Abstract
Background
Mycoplasmal pneumonia of sheep and goats (MPSG) is an important infectious disease that threatens sheep and goat production worldwide, and Mycoplasma ovipneumoniae (Movi) is one of the major aetiological agents causing MPSG. The aim of this study was to investigate the immunological activity of the Hsp70‒P113 fusion protein derived from Movi and to develop a serological assay for the detection of Movi.
Methods
This study involved codon optimization of the dominant antigenic regions of Movi heat shock protein 70 (Hsp70) and adhesin P113. Afterwards, the optimized sequences were inserted into the prokaryotic expression vector pET-30a( +) through tandem linking with the aid of a linker. Once a positive recombinant plasmid (pET-30a-rHsp70-P113) was successfully generated, the expression conditions were further refined. The resulting double gene fusion target protein (rHsp70‒P113) was subsequently purified using ProteinIso® Ni–NTA resin, and the reactivity of the protein was confirmed via SDS‒PAGE and Western blot analysis. An indirect enzyme-linked immunosorbent assay (i-ELISA) technique was developed to detect Movi utilizing the fusion protein as the coating antigen. The specificity, sensitivity, and reproducibility of all methods were assessed after each reaction parameter was optimized.
Results
The resulting rHsp70-P113 protein had a molecular weight of approximately 51 kDa and was predominantly expressed in the supernatant. Western blot analysis demonstrated its favourable reactivity. The optimal parameters for the i-ELISA technique were as follows: the rHsp70-P113 protein was encapsulated at a concentration of 5 μg/mL; the serum was diluted at a ratio of 1:50; the HRP-labelled donkey anti-goat IgG was diluted at a ratio of 1:6,000. The results of the cross-reactivity assays revealed that the i-ELISA was not cross-reactive with other goat-positive sera against Mycoplasma mycodies subsp. capri (Mmc), Mycoplasma capricolum subsp. capripneumoniae (Mccp), Mycoplasma arginini (Marg), orf virus (ORFV) or enzootic nasal tumour virus of goats (ENTV-2). The sensitivity of this method is high, with a maximum dilution of up to 1:640. The results of the intra- and inter-batch replication tests revealed that the coefficients of variation were both less than 10%, indicating excellent reproducibility. The analysis of 108 clinical serum samples via i-ELISA and indirect haemagglutination techniques yielded significant findings. Among these samples, 43 displayed positive results, whereas 65 presented negative results, resulting in a positivity rate of 39.8% for the i-ELISA method. In contrast, the indirect haemagglutination technique identified 20 positive samples and 88 negative samples, resulting in a positivity rate of 18.5%. Moreover, a comparison between the two methods revealed a conformity rate of 78.7%.
Conclusion
The results obtained in this study lay the groundwork for advancements in the use of an Movi antibody detection kit, epidemiological inquiry, and subunit vaccines.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12917-024-04274-7.
Keywords: Mycoplasma ovipneumoniae, Heat shock protein 70, Adhesin P113, I-ELISA
Introduction
Mycoplasma ovipneumoniae (Movi) belongs to the kingdom Prokaryota, class Mollicutes, order Mycoplasmtales, family Mycoplasmataceae and genus Mycoplasma. Movi have an unusually wide host range, with the pathogen being detected in wildlife, such as musk oxen, caribou, bighorn sheep, as well as goats and sheep [1–3]. Movi are among the most important pathogens causing MPSG. MPSG can occur throughout the year, and the incidence rate in winter and spring is greater than that in summer and fall, causing great economic losses to the sheep and goat industry [4–6]. Pathogen isolation and identification are considered the gold standard for Movi detection but are subject to stringent culture conditions, resulting in low isolation rates. In recent years, various molecular biology assays, including PCR, quantitative PCR (qPCR), loop-mediated isothermal amplification (LAMP), single-stranded DNA probes, and recombinase polymerase amplification (RPA), have been widely used to detect Movi [7–13]. The detection of antibodies serves as a valuable tool in disease diagnosis, epidemiological investigations, and the assessment of vaccine immunization efficacy. Among these applications, the enzyme-linked immunosorbent assay (ELISA) has gained widespread use due to its notable attributes such as heightened sensitivity, commendable specificity, stability, and convenient automated operation and batch testing capabilities.
