Abstract
Epigenetic modifications to DNA and chromatin control oncogenic and tumor-suppressive mechanisms in melanoma. Ezh2, the catalytic component of the Polycomb Repressive Complex 2 (PRC2), which mediates methylation of lysine 27 on histone 3 (H3K27me3), can regulate both melanoma initiation and progression. We previously found that mutant Ezh2Y641F interacts with the immune regulator Stat3 and together they affect anti-tumor immunity. However, given the numerous downstream targets and pathways affected by Ezh2, many mechanisms that determine its oncogenic activity remain largely unexplored. Using genetically engineered mouse models, we further investigated the role of pathways downstream of Ezh2 in melanoma carcinogenesis and identified significant enrichment in several autophagy signatures, along with increased expression of autophagy regulators, such as Atg7. In this study, we investigated the effect of Atg7 on melanoma growth and tumor immunity within the context of a wild-type or Ezh2Y641F epigenetic state. We found that the Atg7 locus is controlled by multiple Ezh2 and Stat3 binding sites, Atg7 expression is dependent on Stat3 expression, and that deletion of Atg7 slows down melanoma cell growth in vivo, but not in vitro. Atg7 deletion also results in increased CD8 + T cells in Ezh2Y641F melanomas and reduced myelosuppressive cell infiltration in the tumor microenvironment, particularly in Ezh2WT melanomas, suggesting a strong immune system contribution in the role of Atg7 in melanoma progression. These findings highlight the complex interplay between genetic mutations, epigenetic regulators, and autophagy in shaping tumor immunity in melanoma.
Keywords: Melanoma, Atg7, Autophagy, Tumor-immune response
Introduction
Epigenetic alterations contribute to oncogenesis through multiple mechanisms, from repression of tumor suppressor genes or activation of oncogenes to tumor cell-extrinsic mechanisms such as angiogenesis, invasion, and anti-tumor immunity [1–4]. Epigenetic regulators have thus become effective therapeutic targets in multiple solid tumors. One epigenetic complex that is frequently mutated in many solid tumors and directly implicated in anti-tumor immunity is the Polycomb Repressive Complex 2 and particularly its enzymatic subunit, Ezh2 [5, 6]. Ezh2 possesses histone methyltransferase activity and mediates methylation of histone 3 on lysine 27 (H3K27me). Genetic alterations in Ezh2 include both loss- and gain-of-function events, and it can function both as a tumor suppressor [7–11] and as an oncogene [12–16]. A unique point mutation in the methyltransferase domain of Ezh2 (SET domain) at tyrosine 641 (Y641) alters its methyltransferase activity and may confer neomorphic functions by promoting unconventional changes to the distribution of H3K27me3 across the genome [12, 17], with complicated effects on gene expression.
In previous studies, using a genetically engineered mouse model, we found that expression of mutant Ezh2Y641F is oncogenic and cooperates with BrafV600E mutations and Pten loss to accelerate melanoma formation [12]. Furthermore, we found that mutant Ezh2Y641F co-immunoprecipitates with Stat3, and together they activate expression of several common target genes. One class of genes co-regulated by Ezh2 and Stat3 in Ezh2Y641F mutant melanomas were MHC class I antigen processing genes in the H2-Q cluster, which are directly implicated in anti-tumor immunity [18]. In addition to these MHC class I genes, chromatin immunoprecipitation, followed by sequencing (ChIP-seq), suggests that Ezh2 and Stat3 are also found at the same promoter regions of the autophagy regulator, Atg7. Atg7 is a critical protein for autophagy initiation, as it facilitates an intermediate step in LC3 lipidation through its E1-like enzymatic activity [19]. Atg7 conjugates with and adenylates LC3 (a ubiquitin-like protein also known as Atg8) and then transfers LC3 to the E2-like enzyme Atg3, which catalyzes the conjugation of LC3 to phosphatidylethanolamine (PE) on the autophagosome membrane [20, 21]. LC3 lipidation and, therefore, Atg7 are necessary for normal autophagosome formation, and Atg7 deficient cells are also autophagy-deficient [19, 22]. Autophagy plays a significant role in many different cellular functions, both cell- intrinsically and extrinsically. In cancer, numerous autophagy regulators are mutated or deregulated [23–25], but given autophagy’s role in many cellular mechanisms, its contribution during different phases of carcinogenesis is not entirely understood. In melanoma, previous studies have shown that deletion of Atg7 in a mouse model driven by the oncogenic BrafV600E and deletion of the tumor suppressor Pten significantly slowed down melanoma growth, suggesting that Atg7 functions as an oncogene [26]. Mechanistically, the study showed that deletion of Atg7 resulted in increased oxidative stress and cellular senescence, which served as a barrier to melanomagenesis [26]. Carcinogenesis, however, involves many different steps, from initial melanocyte transformation and immortalization to angiogenesis and immune evasion. The latter is particularly important in melanoma since checkpoint inhibitors have dramatically increased melanoma survival in the last ten years [27–30]. Despite this improvement, many patients do not respond to treatment or experience severe toxicity, necessitating better understanding of anti-tumor immune mechanisms. Many autophagy components have been implicated in tumor immunity in multiple solid tumors [31–35], partially driven by their role in recycling unwanted cellular components and processing peptides, and may therefore play an important role in immunotherapy approaches. Several studies investigated the underlying mechanisms of immunotherapy resistance and identified very complex interplay between many biological mechanisms. These studies also identified increased expression of both STAT and ATG genes, including Stat3 and Atg7, in patients that did not respond to immune checkpoint blockade therapies [36]. Additionally, post-treatment samples had also acquired mutations in numerous autophagy-related ATG genes [37], suggesting that dysregulation of autophagy mechanisms may be important in the immune system’s ability to clear melanoma in response to immune checkpoint blockade.
Given our prior findings that Ezh2Y641F mutant melanomas have a significantly altered tumor immunity and the fact that Ezh2 and Stat3 can both be found at the Atg7 locus, we hypothesized that Atg7 may contribute to the altered tumor immune response in Ezh2Y641F melanomas. In this study, we investigated the role of Atg7 in both Ezh2WT and Ezh2Y641F melanoma tumor growth and its effect on anti-tumor immunity.
