Abstract

Thermal cycling-based quantitative polymerase chain reaction (qPCR) represents the gold standard method for accurate and sensitive nucleic acid quantification in laboratory settings. However, its reliance on costly thermal cyclers limits the implementation of this technique for rapid point-of-care (POC) diagnostics. To address this, isothermal amplification techniques such as rolling circle amplification (RCA) have been developed, offering a simpler alternative that can operate without the need for sophisticated instrumentation. This study focuses on the development and optimization of toehold-mediated RCA (TRCA), which employs a conformationally switchable dumbbell DNA template for the sensitive and selective detection of cancer-associated miRNAs, specifically miR-21. In addition, we developed variants of hyperbranched TRCA (HTRCA), nicking-assisted TRCA (NTRCA), and hyperbranched NTRCA (HNTRCA) to facilitate exponential amplification by enhancing TRCA through the sequential incorporation of reverse primer (Pr) and nicking endonuclease (nE). By conducting a systematic kinetic analysis of the initial rate and end point signals for varying concentrations of key reaction components, we could identify optimal conditions that markedly enhanced the sensitivity and specificity of the TRCA variants. In particular, HNTRCA, which exploits the synergistic effect of Pr and nE, demonstrated an approximately 3000-fold improvement in the detection limit (260 fM) and a wider dynamic range of more than 4 log orders of magnitude compared to TRCA, thereby evidencing its superior performance. Also, we established a mechanistic model for TRCA that includes the roles of Pr and nE in different amplification processes. Model parameters were fitted to the experimental data, and additional simulations were conducted to compare the four amplification methods. Further tests with real biological samples revealed that this technique showed a good correlation with qPCR in quantifying miR-21 expression in various cell lines (0.9510 of Pearson’s r), confirming its potential as a robust and rapid tool for nucleic acid detection. Therefore, the simplicity, high sensitivity, and potential for integration with POC diagnostic platforms make the HNTRCA system suitable for field deployment in resource-limited environments.
Keywords: isothermal nucleic acid amplification, rolling circle amplification, microRNA, systematic optimization, mechanistic modeling
Gene amplification system represents a fundamental tool in the fields of molecular biology and diagnostics, facilitating the detection and analysis of nucleic acids. In particular, amplifying target nucleic acid biomarkers in the background greatly simplifies multiple downstream signaling and detection processes. First developed in the 1990s, quantitative polymerase chain reaction (qPCR) has experienced rapid growth and is now recognized as the gold standard for laboratory-based nucleic acid analysis and detection.1 However, a number of limitations have hindered its application in the field of rapid point-of-care (POC) diagnostics.2,3 The biggest barrier is the reliance on costly thermal cyclers equipped with optics to read the optical signals generated by the highly temperature-controlled thermal cycling system for amplification. This nature necessitates relatively large, reliable power supplies and expensive equipment, which constrains the portability of the technology and excludes it from various POC applications. To address this issue, considerable effort has been made to develop isothermal technologies that can operate at a single optimal temperature, thereby greatly simplifying the thermal cycling requirements of the system.4−6
An exemplar of an isothermal amplification technique is rolling circle amplification (RCA), which employs DNA polymerase (DNAP) with strong strand displacement activity to amplify DNA by rolling along a circular DNA template at a constant temperature.7−9 The RCA reaction generates DNA amplicons with sequences complementary and repetitive to the circular template, which can be customized with functional sequences, including DNA aptamers,10−13 DNAzyme,14−16 restriction or nicking enzyme cleavage sites,17,18 probe binding sites,19−23etc. Notably, RCA reactions can be performed not only in solution but also on a variety of solid substrates (e.g., glass, plastic, or gold substrates, nano/microparticle surfaces)24−28 and in complex biological matrices (e.g., cell membranes, intracellular compartments).29−32 Thus, the robustness, versatility, and programmability of this RCA technique have facilitated its extensive utilization in the fields of in vitro diagnostics, biosensors, and bioanalysis, enabling the sensitive detection and analysis of various biomarkers, including DNA, RNA, proteins, small molecules, and cells.9,33,34 However, although the RCA technique shows great potential in biosensors and bioanalysis, its translation into commercial and field settings remains limited due to challenges, including lack of validated assays, technical optimization, standardization, and nonspecific amplification. Despite these hurdles, RCA-based products such as TempliPhi DNA amplification kit (Cytiva),35 TruePrime RCA kit, (SYGNIS Biotech)36 and DNBSEQ platform (MGI Tech)37 demonstrate that the RCA technique is gradually being adopted in real-world applications, particularly in sequencing and genomic analysis. Still, to fully integrate RCA into clinical practice and POC diagnostics, these limitations—especially the risk of false-positive results, high background noise, and reduced sensitivity and selectivity due to nonspecific amplification—should be addressed. While advantageous for simplifying instrumentation, the isothermal nature of RCA presents challenges that need further resolution to ensure its successful implementation in clinical and field settings.
In addition to the classical RCA, which exhibits linear amplification, several RCA techniques have been developed to allow for exponential amplification through a single binding event. One of the well-established examples is hyperbranched RCA (HRCA), which amplifies DNA by initiating multiple branching events through the hybridization of multiple primers with the amplicon.38 This results in a highly branched and extensive DNA network. Multiprimed RCA also employs multiple primers that hybridize into different sites on the circular template, allowing simultaneous DNA amplifications at various sites from the template.39 Another type of RCA is a combination of RCA with a nicking reaction (nicking-assisted RCA or NRCA), which employs a nicking endonuclease (nE) to introduce nicks in the amplicon at a specific site, enabling the continuous displacement and synthesis of new strands by DNAP in an exponential manner.40 These modified RCA techniques are employed individually or in conjunction with one another and/or other isothermal amplification approaches to enhance sensitivity, particularly when the target molecules are present at low abundance.
Despite the extensive use of RCA techniques in biosensor and bioanalytical applications, the optimization of experimental conditions for different variants of RCA remains a significant challenge. A typical RCA requires five key components: a circular DNA template, primers, a DNAP (e.g., phi29, Bst, Bsu, and Vent DNAPs, etc.), deoxynucleoside triphosphate (dNTP), and a reaction buffer (typically comprising Mg ions). The concentrations of those components affect the overall amplification efficiency. Although the majority of studies have focused on empirically optimizing these components based on the experimental results,41−43 a systematic and comprehensive investigation of the underlying real-time kinetics of the RCA process has frequently been lacking. In order to optimize RCA and its variants for specific applications such as POC diagnostics and quantitative analysis, it is essential to gain a deep understanding of the underlying kinetic parameters, including the initial rate and final yield.44−46 It is of particular importance to optimize the initial amplification rate to reduce detection time and associated costs. In addition, in the context of detecting targets present at very low abundances, it is crucial to comprehend and refine the parameters influencing the final yield to achieve robust and reliable amplification results. This is particularly the case for modified RCAs, such as HRCA and NRCA, where the amplification process is initiated simultaneously at multiple sites, resulting in a more complex kinetic profile than conventional RCA. A thorough understanding of these kinetics enables the design of optimal reaction conditions, taking into account the concentrations and binding efficiencies of the various components that affect the initial rate and final yield. Therefore, kinetic analysis and subsequent optimization of RCA and its variants are essential to facilitate rapid and efficient DNA amplification, enabling reliable detection of target molecules.
In this study, we constructed a toehold-mediated RCA (TRCA) using a conformationally switchable dumbbell DNA template (DBTP) for sensitive and selective detection of cancer-associated microRNAs (miRNAs) without labels. The DBTP consists of two loops and a stem, each containing a toehold region for target miRNA binding and a specific sequence for recognition by a nE. This rationally designed template maintains a stable dumbbell structure and is then activated for TRCA initiation by triggering toehold-mediated strand displacement specific to the target miRNA.47,48 To further construct modified TRCA, we introduced reverse primers (Pr) that bind to TRCA amplicons and a nE to induce hyperbranched TRCA (HTRCA), nicking-assisted TRCA (NTRCA), and hyperbranched NTRCA (HNTRCA). We chose miR-21 as a target miRNA, which is commonly overexpressed in several types of cancer, including breast, lung, and colon cancer.49 In particular, miR-21 is one of the cancer-related miRNAs that has been extensively studied as a potential biomarker for cancer diagnosis and prognosis and as a target for therapeutic intervention, as it plays an important role in tumorigenesis by regulating the genes involved in cell proliferation, apoptosis, and metastasis.50
To optimize the experimental conditions and obtain high amplification yields of the four TRCAs used in this study, we analyzed the time-dependent amplification behavior of each TRCA by varying the concentrations of dominant reaction factors, including DBTP, dNTP, Mg, DNAP, Pr, and nE. Two kinetic parameters, such as initial rate and end point signal, were extracted and examined from the amplification curve to gain insight into the kinetics of the TRCA and its variants. In addition to the experimental kinetic analysis, we developed a mechanistic model for TRCA, HTRCA, NTRCA, and HNTRCA to explore the complex interplay of various species on DNA amplification efficiency. This mechanistic model, with parameters fitted to the experimental data, can also be used to understand the influence of input species concentrations on the temporal evolution of substrates, enzymes, DNA amplicons, and other ionic species.
