Abstract
The only cure of HIV has been achieved in a small number of people who received a hematopoietic stem cell transplant (HSCT) comprising allogeneic cells carrying a rare, naturally occurring, homozygous deletion in the CCR5 gene. The rarity of the mutation and the significant morbidity and mortality of such allogeneic transplants precludes widespread adoption of this HIV cure. Here, we show the application of CRISPR/Cas9 to achieve >90% CCR5 editing in human, mobilized hematopoietic stem progenitor cells (HSPC), resulting in a transplant that undergoes normal hematopoiesis, produces CCR5 null T cells, and renders xenograft mice refractory to HIV infection. Titration studies transplanting decreasing frequencies of CCR5 edited HSPCs demonstrate that <90% CCR5 editing confers decreasing protective benefit that becomes negligible between 54% and 26%. Our study demonstrates the feasibility of using CRISPR/Cas9/RNP to produce an HSPC transplant with high frequency CCR5 editing that is refractory to HIV replication. These results raise the potential of using CRISPR/Cas9 to produce a curative autologous HSCT and bring us closer to the development of a cure for HIV infection.
Subject terms: HIV infections, CRISPR-Cas9 genome editing, Retrovirus, Translational research
Mutations of CCR5 have been linked to improved outcome to HIV infection. Here the authors use CRISPR-Cas9 genome editing of CCR5 in mobilized hematopoietic stem progenitor cells and show protection in a murine xenograft HIV infection model.
Introduction
Although highly effective in managing disease, modern antiretroviral therapy (ART) for Human Immunodeficiency Virus (HIV) infection is not curative1–5, thus warranting the continued effort to develop strategies to achieve a functional cure for HIV infection6,7. The long-term remission of HIV achieved in the “Berlin”, “London”, and “Düsseldorf” patients following allogeneic, CCR5Δ32/Δ32 hematopoietic stem cell transplant (HSCT) demonstrates that an HIV cure is possible8–10. The complex nature of their clinical histories and treatment regimens has made it challenging to identify the minimum intervention(s) required for durable prevention of HIV rebound, but several studies have demonstrated that pre-transplant conditioning and full allogeneic donor chimerism11, post-transplant graft versus host response12,13, and low-frequency disruption of the CCR5 gene in transplanted hematopoietic stem and progenitor cells (HSPCs)14 are themselves insufficient to achieve a lasting cure15. These studies and others suggest that the critical element of an HIV cure is the transplant with CCR5Δ32/Δ32 HSPCs and the resultant hematopoietic reconstitution of an immune system dominated by HIV-resistant CD4+ T cells. Unfortunately, the low frequency of CCR5Δ32/Δ32 homozygosity in the population16, the constraints of donor-recipient HLA-matching, and the morbidity and mortality associated with allogeneic HSCT combine to form a formidable barrier to the broader application of this effective approach.
The continued refinement and clinical application of genetic engineering using the CRISPR/Cas9 system provides the potential for lowering this barrier by allowing recapitulation of the CCR5Δ32/ Δ32 genotype in a patient’s own HSPCs, thereby capitalizing on the reduced clinical challenges of an autologous HSCT14,17,18. However, achieving high-frequency editing by CRISPR/Cas9 can be challenging, especially in the context of maintaining the engraftment potential and pluripotency of HSPCs19. Furthermore, the failure of allogeneic HSCT with CCR5WT/Δ32 heterozygous HSPCs to prevent post-transplant HIV rebound11 suggests that the minimum frequency of disrupted CCR5 disruption necessary to achieve a cure is high. Indeed, prior studies modeling HIV dynamics have predicted that the reduction in CCR5-expressing cells would have to approach 90% to eliminate post-ART rebound, although modest clinical benefits including delayed viral rebound and reduced set-point viral load were predicted to result from lesser reductions in the frequency of CCR5-expressing cells20.
Here, we describe reproducible, and clinically scalable (NCT03745287, NCT03655678)21, high frequency (>90%) CRISPR/Cas-mediated editing of CCR5 in cryopreserved, adult, mobilized, HSPCs that retained normal pluripotency and hematopoietic potential both in vitro and in a mouse xenograft model. This highly CCR5-edited transplant reconstituted a human immune system that contained few CCR5-expressing cells and was refractory to infection by HIV following high dose challenge with a CCR5-tropic virus. We also evaluated the protective threshold of CCR5 editing frequency by titrating the percentage of CCR5 edited HSPCs present in the stem cell transplant infusion followed by repeated HIV challenges. This study provides a pre-clinical, proof-of-concept application of CRISPR/Cas9 editing in the development of an autologous, CCR5-knockout, HSCT to reduce the barriers to more widespread application of this HIV cure strategy.
Results
Discovery pipeline identifies optimal CCR5-specific guide RNAs
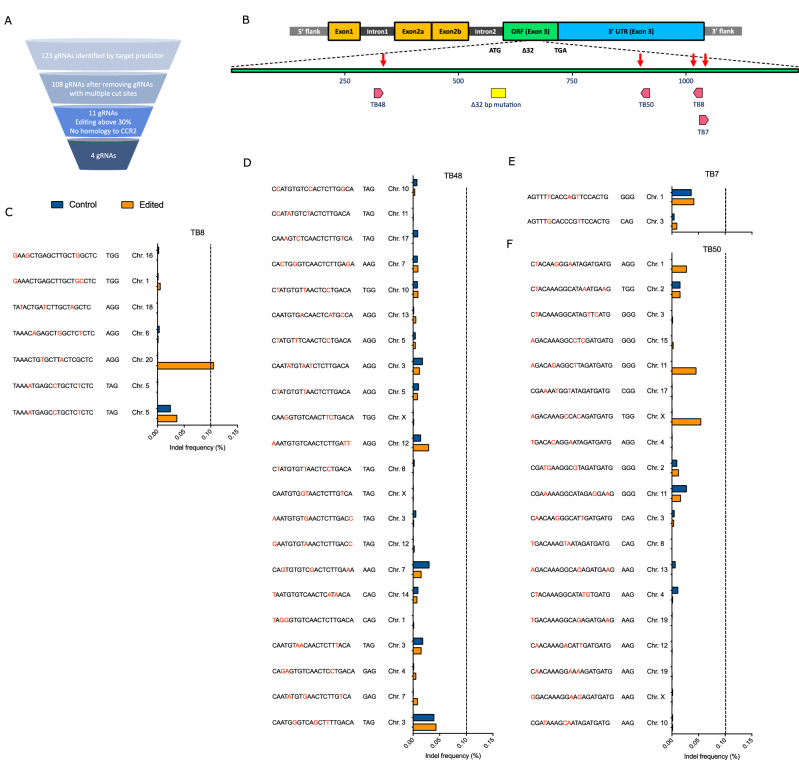
A combination of in silico prediction software and in vitro screening methods were used to identify guide RNAs (gRNAs) capable of high-frequency editing of the human CCR5 gene (Fig. 1A). In silico target prediction identified 123 gRNAs with the ability to cause double-stranded DNA breaks in the open reading frame (exon 3) of CCR5. In silico off-target site prediction excluded 15 gRNAs with multiple potential binding sites in the human genome. The remaining 108 gRNAs were transcribed in vitro and evaluated for editing efficiency when complexed with SpCas9 protein (Cas9) in adult human, mobilized, CD34+ hematopoietic stem progenitor cells (HSPCs) (Supplementary Fig. 1A). The 11 gRNAs displaying the highest editing frequency (>30%), and lacking homology to the CCR2 gene, were further evaluated in an optimized chemically synthesized format at increasing dosages of gRNA (Supplementary Fig. 1B). Four gRNAs exhibiting the highest editing frequency and lacking predicted sequence homology to other human genes (TB7, TB8, TB48, TB50) were selected for more stringent off-target editing evaluation (Fig. 1B). To evaluate off-target editing, HSPCs were either mock edited with Cas9 electroporation only or were electroporated with Cas9 complexed to each of the four gRNAs. Putative off-target gene regions containing sites with <4 base pair mismatches were amplified and deep sequenced to quantify the frequency of indel formation. Off-target editing events were rare for all four gRNAs (Fig. 1C–F), with a single instance of off-target editing observed for gRNA TB8, in which the off-target editing frequency (orange bars), but not mock editing (blue bars), exceeded the background indel threshold (set at 0.1%) (Fig. 1C). Based on the high-frequency editing of CCR5 in primary human HSPCs, and the rarity of off-target editing events, these four gRNAs were taken forward for further evaluation.
Fig. 1. Screening, location, and off-target analysis of optimal CCR5-targeting gRNAs.
A Diagram of the screening process used to identify optimal gRNAs from the initial pool of 123 gRNAs predicted to induce double-stranded breaks in CCR5 exon 3. B Genome organization of the CCR5 locus, and the relative positions of each of the 4 optimal gRNAs within the CCR5 exon 3 open reading frame and in relation to the naturally occurring Δ32 deletion. C–F CD34+ HSPCs were edited with each lead gRNA using a concentration of 150 µg/mL or mock edited. Predicted off-target sites were amplified, sequenced, and analyzed for indel formation. The blue bars represent indel frequency at the off-target sites for gRNAs (C) TB8, (D) TB48, (E) TB7, and (F) TB50 in mock edited CD34+ HSPCs. The orange bars represent indel frequency at off-target sites in CD34+ HSPCs edited with the respective gRNA and Cas9 protein. An indel frequency level of ≥0.10%, represented by the dashed line, was considered the threshold for off-target editing.
