Abstract
Background
Type 1 diabetes, an autoimmune disorder leading to the destruction of pancreatic β-cells, requires lifelong insulin therapy. Islet transplantation offers a promising solution but faces challenges such as limited availability and the need for immunosuppression. Induced pluripotent stem cells (iPSCs) provide a potential alternative source of functional β-cells and have the capability for large-scale production. However, current differentiation protocols, predominantly conducted in hybrid or 2D settings, lack scalability and optimal conditions for suspension culture.
Methods
We examined a range of bioreactor scaleup process parameters and quality target product profiles that might affect the differentiation process. This investigation was conducted using an optimized High Dimensional Design of Experiments (HD-DoE) protocol designed for scalability and implemented in 0.5L (PBS-0.5 Mini) vertical wheel bioreactors.
Results
A three stage suspension manufacturing process is developed, transitioning from adherent to suspension culture, with TB2 media supporting iPSC growth during scaling. Stage-wise optimization approaches and extended differentiation times are used to enhance marker expression and maturation of iPSC-derived islet-like clusters. Continuous bioreactor runs were used to study nutrient and growth limitations and impact on differentiation. The continuous bioreactors were compared to a Control media change bioreactor showing metabolic shifts and a more β-cell-like differentiation profile. Cryopreserved aggregates harvested from the runs were recovered and showed maintenance of viability and insulin secretion capacity post-recovery, indicating their potential for storage and future transplantation therapies.
Conclusion
This study demonstrated that stage time increase and limited media replenishing with lactate accumulation can increase the differentiation capacity of insulin producing cells cultured in a large-scale suspension environment.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13287-024-03973-0.
Keywords: Diabetes, Human induced pluripotent stem cell, Insulin producing cells, Bioreactor, DoE, β-cells, Pancreatic cells, Bioprocess development, Optimization, Islets, iPSCs
Background
Type I Diabetes Mellitus (T1DM) is a chronic autoimmune destruction disorder of the pancreatic β-cells. Latest data collected shows it affected ~ 8.7 million patients worldwide and is expected to rise to 17.4 million in the next two decades with a projected health expenditures of 1,054 billion USD annually [1–3]. Life-long insulin therapy is needed for patients suffering from this disease. Islet transplantation that replaces lost insulin secreting cells is a promising therapy solution [4, 5]. However, it has been tempered by the lack of islets available and the requirement of life-long immunosuppression. This has led to the investigation of iPSCs as an available source of functional β-cells [6]. Cell therapies derived from iPSCs hold promise in treating a variety of clinical indications [7]. They have the ability of replacing damaged or lost cells and are capable of high in vitro proliferation and differentiation into all three germ layers [8, 9]. This makes them ideal for applications that require a large sustainable source of clinical grade cells. To meet the required clinical cell dosage, a large-batch manufacturing system must be established. Thus, a scaleup from a planar adherent culture to a 3D culture that generates aggregates is required. However, most research and established differentiation protocols are done in a 2D environment that lack suspension environment factors and considerations [10, 11].
Adherent culture methods are impractical for scaling due to the limitation of surface area. To increase production beyond these constraints, suspension culture becomes necessary. Recent research indicates that a bioreactor scale-up approach can effectively expand and differentiate iPSCs while reducing the need for manual interventions in static cultures [11–17]. Already established differentiation protocols were conducted side by side in adherent and suspension environments. The results indicated that the cells produced from both methods are different. In our novel HD-DoE protocol for pancreatic β-cell differentiation optimized on adherent plates [18], optimal conditions required BMP antagonism and retinoid input, resulting in the induction of dorsal foregut endoderm (DFE). The study demonstrated that pancreatic identity can be rapidly and robustly induced from DFE, and these cells exhibit a dorsal pancreatic identity [18].
Other published methods for differentiating human iPSCs into β-cells involve a series of developmental stages, including definitive endoderm, primitive gut tube, pancreatic progenitor, endocrine progenitor, and insulin-producing β-cells [19]. These methods typically entail the sequential addition of growth factors and small molecules to mimic pancreatic development [18, 20–29]. The initial step in differentiating iPSCs into pancreatic cells often begins with definitive endoderm commitment. D'Amour et al. established an efficient method using activin A and low serum to direct up to 80% of iPSCs towards the definitive endoderm lineage [30]. Subsequent protocols have further specified and differentiated iPSCs into PDX1 and NKX6.1 positive pancreatic progenitors, leading to the generation of β-cells [21, 31, 32]. For instance, Rezania et al. and Pagliuca et al. successfully generated iPSC-derived β-cells with varying functionality [20, 23]. These cells, while exhibiting reduced glucose-stimulated insulin secretion (GSIS) functionality compared to human islets, have shown promise in reversing diabetes in diabetic mice post-transplant.
Recent advancements have led to the development of methods capable of producing cells with more robust and dynamic GSIS [22, 33, 34]. Nair et al. demonstrated that isolating INS positive cells at an early immature stage followed by reaggregation into clusters enhances β-cell maturation, although sustained second-phase response remains a challenge [19]. Velazco-Cruz et al. reported that selective modulation of transforming growth factor-β signaling, combined with resizing of cell clusters, improves dynamic GSIS with a more sustained second-phase response [34].
While GSIS functionality is of great importance, in vivo transplantation has been the most favorable method for gaining full β-cell maturation and function. A clinical trial presented at the 83rd Scientific Sessions of the American Diabetes Association showcased VX-880, a stem cell-derived islet cell therapy, as a potential treatment for T1DM. Results demonstrated restored insulin secretion, improved glycemic control, and elimination of severe hypoglycemic events in all six treated patients [35].
Despite these advancements, a significant limitation of current differentiation methods is the scalability recovery yield and the use of serum-containing or xeno-containing components, which may pose translational challenges in the future. Efforts to address these limitations are crucial for the clinical development of iPSC-derived β-cells.
Methods
Differentiation of human induced pluripotent stem cell culture
Throughout this study two iPSC lines were used the RCP5005N (Reprocell # 771-3G) and the NCRM-1 (iXCell #CR0000001). Cells were maintained at 37 °C and 5% CO2. Prior to the experiment, cells were grown to 70–80% confluency on vitronectin (ThermoFisher, USA #A14700) coated flasks. Cells were dissociated with TrypLE (ThermoFisher, USA 12605010) for 3 min at 37 °C, resuspended in E8 medium (ThermoFisher, USA #A1517001), transferred to 50 ml conical tubes, and centrifuged at 400 × g for 6 min. The pellet was resuspended in E8 and 10 µM Y-27632 Rho-Kinase (ROCK) inhibitor (Peprotech, USA #1293823). 90 million cells were seeded in a 500 ml PBS vertical wheel bioreactor (PBS Biotech, USA #FA-0.5-D-001) with the TB10 media (Table 1). After 1 day of culture, 50% of the media was replenished. After an additional day, differentiation was started using the differentiation medium as indicated in (Table 1). All reagents used are listed in (Table 2).
Table 1.
Composition and concentration of basal medium and differentiation factors used
| TB10 | TB2 | Stage 1 | Stage 2 | Stage 3 | Concentration | |
|---|---|---|---|---|---|---|
| Basal Media | E8™ Medium | IMDM/F12 (1:1) | IMDM/F12 (1:1) | IMDM/F12 (1:1) | IMDM/F12 (1:1) | |
| Additives | E8 Supplement | 1X | ||||
| Pen/Strep | Pen/Strep | Pen/Strep | Pen/Strep | Pen/Strep | 1X | |
| PEG | PEG | PEG | PEG | PEG | 1% w/v | |
| Heparin sodium Heparin Sodium | Heparin Sodium | Heparin Sodium | Heparin Sodium | |||
| salt | Salt | Salt | Salt | Salt | 1 mg/ml | |
| Albumax | Albumax | Albumax | Albumax | 1X | ||
| Ethanolamine | Ethanolamine | Ethanolamine | Ethanolamine | 10 μM | ||
| Monothioglycerol | Monothioglycerol | Monothioglycerol | Monothioglycerol | |||
| Cholesterol | Cholesterol | Cholesterol | Cholesterol | 150 μM | ||
| Sulfate | Sulfate | Sulfate | Sulfate | 409 nM | ||
| Trolox | Trolox | Trolox | Trolox | 10 μM | ||
| Linoleic Acid | Linoleic Acid | Linoleic Acid | Linoleic Acid | 14.26uM | ||
| Oleic Acid | Oleic Acid | Oleic Acid | Oleic Acid | 10 μM | ||
| Insulin | Insulin | Insulin | Insulin | 10 μg/ml | ||
| Holo-Transferrin | Holo-Transferrin | Holo-Transferrin | Holo-Transferrin | 5 μg/ml | ||
| Differentiation | Retinoic Acid | Retinoic Acid | 2 μM | |||
| Reagents | LDN193189 | LDN193190 | 250 nM | |||
| PD0325 | 250 nM | |||||
| A8301 | A8301 | A8301 | 500 nM | |||
| (5Z)-7-Oxozeaenol | 500 nM | |||||
| SUN11602 | 300 nM | |||||
| AT7867 | 250 nM | |||||
| g-XX | 100 nM | |||||
Table 2.
List of reagents along with cell lines used in the protocol
| Compound | Supplier | Catalogue Number |
|---|---|---|
| Trolox | cayman chemical co | 10,011,659 |
| Accutase | Millipore Sigma | A6964 |
| Gamma-Secretase | Millipore Sigma | 565789 |
| Insulin | Millipore Sigma | 11376497001 |
| Holo transferrin | Millipore Sigma | T0665-500MG |
| Ethanolamine | Millipore Sigma | E0135-100ML |
| Linoleic acid | Millipore Sigma | L1012-1G |
| Oleic acid | Millipore Sigma | O1383-5G |
| Cholesterol Sulfate | Millipore Sigma | C9523-25MG |
| All-trans retinoic acid | Millipore Sigma | R2625-50MG |
| PD0325901 | Millipore Sigma | PZ0162-5MG |
| Fluorescein diacetate | Millipore Sigma | F7378-5G |
| Propidium iodide | Millipore Sigma | P4170 |
| Polyethylene Glycol (PEG) | Millipore Sigma | 89510 |
| Y-27632 (ROCK inhibitor) | Peprotech | 1293823-1 mg |
| LDN193189 | Peprotech | 1066208-5MG |
| A8301 | Peprotech | 9094360-10MG |
| AT7867 | Selleck Chem | S1558 |
| SUN11602 | Selleck Chem | S8192 |
| Glucose solution | Thermofisher Scientific | A2494001 |
| Albumax | Thermofisher Scientific | 11020021 |
| CryoStor® CS10 | STEMCELL Technologies | 100-1061 |
| REAGENTS | ||
| Essential 8™ Flex | ThermoFisher Scientific | A2858501 |
| Vitronectin | ThermoFisher Scientific | A31804 |
| DPBS (without calcium and magnesium) | ThermoFisher Scientific | 14190144 |
| TrypLE | ThermoFisher Scientific | 12563-029 |
| CELL LINES | ||
| RCRP5005N* | REPROCELL | StemRNA™ Human iPSC 771-3G |
| NCRM-1** | iXCells | CR0000001 |
* Established from endothelial progenitor cells from peripheral blood. Reprogrammed with the Stemgent StemRNA 3rd Gen Reprogramming technology
** Established from CD34 + cord blood cells. Reprogrammed with Episomal plasmid method
Cell counting, aggregate size and islet equivalency (IEQ) estimates
Bioreactor samples were taken after mixing using a pipette to avoid sampling bias and gradient formation after settling. 3 ml total was sampled daily for 3 samples of 1 ml for cell count. The cells were dissociated using Accutase (Sigma, USA #A6964) and incubated for 10 min. Cells were then quenched with E8 then centrifuged at 400 × g for 6 min. Pellets were resuspended in an equivalent volume of PBS and analyzed for total cell count using an Attune™ Flow Cytometry (ThermoFisher, USA). Triplicate counts were collected for each bioreactor at each stage of culture. Duplicate 500 μl samples were collected for aggregate imaging on a 24 well plate. An inverted automated microscope, EVOS M7000 imaging system (ThermoFisher, USA #AMF7000), was used for bright field images of the aggregates. ImageJ was used to analyze aggregate size and distribution. IEQ estimates of aggregates were performed using Prodo Laboratories, Inc protocol. Briefly, aggregates were divided into groups based on size and were normalized to a theoretical islet size of 150 μm using the provided conversion factors. Total IEQ was then estimated for the entire bioreactor volume and presented for the individual runs.
