Abstract
Histone acetyltransferases KAT2A and KAT2B are paralogs highly expressed in the intestinal epithelium, but their functions are not well understood. In this study, double knockout of murine Kat2 genes in the intestinal epithelium was lethal, resulting in robust activation of interferon signaling and interferon-associated phenotypes including the loss of intestinal stem cells. Use of pharmacological agents and sterile organoid cultures indicated a cell-intrinsic double-stranded RNA trigger for interferon signaling. Acetyl-proteomics and sequencing of immunoprecipitated double-stranded RNA were used to interrogate the mechanism behind this response, which identified mitochondria-encoded double-stranded RNA as the source of intrinsic interferon signaling. Kat2a and Kat2b therefore play an essential role in regulating mitochondrial functions and maintaining intestinal health.
Histone acetyltransferases unexpectedly suppress interferon signaling induced by mitochondrial dsRNA.
INTRODUCTION
The intestinal epithelium not only serves absorptive functions but also acts as a barrier against pathogens and microbiota within the lumen. Following an infection, nucleotides of pathogenic origin can be detected by pattern recognition sensors, such as Toll-like receptors (TLRs), stimulator of interferon (IFN) genes (STING), melanoma differentiation–associated gene 5, and retinoic acid–inducible gene I. Besides these pathogen-associated molecular patterns, host-derived danger-associated molecular patterns from DNA damage and apoptosis can trigger an epithelial response. Host-derived double-stranded RNA (dsRNA) from the nuclear genome via endogenous retrovirus reactivation, or via leaked mitochondrial transcripts, may activate these pattern recognition sensors (1–9). In response to detection of these nucleotides, type I and type III IFNs are produced in the intestinal epithelium to activate transcription of IFN-stimulated genes (ISGs) that mount a defense against threats (10–12). Furthermore, specialized types of intestinal epithelial cells (IECs) partake in innate immunity, such as Paneth cells via secretion antimicrobial peptides and cytokines (13, 14). Activation of IFN signaling can also have negative consequences on intestinal function, such as with the loss of intestinal stem cells via Irf2 deletion or polyinosinic:polycytidylic acid [poly(I:C)] treatment (15, 16).
DNA, as the carrier of genetic instructions for all cells, must also be carefully regulated to allow or prohibit transcription of genes as needed. DNA is compacted into chromatin by wrapping approximately 146 base pairs around a nucleosome with each nucleosome comprised of an octomer of histones (17). Histones contain unstructured N-terminal “tails” that are the substrate for a variety of posttranslational modifications (PTMs). Histone PTMs serve as a means for controlling formation of euchromatin (open chromatin) and heterochromatin (compacted chromatin) and alter the recruitment of transregulating proteins to the chromatin. Histone PTMs do not necessarily behave independently but rather cross-talk among one another and collectively form a “histone code” as proposed by Strahl and Allis (18). Many histone readers, writers, and erasers maintain this histone code, allowing or disabling transcription of specific genes based on the needs of the cell.
KAT2A and KAT2B encode for proteins general control nonderepressible protein 5 (GCN5, also GCN5L2) and p300/CREB-binding protein (CBP)–associated factor (PCAF), respectively. KAT2A and KAT2B are paralogous genes and have roughly 70% amino acid sequence homology (19, 20). Both KAT2A and KAT2B are reported to redundantly catalyze acetylation of lysine residues on histone H3 tails, particularly lysines 9 and 14 (H3K9ac and H3K14ac, respectively). In addition, these acetyltransferases also catalyze other acetyl and nonacetyl marks (21–34).
Previous studies suggest that KAT2A and KAT2B function together in chromatin remodeling, DNA replication, DNA repair, cell cycle, cell death, and transcriptional regulation (35–40). Other studies also point to their roles in the Notch signaling pathway, heart development, limb development, the immune response, cell adhesion, brown adipogenesis, and keratinocyte differentiation (41–48). However, KAT2A-specific functions include in utero embryonic development, neural tube closure, synaptic plasticity, long-term memory, T cell regulation, and regulatory T cell differentiation (49–51).
We sought to define the functions of KAT2 family members in the intestine. In this study, we find that double knockout (DKO) of lysine acetyltransferases 2A (Kat2a) and 2B (Kat2b) in the intestinal epithelium triggers an intrinsic IFN response and critically compromises intestinal function. Upon inactivation of Kat2 paralogs in the intestinal epithelium, the IFN pathway is activated, and intestinal stem cells are lost. Kat2 DKO in IECs leads to compromised mitochondrial function and the accumulation of mitochondrial dsRNA, which serves as the trigger for IFN signaling. As a result, key epithelial functions such as intestinal stem cell renewal are compromised, and the animal can no longer survive.
RESULTS
To study the functions of KAT2A and KAT2B genes in intestinal health, we generated genetic mouse models that ablate both Kat2 paralogs in the intestinal epithelium (henceforth referred as Kat2 DKO). Specifically, we integrated the Villin-CreERT2 transgenic allele (52) with Kat2aflox/flox alleles (52, 53) and with Kat2b germline KO alleles (Kat2b−/−) (36). Induction of Kat2 DKO via tamoxifen treatment of adult mice was efficient, and loss of Kat2 paralogs was confirmed at the RNA and protein level (Fig. 1, A, B, and F). Induced Kat2 DKO led to rapid and statistically significant weight loss, with animals typically reaching a humane end point and requiring euthanasia within a week after the start of tamoxifen injections (Fig. 1C). These results indicate a vital intestinal function for Kat2 paralogs.
Fig. 1. Intestinal epithelium–specific DKO of Kat2a and Kat2b depletes H3K9ac and is required for survival.
(A) Timeline of induction of genetic KOs and harvest. Vertical arrows indicate the days of tamoxifen injection. Horizontal arrows indicate the average time range for tissue collection. (B) Transcript levels of Kat2a and Kat2b from Kat2 DKO mice in vivo, normalized to housekeeping gene controls [two-way analysis of variance (ANOVA) with Bonferroni’s multiple comparisons test, *adj. P < 0.05, n = 12 to 13 per group]. (C) Average weights over time in percentage, relative to first day of tamoxifen injection, of DKO and control mice show that loss of Kat2 in the intestine leads to rapid weight loss and death (n = 12 to 15 per group, mixed-effects ANOVA analysis, *adj. P < 0.05). (D) Schematic of the pipeline for histone PTM MS. Created with Biorender.com. (E) Relative abundance (%) of H3K9ac and H45acK12acK16ac from histone PTM MS of histones from Kat2 DKO IECs (n = 4 to 5 per group, Welch’s t test). (F) Immunohistochemistry (IHC) of jejunal sections using the indicated antibodies in control and DKO mice (n = 3 to 4 per group). Scale bars, 100 μm. Note efficient depletion of GCN5, PCAF, and H3K9ac in DKO tissues. (G) Additional H3K9ac immunostaining; insets outline the intestinal epithelium to highlight the epithelial-specific reduction of H3K9ac immunoreactivity. Scale bars, 100 μm (center images) and 50 μm (insets). Immunostaining is representative of four biological replicates. (H) Left: Immunoblots of two representative samples for H3K9ac and histone H3 expression on purified histones. Right: Ratios of H3K9ac to histone H3 in DKOs from densitometric analysis using ImageJ (n = 5 per group, unpaired t test, *P < 0.01). All graphs show means with error bars in SEM.
KAT2A and KAT2B are reported to catalyze histone PTMs H3K9ac and H3K14ac (23, 42, 54) but could more broadly affect the histone code. To comprehensively screen changes in histone modifications from Kat2 DKOs, we conducted histone PTM mass spectrometry (MS) (Fig. 1D) (55). Consistent with previous literature linking KAT2 to H3K9ac, the relative abundance of H3K9ac in Kat2 DKO mutants was decreased (Fig. 1E). Other changes were observed in histone PTMs, such as nearly complete loss of detection of multiple acetyl-lysine marks on histone H4 involving H4K5ac, H4K8ac, H4K12ac, and H4K16ac in various combinations (Fig. 1E and table S2). Total acetylation across all histone tails did not significantly change, suggesting the specificity of Kat2a and Kat2b to these specific lysine residues (fig. S1A). Additional changes to histone marks, such as statistically significant increases in methyl marks H3K18me1K23me1 and H3K56me2 (table S2), are not expected to be catalyzed by Kat2, which, we assume, are likely indirect effects of Kat2 loss. Immunohistochemistry (IHC) confirmed notable depletion of H3K9ac immunoreactivity in the intestinal epithelial nuclei and in expanded crypt regions in Kat2 DKO mice compared to controls (Fig. 1, F and G). This loss was not observed in either single Kat2 KO (fig. S1, B and C), suggesting redundant roles of the paralogs in H3K9ac modification. The absence of H3K9ac was additionally quantified by immunoblotting of purified histones from IECs of control and DKO mice (Fig. 1H). Conversely, competing or neighboring marks, such as H3K9me3 and H3K14ac, did not exhibit a qualitative difference between controls and DKO in IHC nor histone PTM MS (fig. S1, D and E). These data suggest efficient depletion of KAT2 activity in an in vivo model.
To further investigate the effects of intestinal Kat2 DKO on intestinal health, we conducted immunostaining for stem cell marker olfactomedin 4 (OLFM4) and proliferation marker protein Ki-67 (KI67) (Fig. 2A). Both protein and transcript levels were notably diminished in Kat2 DKO intestinal epithelium, suggesting that Kat2 function is necessary for maintaining stemness in the intestinal epithelium. In addition, fewer numbers of crypt cells expressed the proliferation marker KI67 and stem cell marker OLFM4 in Kat2 DKO with statistical significance (fig. S2, A to D). To assay for stem cell function, we isolated murine intestine epithelial crypts and cultured them as organoids (56, 57). Of 250 crypts plated from each condition, just over 100 gave rise to organoids from control mice, while, virtually, none were observed from in vivo DKO mice (Fig. 2B), further suggesting compromised stem cell activity upon loss of Kat2 function.
Fig. 2. Intestinal epithelium–specific Kat2a/Kat2b DKO leads to loss of stem cell renewal and skews expression of distinct intestinal lineage markers.
