Summary
Chromothripsis, a hallmark of cancer, is characterized by extensive and localized DNA rearrangements involving one or a few chromosomes. However, its genome-wide frequency and characteristics in urothelial carcinoma (UC) remain largely unknown. Here, by analyzing single-regional and multi-regional whole-genome sequencing (WGS), we present the chromothripsis blueprint in 488 UC patients. Chromothripsis events exhibit significant intertumoral heterogeneity, being detected in 41% of UC patients, with an increase from 30% in non-muscle-invasive disease (Ta/1) to 53% in muscle-invasive disease (T2-4). The presence of chromothripsis correlates with an unstable cancer genome and poor clinical outcomes. Analysis of multi-regional WGS data from 52 patients revealed pronounced intratumoral heterogeneity with chromothripsis events detectable only in specific tumor regions rather than uniformly across all areas. Chromothripsis events evolve under positive selection and contribute to tumor dissemination. This study presents a comprehensive genome-wide chromothripsis landscape in UC, highlighting the significance of chromothripsis in UC development.
Subject areas: Genomics, Genomic analysis, Cancer systems biology, Cancer
Graphical abstract

Highlights
-
•
A landscape of chromothripsis in urothelial carcinoma (UC)
-
•
Chromothripsis is highly heterogeneous in UC
-
•
Chromothripsis correlates with UC clinical outcome
Genomics; Genomic analysis; Cancer systems biology; Cancer
Introduction
Urothelial carcinoma (UC) is a highly invasive malignancy within the urinary tract. Most UC cases, approximately 90%–95%, originate in the bladder, while tumors located in the renal pelvis and ureter are less common, accounting for 5%–10% of all UCs. Depending on the degree of tumor invasion, UC can be classified into non-muscle-invasive (pTa/1) and muscle-invasive (pT2-T4) subtypes. A long-standing challenge in the clinical management of UC is the absence of effective targeted therapeutic drugs, especially for patients with muscle-invasive disease.1,2,3 A comprehensive understanding of the molecular mechanisms underlying the onset of UC is crucial for effectively managing the disease.
Chromosomal instability has been implicated in UC development.4,5,6 This instability can lead to chromothripsis, a phenomenon characterized by extensive and localized DNA rearrangements involving one or a few chromosomes.7,8,9,10 This intricate genomic rearrangement mosaic arises from the continuous, inaccurate repair of shattered DNA fragments, leading to the swift accumulation of numerous rearrangements within only a few cell divisions. Chromothripsis has been observed across a wide spectrum of tumor types and is associated with aggressive biological behavior in certain entities.11,12,13 Notably, chromothripsis has been identified to be the major driver of tumor-suppressor gene inactivation and oncogenes amplification, suggesting a pivotal role in both driving and potentially initiating cancer development. One molecular mechanism of chromothripsis-dependent oncogene amplification is through extrachromosomal DNA (ecDNA).7,11
The breakthrough in unraveling chromothripsis in cancer is facilitated by the development of computational tools for identifying chromothripsis in cancer using whole-genome sequencing (WGS) data.11 The landscape of chromothripsis, including its frequency, distribution, intratumoral diversity/heterogeneity, and clinical significance, is not well understood in UC. Moreover, it is unclear how chromothripsis shapes the genomic heterogeneity within cancer cells. To address this, this study provides a comprehensive analysis of the chromothripsis landscape in UC using single-regional and multi-regional WGS data, combined with bulk transcriptome, from a large cohort of UCs (N = 488 patients).
Results
Chromothripsis is a major driver in UC development
To explore the chromothripsis landscape in UC, we performed chromothripsis analysis using ShatterSeek11 on paired tumor-germline WGS data from our in-house dataset (N = 465 patients) and the Beijing Institute of Genomics (BIG) dataset (N = 3 patients).14 In addition, we performed multi-regional WGS on 20 UC patients. The entire cohort includes the WGS data from 629 tumors, 491 germline samples, and 166 time-matched urine samples from 488 UC patients (Figure 1A; Tables S1 and S2). Among these patients, 52 (11%) had multi-regional tumor WGS data. The cohort included 362 (74%) males and 126 (26%) females, with a median age of 68 years. Muscle-invasive diseases (pT2-T4) were diagnosed in 232 (48%) patients. Clinical metadata—including age, gender, stage, and survival, as well as sequencing information—are provided in Tables 1 and S1.
Figure 1.
Chromothripsis landscape in UC
(A) Overview of the experimental workflow and a brief model illustrating the process of chromothripsis (created with BioRender.com).
(B) Classification of chromothripsis events into high-confidence (HC) and low-confidence (LC) categories.
(C) Patterns and prevalence of chromothripsis across the entire cohort (488 UC patients).
(D) Percentage of canonical and non-canonical chromothripsis events at the chromosome level (N = 402).
(E) Percentage of patients with single and multiple (2–7) chromothriptic chromosomes.
(F) Presence of chromothripsis by clinical subgroup across 488 UC patients (Fisher’s exact test, two-sided).
(G) Kaplan-Meier survival curves illustrating the overall survival of UC patients stratified by chromothripsis status (log rank test).
(H) Hazard ratios with ±95% confidence intervals and p values for chromothripsis status, gender, age, and tumor location calculated using a Cox regression on overall survival.
Table 1.
Clinical and WGS information of patients included in the present study
| Category | No. (%) |
|---|---|
| No. of patients | 488 |
| Age, yr-no. (%) | |
| ≤65 | 200 (41) |
| >65 | 288 (59) |
| Gender-no. (%) | |
| Male | 362 (74) |
| Female | 126 (26) |
| Tumor site-no. (%) | |
| Pelvis | 107 (22) |
| Ureter | 80 (16) |
| Bladder | 301 (62) |
| Histologic grading-no. (%) | |
| Low | 106 (22) |
| High | 382 (78) |
| T-category-no. (%) | |
| Muscle-invasive (pT2-T4) | 232 (48) |
| Non-muscle-invasive (pTa/1) | 256 (52) |
| Multi-regional WGS-no. (%) | |
| Yes | 52 (11) |
| No | 436 (89) |
The widely accepted model of chromothripsis suggests that, during DNA shattering, some fragments are lost (Figure 1A).7,11,15 As a result, the oscillation of copy number (CN) between two states serves as a primary indicator of chromothripsis events. We utilized the ShatterSeek11 method to identify chromothripsis events from paired tumor-germline WGS data (Figure 1A). Based on established criteria,9,11 chromothripsis events were categorized into high-confidence (HC) and low-confidence (LC) groups. The LC calls involved 4–6 segments between two states, whereas HC calls comprised a minimum of 7 adjacent segments (Figure 1B). Chromothripsis events, including both HC and LC calls, were identified in 41% (199 out of 488) of UC patients (Figure 1C; Table S3), indicating that chromothripsis is a common mutational event in UC. Moreover, about 21% of these events were canonical (Figure 1C), in which more than 60% of CN segments in the affected region exhibited oscillation between the two states. In approximately 86% of tumors, the centromere was included in the segment affected by chromothripsis (Figure S1A). Notably, nearly half (49%, 97 out of 199) of chromothripsis+ UC patients exhibited multiple chromothriptic chromosomes (Figures 1E, S1B, and S1C).
