Abstract
Introduction
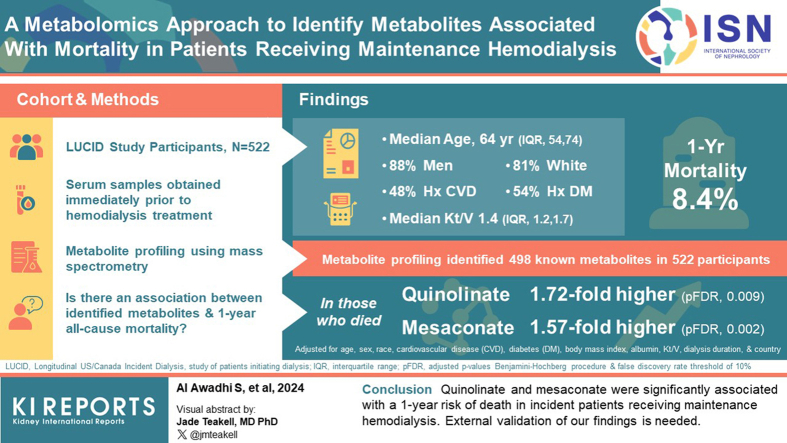
Uremic toxins contributing to increased risk of death remain largely unknown. We used untargeted metabolomics to identify plasma metabolites associated with mortality in patients receiving maintenance hemodialysis.
Methods
We measured metabolites in serum samples from 522 Longitudinal US/Canada Incident Dialysis (LUCID) study participants. We assessed the association between metabolites and 1-year mortality, adjusting for age, sex, race, cardiovascular disease, diabetes, body mass index, serum albumin, Kt/Vurea, dialysis duration, and country. We modeled these associations using limma, a metabolite-wise linear model with empirical Bayesian inference, and 2 machine learning (ML) models: Least absolute shrinkage and selection operator (LASSO) and random forest (RF). We accounted for multiple testing using a false discovery rate (pFDR) adjustment. We defined significant mortality-metabolite associations as pFDR < 0.1 in the limma model and metabolites of at least medium importance in both ML models.
Results
The mean age of the participants was 64 years, the mean dialysis duration was 35 days, and there were 44 deaths (8.4%) during a 1-year follow-up period. Two metabolites were significantly associated with 1-year mortality. Quinolinate levels (a kynurenine pathway metabolite) were 1.72-fold higher in patients who died within year 1 compared with those who did not (pFDR, 0.009), wheras mesaconate levels (an emerging immunometabolite) were 1.57-fold higher (pFDR, 0.002). An additional 42 metabolites had high importance as per LASSO, 46 per RF, and 9 per both ML models but were not significant per limma.
Conclusion
Quinolinate and mesaconate were significantly associated with a 1-year risk of death in incident patients receiving maintenance hemodialysis. External validation of our findings is needed.
Keywords: artificial intelligence, hemodialysis, metabolomics, mortality
Graphical abstract
The prevalence of patients with kidney failure treated with dialysis is increasing worldwide, with the majority treated with hemodialysis.1 Patients who undergo hemodialysis are at substantial mortality risk, with a 20% mortality rate in the first year of dialysis and a staggering 60% mortality rate over 5 years.2 Currently, the main causes of mortality in patients undergoing hemodialysis are cardiovascular (53%), infections (18%), and withdrawal from dialysis (16%).2 Despite notable advances in dialysis treatment technologies and strategies to manage kidney failure related conditions such as anemia and bone mineral disorder, there have been limited improvements in survival for dialysis patients.2 The factors contributing to this persistently high mortality are complex, multidimensional and remain unkown.3, 4, 5 Among these factors, the accumulation of uremic toxins, solutes that accumulate in the body due to impaired kidney function, may play an important role.6, 7, 8 However, the identification of specific uremic toxins that impact mortality in patients receiving maintenance hemodialysis has not yet been systematically examined.
Metabolomics enables extensive profiling of small molecules in biological specimens, providing an unbiased snapshot of the metabolic landscape for an individual.9,10 Metabolomics has been applied to identify markers associated with chronic kidney disease (CKD) and CKD progression,11, 12, 13, 14 as well as to describe the uremic milieu and the effect of the hemodialysis procedure.15, 16, 17, 18 Two previous studies examined the association between serum metabolites and subsequent mortality in patients with kidney failure, however, they only focused on cardiovascular mortality.19,20
Our study aimed to determine the association between plasma metabolite levels measured using a well-established liquid chromatography-mass spectrometry-based metabolomics platform and 1-year all-cause mortality in patients undergoing hemodialysis. We used stored samples from 522 incident patients with kidney failure treated with hemodialysis in the United States and Canada participating in the LUCID and evaluated the association between 498 plasma metabolites and 1-year mortality using state-of-the-art high-dimensional prognostic data methods.