Results
Ezh2 and Stat3 regulate Atg7 expression in melanoma cells
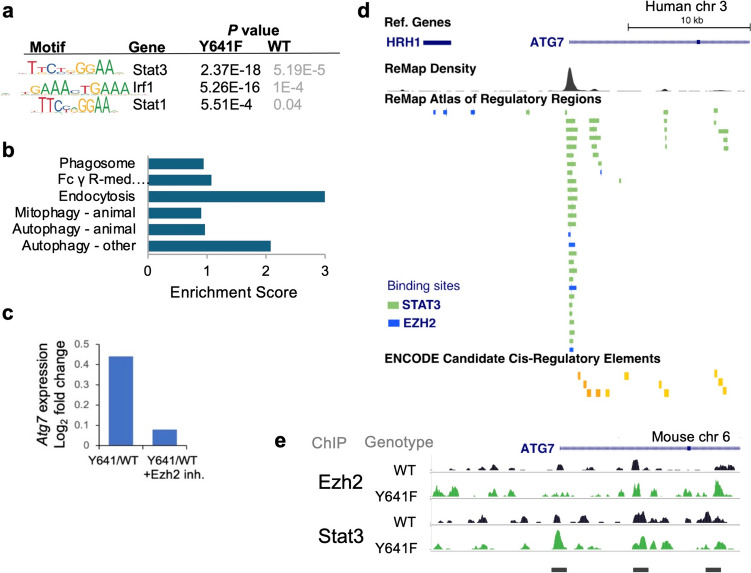
Previously, we investigated the role of Ezh2Y641F mutations in melanoma and found a direct interaction of Ezh2Y641F with Stat3, with direct effects on tumor immunity [18]. We also identified several loci directly bound by both Ezh2 and Stat3 in melanoma cells. Here, we expanded that study to additional cell lines to gain a more comprehensive understanding of genes regulated by both Ezh2 and Stat3 in melanoma in a BrafV600E/PtenF/F background, with or without the Ezh2Y641F mutation. First, using Stat3 ChIP-seq, we confirmed enrichment of Stat3 binding motifs in Ezh2Y641F melanoma cells compared to Ezh2WT cells and identified enriched representation of motifs of other immune regulators, such as Stat1 and Irf1 (Fig. 1a). Gene Set Enrichment Analysis (GSEA) [38] of Stat3 peaks enriched in Ezh2Y641F mutant melanoma cells identified several oncogenic signatures. Interestingly, we also identified several gene expression signatures that implicate autophagy or related cellular processes (Fig. 1b). We next assessed whether autophagy regulators were differentially expressed in Ezh2WT vs. Ezh2Y641F melanoma [12]. We found that Atg7, an important autophagy regulator, was upregulated in Ezh2Y641F melanomas compared to Ezh2WT and its expression was downregulated upon treatment with a pharmacological Ezh2 inhibitor (Fig. 1c). Chromatin immunoprecipitation, followed by sequencing (ChIP-seq) analysis, identified several Stat3 and Ezh2 peaks at the Atg7 gene promoter and the first intron (Fig. 1e). To confirm the relevance of these data in human patients, we analyzed data from the ReMap Atlas of Regulatory Regions (a collection of all public ChIP-seq data for transcriptional regulators from GEO, ArrayExpress, and ENCODE databases) [39] for Ezh2 and Stat3 in various cell types and the ENCODE registry of candidate cis-regulatory elements [40]. We identified several cis-regulatory elements that coincide with mouse experimental Ezh2 and Stat3 binding sites (Fig. 1d), suggesting that our findings in mouse models are conserved and potentially relevant to human disease. Consistent with these observations, expression of Stat3 correlates with increased expression of Atg7 in human melanoma patient samples [41].
Fig. 1.
Regulation of Atg7 expression by Ezh2 and Stat3. a Enriched motifs in Ezh2WT and Ezh2Y641F melanoma cells. b Gene Set Enrichment Analysis (GSEA) of Stat3 ChIP-seq peaks identifies several signatures associated with autophagy mechanisms (FDR < 0.05). c Transcript expression of Atg7 in Ezh2Y641F vs. Ezh2WT melanoma cells measured by RNA-sequencing, in the absence or presence of the Ezh2 inhibitor JQEZ5. d Human ChIP-seq data in various cell lines showing direct binding of both Stat3 (green) and Ezh2 (blue) at the Atg7 promoter and intronic regions that correspond to cis-regulatory elements. Image modified from UCSC Genome Browser. e ChIP-seq tracks for Ezh2 and Stat3 in Ezh2WT and Ezh2Y641F melanoma cells at the mouse Atg7 locus indicating binding at the Atg7 promoter and the first intron
Loss of Atg7 inhibits in vitro and in vivo cell growth
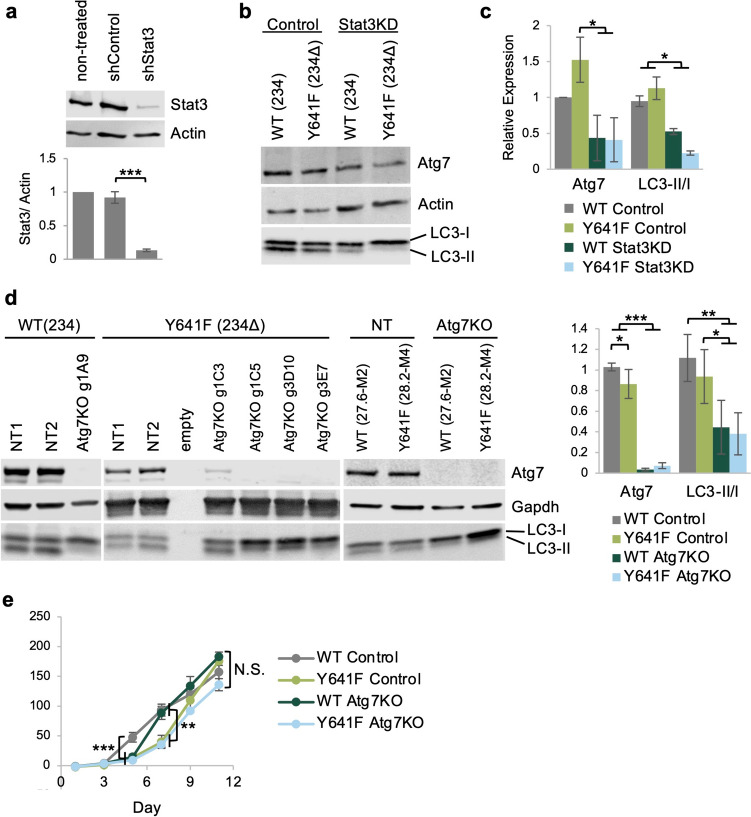
We first assessed the protein levels of Atg7 in the presence or absence of Ezh2Y641F mutations, with or without Stat3 expression. We found that the effect of mutant Ezh2Y641F expression on the protein levels of Atg7 was marginally different, suggesting that perhaps the role of Ezh2 is to fine-tune Atg7 expression and control accessibility by transcription factors, such as Stat3. To determine whether Stat3 controls expression of Atg7, we generated stable Stat3 knockdown melanoma cell lines using shRNA (Fig. 2a). We found that Stat3 knockdown in at least two independent mouse melanoma cell lines resulted in lower Atg7 protein levels, consistent with the hypothesis that Stat3 positively regulates Atg7 expression (Fig. 2b–c). Since Atg7 is an important regulator of autophagy initiation, we