Supplemental Digital Content is Available in the Text.
Back pain is common in idiopathic scoliosis. Thirty-seven known selected pain variants are not associated with back pain in individuals with idiopathic scoliosis.
Keywords: Back pain, Pain, Idiopathic scoliosis, Genetics
Abstract
Introduction:
Back pain is common in idiopathic scoliosis. The aim of this study was to study known genetic variants associated with pain in individuals with idiopathic scoliosis.
Methods:
We included 1442 individuals with juvenile or adolescent idiopathic scoliosis from Sweden and Denmark. Single nucleotide variants (SNV) genotyping was performed on 37 SNVs. Pain was assessed using 2 questionnaires. The mean pain domain score on the Scoliosis Research Society 22 revised questionnaire (SRS-22r) ranging between 1 (worst) and 5 (best) was dichotomized into a “back pain group” (score <4) and a “no back pain group” (score ≥4). The EuroQol 5-dimensions (EQ-5D) 3 level pain domain was dichotomized into a “no pain group” and a “pain group.” Odds ratios were used to describe the associations.
Results:
Based on the SRS-22r pain domain scores, 456 individuals (32%) reported back pain. Based on the EQ-5D questionnaire, 813 individuals (56%) reported moderate or extreme pain/discomfort. The odds ratio for the associations between the selected genetic variants and back pain or pain in general as measured with SRS-22r and EQ-5D-3L ranged between 0.88 to 1.17 and 0.86 to 1.16, with P-values ranging between 0.08 to 0.99 and 0.08 to 0.95.
Conclusion:
This study suggests that known genetic variants associated with pain do not play a significant role in the development of pain in individuals with idiopathic scoliosis.
1. Introduction
Idiopathic scoliosis is the most common spinal deformity affecting children and adolescents, with a prevalence of 1%-3%.7,30,57 Scoliosis is defined as a 3-dimensional spinal deformity with an angle of ≥10° on the coronal plane of a standing full-spine radiograph, determined using the Cobb method.9 Scoliosis can progress rapidly during the final growth spurt, leading to decreased respiratory function, cosmetic issues, and back pain,56 and is therefore treated with bracing, or surgery.7
Back pain is a common and complex condition, and individuals with idiopathic scoliosis are known to have a high prevalence of back pain.46,52,58 Recent genetic studies have suggested several genetic variants associated with back pain phenotypes, including the risk of back pain, back pain intensity, and disability.17,20,27,54,59
The authors are unaware of studies examining the association of genetic variants associated with back pain in individuals with idiopathic scoliosis. The aim of this study was to study the association between a set of previously described pain candidate variants and pain in individuals with idiopathic scoliosis.
2. Methods
2.1. Subjects
Individuals diagnosed with idiopathic scoliosis, with a Cobb angle greater than 10° and onset between ages 4 and 9 years (juvenile) or from the age of 10 years onward (adolescent), were invited to participate. Recruitment took place in 5 Swedish and one Danish hospital between 2004 and 2013, as described earlier.21–23 Exclusion criteria included individuals with an onset or diagnosis less than the age of 4 years, nonidiopathic scoliosis, neural abnormalities on magnetic resonance imaging (MRI) of the spine, incomplete medical records, unsuccessful DNA sampling, or extraction or failure to respond to at least one of the health-related quality-of-life (HRQoL) questionnaires. In total, 1442 individuals were included (Fig. 1).
Figure 1.
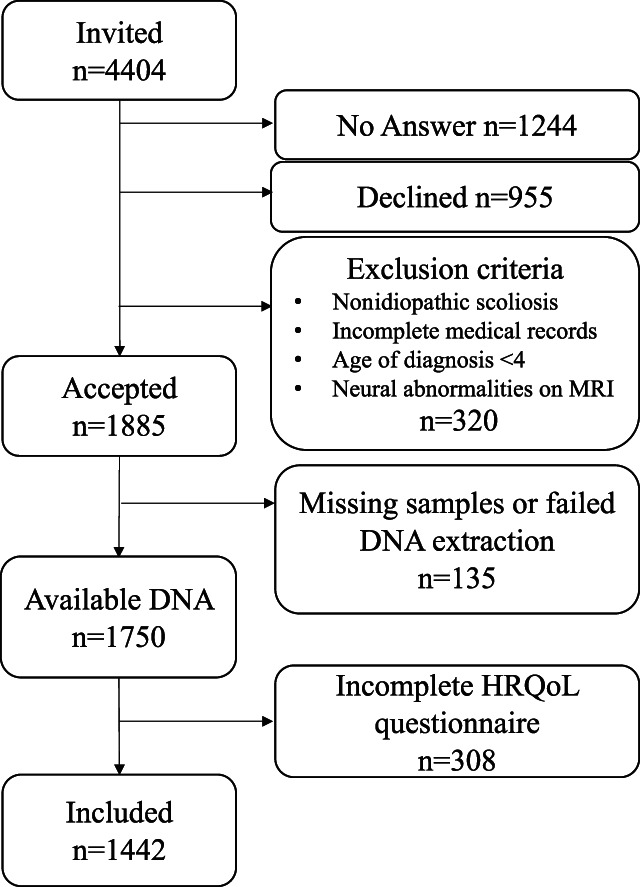
Flow chart of the study participants.
2.2. Questionnaires
We assessed back pain with the validated Swedish and Danish versions of Scoliosis Research Society 22 revised questionnaire (SRS-22r), which stands as the most widely used disease-specific outcome questionnaire for idiopathic scoliosis for all ages.1,14,48 We assessed general pain with the EuroQol 5-dimensions 3 level (EQ-5D-3L) pain/discomfort domain.41
The SRS-22r comprises 5 domains: pain, function, self-image, mental health, and satisfaction with treatment. The pain domain consists of 5 questions, with the first 3 questions focusing on the extent of back pain experienced in the last 6 months, one month, and at rest. The fourth question concerns the use of pain medication for back pain, while the fifth question addresses whether the patient has taken any days off work due to back pain in the last 3 months. Each question offers 5 answer alternatives, each scored from 1 (worst) to 5 (best). The mean pain domain score was calculated based on the scores from these 5 questions.29
In the EQ-5D-3L questionnaire, the pain or discomfort question asked individuals to describe their pain or discomfort at the time of the questionnaire, with 3 available answer alternatives; “no pain/discomfort,” “moderate pain/discomfort,” or “extreme pain/discomfort.” The participants provided demographic information about sex, body height, and weight. Age at the time of the questionnaire assessment was registered. Participants were asked to report their own and their parents' country of birth. Scandinavian ancestry was defined as having both parents born in Sweden, Norway, or Denmark.
2.3. Radiology
The largest available curve for untreated individuals was used.9 For brace or surgically treated individuals, the largest available curve before treatment was used.21–23
2.4. DNA extraction and genotyping
The candidate pain variants were selected based on the existing literature, and a total of 37 single nucleotide variants (SNVs) distributed across 21 genes were included in this study (Table 1). Twenty-two SNVs were directly genotyped, and the genotypes for 15 SNVs were imputed.
Table 1.
