Abstract
Clinical, neuroimaging and genomics evidence have increasingly underscored a degree of overlap between autism and attention-deficit/hyperactivity disorder (ADHD). This study explores the specific contribution of their core symptoms to shared biology in a sample of N=166 verbal children (6–12 years) with rigorously-established primary diagnoses of either autism or ADHD (without autism). We investigated the associations between inter-individual differences in clinician-based dimensional measures of autism and ADHD symptoms and whole-brain low motion intrinsic functional connectivity (iFC). Additionally, we explored their linked gene expression patterns in silico. Whole-brain multivariate distance matrix regression revealed a transdiagnostic association between autism severity and iFC of two nodes: the middle frontal gyrus of the frontoparietal network and posterior cingulate cortex of the default mode network. Across children, the greater the iFC between these nodes, the more severe the autism symptoms, even after controlling for ADHD symptoms. Results from segregation analyses were consistent with primary findings, underscoring the significance of internetwork iFC interactions for autism symptom severity across diagnoses. No statistically significant brain-behavior relationships were observed for ADHD symptoms. Genetic enrichment analyses of the iFC maps associated with autism symptoms implicated genes known to: (i) have greater rate of variance in autism and ADHD, and (ii) be involved in neuron projection, suggesting shared genetic mechanisms for this specific brain-clinical phenotype. Overall, these findings underscore the relevance of transdiagnostic dimensional approaches in linking clinically-defined phenomena to shared presentations at the macroscale circuit- and genomic-levels among children with diagnoses of autism and ADHD.
INTRODUCTION
The growing awareness of frequent comorbidities and symptom co-occurrence among neuropsychiatric conditions has motivated a shift from case-control categorical comparisons to transdiagnostic investigations of the biology underlying symptom dimensions.[1] This is particularly relevant for autism spectrum disorder (henceforth autism) and attention-deficit/hyperactivity disorder (ADHD), two common childhood-onset neurodevelopmental conditions. While autism and ADHD are conceptualized as distinct categorical diagnoses, abundant clinical evidence has documented their overlapping symptoms.[2–6] Some degree of shared genetic variance,[7–9] as well as both brain structural[10–13] and functional[14–17] atypicalities, have also been reported. Yet, despite initial insights from the clinical, neuroimaging and genomics evidence summarized below, the extent to which co-occurring symptom profiles reflect shared biological features is largely unknown.
Clinically, the co-occurrence of ADHD symptoms in children with autism has been recognized, with frequencies ranging from 28% to 80%.[18–20] More recently, the presence of symptoms qualitatively similar to autism (i.e., autistic traits) has been acknowledged in a large proportion of those with a primary ADHD diagnosis. Indeed, when examined, autistic traits have been documented in up to 32% of ADHD children.[2–6] Both patterns of symptom co-occurrence exacerbate clinical impairment, challenge accurate diagnoses, and impede individualized care.[21–23] This clinical evidence supports using transdiagnostic dimensional approaches to investigate the neurobiological underpinnings of inter-individual symptomatology differences in both symptom domains.
From a neuroimaging perspective, accumulating evidence points towards atypical widespread large-scale functional networks in both autism and ADHD.[24–28] As such, intrinsic functional connectivity (iFC) measured with resting-state functional MRI (R-fMRI) is useful to investigate shared atypical neural features. To date, only a few R-fMRI studies have directly examined associations between iFC and dimensional measures of autism and/or ADHD symptoms across individuals with categorical diagnoses of autism or ADHD in the same study.[15–17] However, findings have been inconsistent. One study did not find significant iFC associations specific to either autism or ADHD symptoms.[16] The other two studies revealed significant transdiagnostic iFC associations,[15, 17] but the relative contribution of each targeted symptom domain remains unspecified. For example, the iFC patterns predicted by parent-reported autism severity in an autism dataset were found to be associated with parent ratings of inattention in an independent ADHD sample.[17] However, the lack of autism measures in the independent ADHD dataset prevented assessing the degree to which the identified iFC pattern was uniquely related to the autism domain. As a result, the specific contribution of core autism or ADHD symptoms to the iFC atypicalities shared across children with categorical diagnoses of autism and/or ADHD remains uncertain.
At the genomic level, accumulating evidence also converges on shared genetic mechanisms across autism and ADHD.[8, 9] These highly heritable conditions[29, 30] have been found to co-aggregate in families with a parent or a child with either of these diagnoses,[31, 32] to share the same comorbidity patterns among monozygotic twins,[32, 33] and to be genetically correlated (rG=0.36).[34] For both autism and ADHD, genetic liability is thought to involve polygenic effects of common variants — most still undiscovered — and the role of rare — heritable and de novo — variants.[35, 36] Among those identified, many genes implicated in autism and ADHD are highly expressed in the brain and encode for proteins involved in transcription regulation, synaptic transmission, intracellular signaling, and nervous system development.[37, 38] Whether the expression pattern of such genes is linked to transdiagnostic brain connectivity associated with core symptoms of autism and/or ADHD is unknown.
To clarify the specific contribution of symptom dimensions to shared biological features across autism and ADHD, the present study aims to identify associations between inter-individual differences in core autism and ADHD symptoms, iFC, and their related gene cortical expression patterns. We examined iFC relationships with clinicians’ expert ratings of autism and ADHD symptom severity in school-aged children without intellectual disability and rigorous diagnoses of either autism or ADHD without autism. To assess the unique relative contribution of each symptom domain, all children underwent the same phenotypic protocol. Given that multiple brain networks have been implicated in autism and ADHD,[24, 25, 27, 28] we conducted a whole-brain unbiased investigation using multivariate connectome-based analyses.[39] Further, to address reproducibility concerns in the field, we assessed robustness of iFC-behavioral findings to different methodological choices in MRI processing, data collection, and behavioral measures. Finally, building on the notion that genetic variation contributes to iFC,[40, 41] we explored if genes reported to harbor variants associated with ADHD and autism[42] were significantly enriched among those expressed within symptom-related iFC maps.
METHODS
Sampling
Data were collected prior to the WHO COVID-19 pandemic. Children were mostly recruited from the New York metropolitan area from clinical and/or community sources aiming to reach parents with concerns related to autism and/or ADHD, regardless of prior diagnostic history. We included participants aged 6-to-12 years who were identified as ASD or as ADHD without ASD (i.e., ADHDw/oASD) and completed at least one structural T1-weighted (T1w) and one resting-state functional MRI (R-fMRI) scan passing quality assurance. Inclusion in the ASD group required clinician’s best estimate of ASD per the Diagnostic and Statistical Manual of Mental Disorders, 5th Edition (DSM-5)[43] with or without any psychiatric comorbidities, including ADHD, given its high prevalence in ASD.[18, 20] Inclusion in the ADHDw/oASD group required clinician’s best estimate of meeting ADHD DSM-5 criteria in the absence of meeting DSM-5 criteria for ASD; other comorbid diagnoses for this group were not exclusionary. Inclusion also required a full IQ > 65; a complete list of inclusion/exclusion criteria is in Supplementary material. Over the course of the study, the enrolling and phenotyping site transferred from the NYU Grossman School of Medicine Child Study Center to the Child Mind Institute (CMI). As shown in Table S1, sample characteristics did not differ by enrollment site. The study was conducted in compliance with the Institutional Review Boards at NYU and Advarra Inc. at CMI. Written parental informed consent and verbal assent were provided for all children; children seven years or older also assented in writing.
Clinical and socio-demographics
Phenotyping included direct child assessment and parent(s) interviews, as well as review of available parent questionnaires, teacher and/or prior child’s records. Child assessments included the Differential Ability Scales-2nd Edition (DAS-II)[44] and research-reliable administration/scoring of the Autism Diagnostic Observation Schedule-2nd Edition (ADOS-2)[45] by an evaluator blind to the child’s presumptive diagnosis, prior history and records. For children treated with stimulants, assessments occurred after their discontinuation for at least 24 hours. The clinician-based parent interviews included the Kiddie-Schedule for Affective Disorders and Schizophrenia for School-Age Children–Present and Lifetime Version (KSADS)[46] and the Autism Symptom Interview for verbal children,[47] conducted by an evaluator unblind to current and past concerns/records. Whenever feasible, to further characterize the sample, the Vineland Adaptive Behavior Scales-2nd Edition (VABS-II)[48] parent interview was collected along with parent questionnaires assessing symptoms of autism, ADHD, and general psychopathology using the Social Responsiveness Scale-2nd Edition (SRS-2),[49] the Social Communication Questionnaire-Lifetime (SCQ-L),[50] the Strengths and Weaknesses of ADHD Symptoms and Normal Behavior (SWAN),[51] and the Child Behavior Checklist (CBCL).[52] Information about ADHD-related symptoms in school were obtained via SWAN teacher questionnaires, whenever feasible. Socioeconomic status, indexed by the Hollingshead scale,[53] and parent-reported ethnic/racial backgrounds were also collected.
Diagnostic protocol
Best estimate clinician-based DSM-5 psychiatric diagnoses were established with a rigorous multistep team-based approach. First, following their assessments, the blind and unblind evaluators (i.e., child and parent evaluator, respectively) met to share their impressions and records to reach an initial diagnostic consensus in the primary diagnostic groups. The evaluators provided ratings of diagnostic certainty for the presence or absence of ASD (with or without ADHD) or of ADHDw/oASD. Then, a multidisciplinary conference reviewed all information to confirm best-estimate diagnosis of either ASD (with or without ADHD and other comorbid diagnoses) or ADHDw/oASD (with or without other comorbid diagnoses), and to resolve potential diagnostic disagreements; see Supplementary material.
MRI
Following MRI simulator training, children were invited to at least one MRI session at the NYU Center for Brain Imaging as detailed elsewhere.[54] All MRI images were collected using the same Siemens Prisma 3.0T MRI scanner with a 32-channel head coil (Siemens; Erlangen). T1w images were obtained using a magnetization prepared gradient echo sequence, followed by at least a 6.33 min R-fMRI scan (Table S2). During R-fMRI acquisition, participants were instructed to keep their eyes open, relax and remain still. Whenever feasible, children completed an additional 4.65 min R-fMRI scan. Children treated with stimulant medications discontinued them at least 24 hours prior to MRI.