We observed a significant increase in chromothripsis events in patients with muscle-invasive or high-grade tumors compared to those with non-muscle-invasive or low-grade tumors (Figure 1F; grade, p = 3.48e−10; stage, p = 2.11e−07; Fisher’s exact test, two-sided). This suggests that chromothripsis occurs during tumor development and might be linked to increased tumor malignancy. Moreover, patients with chromothripsis+ tumors have a poorer prognosis compared to those with chromothripsis− tumors (Figure 1G; log rank test, p = 3.558e−04). To further assess the prognostic significance of chromothripsis, we performed a Cox proportional hazards regression analysis, adjusting for age, gender, and tumor location. Patients who carried chromothripsis+ tumors exhibited a significantly higher risk of mortality (Figure 1H; hazard ratio = 1.82; p < 0.001) compared to patients with chromothripsis− tumors. These findings highlight chromothripsis as a key driver of UC progression.
Chromothripsis+ tumors have distinct genetic and transcriptional features
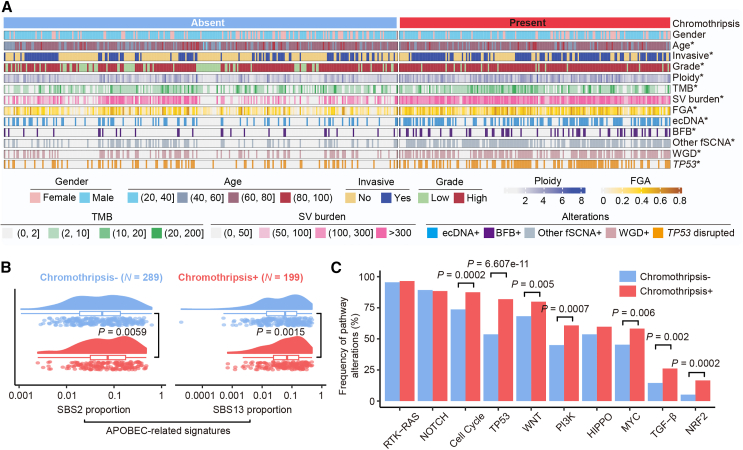
Given the significant impact of chromothripsis on UC prognosis, we further investigated the molecular characteristics associated with chromothripsis. Comparative analysis revealed that chromothripsis+ tumors exhibited higher tumor mutation burden (TMB) and structural variants (SVs) compared to chromothripsis− tumors. Consistently, chromothripsis+ tumors were significantly more chromosomally unstable, as indicated by increased ploidy and a higher fraction of genome altered (FGA) (Figures 2A and S2A; Wilcoxon rank-sum test; TMB, p = 1.5e-05; SV, p = 1.24e−27; ploidy, p = 1.2e−12; FGA, p = 2.3e−16). Genome instability is closely linked to focal amplifications and whole-genome duplication (WGD), both of which were significantly enriched in chromothripsis+ tumors (Figure S2B; Fisher’s exact test, two-sided; WGD, p = 1.53e−13; focal amplification, p = 2.72e−21). Additionally, Apolipoprotein B mRNA editing enzyme catalytic polypeptide (APOBEC) mutagenesis, a well-established driver in UC development, was more prevalent in chromothripsis+ tumors, with higher proportions of APOBEC-related signatures observed compared to chromothripsis− tumors (Figure 2B; Wilcoxon rank-sum test; Catalogue Of Somatic Mutations In Cancer (COSMIC) SBS2, p = 0.0059; COSMIC SBS13, p = 0.0015). Moreover, chromothripsis+ tumors exhibited a significantly higher frequency of alterations in pathways related to the cell cycle (87% vs. 74%; p = 0.0002), TP53 (82% vs. 54%; p = 6.607e−11), WNT signaling pathway (80% vs. 68%; p = 0.005), PI3K signaling pathway (61% vs. 45%; p = 0.0007), MYC signaling pathway (58% vs. 45%; p = 0.006), transforming growth factor β (26% vs. 15%; p = 0.002), and NRF2 (17% vs. 5%; p = 0.0002) pathways16 (Figure 2C; Fisher’s exact test, two-sided). Overall, these data suggested that chromothripsis is linked to an unstable cancer genome in UC.
Figure 2.
Hallmarks of chromothripsis on UC genome instability, mutation, and transcription
(A) Patient classification by chromothripsis status. The asterisk denotes a statistically significant association with the presence of chromothripsis (Wilcoxon rank-sum test for age, ploidy, TMB, SV burden, and FGA; two-sided Fisher’s exact test for other factors). FGA, the fraction of genome altered; TMB, tumor mutation burden; SV, structural variation; fSCNA, focal somatic copy number amplification. BFB, breakage-fusion-bridge; WGD, whole-genome duplication.
(B) Proportion of APOBEC-related mutations (COSMIC SBS2 and SBS13) between chromothripsis− and chromothripsis+ tumors (Wilcoxon rank-sum test).
(C) Percentage of patients with alterations for 10 canonical oncogenic pathways (Fisher’s exact test, two-sided).
To further investigate the impact of chromothripsis on transcriptional changes, we compared the transcriptome between chromothripsis− and chromothripsis+ tumors. Differentially expressed gene (DEG) analysis revealed that 206 genes were specifically up-regulated in chromothripsis− tumors, while 20 genes showed specific up-regulation in chromothripsis+ tumors (Figure S3A). Enrichment analysis indicated that these genes were predominantly involved in endocytosis, focal adhesion, and mitogen-activated protein kinase signaling pathways (Figure S3B; Table S4). We also examined a curated set of well-defined functional pathways related to UC5 (Table S5). Notably, chromothripsis− tumors exhibited remarkably elevated expression of fibroblast growth factor receptor 3 (FGFR3)-associated markers (Figure S3), implying a potential therapeutic benefit from FGFR3 inhibitors. In contrast, chromothripsis+ tumors demonstrated enhanced expression of markers associated with “epithelial-mesenchymal transition (EMT),” “extracellular matrix (ECM),” “smooth muscle,” “squamous,” and “immune checkpoint” markers (Figure S3). Collectively, these data suggest that chromothripsis is associated with increased genome instability, APOBEC-related mutation burden, and distinct transcriptomic signatures.