Quantitative polymerase chain reaction (qPCR)
Triplicate samples were taken from each bioreactor for qPCR testing at different stages throughout the culture time. RNA samples were dissolved in Lysis Buffer and extracted using MagMAX™-96 Total RNA Isolation Kit (ThermoFisher, USA cat #AM1830) according to manufacturer’s protocol. Quantification of RNA was performed on epoch reader (ThermoFisher Scientific, USA #A51119500C). A high-Capacity cDNA RT Kit (cat #4368813) was used for reverse transcription of RNA triplicate samples.
Flow cytometry
A 10 ml sample was taken from each bioreactor at the end of each stage of differentiation for flow testing. The aggregates were dissociated with Accutase for 10 min into single cells. Cells were resuspended in PBS and divided into samples for intracellular staining and cells for extracellular staining. Samples for intracellular staining were fixed with a live/dead stain FVS 780 (BD Biosciences, USA #565388), then permeabilized according to manufacturer’s protocol of overnight incubation in permeabilization buffer at − 20 °C using Perm Buffer III (BD Biosciences, USA #558050). Samples were then stained for antibodies corresponding to each stage of the protocol and flow cytometry was performed using Attune™ Flow Cytometry (ThermoFisher Scientific, USA) (Table 3).
Table 3.
List of antibodies used for immunostaining and flow cytometry
| Antigen | Host | Supplier | Catalogue number | Dilution |
|---|---|---|---|---|
| Primary antibodies | ||||
| C-peptide | Rat | DSHB | GN-ID4 | 1:500 |
| FOXA2 | Goat | Santa Cruz Biotechnologies | sc-6554 | 1:200 |
| Glucagon | Mouse | Sigma | G2654 | 1:500 |
| HNF1B | Rabbit | Santa Cruz Biotechnologies | sc-22840 | 1:200 |
| Insulin | Guinea pig | Dako | A0564 | 1:200 |
| PDX1 | Goat | R&D | AF2419 | 1:200 |
| SST | Rabbit | Dako | A0566 | 1:500 |
| Chromogranin A | Mouse | Agilent | M086901-2 | 1:500 |
| Secondary antibodies | ||||
| Antibody | Supplier | Catalogue number | Dilution | |
| 488-Donkey-a-Goat | Jackson Immuno Research | 705-546-147 | 1:200 | |
| 488-Donkey-a-Mouse | Jackson Immuno Research | 715-546-151 | 1:200 | |
| 488-Donkey-a-Rabbit | Jackson Immuno Research | 711-546-152 | 1:200 | |
| 488-Donkey-a-Rat | Jackson Immuno Research | 712-545-150 | 1:200 | |
| 594-Donkey-a-Goat | Jackson Immuno Research | 705-515-003 | 1:200 | |
| 594-Donkey-a-Mouse | Jackson Immuno Research | 715-585-150 | 1:200 | |
| 594-Donkey-a-Rabbit | Jackson Immuno Research | 711-586-152 | 1:200 | |
| 594-Donkey-a-Rat | Jackson Immuno Research | 712-586-153 | 1:200 | |
| Flow Antibodies | ||||
| Antibody | Supplier | Catalogue number | Dilution | |
| PE Mouse anti-PDX-1 | BD Biosciences | 562161 | 1:33 | |
| Fixable Viability Stain 780 | BD Biosciences | 565388 | 1:10,000 | |
| PE Mouse anti-Human FoxA2 | BD Biosciences | 561589 | 1:100 | |
| BD Pharmingen™Alexa Fluor® 647 Mouse Anti-Nkx2.2 | BD Biosciences | 564729 | 1:33 | |
| Alexa Fluor® 647 Mouse Anti-C-Peptide | BD Biosciences | 565831 | 1:2000 | |
|
Alexa Fluor® 488 Mouse Anti-Human Somatostatin |
BD Biosciences | 566032 | 1:2000 | |
|
Alexa Fluor® 647 Mouse anti-Human TRA-1- 60 Antigen |
BD Biosciences | 560850 | 1:50 | |
| BV605 Mouse Anti-SSEA-4 | BD Biosciences | 563119 | 1:50 | |
| Alexa Fluor® 488 Mouse anti-Oct3/4 | BD Biosciences | 560253 | 1:50 | |
| SYTOX Green Dead Cell Stain for flow cytometry | Thermo Fisher Scientific | S34860 | 1:2000 | |
|
Anti-GP2 (Glycoprotein 2) (Human) mAb- Alexa Fluor® 488 |
MBL | D277-A48 | 1:1000 | |
| Alexa Fluor® 647 Mouse Anti-Sox9 | BD Biosciences | 562161 | 1:33 | |
QuantStudio data analysis
Data collection was performed using an Open Array (QuantStudio, Life Technologies) with custom design gene cards (Table 4). The design focused on endodermal lineages and pancreatic fates. cDNA samples were loaded onto a custom design Quant Studio Card (ThermoFisher, #4471125) using an Open Array AccuFill System (ThermoFisher, USA #4471021) and ran on a QuantStudio 12 k Flex Real-Time qPCR System (ThermoFisher, USA #4471086). QuantStudio runs were performed according to manufacturer’s protocol. The resulting QuantStudio gene expression data was analyzed using Expression Suite™ software (Life Tech). The data set was then exported to Excel and normalized against three internal standard housekeeping genes present on the QS card GAPDH, EEF1A1 and 18S. Final expression levels were expressed as 1/(2ΔCrt) × 10,000 where Crt is the relative threshold cycle.
Table 4.
List of oligonucleotides used on the QuantStudio chip for gene expression testing
| Oligonucleotides | Source | Identifier |
|---|---|---|
| NKX6-1 Hs00232355 ml | Life Technologies | Cat# 4331182 |
| CLOCK Hs00231857 m1 | Life Technologies | Cat# 4331182 |
| PER3_Hs00213466_m1 | Life Technologies | Cat# 4331182 |
| SLC16A1_Hs01560299_m1 | Life Technologies | Cat# 4331182 |
| LDHA_Hs01378790_g1 | Life Technologies | Cat# 4331182 |
| GCG_Hs01031536_m1 | Life Technologies | Cat# 4331182 |
| MFN2 Hs00208382 m1 | Life Technologies | Cat# 4331182 |
| MAFA_Hs01651425_s1 | Life Technologies | Cat# 4331182 |
| PDX1 Hs00236830 m1 | Life Technologies | Cat# 4331182 |
| NEUROD1 Hs01922995 s1 | Life Technologies | Cat# 4331182 |
| NEUROG3 Hs01875204 s1 | Life Technologies | Cat# 4331182 |
| HNF1B Hs01001602 m1 | Life Technologies | Cat# 4331182 |
| MFN1 Hs00250475 m1 | Life Technologies | Cat# 4331182 |
| PER1 Hs00242988 m1 | Life Technologies | Cat# 4331182 |
| HK1 Hs00175976 m1 | Life Technologies | Cat# 4331182 |
| INS Hs02741908 m1 | Life Technologies | Cat# 4331182 |
| 18S Hs03003631 g1 | Life Technologies | Cat# 4331182 |
| NKX2-2_Hs00159616_m1 | Life Technologies | Cat# 4331182 |
| CRY1 Hs00172734 m1 | Life Technologies | Cat# 4331182 |
| GAPDH Hs99999905 m1 | Life Technologies | Cat# 4331182 |
| FOXA2 Hs05036278 s1 | Life Technologies | Cat# 4331182 |
| EEF1A1 Hs00742749 s1 | Life Technologies | Cat# 4331182 |
| PER2 Hs00256143 m1 | Life Technologies | Cat# 4331182 |
| HK2 Hs00606086 m1 | Life Technologies | Cat# 4331182 |
| CRY2 Hs00323654 m1 | Life Technologies | Cat# 4331182 |
| SST Hs00356144 m1 | Life Technologies | Cat# 4331182 |
| CIART Hs00328968 m1 | Life Technologies | Cat# 4331182 |
Immunofluorescent staining
iPSC and differentiated bioreactor aggregates were dissociated with Accutase and plated onto a vitronectin treated 24 well plate and grown for 1 day in TB2 media. Cells were fixed with 5% formalin (Sigma, USA #HT501128) in DPBS for 15 min at room temperature, permeabilized and blocked with a blocking buffer solution composed of 1% BSA (Sigma, USA #A8806) and 0.1% Tween-20 (Sigma, USA #P1379) for 60 min at room temperature and stained with primary antibodies at 1:250 (Table 3) overnight. Cells were then washed with DPBS and stained with secondary antibodies for 60 min. Cells were washed three times with DPBS (5 min each) and stained with DAPI. Plates were imagined using EVOS M7000 Imaging System microscope (ThermoFisher Scientific, USA #AMF7000).
RNA isolation, library preparation and RNA-seq
The RNA was qualified and quantified using Qubit™ RNA BR Assay Kit (ThermoFisher Scientific, USA #Q10211). Total RNA was isolated using MagMAX™-96 Total RNA Isolation Kit (ThermoFisher Scientific #AM1830) according to the manufacturer’s instructions. RNA quality was validated using 4200 TapeStation System (Agilent Technologies, USA #G2991BA). Enrichment of polyadenylated RNA and library preparation were performed using Illumina Stranded mRNA Prep (illumina) using the reagents provided in an Illumina® TruSeq® Stranded mRNA library prep workflow. The library underwent a final cleanup using the Agencourt AMPure XP system (Beckman Coulter, #A63880) after which the library’s quality was assessed using a 4200 TapeStation System.
For all samples, the sequencing was done at Genewiz from Azenta Life Sciences. The quality trimming and alignment of the samples were conducted using the nextflow nf-core/rnaseq pipeline (version 3.6). The pipeline incorporated Trim Galore (v.0.6.7) for adaptor trimming and quality control. The trimmed RNAseq reads were then mapped to the Homo sapiens GRCh38 genome annotation utilizing STAR (v 2.6.1). Datasets underwent filtration to eliminate low counts (< 10 reads). The heatmap was created using the package “pheatmap” (v1.0.12).
Dithizone staining
Dithizone (DTZ) (Sigma, USA Cat# 194832) powder was reconstituted in DMSO (ThermoFisher, USA #J66650.AP), diluted in PBS and filtered to eliminate sediments. Clusters were stained in 5 mg/mL DTZ for 2–3 min and washed with PBS. Images were acquired using an EVOS XL Core (Thermo Fisher Scientific, USA #AMEX1000).
Human C‑peptide content
Stage 3 endocrine clusters were lysed in Tissue Protein Extraction Reagent (ThermoFisher, USA #78510). The cell suspension was centrifuged (1 min, 1000 rcf, 4 °C) to remove cell debris. Cell lysates were stored at − 20 °C until assayed using the human Ultrasensitive C-peptide ELISA kit (Mercodia, SWE #10-1141-01). Content was normalized to aggregate number or protein content using a Bradford kit (ThermoFisher, USA #23200) per the manufacture’s instruction.
