Abstract
Objective:
The main objective of the study is to explore the potential molecular benefits of Nab-paclitaxel as an effective advanced chemotherapeutic agent for HER2-positive breast cancer patients. Specifically, the study aims to assess Nab-paclitaxel as a potential drug candidate for breast cancer treatment.
Methods:
This study used bioinformatics and cheminformatics to analyze the HER2 signaling pathway and its possible interactions with Nab-Paclitaxel. This involves using pharmacokinetic profiling software to evaluate its physicochemical properties, analyzing its potential impact on gene expression modulation, and assessing its binding affinity to the HER2 receptor through molecular docking.
Result:
The results indicate that the most favorable docking pose occurs between chain B of the HER-2 receptor and Paclitaxel, with a binding energy of -9.4 kcal/mol. Notably, a hydrogen bond is observed in ARG849, with 3.0 Angstrom (Å). Previous research highlights Paclitaxel’s impact on breast cancer patients’ genes, particularly the ABCB1 gene responsible for P-glycoprotein production, contributing to drug resistance in chemotherapy. Nab-paclitaxel exhibits potential ease of metabolism, as it minimally inhibits drug-metabolizing cytochrome P450 enzymes. Additionally, despite initial concerns related to drug-likeness parameters and molecular weight discrepancies, the pharmacokinetic profile of Nab-Paclitaxel suggests improvements in delivery facilitated by an albumin-supported nanoparticle delivery mechanism.
Conclusion:
The binding energy confirms the secure docking of ligands to receptors, suggesting the stability of the interaction between them. Nevertheless, prolonged administration of Paclitaxel poses the risk of inducing drug resistance, a significant factor contributing to treatment failure. This emphasizes the need to explore new candidate drug combinations or identify alternative drug-binding interaction sites. Such endeavors hold the potential to enhance the effectiveness of drug treatments and address challenges associated with prolonged Paclitaxel use.
Key Words: HER2- Nab, Paclitaxel, cytochrome P450, ARG849, ABCB1, P-glycoprotein
Introduction
The world’s leading cause of death, according to the World Health Organization (2022)[1], is cancer. The cellular basis of cancer is the uncontrolled proliferation and division of cells due to genetic mutations that disrupt normal mechanisms regulating cell growth, division and behavior leading to the development of tumors [2, 3]. In 2020, cancer contributed to almost ten million global deaths [1]. The Breast Cancer Organization [3] projects that approximately 287,850 new cases of invasive breast cancer and 51,400 new cases of non-invasive (in situ) breast cancer are anticipated to be diagnosed in women alone.
Breast cancer is caused by the hyperproliferation of cells in the body. This involves a rapid division process, wherein the cell loses its ability to regulate and halt its division [4, 5]. The lobular and ductal epithelium of the breast may develop malignant possibilities [6]. Malignant cells can originate from the lobular or milk-producing glands; however, the majority of breast cancers arise from the ductal epithelium. Extensive research has revealed that the primary cause of death in breast cancer patients is often not the cancer located in the initial foci but rather complications associated with secondary metastasis [7, 8]. The insufficient production of the tumor-suppressing gene “E-cadherin” leads to the growth of cells in the ducts and lobes of the breast in non-cohesive clusters. In breast cancer patients, the loss of E-cadherin is attributed to the development of allele homozygosity in the gene responsible for E-cadherin expression [9]. This aberrant gene expression, resulting in the loss of E-cadherin, heightens the risk of tumor invasion by the already malignant neoplasm [10].
HER2-positive breast cancer is one of the major subtypes characterized by its rapid growth and higher propensity for spreading compared to other subtypes [11]. This aggressive form of cancer, marked by HER-2 overexpression, affects approximately 1 in 5 women diagnosed with breast cancer. Particularly, around 20% of all invasive breast cancers fall into the category of HER2-positive breast cancer [12]. HER2-positive cancers exhibit elevated levels of the HER2 protein. When overexpressed, this protein stimulates tumor cells, promoting increased proliferation and reduced susceptibility to pro-apoptotic signals, allowing them to evade cell death [13]. Conversely, treatments targeting HER2-positive breast cancer can be highly effective. Notably, Herceptin is a promising compound with significant results in HER2 inhibition. This specific compound leverages the interception of cell signaling, targeting the anti-apoptotic properties associated with the HER2 protein [14].
HER-2 overexpression is frequently linked to advanced disease stages and an unfavorable prognosis [15]. Currently, the primary therapeutic agents for HER2-positive breast cancer are trastuzumab and lapatinib [16]. However, tumors often develop resistance to these medications. A deeper understanding of the mechanisms underlying HER-2-positive breast cancer is crucial for developing novel therapeutic approaches targeting the root cause of signaling dysregulation. Consequently, chemotherapy is recommended for patients with breast cancer and is the only treatment option for patients with triple-negative breast cancer [12, 17]. This treatment may also be used in patients with HR+ illness who have developed resistance to hormone treatment, or when symptoms are severe enough to merit the use of chemotherapy. Taxanes, such as paclitaxel and Nab-paclitaxel, are a common type of chemotherapy for breast cancer, including recurrent disease following adjuvant treatment [18, 19].
Among the various cancer treatment modalities, including surgery, radiation therapy, immunotherapy, chemotherapy, and more, our focus is on investigating the effectiveness of the chemotherapeutic agent Paclitaxel in conjunction with the Nab molecule. Paclitaxel is an FDA-approved adjuvant treatment for node-positive breast cancers, metastatic breast cancers, and ovarian cancers [20]. However, it is important to note that paclitaxel carries a black box warning due to potential hypersensitivity reactions and adverse effects such as bone marrow suppression, peripheral neurotoxicity, and mucositis [21, 22]. Pharmacokinetic profiling software, including Swiss ADME and ADMET Lab 2.0, is employed to gain insights into the nature of paclitaxel as a raw material. This approach allows for a comprehensive assessment of the drug’s physicochemical properties and metabolic characteristics. Subsequent modifications can be considered to enhance the drug’s bioavailability, guided by a thorough understanding of its properties and metabolic behavior [23].
