Abstract
Purpose
The study evaluates the expression and functional significance of the Small RNA Binding Exonuclease Protection Factor La (SSB) gene in lung adenocarcinoma (LUAD). By utilizing 18F-fluorodeoxyglucose (18F-FDG) positron emission tomography/computed tomography (PET/CT) machines, we correlated SSB gene expression with PET/CT parameters, as well as its value in LUAD diagnosis.
Methods
Fifty-five patients with LUAD underwent 18F-FDG PET/CT imaging prior to pulmonary surgery. Metabolic parameters such as maximum standardized uptake values (SUVmax) were quantitatively calculated from the 18F-FDG PET/CT imaging data. The diagnostic value was compared with that of thyroid transcription factor 1 (TTF1, the current standard-of-care). Publicly procurable datasets from The Cancer Genome Atlas (TCGA) and Gene Expression Omnibus (GEO) were used to establish SSB gene expression patterns across diverse cancer types and specifically in LUAD, along with its associations with glycolysis and N6-methyladenosine (m6A) modification.
Results
SSB was highly expressed in LUAD compared to adjacent non-cancerous tissues. SSB additionally demonstrated superior diagnostic utility for LUAD compared to TTF1. The correlation between SSB and SUVmax as well as average standardized uptake values (SUVmean) was positive (P < 0.001), while TTF1 displayed a negative correlation with metabolic tumor volume (MTV) and total lesion glycolysis (TLG) (P < 0.05).
Conclusion
In LUAD, SSB expression correlated with high metabolic activity (SUV) on 18F-FDG PET/CT imaging. SSB is not only an important prognostic marker for lung cancer metastases, but may also represent a novel therapeutic target.
Keywords: SSB, LUAD, Glucose, Metabolic parameter, 18F-FDG PET/CT
1. Background
Lung cancer is the second most prevalent type of cancer worldwide, with a global incidence of 2.2 million cases per year and representing for 11.4 % of all cancer cases [1]. The primary histological subtypes of lung cancer comprise lung squamous cell carcinoma (LUSC), small cell carcinoma, lung adenocarcinoma (LUAD) and large cell carcinoma [2]. Recent investigations have revealed that LUAD has emerged as the most prevalent subtype of lung cancer worldwide in 2020, particularly in Eastern Asia (including China) among both men and women [3]. Despite the availability of treatment modalities such as chemotherapy, radiation therapy, and curative surgery, patients with LUAD face high mortality rates and have a poor prognosis [4,5]. The initiation and progression of LUAD involve a complex, multistep process involving aberrant genes expression. Hence, by comprehensively understanding of the molecular mechanisms underlying LUAD can unveil better biomarkers for the diagnosis and treatment [[6], [7], [8]].
18F-fluorodeoxyglucose (18F-FDG) positron emission tomography/computed tomography scan (PET/CT) is a non-invasive early diagnostic tool that is frequently utilized for cancer diagnosis, staging, treatment planning, and treatment monitoring [9,10]. PET/CT-related parameters are crucial in the assessment of tumor characteristics. These parameters, including maximum and average standardized uptake values (SUVmax and SUVmean), metabolic tumor volume (MTV), and total lesion glycolysis (TLG), provide valuable insights into the biological factors at play in the tumor microenvironment (TME) [11]. Previous research has identified significant correlations between the uptake of FDG and various biological characteristics of cancer, such as proliferation [12,13], tissue type [14], tumor differentiation [15], and hypoxia [16,17]. PET/CT imaging allows for visualization of the “Warburg effect” in which tumor cells exhibit a preference for aerobic glycolysis as their primary energy-producing pathway, even when there is no adequate oxygen supply [18]. Our previous data has highlighted the clinical significance of 18F-FDG PET/CT parameters in predicting clinical phenotypes and novel targeted molecular of malignancies. These phenotypes comprise the overexpression of NPM1 in lung cancer [19], eIF6 expression in esophageal carcinoma [20], and EIF2S2 expression in colorectal cancer [21].
The thyroid transcription factor 1 (TTF1) is a nuclear protein expressed in thyroid and lung tissues [22]. Pathologists use TTF1 to identify tissue origin. TTF1 expression is positively associated with tumor differentiation and may predict lung cancer survival [23]. Lupus La Protein, also known as Small RNA Binding Exonuclease Protection Factor La (SSB), is a nucleolar ribonucleoprotein that is abundantly expressed in dead tumor cells, essential for cellular function, and widely distributed [24]. The gene has been approved by the HUGO Gene Nomenclature Committee. SSB is preferentially expressed in apoptotic tumor cells and is highly expressed in diseases such as lung cancer [24], cervical cancer [25], head and neck squamous cell carcinoma [26,27], chronic myelogenous leukemia (CML), polycythemia vera, and primary myelofibrosis [28]. It has also been found that SSB causes cancer cells to proliferate, migrate, and invade. Nevertheless, the role and molecular mechanisms of SSB in LUAD have yet not to been excluded. Previous research has suggested that upregulation of glycolysis is associated with significant increase in FDG uptake, which has been associated with increased tumor growth and metastasis [13]. Therefore, this study investigates the relationship between SSB gene expression and metabolism parameters derived from PET/CT imaging. Additionally, the study examines SSB expression in patients with LUAD as a prognostic factor.
2. Material and methods
2.1. Patient samples
55 LUAD patients (17 males and 38 females) with a mean age of 55 ± 10.4 years who underwent preoperative 18F-FDG PET/CT scans at Taihe Hospital from August 2018 to July 2020 were analyzed retrospectively. The selection criteria are as follows: (a) LUAD confirmed by pathology; (b) No biopsy, radiotherapy or chemotherapy before PET/CT; (c) Surgery was performed within 2 weeks of PET/CT imaging; (d) Tissue samples can be used for IHC staining; (e) The medical records are complete. This retrospective study conducted at Taihe Hospital affiliated with Hubei Medical College has been approved by the Ethics Committee.
2.2. Collecting and analyzing raw data for SSB from public databases
Our study utilized the TCGA_GTEx-ALL dataset, which was processed by UCSC XENA with RNAseq data in TPM format from both TCGA and GTEx, processed through the Toil pipeline. Analyzing different tumor types' expression of SSB was based on this dataset (n = 18102). Specifically, we utilized the RNAseq data processed through the STAR pipeline from the TCGA database (https://portal.gdc.cancer.gov) [29] for use in the TCGA-LUAD project and extracted the formatted data in TPM format. This dataset was utilized for the comparative analysis of SSB expression levels in cancerous tissues compared to their corresponding normal tissues (n = 598). We obtained and examined the GSE40791 dataset from the Gene Expression Omnibus (GEO, https://www.ncbi.nlm.nih.gov/geo/) [30] database in order to further confirm the variations in SSB expression between LUAD and normal samples.
The study utilized the Receiver Operating Characteristic (ROC) curves to assess the diagnostic utility of SSB in LUAD patients. A Kaplan-Meier plot was used to evaluate the association between SSB gene expression and LUAD survival (http://kmplot.com/analysis/) [31]. Additionally, we conducted sub-analysis on the relationship between the levels of SSB and the clinicopathological characteristics in patients diagnosed with LUAD, utilizing the TCGA LUAD dataset.
2.3. Immunohistochemistry
IHC staining was performed on lung tumor tissue sections measuring 5 μm in thickness and embedded in paraffin. The sections were stained according to previously established protocols [19,20,32]. The sections were incubated with rabbit anti-SSB antibody (1:150; ab124932; Abcam) and TTF1 (1:250; ab76013; Abcam) antibody during the primary incubation.
SSB and TTF1 expression was evaluated, and nuclear reactivity characterized positive expression. The IHC results were assessed by two experienced pathologists who were fully blinded to the patient's clinical status. Any disparities were resolved through deliberations to reach a consensus.
According to the tumor staining intensity (I score), the scoring was performed as follows: from 0 (negative) to 3+ (strong positive), with intermediate levels of 1+ (weak positive) and 2+ (moderate positive) included in between, analyzing protein expression and scoring from low to high as 0, 1, 2, 3. The percentage of positive cell numbers (P score) was scored as follows: 0 for absence of display, 1 for a display representing 1%–30 %, 2 for a display representing 31%–70 %, and 3 for a display exceeding 71 %. Total score was calculated by multiplying the individual score (I score) with the percentage score (P score), denoted as S score = I score × P score [33]. We categorized samples with S score ≤1 as the “relatively negative group”, 1 < S score ≤4 as the “slightly positive group”, 4 < S score ≤6 as the “moderately positive group”, and 6 < S score ≤9 as the “strongly positive group” [34,35]. Samples in the moderately and strongly positive groups were classified as having high expression, while those in the relatively negative and slightly positive groups as having low expression. All cases in slightly, moderately, and strongly positive groups were regarded as positive expression.
