Abstract
Although, several studies have assessed the association of HLA Class II and genes with bullous pemphigoid (BP) and mucous membrane pemphigoid (MMP), results were inconsistent and between-studies heterogeneity needs to be investigated. An electronic literature search for eligible studies among all papers published prior to May 31, 2024, was conducted through PubMed, EMBASE, Web of science and Scopus databases. Meta-analyses together with subgroup analyses and meta-regressions were performed for the three following HLA genes: DRB1, DQA1 and DQB1. Combined analyses revealed a significant increase in pemphigoid risk conferred by the following alleles: DQB1*0301, DRB1*11, DRB1*1101 subtype and DQA1*0505, all p-values <.001. However, there was a moderate to high level of between-studies heterogeneity. Subgroup analyses revealed that the risk conferred by the aforementioned alleles was significantly higher in case of dipeptidyl peptidase-4 inhibitors induced BP (DBP) comparatively to idiopathic BP and MMP. In addition, the risk conferred by the DQB1*0301 was significantly higher in MMP (OR [95% CI] = 5.25 [4.03–6.84]) than in BP (OR [95% CI] = 2.22 [1.87–2.65]), p = .007. Besides, the DRB1*1101-DQB1*0301 and DRB1*11-DQA1*05-DQB1*0301 haplotypes were significantly associated with an increased pemphigoid risk, both p-values <.001. Conversely, the DQA1*0201 allele was significantly associated with reduced pemphigoid risk (OR [95% CI] = 0.3 [0.17–0.52]), with no between-studies heterogeneity (I2 = 0%, p = .76). This meta-analysis demonstrated that the DRB1*1101, DQA1*0505 and DQB1*0301 were significantly associated with increased pemphigoid risk. These associations were found to be significantly stronger in case of DBP comparatively to idiopathic pemphigoid. The DQA1*0201 allele seems to play a protective role against pemphigoid. Registration: This review has been registered on PROSPERO: CRD42024552821, Available from: https://www.crd.york.ac.uk/prospero/display_record.php?ID=CRD42024552821.
Keywords: bullous pemphigoid, mucous membrane pemphigoid, HLA class II, haplotype, meta-analysis, meta-regression
Introduction
The pemphigoid diseases group includes several distinct conditions such as bullous pemphigoid (BP), mucous membrane pemphigoid (MMP), pemphigoid gestationis, linear IgA disease, epidermolysis bullosa acquisita, anti-p200-pemphigoid and lichen planus pempigoides. 1 Among this group, BP and MMP are the most prevalent disorders and share the BP230 (BPAg1) and BP180 (BPAg2) antigens as main targets of pathogenic autoantibodies. 1 Unlike BP in which the separation of epidermis from dermis results in cutaneous blisters, MMP is distinguishable by predominant involvement of mucosal surfaces. 2 Diagnosis of BP and MMP is based on a combination of clinical criteria, direct immunofluorescence (DIF) microscopy of a perilesional sample and serology including indirect immunofluorescence (IIF) microscopy using salt-split skin and ELISA measuring anti-BP230 and anti-BP180 antibodies.1,2 Disease severity assessment is based on BP and MMP disease area indices (BPDAI and MMPDAI).1,2
The exact etiology and pathogenesis of pemphigoid remain to be determined. However, the disease is believed to result from the interplay of various predisposing and precipitating factors, including genetic factors, comorbidities, drugs such as dipeptidyl peptidase-4 inhibitors (DPP-4), ultraviolet radiation, infections and aging.3,4 Among the environmental factors, DPP-4 was associated with about 7.38-fold increased BP risk. 5 Interestingly it has been shown that sera from DPP-4 induced BP (DBP) patients targeted epitopes of BP180 other than NC16A domain, which is one of the main targets in idiopathic BP. In addition, DBP may present as a unique phenotype with fewer inflammatory lesions and less eosinophilic infiltration comparatively to idiopathic BP.1,3,4 Hence, DBP may have a distinct immunopathological mechanism from the classical idiopathic BP, which could justify its separation as a distinct entity.
Besides, familial clustering of pemphigoid is very rare, suggesting that it is not a typical hereditary disease. 4 Instead, susceptibility alleles may modulate immune responses to target autoantigens only when individuals are exposed to appropriate environmental triggers. 4 The genetic susceptibility is thought to be involved in BP230 and BP180 autoantigen presentation to autoreactive T cells. 4 Indeed, pathogenic autoantibodies production by autoreactive B cells requires the presentation of BP230 and BP180 epitopes to T cells by class II human leukocyte antigens (HLA) including HLA-DR, -DP and -DQ. Hence, numerous studies have investigated HLA class II polymorphisms in pemphigoid risk. 4
A previous meta-analysis assessing HLA-DQA1 polymorphism role in BP genetic risk has already been published in 2023. 6 Nevertheless, this meta-analysis included only five studies among which only four were included for assessing the HLA-DQA1*0505 and HLA-DQA1*0201 alleles role in susceptibility to pemphigoid. 6 The authors concluded that HLA-DQA1*0505 allele conferred a 2.25-fold increased BP risk while the DQA1*0201 allele was associated with roughly a 3-fold decrease in BP risk. 6 Hence, the risk conferred by the HLA DQA1 gene represents only a small part of the genetic component predisposing to BP. Indeed, other HLA-DQB1 and DRB1 alleles could contribute to pemphigoid susceptibility, and thus require further investigation to better characterize the predisposing genetic landscape.
The aim of this review was to summarize existing data on the contribution of HLA-DRB1, DQA1 and DQB1 alleles and haplotypes to BP and MMP susceptibility and to investigate the between-studies heterogeneity by subgroup analyses and meta-regressions.
Material and methods
Search strategy
This study was performed according to the PRISMA guidelines for systematic reviews and meta-analyses. 7 An electronic literature search for eligible studies among all papers published prior to May 31, 2024, was conducted through PubMed, EMBASE, Web of science and Scopus databases. The following search string was used: (((HLA) OR (6p21) OR (Human Leukocyte Antigen) OR (MHC) OR (Major Histocompatibility Complex)) AND (pemphigoid)). The literature search was carried out without any language restriction.
Selection criteria
All studies were independently assessed and evaluated by two reviewers (Tarak Dhaouadi, T.D. and Imen Sfar, I.S.) for the inclusion and the exclusion. The following selection criteria were adopted:
Inclusion criteria:
• Studies of case-control (retrospective) or cohort (prospective) design.
• Studies assessing the association between HLA-DQB1, DRB1 and DQA1 alleles and BP and/or MMP risk.
• Studies providing precise results with alleles and or haplotypes frequencies.
Exclusion criteria:
• Studies carried out in patients with pemphigoid gestationis, linear IgA disease, epidermolysis bullosa acquisita, anti-p200-pemphigoid and lichen planus pempigoides.
• Case series of subjects, narrative or systematic review, comments, or meta-analysis.
• If many studies have been carried out using duplicate cases, only the study with complete data and the largest sample size was included.
Definition of pemphigoid
BP and MMP were defined respectively as cutaneous or mucosal lesions including blisters, erythema, erosions associated with a positive perilesional DIF and/or a positive IIF on salt-split skin and/or positive anti-BP230/BP180 antibodies (Abs) detected by ELISA. DPP-4-induced BP (DBP) was defined as a BP attributed to a DDP-4 use.
Data extraction
Data were extracted using a predeveloped form and entered in an Excel datasheet. Two investigators (T.D. and I.S.) independently extracted the following information: first author, year of publication, country, ethnicity, mean or median age, gender ratio (M/F), number of patients, number of controls, DIF result, serology (IIF, ELISA) results, and HLA-DQB1, DRB1 and DQA1 genotyping method (Table 1). A third investigator (Awatef Riahi) compared the results of the extracted data for potential discrepancies.
Table 1.
