Abstract
Background: Distinguishing latent tuberculosis infection (LTBI) from active tuberculosis (ATB) is very important. This study aims to analyze cases from multiple cohorts and get the signature that can distinguish LTBI from ATB. Methods: Thirteen datasets were downloaded from the gene expression omnibus (GEO) database. Three datasets were selected as discovery datasets, and the hub genes were discovered through WGCNA. In the training cohort, we use machine learning to establish the signature, verify the authentication ability of the signature in the remaining datasets, and compare it with other signatures. Cluster analysis was carried out on ATB cases, immune cell infiltration analysis, GSVA analysis, and drug sensitivity analysis were carried out on different clusters. Results: In the discovery datasets, we discovered five hub genes. A signature (SLC26A8, ANKRD22, and FCGR1B) is obtained in the training cohort. In the total cohort, the three-gene signature can separate LTBI from ATB (the total area under ROC curve (AUC) is 0.801, 95% CI 0.771–0.830). Compared with other author’s signatures, our signature shows good identification ability. Immunological analysis showed that SLC26A8, ANKRD22, and FCGR1B were closely related to the infiltration of immune cells. According to the expression of the three genes, ATB can be divided into two clusters, which are different in immune cell infiltration analysis, gene set variation, and drug sensitivity. Conclusion: Our study produced an immune-related three-gene signature to distinguish LTBI from ATB, which may help us to manage and treat tuberculosis patients.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-80072-3.
Keywords: Latent tuberculosis infection, Active tuberculosis, Molecule subtypes, Immune infiltration, Signature
Subject terms: Diagnostic markers, Tuberculosis
Introduction
It is estimated that about a quarter of the world’s population is infected with Mycobacterium tuberculosis (MTB). After being infected with MTB, the risk of tuberculosis is the highest in the first two years (about 5%), and then it is much lower1. Latent tuberculosis infection (LTBI) refers to the persistent immune response to MTB antigen, but there is no clinical evidence of ATB. The diagnosis of LTBI lacks the gold standard2,3.
In the Stop TB Strategy, the World Health Organization proposes to “stop the global tuberculosis epidemic” in 2035 4. However, the current tuberculosis epidemic has not shown any trend of being contained. Developing a new tuberculosis vaccine, finding biomarkers to distinguish LTBI from ATB, and developing drugs to treat drug-resistant tuberculosis are crucial in controlling the epidemic5.
MTB can survive in the host in a dormant state for a long time, and it can also be reactivated and progress to ATB. The development from LTBI to ATB is a series of heterogeneous processes with specific microbiological and immunological characteristics6. Among them, the change of host immune state is the initial factor. Studying the host immune mechanism and then implementing preventive anti-tuberculosis treatment for high-risk populations is of great significance for reducing the incidence of tuberculosis. In addition, understanding the characteristics of immune function induced by MTB is also helpful in finding biomarkers to predict the progress of LTBI to ATB and developing immunotherapy drugs for tuberculosis7.
Although the immune response to MTB is mainly concentrated in the lungs, the circulating immune cells in peripheral blood determine the pathological state of tuberculosis infection. Whole blood transcriptome analysis can not only help us to determine the molecular components of potential infection but also help us to understand the host immune response in tuberculosis8. The developed signature of blood transcriptome, which can distinguish LTBI from ATB, and analyze the immune status of different ATB may help us to evaluate the risk of tuberculosis reactivation, and then carry out preventive treatment for high-risk infected people, which is very important for reducing the incidence of tuberculosis and controlling the epidemic situation of tuberculosis.
The datasets in the public database provide us with the possibility of investigating molecular patterns through bioinformatics. Studies have shown that the ability of single-gene biomarkers to predict diseases is insufficient, and a signature containing several genes may be a better choice9. Therefore, we use meta-analysis to integrate transcriptome datasets from different studies and screen biomarkers that distinguish LTBI from ATB.
Materials and methods
Experimental design
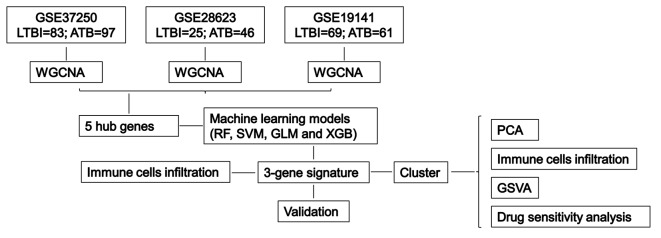
The experimental design flow chart is shown in Fig. 1.
Fig. 1.
Flow chart of finding immune-related signature through multi-cohort study.
Data source
The dataset included in this study was downloaded from GEO database (http://www.ncbi.nlm.nih.gov/geo). The downloaded dataset meets the following criteria: (1) The cases contained in the dataset are mRNA expression data of blood. (2) The specimens contain cases of LTBI and ATB. (3) ATB cases were collected before treatment. (4) The patient’s age is over 15 years old. (5) HIV negative. Based on these four standards, 13 datasets were included, and we excluded the GSE19444 dataset because it is a subset of the GSE19491 dataset. To better understand racial disparities in TB, the specimens of these datasets come from eight different countries and regions, including high- and medium-incidence regions. The details of datasets are shown in Table 1 and supplement Table 1.