2.4. 18F-FDG PET/CT imaging and delineation
Each participant underwent a minimum 6-h fasting period prior to the administration of 18F-FDG in order to maintain blood glucose levels below 7.1 mmol/L. Intravenous injection of the corresponding dose of 18F-FDG (3.7–4.1 MBq/kg) was followed by a rest period of 50–60 min. All imaging scans were conducted using a 64-detector PET/CT scanner (Biograph mCT-S 64 PET/CT, Siemens, Hoffman Estates, IL, USA) according to standard clinical protocols. Urination was required before the PET/CT examination. A CT scan was conducted with parameters encompassing the region from the thighs to the base of the skull: tube current of 100 mA, voltage of 120 kV, and slice thickness of 5 mm. Subsequently, PET images were obtained at 7–9 bed positions, with each position acquiring data for 2 min. After image acquisition, PET images were attenuated using CT data and reconstructed using Siemens TOF PET software with iterative reconstruction. Additionally, each patient underwent a high-resolution breath-hold CT scan. Finally, all PET data were subjected to semi-quantitative analysis using semi-automatic software (Power Imager NM, Mozi Healthcare, Beijing, China), with the involvement of two nuclear medicine experts each with over 10 years of experience. Discrepancies were resolved through consensus.
Briefly, a rectangular three-dimensional region of interest (ROI) was inserted onto the PET image to encompass the entire tumor. Adjust the ROI to exclude surrounding non-tumor activity to measure SUVmax and SUVmean. MTV (cm3) was defined by SUVmax threshold of 2.5, and TLG was calculated by multiplying SUVmean by MTV (TLG = ΣMTV × SUVmean).As previously described, the MTV and TLG were defined semi-automatically using an SUV-based platform [36].
2.5. Quantitative RT-PCR (qRT-PCR)
The Trizol reagent (Ambion, USA) was utilized for extracting RNA from both HBE normal lung cells and 1299 lung cancer cells. Real-time polymerase chain reaction (RT-PCR) was performed utilizing the PrimeScript™ RT Master Mix (Takara, Japan). The mRNA expression was quantitatively analyzed using the SYBR Green Real-time PCR Master Mix (Takara, Japan). The relative expression fold change of candidate genes was calculated using the 2−ΔΔCT formula. The following primers were used:
SSB, forward primer (5′-3′): CCCTGGAGGCCAAAATCTGT; reverse primer (5′-3′): TTGCATCAGTTGGGAAGCCT.
ACTB, forward primer (5′-3′): TGGCACCCAGCACAATGAA; reverse primer (5′-3′): CTAAGTCATAGTCCGCCTAGAAGCA.
2.6. Enrichment of SSB co-expression network in LUAD
We investigated into co-expressed genes that were linked to the expression of SSB, and performed an analysis of the TCGA LUAD dataset utilizing the R software. A statistical correlation was evaluated using the Pearson correlation coefficient. The ggplot2 package in R was employed to create visual representations of the data, which included the volcano plot and heat map. After GO KEGG pathway enrichment analyses, the data was visualized.
2.7. Gene set enrichment analysis (GSEA) synergizes with bioinformatics
In order to explore the involvement of SSB in the biological processes of LUAD, we assessed SSB gene expression profiles with clinical outcomes in the TCGA LUAD dataset using GSEA. The reference genes were designated as h.all.v7.5.1.symbols.gmt [Hallmarks] and c2.cp.all.vi7.5.1.symbols.gmt [All Canonical Pathways], and the analysis was performed for 100000 iterations.
2.8. m6A and SSB expression in LUAD
By using 20 m6A related genes, we examined SSB gene expression in TCGA and GSE40791, and compared differential expression between high and low expression cohorts [37].
2.9. Glycolysis and SSB expression in LUAD
Based on the TCGA and GSE40791 databases, expression levels of glycolysis and SSB genes were determined [38].
2.10. Statistical analysis
In this retrospective study, all test results were expressed as median ± interquartile range. The Shapiro Wilk test method was used to evaluate the normality of data distribution. Using the Mann Whitney U test (Wilcoxon rank sum test) and the Chi square test, we compared quantitative and categorical data with non-normal distributions. We calculated the correlation coefficient between two variables using Spearman rank correlation analysis. By analyzing ROC curves, PET/CT metabolic parameters and model scores were evaluated for their sensitivity and specificity. We then calculated the AUC value to represent the ROC effect. We used the DeLong test to compare the performance of the ROC curves described above to estimate the statistical significance of the difference. We used R version 4.2.1 and Xiantao to conduct statistical analysis. The value of p < 0.05 was considered statistically significant.
3. Results
3.1. The expression of SSB in LUAD
Using the TCGA_GTEx-ALL database, we compiled a comprehensive overview of the variation in SSB gene expression among different types of tumor tissues and their corresponding normal tissues. SSB expression levels were significantly elevated in multiple cancer types, including bladder urothelial carcinoma (BLCA), invasive breast carcinoma (BRCA), cholangiocarcinoma (CHOL), colon adenocarcinoma (COAD), diffuse large B-cell lymphoma (DLBC), esophageal carcinoma (ESCA), glioblastoma multiforme (GBM), head and neck squamous cell carcinoma (HNSC), lower grade glioma (LGG), hepatocellular carcinoma (LIHC), lung adenocarcinoma (LUAD), lung squamous cell carcinoma (LUSC), ovarian serous cystadenocarcinoma (OV), pancreatic adenocarcinoma (PAAD), prostate adenocarcinoma (PRAD), rectal adenocarcinoma (READ), stomach adenocarcinoma (STAD), testicular germ cell tumors (TGCT), thymoma (THYM), uterine corpus endometrial carcinoma (UCEC), and uterine carcinosarcoma (UCS). While SSB expression was increased in the aforementioned malignancies, its expression was decreased in kidney chromophobe (KICH), acute myeloid leukemia (LAML), skin cutaneous melanoma (SKCM), and thyroid carcinoma (THCA) (Fig. 1A).
Fig. 1.
Overexpression of SSB was observed in both LUAD and various types of cancer. (A) SSB expression is upregulated in various types of cancers. (B–C) Compared to normal tissues, there was upregulation of SSB expression in LUAD cancer tissues. (D) In the GSE40791 dataset, SSB was highly expressed in LUAD. (E) Differential expression of SSB mRNA was detected in the HBE normal lung cells and H1299 lung cancer cells using qPCR experiments. (F) The IHC scoring of SSB and TTF1 in both para-cancerous and cancerous tissues from patients diagnosed with LUAD were assessed using IHC. (G) IHC staining of SSB in LUAD. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001.
The TCGA LUAD dataset detected higher levels of SSB than the normal control samples (Fig. 1B). Further analysis of the paired TCGA LUAD data unveiled a notable elevation in the levels of SSB in the LUAD samples as opposed to their corresponding normal samples (Fig. 1C). Analysis of the GSE40791 dataset demonstrated a significant increase in the expression levels of SSB in the LUAD samples compared to the control group (Fig. 1D).
qRT-PCR (Fig. 1E) and IHC experiments (Fig. 1G) confirmed that SSB was expressed at both the RNA and protein levels in LUAD. IHC staining levels of SSB and TTF1 in tumor tissues were significantly higher than those in adjacent normal lung tissues (P < 0.001, Fig. 1F). Although the expression of SSB and TTF1 was meaningful in both tumor and para cancer, the expression of SSB (P = 2.63e-18) was more significant than that of TTF1 (P = 2.58e-06) according to the Wilcoxon rank sum test. The findings suggested a significant increase in both mRNA and protein levels of SSB in adjacent cancer tissue compared to normal tissue, particularly in the context of IHC experiments.
3.2. LUAD patients with high SSB expression have a poorer chance of surviving
Poor survival was predicted by high expression of SSB in LUAD [HR = 1.57 (1.15–2.14), P = 0.0046] (Fig. 2A). In order to evaluate the diagnostic efficacy of the SSB ratio in patients with LUAD, its diagnostic performance was evaluated using ROC curve analysis. There was significant accuracy of SSB in predicting LUAD diagnosis, as evidenced by an AUC value of 0.737 (95 % confidence interval: 0.693–0.782) (Fig. 2B). We analyzed the clinical outcomes of TCGA LUAD samples to determine the clinical significance of SSB expression. SSB expression was significantly associated with factors including gender, smoker, pathologic stage, Overall survival (OS), and TMN staging in LUAD (Fig. 2C–I).