(A) Left: IHC on mouse jejunum for antibodies against OLFM4 and KI67 at 4 to 7 days after onset of tamoxifen treatment (n = 3 per group). Scale bars, 100 μm. Right: RT-qPCR of Olfm4 and Ki67 transcript changes in DKO. Srsf6 was used as an additional reference gene, and all transcript expression was normalized to housekeeping gene control (n = 3 per group, unpaired t test, *P < 0.01). ns, not significant. (B) Left: Crypts were collected from mice 4 to 5 days after the onset of tamoxifen treatment and then seeded in Matrigel for organoid culture. Ex vivo primary epithelium-derived intestinal organoids grown for 6 days (n = 3 per group). Scale bars, 500 μm. Right: Survival graph of intestinal organoids (n = 3 per group, unpaired t test, *P < 0.01). (C) Experimental design for RNA-seq. Arrows indicate the days of tamoxifen injection, with tissue collection occurring on day 3. (D) Heatmap of significantly up-regulated or down-regulated RNA transcripts in Kat2a/Kat2b DKO mice (n = 4 per group, average FPKM > 1, DESeq2 adj. P < 0.05, fold change > 2 or < −2). (E) FPKM values of Olfm4, Mki67, and select intestinal cell lineage markers from RNA-seq (n = 4 per group, *DESeq2 adj. P < 0.05). GSEA enrichment profiles from RNA-seq that are (F) up-regulated or (G) down-regulated (n = 4 per group). FDR, false discovery rate; NES, normalized enrichment score. (H) PAS staining and IHC for lysozyme of intestinal tissue (n = 3 per group). Scale bars, 10 μm (middle) or 100 μm (top and bottom). All graphs show means with error bars in SEM.
To more comprehensively characterize the functions of Kat2a and Kat2b in the intestine epithelium, we used RNA sequencing (RNA-seq). Epithelium was harvested from control and mutant mice before the onset of overt symptoms (Fig. 2C) to understand how the Kat2 paralogs affect the transcriptome before secondary changes of declining mouse health. RNA-seq from Kat2 DKO IECs and controls resulted in 483 up-regulated and 362 down-regulated genes [Fig. 2D; average fragments per kilobase of transcript per million mapped reads (FPKM) > 1, fold change > 2 or < −2, DESeq2 adjusted (adj.) P < 0.05, n = 4]. Consistent with reverse transcription quantitative polymerase chain reaction (RT-qPCR) data (Fig. 2A), RNA-seq transcript levels for Olfm4 and Mki67 decreased with Kat2 DKO (Fig. 2E). In addition, RNA-seq transcript levels for another stem cell marker, Lgr5, were lower with statistical significance in the intestinal epithelium of Kat2 DKO (fig. S2I). Most other key transcripts used as lineage markers of various intestinal cell types did not show statistically significant changes in Kat2 DKO mutants (Fig. 2E). However, use of broader gene sets associated with intestinal lineages using gene set enrichment analysis (GSEA) (58) suggested higher expression of transcripts associated with secretory lineages (enteroendocrine, goblet, and Paneth cell signatures) and reduced expression of transcripts associated with stem cell, differentiation, and enterocyte progenitor signatures (16, 59, 60) in Kat2 DKOs (Fig. 2, F and G). The decrease in stem cell transcripts corroborated the lack of organoid forming ability from the DKO crypts (Fig. 2B). Several of these phenotypes have been associated with a reduction in Notch signaling, and a gene set associated with Notch signaling in the intestine (16) was also reduced in the transcriptome of the Kat2 DKO (fig. S2J). Consistent with an increase in Paneth cell gene signature transcripts, Kat2 DKO mutants also demonstrated increased periodic acid–Schiff (PAS) staining for Paneth cells and disruption in lysozyme protein localization in the intestine (Fig. 2H and fig. S2, E and F). Overall, intestinal KAT2 appears to be required for intestinal stem cell transcript expression and function, and disruption of KAT2 expression leads to changes in intestinal homeostasis and a rapid decline in the health of the mouse.
To understand how KAT2 could function to control these aspects of intestinal homeostasis, we further interrogated RNA-seq data. Database for Annotation, Visualization and Integrated Discovery (DAVID) gene ontology (GO) analysis revealed that the most enriched category of genes is associated with viral infection and related immune responses, while down-regulated genes are associated with oxidoreductase and mitochondrial functions (Fig. 3, A to C, and table S3). Genes induced after viral infection are often linked to an IFN signaling response. We therefore queried whether the genes induced upon Kat2 DKO are similar to genes induced upon IFN-λ treatment of intestinal organoids or upon treatment of neonatal mice with IFN-β in the intestinal epithelium. We found very robust correlations between these datasets using GSEA (Fig. 3C), indicating that the IFN signaling pathway is activated upon Kat2 DKO (61). These findings are consistent with previous literature in which active IFN signaling disturbed intestinal stem cell maintenance and health (15, 16).
Fig. 3. Kat2a/Kat2b DKO induces epithelial-intrinsic IFN signaling, which can be suppressed with pharmacological inhibitors of cytoplasmic double-stranded nucleic acid–sensing pathways.
(A) Top up-regulated (red) and top down-regulated (blue) GO terms in DKO samples (483 and 362 genes, respectively, from RNA-seq Fig. 2D) using DAVID GO. (B) Expression profile of immune-related genes populated from RNA-seq and GO analysis (from five GO terms: GO:0051607, GO:0009615, GO:0002376, GO:0045087, and GO:0045071). (C) GSEA profiles demonstrate enrichment of IFN-β and IFN-λ targets from GSE142166 and down-regulation of mitochondrial complex V in Kat2a/Kat2b DKO. (D) Timeline of events for ex vivo epithelium-derived intestinal organoid experiments, where mice are harvested on day 0 for three-dimensional organoid culture containing antimicrobials [Primocin (0.1 mg/ml) and penicillin-streptomycin (100 U/ml)]. Induction of DKO with 16-hour tamoxifen (1 μM) treatment in ex vivo culture begins on day 4. Inhibitor treatment also begins on day 4 and ends on day 8 at the time of organoid collection. (E) Schematic of IFN signaling pathway activation via double-stranded nucleic acids. Targets of select inhibitors are shown in red. MAVS, mitochondrial antiviral-signaling protein. (F) RT-qPCR of ISGs from epithelium-derived intestinal organoids with DKO induced in culture, which were treated with pharmacological inhibitors targeting TBK1 (GSK8612; 10 μM), JAK (ruxolitinib; 2 μM), and STING (H-151; 2 μM) for 4 days. Inhibitors were added during and after induction of DKO in culture, with media and inhibitors refreshed daily (n = 3 per group, two-way ANOVA with Tukey post hoc analysis, *adj. P < 0.05 for the indicated gene versus 0.2% DMSO vehicle-treated DKO, normalized to Hprt). Mean shown with error bars in SEM.
Activation of IFN signaling in the Kat2 DKO mice could be either dependent on a microbial agent or initiated intrinsically in the mouse cells. To discern whether activation of IFN signature genes was dependent on the microbiome, we induced Kat2 DKO in the context of established intestinal organoid cultures, which are grown under sterile conditions. Organoids were grown for 4 days in the presence of antimicrobials (primocin, 0.1 mg/ml) before DKO induction (Fig. 3D) and then monitored for ISG expression. Compared to control organoids, DKO organoids exhibited statistically significant elevation of a panel of ISGs (Fig. 3F and fig. S3A). Given the absence of microbial populations and other host-derived cell populations in these epithelial organoid cultures, this IFN response upon Kat2 DKO appears intrinsic to the intestinal epithelium.
Kat2s suppress a self-derived dsRNA-induced activation of IFN signaling
Cell-intrinsic IFN responses can come from several sources, including self-derived double-stranded DNA (dsDNA) and dsRNA. dsDNA and dsRNA sensors initially activate different signaling proteins in the IFN response but later converge and share the same downstream pathway to trigger transcription of ISGs (Fig. 3E). In the ex vivo DKO organoid model, the IFN response can be suppressed using a TANK binding kinase 1 (TBK1)–specific inhibitor (62) (GSK8612) or even more potently suppressed with a Janus kinase (JAK) inhibitor (ruxolitinib; Fig. 3F). However, inhibition of STING, which is typically activated by dsDNA, failed to repress DKO-induced ISG activation (H-151; Fig. 3F). To confirm efficacy of the STING inhibitor, we transfected wild-type intestinal organoids with a dsDNA mimetic. Poly(deoxyadenylic-deoxythymidylic) acid [poly(dA:dT)] treatment successfully induced ISG expression, as expected, and addition of H-151 effectively repressed this dsDNA response (fig. S3B). Last, to further test the hypothesis that the Kat2 response was due to dsRNA detection, we challenged organoids with the dsRNA mimetic, poly(I:C). Poly(I:C) induced the same ISG profile induced by Kat2 DKOs, and the induction of ISGs by poly(I:C) could be suppressed with the same pharmacological agents that blocked Kat2 DKO-induced ISG expression (fig. S3, C to F). Thus, dsRNA, and not dsDNA, appears to drive the intrinsic IFN response in Kat2a/Kat2b DKO.
ISGs are not enriched with nucleosomes containing H3K9ac
To understand whether ISGs are induced in response to dysregulation of transcription associated with loss of H3K9ac in the Kat2 DKO, we interrogated H3K9ac chromatin immunoprecipitation sequencing (ChIP-seq) from murine jejunal epithelial cells (GSE86996) (63). H3K9ac signal was aligned at all annotated transcription start sites (TSSs) and enriched immediately upstream of the nucleosome-free region associated with the TSS and immediately downstream of the TSS, as expected on the basis of previous profiles of H3K9ac marked histones (GSE86996; Fig. 4A) (63). K-means clustering resolved five distinct H3K9ac patterns at intestinal genes: high signal downstream the TSS (cluster 1), moderate and bidirectional signal from the TSS (cluster 2), moderate signal downstream the TSS (cluster 3), weak signal downstream the TSS (cluster 4), and negligible to no signal (cluster 5; Fig. 4B). We sought to identify what patterns of H3K9ac signal were associated with ISGs differentially expressed in Kat2 DKO versus controls from RNA-seq analysis (listed in Fig. 3B). Most ISGs that are up-regulated in Kat2 DKO were found in clusters with moderate or low H3K9ac-marked promoters (clusters 3 to 5), suggesting no clear link between ISG induction and epigenomic regulation by KAT2A- and KAT2B-mediated H3K9ac. ISGs cluster similarly to all differentially expressed genes (DEGs) up-regulated in Kat2 DKO (Fig. 4C). DEGs that are down-regulated have a slightly greater proportion of genes in clusters 1 and 3 (Fig. 4C). However, DEGs and even all genes from Kat2 DKO RNA-seq generally do not have a strong correlation with H3K9ac ChIP-seq signal (Fig. 4D), which implies that a non-H3K9ac mechanism is likely activating the IFN response upon Kat2 DKO. Notably, this should not diminish the implications that other biological processes unrelated to IFN signaling are controlled, at least in part, by Kat2 DKO-mediated H3K9ac of TSSs, especially given that ~48% of all Kat2 DKO DEGs that are down-regulated (146 genes) reside in clusters 1 to 3 with moderate to high H3K9ac signal.