TP53 alterations are more frequently detected in chromothripsis+ UC
The tumor suppressor p53, encoded by TP53, is crucial for maintaining genome integrity through its roles in DNA damage response, cell-cycle arrest, apoptosis, and inhibition of angiogenesis. Disruption of TP53 facilitates genomic instability.17,18 Previous research has shown that TP53 is frequently subjected to somatic mutations in UC.2,4,5 To explore the potential relationship between TP53 alterations and chromothripsis, we analyzed somatic TP53 mutations in 488 UC cases. We found that TP53 alterations were significantly more common in chromothripsis+ tumors (52%, 104 out of 199) compared to the chromothripsis− tumors (25%, 73 out of 289; Fisher exact test, p = 1.31e−09, two-sided) (Figures 2C and S2B), implying a potential mechanism of co-evolution between TP53 mutation and chromothripsis.
Frequent co-localization of chromothripsis with ecDNA and kataegis
Chromothripsis, followed by circular recombination, contributes to increased gene dosage in cancer.7,19 To study the relationship between chromothripsis and ecDNA, we applied the AmpliconArchitect20 method to reconstruct the structure of ecDNA and focal amplification from WGS data. Overall, we identified ecDNA and focal amplification signatures within chromothripsis regions in 9% (18 out of 199) and 29% (57 out of 199) of chromothripsis+ UC patients (Figure S4A; Table S6), suggesting that chromothripsis serves as an initiating event for some ecDNAs or focal amplifications. For example, in CCGA-UBC-075T, we detected CCND1 (a well-known driver in UC)-containing ecDNA in chromothripsis regions (Figures 3A–3C). CCND1 amplification was validated by DNA fluorescent in situ hybridization (FISH) of interphase spread chromosomes. It is noteworthy that once ecDNA and focal amplification form, the occurrence of chromothripsis phenomena may diminish in the absence of sustained drivers promoting chromosomal instability,11 which might explain the relatively low co-occurrence frequency of ecDNA (9%)/focal amplification (29%), and chromothripsis may be underestimated. Nevertheless, our results prove that chromothripsis is associated with the genome-wide formation of ecDNA and focal amplification in UC.
Figure 3.
Co-occurrence of chromothripsis with ecDNA and focal amplification
(A) Chromothripsis event (chromosome 11) detected in CCGA-UBC-75T.
(B and C) Structural variant and breakpoint graph (B), along with the structure (C) of the CCND1-containing ecDNA identified in CCGA-UBC-75T. The CCND1-containing ecDNA is localized within chromothripsis regions.
(D) CCND1 amplification validated through interphase fluorescent in situ hybridization (FISH). Scale bars, 10 μm.
Kataegis is a genomic phenomenon characterized by localized clusters of point mutations, often linked to specific mutagenic processes and associated with cancer development. We identified kataegis in 94% of UC patients (458 out of 488). Patients with kataegis mutations exhibited a significantly higher frequency of chromothripsis events compared to those without kataegis (Figure S4B; two-sided Fisher’s test, p = 2.933e−06). Furthermore, we examined kataegis occurrences within chromothripsis regions and found that kataegis was present in 57% of these regions (229 out of 402) (Figure S4C). These findings corroborate previous observations that chromothripsis frequently co-localizes with kataegis.
The landscape of intratumoral chromothripsis heterogeneity
Chromothripsis represents a significant contributor to intratumor heterogeneity,21 yet its intertumoral diversity is largely unexplored. We performed multi-regional WGS analysis (two to six tumor regions per patient) from 193 tumors and 52 germline samples, collected across 52 UC patients (Figure 4A). Chromothripsis events were identified in 67% of (35 out of 52) UC cases (Figure 4A). In most chromothripsis+ UC patients (86%, 30 out of 35), chromothripsis was selectively detected in specific tumor regions, a phenomenon of intertumoral heterogeneity and referred to as branch chromothripsis hereafter (Figure 4A). In five cases (CUGA-MR-001/023/033/035; CUGA-006), chromothripsis was detected in all analyzed tumor regions (Figures 4A and 4B). For instance, in CUGA-MR-001, we observed chromothripsis events involving chromosomes 6 and 13 were detected only in regions tumor-R3 and lymph node metastasis (LNM) tissues (Figure 4D). The identical chromothripsis event in both primary and metastatic tumor tissue also indicated the involvement of chromothripsis in the process of tumor dissemination. These data suggest that most chromothripsis events are likely a later manifestation of genomic instability and provide a strong selective advantage (Figure 4E).
Figure 4.
Intratumoral chromothripsis heterogeneity in UC
(A) Oncoprint tables of samples from the multi-regional WGS cohort (N = 193 tumors from 52 patients are shown). Each column represents one tumor. Genetic alterations, including chromothripsis, WGD, ecDNA, BFB, other fSCNA, and TP53 disrupted, as well as the number of chromosomes affected by chromothripsis, are indicated. fSCNA, focal somatic copy number amplification. BFB, breakage-fusion-bridge; WGD, whole-genome duplication.
(B) Proportion of trunk and branch chromothripsis events across 35 patients with chromothripsis+ tumors. A trunk chromothripsis event was defined as chromothripsis detectable across all tumor regions in the same patient, while a branch chromothripsis event was specific to specific tumor regions.
(C) Representative images illustrating the multi-regional sampling sites of patient CUGA-MR-001.
(D) Example of heterogeneous chromothripsis events involving chromosomes 6 and 13, detected in regions R3 and LNM from patient CUGA-MR-001.
(E) A schematic illustration of chromothripsis evolution in cancer. Initiating driver instigates the initiation of tumors. After genome doubling of cancer cells, a subset of cancer cells experiences chromothripsis events, while others do not. As the tumor progresses, cells with chromothripsis undergo parallel evolution, whereas another subset, under the pressure of natural selection, generates new chromothripsis events and actively contributes to tumor metastasis. These dynamic processes collectively contribute to the subclonal evolution of the tumor.
Discovery of chromothripsis events in the urine of UC patients
We previously demonstrated that extrachromosomal circular DNA with size less than 1 kb (eccDNA) and ecDNA could be detected in urine sediment-derived DNA.22,23 Therefore, we next sought to investigate if chromothripsis could be detected in the urine, a reservoir of biological waste from the urinary tract. To achieve this, we analyzed WGS data from 166 matched tumor-urine samples. Due to the significantly lower tumor purity in urine sediments (Figure 5B; paired t test, p = 3.79e−23), chromothripsis events were detected in the urinary genomic DNA but less frequently (25%, 42 out of 166) compared to tumor tissues (42%, 70 out of 166) (Figures 5A and 5C). Furthermore, the incidence of chromothripsis in urine was higher in patients with bladder tumors (36%, 33 of 92) than in patients with pelvis (13%, 6 out of 46) or ureter tumors (11%, 3 out of 28) (Figure 5D; Fisher exact test, p < 0.05, two-sided). Among 21 patients, chromothripsis was detected in both urine and tumor samples, but the regions of chromothripsis events differed significantly between the two types of samples (Figure S5). Notably, in 13% (21 out of 166) of cases (Figure 5C), chromothripsis was exclusively detected in urine samples, further emphasizing the need to consider intratumoral heterogeneity of chromothripsis events in UC.
Figure 5.