Table 1.
The detailed information of 13 datasets.
| Dataset | Platforms | Country | Submission date | Last update date | Source name | LTBI | ATB | |
|---|---|---|---|---|---|---|---|---|
| Train | GSE19491 | GPL6947 | USA | 2009.02.15 | 2018.08.16 | whole blood | 69 | 61 |
| GSE28623 | GPL4133 | Germany | 2011.04.14 | 2018.02.22 | whole blood | 25 | 46 | |
| GSE37250 | GPL10558 | United Kingdom | 2012.04.13 | 2020.11.30 | whole blood | 83 | 97 | |
| Test | GSE144127 | GPL10558 | United Kingdom | 2021.10.23 | 2021.03.10 | whole blood | 13 | 101 |
| GSE152532 | GPL10558 | USA | 2020.06.18 | 2021.12.09 | whole blood | 111 | 25 | |
| GSE39939 | GPL10558 | United Kingdom | 2012.08.07 | 2018.08.13 | whole blood | 79 | 14 | |
| GSE39940 | GPL10558 | United Kingdom | 2012.08.07 | 2018.08.13 | whole blood | 54 | 70 | |
| GSE40553 | GPL10558 | United Kingdom | 2012.09.03 | 2018.08.13 | whole blood | 38 | 35 | |
| GSE54992 | GPL570 | China | 2014.02.13 | 2020.11.30 | whole blood | 6 | 9 | |
| GSE62525 | GPL16951 | Taiwan | 2014.10.20 | 2019.04.10 | whole blood | 14 | 14 | |
| NO FCGR1B | GSE25534 | GPL1708 | Germany | 2010.11.22 | 2014.05.14 | whole blood | 21 | 23 |
| Not enough genes | GSE74092 | GPL21040 | Germany | 2015.10.16 | 2015.12.31 | whole blood | 56 | 113 |
| subset | GSE19444 | GPL6947 | USA | 2009.12.12 | 2018.08.16 | whole blood | 21 | 21 |
We selected GSE19491, GSE28623, and GSE37250 for WGCNA analysis and merged three datasets into a training cohort, in which there were 177 LTBI and 204 cases of ATB. GSE144127, GSE152532, GSE39939, GSE39940, GSE40553, GSE54992, and GSE62525 were merged into testing cohort. There are 315 cases of LTBI and 268 cases of ATB in testing cohort. The training and testing cohorts merged into the total cohort, including 492 LTBI and 490 cases of ATB.
There is no expression of FCGR1B in GSE25534 dataset, and there is no expression of FCGR1B and SLC26A8 in GSE74092 dataset, so these two datasets are not merged into the total cohort, but verified separately.
Weighted correlation network analysis (WGCNA)
WGCNA can be used to find modules with highly correlated genes, and the characteristic gene network method is used to associate modules with each other, correlate the module with the external sample traits, and calculate the module member metrics. The relevance network promotes the network-based gene screening method, which can be used to identify candidate biomarkers or therapeutic targets10. In the datasets of GSE19491, GSE28623, and GSE37250, the top 25% genes with the highest variance are used for WGCNA analysis to ensure the accuracy of quality results, and the “pickSoftThreshold” function is used to select the best soft threshold (β). Then a weighted adjacency matrix is constructed. Using the TOM dissimilarity measure (1-TOM) based on a hierarchical clustering tree algorithm, the module has more than 50 genes. Each module is assigned a random color. Modular characteristic genes represent the overall gene expression profile of each module. Gene significance (GS) indicates the correlation between gene and clinical phenotype. The relationship between module and disease state is reflected by module significance (MS). Genes with critical values of MS > 0.8 and GS > 0.5 in the significant module were identified as key genes.
Determination of key genes
We use the “veen” package of R language to analyze the intersection genes of key genes in GSE19491, GSE28623 and GSE37250, and identify the intersection genes as hub genes. We use the “sva” software package to merge the datasets and conduct standardized analysis.
Construction of predictive model based on multiple machine learning algorithms
In the training cohort, we use the R language package “caret” to build machine learning models, including random forest (RF), support vector machine (SVM), generalized linear model (GLM), and extreme gradient lifting (XGB). We use “pROC” to visualize the AUC; then, we use “DALEX” to explain these four models. Finally, we draw the residual distribution of each model11.
To verify whether the machine learning model can distinguish between ATB and LTBI, we analyzed the ROC curves of signatures in each dataset: training cohort, testing cohort, and total cohort. We also compared our signature with other signatures in the total cohort 12–23.
In order to verify whether the machine learning model can distinguish LTBI from ATB in HIV-positive patients, we further verify it in the GSE37250 dataset. In the GSE37250 dataset, there are 84 cases of HIV-positive LTBI patients and 98 cases of HIV-positive ATB patients.
