Abstract
Blood vessel endothelial cells (EC) display heterogeneity across vascular beds, which is anticipated to drive site-specific vascular pathology. This heterogeneity is assessed using transcriptomics in vivo, and functional assays in vitro, but how proteomes compare across human in vitro cultured ECs remains incompletely characterized. We generated an in-depth human EC proteomic landscape (>8000 proteins) across six organs and two in vitro models in steady-state and upon IFNγ-induced inflammation. EC proteomes displayed a high similarity and organ-specific proteins were limited. Variation between ECs was mainly based on proliferation and differentiation processes in which Blood outgrowth endothelial cells (BOEC) and Human umbilical vein cells (HUVEC) represented the extremes of proteomic phenotypes. The IFNγ response was highly conserved across all samples. Harnessing dynamics in protein abundances we delineated VWF and VE-Cadherin correlation networks. This EC landscape provides an extensive proteomic addition in studying EC biology and heterogeneity from an in vitro perspective.
Subject terms: Systems biology, Cell biology
Analysis of proteomic signatures of commercially acquired endothelial cells from different organs in steady state and after IFNγ inflammation in the context of understanding vascular heterogeneity.
Introduction
Vascular endothelial cells (ECs) form a dynamic interface between blood and tissue. At this interface ECs regulate blood vessel dilation and permeability (e.g. through regulation of tight junctions) and play a crucial role in hemostasis (e.g. the release of von Willebrand factor (VWF)) and inflammation (e.g. release of chemokines)1–4. The endothelial monolayer is diverse and consists of a spectrum of ECs which are specialized based on the organ or vascular bed they reside in. This heterogeneity is dependent on various factors such as vessel size (macro- versus micro-vascular), vessel type (arterial versus venous) and their respective microenvironment5–7. EC heterogeneity is anticipated to contribute to the site-specificity of various disorders, such as venous thrombosis, atherosclerosis and pulmonary arterial hypertension8–11. Therefore, vascular bed heterogeneity is an important topic in the context of organ-specific vascular disorders and drug-targeting/metabolism.
Our knowledge on EC heterogeneity is mostly based on single-cell mRNA sequencing using murine models8–13. These showed EC adaptations to their physiological roles such as increased mRNA levels of scavenger receptors in liver sinusoidal ECs, tight junctions in brain ECs in the blood brain barrier and the expression of metabolic genes in cardiac ECs8–13. However, it is unclear how these predominantly murine insights translate to human ECs. In addition, studies that assess EC heterogeneity that include unbiased proteomics are limited14, thus how transcriptomic observations translate to differences in protein levels is unresolved. Finally, human ECs are used extensively in vitro in both fundamental and translational settings to investigate functional outcomes and responses to inflammatory challenges15,16–18. Therefore it is important to assess how proteomes compare across ECs in in vitro culture.
To understand how organ-specificity, donor variation and inflammation facet into the context of studying EC biology and heterogeneity, we set out to characterize proteomes across a range of in vitro cultured human ECs. We analyzed the proteomes of cultured primary human ECs originating from aorta, brain, cardiac, kidney, liver and lung vascular beds (all microvascular ECs expect for aorta), and the two most-commonly used in vitro models: Blood-outgrowth ECs (BOECs or Endothelial colony forming ECs (ECFCs)) and human umbilical vein ECs (HUVECs) in both steady state and in response to IFNγ stimulation.
We generated an in-depth human EC proteomic landscape (>8000 proteins) across ECs, and highlight the proteomic overlap and differences between EC types. Moreover, we analyzed IFNγ induced responses, which were highly conserved across all samples. Due to the extensive variation of ECs included in this study, combined with the proteomic depth, we could harness the dynamics in protein abundances to highlight known and putative associations of core EC proteins VWF and VE-Cadherin. Taken together, this EC proteomic landscape, across a wide variety of EC-types, donors and inflammation, provides an extensive source in studying basic EC biology and heterogeneity.
Results
In vitro EC proteomic landscapes across vascular beds
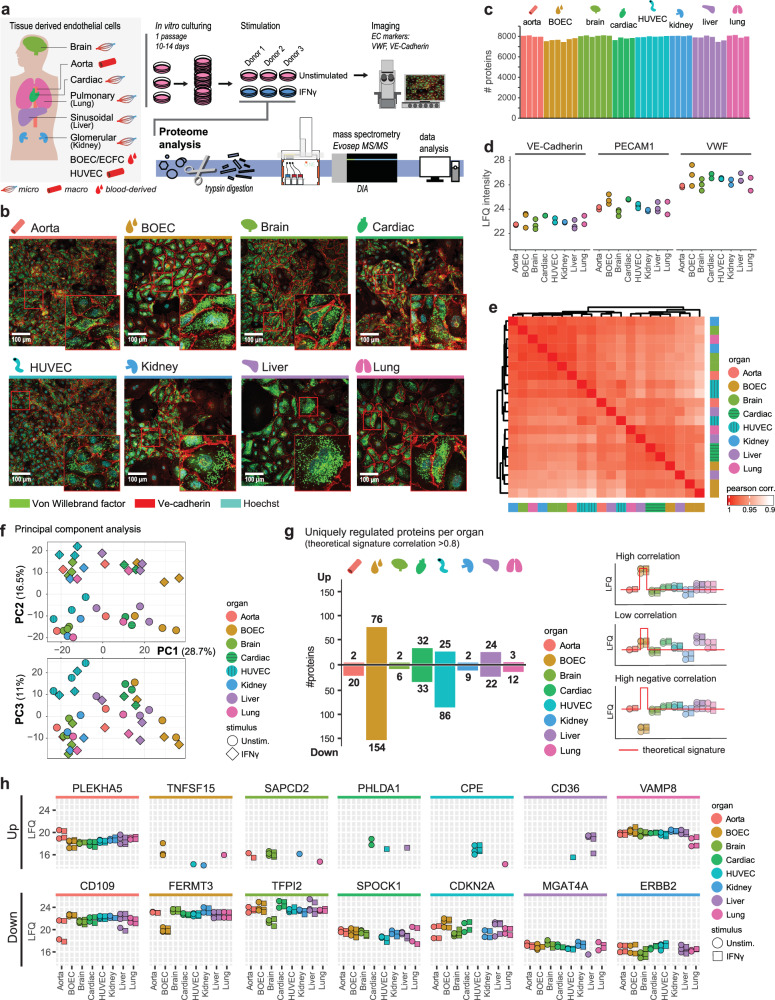
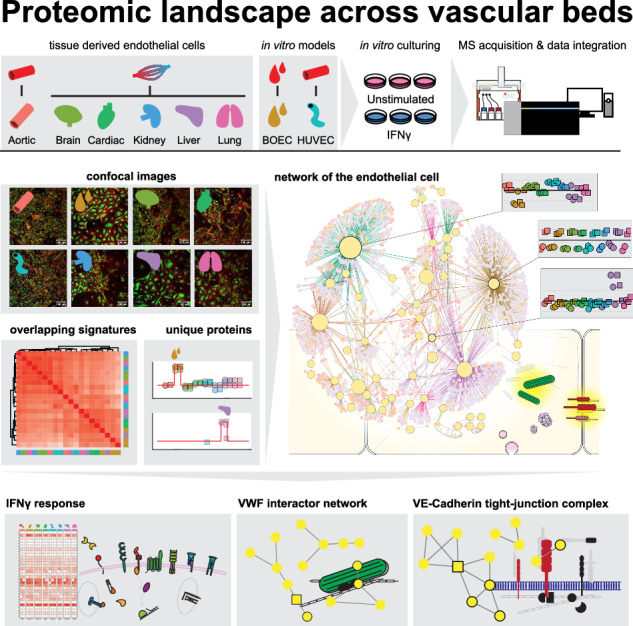