Results obtained from both experiments and simulations demonstrated that the reaction rate and amplification gain could be finely tuned by modulating the concentrations of reaction components. The optimal conditions were then employed to assess the sensitivity and specificity of the four TRCAs. The HNTRCA reaction was found to exhibit superior performance in terms of initial rate, final yield, sensitivity, and specificity compared to the other TRCAs. Subsequently, we conducted the HNTRCA using known concentrations of synthetic miRNA by spiking them into total RNA extracted from cells. This resulted in 90–150% recoveries, confirming the utility of the HNTRCA technique for quantitative detection of the target miRNA in complex biological matrices. Finally, we demonstrated that HNTRCA could identify differences in relative miR-21 expression levels in total RNA extracts derived from five different cell lines, showing its consistency with qPCR analysis. Overall, we developed and optimized a single-step, isothermal TRCA reaction with performance competitive with qPCR, thereby presenting a quantitative molecular diagnostic tool suitable for rapid and sensitive nucleic acid testing in the field.
Materials and Methods
Materials
All DNA and RNA oligonucleotides used in this study were purchased from Integrated DNA Technology (IA, USA) and used without further purifications (see Table S1 for details of oligonucleotide sequences). T4 DNA ligase, exonuclease I and III (Exo I and Exo III), Bst 2.0 DNAP, Nt.BstNBI nE, dNTP solution mix, and 100 mM MgSO4 were purchased from New England Biolabs (MA, USA). SYBR Green I, SYBR Gold nucleic acid stain, 10× TBE buffer (1 M Tris, 900 mM boric acid, 10 mM EDTA, pH 8.3), and nuclease-free deionized water were supplied from ThermoFisher Scientific (MA, USA).
Preparation of DBTP
To synthesize DBTP, a 5′ phosphorylated linear single-stranded DNA template was enzymatically self-ligated in the absence of a splint DNA. First, the linear DNA template (4 μM) was annealed by heating at 80 °C for 5 min and gradually cooling to 25 °C in 1× ligation buffer (50 mM Tris-HCl, 10 mM MgCl2, 1 mM ATP, 10 mM DTT, pH 7.5) using a PCR thermocycler (Eppendorf, Germany). After annealing, the nick site was intramolecularly connected by adding 2 μL of T4 DNA ligase (16 U/μL), and the ligation reaction was performed at 25 °C for 2 h before inactivation at 65 °C for 10 min. The ligated product was followed by treatment with Exo I (0.4 U/μL) and Exo III (2 U/μL) in 1× NEBuffer 1 (10 mM Bis-Tris-Propane-HCl, 10 mM MgCl2, 1 mM DTT, pH 7) at 37 °C for 1 h to digest any residual unsealed template DNA. After the inactivation of Exo I and Exo III at 80 °C for 20 min, the resulting DBTP (2 μM) was stored at −20 °C for further use. To verify the ligation of the linear template and hybridization of DBTP with a target miRNA, 15% native polyacrylamide gel electrophoresis (PAGE) and 15% denatured urea-PAGE were performed in 0.5× TBE at 110 and 120 V for 1 h, respectively. The gels were stained with 1× SYBR Gold nucleic acid stain at 25 °C for 30 min and observed by using the ChemiDoc MP imaging system (Bio-Rad, USA).
Melting Temperature Analysis
To ascertain the melting temperature (Tm) of DBTP, a mixture (20 μL) containing DBTP (200 nM), dNTP (1.5 mM), MgSO4 (6 mM), and 50× SYBR Green I was prepared in 1× isothermal amplification buffer (20 mM Tris-HCl, 10 mM (NH4)2SO4, 50 mM KCl, 2 mM MgSO4, 0.1% Tween 20, pH 8.8). The temperature was increased at a rate of 0.3 °C/s from 35 to 95 °C. The fluorescence intensity (FI) was measured using QuantStudio 5 Real-Time PCR system (ThermoFisher Scientific) with an excitation wavelength of 480 nm and an emission wavelength of 520 nm. The Tm was calculated by converting the fluorescence data into the negative derivative of fluorescence as a function of temperature.
TRCA and Modified TRCA Reactions
In a typical TRCA reaction, a mixture (20 μL) containing miRNA (50 nM), DBTP (200 nM), dNTP (1.5 mM), MgSO4 (6 mM), Bst2.0 DNAP (0.6 nM), and 50× SYBR Green I was prepared in 1× isothermal amplification buffer. The modified RCA reactions involve the addition of the Pr and/or nE to the TRCA reaction. Specifically, for HTRCA and NTRCA, Pr (200 nM) and nE (14.7 nM) were added to the TRCA reaction mixture, respectively, resulting in a final volume of 20 μL. The HNTRCA mixture was prepared by the addition of both Pr (200 nM) and nE (14.7 nM) to the TRCA mixture. All reaction mixtures were prepared on ice and reacted at 50 °C for 60 or 90 min, followed by incubation at 80 °C for 20 min to inactivate Bst2.0 DNAP.
Real-Time Kinetics of TRCA and Modified TRCA Reactions
For the kinetic analysis of TRCA and modified TRCA, real-time fluorescence measurements were conducted at a constant temperature of 50 °C every 1 min using QuantStudio 5 Real-Time PCR system. Fluorescence emission was quantified at 520 nm following excitation at 480 nm. The net signal change (referred to as normalized FI) was calculated by subtracting the initial FI from the FI of the sample during the measurement period. To facilitate a comparison of the relative FI changes between the test groups, the average FI of three independent experimental replicates after 60 min reaction was used. The analysis and plot fitting of the obtained results were conducted using Origin Pro2023b (OriginLab Corporation, USA). The calculations of the initial rate and end point signal were standardized across all amplification curves. The reported initial rate and end point signal refer to the rate of increase in normalized FI within the initial phase of the reaction and the normalized FI after 60 min-reaction (which indicates the overall yield), respectively.
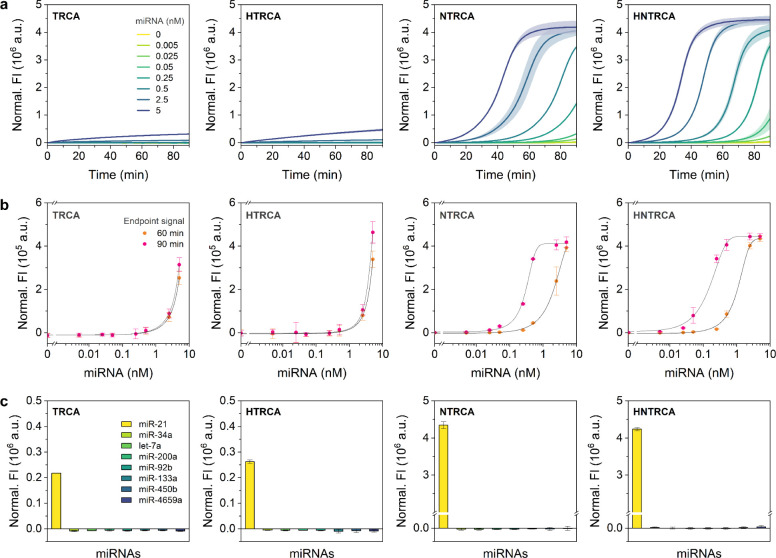
Analytical Sensitivity and Specificity
To ascertain the analytical sensitivity of the TRCA and modified TRCA reactions, amplification curves were generated for varying concentrations of the target miRNA, ranging from 0 to 5 nM. Subsequently, the FIs obtained after 60 and 90 min of amplification were used to derive sigmoid dose–response curves. Specifically, the time required to reach the threshold value (referred to as amplification time) was used to derive linear dose–response curves for the NTRCA and HNTRCA reactions. The curves were plotted and fitted using the dose–response or linear function in OriginPro 2023b. The limit of detection (LOD) was calculated as the target concentration obtained by adding three times the standard deviation (s.d.) from the mean FI signal of 0 nM target and extrapolating the resulting value to the dose–response curve. To assess specificity, each TRCA and modified TRCA was tested in the presence of eight different miRNAs (5 nM), including miR-21, miR-34a, let-7a, miR-200a, miR-92b, miR-133a, miR-450b, and miR-4659a. The TRCA and modified TRCA reactions were performed in the same procedure as described above.
Cell Culture and RNA Extraction
Human breast cancer cells MDA-MB-231 (KCLB no. 30026), HCC1143 (KCLB no. 9S1143), HCC1954 (KCLB no. 9S1954), and SK-BR-3 (KCLB no. 30030) and human embryonic kidney cells HEK 293T (KCLB no. 21573) were obtained from the Korean Cell Line Bank (KCLB). MDA-MB-231 and HEK293T cells were maintained in the DMEM medium (Gibco), and HCC1143, HCC1954, and SK-BR-3 cells were maintained in the RPMI 1640 medium (Gibco). All complete culture media were supplemented with 10% v/v fetal bovine serum (Merck) and 1% v/v penicillin/streptomycin (Gibco), and the cells were kept at 37 °C in a humid atmosphere with 5% CO2. Total RNA containing miRNAs was extracted using miRNeasy kit (Qiagen, Germany) according to the manufacturer’s instructions. Cells were lysed with the Qiazol reagent, mixed with chloroform (1:5 volume ratio of chloroform to Qiazol), and separated by centrifugation at 12 000g for 15 min at 4 °C. Total RNA was then isolated and purified with miRNeasy columns. The RNA concentration and purity were confirmed with a DS-11 FX+ spectrometer (DeNovis Inc., USA). The extracted RNA samples were kept at −80 °C until use.