Included studies characteristics.
| Ref | Study | Pemphigoid | Country | Ethnicity | Age | Gender-ratio (Female/Male) | Anti-BPAg1 (BP230) Ab | Anti-BPAg2 (BP180) Ab | DIF + | IIF + | Genotyping/Subtyping method | Patients | Controls | NOS score |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| [12] | Ahmed 1991 | MMP⁂ | USA | American | NS⁋ | NS | NS | NS | NS | NS | MLC / PCR-RFLP | 23 | 46 | 7 |
| [13] | Banfield 1998 | BP | UK | Caucasian | 74 | 1.18 (40/34) | 35.1% | 16.2% | 71.6% | 73% | MLC / PCR | 74 | 604 | 8 |
| [14] | Büdinger 1998 | BP | Germany | Caucasian | 69.1 | NS | NS | 100% | NS | NS | PCR-SSP | 15 | 24 | 8 |
| [15] | Carrozzo 2001 | MMP | Italy | Caucasian | 65.6 ± 15.14 | 2.11 (19/9) | NS | NS | 100% | 48% | PCR-SSP | 28 | 97 | 7 |
| [16] | Chagury 2018 | BP | Brazil | American | 63.71 | 1.125 (9/8) | NS | NS | NS | NS | PCR-SSO | 17 | 297 | 7 |
| [17] | Chan 1997 | MMP | USA | American | NS | NS | NS | NS | 100% | NS | PCR / dot-blot | 21 | 42 | 7 |
| [18] | Chanprapraph 2021 | DBP | Thailand | Asian | 79.6 | 0.92 (11/12) | 43.5% | 47.8% | NS | NS | PCR-SSO | 20 | 33 | 7 |
| [19] | Delgado 1996 (BP*) | BP | USA | American | NS | NS | NS | NS | NS | NS | PCR / dot-blot | 42 | 218 | 8 |
| [19] | Delgado 1996 (OCP†) | MMP | USA | American | NS | NS | NS | NS | NS | NS | PCR / dot-blot | 34 | 218 | 8 |
| [19] | Delgado 1996 (OP‡) | MMP | USA | American | NS | NS | NS | NS | NS | NS | PCR / dot-blot | 44 | 218 | 8 |
| [20] | Drouet 1998 | MMP | France | Caucasian | NS | NS | NS | NS | NS | NS | PCR-SSP | 25 | 106 | 7 |
| [21] | Esmaili 2013 | BP | Iran | Asian | NS | 0.786 (22/28) | NS | NS | NS | NS | PCR-SSP | 50 | 180 | 8 |
| [22] | Fang 2018 | BP | China | Asian | 63.5 ± 13.5 | 1.234 (58/47) | NS | NS | NS | NS | Sequence-based typing | 105 | 420 | 8 |
| [23] | Gao 2002 | BP | China | Asian | NS | NS | NS | NS | NS | NS | PCR-SSP | 25 | 57 | 7 |
| [24] | Hübner 2018 | MMP | Germany | Caucasian | NS | NS | 31.5% | 18.5% | 72.7% | 44.4% | Allele-specific sequencing | 54 | 39 | 8 |
| [25] | Lindgren 2019 (BP) | BP | Finland | Caucasian | 89.2 | 1.5 | NS | 82.4% | 82.4% | NS | PCR-SSO | 9 | 5982 | 7 |
| [25] | Lindgren 2019 (DBP⁑) | DBP | Finland | Caucasian | 77.1 | 1.125 (9/8) | NS | 77.7% | 100% | NS | PCR-SSO | 14 | 5982 | 7 |
| [26] | Okazaki 2000 | BP | Japan | Asian | 77 | 2.286 (16/7) | NS | NS | 100% | NS | PCR-RFLP | 23 | 525 | 7 |
| [27] | Setterfield 2001 | MMP | UK | Caucasian | 56.3 | 1.98 (87/44) | 33.3% | 57.7% | 84.7% | 80.3% | MLC / PCR-SSP | 128 | 177 | 8 |
| [28] | Sun 2018 | BP | China | Asian | 68.9 | 0.76 (248/327) | 54.4% | 94.7% | 100% | NS | NGS | 575 | 976 | 8 |
| [29] | Ujiie 2018 (BP) | BP | Japan | Asian | 70 | 1.32 (41/31) | NS | NS | NS | NS | Luminex | 72 | 873 | 7 |
| [29] | Ujiie 2018 (DBP) | DBP | Japan | Asian | 77.1 | 0.4 (6/15) | 5.6% | 71.4% | 100% | 100% | Luminex | 21 | 873 | 8 |
| [30] | Walton 2020 | MMP | UK | Caucasian | 66.8 | 3.5 (21/6) | 36% | 24% | 71.4% | 28% | NGS | 27 | 10,000 | 7 |
| [31] | Yunis 1994 (OCP) | MMP | USA | American | NS | NS | NS | NS | 100% | NS | PCR-SSO | 17 | 65 | 7 |
| [31] | Yunis 1994 (OP) | MMP | USA | American | NS | NS | NS | NS | 100% | NS | PCR-SSO | 22 | 65 | 7 |
*BP: Bullous pemphigoid, †OCP: Ocular cicatricial pemphigoid, ‡OP: Oral pemphigoid, ⁑DBP: Dipeptidyl peptidase-4 inhibitor induced BP, ⁂MMP: Mucus membrane pemphigoid, ⁋NS: Not specified, Ab: Antibody, DIF: Direct immunofluorescence, IIF: Indirect immunofluorescence, MLC: Microlymphocytotoxicity.
Quality assessment
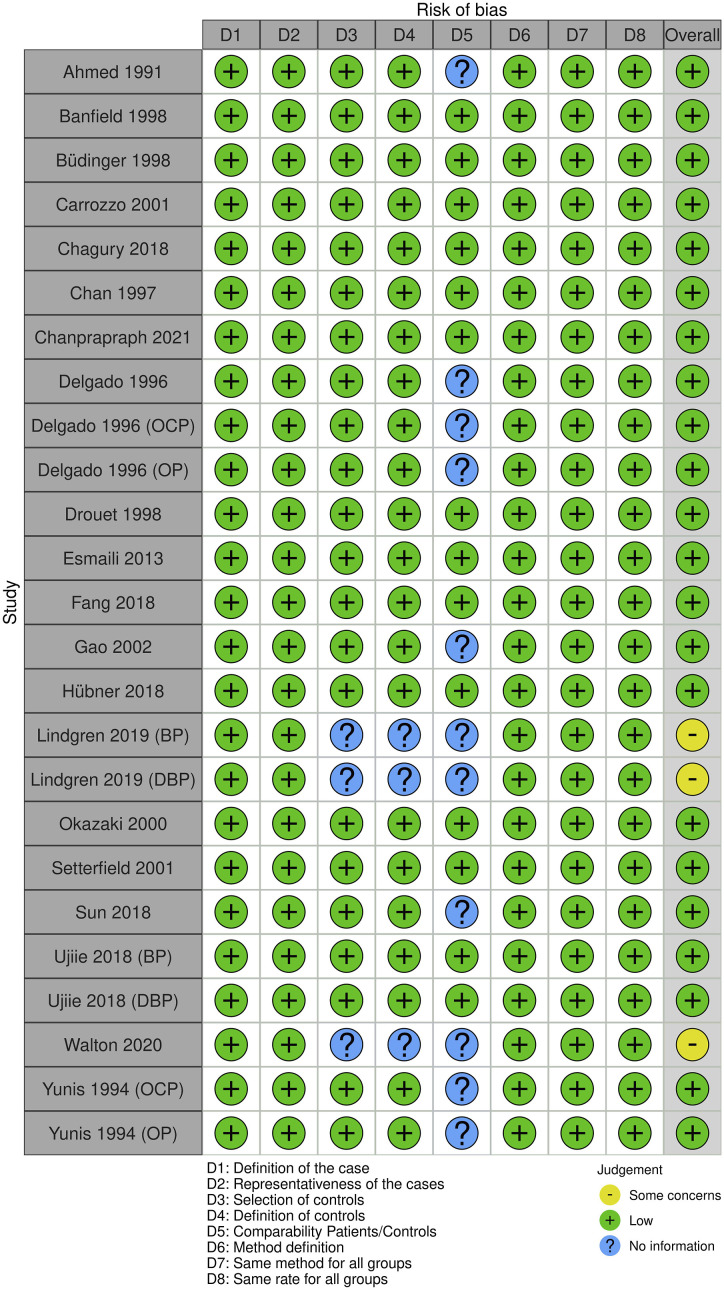
The quality of eligible studies was assessed independently by two reviewers (T.D. and I.S.) using the Newcastle-Ottawa Scale (NOS) 8 which is based on the following three general categories: selection (4 points), Comparability of the study groups (2 points) and ascertainment of outcome (3 points). Studies with a score ≥7 were classified as high-quality reports. Additionally, risk of bias was assessed for each included study through a generic form (Excel spreadsheet) and visualized via the Cochrane ROBVIS online tool (https://mcguinlu.shinyapps.io/robvis/). Two additional independent reviewers (Taieb Ben Abdallah and Yousr Gorgi) examined the quality-assessment results.
Study endpoints
The primary endpoint of this meta-analysis was to estimate the strength of the association HLA-DQB1, DRB1 and DQA1 alleles and haplotypes with BP, MMP and DBP risk. The secondary endpoint was to evaluate potential confounding factors that might influence the impact of the HLA class II alleles and haplotypes on pemphigoid risk in order to identify the sources of heterogeneity.
