Abstract
Aims/hypothesis
Wolfram syndrome 1 (WS1) is an inherited condition mainly manifesting in childhood-onset diabetes mellitus and progressive optic nerve atrophy. The causative gene, WFS1, encodes wolframin, a master regulator of several cellular responses, and the gene’s mutations associate with clinical variability. Indeed, nonsense/frameshift variants correlate with more severe symptoms than missense/in-frame variants. As achieving a genotype–phenotype correlation is crucial for dealing with disease outcome, works investigating the impact of transcriptional and translational landscapes stemming from such mutations are needed. Therefore, we sought to elucidate the molecular determinants behind the pathophysiological alterations in a WS1 patient carrying compound heterozygous mutations in WFS1: c.316-1G>A, affecting the acceptor splice site (ASS) upstream of exon 4; and c.757A>T, introducing a premature termination codon (PTC) in exon 7.
Methods
Bioinformatic analysis was carried out to infer the alternative splicing events occurring after disruption of ASS, followed by RNA-seq and PCR to validate the transcriptional landscape. Patient-derived induced pluripotent stem cells (iPSCs) were used as an in vitro model of WS1 and to investigate the WFS1 alternative splicing isoforms in pancreatic beta cells. CRISPR/Cas9 technology was employed to correct ASS mutation and generate a syngeneic control for the endoplasmic reticulum stress induction and immunotoxicity assays.
Results
We showed that patient-derived iPSCs retained the ability to differentiate into pancreatic beta cells. We demonstrated that the allele carrying the ASS mutation c.316-1G>A originates two PTC-containing alternative splicing transcripts (c.316del and c.316–460del), and two open reading frame-conserving mRNAs (c.271–513del and c.316–456del) leading to N-terminally truncated polypeptides. By retaining the C-terminal domain, these isoforms sustained the endoplasmic reticulum stress response in beta cells. Otherwise, PTC-carrying transcripts were regulated by the nonsense-mediated decay (NMD) in basal conditions. Exposure to cell stress inducers and proinflammatory cytokines affected expression levels of the NMD-related gene SMG7 (>twofold decrease; p<0.001) without eliciting a robust unfolded protein response in WFS1 beta cells. This resulted in a dramatic accumulation of the PTC-containing isoforms c.316del (>100-fold increase over basal; p<0.001) and c.316–460del (>20-fold increase over basal; p<0.001), predisposing affected beta cells to undergo apoptosis. Cas9-mediated recovery of ASS retrieved the canonical transcriptional landscape, rescuing the normal phenotype in patient-derived beta cells.
Conclusions/interpretation
This study represents a new model to study wolframin, highlighting how each single mutation of the WFS1 gene can determine dramatically different functional outcomes. Our data point to increased vulnerability of WFS1 beta cells to stress and inflammation and we postulate that this is triggered by escaping NMD and accumulation of mutated transcripts and truncated proteins. These findings pave the way for further studies on the molecular basis of genotype–phenotype relationship in WS1, to uncover the key determinants that might be targeted to ameliorate the clinical outcome of patients affected by this rare disease.
Data availability
The in silico predicted N-terminal domain structure file of WT wolframin was deposited in the ModelArchive, together with procedures, ramachandran plots, inter-residue distance deviation and IDDT scores, and Gromacs configuration files (doi/10.5452/ma-cg3qd). The deep-sequencing data as fastq files used to generate consensus sequences of AS isoforms of WFS1 are available in the SRA database (BioProject PRJNA1109747).
Graphical Abstract
Supplementary Information
The online version of this article (10.1007/s00125-024-06307-0) contains peer-reviewed but unedited supplementary material.
Keywords: Cell stress, Inflammation, IPSCs, Nonsense-mediated decay, Wolfram syndrome, Wolframin
Introduction
Wolfram syndrome 1 (WS1) is a progressive neurodegenerative disorder mainly characterised by comorbidity of early-onset diabetes mellitus and optic nerve atrophy, with or without other endocrinological, urological and neurological abnormalities, with symptoms of highly variable onset, progression and severity [1, 2]. Disease manifestations are attributable to pathogenetic variants of the WFS1 gene, spanning 33.4 kb of genomic DNA within the short arm of chromosome 4 (4p16). WFS1 consists of eight exons, of which the first one is non-coding. The 3.6 kb WFS1 mRNA encodes a polypeptide named wolframin, which is 890 amino acids in length and has an apparent molecular mass of 100 kDa. The protein displays subcellular localisation to the endoplasmic reticulum (ER), where it plays a pivotal role in the control of the unfolded protein response (UPR), Ca2+ homeostasis, ER–mitochondria crosstalk and autophagy [3–6].
Wolframin’s structure and its interactome have not been entirely elucidated but it has been reported to have protein–protein interaction domains at both N- and C-terminals, separated by nine transmembrane domains (TMDs), allowing its insertion in the ER membrane [7]. Furthermore, the N-terminal domain is involved in the stabilisation of a tetrameric form, which is thought to be the most common and functional conformation of wolframin [8]. The 5′-untranslated region (UTR) of WFS1 mRNA exhibits heterogeneity due to naturally occurring alternative splicing (AS) events [9, 10]. More precisely, AS at the acceptor site of exon 2 causes a 4-bp deletion, enabling the differentiation of isoform 1 (aligned with the initial published sequence) from isoform 2, the latter starting four bases before the canonical ATG translation codon [9]. Despite isoforms 1 and 2 generating the same protein, their biological significance remains unclear [11, 12].
To date, over 200 disease-causing mutations of WFS1 have been identified along the entire sequence, with no evidence of mutational hotspots (https://www.ncbi.nlm.nih.gov/clinvar/?term=WFS1[gene], accessed 16 September 2024). As different WFS1 variants mirror a wide range of clinical phenotypes, several genotype–phenotype association studies have been performed to aid clinicians in predicting more accurate prognoses and pave the way for personalised treatments [13–18]. However, works investigating the molecular effects of specific WFS1 variants on therapeutics are rare [19] and those proposing in-depth characterisation of the transcriptional/translational outcomes stemming from the disruption of splice sites and/or introduction of premature termination codon (PTC) have not been performed yet.
It is well established that AS plays an important role in beta cell function and viability [20, 21]. Several AS variants contain PTC or intronic sequence after the stop codon that can elicit nonsense-mediated decay (NMD) to prevent the accumulation of aberrant transcripts or not-functional proteins [22]. In pancreatic beta cells, homeostatic pathways, including NMD and UPR, are involved in controlling and degradation of AS-deriving aberrant transcripts. Of note, several monogenic diabetes-associated genes, such as HNF1A [23, 24], GCK [24] and TCF7L2 [25], have AS variants, suggesting the importance of understanding the RNA surveillance mechanisms in pancreatic beta cells to better explain disease phenotype [26].
In our work, taking advantage of both induced pluripotent stem cells (iPSCs) and CRISPR/Cas9 technologies, we sought to decipher the consequences at the transcriptional and translational level of two recently discovered pathogenic WFS1 mutations: c.316-1G>A, affecting the acceptor splice site (ASS) upstream of exon 4; and c.757A>T, introducing a PTC in exon 7 [27, 28]. Finally, we explored the maturation of WFS1 mRNA in patient-derived iPSCs and after their differentiation into pancreatic beta cells, we evaluated the impact of WFS1 PTC-carrying transcript accumulation upon exogenous stress and inflammation-dependent NMD inhibition on survival of the insulin-producing beta cells.
Methods
Peripheral blood mononuclear cell isolation, reprogramming and cell culture
The healthy donor and patient-derived peripheral blood mononuclear cells (PBMCs) were obtained following informed consent and collected using Ficoll-Paque separation method on whole blood. WFS1 iPSCs were generated by reprogramming the CD34+ fraction from the patient-derived PBMCs using CytoTune-iPS 2.0 Sendai Reprogramming Kit (Thermo Fisher, USA) according to the manufacturer’s indications. The iPSC lines were routinely tested for mycoplasma contamination by using MycoAlert Mycoplasma Detection Kit by Lonza, Switzerland, according to the manufacturer’s instructions. Karyotyping was performed by the ISENET Biobanking service unit in Milan, Italy. All WFS1-derived iPSC clones and their gene-edited counterparts used in the present study had normal karyotype. Control iPSCs with wild-type (WT) genotype (CGTRCiB10) were already available and routinely employed in the laboratory. This WT iPSC line was used as an unrelated healthy control and its in-depth characterisation has been reported previously [29, 30]. Differentiation into pancreatic beta cells was performed in adhesion following an in vitro protocol published by our group [31] on subclones of WT iPSCs (#05, #B2, #T3), two clones of WFS1 iPSCs (#42 and #43) and three clones of WFS1wt/757A>T iPSCs (#B10, #C6 and #F4). At the end of differentiation (day 24), iPSC-derived beta cells were detached using 0.5 mmol/l EDTA and displaced in 35 mm or 60 mm petri dishes on an orbital shaker at 55 rev/min to aggregate into clusters (pseudo-islets). The pseudo-islets were maintained in suspension culture at 55 rev/min until their use, using the following medium: CMRL 1066 (Mediatech, USA) supplemented with 10% vol./vol. FBS (Lonza), 1% penicillin/streptomycin, 1% vol./vol. l-glutamine (Euroclone, Italy), 10 μmol/l Alk5i II, 1 μmol/l T3, 10 mmol/l nicotinamide (Sigma, USA), 10 μmol/l H1152 (Euroclone) and 100 U/ml DNAse I (Sigma). Skin fibroblasts from a WS1 patient with genotype W189X/W189X and an unrelated healthy control individual were provided by V. Broccoli’s lab at IRCCS San Raffaele Hospital. Fibroblasts were kept in the DMEM with high glucose, supplemented with 10% vol./vol. FBS, 1% vol./vol. penicillin/streptomycin and 1% vol./vol. l-glutamine.
RNA extraction, retrotranscription and PCR/RT-quantitative PCR
Total RNA was extracted by using mirVana Isolation Kit (Ambion, USA) and quantified by BioTek Epoch Microplate Spectrophotometer (Agilent, USA). About 1 µg of TURBO DNAse (Thermo Fisher)-treated RNA was reverse transcribed using SuperScript IV First-Strand Synthesis System (Thermo Fisher), with oligo(dT) primers, according to the manufacturer’s instructions. For the identification of WFS1 AS isoforms, 10 ng of cDNA was pre-amplified using WFS1 primers listed in the electronic supplementary material (ESM) Table 1 (0.4 μmol/l forward/reverse) and the 1.25 U DreamTaq HS DNA Polymerase (Thermo Fisher), with 3.5 mmol/l MgCl2 and 0.2 mmol/l dNTPs, in 50 μl final volume reaction. Pre-amplification consisted of the following cycling conditions: 1× 95°C for 3 min; 15× (95°C for 30 s, 60°C for 30 s, 72°C for 45 s); 1× 72°C for 5 min. A total of 1 μl of pre-amplification reaction was used as a template for further amplification. A second PCR was performed using 1.25 U DreamTaq HS DNA Polymerase, the same primers of pre-amplification PCR (0.4 μmol/l each), 2.0 mmol/l MgCl2 and 0.2 mmol/l dNTPs, with the following cycling conditions: 1× 95°C for 3 min; 35× (95°C for 30 s, 60°C for 30 s, 72°C for 45 s); 1× 72°C for 5 min. The PCR products were run on analytical pre-cast E-Gel Agarose Gels with SYBR Safe DNA Gel Stain, 2% (Thermo Fisher), using the GeneRuler 100 bp Plus DNA Ladder (Thermo Fisher) as size markers. The spliced/unspliced (s/u) XBP1 ratio was measured by agarose gel electrophoresis and densitometric analysis. For RT-quantitative PCR (qPCR) experiments, PowerUp Green Master Mix (Applied Biosystems, USA) and TaqMan Universal PCR Master Mix (Applied Biosystems) were used according to the manufacturer’s protocol, and amplification with an annealing temperature of 60°C was performed on a 7900HT Fast Real-Time PCR System (Applied Biosystems) with a Fast 96-Well Block Module. Expression levels of analysed genes were normalised by applying the method, using GAPDH as a housekeeping gene. A complete list of primers is shown in ESM Table 1 and TaqMan assays are reported in ESM Table 2.
