Abstract
Small cell neuroendocrine cervical carcinoma is a highly aggressive tumor characterized by early metastasis, a high recurrence rate, and poor prognosis. This study represents the first instance of single-cell sequencing conducted on small cell neuroendocrine carcinoma of the cervix worldwide. Analysis of gene expression regulatory networks revealed that the transcription factor TFF3 drived up-regulation of ELF3. Furthermore, our findings indicated that the neuroendocrine marker genes and gene regulatory networks associated with small cell neuroendocrine cervical carcinoma differed from those observed in lung, small intestine, and liver neuroendocrine carcinoma within the GEO database, suggesting tissue-specific origins for these malignancies. Overall, this study addresses a significant research in understanding small cell neuroendocrine cervical carcinoma in vivo and provides valuable insights for guiding radiotherapy, chemotherapy, and targeted therapy.
Graphical abstract
Supplementary Information
The online version contains supplementary material available at 10.1186/s12967-024-05977-z.
Keywords: Neuroendocrine, Cervical cancer, Single-nucleus RNA sequencing, Tumor microenvironment
Introduction
Neuroendocrine carcinoma (NECs) [1] are a rare type of tumor originating from mutated peptidergic neurons and neuroendocrine cells. They exhibit neuroendocrine differentiation and express neuroendocrine markers, most commonly found in the lungs, gastrointestinal tract, and pancreas [2]. The incidence of cervical neuroendocrine carcinoma (NECC) is even lower, accounting for about 1–1.5% of cervical cancers [3, 4]. Vascular invasion, lymph node or visceral metastasis are found at an early stage in NECC. Therefore, NECC shows extremely high invasiveness and poor prognosis. The 5-year survival rate for early stage is about 36%, Only 4% of stage III SCNECC patients survive beyond five years [4], and the recurrence rate exceeds 90% in NECC [3]. Therefore, a deep understanding of the biological behavior of this malignant tumor is crucial for clinical treatment.
A study [5] evaluated the clinicopathological characteristics and prognostic factors of patients with high-grade neuroendocrine carcinoma of the cervix (NECC) treated surgically. It was found that patients with mixed histological types of high-grade NECC had better prognoses. Ovarian preservation, age > 45 years, tumor size > 4 cm, parity > 3, and perineural invasion were identified as predictors of poor prognosis [5]. A study employing multi-omics sequencing and analysis of the tumor immune microenvironment identified three subtypes of small cell neuroendocrine carcinoma of the cervix. The SCCC-I subtype, characterized by an inflammatory phenotype and high expression of MHC-II and IFN-α/β-related genes, was associated with better prognosis and potential responsiveness to immune checkpoint inhibitor therapy, suggesting that patients with this subtype may benefit from immunotherapy [6]. Another study [7] analyzed somatic copy number variations (CNVs) in SCNECC using whole-exome sequencing (WES), identifying two copy number gains (3q27.1 and 19q13.12) and five copy number losses (1p36.21, 5q31.3, 6p22.2, 9q21.11, and 11p15.5). However, the accuracy of WES in detecting CNVs may be inferior to that of whole-genome sequencing. To date, there was few publications about exploring the small cell cervical neuroendocrine carcinoma (SCNECC) at the single-cell level. Due to the scarcity of samples, most NEC-related research is based on virus-induced in vitro models, but there are huge differences between in vitro models and in vivo tumors because of tumor origin process, tumor microenvironment and so on. Almost neither single-cell RNA (scRNA) sequencing data nor bulk RNA sequencing data of SCNECC is available in the public database. In an effort to fill this gap, we utilized single-cell sequencing techniques to compare the tumor and adjacent tissue of SCNECC. By dissecting the tumor microenvironment, we identified genes abnormally expressed during the carcinogenic process and pinpointed the associated key transcription factors.
Our research addressed the gap in the understanding of the in vivo tumor microenvironment of SCNECC, and provided valuable insights for the discovery of new potential therapeutic targets for not only cervical carcinoma, but also other neuroendocrine carcinomas.
We hope that this study can pave new avenues for future research on small cell cervical neuroendocrine carcinoma and provide more possibilities for future clinical treatment.
Materials and methods
Patients and collection of samples
This study has been approved by the Ethics Committee of the Obstetrics and Gynecology Hospital affiliated with Fudan University. The patient provided written informed consent and was preoperatively diagnosed with SCNECC. During the surgery, tumor lesions and adjacent tissues with a diameter of 1 cm surrounding the cancer were collected for single-cell sequencing.
Sample processing and cell sorting
Fresh tissue samples were immediately placed in a sample preservation bag with an ice bag for thermal insulation and quickly sent to “Jizhiyixue” company for single-cell sequencing. Single-cell suspensions were prepared using enzymatic digestion, and single-cell sequencing was strictly performed according to the instructions of the 10 × Genomics platform.
Preprocessing of scRNA-seq data
Unless specifically mentioned, this article utilizes R 4.1.2 [8] for single-cell data processing, using the default parameters of the functions in the regular process. A two-tailed P value < 0.05 is considered statistically significant. Adobe Illustrator 2020 is used for the final layout and combination of pictures. Cytoscape (v3.10.1) is used to draw netplots [9].
Data quality control, normalization and standardization
This study uses the DoubletFinder (2.0.3) [10] R package to predict and filter double cells for each sample.
The distribution of unique molecular identifiers (UMIs), genes, and mitochondria for each project and each sample is statistically analyzed to calculate the specific filtering threshold for each sample. The number of cells before and after filtering for each sample can be found in (Table S1). The single-cell sequencing data downloaded from the GEO database is filtered based on the number of genes and the proportion of mitochondria, with the parameter settings: minGene = 500, maxGene = 4000, mitochondrial gene count pctMT = 10.
In order to centralize the dataset with a high degree of dispersion, data normalization is performed: the gene expression of each cell is divided by the total number of UMIs (Unique Molecular Identifiers) of the cell, then divided by the scaling factor 10,000, and finally log-transformed.
Considering the impact of different samples on expression, in order to eliminate the influence of the average level of expression and deviation, the single-cell data is standardized: the Z-score is calculated, and the data after normalization is converted to data with a mean of 0 and a standard deviation of 1.
Batch effect correction and clustering
Batch effect correction is performed using the Harmony method. The FindNeighbors function is set with dims = 1:20, pc.num = 1:20. Dimensionality reduction and clustering are performed using PCA and UMAP methods, respectively. Clustering is performed using the FindClusters function, with resolution = 0.1 to obtain suitable cell clusterings for subsequent cell type identification. The R dittoSeq [11] package is used to visualize the proportion of cell composition in each sample.
Marker gene identification
The Wilcoxon algorithm is used to analyze the marker genes of each subgroup, and the group one vs rest method is used to score the marker genes. The FindAllMarkers function is used to select genes that are expressed in at least 25% of the cells in each cell subgroup after clustering and have a logfc > 0.25 as the Marker genes for that subgroup.
Cell type identification
First, each cell group is pre-annotated according to the universal standard cell type marker (Table S2), and the results are grouped into AllTypes. Then, when the resolution in FindClusters is 0.1, SCNECC single-cell data is clustered into 11 major celltypes. SingleR (1.8.1) [12] is first used for preliminary cell type identification. Then, the EasyCellType [13] package is loaded (R 4.3.0), and three databases (cellmarker, panglao, clustermole) are used to further identify cell subtypes using two methods (GSEA, Fisher's exact test). Then, the marker genes of cell types in high-impact literature [3, 14–17] are verified. For undetermined cell types, The top10 Marker genes [18] of each population were searched in CellMarker 2.0 [19] database and consulted with human experts.
The final determination of cell types is made by integrating SingleR, EasyCellType (Figure S2B-D), marker genes in high-impact literature, CellMarker 2.0 database, and the opinions of human experts.
Differentiation of tumor cells in epithelial cells by infercnv
The epithelial cells in the paracancerous samples were set as the reference cell population, and infercnv (1.10.1) [20] was employed to analyze the gene copy number of the epithelial cell population in the sample of cervical neuroendocrine carcinoma.