Statistical analysis
Statistical analysis was carried out using the Cochrane Review Manager 5.4 software, the OpenMeta-Analyst software and the online available software MetaGenyo (https://metagenyo.genyo.es/). The associations of DQB1, DRB1 and DQA1 alleles and haplotypes with pemphigoid risk were assessed using pooled Odds Ratios (ORs) with the 95% confidence interval (95% CI). The statistical significance of pooled ORs was tested by Z-test with a threshold of significance set at 0.05. 9 Random-effects models (DerSimonian-Laird 10 ) were used as recommended by Borenstein et al. 9 Indeed, as long as the eligible studies were carried out in genetically diverse populations, the random-effects model applies. 9 Forest-plots were generated to display the distribution of effect size (OR) across included studies. Sensitivity analyses were carried out to test the results stability by omitting sequentially each individual study. The heterogeneity of between-studies was tested by Q test (significance threshold: 0.1), quantified via I2 calculation (proportion of true effects variance) and analyzed through the determination of 95% prediction intervals (PI). PI were obtained through the CMA Prediction Intervals free software. The calculation of the 95% PI was based on the following four items: OR, upper bound of 95% CI, Tau2 and number of included studies. Of note, 95% PI computation requires at least three included studies in a meta-analysis. Permission to use the CMA prediction intervals has been obtained since March 21, 2023 (Figure S1). Subsequently, the heterogeneity was explored for potential sources by subgroup analyses and meta-regressions. Briefly, studies were stratified by ethnicity (American, Asian and Caucasian) and pemphigoid subtype (BP, MMP and DBP). Meta-regressions were performed using age, gender ratio (Female/Male), the positivity rate of DIF, IIF and ELISA, the risk allele/haplotype frequency in control groups as independent variables. Both univariate and multivariate models of meta-regression were generated in order to assess the presence of potential confounding factors. Publication bias were assessed by Egger’s test 11 and visualized through the generation of funnel plots. Of note, Egger’s test p-value computation requires at least three included studies in a meta-analysis.
A supplemental file 1 with additional tables and figures is available with the full-manuscript. A PRISMA checklist is available as a supplemental file 2.
Systematic review registration
This review has been registered on PROSPERO: CRD42024552821, Available from: https://www.crd.york.ac.uk/prospero/display_record.php?ID=CRD42024552821.
Results
Search results and study characteristics
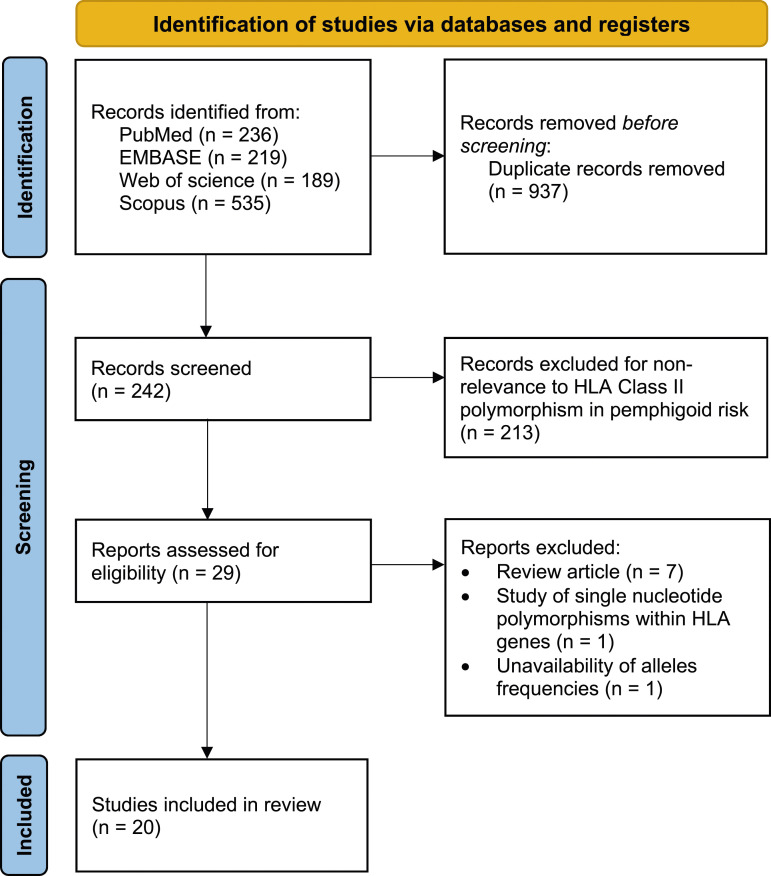
A PRISMA flow diagram was generated to depict the study selection process (Figure 1). Overall, 20 studies with a total of 1485 pemphigoid cases (BP: n = 1007, MMP: n = 423, and DBP: n = 55) and 28,117 controls were included in the present study.12–31 The characteristics of included studies are summarized in Table 1. The frequencies of the following HLA class II alleles and haplotypes in patients and controls are shown in Tables S1, S2, S3, S4, S5, S6 and S7: DQB1*0301, DRB1*11, DRB1*1101, DQA1*0505, DQA1*0201, DRB1*1101-DQB1*0301 and DRB1*11-DQA1*05-DQB1*0301. Twenty studies were included for the DQB1*0301 allele,12–31 11 for the HLA-DB1*11 allele,14,19–24,26,27,29,31 five for the DRB1*1101,14,20,24,26,28 four for the DQA1*0505 allele,16,22,26,29 four for the DQA1*0201 allele,16,21,22,31 eight for the DRB1*1101-DQB1*0301 haplotype14,19–22,26,27,31 and four for the DRB1*11-DQA1*05-DQB1*0301 haplotype.21,22,26,31 The NOS quality score results for each included study are shown in Table 1. Risks of bias are summarized in Figure 2.
Figure 1.
PRISMA flow diagram for study selection.
Figure 2.
Summary of study risk of bias.
HLA-DQB1*0301 allele meta-analysis
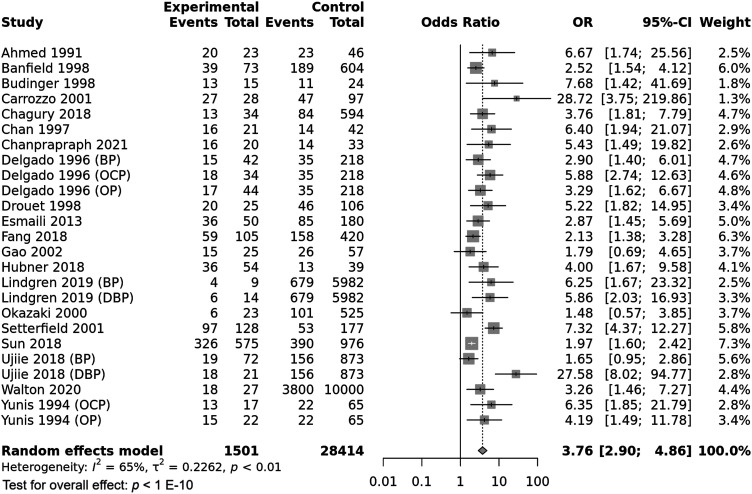
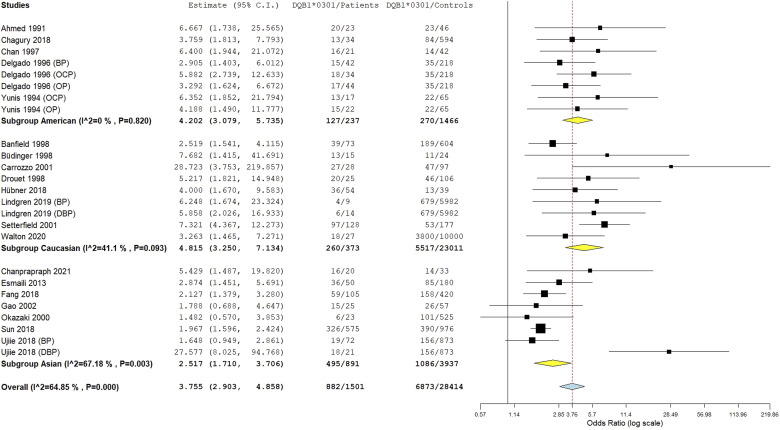
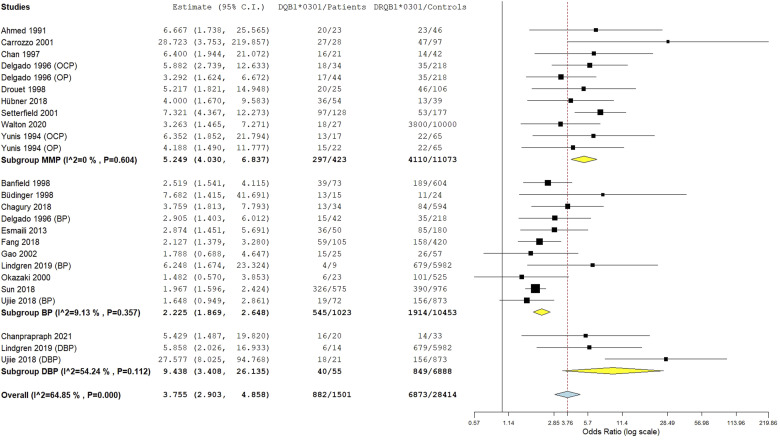
The HLA-DQB1*0301 wild allele was significantly associated with increased pemphigoid risk, OR [95% CI] = 3.76 [2.9–4.86], p < 1 E−10 (Table 2 and Figure 3). Nevertheless, there was a high level of between-studies heterogeneity, I2 = 65%, Tau2 = 0.2262, 95% PI = [1.36–10.43] and p < .01. Subgroup analysis by ethnicity revealed that if the association of the DQB1*0301 allele with pemphigoid susceptibility remained significant in all ethnic groups, the increase of pemphigoid risk was significantly higher in American and Caucasian populations comparatively to Asian (Tables S8 and S9 and Figure 4). Moreover, the risk conferred by the DQB1*0301 allele was significantly higher in case of DBP comparatively to BP and MMP; and in MMP patients than in BP cases (Tables S8 and S9 and Figure 5). Subsequent meta-regression revealed a significant positive correlation of the effect size with DIF positivity rate, p < .0001 (Table S9 and Figure S2). Conversely, the effect size was not correlated with age, gender, or positivity rate of IIF, anti-BP230/BP180 Abs and DQB1*0301 allele frequency in control groups (Table S9 and Figure S3).