Optimal CCR5-targeting gRNAs induce robust ablation of CCR5 expression on human T cells
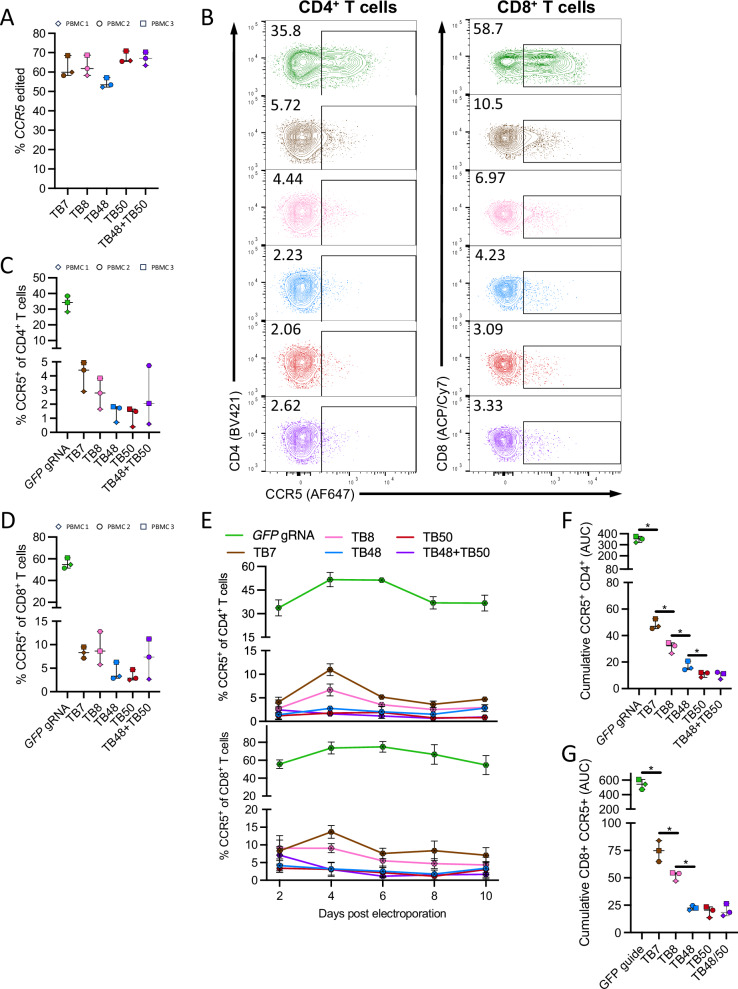
Next, we sought to confirm that CCR5 editing with the four optimal gRNAs (TB7, TB8, TB48, and TB50) could functionally impact CCR5 protein expression on the surface of human T cells. All four optimal gRNAs induced robust CCR5 gene editing in primary T cells, ranging from 52% to 70% (Fig. 2A), and all resulted in effective reduction in the frequency of both CD4+ and CD8+ T cells that exhibited cell surface CCR5 expression (CCR5+) in comparison to mock “editing” using a guide RNA specific to the green fluorescent protein gene (GFP) (Fig. 2B–D). In addition, we tested guides TB48 and TB50 in combination in a “dual guide” approach intended to cause a small gene deletion and thereby approximate the effect of the naturally occurring CCR5Δ32 mutation. We observed similarly robust CCR5 gene editing and reduction in expressed CCR5 with this “dual guide” approach (Fig. 2A–D; purple). The impact on the frequency of CCR5+ cells was maintained over 10 days in culture, with no apparent outgrowth of unedited cells or alteration in CCR5 expression among unedited cells (Fig. 2E). Importantly, quantification of the cumulative reduction in the frequency of CCR5+ cells over time by area under the curve (AUC) analysis revealed a clear hierarchy in gRNA potency that was not observed by simply quantifying the percent of gene editing, with TB48, TB50, and the TB48 + TB50 dual guides demonstrating superior reduction in the frequency of both CCR5+ CD4+ and CD8+ T cells compared to either the TB7 or TB8 gRNA (Fig. 2F, G).
Fig. 2. Robust and durable ablation of CCR5 surface expression in primary human T cells.
A–F Human PBMCs from 3 donors were stimulated with phytohemagglutinin (PHA) for 3 days, electroporated with either a “mock” non-specific gRNA targeting GFP (green), single gRNAs targeting CCR5 (TB7-pink, TB8-brown, TB48-blue, TB50-red), or a dual gRNA approach comprising TB48 + TB50 (purple). Each symbol represents a distinct donor. A 48-h post electroporation, gene editing was measured by Sanger sequencing of CCR5 amplified from genomic DNA. B FACS plots depict CCR5 surface expression on mock edited (GFP gRNA) or CCR5 edited CD4+ (left) and CD8+ (right) T cells in a representative donor 48-h post electroporation. CCR5 expression levels 48-h after electroporation with various gRNAs on (C) CD4+ and (D) CD8+ T cells. E Longitudinal expression of CCR5 in vitro on CD4+ (top) and CD8+ (bottom) T cells up to 10 days post electroporation. Data from 3 independent PBMC donors are averaged, and error bars show ± SD. Area-under-the-curve (AUC) analysis of total CCR5 expression on (F) CD4+ and (G) CD8+ T cells throughout 10 days in culture. Each data point represents the average of 3 technical replicates, and each data point represents a distinct biological replicate using different donor PBMCs. Error bars denote minimum to maximum values. *denotes p < 0.05 and statistics generated from the Mann–Whitney test and p values are one-tailed.
CCR5-editing confers HIV resistance to human CD4± T cells in vitro
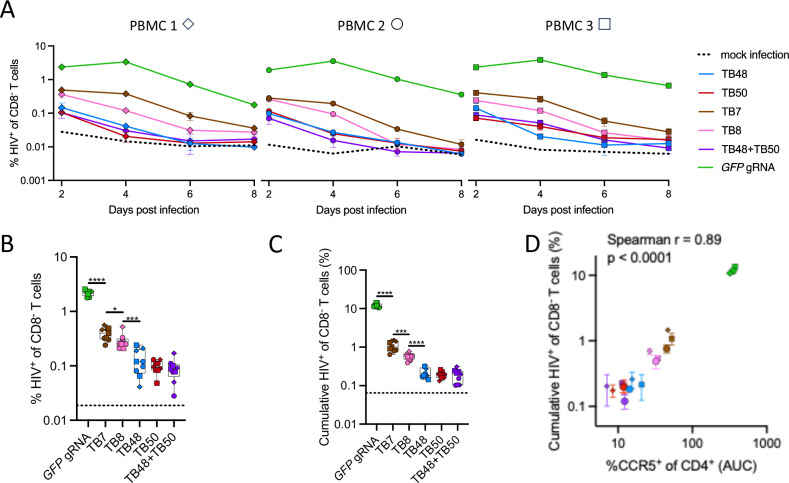
As a validation of this approach to prevent HIV replication, we quantified the impact of CCR5 editing on the susceptibility of CD4+ T cells to HIV infection. After editing by Cas9/gRNA electroporation with either the CCR5-specific gRNAs or the mock GFP-specific gRNA, stimulated human peripheral blood mononuclear cells (PBMCs) from 3 independent donors were challenged with a high dose of CCR5-tropic HIVJRCSF. In contrast to the mock GFP edited CD4+ T cells, all CCR5 edited CD4+ T cells exhibited a reduced frequency of infection that was sustained through the course of the assay (Fig. 3A). Notably, in two of the three donors tested, CCR5 editing with the TB48, TB50, and TB48 + TB50 dual guides resulted in decay of the HIV infection to that of mock infected control cells by 6 days post challenge (Fig. 3A). Indeed, both comparison of initial frequency of HIV infected cells at 2 days post challenge (Fig. 3B) and calculation of the cumulative burden of infection (days 2–8) by area under the curve (AUC) analysis (Fig. 3C) demonstrate a significantly reduced frequency of HIV infection in the TB48, TB50, or TB48 + TB50 dual guide edited cells compared to those edited with gRNAs TB7 or TB8. As expected, the reduction in HIV infection conferred by different gRNAs was well correlated with the reduction in frequency of CCR5+ CD4 + T cells (Fig. 3D). These results validated the potential of this CCR5 editing approach to confer resistance to HIV infection to human CD4+ T cells and indicated that the TB48 + TB50 dual guide approach merited further evaluation.
Fig. 3. CCR5 ablation in primary CD4+ T cells prevents HIV spread in vitro.
PBMCs from 3 healthy adult donors were stimulated with PHA for 3 days, electroporated with indicated gRNAs complexed with Cas9, and challenged with HIVJRCSF at a 0.5 multiplicity of infection 2 days after electroporation and in triplicate for each donor. HIV spread in culture was assessed at 2-day intervals by intracellular staining for Gag p24 antigen. A Kinetics of HIV spread following HIVJRCSF infection of either mock edited (GFP gRNA, green) or CCR5 edited CD4+ T cells. Error bars represent ± SD for 3 technical replicates. B A box and whisker plot depicts the average initial infection rate 2 days after infection for all biological and technical replicates (n = 9) for each gRNA. Error bars denote minimum and maximum values, the line denotes median, and box bounds the 25th and 75th percentiles. C A box and whisker plot depicts the cumulative infection rate calculated as the area-under-the-curve (AUC) for all biological and technical replicates (n = 9) for each gRNA. Error bars denote minimum and maximum values, the line denotes median, and box bounds the 25th and 75th percentiles. Dotted horizontal lines represent the average background Gag p24 staining in mock-infected CD4+ T cells. D Plot depicting the association between the log10-transformed total AUC percentage of CCR5+ CD4+ T cells and the log10-transformed total AUC percentage of p24+ CD8- T cells per PBMC donor and gRNA condition. Different symbols represent distinct PBMC donors and represent the mean of 3 technical replicates. Error bars denote ± SD. B, C ****denotes p < 0.0001, ***denotes p < 0.001, **denotes p < 0.01, *denotes p < 0.05. Statistics were generated from the Mann-Whitney test and p values are two-tailed. D Statistics were generated from a two-tailed Spearman rank correlation (p < 0.0001).