Genes with low expression are filtered with cutoff = 0.1, noise_filter = NA [21]. In the GEO dataset, immune cells (T, B cells) with very low expression of SCNE markers in the sample were used as the reference cell group.
Based on the results of infercnv, the distribution of highly malignant cell groups is preliminarily judged, and then the expr.data in the calculation results of infercnv is re-clustered using the kmeans method (kmeans = 3), and a heatmap is drawn using the ComplexHeatmap [22] package. Cells with obvious malignancy are screened out based on the CNV signal [21]. The CNV level of each cell is quantified, and the following reference thresholds for judging the correlation of benign and malignant are given based on the CNV results: highly malignant cells: CNV mean > 0.004; non-malignant cells: overall CNV mean < 0.002.
Trajectory analysis by monocle 2 analysis and gene enrichment
Cell trajectory analysis is performed using Monocle2 (2.22.0) [23]. The gene expression matrix is extracted from the results obtained from infercnv, and the CNV score and cluster subgroup labels (kmeans_class) are added to the meta.data dataframe of the seurat object. The Monocle2 process is then run, with the fullModelFormulaStr of the differentialGeneTest function set to ~ sm.ns(CNV_score) (Figure S6). The starting state is determined based on the biological process. All differential genes are shown in Table S3. Genes with a q value < 0.001 and expressed in > 50 cells are used for cell sorting. Genes with q value < 0.0001 and expressed in more than 100 cells are selected, sorted by q value from small to large, and the top 20 genes are selected for the graph. Branch point 1 is selected for the gene heatmap related to branch height. The gene cluster is shown in Table S4. The heatmap plotting data is extracted and GO enrichment analysis [24] is performed on the genes in each cluster subgroup, and the results are plotted as a Polar bar Plot using EnrichVisBox [25].
Analysis of other cell subgroups
In addition to the epithelial cell subgroup, there are also stromal cells, monocyte cell subgroups, and T cell subgroups. Further dimensionality reduction clustering is performed using PCA, with the pc.dim parameter uniformly set to 30. Clustree [26] is used to determine the best resolution for each group, and the top 10 Markers of each cell group are extracted for further identification.
Stromal cells typically include fibroblasts, smooth muscle cells, and endothelial cells. In this study, fibroblast groups 1 and 2, smooth muscle cell groups 1 and 2, and endothelial cells are combined into stromal cells for further analysis. The best resolution for re-dimensionality reduction clustering of T cell group subsets is 0.3, and the marker genes in the study [27, 28] are used to distinguish each subgroup. The best resolution for the Monocyte cell group was 0.3, and it was found that group 4 is a doublet and was removed. The marker genes in the paper [15, 29] are used to distinguish various CAFs. Finally, gene sets are downloaded and screened from msigdb to perform enrichment analysis [30–32] on each subgroup and scored using AddModuleScore.
Single-cell regulatory network analysis by SCENIC
SCENIC (1.3.1) [33] is used for gene regulatory network analysis and visualization. The reference databases are hg38__refseq-r80__500bp_up_and_100bp_down_tss.mc9nr.feather and hg38__refseq-r80__10kb_up_and_down_tss.mc9nr.feather. The gene selection criteria are: the minimum count (minCountsPerGene) of each gene in all cells is at least 5% of the total number of cells, and each gene is detected in at least 5% of cells (minSamples = 5%). The GENIE3 method is used to construct a co-expression network. Each gene retains the top 5 weight values of TF to obtain a simplified TF-Target association table, and then genes are assigned to TF to construct co-expression modules. RcisTarget is used to identify motifs directly bound by transcription factors based on DNA-motif analysis, and indirect targets lacking motif support are pruned. After pruning, each TF and its potential direct acting target genes form a regulon. Run steps 1 to 4 of runSCENIC with default parameters, compare CNV score re-clustered groups 2 and 3, and also compare epithelial cell groups 1–3 from the perspective of cell subgroups, calculate and visualize CSI (Cell Specificity Index). The relationships between important TFs and regulatory networks were screened with high confidence. Regulons with significant expression in each group are further analyzed.
Comparison analysis of cell–cell communication analysis by CellChat
This study uses all human databases (Secreted Signaling, ECM-Receptor, Cell–Cell Contact) in CellChat (1.6.1) [34]. The seurat object is divided into two objects based on orig.ident and CellChat is used to calculate communication probabilities and communication networks based on ligand-receptor pairs, and filter out communications between cell groups with < 10 cells. The aggregated cell communication network and network centrality scores are calculated and the object is integrated for further analysis.
Survival data validates gene expression and prognosis
Data from the small cell lung cancer study [35] published in Nature in 2015 is downloaded from cBioPortal [36–38]. With a q value < 1*10–10 and num_cells_expressed > 100 as the standard, differential genes analyzed by Monocle from CNV-H kmean groups 2 and 3 are selected.
The differential gene expression obtained from Monocle is selected in the mRNA expression matrix (data_mrna_seq_fpkm.txt) and merged with survival data (data_clinical_patient.txt), and a survival object is created with the Overall Survival (Months) and Overall Survival Status. The surv_cutpoint function in the survminer [39] package is used to select the best cutoff for survival analysis, and the population is divided into high and low expression groups for each gene. The survival curve is plotted using the survival [40] package, and the log-rank test is used to calculate the p-value for prognosis differences.
Download of GEO dataset
Data from lung neuroendocrine carcinoma (GSM5870250, GSM5870256, GSM5870258), small intestine neuroendocrine carcinoma primary lesion (GSM4159164) and its liver metastatic lesion (GSM4159165) [41] downloaded from GEO. Among them, GSM5870250 and GSM5870258 are histologically atypical carcinoids, and GSM5870256 is histologically typical carcinoid, all of which are well differentiated. The data downloaded from GEO is analyzed with Seurat_4.3.0 [42], with min.cells set to 3 and min.features set to 200 to filter out low-quality cells. The analysis is carried out according to the previous process.
Data availability statement
The data and scripts supporting the findings of this study will be provided during review. All data in the study are available from the corresponding author upon reasonable request.
Results
ScRNA-seq data revealed the cellular composition of SCNECC
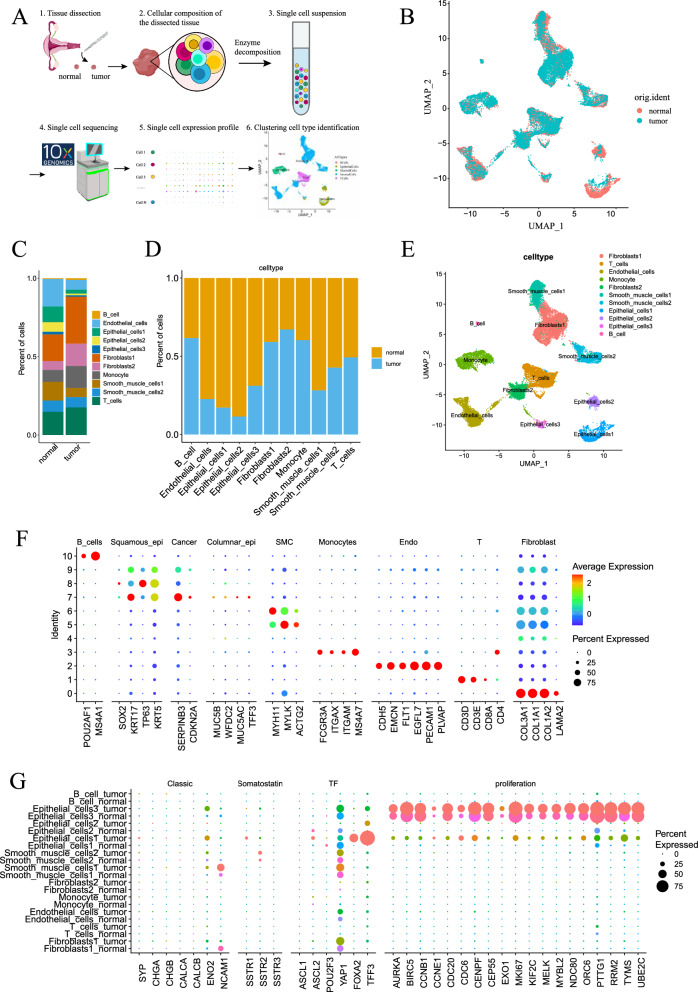
To fully characterize the cell composition of the tissues of SCNECC, we collected SCNECC samples from one patient for single-cell RNA sequence (Fig. 1A, B). A total of 25,264 cells and 23,124 genes passed quality control. The cells were pre-annotated by standard cell type markers and grouped into AllTypes (Figure S1).