The heat shock protein Hsp70 is one of the most important membrane proteins of Movi, and its C-terminal antigenicity advantage is obvious; this advantage can induce humoral and cellular immune responses [14]. Adhesin is an important virulence factor of Movi [15, 16]. A study of the adhesin P113 protein revealed that it can specifically bind to Movi antibodies and has good immunogenicity [17, 18]. Both proteins have the potential to be used as subunit vaccines to provide technical support for the later development of subunit vaccines with Hsp70–p113 fusion proteins. In this study, a two-gene fusion expression plasmid was constructed by connecting the dominant antigenic epitope region of the P113 gene and Hsp70 via a linker. The recombinant protein rHsp70-P113 was obtained through prokaryotic expression, and its reactivity was confirmed through SDS‒PAGE and Western blot analysis. An i-ELISA method was established using the recombinant protein rHsp70-P113 as the coating antigen for the detection of Movi. This method serves as the basis for the development of a Movi antibody detection kit, epidemiological investigations, and subunit vaccine development.
Results
Protein expression and purification
SDS‒PAGE revealed that the expressed target protein (rHsp70‒P113) was, as expected, approximately 51 kDa in size and was present mainly in the supernatant (Fig. 1A). The purified protein exhibited a single electrophoretic band (Fig. 1B) at a concentration of 1,530 μg/mL.
Fig. 1.

SDS‒PAGE detection of recombinant Hsp70‒P113 protein expression and purification. M: protein marker; 1: uninduced pET-30a/BL21(DE3); 2: uninduced pET-30a-rHsp70-P113/BL21(DE3); 3: induced pET-30a-rHsp70-P113/BL21(DE3); 4: supernatant of the induced pET-30a-rHsp70-P113/BL21(DE3); 5: precipitate of induced pET-30a-rHsp70-P113/BL21(DE3); 6: purified recombinant protein Hsp70-P113
Identification of the reactogenicity
Western blot analysis with an anti-His-tag antibody confirmed the expression of rHsp70-P113 (Fig. 2A). The protein showed a specific 51 kDa band when hybridized with immune serum and serum from mice immunized with Movi whole bacterial antigen but not with serum from unimmunized mice (Fig. 2B), indicating the strong reactogenicity of rHsp70-P113.
Fig. 2.
Western blot analysis of the reactogenicity of the recombinant protein Hsp70-P113. 1: Anti-His tag antibody as the primary antibody; 2: recombinant protein hyperimmune serum as the primary antibody (hyperimmune serum prepared by immunizing BALB/c mice with purified Hsp70-P113 recombinant protein); 3: Movi hyperimmune serum as the primary antibody (hyperimmune serum prepared by immunizing BALB/c mice with Movi whole bacterium antigen); 4. The serum of nonimmunized mice (blank control) was used as the primary antibody. M: protein marker. 1–4:HRP-labelled goat anti-mouse IgG was used as the secondary antibody
Optimization of working conditions
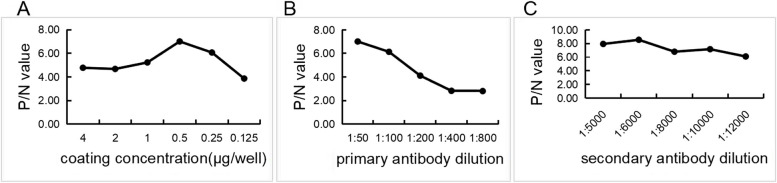
The dilution with the highest OD450nm ratio between the positive and negative samples (P/N values) was chosen as the optimal working concentration. The results revealed that the optimal concentration for antigen encapsulation was 5 µg/mL (Fig. 3A), the optimal dilution of serum (primary antibody) was 1:50 (Fig. 3B), and the optimal dilution of secondary antibody (HRP-labelled donkey anti-goat IgG) was 1:6000 (Fig. 3C).
Fig. 3.