Pain variants included.
| CHR | SNV | Method | Gene | Pain phenotype |
|---|---|---|---|---|
| 1 | rs4645978 | Imputation | CASP9 | Risk |
| 1 | rs932816 | Genotyping | FAAH | Intensity, disability |
| 1 | rs4141964 | Genotyping | FAAH | Intensity, disability |
| 1 | rs324420 | Genotyping | FAAH | Intensity, disability |
| 2 | rs1800587 | Genotyping | IL1A | Intensity, disability |
| 2 | rs1143634 | Genotyping | IL1B | Risk, intensity, disability |
| 2 | rs2234677 | Imputation | IL1RN | Risk, intensity, disability |
| 2 | rs6746030 | Genotyping | SCN9A | Intensity |
| 3 | rs6795970 | Genotyping | SCN10A | Intensity |
| 5 | rs2053044 | Genotyping | ADRB2 | Risk |
| 6 | rs2234693 | Genotyping | ESR1 | Intensity |
| 6 | rs9340799 | Genotyping | ESR1 | Intensity |
| 6 | rs1799971 | Genotyping | OPRM1 | Intensity |
| 8 | rs7814941 | Genotyping | GSDMC/CCDC26 | Risk |
| 10 | rs3180 | Genotyping | SPOCK2/CHST3 | Risk |
| 11 | rs6265 | Genotyping | BDNF | Intensity |
| 11 | rs1799750 | Imputation | MMP1 | Intensity, disability |
| 12 | rs731236 | Genotyping | VDR | Intensity |
| 12 | rs2228570 | Imputation | VDR | Risk |
| 14 | rs998259 | Imputation | GCH1 | Intensity |
| 20 | rs143383 | Genotyping | GDF5 | Risk |
| 20 | rs734784 | Imputation | KCNS1 | Intensity |
| 20 | rs13043825 | Imputation | KCNS1 | Intensity |
| 20 | rs17576 | Imputation | MMP9 | Intensity |
| 21 | rs9981629 | Imputation | KCNJ6 | Intensity |
| 21 | rs928723 | Genotyping | KCNJ6 | Intensity |
| 21 | rs1543754 | Imputation | KCNJ6 | Intensity |
| 21 | rs858035 | Genotyping | KCNJ6 | Intensity |
| 21 | rs2835925 | Imputation | KCNJ6 | Intensity |
| 21 | rs2835930 | Imputation | KCNJ6 | Intensity |
| 21 | rs1787337 | Imputation | KCNJ6 | Intensity |
| 21 | rs2211843 | Genotyping | KCNJ6 | Intensity |
| 22 | rs2075507 | Imputation | COMT | Disability |
| 22 | rs6269 | Genotyping | COMT | Disability |
| 22 | rs4633 | Genotyping | COMT | Disability |
| 22 | rs4818 | Imputation | COMT | Disability |
| 22 | rs4680 | Genotyping | COMT | Risk, intensity, disability |
ADRB2, adrenoceptor beta 2; BDNF, brain derived neurotrophic factor; CASP9, caspase 9; CCDC26; SPOCK2; CHST3, carbohydrate sulfotransferase 3 gene; CHR, chromosome; COMT, catechol-O-methyltransferase; ESR1, estrogen receptor 1; FAAH, fatty acid amide hydrolase; GCH1, GTP cyclohydrolase 1; GDF5, growth differentiation factor 5; GSDMC, Gasdermin C; IL1A, interleukin 1 alpha; IL1B, interleukin 1 beta; IL1RN, interleukin 1 receptor antagonist; KCNJ6, potassium inwardly rectifying channel, subfamily J, member 6; KCNS1, potassium voltage-gated channel modifier subfamily S member; MMP1, matrix metallopeptidase 1; MMP9, matrix metallopeptidase 9; OPRM1, opioid receptor mu 1; SCN10A, sodium voltage-gated channel alpha subunit 10; SCN9A, sodium voltage-gated channel alpha subunit 9; SNV, single nucleotide variant; VDR, vitamin D receptor.
DNA was extracted from blood or saliva using a salt precipitation method on the Autopure LS system (Qiagen, Hilden, Germany) or the QIAamp 96 DNA Blood kit (Qiagen), following the manufacturer's instructions. For the following variants, rs6746030, rs6795970, rs7814941, rs3180, and rs4680, genotyping was conducted at the Mutation Analysis Facility at Karolinska University Hospital Huddinge in Stockholm, Sweden. Genotyping was performed using iPLEX Gold chemistry on the MassARRAY mass spectrometry system (Agena BioScience, San Diego, CA). For the remaining variants, genotyping was performed at the SNP&SEQ Technology Platform at Uppsala University, Uppsala, Sweden (part of NGI and SciLifeLab). The method used was the Illumina Global Screening Array-Multi Disease version 3 (GSAMD-24v3-0-EA_20034606_A1) (Illumina Inc., San Diego, CA).
2.5. Sample quality control
All samples with missing data >5% for each SNV were removed. Duplicate samples were identified using KING v. 2.1.6,31 and one of each pair was removed. Sex discordance was calculated using PLINK.40 All remaining samples were merged with samples from the 1000 genomes (1000 g) database,2 and a principal component analysis (PCA) to identify outliers was performed using PLINK. Study samples deviating >3 standard deviations from the mean of PCA1 or PCA2 for European samples from 1000 g were removed. Finally, KING31 was used to create a set of unrelated samples.
2.6. Imputation
Imputation was performed where missing genotypes were inferred from high-quality reference data and allowed us to obtain genotypes for the SNVs not directly included on the genotyping array. With robust reference genotypes and stringent quality control, imputation is a well-established and reliable bioinformatic approach in genetic analysis.32 Single nucleotide variants with a missingness <1%, not deviating from Hardy–Weinberg Equilibrium (HWE) (P < 1e-6), were used for imputation. Beagle v5.2 (https://faculty.washington.edu/browning/conform-gt.html) was used for phasing and imputation. Before imputation, the conform-gt tool provided with Beagle was used to verify allele consistency with the reference, using European samples only.
2.7. Statistical analysis
Baseline descriptive statistics are presented as number (%) and mean (SD). The allelic case–control association by the Pearson χ2 test8 was performed. Other data were analysed using the Pearson χ2 test for categorical data and Student t-test for continuous data on IBM SPSS Statistics version 27.0 (IBM Corp., Armonk, NY). Effect sizes are presented as odds ratios with corresponding P-values. All P-values are uncorrected.
Based on the mean pain domain score on SRS-22r, individuals were subsequently categorized into a “back pain” group (score less than 4) and a “no back pain” group (score of 4 or more). In additional analyses, each of the 5 SRS-22r pain questions was analysed with and without dichotomization.
Based on the generic EQ-5D-3L pain/discomfort domain, individuals were divided into 2 groups: a “no pain” group that answered, “no pain/discomfort” and a “pain” group that answered, “moderate pain/discomfort” or “extreme pain/discomfort.” In addition, the EQ-5D-3L pain/discomfort domain was also analysed without dichotomization.
In the subanalyses, the 5 questions of the SRS-22r pain domain were dichotomized similarly to the SRS-22r pain domain index score. The allelic case–control association by the Pearson χ2 test was performed in the subanalysis.
2.8. Ethical approval
The study received approval from the Stockholm and Lund ethical boards in Sweden (LU 363-02, 290/2006, 2009/1124-31/2, 2017/2374-31), and the Regional Committee on Health Research Ethics for Southern Denmark (S-2011002).
3. Results
Baseline characteristics are presented in Table 2. Based on the SRS-22r pain domain scores, 456 individuals (32%) were in the back pain group. Based on the EQ-5D-3L questionnaire, 813 individuals (56%) were in the pain group, reporting moderate or extreme pain/discomfort (Table 2). The group of individuals with pain in either EQ-5D-3L or SRS-22r was characterized by a higher proportion of female patients, a larger mean Cobb angle, and an older age at questionnaire response (Table 2). The median age at the questionnaire response for the included 1442 individuals was 40.0 (25th percentile 22.9, 75th percentile 47.8) years. The number of individuals for each SNV is presented in Table 3. Failed extraction, genotyping, or QC occurred in 132 samples in the Illumina GSAMD-24v3 analysis (Table 3).
