Abstract
Peribunyavirids produce enveloped virions with three negative-sense RNA segments comprising 10.7–12.5 kb in total. The family includes globally distributed viruses in multiple genera. While most peribunyavirids are maintained in geographically restricted vertebrate–arthropod transmission cycles, others are arthropod-specific or do not have a known vector. Arthropods can be persistently infected. Human and other vertebrate animal infections occur through blood feeding by an infected vector arthropod, resulting in diverse human and veterinary clinical outcomes in a strain-specific manner. Reassortment can occur between members of the same genus. This is a summary of the International Committee on Taxonomy of Viruses (ICTV) Report on the family Peribunyaviridae, which is available at ictv.global/report/peribunyaviridae.
Keywords: bunyavirus, ICTV Report, Orthobunyavirus, Peribunyaviridae, taxonomy
Virion
Peribunyavirids produce virions that are spherical or pleomorphic, 80–120 nm in diameter [1], with glycoprotein surface projections (5–18 nm) embedded in a lipid bilayer envelope (about 5 nm) (Table 1, Fig. 1). Ribonucleoprotein complexes are associated with the L protein and are made of non-covalently closed, circular genomic RNAs that are individually encapsidated [2].
Table 1. Characteristics of members of the family Peribunyaviridae.
| Example | Bunyamwera virus (S, D00353; M, M11852 and L, X14383), species Orthobunyavirus bunyamweraense, genus Orthobunyavirus |
| Virion | Enveloped, spherical or pleomorphic virions, 80–120 nm in diameter |
| Genome | Three single-stranded, negative-sense RNA molecules, small (S: 0.7–1 kb), medium (M: 2.6–4.6 kb) and large (L: 6.6–7.4 kb) |
| Replication | Cytoplasmic; primary transcription is primed by ‘cap snatching’ of host RNAs |
| Translation | On ER-bound ribosomes for GN and GC and on free ribosomes in the cytoplasm for N and L |
| Host range | Vertebrates and invertebrates (including mammals, birds, fish, mosquitoes, culicoids and psychodid sandflies) |
| Taxonomy | Realm Riboviria, kingdom Orthornavirae, phylum Negarnaviricota, subphylum Polyploviricotina, class Bunyaviricetes, order Elliovirales: the family includes ≥8 genera and ≥148 species |
Fig. 1. Peribunyaviridae family. Representation of a virion in cross-section (top). Generalized coding strategy of a peribunyavirid (bottom).
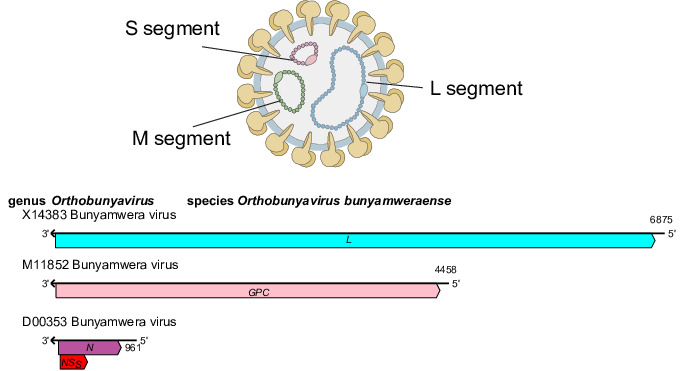
Genome
Peribunyavirid genomes comprise three single-stranded negative-sense RNAs (designated S, small; M, medium and L, large) (Fig. 1), each with complementary terminal nucleotide sequences that base pair to form non-covalently closed, circular genomic RNAs. The S segment encodes the nucleoprotein (N), which is abundant in infected cells and facilitates genomic RNA encapsidation and virus replication. In some viruses, an overlapping reading frame encodes the non-structural protein NSS [3]. The M segment encodes two structural glycoproteins (GN and GC) responsible for viral entry into host cells. Some members also encode a non-structural protein (NSM) between the GN and GC coding regions [4]. The L segment encodes the L protein, which has RNA-directed RNA polymerase and endonuclease functions essential for viral replication.
Replication
Virions attach via surface glycoproteins, entering the cell through clathrin-mediated endocytosis. Fusion of the viral GC protein fusion peptide with endosomal membranes facilitates the release of ribonucleocapsids into the cytoplasm. The complementary 5′- and 3′-termini are promoters for mRNA and antigenome synthesis. Viral mRNAs are not polyadenylated and are truncated relative to the viral (genomic) RNA (vRNA); a 5′-methylated cap is derived from host mRNA via ‘cap snatching’ mediated by the endonuclease function of the L protein. Proteins are translated on free ribosomes (S and L segment mRNAs) or membrane-bound ribosomes (M segment mRNA). The GN and GC proteins are generated by co-translational cleavage and are targeted to and retained in the Golgi complex. Ribonucleoproteins are targeted near the Golgi complex. Genomes are packaged by signals from non-conserved sequences in the terminal untranslated regions. Virions bud into Golgi cisternae and are transported to the cell surface by the secretory pathway [5].