Methods
Study Population
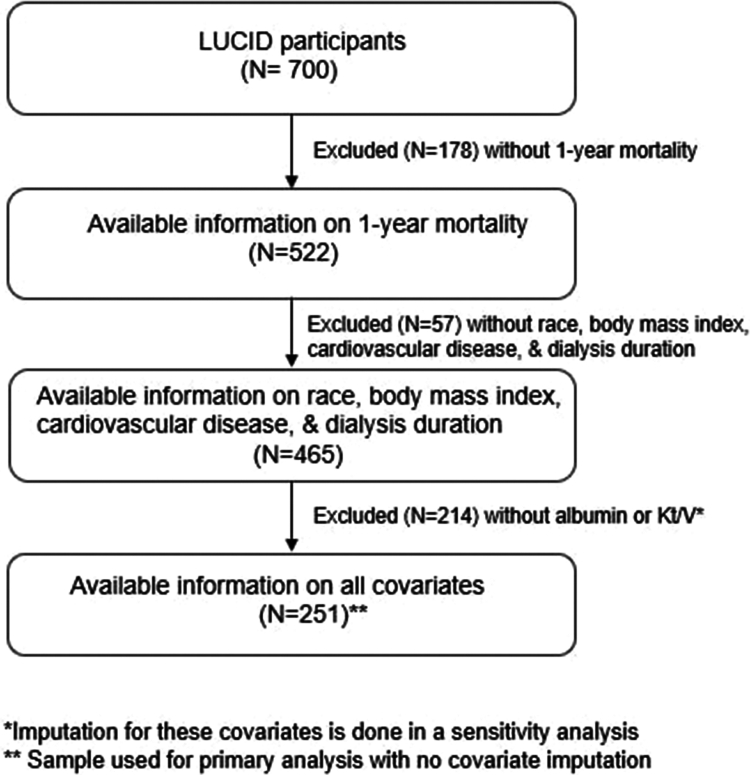
We used registry data from the LUCID study, which was a prospective, multicenter observational study of patients initiating hemodialysis and peritoneal dialysis in the United States and Canada. A total of 823participants were enrolled in 3 centers in the US (New England, Washington, and Indiana) and Canada between May 2011 and December 2017.21 Eligibility criteria were initiation of maintenance dialysis within 6 months, age >18 years, and ability to provide informed consent. For this study, we profiled a subset of 700 participants and analyzed 522 patients on maintenance hemodialysis (3 times per week) who had available stored samples and information on all-cause mortality at one year after dialysis initiation (Figure 1).
Figure 1.
Consort diagram of evaluated patients. LUCID, Longitudinal US/Canada Incident Dialysis.
The LUCID study was approved by the human subjects’ review boards of the participating institutions and all participants provided informed consent.
Samples
Samples from the LUCID participants were obtained immediately predialysis at each individual’s outpatient hemodialysis facility at the time of enrollment. Date of the samples was only available for the US participants, and 58% of samples were collected after a long intradialytic interval (Monday or Tuesday). The samples were collected in red and lavender top- tubes and centrifuged at 3000 revolutions per minute for 10 minutes. They were aliquoted on the same day and then stored at −80 °C at each site. Aliquoted samples were shipped to the Broad Institute on dry ice for analysis.
Metabolomics
Detailed methods, including characterization of technical and intraindividual analyte variation among individuals with CKD for the Broad Institute Metabolomics Platform have been published.22 In brief, 3 liquid chromatography-tandem mass spectrometry (LC-MS) injections were used to profile blood samples. The LC-MS systems consist of Q Exactive/Exactive Plus orbitrap mass spectrometers (Themo Fisher Scientific) and Nexera X2 U-HPLC systems (Shimadzu Scientific Instruments). Measurements of positively charged polar metabolites were performed in samples extracted with acetonitrile/methanol/formic acid and separated by hydrophilic interaction LC (HILIC). Positive ion mode electrospray ionization (ESI) was used for mass spectrometry (MS) analyses. Measurements of positively charged lipids were performed in samples extracted with isopropanol and separated on a C8 column. Similarly, positive ion mode electrospray ionization (ESI) was used for MS analyses. Measurements of negatively charged polar metabolites were performed in samples extracted with methanol and separated by an NH2 column. Negative ion mode ESI was used for MS analyses. Raw metabolomics data were processed using TraceFinder (Thermo Fisher Scientific). Metabolite peaks were identified by matching observed retention times and masses to reference metabolites tested in each batch, and to an internal database of previously identified compounds.12
The analytical performance of the LC-MS systems and the quality of metabolomics data were assured using several strategies. A mixture of synthetic reference standards was analyzed periodically during the analysis queue to assure reproducibility of chromatographic retention times, quality of peak shapes, and the sensitivity of the MS system. Further, internal standard signals were monitored in each sample to ensure that each sample was injected properly and to monitor MS sensitivity. Last, reference samples were inserted in the analysis queue after sets of 20 study samples to determine the reproducibility for all metabolites over the run.
Metabolite profiling identified 507 known metabolites in 522 participants. Metabolites below the level of detection in 95% of samples (n = 9) were excluded, leaving 498 metabolites for evaluation.
Statistical Analysis
We summarized participant characteristics at baseline using medians and interquartile ranges for continuous variables and frequencies for categorical variables. We used STROBE (Strengthening the reporting of observational studies in epidemiology) guidelines for reporting (Supplementary Table S8).23
Metabolite Data Processing
Two key elements of processing high dimensional data are addressing the variability and missingness in the metabolomics data. The potential sources of variability can be preanalytical, for example, due to sample collection, processing, storage, and transportation methods. The variability can also be analytical (instrument drift), induced by the technique used and reflected by retention time and signal intensity drifts.24,25 To assess instrument drift, we evaluated correlations between the quality control samples and performed principal components analysis (PCA) on the metabolite abundances (Supplementary Methods S1, Supplementary Table S1, and Supplementary Figure S1). We then adjusted for instrument drift using the removal of unwanted variation (RUV) method (Supplementary Methods and Supplementary Figure S2).26 In sensitivity analyses we explored the impact of eliminating the RUV correction.
