Abstract
Atherosclerosis (AS) is the leading cause of coronary heart disease, which is the primary cause of death worldwide. Recent studies have identified disulfidptosis as a new type of cell death that may be involved in onset and development of many diseases. However, the role of disulfidptosis in AS is not clear. In this study, bioinformatics analysis and experiments in vivo and in vitro were performed to evaluate the potential relationship between disulfidptosis and AS. AS-related sequencing data were obtained from the Gene Expression Omnibus (GEO). Bioinformatics techniques were used to evaluate differentially expressed genes (DEGs) associated with disulfidptosis-related AS. Hub genes were screened using least absolute shrinkage and selection operator (LASSO) and random forests (RF) methods. In addition, we established a foam cell model in vitro and an AS mouse model in vivo to verify the expressions of hub genes. In addition, we constructed a diagnostic nomogram with hub genes to predict progression of AS. Finally, the consensus clustering method was used to establish two different subtypes, and associations between subtypes and immunity were explored. As the results, 9 disulfidptosis-related AS DEGs were identified from GSE28829 and GSE43292 datasets. Evaluation of DEGs using LASSO and RF methods resulted in identification of 4 hub genes (CAPZB, DSTN, MYL6, PDLIM1), which were analyzed for diagnostic value using ROC curve analysis and verified in vitro and in vivo. Furthermore, a nomogram including hub genes was established that accurately predicted the occurrence of AS. The consensus clustering algorithm was used to separate patients with early atherosclerotic plaques and patients with advanced atherosclerotic plaques into two disulfidptosis subtypes. Cluster B displayed higher levels of infiltrating immune cells, which indicated that patients in cluster B may have a positive immune response for progression of AS. In summary, disulfidptosis-related genes including CAPZB, DSTN, MYL6, and PDLIM1 may be diagnostic markers and therapeutic targets for AS. In addition, these genes are closely related to immune cells, which may inform immunotherapy for AS.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-78392-5.
Keywords: Atherosclerosis, Disulfidptosis, Bioinformatics, Consensus cluster, Immune cell infiltration
Subject terms: Biochemistry, Cell biology, Cardiology, Diseases
Introduction
Atherosclerosis (AS) is a progressive arterial disease in which closely packed endothelial cells gradually deteriorate due to prolonged exposure to chronic inflammation, lipid deposition, and other deleterious stimuli, resulting in thickening of the arterial intima-media, and subsequent plaque formation1. During progression of atherosclerosis inflammation and immune responses affect endothelial cells, and the imbalance between pro-inflammatory and anti-inflammatory mechanisms dictates the final clinical outcome2. Development of effective preventive and treatment measures is critical for acute ischemic events caused by erosion or rupture of atherosclerotic plaques, such as myocardial infarction and stroke, which are fatal or disabling diseases3. Extensive research has been performed to elucidate the effects of anti-inflammatory therapy on AS, but translation into clinical practice remains lacking4,5. Reducing plasma low-density lipoprotein cholesterol (LDL-C) levels is an established method for reducing risk for development and progression of AS, but some patients continue to develop adverse cardiovascular events due to residual inflammatory risk6,7. Therefore, discovery of effective diagnostic tools and therapeutic targets for AS is critical. Disulfidptosis is mediated by abnormal disulfide bond formation of actin cytoskeleton proteins and is new form of cell death. This process results from an imbalance in nicotinamide adenine dinucleotide phosphate (NADPH) and cystine metabolism8. Disulfidptosis represents a novel mechanistic and therapeutic target for AS, but the association between process and disease requires further study, including identification of potential biomarkers.
Cells respond to damaging stimuli by initiating apoptosis, pyroptosis, ferroptosis, autophagy, and other specific programs as adaptive mechanisms. These cell death forms can independently or jointly regulate a wide range of pathophysiological processes, and characterization of these processes provides insights into disease mechanisms and development of therapeutic targets9,10. Chemokine C-C-Motif Receptor 2 (CCR2) has been identified as a critical regulator of neuronal apoptosis following subarachnoid hemorrhage, and suppression of CCR2 reduced uncontrolled apoptosis and alleviated inflammation through reduced PI3K/AKT signaling11. Ferritinophagy aggravates myocardial injury by promoting cardiomyocyte ferroptosis. Inhibition of nuclear receptor co-activator 4 (NCOA4) has been shown to reverse ferritinophagy, providing a potential therapeutic target12. Cuproptosis was discovered within the last year and numerous research have examined how it is associated with pathophysiology of diseases. Cuproptosis represents a promising target for blocking diseases13,14. Further characterization of disulfidptosis may allow for discovery of novel molecular mechanisms and effective interventions for diseases15. However, the effects of disulfidptosis on development of AS has not been studied.
Bioinformatics methods are powerful data processing tools for analysis of complex disease mechanisms and the development of ideal targets for diagnosis or treatment16, which have received wide-spread attention and comprehensive application in recent years. Researchers could predict the risk of AS with systemic lupus erythematosus using various bioinformatic analyses and machine learning algorithms17. By integrating network pharmacology and experimental validation, the bioactive compounds and potential mechanisms of scutellariae radix-coptidis rhizoma have been identified in the treatment of AS18. Another study provided new sights into shared molecular mechanisms between osteoporosis and AS based on bioinformatics analysis19. In this study, we evaluated disulfidptosis-related genes in AS based on GSE28829 and GSE43292, two publicly available AS microarray datasets. We identified four disulfidptosis-related genes using two machine learning algorithms, and a diagnostic signature was established to predict development of AS based on these 4 genes. Furthermore, we showed that patients could benefit from clinical decisions made using this diagnostic signature. In addition, we clustered AS samples according to the expression levels of disulfidptosis-related genes and found two different subtypes with different immune characteristics, which suggested that the subtypes of disulfidptosis-related genes may distinguish the degree of progress of AS. Moreover, regulators of disulfidptosis induce significant effects on the immune microenvironment during development of AS. The workflow of the study is shown in Fig. 1.
Fig. 1.
Workflow of the study.
