Summary
Substantial changes in energy metabolism are a hallmark of pancreatic cancer. To adapt to hypoxic and nutrient-deprived microenvironments, pancreatic cancer cells remodel their bioenergetics from oxidative phosphorylation to glycolysis. This bioenergetic shift makes mitochondria an Achilles' heel. Since mitochondrial function remains essential for pancreatic cancer cells, further depleting mitochondrial energy production is an appealing treatment target. However, identifying effective mitochondrial targets for treatment is challenging. Here, we developed an approach, mitochondria-targeted cancer analysis using survival and expression (mCAUSE), to prioritize target proteins from the entire mitochondrial proteome. Selected proteins were further tested for their impact on pancreatic cancer cell phenotypes. We discovered that targeting a dynamin-related GTPase, OPA1, which controls mitochondrial fusion and cristae, effectively suppresses pancreatic cancer activities. Remarkably, when combined with a mutation-specific KRAS inhibitor, OPA1 inhibition showed a synergistic effect. Our findings offer a therapeutic strategy against pancreatic cancer by simultaneously targeting mitochondria dynamics and KRAS signaling.
Subject areas: Molecular biology, Cell biology, Cancer
Graphical abstract

Highlights
-
•
We developed a method to identify mitochondrial targets for mitigating cancer
-
•
OPA1 inhibition suppresses pancreatic cancer phenotypes
-
•
Cristae integrity is crucial for pancreatic cancer cell proliferation
-
•
OPA1 and KRAS inhibitors synergistically inhibit pancreatic cancer cells
Molecular biology; Cell biology; Cancer
Introduction
Pancreatic cancer is the fourth leading cause of cancer-associated death, with high levels of metastasis.1,2,3,4,5,6 Its average 5-year survival rate is approximately 10%, and with predicted increases in its occurrence, pancreatic cancer is estimated to become the second leading cause of cancer-associated death by 2030.7 Current therapies are not effective enough for patients with pancreatic cancer. There is an urgent need to identify therapeutic interventions for this devastating cancer.
Genetic mutations predominantly drive pancreatic cancer tumorigenesis and progression, especially in the four major oncogenes and tumor suppressors: KRAS, p16/CDKN2A, TP53, and DPC4/SMAD4.1,2,3,4 KRAS mutations are one of the earliest critical events in the development of pancreatic cancer and occur in more than 80% of cases.1,2,3,4 Downstream signaling of KRAS is mediated by two major protein kinases, ERK and AKT, which control cell proliferation and metastasis. In addition to intracellular signal transduction, during the development of pancreatic cancer, cellular energy metabolism shifts from oxidative phosphorylation to glycolysis to fuel rapid and massive cancer cell proliferation.8,9,10 Notably, even after an increase in glycolysis, oxidative phosphorylation remains essential for generating ATP in pancreatic cancer, albeit at a reduced rate, to ensure the growth and viability of cancer cells.9,10,11,12 These unique bioenergetic characteristics present an excellent opportunity to target mitochondria as a therapeutic strategy against pancreatic cancer.9,10,11,12 Since the mitochondrial proteome contains more than 1000 proteins,13 identifying effective targets poses a significant challenge to developing mitochondria-focused strategies to alleviate pancreatic cancer.
In this study, we address this challenge by introducing a method termed Mitochondria-targeted Cancer Analysis Using Survival and Expression (mCAUSE). This method prioritizes target proteins based on their gene expression levels and correlation with the survival rates of patients with pancreatic cancer.
Results
Development of mitochondria-targeted cancer analysis using survival and expression
To understand how the survival of patients with pancreatic cancer is correlated with the expression of genes that encode mitochondrial proteins (termed mito-genes), we first retrieved all 1136 mito-genes listed in human MitoCarta3.013 (Figure 1A). Using the Cancer Genome Atlas (TCGA)14 and OncoDB,15 we divided the cancer cases into a “high” or “low” expression group for each mito-gene based on a median 50% threshold of mRNA levels (Table S1, Total analyzed). The analysis excluded 13 mitochondrial DNA-encoded genes that were not included in the NCBI RefSeq curated database, as well as 7 nuclear DNA-encoded genes that were not part of OncoDB. We then utilized a log rank test to compare the survival between these two groups. This analysis revealed that 219 mito-genes are linked to significant changes in patient survival (Table S1, HR altered). Of these 219, higher levels of 45 mito-genes were associated with compromised survival (Hazard Ratio >1, Blue line, Figure 1A) (Table S1, HR > 1). In contrast, higher levels of 174 mito-genes were associated with better survival (Hazard Ratio <1, Red line, Figure 1A) (Table S1, HR < 1).
Figure 1.
Mitochondria-targeted cancer analysis using survival and expression
(A) Out of 25,000 proteins in the human proteome, MitoCarta3.0 identified 1136 mitochondrial proteins. Of these, 219 mito-genes exhibited a significant difference in hazard ratio (HR). Specifically, 45 mito-genes were associated with decreased survival in patients with pancreatic cancer when expressed at increased levels. Among these, category A includes 39 genes expressed at higher levels in tumors than in normal tissues. In contrast, when expressed at increased levels, 174 mito-genes were associated with better survival. Category C includes 72 of these genes that are also expressed at higher levels in tumors, while category D comprises 17 genes that are expressed at lower levels in tumors. GO enrichment analysis was performed by PANTHER with Bonferroni correction. Biological processes with a p-value less than 0.05 and greater than 20-fold enrichment are presented.
(B and C) PANC-1 cells (B) and MIA PaCa-2 cells (C) were cultured in the presence of the indicated inhibitors for 72 h. Cell density was determined using a crystal violet assay (mean ± SD, n≥ 10). ANOVA with post-hoc Tukey: ∗p < 0.05, ∗∗∗p < 0.001.
To determine whether and how the expression of these 219 genes is altered in pancreatic cancer compared to normal tissues, we analyzed individual mRNA levels and grouped them into four categories using TCGA and OncoDB (Table S2). In the group of 45 mito-genes with a hazard ratio greater than 1, the expression of 39 of these genes was significantly altered, showing a more than 2-fold increase compared to normal tissues. All of these genes were upregulated in tumors (Figure 1A, Category A) (Table S3). We were most interested in this first category (A); these genes potentially exhibit oncogenic properties, and lowering their expression or inhibiting their function might mitigate pancreatic cancer. Therefore, these mito-genes are strong candidates for drug targets to treat pancreatic cancer. However, it should also be noted that such increased levels may be the consequence of other changes in tumors. Gene ontology (GO) analysis showed that two pathways are significantly enriched, including mitochondrial translation (nine genes) and mitochondrial membrane organization (five genes) (Figure 1A) (Table S4). The second category (B) describes genes that are significantly decreased in their expression levels by more than half, yet higher expression levels are associated with poor survival. We expected to find fewer genes in this category as the expression-survival relationship appears somewhat conflicting. Indeed, we found no mito-genes in this category (Figure 1A).
In the group of 174 mito-genes with a hazard ratio smaller than 1, the expression of 89 mito-genes was significantly altered by greater than 2-fold compared to normal tissues. The third category (C) contains 72 genes that are higher in their expression in pancreatic cancer (Figure 1A) (Table S4). GO analysis indicated that three pathways are significantly enriched, including the regulation of mitochondrial translation (four genes), mitochondrial electron transport chain complex I assembly (nine genes), and mitochondrial RNA metabolic processes (five genes) (Table S4). The fourth category (D) has genes that are lower in expression, but increased levels are linked to better survival (Figure 1A) (Table S4). Seventeen mito-genes were found in this category. We consider that these genes might function as tumor suppressors. While these correlations may be secondary consequences of pancreatic cancer, it is also possible that boosting their function would decrease pancreatic cancer growth or improve patient survival. GO analysis revealed one pathway significantly enriched in this category: mitochondrial gene expression (four genes). Thus, mCAUSE divided mito-genes into four categories, identifying 39 genes with potential oncogenic features (category A) and 17 genes that may function such as tumor suppressors (category D).
Evaluation of mitochondria-targeted cancer analysis using survival and expression
To evaluate the usefulness of mCAUSE, we tested the impact of the identified genes on pancreatic cancer cells. Specifically, we focused on the genes in category (A), which show potential oncogenic characteristics, using two pancreatic cancer cell lines with different genetic makeups: PANC-1 and MIA PaCa-2 cells (Figures 1B and 1C). We blocked two enriched pathways — mitochondrial translation by chloramphenicol and mitochondrial membrane organization by MYLS2216,17 (which inhibits OPA1, a mitochondrial dynamin-related GTPase that controls mitochondrial fusion and cristae structure). We also inhibited gene products in category (A) that available drugs can pharmacologically target. These inhibitors include R16218 to inhibit the glutamate dehydrogenase GLUD1, YMU119 to inhibit thymidylate kinase DTYMK, Bl-6C920 to inhibit the apoptotic protein BID, AP5A to inhibit adenylate kinase AK4, Fidarestat to inhibit the aldo-keto reductase AKR1B10, ML348 to inhibits the lysophospholipase LYPLA1, and FC9402 to inhibit the sulfide quinone oxidoreductase SQOR. We used concentrations commonly employed for each chemical. PANC-1 and MIA PaCa-2 cells were cultured for 48 h in the presence of these inhibitors and analyzed for their cell density using crystal violet staining (Figures 1B and 1C). We found that the OPA1 inhibitor MYLS22 induces a significant and large decrease in cell density in both PANC-1 and MIA PaCa-2 cells. In contrast, the mitochondrial translation inhibitor chloramphenicol induced modest reductions in cell density only in MIA PaCa-2 cells. Other inhibitors did not decrease cell density in either cell line.
