Abstract
Viruses constitute the most diverse and abundant biological entities on Earth. However, our understanding of this tiniest life form in complex ecosystems remains limited. Here, we recover 20,102 viral OTUs from twelve intertidal zones along the Chinese coasts. Our analysis demonstrates high viral diversity and functional potential in intertidal zones, encoding important functional genes that can be potentially transferred to microbial hosts and mediate elemental biogeochemical cycles, especially carbon, phosphate and sulfur. Virus-host abundance dynamics vary among different microbial lineages. Viral community composition is closely associated with environmental conditions, including dissolved organic matter. Concordant biogeographic patterns are observed for viruses and microbes. Viral communities are generally habitat specific with low overlaps between intertidal and other habitats. Environmental factors and geographic distance dominate the compositional variation of intertidal viromes. Overall, these findings expand our understanding of intertidal viromes within an ecological framework, providing insights into the virus-host coevolutionary arms race.
Subject terms: Microbial ecology, Metagenomics, Wetlands ecology
The dynamic intertides, located between marine and terrestrial ecosystems, serve as a favorable habitat for exploring virus-host relationships. Here, the authors recover 20,102 viral OTUs from twelve intertidal zones along the Chinese coasts, with further analyses expanding our understanding of intertidal viromes within an ecological framework.
Introduction
Microbes are ubiquitous in the Earth biosphere, forming the most abundant and diverse group of living organisms1. Although tiny in size, they are central to driving the biogeochemical cycling of various elements, and maintaining ecosystem stability2. Over the past decades, great progress has been made to unravel the diversity, function, biogeography and underlying mechanisms of microbes in various ecosystems, largely advancing our knowledge of the unseen majority on the Earth3,4. Compared to the breakthroughs achieved for prokaryotes, much less attention has been paid for the tiniest viral communities5. Progress in viral ecology has been relatively slow for a long period owing to the absence of effective technologies, such as the lack of universal marker genes5,6. More recently, the development of high-throughput sequencing technologies and meta-omics approaches have greatly facilitated viral ecology studies, allowing scientists to more efficiently unravel the mysterious viral communities and their carrying functional potential7,8.
Viruses inhabit almost all environments, shaping the composition and assembly of microbial communities through lysis9. Furthermore, they can also reprogram host metabolism via encoding auxiliary metabolic genes (AMGs) and regulate microbial dynamics by releasing host nutrients, thereby affecting elemental biogeochemical cycles in various ecosystems8,10. Viruses are also important drivers of microbial evolution, owing to their specific mechanisms including lysogenic conversion and transduction9. Similar to what has been observed for macro-organisms and microbes (e.g., prokaryotes and microeukaryotes), large-scale metagenomic studies have demonstrated that viral communities in natural ecosystems such as soil and marine also follow typical biogeographic patterns and are influenced by multiple environmental factors11,12. Despite the rapid development of viral ecology, our comprehension of the immense viral world in the Earth’s biosphere still remains very limited, especially for viral diversity and functional potential in complex ecosystems9,13.
Intertidal zones are critical ecological ecotones located between marine and terrestrial ecosystems, constituting one of the most widespread coastal ecosystems14. Tidal oscillations induce frequent switches between aerobic and anoxic/suboxic conditions in intertidal sediments, representing a major stressor for microbial communities therein15. In response, intertidal microbes have developed flexible and diverse adaptive strategies (e.g., adjusting competitive and symbiotic relationships), resulting in more variable community structures compared to other natural ecosystems16. The dynamic environmental conditions and host communities are expected to significantly influence viral communities, as previously observed in acid mines and high-altitude watershed soil7,17. In turn, viruses can also drive the population and evolutionary diversity of microbial hosts via their unique life strategies9,18. Therefore, intertidal zones serve as a favorable habitat for exploring the virus-host ecological and evolutionary dynamics. However, the restricted viral genomic datasets available in current intertidal studies impede comprehensive understanding of viral biodiversity, both taxonomic and functional. Large scale sampling and investigation of intertidal viromes are desired to resolve their ecological mechanisms, linkages with microbial hosts, and contributions to different elemental biogeochemical cycles.
In this study, viral communities from mudflat intertidal zones spanning from the southmost to the northmost of the Chinese coasts are recovered and analyzed using shotgun metagenomic sequencing and state-of-the-art bioinformatics approaches, aiming to address the following questions related with the diversity, function and ecological mechanisms of mudflat intertidal viromes: (i) How diverse are mudflat intertidal viromes? (ii) How do they potentially contribute to the biogeochemical cycling of various elements? (iii) Do viruses follow typical biogeographic patterns as microbes? (iv) What is the relative importance of deterministic vs stochastic processes in structuring mudflat intertidal viromes? The results demonstrate high viral diversity in the intertidal zones, with significant associations with elemental biogeochemical cycling processes, and comparable biogeographic patterns and assembly mechanisms with their host microbes. The findings expand our understanding of the viral communities in complex natural ecosystems, providing mechanistic insights into the diversity and ecology of mudflat intertidal viromes.
Results
An overview of the intertidal viromes and microbiomes
In this study, 96 mudflat intertidal sediment samples were subjected to shotgun metagenomic sequencing, covering twelve coastal zones in China (Fig. 1a and Supplementary Data 1). The sampling sites spanned from the southernmost (Sanya) to the northernmost (Dandong), representing typical mudflat intertidal zones in the Chinese coasts. High quality contigs (minimum length of 5 kb) generated from multi-sample-assembly were screened by Virsorter219, DeepVirFinder20, and VIBRANT21, resulting in 21,964 viral contigs. By clustering at 95% average nucleotide identity, approximately corresponding to the species delineation cutoff of prokaryotes22, a total of 20,102 viral operational taxonomic units (vOTUs) were obtained. The genome size of vOTUs ranged from 5 kb to 230 kb, and ~96% of them were between 5 kb and 50 kb (Supplementary Data 2). Notably, 239 complete viral genomes were recovered in the intertidal dataset, with a minimum size of only 5.1 kb (Supplementary Data 2). In addition, the 302,424 protein-coding genes carried by 21,964 viral genomes were clustered at 80% coverage and 60% identity7, generating 238,445 viral protein clusters (vPCs).
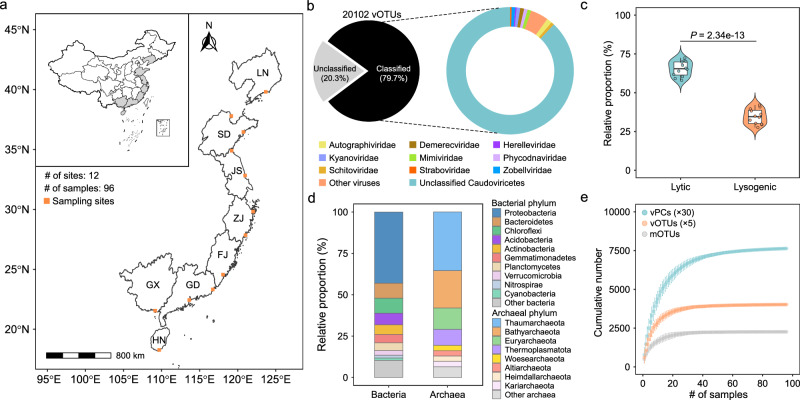
Fig. 1. Overview of viruses and microbes in mudflat intertidal sediments.
a Geographic distribution of collected intertidal sediment samples. The sampling provinces of China are colored in gray, including Liaoning (LN), Shandong (SD), Jiangsu (JS), Zhejiang (ZJ), Fujian (FJ), Guangxi (GX), Guangdong (GD), and Hainan (HN). The sampling regions are marked with orange squares. For each sampling region, eight sedimental cores (0–15 cm) were subjected to shotgun metagenome sequencing. b The proportions of classified and unclassified viruses (left) and the proportions of viral lineages at family level (right). c Relative proportions of lytic and lysogenic viruses in the intertidal sediments. The boxplots show the differences in the relative proportions of viruses with different lifestyles. The points in boxplot represent the average values of eight samples across each sampling region. The center lines of the boxes indicate the median value of 12 sampling regions. The bounds of the box represent the interquartile range, with the lower bound corresponding to the first quartile and the upper bound to the third quartile. The whiskers denote the lowest and highest values within 1.5 times the range of the first and third quartiles. Statistical significance in difference was determined using a two-tailed t test. d The relative proportions of bacterial and archaeal taxa at phylum level. Only the top ten phyla were presented, while the remaining phyla were categorized as others. e Accumulation curves of viral operational taxonomic units (vOTUs, orange), microbial operational taxonomic units (mOTUs, gray), and viral protein clusters (vPCs, cyan). The mean ± SEM values are plotted. Dots represent the average number of vPCs, vOTUs, and mOTUs, and the error bars represent the SEM. The numbers of vPCs and vOTUs were respectively divided by 30 and 5 for better visualization. Source data are provided as a Source Data file. Figure 1a is plotted using the ‘geom_sf’ function in ggplot2. The geojson files of base map were obtained from the public source http://xzqh.mca.gov.cn/map.