Table 2.
Study population demographics and clinical outcomes.
| SRS-22r pain domain score | EQ-5D-3L pain question | |||||
|---|---|---|---|---|---|---|
| No back pain | Back pain | P | No pain | Pain | P | |
| n = 985 (68%) | n = 456 (32%) | n = 627 (44%) | n = 813 (56%) | |||
| Height (cm) | 168.4 (8.0) | 168.2 (7.9) | 0.73 | 168.4 (8.3) | 168.2 (7.7) | 0.62 |
| Weight (kg) | 64.7 (13.0) | 67.8 (13.9) | <0.001 | 64.1 (12.9) | 66.9 (13.6) | <0.001 |
| Age at questionnaire (y) | 36.6 (13.9) | 38.7 (13.3) | 0.006 | 35.7 (13.9) | 38.4 (13.4) | <0.001 |
| Largest Cobb angle (degrees) | 39.3 (16.9) | 42.9 (18.0) | <0.001 | 38.6 (16.7) | 41.9 (17.7) | <0.001 |
| Sex | 0.08 | 0.001 | ||||
| Female | 853 (68%) | 410 (32%) | 530 (42%) | 733 (58%) | ||
| Male | 132 (74%) | 46 (26%) | 97 (55%) | 80 (45%) | ||
| Country of inclusion | 0.06 | 0.28 | ||||
| Denmark | 89 (61%) | 56 (39%) | 57 (39%) | 88 (61%) | ||
| Sweden | 896 (69%) | 400 (31%) | 570 (44%) | 725 (56%) | ||
| Type of scoliosis | 0.07 | 0.73 | ||||
| Adolescent idiopathic scoliosis | 872 (69%) | 388 (31%) | 546 (43%) | 713 (57%) | ||
| Juvenile idiopathic scoliosis | 113 (62%) | 68 (38%) | 81 (45%) | 100 (55%) | ||
| Convexity of major curve | 0.44 | 0.30 | ||||
| Left | 282 (70%) | 121 (30%) | 184 (46%) | 218 (54%) | ||
| Right | 697 (68%) | 330 (32%) | 439 (43%) | 588 (57%) | ||
| Type of curve | 0.81 | 0.29 | ||||
| Thoracal | 607 (69%) | 269 (31%) | 384 (44%) | 492 (56%) | ||
| Thoracolumbar | 153 (68%) | 73 (32%) | 108 (48%) | 118 (52%) | ||
| Lumbar | 100 (68%) | 47 (32%) | 58 (40%) | 88 (60%) | ||
| Double primary | 107 (66%) | 56 (34%) | 64 (39%) | 99 (61%) | ||
| Type of treatment | 0.02 | 0.16 | ||||
| Brace or untreated | 647 (71%) | 271 (29%) | 412 (45%) | 505 (55%) | ||
| Surgery | 338 (65%) | 185 (35%) | 215 (41%) | 308 (59%) | ||
| Ancestry | 0.89 | 0.80 | ||||
| Scandinavian | 858 (68%) | 396 (32%) | 544 (43%) | 709 (57%) | ||
| Non-Scandinavian | 127 (68%) | 60 (32%) | 83 (44%) | 104 (56%) | ||
Data are presented as number of individuals (percentage) or mean (standard deviation).
Table 3.
Association between back pain in the scoliosis research society 22 revised questionnaire domain and pain in the EuroQol 5-dimensions 3 level domain and the included single nucleotide variants.
| CHR | SNV | Samples genotyped & Passed QC | Gene | Minor allele | Major allele | SRS22r back pain index | EQ-5D-3L | SRS22r Back pain 6 mo | SRS22r Back pain 1 mo | SRS22r Back pain rest | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MAF case | MAF control | P | OR | MAF case | MAF control | P | OR | MAF case | MAF control | P | OR | MAF case | MAF control | P | OR | MAF case | MAF control | P | OR | ||||||
| 1 | rs4645978 | 1310 | CASP9 | C | T | 0.45 | 0.45 | 0.89 | 1.01 | 0.45 | 0.45 | 0.82 | 1.02 | 0.45 | 0.45 | 0.91 | 0.99 | 0.44 | 0.46 | 0.5 | 0.94 | 0.46 | 0.45 | 0.4 | 1.07 |
| 1 | rs932816 | 1310 | FAAH | A | G | 0.31 | 0.31 | 0.96 | 1 | 0.32 | 0.3 | 0.27 | 1.1 | 0.32 | 0.3 | 0.37 | 1.08 | 0.31 | 0.31 | 0.76 | 1.03 | 0.31 | 0.31 | 0.82 | 1.02 |
| 1 | rs4141964 | 1310 | FAAH | T | C | 0.38 | 0.39 | 0.54 | 0.95 | 0.39 | 0.38 | 0.55 | 1.05 | 0.38 | 0.39 | 0.65 | 0.96 | 0.38 | 0.39 | 0.4 | 0.93 | 0.39 | 0.39 | 0.84 | 0.98 |
| 1 | rs324420 | 1310 | FAAH | A | C | 0.25 | 0.22 | 0.19 | 1.14 | 0.24 | 0.22 | 0.34 | 1.09 | 0.24 | 0.22 | 0.39 | 1.08 | 0.24 | 0.22 | 0.32 | 1.1 | 0.23 | 0.23 | 0.95 | 0.99 |
| 2 | rs1800587 | 1310 | IL1A | A | G | 0.32 | 0.33 | 0.39 | 0.93 | 0.32 | 0.34 | 0.34 | 0.92 | 0.32 | 0.34 | 0.3 | 0.92 | 0.32 | 0.33 | 0.35 | 0.92 | 0.33 | 0.33 | 0.78 | 0.98 |
| 2 | rs1143634 | 1310 | IL1B | A | G | 0.25 | 0.27 | 0.4 | 0.92 | 0.26 | 0.26 | 0.8 | 0.98 | 0.26 | 0.26 | 0.66 | 0.96 | 0.26 | 0.26 | 0.95 | 0.99 | 0.26 | 0.26 | 0.97 | 1 |
| 2 | rs2234677 | 1310 | IL1RN | A | G | 0.27 | 0.26 | 0.57 | 1.05 | 0.28 | 0.25 | 0.1 | 1.16 | 0.28 | 0.25 | 0.08 | 1.17 | 0.27 | 0.26 | 0.84 | 1.02 | 0.27 | 0.26 | 0.82 | 1.02 |
| 2 | rs6746030 | 1439 | SCN9A | A | G | 0.11 | 0.11 | 0.68 | 1.05 | 0.11 | 0.11 | 0.85 | 1.02 | 0.11 | 0.11 | 0.62 | 1.06 | 0.11 | 0.11 | 0.58 | 1.07 | 0.11 | 0.11 | 0.61 | 1.07 |
| 3 | rs6795970 | 1439 | SCN10A | A | G | 0.36 | 0.39 | 0.19 | 0.9 | 0.38 | 0.38 | 0.81 | 0.98 | 0.37 | 0.39 | 0.22 | 0.91 | 0.36 | 0.39 | 0.07 | 0.86 | 0.37 | 0.39 | 0.46 | 0.94 |
| 5 | rs2053044 | 1310 | ADRB2 | A | G | 0.41 | 0.43 | 0.27 | 0.91 | 0.43 | 0.43 | 0.73 | 0.97 | 0.4 | 0.44 | 0.02 | 0.83 | 0.41 | 0.44 | 0.23 | 0.9 | 0.43 | 0.43 | 0.88 | 1.01 |
| 6 | rs2234693 | 1310 | ESR1 | C | T | 0.48 | 0.45 | 0.19 | 1.12 | 0.46 | 0.46 | 0.75 | 1.03 | 0.48 | 0.45 | 0.17 | 1.12 | 0.48 | 0.45 | 0.15 | 1.13 | 0.48 | 0.44 | 0.06 | 1.16 |
