Summary
Tumors growing in metabolically challenged environments, such as glioblastoma in the brain, are particularly reliant on crosstalk with their tumor microenvironment (TME) to satisfy their high energetic needs. To study the intricacies of this metabolic interplay, we interrogated the heterogeneity of the glioblastoma TME using single-cell and multi-omics analyses and identified metabolically rewired tumor-associated macrophage (TAM) subpopulations with pro-tumorigenic properties. These TAM subsets, termed lipid-laden macrophages (LLMs) to reflect their cholesterol accumulation, are epigenetically rewired, display immunosuppressive features, and are enriched in the aggressive mesenchymal glioblastoma subtype. Engulfment of cholesterol-rich myelin debris endows subsets of TAMs to acquire an LLM phenotype. Subsequently, LLMs directly transfer myelin-derived lipids to cancer cells in an LXR/Abca1-dependent manner, thereby fueling the heightened metabolic demands of mesenchymal glioblastoma. Our work provides an in-depth understanding of the immune-metabolic interplay during glioblastoma progression, thereby laying a framework to unveil targetable metabolic vulnerabilities in glioblastoma.
Keywords: glioblastoma, macrophages, cholesterol, myelin recycling, lipid metabolism, cancer immunity, tumor microenvironment
Graphical abstract

Highlights
-
•
TAMs recycle cholesterol-rich myelin, acquiring lipid-laden features in glioblastoma
-
•
Epigenetic rewiring induced by myelin debris scavenging underlies LLM immunosuppressive features
-
•
LLMs transfer myelin-derived lipids to glioblastoma cells to fuel disease progression
-
•
LLMs correlate with aggressive mesenchymal subtype and poor prognosis in patients
Recycling of cholesterol-rich myelin debris by macrophages leads to the emergence of lipid-laden macrophages (LLMs) in glioblastoma. LLMs support glioblastoma malignancy through transfer of myelin-derived lipids to tumor cells, fueling cancer cell proliferation and recurrence.
Introduction
Tumors arising in the brain benefit from a unique microenvironment that is dynamically harnessed as malignant features expand.1 Glioblastoma, the most aggressive primary brain tumor, is an incurable disease with a dismal prognosis and displays high heterogeneity with regards to tumor subtype, microenvironmental niches, and immune landscape.2,3,4 Besides recapitulating a neurodevelopmental hierarchy,5 glioblastoma subtypes possess distinct metabolic features affecting macrophage polarisation.6,7 As macrophages significantly contribute to gliomagenesis and treatment response, these cells have been explored as potent therapeutic targets.8,9,10,11 However, the high intra-tumoral and inter-patient heterogeneity displayed in glioblastoma challenges a “one-size-fits-all” therapeutic approach targeting pan-macrophages in brain cancer,12 calling for an in-depth appreciation of their immense plasticity.12
Indeed, macrophages are the most abundant immune cells in the glioblastoma tumor microenvironment (TME) and are ontogenically diverse, including brain-resident microglia (MG) and infiltrating monocyte-derived macrophages (MDMs). While recent single-cell transcriptomic analyses reported tumor-associated macrophages (TAMs) subpopulations could predict survival13,14 or displayed altered metabolic features,15,16 the molecular bases underpinning glioblastoma TAM diversity and functions remain poorly understood.
In this study, we employ a multi-omics approach to resolve the heterogeneity and spatial diversity of TAM subsets, revealing their niche-specific interactions with distinct glioblastoma subtypes and impact on disease recurrence post-radiotherapy (RT). Utilizing complementary model systems and functional assays, we uncover TAM-mediated mechanisms sanctioning glioblastoma cells to overcome challenges peculiar to the metabolically autonomous brain environment. We show that lipids play a central role in a metabolic interplay established between mesenchymal-like (MES-like) glioblastoma cancer cells and TAM subsets presenting a lipid-laden phenotype. As TAMs evolve into acquiring this rewired metabolic education, they gain enhanced pro-tumorigenic and immunosuppressive features fueling glioblastoma malignancy. Our study reveals in-depth mechanisms underlying TAM subset specification in the TME and identifies TAM metabolic rewiring as an exploitable therapeutic vulnerability in glioblastoma.
Results
Murine glioblastoma displays dynamic cellular subtype heterogeneity in response to RT
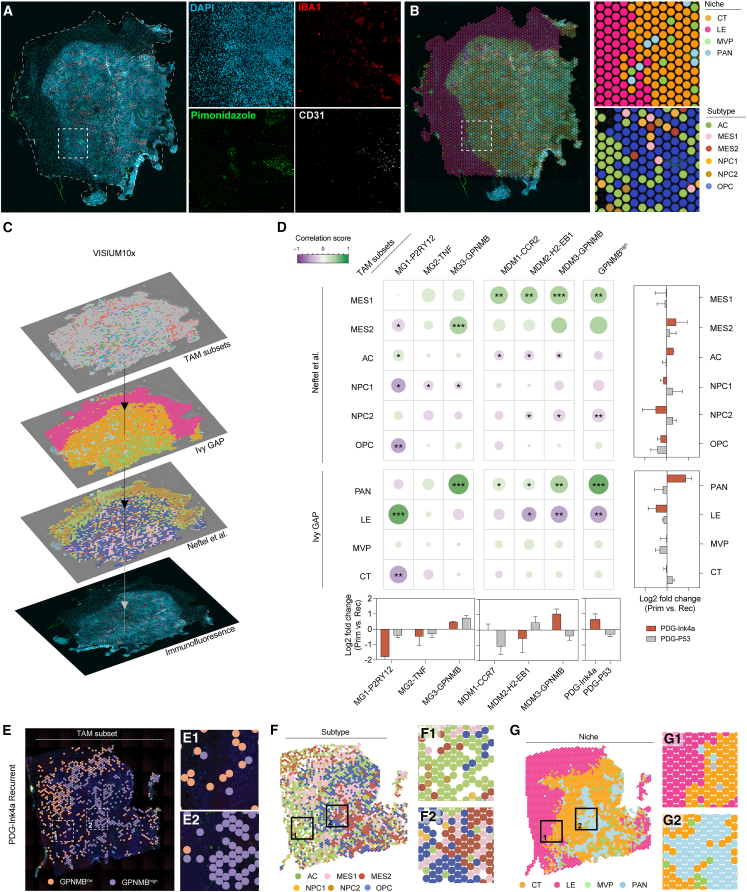
To reveal the reciprocal interactions between macrophages and tumor cells that fuel glioblastoma malignancy, we investigated the dynamic contexture of the glioblastoma TME at the single-cell and spatial transcriptomic levels, in two genetically engineered mouse models (GEMMs) that recapitulate key characteristics of human gliomagenesis and therapeutic response.8,9,10,17 Both GEMMs involve enforced expression of platelet-derived growth factor-β in Nestin-positive progenitor cells, either in an Ink4a/ArfKO background (PDG-Ink4a) or in combination with a short-hairpin-mediated knockdown of p53 in tumor cells (PDG-p53). Orthogonally, publicly available datasets of glioblastoma patients enabled the validation of our murine models’ clinical relevance. We explored the heterogeneity of macrophage subsets in relation to their local microenvironment, employing multi-omics approaches and a wide range of functional ex vivo and in vitro assays to unveil pro-tumorigenic interactions underlying macrophage and tumor cell co-evolution in the glioblastoma TME (Figure 1A).
Figure 1.
Single-cell analyses reveal co-occurrence of glioblastoma mesenchymal subtype and GPNMBhigh TAMs
(A) Schematic diagram of experimental design assessing macrophage subsets and glioblastoma subtype/niche heterogeneity and interactions using glioblastoma GEMM and public datasets (see STAR Methods).
(B) Uniform manifold approximation and projection (UMAP) representation of major cell populations from scRNA-seq glioblastoma mouse dataset (DCs, dendritic cells; ECs, endothelial cells).
(C) Visual representation of glioblastoma subtype heterogeneity subjected to scRNA-seq.18 The module scores of each individual cell are depicted on the axes.
(D) Average fraction of the glioblastoma cell subtypes (outer circle) per model. Pseudo-location2 assignment of glioblastoma cells (inner circle) displayed as a percentage of cellular subtype.
(E) UMAP representation of MG and MDM sub-clusters.
(F) Gene set enrichment analysis (GSEA) scores of depicted pathways in MG and MDM clusters identified in (E).
(G) Pseudo-location niche assignment of TAM subsets from clusters identified in (E); represented as mean ± SEM.
(H) Left and middle: differential abundance of TAM subsets upon recurrence in glioblastoma models. Right: correlation matrix between TAM subset and glioblastoma cellular subtype abundance.
(I) Top 25 differentially expressed genes between GPNMBhigh and GPNMBlow TAM clusters (Table S1C).
(J) Correlation between GPNMBhigh TAMs and MES glioblastoma cells in murine and human glioblastoma scRNA-seq datasets.5,19
Statistics: Kendall trend test (H) or simple linear regression (J) (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001).
P2RY12, Purinergic Receptor P2Y12; TNF, Tumor Necrosis Factor; GPNMB, Glycoprotein Nmb; CCR2, C-C Chemokine Receptor type 2; H2-EB1, Histocompatibility 2, class II antigen E Beta.
See also Figure S1 and Table S1. Relevant gene signatures used for computational analysis of single-cell RNA sequencing and VISIUM10x dataset, related to Figures 1 and 2, Table S2. Gene set over-representation analyses of macrophage subclusters, related to Figure 1, Table S3. Gene set over-representation analysis of GPNMBlow and GPNMBhigh TAMs, related to Figure 1.
Single-cell RNA sequencing (scRNA-seq) of CD45+ and CD45− fluorescence-activated cell sorting (FACS)-purified cells from primary (treatment-naive) and recurrent (relapsed after 5 × 2 Gy fractionated RT) tumors from both glioblastoma GEMMs highlighted the cellular heterogeneity of the glioblastoma TME (Figure 1B). To appreciate the impact of glioblastoma tumor cell heterogeneity on macrophage education, we first classified tumor cells into molecular subtypes associated with distinct transcriptional signatures previously reported at the single-cell level.18 In patients, each of these cellular subtypes—neural progenitor-like (NPC-like), oligodendrocyte-progenitor-like (OPC-like), astrocyte-like (AC-like), and MES-like—is represented within individual tumors20,21 and this heterogeneity was recapitulated in murine glioblastoma models (Figure 1C). We utilized the Ivy Glioblastoma Atlas Project (GAP) dataset2 defining niche-specific transcriptional signatures of major anatomic features of glioblastoma (LE, leading edge; CT, cellular tumor; MVP, micro-vascular proliferation; PAN, pseudo-palisading cells around necrosis), to assign individual tumor cells to a pseudo-spatial location, and found that glioblastoma cellular subtypes were heterogeneously distributed across these niches (Figure 1D).
We next analyzed the intra-tumoral heterogeneity of cancer cells in primary and recurrent murine glioblastoma and addressed whether, as seen in human tumors,20 subtype shifts occurred in relapsed disease post-RT. The PDG-p53 model displayed a dominant AC-like primary tumor, which shifted toward an OPC-like dominant composition upon recurrence, despite possessing high inter-tumor variation (Figure 1D). Distinctively, PDG-Ink4a glioblastoma, characterized by a dominant population of OPC-like tumor cells, underwent the predicted shift toward a more MES-enriched phenotype at recurrence (Figure 1D). Altogether, these results show that heterogeneous and dynamic cancer cell subtype changes occur in murine PDG-driven tumors in a stage and niche-dependent manner, mirroring results reported in glioblastoma patients.18,20
Macrophage subset dynamics correlate with glioblastoma stage, cellular subtype composition, and local niches
We next explored the heterotypic interactions between cancer cell subtype dynamics and macrophage heterogeneity.18,22 Unsupervised clustering of MDMs and MG segregated these cells into 8 states, consisting of 4 MG and 4 MDM subsets (Figures 1E and S1A). We examined the transcriptional phenotypes of these different TAM identities in relation to glioblastoma microanatomical niches, disease stage, and molecular subtype. In both TAM subpopulations, we observed a spectrum of phenotypes ranging from pre-activated (MG1-P2RY12 and MDM1-CCR2) to inflammatory (MG2-TNF and MDM2-H2-EB1), metabolically active (MG3-GPNMB and MDM3-GPNMB), and proliferating states (MG4-MKI67 and MDM4-MKI67) (Figures 1F and S1B–S1D; Tables S1A, S1B, and S2A–S2H). In line with previous reports of increased MDM content in the recurrent glioblastoma TME,8 there was a notable increase of MDM4-MKI67 abundance in recurrent tumors, and MG4-MKI67 and MDM4-MKI67 clusters both positively correlated with the presence of MES-like glioblastoma cells (Figure 1H). Timestamp transcriptional signatures23 revealed a gradual enrichment of the 48-h timestamp from MDM1 to MDM3, indicative of a differentiation trajectory taking place in MDMs rather than expansion of subsets in the TME (Figure S1E).
Figure S1.
In-depth transcriptional analysis of macrophage landscape in glioblastoma, related to Figure 1
(A) Stacked violin plots depicting the expression of representative genes defining MG1–4 and MDM1–4 clusters identified in the MG and MDM population (52,688 cells) represented in Figures 1B and 1E; Table S1A. MG and MDM from all PDG-Ink4a primary (n = 3) and recurrent (n = 3) and PDG-p53 primary (n = 4) and recurrent (n = 3) tumor samples are represented. Colors correspond to cell clusters presented in Figure 1E.
(B) Heatmap of the averaged enrichment scores derived from GSEA for the depicted relevant published signatures per macrophage cluster, data are scaled between 1 and 0 per column. The displayed signatures were derived from published studies referenced in Table S1B.
(C and D) Bar plots depicting the adjusted p values of relevant pathways specific to each MG (C) and MDM (D) clusters. Results were generated by gene set enrichment analyses of the top 30 significantly DE genes for each cluster (padjusted < 0.05) (see Table S2).
(E) Line plot depicting the averaged enrichment scores of the 12- and 48-h glioblastoma MDM timestamp modules23 in each MDM cluster.
(F) Pie chart representing each MG subset present as a percentage of total macrophages of a healthy brain captured by scRNA-seq.
(G) Bar plot showing the adjusted p value of relevant significantly enriched pathways related to the top differentially expressed 30 genes increased in GPNMBlow or GPNMBhigh clusters (Table S3). Statistics: Fisher’s exact test in combination with the Benjamini-Hochberg method for correction of multiple hypotheses testing.
(H) Violin plot of the GPNMBhigh signature enrichment score per MG and MDM cluster, showing percentages of macrophages assigned as GPNMBhigh per cluster.
(I) Pie chart representing the macrophage subset composition assigned as GPNMBhigh TAMs captured by scRNA-seq of all tumors sequenced.
(J) Scatter plot depicting the correlation between T cell content (as percentage of CD45+ cells) and the abundance of GPNMBhigh TAMs (as percentage of total TAMs) in glioblastoma patient sample scRNA-seq dataset.5,19 Each data point represents a single glioblastoma tumor (n = 22). Red line represents simple linear regression. The Pearson’s correlation coefficient and p value are depicted at the top right corner.
As expected, the pre-activated MG1-P2RY12 cluster, closely representing MG FACS-purified from healthy brains, was predicted to be over-represented at the tumor LE compared with other MG subsets, indicative of tumor invasion into the brain parenchyma from which MG originate (Figures 1G and S1F). Similarly, the MDM1-CCR2 cluster resembling pre-activated monocytes infiltrating the tumor site (Figures S1A and S1E) was predicted to be enriched in the MVP area, as expected from peripheral recruitment (Figure 1G).
Budding from these ontogeny-specific clusters, we identified macrophages with inflammatory features. In the MG compartment, cells comprising the MG2-TNF cluster similarly shared transcriptional education reported in neuroinflammation,24 therefore displaying high expression of pro-inflammatory genes (Tnf, Ccl4, and Ccl3) and neuro-inflammatory response pathways (Figures 1F, S1B, and S1C; Table S1B). The counterpart cluster MDM2-H2-EB1 similarly presented increased inflammatory response gene expression, albeit of different nature and distinguished by increased antigen processing signatures (H2-Eb1, H2-Aa, and Cd74) (Figures S1B and S1D; Table S1A). Interestingly, the MG2-TNF and MDM2-H2-EB1 macrophage clusters negatively correlated with the MES2 cellular subtype (Figure 1H), showing that MES-enriched glioblastoma present overall lower inflammatory TAMs, consistent with their aggressive phenotype in patients.25
The MDM3-GPNMB cluster, located at the apex of MG and MDM convergence and MDM differentiation trajectory (Figures 1E and S1E), was specifically increased in MES-enriched PDG-Ink4a recurrent tumors (Figure 1H). Together with MG3-GPNMB cells, both clusters were predicted to be largely located within the PAN (hypoxic) glioblastoma niche, where MES2 tumor cells preferentially reside (Figures 1D and 1G). Functionally, macrophages from the MDM3-GPNMB and MG3-GPNMB clusters shared a transcriptional education reminiscent of “lipid-associated” macrophages (Figure S1B; Tables S2C and S2G).24 Interestingly, these macrophages were transcriptionally akin to TAMs previously ascribed a lipid/phagocytic education or possessing pro-tumorigenic immunosuppressive functions in glioblastoma murine models and patient samples.15,26 Comparably, these clusters displayed overall the least inflammatory phenotype of all TAM subsets while presenting increased activation of cellular pathways involved in foam cell differentiation (Figure 1F).27 Hence, these unbiased analyses highlight the putative lipid-associated or lipid-laden phenotype of MDM3-GPNMB and MG3-GPNMB TAM subsets. While lipid-laden macrophages (LLMs) were previously observed in the context of obesity28 or atherosclerosis,29 and recently in a handful of cancers as a pro-tumorigenic TAM subset,16,30,31 their presence in the brain has only been reported in the course of aging and neurodegenerative diseases.32,33 Altogether, our findings indicate that GPNMBhigh TAMs transcriptionally resembling LLMs are increased in MES-enriched glioblastoma as PDG-Ink4a glioblastoma shifts toward this subtype in recurrent disease and are associated with MES-like cancer cells in hypoxic microanatomical niches. Hypothesizing that this heterotypic crosstalk may fuel glioblastoma progression and the OPC-to-MES transition, we sought to further validate the presence and significance of LLMs in glioblastoma.
Using the differentially expressed genes characterizing the MG3-GPNMB and MDM3-GPNMB TAM clusters, we generated a GPNMBhigh signature to robustly quantify the percentage of TAMs presenting a transcriptional phenotype reminiscent of LLMs in other diseases (Figures 1I and S1G; Tables S3A and S3B).27 Interestingly, aside from the MG3/MDM3 clusters, TAMs classified as GPNMBhigh also encompassed cells belonging to other TAM subsets (Figures S1H and S1I). In line with previous studies,34 correlation analyses revealed a strong association between MES-like glioblastoma cells and GPNMBhigh macrophages in murine and human glioblastoma, independently of the primary/recurrent disease stage (Figure 1J). As expected from their anti-inflammatory profile (Figure 1F), GPNMBhigh macrophages negatively correlated with lymphoid infiltration in patient samples, supporting the notion that macrophages with a lipid-laden phenotype underly the immunosuppressive nature of MES-enriched glioblastoma (Figure S1J). Altogether, our results allude to a functional significance of GPNMBhigh TAMs through acquisition of a lipid-laden, anti-inflammatory phenotype and association with the aggressive MES-like subtype.
Spatial co-localization of GPNMBhigh macrophages with MES-like cells in hypoxic niches
To deepen our understanding of the spatial interactions between macrophage subsets and glioblastoma cellular subtypes, we performed spatial transcriptomics analyses on murine glioblastoma tissue sections. Immunofluorescent (IF) staining of markers representative of macrophages (IBA1), MVP (CD31), PAN (hypoxia marker pimonidazole), LE (low-density DAPI), and CT (high-density DAPI) were used to visualize TAM presence in the various microanatomical glioblastoma niches (Figure 2A).1 Importantly, these staining overlapped with the classification defining each of the Ivy GAP transcriptional niche signature (Figure 2B). Furthermore, curated gene lists specific to each glioblastoma cellular subtype were used to classify spots according to the subtype most represented within that location (Figure 2B). We used a 3-step classification strategy to allocate macrophage subset distribution in relation with niches and glioblastoma cellular subtype (Figure S2A). This approach revealed the abundance and spatial co-localization of TAM subsets and glioblastoma cell subtypes within primary and recurrent tumors from both glioblastoma GEMMs (Figures 2C and 2D).
Figure 2.
MES-like cancer cells and GPNMBhigh TAMs co-localize in hypoxic niches
(A) Representative immunofluorescence staining on fresh-frozen brain tumor sections used in VISIUM 10X spatial transcriptomics.
(B) Visualization of the classification of dominant niche and glioblastoma cellular subtype transcriptional activity per VISIUM 10X sequenced spot (see STAR Methods).
(C) Spatial co-occurrence workflow: transcriptional modules from TAM subsets, glioblastoma cellular subtypes, and niches were assigned to each spot to infer correlation coefficients.
(D) Central: correlation matrix of TAM subset transcriptional modules and either the glioblastoma cellular subtype (top) or microanatomical niche transcriptional modules (bottom) across all VISIUM 10X samples; dot color and size correspond to the correlation coefficient. LogFC of TAM subset, tumor niche, or glioblastoma cell subtype occurrence in recurrent compared with primary glioblastoma are shown in the bottom and right side of the matrix, respectively; data represented as mean + S.D.
(E–G) Representative visualization of spatial transcriptomic analyses, highlighting GPNMBhigh deserted (1) and enriched (2) areas. Spots assigned as GPNMBlow or GPNMBhigh are overlayed onto the corresponding IF image (E) and paralleled to glioblastoma cellular subtypes (F) or microanatomical niches (G) as defined in (B).
Statistics: Kendall trend test (D) (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001).
Figure S2.
Spatial analyses of macrophage subsets and validation that lipid-laden macrophages transcriptionally represent GPNMBhigh TAMs in glioblastoma, related to Figures 2 and 3
(A) Schematic of the 3-step classification strategy to identify macrophage subset spatial distribution on a representative Visium 10X dataset from a primary PDG-Ink4a tumor. After assigning spots positive for a pan-macrophage signature (Aif1, Lgals3, Itga1, Trem2, P2ry12, and Siglech), spots are either classified as MG or MDMs based on microglia-specific gene expression (P2ry12 and Siglech). Curated gene signatures, specific to each MG or MDM clusters (see Table S1H), were used to assign a subset score. The highest subset score classified each respective spot to a specific TAM subset in the TME (right).
(B) Representative IF images of fresh-frozen primary murine glioblastoma tissue sections (PDG-Ink4a). DAPI: nuclear stain (blue); IBA1: pan-macrophage (magenta); PLIN2: lipid droplets (LDs) (white); BODIPY: neutral lipid marker (yellow). Cells positive for IBA1, but negative for PLIN2 and BODIPY were identified as non-LLMs (left magnified). Cells positive for IBA1, PLIN2, and BODIPY were identified as LLMs (right magnified).
(C) IF quantification of LLMs (IBA1+ cells with a surface area of ≥500 pixels [0.325 μm/pixel] and containing ≥5 PLIN2+ spots [defined as LDs]) as a percentage of total TAMs in primary and recurrent tumors isolated from PDG-Ink4a tumor-bearing mice.
(D) Quantification of LLMs in hypoxic and non-hypoxic areas in primary and recurrent PDG-Ink4a glioblastoma. Linked pairs of dots represent the same sample.
(E) Representative flow cytometry plots depicting the LLM FACS-purification gating strategy in a recurrent PDG-Ink4a glioblastoma. Live macrophages (CD45+CD11b+Ly6ClowLy6Glow) of each TAM subpopulations (MG; CD49dlow) and MDMs (CD49dhigh) were sorted as LLMs or non-LLMs (independently of their ontogeny) based on SSC-A (granularity) and BODIPY (neutral lipids) intensity.
(F) Heatmap depicting the logFC of normalized gene expression between LLMs and non-LLMs from the RNA-seq analysis. Genes shown in heatmap are commonly upregulated in lipid-laden MG and MDMs represented in Figure 3B.
Statistics: two-stage step-up unpaired t test (C) or two-tailed paired t test (D). Data are represented as mean + SEM (C).