Table 2.
Main results of HLA-DRB1, DQB1 and DQA1 associations with pemphigoid risk.
| Allele / Haplotype | Pemphigoid risk | Heterogeneity | |||
|---|---|---|---|---|---|
| OR [95% CI] | p-value | I2(%) | p-value | 95% PI | |
| DQB1*0301 | 3.76 [2.9–4.86] | <1 E−10 | 65 | <.001 | 1.36–10.43 |
| DRB1*11 | 3.17 [2.14–4.7] | 1 E-8 | 52 | .02 | 1.06–9.46 |
| DRB1*1101 | 4.35 [2.44–7.76] | 6.3 E-7 | 38 | .15 | 1.45–13.06 |
| DQA1*0505 | 4.39 [2.4–8.04] | 1.6 E-6 | 68 | .01 | 0.57–33.55 |
| DQA1*0201 | 0.3 [0.17–0.52] | 1.6 E-5 | 0 | .76 | 0.17–0.52 |
| DRB1*1101-DQB1*0301 | 2.8 [2.01–3.89] | 1.1 E-9 | 16 | .3 | 1.5–5.23 |
| DRB1*11-DQA1*05-DQB1*0301 | 2.13 [1.44–3.16] | .00016 | 0 | .48 | 1.44–3.16 |
Figure 3.
Forest plot for the association between the HLA-DQB1*0301 allele and pemphigoid risk.
Figure 4.
Subgroup analysis by ethnicity for the HLA-DQB1*0301 allele.
Figure 5.
Subgroup analysis by pemphigoid subtype for the HLA-DQB1*0301 allele.
HLA-DRB1*11 allele meta-analysis
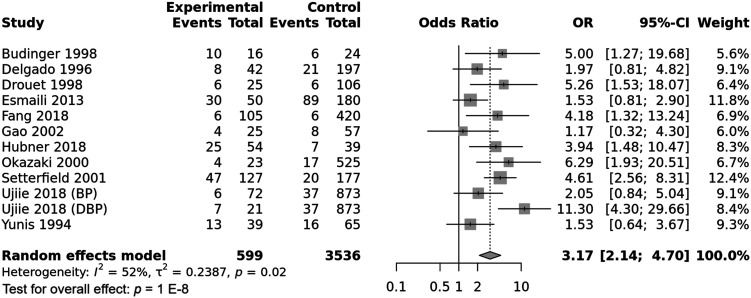
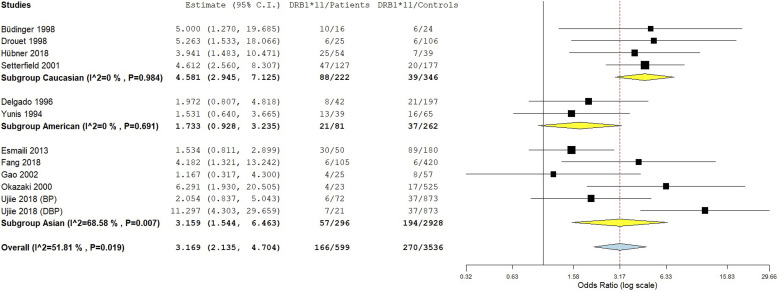
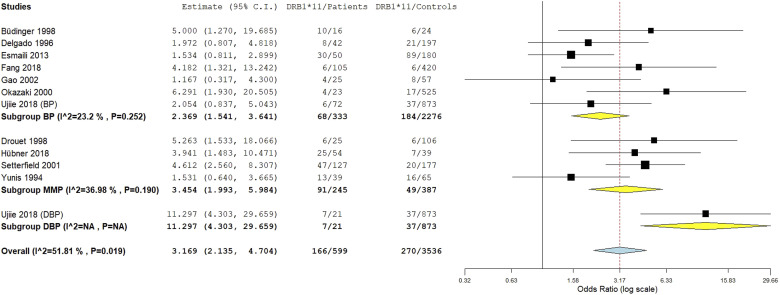
The integrated analysis revealed a significant association between the DRB1*11 allele and susceptibility to pemphigoid, OR [95% CI] = 3.17 [2.14–4.7], p = 1 E-8 (Table 2 and Figure 6). However, the heterogeneity between included studies was substantial, I2 = 52%, Tau2 = 0.2387, 95% PI = [1.18–3.83], p < .01. Subgroup analysis by ethnicity revealed that while the association of the DRB1*11 allele with pemphigoid susceptibility remained significant in all ethnic groups, the increase of pemphigoid risk was significantly greater in Asian and Caucasian populations comparatively to American (Tables S10 and S11 and Figure 7). Besides, subgroup analysis by pemphigoid subtype revealed a significant higher risk conferred by the DRB1*11 allele in DBP patients comparatively to BP and MMP cases (Tables S10 and S11 and Figure 8). Meta-regression revealed a significant negative correlation of the effect size with the DRB*11 allele frequency in control groups, p = .01 (Table S11 and Figure S4). Conversely, the strength of the association between the DRB1*11 allele and pemphigoid risk was not correlated with patients age and gender ratio (M/F) (Table S11 and Figures S5 and S6).
Figure 6.
Forest plot for the association between the HLA-DRB1*11 allele and pemphigoid risk.
Figure 7.
Subgroup analysis by ethnicity for the HLA-DRB1*11 allele.
Figure 8.
Subgroup analysis by pemphigoid subtype for the HLA-DRB1*11 allele.
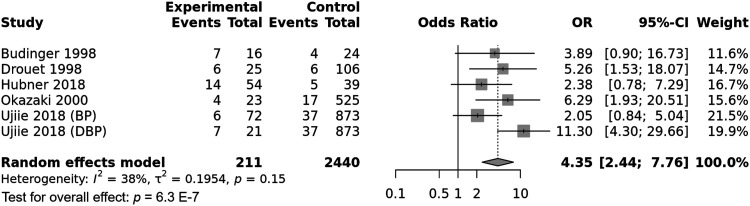
HLA-DRB1*1101 allele meta-analysis
The DRB1*1101 allele was significantly associated with pemphigoid susceptibility, OR [95% CI] = 4.35 [2.44–7.76], p = 6.3 E-7 (Table 2 and Figure 9). Of note, there was a moderate amount of between-studies heterogeneity, I2 = 38%, Tau2 = 0.1954, 95% PI = [1.45–13.06] and p = .15. Subgroup analysis by ethnicity did not provide the present study with information on potential sources of heterogeneity as there was no difference regarding the effect size between Asian and Caucasian populations (Tables S12 and S13 and Figure S7). Conversely, the risk of pemphigoid conferred by the DRB1*1101 allele was significantly higher in case of DBP comparatively to BP and MMP, p = .006 (Tables S12 and S13 and Figure 10). Meta-regression revealed a significant positive correlation between the effect size and patients age, p = .02 (Table S13 and Figure S8). Of note, gender ratio (F/M) and DRB1*1101 allele frequency in control group were not found to be correlated with the effect size (Table S13 and Figures S9 and S10).
Figure 9.
Forest plot for the association between the HLA-DRB1*1101 subtype and pemphigoid risk.
Figure 10.
Subgroup analysis by pemphigoid subtype for the HLA-DRB1*1101 subtype.