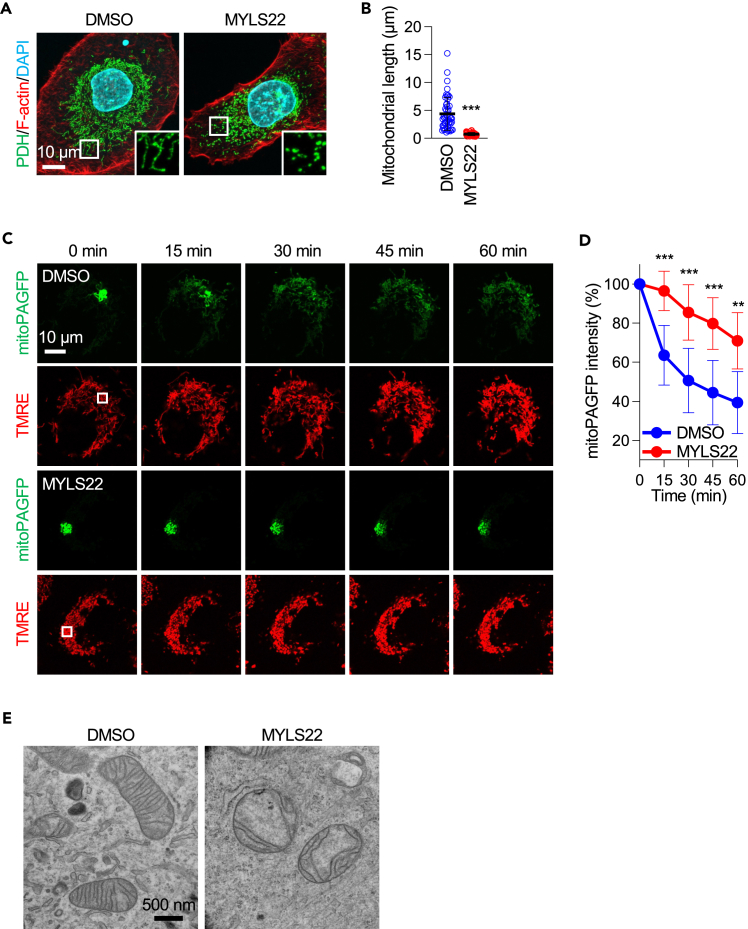
MYLS22 blocks mitochondrial fusion and disorganizes cristae
As described above, OPA1 controls mitochondrial fusion and cristae formation.21 To confirm the effect of MYLS22 on these processes in PANC-1 cells, we analyzed mitochondrial morphology, dynamics, and ultrastructure. First, PANC-1 cells were treated with MYLS22 for 24 h and subjected to laser confocal immunofluorescence microscopy using antibodies against the mitochondrial protein pyruvate dehydrogenase (PDH) (Figures 2A and 2B). In control cells, mitochondria displayed short tubular structures with occasional branches. In contrast, MYLS22-treated cells contained highly fragmented, small mitochondria (Figures 2A and 2B). Mitochondrial fragmentation likely results from decreased mitochondrial fusion, leading to unopposed, excess mitochondrial division. Second, to more directly test whether MYLS22 suppresses mitochondrial fusion in PANC-1 cells, we utilized a mitochondrial fusion assay involving live-cell imaging with matrix-targeted photoactivatable GFP (mitoPAGFP), as we performed22 (Figures 2C and 2D). PANC-1 cells were infected with lentiviruses carrying mitoPAGFP and treated with MYLS22 for 24 h. Following treatment, the cells were stained with tetramethylrhodamine ethyl ester (TMRE) to visualize the entire mitochondrial network. After the photoactivation of mitoPAGFP in a small portion of the mitochondria, we monitored the mixing of the fluorescent matrix marker through mitochondrial fusion. In control PANC-1 cells, the fluorescence intensity of mitoPAGFP gradually decreased over 60 min to approximately 40% of the initial intensity (Figures 2C and 2D). In contrast, treatment with MYLS22 significantly slowed the diffusion of the photoactivated mitoPAGFP among mitochondria. These data show that MYLS22 effectively decreases mitochondrial fusion. Third, to determine if MYLS22 affects cristae structure in PANC-1 cells, we performed transmission electron microscopy. In control cells, the mitochondrial inner membrane displayed well-developed cristae structures (Figure 2E). In contrast, cells treated with MYLS22 showed reduced and disorganized cristae (Figure 2E). Taken together, these data indicate that MYLS22 effectively inhibits mitochondrial fusion and cristae formation.
Figure 2.
MYLS22 blocks mitochondrial fusion and disorganizes inner membrane cristae
(A) Mitochondrial morphology. PANC-1 cells were treated with DMSO or MYLS22 for 24 h and subjected to laser confocal immunofluorescence microscopy with anti-PDH antibodies. The boxed regions are magnified.
(B) Quantification of mitochondrial length (mean ± SD, n = 50).
(C) Mitochondrial fusion. PANC-1 cells expressing mitoPAGFP were incubated with DMSO or MYLS22 for 24 h. Subsequently, cells were stained with 5 nM TMRE. MitoPAGFP was photoactivated using a 405 nm laser in a small region indicated by a square at 0 min. Observations were made at 15-min intervals over 60 min.
(D) The fluorescence intensity of mitoPAGFP in the photoactivated region was quantified (mean ± SD, n > 9).
(E) PANC-1 cells were treated with DMSO or MYLS22 for 24 h and subjected to transmission electron microscopy. Student’s t-test in (B) and ANOVA followed by Šídák’s test in (D): ∗∗p < 0.01, ∗∗∗p < 0.001.
Inhibiting normal cristae formation, but not mitochondrial fusion, suppresses cell proliferation
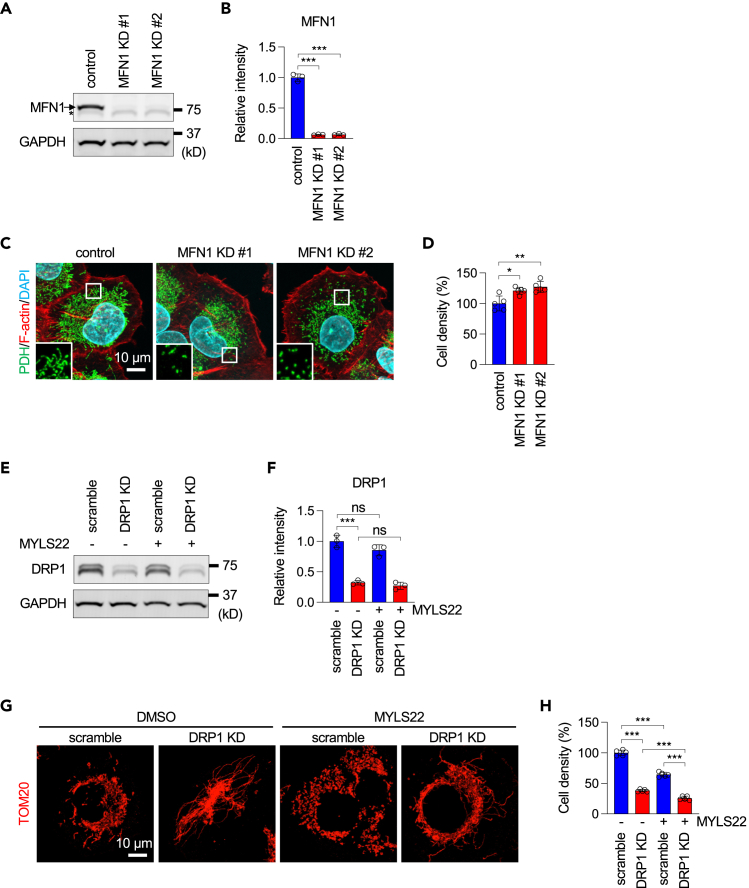
To determine which function of OPA1 is critical for suppressing cell proliferation in PANC-1 cells, we knocked down another dynamin-related GTPase, MFN1, which mediates mitochondrial fusion but not cristae formation, using two different siRNAs. Western blotting showed a significant reduction in MFN1 protein levels by these siRNAs (Figures 3A and 3B). Immunofluorescence microscopy showed the fragmentation of mitochondria due to the inhibition of mitochondrial fusion (Figure 3C). However, MFN1 knockdown did not reduce PANC-1 cell proliferation (Figure 3D). These data suggest that mitochondrial fusion is dispensable for the proliferation of PANC-1 cells.
Figure 3.