Fig. 4. Determination of H3K9ac status at ISGs induced in the Kat2 DKO.
(A) Mean H3K9ac coverage of all murine genes in the jejunal epithelium is shown at a metagene [TSS to transcription end site (TES) ± 2 kb (left) and only the TSS ± 2 kb (right)]. Positive and negative sense strands were reoriented into the same direction for analysis. Data visuals were generated with deepTools (v3.5.0) using ChIP-seq of H3K9ac from wild-type mice in GSE86996. (B) Left: K-means clustering of H3K9ac signal within 2 kb of the TSS forms five distinct patterns in the murine jejunal epithelium. Positive and negative sense strands were reoriented into the same direction for analysis. Heatmap visual was generated with deepTools 3.5.0. Right: Pie charts indicating the percentage of up-regulated ISGs from Kat2 DKO RNA-seq belonging to each cluster, highlighting that the presence of H3K9ac in wild-type mice does not coincide with ISG expression upon Kat2 DKO. bp, base pairs. (C) Bar charts indicating the percentage of ISGs, all DEGs up-regulated, and all DEGs down-regulated from Kat2 DKO RNA-seq in each H3K9ac ChIP-seq cluster from (B). (D) Scatter plots showing the log fold change of Kat2 DKO FPKM values versus control FPKM values from RNA-seq versus the H3K9ac counts from ChIP-seq (GSE86996) for DEGs (top) or all genes (bottom) from Kat2 DKO RNA-seq. There does not appear to be a strong correlation between H3K9ac levels and changes in RNA levels in the Kat2 DKO.
Identification of Kat2-dependent acetylation targets reveals that mitochondrial protein acetylation and mitochondrial function are compromised in Kat2 DKO intestinal epithelium
We next aimed to understand whether KAT2A and KAT2B regulate the IFN response through means beyond transcriptional changes associated with H3K9ac loss. KAT2A and KAT2B are reported to directly acetylate nonhistone proteins in controlling various biological functions in the cell (34, 35, 37, 38, 42, 43, 64, 65). To investigate what proteins may be under Kat2-dependent acetylation, we used acetylomic MS of intestinal epithelial lysates from control versus Kat2 DKO mice (Fig. 5A and table S4). In agreement with our findings at the transcript level, the Kat2 DKO proteome exhibited enrichment in terms related to immune and viral response and suppression of oxidoreductase activity (Fig. 5B and table S5). Acetylomic MS identified more peptides with diminished acetylation than increased acetylation, as expected upon loss of the Kat2 acetyltransferases (Fig. 5C). Among the proteins harboring Kat2-dependent acetylated residues were those associated with mitochondrial functions (Fig. 5, B and C). These included members of the solute carrier family 25 (SLC25) family, which are mitochondrial transport carriers of solutes such as adenosine 5′-diphosphate (ADP)/adenosine 5′-triphosphate (ATP), amino acids, and fatty acids. In light of these changes to the acetylome and diminished mitochondrial transcripts (Fig. 3A) and proteins (Fig. 5, B and C), Kat2 DKO intestinal tissue was stained for activities of enzymes from mitochondrial complexes I and IV to assess disruptions in mitochondrial function. Both reduced form of nicotinamide adenine dinucleotide (NADH) oxidation (complex I) and cytochrome c oxidase (COX; complex IV) activities were weaker in Kat2 DKO (Fig. 5, D and E). In addition, colorimetric-based COX activity was lower with statistical significance in Kat2 DKO intestinal epithelium cells (Fig. 5F), implying that mitochondrial function is disrupted in the Kat2 DKO epithelium. These results provide an intriguing look at the acetyl-proteome and link Kat2 as a potential regulator of mitochondrial function.
Fig. 5. Mitochondrial proteins are differentially acetylated, and mitochondrial enzyme activity is compromised in Kat2 DKO.
(A) Schematic of the pipeline for acetylomics MS. Created with Biorender.com. (B) GO term analysis of the proteome (top) and nonhistone proteins with down-regulated acetyl-lysine residues (bottom) from Kat2 DKO IECs (n = 4 controls and 6 DKOs). (C) Volcano plot of acetyl-lysine residues from acetylomics of Kat2 DKO IECs. Orange points indicate lysine residues on mitochondrial proteins (n = 4 controls and 6 DKOs). Stains for activity of mitochondrial enzymes (D) NADH and (E) COX in jejunal intestine sections (n = 2 to 4 per group). Scale bars, 500 μm. (F) COX activity in the IECs from control and Kat2 DKO mice (n = 3 mice, Welch’s t test) using spectrophotometry. ***P < 0.001.
Mitochondria-derived dsRNA accumulates in Kat2 DKO intestinal epithelium
It has previously been demonstrated that compromised mitochondrial function can lead to the generation of mitochondria-derived dsRNA (4, 66, 67) and activation of IFN signaling. For example, knockdown of polynucleotide phosphorylase, an exoribonuclease responsible for degrading mitochondrial transcripts, results in accumulation of cytoplasmic mitochondrial dsRNA and induces transcription of IFNB in HeLa cells (4). Inhibition of ATP synthase from the mitochondrial electron transport chain with oligomycin A also disrupts mitochondrial membrane potential and structure, promoting mitochondrial dsRNA efflux and activating IFN-β and ISGs in chondrosarcoma cells (67).
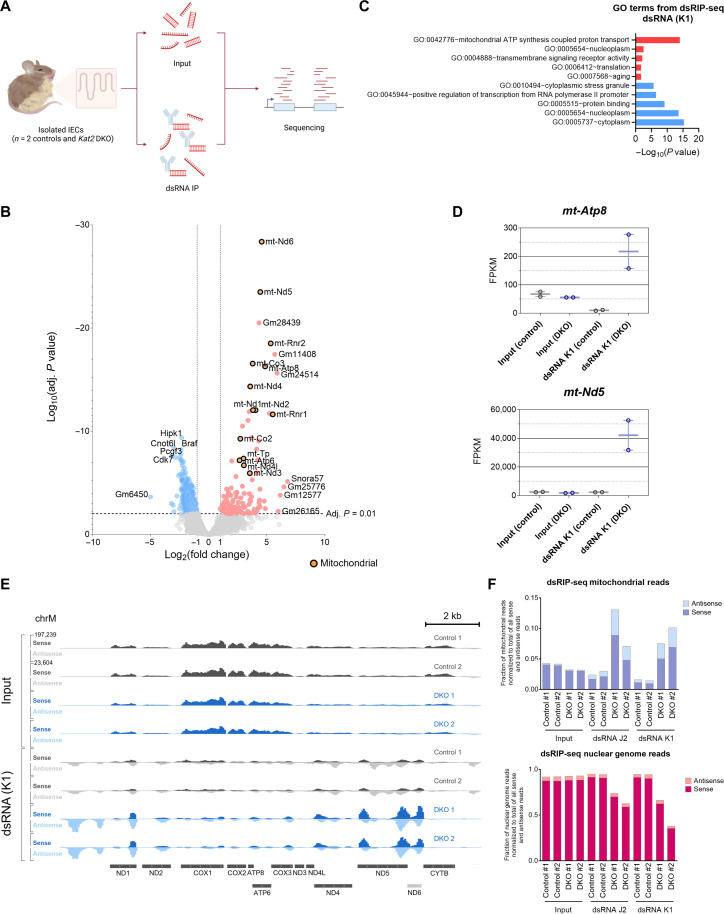
To identify the source of dsRNA in Kat2 DKO epithelium, we conducted sequencing of immunoprecipitated dsRNA (dsRIP-seq) as previously described (68) using two different monoclonal antibodies designed to recognize dsRNA (anti-dsRNA antibody clones K1 and J2; Fig. 6A and fig. S4). Stranded RNA libraries were generated to allow for detection of both sense and antisense transcripts that could contribute to dsRNA generation. Differential enrichment and DAVID GO analyses illustrated an up-regulation of dsRNA in dsRIP-seq originating from mitochondria-encoded genes in the Kat2 DKO samples (Fig. 6, B and C; fig. S4, A and B; and table S6). The highest GO enrichment category of genes associated with dsRNA production is from the mitochondrial genome, regardless of whether the dsRIP-seq was conducted with the K1 or J2 anti-dsRNA antibody. For example, mitochondrial genes mt-Atp8 and mt-Nd5 were prominent in dsRNA pull-downs from DKOs but not in control pull-downs nor any input samples (Fig. 6, D and E, and fig. S4, C and D). Notably, mt-Nd5 and mt-Nd6 are located on opposite strands at adjacent positions in the mitochondrial genome. These two mitochondrial genes are most susceptible to dsRNA generation due to transcription in opposing directions (69) and are among the most abundant sources of dsRNA in our dsRIP-seq results (Fig. 6E and fig. S4D). Furthermore, dsRNA pull-downs in DKOs presented with a higher fraction of unique mitochondrial reads (Fig. 6F). Consistent with successful enrichment of dsRNA in pull-down samples, antisense transcripts were enriched in the immunoprecipitation samples compared to input RNA (fig. S4E), and total antisense transcripts were higher in mutant samples compared to controls (Fig. 6F). Last, we note that antisense transcript enrichment in the Kat2 DKOs was specific to the mitochondrial genome, as the nuclear genome had a smaller and consistent fraction of antisense reads across all samples (Fig. 6F and fig. S4F). Collectively, these findings imply that Kat2 DKO-generated dsRNA originates from the mitochondrial genome rather than the nuclear genome and that mitochondrial dsRNA then triggers an IFN response and subsequent imbalances in intestinal homeostasis.
Fig. 6. Mitochondrial dsRNA is a likely source of ISG induction in Kat2a/Kat2b DKO.