| 6 | rs9340799 | 1310 | ESR1 | G | A | 0.35 | 0.31 | 0.08 | 1.17 | 0.32 | 0.33 | 0.86 | 0.99 | 0.34 | 0.32 | 0.22 | 1.11 | 0.35 | 0.31 | 0.1 | 1.15 | 0.35 | 0.31 | 0.02 | 1.22 |
| 6 | rs1799971 | 1310 | OPRM1 | G | A | 0.09 | 0.1 | 0.52 | 0.91 | 0.1 | 0.1 | 0.88 | 1.02 | 0.1 | 0.1 | 0.69 | 1.06 | 0.09 | 0.1 | 0.26 | 0.85 | 0.09 | 0.1 | 0.4 | 0.89 |
| 8 | rs7814941 | 1439 | GSDMC/CCDC26 | G | A | 0.2 | 0.19 | 0.93 | 1.01 | 0.18 | 0.21 | 0.12 | 0.86 | 0.19 | 0.2 | 0.5 | 0.94 | 0.2 | 0.19 | 0.85 | 1.02 | 0.19 | 0.2 | 0.76 | 0.97 |
| 10 | rs3180 | 1435 | SPOCK2/CHST3 | A | G | 0.42 | 0.43 | 0.57 | 0.95 | 0.42 | 0.44 | 0.23 | 0.91 | 0.42 | 0.43 | 0.72 | 0.97 | 0.41 | 0.43 | 0.34 | 0.92 | 0.42 | 0.43 | 0.63 | 0.96 |
| 11 | rs6265 | 1310 | BDNF | T | C | 0.18 | 0.18 | 0.72 | 0.96 | 0.18 | 0.18 | 0.8 | 1.03 | 0.18 | 0.18 | 0.7 | 0.96 | 0.19 | 0.18 | 0.7 | 1.04 | 0.18 | 0.18 | 0.73 | 1.04 |
| 11 | rs1799750 | 1310 | MMP1 | TC | T | 0.51 | 0.49 | 0.31 | 1.09 | 0.49 | 0.49 | 0.91 | 0.99 | 0.5 | 0.49 | 0.44 | 1.06 | 0.5 | 0.49 | 0.68 | 1.04 | 0.49 | 0.49 | 0.99 | 1 |
| 12 | rs731236 | 1310 | VDR | G | A | 0.41 | 0.39 | 0.25 | 1.1 | 0.41 | 0.38 | 0.16 | 1.12 | 0.41 | 0.39 | 0.26 | 1.1 | 0.41 | 0.39 | 0.49 | 1.06 | 0.41 | 0.38 | 0.11 | 1.14 |
| 12 | rs2228570 | 1310 | VDR | A | G | 0.39 | 0.4 | 0.68 | 0.97 | 0.41 | 0.38 | 0.14 | 1.12 | 0.39 | 0.4 | 0.56 | 0.95 | 0.4 | 0.39 | 0.52 | 1.06 | 0.37 | 0.41 | 0.07 | 0.86 |
| 14 | rs998259 | 1310 | GCH1 | T | C | 0.25 | 0.25 | 0.94 | 0.99 | 0.24 | 0.25 | 0.36 | 0.92 | 0.24 | 0.25 | 0.36 | 0.92 | 0.24 | 0.25 | 0.86 | 0.98 | 0.24 | 0.25 | 0.47 | 0.94 |
| 20 | rs143383 | 1310 | GDF5 | G | A | 0.36 | 0.36 | 0.96 | 1 | 0.36 | 0.36 | 0.86 | 1.01 | 0.37 | 0.35 | 0.53 | 1.05 | 0.35 | 0.36 | 0.8 | 0.98 | 0.36 | 0.35 | 0.64 | 1.04 |
| 20 | rs734784 | 1310 | KCNS1 | C | T | 0.45 | 0.44 | 0.41 | 1.07 | 0.45 | 0.44 | 0.68 | 1.03 | 0.45 | 0.44 | 0.73 | 1.03 | 0.46 | 0.44 | 0.28 | 1.1 | 0.45 | 0.44 | 0.55 | 1.05 |
| 20 | rs13043825 | 1310 | KCNS1 | T | C | 0.28 | 0.28 | 0.7 | 1.04 | 0.29 | 0.27 | 0.35 | 1.08 | 0.28 | 0.28 | 0.93 | 0.99 | 0.28 | 0.28 | 0.66 | 1.04 | 0.27 | 0.28 | 0.62 | 0.96 |
| 20 | rs17576 | 1310 | MMP9 | G | A | 0.34 | 0.36 | 0.15 | 0.88 | 0.35 | 0.36 | 0.63 | 0.96 | 0.35 | 0.36 | 0.61 | 0.96 | 0.33 | 0.37 | 0.04 | 0.83 | 0.34 | 0.36 | 0.36 | 0.93 |
| 21 | rs9981629 | 1310 | KCNJ6 | C | G | 0.48 | 0.46 | 0.24 | 1.1 | 0.47 | 0.46 | 0.45 | 1.06 | 0.49 | 0.45 | 0.07 | 1.16 | 0.49 | 0.45 | 0.14 | 1.13 | 0.47 | 0.46 | 0.38 | 1.07 |
| 21 | rs928723 | 1310 | KCNJ6 | A | C | 0.47 | 0.47 | 0.99 | 1 | 0.46 | 0.48 | 0.55 | 0.95 | 0.46 | 0.47 | 0.74 | 0.97 | 0.46 | 0.47 | 0.78 | 0.98 | 0.48 | 0.46 | 0.38 | 1.07 |
| 21 | rs1543754 | 1310 | KCNJ6 | C | G | 0.48 | 0.5 | 0.38 | 0.93 | 0.48 | 0.51 | 0.16 | 0.9 | 0.49 | 0.5 | 0.63 | 0.96 | 0.49 | 0.49 | 0.83 | 0.98 | 0.5 | 0.49 | 0.47 | 1.06 |
| 21 | rs858035 | 1310 | KCNJ6 | G | A | 0.33 | 0.33 | 0.79 | 0.98 | 0.34 | 0.32 | 0.26 | 1.1 | 0.32 | 0.33 | 0.42 | 0.93 | 0.31 | 0.34 | 0.19 | 0.89 | 0.32 | 0.33 | 0.44 | 0.94 |
| 21 | rs2835925 | 1310 | KCNJ6 | G | A | 0.22 | 0.2 | 0.42 | 1.09 | 0.21 | 0.2 | 0.27 | 1.11 | 0.2 | 0.21 | 0.55 | 0.94 | 0.2 | 0.21 | 0.77 | 0.97 | 0.21 | 0.21 | 0.99 | 1 |
| 21 | rs2835930 | 1310 | KCNJ6 | A | C | 0.23 | 0.24 | 0.54 | 0.94 | 0.23 | 0.25 | 0.23 | 0.9 | 0.23 | 0.25 | 0.47 | 0.94 | 0.24 | 0.24 | 0.81 | 1.02 | 0.23 | 0.25 | 0.28 | 0.9 |
| 21 | rs1787337 | 1310 | KCNJ6 | A | G | 0.43 | 0.42 | 0.42 | 1.07 | 0.44 | 0.4 | 0.08 | 1.15 | 0.42 | 0.42 | 0.87 | 1.01 | 0.42 | 0.42 | 0.82 | 0.98 | 0.43 | 0.42 | 0.47 | 1.06 |
| 21 | rs2211843 | 1310 | KCNJ6 | T | G | 0.26 | 0.26 | 0.91 | 0.99 | 0.26 | 0.26 | 0.76 | 1.03 | 0.27 | 0.25 | 0.25 | 1.11 | 0.27 | 0.26 | 0.57 | 1.06 | 0.26 | 0.26 | 0.71 | 0.97 |
| 22 | rs2075507 | 1310 | COMT | G | A | 0.51 | 0.49 | 0.31 | 1.09 | 0.5 | 0.5 | 0.95 | 0.99 | 0.51 | 0.49 | 0.51 | 1.05 | 0.51 | 0.5 | 0.54 | 1.05 | 0.5 | 0.5 | 0.69 | 1.03 |
| 22 | rs6269 | 1310 | COMT | G | A | 0.37 | 0.38 | 0.5 | 0.94 | 0.37 | 0.38 | 0.53 | 0.95 | 0.36 | 0.39 | 0.25 | 0.91 | 0.36 | 0.38 | 0.27 | 0.91 | 0.36 | 0.38 | 0.31 | 0.92 |
| 22 | rs4633 | 1310 | COMT | C | T | 0.43 | 0.44 | 0.54 | 0.95 | 0.44 | 0.44 | 0.73 | 0.97 | 0.43 | 0.45 | 0.37 | 0.93 | 0.43 | 0.44 | 0.43 | 0.94 | 0.42 | 0.45 | 0.17 | 0.9 |
| 22 | rs4818 | 1310 | COMT | G | C | 0.36 | 0.38 | 0.45 | 0.94 | 0.37 | 0.38 | 0.45 | 0.94 | 0.36 | 0.38 | 0.24 | 0.91 | 0.36 | 0.38 | 0.23 | 0.9 | 0.36 | 0.38 | 0.34 | 0.93 |
| 22 | rs4680 | 1433 | COMT | G | A | 0.44 | 0.44 | 0.76 | 0.98 | 0.44 | 0.44 | 0.83 | 0.98 | 0.43 | 0.45 | 0.39 | 0.94 | 0.43 | 0.44 | 0.5 | 0.95 | 0.42 | 0.45 | 0.17 | 0.9 |
CHR, chromosome; MAF, minor allele frequency; SNV, single nucleotide variant.
3.1. Genetic associations