CCR5 editing of human CD34± HSPCs does not impair long-term engraftment or hematopoiesis
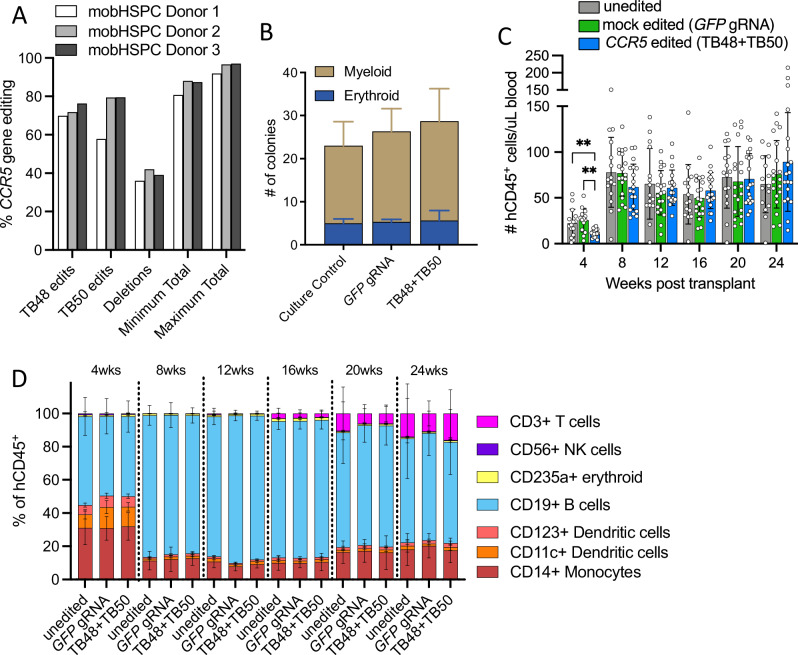
To assess the impact of high-frequency CCR5 editing on HSPC viability and pluripotency, mobilized HSCs from three healthy adult human donors were electroporated with the TB48 and TB50 gRNAs (dual guide) complexed with Cas9. Two days after electroporation, the frequencies of indel editing caused by each single gRNA and of larger deletions caused by synchronous dual gRNA editing were quantified in genomic DNA and used to calculate total CCR5 editing, which ranged from 91% to 97% at maximum across the 3 donors (Fig. 4A). Cell viability 48 h post electroporation was comparable among editing conditions, and the percent recovery was greater than 95% for all donors (Supplementary Table 1). The post-editing colony formation and lineage potential of each of the donor HSPCs was characterized in a single-cell methylcellulose assay, and we observed no significant differences in either the proportion of lineages or the absolute number of colonies between mock edited (GFP gRNA) and TB48 + TB50 dual gRNA CCR5 edited HSPCs (Fig. 4B). We next assessed the long-term engraftment and hematopoiesis potential of edited HSPCs by xenograft transplant into immunocompromised mice (NBSGW) of either 1 × 106 freshly thawed/uncultured HSPCs, mock edited (GFP gRNA) HSPCs, or the TB48 + TB50 dual gRNA CCR5 edited HSPCs. Although the mice transplanted with CCR5 edited HSPCs exhibited fewer total human CD45+ cells in the blood at 1-month post-transplant than the mock edited and untouched control mice, the difference resolved and no significant differences in total human CD45+ cell counts were observed between experimental groups at any subsequent time points (Fig. 4C). Importantly, the timing and proportion of human cells of erythroid, myeloid, and lymphoid lineages were comparable between mice transplanted with untouched, mock edited (GFP gRNA), or CCR5 edited HSPCs, suggesting that hematopoietic potential was unaltered by either the electroporation process or the high-frequency editing of CCR5 (Fig. 4D).
Fig. 4. CCR5 editing in human HSPCs does not significantly alter longitudinal engraftment or hematopoiesis.
A Mobilized CD34+ HSPCs derived from three healthy adult human donors were electroporated with Cas9 complexed to gRNAs TB48 and TB50. Single gRNA gene disruptions were measured by Sanger sequencing of CCR5 amplified from genomic DNA isolated 48-h after electroporation. Dual gRNA deletions were measured via droplet-digital PCR. Minimum and maximum editing was calculated using single and dual gRNA editing frequencies (see Methods for calculation details). B Number and type of colonies derived from 3 donor HSPCs for each condition culture control, mock edited (GFP gRNA), and CCR5 edited (TB48 + TB50 dual gRNA) cultured in a methylcellulose-based colony forming unit assay. Bars represent the mean and error bars represent the standard deviation for the number of colonies of each type for the 3 distinct HSPC donors. C, D The absolute number and lineage distribution of human CD45+ cells in the blood were measured at 1-month intervals by multiparameter flow cytometry in NBSGW immunodeficient mice engrafted with 1 × 106 untouched (n = 15), mock edited (GFP gRNA) (n = 19), or CCR5 edited (n = 19) HSPCs. C The absolute number of human CD45+ cells per uL of blood in NBSGW mice engrafted with untouched (gray bars), mock edited (green bars), or CCR5 edited (blue bars) HSPCs. Bars represent mean and error bars denote ± SD. Statistics generated from the Student’s t-test and p values are two-tailed. **denotes p < 0.01. D Stacked bar graphs depict the average frequency of distinct human hematopoietic cell lineages in mice transplanted with untouched, mock edited, or CCR5 edited HSPCs at monthly intervals. Error bars represent the standard deviation in each lineage frequency.
Stable engraftment of a CCR5-deficient human hematopoietic xenograft
Next, we assessed the stability and functional impact of CCR5 editing in transplanted HSPCs. Here we quantified the longitudinal, post-transplant frequency of CCR5+ human myeloid and T cell subsets, the cells that naturally express CCR5, in xenograft mice transplanted with >90% CCR5 edited HSPCs. The frequency of myeloid (CD33+) cells and T cells (CD3+) expressing CCR5 was found to be negligible compared to mice transplanted with untouched and mock GFP edited HSPCs (Fig. 5A). Overall, the frequency of myeloid lineage cells expressing surface CCR5 was <2% on average across the entire 6-month post-transplant follow-up, which was significantly lower than the ~30% of CCR5+ cells observed in control mice (Fig. 5B). Importantly, T cells (CD3+), which develop at later stages post-transplant but are the critical target cell of HIV, exhibited similarly negligible frequencies of CCR5 expression in the mice transplanted with CCR5 edited HSPCs (Fig. 5C). Consistent results were observed in a second, independent xenograft study comprising editing and transplant of human, adult mobilized HSPCs from a different anonymous, healthy donor (Supplementary Fig. 2). Collectively, these results support that the transplant of human HSPCs highly edited at CCR5 using the dual gRNA approach confer a human hematopoietic graft that is virtually devoid of CCR5 expression, thereby replicating, in principle, a CCR5Δ32/Δ32 HSCT.
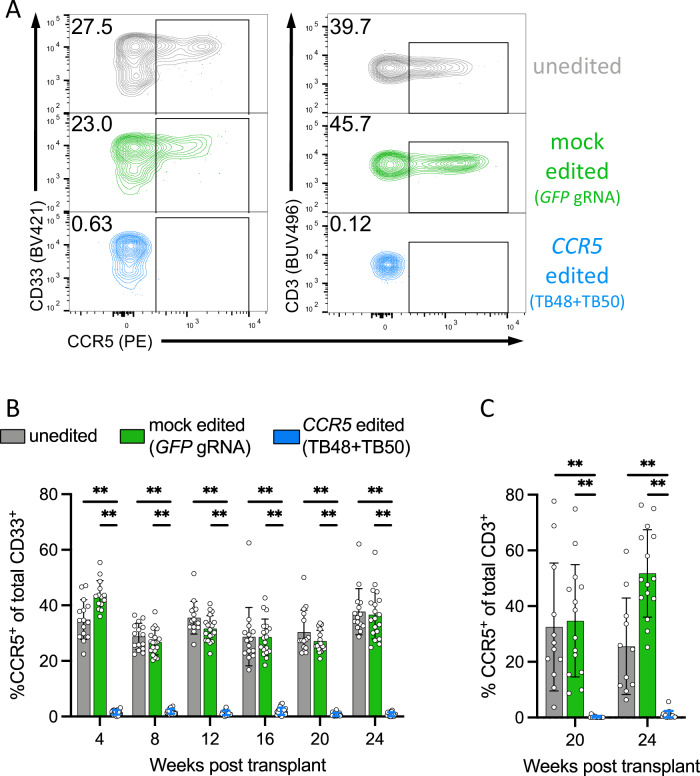
Fig. 5. Robust CCR5 editing in HSPCs is stable in vivo and gives rise to differentiated myeloid and lymphoid cell lineages lacking CCR5.
A Representative FACS plots demonstrating ablation of surface CCR5 protein in CD33+ myeloid cells (left panel) and CD3+/CD4+ T cells (right panel) in the peripheral blood of xenograft mice 20 weeks post-transplant. B The percentage of CCR5-expressing myeloid cells as measured by flow cytometry in the peripheral blood of mice transplanted with 1 × 106 untouched (n = 15, gray bars), mock edited (GFP gRNA) (n = 19, green bars), or dual gRNA CCR5 edited (n = 19) HSPCs at monthly intervals. **denotes p < 0.01 and statistics generated from the Student’s t-test and p values are two-tailed. Bars represent mean and error bars denote ± SD. C The percentage of CCR5-expressing CD3+ T cells at 20 and 24 weeks post-transplant in xenograft mice with appreciable human T cell numbers from different HSPC transplant groups. **denotes p < 0.01 and statistics generated from the Student’s t-test and p values are two-tailed. Bars represent mean and error bars denote ± SD.
