Abstract
Background
Alzheimer’s disease (AD) has a high heritable component characteristic of complex diseases, yet many of the genetic risk factors remain unknown. We combined genome-wide association studies (GWAS) on amyloid endophenotypes measured in cerebrospinal fluid (CSF) and positron emission tomography (PET) as surrogates of amyloid pathology, which may be helpful to understand the underlying biology of the disease.
Methods
We performed a meta-analysis of GWAS of CSF Aβ42 and PET measures combining six independent cohorts (n=2,076). Due to the opposite effect direction of Aβ phenotypes in CSF and PET measures, only genetic signals in the opposite direction were considered for analysis (n=376,599). Polygenic risk scores (PRS) were calculated and evaluated for AD status and amyloid endophenotypes. We then searched the CSF proteome signature of brain amyloidosis using SOMAscan proteomic data (Ace cohort, n=1,008) and connected it with GWAS results of loci modulating amyloidosis. Finally, we compared our results with a large meta-analysis using publicly available datasets in CSF (n=13,409) and PET (n=13,116). This combined approach enabled the identification of overlapping genes and proteins associated with amyloid burden and the assessment of their biological significance using enrichment analyses.
Results
After filtering the meta-GWAS, we observed genome-wide significance in the rs429358-APOE locus and nine suggestive hits were annotated. We replicated the APOE loci using the large CSF-PET meta-GWAS and identified multiple AD-associated genes as well as the novel GADL1 locus. Additionally, we found a significant association between the AD PRS and amyloid levels, whereas no significant association was found between any Aβ PRS with AD risk. CSF SOMAscan analysis identified 1,387 FDR-significant proteins associated with CSF Aβ42 levels. The overlap among GWAS loci and proteins associated with amyloid burden was very poor (n=35). The enrichment analysis of overlapping hits strongly suggested several signalling pathways connecting amyloidosis with the anchored component of the plasma membrane, synapse physiology and mental disorders that were replicated in the large CSF-PET meta-analysis.
Conclusions
The strategy of combining CSF and PET amyloid endophenotypes GWAS with CSF proteome analyses might be effective for identifying signals associated with the AD pathological process and elucidate causative molecular mechanisms behind the amyloid mobilization in AD.
Keywords: Aβ42, CSF biomarkers, PET tomography, GWAS, Proteome
Background
Alzheimer’s disease (AD) is the most common cause of dementia. AD is a growing epidemic with an expected doubling of annual new diagnosis in the next 20 years prevalence and a major socioeconomic impact with a projected direct economic cost of $2 trillion by 20301–3. In this sense, increasing the knowledge of AD aetiology and biomarker development would be an interesting approach to developing a clear understanding of the disease physiopathology and future drug developments. Genome wide association studies (GWAS) have permitted the discovery of more than 80 genetic variants associated with AD risk4,5. Despite the continued efforts led by international consortia, a large fraction of AD heritability remains to be elucidated since only 31% of AD genetic variance is explained by single-nucleotide polymorphisms (SNPs)6.
The analysis of heritable quantitative traits tightly linked to disease pathology, called endophenotypes, has become a promising approach in genetic studies7–9. These intermediate phenotypes might be influenced by the same genetic factors that confer risk to AD development and might have low genetic complexity. Compared to disease phenotypes, there are fewer genes or environmental influences affecting the genetic components of endophenotypes which facilitate finding a genuine association between these phenotypes and AD7,8,10.
The most common endophenotypes for AD are levels of amyloid beta (Aβ42), total tau (t-tau) and phosphorylated tau in threonine 181 (p-tau) in CSF10–12. Moreover, Positron Emission Tomography (PET) using several radiotracers for measuring amyloid and tau burden has been used as AD endophenotypes11,13,14. These biomarkers are surrogates of AD brain pathology and understanding their biology might provide insights into novel mechanisms of AD15,16. To date, relatively few AD loci have been identified using the endophenotype approach9,13. Moreover, GWAS analyses of PET and CSF endophenotypes are commonly analysed separately and comparisons between them have been overlooked.
In this study, we combined GWAS of Aβ CSF levels from four different AD cohorts with two GWAS of Aβ-burden measured using PET radiotracers. We used this strategy of combining both Aβ endophenotypes (CSF and PET) to identify novel genetic variants associated with AD and to replicate known AD signals. We then tested polygenic risk scores (PRS) derived from large studies in our datasets, dissected the CSF proteome signature associated with brain amyloidosis in a sizable CSF collection, and checked the overlapping of genomic meta-analyses and proteomic results.
Materials and methods
GWAS Cohorts
This study comprised a total of 2,076 individuals from Ace, Valdecilla and ADNI cohorts and had data for different Aβ CSF or PET endophenotypes (Supplementary Fig. 1 and Supplementary Table 1). To avoid overlap of subjects between the CSF and PET cohorts, we used only datasets with genotype-level information available.
a. Ace Alzheimer Center Barcelona
The Ace cohort comprised 1,189 individuals with brain amyloidosis measurements obtained using CSF or PET imaging, divided into three independent and non-overlapping cohorts. Because we used different methods to quantify CSF AB42, we decided to analyse the GWAS in two independent groups (536 individuals tested using Innotest ELISA kits and 472 individuals tested using the Lumipulse automated platform17). We included a third dataset of 181 individuals with subjective cognitive decline (SCD) tested using PET Florbetaben measures from the Fundació ACE Healthy Brain Initiative (FACEHBI) study18. The clinical protocols of the Ace Alzheimer Center have been previously published17–19. Briefly, syndromic diagnosis of all subjects was established by a multidisciplinary group of neurologists, neuropsychologists, and social workers. We assigned to healthy controls (HCs) including SCD diagnosis to Clinical Dementia Rating (CDR) of 0 individuals, and mild cognitive impairment (MCI) individuals a CDR of 0.5. For MCI diagnosis, the classification of López et al., and Petersen’s criteria were also used20–23. We employed the 2011 National Institute on Aging and Alzheimer’s Association (NIA-AA) guidelines for AD diagnosis24. We performed a lumbar puncture (LP) to obtain CSF following consensus recommendations25. The CSF obtained was centrifuged (2000 × g for 10 min at 4°C) and stored at −80°C. For Aβ42 analysis, CSF was defrosted at room temperature (20°C), vortexed and protein levels measured using the commercial ELISA kit Innotest β-AMYLOID (1–42) in 536 individuals and the chemiluminescent enzyme-immunoassay LUMIPULSE G600II automated platform (Fujirebio Europe, Belgium) in 472 individuals17. FACEHBI patients were assessed for brain amyloid deposition by PET imaging using the florbetaben [18F] radiotracer (FBB) (NeuraCeq©). A single slow intravenous bolus of 300 Mbq of FBB (6 sec/mL) (>10 mL during 20 min) was administered. After 90 min, PET images were acquired18.
b. Valdecilla cohort for the study of memory and brain aging
The Valdecilla cohort comprised 97 individuals who were older than 55 years and extensively phenotyped. Biological samples were collected at baseline and several tests were performed to evaluate early signs of AD. Moreover, core biomarkers in CSF were analysed and a neuropsychological battery including The Free and Cued Selective Reminding Test26, the Spanish version of the Face-Name Associative Memory Exam27, and the Logical Memory Test of the Wechsler Memory Scale-III28 and depression symptoms by the Geriatric Depression Scale29. HC (CDR=0), MCI (CDR=0.1) and dementia individuals (NIA-AA guidelines) were included in this analysis30. In the Valdecilla cohort, the Aβ42 biomarker was quantified by Lumipulse G600II which were interpreted according to previously established cut-off points31. Further information about phenotype assessment was presented elsewhere32.
c. Alzheimer’s Disease Neuroimaging Initiative (ADNI) cohort
Launched in 2003, ADNI is a longitudinal multicentre cohort for AD research based on United States and Canada33,34. The primary goal of ADNI has been to test whether biological markers, clinical and neuropsychological assessments can be combined to study the progression of MCI and early AD. We selected individuals from two separate ADNI databases: 1) the ADNI1 cohort with 378 individuals with available Aβ42 in CSF and 2) the ADNI2GO cohort with 412 individuals with available PET centiloid measures. In ADNI, syndromic diagnoses were based on a specific cut-off in the WMS-II LM test, education attainment, the Mini-Mental State Exam (MMSE) and CDR score. For HC and those with SCD, an MMSE score of 24–30 and a CDR of 0 were used. For those with MCI, a CDR of 0.5 and MMSE score of 24–30 were used. For those with AD, a CDR of 0.5–1 and an MMSE score of 20–26 were used. For the AD diagnosis, the National Institute of Neurological and Communicative Disorders and Stroke and Alzheimer’s Disease and Related Disorders Association (NINCDS/ADRDA) criteria for probable AD were considered35. In ADNI individuals, Aβ42 CSF biomarker was measured using the Luminex xMAP platform (Luminex Corp, Austin, TX) for multiplexing with Innogenetics immunoassay reagents (INNO-BIA AlzBio3, Ghent, Belgium)36,37. ADNI2GO patients were screened for brain amyloid deposits using the Florbetapir [18F] (AV45) radiotracer. After the injection of 370 MBq (10 mCi), four 5 min scans were acquired 50–70 min after the injection36. Further information about PET data acquisition can be found elsewhere38.
PET imaging acquisition, harmonization and analysis
As FACEHBI and ADNI cohorts had different radiotracers, PET centiloid measures were used to perform a meta-analysis. Centiloids were calculated using equation (1), which was described for the conversion of FBB measures in the FACEHBI cohort39 and equation (2), which was described for the conversion of AV45 in ADNI40.
| (1) |
| (2) |
Genotyping, quality control and imputation
Ace and Valdecilla DNA samples were genotyped using the Axiom 815K Spanish Biobank Array (Thermo Fisher). The genotyping was performed by the Spanish National Center for Genotyping (CeGen, Santiago de Compostela, Spain). Genotyping procedures have been previously published elsewhere5,19. For the ADNI samples, the Illumina Human610-Quad BeadChip platform was used for genotyping in ADNI1, and the Illumina HumanOmniExpress BeadChip was used for ADNI2GO41.
Common quality control was applied to all GWAS datasets. Briefly, individuals with low-quality samples, excess of heterozygosity, sample call rate below 97%, sex discrepancies, variants call rate below 95% or a deviation from the Hardy–Weinberg equilibrium (P>1e-06) were excluded from the analysis. In addition, familial relatedness (PI-HAT>0.1875) or ancestry outliers based on principal component analysis (PCA) were also removed. The imputation was performed using the Haplotype Reference Consortium panel in the Michigan Imputation Server42. Only the common markers (MAF>1%; MAC=20) and high imputation quality (R2>0.3) were selected for the subsequent analyses (GRCh37/hg19 reference assembly).
SOMAscan Proteomic Assay
A subset of 1,008 CSF samples (Ace CSF cohort) was analysed using the SOMAscan panel measuring more than 7,000 proteins (SomaLogic, Boulder, Colorado). Briefly, this multiplex proteomic assay uses a 50 μL CSF sample and modified DNA aptamers to measure protein abundance. First, proteins are bound to immobilized aptamers using streptavidin beads and tagged with fluorescent markers. After washing unbounded proteins, the streptavidin beads are released using ultraviolet light, and the protein–aptamer complex is re-captured by monomeric avidin. To select only specific complexes, the protein–aptamers are exposed to an anionic competitor and then, hybridized in a conventional DNA array for analysis as described in Gold et al43. Finally, the protein level measures expressed in relative fluorescent units (RFU) are normalized using the adaptive normalization by maximum likelihood method further described in Candia et al44.
Statistical analysis
a. GWAS and meta-GWAS
We harmonized CSF and PET endophenotypes measures performing a log10 transformation to adjust to a normal distribution, and Z-score values were determined using the scale R function (center=TRUE, scale=TRUE) (Supplementary Fig. 2). We used R software version 4.1.1.