Discovery of chromothripsis in urinary genomic DNA from UC
(A) Chromothripsis and other genetic alterations in paired tumor and urine samples (N = 166). Each column represents one patient. Each row indicates genetic alterations detected in preoperative urine sediment (indicated by ‘‘U”) and paired tumors (indicated by ‘‘T”).
(B) Comparison of the tumor purity between urine and paired tumor samples (paired t test). Tumor purity was calculated from WGS data using the FACETS algorithm. Values are represented as median with 25th and 75th percentile.
(C) Proportion of patients with indicated events: T+ & U+: chromothripsis detected in both tumor and urine samples. T+ & U−: chromothripsis detected only in tumors. T− & U+: chromothripsis detected only in urine. T− & U−: chromothripsis undetected in both tumor and urine samples.
(D) Proportion of urine samples with chromothripsis, stratified by tumor location (Fisher’s exact test, two-sided).
Discussion
A long-standing challenge in the clinical management of UC is the absence of effective targeted therapeutic drugs, particularly for patients with muscle-invasive disease. Among the limited drugs with Food and Drug Administration-approved usage, FGFR inhibitors such as pemigatinib or erdafitinib currently stand out. By integrating single-regional and multi-regional WGS data from a large cohort of UC patients, we revealed the frequency and diversity of chromothripsis in UC and demonstrated that chromothripsis is a major contributor to UC heterogeneity. Survival analysis revealed that, in comparison to patients with chromothripsis− tumors, those with chromothripsis+ tumors are more likely to succumb during the follow-up period. Detecting chromothripsis in UC tumors is crucial in laying the foundation for precision medicine interventions specifically designed to target chromothripsis.
At the genomic level, chromothripsis in UC is associated with extensive CN variations, which are evident in the amplification and deletion patterns observed across multiple chromosomes in our cohort. The presence of multiple chromothriptic chromosomes in nearly half of the chromothripsis+ UC patients suggests a recurrent and potentially orchestrated process of genomic disruption. This widespread chromosomal fragmentation likely results from a combination of factors, including DNA replication stress and defective DNA repair mechanisms. To further explore the transcriptional impact of chromothripsis in UC, we found that chromothripsis− tumors had higher expression of FGFR3-related markers, suggesting potential responsiveness to FGFR3 inhibitors. In contrast, chromothripsis+ tumors were enriched for markers of EMT, ECM, squamous features, and immune checkpoints, highlighting their distinct transcriptomic profiles and increased genomic instability.
We show the spatial intratumoral heterogeneity of chromothripsis in UC evolution. Even within different tumor regions of the same patient, some tumor regions manifest detectable chromothripsis, while others do not. Previous studies and our data have indicated an association between chromothripsis and drug resistance and tumor metastasis,19,24 underscoring that targeting trunk driver events (such as FGFR3 mutations) in cancer treatment may be affected by the occurrence of chromothripsis events in the later stages of tumor evolution. This observation supports ongoing efforts to enhance early detection methods for UC. Moreover, our data emphasize the importance of multi-region tumor sampling for comprehensive chromothripsis profiling.
Considering the clinical implications of chromothripsis, it is crucial to employ non-invasive methods for detecting chromothripsis-driven tumors. Urothelial tumor tissue directly contacts urine, potentially resulting in the presence of tumor cells in the urine. In this study, we decode chromothripsis events from urine samples based on genome sequencing data. We revealed a significantly lower detection rate of chromothripsis in urine (25%) compared to that in tumor tissue (42%). This phenomenon may be explained by the presence of a higher proportion of non-cancerous cells, such as white blood cells, in the urine of UC patients.25,26 Further studies could explore the potential of initially enriching tumor cells from the urine of cancer patients, followed by chromothripsis detection. Identifying chromothripsis in urine samples opens a wide range of diagnostic opportunities in genitourinary tumors.
Chromothripsis has been implicated in intratumoral heterogeneity. In this study, we have shown that chromothripsis is an independent prognostic factor for UC patients. It is associated with increased mutational load, higher genome instability, oncogene amplification, and tumor dissemination. Further studies of the mechanisms of chromothripsis formation and maintenance may uncover new combinatorial therapies for UC patients.
Limitations of the study
The study primarily focused on the frequency and characteristics of chromothripsis but did not investigate the underlying mechanisms or potential therapeutic implications. Future studies should address these limitations and explore the broader implications of chromothripsis in cancer development and treatment.
Resource availability
Lead contact
Requests for further information and resources should be directed to and will be fulfilled by the lead contact, Yonglun Luo (alun@biomed.au.dk).
Materials availability
This study did not generate new unique reagents.
Data and code availability
-
•
WGS and RNA sequencing data have been deposited at Genome Sequence Archive (GSA) as HRA005963, HRA003461, HRA004718, and HRA005001 and China National GeneBank as CNP0003498 and are publicly available as of the date of publication.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Acknowledgments
We gratefully acknowledge the contributions of our patients who participated in the study and their families. This work was jointly supported by IBMC-BGI Center, Qingdao-Europe Advanced Institute for Life Sciences, Taishan Scholar Program (no. Tsqn202103198), the Shandong Provincial Medical Association Clinical Research Funds Qilu Special (YXH2022ZX02181), the Jinan Science and Technology Plan-Clinical Medical Science and Technology Innovation Program (no. 202134003 and no. 202225057), and the First Affiliated Hospital of Shandong First Medical University National Natural Science Foundation Cultivation Program (QYPY2020NSFC0616). W.L. was supported by the China Scholarship Council (CSC). We thank the support from China National GeneBank. Y. Luo is supported by the Danish Research Council (9041-00317B), European Union’s Horizon 2020 research and innovation program under grant agreement no. 899417, the Novo Nordisk Foundation (NNF21OC0068988; NNF21OC0071031), M-ERA.Net and the Innovation Fund Denmark (9355 PIECRISCI), and the Lundbeck Foundation Ascending Investigator award (R396-2022-350).
Author contributions
W.L., P.H., Y. Luo, and C. Lin contributed to the study design and conceptualization. P.H., Y. Luo, C. Lin, W.L., X.J., and X.S. co-supervised the project. P.H., C. Lin, Y. Luo, and X.S. acquired funding. C. Lin, Y.L., T.Y., S.W., N.L., W.W., J.M., and X.C. contributed to the sample and information collection. Y.L. provided WGS and clinical data for the BIG cohort. P.H. and W.L. performed WGS production. W.D., H.Y., and M.Z. provided computational resources. W.L., Y.Z., H. Tao, C.L., and S.J. contributed to computational data analysis. Y.L., H. Tao, X.C., T.F., K.C., K.M., and P.W. assisted with data analysis. Y.Z., H.T., C.C., and Z.X. uploaded the raw sequencing data to GSA. Y. Luo., B.L., L.L., and W.L. supervised the data analysis, data integration, and interpretation. W.L., Y.Z., C.L., and H. Tao. prepared and modified the figures. W.L. wrote the manuscript with input from Y.Z., H.T., and C.L. Y. Luo, P.H., and W.L. revised the manuscript. All authors discussed the results and contributed to the review and editing of the manuscript.