The association between back pain in the SRS-22r domain and pain in the EQ-5D-3L domain and the included 37 SNVs are presented in Table 3. The odds ratio for the associations between the selected genetic variants and back pain in the SRS-22r domain ranged between 0.88 and 1.17, with P-values ranging between 0.08 and 0.99. The odds ratio for the associations between the selected genetic variants and pain in the EQ-5D-3L domain ranged between 0.86 and 1.16, with P-values ranging between 0.08 and 0.95.
Box plot diagrams depicting the associations in the subanalyses of individual questions on back pain from the SRS-22r and pain from EQ-5D-3L are shown in supplementary Figure 1, http://links.lww.com/PR9/A274. In these analyses, the odds ratio between rs2053044 in ADRB2 and back pain in the last 6 months was 0.83; P = 0.02. The odds ratio between rs17576 in MMP9 and back pain during the last month was 0.83; P = 0.04. The odds ratio between rs9340799 in ESR1 and back pain at rest was 1.22; P = 0.02.
3.2. Nonresponder analysis
Baseline data were available for the 1750 participants who had accepted to participate in the study with available DNA samples. Nonresponder analysis was performed between the included 1442 participants and 308 participants excluded due to incomplete HRQoL questionnaire.
The excluded group were marginally older, had slightly smaller Cobb angle, and more male participants. Only 3 of 145 participants included from Denmark failed to complete the questionnaire. There were no participants with double major curvature in that group. The proportion of participants who were surgically treated were slightly higher in the included group. No statistically significant differences were found for height, weight, type of scoliosis, convexity, and Scandinavian ancestry (Supplementary Table 1, http://links.lww.com/PR9/A274).
4. Discussion
Our study found no significant associations between the 37 selected SNVs and the SRS-22r back pain domain score or EQ-5D-3L pain/discomfort domain. Weak associations were found between some of the back pain questions in SRS-22r and variants in ADRB2, MMP9, and ESR1.
While association has been found between several SNVs and back pain, the mechanism behind the contribution of these variants to back pain is poorly understood. The COMT gene is one of the few well-studied and replicated genes in the modulation of pain. This gene encodes catechol-O-methyltransferase (COMT) that regulates the levels of catecholamines through β2 and β3 adrenergic receptors.36 A reduced enzyme activity results in elevated catecholamine levels and is associated with back pain, degenerative disk disease, lumbar disk herniation, sciatica, and chronic back pain.5,11,24,27,38,39,45,55 Both back pain and lumbar degeneration are common findings in adults with idiopathic scoliosis,12 but our study did not support idiopathic scoliosis individuals with back pain having a higher frequency of the COMT Val158Met genotype or any of the other 4 selected SNVs.39
Two other genes potentially involved in pain perception are the SCN9A and SCN10A genes, which encode for 2 different voltage-gated sodium ion channels, Nav1.7 and Nav1.8, respectively.59 We genotyped a common SNV within the SCN9A gene, rs6746030 (G > A; R1150W), previously found to be associated with levels of pain nociception in amputees and individuals with a variety of medical conditions such as osteoarthritis, sciatica, phantom pain, and higher presurgical pain scores in individuals with lumbar intervertebral disk herniation.19,28,43 For the SCN10A gene, we included a common SNV, rs6795970 (G>A; A1073V) which has been demonstrated to be associated with reduced mechanical pain sensitivity.17 None of the SCN9A and SCN10A variants were associated with back pain or pain/discomfort in our scoliosis cohort.
Two SNVs included in this study, rs781491 and rs3180, have previously been associated with the risk of chronic back pain in a large genome-wide association study (GWAS) for back pain.20 Our study failed to identify association of these variants with back pain in idiopathic scoliosis individuals.
Several of the other genotyped back pain SNVs have been previously associated with discogenic or degenerative back pain in the literature.3,6,18,25,26,35,37,44,50 Bjorland et al.3 reported that there were significant associations for rs17576 in MMP9 and rs1799971 in OPRM1 and pain recovery. A weak association between rs17576 in MMP9 and back pain during the last month was found in our cohort. Roh et al.44 showed that ESR1 polymorphisms were associated with pain intensity in patients with degenerative lumbar spondylolisthesis. In our cohort, a weak association was found between rs9340799 in ESR1 and back pain at rest.