The GWAS on each dataset was run using a generalized linear model in the software PLINK245. The statistical model considered population microstratification (four PCs), sex, age, and dementia status for the association analysis. We then performed an inverse-variance weighted meta-analysis on each amyloid burden endophenotype separately, Aβ42 (n=1,483) in CSF and amyloid PET imaging (n=593).
Thereafter, both Aβ endophenotypes were further combined into a single meta-analysis (n=2,076) using the sample size weighted method in METAL software. This approach integrates p-values from different studies, weighting them by the sample size of each cohort, which provides a way to combine evidence across studies without relying on the effect size direction46. This is particularly useful when dealing with datasets where the effect sizes are not directly comparable or when different methods are used to measure the same biological outcome, as is the case with PET and CSF amyloid measurements.
We chose this meta-analysis of p-values approach because the effect directions and methods applied to measure amyloid burden differ between PET and CSF assessments. Specifically, in AD, the two measurements exhibit opposite biological directions: decreased levels of CSF Aβ42 are associated with increased amyloid plaque deposition in the brain, as observed through PET imaging. PET measures amyloid burden through radiotracer retention, while CSF measures it through soluble Aβ42 levels, which decrease as amyloid plaques accumulate in the brain. Thus, directly comparing effect sizes across these methods could be misleading46,47.
By combining p-values, focusing on the statistical significance and opposite effect size direction, this approach accounts for the differing biological contexts and measurement techniques, enabling a more robust and generalized analysis of amyloid burden across different datasets. The genetic markers evaluated in the meta-analysis were filtered considering the opposite effect direction in each CSF and PET endophenotype-independent GWAS and its presence in at least half of the datasets to select SNPs for further analysis.
Additionally, we performed another CSF-PET meta-analysis considering the largest publicly available datasets for CSF Aβ42 (n=13,116)9 and amyloid PET (n=13,409)48 (publicly available datasets; PAD analysis). To homogenize the results with our primary analysis, those datasets were converted to the GRCh37 assembly using the UCSC LiftOver software49. Because we did not have access to genotype-level information for all cohorts included in these studies, we were unable to prune potential overlapping subjects between both meta-analyses. Therefore, the results of combining these large meta-GWAS should be interpreted cautiously and are considered primarily for generating additional evidence about the pathways observed in our main analysis, where subject overlap was checked at the genotype level and removed to create two genuinely independent datasets (CSF and PET).
Finally, we attempted to replicate previously published genes for AD described in Bellenguez et al4, the significant signals associated with amyloid burden reported by the EADB consortium9 and significant markers associated with neuropathological features described in Beecham et al50 (Supplementary Table 2, Supplementary Table 3, Supplementary Table 4, and Supplementary Material 1).
b. Functional examination of identified sentinel SNPs and linked genomic regions
Clumping and annotation of suggestive signals (P<1e-05) were performed using the software PLINK1.951. Additional annotations of biological function and gene-mapping were performed using meta-analysis summary statistics using the online tool Functional Mapping and Annotation of Genome-Wide Association Studies (FUMA)52. We set the threshold for independent significant SNPs at P<1e-05, R2<0.05, separated by over 250 kb, and we used the 1000G Phase3 reference panel to analyse suggestive signals in European population. For functional annotation, SNPs were matched to available databases such as ANNOVAR, Combined Annotation Dependent Depletion (CADD) scores, RegulomeDB and chromatin states based on a hidden Markov model from the Roadmap Epigenomics Project. Significant hits were mapped to genes according to 3 methods: 1) Physical distance with a maximum of 10 kb from nearby genes in the reference assembly, 2) expression quantitative trait loci (eQTL) associations assigned to SNP in blood, vascular, heart, brain tissues and embryonic stem cell derived cells, and 3) three-dimensional DNA interactions with SNPs and other gene regions where promoters were considered to be 250 bp upstream and 500 bp downstream of the transcription starting site for chromatin interaction. Moreover, a gene-based analysis was performed using MAGMA v1.08 that assigned exclusively protein-coding genes (Ensembl build 85) to the top SNPs found. Only 11,807 genes were mapped, and the gene-wide significance was defined at P=0.05/11,807=4.235e-06. We also conducted FUMA annotations in the amyloid burden meta-analysis considering the largest meta-GWAS for amyloid PET and CSF reported to date9,48.
c. Polygenic Risk Scores (PRS)
We computed the AD PRS described in Bellenguez et al that considered 83 loci. However, some SNPs were not imputed or had a low imputation quality (R2<0.3), and we decided to calculate the AD PRS including genetic variants found in all imputed datasets (n=76; Supplementary Table 5). For PRS calculation, we added the gene dosages of these SNPs weighting by their effect size (beta coefficients); the allele analysed was matched to the reported allele (A1) by Bellenguez et al4. Because some control samples were included in the first stage of the AD GWAS, we considered the independent effects reported in the second stage for the PRS calculation. Additionally, due to the large effect on AD risk and its well-established association with most AD endophenotypes, the APOE locus was excluded from all these PRS. We then tested its association with AD case-control status, CSF Aβ42 and p-tau endophenotypes, and PET amyloid burden measurements. We considered as a covariate the age, sex, and disease status only in associations with biomarkers. These analyses were performed separately in each cohort except for Valdecilla which was excluded due to reduced sample size, while Ace PET cohort was excluded in the case-control analysis because all individuals were cognitively unimpaired. Additionally, we considered the fixed effect meta-analysis model considering the heterogeneity threshold (I2) of 75% as high53.
We also calculated another PRS for A) AD4 (n=76 SNPs; Supplementary Table 5), B) CSF Aβ42 levels (n=30 SNPs; Supplementary Table 6) considering the genetic variants with a P<1e-05 described in Jansen et al9, and C) an amyloid burden PRS considering suggestive variants found in our meta-analysis (combining endophenotypes filtering according to the effect size direction; n=9 SNPs; Supplementary Table 7) in GR@ACE cohort individuals, including 8,110 cases and 9,640 controls the same way as described above. Further information about the cohort has been previously published5,19. For PRS computation, the effect (beta coeffcients) and standard errors were estimated using the equations described by Zhu et al54. Again, we associated these scaled PRS with case-control data in non-overlapping individuals to assess if Aβ genetic determinants are also related to disease risk.
d. Association between biomarker levels and SOMAscan proteomics
We assessed the association between SOMAscan 7k proteomic panel and CSF Aβ42 levels (n=1,008) in the Ace CSF cohort. Briefly, SOMAscan proteomic measures were log10 transformed, outliers were removed at ±3 standard deviations from the mean and standardized using the scale R function with centring and scaling. For further analysis, we selected 2,682 proteins based on correlations between: 1) two independent SOMAscan assay analysing the same samples, and 2) comparing aptamer- and antibody-based proteomic platforms55,56. To identify proteins associated to CSF Aβ42, a linear regression model was performed on scaled CSF Aβ42 levels and proteomic measures. We considered disease status, sex, age, and the CSF biomarker technique as covariates. Subsequently, the top 500 ranking of significant proteins associated with CSF Aβ42 (False discovery rate; FDR<1.864e-05) was analysed in the WEB-based GEne SeT AnaLysis Toolkit (WebGestalt)57 to perform an over-representation analysis (ORA) considering several functional databases and the whole genome as built-in reference gene list following the idea of investigating the complete genome (GWAS and gene-based analyses) (Supplementary Table 8). We also performed an enrichment analysis on the complete subset of valid SOMAscan proteins (n=2,682) to evaluate the impact of platform analyte preselection and quality control process on the results obtained.
To explore the biological significance of the GWAS results, we displayed the overlap between loci controlling amyloidosis and the proteins significantly associated with CSF Aβ42 in the Ace CSF cohort using Venn diagrams. The top 500 proteome and genome hits selected from the CSF Aβ42 meta-GWAS described in Jansen et al, the meta-analysis of CSF-PET endophenotypes filtered by opposite effect size direction, and the gene-based MAGMA analysis performed by FUMA were identified and annotated. The top rankings were reduced to 345, 339, 457, 361 and 465 loci for the meta-GWAS by Jansen et al, our current CSF-PET meta-analysis and its gene-based MAGMA analysis, the PAD CSF-PET meta-GWAS and its gene-based MAGMA analysis9,48, respectively. These reductions were due to the presence of SNPs that were not annotated and could not be matched to UniProt codes (Supplementary Table 9, Supplementary Table 10, Supplementary Table 11, Supplementary Table 12 and Supplementary Table 13). The top rankings were compared and the overlap between genomic and proteomic analysis was identified and evaluated using WebGestalt tool as described above.
Results
Meta-analysis of Aβ endophenotypes
The genome-wide meta-analysis of CSF endophenotypes involved 4 independent AD cohorts with Aβ42 measures (n=1,483; λ=1.009). The genomic inflation factor (λ) suggested no gross bias or stratification. As it was expected, we observed a consistent genome-wide significant association with rs429358-APOE locus as a sentinel variant (Effect=−0.58 [−0.658, −0.503]; P=8.36e-49). We detected 19 additional suggestive pQTL signals for Aβ42 levels in CSF (Supplementary Table 14). Similarly, the meta-analysis of amyloid PET endophenotype (n=593; λ=1.013), revealed a genome-wide significant association in the same sentinel variant in the opposite direction (rs429358-APOE locus; Effect=0.684 [0.555, 0.813]; P = 2.00e-25). An additional novel hit at rs72737013 close to the ANXA1 gene (Effect=0.813 [0.528, 1.099]; P=2.39e-08) was detected. This gene is related to anti-inflammatory reactions, innate immune response, and inflammatory processes58, psychiatric disorders, brain volume59,60, and the degradation of Aβ species61. Additionally, there were 43 additional independent suggestive signals annotated for amyloid burden measured using PET (Supplementary Table 15). We combined the summary statistics from both CSF and PET Aβ meta-analyses without considering the effect direction (n=2,076). Again, we confirmed the sentinel variant rs429358 to be the most significant locus in the APOE region. Other genetic variants emerged as GWAS-significant in this new meta-analysis. However, none of them were inversely associated with CSF and PET endophenotypes in all studies except for the rs429358-APOE marker. We considered these hits as false positive signals (Supplementary Table 16).
In looking for new suggestive signals beyond APOE, we extracted the subset of 376,599 SNPs with consistent opposite effect in CSF and PET analyses. After the SNP selection in the combined Aβ meta-analysis, the rs429358-APOE variant (P=9.50e-67) remained as the only GWAS-significant hit (Fig. 1A, upper) but nine additional suggestive consistent variants were identified in genes such as NPY5R, TIAM2 or MAGI2, among others (Table 1). Additionally, the combination of Aβ endophenotypes enhanced the significant replication of several genetic markers previously described for AD4, CSF Aβ42 levels9 and neuropathological features50 (Supplementary Material 1, Supplementary Table 2, Supplementary Table 3, Supplementary Table 4).
Figure 1. Plots of the Aβ burden meta-analysis combining data of CSF-PET endophenotypes.
A) (upper) Manhattan plot of our CSF-PET meta-analysis (n=2,076). Results were filtered according effect size direction and dataset missingness. Suggestive independent markers were annotated with the nearest gene name. Mapped genes coloured in grey represent those that were not replicated in the PAD CSF-PET meta-GWAS. (lower) Manhattan plot of the PAD CSF-PET meta-analysis filtered (n=23,532). Genome-wide significant independent markers were annotated with the nearest gene name. The Y-axis was restricted to visualize suggestive signals. The genome-wide significance threshold was set to P<5e-08 (red line) and the suggestive threshold was set to P<1e-05 (blue line). B) Venn diagram representing the overlap between the top 500 ranking of independent genetic markers comparing the PAD and our amyloid burden meta-analysis. C) Venn diagram representing the overlap between the top 500 ranking of independent genes in the PAD and our gene-based analysis.