PCR product purification, cloning and Sanger sequencing
WFS1-derived PCR products obtained following amplification with the Ex3fw-Ex5rev primer pair were separated on 2% agarose extraction gel prepared by dissolving low-melt-temperature agarose in 1× TBE. Both 324 bp- and 180 bp-long amplicons were purified separately by NucleoSpin PCR & Gel Clean-up (Macherey Nagel, Germany) and cloned into pCR4-TOPO TA vector using TOPO TA Cloning Kits for Sequencing (Thermo Fisher), following manufacturer’s instructions. The pCR4-TOPO construct was transformed into One Shot TOP10 (Thermo Fisher) chemically competent cells, then using LB-agar plates containing 100 µg/ml ampicillin as selection medium. The bacterial colonies were picked and analysed for the presence of insert by following amplification with the T3/T7 primers, and agarose gel electrophoresis. The pCR4-TOPO plasmid-containing insert was purified using NucleoSpin Plasmid, Mini kit for plasmid DNA (Macherey Nagel). The Sanger sequencing was performed by the EZ-Seq service at Macrogen Europe, the Netherlands. Electropherograms were visualised and sequence analysed by using SnapGene v4.3.2 (Dotmatics, USA).
Next generation sequencing
Following the patient’s referral to a geneticist for further examination, next generation sequencing (NGS) was performed on a panel of 31 monogenic diabetes-related genes, including WFS1 (NM_006005). Enrichment of fragments was performed with the TruSight One Sequencing kit (Illumina, USA). Sequencing of coding regions and exon–intron junctions was done on NextSeq Illumina platform. Diagnosis of WS1 was confirmed when mutations in the WFS1 gene were reported in the medical record of the patients, after comparison with variants deposited in the following databases: NCBI dbSNP; 1000 Genomes; dbNSFP; ClinVar; and Leiden Open Variation Database (LOVD). We classified the genetic variants of WFS1 gene sequence according to variants reported in ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/, accessed 16 September 2024) in compliance with Human Genome Variation Society (HGVS) Nomenclature (https://hgvs-nomenclature.org/stable/, version 20.0.4, August 2024). The deep-sequencing analysis on PCR products obtained by amplifying WFS1 sequence comprised between exon 3 and exon 5 was performed to confirm the sequence of the alternative splice variants deriving from c.316-1G>A-carrying allele in the WFS1 cells. PCR-derived fragments were purified by using NucleoSpin PCR & Gel Clean-up (Macherey Nagel). Libraries were obtained following the TruSeq Nano WGS (Illumina) protocol, deriving 250 nucleotides-long 4 × 106 fragments per sample. Paired-end sequencing was performed through MiSeq_500_v2 Illumina platform on two WFS1 iPSCs clones (#01 and #42) and one WT iPSCs clone (#05). The reads in R1 and R2 FASTQ files generated at the end of the sequencing were analysed for the base quality. Adaptors were removed by using Trimmomatic, v.0.32 (https://github.com/usadellab/Trimmomatic/releases, accessed 3 October 2024). Preliminary, explorative alignment was performed by STAR v2.5.3a (Illumina, USA), mapping reads to the reference human genome hg38, Gencode version 28. Percentages of uniquely mapped reads were about 70–77% for the two WFS1 samples and 85% for the WT sample. More than 99.99% of reads were assigned to the WFS1 gene for all the three samples.
CRISPR/Cas9-mediated gene editing
CRISPR/Cas9-mediated correction of the point mutation located upstream of exon 4 of the WFS1 gene was performed in patient-derived iPSCs. GeneArt Precision gRNA Synthesis Kit (Thermo Fisher) was employed for in vitro synthesis of the gRNAs (sequences are reported in ESM Table 1). As donor templates, a single-strand oligodeoxynucleotide (ssODN) homologous to the region encompassing the WFS1 intron 3–exon 4 boundary junction was used. Twenty micrograms of Cas9 protein (TrueCut Cas9 Protein v2, ThermoFisher) and 4 µg of in vitro transcribed gRNAs were pre-incubated for 20 min at room temperature to form ribonucleoprotein complex. A ribonucleoprotein/ssODN ratio of 1:1.5 was used by adding 36 µg of ssODN. Finally, 2 µg of pmaxGFP plasmid (Lonza) was added to the mix as a selection marker for cell sorting. For Cas9-mediated gene editing, 1×105 iPSCs were electroporated by using the 4D-nucleofector with P3 electroporation solution (Lonza) and setting the CB-150 program, according to the manufacturer’s instructions. The electroporated cells were replated and cultured in complete fresh media for 5 days. Single-cell cloning was performed following cell sorting of GFP-positive cells.
Cell sorting and FACS analysis
Electroporated cells were analysed with FACSAria Fusion Flow Cytometer (BD) and GFP-positive cells were sorted in vitronectin-coated 96-well plate for single-cell cloning. Sorted cells were cultured in Essential 8 Flex supplemented with CloneR (Stemcell Tech., Canada) until colonies emerged. Single-clone-derived colonies were screened by restriction enzyme as described below. For intracellular staining, cells were fixed by using Cytofix/Cytoperm (Becton Dickinson, BD), then permeabilised with Phosflow Perm Buffer III (BD, USA), according to the manufacturer’s instructions. Staining with conjugated antibody was performed for 30 min at 4°C. The list of antibodies used for FACS analysis is reported in ESM Table 3. For the ER stress induction and immunotoxicity assay, early and late apoptosis were evaluated by using the FITC Annexin V Apoptosis Detection Kit I (BD), following the manufacturer’s instructions. Cells were inspected on a CytoFLEX LX (Beckman Coulter, USA) cytometer, and analysed by FlowJo Software V.10 (FlowJo, Ashland, OR, USA).
Homologous-directed repair screening
Single colonies of growing clones were picked and transferred into direct PCR lysis buffer containing 150 mmol/l NaCl, 10 mmol/l Tris-HCl pH7.5, 5% Tween-20 and 0.4 mg/ml proteinase K (Promega, USA), then incubated for 6 h at 55°C until complete lysis was achieved. Crude lysate was incubated for 45 min at 85°C to inactivate proteinase K. After centrifugation to collect debris, 1 μl of lysate was used per 50 μl genotyping PCR reaction with the homologous-directed repair (HR) forward (fw)/reverse (rev) primers reported in ESM Table 1. The PCR product was purified using NucleoSpin PCR & Gel Clean-up (Macherey Nagel) and quantified by BioTek Epoch Microplate Spectrophotometer (Agilent). Up to 1 μg of DNA was digested with 10 U of SacI enzyme (NEB, USA) in NEBuffer r1.1 for 1 h at 37°C. Digested DNA derived from each clone was resolved on 2% agarose gel. The amplified sequence was 272 bp in length. Enzymatic digestion of the recombined sequence produced two bands of 152 bp and 119 bp. If homologous recombination with donor DNA involved one allele, the 272 bp-long and the 152/119 bp-long pair amplicons were observed (50% chance that the c.316-1G>A-carrying allele had been corrected). If homologous recombination occurred in both WFS1 alleles, only digested products were displayed. For subsequent experiments, only clones incurred in bi-allelic recombination were selected and evaluated for WFS1 expression.
Immunofluorescence
For indirect immunofluorescence, cells were cultured on coated Falcon Chambered Cell Culture Slides. Cell monolayers were washed with PBS, fixed with 4% wt/vol. paraformaldehyde in PBS for 20 min at room temperature and then treated with 15 mmol/l glycine for 5 min at room temperature. After blocking and permeabilisation with PBS supplemented with 0.4% vol./vol. Triton X-100, 2% wt/vol. BSA, 5% vol./vol. FBS for 45 min at room temperature, cells were washed twice with PBS and then incubated overnight at 4°C with appropriate primary antibody (ESM Table 3) diluted in PBS supplemented with 2% wt/vol BSA. Cells were incubated with appropriate secondary antibody for 1 h at room temperature. Nuclei were counterstained with Hoechst 33342. Images were acquired using Olympus FluoVIEW FV 3000 (VivaScope Research, Germany) confocal microscope and analysed using Fiji software v.1.52p [32].
Immunoblot analysis
Samples were lysed in M-PER and proteins were quantified by Pierce Rapid Gold BCA Protein Assay Kit. Protein extracts were resolved on SDS-PAGE (4–20%) using Novex Tris-Glycine Mini Protein Gels and then electro-transferred onto a polyvinylidene difluoride (PVDF) membrane. Transfer efficiency was evaluated using Ponceau S (Sigma). After 1 h of incubation in blocking buffer (TBS, 0.1% vol./vol. Tween 20, 5% wt/vol. skimmed milk) at room temperature, the membranes were probed overnight at 4°C with the primary antibodies reported in ESM Table 3 diluted in TBS supplemented with 0.1% vol./vol. Tween 20 and 5% wt/vol. skimmed milk or BSA. Horseradish peroxidase-labelled antibodies (diluted 1:1000 in the same dilution buffer as the primary antibody) were used as secondary antibody, and revelation was performed using the SuperSignal West Pico PLUS chemiluminescent detection system according to the manufacturer’s instructions by ChemiDoc MP (Biorad, USA). Quantification of protein levels was performed using Fiji software [32]. GADPH was used as a normaliser for protein quantification. All the listed reagents were purchased from Thermo Fisher unless otherwise specified. Frozen EndoC-βH1 cell pellets were available in the laboratory [29] and were employed as controls for protein expression in beta cells. They were resuspended in lysis buffer and subsequently processed as the other samples.
NMD inhibition
To determine whether NMD inhibition could increase the PTC-carrying WFS1 transcript levels, iPSCs were cultured in six-well dishes. At ~70% confluence, cells were treated with 5 μmol/l NMDI-14 (MedChemExpress) or with vehicle (DMSO 0.1%) for 16 h. RNA was collected at the end of the treatment and processed following the aforementioned methods.
ER stress induction and immunotoxicity assay
For the thapsigargin (TG; Sigma)-induced ER stress experiments, iPSCs were treated for 2, 4, 8, 16 or 24 h with 100 nmol/l TG. For the cytotoxicity assay, they were treated for 8 h followed by 24 or 48 h of recovery in fresh medium without TG. For iPSC-derived beta cells, TG treatment lasted for a total of 8 or 16 h with 50 nmol/l TG before further analysis. Conditioning of iPSC-derived beta cells with inflammatory cytokines was carried out by adding 50 U/ml rhIL-1β, 1000 U/ml rhIFN-ɣ and 10 ng/ml rhTNF-α (PeproTech, USA) to the culture medium for up to 48 h. Cells were analysed by FACS for positivity to Annexin-V/propidium iodide (PI) at 8, 16 or 48 h, according to the protocol described above.
Hormone secretion
Ghrelin, glucagon and C-peptide levels were measured in culture supernatant fractions by Bio-Plex Pro human diabetes kit (BioRad), following the manufacturer’s instructions; processed samples were read on a Luminex xMAP (BioRad) and analysed with the software Bio-Plex Manager 6.0 (BioRad). Dynamic stimulation of iPSC-derived beta cells was instead performed on an automated perifusion system (BioRep Perifusion V2.0.0, USA). For each line, 100 clusters were picked and stimulated with HEPES-buffered solution (125 mmol/l NaCl, 5.9 mmol/l KCl, 2.56 mmol/l CaCl2, 1 mmol/l MgCl2, 25 mmol/l HEPES, 0.1% wt/vol BSA, pH 7.4) supplemented as follows: 0.5 mmol/l glucose; 11 mmol/l glucose plus 50 µmol/l IBMX (Gibco, USA) or 30 mmol/l KCl. Insulin content was quantified by ultrasensitive ELISA kit (Mercodia, Sweden).
