Summary
Blood cell aggregates are clinically useful biomarkers in a number of medical disorders. This protocol provides accurate and quantitative analysis of cell aggregates using a small volume of whole blood and imaging flow cytometry. We describe steps for sample collection, staining, and measurement. We then detail gating procedures and analysis of cell morphology. Sample preparation artifacts, activation, and morphological changes of cells are mitigated by omitting erythrocyte lysis and leukocyte isolation while maintaining high-throughput accurate imaging of leukocytes and platelets.
Subject areas: Clinical Protocol, Flow Cytometry, Immunology, Microscopy
Graphical abstract

Highlights
-
•
Instructions for the automated analysis of cell aggregates via imaging flow cytometry
-
•
Guidance on sample processing to reduce aggregation and activation of platelets
-
•
Steps for processing 50 μL of whole blood and gaining results in under 1 h
-
•
Directions for anticoagulant selection and comparison to a standard lysis protocol
Publisher’s note: Undertaking any experimental protocol requires adherence to local institutional guidelines for laboratory safety and ethics.
Blood cell aggregates are clinically useful biomarkers in a number of medical disorders. This protocol provides accurate and quantitative analysis of cell aggregates using a small volume of whole blood and imaging flow cytometry. We describe steps for sample collection, staining, and measurement. We then detail gating procedures and analysis of cell morphology. Sample preparation artifacts, activation, and morphological changes of cells are mitigated by omitting erythrocyte lysis and leukocyte isolation while maintaining high-throughput accurate imaging of leukocytes and platelets.
Before you begin
Microaggregates as clinical biomarkers across numerous diseases are being linked to a spectrum of acute and chronic thrombo-inflammatory disorders, including cardiovascular diseases,1,2 Acute Respiratory Distress Syndrome,3,4,5 sepsis,6 stroke,7 and hemodialysis.8 Especially platelet-leukocyte (PLT-WBC) aggregates frequently occur as possible biomarkers in the prediction of microvascular injury in diabetes mellitus.9 We measured the most important surface markers for direct PLT-WBC interactions, namely p-selectin (CD62P), platelet glycoprotein IIb (CD41a), platelet endothelial cell adhesion molecule (CD31), and CD45.
This protocol outlines procedural guidelines for the rapid assessment of platelet activation, platelet-platelet (PLT-PLT), PLT-WBC, and leukocyte-leukocyte aggregates from just 50 μL of whole blood (WB). Additionally, we assess the most suitable anticoagulants for studying cell aggregates and platelet activation. We also report the effects of ammonium chloride-based red blood cell (RBC) lysis buffer on the morphological characteristics of platelets and aggregates of blood cells, as literature on the effects of RBC lysis on blood cell aggregates is limited. Furthermore, a gating strategy was developed to differentiate blood cell aggregates and accurately measure the sizes of individual cells or cell aggregates in peripheral blood using imaging flow cytometry (IFC). The implemented gating method effectively eliminates most undesired coincidental events from the aggregate analysis.
Institutional permissions
To conduct research with patient-derived materials according to this protocol, it is necessary to get ethical permission from the appropriate institutional or regulatory agencies.
Informed consent was obtained from all subjects.
Our study has been approved by the NHG DOMAIN SPECIFIC REVIEW BOARD (DSRB).
Study Reference Number: 2021/00930 and 2021/01130 NHG DSRB – Singapore.
ImageStreamX Mk II imaging flow cytometry setup
The ImageStreamX Mk II used in this protocol is equipped with a total of five lasers, a violet laser with a wavelength of 405 nm, a blue laser with a wavelength of 488 nm, a green laser with a wavelength of 561 nm, a red laser with a wavelength of 642 nm, as well as a side scatter laser with a wavelength of 785 nm. Additionally, the instrument also includes a bright field. As the size of platelets is sufficient, no additional “High Gain” setting is necessary.
-
1.
Configure the camera to operate at a magnification of 60x.
-
2.
The laser intensities should be adjusted in accordance with the signal intensities specific to the given channel.
-
3.
To establish the compensation, single stain controls with either beads or cells are required, isotype controls are optional.
Note: Acquisition Software: INSPIRE v. 200.1.681.0.
Laser settings
| Laser | Proposed Watts |
|---|---|
| 405 nm | 120 mW |
| 488 nm | 100 mW |
| 561 nm | 100 mW |
| 642 nm | 150 mW |
Note: Ensuring appropriate panel design is crucial to avoid spectral overlap. It is advisable to verify the instrument configuration and formulate the panel design in accordance with the filters accessible for each laser.
Note: For a detailed explanation on how to use the instrument and analysis software, please refer to the user manual.