Optimization of the ELISA. The concentration of the coating antigen (A), the dilution of the serum (B), and the dilution of the secondary antibody (C) were determined via the Checkerboard method
Determination of the cut-off value for i-ELISA
Forty-six Movi-negative serum samples that were collected from goats without a history of Movi infection were tested via i-ELISA as described above to determine the cut-off value. Each sample was tested three times, and the frequency statistics are shown in the graph (Fig. 4). The statistical analysis revealed that the OD450nm ranged from 0.195 to 0.499. The mean OD450nm of the Movi-negative serum samples was 0.355, with a standard deviation (SD) of 0.071. When the OD450nm was < 0.497 ( + 2SD), the sample was determined to be Movi negative. Values between 0.497 ( + 2SD) and 0.569 ( + 3SD) were considered suspicious. The analysis of the suspicious samples was repeated once, and if the OD450nm value was still < 0.569 ( + 3SD), the sample was determined to be Movi negative.
Fig. 4.

Statistical frequency of negative sample results
The evaluation of specificity and sensitivity
To determine the specificity of the i-ELISA, goat-positive sera against common pathogens were examined. As shown in Fig. 5, the average OD450nm values of positive serum samples for Mycoplasma mycoides subsp. Capri (Mmc), Mycoplasma capricolum subsp. capripneumoniae (Mccp), Mycoplasma arginini (Marg), Enzootic nasal tumour virus of goats (ENTV-2), and orf virus (ORFV) were 0.225, 0.152, 0.205, 0.393, and 0.310, respectively. These values are all less than 0.497 ( + 2SD), indicating that these serum samples were Movi seronegative and non-cross-reactive according to the results of the i-ELISA; this finding indicates that the established i-ELISA has good specificity. The Movi-positive serum was diluted twofold from 1:40 to 1:2,560 and detected by i-ELISA. The results revealed that the serum was positive, with an OD450nm value of 0.60 (> + 3SD) at a dilution of 1:640 (Table 1), indicating the high sensitivity of the i-ELISA.
Fig. 5.

Specificity of the ELISA
Table 1.
Sensitivity of the antigen capture ELISA
| serum dilution | OD450nm ( ± SD) | Results |
|---|---|---|
| 1:40 | 1.68 ± 0.04 | + |
| 1:80 | 1.50 ± 0.06 | + |
| 1:160 | 1.31 ± 0.02 | + |
| 1:320 | 0.98 ± 0.03 | + |
| 1:640 | 0.60 ± 0.03 | + |
| 1:1280 | 0.41 ± 0.01 | - |
| 1:2560 | 0.28 ± 0.05 | - |
| Negative control | 0.31 ± 0.04 | - |
“ + ” indicates a positive result; “-” indicates a negative result
Evaluation of repeatability
In the repeatability experiment, 10 serum samples were used to determine the intra- and inter-assay coefficients of variation (CVs) of the i-ELISA, which were 0.97% ~ 9.78% and 1.39% ~ 9.53%, respectively (Table 2). The CVs were all less than 10%, which indicates that the established i-ELISA had good repeatability.
Table 2.
Repeatability assay for i-ELISA
| Intra batch | Inter batch | |||
|---|---|---|---|---|
| Sample No | ± SD | CV(%) | ± SD | CV(%) |
| 1 | 1.10 ± 0.04 | 4.07 | 1.03 ± 0.08 | 7.65 |
| 2 | 0.77 ± 0.01 | 0.97 | 0.73 ± 0.06 | 8.07 |
| 3 | 0.60 ± 0.02 | 3.22 | 0.60 ± 0.01 | 1.39 |
| 4 | 0.76 ± 0.04 | 5.51 | 0.73 ± 0.03 | 3.65 |
| 5 | 0.33 ± 0.03 | 9.78 | 0.31 ± 0.02 | 5.50 |
| 6 | 0.83 ± 0.03 | 4.12 | 0.81 ± 0.02 | 2.22 |
| 7 | 0.80 ± 0.02 | 2.65 | 0.75 ± 0.06 | 8.10 |
| 8 | 1.13 ± 0.03 | 2.51 | 1.04 ± 0.08 | 8.13 |
| 9 | 0.27 ± 0.02 | 8.91 | 0.44 ± 0.04 | 9.53 |
| 10 | 1.57 ± 0.06 | 4.06 | 1.56 ± 0.12 | 7.45 |
Comparison of the i-ELISA with a commercial indirect haemagglutination kit
A total of 108 serum samples obtained from clinical sheep and goats were subjected to testing using both i-ELISA and an indirect haemagglutination kit (Table 3). The i-ELISA analysis revealed that 43 of the 108 samples tested positive for Movi, whereas the remaining 65 samples tested negative, resulting in a calculated positivity rate of 39% (43/108). On the other hand, the indirect haemagglutination assay demonstrated that among the same set of samples, 20 were identified as positive for Movi, whereas the remaining 88 samples tested negative, yielding a positivity rate of 18.5% (20/108). The serum samples that tested positive for Movi according to the indirect haemagglutination method were also positive according to the i-ELISA. The overall concordance rate between the two methods was 78.7% (85/108).