assessed the ratio of type I cytosolic LC3 (LC3-I) and the type II lipid-conjugated form that is present on autophagosome membranes (LC3-II), a standard assay for assessing autophagy [42, 43]. We found that after Stat3 knockdown, cells exhibited a lower LC3-II/I ratio, indicating reduced levels of autophagy (Fig. 2b–c), consistent with depletion of Atg7 protein levels. We next investigated whether Atg7 is required for in vitro melanoma growth. We used a lentiviral CRISPR/Cas9 system to inactivate Atg7 expression in two Ezh2WT (234 and 27.6-M2) and two Ezh2Y641F (234Δ and 28.2-M4) melanoma cell lines. The lentiviral system is a single vector delivery of the single guide RNA (sgRNA) targeting Atg7, Cas9, puromycin for selection, and GFP for cell sorting [44]. For controls, we generated stable cell lines using two non-specific sgRNAs. After puromycin selection, GFP + transfected cells were sorted by FACS to generate single-cell clones and tested for knockout efficiency by western blot. We identified multiple clones that exhibited complete loss of Atg7 protein expression (Fig. 2d). We further tested these clones for autophagy activity, and they exhibited a decreased LC3-II/I ratio, verifying disruption of Atg7 function and lower autophagic activity (n = 4, p < 0.01). To determine whether the absence of Atg7 affects cell-intrinsic melanoma growth in vitro, we monitored cell growth by staining with Alamar Blue, a cell-permeable dye (resazurin), which serves as a redox indicator in response to cellular metabolic activity [45]. We found that deletion of Atg7 only transiently slowed the growth of Ezh2WT cells but did not have a significant overall effect during the duration of the in vitro assay (Fig. 2e) or an effect on the growth rate of Ezh2Y641F melanoma cells. These results suggest that the effect of Atg7 deletion on melanoma cell growth may depend not only on increased cellular stress and senescence, as previously suggested [26], but also on specific in vivo variables and cell-extrinsic factors such as the tumor microenvironment and anti-tumor immunity.
Fig. 2.
Deletion of Atg7 in melanoma cells has no significant effect on cell-intrinsic cell growth in vitro. a Top: Protein expression of Stat3 measured by western blot after shRNA-mediated stable gene knockdown in melanoma cell line 234Δ (Y641F). Bottom: Quantification of protein expression, N = 3 independent experiments. b Expression of Atg7 and LC3 after Stat3 knockdown in Ezh2WT and Ezh2Y641F melanoma cell lines 234 and 234Δ. c Quantification of western blot in b, N = 2. d Immunoblotting for Atg7 and LC3 in control and Atg7 knockout clones in the 234, 234Δ, 27.6-M2, and 28.2-M4 cell lines. NT = non-targeted sgRNA. Quantification of the Atg7/GAPDH, N = 4, and LC3-II/I, N = 5. e In vitro growth curve of Ezh2WT and Ezh2Y641F melanoma cell lines 27.6-M2 and 28.2-M4 with and without Atg7 deletion. N.S. = not statistically significant. For all graphs, error bars are standard deviation; *** p value < 0.001, ** p value < 0.01, and * p value < 0.05
Atg7 deletion suppresses in vivo tumor growth and results in increased CD8 + T cells and NK cells in the tumor microenvironment
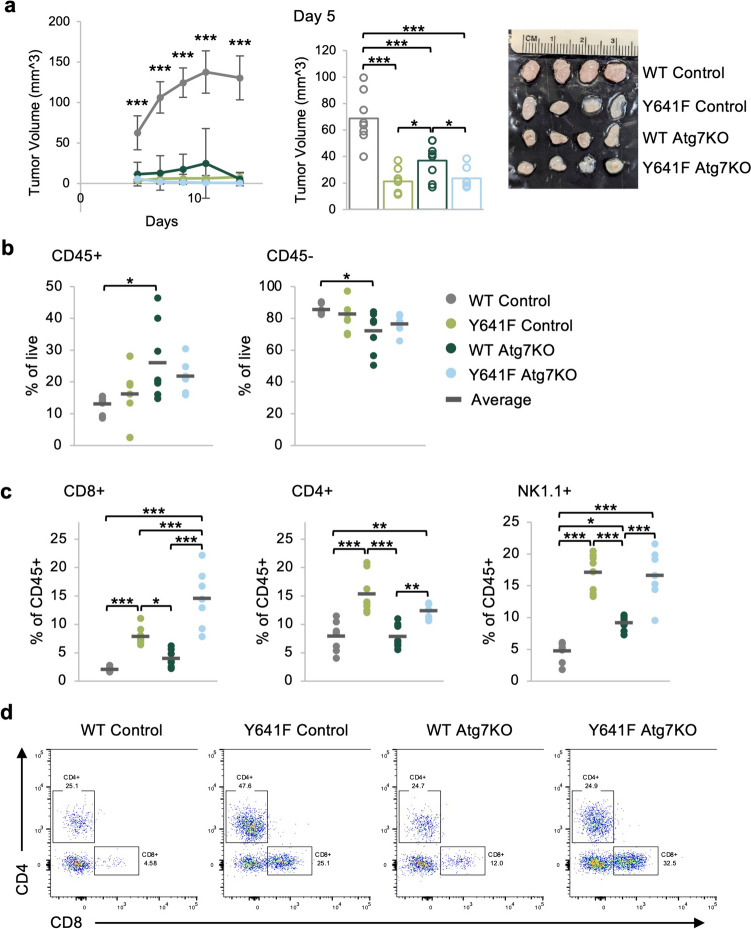
To test whether Atg7 deletion differentially affects in vivo growth of Ezh2WT or Ezh2Y641F mutant melanomas, we adaptively transferred five hundred thousand Atg7 knockout or non-targeted sgRNA, Ezh2WT, and Ezh2Y641F melanoma cells into the left and right flank of wild-type recipient mice. These cells formed tumors, which we monitored for growth over time. Consistent with our prior finding, tumors expressing Ezh2Y641F grew more slowly than Ezh2WT [18], and deletion of Atg7 resulted in slower tumor growth, particularly in Ezh2WT tumors (n = 8, p < 0.001 for WT Control vs. all other groups at every time point) (Fig. 3a). These results are consistent with a prior study that demonstrated the oncogenic activity of Atg7 in a BrafV600E/PtenF/F background [26], which was attributed to a cell-intrinsic