Fig. 1.
Single-cell data annotation and small cell neuroendocrine tumor cell identification. A A schematic diagram showing the single-cell sequencing process. B UMAP dimensionality reduction plot of single-cell sequencing data from normal and tumor samples, where each point represents a single cell. C, D Bar plots showing the proportion and difference of different cell types in normal and tumor samples. E At UMAP resolution of 0.1, the samples were clustered into 11 cell subgroups. F The correspondence between the 11 cell subgroups obtained from UMAP dimensionality reduction clustering and the classic cell marker genes. G Comparison of the expression of classic neuroendocrine markers, growth inhibitory receptors, neuroendocrine-related transcription factors, and cell proliferation markers in each cell subgroup by sample
All cells were clustered into 11 classes at a resolution of 0.1 by UMAP dimensionality reduction (Fig. 1D).We utilized marker genes specific to different cell subsets, as described in the classical literature, for the purpose of identifying and distinguishing various cell types in our study. (Fig. 1F).
We identified 11 cell types based on canonical cell markers (Fig. 1B), including Fibroblasts1 (5754), T cells (4037), endothelial cells (3145), monocytes (2652), Fibroblasts2 (2408), smooth muscle cells1 (2322), smooth muscle cells2 (1749), epithelial cells1 (1690), epithelial cells2 (960), epithelial cells3 (344) and B cells (203) (Table S5).
Identification of neuroendocrine tumor cells
First, the marker genes of neuroendocrine were used to locate neuroendocrine tumor cells in the 5 major cell groups pre-annotated by AllTypes, and we found that the epithelial cell group had significant expression of neuroendocrine cell marker genes SYP, ASCL2, POU2F3, FOXA2 (Figure S1G). The Epithelial_cells1 cell group of tumor samples has higher expression of neuroendocrine marker genes SYP, SSTR1, SSTR2, ENO2, ASCL2, FOXA2, and proliferation markers (such as MKI67) than normal samples (Figure S1H).
Then, the related genes of neuroendocrine carcinoma were used to further locate neuroendocrine tumor cells in the 11 cell groups, and we found that in the epithelial cell group, classic neuroendocrine markers (SYP, CHGA, CHGB, CALCA, CALCB, ENO2, NCAM1), somatostatin receptors (SSTR1-3), neuroendocrine-related transcription factors (ASCL1-2, POU2F3, YAP1, FOXA2, TFF3), and cell proliferation markers (AURKA, BIRC5, CCNB1, CCNE1, CDC20, CDC6, CENPF, CEP55, EXO1, MKI67, KIF2C, MELK, MYBL2, NDC80, ORC6, PTTG1, RRM2, TYMS, UBE2C) all have significant expression compared with other groups (Fig. 1G). The epithelial cell group of tumor samples had higher expression of neuroendocrine marker genes SYP, SSTR1, SSTR2, ENO2, ASCL2, FOXA2, and proliferation markers (such as MKI67) than normal samples.
Copy number variation in cervical neuroendocrine epithelial cell populations
Preliminary results of InferCNV
Because the marker genes of neuroendocrine carcinoma were highly expressed in the Epithelial_cells 1 (Fig. 2A), it inferred that there might be highly malignant neuroendocrine tumor cells in the epithelial cell groups. Therefore, with the help of inferCNV, the Epithelial_cells1, 2, 3 cell groups of normal samples were used as the reference cell group, and the Epithelial_cells1, 2, 3 cell groups of tumor samples were used as the observation cell group to further analyze the copy number variation, identify and locate highly malignant cell groups. The results showed that when compared with normal epithelial cell groups, Epithelial_cells1 in tumor samples had obvious CNV changes (Fig. 2B). As can be seen in the figure, Epithelial_cells1 of tumor samples showed large segment copy number increases and decreases in multiple chromosomal regions, while the copy number changes of Epithelial_cells2, 3 were lighter.
Fig. 2.
CNV analysis and re-clustering of epithelial cells. A Expression proportion and intensity of classic neuroendocrine marker genes (SYP, CHGA) and transcription factors (ASCL1, ASCL2, POU2F3) in each cell subgroup. B Using the Epithelial_cells1, 2, 3 of normal samples as the reference cell group (upper heat map), and the Epithelial_cells1, 2, 3 cell groups of tumor samples (lower heat map) as the observation cell group for CNV analysis and the results of inferCNV. The horizontal axis represents the 22 chromosomes arranged in order, and the different colors on the vertical axis correspond to different cell subgroups. C According to the results of inferCNV, the heat map of all epithelial cells in normal and tumor samples after re-clustering (kmeans_class = 3). The horizontal axis represents the genes on chromosomes 1–22. D Share the same legend as C, it shows the CNV quantification of the 3 cell subgroups after re-clustering. E UMAP reduction plot showing the distribution of different CNV score cells by sample. F UMAP reduction plot showing the distribution of class 2 and class 3 cell after CNV re-clustering by sample
Epithelial cells re-cluster by CNV scores
Because the CNV heterogeneity of Epithelial_cells1 cells in tumor samples was obvious, all epithelial cell CNV score could be re-clustered into 3 classes (Fig. 2C). After re-clustering, the CNV changes were most significant in the second cell subgroup identified by k-means clustering, while the first subgroup showed no significant changes, and the third subgroup exhibited changes that were intermediate between the other two.
Since the more obvious the tumor cell chromosome copy number variation, the greater the difference between them and normal cells, the higher the malignancy of the them [43], we speculated that the second class of cells had the highest malignancy. According to the source of epithelial cells, the second class mainly corresponded to the Epithelial_cells1 in tumor.
CNV quantification
In order to compare the degree of CNV changes, the cells after re-clustering were further quantitatively analyzed for CNV score (Fig. 2D), and we found that the CNV score was highest in the second class, lowest in the first class, and the third class was in the middle transition state. The UMAP dimensionality reduction plot showed the distribution of CNV score in epithelial cell and kmeans_class (2, 3 cell classes) after re-clustering by sample (Fig. 2E, F). The CNV score of epithelial cells in normal samples was lower, while the score in tumor samples was higher, and the kmeans_class 2 cells with the highest CNV score all appeared in tumor samples.
Pseudotime analysis of malignant cells
Pseudotime analysis
To infer the origin and development process of neuroendocrine tumor cells, we performed the pseudotime analysis of the second class (CNV high cell group) and the third class (CNV medium transition cell group) after re-clustering (Fig. 3A, B). In the pseudotime analysis, normal cells gradually transformed into tumor cells (Fig. 3C), the CNV score value of cells gradually increased (Fig. 3D), and the cell groups gradually became malignant. Most of the cells in the right side of Figure E were the third class of cells after re-clustering (green), and most of the cells on the left side were highly malignant second class cells (red), indicating that the CNV score changes from low to high during pseudotime. Figure F showed the classification of cell subgroups, and there were more Epithelial_cells 2, 3 groups on the right side, and gradually differentiate into the Epithelial_cells1 group (SCNE classic marker positive). In summary, it was speculated that the occurrence of tumors might evolve from normal Epithelial_cells1,2 to normal sample's Epithelial_cells3, gradually transform into lesion Epithelial_cells 2,3 group, and finally become neuroendocrine carcinoma (Epithelial_cells1).
Fig. 3.