RT-qPCR
For quantification of miR-21 by reverse transcription-quantitative polymerase chain reaction (RT-qPCR), complementary DNA (cDNA) was prepared in a 10-μL reaction using a miRCURY LNA RT kit (Qiagen) from 100 ng of RNA extracts according to the manufacturer’s protocols. The RT reaction was incubated at 42 °C for 60 min and then inactivated at 95 °C for 5 min before immediately cooling to 4 °C. qPCR was performed on a QuantStudio 5 Real-Time PCR System using a miRCURY LNA SYBR Green PCR kit (Qiagen) and miRCURY LNA miRNA PCR Assays (Qiagen; #YP00204230 for hsa-miR-21-5p) with 0.5 ng input cDNA reaction. The qPCR reaction was as follows: 95 °C for 2 min, followed by 40 cycles of denaturation at 95 °C for 10 s, annealing and extension at 56 °C for 60 s. The cycles to threshold (Ct) values for miR-21 in cell-derived RNA samples were compared with the amplification time values of HNTRCA.
HNTRCA Using Total RNA Extracted from Cell Lines
To construct the standard curve, serial dilutions of synthetic miR-21 (0–5 nM) were spiked into total RNA (100 ng) extracted from HEK 293T cells. The HNTRCA was then performed at 50 °C for 90 min under optimal conditions. In a parallel experiment, four concentrations of synthetic miR-21 (0.1, 0.2, 1, and 2 nM) were spiked into 100 ng of total RNA extracts, and HNTRCA was carried out under the same conditions. The measured miR-21 concentration in the spiked samples was calculated by extrapolation a threshold of 105 based on a linear calibration curve derived from the obtained amplification time. The recovery (%) and relative s.d. (RSD, %) were calculated using the following equations: recovery (%) = (measured miR-21 concentration/spiked miR-21 concentration) × 100; RSD (%) = (s.d./mean) × 100. To ascertain the expression of miR-21 in cell lines, HNTRCA was further conducted under optimal conditions on total RNA extracts (100 ng) derived from five cell lines (MDA-MB-231, HCC1143, HCC1954, SK-BR-3, and HEK 293T). For the correlation study between HNTRCA and RT-qPCR, the amplification times and Ct values of the two assays were compared.
Mechanistic Modeling
Four types of DNA amplification—TRCA, HTRCA, NTRCA, and HNTRCA—were modeled using simple enzymatic reaction kinetics, which resulted in dual Michaelis–Menten-type reaction kinetics equations. The DNA synthesis process begins with the formation of the miRNA-DBTP complex, followed by the involvement of DNAP, which are common steps across all amplification methods. In the presence of Pr, additional DNA synthesis occurs, referred to as branched DNA. The addition of nE leads to the cleavage of the prior DNA into the initial miRNA-DBTP complex and replicon DNA. Variations in the concentrations of ionic species throughout the DNA amplification process were also considered, which were described by a set of algebraic equations involving equilibrium relations and component balances, some of which were necessary for calculating the reaction rates.43,45 Additional mechanisms were incorporated to provide a more realistic representation of the amplification process. First, the precipitation of Mg2PPi was accounted for, based on its solubility limit.44−46 Second, DBTP was assumed to be taken by DNAP in addition to miRNA. Third, the enzymes DNAP and nE could undergo partial deactivation. Finally, the degradation of produced DNA was included.44,45 The temporal concentrations of key species—miDNA, DBTP, dNTP, DNAP, Pr, nE, and DNA—as well as ionic species, PPi4–, H+, Mg2+, and buffer (Tris) were described by a system of ordinary differential equations.44−46 As a result, temporal variations in species concentrations and DNA synthesis rates could be obtained by solving a system of differential and algebraic equations composed of the aforementioned reaction rates. The hypothesis for the DNA amplification steps, the formulation of DNA synthesis rate, and other model equations can be found in the Supporting Information.
Results and Discussion
Reaction Pathways in TRCA and Modified TRCAs
Both conventional toehold-mediated rolling circle amplification (TRCA) and modified TRCA amplify short target microRNA (miRNA) sequences (22 nt) at a single reaction temperature of 50 °C through the action of a thermophilic DNA polymerase (DNAP) and/or nicking endonuclease (nE) on dumbbell-shaped template DNA (DBTP). The representative amplification reaction pathways are shown in Scheme 1. The DBTP strategically designed for target-specific TRCA comprises one stem (12 bp) and two loops (16 bp each) with sealed ends and contains three major sites: (i) a toehold site for association with the target miRNA, (ii) a reverse primer (Pr) binding site, and a nE recognition site. The opening of the DBTP is initiated by a toehold recognition of the target and a toehold-mediated strand displacement process, which is accompanied by penetration into the stem sequence. This results in a conformational change to the activated circular template.47,48 The target, which is bound to the template, serves as a primer and is then elongated to the repeated sequences complementary to the template with the addition of deoxynucleoside triphosphate (dNTP) by DNAP with strong strand displacement activity. In this context, a divalent cation such as magnesium (Mg) ions acts as an essential cofactor for the DNAP-catalyzed formation of phosphodiester bonds, while interacting with incoming dNTP to ensure proper conformational adjustments for catalysis and stabilize the reaction intermediate.51,52 The presence of a Pr binding site and a nE recognition site on the resulting amplicon of TRCA can induce three additional amplifications, including hyperbranched TRCA (HTRCA), nicking-assisted TRCA (NTRCA), and hyperbranched NTRCA (HNTRCA). In HTRCA, hybridization of the amplicon with Pr leads to the extension of Pr by DNAP, which in turn facilitates further DNA amplification. Moreover, during NTRCA, sequence-specific cleavage by nE of the growing amplicon results in sequence extension by DNAP at the nick site and subsequent amplification through displacement of the downstream strand that has not yet dissociated from the template. Ultimately, the incorporation of Pr and nE in HNTRCA initiates both the HTRCA and NTRCA reaction pathways simultaneously, leading to the further elongation of multiple strands. Given that the resulting amplicons adopt repeating hairpin structures, we employed SYBR Green I, which binds specifically to the double-stranded DNA (dsDNA) and emits fluorescence, to monitor the real-time kinetics of these amplification reactions.53
Scheme 1. Schematic Illustration of the Conventional TRCA and Modified TRCAs (HTRCA, NTRCA, and HNTRCA) Reaction Pathways Initiated by Toehold-Mediated Displacement and Subsequent Circularization of DBTP in the Presence of a Target miRNA.
DBTP, dumbbell DNA template; DNAP, DNA polymerase; DNAP–Mg, Mg-bound DNAP; miRNA, microRNA; dNTP, deoxynucleoside triphosphates; dNTP–Mg, Mg-bound dNTP; Pr, reverse primer; nE, nicking endonuclease; Mg, magnesium ion
We first prepared DBTP by enzymatic self-ligation of 5′ phosphorylated linear template DNA in the absence of a splint using T4 DNA ligase. This was followed by subsequent treatments with exonuclease I and III, which served to remove any residual unsealed DBTP. The results of the 15% native and denatured polyacrylamide gel electrophoresis showed that both the 5′ and 3′ ends of DBTP were successfully ligated (Figure S1a). In the presence of a target miRNA, a new band of higher molecular weight appeared, indicating that the hybridization between DBTP and miRNA induced a conformational shift to the circular template (Figure S1b). We then measured the melting temperature (Tm) of DBTP to confirm that its secondary structure remained without denaturation at the TRCA reaction temperature (50 °C). To achieve this, the reaction mixture was prepared in the same manner as that used for the TRCA reaction, with the exception of the inclusion of DNAP. The calculated Tm of DBTP was 76.8 °C, implying that the DBTP maintained a stable dumbbell structure under the reaction conditions (Figure S2).
Real-Time Kinetics of TRCA under Various Conditions
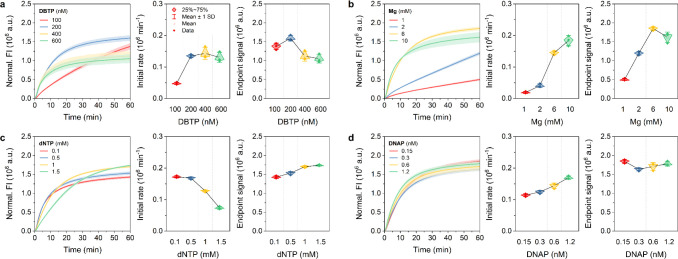
To investigate the effect of key reaction components on the kinetics of TRCA, we first carried out TRCA by varying the concentration of four dominant factors (DBTP, Mg, dNTP, and DNAP) in the reaction. The standard conditions were set at 200 nM DBTP, 6 mM Mg, 1.5 mM dNTP, and 0.6 nM DNAP. When the concentration of one component was altered, the remaining three components were kept at the standard conditions. The fluorescence intensity (FI) was then monitored in real-time fluorescence for 60 min, yielding an amplification curve. The amplification curves for varying concentrations of each component were compared to determine the relationship between the concentration of each factor and the kinetic variables (initial rate and end point signal). Based on these experiments, we sought to identify the optimal reaction conditions for obtaining high-gain DNA products and to elucidate the mechanisms underlying the kinetic differences of TRCA among conditions.