(A) Schematic of the workflow for dsRIP-seq. Created with Biorender.com. IP, immunoprecipitation. (B) Volcano plot of dsRIP-seq for dsRNA (K1 antibody) pull-down (n = 2 per group, DESeq2 adj. P < 0.01, likelihood ratio test, average FPKM > 1) shows enrichment in dsRNA from mitochondrial transcripts that are specific to anti-dsRNA antibodies in the DKO versus controls and normalized for baseline levels of RNA in input samples. Orange points indicate mitochondrial genes. (C) Top up-regulated (red) and top down-regulated (blue) GO terms for DKO from dsRIP-seq using dsRNA (K1) antibody and DAVID GO. (D) FPKM values of select dsRNA species up-regulated in DKO. Minimum values, maximum values, and mean of the values are shown. (E) IGV tracks for input and dsRNA (K1) pull-down samples from dsRIP-seq along the mitochondrial genome. Sense reads are on the same scale (0 to 197,239) in the top half of the track and antisense reads on the same scale (0 to 23,604) in the bottom half for each sample. (F) Number of unique mitochondrial (top) or nuclear (bottom) sense or antisense reads from each dsRIP-seq sample, normalized to the total number of all dsRIP-seq reads (including both sense and antisense transcripts).
DISCUSSION
IEC-specific Kat2 DKO mutants presented with both an IFN response and stem cell loss. Links between the IFN signaling pathways inhibiting intestinal stemness have been previously documented. Transcription factor interferon regulatory factor 2 (IRF2) negatively regulates type I IFN signaling, and deletion of Irf2 impairs regeneration and maintenance of stem cells in the intestine (15, 16). Furthermore, wild-type colonic organoids from mice treated with poly(I:C) to induce chronic type I IFN signaling fail to thrive in culture; organoids from Ifnar1−/− mice undergoing the same treatment, but lacking type I IFN signaling, are capable of growing (15). It is likely that mice with Kat2 DKO in IECs follow a similar trajectory from induction of ISGs to suppression in stemness, ultimately leading to the rapid decline in health.
The IFN response is also connected to changes in intestinal homeostasis as the presence of excess IFN signaling induces stem cells to prematurely differentiate into secretory progenitors (16). An accumulation of immature Paneth cells is observed in Irf2−/− intestines, with PAS+ granule morphology much similar to the IEC Kat2 DKO mutants (16). Deletion of SET domain bifurcated histone lysine methyltransferase 1 (SETDB1) in the intestine also activates ISGs and results in mislocalization of lysozyme expression (8).
Abolishment of histone acetyltransferases such as KAT2A and KAT2B and the resulting decrease in histone acetylation might be expected to repress transcription on a global level, but many genes including ISGs were up-regulated in IEC-specific Kat2 DKO mutants. Therefore, KAT2A and KAT2B may be important components of functioning transcriptional complexes and machinery from initiation to termination, or these histone modulators catalyze acetyl marks on RNA that would confer posttranscriptional stability. For example, KAT2A and KAT2B have been previously described to acetylate proteins recruited for nucleotide excision repair, such as replication protein A1 (RPA1), and catalyze H3K9ac at sites of DNA damage to mark them for repair (35, 70). In the absence of KAT2A/KAT2B, these DNA repair pathways may become inefficient or defective. Future studies should focus their efforts on obtaining a comprehensive understanding of the diverse roles that the KAT2 genes play both inside and outside the nucleus.
Kat2 DKO-induced ISG up-regulation has been previously documented (42, 48). A study found that knockdown of KAT2A and KAT2B in human keratinocyte cell lines induced transcription of ISGs at both subconfluence and confluence (48). In another study, Kat2 DKO in mouse embryonic fibroblasts also identified up-regulation in viral and immune response from RNA-seq, which was attributed to GCN5 directly suppressing TBK1 activity (42). However, the mechanisms behind this response in vivo have not yet been investigated. The intestine-specific KO in the current study suggests a mechanism driving this intrinsic IFN response, using novel techniques including acetylomics and dsRIP-seq to elucidate dsRNA from the mitochondria as the instigator in the intestinal epithelium. How KAT2A and KAT2B modulate mitochondrial function is still uncertain but may relate to acetyl–coenzyme A as a substrate for both posttranslational acetylation and oxidative phosphorylation or the acetylation status of KAT2-targeted mitochondrial proteins. Notably, Kat2 DKO diminished acetylation of solute transporter SLC25 family members at multiple lysines. Modeling simulations of human SLC25A4 determined that acetylation of lysine-23 within the ADP-binding pocket reduced ADP-binding affinity (71), although this particular residue was not identified in our acetylomics data. While published evidence for other acetyl sites is limited, the SLC25 carrier family is closely tied to mitochondrial pathologies and diseases (72–76). Kat2-catalyzed acetylation may be crucial for conformational changes required for normal carrier functions and providing essential solutes for mitochondrial respiratory complex activity. Notably, formation of mitochondrial dsRNA has been reported to activate type I IFN signaling in HeLa cells upon depletion of polynucleotide phosphorylase and in patients with hypomorphic mutations in the corresponding PNPT1 gene (4). Overall, these mitochondrial defects may cause mitochondrial dsRNA to escape into the cytoplasm and trigger IFN signaling.
MATERIALS AND METHODS
Experimental design
The objective of this study is to investigate the functions of Kat2a and Kat2b in the intestinal epithelium using mouse models and inducible KO systems as described below.
Mouse strains
Mouse strains are an outbred mix of 129SvEv and C57BL/6J strains. For Kat2aflox/flox, loxP sites were inserted into the gene locus before exon 3 and after exon 19 (36, 49, 77). Whole-body Kat2b−/− mice with the neomycin construct were generated as previously described (36, 49, 77). Kat2 mice were provided to our laboratory by S. Dent (The University of Texas MD Anderson Cancer Center).
Mouse intestinal epithelium harvesting
Mice taken for harvest were euthanized by cervical dislocation. Mouse intestine was removed and unraveled before flushing with phosphate-buffered saline (PBS) and then cut longitudinally. Intestines were further cut into ~2.54-cm pieces and then incubated in 0.5 M EDTA in a series of 5-, 10-, and 25-min intervals at 4°C on a rotator. After EDTA incubations, intestinal epithelium was isolated by aggressive shaking. For separating crypt and villus preparations, the epithelium-containing EDTA was passed through a 70-μm filter. Isolated crypts and villi were further cleansed in PBS and centrifuged at 4°C for 3 min at 200 rpm to make pellets for organoid culture, RNA, and RT-qPCR. Animal protocols are approved by the Rutgers Institutional Animal Care and Use Committee. For mouse breeding colonies, mice were fed with special breeder diet (PicoLab Mouse Diet 20, 5058). For weaned mice and other nonbreeding adult mice, the standard mouse diet was provided (PicoLab Rodent Diet 20, 5053).
For conditional and inducible KO of Kat2a using intestinal epithelium-specific Villin-CreERT2 in mice (52), tamoxifen [100 mg/kg; 10% (w/v) of ethanol and 90% corn oil] was administered by intraperitoneal injections for 4 days daily; for RNA-seq, the same treatment regimen was implemented except for 3 days daily. Control mice received vehicle intraperitoneal injections (10% ethanol and 90% corn oil).
Intestinal organoid culture
Crypts isolated from mouse intestine were seeded at a density of 250 crypts per 25 μl of Matrigel (Cultrex reduced growth factor basement membrane extract, type R1, R&D Systems, catalog no. 3433-010-R1) in 48-well plates. Organoids were fed with 250 μl of media [N-2, B-27, 0.5 M N-acetylcysteine, mouse epidermal growth factor (50 μg/ml), Noggin (100 μg/ml), R-spondin, and primocin (0.1 mg/ml) in advanced Dulbecco’s modified Eagle’s medium/F-12 supplemented with GlutaMAX, Hepes, and penicillin/streptomycin (100 U/ml)], typically grown until mature, and differentiated at 6 to 7 days. For ex vivo KO, 3- to 4-day-old primary organoids were treated with either 1 μM tamoxifen in ethanol or an equivalent volume of ethanol vehicle (100% ethanol) for 16 hours and then collected 72 hours later in TRIzol for RNA.
For small-molecule inhibitor studies in organoids, all compounds [GSK8612, MedChemExpress, catalog no. HY-111941 (62); H-151, MedChemExpress, catalog no. HY-112693; ruxolitinib, Cayman Chemical, item #11609] were reconstituted in dimethyl sulfoxide (DMSO). Stock concentrations and dilutions were prepared to attain a 0.2% DMSO concentration across all groups and treatments. DKO was induced on 4-day-old primary organoids (tamoxifen treated for 16 hours), with collection in TRIzol for RT-qPCR on day 8 of the same passage. Compound treatment overlapped with DKO induction and was changed every 24 hours for 3 days for a total treatment time of 88 hours. Three wells of a 48-well plate were collected in 1 ml TRIzol per group and treatment.
For poly(dA:dT) (InvivoGen, catalog code tlrl-patn) studies in organoids, the compound was reconstituted in water as instructed by the manufacturer. Transfection with poly(dA:dT) was performed with Lipofectamine 2000 (Thermo Fisher Scientific, catalog no. 11668019) following the instructions from the manufacturer for 6 hours on 10-day-old primary intestinal organoids, which were separated from Matrigel by pipetting in ice-cold PBS. H-151 (2 μM) or 0.2% DMSO vehicle was given concomitantly with poly(dA:dT). Concentrations for poly(dA:dT) was chosen on the basis of the manufacturer’s recommendations.
RNA extraction and cDNA synthesis
For primary mouse intestinal epithelium, ~20 to 50 μl of packed epithelial cell pellet was resuspended in 1 ml of TRIzol, and RNA was isolated according to the manufacturer’s instructions. Mouse intestinal epithelium–derived organoids were also resuspended in TRIzol (3 wells from a 48-well plate in 1 ml of TRIzol) and extracted for RNA using chloroform. RNA-containing aqueous phase was processed following the instructions from QIAGEN’s RNeasy micro kit (QIAGEN, catalog no. 74004). For cDNA synthesis, 500 to 1000 ng of RNA was measured using Nanodrop ND-1000 spectrophotometer (Thermo Fisher Scientific) and then processed per the SuperScript III first-strand synthesis protocol provided by Invitrogen (Thermo Fisher Scientific, catalog no. 18080051).