Taxonomic assignment of vOTUs showed that 16,027 of 20,102 vOTUs (~79.7%) could be classified to known taxa using the Lowest Common Ancestor algorithm (Fig. 1b). Among these, intertidal viral taxonomy covered four major DNA viral realms, including Duplodnaviria (15,543 vOTUs), Monodnaviria (14 vOTUs), Varidnaviria (465 vOTUs), and Adnaviria (1 vOTUs) (Supplementary Data 2). Caudoviricetes (15,540 vOTUs) belonging to Duplodnaviria largely represented the taxonomic diversity of DNA viruses in the intertidal zones (Fig. 1b). The majority of classified vOTUs (14,154) could only be resolved at class level (Fig. 1b). Only 8.5% (1367) vOTUs can be resolved at family level, such as Autographiviridae, Kyanoviridae, Demerecviridae, and Zobellviridae (Fig. 1b). Notably, we observed 409 vOTUs assigned to nucleocytoplasmic large DNA viruses (NCLDVs), primarily comprising the clades of Mimiviridae and Phycodnaviridae (Fig. 1b). Moreover, 16 vOTUs were classified as Lavidaviridae (virophage) (Supplementary Data 2), which act as parasites of NCLDVs and hold the capability to inhibit their replication23.
The potential lifestyles associated with vOTUs were determined by identifying lysogenic hallmark genes (e.g., integrase) and employing a deep-learning model24. As a result, 6680 vOTUs were determined as lysogenic viruses, of which 5503 were temperate viruses and 1177 were proviruses (integrated temperate viruses) (Supplementary Data 2). The relative proportion of lytic viruses (65.4 ± 4.64%) was significantly higher than that of lysogenic viruses (34.6 ± 4.64%) across the sampling sites (Student’s t test, df = 22, t = 15.55, P = 2.34e-13, 95% CI = [26.7, 34.9]) (Fig. 1c).
To establish the virus-host linkages, metagenome-assembled genomes (MAGs) were also recovered from the assembled contigs. A total of 2703 MAGs were obtained and further clustered at 95% average nucleotide identity, yielding 2259 microbial operational taxonomic units (mOTUs) (Supplementary Data 3). Taxonomic assignment showed that these mOTUs were classified as 2228 bacteria and 31 archaea (Supplementary Data 3). The bacterial mOTUs covered 48 phyla, of which Proteobacteria was the most abundant (960 mOTUs), followed by Bacteroidetes (202 mOTUs) and Chloroflexi (201 mOTUs) (Fig. 1d). The archaeal mOTUs belonged to 8 phyla, dominated by Thaumarchaeota (11 mOTUs) and Bathyarchaeota (7 mOTUs) (Fig. 1d). The numbers of vOTUs, mOTUs, and vPCs saturated between 20 and 40 samples, suggesting that more sequencing data and samples tended not to substantially improve the quantity of assembled contigs (Fig. 1e).
Functional potential encoded by intertidal viromes
To investigate the functional potential encoded by intertidal viromes, functional assignment of 302,424 predicted viral genes was carried out by searching against the eggNOG database (Supplementary Data 4). As a result, 69,661 viral genes were assigned to known orthologous groups, of which 46.9% were functionally unknown (Fig. 2a). Functional genes closely related with viral reproduction and transcription, such as “replication, recombination and repair (L)”, “cell wall/membrane/envelope biogenesis (M)”, and “transcription (K)”, were found with high relative proportions (Fig. 2a). Remarkably, a number of viral genes (1,583 of 69,661) were related with “carbohydrate transport and metabolism (G)” (Fig. 2a), which has previously also been noted in mangrove sediment25.
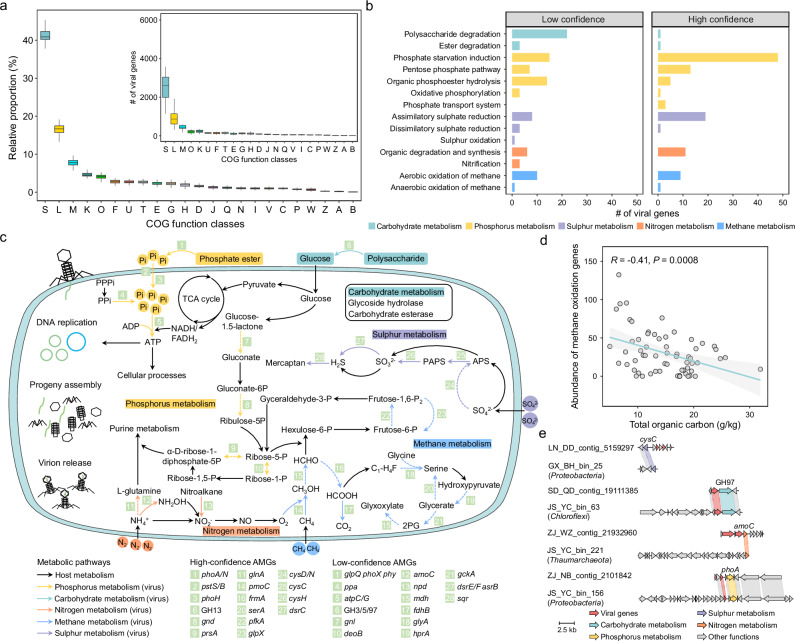
Fig. 2. Functional genes and auxiliary metabolic genes (AMGs) encoded by intertidal viruses.
a The relative proportions (abundances of specific functional class/abundances of all functional genes) of viral functional genes categorized by COG classes. The center lines of the boxes indicate the median values of 96 intertidal samples. The bounds of the box represent the interquartile range, with the lower and upper bounds respectively corresponding to the first and third quartiles. The whiskers denote the lowest and highest values within 1.5 times the interquartile range. The detailed descriptions of COG function classes were: ‘S’ (Function unknown), ‘L’ (Replication, recombination and repair), ‘M’ (Cell wall/membrane/envelope biogenesis), ‘K’ (Transcription), ‘O’ (Posttranslational modification, protein turnover, chaperones), ‘F’ (Nucleotide transport and metabolism), ‘U’ (Intracellular trafficking, secretion, and vesicular transport), ‘T’ (Signal transduction), ‘E’ (Amino acid transport and metabolism), ‘G’ (Carbohydrate transport and metabolism), ‘H’ (Coenzyme transport and metabolism), ‘D’ (Cell cycle control, cell division, chromosome partitioning), ‘J’ (Translation), ‘Q’ (Secondary metabolites biosynthesis, transport and catabolism), ‘N’ (Cell motility), ‘I’ (Lipid transport and metabolism), ‘V’ (Defense mechanisms), ‘C’ (Energy production and conversion), ‘P’ (Inorganic ion transport and metabolism), ‘W’ (Extracellular structures), ‘Z’ (Cytoskeleton), ‘A’ (RNA processing and modification), ‘B’ (Chromatin structure and dynamics). b The number of viral AMGs involved in different metabolic functions. Different colors represent the metabolic pathways of these functions. c A conceptual diagram depicting the potential viral regulation of host metabolism via AMGs. Different colors (excluding black) of arrows indicate different metabolic pathways that viral AMGs may participate in. The solid and dashed arrows were used to distinguish whether viral AMGs were exclusively involved in that metabolic pathway. d The relationship between total organic carbon and normalized abundance of viral AMGs related to methane oxidation. The shaded gray region reflects 95% confidence intervals of the fitted regression line. The Pearson’s correlation coefficient of the linear regression is presented. Statistical significance of the model was evaluated using a two-sided F test. e Comparative genomic analysis of viral and microbial AMGs. Only genes with > 90% identity and > 90% coverage were concatenated. Source data are provided as a Source Data file.
To disentangle the potential roles of viruses in contributing to important biogeochemical cycles, 209 viral AMGs (vAMGs) associated with targeted processes (e.g., carbon, nitrogen, sulfur, and phosphorous cycles) were further identified (Supplementary Data 5). Of these, 113 vAMGs with viral hallmark genes on both flanks were categorized as high confidence, while the remaining were of low confidence (Fig. 2b and Supplementary Data 5). Overall, intertidal vAMGs that participated in 14 important metabolic functions belonging to different biogeochemical cycles were identified (Fig. 2b, c), with the most abundant being associated with phosphate starvation induction (63 vAMGs). These phosphate starvation induction genes (phoH), along with the vAMGs caring organic phosphoester hydrolysis (glpQ, phoA/N/X, and phy), phosphate transport system (pstS/B), and oxidative phosphorylation (atpC/G), may help provide ATP for viral DNA replication and host cellular processes. In addition, we found that 20 vAMGs (gnl, gnd, prsA, and deoB) were involved in pentose phosphate pathway and its close relatives. Of these, deoB exclusively participated in this metabolic pathway. A total of 27 vAMGs were identified as carbohydrate-active enzymes (CAZymes), including one carbohydrates esterase (CE) family (4 vAMGs) and eight glycoside hydrolase (GH) families (23 vAMGs). In particular, vAMGs encoding β-glucosidase (GH3), cellulase (GH5), α-amylase (GH13), and α-glucosidase (GH97) were found, with the potential to help degrade complex polysaccharides into glucose10. This process may further affect the TCA cycle and pentose phosphate pathway within the hosts. Similar to previous findings in marine26, multiple vAMGs encoding sulfate reduction genes were also observed, of which assimilatory sulfate reduction genes (cysC/D/N/H) were the most abundant, followed by dissimilatory sulfate reduction genes (dsrC/E/F and asrB). We also found some virus-encoded nitrogen metabolism AMGs, including glnA and npd genes related to organic nitrogen degradation and synthesis and amoC genes involved in ammonia oxidation. Among these, glutamine synthase (glnA) can provide the substrate for host purine metabolism. Notably, multiple gene families related with aerobic methane oxidation and its close relatives were carried by vAMGs, potentially contributing to energy acquisition and carbon metabolism for host life activities.