To determine the proteomic landscape across different vascular beds, we cultured BOECs, HUVECs and ECs from aorta, brain, cardiac, kidney, liver and lung origin, acquired from different suppliers and various donors (Supplementary Table 1). To keep culturing time to a minimum, all cells were passaged once and subjected to the same criteria with respect to passage, stimulation and harvest conditions (Supplementary Fig. S1). EC markers VWF and VE-Cadherin were assessed in unstimulated conditions through confocal microscopy, and both unstimulated and IFNy-treated cells were analyzed using Label Free Quantification (LFQ) mass spectrometry (Fig. 1a). Notably, all cells sourced from a single supplier exhibited divergent proteomes, irrespective of cell type (Supplementary Fig. S2a). Although showing similar abundances of EC hallmark proteins (VE-Cadherin, PECAM1, VWF) (Supplementary Fig. S2b), these cells showed higher abundances (p value < 0.05, LFC > 1) of LYVE1, a lymphatic EC marker19, and CD68, a classical monocyte/macrophage marker20, compared to other cell types (Supplementary Fig. S2c). Therefore, these samples were omitted from further analyses. Microscopic observation showed presence of endothelial cell markers VWF and VE-Cadherin (CDH5) throughout all other samples, although VWF was heterogeneously expressed between cells within a monolayer (Fig. 1b). Employing data-independent MS acquisition, we generated an in-depth EC proteome, quantifying 7000–8000 proteins per sample (Fig. 1c). Protein abundances of VWF, VE-Cadherin and PECAM1 were stable through all cell types, although VWF showed more heterogeneous abundances between donors (Fig. 1d). Overall correlation between datasets was high (mean Pearson correlation >0.96) indicating a large overlap between EC proteomes. The lowest correlations were observed between BOEC and HUVEC samples (min. Pearson correlation = 0.91) (Fig. 1e). To visualize the variation between samples, we performed a principal component analysis (PCA). This showed clear separation of IFNγ stimulation over PC2, while PC1 and PC3 separated cell types and donors (Fig. 1f). This analysis highlighted a large donor-to-donor variation, most notable for lung, aorta and liver ECs. To assess whether we could identify proteins uniquely regulated per EC type despite the observed variation, we determined protein correlations to theoretical specific profiles per organ. This indicated various vascular bed associated proteins (correlation >0.8), containing both increased as decreased abundances: BOEC—230 proteins, HUVEC—111, cardiac—65, liver—46, aorta—22, brain, kidney and lung—<20 (Fig. 1g). We plotted proteins with an evident unique regulation for aorta (PLEKHA5, CD109), BOEC (TNFSF15, FERMT3), brain (SAPCD2, TFPI2), cardiac (PHLDA1, SPOCK1), HUVEC (CPE, CDKN2A), liver (CD36, MGAT4A), lung (VAMP8) and kidney (ERBB2) (Fig. 1h). We assessed two of these proteins by immunostaining. CD36, a fatty acid uptake receptor in ECs21,22, which was uniquely quantified by proteomics in liver ECs in this study. The immunolabeling revealed fluorescent foci to be predominantly present in livers ECs, with a lower expression in other cell types (Supplementary Fig. S3). In addition, we assessed FERMT3 (KINDLIN-3), which was reduced in BOECs compared to other cell types. FERMT3 is in particular interest in the endothelium as mutations in this gene result in a bleeding phenotype and impaired immune cell adherence to the vessel wall23. Immunostaining showed similar intensities and patterns across cell types, and we could not discern differences in abundance between ECs (Supplementary Fig. S4).
Fig. 1. Proteomic signatures of ECs are dependent on organ-source and donor variations.
a Schematic overview of experimental workflow. Mass spectrometry acquisition was performed in one combined analysis with cells derived from three different donors per organ. b Confocal images per EC-type of VWF (green) and VE-Cadherin (red) immunostaining in unstimulated ECs, Hoechst staining in cyan. Representative experiment shown (n = 2 biologically independent experiments). Upper limit of the display range were adjusted equally across images for visualization purposes. c Number of quantified proteins across samples. d LFQ intensities of EC markers across unstimulated ECs. e Heatmap showing Pearson correlation between unstimulated samples. Color gradient (white to red) indicates Pearson correlation (0.9–1.0). f Principal component analysis (PCA) of proteomes across PC1, PC2 and PC3. Colors indicate EC-type. Unstimulated (circles) and IFNγ stimulated (diamonds) conditions are indicated. g Number of proteins with a unique down- or upregulation per EC-type (absolute correlation to theoretical signature >0.8). Examples of high, low and high negative correlation to one EC-type theoretical signature (red line) are shown. h LFQ intensities of top high and high negative correlating proteins shown per organ. Unstimulated (circles) and IFNγ stimulated (squares) are indicated. EC-types are indicated per color in all panels as follows: Aorta (red), BOEC (brown), Brain (light green), Cardiac (dark green), HUVEC (cyan), Kidney (blue), Liver (purple) and Lung (pink).
To assess whether previously identified organ-specific regulated genes found in murine studies were in line with our study, we compared our protein data with two murine transcriptomics studies8,12 (Supplementary Fig. S5). Using the top-10 genes per organ as reported by Kalucka et al. we observed that most specifically expressed transcripts were equally abundant on proteome level. On the other hand proteins that were uniquely regulated in this study such as TSPAN7 and CPE in HUVECs were reported by Kalucka et al. as specific to lung and heart respectively. Moreover, although Kalucka and Cleueren et al. did show overlapping signatures for brain, lung and liver, many transcripts were not uniquely expressed in these studies either, highlighting the challenges in determining organ-specific genes.
