Abstract
Over the past few decades, the development of potent and safe immune-activating adjuvant technologies has become the heart of intensive research in the constant fight against highly mutative and immune evasive viruses such as influenza, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), and human immunodeficiency virus (HIV). Herein, we developed a highly modular saponin-based nanoparticle platform incorporating Toll-like receptor agonists (TLRas) including TLR1/2a, TLR4a, and TLR7/8a adjuvants and their mixtures. These various TLRa–saponin nanoparticle adjuvant constructs induce unique acute cytokine and immune-signaling profiles, leading to specific T helper responses that could be of interest depending on the target disease for prevention. In a murine vaccine study, the adjuvants greatly improved the potency, durability, breadth, and neutralization of both COVID-19 and HIV vaccine candidates, suggesting the potential broad application of these adjuvant constructs to a range of different antigens. Overall, this work demonstrates a modular TLRa-SNP adjuvant platform that could improve the design of vaccines and affect modern vaccine development.
Saponin-TLRa nanoadjuvants provide distinct immune signatures and drive potent, broad, and durable COVID-19 and HIV vaccine responses.
INTRODUCTION
The development of prophylactic vaccines against infectious diseases has been at the heart of intensive scientific research for the past hundred years, but the emergence of global pandemics such as severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), human immunodeficiency virus (HIV), and influenza has further catalyzed their development (1). Yet, many commercial vaccines comprise traditional live-attenuated or inactivated virus technologies, both of which suffer from complex manufacturing processes and potential safety hazards (2). Over the past few decades, protein-based subunit vaccine approaches have been shown to offer desirable characteristics in terms of safety, cost-effectiveness, scalability, and manufacturability. Subunit vaccines leverage the use of viral protein antigens paired with immune stimulating molecules, also known as adjuvants, that are essential in promoting the magnitude and durability of the immune response.
To date, only a handful of adjuvants are clinically licensed in vaccine formulations and approved by the Food and Drug Administration (FDA), including more traditional insoluble aluminum salts (Alum) and oil-in-water emulsions such as MF59 and AS03 (3). Numerous adjuvant candidates targeting specific molecular pathways have been in development recently; many of which offer promising improvements in the overall efficacy of subunit vaccines. A specific class of adjuvants that has been gaining attention is Toll-like receptor agonists (TLRas) (4, 5). These agonists include Pam3CysSerLys4 (Pam3CSK4; TLR1/2a), polyinosinic:polycytidylic acid [poly(I:C); TLR3a], monophosphoryl lipid (MPL; TLR4a), imidazoquinolines such as resiquimod or 3M-052 (TLR7/8a), and CpG-ODN (TLR9a). While these molecules differ in their physical and chemical properties and include lipids, small molecules, single-stranded DNA, and double-stranded RNA, they all activate different Toll-like receptors (TLRs) commonly expressed by antigen-presenting cells to drive distinct immune signaling pathways. Recent FDA approvals of vaccines comprising CpG-ODN 1018 (Dynavax), AS04 (GSK; MPL/Alum), AS01 (GSK; liposomes comprising MPL), along with numerous ongoing clinical trials using other promising TLRas such as 3M-052 (3M/AAHI), reinforce their promise as adjuvants to improve the efficacy of vaccines.
While TLRa adjuvants have been the primary focus in vaccine design, other non–TLR-based adjuvants have also been of interest. For example, saponins such as Quil-A, natural triterpene glycosides from Quillaja, are leading adjuvant molecules in development. Although saponins can be toxic when delivered in a soluble form, they are well tolerated when delivered in nanoparticles (NPs) such as the well-known honeycomb-like structure ISCOMATRIX (6–12). These well-defined nanostructures are 30 to 70 nm in size and are generated by spontaneous self-assembly of Quil-A saponins with cholesterol and phospholipids. These adjuvants have been successfully used for decades in vaccine formulations and have been demonstrated to be highly potent and safe. Matrix-M (Novavax), a self-assembled saponin-based nanostructure adjuvant comparable to ISCOMATRIX, was approved by the FDA in October 2022 as part of the Novavax COVID-19 vaccine. While the mechanism of immune stimulation of these self-assembled saponin-based nanostructure adjuvants has not yet been fully elucidated, recent reports suggest that they increase lymph flow and lymph node (LN) permeability to enhance antigen acquisition by B cells in the draining LNs (dLNs) (7, 12). More generally, intensive research has demonstrated the advantage of formulating adjuvants as NPs to improve their stability, solubility, cellular uptake, immunogenicity, and safety, all ultimately augmenting vaccine immune responses favorably (13–19).
Few attempts have been made to combine both TLRas and saponin adjuvants into the same nanostructure (7, 20–24). For example, AS01 adjuvant, which is part of the licensed Shingrix and Mosquirix vaccines by GSK, is a liposome containing TLR4a MPL and QS-21 saponin (21). Similarly, Irvine and coworkers recently designed an ISCOMATRIX analog incorporating TLR4a MPLA, named SMNP (7). Despite the potential for synergistic effects associated with combining saponins with a broader array of TLRas, saponin constructs incorporating a variety of TLRas have not yet been reported. Here, we report a library of saponin-based NP adjuvants incorporating various single or multiple clinically relevant TLRas (TLRa-SNPs) to generate a modular platform that could be broadly applicable. We generated four formulations of TLRa-SNPs incorporating Pam3CSK4 (TLR1/2a-SNP), MPLA (TLR4a-SNP), an imidazoquinoline derivative (TLR7/8a-SNP), as well as a mixture of both MPLA and imidazoquinoline derivative (TLR4a-TLR7/8a-SNP). Compared to plain saponin NPs (SNPs), comparable to Matrix-M and ISCOMATRIX, all four TLRa-SNPs elicited improved humoral immune responses when used as adjuvants in SARS-CoV-2 and HIV vaccines in mice. TLRa-SNPs led to more potent antibody titers, improved antibody durability, enhanced breadth against variants of concern (VOCs), and induced better neutralizing responses when compared to SNPs or another clinically relevant control adjuvant CpG/Alum. Notably, TLRa-SNPs induced unique acute cytokine profiles, leading to distinct T helper (TH) skewing depending on the TLRa incorporated. Overall, we describe the generation of a potent and modular TLRa-SNP platform enabling tuning of TH-skewed responses, a pertinent feature in preventing diseases for which different TH-skewed responses lead to better clinical outcomes, as well as enhancing over humoral immune responses to vaccines.
RESULTS
Formulation and characterization of TLRa-SNPs
We sought to create a platform of lipid-based SNPs incorporating various TLRa adjuvants. The formulation of plain SNPs, traditionally known as ISCOMATRIX, has been widely reported in the literature: mixing Quil-A saponin, dipalmitoylphosphatidylcholine (DPPC), and cholesterol at a molar ratio of 10:10:5 in aqueous medium triggers their spontaneous self-assembly into honeycomb-like NPs 30 to 70 nm in diameter (6, 8, 9). We hypothesized that lipid-based TLRas could be readily incorporated with SNPs at a specific stoichiometry via hydrophobic effects, while nonlipidated TLRas could be conjugated with a cholesterol motif to specifically interact with saponins. In this way, we successfully incorporated TLR1/2a Pam3CSK4, TLR4a synthetic MPLA (MPLAs), and cholesteryl TLR7/8a imidazoquinoline derivative with SNPs by mixing the desired TLRa adjuvant, Quil-A saponin, DPPC, and total cholesterol content at a molar ratio of 1:10:2.5:10 across all formulations (Fig. 1A, table S1, figs. S1 to S3, and the Supplementary Materials for synthesis). The resulting NPs showed monodisperse populations of hydrodynamic diameters between 40 and 60 nm and negative surface charges equal to or below −30 mV (Fig. 1, B and C). Cryo–electron microscopy (cryo-EM) confirmed that all NPs conserved the signature honeycomb-like structure after introduction of TLRa adjuvants (Fig. 1D). Moreover, TLRa-SNPs maintained their colloidal stability for at least 6 weeks when stored at 4°C and for several months when stored at −20°C (fig. S4).
Fig. 1. Design and characterization of TLRa-SNPs.
(A) Schematic representation of SNPs and different formulation of TLRa-SNPs: TLR1/2a-SNP incorporating Pam3CSK4, TLR4a-SNP incorporating MPLA, and TLR7/8a-SNP incorporating imidazoquinoline derivative. Hydrodynamic diameter (B) and surface charge (C) of SNPs and TLRa-SNPs measured by dynamic light scattering and Zetasizer. (D) Cryo-EM of (i) SNP, (ii) TLR1/2a-SNP, (iii) TLR4a-SNP, and (iv) TLR7/8a-SNP, demonstrating the maintenance of the SNP structure after introduction of the TLRas. Scale bars, 50 nm. RAW-Blue macrophage cells incubated with SNPs or TLRa-incorporated SNPs (TLRa-SNPs). Activation curves of (E) soluble TLR1/2a and TLR1/2a-SNP, (F) soluble TLR4a and TLR4a-SNP, and (G) soluble cholesteryl-modified TLR7/8a and TLR7/8a-SNP across a range of TLRa concentrations (0.0091 to 2.22 μg/ml) with 100,000 RAW-Blue cells (n = 3). (H) Schematic of in vivo evaluation of antigen (AF647-NP) accumulation with different TLRa-SNPs and CpG/Alum control. (I) Fluorescence IVIS imaging of the inguinal dLNs 48 hours after subcutaneous injection at the tail base. (J) Quantification of the LN accumulations at 48 hours. Data are shown as means ± SEM. P values were determined with one-way analysis of variance (ANOVA) with Tukey’s test of the absorbance values at the highest TLRa concentration or logged radiant efficiency values. Complete P values for comparisons are shown in tables S3 and S4. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
In vitro activation of TLRa-SNPs and in vivo LN accumulation of NP antigen with TLRa-SNPs
To further confirm that TLRas had been incorporated with SNPs and maintained their bioactivities, we performed a RAW-Blue transgenic mouse macrophage in vitro cell assay to confirm TLR activation. RAW-Blue cells were incubated with different formulations of TLRa-SNPs (SNPs, TLR1/2a-SNPs, TLR4a-SNPs, or TLR7/8a-SNPs) or the corresponding soluble TLRa adjuvants. Activation of the TLRs by the TLRas would lead to downstream activation of nuclear factor κB (NF-κB) and activating protein 1 (AP-1) pathways, which can be assessed with QUANTI-Blue for a colorimetric output. Concentration-dependent NF-κB and AP-1 activation curves were generated by stimulating the cells with a range of soluble TLRa or TLRa-SNP concentrations (n = 3; equivalent TLRa concentration of 0.0091 to 2.22 μg/ml; Fig. 1, E to G, and fig. S5; P values in table S3). No activation with plain SNPs or relevant controls was observed. In contrast, all TLRa-SNPs generated concentration-dependent NF-κB and AP-1 activation. Notably, TLRa-SNPs produced more potent activation compared to their soluble counterparts, most likely due to the ability of the NP structure to multivalently display the TLRas and to enhance cellular uptake.
Because saponin-based adjuvants have been found to improve lymphatic drainage (7), we evaluated the accumulation of antigens in the LNs with different TLRa-SNPs. Mice received 5 μg of an Alexa Fluor 647–labeled I53-50 NP antigen (AF647-NP) and 10 μg of SNP or TLRa-SNPs via subcutaneous injections at the tail base (Fig. 1H). I53-50 is a 28-nm-wide, 120-subunit complex that can be assembled in vitro by simply mixing trimeric and pentameric components forming a NP platform to allow for multivalent antigen presentation (25–27). This platform allows us to use AF647-labeled I53-50 NPs for imagining and use the vaccine antigen-tethered I53-50 NPs for immunogenicity studies. CpG/Alum was used as a clinically relevant positive control adjuvant. Forty-eight hours after administration, the inguinal dLNs (n = 4) were harvested for imaging using an in vivo imaging system (IVIS) (Fig. 1I). Compared to CpG/Alum, vaccines comprising SNP or TLRa-SNPs elicited significantly higher accumulation of AF647-NPs in the dLNs (Fig. 1J; P values in table S4). TLRa-SNPs also resulted in higher accumulation compared to plain SNPs, suggesting that the incorporation of TLRas improve antigen accumulation in the dLNs. Overall, the TLRa-SNP constructs maintained the bioactivity of the encapsulated TLRas in vitro and preserved the saponin’s ability to improve lymphatic drainage and LN accumulation in vivo.
RBD-NP vaccines adjuvanted with TLRa-SNPs generate potent and broad humoral responses
The immune responses generated by different TLRa-SNPs were investigated using SARS-CoV-2 vaccines formulated with a receptor binding domain NP (RBD-NP) antigen and TLRa-SNPs as NP adjuvants. The RBD-NP used in this study, also called RBD-16GS-I53-50, has been extensively studied in mice, nonhuman primates (NHPs), and human clinical trials (27–31). It has been shown to generate potent neutralizing humoral responses when formulated with AS03 or CpG/Alum adjuvants, leading to licensure in several countries of RBD-NP adjuvanted with AS03 as a COVID-19 vaccine for adults 18 years or older (32). Following the same immunization regime as previous studies (27, 30), 7- to 8-week-old C57BL/6 mice (n = 5) were subcutaneously immunized with vaccines comprising 1.5 μg of RBD-NP and 10 μg of TLRa-SNP formulations (SNP, TLR1/2a-SNP, TLR4aSNP, or TLR7/8a-SNP). For comparison, a clinical control containing CpG/Alum (20 and 100 μg, respectively) was evaluated. Mice were immunized at week 0 (prime) followed by a boost at week 3, and sera were collected from week 0 to week 11 (Fig. 2A).