Interestingly, vAMGs associated with methane oxidation were more abundantly found in intertidal sediments with low total organic carbon (TOC) content, showing significant negative associations with TOC concentrations (R = −0.41, P = 0.0008) (Fig. 2d). We also observed similar negative correlations between virus-encoded sulfate reduction genes and sulfate (SO42-) concentrations (R = −0.31, P = 0.004), as well as between phosphate starvation induction genes and total phosphorus (TP) concentrations (R = −0.29, P = 0.004) (Fig. S1a, b). These results indicated that viruses may contribute to host metabolism by encoding abundant AMGs under oligotrophic conditions, serving as a potential strategy for viral survival and propagation. In addition, 22 vAMGs involved in different metabolic pathways could also be detected in microbial genomes by mapping vAMGs to the contigs (Supplementary Data 5). Among these, one cysC gene and one phoA gene were mapped to Proteobacteria contigs, one glucosidase (GH97) was mapped to Chloroflexi contigs, and one amoC gene was mapped to Thaumarchaeota contigs (Fig. 2e).
Virus-host relationships in intertidal zones
To explore the potential effects of viruses on microbial taxa, mOTUs were linked to vOTUs through genomic features to assign prokaryotic host information for viruses. As a result, 2233 out of 20,102 vOTUs and 1078 out of 2259 prokaryotic mOTUs were determined to be potentially linked (Supplementary Data 6). Among these, Proteobacteria (~45.5%) was the most frequently predicted viral host phylum (Fig. 3a). Notably, a strong virus-host correlation was observed between the normalized abundances of viruses and prokaryotic hosts at the phylum level (R2 = 0.87, P < 2.2e-16) (Fig. S2a). Significant associations (P < 0.05) were also observed between the normalized abundances of viruses and their corresponding hosts for most specific lineages (34 of 41 phyla), demonstrating high confidence for the predicted prokaryotic hosts (Fig. S2b). For intertidal lineages, higher normalized viral abundances than host abundances were observed for 28 phyla (Fig. 3a and Fig. S2c). Of these, Thermoplasmatota was found with the highest virus/host abundance ratio (VHR), whereas Thaumarchaeota was the lowest (Fig. 3a).
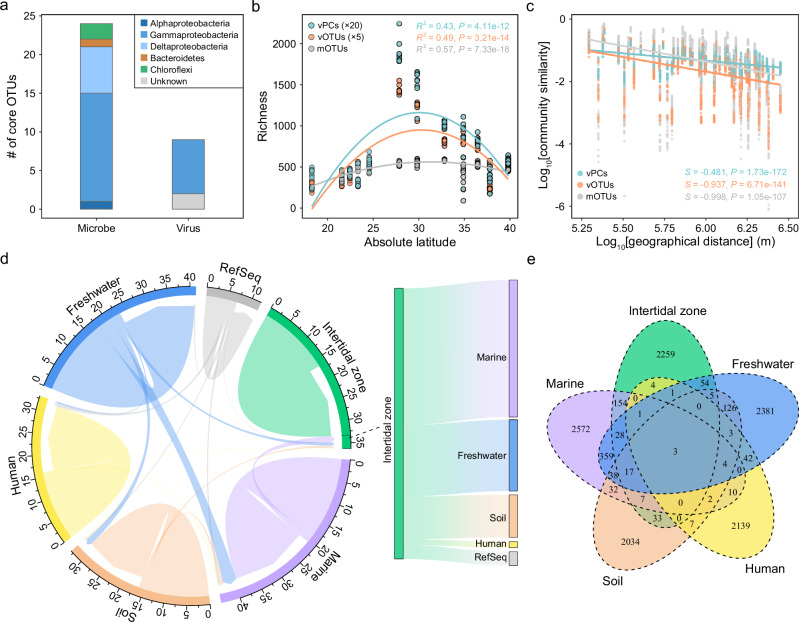
Fig. 3. Virus-host relationships and their linkages with environmental factors and dissolved organic matter (DOM) in intertidal zones.
a Virus/host abundance ratios (VHRs) of specific lineages. The bar graph represents the relative abundances of hosts, the dot (orange) represents the VHRs, and the cyan vertical line represents the 1:1 ratio. Values in the brackets indicate the numbers of predicted viruses belonging to that lineage. Only viral lineages with ≥ 0.2% relative abundances are shown. b Histograms of the frequency of Pearson’s correlation coefficient (r) between normalized abundances of virus-host pairs. c Histograms of the frequency of Pearson’s correlation coefficient (r) between normalized host abundances (log-transformed) and VHRs (log-transformed). d The relationship between normalized host abundances, VHRs and environmental factors. The metabolic pathways of host microbial operational taxonomic units (mOTUs) belonging to Deltaproteobacteria, Thermodesulfobacteria, and Thaumarchaeota were assessed using the KEGG-Decoder module. Only the Deltaproteobacteria and Thermodesulfobacteria mOTUs involved in the sulfate reduction, and the Thaumarchaeota mOTUs involved in the ammonia oxidation were used for correlation analysis. The color gradient in heatmap represents the values of Pearson’s correlation coefficient (r). The stars in heatmap represent the significance levels: * (P < 0.05), ** (P < 0.01), and *** (P < 0.001). e The consensus network representing the correlations between DOM and viral operational taxonomic units (vOTUs) of different lineages. The correlations between DOM and vOTUs were determined using Spearman’s correlation coefficients (ρ), and the statistical significance was adjusted using Bonferroni correction for multiple comparisons (P-adjust). Only the correlations with absolute Spearman’s correlation coefficient (ρ) > 0.7 and P-adjust <0.5 were retained for network construction. The vOTUs were classified according to their host lineages, and only the top ten lineages were displayed. All statistical tests were two-tailed. Source data are provided as a Source Data file.
The relative quantification of metagenomic data revealed a significant correlation between normalized viral abundances and host abundances across the intertidal samples (R2 = 0.41, P = 1.54e-11) (Fig. S2d). However, viral abundance did not increase linearly as host abundance increased. Rather, a trend of decreasing VHRs with increasing host abundances was observed for samples with high host abundances (Fig. S2d). As previously described7, this pattern might be due to the differences in the virus-host abundance dynamics of some specific lineages. For example, Thaumarchaeota and Planctomycetes were similar in host relative abundances (Fig. 3a), but the virus-host abundance dynamics of Thaumarchaeota was significantly weaker than that of Planctomycetes (two-way ANOVA, df = 188, F = 144.38, P < 2.2e-16) (Fig. S2e). To further investigate the effects of host density on viruses, we assessed the relationships between the normalized abundances of each virus-host pair (Fig. 3b). As a result, 1419 virus-host pairs exhibited significant correlations (P < 0.05) (Fig. 3b). The VHRs of ~81.4% virus-host pairs were negatively correlated with host abundances (Fig. 3c), which may contribute to the overall virus-host abundance pattern.
As critical components of the ecosystem, the relationships between viruses and their hosts are not self-driven, but may also be affected by the environmental conditions10. For example, we found that the VHRs of several different lineages exhibited significant associations with the changes of surrounding environmental conditions, such as NO2--N, pH, salinity, and moisture (Fig. 3d). Specifically, the abundances and VHRs of sulfate-reducing bacteria within Deltaproteobacteria and Thermodesulfobacteria, as well as ammonia-oxidizing archaea within Thaumarchaeota, demonstrated significant correlations with SO42- and ammonium nitrogen (NH4+-N) concentrations, respectively (Fig. 3d). Notably, opposite patterns with SO42- were observed for Deltaproteobacteria and Thermodesulfobacteria (Fig. 3d and Fig. S3a), possibly due to their different ecological niches in the intertidal ecosystem, as also supported by the fact that Thermodesulfobacteria are generally more adapted to high temperature environments, whereas Deltaproteobacteria have better sulfate reduction efficiency at ambient temperatures27,28. Accordingly, significant associations were also observed for the VHRs of these lineages with environmental factors (Fig. 3d and Fig. S3b). Strikingly, the patterns with environmental parameters for VHRs and hosts may not always the same, and sometimes opposite (Fig. S3a, b). Such different patterns were partially linked to the relationships of VHRs with their host abundance (Fig. S3c), but also reflected a complex biological and ecological procedure that environmental factors such as SO42- and NH4+-N not only affected microbes, but also their viruses and the virus-host relationships.
Significant associations were also observed between viral communities and dissolved organic matter (DOM) components (Mantel’s r = 0.34, P < 0.001). Pairwise correlation analyses showed that 12,995 of 20,102 vOTUs and 8436 of 20,980 DOM molecules were significantly correlated (absolute Spearman’s ρ > 0.7 and P < 0.05), generating a network with 304,760 correlations (Fig. S4). Aliphatic- (~33.8%) and lignin-like (~20.3%) compounds were the dominant classes associated with vOTUs (Fig. S4). Further analysis of viruses assigned with host and DOM molecules showed that the proportions of DOM classes associated with different viral lineages were generally consistent (Fig. 3e). Although the vOTUs infecting Cyanobacteria were not abundant, they were found to be the most relevant viral lineage to DOM molecules (7563 of 31,717 correlations) (Fig. 3a, e). Mantel test suggested that the VHRs of 15 lineages were significantly correlated with DOM components (Fig. S5a), and most of them (12 of 15 lineages) exhibited high VHRs (Fig. 3a and Fig. S5a). Interestingly, 9.6% of the DOM compositional variations could be purely explained by VHRs, much higher than that by environmental factors (2.5%) and geographic distance (3.1%) (Fig. S5b). Such results suggested that viral lysis of host microbes may have largely contributed to the DOM pools in intertidal zones.