A global network analysis reveals separation between proliferative and EC differentiation processes
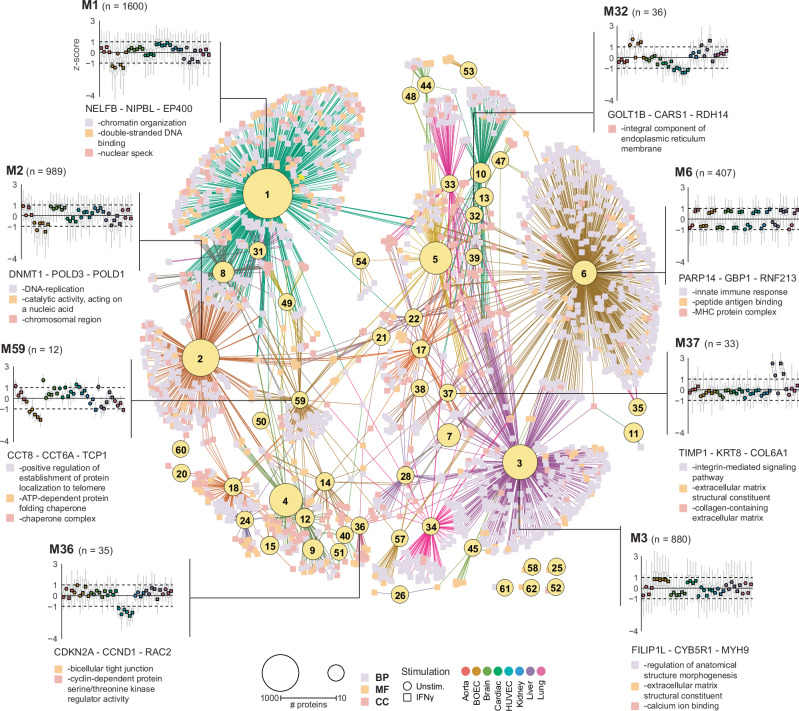
As we identified limited specific marker proteins per organ, we wanted to understand the main drivers underlying the variation between samples. To exploit the observed variation and assess (shared) proteomic signatures between ECs from different origins and upon IFNγ-induced inflammation, we performed weighted gene correlation network analysis (WGCNA) on all quantified proteins which resulted in 62 modules (Supplementary Fig. S6). To visualize the biological processes associated with these modules, we performed GO enrichment for “Biological Process” (BP), “Molecular Function” (MF) and “Cellular Compartment” (CC) per module and generated an interaction network (Fig. 2). Initially, we investigated the three modules containing the most proteins. These modules described the main separation within the network. Module 1 (1600 proteins) and module 2 (989 proteins), enriched for DNA-binding and -replication processes. On the other side module 3 (880 proteins) contained mainly structural and extracellular matrix proteins. Next we analyzed module 6 (407 proteins), this module reflected IFNγ stimulation, enriching for upregulation of innate immune response and MHC-protein complexes and was consistent across all cell types. Finally, we investigated modules which were uniquely regulated in one EC-type. Among these were module 36 (35 proteins), containing tight junction proteins and cell-cycle inhibitors, which were lower abundant in HUVECs. Module 37 (33 proteins), which was upregulated in two liver samples and enriched for extracellular matrix proteins. Among the top three highest module membership proteins in this module was TIMP1, a metalloproteinase inhibitor known to mediate the extracellular matrix in the liver24. Finally, module 59 was uniquely downregulated in BOECs and contained 12 proteins of the chaperone tailless complex polypeptide-1-ring complex (TRiC) (e.g. CCT8, CCT6A, TCP1)25.
Fig. 2. Interaction network shows varied trends in regulation of biological processes across EC-types.
Interaction network of WGCNA analysis modules and enriched GO-enrichment terms. Yellow nodes indicate modules, size shows number of proteins in the respective module. Squares indicate enriched GO-terms, Biological process (BP, blue), Molecular function (MF, yellow), Cellular component (CC, red). Edge colors indicate connected module. Examples of node regulation are shown as z-score means of LFQ intensities of the proteins in the module across EC-types and stimulation. Unstimulated (circles) and IFNγ stimulated (squares) are indicated. Mean z-score per sample per EC-type are indicated per color as follows: Aorta (red), BOEC (brown), Brain (light green), Cardiac (dark green), HUVEC (cyan), Kidney (blue), Liver (purple) and Lung (pink). Upper and lower parts of boxplots indicate the 25th and 75th percentiles of all protein z-scores per sample, outliers are not shown. Per module example, the top three highest correlating protein to respective modules, and top enriched GO-term per type are shown.
The IFNγ response is highly conserved in in vitro cultured ECs
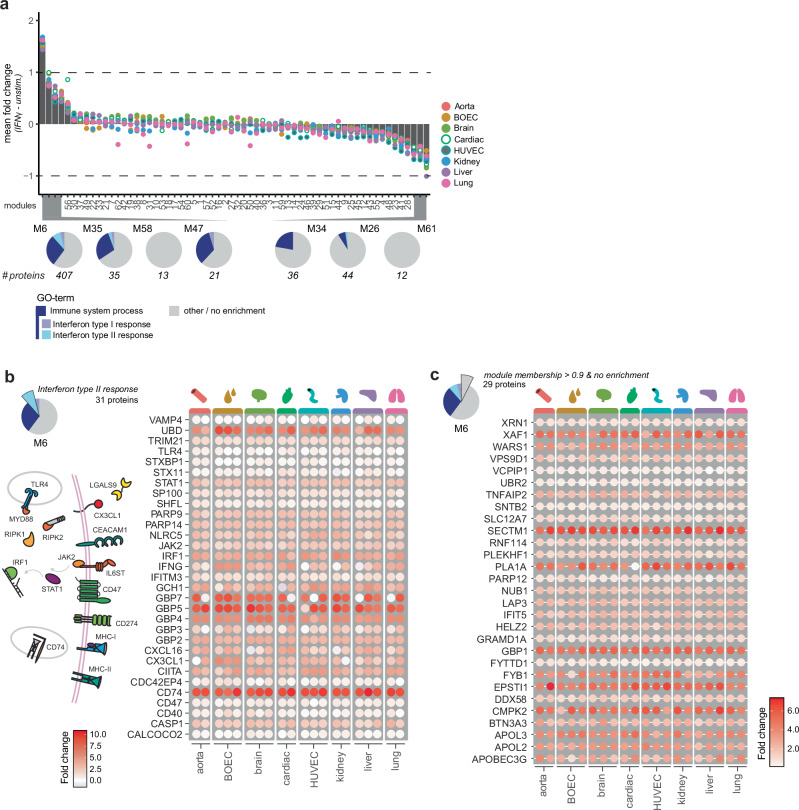
To determine if inflammatory responses are specific or shared between ECs from different vascular beds, we next investigated IFNγ responses across endothelial cell types. To assess which modules described an IFNγ response, we determined the mean log fold changes between IFNγ-stimulated samples versus unstimulated controls per module (Fig. 3a). Several modules showed minor changes, while only the afore mentioned module 6 had a mean LFC > 1. To assess if proteins were equally induced across cell types we first plotted individual fold changes per sample for the “IFN type II response” annotated proteins in module 6. No general trend in either specific organ nor donor regulation was observed and proteins were induced similarly across samples among which hallmark proteins STAT1, JAK2, IRF1 and CD74 (Fig. 3b). Notably, among proteins resembling the module regulation most closely (module membership >0.9), 29 had no direct annotation with “Interferon response type I and II” or the broader “Immune system process” term (Fig. 3c). Among these proteins were known interferon inducible genes (ISGs) such as SECTM126, XAF127, PLA1A28 and EPSTI129, which again showed highly similar increases in protein abundances. Based on these protein dynamics we determined a remarkable similar induction of proteins by IFNγ regardless of EC-type and donor.
Fig. 3. The IFNγ response is highly conserved.
a Bar graph of mean LFQ difference between unstimulated and stimulated proteins per module. Circles indicate mean LFQ difference per EC-type. EC-types are indicated per color as follows: Aorta (red), BOEC (brown), Brian (light green), Cardiac (dark green), HUVEC (cyan), Kidney (blue), Liver (purple) and Lung (pink). Pie charts of modules (mean LFC > 0.5) show relative enrichment of proteins per module of GO-terms “Immune system process” (blue), “Interferon type I response” (purple), “Interferon type II response” (cyan) and other/no enrichment (gray). Number of proteins per module are indicated. Heatmaps showing LFC change between unstimulated and stimulated per sample of proteins enriching for (b) “Interferon type II response” and (c) non-enriching proteins highly correlating to module 6 (Membership score >0.9). Color gradient (white to red) indicates LFC per sample (0–10 or 0–7 respectively). Schematic overview of “Interferon type II response” proteins is shown.