Oxygen consumption rate (OCR)
OCR and Extracellular Acidification Rate (ECAR) were measured using a Seahorse XFe96 analyzer (Agilent, USA #S7800BR). For stage 3, the cells were tested as clusters in the test plate. The cells were lysed and tested using a Bradford kit for protein normalization. The seeded plates were left for 24 h in the incubator. For the Mito Stress Test, cells were incubated in a non-CO2 incubator for 1 h in serum free Seahorse XF Base minimal DMEM media (Agilent, USA#103335-100) supplemented with 3 mM glucose (Agilent, USA #103577-100), 1 mM sodium pyruvate (Agilent, USA #103578-100) and 2 mM L-glutamine (Agilent, USA #103579-100). Following measurement of basal respiration, the cells were treated with sequential injections of 14.5 µM glucose, 1.5 µM oligomycin, 0.5 µM carbonyl cyanide-4-(trifuoromethoxy) phenyl hydrazone (FCCP) and 0.5 µM rotenone/antimycin A (RotA) (Agilent, USA Seahorse XF Cell Mito Stress Test Kit #103015-100).
Metabolite assessment of spent media
Spent media collected from several time points were centrifuged to remove cell debris and frozen at − 80 °C until being assayed. Glucose, lactate, Gln, Glu, NH4 +, Na +, K + , Ca + +, pH, PCO2, and PO2 were measured using a Flex2 analyzer (Nova Biomedical, USA #57900). Amino acid concentrations were measured using REBEL cell culture media analyzer (908devices, USA) from spent media samples loaded onto a round bottom plate and diluted 1:10 with the provided REBEL diluent.
Glucose response
For the glucose stimulated insulin secretion assay, approximately 15–20 stem cell-derived clusters were collected in a 24 well plate. The clusters were equilibrated in 2.8 mM glucose in RPMI medium (ThermoFisher, USA#11879020) for 30 min at 37 °C, and then they were washed and incubated again for 1 h. Aggregates were washed again and were exposed to 2.8 mM glucose for 1 h and then challenged for another hour with 17.5 mM glucose, and then tested for depolarization with KCL. Media samples were collected after each incubation of 1 h and tested using the human Ultrasensitive C-peptide ELISA kit (Mercodia, SWE#10-1141-01). The aggregates were counted and used for normalization.
Cryopreservation and recovery
Harvest aggregates were pelleted and resuspended in 1 ml of CS10 (Stem Cell Technologies, CAN #07930) in cryotubes to achieve a density from 1 to 5 million/ml. Tubes are then placed in a 9X9 freezer box and placed into the 4 °C (pre-chilled) CryoMed Controlled-Rate Freezer (ThermoFisher Scientific, USA #TSCM17PA). After the controlled rate freezing was completed, cells are moved directly into liquid nitrogen. Aggregates are recovered in a TB2 solution with 10 µM Y-27632 ROCK inhibitor and washed to be resuspended again in the same solution.
Generating DoE designs
All experimental designs based on HD-DoE were computer-generated using D-optimal interaction designs in MODDE software (Sartorius Stedim Data Analytical Solutions, SSDAS, FRA). In the Design Wizard within MODDE software, all factors tested and genes measured were manually input, and the screening option was selected within the Objective window. Factors known to initiate insulin secretion were incorporated into our design. The design runs were set to include up to 93 reaction conditions, along with the addition of 3 center point conditions. Following the generation of HD-DoE designs, the design with the highest G-efficiency was selected. This chosen HD-DoE design served as a template for the creation of perturbation media matrices. The perturbation matrices, consisting of 96 independent experimental runs, were generated using a Freedom Evo150 liquid handling robot (TECAN, CHE).
Generating computer gene models
The differentiation space was modeled using MODDE software. After importing the data into MODDE, primary 'Summary of Fit Plots' were automatically generated, providing R2 and Q2 measurements for each gene model. R2 assesses how well the data fits the gene model, while Q2 estimates the precision of the model's prediction. Both metrics range from 0 to 1, with values above 0.5 indicating a significant gene model.
Statistical analysis
Data were analyzed and graphed in Excel and GraphPad Prism9. Comparisons were conducted via ANOVA and T-Test with a significant difference defined as P < 0.05.
Results
Impact of culture system (2D vs. 3D) on differentiation and metabolism
The protocol was originally developed using adherent cultures and derived from design of experiments [18]. Suspension protocol was initiated in TB10 media (Table 1) to support aggregate formation [36]. This media was designed to stabilize aggregate formation while limiting aggregate growth. CDM2 medium [37, 38] poorly sustained aggregate growth during iPSC differentiation in bioreactors (Data not shown), so a descendent media was developed (TB2) specifically for sustaining iPSC differentiation in bioreactor suspension conditions (Table 1). Initial evaluation of the protocol transfer was performed in two culture conditions: adherent and in suspension bioreactors using the NCRM1 iPSC-line. Stage 3 cells from a suspension environment and adherent culture were compared and evaluated through RNA sequencing (Figure S1). RNA sequencing data in heatmap (Figure S1-A) and volcano plot (Figure S1-B-C) evaluation of core endocrine genes showed that expression levels in adherent cells were lower than those expressed in 3D environment (Figure S1-A). Several genes that are essential for insulin transcription such as NEUROD1, MAFA, PPARGC1A and NKX2.2 [19] along with INS were expressed with higher transcript levels in bioreactor samples. It was observed that genes usually expressed in earlier stages such as FOXA2 and HNF1β had higher expression levels in adherent culture indicating that 2D environment differentiation might be lagging. Since the cell architecture of human islets is in 3D and not 2D, dissociating and reaggregating the cells can more closely mimic conditions during embryonic islet development [22]. Thus, endocrine stage adherent cells were dissociated and aggregated in flasks (Figure S1-D) as a means of additional comparison of the final material as aggregates. ELISA assay was used for quantifying C-peptide content per aggregate in this comparison. The results indicated that the C-peptide per aggregate content observed in suspension environment was significantly higher compared to pseudoiselets generated from the adherent cultures (Figure S1-E). These results were consistent with the data from RNA sequencing despite reaggregation. Thus, there are discrepancies between culture characteristics requiring custom differentiation optimization. Aggregate size and density might be potential factors impacting the differentiation process and cell fate [36].
Expansion and differentiation of iPSCs into insulin-producing cells
To develop and solidify the protocol transferred from adherent to suspension environment, an iterative strategy of stage-wise optimization was used to determine and control critical process parameters using two cell lines. Prior to initiating the differentiation protocol, 90 million adherent cells were collected from flasks of two different cell lines, RCRP5005N and NCRM-1. Each batch was then reseeded into a 500 ml vertical wheel bioreactor at a density of approximately 1.8 × 10^5 cells/ml in TB10 media which consists of Essential 8 (E8) medium supplemented with polyethylene glycol (PEG) and Heparin Sodium Salt (HS). On the 3rd day, differentiation was initiated as previously described [18]. The protocol, originally optimized for adherent culture using a HD-DoE approach, aimed to induce dorsal foregut endoderm from pluripotent stem cells. This protocol (TB-beta) was modified as depicted in (Fig. 1A) with the basal media TB2 (Table 1). Modifications in the basal media supplements were made to enhance aggregate stability and limit aggregate fusion events, thus minimizing cluster diameter.
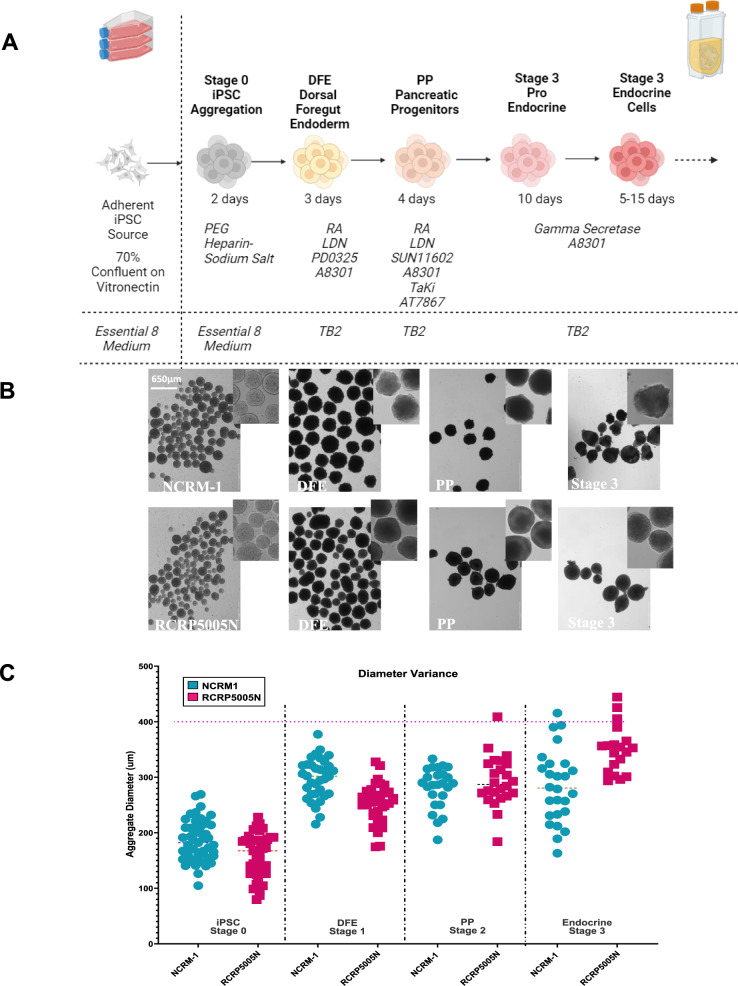
Fig. 1.
Bioreactor-based differentiation protocol timeline. A Schematic of the 3-stage differentiation pancreatic protocol used in bioreactors. Figure is created with BioRender.com B Images of bioreactor aggregates sampled throughout the different protocol stages. C Average aggregate size measurements on two different cell lines used throughout the protocol
During the initial 3 days of differentiation toward definitive endoderm, cells were optimized for FOXA2 expression. The morphology and growth of the clusters were characterized at all stages of differentiation, and the aggregate diameter measured in the range of (300 ± 100) µm for both cell lines (Fig. 1B–C). Retinoic acid, LDN3189, A8301, and PD0325901 were identified as differentiation factors for stage 1 (DFE) based on previous optimization efforts for HNF1β and FOXA2 [18]. By the end of this stage, bioreactors contained approximately 325 million cells of NCRM-1 and 420 million cells of RCRP5005N, representing ~ 4 × fold expansion throughout the process. Following the DFE stage, aggregates from each bioreactor were split into two additional 500 ml bioreactors to control cellular density. One half of the NCRM-1 cells remained in their original spent medium in a continuous bioreactor run with additional Pancreatic Progenitor (PP) stage factors being spiked in (Fig. 1A). It was also noted that aggregate diameter remained consistent for both cell lines throughout PP induction (Fig. 1B–C).
Subsequently, cells were cultured in endocrine inducing media for 10–30 days, consisting of TB2 media supplemented with gamma secretase and A8301. Media changes occurred for all bioreactors except the NCRM-1 cells that had never received a media exchange. NCRM-1 kept the same medium with spiking in additional factors needed for endocrine induction. In this study, several parameters and changes were identified and deemed essential for optimizing efficient large-scale manufacturing of islet-like clusters. Some of these parameters include controlling aggregate size without dissociation, altering basal media to support aggregation, conducting a 100% suspension protocol, extending the PP induction and endocrine induction stages differentiation periods, and limiting basal media changes throughout the process.
Optimization of time through a stage-wise approach
Cell evaluation occurred at the end of each differentiation stage. Prior to bioreactor seeding, cells were assessed for pluripotency markers SSEA4 and TRA-1–60 (Figure S2A). Populations exceeded 90% for both markers and over 85% co-expression. At the conclusion of DFE stage, FOXA2 expression ranged from 50 to 85%, with no detectable OCT4 expression, signifying the absence of undifferentiated cells (Figure S2B). Immunostaining on bioreactor clusters, which were plated on a vitronectin coated 24 well plate, revealed co-expression of HNF1β and FOXA2 (Figure S2C). HNF1β was originally optimized as a marker in the adherent protocol for directing dorsal foregut endoderm differentiation and subsequent endocrine cell fates [18].