increase in oxidative stress and senescence of the tumor cells, without consideration of cell-extrinsic variables. Since we previously showed that tumor immunity is an important factor in the progression of Ezh2Y641F melanomas in vivo, we investigated how deletion of Atg7 affected infiltration of immune cells in Ezh2WT and Ezh2Y641F melanomas. We harvested tumors seven days after injection and analyzed tumor immune cell infiltration by flow cytometry. We found that the overall amount of CD45 + tumor-infiltrating cells, while somewhat variable, tended to be higher after Atg7 deletion, particularly in Ezh2WT melanoma tumors (n = 8, p = 0.024) (Fig. 3b). Nevertheless, we observed more significant differences in the type of immune cells that infiltrated these tumors. In the Ezh2Y641F control group, we detected increased CD8 + T cell infiltration compared to Ezh2WT (n = 8, p < 0.001), confirming our prior findings [18]. Deletion of Atg7 resulted in no change to CD8 + T cell infiltration in Ezh2WT; however, Atg7 deletion in Ezh2Y641F tumors resulted in an approximately twofold increase in the CD8 + population (n = 7–8, p < 0.001) (Fig. 3c–d). Interestingly, we found that expression of Ezh2Y641F, regardless of Atg7 expression, dramatically increased infiltration of natural killer (NK) cells, a population that we had not previously assessed in this model (n = 7–8, p < 0.001) (Fig. 3c). Deletion of Atg7 in Ezh2WT tumors also led to increased NK cells (n = 8, p = 0.0107) (Fig. 3c). Other lymphoid populations such as CD4 + cells were elevated in Ezh2Y641F compared to Ezh2WT, but deletion of Atg7 had no significant effect compared to controls in either Ezh2 genotype (Fig. 3c–d).
Fig. 3.
Deletion of Atg7 in melanoma cells results in slower in vivo tumor growth and increased presence of tumor infiltration of lymphocytes. a (Left) In vivo tumor growth in Ezh2WT (27.6-M2) and Ezh2Y641F (28.2-M4) melanomas, with and without Atg7 deletion. The group average is displayed, and error bars indicate the standard deviation. Control = non-targeted sgRNA, N = 8 per group, representative of two independent experiments. (Right) Tumor volume at day 5 post-injection. The bars indicate the group mean, and the circles are individual tumor sizes. (Far right) Image of tumors at day 7. The image has been cropped, and the brightness and contrast have been increased to improve viewing. b Flow cytometric analysis of tumor-infiltrating CD45 + hematopoietic cells and CD45- cells. N = 6–8 tumors per group. c Flow cytometric analysis of tumor-infiltrating CD8 + , CD4 +, and NK1.1 + cells. N = 7–8 tumors per group. d Representative flow cytometry plots of the CD4 + and CD8 + data shown in panel c. For the graphs in b and c, each dot on the graph represents an individual tumor, and the bar marks the average for the group. *p < 0.05, **p < 0.01, and ***p < 0.001
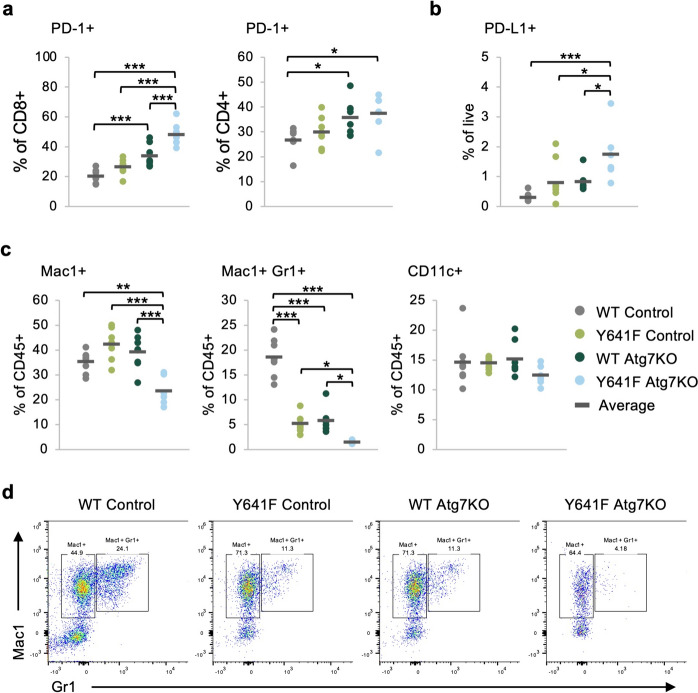
While the number of cytotoxic CD8 + T cells significantly increased with Atg7 deletion in Ezh2Y641F melanoma, it is possible that these T cells are not functionally competent killer cells. T cells have evolved mechanisms to prevent autoreactivity through receptor–ligand interactions, also known as immune checkpoints. These interactions are particularly important in cancer, as ligands expressed on tumors may interact with receptors on T cells to inhibit anti-tumor activity. One such immune checkpoint pair is PD-1 and PD-L1. We thus assessed the presence of PD-1 on T cells in the tumor microenvironment and PD-L1 on the melanoma cells. We found increased expression of PD-1 in CD8 + T cells after Atg7 knockout (n = 7–8, p < 0.001) and to a lesser degree in CD4 + cells (Fig. 4a). Ezh2Y641F Atg7 knockout tumors also exhibited increased expression of PD-L1 compared to all other groups (p < 0.05) (Fig. 4b). These data suggest that while loss of Atg7 results in slower tumor growth, likely partially mediated by the increased presence of CD8 + T cells, it may also eventually lead to T cell inhibition.