Immune infiltration analysis
CIBERSORT is an immunoassay tool developed by Stanford University. It was published in Nature Methods in 2015 and has been cited nearly 6000 times24. CIBERSORT performs deconvolution analysis based on the linear support vector regression principle and provides gene expression feature sets of 22 immune cell subtypes25.
Clustering analysis of ATB cases
Based on the expression of genes in the machine learning model, we used the R package “ConsensusClusterPlus” for unsupervised clustering analysis26. We use the k-means algorithm to classify 472 ATB cases into clusters and make 1,000 iterations on the clusters. The value of k is defined from 1 to 9 to generate different subtypes. We choose the best number of clusters according to the clustering consensus score. We use principal component analysis (PCA) to visualize the distribution of clusters.
Gene set variation analysis (GSVA)
We use the R package “GSVA” to analyze GSVA enrichment and clarify the differences in enrichment gene sets among different clusters. Gene matrix transposition (gmt) files including “c2.cp.kegg.symbols.gmt” and “c5.go.symbols.gmt” were downloaded from the database of MSigDB website for further GSVA analysis27. We use the R package “limma” to identify the pathways and biological functions of differential expression by comparing GSVA scores among different clusters. P value < 0.05 was statistically significant.
Screening of potential therapeutic drugs
We then used the “pRRophetic” R package to assess the treatment response, as indicated by the IC50 of targeted and immunotherapy drugs28.
Statistical analysis
We use R language (version 4.1.3) for statistical analysis. Spearman correlation calculated by the “cor.test” function was used to analyze the relationship between the expression genes and immune cells. P value < 0.05 was statistically significant.
Results
Identification of key gene by WGCNA
In GSE37250 dataset, there were 83 cases of LTBI and 97 cases of ATB (HIV negative). We selected the top 25% genes (7853 genes) with significant fluctuation for WGCNA analysis. When the soft threshold is 2, we establish a scale-independent topological network. The correlation analysis between MS value and sample traits found that the red module was most closely related to the progress from LTBI to ATB (cor = 0.89, p = 3e-33). Under the precise cut-off criteria (MS > 0.8 and GS > 0.5), seven genes from the central module were identified as important genes (Fig. 2A and B; Supplement Fig. 1).
Fig. 2.
Evaluating the module-trait association through the correlation between the module characteristic genes and the sample traits in GSE37250 (A), GSE28623 (C), and GSE19141 (E) dataset. B. The gene scatter plot in the red module in the GSE37250 dataset. D. The gene scatter plot in the yellow module in the GSE28623 dataset. F. The gene scatter plot in the brown module in the GSE19141 dataset. G. The intersection gene of important genes in three datasets. H. The expression of hub genes in LTBI and ATB cases in the training cohort. I. The expression of hub genes in LTBI and ATB cases in testing cohort. J. ROC curve analyses for SLC26A8, CD274, FCGR1B, SERPING1, and ANKRD22 in the training cohort. K. Residual distribution of each machine learning model. L. ROC analysis of four machine learning models in the training cohort.
In GSE28623 dataset, there are 25 cases of LTBI and 46 cases of ATB. The top 25% genes (4924 genes) with significant fluctuation were selected for WGCNA analysis. We establish a scale-independent topological network when the soft threshold is 4. The analysis shows that the yellow module is most closely related to the progress from LTBI to ATB (cor = 0.76, p = 1.9e-104). 113 genes from the central module were identified as important genes (Fig. 2C and D; Supplement Fig. 2).
In GSE19141 dataset, there are 69 cases of LTBI and 61 cases of ATB, and there are 6228 genes in the top 25% with significant fluctuation. WGCNA analyzes these genes. When the soft threshold is 1, we establish a scale-independent topological network. It is found that the brown module is most closely related to the progress from LTBI to ATB (cor = 0.92, p = 1.8e-168). According to the criteria of MS > 0.8 and GS > 0.5, we selected 16 genes from the central module as important genes (Fig. 2E and F; Supplement Fig. 3).
Determination of hub genes
We used the “veen” package to analyze whether the important genes of GSE19491, GSE28623, and GSE37250 intersect. We found that SLC26A8, CD274, FCGR1B, SERPING1, and ANKRD22 are the intersection genes of three datasets, and we identified these genes as hub genes. (Fig. 2G). In the training cohort, compared with LTBI patients, the expression of hub genes in ATB patients increased significantly (Fig. 2H), and similar findings were also observed in the testing dataset (Fig. 2I). ROC curve analysis of training datasets shows that the AUC values of SLC26A8, CD274, FCGR1B, SERPING1, and ANKRD22 are 0.745, 0.750, 0.798, 0.769, and 0.783, respectively (Fig. 2J).
Construction of machine learning models
In training cohort, we constructed four machine learning models based on the expression of hub genes, including RF, SVM, GLM and XGB. It is found that the residuals of SVM and RF machine learning models are low (Fig. 2K). The AUC of the four models is 0.918 for RF, 0.924 for SVM, 0.905 for XGB and 0.904 for GLM, respectively (Fig. 2L). Based on residual and AUC, the SVM machine learning model best distinguishes LTBI from ATB. We select the three-gene signature in the SVM model (SLC26A8, ANKRD22, and FCGR1B) for further analysis.