Pseudotime analysis and GO enrichment of re-clustered epithelial cells 2 and 3. A–F Pseudo-temporal plot of the Epithelial Cells 2 and 3 cell group after re-clustering, where each point represents a single cell. A The color gradient from dark to light indicates the progression of pseudotime. B Branch node 1 divides the cell group into 3 states. C The origin of samples. D The distribution of CNV scores. E The classification of each cell after CNV re-clustering. F The group of epithelial cells to which each cell belongs. G Heatmap showing the differential gene expression patterns of cells with different cell destinies at branch point 1. Rows represent genes, and columns represent cells. In the heatmap, the redder (bluer) the color, the higher (lower) the gene expression level. H Stacked polar bar chart showing the results of GO enrichment of cluster1 genes in G. The color gradient of the central column represents the p-value of the GO term; the height of the column represents the number of differential genes enriched to the GO term. I Stacked polar bar chart showing the results of GO enrichment of cluster2 genes in G. J, K, L, M, N Showing the changes in the expression levels of important differential genes (ELF3, CALD1, COL1A1, COL1A2, MKI67) as pseudotime progresses. Shared legend and horizontal axis: Each point represents the expression level of the gene in a cell. The horizontal axis represents pseudotime, and the vertical axis represents gene expression level. O, P, Q, R, S Survival curve plots of small cell lung cancer patients grouped by high and low gene expression, corresponding to J, K, L, M, N. The dashed lines on the horizontal and vertical axes represent the median survival time and median survival rate, respectively
Analysis of gene transformation during cell malignancy and GO enrichment
Previously, we discovered and demonstrated the process of cervical neuroendocrine carcinoma transforming from normal tissue to malignant tumor. Next, we further explored the transformation in genes during the process of cell malignancy. Heat map (Fig. 3G) showed the changes in gene expression among cells with distinct fatesat branch point 1 during the process of cell malignancy, and these genes were clustered into 3 classes. In the pseudotime trajectory, from the middle to both sides, the genes of C1 decreased in state1 and slightly increased in state2 cells, while the genes of C2 slightly decreased in state2 but significantly increased in state1 cells. The expression pattern of C3 group genes was not obvious. In order to explore the biological function of these changing gene sets in small cell cervical neuroendocrine carcinoma, the genes in C1 and C2 classes were extracted and enriched by GO. The C2 gene set enrichment primarily focued on pathways such as cell–cell junction, wound healing, and actin binding. These pathways were mainly related to cell–cell connections. The C1 gene set enrichment primarily emphasied on pathways such as ribosomal subunit, structural constituent of ribosome, and cytoplasmic translation. These pathways were mainly related to ribosome synthesis within the cell (Tables S6–8).
Gene expression changes and prognosis
In the process of malignant transformation of tumor cells, ELF3 was a significantly up-regulated gene in the epithelial cell population of small cell cervical neuroendocrine carcinoma during (state 1) malignant transformation, and showed a downward trend in the transitional cell population (state 2) (Fig. 3J–N).While other genes such as CALD1, COL1A1, COL1A2, COL3A1, DCN, TAGLN, TIMP1, VIM, LGALS1, MT2A showed an opposite trend (Figure S3). In smallcell lung neuroendocrine carcinoma, there was no significant difference in the prognosis between ELF3 high expression group and low expression group (p = 0.16). There were significant differences between the high and low expression groups of CALD1 (p = 0.0039), COL1A1(p = 0.0001), COL1A2(p = 0.0018), MKI67(p = 0.0041), COL3A1(p = 0.002), IGFBP7 (p = 0.00079), COL1A1 (p = 0.00079) and so on. (Fig. 3O–S, Figure S3).
Gene expression and regulatory networks in malignant cells
Gene expression and regulatory networks
Pseudotime analysis exhibited differences in gene expression during the malignant transformation of small cell cervical neuroendocrine carcinoma cells. Therefore, further exploration of the gene regulatory expression network was necessary (Table S9, Fig. 4A, B). By analyzing the gene expression regulatory network from the perspective of epithelial cell subgroup and CNV re-clustering, we found that TFF3 was more specific in Epithelial cells1/re-clustered 2 cell group (which represented high copy number variation, CNV-H), and CEBPB was very specific in Epithelial cells3/re-clustered 3 cells (which represented middle copy number variation, CHV-M) (Fig. 4C–L, N–P). TFF3 mainly regulated EPCAM, ELF3 (Table S10), while CEBPB mainly regulated FTL, LITAF (Table S11).
Fig. 4.
Analysis of gene expression regulatory network of tumor neuroendocrine cell subsets. A Heat map of comparing gene expression regulatory networks in different cell populations after CNV re-clustering, showing regulon activity in different cell subsets. B Heat map of comparing the regulatory network of gene expression in epithelial cell. C Epithelial cell Regulon Specificity Score (RSS) plot showes the cell type-specific regulon in each epithelial cell groups. The size of the dot represents the RSS score, and the yellower color of the dot, the higher specificity of this regulon in this cell type. D RSS plot of cell groups after CNV re-clustering shows the specific regulon of each cell type after CNV re-clustering. The legend is as same as Fig. 4C. E, F, G, H, I The top 3 regulon in each cell group of epithelial cells and CNV re-clustered cells. J Distribution of regulon TFF3_173g in UMAP dimensionality reduction results; K The upper ridge plot and the lower violin plot show the distribution of TFF3_173g in different epithelial cell subsets. L Ridge plot and violin plot below show the distribution difference of TFF3_173g among cell groups after CNV re-clustering. M K-M survival curve of TFF3 high expression group and low expression group in small cell lung cancer patients. The N, O, P and Q plots are similar to J, K, L and M, respectively, showing the distribution of regulon CEBPB_117g in UMAP dimension reduction results, the distribution difference among different cell subsets and the K-M survival curve of small cell lung cancer
Transcription factors and prognosis
To further investigate the impact of the expression levels of significant transcription factor genes on patient prognosis, patients with small cell lung cancer were categorized into high/low expression groups based on mRNA expression levels. It was observed that the prognosis of patients with TFF3 high expression group was significantly better than those of the low expression group (p = 0.004) (Fig. 4M). No significant difference was noted for CEBPB (p = 0.081) (Fig. 4Q).
Verification of GEO neuroendocrine carcinoma
In the comparison of single-cell sequencing data from pulmonary neuroendocrine tumors, primary small intestine lesions, and liver metastases of neuroendocrine tumors on GEO, the results revealed tissue heterogeneity in cellular composition and proportions across the samples (Fig. 5A, B). CHGA and CHGB were found to be more sensitive in cancer cells of pulmonary, small intestine, and liver neuroendocrine tumors, while the expression ratio of SYP was slightly lower. ENO2 expressionin pulmonary, small intestine, and liver neuroendocrine tumors was partial and not universal (Fig. 5C). The gene expression regulatory networks of neuroendocrine tumors from different samples exhibited heterogeneity, and transcription factors showed sample specificity. ETV1 was more specific in GSM5870258, while JUND_extended (2126 g) was more specific in GSM4159165. The network diagram displayed transcription factors were common in both pulmonary and cervical neuroendocrine tumors (Fig. 5J). The key transcription factors that were common to these two neuroendocrine carcinomas primarily include CEBPB, POLR2A, SNAI1, YY1, PBX1, ELK3, SOX4, ELF1, ATF3, XBP1, NR2F6.
Fig. 5.