Declaration of interests
The authors declare no competing interests.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Biological samples | ||
| 71 tumors and 20 adjacent tumor samples from 20 UC patients | This paper | |
| Critical commercial assays | ||
| MagAttract HMW DNA kit | Qiagen | 67563 |
| Qubit dsDNA HS Assay kit | Invitrogen | Q32851 |
| MGIEasy DNA Library Preparation Kit | MGI | 1000006985 |
| ZytoLight® SPEC CCND1/CEN11 Dual Color Probe | ZytoVision | Z-2071-200 |
| Deposited data | ||
| In-house-This study Data | This paper, GSA-Human: HRA005963 | https://ngdc.cncb.ac.cn/gsa-human/s/CGL69Tjc |
| Raw sequencing data of three paired tumor-germline WGS-data from the BIG dataset | GSA-Human: HRA000029; GVM: GVM000054 |
https://ngdc.cncb.ac.cn/gsa-human/browse/HRA000029; https://ngdc.cncb.ac.cn/gvm/getProjectDetail?project=GVM000054 |
| In-house Data | GSA-Human: HRA003461, HRA004718, HRA005001; CNGB: CNP0003498 |
https://ngdc.cncb.ac.cn/gsa-human/browse/HRA003461, https://ngdc.cncb.ac.cn/gsa-human/s/Mbxq9RV8, https://ngdc.cncb.ac.cn/gsa-human/s/qTvrKj89, https://db.cngb.org/search/project/CNP0003498/ |
| Software and algorithms | ||
| R (v4.2.2) | R Foundation | https://www.r-project.org/ |
| ShatterSeek (version1.1) | Cortés-Ciriano et al.11 | https://github.com/parklab/shatterseek/ |
| bwa (version 0.7.17) | Li et al.27 | http://bio-bwa.sourceforge.net/bwa.shtml |
| samtools (version 1.11) | Li et al.28 | http://samtools.sourceforge.net/ |
| fastQC (version 0.11.7) | Babraham Bioinformatics | https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ |
| fastp (version 0.23.2) | Chen et al.29 | https://github.com/OpenGene/fastp |
| Perl (version 5.16.3) | Larry Arnold Wall | http://www.perl.org |
| Java (version 1.8.0_262) | / | https://www.oracle.com/java |
| Mutect2 (version 4.2.5.0) | Cibulskis et al.30 | https://gatk.broadinstitute.org/ |
| ANNOVAR (version 2020.06.07) | Wang et al.31 | http://www.openbioinformatics.org/annovar/ |
| AmpliconSuite-pipeline (version 1.0.0) | Deshpande et al.20 | https://github.com/AmpliconSuite/AmpliconSuite-pipeline |
| AmpliconArchitect (version 1.3.r6) | Deshpande et al.20 | https://github.com/AmpliconSuite/AmpliconArchitect |
| AmpliconClassifier (version 1.0.0) | Deshpande et al.20 | https://github.com/jluebeck/AmpliconClassifier |
| CycleViz (version 0.1.5) | Deshpande et al.20 | https://github.com/jluebeck/CycleViz |
| MutationalPatterns (version 3.8.1) | Blokzijl et al.32 | https://bioconductor.org/packages/release/bioc/html/MutationalPatterns.html |
| Bedtools (version 2.30.0) | Quinlan laboratory | https://bedtools.readthedocs.io/en/latest/ |
| gistic2 (version 2.0.23) | Mermel et al.33 | https://broadinstitute.github.io/gistic2/ |
| CNVkit (version 0.9.9) | Talevich et al.34 | https://cnvkit.readthedocs.io/en/stable/ |
| Delly2 (version 1.1.3) | Rausch et al.35 | https://tobiasrausch.com/delly/ |
| Manta (version 1.6.0) | Chen et al.36 | https://github.com/Illumina/manta |
| SvABA (version 1.1.0) | Wala et al.37 | https://github.com/walaj/svaba |
| Lumpy (version 0.2.13) | Layer et al.38 | https://github.com/arq5x/lumpy-sv |
| SURVIVOR (version 1.0.7) | Jeffares et al.39 | https://github.com/fritzsedlazeck/SURVIVOR |
| FACETS (version 0.5.14) | Shen et al.40 | https://github.com/mskcc/facets |
| TrimGalore (version 0.6.7) | / | https://github.com/FelixKrueger/TrimGalore |
| STAR (version 2.7.10a) | Dobin et al.41 | https://github.com/alexdobin/STAR |
| RSEM (version 1.3.3) | Li et al.42 | https://github.com/deweylab/RSEM |
| DESeq2 (version 1.40.2) | Love et al.43 | https://github.com/thelovelab/DESeq2 |
| ClusterProfiler (version 4.6.2) | Wu et al.44 | https://github.com/YuLab-SMU/clusterProfiler |
| GSVA (version 1.46.0) | Hänzelmann et al.45 | https://github.com/rcastelo/GSVA |
| maftools (version 2.14.0) | Mayakonda et al.46 | https://github.com/PoisonAlien/maftools |
| survminer (version 0.4.9) | Alboukadel Kassambara et al. | https://rpkgs.datanovia.com/survminer/index.html |
Experimental model and study participant details
Human samples
We collected 71 tumors and 20 adjacent tumor samples from 20 UC patients, who had not undergone systemic therapy before surgery. There was no selection of patients for specific clinical features. For multifocal urothelial tumors, we sampled tissues from different sites, while for solitary urothelial tumors, we collected samples from distinct tumor regions that were at least 5 mm apart and located away from the tumor margin. Paired adjacent tumor tissues (ATs) were collected at a minimum distance of 2 cm from the tumor margin. To ensure privacy, the identifiers of patients were reassigned. Tissue samples were promptly collected within 30 minutes post-surgery and immediately snap-frozen in liquid nitrogen for subsequent processing. The histology of tumors was evaluated by three pathologists. The study adhered to both national and local laws and regulations and received approval from the Research Ethics Committee of Yantai Yuhuangding Hospital. Before enrollment, all participating patients provided written informed consent, and no compensation was provided for their participation.
Method details
Datasets
We collected WGS data from 558 tumors, 471 germline samples, and 166 urine samples, along with RNA-seq data from 328 tumors sourced from both our in-house and BIG datasets.14 Clinical and sequencing information of included patients is provided in Tables S1 and S2.
Genomic DNA isolation
Genomic DNA was isolated from fresh frozen tissues using the MagAttract HMW DNA kit (Qiagen) according to the manufacturer’s instructions. The concentration and integrity of DNA were assessed using Qubit dsDNA BR (Invitrogen) and electrophoresis on a 0.8% agarose gel, respectively.