For the TRCA reaction, the initial rate of DNA amplification was found to be proportional to the DBTP concentration, with the maximum rate and end point signal at 200 nM of DBTP. At higher concentrations, the initial rate was negligible, while the end point signal decreased, indicating that the excess of DBTP exerted an inhibitory effect on the TRCA reaction (Figure 1a). This may be attributed to the competitive reaction of miRNA-bound DBTP and free DBTP for DNAP, as evidenced by the observation of a high DNA binding rate of DNAP in the presence of high concentrations of dsDNA.54 This subsequently reduces the availability of DNAP to bind and extend the miRNA-template complex, thereby lowering the amplification efficiency.55 An increase in Mg concentration resulted in a discernible initial rate increase, with the maximum end point signal at 6 mM, suggesting a pronounced Mg ion dependence of DNAP activity (Figure 1b). In contrast, the initial rate was observed to decrease continuously with increasing dNTP concentration (Figure 1c). These results are consistent with previous studies demonstrating that during RNA polymerase-catalyzed RNA polymerization, the binding of nucleoside triphosphate (NTP) ions with Mg ions resulted in the formation of several complexes, including Mg2NTP, MgNTP, and MgHNTP. This process leads to a reduction in the concentration of free Mg ions and an NTP concentration-dependent decrease in the initial rate of RNA amplification.44,46 Additionally, an alternative mechanism involves the formation of insoluble magnesium pyrophosphate (Mg2PPi) crystal precipitates, derived from the binding of Mg ions with pyrophosphate (PPi) ions released from dNTP during DNA synthesis. The generation of these crystals competes with dNTP for Mg ions, which ultimately leads to a decline in the concentration of free Mg. This is supported by the observation that the addition of PPi to an aqueous Mg solution in the absence of a polymerization reaction resulted in a decrease in Mg concentration due to the formation of Mg2PPi precipitates, while the addition of NTP increased the Mg concentration in solution.46 Moreover, in contrast to the initial rate trend, the end point signal displayed a gradual increase with increasing dNTP concentration. This result can be explained by the fact that the production of Mg2PPi precipitates is suppressed at elevated dNTP concentrations over time, which prevents the termination of the polymerization reaction and contributes to enhanced yields. Finally, increasing DNAP concentrations increased the initial rate of amplification without any noticeable effect on DNA yield (Figure 1d).
Figure 1.
Kinetics of TRCA under four different conditions. Real-time fluorescence curves (left), initial rates (middle), and end point signals (right) of TRCA performed in the presence of a target miRNA (50 nM) at 50 °C for 1 h with various concentrations of a) DBTP, b) Mg, c) dNTP, and d) DNAP, respectively. The initial rate and end point signal represent the rate of increase in normalized fluorescence intensity (FI) within the initial phase of the reaction and the normalized FI after 1-h reaction. Data represent mean ± s.d. for three independent experiments.
Real-Time Kinetics of Modified TRCAs under Various Conditions
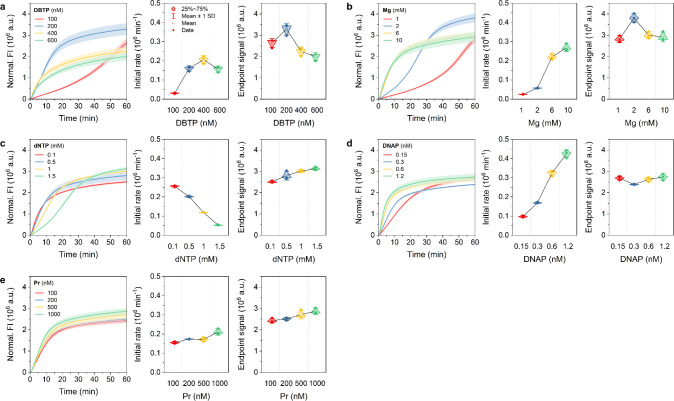
Subsequently, we conducted modified TRCAs, including HTRCA, NTRCA, and HNTRCA, and compared their initial rate and end point signals with TRCA in order to gain insight into the kinetic differences and product yields. In comparison with TRCA, a prominent feature of the amplification curve in HTRCA was the overall enhancement of the FI values and the sigmoidal profile at low concentration conditions. The introduction of Pr into the reaction mixture initiates a secondary polymerization event, resulting in a hyperbranched structure with multiple replication forks.38 This differed from the amplification curve for TRCA, which showed an initial rapid amplification followed by saturation (Figure 1). Instead, HTRCA produced a sigmoidal curve with an initial lag phase followed by an exponentially rapid increase in amplification to saturation (Figure 2).
Figure 2.
Kinetics of HTRCA under five different conditions. Real-time fluorescence curves (left), initial rates (middle), and end point signals (right) of HTRCA performed in the presence of a target miRNA (50 nM) at 50 °C for 1 h with various concentrations of a) DBTP, b) Mg, c) dNTP, d) DNAP, and e) Pr, respectively. Data represent mean ± s.d. for three independent experiments.
In HTRCA, DBTP concentrations exhibited behavior comparable to that of TRCA in terms of initial rate and end point signal (Figure 2a). An increase in Mg concentrations resulted in a steeper increase in the initial rate compared to TRCA (1.83 × 104 min–1 mM–1 for TRCA vs 2.70 × 104 min–1 mM–1 for HTRCA), with the maximum end point signal obtained at 2 mM (Figure 2b). The dNTP concentrations had a comparable impact on both TRCA and HTRCA in terms of initial rate and end point signal, indicating that the concentration for optimal yield was 1.5 mM (Figure 2c). Notably, the initial rate inhibition was more pronounced for HTRCA (−7.04 × 104 min–1 mM–1 for TRCA vs −1.45 × 105 min–1 mM–1 for HTRCA), suggesting that HTRCA is more sensitive to changes in dNTP concentrations than TRCA. As with the Mg ion, the increase in DNAP concentration also increased more rapidly than that observed for TRCA (5.30 × 104 min–1 mM–1 for TRCA vs 3.12 × 105 min–1 mM–1 for HTRCA), with the maximum reaction yield being reached at 1.2 nM (Figure 2d). An increase in Pr concentration showed a gradual enhancement of both the initial rate and reaction yield (Figure 2e). These comparisons demonstrate the subtle differences in reaction kinetics between TRCA and HTRCA, with HTRCA displaying more sensitive kinetics and exhibiting improved DNA yields.
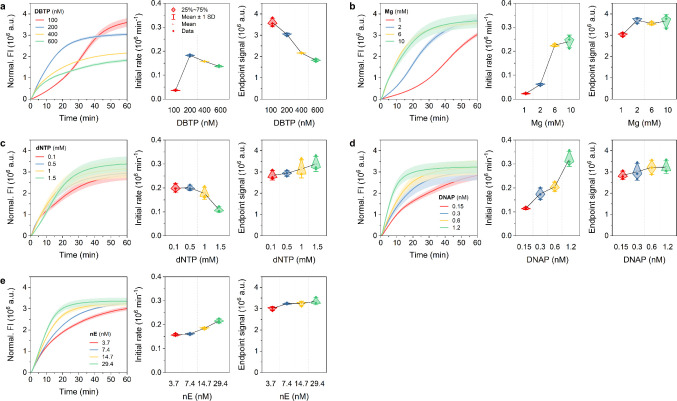
The NTRCA, which is initiated by the introduction of nE, creates a nick at a specific site on the newly synthesized DNA strand, thereby providing an additional starting point for a new polymerase reaction. This leads to exponential amplification, comparable to HTRCA, but driven by the nick rather than the primer.40 This induces an initial delay during the nick formation step, followed by a rapid signal increase before reaching a plateau and a higher end point signal than that observed in TRCA. Consequently, the amplification curves for NTRCA also showed an overall increase in FI signals, analogous to that observed in HTRCA, and more sensitive initial reaction rates than TRCA (Figure 3). The initial response rate of NTRCA presented greater sensitivity to changes in DBTP concentrations compared to TRCA, demonstrating a more rapid increase between 100 nM and 200 nM. However, the end point signal showed a definite decline (Figure 3a). This level of sensitivity can be attributed to the distinctive mechanism of NTRCA, whereby nE rapidly induces additional amplification sites of target miRNA-bound DBTP, necessitating immediate and efficient utilization of DNAP. This process can be significantly disrupted when DNAP is sequestered by high concentrations of free DBTP, resulting in a more substantial decrease in the end point signal of NTRCA compared to TRCA or HTRCA. The initial rate and the responsiveness of the end point signal to changes in Mg ions, dNTP, and DNAP concentrations were found to be generally similar to those observed for HTRCA (Figure 3b–d). Nevertheless, the inhibition and enhancement of the initial rate by dNTP (−1.45 × 105 min–1 mM–1 for HTRCA vs −6.48 × 104 min–1 mM–1 for NTRCA) and DNAP (3.12 × 105 min–1 mM–1 for HTRCA vs 1.96 × 105 min–1 mM–1 for NTRCA) showed fewer sensitive changes when compared to those of HTRCA. This suggests that dNTP consumption and DNAP activity by nE provided less rapid and controlled amplification than HTRCA, which induced rapid and extensive amplification in the presence of Pr. Finally, increasing nE concentration slightly increased the initial rate but did not improve the amplification yield (Figure 3e).
Figure 3.
Kinetics of NTRCA under five different conditions. Real-time fluorescence curves (left), initial rates (middle), and end point signals (right) of NTRCA performed in the presence of a target miRNA (50 nM) at 50 °C for 1 h with various concentrations of a) DBTP, b) Mg, c) dNTP, d) DNAP, and e) nE, respectively. Data represent mean ± s.d. for three independent experiments.