Methods and materials
Data collection and process
The microarray datasets GSE28829 and GSE43292 were downloaded from Gene Expression Omnibus (GEO, https://www.ncbi.nlm.nih.gov/geo/). There are 13 early carotid atherosclerotic plaque specimens and 16 advanced carotid atherosclerotic plaque specimens in the GSE28829 dataset. There are 32 early stage and 32 advanced stage carotid atherosclerotic plaque specimens in the GSE43292 dataset. Based on platform annotation information, we converted the probes into gene symbols and excluded probes containing multiple genes. Furthermore, we removed batch effects using “sva” package in R. Utilizing the empirical Bayesian framework, the comBat function within the “sva” package models the matrix data. After inputting the batch covariates and grouping information, the data after removing the batch effect is obtained.
Identification of disulfidptosis-related DEGs
We applied “limma” package to identify differentially expressed genes (DEGs) in GSE28829 and GSE43292 with adjusted p-value < 0.05 and |log2FC| > 1 set as thresholds based on previous studies20,21. A heatmap was generated to present the DEGs. Fourteen genes related to disulfidptosis were identified from a previous study15. Supplementary Table S1 shows disulfidptosis-related genes, and a heatmap was generated to present the disulfidptosis-related DEGs.
Hub genes screening and verification via GEO
We performed least absolute shrinkage and selection operator (LASSO) regression using the R package “glmnet” and random forest (RF) using the R package “randomForest” to screen for candidate hub genes. The LASSO regression model was trained using ten-fold cross-validation to improve performance, setting the response type to binomial and alpha to 1. The importance of each variable in the Random Forest model was calculated and the Random Forest model was evaluated using five-fold cross-validation. The differentially expressed candidate hub genes were visualized using the R package “ggpubr” with P < 0.05 indicating statistical significance.
In-depth analysis of the efficacy of candidate hub genes was performed using receiver operating characteristic (ROC) with the R package “pROC”. The area under the curve (AUC) was calculated to evaluate the predictive value of the algorithms. The value of AUC ranges from 0 to 1, with higher values indicating better classification performance of the model. An AUC value of 0.7 or more indicates an acceptable level of model performance capability. Two-sided P < 0.05 indicated statistical significance. In addition, we used the “corrplot” package to show the correlation among hub genes.
Verification in vivo
Twenty-four male 8-week-old ApoE-/- mice (Purchased from Jiangsu Jicui Pharmachem Biotechnology Co., Ltd) were housed in an SPF animal laboratory in the Experimental Center of First Affiliated Hospital of Guangzhou University of Chinese Medicine. All animal experiments were conducted in accordance with the guidelines outlined in the National Institutes of Health Guide for the Care and Use of Laboratory Animals. The animals were maintained at a temperature of 23 ± 1 °C under a 12-hour light-dark cycle and were provided with standard chow and water ad libitum. The mice were acclimatized and fed for 7 days, then divided into Control and Model groups (12 mice per group) according to the random number table method. The model group was given a high-fat diet to induce AS (high-fat diet formula: 21% fat, 0.15% cholesterol, 15.5% protein, 62% common feed, purchased from Guangdong Medical Laboratory Animal Center) for 16 weeks22,23. The mice were then placed under anesthesia using sodium pentobarbital (50 mg/kg intraperitoneally). The mice were sacrificed via abdominal aorta exsanguination, blood specimens were collected, and aortic tissue was placed in 4% paraformaldehyde and fixed at room temperature for 48 h.
Blood samples were incubated at room temperature for 1 h, centrifuged at 3,000 rpm for 15 min, and analyzed for serum total cholesterol (Tc), triglycerides (TG), low-density lipoprotein (LDL-c), and high-density lipoprotein (HDL-c) using a fully automated biochemical analyzer (Myriad, DC-220, USA).
For oil red O staining, the isolated aorta were rinsed with distilled water for 1 min, then washed with 60% isopropanol for 30 s. The aortas were then placed in modified oil red O staining solution for 10 min for closed staining, then placed in 60% isopropanol for 1 min, rinsed with distilled water for 1 min, and blotted dry on filter paper. Lipid deposition was assessed by taking pictures under the microscope.
For hematoxylin and eosin (H&E) staining, aortic tissues were fixed in 4% paraformaldehyde, embedded in paraffin, cut into 4-µm sections using a microtome (Leica, Germany), spread into a water bath at 42 ˚C to fully unfold the sections, placed on slides to dry, then placed in an oven at 65 ˚C. The sections were then dewaxed, dehydrated, stained with H&E, neutral resin sealed, and visualized using a microscope.
Total RNA in aortic samples was obtained for PCR assay using a SteadyPure Universal RNA Extraction Kit (AG, China) according to the manufacturer’s instructions. Real time qPCR was performed using a QuantStudio™ 5 System (ABI). Glyceraldehyde phosphate dehydrogenase (GAPDH) was used as an endogenous control. The primer sequences are listed in Table 1.
Table 1.
Primer sequences used for RT-qPCR.