Elucidating the VWF correlation network
Next to investigating variation in biological processes from an unbiased perspective, the proteome dynamics based analysis can be harnessed to study processes or complexes from a protein-centric angle. A hallmark EC protein is VWF, which is crucial in mediating platelet interactions important for hemostasis and is the main constituent of Weibel-Palade bodies (WPBs). VWF was a member of module 25, which showed a general increased abundance for BOEC/cardiac/HUVEC/liver versus aorta/brain/kidney/lung ECs and enriched predominantly for actin- cytoskeleton and binding processes (Fig. 4a, b). To assess which proteins in this module were most associated with the abundance of VWF, we determined the Pearson correlation of proteins in module 25 to VWF and observed 16 highly correlating proteins (Pearson correlation >0.7) which were plotted in a STRING-DB protein-protein interaction network (confidence interactions score >0.4) (Fig. 4c). The highest correlations were between VWF and RAB3D, MYO5C, LIMCH1 and ABLIM1, which are known interactors of VWF30–35 (Fig. 4d and Supplementary Table 2). Interestingly, there were several other highly correlating proteins in this module, such as AFDN, EHD3, LRATD2 and PALD1, which had no STRING-db protein-interaction and have previously not been characterized as VWF-interacting proteins. Next, we determined the correlation between well-established proteins present in- or interacting with- WPBs that were not in module 25 (Fig. 4e). Of these SELP and ANGPT2, both present in WPBs, showed minor correlation with VWF (Pearson correlation 0.52 and 0.36 respectively), while CD63 and IGFBP7 had correlation values close to 0 (0.03 and 0.11 respectively). This low correlation was also observed for other known WPB-associated proteins, including RAB27A, RAB3B and SEC22B.
Fig. 4. Protein dynamics reveal VWF correlation network.
a Module protein dynamics is shown as means of z-scores of LFQ intensities across EC-types and stimulation. Upper and lower parts of boxplots indicate the 25th and 75th percentiles of all protein z-scores per sample, outliers are not shown. b Top five significantly enriched GO-terms are shown (p < 0.05). Node size indicates number of enriched proteins per GO-term. Color indicates GO-term type, Biological process (BP, blue), Molecular function (MF, yellow), Cellular component (CC, red). c STRING-DB protein-protein interaction network of proteins with a reported interaction (confidence interactions score >0.4) and proteins with a high correlation to VWF (Pearson correlation >0.7, black border). Color gradient (white to red) indicates Pearson correlation (0.2–1.0). d LFQ intensities of top 4 VWF correlating proteins shown per organ. Pearson correlation is shown in top right per protein. e Line plots of mean LFQ intensities per organ and stimulation of known WPB cargo or interactors (black). Pearson correlation to VWF is shown in top right and mean VWF LFQ intensity is shown per protein (gray) For all panels: unstimulated (circles) and IFNγ stimulated (squares) are indicated. EC-types are indicated per color as follows: Aorta (red), BOEC (brown), Brain (light green), Cardiac (dark green), HUVEC (cyan), Kidney (blue), Liver (purple) and Lung (pink).
The VE-Cadherin tight junction complex
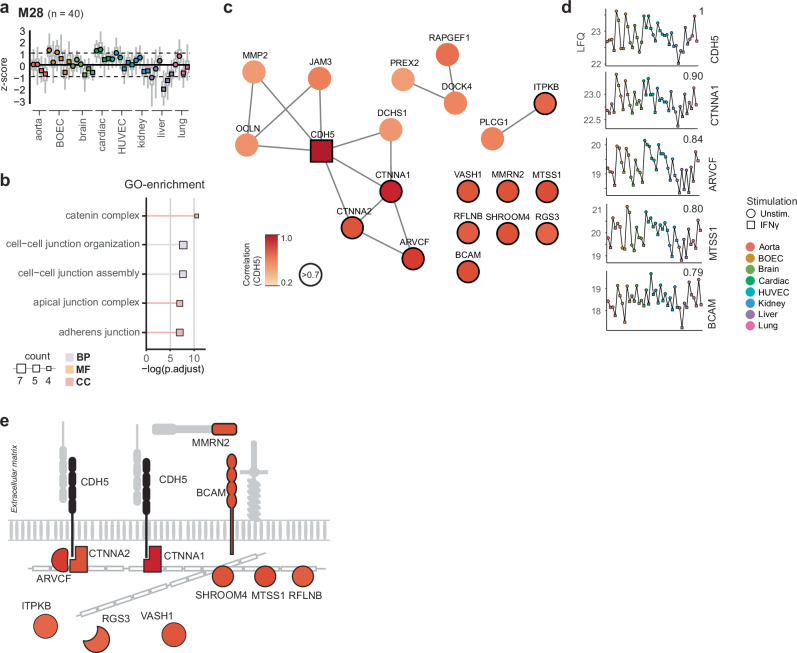
It is well established that EC activation impacts tight junctions and EC permeability. CDH5 (VE-Cadherin) is a pivotal protein in endothelial cell-cell junctions. This protein was in module 28, which enriched for cell-cell junction processes and was minorly downregulated after IFNγ stimulation in all samples (Fig. 5a, b). To assess closely associated proteins, we determined the Pearson correlation of proteins in this module to CDH5 and plotted the ones with high correlation (>0.7) (Fig. 5c). Among the highest correlation were actin anchors CTNNA1 and ARVCF known to interact with VE-Cadherin36,37, as well as laminin binding receptor BCAM (Fig. 5d). Interestingly we observed several inhibitors of angiogenesis including Vasohibin (VASH1), Multimerin-2 (MMRN2), Metastasis suppressor (MTSS1), Refelin-B (RFLNB) to be highly correlated to CDH5 as well (Fig. 5e).
Fig. 5. The VE-Cadherin complex network.
a Module protein dynamics is shown as means of z-scores of LFQ intensities across EC-types and stimulation. Upper and lower parts of boxplots indicate the 25th and 75th percentiles of all protein z-scores per sample, outliers are not shown. b Top five significantly enriched GO-terms are shown (p < 0.05). Node size indicates number of enriched proteins per GO-term. Color indicates GO-term type, Biological process (BP, blue), Molecular function (MF, yellow), Cellular component (CC, red). c STRING-DB protein-protein interaction network of proteins with a reported interaction (confidence interactions score >0.4) and proteins with a high correlation to VE-Cadherin (Pearson correlation >0.7, black border). Color gradient (white to red) indicates Pearson correlation (0.2–1.0). d LFQ intensities of top VE-Cadherin correlating proteins shown per organ. Pearson correlation is shown in top right per protein. e Schematic overview of VE-Cadherin (black) and highly correlating proteins (Pearson correlation >0.7). Color gradient (white to red) indicates Pearson correlation (0.2–1.0). For all panels: unstimulated (circles) and IFNγ stimulated (squares) are indicated. EC-types are indicated per color as follows: Aorta (red), BOEC (brown), Brain (light green), Cardiac (dark green), HUVEC (cyan), Kidney (blue), Liver (purple) and Lung (pink).
Discussion
This study provides a human proteomic landscape across a broad range of in vitro cultured EC-types. We highlight that all ECs have similar proteomic makeup, with limited specific signatures between organs and donors. Notably, IFNy stimulation resulted in a homogeneous proteomic response across all ECs, indicating this is a highly conserved cellular process. The proteomic depth across multiple varying endothelial samples, allowed to discern protein networks of known and unknown proteins in the context of fundamental EC processes, including VWF and VE-Cadherin.