Paclitaxel functions by promoting the assembly of microtubules through tubulin dimers while concurrently stabilizing existing microtubules, and it inhibits their disassembly [22, 24]. This microtubule stabilization then inhibits the G2 phase of cell replication, and in some cases, it causes the breakage of the chromosomes by distorting their mitotic spindles [25]. Utilizing software tools such as Swiss Target and Comparative Toxicogenomic Data offers foundational insights into the general targets of the drug and the associated diseases that may benefit from favorable interactions. On the other hand, molecular docking provides a more in-depth understanding by revealing specific residues or binding sites occupied by the drug and elucidating the clinical significance inherent in the ligand-receptor interaction [26].
Nab-paclitaxel or Albumin-bound paclitaxel is a novel solvent-free taxane [27]. Taxanes play a key role in chemotherapy in malignant cancers such as metastatic breast cancer, ovarian cancer, and advanced non-small cell lung cancer [28]. Nab-paclitaxel demonstrated elevated response rates and enhanced tolerability in patients with metastatic breast cancer compared to solvent-based formulations [29, 30]. Its solvent-free formulation also enables Nab-paclitaxel to circumvent specific toxicities associated with existing solvent-based formulations, such as sb-paclitaxel and docetaxel [30, 31]. Further research is essential to comprehensively explore the diverse mechanisms through which Nab-paclitaxel can interact with tumor cells.
This study has looked at the effectiveness of Nab-paclitaxel as an advanced chemotherapeutic agent for HER2-positive breast cancer patients. However, little attention has been given to the undesirable toxicities and a few challenges of the conventional formulation. Nevertheless, molecular docking and pharmacokinetic profiling showed high-yield progress in providing not only utilizable binding sites from target compounds but also modifiable properties of the chemotherapeutic agent. Furthermore, according to Shi et al. [32], molecular dynamics-based screening aids in hypothesizing relevant therapeutic candidates’ efficacy, stability, and toxicity.
Materials and Methods
Acquisition of PDB Files of Receptors and Ligands
The ligand and macromolecule samples have structures important for in-silico analysis since these structures have components vital for ligand-receptor binding. The collection of the ligand structure, Paclitaxel, was provided by PubChem while the crystallized structure of the Kinase Domain of HER2 molecule (PDB ID: 3PP0) was accessed from the Protein Data Bank. For pharmacokinetic profiling only the canonical SMILES of the ligand, CC1=C2C(C(=O)C3(C(CC4C(C3C(C(C2(C)C)(CC1OC(=O)C(C(C5=CC=CC=C5)NC(=O)C6=CC=CC=C6)O)O)OC(=O)C7=CC=CC=C7)(CO4)OC(=O)C)O)C)OC(=O)C, was necessary for the data gathering while its SDF file needs conversion to PDB file via open babel for it to be utilized during the molecular docking phase of the in silico experiment- the macromolecule’s PDB file was only utilized for molecular docking with structural preparations to be discussed in the succeeding sections.
Swiss Target
The Nab-paclitaxel canonical SMILES downloaded from PubChem were uploaded to the query bar of the Swiss Institute of Bioinformatics’ SwissTargetPrediction section. The time to run the canonical SMILES within the SwissTargetPrediction took almost 20-30 seconds. The Top 15 targets were identified and analyzed in a chemotherapeutic context.
Comparative Toxicogenomic Database (CTD)
The Nab-paclitaxel was searched in the query search bar of the main CTD website. The target genes, gene interactions, and disease targets were all analyzed to summarize the theoretical action of the ligand inside the body.
SwissADME
The canonical SMILES of Nab-paclitaxel gathered from PubChem were loaded in the search query of the website- this ran for at least 20 seconds. The data regarding Physicochemical Properties, Lipophilicity, Water Solubility, Pharmacokinetics, Druglikeness, and Medicinal Chemistry were collected and analyzed accordingly.
ADMET Lab 2.0
The canonical SMILES of Nab-paclitaxel were loaded in ADMET Lab 2.0 to evaluate its physicochemical properties, medicinal chemistry, absorption, distribution, metabolism, excretion, and toxicity. The obtained results were recorded and analyzed.
ADVER-Pred
The canonical SMILES of Nab-paclitaxel were used for the prediction of its adverse effects. The resulting Pa and Pi values and their corresponding side effects were recorded and analyzed.
CLC-Pred
The canonical SMILES of Nab-paclitaxel were loaded in CLC-Pred to predict its cytotoxicity against tumor and non-tumor cell lines. The results obtained for cancer and non-tumor cell line prediction will be recorded and analyzed. The results include the Pa and Pi values, the name of the cell line, and the type of tissue. Moreover, the type of tumor was included in the cancer cell line prediction.
Protein and Ligand Preparation
The 3D crystal structure of the kinase domain of human HER2 (PDB ID: 3PP0) was retrieved from Protein Data Bank (PDB) and was optimized through Discovery Studio and Chimera. All the 3-dimensional structures of the ligands were obtained from PubChem in the SDF format. Afterward, the molecular geometry of ligands was optimized using Avogadro and saved in PDB format.
Molecular Docking
For the screening of the compounds, Chimera-Autodock Vina was used to dock Paclitaxel against HER2. The active site of the receptor was determined through the use of a Protein Data Bank where the X, Y, and Z coordinates of chains A (Ligand 1) and B (Ligand 2) were stated to be 17.37, 17.58, 27.00, and 34.28, 45.23, -11.01, respectively. After setting the parameters required for the docking procedure, such as the selected active site, energy minimization, and H-bond optimization, the one with the lowest binding affinity and RMSD value was selected for further analysis.
Receptor-Ligand Interactions
After the docking process, the Receptor-Ligand Interactions will be studied using Discovery Studio Visualizer. The amino acids present in chains A and B will be identified. Also, the different non-bond interactions, such as hydrogen, electrostatic, and hydrophobic bonds, will be classified and their distances determined
Results
HER-2 Ligand interactions
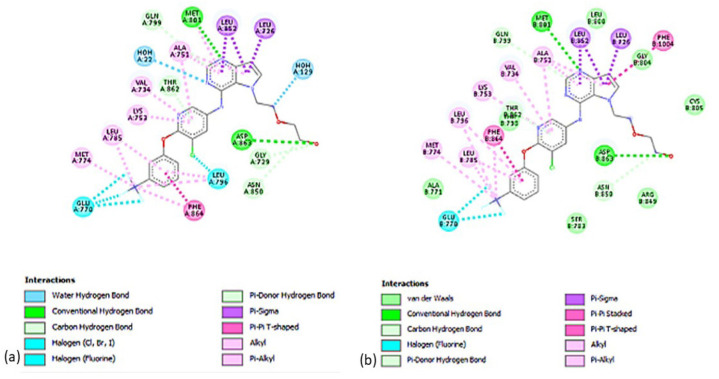
Figure 1 shows all possible interactions between the anticancer drug, Paclitaxel, and the active-site pocket of HER-2 protein which has two identical structural chains. Figure 1a shows a water hydrogen bond of Paclitaxel with Chain A of HER-2. Figure 1b indicates a potential van der Waals interaction with hydrogen bonding and pi-alkyl interactions are present between Chain B and Paclitaxel.