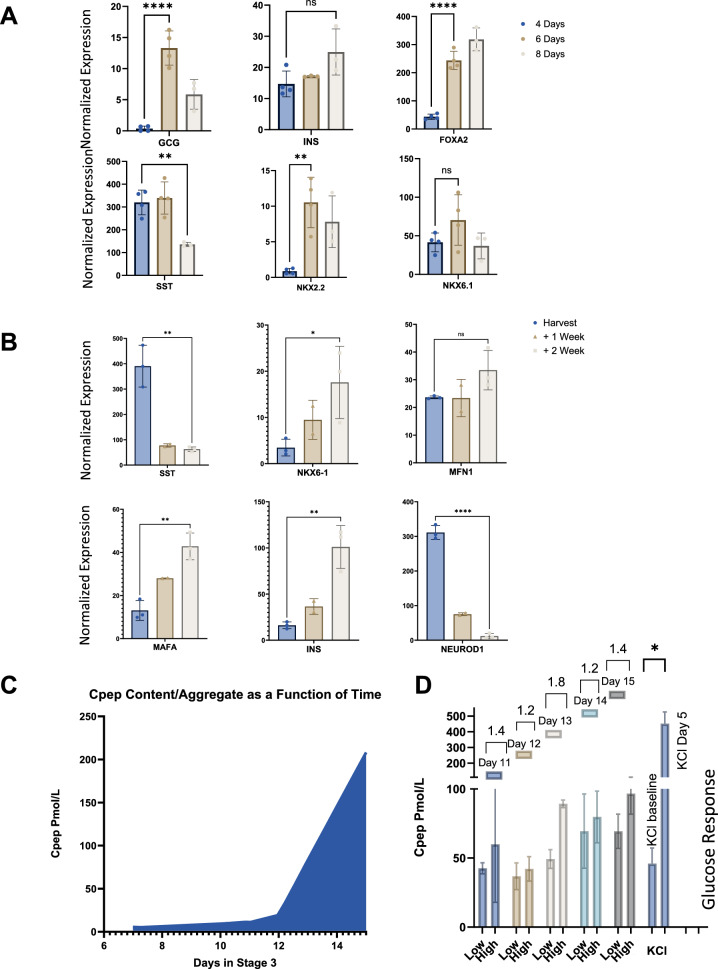
To optimize PDX1 expression, cells remained as clusters in PP stage media for 4–8 days. This time study on PP stage cells determined the optimal duration for differentiation in a suspension environment. Bioreactor PP stage cells were sampled at various time points. While the control sample remained in stage 2 (PP) for only 4 days, two additional samples were kept in PP induction stage media for 6 and 8 days, respectively. Subsequently, the cells were subjected to endocrine induction medium for 10 days and then analyzed for the expression of several endocrine specific markers including GCG, INS, SST, NKX6.1, NKX2.2, and FOXA2 (Fig. 2A). Prolonging the exposure of the PP inducing media led to increased expression of all the afore-mentioned markers except for SST. Notably, the extra two days in the PP inducing media resulted in increased expression of GCG, SST, NKX6.1, and NKX2.2 as compared to both the 4- and 8-day induction periods. It was noted that the expression of INS and FOXA2 continued to increase over time. During the endocrine induction period, marker expression analysis was performed weekly. Prolonging the exposure to the endocrine inducing medium resulted in increased expression of INS, MAFA, MFN1, NKX6.1, and PDX1.Only SST and NEUROD1 expression decreased over time (Fig. 2B). Additionally, daily sampling after 10 days in endocrine induction media revealed that extending this time by at least an additional five days resulted in a 20-fold increase in C-PEP expression. Glucose stimulated insulin release assays conducted over this 5-day time-period showed a minimal increase in C-PEP on day 10, and the largest increase was observed on day 13. Further increases following depolarization with KCL on day 15 showed a significantly higher C-PEP content (~ tenfold) as compared to the control samples performed on day 10 of endocrine inducing stage 3 media (Fig. 2D).
Fig. 2.
Time study evaluating differentiation and function. A Gene expression profile from the end of stage 3 of harvested cells that had different prolonged PP induction timing of 4 days, 6 days, and 8 days B Gene expression profile of harvested endocrine cells with increasing 1-week intervals. C C-Peptide concentration per aggregate count as a function of time in endocrine induction media. D Glucose-Stimulated Insulin Secretion of endocrine aggregates as a function of time. The stimulation index for day 11, 12, 13, 14, 15 after the end of endocrine induction (10 days), was 1.4, 1.2, 1.8, 1.2 and 1.4 respectively. All bar charts show individual points with mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001
Characterization of stage-specific markers throughout the protocol
The transition of β-cells during maturation involves notable alterations in the expression levels of HK2 (Hexokinase 2), LDHA (Lactate Dehydrogenase A), and SLC16A1 (Monocarboxylate Transporter 1) [19, 39, 40]. These genes play crucial roles in glucose metabolism and lactate production within cells. As β-cells mature, LDHA expression declines, leading to decreased lactate production. Additionally, SLC16A1 expression decreases, limiting the capacity for lactate transport further contributing to the metabolic shift from lactate generation to pyruvate utilization. This causes a β-cell maturation event by facilitating functional adaptation to full glucose utilization [19].
Comparing the original adherent protocol to the bioreactor production of β-cells over time focusing on these three genes (Fig. 3A) showed that these ‘functional disallowed genes’ are repressed throughout differentiation process within bioreactors. The difference between adherent and bioreactor culture becomes especially significant during the PP-induction stage and throughout the endocrine induction stage, where suspension cultures show significant repression of these glycolytic genes. These results match the findings shown in our RNA sequencing analysis presented in (Figure S1) which suggested that adherent culture lags in differentiation. To determine the shift in gene expression occurring throughout the differentiation protocol, key insulin transcription factors such as NEUROD1, NEUROG3, NKX6.1, NKX2.2, and MAFA, which play crucial roles in β-cell maturation [19, 39, 41–43], were monitored throughout the entire process (Fig. 3B). NKX6.1 expression increased after the media changed into the endocrine induction media. Conversely, NEUROD1, NEUROG3, NKX2.2, and MAFA expression began to significantly increase as the duration of the endocrine induction media was lengthened, suggesting a maturation benefit for extended differentiation time. Throughout the culture, FOXA2 and HNF1β were also tracked. FOXA2 regulates gene expression crucial for β-cell development during embryonic pancreatic development [18], and HNF1β dives exocrine development [18]. HNF1β exhibited a significant increase in expression levels between the differentiation from DFE to a PP but had a diminishing expression after endocrine induction. This was expected since its continued expression leads to exocrine development. In contrast, FOXA2 continued to rise post endocrine induction since it governs the expression of key genes like PDX1 and maintains the differentiated state and functionality of mature β-cells. Additionally, the expression levels of INS and GCG were mapped throughout the process, both showing significant increases during endocrine induction, in agreement with the discussed results.
Fig. 3.
Characterization of glycolytic and stage specific markers throughout the differentiation protocol. A Glycolytic genes expression comparison between adherent and suspension culture over time. Comparisons are made at the end of each differentiation stage. B Gene expression profile of cells at different stages of the protocol. The stage being assayed is indicated below the graph. All bar charts show individual points with mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001
The iPSC-derived islet-like clusters generated from the bioreactors were assessed for endocrine marker expression through immunostaining of the clusters. It was revealed that the aggregates expressed several known pancreatic markers, including PDX1, C-PEP, CGA, GCG, and SST, indicating the induction of a genuine pancreatic endocrine state (Figure S3A). Some co-expression of endocrine products was noted with the presence of CPEP + /SST + and CPEP + /GCG + cells being observed.
Continuous culture dramatically impacts the differentiation towards pancreatic progenitors
Maintaining stable cellular homeostasis, characterized by minimal fluctuations, is a prerequisite for optimal cellular function and environment understanding which leads to better system control [44]. This stability ensures that processes such as metabolism, signaling, and gene expression remain finely regulated within narrow ranges conducive to cellular function. Multiple factors contribute to the fluctuation and homeostasis and not just a small number of regulatory enzymes [45–47]. Factors include growth conditions, glucose concentration, cellular signaling pathways like AKT1, enzymatic regulation by LDH, PFK, and PEP, oxygen availability, cellular metabolism, tissue-specific functions, metabolic shifts, all collectively shaping the balance between lactate production and consumption [44]. To better understand the full bioprocess and nutrient limitations in maintaining a cellular homeostatic environment within the culture, a continuous media study was conducted throughout the differentiation protocol (Figure S3B). The stage-specific utilization of glucose and lactate generation throughout the culture time was assessed. A comparison was made between the Control media change bioreactor and the continuous bioreactor at the end of each stage and every week after endocrine induction. Spent media samples collected throughout the differentiation protocol were evaluated (Fig. 4A). Initially, the glucose concentration in the Control media change bioreactor decreased by approximately 30%. By around day 17 of the protocol, this decrease became less than approximately 16%, eventually reaching around 11% at harvest. With each media change, a steady increase in lactate concentration in the basal medium was observed. Toward the end of the control protocol which received full media changes throughout the process, an accumulation of (6 mM) lactate was measured. Comparing the profile of glucose and lactate change over time between the Control media change bioreactor and the continuous condition revealed significant differences. The glucose consumption rate decreased from approximately 30% before the initiation of differentiation to negligible levels toward the end of the protocol, stabilizing at around 7 mM after approximately 5 days from the start of differentiation (Fig. 4A). Similarly, lactate accumulation increased only during the first 3–5 days of the protocol, reaching a steady state after 5 days in culture at around 17 mM.
Fig. 4.
Impact of glucose consumption and lactate accumulation and profile on the differentiation and growth of cells A. Glucose and lactate concentration profile throughout the differentiation protocol on two bioreactors: the control which has frequent media changes at the different stages and the continuous which has no media changes but only spike in of differentiation factors at each stage. B Gene expression profile of cells for stage specific markers on both bioreactors after 20 days in stage 3 endocrine induction media. C Cell growth profile throughout the differentiation period on both control and continuous bioreactors. D The change of glucose over lactate concentration over time for the continuous bioreactor. All bar charts show individual points with mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001
The impact of not changing the basal medium and spiking differentiation factors in a continuous culture of cells was assessed alongside the Control media change bioreactors that underwent the process previously described (Fig. 1A). A comparison of expression levels at late endocrine induction stage revealed significant differences in the levels of PDX1, NEUROD1, and SST between the control cells and the cells that remained in the same medium with only spiking in stage-specific differentiation factors (Fig. 4B). The cells that were not subjected to basal media change exhibited higher expressions of PDX1, NEUROD1, INS, and NKX2.2 but lower expression of SST and GCG. This suggests an increased preference for β-cells throughout the differentiation process.
To explore the metabolic phenotype of the islet aggregates generated by our suspension protocol, we examined the growth rate of cells in both cultures (Fig. 4C) along with the consumption rate of glucose relative to lactate production over the culture period (Fig. 4D). The results further suggest that the cells are potentially primarily relying on glycolysis during the early stages of differentiation (S1-2) and transition to oxidative phosphorylation towards endocrine induction during the protocol (Fig. 4D). This transition occurs earlier in the continuous bioreactor because the glucose consumption rate reached a steady state faster. Endocrine cells produced from a continuous bioreactor run were assessed for INS expression using a HD-DoE assay that included multiple secretagogues (Figure S4A). This demonstrated that a fivefold increase in insulin levels can be attained within a 3-h incubation using the combinatorial influences of high glucose Rapamycin, DCA, oxytocin, and arginine (Figure S4B). The baseline medium on the cells was at low glucose whereas high glucose (17.5 mM) was incorporated as an additive in the design.