| Gene | Forward | Reverse | |
|---|---|---|---|
| CAPZB | Human | GCACCGCCCATTACAAGTTGAC | GGCTGCCTCCGAGGTTCATG |
| Mouse | AAGTTGACCTCCACGGTGATGC | GGCTGCCTCCCAGGTTCATG | |
| DSTN | Human | TGGGCACCAGAACTAGCACCTC | CACAGGGCATCCTTCAAAGGCTAC |
| Mouse | TGACGCCAGCTTTGAGACCAAG | TTGTTCTGGTGCCCACAGGAAG | |
| MYL6 | Human | CCAGACCGCAGAGTTCAAGGAG | CTCGGCGTTGGTAGGGTTCTG |
| Mouse | CCTAGTCACACTGGGCGAGAAG | TGGCAGGAGGAGAGGTCAGC | |
| PDLIM1 |
Human Mouse |
CGGAGGAAGGGAAGCGTCATC ACCATCGCCACCCTGAATGC |
GCAGGCGAGGCGGTAAAGG ACACTGTGACCACATCGTAGCC |
| GAPDH |
Human Mouse |
CAGGAGGCATTGCTGATGAT GGTTGTCTCCTGCGACTTCA |
GAAGGCTGGGGCTCATTT TGGTCCAGGGTTTCTTACTCC |
Verification in vitro
RAW264.7 cells were cultured in high-sugar Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin in an incubator at 37 ˚C and 5% CO2. RAW264.7 (1 × 106 cells/well) were cultured in 6-well plates and treated with ox-LDL (80 µg/ml) for 24 h to establish a foam cell model24. THP-1 cells were cultured in Roswell Park Memorial Institute (RPMI) 1640 medium containing 10% FBS and 1% penicillin-streptomycin in an incubator at 37 ˚C and 5% CO225. Then, THP-1 (1 × 106 cells/well) were plated in 6-well plates, pre-intervention with PMA (100 ng/ml) for 24 h and stimulated with ox-LDL (40 µg/ml) for 24 h. Total RNA in RAW264.7 and THP-1 cells was collected for PCR assay using a SteadyPure Universal RNA Extraction Kit (AG, China) according to the manufacturer’s instructions. Quantitative PCR was performed using a QuantStudio™ 5 System (ABI). Glyceraldehyde phosphate dehydrogenase was used as an endogenous control. The primer sequences are listed in Table 1.
Nomogram construction
Characteristic genes were incorporated into a nomogram using the “rms” package. A calibration curve was used to evaluate the accuracy of the nomogram. A calibration curve close to 45° indicates strong agreement between the model’s predicted probabilities and the actual observed probabilities. The clinical usefulness of the nomogram was evaluated using the decision curve analysis (DCA) and clinical impact curve (CIC). DCA was used to assess the clinical benefit of the model, with the portion of the model curve above the all and none curves corresponding to the risk threshold on the x-axis having a higher benefit. The CIC provides a more visual representation of the difference between the model’s predicted and actual values at different risk thresholds. The diagnostic ability of the nomogram was evaluated using ROC analysis using the R package “pROC”.
Identification of disulfidptosis subtypes
Consensus clustering with K-means algorithms was used to identify disulfidptosis-related subtypes that correlated with gene expression. We used “Consensus Cluster Plus” package to determine the quantity and robustness of clusters.
Establishment of the disulfidptosis gene signature
Principal component analysis (PCA) was conducted to obtain the disulfidptosis score for individual specimens, which allowed for quantitation of the disulfidptosis subtypes. We used PCA to identify disulfidptosis subgroups, and disulfidptosis score was determined as follows: disulfidptosis score = PC1i, in which PC1 refers to principal component 1 and i to DEG expression.
Evaluation of immune cell infiltration using ssGSEA
Single sample gene set enrichment analysis (ssGSEA) was performed using “GSVA” package in R software to assess infiltration of 28 immune cells between patients with early atherosclerotic plaques and patients with advanced atherosclerotic plaques. The gene set of 28 immune cells used for analysis was obtained from the TISIDB dataset26.
Statistical analysis
SPSS 23.0 software was used for data processing and analysis. Data were normally distributed within the groups and were expressed as the mean ± standard deviation (SD). Intergroup comparisons were analyzed using independent-sample t-tests. P < 0.05 indicated statistically significant differences.
Results
Disulfidptosis-related genes for the progress of AS
GSE28829 and GSE43292 were downloaded from GEO and normalized using “sva” package in R. Disulfidptosis-related genes were extracted from the matrix. Eleven disulfidptosis-related DEGs including ACTN4, CD2AP, CAPZB, DSTN, FLNA, FLNB, MYH10, MYL6, MYH9, PDLIM1, and TLN1 were identified between patients with early atherosclerotic plaques and patients with advanced atherosclerotic plaques (Fig. 2A). The expression levels of the 11 disulfidptosis-related DEGs in each sample were visualized using a heatmap as shown in Fig. 2B. The correlation among the 11 disulfidptosis-related DEGs is presented in Fig. 2C.
Fig. 2.
Disulfidptosis-related DEGs between patients with early atherosclerotic plaques and patients with advanced atherosclerotic plaques in GSE28829 and GSE43292 databases. (A) Differential expression analysis of the 14 disulfidptosis-related genes. (B) Heatmap of 11 disulfidptosis-related DEGs. (C) The correlation among the 11 disulfidptosis-related DEGs.
Candidate hub disulfidptosis-related genes screening
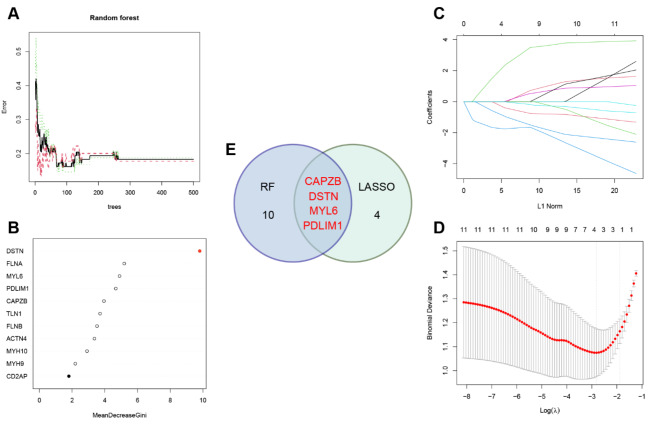
Machine learning was used to identify optimal disulfidptosis-related genes to predict progression of AS. We constructed an RF model to identify optimal disulfidptosis-related genes from the 11 identified DEGs. The error of the model was deemed acceptable based on the overall model error and number of decision trees (Fig. 3A). We ranked the 11 disulfidptosis-related DEGs based on their respective gene importance using the RF model. The results showed that DSTN was a high priority gene (Fig. 3B). Furthermore, LASSO logistic regression was used to identify 4 disulfidptosis-related genes (Fig. 3C-D). Four key disulfidptosis-related genes (CAPZB, DSTN, MYL6, and PDLIM1) were obtained by determining the intersection between the LASSO and RF algorithms, as visualized in a Venn diagram (Fig. 3E).
Fig. 3.