MRI data quality assurance (Q/A) and preprocessing
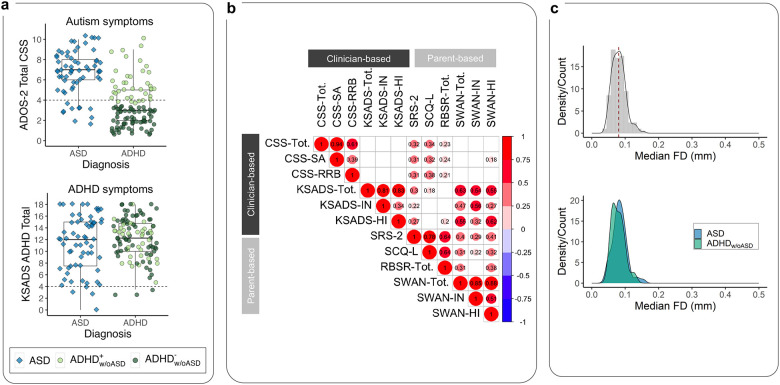
Structural T1w scans were visually inspected for artifacts (e.g., severe ringing, blurring, ghosting, or gross anatomical abnormalities) by one of two raters with excellent interrater reliability established on 84 study images (ICC(3,1): 0.96; 95% CI: 0.94–0.97). For R-fMRI data, following visual inspection for signal dropouts/artifacts, only scans with a median framewise displacement[55] (FD) ≤0.2mm were analyzed. Using this cutoff, of the 194 children meeting inclusion criteria and completing at least one MRI session, 28 did not pass Q/A, and thus were excluded from analyses. No differences were seen between those excluded and included with respect to demographics, intelligence, or key symptom ratings (Table S3). For further characterization of Q/A data, we computed individual-level Euler index[56] for T1w data and the number of R-fMRI peaks with FD≥1mm. Overall, motion was low across the sample analyzed, with no differences by primary diagnosis (Table 1, Fig. 1c).
Table 1.
Characteristics of the sample combined and by primary diagnostic groups.
| Variable | Total N=166 |
ASD n=63 |
ADHD w/o ASDl n=103 |
Statistics | |||
|---|---|---|---|---|---|---|---|
| df | W χ2 | p value | FDR-p value | ||||
| Age, years, M (SD) | 8.9 (1.7) | 8.8 (1.9) | 8.9 (1.7) | - | 3419.0 | 0.56 | 0.67 |
| Sex assigned at birth, males #, (%) | 125 (75) | 53 (84) | 72 (70) | 1 | 3.5 | 0.06 | 0.11 |
| Ethnicity, Hispanic #, (%)a | 39 (24) | 16 (26) | 23 (22) | 1 | 0.1 | 0.75 | 0.79 |
| Race, #, (%)a,b | 4 | 5.3 | 0.26 | 0.34 | |||
| White | 95 (58) | 32 (52) | 63 (61) | ||||
| Mixed | 26 (16) | 8 (13) | 18 (17) | ||||
| Black/African American | 19 (12) | 9 (15) | 10 (10) | ||||
| Asian | 14 (8) | 6 (10) | 8 (8) | ||||
| Other | 11 (7) | 7 (11) | 4 (4) | ||||
| Socio-economic status, class #, (%)c | 1 | 0.4 | 0.53 | 0.66 | |||
| Classes 1–3 | 45 (28) | 19 (32) | 26 (26) | ||||
| Classes 4–5 | 116 (72) | 41 (68) | 75 (74) | ||||
| Psychoactive Medication Status, #, (%)d | 2 | 5.1 | 0.08 | 0.14 | |||
| Med naïve | 117 (72) | 39 (65) | 78 (76) | ||||
| Currently on medication | 34 (21) | 18 (30) | 16 (16) | ||||
| Not Naïve but current off treatment | 13 (7) | 4 (5) | 9 (9) | ||||
| Psychiatric Comorbidity (yes), # (%) | 108 (65) | 59 (94) | 49 (48) | 1 | 34.5 | p<0.0001 | p<0.0001 |
| DAS-II IQ Standard scores, M (SD) | |||||||
| Verbal IQ | 107 (16) | 105 (17) | 108 (16) | - | 3565.5 | 0.29 | 0.40 |
| Non-verbal VIQ | 104 (18) | 103 (19) | 104 (18) | - | 3294.5 | 0.87 | 0.87 |
| Full-scale IQ | 105 (17) | 103 (19) | 105 (15) | - | 3450.5 | 0.49 | 0.65 |
| ADOS-2, M (SD)e | |||||||
| Expressive Language scoref | 7.9 (0.4) | 7.8 (0.6) | 7.9 (0.3) | - | 3604.5 | 0.03 | 0.06 |
| SA CSS | 5.1 (2.6) | 6.9 (2.1) | 4.1 (2.3) | - | 1230.5 | p<0.0001 | p<0.0001 |
| RRB CSS | 4.6 (3.2) | 6.8 (2.9) | 3.3 (2.7) | - | 1297.5 | p<0.0001 | p<0.0001 |
| Total CSS | 4.7 (2.7) | 6.7 (2.2) | 3.4 (2.1) | - | 992.5 | p<0.0001 | p<0.0001 |
| SRS-2 Parent Total T scores, M (SD) | 62 (12.2) | 70.2 (9.7) | 57 (10.8) | - | 1184.5 | p<0.0001 | p<0.0001 |
| SCQ-Lifetime Total, M (SD)g | 9.7 (7.7) | 15.5 (6.7) | 6.2 (5.9) | - | 783.5 | p<0.0001 | p<0.0001 |
| ASI Total, M (SD) | 35.8 (14.9) | 45.5 (11.3) | 30 (13.9) | - | 1195.0 | p<0.0001 | p<0.0001 |
| Vineland Adaptive Functioning-II, M | |||||||
| Communication | 81.9 (10.6) | 79.9 (11.2) | 83.2 (10) | - | 3600.5 | 0.04 | 0.07 |
| Socialization | 83.4 (14.4) | 76.6 (12.9) | 87.8 (13.6) | - | 4323.5 | p<0.0001 | p<0.0001 |
| Daily Living | 81.1 (10.9) | 77.4 (11) | 83.5 (10.2) | - | 4013.0 | p<0.0001 | 0.001 |
| ABC Composite scores | 80 (10.9) | 75.4 (11.7) | 83 (9.3) | - | 4195.5 | p<0.0001 | p<0.0001 |
| K-SADS ADHD severity, M (SD) | |||||||
| Inattentive | 6.8 (2.2) | 6.4 (2.5) | 7.1 (2) | - | 3668.0 | 0.16 | 0.24 |
| Hyperactive | 5.6 (2.4) | 5.5 (2.6) | 5.7 (2.4) | - | 3332.5 | 0.77 | 0.80 |
| Total | 12.4 (3.8) | 11.9 (4.4) | 12.8 (3.3) | - | 3541.0 | 0.32 | 0.44 |
| SWAN Parent Averaged scores, M (SD) | |||||||
| Inattentive | 1.1 (0.9) | 1.1 (0.9) | 1.2 (0.9) | - | 3397.5 | 0.61 | 0.69 |
| Hyperactive | 0.9 (1) | 1.1 (1) | 0.8 (1) | - | 2642.0 | 0.05 | 0.09 |
| Total | 1 (0.8) | 1.1 (0.8) | 1 (0.8) | - | 2982.0 | 0.38 | 0.52 |
| SWAN Teacher Averaged scores, M (SD)i | |||||||
| Inattentive | 1.1 (0.83) | 1.09 (0.723) | 1.12 (0.9) | - | 582.5 | 0.65 | 0.72 |
| Hyperactive | 0.86 (0.89) | 0.76 (0.83) | 0.92 (0.9) | - | 587.0 | 0.61 | 0.71 |
| Total | 0.98 (0.72) | 0.92 (0.65) | 1.02 (0.77) | - | 584.0 | 0.64 | 0.72 |
| CBCL T scores, M (SD)j | |||||||
| Internalizing | 58.5 (10.6) | 62.4 (9.3) | 56.2 (10.7) | - | 2129.0 | p<0.0001 | 0.001 |
| Externalizing | 58.8 (10.2) | 60.4 (10.6) | 57.9 (9.9) | - | 2770.5 | 0.16 | 0.25 |
| Total Problems | 62.3 (8.9) | 65.8 (8.1) | 60.2 (8.8) | - | 2059.5 | p<0.0001 | p<0.0001 |
| In-scanner motion, median FD, M (SD) | |||||||
| R-fMRI-1 6’20” | 0.08 (0.02) | 0.08 (0.02) | 0.08 (0.02) | - | 2784.5 | 0.13 | 0.21 |
| R-fMRI-2 4’39”k | 0.14 (0.04) | 0.1 (0) | 0.1 (0) | - | 967.0 | 0.15 | 0.23 |
| # Peaks ≥ 1 mm, M (SD) | |||||||
| R-fMRI-1 6’20” | 2.4 (6.8) | 3.4 (8.6) | 1.8 (5.3) | - | 2794.0 | 0.07 | 0.12 |
| R-fMRI-2 4’39”k | 2.8 (7.4) | 4.6 (11.2) | 2.1 (4.6) | - | 1003.5 | 0.18 | 0.24 |
| Euler index | −112 (76.6) | −117 (78.1) | −110 (75.9) | - | 3301.0 | 0.77 | 0.79 |
Diagnostic group differences (i.e., Autism Spectrum Disorder [ASD] vs Attention Deficit Hyperactivity Disorder without ASD [ADHDw/oASD]) were tested with Wilcoxon Rank sum tests for continous variables and with Pearson Chi-square tests for categorical variables. p-values were corrected with False Discovery Rate (FDR) at q>0.05; those remained significant after correction are shown in bold.
Missing data for one child with ASD;
“Other” includes American Indian/Alaskan Native, Native Hawaiian/Pacific Islander, mixed race or other not specified. Of the n=7 children with Autism Spectrum disorder (ASD) under “Other,” 5 identified as ‘other’, 1 as American Indian/Alaskan Native, and 1 as Native Hawaiian/other Pacific Islander. All 4 children with (Attention Deficit Hyperactivity disorder (ADHD) under “Other”, identified as ‘other’;
Missing data for n=3 children with ASD and n=2 with ADHD. The ASD group included: n=4 children in socioeconomic status (SES) Class 1, n=4 in SES Class 2, n=11 in SES Class 3, n=17 in SES Class 4, and n=24 in SES Class 5. The ADHD group included n=5 children in SES Class 1, n=8 children in SES Class 2, n=13 in SES Class 3, n=20 in SES Class 4 and n=55 SES Class 5;
medication status was missing for n=2 children with ASD.
Most children were administered Autism Diagnostic Observation Schedule-second edition (ADOS-2) module 3 (n=162, 98%; including n=59 ASD and n=103 ADHD), n=4 (2%) children with ASD were administered ADOS-2 module 2;
ADOS-based measure of expressive language validated in Mazurek et al. 2019, it ranges from 1 to 8 (“1” corresponds to “No spontaneous words or word approximations” and “8” to “Uses sentences in a largely correct fashion (must use some complex speech)”.