The missing data in metabolomics studies are often because of limit of detection issues (a type of left censoring), which we addressed by using the quantile regression imputation of left-censored data (QRILC) method.27 We evaluated the quality of metabolomics imputation by first manually left censoring the metabolomics data, dropping the 20 lowest abundances for metabolites that did not already have a high rate of missingness (<60% missing values) and then using root mean squared error to quantify imputation error (See Supplementary Methods and Supplementary Figure S3). We tested additional imputation methods such as half-minimum (uniformly distributed random values from 0 to half of the minimum observed abundance) and k-nearest neighbors’ methods.28 QRILC showed the lowest imputation error of all methods and was used for the subsequent analysis.
Covariates
A priori, we determined covariates to be included in adjusted models based on biological plausibility, and included age, sex, race, cardiovascular disease, diabetes, body mass index (BMI), albumin, Kt/Vurea (K represents urea clearance by the dialyzer, t represents treatment time, and V represents urea distribution volume), dialysis duration, and country (USA and Canada). We excluded 57 patients with missing information on race, BMI, cardiovascular disease, and dialysis duration. To account for missing albumin (n = 208) and Kt/Vurea (n = 215), we used a RF imputation approach as implemented in the missForest R package.29 RF imputation has been found to have good performance for categorical variables following a missing at random mechanism but poorer performance for continuous variables that are skewed.30 To address this limitation of RF imputation, we imputed log-transformed versions of albumin and Kt/V (See Supplementary methods and Supplementary Table S2). Analyses that used imputed albumin and Kt/V had a sample size of 465 patients. Additionally, we performed sensitivity analyses where we repeated our analyses on the 251 patients with complete records on all covariates.
Outcome
The primary outcome of the study was all-cause mortality within one year of dialysis initiation. The information on mortality was ascertained from the electronic health records of the dialysis providers for the US participants and by research physician adjudication for the Canadian participants.
Because there is no single best statistical method to analyze high dimensional metabolomic data,31,32 we a priori assumed that metabolites identified by >1 statistical model would be more likely associated with the study outcome. We applied 3 statistical methods to identify metabolites associated with one-year all-cause mortality: linear models with empirical Bayesian inference, ML models using LASSO, and random forests.
Linear models with empirical Bayesian inference use a differential analysis framework modeling strategy, which shrinks a sample’s variance towards a pooled estimate.33 The advantage over the standard linear regression estimator is that Bayesian Linear Regression determines a probability distribution instead of a single “best” value.34 This leads to a more powerful and stable inference to detect significant changes in metabolite abundance and association with outcomes.35 To study the association between metabolite abundance and mortality, we regressed log2-metabolite levels as the dependent variable on mortality (yes/no) and covariates as the independent variables using the limma software package.36 We used metabolite levels as the dependent variable instead of mortality because the theory underlying the limma method makes assumptions about the distribution of the biological measurement (metabolite levels) rather than assumptions about the grouping variable (mortality).33 The covariates were determined a priori and included baseline age, sex, race, cardiovascular disease, diabetes, BMI, albumin, Kt/Vurea, and dialysis duration. BMI, albumin, Kt/Vurea, and dialysis duration were modeled with natural splines. When estimating moderated variances, we retained unidentified metabolites to improve estimation accuracy but removed them after completion of modeling, resulting in 498 metabolites. We adjusted P-values for multiple testing using the Benjamini-Hochberg procedure and used a false discovery rate threshold of 10% (pFDR <0.10) for the association between mortality and metabolites.37,38 We used the coefficients on mortality indicators as our measures of association. These coefficients give the adjusted log2-fold change in metabolite levels comparing patients who died within 1 year of dialysis initiation and those who did not.
In addition to empirical Bayesian inference, we used LASSO and RF models to crosscheck metabolites predictive of one-year mortality as a binary variable. In these models, covariates and all known metabolites were included as predictors, and variable importance measures were used to quantify the predictive ability of metabolites. In the LASSO models, variable importance was quantified as the number of times a variable had a nonzero coefficient across different values of the regularization parameter (lambda). In the RF models, variable importance was quantified as the mean decrease in Gini index resulting from splits on that variable, averaged over the trees in which that variable appeared. We categorized variable importance as high (≥90th percentile), medium (70th–90th percentile), or low (<70th percentile). Overall, we considered a metabolite significantly associated with mortality if the pFDR was <0.1 and showed medium or high variable importance by LASSO and RF models. Additionally, we explored metabolites that were considered significant by any one of the 3 statistical models.
Sensitivity Analyses
Our primary analysis included RUV correction and no covariate imputation. In sensitivity analyses, we explored the impact of imputing albumin and Kt/V covariates (imputation = yes) and eliminating RUV correction (RUV correction = no).
All analyses were done using Stata SE 17.0 (StataCorp, College Station, TX, USA) and R (R Core Team, 2023).