Starting and priming the imaging flow cytometer prior to protocol start
Timing: 1 h
-
4.
Refill consumables: Speedbeads, PBS 1X, detergent and empty waste container.
-
5.
Switch on the imaging flow cytometry equipment.
-
6.
Open the pre-installed INSPIRE data acquisition software on the computer.
-
7.
Execute the automatic startup process of the instrument and view the diagnostic plots.
-
8.
Turn on the lasers and bright field corresponding to the fluorochromes used for the analysis.
-
9.First time use only: Design an acquisition template that aligns with the desired objective and focus on the immune cell population of interest (Figure 1A) and establish a compensation matrix.Note: We focus on excluding erythrocytes. For the analysis of PLT-WBC and PLT-PLT aggregates we recommend a CD41a - CD45 biplot.
-
a.Save all events excluding the double negative population.
-
b.Load the compensation matrix.
-
c.Set the channel names and optimize image quality via the histogram tool.
-
d.Save the template for future use.
-
a.
-
10.
Open the acquisition template.
-
11.
Make sure appropriate storage space is available on the PC.
Note: Always switch on the instrument in time to address possible QC issues in the automatic startup.
Figure 1.
Gating of IFC data, WB-Protocol, unstimulated and unfixed
(A) The acquisition template is designed to gate out most RBCs via negative selection. The left figure displays the available populations for reference, the right figure displays the actual acquisition gate used for the experiments. Only events in this gate are saved for future analysis.
(B) The analysis is done in Ideas 6.3, proprietary software which combines bright-field imaging and fluorescent intensities to provide high throughput image-based analysis. First, small particles and focusing beads (Amnis SpeedBead) were excluded. After gating on in-focus events, we analyze aggregates containing WBCs or only PLTs.
(B I) To detect WBC-PLT aggregates and WBC-WBC aggregates, we use following gating strategy: WBC-WBC aggregates are detected via symmetry and circularity analysis. For WBC-PLT aggregates, double positive events for CD45+ and CD41a+ were selected. To exclude coincidental events (platelets and WBCs share one frame but are not adherent to each other) we used the internalization mask. The actual aggregates can now be analyzed using various morphological features.
(B II) To detect PLT-PLT aggregates, PLT activation and shape following gating strategy was used: All WBC-PLT aggregates were excluded. To remove remaining RBCs we gate on CD31-CD41a double positive events. This population is analyzed for PLT-PLT aggregates, platelet activation and platelet morphology.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| BV421 mouse anti-human CD45 (clone: HI30) dilution 1:100 |
BD Biosciences | Cat # 563880; RRID AB_2744402 |
| APC mouse anti-human CD41a (HIP8) dilution 1:50 |
BD Biosciences | Cat # 559777; RRID AB_398671 |
| BV510 mouse anti-human CD62p (clone: AK-4) dilution 1:67 |
BD Biosciences | Cat # 744225; RRID AB_2742072 |
| RY586 mouse anti-human CD31 (clone: L133.1) dilution 1:50 |
BD Biosciences | Cat # 753147; RRID AB_3096285 |
| RY586 mouse IgG1, k isotype control | BD Biosciences | Cat # 568097; RRID AB_3102022 |
| BV421 mouse IgG1, κ isotype control (clone: X40) | BD Biosciences | Cat # 562438; RRID AB_11207319 |
| APC mouse IgG1, κ isotype control | BD Biosciences | Cat # 555751; RRID AB_398576 |
| BV 510 mouse IgG1, k isotype control | BD Biosciences | Cat # 562946; RRID AB_2784530 |
| BD Pharmingen human BD Fc Block clone FC1 RUO |
BD Biosciences | Cat #564219; RRID AB_2728082 |
| Chemicals, peptides, and recombinant proteins | ||
| HEPES buffer solution | Gibco | 15630-106 |
| NaCl | Sigma-Aldrich | S7653-1KG |
| KCl | Sigma-Aldrich | P9333-1KG |
| MgCl2 | Sigma-Aldrich | M8266-1KG |
| NaHCO3 | Sigma-Aldrich | S6014-1KG |
| Na2HPO4 | Sigma-Aldrich | S9763-1KG |