Diversity and biogeographic patterns of intertidal viromes along the latitudes
The composition of both viral (R = 0.982, P = 0.001) and microbial (R = 0.97, P = 0.001) communities from different sampling regions clearly differed from each other (Fig. S6a and b). Only 9 core viruses (mean relative abundance > 0.1% and existed in more than 80% of samples) were detected (1.64% in relative abundance), corresponding to the situation of core prokaryotes (Fig. S6c and Supplementary Data 7). Of the 9 core viruses detected, 7 were found to infect Gammaproteobacteria (Fig. 4a).
Fig. 4. Biogeographic patterns of intertidal viromes and their linkages with other habitats.
a The taxonomic diversity of core vOTUs and mOTUs in the intertidal sediments. The taxonomy of viruses was determined according to their predicted hosts. b Latitudinal diversity patterns for vOTUs, mOTUs, and vPCs. The relationship between absolute latitude and richness was analyzed. The numbers of vPCs and vOTUs were respectively divided by 20 and 5 for better visualization. The best polynomial fit was determined between first- and second-order polynomial fits based on the corrected Akaike Information Criterion (AICc). The R2 values represent the proportion of variance explained by the polynomial regression model. c Distance-decay relationships for vOTUs, mOTUs, and vPCs. The relationships between geographic distance (log-transformed) and community similarity (log-transformed) were analyzed. The Pearson’s correlation coefficients of the linear regression model are presented. d The consensus network representing shared viruses across different habitats. The circos illustration was employed to show the proportion of shared viral clusters (VCs) across different habitats. The Sankey diagram was used to show the proportions of VCs shared between intertidal zones and other habitats. e The numbers of shared VCs among intertidal zones, marine, soil, freshwater, and human. Statistical significance of each regression model was evaluated using a two-sided F test. Source data are provided as a Source Data file.
We also investigated whether intertidal viromes followed similar biogeographic patterns as microbes, especially their hosts. Two typical biogeographic distribution patterns, including latitudinal diversity gradients (LDGs) and distance-decay relationships (DDRs), were investigated. The richness of viruses (vOTUs), viral genes (vPCs) and prokaryotes (mOTUs) all peaked at midlatitude (Fig. 4b and Fig. S6d), showing a latitudinal pattern different from conventional LDG patterns. At the meanwhile, strong DDR patterns were observed for the community similarity of viruses, viral genes and prokaryotes (Fig. 4c). The slope coefficients of DDRs were similarly steep for vOTUs (S = −0.937, P < 0.001) and mOTUs (S = −0.998, P < 0.001) (Fig. 4c). Such similarly strong DDR patterns were also observed for viruses and their hosts (Fig. S7). In contrast, much weaker slope coefficient of vPCs was observed (Fig. 4c), reflecting more similar functional gene composition than taxonomy across geographic distance.
Linkages of intertidal viromes with other habitats
As one of the most complex ecosystems, intertidal zones are located between terrestrial and marine environments and are often under strong pressure from human activities14. Here, we analyzed the linkages between intertidal viromes and the viromes in other habitats using a weighted network that can cluster viruses approximately at genus level (Fig. S8). In the network, the shared viral genes from different habitats were closely linked to generate 13,008 viral clusters (VCs), including 7363 viruses from intertidal zones, 15,746 viruses from marine, 13,828 viruses from soil, 15,105 viruses from freshwater, 17,506 viruses from human, and 3807 known viruses from RefSeq (Fig. S8 and Supplementary Data 8).
Of the 2566 VCs present in intertidal zones, 2259 ( ~ 88%) were intertidal exclusive, showing a high degree of habitat specificity of viromes (Fig. 4d and e). Of the VCs co-occurred in intertidal zones and other habitats, ~50.6% coexisted in the marine habitat, followed by freshwater (26.3%), soil (15.7%), and human (2.2%) (Fig. 4d, e). Importantly, 3 VCs classified as Caudoviricetes and may infect Proteobacteria taxa were detected in all habitats/sources (Fig. 4e), suggesting that some viral taxa might be widely distributed across various habitats.
Deterministic vs stochastic processes in structuring intertidal viromes
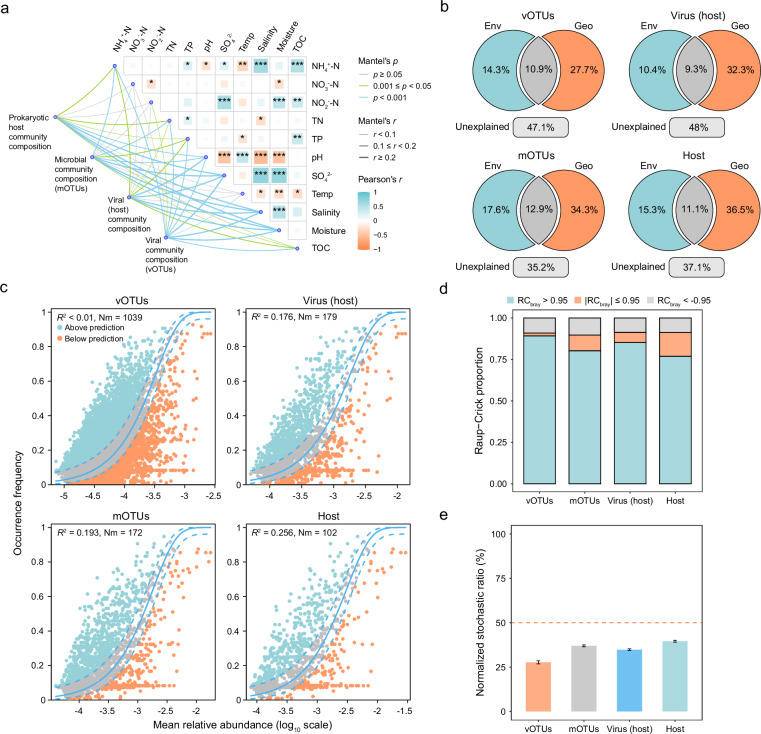
Both deterministic and stochastic processes mediate the compositional variation of microbial communities29,30. Here, multiple statistical approaches were employed to explore the relative importance of deterministic vs stochastic processes in structuring the intertidal viromes. First, partial mantel test demonstrated significant associations between multiple environmental factors and the viral communities (Mantel’s r = 0.27, P < 0.001), as well as viral hosts (Mantel’s r = 0.33, P < 0.001) and microbial communities (Mantel’s r = 0.31, P < 0.001) (Supplementary Data 9). In general, the same set of environmental factors, including NH4+-N, total nitrogen (TN), total phosphorus (TP), pH, TOC, SO42-, salinity, and moisture were significantly associated with the compositional variations of viral, prokaryotic host, and microbial communities (Fig. 5a). Second, the effects of environmental factors on viral communities were further assessed by linking community similarity with environmental heterogeneity, showing significant decay pattern with increasing environmental heterogeneity (Fig. S9). Third, variation partitioning analyses suggested that environmental factors and geographic distance together explained the major compositional variation of viral, prokaryotic host, and microbial communities (Fig. 5b). Fourth, the neutral community model only explained a small fraction (R2 < 0.5) of the relationships between the occurrence frequency and relative abundances of viral, prokaryotic host, and microbial communities, demonstrating weak neutral processes on the viral and host communities (Fig. 5c). Fifth, the RCbray metric (Raup-Crick index based on Bray–Curtis dissimilarity) showed greater community turnover than null expectations, suggesting that deterministic processes mainly accounted for the compositional variations (Fig. 5d). Finally, stochastic ratio analysis was also carried out, showing that the assembly of both viral and microbial communities were highly deterministic (Fig. 5e), consistent with the above results. Such results demonstrated that the intertidal viral communities and their corresponding host microbiome were similarly mainly structured by deterministic processes.
Fig. 5. Ecological mechanisms driving intertidal viromes and their hosts.
Analyses were performed for vOTUs, mOTUs, vOTUs assigned with hosts, and microbial hosts. a Partial mantel tests showing the relationship between environmental factors and viral/microbial communities. The edge color and width represent the Mantel’s r and p value, respectively. The color gradient in heatmap represents the Pearson’s correlation coefficients between different environmental factors. The stars in the heatmap indicate significance levels: * (P < 0.05), ** (P < 0.01), and *** (P < 0.001). All statistical tests were two-tailed. b Variation partitioning analysis showing the contributions of geographic distance and environmental factors to the compositional variations of viral and microbial communities. c Neutral community model analyses based on the predicted occurrence frequencies and their relative abundances. The solid blue lines indicate the best fit to the neutral community model and the dashed blue lines represented 95% confidence intervals. Nm represents the community size times immigration, R2 represents the fit strength to this model. d The Raup-Crick proportion of viral and microbial communities showing the contribution of different processes in community assembly, including deterministic processes (|RCbray | > 0.95), and stochastic processes (|RCbray | ≤ 0.95). e Stochastic ratio analyses of viral and microbial communities. The normalized stochastic ratios were calculated based on 1000 null models, using a threshold of 50% to distinguish between deterministic and stochastic processes. The mean ± SEM of stochastic ratio values calculated from 4560 pairwise comparisons among 96 intertidal samples were plotted. Source data are provided as a Source Data file.