Fig. 2. In vivo humoral response to RBD-NP vaccines adjuvanted with TLRa-SNPs.
(A) Timeline of immunization and blood collection to determine IgG titers. Mice were immunized on week 0 and boosted on week 3 with RBD-NP vaccines adjuvanted with CpG/Alum, SNP, or TLRa-SNPs. IgG1, IgG2c, neutralization, and variants titers were determined on week 5. (B) Anti-RBD IgG binding endpoint titers of RBD-NP vaccines adjuvanted with CpG/Alum, SNP, or TLRa-SNPs. (C) AUCs of anti-RBD IgG endpoint antibody titers from week 0 to week 11 of the different RBD-NP vaccines. (D) EC50 of anti-RBD IgG endpoint titers on week 5 of the different RBD-NP vaccines. (E) Anti-RBD IgG binding endpoint titers a year (D365) after immunization. (F) Anti-spike IgG binding endpoint titers from sera collected on week 5 after the initial immunization. Titers were determined for wild-type (WT) spike as well as Alpha (B.1.1.7), Beta (B.1.351), Delta (B.1.617.2), and Omicron (B.1.1.529) variants of the spike protein. Data (n = 4 to 5) are shown as means ± SEM. P values were determined using the GLM followed by Tukey’s post hoc comparison procedure on the logged titer values for IgG titer comparisons. Complete P values for comparisons are shown in tables S5 to S9. ns, not significant; *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
We first evaluated anti-RBD total immunoglobulin G (IgG) titers over time by measuring the endpoint binding antibody titers using enzyme-linked immunosorbent assays (ELISAs) (Fig. 2B). At week 3, prior to boosting, mice immunized with RBD-NP vaccines adjuvanted with CpG/Alum or SNP elicited inconsistent antibody titers with around half the mice’s anti-RBD IgG endpoint below the limit of detection. In contrast, on week 3, all mice immunized with vaccines adjuvanted with TLRa-SNPs (TLR1/2a-SNP, TLR4a-SNP, and TLR7/8aSNP) seroconverted and generated antibody titers significantly higher than the CpG/Alum and SNP controls (P values in table S5). Two weeks after boost (week 5), no significant difference in titers was observed between the two controls CpG/Alum and SNP, with an average endpoint titer of 2.1 × 105 and 9.6 × 104, respectively. However, TLRa-SNPs induced significantly higher titers for which the average endpoint titers were 9.1 × 105 (TLR1/2a-SNP), 5.2 × 105 (TLR4a-SNP), and 4.3 × 105 (TLR7/8a-SNP). In addition, TLRa-SNP vaccinated mice maintained significantly higher anti-RBD IgG endpoint titers at all time points after boost and produced robust titers with little deviations across animals (P values in table S5). In contrast, mice vaccinated with CpG/Alum resulted in variable titer responses spanning up to two orders of magnitude, leading to an SEM measured to be at least twice that of the rest of the treatments after boosting.
We calculated the area under the curves (AUCs) of the endpoint titers over the 11-week period to estimate the mean IgG production. No significant difference in AUC titers between CpG/Alum and SNP was observed (Fig. 2C; P values in table S6). While all TLRa-SNPs resulted in significantly higher AUCs than CpG/Alum, only TLR1/2aSNP produced a significantly higher AUC than SNP. EC50 titers (halfmaximal binding dilution) were also measured 2 weeks after boost (week 5) to ensure that the endpoint titers reported correlated with the functional potency of the antibody (Fig. 2D). Similar to comparisons of endpoint titers, CpG/Alum and SNP induced low and variable EC50 titers compared to TLRa-SNPs, which all generated significantly higher average EC50 (P values in table S7). Furthermore, we assessed the endpoint titers of the vaccinated mice a year after immunization (D365; Fig. 2E). While the average endpoint titers from week 11 to D365 drastically dropped by 24-fold for CpG/Alum (2.6 × 103) and 13-fold for SNP (3.9 × 103), TLRa-SNPs’ endpoint titers remained high with only a 7-fold (6.7 × 104) and 5-fold (5.4 × 104) decrease for TLR1/2a-SNP and TLR4a-SNP, respectively. TLR7/8aSNP vaccinated mice were excluded from the comparison due to an insufficient sample size that remained (n = 2 at D365 due to murine dermatitis leading to euthanasia criteria; statistical power calculated using Mead’s resource equation) (33, 34). Overall, the titers measured at D365 from mice receiving vaccines adjuvanted with TLRaSNPs were significantly higher than mice that received vaccines adjuvanted with either SNP or CpG/Alum controls. These titers were at the same order of magnitude as the endpoint titers measured for the SNP and CpG/Alum controls on week 11, demonstrating the durability of RBD-NP vaccines adjuvanted with TLRa-SNPs (fig. S6; P values in table S8).
We further determined anti-spike antibody titers to confirm that the antibodies produced from RBD-NP adjuvanted with TLRa-SNP vaccines could bind to native spike proteins. Moreover, the breadth of the antibody response was assessed by measuring the endpoint titers against different SARS-CoV-2 VOCs including Alpha (B.1.1.7), Beta (B.1.351), Delta (B.1.617.2), and Omicron (B.1.1.529) spike proteins 2 weeks after boost (week 5) compared to CpG/Alum (Fig. 2F). While no differences in endpoint titers were observed between SNP and CpG/Alum regardless of variants (P values in table S9), TLR1/2a-SNP led to significantly higher titers against all variants. Overall improved titers against variants were also observed for TLR4a-SNP and TLR7/8a-SNP. Notably, the ratio of titers against different variants compared to wild-type spike titer (percent drop) showed more consistent and higher breadth for TLRa-SNPs compared to CpG/Alum and SNP controls, for which more variable and larger interquartile ranges were observed (fig. S7). Overall, these findings suggested that SNP as an adjuvant did not perform better than the clinical control CpG/Alum in generating humoral responses. Only TLRa-containing SNPs elicited more potent and durable titer responses with better breadth.
RBD-NP vaccines adjuvanted with TLRa-SNPs produce strong neutralizing antibodies
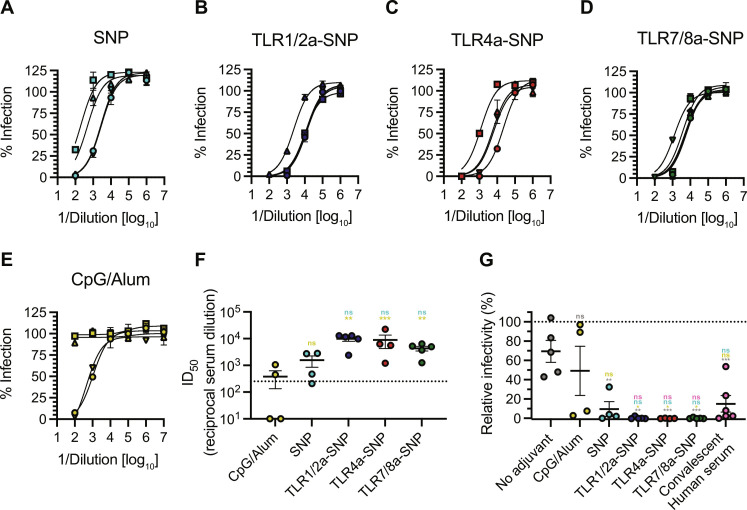
Because mice immunized with RBD-NP vaccines adjuvanted with TLRa-SNPs generated potent, durable, and broad antibody responses, we sought to measure the neutralizing activities of the sera. Week 5 sera neutralization was evaluated by using a SARS-CoV-2 spike pseudotyped lentivirus to measure serum-mediated inhibition of viral entry into HeLa cells overexpressing angiotensin-converting enzyme 2 (ACE2) and transmembrane serine protease 2 (TMPRSS2) (Fig. 2A). Serum neutralization ID50 was measured through neutralizing activities of a range of sera concentrations to determine the half-maximal inhibition of infectivity (ID50) (Fig. 3, A to F). Consistent with previous findings (31), mice immunized with RBD-NP adjuvanted with CpG/Alum resulted in highly variable neutralizing responses, with two mice having serum neutralizing activity below the limit of detection. In contrast to CpG/Alum, RBD-NP vaccines adjuvanted with SNP and TLRa-SNPs induced significantly higher neutralizing ID50 (P values in table S10). Notably, TLRa-SNPs elicited higher ID50 than the “high titer” classification according to FDA’s recommendation (ID50 ~ 102.4) (35). Moreover, mice sera neutralization activities were compared with human patients’ convalescent plasma by testing the pseudoviruses’ infectivity at the lowest sera dilution (1:100 dilution; Fig. 3G). Variable results were observed for SNP and CpG/Alum controls, where not all mice receiving vaccines with these two adjuvants reached 0% infectivity. Mice immunized with vaccines adjuvanted with CpG/Alum resulted in significantly higher infectivity than those immunized with vaccines adjuvanted with TLRa-SNPs, with an average comparable to mice that received nonadjuvanted RBD-NP vaccine (P values in table S11). On the contrary, we measured 0% infectivity from all mice receiving vaccines adjuvanted with TLRa-SNPs, indicating neutralization of all lentivirus and prevention of their entry into and infection of HeLa cells. Notably, the sera were even more neutralizing than the human patients’ convalescent plasma, for which the average infectivity was 15%. Overall, TLRa-SNPs as vaccine adjuvants generated superior humoral responses in potency, durability, breadth, and neutralization compared to both SNP and CpG/Alum controls.
Fig. 3. Neutralizing antibodies in mice after RBD-NP vaccines adjuvanted with TLRa-SNPs.
(A to E) Percent infectivity for all vaccine treatments at a range of week 5 serum dilutions as determined by a SARS-CoV-2 spike-pseudotyped viral neutralization assay. (F) Comparison of ID50 values determined from neutralization curves. Dotted line denotes the threshold for which the FDA considers as high titer (35). (G) Relative percent infectivity of all vaccine formulations compared to convalescent human serum at 1:100 dilution. Data (n = 4 to 5) are shown as means ± SEM. P values were determined using the GLM followed by Tukey’s post hoc multiple comparisons procedure on the logged neutralization ID50 values or relative infectivity comparisons. Complete P values for comparisons are shown in table S10 and S11. **P < 0.01 and ***P < 0.001.
TLRa-SNPs elicit tunable TH-skewed responses
After assessing the potency of TLRa-SNPs in inducing durable and broad antibody responses, we further evaluated if the nature of TLRa adjuvants affected the immune response of the TLRa-SNPs because TLRas have been shown to generate unique immune signaling and TH responses (36, 37). We hypothesized that different TLRa-SNPs could elicit unique levels of IgG1 and IgG2c titers as these isotypes are respectively strong indicators of TH2- and TH1-skewed responses. IgG isotypes were therefore measured across the different vaccine treatments. Even though all TLRa-SNPs led to improved total IgG antibody responses compared to SNP and CpG/Alum controls, they generated different levels of IgG isotypes. While TLR4a-SNP and TLR7/8a-SNP generated similar IgG1 responses compared to SNP and CpG/Alum, TLR1/2a-SNP induced significantly higher IgG1 titers compared to all other groups, with average titers an order of magnitude higher compared to those generated by SNPs (Fig. 4A; P values in table S12). On the other hand, all TLRa-SNPs elicited higher IgG2c titers, with at least a fivefold increase in average titers compared to the controls (Fig. 4B; P values in table S13). The differences in IgG1 and IgG2c titers suggested that different TLRa-SNPs resulted in different “flavors” of antibody responses, which we measured by calculating the ratio of IgG2c to IgG1 titers (Fig. 4C). The ratio suggested that TLR1/2a-SNP elicited a TH2-skewed response whereas TLR4a-SNP and TLR7/8a-SNP induced a TH1-skewed response. These results are drastically different from the balanced TH1/TH2 responses generated by SNP and CpG/Alum controls. While TH1skewed responses may lead to better disease outcomes for SARSCoV-2 infections (38–41), the tailored TH responses induced from the different TLRa-SNPs could position them as a potent and modular adjuvant platform for inducing immunity against other challenging viruses by enabling the generation of bespoke adjuvant responses with distinct immune signatures.
Fig. 4. Antibody subtype response to RBD-NP vaccines adjuvanted with TLRa-SNPs.
(A) Anti-RBD IgG1 and (B) IgG2c titers from sera collected on week 5, 2 weeks after boost. (C) Ratio of anti-RBD IgG2c to IgG1 titers. Lower values (below 1) suggest a TH2 response or humoral response, and higher values (above 1) imply a TH1 response or cellular response. Data (n = 4 to 5) are shown as means ± SEM. P values listed were using the GLM followed by Tukey’s post hoc multiple comparison procedure on the logged titer values. Complete P values for comparisons are shown in tables S12 and S13. *P < 0.05 and **P < 0.01.