Mitochondrial fusion is dispensable for cell proliferation
(A) Western blotting of PANC-1 cells treated with non-targeting control or two different MFN1 siRNAs for 5 days. An arrow and an asterisk indicate MFN1 and non-specific bands, respectively.
(B) Quantification of band intensity (mean ± SD, n = 3).
(C) PANC-1 cells were transfected with the indicated siRNAs and subjected to laser confocal immunofluorescence microscopy with anti-PDH antibodies. The boxed regions are magnified.
(D) Cell proliferation of the transfectants was examined by a crystal violet assay (mean ± SD, n = 5).
(E) PANC-1 cells were infected with lentiviruses expressing scramble or DRP1 shRNAs and then treated with DMSO or MYLS22 for 24 h. Knockdown of DRP1 was confirmed by Western blotting.
(F) Quantification of band intensity (mean ± SD, n = 3).
(G) Mitochondria were analyzed by immunofluorescence microscopy with anti-TOM20 antibodies.
(H) Cell proliferation was analyzed by a crystal violet assay (mean ± SD, n = 5). ANOVA with post-hoc Tukey in (B, D, F, H): ∗p < 0.05, ∗∗∗p < 0.01, ∗∗∗p < 0.001.
To further confirm this notion, we blocked mitochondrial division by using shRNA knockdown of DRP1 in PANC-1 cells treated with MYLS22 (Figures 3E and 3F). We found that mitochondrial fragmentation is rescued by DRP1 knockdown (Figure 3G). However, rescuing mitochondrial fragmentation failed to rescue cell proliferation defects caused by MYLS22: knockdown of DRP1 decreased cell proliferation, consistent with a previous study,23 and the combination of DRP1 knockdown and MYLS22 showed further reduction (Figure 3H).
To test the effect of cristae formation inhibition, we knocked down MIC60, a vital component of the MICOS complex responsible for cristae formation,24,25 using two different siRNAs. We confirmed the knockdown using Western blotting (Figures 4A and 4B). Electron microscopy showed the loss of normal cristae structure in MIC60 knockdown cells, consistent with previous reports24,25 (Figure 4C). MIC60 knockdown resulted in significant decreases in cell proliferation for both siRNAs (Figure 4D). Furthermore, we treated PANC-1 cells with a MIC60 inhibitor, miclxin.26 Results showed that miclxin disorganizes inner membrane cristae (Figure 4E) and blocks cell proliferation in a dose-dependent manner (Figure 4F). These data suggest that cristae morphology plays a crucial role in the proliferation of PANC-1 cells.
Figure 4.
Cristae are important for cell proliferation
(A) Western blotting of PANC-1 cells treated with non-targeting control or two different MIC60 siRNAs for 5 days.
(B) Quantification of band intensity (mean ± SD, n = 3).
(C) Mitochondria were analyzed in PANC-1 cells transfected with the indicated siRNAs by electron microscopy.
(D) PANC-1 cells were transfected with the indicated siRNAs. Cell proliferation was assessed using a crystal violet assay (mean ± SD, n = 5).
(E) Mitochondria were analyzed in PANC-1 cells treated with DMSO or 10 μM miclxin by electron microscopy.
(F) PANC-1 cells were cultured with 0–50 μM miclxin for 72 h. Cell density was assessed using a crystal violet assay (mean ± SD, n = 5). ANOVA with post-hoc Tukey in (B, D): ∗∗∗p < 0.001.
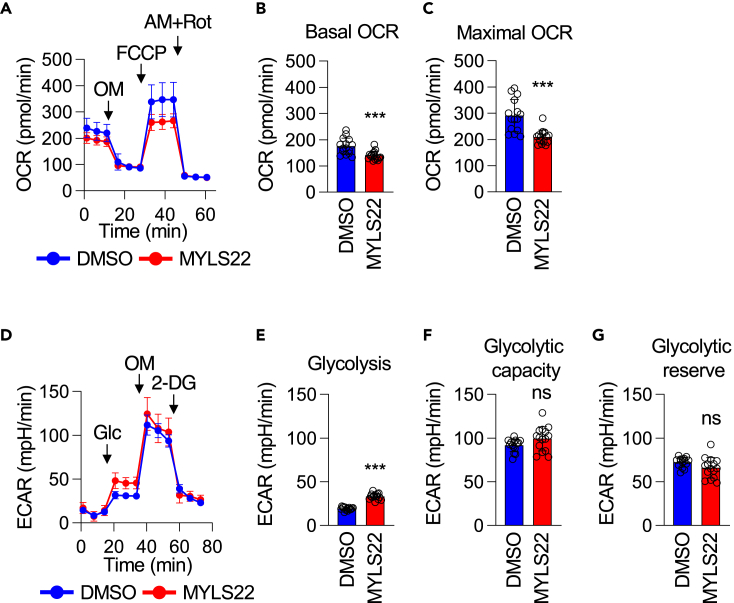
MYLS22 decreases mitochondrial respiration
Since cristae are important for efficient mitochondrial oxidative phosphorylation, we investigated how MYLS22 affects oxygen consumption rates (OCRs) in PANC-1 cells. We found that MYLS22 strongly decreases both basal OCRs and respiratory capacity (Figures 5A–5C). Since reduced mitochondrial energy production can lead to a compensatory upregulation of glycolysis, we also measured the extracellular acidification rates (ECARs) for glycolysis. Indeed, we observed an increase in glycolysis in MYLS22-treated PANC-1 cells (Figures 5D and 5E). This increase did not affect glycolytic capacity or reserve (Figures 5F and 5G). Therefore, MYLS22 decreases mitochondrial respiration but not cytosolic glycolysis in PANC-1 cells.
Figure 5.
MYLS22 alters energy metabolism
(A) Mitochondrial respiration was analyzed by measuring the OCRs in PANC-1 cells treated with MYLS22.
(B and C) Both basal and maximal OCRs are presented (mean ± SD, n = 15).
(D–G) Glycolysis was assessed by measuring the ECARs in the same set of cells. Basal glycolysis (E), glycolytic capacity (F), and glycolytic reserve (G) are shown (mean ± SD, n = 13). Student’s t-test in (B, C, E, F, G): ∗∗∗p < 0.001.
MYLS22 decreases spheroid growth and cell motility
To further analyze the impact of MYLS22 on pancreatic cell phenotypes, we utilized a three-dimensional spheroid growth model, which closely represents physiological conditions.27 We generated spheroids of PANC-1 cells using the hanging drop method and placed the spheroids in a 3D meshwork of the extracellular matrix using Geltrex.27 Control spheroids showed substantial growth at 5 days and invasion at 7 days (Figures 6A and 6B). In contrast, spheroids treated with MYLS22 displayed significantly decreased growth and no invasion. Inhibition of invasion in spheroids by MYLS22 (Figures 6A and 7 days) suggested that MYLS22 suppresses migratory activities of PANC-1 cells. To directly test this notion, we examined cell motility using a wound-healing assay (Figures 6C and 6D). In this assay, we plated cells and cultured them to 100% confluency. Then, a line of scratches was made with small pipette tips. The migration of cells into the scratched width was monitored over 72 h in the presence or absence of 50 μM MYLS22. MYLS22 significantly decreased cell migration (Figures 6C and 6D).
Figure 6.
OPA1 inhibition suppresses spheroid growth and cell migration
(A) Spheroid growth. Spheroids of PANC-1 cells were cultured in Geltrex for 7 days in the presence of DMSO or 50 μM MYLS22.
(B) Quantification of the total and invasion areas (mean ± SD, n > 20).
(C) Cell migration. Motility of PANC-1 cells was analyzed using a wound-healing assay. Representative images at 0 h and 72 h are shown.
(D) Wound closure was assessed by determining the relative width over the initial width (mean ± SD, n = 5). Student’s t-test in (B, D): ∗∗p < 0.01, ∗∗∗p < 0.001.
Figure 7.
MYLS22 promotes the phosphorylation of AKT and ERK in a KRAS-dependent manner
(A) PANC-1 cells were treated with DMSO, MRTX1133, MYLS22, or both for 24 h. Whole-cell lysates were subjected to Western blotting using the indicated antibodies.
(B) Quantification of band intensity (mean ± SD, n = 3). ANOVA with post-hoc Tukey: ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
MYLS22 stimulates the phosphorylation of AKT and ERK
Mutated, constitutively active KRAS mutations are the primary driver of pancreatic cancer.1,2,3,4 Activated KRAS signaling promotes mitochondrial division by phosphorylating the division GTPase DRP1 through one of the major downstream kinases, ERK1/2.28,29 However, the functional relationships between the fusion GTPase OPA1 and the KRAS pathway are not well understood. Therefore, we were interested in determining if MYLS22 affects this oncogenic pathway. Since the phosphorylation of ERK1/2 and AKT are two major downstream events in the KRAS signaling pathway, we tested whether these phosphorylation events are affected by MYLS22. Specifically, we examined ERK 1/2 phosphorylation at Thr202/Tyr204 and AKT phosphorylation at Ser473 and Thr30830,31,32,33 (Figures 7A and 7B). We found a significant increase in the phosphorylation of both ERK1/2 and AKT following MYLS22 treatment (Figures 7A and 7B).
