EM, et al. T-cell transfer therapy targeting mutant KRAS in cancer. N Engl J Med. 2016;375:2255–2262. doi: 10.1056/NEJMoa1609279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ma R, Rei M, Woodhouse I, Ferris K, Kirschner S, Chandran A, Gileadi U, Chen JL, Pereira Pinho M, Ariosa-Morejon Y, et al. Decitabine increases neoantigen and cancer testis antigen expression to enhance T-cell-mediated toxicity against glioblastoma. Neuro Oncol. 2022;24:2093–2106. doi: 10.1093/neuonc/noac107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Okemoto K, Kasai K, Wagner B, Haseley A, Meisen H, Bolyard C, Mo X, Wehr A, Lehman A, Fernandez S, et al. DNA demethylating agents synergize with oncolytic HSV1 against malignant gliomas. Clin Cancer Res. 2013;19:5952–5959. doi: 10.1158/1078-0432.CCR-12-3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Nebhan CA, Johnson DB. Pembrolizumab in the adjuvant treatment of melanoma: Efficacy and safety. Expert Rev Anticancer Ther. 2021;21:583–590. doi: 10.1080/14737140.2021.1882856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Allen PB, Savas H, Evens AM, Advani RH, Palmer B, Pro B, Karmali R, Mou E, Bearden J, Dillehay G, et al. Pembrolizumab followed by AVD in untreated early unfavorable and advanced-stage classical Hodgkin lymphoma. Blood. 2021;137:1318–1326. doi: 10.1182/blood.2020007400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Nie J, Wang C, Liu Y, Yang Q, Mei Q, Dong L, Li X, Liu J, Ku W, Zhang Y, et al. Addition of low-dose decitabine to anti-PD-1 antibody camrelizumab in relapsed/refractory classical hodgkin lymphoma. J Clin Oncol. 2019;37:1479–1489. doi: 10.1200/JCO.18.02151. [DOI] [PubMed] [Google Scholar]
- 99.Merseburger AS, Apolo AB, Chowdhury S, Hahn NM, Galsky MD, Milowsky MI, Petrylak D, Powles T, Quinn DI, Rosenberg JE, et al. SIU-ICUD recommendations on bladder cancer: Systemic therapy for metastatic bladder cancer. World J Urol. 2019;37:95–105. doi: 10.1007/s00345-018-2486-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Chowdhury S, Infante JR, Hawkins R, Voss MH, Perini R, Arkenau T, Voskoboynik M, Aimone P, Naeije I, Reising A, McDermott DF. A phase I/II study to assess the safety and efficacy of pazopanib and pembrolizumab combination therapy in patients with advanced renal cell carcinoma. Clin Genitourin Cancer. 2021;19:434–446. doi: 10.1016/j.clgc.2021.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ghoneim HE, Fan Y, Moustaki A, Abdelsamed HA, Dash P, Dogra P, Carter R, Awad W, Neale G, Thomas PG, Youngblood B. De novo epigenetic programs inhibit PD-1 blockade-mediated T cell rejuvenation. Cell. 2017;170:142–157.e19. doi: 10.1016/j.cell.2017.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Li X, Li Y, Dong L, Chang Y, Zhang X, Wang C, Chen M, Bo X, Chen H, Han W, Nie J. Decitabine priming increases anti-PD-1 antitumor efficacy by promoting CD8+ progenitor exhausted T cell expansion in tumor models. J Clin Invest. 2023;133(e165673) doi: 10.1172/JCI165673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wang Y, Tong C, Dai H, Wu Z, Han X, Guo Y, Chen D, Wei J, Ti D, Liu Z, et al. Low-dose decitabine priming endows CAR T cells with enhanced and persistent antitumour potential via epigenetic reprogramming. Nat Commun. 2021;12(409) doi: 10.1038/s41467-020-20696-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Papadatos-Pastos D, Yuan W, Pal A, Crespo M, Ferreira A, Gurel B, Prout T, Ameratunga M, Chénard-Poirier M, Curcean A, et al. Phase 1, dose-escalation study of guadecitabine (SGI-110) in combination with pembrolizumab in patients with solid tumors. J Immunother Cancer. 2022;10(e004495) doi: 10.1136/jitc-2022-004495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wei SC, Duffy CR, Allison JP. Fundamental mechanisms of immune checkpoint blockade therapy. Cancer Discov. 2018;8:1069–1086. doi: 10.1158/2159-8290.CD-18-0367. [DOI] [PubMed] [Google Scholar]