Screening for hub disulfidptosis-related genes using multiple strategies. (A) The influence of the number of decision trees on the error rate. (B) The importance of the 11 disulfidptosis-related DEGs based on the RF model. (C) Ten-fold cross-verification for tuning parameter selection in the LASSO model. Each curve corresponds to a single gene. (D) LASSO coefficient profiling. The dotted vertical line is drawn at the optimal lambda. (E) Venn diagram of LASSO and RF algorithms.
Hub genes verification and enrichment analyses
The expression levels of DSTN (P < 0.001), MYL6 (P < 0.001), and PDLIM1 (P < 0.001) were down-regulated in patients with advanced atherosclerotic plaques, and CAPZB (P < 0.001) was up-regulated in patients with advanced atherosclerotic plaques (Fig. 4A). To validate the diagnostic ability of the candidate hub genes, ROC curves were generated and the AUC values were used to determine the diagnostic efficacy for distinguishing between patients with early atherosclerotic plaques and advanced atherosclerotic plaques. Receiver operating characteristic curves were generated for CAPZB, DSTN, MYL6, and PDLIM1 with AUCs of 0.758, 0.850, 0.753, and 0.759, respectively, which indicated that the hub genes had high diagnostic capability (Fig. 4B). Oil Red O and H&E staining allowed for visualization of plaques. Compared with the Control group, Tc, TG, and LDL-c levels were up-regulated in the Model group, and HDL-c level was downregulated. These results indicated that the AS model was successfully generated (Fig. 4C-E).
Fig. 4.
Hub genes verification. (A) The expression levels of CAPZB, DSTN, MYL6, and PDLIM1 in GSE28829 and GSE43292 datasets. (B) Receiver operating characteristic curves for CAPZB, DSTN, MYL6, and PDLIM1 in GSE28829 and GSE43292 datasets. (C) H&E staining of mouse aorta. (D) Oil red O staining of mouse aorta. (E) Tc, TG, LDL-c and HDL-c levels in ApoE−/− mice. (F) The expression levels of CAPZB, DSTN, MYL6, and PDLIM1 in ApoE−/− mice. (G) The expression levels of CAPZB, DSTN, MYL6, and PDLIM1 in a foam cell model derived from THP-1 cells. (H) The expression levels of CAPZB, DSTN, MYL6, and PDLIM1 in a foam cell model derived from RAW264.7 cells.
We then investigated total mRNA expression levels of hub genes in aortic tissue (Fig. 4F). Then, we constructed a foam cell model in RAW264.7 and THP-1 cells to further evaluate the expression levels of hub genes in vitro (Fig. 4G-H). The results showed that the expression trends of hub genes were consistent across the different samples. Compared with the Control group, the expression level of CAPZB was significantly upregulated in the Model group. In addition, the expression levels of DSTN, MYL6, and PDLIM1 were downregulated in the Model group, which was consistent with the trends in the sequencing results. These results further supported our hub genes as potential diagnostic and therapeutic markers.
Nomogram of hub genes construction
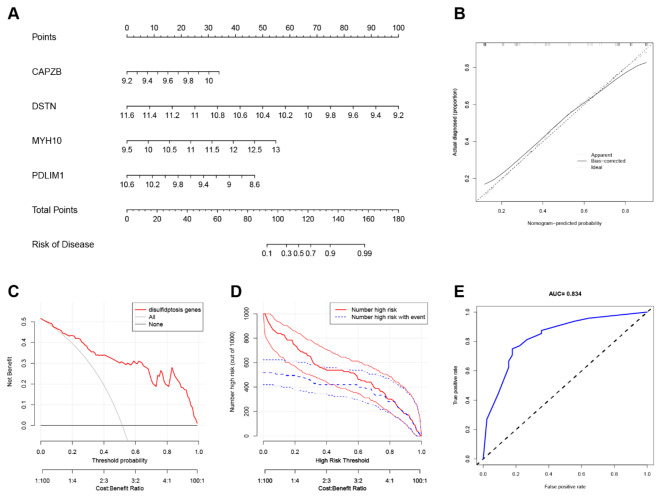
A nomogram constructed from AS diagnostic genes was used as a diagnostic tool for AS (Fig. 5A). In the nomogram, each characteristic gene corresponded to a score, and the total score was calculated by adding the scores for all diagnostic genes. The total points corresponded to different risks of AS. The calibration curve showed that the nomogram enabled an accurate estimate of the progression of AS (Fig. 5B). As shown in the DCA, patients diagnosed with AS could clinically benefit from the nomogram (Fig. 5C). The clinical effectiveness of the nomogram was demonstrated by the CIC. The number of subjects who were at a high risk (the number of positive cases predicted by the nomogram) matched strongly with the number of subjects with high risk outcomes (the number of true-positive cases) when the threshold probability was above 80% (Fig. 5D). In addition, ROC curves were constructed and AUC values were used to determine the accuracy of nomogram model (Fig. 5E).
Fig. 5.
Nomogram construction (A) Nomogram for prediction of MI. (B) Calibration curve analysis of the nomogram. The dashed line represents perfect prediction. The dotted line represents apparent estimates of predicted vs. observed values, and the solid line (bias) shows the corrected estimates. (C) The DCA of the proposed nomogram for predicting MI in the training datasets. (D) The CIC curve of the proposed nomogram. The red curve indicates the number of people classified as positive (high risk) by the prediction model at each threshold probability. The blue curve is the number of true positives at each threshold probability. (E) ROC curves showing the accuracy of the nomogram.
Analysis of disulfidptosis subtypes
We performed a consensus clustering algorithm to identify different subtypes using disulfidptosis-related DEGs. The optimal number of subtypes was 2, including cluster A and cluster B, which was determined using a consensus matrix plot and a CDF plot (Fig. 6A-C). Then, we found that the expression levels of ACTN4, DSTN, FLNA, MYH10, MYL6, MYH9, PDLIM1, and TLN1 in cluster A were lower than those in cluster B. In addition, CD2AP, CAPZB, and FLNB did not differ significantly between the two clusters (Fig. 6D-E). Principal component analysis showed that the 11 disulfidptosis-related genes were able to classify the two subtypes (Fig. 6F).