Figure 1.
Two-Dimensional Illustration of the Amino Acid Residues Involved in Binding between the Paclitaxel and Active Site of HER-2. 1a shows the conventional and Pi-donor H-bonds of Paclitaxel with Chain A of HER-2, and 2b depicts the hydrophobic interactions between Chain B of HER-2 and Paclitaxel.
Summary of Binding Interactions of Potential Anti-Breast Cancer Compounds on HER-2 Protein
Table 1 displays the results of the molecular docking and docking scores for each Ligand interaction with the 3PP0 Receptor using Chimera. The results show that chain B of HER-2 receptor has the greatest binding affinity with Paclitaxel having a general docking energy of about -9.4 kcal/mol with a total of 8 sites of interactions, whereas the binding affinity of chain A of HER-2 receptor showed to have a general docking energy of about -8.3 kcal/mol with 15 sites of interactions. The binding affinities of the other chemotherapeutic agents used in Nab-paclitaxel were also elucidated showed for reference which showed to be -8.9 kcal/mol for Epirubicin, -8.7 kcal/mol for Capecitabine, -6.8 kcal/mol for Gemcitabine, and -5.6 kcal/mol for Carboplatin. It must be noted that values presented from the molecular docking results represent the highest protein-ligand affinity and the lowest RMSD values obtained.
Table 1.
Binding Energies of Conventional Anti-Cancer Drugs against Breast Cancer on HER-2 Protein.
| Compound | Docking Score | Ranking |
| Paclitaxel bound to Chain B | -9.4 | 1 |
| Epirubicin | -8.9 | 2 |
| Capecitabine | -8.7 | 3 |
| Paclitaxel bound to Chain A | -8.3 | 4 |
| Gemcitabine | -6.8 | 5 |
| Carboplatin | -5.6 | 6 |
| Paclitaxel bound to Chain B | -9.4 | 1 |
Predicted Protein Targets of Nab-Paclitaxel
The top 2 significant target classes of the ligand are Family A G protein-coupled receptors with 26.7% target likelihood and a tie between kinases and other systolic proteins with 20.0% target proteins with 20.0% target likelihood. The rest of the target classes obtained 6.7% as shown in Figure 3. The top 10 specific target proteins in various target classes are summarized in Table 2 with Neurokinin 2 Receptor being the most probable target for Paclitaxel with a probability of 0.81700. Table 2 shows the top 10 specific target proteins of paclitaxel with varying probability. It also shows the target class of each protein receptor. Neurokinin 2 receptor, which belongs to the target class family - G protein-coupled receptor, had the highest probability of 0.817, whereas Endothelin receptor ET-A, which belongs to the same family had the lowest probability of 0.0642.
Figure 3.
Predicted Classes of Proteins that Interact with Paclitaxel as Determined by Swiss Target
Table 2.
Top 10 Specific Target Proteins of Nab-Paclitaxel with Varying Probability.
| Target Number | Target | Target Class | Probability |
|---|---|---|---|
| 1 | Neurokinin 2 receptor | Family A G protein-coupled receptor | 8.17004E+11 |
| 2 | Cholecystokinin A receptor | Family A G protein-coupled receptor | 7.59417E+11 |
| 3 | Delta opioid receptor | Family A G protein-coupled receptor | 7.59417E+11 |
| 4 | Tubulin beta-3 chain | Structural Protein | 7.59417E+11 |
| 5 | P-glycoprotein 1 | Primary active transporter | 7.59417E+11 |
| 6 | Proto-oncogene tyrosine-protein kinase MER | Kinase | 7.59417E+11 |
| 7 | Growth hormone-releasing | Family B G protein-coupled receptor | 0.208708525 |
| hormone receptor | |||
| 8 | Epidermal growth factor | Kinase | 0.208708525 |
| receptor erbB1 | |||
| 9 | Cytochrome P450 3A4 | Cytochrome p450 | 0.208708525 |
| 10 | Endothelin receptor ET-A | Family A G protein-coupled receptor | 0.06423878 |
Predicted Gene Interactions of Nab-Paclitaxel and their disease targets
The administration of the Nab-paclitaxel to the CT database yielded the top 10 affected genes, participating gene interactions, and target diseases, summarized in Table 3. The top 3 most likely to be subjected to toxicogenomic effects in the affected genes are ATP binding cassette subfamily B member 1 gene, caspase 3 gene, and Bcell-lymphoma-2 gene. The gene interactions with regards to Nab-paclitaxel were a set of increased, decreased, inhibited, and affected activity. Eight gene interactions are projected to have increased expression, susceptibility, uptake, or export when interacting with NAB-paclitaxel, while four gene interactions showed decreased expression, susceptibility, uptake, or export. Three of 10 gene interactions were identified to be affected by the drug, but synergistic or antagonistic effects were not indicated. The most probable diseases to be targeted by NAB-paclitaxel were also identified in the CT database. Five diseases were cardiovascular system-related, another five were nervous system-related, three were pulmonary system-related, and the last two were sepsis-related and side-effects-related.
Table 3.