Results
Descriptive Characteristics
Baseline characteristics of the 522 included patients are shown in Table 1. The median age (interquartile range, IQR) was 64 (54–74) years, 88% were men, and 81% self-identified as White. The median BMI (IQR) was 27 (23–31) kg/m2, 48% of patients had a history of cardiovascular disease, and 54% of patients had a history of diabetes mellitus. Median serum albumin was 3.3 (2.7–3.7) g/dl, and median Kt/Vurea was 1.4 (1.2–1.7). The median dialysis duration was 35 (15–66) days. The characteristics of all LUCID participants (n = 700), including those who were excluded from this study due to lack of mortality information, are shown in Supplementary Table S6.
Table 1.
Characteristics of 522 Longitudinal US/Canada Incident Dialysis study participants included in present study
| Characteristics | Overall |
|---|---|
| Median age (IQR), yr | 64 (54–74) |
| Male, n (%) | 457 (88%) |
| Race/ethnicity, n (%) | |
| Non-Hispanic White | 424 (81) |
| Non-Hispanic Black | 22 (4) |
| Other | 75 (15) |
| Median BMI (IQR), kg/m2 | 27 (23–31) |
| Diabetes, n (%) | 280 (54) |
| Cardiovascular disease, n (%) | 248 (48) |
| Median albumin (IQR), g/dl | 3.3 (2.7– 3.7) |
| Median Kt/Vurea (IQR) | 1.4 (1.2–1.7) |
| Median dialysis duration (IQR), days | 35 (15–66) |
| 1-year mortality, n (%) | 44 (8.4) |
BMI, body mass index; IQR, interquartile range.
Missingness: Race (n = 1), BMI (n = 51), cardiovascular disease (n = 3), albumin (n = 208), Kt/Vurea (n = 215), dialysis duration (n = 8).
Metabolites Associated With Mortality
The cumulative 1-year mortality in the cohort was 8.4%. Two metabolites, quinolinate and mesaconate, were significantly associated with one-year posthemodialysis-initiation mortality (pFDR<0.1 and ranked high importance using RF and LASSO) (Table 2). The levels of quinolinate were 1.72-fold higher in patients who died by year 1 compared with those who did not (pFDR, 0.009), while the levels of mesaconate were 1.57-fold higher in those who died compared to those who did not (pFDR, 0.002).
Table 2.
Metabolites associated with one-year posthemodialysis mortality in Longitudinal US/Canada Incident Dialysis cohort
| Analysis | Metabolite | 1-year mortality |
|||
|---|---|---|---|---|---|
| Adjusted fold change | pFDR | LASSO | Random forest | ||
| Primary analysis | |||||
| RUV correction; no covariate imputation (n = 251) | Quinolinate | 1.72 | 0.009 | High | High |
| Mesaconate | 1.57 | 0.002 | High | High | |
| Sensitivity analyses | |||||
| No RUV correction; no covariate imputation (n = 251) | Quinolinate | 1.72 | 0.008 | High | High |
| Mesaconate | 1.53 | 0.003 | High | High | |
| No RUV correction; covariate imputation (n = 465) | Quinolinate | 1.66 | 0.001 | High | High |
| Mesaconate | 1.30 | 0.073 | High | High | |
| RUV correction & covariate imputation (n = 465) | Quinolinate | 1.66 | 0.001 | High | High |
| Mesaconate | 1.36 | 0.012 | High | High | |
FDR, false discovery rate; LASSO, Least absolute shrinkage and selection operator; RUV, removal of unwanted variation.
Other than quinolinate and mesaconate, a total of 42 metabolites were identified as high importance by LASSO (Supplementary Table S3), 46 by RF (Supplementary Table S4) and 9 by both these models (Supplementary Table S5). However, the pFDR was >0.1 for all. The 9 metabolites included 2-aminoheptanoate, C22:5 cholesteryl eicosapentaenoic acid, C54:6 monoarachidonic acid triglyceride, C54:4 monoarachidonic acid triglyceride, citrate/isocitrate, N1-methyl-2-pyridone-5-carboxamide, C34:2 phosphatidylethanolamine, N-acetylornithine, and guanidinoacetic acid (Supplementary Table S5). Accuracy measures for the ML models are available (Supplementary Table S7).
A comprehensive list of all identified metabolites and their degree of association with one-year mortality using limma, LASSO and RF is available in Supplementary Data S2.
Sensitivity and Additional Analyses
Sensitivity analyses showed that quinolinate and mesaconate remained significantly associated with one-year mortality under all permutations accounting for RUV and missing covariate imputation (Table 2). In exploratory analyses, the associations remained unchanged after adjusting for indoxyl sulfate, another protein-bound uremic solute (data not shown).
Discussion
In this cohort of incident patients with kidney failure treated with hemodialysis in the United States and Canada, we identified 498 known plasma metabolites and evaluated their association with 1-year mortality. Using state-of-the-art high-dimensional data methods, we found 2 metabolites, quinolinate and mesaconate, were significantly associated with 1-year mortality.
Quinolinate (HMDB0000232) is a 167 Da metabolite generated in humans by the kynurenine pathway of tryptophan metabolism. It is also present in a variety of foods, including fruits, nuts, and meats.39 Quinolinate is considered a putative protein-bound uremic toxin, and its plasma concentrations in patients with kidney failure are 15 times higher than in people with normal kidney function.40 More frequent hemodialysis (6 times per week) compared to conventional thrice-weekly hemodialysis lowers quinolinate levels by 35%.17 In our study, quinolinate levels were 72% higher in the patients that died during year 1 compared with those who survived. However, we did not find an association between mortality and other metabolites of the kynurenine pathway, including kynurenic acid (P = 0.98), tryptophan (P = 0.98), and xanthurenate (P = 0.98).