Spatial transcriptomic analyses confirmed that MG belonging to the pre-activated MG1-P2RY12 cluster were indeed enriched in the tumor LE (Figure 2D). Importantly, while all MDM clusters correlated with the MES1 subtype, GPNMBhigh TAMs were specifically associated with the presence of MES2 glioblastoma cells, particularly in PAN niches where both cell types are enriched and their interaction is heightened (Figures 2D–2G). We identified TAMs expressing the lipid droplet marker perilipin-2 (PLIN2) and neutral lipid stain BODIPY in the TME, confirming the accumulation of lipid droplets within specific TAM subsets (Figure S2B). Consistent with the GPNMBhigh subset enrichment occurring during OPC-to-MES transition post-RT in the PDG-Ink4a model (Figures 1D and 2D), a marked increase in TAMs displaying a lipid-laden phenotype (high levels of PLIN2+ lipid droplets and increased size) was observed upon recurrence (Figure S2C). Additionally, combining PLIN2 staining and pimonidazole confirmed that LLMs accumulate in hypoxic areas, where MES2 glioblastoma cells reside, particularly in the recurrent setting (Figures 1D and S2D). These findings highlight the intricate reciprocal communication between GPNMBhigh TAMs and MES-like tumor cells in hypoxic niches and demonstrate that GPNMBhigh macrophages display a characteristic lipid-laden phenotype.
LLMs display metabolic and immunosuppressive programs associated with altered chromatin landscapes and promote glioblastoma relapse post-RT
To explore the origin of lipid-laden GPNMBhigh TAM formation, we first validated that GPNMBhigh macrophages were indistinguishable from LLMs and performed RNA-seq analysis on FACS-purified lipid-enriched (BODIPYhigh, SSC-Ahigh) MG and MDMs from PDG-Ink4a tumors (Figures 3A and S2E). Comparing differentially expressed genes between LLMs and non-LLMs from MG and MDM subpopulations, we identified gene signatures specific and shared between LLMs and non-LLMs from distinct ontogeny (Figure 3B; Tables S3A and S3B). Genes commonly upregulated in FACS-purified LLMs from tissue-resident or monocytic origin overlapped with several genes from the GPNMBhigh TAM transcriptional signature (Figures 1I and S2F). Indeed, GPNMBhigh macrophages transcriptionally overlap with LLMs, confirming that GPNMBhigh macrophages are indeed lipid-laden in glioblastoma (Figure 3C). These findings prompted us to refer to GPNMBhigh TAMs as LLMs and use the GPNMBhigh signature to transcriptionally identify LLMs.
Figure 3.
Lipid-laden macrophage pro-tumorigenic phenotype correlates with loss of chromatin accessibility
(A) Schematic overview of lipid-laden macrophage (LLM) multi-omics analyses.
(B) Venn diagram of differentially expressed genes identified by RNA-seq in non-LLMs and LLMs from FACS-isolated MG and MDM subpopulations.
(C) Stacked violin plots depicting TAM subset (from Figure 1E) transcriptional enrichment for gene signatures identified in RNA-seq analyses from FACS-purified macrophage subpopulations (Table S1D).
(D) Average peak profiles (top) and heatmaps (bottom) depicting the normalized ATAC-seq (assay for transposase-accessible chromatin with sequencing) signals at differentially accessible chromatin regions in macrophage subpopulations.
(E) LogFC of gene expression associated with higher and lower accessibility in LLM promoter regions, compared with expression of the same genes in non-LLMs from MG and MDM subpopulations.
(F) Log10(FDR, false discovery rate) of significantly enriched gene sets based on gene expression associated with higher or lower promoter accessibility in MDM-LLMs compared with non-LLMs.
(G) Normalized relevant ATAC-seq peak signals from MDM subpopulations.
(H) Flow cytometry MFI of H3K27me3 in macrophage subpopulations.
(I) Log2FC of MFI for the indicated cell surface marker expression in macrophage subpopulations. Statistics shown in Figure S3J.
(J) Flow cytometry quantification of LLMs (BODIPYhigh, SSC-Ahigh).
(K) Kaplan-Meier curve showing animal survival over time in control (DMSO); RT (5x2 Gy) and DMSO; SSO (30 mg/kg daily treatment); or RT + SSO treatment groups.
(L) Flow cytometry quantification of LLMs (BODIPYhigh, SSC-Ahigh) from treatment groups in (K).
Statistics: Wilcoxon signed-rank test (E), two-way ANOVA with Sídák correction for multiple comparisons (H and L), two-stage step-up unpaired t test with Benjamini, Krieger, and Yekutiel correction for multiple comparisons (J), or log-rank test (K). Data are represented as mean ± SEM (H–J and L).
See also Figures S2, S3, and S4 and Tables S3 and S4.
To deepen our understanding of the underlying mechanisms driving LLM formation, we examined the chromatin state and accessibility of FACS-purified lipid- and non-lipid-laden MG and MDMs (Figure 3A). We confirmed that MG and MDMs displayed ontogeny-specific peaks reminiscent of gene expression underlying their embryological origins (Sall135 and Itga4,10 respectively) (Figure S3A). MDMs displayed more accessible regulatory elements compared with MG, supporting the notion that tissue imprinting restricts macrophage plasticity toward functional specification (Figure S3B).36 Peaks enriched in non-lipid-laden MDMs were identified in genes regulating inflammatory responses and cytokine-mediated signaling pathways, while non-lipid-laden MG-specific peaks related to gene expression regulating tissue-resident functions (Figures S3C and S3D; Tables S4A–S4H).
Figure S3.
Extended ontogeny-specific lipid-laden macrophage chromatin landscape analysis, related to Figure 3
(A) Normalized ATAC-seq signal at gene regions related to MG specific (Sall1) or MDM specific (Itga4) accessibility in LLMs and non-LLMs from MG and MDM subpopulations.
(B) Venn diagram depicting the number of consensus peaks from the ATAC-seq in non-lipid-laden MG and MDMs.
(C and D) Histogram showing the −log10(FDR) of significantly enriched gene sets in non-lipid laden MDMs (C) and MG (D) based on differentially expressed genes depicted in (B). Vertical line at −log10(FDR) = 2 represents the threshold for significance. Colors indicate the overlap between the differentially accessible genes and gene sets.
(E) Venn diagram depicting the consensus peaks in LLMs versus non-LLMs from MG (top) and MDMs (bottom) determined from ATAC-seq analyses.
(F) Dot plots depicting the −log10(FDR) of the motif enrichment analyses performed for all differentially accessible peaks in LLMs (left) and non-LLMs (right) from monocytic origin. Colors represent the logFC of the motif enrichment from the differentially accessible peaks compared with all peaks detected (background). Relevant motif clusters are highlighted in blue, and TFs related to these motifs are given in the bottom panels.
(G) Heatmap depicting the logFC (comparison based on variable on x axis) of the gene expression of chromatin regulators determined from bulk RNA-seq analyses (shown in Figure 3B).
(H) Histogram depicting the mean fluorescent intensity (MFI) of H3K27me3 in LLMs and non-LLMs from MG (purple) and MDM (blue) subpopulations from recurrent PDG-Ink4a tumor-bearing mice.
(I) Quantification of mean fluorescent intensity (MFI) of EZH2 in LLMs and non-LLMs from MG and MDM subpopulations from primary (left) and recurrent (right) PDG-Ink4a tumor-bearing mice.
(J) MFI of depicted cell surface markers in lipid-laden MG (left) and MDMs (right) compared with non-lipid-laden MG and MDMs from the same primary PDG-Ink4a glioblastoma samples.
Statistics: two-stage step-up multiple paired t test with Benjamini, Krieger, and Yekutieli correction for multiple testing (H–J). Data shown as mean + SEM (H–J).
We next assessed the chromatin landscape changes associated with the lipid-laden phenotype. Independent of their ontogeny, LLMs lost access to numerous chromatin regions compared with their non-LLM counterparts, while substantially less peaks, displaying more subtle changes, were gained (Figures 3D and S3E). Interestingly, chromatin accessibility to promotor regions correlated with gene expression only in lipid-laden MDMs but not in lipid-laden MG (Figure 3E). Low chromatin accessibility in lipid-laden MDMs was associated with downregulated expression of genes involved in inflammatory pathways, such as H2-Eb1 and Cd74 (Figures 3F and 3G). Importantly, lipid-laden MDMs gained specific peaks in genes related to lipid transport (Abca1), phagocytosis, and synapse pruning while losing accessibility to regions in genes associated with cholesterol biosynthesis (Dhcr24), compared with non-lipid laden MDMs (Figures 3F and 3G). We next performed motif analysis of these peaks, which revealed an enrichment of NF-κB (Nuclear factor kappa-light-chain-enhancer of activated B cells) and RUNX (Runt-Related Transcription Factor) in non-lipid laden MDMs, two transcription factors (TFs) known to mediate inflammatory cascades (Figure S3F).37,38 By contrast, lipid-laden MDMs displayed enrichment of TFs known to be involved in macrophage immunosuppressive phenotype, such as CCAAT/enhancer-binding protein beta (C/EBPβ) and Activating transcription factor 3 (ATF3) (Figure S3F).39,40
Interestingly, the H3K27me3 histone demethylase Kdm6b was significantly downregulated in both lipid-laden MG and MDMs (Figure S3G). These findings suggest that a loss of Kdm6b-mediated decrease in the H3K27me3 repressive histone mark underlies the reduced chromatin accessibility of LLM inflammatory genes. Flow cytometry analyses confirmed that H3K27me3 levels were heightened in lipid-laden MDMs compared with their non-lipid-laden counterpart. The elevated H3K27me3 levels in MG correlated with their less accessible chromatin status, potentially hindering the integration of extrinsic cues from the TME in both primary and recurrent tumors (Figures 3H and S3H). In addition to downregulating Kdm6b, LLMs displayed increased EZH2 (Enhancer Of Zeste Homolog 2) levels, the major catalyzer of H3K27me3.41 The higher EZH2 level in MG compared with MDMs independent of their lipid-laden phenotype potentially underlies the heightened H3K27me3 base level in tissue-resident MG (Figure S3I).
Both lipid-laden MG and MDMs displayed enhanced immunosuppressive features compared with their non-LLM counterpart, including increased levels of CD39 and PD-L1 (Programmed death-ligand 1) (Figures 3I and S3J). Lipid-laden MDMs specifically downregulated major histocompatibility complex (MHC)-II expression, suggesting that MDM antigen presentation is compromised when acquiring an LLM phenotype (Figure 3I). Interestingly, expression of the lipid receptor CD36 was upregulated in LLMs (Figure 3I), corroborating our previous observations of the lipid droplet accumulation characteristic of LLMs (Figure S2B). The content of lipid-laden MDM was increased in recurrent PDG-Ink4a glioblastoma, in line with the OPC-to-MES transition revealed by scRNA-seq (Figures 1D and 3J). Importantly, the association between lipid-laden MDM enrichment and MES-like glioblastoma was confirmed in the treatment-naive NF1-driven MES-dominant glioblastoma model42 (Figure 3J). Altogether, our findings consolidate the association between MES-like glioblastoma and LLMs, which immunosuppressive and lipid-associated phenotype is partly driven by downregulation of Kdm6b and upregulation of EZH2. Consequently, heightened levels of the H3K27me3 repressive mark in lipid-laden MDMs correlates with loss of chromatin accessibility underlying inflammatory gene downregulation.
We next assessed the functional significance of LLMs in glioblastoma progression by targeting CD36, a lipid receptor preferentially expressed by lipid-laden MDMs and MG (Figures 3I, S4A, and S4B). Based on the kinetics of LLM frequency in the PDG-Ink4a model (Figure S4C), we initiated daily longitudinal treatment with the blood-brain barrier permeable CD36 inhibitor sulfosuccinimidyl oleate (SSO)43 2 days post-RT completion (Figure S4D). Importantly, SSO treatment in combination with RT resulted in a significant survival benefit in glioblastoma-bearing mice (Figure 3K). A moderate decrease in LLM frequency was observed in RT + SSO compared with the RT + vehicle, which was only significant in the MDM-TAM subset (Figures 3L and S4E). SSO-treated tumors displayed a significantly larger population of MHC-II+ MDMs (Figure S4F), while lymphoid immune composition and activation remained unchanged (Figures S4G–S4I). These experiments establish the proof-of-concept potential of targeting LLMs, prompting us to further examine the mechanistic bases underlying their emergence and pro-tumoral functions in the glioblastoma TME.
Figure S4.
Validation of myelin-derived lipids as modulators of TAM phenotype, which can be altered by CD36 inhibition in combination with radiotherapy, related to Figures 3 and 4
(A) Dot plot displaying CD36 RNA expression levels in the most abundant cell types identified in the murine scRNA-seq dataset. Colors represent the average expression; size depicts the percentage of cells that express Cd36.
(B) Quantification of mean fluorescent intensity (MFI) of CD36 in various cell populations in recurrent PDG-Ink4a tumors post RT at the trial endpoint. Tumor cells: CD45−CD11b−CD31− cells; endothelial cells: CD45−CD11b−CD31+ cells. All macrophages and LLM-macrophages: see Figure S2E.
(C) Flow cytometry quantification of lipid-laden (BODIPYhighSSC-Ahigh) MG (left) and MDMs (right) in primary, 2 days post-5x2 Gy RT, and recurrent PDG-Ink4a glioblastoma.
(D) Schematic overview of the experimental design and treatment schedules. Tumors were initiated as described in STAR Methods to generate PDG-Ink4a tumor-bearing mice. At 4–6 weeks post tumor initiation, tumor size was quantified by MRI, and block randomization was used to evenly distribute mice between treatment groups based on tumor volume (20–90 mm3). Treatment groups were control (DMSO), fractionated ionizing radiation (RT, 5 × 2 Gy), and DMSO, SSO (30 mg/kg daily treatment), or RT + SSO. DMSO and SSO treatments were initiated 48 h post last dose of RT in the combination groups. Mice were euthanized when symptomatic or at the indicated time points.
(E) Graph depicting the mean fluorescent intensity (MFI) of lipid metabolism markers in MG (left) and MDMs (right) isolated from recurrent PDG-Ink4a tumors post RT + DMSO and RT + SSO treatment at the trial endpoint.
(F) Flow cytometry quantification of MHC-II+ MDMs as a percentage of total MDMs in recurrent PDG-Ink4a tumors post RT + DMSO and RT + SSO treatment at the trial endpoint.
(G) Quantification of CD3+ lymphoid cell populations (as percentage of CD45+ cells) in recurrent PDG-Ink4a glioblastoma post RT + DMSO or RT + SSO treatment at the trial endpoint. B cells: NK1.1−CD19+; CD8T cells: NK1.1−CD19−CD3+CD8+; CD4T cells: NK1.1−CD19−CD3+CD4+.
(H and I) Mean fluorescent intensity (MFI) of depicted activation markers in CD8 T cells (H) and CD4 T cells (I) from recurrent PDG-Ink4a glioblastoma post RT + DMSO or RT + SSO treatment at the trial endpoint.
(J) Schematic of cholesterol de novo biosynthesis pathway, depicting cholesterol precursors and derivatives, as well as enzymes (in italics) involved. Enzymes and intermediates that are increased in LLMs are marked in green, decreased in red, as determined by lipidomics (Figures 4A–4C) and RNA-seq (Figure 4D).
(K) Representative IF images of fresh-frozen recurrent PDG-Ink4a glioblastoma tissue sections illustrating the LLM location in relation to myelin basic protein. DAPI: nuclear stain (blue); IBA1: pan-macrophage (magenta); PLIN2: lipid droplets (LDs) (white); myelin basic protein (MBP) (yellow). T, tumor; H, adjacent healthy brain.
(L) Pie chart depicting the average fraction of each major lipid species found in myelin isolated from healthy mouse brains. Data for this graph was extracted from NMR to quantify cholesterol (red) and Lipidyzer analysis quantifying other lipid classes (gray).
Statistics: mixed-effects analysis (B and C) or two-way ANOVA with Sídák correction for multiple comparisons (E–I). Data are represented as mean + SEM (B and G–I), ± SEM (C, E, and F), or ± SD (L).
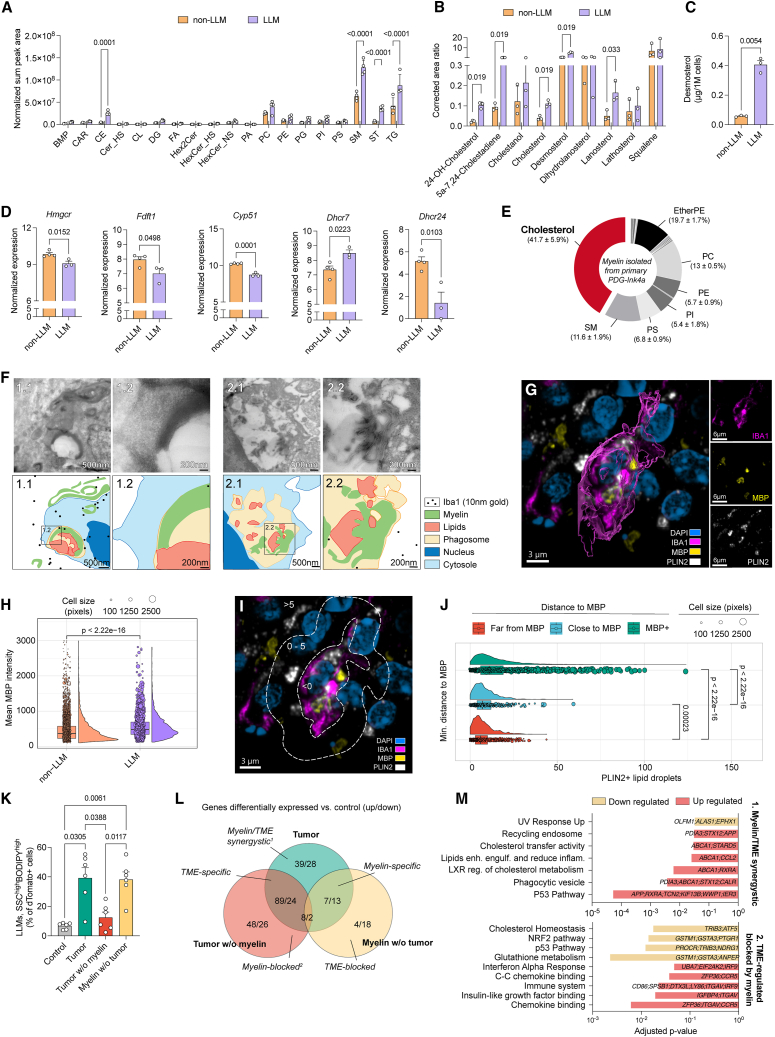
Myelin debris in the TME drives cholesterol accumulation and LXR activation in TAMs
We investigated the lipidome landscape specific to LLMs versus their non-LLM counterpart in vivo that may underlie their pro-tumorigenic features. Lipidomics analyses showed increased levels of sterols, sphingomyelin, cholesteryl esters (CEs), and triacylglycerols in LLMs (Figure 4A). Strikingly, LLMs displayed increased levels of cholesterol and cholesterol precursors, such as 5α-7,24-cholestadiene, lanosterol, and desmosterol (Figures 4B and 4C). Interestingly, downregulation of multiple enzymes involved in the cholesterol de novo biosynthesis pathway (Figures 4D and S4J) indicated that the observed cholesterol accumulation in LLMs originates from extrinsic sources in the TME rather than from de novo biosynthesis. These findings resonate with the phenotype of foamy macrophages in neurodegenerative diseases, in which phagocytosis of cholesterol-rich myelin debris triggers cholesterol accumulation and cholesterol biosynthesis shutdown. This negative feedback results in build-up of cholesterol precursors such as desmosterol, which consequently activates liver X receptor (LXR) to overcome cholesterol overloading by increasing cholesterol efflux.29,44
Figure 4.
LLM formation relies on myelin phagocytosis and subsequent sterol accumulation
(A and B) Normalized levels of depicted lipid classes (abbreviations in Figure S6J) and corrected area ratio of sterols in FACS-purified LLMs and non-LLMs.
(C) Desmosterol concentration in FACS-purified LLMs and non-LLMs, assessed from the lipidomic analyses depicted in (B).
(D) Normalized expression of cholesterol synthesis pathway genes in FACS-purified LLMs and non-LLMs from Figure 3B.
(E) Average fraction of major lipid species present in myelin isolated from tumor.
(F) Representative electron microscopy images of primary PDG-Ink4a tumor sections. Boxes 1.2 and 2.2 are high-magnification images from boxes 1.1 and 2.1, respectively.
(G) Representative IF image of primary PDG-Ink4a tumor sections. DAPI: nuclear stain (blue); IBA1: pan-macrophage (magenta); PLIN2: lipid droplets (white); MBP: myelin basic protein (yellow).
(H) Quantification of mean MBP intensity in TAM subsets in primary PDG-Ink4a tumors (as presented in Figure S4K). Each dot represents individual TAMs (IBA1+), size corresponds to cell area.
(I) Visual IF image representation of an IBA1+PLIN2+ LLM and distance to myelin used to annotate individual TAMs (IBA1+) as MBP+ (minimal distance to MBP < 0 pixels), close to MBP (0–5 pixels), or far from MBP (>5 pixels).
(J) Quantification of the total number of lipid droplets present in each TAM correlated to their minimal distance to MBP+ staining as described in (I).
(K) Flow cytometry quantification of ex vivo LLMs (experiments depicted in Figure S5C).
(L) Differentially expressed genes (Table S5A) identified in RNA-seq from FACS-purified dTomato+ BMDMs used in (K).
(M) p values of relevant pathways enriched based on GSEA of signatures derived from (L) (Tables S5B–S5J).
Statistics: two-way ANOVA with Sídák correction for multiple comparisons (A and B), two-tailed paired t test (C and D), pairwise comparison for analyzing multiple population means in pairs (H), pairwise comparison for analyzing multiple population means in pairs (J), one-way ANOVA with Sídák correction for multiple comparisons (K), Fisher’s exact test in combination with the Benjamini-Hochberg correction (M). Data are represented as mean + SEM (A–D and K) or ± SD (E).
See also Figures S4 and S5 and Table S5.
In order to address the potential of myelin as a key source of lipids conferring TAMs an LLM phenotype, we first probed the presence of myelin within tumor tissue using myelin basic protein (MBP) as a marker. Within the TME, structures reminiscent of myelin debris, presenting a distorted configuration compared with myelin in adjacent healthy brain tissue, were identified in the proximity of PLIN2+LLMs (Figure S4K). Importantly, lipidomics analyses of myelin isolated from glioblastoma tumors confirmed the over-representation of cholesterol, similarly to myelin from non-tumor-bearing brains (Figures 4E and S4L). Probing de- and remyelination transcriptional signatures45 revealed a positive correlation with glioblastoma tumors but not with adjacent normal brain tissue, corroborating the accumulation of myelin debris within glioblastoma (Figures S5A and S5B). These findings imply that de- and remyelination pathways are triggered within glioblastoma, potentially underlying the presence of myelin debris within the TME. Importantly, electron microscopy (EM) imaging confirmed the presence of myelin debris within macrophage phagosomes in vivo (Figure 4F). Moreover, myelin engulfment and MBP accumulation in TAMs were contiguous to size increase and lipid accumulation, suggesting that macrophages phagocytose and recycle myelin into lipids (Figures 4G–4J). These findings highlight the requirement for macrophages to scavenge myelin debris to acquire an LLM phenotype in the glioblastoma TME.
Figure S5.
Assessment of the impact of TME-mediated education and myelin uptake on TAM lipid metabolism and inflammation activity, related to Figures 4 and 5
(A) Representative spatial expression plot of de- and remyelination transcriptional modules45 overlayed on recurrent PDG-Ink4a tumor tissue used in VISIUM 10x spatial transcriptomics.
(B) Dot plot representing the correlation matrix between de- or remyelination transcriptional signatures45 and healthy brain, tumor, glioblastoma cellular subtypes, glioblastoma niches, or TAM subsets transcriptional modules across all VISIUM 10x samples (n = 13, n = 4 PDG-Ink4a primary, n = 3 PDG-Ink4a recurrent, n = 3 PDG-p53 primary, n = 3 PDG-p53 recurrent). Dot color and size correspond to the correlation coefficient.
(C) Schematic of ex vivo assay used for LLM quantification in Figure 4K and in (D). Tumors were extracted from PDG-Ink4a glioblastoma-bearing mice and dissociated as a single cell preparation prior to magnetic beads-isolation of myelin. The dissociated glioblastoma (with or without myelin) or myelin only (extracted from the tumor) was then added to dTomato+ BMDMs for 48 h, after which LLM formation was assessed by flow cytometry based on dTomato, SSC-A (granularity), and BODIPY staining (as described in Figure S2E).
(D) Flow cytometry quantification of BODIPY (left) or SSC-A (right) mean fluorescence intensity (MFI) in dTomato+ BMDMs incubated with control media (gray); dissociated PDG-Ink4a primary tumors containing myelin (green) or not (red); tumor-extracted myelin (yellow).
(E) Bar plots depicting the p values of relevant pathways related to TME-driven and Myelin-regulated genes depicted in Figure 4L. Genes driving pathway enrichment are stated within each respective bar. Colors represent pathways that belong to genes that are either down- (beige) or upregulated (red) within that signature (Tables S5F, S5G, S5K, and S5L).