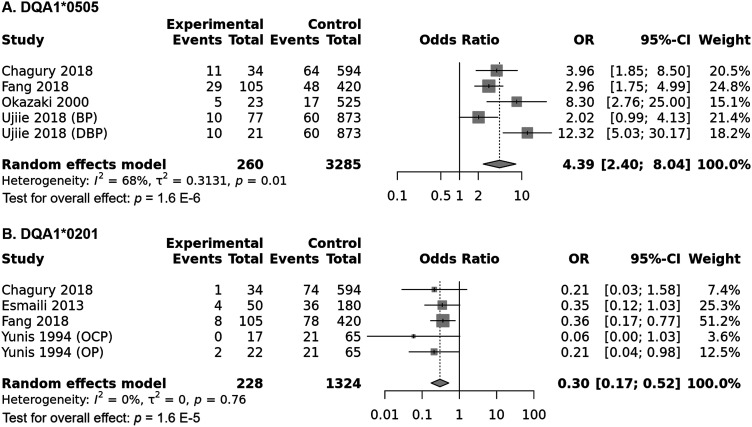
HLA-DQA1*0505 allele meta-analysis
Combined analysis showed a significant association of the DQA1*0505 allele with increased pemphigoid risk, OR [95% CI] = 4.39 [2.4–8.04], p < 1 E−10 (Table 2 and Figure 11(a)). Nonetheless, there was a considerable amount of between-studies heterogeneity, I2 = 68%, Tau2 = 0.3131, 95% PI = [0.57–33.55], p = .01. Subgroup analysis by ethnicity did not reveal any significant difference in pemphigoid risk between Asian and Caucasian populations, p = .567 (Tables S14 and S15 and Figure S11). Inversely, the association between the DQA1*0505 and pemphigoid was significantly stronger in case of DBP comparatively to BP, p = .006 (Tables S14 and S15 and Figure S12). Subsequent meta-regression model showed that patients age was positively and significantly correlated with pemphigoid risk, while gender ratio (F/M) and the DQA1*0505 allele frequency in control groups were not (Table S15 and Figures S13, S14 and S15).
Figure 11.
(a) Forest plot for the association between the HLA-DQA1*0505 allele and pemphigoid risk. (b) Forest plot for the association between the HLA-DQA1*0201 and pemphigoid risk.
HLA-DQA1*0201 allele meta-analysis
The DQA1*0201 allele was significantly associated with decreased pemphigoid risk, OR [95% CI] = 0.3 [0.17–0.52], p = 1.6 E-5 (Table 2 and Figure 11(b)). Besides there was no between-studies heterogeneity, I2 = 0%, Tau2 = 0, 95% PI = [0.17–0.52], p = .76. Subgroup analysis by ethnicity revealed that the association remained significant in all ethnic groups (Table S16 and S17 and Figure S16). Similarly, the decrease of pemphigoid risk conferred by the DQB1*0201 did not differ between BP and MMP, p = .686 (Tables S16 and S17 and Figure S17). In addition, meta-regression did not reveal any significant correlation of the DQA1*0201 allele frequency in control groups and the effect size (Table S17 and Figure S18).
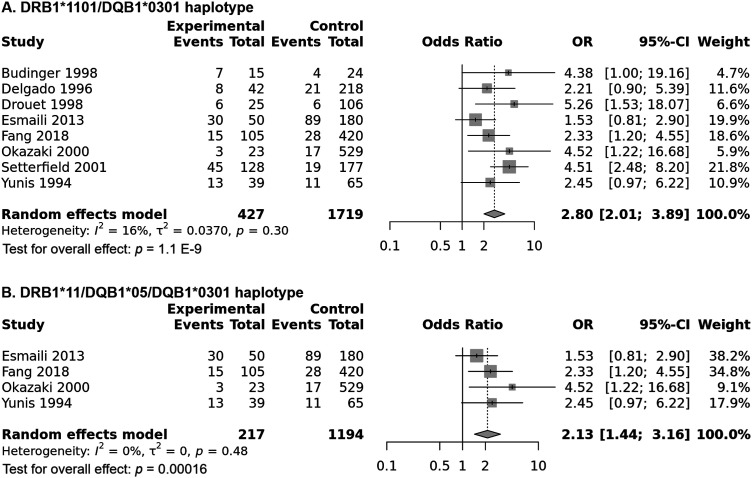
HLA-DRB1*1101-DQB1*0301 haplotype meta-analysis
Combined analysis showed a significant association of the DRB1*1101-DQB1*0301 haplotype with increased pemphigoid risk, OR [95% CI] = 2.8 [2.01–3.89], p = 1.1 E-9 (Table 2 and Figure 12(a)). In addition, there was a low level of between-studies heterogeneity, I2 = 16%, Tau2 = 0.037, 95% PI = [1.5–5.23], p = .3. Subgroup analysis by ethnicity revealed that if the association remained significant among all ethnic groups, the risk conferred by the DRB1*1101-DQB1*0301 haplotype was significantly higher in Caucasians (OR [95% CI] = 4.61 [2.78–7.65]) than in Asians (OR [95% CI] = 2.1 [1.3–3.41] and American (OR [95% CI] = 2.32 [1.22 – 4.42]; p = .041 and p = .026, respectively (Tables S18 and S19 and Figure 13). Moreover, the strength of the association between the DRB1*1101-DQB1*0301 haplotype with pemphigoid was significantly higher MMP (OR [95% CI] = 3.96 [2.48–6.3]) than in BP (OR [95% CI] = 2.19 [1.5–3.2]), p = .047 (Tables S18 and S19 and Figure 14). Besides, meta-regression showed while patients age was not correlated with the effect size (Figure S19), gender-ratio (F/M) was positively correlated with the pemphigoid risk conferred by the DRB1*1101-DQB1*0301 haplotype, p = .011 (Table S19 and Figure S20). In addition, this haplotype frequency in controls was negatively correlated with the effect size, p = .03 (Table S19 and Figure S21).
Figure 12.
(a) Forest plot for the association between the HLA-DRB1*1101-DQB1*0301 haplotype and pemphigoid risk. (b) Forest plot for the association between the HLA-DRB1*11-DQA1*05-DQB1*0301 haplotype and pemphigoid risk.
Figure 13.
Subgroup analysis by ethnicity for the HLA-DRB1*1101-DQB1*0301 haplotype.
Figure 14.
Subgroup analysis by pemphigoid subtype for the HLA-DRB1*1101-DQB1*0301 haplotype.
HLA-DRB1*11-DQA1*05-DQB1*0301 haplotype meta-analysis
Only four studies investigated the influence of the HLA-DRB1*11-DQA1*05-DQB1*0301 haplotype on susceptibility to pemphigoid. This peculiar haplotype was significantly associated with increased pemphigoid risk, OR [95% CI] = 2.13 [1.44–3.16], p = .00016 (Table 2 and Figure 12(b)). In addition, there was no evidence of heterogeneity between included studies, I2 = 0%, Tau2 = 0, 95% PI = [1.44–3.16], p = .48. Subgroup analyses by ethnicity and pemphigoid subtype did not reveal any significant difference (Tables S20 and S21 and Figures S22 and S23). Meta-regressions revealed a positive correlation of the effect size with gender ratio (F/M), p = .042 (Table S21 and Figure S24), and a negative correlation with the DRB1*11-DQA1*05-DQB1*0301 haplotype frequency in control groups, p = .039 (Table S21 and Figure S25).
Sensitivity analysis
The sensitivity analysis revealed that association results were stable for all performed meta-analyses (Figures S26, S27, S28, S29, S30, S31 and S32), suggesting a high level of integrity with reliable results.
Publication bias
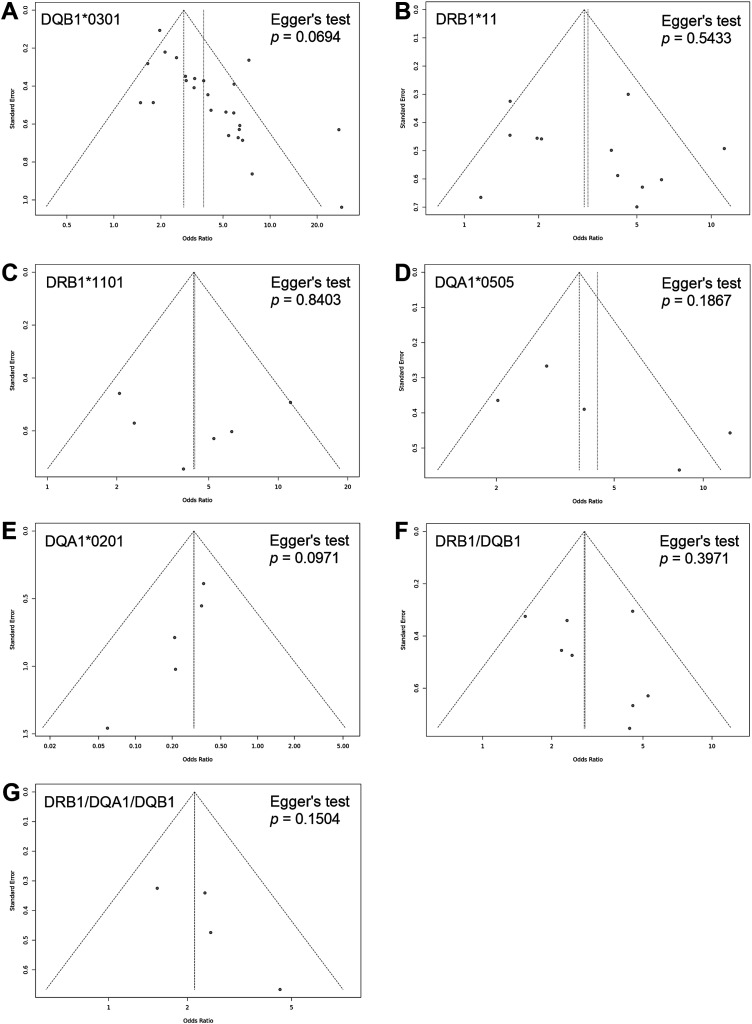
Generated funnel plots (Figure 15) were found to be overall symmetrical and Egger’s tests confirmed these findings with non-significant p-values for all investigated HLA class II alleles and haplotypes (.0694, .5433, .8403, .1867, .0971, .3971, and .1504), which indicated that results were not weakened by publication biases.