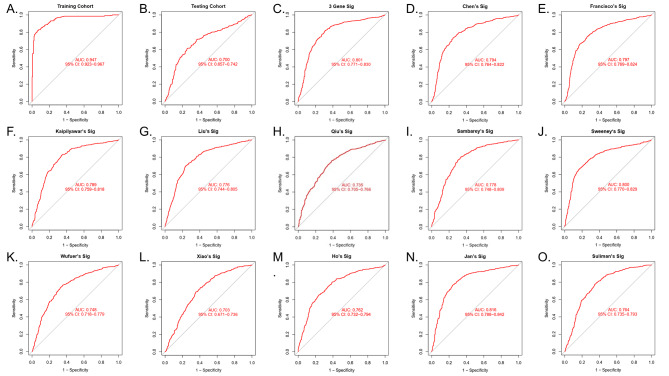
ROC analysis showed that the AUC of the three-gene signature was 0.947 in the training cohort (Fig. 3A), 0.700 in the testing cohort (Fig. 3B), and 0.801 (Fig. 3C) in the total cohort.
Fig. 3.
To verify the diagnostic value of the three-gene signature by ROC analysis in the training set (A), testing cohort (B), and total cohort (C). D-O. In the total cohort, the ability of other signatures to distinguish LTBI from ATB.
In the total cohort, we compared the identification potential of the three-gene signature with other signatures. The AUC curve shows that except for Jan’s Sig (22-gene signature), the ability of the three-gene signature to distinguish LTBI from ATB is not inferior to other models (Fig. 3D-O).
Table 2 shows the AUC of the three-gene signature in each dataset. The GSE25534 dataset does not have FCGR1B gene expression, and the AUC of SLC26A8 and ANKRD22 in differentiating LTBI from ATB is 0.988 (95% CI: 0.961-1.000). The platform file GPL21040 of GSE74092 data set only has 281 genes expressed (excluding FCGR1B and SLC26A8), so no further analysis was conducted.
Table 2.
In each dataset, the diagnostic value of the three-gene signature was verified by ROC analysis.
| Dateset | AUC | 95% CI |
|---|---|---|
| GSE19491 | 0.945 | 0.899–0.979 |
| GSE28623 | 0.910 | 0.832–0.970 |
| GSE37250 | 0.972 | 0.947–0.992 |
| GSE152532 | 0.685 | 0.547–0.817 |
| GSE39940 | 0.879 | 0.815–0.934 |
| GSE40553 | 0.973 | 0.938–0.995 |
| GSE39939 | 0.847 | 0.740–0.932 |
| GSE144127 | 0.851 | 0.743–0.931 |
| GSE62525 | 0.980 | 0.918-1.000 |
| GSE54992 | 1.000 | 1.000–1.000 |
| GSE25534 | 0.990 | 0.961-1.000 |
| GSE19444 | 0.943 | 0.886–0.995 |
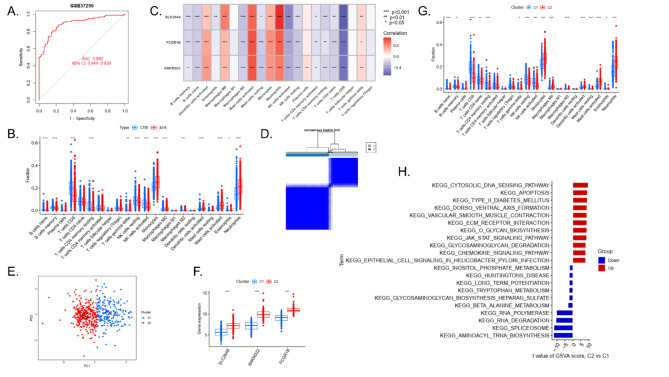
Among HIV-positive cases in GSE37250 dataset, the AUC of identifying LTBI and ATB by the three-gene signature is 0.892 (95% CI: 0.844–0.938) (Fig. 4A).
Fig. 4.
A. The AUC curve of the three-gene signature in HIV positive cases. B. Difference of immune cell infiltration between LTBI and ATB. C. Correlation analysis between SLC26A8, ANKRD22, FCGR1B, and infiltrated immune cells. D. The cluster number is most stable when the k value is set to 2. E. PCA showed significant differences between the two clusters. F. SLC26A8, ANKRD22, and FCGR1B expression in C1 and C2 cases in total cohort. G. Difference of immune cell infiltration between C1 and C2. H. The difference in biological function between C1 and C2 was analyzed by the GSVA method.
Immune cell infiltration in LTBI and ATB
CIBERSORT was used to analyze the difference of immune cell infiltration between LTBI and ATB. It was found that the expressions of B cells naïve, B cells memory, T cells CD8, T cells CD4 memory resting, NK cells resting, NK cells activated, macrophages M2 and eosinophils increased in LTBI. The expression of T cells gamma delta, monocytes, macrophages M0, dendritic cells activated, mast cells activated and neutrophils increased in ATB (Fig. 4B).