Top 10 Affected Genes in the Presence of Nab-Paclitaxel with Interactions and Disease Targets
| Rank | Genes | Gene Interactions | Diseases | |
|---|---|---|---|---|
| 1 | ABCB1 | ↑ | ABCA5 mRNA- expression | Myocardial Ischemia |
| 2 | CASP3 | ↑ | ABCA7 mRNA - expression | Pulmonary Fibrosis |
| 3 | BCL2 | ↓ | ABCB1 - 6-OH-BDE-47 | Hypertension |
| 4 | BAX | ∞ | ABCB1 gene polymorphism | Cardiomyopathy |
| 5 | TP53 | ∞ | ABCB1 gene SNP | Myocardial Infarction |
| 6 | CASP9 | ↓ | ABCB1 mRNA | Heart Failure |
| 7 | PARP1 | ↑ | ABCB1 mRNA | Sepsis |
| 8 | MAPK3 | ↑ | ABCB1 protein - acetochlor | Hyperalgesia |
| 9 | MAPK1 | ↑ | ABCB1 protein - alachlor | Drug-Related Side Effects And Adverse Reaction |
| 10 | CYP3A4 | ↑ | ABCB1 protein - metolachlor | Nerve Degeneration |
Legends: ↑, increased expression, susceptibility, uptake, or export; ↓, decrease expression, susceptibility, uptake, or export; ∞, affects the expression, susceptibility, uptake, or export in an unidentified way
Pharmacokinetic Profiling of Nab-Paclitaxel
Drug Likeness was also included in the parameters of SWISS ADME to determine any violations that may come across in the candidacy of Nab-paclitaxel in drug discovery. In Lipinski’s standards, there were two violations in MW>500 and NorO>10 categories. In Ghose’s standard, NAB-paclitaxel garnered three violations in MW>480, MR>130, and #atoms>70 categories. Muegge’s standards also depicted three violations for Nab-paclitaxel in the MW>600, TPSA>150, and H-acc>10 categories The least violated standard was that of Egan’s with only the TPSA>140 categories violated in the analysis of the drug candidate. All these standards were also considered in the determination of the bioavailability score of the drug candidate measured as 0.17. For the medicinal chemistry parameter, zero alerts were detected for the Pan Assay Interference Structures while two alerts were detected for the Brenk standard which also acts as a structural alert (alert for the presence of isolated alkene and more than two esters detected). The highest number of violations detected was those of Lead likeness which breached MW>350, rotors>7, and XLOGP3>3.5 categories. The synthetic accessibility or the capability of the compound to be easily synthesized was identified to be equal to 8.34.
Table S1 shows some important physicochemical properties of Nab-paclitaxel which includes its molecular weight (MW), volume, density, nHA, nHD, nRot, nRing, MaxRing, nHet, fChar, nRig, Flexibility, number of stereocenters, Topological polar surface area (TPSA), logS, logP, and logD. TPSA is an essential physicochemical property as it indicates if a compound can be absorbed in the intestine which is a factor to consider in the administration of drugs. In this case, the TPSA value of Paclitaxel satisfied the criteria of having a value of greater than 140 Ų which signifies good intestinal absorption. Table 4 shows the vital pharmacokinetics and medicinal chemistry of Paclitaxel, including whether a compound has satisfied the criteria of the Lipinski Rule, Pfizer Rule, GSK Rule, and Golden Triangle. Results showed that the Lipinski rule, GSK rule, and Golden triangle rejected validation. At the same time, the only criteria that were satisfied were the Pfizer rule due to having a >3 log P value and a TPSA value of 75. These ranges are essential in the pharmacokinetic property of a potential drug since compounds with a higher logP and lower TPSA than the indicated values are susceptible to toxicity [33].
Table 4.
Summary of SWISS ADME Pharmacokinetic Parameters
| SWISS ADME of Nab-paclitaxel (MW: 853.91) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Lipophilicity | Water Solubility | Pharmacokinetics | Drug likeness | Medicinal Chemistry | |||||
| Parameter | Result | Parameter | Result | Parameter | Result | Parameter | Result | Parameter | Result |
| LogPo/w (iLOGP) | 4.51 | Log S (ESOL) | -6.66 | GI absorption | Low | Lipinski | 2 Violations MW>500 NorO>10 |
PAINS | 0 alert |
| LogPo/w (XLOGP3) | 3.66 | Solubility | 1.85e-04 mg/mL | BBB permeant | No | Ghose | 3 violations MW>480 MR>130 #atoms>70 |
Brenk | 2 alerts Isolated alkene No ester>2 |
| LogPo/w (WLOGP) | 3.41 | Class | Poorly soluble | P-gp substrate | Yes | Veber | 2 violations Rotors>10 TPSA> 140 |
Leadlikeness | 3 violations MW>350 Rotors>7 XLOGP3>3.5 |
| LogPo/w (MLOGP) | 1.7 | Log S (Ali) | -8 | CYP1A2 inhibitor | No | Egan | 1 violation TPSA>140 |
Synthetic accessibility | 8.34 |
| LogPo/w (SILICOS-IT) | 4.59 | Solubility | 8.61e-06 mg/mL | CYP2C19 inhibitor | No | Muegge | 3 violations MW>600 TPSA>150 H-acc>10 |
||
| Consensus LogPo/w | 3.58 | Class | Poorly soluble | CYP2C9 inhibitor CYP2D6 inhibitor |
No No |
Bioavailability score | 0.17 | ||
Predicted Adverse Effects of Nab Paclitaxel
Table 5 depicts the evaluation of the adverse effects of Nab-paclitaxel. The values for Pharmacologically active (Pa) and pharmacologically inactive (Pi) were also shown. Hepatotoxicity yielded a Pa = 0.979 and Pi = 0.006, while Nephrotoxicity yielded a Pa = 0.470 and Pi = 0.060.
Table 5.
Adverse Effects of Nab-paclitaxel determined by ADVER-Pred
| Pa | Pi | Side Effect |
|---|---|---|
| 0.979 | 0.006 | Hepatotoxicity |
| 0.47 | 0.06 | Nephrotoxicity |
Predicted Cytotoxicity of Nab-Paclitaxel as determined by CLC-Pred
Table 6 depicts the pharmacological activity and inactivity of Nab-paclitaxel against cancer cell lines. MCF-7 Breast Carcinoma exhibited the highest Pa value with the cytotoxicity of Nab-paclitaxel against this cell line. This observation is corroborated by the medication’s frequent use as a first-line treatment for patients diagnosed with early-stage breast cancer (BC) [20]. Table S2 shows the cytotoxicity of Nab-paclitaxel against cancer cell lines, along with the pharmacological active and inactive value indicators. The appropriate and well-studied cells which were WI-38 VA13, HEK293, and RPTEC correspond to the name, tissue location, and tumor type of the cancer.
Table 6.