The most profound proteomic differences between EC types was observed between HUVECs and BOECs, mirroring their distinctly different origins from the other cell types in this study, i.e., the umbilical vein and blood resident progenitor cells. Notably, these two in vitro models captured the entire proteomic range of in vitro cultured ECs in our study, emphasizing their applicability for studying EC biology. The main axis of separation was between proliferative processes (DNA-binding, histone modulation, ribosome complexes) versus differentiation (extracellular matrix and structural constituents), a relationship which has been described in other (developmental) studies13,38,39. BOECs enriched for upregulation of differentiation processes, which was also observed for cardiac and liver ECs. On the other hand, HUVECs mostly enriched for proliferative processes, together with brain and kidney samples. HUVECs also had an uniquely downregulated module, containing cell-cycle inhibitors CDKN2A and CCND1 among the highest correlating, as well as several tight-junction proteins. These ECs were the only non-adult derived cells in this study, which may be at the basis of this unique regulation.
Our observations on IFNγ responses showed that regardless of vascular bed, ECs contain the molecular machinery to respond to IFNγ. Moreover, the fact that newly synthesized IFNγ-induced proteins result in highly similar protein abundances across all samples, marks a tightly controlled process of transcriptomic induction and subsequent proteomic translation. This similarity is remarkable in comparison to reports on TNFα-induced inflammation which describe organ-specific EC responses11,40. Moreover, this EC IFNγ response is similar to that of other (immune) cell types, highlighting a pivotal conserved and regulated IFNγ response throughout our cells41–43.
A strength of our in-depth proteomic dataset over a wide variety of donors, EC-types and inflammation is that it provides a basis for studying fundamental endothelial biology on a molecular level. Given their central role in EC biology, we examined VWF-containing WPBs and VE-Cadherin mediated tight-junctions. VWF was in a module which highly enriched for actin binding processes, which is as expected since WPBs are tethered to and transported across the actin-network44. RAB3D showed a very high correlation to VWF and is also known to, not only localize to, but also necessary in the formation of WPBs34,35. On the other hand other well-described RABs such as RAB27A and RAB3B showed limited correlation. This is surprising, as both RAB27A and RAB3B are reported to be recruited to WPBs35,45,46. Of all other proteins with a high correlation to VWF, several have previously been detected in pull-downs with VWF-interactors (ABLIM1 and LIMCH1)30,33,47, while others, AFDN and EHD3 are actin regulating proteins and thus can be expected to play a role in anchoring VWF to the actin-network. Interestingly, SLC44A2 has been shown to interact with VWF in platelets, but not in ECs48. Moreover, another protein with a high correlation, PALD1, is poorly characterized both in function and in relation to VWF. It has been reported as an important protein in endothelial development49, but a relation to VWF has previously not been made.
Concerning VE-Cadherin, we observed that known complex member CTNNA1, CTNNA2 and ARVCF highly correlated to VE-Cadherin36,37. Interestingly, BCAM, an extracellular laminin binding protein, showed a similar correlation to CDH5, suggesting this protein is associated with VE-Cadherin. Moreover, several inhibitors of angiogenesis VASH1, MMRN2, MTSS1 and RFLNB50–53, were tightly correlated with VE-Cadherin levels. Of note is that these proteins were all downregulated by IFNγ, in line with IFNy-induced inhibition of proliferation41, albeit fold changes were limited. As such, utilizing dynamic protein levels allows for investigating of known and potential WBP binding- and tight-junction- proteins in steady state condition and this analysis could be used to investigate other EC biological processes of interest. However, it should be noted this remains an explorative analysis indicating co-regulation of proteins. Therefore the nature of these associations should be validated through dedicated experiments.
A major challenge of in vitro endothelial cultures, is the adaptation of ECs to the in vitro microenvironment which lacks factors such as flow, vessel size or contact with other cell types. This loss of organ specificity during cell culture has been highlighted by studies comparing in vitro grown and in vivo isolated ECs14,53–57, which could contribute to the observed proteomic overlap. Although this adaptation of transcriptional expression is rapid and can occur within 1 passage, cells in this study maintained separation between organs and especially donors over several passages on the proteome level. In contrast to in vivo single-cell transcriptomics studies we also observed less defined separation of cell types8,10–12. We do find EC specific regulation for some proteins, which was highlighted in uniquely regulated modules for BOECs, HUVECs and Liver ECs. Especially liver ECs showed a cluster of uniquely upregulated proteins. These included CD36 and TIMP1, which have been reported as liver specific24,58–60. However, CD36 has also been reported as specifically expressed in muscle (Soleus) ECs8, while others observed CD36 expression predominantly in lung and heart21. Moreover, various studies found CD36 in microvasculature ECs22,61, as well as in lymphatic endothelium62. Furthermore, BOECs derived from different human donors showed varied transcript levels within the same study63. Finally in our study, CD36 protein expression in liver ECs was only prevalent in two out of three donors. These diverse observations exemplify the challenges in interpreting vascular heterogeneity. Whether uniquely regulated proteins are specific to an organ and how transcript levels in mice translates to protein levels in humans remain incompletely resolved. As the robustness, throughput and proteomic depth of single-cell proteomics workflows advances, we envision that a combined study of single-cell -transcriptomics and -proteomics on ECs isolated directly from human tissues, would provide the next level in understanding the nature of EC heterogeneity, bridging the gaps of murine-to-human, in vitro-to-in vivo and RNA-to-protein. Our in-depth proteomic characterization representing eight EC-types highlights the opportunities and challenges for comprehensively understanding human vascular specificity from an in vitro perspective. EC heterogeneity from vessel to vessel has been firmly established and some of these archetypes were indeed observed on the proteome level in vitro, however variation in the cell of origin and de-phenotypization should be seriously considered in translation of organ-specific in vitro findings.
In conclusion, we provide a human proteomic landscape of in vitro cultured organ-specific ECs that gives a proteomics perspective to both basic EC biology and EC heterogeneity.
Methods
Cell culture and stimulation
Endothelial cells from different sources were acquired from various suppliers specified in Supplementary Table 1. Blood outgrowth endothelial cells (BOECs) were isolated from healthy donors as described previously64. For all cell types culture flasks and dishes were coated with collagen type I (50 µg/ml, BD biosciences) for 1 h prior to use. Cells were cultured in Endothelial Cell Basal Medium w/o Glutamine (Cellovations) supplemented with Microvascular Endothelial Cell Growth Supplements Enhanced (Cellovations). Cells were passaged when reaching 100% confluency, stimulations were performed 4 days after reaching 100% confluency in the following passage (Supplementary Fig. 1). For IFNγ stimulated conditions, cells were stimulated with 10 ng/ml IFNγ (Peprotech) for 24 h.
Immunostaining and imaging
Cells were grown to confluence on collagen-coated glass coverslips, and fixed after 4 days with 4% PFA (Thermo Scientific), washed 3x with PBS and quenched with 50 mM ammonium chloride (Sigma-Aldrich). Antibody staining steps were performed in 1% BSA (Serva), 0.1% Saponin (Sigma-Aldrich) to permeabilize cells. VWF was stained using mouse-monoclonal RAG2065 (IL262, 10 µg/ml), and VE-cadherin using goat polyclonal anti-Ve-Cadherin (sc-6458, #H1814) 1 ug/ml. Alexa Fluor 488 chicken-anti-mouse (Invitrogen, #A21200), and Alexa fluor 568 donkey anti-goat (Invitrogen, #A11057) conjugated secondary antibodies were used for respective stainings. CD36 was stained using rabbit-polyclonal anti-CD36 antibody (Invitrogen, PA1-16813, #ZA4176203, 20 ug/ml) and FERMT3 using rabbit polyclonal anti-Kindlin-3 antibody (Invitrogen, PA5-116109. #ZC4247225B, 10 ug/ml). For these stainings, Alexa Fluor 488 chicken-anti-rabbit (Invitrogen, A21441, #1512063), and Alexa fluor 568 donkey anti-rabbit (A10042, #1964370) conjugated secondary antibodies were used respectively. Slides were fixed in Mowiol 4-88 (Polysciences). Images were acquired on a SP8 Confocal Laser Scanning Microscope (Leica) with a 40x/1.30 oil objective (Leica, 11506359) and processed using Fiji software. Staining was performed in two independent experiments.