To determine whether the increased phosphorylation of ERK1 and AKT is dependent on KRAS, we added MRTX1133, an allele-specific KRAS inhibitor targeting the oncogenic G12D mutation, along with MYLS22, as PANC-1 cells carry this KRAS mutation.34 The G12D mutation is the most frequent KRAS mutation, accounting for approximately 40% of pancreatic cancer cases. MRTX1133 is currently undergoing Phase 2 clinical trials for the treatment of solid tumor malignancies harboring this mutation, including pancreatic cancer. We found that MRTX1133 strongly blocks ERK1/2 phosphorylation induced by MYLS22 (Figures 7A and 7B). AKT phosphorylation was also suppressed by MRTX1133 (Figures 7A and 7B). These results indicate that MYLS22 affects KRAS signaling in PANC-1 cells. The elevated levels of ERK1/2 and AKT phosphorylation by MYLS22 may be an adoptive response to the decreased cell proliferation and migration resulting from mitochondrial deficits.
Effects of combined treatments with MYLS22 and MRTX1133 on cell proliferation and spheroid growth
If increased KRAS signaling is a compensatory mechanism in the presence of OPA1 inhibition, concurrently inhibiting OPA1 and KRAS signaling could be an effective strategy against pancreatic cancer cells. To test our hypothesis, we treated PANC-1 cells with MRTX1133 in the presence or absence of MYLS22. First, we confirmed that MRTX1133 inhibits the proliferation of PANC-1 cells in a dose-dependent manner (Figure 8A) and determined its IC50 to be approximately 30 μM (Figure 8C). To test whether the presence of MYLS22 changes the IC50 of MRTX1133, we included a sub-effective concentration of MYLS22 (12.5 μM, Figure 8B, arrow) and found that 12.5 μM MYLS22 significantly decreases the IC50 of MRTX1133 in the proliferation of PANC-1 cells (Figure 8C). These data suggest that MRTX1133 and MYLS22 have a synergistic effect on cell proliferation. To further explore the combined effects of MRTX1133 and MYLS22, we incubated the spheroids with low doses of MRTX1133 (4 nM) and MYLS22 (1 μM). At these concentrations, individual drug treatments showed no significant effects on the growth or invasion of spheroids (Figures 8D and 8E). In contrast, the combination of these drugs demonstrated synergistic effects in inhibiting spheroid growth and dissemination (Figures 8D and 8E). These data taken together suggest that combining the inhibition of KRAS and OPA1 provides an effective strategy to counteract pancreatic cancer phenotypes.
Figure 8.
Synergistic effects of MRTX1133 and MYLS22 on cell proliferation and spheroid growth
(A and B) PANC-1 cells were cultured with 0–50 μM of MRTX1133 (A) or MYLS22 (B) for 72 h. Cell density was assessed using a crystal violet assay (mean ± SD, n = 5).
(C) The IC50 of MRTX1133 for cell proliferation was evaluated in the presence or absence of a sub-effective concentration of MYLS22 (12.5 μM, indicated by an arrow in panel B) (mean ± SD, n = 5).
(D) Spheroid growth. Spheroids of PANC-1 cells were cultured in Geltrex for 7 days in the presence or absence of 4 nM MRTX1133 and/or 1 μM MYLS22.
(E) Quantification of the total and invasion areas (mean ± SD, n > 20). Student’s t-test in (C) and ANOVA with post-hoc Tukey in (E): ∗p < 0.05, ∗∗∗p < 0.001.
Discussion
In the current work, we have developed mCAUSE, a method to identify mitochondrial proteins that could be targeted to mitigate phenotypes in cancer cells. This identification is based on a correlation analysis between patient survival and gene expression using publicly available datasets. We found 39 genes with potential oncogenic features (category A) and 17 genes that may function such as tumor suppressors (category D). We showed that pharmacologically targeting the dynamin-related GTPase OPA1 identified in category A successfully suppresses various pancreatic cancer phenotypes. These phenotypes include cell proliferation in both 2D and 3D cultures, cell migration, mitochondrial bioenergetics, and alterations in mitochondrial morphology and dynamics. Our data support and extend a previous study that reported genetic OPA1 knockdown using siRNAs suppresses spheroid growth and anchorage-independent growth in PANC-1 cells.35 Furthermore, we show that the inhibition of OPA1 produces a synergistic effect when combined with a mutation-specific KRAS inhibitor, significantly reducing cell proliferation and spheroid growth. Our findings propose a therapeutic approach for treating cancer by simultaneously targeting mitochondrial function and KRAS signaling.
Mitochondria are dynamic organelles that actively fuse and divide to regulate their structure.21,36,37,38 Mitochondrial fusion is controlled by inner-membrane-located OPA1 and outer-membrane-located GTPases, mitofusins.21 In contrast, division is mediated by another GTPase, DRP1. During KRAS-driven oncogenic transformation, the mitochondrial division is upregulated, resulting in smaller mitochondria.28,29 This may suggest that further tilting the balance between fusion and division against fusion critically decreases mitochondrial functional competence. Interestingly, however, we found that the inhibition of mitochondrial fusion by knocking down MFN1 causes the fragmentation of mitochondria but does not inhibit cell proliferation in PANC-1 cells. Therefore, PANC-1 cells appear to be independent of mitochondrial fusion for their proliferation. Instead, our data suggest that inner membrane cristae are important for pancreatic cancer cell proliferation. Since cristae are involved in multiple processes in mitochondria including ATP production, metabolic reprogramming, and ROS production, it would be of great interest to further define the role of cristae in pancreatic cancer. Supporting this notion, we found that MYLS22 inhibits OCRs in PANC-1 cells.
Another pathway enriched in category A is mitochondrial translation. Mitochondrial energy production is critical for the proliferation of pancreatic cancer, even though glycolysis is enhanced. Therefore, enhanced mitochondrial translation may help produce cellular ATP to support the high energy demands of rapidly proliferating cancer cells, even under conditions where glycolysis is also upregulated. However, we found that chloramphenicol, which inhibits mitochondrial translation, suppresses the growth of MIA PaCa-2 cells but not PANC-1 cells. Our findings suggest that the potential oncogenic roles of mitochondrial translation may be specific to subpopulations of pancreatic cancer cells.
Components associated with mitochondrial gene expression are enriched as potential tumor suppressors in category D. These components can also affect mitochondrial energy production and metabolism. One possibility that explains this enrichment is that specific metabolic pathways or bioenergetic intermediates play a crucial role in tumor suppression. Alternatively, mitochondrial gene expression can affect nuclear gene expression for tumor suppressor genes through their inter-organelle crosstalk.39 In future studies, we are interested in defining specific mechanisms that could affect pancreatic cancer through this pathway.
The potential of mCAUSE extends beyond pancreatic cancers. It has the potential to map the landscape of mitochondrial proteins that may be inhibited to control cancer cell phenotypes in vitro and in vivo across various types of cancers. Considering the existence of around 30 different cancer types, mCAUSE offers a promising navigational tool. It can effectively prioritize potential targets for cancer treatment, filtering through the extensive combinations of mitochondrial proteins and cancer types. This method, therefore, represents a significant stride forward in personalized cancer therapy and offers a wide array of future research and clinical applications.
Limitations of the study
In the current work, our investigation was limited to an in vitro culture system. Extending our findings to animal models to explore tumor mitigation in vivo would be of great interest. Additionally, testing the efficacy of targeting mitochondrial structure and dynamics alongside KRAS in human primary tumors represents another crucial future direction. Such investigations could potentially lead to the development of therapeutic strategies that control mitochondrial bioenergetics and intracellular signaling in this devastating cancer. Beyond pancreatic cancer, we aim to extend mCAUSE to other, and possibly all, cancer types to identify effective mitochondrial targets for treating each one.
Resource availability
Lead contact
Further information and requests for resources should be directed to and will be fulfilled by the lead contact, Hiromi Sesaki (hsesaki@jhmi.edu).
Materials availability
Cell lines and plasmids constructed in this study are available from the lead contact upon request.
Data and code availability
-
•
All data reported in this article will be shared by the lead contact upon request.
-
•
This article does not report original code.
-
•
Any additional information required to reanalyze the data reported in this article is available from the lead contact upon request.
Acknowledgments
We thank members of the Iijima and Sesaki labs for invaluable discussions and technical assistance. We are also grateful to Dr. Lei Zheng for his insightful and valuable comments on the article. This work was supported by NIH grants to MI (GM131768), AJE (U54CA26808), and HS (GM144103) by a Black in Cancer/Emerald Foundation Postdoctoral Fellowship to JJW, a Breast Cancer Research Foundation (BCRF-22-048) grant to AJE, and a Sol Goldman Pancreatic Cancer Research Grant to HS.
Author contributions
DM, FI, WI, MI, and HS designed the study. DM, FI, GT, and WI performed experiments. DM, FI, WI, NY, XW, JJW, AJE, MI, and HS analyzed and discussed data. DM, FI, WI, GT, NY, MI, and HS wrote and reviewed the article.