Fig. 2.
The correlation between the expression of SSB and clinicopathological parameters in LUAD patients. (A) The KM curves presented the survival outcomes of LUAD according to SSB expression levels. (B) LUAD ROC curve for SSB diagnosis. The SSB expression level was analyzed in relation to (C)gender, (D)smoker, (E)Pathologic stage, (F)OS event, (G–I) Pathologic T.M.N stage. ∗P < 0.05, ∗∗∗P < 0.001.
3.3. The expression and metabolic parameters of SSB and TTF1 in PET
The ROC curves of the samples we collected from clinical practice were analyzed to examine the diagnostic capacity of SSB and TTF1 for LUAD (Fig. 3A). Compared to TTF1 (AUC = 0.754), SSB (AUC = 0.964) had higher diagnostic efficacy. In addition, we found that 94.5 % and 90.9 % of tumor tissues had positive SSB and TTF1 levels, respectively, while 5.5 % and 89.1 % of adjacent normal tissues contained positive expression (Table 1). There was a significant difference in the expression of SSB between LUAD tissue and adjacent lung tissue (P < 0.001), while TTF1 expression was not significantly different (P = 0.751). We concluded that SSB is superior to TTF1 in diagnosing LUAD, and may be used as a new target for lung cancer treatment.
Fig. 3.

Diagnostic efficacy of SSB and TTF1 in LUAD. (A)The ROC curve of SSB and TTF1.
Table 1.
The expression levels of SSB and TTF1 in LUAD and adjacent tissues.
| Tissue | Number | SSB expression |
TTF1 expression |
||||||
|---|---|---|---|---|---|---|---|---|---|
| All Positive (%) | Relatively Negative (%) | Χ2 | P value | All Positive (%) | Relatively Negative (%) | Χ2 | P value | ||
| Tumor | 55 | 52 (94.5 %) | 3 (5.5 %) | 87.309 | <0.001 | 50 (90.9 %) | 5 (9.1 %) | 0.10101 | 0.751 |
| Para-cancerous | 55 | 3 (5.5 %) | 52 (94.5 %) | 49 (89.1 %) | 6 (10.9 %) | ||||
To confirm the predictive accuracy of SSB and TTF1 in clinical samples, we categorized a total of 55 SSB cases into low (n = 22) and high expression group (n = 33) based on the median IHC score. The median IHC score was used to categorize TTF1 into low (n = 26) and high expression groups (n = 29). It was found that there was a significant association between SSB and T stage, as well as between TTF1 and gender expression levels (Table 2). In 55 LUAD patients, we also examined the expression of SSB using 18F-FDG PET/CT metabolic parameters (Table 3). High SSB expression was associated with higher SUVmax and SUVmean values compared to low SSB expression (P < 0.05). However, the expression of SSB was not associated with TLG and MTV in a statistically significant manner. Fig. 4A illustrated PET/CT images of patients with high and low SUVmax for LUAD. Compared to TTF1, the result of SSB in comparing high and low SUVmax values were significant (P < 0.05, Fig. 4B). Patients with high SUVmax values in the primary lesion exhibited increased expression of SSB and TTF1 in comparison to patients with lower SUVmax values (P < 0.05, Fig. 4C). Furthermore, the protein levels of SSB were significantly correlated with the SUVmax and SUVmean (cor = 0.477 and 0.466, respectively, P < 0.001, Fig. 5A). However, there was no statistically significant correlation between the intensity of SSB on IHC and MTV or TLG. The IHC score of TTF1 also showed no statistically significant correlation with SUVmax or SUVmean (Fig. 5B). These results suggested a significant relationship between SSB and glucose metabolism in LUAD, suggesting that SSB may regulate glycolytic pathways to play a role in LUAD tumor development and occurrence.
Table 2.
Clinic-pathological characteristics of 55 patients.
| Characteristics | SSB expression |
TTF1 expression |
||||
|---|---|---|---|---|---|---|
| Low | High | P value | Low | High | P value | |
| Number | 22 | 33 | 26 | 29 | ||
| Age (years) | 0.604 | 0.587 | ||||
| ≤ 60 | 16 (29.1 %) | 26 (47.3 %) | 19 (34.5 %) | 23 (41.8 %) | ||
| > 60 | 6 (10.9 %) | 7 (12.7 %) | 7 (12.7 %) | 6 (10.9 %) | ||
| Gender | 0.905 | 0.021 | ||||
| Female | 15 (27.3 %) | 23 (41.8 %) | 14 (25.5 %) | 24 (43.6 %) | ||
| Male | 7 (12.7 %) | 10 (18.2 %) | 12 (21.8 %) | 5 (9.1 %) | ||
| pT stage | 0.010 | 0.201 | ||||
| T1 | 22 (40 %) | 22 (40 %) | 20 (36.4 %) | 24 (43.6 %) | ||
| T2 | 0 (0 %) | 9 (16.4 %) | 6 (10.9 %) | 3 (5.5 %) | ||
| T3 | 0 (0 %) | 2 (3.6 %) | 0 (0 %) | 2 (3.6 %) | ||
| pN stage | 0.353 | 0.745 | ||||
| N0 | 21 (38.2 %) | 26 (47.3 %) | 22 (40 %) | 25 (45.5 %) | ||
| N1 | 0 (0 %) | 2 (3.6 %) | 1 (1.8 %) | 1 (1.8 %) | ||
| N2 | 1 (1.8 %) | 4 (7.3 %) | 2 (3.6 %) | 3 (5.5 %) | ||
| N3 | 0 (0 %) | 1 (1.8 %) | 1 (1.8 %) | 0 (0 %) | ||
| Pathological stage | 0.128 | 0.940 | ||||
| I | 21 (38.2 %) | 25 (45.5 %) | 22 (40 %) | 24 (43.6 %) | ||
| II | 0 (0 %) | 4 (7.3 %) | 2 (3.6 %) | 2 (3.6 %) | ||
| III | 1 (1.8 %) | 4 (7.3 %) | 2 (3.6 %) | 3 (5.5 %) | ||
Table 3.
PET/CT metabolic parameters of 55 patients.
| Characteristics | SSB expression |
TTF1 expression |
||||
|---|---|---|---|---|---|---|
| Low | High | P value | Low | High | P value | |
| SUVmax, median (IQR) | 3.04 (2.425, 4.67) | 5.63 (3.14, 12.76) | 0.018 | 5.185 (2.6125, 9.8272) | 3.61 (2.86, 7.16) | 0.533 |
| SUVmean, median (IQR) | 1.86 (1.4225, 2.7475) | 3.31 (1.78, 7.93) | 0.019 | 2.825 (1.4225, 5.325) | 2.1 (1.59, 3.93) | 0.692 |
| TLG, median (IQR) | 6 (4.2775,13.457) | 5.43 (3.32, 36.59) | 0.372 | 7.895 (5.57, 31.977) | 4.13 (2.56, 13.4) | 0.047 |
| MTV, median (IQR) | 3.405 (1.8775, 4.5175) | 2.54 (1.44, 6.12) | 0.884 | 3.605 (2.34, 6.795) | 1.89 (1.29, 3.78) | 0.023 |
Fig. 4.
The expression of TTF1 and SSB in LUAD patients. (A) Two PET/CT images of patients with different SUVmax. The left image displayed an SUVmax of 19.11, while the right image showed an SUVmax of 3.13. (B) The expression of SSB and TTF1 differed between SUVmax high and low. (C) IHC staining of SSB and TTF1 in LUAD. ∗P < 0.05.
Fig. 5.
SSB (A) and TTF1(B) IHC scores and PET/CT metabolic parameters: SUVmax, SUVmean, TLG, and MTV were correlated.
3.4. Enrichment analysis of SSB gene co-expression network in LUAD
In order to identify genes co-expressed with SSB, the TCGA LUAD dataset was analyzed using the R package ggplot2. In this study, only protein-coding genes were retained. Fig. 6A demonstrated that 14945 genes exhibited a positive correlation with SSB expression, whereas 16086 genes showed a significant negative correlation with SSB expression (P < 0.05). When cor >0.7 was chosen as the threshold, three genes exhibited the highest correlation: HAT1 (cor = 0.763, P = 3.27463e-99), ZC3H15 (cor = 0.728, P = 6.09671e-86), and CWC22 (cor = 0.727, P = 1.1591e-85). Notably, RPS23P6 (cor = −0.377, P = 2.58637e-18) and AL031733.2 (cor = −0.351, P = 8.73374e-16) exhibited the most pronounced inverse relationship with SSB (Fig. 6A). The heatmap revealed that the 35 most important genes had both positive and negative correlations with SSB expression (Fig. 6B).