Neuroendocrine markers and gene expression regulatory network analysis of lung, small intestine, and liver neuroendocrine carcinoma in GEO data. A Bar graphs: the bar graphs display the composition of major cell types in different GEO single-cell sequencing datasets. GSM5870250, GSM5870256, GSM5870258 represent lung adenocarcinoma; GSM4159164 indicates small intestine neuroendocrine carcinoma (primary lesion); GSM4159165 denotes liver neuroendocrine carcinoma (metastatic lesion). B Cell type proportions: the bar graphs show the proportions of different cell types across various GEO single-cell sequencing datasets (neurons: small cell lung cancer cells, Neurons1/2: small intestine/liver neuroendocrine tumor cells). C Dot plots: the dot plots of GEO data illustrate the expression levels of classic neuroendocrine markers, somatostatin receptors, and transcription factors across different cell subgroups; D RSS plots: the RSS plots reveal regulons specific to neuroendocrine tumor cells (Neurons) in lung neuroendocrine carcinoma. The size of the dots represents the RSS score, and the more yellow the color, the higher the specificity of the regulon in that cell type. E The RSS plots reveal regulons specific to neuroendocrine tumor cells (Neurons1/2) in small intestine and liver neuroendocrine carcinoma. Legends are as same as decribed in D. F Distribution of CEBPD_13g: the distribution of CEBPD_13g within the UMAP dimensionality reduction results is depicted (left) while the ridge plot (top right) and violin plot (bottom) both illustrate the distribution differences of CEBPD_13g across various samples. G, H, I TFs’ Distribution: G, H, and I display the distribution differences of NFIB_39g, ETV1_20g, and FOS_60g across different samples, with legends corresponding to F. J Network diagram: the network diagram reveals the intersection of gene expression regulatory networks between small cell neuroendocrine cervical carcinoma and small cell neuroendocrine lung carcinoma. Blue represents transcription factors common to both; green denotes transcription factors or target genes shared by both
Features of other cell groups in SCNECC
T cell subgroups
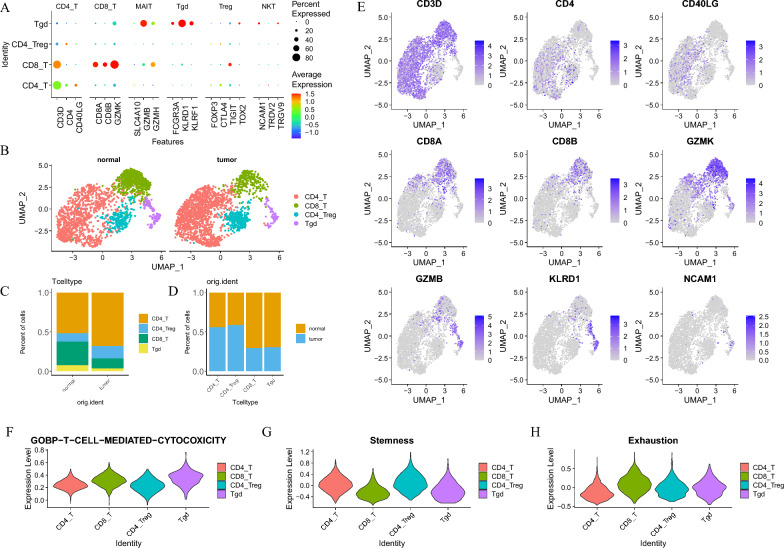
In SCNECC, T cells were divided into four subgroups, including conventional CD4 T cells (CD4_T), CD8 T cells (CD8_T), T regulatory cells (CD4_Treg), and γδT (Tgd) (Fig. 6). CD4_T was featured by CD3D and CD4, CD8 T cells was featured by CD8A and CD8B, CD4_Treg was featured by TIGHT, and Tgd was featured by KLRD1. The γδT subgroup played a major role in cytotoxic tumor cell killing, yet it was the least abundant. The CD8 subgroup, typically associated with cytotoxic functions, exhibited the poorest stemness and highest exhaustion scores.
Fig. 6.
T cell subgroups. A The dot plot illustrates the expression of cellular markers corresponding to T cells after re-dimensionality reduction clustering. B Comparative UMAP dimensionality reduction plots reveal the distributional differences between normal and tumor T cells in two-dimensional space. C Stacked bar plots depict the percentage differences in the distribution of T cell subpopulations between normal and tumor samples. D Histograms show the percentage differences in the distribution of T cell subpopulations between normal and tumor samples. E Feature plots demonstrate the expression patterns of various marker genes on the UMAP dimensionality reduction plots. F, G, H Violin plots present the scoring of each T cell subpopulation on gene sets associated with GOBP_T_CELL_MEDIATED_CYTOTOXICITY, Stemness, and Exhaustion
Monocytes subgroups
Monocytes were divided into three subgroups: macrophages, MDSCs, and DCs. Macrophages was featured by C1QA, C1QB, C1QC, APOE. MDSCs was featured by FCN1, and DCs was featured by CLEC9A (conventional dendritic cells1) and CD1C (conventional dendritic cells2). Macrophages were more prevalent in tumor samples and had been found to possess a high potential for phagocytosis, while MDSCs exhibited high angiogenesis and MDSC scores, suggesting their primary role in promoting angiogenesis and assisting tumor metastasis within the tumor. DCs possessed a relatively high antigen-presenting function but a low phagocytic function (Fig. 7).
Fig. 7.
Subgroups of monocytes. A Dot plot illustrating the expression of cell markers corresponding to monocytes subgroups after further dimensionality reduction clustering. B UMAP dimensionality reduction plot comparing the distribution differences of normal and tumor monocytes in two-dimensional space. C Stacked bar chart showing the percentage distribution differences among monocyte subgroups between normal and tumor samples. D Bar chart displaying the percentage distribution differences of monocyte subgroups between normal and tumor samples. E, F, G, H Violin plots revealing the scores of each monocyte subgroup in angiogenesis, antigen processing, phagocytosis, and MDSC gene sets
Stromal cells
Previous studies indicated that cancer-associated fibroblasts (CAFs) exhibited significant heterogeneity within the tumor microenvironment and played a pivotal role. This study identified several CAF subgroups in SCNECC, including inflammatory CAFs (i_matrix_CAF), RAMP1 positive soomth musscle cells (RAMP1 + _SMC), antigen-presenting CAFs (apCAF), IGKC positive CAF (IGKC + _CAF), inflammatory matrix CAFs with a highly activated metabolic state (i_matrix_me_CAF), iCAF or SMC (iCAF_SMC), vascular CAFs (vCAF), GNG11 positive apCAF (GNG11 + _apCAF) (Figure S4). In tumor samples, the proportions of apCAF, SMC, and vCAF were notably reduced, while iCAF, matrix CAF, and meCAF subgroups were more prevalent.
Cell–cell communication among subgroups of SCNECC
After analyzing individual cell subgroups, we sought to further explore the interactions among SCNECC cells. Given the critical role of T cells in tumor immunity, T cell subgroups were renamed based on previous identification results. Cell communication analysis revealed that while the overall intensity of communication in SCNECC tumor samples remained unchanged compared to normal cell samples, the total number of interactions increased. Normal and tumor cells shared 65 signaling pathways, of which 24 were unique to tumors and some were associated with the nervous system and tumorigenesis. Normal tissues exhibited high levels of THBS and CCL signals and a high proportion of IL-I signals. In tumors, MHC-II signal flow was strong, and SPP1 was most specific. Cell-autonomous signaling in small cell cervical neuroendocrine tumor cells (Epithelial_cells1) differed from normal cells, including DESMOSOME, MPZ, CDH, and EPHA signals (Fig. 8).
Fig. 8.
Comparison analysis of cell–cell communication analysis in SCNECC. A–F Network diagrams: nodes of different colors represent various cell subgroups. Each line indicates a signal from the sender to the receiver, with thickness representing the number or strength of interactions between subgroups. C and F show differences in cell communication networks between samples, with red (or blue) lines indicating increased (or decreased) signals in small cell neuroendocrine cervical tumor compared to normal samples. C (/F) represents the difference between panels A and B (/D and E). G, H Differential network analysis heatmaps: Display variations in the number or strength of interactions between different cell groups. The colored bar at the top indicates the sum of incoming signals (columns), and the colored bar on the right indicates the sum of outgoing signals (rows). Red (or blue) squares indicate increased (or decreased) signals in tumors compared to normal samples. I, J Ranking of important signaling pathways based on inferred overall information flow differences between Normal and Tumor within the network. The top signaling pathways colored red were enriched in Normal samples, while the bottom signaling pathways colored green were enriched in Tumor samples. K, L, M, N Comparisons of DESMOSOME and EPHA signaling pathways between normal and tumor samples; O, P Comparative distribution of gene expression related to DESMOSOME and EPHA signaling pathways between normal and tumor samples
Discussion
This single-cell study discovered that ELF3 gene upregulated in SCNECC tumor cells during the malignant transformation process. And TFF3 regulated the transcription of the ELF3 gene.