Fig. 6.
Consensus clustering of the 11 significant disulfidptosis-related genes in GSE28829 and GSE43292 datasets. (A) Consensus matrix of the 11 significant disulfidptosis-related genes when k = 2. (B) Consensus CDF when k = 2–9. (C) Relative alterations in the area under the CDF curve. (D) Expression of the 11 significant disulfidptosis-related genes in disulfidptosis cluster A and cluster B via heatmap. (E )Differential expression of the 11 significant disulfidptosis-related genes in disulfidptosis cluster A and cluster B. (F) Principal component analysis of the expression data of the 11 significant disulfidptosis-related genes showed differences between the transcriptomes of the two disulfidptosis subtypes.
Analysis of Immune cell infiltration
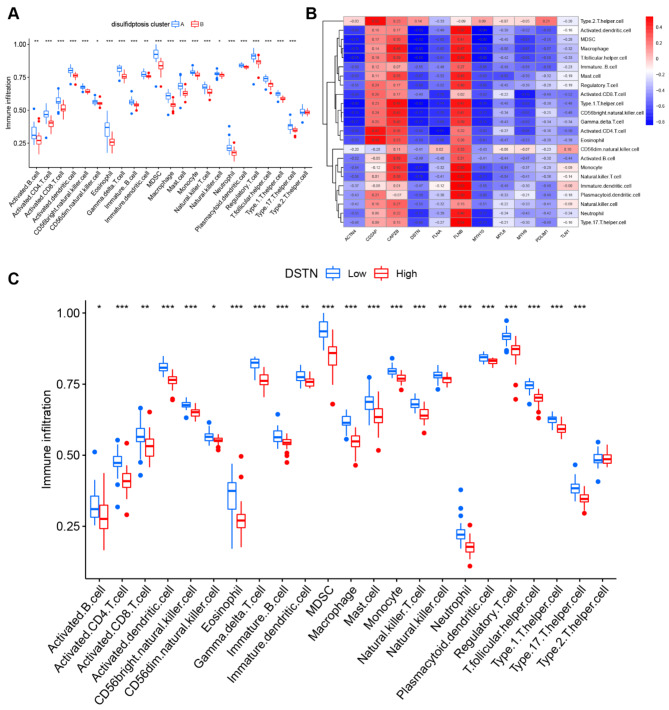
The ssGSEA algorithm was used to analyze immune cell infiltration between patients with early atherosclerotic plaques and patients with advanced atherosclerotic plaques, and to evaluate the relationship between immune cells and disulfidptosis-related genes (Fig. 7A). The results showed enrichment of immune cells in patients with high- or low-disulfidptosis-related genes, including the 4 key disulfidptosis-related genes (CAPZB, DSTN, MYL6, and PDLIM1), as shown in Fig. 7B. Our findings showed that patients with high CAPZB expression had higher levels of immune cells, which indicated this may be a therapeutic target for AS. Patients with low DSTN, MYL6, and PDLIM1 expression had increased numbers of immune cells, which protect against progression of AS. Furthermore, we explored differential immune cell enrichment between disulfidptosis cluster A and cluster B. We found that cluster B displayed greater infiltration of immune cells than that observed in cluster A, which indicated that patients in cluster B may have a positive immune response for progression of AS (Fig. 7C).
Fig. 7.
Immune cell infiltration analysis. (A) Heatmap of correlations between immune cells and the 11 significant disulfidptosis-related genes. (B) Difference in the abundance of infiltrating immune cells between the high and low expression groups, including CAPZB, DSTN, MYL6, and PDLIM1. (C) Differential immune cell infiltration between disulfidptosis cluster A and cluster B.
Evaluation of the disulfidptosis gene signature
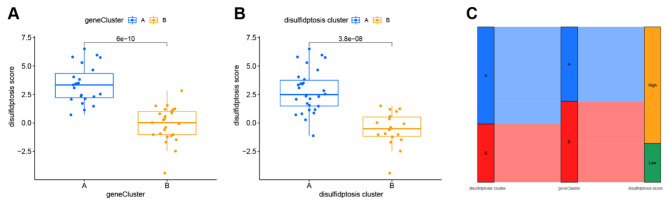
A consensus clustering algorithm was used to determine the two disulfidptosis subtypes. Patients were divided into different gene subgroups based on 47 disulfidptosis-related DEGs. The optimal number of subtypes was 2 (cluster A and cluster B), which was determined using a consensus matrix plot and a CDF plot (Fig. 8A-C). We observed that two gene subtypes agreed with the disulfidptosis subtypes. The differential expression of the 47 disulfidptosis-related DEGs is shown in Fig. 8D. In addition, the differential expression of the 11 disulfidptosis-related genes and immune cells between different gene clusters showed similar to results to those observed between the disulfidptosis subtypes (Fig. 8E-F), which verified the disulfidptosis clustering algorithm. Furthermore, PCA was used to obtain disulfidptosis scores to quantify the disulfidptosis subtypes. We compared the disulfidptosis scores in the disulfidptosis clusters and gene clusters. The results showed that the disulfidptosis score in cluster A was significantly higher than that in cluster B in the disulfidptosis and gene clusters (Fig. 9A-B). The correlation between the disulfidptosis cluster, disulfidptosis gene clusters, and disulfidptosis scores are shown in Fig. 9C.
Fig. 8.
Consensus clustering of the 47 disulfidptosis-related DEGs in GSE28829 and GSE43292 datasets. (A) Consensus matrix of the 47 m6A-related DEGs when k = 2. (B) Consensus CDF when k = 2–9. (C) Relative alterations in the area under the CDF curve. (D) Expression of the 47 disulfidptosis-related DEGs in gene cluster A and cluster B using a heatmap. (E) Differential expression of the 11 significant disulfidptosis-related genes in gene cluster A and cluster B. (F) Differential enrichment of immune cells between gene cluster A and cluster B.
Fig. 9.