Fig. 6.
SSB gene was co-expression and enriched in LUAD. (A) The volcano plot depicted genes co-expressed with SSB in the TCGA LUAD datasets. (B) The heat map showed a significant positive and negative correlation between the first 35 co-expressed genes and the expression levels of SSB in the LUAD dataset. (C–F) Enrichment analysis was conducted to elucidate the functional significance of GO terms associated with SSB co-expression genes. Furthermore, KEGG terms enrichment analyses were conducted to gain insights into the molecular pathways linked to SSB co-expression genes.
Using R software, 724 co-expressed genes positively correlated with SSB expression were analyzed for GO function and KEGG pathway enrichment. SSB co-expressed genes were associated with 276 biological processes (GO-BP), 164 cellular components (GO-CC), 141 molecular functions (GO-MF), and 101 KEGGs when P < 0.05. Bubble charts show the top 5 messages for GO-BP, GO-CC, GO-MF, and KEGG. According to the GO functional annotation, the genes co-expression with SSB play a major role in ribonucleoprotein complex biogenesis, chromosomal region, ATP hydrolysis activity and spliceosome (Fig. 6C–F).
3.5. The expression of m6A and SSB in LUAD
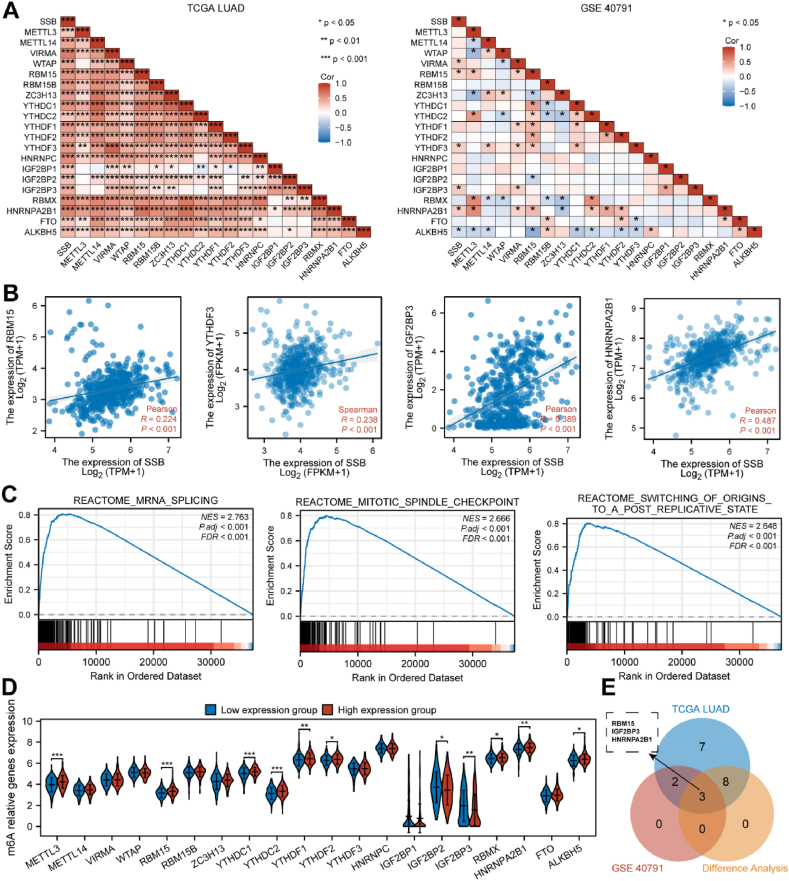
There was a positive correlation between SSB and 20 m6A-related genes in the TCGA dataset (P < 0.05). As seen in the GEO dataset, SSB was significantly and positively related to RBM15, YTHDF3, IGF2BP3, and HNRNPA2B1 (Fig. 7A–B). In addition, multiple m6A pathways in which SSB are involved were identified (Fig. 7C). A notable disparity was observed between the low- and high-expressing groups of SSB in the expression of multiple m6A-associated genes within LUAD (Fig. 7D). From the intersection of Figure A and D, we obtained four genes: RBM15, IGF2BP3 and HNRNPA2B1 (Fig. 7E).
Fig. 7.
Correlation of SSB expression levels with m6A-related genes in LUAD. (A) m6A-gene-SSB correlation was derived from TCGA and GSE40791 dataset analysis. (B) Graph showed the correlation between four highly correlated genes with SSB. (C) SSB participated in pathways related to m6A. (D) High and low SSB groups expressed m6A-related genes differently in LUAD. (E) The Venn plot showed the expression association and differential expression of RBM15, IGF2BP3 and HNRNPA2B1 genes. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001.
3.6. Glycolysis and expression of SSB in LUAD
The correlation between the SSB ratio and genes related to glycolysis was investigated utilizing the TCGA and GEO databases. SSB exhibited positive correlations with PGAM1 (Fig. 8A). SSB is involved in numerous glycolysis-related pathways (Fig. 8B). In LUAD, there were differences in the expression of several glycolytic related genes between the normal and tumor groups, as well as between the high and low expression groups of SSB (Fig. 8C). From the intersection of Figure A and C, we obtained one gene: PGAM1 (Fig. 8D).
Fig. 8.
Glycolysis-related genes in LUAD were correlated with SSB expression. (A) Correlation between glycolysis-related genes and SSB in TCGA and GSE40791. (B) SSB and pathways associated with glycolysis. (C) LUAD displayed differential expression of glycolytic related genes between normal and tumor groups, as well as between SSB high and low expression groups. (D) The Venn plot showed the expression association and differential expression of PGAM1. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001.
4. Discussion
LUAD, is the most prevalent subtype of lung cancer, with often subclinical manifestations in its early stages, that may delay diagnosis. The mainstay treatment for lung cancer involves surgical intervention, chemotherapy, or a combination of both modalities. Unfortunately, a considerable proportion of LUAD patients receive their diagnosis at an advanced stage, leading to an unfavorable prognosis characterized by high mortality and recurrence rates (30–32 % higher than the overall average) [[39], [40], [41]]. Elucidating the molecular mechanisms behind carcinogenesis may lead to the development of better therapeutics to improve survival in the LUAD population.
TTF1, a transcription factor composed of homologous domains [42], is a highly sensitive and specific molecular marker for LUAD [43]. Nevertheless, Myong et al. discovered that TTF1 expression correlates negatively with cell proliferation antigen (Ki67), indicating that higher TTF1 expression associated with lower tumor cell proliferation rate [44]. In contrast, some scientists believe that lung cancer patients with positive expression of TTF1 have a poor prognosis. A study by Lee et al. found that EGFR or TTF1 gene amplification is a poor prognostic factor for disease-free survival in patients with primary lung adenocarcinoma [45]. According to these previous literatures, it appears that TTF1 plays a double role in lung cancer pathogenesis, but the specific mechanism of action is not well understood. About 15 %–20 % of primary LUAD cases lack TTF1 expression, which poses challenges to treatment [46,47]. Therefore, the identification of a biomarker capable of enhancing the diagnostic efficacy of LUAD is of utmost importance.
SSB antigen, also referred to as La antigen, is a nuclear phosphoprotein consisting of two distinct domains of 23 and 28 kDa. The SSB antigen is prone to protein hydrolysis, resulting in the formation of numerous smaller peptides, which still retain immunoreactivity. SSB mainly resides in the cellular nucleus and is associated with transcription by RNA polymerase III. Antibodies against SSB belong to a class of anti-nuclear antibodies associated with various autoimmune diseases, particularly Sjögren's syndrome (SS) [48] and systemic lupus erythematosus (SLE) [49]. These antibodies can be detected in 40%–90 % of primary SS patients and 9%–35 % of SLE patients. Recent work on SSB has unveiled its upregulation in several disease, including lung cancer [24], cervical cancer [25], head and neck squamous cell carcinoma [26,27], chronic myelogenous leukemia, polycythemia vera, and primary myelofibrosis [28]. Through bioinformatics analysis, we identified high SSB expression in lung adenocarcinoma, which was further validated through cellular assays in both HBE normal lung cells and H1299 lung cancer cells. Research on SSB may lay the foundation for therapeutic targeting of LUAD in the future.