Declaration of interests
AJE has unlicensed patents related to keratin 14 as a prognostic marker and antibody strategy for anti-cancer therapeutics. AJE is a consultant for BioNTech. AJE’s spouse is an employee of ImmunoCore.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-PDH | Abcam | Cat# ab110333; RRID: AB_10862029 |
| Rabbit polyclonal anti-MFN1 | Proteintech | Cat# 13798-1-AP; RRID: AB_2266318 |
| Mouse monoclonal anti-GAPDH | Thermo Fisher Scientific | Cat# MA5-15738; RRID: AB_10977387 |
| Mouse monoclonal anti-DRP1 | BD Biosciences | Cat# 611113; RRID:AB_398424 |
| Rabbit polyclonal anti-TOM20 | Proteintech | Cat# 11802-1-AP; RRID: AB_2207530 |
| Rabbit polyclonal anti-MIC60 | Proteintech | Cat# 10179-1-AP; RRID: AB_2127193 |
| Rabbit polyclonal anti-AKT | Cell Signaling Technology | Cat# 9272; RRID: AB_329827 |
| Rabbit monoclonal anti-pAKT(S473) | Cell Signaling Technology | Cat# 4060; RRID: AB_2315049 |
| Rabbit monoclonal anti-pAKT(T308) | Cell Signaling Technology | Cat# 2965; RRID: AB_2255933 |
| Mouse monoclonal anti-ERK1/2 | Cell Signaling Technology | Cat#9107; RRID: AB_10695739 |
| Rabbit monoclonal anti-pERK1/2 | Cell Signaling Technology | Cat#4370; RRID: AB_2315112 |
| Alexa 488-conjugated anti-mouse IgG | Thermo Fisher Scientific | Cat# A21202; RRID: AB_141607 |
| Alexa 488-conjugated anti-rabbit IgG | Thermo Fisher Scientific | Cat# A21206; RRID: AB_2535792 |
| Alexa 568-conjugated anti-rabbit IgG | Thermo Fisher Scientific | Cat# A10042; RRID: AB_2534017 |
| Alexa 647-conjugated anti-rabbit IgG | Thermo Fisher Scientific | Cat# A31573; RRID: AB_2536183 |
| Chemicals, peptides, and recombinant proteins | ||
| Dulbecco’s Modified Eagle’s Medium (DMEM) – high glucose | Sigma-Aldrich | Cat# D5796 |
| Fetal Bovine Serum (FBS) | Corning | Cat# 35-010-CV |
| DPBS | Corning | Cat# 21-031-CV |
| 0.05% Trypsin-EDTA | Gibco | Cat# 25300-054 |
| DMSO | Corning | Cat# 25-950-CQC |
| Chloramphenicol | Sigma-Aldrich | Cat# C0378-5G |
| MYLS22 | Med Chem Express | Cat# HY-136446 |
| R162 | Caymen | Cat# 30922 |
| YMU1 | Caymen | Cat# 21981 |
| BI-6C9 | Santa Cruz Biotechnology | Cat# sc-210915 |
| AP5A | Santa Cruz Biotechnology | Cat# sc-204156 |
| Fidarestat | Med Chem Express | Cat# HY-105185 |
| ML348 | Med Chem Express | Cat# HY-100736 |
| FC9402 | Med Chem Express | Cat# HY-141552 |
| Miclxin | Med Chem Express | Cat# HY-138301 |
| MRTX1133 | Fisher Scientific | Cat# NC2083191 |
| Crystal violet | Sigma-Aldrich | Cat# 65092A-95 |
| Acetic acid | Thermo Fisher Scientific | Cat# A38-212 |
| Paraformaldehyde | Sigma-Aldrich | Cat# P6148 |
| Triton X-100 | Sigma-Aldrich | Cat# T9284 |
| BSA | Sigma-Aldrich | Cat# A7906 |
| Alexa 568-conjugated phalloidin | Thermo Fisher Scientific | Cat# A12380 |
| DAPI | Roche | Cat# 10236276001 |
| Phusion High-Fidelity DNA Polymerase | NEB | Cat# M0530 |
| Quick Ligation Kit | NEB | Cat# M2200 |
| Lipofectamine 2000 Transfection Reagent | Thermo Fisher Scientific | Cat# 11668019 |
| Lipofectamine RNAiMAX Transfection Reagent | Thermo Fisher Scientific | Cat# 13778150 |
| Polybrene Infection/Transfection Reagent | MilliporeSigma | Cat# TR-1003-G |
| Tetramethylrhodamine, ethyl ester (TMRE) | Thermo Fisher Scientific | Cat# T669 |
| Sodium cacodylate, trihydrate | Electron Microscopy Sciences | Cat# 12300 |
| 25% glutaraldehyde | Electron Microscopy Sciences | Cat# 16220 |
| 4% Osmium Tetroxide (OsO4) | Electron Microscopy Sciences | Cat# 19190 |
| Potassium hexacyanoferrate(II), trihydrate | Sigma-Aldrich | Cat# P3289 |
| Uranyl acetate | Ted Pella, Inc. | Cat# 19481 |
| EMBED 812 RESIN | Electron Microscopy Sciences | Cat# 14900 |
| DDSA | Electron Microscopy Sciences | Cat# 13710 |
| NMA | Electron Microscopy Sciences | Cat# 19000 |
| DMP-30 | Electron Microscopy Sciences | Cat# 13600 |
| RIPA Buffer (10X) | Cell Signaling Technology | Cat# 9806S |
| cOmplete, Mini, EDTA-free Protease Inhibitor Cocktail | Roche | Cat# 11836170001 |
| Phosphatase Inhibitor Cocktail 2 | Sigma-Aldrich | Cat# P5726 |
| Phosphatase Inhibitor Cocktail 3 | Sigma-Aldrich | Cat# P0044 |
| Tween 20 | Sigma-Aldrich | Cat# P7949 |
| 4–20% Criterion TGX Precast Gels | Bio-Rad Laboratories | Cat# 5671095 |
| Seahorse XF Base Medium Minimal DMEM | Agilent | Cat# 102353-100 |
| L-Glutamine (200 mM) | Gibco | Cat# 25030081 |
| Sodium pyruvate (100 mM) | Sigma-Aldrich | Cat# S8636 |
| D-Glucose | Sigma-Aldrich | Cat# G7021 |
| Oligomycin | Sigma-Aldrich | Cat# O4876 |
| FCCP | Sigma-Aldrich | Cat# C2920 |
| Antimycin A | Sigma-Aldrich | Cat# A8674 |
| Rotenone | Sigma-Aldrich | Cat# R8875 |
| 2-Deoxy-D-glucose | MilliporeSigma | Cat# 25972-1GM |
| Methylcellulose | Sigma-Aldrich | Cat# M0512 |
| Geltrex | Thermo Fisher Scientific | Cat# A1413202 |
| Critical commercial assays | ||
| Seahorse XFe96/XF Pro FluxPak | Agilent | Cat# 103792-100 |
| Experimental models: Cell lines | ||
| Human: Panc-1 cells | ATCC | CRL-1469 |
| Human: HEK293T cells | ATCC | CRL-3216 |
| Oligonucleotides | ||
| siRNA: Negative control | Thermo Fisher Scientific | Cat# 4390843 |
| siRNA: MFN1 #1 | Thermo Fisher Scientific | Assay ID: s31218 |
| siRNA: MFN1 #2 | Thermo Fisher Scientific | Assay ID: s31220 |
| siRNA: MIC60 #1 | Thermo Fisher Scientific | Assay ID: s21633 |
| siRNA: MIC60 #2: | Thermo Fisher Scientific | Assay ID: s21634 |
| shRNA targeting sequence: scramble: CCTAAGGTTAAGTCGCCCTCG | Sarbassov et al.40 | Addgene Plasmid #1864 |
| shRNA targeting sequence: DRP1: GCTACTTTACTCCAACTTATT | Sigma-Aldrich | TRCN0000001097 |
| Recombinant DNA | ||
| pHR-CMV8.2ΔR | Stewart et al.41 | Addgene Plasmid #8455 |
| pCMV-VSVG | Stewart et al.41 | Addgene Plasmid #8454 |
| pPAGFP-N1 | Patterson et al.42 | Addgene Plasmid #11909 |
| pHR-SIN Su9-PAGFP | This paper | N/A |
| pLKO.1 scrambled shRNA | This paper | N/A |
| pLKO.1 DRP1 shRNA | This paper | N/A |
| Software and algorithms | ||
| MitoCarta3.0 | Rath et al., 202113 | www.broadinstitute.org/mitocarta |
| OncoDB | Tang et al., 202215 | https://oncodb.org |
| GTEx database | Consortium, 201343 | https://gtexportal.org/home/ |
| FIJI | NIH | https://imagej.net/software/fiji/downloads |
| Prism | GraphPad | https://www.graphpad.com/features |
| Seahorse Wave Controller Software | Agilent | https://www.agilent.com/en/product/cell-analysis/real-time-cell-metabolic-analysis/xf-software/seahorse-wave-controller-software-2-6-1-740904 |
Experimental model and study participant details
Cells
PANC-1 and MIA PaCa-2 cells were obtained from the American Type Culture Collection and were cultured in Dulbecco's Modified Eagle Medium (DMEM) containing 10% fetal bovine serum. For siRNA knockdown, PANC-1 cells were plated at a density of 100,000 cells/well in a 6-well plate and cultured for 24 h. The cells were transfected with siRNAs at 10 nM using Lipofectamine RNAiMAX (Invitrogen). The following Silencer Select siRNAs from Thermo Fisher were used: negative control (4390843), MFN1 #1 (s31218), MFN1 #2 (s31220), MIC60 #1 (s21633), and MIC60 #2 (s21634).