Different TLRa-SNPs induced unique acute cytokine induction in the dLN and robust germinal center responses
We hypothesized that the differences in TH-skewed responses from different TLRa-SNP formulations were due to different TLRas leading to differences in immune signaling. To measure the acute immune response postvaccination, we assessed dLN cytokine profiles 1 day (24 hours) after immunization through a Luminex assay (Fig. 5A). Instead of reporting the results in pg/ml, which can suffer from plate/batch/lot inconsistencies, nonspecific binding, differences between standards and real biological samples, and other experimental artifacts, we analyzed the raw fluorescence signal by correcting for nonspecific binding as a covariate in the regression analysis to better infer the true biological patterns in Luminex data (42–45) (fig. S8). Sufficient levels of cytokines in the dLN are necessary to mount a successful vaccine response, and different cytokines induced can result in different TH responses. Comparing to mice adjuvanted with SNP, those that received TLRa-SNPs (TLR1/2a-SNP, TLR4a-SNP, and TLR7/8a-SNP) all induced significantly higher median fluorescence intensities (MFIs) of interferon-γ (IFN-γ), interferon gammainduced protein 10 (IP10 or CXCL10), interleukin (IL)-3, and IL-15 (Fig. 5B; P values in table S14). These are critical proinflammatory cytokines for initiating strong immune responses and overall improved TH response (46). Additionally, elevated levels of IL-6, IL-25, and IL-31 observed with TLR1/2a-SNP–adjuvanted vaccine could explain the observed TH2-skewed response (fig. S8). On the other hand, elevated levels of cytokines such as IFN-γ, IP10, IL-18, IL-27, and granulocyte-macrophage colony-stimulating factor (GM-CSF) observed with TLR4a-SNP could be the cause of the observed TH1-skewed response (fig. S8). Notably, of the 48 cytokines measured in the Luminex assay, only 11 did not show significant differences between SNP and at least one of the TLRa-SNPs (fig. S8). Thus, to better understand the overall cytokine induction profile of each adjuvant treatment, we performed a dimensional reduction analysis by creating a penalized supervised star (PSS) plot (Fig. 5C). We were successful in generating distinct and separate cluster for each adjuvant. Not only did TLRa-SNPs induce higher effector cytokine production in the dLNs, but different formulations of TLRa-SNPs also led to unique acute cytokine profiles postvaccination.
Fig. 5. dLN analysis postimmunization with RBD-NP vaccines adjuvanted with SNP or TLRa-SNPs.
(A) Timeline of immunization and dLN analysis. Luminex analysis was performed 1 day (24 hours) after vaccination, and flow cytometry was performed to measure GCBC and TFH population 12 days after vaccination. (B) All three TLRa-SNPs induced significantly higher beneficial proinflammatory cytokine levels of IFN-γ, IP10, IL-3, and IL-15 in the dLNs compared to SNPs, reported in MFI. (C) Dimensional reduction analysis of the full murine Luminex 48-plexed cytokine assay (fig. S8) by performing a PSS plot of the RBD-NP vaccines. The result shows a clear separation of vaccines containing different adjuvants, with each number signifying an individual sample from the treatment groups. Vectors are the projection coefficients for each individual cytokine. (D) Total GCBC count and (E) frequency of GCBC from all B cells from RBD-NP vaccines. (F) GCBC-to-TFH cell ratio. Data (n = 4 to 5) are shown as means ± SEM. P values for the Luminex assay were from a generalized maximum entropy estimation regression adjusted to control the false discovery rate, and P values for flow cytometry were determined using the GLM followed by Tukey’s post hoc comparison procedure on the logged values. Complete P values for comparisons are shown in tables S14 to S16 and S18.**P < 0.01, ***P < 0.001, and ****P < 0.0001.
We also assessed germinal center (GC) activity in the dLN 12 days after immunizing with RBD-NP vaccines adjuvanted with TLRaSNPs using flow cytometry (Fig. 5A; sample gating scheme in fig. S9). While mice treated with CpG/Alum-adjuvanted vaccines had the highest count of GC B cells (GCBCs), they also had the lowest percent of GCBCs of total B cells, indicating CpG/Alum’s inability to convert B cells to GCBCs (Fig. 5, D and E; P values in tables S15 and S16). On the contrary, mice immunized with vaccines adjuvanted with TLRa-SNPs resulted in higher percentages of GCBCs as well as higher total GCBC counts compared to mice receiving vaccines adjuvanted with SNP, suggesting robust GC activities. Moreover, compared to TLRa-SNP groups, mice receiving vaccines adjuvanted with SNP resulted in the lowest count of T follicular helper cells (TFHs) (fig. S10; P values in table S17). We then assessed the ratio of GCBCs to TFHs because this ratio has been found to correlate with the quality of T cell help (47–49). SNP and TLRa-SNP groups all resulted in a higher GCBC:TFH ratio compared to the CpG/Alum group (Fig. 5F; P values in table S18). The overall improvements in TFH count and the higher ratio of GCBC:TFH suggest that TLRa-SNPs can improve the quality of T cell help compared to SNP and CpG/Alum adjuvants. Because robust GC activity is important for generating better neutralizing and higher affinity antibodies, these data are consistent with the vaccination results that demonstrated increased magnitude and higher neutralizing antibodies with the TLRa-SNP groups. These experiments suggest TLRa-SNPs are superior in generating unique early proinflammatory responses and robust GC activity compared to SNP and CpG/Alum, resulting in an overall improved potency, durability, and breadth of humoral responses.
SNP incorporating both TLR4a and TLR7/8a generated synergistic immune stimulation
Prior research has shown that simultaneously triggering TIR domain-containing adaptor inducing interferon-β (TRIF)-coupled TLR and endosomal TLR can synergistically activate the immune system (5, 50, 51). As such, activating both TRIF-coupled TLR4 and endosomal TLR7/8 has been found to increase antigen-specific, neutralizing antibodies by enhancing the persistence of GCs and plasma-cell responses compared to activating single TLR ligands (50). We therefore created an additional formulation of TLRa-SNP, TLR4a-TLR7/8a-SNP, in which we incorporated equimolar amounts of TLR4a MPLA and cholesteryl TLR7/8a imidazoquinoline derivative in SNPs (Fig. 6A). The total molar ratio of TLRa to saponin was kept identical to the previously described TLRa-SNP formulations (table S1). The average hydrodynamic diameter of TLR4a-TLR7/8a-SNP, 47.9 nm, is consistent with the diameters measured from SNPs and other TLRaSNPs (Fig. 1B and fig. S11, A and B). Similar to the other TLRa-SNPs evaluated, TLR4a-TLR7/8a-SNP significantly enhanced AF647-NP accumulation in the dLNs in mice compared to CpG/Alum (fig. S11, C to E; P values in table S4). To confirm if coincorporating TLR4a and TLR7/8a in the same SNP is synergistic, we immunized C57BL/6 mice (n = 5) with vaccines formulated with 1.5 μg of RBD-NP adjuvanted with either 10 μg of TLR4a-TLR7/8a-SNP or a 1:1 mixture of TLR4a-SNP and TLR7/8a-SNP (5 μg each; referred as TLR4aSNP + TLR7/8a-SNP). Mice were vaccinated on week 0 and boosted on week 3 matching the previous schedule. Endpoint binding titers were measured to compare with those from SNP, TLR4a-SNP, and TLR7/8a-SNP formulations (Fig. 6B). We observed no significant difference of anti-RBD total IgG titers over time in mice adjuvanted with TLR4a-SNP + TLR7/8a-SNP compared to mice adjuvanted with either TLR4a-SNP or TLR7/8a-SNP (Fig. 6C; P values in table S19). In contrast, mice adjuvanted with TLR4a-TLR7/8a-SNP resulted in overall higher titers than all other groups, with a significant increase observed after the boost. The coincorporation of both TLR4a and TLR7/8a on the same SNP is therefore pivotal for their synergistic effect. Consistent with other TLRa-SNP formulations, mice receiving vaccines adjuvanted with either TLR4a-SNP + TLR7/8a SNPs or TLR4a-TLR7/8a-SNP maintained robust binding titers against wild-type spike and different SARS-CoV-2 VOCs (fig. S11, F and G). Last, both the TLR4a-SNP + TLR7/8a-SNP and TLR4a-TLR7/8a-SNP formulations maintained similar IgG1 and IgG2c titers compared to mice adjuvanted with either TLR4a-SNP or TLR7/8a-SNP, resulting in overall equally TH1-skewed responses (Fig. 6, D to F; P values in tables S20 and S21).
Fig. 6. TLRa-SNP incorporating both TLR4a and TLR7/8a generated synergistic effect.
(A) Schematic representation of TLR4a-TLR7/8a-SNP for which SNPs coincorporate TLR4a MPLA and TLR7/8a imidazoquinoline derivative. (B) Timeline of RBD-NP vaccines immunization and blood collection to determine IgG titers. Mice were immunized on week 0 and boosted on week 3 with RBD-NP vaccines adjuvanted with a 1:1 mixture of TLR4a-SNP and TLR7/8a-SNP or TLR4a-TLR7/8a-SNP. IgG1, IgG2c, and variants titers were measured on week 5. (C) Anti-RBD IgG binding endpoint titers of RBD-NP vaccines. (D) IgG1 titers and (E) IgG2c titers. (F) Ratio of anti-RBD IgG2c to IgG1 titers. Lower values (below 1) suggest a TH2 response or humoral response, and higher values (above 1) imply a TH1 response or cellular response. (G) Week 7 competitive binding curves for different RBD-NP vaccines compared to an mAb reference competing with the same mAb. OD, optical density. (H) Calculated KD values from fitted binding curves. Data (n = 4 to 5) are shown as means ± SEM. P values were determined using the GLM followed by Tukey’s post hoc comparison procedure on the logged titer values for IgG titer comparisons. Complete P values for comparisons are shown in tables S19 to S22. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
To determine if RBD-NP vaccines adjuvanted with TLRa-SNPs resulted in improved antibody quality, we investigated the post-boost serum antibody affinities toward RBD using a competitive binding ELISA (Fig. 6, G and H). The polyclonal populations of antibodies produced by mice that received vaccines adjuvanted with SNP and CpG/Alum were found to have the highest dissociation constant (KD) with values of 134 and 60 nM, respectively. All antibodies from mice that received vaccines adjuvanted with TLRa-SNPs exhibit KD values almost an order of magnitude lower than the vaccine adjuvanted with SNP, with the TLR4a-TLR7/8a-SNP group eliciting a significantly lower KD of 5.5 nM (P values in table S22). These results are consistent with the findings that TLRa-SNPs enhance GC responses and improve antibody neutralization.
TLRa-SNPs as gp120 vaccine adjuvants induce robust humoral responses
Last, to confirm that TLRa-SNPs can elicit robust immune responses against other antigens, we vaccinated C57BL/6 mice (n = 5) with HIV gp120 antigen adjuvanted with TLR1/2a-SNP, TLR4a-SNP, TLR7/8a-SNP, and TLR4a-TLR7/8a-SNP. Titer responses were compared to the two following controls: gp120 vaccines adjuvanted with SNP or Alum. Mice were immunized on week 0 and boosted on week 4, and sera were collected from week 0 to week 6 to assess total anti-gp120 IgG and subtype titers (Fig. 7A). Before boosting, most mice that received Alum- or SNP-adjuvanted vaccines did not have detectable titers, but all mice that received vaccines adjuvanted with TLRa-SNPs seroconverted and elicited significantly higher titers (Fig. 7B; P values in table S23). Two weeks after boost (week 6), vaccines containing TLRa-SNP adjuvants generated significantly higher antibody titers compared to those with Alum, with average endpoint titers around 2000-fold higher. Notably, mice adjuvanted with TLR4a-TLR7/8a-SNP elicited the strongest response. Moreover, TLRaSNPs also led to over threefold higher titers than SNPs. Similarly, we observed significantly higher overall antibody production from mice adjuvanted with TLRa-SNPs compared to the controls by determining the AUCs of titers over the 6-week period (Fig. 7C; P values in table S24).
Fig. 7. In vivo humoral response to HIV gp120 vaccines adjuvanted with TLRa-SNPs.