Reverse transcription quantitative polymerase chain reaction
RT-qPCR was conducted with the ABI PRISM 7900HT Sequence Detection System for a 384-well plate loaded with SYBR green (Thermo Fisher Scientific, catalog no. 4309155), cDNA, and reaction mix containing a 0.5 μM final primer concentration. For analysis, Ct values were normalized to Hprt, Cyb561d1, or Srsf6, which were chosen because of Kat2 DKO-induced expression changes in typical housekeeping genes and based on RNA-seq.
RNA sequencing
Whole mouse duodenal epithelial cells were isolated and extracted for RNA using TRIzol (Invitrogen) and chloroform. RNA samples were sent to BGI for transcriptome library preparation, paired-end RNA-seq, and quality control check using their DNBSEQ platform (40 million reads per sample). Subsequent fastq files were processed using Kallisto (version 0.46.1) with 100 bootstraps for raw read counts and indexed to the mouse (mm9) genome. Transcript abundance HDF5 binary files were imported with tximport and then processed using DESeq2 (version 1.32) for calculating FPKM normalization and for differential expression analysis in R. Genes for analysis were filtered for an average FPKM of >1. DESeq2 was used to generate a preranked gene list based on signal-to-noise ratios for conducting GSEA (58).
Sequencing of immunoprecipitated dsRNA
Whole IECs from the proximal half of the small intestine of two control and two Kat2a/Kat2b DKO mice were collected for dsRIP-seq, following Gao et al. (68). Briefly, ~20 to 30 μl of epithelial cell pellet per sample was resuspended in TRIzol (Invitrogen) as inputs. A total of 200 μl of pellets per sample were also resuspended in 3 ml of dsRIP buffer [100 mM NaCl, 50 mM tris-HCl (pH 7.4), 3 mM MgCl2, and 0.5% IGEPAL CA-630 with 1× mammalian ProteaseArrest protease inhibitor (G-Biosciences, catalog no. 786-433) and SUPERase·In ribonuclease inhibitor (40 U/ml; Thermo Fisher Scientific, catalog no. AM2696) in water], incubated for 5 min on ice, and then centrifuged at maximum speed (13,000g) for 10 min at 4°C. Supernatant was divided into three aliquots for incubation in normal mouse immunoglobulin G (IgG) (MilliporeSigma, catalog no. 12-371), dsRNA K1 (Cell Signaling Technology, #28764), or dsRNA J2 (Cell Signaling Technology, #76651) at 5 μl per 300 μl of supernatant. Samples in normal mouse IgG, dsRNA K1, or dsRNA J2 were incubated with rotation at 4°C for 2 hours and then incubated along with pre-washed Protein G Dynabeads (Thermo Fisher Scientific, catalog no. 10003D), using 25 μl of beads per 300 μl of supernatant, for 1 hour at 4°C with rotation. Beads were collected with a magnetic rack and washed four times in a cold room before addition of TRIzol.
RNA extraction for dsRIP was performed using an miRNeasy micro kit (QIAGEN, catalog no. 217084) following the manufacturer’s protocol and two rounds of chloroform incubations to reduce phenol contamination. RNA was measured using Nanodrop ND-1000 spectrophotometer (Thermo Fisher Scientific) before shipping to Azenta (GeneWiz) for next-generation sequencing. Similar to previous reports, IgG samples yielded insufficient amounts of RNA for sequencing and were not included for further analysis (68). Library prep included ribosomal RNA–depleted total RNA, followed by cDNA synthesis using random hexamers. FastQC (version 0.12.0) was used for quality checking fastq files. Unique molecular identifiers (UMIs) were provided by Azenta in a separate fastq file. UMIs were linked to their respective sequencing read with umis (version 1.0.3) (78), and strand-specific reads were aligned to the mm10 genome (from GENCODE, GRCm38.p4) using STAR (version 2.7.10b) for splice awareness. The resulting BAM file was then indexed with SAMtools (version 1.17) and deduplicated with UMI-tools (version 1.1.4) (79–82). Per-gene raw reads counts were obtained using HTSeq (version 2.0.2), and differential expression analyses were conducted with DESeq2 (version 1.40.1) by calculating ratios of ratios (83, 84). FPKM values were determined using edgeR (version 3.42.2) following the script provided by Gao et al. (68) and Robinson et al. (85). Bigwig files for Integrative Genomics Viewer (IGV) tracks were generated using BAM files from STAR and deepTools (version 3.5.1) (86).
ChIP-seq analysis
Analysis of H3K9ac ChIP-seq from GSE86996 (63) was conducted using Bowtie (v1.3.0) (87) and deepTools (v3.5.0) (86) with alignment to the mm9 mouse genome. Positive and negative sense strands were reoriented into the same direction for analysis. Counts from BAM files were used to compare H3K9ac signal from ChIP-seq to the log fold change of FPKM values between Kat2 DKOs and controls from RNA-seq.
Histology
Harvested mouse intestine used for paraffin embedding was flushed with PBS and cut longitudinally, then pinned, and fixed in 4% paraformaldehyde in PBS for 10 min before Swiss rolling. After overnight incubation in additional 4% paraformaldehyde, intestine Swiss rolls were briefly rinsed in PBS and dehydrated in a series of 70 to 100% ethanol incubations. Rolls were transferred into xylene for 2 hours, then submerged in melted paraffin for 4 hours, and embedded for sectioning. Paraffin-embedded tissues were sectioned at a thickness of 5 μm.
Immunohistochemistry
Five-micrometer paraffin-embedded tissue sections were heated at 65°C and incubated in xylene before processing in increasing concentrations of ethanol. Sections were rehydrated and underwent antigen retrieval with 10 mM sodium citrate buffer (pH 6) along with peroxidase quenching and permeabilized with 0.5% Triton X-100. Samples were blocked in 5% fetal bovine serum and incubated overnight with primary antibody diluted in fetal bovine serum (see table S1 for antibody list). After biotinylated secondary antibody incubation following the instructions from VECTASTAIN Elite ABC (avidin/biotin complexing) HRP (horseradish peroxidase) Kit (Vector Laboratories, catalog no. PK-6101), antigen signal was amplified with ABC for 1 hour and developed with 0.05% 3,3′-diaminobenzidine [Chemical Abstracts Service (CAS), #868272-85-9] and 0.015% hydrogen peroxide in 0.1 M tris. Tissue was counterstained with hematoxylin and then processed again in increasing ethanol concentrations and xylene before mounting.
Mitochondrial enzymatic activity staining
Four-micrometer sections of unfixed, Optimal Cutting Temperature (OCT)–embedded jejunal tissue were incubated in NADH-containing buffer [nitro blue tetrazolium chloride (1 mg/ml), 5 mM MgCl2, 25 mM CoCl2, NADH (2 mg/ml), and 0.2 M tris-HCl buffer (pH 7.4)] at room temperature until the tissue developed a blue color or buffer containing cytochrome c [5 mg of 3,3′-diaminobenzidine, 10 mg of cytochrome c, 20 μg of catalase, 750 mg of sucrose, and 2.5 ml of 0.2 M sodium phosphate buffer, (pH 7.6) in 7.5 ml of deionized water] at 37°C until the tissue developed a brown color. For mitochondrial complex I activity staining, tissue was then fixed in 4% paraformaldehyde for 10 to 15 min, washed two times in water, and then mounted in water-based mounting medium (88). For COX activity staining, tissue was washed in water before dehydration with increasing concentrations of ethanol, clearance with xylene, and mounting with a xylene-based medium (89).
COX activity assay
IEC homogenates from control and Kat2 DKO mice were used to quantify the activity of COX by spectrophotometry using the COX Assay Kit (Abcam, #ab239711) according to the instructions of the manufacturer. After protein quantification, 25 μg of cell homogenate protein was used to measure COX activity. To characterize the enzyme activity over a 30-min period, the oxidation of reduced cytochrome c with an absorbance decrease at 550 nm was measured. The rate of the enzyme reaction was calculated in the linear range (n = 3 mice per group) and normalized per protein amount, according to the manufacturer.
Cell lysis and protein extraction
Primary IECs were processed in radioimmunoprecipitation assay (RIPA) buffer [150 mM sodium chloride, 50 mM tris (pH 8.0), 1% NP-40, 0.5% sodium deoxycholate, and 0.1% SDS in water] to extract total protein. RIPA buffer was spiked with inhibitors to achieve final concentrations of 1× protease inhibitor, 1 mM phenylmethylsulfonyl fluoride, 1 mM sodium orthovanadate, 10 mM sodium fluoride, and 10 mM sodium butyrate. Samples were sonicated for 2 to 3 min before incubating at 4°C with rotation for 30 min. After spin down at maximum speed in a mini centrifuge for 15 min at 4°C, the resulting supernatant containing protein was saved for additional assays described below. Protein quantification was performed following the instructions from the Pierce BCA Protein Assay Kit (Thermo Fisher Scientific, catalog no. 23225), using the SpectraMax M2 (Molecular Devices) plate reader and SoftMax Pro software (Molecular Devices).
Histone purification and PTM MS
Histones were purified from isolated IECs using acid-based methodology for MS of histone PTMs, as described by Sidoli et al. (55) and Yuan et al. (90). Briefly, for histone extraction and digestion, histone proteins were extracted from pelleted IECs to ensure good-quality identification and quantification of single histone marks (55). Briefly, histones were acid-extracted with chilled 0.2 M sulfuric acid (5:1, sulfuric acid:pellet) and incubated with constant rotation for 4 hours at 4°C, followed by precipitation with 33% trichloroacetic acid overnight at 4°C. Then, the supernatant was removed, and the tubes were rinsed with ice-cold acetone containing 0.1% HCl, centrifuged, and rinsed again using 100% ice-cold acetone. After the final centrifugation, the supernatant was discarded, and the pellet was dried using a vacuum centrifuge. The pellet was dissolved in 50 mM ammonium bicarbonate (pH 8.0), and histones were subjected to derivatization using 5 μl of propionic anhydride and 14 μl of ammonium hydroxide (all Sigma-Aldrich) to balance the pH at 8.0. The mixture was incubated for 15 min, and the procedure was repeated. Histones were then digested with 1 μg of sequencing grade trypsin (Promega) diluted in 50 mM ammonium bicarbonate (1:20, enzyme:sample) overnight at room temperature. Derivatization reaction was repeated to derivatize peptide N termini. The samples were dried in a vacuum centrifuge.