Table1.
Results of the genome wide meta-analysis filtered combining CSF and PET endophenotypes (4.9% of total SNPs).
| rsID | SNP | Nearest Gene | REF | ALT | A1 | FreqA1 | Call Rate | n | Zscore | P-value | Direction |
|---|---|---|---|---|---|---|---|---|---|---|---|
| rs429358 | 19:45411941 | APOE (0 kb) | T | C | C | 0.211 | 1.000 | 2076 | −17.259 | 9.50E-67 | ---++ |
| rs78402940 | 2:59125664 | FLJ30838 (0 kb) | A | G | G | 0.089 | 0.999 | 1979 | −4.849 | 1.24E-06 | --?-++ |
| rs62354504 | 5:36361976 | RANBP3L (59.96 kb) | G | A | A | 0.027 | 0.999 | 1262 | −4.807 | 1.53E-06 | ?-?-+? |
| rs62340552 | 4:180023617 | NA | C | T | T | 0.041 | 0.993 | 1798 | −4.582 | 4.61E-06 | --?-+? |
| rs2902373 | 11:43637563 | MIR129-2 (34.53 kb) | C | T | T | 0.222 | 0.993 | 2076 | −4.571 | 4.84E-06 | ----++ |
| rs73141455 | 7:78648931 | MAGI2 (0 kb) | G | T | T | 0.038 | 0.995 | 1798 | −4.555 | 5.24E-06 | --?-+? |
| rs115822934 | 4:164308011 | NPY5R (34.92 kb) | G | T | T | 0.032 | 0.998 | 1798 | −4.448 | 8.65E-06 | ++?+-? |
| rs1523589 | 18:30242590 | KLHL14 (−10.04 kb) | C | A | A | 0.201 | 0.996 | 2076 | −4.440 | 8.99E-06 | ++++-- |
| rs4395536 | 4:168504316 | NA | A | G | G | 0.914 | 0.987 | 1979 | −4.435 | 9.23E-06 | ++?+-- |
| rs11963901 | 6:155436138 | TIAM2 (0 kb) | C | T | T | 0.106 | 0.999 | 1979 | −4.434 | 9.23E-06 | --?-++ |
Bold: significant results P < 5e-08 with consistent effect direction.
Note: Meta-analysis was performed using a sample size weighted method without considering effect size direction and filtered by effect size and data availability in at least half of each endophenotypes cohorts. Reference SNP (rs) code for the SNP: rsID, Genetic markers in GRCh37/hg19 genomic assembly: SNP, Reference allele: REF, Alternative allele: ALT, Effect allele: A1, Sample size: n.
However, the PAD CSF-PET meta-analysis9,48 (effective sample size n=23,532) identified several markers previously associated with AD and its endophenotypes. These significant markers were identified on chromosome 19 including the rs429358-APOE (P=5.94e-601), as well as, the rs4844610-CR1 (P=5.76e-18), rs7982-CLU (P=7.81e-11), rs12151021-ABCA7 (P=3.92e-10), rs6733839-BIN1 (P=1.02e-08), rs117834516-FERMT2 (P=4.82e-08) and the novel rs4955351-GADL1 (P=3.19e-08) which was not previously associated to AD or amyloid levels (Fig. 1A lower, Supplementary Table 17). Additionally, the PAD analysis replicated the rs115822934-NPY5R variant (P=3.21e-04) originally found suggestive in our CSF-PET meta-analysis (Supplementary Table 18). Importantly, we also observe concordances between our local effort (amyloid burden CSF-PET meta-GWAS) and the PAD. Specifically, we detected 15 overlapping sentinel markers in the top 500 ranking of the amyloid burden meta-GWAS from both the PAD and our current meta-analysis (Fig.1B), as well as 67 overlapping genes in the PAD and our gene-based top 500 ranking (Fig.1C).
To link the variants of interest to specific genes and obtain relevant functional information about these loci, we applied FUMA to the suggestive signals from the Aβ meta-analysis that were filtered based on opposite direction in CSF and PET (Table 1). There were 125 prioritized genes mapped using at least two strategies (positional mapping, eQTL or chromatin interactions) and 45 genes were selected based on the three strategies described in methods. As expected, the majority of the prioritized genes were related to the rs429358-APOE. Excluding chromosome 19, we prioritized 23 genes mapped (6 SNPs) with a CADD score > 12.37 suggesting a potential deleterious effect (Supplementary Table 19)62. In contrast to the univariate SNP analysis, the gene-based analysis performed using MAGMA revealed 15 study-wise significant loci (P<4.235e-06) excluding the APOE region (Table 2, Supplementary Fig. 3). Interestingly, the identified genetic variants in some of these genes (TENM3, TMEM132D, PTPRD, CNTN5, RBFOX1, CSMD1, TIAM2, RORA and WWOX) have been previously related to neuroimaging endophenotypes63–65, extreme AD PRS measures66, AD endophenotypes (CSF Aβ42 or p-tau levels13,67–70), mental disorders71,72 and cognitive decline in AD64,73,74. Additionally, the gene-based analysis of the PAD amyloid burden meta-GWAS revealed genes previously associated to AD such as APOE (P=2.09e-13), CLU (P=2.13e-07), FERMT2 (P=3.49e-07) and the CR1 locus (P=3.64e-06), which reached borderline gene-wide significance threshold at P<2.717e-06 (Supplementary Table 20).
Table 2.
Gene-based MAGMA results from FUMA analysis considering genome-wide significant results P<4.235e-06.
| Gene | EntrezID | UniProtID | CHR | Start | Stop | n SNPS | Zscore | P-value |
|---|---|---|---|---|---|---|---|---|
| NECTIN2 | 5819 | Q92692 | 19 | 45349432 | 45392485 | 37 | 7.787 | 3.442E-15 |
| APOE | 348 | P02649 | 19 | 45409011 | 45412650 | 4 | 7.282 | 1.648E-13 |
| APOC1 | 341 | P02654 | 19 | 45417504 | 45422606 | 5 | 6.774 | 6.278E-12 |
| CSMD1 | 64478 | Q96PZ7 | 8 | 2792875 | 4852494 | 929 | 6.437 | 6.076E-11 |
| TOMM40 | 10452 | O96008 | 19 | 45393826 | 45406946 | 24 | 6.109 | 5.000E-10 |
| WWOX | 51741 | Q9NZC7 | 16 | 78133310 | 79246564 | 323 | 5.566 | 1.307E-08 |
| LHPP | 64077 | Q9H008 | 10 | 126150403 | 126306457 | 153 | 5.507 | 1.822E-08 |
| BCL3 | 602 | P20749 | 19 | 45250962 | 45263301 | 6 | 5.449 | 2.540E-08 |
| CNTN5 | 53942 | O94779 | 11 | 98891683 | 100229616 | 556 | 5.358 | 4.217E-08 |
| TENM3 | 55714 | Q9P273 | 4 | 183065140 | 183724177 | 114 | 5.312 | 5.432E-08 |
| PTPRD | 5789 | P23468 | 9 | 8314246 | 10612723 | 332 | 5.258 | 7.272E-08 |
| TIAM2 | 26230 | Q8IVF5 | 6 | 155153831 | 155578857 | 60 | 5.243 | 7.904E-08 |
| RBFOX1 | 54715 | Q9NWB1 | 16 | 6069095 | 7763340 | 382 | 5.079 | 1.900E-07 |
| GPC5 | 2262 | P78333 | 13 | 92050929 | 93519490 | 275 | 4.956 | 3.604E-07 |
| RORA | 6095 | P35398 | 15 | 60780483 | 61521518 | 185 | 4.827 | 6.948E-07 |
| MACROD2 | 140733 | A1Z1Q3 | 20 | 13976015 | 16033842 | 354 | 4.787 | 8.471E-07 |
| TMEM132D | 121256 | Q14C87 | 12 | 129556270 | 130388211 | 205 | 4.759 | 9.717E-07 |
| CDH13 | 1012 | P55290 | 16 | 82660408 | 83830204 | 315 | 4.592 | 2.201E-06 |
| LRP1B | 53353 | Q9NZR2 | 2 | 140988992 | 142889270 | 318 | 4.549 | 2.696E-06 |
| KIR3DX1 | 90011 | Q9H7L2 | 19 | 55043909 | 55057053 | 9 | 4.487 | 3.616E-06 |
| FAM53B | 9679 | Q14153 | 10 | 126307861 | 126432838 | 42 | 4.479 | 3.751E-06 |
Chromosome: CHR, Number of SNPs: NSNP, Sample size: n, Entrez Gene Identifier: EntrezID, UniProt Swiss Protein Identifier: UniProtID.
Association between AD PRS with AD endophenotypes and other clinical features
We observed a significant result in the meta-analysed associations between the AD PRS and Aβ levels (CSF Effect = −0.05 [−0.10, −0.00]; P=3.43e-02 and PET Effect = 0.10 [0.02, 0.17]; P=1.30e-02). These results suggest that genes involved in AD risk indeed modulate amyloid levels (Fig. 2A, Fig. 2B).
Figure 2. Forest plot of the meta-analysis association between the AD PRS.
A) CSF Aβ42, and B) Aβ PET endophenotypes. The significance threshold was set to 0.05.
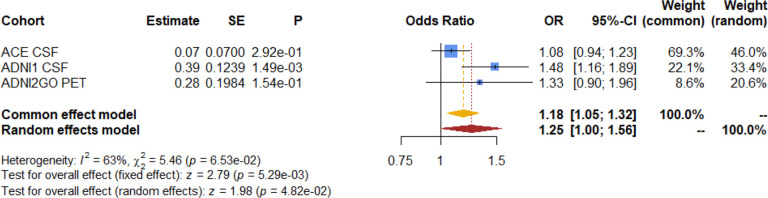
As expected, we observed a significant result in the association meta-analysis of the AD PRS with case-control dementia status in all 3 endophenotype datasets (OR=1.18 [1.05, 1.32]; P=5.29e-03). These results suggest that these genes modulate the disease status as previously reported4 (Fig. 3). Even though ADNI2GO did not reach statistical significance, it had a similar effect size and direction, possibly due to the low proportion of AD cases in this cohort (6.55 %).
Figure 3. Forest plot of the meta-analysis association between the AD PRS and dementia status as case-control.
In ACE (305 cases and 703 controls, 30.25%), ADNI1 (94 cases and 285 controls, 24.80%) and ADNI2GO cohorts (27 cases and 385 controls, 6.55%).
Association between genetic variants of amyloid endophenotypes with case-control status
To assess whether CSF Aβ42 genetic modulators are also related to AD risk, we constructed different PRS including variants detected in our study and previous meta-GWAS4. We then checked the association of calculated PRS in the GR@ACE case-control study5. We did not observe any significant association for any calculated PRS for amyloid (Fig. 4) which could be due to the reduced set of independent markers reported for these phenotypes (P<1e-05) or not having an impact on AD pathology. As expected, the AD PRS was highly associated with the case-control (OR=1.35 [1.30, 1.40]; P=3.02e-49), thus confirming that the AD genes previously described by us and the EADB consortium4,5 truly modulate disease risk in the GR@ACE/DEGESCO cohort.
Figure 4. Forest plot of the association between the AD, Aβ PRS and case-control status.
PRS for AD (76 SNPs from Bellenguez et al., 2022) and Aβ42 (30 SNPs from Jansen et al., 2022, 9 SNPs from our meta-analysis). The GR@ACE cohort included 8110 cases and 9640 controls.