Method details
Data analysis
We selected 1136 mito-genes that encode mitochondrial proteins based on MitoCarta3.0.13 Using OncoDB,15 we retrieved gene expression profiles of these genes from pancreatic cancer samples in TCGA14 and normal pancreas tissues in the GTEx database.43 We employed Student's t-test followed by the Benjamini-Hochberg procedure to calculate adjusted p-values. For the survival analysis of pancreatic cancer, we applied the Kaplan-Meier model to estimate the survival probabilities along with the survival time. Based on RNA expression, we categorized the cancer cases into 'high' or 'low' expression groups using a median 50% threshold. We then used the log-rank test to compare the survival curves between these two groups. Additionally, we employed the Cox regression analysis to determine the hazard ratio associated with the expression levels of the selected genes. GO enrichment analysis of differentially expressed genes was carried out using PANTHER, and statistical significance was adjusted using Bonferroni correction. Biological processes that met the criteria of a p-value less than 0.05 and showed gene enrichment greater than 20-fold are presented.
Inhibitors
Chloramphenicol (C0378-5G, Sigma), MYLS22 (HY-136446, Med Chem Express), R162 (30922, Cayman), YMU1 (21981, Cayman), BI-6C9 (sc-210915, Santa Cruz Biotechnology), AP5A (sc-204156, Santa Cruz Biotechnology), Fidarestat (HY-105185, Med Chem Express), ML348 (HY-100736, Med Chem Express), FC9402 (HY-141552, Med Chem Express), MRTX1133 (NC2083191, Fisher Scientific), and Miclxin (HY-138301, Med Chem Express) were used.
Cell proliferation
PANC-1 and MIA PaCa-2 cells were plated at 3,000 cells/well in a 96-well plate and incubated at 37°C with 5% CO2 for 24 h. Cells were then treated with various inhibitors for 48 h. To determine IC50, PANC-1 cells were plated at 3,000 cells/well at in a 96-well plate and incubated at 37°C with 5% CO2 for 24 h. Cells were then treated with various inhibitors for 72 h. To examine the combined effect of DRP1 knockdown and MYLS22, PANC-1 cells were seeded at 100,000 cells/well in a 6-well plate and cultured for 24 h. The cells were then incubated with lentiviruses expressing either scramble or DRP1 shRNA in DMEM containing 10% FBS and 8 μg/ml polybrene for 24 h. The cells were subsequently cultured in DMEM containing 10% FBS for an additional 3 days. The cells were then replated at 10,000 cells/well in a 96-well plate and cultured for 24 h before being treated with 50 μM MYLS22 for an additional 2 days. For the siRNA knockdown of MFN1 and MIC60, PANC-1 cells were plated at a density of 100,000 cells/well in a 6-well plate and cultured for 24 h. The cells were transfected with siRNAs at 10 nM using Lipofectamine RNAiMAX (Invitrogen). After 3 days, the cells were replated at a density of 10,000 cells/well in a 96-well plate and cultured for 24 h. The cells were then transfected with siRNAs again and cultured for an additional 2 days. For crystal violet staining, the cells were fixed with 4% paraformaldehyde in PBS at room temperature for 20 min. After washing with MilliQ water, cell densities were assessed by staining with 0.35% crystal violet (65092A-95, Sigma Aldrich) for 20 min. After washing three times with water and air-drying, 150 μl of 10% acetic acid was added, and the absorbance was measured at 595 nm using a multimode microplate reader (CLARIOstar, BMG Labtech).
Spheroid growth
PANC-1 cells were cultured in a 10-cm dish in DMEM containing 10% FBS. The cells were washed with PBS and detached from the dish using 2 ml of pre-warmed 0.05% trypsin-EDTA for 5 min. After adding 5 ml of the culture medium, the cells were centrifuged twice in a 15 ml conical tube at 1500 rpm for 3 min and then pelleted. The cells were resuspended at 10,000 cells/ml in 10 ml of the culture medium containing 0.24% methylcellulose. A 20 μl aliquot of the cell suspension was placed on the bottom of the lid of a 10-cm dish filled with PBS and incubated for 2–4 days in a cell culture incubator at 37°C with 5% CO2 to allow spheroid formation.27 The spheroids were suspended in 2–3 ml of culture medium and transferred to a 15 ml conical tube coated with 3% BSA in PBS, using a 10 ml pipette coated with 3% BSA in PBS. The spheroids were washed three times in the culture medium at 1500 rpm for 3 s each time and resuspended at a concentration of 1 spheroid/μl in Geltrex (A1413202, Thermo Fisher). A 50-μl volume of the spheroid suspension was placed in 24-well plates and incubated at 37°C for 30 min to allow the Geltrex to solidify. Subsequently, 1 ml of the culture medium was added. The spheroids were then cultured for 7 days in a cell culture incubator at 37°C with 5% CO2. The culture medium was changed on day 3 or 4. Images of the spheroids were captured using phase-contrast microscopy (AXIO Observer Z1, Zeiss). The size of the total and their invasion area were calculated using NIH Fiji software.
Wound-healing assay
PANC-1 cells were seeded in a 12-well plate and cultured until they reached 100% confluence. A wound was created by scratching a line across the bottom of the dish through the confluent cell monolayer using a sterile P-200 pipette tip. The cells were then gently rinsed with PBS and treated with DMEM containing either DMSO or 50 μM MYLS22 for 72 h. Images were captured using phase-contrast microscopy (AXIO Observer Z1, Zeiss). The width of the scratches was measured using NIH Fiji software.
Western blotting
PANC-1 cells were harvested and lysed in RIPA buffer (9806S, Cell Signaling), supplemented with cOmplete, Mini, EDTA-free Protease Inhibitor Cocktail (11836170001, Roche) and Phosphatase Inhibitor Cocktail 2 and 3 (P5726 and P0044, Sigma Aldrich), while on ice.44 The lysates were centrifuged at 16,000 g for 10 min at 4°C, and the supernatants were collected. Proteins were separated using SDS-PAGE and subsequently transferred onto Immobilon-FL Transfer Membranes (Millipore). These membranes were blocked with PBS-T (PBS containing 0.05% Tween 20) and 3% BSA at room temperature for 1 h and were incubated with primary antibodies in PBS-T containing 3% BSA at 4°C overnight. The antibodies used were MFN1 (13798-1-AP; Proteintech), DRP1 (611113; BD Biosciences), MIC60 (10179-1-AP; Proteintech), AKT (9272, Cell Signaling), phospho-AKT at S473 (4060, Cell Signaling), phospho-AKT at T308 (13038, Cell Signaling), ERK1/2 (9107, Cell Signaling), phospho-ERK1/2 (4370, Cell Signaling), and GAPDH (MA5-15738, Invitrogen). After washing the membranes three times in PBS-T, they were incubated with appropriate fluorescently labeled secondary antibodies at room temperature for 1 h. Following another three washes in PBS-T, fluorescence signals were detected using a Typhoon laser-scanner platform (Amersham).
Laser confocal immunofluorescence microscopy
PANC-1 cells were seeded at 20,000 cells/well in an 8-well chamber and cultured for 24 h. The cells were then incubated with 50 μM MYLS22 for an additional 24 h. Subsequently, the cells were fixed in pre-warmed (37°C) PBS containing 4% paraformaldehyde for 20 min.44,45 After washing three times with PBS, the cells were permeabilized with PBS containing 0.1% Triton X-100 for 8 min. The cells were then washed three more times with PBS and blocked in PBS containing 0.5% BSA at room temperature for 1 h.44,45 Next, the cells were incubated with anti-PDH antibody (1:300 dilution in PBS containing 0.5% BSA; ab110333, Abcam) or TOM20 (1:1,000 dilution in PBS containing 0.5% BSA; 11802-1-AP; Proteintech) at 4°C overnight. The cells were washed three times with PBS and incubated with Alexa 488-conjugated anti-mouse IgG (1:400 dilution in PBS; A21202, Thermo Fisher) and Alexa 568-conjugated phalloidin (1:500 dilution in PBS; A12380, Thermo Fisher) at room temperature for 1 h. Finally, the cells were again washed three times with PBS and stained with 1 μg/ml DAPI. Samples were observed using a Zeiss LSM800 GaAsP laser scanning confocal microscope.44,45 Mitochondrial length was measured using NIH Fiji software.44,45
Mitochondrial fusion
Mitochondrial fusion was examined using matrix-targeted photoactivatable GFP (mitoPAGFP).22 The matrix-targeted presequence from Su9 was fused to the N-terminus of photoactivatable GFP (Addgene #11909) and cloned into the lentiviral vector pHR-SIN.22 Cells were infected with lentiviral particles carrying mitoPAGFP. Fifteen minutes before observation, cells were stained with 5 nM TMRE to visualize the mitochondria. MitoPAGFP was photoactivated using 405 nm light at 50% power, with 25 repetitions, for a total of 2.5 seconds, in a small region measuring 16 μm2. The imaging was performed using a Zeiss LSM800 GaAsP confocal microscope equipped with an environmentally controlled chamber. Images were captured at 15-min intervals over a 60-min period. The fluorescence intensity of mitoPAGFP in the photoactivated region was quantified using NIH Fiji software.