| D-(+)-Glucose | Sigma-Aldrich | G8270-1KG |
| Bovine serum albumin (BSA) | Sigma-Aldrich | A2153-10G |
| NaOH | Sigma-Aldrich | 1310-73-2 |
| Thrombin receptor activator peptide-6 (TRAP-6) | Sigma-Aldrich | T1573-5MG |
| RBC lysis buffer (10x) | BioLegend | Cat # 420302 |
| Human TruStain FcX | BioLegend | Cat # 422302 |
| 10× phosphate-buffered saline | 1st BASE | BUF-2040-10X4L |
| BD Vacutainer K2 EDTA 7.2 mg | BD | 367856 |
| BD Vacutainer buffered Na citrate 0.109 M, 3.2% | BD | 363083 |
| BD Vacutainer lithium heparin 68 USP units | BD | 367884 |
| Amnis SpeedBead kit for ImageStreamX system, ISX400041 | CYTEK | CN-0440-01 |
| Software and algorithms | ||
| IDEAS 6.3 | CYTEK | CN-SW69-01; https://cytekbio.com/pages/imagestream |
| INSPIRE acquisition software | CYTEK | Version 200.1.681.0 https://cytekbio.com/pages/imagestream |
| Deposited data | ||
| Raw data | this manuscript | upon request |
| Other | ||
| Luminex Amnis ImageStreamX Mk II system | Luminex | RRID:SCR_018589 |
| 10 μL pipette tips | any supplier | NA |
| 200 μL pipette tips | any supplier | NA |
| 1,000 μL pipette tips | any supplier | NA |
| 1.5 mL Eppendorf tubes | any supplier | NA |
| 50 mL Falcon tubes | any supplier | NA |
| B. Braun butterfly Venofix 19G | B. Braun | Cat # 10552-001 |
Materials and equipment
Preparation of reagents
HEPES modified Tyrode’s buffer10
| Reagent | Final concentration | Amount |
|---|---|---|
| HEPES Buffer Solution 1 M | 10 mM | 5 mL |
| NaCl | 137 mM | 4 g |
| KCl | 2.8 mM | 104 mg |
| MgCl2 | 1 mM | 47 mg |
| NaHCO3 | 12 mM | 504 mg |
| Na2HPO4 | 0.4 mM | 28 mg |
| D-(+)-Glucose | 5.5 mM | 495 mg |
| Bovine Serum Albumin (BSA) | 0.35% | 1.75 g |
| ddH2O | N/A | 495 mL |
| Total | N/A | 500 mL |
Note: Store at 4°C for up to 1 month or at −15°C for up to 6 months.
Note: After preparation, the PH should be adjusted to 7.4 using 0.1 M NaOH.
Lysis buffer: 5 mL RBC Lysis Buffer (10x) in 45 mL ddH2O.
Note: Store at 4°C for up to 1 month or at −15°C for up to 6 months.
Preparation before blood draw
Antibody mix
Prepare antibody mix, store at 4°C in darkness until used.
Antibody mix - sample number and lot dependent (50 μL/sample)
| Reagent | Final concentration in staining mix | Amount for one sample | Dilution |
|---|---|---|---|
| BV421 Mouse Anti-human CD45 (clone HI30) | 4 μg/mL | 1 μL | 1:100 |
| APC Mouse Anti-human CD41a (clone HIP8) | 1 μg/mL | 2 μL | 1:50 |
| BV510 Mouse Anti-human CD62p (clone AK-4) | 6 μg/mL | 1.5 μL | 1:67 |
| RY586 Mouse Anti-human CD31 (clone L133.1) | 8 μg/mL | 2 μL | 1:50 |
| HEPES modified Tyrode’s buffer | – | 43.5 μL | – |
Note: Antibody concentration and dilution is Lot Number dependant.
Note: prepare fresh for each day.
Optional: Thrombin Receptor Activator Peptide-6 for platelet stimulation
10x Stock solution Thrombin Receptor Activator Peptide-6 (TRAP-6): To prepare 1.1 mmol/L TRAP-6 10x stock solution dissolve 0.5 mg TRAP-6 in 600 μL 1X PBS (see key resources table).
Note: Store at 4°C for up to 1 month or at −15°C for up to 6 months.
1x Stock solution of TRAP-6: Only immediately before stimulating the platelets: dilute 10x stock solution 1:10 in 1X PBS to obtain 1X working solution TRAP-6 (0.11 mmol/L).
4% Paraformaldehyde solution (PFA)
| Reagent | Final concentration | Amount |
|---|---|---|
| Paraformaldehyde 32% vial | 4% | 10 mL |
| PBS | N/A | 70 mL |
| Total | N/A | 80 mL |
Note: Store at 4°C for 2 weeks or at −20°C for a long-time storage.
Note: Work with paraformaldehyde should be conducted in a well-ventilated area, preferably within a chemical fume hood, to minimize inhalation of formaldehyde vapors. Personal protective equipment (PPE) such as gloves, safety goggles, and lab coats should be worn to prevent skin and eye contact.
Alternative reagents
Antibodies and lasers can be exchanged freely. Make sure that the selected antibodies do not interfere with cell activation or morphology.