CSF proteome signatures associated with the Aβ42 CSF levels
We regressed the CSF Aβ42 peptide levels on CSF SOMAscan aptamer levels to identify the proteomic signature associated with amyloid burden (Fig. 5A). We identified 1,387 study-wide significant proteins in the linear model of CSF Aβ42 (FDR<1.864e-05) (Supplementary Table 21). Notably, we observed a marked asymmetry in the effect of SOMAmers on CSF Aβ42 levels, with the majority showing estimates greater than 0, suggesting a positive correlation contributing to increased CSF Aβ42 levels (Fig. 5A). Thus, the top 100 ranks of significant associations have an estimate range between 0.449 and 0.317, which contributes to an increase of this magnitude in CSF Aβ42 levels, and the variance explained by these highly associated proteins ranges of between 0.202 and 0.297. Importantly, we observed multiple proteins that have been associated with the CSF levels of Aβ species or its mechanisms in previous studies, such as MTMR7, LMOD4, GD3S, SERA/PHGDH, SELS, ATE1, NPTXR, and the 14-3-3 eta protein, among others75–81.
Figure 5. Associations between CSF SOMAscan and CSF Aβ42 levels.
A) Vulcano plot only considering proteins with good inter-assay correlation (n=2,682), significant proteins (FDR < 1.864e-05) were highlighted in red (n=1,387). B) Top 10 results of the enrichment analysis of significant protein associations with CSF Aβ42 levels using the WebGestalt tool.
An enrichment analysis performed for significant proteins associated with CSF Aβ42 levels revealed genes involved in neuronal projection guidance (enrichment ratio=11.034; FDR<2.2e-16), synaptic structure and activity (enrichment ratio=10.868; FDR<2.2e-16), cell–cell adhesion by plasma membrane molecules (enrichment ratio=7.660; FDR<2.2e-16), peptidyl-tyrosine modifications (enrichment ratio=6.174; FDR<2.2e-16), regulation of cell morphogenesis (enrichment ratio=5.786; FDR<2.2e-16) and angiogenesis (enrichment ratio=5.617; FDR<2.2e-16) which are mainly driven by the large proportion of proteins with a positive effect (n=1,300; 93.73%) (Fig. 5B; Supplementary Table 22, Supplementary Table 23). Furthermore, when comparing the enrichment results from the ORA analysis between the entire set of valid SOMAscan proteins and the proteins significantly associated with Aβ42 levels, we observed a complete lack of overlap, reinforcing the validity of our findings (Supplementary Fig. 4).
To identify those genes that were commonly associated with CSF Aβ42 levels in genomic and proteomic analyses, we compared the top 500 common list of signals in the following four analyses: meta-GWAS by EADB4, our meta-analysis of CSF-PET, gene-based MAGMA, and SOMAscan protein analysis. We found three genes/proteins (CHST1, PTPRD and TMEM132D) present in all four analyses, representing only 0.2% of the total loci/proteins analysed (full overlap). In addition, 32 other proteins overlapped between the SOMAscan proteomics and any genomic analysis, including four proteins represented in 3 different analyses (Fig. 6A, Supplementary Table 24). Similar results were obtained in investigating the top rankings of the SOMAscan analysis, the PAD CSF-PET meta-GWAS and its gene-based analysis; only the TMEM132D was represented in all analyses (Fig. 6B). Interestingly, we found that 10 of the 23 loci/proteins observed were also overlapping with the ranking considering our main CSF-PET meta-GWAS results. This overlapping with PAD CSF-PET meta-GWAS support the validity of our approach (Supplementary Fig. 5, Supplementary Table 25). However, there was a reduced consistency between the top 500 SOMAscan proteins associated with CSF Aβ42 and any genomic results with less than 2.5% of overlapping proteins. These results suggest that the Aβ42-related protein signature in CSF might not be closely linked to amyloid genetic modulators, indicating that the proteome signature associated with Aβ42 burden in the brain primarily reflects general disease processes largely unrelated to the genetic elements controlling amyloid production.
Figure 6. Overlapping loci/proteins in genomic and proteomic analysis.
A) Venn diagram of the top 500 ranking of CSF Aβ42-associated proteins in the SOMAscan panel (orange), our gene-based MAGMA analysis (red), GWAS of CSF Aβ42 (Jansen et al., 2022) (dark blue) and our amyloid burden meta-analysis of filtered CSF-PET endophenotypes (light blue). B) Venn diagram of the top 500 ranking of CSF Aβ42-associated proteins in the SOMAscan panel (orange), PAD gene-based MAGMA meta-analysis (red) and PAD amyloid burden meta-analysis of filtered CSF-PET endophenotypes (light blue). C) Top 10 enrichment analysis results of the overlapping proteins between our genomic and proteomic analyses. C) Top 10 enrichment analysis results of the overlapping proteins between proteomic and PAD genomic analyses. The analysis was done using the WebGestalt tool.
Finally, to gain insight into the few commonalities identified by comparing genetic and proteome signatures associated to the amyloid burden in the brain, we conducted a new enrichment analysis. Despite the reduced overlapping hits among proteome and genome studies, several significant mechanisms related to the synthesis of glycosylphosphatidyl inositol (GPI)-anchored proteins by post-translational modifications were identified (enrichment ratio=48.070; FDR=1.86e-04) and the anchored component of the membrane (enrichment ratio=31.778; FDR=1.32e-04), cell–cell adhesion via plasma membrane molecules (enrichment ratio=26.207; FDR=3.42e-06), mental disorders (enrichment ratio=12.853; FDR=3.42e-06) such as autism (enrichment ratio=18.556; FDR=0.002) and anxiety (enrichment ratio=23.524; FDR=0.004), and regulation and development of neuron projections (enrichment ratio=13.035; FDR=0.002) among others (FDR<0.05). Interestingly, six of these mechanisms were also represented in the enrichment analysis of the PAD CSF-PET meta-analysis, which confirms our main results (Fig. 6C, Supplementary Table 26, Supplementary Table 27, Fig.6D, Supplementary Table 28, Supplementary Table 29).
Discussion
For the first time, we have combined meta-GWAS results obtained from analysing amyloid PET and CSF Aβ42 levels. Our innovative experimental approach identified novel genetic variants associated with amyloid burden endophenotypes. This meta-analytic approach benefited from combining endophenotypic information from six cohorts thereby increasing our statistical power. As expected, we identified a genome-wide significant hit at the rs429358-APOE loci. We also observed a novel genome-wide significant hit near the ANXA1 locus exclusively associated with PET amyloid. SNPs in this locus were previously linked to psychiatric disorders, brain volume59,60, and the degradation of Aβ species61. However, neither the large PET meta-GWAS available nor the PAD meta-analysis conducted by us replicated this finding. For these reasons, we believe that this signal could be a false positive. We attribute the lack of additional hits to the relatively small sample size of our CSF-PET meta-GWAS. By repeating this strategy with a larger sample size, we expect to identify more genetic modulators of Aβ42 peptide expression in the brain. Indeed, using a similar approach with currently available summary statistics (PAD study), we were able to detect several sentinel markers surpassing the GWAS significance threshold. Specifically, the PAD CSF-PET meta-analysis identified several significant genes that have been previously related to AD (CR1, BIN1, CLU, ABCA7, FERMT2 and APOE4,82–85) or amyloid proteins (CR1, CLU, APOE and FERMT29,86,87), as well as PICALM and GPC5 suggestive genes84,88. Notably, we identified the novel GADL1 locus, which encodes for a protein from the glutamate decarboxylase family, suggesting that it might have a glutamate decarboxylase activity in the CNS89,90.
In AD, the glutamate excess generates a continuous glutamatergic activity, impairing neuronal plasticity and long-term potentiation leading to excitotoxicity. Therefore, using receptor antagonists such as memantine, which has shown neuroprotective effects, could be a crucial therapeutic intervention91,92.
Importantly, these results should be interpreted with extreme caution because PAD analysis is not entirely independent as various cohorts were represented in both summary statistics of the PAD analysis (Supplementary Table 30). This overlapping samples (11.284%) could lead to overestimated effects and increased proportion of false positive findings. Compared to our local effort, where we eliminated any potential overlap between CSF and PET cohorts, we remain very cautious about the PAD results due to the potential overlap of subjects among studies. Future efforts are necessary to confirm the findings from the PAD analysis. Nevertheless, the PAD analysis replicated the rs115822934-NPY5R marker, alongside the rs429358-APOE, originally identified in our CSF-PET meta-analysis. These results might suggest that NPY5R could be genuinely involved in amyloid pathology, as well as panic disorders93,94. Again, further completely independent studies, expanding the sample size of these analysis, are needed to validate our observation and working hypothesis.
In spite of these limitations, our experimental strategy permitted us to evaluate common pathways potentially associated to CSF-soluble Aβ42 (circulating amyloid)95 and brain amyloid species detected by PET (insoluble species such as amyloid plaques or cerebral amyloid angiopathy)96 and proteome signature associated to CSF Aβ42 peptide levels. To assess the relationship between genetic modulators and protein levels, we analysed the overlap between loci-controlling amyloid levels and significant proteins associated with CSF Aβ42 levels. Importantly, three genes/proteins (CHST1, PTPRD and TMEM132D) were identified and prioritized in all analyses, thus suggesting that these modulators might be key drivers controlling amyloid pathology. Lower TMEM132D levels have been observed in patients with frontotemporal dementia97, and genetic markers in this gene have been related to anxiety, panic disorders and the rate of cognitive decline73,98,99. This locus was the only that also overlapped with all PAD rankings, suggesting that might be a potential modulator of amyloid pathology. The PTPRD gene, which was also represented in the large meta-GWAS gene-based ranking, has been significantly associated to synaptic process in schizophrenia100, AD susceptibility, neurofibrillary tangle and neuritic plaques68. We consider these two loci excellent candidates for further translational research due to their consistent statistical significance and previous literature findings. Nevertheless, there is a possibility that we are not capturing pathological mechanisms occurring similarly in both biofluids due to the opposite direction filtering, which could be contributing to the accumulation or reduction in both CSF and PET amyloid levels. These discordances have been described in previous articles101–103, suggesting that they might be caused due to the differential sensitivity to amyloid species across the AD continuum. Further research is needed to elucidate the role of these common and discordant amyloid mechanisms occurring in brain and their impact on disease development.
In this study, we found a limited overlap between genetic modulators of amyloid burden and the proteins associated with the CSF levels of Aβ42. This could be interpreted as a result of the inherent statistical noise in these multiomic analyses, the lack of power in our main analysis, or it could indicate that the observed discordance is genuine. The poor heritability reported for CSF traits in previous studies9 supports that common SNPs might not strongly modulate the CSF amyloid burden. Moreover, no amyloid PRS showed a significant association with the risk of developing AD, whereas the AD PRS showed a strong association with the AD case-control status and amyloid levels, which is fully consistent with previous studies70,104–106. These results suggest either a lack of statistical power to detect genuine hits associated with amyloid burden or a limited causal role of common genetic modulators of amyloid deposits in the aetiology of clinical AD. Further studies are needed to clarify these discrepancies. Interestingly, we observed a higher number of loci/proteins overlapped with the SOMAscan protein associations with CSF Aβ42 levels and the gene-based analysis than in the sentinel SNP-based GWAS analyses (our meta-analysis n=21). The gene-based approach could be particularly powerful because the genetic markers summarised at (protein-coding) gene level might reduce the statistical noise on a full GWAS dataset52.
We also noted a large number of significant CSF SOMAscan proteins associated with CSF Aβ42 levels. Notably, most of the observed associations were predominantly positive in our study. Interestingly, Bader et al107 reported a correlation map illustrating high correlations between CSF proteomic measures suggesting that these measures might lead to multiple significant associations. The massive abundance of significant proteins might simply reflect a general neurodegenerative signature that occurs as a result of widespread neuronal cell death or reactive gliosis. These changes are likely to be epiphenomenal rather than specific to the AD process. The potential implication of these findings is important for interpreting CSF proteome results. Indeed, only a minority of proteomic markers associated to Aβ42 might be genuine mediators modulating the AD-related amyloid endophenotype. Overall, the lack of overlap between Aβ42 and AD risk GWAS studies suggests that genetic factors modulating amyloid production may represent only a relatively small component of overall AD causality. These findings are also in line with several clinical trials targeting amyloid, that have observed a reduced association between Aβ reduction and AD progression, as well as only modest control of AD progression with these monotherapies. This also suggests that both amyloid-dependent and amyloid-independent mechanisms must be addressed simultaneously to effectively control disease progression108,109.