Nutrient consumption and metabolite measurements
Examining potential alternative fuel and nutrients sources for the cell, the amino acid profile throughout the culture of both the control and the continuous bioreactors was analyzed (Figure S5A-B). Amino acid concentrations were measured using the REBEL Cell Culture Analyzer (908 Devices). Essential amino acids such as histidine, isoleucine, leucine, lysine, methionine, phenylalanine, threonine, tryptophan, and valine were maintained throughout the culture period in both bioreactors. However, some amino acids were completely depleted in both culture mediums, including L-aspartic acid after 5 days and L-glutamic acid after 16 days. Amino acid metabolism is crucial for normal pancreatic β-cell function, and alanine and glutamine are known for their role in regulating β-cell function and insulin secretion [48]. At the end of culture, glutamine and alanine concentrations were at a higher concentration than they occur in fresh media, suggesting they were not growth-limiting (Figure S5A-B). However, the source of their increase remains unknown, unlike previous observations attributing their increase to the GlutaMAX™ additive [49]. This increased level of alanine and glutamine compared to the starting medium was not observed in the Control bioreactor that had frequent media changes between stages and throughout the extended endocrine induction stage. No other significant differences were observed between the two bioreactors. As previously noted, the bioreactor with limited media replenishment showed better differentiation than the Control media change bioreactor. Dietary manipulations of amino acids and serum deprivation have been linked to promoting adult-like traits in pancreas β-cells derived from human stem cells [34, 50–52].
In addition, both culture bio-profiles were assessed using FLEX2 (Nova Biomedical), analyzing Gln, Glu, NH4 +, Na +, K +, Ca + +, pH, PCO2, and PO2 throughout the culture period for both reactors (Figure S6A-B). The osmolarity of the culture medium steadily increased in the continuous bioreactor but remained within the range of 280–320 mOsm/kg. This increase can be attributed to the accumulation of solutes from nutrient metabolism and other waste products. In contrast, the osmolarity of the Control media change bioreactor fluctuated as the media was replenished at different stages of the differentiation process. Glutamine and glutamate levels were also assessed, and both showed depletion over time. This was consistent with measurements taken using the REBEL analyzer. Both bioreactors were comparable in their bio-profile, except for major differences observed in the continuous decrease of pH in the continuous bioreactor, as expected, and the rate of oxygen consumption. The gases measured in the media may have been impacted by the time between collection and measurement, however, the overall impact is the same for all samples. The overall data profile showed that the PO2 level began steadily decreasing after 10 days of culture or PP induction stage of differentiation. Although the interface of the media with the gas in the headspace of both bioreactors is the same since they are both the same size (500 ml), the near-equivalent flux of oxygen into the media may not be sufficient to replenish increased oxygen consumption in the 0.5L continuous vessel as compared to the Control media change bioreactor that simply has all the media replaced at regular intervals.
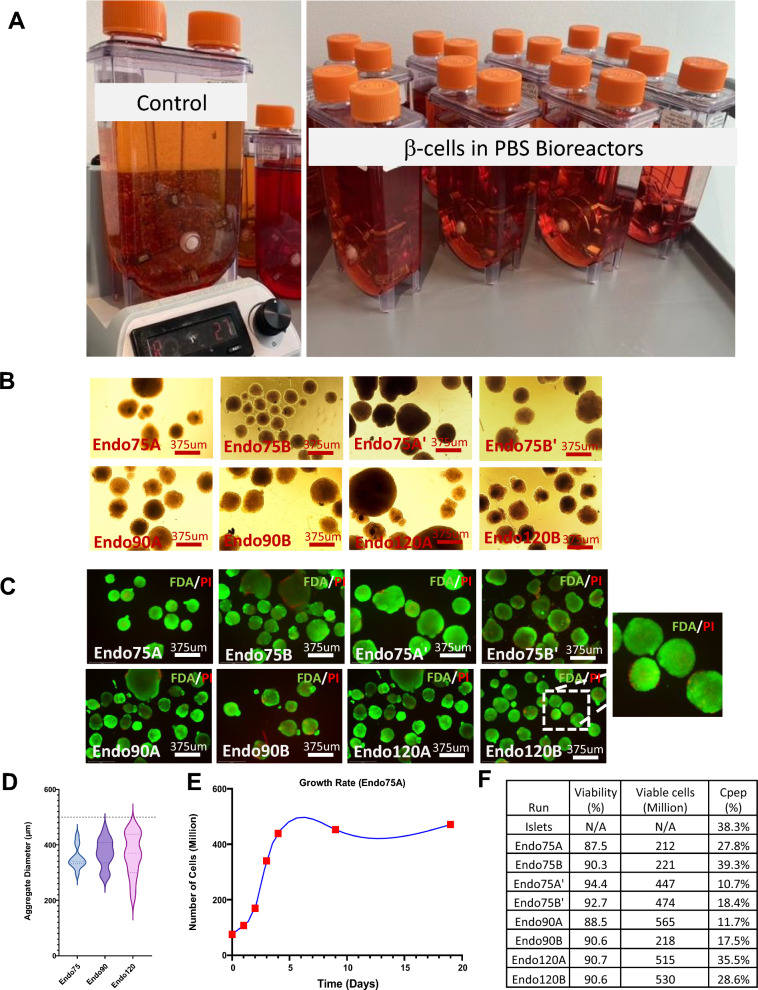
Validation and scaling
After resuming iterative process optimization and improvement efforts and implementing them as needed to achieve the desired culture outcomes, the process was validated using multiple bioreactors at different densities. Three different seeding densities were used to seed 0.5L bioreactors (75 M, 90 M and 120 M). The aggregates generated were monitored throughout the experiment and harvested at the end of the process (Fig. 5A). The cell clusters were evaluated for staining with dithizone (DTZ, which binds zinc within insulin granules), flow cytometry and protein levels of selected markers, viability, and oxygen consumption rate. There was a variable retention of DTZ throughout the different runs with the strongest intensity observed in Endo75A’ while little if any was observed in Endo75B’ (Fig. 5B). The aggregates were then analyzed for viability (Fig. 5C) and size (Fig. 5D). The cells were viable after the induction of endocrine cells, and the culture aggregate diameter average was below 500 µm. The growth rate of the bioreactors followed a profile of rapid proliferation during the early stages of differentiation (Generation of PP) and plateauing after endocrine induction (Fig. 5E). A sample of the clusters was digested to be further tested for counts (Fig. 5F) and flow cytometry (Figure S7–10). As expected, the largest yield was observed in bioreactors with the largest seeding density. These bioreactors also had the largest aggregate diameter; however, this didn’t impact viability (Fig. 5E).
Fig. 5.
Scale-up validation on multiple 0.5L bioreactors with different seeding densities. A An image of all bioreactors runs for this validation. B Dithizone staining of endocrine aggregates products. C Live dead staining on endocrine stage aggregates generated from all bioreactors using fluorescein diacetate for live staining and propidium iodide for dead staining. D Violin plot of the aggregate diameter variance on endocrine stage aggregate from all bioreactors runs including previous control. E The growth rate of cells over time for the Control media change bioreactor Endo75A. F Table summary of the viability of digested endocrine aggregates and their respective total cell count from each 500 ml bioreactor
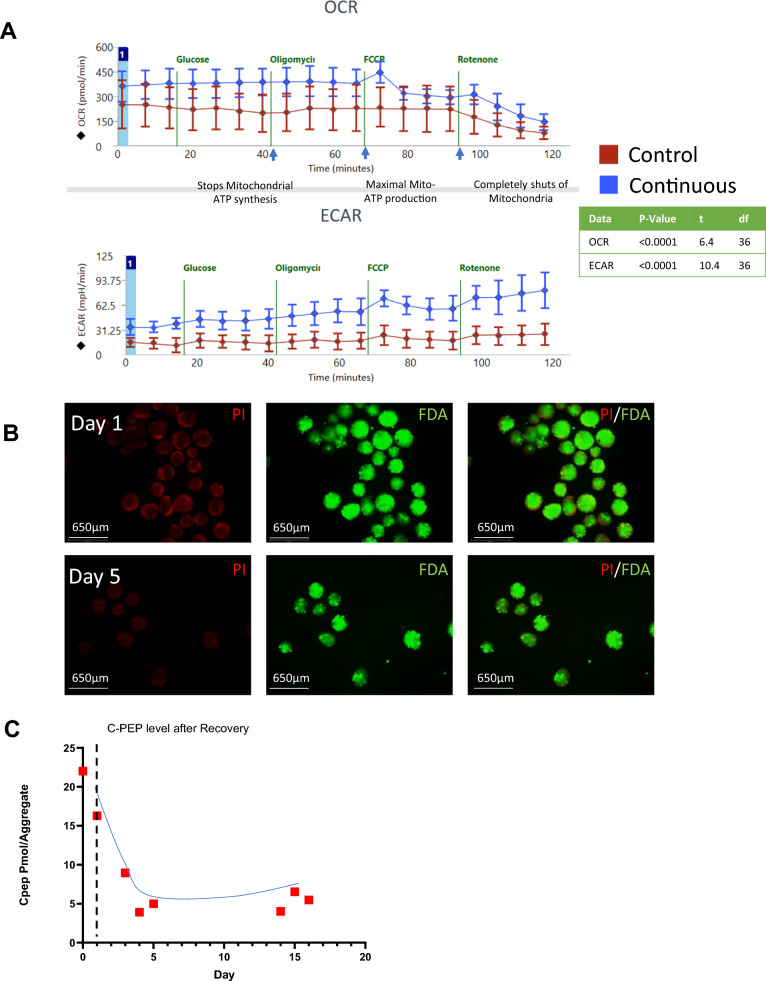
The continuous and Control media change bioreactors were tested for OCR and ECAR (Fig. 6A). Consistent with our findings, iPSCs-derived pancreatic progenitors produced with constant medium changes have lower OCR compared to those produced in a continuous bioreactor. iPSC-islets generated with our continuous bioreactor protocol have a higher aerobic respiration capacity. Mature function is dependent on obligatory aerobic metabolism and an increase in aerobic capacity of iPSC-derived islets is suggestive of an increased functionality, though full function has not been obtained. To assess the robustness of the continuous bioreactor process we next performed a continuous run using three different cell iPSC lines (Figure S11). It was shown that the process itself worked for all cell lines, however the efficacy of the pancreatic induction protocol varied widely. The early endodermal genes FOXA2 and HNF1β were all induced to similar levels, however genes representative of a pancreatic progenitor state showed variance between the cell-lines (Figure S11A). PDX1 induction was detected in all three cell lines, but was significantly reduced in the RCRP5005N lines, whereas NKX6.1 was significantly reduced in the NCRM-4 line. Consequently, endocrine induction was severely reduced with only SST reaching similar induction levels in the NCRM-4 cell line. Immunofluorescent staining was performed on the three continuous bioreactor runs to confirm the reduced induction levels of PDX1 and CPEP in the different iPSC lines (Figure S11B).
Fig. 6.
Energy map profile and cryopreservation recovery on endocrine cells. A Seahorse data on OCR and their respective ECAR profiles on a control run with regular media changes versus a continuous bioreactor run. B Live dead staining using fluorescein diacetate for live staining and propidium iodide for dead staining. On endocrine aggregates that were recovered from cryopreservation. C-peptide concentration profile normalized to the number of aggregates on cryopreserved and recovered cells from bioreactors. OCR and ECAR charts show individual points with mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001
Cryopreservation and recovery
The harvested aggregates were cryopreserved in CS10 solution with 10% DMSO in liquid nitrogen. Some vials were then recovered in TB2 medium + 10 µM Y-27632 ROCK inhibitor as aggregates. The clusters were checked for viability using FDA/PI stain immediately, and after 5 days of recovery, (Fig. 6B) no change was observed. The cells were > 85% viable after recovery. In addition, the cells were evaluated for C-PEP content in high glucose medium (17.5 mM) over time using a C-PEP Elisa kit. The concentration was then normalized per aggregate number (Fig. 6C). The C-PEP content was ~ 22 pmol per aggregate before cryopreservation. It decreased immediately after recovery but then reached a steady state after 5 days and stayed at 5 pmol per aggregate for the next 10 days.