TRAP6 can be exchanged for stimulants like ADP, arachidonic acid, or adrenaline, which can be employed for platelet stimulation. To properly stimulate platelets, it is possible to add calcium gluconate to a final concentration of 2 mmol/L to the buffer. This allows for a more physiological platelet response.
This protocol was also tested on the Luminex Amnis ImageStreamX Mk II System featuring 8 channels. Using this protocol, measurements with a conventional cytometer are possible, but the gating strategy needs to be redesigned, as morphological features can no longer be analyzed.
Step-by-step method details
Note: It is highly recommended to start the IFC instrument prior to blood collection, since the device requires around one hour to prime and start (see above).
Collection of human peripheral blood and transportation to research laboratory
Timing: 20 min
In this step, we summarize our sample collection and handling recommendations.
Note: We recommend buffered Na citrate, 0.109 M, 3.2%, as the anticoagulant of choice as it is commonly available and causes minimal platelet activation.
CRITICAL: Rapid processing of the sample is of critical importance. A vacuum tube or a manually-aspirated syringe with a minimum size of 21G can be used when assessing platelet activation, there is no need for a discard tube.11
-
1.Obtain peripheral blood samples.
-
a.Obtain informed consent from the patient prior to the procedure.Note: Blood collection must be performed exclusively by trained and certified phlebotomists or healthcare professionals who are proficient in venipuncture techniques as repeated or insufficiently performed venipuncture will lead to premature platelet activation. 5 min
-
b.Ensure all necessary equipment (e.g., needles, collection tubes, tourniquets, antiseptics) is sterile and readily available. 5 min
-
c.Collect blood samples by venipuncture into citrated vacutainer tubes (BD, 0.109 M, citrated). Don’t underfill or overfill the tubes. 5 min
-
d.Check that blood flows freely into the collection tube once the vein is punctured, indicating proper needle placement. 5 min
-
e.Gently invert the tubes immediately after collection. Do not shake the tubes.
-
f.Once the blood has been collected, ensure rapid transport to the lab at room temperature.12Optional: If processing the sample within 2 h is not possible, consider following option.
-
i.Fix the WB sample within 1 h in 2% paraformaldehyde (PFA) final concentration for 10 min
-
ii.Wash the sample with 1 mL of HEPES-modified Tyrode’s buffer at 400 g for 6 min.
-
iii.Resuspend pellet in HEPES buffer and store in 4 degrees Celsius for up to 12 h.
 CRITICAL: Sample staining should be initiated in a 30-min timeframe after blood collection to prevent pre-analytic platelet activation, aggregation of blood cells, and modifications in cellular function that may impact the expression of surface receptors.
CRITICAL: Sample staining should be initiated in a 30-min timeframe after blood collection to prevent pre-analytic platelet activation, aggregation of blood cells, and modifications in cellular function that may impact the expression of surface receptors.
-
i.
-
a.
RBC lysis method
Timing: 25 min
This step is only required for the reproduction of our data or if needed for specific research questions.
Note: We advise against performing lysis, especially without fixative. As shown in Figure 5, lysis induces significant platelet activation compared to unlysed whole blood.
-
2.
Mix 50 μL of whole blood with 1 mL of room temperature RBC lysis buffer for a duration of 5–8 min in a 1.5 mL Eppendorf tube. 8 min
-
3.
Gently agitate until the solution is clear. 1 min
-
4.
Remove the lysate solution by centrifugation at 400g for 5 min at RT. 7 min
Note: Select the slowest possible acceleration and deceleration rate.
-
5.
Discard the supernatant.
-
6.
Add 1 mL of HEPES modified Tyrode’s buffer and centrifuge the sample at 400g for 5 min at room temperature. 7 min
-
7.
Carefully remove the supernatant. 1 min
-
8.
Resuspend the pellet in 50 μL of HEPES modified Tyrode’s buffer. 1 min
-
9.
Continue with the cell staining protocol.
CRITICAL: Warm up the lysis buffer to RT while setting up the IFC.