(F) Relative uptake of CFSE-labeled myelin or FluoSpheres based on mean fluorescent intensity (MFI) of BMDMs educated with conditioned media (CM) from ex vivo dissociated primary PDG-Ink4a tumors ± SSO (100 μM). Data normalized per experiment.
(G) Relative expression of genes representative of the OPC and MES glioblastoma subtypes assessed by RT-qPCR in cell lines isolated from PDG-Ink4a primary (OPC-like) or recurrent (MES-like) glioblastoma. Represented genes were obtained from previously reported glioblastoma subtype classification,18 with OPC-like cells resembling the proneural state. Data scaled by columns.
(H–J) Seahorse assay was performed on glioblastoma cell lines generated from primary (blue, n = 4) or recurrent (red, n = 4) PDG-Ink4a mice to assess (H and I) mitochondrial respiration and (J) glycolysis and glycolytic reserve.
(H) Oxygen consumption rate (OCR) over time upon injection of drugs interfering with mitochondrial respiration, as indicated.
(I) Left: OCR measured at start minus OCR after injection of antimycin A and rotenone reflects basal respiration; right: OCR difference measured of basal respiration and maximal OCR reflects maximal respiratory capacity.
(J) Left: ECAR measured after injection of glucose reflects basal glycolytic activity; right: ECAR measured after injection of FCCP reflects glycolytic reserve.
(K) In vitro assay experimental setup used to generate data for (L) and for Figures 5B, 5C, and S6C: bone marrow-derived macrophages (BMDMs) or primary MG were cultured in control media (10% FBS) or tumor-conditioned media (TCM) derived from OPC-like or MES-like glioblastoma cells for 24 h. When indicated, 2-DG was incorporated to the TCM preparation in order to block glycolysis. BMDMs were then exposed either to myelin for 3 h and maintained in culture for an additional 24 h before assessing LLM formation using flow cytometry analyses of intracellular BODIPY staining and SSC-A; or to myelin-CFSE or FluoSpheres for 3 h before assessing phagocytosis activity using flow cytometry analyses of the fluorescent phagocytic source.
(L) Graph depicting the fold-change of CFSE-labeled myelin and FluoSpheres mean fluorescent intensity (MFI) (added 3 h before measurement) in BMDMs previously exposed to MES-like TCM for 24 h. Statistics: two-tailed unpaired t test.
Statistics: Kendall trend test (B), one-way ANOVA Dunnett’s multiple comparisons test (D), Fisher’s exact test in combination with the Benjamini-Hochberg method for correction of multiple hypotheses testing (E), two-way ANOVA with Sídák correction for multiple comparisons (F), two-stage step-up multiple paired t test with Benjamini, Krieger, and Yekutieli method for multiple testing (G), or two-tailed unpaired t test (I, J, and L) (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001). Data are represented as mean + SEM (D, F, H–J, and L).
To confirm the role of local myelin debris uptake in LLM formation, we established an ex vivo co-culture system in which dTomato+ bone-marrow derived macrophages (BMDMs) were exposed to dissociated murine glioblastoma tumors with or without prior myelin depletion (Figure S5C). Interestingly, the whole dissociated tumor or myelin debris alone led to significant LLM formation, whereas this effect was abrogated in absence of myelin (Figures 4K and S5D), highlighting the central role of myelin as a lipid source for TAMs to acquire a lipid-laden phenotype. Interestingly, addition of the whole tumor (containing myelin) led to a 3-fold increase in general lipid content, compared with 2-fold increase with myelin alone (Figure S5D), indicative of the TME contribution in priming macrophage LLM phenotype.
To parse out the contribution of signaling cues originating from the TME or from myelin itself that drive LLM formation, we performed RNA-seq on dTomato+ BMDMs FACS-isolated from ex vivo co-culture assays (Figures 4L and S5C; Tables S5A–S5L). Interestingly, upregulation of pathways involved in phagocytosis and complement activation suggested that the TME bestowed “eat-me” signals, instructing macrophages to enhance their phagocytic capacity (Figure S5E). Importantly, these signals likely regulate the specific uptake of myelin, as CD36 inhibition in conditioned media (CM)-educated macrophages hindered myelin engulfment but did not impair microsphere phagocytosis (Figure S5F). Orthogonally, myelin exposure induced expression of pathways involved in cholesterol metabolism and lipid transport while downregulating cholesterol biosynthesis (Figure S5E), similarly to in vivo LLM features (Figure 4D). Interestingly, the intersect of TME- and myelin-derived signals instructing macrophage education encompassed upregulation of pathways involved in lipid engulfment, phagocytosis, and activation of the LXR pathway, as well as increased expression of genes pertaining to lipid export (Abca1) (Figure 4M). Strikingly, TME-specific education promoting inflammatory responses in macrophages was inhibited in presence of myelin (Figure 4M), inferring that glioblastoma-induced inflammation is hindered by this lipid source, fueling LLM immunosuppressive phenotype (Figure 3I). Taken together, these findings reveal that signals derived from the TME prime macrophages to scavenge myelin debris as a source of lipids. In turn, phagocytosed myelin debris drives the accumulation of cholesterol precursors activating LXR, consequently dampening inflammatory activity and upregulating lipid export in LLMs.
MES-like glioblastoma cells enhance TAM-mediated myelin recycling and lipid export into the TIF
We next sought to investigate the functional significance of myelin recycling in the context of LLM and glioblastoma cell crosstalk. To model this interplay in vitro, we generated OPC-like and MES-like glioblastoma cell lines from primary or recurrent PDG-Ink4a tumors, respectively. Primary/OPC-like and recurrent/MES-like glioblastoma cells expressed genes characteristic of the glioblastoma subtype they represent, and MES-like glioblastoma cells displayed a distinctive increase in glycolytic activity, representative of their in vivo properties (Figures 5A and S5G–S5J).46 Myelin debris phagocytosis, but not general phagocytosis (Figures S5K and S5L), was enhanced when BMDMs were educated with glioblastoma tumor-conditioned media (TCM) (Figure 5B), leading to increased lipid accumulation (Figures 5C and S6A) reminiscent of the in vivo LLM phenotype (Figures S2B and S2E). In a similar fashion, TCM-mediated priming of primary MG increased LLM formation in response to myelin debris (Figure S6B), highlighting the myelin-mediated LLM phenotype is not restricted to MDMs. While OPC-like and MES-like glioblastoma cells both instructed LLM formation in BMDMs, the highly glycolytic features of MES-like glioblastoma cells (Figures 5A and S5J) impacted this process, as shown by the decreased LLM generation upon inhibition of glycolysis (Figure S6C). Additionally, exposure to myelin (but not to TCM) triggered the upregulation of H3K27me3 (Figure 5D), reinforcing the role of myelin in orchestrating gene expression changes underlying the acquisition of macrophage LLM phenotype.
Figure 5.
LXR pathway activation in LLMs stimulates lipid exchange within the TME
(A) Extracellular acidification rate (ECAR) over time in Seahorse glycolytic activity assay on glioblastoma cell lines generated from primary/OPC-like (n = 4) or recurrent/MES-like (n = 4) PDG-Ink4a tumors.
(B) Relative fold-change of myelin-carboxyfluorescein succinimidyl ester (CFSE) uptake (MFI) in BMDMs.
(C) Relative fold-change of BODIPY MFI in myelin-exposed BMDMs over control (no myelin).
(D) Relative fold-change of H3K27me3 MFI in BMDMs in monoculture (±myelin) or in co-culture with MES-like glioblastoma cells.
(E and F) Relative expression LXR pathway genes assessed by quantitative real-time PCR (RT-qPCR) in BMDMs exposed to (E) control media or MES-like TCM, and (F) MES-like TCM ± myelin.
(G) Expression levels of lipid export and lipoprotein receptor genes in the most abundant cell types identified in the murine scRNA-seq dataset (Figure 1B).
(H) Free cholesterol quantified in the supernatant of BMDMs previously exposed to myelin for 24 h in vitro.
(I) Experimental design illustrating the collection of TIF from glioblastoma (see STAR Methods).
(J) Relative levels (sum of peaks) of total lipids detected in the TIF.
(K) Fold-change in cholesteryl ester (CE) levels quantified in the TIF compared with Ntb tissue (control).
Statistics: Friedman test with Dunn’s correction for multiple comparisons (B, C, J, and K), one-way ANOVA with Sídák (D), two-way ANOVA with Sídák correction for multiple comparisons (E and F), or two-tailed paired t test (H). Data are represented as mean + SEM (A–C) or ± SEM (D–F, H, J, and K).
Figure S6.
Analysis of myelin-induced LXR activation in macrophages orchestrating lipid exchanges in the TME, related to Figure 5
(A) Representative flow cytometry plots from BMDMs exposed to MES-like TCM ± myelin as shown in Figure S5K. Gate is set on BODIPYhigh and SSC-Ahigh BMDMs to identify LLMs in vitro.
(B) Flow cytometry quantification of LLMs (BODIPYhigh, SSC-Ahigh) as a percentage of primary microglia (MG) previously exposed to control media or MES-like TCM, 24 h post myelin debris exposure (3 h).
(C) BODIPY MFI fold-change in MES-like TCM-educated BMDMs after exposure to myelin (3 h), compared with no myelin exposure. MES-like TCM was collected from cells treated or not with the glycolysis inhibitor 2-DG (Figure S5K). Lipid accumulation in response to myelin uptake in BMDMs was assessed by flow cytometry using BODIPY staining.
(D and E) Relative expression of genes representative of the LLM signature (as established in Figure 1I) was assessed by qPCR in BMDMs exposed to (D) MES-like TCM ± myelin for 1 or 24 h, (E) control media, or MES-like TCM alone.
(F) Relative expression of Abca1, Abcg1, Dhcr24, and Nr1h3 as assessed by qPCR in BMDMs conditioned with MES-like TCM and exposed to myelin ± LXR inhibitor.
(G and H) Immunofluorescence staining of Filipin (cholesterol stain) was performed on in vitro BMDMs conditioned with MES-like TCM ± LXR inhibitor (LXRi) for 24 h and exposed or not to myelin (3 h). Histogram depicting mean intensity of Filipin per BMDM (G) or per lipid droplet in BMDMs (H). Lines represent median and quartiles.
(I) Violin plots depicting normalized gene expression levels of the cholesterol efflux-related genes Abca1, Abcg1, Apoc1, and Apoe in LLMs and non-LLMs, extracted from the murine scRNA-seq dataset.
(J) Fold-change in the depicted lipid classes quantified by lipidomics analyses in the TIF of primary/OPC-like and recurrent/MES-like PDG-Ink4a tumors compared with non-tumor-bearing brain (Ntb) tissue. BRSE, brassicasterol ester; CASE, campesterol ester; CL, cardiolipin; CE, cholesteryl ester; Cer_BS, ceramide beta-hydroxy fatty acid-sphingosine; Cer_NS, ceramide non-hydroxyfatty acid-sphingosine; CoQ, coenzyme Q; DG, diacylglycerol; FA, free fatty acid; LPC, lysophophatidylcholine; LPI, lysophosphatidylinositol; MGDG, monogalactosyldiacylglycerol; PA, phosphatidic acid; PC, phosphatidylcholine; PE, phosphatidylethanolamine; PE_Cer, ceramide phosphoethanolamine; PG, phosphatidylglycerol; PI, phosphatidylinositol; PI_Cer, ceramide phosphoinositol; PS, phosphatidylserine; SHexCer, sulfatide; SM, sphingomyelin; SSulfate, sterol sulfate; TG, triacylglycerol; CAR, acylcarnitine; ST, sterol.
(K) Relative fold-change of ceramide (Cer) species levels in the interstitial fluid (IF) of Ntb mice, primary/OPC-like and recurrent/MES-like PDG-Ink4a tumors.
(L) Total lipid levels (sum peak area) detected in the IF of Ntb, primary, RT-recurrent, and SSO-treated recurrent PDG-Ink4a tumors. Data points in the Ntb, primary, and recurrent PDG-Ink4a group from Figure 5J.
(M) Relative levels of cholesteryl ester (CE) quantified in the tumor interstitial fluid (TIF) of Ntb, primary, RT-recurrent, and SSO-treated recurrent PDG-Ink4a tumors. Data points in the Ntb, primary, and recurrent PDG-Ink4a group from Figure 5K.
Statistics: two-way ANOVA with Sídák correction for multiple comparisons (B, D–F, J, and K), two-stage step-up ratio paired t test (C), or mixed-effects analysis with Sídák correction for multiple comparisons (G, H, L, and M). Data are represented as mean + SEM (B, C, and F) and ± SEM (D, E, and J–M).
Next, we further investigated the interplay between LLM and MES-like glioblastoma cells, in light of the strong correlation observed between these two cell types at the transcriptional and spatial levels (Figures 1J and 2D). BMDM acquisition of LLM-specific genes was induced upon exposure to MES-like TCM + myelin debris, but not by MES-like TCM alone (Figures S6D and S6E). While MES-like TCM led to increased Nr1h3 gene expression, which encodes for the LXR-alpha protein (Figure 5E), exposure to myelin in this context was required to upregulate the ATP-binding cassette transporters Abca1 and Abcg1 (both downstream LXR activation)44 expression and to downregulate Dhcr24 (Figure 5F), both indicative of intracellular accumulation of cholesterol and desmosterol following myelin uptake, as observed in vivo (Figures 4B–4D and S4J). As expected from the agonistic role of desmosterol in activating LXR during foamy macrophage formation,43 pharmacological inhibition of LXR hindered the expression of the myelin-induced lipid export genes Abca1 and Abcg1 (Figure S6F), resulting in cholesterol accumulation in macrophages (Figures S6G and S6H). These cholesterol efflux transporters44 were found mainly expressed in TAMs (Figure 5G) and upregulated in glioblastoma LLMs (Figure S6I). Consistent with this increased expression of lipid transporters, macrophages were able to efficiently efflux cholesterol in vitro following myelin uptake (Figure 5H). These findings prompted us to assess the lipid content of the tumor interstitial fluid (TIF) in primary/OPC-like and recurrent/MES-like glioblastoma (Figure 5I). Lipidomic analyses revealed that the total amount of lipids detected in the TIF was increased in MES-like, LLM-enriched tumors (Figures 5J and S6J), including CEs (Figure 5K), which were previously found elevated in LLMs (Figure 4A). As CEs are a major component of high-density lipoproteins (HDLs),47 and HDL-mediated lipid transport activity is upregulated in macrophages following myelin exposure (Figure S5E), these results are indicative of increased HDL-mediated cholesterol efflux from LLMs. Furthermore, MES-like enriched recurrent glioblastoma displayed increased levels of ceramides (Figure S6K), a lipid class involved in demyelination,48 altogether exposing a regulatory mechanism during which increased levels of myelin debris underlies LLM accumulation in MES-like glioblastoma. Notably, recurrent tumors in which LLM formation was hindered by SSO treatment (Figures 3K, 3L, and S4E) displayed reduced TIF total lipid levels (Figure S6L), including CEs (Figure S6M).
In sum, these findings reveal that LLMs not only possess increased levels of a wide variety of lipid classes but also export myelin-derived lipids into the TIF through TME-mediated priming of the LXR pathway. These results prompted us to further examine the significance of LLM-mediated cholesterol efflux on the malignant features of glioblastoma cancer cells.
Myelin-induced LLMs fuel MES-like glioblastoma cell malignancy
To determine how glioblastoma cells benefit from LLM-derived lipids subsequent to myelin engulfment and processing, we examined the metabolic features of distinct glioblastoma subtypes that would require such external lipid sources. While tumor cells in general showed high transcriptional activity for cholesterol de novo synthesis pathways (Figure S7A), MES-like cells, and particularly the MES2 subtype, displayed the lowest cholesterol biosynthesis activity. Indeed, MES-like cells showed low activity in multiple lipid biosynthetic pathways compared with other cancer cell subtypes (Figures 6A and 6B), a feature also observed in patient samples (Figures S7B and S7C). Conversely, MES2 cells expressed high levels of Vldlr (Figure 6B), a membrane receptor for HDL, potentially favoring their ability to scavenge these lipoproteins containing LLM-derived cholesterol (Figure S5E).49 This led us to postulate that a metabolic crosstalk centered on lipid uptake and exchange is at the core of MES-like cell and LLM reciprocal interactions. Indeed, lipids stored in myelin sheaths were not readily available to cancer cells (Figure S7D), whose proliferation was reduced when exposed to myelin following macrophage depletion ex vivo (Figures S7E and S7F). Importantly, myelin lipotoxic effect on glioblastoma cancer cells was rescued by reintroducing macrophages—revealing the importance of macrophages in recycling myelin debris to both protect and benefit tumor cells (Figure S7F). Tracing the fate of lipids from BMDM loaded with clickable cholesterol in co-culture (Figure S7G) revealed that direct transfer occurs to MES-like but not to OPC-like glioblastoma cells (Figure 6C). In order to address whether tumor cells scavenge lipids derived from LLM-mediated myelin recycling, we designed a multi-omics co-culture experiment under various conditions of myelin exposure, subsequently applying quantitative lipidomics and transcriptomics analyses on FACS-purified glioblastoma cancer cells (Figure S7H). In monoculture, MES-like cells exposed to myelin did not display altered lipid contents (Figure 6D), corollary to their inability to phagocytose myelin debris (Figure S7D). However, processes associated with cellular stress were upregulated in this condition, while proliferation-associated pathways were decreased (Figures S7I and S7J), emphasizing the lipotoxic features of myelin toward cancer cells (Figure S7F; Tables S6A and S6B). Distinctively, when co-cultured with macrophages and in presence of myelin, multiple lipid classes gradually accumulated in MES-like tumor cells, including cholesterol and triglycerides (TGs) (Figures 6D and 6E). Transcriptionally, MES-like tumor cells upregulated various pathways involved in proliferation and cell cycle (Figures S8A and S8B; Tables S6C–S6F). Altogether, these analyses expose the molecular mechanisms underlying TAM/cancer cell interplay, through which myelin-derived lipids processed by LLMs promote tumor growth.
Figure S7.
Conserved human mesenchymal glioblastoma metabolic properties in murine tumors, which underlie MES-like tumor cells’ requirement to outsource lipidic needs, related to Figure 6
(A) Dot plot displaying the expression levels of de novo cholesterol biosynthesis genes in the most abundant cell types identified in the murine scRNA-seq dataset (Figure 1B). Color represents the average expression; size depicts percentage of cells that express each specific gene.
(B) Dot plot displaying the expression levels of de novo cholesterol biosynthesis and import genes in the glioblastoma cell subtypes identified in patient scRNA-seq datasets.19 Dot color represents the average expression; size depicts percentage of cells expressing each specific gene.
(C) Radar plot depicting normalized GSEA scores (between 0 and 1 based on each pathway) of the represented metabolic pathways (KEGG) between OPC, MES, NPC, and AC glioblastoma cancer cells extracted from patient scRNA-seq datasets.19 Gray area represents the average activity in each metabolic pathway across all tumor cells.
(D) Graph depicting MFI of CFSE-labeled myelin uptake in BMDMs or OPC-like and MES-like glioblastoma cells.
(E) Schematic of ex vivo assay used for EdU and Annexin V staining as depicted in (F) and in Figures 6K and S8G–S8I. Primary tumors were extracted from PDG-Ink4a glioblastoma-bearing mice and dissociated as single cells prior to magnetic beads-mediated myelin removal and/or CD11b+ cell depletion. The dissociated tumors were then placed in culture for 24 h, media was refreshed to remove cellular debris, and LXRi, ABCA1i, or BMDMs were added when specified. Tumor cell proliferation (EdU staining) or viability (Annexin V−, Zombie−) was assessed after 48 h by flow cytometry.
(F) Histogram depicting the fold-change of tumor cells in S phase (EdU+) compared with tumor cells without CD11b+Abca1/Abcg1KO BMDMs ± myelin. Culture conditions are described in (E).
(G) Schematic of in vitro LLM/glioblastoma cell co-culture experimental design: BMDMs educated in OPC-like or MES-like TCM as previously described (Figure S5K) were exposed to (1) clickable cholesterol overnight or (2) myelin for 3 h and placed in co-culture with glioblastoma cells. (1) Uptake/transfer of cholesterol from BMDMs to glioblastoma cells was measured by flow cytometry after 0, 1, and 3 h of co-culture to reveal macrophage-derived cholesterol uptake with a Click-iT reaction (see STAR Methods). (2) Glioblastoma cell proliferation was measured by flow cytometry after 48 h of co-culture (EdU staining).
(H) Schematic of in vitro LLM/MES-like glioblastoma cancer cell co-culture design to study effect of macrophage-mediated myelin recycling on tumor cell lipidome and transcriptome: MES-like tumor cells were grown in 2% lipid-free FBS DMEM before addition of BMDMs (or not) to the co-culture. After 24 h, myelin was added for different durations (0, 6, or 24 h) prior to MES-like tumor cells and BMDMs FACS-isolation and downstream bulk RNA-seq or lipidomics analyses (see STAR Methods).
(I) Volcano plot depicting the differentially expressed genes between MES-like tumor cells cultured with myelin debris for 24 h or not. Colors correspond to significantly increased (red) or decreased (blue) genes. Genes are identified by bulk RNA-seq from FACS-purified MES-like tumor cells as depicted in (H).
(J) Bar plots depicting the adjusted p values of relevant pathways specific to genes downregulated (blue) or upregulated (red) in MES-like tumor cells in response to myelin exposure in monoculture from (I).
Statistics: one-way ANOVA (D) or two-way ANOVA with Sídák correction for multiple comparisons (F) or Fisher’s exact test in combination with the Benjamini-Hochberg method for correction of multiple hypotheses testing (J). Data are represented as mean ± SEM (D and F).
Figure 6.
LLM-mediated lipid export fuels MES-like cell malignancy in the lipid-scarce glioblastoma TME
(A) Normalized GSEA scores in glioblastoma cell subtypes from murine scRNA-seq dataset. Gray areas = average metabolic activity of each pathway.
(B) Expression levels of cholesterol biosynthesis and import genes in glioblastoma cell subtypes from murine scRNA-seq dataset.
(C) Flow cytometry MFI quantification of cholesterol content in glioblastoma cells following co-culture with BMDMs previously loaded with clickable cholesterol (Figure S7G).
(D) Fold-change expression (x axis) and −log10p value (y axis) of depicted lipid classes quantified in MES-like tumor cells in mono- or co-culture with TAMs + myelin, compared with no myelin (Figure S7H).
(E) Cholesterol and triglyceride (TG) levels in MES-like glioblastoma cells exposed to myelin in monoculture or co-culture with BMDMs.
(F) Relative distribution (based on lipid ng/μg protein) of labeled 13C among lipid classes quantifiable 24 h after administration of CE13C-FA in MES-like glioblastoma cells in vitro (n = 3).
(G) Quantification of metabolized CE13C-FA per lipid species in which U13C-FA18:1 was detected.
(H) Percentage of glioblastoma cells in S phase (EdU+) after 48 h co-culture with TCM-conditioned BMDMs ± myelin (Figure S7G). EdU, 5-ethynyl 2′-deoxyuridine.
(I) Electron microscopy representative images depicting the contact points between MES-like tumor cells (T) and myelin-loaded BMDMs (M). LD, lipid droplet; CV, coated vesicle; MD, myelin debris.
(J) Percentage of MES-like glioblastoma cells in S phase (EdU+) after 48 h of co-culture with TCM-conditioned BMDMs ±myelin, ±CD36 inhibitor (SSO) ±ABCA1 inhibitor valspodar (ABCA1i).
(K) Viable glioblastoma cells (ZombieNIR−, Annexin V−) as a percentage of total tumor cells in dissociated glioblastoma where myeloid cells were maintained (+CD11b+) or ex vivo depleted (−CD11b+) (Figure S7E) ± ABCA1i or LXR inhibitor GSK2033 (LXRi).
Statistics: two-way ANOVA (C, E, H, and K) and mixed-effects analysis with Sídák correction for multiple comparisons (J). Data are represented as mean + SEM (C, E, G, H, J, and K).
See also Figures S7 and S8 and Table S6.
Figure S8.
Extended analyses of macrophage-mediated myelin recycling on shielding tumor cells from lipotoxicity and fueling MES-like cells in an LXR-Abca1/Abcg1-dependent manner, related to Figure 6
(A) Venn diagram depicting the differentially expressed genes in MES-like tumor cells co-cultured with BMDMs in the absence (control) or presence of myelin debris (+myelin debris), compared with MES-like tumor cells in monoculture (Table S6). Genes are identified by bulk RNA-seq from FACS-purified MES-like tumor cells as described in Figure S7H.
(B) Bar plots depicting the p values of pathways downregulated (left) and upregulated (right) in MES-like tumor cells in co-culture with BMDMs (±myelin debris) compared with MES-like tumor cells in monoculture from Figure S7H.