Studies of ELF3 in SCNECC
The ELF3 gene, encoding a 371-amino acid protein, is an epithelial cell-specific ETS transcription factor involved in development and disease [44]. ELF3 is an epithelial cell-specific transcription factor [44]. Genes regulated by ELF3 are closely associated with malignant phenotypes, such as tumor initiation, progression, invasion, and metastasis. ELF3 functions dually as a tumor suppressor and an oncogene. ELF3 acts as an oncogene in lung, liver, and breast cancers, and as a tumor suppressor in ovarian and oral cancers. [45]. Currently, there are no studies on ELF3 in small cell neuroendocrine carcinoma of the cervix (SCNECC). Studies have linked ELF3 to super-enhancers in neuroendocrine carcinomas [46]. Certain super-enhancers in malignant tumors may be aberrantly activated, leading to the overexpression of oncogenes that promote tumor cell proliferation, survival, and invasion. The ELF3 gene is highly expressed in cervical cancer tissues but not in normal tissues [47]. ELF3 can promote the proliferation and invasion of cervical cancer cells, resulting in poor prognosis [44]. Increased ELF3 expression during cervical neuroendocrine carcinoma malignancy suggests its potential as a biomarker for early detection and prognosis. Further exploration is needed to elucidate the specific mechanisms of ELF3 aberrant expression in cervical neuroendocrine carcinoma. Inhibition of ELF3 or its downstream effectors could lead to new targeted therapies. Personalized treatment based on ELF3 expression levels could potentially enhance therapeutic efficacy and reduce unnecessary side effects.
In addition to super-enhancers, mutations in the ELF3 gene itself may also be a significant cause of neuroendocrine carcinoma. ELF3 gene frameshift mutations occur in 13% of cervical cancers [48]. Compared to wild-type tumors, cervical adenocarcinomas with ELF3 mutations express higher levels of ELF3 mRNA [44]. Currently, there are no studies on ELF3 mutations in small cell neuroendocrine carcinoma of the cervix (SCNECC). In this SCNECC single-cell study, inferCNV was used to predict changes in DNA copy number, revealing large segmental amplifications and deletions on chromosome 1 in SCNECC. Specific loci require further investigation using specialized mutation detection.
The ELF3 gene is closely associated with high-risk HPV infection in cervical cancer [48]. Neuroendocrine cervical cancers are often linked to HPV. A multicenter study [7] involving 64 SCNECC patients, integrating whole-exome and RNA sequencing analyses, found that HPV DNA was detected in 65.6% (42/64) of the tumors. ELF3 upregulation in HIDDEN cells suggests its role as an HPV-induced transcriptional driver [49]. In our samples, in situ hybridization results of postoperative pathological samples indicated high-risk HPV (+). The patient tested positive for HPV-18 within one month before surgery. Single-cell analysis revealed upregulation of ELF3, mediated by TFF3, during SCNECC transformation, suggesting its potential as an early diagnostic marker. These findings suggest that the expression of the transcription factor ELF3 is potentially closely related to high-risk HPV (types 16 and 18) infection, warranting further exploration. Reviews link SCNECC more closely to HPV-18 than HPV-16 [50].
However, there have been no reports on whether the ELF3 and TFF3 genes undergo mutations or amplifications in SCNECC. Further exploration of the ELF3-HPV relationship in SCNECC is warranted.
A retrospective study analyzed the liquid-based cytology characteristics of small cell neuroendocrine carcinoma of the cervix [51]. The results showed that although the initial cytological screening had a positive rate of 63.6% (7/11), none were accurately diagnosed as small cell carcinoma of the cervix. The potential of using ELF3 as an auxiliary diagnostic marker to improve the initial screening accuracy for small cell neuroendocrine carcinoma of the cervix (SCNECC) warrants further exploration.
Studies of TFF3 in SCNECC
TFF3 is predominantly expressed in the columnar epithelium of the cervical endometrium, not in the squamous epithelium [52]. Upregulation of TFF3 in neuroendocrine cells of cervical cancer suggests a possible origin from columnar epithelium. The columnar epithelium's close association with neuroendocrine carcinoma suggests potential for abnormal differentiation into tumors.
TFF3 is implicated in CTC migration and hematogenous dissemination, with higher expression in CTCs and lung metastases [53]. Our study indicates TFF3's role in early hematogenous metastasis of SCNECC, possibly through intercellular connection remodeling.
TFF3 serves as a marker for epithelial-like CTCs, and its upregulation of ELF3 may enhance CTC detection [53]. Our gene expression regulatory network analysis revealed that TFF3 upregulates ELF3 expression with high confidence and correlation. TFF3 is anticipated to be a marker for epithelial tumor hematogenous metastasis.
Markers of small cell neuroendocrine carcinoma
Identification of small cell neuroendocrine carcinoma cells requires combined analysis of various neuroendocrine markers and tumor proliferation factors.
Chromogranin A (CgA) and Synaptophysin (Syn) are two significant neuroendocrine carcinoma immunohistochemical markers clinically [1, 54]. The encoding genes for CgA and Syn are CHGA and SYP, respectively. In SCNECC, the expression ratio of SYP was low, while that of CHGA was even lower, indicating that these commonly used classical neuroendocrine markers are not sensitive enough. They may not be entirely suitable for identifying rare neuroendocrine carcinomas occurring at non-canonical sites, potentially due to the unique tumor microenvironment at various locations. Therefore, there is a need to discover markers that possess higher sensitivity [55].
Studies have shown that the encoding genes for somatostatin receptors were often highly expressed in neuroendocrine tumor cells [56]. In SCNECC, SSTR1 and SSTR2 were significantly overexpressed in neuroendocrine tumor cell clusters, serving as markers for neuroendocrine tumor identification. This suggested that targeted drugs approved for the treatment of neuroendocrine tumors [56] (such as octreotide and lanreotide) could also be used to treat patients with SCNECC.
ASCL1, NEUROD1, POU2F3, and YAP1 were important transcription factors in neuroendocrine carcinoma, which were used to divide four major subgroups in lung neuroendocrine carcinoma [1]. In the small cell neuroendocrine prostate cancer model, ASCL1 and ASCL2 marked different transdifferentiation trajectories of tumor cells [14]. In our SCNECC smaple, ASCL2 had a higher expression ratio than ASCL1, suggesting that ASCL2 may be more suitable for the identification of SCNECC. A study indicates that the expression of NEUROD1 is an independent prognostic factor for poor outcomes in SCNECC [57]. However, in our sample, NEUROD1 expression was not detected.
FOXA2 was a sensitive and specific marker for small cell prostate neuroendocrine carcinoma [14], and it also had high sensitivity and specificity in SCNECC. In our data, the expression ratio of FOXA2 was high and it was only enriched in tumor epithelial tissue. Therefore, it could clearly distinguish tumor epithelium from normal epithelial cells. Consequently, FOXA2 had great potential and could be attempted for the identification of other rare and complex neuroendocrine carcinomas.
CALCA and CALCB co-encode the neuropeptide CGRP (Calcitonin Gene-Related Peptide) [58], a marker for mature (Neuroendocrine) NE in the lungs. The expression levels of CALCA and CALCB in SCNECC were very low, which was possibly due to significant functional differences between neuroendocrine cells in the cervix and lungs, indicating that the CALCA and CALCB genes were limited by tissue origin for neuroendocrine tumor identification.
Origin and malignant transformation of neuroendocrine tumor cells
This study suggested that small cell neuroendocrine cervical carcinaoma cells may originate from epithelial cells. In SCNECC, proliferation markers in epithelial cell groups were significantly upregulated, while there was no significant expression in stromal cells and immune cells. Pseudotime analysis showed that cell types underwent a gradual transition from normal cells to tumor cells by pseudotime trajectory, rather than an abrupt cutoff, indicating that the malignant transformation of tumor cells was a gradual process. SCNECC single-cell data suggested high heterogeneity and complex origins of neuroendocrine carcinoma tumors.
Overall signaling pathway differences in neuroendocrine carcinoma
The THBS signaling pathway played a protective role in normal tissues, consistent with literatures [59, 60]. However, in small cell cervical neuroendocrine carcinoma tissues, THBS-related signals were significantly weakened, contributing to tumor occurrence, progression, and metastasis. In tumors, MHC-II class signals were abnormally active when compared to normal samples, suggesting the presence of active exogenous antigen presentation activities in the tumor microenvironment. It indicated that infection by exogenous pathogens might be the cause of SCNECC. We conducted an analysis of the relevant signaling pathways, the details of which are not presented here.