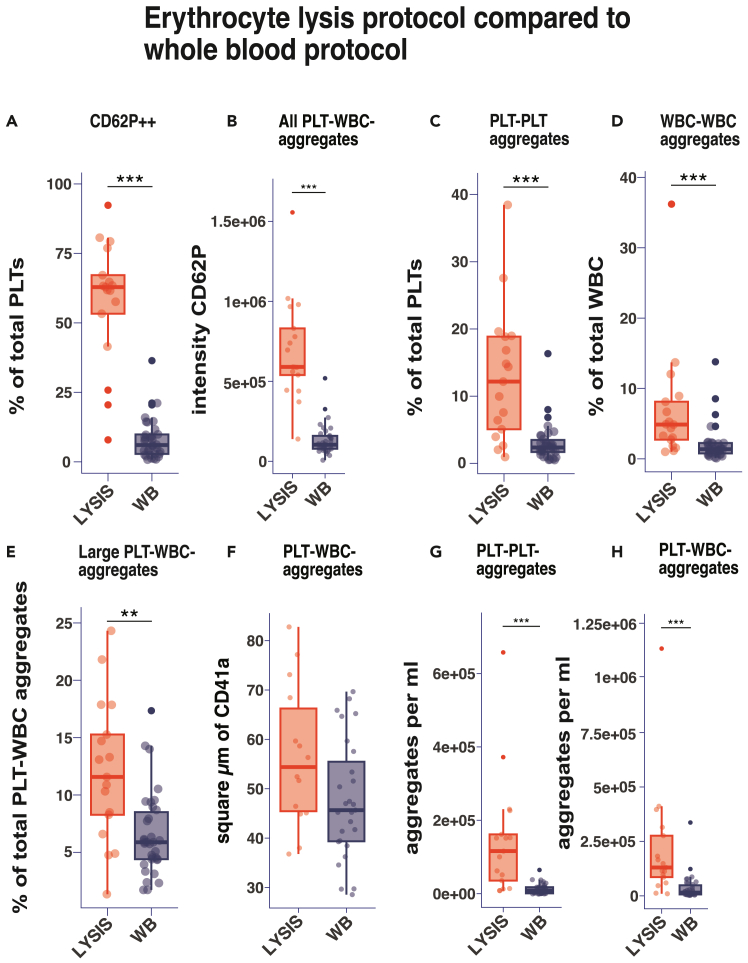
Figure 5.
Comparison of WB and RBC lysis protocols
The WB protocol was compared with an RBC lysis protocol, n = 17. The median intensity or the ratio of the selected population is displayed. We performed a Kruskal-Wallis with Dunn’s Post hoc where ∗ : p < 0.05, ∗∗ :p < 0.005, ∗∗∗: p < 0.0005.
(A) The ratio of positive CD62P expression on the PLT surface based on gate B-II-5b was compared.
(B) The intensity of CD62P on PLT-WBC-aggregates.
(C–E) The ratio of cell aggregates compared to the respective parent populations are displayed. Large PLT-leukocyte aggregates are defined by gate B-I-7.
(F) The size of PLT aggregates determined by the CD41a expression area, based on gate B-I-6 are being compared.
(G and H) The concentration of cell aggregates in WB and lysis are being compared.
Antibody staining (and platelet stimulation)
Timing: 50 min
This step describes the staining and stimulation of the citrated whole blood with the prepared antibody cocktail.
Note: If desired, replace WB with the lysed sample directly.
CRITICAL: Please ensure adherence to all precautions and local regulations outlined in Lab Safety protocols for handling biohazard samples such as human blood in a BSL2-level laboratory. Further, ensure strict measures to prevent spillage and bio-contamination are followed.
CRITICAL: All steps are performed at room temperature (RT).
Note: The following quantities and timing are calculated for 1 sample.
-
10.
Invert from the anticoagulant citrate tube containing WB once, then transfer 50 μL of WB into a 1.5 mL Eppendorf tube. 1 min
-
11.Incubate the cells for 5 min with 2 μL of FC block. 5 minOptional: Incubate cells with a stimulation medium.
-
a.As a positive control for platelet activation we add 1 μL 1x Trap6 solution (of 0.11 mM, see stimulation mix) to the sample for 1 min (2 μM TRAP6 in final volume).
-
b.Control Tubes: Incubate cells with a vehicle control solution (HEPES-modified Tyrode’s buffer). 3 min.
-
a.
-
12.
Add 50 μL antibody cocktail to cells and mix by pipetting up and down twice. 1 min
-
13.
Incubate cells and antibody cocktail for 30 min in the dark at RT. 30 min
-
14.
Wash the cells with 1 mL of HEPES-modified Tyrode’s buffer at 400g for 5 min at RT. 7 min
CRITICAL: Make sure to use the slowest possible acceleration and deceleration rate to minimize shear forces.13,14
-
15.
Carefully discard the supernatant as the pellet is not very firm. Use a pipette to gently remove the supernatant without disturbing the pellet. 2 min
-
16.
Resuspend the pellet in 600 μL of HEPES-modified Tyrode’s buffer. 1 min
CRITICAL: Do not vortex, but pipette up and down twice.
-
17.
Measure the resuspended cells immediately.
Note: The use of Fc receptor blocking is recommended to prevent non-specific binding of antibodies.
Note: The washing step is critical to limit platelet sedimentation and clogging in the fluidics system with patient samples.