We used spearman correlation to analyze the correlation between FCGR1B, SLC26A8, and ANKRD22 and immune cell infiltration. The study showed that the expression of FCGR1B, SLC26A8, and ANKRD22 was positively correlated with the levels of dendritic cells activated, macrophages M0, mast cells activated, monocytes and neutrophils. It was negatively correlated with the expression levels of B cells naïve, B cells memory, macrophages M2, mast cells resting, NK cells activated, NK cells resting, T cells CD4 memory activated, T cells CD4 memory resting and T cells CD4 naïve (Fig. 4C).
Clustering analysis of ATB cases
In the total cohort, based on the expressions of SLC26A8, ANKRD22, and FCGR1B, we use a consensus clustering algorithm to cluster 472 ATB cases. When the value of k is set to 2(k = 2), the optimal number of clusters is observed (Fig. 4D). PCA analysis showed that 472 patients with tuberculosis could be well divided into the cluster 1 (C1, n = 189) and the cluster 2 (C2, n = 283) (Fig. 4E).
Compared with C1, the expression of three genes in C2 is higher (Fig. 4F).
Through the analysis of immune cell infiltration, we found that there were significant differences in immune state between C1 and C2 (Fig. 4G). In C1, B cells naïve, B cells memory, T cells CD8, T cells CD4 naïve, T cells CD4 memory resting, T cells gamma delta, NK cells resting, NK cells activated, macrophages M2, the expressions of mast cells resting and eosinophils increased significantly (Fig. 4G). In C2, the expressions of monocytes, macrophages M0, dendritic cells activated, mast cells activated and neutrophils increased significantly (Fig. 4G).
GSVA analysis results
We identified the pathway activity and biological function related to each cluster by GSVA. Functional enrichment analysis showed that inositol phosphate metabolism, Huntingtons disease, long-term potentiation, tryptophan metabolism, Glycosaminoglycan biosynthesis - heparan sulfate, beta-Alanine metabolism, RNA polymerase, RNA degradation, spliceosome and aminoacyl-tRNA biosynthesis were significantly enriched in C1 (Fig. 4H).
In contrast, Cytosolic DNA-sensing pathway, apoptosis, type II diabetes mellitus, dorsoventral axis formation, vascular smooth muscle cell, EMC receptor interaction, O glycan biosynthesis, JAK-STAT signaling pathway, glycosaminoglycan degradation, chemokine signaling pathway and epidermal cell signaling in helicobacter infection were enriched in C2 (Fig. 4H).
Screening of potential therapeutic drugs
We have identified 35 kinds of immunotherapy or targeted drugs, and the IC50 values of these drugs in C2 are lower than those in C1, including Bicalutamide, CGP.60,474, KU.55,933, Parthenolide and NSC.87,877(Fig. 5A-E, Supplement Table 2). We also identified 62 drugs with IC50 values lower than C2 in C1, including AZD8055, Camptothecin, Lenalidomide, Metformin and Metformin (Fig. 5F-J, Supplement Table 2).
Fig. 5.
Drug sensitivity analysis. A-E showed that the drug had a lower IC50 value in the C2 group. F-J showed that the drug had a lower IC50 value in the C1 group.
Discussion
Although the tuberculin skin test (TST) and the interferon gamma release assays (IGRA) have been widely used in LTBI screening among students and special people with immunosuppression, and in clinical auxiliary diagnosis of ATB disease, TST and IGRA are of limited value in distinguishing ATB cases from LTBI29.
MTB infection is a dynamic process. Host and bacteria are the main factors that induce LTBI to develop into ATB, and the change of host immune state is the most direct reason for LTBI to develop into ATB. The immunological mechanism of tuberculosis infection is very complicated, and innate immunity and adaptive immunity are very important in maintaining the state of latent infection. In innate immunity, alveolar macrophages, dendritic cells, neutrophils, and natural killer cells play an important role in tuberculosis infection. Adaptive immunity includes cellular immunity and humoral immunity. Cellular immunity mediated by T cells is the main immune mechanism of anti-TB6. It was correlated with the expression of many immune cells. However, we should have elaborated on the dynamic changes of the host immune state from normal to LTBI and then to ATB, this is not the main purpose of this study.
Our study found that SLC26A8, CD274, FCGR1B, SERPING1 and ANKRD22 may be hub genes for LTBI to develop into ATB. Among these five genes, the interesting one is the CD274 gene. In some literatures focusing on the study of tuberculosis signature, it is found that CD274 may play an important role in the process of tuberculosis activity 30,31. Many studies show that anti-PD-(L)1(CD274) therapy may lead to ATB32. However, some studies have shown that patients with both malignant tumors and tuberculosis benefit from anti-PD-(L)1 treatment, while the anti-tuberculosis response of ATB patients is not affected. It is worth noting that the combination of anti-PD-(L)1(CD274) and anti-tuberculosis therapy is well tolerated and has no obvious accidental toxicity33. Therefore, the effects of CD274 and anti-PD-(L)1 therapy on ATB need further study. Compared with the normal control, the expression of SERPING1 in tuberculosis patients increased continuously, and its coding protein C1-INH was positively correlated with the level of complement C1q. In tuberculosis patients, the increase of C1-INH and C1q may represent the immune escape mechanism of Mtb34. However, no more research exists on the correlation between SLC26A8, FCGR1B, and ANKRD22 and tuberculosis.