Cell-Line Cytotoxicity of Nab-Paclitaxel as Determined by CLC-Pred
| Pa | Pi | Cell-line | Cell-line full name | Tissue | Tumor Type |
|---|---|---|---|---|---|
| 0.909 | 0.005 | MCF-7 | Breast carcinoma | Breast | Carcinoma |
| 0.788 | 0.003 | NCI-H838 | Non-small cell lung cancer. 3 stages | Lung | Carcinoma |
| 0.73 | 0.018 | A549 | Lung carcinoma | Lung | Carcinoma |
| 0.675 | 0.009 | HT-29 | Colon adenocarcinoma | Colon | Adenocarcinoma |
| 0.662 | 0.005 | A2780 | Ovarian carcinoma | Ovarium | Carcinoma |
| 0.604 | 0.009 | DMS-114 | Lung carcinoma | Lung | Carcinoma |
| 0.596 | 0.005 | SK-MEL-1 | Metastatic carcinoma | Skin | Melanoma |
| 0.588 | 0.004 | CFPAC-1 | Pancreatic carcinoma | Pancreas | Carcinoma |
| 0.575 | 0.004 | NCI-H295R | Adrenal cortex carcinoma | Adrenal cortex | Carcinoma |
| 0.516 | 0.01 | A2058 | Melanoma | Skin | Melanoma |
Discussion
The docking results were generated through the molecular docking of the HER-2 receptor and chosen ligands using Chimera, AutoDock Vina, and BIOVIA Discovery Studio, and the pharmacokinetic profiling of the Top 10 Target Classes of Paclitaxel with varying target likelihood percentages was done using Swiss Target, Swiss ADME, ADMET Lab 2.0, ADVER-Pred, and CLC-Pred.
The 2-D interactions were displayed to detect probable bond interactions between HER-2 protein amino acid residues and the ligands. Both Ligands 1 and 2 created three types of connections, as shown in Figures 1 and 2, including H-bonds, Pi contacts, and halogen interactions. Hydrogen bonds are crucial in protein structure’s overall stability and molecular recognition. Moreover, Ligand 1 has water hydrogen bond interactions while ligand 2 has Van der Waals.
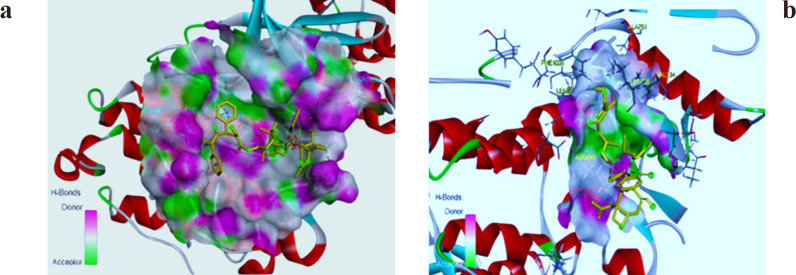
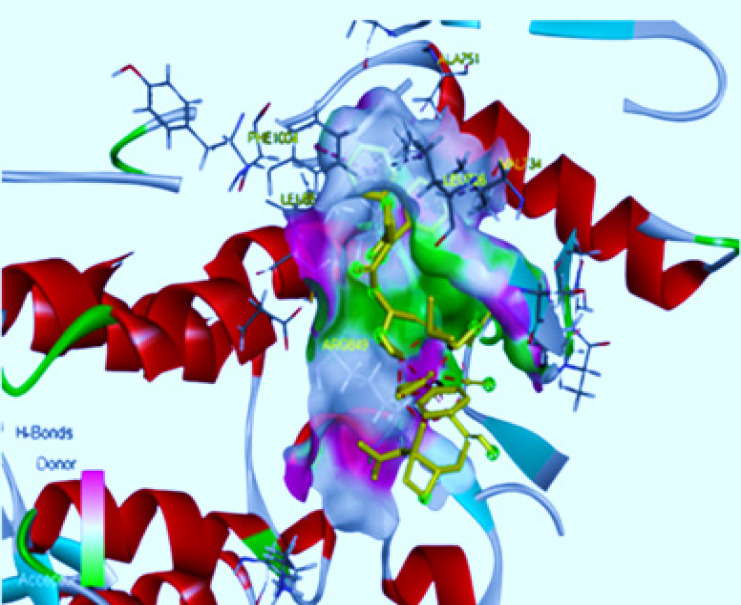
Figure 2.
Three-Dimensional Depiction of Molecular Interaction of Paclitaxel to HER-2 Active Site. 2a indicates the position of Paclitaxel on Chain 1 and 2b shows Paclitaxel interaction on Chain 2 in the HER-2 active site pocket.
In the results of the molecular docking, it shows that the docking pose of chain B with Paclitaxel is at -9.4 kcal/mol binding affinity which has favorable interactions. As presented in Table 1, the strongest hydrogen bond can be seen in Arg849 with 3.0 Angstrom (Å) which is the most common value for protein and water hydrogen bonds, while weak H-bond can be observed at 3.8 Å. Moreover, the docking pose of chain A with Paclitaxel is at -8.3 kcal/mol. The strongest H-bond interaction in Chain A is located in LYS921 at 3.0 Å as shown in Table 1, it has more unfavorable interactions than Chain B which is 18 and 15 respectively. The binding affinity of the best docking pose of other breast chemotherapeutic agents in HER-2, namely Epirubicin, Capecitabine, Gemcitabine, and Carboplatin, were -8.9, -8.7, -6.8, and -5.6 kcal/mol, respectively. In molecular docking results, Paclitaxel had the highest conformation energy at -9.4 kcal/mol with the chain B of the HER-2 receptor. Furthermore, the pharmacokinetic profiling of the physicochemical properties of Paclitaxel was shown where the logP and TPSA values obtained from ADMET Lab satisfied the ranges of a potential drug. The HER-2 inhibitors as anticancer agents showed that the catalytic site with important interacting amino acid (AA) residues are Leu726, Gly727, Ser728, Gly729, Val734, Ala751, Lys753, Glu770, Ala771, Met774, Leu785, Leu796, Val797, Thr798, Gln799, Leu800, Met801, Gly804, Cys805, Arg849, Asn850, Leu852, Thr862, Asp863, and Phe864 of HER2 which is comparable with the results of the current docking studies where the strongest hydrogen bond can be seen in ARG849 [33].