Bioinformatic analysis
Inference of the alternative splicing events potentially occurring after disruption of the ASS at c.316-2 (AG) of WFS1 coding sequence was conducted following preliminary examination of the cryptic splice sites within the exon 4 sequence (ENSE00000701011) ±150 bp flanking intronic regions. Analysis of the canonical splice sites, branch points (BP score), cis-elements (exonic splicing enhancer [ESE] and exonic splicing silencer [ESS]) and trans-acting splicing factor sequences, was performed by the Human Splice Finder (HSF) Pro (https://hsf.genomnis.com, version 3.1, accessed 3 October 2024). The HSF and BP scores were computed according to the weighted matrices as described by Desmet et al [33]. To model the most thermodynamically favoured AS events following the c.316-1G>A transition, the MaxEntScan framework [34] based on the maximum entropy principle was used. For the generation of AS isoform consensus sequences from deep-sequencing raw data, the trimmed paired-end reads were aligned by STAR v2.5.3a to custom reference genome consisting of the WT WFS1 mRNA sequence as deposited in the RefSeq database (NM_006005.3) and the three AS isoform sequences as obtained by Sanger sequencing (c.316del, c.316–456del and c.316–460del). Due to higher probability of sequencing errors associated with read length, -outFilterMismatchNmax parameter was set to 10. Reads that did not align to any sequence of the custom reference genome were processed to identify other AS isoforms, if any, and used to generate the relative consensus sequence using EMBOSS’ Cons software (https://github.com/kimrutherford/EMBOSS, version 6.6, accessed 3 October 2024). Mapping of paired-end reads to the custom reference genome was visualised with Integrative Genomics Viewer (IGV) v2.5. To infer the tridimensional (3D)-structure of wolframin N-terminal domain, the entire amino-acid sequence of the protein was used with Ginzu protocol [35] to predict the functional domains of the protein. The sequence of N-terminal domain (residues 1–402) was then submitted to Robetta server’s RoseTTAFold (https://robetta.bakerlab.org/, accessed 3 October 2024), following ab initio modelling pipeline. The structures of mutated isoforms were generated employing Rosetta’s comparative modelling [36] and using the de novo modelled WT N-terminal structure as reference. The molecular dynamics simulations and conformational change analysis were performed by Gromacs v.2021.7 [37].
Statistical analysis
Prism software version 9.0.1 (GraphPad, USA) was employed to perform statistical analyses. Assuming the normal distribution and according to type of data, Student’s unpaired or paired t test (one- or two-tailed) was used for comparison between two groups, while to evaluate the effects of two independent variables on a dependent variable, two-way ANOVA with Dunn–Šídák correction for multiple comparison test was applied. Data are graphed as mean ± SEM or as mean ± SD. Error bar meaning, number and characteristics of replicates, and statistical analysis are reported in the figure legends.
Results
Analysis of patient’s WFS1 mutations
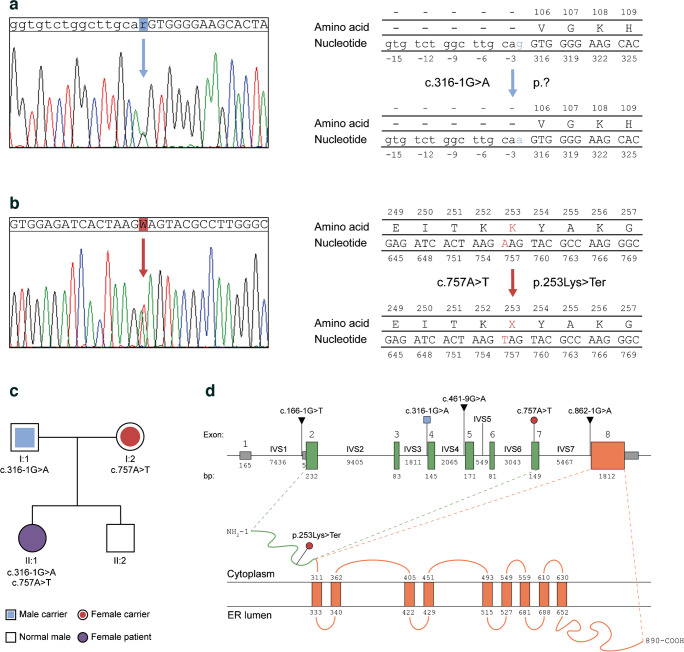
The patient did not show any notable clinical manifestation before type 1 diabetes diagnosis at the age of 5.2 years, with the exception of a transient mild speech delay. At age 8.7 years, WS1 was diagnosed by NGS-based screening for monogenic diabetes-driving mutations. Residual C-peptide at baseline screening was 0.1 ng/ml, with an HbA1c of 50 mmol/mol (6.7%) and an insulin requirement of 0.28 U kg−1 day−1. MRI revealed slight bilateral atrophy of brainstem and optic nerves, as extensively described in our previous paper [38]. Moreover, the patient presented a pubertal delay and the analysis of hormonal values suggested an early stage primary ovarian insufficiency [39]. DNA-sequencing patterns were compared with the sequence of WFS1 transcript variant 1 in GenBank (NCBI Ref. Seq: NM_006005.3; Gene ID: 7466), leading to detection of the heterozygous mutations c.316-1G>A and c.757A>T (Fig. 1a, b), and other homozygous benign variations (Table 1). The c.316-1G>A transition disrupts the acceptor (3′) splice site of exon 4 with uncertain outcome (Fig. 1a) and was classified as likely pathogenic, according to the American College of Medical Genetics and Genomics (ACMG) guidelines [40]. The c.757A>T transversion was classified as pathogenic and located on exon 7, where it gives rise to a PTC at the amino-acid residue 253 (p.Lys253Ter), as shown in Fig. 1b. Sanger sequencing on the PCR products from genomic DNA isolated from the family members confirmed that the patient inherited the c.316-1G>A and c.757A>T mutations from the father and the mother, respectively (Fig. 1c). Since the parents had no symptoms related to the syndrome, we assumed that the mutations did not exert any dominant negative effect and were transmitted as an autosomal recessive trait. By querying the ClinVar archives of NCBI, the Human Gene Mutation Database and the LOVD, we found that splicing-site variations of WFS1 account for 2.9% of the total mutations and, among them, only one out of three involves an ASS or its proximal polypyrimidine (PY) tract. In Fig. 1d we display the genomic position of the mutations carried by our patient and we highlight the previously reported ASS variants mapping to the intervening sequence (IVS)-1, IVS-4 and IVS-7 of the gene [41, 42]. To infer the effects of disruption of canonical AG ASS, we evaluated the cryptic splice sites along the WFS1 exon 4 sequence (ENSE00000701011) ±150 bp flanking intronic regions, by using the Human Splice Finder (HSF) sequence analysis tool (Fig. 2a–c) and we identified a total of 4 ASSs with an HSF score ≥75 (Fig. 2a and ESM Table 4). Among these, the acceptor splice motif located at 6288996 (CACTACCTGCAGTT; HSF Score: 81.29) might cause a 21-bp deletion (c.316–336del) that preserves the open reading frame (ORF) generating a protein lacking a seven-amino-acid fragment (VGKHYLQ) with apparent molecular mass of 99.5 kDa. None of these alternative splice sites were confirmed using inhomogeneous Markov models as shown by plotting the MaxEntScores ≥4 (Fig. 2a and ESM Table 4). Interestingly, we observed that the exonic splicing auxiliary sequences ratio (ESR) was significantly higher (>15) along the cryptic acceptor splice sequence starting at 6289115 (CTTGGCGGACAGAA; HSF Score 78.06) (Fig. 2b). This positive ESR was suggestive of an increased activity of exon splicing enhancer proteins as the analysis displayed the presence at the 5′ of exon 4 of the recognition motifs of the SC35, SRp40 and 9G8 splicing factors (Fig. 2b). We also found a total of six branch points (BP score >65) along the sequence comprising between 6288948 and 6289146 that could be used from the cryptic ASSs (Fig. 2c and ESM Table 4). To infer which ASS could be selected by the spliceosome when the disruption of the WT site occurs, we applied a method based on maximum entropy principle [34]. We expected the abolishment of the WT ASS would drive the splicing machinery to utilise the cryptic sequence at position 6288996. However, since the c.316-1G>A mutation determines the 1 bp shifting of the canonical AG motif, according to the maximum entropy model, this ASS was thermodynamically favoured when compared with the cryptic motif at 6288996 (Fig. 2d). Nevertheless, the generation of the 1 bp-shifted splice site at position c.317 dramatically alters the ORF and results in a PTC. Anyhow, the maximum entropy-based probabilistic model suggested exon 4 skipping as the most probable outcome (MaxEntScore >6) causing ORF breakdown and PTC formation (Fig. 2d).
Fig. 1.
(a, b) Sanger sequencing chromatograms showing the heterozygous c.316-1G>A transition and the c.757A>T transversion in the WFS1 gene of the patient. The modified nucleotides are highlighted in blue and red, respectively, as well as the resulting amino-acid substitution in the protein sequence, if known. (c) Genetic pedigree of the patient’s family. The first line below each symbol represents generation and identification number. The heterozygous mutations found in the patient’s father and mother are reported; the patient’s brother carries no mutated allele. (d) Schematic representation of WFS1 gene and wolframin protein. The exons 1 (non-coding, grey boxes), 2–7 (encoding the N-terminal domain of wolframin, green boxes) and 8 (encoding the transmembrane and C-terminal domains of wolframin, orange box) are reported. The genomic position of the patient’s WFS1 mutations are highlighted and colour-coded as in (a, b). The previously reported ASS variants are reported as well. The N-terminal domain (green) and transmembrane/C-terminal domains (orange) are shown along with the c.757A>T transversion introducing a premature stop codon
Table 1.
List of the WFS1 variants of the WS1 patient as found by NGS
| Classification | Pathogenicity score | Zygosity | Effect | HGVS coding | HGVS protein | ClinVar |
|---|---|---|---|---|---|---|
| Likely pathogenic | 7.0 | Heterozygosity |
splice_acceptor_variant intron_variant |
c.316-1G>A | NA | NA |
| Pathogenic | 7.0 | Heterozygosity | stop_gained | c.757A>T | p.Lys253* | NA |
| Benign | −1.5 | Homozygosity | intron_variant | c.461–15C>T | NA |
RCV000303369 (benign) RCV001513862 (benign) RCV000038663 (benign) RCV000365086 (benign) RCV002464098 (benign) |
| Benign | −1.5 | Homozygosity | intron_variant | c.461–9A>G | NA |
RCV000325882 (benign) RCV001513863 (benign) RCV000038664 (benign) RCV000004785 (pathogenic) RCV000273205 (benign) RCV002225069 (benign) |
| Benign | −1.5 | Homozygosity | synonymous_variant | c.684C>G | p.Arg228Arg |
RCV001155464 (likely benign) RCV000515386 (uncertain significance) RCV000724125 (likely benign) RCV000200201 (uncertain significance) RCV001155463 (uncertain significance) RCV000023512 (pathogenic) RCV002042600 (uncertain significance) RCV000319343 (benign) RCV001513864 (benign) RCV000038665 (benign) RCV000261705 (benign) RCV002464099 (benign) |
| Benign | −1.5 | Homozygosity | missense_variant | c.997G>A | p.Val333Ile |
RCV003039715 (uncertain significance) RCV000155337 (benign) RCV001904627 (uncertain significance) RCV001157248 (benign) RCV002496615 (benign) RCV001523396 (benign) RCV000038668 (benign) RCV000259675 (benign) RCV002464100 (likely benign) RCV001414831 (likely benign) |
| Benign | −1.5 | Homozygosity | synonymous_variant | c.1185C>T | p.Val395 Val |
RCV001858397 (uncertain significance) RCV000825500 (uncertain significance) RCV002509559 (uncertain significance) RCV000344908 (benign) RCV001523397 (benign) RCV000038635 (benign) RCV000380942 (benign) RCV002463627 (uncertain significance) RCV000825690 (benign) |
| Benign | −1.5 | Homozygosity | synonymous_variant | c.1500C>T | p.Asn500Asn |
RCV002765362 (uncertain significance) RCV000294241 (benign) RCV001513865 (benign) RCV000038641 (benign) RCV002285139 (uncertain significance) RCV000330600 (benign) |
| Benign | −1.5 | Homozygosity | missense_variant | c.1832G>A | p.Arg611His |
RCV002149055 (likely benign) RCV002492910 (uncertain significance) RCV000196564 (uncertain significance) RCV003126590 (uncertain significance) RCV001157549 (uncertain significance) RCV002491456 (uncertain significance) RCV001570232 (uncertain significance) RCV001157550 (uncertain significance) RCV000339307 (benign) RCV001523401 (benign) RCV000038646 (benign) RCV000404430 (benign) RCV000987411 (likely benign&benign) RCV003075593 (likely benign) |
| Benign | −1.5 | Homozygosity | synonymous_variant | c.2433G>A | p.Lys811Lys |
RCV001768421 (uncertain significance) RCV001504964 (likely benign&uncertain significance) RCV000352148 (benign) RCV002287353 (benign) RCV001523402 (benign) RCV000038659 (benign) RCV002287352 (benign) RCV000292566 (benign) |
| Benign | −1.5 | Homozygosity | synonymous_variant | c.2565A>G | p.Ser855Ser |
RCV000722519 (uncertain significance) RCV001816793 (likely benign) RCV003148856 (uncertain significance) RCV000664096 (likely benign) RCV000952030 (likely benign&benign) RCV001700444 (benign) RCV002464286 (benign) RCV001893060 (uncertain significance) RCV000329623 (benign) RCV001523403 (benign) RCV000038660 (benign) RCV000272258 (benign) RCV002502581 (likely benign) RCV001721445 (likely benign) |
| Benign | −1.5 | Homozygosity | 3_prime_UTR_variant | c.*47T>C | NA |
RCV000393152 (benign) RCV001643058 (benign) RCV000727866 (benign) RCV000288103 (benign) |
Fig. 2.