Several metabolites of the kynurenine pathway, including quinolinate, are upregulated with oxidative stress and are positively associated with inflammatory markers and carotid intima-media thickness.41 Quinolinate and other metabolites of the kynurenine pathway are also positively associated with hypercoagulability as manifested by elevated fibrinogen, thrombomodulin, and von Willebrand factor levels in patients on dialysis.42,43 In animal models, quinolinate has a direct, dose-dependent, inhibitory effect on endogenous erythropoietin production.44 Although not yet demonstrated in kidney failure, inhibition of erythropoiesis would likely require higher doses of erythropoiesis-stimulating agents, which are also associated with thrombotic events. Thus, the activation of the kynurenine pathway and accumulation of quinolinate could contribute to an increased risk of mortality in patients on dialysis through atherogenic and thrombotic pathways.45,46 Alternatively, elevations of blood quinolinate might indicate a bottleneck in the production of downstream tissue nicotinamide adenine dinucleotide+, as has been observed in other contexts, including acute kidney injury.47, 48, 49 In this case, quinolinate would serve as a marker of NAD+ depletion rather than a functional mediator of mortality risk in kidney failure.
Much less is known about mesaconate, or mesaconic acid (HMDB0000749), the second metabolite associated with mortality in our study. Mesaconate is a 130 Da methyl-branched fatty acid with an unknown protein binding. Recently, mesaconate was shown to be synthesized in inflammatory macrophages from itaconate, itself a macrophage-derived metabolite.50 Microbial components like lipopolysaccharide and fungal cell wall sugars, as well as several cytokines, are known to upregulate macrophage aconitate decarboxylate 1, which synthesizes itaconate from the Krebs cycle intermediate aconitate. Itaconate has complex immunomodulatory effects, although the general view is that it acts as a negative regulator of innate immunity.51
Mesaconate is structurally very similar to itaconate—it differs in the position of one proton and the double bond—with similar immunomodulatory effects in proinflammatory macrophages, including reduced IL-6 and IL-12 secretion and increased CXCL10 production.50 In mice, administration of exogenous mesaconate improved survival following intraperitoneal injection of LPS, a commonly used sepsis model.50 Thus, mesaconate is an emerging “immunometabolite,” although to our knowledge its levels have not been associated with outcomes in humans to date. In our study, mesaconate levels were 57% higher in the patients who died within 1 year compared to those who were alive. There was no association with its precursor aconitate (P = 0.46); our platform did not measure itaconate. Our findings raise the possibility that, like itaconate, mesaconate could be a functional mediator of immunologic dysfunction, thereby increasing the risk of mortality in kidney failure.52
Two previous studies have used an untargeted metabolomics approach to identify metabolites associated with mortality in patients receiving hemodialysis. Kalim et al.19 found an association between long-chain carnitines (oleoylcarnitine, linoleylcarnitine, palmitoleoylcarnitine, stearoylcarnitine), measured on the Broad platform (the same platform as our study) and cardiovascular disease mortality in the Accelerated Mortality on Renal Replacement (ArMORR) study cohort. None of these metabolites were significant in our analysis. Hu et al.20 evaluated the association of metabolites, measured on the Metabolon platform, and cardiovascular mortality in a subsample (n = 94) of the Hemodialysis (HEMO) Study. None of the metabolites were associated with mortality using the false discovery threshold of 10%. Using a raw P-value threshold of <0.005 (unadjusted for multiple comparisons), several metabolites were associated with cardiovascular mortality. Of these metabolites, only palmitoleoylcarnitine and 2-hydroxybutyrate/2-hydroxyisobutyrate were detected in our cohort but were not associated with mortality. This could be due to differences in sample size and statistical methods used for inference or biological differences such as diet, microbiome, variability in dialysis intensity, timing of sample collection relative to the long interdialytic interval, intradialytic weight gain and residual renal function, given that our study included incident dialysis patients with presumably some residual kidney function. In contrast, ArMORR and HEMO studies evaluated prevalent patients. This highlights the need for larger validation studies with rigorous methodologies.
Our study has several strengths, including an incident dialysis cohort, a state-of-the-art metabolomics platform, and our rigorous statistical approach with a priori selection of covariates and statistical models. The limitations include a single timepoint measurement of metabolites and lack of standardization of the day of the week for sample collection. In addition, the observational design of the study precludes the assessment of causality, and our cohort had a mortality rate (8%) that was lower than the reported by the United States Renal Data System (16%), suggesting selection of healthier patients recruited in the study. Therefore, the next steps should include external validation on diverse populations, with a longer follow-up period, and repeated metabolite measurements. In addition, an exploration of the association of metabolites with uremic symptoms in patients receiving maintenance hemodialysis remains lacking and constitutes an important future direction.
Conclusion
We report 2 novel metabolites, quinolinate and mesaconate, are associated with 1-year mortality in incident patients receiving maintenance hemodialysis. Our findings call for further external validation, and if confirmed, exploration of biological pathways of toxicity and search for possible therapeutic targets.
Disclosures
All the authors declared no competing interests.
Funding
The metabolomics investigation in this study was funded by the National Institute of Nursing Research (R01NR017399).