Table 3.
Comparison of a commercial indirect haemagglutination kit and the i-ELISA
| Detection method | No.of samples | No.of positive samples | No.of negative samples | Positive rate/% |
|---|---|---|---|---|
| indirect haemagglutination kit | 108 | 43 | 65 | 39.8 |
| i-ELISA | 108 | 20 | 88 | 18.5 |
Discussion
The sole reliance of the diagnosis of Movi based on the clinical symptoms exhibited by infected sheep can lead to potential confusion with other pathogens, such as Mccp, Mmc, ENTV-2, and others that induce similar respiratory clinical symptoms [19, 20]. Therefore, it is imperative to complement clinical observations with laboratory tests to ensure an accurate diagnosis. There is no universally accepted, gold standard serological test for the detection of antibodies against Movi. A variety of targets can be detected via ELISA. Several ELISA methods have been devised to identify serum antibodies directed against Movi EF-TU, pdhD, P60, enolase, P113, P130, Hsp70, and other proteins [18, 21–26]. Nevertheless, there is presently a dearth of scholarly literature pertaining to the detection of serum antibodies, specifically employing ELISA techniques established for Hsp70-P113 fusion proteins.
Sequencing analysis of the Hsp70 gene of six Movi isolates from the Fujian Province revealed that the nucleotide and amino acid homology between the Hsp70 gene and other reference strains of Movi ranged from 96.0% to 99.4% and 98.0% to 100.0%, respectively [27]. These findings suggest a high level of conservation of the Hsp70 gene within the Movi species. The complete sequences of two Movi P113 genes were published in the NCBI database for strain ATCC 29419 (GenBank accession number: KR021380.1) and strain GZ-QX1 (GenBank accession number: KR270152.1), which presented up to 99.9% gene sequence homology. In the design, the author analysed the dominant antigen regions of the Hsp70 gene and P113 gene through bioinformatics analysis software and selected the conserved regions between different strains for tandem expression. The combination of a variety of highly specific antigens not only avoids the limited detection efficiency of a single antigen, but also provides the possibility for subunit vaccines to offer immune protection to different strains in subsequent development.
In this study, an i-ELISA based on the Hsp70‒P113 fusion protein was established to detect the serum antibody levels of Movi. This method has high specificity and does not cross-react with positive goat sera of Mmc, Mccp, Marg, ORFV, ENTV-2, or other pathogens. It also has high sensitivity, as it shows positive detection results even after a serum dilution of 1:640. Additionally, it exhibits excellent repeatability, with the coefficient of variation of intra- and inter-batch repeated tests being less than 10%. In this study, the i-ELISA method was utilized to test 108 clinical sheep serum samples from the Fujian Province. The findings revealed a Movi positivity rate of 39.8%. Previous reports have indicated the presence of Movi in various provinces in China, including a 40.2% Movi positivity rate in sheep serum samples from the Guizhou Province, a 38.8% Movi positivity rate in sheep serum samples from the Sichuan Province [18, 25], and a 40.8% Movi positivity rate in nasal swab samples from six regions in southern Xinjiang [6]. These results suggest a high prevalence of Movi infection and a wide range of prevalence rates across different regions. Consequently, the advancement of superior quality serological diagnostic reagents can offer technical assistance for conducting epidemiological investigations and facilitating the development of subunit vaccines targeting the Hsp70‒P113 fusion protein.
Conclusion
In this study, we effectively generated a prokaryotic expression vector for the recombinant protein Hsp70-P113 of Movi. The expressed protein in Escherichia coli had a molecular weight of approximately 51 kDa and was predominantly found in a soluble state, resulting in a successful purification process. The recombinant Hsp70-P113 protein displayed favourable reactivity. Moreover, we developed an i-ELISA employing the recombinant Hsp70-P113 protein for the detection of Movi antibodies. The i-ELISA employed in this study exhibited remarkable specificity, sensitivity, and reproducibility. Consequently, this research offers valuable technological assistance for the clinical detection of Movi and the subsequent advancement of a subunit vaccine targeting the Hsp70-P113 fusion protein.