(a) Genomic positions of the natural and cryptic donor (green) and acceptor (red) splice sites along WFS1 exon 4 (145 bp) and its flanking regions (150 bp/each) as predicted by Human Splice Finder. Only the splice sites with an HSF score >75 and a MaxEnt score >4 are plotted. (b) ESR calculated as a ratio between auxiliary splicing signals (ESE and ESS) along the same genomic region. The binding sites for the splicing factors SC35 (dark blue), SRp40 (purple) and 9G8 (light blue) are noted for the region of interest surrounding the cryptic ASSs inside exon 4. (c) Position of the natural and alternative branch points (light blue) along the analysed genomic region. Only the branch points with a BP score >65 are plotted. The coordinates indicate the base number counting from the p-arm telomere of chromosome 4 (according to the UCSC Genome Browser on Human GRCh38/hg38 Assembly). (d) Results of maximum entropy modelling in the presence of the WT ASS and after the c.316-1G>A transition. The two most likely alternatives upon the canonical splice site disruption are shown. ASSs scored by the model are underlined and highlighted in yellow. Exonic bases are in upper case letters; intronic bases in lower case letters. The grey area represents the spliced-out sequence. Coordinates refer to the nucleotide position in the coding sequence
Characterisation of the transcriptional outcomes from the c.316-1 G>A allele
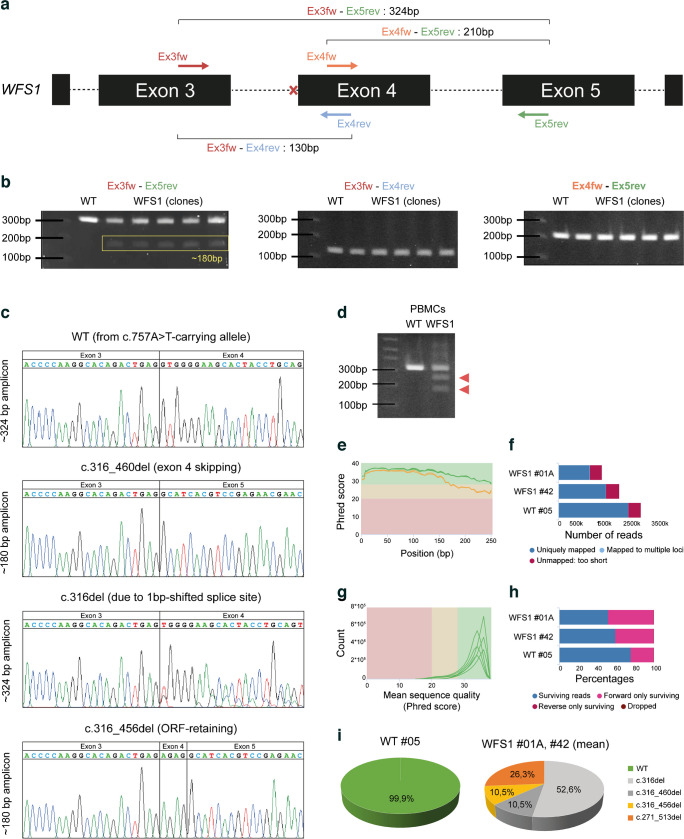
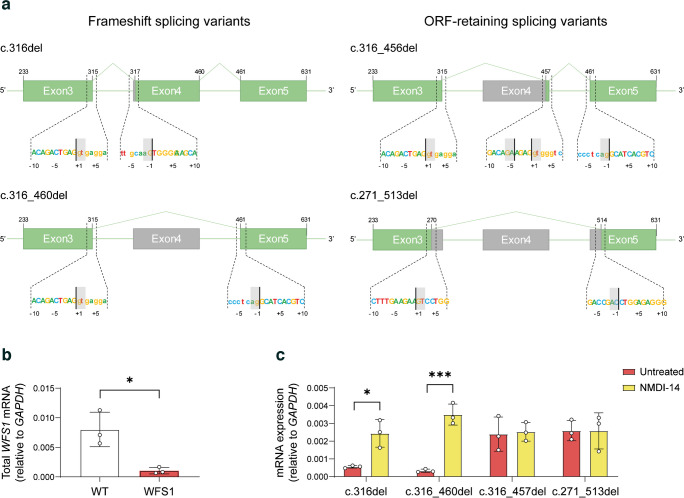
To explore the transcriptional landscape arising from ASS disruption in IVS-3, we decided to exploit stem cell technology. Specifically, we reprogrammed peripheral blood CD34+ cells of the patient into iPSCs, as previously described by our group [31], successfully generating a total of eight iPSC syngeneic clones with normal karyotype and conserved WFS1 heterozygous mutations. Details on genomic integrity and analysis of pluripotency are reported in ESM Fig. 1a–c. We designed a PCR strategy to amplify WFS1 transcripts along exons 3, 4 and 5 by using two pairs of primers (Fig. 3a). When the WFS1 mRNA between exon 3 and 5 was amplified in both WT and WFS1 iPSCs, we highlighted a potential mutation-related AS isoform, as we observed an amplicon of ~180 bp in the WFS1 iPSCs (Fig. 3b). Both cell lines also displayed the expected amplicon of 324 bp, which derives from the normal gene in the WT cells, and likely from the c.757A>T-carrying allele in WFS1. The amplification of the sequence between exons 3 and 4 or between exons 4 and 5 generated a single amplicon in WFS1 iPSCs, supporting the hypothesis that exon 4 skipping was the most probable event occurring in the c.316-1G>A-carrying pre-mRNA (Fig. 3b). To determine the sequences of the observed PCR products, we TA-cloned the WFS1-derived 324 bp- and 180 bp-long bands and >20 clones were analysed by Sanger sequencing. We found both natural and AS variants that we named according to Human Genome Variant Society nomenclature standard. Among the 324 bp-long amplicons, we identified two sequences: the WT (deriving from the c.757A>T-carrying allele); and the c.316del, corresponding to the maximum entropy-based prediction of alternative splicing resulting from the 1 bp shifting of ASS. Analysing the 180 bp-long amplicons, we found two different sequences, one resulting from the skipping of the entire exon 4 (c.316–460del) and the other retaining the last four bases of the exon 4 (c.316–356del). Among the identified c.316-1G>A-carrying allele-derived isoforms, only the c.316–356del preserved the ORF (Fig. 3c). To exclude the possibility that reprogramming could alter the processing of WFS1 pre-mRNA, as well as to rule out the possibility that observed isoforms were a specific characteristic of stem cells, due to a broader expression of splicing enhancer factors, we amplified the WT and WFS1 primary PBMC-derived cDNA by Ex3fw-Ex5rev primer pair, observing a complex pattern of amplicons in patient-derived PBMCs (Fig. 3d). Based on these findings, we generated a sequence library from both WT and WFS1-derived amplification products of the Ex3fw-Ex5rev primer pair and performed deep sequencing to identify all the potential AS isoforms occurring in the mutated cells. We performed paired-end sequencing on the PCR products from one WT subclone and two WFS1 clones. Assessment of read quality showed that R1 (forward) reads had poor quality after 210 bp, while R2 (reverse) reads lost accuracy after 180 bp. Preliminary STAR alignment to reference human genome hg38 revealed a reduced number of reads mapping to the WFS1 gene in the affected cell-derived samples. This mirrored the percentage of trimmed paired-end reads per sample used to individuate the AS isoforms in WFS1 iPSCs (Fig. 3e–h). Interestingly, a total of 4 c.316-1G>A-carrying allele-deriving AS isoforms were identified in WFS1 iPSCs, whereas in WT cells the normal, correctly spliced transcript represented more than 99% of the mapped reads (Fig. 3i). NGS analysis thus confirmed sequences identified by cloning and Sanger sequencing and allowed us to identify an additional splice variant: c.271–513del. Among the detected transcripts, the c.316del and c.316–460del isoforms resulted in frameshift and PTC generation, whereas the c.316–356del and c.271–513del isoforms conserved the ORF, missing part of the WFS1 mRNA. Specifically, c.316–356del lost the entire exon 4 with the exception of the last four nucleotides (AGAG), while c.271–513del included a larger deletion comprising the last 45 bp of exon 3, the entire exon 4 (145 bp) and the first 53 bp of exon 5 (Fig. 4a and ESM Table 5). Relative quantification of all the WFS1 transcripts revealed a significant reduction in WFS1 iPSCs (p<0.05; one-tailed t test) but not the complete absence of stable transcripts (Fig. 4b). We speculated that stable low expression of WFS1 total mRNAs in affected iPSCs might result from post-transcriptional regulation by NMD of the transcript deriving from the c.757A>T-carrying allele and the two out of four transcripts derived from the c.316-1G>A-carrying allele as well. Indeed, the two isoforms c.316del and c.316–460del were significantly upregulated by treatment with NMDI-14, a molecule that disrupts the SMG7 nonsense-mediated MRNA decay factor (SMG7)–UPF1 RNA helicase and ATPase (UPF1) interactions and strongly inhibits NMD (Fig. 4c).
Fig. 3.
(a) Schematic representation of PCR strategy for the amplification of the WFS1 cDNA among exons 3, 4 and 5. Coloured arrows show the mapping of the four primers along the WFS1 gene sequence. The red cross indicates the mutated canonical ASS. The theoretical length of PCR products obtained by performing amplification with different combinations of the indicated primers is shown. (b) Representative PCR results for the indicated primer pairs, performed on WT and five patient-derived clones. The yellow rectangle highlights the ~180 bp-long amplicon obtained with the Ex3fw-Ex5rev primer pair. (c) Electropherograms from Sanger sequencing of WFS1-derived PCR products. (d) Agarose gel electrophoresis of amplification with Ex3fw-Ex5rev from WT donor- and WFS1 patient-derived PBMCs. The additional amplicons, including the 180 bp-long amplicon observed in WFS1 iPSCs, are indicated by red triangles. (e) High-throughput evaluation of RNA-seq data quality and goodness of alignment: the sequence quality graph reports the mean quality value (Phred score) across each base position in the R1 (green line) and R2 (orange line) reads. (f) STAR alignment score, reporting the number of mapped/unmapped reads per sample. (g) Per sequence quality score graph reports the number of reads with average quality scores (red, yellow and green backgrounds highlight the Phred score ranges of bad, poor and good quality, respectively). (h) Percentage of reads per sample after adaptor removal by Trimmomatic. (i) Percentage of NGS reads aligned to the WT or the four alternative splice isoform consensus sequences in WT and WFS1 iPSCs. Alignment data for WFS1 are reported as mean of the two analysed clones
Fig. 4.