It is of great significance to find biomarkers to distinguish LTBI from ATB for the prevention and treatment of tuberculosis35. Relying on a single biomarker to distinguish ATB disease from LTBI may not achieve ideal results, but the combination of multiple biomarkers may bring new hope. The prospective study conducted by Sweeney et al. shows that the mRNA models based on three genes (GBP5, DUSP3, KLF2) have a reasonable correlation with the development of LTBI into ATB, but its exact value needs more research21. In distinguishing the predictive ability of LTBI and ATB, the three-gene signature is not lower than Sweeney’s signature. In addition to Sweeney’s signature, researchers also reported some other signatures. Except for Jan’s signature15, the discrimination ability of the 3-gene model is not lower than that of other models, and the number of genes is also the lowest. At the same time, our gene model also showed stable discrimination performance in HIV-positive patients.
The three-gene model can distinguish LTBI from ATB and is closely related to immune cells infiltration. According to the expression of three genes, ATB can be well divided into two clusters. These two clusters have different immune states. However, whether different immune states are related to the therapeutic effect of anti-tuberculosis drugs, the choice of therapeutic drugs and drug resistance needs further study. GSVA analysis showed that C1 was enriched in metabolism-related pathways. In contrast, C2 was mainly enriched in apoptosis, JAK-STAT signaling pathway and immune-related pathways, suggesting that C1 and C2 might differ in tuberculosis infection, immunity and treatment, and drug sensitivity analysis verified our conjecture.
Although chemotherapy should always be the primary treatment for tuberculosis at present, immunotherapy can only be used as an auxiliary treatment method, and clinicians lack understanding of immunotherapy for ATB. Tuberculosis immunotherapy includes immunoactive substances, TB therapeutic vaccines, chemical agents, cellular therapy, etc. Immunoactive substances include cytokines such as IL-2 and GM-CSF and immune blockers such as IL-4, denileukin diftitox, and PD-1 blockers36. Immunotherapy, such as cytokines and immune blockers, is limited to refractory pulmonary tuberculosis or in vitro experiments, including IL-237, GM-CSF38, and PD-1 blockers39. However, these drugs have not been identified as biomarker indications, which need improvement. Moreover, unlike the pathological diagnosis of tumors, we do not detect pathological immune responses in the pathological diagnosis of tuberculosis patients, which also needs improvement. The only immunotherapy for HIV-positive patients is TB therapeutic vaccines. Whether the immune change process between LTBI and ATB is similar to that of HIV-positive patients needs further study. Generally speaking, immunotherapy for tuberculosis still has great potential, especially for drug-resistant tuberculosis.
In this study, we included all the datasets containing LTBI and ATB in the GEO database. Compared with previous studies, we included the largest number of LTBI cases (492 cases of LTBI). Of course, our research also has limitations. First of all, we did not include the normal sample. There are reliable and economical methods to distinguish normal from LTBI, such as TST, and IGRAs35. The focus of this study is how to identify LTBI and ATB, so we did not include normal cases. Moreover, if too many datasets are included, the expression of some genes may be missing during the batch removal process. A total of 10 datasets were included in the total cohort, while six datasets used the GPL10558 platform, which makes the analysis results more reliable. Secondly, the three-gene signature can distinguish LTBI from ATB, but whether it can identify high-risk LTBI still needs further study. Third, our research results need to be further verified in a larger multi-center study. Because the number of specimens we included is large enough, we have verified it not only in the testing and total cohorts but also in various datasets. To some extent, it makes up for this defect. Finally, the effectiveness of our signature in children, tumors, rheumatic immune system diseases and patients undergoing immunotherapy needs further verification.
Conclusion
Generally speaking, our research has found a three-gene signature that can not only distinguish LTBI from ATB but also be closely related to tuberculosis immunity. The results of this study may be helpful to the clinical management of tuberculosis patients and guide the testing of new drugs in clinical trials, but they still need further verification.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
The script for R language and Perl is provided by biowolf_cn (https://mp.weixin.qq.com/s/kuTGlMFRlbUN63FL4XXXAg).
Abbreviations
- ANKRD22
Ankyrin Repeat Domain 22
- ATB
Active tuberculosis
- AUC
The total area under ROC curve
- C1
Cluster 1
- C2
Cluster 2
- CD274
Programmed Cell Death 1 Ligand 1(PD-L1)
- DUSP3
Dual Specificity Phosphatase 3
- FCGR1B
Fc Gamma Receptor Ib
- GBP5
Guanylate Binding Protein 5
- GEO
The gene expression omnibus
- GLM
Generalized linear model
- GS
Gene significance
- GSVA
Gene set variation analysis analysis
- IGRA
Interferon gamma release assays
- KLF2
KLF Transcription Factor 2
- LTBI
Latent tuberculosis infection
- MS
Module significance
- MTB
Mycobacterium tuberculosis
- RF
Random forest
- SERPING1
Serpin Family G Member 1
- SLC26A8
Solute Carrier Family 26, Member 8
- SVM
Support vector machine
- TST
Tuberculin skin test
- WGCNA
Weighted correlation network analysis
- XGB
Extreme gradient lifting
Author contributions
Qingqing Shan and Yangke Li analyzed and interpreted the data and was a major contributor to writing the manuscript. They contributed equally to this work and should be considered co-first authors. Kun Yuan, Xiao Yang, Li Yang, and Jian-Qing He reviewed the manuscript. All authors read and approved the final manuscript.