(C) Representative IF images of MES-like tumor cells exposed to cholesteryl esters (CEs) containing a click handle on the cholesterol (left, yellow) or fatty acid (right, pink) at various time point (0, 8, and 24 h post addition of Click-CE). DAPI: nuclear stain (blue).
(D) Representative flow cytometry plots depicting the gating strategy for the EdU proliferation assay analyses, in which EdU+ cells are depicting cells in S phase, as shown in (E), (F), and Figures 6H and 6J.
(E) Graph depicting the percentage of live glioblastoma cells in S phase (EdU+) assessed 48 h post co-culture with TCM-preconditioned BMDMs previously exposed to myelin (3 h) or not and separated by transwells.
(F) Graph depicting the percentage of MES-like glioblastoma cells in S phase (EdU+) assessed after 48 h of co-culture with WT or Abca1/Abcg1 KO BMDMs previously educated in MES-like TCM ± myelin (3 h).
(G) Quantification of viable glioblastoma cells (ZombieNIR−, Annexin V−) as a percentage of total tumor cells (CD45−CD11b− cells) in ex vivo assays (Figure S7E). PDG-Ink4a glioblastoma from tumor-bearing mice was dissociated into single cells and cultured in media supplemented with lipid-rich FBS. Myeloid cells were either maintained (+CD11b+) or depleted (−CD11b+) from the dissociated tumor, and the ABCA1 inhibitor valspodar (ABCA1i) or LXR inhibitor GSK2033 (LXRi) were added.
(H and I) Quantification of viable glioblastoma cells (ZombieNIR−, Annexin V−) as a percentage of total tumor cells (CD45−CD11b− cells) in ex vivo assays (Figure S7E). PDG-Ink4a glioblastoma from tumor-bearing mice was dissociated into single cells and cultured in media supplemented with lipid-free FBS. Myeloid cells were depleted (−CD11b+) before addition of WT or Abca1/Abcg1 KO BMDMs (previously exposed to myelin debris or not) to the dissociated tumor ± LXR inhibitor GSK2033 (LXRi). Data points from non-treated (no Tx) controls in (I) are taken from (H).
Statistics: Fisher’s exact test in combination with the Benjamini-Hochberg method for correction of multiple hypotheses testing (B), two-way ANOVA with Sídák correction for multiple comparisons (E and G–I), or mixed-effects analysis with Tukey’s correction for multiple comparisons (F). Data are represented as mean + SEM (E–I).
Macrophages largely transport cholesterol in its esterified form,50 prompting us to trace cholesterol/CE integration within MES-like tumor cells using cholesterol esters with a clickable handle on either the cholesterol component or on the fatty acid (FA) chain.51 Interestingly, cholesterol located into structures resembling lipid droplets early on, in addition to apparent incorporation into membranous structures after 24 h (Figure S8C). Distinctively, the initially diffused distribution of FAs suggested incorporation into the cytoplasm, while over time, FAs accumulated within lipid droplets (Figure S8C). Orthogonally, we used uniformly labeled oleic acid (U13C-FA18:1) as carbon-tracer of the FA group of cholesteryl oleate (CE13C-FA), which showed that U13C-FA18:1 remained mostly unmetabolized after incorporation in MES-like cancer cells with neither elongation nor shortening (Figure 6F). Minute labeling of oleoyl-carnitine (CAR 18:1), an intermediate necessary for directing U13C-FA18:1 toward mitochondrial metabolism, and the undetected labeling of the tricarboxylic acid (TCA) cycle intermediates citrate, malate, and aspartate indicated that CE-derived FAs were not integrated into the TCA cycle for FA oxidation or energy production (Figure 6F). Rather, metabolized CE13C-FA derived FAs were integrated into phospholipids (predominantly phosphatidylcholines [PCs]) (Figure 6G), suggestive of their role as cell membrane building blocks, in line with click-chemistery-based immunofluorescence staining (Figure S8C). Taken together, these results indicate that by metabolizing myelin, LLMs not only shelter cancer cells from lipotoxicity but actively supply them with myelin-derived sterols and FAs, which are either stored or used as building blocks for the biosynthesis of cellular membranes.
We next interrogated the functional consequences of LLM ability to directly transfer myelin-derived lipid sources to glioblastoma MES-like cells. In co-culture, myelin-loaded BMDMs enhanced MES-like cell proliferation (Figures 6H and S8D), in a cell-cell contact-dependent manner (Figure S8E). The requirement for cell-cell contact was further supported by EM imaging, which revealed tight interactions between BMDMs and MES-like glioblastoma cells and release/uptake of coated vesicles between the two cell types (Figure 6I). These results support the notion that both passive and active mechanisms are involved in tumor cell uptake of myelin-derived lipids from LLMs. Indeed, pharmacological inhibition of either the lipid receptor CD36 or the lipid exporter ABCA1, both upregulated in LLMs (Figures 3I and S6I) and regulated by the LXR pathway,44,52 abrogated the pro-proliferative effects bestowed by LLMs on MES-like glioblastoma cells (Figure 6J). These results were genetically validated using BMDMs isolated from LysM-Cre Abca1/Abcg1fl/fl mice (Figure S8F).53
Interestingly, inhibition of either ABCA1 or LXR significantly reduced ex vivo tumor cell viability, specifically in lipid-free media co-cultures, a condition in which tumor cells rely on LLM-supplied lipids subsequent to myelin uptake (Figures 6K and S8G). These results demonstrate that the inhibitory effects of LXR and ABCA1 blockade on tumor cell growth relies on the exchange of lipids between LLMs and tumor cells, as these inhibitors did not hinder glioblastoma cell viability in lipid-rich medium or in absence of macrophages. This mechanism was further validated using LysM-Cre Abca1/Abcg1fl/fl macrophages in ex vivo co-cultures, confirming that the pro-proliferative effect of LLMs toward MES-like cancer cells was dependent on both Abca1/Abcg1 and LXR (Figures S8H and S8I). Altogether, these findings reveal a symbiotic relationship between LLMs and MES-like tumor cells, whereby glioblastoma cells instruct LLM formation through promotion of their lipid-uptake capability. Reciprocally, myelin-loaded LLMs fuel MES-like glioblastoma cell proliferation through ABCA1-mediated cholesterol/cholesterol ester efflux and lipid transfer. Therefore, preventing lipid transfer from LLMs to glioblastoma cells curbs LLMs pro-tumorigenic effects.
LLMs predict glioblastoma survival and response to immunotherapy
To assess the translational potential of our findings, we investigated the prognostic value of the LLM signature (Figure 1I) using The Cancer Genome Atlas54 (TCGA) and the Glioma Longitudinal Analysis (GLASS) Consortium55 datasets. We stratified primary glioblastoma samples into LLMlow and LLMhigh groups and determined that LLMhigh patients presented a significantly worse prognosis (Figure 7A). As expected, most glioblastoma samples included in the LLMhigh signature group were classified as MES-enriched (Figures 7B–7E). Nonetheless, when MES tumors were excluded from these analyses, the LLMhigh classification of glioblastoma patients retained its association with reduced survival (Figure 7F). Altogether, these data highlight the potential of LLMs as a diagnostic and classification tool, eventually applicable to glioblastoma patient stratification into personalized therapy approaches.
Figure 7.
Lipid-laden macrophages predict patients’ clinical outcome
(A) Kaplan-Meier curves of glioblastoma patients54,55 stratified by low and high LLM signature enrichment in primary Isocitrate dehydrogenase wild-type (IDHWT) glioblastoma patients.
(B and C) Boxplots depicting the single-sample (ss) GSEA scores of the LLM signature for all IDHWT glioblastoma samples segregated according to their dominant transcriptional subtype (CL, classical; MES, mesenchymal; PN, proneural) in the (B) TCGA54 and (C) GLASS datasets.55
(D and E) Heatmap of the LLM gene signature expression in primary IDHWT glioblastoma tumors stratified by low/high LLM enrichment scores based on ssGSEA scores.
(F) Kaplan-Meier curves of glioblastoma patients54,55 stratified by low and high LLM signature enrichment in primary IDHWT glioblastoma patients excluding MES glioblastoma.
(G) Left: schematic overview of pan-cancer TAM analysis.5,19,56 Right: violin plot depicting the average LLM signature scores in TAMs.
(H) Percentage of LLMs in scRNA-seq datasets of treatment-naive, recurrent, and neoadjuvant anti-PD-1-treated glioblastoma patient samples.57
(I) Percentage of non-LLM or LLM TAMs in responder and non-responder melanoma patients58 prior to ICB treatment.
(J) Receiver operating characteristic curves comparing the predictive value of tumor mutational burden, LLMs, non-LLMs, and total macrophages as a percentage of CD45+ cells to predict ICB response in melanoma. Statistics: log-rank test (A and F), Kruskal-Wallis test and the pairwise comparisons with Wilcoxon-test (B and C), one-way ANOVA with Sídák correction for multiple comparisons (H), two-tailed paired t test (I), or unpaired two-sided bootstrap test (J). Data are represented as mean ± SEM (H and I).
To place the prevalence of LLMs into a broader perspective and assess the significance of these cells in the scope of different cancer types, we queried the LLM transcriptional gene signature expression in pan-cancer myeloid cell scRNA-seq.56 Notably, the proportion of TAMs enriched for the LLM signature varied greatly between glioblastoma, lung cancer, and ovarian cancer patients, while melanoma patients displayed overall low levels of LLMs, despite a subset of these exhibiting heightened LLM enrichment (Figure 7G). In light of the immunosuppressive features of LLMs (Figures 3I and S1J), we assessed whether the LLM signature could predict T cell-directed immune checkpoint blockade (ICB) response in cancer patients.58 In recent clinical trials, neoadjuvant anti-PD-1 (Programmed cell death protein 1) was incorporated to resectable recurrent glioblastoma and showed significant survival benefit compared with adjuvant treatment.59 We queried the scRNA-seq dataset of patients that underwent such therapies,57 which revealed that neoadjuvant anti-PD-1 treated glioblastoma presented significantly lower percentage of TAM-LLMs compared with treatment-naive or recurrent glioblastoma (Figure 7H). We thus hypothesized that the LLMhigh transcriptional signature found in glioblastoma patients may account for their poor responsiveness to ICB, which remains to be confirmed as established ICB responder and non-responder datasets are not available for glioblastoma patients. We therefore interrogated publicly available datasets of melanoma patients treated with immunotherapy to assess whether LLM enrichment was predictive of a therapeutic benefit in a patient population that favorably respond to ICB.58 Strikingly, the content of LLMs predicted treatment response in melanoma, while total TAM levels did not (Figure 7I). Moreover, using LLMs as biomarkers of ICB response yielded a superior predictive value than either total TAM content or the commonly employed tumor mutational burden (TMB) (Figure 7J). Altogether, these findings support the relevance of LLMs as a predictive cell type for glioblastoma patient survival and response to immunotherapy.
Discussion
The multifaceted phenotype of tissue-resident and infiltrating macrophages is shaped by diverse cues, including the metabolic landscape of the TME in which they evolve.12,60 In this study, we moved beyond multi-omics description of the glioblastoma TME and uncovered an intricate metabolic crosstalk between TAMs and cancer cells that relies on recycling of cholesterol-rich myelin debris. Glioblastoma cells utilize neuronal homeostatic functions to their advantage, for instance through neuronal mimicry.61,62 Comparably, we propose that glioblastoma cells hijack and exacerbate the ability of macrophages to orchestrate de- and remyelination during development and homeostasis44,63 by co-opting cellular pathways that promote engulfment of myelin debris,64 leading to lipid overloading in TAMs as a result of chronic exposure to myelin debris. This metabolic interplay benefits MES-like glioblastoma cells, whereby TAMs’ ability to uptake, recycle, and process myelin into freely available lipids fuel their survival and proliferation. Notably, such lipid exchange between myeloid cells and cancer cells had been observed in prostate and lung cancers.53,65
Given that circulating cholesterol does not cross the blood-brain barrier and that the brain environment is metabolically autonomous,66 our data suggest MES-like glioblastoma cells rely heavily on local cholesterol metabolism29 and are thus required to harness the homeostatic functions of resident cells to advance into malignancy. This metabolic crosstalk is particularly enriched in hypoxic areas where lipid-auxotrophic MES-like glioblastoma cells preferentially home, and scavenging of extracellular lipids is preferred over de novo lipid synthesis.67 The complex glioblastoma environment is a cornerstone of therapy resistance,11 eventually culminating into the generation of a TME that has dynamically adapted in recurrent, often MES tumors. Our findings that LLMs are enriched in this context raise the question of their role as a dominant driver of such transition, orchestrating adaptive mechanisms supportive of glioblastoma relapse post-therapy.
LLMs display immunosuppressive features and negatively correlate with response to ICB, supporting the notion that LLMs actively participate to the glioblastoma immunosuppressive microenvironment. These findings are in line with previous studies in multiple sclerosis, showing myelin-phagocytosis in macrophages directly dampens T cell proliferation and neuroinflammation.43,68 Surprisingly, however, targeting LLMs in vivo did not lead to overt changes in lymphoid populations frequency in end-stage tumors. In light of the controversial roles lipids play in immune activation,69 careful examination of the (in-)direct roles of TAM-mediated myelin recycling on lymphocytes functions will be central to designing combinatorial treatment approaches bolstering anti-tumoral response.
In summary, we present in-depth functional analyses of a pro-tumorigenic LLM subset in glioblastoma, spanning from studying their transcriptional education and chromatin alterations to their re-configured lipidome. We reveal that lipid-laden-TAMs’ pro-tumoral functions are largely driven by the excessive, TME-instructed uptake of myelin debris. The discovery of LLMs’ role in fueling glioblastoma malignancy warrants further investigation into efficient strategies targeting the metabolic co-evolution processes occurring in TAM subsets to acquire specified pro-tumorigenic functions.
Limitations of the study
This study identifies macrophage-mediated myelin recycling as a mechanism underlying their pro-tumorigenic phenotype in glioblastoma, with a primary focus on the role of cholesterol and FAs in this process. However, myelin is composed of a wide range of lipids and proteins, which can also affect macrophage phenotype and functions. Further experiments focused on the exact mechanism behind myelin recycling and transfer will be important to define other important regulators of the lipid-laden phenotype. In addition, mimicking metabolic crosstalk between macrophages and MES-like tumor cells in vitro or ex vivo may not fully capture the complexity of the TME, in which multiple variables, such as oxygen and nutrient availability greatly impact cellular interactions.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| FCM: Annexin V APC, dilution 1:150 | BioLegend | Cat# 640920 |
| FCM: CD11b BV650 (clone M1/70), dilution 1:400 | BD Biosciences | Cat# 563402; RRID:AB_2738184 |
| FCM: CD11b BV786 (clone M1/70), dilution 1:1200 | BioLegend | Cat# 101243; RRID:AB_2561373 |
| FCM: CD11c BV421 (clone N418), dilution 1:400 | BioLegend | Cat# 117329; RRID:AB_10897814 |
| FCM: CD11c PE-Cy5.5 (clone N418), dilution 1:200 | Life Technologies | Cat# 35-0114-82; RRID:AB_469709 |
| FCM: CD19 BV510 (clone 6D5), dilution 1:200 | Biolegend | Cat# 115546; RRID:AB_2562137 |
| FCM: CD25 APC (clone PC61), dilution 1:400 | BioLegend | Cat# 102012; RRID:AB_312861 |
| FCM: CD3 PerCP-eFluor710 (clone 145-500A2), dilution 1:300 | eBioscience | Cat# 46-0081-82 |
| FCM: CD31 BV421 (clone 390), dilution 1:400 | BioLegend | Cat# 102424; RRID:AB_2650892 |
| FCM: CD36 APC (clone HM36), dilution 1:150 | BioLegend | Cat# 102612; RRID:AB_2072639 |
| FCM: CD39 SB702 (clone 24DMS1), dilution 1:100 | eBioscience | Cat# 67-0391-82; RRID:AB_2717143 |
| FCM: CD4 BV711 (clone RM405), dilution 1:100 | BioLegend | Cat# 100550; RRID:AB_2562099 |
| FCM: CD44 BV605 (clone IM7), dilution 1:100 | BioLegend | Cat# 103047; RRID:AB_2562451 |
| FCM: CD45 AF488 (clone 30-F11), dilution 1:200 | BioLegend | Cat# 103122; RRID:AB_493531 |
| FCM: CD45 AF700 (clone 30-F11), dilution 1:200 | BioLegend | Cat# 103128; RRID:AB_493715 |
| FCM: CD49d PE-Cy7 (clone R1-2), dilution 1:150 | BioLegend | Cat# 103618; RRID:AB_2563700 |
| FCM: CD69 PE/Dazzle (clone H1.2F3), dilution 1:200 | BioLegend | Cat# 104536; RRID:AB_2565583 |
| FCM: CD8 FITC (clone 53-6.7), dilution 1:200 | Life Technologies | Cat# 11-0081-82 |
| FCM: EZH2 AF488 (clone D2C9), dilution 1:50 | Cell Signaling Tech. | Cat# 30233S; RRID:AB_2798987 |
| FCM: Fc blocking reagent (aCD16/CD32), dilution 1:400 | BD Biosciences | Cat# 553142; RRID:AB_394657 |
| FCM: FoxP3 PE/Cy5.5 (clone PJK-16s), dilution 1:200 | Life Technologies | Cat# 35-5773-82; RRID:AB_11218094 |
| FCM: GrzB PE (clone QA16A02), dilution 1:200 | BioLegend | Cat# 372208; RRID:AB_2687032 |
| FCM: H3K27me3 AF657 (clone C36B11), dilution 1:1200 | Cell Signaling Tech. | Cat# 12158S; RRID:AB_2797834 |
| FCM: Ki67 BV785 (clone B56), dilution 1:200 | BD Biosciences | Cat# 563756; RRID:AB_2732007 |
| FCM: Ly6C APC-Cy7 (clone HK1.4), dilution 1:400 | BioLegend | Cat# 128026; RRID:AB_10640120 |
| FCM: Ly6C BV605 (clone HK1.4 ), dilution 1:400 | BioLegend | Cat# 128036; RRID:AB_2562353 |
| FCM: Ly6G BV605 (clone 1A8), dilution 1:200 | BD Biosciences | Cat# 563005; RRID:AB_2737946 |
| FCM: Ly6G BV711 (clone 1A8), dilution 1:200 | BioLegend | Cat# 127643; RRID:AB_2565971 |
| FCM: MHC II PerCP-eFluor710 (clone M5/114.15.2), dilution 1:1000 | Invitrogen | Cat# 46-5321-82; RRID:AB_1834439 |
| FCM: NK1.1 PE-Cy7 (clone PK136), dilution 1:200 | Biolegend | Cat# 108714; RRID:AB_389364 |
| FCM: PD-1 BV421 (clone J43), dilution 1:500 | BD Biosciences | Cat# 562584; RRID:AB_2737668 |
| FCM: PD-L1 PE (clone 10F.9G2), dilution 1:400 | BioLegend | Cat# 124308; RRID:AB_2073556 |
| IF: anti-goat AF568 donkey, dilution 1:500 | Thermo Fisher Scientific | Cat# A11057; RRID:AB_2534104 |
| IF: anti-rabbit AF647 donkey, dilution 1:500 | Thermo Fisher Scientific | Cat# A31573; RRID:AB_2536183 |
| IF: anti-rat AF488 donkey, dilution 1:500 | Thermo Fisher Scientific | Cat# A21208; RRID:AB_2535794 |
| IF: CD31 rat, dilution 1:200 | Novus Bio | Cat# NB600-1475; RRID:AB_789108 |
| IF: Hypoxyprobe-RedAPC-Mab mouse, dilution 1:1000 | Hypoxyprobe™ | Cat# HP8-100kit |
| IF: IBA1 goat, dilution 1:1000 | Novus Bio | Cat# NB100-1028; RRID:AB_521594 |
| IF: MBP rat, dilution 1:400 | Sigma-Aldrich | Cat# MAB386; RRID:AB_94975 |
| IF: P2RY12 rabbit, dilution 1:200 | Thermo Fisher Scientific | Cat# 702516; RRID:AB_2689476 |
| IF: PLIN2 rabbit, dilution 1:200 | Novus Bio | Cat# NB110-40877; RRID:AB_787904 |
| Chemicals, peptides, and recombinant proteins | ||
| PBS | Gibco | Cat# 18912014 |
| DMEM | Gibco | Cat# 61965059 |
| RPMI-1640 | Gibco | Cat# 11875093 |
| Penicillin-Streptomycin | Roche | Cat# 110774440001 |
| Trypsin-EDTA | Gibco | Cat# 15400-054 |
| DMSO | Sigma-Aldrich | Cat# 276855 |
| PFA | Sigma-Aldrich | Cat# P6148-500G |
| TRIzol™ | Thermo Fisher Scientific | Cat# 15596026 |
| Triton | Sigma-Aldrich | Cat# 9002-93-1 |
| UltraPure™ Glycine | Invitrogen | Cat# 15527-013 |
| Saponin | Sigma-Aldrich | Cat# S4521 |
| HBSS | Gibco | Cat# 24020-133 |
| Poly-D-Lysine | Thermo Fisher Scientific | Cat# A3890401 |
| RLT Buffer | Qiagen | Cat# 79216 |
| BSA | Roche | Cat# 03116964001 |
| FBS | Capricorn Scientific | Cat#FBS-12A |
| Lipid-depleted FBS | Biowest | Cat# S181L-500 |
| CSF1 | R&D | Cat# 4160ML |
| IL-34 | R&D | Cat# 5195-ML |
| TGF-β | R&D | Cat# 7666-MB |
| Sulfosuccinimidyl oleate (SSO) | Cayman | Cat# Cay11211 |
| Alkyne Cholesterol | Click Chemistry tools | Cat# 1409-5 |
| Alkyne Cholesterol-ester (FA-handle) | This paper | N/A |
| Alkyne Cholesterol-ester (Cholesterol-handle) | This paper | N/A |
| Oleic Acid (U-13C18, 98%) | Cambridge Isotope Laboratories | Cat# CLM-460-0.005 |
| BD brilliant stain buffer | BD Biosciences | Cat# 566349 |
| BODIPY™ 493/503 | ThermoFisher | Cat# D3922 |
| Zombie Aqua | BioLegend | Cat# 423102 |
| Zombie NIR™ | BioLegend | Cat# 423105 |
| DAPI | Thermo Fisher Scientific | Cat# D3571 |
| Pimonidazole | Hypoxyprobe™ | Cat# HP8-100kit |
| Fluorescent mounting medium | Dako | Cat# GM30411-2 |
| Optimal Cutting Temperature (OCT) compound | Tissue-Tek | Cat# SA62550 |
| Critical commercial assays | ||
| Brain Tumor Dissociation Kit | Miltenyi | Cat# 130-095-942 |
| Adult Brain Dissociation Kit | Miltenyi | Cat# 130-107-677 |
| Fixation/Permeabilization Concentrate | Thermo Fisher Scientific | Cat# 00-5123-43 |
| Fixation/Permeabilization Diluent | Thermo Fisher Scientific | Cat# 00-5223-56 |
| Permeabilization buffer | Ebiosciences | Cat# 00-8333-56 |
| BD brilliant stain buffer | BD Biosciences | Cat# 566349 |
| Click-iT™ EdU Alexa Fluor™ 647 Flow Cytometry Assay Kit | Thermo Fisher Scientific | Cat# C10419 |
| Cholesterol Assay Kit | Abcam | Cat# ab133116 |
| Myelin Removal Beads II | Miltenyi | Cat# 130-096-433 |
| CD11b Microbeads | Miltenyi | Cat# 130-093-634 |
| UltraComp eBeads™ Compensation Beads | Ebiosciences | Cat# 01-2222-42 |
| Seahorse XFe24 FluxPaks mini | Agilent | Cat# 102342-100 |
| Seahorse XF Glycolytic Rate Assay Kit | Agilent | Cat# 103344-100 |
| Seahorse XF Cell Mito Stress Test Kit | Agilent | Cat# 103015-100 |
| FluoSpheres™ | Thermo Fisher Scientific | Cat# F8803 |
| Transcriptor First Strand cDNA Synthesis Kit | Roche | Cat# 50-997-552 |
| TaqMan™ Fast Advanced Master Mix | Thermo Fisher Scientific | Cat# 4444557 |
| Deposited data | ||
| Raw and processed data | This paper | GEO: GSE266149 |
| scRNA-seq Myeloid Cell Atlas dataset | Cheng et al.56 | GEO: GSE154763 |
| scRNA-seq Melanoma ICB dataset | Sade-Feldman et al.58 | GEO: GSE120575 |
| scRNA-seq Glioblastoma ICB dataset | Lee et al.57 | GEO: GSE154795 |
| RNAseq and Survival Glioblastoma dataset | TCGA | http://gdac.broadinstitute.org/ |
| RNAseq and Survival Glioblastoma dataset | The GLASS Consortium70 | https://www.synapse.org/Synapse:syn17038081/wiki/585622 |
| Experimental models: Cell lines | ||
| Mouse primary PDG-Ink4a glioblastoma | This paper | N/A |
| Mouse recurrent PDG-Ink4a glioblastoma | This paper | N/A |
| DF1-PDGFB-HA, DF1 chicken fibroblasts (ATCC) with RCAS viral vector expressing PDGFB-HA | Dr Tatsuya Ozawa and Dr Eric Holland | N/A |
| DF1-PDGFA-HA, DF1 chicken fibroblasts (ATCC) with RCAS viral vector expressing PDGFA-HA | Dr Tatsuya Ozawa and Dr Eric Holland | N/A |
| DF1-shP53, DF1 chicken fibroblasts (ATCC) with a short hairpin against murine p53 | Dr Tatsuya Ozawa and Dr Eric Holland | N/A |
| DF1-shNF1, DF1 chicken fibroblasts (ATCC) with a short hairpin against murine NF1 | Dr Tatsuya Ozawa and Dr Eric Holland | N/A |
| Experimental models: Organisms/strains | ||
| Mouse: C57BL/6J mice | The Jackson Laboratory | RRID:IMSR_JAX:000664 |
| Mouse: mT/mG (C57BL/6J background) | The Jackson Laboratory | RRID:IMSR_JAX:007576 |
| Mouse: Nestin-Tv-a;Ink4a/Arf-/- (C57BL/6J background) | Holland et al.71 | N/A |
| Mouse: BL/6 Nestin-Tv-a (C57BL/6J background) | Holland et al.71 | N/A |
| Mouse: LysM-Cre Abca1/Abcg1fl/fl (C57BL/6J background) | El-Kenawi et al.53 | N/A |
| Software and algorithms | ||
| FlowJo, version 10.8.0 | BD Bioscience | https://www.flowjo.com/ |
| MIPAV, 11.0.8 | Center for Information Technology - NIH | https://mipav.cit.nih.gov/ |
| GraphPad Prism, version 10.0.3 (217) | GraphPad Software | https://www.graphpad.com/ |
| ZEN software, version 3.3 | Zeiss | https://www.zeiss.com/microscopy/en/products/software.html |
| Huygens software, version 23.04 | Scientific Volume Imaging | https://svi.nl/Huygens-Software |
| R environment, version 4.3.2 | The R Foundation | https://www.r-project.org |
| RStudio, version 2023.09.1+494 | RStudio | https://www.posit.co/ |
| Other | ||
| Resource website for the code used in this paper | This paper | https://github.com/djkloosterman/Kloosterman_et_al_Cell_2024 |
| gentleMACS Octo Dissociator | Miltenyi | Cat# 130-095-037 |
| gentleMACS C Tubes | Miltenyi | Cat# 130-096-334 |
| LS Columns | Miltenyi | Cat# 130-042-401 |
| LSRFortessa, flow cytometry | BD Biosciences | N/A |
| FACSAria Fusion, flow cytometry & cell sorter | BD Biosciences | N/A |
| Tecnai12G2 Electron Microscope | Thermo Fisher Scientific | N/A |
| Axio Scan Z1 slide scanner | Zeiss | N/A |
| Zeiss Confocal LSM 980 Airyscan 2 | Zeiss | N/A |
| Cryomold | Tissue-Tek | Cat# 4557 |
| Super PAP Pen Liquid Blocker | Science Services | Cat# N71312-N |
| Cover glass | VWR | Cat# 631-0662 |
| IHC Microscope Slides | Dako Agilent | Cat# K8020 |
| Illumina NovaSeq 6000 Sequencer | Illumina | N/A |
| Illumina NextSeq 550 Sequencer | Illumina | N/A |
| QuantStudio™ 6 Flex Real-Time PCR System | Thermo Fisher Scientific | N/A |
| Teflon bags | OriGen | Cat# PL07-PL30 |
| 70μm Cell Strainer | Falcon | Cat# 352350 |
| 40μm Cell Strainer | Falcon | Cat# 352340 |
| 20μm Nylon Net Filters, 47mm | Merck | Cat# NY2004700 |
| Amicon® Ultra-15 Centrifugal Filter Unit | Milllipore | Cat# UFC901096 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Leila Akkari (l.akkari@nki.nl).