Correlation between disulfidptosis cluster, gene cluster, and disulfidptosis score. (A) Differences in disulfidptosis scores between disulfidptosis cluster A and cluster B. (B) Differences in disulfidptosis score between gene cluster A and cluster B. (C) Sankey diagram of the relationship between disulfidptosis cluster, gene cluster, and disulfidptosis score.
Discussion
The view of linear progression of AS driven by inflammation and lipids may underestimate the complexity of AS etiology. Although focused therapeutic strategies have shown some success, they remain insufficient27. Recent studies have shown that programmed cell death regulates the inflammatory response, oxidative stress, and abnormalities in lipid metabolism in development of AS, which may allow for identification of novel therapeutic targets28,29. The role of disulfidptosis and its related regulators has not been evaluated in AS. In this study, several machine learning techniques were used to investigate the transcriptomes and immune infiltration of genes related to disulfidptosis. Furthermore, potential early AS biomarkers were identified and evaluated. An AS risk prediction model was built based on key biomarkers. According to the disulfidptosis-related DEGs, patients with advanced AS were separated into two clusters for analysis of differences in immune infiltration, which may help to determine immunotherapy targets.
We showed that most of the disulfidptosis-related DEGs were closely associated with either early or advanced plaques, which indicated that disulfidptosis may play a significant role in progression of AS. We then selected the most promising potential biomarkers from the 11 DEGs using a machine learning technique. We used random attribute selection in the decision tree training process. The RF algorithm, which is the most representative algorithm in Bagging ensemble learning, is a potent tool for screening the importance ranking of variables30. The LASSO regression model can compress the variable coefficients for choosing the crucial variables to solve the regression problem in machine learning31. Confounding factors were avoided by merging the two methods, and CAPZB, DSTN, MYL6, and PDLIM1 were selected as potential AS biomarkers. The findings of our investigation are consistent with those from previous studies. For example, DSTN, which is primarily expressed in vascular smooth muscle cells, has been reported as a therapeutic target for slowing the progression of AS by controlling the phenotypic differentiation and migration of smooth muscle cells32. When compared with ApoE−/− mice, miR150 and ApoE double knockout mice showed more stable atherosclerotic plaques and lower inflammation levels through increased PDLIM1 expression, while PDLIM1 knockdown reversed these anti-atherosclerotic effects33. Although the precise functions of CAPZB and MYL6 in AS have not been reported, the present study showed the intrinsic relationship between these genes and atherosclerotic disease. The cytoskeletal protein CAPZB is significantly differentially expressed before and after targeted drug treatment in Alzheimer’s disease34, and it has also been identified as a genetically sensitive gene that affects circulating thyroxine35,36. In addition, a Mendelian randomization study combined with experimental validation showed that MYL6 is a hallmark gene for obesity and obesity-related disorders37. Interestingly, anti-MYL6 antibody has been shown to alleviate polyangiitis-related multi-system damage38. Our results provide a theoretical reference for further understanding the pathophysiological mechanism of disulfidptosis in AS and resulted in identification of four novel biomarkers.
According to our results, the expression levels of DSTN, MYL6, and PDLIM1 were significantly downregulated, and the expression of CAPZB was upregulated, in advanced AS compared with those in early AS. Increased expression levels of DSTN and PDLIM1 may be associated with slowed progression of AS33. In addition, ROC curves showed excellent diagnostic value for all four genes. To support this finding, we performed validation in vitro and in vivo. The results agreed with our findings regarding the expression levels of these genes. Therefore, we identified CAPZB, DSTN, MYL6, and PDLIM1 as the signature genes for plaque stability, and we used the expression levels of these genes to generate a nomogram for predicting the risk of AS. Nomograms are preferred over conventional disease prediction models due to their convenience for clinical applications. Nomograms are frequently used in risk assessment, clinical judgment, and prognosis assessment for tumor and non-tumor illnesses39. Based on the four key disulfidptosis-related genes, we built a risk prediction model that was calibrated for clinical decision-making. However, the nomogram’s performance does not necessarily indicate the accuracy of illness risk prediction, and more testing is required to determine the nomogram’s benefits in clinical practice40. However, we developed the first practical AS risk prediction model for genes related to disulfidptosis, which may be beneficial for determination of a causal connection between disulfidptosis and AS.
Previous studies have shown that an imbalance in the immune response and promotion of chronic inflammation in plaque formation is mediated by synergistic interactions between various immune cells and crosstalk between immune cells and lipid metabolism41,42. At different stages, the immune response either promotes or inhibits AS, and determination of the mechanisms of this functional dichotomy may aid in creation of new immunotherapies43. In addition, it has become increasingly clear that cell death, and the synergistic immunological response it triggers, plays a role in AS progression44. Although more evidence is needed, crosstalk between cell death and immunity will allow for better understanding of the mechanisms of development of AS. To further explore the potential role of disulfidptosis-related genes in AS, we divided AS samples into two subgroups based on differential expression of 11 genes, then thoroughly analyzed infiltration of 28 immune cells between the two clusters. The results showed that cluster A had significantly greater immune cell densities than cluster B, with the exception of T helper 2 cells. Furthermore, low levels of DSTN have been shown to increase inflammatory factors and increase immune cell aggregation45. Therefore, we believe that atherosclerotic plaques with decreased DSTN expression may exhibit an enhanced immune response. Based on all DEGs, patients with AS were then divided into two subgroups, and patients with significantly worse immune responses exhibited relatively high levels of disulfidptosis-related gene expression. Therefore, our study was the first to assess the relationship between immune cell infiltration and disulfidptosis-related gene expression, which may inform development of immunotherapies for AS.