Funding
No funding.
Data availability
Data is provided within the manuscript or supplementary information files.
Declarations
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Ethical approval and consent to participate
Not applicable.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Qingqing Shan and Yangke Li contributed equally to this work and should be considered co-first authors.
References
- 1.Floyd, K., Glaziou, P., Zumla, A. & Raviglione, M. The global tuberculosis epidemic and progress in care, prevention, and research: an overview in year 3 of the end TB era. Lancet Respir Med.6, 299–314. 10.1016/s2213-2600(18)30057-2 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Lewinsohn, D. M. et al. Official American Thoracic Society/Infectious Diseases Society of America/Centers for Disease Control and Prevention Clinical Practice Guidelines: diagnosis of tuberculosis in adults and children. Clin. Infect. Dis.64, e1–e33. 10.1093/cid/ciw694 (2016). [DOI] [PubMed] [Google Scholar]
- 3.In WHO consolidated guidelines on tuberculosis: Module 2: screening – systematic screening for tuberculosis disease (World Health Organization© World Health Organization 2021). (2021). [PubMed]
- 4.Goletti, D. et al. World Tuberculosis Day 2024 theme yes! We can end TB can be made a reality through concerted global efforts that advance detection, diagnosis, and treatment of tuberculosis infection and disease. Int. J. Infect. Dis.141s, 106993. 10.1016/j.ijid.2024.106993 (2024). [DOI] [PubMed] [Google Scholar]
- 5.Organization, W. H. Global tuberculosis report 2023. Geneva: World Health Organ. (2023).
- 6.Ernst, J. D. The immunological life cycle of tuberculosis. Nat. Rev. Immunol.12, 581–591. 10.1038/nri3259 (2012). [DOI] [PubMed] [Google Scholar]
- 7.Li, L. S. et al. From immunology to artificial intelligence: revolutionizing latent tuberculosis infection diagnosis with machine learning. Mil Med. Res.10, 58. 10.1186/s40779-023-00490-8 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang, Z., Gerstein, M. & Snyder, M. RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet.10, 57–63. 10.1038/nrg2484 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang, Q. W. et al. Prognostic correlation of autophagy-related gene expression-based risk signature in patients with Glioblastoma. Onco Targets Ther.13, 95–107. 10.2147/ott.S238332 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Langfelder, P. & Horvath, S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinform.9, 559. 10.1186/1471-2105-9-559 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li, S. et al. Identification of immune infiltration and cuproptosis-related molecular clusters in tuberculosis. Front. Immunol.14, 1205741. 10.3389/fimmu.2023.1205741 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen, L., Hua, J. & He, X. Coexpression network analysis-based identification of critical genes differentiating between latent and active tuberculosis. Dis. Markers. 2022 (2090560). 10.1155/2022/2090560 (2022). [DOI] [PMC free article] [PubMed]
- 13.Francisco, N. M. et al. Diagnostic accuracy of a selected signature gene set that discriminates active pulmonary tuberculosis and other pulmonary diseases. J. Infect.75, 499–510. 10.1016/j.jinf.2017.09.012 (2017). [DOI] [PubMed] [Google Scholar]
- 14.Ho, J. et al. A transcriptional blood signature distinguishes early tuberculosis disease from latent tuberculosis infection and uninfected individuals in a Vietnamese cohort. J. Infect.81, 72–80. 10.1016/j.jinf.2020.03.066 (2020). [DOI] [PubMed] [Google Scholar]
- 15.Heyckendorf, J. et al. Prediction of anti-tuberculosis treatment duration based on a 22-gene transcriptomic model. Eur. Respir J.5810.1183/13993003.03492-2020 (2021). [DOI] [PMC free article] [PubMed]
- 16.Kaipilyawar, V. et al. Development and validation of a parsimonious tuberculosis gene signature using the digital NanoString nCounter platform. Clin. Infect. Dis.75, 1022–1030. 10.1093/cid/ciac010 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu, Y. et al. Identification and validation of a pyroptosis-related signature in identifying active tuberculosis via a deep learning algorithm. Front. Cell. Infect. Microbiol.13, 1273140. 10.3389/fcimb.2023.1273140 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qiu, Q. et al. Diagnosis of pulmonary tuberculosis via identification of core genes and pathways utilizing blood transcriptional signatures: a multicohort analysis. Respir Res.23, 125. 10.1186/s12931-022-02035-4 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sambarey, A. et al. Unbiased identification of blood-based biomarkers for pulmonary tuberculosis by modeling and Mining Molecular Interaction Networks. EBioMedicine15, 112–126. 