Potential of immunotherapy in neuroendocrine carcinoma
Immune subgroup analysis revealed CD8 T cell exhaustion and low γδT cell counts in tumors; thus, reversing CD8 exhaustion and enhancing γδT cell recruitment could improve immunotherapy. Additionally, activating tumor-associated macrophages' phagocytosis might augment tumor cell destruction.
Novelty of the research
The novelty of this research lied in the systematic, comprehensive, and in-depth advanced analysis of single-cell sequencing data from fresh SCNECC surgical samples. In this study, we thoroughly revealed the origins, development, gene expression transcriptional regulatory networks, and mechanisms of early metastasis in SCNECC, as well as related potential diagnostic and therapeutic targets, paving ways towards personalized and targeted interventions. This study was also the first to combine CNV analysis and pseudotime analysis to explore the changes in chromosomal copy numbers during tumor development, laying the foundation for the inference of the development of malignant cells.
Limitations of the research
This study faced challenges due to the rarity of cervical neuroendocrine carcinoma, limiting sample availability and complicating identification with low marker gene expression. Future single-cell sequencing on more fresh lesions is recommended to elucidate the biology of this aggressive tumor, guiding clinical treatment and improving patient outcomes.
Future directions
Directions for future research include validation of key findings of this research in independent cohorts, confirmation of the functional relevance of these genes and pathways in neuroendocrine cancer progression, and response to therapy by experimental models. To address the limitations of single-cell RNA sequencing, we will employ bulk RNA sequencing, spatial transcriptomics, epigenomics, and proteomics to provide additional layers of data.
Longitudinal follow-up of the patient
In this study, the patient underwent surgery in January 2023, followed by multiple cycles of EP chemotherapy, pelvic radiotherapy, and maintenance immunotherapy with toripalimab, concluding in December 2023. She remained in good condition with normal tumor markers. MRI (August 2024) revealed enlarging abnormal signal foci in the spleen, necessitating ongoing surveillance.
Conclusions
In conclusion, our study found that the transcription factor TFF3 directly up-regulates ELF3, suggesting a new target for therapy.. Comparing single-cell sequencing data of neuroendocrine cancer among lung, small intestine, and liver, we found that the transcriptional states of neuroendocrine cancer among different origins were highly heterogeneous and should be treated individually.
This study discovered for the first time that the early metastasis characteristics observed in small cell neuroendocrine cancer in clinical circumstances may be related to the dismantling of tight intercellular connections and desmosomes as indicated by GO enrichment analysis. However, proliferation markers, such as MKI67 reflected less proliferative vitality than squamous/adeno-carcinoma (Fig. 1G). While the current study provides meaningful insights, additional studies is necessary in order to fully validate these findings in future.
Supplementary Information
Acknowledgements
This work was supported by the Medical Science Data Center of Fudan University.
Author contributions
Conceptualization, C.W. and T.W.; methodology, T.W. and S.M.; software, T.W. C.W. collected the sample. B.W. sent the sample to “Jizhiyixue” company for single cell RNA (scRNA) sequencing and primary data analysis. T.W. washed the primary scRNA data and performed advanced analysis. T.W. discussed the results with C.W. and S.M. and conceived the topic and wrote the first draft. S.M. guided the analysis of immune cells. C.W., B.W., S.M., L.Z., J.L. and W.Y. reviewed the manuscript, tables, and figures. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by National Natural Science Foundation of China (General Program, 82273233); Natural Science Foundation of Shanghai (22ZR1408900); National Natural Science Foundation of China (General Program, 81772777); Clinical Research Plan of SHDC (No. SHDC2020CR4079); Shanghai "Rising Stars of Medical Talent" Youth Development Program-Outstanding Youth Medical Talents (SHWJRS2021-99), Shanghai Science and technology committee medical guidance program (18411963700).
Availability of data and materials
The datasets analysed during the current study are available at the GEO database under the accession code (GSM5870250, GSM5870256, GSM5870258, GSM4159164, GSM4159165). The raw sequencing data generated in this study have been deposited in the Genome Sequence Archive (GSA) by CNCB under the BioProject accession number PRJCA033235.
Code availability
R scripts for analyzing data are available upon reasonable request.
Declarations
Ethics approval and consent to participate
The study was conducted in accordance with the Declaration of Helsinki, and approved by the Ethics Committee of Obstetrics and Gynecology Hospital of Fudan University(2024-113). Informed consent was obtained from all subjects involved in the study. Written informed consent has been obtained from the patient to publish this paper.
Consent for publication
Not applicable.
Competing interests
The authors declare no conflicts of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Tianyou Wang and Li Zhang have contributed equally to this work.
Contributor Information
Jiongbo Liao, Email: liaojiongbo@163.com.
Chao Wang, Email: wang1980-55@163.com.
References
- 1.Chao A, Wu RC, Lin CY, et al. Small cell neuroendocrine carcinoma of the cervix: from molecular basis to therapeutic advances. Biomed J. 2023;46(5): 100633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Society of Neuroendocrine Neoplasm of China Anti-Cancer Association. China Anti-Cancer Association guideline for diagnosis and treatment of neuroendocrine neoplasm (2022 edition). China Oncol. 2022;32(6):545–79. [Google Scholar]
- 3.Mahdi H, Joehlin-Price A, Elishaev E, et al. Genomic analyses of high-grade neuroendocrine gynecological malignancies reveal a unique mutational landscape and therapeutic vulnerabilities. Mol Oncol. 2021;15(12):3545–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saraei P, Heshmati A, Hosseini S. Small-cell neuroendocrine carcinoma of the female genital tract: a comprehensive overview. J Neuroendocrinol. 2024;36(6): e13394. [DOI] [PubMed] [Google Scholar]
- 5.Lyu YH, Liu HX, Han X, et al. Clinicopathologic characteristics and prognostic factors of patients with surgically treated high-grade neuroendocrine carcinoma of the cervix: a multicenter retrospective study. Int J Gynaecol Obstet. 2024. [DOI] [PubMed]
- 6.Pan B, Yan S, Yuan L, et al. Multiomics sequencing and immune microenvironment characteristics define three subtypes of small cell neuroendocrine carcinoma of the cervix. J Pathol. 2024;263(3):372–85. [DOI] [PubMed] [Google Scholar]
- 7.Bellone S, Jeong K, Halle MK, et al. Integrated mutational landscape analysis of poorly differentiated high-grade neuroendocrine carcinoma of the uterine cervix. Proc Natl Acad Sci U S A. 2024;121(17): e2321898121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. 2021. https://www.R-project.org/.
- 9.Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wolock SL, Lopez R, Klein AM. Scrublet: computational identification of cell doublets in single-cell transcriptomic data. Cell Syst. 2019;8(4):281-291.e289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bunis DG, Andrews J, Fragiadakis GK, Burt TD, Sirota M. dittoSeq: universal user-friendly single-cell and bulk RNA sequencing visualization toolkit. Bioinformatics. 2020;36:5535–6. 10.1093/bioinformatics/btaa1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aran D, Looney AP, Liu L, et al. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat Immunol. 2019;20(2):163–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li R, Zhang J, Li Z. EasyCellType: marker-based cell-type annotation by automatically querying multiple databases. Bioinform Adv. 2023;3(1):vbad029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen CC, Tran W, Song K, et al. Temporal evolution reveals bifurcated lineages in aggressive neuroendocrine small cell prostate cancer trans-differentiation. Cancer Cell. 2023;41(12):2066-2082 e2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ou Z, Lin S, Qiu J, et al. Single-nucleus RNA sequencing and spatial transcriptomics reveal the immunological microenvironment of cervical squamous cell carcinoma. Adv Sci (Weinh). 2022;9(29): e2203040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu C, Zhang M, Yan X, et al. Single-cell dissection of cellular and molecular features underlying human cervical squamous cell carcinoma initiation and progression. Sci Adv. 2023;9(4):eadd8977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baine MK, Hsieh MS, Lai WV, et al. SCLC subtypes defined by ASCL1, NEUROD1, POU2F3, and YAP1: a comprehensive immunohistochemical and histopathologic characterization. J Thorac Oncol. 2020;15(12):1823–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clarke ZA, Andrews TS, Atif J, et al. Tutorial: guidelines for annotating single-cell transcriptomic maps using automated and manual methods. Nat Protoc. 2021;16(6):2749–64. [DOI] [PubMed] [Google Scholar]
- 19.Hu C, Li T, Xu Y, et al. Cell Marker 2.0: an updated database of manually curated cell markers in human/mouse and web tools based on scRNA-seq data. Nucleic Acids Res. 2023;51(D1):D870-d876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.inferCNV of the Trinity CTAT Project. https://github.com/broadinstitute/inferCNV.