(a) Schematic representation of the WFS1 AS variants found by NGS, showing the conserved exonic sequence (green) and spliced-out sequence (grey). Sequences at the bottom of each structure zoom at the donor/acceptor AS sites. (b) RT-qPCR of total WFS1 mRNA, in WT and WFS1 iPSCs. Data are plotted as mean ± SD. n=3. *p<0.05 (by Student’s unpaired one-tailed t test). (c) RT-qPCR of the four WFS1 isoforms in WFS1 iPSCs after treatment with NMDI-14 or vehicle (untreated). Data are plotted as mean ± SD. n=3 independent experiments. *p<0.05, ***p<0.001 (by two-way ANOVA with Dunn–Šídák correction)
Characterisation of the translational outcomes from the c.316-1 G>A allele
The ORF-conserving c.316–356del and c.271–513del isoforms induce an in-frame skipping of 141 bp and 243 bp, resulting in 47 amino acids (Val106–Arg152) and 81 amino acids (Val91–Asp171) losses, respectively. This would lead to internally truncated forms of the protein, lacking part of the N-terminal domain but preserving transmembrane and C-terminal domains. To confirm the presence of mutated wolframin, we performed immunoblot with two antibodies recognising the N-terminal residues surrounding the Ala43 position and the Lys679-Phe783 C-terminal residues, respectively. The WT and WFS1-KO fibroblasts were used as controls of antibody specificity. As shown in Fig. 5a, the C-terminal-recognising antibody confirmed that the amount of wolframin in WFS1 iPSCs was ~50% lower than in the WT counterpart. This finding pointed towards the residual protein in affected cells being derived from the c.316-1G>A-carrying allele only. Of note, the higher expression variability and the stable levels of wolframin in WT and WFS1 iPSC clones, respectively, mirrored that observed at transcriptional level. As expected, the N-terminal-recognising antibody did not detect altered isoforms in WFS1 iPSCs (Fig. 5b). These results confirmed that at least one of the splicing isoforms found in WFS1 iPSCs leads to residual protein production, albeit missing a portion of the N-terminal domain. To uncover the impact of the portions lost upon AS events, we refined the structure of the N-terminal domain (residues 1–402) by following the ab initio modelling pipeline of RoseTTAFold. We deposited the de novo model in the ModelArchive Database (doi: 10.5452/ma-cg3qd) and used it to perform homology-based, comparative modelling of the mutated isoforms p.Val106-Arg152del and p.Val91-Thr170del (Fig. 5c, d). The p.Val106-Arg152del isoform partially preserved the wolframin-calmodulin (CaM) binding site, whereas the p.Val91-Thr171 isoform lost the entire wolframin-CaM domain [43]. Moreover, both isoforms resulted in altered N-terminal structure, potentially affecting interaction with other well-known wolframin interactors such as neuronal calcium sensor 1 (NCS1), which is involved in the mitochondria-associated membranes formation and calcium signalling [44]. The dynamic simulation of predicted structures revealed higher instability of the mutated N-terminal domain (>0.5 root mean SD [RMSD]) compared with the WT domain (<0.3 RMSD) after 10 ns into a salt-equilibrated aqueous solution (Fig. 5e). Finally, a radius of gyration of more than 30Å of the p.Val106-Arg152del isoform suggested a reduced compactness of its N-terminal domain (Fig. 5f). These in silico inferences support the hypothesis that the observed residual proteins in WFS1 cells might display reduced binding with cytoplasmic interactors as well as altered capacity to form homotetramers, which are considered the most stable and functional forms of wolframin in the ER membrane [7, 8].
Fig. 5.
(a, b) Immunoblot analysis of wolframin in WT and WFS1 iPSC clones recognised by polyclonal antibodies targeting the C-terminal (a) and N-terminal (b) domains. GAPDH was used as housekeeping gene; Fibro denotes fibroblasts with the indicated genotype. The densitometric analysis of the wolframin/GAPDH ratio is shown; n=3. ***p<0.001 (by Student’s unpaired two-tailed t test). (c, d) Three-dimensional models of WT (residues 1–402), and mutated p.Val106-Arg152del (residues 1–355) (c), and p.Val91–Asp171del (residues 1–322) (d) N-terminal domains, including the first two transmembrane regions. Proteins are represented as ribbons. Region lost in mutated isoforms (green), the mutation-induced conformational changes (blue) and the unaltered portion of the domain (red) are highlighted. In the images displaying the mutated isoforms, magnifications show the superimposed view of transmembrane region α helices in WT (light blue) and p.Val106–Arg152del (red) (c), and the superimposed view of wolframin-CaM binding domain (yellow) loss in p.Val91–Asp171del (d). In p.Val106–Arg152del, the partially conserved wolframin-CaM binding interface is shown as Connolly surface (c). (e, f) Results of molecular dynamics simulation, reporting RMSD of protein backbone atoms and radius of gyration (Rg) of the WT (grey) and mutated (orange/purple) N-terminal domains. Flattening of the RMSD and Rg plots of protein was observed around 10 ns at 300 K
CRISPR/Cas9-mediated gene correction of the c.316-1 G>A allele
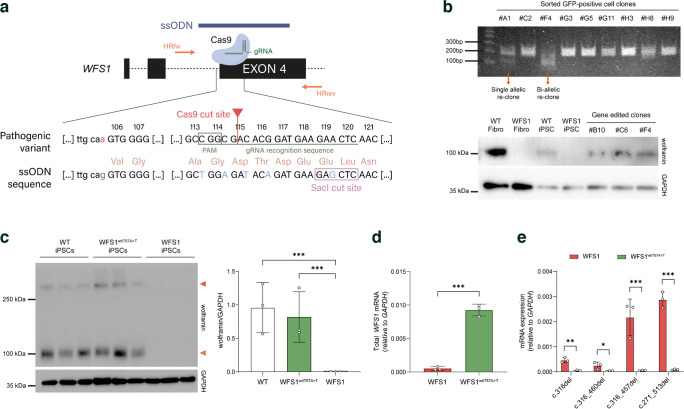
To further confirm that the WFS1 altered isoforms derive from the c.316-1G>A-carrying allele and to obtain a syngeneic allele-specific edited iPSC line to explore the functional impact of mutated transcripts, we performed a CRISPR/Cas9-mediated correction of the 316–1G>A point mutation. We designed single guide RNA (sgRNA) to target exon 4 of the WFS1 gene and trigger HR of the upstream intron–exon junction by using an ssODN, as shown in Fig. 6a. In the ssODN sequence we introduced A>G modification at c.316-1 to restore the natural ASS, and inserted four silent mutations at position p.113, 114, 115 and 116 (indicated in light blue in Fig. 6a) to destroy the protospacer adjacent motif (PAM) sequence and the gRNA recognition motif, without altering amino-acid code, to avoid re-targeting upon homologous recombination. Finally, we also introduced a SacI cut site (GAGCTC) to allow identification of gene engineered cells. Indeed, the gene-edited clones were screened by amplifying the targeted sequence, followed by digestion with the SacI restriction enzyme (Fig. 6b). We obtained a total of six WFS1wt/757A>T iPSC clones in which canonical splicing of WFS1 mRNA and full-length isoform expression of wolframin were restored (Fig. 5c, d); of note, protein levels were similar to those found in WT iPSCs (Fig. 6c). Restoration of the natural ASS was confirmed by RT-qPCR showing the absence of the four alternative splicing isoforms in WFS1wt/757A>T iPSCs (Fig. 6e). All selected gene-edited clones maintained normal morphology and karyotype, and displayed stemness properties as confirmed by expression of the pluripotency markers NANOG, POU class 5 homeobox 1 (OCT4) and SRY-box transcription factor 2 (SOX2) (ESM Fig. 1d–f).
Fig. 6.
(a) Schematic representation of the c.316-1G>A point mutation correction strategy based on the Cas9/gRNA-mediated targeting of WFS1 exon 4. Mapping of the ssODN (blue) and the fw/rev primer pair (orange) used for screening of the homology-directed repair is shown. Magnification highlights the comparison between the pathogenic variant containing the c.316-1G>A mutation with the ssODN sequence re-establishing the natural ASS upstream the exon 4. Protospacer adjacent motif (PAM) sequence and gRNA recognition site are emphasised in dark green. Silent mutations and SacI cut site in ssODN sequence are shown in light blue and framed by a pink rectangle, respectively. (b) Screening by SacI restriction enzyme digestion of GFP-positive iPSC clones and immunoblotting of wolframin in WT and WFS1 fibroblasts (Fibro; as positive and negative controls, respectively), and WT, WFS1 and WFS1wt/757A>T iPSC clones recognised by the polyclonal antibody targeting the N-terminal domain. GAPDH was used as housekeeping gene. (c) Immunoblot analysis of wolframin expression in WT, WFS1wt/757A>T and WFS1 iPSCs. The relative quantification of wolframin/GAPDH ratio is reported and expressed as mean ± SD. n=3 different iPSC clones. ***p<0.001 (by Student’s unpaired two-tailed t test). (d, e) RT-qPCR of total WFS1 mRNA (d) and of AS isoforms (e) in WFS1 and WFS1wt/757A>T iPSCs expressed as mean ± SD; n=3. *p<0.05, **p<0.01, ***p<0.001 (by two-way ANOVA with Dunn–Šídák correction)
Expression of c.316-1 G>A allele-derived WFS1 isoforms in insulin-producing beta cells
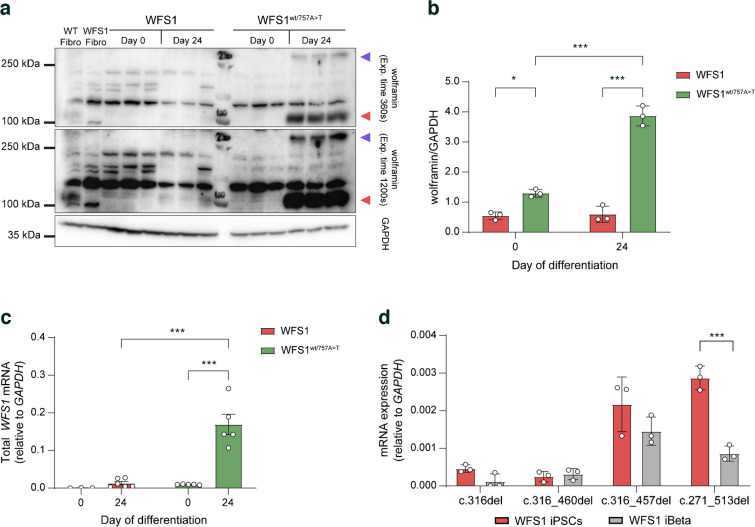
Since pancreatic tissue and in particular insulin-producing beta cells, are affected in WS1, we aimed to explore the impact of aberrant isoforms in iPSC-derived beta cells (iBeta). WFS1 and WFS1wt/757A>T iPSCs were differentiated into pancreatic beta cells following 24 days of in vitro differentiation protocol (ESM Fig. 2a) and WFS1 cells did not display any alterations in the expression of key genes and proteins for the different developmental stages (ESM Fig. 2b, c). Furthermore, pancreatic differentiation efficiency and basal ghrelin, glucagon and insulin secretion did not differ significantly among WT, WFS1 and WFS1wt/757A>T cells at the iBeta stage (ESM Fig. 2c, d). Conversely, the WFS1 iBeta showed a slight, significant decrease in insulin secretion in response to high glucose (11 mmol/l) plus the cAMP inducer IBMX at 15, 20 and 25 min, in comparison with both WT and WFS1wt/757A>T cells (ESM Fig. 2e). Given the pivotal role played by wolframin in maintaining homeostasis and promoting insulin biosynthesis in pancreatic beta cells [1], we expected WFS1 expression to increase during differentiation compared with the iPSC stage. Indeed, in WFS1wt/757A>T cells we observed a 20- to 50-fold increase in WFS1 mRNA levels after 24 days of differentiation, whereas the protein exceeded over threefold the GAPDH levels. Contrariwise, WFS1 cells did not display significant changes in either mRNA or protein levels of wolframin after differentiation (Fig. 7a–c). We confirmed proper upregulation of WFS1 upon pancreatic differentiation in the gene corrected clones by comparing the wolframin levels with WT iBeta. Indeed, overall protein levels did not change between WT and WFS1wt/757A>T iBeta (ESM Fig. 3). Moreover, by evaluating the levels of the c.316-1G>A-carrying allele-derived mRNA isoforms we did not find any difference in c.316del and c.316–460del transcript expression between iPSCs and iBeta, confirming that such isoforms are subjected to NMD also in terminally differentiated cells. Surprisingly, we did not observe any increase in the ORF-maintaining isoforms after differentiation; conversely, WFS1 iBeta displayed a significant reduction in c.271–513del expression (Fig. 7d), suggesting that other mechanisms might be involved in their post-transcriptional regulation. As the wolframin levels did not increase in the WFS1 iBeta, we hypothesised that the residual protein would not be able to guarantee the wolframin-related functions in beta cell stressogenic conditions. Finally, we observed the homotetrameric form of wolframin in WFS1wt/757A>T iBeta only, confirming that the altered residual protein lacked the portion of the N-terminal domain involved in the tetramer formation in WFS1 cells (Fig. 7a).