Methods
Experimental animals and sera
The experimental SPF-grade BALB/c mice were purchased from Beijing Huafukang Bio. Clinical serum samples were collected from several goats and sheep on farms in the Fujian Province. The Movi-negative serum samples were collected from goats and sheep that were confirmed by qPCR to be free of infection and had not received immunization against Movi. The Movi indirect haemagglutination test antigen and Movi-negative and -positive sera were purchased from LanRan Biologicals (Gansu, China). High-immunity sera prepared from Movi whole bacterial antigen-immunized BALB/c mice, high-immunity sera prepared from rHsp70-P113 protein-immunized BALB/c mice, Mycoplasma mycodies subsp. Capri (Mmc)-positive goat sera, Mycoplasma capricolum subsp. capripneumoniae (Mccp)-positive goat sera, Mycoplasma arginini (Marg)-positive goat sera, enzootic nasal tumour virus (ENTV-2)-positive sera, and orf virus (ORFV)-positive sera were preserved by the Institute of Animal Husbandry & Veterinary Medicine, Fujian Academy of Agricultural Sciences (China).
Prokaryotic expression of proteins
Construction of pET-30a-rHsp70-P113
The Protean program in DNAStar 7.0 software was utilized to analyse the secondary structures of the Hsp70 gene (FJ-29 strain, GenBank accession number: OP121620) and the P113 gene (ATCC 29419 strain, GenBank accession number: KR021380) of Movi, with the aim of identifying prominent antigenic regions. Using the ExpOptimizer codon optimization tool, the gene sequences of the two regions were optimized for rare codons based on E. coli codon preference. A linker was then added to the middle of the two gene sequences. The fragment, synthesized by Nanjing Kingsley Co., Ltd., was inserted into the pET-30a( +) vector between the NdeI and HindIII cleavage sites. After screening, the prokaryotic expression recombinant plasmid pET-30a-rHsp70-P113 was obtained. The optimized sequences were as follows: ATG—6*His tag—Hsp70 (red mark)-Linker—P113 (yellow mark)-Stop codon (**).

Protein expression
The recombinant plasmid pET-30a-rHsp70-P113 was introduced into E. coli BL21 (DE3) receptor cells. After reaching an optical density of 0.6 ~ 0.8 at OD600nm, the cells were subjected to induction with 0.5 mmol/L isopropyl-β-D-thiogalactoside (IPTG) at 15 °C for 16 h. Subsequently, the bacterial solution was harvested and subjected to ultrasonic disruption followed by centrifugation at 12,000 rpm/min for 10 min. The resulting bacterial liquid was collected and subjected to further ultrasonication and centrifugation under the same conditions, yielding a supernatant and a precipitate that were separately collected for subsequent analysis via SDS‒PAGE.
Protein purification
The positive bacterial growth medium was collected and induced with IPTG for mass-induced expression of the rHsp70-P113 protein. The bacterial growth fluid was then collected, washed three times with PBS, and resuspended. The mixture was subsequently sonicated and centrifuged to collect the supernatant, which was subsequently purified using ProteinIso® Ni–NTA resin. The concentration of the purified protein was determined following the instructions of the Easy II Protein Quantitative Kit (BCA).
Reactogenicity
The immunogen employed in this study was the purified recombinant Hsp70-P113 protein. BALB/c mice were subjected to a total of three immunizations via multiple subcutaneous injections. During the initial immunization, the recombinant protein was emulsified by incorporating an equal volume of Freund's complete adjuvant. For the subsequent two immunizations, Freund's incomplete adjuvant was added to the recombinant protein. The immunizations were administered at intervals of 14 days. Each immunization involved the administration of a 100 μg dose per mouse. Blood was collected from the mice at 9 days after the third immunization, and the recombinant protein hyperimmune serum was subsequently separated. The purified Hsp70-P113 protein was analysed by SDS‒PAGE, and the protein was transferred to a PVDF membrane using the semi-dry Western blot method. Primary antibodies, including anti-His tag antibodies, recombinant Hsp70-P113 protein hyperimmune serum (1:100), Movi hyperimmune serum (1:100), and blank control mouse serum (1:100), were used. HRP-labelled goat anti-mouse IgG (1:5,000) was used as the secondary antibody. The results were observed through DAB colour development.