Social Communication Questionnaire (SCQ)-lifetime total scores are missing in n=6 children with ASD and n=6 with ADHD.
Missing in n=8 children (1 ASD, 6 ADHD);
The teacher version of the Strengths and Weaknesses of ADHD Symptoms and Normal Behavior Rating scale was available for n=69 children (n=26 with ASD, and n=42 with ADHD).
Missing the Child Behavior Checklist (CBCL) in n=1 child with ASD.
Second R-fMRI data collected for a subset of children from the primary sample (n=118);
Of 103 children designated as ADHDw/oASD, n=53 were identified as ADHD combined, n=36 were classified as predominantly inattentive, n=2 as predominantly hyperactive/impulsive, and n=10 as ADHD otherwise specified.
Abbreviations: DAS-II, Differential Ability Scales-2nd Edition; CSS, calibrated severity scores; SA, Social Affect; RRB, Restricted Repetitive Behaviors; SRS-P 2, Social Responsiveness Scale-second edition parent version; ASI, Autism Symptom Interview; K-SADS; Kiddie Schedule for Affective Disorders and Schizophrenia; df, degrees of freedom; W χ2, Wilcoxon/chi-square statistic.
Fig. 1: Key characteristics of the sample.
a) We included N=166 children comprising n=63 classified with autism and n=103 classified as ADHDw/oASD (blue diamonds and green circles, respectively). The box plots show the distribution of clinician-based ratings of autism and ADHD symptoms, indexed by the ADOS-2 CSS-Total (top) and KSADS total scores (bottom), respectively for the children divided by their primary diagnostic group. As expected, on average, the children in the autism group had significantly elevated CSS-Total scores. While on average, children in the ADHDw/oASD group had lower CSS-Total scores than the autism group; 38 (37%) of these children were at or above the ADOS-2 CSS-Total cutoff for autism spectrum (i.e., CSS-Total=4). This subset of children, labeled ADHDw/oASDAS+, is shown as dark green circles, the remaining (ADHDw/oASDAS−) are marked as light green circles. b) Correlation matrix across autism and ADHD symptom severity measures based on clinician’s ratings (ADOS-2 observation and KSADS parent interview) and parent questionnaire scores (SRS-2, SCQ, and SWAN, respectively). Only correlations surviving FDR-correction q<0.05 are shown in the matrix, with circle size indicating the correlation magnitude. Notably, unlike autism severity based on parent responses (i.e., SCQ and SRS-2), ratings based on clinicians’ observation (i.e., ADOS-2) were not significantly related to any ADHD ratings (i.e., SWAN and KSADS). c) Head micromovement distribution indexed by median framewise displacement (FD) across all 166 children included in analyses (top gray density plot), and by diagnostic group (bottom plots; blue autism and green ADHDw/oASD). Overall head motion was low with no significant differences between diagnostic groups. Abbreviations: ADOS-2 CSS-Total, Autism Diagnostic Observation Scale-2nd Edition Total Calibrated Severity scores; KSADS, Kiddie-Schedule for Affective Disorders and Schizophrenia for School-Age Children–Present and Lifetime Version; SRS-2, Social Responsiveness Scale-Second edition; SCQ-L, Social Communication Questionnairelifetime; SWAN, Strengths and Weaknesses of ADHD Symptoms and Normal Behavior.
Preprocessing was performed using the Configurable Pipeline for the Analysis of Connectomes (Fig S1; supplementary material).[57] Briefly, preprocessing steps for T1w images included intensity non-uniformity correction, skull stripping, tissue segmentation, and spatial normalization using ANTS. For R-fMRI scans, nuisance signal regression followed realignment to anatomical images and skull-stripping and included temporal bandpass filtering (0.01–0.1 Hz), removal of Friston 24-motion parameters, and 5-component-based noise correction (aCompCor);[58] functional-to-anatomical co-registration using FSL FLIRT’s boundary-based registration, spatial smoothing (6mm full-width-at-half-maximum) followed.
Brain connectome-wide association analyses
Overview.
To explore brain-behavior associations in the whole-brain, we employed a multivariate voxel-wise distance matrix regression (MDMR) discovery framework.[39, 59] MDMR identifies brain regions for which full-brain connectivity patterns vary by the behavioral variables of interest. The regions identified are then examined using seed-based voxel-wise correlation analysis (SCA) to delineate the specific iFC associated with that behavioral variable. Analyses focused on total ADOS-2 Calibrated Severity Scores (CSS-Total). We focused on total scores because autism is linked to impairment in both social communication and restricted and repetitive behaviors (RRBs)[60, 61] which have been both observed in children with ADHD.[4–6, 62, 63] Follow-up analyses explored ADOS-2 Social Affect (SA) and RRB domain calibrated subscores, separately. We explored ADHD-related iFC relationships using clinician-based ADHD total severity scores derived from the KSADS ratings, with follow-up exploration of inattention and hyperactivity/impulsivity ratings (see Supplementary material, Fig S2).
MDMR-based discovery framework.
Consistent with the previously developed MDMR approach,[39, 59] we first calculated voxel-wise iFC using Pearson correlations at the subject level. Second, for each voxel, we computed the distances between any possible pairing of participants’ correlation matrices (iFC) to yield a group-level distance matrix. Third, we calculated the distance matrix for each behavioral score across participants and modeled the associations between each behavioral distance matrix and brain-wide connectivity distance matrix at each voxel. We restricted analyses to voxels falling within a ≥25% probability FSL-based gray matter mask common to all participants (Fig S3). The MDMR model included age, sex, and median FD as nuisance covariates. To examine the unique contributions of autism or ADHD symptom severities, both ADOS-2 CSS and ADHD-KSADS ratings were included in the model. Statistical significance was assessed with pseudo F-statistic testing using 5,000 permutations, and was followed by Gaussian random field theory correction at voxel-level thresholds Z>3.1 and cluster level p<0.01. The significant regions identified from MDMR, i.e., those that revealed significant relationships between variation in symptoms ratings and iFC, were selected as seeds for subsequent whole-brain voxel-wise SCA; for each region of interest (ROI, i.e., seed), iFC was computed using the Pearson correlation between the mean time series of the ROI and those of any other voxels included in the study’s gray matter mask. Correlation coefficients were Fisher r-to-z transformed at the individual level. Then, a group-level random effect general linear model via FSL-FEAT was conducted to assess the ROI’s iFC with the behavioral variable(s) of interest, including the same covariates described above along with each participant’s mean iFC of the seed examined. We corrected for multiple comparisons using Gaussian random field theory (Z>3.1, p<0.01). The MDMR code is available at http://connectir.projects.nitrc.org.
Robustness
Secondary analyses investigated the robustness of our cluster-level findings to different methodological decisions. We assessed whether similar iFC-behavior associations identified in primary analyses employing the 24-parameter model and CompCor for nuisance regression, were observable after using either of two alternative MRI denoising pipelines, or longer scan data, or parent behavioral measures. Specifically, for alternative denoising, we selected two validated approaches — global signal regression (GSR)[64] and 36-parameter nuisance regression (36P).[65] Guided by accumulating evidence that longer scans yield more reliable results,[66, 67] we also assessed the robustness of primary findings after concatenating R-fMRI data across two rest scans (6.33+4.65=10.98 min), when available (n=118). For alternative symptom measures, we examined commonly used parent-based ratings including the SRS-2[49] and SCQ-L,[50] for autism, and the SWAN,[51] for ADHD symptoms. Analyses were conducted at the cluster-level, as detailed in Supplementary material. Findings with statistically significant correlations surviving FDR correction across all tests were considered robust.
Genetic decoding and enrichment
We explored whether gene expression topographically associated with the iFC map(s) revealed by our discovery MDMR/SCA approach were significantly enriched for genes found to have greater rate of variants in autism and ADHD.[42] Consistent with previous studies,[68, 69] we carried out gene expression decoding and enrichment analyses. First, using NeuroVault[70] and the Allen Institute Human Brain Gene Expression Atlas,[71] we identified genes topographically expressed in the iFC map(s) of interest, i.e., gene decoding. Next, using hypergeometric-based statistical testing,[72] we assessed if the decoded gene list would include, above chance, genes implicated in autism and ADHD,[42] i.e., gene enrichment. The gene list we tested included 1046 protein-truncating and missense genes that have been identified in the largest exome sequence, to date, of individuals with autism and/or ADHD, (Supplementary material, Table S4).[42] Finally, to explore the biological processes potentially associated with the significantly enriched genes, we performed gene ontology (GO) searches with the Gene Ontology Consortium database[73] retaining terms surviving FDR q<0.05 (Supplementary material).
RESULTS
Sample Characteristics
Table 1 and Fig. 1 summarize the characteristics of the sample which included n=63 children meeting criteria for ASD and n=103 children for ADHDw/oASD with group average intelligence within the typical range. Child and parent evaluators expressed substantial certainty for these primary diagnostic assignments (8.5±1.6 and 8.7±1.6, for ASD and ADHDw/oASD, respectively; 10 indicates full certainty - see Supplementary material) and inter-rater agreement was excellent (ICC(1,1)=0.94 and 0.91 for ASD and ADHDw/oASD, respectively). Consistent with the clinical literature on autism and ADHD,[18, 74] psychiatric comorbidity was frequent (n=108, 65%; see Table S5). Notably, within the autism group, the most represented comorbidity was ADHD, alone or in combination with other diagnoses (84%, n=53). As expected, the autism group showed significantly more severe ratings across all relevant autism measures than ADHDw/oASD (Table 1, Fig. 1). However, consistent with prior reports of elevated autistic traits in ADHD,[3, 5] 37% (n=38) of the ADHDw/oASD group fell at or above the ADOS-2 CSS-Total cutoff for autism spectrum (i.e., CSS=4; Fig. 1a) despite not fully meeting DSM-5 ASD criteria per clinician’s best estimates. Consistent with reports of high prevalence of ADHD in autism,[3, 5, 6] and with the high rate of comorbid ADHD in the autism sample, the two primary diagnostic groups did not differ in ADHD symptom severity across clinician, parent, nor teacher ratings (Fig. 1a, Table 1). Of note, parent-reported autism-related scores were significantly correlated with ADHD ratings derived by clinician’s parent interviews or questionnaires. In contrast, clinician-based ADOS-2-CSS were not significantly related to those ratings (Fig. 1b).