Electron microscopy
Cells were fixed by 2% glutaraldehyde, 3 mM CaCl2 and 0.1 M cacodylate buffer, pH 7.4, for 1 h.46 After washes, samples were post-fixed in 1% OsO4, 1% potassium hexacyanoferrate, and 0.1 M cacodylate, pH 7.4, for 1 h on ice. After washes in water, samples were incubated in 2% uranyl acetate for 30 min on ice. After dehydration using 50, 70, 90, and 100% ethanol, samples were embedded in EPON resin. Ultrathin sections were obtained using a Reichert-Jung ultracut E, stained with 2% uranyl acetate and lead citrate, and viewed using a transmission electron microscope (H-7600; Hitachi) equipped with a dual CCD camera (Advanced Microscopy Techniques).
Plasmids
To generate shRNA plasmids, the following target sequences were cloned into pLKO.1. Scramble: CCTAAGGTTAAGTCGCCCTCGctcgagCGAGGGCGACTTAACCTTAGG, DRP1: GCTACTTTACTCCAACTTATTctcgagAATAAGTTGGAGTAAAGTAGC.
Lentivirus
HEK293T cells were seeded at 1.5 × 106 cells in a 10-cm dish and cultured for 24 h. To produce lentiviruses, 3 μg of pLKO.1 carrying shRNAs was co-transfected into HEK293T cells along with 3 μg of pHR-CMV8.2ΔR and 0.3 μg of pCMV-VSVG using Lipofectamine 2000 (Invitrogen).44,47 After 20–22 h, the culture medium was replaced with fresh medium. After an additional 24 h, the culture medium containing the released viruses was collected. For lentiviral transduction, PANC-1 cells were seeded at 1 × 105 cells/well in a 6-well plate and cultured for 24 h. Cells were then incubated with lentivirus in the cell culture medium containing 10% FBS and 8 μg/ml polybrene for 24 h.
Mitochondrial respiration
Mitochondrial OCRs were measured using an XF96 Extracellular Flux Analyzer (Seahorse Bioscience) as described in previous studies.44,47 PANC-1 cells were seeded at 10,000 cells/well in an XF 96-well culture microplate and cultured for 24 h. The cells were then incubated with 50 μM MYLS22 for an additional 24 h. Subsequently, the cells were washed twice with an XF base medium supplemented with 50 μM MYLS22, 25 mM glucose, and 4 mM L-glutamine. The cells were then incubated at 37°C in a CO2-free incubator for 1 h. OCR measurements were performed according to the manufacturer's instructions. Baseline OCR was recorded three times, after which 1.6 μg/ml oligomycin (OM), 1 μM FCCP, and 0.5 μM rotenone/antimycin A (AM+Rot) were sequentially injected into each well.
Glycolysis
Glycolysis was measured by analyzing ECARs using an XF96 Extracellular Flux Analyzer (Seahorse Bioscience). PANC-1 cells were seeded at 10,000 cells/well in an XF 96-well culture microplate and cultured for 24 h. The cells were then incubated with 50 μM MYLS22 for an additional 24 h. Subsequently, the cells were washed twice with XF base medium supplemented with 50 μM MYLS22 and 4 mM L-glutamine and then incubated at 37°C in a CO2-free incubator for 1 h. ECAR measurements were performed according to the manufacturer's instructions. Non-glycolytic acidification was recorded three times, after which 10 mM glucose, 1.6 μg/ml oligomycin, and 50 mM 2-deoxyglucose (2-DG) were sequentially injected into each well.
Quantification and statistical analysis
Statistical analysis was performed using GraphPad Prism 7. Student's t-test, ANOVA followed by post-hoc Tukey test or Šídák's test were used: ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. Specific statistical tests and significance thresholds are detailed in the figure legends.
Published: September 3, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2024.110880.
Contributor Information
Miho Iijima, Email: miijima@jhmi.edu.
Hiromi Sesaki, Email: hsesaki@jhmi.edu.
Supplemental information
References
- 1.Collisson E.A., Bailey P., Chang D.K., Biankin A.V. Molecular subtypes of pancreatic cancer. Nat. Rev. Gastroenterol. Hepatol. 2019;16:207–220. doi: 10.1038/s41575-019-0109-y. [DOI] [PubMed] [Google Scholar]
- 2.Buscail L., Bournet B., Cordelier P. Role of oncogenic KRAS in the diagnosis, prognosis and treatment of pancreatic cancer. Nat. Rev. Gastroenterol. Hepatol. 2020;17:153–168. doi: 10.1038/s41575-019-0245-4. [DOI] [PubMed] [Google Scholar]
- 3.Hayashi A., Hong J., Iacobuzio-Donahue C.A. The pancreatic cancer genome revisited. Nat. Rev. Gastroenterol. Hepatol. 2021;18:469–481. doi: 10.1038/s41575-021-00463-z. [DOI] [PubMed] [Google Scholar]
- 4.Park W., Chawla A., O'Reilly E.M. Pancreatic Cancer: A Review. JAMA. 2021;326:851–862. doi: 10.1001/jama.2021.13027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hosein A.N., Dougan S.K., Aguirre A.J., Maitra A. Translational advances in pancreatic ductal adenocarcinoma therapy. Nat. Can. (Ott.) 2022;3:272–286. doi: 10.1038/s43018-022-00349-2. [DOI] [PubMed] [Google Scholar]
- 6.Mizrahi J.D., Surana R., Valle J.W., Shroff R.T. Pancreatic cancer. Lancet. 2020;395:2008–2020. doi: 10.1016/S0140-6736(20)30974-0. [DOI] [PubMed] [Google Scholar]
- 7.Siegel R.L., Miller K.D., Fuchs H.E., Jemal A. Cancer Statistics, 2021. CA A Cancer J. Clin. 2021;71:7–33. doi: 10.3322/caac.21654. [DOI] [PubMed] [Google Scholar]
- 8.Encarnación-Rosado J., Kimmelman A.C. Harnessing metabolic dependencies in pancreatic cancers. Nat. Rev. Gastroenterol. Hepatol. 2021;18:482–492. doi: 10.1038/s41575-021-00431-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Camelo F., Le A. The Intricate Metabolism of Pancreatic Cancers. Adv. Exp. Med. Biol. 2021;1311:77–88. doi: 10.1007/978-3-030-65768-0_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vaziri-Gohar A., Zarei M., Brody J.R., Winter J.M. Metabolic Dependencies in Pancreatic Cancer. Front. Oncol. 2018;8:617. doi: 10.3389/fonc.2018.00617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Viale A., Pettazzoni P., Lyssiotis C.A., Ying H., Sánchez N., Marchesini M., Carugo A., Green T., Seth S., Giuliani V., et al. Oncogene ablation-resistant pancreatic cancer cells depend on mitochondrial function. Nature. 2014;514:628–632. doi: 10.1038/nature13611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Siolas D., Morrissey C., Oberstein P.E. The Achilles' Heel of Pancreatic Cancer: Targeting pancreatic cancer's unique immunologic characteristics and metabolic dependencies in clinical trials. J. Pancreatol. 2020;3:121–131. doi: 10.1097/JP9.0000000000000052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rath S., Sharma R., Gupta R., Ast T., Chan C., Durham T.J., Goodman R.P., Grabarek Z., Haas M.E., Hung W.H.W., et al. MitoCarta3.0: an updated mitochondrial proteome now with sub-organelle localization and pathway annotations. Nucleic Acids Res. 2021;49:D1541–D1547. doi: 10.1093/nar/gkaa1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cancer Genome Atlas Research Network. Weinstein J.N., Collisson E.A., Mills G.B., Shaw K.R.M., Ozenberger B.A., Ellrott K., Shmulevich I., Sander C., Stuart J.M. The Cancer Genome Atlas Pan-Cancer analysis project. Nat. Genet. 2013;45:1113–1120. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tang G., Cho M., Wang X. OncoDB: an interactive online database for analysis of gene expression and viral infection in cancer. Nucleic Acids Res. 2022;50:D1334–D1339. doi: 10.1093/nar/gkab970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Herkenne S., Ek O., Zamberlan M., Pellattiero A., Chergova M., Chivite I., Novotná E., Rigoni G., Fonseca T.B., Samardzic D., et al. Developmental and Tumor Angiogenesis Requires the Mitochondria-Shaping Protein Opa1. Cell Metabol. 2020;31:987–1003.e8. doi: 10.1016/j.cmet.2020.04.007. [DOI] [PubMed] [Google Scholar]