Figure 15.
Funnel plots assessing publication bias: Symmetrical funnel plots with no evidence of publication biases. (a) HLA-DQB1*0301 allele. (b) HLA-DRB1*11 allele. (c) HLA-DRB1*11 subtype. (d) HLA-DQA1*0505 allele. (e) HLA-DQA1*0201 allele. (f) HLA-DRB1*1101-DQB1*0301 haplotype. (g) HLA-DRB1*11-DQA1*05-DQB1*0301 haplotype.
Discussion
Pemphigoid is an antibody-mediated cutaneous and/or mucous membrane disease caused by the binding of autoantibodies, mainly anti-BP230 and anti-BP180 Abs, to components of hemidesmosomes.1,32 In addition, to their diagnostic specificity for pemphigoid, these autoantibodies have also been reported to be of a prognostic significance.32,33 In this regard, HLA class II risk alleles showed an increased avidity to targeted antigens by these autoantibodies.32,33 This increased avidity of HLA alleles with respect to target autoantigens favors their presentation to autoreactive T cells, which in turn results in autoantibodies production by specific autoreactive B cells.32,33
Like other autoimmune diseases, BP and MMP etiology is complex including multigenetic background and environmental factors.1–4 Among genetic factors, HLA class II genes were the most associated with pemphigoid risk. In fact, over the past 3 decades several studies focused on HLA-DQB1, DRB1 and DQA1 gene polymorphisms, though with inconsistent findings.3,4 Although, a previous meta-analysis assessing HLA-DQA1 polymorphism role in BP genetic risk has already been published in 2023, 6 HLA-DQB1 and DRB1 gene polymorphisms impact on pemphigoid risk needs to be summarized. Therefore, this study aimed to summarize published results on pemphigoid risk conferred by HLA-DQB1 and DRB1 alleles and haplotypes together with an update on HLA-DQA1 role in pemphigoid susceptibility. In addition, while the previous meta-analysis involved only four studies, our meta-analysis included 20 reports which could provide a more comprehensive and accurate estimation of HLA class II alleles influence on pemphigoid risk. Furthermore, we assessed the between-studies heterogeneity by subgroup analyses and building univariate and multivariate models of meta-regression. Overall, the HLA-DQB1*0301, DRB1*11, DRB1*1101, DQA1*0505 alleles were found to be associated with pemphigoid risk, whereas the DQA1*0201 allele was protective against pemphigoid. The associations with pemphigoid risk were stronger in case of DBP comparatively to BP and MMP. In addition, the risk conferred by the DQB1*0301 was greater in MMP than in BP.
The present study revealed that the HLA-DQB1*0301 allele was associated with roughly a 3.76-fold increase in pemphigoid risk, though with a high level of between-studies heterogeneity. In order to explore this heterogeneity, several subgroup analyses and meta-regressions were carried out. Interestingly, the strength of the association with pemphigoid was greater in American and Caucasian when compared to Asian. It should be noted that the included studies from American populations specified that the patients recruited were mostly of European descent and Caucasian in morphology.12,16,19,31 Observed inter-ethnic difference regarding the association strength could be due to different genetic background as well as disparate environmental factors. Of note, between-studies inconsistency remained significant in the Asian subgroup (I2 = 67%). Asian subgroup included a study performed in DBP patients with a much greater OR, which resulted in a significant heterogeneity. 29 Besides, the DQB1*0301 allele conferred an almost 9.44-fold increase in the risk of DBP which was significantly greater than the conferred risk of BP and MMP. This peculiar finding points to a possible interaction between HLA-DQB1*0301 allele and the use of DPP-4 inhibitors. Nevertheless, it is important to acknowledge that there were only three included studies that have examined the role of the DQB1*0301 allele in DBP risk along with a total number of patients of 55. Hence, further studies in DBP patients are required to confirm this finding. Moreover, in the study of Ujiie, 29 the DQB1*0301 conferred a much higher risk for DBP when compared to the two other included studies (27.58 vs ≈5) which resulted in a significant between-studies heterogeneity (I2 = 54%). These differences could be due to a small number of patients, which could have led to sampling errors. In addition, the risk conferred by the DQB1*0301 allele was significantly higher in case of MMP (≈5.25-fold) comparatively to BP (≈2.22-fold). This indicates that although these two entities share, at least in part, the same genetic background and some immunopathological features, they are two distinct but related conditions. Indeed, while most BP patients raise IgG against epitopes outside the NC16A domain of BP180, this domain is targeted in 50% of MMP patients.1,2 Remarkably, reactivity to NC16A domain in BP patients was found to be associated with mucosal involvement in addition to a more severe skin disease. 1 Therefore, we can hypothesize that the HLA-DQB1*0301 allele may have a higher affinity for NC16A domain epitopes, which could lead to a stronger association with MMP risk than with BP risk. Besides, the present study revealed a significant correlation of DIF positivity rate with the risk of pemphigoid conferred by the DQB1*0301 allele. Hence, the association was rather noted in DIF-positive patients, which demonstrates the link between the DQB1*0301 allele and self-reactivity towards hemidesmosome antigens.
In this meta-analysis, the DRB1*11 allele conferred a roughly 3.17-fold increase in pemphigoid risk. As for the DQB1*0301 allele, the association of the DRB1*11 allele with susceptibility to pemphigoid suffered from a significant amount of between-studies inconsistency. The association remained significant in all ethnic groups, even though the effect size was smaller in American. However, only one study from an American population with two small MMP subgroups (OCP and OP) was included in the meta-analysis. Hence future studies in American populations are needed. Besides, the DRB1*11 allele conferred an approximately 11.3-fold increase in the risk of DBP which was significantly larger than the conferred risk of BP and MMP. As for the DQB1*0301 allele, there might be a gene-drug interaction for the DRB1*11 allele, though there was only one included study of DBP patients. Therefore, more studies in DBP patients are required to confirm this finding. Conversely, no significant difference regarding the effect size between BP and MMP was noted. Of note, no previous study investigated targeted autoantigen epitopes affinity with DRB1*11 allele. Besides, meta-regression revealed a negative correlation of the effect size with the DRB1*11 allele frequency in control groups. Thus, the association between the DRB1*11 allele and pemphigoid risk was rather observed when the aforementioned allele is rare in the general population.
This study showed that the DRB1*1101 subtype was associated with roughly 4.34-fold increased pemphigoid risk. Therefore, Thus, the association of the DRB1*11 allele with the risk of pemphigoid is predominantly carried by the DRB1*1101 subtype. The between-studies heterogeneity was moderate and the predicted risk might fluctuate between 1.45- and 13-fold increase in pemphigoid risk. Almost all included studies were performed in Asians, thus, subgroup analysis by ethnicity could not be useful to determine the sources of heterogeneity. Conversely, the effect size was significantly larger in DBP comparatively to BP and MMP suggesting a drug-gene interaction. Interestingly, meta-regression revealed a significant positive correlation between patients age and the effect size. This finding indicated that the association between the DRB1*1101 subtype and pemphigoid was rather noted in more elderly patients which suggests a possible interaction between age and the DRB1*1101 allele in susceptibility to pemphigoid.
In the current study, the HLA-DQA1*0505 allele was associated with approximately 4.4-fold higher pemphigoid risk. The heterogeneity was substantial and the true effect in 95% of comparable populations could vary from 0.57 to 33.55 times. Subgroup analysis by ethnicity was of no help as most include studies were carried out in Asian populations. Inversely, the effect size was significantly larger in DBP than in BP as the DQA1*0505 allele conferred a 12.32-fold increased risk which is suggestive of a drug-gene interaction. Again, meta-regression showed a significant positive correlation of the effect size with patients age, which is indicative of a stronger association in more elderly patients.
Besides, the combined analysis revealed a 3.33-fold decrease in pemphigoid risk conferred by the HLA-DQA1*0201 allele. In addition, there was no between-studies heterogeneity. Even though there was no inconsistency between included studies, subgroup analyses and meta-regression were performed. Subgroup analyses by ethnicity and pemphigoid subtype did not show any significant difference regarding the effect size. Similarly, meta-regression did not reveal any significant correlation of the effect size with the DQA1*0201 allele frequency in control groups. Hence, it seems that the DQA1*0201 might play a protective role against pemphigoid.