Autism symptom severity is associated with connectivity between nodes of the frontoparietal and default mode networks
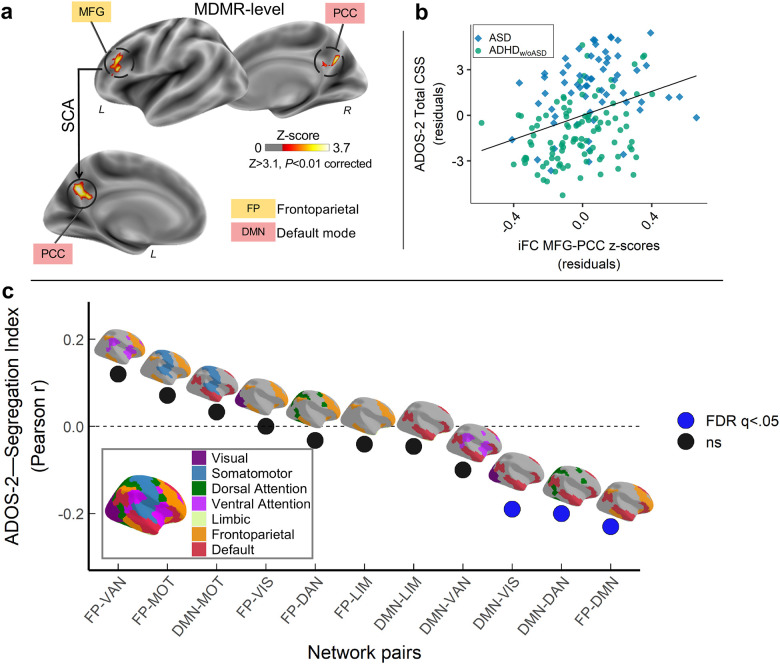
MDMR.
Across children, MDMR discovery whole-brain analyses revealed statistically significant (Z>3.1, p<0.01) transdiagnostic iFC relationships with ADOS-2 CSS-Total in two clusters in the left hemisphere: middle frontal gyrus (MFG), and precuneus/posterior cingulate cortex (PCC), nodes of the frontoparietal (FP) and default mode (DM) networks, respectively (Fig. 2a). Secondary analyses exploring the contribution of ADOS-2 subdomain scores, CSS-SA and CSS-RRB, yielded no connectome-wide level associations surviving our rigorous statistical threshold (Z>3.1; p<0.01; Fig S4a). MDMR did not reveal statistically significant associations with ADHD symptoms indexed by total, inattentive or hyperactivity/impulsivity ratings from clinicians’ parent interviews nor parent questionnaires (Fig S5). A similar pattern of MDMR results for ADOS-2 and KSADS-ADHD total was observed after adding full-scale IQ to the models. Accordingly, the follow-up SCA analyses focused on autism symptoms.
Fig. 2: Internetwork connectivity between frontoparietal and default networks is transdiagnostically associated with autistic symptoms.
a) Overlaid on inflated brain surface maps are the clusters with intrinsic functional connectivity (iFC) significantly associated with Autism Diagnostic Observation Scale-2nd Edition Total Calibrated Severity scores (ADOS-2 CSS-Total) based on multivariate distance matrix regression (MDMR) and follow-up seed-based correlation analyses (SCA): left middle frontal gyrus (MFG: MNI152 x=−45, y=29, z=18) within the frontoparietal network (FP; mustard), and posterior cingulate cortex/precuneus (PCC: MNI152 x=−5, y=−59, z=30) within the default mode network (DMN; red). All analyses were Gaussian Random Field corrected at Z>3.1 and p<0.01. b) Scatter plot showing the relationship between CSS-Total and MFG iFC with PCC across children with autism spectrum disorder (ASD; blue diamonds) and Attention-Deficit/Hyperactivity Disorder without autism (ADHDw/oASD; green circles). Both iFC and CSS-Total are shown after regressing the same covariates used in discovery brain-behavior analyses (i.e., median FD, age, sex and KSADS ADHD total score). c) Correlation values (Pearson r) between ADOS-2 CSS-Total and segregation index (SI) between FP and DMN, as well as the SI for DMN and FP with each of the other networks defined by the 7-network Yeo-Krienen atlas (i.e., visual [VIS; purple], somatomotor [MOT; blue], dorsal attention [DAN; green], ventral attention [VAN; pink], limbic [LIM; khaki]). The SI-CSS correlations surviving FDR correction at q<0.05 are indicated as blue circles whereas black circles indicate non-significant associations. As shown, SI between DMN and FP, as well as between DMN and both DAN and VIS were significantly negatively correlated with CSS.
SCA.
Follow-up SCA for the MDMR-identified clusters in MFG and PCC revealed a statistically significant association with CSS-Total and iFC between these nodes after controlling for KSADS ADHD total, along with the nuisance covariates listed above. Results indicated that children with higher (i.e., more severe) CSS-Totals have stronger iFC between MFG and PCC (Fig. 2b). Although MDMR analyses did not reveal significant iFC associations with CSS-SA or CSS-RRB, exploring these symptom correlations within MFG-PCC iFC indicated a statistically significant correlation with CSS-SA but not with CSS-RRB scores (r(164)=0.23, p=0.003; r(164)=0.08, p=0.290, respectively; Fig S4b).
‘High’ and ‘low’ autistic traits subgroups.
To further summarize findings in the context of transdiagnostic clinical presentations, we identified and compared two subgroups defined as having either ‘high’ or ‘low’ autistic traits. Specifically, regardless of their primary diagnostic status, children falling at or above the ADOS-2 autism cutoff (i.e., CSS-Total≥4) were included in the ‘high’ autistic traits subgroup, the remaining were in the ‘low’ subgroup (n=92, 55% and n=74, 45%, respectively). As expected, the ‘high’ autistic traits subgroup had significantly more positive MFG-PCC iFC. Both subgroups included children from the autism and ADHDw/oASD primary diagnostic groups, though the ‘high’ subgroup included a statistically greater proportion of those with DSM-5 ASD diagnoses (86% of the autism sample; see Table 2). The ‘high’ subgroup had greater prevalence of males, showed elevated parent autism severity ratings and greater impairment in adaptive functioning (Table 2). Importantly, no significant subgroup differences were noted in ADHD severity measures, comorbidity rates, nor micromotion in R-fMRI data (Table 2).
Table 2.
Characteristics of the ‘high’ and ‘low’ autistic traits subgroups.
| Variable | ASD traits | Statistics | ||||
|---|---|---|---|---|---|---|
| High n=92 |
Low n=74 |
df | F χ2 | p value | FDR-p value | |
| Primary Diagnosis, # (%) | 1 | 35.8 | p<0.0001 | p<0.0001 | ||
| ASD | 54 (59) | 9 (12) | ||||
| ADHDw/oASD | 38 (41) | 65 (88) | ||||
| ADHD Presentation, # (%) | 3 | 1.4 | 0.72 | 0.76 | ||
| ADHD-Combined | 39 (57) | 32 (48) | ||||
| ADHD-Inattentive | 21 (30) | 23 (34) | ||||
| ADHD-Hyperactive/Impulsive | 3 (4) | 3 (4) | ||||
| ADHD Not Otherwise Specified | 6 (9) | 9 (13) | ||||
| Age, years, M (SD) | 8.8 (1.8) | 9.0 (1.7) | - | 3044.0 | 0.24 | 0.32 |
| Sex assigned at birth (males) #, (%) | 78 (85) | 47 (64) | 1 | 8.9 | 0.003 | 0.01 |
| Ethnicity (Hispanic) #, (%)a | 22 (24) | 17 (23) | 1 | 0.0 | 1 | 1 |
| Race #, (%)ab | 4 | 2.2 | 0.691 | 0.73 | ||
| White | 50 (55) | 45 (61) | ||||
| Mixed | 16 (18) | 10 (14) | ||||
| Black/African American | 10 (11) | 9 (12) | ||||
| Asian | 7 (8) | 7 (9) | ||||
| Other | 8 (9) | 3 (4) | ||||
| Socio-economic status class #, (%)c | 1 | 3.0 | 0.084 | 0.14 | ||
| Class 1–3 | 30 (34) | 15 (21) | ||||
| Class 4–5 | 58 (66) | 58 (79) | ||||
| Psychiatric Comorbidity (yes), # (%) | 67 (73) | 41 (55) | 1 | 4.7 | 0.03 | 0.06 |
| DAS-II IQ Standard scores, M (SD) | ||||||
| Verbal IQ | 105 (17) | 109 (14) | - | 2852.0 | 0.07 | 0.13 |
| Non-verbal VIQ | 103 (19) | 105 (15) | - | 2954.0 | 0.14 | 0.21 |
| Full-scale IQ | 103 (19) | 106 (13) | - | 2983.0 | 0.17 | 0.23 |
| SRS-2 Parent Total T scores, M (SD) | 64.9 (11) | 58.4 (12.7) | 4509.5 | p<0.0001 | 0.001 | |
| SCQ-Lifetime Total scores, M (SD)d | 11.1 (7.3) | 7.9 (7.8) | - | 3783.0 | 0.002 | 0.01 |
| ASI total scores, M (SD) | 39.9 (13.0) | 30.9 (15.7) | - | 4451.0 | p<0.0001 | p<0.001 |
| Vineland Adaptive Functioning-II, M | ||||||
| Com munication | 81.0 (11.3) | 83.1 (9.6) | - | |||
| Socialization | 81.1 (13.8) | 86.3 (14.6) | - | 2421.5 | 0.02 | 0.04 |
| Daily Living | 79.1 (10.1) | 83.6 (11.4) | - | 2410.0 | 0.013 | 0.03 |
| ABC Composite scores | 77.9 (11.3) | 82.6 (10) | - | 2330.5 | 0.008 | 0.02 |
| K-SADS ADHD severity, M (SD) | ||||||
| Inattentive | 6.46 (2.31) | 6.68 (2.43) | - | 3128.5 | 0.37 | 0.44 |
| Hyperactive | 5.15 (2.39) | 5.43 (2.66) | - | 3175.0 | 0.46 | 0.51 |
| Total | 11.61 (3.96) | 12.11 (4.08) | - | 3122.5 | 0.36 | 0.44 |
| SWAN Parent Averaged scores, M (SD) | ||||||
| Inattentive | 1.23 (0.89) | 1.02 (0.95) | - | 3849.0 | 0.15 | 0.21 |
| Hyperactive | 1.06 (0.9) | 0.79 (1.04) | - | 3850.0 | 0.15 | 0.21 |
| Total | 1.15 (0.77) | 0.91 (0.85) | - | 4006.0 | 0.05 | 0.10 |
| CBCL T scores, M (SD)f | ||||||
| Internalizing | 59.22 (10.09) | 57.68 (11.19) | - | 3631.0 | 0.39 | 0.44 |
| Externalizing | 59.38 (10.21) | 58.19 (10.25) | - | 3658.0 | 0.34 | 0.43 |
| Total Problems | 63.44 (8.68) | 60.93 (9.08) | - | 3934.0 | 0.06 | 0.12 |
| In-scanner motion, median FD, M (SD) | 0.08 (0.02) | 0.08 (0.02) | - | 3784.5 | 0.22 | 0.28 |
| MFG-PCC iFC, M (SD) | 0.13 (0.22) | 0.001 (0.23) | - | 4429.0 | 0.001 | 0.003 |
Subgroup differences were tested with Wilcoxon Rank sum tests for continous variables and with Pearson Chi-square tests for categorical variables. p-values were corrected with False Discovery Rate (FDR) at q>0.05; those remained significant after correction are shown in bold
Missing data for one child from the ‘high’ autistic traits group;
“Other” includes American Indian/Alaskan Native, Native Hawaiian/Pacific Islander, mixed race or other not specified. Of the n=8 children of the ‘high’ group under “Other,” 6 identified as ‘other’, 1 as American Indian/Alaskan Native, and n=1 as Native Hawaiian/other Pacific Islander. All n=3 children in the ‘low’ autistic traits subgroup under “Other”, identified as ‘other’;
The ‘high’ subgroup included: n=4 children in socioeconomic status (SES) Class 1, n=8 in SES Class 2, n=18 in SES Class 3, n=19 in SES Class 4, and n=39 in SES Class 5. The ‘low’ group included n=5 children in SES Class 1, n=4 children in SES Class 2, n=6 in SES Class 3, n=18 in SES Class 4 and n=40 SES Class 5. Missing data for n=4 children from the ‘high’ group and one child from the ‘low’;
Social Communication questionnaire (SCQ)-lifetime total scores were missing for n=5 children from the ‘high’ group and n=7 in the ‘low’ group.