The remaining selected SNVs concerned other pain phenotypes such as pain intensity or pain medication usage. Several studies have reported association between variants in interleukin genes IL1A, IL1RN and pain.34,47,49 Bruehl et al.4 used a tag SNV approach and reported 8 SNVs in the KCNJ6 gene to be significantly associated with the use of opioid analgesic medication in chronic LBP. KCNS1 is one of 3 potassium channel modulatory Kv9 subunits. Costigan et al.10 presented association between the minor allele for rs734784 and rs13043825 and greater pain. Vossen et al.55 reported that the SNV rs6265 (Val66Met) was associated with chronic back pain, similar to previous results on COMT rs4680 (Val158Met). Ramesh et al.42 examined 3 FAAH SNVs and reported significant association with increased pain scores in LBP patients. Skouen et al. reported a significant association between rs2053044 in ADRB2 and chronic disabling comorbid neck and low back pain. We found a weak association between rs2053044 in ADRB2 and back pain in the last 6 months in our cohort.
The risk of developing back pain and the differences in pain perception are likely a complex trait, with several SNVs identified as associated with back pain phenotypes,15 but their role has not been studied previously in the context of back pain in idiopathic scoliosis. The variants in this study were mainly selected due to their association with the risk of back pain in degenerative conditions in the spine. Despite idiopathic scoliosis being reported as associated with an increased risk of degenerative lumbar changes, other factors may be involved in the development of back pain.12,13 Pain in idiopathic scoliosis may be considered more “mechanical”; other studies have suggested deformity severity including curve type, decreased kyphosis, and pelvic asymmetry to be associated with back pain.51,53 Matamalas et al.33 showed that older age and larger Cobb angle associate with back pain, which concurs with the data from the present cohort.
One strength of our study is the inclusion of a large and relatively homogeneous population consisting of individuals with predominantly Scandinavian ancestry and with the non-Scandinavian individuals demonstrating similar pain status. However, our study is not without limitations, and one concerns the questionnaires and the dichotomization of the pain scores. The SRS-22r is a scoliosis-specific questionnaire, and the pain domain questions are not specific to scoliosis-related back pain. This is however the best available and most used and validated tool to measure quality of life including back pain in scoliosis patients. The EQ-5D-3L is a generic quality of life questionnaire that is not focused on back pain but rather pain or discomfort in general. Including patients based on their EQ-5D-3L scores might misinterpret their level of pain, since feeling out of balance or stiff after spinal fusion surgery, might lead to poor scores in EQ-5D-3L questionnaire. Second, to determine the pain status of the patient's scoliosis, we divided the groups as in Djurasovic et al.16 and Teles et al.51 In the “no back pain” group, some individuals are not completely pain-free. However, we believe that occasional mild back pain may be considered normal and categorizing them into the “back pain” group would overestimate the prevalence. Third, as with the design of cross-sectional studies, individuals answered about their pain status on only one occasion, making it difficult to assess chronic pain and pain progression. Individuals were also required to recall their pain status up to 6 months with risk of recall bias. Moreover, our study lacked a numerical scale for quantifying pain, which would have provided additional insight into the pain levels experienced by the individuals. Finally, we only tested a selection of known variants associated with the phenotype pain. As with other candidate variant approaches, other variants in the region associated with certain genes were not analysed. In conclusion, our study suggests that known selected pain variants are not associated with back pain in individuals with idiopathic scoliosis.
Disclosures
The authors have no conflict of interest to declare. All authors have completed the ICMJE disclosure form.
Appendix A. Supplemental digital content
Supplemental digital content associated with this article can be found online at http://links.lww.com/PR9/A274.
Supplementary Material
Acknowledgments
This study was supported by grants from the Swedish Research Council (Dnr 2012-02275 and 2017-0139), the Stockholm County Council, and The Swedish Brain Foundation. The authors would gratefully acknowledge the support of Diana Ekman and Marcin Kierczak at the National Bioinformatics Infrastructure Sweden (NBIS) at Science for Life Laboratory (SciLifeLab) in the statistical analysis. Genotyping was performed by the SNP&SEQ Technology Platform in Uppsala. The facility is part of the National Genomics Infrastructure (NGI) Sweden and SciLifeLab. The SNP&SEQ Technology Platform is also supported by the Swedish Research Council and the Knut and Alice Wallenberg Foundation.
Data availability statement: The authors will make the program codes used for analysis available on request. Anonymized data may be shared on reasonable request. The corresponding authors are to be contacted to obtain the data. In case of data sharing, a data transfer agreement will be needed.
Footnotes
Sponsorships or competing interests that may be relevant to content are disclosed at the end of this article.
Supplemental digital content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal's Web site (www.painrpts.com).
Contributor Information
Elias Diarbakerli, Email: elias.diarbakerli@ki.se.
Ane Simony, Email: ane.simony1@rsyd.dk.
Mikkel Østerheden Andersen, Email: mikkel.andersen2@rsyd.dk.
Aina Danielsson, Email: aina.danielsson@gu.se.
Juha Kere, Email: juha.kere@ki.se.
Elisabet Einarsdottir, Email: elisabet.einarsdottir@scilifelab.se.
Paul Gerdhem, Email: paul.gerdhem@uu.se.
References
- [1].Asher M, Min Lai S, Burton D, Manna B. The reliability and concurrent validity of the scoliosis research society-22 patient questionnaire for idiopathic scoliosis. Spine (Phila Pa 1976) 2003;28:63–9. [DOI] [PubMed] [Google Scholar]
- [2].Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, Marchini JL, McCarthy S, McVean GA, Abecasis GR. A global reference for human genetic variation. Nature 2015;526:68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Bjorland S, Røe C, Moen A, Schistad E, Mahmood A, Gjerstad J. Genetic predictors of recovery in low back and lumbar radicular pain. PAIN 2017;158:1456–60. [DOI] [PubMed] [Google Scholar]
- [4].Bruehl S, Denton JS, Lonergan D, Koran ME, Chont M, Sobey C, Fernando S, Bush WS, Mishra P, Thornton-Wells TA. Associations between KCNJ6 (GIRK2) gene polymorphisms and pain-related phenotypes. PAIN 2013;154:2853–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Cargnin S, Magnani F, Viana M, Tassorelli C, Mittino D, Cantello R, Sances G, Nappi G, Canonico PL, Genazzani AA, Raffaeli W, Terrazzino S. An opposite-direction modulation of the COMT Val158Met polymorphism on the clinical response to intrathecal morphine and triptans. J Pain 2013;14:1097–106. [DOI] [PubMed] [Google Scholar]