Discussion
As the tiniest and most abundant lifeform in natural ecosystems, resolving the mysterious viral communities has been almost impossible, until recent advances in high throughput metagenomic sequencing technologies and associated bioinformatics approaches5,6. Multiple studies have been recently carried out, uncovering the diversity of viral communities in complex natural ecosystems7,8,26. In this study, the viral communities and their prokaryotic hosts in mudflat intertidal sediments were comparatively investigated, largely expanding our understanding of the viral diversity, biogeography, and functional potential in complex ecosystems. A diverse set of intertidal viruses were recovered, potentially infecting a broad range of prokaryotic hosts. Consistent with previous findings in marine and soil ecosystems11,12, Caudoviricetes was the most abundant DNA viruses in the intertidal sediments. As the most widespread, abundant and diverse group of viruses on Earth6, Caudoviricetes are thought to infect hosts from almost all bacterial lineages6, as also supported by the current study. Interestingly, Thermoplasmatota, a globally distributed and ecologically important archaeal phylum31, was found with the highest VHR, despite of its low relative abundance. Also, NCLDVs were frequently detected, demonstrating their wide distribution in intertidal ecosystems. Although these NCLDVs were only genomic fragments, the majority of them could be identified by multiple pipelines, ensuring their accuracy. To offer additional insights into the phylogenetic diversity of intertidal NCLDVs, further approaches like binning are required to obtain more complete viral genomes32.
Previously, the virus-host dynamics are typically estimated using the virus-to-microbe ratio derived from the counts of viruses and cells employing epifluorescence microscopy or flow cytometry33. Recently, shotgun metagenome/metavirome have been utilized as a routine approach for viral ecology studies, in which virus-host relationships are analyzed based on relative abundances11,18,33,34. Comparatively, different limitations are present for these different technologies33,35. For instance, absolute-abundance-based approaches like flow cytometry quantifies the overall virus-to-microbe ratio based on counts, but lacks more detailed information such as the taxonomy of viruses and hosts, hindering further statistical analyses at fine levels, albeit the potential false positive counting of viral particles in soils and sediments33,34. Metaviromes usually recover more viruses than metagenomes36, though with some exceptions37, but require metagenomes in different batches for virus-host relationship analyses, introducing other systematic artifacts. Considering various issues, shotgun metagenomes were employed in this study, aiming to provide insights into the complex virus-host relationships and their potential function in intertidal ecosystems38. Similar to several studies in marine and soil habitats11,39, a sublinear relationship was observed for the relative abundances of viruses against microbial hosts in intertidal sediments, in which the peaked VHRs tended to decrease when microbial host further increased. In addition, we found that the virus-host abundance dynamics significantly differed among different microbial lineages, which may contribute to the observed sublinear relationship. Nevertheless, the virus-host relationships in this study were analyzed based on the relative abundances derived from metagenomic data, absolute quantification approaches at fine taxonomic levels are desired to determine the viral predation models in complex environments.
As the most abundant biological entities in the Earth’s biosphere, viruses also execute critical ecosystem functions and maintain ecosystem stability8,10,26. Although viruses are usually not directly involved in biogeochemical cycles, they can promote the release of nutrients from hosts by lysis and enhance host metabolism by encoding critical functional genes8,26. A number of studies have demonstrated that viruses frequently carry AMGs to regulate the corresponding biogeochemical cycling processes in different habitats7,8,10,26. In this study, vAMGs related to carbon, nitrogen, phosphorus, and sulfur metabolism were detected. Importantly, the results demonstrated that intertidal viruses carried multiple AMGs related with aerobic methane oxidation and its close relatives, as also observed in a recent study40. Among these, pmoC and fdhB genes were exclusively specific to aerobic methane metabolism. We anticipated that viral regulation of host metabolism may have imposed important impacts on intertidal methane metabolism, particularly in terms of methane oxidation40. In addition, the relative abundances of vAMGs involved in specific metabolic pathways and the VHRs for lineages with specific functions were significantly correlated with corresponding environmental factors, providing compelling evidence for the contribution of intertidal viromes in various biogeochemical cycling.
Insights were also gained for the involvement of intertidal viromes in mediating DOM, which is the largest reservoir of organic carbon and is the source of recalcitrant DOM (RDOM), impacting global carbon cycling and climate change41. It is estimated that approximately 96% of DOM is difficult to be degraded and utilized by microbes and is present in the ocean as RDOM42. Viral shunt is reported as an important source of DOM and RDOM production41,43. Significant associations between intertidal viruses and DOM components were observed in this study. Viral lysis of Cyanobacteria is considered a major driving force in shaping DOM components44, as also supported by our findings in intertidal sediments. Aliphatic-like compounds consistently emerged as the most relevant DOM class to intertidal viruses infecting different lineages. Most of these compounds are highly bio-labile45, making them bioavailable and significant contributors to biogeochemical processes in intertidal sediments. In addition, lignin- and phenolic-like compounds were the other two classes that strongly correlated with viruses, suggesting that intertidal viruses may have also contributed to the accumulation of RDOM45. Notably, VHRs explained much higher proportion of the compositional variation of DOM pools than geo-environmental factors. Such results suggested important contribution of viral communities to intertidal DOM composition, providing insights into the role of viral shunt in intertidal DOM pools.
Finally, the biogeographic patterns and community assembly mechanisms for the intertidal viromes revealed several interesting mechanistic patterns along the latitudes. First, similar biogeographic patterns were followed by viral and microbial communities, demonstrating interconnected relationships between them7. Second, much weaker spatial scaling pattern was observed for the viral functional genes than taxonomic groups, suggesting functional redundancy despite of dissimilar communities46,47. Third, deterministic factors mainly mediated the compositional variation of intertidal viromes and microbial communities, as evidenced by multiple statistical approaches. Both environmental heterogeneity and geographic distance strongly influenced the compositional variations of intertidal viromes. Such results demonstrated that patterns followed by microbial communities can also be conveyed to viral communities, or vice versa7.
In conclusion, this study demonstrated the biodiversity, functional potential, biogeography, and ecology of intertidal viromes along the Chinese coasts. An extensive dataset of intertidal viral genomes and their microbial hosts was recovered. A sublinear relationship between viral and microbial host relative abundances was observed, with varied virus-host abundance dynamics among different microbial lineages. Furthermore, diverse AMGs that may potentially enhance host metabolisms were encoded by viruses, which may serve as a complimentary life strategy for intertidal viruses. The results also suggested close interconnected relationships between viral and microbial communities, by sharing similar ecological patterns and mechanistic processes. Our findings contribute a comprehensive understanding of intertidal viromes within an ecological framework, and provide novel perspectives for investigating the interaction and coevolution between viruses and their host microbes.
Methods
Study locations and sample collections
In this study, mudflat intertidal sediments were collected in twelve coastal regions spanning from the southmost (Sanya, 18.27o N, 109.68 o E) to the northmost (Dandong, 39.81 o N, 123.69 o E) in China. Samples were collected from April (South) to June (North) in 2021, balancing the sampling temperature differences in South and North China. For each sampling site, fifteen different homogenized sedimental samples were collected. Of them, eight were subject to shotgun metagenome sequencing. For each sample, five sedimental cores were collected from 0–15 cm within 1 m2 and then homogenized as one sample. The collected samples were stored on ice, and immediately transported to the laboratory. All samples were stored at −80 °C before subject to DNA extraction and environmental factor measurement.
Environmental factors and DOM measurement
A total of 11 environmental factors were measured for each sample (Supplementary Data 9), including temperature, pH, salinity, total organic carbon (TOC), total nitrogen (TN), total phosphorous (TP), ammonia nitrogen (NH4+-N), nitrate nitrogen (NO3--N), nitrite nitrogen (NO2--N), sulfate (SO42-), and moisture. The temperature for each sample was measured in situ using a mercury thermometer (−50 °C ~ 50 °C) when the sediment was collected. The pH of sediments was measured using a pH meter (STARTER 300, OHAUS, Beijing, China). The salinity of sediments was measured using a salinity meter (WS-31, Xudu, Beijing, China). The TOC of sediments was measured using TOC-L CPH meter (Shimadzu, Kyoto, Japan). The moisture of sediments was determined by drying 5.0 g fresh sediment at −80 °C to a constant weight. The concentrations of TN, TP, NH4+-N, NO3--N, NO2--N, and SO42- were measured by spectrophotometry (Cytation5, BioTek, USA). DOM was extracted using a solid-phase method and analyzed using Fourier transform ion cyclotron resonance mass spectrometry (Solarix 15 T, Bruker, USA) at the Research Center for Eco-Environmental Sciences, Chinese Academy of Sciences, Beijing, China. Then, the DOM molecular formulas were assigned by Formularity software48 and the downstream data analysis was performed with FtmsAnalysis (https://github.com/KaiMa-endeavour/DOM-in-MicroEco)49. The DOM table can be obtained from https://zenodo.org/records/10827260.
DNA extraction and bulk metagenomic sequencing
The total DNA of each sediment sample was extracted from 0.5 g homogenized sediment after freeze-drying using a FastDNA SPIN kit for soil (MP Biochemicals, USA) according to the manufacturer’s instructions. Subsequently, DNA quality was assessed based on ratios of 260/280 and 260/230 using Nanodrop ONE (Thermo Fisher Scientific, MA, USA). DNA with good quality was sequenced on the Illumina NovaSeq 6000 platform (paired-end, 2 × 150 bp, Inc., San Diego, CA, USA), and each metagenomic sample had a raw data size of >20 Gb. High throughput sequencing was performed by NovoGene Co., Ltd. (Tianjin, China).