Vineland Adaptive Functioning-2nd edition were not available for n=8 children (5 ‘high’, 3 ‘low’);
Missing CBCL in n=1 child from the ‘high’ group.
Abbreviations: ASD, Autism Spectrum Disorder; ADHDw/oASD, Attention Deficit Hyperactivity Disorder - without ASD; DAS-II, Differential Ability Scale-2nd Edition; SRS-P 2, Social Responsiveness Scale, parent version 2; ASI, Autism Symptom Interview, SWAN-P, Strengths and Weaknesses of ADHD Symptoms and Normal Behavior Rating Scale, parent version; K-SADS; Kiddie Schedule for Affective Disorders and Schizophrenia; df, degrees of freedom; W χ2, Wilcoxon /chi-square statistic.
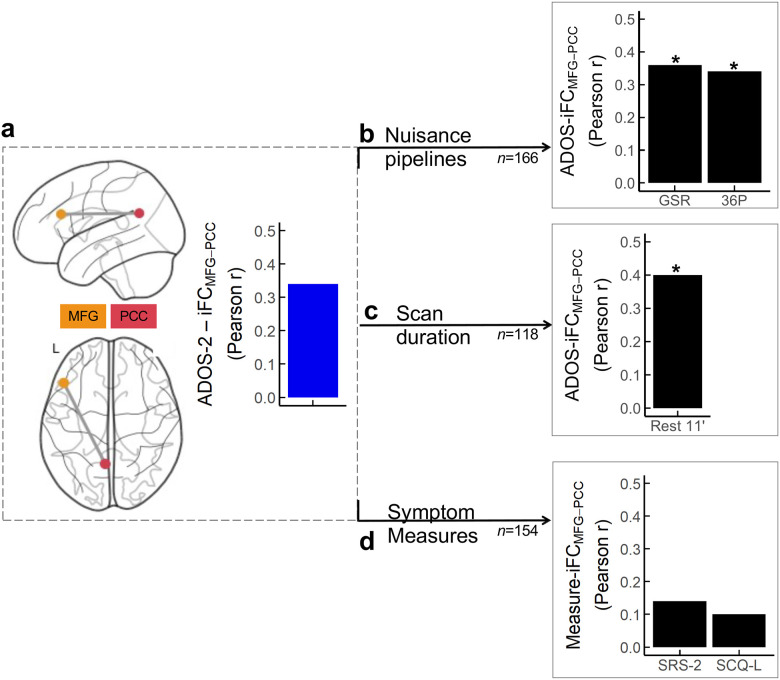
Robustness.
Cluster-level robustness analyses revealed that the association between MFG-PCC iFC and CSS-Total was robust to alternative MRI preprocessing pipelines (r(164)=0.35 and 0.34, for GSR and 36P, respectively, with FDR q<0.0001; Fig. 3) and to longer R-fMRI data (r(116)=0.42; FDR q<0.0001). Findings were not robust to the two parent-based measures of autism symptoms used as alternatives to the clinician-based ADOS-2 scores (i.e., r(152)=0.14 and 0.10, for SRS-2 and SCQ-L, respectively; FDR q=0.08 and q=0.21).
Fig. 3: Robustness of brain-behavior primary findings to alternative MRI nuisance strategies, scan duration, and behavioral measures.
a) Overlaid on Nilearn surface glass brains are the clusters with iFC significantly related to the Autism Diagnostic Observation Scale-second edition (ADOS-2) Total Calibrated Severity Scores (CSS-Total) based on the MDMR/SCA discovery analyses — i.e., the middle frontal gyrus (MFG) of the frontoparietal network (FP; mustard), and the posterior cingulate/precuneus (PCC) of the default mode network (DMN, red). The blue histogram illustrates the magnitude of the correlation between the MFG-PCC iFC and the ADOS-2 CSS-Total (Rest 6 minutes + discovery preprocessing pipeline, aCompCor) from the discovery analyses. The black histograms index the magnitude of the MFG-PCC correlation with ADOS-2 CSS-Total after: b) preprocessing using global signal regression (GSR) or 36 nuisance parameter regression (36P) in the 166 children included in the study; c) concatenating two quality assured resting state fMRI (R-fMRI) scans (6’20”+4’39”=10’59”) in a subset of children completing both R-fMRI scans n=118); d) using parent ratings on the Social Responsiveness Scale-second edition (SRS-2) or the Social Communication Questionnaire-lifetime (SCQ-L) available in n=154 children. Asterisks indicate that correlations were statistically significant following FDR correction.
Reduced segregation between DMN and the FP, dorsal attention and visual networks
Given that the iFC association with CSS-Total involved regions of the FP and DMN, we investigated if these brain-behavior patterns extended more broadly to FP and DMN internetwork patterns. To this end, we computed the segregation index (SI)[75] — i.e., the relative strength of within-network to between-network iFC — of each DMN and FP networks with the other functional networks defined by Yeo-Krienen[76] (see Supplementary material). Consistent with primary findings, we found significant negative associations between SI and CSS-Total such that more severe autism symptoms were associated with lower SI (higher internetwork iFC) between DMN and the FP, visual and dorsal attention networks (FDR q<0.05), but not with other network pairs (Fig. 2c).
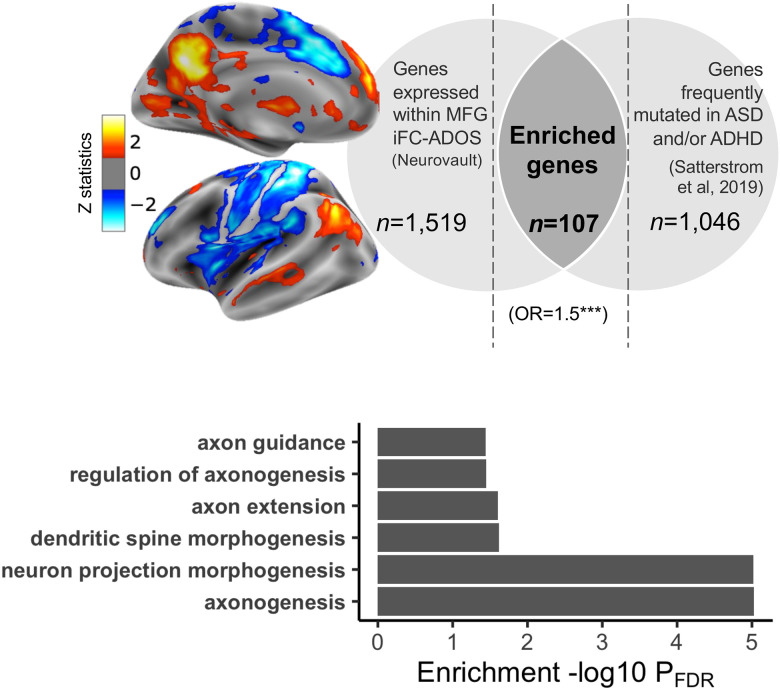
Transdiagnostic in silico gene expression
Given that primary SCA analyses revealed a transdiagnostic association between autism symptom severity and iFC of the MFG, we first ran gene decoding in the MFG-iFC map. This analysis identified n=1,519 genes topographically expressed in the MFG-iFC map (Fig. 4; gene list in GitHub). We then tested for genetic enrichment among rare genetic variants (protein truncating and missense) previously shown to have greater rates in autism and ADHD relative to controls.[42] Analyses identified significant enrichment for n=107 of these genes (OR=1.6, p=0.0002; Supplementary material, Fig. 4, Table S6). Confirming specificity of these findings, control enrichment analyses conducted using an alternative topographic gene expression map, selected to not spatially overlap with the MFG-iFC map, did not yield any significant findings (see Supplementary material). To explore the biological processes related to the 107 enriched genes, we leveraged the Gene Ontology (GO) annotation database.[73] Analyses yielded significant (FDR q<0.05) associations with n=78 GO terms (Table S7); among them most significant terms indexed neurodevelopmental morphogenesis processes involved in axonogenesis (Supplementary Material, Fig. 4b, Fig S6).
Fig. 4: Transdiagnostic transcriptomic signature of iFC maps associated with autism symptom severity.
a. We performed gene decoding analyses on the iFC statistical map of the middle frontal gyrus (MFG) seed associated with the Autism Diagnostic Observation Scale-2nd Edition (ADOS-2) Total Calibrated Severity Scores (CSS-Total). The Z statistic MFG iFC map is overlaid on inflated surface maps. Gene decoding identified N=1,519 genes as having a spatial expression pattern significantly similar to the MFG iFC map. They are shown in the left light gray area of the Venn diagram. The right light gray area of the Venn diagram shows the number of genes (N=1,046 reported to be dysregulated in autism and/or ADHD by Satterstrom et al. 2019.[42] The number of genes resulting from gene enrichment analysis is depicted in the dark gray area of the Venn diagram as the overlap between these genes and those decoded. b. Specific biological GO terms included under the neuron morphogenesis term class associated with genes linked to the MFG iFC map, and related statistics (see also Fig S6 and Table S7).