Fig. 4.
Deletion of Atg7 in melanoma cells results in decreased infiltration of myelosuppressive cells. a Expression of PD-1 on tumor-infiltrating CD8 + and CD4 + T cells in Ezh2WT and Ezh2Y641F melanoma cells, with and without Atg7 deletion. N = 7–8 tumors per group. b Expression of the PD-1 ligand (PD-L1) on the melanoma cells from panel a. N = 6–8 tumors per group. c Flow cytometric analysis of tumor-infiltrated CD11c + , Mac1 +, and double Mac1/Gr1 + cells in Ezh2WT and Ezh2Y641F melanoma tumors, with and without Atg7 deletion. N = 6–8 tumors per group. d Representative flow cytometry plots for the Mac1 + and double Mac1/Gr1 + data in panel c. For the graphs in a–c, each dot on the graph represents an individual tumor, and the bar marks the average for the group. *p < 0.05, **p < 0.01, and ***p < 0.001
Deletion of Atg7 results in a decrease in myelosuppressive cells in the melanoma tumor microenvironment
Another important immune population that plays a critical role in tumor immunity is myeloid-derived suppressor cells (MDSCs). To test whether Atg7 deletion affects infiltration of these cells in the melanoma tumor microenvironment, we measured expression of myeloid markers using flow cytometry. We found a significant decrease in Mac1 +/Gr1 + double-positive cells after Atg7 deletion in both Ezh2WT and Ezh2Y641F cells (n = 6–8, p < 0.001 WT, p < 0.05 Y641F), with a significantly lower frequency in the Ezh2Y641F tumors (p = 0.04), while Mac1 + cells decreased only in the Ezh2Y641F Atg7 knockout tumors (n = 6–8, p < 0.01) (Fig. 4c–d). While tumor size itself has been associated with differences in tumor immunity, the Ezh2Y641F tumors with or without Atg7 deletion were of similar size, suggesting that tumor size is not a confounding variable in the observed phenotypes. Finally, we did not find changes in the dendritic cell population as determined by CD11c expression in any of the groups, regardless of Ezh2 status or Atg7 expression (Fig. 4c).
Overall, these results suggest that deletion of Atg7 significantly suppresses in vivo melanoma tumor growth, particularly in Ezh2WT tumors, which correlated with a significant decrease in myelosuppressive cells in the tumor microenvironment. Atg7 loss also affects the recruitment of lymphoid populations in the melanoma tumor microenvironment, with a more pronounced effect in the presence of Ezh2Y641F, suggesting that some of the effects of Ezh2Y641F on melanoma tumor immunity may be mediated by Atg7. It remains to be seen whether the effects of Atg7 on tumor immunity are mediated through its role in autophagy or whether they are mediated by autophagy-independent, cell-intrinsic mechanisms.
Discussion
In this study, we investigated the role of downstream targets of Ezh2 in the melanoma tumor immune response. Ezh2 regulates many different hallmarks of cancer, from cell-intrinsic cell cycle regulation to tumor immunity. Ezh2 has a complex role in cancer. It is often deleted in some cancers while amplified in others, consequently functioning both as a tumor suppressor and as an oncogene. While typically functioning within the PRC2 complex and mediating methylation of lysine 27 on histone 3, Ezh2 can also function independently of the PRC2 complex, sometimes as a transcriptional activator as we and others have previously shown [18, 46]. Here, we investigated the role of one of its non-canonical targets, Atg7, an autophagy regulator.
Autophagy is a fundamental cellular mechanism required to maintain cellular health. When perturbed, it can result in the onset of different diseases. In antigen-presenting cells, such as dendritic cells, autophagy generates peptides from endogenous antigens, which are presented by MHC class II proteins to CD4 + cells to prime the immune response. In cancer, the role of autophagy is context-dependent. Autophagy in tumor cells can enhance processing of exogenous antigens and MHC-I antigen presentation, inducing CD8 T cell priming and cytotoxic activity [47]. Specifically, ATG genes, such as Atg7, are involved in the internalization and recycling of the MHC-I molecules themselves [47], and dendritic cells deficient in Atg7 have increased cell surface expression of MHC-I molecules [48]. Autophagy, therefore, can stimulate CD8 + T cells, thus functioning in a tumor-suppressive manner [49]. In our melanoma models, it is possible that deletion of Atg7 similarly increases the amount of MHC-I at the cell surface, resulting in the increased CD8 + T cell infiltration that we observe in melanoma tumors. On the other hand, because cancer cells require autophagy for growth, autophagy-regulating genes can also function as oncogenes [26]. Consistent with an oncogenic function, in humans, melanoma patients with a high autophagic index benefit less from chemotherapy, exhibit increased tumor cell proliferation and metastasis, and have poor outcomes [50, 51]. Overall, this dual role of autophagy in cancer is not well understood and may be context-dependent.
Within the context of Ezh2Y641F mutant melanomas, loss of Atg7 does not have a significant effect on cell-intrinsic cell growth or in vivo tumor growth, but it appears to further enhance anti-tumor immunity with the increased presence of cytotoxic CD8 + T cells and decreased MDSCs populations in the tumor microenvironment, a combination that is not conducive to tumor growth. In Ezh2WT melanomas, loss of Atg7 does not affect cell growth in vitro; however, Atg7 loss has a significant effect on tumor growth in vivo. Specifically, Atg7 deletion in Ezh2WT tumors results in more than fivefold smaller tumors than the control group. Atg7 loss in Ezh2WT tumors also affects the anti-tumor immune response, as evidenced by decreased MDSCs and increased NK cell infiltration. Expression of Atg7 does not change dramatically with expression of Ezh2Y641F in vitro, but its expression is regulated by Stat3, as clearly demonstrated with Stat3 knockdown experiments. Ezh2 and Stat3 may, therefore, play a role in sustaining Atg7 expression within the context of a more complicated transcriptional network, and Atg7 may be playing a secondary role in the oncogenic mechanisms of Ezh2Y641 mutations in melanoma.