The scope of molecular docking was further broadened to include drug pharmacokinetics, aiming to elucidate not just the safety and efficacy of the potential drug, but also its effectiveness against other diseases. With the SWISS target, the rhodopsin family or Family A GPCR gained the top target class percentage out of 10 target classes- the rhodopsin family, specifically GPR161 [34,35] were overexpressed in triple-negative breast cancer patients. The action of paclitaxel was predicted to target GPR161 and associated growth factor like HER2 proteins with 26.7% target percentage. In contrast with this, another rhodopsin family member is the neurokinin 2 receptor with approximately 0.82 probability of being a target of the Nab-paclitaxel- this is the receptor, when targeted, which had no apparent antiproliferative effect on the metastasizing breast cancer but this receptor can be targeted in conjunction with a fellow Family A GPCR, delta-opioid receptor, which are highly expressed in breast cancer patients thus can be further used as a biomarker in future drug efficacy studies of nab-paclitaxel.
More to the context of targeting, Nab-paclitaxel has several effects on the genes of breast cancer patients, specifically the ABCB1 gene which codes for the P-glycoprotein (P-gp) responsible for the resistance of breast cancer cells to various chemotherapeutic drugs. Nab-paclitaxel has gene interaction effects on ABCB1, which lowers protein susceptibility and uptake- the drug also affects the ABCB1 mRNA. It lowers its expression, thus further lowering the production of the chemotherapeutic drug resistance protein P-gp. Moreover, to the ABCB1-coded protein, Nab-paclitaxel increases the drug exportation when interacting with the mentioned protein which facilitates a faster drug detoxification process due to decreased stay in the body. The approach of using NAB-paclitaxel as a P-gp inhibitor works well when administering a cocktail of chemotherapeutic drugs to breast cancer patients to maximize drug uptake. Besides breast cancer, NAB-paclitaxel was predicted to affect cardio-pulmonary diseases- the ABCA7 gene interacts with the drug and may potentially be a target of further inhibition assays to prove its potency in lowering the expression of ABCA7 mRNA in ischemic heart disease patients.
Regarding pharmacokinetic parameters, the results gathered positive and negative qualities of Nab-paclitaxel as a drug candidate. The first parameter, lipophilicity, identified that its consensus LogPo/w =3.58 is fairly soluble in non-polar solvents and thus also fairly efficient in binding with nonpolar biological components like lipid membranes and nonpolar proteins like HER-2 protein. The second parameter, water solubility, further supported the first parameter with the values on the Nab-paclitaxel’s poor solubility in water, which increases the possible dosage of the drug to be able to penetrate the plasma membrane therapeutically. The third parameter, pharmacokinetics, identified the difficulty of the drug to pass both the GI tract and the blood-brain barrier but this can be modified to mend the limitations - The drug needs to be administered intravenously so the transit in the GI tract does not affect the drug absorption while limitations brought about by the molecular weight of the drug in the prevention of its passage through the BBB can be averted using receptor-mediated transport systems as suggested by [36, 37]. Upon successfully getting past the limitations of the GI and the BBB, the drug has the advantage of being transported by the P-gp in the cellular target without difficulty in its transit due to guaranteed lipophilicity. The Nab-paclitaxel can also be potentially metabolized without difficulty since it does not inhibit most of the drug-metabolizing cytochrome P450 proteins - increasing more proteins that can metabolize the drug translates to an increased efficacy in the binding of the drug towards the target receptor. The fourth parameter, drug-likeness, indicates that the drug has a low chance of being an oral drug because it violates certain rules or standards. However, it was accepted for the Pfizer rule (or 3/75 rule), which suggests that the drug is unlikely to cause toxicological effects. This rule can be justified and supported by previous studies, where a correlation has been observed between the logP and TPSA values, and the in vivo toxicity results obtained [38]. And lastly, the fifth parameter, medicinal chemistry, indicates that there are no interfering substructures in the drug molecule. Despite this, however, the Brenk filter indicates that there are fragments within the molecule that have been identified as potentially toxic, and reactive, and can cause poor pharmacokinetics.
Predictions on the adverse effects and cytotoxicity of the drug were also obtained. First, hepatotoxicity and nephrotoxicity were shown as side effects because of the ability of Paclitaxel as a microtubule inhibitor [39, 40]. It was also found in a previous study that it can cause an increase in aminotransferase levels, which can lead to liver injury [39]. Moreover, it was found that this drug has cytotoxic effects against cancer cell lines, which can again be attributed to the role of Paclitaxel as one of the most used chemotherapeutic agents, especially against primary breast cancer [20, 41].
In conclusion, the molecular docking results showed that the docking pose of chain B of the HER-2 receptor with Paclitaxel exhibits the highest binding affinity of -9.4 kcal/mol. The strongest hydrogen bond observed is with ARG849, at a distance of 3.0 Angstroms (Å). Studies reported that Paclitaxel affects the genes of breast cancer patients, particularly the ABCB1 gene [42]. This gene encodes P-glycoprotein (P-gp), which has a crucial role in the resistance of breast cancer cells to various chemotherapeutic drugs. NAB-paclitaxel can also be potentially metabolized without difficulty since it does not inhibit most of the drug-metabolizing cytochrome P450 [43]. However, prolonged administration of Paclitaxel may induce drug resistance, a significant factor contributing to treatment failure [44]. This warrants the need for developing new combinations of candidate drugs or identifying novel drug-binding sites that could enhance treatment efficacy.
Acknowledgements
General
The authors would like to thank the academic staff of the internship committee of the Mammalian Tissue Culture Laboratory of the University of Santo Tomas Research Center for Natural and Applied Sciences through the Department of Biochemistry, Faculty of Pharmacy, the University of Santo Tomas for providing the training for the use of in silico tools used in this study.
Data Availability
The model simulations based on this study are too extensive to archive. Instead, we provide all the information needed to replicate the simulations. Due to confidentiality agreements, supporting data can only be made available to bona fide researchers subject to a non-disclosure agreement. Details of the data and how to request access are available from the corresponding author at the University of Santo Tomas, Department of Biochemistry.
Approval
This study is part of an approved undergraduate thesis that has been approved and accepted as partial fulfillment of the requirements for the degree of Bachelor of Science in Biochemistry at the University of Santo Tomas, Manila, Philippines.
Conflict of Interest
The authors declare that they have no competing interests.