Whole genome sequencing (WGS)
500 ng of genomic DNA was subjected to the WGS library construction using the MGIEasy DNA Library Preparation Kit (MGI). Briefly, the genomic DNA was fragmented, ligated with adapters, and subjected to PCR amplification. The constructed libraries were quality-controlled using the Qubit dsDNA HS (Invitrogen), and Agilent 2100 system, and were deep sequenced on DNBSEQ with 150 bp paired-end reads.
WGS processing
FastQC (v0.11.7) was performed to assess the sequencing quality of FASTQ files. Subsequently, fastp27 (v0.23.2) was utilized to trim reads of adapters, N content, and low-quality bases. The processed reads, meeting all filtering criteria, were aligned to the human reference genome GRCh38 using BWA-MEM28 (v0.7.17), resulting in the generation of BAM files. SAMtools29 (v1.11) was then employed to sort and index the BAM files. Coverage analysis was conducted using SAMtools, and the mean coverage for the WGS data used in this study was determined to be 41X (Table S2).
Variant calling
SNVs and indels
Somatic mutations were detected from tumor samples, using matched germline samples as controls. In detail, somatic point mutations were detected using Mutect230 (v4.2.5.0). Somatic small indels were detected using Mutect2,30 and Strelka231 (v2.9.10). Two software were run independently. The indels detected by two callers were merged using BEDTools (v2.30.0). The combined variant format files (VCF) files were split and left trimmed using GATK (v4.2.5.0). The VCF was annotated using ANNOVAR.32 To remove artifacts and false positives, we filtered for variants with > 0.01 allele frequency in 1000 Genomes or Exome Aggregation Consortium (ExAC)33 unless ClinVar34 flags it as pathogenic.
SVs
Somatic SVs were identified from paired tumor-germline WGS data using four callers: Delly235 (v1.1.3), SvABA36 (v 1.1.0), Lumpy37 (v0.2.13), and Manta38 (v1.6.0). Each algorithm was executed independently. The SVs generated by each caller were merged using SURVIVOR39 (v1.0.7), with a 1 Kb allowance for slop at the breakpoints. Only calls generated by at least two algorithms were retained. We calculated the number of SV for each sample.
CNVs
Tumor purity and ploidy were computed for all tumor and urine samples using FACETS40 (v0.5.14). FACETS provides allele-specific estimations of the copy numbers. As previously described,41 if the fraction of the genome with a major copy number ≥ 2 was greater than 50%, samples were classified as exhibiting whole genome duplication (WGD). Somatic CNVs were inferred by CNVkit42 (v0.9.9). GISTIC243 was applied to evaluate the gene copy number status. The somatic variations (including CNV, SNV, and indel) in ten 10 canonical signaling pathways were assessed using maftools44 (2.14.0).
Kataegis detection
We used maftools (v2.14.0)44 to identify kataegis by analyzing mutation clusters across the genome.
Chromothripsis calling
Chromothripsis events were identified through ShatterSeek11 (v1.1) using CNV and SV data as input. These chromothripsis events were classified into high-confidence (HC) and low-confidence (LC) categories based on established criteria outlined by Cortés-Ciriano et al. LC calls encompassed 4-6 segments between two states, while HC calls consisted of a minimum of 7 adjacent segments. Both HC and LC calls were retained for subsequent analysis. Moreover, we defined chromothripsis events in which more than 60% of the CN segments in the affected region oscillated between two states as canonical chromothripsis events.
Focal amplifications calling
To identify the presence of focal amplifications, we applied the AmpliconSuite-pipeline wrapper as previously described.45 For seed detection, the wrapper pipeline integrated CNVKit42 in tumor-normal mode, calling somatic CNVs against matched germline WGS data for each case. The amplified_intervals.py script was then utilized to filter regions with a copy number exceeding 4.5 and a size larger than 50 Kb, yielding a set of seed regions. Subsequently, the wrapper invoked AmpliconArchitect20 (v1.3.r6) in default mode on the WGS BAM files to scrutinize seed regions and profile the architecture of focal amplifications. AmpliconClassifier46 (v0.4.12) was used to predict the types of amplifications present in AmpliconArchitect’s output. Through this approach, we categorized focal amplifications into ecDNA, BFB, and other fSCNA (including complex non-cyclic and linear focal amplifications).
Mutational signature analysis
Point mutations, including non-synonymous and synonymous mutations, were classified into 96 substitution types, based on six base substitutions (C > A, C > G, C > T, T > A, T > C, and T > G) and their neighboring bases. The MutationalPatterns47 R package was employed to assess the activity of APOBEC-related mutational signatures (COSMIC SBS2 and SBS13) for each sample.
CNV signature quantification
The activities of the CNV signature were quantified using sigminer (v2.2.0) software.
Processing of RNA-seq data
The raw RNA-seq data from 328 tumors were evaluated for quality using FastQC (v. 0.11.7). We then used TrimGalore (v.0.6.7) to remove low-quality bases and adapter sequences. The clean reads were aligned to the human reference genome (GRCh38) using STAR48 (v.2.7.10a). Transcript quantification was calculated using RSEM49 (v.1.3.3). We filtered out genes with no expression in all of the samples for downstream GSVA analysis. Single-sample gene set enrichment analysis scores for each sample were calculated using the previously defined gene sets and the GSVA (v.1.46.0)50 R package. DEG analysis was performed by DESeq2 (v.1.40.2).51 ClusterProfiler (v.4.6.2)52 R package was performed for KEGG pathway analysis.
Interphase FISH
Interphase FISH was carried out using the ZytoLight ® SPEC CCND1/CEN 11 Dual Color Probe to confirm CCND1 amplification in four available FFPE tumor samples. Briefly, FFPE samples were deparaffinized, rehydrated, and underwent heat-induced epitope retrieval. Slides were dehydrated, and FISH probes in a hybridization buffer were applied. Denaturation occurred at 72°C for 5 minutes, followed by overnight hybridization at 37°C. Slides were washed in 2×SSC/0.3% NP-40 (pH 7.0-7.5) and stained with 10μl of DAPI buffer. Images were captured using OLYMPUS BX53.
Analysis of clinical outcomes
The prognostic values of chromothripsis were assessed in our cohort. The sample size was N = 436 (272 chromothripsis+; 164 chromothripsis-). We employed the log-rank test to evaluate survival differences and conducted Cox proportional hazards regression analysis to identify independent prognostic factors. Survival curves were generated using the R package Survminer (v.0.4.9).
Quantification and statistical analysis
All statistical tests were performed in R (v4.2.2). Statistical tests, test statistics, and p values are indicated in figure legends and the results section. p-values < 0.05 were considered statistically significant.
Published: December 2, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2024.111510.
Contributor Information
Wei Lv, Email: lvwei@biomed.au.dk.
Jinzhao Song, Email: songjinzhao@ucas.ac.cn.
Xicheng Song, Email: drxchsong@163.com.
Yonglun Luo, Email: alun@biomed.au.dk.
Chunhua Lin, Email: chunhua.lin@qdu.edu.cn.
Peng Han, Email: han.peng@bio.ku.dk.