CCR5 edited xenograft mice are refractory to HIV infection
The significant reduction of CCR5 expressing CD4+ T cells in the mice transplanted with TB48 + TB50 dual gRNA CCR5 edited HSPCs suggested that such a transplant may be resistant to HIV infection. To assess this, we challenged mice with a high dose of CCR5-tropic HIVJRCSF and compared the outcome in CCR5-edited mice to mock-edited (GFP gRNA) control mice. All mock edited control mice were infected by a single HIV challenge and exhibited substantial viremia of >103 HIV RNA copies/mL of plasma at 2-weeks post challenge (Fig. 6A). In contrast, the CCR5 edited mice resisted HIV infection, including a follow-up challenge conducted at a 5-fold higher dose (Fig. 6A). The sustained viremia in control mice was associated with the rapid decline of peripheral human CD4+ T cells, whereas CCR5 edited mice showed no such loss of these target cells of HIV (Fig. 6B). We did not find any evidence of non-productive or low-level HIV infection in the CCR5 edited mice by digital droplet PCR (ddPCR) specific for integrated HIV DNA in human cells derived from lymph node tissue harvested at necropsy 5 weeks post challenge (Fig. 6C). Finally, to confirm that the protective effect was specific to the CCR5 editing and not a byproduct of the editing process, we challenged a CCR5 edited mouse that had resisted both prior HIVJRCSF challenges with the CCR5-independent strain HIVNL43 that utilizes the alternative HIV co-receptor CXCR4. The efficient infection and robust HIV viremia in this mouse supports that the protection against HIV infection in CCR5 edited mice was specific to the ablation of CCR5 expression (Fig. 6D). These results demonstrate that a human HSPC transplant exhibiting high-frequency CCR5 editing can effectively reconstitute peripheral and tissue-resident CD4+ cells that are refractory to infection by CCR5-tropic HIV.
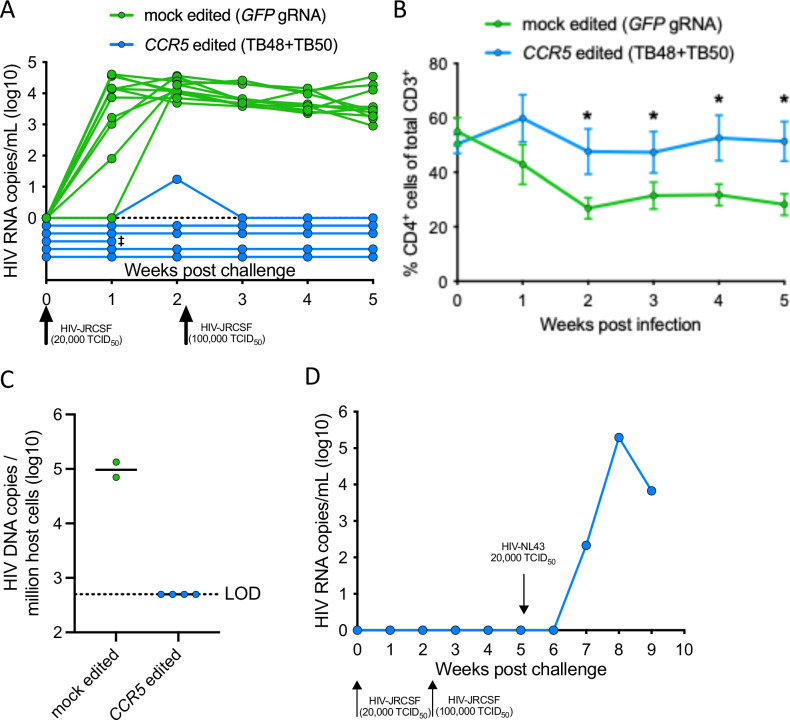
Fig. 6. Xenograft transplant with HSPCs exhibiting high-frequency CCR5 editing confers resistance to CCR5-tropic HIV infection.
NSG mice were transplanted with either 1 × 106 mock edited (GFP gRNA) or CCR5 edited (TB48 + TB50 dual gRNA) HSPCs following sublethal irradiation of 200 cGy. 24 weeks after transplant, mice demonstrating sufficient human T cell reconstitution (>10 CD4+ T cells/uL of blood) were challenged intraperitoneally with 20,000 TCID50 of CCR5-tropic HIVJRCSF. Viral loads and CD4+ T cell counts were assessed weekly by qRT-PCR and flow cytometry, respectively. Following two negative viral loads, uninfected mice were re-challenged with 100,000 TCID50 HIVJRCSF. A Viral load in individual mock edited (n = 11, green) and CCR5 edited (n = 6, blue) mice via qRT-PCR from weekly blood draws following HIVJRCSF challenge. ‡denotes animal euthanized prior to experimental endpoint due to health concerns. B Average frequency of human CD4+ T cells within total T cells in the blood of mock edited (green, n = 11) and CCR5 edited (blue, n = 5) mice following HIVJRCSF challenge. *denotes p < 0.05 and statistics are generated from a one-tailed Student’s t-test. Error bars represent ± SEM. C Log10-transformed number of HIV DNA copies per 1 × 106 human cells as measured in genomic DNA extracted from pooled lymph nodes by a multiplexed droplet-digital PCR assay. The limit of detection (LOD) for the assay is set as the threshold to detect a single HIV DNA copy in the total number of human cells assayed per mouse. Lines represent the mean. D A single CCR5 edited humanized mouse that was resistant to both HIVJRCSF challenges was subsequently challenged with 20,000 TCID50 of HIVNL4-3 via the intraperitoneal route and log10-transformed HIV RNA copies per mL are plotted.
Dose effect of CCR5 edited HSPCs on HIV acquisition and pathogenesis
In the above study, mice receiving a transplant of >90% CCR5 edited human HSPCs developed a human immune graft that was completely protected from CCR5-tropic HIV challenge. However, in a clinical setting, it is possible that lower frequencies of CCR5 editing and/or engraftment may occur. To evaluate the protective benefit that might still be garnered by lower CCR5 editing, we transplanted immunodeficient mice with decreasing numbers of CCR5 edited HSPCs that were mixed with mock edited (GFP gRNA) HSPCs to create the transplant. In this study, NBSGW mice were used to improve life span of the mice as the mutation in c-kit obviates the need for intense whole-body irradiation22, and can also improve the engraftment of human HSCs23. Five transplant groups were created that received either a full (100%) dose of CCR5 edited cells, a 75% dose, a 50% dose, a 25% dose, or a 0% dose comprising mock edited cells only (control mice) (Fig. 7A). CCR5 editing quantification in human, non-T cell, bone marrow cells taken at necropsy revealed that although there was intragroup variability, this “dosing” of transplanted CCR5 edited HSPCs resulted in transplant groups with mean actual CCR5 editing frequencies of 26%, 54%, 69%, and 96% (Fig. 7B). Further, longitudinal CCR5 expression on CD33+ myeloid cells (Fig. 7C) and on T cells (Fig. 7D) in each group as measured in the peripheral blood was consistent with the transplant dose of CCR5 edited HSPCs.
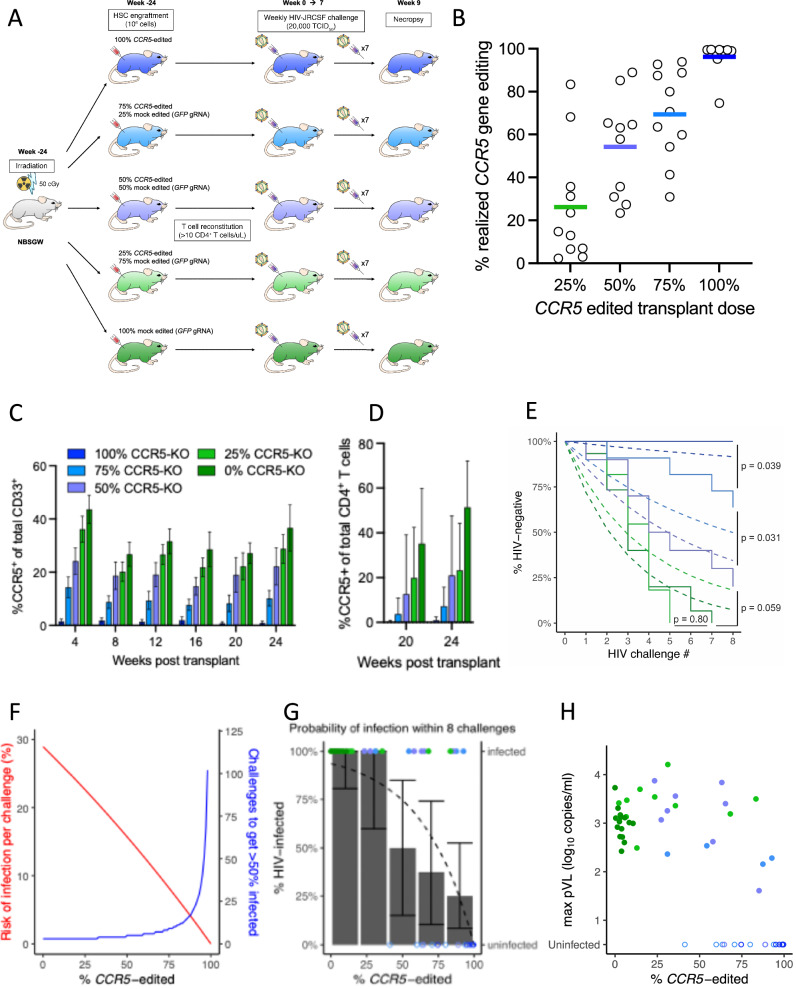
Fig. 7. Frequency of CCR5 editing determines risk of HIV infection.