Our IHC staining results validate an augmented expression of SSB and TTF1 in LUAD, with SSB being specifically localized within the cell nucleus. Our study demonstrated, for the first time, that the positivity rate of SSB in LUAD tissues is significantly higher than that of TTF1. Statistical analysis using Chi-square test analysis demonstrated a significant disparity in the distribution of SSB positive expression between cancerous and non-cancerous tissues, and SSB expression was more significant than TTF1 expression. In conclusion, our findings highlight the remarkable performance of SSB in LUAD diagnosis. Furthermore, experimental evidence has demonstrated that the overexpression of SSB significantly expedites the advancement of the cell cycle and the expression of cyclin D1 (CCND1) in cervical cancer, thereby promoting tumor cell proliferation [25]. This gives us the idea that SSB may also promote cell cycle progression in LUAD. In addition, we performed GO and KEGG enrichment analyses on SSB and co-expressed genes, identifying several significantly enriched categories among the positively correlated group, consisting of cell cycle, ribonucleoprotein complex biogenesis, chromosomal region, ATP hydrolysis activity, and spliceosome. These findings imply that SSB may function as an oncogene in the initiation and progression of lung cancer.
Previous research has shown the association between m6A modification and tumor progression [50]. These aspects encompass proliferation, differentiation, initiation, invasion, and metastasis [51,52], which are mediated by m6A regulators [53]. These m6A genes exhibit a wide range of functions spanning mRNA pre-processing, translation, miRNA biogenesis, and mRNA decay, potentially explaining the underlying mechanisms of tumorigenesis [54]. In addition to their involvement in RNA metabolism, m6A readers are involved in a host of other diverse biological processes, including tumor development, blood cell differentiation, virus replication, immune response, and lipid synthesis [[55], [56], [57]]. This study identified a strong correlation between SSB and 20 m6A-related genes, with SSB expression influencing the expression of these genes in LUAD. Through m6A, RBM15 [58], IGF2BP3 [59] and ALKBH5 [60] have been shown to be tumor oncogenes or biomarkers. It is possible to use RBM15 and IGF2BP3 as pathological diagnostic indicators and potential therapeutic targets for lung cancer. The RNA-binding protein RBM15 modulates multiple signaling pathways, including Notch and Wnt, to regulate cell growth and apoptosis [61]. Colorectal cancer cells proliferate and metastasize when the RBM15 gene is overexpressed [62]. SSB is known to regulate LUAD methylation, mostly m6A, and may affect the methylation level of LUAD and ultimately the progression of LUAD through interactions with RBM15, IGF2BP3, and ALKBH5.Studies investigating the relationship between tumor cell metabolism and SSB expression are lacking. 18F-FDG PET/CT, as a non-invasive imaging modality, has emerged as an effective means to evaluate cellular energy metabolism on a macroscopic scale, particularly in the context of tumor cells [63].Notably, PET/CT is widely used for staging, detection of recurrent lesions, treatment monitoring, and prognostication in LUAD patients [64]. In clinical practice, PET/CT serves as a quantitative indicator for the assessment of tumor glucose affinity. Previous studies have focused on examining the correlation of PET/CT and treatment outcomes, specifically regarding tyrosine kinase inhibitors (TKIs) and immune checkpoint inhibitors (ICIs), in predicting treatment outcomes in non-small cell lung cancer [[65], [66], [67]]. The most commonly used metabolic parameters in PET/CT include SUVmax, TLG, and MTV; they are considered potential indicators for evaluating LUAD PD-L1 expression in individualized treatment [[68], [69], [70]]. Our study was the first of its kind to demonstrate an association between SSB expression and PET/CT in LUAD patients. SSB expression positively correlated with SUVmax and SUVmean in LUAD patients. We observed a robust correlation between conventional metabolic parameters derived from PET/CT scans and the expression of SSB, but not with MTV and TLG. However, consistent with previous studies, there existed a significant negative relationship among levels of TLG, MTV and TTF1 expression. As a composite parameter that combines tumor morphology and functional metabolism, MTV can more accurately reflect the metabolic burden of tumors, which can provide more references for the prognosis evaluation of patients with advanced NSCLC. TLG is also one of the research hotspots in recent years to evaluate the prognosis of PET/CT metabolic parameters, and it is a composite index reflecting the degree of uptake and metabolic volume of tumor lesions. The value of SUV, MTV and TLG in the prognostic evaluation of patients with early-stage NSCLC has been recognized. However, MTV and TLG can reflect systemic metabolism and tumor volume involved, which can be used as a supplement to the prognostic evaluation of patients with advanced NSCLC, and provide a reference for the selection of treatment regimens after accurate evaluation. In our 55 clinical samples, it was shown that the expression of SSB was not related to the metabolic parameters MTV and TLG, which may be related to our small sample size and algorithm, and we need to further explore the large sample size clinical trials. GSEA analysis indicates that SSB participates in glycolysis and is strongly correlated with glycolysis-related genes. Based on our hypothesis, SSB promotes glycolysis in LUAD tissue, thereby promoting LUAD occurrence and development. Therefore, we aim to validate preliminary results regarding the involvement of SSB in LUAD cell transformation.
5. Conclusion
Overexpression of SSB not only affects the clinical manifestations of LUAD patients, but also the severity and prognosis of the disease. Furthermore, SSB expression is correlated with 18F-FDG uptake. 18F-FDG PET/CT has the ability to predict SSB expression in LUAD, showing a positive correlation with SUVmax and SUVmean. SSB is a critical regulator of DNA replication and cell cycle progression. Furthermore, SSB is involved in multiple glycolytic pathways. SSB may influence 18F-FDG uptake by modulating glycolysis. SSB may influence the biological functions of cancer cells by controlling cell cycle and glycolysis. Therefore, SSB may serve as a useful molecular biomarker for the diagnosis of LUAD, in conjunction with 18F-FDG PET/CT imaging. The use of SSB as a potential therapeutic target for the treatment of LUAD may be worthwhile. Nevertheless, it is important to recognize the constraints of this study. One limitation is the absence of basic in vitro experiments to confirm clinical findings. We will plan to further investigate the expression of SSB in lung adenocarcinoma cells in future in vitro experiments.
Ethics approval and consent to participate
The studies involving human participants were reviewed and approved by Ethics Committee of Taihe Hospital Affiliated of Hubei University of Medicine (NO.2022KS010) and was in accordance with the Declaration of Helsinki. Written informed consent for participation was not required for this study in accordance with the Ethics Committee of Taihe Hospital Affiliated with Hubei University of Medicine.
Consent for publication
Not applicable.
Availability of data and materials
No data related to my research has been deposited in a publicly available repository. The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Funding
This work was supported by the Hubei Province's Outstanding Medical Academic Leader program, the Foundation for Innovative Research Team of Hubei Provincial Department of Education (No. T2020025),Freeexploring Foundation of Hubei University of Medicine (grant no. FDFR201903)Innovative Research Program for Graduates of Hubei University of Medicine (grant no. YC2023007, YC2023035), the Shiyan Taihe Hospital level project (2022JJXM006 and 2022JJXM007), and the Key Discipline Project of Hubei University of Medicine.
CRediT authorship contribution statement
Zi-Yue Liu: Writing – original draft, Methodology, Conceptualization. Ling-Ling Yuan: Writing – original draft, Validation, Software. Yan Gao: Validation, Conceptualization. Yu Zhang: Investigation, Formal analysis. Yao-Hua Zhang: Investigation, Formal analysis. Yi Yang: Investigation, Formal analysis. Yu-Xuan Chen: Investigation, Formal analysis. Xu-Sheng Liu: Methodology. Zhi-Jun Pei: Writing – review & editing, Supervision.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors would like to thank editors and reviewers for their insightful suggestions and careful reading of manuscript.
Contributor Information
Xu-Sheng Liu, Email: lxsking@taihehospital.com.
Zhi-Jun Pei, Email: pzjzml1980@taihehospital.com.