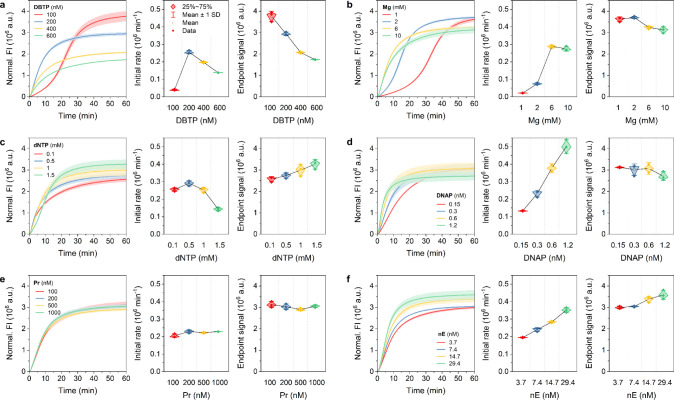
The HNTRCA relies on a synergistic amplification process that combines the nicking of nE with the hyperbranching effect of Pr. The nE generates another initiation site for synthesis on the DNA strand growing on the DBTP, and the Pr anneals to the resulting DNA strand to provide various additional synthesis sites. This synergistic effect resulted in a markedly more dynamic and robust exponential amplification compared to other modified TRCAs and produced higher yields of DNA (Figure 4). It is also noteworthy that across all parameters (DBTP, Mg, dNTP, DNAP, Pr, and nE concentrations), HNTRCA revealed consistently higher initial reaction rates compared to other amplifications. This steep initial increase indicates that the reaction rapidly generates substantial quantities of DNA within a relatively short time frame. This is particularly advantageous for applications that require rapid, high-yield amplification, such as molecular diagnostics, where the detection of low levels of target DNA or RNA is critical.
Figure 4.
Kinetics of HNTRCA under six different conditions. Real-time fluorescence curves (left), initial rates (middle), and end point signals (right) of HNTRCA performed in the presence of a target miRNA (50 nM) at 50 °C for 1 h with various concentrations of a) DBTP, b) Mg, c) dNTP, d) DNAP, e) Pr, and f) nE, respectively. Data represent mean ± s.d. for three independent experiments.
Overall, the initial reaction rates and maximum yields of the four TRCAs were analyzed for four to six reaction factors. The results demonstrated that HNTRCA consistently had the highest initial rate and maximum yield under the test conditions, while TRCA had the lowest performance (Figures S3 and S4). This is to be expected, given the linear amplification mechanism of TRCA and the exponential amplification scenario resulting from the synergistic effect of the roles of Pr and nE in HNTRCA. Furthermore, it was observed that the initial reaction rate was enhanced for all TRCA reactions when the concentration of Mg, DNAP, Pr, and nE was increased. However, the initial reaction rate was inhibited when excess DBTP and dNTP were present due to competitive reactions. These findings indicate that HNTRCA is the optimal approach when utilizing optimized concentrations of each component for high gain of amplification. It is also crucial to avoid the occurrence of plateauing or efficiency reduction observed at high concentrations. Therefore, based on these results, the optimal conditions for achieving maximum amplification yield were identified as 200 nM DBTP, 6 mM Mg, 1.5 mM dNTP, 0.6 nM DNAP, 200 nM Pr, and 14.7 nM nE.
Analytical Sensitivity and Specificity
To compare the analytical sensitivity of the four TRCAs, each amplification reaction was conducted under optimal conditions using various concentrations of miR-21 ranging from 0 to 5 nM. The resulting amplification profiles and dose–response curves based on the end point signal at 60 and 90 min were shown in Figure 5a,b. As expected, the end point signal showed a positive correlation with miRNA concentration. The limit of detection (LOD) was calculated based on the dose–response curve using three times the standard deviation of the negative sample. As summarized in Table S2, the NTRCA and HNTRCA showed approximately 25- and 23-fold lower LODs than TRCA (based on 60 min end point signals). This resulted in approximately 2516- and 3110-fold improvement when using the 90 min end point signals. In contrast, HTRCA did not show a notable increase in sensitivity compared to TRCA. Although quantitative measurements using this single end point readout are straightforward, they produce a relatively narrow linear dynamic range with one or two log orders of magnitude, particularly when an exponentially amplified signal quickly reaches saturation. Therefore, to obtain a broader dynamic range, the time required to reach the threshold of the amplified signal (amplification time) was used as a quantitative metric to generate linear dose–response curves for NTRCA and HNTRCA (Figure S5). Both responses displayed a broader linear relationship with four log orders of magnitude (5 pM to 5 nM).
Figure 5.
Comparison of analytical sensitivity and specificity of TRCAs. a) Real-time fluorescence curves, b) target dose–response curves, and c) target specificity results of TRCA, HTRCA, NTRCA, and HNTRCA. For sensitivity tests, each reaction was performed with various concentrations of target miRNA, ranging from 0 to 5 nM, at 50 °C for 90 min. The end point signals after 60- and 90-min reactions were used to construct dose–response curves. For specificity tests, each reaction was conducted with eight different miRNAs (5 nM) at 50 °C for 1 h. All reactions were carried out under the optimized conditions, including 200 nM DBTP, 6 mM Mg, 1.5 mM dNTP, 1.5 mM DNAP, 200 nM Pr, and 14.7 nM nE. Data represent mean ± s.d. for three independent experiments.
Next, in order to evaluate the specificity of the amplification system, each TRCA was performed against eight miRNAs (miR-21, miR-34a, let-7a, miR-200a, miR-92b, miR-133a, miR-450b, and miR-4659a), followed by analysis with the 60 min end point signals. As shown in Figure 5c, all four TRCAs exhibited high fluorescence specific to the target miRNA (miR-21), and the signal difference between target and nontarget miRNAs was significantly higher for NTRCA and HNTRAC, which demonstrated relatively better sensitivity and amplification efficiency. Therefore, based on the amplification rate and yield, sensitivity and specificity results, it can be concluded that HNTRCA is the most suitable for highly sensitive and specific detection of miRNAs.
Model Fitting and Simulation Results
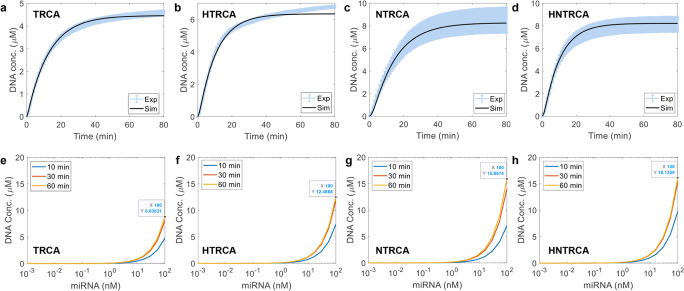
Model parameters were fitted to each individual set of kinetic experiments. In TRCA, a total of 9 parameters were estimated for each experiment set, while an additional 7 parameters were included for the estimation in each HTRCA and NTRCA. As shown in Figure 6a–d, the calculated total DNA concentrations closely matched the experimental values. Although the parameter values should ideally be consistent across different experiments with varied input concentrations, they showed significant variation from one condition to another. It is speculated that these variations in model parameters may be due to diffusion limitations within the reaction volume. Since the participating species need to be effectively delivered to the relevant sites for biochemical reactions to occur, such crowded conditions as those in the DNA amplification setup—where the total reaction volume is minimal and contains numerous substances and growing DNA—can limit diffusion. Additionally, the FI measured during DNA amplification might not be directly proportional to DNA concentration, especially in the presence of numerous ionic and chemical species. Since the dye molecules also need to bind to DNA to emit fluorescence, the measured FI might only reflect a portion of DNA due to their diffusion-limited access.
Figure 6.
Comparison of experimental data with calculation results. a–d) Measured and simulated results of total DNA concentration changes with time. e–h) Simulation results of total DNA concentration changes with respect to varying miRNA concentrations at 10, 30, and 60 min of amplification. Simulations were performed with the data-fitted model parameters for representative conditions, including 200 nM DBTP, 6 mM Mg, 1 mM dNTP, 0.3 mM DNAP, 500 nM Pr, and 7.35 nM nE.
Using the parameters separately identified for TRCA, HTRCA, NTRCA, and HNTRCA, we performed simulations with different amounts of miRNA to compare DNA amplification efficiency across different TRCA methods. Figures 6e–h and S6 illustrate DNA amplification with respect to the amount of miRNA at three-time points (10, 30, and 60 min) for the four TRCA methods. The general trend of increasing DNA synthesis with an increase in miRNA was observed across all methods, consistent with experimental results (Figure 5b). However, the current mechanistic model failed to capture the high nonlinearity in the increase of DNA for NTRCA and HNTRCA. This could be because the model parameters used for this analysis were estimated for much higher miRNA concentration (50 nM) than those simulated here. Nonetheless, the simulations clearly showed that HNTRCA outperforms the other methods in terms of end point signals. Further detailed results on the temporal evolution of biochemical molecules and ionic species can be found in Figures S7–S14.
Quantification of MiR-21 in Total RNA Extracts Using HNTRCA
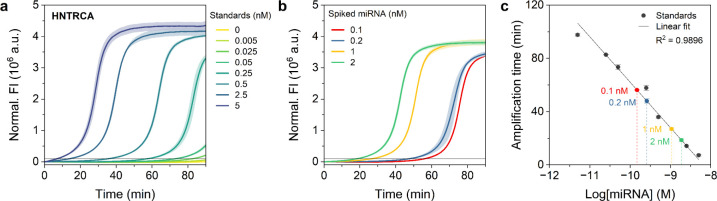
To further test the feasibility of the NHTRCA assay for target miRNA quantification in real biological samples, we adapted this method to quantify miR-21 levels in total RNA extracts of human embryonic kidney cells HEK 293T, which express relatively low levels of miR-21 compared to cancer cells.18,56 To this end, synthetic miR-21 was serially diluted in 100 ng of total RNA extracts, after which the HNTRCA was performed to obtain a standard curve (Figure 7a,c). In a separate experiment, varying concentrations of miR-21 (100, 200, 1000, and 2000 pM) were spiked into RNA extracts. The amplification times from HNTRCA curves were recorded and subsequently analyzed by substituting them into the standard curve, yielding 147.6, 258.7, 1067.1, and 1868.8 pM, respectively (Figure 7b,c). This resulted in recoveries of 147.6, 129.3, 106.7, and 93.44%, as illustrated in Table 1. The observed recoveries above 100% can be attributed to several factors, including potential dilution inaccuracies, minor deviations in the calibration curve, and the possibility of cross-reactivity with other RNAs present in the sample. These factors may result in an overestimation of the measured miR-21 concentration. Therefore, HNTRCA can be a practical and reliable approach for the precise detection of target miRNAs, effectively eliminating the impact of interfering substances that may present in biological samples.