Metagenomic assembly
Trimmomatic v0.39 was used to remove adapters and filter low-quality raw reads with a minimum base quality threshold of 20 and minimum read length of 3650. Then, clean reads from eight sediment samples at each sampling site were co-assembled (i.e., assembly of multiple samples) using MEGAHIT v1.2.951 with k-mer sizes of 29, 39, 59, 79, 99, 109, 127.
Identification and clustering of viral genomes and genes
Three mainstream pipelines, including Virsorter2 v2.2.319, DeepVirFinder v1.020, and VIBRANT v1.2.121, were first used to identify viruses from co-assembled contigs (≥5 kb). Then, a more precise procedure was used to screen and retain viral genomes according to the following criteria: (i) high confidence level (score ≥ 0.7 and had hallmark genes) of VirSorter2 (parameters: --keep-original-sequence), (ii) identified by VirSorter2 (score ≥ 0.5), DeepVirFinder (score ≥ 0.7 and p ≤ 0.05), and VIBRANT simultaneously. (iii) identified by any two of VirSorter2 (score ≥ 0.5), DeepVirFinder (score ≥ 0.7 and p ≤ 0.05), and VIBRANT, and further screened by CheckV v1.0.152 with at least one viral hallmark gene. The proviral regions were extracted from recovered viral genomes based on the CheckV contamination estimates.
The viral genomes after removing host contamination were de-replicated and clustered into viral operational taxonomic units (vOTUs) at 95% average nucleotide identity and 85% alignment fraction of the shortest genome using the python scripts in CheckV (https://bitbucket.org/berkeleylab/checkv/src/master/scripts/)52. The representative genome of each vOTU was used as input to CheckV to evaluate genome completeness52. The Prodigal 2.6.3 (-p meta)53 software was used to predict viral genes in viral genomes after removing host contamination. The proteins encoded by viral genes were further clustered at 60% identity and 80% coverage using cd-hit v4.8.1 (parameters: -c 0.6 -aS 0.8 -n 4 -g 1) to get viral protein clusters (vPCs)54.
Lifestyles and taxonomy prediction of viruses
Viral lifestyles were first determined by identifying lysogenic signals (including integrase, recombinase, provirus, transposase, and repressor)26. The proteins of 20,102 vOTUs were annotated against eggNOG database v5.0 using DIAMOND (bit score > 50 and e-value < 1e-5) and integrated database in VIBRANT21,55,56. The viruses without lysogenic signals were further detected with PhaTYP (default parameters)24 to distinguish lifestyles (lysogenic or lytic).
To assign taxonomic lineages to viral genomes in accordance with the latest ICTV classification, geNomad v1.7.4 (score ≥ 0.7) was adopted based on the taxonomic rank of annotated proteins57. Subsequently, the viral genomes classified as NCLDVs were further validated using Virsorter2 v2.2.3 (--include-groups NCLDV)19 and Viralrecall v2.158.
Recovery and clustering of microbial genomes
The recovered viral genomes (excluding proviruses) were first removed from co-assembled contigs7. The metaWRAP v1.3.2 pipeline59, an integrative pipeline of multiple binning methods, were used to bin metagenome-assembled genomes (MAGs) from co-assembled contigs (≥1.5 kb) for each sampling site. All MAGs were further consolidated into a final bin set using Bin_refinement module (>50% completeness and <10% contamination) within metaWRAP59. MAGs in the final bin set were de-replicated and clustered into microbial operational taxonomic units (mOTUs) at 95% average nucleotide identity using dRep v3.4.060. CheckM v1.0.1261 was then used to assess the genome quality and features of mOTUs with the lineage_wf module. Taxonomic information of mOTUs was assigned using GTDB-tk v2.1.162 based on the Genome Taxonomy Database R07-RS207 v2. The classification results were further refined by NCBI taxonomy using the GTDB-tk script (https://github.com/Ecogenomics/GTDBTk/tree/master/scripts) for downstream analysis. The KEGG-Decoder module63 was used to evaluate the metabolic pathways of sulfate-reducing bacteria within Deltaproteobacteria and Thermodesulfobacteria, as well as ammonia-oxidizing archaea within Thaumarchaeota.
Identification of viral functional genes and AMGs related to biogeochemical cycles
Viral proteins were searched against eggNOG database v5.0 using DIAMOND with an e-value cutoff of 1e-5 and a bit score of 5055,56. The functions of viral genes were categorized according to COG function classes. Subsequently, viral genes assigned KEGG_ko numbers by eggNOG were considered as potential AMGs and were further searched against various databases targeting specific biogeochemical processes using DIAMOND (identity > 30%, e-value > 1e-5, and bit score > 50), including NCycDB (nitrogen metabolism)64, SCycDB (sulfur metabolism)65, PCycDB (phosphorus metabolism)66, MCycDB (methane metabolism)67, and dbCAN2 (carbohydrate metabolism)68. To ensure the accuracy of AMGs, only the consistent annotations between eggNOG and these specific databases were retained. Further, AMGs were screened based on previously proposed criteria (Supplementary Data 5)69: (i) glycoside hydrolase family (e.g., lysozyme) related to viral infection were removed; (ii) AMGs related to nucleotide or amino acid metabolism (e.g., GlycosylTransferase and multiple phosphorus metabolism families) were removed; (iii) AMGs neighbored to viral hallmark gene (the viral hallmark genes were determined by searching against viral RefSeq and geNomad_db with e-value > 1e-5 and bit score > 50) were retained and only AMGs with viral hallmark gene on both the left and right flanks were considered as of high confidence. For AMGs related to amo/pmo gene families, both database searching against NCycDB and phylogenetic analyses using the maximum likelihood algorithm were conducted to clarify their homology. All proteins encoded by AMGs were then compared to the proteins of mOTUs with identity and coverage > 90% to determine the source of AMGs. The comparative genomic analyses and visualization of viruses were performed using clinker v0.0.2370.
Prokaryotic host prediction for intertidal viruses
The 451 intertidal vOTUs that infected eukaryotes based on viral taxonomy were initially excluded, including vOTUs belonging to Parvoviridae, Nanoviridae, Baculoviridae, Herpesviridae, Malacoherpesviridae, Nucleocytoviricota, Lavidaviridae, and Adintoviridae. Subsequently, 19,651 vOTUs were linked to microbial genomes using five different methods: (i) CRISPR spacers matches. CRISPR spacers were recovered from 2259 microbial genomes using metaCRT71 and PLIERCR72. The recovered spacers were then compared to viral genomes using BLASTn with the following parameters: e-value ≤ 1e-5, percentage identity ≥ 95%, and mismatch ≤ 2; (ii) tRNA sequence matches. tRNA sequences in viral genomes were recovered with tRNAscan-SE v2.0.9 (using general tRNA model)73 and then compared to microbial genomes using BLASTn with identity ≥ 90% and coverage ≥ 90%; (iii) shared genomic sequence homology. Viral genomes were compared to microbial genomes using BLASTn. Only the best matches having ≥2 kb alignment length and ≥ 70% identity were considered8. Short proviruses (≤5 kb) having ≥50% coverage over the genome length and ≥ 70% identity can also be considered as effective hits; (iv) integrated phage host prediction (iPHoP)74. The 2259 recovered intertidal microbial genomes were first added to the iPHoP database. Then, iPHoP v1.3.2 was used to predict microbial hosts for intertidal viruses with a minimum confidence score of 90 (i.e., false discovery rate <10%). Only the predicted hosts that hit intertidal microbes were retained; (v) oligonucleotide frequency. VirHostMatcher v1.0 was employed for calculating the oligonucleotide frequency between viral and microbial genomes75. The best matches with d2* values < 0.25 were retained.
Viral gene sharing network between intertidal zones and other habitats
The predicted viral proteins were clustered with NCBI Viral RefSeq v211 using vConTACT2 v0.11.3 to construct the gene sharing network76. Specifically, 20,102 intertidal viral genomes were compared to the same number of randomly selected high-quality viral genomes from other habitats in IMG/VR v4 database77: (i) 20,102 from soil (grassland, forest, and agricultural land), (ii) 20,102 from freshwater (lake and river), (iii) 20,102 from marine, (iv) 20,102 from human (intestine and oral cavity).
Calculating the normalized abundance of vOTUs, mOTUs, and vPCs
Coverm v0.6.1 (https://github.com/wwood/CoverM) was used to calculate the average read depths of representative genomes or genes. Briefly, read mapping was performed using BWA-MEM78 with nucleotide identity ≥ 95% and coverage ≥ 90% and the average sequencing depths were calculated with ‘trimmed_mean’ (remove the top and bottom 10% depths) coverage mode of Coverm. Then, the average sequencing depths were further normalized to eliminate the differences in the average read length and read number of each sample according to the following function:
Here, the total read number and average read length of each sample were recorded in Supplementary Data 1. The normalized results represented the coverage of genomes/genes in each sample when the sequencing depth is one billion reads and the average read length is 150 bp.