References
- 1.Bray F., Laversanne M., Sung H., Ferlay J., Siegel R.L., Soerjomataram I., Jemal A. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA. Cancer J. Clin. 2024 doi: 10.3322/caac.21834. caac. [DOI] [PubMed] [Google Scholar]
- 2.Lortet-Tieulent J., Soerjomataram I., Ferlay J., Rutherford M., Weiderpass E., Bray F. International trends in lung cancer incidence by histological subtype: adenocarcinoma stabilizing in men but still increasing in women. Lung Cancer. 2014;84:13–22. doi: 10.1016/j.lungcan.2014.01.009. [DOI] [PubMed] [Google Scholar]
- 3.Zhang Y., Vaccarella S., Morgan E., Li M., Etxeberria J., Chokunonga E., Manraj S.S., Kamate B., Omonisi A., Bray F. Global variations in lung cancer incidence by histological subtype in 2020: a population-based study. Lancet Oncol. 2023;24:1206–1218. doi: 10.1016/S1470-2045(23)00444-8. [DOI] [PubMed] [Google Scholar]
- 4.Li Y., Sheng H., Ma F., Wu Q., Huang J., Chen Q., Sheng L., Zhu X., Zhu X., Xu M. RNA m6A reader YTHDF2 facilitates lung adenocarcinoma cell proliferation and metastasis by targeting the AXIN1/Wnt/β-catenin signaling. Cell Death Dis. 2021;12:479. doi: 10.1038/s41419-021-03763-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hirsch F.R., Scagliotti G.V., Mulshine J.L., Kwon R., Curran W.J., Wu Y.-L., Paz-Ares L. Lung cancer: current therapies and new targeted treatments. Lancet. 2017;389:299–311. doi: 10.1016/S0140-6736(16)30958-8. [DOI] [PubMed] [Google Scholar]
- 6.Liu X.-S., Zhou L.-M., Yuan L.-L., Gao Y., Kui X.-Y., Liu X.-Y., Pei Z.-J. NPM1 is a prognostic biomarker involved in immune infiltration of lung adenocarcinoma and associated with m6A modification and glycolysis. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.724741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu X.-S., Zhang Y., Ming X., Hu J., Chen X.-L., Wang Y.-L., Zhang Y.-H., Gao Y., Pei Z.-J. SPC25 as a novel therapeutic and prognostic biomarker and its association with glycolysis, ferroptosis and ceRNA in lung adenocarcinoma. Aging-Us. 2024;16:779–798. doi: 10.18632/aging.205418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu X.-S., Yuan L.-L., Gao Y., Ming X., Zhang Y.-H., Zhang Y., Liu Z.-Y., Yang Y., Pei Z.-J. DARS2 overexpression is associated with PET/CT metabolic parameters and affects glycolytic activity in lung adenocarcinoma. J. Transl. Med. 2023;21:574. doi: 10.1186/s12967-023-04454-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lopci E., Toschi L., Grizzi F., Rahal D., Olivari L., Castino G.F., Marchetti S., Cortese N., Qehajaj D., Pistillo D., Alloisio M., Roncalli M., Allavena P., Santoro A., Marchesi F., Chiti A. Correlation of metabolic information on FDG-PET with tissue expression of immune markers in patients with non-small cell lung cancer (NSCLC) who are candidates for upfront surgery. Eur. J. Nucl. Med. Mol. Imag. 2016;43:1954–1961. doi: 10.1007/s00259-016-3425-2. [DOI] [PubMed] [Google Scholar]
- 10.Chung H.W., Lee K.Y., Kim H.J., Kim W.S., So Y. FDG PET/CT metabolic tumor volume and total lesion glycolysis predict prognosis in patients with advanced lung adenocarcinoma. J. Cancer Res. Clin. Oncol. 2014;140:89–98. doi: 10.1007/s00432-013-1545-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hofheinz F., Li Y., Steffen I.G., Lin Q., Lili C., Hua W., van den Hoff J., Zschaeck S. Confirmation of the prognostic value of pretherapeutic tumor SUR and MTV in patients with esophageal squamous cell carcinoma. Eur. J. Nucl. Med. Mol. Imag. 2019;46:1485–1494. doi: 10.1007/s00259-019-04307-6. [DOI] [PubMed] [Google Scholar]
- 12.Na K.J., Choi H. Tumor metabolic features identified by 18F-FDG PET correlate with gene networks of immune cell microenvironment in head and neck cancer. J. Nucl. Med. 2018;59:31–37. doi: 10.2967/jnumed.117.194217. [DOI] [PubMed] [Google Scholar]
- 13.Nair V.S., Gevaert O., Davidzon G., Napel S., Graves E.E., Hoang C.D., Shrager J.B., Quon A., Rubin D.L., Plevritis S.K. Prognostic PET 18F-FDG uptake imaging features are associated with major oncogenomic alterations in patients with resected non–small cell lung cancer. Cancer Res. 2012;72:3725–3734. doi: 10.1158/0008-5472.CAN-11-3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vesselle H., Salskov A., Turcotte E., Wiens L., Schmidt R., Jordan C.D., Vallières E., Wood D.E. Relationship between non-small cell lung cancer FDG uptake at PET, tumor histology, and ki-67 proliferation index. J. Thorac. Oncol. 2008;3:971–978. doi: 10.1097/JTO.0b013e31818307a7. [DOI] [PubMed] [Google Scholar]
- 15.Song J.-Y., Lee Y.N., Kim Y.S., Kim S.G., Jin S.J., Park J.M., Choi G.S., Chung J.C., Lee M.H., Cho Y.H., Choi M.H., Kim D.C., Choi H.J., Moon J.H., Lee S.H., Jeong S.W., Jang J.Y., Kim H.S., Kim B.S. Predictability of preoperative 18F-FDG PET for histopathological differentiation and early recurrence of primary malignant intrahepatic tumors. Nucl. Med. Commun. 2015;36:319–327. doi: 10.1097/MNM.0000000000000254. [DOI] [PubMed] [Google Scholar]
- 16.Kaira K., Serizawa M., Koh Y., Takahashi T., Yamaguchi A., Hanaoka H., Oriuchi N., Endo M., Ohde Y., Nakajima T., Yamamoto N. Biological significance of 18F-FDG uptake on PET in patients with non-small-cell lung cancer. Lung Cancer. 2014;83:197–204. doi: 10.1016/j.lungcan.2013.11.025. [DOI] [PubMed] [Google Scholar]
- 17.Zegers C.M.L., van Elmpt W., Reymen B., Even A.J.G., Troost E.G.C., Öllers M.C., Hoebers F.J.P., Houben R.M.A., Eriksson J., Windhorst A.D., Mottaghy F.M., De Ruysscher D., Lambin P. In vivo quantification of hypoxic and metabolic status of NSCLC tumors using [18F]HX4 and [18F]FDG-PET/CT imaging. Clin. Cancer Res. 2014;20:6389–6397. doi: 10.1158/1078-0432.CCR-14-1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vander Heiden M.G., Cantley L.C., Thompson C.B. Understanding the warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou L.-M., Yuan L.-L., Gao Y., Liu X.-S., Dai Q., Yang J.-W., Pei Z.-J. Nucleophosmin 1 overexpression correlates with 18F-FDG PET/CT metabolic parameters and improves diagnostic accuracy in patients with lung adenocarcinoma. Eur. J. Nucl. Med. Mol. Imag. 2021;48:904–912. doi: 10.1007/s00259-020-05005-4. [DOI] [PubMed] [Google Scholar]
- 20.Gao Y., Yuan L., Zeng J., Li F., Li X., Tan F., Liu X., Wan H., Kui X., Liu X., Ke C., Pei Z. eIF6 is potential diagnostic and prognostic biomarker that associated with 18F-FDG PET/CT features and immune signatures in esophageal carcinoma. J. Transl. Med. 2022;20 doi: 10.1186/s12967-022-03503-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang J.-W., Yuan L.-L., Gao Y., Liu X.-S., Wang Y.-J., Zhou L.-M., Kui X.-Y., Li X.-H., Ke C.-B., Pei Z.-J. 18F-FDG PET/CT metabolic parameters correlate with EIF2S2 expression status in colorectal cancer. J. Cancer. 2021;12:5838–5847. doi: 10.7150/jca.57926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Berghmans T., Paesmans M., Mascaux C., Martin B., Meert A.-P., Haller A., Lafitte J.-J., Sculier J.-P. Thyroid transcription factor 1—a new prognostic factor in lung cancer: a meta-analysis. Ann. Oncol. 2006;17:1673–1676. doi: 10.1093/annonc/mdl287. [DOI] [PubMed] [Google Scholar]
- 23.Ooi H., Chen C.-Y., Hsiao Y.-C., Huang W.-S., Hsieh B.-T. Influence of thyroid transcription factor-1 on fluorodeoxyglucose uptake and prognosis of non-small cell lung cancer. Anticancer Res. 2014;34:2467–2475. [PubMed] [Google Scholar]