Discussion
The translation of a developed large-scale process for β-cell production into the clinic applications is much needed [53, 54]. In this study, the process development and improvement for transitioning adherent cell culture protocols to a bioreactor suspension culture system involved several sequential steps (Table 5). Firstly, an assessment of the current cell culture parameters, including medium composition, cell seeding density, differentiation factors, and culture vessel, is needed. Next is the identification of key performance indicators and desired process outcomes to establish the goals of the optimization process. Before this study, feasibility studies were performed to evaluate the suitability of bioreactor culture vessels and impeller speed for the specific cell line or type. This was followed by small-scale bioreactor trials (100 ml) to assess cell expression, growth kinetics, and overall culture performance [36]. In this study, adjustments were made to bioreactor operating parameters such as stage time, culture medium change, and nutrient feeding strategies based on the results of these trials. Throughout the process, cell viability, proliferation, metabolite assessment and product quality were monitored and evaluated. A gradual transition from adherent cell culture to bioreactor cell culture was then achieved with careful optimization and monitoring at each step (Table 5).
Table 5.
Process optimization and improvement flowchart
| Process optimization and improvement flowchart | ||
|---|---|---|
| Step | Description | List |
| 1. Initial Adherent Cell Culture Process | Assess current cell culture parameters |
Culture medium Surface coating Stage medium development (HD-DoE) Seeding density optmization |
| 2. Identify Key Performance Indicators | Identify key performance indicators and desired process outcomes |
Gene Expression Glucose response Growth and viability Aggregate diameter oxygen consumption Glucose/Lactate Amino acid consumption |
| 3. Feasibility Studies | Conduct feasibility studies to evaluate the suitability of bioreactor culture for the specific cell line or type |
Direct protocol transfer RPM and shear stress minimization Seeding density optmization |
| 4. Small-Scale Bioreactor Trials | Perform small-scale bioreactor trials | 100 ml Bioreactor validation Differentiation medium optmization Aggregate stability additives |
| 5. Adjust Bioreactor Operating Parameters | Evaluate and adjust bioreactor operating parameters |
RPM Media change frequency Disaggregation between stages Media change method |
| 6. Monitoring and Evaluation | Monitor cell viability, proliferation, and product quality throughout the process |
Sampling frequency Stage timing QC methods and freasibilty |
|
7. Transition to Bioreactor Culture |
Implement gradual transition from adherent cell culture to bioreactor cell culture, with careful optimization and monitoring at each step of the process |
Large scale transition Validation Repeatabilty and reproducability check Stage wise validation |
| 8. Continuous Monitoring and Data Collection | Continuously monitor and collect data on bioreactor performance and cell culture characteristics for further process optimization and refinement | Continuous culture study Nutrient limiation study Environment control limitation Data monitoring methods |
| 9. Analysis and Iteration |
Document and analyze results to identify areas for further improvement and innovation |
Determining needs Process iteration changes QC check repeat with control |
|
10. Iterative Process Optimization and Improvement |
Iterate process optimization and improvement steps as needed to achieve desired culture outcomes |
Contrinuous process improvement Eliminate variables Fix acceptance critera |
| 11. Successful Transition to Bioreactor | Successful Transition to Bioreactor |
Validation Process control |
Developing a manufacturing process for robust and scaled production of iPSC-derived pancreatic islet-like cells produces a renewable source of islets needed both for diabetes investigations as well as diabetes therapies. Making islets available to address these needs will promote discovery of new treatments and increase the accessibility of existing treatments. In this study we hypothesized that lactate accumulation and glucose stability resulting from a limitation of media replenishment may be beneficial, if not necessary, for efficient differentiation. However, further experiments are required to validate this hypothesis. Various compounds constitute the common basal media used in iPSC culture maintenance and differentiation. IMDM and F12 are examples of basal media used in the field, however, there has been limited research on whether all the components that constitute these mediums are needed. Future work assessing the need of medium components and amino acids may play a pivotal role in improving targeted differentiation and reducing production costs. Another method that can contribute to an increased efficiency in manufacturing is assessing the underlying cause of improved differentiation seen in constant medium bioreactor cultures. Identifying the exact parameters allows researchers to build on these process improvements. In addition, defining limiting nutrient parameters and designing a strategy for spiking additives such as essential amino acids, other nutrients or increasing oxygen levels will play an instrumental role in enhancing function of the process and on the differentiated cells. Finally, optimization of bioprocess design and defining new key parameters for monitoring and control represent a promising avenue for enhancing the scalability and efficiency of iPSC expansion and differentiation.
As the realization of implementing cellular replacement therapies for Type 1 diabetes in clinical settings advances, it is crucial to refine the current adapted manufacturing methods, their feasibility, and efficiency. This research focuses on identifying and assessing various bioprocess variables in the production of insulin-producing cells derived from human iPSCs. We examined gene expression profile throughout the process, metabolic behaviors after nutrient limitation, and growth patterns, cell functionalities, and the activation of specific markers associated with maturation and metabolic shifts during differentiation.
Our study has shown that 3D culture systems better mimic the in vivo microenvironment, leading to improved differentiation efficiency and functionality of β-cells. We determined that culture platform can affect cell fate and differentiation efficiency. Understanding the effects of culture morphology on β-cell production is crucial for optimizing differentiation protocols. The differentiation towards endocrine fates was improved when transferring the protocol from adherent platform to suspension platform. Crucial for this improvement was developing a media formulation optimized for suspension cultures and is reflected in the changing of the basal culture medium from CDM2 to TB2 to better sustain aggregate stability. While we believe the general advantage of 3D culture on differentiation over 2D cultures, the differentiation protocol and process have factors that could cause variability in results. Some inherent causes of variability throughout the process that were evaluated here are the cell source used, differentiation media, aggregation method and reagents used. In addition, the impact of time on the differentiation process cannot be understated. We show here that prolonged culture durations lead to increased cell differentiation though they have the potential to increase process cost. Here we propose that a compromise between an increased maturation level of a biologic and the overall process cost needs to be fully addressed during process optimization. In this study, extending stage durations was shown to benefit the differentiation capacity of desired markers associated with our target product profile. To counter the associated cost with this increased production time, we evaluated the need for replenishing media throughout the process and how the overall differentiation process was affected.
Achieving significant cell quantities is essential for producing insulin-producing cells efficiently. Estimates suggest around 1 billion β-cells would be necessary for the treatment of a type 1 diabetic patient [5, 55–58]. By defining critical quality attributes, we can pinpoint stages for enhancing cell differentiation without compromising process quality and yield. Our study showed an approximate production capability of 1 billion cells per liter of production run. The cell yield was enhanced by eliminating cell disruption and digestion throughout the entire process, increasing seeding density and culturing the cells in an aggregate stability enhanced media [36].Though increasing the seeding density was shown to have the direct effect of increasing aggregate size, a balance between initial seeding density and product yield must center on defining the desired aggregate size of the final product.
Despite observing a large increase in cell number and glucose consumption in the first 5 days indicating proliferation, the actual proliferative capacity was limited as the differentiation continued presumably due to at least a partial metabolic shift away from glycolysis. This shift from proliferation to differentiation occurred faster in continuous bioreactors when compared to the control medium change environment. This change in addition to reduced glucose consumption rate can be partially attributed to insulin degradation in medium and inability to facilitate glucose uptake by thus regulating glucose consumption in cell medium. We propose that glucose utilization and lactate accumulation are critical quality attributes that can influence cell fate. We report in our study that while the intention behind running continuous bioreactors was for media nutrition limitation studies, the cells produced in that environment achieved a more desirable differentiation state. This may result from achieving a level of cellular homeostasis and stability not capable when cultures are continuously shocked by drastic environmental changes. Replenishment of media was shown to drastically change both the glucose and lactate levels while process related changes, such as centrifugation and the complete removal from culture media, are suggested to also have negative effects. The continuous culturing process described here was found to be practically amenable to this protocol since none of the differentiation factors used work directly against each other. A situation unlikely to occur in all directed differentiation protocols. Most small molecules are relatively stable as proteins or metabolites that can be used as differentiation reagents. The latter two naturally degrading over time in culture. Recent studies have reported and support the notion that lactate accumulation contribute to a slightly acidic environment and may be beneficial for differentiation efficiency [49]. Lactate production rates in our continuous process media were similar to what is observed in physiological fasting levels [59].Lactate is approximately double glucose concentrations when measured on a molar basis. This equivalence extends to a carbon-atom basis because two lactate molecules equate to one glucose molecule [59]. The clear implication from these findings is that pyruvate, a product of glycolysis, might not enter the tricarboxylic acid (TCA) cycle directly within cells, but may instead be converted into lactate and released into the bloodstream. This conversion process necessitates the activity of lactate dehydrogenase (LDH) [59]. The lactate steady state achieved in the continuous bioreactor could be attributed to a lower glucose consumption rate after a metabolic shift away from glycolysis and towards oxidative phosphorylation during our endocrine induction stage.
Undifferentiated human iPSCs primarily utilize glycolysis for glucose metabolism, whereas during differentiation, they transition to oxidative phosphorylation [60]. Even when maintained in an undifferentiated state, culturing iPSCs in stirred suspension bioreactors prompts a shift from glycolysis accompanied by increased lactate production compared to differentiated cells [61]. Adapting to the dynamic metabolic changes and consequential fate decisions of iPSCs in bioreactors will necessitate a departure from conventional cell culture techniques [62].
Metabolic profiling of the cell culture medium is shown to be an essential parameter. The composition of the medium, including glucose concentration, amino acids, and lactate, influences cellular metabolism and ultimately effects β-cell production. For instance, higher glucose concentrations will stimulate glycolysis, whereas lower concentrations may favor oxidative phosphorylation as supported by data from our continuous bioreactor runs. Understanding these metabolic shifts and their impact on β-cell differentiation is critical for optimizing culture conditions to both enhance cell yield and functionality. These results offer valuable insights for manufacturing processes, though there are limitations to consider such as variations between cell lines and protocols, the need to evaluate robustness and reproducibility of both manufacturing and QC processes.
Data suggests that stem cell derived endocrine cells still lack glucose response function in comparison to human islet cells. While our cells were mildly glucose responsive, the high glucose set point is still lacking insulin increase [17, 37, 39]. Previous studies have shown that less mature β-cells exhibit heightened responsiveness to calcium levels when glucose concentrations are low [19]. This can explain the elevated basal insulin secretion and poor GSIS [19]. Our cells were assessed for function using a basic static GSIS (Fig. 2D) while a more dynamic INS expression assay using our continuous bioreactor endocrine cells incorporated a HD-DoE assay. Mathematical models from MODDE software (Figure S4) have demonstrated an ability to increase insulin secretion levels fivefold. It is reasonable to assume that our endocrine aggregates have the potential for an insulin secretion profile of human islets, but still lack necessary mechanism for achieving this level of insulin response.
Strategic scale-up and design transfer processes are vital for translating laboratory-scale protocols into the large-scale manufacturing needed for commercial and clinical uses. Ensuring scalability, reproducibility, and validation of the manufacturing process is essential for generating consistent high-quality cellular products. This includes properly identifying critical process parameters and defining adequate quality control measures that can establish robust protocols ensuring reproducibility for a manufacturing site.