Statistical analyses
Statistical analyses were performed with multiple packages in R v4.2.0. The cumulative curves were calculated using the ‘specaccum’ function of vegan package79. Bray–Curtis dissimilarity of communities and DOM pools and Euclidean distances of geo-environmental factors and VHRs were calculated using the ‘vegdist’ function of vegan package79. Analysis of similarity was used to test the significance between different groups presented in non-metric multidimensional scaling (NMDS). The geographic distances between different sites were calculated using the ‘geoXY’ function of SoDA package7. The partial mantel tests between communities and environmental factors were performed by controlling the effects of geographic distance with 999 permutations. The mantel tests between DOM pools and VHR of each lineage were performed with the method of Spearman correlation and 999 permutations. The geographic variables used for variation partitioning analysis were calculated by the principal coordinates of neighbor matrices procedure of vegan package79. Then, the variation partitioning analysis was performed with a forward selection procedure using the “ordistep” function of vegan packages to select geographic variables, environmental variables, and VHRs of specific lineages for constructing significant (P < 0.05) canonical correlation analysis models, respectively47. The screened variables were divided into different groups to get their variation using the “varpart” function of vegan package79. The Spearman correlations between the relative abundance of each vOTU and each DOM molecular formula were calculated using the ‘rcorr’ function of Hmisc package80. Then, the statistical significance was adjusted by applying a Bonferroni correction.
To disentangle the relative importance of deterministic and stochastic processes, multiple approaches including neutral community model, RCbray metric, and normalized stochastic ratio were used. For neutral community model, the correlations between occurrence frequency and regional relative abundance were calculated using the Rscript (https://github.com/Weidong-Chen-Microbial-Ecology/Stochastic-assembly-of-river-microeukaryotes) described previously81. The overall fit (R2) of the community to the neutral model was determined. A well-fitted overall neutral model indicated that the community was structured by the neutral theory, whereas a low R2 value suggested the dominance of ecological niche theory. For RCbray metric calculation, the R function “Raup_Crick_Abundance.r” (https://github.com/stegen/Stegen_etal_ISME_2013)29 was used. In the result, |RCbray | ≤ 0.95 suggests comparable community turnover between the observed and null communities, meaning that the compositional variations are dominated by stochastic processes. RCbray values larger than 0.95 or smaller than −0.95 indicate deterministic factors that lead to heterogeneous or homogeneous communities could be the dominant process for the compositional variations. For normalized stochastic ratio calculation, a total of 1000 null models were generated based on Bray–Curtis dissimilarity using the NST package30. A normalized stochastic ratio below 50% indicates that the deterministic factors dominate community variations, while a ratio above 50% suggests the domination of stochasticity.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Source data
Acknowledgements
This study was supported by National Key Research and Development Program of China [2020YFA0607600 (Q.T.), 2019YFA0606700 (Q.T.)], the National Natural Science Foundation of China [32371598 (Q.T.), 92051110 (Q.T.), 31971446 (Q.T.)], the Natural Science Foundations of Shandong Province [2020ZLYS04 (Q.T.)], the Southern Marine Science and Engineering Guangdong Laboratory (Zhuhai) [SML2023SP218 (Q.T.)], the Taishan Young Scholarship of Shandong Province, and the Distinguished Young Scholarship of Shandong University. This study contributes to the science plan of the Ocean Negative Carbon Emissions (ONCE) Program. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Author contributions
The research was conceived by Q.T. and J.Z. (Joe). Sample collection and physicochemical characterization were carried out by K.M., W.S., and Y.L. Data analysis was done by M.J., J.Z., Y.L., and K.M. All data analysis and integration were guided by Q.T. The manuscript and figures were prepared by M.J. and Q.T. All authors have read and approved the submitted version of manuscript.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Data availability
The sequencing data generated in this study have been deposited in NCBI Sequence Read Archive (SRA) database under project ID PRJNA957716 as well as in the NODE under project ID OEP004120 [https://www.biosino.org/node/project/detail/OEP00004120]. The detailed information of sequencing data is provided in the Supplementary Data 1. The representative sequences of vOTUs, mOTUs, and vPCs and DOM tables generated from this work are available at https://zenodo.org/records/10827260. Source data are provided with this paper.
Code availability
R code used for generating figures and performing data statistics in this study are publicly available on GitHub at https://github.com/MengzhiJ/Biodiversity-of-mudflat-intertidal-viromes-along-the-Chinese-coasts.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-52996-x.
References
- 1.Torsvik, V., Øvreås, L. & Thingstad, T. F. Prokaryotic diversity-magnitude, dynamics, and controlling factors. Science296, 1064–1066 (2002). [DOI] [PubMed] [Google Scholar]
- 2.Fuhrman, J. A. Microbial community structure and its functional implications. Nature459, 193–199 (2009). [DOI] [PubMed] [Google Scholar]
- 3.Sunagawa, S. et al. Structure and function of the global ocean microbiome. Science348, 1261359 (2015). [DOI] [PubMed] [Google Scholar]
- 4.Nayfach, S. et al. A genomic catalog of Earth’s microbiomes. Nat. Biotechnol.39, 499–509 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roux, S., Hallam, S. J., Woyke, T. & Sullivan, M. B. Viral dark matter and virus–host interactions resolved from publicly available microbial genomes. elife4, e08490 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dion, M. B., Oechslin, F. & Moineau, S. Phage diversity, genomics and phylogeny. Nat. Rev. Microbiol.18, 125–138 (2020). [DOI] [PubMed] [Google Scholar]
- 7.Gao, S. et al. Patterns and ecological drivers of viral communities in acid mine drainage sediments across Southern China. Nat. Commun.13, 2389 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roux, S. et al. Ecogenomics and potential biogeochemical impacts of globally abundant ocean viruses. Nature537, 689–693 (2016). [DOI] [PubMed] [Google Scholar]
- 9.Chevallereau, A., Pons, B. J., van Houte, S. & Westra, E. R. Interactions between bacterial and phage communities in natural environments. Nat. Rev. Microbiol.20, 49–62 (2022). [DOI] [PubMed] [Google Scholar]
- 10.Emerson, J. B. et al. Host-linked soil viral ecology along a permafrost thaw gradient. Nat. Microbiol.3, 870–880 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ma, B. et al. Biogeographic patterns and drivers of soil viromes. Nat. Ecol. Evol.8, 717–728 (2024). [DOI] [PubMed]
- 12.Gregory, A. C. et al. Marine DNA viral macro-and microdiversity from pole to pole. Cell177, 1109–1123. e1114 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roux, S. & Emerson, J. B. Diversity in the soil virosphere: to infinity and beyond? Trends Microbiol.30, 1025–1035 (2022). [DOI] [PubMed] [Google Scholar]
- 14.Murray, N. J. et al. The global distribution and trajectory of tidal flats. Nature565, 222–225 (2019). [DOI] [PubMed] [Google Scholar]
- 15.Wang, J. et al. Denitrifying anaerobic methane oxidation: a previously overlooked methane sink in intertidal zone. Environ. Sci. Technol.53, 203–212 (2018). [DOI] [PubMed] [Google Scholar]
- 16.Bang, C. et al. Metaorganisms in extreme environments: do microbes play a role in organismal adaptation? Zoology127, 1–19 (2018). [DOI] [PubMed] [Google Scholar]
- 17.Coclet, C. et al. Virus diversity and activity is driven by snowmelt and host dynamics in a high-altitude watershed soil ecosystem. Microbiome11, 237 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Knowles, B. et al. Lytic to temperate switching of viral communities. Nature531, 466–470 (2016). [DOI] [PubMed] [Google Scholar]
- 19.Guo, J. et al. VirSorter2: a multi-classifier, expert-guided approach to detect diverse DNA and RNA viruses. Microbiome9, 1–13 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ren, J. et al. Identifying viruses from metagenomic data using deep learning. Quant. Biol.8, 64–77 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kieft, K., Zhou, Z. & Anantharaman, K. VIBRANT: automated recovery, annotation and curation of microbial viruses, and evaluation of viral community function from genomic sequences. Microbiome8, 1–23 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Konstantinidis, K. T. & Tiedje, J. M. Genomic insights that advance the species definition for prokaryotes. Proc. Natl. Acad. Sci. USA102, 2567–2572 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.La Scola, B. et al. The virophage as a unique parasite of the giant mimivirus. Nature455, 100–104 (2008). [DOI] [PubMed] [Google Scholar]
- 24.Shang, J., Tang, X. & Sun, Y. PhaTYP: predicting the lifestyle for bacteriophages using BERT. Brief. Bioinforma.24, bbac487 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jin, M. et al. Diversities and potential biogeochemical impacts of mangrove soil viruses. Microbiome7, 1–15 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kieft, K. et al. Ecology of inorganic sulfur auxiliary metabolism in widespread bacteriophages. Nat. Commun.12, 3503 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rabus, R. et al. A post-genomic view of the ecophysiology, catabolism and biotechnological relevance of sulphate-reducing prokaryotes. Adv. Microb. Physiol.66, 55–321 (2015). [DOI] [PubMed] [Google Scholar]
- 28.Muyzer, G. & Stams, A. J. The ecology and biotechnology of sulphate-reducing bacteria. Nat. Rev. Microbiol.6, 441–454 (2008). [DOI] [PubMed] [Google Scholar]