Before MS analysis, samples were desalted using a 96-well plate filter (Orochem) packed with 1 mg of Oasis HLB C-18 resin (Waters). Briefly, the samples were resuspended in 100 μl of 0.1% trifluoroacetic acid (TFA) and loaded onto the hydrophilic/lipophilic balanced (HLB) resin, which was previously equilibrated using 100 μl of the same buffer. After washing with 100 μl of 0.1% TFA, the samples were eluted with a buffer containing 70 μl of 60% acetonitrile and 0.1% TFA and then dried in a vacuum centrifuge.
For liquid chromatography–MS/MS acquisition and analysis, samples were resuspended in 10 μl of 0.1% TFA and loaded onto a Dionex RSLC Ultimate 300 (Thermo Fisher Scientific), coupled online with an Orbitrap Fusion Lumos (Thermo Fisher Scientific). Chromatographic separation was performed with a two-column system, consisting of a C-18 trap cartridge (300 μm in inside diameter and 5 mm in length) and a picofrit analytical column (75 μm in inside diameter and 25 cm in length) packed in-house with reversed-phase Repro-Sil Pur C18-AQ 3-μm resin. Peptides were separated using a 30-min gradient from 1 to 30% buffer B (buffer A, 0.1% formic acid; buffer B, 80% acetonitrile + 0.1% formic acid) at a flow rate of 300 nl/min. The mass spectrometer was set to acquire spectra in a data-independent acquisition mode. Briefly, the full MS scan was set to 300 to 1100 mass/charge ratio (m/z) in the orbitrap with a resolution of 120,000 (at 200 m/z) and an automatic gain control (AGC) target of 5 × 105. MS/MS was performed in the orbitrap with sequential isolation windows of 50 m/z with an AGC target of 2 × 105 and a higher-energy collisional dissociation (HCD) collision energy of 30.
Histone peptides raw files were imported into EpiProfile 2.0 software (90). From the extracted ion chromatogram, the area under the curve was obtained and used to estimate the abundance of each peptide. To achieve the relative abundance of PTMs, the sum of all different modified forms of a histone peptide was considered as 100%, and the area of the particular peptide was divided by the total area for that histone peptide in all of its modified forms. The relative ratio of two isobaric forms was estimated by averaging the ratio for each fragment ion with different masses between the two species. The resulting peptide lists generated by EpiProfile were exported to Microsoft Excel and further processed for a detailed analysis.
Acetylomics
Primary IECs in the proximal half of the small intestine from four control and six Kat2a/Kat2b DKO mice were collected. Protein was extracted with RIPA buffer and processed for acetylomic MS as described previously (91) using PTMScan HS Acetyl-Lysine Motif (Ac-K) Kit (Cell Signaling Technology, #46784).
Immunoblotting
Protein preps were mixed with loading dye and a final concentration of 0.1 M dithiothreitol and then heated at 95°C for 10 min. For purified histones, 5 μg was loaded for each sample into a NuPAGE 4 to 12% bis-tris polyacrylamide gel (Thermo Fisher Scientific, catalog no. NP0336BOX) and run in NuPAGE MES SDS running buffer (Thermo Fisher Scientific, catalog no. NP0002). Proteins were wet transferred onto polyvinylidene difluoride membrane, which was subsequently blocked in 5% bovine serum albumin for phosphorylated proteins or 5% milk for nonphosphorylated proteins for 1 hour at room temperature and incubated overnight with primary antibody in blocking buffer (table S1). After washing in Tris-buffered saline with 0.1% Tween 20 (TBS-T), membranes were incubated in secondary HRP antibody for 1 hour. Enhanced chemiluminescence substrate (Ultra Digital-ECL substrate solution, Kindle Biosciences, #R1002) was used to detect and image protein (KwikQuant Imager, Kindle Biosciences, #D1001). Quantification was performed using ImageJ by measuring the mean gray value of each band and subtracting it from the background. Ratios were calculated by dividing the resulting value for the target protein of interest by that of the total protein or control.
Statistical analysis
Statistical analyses were typically conducted with t tests or two-way analysis of variance (ANOVA) with post hoc analyses, unless otherwise specified, as described in the figure legends for each figure.
Acknowledgments
We thank Q. Nguyen and K. Vemuri for bioinformatics assistance and J. Xing for key discussions.
Funding: This research was funded by grants from The New Jersey Commission on Cancer Research, DCHS20PPC023 (to M.-U.N.); National Institutes of Health, F31CA254086 (to M.-U.N.); National Institutes of Health, DK126446 and DK121915 (to M.V.); National Institutes of Health, S10OD025140 (to H.Z. and C.Z.); National Institutes of Health, R35 GM131678 (to S.D.); and National Institutes of Health, P30CA072720.
Author contributions: Conceptualization: M.V. and M.-U.N. Methodology: M.-U.N., M.L., Z.-F.Y., S.D., H.Z., S.S., C.E.E., and M.V. Data generation: M.-U.N., J.I., S.P., W.H., J.P., S.W., C.Z., R.Y., R.R., and Z.-F.Y. Data analysis: M.-U.N. and J.I. Writing—original draft: M.-U.N. and M.V. Key discussions: A.H. and J.B.S.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: RNA-seq and dsRIP-seq data from this publication have been deposited to GEO SuperSeries accession number GSE235796 for both RNA-seq (GSE235789) and dsRIP-seq (GSE235794). MS raw files of histone peptides are publicly available on the repository Chorus (https://chorusproject.org/) at the project number 1828. Acetylomics MS raw files are available on the repository MassIVE (https://massive.ucsd.edu/ProteoSAFe/dataset.jsp?task=ba975beda43f4f9d90de22e7e588b1da).
Supplementary Materials
This PDF file includes:
Figs. S1 to S4
Legends for tables S1 to S6
Other Supplementary Material for this manuscript includes the following:
Tables S1 to S6
REFERENCES AND NOTES
- 1.Li T., Chen Z. J., The cGAS-cGAMP-STING pathway connects DNA damage to inflammation, senescence, and cancer. J. Exp. Med. 215, 1287–1299 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yu L., Liu P., Cytosolic DNA sensing by cGAS: Regulation, function, and human diseases. Signal Transduct. Target. Ther. 6, 170 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sadeq S., Al-Hashimi S., Cusack C. M., Werner A., Endogenous double-stranded RNA. Noncoding RNA 7, 15 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dhir A., Dhir S., Borowski L. S., Jimenez L., Teitell M., Rötig A., Crow Y. J., Rice G. I., Duffy D., Tamby C., Nojima T., Munnich A., Schiff M., de Almeida C. R., Rehwinkel J., Dziembowski A., Szczesny R. J., Proudfoot N. J., Mitochondrial double-stranded RNA triggers antiviral signalling in humans. Nature 560, 238–242 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schlee M., Hartmann G., Discriminating self from non-self in nucleic acid sensing. Nat. Rev. Immunol. 16, 566–580 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kariko K., Buckstein M., Ni H., Weissman D., Suppression of RNA recognition by Toll-like receptors: The impact of nucleoside modification and the evolutionary origin of RNA. Immunity 23, 165–175 (2005). [DOI] [PubMed] [Google Scholar]
- 7.Wang R., Li H., Wu J., Cai Z. Y., Li B., Ni H., Qiu X., Chen H., Liu W., Yang Z. H., Liu M., Hu J., Liang Y., Lan P., Han J., Mo W., Gut stem cell necroptosis by genome instability triggers bowel inflammation. Nature 580, 386–390 (2020). [DOI] [PubMed] [Google Scholar]
- 8.Južnić L., Peuker K., Strigli A., Brosch M., Herrmann A., Häsler R., Koch M., Matthiesen L., Zeissig Y., Löscher B. S., Nuber A., Schotta G., Neumeister V., Chavakis T., Kurth T., Lesche M., Dahl A., von Mässenhausen A., Linkermann A., Schreiber S., Aden K., Rosenstiel P. C., Franke A., Hampe J., Zeissig S., SETDB1 is required for intestinal epithelial differentiation and the prevention of intestinal inflammation. Gut 70, 485–498 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yuan H., Wu X., Wu Q., Chatoff A., Megill E., Gao J., Huang T., Duan T., Yang K., Jin C., Yuan F., Wang S., Zhao L., Zinn P. O., Abdullah K. G., Zhao Y., Snyder N. W., Rich J. N., Lysine catabolism reprograms tumour immunity through histone crotonylation. Nature 617, 818–826 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stanifer M. L., Guo C., Doldan P., Boulant S., Importance of type I and III interferons at respiratory and intestinal barrier surfaces. Front. Immunol. 11, 608645 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sommereyns C., Paul S., Staeheli P., Michiels T., IFN-lambda (IFN-λ) is expressed in a tissue-dependent fashion and primarily acts on epithelial cells in vivo. PLOS Pathog. 4, e1000017 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kotenko S. V., Durbin J. E., Contribution of type III interferons to antiviral immunity: Location, location, location. J. Biol. Chem. 292, 7295–7303 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lueschow S. R., McElroy S. J., The Paneth cell: The curator and defender of the immature small intestine. Front. Immunol. 11, 587 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elphick D. A., Mahida Y. R., Paneth cells: Their role in innate immunity and inflammatory disease. Gut 54, 1802–1809 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Minamide K., Sato T., Nakanishi Y., Ohno H., Kato T., Asano J., Ohteki T., IRF2 maintains the stemness of colonic stem cells by limiting physiological stress from interferon. Sci. Rep. 10, 14639 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sato T., Ishikawa S., Asano J., Yamamoto H., Fujii M., Sato T., Yamamoto K., Kitagaki K., Akashi T., Okamoto R., Ohteki T., Regulated IFN signalling preserves the stemness of intestinal stem cells by restricting differentiation into secretory-cell lineages. Nat. Cell Biol. 22, 919–926 (2020). [DOI] [PubMed] [Google Scholar]