Figure 7.
Quantification of synthetic miR-21 in total RNA extracts using HNTRCA. Real-time fluorescence curves of the HNTRCA with a) serial dilutions of synthetic miR-21 standards (0–5 nM) and b) miR-21 at four different concentrations (0.1, 0.2, 1, and 2 nM) spiked into 100 ng total RNA extracted from HEK 293T cells. The HNTRCA reaction was conducted at 50 °C for 90 min under the optimized conditions. The black solid line of the curves in a) and b) indicates the threshold (105) to determine the amplification time. c) Linear standard curve with serial dilutions of synthetic miR-21 standards to quantify the concentration of spiked miRNA in total RNA extracts. Data represent mean ± s.d. for two or three independent experiments.
Table 1. Quantification of Synthetic miR-21 in Total RNA Extracted from HEK 293T Cells Using HNTRCA.
| Spiked miRNA (pM)a | Calculated miRNA (pM)b | Recovery (%)b | RSD (%)b |
|---|---|---|---|
| 100 | 147.55 ± 0.18 | 147.55 ± 0.18 | 0.12 |
| 200 | 258.72 ± 28.49 | 129.36 ± 14.24 | 11.01 |
| 1000 | 1067.05 ± 59.70 | 106.71 ± 5.97 | 5.59 |
| 2000 | 1868.79 ± 132.97 | 93.44 ± 6.65 | 7.14 |
Synthetic miRNA was spiked into 100 ng of total RNA extracts.
Data represent mean ± s.d. for three independent replicates.
Determination of MiR-21 Expression Levels in Various Cell Lines Using HNTRCA
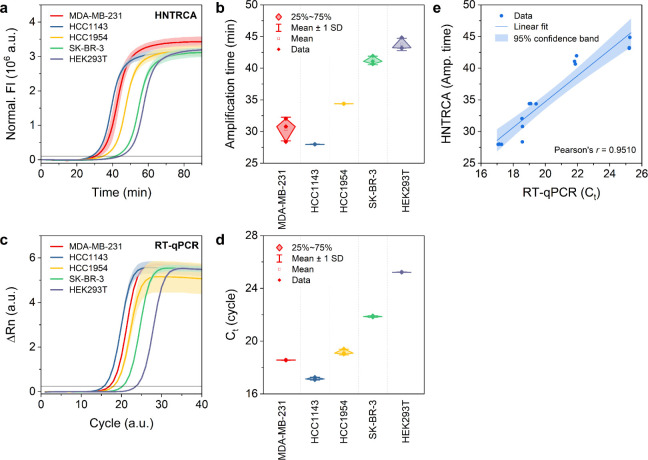
To validate the utility of HNTRCA for identifying the expression of miR-21 in cancer cells, total RNA was extracted from four human breast cancer cell lines (MDA-MB-231, HCC1143, HCC1954, and SK-BR-3) and one normal cell line (HEK 293T). The extracted RNA was subjected to both single-step HNTRCA and two-step reverse transcription-quantitative polymerase chain reaction (RT-qPCR). The HNTRCA analysis demonstrated that cancer cells exhibited relatively higher expression of miR-21 compared to normal cells, indicating the potential of miR-21 as a diagnostic biomarker for breast cancer cells (Figure 8a,b).49 The expression levels of miR-21 were found to be higher in HCC1143, MDA-MB-231, HCC1954, SK-BR-3, and HEK 293T, in that order, which was in good agreement with the results obtained using the gold standard RT-qPCR method (Figure 8c,d). Furthermore, a good correlation was observed between the two methods for detecting miR-21, as indicated by Pearson’s r of 0.9510 (Figure 8e). These results confirm that our HNTRCA approach has the capacity to sensitively detect miRNAs with comparable performance compared to RT-qPCR. Notably, it offers distinct advantages of rapid miRNA detection in complicated biological samples in a single isothermal reaction (<60 min), eliminating the need for an RT step.
Figure 8.
Determination of the presence of miR-21 in total RNA extracted from various cell lines using HNTRCA and RT-qPCR. a) Real-time fluorescence curves and b) amplification time of the HNTRCA reaction with 100 ng total RNA extracted from five different cell lines. The HNTRCA reaction was conducted at 50 °C for 90 min under the optimized conditions. c) Real-time amplification curves and d) cycles to threshold (Ct) of the RT-qPCR assay with 0.5 ng input cDNA prepared from five different cell lines. e) Correlation between the measurements by HNTRCA and RT-qPCR for miR-21 quantification. The black solid line of the curves in a) and c) indicates the threshold to determine the amplification time and Ct value, respectively. Data represent mean ± s.d. for two or three independent experiments.
Conclusions
In this study, we explored modified TRCA techniques with the aim of improving the specificity and sensitivity of a conventional TRCA technique for the detection of disease-associated miRNAs, utilizing the DBTP with the capacity for target-specific conformational switching. The modified TRCA reactions, including HTRCA, NTRCA, and HNTRCA, were implemented by sequentially incorporating Pr and nE, which promoted exponential amplification, into the TRCA system. In order to understand and refine the reaction parameters that affect the real-time kinetics of these four TRCAs, each reaction was performed over a range of concentrations of four to six major components, and two kinetic parameters were subjected to close analysis. Among the TRCA variants, we found that HNTRCA consistently showed the highest amplification rate and maximum yield over a range of concentration conditions. This distinctive kinetics was accompanied by a favorable amplification profile compared to the linear TRCA approach, resulting in a wider dynamic range with a significantly improved detection limit (260 fM) and target specificity. This superior performance of HNTRCA could be attributed to the synergistic effect of Pr and nE, enabling more efficient and broader amplification. Moreover, these results highlight the importance of optimizing biochemical reaction components, including DBTP, Mg, dNTP, DNAP, Pr, and nE concentrations, as well as physical factors such as reaction temperature and time, to circumvent amplification inhibition and maximize efficiency.
We also validated the HNTRCA system with real biological samples under the optimized conditions and demonstrated excellent detection performance and recovery in complex matrices containing different RNA molecules. In addition, the system showed a high correlation with the standard RT-qPCR method for quantifying the expression levels of miR-21 in various cell lines. Consequently, through kinetic analysis and systematic optimization of the four TRCAs, we proposed the HNTRCA system, which features a single-step, easy-to-operate isothermal process for rapid, robust, and efficient DNA amplification.
The kinetic analysis was also performed numerically through mechanistic modeling to better understand the complex interplay between various ionic and biochemical species during different DNA amplification processes. Although further refinement of the mechanistic model is needed to more accurately mimic the fluorescence-based DNA quantification and account for potential diffusion-limited DNA amplification, the current work lays the foundation for in silico platforms aimed at developing and optimizing other RCA variants.
In conclusion, the simplicity and usability of the HNTRCA method make it a promising technique in POC diagnostics, where cost-effective and portable instruments are essential. Integration with POC-friendly systems such as lateral flow assays or smartphone-based signal detection platforms could enhance its applicability. Given that conventional TRCAs are widely used to detect and analyze various types of biomarkers, including nucleic acids, proteins, and small molecules, the developed HNTRCA technique offers considerable potential as a versatile molecular diagnostic tool for developing novel biosensors targeting diverse biomarkers. Furthermore, this study could contribute to improving the accuracy of disease diagnosis, including cancer and rare genetic disorders, and to the rapid and sensitive identification of bacterial or viral pathogens, especially with respect to emerging variants.
Acknowledgments
This work was supported by Incheon National University Research Grant in 2020.
Data Availability Statement
Data requests can be made to e.kim@inu.ac.kr.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsmeasuresciau.4c00063.
Additional tables of oligonucleotide sequences and calculated LODs of four TRCAs and figures, including PAGE analysis of DBTP, melt curve of DBTP, initial rate and end point signal analysis of four TRCAs, target dose–response curves of NTRCA and HNTRCA, and simulation results of four TRCAs (PDF)
Author Contributions
The manuscript was written through the contributions of all authors. All authors have given approval to the final version of the manuscript.
The authors declare no competing financial interest.
Special Issue
Published as part of ACS Measurement Science Auspecial issue “2024 Rising Stars”.