- 24.Staudacher A.H., Al-Ejeh F., Fraser C.K., Darby J.M., Roder D.M., Ruszkiewicz A., Manavis J., Brown M.P. The La antigen is over-expressed in lung cancer and is a selective dead cancer cell target for radioimmunotherapy using the La-specific antibody APOMAB. EJNMMI Res. 2014;4:2. doi: 10.1186/2191-219X-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sommer G., Dittmann J., Kuehnert J., Reumann K., Schwartz P.E., Will H., Coulter B.L., Smith M.T., Heise T. The RNA-binding protein La contributes to cell proliferation and CCND1 expression. Oncogene. 2011;30:434–444. doi: 10.1038/onc.2010.425. [DOI] [PubMed] [Google Scholar]
- 26.Sommer G., Rossa C., Chi A.C., Neville B.W., Heise T. Implication of RNA-binding protein La in proliferation, migration and invasion of lymph node-metastasized hypopharyngeal SCC cells. PLoS One. 2011;6 doi: 10.1371/journal.pone.0025402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heise T., Kota V., Brock A., Morris A.B., Rodriguez R.M., Zierk A.W., Howe P.H., Sommer G. The La protein counteracts cisplatin-induced cell death by stimulating protein synthesis of anti-apoptotic factor Bcl2. Oncotarget. 2016;7:29664–29676. doi: 10.18632/oncotarget.8819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trotta R., Vignudelli T., Candini O., Intine R.V., Pecorari L., Guerzoni C., Santilli G., Byrom M.W., Goldoni S., Ford L.P., Caligiuri M.A., Maraia R.J., Perrotti D., Calabretta B. BCR/ABL activates mdm2 mRNA translation via the La antigen. Cancer Cell. 2003;3:145–160. doi: 10.1016/s1535-6108(03)00020-5. [DOI] [PubMed] [Google Scholar]
- 29.Tomczak K., Czerwińska P., Wiznerowicz M. The Cancer Genome Atlas (TCGA): an immeasurable source of knowledge. Współczesna Onkol. 2015;1A:68–77. doi: 10.5114/wo.2014.47136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barrett T., Wilhite S.E., Ledoux P., Evangelista C., Kim I.F., Tomashevsky M., Marshall K.A., Phillippy K.H., Sherman P.M., Holko M., Yefanov A., Lee H., Zhang N., Robertson C.L., Serova N., Davis S., Soboleva A. NCBI GEO: archive for functional genomics data sets--update. Nucleic Acids Res. 2012;41:D991–D995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Győrffy B., Surowiak P., Budczies J., Lánczky A. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS One. 2013;8 doi: 10.1371/journal.pone.0082241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu X.-Y., Gao Y., Kui X.-Y., Liu X.-S., Zhang Y., Zhang Y., Ke C.-B., Pei Z.-J. High expression of HNRNPR in ESCA combined with 18F-FDG PET/CT metabolic parameters are novel biomarkers for preoperative diagnosis of ESCA. J. Transl. Med. 2021;20 doi: 10.1186/s12967-022-03665-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang R., Liang X., Wang H., Guo M., Shen H., Shi Y., Liu Q., Sun Y., Yang L., Zhan M. The RNA methyltransferase NSUN6 suppresses pancreatic cancer development by regulating cell proliferation. EBioMedicine. 2021;63 doi: 10.1016/j.ebiom.2020.103195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang T.-W., Lin K.-F., Lee C.-H., Chang H., Lee S.-C., Shieh Y.-S. The role of thyroid transcription factor-1 and tumor differentiation in resected lung adenocarcinoma. Sci. Rep. 2021;7 doi: 10.1038/s41598-017-14651-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Luna H. Gloriane C., Severino Imasa M., Juat N., Hernandez K.V., May Sayo T., Cristal-Luna G., Marie Asur-Galang S., Bellengan M., John Duga K., Brian Buenaobra B., De los Santos M.I., Medina D., Samo J., Minerva Literal V., Andrew Bascos N., Sy-Naval S. Expression landscapes in non-small cell lung cancer shaped by the thyroid transcription factor 1. Lung Cancer. 2023;176:121–131. doi: 10.1016/j.lungcan.2022.12.015. [DOI] [PubMed] [Google Scholar]
- 36.Liu X.-S., Yuan L.-L., Gao Y., Zhou L.-M., Yang J.-W., Pei Z.-J. Overexpression of METTL3 associated with the metabolic status on 18F-FDG PET/CT in patients with Esophageal Carcinoma. J. Cancer. 2020;11:4851–4860. doi: 10.7150/jca.44754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li Y., Xiao J., Bai J., Tian Y., Qu Y., Chen X., Wang Q., Li X., Zhang Y., Xu J. Molecular characterization and clinical relevance of m6A regulators across 33 cancer types. Mol. Cancer. 2019;18:137. doi: 10.1186/s12943-019-1066-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li L., Liang Y., Kang L., Liu Y., Gao S., Chen S., Li Y., You W., Dong Q., Hong T., Yan Z., Jin S., Wang T., Zhao W., Mai H., Huang J., Han X., Ji Q., Song Q., Yang C., Zhao S., Xu X., Ye Q. Transcriptional regulation of the warburg effect in cancer by SIX1. Cancer Cell. 2018;33:368–385.e7. doi: 10.1016/j.ccell.2018.01.010. [DOI] [PubMed] [Google Scholar]
- 39.Thai A.A., Solomon B.J., Sequist L.V., Gainor J.F., Heist R.S. Lung cancer. Lancet. 2021;398:535–554. doi: 10.1016/S0140-6736(21)00312-3. [DOI] [PubMed] [Google Scholar]
- 40.Seguin L., Durandy M., Feral C.C. Lung adenocarcinoma tumor origin: a guide for personalized medicine. Cancers. 2021;14:1759. doi: 10.3390/cancers14071759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gao S., Li N., Wang S., Zhang F., Wei W., Li N., Bi N., Wang Z., He J. Lung cancer in people's Republic of China. J. Thorac. Oncol. 2020;15:1567–1576. doi: 10.1016/j.jtho.2020.04.028. [DOI] [PubMed] [Google Scholar]
- 42.Bingle C.D. Thyroid transcription factor-1. Int. J. Biochem. Cell Biol. 1997;29:1471–1473. doi: 10.1016/s1357-2725(97)00007-1. [DOI] [PubMed] [Google Scholar]
- 43.Tang X., Kadara H., Behrens C., Liu D.D., Xiao Y., Rice D., Gazdar A.F., Fujimoto J., Moran C., Varella-Garcia M., Lee J.J., Hong W.K., Wistuba I.I. Abnormalities of the TITF-1 lineage-specific oncogene in NSCLC: implications in lung cancer pathogenesis and prognosis. Clin. Cancer Res. 2011;17:2434–2443. doi: 10.1158/1078-0432.CCR-10-1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Myong N.H. Thyroid transcription factor-1 (TTF-1) expression in human lung carcinomas: its prognostic implication and relationship with expressions of p53 and ki-67 proteins. J. Kor. Med. Sci. 2003;18:494. doi: 10.3346/jkms.2003.18.4.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee J.S., Kim H.R., Lee C.Y., Shin M., Shim H.S. EGFR and TTF-1 gene amplification in surgically resected lung adenocarcinomas: clinicopathologic significance and effect on response to EGFR-tyrosine kinase inhibitors in recurred cases. Ann. Surg Oncol. 2013;20:3015–3022. doi: 10.1245/s10434-013-2937-2. [DOI] [PubMed] [Google Scholar]
- 46.Tanaka I., Dayde D., Tai M.C., Mori H., Solis L.M., Tripathi S.C., Fahrmann J.F., Unver N., Parhy G., Jain R., Parra E.R., Murakami Y., Aguilar-Bonavides C., Mino B., Celiktas M., Dhillon D., Casabar J.P., Nakatochi M., Stingo F., Baladandayuthapani V., Wang H., Katayama H., Dennison J.B., Lorenzi P.L., Do K.-A., Fujimoto J., Behrens C., Ostrin E.J., Rodriguez-Canales J., Hase T., Fukui T., Kajino T., Kato S., Yatabe Y., Hosoda W., Kawaguchi K., Yokoi K., Chen-Yoshikawa T.F., Hasegawa Y., Gazdar A.F., Wistuba I.I., Hanash S., Taguchi A. SRGN-triggered aggressive and immunosuppressive phenotype in a subset of TTF-1–negative lung adenocarcinomas. JNCI: Journal of the National Cancer Institute. 2022;114:290–301. doi: 10.1093/jnci/djab183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Anagnostou V.K., Syrigos K.N., Bepler G., Homer R.J., Rimm D.L. Thyroid transcription factor 1 is an independent prognostic factor for patients with stage I lung adenocarcinoma. J. Clin. Orthod. 2009;27:271–278. doi: 10.1200/JCO.2008.17.0043. [DOI] [PubMed] [Google Scholar]