- [6].Cauci S, Migliozzi F, Trombetta CS, Venuto I, Saccheri P, Travan L, Chiriacò G. Low back pain and FokI (rs2228570) polymorphism of vitamin D receptor in athletes. BMC Sports Sci Med Rehabil 2017;9:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Cheng JC, Castelein RM, Chu WC, Danielsson AJ, Dobbs MB, Grivas TB, Gurnett CA, Luk KD, Moreau A, Newton PO, Stokes IA, Weinstein SL, Burwell RG. Adolescent idiopathic scoliosis. Nat Rev Dis Primers 2015;1:15030. [DOI] [PubMed] [Google Scholar]
- [8].Clarke GM, Anderson CA, Pettersson FH, Cardon LR, Morris AP, Zondervan KT. Basic statistical analysis in genetic case-control studies. Nat Protoc 2011;6:121–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Cobb J. Outline for the study of scoliosis. Instructional Course Lecture 1948;5:261–75. [Google Scholar]
- [10].Costigan M, Belfer I, Griffin RS, Dai F, Barrett LB, Coppola G, Wu T, Kiselycznyk C, Poddar M, Lu Y, Diatchenko L, Smith S, Cobos EJ, Zaykin D, Allchorne A, Gershon E, Livneh J, Shen PH, Nikolajsen L, Karppinen J, Männikkö M, Kelempisioti A, Goldman D, Maixner W, Geschwind DH, Max MB, Seltzer Z, Woolf CJ. Multiple chronic pain states are associated with a common amino acid-changing allele in KCNS1. Brain 2010;133:2519–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Dai F, Belfer I, Schwartz CE, Banco R, Martha JF, Tighioughart H, Tromanhauser SG, Jenis LG, Kim DH. Association of catechol-O-methyltransferase genetic variants with outcome in patients undergoing surgical treatment for lumbar degenerative disc disease. Spine J 2010;10:949–57. [DOI] [PubMed] [Google Scholar]
- [12].Danielsson AJ, Cederlund CG, Ekholm S, Nachemson AL. The prevalence of disc aging and back pain after fusion extending into the lower lumbar spine. A matched MR study twenty-five years after surgery for adolescent idiopathic scoliosis. Acta Radiol 2001;42:187–97. [DOI] [PubMed] [Google Scholar]
- [13].Danielsson AJ, Nachemson AL. Back pain and function 22 years after brace treatment for adolescent idiopathic scoliosis: a case-control study-part I. Spine (Phila Pa 1976) 2003;28:2078–86; discussion 2086. [DOI] [PubMed] [Google Scholar]
- [14].Danielsson AJ, Romberg K. Reliability and validity of the Swedish version of the Scoliosis Research Society-22 (SRS-22r) patient questionnaire for idiopathic scoliosis. Spine (Phila Pa 1976) 2013;38:1875–84. [DOI] [PubMed] [Google Scholar]
- [15].Diatchenko L, Nackley AG, Tchivileva IE, Shabalina SA, Maixner W. Genetic architecture of human pain perception. Trends Genet 2007;23:605–13. [DOI] [PubMed] [Google Scholar]
- [16].Djurasovic M, Glassman SD, Sucato DJ, Lenke LG, Crawford CH, III, Carreon LY. Improvement in scoliosis research society-22r pain scores after surgery for adolescent idiopathic scoliosis. Spine (Phila Pa 1976) 2018;43:127–32. [DOI] [PubMed] [Google Scholar]
- [17].Duan G, Han C, Wang Q, Guo S, Zhang Y, Ying Y, Huang P, Zhang L, Macala L, Shah P, Zhang M, Li N, Dib-Hajj SD, Waxman SG, Zhang X. A SCN10A SNP biases human pain sensitivity. Mol Pain 2016;12:1744806916666083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Eskola PJ, Lemmelä S, Kjaer P, Solovieva S, Männikkö M, Tommerup N, Lind-Thomsen A, Husgafvel-Pursiainen K, Cheung KM, Chan D, Samartzis D, Karppinen J. Genetic association studies in lumbar disc degeneration: a systematic review. PLoS One 2012;7:e49995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Estacion M, Harty TP, Choi JS, Tyrrell L, Dib-Hajj SD, Waxman SG. A sodium channel gene SCN9A polymorphism that increases nociceptor excitability. Ann Neurol 2009;66:862–6. [DOI] [PubMed] [Google Scholar]
- [20].Freidin MB, Tsepilov YA, Palmer M, Karssen LC, Suri P, Aulchenko YS, Williams FMK, CHARGE Musculoskeletal Working Group. Insight into the genetic architecture of back pain and its risk factors from a study of 509,000 individuals. PAIN 2019;160:1361–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Grauers A, Danielsson A, Karlsson M, Ohlin A, Gerdhem P. Family history and its association to curve size and treatment in 1,463 patients with idiopathic scoliosis. Eur Spine J 2013;22:2421–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Grauers A, Topalis C, Möller H, Normelli H, Karlsson MK, Danielsson A, Gerdhem P. Prevalence of back problems in 1069 adults with idiopathic scoliosis and 158 adults without scoliosis. Spine (Phila Pa 1976) 2014;39:886–92. [DOI] [PubMed] [Google Scholar]
- [23].Grauers A, Wang J, Einarsdottir E, Simony A, Danielsson A, Åkesson K, Ohlin A, Halldin K, Grabowski P, Tenne M, Laivuori H, Dahlman I, Andersen M, Christensen SB, Karlsson MK, Jiao H, Kere J, Gerdhem P. Candidate gene analysis and exome sequencing confirm LBX1 as a susceptibility gene for idiopathic scoliosis. Spine J 2015;15:2239–46. [DOI] [PubMed] [Google Scholar]
- [24].Gruber HE, Sha W, Brouwer CR, Steuerwald N, Hoelscher GL, Hanley EN, Jr. A novel catechol-O-methyltransferase variant associated with human disc degeneration. Int J Med Sci 2014;11:748–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Guo TM, Liu M, Zhang YG, Guo WT, Wu SX. Association between Caspase-9 promoter region polymorphisms and discogenic low back pain. Connect Tissue Res 2011;52:133–8. [DOI] [PubMed] [Google Scholar]
- [26].Jacobsen LM, Schistad EI, Storesund A, Pedersen LM, Espeland A, Rygh LJ, Røe C, Gjerstad J. The MMP1 rs1799750 2G allele is associated with increased low back pain, sciatica, and disability after lumbar disk herniation. Clin J Pain 2013;29:967–71. [DOI] [PubMed] [Google Scholar]
- [27].Jacobsen LM, Schistad EI, Storesund A, Pedersen LM, Rygh LJ, Røe C, Gjerstad J. The COMT rs4680 Met allele contributes to long-lasting low back pain, sciatica and disability after lumbar disc herniation. Eur J Pain 2012;16:1064–9. [DOI] [PubMed] [Google Scholar]
- [28].Kurzawski M, Rut M, Dziedziejko V, Safranow K, Machoy-Mokrzynska A, Drozdzik M, Bialecka M. Common missense variant of SCN9A gene is associated with pain intensity in patients with chronic pain from disc herniation. Pain Med 2018;19:1010–4. [DOI] [PubMed] [Google Scholar]
- [29].Lai SM, Burton DC, Asher MA, Carlson BB. Converting SRS-24, SRS-23, and SRS-22 to SRS-22r: establishing conversion equations using regression modeling. Spine (Phila Pa 1976) 2011;36:E1525–33. [DOI] [PubMed] [Google Scholar]
- [30].Luk KD, Lee CF, Cheung KM, Cheng JC, Ng BK, Lam TP, Mak KH, Yip PS, Fong DY. Clinical effectiveness of school screening for adolescent idiopathic scoliosis: a large population-based retrospective cohort study. Spine (Phila Pa 1976) 2010;35:1607–14. [DOI] [PubMed] [Google Scholar]
- [31].Manichaikul A, Mychaleckyj JC, Rich SS, Daly K, Sale M, Chen WM. Robust relationship inference in genome-wide association studies. Bioinformatics 2010;26:2867–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Marchini J, Howie B. Genotype imputation for genome-wide association studies. Nat Rev Genet 2010;11:499–511. [DOI] [PubMed] [Google Scholar]