Tumor immunobiology is very complex and is affected by a multitude of factors, including cell-intrinsic variables as well as cell-extrinsic factors such as the stroma, fibrosis, tumor tissue location, tumor vascularity, tumor burden, and signals or cytokines secreted by tumor cells, and others. It is possible that deletion of Atg7 affects any of these factors, whether via autophagy-dependent or -independent functions. Regardless of the mechanisms, our results indicate the relevance of tumor immunity in melanoma tumors lacking expression of Atg7. Future studies are needed to further delineate mechanistically how Atg7 deletion results in such significant changes to the tumor immune response in melanoma and how it cooperates with mutations in Ezh2. With the availability of several pharmacological inhibitors of autophagy mechanisms, our study suggests that targeting autophagy-related pathways could be a viable strategy to modulate anti-tumor immunity, offering potential for therapeutic advancements in melanoma treatment.
Materials and methods
Genomic analysis
ChIP-seq and RNA-seq were performed on Ezh2WT and Ezh2Y641F mouse melanoma cells with or without treatment with the Ezh2 inhibitor JQEZ5 as described previously [18]. Analysis of transcription factor motif enrichment was carried out using HOMER [52]. Functional significance of Ezh2 and Stat3 binding sites/peaks was evaluated using the Genomic Regions Enrichment of Annotations Tool (GREAT) [53], and Gene Set Enrichment Analysis was performed as described here [38]. The UCSC Genome Browser was used to visualize Ezh2 and Stat3 binding sites at the Atg7 locus (human GRCh38/hg38) using tracks for the ReMap Atlas of Regulatory Regions and the ENCODE Candidate Cis-Regulatory Elements (cCREs) [39].
Cell culture and CRISPR knockouts
Eight mouse melanoma cell lines were used: 234, 480, and 855 (Ezh2WT Tyr-CREERT2 BrafV600E/+ Ptenflox/flox); 234Δ, 480Δ, and 855Δ (Ezh2Y641F Tyr-CREERT2 BrafV600E/+ Ptenflox/flox); 27.6-M2 (Ezh2WT Tyr-CREERT2 BrafV600E/+ Ptenflox/+); and 28.2-M4 (Ezh2Y641F Tyr-CREERT2 BrafV600E/+ Ptenflox/+). Cell lines 234, 480, and 855 were previously characterized [12, 18]. Cells were cultured in DMEM (Sigma D6429) with 10% FBS (Corning Cat# MT35010CV) and 1% penicillin–streptomycin (Genesee Scientific Cat# 25–512). Atg7 knockout cell lines were generated by transducing cells with lentiviral CRISPR/Cas9 (TLCV2 Addgene plasmid #87360). Lentiviruses were generated using 293T cells via transfection with PEI. Stable cell lines were selected by treating with puromycin for seven days (3 µg/ml, refreshed every other day), and Cas9 expression was induced with 3–5 doses of doxycycline at 1 µg/ml. To generate single clones, GFP-positive and propidium iodide (PI)-negative cells were single-cell sorted into 96-well plates on the MoFlo sorter (Beckman Coulter) at the Siteman Flow Cytometry Core Facility. The clones were tested for Atg7 knockout by immunoblotting. For the in vitro cell growth assay, cells were plated at 500 cells/well in a 24-well plate in triplicate, one set of triplicates for each time point. For each measurement, the growth media were aspirated and replaced with media containing Alamar Blue (Invitrogen #A50100) cell viability reagent at 1:10 dilution [45]. The cells were returned to the incubator for 1 h, after which 100 µl of supernatant was transferred from the 24-well plate to a clean 96-well plate. The samples were scanned on a BioTek Synergy HT plate reader using fluorescent excitation at 485/20 nm and detection at 590/35 nm. Data analysis was performed in Excel, and statistically significant differences were determined by one-way ANOVA.
Immunoblotting
Samples were prepared in Laemmli buffer with beta-mercaptoethanol, run on 4–20% pre-cast gels (BioRad Mini-PROTEAN TGX Gels Cat# 4561095) using the BioRad Mini-PROTEAN system, and then transferred onto nitrocellulose membranes. The membranes were blocked for 1 h in 5% milk in TBS-T and then incubated with primary antibodies overnight at 4°C. Primary antibodies: anti-ATG7 (Cell Signaling #8558 at 1:500), anti-ACTIN (Abcam ab213262 at 1:1000), anti-GAPDH (Cell Signaling #5174 at 1:1000), and anti-LC3A/B (Cell Signaling #12741 at 1:1000). Membranes were washed with TBS-T before staining with secondary anti-rabbit IgG (H + L) DyLight 800 4X PEG Conjugate (Cell Signaling #5151) at 1:20,000 at room temperature for 1 hour. Membranes were imaged using a Licor Odyssey Infrared Imager, and Image Studio software was used for densitometry analysis. Statistically significant differences were detected using one-way ANOVA.
Animals
Animals were housed in an Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC)-accredited facility and treated in accordance with protocols approved by the Institutional Animal Care and Use Committee (IACUC) for animal research at Washington University in St. Louis.
In vivo tumor models
Wild-type C57BL/6 mice were purchased from Charles River laboratories or bred in house. Tumor cells suspended in HBSS were mixed 1:1 with Matrigel (Corning 354234) and injected subcutaneously in the flank at 0.5 × 106 cells per injection, two injections per mouse. Both male and female mice were used as tumor recipients. Mice were of similar age (4–6 months old) and size (> 20 g) and were randomized during injections. Eight to ten tumors were generated per group, which was based on prior preliminary data that reach statistical significance between groups. Tumor growth was measured in a blinded manner using digital calipers on day 5 post-injection and then every other day. For the flow cytometry analysis of tumor-infiltrating lymphocytes, tumors were harvested on day 7. Tumors were dissociated in HBSS media, dispersed using a syringe with 18G needle, and filtered through a 0.40 µm filter.