Besides, considering the universal strong linkage disequilibrium between HLA-alleles, 34 haplotype meta-analyses were performed. In this study, the HLA-DRB1*1101-DQB1*0301 haplotype was associated with approximately 2.8 times increased pemphigoid risk with a low level of between-studies heterogeneity (I2 = 16%). Subgroup analysis revealed that the DRB1*1101-DQB1*0301 haplotype conferred a twofold higher risk of MMP than BP (3.96 vs 2.19). Moreover, the risk of pemphigoid conferred by this haplotype was significantly larger in Caucasian comparatively to American and Asian populations. These differences could be due to disparate genetic background and additional environmental factors. Besides, there was a significant correlation between gender ratio (F/M) and the effect size, which is suggestive that the risk of pemphigoid conferred by this haplotype is higher in female patients. In addition, meta-regression revealed a negative correlation of the effect size with the DRB1*1101-DQB1*0301 haplotype frequency in control groups. Thus, the association between this haplotype and pemphigoid risk was rather observed when it is rare in the general population.
Only four studies reported the HLA-DRB1*11-DQA1*05-DQB1*0301 haplotype frequency in pemphigoid patients. Integrated analysis showed a 2.13-fold increase in pemphigoid risk conferred by this haplotype without any between-studies heterogeneity (I2 = 0%). Despite the absence of inconsistency between included studies, subgroup analyses and meta-regression have been performed. Subgroup analyses by ethnicity and pemphigoid subtype did not show any significant difference. Conversely, the risk of pemphigoid conferred by the DRB1*11-DQA1*05-DQB1*0301 haplotype was positively correlated with gender ratio (F/M) which is suggestive of increased effect in females comparatively to males. Moreover, there was a significant negative correlation between this haplotype frequency in control groups and the effect size which indicates an increased risk when it is rare in the general population.
In summary, the present meta-analysis revealed that the HLA-DQB1*0301, DRB1*11/DRB1*1101 and the DQA1*0505 alleles could play a significant role in susceptibility to pemphigoid. However, it is noteworthy to admit the substantial between studies heterogeneity for some studied alleles. Indeed, most effect sizes were larger in DBP than in BP and MMP; and in MMP comparatively to BP. In addition, there were inter-ethnic differences regarding the associations between the aforementioned alleles and pemphigoid risk. Likewise, haplotypes bearing these risk alleles were significantly associated with pemphigoid risk. Nevertheless, it is important to acknowledge that only the DQB1*0301 allele has previously been shown to have increased affinity to epitopes from BP180, BP230 and other components of hemidesmosomes.32,33 Hence, the association of the DRB1*1101 and DQA1*0505 could be accounted for by the linkage disequilibrium with the DQB1*0301 allele. computer modeling could help determine whether there are T-cell epitopes from target autoantigens that can be presented by the DRB1*1101 and DQA1*0505 alleles.
Besides, to the best of our knowledge, this study was the first to perform several meta-regressions in order to explore the between-studies heterogeneity. Meta-regressions provided the present study with some interesting findings such as significant correlations with DIF positivity rate, age, gender ratio (F/M) and allele/haplotype frequencies in control groups. However, there are some limitations that need to be acknowledged. Firstly, like other autoimmune disease, pemphigoid risk depends on several factors such as environmental, infectious, psychological and genetic factors, and as the present study analyses derived from pooling HLA class II aggregate findings without any access to raw data, there was a lack of further adjustment for baseline characteristics. Secondly, the majority of included studies were performed in Caucasians, American of European descent and Asians. Hence, the present meta-analysis results cannot be generalized to sub-Saharan-African, North-African and indigenous American populations. Thirdly, there were only a few numbers of included studies in the HLA-DQA1*0505 and DQA1*0201 alleles meta-analyses, and consequently our results require replication in independent large cohorts. Fourthly, the included studies did not specify BPDAI or MMPDAI in patients and response to treatment. This issue prevented us from analyzing the impact of HLA class II alleles and haplotypes on disease severity and progression under treatment.
Conclusions
This meta-analysis demonstrated that the DRB1*1101, DQA1*0505 and DQB1*0301 were significantly associated with increased pemphigoid risk. These associations were found to be significantly stronger in case of DBP comparatively to idiopathic pemphigoid. The DQA1*0201 allele seems to play a protective role against pemphigoid.
Supplemental Material
Supplemental Material for Association of HLA class II alleles and haplotypes with bullous and mucus membrane pemphigoid risk: A systematic review, a meta-analysis and a meta-regression by Tarak Dhaouadi, Awatef Riahi, Taïeb Ben Abdallah, Yousr Gorgi and Imen Sfar in International Journal of Immunopathology and Pharmacology
Supplemental Material for Association of HLA class II alleles and haplotypes with bullous and mucus membrane pemphigoid risk: A systematic review, a meta-analysis and a meta-regression by Tarak Dhaouadi, Awatef Riahi, Taïeb Ben Abdallah, Yousr Gorgi and Imen Sfar in International Journal of Immunopathology and Pharmacology
Appendix.
Abbreviations
- BP
Bullous pemphigoid
- MMP
Mucous membrane pemphigoid
- OCP
Ocular cicatricial pemphigoid
- OP
Oral pemphigoid
- BPDAI
BP disease area index
- MMPDAI
MMP disease are index
- DPP-4
Dipeptidyl peptidase-4 inhibitors
- DBP
DPP-4-induced bullous pemphigoid
- Abs
antibodies
- HLA
Human Leukocyte Antigen
- NOS
Newcastle-Ottawa Scale
- PI
prediction interval
- NC16A
16th non-collagenous domain of BP180
Author contributions: Conceptualization: Tarak Dhaouadi, Aouatef Riahi, Yousr Gorgi, Taieb Ben Abdallah, Imen Sfar. Data curation: Tarak Dhaouadi, Awatef Riahi. Formal analysis: Tarak Dhaouadi, Imen Sfar. Investigation: Tarak Dhaouadi, Awatef Riahi, Imen Sfar. Methodology: Tarak Dhaouadi, Imen Sfar, Taieb Ben Abdallah, Yousr Gorgi. Project administration: Yousr Gorgi, Taieb Ben Abdallah, Imen Sfar. Supervision: Yousr Gorgi, Taieb Ben Abdallah, Imen Sfar. Writing - original draft: Tarak Dhaouadi. Writing - review & editing: Tarak Dhaouadi.
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: This study was granted and supported by the Research Laboratory in Immunology of Renal Transplantation and Immunopathology (LR03SP01), Charles Nicolle Hospital, Tunis El Manar University, Tunisia.
Supplemental Material: Supplemental material for this article is available online.
ORCID iD
Tarak Dhaouadi https://orcid.org/0000-0002-6376-0950
Data availability statement
The authors confirm that the data supporting the findings of this study are available within the article [and/or] its supplementary materials.
References
- 1.Schmidt E, Zillikens D. (2013) Pemphigoid diseases. Lancet 381(9863): 320–332. DOI: 10.1016/S0140-6736(12)61140-4. [DOI] [PubMed] [Google Scholar]
- 2.Du G, Patzelt S, van Beek N, et al. (2022) Mucous membrane pemphigoid. Autoimmunity Reviews 21(4): 103036. DOI: 10.1016/j.autrev.2022.103036. [DOI] [PubMed] [Google Scholar]
- 3.Moro F, Fania L, Sinagra JLM, et al. (2020) Bullous pemphigoid: trigger and predisposing factors. Biomolecules 10(10): 1432. DOI: 10.3390/biom10101432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang J, Wang G. (2020) Genetic predisposition to bullous pemphigoid. Journal of Dermatological Science 100(2): 86–91. DOI: 10.1016/j.jdermsci.2020.05.010. [DOI] [PubMed] [Google Scholar]
- 5.Yang W, Cai X, Zhang S, et al. (2021) Dipeptidyl peptidase-4 inhibitor treatment and the risk of bullous pemphigoid and skin-related adverse events: a systematic review and meta-analysis of randomized controlled trials. Diabetes/ Metabolism Research and Reviews 37(3): e3391. DOI: 10.1002/dmrr.3391. [DOI] [PubMed] [Google Scholar]
- 6.Hesari R, Thibaut D, Schur N, et al. (2023) Bullous pemphigoid and human leukocyte antigen (HLA)-DQA1: a systematic review. Cureus 15(6): e39923. DOI: 10.7759/cureus.39923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McLeroy KR, Northridge ME, Balcazar H, et al. (2012) Reporting guidelines and the American Journal of Public Health’s adoption of preferred reporting items for systematic reviews and meta-analyses. American Journal of Public Health 102(5): 780–784. DOI: 10.2105/AJPH.2011.300630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wells GA, Shea B, O’Connell D, et al. The Newcastle-Ottawa Scale (NOS) for assessing the quality if nonrandomized studies in meta-analyses. Available from: https://www.ohri.ca/programs/clinical_epidemiology/oxford.htm [cited 2024 January 05].