Standardization of i-ELISA methods
Optimization of the working conditions
The checkerboard method was used to optimize the concentration of Hsp70-P113 protein (40 µg/mL, 20 µg/mL, 10 µg/mL, 5 µg/mL, 2.5 µg/mL, and 1.25 µg/mL), and 100 µL was added transversely to each well and incubated at 4 °C for 16 h. The plates were then washed five times with 350 µL of PBST (1 × PBS containing 0.05% Tween-20)/well/each time and blotted dry. The plates were blocked with 3% bovine serum albumin (BSA) for 2 h at 37 °C. After the same washing step, the Movi-positive and -negative sera were diluted 1:50, 1:100, 1:200, 1:400, and 1:800, respectively, added lengthwise to the plate and incubated at 37 °C for 45 min. After the same washing step, the plates were reacted with 100 µL of diluted secondary antibody (HRP-labelled donkey anti-goat IgG) and diluted 1:10,000 for 45 min at 37 °C. The plates were then washed five times, and the peroxidase reaction was visualized using 100 µL of 3,3’,5,5′-tetramethylbenzidine (TMB) as the substrate of HRP for 10 min at 37 °C in complete darkness. The reaction was stopped by adding 50 µL of TMB termination solution to each well. Finally, the OD450nm of each well was measured and recorded immediately. The conditions that gave the highest OD450nm ratio between the positive and negative sera (P/N value) were considered optimal. Further optimization of donkey anti-goat IgG HRP dilutions (1:5,000, 1:6,000, 1:8,000, 1:10,000, and 1:12,000) was performed to determine the optimal conditions.
Determination of the cut-off value
Movi-negative serum (N = 46) was tested following the i-ELISA procedure mentioned above. Statistical analysis was performed using SPSS 23.0 software, and the average () and standard deviation (SD) were calculated. According to statistical principles, when the OD450nm of a sample was ≥ + 3SD, the sample was considered positive with an accuracy of 99.9%. When the OD450nm was < + 2SD, the sample was determined to be Movi negative, and when + 2SD ≤ OD450nm < + 3SD, the sample was determined to be suspicious. The analysis of the suspicious samples was repeated once, and when the OD450nm was still < + 3SD, the sample was determined to be Movi negative.
Specificity and sensitivity of the i-ELISA
Goat sera positive for Mmc, Mccp, Marg, Movi, ORFV and ENTV-2 (three replicates of each sample) were tested using an established i-ELISA to assess specificity.
Sensitivity analysis, also known as the lower detection limit, was estimated by endpoint titration and was defined as the maximum dilution of the sample detected just above the cut-off value [28]. Therefore, Movi-positive goat sera were diluted 1:40, 1:80, 1:160, 1:320, 1:640, 1:1,280 and 1:2,560 in PBST (100 µL/well) and tested by i-ELISA to determine the sensitivity. The sensitivity of the i-ELISA was evaluated on the basis of the cut-off. Each dilution was tested in triplicate, and the Movi-negative serum samples served as controls.
Determination of the repeatability of i-ELISA
Serum samples (N = 10) were randomly selected. Three replicates of each sample were assayed on the same plate for intra-assay analysis and on three different plates for inter-assay analysis. The mean () and standard deviation (SD) of the intra- and inter-batch tests were calculated, and the coefficient of variation of the intra- and inter-batch reproducibility tests was calculated according to the following formula: coefficient of variation (CV) = standard deviation (SD)/mean ().
Comparison of the i-ELISA with a commercial indirect haemagglutination kit
A total of 108 clinical serum samples were diluted 1:50 in PBS and tested via i-ELISA and an indirect haemagglutination kit.
Supplementary Information
Acknowledgements
Not applicable.
Authors’ contributions
Jinxiu JIANG and Yongliang CHE conceived and designed the study. Jinxiu JIANG: performed the test, collected and analysed the data, and writing the manuscript. Yongliang CHE and Qilin HU: edited and revised the manuscript. Yusheng Lin and Jingpeng Zhang: Analysed the data and editing the manuscript. Weiwei Liu and Lina HUANG collected the serum and analysed the data. All the authors reviewed the manuscript.