(A) Timeline of immunization and blood collection to measure IgG titers. Mice were immunized on week 0 and boosted on week 4 with gp120 vaccines adjuvanted with Alum, SNP, or TLRa-SNPs. IgG1 and IgG2c titers were determined on week 6. (B) Anti-gp120 IgG binding endpoint titers of gp120 vaccines adjuvanted with Alum, SNP, or TLRa-SNPs. (C) AUCs of anti-gp120 IgG endpoint antibody titers from week 0 to week 6 of different gp120 vaccines. (D) Anti-gp120 IgG1 and (E) IgG2c titers from sera collected on week 6, 2 weeks after boost. (F) Ratio of anti-gp120 IgG2c to IgG1 titers. Lower values (below 1) suggest a TH2 response or humoral response, and higher values (above 1) imply a TH1 response or cellular response. (G) Total GCBC count from gp120 vaccines and (H) frequency of GCBC from all B cells. Data (n = 5 to 6) are shown as means ± SEM. P values were determined using the GLM followed by Tukey’s post hoc comparison procedure. Complete P values for comparisons are shown in tables S23 to S28. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
To confirm that the unique TH-skewed responses observed from different TLRa-SNPs in RBD-NP vaccines were due to influences of TLRas in immune signaling, we measured week 6 IgG1 and IgG2c titers from mice immunized with gp120 vaccines. Consistent with our previous observations, mice adjuvanted with TLR1/2a-SNP, TLR4a-SNP, and TLR7/8a-SNP all induced robust IgG2c responses while only TLR1/2a-SNP generated significantly higher IgG1 titers compared to SNP (Fig. 7, D and E; P values in tables S25 and S26). On the other hand, TLR4a-TLR7/8a-SNP elicited significantly higher IgG1 and IgG2c titers compared to SNP. This resulted in a balanced TH1/TH2 response from mice adjuvanted with TLR4a-TLR7/8a-SNP, a more TH2-skewed response from mice adjuvanted with TLR1/2aSNP, and a more TH1-skewed response from mice adjuvanted with TLR4a-SNP and TLR7/8a-SNP (Fig. 7F). In addition, we assessed GC activity in the dLN 14 days after immunizing with gp120 adjuvanted with TLRa-SNPs (Fig. 7A). Mice immunized with vaccines adjuvanted with TLRa-SNPs resulted in overall higher count and percent of GCBCs compared to mice adjuvanted with SNP or Alum (Fig. 7, G and H; P values in tables S27 and S28).
Last, we immunized male C57BL/6 mice with gp120 vaccines adjuvanted with either SNP or TLRa-SNPs to confirm that TLRa-SNPs have similar potent vaccine adjuvanting effects in both sexes (fig. S12A). We observed similar antibody titers between the male and female mice after immunizing them with gp120 vaccines adjuvanted with TLRa-SNPs (fig. S12, B and C). Consistent with previous findings in female mice, mice adjuvanted with TLR1/2a-SNP, TLR4a-SNP, TLR7/8a-SNP, or TLR4a-TLR7/8a-SNP all elicited significantly higher titers both pre-and post-boost compared to the SNP control (fig. S12, B and C; P values in table S29). Together, these results confirmed TLRa-SNPs’ ability to generate potent and robust antibody responses with distinctive TH-skewed responses regardless of the antigen or gender of the animal.
DISCUSSION
Over the past few decades, modern vaccinology has moved toward the use of subunit protein-based vaccines over more traditional vaccine formats on account of their increased affordability, ease of manufacturing, and high safety profile (2). Unfortunately, the poor immunogenicity of many protein antigens reinforces the need for developing more potent and modular adjuvants. In this study, we designed an adjuvant platform combining three approaches in adjuvant design and selection: (i) molecular TLRa adjuvants resembling pathogen-associated molecular patterns found on bacteria, viruses, and other foreign invaders our immune system is trained to recognize; (ii) saponin adjuvants from natural sources that improve lymphatic flow; and (iii) particulate design displaying both adjuvants together with improved trafficking kinetics and cellular uptake. Previous studies have reported the design of combined TLR4a MPLA and saponins into NP constructs, such as AS01 and SMNP. Nonetheless, a tunable platform approach must consider incorporating other clinically relevant TLRas. In this regard, we demonstrated that lipid TLRas, such as MPLA and Pam3CSK4, could be readily incorporated with clinically relevant stoichiometric ratios due to the hydrophobic nature of SNPs. Nonlipid TLRas, such as TLR7/8a resiquimod, could be incorporated by conjugating a cholesterol motif that hydrophobically interacts with SNPs. The facile assembly and modularity of the SNP platform also allow the design and investigation of SNPs containing multiple TLRas in the same particle such as TLR4a and TLR7/8a (TLR4a-TLR7/8a-SNP). As such, consistent with previous reports suggesting that TLR4a and TLR7/8a synergistically activate the innate immune system (5, 50), we observed improved antibody responses from mice receiving vaccines adjuvanted with TLR4aTLR7/8a-SNP. While prior research supported synergistic responses by activating both TRIF-coupled TLR4 and endosomal TLR7/8, examining TLRa-SNPs combining TLR1/2a and TLR4a or TLR1/2a with TLR7/8a may be of interest for future studies.
A recent report by Irvine and coworkers provided detailed mechanistic analyses of the improved humoral response induced by ISCOMATRIX and SMNP, similar to SNP and TLR4a-SNP in the present study (7). Mainly, both adjuvants enhance lymph flow in a mast cell–dependent manner, which promotes antigen delivery into the dLNs. Because this mechanism was determined to be saponin-dependent, we assumed that all TLRa-SNPs improved vaccine immune responses in a similar manner. Furthermore, because mast cells were shown to be essential in saponins’ ability to improve lymph flow and are effector cells of TH2 response, we hypothesized that TLR1/2a-SNP conferred superior humoral responses due to its ability to generate a TH2 skewed response. Future investigation could determine other synergistic effects of different TLRas with SNPs. Moreover, no significant elevation in proinflammatory cytokines or chemokines in the dLNs was observed when mice received soluble MPLA mixed with ISCOMATRIX compared to mice receiving ISCOMATRIX alone, suggesting that it is necessary to deliver both adjuvants in the same particle for their synergistic effects. In addition, previous reports have also suggested that delivering soluble TLRas can lead to potentially toxic levels of systemic inflammation due to their rapid clearance into the bloodstream (7, 52, 53). Hence, delivering soluble TLRas without carriers such as Alum or various NP constructs poses safety risks that may hinder clinical translation (54). As such, soluble TLRas mixed with SNP matching TLRa-SNP formulations were not tested as controls in this study. Furthermore, Irvine and coworkers reported no signs of toxicity of their SMNP design. The present study includes the use of clinically safe TLRas dosed in mice in lower masses than previously reported (7, 31, 55, 56). We therefore assumed that the TLRa-SNPs were nontoxic as well, an assumption supported by the absence of any toxicity signs in mice such as weight loss or injection site reactogenicity.
Numerous preclinical and clinical studies have used the RBD-NP antigen RBD-16GS-I53-50 developed by King and coworkers (27–30). In this study, we used the same RBD-NP antigen in a dose- and schedule-matched study design to allow for comparison of the potency of TLRa-SNPs with previously screened adjuvants (27–30). From those reports, AS03 and CpG/Alum have been reported to induce comparable humoral responses, including robust neutralizing responses, and both outperformed other clinically relevant adjuvants such as AS37, AddaVax, and Essai O/W 1849101 (30). The use of CpG/Alum as a control in the present study therefore highlights the superiority of all TLRa-SNPs (i.e., TLR1/2a-SNP, TLR4a-SNP, TLR7/8a-SNP, and TLR4a-TLR7/8a-SNP) in generating potent, durable, broad, and neutralizing antibody responses (30). This observation is even more notable considering that the dosage of CpG/Alum was doubled compared to that used in the literature (30). In addition, mice receiving vaccines adjuvanted with TLRa-SNPs demonstrated complete neutralization in pseudovirion neutralization assays, in contrast to convalescent human plasma, which resulted in a quantifiable level of infectivity. This observation is particularly notable considering all human patients had previously received up-to-date COVID-19 vaccines, including an original prime and boost regimen plus an additional booster of either Moderna or Pfizer COVID-19 mRNA vaccines, prior to contracting COVID-19 only 8 to 12 weeks before sample collection. Previous reports have demonstrated that patients with such hybrid immunity have superior neutralizing antibodies than patients who had been infected but not previously vaccinated (57–61). These data show great promise for further investigation of the TLRa-SNPs adjuvant technology in larger animal models. This would be especially of interest in assessing neutralizing antibody responses postvaccination with HIV vaccines adjuvanted with TLRa-SNPs because there are generally no detectable neutralizing antibodies toward HIV in mice.
In addition, in an NHP study, treatment groups receiving AS03-adjuvanted RBD-NP vaccines had extended antibody titers and more durable Omicron variant protection when compared to values previously reported for commercial Pfizer-BioNTech and Moderna mRNA vaccines (28). Nonetheless, antibody titers in NHPs waned to pre-boost magnitudes by 6 months after boost, necessitating additional boosters to maintain robust protection (28). We showed that mice receiving RBD-NP vaccines adjuvanted with TLRa-SNPs maintained antibody endpoint titers for an entire year following vaccination that were of similar magnitude to mice receiving vaccines adjuvanted with SNP or CpG/Alum at week 11 following vaccination. These astonishing results were further emphasized by the rapid seroconversion of all mice after a single immunization (i.e., the prime immunization), thereby demonstrating the potential of single-immunization vaccines adjuvanted with these TLRa-SNPs. The rapid and complete seroconversion was further verified with HIV gp120 protein antigen, suggesting the superiority and potential broad application of TLRa-SNPs compared to SNPs and clinically relevant control adjuvants. Notably, we did not observe any substantial differences between the two clinical controls CpG/Alum (mimicking Dynavax’s CpG 1018) and SNP (mimicking Novavax’s Matrix-M). Although the adjuvant potencies remained similar for these two control adjuvants, the differences in their immune-stimulating mechanism might create synergistic effects when combined, illustrated by the potent, durable, broad, and neutralizing humoral responses induced by TLRa-SNPs, which take advantage of multiple immune-stimulating mechanisms. Overall, TLRa-SNP adjuvants can rapidly lead to protective levels of neutralizing antibodies and prolong the durability of neutralizing responses, therefore strongly decreasing the need for costly booster shots, which is crucial for vaccines capable of rapidly and broadly protecting worldwide populations during a pandemic.
The unique acute cytokine induction profiles and TH-skewed responses observed in this study for each TLRa-SNP formulation are consistent with prior literature (3, 5, 62). TLR1/2a-SNP, TLR4a-SNP, and TLR7/8a-SNP generated robust IgG2c titers, thereby suggesting a strong TH1 response. All three TLRas can induce TH1 response by activating the NF-κB pathway. Elevated levels of IL-12 and IL-1β, hallmark cytokines of NF-κB activation, in the Luminex assay further verify the incorporation of these three TLRas and their impact on immune signaling (3). In addition, only TLR1/2a-SNP elicited higher IgG1 titer, indicating an overall TH2-skewed response. The activation of TLR1 and TLR2 triggers the ERK1 and ERK2 pathways, which could result in higher TH2 responses (3). Similarly, the initiation of endosomal TLRs such as TLR7 and TLR8 activates the interferon regulatory factor (IRF) pathway, which, in turn, induces strong type I interferon response such as the secretion of IFN-α and IFN-β. The PSS plot clearly indicates a stronger expression of IFN-α (demonstrated by the vector) in the TLR7/8a-SNP cluster (cluster 4; Fig. 5C). While the Luminex panel did not include IFN-β, we nonetheless observed other strong interferon responses, such as high IFN-γ, IP10, and IL-28 production from mice adjuvanted with TLR7/8a-SNP. The overall balanced but slightly TH1-skewed response is consistent with prior findings (3, 53, 62). Likewise, the TH1-skewed response observed with TLR4a-SNP is consistent with the previously reported SMNP, for which only IgG2a titers, but not IgG1, were significantly higher than the SNP control (7). Overall, the Luminex assay helped to further verify the success of TLRas incorporation to the SNPs as well as the unique TH responses induced from the library of TLRacontaining saponin-based NPs. In this regard, literature reports have shown that, while Th2-skewed responses resulted in more favorable clinical outcomes for some infectious diseases such as rabies virus (63, 64), TH1-skewed responses were favorable for diseases such as COVID-19 (38–41). Thus, having a library of TLRa-SNPs with tailored TH-skewed responses may have profound clinical implications for which a specific adjuvant formulation could be selected during the early stage of vaccine development depending on the disease the vaccine is designed to prevent.
Additional characterization of the TLRa-SNPs, including evaluation of how these NPs could influence antigen stability and conformation when combined, could be of interest to further extend the broad use of this NP platform. Moreover, protein-based vaccines have been previously reported to not induce strong cellular immunity. RBD-NP, the primary antigen used in this study, was found to not generate a significant CD8+ T cell response (29). We observed comparable results when assessing antigen-specific IFN-γ+CD8+ T cells by ELISpot following activation of splenocytes 2 weeks after boost with pan-RBD peptides (fig. S13; P values in table S30). While most clinical vaccines focus on inducing humoral responses (65), further insights into TLRa-SNPs’ ability to generate cellular responses could make them promising candidates as T cell or cancer vaccines. In addition, other than generating different TH-skewed responses, different TLRas have also been found to induce differences in other immune cell populations, such as different levels of longlived plasma cells or memory cells. Studying the activation and induction of innate cells and effector cells by different TLRa-SNPs could be of interest for developing a comprehensive understanding of the immune-stimulating nature of these adjuvant formulations.