- 17.Bowman G. D., Poirier M. G., Post-translational modifications of histones that influence nucleosome dynamics. Chem. Rev. 115, 2274–2295 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Strahl B. D., Allis C. D., The language of covalent histone modifications. Nature 403, 41–45 (2000). [DOI] [PubMed] [Google Scholar]
- 19.Xu W., Edmondson D. G., Roth S. Y., Mammalian GCN5 and P/CAF acetyltransferases have homologous amino-terminal domains important for recognition of nucleosomal substrates. Mol. Cell. Biol. 18, 5659–5669 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nagy Z., Tora L., Distinct GCN5/PCAF-containing complexes function as co-activators and are involved in transcription factor and global histone acetylation. Oncogene 26, 5341–5357 (2007). [DOI] [PubMed] [Google Scholar]
- 21.Huang H., Sabari B. R., Garcia B. A., Allis C. D., Zhao Y., SnapShot: Histone modifications. Cell 159, 458–458.e1 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Y., Guo Y. R., Liu K., Yin Z., Liu R., Xia Y., Tan L., Yang P., Lee J. H., Li X. J., Hawke D., Zheng Y., Qian X., Lyu J., He J., Xing D., Tao Y. J., Lu Z., KAT2A coupled with the α-KGDH complex acts as a histone H3 succinyltransferase. Nature 552, 273–277 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jin Q., Yu L. R., Wang L., Zhang Z., Kasper L. H., Lee J. E., Wang C., Brindle P. K., Dent S. Y. R., Ge K., Distinct roles of GCN5/PCAF-mediated H3K9ac and CBP/p300-mediated H3K18/27ac in nuclear receptor transactivation. EMBO J. 30, 249–262 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bai A. H., Wu W. K. K., Xu L., Wong S. H., Go M. Y., Chan A. W. H., Harbord M., Zhang S., Chen M., Wu J. C. Y., Chan M. W. Y., Chan M. T. V., Chan F. K. L., Sung J. J. Y., Yu J., Cheng A. S. L., Ng S. C., Dysregulated lysine acetyltransferase 2B promotes inflammatory bowel disease pathogenesis through transcriptional repression of interleukin-10. J. Crohns Colitis 10, 726–734 (2016). [DOI] [PubMed] [Google Scholar]
- 25.Grant P. A., Eberharter A., John S., Cook R. G., Turner B. M., Workman J. L., Expanded lysine acetylation specificity of Gcn5 in native complexes. J. Biol. Chem. 274, 5895–5900 (1999). [DOI] [PubMed] [Google Scholar]
- 26.Marmorstein R., Zhou M. M., Writers and readers of histone acetylation: Structure, mechanism, and inhibition. Cold Spring Harb. Perspect. Biol. 6, a018762 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Noberini R., Restellini C., Savoia E. O., Raimondi F., Ghiani L., Jodice M. G., Bertalot G., Bonizzi G., Capra M., Maffini F. A., Tagliabue M., Ansarin M., Lupia M., Giordano M., Osti D., Pelicci G., Chiocca S., Bonaldi T., Profiling of epigenetic features in clinical samples reveals novel widespread changes in cancer. Cancers 11, 723 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Riss A., Scheer E., Joint M., Trowitzsch S., Berger I., Tora L., Subunits of ADA-two-A-containing (ATAC) or Spt-Ada-Gcn5-acetyltrasferase (SAGA) coactivator complexes enhance the acetyltransferase activity of GCN5. J. Biol. Chem. 290, 28997–29009 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schiltz R. L., Mizzen C. A., Vassilev A., Cook R. G., Allis C. D., Nakatani Y., Overlapping but distinct patterns of histone acetylation by the human coactivators p300 and PCAF within nucleosomal substrates. J. Biol. Chem. 274, 1189–1192 (1999). [DOI] [PubMed] [Google Scholar]
- 30.Klein B. J., Simithy J., Wang X., Ahn J. W., Andrews F. H., Zhang Y., Côté J., Shi X., Garcia B. A., Kutateladze T. G., Recognition of histone H3K14 acylation by MORF. Structure 25, 650–654.e2 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuo M. H., Brownell J. E., Sobel R. E., Ranalli T. A., Cook R. G., Edmondson D. G., Roth S. Y., Allis C. D., Transcription-linked acetylation by Gcn5p of histones H3 and H4 at specific lysines. Nature 383, 269–272 (1996). [DOI] [PubMed] [Google Scholar]
- 32.Leemhuis H., Packman L. C., Nightingale K. P., Hollfelder F., The human histone acetyltransferase P/CAF is a promiscuous histone propionyltransferase. Chembiochem 9, 499–503 (2008). [DOI] [PubMed] [Google Scholar]
- 33.Cieniewicz A. M., Moreland L., Ringel A. E., Mackintosh S. G., Raman A., Gilbert T. M., Wolberger C., Tackett A. J., Taverna S. D., The bromodomain of Gcn5 regulates site specificity of lysine acetylation on histone H3. Mol. Cell. Proteomics 13, 2896–2910 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xia L., Liu Y., Zhang Z., Gong Y., Yu T., Zhao D., Qiu W., Wang Y., Zhang J., Modulation of IL-6 expression by KLF4-mediated transactivation and PCAF-mediated acetylation in sublytic C5b-9-induced rat glomerular mesangial cells. Front. Immunol. 12, 779667 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao M., Geng R., Guo X., Yuan R., Zhou X., Zhong Y., Huo Y., Zhou M., Shen Q., Li Y., Zhu W., Wang J., PCAF/GCN5-mediated acetylation of RPA1 promotes nucleotide excision repair. Cell Rep. 20, 1997–2009 (2017). [DOI] [PubMed] [Google Scholar]
- 36.Xu W., Edmondson D. G., Evrard Y. A., Wakamiya M., Behringer R. R., Roth S. Y., Loss of Gcn5l2 leads to increased apoptosis and mesodermal defects during mouse development. Nat. Genet. 26, 229–232 (2000). [DOI] [PubMed] [Google Scholar]
- 37.Fournier M., Tora L., KAT2-mediated PLK4 acetylation contributes to genomic stability by preserving centrosome number. Mol. Cell. Oncol. 4, e1270391 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fournier M., Orpinell M., Grauffel C., Scheer E., Garnier J. M., Ye T., Chavant V., Joint M., Esashi F., Dejaegere A., Gönczy P., Tora L., KAT2A/KAT2B-targeted acetylome reveals a role for PLK4 acetylation in preventing centrosome amplification. Nat. Commun. 7, 13227 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee S. Y., Kim J. J., Miller K. M., Single-cell analysis of histone acetylation dynamics at replication forks using PLA and SIRF. Methods Mol. Biol. 2589, 345–360 (2023). [DOI] [PubMed] [Google Scholar]
- 40.Fournier M., Rodrigue A., Milano L., Bleuyard J. Y., Couturier A. M., Wall J., Ellins J., Hester S., Smerdon S. J., Tora L., Masson J. Y., Esashi F., KAT2-mediated acetylation switches the mode of PALB2 chromatin association to safeguard genome integrity. eLife 11, e57736 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jin Q., Wang C., Kuang X., Feng X., Sartorelli V., Ying H., Ge K., Dent S. Y. R., Gcn5 and PCAF regulate PPARγ and Prdm16 expression to facilitate brown adipogenesis. Mol. Cell. Biol. 34, 3746–3753 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jin Q., Zhuang L., Lai B., Wang C., Li W., Dolan B., Lu Y., Wang Z., Zhao K., Peng W., Dent S. Y. R., Ge K., Gcn5 and PCAF negatively regulate interferon-β production through HAT-independent inhibition of TBK1. EMBO Rep. 15, 1192–1201 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ghosh T. K., Aparicio-Sánchez J. J., Buxton S., Ketley A., Mohamed T., Rutland C. S., Loughna S., Brook J. D., Acetylation of TBX5 by KAT2B and KAT2A regulates heart and limb development. J. Mol. Cell. Cardiol. 114, 185–198 (2018). [DOI] [PubMed] [Google Scholar]
- 44.Kurooka H., Honjo T., Functional interaction between the mouse notch1 intracellular region and histone acetyltransferases PCAF and GCN5. J. Biol. Chem. 275, 17211–17220 (2000). [DOI] [PubMed] [Google Scholar]
- 45.Brown K., Chen Y., Underhill T. M., Mymryk J. S., Torchia J., The coactivator p/CIP/SRC-3 facilitates retinoic acid receptor signaling via recruitment of GCN5. J. Biol. Chem. 278, 39402–39412 (2003). [DOI] [PubMed] [Google Scholar]
- 46.Gonçalves S., Patat J., Guida M. C., Lachaussée N., Arrondel C., Helmstädter M., Boyer O., Gribouval O., Gubler M. C., Mollet G., Rio M., Charbit M., Bole-Feysot C., Nitschke P., Huber T. B., Wheeler P. G., Haynes D., Juusola J., Billette de Villemeur T., Nava C., Afenjar A., Keren B., Bodmer R., Antignac C., Simons M., A homozygous KAT2B variant modulates the clinical phenotype of ADD3 deficiency in humans and flies. PLOS Genet. 14, e1007386 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li J., Yan C., Wang Y., Chen C., Yu H., Liu D., Huang K., Han Y., GCN5-mediated regulation of pathological cardiac hypertrophy via activation of the TAK1-JNK/p38 signaling pathway. Cell Death Dis. 13, 421 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Walters B. W., Tan T. J., Tan C. T., Dube C. T., Lee K. T., Koh J., Ong Y. H. B., Tan V. X. H., Jahan F. R. S., Lim X. N., Wan Y., Lim C. Y., Divergent functions of histone acetyltransferases KAT2A and KAT2B in keratinocyte self-renewal and differentiation. J. Cell Sci. 136, jcs260723 (2023). [DOI] [PubMed] [Google Scholar]
- 49.Yamauchi T., Yamauchi J., Kuwata T., Tamura T., Yamashita T., Bae N., Westphal H., Ozato K., Nakatani Y., Distinct but overlapping roles of histone acetylase PCAF and of the closely related PCAF-B/GCN5 in mouse embryogenesis. Proc. Natl. Acad. Sci. U.S.A. 97, 11303–11306 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lin W., Zhang Z., Chen C. H., Behringer R. R., Dent S. Y., Proper Gcn5 histone acetyltransferase expression is required for normal anteroposterior patterning of the mouse skeleton. Dev. Growth Differ. 50, 321–330 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ashburner M., Ball C. A., Blake J. A., Botstein D., Butler H., Cherry J. M., Davis A. P., Dolinski K., Dwight S. S., Eppig J. T., Harris M. A., Hill D. P., Issel-Tarver L., Kasarskis A., Lewis S., Matese J. C., Richardson J. E., Ringwald M., Rubin G. M., Sherlock G., Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25, 25–29 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.el Marjou F., Janssen K. P., Hung-Junn Chang B., Li M., Hindie V., Chan L., Louvard D., Chambon P., Metzger D., Robine S., Tissue-specific and inducible Cre-mediated recombination in the gut epithelium. Genesis 39, 186–193 (2004). [DOI] [PubMed] [Google Scholar]