Mass spectrometry sample preparation
For mass spectrometry analysis of EC proteomes, cells were lysed in 1% sodium deoxycholate (Bioworld), 10 mM TCEP (Thermo Scientific), 40 mM chloroacetamide (Sigma-Aldrich), 100 mM Tris-HCl pH 8.0 (Gibco). Lysates were incubated for 5 min at 95 °C and sonicated for 10 min in a sonifier bath (Branson model 2510), after which trypsin gold (Promega) was added in a 1:50 (w/w) protein ratio and digested overnight at room temperature.
Mass spectrometry acquisition and analysis
Tryptic digests were transferred to an Evotip Pure (Evosep) according to manufactures guidelines and separated on a 15 cm × 150 μm, 1.5 μm Performance Column (EV1137 from EvoSep) with an Evosep One liquid chromatography (LC) system (Evosep) with the 15 samples per day program. Buffer A was composed of 0.1% formic acid, buffer B of 0.1% formic acid in acetonitrile (Biosolve, NLD). Peptides were ionized and introduced electrosprayed into the Orbitrap Fusion Lumos Tribrid mass spectrometer (Thermo Fisher scientific, USA). Data were acquired in Data Independent Acquisition mode (DIA), consisting of a MS1 scan from 390 to 1010 m/z, performed at 60 K resolution (AGC target of 4 * 105) with a maximum injection time of 100 ms. This was followed by MS2 data acquisition in centroid mode by, using 75 isolation windows of 8 m/z, with an overlap of 1 m/z. DIA segments/spectra were acquired at 30 K resolution (with an AGC target 4 * 105) with a maximum injection time set on 54 ms. HCD fragmentation was used in MS2 with a normalized collision energy of 23%. A default charge state of 2 for MS2 was used. Spectra were recorded in centroid mode. The setting “inject ions for all available parallelizable time” was set.
Data analysis
The raw mass spectrometry data files were processed using the DIANN software (version 1.8), proteins and peptides were detected by querying the filtered human Swissprot database (release 2021.22.04). Standard settings were used, using a generated library based spectra search. Maximum number of variable modifications was set at 2, Protein Interference used was “Protein names (from FASTA)” and quantification strategy “Robust LC (high accuracy)”.
Data were analyzed using R 4.2.3/Rstudio (2022.07.02). Detected proteins were filtered for proteotypic and ≥2 unique peptides per protein and proteins should be quantified in 100% of samples in at least one cell type and condition. LFQ values were transformed in log2 scale. Missing values were imputed by normal distribution (width = 0.3, shift = 2.5), assuming these proteins were close to the detection limit. Organ unique regulated proteins were determined by supervised classification, generating theoretical profiles for each organ in which intensities are high for a given organ and low in all other samples. These profiles were correlated with all quantified proteins, an absolute Pearson correlation coefficient of 0.8 or higher was used as a threshold for an organ-unique regulation. Analysis of protein dynamics was performed using the weighted gene co-expression network analysis (WGCNA)66 using signed network and a soft power of 9, minClusterSize was set to 10. Gene ontology term enrichment and pathway analyses were performed using the clusterPofiler67 package, enrichments with a BH adjusted p value < 0.05 were considered significant. Connections between GO-terms and modules were visualized in Cytoscape 3.8.0. We first obtained the “EdgeBetweenness” using the “Analyse Network” function, after which we used “Edge-weighted Spring Embedded Layout” to visualize the network.
Statistics and reproducibility
Statistical tests employed and significance cut-off values are indicated per experiment in the “Methods” section. ECs from three different donors per organ were used as biological replicates as indicated in Supplementary Table 1.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Acknowledgements
This work was supported by the Landsteiner Foundation for Blood Transfusion Research grants LSBR-1517 awarded to M.v.d.B. and LSBR-1923 awarded to A.J.H. and M.v.d.B.
Author contributions
S.A.G. performed experiments, analyzed data, and drafted the manuscript. E.R.S. developed and contributed in mass-spec data acquisition. M.v.d.B. and A.J.H. designed the study, supervised research and drafted the manuscript. All authors read and approved the final version of the manuscript.
Peer review
Peer review information
Communications Biology thanks Hisamichi Naito and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editor: Ophelia Bu.
Data availability
The raw mass spectrometry acquisition files and DIA-NN search files have been deposited to the ProteomeXchange Consortium via the PRIDE68 partner repository with the dataset identifier PXD045899. Source data used for all figures in this study can be found in Supplementary Data 1. Any remaining information can be obtained from the authors upon reasonable request.
Code availability
No original code was generated in this study. In-house written scripts are available from the corresponding author upon reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Maartje van den Biggelaar, Arie J. Hoogendijk.
Supplementary information
The online version contains supplementary material available at 10.1038/s42003-024-06649-w.