Fig. 7.
(a) Representative immunoblot showing wolframin protein recognised by the anti-wolframin C-terminal antibody before (day 0) and after (day 24) iPSC differentiation into beta cells. The monomeric form (100 kDa) and homotetrameric form (>250 kDa) are indicated by red and violet triangles, respectively. The signal was acquired at different exposure times to detect homotetramers. (b) Relative quantification of wolframin in WFS1 and WFS1wt/757A>T iPSCs (day 0) and iBeta (day 24), expressed as mean ± SD. n=3 independent in vitro differentiation experiments. *p<0.05, ***p<0.001 (by two-way ANOVA with Dunn–Šídák correction). (c) RT-qPCR analysis of total WFS1 mRNA in WFS1 and WFS1wt/757A>T iPSCs (day 0) vs iBeta (day 24), presented as mean ± SD. n=3–6 independent in vitro differentiation experiments. ***p<0.001 (by two-way ANOVA with Dunn–Šídák correction). (d) Quantification of the AS isoforms in iPSCs and iBeta expressed as mean ± SD. n=3. ***p<0.001 (by two-way ANOVA with Dunn–Šídák correction)
Impact of WFS1 aberrant isoforms on iBeta survival upon stress and inflammation
From a functional point of view, WS1 is characterised by aberrant ER stress responses and overall upregulation of ER stress-related genes in basal conditions [3]. However, our data are in contrast with those reported in the literature, as key ER stress-related markers were comparable in both WFS1 and WFS1wt/757A>T iPSCs at the protein and transcriptional levels. Indeed, we did not find statistically significant differences in the protein levels of markers of the three ER stress-related pathways in WFS1 and WFS1wt/757A>T iPSCs in basal conditions, with the exception of activating transcription factor 4 (ATF4) and endoplasmic reticulum oxidoreductase 1α (ERO1α) (ESM Fig. 4a). More interestingly, no differences in the transcription levels of the ER stress-related genes ATF4, HSPA5 and DDIT3 were detected after 2, 4, 8, 16 or 24 h upon TG treatment in WFS1 and WFS1wt/757A>T iBeta, probably because residual protein activity was sufficient to control ER stress response (ESM Fig. 4b). Of note, WFS1 cells showed a significant decrease in sXBP-1/uXBP-1 ratio in comparison with the WFS1wt/757A>T counterpart after 24 h following TG treatment (ESM Fig. 4b), supporting the hypothesis that the Inositol-requiring enzyme 1 (IRE1)-dependent UPR may not be effective in controlling stress long term. However, reduced sXBP-1 levels did not correlate with higher apoptotic rate in WFS1 cells, supporting the hypothesis that cell death may be a consequence of ineffective short-term ER stress response. Although TG treatment induced the same magnitude of cell death in WFS1 and WFS1wt/757A>T iPSCs after 8 h, we observed that recovery from TG-induced apoptosis occurred in the WFS1wt/757A>T cells only. Indeed, the genetically corrected cells were able to reverse TG-induced apoptosis after 24 h, while the percentage of Annexin-V-positive WFS1 cells remained around 40% up to 48 h after stress induction (ESM Fig. 4c). We further examined ATF4, HSPA5 and DDIT3 expression in WFS1 and WFS1wt/757A>T iBeta 8 h and 16 h after TG treatment, confirming that TG stimulation induced a similar increase in UPR markers in both the iBeta lines at early time points. However, as observed in iPSCs, the UPR in WFS1 cells was less persistent over time when compared with WFS1wt/757A>T cells (Fig. 8a, d, g), as we reported a decrease in mRNA expression for ATF4 (relative to GAPDH: 0.24 ± 0.11 at 8 h vs 0.12 ± 0.10 at 16 h; p=0.12), HSPA5 (relative to GAPDH: 1.46 ± 0.48 at 8 h vs 0.82 ± 0.61 at 16 h; p=0.41) and DDIT3 (relative to GAPDH: 0.35 ± 0.11 at 8 h vs 0.07 ± 0.04 at 16 h; p<0.01) in WFS1 iBeta after 16 h. Both ATF4 and HSPA5 expression levels remained significantly higher compared with the baseline in WFS1wt/757A>T cells at the same time point (over fivefold for ATF4 and over twofold for HSPA5 after 16 h; p<0.01). We then explored the impact of inflammation on the UPR in both WFS1 and WFS1wt/757A>T iBeta. The exposure to inflammatory cytokines induced a significant upregulation of the UPR in WFS1wt/757A>T iBeta, but not in WFS1 after 48 h of treatment (Fig. 8b,c,e,f,h,i). Both TG and cytokine treatments triggered statistically significant higher rates of early and late apoptotic cell death in WFS1 compared with WFS1wt/757A>T iBeta, as revealed by the frequency of Annexin-V+ and PI+ cells, respectively (Fig. 9a–d). In particular, the WFS1 iBeta displayed a significant increase in Annexin-V+ cells after exposure to inflammatory cytokines and overall higher susceptibility to inflammatory signals compared with the gene-edited counterpart. Of note, as it has been shown that both ER stress induction and inflammation are able to inhibit NMD and increase PTC-carrying mRNAs stability [45, 46], we wondered whether this might affect the PTC-carrying WFS1 isoforms, resulting in toxic accumulation of the aberrant transcripts. Therefore, we evaluated the transcription levels of UPF1 and SMG7 genes, which encode for the main components of the NMD complex. As expected, treatment with both TG and inflammatory cytokines significantly downregulated SMG7 more than twofold compared with baseline, but did not affect UPF1, in both WFS1 and WFS1wt/757A>T iBeta (Fig. 9e, f). We further detected a dramatic increase in the PTC-carrying splicing isoforms c.316del and c.316–460del after exposure to cell stress inducer or proinflammatory cytokines (c.316del: >60-fold and 100-fold after 16 h TG and 48 h cytokine treatments, respectively; c.316–460del: >20-fold after either 16 h TG or 48 h cytokine treatments) (Fig. 9g, h). These findings suggest that exogenous stress strongly affected the NMD-related gene SMG7 and induced accumulation of PTC-carrying isoforms, correlating with increased cell death in WFS1 iBeta, without UPR upregulation. A slight but non-significant increase in the ORF-conserving isoforms c.316–356del and c.271–513del was also detected.
Fig. 8.
(a, d, g) Relative expression of ATF4, HSPA5 and DDIT3 in WFS1 and WFS1wt/757A>T beta cells at 8 h or 16 h post-TG exposure. Data are plotted as mean ± SEM. n=4–9 independent replicates. *p<0.05, **p<0.01, ***p<0.001 (by two-way ANOVA with Dunn–Šídák correction). (b, c, e, f, h, i) Estimation plot including the mean of difference in ATF4, HSPA5 and DDIT3 mRNAs calculated at 48 h after cytokine treatment (50 U/ml IL-1β, 1000 U/ml IFN-γ and 10 ng/ml TNF-α) in WFS1 (red) and WFS1wt/757A>T (green) beta cells. n=6 for WFS1, n=5 for WFS1wt/757A>T iBeta. *p<0.05 (by paired Student’s t test)
Fig. 9.
(a–d) Early and late apoptosis were measured as percentage (%) of Annexin-V and PI-positive cells, respectively, in WFS1 and WFS1wt/757A>T iBeta after 50 nmol/l TG or inflammatory cytokine exposure. Data are plotted as mean ± SD. n=3–5. **p<0.01, ***p<0.001 (by two-way ANOVA with Dunn–Šídák correction). (e, f) Gene expression of the splicing factors UPF1 and SMG7 in WFS1 and WFS1wt/757A>T iBeta upon TG or inflammatory cytokine treatment at the indicated times. Data are expressed as fold change over the untreated control. Data are expressed as mean ± SD; n=3. **p<0.01, ***p<0.001 (by two-way ANOVA with Dunn–Šídák correction). (g, h) Relative quantification of AS isoforms in WFS1 iBeta upon treatment for 8 h and 16 h with TG (g) or treatment for 48 h with inflammatory cytokines (h). Data are plotted as mean ± SD. n=3 independent experiments. **p<0.01, ***p<0.001 (by two-way ANOVA with Dunn–Šídák correction)
Discussion
Over the last few years, WS1 has assumed the connotation of a spectrum of disorders, due to the strong variability in terms of clinical manifestations and progression [47]. Because of the role that wolframin plays in a plethora of cellular processes, exploring the impact of WFS1 variants on cell function and survival may represent a crucial step for their association with clinical phenotype.
Previous attempts to discover genotype and phenotype correlations led to the classification of WFS1 mutations according to their impact on the protein expression and stability [16, 48, 49]. A robust classification of WFS1 genetic variants by disease onset, and spectrum and severity of symptoms, has been recently published [18]. Studying the relationship between disease severity score reported by the Wolfram Unified Rating Scale and WFS1 variants, the metanalysis performed by Urano’s group highlighted the importance of characterisation of the classes of mutations for providing a more accurate prognoses of outcomes and a reliable therapy for patients with WS1.
The patient characterised in this work presents clinical symptoms with a mild progression of the disease in relation to age, with marginal optic nerve and hearing involvement [27], and residual circulating C-peptide [38]. The mutation in exon 7 (c.757A>T) introduces a PTC; the other (c.316-1G>A) gives rise to four splice isoforms, two of which are frameshift and two preserve the ORF. According to the classification offered by de Heredia et al [16], the introduction of a PTC before exon 8 causes NMD intervention, transcript degradation and therefore protein absence. By confirming that NMD induces complete degradation of PTC-carrying isoforms, we can state that both mutations c.757A>T and c.316-1G>A can be classified as type 1 [16]. However, the isoforms that preserve the ORF generate a functional residual protein, which would allow us to associate the patient genotype with an intermediate, mild–moderate phenotype, according to the classification reported by Urano’s work [18]. In light of this consideration, the c.316-1G>A mutation has a transcriptional and translational phenotype more similar to that deriving from missense mutations, which maintain partial protein structure and, therefore, function.