Supplemental information
References
- 1.Flaig T.W., Spiess P.E., Abern M., Agarwal N., Bangs R., Buyyounouski M.K., Chan K., Chang S.S., Chang P., Friedlander T., et al. NCCN Guidelines® Insights: Bladder Cancer, Version 3.2024. J. Natl. Compr. Cancer Netw. 2024;22:216–225. doi: 10.6004/jnccn.2024.0024. [DOI] [PubMed] [Google Scholar]
- 2.Tran L., Xiao J.F., Agarwal N., Duex J.E., Theodorescu D. Advances in bladder cancer biology and therapy. Nat. Rev. Cancer. 2021;21:104–121. doi: 10.1038/s41568-020-00313-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Glaser A.P., Fantini D., Shilatifard A., Schaeffer E.M., Meeks J.J. The evolving genomic landscape of urothelial carcinoma. Nat. Rev. Urol. 2017;14:215–229. doi: 10.1038/nrurol.2017.11. [DOI] [PubMed] [Google Scholar]
- 4.Sfakianos J.P., Cha E.K., Iyer G., Scott S.N., Zabor E.C., Shah R.H., Ren Q., Bagrodia A., Kim P.H., Hakimi A.A., et al. Genomic Characterization of Upper Tract Urothelial Carcinoma. Eur. Urol. 2015;68:970–977. doi: 10.1016/j.eururo.2015.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fujii Y., Sato Y., Suzuki H., Kakiuchi N., Yoshizato T., Lenis A.T., Maekawa S., Yokoyama A., Takeuchi Y., Inoue Y., et al. Molecular classification and diagnostics of upper urinary tract urothelial carcinoma. Cancer Cell. 2021;39:793–809.e8. doi: 10.1016/j.ccell.2021.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Robertson A.G., Kim J., Al-Ahmadie H., Bellmunt J., Guo G., Cherniack A.D., Hinoue T., Laird P.W., Hoadley K.A., Akbani R., et al. Comprehensive Molecular Characterization of Muscle-Invasive Bladder Cancer. Cell. 2017;171:540–556.e25. doi: 10.1016/j.cell.2017.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stephens P.J., Greenman C.D., Fu B., Yang F., Bignell G.R., Mudie L.J., Pleasance E.D., Lau K.W., Beare D., Stebbings L.A., et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell. 2011;144:27–40. doi: 10.1016/j.cell.2010.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang C.Z., Spektor A., Cornils H., Francis J.M., Jackson E.K., Liu S., Meyerson M., Pellman D. Chromothripsis from DNA damage in micronuclei. Nature. 2015;522:179–184. doi: 10.1038/nature14493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Korbel J.O., Campbell P.J. Criteria for inference of chromothripsis in cancer genomes. Cell. 2013;152:1226–1236. doi: 10.1016/j.cell.2013.02.023. [DOI] [PubMed] [Google Scholar]
- 10.Leibowitz M.L., Zhang C.Z., Pellman D. Chromothripsis: A New Mechanism for Rapid Karyotype Evolution. Annu. Rev. Genet. 2015;49:183–211. doi: 10.1146/annurev-genet-120213-092228. [DOI] [PubMed] [Google Scholar]
- 11.Cortés-Ciriano I., Lee J.J.K., Xi R., Jain D., Jung Y.L., Yang L., Gordenin D., Klimczak L.J., Zhang C.Z., Pellman D.S., et al. Comprehensive analysis of chromothripsis in 2,658 human cancers using whole-genome sequencing. Nat. Genet. 2020;52:331–341. doi: 10.1038/s41588-019-0576-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Voronina N., Wong J.K.L., Hübschmann D., Hlevnjak M., Uhrig S., Heilig C.E., Horak P., Kreutzfeldt S., Mock A., Stenzinger A., et al. The landscape of chromothripsis across adult cancer types. Nat. Commun. 2020;11:2320. doi: 10.1038/s41467-020-16134-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bolkestein M., Wong J.K.L., Thewes V., Körber V., Hlevnjak M., Elgaafary S., Schulze M., Kommoss F.K.F., Sinn H.P., Anzeneder T., et al. Chromothripsis in Human Breast Cancer. Cancer Res. 2020;80:4918–4931. doi: 10.1158/0008-5472.Can-20-1920. [DOI] [PubMed] [Google Scholar]
- 14.Lu H., Liang Y., Guan B., Shi Y., Gong Y., Li J., Kong W., Liu J., Fang D., Liu L., et al. Aristolochic acid mutational signature defines the low-risk subtype in upper tract urothelial carcinoma. Theranostics. 2020;10:4323–4333. doi: 10.7150/thno.43251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Y., Schwab C., Ryan S., Papaemmanuil E., Robinson H.M., Jacobs P., Moorman A.V., Dyer S., Borrow J., Griffiths M., et al. Constitutional and somatic rearrangement of chromosome 21 in acute lymphoblastic leukaemia. Nature. 2014;508:98–102. doi: 10.1038/nature13115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sanchez-Vega F., Mina M., Armenia J., Chatila W.K., Luna A., La K.C., Dimitriadoy S., Liu D.L., Kantheti H.S., Saghafinia S., et al. Oncogenic Signaling Pathways in The Cancer Genome Atlas. Cell. 2018;173:321–337.e10. doi: 10.1016/j.cell.2018.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hanel W., Moll U.M. Links between mutant p53 and genomic instability. J. Cell. Biochem. 2012;113:433–439. doi: 10.1002/jcb.23400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eischen C.M. Genome Stability Requires p53. Cold Spring Harbor Perspect. Med. 2016;6 doi: 10.1101/cshperspect.a026096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shoshani O., Brunner S.F., Yaeger R., Ly P., Nechemia-Arbely Y., Kim D.H., Fang R., Castillon G.A., Yu M., Li J.S.Z., et al. Chromothripsis drives the evolution of gene amplification in cancer. Nature. 2021;591:137–141. doi: 10.1038/s41586-020-03064-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deshpande V., Luebeck J., Nguyen N.P.D., Bakhtiari M., Turner K.M., Schwab R., Carter H., Mischel P.S., Bafna V. Exploring the landscape of focal amplifications in cancer using AmpliconArchitect. Nat. Commun. 2019;10:392. doi: 10.1038/s41467-018-08200-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vitale I., Shema E., Loi S., Galluzzi L. Intratumoral heterogeneity in cancer progression and response to immunotherapy. Nat. Med. 2021;27:212–224. doi: 10.1038/s41591-021-01233-9. [DOI] [PubMed] [Google Scholar]
- 22.Lv W., Pan X., Han P., Wang Z., Feng W., Xing X., Wang Q., Qu K., Zeng Y., Zhang C., et al. Circle-Seq reveals genomic and disease-specific hallmarks in urinary cell-free extrachromosomal circular DNAs. Clin. Transl. Med. 2022;12 doi: 10.1002/ctm2.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lv W., Pan X., Han P., Wu S., Zeng Y., Wang Q., Guo L., Xu M., Qi Y., Deng L., et al. Extrachromosomal circular DNA orchestrates genome heterogeneity in urothelial bladder carcinoma. Theranostics. 2024;14:5102–5122. doi: 10.7150/thno.99563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rode A., Maass K.K., Willmund K.V., Lichter P., Ernst A. Chromothripsis in cancer cells: An update. Int. J. Cancer. 2016;138:2322–2333. doi: 10.1002/ijc.29888. [DOI] [PubMed] [Google Scholar]