Despite the poor overlap, we detected 35 overlapping genes and proteins pointing to a few enriched mechanisms in our CSF-PET meta-GWAS. We consider these overlapping signals of special importance because they could point to genuine amyloid-related mechanisms involved in AD causality and development. We found Aβ burden significantly associated with pathways controlling the anchored proteins in the membrane, which had also been represented in the PAD enriched analysis (n=23 loci/proteins). Interestingly, six enriched mechanisms were represented in both overlapping loci/protein rankings of the PAD and our CSF-PET meta-analysis. These results validate our findings and suggest that the enrichment analysis is more powerful in detecting genuine associations than analysing individual genes, particularly in the context of reduced statistical power.
Additionally, the enrichment analysis pointed to synapse molecules and cell adhesion mechanisms. Neuronal cadherins and integrins have been linked to the synaptic process, plasticity and long-term potentiation and modulation of Aβ levels110, while their loss has been correlated to cognitive decline111–113. Furthermore, we detected a link between amyloid levels and mental disorders, such as anxiety which has been associated with high Aβ deposition across the AD continuum114–116. On the contrary, autism spectrum disorder (ASD) has been associated with Aβ processing via the non-amyloidogenic pathway leading to reduced Aβ levels in ASD patients117. Other overlapping loci and proteins such as ROBO2, CNTN5, OPCML, NRG3, NGFR or CACNA2D3, have been associated with cognitive performance118,119, age at onset120,121, schizophrenia122–124 or ASD125,126, AD127,128 and its endophenotypes129,130.
Considering all these observations, it is difficult to conceive that all of them can be explained by pure random chance. However, our analysis had important limitations. First, we use a suboptimal p-value-based meta-analysis method, however, this strategy becomes highly valuable for integrating diverse studies reporting different estimate metrics and combining endophenotypes measured by various techniques131,132. Also, the CSF-PET meta-analysis did not report effect size which were estimated. The restrictive SNP filtering allowed the evaluation of only 4.9% of genomic markers, likely due to meta-analysing multiple datasets and reducing marker identification involved in common mechanisms between soluble-CSF and insoluble-PET amyloid species. Moreover, as mentioned earlier, the PAD analysis was not completely independent, with an 8.272% and 3.073% of overlapping samples between our main meta-analysis, the CSF9 and PET48 summary statistics, respectively. The PAD CSF-PET meta-analysis should be interpreted with extreme caution due to these overlapping samples among summary statistics. Because we used publicly available results, we could not confirm the presence of additional overlapping samples, potentially leading to overfitting. The Ali et al meta-GWAS conducted a different data harmonisation process, potentially introducing variability. Furthermore, neuropathological information was not available for these samples, leaving us unaware of other concomitant pathological changes. Finally, the lack of significant findings for several PRS associations may suggest that there is insufficient statistical power to find genetic variants that affect the amyloid endophenotype. These concerns should be addressed in future research.
In summary, our results demonstrate the feasibility of combining Aβ endophenotypes in CSF and PET, along with proteome analysis, to gain novel insights into the fundamental biology of AD. The strong proteomic associations with Aβ endophenotypes could help identify signalling pathways and molecular mechanisms involved in Aβ and AD pathology, as well as the overlapping pathways that control the amyloidotic process. Further studies are needed to refine these observed associations, connecting AD loci and proposed causal pathways with brain amyloidogenesis.
Supplementary Material
Acknowledgments
We would like to thank patients and controls who participated in this project. The present work has been performed as part of the doctoral thesis of RPF at the University of Barcelona (Barcelona, Spain). Some control samples and data from patients included in this study were provided in part by the National DNA Bank Carlos III (www.bancoadn.org, University of Salamanca, Spain) and Hospital Universitario Virgen de Valme (Sevilla, Spain); they were processed following standard operating procedures with the appropriate approval of the Ethical and Scientific Committee. Data used in this article were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (adni.loni.usc.edu). As such, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in analysis or writing of this report. A complete listing of ADNI investigators can be found at: http://adni.loni.usc.edu/wp-content/uploads/how_to_apply/ADNI_Acknowledgement_List.pdf. Data was used for this project of which collection and sharing was funded by the Alzheimer’s Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12-2-0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: AbbVie, Alzheimer’s Association; Alzheimer’s Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Biogen; Bristol-Myers Squibb Company; CereSpir, Inc.; Cogstate; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; EuroImmun; F. Hofmann-La Roche Ltd and its afliated company Genentech, Inc.; Fujirebio; GE Healthcare; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Lumosity; Lundbeck; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfzer Inc.; Piramal Imaging; Servier; Takeda Pharmaceutical Company; and Transition Therapeutics. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Therapeutic Research Institute at the University of Southern California. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California.
Funding
The Genome Research @ Ace Alzheimer Center Barcelona project (GR@ACE) is supported by Grifols SA, Fundación bancaria ‘La Caixa’, Ace Alzheimer Center Barcelona and CIBERNED. Ace Alzheimer Center Barcelona is one of the participating centers of the Dementia Genetics Spanish Consortium (DEGESCO). The FACEHBI study is supported by funds from Ace Alzheimer Center Barcelona, Grifols, Life Molecular Imaging, Araclon Biotech, Alkahest, Laboratorio de análisis Echevarne and IrsiCaixa. Authors acknowledge the support of the Spanish Ministry of Science and Innovation, Proyectos de Generación de Conocimiento grants PID2021-122473OA-I00, PID2021-123462OB-I00 and PID2019-106625RB-I00. ISCIII, Acción Estratégica en Salud, integrated in the Spanish National R+D+I Plan and financed by ISCIII Subdirección General de Evaluación and the Fondo Europeo de Desarrollo Regional (FEDER “Una manera de hacer Europa”) grants PI13/02434, PI16/01861, PI17/01474, PI19/00335, PI19/01240, PI19/01301, PI22/01403, PI22/00258 and the ISCIII national grant PMP22/00022, funded by the European Union (NextGenerationEU). The support of CIBERNED (ISCIII) under the grants CB06/05/2004 and CB18/05/00010. The support from the ADAPTED and MOPEAD projects, European Union/EFPIA Innovative Medicines Initiative Joint (grant numbers 115975 and 115985, respectively); from PREADAPT project, Joint Program for Neurodegenerative Diseases (JPND) grant N° AC19/00097; from HARPONE project, Agency for Innovation and Entrepreneurship (VLAIO) grant N° PR067/21 and Janssen. DESCARTES project is funded by German Research Foundation (DFG). Additionally, IdR is supported by the Instituto de Salud Carlos III (ISCIII) under grant FI20/00215. PGG is supported by CIBERNED employment plan (CNV-304-PRF-866). ACF received support from the ISCIII under the grant Sara Borrell (CD22/00125). JEC received support from National Institute of Health award P30AG066546.
Abbreviations:
- AD
Alzheimer’s disease
- ADNI
Alzheimer’s Disease Neuroimaging Initiative
- ASD
autism spectrum disorder
- Aβ
amyloid
- Aβ42
amyloid beta 42
- AV45
Florbetapir
- CADD
Combined Annotation Dependent Depletion
- CDR
Clinical Dementia Rating
- CI
Confidence interval
- eQTL
expression quantitative trait loci
- FBB
Florbetaben
- FDR
false discovery rate
- FUMA
Functional Mapping and Annotation of Genome-Wide Association Studie
- GPI
glycosylphosphatidyl inositol
- GWAS
Genome-wide association studie
- HC
Healthy Control
- LP
lumbar puncture
- MAC
minor allele count
- MAF
minor allele frequency
- MCI
Mild Cognitive impairment
- MMSE
Mini-Mental State Exam
- n
Sample size
- NIA-AA
National Institute on Aging and Alzheimer’s Association
- NINCDS/ADRDA
National Institute of Neurological and Communicative Disorders and Stroke and Alzheimer’s Disease and Related Disorders Association
- OR
Odds ratio
- P
p-value
- p-tau
phosphorylated tau in Thr 181
- PAD
publicly available datasets
- PET
Positron Emission Tomography
- PCA
principal component analysis
- PCs
Principal components
- pQTL
protein quantitative trait loci
- PRS
Polygenic risk scores
- RFU
relative fluorescent units
- SCD
subjective cognitive decline
- SNP
single nucleotide polymorphisms
- t-tau
total tau
- λ
genomic inflation facto
Funding Statement
The Genome Research @ Ace Alzheimer Center Barcelona project (GR@ACE) is supported by Grifols SA, Fundación bancaria ‘La Caixa’, Ace Alzheimer Center Barcelona and CIBERNED. Ace Alzheimer Center Barcelona is one of the participating centers of the Dementia Genetics Spanish Consortium (DEGESCO). The FACEHBI study is supported by funds from Ace Alzheimer Center Barcelona, Grifols, Life Molecular Imaging, Araclon Biotech, Alkahest, Laboratorio de análisis Echevarne and IrsiCaixa. Authors acknowledge the support of the Spanish Ministry of Science and Innovation, Proyectos de Generación de Conocimiento grants PID2021-122473OA-I00, PID2021-123462OB-I00 and PID2019-106625RB-I00. ISCIII, Acción Estratégica en Salud, integrated in the Spanish National R+D+I Plan and financed by ISCIII Subdirección General de Evaluación and the Fondo Europeo de Desarrollo Regional (FEDER “Una manera de hacer Europa”) grants PI13/02434, PI16/01861, PI17/01474, PI19/00335, PI19/01240, PI19/01301, PI22/01403, PI22/00258 and the ISCIII national grant PMP22/00022, funded by the European Union (NextGenerationEU). The support of CIBERNED (ISCIII) under the grants CB06/05/2004 and CB18/05/00010. The support from the ADAPTED and MOPEAD projects, European Union/EFPIA Innovative Medicines Initiative Joint (grant numbers 115975 and 115985, respectively); from PREADAPT project, Joint Program for Neurodegenerative Diseases (JPND) grant N° AC19/00097; from HARPONE project, Agency for Innovation and Entrepreneurship (VLAIO) grant N° PR067/21 and Janssen. DESCARTES project is funded by German Research Foundation (DFG). Additionally, IdR is supported by the Instituto de Salud Carlos III (ISCIII) under grant FI20/00215. PGG is supported by CIBERNED employment plan (CNV-304-PRF-866). ACF received support from the ISCIII under the grant Sara Borrell (CD22/00125). JEC received support from National Institute of Health award P30AG066546.
Footnotes
Ethics approval and consent to participate
In accordance with Spanish regulations for the biomedical research field, all the protocols of this study were approved by the Clinical Research Ethics Commission of the Hospital Clinic (Barcelona, Spain) for Ace cohort and the Clinical Research Ethics Commission of Cantabria (Spain) for Valdecilla cohort. This research followed the Declaration of Helsinki. All participants were informed about the procedures and objectives of this study by a neurologist before signing an informed consent. Moreover, data confidentiality and privacy of patients were protected as specified in applicable laws.
Competing interests
All authors declare that the research was conducted in the absence of any conflict of interest.
Availability of data and materials
The data that support the findings of this study are publicly available from the corresponding authors upon reasonable request. Additionally, the raw SOMAscan proteomic data is publicly accessible through the Alzheimer’s Disease Data Initiative (ADDI) community.