DISCUSSION
Recognizing the frequent co-occurrence of autism- and ADHD-related symptoms,[2–6] we investigated their specific contribution to iFC across school-aged verbal children with rigorously established DSM-5 diagnoses of ASD or of ADHDw/oASD. Whole-brain voxel-wise multivariate analyses of low motion data revealed transdiagnostic brain-behavior relationships specific to autism symptom severity, even after accounting for ADHD symptoms. Across children, inter-individual variability in symptom severity, indexed by gold-standard clinicians’ observation ratings (i.e., ADOS-2), was associated with inter-individual variability in iFC between nodes of the DMN and FP networks. Complementing results from network segregation analyses showed that autism symptom severity was inversely related to the segregation of the DMN with the FP, dorsal attention, and visual networks. In silico analyses revealed that gene expression within the iFC map associated with autism symptoms was significantly enriched for genes previously shown to have a higher rate of variants in individuals with autism and ADHD diagnoses without intellectual disability. Overall, our results point towards transdiagnostically shared biological underpinnings of autism symptom severity that may inform biomarker research, as well as models of vulnerability, for these neurodevelopmental conditions.
The present study underscores the role of internetwork iFC for autism-related behaviors across diagnoses. Autism-related increases in internetwork iFC have been reported in both traditional case-control comparisons[77–85] and dimensional designs within autistic samples.[77–81, 86] Our findings not only support their role among children with autism, but also elucidate their specific contribution to autism symptoms among children with ADHDw/oASD. Although R-fMRI studies of samples combining autism and ADHD are increasing,[87] only three studies have examined a priori dimensional associations of inter-individual differences in iFC with autism symptoms.[15–17] Two of them identified autism symptom associations with internetwork iFC.[15, 17] However, the autism-related iFC patterns were also associated with ADHD, thus neither study established specificity to autistic symptoms. In contrast, our phenotyping strategy of meticulously assessing both autism and ADHD symptoms in all participants allowed us to examine their relative contribution to iFC.
Additionally, prior MRI studies have primarily used parent questionnaires to index symptom severity. Although valid, these measures are known to covary with more general ratings of psychopathology (i.e., not specific to autism),[88] a finding also observed in our data. Thus, unlike prior efforts, to index variation more specific to autism we leveraged expert-based observational measures of autism severity (i.e., ADOS-2) obtained blindly to diagnostic concerns across all children.[88] Relatedly, brain-behavioral associations were weaker and statistically non-significant for parent-based ratings. In contrast, internetwork connectivity associations with ADOS-2 scores were robust to different R-fMRI analytical strategies. These findings underscore that the behavioral measures employed for brain association analyses matter, as much as the MRI metrics examined. This is in line with recent work highlighting that selecting clinical measures with high test-retest reliability increases the ability to detect relationships with brain functional metrics.[89]
Our internetwork connectivity findings involving the DMN shed new light on the role of DMN atypical connectivity previously reported in studies of either autism or ADHD.[87] By focusing on symptom domains rather than diagnostic status, our results underscore that the DMN’s interaction with other functional networks is relevant to autism symptom severity across both conditions. In typical children, internetwork iFC decreases significantly during childhood.[28, 83] These maturational processes are thought to support greater functional network segregation/specialization. Accordingly, mechanisms involved in functional network maturation may play a significant role in the emergence of autistic symptoms in children with diagnoses of autism, and, at least a subset of those with diagnosis of ADHD. The present work primarily focused on total scores of autism severity given that autism is linked to impairment in both social communication and restricted and repetitive behaviors; it was however, limited in its ability to differentiate the contribution of specific symptom subdomains. Future larger studies would benefit from identifying and leveraging finer grained autism symptom metrics to clarify whether reduced segregation of the DMN indexes a general vulnerability to autism severity or whether it has a specific role for symptom subdomains.
To our knowledge this is the first study examining, albeit in silico, gene expression associations with iFC in the context of brain-behavior dimensional relationships in a transdiagnostic sample including children with autism and/or ADHD. We leveraged a gene list derived from the largest exome sequencing study of autism and ADHD including protein truncating and missense genetic variants[42]. Analyses revealed that genes with a greater burden of pathogenic variants in these conditions are topographically associated with iFC patterns relevant to autism symptom severity across diagnostic groups. While this finding is consistent with models suggesting that shared genetic factors underlie disorder overlaps,[90–92] it also extends this line of work one step further by revealing that aspects of shared genetic variance among these conditions may mark vulnerability specifically for autism symptom severity rather than categorical diagnoses. This provides a more specific framework to explore previously suggested pleiotropic effects. Notably, gene ontology analyses involved terms associated to neuron morphogenesis including axonogenesis, dendritic spine morphogenesis, that have previously implicated in autism and ADHD.[93, 94] Such in silico associations provide hypotheses for future large-scale genome-wide association studies that can investigate shared and distinct genetic vulnerabilities of both rare and common variants.
Beyond providing biological insights, our systematic administration of clinician-based autism assessments in all children, regardless of their diagnostic concerns, detected elevated autistic traits in a substantial group of children identified as ADHDw/oASD. This is consistent with an emerging clinical literature recognizing that autism symptoms can be expressed in varying degrees of severity across individuals — both within and beyond diagnostic boundaries.[2–6] Recognizing children with ADHD and co-occurring autistic traits is clinically relevant. Consistent with prior work,[95] we found greater impairment in adaptive functioning in the ‘high’ autistic traits subgroup, regardless of diagnostic status (autism, ADHDw/oASD). Taken together, the present work highlights the importance of accounting for autistic traits when attempting to explain both the clinical and biological variance in ADHD samples.
Finally, we note that we did not observe significant connectome-wide associations specific to ADHD parent-reported severity indices, even after controlling for autism severity. Given the absence of standardized ADHD observational measures, clinician ratings were necessarily based on semi-structured parent interviews. Additionally, the relatively restricted (albeit in the clinical range) distribution of ADHD severity in the sample, may have limited the ability to detect brain-behavior associations. These observations must be placed in the context of our sample size, which, although considerably larger than the median of N~23 in the literature,[96] is nonetheless only moderately large. At the same time, our findings underscore that observational behavioral measures of ADHD are needed for brain behavior associations along with a finer characterization of ADHD presentations.
Our results should be interpreted in light of limitations. First, reflecting the known preponderance of boys with these conditions, most participants were male. Thus, although we found greater prevalence of males in the ‘high’ autistic traits subgroup, this observation needs to be replicated in larger female samples. Second, as already mentioned,[96] our sample size is only moderately large and thus future efforts with better powered samples are needed. Larger transdiagnostic rigorously characterized samples will allow studies to represent, and thus assess the role of, a greater range of demographics, cognitive, as well as autism and ADHD presentations that were not assessed here.
In sum, the findings from this study enhance our understanding of the significance of internetwork iFC in autism symptom severity in children with autism and/or ADHD. They also point towards shared molecular pathways involving morphogenetic developmental processes linked to neuron projection and regulation. This work underscores the promise of exploring symptom dimensions across diagnostic categories to identify children with linked clinical and biological phenotypes and their underlying vulnerabilities. It also highlights the importance of utilizing more objective markers of behavioral phenotypes in these efforts. To realize the full potential of the framework presented in this study, future work needs to extend it to multiple dimensions across diagnoses as in ongoing efforts.[97] To fully accomplish this goal, collaborative transdiagnostic efforts using harmonized phenotyping and neuroimaging protocols are needed to gather sufficiently large samples. Such efforts will facilitate the assessment of diagnostic-specific dimensional relationships that may accompany diagnostically-shared brain-behavior associations, as shown in prior transdiagnostic studies,[98, 99] as well as enable brain-behavior subtyping to account for ADHD/ASD heterogeneity.[100]
Supplementary Material
ACKNOWLEDGEMENTS
We are deeply grateful to the children and their families participating in the study, as well as the clinical and research staff members at the Child Mind Institute and at the Child Study Center at NYU Langone Health, who supported data collection and curation. We would like to thank Steven Giavasis and the CPAC team for providing technical support for setting up the MRI preprocessing pipelines in CPAC, and to Jessica Cloud for her assistance in some aspects of MRI data quality assurance. We would also like to acknowledge the Neurovalt’s development team and the Allen Institute for their open science initiatives that enabled our in silico gene expression analyses. This work was partially funded by NIH R01MH105506 and R01MH115363 to ADM, R01MH091864 and R01MH120482 to MPM; the Korean Ministry of Science and ICT of the National Research Foundation RS-2023-00265410 to SHK, as well as generous gifts to the Child Mind Institute from the Dr. John and Consuela Phelan (to ADM), and from the Phyllis Green and Randolph Cowen Foundation (to MPM).
Footnotes
CONFLICT OF INTEREST
Drs. Lord and Bishop receive royalties for the sale of diagnostic instruments they have co-authored (ADOS-2 and/or SCQ); profits generated from any of their own research or clinical activities are donated to charity. Dr. Di Martino is coauthor of the Italian version of the Social Responsiveness Scale — child version distributed in Italy by Organizzazioni Speciali, Italy. All other co authors report no financial interests or potential conflicts of interest.