- 9.Borenstein M, Hedges LV, Higgins JP, et al. (2010) A basic introduction to fixed-effect and random-effects models for meta-analysis. Research Synthesis Methods 1(2): 97–111. DOI: 10.1002/jrsm.12. [DOI] [PubMed] [Google Scholar]
- 10.DerSimonian R, Laird N. (2015) Meta-analysis in clinical trials revisited. Contemporary Clinical Trials 45(Pt A): 139–145. DOI: 10.1016/j.cct.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Egger M, Davey Smith G, Schneider M, et al. (1997) Bias in meta-analysis detected by a simple, graphical test. BMJ 315(7109): 629–634. DOI: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ahmed AR, Foster S, Zaltas M, et al. (1991) Association of DQw7 (DQB1*0301) with ocular cicatricial pemphigoid. Proceedings of the National Academy of Sciences of the United States of America 88(24): 11579–11582. DOI: 10.1073/pnas.88.24.11579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Banfield CC, Wojnarowska F, Allen J, et al. (1998) The association of HLA-DQ7 with bullous pemphigoid is restricted to men. British Journal of Dermatology 138(6): 1085–1090. DOI: 10.1046/j.1365-2133.1998.02350.x. [DOI] [PubMed] [Google Scholar]
- 14.Büdinger L, Borradori L, Yee C, et al. (1998) Identification and characterization of autoreactive T cell responses to bullous pemphigoid antigen 2 in patients and healthy controls. The Journal of Clinical Investigation 102(12): 2082–2089. DOI: 10.1172/JCI3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carrozzo M, Fasano ME, Broccoletti R, et al. (2001) HLA-DQB1 alleles in Italian patients with mucous membrane pemphigoid predominantly affecting the oral cavity. British Journal of Dermatology 145(5): 805–808. DOI: 10.1046/j.1365-2133.2001.04448.x. [DOI] [PubMed] [Google Scholar]
- 16.Chagury AA, Sennes LU, Gil JM, et al. (2018) HLA-C*17, DQB1*03:01, DQA1*01:03 and DQA1*05:05 alleles associated to bullous pemphigoid in Brazilian population. Annals of Dermatology 30(1): 8–12. DOI: 10.5021/ad.2018.30.1.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chan LS, Hammerberg C, Cooper KD. (1997) Significantly increased occurrence of HLA-DQB1*0301 allele in patients with ocular cicatricial pemphigoid. Journal of Investigative Dermatology 108(2): 129–132. DOI: 10.1111/1523-1747. [DOI] [PubMed] [Google Scholar]
- 18.Chanprapaph K, Pratumchart N, Limtong P, et al. (2021) Dipeptidyl peptidase-4 inhibitor-related bullous pemphigoid: a comparative study of 100 patients with bullous pemphigoid and diabetes mellitus. The Journal of Dermatology 48(4): 486–496. DOI: 10.1111/1346-8138.15778. [DOI] [PubMed] [Google Scholar]
- 19.Delgado JC, Turbay D, Yunis EJ, et al. (1996) A common major histocompatibility complex class II allele HLA-DQB1* 0301 is present in clinical variants of pemphigoid. Proceedings of the National Academy of Sciences of the United States of America 93(16): 8569–8671. DOI: 10.1073/pnas.93.16.8569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Drouet M, Delpuget-Bertin N, Vaillant L, et al. (1998) HLA-DRB1 and HLA-DQB1 genes in susceptibility and resistance to cicatricial pemphigoid in French Caucasians. Eur The Journal of Dermatology 8(5): 330–333. [PubMed] [Google Scholar]
- 21.Esmaili N, Mortazavi H, Chams-Davatchi C, et al. (2013) Association between HLA-DQB1*03:01 and Bullous pemphigoid in Iranian patients. Iranian Journal Immunology 10(1): 1–9. [PubMed] [Google Scholar]
- 22.Fang H, Shen S, Zheng X, et al. (2018) Association of HLA class I and class II alleles with bullous pemphigoid in Chinese Hans. Journal of Dermatological Science 89(3): 258–262. DOI: 10.1016/j.jdermsci.2017.11.014. [DOI] [PubMed] [Google Scholar]
- 23.Gao XH, Winsey S, Li G, et al. (2002) HLA-DR and DQ polymorphisms in bullous pemphigoid from northern China. Clinical and Experimental Dermatology 27(4): 319–321. DOI: 10.1046/j.1365-2230.2002.01037.x. [DOI] [PubMed] [Google Scholar]
- 24.Hübner F, Setterfield J, Recke A, et al. (2018) HLA alleles in British Caucasians with mucous membrane pemphigoid. Eye (London, England) 32(9): 1540–1541. DOI: 10.1038/s41433-018-0092-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lindgren O, Varpuluoma O, Tuusa J, et al. (2019) Gliptin-associated bullous pemphigoid and the expression of dipeptidyl peptidase-4/CD26 in bullous pemphigoid. Acta Dermato-Venereologica 99(6): 602–609. DOI: 10.2340/00015555-3166. [DOI] [PubMed] [Google Scholar]
- 26.Okazaki A, Miyagawa S, Yamashina Y, et al. (2000) Polymorphisms of HLA-DR and -DQ genes in Japanese patients with bullous pemphigoid. The Journal of Dermatology 27(3): 149–156. DOI: 10.1111/j.1346-8138.2000.tb02141.x. [DOI] [PubMed] [Google Scholar]
- 27.Setterfield J, Theron J, Vaughan RW, et al. (2001) Mucous membrane pemphigoid: HLA-DQB1*0301 is associated with all clinical sites of involvement and may be linked to antibasement membrane IgG production. British Journal of Dermatology 145(3): 406–414. DOI: 10.1046/j.1365-2133.2001.04380.x. [DOI] [PubMed] [Google Scholar]
- 28.Sun Y, Liu H, Wang Z, et al. (2018) The HLA-DQB1*03:01 is associated with bullous pemphigoid in the Han Chinese population. Journal of Investigative Dermatology 138(8): 1874–1877. DOI: 10.1016/j.jid.2018.02.021. [DOI] [PubMed] [Google Scholar]
- 29.Ujiie H, Muramatsu K, Mushiroda T, et al. (2018) HLA-DQB1*03:01 as a biomarker for genetic susceptibility to bullous pemphigoid induced by DPP-4 inhibitors. Journal of Investigative Dermatology 138(5): 1201–1204. DOI: 10.1016/j.jid.2017.11.023. [DOI] [PubMed] [Google Scholar]
- 30.Walton R, Robinson M, Carrozzo M. (2020) A service evaluation of the diagnostic testing for mucous membrane pemphigoid in a UK oral medicine unit. Journal of Oral Pathology & Medicine 49(7): 687–692. DOI: 10.1111/jop.13054. [DOI] [PubMed] [Google Scholar]
- 31.Yunis JJ, Mobini N, Yunis EJ, et al. (1994) Common major histocompatibility complex class II markers in clinical variants of cicatricial pemphigoid. Proceedings of the National Academy of Sciences of the United States of America 91(16): 7747–7751. DOI: 10.1073/pnas.91.16.7747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Amber KT, Zikry J, Hertl M. (2017) A multi-hit hypothesis of bullous pemphigoid and associated neurological disease: is HLA-DQB1*03:01, a potential link between immune privileged antigen exposure and epitope spreading? HLA 89(3): 127–134. DOI: 10.1111/tan.12960. [DOI] [PubMed] [Google Scholar]
- 33.Oyama N, Setterfield JF, Powell AM, et al. (2006) Bullous pemphigoid antigen II (BP180) and its soluble extracellular domains are major autoantigens in mucous membrane pemphigoid: the pathogenic relevance to HLA class II alleles and disease severity. British Journal of Dermatology 154(1): 90–98. DOI: 10.1111/j.1365-2133.2005.06998.x. [DOI] [PubMed] [Google Scholar]
- 34.Robinson MA. (1998) Linkage disequilibrium. In: Encyclopedia of Immunology. 2nd ed. Amsterdam: Elsevier, 1586–1588. DOI: 10.1006/rwei.1999.0406. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Material for Association of HLA class II alleles and haplotypes with bullous and mucus membrane pemphigoid risk: A systematic review, a meta-analysis and a meta-regression by Tarak Dhaouadi, Awatef Riahi, Taïeb Ben Abdallah, Yousr Gorgi and Imen Sfar in International Journal of Immunopathology and Pharmacology
Supplemental Material for Association of HLA class II alleles and haplotypes with bullous and mucus membrane pemphigoid risk: A systematic review, a meta-analysis and a meta-regression by Tarak Dhaouadi, Awatef Riahi, Taïeb Ben Abdallah, Yousr Gorgi and Imen Sfar in International Journal of Immunopathology and Pharmacology
Data Availability Statement
The authors confirm that the data supporting the findings of this study are available within the article [and/or] its supplementary materials.