References
- 1.Yau, J. W., Teoh, H. & Verma, S. Endothelial cell control of thrombosis. BMC Cardiovasc. Disord.15, 130 (2015). 10.1186/s12872-015-0124-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Galley, H. F. & Webster, N. R. Physiology of the endothelium. Br. J. Anaesth.93, 105–113 (2004). 10.1093/bja/aeh163 [DOI] [PubMed] [Google Scholar]
- 3.Pober, J. S. & Sessa, W. C. Evolving functions of endothelial cells in inflammation. Nat. Rev. Immunol.7, 803–815 (2007). 10.1038/nri2171 [DOI] [PubMed] [Google Scholar]
- 4.Ashina, K. et al. Histamine induces vascular hyperpermeability by increasing blood flow and endothelial barrier disruption in vivo. PLoS ONE10, e0132367 (2015). [DOI] [PMC free article] [PubMed]
- 5.Aird, W. C. Phenotypic heterogeneity of the endothelium. Circ. Res.100, 158–173 (2007). 10.1161/01.RES.0000255691.76142.4a [DOI] [PubMed] [Google Scholar]
- 6.Garlanda, C. & Dejana, E. Heterogeneity of endothelial cells. Arterioscler. Thromb. Vasc. Biol.17, 1193–1202 (1997). 10.1161/01.ATV.17.7.1193 [DOI] [PubMed] [Google Scholar]
- 7.Potente, M. & Mäkinen, T. Vascular heterogeneity and specialization in development and disease. Nat. Rev. Mol. Cell Biol.18, 477–494 (2017). 10.1038/nrm.2017.36 [DOI] [PubMed] [Google Scholar]
- 8.Kalucka, J. et al. Single-cell transcriptome atlas of murine endothelial cells. Cell180, 764–779.e20 (2020). 10.1016/j.cell.2020.01.015 [DOI] [PubMed] [Google Scholar]
- 9.Becker, L. M. et al. Deciphering endothelial heterogeneity in health and disease at single-cell resolution: progress and perspectives. Cardiovasc. Res.119, 6–27 (2023). 10.1093/cvr/cvac018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Serra, A. et al. Vascular bed molecular profiling by differential systemic decellularization in vivo. Arterioscler. Thromb. Vasc. Biol.38, 2396–2409 (2018). 10.1161/ATVBAHA.118.311552 [DOI] [PubMed] [Google Scholar]
- 11.Jambusaria, A. et al. Endothelial heterogeneity across distinct vascular beds during homeostasis and inflammation. Elife9, e51413 (2020). [DOI] [PMC free article] [PubMed]
- 12.Cleuren, A. C. A. et al. The in vivo endothelial cell translatome is highly heterogeneous across vascular beds. Proc. Natl Acad. Sci.116, 23618–23624 (2019). 10.1073/pnas.1912409116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Trimm, E. & Red-Horse, K. Vascular endothelial cell development and diversity. Nat. Rev. Cardiol.20, 197–210 (2023). 10.1038/s41569-022-00770-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Durr, E. et al. Direct proteomic mapping of the lung microvascular endothelial cell surface in vivo and in cell culture. Nat. Biotechnol.22, 985–992 (2004). 10.1038/nbt993 [DOI] [PubMed] [Google Scholar]
- 15.Bachetti, D. & Morbidelli, L. Endothelial cells in culture: a model for studying vascular functions. Pharm. Res.42, 9–19 (2000). 10.1006/phrs.1999.0655 [DOI] [PubMed] [Google Scholar]
- 16.Nachman, R. L. & Jaffe, E. A. Endothelial cell culture: beginnings of modern vascular biology. J. Clin. Invest.114, 1037–1040 (2004). 10.1172/JCI23284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Helms, H. C. et al. In vitro models of the blood–brain barrier: an overview of commonly used brain endothelial cell culture models and guidelines for their use. J. Cereb. Blood Flow. Metab.36, 862–890 (2016). 10.1177/0271678X16630991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bouïs, D., Hospers, G. A. P., Meijer, C., Molema, G. & Mulder, N. H. Endothelium in vitro: a review of human vascular endothelial cell lines for blood vessel-related research. Angiogenesis4, 91–102 (2001). 10.1023/A:1012259529167 [DOI] [PubMed] [Google Scholar]
- 19.Banerji, S. et al. LYVE-1, a new homologue of the CD44 glycoprotein, is a lymph-specific receptor for hyaluronan. J. Cell Biol.144, 789–801 (1999). 10.1083/jcb.144.4.789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Micklem, K. et al. A human macrophage-associated antigen (CD68) detected by six different monoclonal antibodies. Br. J. Haematol.73, 6–11 (1989). 10.1111/j.1365-2141.1989.tb00210.x [DOI] [PubMed] [Google Scholar]
- 21.Nolan, D. J. et al. Molecular signatures of tissue-specific microvascular endothelial cell heterogeneity in organ maintenance and regeneration. Dev. Cell26, 204–219 (2013). 10.1016/j.devcel.2013.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Son, N.-H. et al. Endothelial cell CD36 optimizes tissue fatty acid uptake. J. Clin. Investig.128, 4329–4342 (2018). 10.1172/JCI99315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fagerholm, S. C., Lek, H. S. & Morrison, V. L. Kindlin-3 in the immune system. Am. J. Clin. Exp. Immunol.3, 37–42 (2014). [PMC free article] [PubMed] [Google Scholar]
- 24.Hemmann, S., Graf, J., Roderfeld, M. & Roeb, E. Expression of MMPs and TIMPs in liver fibrosis—a systematic review with special emphasis on anti-fibrotic strategies. J. Hepatol.46, 955–975 (2007). 10.1016/j.jhep.2007.02.003 [DOI] [PubMed] [Google Scholar]
- 25.Grantham, J. The molecular chaperone CCT/TRiC: an essential component of proteostasis and a potential modulator of protein aggregation. Front. Genet.11, 172 (2020). [DOI] [PMC free article] [PubMed]
- 26.Huyton, T., Göttmann, W., Bade-Döding, C., Paine, A. & Blasczyk, R. The T/NK cell co-stimulatory molecule SECTM1 is an IFN “early response gene” that is negatively regulated by LPS in Human monocytic cells. Biochim. Biophys. Acta Gen. Subj.1810, 1294–1301 (2011). 10.1016/j.bbagen.2011.06.020 [DOI] [PubMed] [Google Scholar]
- 27.Leaman, D. W. et al. Identification of X-linked inhibitor of apoptosis-associated factor-1 as an interferon-stimulated gene that augments TRAIL Apo2L-induced apoptosis. J. Biol. Chem.277, 28504–28511 (2002). 10.1074/jbc.M204851200 [DOI] [PubMed] [Google Scholar]
- 28.Hölzer, M. et al. Virus- and interferon alpha-induced transcriptomes of cells from the microbat myotis daubentonii. iScience19, 647–661 (2019). 10.1016/j.isci.2019.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ishii, T. et al. Isolation and expression profiling of genes upregulated in the peripheral blood cells of systemic lupus erythematosus patients. DNA Res.12, 429–439 (2005). 10.1093/dnares/dsi020 [DOI] [PubMed] [Google Scholar]
- 30.Holthenrich, A., Drexler, H. C. A., Chehab, T., Naß, J. & Gerke, V. Proximity proteomics of endothelial Weibel-Palade bodies identifies novel regulator of von Willebrand factor secretion. Blood134, 979–982 (2019). 10.1182/blood.2019000786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Holthenrich, A. et al. Spire1 and Myosin Vc promote Ca2+-evoked externalization of von Willebrand factor in endothelial cells. Cell. Mol. Life Sci.79, 96 (2022). 10.1007/s00018-021-04108-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.El-Mansi, S. et al. Proximity proteomics identifies septins and PAK2 as decisive regulators of actomyosin-mediated expulsion of von Willebrand factor. Blood141, 930–944 (2023). 10.1182/blood.2022017419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van Breevoort, D. et al. STXBP1 promotes Weibel-Palade body exocytosis through its interaction with the Rab27A effector Slp4-a. Blood123, 3185–3194 (2014). 10.1182/blood-2013-10-535831 [DOI] [PubMed] [Google Scholar]
- 34.Knop, M., Aareskjold, E., Bode, G. & Gerke, V. Rab3D and annexin A2 play a role in regulated secretion of vWF, but not tPA, from endothelial cells. EMBO J.23, 2982–2992 (2004). 10.1038/sj.emboj.7600319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zografou, S. et al. Rab-genome analysis reveals novel insights in Weibel-Palade body exocytosis. J. Cell Sci.10.1242/jcs.104174 (2012). [DOI] [PubMed]
- 36.Huveneers, S. et al. Vinculin associates with endothelial VE-cadherin junctions to control force-dependent remodeling. J. Cell Biol.196, 641–652 (2012). 10.1083/jcb.201108120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kaufmann, U. et al. The armadillo repeat region targets ARVCF to cadherin-based cellular junctions. J. Cell Sci.113, 4121–4135 (2000). 10.1242/jcs.113.22.4121 [DOI] [PubMed] [Google Scholar]