REFERENCES
- 1.Cuthbert BN. The role of RDoC in future classification of mental disorders. Dialogues Clin Neurosci. 2020;22:81–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kern JK, Geier DA, Sykes LK, Geier MR, Deth RC. Are ASD and ADHD a Continuum? A Comparison of Pathophysiological Similarities Between the Disorders. J Atten Disord. 2015;19:805–827. [DOI] [PubMed] [Google Scholar]
- 3.Hollingdale J, Woodhouse E, Young S, Fridman A, Mandy W. Autistic spectrum disorder symptoms in children and adolescents with attention-deficit/hyperactivity disorder: a meta-analytical review. Psychol Med. 2020;50:2240–2253. [DOI] [PubMed] [Google Scholar]
- 4.Martin J, Hamshere ML, O’Donovan MC, Rutter M, Thapar A. Factor structure of autistic traits in children with ADHD. J Autism Dev Disord. 2014;44:204–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grzadzinski R, Di Martino A, Brady E, Mairena MA, O’Neale M, Petkova E, et al. Examining autistic traits in children with ADHD: does the autism spectrum extend to ADHD? J Autism Dev Disord. 2011;41:1178–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grzadzinski R, Dick C, Lord C, Bishop S. Parent-reported and clinician-observed autism spectrum disorder (ASD) symptoms in children with attention deficit/hyperactivity disorder (ADHD): implications for practice under DSM-5. Mol Autism. 2016;7:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roelfs D, Frei O, van der Meer D, Tissink E, Shadrin A, Alnaes D, et al. Shared genetic architecture between mental health and the brain functional connectome in the UK Biobank. BMC Psychiatry. 2023;23:461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rommelse NNJ, Franke B, Geurts HM, Hartman CA, Buitelaar JK. Shared heritability of attention-deficit/hyperactivity disorder and autism spectrum disorder. Eur Child Adolesc Psychiatry. 2010;19:281–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoogman M, van Rooij D, Klein M, Boedhoe P, Ilioska I, Li T, et al. Consortium neuroscience of attention deficit/hyperactivity disorder and autism spectrum disorder: The ENIGMA adventure. Hum Brain Mapp. 2022;43:37–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baribeau DA, Dupuis A, Paton TA, Hammill C, Scherer SW, Schachar RJ, et al. Structural neuroimaging correlates of social deficits are similar in autism spectrum disorder and attention-deficit/hyperactivity disorder: analysis from the POND Network. Translational Psychiatry. 2019;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ameis SH, Lerch JP, Taylor MJ, Lee W, Viviano JD, Pipitone J, et al. A diffusion tensor imaging study in children with ADHD, autism spectrum disorder, OCD, and matched controls: Distinct and non-distinct white matter disruption and dimensional brain-behavior relationships. Am J Psychiatry. 2016;173:1213–1222. [DOI] [PubMed] [Google Scholar]
- 12.Kushki A, Cardy RE, Panahandeh S, Malihi M, Hammill C, Brian J, et al. Cross-Diagnosis Structural Correlates of Autistic-Like Social Communication Differences. Cereb Cortex. 2021;31:5067–5076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aoki Y, Yoncheva YN, Chen B, Nath T, Sharp D, Lazar M, et al. Association of White Matter Structure With Autism Spectrum Disorder and Attention-Deficit/Hyperactivity Disorder. JAMA Psychiatry. 2017;74:1120–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Di Martino A, Zuo X-N, Kelly C, Grzadzinski R, Mennes M, Schvarcz A, et al. Shared and distinct intrinsic functional network centrality in autism and attention-deficit/hyperactivity disorder. Biol Psychiatry. 2013;74:623–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Itahashi T, Fujino J, Sato T, Ohta H, Nakamura M, Kato N, et al. Neural correlates of shared sensory symptoms in autism and attention-deficit/hyperactivity disorder. Brain Commun. 2020;2:fcaa186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Choi EJ, Vandewouw MM, Taylor MJ, Arnold PD, Brian J, Crosbie J, et al. Beyond diagnosis: Cross-diagnostic features in canonical resting-state networks in children with neurodevelopmental disorders. Neuroimage Clin. 2020;28:102476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lake EMR, Finn ES, Noble SM, Vanderwal T, Shen X, Rosenberg MD, et al. The Functional Brain Organization of an Individual Allows Prediction of Measures of Social Abilities Transdiagnostically in Autism and Attention-Deficit/Hyperactivity Disorder. Biological Psychiatry. 2019;86:315–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Simonoff E, Pickles A, Charman T, Chandler S, Loucas T, Baird G. Psychiatric disorders in children with autism spectrum disorders: prevalence, comorbidity, and associated factors in a population-derived sample. J Am Acad Child Adolesc Psychiatry. 2008;47:921–929. [DOI] [PubMed] [Google Scholar]
- 19.Antshel KM, Russo N. Autism Spectrum Disorders and ADHD: Overlapping Phenomenology, Diagnostic Issues, and Treatment Considerations. Curr Psychiatry Rep. 2019;21:34. [DOI] [PubMed] [Google Scholar]
- 20.Rong Y, Yang C-J, Jin Y, Wang Y. Prevalence of attention-deficit/hyperactivity disorder in individuals with autism spectrum disorder: A meta-analysis. Res Autism Spectr Disord. 2021;83:101759. [Google Scholar]
- 21.Antshel KM, Zhang-James Y, Wagner KE, Ledesma A, Faraone SV. An update on the comorbidity of ADHD and ASD: a focus on clinical management. Expert Rev Neurother. 2016;16:279–293. [DOI] [PubMed] [Google Scholar]
- 22.Taurines R, Schwenck C, Westerwald E, Sachse M, Siniatchkin M, Freitag C. ADHD and autism: differential diagnosis or overlapping traits? A selective review. Atten Defic Hyperact Disord. 2012;4:115–139. [DOI] [PubMed] [Google Scholar]
- 23.Kentrou V, de Veld DM, Mataw KJ, Begeer S. Delayed autism spectrum disorder recognition in children and adolescents previously diagnosed with attention-deficit/hyperactivity disorder. Autism. 2019;23:1065–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hernandez LM, Rudie JD, Green SA, Bookheimer S, Dapretto M. Neural signatures of autism spectrum disorders: insights into brain network dynamics. Neuropsychopharmacology. 2015;40:171–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Castellanos FX, Aoki Y. Intrinsic Functional Connectivity in Attention-Deficit/Hyperactivity Disorder: A Science in Development. Biol Psychiatry Cogn Neurosci Neuroimaging. 2016;1:253–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Anagnostou E, Taylor MJ. Review of neuroimaging in autism spectrum disorders: what have we learned and where we go from here. Mol Autism. 2011;2:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Konrad K, Eickhoff SB. Is the ADHD brain wired differently? A review on structural and functional connectivity in attention deficit hyperactivity disorder. Hum Brain Mapp. 2010;31:904–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Picci G, Gotts SJ, Scherf KS. A theoretical rut: revisiting and critically evaluating the generalized under/over-connectivity hypothesis of autism. Dev Sci. 2016;19:524–549. [DOI] [PubMed] [Google Scholar]
- 29.Larsson H, Chang Z, D’Onofrio BM, Lichtenstein P. The heritability of clinically diagnosed attention deficit hyperactivity disorder across the lifespan. Psychol Med. 2014;44:2223–2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sandin S, Lichtenstein P, Kuja-Halkola R, Hultman C, Larsson H, Reichenberg A. The Heritability of Autism Spectrum Disorder. JAMA. 2017;318:1182–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Musser ED, Hawkey E, Kachan-Liu SS, Lees P, Roullet J-B, Goddard K, et al. Shared familial transmission of autism spectrum and attention-deficit/hyperactivity disorders. J Child Psychol Psychiatry. 2014;55:819–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ghirardi L, Brikell I, Kuja-Halkola R, Freitag CM, Franke B, Asherson P, et al. The familial co-aggregation of ASD and ADHD: a register-based cohort study. Mol Psychiatry. 2018;23:257–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ronald A, Simonoff E, Kuntsi J, Asherson P, Plomin R. Evidence for overlapping genetic influences on autistic and ADHD behaviours in a community twin sample. J Child Psychol Psychiatry. 2008;49:535–542. [DOI] [PubMed] [Google Scholar]
- 34.Grove J, Ripke S, Als TD, Mattheisen M, Walters RK, Won H, et al. Identification of common genetic risk variants for autism spectrum disorder. Nat Genet. 2019;51:431–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Faraone SV, Larsson H. Genetics of attention deficit hyperactivity disorder. Mol Psychiatry. 2019;24:562–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Willsey HR, Willsey AJ, Wang B, State MW. Genomics, convergent neuroscience and progress in understanding autism spectrum disorder. Nat Rev Neurosci. 2022;23:323–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cristino AS, Williams SM, Hawi Z, An J-Y, Bellgrove MA, Schwartz CE, et al. Neurodevelopmental and neuropsychiatric disorders represent an interconnected molecular system. Mol Psychiatry. 2014;19:294–301. [DOI] [PubMed] [Google Scholar]
- 38.Martin J, Cooper M, Hamshere ML, Pocklington A, Scherer SW, Kent L, et al. Biological overlap of attention-deficit/hyperactivity disorder and autism spectrum disorder: evidence from copy number variants. J Am Acad Child Adolesc Psychiatry. 2014;53:761–70.e26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shehzad Z, Kelly C, Reiss PT, Cameron Craddock R, Emerson JW, McMahon K, et al. A multivariate distance-based analytic framework for connectome-wide association studies. Neuroimage. 2014;93 Pt 1:74–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao B, Li T, Smith SM, Xiong D, Wang X, Yang Y, et al. Common variants contribute to intrinsic human brain functional networks. Nat Genet. 2022;54:508–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miranda-Dominguez O, Feczko E, Grayson DS, Walum H, Nigg JT, Fair DA. Heritability of the human connectome: a connectotyping study. Netw. Neurosci. 2, 175−−199. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Satterstrom FK, Walters RK, Singh T, Wigdor EM, Lescai F, Demontis D, et al. Autism spectrum disorder and attention deficit hyperactivity disorder have a similar burden of rare protein-truncating variants. Nat Neurosci. 2019;22:1961–1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Edition. Diagnostic and statistical manual of mental disorders. Am Psychiatric Assoc. 2013. 2013. [Google Scholar]
- 44.Elliott Murray, Pearson. Differential ability scales. San Antonio, Texas. [Google Scholar]
- 45.Lord C. Autism Diagnostic Observation Schedule: ADOS-2. Western Psychological Services; 2012. [Google Scholar]
- 46.Kaufman J, Birmaher B, Brent D, Rao U, Flynn C, Moreci P, et al. Schedule for Affective Disorders and Schizophrenia for School-Age Children-Present and Lifetime Version (K-SADS-PL): initial reliability and validity data. J Am Acad Child Adolesc Psychiatry. 1997;36:980–988. [DOI] [PubMed] [Google Scholar]
- 47.Bishop SL, Huerta M, Gotham K, Alexandra Havdahl K, Pickles A, Duncan A, et al. The autism symptom interview, school-age: A brief telephone interview to identify autism spectrum disorders in 5-to-12-year-old children. Autism Res. 2017;10:78–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sparrow SS. VINELAND-II: Vineland adaptive behavior Scales. Giunti O.S.; 2005. [DOI] [PubMed] [Google Scholar]
- 49.Constantino JN, Gruber CP. Social responsiveness scale: SRS-2. Western Psychological Services Torrance, CA; 2012. [Google Scholar]
- 50.Rutter M. The Social Communication Questionnaire: Manual. Western Psychological Services; 2003. [Google Scholar]