Flow cytometric analysis
Single-cell suspensions from tumors were washed with HBSS containing 2% FBS and 1 mM EDTA and stained with the following antibody cocktails for detecting lymphoid populations: anti-CD45-PerCP/Cy5.5 (BioLegend 103132), anti-NK1.1-FITC (BioLegend 108706), anti-CD3-PB (BioLegend 100214), anti-CD4-APC (BioLegend 100412), anti-CD8-AF700 (BioLegend 100730), and anti-PD-1 (CD279)-PE/Cy7 (BioLegend 135216) and myeloid populations: anti-CD45-PerCP/Cy5.5 (BioLegend 103132), anti-CD19-FITC (BioLegend 115506), anti-B220-FITC (BioLegend 103206), anti-CD3-FITC (BioLegend 100204), anti-CD11b (Mac1)-PB (BioLegend 101224), anti-CD11c-PE/Cy7 (BioLegend 117318), and anti-Ly-6G (Gr1)-AF700 (BioLegend 127622). Propidium iodide was used to exclude dead cells. Samples were run on an Attune NxT Flow Cytometer (ThermoFisher Scientific) at the Siteman Flow Cytometry Core Facility, analysis was done in FlowJo v10, and statistically significant differences were identified using one-way ANOVA.
Acknowledgments
We thank the Siteman Flow Cytometry Facility and the Department of Comparative Medicine for animal expertise. We also thank all members of the Souroullas Lab for critical input on the manuscript. This work was supported by the Alvin J. Siteman Cancer Center, The Harry J. Lloyd Charitable Trust (GPS), and T32 CA113275-10 (SZ),
Author contributions
GPS and SMZ designed the experiments and wrote the manuscript. GPS, SMZ, ES, and SS performed the experiments, analyzed, and interpreted the data. ES and SS performed the experiments. GPS conceived of and supervised the study.
Funding
This work was funded by National Institutes of Health, T32 CA113275-10, Alvin J. Siteman Cancer Center and Harry J. Lloyd Charitable Trust.
Data Availability
Sequence data that support the findings have been deposited in Gene Expression Omnibus (GSE183819ID).
Declarations
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Villanueva L, Álvarez-Errico D, Esteller M (2020) The contribution of epigenetics to cancer immunotherapy. Trends Immunol 41:676–691 [DOI] [PubMed] [Google Scholar]
- 2.Hogg SJ, Beavis PA, Dawson MA, Johnstone RW (2020) Targeting the epigenetic regulation of antitumour immunity. Nat Rev Drug Discov 19:776–800 [DOI] [PubMed] [Google Scholar]
- 3.Feinberg AP, Koldobskiy MA, Göndör A (2016) Epigenetic modulators, modifiers and mediators in cancer aetiology and progression. Nat Publ Gr 17:284–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jones PA, Baylin SB (2002) The fundamental role of epigenetic events in cancer. Nat Rev Genet 3:415–428 [DOI] [PubMed] [Google Scholar]
- 5.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO et al (2013) Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 10.1126/scisignal.2004088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA et al (2012) The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data: figure 1. Cancer Discov 2:401–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ntziachristos P, Tsirigos A, Vlierberghe PV, Nedjic J, Trimarchi T, Flaherty MS et al (2012) Genetic inactivation of the polycomb repressive complex 2 in T cell acute lymphoblastic leukemia. Nat Med 18:298–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Muto T, Sashida G, Oshima M, Wendt GR, Mochizuki-Kashio M, Nagata Y et al (2013) Concurrent loss of Ezh2and Tet2cooperates in the pathogenesis of myelodysplastic disorders. J Exp Med 210:2627–2639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clair JM-S, Soydaner-Azeloglu R, Lee KE, Taylor L, Livanos A, Pylayeva-Gupta Y et al (2012) EZH2 couples pancreatic regeneration to neoplastic progression. Genes & Dev 26:439–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Y, Hou N, Cheng X, Zhang J, Tan X, Zhang C et al (2017) Ezh2 acts as a tumor suppressor in kras-driven lung adenocarcinoma. Int J Biol Sci 13:652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mochizuki-Kashio M, Aoyama K, Sashida G, Oshima M, Tomioka T, Muto T et al (2015) Ezh2 loss in hematopoietic stem cells predisposes mice to develop heterogeneous malignancies in an Ezh1-dependent manner. Blood 126:1172–1183 [DOI] [PubMed] [Google Scholar]
- 12.Souroullas GP, Jeck WR, Parker JS, Simon JM, Liu J-Y, Paulk J et al (2016) An oncogenic Ezh2 mutation induces tumors through global redistribution of histone 3 lysine 27 trimethylation. Nat Med 22:632–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zingg D, Debbache J, Schaefer SM, Tuncer E, Frommel SC, Cheng P et al (2015) The epigenetic modifier EZH2 controls melanoma growth and metastasis through silencing of distinct tumour suppressors. Nat Commun 6:6051 [DOI] [PubMed] [Google Scholar]
- 14.Béguelin W, Popovic R, Teater M, Jiang Y, Bunting KL, Rosen M et al (2013) EZH2 Is required for germinal center formation and somatic EZH2 mutations promote lymphoid transformation. Cancer Cell 23:677–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Varambally S, Dhanasekaran SM, Zhou M, Barrette TR, Kumar-Sinha C, Sanda MG et al (2002) The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature 419:624–629 [DOI] [PubMed] [Google Scholar]
- 16.Zhang H, Qi J, Reyes JM, Li L, Rao PK, Li F et al (2016) Oncogenic deregulation of EZH2 as an opportunity for targeted therapy in lung cancer. Cancer Discov 6:1006–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Romero P, Richart L, Aflaki S, Petitalot A, Burton M, Michaud A et al (2024) EZH2 mutations in follicular lymphoma distort H3K27me3 profiles and alter transcriptional responses to PRC2 inhibition. Nat Commun 15:3452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zimmerman SM, Nixon SJ, Chen PY, Raj L, Smith SR, Paolini RL et al (2022) Ezh2Y641F mutations co-operate with Stat3 to regulate MHC class I antigen processing and alter the tumor immune response in melanoma. Oncogene. 10.1038/s41388-022-02492-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Collier JJ, Suomi F, Oláhová M, McWilliams TG, Taylor RW (2021) Emerging roles of ATG7 in human health and disease. EMBO Mol Med 13(12):e14824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ichimura Y, Kirisako T, Takao T, Satomi Y, Shimonishi Y, Ishihara N et al (2000) A ubiquitin-like system mediates protein lipidation. Nature 408:488–492 [DOI] [PubMed] [Google Scholar]