Following our efforts to develop broadly applicable vaccine technologies, TLRa-SNPs showed great promise in improving the potency, durability, breadth, and neutralization of vaccines with the potential of preventing infection by complex immune evasive viruses such as SARS-CoV-2 and HIV. Notably, the different adjuvant formulations induced unique acute cytokine and immune-signaling profiles, leading to different TH responses. This robust and tunable adjuvant library reinforces the global effort of the Coalition for Epidemic Preparedness Innovations and the World Health Organization to develop and manufacture vaccines within 100 days in response to “Disease X,” an infectious agent currently unknown to cause human disease.
MATERIALS AND METHODS
Materials
Cholesterol (ovine wool, >98%) and 1,2-dipalmitoyl-sn-glycero-3phosphocholine (DPPC) were purchased from Avanti Polar Lipids. Quil-A adjuvant, MPLA (from S. minnesota R595, MPLA-SM VacciGrade), and Pam3CSK4 (VacciGrade) were purchased from Invivogen. Cholesteryl imidazoquinoline derivative was synthetized in a one-step reaction from 4-amino-2-(ethoxymethyl)-1H-imidazo[4,5-c]quinoline-1-butylamine (Career Henan Chemical Co., Henan Province, China; see the Supplementary Materials). SARS-CoV-2 proteins and antibodies were purchased from Sino Biological including SARS-CoV-2 RBD protein (40592-V08H), SARS-CoV-2 spike protein (40589-V08H4), Alpha B.1.1.7 spike (40591-V08H10), Beta B.1.351 spike (40591-V08H12), Delta B.1.617.2 spike (40591-V08H23), Omicron B.1.1.529 spike (40591-V08H41), SARS CoV-2 spike RBD monoclonal antibody (mAb) (40150-D001), and SARS CoV-2 spike RBD mAb with horseradish peroxidase (HRP) (40150-D001-H). HIV antigen gp120 was purchased from Immune Technology Corporation (IT-001-022p). CpG1826 (Vac-1826) and Alum (Alhydrogel 2%, vac-alu) were purchased from Invivogen. Maxisorp plates were purchased from Thermo Fisher Scientific. Goat anti-mouse IgG Fc secondary antibody (A16084) HRP was purchased from Invitrogen. Goat antimouse IgG1 and IgG2c Fc secondary antibodies with HRP (ab97250 and ab97255) and 3,3′,5,5′-tetramethylbenzidine (TMB) high sensitivity ELISA Substrate were purchased from Abcam. Single-color mouse IFN-γ ELISPOT kit was purchased from Cellular Technology Limited. Antigen-specific cell stimulating peptides (Pan-SARS-CoV-2 S-RBD peptides) were purchased from JPT Peptide Technologies (PM-WCPV-S-RBD-1).
Formulation of SNPs and TLRa-SNPs
The formulation of lipid-based NPs has been previously reported (6–11). Briefly, cholesterol and DPPC (20 mg/ml) were each dissolved in a solution of 20% (w/v) of MEGA-10 detergent in Milli-Q water. Quil-A saponin (20 mg/ml) was dissolved in pure Milli-Q water. All solutions were heated at 50°C until complete dissolution before use. When required, cholesteryl imidazoquinoline, MPLA, and Pam3CSK4 (2 mg/ml) were each dissolved in a solution of 20% (w/v) of MEGA-10 detergent in Milli-Q water and heated at 70°C under sonication until dissolution. All formulations were obtained after quickly mixing the appropriate volume of each compound at 60°C without stirring in this order: cholesterol/DPPC/adjuvant/Quil-A saponin followed by its dilution with phosphate-buffered saline (PBS) 1X to reach a final concentration of 1 mg/ml in cholesterol (table S1). Solutions were left for equilibration overnight at room temperature and dialyzed against PBS 1X [molecular weight cut-off (MWCO) of 10 kDa] for 5 days. Solutions were then filtered using 0.2-μm Acrodisc Syringe Filters and concentrated using Centricon spin filters (MWCO 50 kDa, 3000 relative centrifugal force (RCF), 40 min) in stock solutions of 5 mg/ml in Quil-A saponin.
Dynamic light scattering and zeta potential
The hydrodynamic diameter and surface charge of the NPs were respectively measured on a DynaPro II plate reader (Wyatt Technology) and a Zetasizer Nano Zs (Malvern Instruments). Three independent measurements were performed for each sample.
Cryo-EM and cryo-ET sample preparation and data collection
Cryo-EM and cryo–electron tomography (cryo-ET) samples were prepared using Vitrobot Mark IV. First, 1.2/1.3 Quantifoil grids (Ted Pella Inc., 658-200-CU-100) were glow discharged using PELCO easiGlow for 30 s. For cryo-ET, 6-nm gold fiducial was mixed with the samples in a 1:10 volume ratio. Then, the sample (2 mg/ml) was applied to the glow discharged grid (Electron Microscopy Sciences, 25486), followed by removing the extra sample amount using filter paper and quickly plunging into liquid ethane. Cryo-EM and cryo-ET data were acquired by a Titan Krios G3 electron microscope equipped with a K3 direct detector and energy filter with an accelerated voltage of 300 kV using SerialEM (66). Cryo-EM micrographs were collected on a pixel size of 1.13 Å per pixel with and −2-μm defocus. Cryo-ET data was collected from −60° to +60°C with a 2°C increment. The total electron dose was 80 e−/Å2 with −3-μm defocus, and the pixel size was 1.13 Å. Reconstruction was done by IMOD (67) and visualized by Chimera (68).
In vitro RAW-Blue reporter assay
RAW-Blue [NF-κB-SEAP (secreted embryonic alkaline phosphatase)] reporter cell line (Invivogen, raw-sp) was used to confirm the proper incorporation of TLRas with the SNPs and their potencies compared to soluble TLRas. RAW-Blue cells were cultured at 37°C with 5% CO2 in Dulbecco’s modified Eagle’s medium supplemented with l-glutamine (2 mM), d-glucose (4.5 g/liter), 10% heat-inactivated fetal bovine serum (FBS), and penicillin (100 U/ml)/streptomycin (100 μg). Every other passage, zeocin (100 μg/ml) was added to the culture medium. Serial dilutions of solvent controls, soluble SNP components (cholesterol, DPPC, and saponin), soluble TLRas (TLR1/2 Pam3CSK4, TLR4 MPLAs, and TLR7/8 cholesteryl imidazoquinoline derivative), SNP, and TLRa-SNPs were added to a 96-well tissue culture treated plate to achieve final concentrations between 40 and 0.01 μg/ml of TLRas (or an equivalent concentration of the individual component based on TLRa-SNPs added for the negative controls). We assumed 100% TLRa incorporation during TLRa-SNP formulations to determine the TLRa concentrations. About 100,000 cells were added to each well in 180 μl of medium and were incubated for 24 hours at 37°C in a 5% CO2 incubator. Manufacturer’s instructions were followed for SEAP quantification, and absorbance levels were detected at 655 nm after 2-hour incubation with QUANTI-Blue Solution (Invivogen). Nonlinear regression fits were estimated using the “Log(agonist) versus response–EC50” function in GraphPad Prism 8.4 software.
In vivo LN accumulation of NP antigen with TLRa-SNPs
C57BL/6 mice were injected subcutaneously at the tail base with 100 μl of PBS 1X buffer containing 10 μg of AF647-NPs and 10 μg of SNP, 10 μg of TLRa-SNPs, or 20 μg of CpG and 100 μg of Alum. Mice were euthanized 48 hours after injection with CO2, and their inguinal dLNs were imaged using an IVIS (IVIS Lago). Imaging procedures and data analysis methods were identical to those thoroughly described in previously published works (69–71). AF647-NPs were imaged using a 30-s exposure time, an excitation wavelength of 660 nm, and an emission wavelength of 710 nm, medium binning, and an F/stop of 4. Total radiant efficiency was quantified using the Living Image software version 4.2 with the region of interest defined as an oval of consistent size around the entire LN.
Preparation of RBD-NPs
The NP immunogen (RBD-NP) components and the NP formulation methodology have been previously described (27). RBD-NP was kept under the following buffer conditions: 50 mM Tris (pH 8), 150 mM NaCl, 100 mM l-arginine, and 5% sucrose.
Vaccine formulation
The model vaccines included an antigen of either 1.5 μg dose of RBD-NP (University of Washington) or 10 μg dose of gp120 (SIV/mac239, Immune Technology Corporation). Depending on the formulations, vaccines also contained an adjuvant of CpG1826/Alum (20 μg + 100 μg, respectively, Invivogen), SNP, or one of the four TLRa-SNPs with an equivalent dose of 10 μg of Quil-A saponins (TLR1/2a-SNP, TLR4a-SNP, TLR7/8a-SNP, or TLR4a-TLR7/8a-SNP). An additional vaccine was formulated with an equivalent dose of 5 μg saponins of TLR4a-SNP and 5 μg saponins of TLR7/8a-SNP to test their synergistic effects. The CpG/Alum group adjuvanted with RBD-NP was prepared similarly as described previously by Pulendran and coworkers as the clinical control (29, 30), although both the CpG and Alum dosages were doubled in the present study. Vaccines were prepared in PBS 1X to a volume of 100 μl per dose and loaded into syringes with a 26-gauge needle for subcutaneous injection. Mice were boosted on week 3 for RBD-NP vaccines or week 4 for gp120 vaccines.
Mice and vaccination
Six-to 7-week-old female (except for one experiment where male mice were vaccinated with gp120 vaccines) C57BL/6 (B6) mice were purchased from Charles River and housed in the animal facility at Stanford University. Several days before vaccine administration, mice were shaved on their right flank. On the day of injection, 100 μl of soluble vaccines was subcutaneously injected on the right side of their flank under brief isoflurane anesthesia. Mouse blood was collected from the tail vein following the schematic shown in Figs. 2 and 7. The inguinal dLNs were collected for GC analysis and Luminex after euthanasia, and spleens were collected for splenocyte ELISpot analysis after euthanasia.
Mouse serum antibody concentration and affinity with ELISAs
Serum anti-RBD or anti-gp120 IgG antibody endpoint titers were measured using an endpoint ELISA. Maxisorp plates were coated with SARS-CoV-2 RBD protein, spike protein, a spike protein variant (B.1.1.7, B.1.351, B.1.617.2, or B.1.529), or gp120 at 2 μg/ml in PBS 1X overnight at 4°C and subsequently blocked with PBS 1X containing 1 wt % bovine serum albumin (BSA) for 1 hour at 25°C. Plates were washed 5x in between each step with PBS 1X containing 0.05% of Tween 20. Serum samples were diluted in diluent buffer (PBS 1X containing 1% BSA) starting at 1:100 followed by a fourfold serial dilution and incubated in the coated plates for 2 hours at 25°C. Then, goat anti-mouse IgG Fc-HRP (1:10,000), IgG1 Fc-HRP (1:10,000), or IgG2c Fc-HRP (1:10,000) was incubated for 1 hour at 25°C. Plates were developed with TMB substrate for 6 min, and the reaction was stopped with 1Ν HCl aqueous solution. Plates were analyzed using a Synergy H1 microplate reader (BioTek Instruments) at 450 nm in absorbance. Total IgG, subtypes, and variants were imported into GraphPad Prism 8.4.1 to determine serum titers by fitting the curves (absorbance versus dilution) with a three-parameter nonlinear logistic regression (baseline constrained to 0.054, the negative control average). The dilution titer value at which the endpoint threshold (0.2 and 0.1 for RBD-NP and gp120 vaccines, respectively) was crossed for each curve was reported as the endpoint titer. Samples failing to meet the endpoint threshold at a 1:100 dilution were set to below the limit quantitation for the assay (which we plotted as 1:25 dilution).
For affinity measurements, competitive ELISAs were performed in a similar manner to previously published procedures (72). In these assays, 2X dilutions of the serum (starting at 1:50) were mixed with a constant 0.07 nM concentration of an HRP-conjugated anti-RBD mAb (Sino Biological) and incubated for 2 hours at room temperature. Plates were developed with TMB substrate for 6 min, and the reaction was stopped with 1 N HCl aqueous solution. Plates were analyzed using a Synergy H1 microplate reader (BioTek Instruments) at 450 nm in absorbance. Data were fitted with a one-site competitive binding model using GraphPad Prism 10. The control mAb reference antibody was assumed to have a KD value of 1 nM based on common affinities found in the industry. This assumption only affects the absolute KD values reported and not the relative differences between treatment groups.