Materials availability
Materials Availability Materials and reagents used in this study are listed in the key resources table. Reagents generated in our laboratory in this study or previous studies are available upon request. Extended figure legends have been made available on at https://github.com/djkloosterman/Kloosterman_et_al_Cell_2024.
Data and code availability
-
•
Data generated in this study was aligned using the mouse reference genome mm10 and is deposited on GSE266149. The glioblastoma niche and subtype meta-module gene lists used in this study can be obtained from the Ivy Glioblastoma Atlas Project (GAP) or Neftel et al.18 (Table S2), respectively. The glioblastoma patient scRNA-seq datasets were kindly provided to us by Dr. Kevin Petrecca, McGill University.5,19 The publicly available single-cell RNA sequencing datasets which have been used in this study, including the myeloid cell atlas,56 the melanoma treatment-related dataset,58 and the response of glioblastoma patients to neoadjuvant aPD157 can be obtained from GEO: GSE154763, GSE120575, and GSE154795, respectively. TCGA-glioblastoma data was obtained from http://gdac.broadinstitute.org/ using the package UCSCXenaTools.72 The raw counts were processed using EdgeR.73 The GLASS dataset70 was downloaded from https://www.synapse.org/#!Synapse:syn17038081/wiki/585622, where patients for glioblastoma and IDH-wild-type were selected.
-
•
The original codes for the murine scRNA-seq and Visium spatial transcriptomic analyses are made available at https://github.com/djkloosterman/Kloosterman_et_al_Cell_2024.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Experimental model and study participant details
Glioblastoma mouse model and treatment
Nestin-TV-a;Ink4a/Arf-/- mice (BL/6 background) and Nestin-Tv-a mice (BL/6 background) have been previously described8,9,10,11,74 and were bred within the animal facilities of the Netherlands Cancer Institute (NKI). Briefly, gliomas were initiated by injection of DF-1 cells expressing an RCAS (Replication-Competent ASLV long terminal repeat (LTR) with a Splice Acceptor) vector encoding PDGF-B HA in male and female Nestin-Tv-a;Ink4a/Arf-/- mice (PDG-Ink4a model) aged between 5-7 weeks, or DF-1 cells expressing PDGF-B HA and a short hairpin RNA targeting TP53 in Nestin-Tv-a mice (PDG-p53 model) or DF-1 expressing PDGF-A HA, a short hairpin RNA targeting NF1 and a short hairpin RNA targeting TP53 in male and female Nestin-Tv-a mice (PDGFA-NF1-P53 model) aged between 5-7 weeks.
Mice underwent weekly or bi-weekly MRI scans which were measured as previously described.11 Following emergence of brain tumors (tumor volume 30-90 mm3), mice were sedated with isoflurane and head was irradiated using a X-RAD 320 (Precision X-Ray) at 115 cGy/min. Radiation was dosed at 2 Gy/day for five days. Animals were euthanized upon recurrence of the tumor as monitored by regular MRI imaging, or by neurological symptoms.
Sulfosuccinimidyl oleate (SSO) obtained from Cayman (cat.no. Cay11211) was dissolved in DMSO (66mg/ml) and diluted 1:20 in PBS prior to intraperitoneal (i.p.) administration. SSO (30mg/kg) or vehicle control (5% DMSO in PBS) was injected daily. All mouse procedures were approved by the animal ethics committee of the Netherlands Cancer Institute and performed in accordance with institutional, national and European guidelines for animal care and use. Mice were euthanized at defined time points as indicated, or when symptomatic. Euthanasia was performed by carbon dioxide asphyxiation.
Method details
Experimental design
Glioblastoma were initiated in the PDGFB-Ink4a/Arf KO (PDG-Ink4a) and PDGFB-p53 KD (PDG-p53) GEMM models (as described in STAR Methods). Mice were assigned to primary control (treatment-naïve) or fractionated ionizing radiotherapy (RT- 5 x 2 Gy) groups. Tumors of comparable volumes (determined by MRI) were collected when animals reached the humane endpoint in the primary (n = 3 PDG-Ink4a, n = 4 PDG-p53) or recurrence post-RT (n = 3 PDG-Ink4a, n = 3 PDG-p53) treatment groups and processed into single-cell suspensions. CD45+ immune cells and CD45- cells were FACS-isolated and subjected to 10X Chromium single-cell RNA sequencing (scRNA-seq). Microglia (MG gated on CD45+, CD11b+, Ly6Clow, Ly6Glow, CD49dlow) and monocyte-derived macrophages (MDMs, gated on CD45+, CD11b+, Ly6Clow, Ly6Glow, CD49d+) were FACS-purified from independent cohorts of non-tumor bearing animals (only MG, n = 1 Ntv-a), primary (n = 1 PDG-Ink4a, n = 1 PDG-p53) or recurrent (n = 1 PDG-Ink4a, n = 1 PDG-p53) and subjected to 10X Chromium scRNA-seq to complement the pool of CD45+ cells. Independent cohort of PDG-Ink4a animals were sacrificed, and snap-frozen sections were processed for Visium 10X spatial transcriptomics (n = 4 PDG-Ink4a primary; n = 3 PDG-Ink4a recurrent, n = 3 PDG-Ink4a primary; n = 3 PDG-Ink4a recurrent). Published patient datasets, including Ivy GAP glioblastoma niche signatures2 and glioblastoma patient scRNA-seq,19 were included in downstream analyses to assess the translational value of the mouse sequencing results.
Flowcytometry-based cell surface marker analysis
Flow cytometry procedures were performed as previously described.10,11 Briefly, following euthanasia, mouse brain tumors were macrodissected and dissociated using the Tumor Dissociation Kit (Miltenyi, cat.no. 130-096-730) and a single cell suspension was generated using the OctoMACS dissociator (Miltenyi). All antibodies used for flow cytometry were titrated in a lot-dependent manner and are listed in the key resources table. To measure neutral lipid levels, cells were stained with 2.5ng/mL BODIPY 493/503 (Thermofisher, cat.no. D3922) for 20 minutes at 37 degrees. Analyses were completed on a BD Fortessa instrument at the Flow Cytometry Core Facility of NKI. Data were analyzed using the FlowJo v10 software.
Fluorescence-activated cell sorting
To isolate macrophages populations from murine glioblastoma, single cell suspensions were stained with antibodies as listed in the key resources table. Viable cells (NIRlow) were sorted based on CD11bhighCD45highLy6ClowLy6GlowBODIPYhighSSC-Ahigh and CD11bhighCD45highLy6ClowLy6GlowBODIPYlowSSC-Alow to isolate LLMs and non-LLMs respectively, as described in Figure S2E.
For single-cell RNA sequencing (scRNA-seq), single cell suspensions were stained with live/dead staining using Zombie NIR (Biolegend, cat.no. 423106) and CD45-AF488 as listed in the key resources table. Single viable cells (NIRlow) were sorted based on CD45 expression (negative and positive) in 2% FBS PBS. All sorting was performed on a Fusion II instrument (BD) using a 100μm nozzle. Single cell suspensions were centrifuged at 300rcf at 4 degrees for 5 minutes and washed with PBS with 0.04% BSA, before resuspension at a concentration of 1000 cells per μl in PBS with 0.04% BSA for 10x Chromium scRNA-seq purpose. When prepared for mass spectrometry, single cell suspensions were washed with PBS and snap-frozen on dry ice.
Tissue and cell imaging
For immunofluorescence (IF) staining, mice were intracardially perfused with 20 ml of PBS. Whole brains were collected, sectioned in 4 slices, embedded and frozen in optimal cutting temperature (OCT) compound (Tissue-Tek, cat.no. SA62550), and 10 μm cryostat tissue sections were cut. When indicated, mice were injected i.p. with 1.5mg of pimonidazole (Hypoxyprobe™, cat.no. HP8-100kit) 1h prior to euthanasia. Tissue sections or plated cells were thawed and dried at room temperature, fixed in 3% paraformaldehyde for 10min at room temperature, washed in PBS, and blocked in PBS supplemented with 2% BSA, 0.1% saponin and 0,.2M glycine for 1h at room temperature or permeabilized with 0.01% Triton in PBS for 10min at room temperature when stained for clickable lipids. Sections were then incubated with primary antibodies overnight at 4°C, followed by PBS washes and incubation with fluorescent secondary antibodies for 45min at room temperature. When indicated, neutral lipids were stained with 1:500 BODIPY 493/503. When specified, a click reaction (Click-iT™ EdU Alexa Fluor™ kit, Thermofisher, cat.no. C10337) mix was prepared to stain the clickable lipids for 30min at RT, followed by two washes with 2% BSA in PBS and one wash with PBS alone. All antibodies and dilution factors are listed in the key resources table. Antibodies were diluted in PBS + 2% BSA + 0.1% saponin. DAPI (Thermofisher, cat.no. D3571) was used as a counterstain prior to mounting with fluorescent mounting medium (Dako, cat.no. GM30411-2).
3D Whole slide immunofluorescent images were acquired with a Zeiss AxioScan Z1 slide scanner, equipped with a high-resolution Hamamatsu Orca FLASH 4.0 V3 camera and a 20x/0.8 objective (Zen 2.6 Zeiss software) with a pixel size 0.325x0.325x1μm. After acquisition, z-stack tiled images were deconvoluted and stitched in Huygens (SVI). Confocal images were made using the super-resolution Airyscan setting on a Zeiss Confocal LSM 980 Airyscan 2, equipped with a 63x / 1.40 oil Plan-ApoChromat (Zeiss 440756) objective (Zen 3.3 Zeiss software) and images were processed with Airyscan processing in ZEN software (Zeiss).
Visium10x spatial transcriptomics
Samples were collected as described above for IF staining. Fresh-frozen samples were cryo-sectioned and placed on Visium Tissue Optimization Slides and Spatial Gene Expression Slides (10X Genomics) that were pre-equilibrated at -20°C. Tissue sections were then fixed in chilled methanol and stained with rat anti-CD31 followed by donkey anti-rat AF488 (as listed in the key resources table), according to the Visium Tissue Fixation & Immunofluorescence staining guide (CG000312 Rev C, 10X Genomics). Optimal permeabilization time was determined using the Visium Tissue Optimization slide according to the manufacturer’s protocol (CG000238 Rev D), and tissue was permeabilized for 12min for gene expression samples. Immunofluorescent images were taken using a 10X objective on a Zeiss observer Z1 (Carl Zeiss Microscopy). Raw images were stitched using the Zen 2.6 software (Zeiss), and image was optimized by removal of background autofluorescence with Fiji software.
Interstitial fluid collection
To collect tumor interstitial fluid, tumors were macrodissected and weighed, then rinsed in PBS and placed on top of a 20μm pore sized nylon net filter (Merck Millipore, cat.no. NY2004700) in an Eppendorf tube. After centrifugation for 15min at 400g, 4°C, TIF was collected from the bottom of the Eppendorf tube and snap-frozen on dry ice.
Sample preparation for lipidomics analysis
Lipid extraction was conducted by adding 250 or 750 μL of 2-propanol to the tumor interstitial fluid (TIF) and isolated myelin or macrophage samples, respectively. Samples were then vortexed for 10 s and sonicated in an ultrasound bath for 5 min at room temperature. Subsequently, samples were centrifuged at 18,000g for 3 min at 20 °C and the resulting supernatants were transferred to an HPLC glass vial. The extraction was repeated with 250 μL of 2-propanol and organic extracts were combined. Samples were dried under a gentle stream of nitrogen and reconstituted in 50 μL of 2-propanol, vortexed for 10 s and sonicated for 5 min. Next, 50 μL of LC-MS grade water were added and samples were again vortexed for 10 s and sonicated for 5 min. Finally, samples were transferred to micro-vial inserts and placed in the autosampler.
Untargeted lipidomic analysis by LC-MS/MS
Lipidomic analysis of the lipid extracts was performed using a LC-MS/MS based lipid profiling method.75 A Shimadzu Nexera X2 (Shimadzu, ‘s Hertogenbosch, The Netherlands) was used to deliver a gradient of water:acetonitrile 80:20 (eluent A) and water:2-propanol:acetonitrile 1:90:9 (eluent B). Both eluents contained 5 mM ammonium formate and 0.05% formic acid. The applied gradient, with a column flow of 300 μL/min, was as follows: 0 min 40% B, 10 min 100% B, 12 min 100% B. A Phenomenex Kinetex C18, 2.7 μm particles, 50 × 2.1 mm (Phenomenex, Utrecht, The Netherlands) was used as column with a Phenomenex SecurityGuard Ultra C8, 2.7 μm, 5 × 2.1 mm cartridge (Phenomenex, Utrecht, The Netherlands) as guard column. The column was kept at 50 ˚C. The injection volume was 10 μL. The MS was a Sciex TripleTOF 6600 (AB Sciex Netherlands B.V., Nieuwerkerk aan den Ijssel, The Netherlands) operated in positive (ESI+) and negative (ESI-) ESI mode, with the following conditions: ion source gas 1 45 psi, ion source gas 2 50 psi, curtain gas 35 psi, temperature 350 ˚C, acquisition range m/z 100-1800, ion spray Voltage 5500 V (ESI+) and -4500 V (ESI-), declustering potential 80 V (ESI+) and -80 V (ESI-). An information dependent acquisition (IDA) method was used to identify lipids, with the following conditions for MS analysis: collision energy ±10, acquisition time 250 ms and for MS/MS analysis: collision energy ±45, collision energy spread 25, ion release delay 30, ion release width 14, acquisition time 40 ms. The IDA switching criteria were set as follows: for ions greater than m/z 300, which exceed 200 cps, exclude former target for 2 seconds, exclude isotopes within 1.5 Da, max. candidate ions 20. MS-DIAL (v4.70), with lipid database version Msp20210527163602, was used to align the data and identify the different lipids.76
Quantitative lipidomics analysis (NMR)
For in vitro isolated tumor cells, lipid extraction using 2-propranol was conducted until the drying step with N2. Then, 750 μL of 99.8% deuterated chloroform (Cortecnet) was added to the dried lipid extract. In the case of the myelin samples, 25 μL of the remaining lipid extract coming from the untargeted lipidomics analysis were pipetted into glass vials and dried under a N2 gas stream. 250 μL of 99.8% deuterated chloroform (Cortecnet) was added. In both cases (tumor cells and myelin samples), the vials were sonicated for 30 min after which 200 μL of the solutions were transferred to 3 mm Bruker SampleJet NMR tubes. 1H NMR data were collected using a Bruker 600 MHz Avance Neo/IVDr spectrometer equipped with a 5 mm TCI cryogenic probe head and a z-gradient system. The analysis of the NMR spectra was performed in KNIME77 using a customized KIMBLE workflow.78 The cholesterol component of the total lipid content was calculated by converting the areas to masses and taking the ratio of the cholesterol mass to sum of the cholesterol mass and the alkyl chain masses.
Quantitative lipidomics analysis (Lipidyzer)
The remaining 550 μL deuterated chloroform-sample from the previous section was dried under a stream of N2. 25 μL Lipidyzer internal standard mix, 600 μL methyl-tert-butyl ether (MTBE) and 150 μL methanol were added. The mixture was vortexed and left for 30 min at room temperature. Then, the sample was centrifuged at 18,000g for 5 min. From each sample, 750 μL of supernatant was transferred to a new Eppendorf tube. Extraction was repeated on the original sample tubes with 300 μL MTBE and 100 μL methanol. The samples were vortexed and centrifuged at 18,000g for 5 min. Then supernatants were combined and 300 μL water was added and sample was further centrifuged at 18,000g for 5 min. The upper organic layer was transferred to a glass vial, then, dried under a stream of N2. Finally, the samples were reconstituted with 250 μL of Lipidyzer running buffer (10 mM ammonium acetate in dichloromethane: methanol (50:50 v/v)). 75 μL of these samples were injected on the Lipidyzer.79
Sterol analysis by GC-MS
Sterols and hydroxysterols were analyzed by GC-MS as described previously with some modifications.80 To each sample 5 μL of an internal standard mixture consisting of 1 mg/mL cholesterol-d7, 20 μg/mL desmosterol-d6 and 20 μg/mL 25-OH-cholesterol-d6 was added. Lipids were extracted twice into MTBE after hydrolysis in 80% (v/v) ethanolic sodium hydroxide (1 M). Samples were dried under N2 and derivatized with N-methyl-N-(trimethylsilyl)trifluoroacetamide before GC-MS.
Samples were analyzed on an Agilent GC-MS 5977B gas chromatograph coupled to a single quadrupole mass analyzer. For sterol quantification the following ion traces were monitored: m/z 444.4, 465.5, 327.2 and 333.3 for cholesterol, cholesterol-d7, desmosterol and desmosterol-d6, respectively. Quantification was achieved with external calibration lines.
Metabolite and lipid extraction for isotope tracing experiments
A dual phase extraction enabling to collect lipids and water-soluble metabolites was applied. Briefly, 500 μL methanol and 250 μL chloroform were added to each cell pellet. After mixing thoroughly, 250 μL chloroform and 400 μL LC-MS quality water were added. Samples were vortexed and placed on ice for 10 min followed by a 10,000g centrifugation step at 4°C for 10 min. The upper polar phase was collected and dried under a gentle stream of N2, containing the water-soluble fraction. The lower lipid phase was collected, split in two equal aliquots and dried likewise. The dried polar extracts were reconstituted in 200 μL water, vortexed, and sonicated for 1 min and transferred to glass inserts placed in LC vials for further LC-MS/MS metabolomics analysis. One of the dried lipid aliquots, was used for untargeted lipidomics analysis, as described above. The extract was reconstituted in 100 μL 2-propanol, vortexed and sonicated for 5 min. Then 100 μL of LC-MS water were added, vortexed, sonicated for 5 min, and transferred to glass inserts placed in LC vials for further LC-MS/MS lipidomics analysis (see above). For GC-MS analysis, 80 μL ethanol and 10 μL LC-MS grade water were added to the remaining dried lipid aliquot. The sample was saponified by adding 10 μL 10 M sodium hydroxide. Nitrogen was flushed on the sample and the sample was incubated for 1 h at 70°C. Subsequently, liquid-liquid extraction of fatty acids and sterols was accomplished by adding 700 μL MTBE and 550 μL LC-MS grade water after the samples had been neutralized using 35 μL 6 M hydrochloric acid. The MTBE phase was dried and then reconstituted in 25 μL N-tert-butyldimethylsilyl-N-methyltrifluoroacetamide and incubated for 10 min to derivatize fatty acids. Thereafter sterols were derivatized by adding 25 μL N-methyl-N-trimethylsilyltrifluoroacetamide containing 1% trimethylchlorosilane and 10% N-trimethylsilyl-imidazole and incubating for 15 min at 50°C. Finally, 150 μL n-hexane were added and samples were vortexed and transferred to glass inserts for further GC-MS analysis.
Fatty acid analysis
The method was based on a chromatography-electron ionisation mass spectrometric analysis method previously described.81 Briefly, samples (1 μL) were injected splitless at 280°C on a Gas Chromatograph (Agilent 8890 GC system) - Mass Spectrometer (Agilent 5977B GC/MSD) equipped with an electron ionization source. Analytes were separated on a 5% phenyl-95% methylpolysiloxane column (30m × 0.25mm i.d. × 0.25μm film thickness; Agilent) using Helium as carrier gas at a flow of 1 mL/min. The oven was set as follows: 1.5 min constant at 50°C, increase at 30°C/min to 210°C, increase at 5°C/min to 310°C, 4 min constant at 310°C. The transfer line, source and quadrupole temperature were set at 280, 230 and 150°C, respectively.
Untargeted metabolomics analysis by LC-MS/MS
Metabolomics analysis of the polar extracts was performed using a LC-MS/MS-based in-house library.82 A Shimadzu Nexera X2 (consisting of two LC30AD pumps, a SIL30AC autosampler, a CTO20AC column oven and a CBM20A controller) (Shimadzu, ‘s Hertogenbosch, The Netherlands) was used to deliver a programmed gradient of water (eluent A) and methanol (eluent B), both having 0.1 % formic acid. The gradient was 0% B at 0 min, 0% B at 1.5 min, 97% B at 9.9 min, 97% B at 12.9 min, 0% B at 13.0 min and 0% B at 13.8 min using a flow rate of 0.4 mL/min. A Synergi Hydro-RP, 2.5 μm, 100 × 2 mm column was used with a Phenomenex SecurityGuard Ultra C8, 2.7 μm, 5 × 2.1 mm cartridge as a guard column. The column oven was set at 40°C. The injection volume was 10 μL. The MS was a Sciex TripleTOF 6600 (AB Sciex Netherlands B.V., Nieuwerkerk aan den Ijssel, The Netherlands) operated in positive and negative ESI mode, with the following conditions: ion source gas 1 50 psi, ion source gas 2 50 psi, curtain gas 30 psi, temperature 500°C, acquisition range m/z 75-650, ion spray voltage 5500 V (ESI+) and -4500 V (ESI-), and declustering potential 80 V (ESI+) and -80 V (ESI-). An information dependent acquisition (IDA) method was used to identify the different metabolites, with the following conditions for MS analysis: collision energy ±10, acquisition time 80 ms and for MS/MS analysis: collision energy ±30, collision energy spread 15, ion release delay 30, ion release width 15 and acquisition time 40 ms. The IDA switching criteria was to exclude isotopes within 4 Da for a maximum of 10 (ESI+) or 18 (ESI-) candidate ions to monitor per cycle.