CRITICAL: Do not vortex, rapid agitation leads to activation of platelets and subsequent aggregation with other cells.
CRITICAL: It is recommended to use HEPES modified Tyrode’s buffer for the investigation of platelets and blood cell aggregates.10
Sample acquisition
Timing: 10 min per sample
This step describes how to efficiently aquire cell aggregates with the IFC.
-
18.
Load the stained cell suspension into the IFC sample chamber.
-
19.
Validate your target population to record in the acquisition template.
Note: Don’t save all events, as most will be erythrocytes. We are interested in CD41a and CD45 positive events (PLTs and WBCs).
-
20.
In healthy controls, collect images of approximately 1000–2000 leukocytes and 20000 in-focus platelet events per sample in 10 min.
Note: If the raw data files (.rif files) are too large, system stability degrades, and file loading times become excessive.
CRITICAL: It is recommended to carry out a pre-acquisition cleaning procedure with 10% bleach and Millipore distilled water for every 5 samples to avoid any potential obstruction of the fluidics system and to prevent clogging-associated focusing issues when measuring unfixated samples.
Image analysis with IDEAS 6.3
This step first describes how to gate the acquired data and then how to export this data for statistical analysis.
Gating strategy
Timing: 20 min for initial setup; 5 min using established template
-
21.Acquisition gate (Figure 1A).
-
a.Intensity CD41a – Intensity CD45 biplot: Set the gate in the IFC at acquisition. Use the INSPIRE software for acquisition.
-
a.
Note: As our target population is either CD41a and/or CD45 positive, we can exclude all double negative events. This gate eliminates most RBCs from our total events.
-
22.Gating strategy for analysis (Figure 1B).
-
a.Initial gating on cells: Aspect ratio vs. area (gate 1).
-
i.Use the aspect ratio vs. area of cells in the bright-field channel to exclude the focusing beads (Amnis SpeedBead) and cell debris.
-
ii.To find the correct cutoff in the cell-area variable check the bright-field channel until platelets are the smallest events and beads are excluded.
-
i.
-
b.Select in-focus events: Root mean square (rms) gradient histogram (gate 2).
-
i.Set the threshold for in-focus events by looking at the bright-field images. Exclude cells with an RMS-gradient below the threshold.
-
i.
-
c.Select all cells that are CD45 positive: CD45 histogram (gate I3).
-
d.Investigation of WBC-WBC aggregates: Circularity-CD45 vs. Symmetry–CD45 biplot (gate I4a).
-
i.Separate single and aggregated WBCs by comparing the shapes of CD45+ events.
-
i.
-
a.
-
23.Investigation of WBC-PLT aggregates (gating I).
-
a.Select for PLT-WBC aggregates by gating on events which are positive for CD41a: CD41a vs. CD62P biplot (gate I4b).
-
i.This gate still includes coincidental events. Use CD62P to exclude large PLT-PLT aggregates which are extremely bright in the CD62P channel (see troubleshooting).
-
i.
-
b.Remove coincidental events: Internalization mask vs. CD41a bright detail intensity biplot (gate I5).
-
i.To exclude most platelets which are on the same image section as WBCs but not actually attached to any cell we recommend to set up masks on the channels for CD45 and CD41a.Optional: These masks are usually set automatically by the software. It is possible to edit these masks to better fit the cell shapes in the “masks” tool. These masks trace the outlines of the fluorescence signal in the matching channels. The internalization mask measures a score depending on the overlap between the selected masks. To find the correct threshold check the images of the relevant merged channels.
-
ii.Exclude events where CD41a intensity is too low, as these events are mostly debris.Optional: Remove coincidental events caused by artefacts (gate I6).Sometimes large platelet aggregates have intense fluorescence signals (artefacts) which bleed into all channels. To exclude these events inverse the internalization mask and set the threshold to exclude the relevant events.
-
i.
-
c.Analyze the aggregates: Spot-count CD41a vs. area CD41a biplot (gate I7).
-
i.To gain statistical data about the size and composition of the aggregates, plot the masked area of CD41a vs. the spot count of CD41a events.Note: The spot count detects the approximate amount of isolated CD41a positive events in the analyzed image. This allows to gate for an approximate amount of platelets attached to a leukocyte as well as the size of these platelet aggregates.
-
ii.The size value measures the area of the CD41a positive mask.
-
i.
-
a.
-
24.Investigation of PLT-PLT aggregates (gating II).
-
a.Detect platelets not attached to WBCs: Intensity CD41a vs. Intensity CD45 biplot (gate II3)
-
i.Select the population which is positive for CD41a but negative for CD45. This gate excludes the PLT-WBC aggregates.