SARS-CoV-2 spike-pseudotyped viral neutralization assay
Neutralization assays were conducted as described previously (73). Briefly, SARS-CoV-2 spike-pseudotyped lentivirus was produced in human embryonic kidney (HEK) 293T cells. Six million cells were seeded 1 day prior to transfection. A five-plasmid system (74) and BioT (Bioland Scientific) were used for viral production per manufacturer’s protocols with the full-length wild-type spike sequence from the Wuhan-Hu-1 strain of SARS-CoV-2. Virus-containing culture supernatants were harvested ≈72 hours after transfection by centrifugation and filtered through a 0.45-μm syringe filter. Stocks were stored at −80°C prior to use. For the neutralization assay, ACE2/HeLa cells were plated 1 to 2 days prior to infection (75). Mouse and human sera were heat inactivated at 56°C for 30 min prior to use. Sera and viruses were diluted in cell culture medium and supplemented with a polybrene at a final concentration of 5 μg/ml. Serum/virus dilutions were incubated at 37°C for 1 hour. After incubation, media was removed from cells and replaced with serum/virus dilutions and incubated at 37°C for 2 days. Cells were then lysed using BriteLite (PerkinElmer) luciferase readout reagent, and luminescence was measured with a BioTek plate reader. Each plate was normalized by wells with cells or virus only, and curves were fit with a threeparameter nonlinear logistic regression inhibitor curve to obtain IC50 values. Serum dilution curves display mean infectivity ± SEM for each individual mouse at each serum dilution. Normalized values were fit with a three-parameter nonlinear logistic regression inhibitor curve in GraphPad Prism 8.4.1 to obtain IC50 values. Fits were constrained to have a value of 0% at the bottom of the fit.
Luminex cytokine assay
Mice were immunized with RBD-NP and different TLRa-SNPs as described in the immunization section. Twenty-four hours after injection, mice were euthanized and the injection inguinal dLNs were excised and processed for analysis (76). Briefly, the dLNs were weighed and 10 ml of lysis buffer (Cell Extraction Buffer FNN0011 with 1x Halt protease inhibitor, Thermo Fisher Scientific) was added per 1 g of tissue. Tissues were then grinded using a Dounce tissue grinder and centrifuged at 10,000 RCF for 5 min using protein LoBind Eppendorf tubes to pellet tissue debris. Supernatants were collected into new protein LoBind Eppendorf tubes. Meanwhile, a Pierce BCA (bovine serum albumin) assay (Thermo Fisher Scientific) was set up to analyze the total protein content of the samples. When possible, samples were diluted and normalized to 2 mg/ml of the total sample protein content per sample using additional lysis buffer. Tissues/homogenates were kept on ice throughout processing and normalized samples were stored at −80°C. Injection dLN homogenates were provided to the Human Immune Monitoring Center (HIMC) for analysis by a 48-plex murine Luminex cytokine panel.
Dimensional reduction analysis via PSS plot
Cytokine MFI data were detrended for cage and nonspecific binding effects using ordinary least squares (77). Detrended data were used to construct a PSS plot (78). Analyses were conducted in SAS v.9.4 (SAS Institute, Cary, North Carolina, United States) and R (www.r-project.org). Star plots were constructed with R packages maptools (79), matrixcalc (80), plotrix (81), JPEN (82), and VCA (83).
Immunophenotyping in LNs by flow cytometry
According to a previously reported methodology (72), mice were euthanized using CO2 12 (RBD-NP vaccines) or 14 (gp120 vaccines) days after immunization. Inguinal LNs were collected and dissociated into single cell suspensions. Cells were stained for viability using Ghost Dye Violet 510 (Tonbo Biosciences, catalog no. 13-0870-T100) for 5 min on ice and washed with fluorescence-activated cell sorting (FACS) buffer (PBS 1X with 2% FBS and 1 mM EDTA). Fc receptor was blocked using anti-CD16/CD38 (clone: 2.4G2, BD Biosciences, catalog no. 553142) for 5 min on ice and then stained with the following fluorochrome conjugated antibodies: CD19 (PerCP-Cy5.5, clone: 1D3, BioLegend, catalog no. 152406), CD95 (PE-Cy7, clone: Jo2, BD Biosciences, catalog no. 557653), CD38 (BUV395, clone: 90, BD Biosciences, catalog no. 740245), CXCR4 (BV421, clone: L276F12, BioLegend, catalog no. 146511), CD86 (BV785, clone: GL1, BioLegend, catalog no. 105043), GL7 (AF488, clone: GL7, BioLegend, catalog no. 144613), CD3 (AF700, clone: 17A2, BioLegend, catalog no. 100216), CD4 (BV650, clone: GK1.5, BioLegend, catalog no. 100469), CXCR5 (BV711, clone: L138D7, BioLegend, catalog no. 145529), and PD1 (PE-Dazzle594, clone: 29F.1A12, BioLegend, catalog no. 135228) for 30 min on ice (table S2). Cells were washed, fixed with 4% paraformaldehyde on ice, washed again, and analyzed on an LSRII flow cytometer (BD Biosciences). Data were analyzed with FlowJ 10 (FlowJo, LLC; representative gating scheme in fig. S9).
ELISpot
The number of antigen-specific IFN-γ producing splenocytes was determined to correlate with antigen-specific IFN-γ producing CD8+ T cells using a mouse IFN-γ Single-Color ELISPOT kit (CTL ImmunoSpot). Spleen cells were harvested 2 weeks after boost (D35) following the immunization outlined in fig. S13A. A total of 800,000 splenocytes per sample were pipetted into the well and were stimulated with Pan-SARS-CoV-2-RBD peptides (1 μg/ml; JPT) for 24 hours at 37°C. Spots were then developed following the manufacturer’s instructions (Immunospot S6 Ultra M2).
Animal protocol
Mice were cared for following the Institutional Animal Care and Use guidelines. All animal studies were performed in accordance with the National Institutes of Health guidelines and the approval of Stanford Administrative Panel on Laboratory Animal Care (protocol APLAC-32109).
Collection of serum from human patients
Convalescent COVID-19 blood was collected from donors 8 to 12 weeks after the onset of symptoms; all donors had previously been immunized with COVID vaccines authorized by the FDA (total of three immunizations, excluding the omicron bivalent booster). Blood was collected in microtubes with serum gel for clotting (Starstedt) and centrifuged for 5 min at 10,000g, and then serum was stored at −80°C until used. Blood collection was performed by fingerprick in accordance with the National Institutes of Health guidelines, the approval of the Stanford Human Subjects Research and IRB Compliance Office (IRB-58511), and the consent of the individuals.
Statistical analysis
For in vivo experiments, animals were caged blocked. Mead’s resource equation was used to calculate the sample size above which additional subjects (n) will not have significant impact on power (33, 34). For most experiments, a sample size of n = 5 per group was used. A few mice died due to reasons external to the treatment effects as evaluated by the veterinarian staff. Groups resulting with n = 4 still had enough power for statistical analyses using Mead’s resource equation. No animals were considered outliers.
All results except for Luminex are expressed as means ± SEM. Unless otherwise specified, comparison between multiple groups and P values were conducted with the general linear model (GLM) followed by Tukey’s post hoc procedure test in JMP and we accounted and adjusted for unequal variances, cage blocking, and multiple comparison. Statistical significance was considered as P < 0.05. Selected P values are shown in the text, and the figures and remaining P values are in the Supplementary Materials.
For the Luminex assay, statistical analyses were provided by the Statistical Consultation Service of the HIMC at the Institute for Immunity, Transplantation and Infection, Stanford University School of Medicine. The logarithm of MFI was regressed on treatment group, cage, and logarithm of nonspecific binding MFI using generalized maximum entropy estimation regression (84). Within each treatment group, P values were adjusted to control the false discovery rate (85, 86) at 5% across all cytokines. Analyses and plotting were done in SAS (SAS Institute, Cary, North Carolina, United States).
Acknowledgments
We would like to thank former and current members of the Appel lab for their technical expertise and scientific discussion, especially C. S. Liong, C. M. Kasse, and A. Nejatfard for help in animal work and advice. We acknowledge W. Chiu and P. Kim for their assistance and advice. We extend our gratitude to T. H. Holmes and Y. Rosenberg-Hasson at the Stanford HIMC as well as D. Xu for invaluable advice in implementing the spike-pseudotyped lentiviral neutralization assay (to M.V.F.I.). We also would like to thank the human patients for donating their blood, the staff of the BioE/ChemE Animal Facility who cared for the mice, and the Stanford Core Facilities that were central to the completion of this work. Specifically, we thank the Vincent Coates Foundation Mass Spectrometry Laboratory (Stanford University Mass Spectrometry, RRID:SCR_017801) and the Department of Chemistry Nuclear Magnetic Resonance facility (Stanford University). Cryo-EM was performed at the Stanford-SLAC facility. FACS sorting was performed on an instrument in the Shared FACS Facility.
Funding: This work was financially supported by the Center for Human Systems Immunology with the Bill & Melinda Gates Foundation (OPP1113682, OPP1211043, INV027411, and INV-010680) and the National Institute of Allergy and Infectious Disease (P01AI167966), Eastman Kodak Fellowship (to B.S.O.); the National Institutes of Health (award numbers T32GM007365 and F30AI152943 to M.V.F.I.), the National Science Foundation Graduate Research Fellowship (to O.M.S., J.Y., and C.K.L.), the Hancock Fellowship of the Stanford Graduate Fellowship in Science and Engineering (to O.M.S.), the Stanford ChEM-H COVID-19 Drug and Vaccine Prototyping Seed Grant (to X.Z.), the NIH Biotechnology Training Program (T32 GM008412 to E.L.M.), the NIH High End Instrumentation grant (1 S10 OD028697-01), the NIH shared instrumentation grant (S10OD021600), and the NIH S10 Shared Instrument Grant (S10RR027431-01).
Author contributions: Conceptualization: B.S.O., J.B., O.M.S., J.Y., B.P., and E.A.A. Methodology: B.S.O., J.B., J.Z.A., O.M.S., J.Y., A.S.V., and E.A.A. Investigation: B.S.O., J.B., M.V.F.I., J.Z.A., X.Z., O.M.S., J.Y., J.H.K., C.K.J., E.L.M., L.C., and B.P. Visualization: B.S.O. and J.B. Supervision: B.S.O., J.B., N.P.K., and E.A.A. Resources: B.S.O., J.B., M.V.F.I., X.Z., J.Y., A.S.V., L.C., B.P., and N.P.K. Data curation: B.S.O. Validation: B.S.O., J.B., J.Z.A., X.Z., O.M.S., N.P.K., and E.A.A. Formal analysis: B.S.O., J.B., M.V.F.I., J.Z.A., and C.K.J. Project administration: B.S.O., J.B.,. A.S.V., N.P.K., and E.A.A. Funding acquisition: B.P., N.P.K., and E.A.A. Writing—original draft: B.S.O. and J.B. Writing—review and editing: B.S.O., J.B., X.Z., O.M.S., J.H.K., N.P.K., and E.A.A. All authors have given approval to the final version of the manuscript.