- 21.Taherbhoy AM, Tait SW, Kaiser SE, Williams AH, Deng A, Nourse A et al (2011) Atg8 transfer from Atg7 to Atg3: a distinctive E1–E2 architecture and mechanism in the autophagy pathway. Mol Cell 44:451–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Komatsu M, Waguri S, Ueno T, Iwata J, Murata S, Tanida I et al (2005) Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol 169:425–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wen X, Klionsky DJ (2020) At a glance: a history of autophagy and cancer. Semin Cancer Biol 66:3–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li X, He S, Ma B (2020) Autophagy and autophagy-related proteins in cancer. Mol Cancer 19:12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Debnath J, Gammoh N, Ryan KM (2023) Autophagy and autophagy-related pathways in cancer. Nat Rev Mol Cell Biol 24:560–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xie X, Koh JY, Price S, White E, Mehnert JM (2015) Atg7 Overcomes Senescence and Promotes Growth of BrafV600E-Driven Melanoma. Cancer Discov 5:410–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Js W, Kc K, A H, (2012) Management of immune-related adverse events and kinetics of response with ipilimumab. J clin oncol off J Am Soc Clin Oncol 30(21):2691 [DOI] [PubMed] [Google Scholar]
- 28.Robert C, Long GV, Brady B, Dutriaux C, Maio M, Mortier L et al (2015) Nivolumab in previously untreated melanoma without BRAF mutation. N Engl J Med 372:320–330 [DOI] [PubMed] [Google Scholar]
- 29.Linardou H, Gogas H (2016) Toxicity management of immunotherapy for patients with metastatic melanoma. Ann Transl Med 4:272–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.PatelOthusChenWrightYostHyngstrom SPMYGPKJJR et al (2023) Neoadjuvant-adjuvant or adjuvant-only pembrolizumab in advanced melanoma. New Engl J Med 388:813–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xia H, Green DR, Zou W (2021) Autophagy in tumour immunity and therapy. Nat Rev Cancer 21:281–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Van Kaer L, Parekh VV, Postoak JL, Wu L (2019) Role of autophagy in MHC class I-restricted antigen presentation. Mol Immunol 113:2–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chemali M, Radtke K, Desjardins M, English L (2011) Alternative pathways for MHC class I presentation: a new function for autophagy. Cell Mol Life Sci 68:1533–1541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yamamoto K, Venida A, Yano J, Biancur DE, Kakiuchi M, Gupta S et al (2020) Autophagy promotes immune evasion of pancreatic cancer by degrading MHC-I. Nature 581:100–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Valečka J, Almeida CR, Su B, Pierre P, Gatti E (2018) Autophagy and MHC-restricted antigen presentation. Mol Immunol 99:163–170 [DOI] [PubMed] [Google Scholar]
- 36.Lauss M, Phung B, Borch TH, Harbst K, Kaminska K, Ebbesson A et al (2024) Molecular patterns of resistance to immune checkpoint blockade in melanoma. Nat Commun 15:3075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Freeman SS, Sade-Feldman M, Kim J, Stewart C, Gonye ALK, Ravi A et al (2022) Combined tumor and immune signals from genomes or transcriptomes predict outcomes of checkpoint inhibition in melanoma. Cell Rep Med 3:100500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA et al (2005) Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci 102:15545–15550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hammal F, de Langen P, Bergon A, Lopez F, Ballester B (2022) ReMap 2022: a database of human, mouse, drosophila and arabidopsis regulatory regions from an integrative analysis of DNA-binding sequencing experiments. Nucleic Acids Res 50:D316–D325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.The ENCODE Project Consortium, Abascal F, Acosta R, Addleman NJ, Adrian J, Afzal V et al (2020) Expanded encyclopaedias of DNA elements in the human and mouse genomes. Nature 583:699–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Network TCGA, Akbani R, Akdemir KC, Aksoy BA, Albert M, Ally A et al (2015) Genomic classification of cutaneous melanoma. Cell 161:1681–1696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mizushima N, Yoshimori T, Levine B (2010) Methods in mammalian autophagy research. Cell 140:313–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Singh B, Bhaskar S (2019) Methods for detection of autophagy in mammalian cells. Methods Mol Biol 2045:245–258 [DOI] [PubMed] [Google Scholar]
- 44.Barger CJ, Branick C, Chee L, Karpf AR (2019) Pan-cancer analyses reveal genomic features of FOXM1 overexpression in cancer. Cancers (Basel) 11:251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kumar P, Nagarajan A, Uchil PD (2018) Analysis of cell viability by the alamarblue assay. Cold Spring Harb Protoc. 10.1101/pdb.prot095489 [DOI] [PubMed] [Google Scholar]
- 46.Zimmerman SM, Lin PN, Souroullas GP (2023) Non-canonical functions of EZH2 in cancer. Front Oncol 13:1233953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fonderflick L, Adotévi O, Guittaut M, Adami P, Delage-Mourroux R (2020) Role of autophagy in antigen presentation and its involvement on cancer immunotherapy. In: Autophagy in immune response: impact on cancer immunotherapy. Elsevier, pp 175–196
- 48.Loi M, Müller A, Steinbach K, Niven J, Barreira da Silva R, Paul P et al (2016) Macroautophagy proteins control MHC class I levels on dendritic cells and shape anti-viral CD8 T cell responses. Cell Rep 2016(15):1076–1087 [DOI] [PubMed] [Google Scholar]
- 49.Liang C, Feng P, Ku B, Dotan I, Canaani D, Oh B-H et al (2006) Autophagic and tumour suppressor activity of a novel Beclin1-binding protein UVRAG. Nat Cell Biol 8:688–698 [DOI] [PubMed] [Google Scholar]
- 50.Ma X-H, Piao S, Wang D, Mcafee QW, Nathanson KL, Lum JJ et al (2011) Measurements of tumor cell autophagy predict invasiveness, resistance to chemotherapy, and survival in melanoma. Clin Cancer Res 17:3478–3489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lazova R, Camp RL, Klump V, Siddiqui SF, Amaravadi RK, Pawelek JM (2012) Punctate LC3B expression Is a common feature of solid tumors and associated with proliferation, metastasis, and poor outcome. Clin Cancer Res 18:370–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P et al (2010) Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38:576–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB et al (2010) GREAT improves functional interpretation of cis -regulatory regions. Nat Biotechnol 28:495–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Sequence data that support the findings have been deposited in Gene Expression Omnibus (GSE183819ID).