- 29.Stegen, J. C. et al. Quantifying community assembly processes and identifying features that impose them. ISME J.7, 2069–2079 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ning, D., Deng, Y., Tiedje, J. M. & Zhou, J. A general framework for quantitatively assessing ecological stochasticity. Proc. Natl. Acad. Sci. USA116, 16892–16898 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sheridan, P. O., Meng, Y., Williams, T. A. & Gubry-Rangin, C. Recovery of Lutacidiplasmatales archaeal order genomes suggests convergent evolution in Thermoplasmatota. Nat. Commun.13, 4110 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schulz, F., Abergel, C. & Woyke, T. Giant virus biology and diversity in the era of genome-resolved metagenomics. Nat. Rev. Microbiol.20, 721–736 (2022). [DOI] [PubMed] [Google Scholar]
- 33.Roux, S. & Brum, J. R. Counting dots or counting reads? Complementary approaches to estimate virus-to-microbe ratios. ISME J.17, 1521–1522 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.López-García, P. et al. Metagenome-derived virus-microbe ratios across ecosystems. ISME J.17, 1552–1563 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heinrichs, M. E., De Corte, D., Engelen, B. & Pan, D. An advanced protocol for the quantification of marine sediment viruses via flow cytometry. Viruses13, 102 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Santos-Medellin, C. et al. Viromes outperform total metagenomes in revealing the spatiotemporal patterns of agricultural soil viral communities. ISME J.15, 1956–1970 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bi, L., He, J.-Z. & Hu, H.-W. Total metagenomes outperform viromes in recovering viral diversity from Sulfuric soils. ISME Commun.4, ycae017 (2024). [DOI] [PMC free article] [PubMed]
- 38.Alrasheed, H., Jin, R. & Weitz, J. S. Caution in inferring viral strategies from abundance correlations in marine metagenomes. Nat. Commun.10, 501 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wigington, C. H. et al. Re-examination of the relationship between marine virus and microbial cell abundances. Nat. Microbiol.1, 1–9 (2016). [DOI] [PubMed] [Google Scholar]
- 40.Zhong, Z.-P. et al. Viral potential to modulate microbial methane metabolism varies by habitat. Nat. Commun.15, 1857 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jiao, N. et al. Microbial production of recalcitrant dissolved organic matter: long-term carbon storage in the global ocean. Nat. Rev. Microbiol.8, 593–599 (2010). [DOI] [PubMed] [Google Scholar]
- 42.Osterholz, H., Niggemann, J., Giebel, H.-A., Simon, M. & Dittmar, T. Inefficient microbial production of refractory dissolved organic matter in the ocean. Nat. Commun.6, 7422 (2015). [DOI] [PubMed] [Google Scholar]
- 43.Suttle, C. A. Marine viruses—major players in the global ecosystem. Nat. Rev. Microbiol.5, 801–812 (2007). [DOI] [PubMed] [Google Scholar]
- 44.Zhao, Z. et al. Microbial transformation of virus-induced dissolved organic matter from picocyanobacteria: coupling of bacterial diversity and DOM chemodiversity. ISME J.13, 2551–2565 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen, X. et al. Niche differentiation of microbial community shapes vertical distribution of recalcitrant dissolved organic matter in deep-sea sediments. Environ. Int.178, 108080 (2023). [DOI] [PubMed] [Google Scholar]
- 46.Louca, S. et al. Function and functional redundancy in microbial systems. Nat. Ecol. Evol.2, 936–943 (2018). [DOI] [PubMed] [Google Scholar]
- 47.Song, W. et al. Functional traits resolve mechanisms governing the assembly and distribution of nitrogen-cycling microbial communities in the global ocean. MBio13, e03832–03821 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tolić, N. et al. Formularity: software for automated formula assignment of natural and other organic matter from ultrahigh-resolution mass spectra. Anal. Chem.89, 12659–12665 (2017). [DOI] [PubMed] [Google Scholar]
- 49.Ma, K. et al. Disentangling drivers of mudflat intertidal DOM chemodiversity using ecological models. Nat. Commun.15, 6620 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics30, 2114–2120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li, D., Liu, C.-M., Luo, R., Sadakane, K. & Lam, T.-W. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics31, 1674–1676 (2015). [DOI] [PubMed] [Google Scholar]
- 52.Nayfach, S. et al. CheckV assesses the quality and completeness of metagenome-assembled viral genomes. Nat. Biotechnol.39, 578–585 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hyatt, D. et al. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinforma.11, 1–11 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fu, L., Niu, B., Zhu, Z., Wu, S. & Li, W. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics28, 3150–3152 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Huerta-Cepas, J. et al. eggNOG 5.0: a hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res.47, D309–D314 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Buchfink, B., Xie, C. & Huson, D. H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods12, 59–60 (2015). [DOI] [PubMed] [Google Scholar]
- 57.Camargo, A.P. et al. Identification of mobile genetic elements with geNomad. Nat. Biotechnol.42, 1303–1312 (2024). [DOI] [PMC free article] [PubMed]
- 58.Aylward, F. O. & Moniruzzaman, M. ViralRecall—a flexible command-line tool for the detection of giant virus signatures in ‘Omic Data. Viruses13, 150 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Uritskiy, G. V., DiRuggiero, J. & Taylor, J. MetaWRAP—a flexible pipeline for genome-resolved metagenomic data analysis. Microbiome6, 1–13 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Olm, M. R., Brown, C. T., Brooks, B. & Banfield, J. F. dRep: a tool for fast and accurate genomic comparisons that enables improved genome recovery from metagenomes through de-replication. ISME J.11, 2864–2868 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Parks, D. H., Imelfort, M., Skennerton, C. T., Hugenholtz, P. & Tyson, G. W. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res.25, 1043–1055 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chaumeil, P.-A., Mussig, A. J., Hugenholtz, P. & Parks, D. H. GTDB-Tk v2: memory friendly classification with the Genome Taxonomy Database. Bioinformatics38, 5315–5316 (2022). [DOI] [PMC free article] [PubMed]
- 63.Graham, E., Heidelberg, J. & Tully, B. Potential for primary productivity in a globally-distributed bacterial phototroph. ISME J.12, 1861–1866 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tu, Q., Lin, L., Cheng, L., Deng, Y. & He, Z. NCycDB: a curated integrative database for fast and accurate metagenomic profiling of nitrogen cycling genes. Bioinformatics35, 1040–1048 (2019). [DOI] [PubMed] [Google Scholar]
- 65.Yu, X. et al. SCycDB: a curated functional gene database for metagenomic profiling of sulphur cycling pathways. Mol. Ecol. Resour.21, 924–940 (2021). [Google Scholar]
- 66.Zeng, J. et al. PCycDB: a comprehensive and accurate database for fast analysis of phosphorus cycling genes. Microbiome10, 101 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Qian, L. et al. MCycDB: a curated database for comprehensively profiling methane cycling processes of environmental microbiomes. Mol. Ecol. Resour.22, 1803–1823 (2022). [DOI] [PubMed] [Google Scholar]
- 68.Zhang, H. et al. dbCAN2: a meta server for automated carbohydrate-active enzyme annotation. Nucleic Acids Res.46, W95–W101 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pratama, A. A. et al. Expanding standards in viromics: in silico evaluation of dsDNA viral genome identification, classification, and auxiliary metabolic gene curation. PeerJ9, e11447 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gilchrist, C. L. & Chooi, Y.-H. Clinker & clustermap. js: Automatic generation of gene cluster comparison figures. Bioinformatics37, 2473–2475 (2021). [DOI] [PubMed] [Google Scholar]
- 71.Rho, M., Wu, Y.-W., Tang, H., Doak, T. G. & Ye, Y. Diverse CRISPRs evolving in human microbiomes. PLoS Genet.8, e1002441 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Edgar, R. C. PILER-CR: fast and accurate identification of CRISPR repeats. BMC Bioinforma.8, 1–6 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chan, P. P., Lin, B. Y., Mak, A. J. & Lowe, T. M. tRNAscan-SE 2.0: improved detection and functional classification of transfer RNA genes. Nucleic Acids Res.49, 9077–9096 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Roux, S. et al. iPHoP: an integrated machine learning framework to maximize host prediction for metagenome-derived viruses of archaea and bacteria. PLoS Biol.21, e3002083 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ahlgren, N. A., Ren, J., Lu, Y. Y., Fuhrman, J. A. & Sun, F. Alignment-free oligonucleotide frequency dissimilarity measure improves prediction of hosts from metagenomically-derived viral sequences. Nucleic Acids Res.45, 39–53 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bin Jang, H. et al. Taxonomic assignment of uncultivated prokaryotic virus genomes is enabled by gene-sharing networks. Nat. Biotechnol.37, 632–639 (2019). [DOI] [PubMed] [Google Scholar]
- 77.Camargo, A. P. et al. IMG/VR v4: an expanded database of uncultivated virus genomes within a framework of extensive functional, taxonomic, and ecological metadata. Nucleic Acids Res.51, D733–D743 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv preprint arXiv:1303.3997 (2013).
- 79.Oksanen, J. et al. Package ‘vegan’. Community ecology package. version2, 1–295 (2013). [Google Scholar]
- 80.Harrell, F. E. Jr & Harrell, M. F. E. Jr Package ‘hmisc’. CRAN20182019, 235–236 (2019). [Google Scholar]
- 81.Chen, W. et al. Stochastic processes shape microeukaryotic community assembly in a subtropical river across wet and dry seasons. Microbiome7, 1–16 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
The sequencing data generated in this study have been deposited in NCBI Sequence Read Archive (SRA) database under project ID PRJNA957716 as well as in the NODE under project ID OEP004120 [https://www.biosino.org/node/project/detail/OEP00004120]. The detailed information of sequencing data is provided in the Supplementary Data 1. The representative sequences of vOTUs, mOTUs, and vPCs and DOM tables generated from this work are available at https://zenodo.org/records/10827260. Source data are provided with this paper.
R code used for generating figures and performing data statistics in this study are publicly available on GitHub at https://github.com/MengzhiJ/Biodiversity-of-mudflat-intertidal-viromes-along-the-Chinese-coasts.