- 38.Chavkin, N. W. et al. Endothelial cell cycle state determines propensity for arterial-venous fate. Nat. Commun.13, 5891 (2022). 10.1038/s41467-022-33324-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Luo, W. et al. Arterialization requires the timely suppression of cell growth. Nature589, 437–441 (2021). 10.1038/s41586-020-3018-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gunawardana, H. et al. Tissue-specific endothelial cell heterogeneity contributes to unequal inflammatory responses. Sci. Rep.11, 1949 (2021). 10.1038/s41598-020-80102-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schroder, K., Hertzog, P. J., Ravasi, T. & Hume, D. A. Interferon-γ: an overview of signals, mechanisms and functions. J. Leukoc. Biol.75, 163–189 (2004). 10.1189/jlb.0603252 [DOI] [PubMed] [Google Scholar]
- 42.Grasso, C. S. et al. Conserved interferon-γ signaling drives clinical response to immune checkpoint blockade therapy in melanoma. Cancer Cell38, 500–515.e3 (2020). 10.1016/j.ccell.2020.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Castro, F., Cardoso, A. P., Gonçalves, R. M., Serre, K. & Oliveira, M. J. Interferon-gamma at the crossroads of tumor immune surveillance or evasion. Front. Immunol.9, 847 (2018). [DOI] [PMC free article] [PubMed]
- 44.Manneville, J.-B. et al. Interaction of the actin cytoskeleton with microtubules regulates secretory organelle movement near the plasma membrane in human endothelial cells. J. Cell Sci.116, 3927–3938 (2003). 10.1242/jcs.00672 [DOI] [PubMed] [Google Scholar]
- 45.Nightingale, T. D., Pattni, K., Hume, A. N., Seabra, M. C. & Cutler, D. F. Rab27a and MyRIP regulate the amount and multimeric state of VWF released from endothelial cells. Blood113, 5010–5018 (2009). 10.1182/blood-2008-09-181206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bierings, R. et al. The interplay between the Rab27A effectors Slp4-a and MyRIP controls hormone-evoked Weibel-Palade body exocytosis. Blood120, 2757–2767 (2012). 10.1182/blood-2012-05-429936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kat, M. et al. GDP/GTP exchange factor MADD drives activation and recruitment of secretory Rab GTPases to Weibel-Palade bodies. Blood Adv.5, 5116–5127 (2021). 10.1182/bloodadvances.2021004827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bennett, J. A. et al. The choline transporter Slc44a2 controls platelet activation and thrombosis by regulating mitochondrial function. Nat. Commun.11, 3479 (2020). 10.1038/s41467-020-17254-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Egaña, I. et al. Female mice lacking Pald1 exhibit endothelial cell apoptosis and emphysema. Sci. Rep.7, 15453 (2017). 10.1038/s41598-017-14894-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Watanabe, K. et al. Vasohibin as an endothelium-derived negative feedback regulator of angiogenesis. J. Clin. Investig.114, 898–907 (2004). 10.1172/JCI200421152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.del Valle-Pérez, B. et al. Filamin B plays a key role in vascular endothelial growth factor-induced endothelial cell motility through its interaction with Rac-1 and Vav-2. J. Biol. Chem.285, 10748–10760 (2010). 10.1074/jbc.M109.062984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dawson, J. C., Bruche, S., Spence, H. J., Braga, V. M. M. & Machesky, L. M. Mtss1 promotes cell-cell junction assembly and stability through the small GTPase Rac1. PLoS ONE7, e31141 (2012). 10.1371/journal.pone.0031141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Galvagni, F. et al. Dissecting the CD93-Multimerin 2 interaction involved in cell adhesion and migration of the activated endothelium. Matrix Biol.64, 112–127 (2017). 10.1016/j.matbio.2017.08.003 [DOI] [PubMed] [Google Scholar]
- 54.Amatschek, S. et al. Blood and lymphatic endothelial cell-specific differentiation programs are stringently controlled by the tissue environment. Blood109, 4777–4785 (2007). 10.1182/blood-2006-10-053280 [DOI] [PubMed] [Google Scholar]
- 55.Géraud, C. et al. Liver sinusoidal endothelium: a microenvironment-dependent differentiation program in rat including the novel junctional protein liver endothelial differentiation-associated protein-1. Hepatology52, 313–326 (2010). 10.1002/hep.23618 [DOI] [PubMed] [Google Scholar]
- 56.Calabria, A. R. & Shusta, E. V. A genomic comparison of in vivo and in vitro brain microvascular endothelial cells. J. Cereb. Blood Flow. Metab.28, 135–148 (2008). 10.1038/sj.jcbfm.9600518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lacorre, D.-A. et al. Plasticity of endothelial cells: rapid dedifferentiation of freshly isolated high endothelial venule endothelial cells outside the lymphoid tissue microenvironment. Blood103, 4164–4172 (2004). 10.1182/blood-2003-10-3537 [DOI] [PubMed] [Google Scholar]
- 58.Pandey, E., Nour, A. S. & Harris, E. N. Prominent receptors of liver sinusoidal endothelial cells in liver homeostasis and disease. Front. Physiol.11, 873 (2020). [DOI] [PMC free article] [PubMed]
- 59.Zhao, L. et al. CD36 palmitoylation disrupts free fatty acid metabolism and promotes tissue inflammation in non-alcoholic steatohepatitis. J. Hepatol.69, 705–717 (2018). 10.1016/j.jhep.2018.04.006 [DOI] [PubMed] [Google Scholar]
- 60.Reventun, P. et al. CD36 regulates factor VIII secretion from liver endothelial cells. Blood Adv.8, 143–149 (2024). 10.1182/bloodadvances.2023010023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Peche, V. S. et al. Endothelial cell CD36 regulates membrane ceramide formation, exosome fatty acid transfer and circulating fatty acid levels. Nat. Commun.14, 4029 (2023). 10.1038/s41467-023-39752-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cifarelli, V. et al. Visceral obesity and insulin resistance associate with CD36 deletion in lymphatic endothelial cells. Nat. Commun.12, 3350 (2021). 10.1038/s41467-021-23808-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Laan, S. N. J. et al. Transcriptional and functional profiling identifies inflammation and endothelial-to-mesenchymal transition as potential drivers for phenotypic heterogeneity within a cohort of endothelial colony forming cells. J. Thromb. Haemost.10.1016/j.jtha.2024.03.018 (2024). [DOI] [PubMed]
- 64.Martin-Ramirez, J., Hofman, M., van den Biggelaar, M., Hebbel, R. P. & Voorberg, J. Establishment of outgrowth endothelial cells from peripheral blood. Nat. Protoc.7, 1709–1715 (2012). 10.1038/nprot.2012.093 [DOI] [PubMed] [Google Scholar]
- 65.Stel, H. V. et al. Characterization of 25 monoclonal antibodies to factor VIII-von Willebrand factor: relationship between ristocetin-induced platelet aggregation and platelet adherence to subendothelium. Blood63, 1408–1415 (1984). 10.1182/blood.V63.6.1408.1408 [DOI] [PubMed] [Google Scholar]
- 66.Langfelder, P. & Horvath, S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinform.9, 559 (2008). [DOI] [PMC free article] [PubMed]
- 67.Yu, G., Wang, L. G., Han, Y. & He, Q. Y. ClusterProfiler: an R package for comparing biological themes among gene clusters. OMICS16, 284–287 (2012). 10.1089/omi.2011.0118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Perez-Riverol, Y. et al. The PRIDE database resources in 2022: a hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res.50, D543–D552 (2022). 10.1093/nar/gkab1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
The raw mass spectrometry acquisition files and DIA-NN search files have been deposited to the ProteomeXchange Consortium via the PRIDE68 partner repository with the dataset identifier PXD045899. Source data used for all figures in this study can be found in Supplementary Data 1. Any remaining information can be obtained from the authors upon reasonable request.
No original code was generated in this study. In-house written scripts are available from the corresponding author upon reasonable request.