- [33].Matamalas A, Figueras C, Pizones J, Moreno-Manzanaro L, Betegón J, Esteban M, Pellisé F, Sanchez-Raya J, Sanchez-Marquez JM, Bagó J. How back pain intensity relates to clinical and psychosocial factors in patients with idiopathic scoliosis. Eur Spine J 2022;31:1006–12. [DOI] [PubMed] [Google Scholar]
- [34].Moen A, Schistad EI, Rygh LJ, Røe C, Gjerstad J. Role of IL1A rs1800587, IL1B rs1143627 and IL1RN rs2234677 genotype regarding development of chronic lumbar radicular pain; a prospective one-year study. PLoS One 2014;9:e107301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Mu J, Ge W, Zuo X, Chen Y, Huang C. Analysis of association between IL-1β, CASP-9, and GDF5 variants and low-back pain in Chinese male soldier: clinical article. J Neurosurg Spine 2013;19:243–7. [DOI] [PubMed] [Google Scholar]
- [36].Nackley AG, Tan KS, Fecho K, Flood P, Diatchenko L, Maixner W. Catechol-O-methyltransferase inhibition increases pain sensitivity through activation of both beta2- and beta3-adrenergic receptors. PAIN 2007;128:199–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Olsen MB, Jacobsen LM, Schistad EI, Pedersen LM, Rygh LJ, Røe C, Gjerstad J. Pain intensity the first year after lumbar disc herniation is associated with the A118G polymorphism in the opioid receptor mu 1 gene: evidence of a sex and genotype interaction. J Neurosci 2012;32:9831–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Omair A, Lie BA, Reikeras O, Holden M, Brox JI. Genetic contribution of catechol-O-methyltransferase variants in treatment outcome of low back pain: a prospective genetic association study. BMC Musculoskelet Disord 2012;13:76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Omair A, Mannion AF, Holden M, Fairbank J, Lie BA, Hägg O, Fritzell P, Brox JI. Catechol-O-methyltransferase (COMT) gene polymorphisms are associated with baseline disability but not long-term treatment outcome in patients with chronic low back pain. Eur Spine J 2015;24:2425–31. [DOI] [PubMed] [Google Scholar]
- [40].Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007;81:559–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Rabin R, de Charro F. EQ-5D: a measure of health status from the EuroQol Group. Ann Med 2001;33:337–43. [DOI] [PubMed] [Google Scholar]
- [42].Ramesh D, D'Agata A, Starkweather AR, Young EE. Contribution of endocannabinoid gene expression and genotype on low back pain susceptibility and chronicity. Clin J Pain 2018;34:8–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Reimann F, Cox JJ, Belfer I, Diatchenko L, Zaykin DV, McHale DP, Drenth JP, Dai F, Wheeler J, Sanders F, Wood L, Wu TX, Karppinen J, Nikolajsen L, Männikkö M, Max MB, Kiselycznyk C, Poddar M, Te Morsche RH, Smith S, Gibson D, Kelempisioti A, Maixner W, Gribble FM, Woods CG. Pain perception is altered by a nucleotide polymorphism in SCN9A. Proc Natl Acad Sci U S A 2010;107:5148–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Roh HL, Lee JS, Suh KT, Kim JI, Lee HS, Goh TS, Park SH. Association between estrogen receptor gene polymorphism and back pain intensity in female patients with degenerative lumbar spondylolisthesis. J Spinal Disord Tech 2013;26:E53–7. [DOI] [PubMed] [Google Scholar]
- [45].Rut M, Machoy-Mokrzyńska A, Ręcławowicz D, Słoniewski P, Kurzawski M, Droździk M, Safranow K, Morawska M, Białecka M. Influence of variation in the catechol-O-methyltransferase gene on the clinical outcome after lumbar spine surgery for one-level symptomatic disc disease: a report on 176 cases. Acta Neurochir (Wien) 2014;156:245–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Sato T, Hirano T, Ito T, Morita O, Kikuchi R, Endo N, Tanabe N. Back pain in adolescents with idiopathic scoliosis: epidemiological study for 43,630 pupils in Niigata City, Japan. Eur Spine J 2011;20:274–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Schistad EI, Jacobsen LM, Røe C, Gjerstad J. The interleukin-1α gene C>T polymorphism rs1800587 is associated with increased pain intensity and decreased pressure pain thresholds in patients with lumbar radicular pain. Clin J Pain 2014;30:869–74. [DOI] [PubMed] [Google Scholar]
- [48].Simony A, Carreon LY, Andersen MO. Reliability and validity testing of a Danish translated version of the scoliosis research society instrument-22 revised (SRS-22R). Spine Deform 2016;4:16–21. [DOI] [PubMed] [Google Scholar]
- [49].Solovieva S, Leino-Arjas P, Saarela J, Luoma K, Raininko R, Riihimäki H. Possible association of interleukin 1 gene locus polymorphisms with low back pain. PAIN 2004;109:8–19. [DOI] [PubMed] [Google Scholar]
- [50].Tegeder I, Costigan M, Griffin RS, Abele A, Belfer I, Schmidt H, Ehnert C, Nejim J, Marian C, Scholz J, Wu T, Allchorne A, Diatchenko L, Binshtok AM, Goldman D, Adolph J, Sama S, Atlas SJ, Carlezon WA, Parsegian A, Lötsch J, Fillingim RB, Maixner W, Geisslinger G, Max MB, Woolf CJ. GTP cyclohydrolase and tetrahydrobiopterin regulate pain sensitivity and persistence. Nat Med 2006;12:1269–77. [DOI] [PubMed] [Google Scholar]
- [51].Teles AR, St-Georges M, Abduljabbar F, Simões L, Jiang F, Saran N, Ouellet JA, Ferland CE. Back pain in adolescents with idiopathic scoliosis: the contribution of morphological and psychological factors. Eur Spine J 2020;29:1959–71. [DOI] [PubMed] [Google Scholar]
- [52].Théroux J, Le May S, Fortin C, Labelle H. Prevalence and management of back pain in adolescent idiopathic scoliosis patients: a retrospective study. Pain Res Manag 2015;20:153–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Théroux J, Le May S, Hebert JJ, Labelle H. Back pain prevalence is associated with curve-type and severity in adolescents with idiopathic scoliosis: a cross-sectional study. Spine (Phila Pa 1976) 2017;42:E914–9. [DOI] [PubMed] [Google Scholar]
- [54].Vlaeyen JWS, Maher CG, Wiech K, Van Zundert J, Meloto CB, Diatchenko L, Battié MC, Goossens M, Koes B, Linton SJ. Low back pain. Nat Rev Dis Primers 2018;4:52. [DOI] [PubMed] [Google Scholar]
- [55].Vossen H, Kenis G, Rutten B, van Os J, Hermens H, Lousberg R. The genetic influence on the cortical processing of experimental pain and the moderating effect of pain status. PLoS One 2010;5:e13641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Weinstein SL, Dolan LA, Spratt KF, Peterson KK, Spoonamore MJ, Ponseti IV. Health and function of patients with untreated idiopathic scoliosis: a 50-year natural history study. JAMA 2003;289:559–67. [DOI] [PubMed] [Google Scholar]
- [57].Willner S, Udén A. A prospective prevalence study of scoliosis in Southern Sweden. Acta Orthop Scand 1982;53:233–7. [DOI] [PubMed] [Google Scholar]
- [58].Wong AYL, Samartzis D, Cheung PWH, Cheung JPY. How common is back pain and what biopsychosocial factors are associated with back pain in patients with adolescent idiopathic scoliosis? Clin Orthop Relat Res 2019;477:676–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Xue Y, Chidiac C, Herault Y, Gaveriaux-Ruff C. Pain behavior in SCN9A (Nav1.7) and SCN10A (Nav1.8) mutant rodent models. Neurosci Lett 2021;753:135844. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental digital content associated with this article can be found online at http://links.lww.com/PR9/A274.