Identification and estimation of lipid labeling
Given the fact that GC-MS analysis revealed no conversion of U13C-FA18:1 into shortened or elongated FA, we calculated the expected molecular ions for several U13C-FA18:1 containing lipids from various classes (i.e. PC, PE, TG, DG, CE, CAR, FA) using a characteristic mass shift of 18.0604 Da when compared to their unlabelled counterparts and investigated our untargeted lipidomics data for these signals. As additional validation we confirmed the presence of the characteristic m/z 299.3 fragment (coming from U13C-FA18:1) in negative ESI mode in the collected MS/MS spectra. Moreover, to allow estimating the quantities of the most intense labelled lipids, external calibration lines for PC 18:1/18:1, FA 18:1, CE 18:1 and CAR 18:1 were constructed between 5 -1000 ng/mL. Quantification was carried out in MS1 mode using the [M+HCOO]- ion for PC 18:1/18:1, the [M-H]- ion for FA 18:1, the [M+NH4]+ ion for CE 18:1, and the [M+H]+ ion for CAR 18:1 using high resolution mass spectrometry with a mass accuracy set to ±0.025 Da. Both LC and MS parameters were identical as the ones employed in the untargeted lipidomics analysis described above, with the exception that only MS1 analysis were performed. The so obtained lipid specific responses were then used to estimate the quantities of labelled lipids, subsequently correcting them for the protein content of the analyzed cell pellet using the bicinchoninic acid (BCA) assay according to the manufacturers instructions.
Preparation of reagents for in vitro experiments
BMDMs were incubated for 3h at 37°C with 1mg/mL myelin debris. Myelin debris were obtained from healthy C57BL/6 mice brains, as previously described.83 Briefly, dissected brains were homogenized in 0.32M glucose using a hand-held rotary homogenizer and added to 0.82M glucose in an ultracentrifuge tube to create a density gradient. After ultracentrifugation (100.000G, 45 min at 4°C), crude myelin debris was collected from the interphase, homogenized again and the ultracentrifugation step was repeated. Next, the myelin debris pellet was resuspended in Tris-Cl-EDTA before repeating the ultracentrifugation step. Hereafter, purified myelin debris was collected and labelled with carboxyfluorescein succinimidyl ester (CFSE) (Invitrogen, cat.no. C34554) when indicated, as previously described.83
Cell culture experiments
Glioblastoma cells were isolated from primary, treatment-naïve or recurrent PDG-Ink4a mice post RT and maintained in culture in DMEM (Gibco, cat.no. 61965059) supplemented with 10% FBS and 1x penicillin/streptomycin (Roche, cat.no. 110774440001) (referred to as complete medium) as previously described.8 To generate tumor-conditioned media (TCM), 2x106 glioblastoma cells were seeded in T75 flasks in complete medium, and medium was replaced by DMEM. After overnight incubation, TCM was collected, filtered and concentrated in Amicon® Ultra-15 Centrifugal Filter Unit (Merck, cat.no. C7715), then resuspended in fresh complete medium.
Generation of primary macrophages
Bone marrow-derived macrophages (BMDMs) were generated as previously described.10 Briefly, both femurs and tibiae from C57BL/6 or LysM-Cre Abca1/Abcg1fl/fl mice (kindly provided by Brian Ruffell, Moffit Cancer Center) were flushed in complete medium. The cell suspension obtained was filtered through a 40μm strainer and cultured in 30mL Teflon bags (OriGen, cat.no. PL07 and PL30) in complete medium + 10ng/ mL recombinant mouse CSF-1 (R&D systems, cat.no. 4160ML). Primary microglia (MG) were isolated from brains of C57BL/6 mice which were macrodissected and dissociated using the Adult Brain Dissociation Kit (Miltenyi, cat.no. 130-107-677) and a single cell suspension was generated using the OctoMACS dissociator (Miltenyi). The single cell suspension was incubated with CD11b+ selection beads to effectively purify for MG when using LS Columns and a MidiMACS Separator. Cells were maintained in culture for a total of 5 days, with change of medium 2 and 4 days after collection, before use in in vitro experiments. Cells were then educated in control complete medium or TCM (both supplemented with either 10ng/mL CSF-1 for BMDM differentiation or 20ng/ml IL-34 (R&D systems, cat.no. 5195-ML) and 5ng/ml TGF-β (R&D systems, cat.no. 7666-MB) for MG differentiation) on day 5, either in 6-well plates for monoculture lipid uptake assays (BMDMs seeded at 3x105 cells per well, MG seeded at 1x105 cells per well) or in 3mL Teflon bags for co-culture assays (at a density of 1x106 cells/mL). On day 6, BMDMs or MG were incubated with myelin as indicated, then washed in PBS.
BMDM flow cytometry
BMDMs educated in control media or glioblastoma cancer cell TCM and exposed to different conditions ± lipids were assessed for LLM formation by flow cytometry with BODIPY staining as described above. Alternatively, myelin or general phagocytosis was assessed by measuring CFSE or FluoSpheres™ (ThermoFisher, cat.no. F8803) mean fluorescence intensity (MFI), respectively, by flow cytometry.
Lipid tracing in tumor cell mono-culture
MES-like tumor cells were seeded at 5x104 cells per well of a 100mm Petri dishes or 8 well glass chamber slide (Ibidi, cat.no. 80841) previously coated with Poly-D-lysin (Thermofisher, cat.no. A3890401), in DMEM + 10% FBS for 12 hours. Traceable CE, either carbon (250μM) or click labeled (250μM), was synthesized and conjugated with 0.5% lipid-free BSA in PlasmaX media (Ximbio) for 30min before added to the tumor cells and incubated for various durations (0h, 8h or 24h) before cells were isolated for metabolic or IF analysis (as described above) to trace the fate of the CE.
Cholesterol staining in BMDM monoculture
BMDMs were seeded at 5x104 cells per well of an 8 well glass chamber slide (Ibidi, cat.no. 80841) previously coated with Poly-D-lysin (Thermofisher, cat.no. A3890401) in tumor conditioned media generated from MES-like cells as described above. The next day, 5μM LXR antagonist (GSK2033) was added for 3h prior to addition of 10mg/mL myelin for another 3h. Cholesterol content was analyzed 24h later with a Filipin III staining (Cholesterol Assay Kit, Abcam, cat. no. ab133116) according to the manufacturer’s instructions. The slides were imaged with a Zeiss AxioScan Z1 slide scanner as described above.
Co-culture and proliferation assays
BMDMs (myelin-loaded or not) were seeded at a 1:1 ratio with 1x105 serum-starved (for 24h) glioblastoma cells in 6-well plates for a further 48 hours, in DMEM + 2%FBS + 10ng/mL CSF-1. Cell proliferation was measured by flow cytometry after 48h co-culture using the Click-iT™ EdU Alexa Fluor™ kit (Thermofisher, cat.no. C10337) according to the manufacturer’s instructions, before resuspension in 100μl saponin azide buffer (SAB) with 0.01mg/ml propidium iodide (Invitrogen, P3566). BMDM mediated cholesterol transfer to tumor cells was measured using flow cytometry as previously described.84 BMDMs were seeded in RPMI + 0.1% lipid-free BSA + 10ng/mL CSF-1 ± 50μM of traceable alkyne (“clickable”) cholesterol, and incubated overnight. Next, cholesterol-loaded BMDMs were extensively washed and co-cultured with tumor cells for 0, 1 or 3 hours, before flow cytometry analysis. For all assays, 6-well plates were previously coated with poly-D-lysine (Thermofisher, cat.no. A3890401) to facilitate BMDM adhesion.
Co-culture assay multi-omics analysis
MES-like glioblastoma cells and dTom+ BMDMs were seeded (1x106:3 x106, MES-like:BMDM ratio) in 100mm PDL coated Petri dishes in DMEM + 10%FBS + 10ng/mL CSF-1. After 4h, media was refreshed with DMEM + 2% lipid-free FBS + 10ng/mL CSF-1 and cultured for 12h. Hereafter, myelin debris was added (1mg/mL) and cells were collected at various timepoints (0h, 6h and 24h) for FACS-purification of tumor cells and BMDMs based on dTomato signal. Sorted cell suspensions were washed with PBS and were homogenized in RLT buffer (79216, Qiagen), or resuspended in PBS for preparation of downstream RNA-seq and lipidomics analysis, respectively.
Ex vivo assays
To assess the functional significance of macrophages and myelin in the glioblastoma TME, mouse brain tumors were macrodissected and dissociated as described above. The single cell suspension was incubated with myelin removal and/or CD11b+ selection beads to effectively remove myelin debris and/or macrophages respectively when using LS Columns and a MidiMACS Separator. After negative selection, CD11b- tumor cells were plated in various conditions (mono-culture or co-culture, with CD11b+ cells, WT or Abca1/Abcg1 KO BMDMs pre-loaded, with myelin debris, or not, as described above) in PDL-coated 6-well plates in 10% FBS DMEM. Following 24 hours of incubation in 37C, media was refreshed with 10% FBS DMEM or 2% lipid-free FBS DMEM ± 5μM LXR antagonist (GSK2033) or 1μM Abca1 inhibitor (Valspodar) and maintained for 48 hours for functional studies, or with 10% FBS DMEM with dTomato+ BMDMs, generated as described previously, for 48 hours for LLM formation studies.
Seahorse assay
To characterize the glycolytic activity and mitochondrial respiration of the glioblastoma cell lines, a Seahorse assay (Seahorse Bioscience) was performed according to the manufacturer’s protocol. Briefly, glioblastoma cells were seeded at 1x104 cells per well of a XFe24 Cell Culture microplate in complete medium. The next day, media was replaced with RPMI-1640 (Sigma, cat.no. R1383) supplemented with 4mM glutamine, and 10mM glucose for the mitochondrial respiration assay only. The culture microplate was loaded into Seahorse XF24 analyzer (Seahorse Bioscience). For glycolytic activity, extracellular acidification rate (ECAR) was measured for basal value and after each sequential injection of D-(+)-glucose (10mM final), oligomycin (1μM final), and 2-Deoxy-D-glucose (2-DG) (50mM final). For mitochondrial respiration, oxygen consumption rate (OCR) was measured for basal value and after each sequential addition of oligomycin, carbonyl cyanide-4 trifluoromethoxy phenylhydrazone (FCCP), rotenone + antimycin A (all 1μM final). Assay injection compounds were all from Sigma-Aldrich.
RNA extraction, cDNA synthesis and RT-qPCR
RNA was extracted using TRIzol (Thermofisher, cat.no. 15596026). For cDNA synthesis, the Transcriptor First Strand cDNA Synthesis Kit (Roche, cat.no. 50-997-552) was used with 0.2μg of RNA. The following TaqMan probes (Life Technologies) were used for qPCR: Ubc (Mm01201237_m1), Lgals3 (Mm00802901), Slc1a1 (Mm00436590), Ascl1 (Mm00484217), Sox2 (Mm03053810), Pdgfra (Mm0040701), Tgfbr2 (Mm03024091), Col3a1 (Mm00802300), Col1a1 (Mm00801666) and Scd1 (Mm00772290). Assays were performed in triplicate and relative expression was calculated after normalization to the housekeeping gene Ubc for each sample.
Electron microscopy (EM)
For EM on in vitro samples, samples were fixed in Karnovsky’s fixative. Postfixation was performed with 1% Osmium tetroxide in 0,1 M cacodylate buffer. After washing, cells were stained with Ultrastain 1 (Leica, Vienna, Austria), followed by ethanol dehydration series. Samples were embedded in a mixture of DDSA/NMA/Embed-812 (EMS, Hatfield, U.S.A), sectioned from the basal side upwards parallel to the petri dish and analyzed with a Tecnai12G2 electron microscope (Thermo Fischer, Eindhoven, the Netherlands).
For EM on in vivo brain tissues, samples were fixed with 4% paraformaldehyde, 0.4% glutaraldehyde in 0.1M PHEM buffer. After imbedding in increasing amounts of gelatin, 3-5 mm blocks were infiltrated in 2.3 M sucrose and after overnight incubation, plunge frozen in liquid nitrogen. Then 60-90 nm sections were generated on a Leica EM UC-7 ultra-cryomicrotome at -120 C. Sections were transferred to copper grids and immuno-gold labelled with anti-IBA1 primary antibody, 10 nm gold conjugated to protein A (Utrecht University) and stained with Uranyl acetate. Imaging was performed at a Tecnai T12 with Xarosa and Velata camera.
Single cell sequencing library preparation
The Chromium Controller platform of 10X Genomics was used for single cell partitioning and barcoding. Each cell’s transcriptome was barcoded during reverse transcription, pooled cDNA was amplified and Single Cell 5’ Gene expression libraries were prepared according to the manufacturer’s protocol (CG000207 Rev E and CG000331 Rev A, 10X Genomics). The Single Cell 5’ Gene Expression libraries were pooled to create three sequencing pools. The sequencing pools were quantified by qPCR, according to the KAPA Library Quantification Kit Illumina® Platforms protocol (KR0405, KAPA Biosystems). The Single Cell 5’ Gene Expression libraries were sequenced on a NextSeq550 or NovaSeq 6000 system (Illumina) using a NextSeq 500/550 High Output Kit v2.5 (cat. no. 20024906, Illumina) and NovaSeq SP Reagent Kit v1.5 (cat.no. 20028401, Illumina), at a sequencing depth of 20.000 reads pairs/cell. NextSeq paired-end sequencing was performed using 28 cycles for Read 1, 8 cycles for Read i7 and 56 cycles for Read 2. Novaseq paired-end sequencing was performed using 26 cycles for Read 1, 10 cycles for each Read i7 and i5 and 90 cycles for Read 2.
Visium 10x library preparation
For the Visium samples, libraries were prepared according to the Visium Spatial Gene Expression User Guide (CG000239 RevD). The Visium Spatial Gene Expression libraries of all 8 samples were pooled to create one sequencing pool. The sequencing pool was quantified by qPCR, according to the KAPA Library Quantification Kit Illumina® Platforms protocol (KR0405, KAPA Biosystems). Libraries were loaded at a concentration of 250 pM (1% PhiX) and sequenced on a NovaSeq 6000 System (Illumina) using a NovaSeq SP Reagent Kit v1.5 (cat no. 20028401, Illumina), at a sequencing depth of approximately 40.000 to 50.000 read pairs per tissue covered spot. NovaSeq sequencing was performed using 28 cycles for Read 1, 10 cycles for each Read i7 and i5 and 90 cycles for Read 2. The Space Ranger software v1.0.0 (10X Genomics) was used to align the barcoded reads with the imaged tissue.
Bulk RNA-seq library preparation
For RNA-seq samples, FACS-purified (non-) LLMs from MG and MDMs from primary PDG-Ink4a tumors, dTomato+ BMDMs from ex vivo assays (4.000-20.000 cells) or MES-like tumor cells from in vitro assays (20.000-100.000 cells) were homogenized in RLT buffer (79216, Qiagen), total RNA was isolated using the RNeasy Micro Kit (74004, Qiagen), including an on-column DNase digestion (79254, Qiagen), according to the manufacturer’s instructions. Quality and quantity of the total RNA was assessed on the 2100 Bioanalyzer instrument following manufacturer’s instructions “Agilent RNA 6000 Pico” (G2938-90046, Agilent Technologies). In general RNA yields of 3-135 ng total RNA and RNA integrity numbers (RIN) above 8 were obtained. RNA samples were concentrated with Agencourt RNAClean XP beads (A63987), according to manufacturer’s instructions (Protocol 001298v001, Agencourt) with modifications. In short, 2.5-5 ng of total RNA for each sample was prepared in a volume of 30 μl by adding nuclease free water (AM9937, Ambion). 1.5 μl of Recombinant RNase inhibitor (2313A, Clontech) was added to each RNA sample. A 1.8x reaction volume RNAClean XP bead cleanup was performed, instead of adding 54 ul RNAClean XP bead suspension, 18 μl RNAClean XP bead suspension and 36 μl RNAClean XP bead buffer (same solution without the beads) were added to each RNA sample. After 3 times wash with 70% ethanol, the bead pellets were dried at 37 degrees Celsius for 3-5 minutes, untill bead pellet begins to show cracks. RNA was eluted by adding 4 μl of NF-H20 and mixing by vortexing. RNA library preparation was performed as previously described with modifications.85 In short, oligo dT primer hybridization was performed by the addition of Oligo dT mix (0.7ul H2O, 0.1 ul RNAse inhibitor (40U/ul), 0.1 ul dNTP mix (100mM) and 0.1 ul Oligo-dT30VN primer (100uM)) to the 4 ul sample-RNA bead suspension. Reverse transcription was performed as described but the MgCl2 concentration was adjusted to 10 mM. Template switching and 7-12 cycles pre-amplification of full length cDNAs with template switching oligo’s was performed using ISPCR primer at a final concentration of 0.08 uM. The amplified full length cDNA was used for NGS library construction by Tagmentation for Illumina sequencing, using the Illumina Nextera XT DNA sample preparation kit (FC-131-1096, Illumina) and IDT® for Illumina Nextera DNA Unique Dual Indexes (20027216, Illumina). RNA sequencing libraries were quantified and normalized based library QC data generated on the Bioanalyzer system according to manufacturer’s protocols (G2938-90321, Agilent Technologies). A multiplex sequencing pool of all uniquely dual indexed RNA libraries was composed by equimolar pooling before sequencing on the NovaSeq 6000 Illumina sequencing platform. Paired-end sequencing was performed using 54 cycles for Read 1, 10 cycles for Read i7, 10 cycles for Read i5 and 54 cycles for Read 2, using a NovaSeq SP Reagent Kit v1.5 (cat no. 20028401, Illumina). This resulted in, on average, 20 M passing filter (PF) reads per sample.
Bulk ATAC-seq library preparation
ATAC-seq libraries were prepared using an optimized protocol.86 Nuclei of the FACS sorted cells were isolated, permeabilized and tagmented by using in-house-generated Tn5 transposase. The resulting DNA fragments underwent a first 9-cycle PCR amplifications using KAPA HiFi HotStart ReadyMix (7958935001, Roche), and we selected the fragments between 150 and 700 bp in size using with SPRI beads (A63881, Backman Coulter). The size-selected fragments were amplified for a second 9-cycle PCR reactions, and the resulted libraries were purified using SPRI beads. The ATAC-seq libraries were sequenced in a paired-end mode (2×38 bp) on a NextSeq 550.
The recorded ATAC-seq data were mapped to the mm10 reference genome with BWA-MEM (version 0.7.17-r1188).87 SAMtools (version 1.10)88 was used to discard the mapped reads with a mapping quality score <15, and PICARD tools (version 2.12.0) was applied to remove optical PCR duplicates. The coverage files were generated with the ‘normalize to 1× genome coverage’ methods in deepTools (version 3.5.1).89
Quantification and statistical analysis
Image analysis
LLMs were identified by segmentation of IBA1+ clusters using a customized surface detection workflow with smoothing, manual thresholding and split touching objects in Imaris software (Oxford instruments v9.8). Objects smaller than 100μm and large patches in necrotic areas (>2000μm) were excluded from analysis. PLIN2+ droplets were detected with adapted spot algorithm and counted inside the IBA1 positive cells. Distances of the LLMs to myelin (MBP surfaces) were calculated using a distance transform based on MBP positive surfaces. Data was further analyzed in a R environment.
To quantify the mean intensity Filipin per lipid droplet we first determined the number of lipid droplets per cell in Fiji using the macro ‘Foci Analyzer’ (https://github.com/BioImaging-NKI/Foci-analyzer/), v1.4. Briefly, after maximum intensity projection of the 3D data, cells are segmented in 2D using Cellpose.90 For droplet detection the signal is background-subtracted using a difference-of-Gaussians filter. Droplets are identified as local maxima, which are then used as seeds for the marker-controlled watershed algorithm of MopholibJ,91 after which detected droplets are size-filtered to exclude unrealistically small detections. Many operations in this macro are executed using CLIJ2/CLIJX libraries.92
scRNA-seq processing
The single cell expression data was loaded in R version 4.1.1 using Seurat v4.4. and SeuratObject v4.0.93 To eliminate batch effect, samples were integrated using the IntegrateData() function before log normalization. As quality control, only cells that had between 1000 and 5500 genes detected and had less than 30.000 counts were included. Among the remaining cells, we calculated the aggregated expression of mitochondrial genes to exclude apoptotic cells (dead cells > 0.2). A total of 96,135 cells remained after filtering. Unsupervised clusters (Louvain algorithm, 0.3 resolution) were annotated as tumor cells, astrocytes, pericytes, endothelial, T-cells, DCs, MDMs and MG based on the expression of cell specific markers, respectively Olig2, Gfap, Des, Pecam1, Cd3e, Ccr7, Itga4 and P2ry12. The dataset was split in a tumor cell and macrophage (MDMs and Mgs) dataset for further downstream analysis. Unsupervised clustering (Louvain algorithm, 0.3 resolution) of macrophages gave rise to 4 MDM and 4 MG clusters, from which each the top 30 differentially expressed (DE) genes were calculated using the FindMarkers() function.
Visium 10x processing
As described for the scRNA-seq, Seurat v3.0 was used merge and normalize the gene count matrices derived from the Visium 10x pipeline using SCT normalization to take into account RNA count related to number of cells per spot.
Integration of macrophage clusters data into a spatial context
In order to integrate the identified macrophage clusters into the coordinates of the spatially resolved transcriptomic data, we first interrogated the presence of MDMs and MG within each spot by calculating a module score with AddModuleScore() for general macrophage (Aif1, Lgals3, Itga1, Trem2, P2ry12, Siglech)94 and microglia (P2ry12, Siglech)24 specific genes. Spots classified as pan-macrophage (scorepan-macrophage > 0.1) and MG (scoremicroglia > 0.1) were assigned as MG, whereas spots classified as pan-macrophage (scorepan-macrophage > 0.1) and not MG (scoremicroglia < 0.1) were assigned as MDMs. Next, we used the method used to predict cell cycle stage95 and interchanged the gene sets with the top 30 DE genes for each MG clusters and MDM clusters, respectively, to infer the overrepresentation of macrophage subsets (Table S2). As tumor cells are overrepresented within most spots, the proliferating macrophages MDM4 and MG4 clusters were excluded from the analyses, to avoid that overlap of these clusters’ gene signatures with tumor cell signatures to lead to skewing of most spots towards MG/MDM4 when added to subset classification.
Pseudo-location and subtype assignment
The pseudo-location and cellular subtype of single cells (scRNA-seq) or spots (Visium 10x) were inferred by using gene sets generated by the Ivy GAP associated with glioblastoma anatomical features (CT, cellular tumor; LE, leading edge; MVP: microvascular proliferation; PAN, pseudopalisading cells around necrosis) to predict location,2 gene sets generated to identify glioblastoma cellular subtypes18 or de- and remyelination modules45 using a previously described prediction methods.95
Before interrogation of the pseudo-location in the scRNA-seq, the gene sets associated to their anatomical features were curated for tumor cells and macrophage specific gene markers using a-priori knowledge (Tables S1E and S1F). First, we generated an average expression table for genes of all gene sets that were expressed by tumor cells or MDMs. The tumor-specific curated CT, LE and PAN gene lists consisted of genes with an average expression > 0.5 in tumor cells; whereas we were prompted to use genes from the MVP gene list with an average expression > 0.1 in tumor cells, to account for the overall low expression of genes that compromise this list expressed by tumor cells.
The macrophage-specific curated anatomical features gene lists were determined as followed: as the CT gene list contained overall many tumor-specific genes due to the over-representation of tumor cells in this area, many proliferation-related genes are listed which skews the pseudo-location determination of proliferating macrophages to the CT area. To account for this, we decided to use non-tumor specific genes (average expression < 0.25 in tumor cells) in the macrophage specific CT gene list. In similar fashion, the PAN gene list contained overall many macrophage-specific genes due to over-representation of macrophages in this area, for which we accounted by including only genes highly expressed by macrophages (average expression > 1.5 by MDMs). The LE and MVP macrophage curated gene lists consisted of genes with an average expression higher than 0.05 and 0.6 in MDMs, respectively.