- 53.Lin W., Zhang Z., Srajer G., Chen Y. C., Huang M., Phan H. M., Dent S. Y. R., Proper expression of the Gcn5 histone acetyltransferase is required for neural tube closure in mouse embryos. Dev. Dyn. 237, 928–940 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tjeertes J. V., Miller K. M., Jackson S. P., Screen for DNA-damage-responsive histone modifications identifies H3K9Ac and H3K56Ac in human cells. EMBO J. 28, 1878–1889 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sidoli S., Bhanu N. V., Karch K. R., Wang X., Garcia B. A., Complete workflow for analysis of histone post-translational modifications using bottom-up mass spectrometry: From histone extraction to data analysis. J. Vis. Exp., 54112 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sato T., Clevers H., Primary mouse small intestinal epithelial cell cultures. Methods Mol. Biol. 945, 319–328 (2012). [DOI] [PubMed] [Google Scholar]
- 57.Sato T., Vries R. G., Snippert H. J., van de Wetering M., Barker N., Stange D. E., van Es J. H., Abo A., Kujala P., Peters P. J., Clevers H., Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 459, 262–265 (2009). [DOI] [PubMed] [Google Scholar]
- 58.Subramanian A., Tamayo P., Mootha V. K., Mukherjee S., Ebert B. L., Gillette M. A., Paulovich A., Pomeroy S. L., Golub T. R., Lander E. S., Mesirov J. P., Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Merlos-Suárez A., Barriga F. M., Jung P., Iglesias M., Céspedes M. V., Rossell D., Sevillano M., Hernando-Momblona X., da Silva-Diz V., Muñoz P., Clevers H., Sancho E., Mangues R., Batlle E., The intestinal stem cell signature identifies colorectal cancer stem cells and predicts disease relapse. Cell Stem Cell 8, 511–524 (2011). [DOI] [PubMed] [Google Scholar]
- 60.Haber A. L., Biton M., Rogel N., Herbst R. H., Shekhar K., Smillie C., Burgin G., Delorey T. M., Howitt M. R., Katz Y., Tirosh I., Beyaz S., Dionne D., Zhang M., Raychowdhury R., Garrett W. S., Rozenblatt-Rosen O., Shi H. N., Yilmaz O., Xavier R. J., Regev A., A single-cell survey of the small intestinal epithelium. Nature 551, 333–339 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Van Winkle J. A., Constant D. A., Li L., Nice T. J., Selective interferon responses of intestinal epithelial cells minimize tumor necrosis factor alpha cytotoxicity. J. Virol. 94, e00603–e00620 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Thomson D. W., Poeckel D., Zinn N., Rau C., Strohmer K., Wagner A. J., Graves A. P., Perrin J., Bantscheff M., Duempelfeld B., Kasparcova V., Ramanjulu J. M., Pesiridis G. S., Muelbaier M., Bergamini G., Discovery of GSK8612, a highly selective and potent TBK1 inhibitor. ACS Med. Chem. Lett. 10, 780–785 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen S., Lu W., Yueh M. F., Rettenmeier E., Liu M., Paszek M., Auwerx J., Yu R. T., Evans R. M., Wang K., Karin M., Tukey R. H., Intestinal NCoR1, a regulator of epithelial cell maturation, controls neonatal hyperbilirubinemia. Proc. Natl. Acad. Sci. U.S.A. 114, E1432–E1440 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bondy-Chorney E., Denoncourt A., Sai Y., Downey M., Nonhistone targets of KAT2A and KAT2B implicated in cancer biology. Biochem. Cell Biol. 97, 30–45 (2019). [DOI] [PubMed] [Google Scholar]
- 65.Cai K., Wan Y., Wang Z., Wang Y., Zhao X., Bao X., C5a promotes the proliferation of human nasopharyngeal carcinoma cells through PCAF-mediated STAT3 acetylation. Oncol. Rep. 32, 2260–2266 (2014). [DOI] [PubMed] [Google Scholar]
- 66.Chen Y. G., Hur S., Cellular origins of dsRNA, their recognition and consequences. Nat. Rev. Mol. Cell Biol. 23, 286–301 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kim S., Lee K., Choi Y. S., Ku J., Kim H., Kharbash R., Yoon J., Lee Y. S., Kim J. H., Lee Y. J., Kim Y., Mitochondrial double-stranded RNAs govern the stress response in chondrocytes to promote osteoarthritis development. Cell Rep. 40, 111178 (2022). [DOI] [PubMed] [Google Scholar]
- 68.Gao Y., Chen S., Halene S., Tebaldi T., Transcriptome-wide quantification of double-stranded RNAs in live mouse tissues by dsRIP-seq. STAR Protoc 2, 100366 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mercer T. R., Neph S., Dinger M. E., Crawford J., Smith M. A., Shearwood A. M. J., Haugen E., Bracken C. P., Rackham O., Stamatoyannopoulos J. A., Filipovska A., Mattick J. S., The human mitochondrial transcriptome. Cell 146, 645–658 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Guo R., Chen J., Mitchell D. L., Johnson D. G., GCN5 and E2F1 stimulate nucleotide excision repair by promoting H3K9 acetylation at sites of damage. Nucleic Acids Res. 39, 1390–1397 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mielke C., Lefort N., McLean C. G., Cordova J. M., Langlais P. R., Bordner A. J., te J. A., Ozkan S. B., Willis W. T., Mandarino L. J., Adenine nucleotide translocase is acetylated in vivo in human muscle: Modeling predicts a decreased ADP affinity and altered control of oxidative phosphorylation. Biochemistry 53, 3817–3829 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kunji E. R. S., King M. S., Ruprecht J. J., Thangaratnarajah C., The SLC25 carrier family: Important transport proteins in mitochondrial physiology and pathology. Physiology (Bethesda) 35, 302–327 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Palmieri F., Monne M., Discoveries, metabolic roles and diseases of mitochondrial carriers: A review. Biochim. Biophys. Acta 1863, 2362–2378 (2016). [DOI] [PubMed] [Google Scholar]
- 74.Chen Y. J., Hong W. F., Liu M. L., Guo X., Yu Y. Y., Cui Y. H., Liu T. S., Liang L., An integrated bioinformatic investigation of mitochondrial solute carrier family 25 (SLC25) in colon cancer followed by preliminary validation of member 5 (SLC25A5) in tumorigenesis. Cell Death Dis. 13, 237 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Iacobazzi V., Infantino V., Castegna A., Menga A., Palmieri E. M., Convertini P., Palmieri F., Mitochondrial carriers in inflammation induced by bacterial endotoxin and cytokines. Biol. Chem. 398, 303–317 (2017). [DOI] [PubMed] [Google Scholar]
- 76.Mosaoa R., Kasprzyk-Pawelec A., Fernandez H. R., Avantaggiati M. L., The mitochondrial citrate carrier SLC25A1/CIC and the fundamental role of citrate in cancer, inflammation and beyond. Biomolecules 11, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lin W., Srajer G., Evrard Y. A., Phan H. M., Furuta Y., Dent S. Y. R., Developmental potential of Gcn5−/− embryonic stem cells in vivo and in vitro. Dev. Dyn. 236, 1547–1557 (2007). [DOI] [PubMed] [Google Scholar]
- 78.Svensson V., Natarajan K. N., Ly L. H., Miragaia R. J., Labalette C., Macaulay I. C., Cvejic A., Teichmann S. A., Power analysis of single-cell RNA-sequencing experiments. Nat. Methods 14, 381–387 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Frankish A., Diekhans M., Jungreis I., Lagarde J., Loveland J. E., Mudge J. M., Sisu C., Wright J. C., Armstrong J., Barnes I., Berry A., Bignell A., Boix C., Carbonell Sala S., Cunningham F., di Domenico T., Donaldson S., Fiddes I. T., García Girón C., Gonzalez J. M., Grego T., Hardy M., Hourlier T., Howe K. L., Hunt T., Izuogu O. G., Johnson R., Martin F. J., Martínez L., Mohanan S., Muir P., Navarro F. C. P., Parker A., Pei B., Pozo F., Riera F. C., Ruffier M., Schmitt B. M., Stapleton E., Suner M. M., Sycheva I., Uszczynska-Ratajczak B., Wolf M. Y., Xu J., Yang Y. T., Yates A., Zerbino D., Zhang Y., Choudhary J. S., Gerstein M., Guigó R., Hubbard T. J. P., Kellis M., Paten B., Tress M. L., Flicek P., GENCODE 2021. Nucleic Acids Res. 49, D916–D923 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dobin A., Davis C. A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T. R., STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., 1000 Genome Project Data Processing Subgroup , The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Smith T., Heger A., Sudbery I., UMI-tools: Modeling sequencing errors in unique molecular identifiers to improve quantification accuracy. Genome Res. 27, 491–499 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Anders S., Pyl P. T., Huber W., HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Robinson M. D., McCarthy D. J., Smyth G. K., edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ramírez F., Ryan D. P., Grüning B., Bhardwaj V., Kilpert F., Richter A. S., Heyne S., Dündar F., Manke T., deepTools2: A next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44, W160–W165 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Langmead B., Trapnell C., Pop M., Salzberg S. L., Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Christensen T., Diemer N. H., Reduction of mitochondrial electron transport complex activity is restricted to the ischemic focus after transient focal cerebral ischemia in rats: A histochemical volumetric analysis. Neurochem. Res. 28, 1805–1812 (2003). [DOI] [PubMed] [Google Scholar]
- 89.Hench J., Hench I. B., Pujol C., Ipsen S., Brodesser S., Mourier A., Tolnay M., Frank S., Trifunović A., A tissue-specific approach to the analysis of metabolic changes in Caenorhabditis elegans. PLOS ONE 6, e28417 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yuan Z. F., Sidoli S., Marchione D. M., Simithy J., Janssen K. A., Szurgot M. R., Garcia B. A., EpiProfile 2.0: A computational platform for processing epi-proteomics mass spectrometry data. J. Proteome Res. 17, 2533–2541 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lancho O., Singh A., da Silva-Diz V., Aleksandrova M., Khatun J., Tottone L., Nunes P. R., Luo S., Zhao C., Zheng H., Chiles E., Zuo Z., Rocha P. P., Su X., Khiabanian H., Herranz D., A therapeutically targetable NOTCH1-SIRT1-KAT7 axis in T-cell leukemia. Blood Cancer Discov. 4, 12–33 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S4
Legends for tables S1 to S6
Tables S1 to S6