References
- 1.El-Hayek YH, Wiley RE, Khoury CP, et al. Tip of the Iceberg: Assessing the Global Socioeconomic Costs of Alzheimer’s Disease and Related Dementias and Strategic Implications for Stakeholders. Journal of Alzheimer’s Disease. 2019;70:323–341. doi: 10.3233/JAD-190426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.2022 Alzheimer’s disease Facts and Figures: More Than Normal Aging: Understanding Mild Cognitive Impairment. Alzheimer’s & Dementia. 2022;18(4):700–789. [DOI] [PubMed] [Google Scholar]
- 3.Nandi A, Counts N, Chen S, et al. Global and regional projections of the economic burden of Alzheimer’s disease and related dementias from 2019 to 2050: A value of statistical life approach. EClinicalMedicine. 2022;51(101580):1–10. doi: 10.1016/j.eclinm.2022.101580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bellenguez C, Küçükali F, Jansen I, et al. New insights on the genetic etiology of Alzheimer’s and related dementia. Nat Genet. 2022;54:412–436. doi: 10.1038/s41588-022-01024-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Rojas I, Moreno-Grau S, Tesi N, et al. Common variants in Alzheimer’s disease and risk stratification by polygenic risk scores. Nat Commun. 2021;12(1):1–16. doi: 10.1038/s41467-021-22491-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ridge P, Adams PM, Albert MS, et al. Assessment of the genetic variance of late-onset Alzheimer’s disease. Neurobiol Aging. 2016;41:200.e13–200.e20. doi: 10.1016/j.neurobiolaging.2016.02.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Glahn DC, Bearden CE, Niendam TA, Escamilla MA. The feasibility of neuropsychological endophenotypes in the search for genes associated with bipolar affective disorder. Bipolar Disord. 2004;6(3):171–182. doi: 10.1111/j.1399-5618.2004.00113.x [DOI] [PubMed] [Google Scholar]
- 8.Zhang Q, Cai Z, Lhomme M, et al. Inclusion of endophenotypes in a standard GWAS facilitate a detailed mechanistic understanding of genetic elements that control blood lipid levels. Scientific Report Nature. 2020;10(18434):1–14. doi: 10.1038/s41598-020-75612-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jansen IE, van der Lee SJ, Gomez-Fonseca D, et al. Genome-wide meta-analysis for Alzheimer’s disease cerebrospinal fluid biomarkers. Acta Neuropathol. 2022;144(5):821–842. doi: 10.1007/S00401-022-02454-Z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gottesman II, Todd Gould HD, FRCPsych. The Endophenotype Concept in Psychiatry: Etymology and Strategic Intentions. American Journal of Psychiatry. 2003;160:637–645. doi: 10.1176/appi.ajp.160.4.636 [DOI] [PubMed] [Google Scholar]
- 11.Cruchaga C, Kauwe JS, Harari O, et al. GWAS of cerebrospinal fluid tau levels identifies novel risk variants for Alzheimer’s disease The Alzheimer Disease Genetic Consortium (ADGC), and. Neuron. 2013;78(2):256–268. doi: 10.1016/j.neuron.2013.02.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hall MH, Smoller JW. A New Role for Endophenotypes in the GWAS Era: Functional Characterization of Risk Variants. Harv Rev Psychiatry. 2010;18(1):67–74. doi: 10.3109/10673220903523532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deming Y, Li Z, Kapoor M, et al. Genome-wide association study identifies four novel loci associated with Alzheimer’s endophenotypes and disease modifiers. Acta Neuropathol. 2017;17(5):839–856. doi: 10.1007/s00401-017-1685-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kauwe JSK, Wang J, Mayo K, et al. Alzheimer’s disease risk variants show association with cerebrospinal fluid amyloid beta. Neurogenetics. 2009;10(1):13–17. doi: 10.1007/s10048-008-0150-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hardy J, Higgins G. Alzheimer’s disease: The amyloid cascade hypothesis. Science (1979). 1992;256(5054):184–185. doi: 10.1126/science.1566067 [DOI] [PubMed] [Google Scholar]
- 16.Hardy J, Selkoe D. The Amyloid Hypothesis of Alzheimer’s Disease: Progress and Problems on the Road to Therapeutics. Science (1979). 2002;297:353–356. [DOI] [PubMed] [Google Scholar]
- 17.Orellana A, García-gonzález P, Valero S, et al. Establishing In-House Cutoffs of CSF Alzheimer’s Disease Biomarkers for the AT(N) Stratification of the Alzheimer Center Barcelona Cohort. Int J Mol Sci. 2022;23(13):1–18. doi: 10.3390/IJMS23136891/S1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rodriguez-Gomez O, Sanabria A, Perez-Cordon A, et al. FACEHBI: A Prospective Study of Risk Factors, Biomarkers and Cognition in a Cohort of Individuals with Subjective Cognitive Decline. Study Rationale and Research Protocols. J Prev Alzheimers Dis. 2017;4(2):100–108. doi: 10.14283/jpad.2016.122 [DOI] [PubMed] [Google Scholar]
- 19.Moreno-Grau S, de Rojas I, Hernández I, et al. Genome-wide association analysis of dementia and its clinical endophenotypes reveal novel loci associated with Alzheimer’s disease and three causality networks: The GR@ACE project. Alzheimer’s and Dementia. 2019;15(10):1333–1347. doi: 10.1016/j.jalz.2019.06.4950 [DOI] [PubMed] [Google Scholar]
- 20.Jessen F, Amariglio RE, van Boxtel M, et al. A conceptual framework for research on subjective cognitive decline in preclinical Alzheimer’s disease. Alzheimer’s Dementia. 2014;10(6):844–852. doi: 10.1016/j.jalz.2014.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lopez OL, Jagust WJ, Dulberg C, et al. Risk factors for mild cognitive impairment in the cardiovascular health study cognition study. Arch Neurol. 2003;60(10):1394–1399. doi: 10.1001/archneur.60.10.1394 [DOI] [PubMed] [Google Scholar]
- 22.Petersen RC, Caracciolo B, Brayne C, Gauthier S, Jelic V, Fratiglioni L. Mild cognitive impairment: a concept in evolution. Journal of Intern Med. 2014;275(3):214–228. doi: 10.1111/joim.12190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Petersen RC, Smith GE, Waring SC, Ivnik RJ, Tangalos EG, Kokmen E. Mild Cognitive Impairment: Clinical Characterization and Outcome. American Medical Association. 1999;56:303–308. [DOI] [PubMed] [Google Scholar]
- 24.Jack CR, Bennett DA, Blennow K, et al. NIA-AA Research Framework: Toward a biological definition of Alzheimer’s disease. Alzheimer’s & Dementia. 2018;14(4):535–562. doi: 10.1016/j.jalz.2018.02.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vanderstichele H, Bibl M, Engelborghs S, et al. Standardization of preanalytical aspects of cerebrospinal fluid biomarker testing for Alzheimer’s disease diagnosis: A consensus paper from the Alzheimer’s Biomarkers Standardization Initiative. Alzheimer’s & Dementia. 2012;8(1):65–73. doi: 10.1016/j.jalz.2011.07.004 [DOI] [PubMed] [Google Scholar]
- 26.Buschke H. Cued recall in Amnesia. J Clin Neuropsychol. 1984;6(4):433–440. doi: 10.1080/01688638408401233 [DOI] [PubMed] [Google Scholar]
- 27.Alegret M, Valero S, Ortega G, et al. Validation of the Spanish Version of the Face Name Associative Memory Exam (S-FNAME) in Cognitively Normal Older Individuals. Arch Clin Neuropsychol. 2015;30(7):712–720. doi: 10.1093/ARCLIN/ACV050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wechsler D. A Standardized Memory Scale for Clinical Use. The Journal of Psychology: Interdisciplinary and Applied. 1945;19(1):87–95. doi: 10.1080/00223980.1945.9917223 [DOI] [Google Scholar]
- 29.Sheikh JI, Yesavage JA. Geriatric Depression Scale (GDS): Recent evidence and development of a shorter version. Clinical Gerontologist: The Journal of Aging and Mental Health. 1986;5(1–2):165–173. doi: 10.1300/J018V05N01_09 [DOI] [Google Scholar]
- 30.Morris JC. The Clinical Dementia Rating (CDR). Neurology. 1993;43(11):2412–2414. doi: 10.1212/wnl.43.11.2412-a [DOI] [PubMed] [Google Scholar]
- 31.Alcolea D, Pegueroles J, Muñoz L, et al. Agreement of amyloid PET and CSF biomarkers for Alzheimer’s disease on Lumipulse. Ann Clin Transl Neurol. 2019;6(9):1815–1824. doi: 10.1002/acn3.50873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.López-García S, Lage C, Pozueta A, et al. Sleep Time Estimated by an Actigraphy Watch Correlates With CSF Tau in Cognitively Unimpaired Elders: The Modulatory Role of APOE. Front Aging Neurosci. 2021;13:1–9. doi: 10.3389/FNAGI.2021.663446/FULL [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mueller SG, Weiner MW, Thal LJ, et al. Ways toward an early diagnosis in Alzheimer’s disease: The Alzheimer’s Disease Neuroimaging Initiative (ADNI). Alzheimer’s & Dementia. 2005;1(1):55–66. doi: 10.1016/j.jalz.2005.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.ADNI | Alzheimer’s Disease Neuroimaging Initiative. https://adni.loni.usc.edu/
- 35.Petersen RC, Aisen PS, Beckett LA, et al. Alzheimer’s Disease Neuroimaging Initiative (ADNI): Clinical characterization. Neurology. 2010;74(3):201–209. doi: 10.1212/WNL.0b013e3181cb3e25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Landau SM, Lu M, Joshi AD, et al. Comparing positron emission tomography imaging and cerebrospinal fluid measurements of β-amyloid. Ann Neurol. 2013;74(6):826–836. doi: 10.1002/ana.23908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shaw LM, Vanderstichele H, Knapik-Czajka M, et al. Cerebrospinal fluid biomarker signature in alzheimer’s disease neuroimaging initiative subjects. Ann Neurol. 2009;65(4):403–413. doi: 10.1002/ana.21610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.PET Acquisition Methods ADNI. Accessed May 29, 2023. https://adni.loni.usc.edu/methods/pet-analysis-method/pet-analysis/
- 39.Rowe CC, Doré V, Jones G, et al. 18F-Florbetaben PET beta-amyloid binding expressed in Centiloids. Eur J Nucl Med Mol Imaging. 2017;44(12):2053–2059. doi: 10.1007/S00259-017-3749-6/FIGURES/4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Klunk WE, Koeppe RA, Price JC, et al. The Centiloid Project: Standardizing quantitative amyloid plaque estimation by PET. Alzheimer’s & Dementia. 2015;11(1):1–15. doi: 10.1016/J.JALZ.2014.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saykin AJ, Shen L, Foroud TM, et al. Alzheimer’s Disease Neuroimaging Initiative biomarkers as quantitative phenotypes: Genetics core aims, progress, and plans. Alzheimers Dement. 2010;6(3):265–273. doi: 10.1016/j.jalz.2010.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Michigan Imputation Server. Accessed June 9, 2023. https://imputationserver.sph.umich.edu/index.html#!