A NBSGW mice were transplanted with 1 × 106 HSPCs in five “doses” of CCR5 edited HSPCs with the balance of HSPCs comprising mock edited (GFP gRNA) cells ranging from 100% to 0%. Animals were monitored for frequency of CCR5 expression on relevant cell populations via flow cytometry at 1-month intervals for 6 months, then challenged weekly with 20,000 TCID50 of HIVJRCSF via the intraperitoneal route for 8 weeks. B CD19+ B cells were isolated from the bone marrow of HIV-challenged mice at necropsy and CCR5 gene editing in the human graft was quantified for each CCR5 edited HSPC transplant dose. Group means indicated by horizontal lines. C CCR5 expression levels in human CD33+ myeloid cells at monthly intervals following transplant for each HSPC titration group (n = 19 mice per group) and before HIV challenge. Bars represent mean and error bars denote ± SD. D CCR5 expression levels in CD3+CD4+ T cells 20 and 24 weeks after transplant and before HIV challenge (n = 15 mice per group). Bars represent mean and error bars denote + SD. E Plasma viral loads were monitored weekly for all mice via qRT-PCR. Kaplan–Meier curves for each study arm (solid lines) and corresponding expected survival curves based on the best fit mechanistic model assuming the “barrier at inoculation” mechanism (dashed curves). Statistics were generated by comparing each pair of transplant groups using the log-rank test. F Mechanistic model of the risk of infection per HIV challenge (red line) and the corresponding number of inoculations required to infect 50% of animals (blue line) as functions of the quantified frequency of CCR5 editing. G Observed percentage of animals infected after 8 HIV challenges for each quintile of frequency of CCR5 editing (bars with 95% confidence intervals) and the expected percentage of infection (dashed curve) based on the model in (F). Infection status of each animal after 8 challenges (solid or open points for infected or uninfected, respectively) indicated by the right-hand vertical axis. H Maximum viral load vs. %CCR5 edited.
These 5 groups of mice were then challenged weekly with high-dose, HIVJRCSF (CCR5-tropic) for 8 weeks to determine the protective benefit against HIV infection of the different doses of transplanted CCR5 edited cells (Fig. 7A). The group that had the lowest mean realized CCR5 editing (26%) was as susceptible to HIV infection as the controls (0% CCR5 editing) with all mice becoming infected. However, the protective benefit of CCR5 editing increased with transplant dose, and as in the first study, none of the mice in the highest editing group (96% editing) was infected after 8 consecutive challenges, supporting the potential for complete protection. The survival curves (Fig. 7E, solid lines) were used to mathematically quantify the HIV infection risk for each transplant group. From this, the lowest level of CCR5 editing (26%) exhibited an HIV infection risk of 0.28 per challenge dose, no different from the baseline risk of infection in the absence of any CCR5 editing (0.28 per challenge dose). The next group of mice with modest levels of CCR5 editing (54%) had a reduced infection risk of 0.16 per challenge dose (45% reduction in risk), albeit not statistically significant. However, the last two groups of mice with the highest levels of CCR5 editing (69% and 96%) conferred a statistically significant protective benefit with 0.051 risk per challenge (82% reduction) and 0 risk per challenge (100% reduction), respectively.
Next, we considered the HIV infection data and distribution of CCR5 editing across all mice, regardless of transplant dose group, and developed a mechanistic model (see Supplementary Text S1.5 for details) to estimate the risk of infection per challenge as a function of the frequency of CCR5 editing (Fig. 7F, red curve). To demonstrate the implication of decreasing risk of infection, the number of inoculations per animal required to obtain infection in 50% of animals is plotted in blue. Based on the model fit here, greater than 100 inoculations would be required for infection in animals with greater than 98% CCR5-gene-disruption (Fig. 7F).
Given the heterogeneity in actual %CCR5-gene-disruption among animals within a study arm, it may be more illustrative to consider animal-specific probabilities of infection than study arm survival curves. Thus, we binned animals into quintiles of actual CCR5 editing frequency. As expected, independent of transplant arm, the fraction of mice infected (bars) was highest in the two lowest quintiles of actual CCR5 editing frequency (0–20% and 21–40%) and decreased across the three higher quintiles (41–60%, 61–80%, 81–100 (Fig. 7G)). The model estimated probability of infection at the end of 8 challenges is shown by the black curve.
In addition to protection from HIV infection, we considered the benefit that a reduction in CCR5-expressing cells could have on viral load. We have previously predicted that increasing the proportion of HIV-resistant CD4+ T cells, e.g., CCR5 negative, would ultimately result in the target cell frequency dropping below the level required to sustain HIV dissemination, i.e., causing the basic reproductive number to drop below one, R0 < 120. Hence, the animal would be protected from long-term HIV infection even if one or more cells were infected by the inoculation. We refer to this potentially protective mechanism as a “barrier at dissemination,” as opposed to the assumption (as in the model presented above) that infection is purely determined by if the inoculation infects one or more target cells, i.e., a “barrier at inoculation.” To investigate if the barrier at dissemination is relevant in the current study, we analyzed the relationship between the actual frequency of CCR5 editing and the peak viral load (Fig. 7H). This showed a trend to lower peak VL at higher levels of CCR5 gene disruption and an increasing number of uninfected animals at high gene disruption. However, when we expanded our mechanistic model of infection probability to incorporate the barrier at dissemination mechanism (and the resultant CCR5 editing threshold above which sustained infection cannot occur), this did not improve the fit (unsurprisingly, since so few animals are expected to be infected at high CCR5-KO in either model) (see Supplementary Text S1.6).
Discussion
The efficacy of HSCT with CCR5Δ32/ Δ32 HSPCs in the Berlin Patient and several subsequent individuals provided powerful proof of concept for an HIV cure, but the challenges of allogeneic transplant with a low-frequency genetic variant pose a major hurdle to more widespread adoption of this approach15,16. In this study, we explored the possibility that CRISPR/Cas9 editing of CCR5 in mobilized CD34+ HSPCs could replicate the protective value of naturally occurring CCR5Δ32/ Δ32 allogeneic donors, thereby providing an autologous HSCT approach that could be more widely applied. Our comprehensive screen identified two guide RNAs with efficient on-target editing of the human CCR5 gene with no detectable off-target editing in related gene regions. Using these two guide RNAs, we achieved high frequency (>90%) editing of CCR5 in mobilized CD34+ HSPCs that engrafted with no notable deficit in hematopoietic potential enabling reconstitution of a human immune graft that was refractory to infection despite repeat high dose HIV challenges. These results support a cure strategy that leverages CRISPR/Cas9 editing to replicate the successes of the Berlin, London, and Dusseldorf patients8–10 towards a safer and more tractable, autologous HSCT approach.
The finding that it is possible to achieve a sufficiently high frequency of CCR5 editing of human HSPCs to replicate the protective benefit of the CCR5Δ32/ Δ32 homozygous genotype, and thereby prevent productive infection despite repeated HIV challenges, is a critical proof of concept for the application of gene editing towards a functional cure for HIV. In addition, the dose of delivered CCR5 edited HSPCs well reflected the average actual level of CCR5 editing in the graft (Fig. 7B). This suggested there were no long-term deficits in engraftment potential of the edited versus unedited HSPCs in our model system, which is a frequent concern for edited HSPCs24,25. However, we did observe a transient reduction in human cell engraftment in HSPC groups with high levels of editing (Fig. 4C), suggesting that high frequencies of editing and subsequent DNA repair mechanisms may have impeded engraftment of a subset of edited HSPCs. Notably, this difference in engraftment resolved by 8 weeks post-transplant, suggesting that long-term progenitors were largely unaffected by high-frequency editing and further supporting that high levels of CCR5 editing did not disrupt long-term engraftment potential. However, in the transplants comprising mixtures of high-frequency edited and unedited HSPCs, we observed a range in the measured frequency of CCR5 editing in the resulting human graft for each CCR5 edited HSPC transplant dose (Fig. 7B), and this warrants follow-up studies to identify factors that may influence HSPC engraftment potential. Similarly, we observed no reduction in lineage potential of edited HSPCs, which would otherwise represent a significant hurdle to achieving high-frequency peripheral editing in the clinic. Perhaps equally important, our experimental titration of the dose of CCR5 edited HSPCs in the transplant provides the opportunity to explore the minimum CCR5 editing frequency necessary to confer a protective benefit. Our results suggest that a frequency of CCR5 editing in the mature human graft of >90% is necessary for complete abrogation of productive HIV infection, although we did observe some reduced risk of infection even at lower CCR5 editing frequencies. Notably, the lack of protective benefit that we observed at the lowest quintiles of editing frequencies (Fig. 7G) is consistent with the outcome of prior studies of HSCT with low-frequency CCR5 edited HSPCs in immunodeficient mice26, non-human primates27,28, and humans14.