- 106.Reardon DA, Brandes AA, Omuro A, Mulholland P, Lim M, Wick A, Baehring J, Ahluwalia MS, Roth P, Bähr O, et al. Effect of nivolumab vs bevacizumab in patients with recurrent glioblastoma: The CheckMate 143 phase 3 randomized clinical trial. JAMA Oncol. 2020;6:1003–1010. doi: 10.1001/jamaoncol.2020.1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Omuro A, Brandes AA, Carpentier AF, Idbaih A, Reardon DA, Cloughesy T, Sumrall A, Baehring J, van den Bent M, Bähr O, et al. Radiotherapy combined with nivolumab or temozolomide for newly diagnosed glioblastoma with unmethylated MGMT promoter: An international randomized phase III trial. Neuro Oncol. 2023;25:123–134. doi: 10.1093/neuonc/noac099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lim M, Weller M, Idbaih A, Steinbach J, Finocchiaro G, Raval RR, Ansstas G, Baehring J, Taylor JW, Honnorat J, et al. Phase III trial of chemoradiotherapy with temozolomide plus nivolumab or placebo for newly diagnosed glioblastoma with methylated MGMT promoter. Neuro Oncol. 2022;24:1935–1949. doi: 10.1093/neuonc/noac116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hilf N, Kuttruff-Coqui S, Frenzel K, Bukur V, Stevanović S, Gouttefangeas C, Platten M, Tabatabai G, Dutoit V, van der Burg SH, et al. Actively personalized vaccination trial for newly diagnosed glioblastoma. Nature. 2019;565:240–245. doi: 10.1038/s41586-018-0810-y. [DOI] [PubMed] [Google Scholar]
- 110.Keskin DB, Anandappa AJ, Sun J, Tirosh I, Mathewson ND, Li S, Oliveira G, Giobbie-Hurder A, Felt K, Gjini E, et al. Neoantigen vaccine generates intratumoral T cell responses in phase Ib glioblastoma trial. Nature. 2019;565:234–239. doi: 10.1038/s41586-018-0792-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Rojas LA, Sethna Z, Soares KC, Olcese C, Pang N, Patterson E, Lihm J, Ceglia N, Guasp P, Chu A, et al. Personalized RNA neoantigen vaccines stimulate T cells in pancreatic cancer. Nature. 2023;618:144–150. doi: 10.1038/s41586-023-06063-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Weller M, Butowski N, Tran DD, Recht LD, Lim M, Hirte H, Ashby L, Mechtler L, Goldlust SA, Iwamoto F, et al. Rindopepimut with temozolomide for patients with newly diagnosed, EGFRvIII-expressing glioblastoma (ACT IV): A randomised, double-blind, international phase 3 trial. Lancet Oncol. 2017;18:1373–1385. doi: 10.1016/S1470-2045(17)30517-X. [DOI] [PubMed] [Google Scholar]
- 113.Zhao T, Cai Y, Jiang Y, He X, Wei Y, Yu Y, Tian X. Vaccine adjuvants: Mechanisms and platforms. Signal Transduct Target Ther. 2023;8(283) doi: 10.1038/s41392-023-01557-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Palucka K, Banchereau J. Cancer immunotherapy via dendritic cells. Nat Rev Cancer. 2012;12:265–277. doi: 10.1038/nrc3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Liau LM, Ashkan K, Brem S, Campian JL, Trusheim JE, Iwamoto FM, Tran DD, Ansstas G, Cobbs CS, Heth JA, et al. Association of autologous tumor lysate-loaded dendritic cell vaccination with extension of survival among patients with newly diagnosed and recurrent glioblastoma: A phase 3 prospective externally controlled cohort trial. JAMA Oncol. 2023;9:112–121. doi: 10.1001/jamaoncol.2022.5370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Everson RG, Antonios JP, Lisiero DN, Soto H, Scharnweber R, Garrett MC, Yong WH, Li N, Li G, Kruse CA, et al. Efficacy of systemic adoptive transfer immunotherapy targeting NY-ESO-1 for glioblastoma. Neuro Oncol. 2016;18:368–378. doi: 10.1093/neuonc/nov153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Zhang X, Rao A, Sette P, Deibert C, Pomerantz A, Kim WJ, Kohanbash G, Chang Y, Park Y, Engh J, et al. IDH mutant gliomas escape natural killer cell immune surveillance by downregulation of NKG2D ligand expression. Neuro Oncol. 2016;18:1402–1412. doi: 10.1093/neuonc/now061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zhang X, Kim WJ, Rao AV, Jaman E, Deibert CP, Sandlesh P, Krueger K, Allen JC, Amankulor NM. In vivo efficacy of decitabine as a natural killer cell-mediated immunotherapy against isocitrate dehydrogenase mutant gliomas. Neurosurg Focus. 2022;52(E3) doi: 10.3171/2021.11.FOCUS21489. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.