- 48.Liang C., Xiong K., Szulwach K.E., Zhang Y., Wang Z., Peng J., Fu M., Jin P., Suzuki H.I., Liu Q. Sjögren syndrome antigen B (SSB)/La promotes global MicroRNA expression by binding MicroRNA precursors through stem-loop recognition. J. Biol. Chem. 2013;288:723–736. doi: 10.1074/jbc.M112.401323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tarter L., Bermas B.L. Expert perspective on a clinical challenge: lupus and pregnancy. Arthritis Rheumatol. 2024;76:321–331. doi: 10.1002/art.42756. [DOI] [PubMed] [Google Scholar]
- 50.Li Y., Gu J., Xu F., Zhu Q., Chen Y., Ge D., Lu C. Molecular characterization, biological function, tumor microenvironment association and clinical significance of m6A regulators in lung adenocarcinoma. Briefings Bioinf. 2021;22 doi: 10.1093/bib/bbaa225. [DOI] [PubMed] [Google Scholar]
- 51.Yang X., Bai Q., Chen W., Liang J., Wang F., Gu W., Liu L., Li Q., Chen Z., Zhou A., Long J., Tian H., Wu J., Ding X., Zhou N., Li M., Yang Y., Cai J. m6A‐Dependent modulation via IGF2BP3/MCM5/notch Axis promotes partial EMT and LUAD metastasis. Adv. Sci. 2023;10 doi: 10.1002/advs.202206744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fang H., Sun Q., Zhou J., Zhang H., Song Q., Zhang H., Yu G., Guo Y., Huang C., Mou Y., Jia C., Song Y., Liu A., Song K., Lu C., Tian R., Wei S., Yang D., Chen Y., Li T., Wang K., Yu Y., Lv Y., Mo K., Sun P., Yu X., Song X. m6A methylation reader IGF2BP2 activates endothelial cells to promote angiogenesis and metastasis of lung adenocarcinoma. Mol. Cancer. 2021;22 doi: 10.1186/s12943-023-01791-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Delaunay S., Frye M. RNA modifications regulating cell fate in cancer. Nat. Cell Biol. 2019;21:552–559. doi: 10.1038/s41556-019-0319-0. [DOI] [PubMed] [Google Scholar]
- 54.Mauer J., Luo X., Blanjoie A., Jiao X., Grozhik A.V., Patil D.P., Linder B., Pickering B.F., Vasseur J.-J., Chen Q., Gross S.S., Elemento O., Debart F., Kiledjian M., Jaffrey S.R. Reversible methylation of m6Am in the 5′ cap controls mRNA stability. Nature. 2017;541:371–375. doi: 10.1038/nature21022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen X.-Y., Zhang J., Zhu J.-S. The role of m6A RNA methylation in human cancer. Mol. Cancer. 2019;18 doi: 10.1186/s12943-019-1033-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang B., Wu Q., Li B., Wang D., Wang L., Zhou Y.L. m6A regulator-mediated methylation modification patterns and tumor microenvironment infiltration characterization in gastric cancer. Mol. Cancer. 2020;19 doi: 10.1186/s12943-020-01170-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lin X., Ye R., Li Z., Zhang B., Huang Y., Du J., Wang B., Meng H., Xian H., Yang X., Zhang X., Zhong Y., Huang Z. KIAA1429 promotes tumorigenesis and gefitinib resistance in lung adenocarcinoma by activating the JNK/MAPK pathway in an m6A-dependent manner. Drug Resist. Updates. 2023;66 doi: 10.1016/j.drup.2022.100908. [DOI] [PubMed] [Google Scholar]
- 58.Feng J., Li Y., He F., Zhang F. RBM15 silencing promotes ferroptosis by regulating the TGF-β/Smad2 pathway in lung cancer. Environ. Toxicol. 2023;38:950–961. doi: 10.1002/tox.23741. [DOI] [PubMed] [Google Scholar]
- 59.Lin Z., Li J., Zhang J., Feng W., Lu J., Ma X., Ding W., Ouyang S., Lu J., Yue P., Wan G., Liu P., Zhang X. Metabolic reprogramming driven by IGF2BP3 promotes acquired resistance to EGFR inhibitors in non–small cell lung cancer. Cancer Res. 2023;83:2187–2207. doi: 10.1158/0008-5472.CAN-22-3059. [DOI] [PubMed] [Google Scholar]
- 60.Shen W., Pu J., Zuo Z., Gu S., Sun J., Tan B., Wang L., Cheng J., Zuo Y. The RNA demethylase ALKBH5 promotes the progression and angiogenesis of lung cancer by regulating the stability of the LncRNA PVT1. Cancer Cell Int. 2022;22 doi: 10.1186/s12935-022-02770-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hu M., Yang Y., Ji Z., Luo J. RBM15 functions in blood diseases. Curr. Cancer Drug Targets. 2016;16:579–585. doi: 10.2174/1568009616666160112105706. [DOI] [PubMed] [Google Scholar]
- 62.Zhang Z., Mei Y., Hou M. Knockdown RBM15 inhibits colorectal cancer cell proliferation and metastasis via N6-methyladenosine (m6A) modification of MyD88 mRNA. Cancer Biother. Radiopharm. 2022;37:976–986. doi: 10.1089/cbr.2021.0226. [DOI] [PubMed] [Google Scholar]
- 63.Greenspan B.S. Role of PET/CT for precision medicine in lung cancer: perspective of the society of nuclear medicine and molecular imaging, transl. Lung Cancer Res. 2017;6:617–620. doi: 10.21037/tlcr.2017.09.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kandathil A., Kay F.U., Butt Y.M., Wachsmann J.W., Subramaniam R.M. Role of FDG PET/CT in the eighth edition of TNM staging of non–small cell lung cancer. Radiographics. 2018;38:2134–2149. doi: 10.1148/rg.2018180060. [DOI] [PubMed] [Google Scholar]
- 65.Chang C., Zhou S., Yu H., Zhao W., Ge Y., Duan S., Wang R., Qian X., Lei B., Wang L., Liu L., Ruan M., Yan H., Sun X., Xie W. A clinically practical radiomics-clinical combined model based on PET/CT data and nomogram predicts EGFR mutation in lung adenocarcinoma. Eur. Radiol. 2021;31:6259–6268. doi: 10.1007/s00330-020-07676-x. [DOI] [PubMed] [Google Scholar]
- 66.Zhang J., Zhao X., Zhao Y., Zhang J., Zhang Z., Wang J., Wang Y., Dai M., Han J. Value of pre-therapy 18F-FDG PET/CT radiomics in predicting EGFR mutation status in patients with non-small cell lung cancer. Eur. J. Nucl. Med. Mol. Imag. 2020;47:1137–1146. doi: 10.1007/s00259-019-04592-1. [DOI] [PubMed] [Google Scholar]
- 67.Chetan M.R., Gleeson F.V. Radiomics in predicting treatment response in non-small-cell lung cancer: current status, challenges and future perspectives. Eur. Radiol. 2021;31:1049–1058. doi: 10.1007/s00330-020-07141-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang L., Ruan M., Lei B., Yan H., Sun X., Chang C., Liu L., Xie W. The potential of 18F-FDG PET/CT in predicting PDL1 expression status in pulmonary lesions of untreated stage IIIB-IV non-small-cell lung cancer. Lung Cancer. 2020;150:44–52. doi: 10.1016/j.lungcan.2020.10.004. [DOI] [PubMed] [Google Scholar]
- 69.Hashimoto K., Kaira K., Yamaguchi O., Mouri A., Shiono A., Miura Y., Murayama Y., Kobayashi K., Kagamu H., Kuji I. Potential of FDG-PET as prognostic significance after anti-PD-1 antibody against patients with previously treated non-small cell lung cancer. JCM. 2021;9:725. doi: 10.3390/jcm9030725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wu X., Huang Y., Zhao Q., Wang L., Song X., Li Y., Jiang L. PD-L1 expression correlation with metabolic parameters of FDG PET/CT and clinicopathological characteristics in non-small cell lung cancer. EJNMMI Res. 2020;10:51. doi: 10.1186/s13550-020-00639-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No data related to my research has been deposited in a publicly available repository. The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.