The four novel isoforms of WFS1 mRNA result from alternative splicing due to loss of the canonical ASS upstream of exon 4. In particular, two out of the four isoforms were correctly predicted by the use of a probabilistic model based on the maximum entropy principle. Indeed, we inferred that the two most probable alternative splicing isoforms could be c.316del (deriving from 1-bp shifting of the ASS following the mutation in c.316-1) and c.316–460del (losing exon 4 due to the direct skipping to the ASS of exon 5). Otherwise, the other two isoforms c.316–356del and c.271–513del were not predicted by the in silico analysis but we found that the sequence surrounding the cryptic uncanonical splicing site generating the c.316–356del isoform was particularly enriched in ESEs. We excluded that the AS events occurring in the patient were a consequence of reprogramming of the somatic cells into iPSCs, refuting previous evidence reporting that PBMCs from the same patient displayed WFS1 transcript originating from the c.757A>T-carrying allele only [28]. Despite the design of primers for amplifying WFS1 mRNA being similar to that described by Panfili et al [28], our protocol allows us to identify the AS isoforms deriving from the c.316-1G>A-carrying allele also in patient-derived primary PBMCs. Deep sequencing further confirmed the four isoforms, which we have chosen to classify based on the maintenance of the ORF and, therefore, on their ability to originate a protein, although with altered or reduced function.
Interestingly, the two ORF-maintaining isoforms, c.316–356del and c.271–513del, conserved the ability to produce a protein with intact transmembrane and C-terminal domains, thus potentially preserving its subcellular localisation and intraluminal interactions. In WFS1 iPSCs, we confirmed expression of wolframin lacking part of N-terminal domain but still maintaining the integrity of the C-terminal domain; this could explain the absence of significant alterations in ER stress markers upon ER stress induction and of the activating transcription factor 6 (ATF6) cleaved form. However, we found that after differentiation into pancreatic beta cells, the WFS1 gene in affected cells was unable to reach the expression levels of the gene corrected clones. This resulted in low levels of residual proteins.
Our work highlights another important aspect concerning the physiological mechanisms involved in pancreatic beta cells: NMD. Indeed, the integrity and accuracy of transcript processing is pivotal for beta cells to meet the physiological demands and pathophysiological challenges they face [21]. The disruption of canonical splice sites and AS events introducing PTC may shift the balance towards the accumulation of aberrant transcripts during stress conditions, potentially affecting beta cell survival [50].
We report that the PTC-carrying c.316del and c.316–460del isoforms are regulated by NMD, as by inhibiting the UPF1/SMG7 interaction the degradation of such transcripts was prevented. Indeed, NMD orchestrates the regulated unproductive splicing and translation (RUST) mechanism to eliminate the PTC-containing transcript isoforms generated due to perturbed AS [51]. Although the role of RUST in pancreatic beta cells is still largely unknown, deregulation of NMD induced by islet stress or inflammation influences RUST, leading to accumulation of unproductive transcript isoforms, which are implicated in beta cell dysfunction, vulnerability and death [50]. Interestingly, we found that patient-derived cells were particularly susceptible to the TG-induced cell death due to impairment in recovery from sarco-ER calcium ATPase (SERCA) inhibition. In this context, we observed the early upregulation of UPR genes by both WFS1 and WFS1wt/757A>T iBeta, but at later times (16 h) WFS1 cells did not maintain the upregulation and, as a consequence, displayed increased apoptosis. A similar process seems to be involved in inflammatory cytokine-dependent apoptosis, which is higher in WFS1 iBeta compared with their genetically corrected counterpart. This result is driven by a lack of UPR upregulation upon cytokine exposure, which is, in contrast, efficiently achieved by WFS1wt/757A>T iBeta. Our results point to lack of UPR upregulation as the cause of cell death following ER stress induction by inflammation, as already reported by other groups [52, 53]. A previous study on the WS1 patient presented in this work reported a state of chronic inflammation associated with high levels of inflammatory cytokines, including IL-1β, TNF-α, IFN-γ and IL-6 [28]. Increased secretion of inflammatory cytokines in the serum of the patient and susceptibility of WFS1-deficient beta cells to exposure to proinflammatory factors could represent a new aetiopathogenetic model for WS1-related diabetes. Both inflammation and other exogenous stress could be the cause of NMD inhibition and consequently of accumulation of aberrant transcripts, as shown by the increase in all AS isoforms upon stress induction. Indeed, after exogenous stimulus, we observed a decrease in SMG7 expression and a critical increase in WFS1 PTC-containing isoforms. Accumulation of these transcripts could be the cause of beta cell dysfunction and apoptosis.
The case study we report represents a new model for studying wolframin from a functional point of view, highlighting how each single mutation of the WFS1 gene can determine dramatically different outcomes from a clinical point of view. We underline the need for a better classification of mutations in the WFS1 gene in order to explain the penetrance and expressivity of the clinical characteristics of the disease. Further understanding of the mechanisms by which WFS1 variants functionally affect ER stress or inflammatory responses may aid in the identification of other potential therapeutic targets.
Finally, the role of NMD in WS1 has not been considered until now, despite the majority of mutations in the WFS1 gene introducing a PTC. Accumulation of WFS1 unproductive transcripts whose translation into unfolded polypeptides overwhelms ER capacity may drive unresolved ER stress. The role of NMD among different cell contexts may explain susceptibility to WFS1 mutations of specific cell types. Although further research is needed to validate this hypothesis, NMD inhibition and PTC-carrying mRNA accumulation in WS1 could represent targetable mechanisms for the development of new therapeutic strategies to lengthen the t½ of the residual protein or compensate for missing functions. A similar rationale is highlighted in the field of cystic fibrosis, another autosomal recessive disorder, where knowledge acquired from genotype–phenotype association studies has helped in the identification of effective therapeutic strategies and improved the prognosis [54–56].
Supplementary Information
Below is the link to the electronic supplementary material.
Abbreviations
- AS
Alternative splicing
- ASS
Acceptor splice site
- BP score
Branch point score
- CaM
Calmodulin
- ER
Endoplasmic reticulum
- ESE
Exonic splicing enhancer
- ESR
Exonic splicing auxiliary sequences ratio
- ESS
Exonic splicing silencer
- fw
Forward
- HR
Homologous-directed repair
- iBeta
iPSC-derived beta cell
- iPSC
Induced pluripotent stem cells
- IVS
Intervening sequence
- LOVD
Leiden Open Variation Database
- NGS
Next generation sequencing
- NMD
Nonsense-mediated decay
- ORF
Open reading frame
- PBMC
Peripheral blood mononuclear cell
- PTC
Premature termination codon
- qPCR
Quantitative PCR
- rev
Reverse
- RMSD
Root mean SD
- RUST
Regulated unproductive splicing and translation
- sgRNA
Single guide RNA
- ssODN
Single-strand oligodeoxynucleotide
- TG
Thapsigargin
- TMD
Transmembrane domain
- UPR
Unfolded protein response
- UTR
Untranslated region
- WS1
Wolfram syndrome 1
- WT
Wild-type
Acknowledgements
We thank the Advanced Light and Electron Microscopy BioImaging Center (ALEMBIC) at San Raffaele Scientific Institute, Milan (Italy), for confocal immunofluorescence images and the Flow cytometry Resource, Advanced Cytometry Technical Applications Laboratory (FRACTAL), at San Raffaele Scientific Institute, Milan (Italy), for cell sorting experiments. We also thank F. Giannese and D. Lazarevic at the Center for Omics Sciences (COSR) of San Raffaele Scientific Institute, Milan (Italy), for providing support in library preparation, deep sequencing and bioinformatic analysis. We are grateful to A. Lombardo at the San Raffaele Telethon Institute for Gene Therapy (SR-TIGET), San Raffaele Scientific Institute, Milan (Italy), for the access to BLS2 work areas and the use of the 4D-Nucleofector System. S. Torchio conducted this study as partial fulfilment of an international PhD in Molecular Medicine at Vita-Salute San Raffaele University.
Data availability
The in silico predicted N-terminal domain structure file of WT wolframin was deposited in the ModelArchive, together with procedures, ramachandran plots, inter-residue distance deviation and IDDT scores, and Gromacs configuration files (doi/10.5452/ma-cg3qd). The deep-sequencing data as fastq files used to generate consensus sequences of AS isoforms of WFS1 are available in the SRA database (BioProject PRJNA1109747). All raw data that were not directly included in the manuscript or that have not been deposited in online repositories, are available on request from the corresponding authors.
Funding
This study was supported by a private family donation financing investigation on Wolfram syndrome 1 at the Diabetes Research Institute (DRI) of the IRCCS San Raffaele Hospital. Part of the activities were also supported through the funds from the European Union - Next Generation EU - PNRR M6C2 - Investment 2.1 Enhancement and strengthening of NHS biomedical research (PNRR-MR1-2022-12375914).
Authors’ relationships and activities
The authors declare that there are no relationships or activities that might bias, or be perceived to bias, their work.
Contribution statement
RC, GF and LP were responsible for the conception and design of the study. ST contributed to the experimental design and to the collection and analysis of the data, together with RC and GS. RC, ST, GS, VZ, LM, MTL, FM and FC conducted wet experiments. SP supported collection of data during revision activities. RC performed bioinformatic analysis. RC, ST, GS and LP interpreted the results. PC followed genetic testing in clinical practice. Medical evaluations and patient specimen collection was performed by GF and RB. GR and VB supported cell reprogramming activities. VZ, LM, MTL, SP and VS performed stem cell differentiation and beta cell function analysis. GC contributed to data interpretation. RC and ST wrote the original draft, and all authors contributed to the critical review and editing of the manuscript. All authors approved the final version to be published. LP takes responsibility for the integrity of the data and is the guarantor of this work.
Footnotes
Raniero Chimienti and Silvia Torchio are joint first authors and Giulio Frontino and Lorenzo Piemonti are joint last authors.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Raniero Chimienti, Email: chimienti.raniero@hsr.it.
Silvia Torchio, Email: silvia.torchio@curie.fr.
Lorenzo Piemonti, Email: piemonti.lorenzo@hsr.it.