Author Contribution Statement
NJDJM, ABSA, KMDLB, JMVE, JLDD, JGN, MSAE, AML, JPP, and MRSB conceptualized the in silico experimental design and computational framework, and analysis and interpretation of data. NJDJM, ABSA, KMDLB, JMVE, JLDD, and JGN carried out the data collection and wrote the manuscript with input and guidance from MSAE, AML, JPP, and MRSB. MSAE, AML, JPP, and MRSB were responsible for the paper’s overall concept, development, revision, proofreading, and approval of the final manuscript version.
Supplementary Materials
References
- 1.World Health Organization. [(2022, February 3)]. Retrieved from https://www.who.int/news-room/fact-sheets/detail/cancer?fbclid=
- 2.Cooper gm, hausman re. The development and causes of cancer. The cell: A molecular approach. 2000;2:725–66. [Google Scholar]
- 3.El Nachef L, Bouchet A, Bourguignon M, Foray N. When DNA mutations interplay with cellular proliferation: A narrative history of theories of carcinogenesis. Cancers. 2024;16:2104. doi: 10.3390/cancers16112104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ochwang’i DO, Kimwele CN, Oduma JA, Gathumbi PK, Mbaria JM, Kiama SG. Medicinal plants used in treatment and management of cancer in kakamega county, kenya. J Ethnopharmacol. 2014;151(3):1040–55. doi: 10.1016/j.jep.2013.11.051. [DOI] [PubMed] [Google Scholar]
- 5.Mohamed El-Tanani, Arwa Omar Al Khatib, Belal O, Ashok K, Yahia El-Tanani, Yin-Fai Lee, et al. Tambuwala, Cellular and molecular basis of therapeutic approaches to breast cancer. Cell Signal. 2023;101:110492. doi: 10.1016/j.cellsig.2022.110492. [DOI] [PubMed] [Google Scholar]
- 6.Trapani D, Gandini S, Corti C, Crimini E, Bellerba F, Minchella I, et al. Benefit of adjuvant chemotherapy in patients with lobular breast cancer: A systematic review of the literature and metanalysis. Cancer Treat Rev. 2021;97:102205. doi: 10.1016/j.ctrv.2021.102205. [DOI] [PubMed] [Google Scholar]
- 7.Weigelt B, Peterse JL, van ‘t Veer LJ. Breast cancer metastasis: Markers and models. Nat Rev Cancer. 2005;5(8):591–602. doi: 10.1038/nrc1670. [DOI] [PubMed] [Google Scholar]
- 8.Wicker MN, Wagner KU. Cellular plasticity in mammary gland development and breast cancer. Cancers (Basel) 2023;15:23. doi: 10.3390/cancers15235605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ter Steege EJ, Sijnesael T, Enserink L, Klarenbeek S, Haakma WE, Bakker ERM, et al. Lgr6-dependent conditional inactivation of e-cadherin and p53 leads to invasive skin and mammary carcinomas in mice. Neoplasia. 2023;35:100844. doi: 10.1016/j.neo.2022.100844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaszak I, Witkowska-Piłaszewicz O, Niewiadomska Z, Dworecka-Kaszak B, Ngosa Toka F, Jurka P. Role of cadherins in cancer-a review. Int J Mol Sci. 2020;21:20. doi: 10.3390/ijms21207624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yersal O, Barutca S. Biological subtypes of breast cancer: Prognostic and therapeutic implications. World J Clin Oncol. 2014;5(3):412–24. doi: 10.5306/wjco.v5.i3.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shagufta S, Ahmad I. Therapeutic significance of molecular hybrids for breast cancer research and treatment. RSC Med Chem. 2023;14(2):218–38. doi: 10.1039/d2md00356b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brasó-Maristany F, Griguolo G, Pascual T, Paré L, Nuciforo P, Llombart-Cussac A, et al. Phenotypic changes of her2-positive breast cancer during and after dual her2 blockade. Nat Commun. 2020;11(1):385 . doi: 10.1038/s41467-019-14111-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Patel A, Unni N, Peng Y. The changing paradigm for the treatment of her2-positive breast cancer. Cancers (Basel) 2020;12(8) doi: 10.3390/cancers12082081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang J, Xu B. Targeted therapeutic options and future perspectives for her2-positive breast cancer. Signal Transduct Target Ther. 2019;4:34. doi: 10.1038/s41392-019-0069-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang X, Wang L, Yu Q, Liu Z, Li C, Wang F, et al. The effectiveness of lapatinib in her2-positive metastatic breast cancer patients pretreated with multiline anti-her2 treatment: A retrospective study in china. Technol Cancer Res Treat. 2021;20:15330338211037812. doi: 10.1177/15330338211037812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wahba HA, El-Hadaad HA. Current approaches in treatment of triple-negative breast cancer. Cancer Biol Med. 2015;12(2):106–16. doi: 10.7497/j.issn.2095-3941.2015.0030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yao H, He G, Yan S, Chen C, Song L, Rosol TJ, et al. Triple-negative breast cancer: Is there a treatment on the horizon? Oncotarget. 2017;8(1):1913–24. doi: 10.18632/oncotarget.12284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim JS, Suh KJ, Lee DW, Woo GU, Kim M, Kim SH, et al. A real-world efficacy of nab-paclitaxel monotherapy in metastatic breast cancer. Cancer Res Treat. 2022;54(2):488–96. doi: 10.4143/crt.2021.394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abu Samaan TM, Samec M, Liskova A, Kubatka P, Büsselberg D. Paclitaxel’s mechanistic and clinical effects on breast cancer. Biomolecules. 2019;9:12. doi: 10.3390/biom9120789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kloover JS, den Bakker MA, Gelderblom H, van Meerbeeck JP. Fatal outcome of a hypersensitivity reaction to paclitaxel: A critical review of premedication regimens. Br J Cancer. 2004;90(2):304–5. doi: 10.1038/sj.bjc.6601303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tuyelee Das, Samapika Nandy, Devendra Kumar Pandey, Abdel Rahman Al-Tawaha, Mallappa Kumara Swamy, Vinay Kumar, Potshangbam Nongdam, Abhijit Dey. In: 12 - An update on paclitaxel treatment in breast cancer. Mallappa Kumara Swamy., editor. Academic Press; 2022. [Google Scholar]