- 43.Gold L, Ayers D, Bertino J, et al. Aptamer-Based Multiplexed Proteomic Technology for Biomarker Discovery. Gelain F, ed. PLoS One. 2010;5(12):e15004. doi: 10.1371/journal.pone.0015004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Candia J, Cheung F, Kotliarov Y, et al. Assessment of Variability in the SOMAscan Assay. Scientific Reports 2017 7:1. 2017;7(1):1–13. doi: 10.1038/s41598-017-14755-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chang CC, Chow CC, Tellier LCAM, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: Rising to the challenge of larger and richer datasets. Gigascience. 2015;4(1):1–16. doi: 10.1186/S13742-015-0047-8/2707533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Willer CJ, Li Y, Abecasis GR. METAL: Fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26(17):2190–2191. doi: 10.1093/bioinformatics/btq340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stouffer SA, Suchman EA, DeVinney LC, Star SA, Williams RM. American soldier: Adjustment during army life (Vol. 1). Princeton University Press. 1949;28(1):87–90. doi: 10.2307/2572105 [DOI] [Google Scholar]
- 48.Ali M, Archer DB, Gorijala P, et al. Large multi-ethnic genetic analyses of amyloid imaging identify new genes for Alzheimer disease. Acta Neuropathol Commun. 2023;11(1):1–20. doi: 10.1186/S40478-023-01563-4/FIGURES/4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lift Genome Annotations. Accessed November 20, 2023. https://genome.ucsc.edu/cgi-bin/hgLiftOver
- 50.Beecham GW, Hamilton K, Naj AC, et al. Genome-Wide Association Meta-analysis of Neuropathologic Features of Alzheimer’s Disease and Related Dementias. PLoS Genet. 2014;10(9):e1004606. doi: 10.1371/JOURNAL.PGEN.1004606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Purcell S, Neale B, Todd-Brown K, et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. doi: 10.1086/519795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Watanabe K, Taskesen E, Van Bochoven A, Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat Commun. 2017;8(1):1–11. doi: 10.1038/s41467-017-01261-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Higgins JPT, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327(7414):557–560. doi: 10.1136/BMJ.327.7414.557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhu Z, Zhang F, Hu H, et al. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat Genet. 2016;48(5):481–487. doi: 10.1038/ng.3538 [DOI] [PubMed] [Google Scholar]
- 55.Puerta R, Cano A, García-González P, et al. Head-to-head comparison of aptamer- and antibody-based proteomic platforms in human cerebrospinal fluid samples from a real-world memory clinic cohort. medRxiv. Published online July 18, 2024:2024.07.18.24310563. doi: 10.1101/2024.07.18.24310563 [DOI] [PubMed] [Google Scholar]
- 56.Frick EA, Emilsson V, Jonmundsson T, et al. Serum proteomics reveal APOE-ε4-dependent and APOE-ε4-independent protein signatures in Alzheimer’s disease. Nature Aging 2024. Published online August 21, 2024:1–19. doi: 10.1038/s43587-024-00693-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liao Y, Wang J, Jaehnig EJ, Shi Z, Zhang B. WebGestalt 2019: gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res. 2019;47(W1):W199–W205. doi: 10.1093/NAR/GKZ401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shen X, Zhang S, Guo Z, Xing D, Chen W. The crosstalk of ABCA1 and ANXA1: A potential mechanism for protection against atherosclerosis. Molecular Medicine. 2020;26(1):1–8. doi: 10.1186/S10020-020-00213-Y/FIGURES/1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhao B, Luo T, Li T, et al. Genome-wide association analysis of 19,629 individuals identifies variants influencing regional brain volumes and refines their genetic co-architecture with cognitive and mental health traits. Nat Genet. 2019;51(11):1637–1644. doi: 10.1038/S41588-019-0516-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Suhre K, Arnold M, Bhagwat AM, et al. Connecting genetic risk to disease end points through the human blood plasma proteome. Nat Commun. 2017;8:1–14. doi: 10.1038/NCOMMS14357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ries M, Loiola R, Shah UN, Gentleman SM, Solito E, Sastre M. The anti-inflammatory Annexin A1 induces the clearance and degradation of the amyloid-β peptide. J Neuroinflammation. 2016;13(234):1–15. doi: 10.1186/S12974-016-0692-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kircher M, Witten DM, Jain P, et al. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46(3):310–315. doi: 10.1038/ng.2892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Brouwer RM, Klein M, Grasby KL, et al. Genetic variants associated with longitudinal changes in brain structure across the lifespan. Nature Neuroscience 2022 25:4. 2022;25(4):421–432. doi: 10.1038/s41593-022-01042-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Homann J, Osburg T, Ohlei O, et al. Genome-Wide Association Study of Alzheimer’s Disease Brain Imaging Biomarkers and Neuropsychological Phenotypes in the European Medical Information Framework for Alzheimer’s Disease Multimodal Biomarker Discovery Dataset. Front Aging Neurosci. 2022;14:1–14. doi: 10.3389/FNAGI.2022.840651/BIBTEX [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Furney SJ, Simmons A, Breen G, et al. Genome-wide association with MRI atrophy measures as a quantitative trait locus for Alzheimer’s disease. Mol Psychiatry. 2010;16(11):1130–1138. doi: 10.1038/mp.2010.123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gouveia C, Gibbons E, Dehghani N, Eapen J, Guerreiro R, Bras J. Genome-wide association of polygenic risk extremes for Alzheimer’s disease in the UK Biobank. Sci Rep. 2022;12:1–12. doi: 10.1038/S41598-022-12391-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hong S, Prokopenko D, Dobricic V, et al. Genome-wide association study of Alzheimer’s disease CSF biomarkers in the EMIF-AD Multimodal Biomarker Discovery dataset. Translational Psychiatry 2020 10 :1. 2020;10(1):1–12. doi: 10.1038/s41398-020-01074-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chibnik LB, White CC, Mukherjee S, et al. Susceptibility to neurofibrillary tangles: role of the PTPRD locus and limited pleiotropy with other neuropathologies. Mol Psychiatry. 2018;23(6):1521–1529. doi: 10.1038/MP.2017.20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Raghavan NS, Dumitrescu L, Mormino E, et al. Association Between Common Variants in RBFOX1, an RNA-Binding Protein, and Brain Amyloidosis in Early and Preclinical Alzheimer Disease. JAMA Neurol. 2020;77(10):1288–1298. doi: 10.1001/JAMANEUROL.2020.1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kunkle BW, Grenier-Boley B, Sims R, et al. Genetic meta-analysis of diagnosed Alzheimer’s disease identifies new risk loci and implicates Aβ, tau, immunity and lipid processing. Nat Genet. 2019;51:414–430. doi: 10.1038/s41588-019-0358-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bigdeli TB, Fanous AH, Li Y, et al. Genome-Wide Association Studies of Schizophrenia and Bipolar Disorder in a Diverse Cohort of US Veterans. Schizophr Bull. 2021;47(2):517–529. doi: 10.1093/SCHBUL/SBAA133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Luciano M, Hagenaars SP, Davies G, et al. Association analysis in over 329,000 individuals identifies 116 independent variants influencing neuroticism. Nat Genet. 2018;50(1):6–11. doi: 10.1038/S41588-017-0013-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sherva R, Gross A, Mukherjee S, et al. Genome-wide association study of rate of cognitive decline in Alzheimer’s disease patients identifies novel genes and pathways. Alzheimers Dement. 2020;16(8):1134–1145. doi: 10.1002/ALZ.12106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lee E, Giovanello KS, Saykin AJ, et al. Single-nucleotide polymorphisms are associated with cognitive decline at Alzheimer’s disease conversion within mild cognitive impairment patients. Alzheimer’s & Dementia: Diagnosis, Assessment & Disease Monitoring. 2017;8(1):86–95. doi: 10.1016/J.DADM.2017.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ramanan VK, Lesnick TG, Przybelski SA, et al. Coping with brain amyloid: genetic heterogeneity and cognitive resilience to Alzheimer’s pathophysiology. Acta Neuropathol Commun. 2021;9(1):1–14. doi: 10.1186/S40478-021-01154-1/FIGURES/4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Leuba G, Vernay A, Vu D, et al. Differential expression of LMO4 protein in Alzheimer’s disease. Neuropathol Appl Neurobiol. 2004;30(1):57–69. doi: 10.1046/J.0305-1846.2003.00511.X [DOI] [PubMed] [Google Scholar]
- 77.Bernardo A, Harrison FE, McCord M, et al. Elimination of GD3 synthase improves memory and reduces amyloid-β plaque load in transgenic mice. Neurobiol Aging. 2009;30(11):1777–1791. doi: 10.1016/J.NEUROBIOLAGING.2007.12.022 [DOI] [PubMed] [Google Scholar]
- 78.Hamano M, Ichinose T, Yasuda T, et al. Bioinformatics Analysis of the Molecular Networks Associated with the Amelioration of Aberrant Gene Expression by a Tyr–Trp Dipeptide in Brains Treated with the Amyloid-β Peptide. Nutrients. 2023;15(12):2731. doi: 10.3390/NU15122731/S1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Galiano MR, Goitea VE, Hallak ME. Post-translational protein arginylation in the normal nervous system and in neurodegeneration. J Neurochem. 2016;138(4):506–517. doi: 10.1111/JNC.13708 [DOI] [PubMed] [Google Scholar]
- 80.Wesenhagen KEJ, Gobom J, Bos I, et al. Effects of age, amyloid, sex, and APOE ε4 on the CSF proteome in normal cognition. Alzheimer’s & Dementia: Diagnosis, Assessment & Disease Monitoring. 2022;14(1):e12286. doi: 10.1002/DAD2.12286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Do AN, Ali M, Timsina J, et al. CSF proteomic profiling with amyloid/tau positivity identifies distinctive sex-different alteration of multiple proteins involved in Alzheimer’s disease. medRxiv. Published online March 16, 2024:2024.03.15.24304164. doi: 10.1101/2024.03.15.24304164 [DOI] [Google Scholar]
- 82.Lambert JC, Ibrahim-Verbaas CA, Harold D, et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nature Genetics 2013 45:12. 2013;45(12):1452–1458. doi: 10.1038/ng.2802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lambert JC, Heath S, Even G, et al. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer’s disease. Nat Genet. 2009;41(10):1094–1100. doi: 10.1038/ng.439 [DOI] [PubMed] [Google Scholar]
- 84.Harold D, Abraham R, Hollingworth P, et al. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer’s disease. Nat Genet. 2009;41(10):1088–1093. doi: 10.1038/ng.440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Corder EH, Saunders AM, Gaskell PC, et al. Gene Dose of Apolipoprotein E Type 4 Allele and the Risk of Alzheimer’s Disease in Late Onset Families. Science (1979). 1993;261:921–923. doi: 10.1126/science.8346443 [DOI] [PubMed] [Google Scholar]
- 86.Chapuis J, Flaig A, Grenier-Boley B, et al. Genome-wide, high-content siRNA screening identifies the Alzheimer’s genetic risk factor FERMT2 as a major modulator of APP metabolism. Acta Neuropathol. 2017;133(6):955–966. doi: 10.1007/S00401-016-1652-Z/FIGURES/5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Calero M, Rostagno A, Matsubara E, Zlokovic B, Frangione B, Ghiso J. Apolipoprotein J (Clusterin) and Alzheimer’s Disease. Microsc Res Tech. 2000;50:305–315. [DOI] [PubMed] [Google Scholar]