Bioinformatics analysis of immune cell infiltration showed that myeloid-derived suppressor cells (MDSC) and regulatory T cells (Tregs) differed significantly between the AS subtypes. Myeloid-derived suppressor cells are associated with immunosuppression, and studies have focused on biological functions of MDSC due to the lack of an established phenotype, particularly in the field of tumor immunotherapy46. Recent studies of MDSCs have challenged MDSC cell types, whether they are anti-inflammatory or pro-inflammatory, and potential phenotypes. An interesting area of research has focused on the pathophysiological connection between MDSCs and non-tumor disorders47. Atherosclerosis model mice were shown to have higher levels of polymorphonuclear myeloid-derived suppressor cells (PMN-MDSC) than control mice, which may be involved in development of neutrophil extracellular traps to exacerbate AS. In addition, Tregs buildup in atherosclerotic plaques can promote macrophage M2 polarization and produce inflammatory regulators to induce atherosclerotic plaque regression48. In contrast, AS development and dyslipidemia have been shown to accelerate in response to Treg depletion49. Examination of differential immune cell infiltration showed the immune heterogeneity of atherosclerotic plaques and showed the importance of disulfidptosis-related genes in immune control.
Our study was subject to the following limitations, either. First, although minority disulfidptosis markers can be directly evaluated by specific inhibitors or co-staining, this method is challenging in large-scale AS patient samples. Similarly, the sample sizes of the AS datasets are relatively small, and it is challenging to maintain consistent plaque samples, which may have negatively impacted this study. Second, as disulfidptosis is a recent research advancement, constructing appropriate mouse or cell models for functional experiments could been further established and validated. Although the disulfidptosis-related genes included in the study were validated in vitro using multiple bioinformatics algorithms, future studies in vivo and clinical trials are required for verification. Therefore, more work is needed to precisely define the role of disulfidptosis in AS.
Conclusions
The model constructed using CAPZB, DSTN, MYL6, and PDLIM1 had promising diagnostic capability for AS. In addition, our study identified two disulfidptosis subtypes and cluster B may have a positive immune response for progression of AS. These findings may contribute to better treatment of AS. However, additional clinical samples and additional basic experiments are needed to verify the AS diagnostic model in this study.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
Thank the GEO database for providing the data.
Author contributions
Wei Wu designed the study. Huanyi Zhao carried out the data acquisition and bioinformatics analysis. Zheng Jin drafted the manuscript. Junfeng Fang analysed and prepared the charts. Junlong Li contributed to the writing of the manuscript. All authors read and approved the final manuscript and agreed to the publication of this study.
Funding
This study was supported by grants from the National Natural Science Foundation of China (No. 82204984, 82204987).
Data availability
The datasets in our study are available in the GEO database (https:// www. ncbi. nlm. nih. gov/ geo).
Declarations
Ethics approval and consent to participate
This study was approved by the Ethics Committee of the First Affiliated Hospital of Guangzhou University of Chinese Medicine (No.GZTCMF1-2022016). All the methods are reported in accordance with ARRIVE guidelines.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Ethical Statement
This study is performed in accordance with relevant guidelines and regulations. All methods are reported in accordance with ARRIVE guidelines.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Wei Wu, Email: wwhrlz@163.com.
J. F. Fang, Email: gzyyfyfjf@163.com
References
- 1.Legein, B. et al. Inflammation and immune system interactions in atherosclerosis. Cell. Mol. Life Sci.70(20), 3847–3869 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roth, G. A. et al. Global Burden of Cardiovascular diseases and Risk factors, 1990–2019: Update from the GBD 2019 study. J. Am. Coll. Cardiol.76(25), 2982–3021 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Björkegren, J. L. M. & Lusis, A. J. Atherosclerosis: recent developments. Cell. 185(10), 1630–1645 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Libby, P. Inflammation in Atherosclerosis-No longer a theory. Clin. Chem.67(1), 131–142 (2021). [DOI] [PubMed] [Google Scholar]
- 5.Weber, C. & Noels, H. Atherosclerosis: current pathogenesis and therapeutic options. Nat. Med.17(11), 1410–1422 (2011). [DOI] [PubMed] [Google Scholar]
- 6.Visseren, F. L. J. et al. 2021 ESC guidelines on cardiovascular disease prevention in clinical practice. Eur. Heart J.42(34), 3227–3337 (2021). [DOI] [PubMed] [Google Scholar]
- 7.Riaz, H. et al. Residual inflammatory risk after contemporary lipid lowering therapy. Eur. Heart J. Qual. Care Clin. Outcomes. 6(2), 105–111 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu, X. et al. Actin cytoskeleton vulnerability to disulfide stress mediates disulfidptosis. Nat. Cell. Biol.25(3), 404–414 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tang, D. et al. The molecular machinery of regulated cell death. Cell. Res.29(5), 347–364 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peng, F. et al. Regulated cell death (RCD) in cancer: key pathways and targeted therapies. Signal. Transduct. Target. Ther.7(1), 286 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tian, Q. et al. Inhibition of CCR2 attenuates neuroinflammation and neuronal apoptosis after subarachnoid hemorrhage through the PI3K/Akt pathway. J. Neuroinflammation. 19(1), 312 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu, M. et al. STAT3 signaling promotes cardiac injury by upregulating NCOA4-mediated ferritinophagy and ferroptosis in high-fat-diet fed mice. Free Radic Biol. Med.201, 111–125 (2023). [DOI] [PubMed] [Google Scholar]
- 13.Chen, X. et al. Copper homeostasis and copper-induced cell death in the pathogenesis of cardiovascular disease and therapeutic strategies. Cell. Death Dis.14(2), 105 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen, L., Min, J. & Wang, F. Copper homeostasis and cuproptosis in health and disease. Signal. Transduct. Target. Ther.7(1), 378 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Machesky, L. M. Deadly actin collapse by disulfidptosis. Nat. Cell. Biol.25(3), 375–376 (2023). [DOI] [PubMed] [Google Scholar]