- 21.Peng J, Sun BF, Chen CY, et al. Single-cell RNA-seq highlights intra-tumoral heterogeneity and malignant progression in pancreatic ductal adenocarcinoma. Cell Res. 2019;29(9):725–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gu, Z. Complex Heatmap Visualization. iMeta 2022. [DOI] [PMC free article] [PubMed]
- 23.Qiu X, Hill A, Packer J, et al. Single-cell mRNA quantification and differential analysis with Census. Nat Methods. 2017;14(3):309–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25(1):25–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang S, Zhong Y, Cheng J, et al. EnrichVisBox: a versatile and powerful web toolbox for visualizing complex functional enrichment results of omics data. J Comput Biol. 2021;28(9):922–30. [DOI] [PubMed] [Google Scholar]
- 26.Zappia L, Oshlack A. Clustering trees: a visualization for evaluating clusterings at multiple resolutions. Gigascience. 2018. 10.1093/gigascience/giy083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chu Y, Dai E, Li Y, et al. Pan-cancer T cell atlas links a cellular stress response state to immunotherapy resistance. Nat Med. 2023;29(6):1550–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang L, Yu X, Zheng L, et al. Lineage tracking reveals dynamic relationships of T cells in colorectal cancer. Nature. 2018;564(7735):268–72. [DOI] [PubMed] [Google Scholar]
- 29.Han C, Liu T, Yin R. Biomarkers for cancer-associated fibroblasts. Biomark Res. 2020;8(1):64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liberzon A, Birger C, Thorvaldsdóttir H, et al. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1(6):417–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liberzon A, Subramanian A, Pinchback R, et al. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 2011;27(12):1739–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aibar S, González-Blas CB, Moerman T, et al. SCENIC: single-cell regulatory network inference and clustering. Nat Methods. 2017;14(11):1083–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jin S, Guerrero-Juarez CF, Zhang L, et al. Inference and analysis of cell-cell communication using Cell Chat. Nat Commun. 2021;12(1):1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.George J, Lim JS, Jang SJ, et al. Comprehensive genomic profiles of small cell lung cancer. Nature. 2015;524(7563):47–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.de Bruijn I, Kundra R, Mastrogiacomo B, et al. Analysis and visualization of longitudinal genomic and clinical data from the AACR Project GENIE biopharma collaborative in cBioPortal. Cancer Res. 2023;83(23):3861–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kassambara A, Kosinski M, Biecek P. survminer: Drawing Survival Curves using 'ggplot2'. R package version 0.4.9. 2021. https://CRAN.R-project.org/package=survminer.
- 40.Therneau T. A Package for Survival Analysis in R. R package version 3.5–7, 2023. https://CRAN.R-project.org/package=survival.
- 41.Rao M, Oh K, Moffitt R, et al. Comparative single-cell RNA sequencing (scRNA-seq) reveals liver metastasis-specific targets in a patient with small intestinal neuroendocrine cancer. Cold Spring Harb Mol Case Stud. 2020. 10.1101/mcs.a004978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hao Y, Hao S, Andersen-Nissen E, et al. Integrated analysis of multimodal single-cell data. Cell. 2021;184(13):3573-3587.e3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang L, Li Z, Skrzypczynska KM, et al. Single-cell analyses inform mechanisms of myeloid-targeted therapies in colon cancer. Cell. 2020;181(2):442–459429. [DOI] [PubMed] [Google Scholar]
- 44.Ju Y, Fang S, Liu L, et al. The function of the ELF3 gene and its mechanism in cancers. Life Sci. 2024;346: 122637. [DOI] [PubMed] [Google Scholar]
- 45.Luk IY, Reehorst CM, Mariadason JM. ELF3, ELF5, EHF and SPDEF transcription factors in tissue homeostasis and cancer. Molecules. 2018. 10.3390/molecules23092191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Horie M, Tanaka H, Suzuki M, et al. An integrative epigenomic approach identifies ELF3 as an oncogenic regulator in ASCL1-positive neuroendocrine carcinoma. Cancer Sci. 2023;114(6):2596–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lu TONG, Shan LI, Zheng P. Expression of ELF3 in cervical cancer and its effect on the proliferation of HeLa cells. J Shanxi Med Univ. 2022;53(8):941–9. [Google Scholar]
- 48.Ojesina AI, Lichtenstein L, Freeman SS, et al. Landscape of genomic alterations in cervical carcinomas. Nature. 2014;506(7488):371–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bedard MC, Chihanga T, Carlile A, et al. Single cell transcriptomic analysis of HPV16-infected epithelium identifies a keratinocyte subpopulation implicated in cancer. Nat Commun. 2023;14(1):1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang Y, Qiu H, Lin R, et al. Advancements in the understanding of small-cell neuroendocrine cervical cancer: where we stand and what lies ahead. J Pers Med. 2024. 10.3390/jpm14050462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu Y, Li M, Liu Y, et al. Liquid-based cytology of small cell carcinoma of the cervix: a multicenter retrospective study. Onco Targets Ther. 2024;17:557–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Laskou A, Znalesniak EB, Harder S, et al. Different forms of TFF3 in the human endocervix, including a complex with IgG Fc binding protein (FCGBP), and further aspects of the cervico-vaginal innate immune barrier. Int J Mol Sci. 2024. 10.3390/ijms25042287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fina E, Cleris L, Dugo M, et al. Gene signatures of circulating breast cancer cell models are a source of novel molecular determinants of metastasis and improve circulating tumor cell detection in patients. J Exp Clin Cancer Res. 2022;41(1):78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lorenz A, Lenkiewicz S, Kozlowski M, et al. Neuroendocrine neoplasms of the gastrointestinal tract versus neuroendocrine neoplasms of the gynaecological tract-comparison of the risk factors and non-surgical treatment efficacy. Int J Mol Sci. 2023. 10.3390/ijms24076853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Oberg K, Califano A, Strosberg JR, et al. A meta-analysis of the accuracy of a neuroendocrine tumor mRNA genomic biomarker (NETest) in blood. Ann Oncol. 2020;31(2):202–12. [DOI] [PubMed] [Google Scholar]
- 56.Ichikawa Y, Kobayashi N, Takano S, et al. Neuroendocrine tumor theranostics. Cancer Sci. 2022;113(6):1930–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kim G, Kim M, Nam EJ, et al. Application of small cell lung cancer molecular subtyping markers to small cell neuroendocrine carcinoma of the cervix: NEUROD1 as a poor prognostic factor. Am J Surg Pathol. 2024;48(3):364–72. [DOI] [PubMed] [Google Scholar]
- 58.Ouadah Y, Rojas ER, Riordan DP, et al. Rare pulmonary neuroendocrine cells are stem cells regulated by Rb, p53, and Notch. Cell. 2019;179(2):403–416423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kaur S, Roberts DD. Emerging functions of thrombospondin-1 in immunity. Semin Cell Dev Biol. 2024;155(Pt B):22–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kaur S, Bronson SM, Pal-Nath D, et al. Functions of thrombospondin-1 in the tumor microenvironment. Int J Mol Sci. 2021. 10.3390/ijms22094570. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data and scripts supporting the findings of this study will be provided during review. All data in the study are available from the corresponding author upon reasonable request.
The datasets analysed during the current study are available at the GEO database under the accession code (GSM5870250, GSM5870256, GSM5870258, GSM4159164, GSM4159165). The raw sequencing data generated in this study have been deposited in the Genome Sequence Archive (GSA) by CNCB under the BioProject accession number PRJCA033235.
R scripts for analyzing data are available upon reasonable request.