Data Sharing Statement
The data (metabolites, deidentified clinical data, outcomes) supporting the findings of this study will be made publicly available in the Metabolomics Workbench repository at https://www.metabolomicsworkbench.org/ after the acceptance of the manuscript. The code used in the analyses is available in GitHub at https://github.com/lmyint/metabolomics_mortality_hemodialysis.
Footnotes
Supplementary Methods S1.
Assessment of risk of instrument drift.
Adjustment for risk of instrument drift.
Imputation of missing metabolite data.
Imputation of missing covariate data.
Figure S1. Principal components analysis (PCA) on the metabolite abundances before adjustment for instrument drift.
Figure S2. Principal components analysis (PCA) on the metabolite abundances after adjustment for instrument drift bias.
Figure S3. Evaluation of imputation methods for missing metabolite data.
Table S1. Coefficients of variation for metabolomics data in the LUCID study.
Table S2. Misclassification rate of covariate imputation modeling.
Table S3. Risk of one-year posthemodialysis mortality associated with metabolites classified as high importance using LASSO machine learning model.
Table S4. Risk of one-year posthemodialysis mortality associated with metabolites classified as high importance using random forest machine learning model.
Table S5. Risk of one-year posthemodialysis mortality associated with metabolites classified as high importance using both LASSO and random forest machine learning models.
Table S6. Descriptive characteristics comparison between those in the mortality analysis and the remaining LUCID participants.
Table S7. Accuracy measures for machine learning models.
Table S8. STROBE statement checklist.
Supplementary Data S2.
Degree of association of all identified metabolites with 1-year mortality.
Supplementary Material
Supplementary Methods S1. Assessment of risk of instrument drift. Adjustment for risk of instrument drift. Imputation of missing metabolite data. Imputation of missing covariate data. Figure S1. Principal components analysis (PCA) on the metabolite abundances before adjustment for instrument drift. Figure S2. Principal components analysis (PCA) on the metabolite abundances after adjustment for instrument drift bias. Figure S3. Evaluation of imputation methods for missing metabolite data. Table S1. Coefficients of variation for metabolomics data in the LUCID study. Table S2. Misclassification rate ocf covariate imputation modeling. Table S3. Risk of one-year posthemodialysis mortality associated with metabolites classified as high importance using LASSO machine learning model. Table S4. Risk of one-year posthemodialysis mortality associated with metabolites classified as high importance using random forest machine learning model. Table S5. Risk of one-year posthemodialysis mortality associated with metabolites classified as high importance using both LASSO and random forest machine learning models. Table S6. Descriptive characteristics comparison between those in the mortality analysis and the remaining LUCID participants. Table S7. Accuracy measures for machine learning models. Table S8. STROBE statement checklist.
Supplementary Data S2. Degree of association of all identified metabolites with 1-year mortality.
References
- 1.Kidney disease statistics for the United States. National Institute of Diabetes and Digestive and Kidney Diseases. https://www.niddk.nih.gov/health-information/health-statistics/kidney-disease
- 2.2023 Annual data report. USRDS. https://adr.usrds.org/
- 3.Foley R.N., Hakim R.M. Why is the mortality of dialysis patients in the United States much higher than the rest of the world? J Am Soc Nephrol. 2009;20:1432–1435. doi: 10.1681/ASN.2009030282. [DOI] [PubMed] [Google Scholar]
- 4.Msaad R., Essadik R., Mohtadi K., et al. Predictors of mortality in hemodialysis patients. Pan Afr Med J. 2019;33:61. doi: 10.11604/pamj.2019.33.61.18083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ferreira Ede S., Moreira T.R., da Silva R.G., et al. Survival and analysis of predictors of mortality in patients undergoing replacement renal therapy: a 20-year cohort. BMC Nephrol. 2020;21:502. doi: 10.1186/s12882-020-02135-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Falconi C.A., Junho C.V.D.C., Fogaça-Ruiz F., et al. Uremic toxins: an alarming danger concerning the cardiovascular system. Front Physiol. 2021;12 doi: 10.3389/fphys.2021.686249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arai Y., Shioji S., Tanaka H., Katagiri D., Hinoshita F. A novel uremic score reflecting accumulation of specific uremic toxins more precisely predicts one-year mortality after hemodialysis commencement: a retrospective cohort study. Toxins (Basel) 2020;12:634. doi: 10.3390/toxins12100634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sapa H., Gutiérrez O.M., Shlipak M.G., et al. Association of uremic solutes with cardiovascular death in diabetic kidney disease. Am J Kidney Dis. 2022;80:502–512.e1. doi: 10.1053/j.ajkd.2022.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beger R.D., Dunn W., Schmidt M.A., et al. Metabolomics enables precision medicine: “A White Paper, Community Perspective.”. Metabolomics. 2016;12:149. doi: 10.1007/s11306-016-1094-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buergel T., Steinfeldt J., Ruyoga G., et al. Metabolomic profiles predict individual multidisease outcomes. Nat Med. 2022;28:2309–2320. doi: 10.1038/s41591-022-01980-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhu S., Zhang F., Shen A.W., et al. Metabolomics evaluation of patients with Stage 5 chronic kidney disease before dialysis, maintenance hemodialysis, and peritoneal dialysis. Front Physiol. 