-
i.
-
b.Remove remaining RBCs: Intensity CD41a vs. Intensity CD31 biplot (gate II4).
-
i.Remaining RBCs are excluded by gating on CD41a - CD31 double positive events.Note: This is important as the RBCs display weak autofluorescence which can intervene with the analysis.
-
i.
-
c.Detect PLT-PLT aggregates: Aspect ratio vs. area of CD41a biplot (gate II5a).
-
i.Single platelets and larger platelet aggregates are separated by the size and shape of the CD41a mask in the analyzed picture. Set the threshold according to the CD41a and bright-field channel to separate single platelets and PLT-PLT aggregates.
-
ii.Exclude events with the aspect ratio of 1 as these are mostly remaining erythrocytes or debris.Note: This method can only approximate the number of platelets in the aggregate based on the masked area.
-
i.
-
d.Measure platelet activation: Bright detail intensity CD41a vs. CD62P intensity biplot (gate II5b).
-
i.To set the threshold for CD62P positivity compare the sample to the FMO (Fluorescence Minus One) of the relevant activation marker and the positive control stimulated with TRAP6.
-
i.
-
e.Exclude blurry events: H-Entropy in CD41a vs. Standard deviation in CD41a biplot (gate II5c).
-
i.Select small focused events. The analysis relies on sharp contrast.
-
i.
-
f.Investigate PLT morphology: Circularity Mask vs. Shape Ratio Mask of CD41a biplot (gate II6).
-
i.To investigate the distribution of round vs. spiky/elongated platelets we compare the circularity vs. the shape ratio of the analyzed image.
-
i.
-
a.
-
25.Export data.
-
a.Define your statistical parameters of interest for each gated population under the “Edit Statistic Table” tab.
-
b.Save the file as analysis template.
-
c.Apply analysis template to samples.
-
a.
Note: Batch processing allows you to automatically analyze a group of files with one template when a compensation matrix has already been generated for the experiment.
Note: To check if the exported statistics table is correct, check “Preview Statistics” in the batch window. This will output a .txt file which can be imported into any data analysis software.
Expected outcomes
In this protocol, we have refined existing flow cytometry techniques15 for our application in imaging cytometry, leveraging the latter’s capacity to incorporate morphological and spatial information into the analysis. This enhancement allows for a more precise understanding of cellular interactions.16 Focusing on platelets, our methodology uses fewer steps in sample preparation to preserve the physiological state. Alterations in cell morphology and activation status are avoided by omitting fixation,17 if possible. Figure 2 and Figure S1 feature representative images using our protocol. To establish a benchmark for platelet activation, we stimulated whole blood samples for 1 min with 2 μM of TRAP-6, serving as a positive control. In healthy controls (individuals with normal cell counts), we could image between 1000-2000 leukocytes and more than 20,000 in-focus PLTs in 10 min of measuring time.
Figure 2.
Expected results of the WB protocol, IFC pictures of various cell aggregates
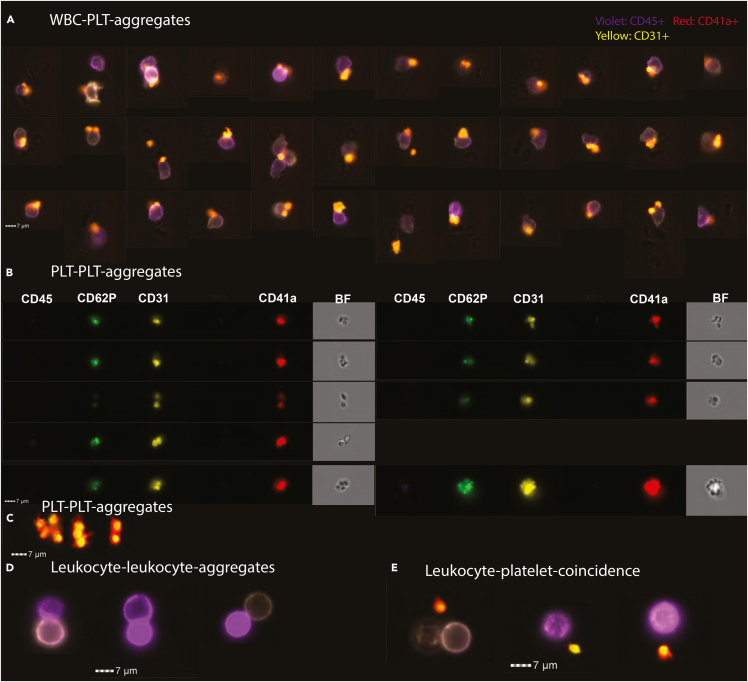
(A, C, D, and E) Depict the composite images of the channels for CD41a (red), CD31 (yellow) and CD45 (violet). (A) Features PLT-WBC-aggregates (Figure 1, gate BI6). The scale bar measures 7 micrometers.