Funding
This work was supported by the Basic Research Special Project for Public Welfare Research Institutes in Fujian Province (grant number 2020R1026009 and grant number 2023R1077), Fujian Province Agricultural High-quality Development Beyond "5511" Collaborative Innovation Project (grant number XTCXGC2021008) and Science and Technology Innovation Team Building Project of Fujian Academy of Agricultural Sciences (grant number CXTD2021007-2).
Availability of data and materials
All the data generated or analysed during this study are included in this manuscript. The sequence of the recombinant protein of the Hsp70-P113 double gene was deposited in the National Microbiology Data Center (NMDC) with accession number NMDCP0000170. URL is https://nmdc.cn/resource/genomics/protein/detail/NMDCP0000170.
Declarations
Ethics approval and consent to participate
Animal studies were conducted in strict accordance with the Guide for the Care and Use of Laboratory Animals, Fujian Province, China. The goat and sheep sera used in this study were collected from goats and sheep husbandry farms. Written consent for the use of the samples before participation in the study was obtained from the farmers. This study was approved by the Institute of Animal Husbandry & Veterinary Medicine, Fujian Academy of Agricultural Sciences (approval no. 202307FJ014).
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Besser TE, Cassirer EF, Potter KA, et al. Association of Mycoplasma ovipneumoniae Infection with Population-Limiting Respiratory Disease in Free-Ranging Rocky Mountain Bighorn Sheep (Ovis canadensis). J Clin Microbiol. 2008;46:423–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Handeland K, Tengs T, Kokotovic B, et al. Mycoplasma ovipneumoniae-A Primary Cause of Severe Pneumonia Epizootics in the Norwegian Muskox (Ovibos moschatus) Population. PLoS ONE. 2014;9: e106116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Highland M, Herndon D, Bender S, et al. Mycoplasma ovipneumoniae in Wildlife Species beyond Subfamily Caprinae. Emerg Infect Dis. 2018;24:2384–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rong G, Zhao JM, Hou GY, et al. Seroprevalence and molecular detection of Mycoplasma ovipneumoniae in goats in tropical China. Trop Anim Health Prod. 2014;46:1491–5. [DOI] [PubMed] [Google Scholar]
- 5.Spaan RS, Epps CW, Crowhurst R, et al. Impact of Mycoplasma ovipneumoniae on juvenile bighorn sheep (Ovis canadensis) survival in the northern Basin and Range ecosystem. Peer J. 2021;9:e10710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhao JY, Du YZ, Song YP, et al. Investigation of the Prevalence of Mycoplasma ovipneumoniae in Southern Xinjiang, China. J Vet Res. 2021;65(2):155–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Noll Lance W, Highland Margaret A, Hamill Vaughn A, et al. Development of a real-time PCR assay for detection and differentiation of Mycoplasma ovipneumoniae and a novel respiratory-associated Mycoplasma species in domestic sheep and goats. Transbound Emerg Dis. 2022;69(5). [DOI] [PMC free article] [PubMed]
- 8.Yang F, Han X, Tang C, et al. A Hsp70 Gene-Based PCR for Detection of Mycoplasma ovipneumoniae. J Anim Vet Adv. 2013;12:1495–7. [Google Scholar]
- 9.Yang F, Dao X, Rodriguez-Palacios A, et al. A Real-Time PCR for Detection and Quantification of Mycoplasma ovipneumoniae. J Vet Med Sci. 2014;76:1631–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Noll LW, Highland MA, Hamill VA, et al. Development of a Real-time PCR Assay for Detection and Differentiation of Mycoplasma ovipneumoniae and a Novel Respiratory-associated Mycoplasma Species in Domestic Sheep and Goats. Transbound Emerg Dis. 2022;69:1460–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang J, Cao J, Zhu M, et al. Loop-Mediated Isothermal Amplification-Lateral-Flow Dipstick (LAMP-LFD) to Detect Mycoplasma ovipneumoniae. WorldJ Microbiol Biotechnol. 2019;35:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen Q, Zhang L, Jiang F, et al. MnO2 Microsphere Absorbing Cy5-Labeled Single Strand DNA Probe Serving as Powerful Biosensor for Effective Detection of Mycoplasma ovipneumoniae. Sens Actuators B Chem. 2017;244:1138–44. [Google Scholar]