2020;11 doi: 10.3389/fphys.2020.630646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wen D., Zheng Z., Surapaneni A., et al. Metabolite profiling of CKD progression in the chronic renal insufficiency cohort study. JCI Insight. 2022;7 doi: 10.1172/jci.insight.161696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen D.Q., Cao G., Chen H., et al. Identification of serum metabolites associating with chronic kidney disease progression and anti-fibrotic effect of 5-methoxytryptophan. Nat Commun. 2019;10:1476. doi: 10.1038/s41467-019-09329-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kwan B., Fuhrer T., Zhang J., et al. Metabolomic markers of kidney function decline in patients with diabetes: evidence from the chronic renal insufficiency cohort (CRIC) study. Am J Kidney Dis. 2020;76:511–520. doi: 10.1053/j.ajkd.2020.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rhee E.P., Souza A., Farrell L., et al. Metabolite profiling identifies markers of uremia. J Am Soc Nephrol. 2010;21:1041–1051. doi: 10.1681/ASN.2009111132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tanaka H., Sirich T.L., Plummer N.S., Weaver D.S., Meyer T.W. An enlarged profile of uremic solutes. PLoS One. 2015;10 doi: 10.1371/journal.pone.0135657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sirich T.L., Fong K., Larive B., et al. Limited reduction in uremic solute concentrations with increased dialysis frequency and time in the Frequent Hemodialysis Network Daily Trial. Kidney Int. 2017;91:1186–1192. doi: 10.1016/j.kint.2016.11.002. [DOI] [PubMed] [Google Scholar]
- 18.Kalim S., Wald R., Yan A.T., et al. Extended duration nocturnal hemodialysis and changes in plasma metabolite profiles. Clin J Am Soc Nephrol. 2018;13:436–444. doi: 10.2215/CJN.08790817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kalim S., Clish C.B., Wenger J., et al. A plasma long-chain acylcarnitine predicts cardiovascular mortality in incident dialysis patients. J Am Heart Assoc. 2013;2 doi: 10.1161/JAHA.113.000542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hu J.R., Grams M.E., Coresh J., et al. Serum metabolites and cardiac death in patients on hemodialysis. Clin J Am Soc Nephrol. 2019;14:747–749. doi: 10.2215/CJN.12691018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rhee E.P., Guallar E., Hwang S., et al. Prevalence and persistence of uremic symptoms in incident dialysis patients. Kidney360. 2020;360:86–92. doi: 10.34067/KID.0000072019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rhee E.P., Waikar S.S., Rebholz C.M., et al. Variability of two metabolomic platforms in CKD. Clin J Am Soc Nephrol. 2019;14:40–48. doi: 10.2215/CJN.07070618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.STROBE checklists. STROBE. https://www.strobe-statement.org/checklists/
- 24.Fernández-Albert F., Llorach R., Garcia-Aloy M., Ziyatdinov A., Andres-Lacueva C., Perera A. Intensity drift removal in LC/MS metabolomics by common variance compensation. Bioinformatics. 2014;30:2899–2905. doi: 10.1093/bioinformatics/btu423. [DOI] [PubMed] [Google Scholar]
- 25.Märtens A., Holle J., Mollenhauer B., Wegner A., Kirwan J., Hiller K. Instrumental drift in untargeted metabolomics: optimizing data quality with intrastudy QC samples. Metabolites. 2023;13:665. doi: 10.3390/metabo13050665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Livera A.M.D., Sysi-Aho M., Jacob L., et al. Statistical methods for handling unwanted variation in metabolomics data. Anal Chem. 2015;87:3606–3615. doi: 10.1021/ac502439y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cao W., Wang D., Li J., Zhou H., Li L., Li Y. Cornell University; Published May 26, 2018. BRITS: Bidirectional Recurrent Imputation for Time Series.https://arxiv.org/abs/1805.10572 [Google Scholar]
- 28.Wei R., Wang J., Su M., et al. Missing value imputation approach for mass spectrometry-based metabolomics data. Sci Rep. 2018;8:663. doi: 10.1038/s41598-017-19120-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stekhoven D.J., Bühlmann P. MissForest—non-parametric missing value imputation for mixed-type data. Bioinformatics. 2012;28:112–118. doi: 10.1093/bioinformatics/btr597. [DOI] [PubMed] [Google Scholar]
- 30.Shah A.D., Bartlett J.W., Carpenter J., Nicholas O., Hemingway H. Comparison of random forest and parametric imputation models for imputing missing data using MICE: a CALIBER study. Am J Epidemiol. 2014;179:764–774. doi: 10.1093/aje/kwt312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Antonelli J., Claggett B.L., Henglin M., et al. Statistical workflow for feature selection in human metabolomics data. Metabolites. 2019;9:143. doi: 10.3390/metabo9070143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ogutu J.O., Schulz-Streeck T., Piepho H.P. Genomic selection using regularized linear regression models: ridge regression, lasso, elastic net and their extensions. BMC Proc. 2012;6(Suppl 2) doi: 10.1186/1753-6561-6-S2-S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smyth G.K. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 34.Koehrsen W. Introduction to Bayesian linear regression. Medium. Published 2018 https://towardsdatascience.com/introduction-to-bayesian-linear-regression-e66e60791ea7 [Google Scholar]