The characterization of HIV infection across a range of CCR5 editing frequencies in this study also provided the opportunity to model how an HSCT-based treatment at varying CCR5 editing frequencies could impact the risk of HIV reactivation following post-transplant cessation of ART. First, to model the relationship between frequency of CCR5 editing and risk of post-ART reactivation of HIV infection, we consider reactivations of latent HIV provirus to be analogous to sequential infectious challenges, as used in this study. That is, we assume CCR5 editing will reduce the risk of successful infection following HIV reactivation from latency in humans in a similar manner to the observed reduction of infection from HIV challenge in the xenograft mice (Barrier at Inoculation; illustrated in red panel of Fig. 8A). We previously estimated that the frequency of successful HIV reactivation from latency was around once a week29. Therefore, we can estimate the impact of CCR5 editing frequency on post-transplant time-to-rebound following cessation of ART (red line in Fig. 8B). We estimate that a 90% CCR5 editing frequency in the transplant would delay rebound post-ART interruption by less than a week and that an editing frequency of greater than 98% would be required to achieve a 2-month delay (see Supplementary Text S1.8 for details). However, modeling predicts that the relative frequency of CCR5-null CD4+ T cells not only impacts the “binary” parameter of “infected” vs. “not infected” measured in this study, but also the subsequent steady-state, or set-point viral load that results from infection. By reducing the frequency of available target cells for HIV, CCR5 editing is predicted to also lower the set-point viral load (Barrier at Dissemination; illustrated in blue panel of Fig. 8A)20. Based on published estimates of HIV replication capacity (R0) in humans, we estimate the fold change in set-point viral load in response to the frequency of CCR5 editing (Fig. 8B blue line; see Supplementary Text S1.8 for details). Of particular note, we have previously predicted that a CCR5 editing frequency of 87.5% would reduce the effective target cell population to such an extent that the virus would not be able to replicate sufficiently to sustain a productive, viremic infection (illustrated in blue panel of Fig. 8A). This suggests that the high CCR5 editing frequency achieved in these studies may be approaching the thresholds necessary to prevent post-ART reactivating HIV from establishing a productive infection and viremic expansion, especially in the presence of contributing HIV-specific immunity which is absent in this model system.
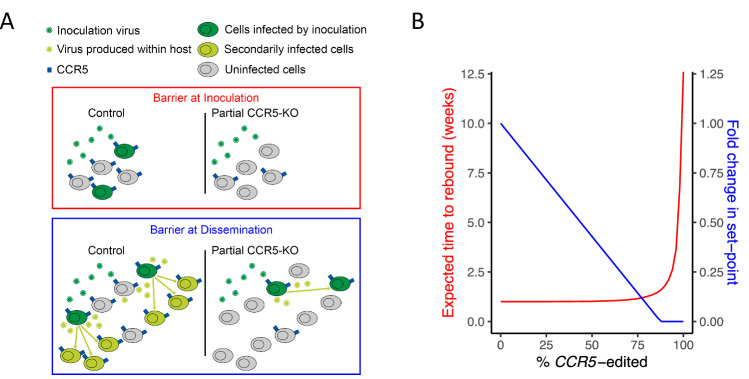
Fig. 8. Potential protective mechanisms of CCR5 editing in the human clinical context.
A Schematics of mechanisms of action by which partial CCR5 editing may confer protection from post-transplant, post-ART HIV rebound. In this study, we have shown that protection from HIV challenge is conferred by CCR5 editing in proportion to the frequency of CCR5 edited target cells. The same mechanism of reducing the available target cell concentration may also reduce the chance of successful HIV reactivation from latency during a post-transplant cessation of ART as reactivating virions will not encounter sufficient target cells (creating a “Barrier at Inoculation”; red panel). Additionally, a sub-sterilizing reduction of target cell concentration may still reduce subsequent HIV replication capacity (R0) by reducing target cell availability. If the frequency of target cells is sufficiently reduced, by virtue of a high enough frequency of CCR5 edited cells, post-ART viral replication, and viremia may be blunted (“Barrier at Dissemination”, blue panel). B Projected post-transplant, post-ART time-to-rebound (red; corresponding to “Barrier at Inoculation” mechanism) and fold change in set-point viral load (blue; corresponding to “Barrier at Dissemination” mechanism), shown as functions of theoretical frequency of CCR5 editing, based on the observed relationship between HIV infection risk and frequency of CCR5 editing in this study (see Supplementary Text S1.8 for details).
This study builds upon a growing list of efforts to achieve clinically meaningful outcomes against HIV through the targeting of CCR5, in both CD4+ T cells30,31, HSPCs14,26–28,32, and iPSCs33, with varying degrees of efficacy observed in humanized mice26,32,33, non-human primates27,28, as well as patients14,30,31. With the ever-increasing efficiency and appreciated safety profile of gene editing in the clinic, this opens the door to the potential for autologous transplants which exhibit reduced rates of complications, more rapid immune reconstitution, and lower risks of opportunistic infections due to the absence of immunosuppression requirements as compared to allogeneic HSCT with CCR5Δ32/ Δ32stem cells15. Moreover, the recent demonstration of direct in vivo HSPC editing of the β-globin gene holds promise for yet further streamlining the delivery of HSPC editing34. The clinical potential of CRISPR/Cas9 editing of autologous human HSPCs prior to HSCT is currently being tested in a series of clinical trials for the treatment of sickle cell disease and β-thalassemia (NCT03655678, NCT03745287, NCT04443907, NCT04925206, NCT04853576). In the first of these trials, an average editing frequency of approximately 80% has been associated with an effective treatment for these genetic disorders21. This provides support for the clinical potential of CRISPR/Cas9 editing of HSPCs for use in HSCT and suggests a viable pathway towards an autologous HSCT-based treatment for HIV.
Methods
Ethics statement
The animal studies in this work were conducted according to protocols that were reviewed, approved, and monitored by the Massachusetts General Hospital Institutional Animal Care and Use Committee (IACUC). The use of anonymous, adult human CD34+ cells was reviewed by the Massachusetts General Brigham Institutional Review Board (IRB) and determined to be not human subjects research.
gRNA selection and predicted off-target site identification
CRISPR Therapeutics (Boston, MA, USA) proprietary in silico predictor software was used to identify gRNAs targeting exon 3 of the CCR5 gene. The software was used to search the human genome for potential off-target sites with up to 3 nucleotide mismatches relative to each gRNA that also lies adjacent to an NNG PAM sequence.
Cell sourcing and culturing
CD34+ HSPCs used for the initial gRNA screening were isolated from leukopaks from mobilized peripheral blood collected from normal, healthy donors injected with G-CSF and Mozobil®. HSPCs used for all other experiments were purchased from AllCells (Alameda, CA, USA) and were isolated from leukopaks from mobilized peripheral blood collected from normal, healthy donors injected with G-CSF. HSPCs were cultured at a density of 750,000 cells/mL at 37 °C/5% CO2 in CellGenix (Breisgau, DE) SCGM medium supplemented with the following cytokines: SCF, FLT3L, and TPO (R&D Systems, Minneapolis, MN, USA). HSPCs were cultured for 48 h before and after electroporation. PBMCs were isolated from whole blood by density gradient centrifugation over Histopaque-1077 (Sigma-Aldrich, St. Louis, MO, USA). The mononuclear layer was washed with PBS, cryopreserved in freezing media (10% DMSO, 90% fetal bovine serum), and stored in liquid nitrogen. For experiments, PBMCs were thawed, washed, and cultured 1 × 106 cells/mL at 37 °C/5% CO2 in R10 media (RPMI 1640, 10% fetal bovine serum, 1% L-glutamine, 1% penicillin-streptomycin) supplemented with 50 U/mL of IL-2. Prior to electroporation and HIV infection assays, PBMCs were stimulated with PHA (phytohemagglutinin, Sigma-Aldrich, cat# 11249738001) for 72 h in R10 supplemented with 50 U/mL of IL-2. After 72 h of stimulation, PHA-containing media was washed out, replaced with fresh R10 supplemented with IL-2, and cultured for 24 h before electroporation.
Editing of primary human cells with RNP
Initial gRNA screening in HSPCs used gRNAs produced through in vitro transcription by Thermo Scientific (Waltham, MA, USA) and Cas9 protein (sNLS-SpCas9-sNLS nuclease) (Aldevron, Fargo, ND, USA). Chemically synthesized gRNAs (AxoLabs, Kulmbach, DE) and the sNLS-SpCas9-sNLS Cas9 were used for all other experiments. The Lonza 4D Nucleofector or Maxcyte ATx instruments were used to electroporate the gRNAs complexed with Cas9 into the HSPCs and T cells. After 48 h, genomic DNA was extracted using prepGEM Universal Kit (MicroGem, Charlottesville, VA, USA), the CCR5 gene was amplified using primers (IDT) for each gRNA (Supplementary Table 2), and editing of the CCR5 gene was measured using Sanger sequencing analyzed using a CRISPR Therapeutics proprietary software to quantify indels. For dual gRNA editing, a droplet-digital PCR (ddPCR) assay (Bio-Rad, Waltham, MA, USA; Supplementary Table 3) was also used to quantify deletions in the CCR5 gene.
Off-target editing analysis via amplicon sequencing
For each gRNA, edited HSPC pellets, along with unedited HSPC pellets, were sent to GeneWiz (Azenta Life Sciences, Burlington, MA, USA) for DNA extraction and amplicon sequencing. Off-target sites with ≤3 mismatches were identified based on in silico homology predictions and screened using multiplex PCR on the predicted sites (Supplementary Table 4). The amplified regions were 270 base pairs in length and the distance between the off-target cut site and the primer was between 70 and 150 base pairs. Off-target amplicon products were sequenced with the Illumina Hi-Seq 2x250 method and analyzed for indel formation. Each off-target site had a minimum of 1000 reads and a maximum of 10,000 reads across each off-target cut site. All raw reads were analyzed using an amplicon sequencing pipeline developed by CRISPR Therapeutics to characterize the indels in each sample.