Taxonomy
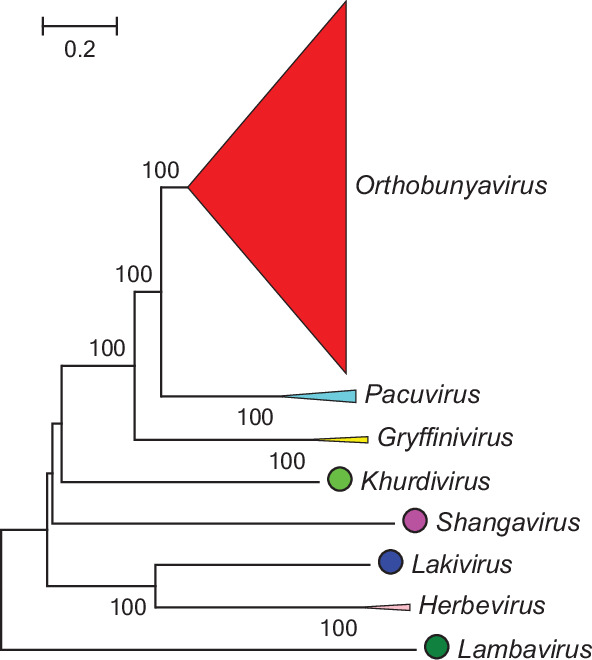
Current taxonomy: ictv.global/taxonomy. Genera are monophyletic based on the analysis of the virus L protein (Fig. 2); members of a genus have similar genomic organizations and transmission cycles. Peribunyavirids share some of the following characteristics: (i) enveloped spherical or pleomorphic virions; (ii) three segments of single-stranded, negative-sense RNA, with all proteins encoded in the same sense and (iii) capped but not polyadenylated viral mRNA.
Fig. 2. Phylogenetic analysis of the L protein of peribunyavirids.
Resources
Full ICTV Report on the family Peribunyaviridae: ictv.global/report/peribunyaviridae.
Acknowledgements
We thank Evelien Adriaenssens, Mart Krupovic, Jens H. Kuhn, Elliot J. Lefkowitz, Luisa Rubino, Sead Sabanadzovic, Peter Simmonds, Dann Turner, F. Murilo Zerbini, Arvind Varsani (ICTV Report Editors) and Donald B. Smith (Managing Editor, ICTV Report). We thank Jacqueline A. Tida (www.plotmyscience.com) for figure editing. The findings and conclusions in this report are the opinions of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
Abbreviations
- ICTV
International Committee on Taxonomy of Viruses
Footnotes
Funding: Production of this summary, the online chapter, and associated resources was supported by the Microbiology Society.
Contributor Information
William M. de Souza, Email: wmdesouza@uky.edu.
Charles H. Calisher, Email: calisher6920@gmail.com.
Jean Paul Carrera, Email: jpcarrera@gorgas.gob.pa.
Holly R. Hughes, Email: ltr8@cdc.gov.
Marcio R. T. Nunes, Email: marcionunesbrasil@yahoo.com.br.
Brandy Russell, Email: bmk8@cdc.gov.
Natasha L. Tilson-Lunel, Email: ntilston@iu.edu.
Marietjie Venter, Email: marietjie.venter1@wits.ac.za.
References
- 1.Bowden TA, Bitto D, McLees A, Yeromonahos C, Elliott RM, et al. Orthobunyavirus ultrastructure and the curious tripodal glycoprotein spike. PLoS Pathog. 2013;9:e1003374. doi: 10.1371/journal.ppat.1003374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barr JN, Elliott RM, Dunn EF, Wertz GW. Segment-specific terminal sequences of Bunyamwera bunyavirus regulate genome replication. Virology. 2003;311:326–338. doi: 10.1016/s0042-6822(03)00130-2. [DOI] [PubMed] [Google Scholar]
- 3.Fuller F, Bhown AS, Bishop DHL. Bunyavirus nucleoprotein, N, and a non-structural protein, NSS, are coded by overlapping reading frames in the S RNA. J Gen Virol. 1983;64 (Pt 8):1705–1714. doi: 10.1099/0022-1317-64-8-1705. [DOI] [PubMed] [Google Scholar]
- 4.Fazakerley JK, Gonzalez-Scarano F, Strickler J, Dietzschold B, Karush F, et al. Organization of the middle RNA segment of snowshoe hare bunyavirus. Virology. 1988;167:422–432. [PubMed] [Google Scholar]
- 5.Shi X, Lappin DF, Elliott RM. Mapping the Golgi targeting and retention signal of Bunyamwera virus glycoproteins. J Virol. 2004;78:10793–10802. doi: 10.1128/JVI.78.19.10793-10802.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]