- 23.Caasi JMN, Baldoza R, Bauzon MSC, Odtohan MAF, Santiago LA, Santiago-Bautista MR. In silico prediction of selected bioactive compounds present in alpinia elegans (c Presl) k Schum seed oil as potential drug candidates against human cancer cell lines. Asian Pac J Cancer Prev. 2023;24(8):2601–14. doi: 10.31557/APJCP.2023.24.8.2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kampan NC, Madondo MT, McNally OM, Quinn M, Plebanski M. Paclitaxel and its evolving role in the management of ovarian cancer. Biomed Res Int. 2015;2015:413076. doi: 10.1155/2015/413076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peronne L, Denarier E, Rai A, Prudent R, Vernet A, Suzanne P, et al. Two antagonistic microtubule targeting drugs act synergistically to kill cancer cells. Cancers (Basel) 2020;12(8) doi: 10.3390/cancers12082196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Burayag LJ, Acmad YH, Alberto C, Ang BN, Arona ME, Balajadia VR, et al. Predicted inflammatory protein targets of tinospora cordifolia secondary metabolites: Admet and molecular docking studies. Acta manilana. 2024;72: 33–54. [Google Scholar]
- 27.Li Y, Lu X, Lin Q, Li W. Is nab-paclitaxel better than conventional taxanes as neoadjuvant therapy for breast cancer? A meta-analysis. J Int Med Res. 2020;48(8):300060520943473. doi: 10.1177/0300060520943473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tian Z, Yao W. Albumin-bound paclitaxel: Worthy of further study in sarcomas. Front Oncol. 2022;12:815900. doi: 10.3389/fonc.2022.815900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yamamoto Y, Kawano I, Iwase H. Nab-paclitaxel for the treatment of breast cancer: Efficacy, safety, and approval. Onco Targets Ther. 2011;4:123–36. doi: 10.2147/OTT.S13836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu M, Liu S, Yang L, Wang S. Comparison between nab-paclitaxel and solvent-based taxanes as neoadjuvant therapy in breast cancer: A systematic review and meta-analysis. BMC Cancer. 2021;21(1):118 . doi: 10.1186/s12885-021-07831-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vishnu P, Roy V. Safety and efficacy of nab-paclitaxel in the treatment of patients with breast cancer. Breast Cancer (Auckl) 2011;5:53–65. doi: 10.4137/BCBCR.S5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shi M, Chen T, Wei S, Zhao C, Zhang X, Li X, et al. Molecular docking, molecular dynamics simulations, and free energy calculation insights into the binding mechanism between vs-4718 and focal adhesion kinase. ACS Omega. 2022;7(36):32442–56. doi: 10.1021/acsomega.2c03951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Singh G, Al-Fahad D, Al-Zrkani MK, Chaudhuri TK, Soni H, Tandon S, et al. Identification of potential inhibitors of HER2 targeting breast cancer—a structure-based drug design approach. J Biomol Struct Dyn. 2024;42(15):8184–8201. doi: 10.1080/07391102.2023.2246576. [DOI] [PubMed] [Google Scholar]
- 34.Feigin ME, Xue B, Hammell MC, Muthuswamy SK. G-protein-coupled receptor gpr161 is overexpressed in breast cancer and is a promoter of cell proliferation and invasion. Proc Natl Acad Sci U S A. 2014;111(11):4191–6. doi: 10.1073/pnas.1320239111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chaudhary PK, Kim S. An insight into gpcr and g-proteins as cancer drivers. Cells. 2021;10(12) doi: 10.3390/cells10123288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pardridge WM. Drug transport across the blood-brain barrier. J Cereb Blood Flow Metab. 2012;32(11):1959–72. doi: 10.1038/jcbfm.2012.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Terceiro LEL, Ikeogu NM, Lima MF, Edechi CA, Nickel BE, Fischer G, et al. Navigating the blood-brain barrier: Challenges and therapeutic strategies in breast cancer brain metastases. Int J Mol Sci. 2023;24(15) doi: 10.3390/ijms241512034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yukawa T, Naven R. Utility of Physicochemical Properties for the Prediction of Toxicological Outcomes: Takeda Perspective. ACS Med Chem Lett. 2020;11:2–9. doi: 10.1021/acsmedchemlett.9b00536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vincenzi B, Armento G, Spalato Ceruso M, Catania G, Leakos M, Santini D, et al. Drug-induced hepatotoxicity in cancer patients - implication for treatment. Expert Opin Drug Saf. 2016;15(9):1219–38. doi: 10.1080/14740338.2016.1194824. [DOI] [PubMed] [Google Scholar]
- 40.Santos MLC, de Brito BB, da Silva FAF, Botelho A, de Melo FF. Nephrotoxicity in cancer treatment: An overview. World J Clin Oncol. 2020;11(4):190–204. doi: 10.5306/wjco.v11.i4.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Repotente E, Carreon A, Devanadera MK, Esmalla MS, Santiago M. Cytotoxic potential on human breast and lung cancer cells of the biosynthesized gold nanoparticles from the reduction of chloroauric acid by lactic acid isolated from lactobacillus acidophilus. Front Mater. 2022;9:933749. [Google Scholar]
- 42.Lopez-Gonzalez L, Sanchez Cendra A, Sanchez Cendra C, Roberts Cervantes ED, Espinosa JC, Pekarek T, et al. Exploring Biomarkers in Breast Cancer: Hallmarks of Diagnosis, Treatment, and Follow- Up in Clinical Practice. Medicina (Kaunas) 2024;60(1):168. doi: 10.3390/medicina60010168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yardley DA. Nab-paclitaxel mechanisms of action and delivery. J Control Release. 2013;170(3):365–72. doi: 10.1016/j.jconrel.2013.05.041. [DOI] [PubMed] [Google Scholar]
- 44.Smith E, Chen Z, Xu X. Paclitaxel and cancer treatment: Non-mitotic mechanisms of paclitaxel action in cancer therapy. Paclitaxel: Sources, Chemistry, Anticancer Actions, and Current Biotechnology. 2022;1:269–286. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The model simulations based on this study are too extensive to archive. Instead, we provide all the information needed to replicate the simulations. Due to confidentiality agreements, supporting data can only be made available to bona fide researchers subject to a non-disclosure agreement. Details of the data and how to request access are available from the corresponding author at the University of Santo Tomas, Department of Biochemistry.