There are several limitations to this study. Although, two iPSC lines are tested in this study and two embryonic cell lines were used originally for the protocol [18], most differentiation protocols whether adherent or in suspension have different efficiencies and preferences when used on different cell lines. Certain cell lines might need slight process parameter changes to better fit the protocol, as an example the repro cell-line was previous shown to require lower RPM when grown in vertical wheel bioreactors [36]. While we expect that the insights gained could be relevant to other cell lines, factors such as seeding density, dissociation enzyme and time, cell passage number, media change method, basal media and human variables can also impact the differentiation efficiency. The fact that the additional cell lines did not perform as well in the continuous process may be attributed to some of these reasons and because the process was developed using the NCRM-1 line. It is likely that additional process parameter optimization is required for different cell lines as it is readily accepted that iPSC-lines often behave differently. Some notable difference we observed in this series of experiments with the RCRP5005P cell line is a greater growth rate, the requirement for a lower rotational rate in the bioreactors and a greater capacity to attach to TC-plates. Differences like these would need to be addressed before a continuous run could be optimized. Altogether this suggests that different CPPs will be identified for different cell lines and manufacturing processes should be built around a single dedicated cell line.
Ultimately, the successful translation of iPSC-derived β-cells into manufacturing and production relies on a comprehensive understanding of various factors, including culture morphology, differentiation time, culture medium metabolic profile, cryopreservation, and scalability of the manufacturing process. By addressing these considerations, researchers can optimize protocols for efficient β-cell production and pave the way for successful clinical trials and ultimately, the treatment of Type 1 Diabetes (Table 6).
Table 6.
Experimental data generated during study
| P-values and fold changes for all significant changes | ||||
|---|---|---|---|---|
| Figure | Comparison | Mesurement | Fold change | P-value |
| Fig. S1 | Bioreactor vs Adhernet | Cpep Content | 24.9 | < 0.0001 |
| Figure 2A | 6 days vs 4 days | GCG Expression | 37.6 | 0.0026 |
| Figure 2A | 6 days vs 4 days | FOXA2 Expression | 5.6 | < 0.0001 |
| Figure 2A | 4 days vs 8 days | SST Expression | 2.4 | 0.0023 |
| Figure 2A | 6 days vs 4 days | NKX2.2 Expression | 12.1 | 0.0016 |
| Figure 2B | 2 weeks vs harvest | SST Expression | 6.2 | 0.0034 |
| Figure 2B | 2 weeks vs harvest | NKX6.1 Expression | 5.1 | 0.0381 |
| Figure 2B | 2 weeks vs harvest | MAFA Expression | 3.2 | 0.0026 |
| Figure 2B | 2 weeks vs harvest | INS Expression | 6.3 | 0.0033 |
| Figure 2B | Harvest vs 2 weeks | NEUROD Expression | 26.0 | < 0.0001 |
| Figure 2D | 30 mM KCl vs 4.8 mM | C-peptide release | 9.8 | 0.0168 |
| Figure 3A | Adherent PP vs Bioreactor PP | HK2 Expression | 1.8 | 0.002 |
| Figure 3A | Bioreactor PP vs Bioreactor Endo | HK2 Expression | 22.5 | < 0.0001 |
| Figure 3A | Adherent Endo vs Bioreactor Endo | HK2 Expression | 3.3 | 0.0055 |
| Figure 3A | Adherent PP vs Bioreactor PP | LDHA Expression | 2.7 | 0.0016 |
| Figure 3A | Bioreactor PP vs Bioreactor Endo | LDHA Expression | 3.5 | 0.0166 |
| Figure 3A | Adherent PP vs Bioreactor PP | SLC16A1 Expression | 1.5 | 0.012 |
| Figure 3A | Bioreactor PP vs Bioreactor Endo | SLC16A1 Expression | 8.5 | 0.0005 |
| Figure 3A | Adherent Endo vs Bioreactor Endo | SLC16A1 Expression | 2.8 | 0.0005 |
| Figure 4B | No media change vs media change | PDX1 Expression | 1.7 | 0.0031 |
| Figure 4B | No media change vs media change | NEUROD1 Expression | 3.8 | 0.0103 |
| Figure 4B | Media change vs no media change | SST Expression | 4.9 | 0.036 |
| Fig. S11 | NCRM1 vs NCRM4 | NKX6.1 Expression | 5.4 | 0.0080 |
| Fig. S11 | NCRM1 vs RCRP5005N | PDX1 Expression | 32.1 | 0.0024 |
| Fig. S11 | NCRM1 vs RCRP5005N | SST Expression | 2.9 | 0.0095 |
| Fig. S11 | NCRM1 vs RCRP5005N | INS Expression | 146.8 | 0.0005 |
| Fig. S11 | NCRM1 vs NCRM4 | INS Expression | 33.2 | 0.0005 |
| Bioreactor conditions from Fig. 5 | |
|---|---|
| Abbreviation | Seeding (per 500 ml Bioreactor) |
| Endo75A | 7.50E + 07 |
| Endo75B | Split from Endo75A at stage 2 |
| Endo75A' | 7.50E + 07 |
| Endo75B' | Split from Endo75A at stage 2 |
| Endo90A | 9.00E + 07 |
| Endo90B | Split from Endo75A at stage 2 |
| Endo120A | 1.20E + 08 |
| Endo120B | Split from Endo75A at stage 2 |
Conclusion
Our study outlines a large-scale differentiation protocol for generating insulin-producing cells from human iPSCs, emphasizing the need for extended differentiation periods and minimal culture interventions. We highlight the impact of reducing media changes on process efficiency and differentiation, underscoring the importance of refining manufacturing protocols. These insights advance cell replacement therapy initiatives by iPSC-derived islet-like cluster production, offering valuable insights to the field's understanding and practices.
Supplementary Information
Acknowledgements
We would like to thank Trailhead Biosystems for funding this research. The authors also thank Cleveland State University for their support.
Abbreviations
- HS
Heparin sodium salt
- PEG
Polyethylene glycol
- E8
Essential 8
- DMSO
Dimethyl Sulfoxide
- HD-DoE
High Dimensional Design of Experiments
- 3D
Three-dimensional
- iPSC
Human Induced pluripotent stem cell
- PBS
Phosphate buffer solution
- qPCR
Quantitative PCR
- RPM
Revolution per minute
- ROCK
Rho-Kinase
- PP
Pancreatic Progenitor
- DFE
Dorsal Foregut Endoderm
- Crt
Relative threshold cycle
- OCR
Oxygen consumption rate
- ECAR
Extracellular Acidification Rate
- FCCP
Carbonyl Cyanide-4-(trifuoromethoxy) Phenyl Hydrazone
- RotA
Rotenone/antimycin A
- 2D
Two-dimensional
- TCA
Tricarboxylic acid
- T1DM
Type 1 Diabetes Mellitus
- LDH
Lactate Dehydrogenase
- GSIS
Glucose-stimulated insulin secretion
- PI
Propidium iodide
- FDA
Fluorescein diacetate
Author contributions
M.B conceived and supervised the project. H.Y. designed, conducted all experiments, data & samples collection, and analysis, and wrote the full manuscript. A.W. cultured the starting iPSC material for all the experiments and conducted ELISA testing. F.S. assisted in QuantStudio QC. M.M. and D.N. ran the validation bioreactors for large-scale production. R.K. assisted with amino acids testing and cryopreservation. R.P. conducted the RNA Seq analysis and figures. J.J. provided review feedback. All authors approved the manuscript.
Funding
The work was supported and funded by Trailhead Biosystems Inc. No external funding was used.
Availability of Data and Materials
Requests for further information or more detailed protocols should be directed to and will be fulfilled by the corresponding author. This study did not generate new unique reagents. All the RNA-Seq data are deposited in NCBI’s Gene Expression Omnibus (GEO) dataset with the accession number GSE264176. All the sequencing data sets are publically availiable.
Declarations
Ethics approval and consent to participate
The study does not include a clinical trial, therefore a consent to participate and the declaration of Helsinki—Ethical principles for medical research involving human subjects are not applicable. Experiments used human induced pluripotent stem cells purchased from REPROCELL (Cat#RCRP005N) and iXCells (Cat#NCRM-1, & NCRM-4) that have had their IRB approval documents and informed consents examined by National Institute of Neurological Disorders and Stroke to ensure that voluntary informed consent was obtained from the donor. The title of the project is, “Identifying and Optimizing Critical Process Parameters for Large-Scale Manufacturing of iPSC Derived Insulin-Producing β-cells”. All experiments were performed at Trailhead Biosystems. Approval and oversight of the project were performed by PhD committee at The Cleveland State University and approved on the 19th day of July 2022. No formal oversight committee was used for these reasons, so no approval number is associated with the study. However, full institutional approval was given by Trailhead Biosystems for all aspects of the study and Trailhead Biosystems does abide by all the ethical principles and guidelines established by the International Society of Stem Cell Research (ISSCR).
Consent for publication
Not applicable.
Competing interests
J.J. is founder of and shareholder of Trailhead Biosystems, Inc., Beachwood, OH, USA. M. B. is a shareholder in Trailhead Biosystems, Inc., Beachwood OH. This work has been filed as US Provisional Application No. application pending.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Roep BO, Thomaidou S, van Tienhoven R, Zaldumbide A. Type 1 diabetes mellitus as a disease of the β-cell (do not blame the immune system?). Nat Rev Endocrinol. 2021;17:150–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sun H, Saeedi P, Karuranga S, Pinkepank M, Ogurtsova K, Duncan BB, et al. IDF Diabetes Atlas: Global, regional and country-level diabetes prevalence estimates for 2021 and projections for 2045. Diabetes Res Clin Pract. 2022;183. [DOI] [PMC free article] [PubMed]
- 3.T1D Index Website. 2024.