Supplementary Material
References
- Heid C. A.; Stevens J.; Livak K. J.; Williams P. M. Real Time Quantitative PCR. Genome Res. 1996, 6 (10), 986–994. 10.1101/gr.6.10.986. [DOI] [PubMed] [Google Scholar]
- Nayak S.; Blumenfeld N. R.; Laksanasopin T.; Sia S. K. Point-of-Care Diagnostics: Recent Developments in a Connected Age. Anal. Chem. 2017, 89 (1), 102–123. 10.1021/acs.analchem.6b04630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petralia S.; Conoci S. PCR Technologies for Point of Care Testing: Progress and Perspectives. ACS Sens. 2017, 2 (7), 876–891. 10.1021/acssensors.7b00299. [DOI] [PubMed] [Google Scholar]
- Craw P.; Balachandran W. Isothermal Nucleic Acid Amplification Technologies for Point-of-Care Diagnostics: A Critical Review. Lab Chip 2012, 12 (14), 2469–2486. 10.1039/c2lc40100b. [DOI] [PubMed] [Google Scholar]
- Zhao Y.; Chen F.; Li Q.; Wang L.; Fan C. Isothermal Amplification of Nucleic Acids. Chem. Rev. 2015, 115 (22), 12491–12545. 10.1021/acs.chemrev.5b00428. [DOI] [PubMed] [Google Scholar]
- Kang T.; Lu J.; Yu T.; Long Y.; Liu G. Advances in Nucleic Acid Amplification Techniques (NAATs): COVID-19 Point-of-Care Diagnostics as An Example. Biosens. Bioelectron. 2022, 206, 114109. 10.1016/j.bios.2022.114109. [DOI] [PubMed] [Google Scholar]
- Ali M. M.; Li F.; Zhang Z.; Zhang K.; Kang D.-K.; Ankrum J. A.; Le X. C.; Zhao W. Rolling Circle Amplification: A Versatile Tool for Chemical Biology, Materials Science And Medicine. Chem. Soc. Rev. 2014, 43 (10), 3324–3341. 10.1039/c3cs60439j. [DOI] [PubMed] [Google Scholar]
- Mohsen M. G.; Kool E. T. The Discovery of Rolling Circle Amplification and Rolling Circle Transcription. Acc. Chem. Res. 2016, 49 (11), 2540–2550. 10.1021/acs.accounts.6b00417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bialy R. M.; Mainguy A.; Li Y.; Brennan J. D. Functional Nucleic Acid Biosensors Utilizing Rolling Circle Amplification. Chem. Soc. Rev. 2022, 51 (21), 9009–9067. 10.1039/D2CS00613H. [DOI] [PubMed] [Google Scholar]
- Zhao W.; Cui C. H.; Bose S.; Guo D.; Shen C.; Wong W. P.; Halvorsen K.; Farokhzad O. C.; Teo G. S. L.; Phillips J. A.; et al. Bioinspired Multivalent DNA Network for Capture and Release of Cells. Proc. Natl. Acad. Sci. U. S. A. 2012, 109 (48), 19626–19631. 10.1073/pnas.1211234109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S.; Yang S.; Hu X.; Zheng C.; Song H.; Wang L.; Shen Z.; Wu Z.-S. Combination of Immunomagnetic Separation with Aptamer-Mediated Double Rolling Circle Amplification for Highly Sensitive Circulating Tumor Cell Detection. ACS Sens. 2020, 5 (12), 3870–3878. 10.1021/acssensors.0c01082. [DOI] [PubMed] [Google Scholar]
- Kim N.; Kim E.; Kim H.; Thomas M. R.; Najer A.; Stevens M. M. Tumor-Targeting Cholesterol-Decorated DNA Nanoflowers for Intracellular Ratiometric Aptasensing. Adv. Mater. 2021, 33 (11), 2007738. 10.1002/adma.202007738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J.; Jia X.; Li Q.; Cui Z.; Liang A.; Ke B.; Yang D.; Yao C. A DNA-Based Hydrogel for Exosome Separation and Biomedical Applications. Proc. Natl. Acad. Sci. U. S. A. 2023, 120 (28), e2303822120 10.1073/pnas.2303822120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F.; Lu C.-H.; Liu X.; Freage L.; Willner I. Amplified and Multiplexed Detection of DNA Using the Dendritic Rolling Circle Amplified Synthesis of DNAzyme Reporter Units. Anal. Chem. 2014, 86 (3), 1614–1621. 10.1021/ac4033033. [DOI] [PubMed] [Google Scholar]
- Kim E.; Howes P. D.; Crowder S. W.; Stevens M. M. Multi-Amplified Sensing of MicroRNA by a Small DNA Fragment-Driven Enzymatic Cascade Reaction. ACS Sens. 2017, 2 (1), 111–118. 10.1021/acssensors.6b00601. [DOI] [PubMed] [Google Scholar]
- Chai H.; Wang M.; Zhang C.; Tang Y.; Miao P. Highly Sensitive Genosensing Coupling Rolling Circle Amplification with Multiple DNAzyme Cores for DNA Walking. Bioconjugate Chem. 2020, 31 (3), 764–769. 10.1021/acs.bioconjchem.9b00861. [DOI] [PubMed] [Google Scholar]
- Mahmoudian L.; Kaji N.; Tokeshi M.; Nilsson M.; Baba Y. Rolling Circle Amplification and Circle-to-circle Amplification of a Specific Gene Integrated with Electrophoretic Analysis on a Single Chip. Anal. Chem. 2008, 80 (7), 2483–2490. 10.1021/ac702289j. [DOI] [PubMed] [Google Scholar]
- Tian B.; Fock J.; Minero G. A. S.; Hansen M. F. Nicking-Assisted On-Loop and Off-Loop Enzymatic Cascade Amplification for Optomagnetic Detection of a Highly Conserved Dengue Virus Sequence. Biosens. Bioelectron. 2020, 160, 112219. 10.1016/j.bios.2020.112219. [DOI] [PubMed] [Google Scholar]
- Nilsson M. Real-Time Monitoring of Rolling-Circle Amplification Using a Modified Molecular Beacon Design. Nucleic Acids Res. 2002, 30 (14), e66 10.1093/nar/gnf065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali M. M.; Li Y. Colorimetric Sensing by Using Allosteric-DNAzyme-Coupled Rolling Circle Amplification and a Peptide Nucleic Acid–Organic Dye Probe. Angew. Chem., Int. Ed. 2009, 48 (19), 3512–3515. 10.1002/anie.200805966. [DOI] [PubMed] [Google Scholar]
- Hu J.; Zhang C.-Y. Sensitive Detection of Nucleic Acids with Rolling Circle Amplification and Surface-Enhanced Raman Scattering Spectroscopy. Anal. Chem. 2010, 82 (21), 8991–8997. 10.1021/ac1019599. [DOI] [PubMed] [Google Scholar]
- Liu M.; Song J.; Shuang S.; Dong C.; Brennan J. D.; Li Y. A Graphene-Based Biosensing Platform Based on the Release of DNA Probes and Rolling Circle Amplification. ACS Nano 2014, 8 (6), 5564–5573. 10.1021/nn5007418. [DOI] [PubMed] [Google Scholar]
- Zhang Z.; Wang S.; Ma J.; Zhou T.; Wang F.; Wang X.; Zhang G. Rolling Circle Amplification-Based Polyvalent Molecular Beacon Probe-Assisted Signal Amplification Strategies for Sensitive Detection of B16 Cells. ACS Biomater. Sci. Eng. 2020, 6 (5), 3114–3121. 10.1021/acsbiomaterials.0c00288. [DOI] [PubMed] [Google Scholar]
- Nallur G. Signal Amplification by Rolling Circle Amplification on DNA Microarrays. Nucleic Acids Res. 2001, 29 (23), e118 10.1093/nar/29.23.e118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell C.; Welch K.; Jarvius J.; Cai Y.; Brucas R.; Nikolajeff F.; Svedlindh P.; Nilsson M. Gold Nanowire Based Electrical DNA Detection Using Rolling Circle Amplification. ACS Nano 2014, 8 (2), 1147–1153. 10.1021/nn4058825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S.; Bi S.; Wang Z.; Xia J.; Zhang F.; Yang M.; Gui R.; Li Y.; Xia Y. A Plasmonic Aptasensor for Ultrasensitive Detection of Thrombin via Arrested Rolling Circle Amplification. Chem. Commun. 2015, 51 (37), 7927–7930. 10.1039/C5CC02069G. [DOI] [PubMed] [Google Scholar]
- Qu X.; Bian F.; Guo Q.; Ge Q.; Sun Q.; Huang X. Ligation-Rolling Circle Amplification on Quantum Dot-Encoded Microbeads for Detection of Multiplex G-Quadruplex-Forming Sequences. Anal. Chem. 2018, 90 (20), 12051–12058. 10.1021/acs.analchem.8b02820. [DOI] [PubMed] [Google Scholar]
- Tian B.; Qiu Z.; Ma J.; Donolato M.; Hansen M. F.; Svedlindh P.; Strömberg M. On-Particle Rolling Circle Amplification-Based Core–Satellite Magnetic Superstructures for MicroRNA Detection. ACS Appl. Mater. Interfaces 2018, 10 (3), 2957–2964. 10.1021/acsami.7b16293. [DOI] [PubMed] [Google Scholar]
- Zhang X.; Li R.; Chen Y.; Zhang S.; Wang W.; Li F. Applying DNA Rolling Circle Amplification in Fluorescence Imaging of Cell Surface Glycans Labeled by a Metabolic Method. Chem. Sci. 2016, 7 (9), 6182–6189. 10.1039/C6SC02089E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng R.; Zhang K.; Wang L.; Ren X.; Sun Y.; Li J. DNA-Sequence-Encoded Rolling Circle Amplicon for Single-Cell RNA Imaging. Chem 2018, 4 (6), 1373–1386. 10.1016/j.chempr.2018.03.003. [DOI] [Google Scholar]