- 13.Wang JF, Li RW, Sun XX, et al. Development and validation of the isothermal recombinase polymerase amplification assays for rapid detection of Mycoplasma ovipneumoniae in sheep. BMC Vet Res. 2020;16(1):172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang H, Zhou BJ, Luo CB, et al. Cloning and partial prokaryotic expression of the gene heat shock protein Hsp70 of Mycoplasma ovipneumoniae. Acta Agriculturae Boreali-Occidentalis Sinica. 2014;23(04):130–4. [Google Scholar]
- 15.Aparicio D, Torres-puig S, Ratera M, et al. Mycoplasma genitalium adhesin P110 binds sialic-acid human receptors. Nat Commun. 2018;9(1):4471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xue GH, Cao L, Wang LP, et al. Evaluation of P1 adhesin epitopes for the serodiagnosis of Mycoplasma pneumoniae infections. FEMS Microbiol Lett. 2013;340(2):86–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu Y, Wang Q, Zhou B, et al. Sequence analysis of P113 gene of Mycoplasma ovipneumoniae strains epidemic in Guizhou province. Chinese J Vet Sci. 2016;36(05):756–62. [Google Scholar]
- 18.Zhang HR, Zhang XY, Yang FL, et al. Development of an Indirect ELISA for detection of Mycoplasma ovipneumoniae antibody in goats based on recombinant protein P113. Chinese J Prevent Vet Med. 2020;42(12):1244–9. [Google Scholar]
- 19.Rifatbegovic M, Maksimovic Z, Hulaj B. Mycoplasma ovipneumoniae associated with severe respiratory disease in goats. Vet Rec. 2011;168(21):565. [DOI] [PubMed] [Google Scholar]
- 20.Yi G, Kaiyu W, Qigui Y, et al. Descriptive study of enzootic nasal adenocarcinoma in goats in southwestern China. Transbound Emerg Dis. 2010;57(3):197–200. [DOI] [PubMed] [Google Scholar]
- 21.Chen S Y, Zhang M J, Tian R, et al. Establishment of An Indirect ELISA Method for Antibody Detection of Mycoplasma ovnipneumoniae Based on EF-Tu Protein. Chinese J Animal Infectious Dis. 2023:1–10.
- 22.Li QQ, Gao PC, Ge JZ, et al. Screening of specific diagnostic targets of Mycoplasma Ovnipneumoniae and establishment of the indirect ELISA. Chinese Veterinary Science. 2023;53(04):439–47. [Google Scholar]
- 23.Tian GY, Wang YC, Zhou YP, et al. Prokaryotic Expression and Indirect ELISA Establishment of P60_(58aa-928aa) Protein of Mycoplasma ovnipneumoniae. China Animal Husb Vet Med. 2022;49(05):1960–9. [Google Scholar]
- 24.Zhang MJ, Wang JJ, He MF, et al. Establishment of an indirect ELISA method for antibody detection based on Mycoplasma ovipneumoniae Enolase protein. Chinese Vet Sci. 2021;51(12):1490–7. [Google Scholar]
- 25.Yang Y, Wang H, Wang J, et al. Establishment of indirect ELISA method based on Mo Hsp70C terminal protein. Chinese J Vet Sci. 2018;38(11):2107–13. [Google Scholar]
- 26.Wang X, Zhang XY, Zhang JH, et al. Development of an indirect ELISA with the main antigentic region of Mycoplasma ovipneumoniae P130 protein expressed in E.coli. Chinese Vet Sci. 2018;48(03):281–7. [Google Scholar]
- 27.Jiang JX, Zhang JP, Lin YS, et al. Establishment of a TaqMan Assay for Mycoplasma ovipneumoniae based on the Hsp70 Gene and Analysis of Its Genetic Evolution. Acta Veterinaria et Zootechnica Sinica. 2024;55(04):1684–95. [Google Scholar]
- 28.Chen RC, Shang HQ, Niu XY, et al. Establishment and evaluation of an indirect ELISA for detection of antibodies to goat Klebsiella pneumonia. BMC Vet Res. 2021;17(1):107–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the data generated or analysed during this study are included in this manuscript. The sequence of the recombinant protein of the Hsp70-P113 double gene was deposited in the National Microbiology Data Center (NMDC) with accession number NMDCP0000170. URL is https://nmdc.cn/resource/genomics/protein/detail/NMDCP0000170.