- 17.Zamberlan M., Boeckx A., Muller F., Vinelli F., Ek O., Vianello C., Coart E., Shibata K., Christian A., Grespi F., et al. Inhibition of the mitochondrial protein Opa1 curtails breast cancer growth. J. Exp. Clin. Cancer Res. 2022;41:95. doi: 10.1186/s13046-022-02304-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jin L., Li D., Alesi G.N., Fan J., Kang H.B., Lu Z., Boggon T.J., Jin P., Yi H., Wright E.R., et al. Glutamate dehydrogenase 1 signals through antioxidant glutathione peroxidase 1 to regulate redox homeostasis and tumor growth. Cancer Cell. 2015;27:257–270. doi: 10.1016/j.ccell.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu C.M., Yeh M.T., Tsao N., Chen C.W., Gao Q.Z., Chang C.Y., Lee M.H., Fang J.M., Sheu S.Y., Lin C.J., et al. Tumor cells require thymidylate kinase to prevent dUTP incorporation during DNA repair. Cancer Cell. 2012;22:36–50. doi: 10.1016/j.ccr.2012.04.038. [DOI] [PubMed] [Google Scholar]
- 20.Becattini B., Sareth S., Zhai D., Crowell K.J., Leone M., Reed J.C., Pellecchia M. Targeting apoptosis via chemical design: inhibition of bid-induced cell death by small organic molecules. Chem. Biol. 2004;11:1107–1117. doi: 10.1016/j.chembiol.2004.05.022. [DOI] [PubMed] [Google Scholar]
- 21.Murata D., Arai K., Iijima M., Sesaki H. Mitochondrial division, fusion and degradation. J. Biochem. 2020;167:233–241. doi: 10.1093/jb/mvz106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bannwarth S., Ait-El-Mkadem S., Chaussenot A., Genin E.C., Lacas-Gervais S., Fragaki K., Berg-Alonso L., Kageyama Y., Serre V., Moore D.G., et al. A mitochondrial origin for frontotemporal dementia and amyotrophic lateral sclerosis through CHCHD10 involvement. Brain. 2014;137:2329–2345. doi: 10.1093/brain/awu138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nagdas S., Kashatus J.A., Nascimento A., Hussain S.S., Trainor R.E., Pollock S.R., Adair S.J., Michaels A.D., Sesaki H., Stelow E.B., et al. Drp1 Promotes KRas-Driven Metabolic Changes to Drive Pancreatic Tumor Growth. Cell Rep. 2019;28:1845–1859.e5. doi: 10.1016/j.celrep.2019.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stephan T., Brüser C., Deckers M., Steyer A.M., Balzarotti F., Barbot M., Behr T.S., Heim G., Hübner W., Ilgen P., et al. MICOS assembly controls mitochondrial inner membrane remodeling and crista junction redistribution to mediate cristae formation. EMBO J. 2020;39 doi: 10.15252/embj.2019104105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kozjak-Pavlovic V. The MICOS complex of human mitochondria. Cell Tissue Res. 2017;367:83–93. doi: 10.1007/s00441-016-2433-7. [DOI] [PubMed] [Google Scholar]
- 26.Ikeda H., Muroi M., Kondoh Y., Ishikawa S., Kakeya H., Osada H., Imoto M. Miclxin, a Novel MIC60 Inhibitor, Induces Apoptosis via Mitochondrial Stress in beta-Catenin Mutant Tumor Cells. ACS Chem. Biol. 2020;15:2195–2204. doi: 10.1021/acschembio.0c00381. [DOI] [PubMed] [Google Scholar]
- 27.Padmanaban V., Krol I., Suhail Y., Szczerba B.M., Aceto N., Bader J.S., Ewald A.J. E-cadherin is required for metastasis in multiple models of breast cancer. Nature. 2019;573:439–444. doi: 10.1038/s41586-019-1526-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Serasinghe M.N., Wieder S.Y., Renault T.T., Elkholi R., Asciolla J.J., Yao J.L., Jabado O., Hoehn K., Kageyama Y., Sesaki H., Chipuk J.E. Mitochondrial Division Is Requisite to RAS-Induced Transformation and Targeted by Oncogenic MAPK Pathway Inhibitors. Mol. Cell. 2015;57:521–536. doi: 10.1016/j.molcel.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kashatus J.A., Nascimento A., Myers L.J., Sher A., Byrne F.L., Hoehn K.L., Counter C.M., Kashatus D.F. Erk2 phosphorylation of Drp1 promotes mitochondrial fission and MAPK-driven tumor growth. Mol. Cell. 2015;57:537–551. doi: 10.1016/j.molcel.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoxhaj G., Manning B.D. The PI3K-AKT network at the interface of oncogenic signalling and cancer metabolism. Nat. Rev. Cancer. 2020;20:74–88. doi: 10.1038/s41568-019-0216-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Caunt C.J., Sale M.J., Smith P.D., Cook S.J. MEK1 and MEK2 inhibitors and cancer therapy: the long and winding road. Nat. Rev. Cancer. 2015;15:577–592. doi: 10.1038/nrc4000. [DOI] [PubMed] [Google Scholar]
- 32.Quirós P.M., Mottis A., Auwerx J. Mitonuclear communication in homeostasis and stress. Nat. Rev. Mol. Cell Biol. 2016;17:213–226. doi: 10.1038/nrm.2016.23. [DOI] [PubMed] [Google Scholar]
- 33.Pakos-Zebrucka K., Koryga I., Mnich K., Ljujic M., Samali A., Gorman A.M. The integrated stress response. EMBO Rep. 2016;17:1374–1395. doi: 10.15252/embr.201642195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hallin J., Bowcut V., Calinisan A., Briere D.M., Hargis L., Engstrom L.D., Laguer J., Medwid J., Vanderpool D., Lifset E., et al. Anti-tumor efficacy of a potent and selective non-covalent KRASG12D inhibitor. Nat. Med. 2022;28:2171–2182. doi: 10.1038/s41591-022-02007-7. [DOI] [PubMed] [Google Scholar]
- 35.Carmona-Carmona C.A., Dalla Pozza E., Ambrosini G., Cisterna B., Palmieri M., Decimo I., Cuezva J.M., Bottani E., Dando I. Mitochondrial Elongation and OPA1 Play Crucial Roles during the Stemness Acquisition Process in Pancreatic Ductal Adenocarcinoma. Cancers. 2022;14 doi: 10.3390/cancers14143432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Friedman J.R., Nunnari J. Mitochondrial form and function. Nature. 2014;505:335–343. doi: 10.1038/nature12985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Youle R.J., van der Bliek A.M. Mitochondrial fission, fusion, and stress. Science. 2012;337:1062–1065. doi: 10.1126/science.1219855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mishra P., Chan D.C. Metabolic regulation of mitochondrial dynamics. J. Cell Biol. 2016;212:379–387. doi: 10.1083/jcb.201511036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Walker B.R., Moraes C.T. Nuclear-Mitochondrial Interactions. Biomolecules. 2022;12 doi: 10.3390/biom12030427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sarbassov D.D., Guertin D.A., Ali S.M., Sabatini D.M. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307:1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- 41.Stewart S.A., Dykxhoorn D.M., Palliser D., Mizuno H., Yu E.Y., An D.S., Sabatini D.M., Chen I.S., Hahn W.C., Sharp P.A., et al. Lentivirus-delivered stable gene silencing by RNAi in primary cells. RNA. 2003;9:493–501. doi: 10.1261/rna.2192803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Patterson G.H., Lippincott-Schwartz J. A photoactivatable GFP for selective photolabeling of proteins and cells. Science. 2002;297:1873–1877. doi: 10.1126/science.1074952. [DOI] [PubMed] [Google Scholar]
- 43.GTEx Consortium The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Murata D., Yamada T., Tokuyama T., Arai K., Quirós P.M., López-Otín C., Iijima M., Sesaki H. Mitochondrial Safeguard: a stress response that offsets extreme fusion and protects respiratory function via flickering-induced Oma1 activation. EMBO J. 2020;39 doi: 10.15252/embj.2020105074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Adachi Y., Itoh K., Yamada T., Cerveny K.L., Suzuki T.L., Macdonald P., Frohman M.A., Ramachandran R., Iijima M., Sesaki H. Coincident Phosphatidic Acid Interaction Restrains Drp1 in Mitochondrial Division. Mol. Cell. 2016;63:1034–1043. doi: 10.1016/j.molcel.2016.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Murata D., Roy S., Lutsenko S., Iijima M., Sesaki H. Slc25a3-dependent copper transport controls flickering-induced Opa1 processing for mitochondrial safeguard. Dev. Cell. 2024 doi: 10.1016/j.devcel.2024.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kageyama Y., Hoshijima M., Seo K., Bedja D., Sysa-Shah P., Andrabi S.A., Chen W., Höke A., Dawson V.L., Dawson T.M., et al. Parkin-independent mitophagy requires Drp1 and maintains the integrity of mammalian heart and brain. EMBO J. 2014;33:2798–2813. doi: 10.15252/embj.201488658. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
All data reported in this article will be shared by the lead contact upon request.
-
•
This article does not report original code.
-
•
Any additional information required to reanalyze the data reported in this article is available from the lead contact upon request.