Competing interests: E.A.A., B.S.O., and J.B. are listed as inventors on a pending patent application. N.P.K. is a cofounder, shareholder, paid consultant, and chair of the scientific advisory board of Icosavax Inc. The King lab has received unrelated sponsored research agreements from Pfizer and GSK. All other authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Supplementary Methods
Figs. S1 to S13
Tables S1 to S30
Legend for data file S1
Other Supplementary Material for this manuscript includes the following:
Data file S1
REFERENCES AND NOTES
- 1.Mascola J. R., Fauci A. S., Novel vaccine technologies for the 21st century. Nat. Rev. Immunol. 20, 87–88 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moyle P. M., Toth I., Modern subunit vaccines: Development, components, and research opportunities. ChemMedChem 8, 360–376 (2013). [DOI] [PubMed] [Google Scholar]
- 3.Pulendran B., Arunachalam P. S., O’Hagan D. T., Emerging concepts in the science of vaccine adjuvants. Nat. Rev. Drug Discov. 20, 454–475 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maisonneuve C., Bertholet S., Philpott D. J., De Gregorio E., Unleashing the potential of NOD- and Toll-like agonists as vaccine adjuvants. Proc. Natl. Acad. Sci. U.S.A. 111, 12294–12299 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Duthie M. S., Windish H. P., Fox C. B., Reed S. G., Use of defined TLR ligands as adjuvants within human vaccines. Immunol. Rev. 239, 178–196 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morein B., Sundquist B., Höglund S., Dalsgaard K., Osterhaus A., Iscom, a novel structure for antigenic presentation of membrane proteins from enveloped viruses. Nature 308, 457–460 (1984). [DOI] [PubMed] [Google Scholar]
- 7.Silva M., Kato Y., Melo M. B., Phung I., Freeman B. L., Li Z., Roh K., Van Wijnbergen J. W., Watkins H., Enemuo C. A., Hartwell B. L., Chang J. Y. H., Xiao S., Rodrigues K. A., Cirelli K. M., Li N., Haupt S., Aung A., Cossette B., Abraham W., Kataria S., Bastidas R., Bhiman J., Linde C., Bloom N. I., Groschel B., Georgeson E., Phelps N., Thomas A., Bals J., Carnathan D. G., Lingwood D., Burton D. R., Alter G., Padera T. P., Belcher A. M., Schief W. R., Silvestri G., Ruprecht R. M., Crotty S., Irvine D. J., A particulate saponin/TLR agonist vaccine adjuvant alters lymph flow and modulates adaptive immunity. Sci. Immunol. 6, eabf1152 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.K. Lövgren-Bengtsson, B. Morein, Vaccine Adjuvants: Preparation Methods and Research Protocols, D. T. O’Hagan, Ed., Methods in Molecular Medicine (Humana Press, 2000). [Google Scholar]
- 9.Pedersen J. S., Oliveira C. L., Hübschmann H. B., Henriette L., Arleth L., Manniche S., Kirkby N., Nielsen H. M., Structure of immune stimulating complex matrices and immune stimulating complexes in suspension determined by small-angle x-ray scattering. Biophys. J. 102, 2372–2380 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sanders M. T., Brown L. E., Deliyannis G., Pearse M. J., ISCOM-based vaccines: The second decade. Immunol. Cell Biol. 83, 119–128 (2005). [DOI] [PubMed] [Google Scholar]
- 11.Pearse M. J., Drane D., ISCOMATRIX adjuvant for antigen delivery. Adv. Drug Deliv. Rev. 57, 465–474 (2005). [DOI] [PubMed] [Google Scholar]
- 12.Maraskovsky E., Schnurr M., Wilson N. S., Robson N. C., Boyle J., Drane D., Development of prophylactic and therapeutic vaccines using the ISCOMATRIX adjuvant. Immunol. Cell Biol. 87, 371–376 (2009). [DOI] [PubMed] [Google Scholar]
- 13.Ledesma F., Ozcan B., Sun X., Medina S. M., Landry M. P., Nanomaterial strategies for delivery of therapeutic cargoes. Adv. Funct. Mater. 32, 2107174 (2022). [Google Scholar]
- 14.Wu S., Xia Y., Hu Y., Ma G., Bio-mimic particles for the enhanced vaccinations: Lessons learnt from the natural traits and pathogenic invasion. Adv. Drug Deliv. Rev. 176, 113871 (2021). [DOI] [PubMed] [Google Scholar]
- 15.Ou B. S., Baillet J., Picece V. C. T. M., Gale E. C., Powell A. E., Saouaf O. M., Yan J., Nejatfard A., Lopez Hernandez H., Appel E. A., Nanoparticle-conjugated toll-like receptor 9 agonists improve the potency, durability, and breadth of COVID-19 vaccines. ACS Nano 18, 3214–3233 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ou B. S., Saouaf O. M., Baillet J., Appel E. A., Sustained delivery approaches to improving adaptive immune responses. Adv. Drug Deliv. Rev. 187, 114401 (2022). [DOI] [PubMed] [Google Scholar]
- 17.Sun B., Xia T., Nanomaterial-based vaccine adjuvants. J. Mater. Chem. B 4, 5496–5509 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Irvine D. J., Aung A., Silva M., Controlling timing and location in vaccines. Adv. Drug Deliv. Rev. 158, 91–115 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Irvine D. J., Hanson M. C., Rakhra K., Tokatlian T., Synthetic nanoparticles for vaccines and immunotherapy. Chem. Rev. 115, 11109–11146 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Van Hoeven N., Wiley S., Gage E., Fiore-Gartland A., Granger B., Gray S., Fox C., Clements D. E., Parks D. E., Winram S., Stinchcomb D. T., Reed S. G., Coler R. N., A combination of TLR-4 agonist and saponin adjuvants increases antibody diversity and protective efficacy of a recombinant West Nile Virus antigen. NPJ Vaccines 3, 39 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Didierlaurent A. M., Laupèze B., Di Pasquale A., Hergli N., Collignon C., Garçon N., Adjuvant system AS01: Helping to overcome the challenges of modern vaccines. Expert Rev. Vaccines 16, 55–63 (2017). [DOI] [PubMed] [Google Scholar]
- 22.Silva A., Mount A., Krstevska K., Pejoski D., Hardy M. P., Owczarek C., Scotney P., Maraskovsky E., Baz Morelli A., The combination of ISCOMATRIX adjuvant and TLR agonists induces regression of established solid tumors in vivo. J. Immunol. 194, 2199–2207 (2015). [DOI] [PubMed] [Google Scholar]
- 23.Martinez P., Sundling C., O'Dell S., Mascola J. R., Wyatt R. T., Karlsson Hedestam G. B., Primate immune responses to HIV-1 Env formulated in the saponin-based adjuvant AbISCO-100 in the presence or absence of TLR9 co-stimulation. Sci. Rep. 5, 8925 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Raaijmakers T. K., Van Den Bijgaart R. J. E., Den Brok M. H., Wassink M., De Graaf A., Wagenaars J. A., Nierkens S., Ansems M., Scheffer G. J., Adema G. J., Tumor ablation plus co-administration of CpG and saponin adjuvants affects IL-1 production and multifunctional T cell numbers in tumor draining lymph nodes. J. Immunother. Cancer 8, e000649 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brouwer P. J. M., Antanasijevic A., Berndsen Z., Yasmeen A., Fiala B., Bijl T. P. L., Bontjer I., Bale J. B., Sheffler W., Allen J. D., Schorcht A., Burger J. A., Camacho M., Ellis D., Cottrell C. A., Behrens A. J., Catalano M., Del Moral-Sanchez I., Ketas T. J., LaBranche C., van Gils M. J., Sliepen K., Stewart L. J., Crispin M., Montefiori D. C., Baker D., Moore J. P., Klasse P. J., Ward A. B., King N. P., Sanders R. W., Enhancing and shaping the immunogenicity of native-like HIV-1 envelope trimers with a two-component protein nanoparticle. Nat. Commun. 10, 4272 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marcandalli J., Fiala B., Ols S., Perotti M., de Van der Schueren W., Snijder J., Hodge E., Benhaim M., Ravichandran R., Carter L., Sheffler W., Brunner L., Lawrenz M., Dubois P., Lanzavecchia A., Sallusto F., Lee K. K., Veesler D., Correnti C. E., Stewart L. J., Baker D., Lore K., Perez L., King N. P., Induction of potent neutralizing antibody responses by a designed protein nanoparticle vaccine for respiratory syncytial virus. Cell 176, 1420–1431.e17 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Walls A. C., Fiala B., Schäfer A., Wrenn S., Pham M. N., Murphy M., Tse L. V., Shehata L., O’Connor M. A., Chen C., Navarro M. J., Miranda M. C., Pettie D., Ravichandran R., Kraft J. C., Ogohara C., Palser A., Chalk S., Lee E.-C., Guerriero K., Kepl E., Chow C. M., Sydeman C., Hodge E. A., Brown B., Fuller J. T., Dinnon K. H., Gralinski L. E., Leist S. R., Gully K. L., Lewis T. B., Guttman M., Chu H. Y., Lee K. K., Fuller D. H., Baric R. S., Kellam P., Carter L., Pepper M., Sheahan T. P., Veesler D., King N. P., Elicitation of potent neutralizing antibody responses by designed protein nanoparticle vaccines for SARS-CoV-2. Cell 183, 1367–1382.e17 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arunachalam P. S., Feng Y., Ashraf U., Hu M., Walls A. C., Edara V. V., Zarnitsyna V. I., Aye P. P., Golden N., Miranda M. C., Green K. W. M., Threeton B. M., Maness N. J., Beddingfield B. J., Bohm R. P., Scheuermann S. E., Goff K., Dufour J., Russell-Lodrigue K., Kepl E., Fiala B., Wrenn S., Ravichandran R., Ellis D., Carter L., Rogers K., Shirreff L. M., Ferrell D. E., Deb Adhikary N. R., Fontenot J., Hammond H. L., Frieman M., Grifoni A., Sette A., O’Hagan D. T., Van Der Most R., Rappuoli R., Villinger F., Kleanthous H., Rappaport J., Suthar M. S., Veesler D., Wang T. T., King N. P., Pulendran B., Durable protection against the SARS-CoV-2 Omicron variant is induced by an adjuvanted subunit vaccine. Sci. Transl. Med. 14, eabq4130 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arunachalam P. S., Walls A. C., Golden N., Atyeo C., Fischinger S., Li C., Aye P., Navarro M. J., Lai L., Edara V. V., Röltgen K., Rogers K., Shirreff L., Ferrell D. E., Wrenn S., Pettie D., Kraft J. C., Miranda M. C., Kepl E., Sydeman C., Brunette N., Murphy M., Fiala B., Carter L., White A. G., Trisal M., Hsieh C.-L., Russell-Lodrigue K., Monjure C., Dufour J., Spencer S., Doyle-Meyers L., Bohm R. P., Maness N. J., Roy C., Plante J. A., Plante K. S., Gorman M. J., Shin S., Shen X., Fontenot J., Gupta S., O’Hagan D. T., Van Der Most R., Rappuoli R., Coffman R. L., Novack D., Mclellan J. S., Subramaniam S., Montefiori D., Boyd S. D., Flynn J. L., Alter G., Villinger F., Kleanthous H., Rappaport J., Suthar M. S., King N. P., Veesler D., Pulendran B., Adjuvanting a subunit COVID-19 vaccine to induce protective immunity. Nature 594, 253–258 (2021). [DOI] [PubMed] [Google Scholar]
- 30.Grigoryan L., Lee A., Walls A. C., Lai L., Franco B., Arunachalam P. S., Feng Y., Luo W., Vanderheiden A., Floyd K., Wrenn S., Pettie D., Miranda M. C., Kepl E., Ravichandran R., Sydeman C., Brunette N., Murphy M., Fiala B., Carter L., Coffman R. L., Novack D., Kleanthous H., O’Hagan D. T., Van Der Most R., Mclellan J. S., Suthar M., Veesler D., King N. P., Pulendran B., Adjuvanting a subunit SARS-CoV-2 vaccine with clinically relevant adjuvants induces durable protection in mice. NPJ Vaccines 7, 55 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ou B. S., Saouaf O. M., Yan J., Bruun T. U. J., Baillet J., Zhou X., King N. P., Appel E. A., Broad and durable humoral responses following single hydrogel immunization of SARS-CoV-2 subunit vaccine. Adv. Healthc. Mater. 12, e2301495 (2023). [DOI] [PubMed] [Google Scholar]
- 32.“SK bioscience’s COVID-19 vaccine approved for use by Republic of Korea,” International Vaccine Institute, 2022.