In the scRNA-seq dataset, tumor cell subtype scores and classification were calculated using the AddModuleScore() function of previously described subtype signatures18 in both murine and patient samples. To validate this method accurately classified the glioblastoma cellular subtype, which functionally represent clinically validated glioblastoma subtype characteristics, the FindAllMarkers() function was used to curate a list of genes differently expressed by tumor cells subtype and niches. These genes were clustered based on a hierarchical clustering analysis (threshold = 4.8), which resulted in 6 groups of genes that behave similarly. Gene-set overrepresentation analyses of these gene groups was used to extrapolate and validate their involvement in subtype specific characteristics.
Before interrogation of the cellular subtype presence in the VISIUM10x spots, the gene sets associated to their cellular subtype were curated for tumor cells using a-priori knowledge of the cellular subtype distribution observed in the scRNA-seq (Tables S1G and S1H).
LLM (GPNMBhigh) classification
A curated list of 5 genes (Table S1C) specifically expressed by LLMs was used to calculate a LLM score for each macrophages using the AddModuleScore(). The average LLM score of MDM4 and MG4 ((averageLLMMDM3 + averageLLMMG3)/2) was used as threshold to classify macrophages as LLMs or non-LLM.
To classify macrophages as LLMs in glioblastoma patient scRNA-seq samples, the LLM signature genes were converted to their human homologue (Table S1C) and were used to calculate a LLM score. We used the following equation to classify macrophages as an LLM in an unbiased manner in glioblastoma patient samples; Gpnmbhigh -Score > median(Gpnmbhigh -ScoreCluster with highest GPNMB score), and another equation to classify macrophages as an LLM in melanoma patients; Gpnmbhigh -Score > 0.
RNA-seq and ATAC-seq analyses
For RNA-seq analyses, a raw count matrix was produced and loaded within the R environment (v.4.1.1, 2021-08-10), unless mentioned otherwise. For ATA-seq, macs2 (v.2.2.7.1) was applied with default parameters to identify peaks. Using this initial estimation, ChIPQC (R v.1.30.0) was used to optimise and calculate extension sizes. Using --no-model and --extsize peaks were called again using the new extension sizes estimates. Peaks existent in all replicates of a condition were defined as consensus peaks per sample. THOR96 was used to normalise (TMM) ATAC-seq signal and create Bigwig file format for visualization in the IGV genome browser.97 The parameters used are: --no-gc-content --merge --rmdup --report --deadzones blacklist.bed --pvalue 1 --exts extension sizes. DESeq2 (version 3.14) and EdgeR (v.3.36.0) was used to assess the differential gene expression/binding between grouped samples. All samples were trimmed-mean-of-M-values (TMM) normalized to account for library size. To test for differential expression, the generalized linear model method using the glmLRT function was used with a significance cutoff of FDR 1%. Plots were generated using the package ggplot (v.3.3.6) or ggpubr (v.0.4.0), except where mentioned. Peaks were annotated to different genomic regions using the function annotatePeak from the ChIPseeker R package (v.1.30.3). To create the heatmaps and average profiles,the functions functions computeMatrix and plotHeatmap from deepTools89 were used with default parameters.
Gene set-centered analyses
KEGG98 and GO99 databases, supplemented with internally created gene sets and literature-derived signatures (Table S1B) were used as a primary source for gene set-centered analyses (Tables S2, S3, S4, S5, and S6). Over-representation was assessed with the g:GOSt function of the gProfiler R package100 to check whether an input set of genes significantly overlaps with annotated gene sets using a false discovery rate equal to 0.05. Geneset enrichment for RNA- and ATAC-seq analyses was performed with the enrichR (v.3.0.0) using the GO: biological Processes database for gene set over-representation analyses genes of genes with altered accessibility at the promoters and differential expression in LLMs relative to non-LLMs (Table S4). Significant overlaps of genes with annotated gene sets were defined a false discovery rate of 0.005-0.01. Gene set enrichment analysis (GSEA) was assessed with the GAGE package,101 which uses the average of the absolute values of the per gene test statistics to account for both up- and down-regulation of the curated pathways. GSEA data for visualization in heatmaps was further transformed to z-scores ranging from 1 to 0. For the analysis of the GLASS and TCGA datasets, the single sample gene set enrichment analysis102 for the LLM signature (Table S1C) was performed using GenePattern (ssGSEA 10.0.11).103 The input was TMM-normalised expression in log2(TPMs) for each of the glioblastoma samples. To define the tumors with high enrichment, the cut-off was a ssGSEA score ≥ the 3rd quantile of the of the data, whereas the low enrichment tumors were chosen based on a ssGSEA score ≤ the 1st quantile. For the GLASS data, after the selection of the high/low enrichment tumors, they were further filtered for glioblastoma and IDH-wild-type.
Motif analysis
Motif enrichment analysis was performed using a published pipeline with some small modifications.104 For each of the differential peak lists identified in the DESeq2 analysis, we searched for transcription factor motifs presented in the peak sets in the nonredundant cis-bp database (version 3.0) using the “gimme scan” function with the default settings (GimmeMotifs, version 0.13.1).105 Next, we calculated for each motif the frequency in a differential peak set (foreground) and the corresponding peak set used to call the differential peaks (background). We normalized the motif frequency by dividing the individual motif frequency by the total number of identified motifs (relative motif frequency). We calculated the log2-transformed enrichment score by computing the ratio of the enriched relative motif frequency in the foreground to the background motif frequency. P values were calculated by Fisher exact test on the following 2 × 2 table: for each motif M, we determined the number of peaks enriched in the foreground with or without M and for the background with or without M.
Survival plots
For the calculation of the survival curves in Figures 7A and 7F, the Kaplan-Meier estimator was obtained with the survfit function from the package survival.106 The statistical test applied was the log-rank test using the survminer package.107
Quantification and statistical analysis
Summary of data are presented as mean ±/+ standard error of mean (SEM) using PRISM v9 or “ggplot2” package in R. For boxplots in Figures 3E, 7B, and 7C, the boxes represent data from the first to the third quartile, the middle line represents the median, the whiskers denote a minimum (first quartile – 1.5∗(first - third quartile)) and a maximum (third quartile + 1.5∗(first - third quartile)). Comparisons between groups were analyzed using statistical tests noted in the corresponding figure legends.
Acknowledgments
We thank members of the Akkari lab for insightful comments and discussion and Marnix de Groot, Cankat Ertekin, Naz Kocabay, Lesley Cornet, Lotte van Mil, and Jeremy Tessier for excellent technical support. We thank Rico J.E. Derks, Kevin A.J. Brewster, Mohan Ghorasaini, Aswin Verhoeven, and Marieke Heijink for technical assistance with lipidomics analysis. We are grateful to Drs. Eric Holland and Tatsuya Ozawa for providing the RCAS vectors and Nestin-Tv-a mice. We thank the animal facilities, imaging cores, experimental animal pathology facility, and flow cytometry cores at the Netherlands Cancer Institute (NKI) for excellent technical assistance. Particularly, we are grateful to Martijn van Baalen for technical flow cytometry support, Jelrik van der Meer for assistance with Visium 10X sample preparation, Amalie Dick and Bram van den Broek for support with Visium 10X imaging acquisition and analysis, Iris de Rink for support with pre-processing the Visium 10X data, and Hans Janssen for performing EM experiments. We thank Drs. Daniela Quail, Oakley Olson, Karin de Visser, and Logan Walsh for critically reading the manuscript. This research was supported by the Dutch Cancer Society (L.A., 2017-10658, 2021-14339, and 2022-14530), the European Research Council (L.A. and A.K., ERC SYN 101118999), the Oncode Institute and Cancer Genomics Center (L.A.), and NWO (L.A., Vidi 91719355; L.A. and A.K., ENW-XL 21-XL21-071).
Author contributions
L.A., D.J.K., and J.E. conceived the study, designed experiments, interpreted data, and wrote the manuscript. D.J.K., J.E., M.B., M.F., S.M.H., M.A.-K., M.N., M.M., B.F., E.d.W., N.Q.L., and G.F.-C. performed and analyzed experiments. D.J.K., M.A.-K., and N.Q.L. performed all computational analyses. B.R. provided LysM-Cre Abca1/Abcg1fl/fl. D.B. and K.P. provided patient samples/datasets. E.S.-L. and M.G. performed and interpreted lipid and lipidomics analyses. L.R. and S.I.v.K. synthesized clickable lipids. N.N.v.d.W. and A.E.G. performed and analyzed EM images. A.K. and E.S.-J. contributed by providing relevant reagents. All authors edited or commented on the manuscript.
Declaration of interests
L.A., D.J.K., A.K., and J.E. are inventors on European patents P091147NL and P099572EP describing relevant claims and their therapeutic potential.
Published: August 12, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.cell.2024.07.030.
Supplemental information
(A) Macrophage subset signatures identified scRNA-seq dataset of GEMMs of glioblastoma.
(B) Relevant gene signatures for TAMs in glioblastoma from cited literature.
(C) GPNMBhigh signatures for analysis of murine or patient datasets.
(D) Gene signatures derived from FACS-purified (non-) lipid-laden MG and MDMs generated by RNA-seq.
(E) Curated Ivy GAP niche gene sets for scRNA-seq analysis of tumor cells.
(F) Curated Ivy GAP niche gene sets for scRNA-seq analysis of macrophages.
(G) Curated Neftel et al. cellular subtype gene sets for assessing subtype localisation in VISIUM10x samples.
(H) Curated TAM subset gene sets for assessing localisation in VISIUM10x samples.
(A) Gene set over-representation analysis, MG1-P2RY12.
(B) Gene set over-representation analysis, MG2-TNF.
(C) Gene set over-representation analysis, MG3-GPNMB.
(D) Gene set over-representation analysis, MG4-MKI67.
(E) Gene set over-representation analysis, MDM1-CCR2.
(F) Gene set over-representation analysis, MDM2-H2-EB1.
(G) Gene set over-representation analysis, MDM3-GPNMB.
(H) Gene set over-representation analysis, MDM4-MKI67.
(A) Gene set over-representation analysis, GPNMBlow.
(B) Gene set over-representation analysis, GPNMBhigh.
(A) GSEA on genes associated with increased accessible promoter regions in MG versus MDMs.
(B) GSEA on genes associated with increased accessible promoter regions in MDMs versus MG.
(C) GSEA on genes associated with increased accessible chromatin regions in lipid-laden MDMs versus non-lipid-laden MDMs.
(D) GSEA on genes associated with decreased accessible chromatin regions in lipid-laden MDMs versus non-lipid-laden MDMs.
(E) GSEA on genes associated with increased accessible chromatin regions in lipid-laden MG versus non-lipid-laden MG.
(F) GSEA on genes associated with decreased accessible chromatin regions in lipid-laden MG versus non-lipid-laden MG.
(G) GSEA on genes associated with increased accessible chromatin regions in lipid-laden TAMs versus non-lipid-laden TAMs.
(H) GSEA on genes associated with decreased accessible chromatin regions in lipid-laden TAMs versus non-lipid-laden TAMs
(A) Gene signatures from FACS-purified dTom+ macrophages from ex vivo LLM formation assay analysed by RNA-seq.
(B) Gene set enrichment analysis on gene signature "up.all".
(C) Gene set enrichment analysis on gene signature "up.full".
(D) Gene set enrichment analysis on gene signature "up.tumor".
(E) Gene set enrichment analysis on gene signature "up.myelin".
(F) Gene set enrichment analysis on gene signature "up.full.myelin".
(G) Gene set enrichment analysis on gene signature "up.full.tumor".
(H) Gene set enrichment analysis on gene signature "down.full".
(I) Gene set enrichment analysis on gene signature "down.tumor".
(J) Gene set enrichment analysis on gene signature "down.myelin".
(K) Gene set enrichment analysis on gene signature "down.full.tumor".
(L) Gene set enrichment analysis on gene signature "down.full.myelin".
(A) Gene set enrichment analysis on gene signature "down.hpm0_vs_hpm24_mono_up_tumor".
(B) Gene set enrichment analysis on gene signature "down.hpm0_vs_hpm24_mono_down_tumor".
(C) Gene set enrichment analysis on gene signature "down.hpm0_movo_vs_co_up_tumor".
(D) Gene set enrichment analysis on gene signature "down.hpm0_movo_vs_co_down_tumor".
(E) Gene set enrichment analysis on gene signature "down.hpm24_movo_vs_co_up_tumor".
References
- 1.Erbani J., Boon M., Akkari L. Therapy-induced shaping of the glioblastoma microenvironment: macrophages at play. Semin. Cancer Biol. 2022;86:41–56. doi: 10.1016/j.semcancer.2022.05.003. [DOI] [PubMed] [Google Scholar]
- 2.Puchalski R.B., Shah N., Miller J., Dalley R., Nomura S.R., Yoon J.-G., Smith K.A., Lankerovich M., Bertagnolli D., Bickley K., et al. An anatomic transcriptional atlas of human glioblastoma. Science. 2018;360:660–663. doi: 10.1126/science.aaf2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Klemm F., Maas R.R., Bowman R.L., Kornete M., Soukup K., Nassiri S., Brouland J.-P., Iacobuzio-Donahue C.A., Brennan C., Tabar V., et al. Interrogation of the microenvironmental landscape in brain tumors reveals disease-specific alterations of immune cells. Cell. 2020;181:1643–1660.e17. doi: 10.1016/j.cell.2020.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Friebel E., Kapolou K., Unger S., Núñez N.G., Utz S., Rushing E.J., Regli L., Weller M., Greter M., Tugues S., et al. Single-cell mapping of human brain cancer reveals tumor-specific instruction of tissue-invading leukocytes. Cell. 2020;181:1626–1642.e20. doi: 10.1016/j.cell.2020.04.055. [DOI] [PubMed] [Google Scholar]
- 5.Couturier C.P., Ayyadhury S., Le P.U., Nadaf J., Monlong J., Riva G., Allache R., Baig S., Yan X., Bourgey M., et al. Single-cell RNA-seq reveals that glioblastoma recapitulates a normal neurodevelopmental hierarchy. Nat. Commun. 2020;11 doi: 10.1038/s41467-020-17186-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kesarwani P., Prabhu A., Kant S., Chinnaiyan P. Metabolic remodeling contributes towards an immune-suppressive phenotype in glioblastoma. Cancer Immunol. Immunother. 2019;68:1107–1120. doi: 10.1007/s00262-019-02347-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garofano L., Migliozzi S., Oh Y.T., D’Angelo F., Najac R.D., Ko A., Frangaj B., Caruso F.P., Yu K., Yuan J., et al. Pathway-based classification of glioblastoma uncovers a mitochondrial subtype with therapeutic vulnerabilities. Nat. Cancer. 2021;2:141–156. doi: 10.1038/s43018-020-00159-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Akkari L., Bowman R.L., Tessier J., Klemm F., Handgraaf S.M., de Groot M., Quail D.F., Tillard L., Gadiot J., Huse J.T., et al. Dynamic changes in glioma macrophage populations after radiotherapy reveal CSF-1R inhibition as a strategy to overcome resistance. Sci. Transl. Med. 2020;12 doi: 10.1126/scitranslmed.aaw7843. [DOI] [PubMed] [Google Scholar]
- 9.Pyonteck S.M., Akkari L., Schuhmacher A.J., Bowman R.L., Sevenich L., Quail D.F., Olson O.C., Quick M.L., Huse J.T., Teijeiro V., et al. CSF-1R inhibition alters macrophage polarization and blocks glioma progression. Nat. Med. 2013;19:1264–1272. doi: 10.1038/nm.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bowman R.L., Klemm F., Akkari L., Pyonteck S.M., Sevenich L., Quail D.F., Dhara S., Simpson K., Gardner E.E., Iacobuzio-Donahue C.A., et al. Macrophage ontogeny underlies differences in tumor-specific education in brain malignancies. Cell Rep. 2016;17:2445–2459. doi: 10.1016/j.celrep.2016.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Quail D.F., Bowman R.L., Akkari L., Quick M.L., Schuhmacher A.J., Huse J.T., Holland E.C., Sutton J.C., Joyce J.A. The tumor microenvironment underlies acquired resistance to CSF-1R inhibition in gliomas. Science. 2016;352 doi: 10.1126/science.aad3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kloosterman D.J., Akkari L. Macrophages at the interface of the co-evolving cancer ecosystem. Cell. 2023;186:1627–1651. doi: 10.1016/j.cell.2023.02.020. [DOI] [PubMed] [Google Scholar]
- 13.Abdelfattah N., Kumar P., Wang C., Leu J.-S., Flynn W.F., Gao R., Baskin D.S., Pichumani K., Ijare O.B., Wood S.L., et al. Single-cell analysis of human glioma and immune cells identifies S100A4 as an immunotherapy target. Nat. Commun. 2022;13 doi: 10.1038/s41467-022-28372-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karimi E., Yu M.W., Maritan S.M., Perus L.J.M., Rezanejad M., Sorin M., Dankner M., Fallah P., Doré S., Zuo D., et al. Single-cell spatial immune landscapes of primary and metastatic brain tumours. Nature. 2023;614:555–563. doi: 10.1038/s41586-022-05680-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pombo Antunes A.R., Scheyltjens I., Lodi F., Messiaen J., Antoranz A., Duerinck J., Kancheva D., Martens L., De Vlaminck K., Van Hove H., et al. Single-cell profiling of myeloid cells in glioblastoma across species and disease stage reveals macrophage competition and specialization. Nat. Neurosci. 2021;24:595–610. doi: 10.1038/s41593-020-00789-y. [DOI] [PubMed] [Google Scholar]
- 16.Masetti M., Carriero R., Portale F., Marelli G., Morina N., Pandini M., Iovino M., Partini B., Erreni M., Ponzetta A., et al. Lipid-loaded tumor-associated macrophages sustain tumor growth and invasiveness in prostate cancer. J. Exp. Med. 2022;219 doi: 10.1084/jem.20210564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hambardzumyan D., Amankulor N.M., Helmy K.Y., Becher O.J., Holland E.C. Modeling adult gliomas using RCAS/t-va technology. Transl. Oncol. 2009;2:89–95. doi: 10.1593/tlo.09100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Neftel C., Laffy J., Filbin M.G., Hara T., Shore M.E., Rahme G.J., Richman A.R., Silverbush D., Shaw M.L., Hebert C.M., et al. An integrative model of cellular states, plasticity, and genetics for glioblastoma. Cell. 2019;178:835–849.e21. doi: 10.1016/j.cell.2019.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Couturier C.P., Baig S., Nadaf J., Li Z., Riva G., Le P., Kloosterman D.J., Monlong J., Allache R., Degenhard T., et al. scRNAseq shows treatment-induced, immune-dependent rise in mesenchymal cancer cells, and structural variants in distal neural stem cells. Neuro Oncol. 2022;24:1494–1508. doi: 10.1093/neuonc/noac085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Varn F.S., Johnson K.C., Martinek J., Huse J.T., Nasrallah M.P., Wesseling P., Cooper L.A.D., Malta T.M., Wade T.E., Sabedot T.S., et al. Glioma progression is shaped by genetic evolution and microenvironment interactions. Cell. 2022;185:2184–2199.e16. doi: 10.1016/j.cell.2022.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang J., Cazzato E., Ladewig E., Frattini V., Rosenbloom D.I.S., Zairis S., Abate F., Liu Z., Elliott O., Shin Y.-J., et al. Clonal evolution of glioblastoma under therapy. Nat. Genet. 2016;48:768–776. doi: 10.1038/ng.3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hara T., Chanoch-Myers R., Mathewson N.D., Myskiw C., Atta L., Bussema L., Eichhorn S.W., Greenwald A.C., Kinker G.S., Rodman C., et al. Interactions between cancer cells and immune cells drive transitions to mesenchymal-like states in glioblastoma. Cancer Cell. 2021;39:779–792.e11. doi: 10.1016/j.ccell.2021.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kirschenbaum D., Xie K., Ingelfinger F., Katzenelenbogen Y., Abadie K., Look T., Sheban F., Phan T.S., Li B., Zwicky P., et al. Time-resolved single-cell transcriptomics defines immune trajectories in glioblastoma. Cell. 2024;187:149–165.e23. doi: 10.1016/j.cell.2023.11.032. [DOI] [PubMed] [Google Scholar]
- 24.Masuda T., Sankowski R., Staszewski O., Prinz M. Microglia heterogeneity in the single-cell era. Cell Rep. 2020;30:1271–1281. doi: 10.1016/j.celrep.2020.01.010. [DOI] [PubMed] [Google Scholar]
- 25.Behnan J., Finocchiaro G., Hanna G. The landscape of the mesenchymal signature in brain tumours. Brain. 2019;142:847–866. doi: 10.1093/brain/awz044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sa J.K., Chang N., Lee H.W., Cho H.J., Ceccarelli M., Cerulo L., Yin J., Kim S.S., Caruso F.P., Lee M., et al. Transcriptional regulatory networks of tumor-associated macrophages that drive malignancy in mesenchymal glioblastoma. Genome Biol. 2020;21 doi: 10.1186/s13059-020-02140-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Eijk M., Aerts J.M.F.G. The unique phenotype of lipid-laden macrophages. Int. J. Mol. Sci. 2021;22 doi: 10.3390/ijms22084039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jaitin D.A., Adlung L., Thaiss C.A., Weiner A., Li B., Descamps H., Lundgren P., Bleriot C., Liu Z., Deczkowska A., et al. Lipid-associated macrophages control metabolic homeostasis in a Trem2-dependent manner. Cell. 2019;178:686–698.e14. doi: 10.1016/j.cell.2019.05.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Spann N.J., Garmire L.X., McDonald J.G., Myers D.S., Milne S.B., Shibata N., Reichart D., Fox J.N., Shaked I., Heudobler D., et al. Regulated accumulation of desmosterol integrates macrophage lipid metabolism and inflammatory responses. Cell. 2012;151:138–152. doi: 10.1016/j.cell.2012.06.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu H., Han Y., Rodriguez Sillke Y., Deng H., Siddiqui S., Treese C., Schmidt F., Friedrich M., Keye J., Wan J., et al. Lipid droplet-dependent fatty acid metabolism controls the immune suppressive phenotype of tumor-associated macrophages. EMBO Mol. Med. 2019;11 doi: 10.15252/emmm.201910698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Casanova-Acebes M., Menéndez-Gutiérrez M.P., Porcuna J., Álvarez-Errico D., Lavin Y., García A., Kobayashi S., Le Berichel J., Núñez V., Were F., et al. RXRs control serous macrophage neonatal expansion and identity and contribute to ovarian cancer progression. Nat. Commun. 2020;11 doi: 10.1038/s41467-020-15371-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boven L.A., Van Meurs M., Van Zwam M., Wierenga-Wolf A., Hintzen R.Q., Boot R.G., Aerts J.M., Amor S., Nieuwenhuis E.E., Laman J.D. Myelin-laden macrophages are anti-inflammatory, consistent with foam cells in multiple sclerosis. Brain. 2006;129:517–526. doi: 10.1093/brain/awh707. [DOI] [PubMed] [Google Scholar]
- 33.Shimabukuro M.K., Langhi L.G.P., Cordeiro I., Brito J.M., Batista C.M.d.C., Mattson M.P., de Mello Coelho V. Lipid-laden cells differentially distributed in the aging brain are functionally active and correspond to distinct phenotypes. Sci. Rep. 2016;6 doi: 10.1038/srep23795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xiong A., Zhang J., Chen Y., Zhang Y., Yang F. Integrated single-cell transcriptomic analyses reveal that GPNMB-high macrophages promote PN-MES transition and impede T cell activation in GBM. EBiomedicine. 2022;83 doi: 10.1016/j.ebiom.2022.104239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lavin Y., Winter D., Blecher-Gonen R., David E., Keren-Shaul H., Merad M., Jung S., Amit I. Tissue-resident macrophage enhancer landscapes are shaped by the local microenvironment. Cell. 2014;159:1312–1326. doi: 10.1016/j.cell.2014.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guilliams M., Svedberg F.R. Does tissue imprinting restrict macrophage plasticity? Nat. Immunol. 2021;22:118–127. doi: 10.1038/s41590-020-00849-2. [DOI] [PubMed] [Google Scholar]