- 88.Harper JD, Fan KH, Aslam MM, et al. Genome-Wide Association Study of Incident Dementia in a Community-Based Sample of Older Subjects. Journal of Alzheimer’s Disease. 2022;88(2):787–798. doi: 10.3233/JAD-220293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Goudet C, Magnaghi V, Landry M, Nagy F, Gereau IV RW, Pin JP. Metabotropic receptors for glutamate and GABA in pain. Brain Res Rev. 2009;60(1):43–56. doi: 10.1016/J.BRAINRESREV.2008.12.007 [DOI] [PubMed] [Google Scholar]
- 90.Chen CH, Lee CS, Lee MTM, et al. Variant GADL1 and Response to Lithium Therapy in Bipolar I Disorder. New England Journal of Medicine. 2014;370(2):119–128. doi: 10.1056/NEJMOA1212444 [DOI] [PubMed] [Google Scholar]
- 91.Li S, Mallory M, Alford M, Tanaka S, Masliah E. Glutamate Transporter Alterations in Alzheimer Disease Are Possibly Associated with Abnormal APP Expression. J Neuropathol Exp Neurol. 1997;56(8):901–911. doi: 10.1097/00005072-199708000-00008 [DOI] [PubMed] [Google Scholar]
- 92.Molinuevo JL, Lladó A, Rami L. Memantine: Targeting glutamate excitotoxicity in Alzheimer’s disease and other dementias. http://dx.doi.org/101177/153331750502000206. 2005;20(2):77–85. doi: 10.1177/153331750502000206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Domschke K, Hohoff C, Jacob C, et al. Chromosome 4q31–34 panic disorder risk locus: Association of neuropeptide Y Y5 receptor variants. American Journal of Medical Genetics Part B: Neuropsychiatric Genetics. 2008;147B(4):510–516. doi: 10.1002/AJMG.B.30629 [DOI] [PubMed] [Google Scholar]
- 94.Otowa T, Kawamura Y, Nishida N, et al. Meta-analysis of genome-wide association studies for panic disorder in the Japanese population. Transl Psychiatry. 2012;2(11):1–8. doi: 10.1038/TP.2012.89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Strozyk D, Blennow K, White LR, Launer LJ. CSF Aß 42 levels correlate with amyloid-neuropathology in a population-based autopsy study. Neurology. 2003;60(4):652–656. doi: 10.1212/01.WNL.0000046581.81650.D0 [DOI] [PubMed] [Google Scholar]
- 96.Sabri O, Sabbagh MN, Seibyl J, et al. Florbetaben PET imaging to detect amyloid beta plaques in Alzheimer’s disease: Phase 3 study. Alzheimer’s & Dementia. 2015;11(8):964–974. doi: 10.1016/J.JALZ.2015.02.004 [DOI] [PubMed] [Google Scholar]
- 97.Remnestål J, Öijerstedt L, Ullgren A, et al. Altered levels of CSF proteins in patients with FTD, presymptomatic mutation carriers and non-carriers. Transl Neurodegener. 2020;9(1):1–13. doi: 10.1186/S40035-020-00198-Y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Erhardt A, Akula N, Schumacher J, et al. Replication and meta-analysis of TMEM132D gene variants in panic disorder. Transl Psychiatry. 2012;2(9):1–6. doi: 10.1038/TP.2012.85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Haaker J, Lonsdorf TB, Raczka KA, Mechias ML, Gartmann N, Kalisch R. Higher anxiety and larger amygdala volumes in carriers of a TMEM132D risk variant for panic disorder. Transl Psychiatry. 2014;4(2):1–2. doi: 10.1038/TP.2014.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Trubetskoy V, Pardiñas AF, Qi T, et al. Mapping genomic loci implicates genes and synaptic biology in schizophrenia. Nature 2022 604:7906. 2022;604(7906):502–508. doi: 10.1038/s41586-022-04434-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Mattsson N, Insel PS, Donohue M, et al. Independent information from cerebrospinal fluid amyloid-β and florbetapir imaging in Alzheimer’s disease. Brain. 2015;138(3):772–783. doi: 10.1093/BRAIN/AWU367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Blennow K, Mattsson N, Schöll M, Hansson O, Zetterberg H. Amyloid biomarkers in Alzheimer’s disease. Trends Pharmacol Sci. 2015;36(5):297–309. doi: 10.1016/J.TIPS.2015.03.002 [DOI] [PubMed] [Google Scholar]
- 103.Reimand J, Boon BDC, Collij LE, et al. Amyloid-β PET and CSF in an autopsy-confirmed cohort. Ann Clin Transl Neurol. 2020;7(11):2150–2160. doi: 10.1002/ACN3.51195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kumar A, Janelidze S, Stomrud E, Palmqvist S, Hansson O, Mattsson-Carlgren N. β-Amyloid-Dependent and -Independent Genetic Pathways Regulating CSF Tau Biomarkers in Alzheimer Disease. Neurology. 2022;99(5):E476–E487. doi: 10.1212/WNL.0000000000200605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Jansen IE, Savage JE, Watanabe K, et al. Genome-wide meta-analysis identifies new loci and functional pathways influencing Alzheimer’s disease risk. Nat Genet. 2019;51(3):404–413. doi: 10.1038/S41588-018-0311-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hong S, Prokopenko D, Dobricic V, et al. Genome-wide association study of Alzheimer’s disease CSF biomarkers in the EMIF-AD Multimodal Biomarker Discovery dataset. Transl Psychiatry. 17:41. doi: 10.1038/s41398-020-01074-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bader JM, Geyer PE, Müller JB, et al. Proteome profiling in cerebrospinal fluid reveals novel biomarkers of Alzheimer’s disease. Mol Syst Biol. 2020;16(6):e9356. doi: 10.15252/MSB.20199356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hyman BT. Amyloid-Dependent and Amyloid-Independent Stages of Alzheimer Disease. Arch Neurol. 2011;68(8):1062–1064. doi: 10.1001/ARCHNEUROL.2011.70 [DOI] [PubMed] [Google Scholar]
- 109.Leonenko G, Shoai M, Bellou E, et al. Genetic risk for alzheimer disease is distinct from genetic risk for amyloid deposition. Ann Neurol. 2019;86(3):427–435. doi: 10.1002/ANA.25530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Asada-Utsugi M, Uemura K, Noda Y, et al. N-cadherin enhances APP dimerization at the extracellular domain and modulates Aβ production. J Neurochem. 2011;119(2):354–363. doi: 10.1111/J.1471-4159.2011.07364.X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Terry RD, Masliah E, Salmon DP, et al. Physical basis of cognitive alterations in Alzheimer’s disease: synapse loss is the major correlate of cognitive impairment. Ann Neurol. 1991;30(4):572–580. doi: 10.1002/ANA.410300410 [DOI] [PubMed] [Google Scholar]
- 112.Uemura T. The cadherin superfamily at the synapse: More members, more missions. Cell. 1998;93(7):1095–1098. doi: 10.1016/S0092-8674(00)81452-X [DOI] [PubMed] [Google Scholar]
- 113.Wu X, Reddy DS. Integrins as receptor targets for neurological disorders. Pharmacol Ther. 2012;134(1):68–81. doi: 10.1016/J.PHARMTHERA.2011.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Bensamoun D, Guignard R, Furst AJ, et al. Associations between Neuropsychiatric Symptoms and Cerebral Amyloid Deposition in Cognitively Impaired Elderly People. Journal of Alzheimer’s Disease. 2016;49(2):387–398. doi: 10.3233/JAD-150181 [DOI] [PubMed] [Google Scholar]
- 115.Kuo HC, Yen HC, Huang CC, Hsu WC, Wei HJ, Lin CL. Cerebrospinal fluid biomarkers for neuropsychological symptoms in early stage of late-onset Alzheimer’s disease. http://dx.doi.org/103109/002074542014971787. 2014;125(10):747–754. doi: 10.3109/00207454.2014.971787 [DOI] [PubMed] [Google Scholar]
- 116.Cai WJ, Tian Y, Ma YH, Dong Q, Tan L, Yu JT. Associations of Anxiety with Amyloid, Tau, and Neurodegeneration in Older Adults without Dementia: A Longitudinal Study. Journal of Alzheimer’s Disease. 2021;82(1):273–283. doi: 10.3233/jad-210020 [DOI] [PubMed] [Google Scholar]
- 117.Lahiri DK, Maloney B, Wang R, Sokol DK, Rogers JT, Westmark CJ. How autism and Alzheimer’s disease are TrAPPed. Molecular Psychiatry 2020 26:1. 2020;26(1):26–29. doi: 10.1038/s41380-020-00928-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Liu F, Arias-Vásquez A, Sleegers K, et al. A genomewide screen for late-onset Alzheimer disease in a genetically isolated Dutch population. Am J Hum Genet. 2007;81(1):17–31. doi: 10.1086/518720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Li Z, Liu L, Lin W, et al. NRG3 contributes to cognitive deficits in chronic patients with schizophrenia. Schizophr Res. 2020;215:134–139. doi: 10.1016/J.SCHRES.2019.10.060 [DOI] [PubMed] [Google Scholar]
- 120.Hollingworth P, Sweet R, Sims R, et al. Genome-wide association study of Alzheimer’s disease with psychotic symptoms. Molecular Psychiatry 2012 17:12. 2011;17(12):1316–1327. doi: 10.1038/mp.2011.125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Wang KS, Xu N, Wang L, et al. NRG3 gene is associated with the risk and age at onset of Alzheimer disease. J Neural Transm. 2014;121(2):183–192. doi: 10.1007/S00702-013-1091-0 [DOI] [PubMed] [Google Scholar]
- 122.Morar B, Dragović M, Waters FAV, Chandler D, Kalaydjieva L, Jablensky A. Neuregulin 3 (NRG3) as a susceptibility gene in a schizophrenia subtype with florid delusions and relatively spared cognition. Molecular Psychiatry 2011 16:8. 2010;16(8):860–866. doi: 10.1038/mp.2010.70 [DOI] [PubMed] [Google Scholar]
- 123.Zhang T, Zhu L, Ni T, et al. Voltage-gated calcium channel activity and complex related genes and schizophrenia: A systematic investigation based on Han Chinese population. J Psychiatr Res. 2018;106:99–105. doi: 10.1016/J.JPSYCHIRES.2018.09.020 [DOI] [PubMed] [Google Scholar]
- 124.Potkin SG, Turner JA, Guffanti G, et al. A Genome-Wide Association Study of Schizophrenia Using Brain Activation as a Quantitative Phenotype. Schizophr Bull. 2009;35(1):96–108. doi: 10.1093/SCHBUL/SBN155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Suda S, Iwata K, Shimmura C, et al. Decreased expression of axon-guidance receptors in the anterior cingulate cortex in autism. Mol Autism. 2011;2(1):1–5. doi: 10.1186/2040-2392-2-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Zuko A, Kleijer KTE, Oguro-Ando A, et al. Contactins in the neurobiology of autism. Eur J Pharmacol. 2013;719(1–3):63–74. doi: 10.1016/J.EJPHAR.2013.07.016 [DOI] [PubMed] [Google Scholar]
- 127.Tosto G, Fu H, Vardarajan BN, et al. F-box/LRR-repeat protein 7 is genetically associated with Alzheimer’s disease. Ann Clin Transl Neurol. 2015;2(8):810–820. doi: 10.1002/ACN3.223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Dauar MT, Labonté A, Picard C, et al. Characterization of the contactin 5 protein and its risk-associated polymorphic variant throughout the Alzheimer’s disease spectrum. Alzheimer’s & Dementia. Published online 2022:1–15. doi: 10.1002/ALZ.12868 [DOI] [PubMed] [Google Scholar]
- 129.Cano A, Esteban-de-Antonio E, Bernuz M, et al. Plasma extracellular vesicles reveal early molecular differences in amyloid positive patients with early-onset mild cognitive impairment. J Nanobiotechnology. 2023;21(1):1–19. doi: 10.1186/S12951-023-01793-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Van Der Meer D, Kaufmann T, Shadrin AA, et al. The genetic architecture of human cortical folding. Sci Adv. 2021;7(51):9446. doi: 10.1126/SCIADV.ABJ9446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Borenstein M, Hedges L V., Higgins JPT, Rothstein HR. Introduction to meta-analysis. Introduction to Meta-Analysis. Published online January 1, 2009:1–421. doi: 10.1002/9780470743386 [DOI] [Google Scholar]
- 132.Yoon S, Baik B, Park T, Nam D. Powerful p-value combination methods to detect incomplete association. Scientific Reports 2021 11:1. 2021;11(1):1–11. doi: 10.1038/s41598-021-86465-y [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are publicly available from the corresponding authors upon reasonable request. Additionally, the raw SOMAscan proteomic data is publicly accessible through the Alzheimer’s Disease Data Initiative (ADDI) community.