- 33.Charan J., Kantharia N. D., How to calculate sample size in animal studies? J. Pharmacol. Pharmacother. 4, 303–306 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arifin W. N., Zahiruddin W. M., Sample Size calculation in animal studies using resource equation approach. Malays. J. Med. Sci. 24, 101–105 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.D. Hinton, “Convalescent plasma EUA letter of authorization” (US Food and Drug Administration, 2021). [Google Scholar]
- 36.Fornefett J., Krause J., Klose K., Fingas F., Hassert R., Benga L., Grunwald T., Muller U., Schrodl W., Baums C. G., Comparative analysis of humoral immune responses and pathologies of BALB/c and C57BL/6 wildtype mice experimentally infected with a highly virulent Rodentibacter pneumotropicus (Pasteurella pneumotropica) strain. BMC Microbiol. 18, 45 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Coffman R. L., Sher A., Seder R. A., Vaccine adjuvants: Putting innate immunity to work. Immunity 33, 492–503 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gil-Etayo F. J., Garcinuño S., Utrero-Rico A., Cabrera-Marante O., Arroyo-Sanchez D., Mancebo E., Pleguezuelo D. E., Rodriguez-Frias E., Allende L. M., Morales- Pérez P., Castro-Panete M. J., Lalueza A., Lumbreras C., Paz-Artal E., Serrano A., An early Th1 response is a key factor for a favorable COVID-19 evolution. Biomedicines 10, 296 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gil-Etayo F. J., Suàrez-Fernández P., Cabrera-Marante O., Arroyo D., Garcinuño S., Naranjo L., Pleguezuelo D. E., Allende L. M., Mancebo E., Lalueza A., Díaz-Simón R., Paz-Artal E., Serrano A., T-helper cell subset response is a determining factor in COVID-19 progression. Front. Cell. Infect. Microbiol. 11, 624483 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Howard F. H. N., Kwan A., Winder N., Mughal A., Collado-Rojas C., Muthana M., Understanding immune responses to viruses—Do underlying Th1/Th2 cell biases predict outcome? Viruses 14, 1493 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Infante-Duarte C., Kamradt T., Th1/Th2 balance in infection. Springer Semin. Immunopathol. 21, 317–338 (1999). [DOI] [PubMed] [Google Scholar]
- 42.Rosenberg-Hasson Y., Hansmann L., Liedtke M., Herschmann I., Maecker H. T., Effects of serum and plasma matrices on multiplex immunoassays. Immunol. Res. 58, 224–233 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Breen E. J., Tan W., Khan A., The statistical value of raw fluorescence signal in luminex xMAP based multiplex immunoassays. Sci. Rep. 6, 26996 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Phillips D. J., League S. C., Weinstein P., Hooper W. C., Interference in microsphere flow cytometric multiplexed immunoassays for human cytokine estimation. Cytokine 36, 180–188 (2006). [DOI] [PubMed] [Google Scholar]
- 45.Maecker H. T., Rosenberg-Hasson Y., Kolstad K. D., Steen V. D., Chung L. S., A novel utility to correct for plate/batch/lot and nonspecific binding artifacts in luminex data. J. Immunol. 204, 3425–3433 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mosmann T. R., Sad S., The expanding universe of T-cell subsets: Th1, Th2 and more. Immunol. Today 17, 138–146 (1996). [DOI] [PubMed] [Google Scholar]
- 47.Phung I., Rodrigues K. A., Marina-Zárate E., Maiorino L., Pahar B., Lee W.-H., Melo M., Kaur A., Allers C., Fahlberg M., Grasperge B. F., Dufour J. P., Schiro F., Aye P. P., Lopez P. G., Torres J. L., Ozorowski G., Eskandarzadeh S., Kubitz M., Georgeson E., Groschel B., Nedellec R., Bick M., Kaczmarek Michaels K., Gao H., Shen X., Carnathan D. G., Silvestri G., Montefiori D. C., Ward A. B., Hangartner L., Veazey R. S., Burton D. R., Schief W. R., Irvine D. J., Crotty S., A combined adjuvant approach primes robust germinal center responses and humoral immunity in non-human primates. Nat. Commun. 14, 7107 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Havenar-Daughton C., Carnathan D. G., Torrents De La Peña A., Pauthner M., Briney B., Reiss S. M., Wood J. S., Kaushik K., Van Gils M. J., Rosales S. L., Van Der Woude P., Locci M., Le K. M., De Taeye S. W., Sok D., Mohammed A. U. R., Huang J., Gumber S., Garcia A., Kasturi S. P., Pulendran B., Moore J. P., Ahmed R., Seumois G., Burton D. R., Sanders R. W., Silvestri G., Crotty S., Direct probing of germinal center responses reveals immunological features and bottlenecks for neutralizing antibody responses to HIV Env trimer. Cell Rep. 17, 2195–2209 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Verstegen N. J. M., Ubels V., Westerhoff H. V., van Ham S. M., Barberis M., System-level scenarios for the elucidation of T cell-mediated germinal center B cell differentiation. Front. Immunol. 12, 734282 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kasturi S. P., Skountzou I., Albrecht R. A., Koutsonanos D., Hua T., Nakaya H. I., Ravindran R., Stewart S., Alam M., Kwissa M., Villinger F., Murthy N., Steel J., Jacob J., Hogan R. J., Garcia-Sastre A., Compans R., Pulendran B., Programming the magnitude and persistence of antibody responses with innate immunity. Nature 470, 543–547 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Napolitani G., Rinaldi A., Bertoni F., Sallusto F., Lanzavecchia A., Selected Toll-like receptor agonist combinations synergistically trigger a T helper type 1-polarizing program in dendritic cells. Nat. Immunol. 6, 769–776 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu H., Moynihan K. D., Zheng Y., Szeto G. L., Li A. V., Huang B., Van Egeren D. S., Park C., Irvine D. J., Structure-based programming of lymph-node targeting in molecular vaccines. Nature 507, 519–522 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Smirnov D., Schmidt J. J., Capecchi J. T., Wightman P. D., Vaccine adjuvant activity of 3M-052: An imidazoquinoline designed for local activity without systemic cytokine induction. Vaccine 29, 5434–5442 (2011). [DOI] [PubMed] [Google Scholar]
- 54.Wu T. Y., Singh M., Miller A. T., De Gregorio E., Doro F., D'Oro U., Skibinski D. A., Mbow M. L., Bufali S., Herman A. E., Cortez A., Li Y., Nayak B. P., Tritto E., Filippi C. M., Otten G. R., Brito L. A., Monaci E., Li C., Aprea S., Valentini S., Calabrό S., Laera D., Brunelli B., Caproni E., Malyala P., Panchal R. G., Warren T. K., Bavari S., O'Hagan D. T., Cooke M. P., Valiante N. M., Rational design of small molecules as vaccine adjuvants. Sci. Transl. Med. 6, 263ra160 (2014). [DOI] [PubMed] [Google Scholar]
- 55.Gale E. C., Powell A. E., Roth G. A., Meany E. L., Yan J., Ou B. S., Grosskopf A. K., Adamska J., Picece V. C. T. M., D'Aquino A. I., Pulendran B., Kim P. S., Appel E. A., hydrogel-based slow release of a receptor-binding domain subunit vaccine elicits neutralizing antibody responses against SARS-CoV-2. Adv. Mater. 33, e2104362 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lynn G. M., Laga R., Darrah P. A., Ishizuka A. S., Balaci A. J., Dulcey A. E., Pechar M., Pola R., Gerner M. Y., Yamamoto A., Buechler C. R., Quinn K. M., Smelkinson M. G., Vanek O., Cawood R., Hills T., Vasalatiy O., Kastenmuller K., Francica J. R., Stutts L., Tom J. K., Ryu K. A., Esser-Kahn A. P., Etrych T., Fisher K. D., Seymour L. W., Seder R. A., In vivo characterization of the physicochemical properties of polymer-linked TLR agonists that enhance vaccine immunogenicity. Nat. Biotechnol. 33, 1201–1210 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Oh S. J., O S. W., Choi Y. J., Kim J. M., Kim D., Kim I. H., Park A. K., Kim H. M., Rhee J. E., Jang Y. R., Yoo C. K., Kim J. Y., Kim E. J., Neutralizing antibody responses in vaccinated and unvaccinated individuals infected with Omicron BA.1 variant. J. Clin. Virol. 155, 105253 (2022). [DOI] [PubMed] [Google Scholar]
- 58.Bates T. A., McBride S. K., Leier H. C., Guzman G., Lyski Z. L., Schoen D., Winders B., Lee J.-Y., Lee D. X., Messer W. B., Curlin M. E., Tafesse F. G., Vaccination before or after SARS-CoV-2 infection leads to robust humoral response and antibodies that effectively neutralize variants. Sci. Immunol. 7, eabn8014 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yu Y., Esposito D., Kang Z., Lu J., Remaley A. T., De Giorgi V., Chen L. N., West K., Cao L., mRNA vaccine-induced antibodies more effective than natural immunity in neutralizing SARS-CoV-2 and its high affinity variants. Sci. Rep. 12, 2628 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jeong H. W., Kim S.-M., Jung M. K., Noh J. Y., Yoo J.-S., Kim E.-H., Kim Y.-I., Yu K., Jang S.-G., Gil J., Casel M. A., Rare R., Choi J. H., Kim H.-S., Kim J. H., Um J., Kim C., Kim Y., Chin B. S., Jung S., Choi J. Y., Song K.-H., Kim Y.-D., Park J.-S., Song J. Y., Shin E.-C., Choi Y. K., Enhanced antibody responses in fully vaccinated individuals against pan-SARS-CoV-2 variants following Omicron breakthrough infection. Cell Rep. Med. 3, 100764 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lechmere T., Snell L. B., Graham C., Seow J., Shalim Z. A., Charalampous T., Alcolea-Medina A., Batra R., Nebbia G., Edgeworth J. D., Malim M. H., Doores K. J., Broad neutralization of SARS-CoV-2 variants, including omicron, following breakthrough infection with delta in COVID-19-vaccinated individuals. mBio 13, e0379821 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kwissa M., Nakaya H. I., Oluoch H., Pulendran B., Distinct TLR adjuvants differentially stimulate systemic and local innate immune responses in nonhuman primates. Blood 119, 2044–2055 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Moore S. M., Wilkerson M. J., Davis R. D., Wyatt C. R., Briggs D. J., Detection of cellular immunity to rabies antigens in human vaccinees. J. Clin. Immunol. 26, 533–545 (2006). [DOI] [PubMed] [Google Scholar]
- 64.Garenne M., Lafon M., Sexist diseases. Perspect. Biol. Med. 41, 176–190 (1998). [DOI] [PubMed] [Google Scholar]
- 65.Plotkin S. A., Correlates of protection induced by vaccination. Clin. Vaccine Immunol. 17, 1055–1065 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mastronarde D. N., Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51 (2005). [DOI] [PubMed] [Google Scholar]
- 67.Kremer J. R., Mastronarde D. N., McIntosh J. R., Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 116, 71–76 (1996). [DOI] [PubMed] [Google Scholar]
- 68.Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E., UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 69.Grosskopf A. K., Labanieh L., Klysz D. D., Roth G. A., Xu P., Adebowale O., Gale E. C., Jons C. K., Klich J. H., Yan J., Maikawa C. L., Correa S., Ou B. S., d’Aquino A. I., Cochran J. R., Chaudhuri O., Mackall C. L., Appel E. A., Delivery of CAR-T cells in a transient injectable stimulatory hydrogel niche improves treatment of solid tumors. Sci. Adv. 8, eabn8264 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Saouaf O. M., Roth G. A., Ou B. S., Smith A. A. A., Yu A. C., Gale E. C., Grosskopf A. K., Picece V. C. T. M., Appel E. A., Modulation of injectable hydrogel properties for slow co-delivery of influenza subunit vaccine components enhance the potency of humoral immunity. J. Biomed. Mater. Res. A 109, 2173–2186 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jons C. K., Grosskopf A. K., Baillet J., Yan J., Klich J. H., Saouaf O. M., Appel E. A., Yield-stress and creep control depot formation and persistence of injectable hydrogels following subcutaneous administration. Adv. Funct. Mater. 32, 2203402, (2022). [Google Scholar]
- 72.Roth G. A., Gale E. C., Alcántara-Hernández M., Luo W., Axpe E., Verma R., Yin Q., Yu A. C., Lopez Hernandez H., Maikawa C. L., Smith A. A. A., Davis M. M., Pulendran B., Idoyaga J., Appel E. A., Injectable hydrogels for sustained codelivery of subunit vaccines enhance humoral immunity. ACS Cent. Sci. 6, 1800–1812 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Powell A. E., Zhang K., Sanyal M., Tang S., Weidenbacher P. A., Li S., Pham T. D., Pak J. E., Chiu W., Kim P. S., A single immunization with spike-functionalized ferritin vaccines elicits neutralizing antibody responses against SARS-CoV-2 in mice. ACS Cent. Sci. 7, 183–199 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Crawford K. H. D., Eguia R., Dingens A. S., Loes A. N., Malone K. D., Wolf C. R., Chu H. Y., Tortorici M. A., Veesler D., Murphy M., Pettie D., King N. P., Balazs A. B., Bloom J. D., Protocol and reagents for pseudotyping lentiviral particles with SARS-CoV-2 spike protein for neutralization assays. Viruses 12, 513 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rogers T. F., Zhao F., Huang D., Beutler N., Burns A., He W. T., Limbo O., Smith C., Song G., Woehl J., Yang L., Abbott R. K., Callaghan S., Garcia E., Hurtado J., Parren M., Peng L., Ramirez S., Ricketts J., Ricciardi M. J., Rawlings S. A., Wu N. C., Yuan M., Smith D. M., Nemazee D., Teijaro J. R., Voss J. E., Wilson I. A., Andrabi R., Briney B., Landais E., Sok D., Jardine J. G., Burton D. R., Isolation of potent SARS-CoV-2 neutralizing antibodies and protection from disease in a small animal model. Science 369, 956–963 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Correa S., Meany E. L., Gale E. C., Klich J. H., Saouaf O. M., Mayer A. T., Xiao Z., Liong C. S., Brown R. A., Maikawa C. L., Grosskopf A. K., Mann J. L., Idoyaga J., Appel E. A., Injectable nanoparticle-based hydrogels enable the safe and effective deployment of immunostimulatory CD40 agonist antibodies. Adv. Sci. 9, e2103677 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Blais J. A. R., Least squares for practitioners. Math. Probl. Eng. 2010, 1–19 (2010). [Google Scholar]
- 78.Holmes T. H., Subrahmanyam P. B., Wang W., Maecker H. T., Penalized supervised star plots: Example application in influenza-specific CD4+ T cells. Viral Immunol. 32, 102–109 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.R. Bivand, N. Lewin-Koh, maptools: Tools for handling spatial objects. R package version 1.1-4 (The Comprehensive R Archive Network, 2022); https://CRAN.R-project.org/package=maptools.
- 80.F. Novomestky, matrixcalc: Collection of functions for matrix calculations. R package version 1.0-5 (The Comprehensive R Archive Network, 2021); https://CRAN.R-project.org/package=matrixcalc.
- 81.Lemon J., Plotrix: A package in the red light district of R. R-News 6, 8–12 (2006). [Google Scholar]
- 82.A. Maurya, JPEN: Covariance and inverse covariance matrix estimation using joint penalty. R package version 1.0 (The Comprehensive R Archive Network, 2015); https://CRAN.R-project.org/package=JPEN.
- 83.A. Schuetzenmeister, F. Dufey, VCA: Variance component analysis. R package version 1.4.4 (The Comprehensive R Archive Network, 2022); https://CRAN.R-project.org/package=VCA.
- 84.A. Golan, G. Judge, D. J. Miller, Maximum Entropy Econometrics: Robust Estimation with Limited Data (Wiley, 1996). [Google Scholar]
- 85.Benjamini Y., Krieger A. M., Yekutieli D., Adaptive linear step-up procedures that control the false discovery rate. Biometrika 93, 491–507 (2006). [Google Scholar]
- 86.Kim K. I., Van De Wiel M. A., Effects of dependence in high-dimensional multiple testing problems. BMC Bioinformatics 9, 114 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Methods
Figs. S1 to S13
Tables S1 to S30
Legend for data file S1
Data file S1