- 51.Swanson JM, Schuck S, Porter MM, Carlson C, Hartman CA, Sergeant JA, et al. Strengths and Weaknesses of ADHD Symptoms and Normal Behaviors Rating Scale. PsycTESTS Dataset. 2020. [Google Scholar]
- 52.Achenbach, Rescorla. Manual for the ASEBA school-age forms & profiles: an integrated system of multi-informant assessment Burlington, VT: University of Vermont. Research Center for Children, Youth, & Families. [Google Scholar]
- 53.Hollingshead AB, Others. Four factor index of social status. 1975. 1975. [Google Scholar]
- 54.Simhal AK, Filho JOA, Segura P, Cloud J, Petkova E, Gallagher R, et al. Predicting multiscan MRI outcomes in children with neurodevelopmental conditions following MRI simulator training. Developmental Cognitive Neuroscience. 2021;52:101009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jenkinson M, Bannister P, Brady M, Smith S. Improved optimization for the robust and accurate linear registration and motion correction of brain images. Neuroimage. 2002;17:825–841. [DOI] [PubMed] [Google Scholar]
- 56.Rosen AFG, Roalf DR, Ruparel K, Blake J, Seelaus K, Villa LP, et al. Quantitative assessment of structural image quality. Neuroimage. 2018;169:407–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Craddock C, Sikka S, Cheung B, Khanuja R, Ghosh SS, Yan C, et al. Towards automated analysis of connectomes: The configurable pipeline for the analysis of connectomes (c-pac). Front Neuroinform. 2013;42:10–3389. [Google Scholar]
- 58.Behzadi Y, Restom K, Liau J, Liu TT. A component based noise correction method (CompCor) for BOLD and perfusion based fMRI. Neuroimage. 2007;37:90–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bennett RH, Somandepalli K, Roy AK, Di Martino A. The Neural Correlates of Emotional Lability in Children with Autism Spectrum Disorder. Brain Connect. 2017;7:281–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Grzadzinski R, Huerta M, Lord C. DSM-5 and autism spectrum disorders (ASDs): an opportunity for identifying ASD subtypes. Mol Autism. 2013;4:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Frazier TW, Ratliff KR, Gruber C, Zhang Y, Law PA, Constantino JN. Confirmatory factor analytic structure and measurement invariance of quantitative autistic traits measured by the social responsiveness scale-2. Autism. 2014;18:31–44. [DOI] [PubMed] [Google Scholar]
- 62.Cooper M, Martin J, Langley K, Hamshere M, Thapar A. Autistic traits in children with ADHD index clinical and cognitive problems. Eur Child Adolesc Psychiatry. 2014;23:23–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dellapiazza F, Michelon C, Vernhet C, Muratori F, Blanc N, Picot M-C, et al. Sensory processing related to attention in children with ASD, ADHD, or typical development: results from the ELENA cohort. Eur Child Adolesc Psychiatry. 2021;30:283–291. [DOI] [PubMed] [Google Scholar]
- 64.Murphy K, Birn RM, Handwerker DA, Jones TB, Bandettini PA. The impact of global signal regression on resting state correlations: are anti-correlated networks introduced? Neuroimage. 2009;44:893–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ciric R, Wolf DH, Power JD, Roalf DR, Baum GL, Ruparel K, et al. Benchmarking of participant-level confound regression strategies for the control of motion artifact in studies of functional connectivity. Neuroimage. 2017;154:174–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cho JW, Korchmaros A, Vogelstein JT, Milham MP, Xu T. Impact of concatenating fMRI data on reliability for functional connectomics. Neuroimage. 2021;226:117549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Milham MP, Vogelstein J, Xu T. Removing the Reliability Bottleneck in Functional Magnetic Resonance Imaging Research to Achieve Clinical Utility. JAMA Psychiatry. 2021;78:587–588. [DOI] [PubMed] [Google Scholar]
- 68.Lombardo MV, Auyeung B, Pramparo T, Quartier A, Courraud J, Holt RJ, et al. Sex-specific impact of prenatal androgens on social brain default mode subsystems. Mol Psychiatry. 2020;25:2175–2188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pagani M, Barsotti N, Bertero A, Trakoshis S, Ulysse L, Locarno A, et al. mTOR-related synaptic pathology causes autism spectrum disorder-associated functional hyperconnectivity. Nat Commun. 2021;12:6084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gorgolewski KJ, Varoquaux G, Rivera G, Schwarz Y, Ghosh SS, Maumet C, et al. Neurovault.org: a web-based repository for collecting and sharing unthresholded statistical maps of the human brain. Front Neuroinform. 2015;9:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hawrylycz MJ, Lein ES, Guillozet-Bongaarts AL, Shen EH, Ng L, Miller JA, et al. An anatomically comprehensive atlas of the adult human brain transcriptome. Nature. 2012;489:391–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rivals I, Personnaz L, Taing L, Potier M-C. Enrichment or depletion of a GO category within a class of genes: which test? Bioinformatics. 2006;23:401–407. [DOI] [PubMed] [Google Scholar]
- 73.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yoshimasu K, Barbaresi WJ, Colligan RC, Voigt RG, Killian JM, Weaver AL, et al. Childhood ADHD is strongly associated with a broad range of psychiatric disorders during adolescence: a population-based birth cohort study. J Child Psychol Psychiatry. 2012;53:1036–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chan MY, Park DC, Savalia NK, Petersen SE, Wig GS. Decreased segregation of brain systems across the healthy adult lifespan. Proc Natl Acad Sci U S A. 2014;111:E4997–5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yeo BTT, Krienen FM, Sepulcre J, Sabuncu MR, Lashkari D, Hollinshead M, et al. The organization of the human cerebral cortex estimated by intrinsic functional connectivity. J Neurophysiol. 2011;106:1125–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fishman I, Datko M, Cabrera Y, Carper RA, Müller R-A. Reduced integration and differentiation of the imitation network in autism: A combined functional connectivity magnetic resonance imaging and diffusion-weighted imaging study. Ann Neurol. 2015;78:958–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fishman I, Keown CL, Lincoln AJ, Pineda JA, Müller R-A. Atypical cross talk between mentalizing and mirror neuron networks in autism spectrum disorder. JAMA Psychiatry. 2014;71:751–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rudie JD, Brown JA, Beck-Pancer D, Hernandez LM, Dennis EL, Thompson PM, et al. Altered functional and structural brain network organization in autism. Neuroimage Clin. 2012;2:79–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Shih P, Keehn B, Oram JK, Leyden KM, Keown CL, Müller R-A. Functional differentiation of posterior superior temporal sulcus in autism: a functional connectivity magnetic resonance imaging study. Biol Psychiatry. 2011;70:270–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yerys BE, Gordon EM, Abrams DN, Satterthwaite TD, Weinblatt R, Jankowski KF, et al. Default mode network segregation and social deficits in autism spectrum disorder: Evidence from non-medicated children. Neuroimage Clin. 2015;9:223–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yang B, Wang M, Zhou W, Wang X, Chen S, Potenza MN, et al. Disrupted network integration and segregation involving the default mode network in autism spectrum disorder. J Affect Disord. 2023;323:309–319. [DOI] [PubMed] [Google Scholar]
- 83.Rudie JD, Shehzad Z, Hernandez LM, Colich NL, Bookheimer SY, Iacoboni M, et al. Reduced functional integration and segregation of distributed neural systems underlying social and emotional information processing in autism spectrum disorders. Cereb Cortex. 2012;22:1025–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Haghighat H, Mirzarezaee M, Araabi BN, Khadem A. Functional Networks Abnormalities in Autism Spectrum Disorder: Age-Related Hypo and Hyper Connectivity. Brain Topogr. 2021;34:306–322. [DOI] [PubMed] [Google Scholar]
- 85.Lombardo MV, Eyler L, Moore A, Datko M, Carter Barnes C, Cha D, et al. Default mode-visual network hypoconnectivity in an autism subtype with pronounced social visual engagement difficulties. Elife. 2019;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Xiao J, Chen H, Shan X, He C, Li Y, Guo X, et al. Linked Social-Communication Dimensions and Connectivity in Functional Brain Networks in Autism Spectrum Disorder. Cereb Cortex. 2021;31:3899–3910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Harikumar A, Evans DW, Dougherty CC, Carpenter KLH, Michael AM. A Review of the Default Mode Network in Autism Spectrum Disorders and Attention Deficit Hyperactivity Disorder. Brain Connect. 2021;11:253–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hus V, Bishop S, Gotham K, Huerta M, Lord C. Factors influencing scores on the social responsiveness scale. J Child Psychol Psychiatry. 2013;54:216–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Nikolaidis A, Chen AA, He X, Shinohara R, Vogelstein J, Milham M, et al. Suboptimal phenotypic reliability impedes reproducible human neuroscience. BioRxiv. 2022:2022.07.22.501193. [Google Scholar]
- 90.Gandal MJ, Haney JR, Parikshak NN, Leppa V, Ramaswami G, Hartl C, et al. Shared Molecular Neuropathology Across Major Psychiatric Disorders Parallels Polygenic Overlap. Focus . 2019;17:66–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cross-Disorder Group of the Psychiatric Genomics Consortium. Electronic address: plee0@mgh.harvard.edu, Cross-Disorder Group of the Psychiatric Genomics Consortium. Genomic Relationships, Novel Loci, and Pleiotropic Mechanisms across Eight Psychiatric Disorders. Cell. 2019;179:1469–1482.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Berg LM, Gurr C, Leyhausen J, Seelemeyer H, Bletsch A, Schaefer T, et al. The neuroanatomical substrates of autism and ADHD and their link to putative genomic underpinnings. Mol Autism. 2023;14:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yadav SK, Bhat AA, Hashem S, Nisar S, Kamal M, Syed N, et al. Genetic variations influence brain changes in patients with attention-deficit hyperactivity disorder. Transl Psychiatry. 2021;11:349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Voineagu I, Wang X, Johnston P, Lowe JK, Tian Y, Horvath S, et al. Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature. 2011;474:380–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Saulnier CA, Klaiman C, McQueen E. Adaptive Behavior Profiles in Autism Spectrum Disorder. Curr Psychiatry Rep. 2022;24:749–756. [DOI] [PubMed] [Google Scholar]
- 96.Marek S, Tervo-Clemmens B, Calabro FJ, Montez DF, Kay BP, Hatoum AS, et al. Publisher Correction: Reproducible brain-wide association studies require thousands of individuals. Nature. 2022;605:E11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Knott R, Johnson BP, Tiego J, Mellahn O, Finlay A, Kallady K, et al. The Monash Autism-ADHD genetics and neurodevelopment (MAGNET) project design and methodologies: a dimensional approach to understanding neurobiological and genetic aetiology. Mol Autism. 2021;12:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Elton A, Di Martino A, Hazlett HC, Gao W. Neural Connectivity Evidence for a Categorical-Dimensional Hybrid Model of Autism Spectrum Disorder. Biol Psychiatry. 2016;80:120–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chabernaud C, Mennes M, Kelly C, Nooner K, Di Martino A, Castellanos FX, et al. Dimensional brain-behavior relationships in children with attention-deficit/hyperactivity disorder. Biol Psychiatry. 2012;71:434–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hong S-J, Vogelstein JT, Gozzi A, Bernhardt BC, Yeo BTT, Milham MP, et al. Towards neurosubtypes in autism. Biol Psychiatry. 2020. 2020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.