- 4.Gamble A, Pepper AR, Bruni A, Shapiro AMJ. The journey of islet cell transplantation and future development. Islets. 2018;10:80–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shapiro AMJ, Lakey JRT, Ryan EA, Korbutt GS, Toth E, Warnock GL, et al. Islet transplantation in seven patients with type 1 diabetes mellitus using a glucocorticoid-free immunosuppressive regimen. N Engl J Med. 2000;343:230–8. [DOI] [PubMed] [Google Scholar]
- 6.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–76. [DOI] [PubMed] [Google Scholar]
- 7.Yamanaka S. Pluripotent stem cell-based cell therapy—promise and challenges. Cell Stem Cell. 2020;27:523–31. [DOI] [PubMed] [Google Scholar]
- 8.Chen G, Gulbranson DR, Hou Z, Bolin JM, Ruotti V, Probasco MD, et al. Chemically defined conditions for human iPSC derivation and culture. Nat Methods. 2011;8:424–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carvalho AM, Nunes R, Sarmento B. From pluripotent stem cells to bioengineered islets: a challenging journey to diabetes treatment. Eur J Pharm Sci. 2022. [DOI] [PubMed]
- 10.Tohyama S, Fujita J, Fujita C, Yamaguchi M, Kanaami S, Ohno R, et al. Efficient large-scale 2D culture system for human induced pluripotent stem cells and differentiated cardiomyocytes. Stem Cell Rep. 2017;9:1406–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lambrechts T, Papantoniou I, Viazzi S, Bovy T, Schrooten J, Luyten FP, et al. Evaluation of a monitored multiplate bioreactor for large-scale expansion of human periosteum derived stem cells for bone tissue engineering applications. Biochem Eng J. 2016;108:58–68. [Google Scholar]
- 12.Amini N, Paluh JL, Xie Y, Saxena V, Sharfstein ST. Insulin production from hiPSC-derived pancreatic cells in a novel wicking matrix bioreactor. Biotechnol Bioeng. 2020;117:2247–61. [DOI] [PubMed] [Google Scholar]
- 13.Nogueira DES, Rodrigues CAV, Carvalho MS, Miranda CC, Hashimura Y, Jung S, et al. Strategies for the expansion of human induced pluripotent stem cells as aggregates in single-use Vertical-Wheel™ bioreactors. J Biol Eng. 2019;13:74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abbasalizadeh S, Larijani MR, Samadian A, Baharvand H. Bioprocess development for mass production of size-controlled human pluripotent stem cell aggregates in stirred suspension bioreactor. Tissue Eng Part C Methods. 2012;18:831–51. [DOI] [PubMed] [Google Scholar]
- 15.Mihara Y, Matsuura K, Sakamoto Y, Okano T, Kokudo N, Shimizu T. Production of pancreatic progenitor cells from human induced pluripotent stem cells using a three-dimensional suspension bioreactor system. J Tissue Eng Regen Med. 2017;11:3193–201. [DOI] [PubMed] [Google Scholar]
- 16.Borys BS, Dang T, So T, Rohani L, Revay T, Walsh T, et al. Overcoming bioprocess bottlenecks in the large-scale expansion of high-quality hiPSC aggregates in vertical-wheel stirred suspension bioreactors. Stem Cell Res Ther. 2021;12:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jeske R, Liu C, Duke L, Canonicco Castro ML, Muok L, Arthur P, et al. Upscaling human mesenchymal stromal cell production in a novel vertical-wheel bioreactor enhances extracellular vesicle secretion and cargo profile. Bioact Mater. 2023;25:732–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bukys MA, Mihas A, Finney K, Sears K, Trivedi D, Wang Y, et al. High-dimensional design-of-experiments extracts small-molecule-only induction conditions for dorsal pancreatic endoderm from pluripotency. iScience. 2020;23:101346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nair GG, Tzanakakis ES, Hebrok M. Emerging routes to the generation of functional β-cells for diabetes mellitus cell therapy. Nat Rev Endocrinol. 2020; 506–18. [DOI] [PMC free article] [PubMed]
- 20.Rezania A, Bruin JE, Arora P, Rubin A, Batushansky I, Asadi A, et al. Reversal of diabetes with insulin-producing cells derived in vitro from human pluripotent stem cells. Nat Biotechnol. 2014;32:1121–33. [DOI] [PubMed] [Google Scholar]
- 21.Kroon E, Martinson LA, Kadoya K, Bang AG, Kelly OG, Eliazer S, et al. Pancreatic endoderm derived from human embryonic stem cells generates glucose-responsive insulin-secreting cells in vivo. Nat Biotechnol. 2008;26:443–52. [DOI] [PubMed] [Google Scholar]
- 22.Nair GG, Liu JS, Russ HA, Tran S, Saxton MS, Chen R, et al. Recapitulating endocrine cell clustering in culture promotes maturation of human stem-cell-derived β cells. Nat Cell Biol. 2019;21:263–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pagliuca FW, Millman JR, Gürtler M, Segel M, Van Dervort A, Ryu JH, et al. Generation of functional human pancreatic β cells in vitro. Cell. 2014;159:428–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Russ HA, Parent AV, Ringler JJ, Hennings TG, Nair GG, Shveygert M, et al. Controlled induction of human pancreatic progenitors produces functional beta-like cells in vitro. EMBO J. 2015;34:1759–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Millman JR, Xie C, Van Dervort A, Gürtler M, Pagliuca FW, Melton DA. Generation of stem cell-derived β-cells from patients with type 1 diabetes. Nat Commun. 2016;7:11463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Velazco-Cruz L, Goedegebuure MM, Millman JR. Advances toward engineering functionally mature human pluripotent stem cell-derived β cells. Front Bioeng Biotechnol. 2020;8. [DOI] [PMC free article] [PubMed]
- 27.Kunisada Y, Tsubooka-Yamazoe N, Shoji M, Hosoya M. Small molecules induce efficient differentiation into insulin-producing cells from human induced pluripotent stem cells. Stem Cell Res. 2012;8:274–84. [DOI] [PubMed] [Google Scholar]
- 28.Ghazizadeh Z, Kao DI, Amin S, Cook B, Rao S, Zhou T, et al. ROCKII inhibition promotes the maturation of human pancreatic beta-like cells. Nat Commun. 2017;8. [DOI] [PMC free article] [PubMed]
- 29.D’Amour KA, Bang AG, Eliazer S, Kelly OG, Agulnick AD, Smart NG, et al. Production of pancreatic hormone–expressing endocrine cells from human embryonic stem cells. Nat Biotechnol. 2006;24:1392–401. [DOI] [PubMed] [Google Scholar]
- 30.D’Amour KA, Agulnick AD, Eliazer S, Kelly OG, Kroon E, Baetge EE. Efficient differentiation of human embryonic stem cells to definitive endoderm. Nat Biotechnol. 2005;23:1534–41. [DOI] [PubMed] [Google Scholar]
- 31.Rezania A, Bruin JE, Riedel MJ, Mojibian M, Asadi A, Xu J, et al. Maturation of human embryonic stem cell-derived pancreatic progenitors into functional islets capable of treating pre-existing diabetes in mice. Diabetes. 2012;61:2016–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Loo LSW, Lau HH, Jasmen JB, Lim CS, Teo AKK. An arduous journey from human pluripotent stem cells to functional pancreatic β cells. Diabetes Obes Metab. 2018;20:3–13. [DOI] [PubMed] [Google Scholar]
- 33.Hogrebe NJ, Maxwell KG, Augsornworawat P, Millman JR. Generation of insulin-producing pancreatic β cells from multiple human stem cell lines. Nat Protoc. 2021;16:4109–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Velazco-Cruz L, Song J, Maxwell KG, Goedegebuure MM, Augsornworawat P, Hogrebe NJ, et al. Acquisition of dynamic function in human stem cell-derived β cells. Stem Cell Rep. 2019;12:351–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Business Wire. Vertex presents positive VX-880 results from ongoing phase 1/2 study in type 1 diabetes at the American diabetes association 83rd scientific sessions. Business Wire. 2023 Jun 23;
- 36.Yehya H, Raudins S, Jensen J, Bukys MA. Addressing bioreactor hiPSC aggregate stability, maintenance and scaleup challenges using a design of experiment approach. Res Sq. 2023. [DOI] [PMC free article] [PubMed]
- 37.Takao T, Yamada D, Takarada T. A protocol to induce expandable limb-bud mesenchymal cells from human pluripotent stem cells. STAR Protoc. 2022;3: 101786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Loh KM, Ang LT, Zhang J, Kumar V, Ang J, Auyeong JQ, et al. Efficient endoderm induction from human pluripotent stem cells by logically directing signals controlling lineage bifurcations. Cell Stem Cell. 2014;14. [DOI] [PMC free article] [PubMed]
- 39.Lemaire K, Thorrez L, Schuit F. Disallowed and allowed gene expression: two faces of mature islet beta cells. Annu Rev Nutr. 2016;36:45–71. [DOI] [PubMed] [Google Scholar]
- 40.Thorrez L, Laudadio I, Van Deun K, Quintens R, Hendrickx N, Granvik M, et al. Tissue-specific disallowance of housekeeping genes: the other face of cell differentiation. Genome Res. 2011;21:95–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Aguayo-Mazzucato C, Zavacki AM, Marinelarena A, Hollister-Lock J, El Khattabi I, Marsili A, et al. Thyroid hormone promotes postnatal rat pancreatic β-cell development and glucose-responsive insulin secretion through MAFA. Diabetes. 2013;62:1569–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gu C, Stein GH, Pan N, Goebbels S, Hörnberg H, Nave K-A, et al. Pancreatic beta cells require NeuroD to achieve and maintain functional maturity. Cell Metab. 2010;11:298–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Taylor BL, Liu F-F, Sander M. Nkx6.1 is essential for maintaining the functional state of pancreatic beta cells. Cell Rep. 2013;4:1262–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mulukutla BC, Gramer M, Hu WS. On metabolic shift to lactate consumption in fed-batch culture of mammalian cells. Metab Eng. 2012;14:138–49. [DOI] [PubMed] [Google Scholar]
- 45.Mulukutla BC, Khan S, Lange A, Hu W-S. Glucose metabolism in mammalian cell culture: new insights for tweaking vintage pathways. Trends Biotechnol. 2010;28:476–84. [DOI] [PubMed] [Google Scholar]
- 46.Levine AJ, Puzio-Kuter AM. The control of the metabolic switch in cancers by oncogenes and tumor suppressor genes. Science. 1979;2010(330):1340–4. [DOI] [PubMed] [Google Scholar]
- 47.Cairns RA, Harris IS, Mak TW. Regulation of cancer cell metabolism. Nat Rev Cancer. 2011;11:85–95. [DOI] [PubMed] [Google Scholar]
- 48.Newsholme P, Brennan L, Bender K. Amino acid metabolism, β-cell function, and diabetes. Diabetes. 2006;55.
- 49.Iworima DG, Baker RK, Ellis C, Sherwood C, Zhan L, Rezania A, et al. Metabolic switching, growth kinetics and cell yields in the scalable manufacture of stem cell-derived insulin-producing cells. Stem Cell Res Ther. 2024;15:1. 10.1186/s13287-023-03574-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Veres A, Faust AL, Bushnell HL, Engquist EN, Kenty JH-R, Harb G, et al. Charting cellular identity during human in vitro β-cell differentiation. Nature. 2019;569:368–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Helman A, Cangelosi AL, Davis JC, Pham Q, Rothman A, Faust AL, et al. A nutrient-sensing transition at birth triggers glucose-responsive insulin secretion. Cell Metab. 2020;31:1004-1016.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Alvarez-Dominguez JR, Melton DA. Cell maturation: hallmarks, triggers, and manipulation. Cell. 2022; 235–49. [DOI] [PMC free article] [PubMed]
- 53.Iworima DG, Rieck S, Kieffer TJ. Process parameter development for the scaled generation of stem cell-derived pancreatic endocrine cells. Stem Cells Transl Med. 2021;10:1459–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ghorbani-Dalini S, Azarpira N, Sangtarash MH, Urbach V, Yaghobi R, Soleimanpour-Lichaei HR, et al. Optimization of 3D islet-like cluster derived from human pluripotent stem cells: an efficient in vitro differentiation protocol. Gene. 2022;845. [DOI] [PubMed]
- 55.Pisania A, Weir GC, O’Neil JJ, Omer A, Tchipashvili V, Lei J, et al. Quantitative analysis of cell composition and purity of human pancreatic islet preparations. Lab Invest. 2010;90:1661–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shapiro AMJ. Islet transplantation in type 1 diabetes: ongoing challenges, refined procedures, and long-term outcome. Rev Diabet Stud. 2012;9:385–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Docherty K, Bernardo AS, Vallier L. Embryonic stem cell therapy for diabetes mellitus. Semin Cell Dev Biol. 2007;18:827–38. [DOI] [PubMed] [Google Scholar]
- 58.Lock LT, Tzanakakis ES. Stem/progenitor cell sources of insulin-producing cells for the treatment of diabetes. Tissue Eng. 2007;13:1399–412. [DOI] [PubMed] [Google Scholar]
- 59.Rabinowitz JD, Enerbäck S. Lactate: the ugly duckling of energy metabolism. Nat Metab. 2020;2:566–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cliff TS, Dalton S. Metabolic switching and cell fate decisions: implications for pluripotency, reprogramming and development. Curr Opin Genet Dev. 2017;46:44–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kropp C, Kempf H, Halloin C, Robles-Diaz D, Franke A, Scheper T, et al. Impact of feeding strategies on the scalable expansion of human pluripotent stem cells in single-use stirred tank bioreactors. Stem Cells Transl Med. 2016;5:1289–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hakim F, Kaitsuka T, Raeed JM, Wei F-Y, Shiraki N, Akagi T, et al. High oxygen condition facilitates the differentiation of mouse and human pluripotent stem cells into pancreatic progenitors and insulin-producing cells. J Biol Chem. 2014;289:9623–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Requests for further information or more detailed protocols should be directed to and will be fulfilled by the corresponding author. This study did not generate new unique reagents. All the RNA-Seq data are deposited in NCBI’s Gene Expression Omnibus (GEO) dataset with the accession number GSE264176. All the sequencing data sets are publically availiable.