Colony forming assay
The MethoCult™ H4034 Optimum kit and protocol from StemCell Technologies (Cambridge, MA, USA) were used for the colony-forming assay according to manufacturer’s instructions. 125 HSPCs were plated and incubated for 14 days. Myeloid and erythroid colonies were determined and counted using the STEMvision system.
In vitro HIV infection assay
After electroporation with Cas9/gRNA combinations, PHA-stimulated T cells were rested for 48 h in R10 supplemented with IL-2. T cells were then plated at 1 × 106 cells per well in a 96-well U-bottom plate. HIVJRCSF virus was added to each well (except mock-infected wells) at a multiplicity of infection (MOI) of 0.5, and plates were centrifuged at 700 × g for 2 h at room temperature. Excess virus was washed from the cells, and then cells were transferred to a 24-well plate at a final concentration of 1 × 106 cells/mL in R10 supplemented with 50 U/mL IL-2. Every two days, cells were resuspended, and 1/10th of the total cells were stained for Gag-p24 expression, and volume replaced with fresh R10 supplemented with IL-2.
Calculation for dual gRNA minimum and maximum editing
When using dual gRNA editing with both TB48 and TB50, large deletions occur or indels are generated at one or both gRNA sites. The ddPCR assay only measures large deletions, while the Sanger Sequencing only measures small indels at each gRNA site. To determine the possible range of editing, both large deletions generated from synchronous dual gRNA editing and indels at each single gRNA site need to be considered.
The calculation for minimal and maximum total editing % for samples is as follows:
Flow cytometry
For surface staining, cells were washed with FACS buffer (PBS with 2 mM EDTA and 2% FBS) and resuspended in a final volume of 50 µL containing fluorochrome-conjugated antibodies and FACS buffer. Cells were incubated for 20 min at room temperature and washed twice with FACS buffer before resuspension in 0.2% paraformaldehyde. For surface staining of whole blood collected from humanized mice, 5 µL of Human TruStain FcX (BioLegend, San Diego, CA, USA) was first added and cells were incubated for 10 min before addition of fluorochrome-conjugated antibodies with an additional 20-min incubation at room temperature. Red blood cell lysis and fixation were achieved using BD FACS lysing solution (BD Biosciences, Woburn, MA, USA). For intracellular Gag-p24 staining, cells were fixed and permeabilized using the FIX & PERM Cell Permeabilization Kit (Invitrogen, Waltham, MA, USA) after staining for surface markers. Cells were resuspended in 100 µL final volume containing anti-p24 antibody (Beckman Coulter, Brea, CA, USA) diluted in permeabilization medium B and incubated for 20 min at room temperature. After incubation for intracellular staining, cells were washed with FACS buffer and resuspended in 0.1% paraformaldehyde. A list of fluorochrome-conjugated antibodies used in all flow-based assays can be found in Supplementary Table 5. Samples were acquired on a BD FACS Symphony instrument using BD FACSDiva Software v.8.0 (BD Biosciences). Flow cytometry data was analyzed using Flowjo software v10.8.2 (TreeStar, Woodburn, OR, USA). Representative flow gating is provided in Supplementary Fig. 3.
Production of HIV stocks
To produce replication-competent HIV stocks, the HIVJRCSF or HIVNL4-3 plasmids were transfected into HEK293T cells using the Takara Bio USA CalPhos transfection kit (San Jose, CA, USA) per manufacturer protocol. Supernatants were harvested at 48- and 72-h post-transfection, supernatant containing virus was filtered through a 0.45 µm membrane, and virus was concentrated using PEG-it (System Biosciences, Palo Alto, CA, USA) per manufacturer protocol. PEG-it concentrated virus stocks were resuspended in R10 media and stored at −80 °C. Virus stocks were titered by TCID50 assay that consisted of plating 105 PBMCs into 24 wells of a 96-well plate, stimulating for 3 days in RPMI supplemented with 20% FBS, 1% penicillin-streptomycin, 5 ug/ml phytohemagglutinin (PHA) (Sigma-Aldrich, St. Louis, MO, USA), and 20 U/ml recombinant human interleukin-2 (rhIL-2) (Roche Applied Sciences, Indianapolis, IN). After 3 days, the medium was removed and replaced with RPMI supplemented with 20% FBS, 1% penicillin-streptomycin, and 20 U/ml rhIL-2, and the mixture was inoculated in triplicate with 4-fold serial dilutions of stock virus ranging from 4−6 to 4−13. On day 4 postinfection (p.i.), 100 µl of medium was removed and replaced. On day 7 p.i., wells were scored for infection by p24 ELISA per the manufacturer protocol’s and TCID50 was calculated by the Spearman-Karber method.
Humanized mouse studies
Mice were housed under a 7p-7a dark/light cycle in a facility that is maintained ambient temperature and humidity of 68–73 F and 30–70%, respectively. To generate humanized mice, 6–8 week old female NSG (NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ) or NBSGW (NOD.Cg-KitW-41J Tyr+ Prkdcscid Il2rgtm1Wjl/ThomJ) mice were purchased from The Jackson Laboratory (Bar Harbor, ME, USA). The mice received a single sublethal doses of total body irradiation of 200 cGy (NSG) or 50 cGy (NBSGW). After 4 h, 1 × 106 HSPCs were transferred via tail vein injection. To monitor reconstitution of human cells, blood was collected from mice monthly via puncture of the retro-orbital sinus. Mice harboring sufficient human T cell reconstitution (>10 T cells/µL of blood) were challenged with HIVJRCSF or HIVNL4-3 in 300 µL of PBS via intraperitoneal injection. After HIV challenge, mice were bled weekly via the retro-orbital sinus, and plasma was separated from whole blood by low-speed centrifugation. Plasma was frozen and stored at −80 °C, and the cell pellet was resuspended in PBS, stained with fluorochrome-conjugated antibodies, and analyzed for human cell frequencies. At necropsy, the spleen, bone marrow, and lymph nodes were harvested. Single-cell suspensions were isolated from the spleen and femurs (bone marrow), cryopreserved in freezing medium (10% DMSO, 90% FBS), and stored in liquid nitrogen. Lymph nodes were added directly to RNAlater Stabilization solution (Invitrogen, Waltham, MA, USA) and stored at −80°C.
HIV viral load quantification
Plasma viral load was quantified by extracting viral RNA from 40 µL of plasma obtained from the peripheral blood of humanized mice using the Qiagen Viral RNA extraction kit (Germantown, MD, USA) per manufacturer protocol. The Quantifast SYBR Green RT-PCR kit (Qiagen, Germantown, MD, USA) was used per manufacturer protocol with HIV-1 gag SK145 (AGTGGGGGGACAT CAAGCAGCCATGCAAAT) and SK431 (TGCTATGTCACTTCCCCTT GGTTCTCT) primers to quantify HIV RNA copies via quantitative RT-PCR35. To measure HIV DNA in the tissue, lymph nodes preserved in RNAlater were first homogenized in buffer RLT using a handheld homogenizer (VWR, Radnor, PA, USA). Genomic DNA was extracted from homogenates using the AllPrep DNA/RNA mini kit (Qiagen, Germantown, MD, USA) according to the manufacturer's protocol. A ddPCR assay which simultaneously measures the presence of the HIV gag gene and the human RPP30 gene was used to quantify HIV viral burden in the extracted genomic DNA isolated from the lymph nodes of humanized mice36. Briefly, total HIV DNA was measured in each sample using a multiplexed ddPCR assay specific for HIV gag and the human RPP30 gene using the gag SK145 and SK431 primers and a 5′ HEX-labeled hydrolysis probe (HEX-CCATCAATGAGGAAGCTGCAGAATGGGA) and RPP30 forward (5′-GATTTGGACCTGCGAGCG) and reverse (5′-GCGGCTGTCTCCACAAGT) primers and a 5′ 6-FAM-labeled hydrolysis probe (6-FAM-CTGACCTGAAGGCTCT).
B cell isolation from the bone marrow
Cryopreserved bone marrow cells were thawed, washed, and incubated with DNase I (STEMCELL, 100-0762) for 15 min at room temperature to prevent cell clumping. Cells were then concentrated to 1 × 108 cells/mL and incubated with FcR blocking reagent (Miltenyi Biotec, CHARLESTOWN, MA, USA) and CD19 microbeads (Miltenyi Biotec, Charlestown, MA, USA) in order to isolate CD19+ cells according to the manufacturer’s protocol. Genomic DNA was isolated from CD19+ cells using the DNeasy Blood and Tissue kit (Germantown, MD, USA).
Statistics and reproducibility
For pairwise analyses of groups of <5 data points, the Mann-Whitney U test was used, and p-values are two-tailed. For pairwise analyses of groups >5 data point, the Student’s t-test was used, and p value are two-tailed. No statistical method was used to predetermine sample sizes. The experiments were not randomized. The investigators were not blinded to allocation during experiments and outcome assessments.
Modeling
To mechanistically model the titration of transplanted CCR5 edited HSPCs experiment (Fig. 7), we assumed that the probability of infection at each inoculation was dependent on the realized % CCR5 gene editing in the animal. Specifically, we assumed that the number of cells infected by inoculating an animal with no CCR5 editing is Poisson distributed with mean b (a free parameter). We then assumed that for an animal in this study with gene-edited cells, the number of cells infected by an inoculation is still Poisson distributed, but the expected number of cells that are infected is proportional to the availability of unedited CCR5+ target cells and therefore is proportional to the percentage of unedited cells in the animal, xa. Hence, we assume that the expected number of cells that are infected following an inoculation of animal a is given by bxa. We define xa as the mean of the minimum and maximum estimated frequencies of intact CCR5 alleles in CD19+ cells taken from bone marrow at necropsy (calculations outlined above). Therefore, the probability of infection of animal a following an inoculation (i.e., the probability that ≥1 cell is infected), is
| 1 |
and the probability of observing animal a becoming infected or censored (annotated by the indicator variable da being equal to 1 or 0, respectively) at inoculation ia is
| 2 |
We fit b using a maximum likelihood estimation (see Supplementary Text S1.6 for more details).