- 16.Oulas, A. et al. Systems Bioinformatics: Increasing precision of computational diagnostics and therapeutics through network-based approaches. Brief. Bioinform. 20(3), 806–824 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu, C. et al. Identification of crucial genes for predicting the risk of atherosclerosis with system lupus erythematosus based on comprehensive bioinformatics analysis and machine learning. Comput. Biol. Med.152, 106388 (2023). [DOI] [PubMed] [Google Scholar]
- 18.Ji, L. et al. Identification of bioactive compounds and potential mechanisms of scutellariae radix-coptidis rhizoma in the treatment of atherosclerosis by integrating network pharmacology and experimental validation. Biomed. Pharmacother. 165, 115210 (2023). [DOI] [PubMed] [Google Scholar]
- 19.Mo, L. et al. Integrated Bioinformatic Analysis of the Shared Molecular mechanisms between osteoporosis and atherosclerosis. Front. Endocrinol. (Lausanne). 13, 950030 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang, X. et al. Identification of a novel immune-related transcriptional regulatory network in Sarcopenia. BMC Geriatr.23(1), 463 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Song, Y. et al. Screening and validation of atherosclerosis PAN-apoptotic immune-related genes based on single-cell sequencing. Front. Immunol.15, 1297298 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Getz, G. S. & Reardon, C. A. Animal models of atherosclerosis. Arterioscler. Thromb. Vasc Biol.32(5), 1104–1115 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao, Y. et al. Small rodent models of atherosclerosis. Biomed. Pharmacother. 129, 110426 (2020). [DOI] [PubMed] [Google Scholar]
- 24.Wang, N. et al. Combination of tanshinone IIA and astragaloside IV attenuate atherosclerotic plaque vulnerability in ApoE(-/-) mice by activating PI3K/AKT signaling and suppressing TRL4/NF-κB signaling. Biomed. Pharmacother. 123, 109729 (2020). [DOI] [PubMed] [Google Scholar]
- 25.Shen, S. et al. The exoprotein gbp of Fusobacterium nucleatum promotes THP-1 cell lipid deposition by binding to CypA and activating PI3K-AKT/MAPK/NF-κB pathways. J. Adv. Res.57, 93–105 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ru, B. et al. TISIDB: an integrated repository portal for tumor-immune system interactions. Bioinformatics. 35(20), 4200–4202 (2019). [DOI] [PubMed] [Google Scholar]
- 27.Libby, P. Inflammation during the life cycle of the atherosclerotic plaque. Cardiovasc. Res.117(13), 2525–2536 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Puylaert, P. et al. Regulated necrosis in atherosclerosis. Arterioscler. Thromb. Vasc Biol.42(11), 1283–1306 (2022). [DOI] [PubMed] [Google Scholar]
- 29.Li, M. et al. Programmed cell death in atherosclerosis and vascular calcification. Cell. Death Dis.13(5), 467 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hu, J. & Szymczak, S. A review on longitudinal data analysis with random forest. Brief. Bioinform, 24(2). (2023). [DOI] [PMC free article] [PubMed]
- 31.Tibshirani, R. The lasso method for variable selection in the Cox model. Stat. Med.16(4), 385–395 (1997). [DOI] [PubMed] [Google Scholar]
- 32.Liao, K. A. et al. The actin depolymerizing factor destrin serves as a negative feedback inhibitor of smooth muscle cell differentiation. Am. J. Physiol. Heart Circ. Physiol.321(5), H893–h904 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gong, F. H. et al. Reduced atherosclerosis lesion size, inflammatory response in miR-150 knockout mice via macrophage effects. J. Lipid Res.59(4), 658–669 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bastrup, J. et al. Anti-Aβ antibody Aducanumab regulates the Proteome of Senile plaques and closely surrounding tissue in a transgenic mouse model of Alzheimer’s Disease. J. Alzheimers Dis.79(1), 249–265 (2021). [DOI] [PubMed] [Google Scholar]
- 35.Panicker, V. et al. A locus on chromosome 1p36 is associated with thyrotropin and thyroid function as identified by genome-wide association study. Am. J. Hum. Genet.87(3), 430–435 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhan, M. et al. Genome-wide association study identifies a novel susceptibility gene for serum TSH levels in Chinese populations. Hum. Mol. Genet.23(20), 5505–5517 (2014). [DOI] [PubMed] [Google Scholar]
- 37.Zhu, Z. et al. Shared genetic and experimental links between obesity-related traits and asthma subtypes in UK Biobank. J. Allergy Clin. Immunol.145(2), 537–549 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yoshinari, M. et al. Low disease activity of microscopic polyangiitis in patients with anti-myosin light chain 6 antibody that disrupts actin rearrangement necessary for neutrophil extracellular trap formation. Arthritis Res. Ther.24(1), 274 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang, X. et al. From past to future: bibliometric analysis of global research productivity on nomogram (2000–2021). Front. Public. Health. 10, 997713 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Balachandran, V. P. et al. Nomograms in oncology: more than meets the eye. Lancet Oncol.16(4), e173–e180 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roy, P., Orecchioni, M. & Ley, K. How the immune system shapes atherosclerosis: roles of innate and adaptive immunity. Nat. Rev. Immunol.22(4), 251–265 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schaftenaar, F. et al. Atherosclerosis: the interplay between lipids and immune cells. Curr. Opin. Lipidol.27(3), 209–215 (2016). [DOI] [PubMed] [Google Scholar]
- 43.Wolf, D. & Ley, K. Immunity and inflammation in atherosclerosis. Circ. Res.124(2), 315–327 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rayner, K. J. Cell death in the Vessel Wall: the Good, the bad, the Ugly. Arterioscler. Thromb. Vasc Biol.37(7), e75–e81 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Verdoni, A. M. et al. Defects in actin dynamics lead to an autoinflammatory condition through the upregulation of CXCL5. PLoS One. 3(7), e2701 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hegde, S., Leader, A. M. & Merad, M. MDSC: markers, development, states, and unaddressed complexity. Immunity. 54(5), 875–884 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yaseen, M. M. et al. Recent advances in myeloid-derived suppressor cell biology. Front. Med.15(2), 232–251 (2021). [DOI] [PubMed] [Google Scholar]
- 48.Sharma, M. et al. Regulatory T cells license macrophage pro-resolving functions during atherosclerosis regression. Circ. Res.127(3), 335–353 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Klingenberg, R. et al. Depletion of FOXP3 + regulatory T cells promotes hypercholesterolemia and atherosclerosis. J. Clin. Invest.123(3), 1323–1334 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets in our study are available in the GEO database (https:// www. ncbi. nlm. nih. gov/ geo).