References
- 1.Abreu D, Asada R, Revilla JMP et al (2020) Wolfram syndrome 1 gene regulates pathways maintaining β-cell health and survival. Lab Invest 100:849–862. 10.1038/s41374-020-0408-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rigoli L, Bramanti P, Di Bella C, De Luca F (2018) Genetic and clinical aspects of Wolfram syndrome 1, a severe neurodegenerative disease. Pediatr Res 83:921–929. 10.1038/pr.2018.17 [DOI] [PubMed] [Google Scholar]
- 3.Fonseca SG, Fukuma M, Lipson KL et al (2005) WFS1 is a novel component of the unfolded protein response and maintains homeostasis of the endoplasmic reticulum in pancreatic β-cells. J Biol Chem 280:39609–39615. 10.1074/jbc.M507426200 [DOI] [PubMed] [Google Scholar]
- 4.Cagalinec M, Liiv M, Hodurova Z et al (2016) Role of mitochondrial dynamics in neuronal development: mechanism for wolfram syndrome. PLoS Biol 14:e1002511. 10.1371/journal.pbio.1002511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.La Morgia C, Maresca A, Amore G et al (2020) Calcium mishandling in absence of primary mitochondrial dysfunction drives cellular pathology in Wolfram Syndrome. Sci Rep 10:4785. 10.1038/s41598-020-61735-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crouzier L, Richard EM, Diez C et al (2022) Morphological, behavioral and cellular analyses revealed different phenotypes in Wolfram syndrome wfs1a and wfs1b zebrafish mutant lines. Hum Mol Genet 31:2711–2727. 10.1093/hmg/ddac065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wilf-Yarkoni A, Shor O, Fellner A et al (2021) Mild phenotype of wolfram syndrome associated with a common pathogenic variant is predicted by a structural model of wolframin. Neurol Genet 7:e578. 10.1212/NXG.0000000000000578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hofmann S, Philbrook C, Gerbitz KD, Bauer MF (2003) Wolfram syndrome: structural and functional analyses of mutant and wild-type wolframin, the WFS1 gene product. Hum Mol Genet 12:2003–2012. 10.1093/hmg/ddg214 [DOI] [PubMed] [Google Scholar]
- 9.Eller P, Föger B, Gander R et al (2001) Wolfram syndrome: a clinical and molecular genetic analysis. J Med Genet 38(11):E37. 10.1136/jmg.38.11.e37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van den Ouweland JMW, Cryns K, Pennings RJE et al (2003) Molecular characterization of WFS1 in patients with Wolfram syndrome. J Mol Diagn 5:88–95. 10.1016/S1525-1578(10)60457-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Inoue H, Tanizawa Y, Wasson J et al (1998) A gene encoding a transmembrane protein is mutated in patients with diabetes mellitus and optic atrophy (Wolfram syndrome). Nat Genet 20:143–148. 10.1038/2441 [DOI] [PubMed] [Google Scholar]
- 12.Strom TM, Hörtnagel K, Hofmann S et al (1998) Diabetes insipidus, diabetes mellitus, optic atrophy and deafness (DIDMOAD) caused by mutations in a novel gene (wolframin) coding for a predicted transmembrane protein. Hum Mol Genet 7:2021–2028. 10.1093/hmg/7.13.2021 [DOI] [PubMed] [Google Scholar]
- 13.Rohayem J, Ehlers C, Wiedemann B et al (2011) Diabetes and neurodegeneration in Wolfram syndrome: a multicenter study of phenotype and genotype. Diabetes Care 34:1503–1510. 10.2337/dc10-1937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chaussenot A, Bannwarth S, Rouzier C et al (2011) Neurologic features and genotype-phenotype correlation in Wolfram syndrome. Ann Neurol 69:501–508. 10.1002/ana.22160 [DOI] [PubMed] [Google Scholar]
- 15.Cano A, Rouzier C, Monnot S et al (2007) Identification of novel mutations in WFS1 and genotype-phenotype correlation in Wolfram syndrome. Am J Med Genet A 143A:1605–1612. 10.1002/ajmg.a.31809 [DOI] [PubMed] [Google Scholar]
- 16.de Heredia ML, Clèries R, Nunes V (2013) Genotypic classification of patients with Wolfram syndrome: insights into the natural history of the disease and correlation with phenotype. Genet Med 15:497–506. 10.1038/gim.2012.180 [DOI] [PubMed] [Google Scholar]
- 17.Delvecchio M, Iacoviello M, Pantaleo A, Resta N (2021) Clinical spectrum associated with Wolfram syndrome type 1 and type 2: a review on genotype-phenotype correlations. Int J Environ Res Public Health 18:4796. 10.3390/ijerph18094796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee EM, Verma M, Palaniappan N et al (2023) Genotype and clinical characteristics of patients with Wolfram syndrome and WFS1-related disorders. Front Genet 14:1198171. 10.3389/fgene.2023.1198171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kitamura RA, Maxwell KG, Ye W et al (2022) Multidimensional analysis and therapeutic development using patient iPSC–derived disease models of Wolfram syndrome. JCI Insight 7(18):e156549. 10.1172/jci.insight.156549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moss ND, Sussel L (2020) mRNA Processing: an emerging frontier in the regulation of pancreatic β cell function. Front Genet 11:983. 10.3389/fgene.2020.00983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ghiasi SM, Rutter GA (2021) Consequences for pancreatic β-cell identity and function of unregulated transcript processing. Front Endocrinol 12:625235. 10.3389/fendo.2021.625235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hug N, Longman D, Cáceres JF (2016) Mechanism and regulation of the nonsense-mediated decay pathway. Nucleic Acids Res 4:1483–95. 10.1093/nar/gkw010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bulman MP, Harries LW, Hansen T et al (2002) Abnormal splicing of hepatocyte nuclear factor 1 alpha in maturity-onset diabetes of the young. Diabetologia 10:1463–7. 10.1007/s00125-002-0919-1 [DOI] [PubMed] [Google Scholar]
- 24.Cappelli A, Tumini S, Consoli A et al (2009) Novel mutations in GCK and HNF1A genes in Italian families with MODY phenotype. Diabetes Res Clin Pract 3:e72-4. 10.1016/j.diabres.2008.12.007 [DOI] [PubMed] [Google Scholar]
- 25.Prokunina-Olsson L, Welch C, Hansson O et al (2009) Tissue-specific alternative splicing of TCF7L2. Hum Mol Genet 20:3795–804. 10.1093/hmg/ddp321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Locke JM, Harries LW (2008) RNA processing and mRNA surveillance in monogenic diabetes. Gene Regul Syst Bio 2:203–12. 10.4137/grsb.s782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Squitti R, Cerchiaro G, Giovannoni I et al (2019) A case of a mild Wolfram Syndrome with concomitant ATP7B mutation. CellR4 Repair Replace Regen Reprogram 7:e2735. 10.32113/cellr4_20198_2735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Panfili E, Mondanelli G, Orabona C et al (2021) Novel mutations in the WFS1 gene are associated with Wolfram syndrome and systemic inflammation. Hum Mol Genet 30:265–276. 10.1093/hmg/ddab040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pellegrini S, Manenti F, Chimienti R et al (2018) Differentiation of Sendai virus-reprogrammed iPSC into β cells, compared with human pancreatic islets and immortalized β cell line. Cell Transpl 27:1548–1560. 10.1177/0963689718798564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chimienti R, Baccega T, Torchio S et al (2022) Engineering of immune checkpoints B7–H3 and CD155 enhances immune compatibility of MHC-I–/– iPSCs for β cell replacement. Cell Rep 40:111423. 10.1016/j.celrep.2022.111423 [DOI] [PMC free article] [PubMed]
- 31.Pellegrini S, Pipitone GB, Cospito A et al (2021) Generation of β cells from iPSC of a MODY8 patient with a novel mutation in the Carboxyl Ester Lipase (CEL) gene. J Clin Endocrinol Metab 106:e2322–e2333. 10.1210/clinem/dgaa986 [DOI] [PubMed] [Google Scholar]
- 32.Schindelin J, Arganda-Carreras I, Frise E et al (2012) Fiji: an open-source platform for biological-image analysis. Nat Methods 9:676–682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Desmet FO, Hamroun D, Lalande M et al (2009) Splicing Finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res 37(9):e67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yeo G, Burge CB (2004) Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J Comput Biol 11:377–394. 10.1089/1066527041410418 [DOI] [PubMed] [Google Scholar]
- 35.Kim DE, Chivian D, Malmström L, Baker D (2005) Automated prediction of domain boundaries in CASP6 targets using Ginzu and RosettaDOM. Proteins 61(Suppl 7):193–200. 10.1002/prot.20737 [DOI] [PubMed] [Google Scholar]
- 36.Kim DE, Chivian D, Baker D (2004) Protein structure prediction and analysis using the Robetta server. Nucleic Acids Res 32:W526. 10.1093/nar/gkh468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Van Der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJC (2005) GROMACS: fast, flexible, and free. J Comput Chem 26:1701–1718. 10.1002/jcc.20291 [DOI] [PubMed] [Google Scholar]
- 38.Frontino G, Raouf T, Canarutto D et al (2021) Off-label Liraglutide use in children with Wolfram Syndrome type 1: extensive characterization of four patients. Front Pediatr 9:755365. 10.3389/fped.2021.755365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Frontino G, Di Tonno R, Stancampiano MR et al (2023) Paediatric wolfram syndrome Type 1: should gonadal dysfunction be part of the diagnostic criteria? Front Endocrinol (Lausanne) 14:1155644. 10.3389/fendo.2023.1155644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Richards S, Aziz N, Bale S et al (2015) Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American college of medical genetics and genomics and the association for molecular pathology. Genet Med Off J Am Coll Med Genet 17:405. 10.1038/gim.2015.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Astuti D, Sabir A, Fulton P et al (2017) Monogenic diabetes syndromes: Locus-specific databases for Alström, Wolfram, and Thiamine-responsive megaloblastic anemia. Hum Mutat 38:764. 10.1002/humu.23233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sandhu MS, Weedon MN, Fawcett KA et al (2007) Common variants in WFS1 confer risk of type 2 diabetes. Nat Genet 39:951–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yurimoto S, Hatano N, Tsuchiya M et al (2009) Identification and characterization of wolframin, the product of the wolfram syndrome gene (WFS1), as a novel calmodulin-binding protein. Biochemistry 48:3946–3955. 10.1021/bi900260y [DOI] [PubMed] [Google Scholar]
- 44.Angebault C, Fauconnier J, Patergnani S et al (2018) ER-mitochondria cross-talk is regulated by the Ca2+ sensor NCS1 and is impaired in Wolfram syndrome. Sci Signal 11:eaaq1380. 10.1126/scisignal.aaq1380 [DOI] [PubMed]
- 45.Li Z, Vuong JK, Zhang M, Stork C, Zheng S (2017) Inhibition of nonsense-mediated RNA decay by ER stress. RNA 23:378–394. 10.1261/rna.058040.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Usuki F, Yamashita A, Fujimura M (2019) Environmental stresses suppress nonsense-mediated mRNA decay (NMD) and affect cells by stabilizing NMD-targeted gene expression. Sci Rep 9:1279. 10.1038/s41598-018-38015-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Urano F (2016) Wolfram syndrome: diagnosis, management, and treatment. Curr Diab Rep 16:6. 10.1007/s11892-015-0702-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hu K, Zatika M, Astuti D et al (2022) WFS1 protein expression correlates with clinical progression of optic atrophy in patients with Wolfram syndrome. J Med Genet 59:65–74. 10.1136/jmedgenet-2020-107257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Majander A, Jurkute N, Burté F et al (2022) WFS1-associated optic neuropathy: genotype-phenotype correlations and disease progression. Am J Ophthalmol 241:9–27. 10.1016/j.ajo.2022.04.003 [DOI] [PubMed] [Google Scholar]
- 50.Dlamini Z, Mokoena F, Hull R (2017) Abnormalities in alternative splicing in diabetes: therapeutic targets. J Mol Endocrinol 2:R93–R107. 10.1530/JME-17-0049 [DOI] [PubMed] [Google Scholar]
- 51.McGlincy NJ, Smith CWJ (2008) Alternative splicing resulting in nonsense-mediated mRNA decay: what is the meaning of nonsense? Trends Biochem Sci 33:385–393. 10.1016/j.tibs.2008.06.001 [DOI] [PubMed] [Google Scholar]
- 52.Akerfeldt MC, Howes J, Chan JY et al (2008) Cytokine-induced beta-cell death is independent of endoplasmic reticulum stress signaling. Diabetes 11:3034–44. 10.2337/db07-1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brozzi F, Nardelli TR, Lopes M et al (2015) Cytokines induce endoplasmic reticulum stress in human, rat and mouse beta cells via different mechanisms. Diabetologia 10:2307–16. 10.1007/s00125-015-3669-6 [DOI] [PubMed] [Google Scholar]
- 54.Cutting GR (2015) Cystic fibrosis genetics: from molecular understanding to clinical application. Nat Rev Genet 16:45–56. 10.1038/nrg3849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Claustres M, Thèzedes Georges CM et al (2017) CFTR-France, a national relational patient database for sharing genetic and phenotypic data associated with rare CFTR variants. Hum Mutat 38:1297–1315. 10.1002/humu.23276 [DOI] [PubMed] [Google Scholar]
- 56.Noel S, Servel N, Hatton A et al (2022) Correlating genotype with phenotype using CFTR-mediated whole-cell Cl- currents in human nasal epithelial cells. J Physiol 600:1515–1531. 10.1113/JP282143 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The in silico predicted N-terminal domain structure file of WT wolframin was deposited in the ModelArchive, together with procedures, ramachandran plots, inter-residue distance deviation and IDDT scores, and Gromacs configuration files (doi/10.5452/ma-cg3qd). The deep-sequencing data as fastq files used to generate consensus sequences of AS isoforms of WFS1 are available in the SRA database (BioProject PRJNA1109747).
The in silico predicted N-terminal domain structure file of WT wolframin was deposited in the ModelArchive, together with procedures, ramachandran plots, inter-residue distance deviation and IDDT scores, and Gromacs configuration files (doi/10.5452/ma-cg3qd). The deep-sequencing data as fastq files used to generate consensus sequences of AS isoforms of WFS1 are available in the SRA database (BioProject PRJNA1109747). All raw data that were not directly included in the manuscript or that have not been deposited in online repositories, are available on request from the corresponding authors.