- 35.Kammers K., Cole R.N., Tiengwe C., Ruczinski I. Detecting significant changes in protein abundance. EuPA Open Proteom. 2015;7:11–19. doi: 10.1016/j.euprot.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Haynes W. In: Encyclopedia of Systems Biology. Dubitzky W., Wolkenhauer O., Cho K.H., Yokota H., editors. Springer; 2013. Benjamini–Hochberg method; p. 78. 78. [DOI] [Google Scholar]
- 38.Chen S.Y., Feng Z., Yi X. A general introduction to adjustment for multiple comparisons. J Thorac Dis. 2017;9:1725–1729. doi: 10.21037/jtd.2017.05.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Friedman M. Analysis, nutrition, and health benefits of tryptophan. Int J Tryptophan Res. 2018;11 doi: 10.1177/1178646918802282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vanholder R., De Smet R., Glorieux G., et al. Review on uremic toxins: classification, concentration, and interindividual variability. Kidney Int. 2003;63:1934–1943. doi: 10.1046/j.1523-1755.2003.00924.x. [DOI] [PubMed] [Google Scholar]
- 41.Pawlak K., Myśliwiec M., Pawlak D. Kynurenine pathway - a new link between endothelial dysfunction and carotid atherosclerosis in chronic kidney disease patients. Adv Med Sci. 2010;55:196–203. doi: 10.2478/v10039-010-0015-6. [DOI] [PubMed] [Google Scholar]
- 42.Pawlak K., Mysliwiec M., Pawlak D. Hypercoagulability is independently associated with kynurenine pathway activation in dialysed uraemic patients. Thromb Haemost. 2009;102:49–55. doi: 10.1160/TH08-10-0696. [DOI] [PubMed] [Google Scholar]
- 43.Pawlak K., Domaniewski T., Mysliwiec M., Pawlak D. The kynurenines are associated with oxidative stress, inflammation and the prevalence of cardiovascular disease in patients with end-stage renal disease. Atherosclerosis. 2009;204:309–314. doi: 10.1016/j.atherosclerosis.2008.08.014. [DOI] [PubMed] [Google Scholar]
- 44.Pawlak D., Koda M., Pawlak S., Wolczynski S., Buczko W. Contribution of quinolinic acid in the development of anemia in renal insufficiency. Am J Physiol Ren Physiol. 2003;284:F693–F700. doi: 10.1152/ajprenal.00327.2002. [DOI] [PubMed] [Google Scholar]
- 45.Mair R.D., Sirich T.L., Meyer T.W. Uremic toxin clearance and cardiovascular toxicities. Toxins (Basel) 2018;10:226. doi: 10.3390/toxins10060226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Benitez T., VanDerWoude E., Han Y., et al. Kynurenine pathway metabolites predict subclinical atherosclerotic disease and new cardiovascular events in chronic kidney disease. Clin Kidney J. 2022;15:1952–1965. doi: 10.1093/ckj/sfac138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Clark A.J., Saade M.C., Vemireddy V., et al. Hepatocyte nuclear factor 4α mediated quinolinate phosphoribosylltransferase (QPRT) expression in the kidney facilitates resilience against acute kidney injury. Kidney Int. 2023;104:1150–1163. doi: 10.1016/j.kint.2023.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mehr A.P., Tran M.T., Ralto K.M., et al. De novo NAD+ biosynthetic impairment in acute kidney injury in humans. Nat Med. 2018;24:1351–1359. doi: 10.1038/s41591-018-0138-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saade M.C., Clark A.J., Parikh S.M. States of quinolinic acid excess in urine: a systematic review of human studies. Front Nutr. 2022;9 doi: 10.3389/fnut.2022.1070435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.He W., Henne A., Lauterbach M., et al. Mesaconate is synthesized from itaconate and exerts immunomodulatory effects in macrophages. Nat Metab. 2022;4:524–533. doi: 10.1038/s42255-022-00565-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Coelho C. Itaconate or how I learned to stop avoiding the study of immunometabolism. PLOS Pathog. 2022;18 doi: 10.1371/journal.ppat.1010361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Winterhoff M., Chen F., Sahini N., et al. Establishment, validation, and initial application of a sensitive LC-MS/MS assay for quantification of the naturally occurring isomers itaconate, mesaconate, and citraconate. Metabolites. 2021;11:270. doi: 10.3390/metabo11050270. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Methods S1. Assessment of risk of instrument drift. Adjustment for risk of instrument drift. Imputation of missing metabolite data. Imputation of missing covariate data. Figure S1. Principal components analysis (PCA) on the metabolite abundances before adjustment for instrument drift. Figure S2. Principal components analysis (PCA) on the metabolite abundances after adjustment for instrument drift bias. Figure S3. Evaluation of imputation methods for missing metabolite data. Table S1. Coefficients of variation for metabolomics data in the LUCID study. Table S2. Misclassification rate ocf covariate imputation modeling. Table S3. Risk of one-year posthemodialysis mortality associated with metabolites classified as high importance using LASSO machine learning model. Table S4. Risk of one-year posthemodialysis mortality associated with metabolites classified as high importance using random forest machine learning model. Table S5. Risk of one-year posthemodialysis mortality associated with metabolites classified as high importance using both LASSO and random forest machine learning models. Table S6. Descriptive characteristics comparison between those in the mortality analysis and the remaining LUCID participants. Table S7. Accuracy measures for machine learning models. Table S8. STROBE statement checklist.
Supplementary Data S2. Degree of association of all identified metabolites with 1-year mortality.