- 37.Liu T., Zhang L., Joo D., Sun S.-C. NF-κB signaling in inflammation. Signal Transduct. Target. Ther. 2017;2 doi: 10.1038/sigtrans.2017.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Luo M.-C., Zhou S.-Y., Feng D.-Y., Xiao J., Li W.-Y., Xu C.-D., Wang H.-Y., Zhou T. Runt-related Transcription Factor 1 (RUNX1) Binds to p50 in Macrophages and Enhances TLR4-triggered Inflammation and Septic Shock. J. Biol. Chem. 2016;291:22011–22020. doi: 10.1074/jbc.M116.715953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ruffell D., Mourkioti F., Gambardella A., Kirstetter P., Lopez R.G., Rosenthal N., Nerlov C. A CREB-C/EBPbeta cascade induces M2 macrophage-specific gene expression and promotes muscle injury repair. Proc. Natl. Acad. Sci. USA. 2009;106:17475–17480. doi: 10.1073/pnas.0908641106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sha H., Zhang D., Zhang Y., Wen Y., Wang Y. ATF3 promotes migration and M1/M2 polarization of macrophages by activating tenascin-C via Wnt/β-catenin pathway. Mol. Med. Rep. 2017;16:3641–3647. doi: 10.3892/mmr.2017.6992. [DOI] [PubMed] [Google Scholar]
- 41.Gan L., Yang Y., Li Q., Feng Y., Liu T., Guo W. Epigenetic regulation of cancer progression by EZH2: from biological insights to therapeutic potential. Biomarker Res. 2018;6 doi: 10.1186/s40364-018-0122-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ozawa T., Riester M., Cheng Y.-K., Huse J.T., Squatrito M., Helmy K., Charles N., Michor F., Holland E.C. Most human non-GCIMP glioblastoma subtypes evolve from a common proneural-like precursor glioma. Cancer Cell. 2014;26:288–300. doi: 10.1016/j.ccr.2014.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Grajchen E., Wouters E., van de Haterd B., Haidar M., Hardonnière K., Dierckx T., Van Broeckhoven J., Erens C., Hendrix S., Kerdine-Römer S., et al. CD36-mediated uptake of myelin debris by macrophages and microglia reduces neuroinflammation. J. Neuroinflammation. 2020;17 doi: 10.1186/s12974-020-01899-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Berghoff S.A., Spieth L., Saher G. Local cholesterol metabolism orchestrates remyelination. Trends Neurosci. 2022;45:272–283. doi: 10.1016/j.tins.2022.01.001. [DOI] [PubMed] [Google Scholar]
- 45.Samtani G., Kim S., Michaud D., Hillhouse A.E., Szule J.A., Konganti K., Li J. Brain region dependent molecular signatures and myelin repair following chronic demyelination. Front. Cell. Neurosci. 2023;17 doi: 10.3389/fncel.2023.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim Y., Varn F.S., Park S.-H., Yoon B.W., Park H.R., Lee C., Verhaak R.G.W., Paek S.H. Perspective of mesenchymal transformation in glioblastoma. Acta Neuropathol. Commun. 2021;9 doi: 10.1186/s40478-021-01151-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Khera A.V., Cuchel M., de la Llera-Moya M., Rodrigues A., Burke M.F., Jafri K., French B.C., Phillips J.A., Mucksavage M.L., Wilensky R.L., et al. Cholesterol efflux capacity, high-density lipoprotein function, and atherosclerosis. N. Engl. J. Med. 2011;364:127–135. doi: 10.1056/NEJMoa1001689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim S., Steelman A.J., Zhang Y., Kinney H.C., Li J. Aberrant upregulation of astroglial ceramide potentiates oligodendrocyte injury. Brain Pathol. 2012;22:41–57. doi: 10.1111/j.1750-3639.2011.00501.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Remmerie A., Scott C.L. Macrophages and lipid metabolism. Cell. Immunol. 2018;330:27–42. doi: 10.1016/j.cellimm.2018.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ho Y.K., Brown M.S., Goldstein J.L. Hydrolysis and excretion of cytoplasmic cholesteryl esters by macrophages: stimulation by high density lipoprotein and other agents. J. Lipid Res. 1980;21:391–398. doi: 10.1016/S0022-2275(20)39788-1. [DOI] [PubMed] [Google Scholar]
- 51.Poulcharidis D., Belfor K., Kros A., van Kasteren S.I. A flow cytometry assay to quantify intercellular exchange of membrane components. Chem. Sci. 2017;8:5585–5590. doi: 10.1039/c7sc00260b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhou J., Febbraio M., Wada T., Zhai Y., Kuruba R., He J., Lee J.H., Khadem S., Ren S., Li S., et al. Hepatic fatty acid transporter CD36 is a common target of LXR, PXR, and PPARγ in promoting steatosis. Gastroenterology. 2008;134:556–567. doi: 10.1053/j.gastro.2007.11.037. [DOI] [PubMed] [Google Scholar]
- 53.El-Kenawi A., Dominguez-Viqueira W., Liu M., Awasthi S., Abraham-Miranda J., Keske A., Steiner K.K., Noel L., Serna A.N., Dhillon J., et al. Macrophage-derived cholesterol contributes to therapeutic resistance in prostate cancer. Cancer Res. 2021;81:5477–5490. doi: 10.1158/0008-5472.CAN-20-4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cancer Genome Atlas Research Network. Weinstein J.N., Collisson E.A., Mills G.B., Shaw K.R.M., Ozenberger B.A., Ellrott K., Shmulevich I., Sander C., Stuart J.M. The Cancer Genome Atlas Pan-Cancer analysis project. Nat. Genet. 2013;45:1113–1120. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.GLASS Consortium Glioma through the looking glass: molecular evolution of diffuse gliomas and the Glioma Longitudinal Analysis Consortium. Neuro-Oncology. 2018;20:873–884. doi: 10.1093/neuonc/noy020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cheng S., Li Z., Gao R., Xing B., Gao Y., Yang Y., Qin S., Zhang L., Ouyang H., Du P., et al. A pan-cancer single-cell transcriptional atlas of tumor infiltrating myeloid cells. Cell. 2021;184:792–809.e23. doi: 10.1016/j.cell.2021.01.010. [DOI] [PubMed] [Google Scholar]
- 57.Lee A.H., Sun L., Mochizuki A.Y., Reynoso J.G., Orpilla J., Chow F., Kienzler J.C., Everson R.G., Nathanson D.A., Bensinger S.J., et al. Neoadjuvant PD-1 blockade induces T cell and cDC1 activation but fails to overcome the immunosuppressive tumor associated macrophages in recurrent glioblastoma. Nat. Commun. 2021;12 doi: 10.1038/s41467-021-26940-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sade-Feldman M., Yizhak K., Bjorgaard S.L., Ray J.P., de Boer C.G., Jenkins R.W., Lieb D.J., Chen J.H., Frederick D.T., Barzily-Rokni M., et al. Defining T cell states associated with response to checkpoint immunotherapy in melanoma. Cell. 2018;175:998–1013.e20. doi: 10.1016/j.cell.2018.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cloughesy T.F., Mochizuki A.Y., Orpilla J.R., Hugo W., Lee A.H., Davidson T.B., Wang A.C., Ellingson B.M., Rytlewski J.A., Sanders C.M., et al. Neoadjuvant anti-PD-1 immunotherapy promotes a survival benefit with intratumoral and systemic immune responses in recurrent glioblastoma. Nat. Med. 2019;25:477–486. doi: 10.1038/s41591-018-0337-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hobson-Gutierrez S.A., Carmona-Fontaine C. The metabolic axis of macrophage and immune cell polarization. Dis. Model. Mech. 2018;11 doi: 10.1242/dmm.034462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Venkatesh H.S., Morishita W., Geraghty A.C., Silverbush D., Gillespie S.M., Arzt M., Tam L.T., Espenel C., Ponnuswami A., Ni L., et al. Electrical and synaptic integration of glioma into neural circuits. Nature. 2019;573:539–545. doi: 10.1038/s41586-019-1563-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Krishna S., Choudhury A., Keough M.B., Seo K., Ni L., Kakaizada S., Lee A., Aabedi A., Popova G., Lipkin B., et al. Glioblastoma remodelling of human neural circuits decreases survival. Nature. 2023;617:599–607. doi: 10.1038/s41586-023-06036-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Brooks L.J., Clements M.P., Burden J.J., Kocher D., Richards L., Devesa S.C., Zakka L., Woodberry M., Ellis M., Jaunmuktane Z., et al. The white matter is a pro-differentiative niche for glioblastoma. Nat. Commun. 2021;12 doi: 10.1038/s41467-021-22225-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stevens B., Allen N.J., Vazquez L.E., Howell G.R., Christopherson K.S., Nouri N., Micheva K.D., Mehalow A.K., Huberman A.D., Stafford B., et al. The classical complement cascade mediates CNS synapse elimination. Cell. 2007;131:1164–1178. doi: 10.1016/j.cell.2007.10.036. [DOI] [PubMed] [Google Scholar]
- 65.Kuhlmann-Hogan A., Cordes T., Xu Z., Kuna R.S., Traina K.A., Robles-Oteíza C., Ayeni D., Kwong E.M., Levy S., Globig A.-M., et al. EGFR-driven lung adenocarcinomas co-opt alveolar macrophage metabolism and function to support EGFR signaling and growth. Cancer Discov. 2024;14:524–545. doi: 10.1158/2159-8290.CD-23-0434. [DOI] [PubMed] [Google Scholar]
- 66.Wang S., Yan W., Kong L., Zuo S., Wu J., Zhu C., Huang H., He B., Dong J., Wei J. Oncolytic viruses engineered to enforce cholesterol efflux restore tumor-associated macrophage phagocytosis and anti-tumor immunity in glioblastoma. Nat. Commun. 2023;14 doi: 10.1038/s41467-023-39683-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kamphorst J.J., Cross J.R., Fan J., de Stanchina E.d., Mathew R., White E.P., Thompson C.B., Rabinowitz J.D. Hypoxic and Ras-transformed cells support growth by scavenging unsaturated fatty acids from lysophospholipids. Proc. Natl. Acad. Sci. USA. 2013;110:8882–8887. doi: 10.1073/pnas.1307237110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bogie J.F.J., Stinissen P., Hellings N., Hendriks J.J.A. Myelin-phagocytosing macrophages modulate autoreactive T cell proliferation. J. Neuroinflammation. 2011;8 doi: 10.1186/1742-2094-8-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yu W., Lei Q., Yang L., Qin G., Liu S., Wang D., Ping Y., Zhang Y. Contradictory roles of lipid metabolism in immune response within the tumor microenvironment. J. Hematol. Oncol. 2021;14 doi: 10.1186/s13045-021-01200-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Barthel F.P., Johnson K.C., Varn F.S., Moskalik A.D., Tanner G., Kocakavuk E., Anderson K.J., Abiola O., Aldape K., Alfaro K.D., et al. Longitudinal molecular trajectories of diffuse glioma in adults. Nature. 2019;576:112–120. doi: 10.1038/s41586-019-1775-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Holland E.C., Hively W.P., DePinho R.A., Varmus H.E. A constitutively active epidermal growth factor receptor cooperates with disruption of G1 cell-cycle arrest pathways to induce glioma-like lesions in mice. Genes Dev. 1998;12:3675–3685. doi: 10.1101/gad.12.23.3675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang S., Liu X.-S. The UCSCXenaTools R package: a toolkit for accessing genomics data from UCSC Xena platform, from cancer multi-omics to single-cell RNA-seq. J. Open Source Software. 2019;4 doi: 10.21105/joss.01627. [DOI] [Google Scholar]
- 73.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yan D., Kowal J., Akkari L., Schuhmacher A.J., Huse J.T., West B.L., Joyce J.A. Inhibition of colony stimulating factor-1 receptor abrogates microenvironment-mediated therapeutic resistance in gliomas. Oncogene. 2017;36:6049–6058. doi: 10.1038/onc.2017.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kong L., Dawkins E., Campbell F., Winkler E., Derks R.J.E., Giera M., Kamp F., Steiner H., Kros A. Photo-controlled delivery of very long chain fatty acids to cell membranes and modulation of membrane protein function. Biochim. Biophys. Acta Biomembr. 2020;1862 doi: 10.1016/j.bbamem.2020.183200. [DOI] [PubMed] [Google Scholar]
- 76.Feuerstein M.L., Kurulugama R.T., Hann S., Causon T. Novel acquisition strategies for metabolomics using drift tube ion mobility-quadrupole resolved all ions time-of-flight mass spectrometry (IM-QRAI-TOFMS) Anal. Chim. Acta. 2021;1163 doi: 10.1016/j.aca.2021.338508. [DOI] [PubMed] [Google Scholar]
- 77.Berthold M.R., Cebron N., Dill F., Gabriel T.R., Kötter T., Meinl T., Ohl P., Sieb C., Thiel K., Wiswedel B. In: Preisach C., Burkhardt H., Schmidt-Thieme L., Decker R., editors. Springer; 2008. KNIME: the Konstanz information miner. held in Berlin, Heidelberg; pp. 319–326. [Google Scholar]
- 78.Verhoeven A., Giera M., Mayboroda O.A. Kimble: A versatile visual NMR metabolomics workbench in KNIME. Anal. Chim. Acta. 2018;1044:66–76. doi: 10.1016/j.aca.2018.07.070. [DOI] [PubMed] [Google Scholar]
- 79.Ghorasaini M., Tsezou K.I., Verhoeven A., Mohammed Y., Vlachoyiannopoulos P., Mikros E., Giera M. Congruence and complementarity of differential mobility spectrometry and NMR spectroscopy for plasma lipidomics. Metabolites. 2022;12 doi: 10.3390/metabo12111030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Müller C., Junker J., Bracher F., Giera M. A gas chromatography-mass spectrometry-based whole-cell screening assay for target identification in distal cholesterol biosynthesis. Nat. Protoc. 2019;14:2546–2570. doi: 10.1038/s41596-019-0193-z. [DOI] [PubMed] [Google Scholar]
- 81.Kloos D.P., Gay E., Lingeman H., Bracher F., Müller C., Mayboroda O.A., Deelder A.M., Niessen W.M.A., Giera M. Comprehensive gas chromatography-electron ionisation mass spectrometric analysis of fatty acids and sterols using sequential one-pot silylation: quantification and isotopologue analysis. Rapid Commun. Mass Spectrom. 2014;28:1507–1514. doi: 10.1002/rcm.6923. [DOI] [PubMed] [Google Scholar]
- 82.Ciurli A., Mohammed Y., Ammon C., Derks R.J.E., Olivier-Jimenez D., Ducarmon Q.R., Slingerland M., Neefjes J., Giera M. Spatially and temporally resolved metabolome of the human oral cavity. iScience. 2024;27 doi: 10.1016/j.isci.2024.108884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rolfe A.J., Bosco D.B., Broussard E.N., Ren Y. In vitro phagocytosis of myelin debris by bone marrow-derived macrophages. J. Vis. Exp. 2017 doi: 10.3791/56322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hofmann K., Thiele C., Schött H.F., Gaebler A., Schoene M., Kiver Y., Friedrichs S., Lütjohann D., Kuerschner L. A novel alkyne cholesterol to trace cellular cholesterol metabolism and localization. J. Lipid Res. 2014;55:583–591. doi: 10.1194/jlr.D044727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Picelli S., Faridani O.R., Björklund A.K., Winberg G., Sagasser S., Sandberg R. Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 2014;9:171–181. doi: 10.1038/nprot.2014.006. [DOI] [PubMed] [Google Scholar]
- 86.Liu N.Q., Ter Huurne M., Nguyen L.N., Peng T., Wang S.Y., Studd J.B., Joshi O., Ongen H., Bramsen J.B., Yan J., et al. The non-coding variant rs1800734 enhances DCLK3 expression through long-range interaction and promotes colorectal cancer progression. Nat. Commun. 2017;8 doi: 10.1038/ncomms14418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Liu T. Use model-based analysis of ChIP-Seq (MACS) to analyze short reads generated by sequencing protein-DNA interactions in embryonic stem cells. Methods Mol. Biol. 2014;1150:81–95. doi: 10.1007/978-1-4939-0512-6_4. [DOI] [PubMed] [Google Scholar]
- 88.Danecek P., Bonfield J.K., Liddle J., Marshall J., Ohan V., Pollard M.O., Whitwham A., Keane T., McCarthy S.A., Davies R.M., et al. Twelve years of SAMtools and BCFtools. GigaScience. 2021;10 doi: 10.1093/gigascience/giab008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ramírez F., Ryan D.P., Grüning B., Bhardwaj V., Kilpert F., Richter A.S., Heyne S., Dündar F., Manke T. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016;44:W160–W165. doi: 10.1093/nar/gkw257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Stringer C., Wang T., Michaelos M., Pachitariu M. Cellpose: a generalist algorithm for cellular segmentation. Nat. Methods. 2021;18:100–106. doi: 10.1038/s41592-020-01018-x. [DOI] [PubMed] [Google Scholar]
- 91.Legland D., Arganda-Carreras I., Andrey P. MorphoLibJ: integrated library and plugins for mathematical morphology with ImageJ. Bioinformatics. 2016;32:3532–3534. doi: 10.1093/bioinformatics/btw413. [DOI] [PubMed] [Google Scholar]
- 92.Haase R., Royer L.A., Steinbach P., Schmidt D., Dibrov A., Schmidt U., Weigert M., Maghelli N., Tomancak P., Jug F., et al. CLIJ: GPU-accelerated image processing for everyone. Nat. Methods. 2020;17:5–6. doi: 10.1038/s41592-019-0650-1. [DOI] [PubMed] [Google Scholar]
- 93.Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M., 3rd, Hao Y., Stoeckius M., Smibert P., Satija R. Comprehensive integration of single-cell data. Cell. 2019;177:1888–1902.e21. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Prinz M., Masuda T., Wheeler M.A., Quintana F.J. Microglia and central nervous system-associated macrophages-from origin to disease modulation. Annu. Rev. Immunol. 2021;39:251–277. doi: 10.1146/annurev-immunol-093019-110159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Scialdone A., Natarajan K.N., Saraiva L.R., Proserpio V., Teichmann S.A., Stegle O., Marioni J.C., Buettner F. Computational assignment of cell-cycle stage from single-cell transcriptome data. Methods. 2015;85:54–61. doi: 10.1016/j.ymeth.2015.06.021. [DOI] [PubMed] [Google Scholar]
- 96.Allhoff M., Seré K., F Pires J., Zenke M., G Costa I. Differential peak calling of ChIP-seq signals with replicates with Thor. Nucleic Acids Res. 2016;44 doi: 10.1093/nar/gkw680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Thorvaldsdóttir H., Robinson J.T., Mesirov J.P. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinform. 2013;14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kanehisa M., Furumichi M., Sato Y., Ishiguro-Watanabe M., Tanabe M. KEGG: integrating viruses and cellular organisms. Nucleic Acids Res. 2021;49:D545–D551. doi: 10.1093/nar/gkaa970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Reimand J., Kull M., Peterson H., Hansen J., Vilo J. g:profiler--a web-based toolset for functional profiling of gene lists from large-scale experiments. Nucleic Acids Res. 2007;35:W193–W200. doi: 10.1093/nar/gkm226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Luo W., Friedman M.S., Shedden K., Hankenson K.D., Woolf P.J. GAGE: generally applicable gene set enrichment for pathway analysis. BMC Bioinformatics. 2009;10 doi: 10.1186/1471-2105-10-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Barbie D.A., Tamayo P., Boehm J.S., Kim S.Y., Moody S.E., Dunn I.F., Schinzel A.C., Sandy P., Meylan E., Scholl C., et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature. 2009;462:108–112. doi: 10.1038/nature08460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Reich M., Liefeld T., Gould J., Lerner J., Tamayo P., Mesirov J.P. GenePattern 2.0. Nat. Genet. 2006;38:500–501. doi: 10.1038/ng0506-500. [DOI] [PubMed] [Google Scholar]
- 104.Liu N.Q., Maresca M., van den Brand T., Braccioli L., Schijns M.M.G.A., Teunissen H., Bruneau B.G., Nora E.P., de Wit E. WAPL maintains a cohesin loading cycle to preserve cell-type-specific distal gene regulation. Nat. Genet. 2021;53:100–109. doi: 10.1038/s41588-020-00744-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.van Heeringen S.J., Veenstra G.J.C. GimmeMotifs: a de novo motif prediction pipeline for ChIP-sequencing experiments. Bioinformatics. 2011;27:270–271. doi: 10.1093/bioinformatics/btq636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Therneau . 2022. A Package for Survival Analysis in R.https://CRAN.R-project.org/package=survival R Package Version 3.3.-1. [Google Scholar]
- 107.Kassambara A., K.M.a.B.P. 2021. Survminer: drawing Survival Curves using 'ggplot2'.https://rpkgs.datanovia.com/survminer/index.html R package version 0.4.9. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Macrophage subset signatures identified scRNA-seq dataset of GEMMs of glioblastoma.
(B) Relevant gene signatures for TAMs in glioblastoma from cited literature.
(C) GPNMBhigh signatures for analysis of murine or patient datasets.
(D) Gene signatures derived from FACS-purified (non-) lipid-laden MG and MDMs generated by RNA-seq.
(E) Curated Ivy GAP niche gene sets for scRNA-seq analysis of tumor cells.
(F) Curated Ivy GAP niche gene sets for scRNA-seq analysis of macrophages.
(G) Curated Neftel et al. cellular subtype gene sets for assessing subtype localisation in VISIUM10x samples.
(H) Curated TAM subset gene sets for assessing localisation in VISIUM10x samples.
(A) Gene set over-representation analysis, MG1-P2RY12.
(B) Gene set over-representation analysis, MG2-TNF.
(C) Gene set over-representation analysis, MG3-GPNMB.
(D) Gene set over-representation analysis, MG4-MKI67.
(E) Gene set over-representation analysis, MDM1-CCR2.
(F) Gene set over-representation analysis, MDM2-H2-EB1.
(G) Gene set over-representation analysis, MDM3-GPNMB.
(H) Gene set over-representation analysis, MDM4-MKI67.
(A) Gene set over-representation analysis, GPNMBlow.
(B) Gene set over-representation analysis, GPNMBhigh.
(A) GSEA on genes associated with increased accessible promoter regions in MG versus MDMs.
(B) GSEA on genes associated with increased accessible promoter regions in MDMs versus MG.
(C) GSEA on genes associated with increased accessible chromatin regions in lipid-laden MDMs versus non-lipid-laden MDMs.
(D) GSEA on genes associated with decreased accessible chromatin regions in lipid-laden MDMs versus non-lipid-laden MDMs.
(E) GSEA on genes associated with increased accessible chromatin regions in lipid-laden MG versus non-lipid-laden MG.
(F) GSEA on genes associated with decreased accessible chromatin regions in lipid-laden MG versus non-lipid-laden MG.
(G) GSEA on genes associated with increased accessible chromatin regions in lipid-laden TAMs versus non-lipid-laden TAMs.
(H) GSEA on genes associated with decreased accessible chromatin regions in lipid-laden TAMs versus non-lipid-laden TAMs
(A) Gene signatures from FACS-purified dTom+ macrophages from ex vivo LLM formation assay analysed by RNA-seq.
(B) Gene set enrichment analysis on gene signature "up.all".
(C) Gene set enrichment analysis on gene signature "up.full".
(D) Gene set enrichment analysis on gene signature "up.tumor".
(E) Gene set enrichment analysis on gene signature "up.myelin".
(F) Gene set enrichment analysis on gene signature "up.full.myelin".
(G) Gene set enrichment analysis on gene signature "up.full.tumor".
(H) Gene set enrichment analysis on gene signature "down.full".
(I) Gene set enrichment analysis on gene signature "down.tumor".
(J) Gene set enrichment analysis on gene signature "down.myelin".
(K) Gene set enrichment analysis on gene signature "down.full.tumor".
(L) Gene set enrichment analysis on gene signature "down.full.myelin".
(A) Gene set enrichment analysis on gene signature "down.hpm0_vs_hpm24_mono_up_tumor".
(B) Gene set enrichment analysis on gene signature "down.hpm0_vs_hpm24_mono_down_tumor".
(C) Gene set enrichment analysis on gene signature "down.hpm0_movo_vs_co_up_tumor".
(D) Gene set enrichment analysis on gene signature "down.hpm0_movo_vs_co_down_tumor".
(E) Gene set enrichment analysis on gene signature "down.hpm24_movo_vs_co_up_tumor".
Data Availability Statement
-
•
Data generated in this study was aligned using the mouse reference genome mm10 and is deposited on GSE266149. The glioblastoma niche and subtype meta-module gene lists used in this study can be obtained from the Ivy Glioblastoma Atlas Project (GAP) or Neftel et al.18 (Table S2), respectively. The glioblastoma patient scRNA-seq datasets were kindly provided to us by Dr. Kevin Petrecca, McGill University.5,19 The publicly available single-cell RNA sequencing datasets which have been used in this study, including the myeloid cell atlas,56 the melanoma treatment-related dataset,58 and the response of glioblastoma patients to neoadjuvant aPD157 can be obtained from GEO: GSE154763, GSE120575, and GSE154795, respectively. TCGA-glioblastoma data was obtained from http://gdac.broadinstitute.org/ using the package UCSCXenaTools.72 The raw counts were processed using EdgeR.73 The GLASS dataset70 was downloaded from https://www.synapse.org/#!Synapse:syn17038081/wiki/585622, where patients for glioblastoma and IDH-wild-type were selected.
-
•
The original codes for the murine scRNA-seq and Visium spatial transcriptomic analyses are made available at https://github.com/djkloosterman/Kloosterman_et_al_Cell_2024.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.