- Wang X.; Allen W. E.; Wright M. A.; Sylwestrak E. L.; Samusik N.; Vesuna S.; Evans K.; Liu C.; Ramakrishnan C.; Liu J.; et al. Three-Dimensional Intact-Tissue Sequencing of Single-Cell Transcriptional States. Science 2018, 361 (6400), eaat5691 10.1126/science.aat5691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao T.; Chen T.; Feng C.; He X.; Mu C.; Anzai J.-I.; Li G. Design and Fabrication of Flexible DNA Polymer Cocoons to Encapsulate Live Cells. Nat. Commun. 2019, 10 (1), 2946. 10.1038/s41467-019-10845-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue S.; Li Y.; Qiao Z.; Song W.; Bi S. Rolling Circle Replication for Biosensing, Bioimaging, and Biomedicine. Trends Biotechnol. 2021, 39 (11), 1160–1172. 10.1016/j.tibtech.2021.02.007. [DOI] [PubMed] [Google Scholar]
- Gao Y.-P.; Huang K.-J.; Wang F.-T.; Hou Y.-Y.; Xu J.; Li G. Recent Advances in Biological Detection with Rolling Circle Amplification: Design Strategy, Biosensing Mechanism, and Practical Applications. Analyst 2022, 147 (15), 3396–3414. 10.1039/D2AN00556E. [DOI] [PubMed] [Google Scholar]
- Nelson J. R.; Cai Y. C.; Giesler T. L.; Farchaus J. W.; Sundaram S. T.; Ortiz-Rivera M.; Hosta L. P.; Hewitt P. L.; Mamone J. A.; Palaniappan C.; et al. TempliPhi, φ29 DNA Polymerase Based Rolling Circle Amplification of Templates for DNA Sequencing. BioTechniques 2002, 32 (sup6), S44–S47. 10.2144/jun0208. [DOI] [PubMed] [Google Scholar]
- Picher Á. J.; Budeus B.; Wafzig O.; Krüger C.; García-Gómez S.; Martínez-Jiménez M. I.; Díaz-Talavera A.; Weber D.; Blanco L.; Schneider A. TruePrime is a Novel Method for Whole-Genome Amplification from Single Cells Based on TthPrimPol. Nat. Commun. 2016, 7 (1), 13296. 10.1038/ncomms13296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drmanac R.; Sparks A. B.; Callow M. J.; Halpern A. L.; Burns N. L.; Kermani B. G.; Carnevali P.; Nazarenko I.; Nilsen G. B.; Yeung G.; Dahl F.; Fernandez A.; Staker B.; Pant K. P.; Baccash J.; Borcherding A. P.; Brownley A.; Cedeno R.; Chen L.; Chernikoff D.; Cheung A.; Chirita R.; Curson B.; Ebert J. C.; Hacker C. R.; Hartlage R.; Hauser B.; Huang S.; Jiang Y.; Karpinchyk V.; Koenig M.; Kong C.; Landers T.; Le C.; Liu J.; McBride C. E.; Morenzoni M.; Morey R. E.; Mutch K.; Perazich H.; Perry K.; Peters B. A.; Peterson J.; Pethiyagoda C. L.; Pothuraju K.; Richter C.; Rosenbaum A. M.; Roy S.; Shafto J.; Sharanhovich U.; Shannon K. W.; Sheppy C. G.; Sun M.; Thakuria J. V.; Tran A.; Vu D.; Zaranek A. W.; Wu X.; Drmanac S.; Oliphant A. R.; Banyai W. C.; Martin B.; Ballinger D. G.; Church G. M.; Reid C. A. Human Genome Sequencing Using Unchained Base Reads on Self-Assembling DNA Nanoarrays. Science 2010, 327 (5961), 78–81. 10.1126/science.1181498. [DOI] [PubMed] [Google Scholar]
- Lizardi P. M.; Huang X.; Zhu Z.; Bray-Ward P.; Thomas D. C.; Ward D. C. Mutation Detection and Single-Molecule Counting Using Isothermal Rolling-Circle Amplification. Nat. Genet. 1998, 19 (3), 225–232. 10.1038/898. [DOI] [PubMed] [Google Scholar]
- Hutchison C. A.; Smith H. O.; Pfannkoch C.; Venter J. C. Cell-Free Cloning Using φ29 DNA Polymerase. Proc. Natl. Acad. Sci. U. S. A. 2005, 102 (48), 17332–17336. 10.1073/pnas.0508809102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami T.; Sumaoka J.; Komiyama M. Sensitive Isothermal Detection of Nucleic-Acid Sequence by Primer Generation–Rolling Circle Amplification. Nucleic Acids Res. 2009, 37 (3), e19–e19 10.1093/nar/gkn1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukal A. C.; Kidanemariam D. B.; Dale J. L.; Harding R. M.; James A. P. Assessment and Optimization of Rolling Circle Amplification Protocols for the Detection and Characterization of Badnaviruses. Virology 2019, 529, 73–80. 10.1016/j.virol.2019.01.013. [DOI] [PubMed] [Google Scholar]
- Li D.; Zhang T.; Yang F.; Yuan R.; Xiang Y. Efficient and Exponential Rolling Circle Amplification Molecular Network Leads to Ultrasensitive and Label-Free Detection of MicroRNA. Anal. Chem. 2020, 92 (2), 2074–2079. 10.1021/acs.analchem.9b04585. [DOI] [PubMed] [Google Scholar]
- Smith L. D.; Nalla S.; Kuo C.-W.; Kohli M.; Smith A. M. Rapid Quantification of MicroRNA-375 Through One-Pot Primer-Generating Rolling Circle Amplification. Analyst 2022, 147 (13), 2936–2941. 10.1039/D2AN00263A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akama S.; Yamamura M.; Kigawa T. A Multiphysics Model of In Vitro Transcription Coupling Enzymatic Reaction and Precipitation Formation. Biophys. J. 2012, 102 (2), 221–230. 10.1016/j.bpj.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Berg D.; Kis Z.; Behmer C. F.; Samnuan K.; Blakney A. K.; Kontoravdi C.; Shattock R.; Shah N. Quality by Design Modelling to Support Rapid RNA Vaccine Production Against Emerging Infectious Diseases. npj Vaccines 2021, 6 (1), 65. 10.1038/s41541-021-00322-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stover N. M.; Ganko K.; Braatz R. D. Mechanistic Modeling of In Vitro Transcription Incorporating Effects of Magnesium Pyrophosphate Crystallization. Biotechnol. Bioeng. 2024, 121 (9), 2636–2647. 10.1002/bit.28699. [DOI] [PubMed] [Google Scholar]
- Deng R.; Tang L.; Tian Q.; Wang Y.; Lin L.; Li J. Toehold-Initiated Rolling Circle Amplification for Visualizing Individual MicroRNAs In Situ in Single Cells. Angew. Chem., Int. Ed. 2014, 53 (9), 2389–2393. 10.1002/anie.201309388. [DOI] [PubMed] [Google Scholar]
- Kim D.; Lee J.; Han J.; Lim J.; Lim E.-K.; Kim E. A Highly Specific and Flexible Detection Assay Using Collaborated Actions of DNA-Processing Enzymes for Identifying Multiple Gene Expression Signatures in Breast Cancer. Analyst 2023, 148 (2), 316–327. 10.1039/D2AN01672A. [DOI] [PubMed] [Google Scholar]
- Bautista-Sánchez D.; Arriaga-Canon C.; Pedroza-Torres A.; De La Rosa-Velázquez I. A.; González-Barrios R.; Contreras-Espinosa L.; Montiel-Manríquez R.; Castro-Hernández C.; Fragoso-Ontiveros V.; Álvarez-Gómez R. M.; et al. The Promising Role of miR-21 as a Cancer Biomarker and Its Importance in RNA-Based Therapeutics. Mol. Ther. -Nucleic Acids 2020, 20, 409–420. 10.1016/j.omtn.2020.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krichevsky A. M.; Gabriely G. MiR-21: A Small Multi-Faceted RNA. J. Cell. Mol. Med. 2009, 13 (1), 39–53. 10.1111/j.1582-4934.2008.00556.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batra V. K.; Beard W. A.; Shock D. D.; Krahn J. M.; Pedersen L. C.; Wilson S. H. Magnesium-Induced Assembly of a Complete DNA Polymerase Catalytic Complex. Structure 2006, 14 (4), 757–766. 10.1016/j.str.2006.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W. -J.; Yang W.; Tsai M.-D. How DNA Polymerases Catalyse Replication and Repair with Contrasting Fidelity. Nat. Rev. Chem. 2017, 1 (9), 0068. 10.1038/s41570-017-0068. [DOI] [Google Scholar]
- Zipper H.; Brunner H.; Bernhagen J.; Vitzthum F. Investigations on DNA Intercalation and Surface Binding by SYBR Green I, Its Structure Determination and Methodological Implications. Nucleic Acids Res. 2004, 32 (12), e103–e103 10.1093/nar/gnh101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oscorbin I.; Filipenko M. Bst Polymerase — A Humble Relative of Taq Polymerase. Comp. Struct. Biotechnol. J. 2023, 21, 4519–4535. 10.1016/j.csbj.2023.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langer A.; Schräml M.; Strasser R.; Daub H.; Myers T.; Heindl D.; Rant U. Polymerase/DNA Interactions and Enzymatic Activity: Multi-Parameter Analysis with Electro-Switchable Biosurfaces. Sci. Rep. 2015, 5 (1), 12066. 10.1038/srep12066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S.; Wu H.; Wu F.; Nie D.; Sheng S.; Mo Y.-Y. MicroRNA-21 Targets Tumor Suppressor Genes in Invasion and Metastasis. Cell Res. 2008, 18 (3), 350–359. 10.1038/cr.2008.24. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data requests can be made to e.kim@inu.ac.kr.