- 25.Rose K.M., Huelster H.L., Meeks J.J., Faltas B.M., Sonpavde G.P., Lerner S.P., Ross J.S., Spiess P.E., Grass G.D., Jain R.K., et al. Circulating and urinary tumour DNA in urothelial carcinoma - upper tract, lower tract and metastatic disease. Nat. Rev. Urol. 2023;20:406–419. doi: 10.1038/s41585-023-00725-2. [DOI] [PubMed] [Google Scholar]
- 26.Zeng Y., Wang A., Lv W., Wang Q., Jiang S., Pan X., Wang F., Yang H., Bolund L., Lin C., et al. Recent development of urinary biomarkers for bladder cancer diagnosis and monitoring. Clin. Transl. Discov. 2023;3 doi: 10.1002/ctd2.183. [DOI] [Google Scholar]
- 27.Chen S., Zhou Y., Chen Y., Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34:i884–i890. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vasimuddin M., Misra S., Li H., Aluru S. 2019 IEEE international parallel and distributed processing symposium (IPDPS) IEEE; 2019. Efficient Architecture-Aware Acceleration of BWA-MEM for Multicore Systems; pp. 314–324. [Google Scholar]
- 29.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cibulskis K., Lawrence M.S., Carter S.L., Sivachenko A., Jaffe D., Sougnez C., Gabriel S., Meyerson M., Lander E.S., Getz G. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat. Biotechnol. 2013;31:213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim S., Scheffler K., Halpern A.L., Bekritsky M.A., Noh E., Källberg M., Chen X., Kim Y., Beyter D., Krusche P., Saunders C.T. Strelka2: fast and accurate calling of germline and somatic variants. Nat. Methods. 2018;15:591–594. doi: 10.1038/s41592-018-0051-x. [DOI] [PubMed] [Google Scholar]
- 32.Wang K., Li M., Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38 doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O'Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B., et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Landrum M.J., Lee J.M., Benson M., Brown G.R., Chao C., Chitipiralla S., Gu B., Hart J., Hoffman D., Jang W., et al. ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 2018;46:D1062–D1067. doi: 10.1093/nar/gkx1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rausch T., Zichner T., Schlattl A., Stütz A.M., Benes V., Korbel J.O. DELLY: structural variant discovery by integrated paired-end and split-read analysis. Bioinformatics. 2012;28:i333–i339. doi: 10.1093/bioinformatics/bts378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wala J.A., Bandopadhayay P., Greenwald N.F., O'Rourke R., Sharpe T., Stewart C., Schumacher S., Li Y., Weischenfeldt J., Yao X., et al. SvABA: genome-wide detection of structural variants and indels by local assembly. Genome Res. 2018;28:581–591. doi: 10.1101/gr.221028.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Layer R.M., Chiang C., Quinlan A.R., Hall I.M. LUMPY: a probabilistic framework for structural variant discovery. Genome Biol. 2014;15:R84. doi: 10.1186/gb-2014-15-6-r84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen X., Schulz-Trieglaff O., Shaw R., Barnes B., Schlesinger F., Källberg M., Cox A.J., Kruglyak S., Saunders C.T. Manta: rapid detection of structural variants and indels for germline and cancer sequencing applications. Bioinformatics. 2016;32:1220–1222. doi: 10.1093/bioinformatics/btv710. [DOI] [PubMed] [Google Scholar]
- 39.Jeffares D.C., Jolly C., Hoti M., Speed D., Shaw L., Rallis C., Balloux F., Dessimoz C., Bähler J., Sedlazeck F.J. Transient structural variations have strong effects on quantitative traits and reproductive isolation in fission yeast. Nat. Commun. 2017;8 doi: 10.1038/ncomms14061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shen R., Seshan V.E. FACETS: allele-specific copy number and clonal heterogeneity analysis tool for high-throughput DNA sequencing. Nucleic Acids Res. 2016;44 doi: 10.1093/nar/gkw520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bielski C.M., Zehir A., Penson A.V., Donoghue M.T.A., Chatila W., Armenia J., Chang M.T., Schram A.M., Jonsson P., Bandlamudi C., et al. Genome doubling shapes the evolution and prognosis of advanced cancers. Nat. Genet. 2018;50:1189–1195. doi: 10.1038/s41588-018-0165-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Talevich E., Shain A.H., Botton T., Bastian B.C. CNVkit: Genome-Wide Copy Number Detection and Visualization from Targeted DNA Sequencing. PLoS Comput. Biol. 2016;12 doi: 10.1371/journal.pcbi.1004873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mermel C.H., Schumacher S.E., Hill B., Meyerson M.L., Beroukhim R., Getz G. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 2011;12:R41. doi: 10.1186/gb-2011-12-4-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mayakonda A., Lin D.C., Assenov Y., Plass C., Koeffler H.P. Maftools: efficient and comprehensive analysis of somatic variants in cancer. Genome Res. 2018;28:1747–1756. doi: 10.1101/gr.239244.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Luebeck J., Ng A.W.T., Galipeau P.C., Li X., Sanchez C.A., Katz-Summercorn A.C., Kim H., Jammula S., He Y., Lippman S.M., et al. Extrachromosomal DNA in the cancerous transformation of Barrett's oesophagus. Nature. 2023;616:798–805. doi: 10.1038/s41586-023-05937-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim H., Nguyen N.P., Turner K., Wu S., Gujar A.D., Luebeck J., Liu J., Deshpande V., Rajkumar U., Namburi S., et al. Extrachromosomal DNA is associated with oncogene amplification and poor outcome across multiple cancers. Nat. Genet. 2020;52:891–897. doi: 10.1038/s41588-020-0678-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Blokzijl F., Janssen R., van Boxtel R., Cuppen E. MutationalPatterns: comprehensive genome-wide analysis of mutational processes. Genome Med. 2018;10:33. doi: 10.1186/s13073-018-0539-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li B., Dewey C.N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinf. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hänzelmann S., Castelo R., Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinf. 2013;14 doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu T., Hu E., Xu S., Chen M., Guo P., Dai Z., Feng T., Zhou L., Tang W., Zhan L., et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation. 2021;2 doi: 10.1016/j.xinn.2021.100141. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
WGS and RNA sequencing data have been deposited at Genome Sequence Archive (GSA) as HRA005963, HRA003461, HRA004718, and HRA005001 and China National GeneBank as CNP0003498 and are publicly available as of the date of publication.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.