Modeling computation was performed in R version 4.0.3 run through RStudio.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Source data
Acknowledgements
The research described here honors the memory of Todd Mackenzie Allen, Ph.D., whose rare blend of scientific acumen and instinct, patience and determination, and above all kindness and compassion, elevated all of those who had the privilege to pursue science with him. We thank the Massachusetts General Hospital Blood Transfusion Service for the provision of leukocytes for in vitro assays. We thank the Ragon Institute Flow Core for supporting data collection. We thank the Ragon Institute Human Immune System Mouse Core for guidance in the planning of humanized mouse experiments. This study was supported by funding through the Ragon Institute of Mass General, MIT, and Harvard, and NIH grants U19 HL129903 (TMA) and U19 HL156247 (TMA). MPD is supported by an NHMRC Investigator grant (1173027) and an NHMRC Program grant (149990). DC is supported by an NHMRC Investigator grant (1173528).
Author contributions
D.T.C, Z.D., S.S.D., T.D.B., M.P.D., T.W.H., C.L.B., and T.M.A. conceived and designed the project and contributed to the interpretation of data. D.T.C, Z.D., T.D.B., D.C., A.S., Y.O., K.O., T.B., T.C., W.H., and R.T. contributed to the acquisition and analysis of data. D.T.C, Z.D., S.S.D., M.P.D., C.L.B., and T.M.A. drafted the manuscript.
Peer review
Peer review information
Nature Communications thanks Ester Ballana, who co-reviewed with Edurne Garcia-Vidal; and the other, anonymous, reviewer for their contribution to the peer review of this work. A peer review file is available.
Data availability
Source data for Figs. 1–6 is available in the Source Data file accompanying the manuscript. Source data for Fig. 7 is available in the mathematical modeling code as outlined in the Code Availability section. Source data are provided with this paper.
Code availability
Code used for the mathematical modeling component of this work is publicly available at the following link: https://github.com/iap-sydney/CCR5_editing_and_HIV_infection.
Competing interests
ZD is a current employee, holds stock and stock options in CRISPR Therapeutics AG. TB and TH are former employees and shareholders of CRISPR Therapeutics AG. The other authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Deceased: Todd M. Allen.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-025-55873-3.
References
- 1.Siliciano, J. D. et al. Long-term follow-up studies confirm the stability of the latent reservoir for HIV-1 in resting CD4+ T cells. Nat. Med.9, 727–728 (2003). [DOI] [PubMed] [Google Scholar]
- 2.Josefsson, L. et al. The HIV-1 reservoir in eight patients on long-term suppressive antiretroviral therapy is stable with few genetic changes over time. Proc. Natl. Acad. Sci. USA110, E4987–E4996 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Besson, G. J. et al. HIV-1 DNA decay dynamics in blood during more than a decade of suppressive antiretroviral therapy. Clin. Infect. Dis.59, 1312–1321 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.von Stockenstrom, S. et al. Longitudinal genetic characterization reveals that cell proliferation maintains a persistent HIV type 1 DNA pool during effective HIV therapy. J. Infect. Dis.212, 596–607 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brodin, J. et al. Establishment and stability of the latent HIV-1 DNA reservoir. Elife5, e18889 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sengupta, S. & Siliciano, R. F. Targeting the latent reservoir for HIV-1. Immunity48, 872–895 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moranguinho, I. & Valente, S. T. Block-and-lock: new horizons for a cure for HIV-1. Viruses12, 1443 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hutter, G. et al. Long-term control of HIV by CCR5 Delta32/Delta32 stem-cell transplantation. N. Engl. J. Med.360, 692–698 (2009). [DOI] [PubMed] [Google Scholar]
- 9.Gupta, R. K. et al. HIV-1 remission following CCR5Delta32/Delta32 haematopoietic stem-cell transplantation. Nature568, 244–248 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jensen, B. O. et al. In-depth virological and immunological characterization of HIV-1 cure after CCR5Delta32/Delta32 allogeneic hematopoietic stem cell transplantation. Nat. Med.29, 583–587 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Henrich, T. J. et al. Antiretroviral-free HIV-1 remission and viral rebound after allogeneic stem cell transplantation: report of 2 cases. Ann. Intern. Med.161, 319–327 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Henrich, T. J. et al. Long-term reduction in peripheral blood HIV type 1 reservoirs following reduced-intensity conditioning allogeneic stem cell transplantation. J. Infect. Dis.207, 1694–1702 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Salgado, M. et al. Mechanisms that contribute to a profound reduction of the HIV-1 reservoir after allogeneic stem cell transplant. Ann. Intern. Med.169, 674–683, (2018). [DOI] [PubMed] [Google Scholar]
- 14.Xu, L. et al. CRISPR-edited stem cells in a patient with HIV and acute lymphocytic leukemia. N. Engl. J. Med.381, 1240–1247 (2019). [DOI] [PubMed] [Google Scholar]
- 15.Kuritzkes, D. R. Hematopoietic stem cell transplantation for HIV cure. J. Clin. Investig.126, 432–437 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martinson, J. J., Chapman, N. H., Rees, D. C., Liu, Y. T. & Clegg, J. B. Global distribution of the CCR5 gene 32-basepair deletion. Nat. Genet.16, 100–103 (1997). [DOI] [PubMed] [Google Scholar]
- 17.Freen-van Heeren, J. J. Closing the door with CRISPR: genome editing of CCR5 and CXCR4 as a potential curative solution for HIV. BioTechnology11, 25 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dash, P. K. et al. CRISPR editing of CCR5 and HIV-1 facilitates viral elimination in antiretroviral drug-suppressed virus-infected humanized mice. Proc. Natl. Acad. Sci. USA120, e2217887120 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Genovese, P. et al. Targeted genome editing in human repopulating haematopoietic stem cells. Nature510, 235–240 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Davenport, M. P. et al. Functional cure of HIV: the scale of the challenge. Nat. Rev. Immunol.19, 45–54 (2019). [DOI] [PubMed] [Google Scholar]
- 21.Frangoul, H. et al. CRISPR-Cas9 gene editing for sickle cell disease and beta-thalassemia. N. Engl. J. Med.384, 252–260 (2021). [DOI] [PubMed] [Google Scholar]
- 22.McIntosh, B. E. et al. Nonirradiated NOD, B6.SCID Il2rgamma-/- Kit(W41/W41) (NBSGW) mice support multilineage engraftment of human hematopoietic cells. Stem Cell Rep.4, 171–180 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Choo, S. et al. Choosing the right mouse model: comparison of humanized NSG and NBSGW mice for in vivo HSC gene therapy. Blood Adv.8, 916–926 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gomez-Ospina, N. et al. Human genome-edited hematopoietic stem cells phenotypically correct Mucopolysaccharidosis type I. Nat. Commun.10, 4045 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Romero, Z. et al. Editing the sickle cell disease mutation in human hematopoietic stem cells: comparison of endonucleases and homologous donor templates. Mol. Ther.27, 1389–1406 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu, L. et al. CRISPR/Cas9-mediated CCR5 ablation in human hematopoietic stem/progenitor cells confers HIV-1 resistance in vivo. Mol. Ther.25, 1782–1789 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peterson, C. W. et al. Differential impact of transplantation on peripheral and tissue-associated viral reservoirs: implications for HIV gene therapy. PLoS Pathog.14, e1006956 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cardozo-Ojeda, E. F. et al. Thresholds for post-rebound SHIV control after CCR5 gene-edited autologous hematopoietic cell transplantation. Elife10, e57646 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pinkevych, M. et al. HIV reactivation from latency after treatment interruption occurs on average every 5–8 days–implications for HIV remission. PLoS Pathog.11, e1005000 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tebas, P. et al. Gene editing of CCR5 in autologous CD4 T cells of persons infected with HIV. N. Engl. J. Med.370, 901–910 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tebas, P. et al. CCR5-edited CD4+ T cells augment HIV-specific immunity to enable post-rebound control of HIV replication. J. Clin. Investig.131, e144486 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Holt, N. et al. Human hematopoietic stem/progenitor cells modified by zinc-finger nucleases targeted to CCR5 control HIV-1 in vivo. Nat. Biotechnol.28, 839–847 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morvan, M. G. et al. Genetically edited CD34(+) cells derived from human iPS cells in vivo but not in vitro engraft and differentiate into HIV-resistant cells. Proc. Natl. Acad. Sci. USA118, e2102404118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li, C. et al. In vivo HSC prime editing rescues sickle cell disease in a mouse model. Blood141, 2085–2099 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boutwell, C. L., Rowley, C. F. & Essex, M. Reduced viral replication capacity of human immunodeficiency virus type 1 subtype C caused by cytotoxic-T-lymphocyte escape mutations in HLA-B57 epitopes of capsid protein. J. Virol.83, 2460–2468 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maldini, C. R. et al. Dual CD4-based CAR T cells with distinct costimulatory domains mitigate HIV pathogenesis in vivo. Nat. Med.26, 1776–1787 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Source data for Figs. 1–6 is available in the Source Data file accompanying the manuscript. Source data for Fig. 7 is available in the mathematical modeling code as outlined in the Code Availability section. Source data are provided with this paper.
Code used for the mathematical modeling component of this work is publicly available at the following link: https://github.com/iap-sydney/CCR5_editing_and_HIV_infection.