10.1016/j.ebiom.2016.12.009 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Suliman, S. et al. Four-gene pan-african blood signature predicts progression to tuberculosis. Am. J. Respir Crit. Care Med.197, 1198–1208. 10.1164/rccm.201711-2340OC (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sweeney, T. E., Braviak, L., Tato, C. M. & Khatri, P. Genome-wide expression for diagnosis of pulmonary tuberculosis: a multicohort analysis. Lancet Respir Med.4, 213–224. 10.1016/s2213-2600(16)00048-5 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wufuer, D., Li, Y., Aierken, H. & Zheng, J. Bioinformatics-led discovery of ferroptosis-associated diagnostic biomarkers and molecule subtypes for tuberculosis patients. Eur. J. Med. Res.28, 445. 10.1186/s40001-023-01371-5 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xiao, S. et al. Identifying autophagy-related genes as potential targets for immunotherapy in tuberculosis. Int. Immunopharmacol.118, 109956. 10.1016/j.intimp.2023.109956 (2023). [DOI] [PubMed] [Google Scholar]
- 24.Newman, A. M. et al. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods. 12, 453–457. 10.1038/nmeth.3337 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen, B., Khodadoust, M. S., Liu, C. L., Newman, A. M. & Alizadeh, A. A. Profiling Tumor infiltrating Immune cells with CIBERSORT. Methods Mol. Biol.1711, 243–259. 10.1007/978-1-4939-7493-1_12 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wilkerson, M. D. & Hayes, D. N. ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics26, 1572–1573. 10.1093/bioinformatics/btq170 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hänzelmann, S., Castelo, R. & Guinney, J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinform.1410.1186/1471-2105-14-7 (2013). [DOI] [PMC free article] [PubMed]
- 28.Geeleher, P., Cox, N. & Huang, R. S. pRRophetic: an R package for prediction of clinical chemotherapeutic response from tumor gene expression levels. PLoS One. 9, e107468. 10.1371/journal.pone.0107468 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pai, M. & Behr, M. Latent < i > Mycobacterium tuberculosis infection and Interferon-Gamma release assays. Microbiol. Spectr.410.1128/microbiolspec.tbtb1122-0023-2016 (2016). [DOI] [PubMed]
- 30.Yang, Y. et al. Screening for diagnostic targets in tuberculosis and study on its pathogenic mechanism based on mRNA sequencing technology and miRNA-mRNA-pathway regulatory network. Front. Immunol.14, 1038647. 10.3389/fimmu.2023.1038647 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shi, T., Huang, L., Zhou, Y. & Tian, J. Role of GBP1 in innate immunity and potential as a tuberculosis biomarker. Sci. Rep.12, 11097. 10.1038/s41598-022-15482-2 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu, K. et al. Increased tuberculosis incidence due to Immunotherapy based on PD-1 and PD-L1 blockade: a systematic review and Meta-analysis. Front. Immunol.13, 727220. 10.3389/fimmu.2022.727220 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Su, S. et al. Assessment of anti-PD-(L)1 for patients with coexisting malignant tumor and tuberculosis classified by active, latent, and obsolete stage. BMC Med.19, 322. 10.1186/s12916-021-02194-z (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lubbers, R. et al. Expression and production of the SERPING1-encoded endogenous complement regulator C1-inhibitor in multiple cohorts of tuberculosis patients. Mol. Immunol.120, 187–195. 10.1016/j.molimm.2020.02.006 (2020). [DOI] [PubMed] [Google Scholar]
- 35.Gong, W. & Wu, X. Differential diagnosis of latent tuberculosis infection and active tuberculosis: a key to a successful tuberculosis control strategy. Front. Microbiol.12, 745592. 10.3389/fmicb.2021.745592 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mi, J. et al. The Research Progress in Immunotherapy of Tuberculosis. Front. Cell. Infect. Microbiol.11, 763591. 10.3389/fcimb.2021.763591 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Johnson, J. L. et al. Randomized trial of adjunctive interleukin-2 in adults with pulmonary tuberculosis. Am. J. Respir Crit. Care Med.168, 185–191. 10.1164/rccm.200211-1359OC (2003). [DOI] [PubMed] [Google Scholar]
- 38.Francisco-Cruz, A. et al. Efficacy of gene-therapy based on adenovirus encoding granulocyte-macrophage colony-stimulating factor in drug-sensitive and drug-resistant experimental pulmonary tuberculosis. Tuberculosis (Edinb). 100, 5–14. 10.1016/j.tube.2016.05.015 (2016). [DOI] [PubMed] [Google Scholar]
- 39.Singh, A., Mohan, A., Dey, A. B. & Mitra, D. K. Inhibiting the programmed death 1 pathway rescues Mycobacterium tuberculosis-specific interferon γ-producing T cells from apoptosis in patients with pulmonary tuberculosis. J. Infect. Dis.208, 603–615. 10.1093/infdis/jit206 (2013). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data is provided within the manuscript or supplementary information files.