(B) Displays PLT-PLT aggregates (Figure 1, gate BII5a). Here each channel and the bright-field image are displayed individually.
(C) Highlights additional composite images of platelet-platelet aggregates. (D) Shows leukocyte-leukocyte aggregates (Figure 1, gate BI4a). (E) Displays coincidental events (Figure 1, gate BI5). Additional populations are shown in Figure S1 e.g., erythrocyte-platelet-aggregates.
For data analysis, we devised a gating strategy to quantify the target aggregates in an unbiased way. Given the challenge of distinguishing between genuine aggregates and coincidental events in conventional flow cytometry,18 we used the high-throughput image analysis capabilities of an IFC. Utilizing the Ideas 6.3 software, we capitalized on its internalization feature, which facilitates the detection of signal overlaps across two channels within a specific masked area on the cells. This approach strongly enhanced our confidence in the data quality, allowing for more precise and automated measurements of cell aggregates. In contrast to microscopy-based approaches, this method enables the measurement of thousands of leukocytes, thereby allowing for the reliable detection of aggregates, which may only be present in a small fraction of the total leukocyte population.19
Reproducibility
As shown in [Figure 3], our measurements respond to platelet stimulation properly with increased CD62P expression and the formation of PLT-PLT aggregates [Figures 3A-3D], and baseline platelet activation, defined by CD62P expression is low. Interestingly we failed to induce PLT-WBC aggregates with TRAP6 in the calcium-depleted whole blood (Figure 3E). To validate our protocol, we conducted a comparative analysis of single leukocyte and platelet concentrations as determined by our method against those measured using a clinically validated diagnostic hemocytometer, the Sysmex XN-350. We detected a strong correlation between both measurements, especially for leukocyte detection [Figures 3F and 3G]. If not stated otherwise, samples are collected from a fever patient cohort. For our data, we usually measured 2 clinical samples with 4 experiments per sample (unstained, WB, stimulated with TRAP6 and lysis) per day.
Figure 3.
Ratios of cell aggregates and surface marker intensities – WB protocol vs. TRAP6-stimulated positive control
(A–E) The unstimulated WB protocol was compared with a WB sample stimulated for 1 min with TRAP6 as positive control, n = 25. The median intensity or the percentage of the selected population is displayed. The platelet marker CD62P is used to assess platelet activation. Additionally, morphological changes of platelets, which are a sign of early platelet activation (pseudopods, see Figure S1F), are being compared, based on gate B-II-6. Cell aggregates were determined using the mentioned gating protocol. A Kruskal-Wallis with Dunn’s Post hoc was performed, where ∗ : p < 0.05, ∗∗ :p < 0.005, ∗∗∗: p < 0.0005.
(F and G) Spearman correlation analysis conducted between cell concentrations of platelets and leukocytes obtained using the WB protocol and those measured by a validated automated hematology analyzer from the routine diagnostics laboratory (Sysmex XN-350). Cell concentrations were automatically determined by the IFC. PLT-WBC aggregate concentrations are based on gate B-I-6, Platelet concentrations are based on gate B-II-4, WBC concentrations on gate B-I-3. The Spearman correlation coefficient (r) and its corresponding p-value are annotated on the plot.
(H) Kernel Density Estimation of absolute WBC counts of CD45+ cells of all measured unstimulated samples. The X-axis depicts the counts, the Y-axis the respective density. The dashed line indicates the mean WBC counts of all samples.
The inclusion criteria for the patient cohort in our study are the following:
Participants must be adults aged 18 years or older who presented with fever (temperature exceeding 38.5°C ) and have been admitted to the emergency department. To avoid potential confounding factors, individuals receiving immunosuppressive therapy or those with immunosuppressive diseases are excluded from the cohort. Similarly, patients with a prior or current diagnosis of cancer are not eligible for inclusion.
Patient characteristics
Our patient cohort is described in Table 1.
Table 1.
Patient data
| Characteristic | Mean | 95% CI | n (of samples measured) | Range |
|---|---|---|---|---|
| Age | 47.9 | 42.3, 53.4 | 30 | (21, 77) |
| Mean arterial pressure (mmHg) | 76.2 | 72.4, 80.0 | 25 | (60.7, 99.0) |
| Heart rate (bpm) | 106.9 | 100.2, 113.7 | 25 | (72.0, 140.0) |
| WBC (1000 per μL) | 12 | 8.5, 15.5 | 25 | (2.5, 32.6) |
| PLT (1000 per μL) | 221.1 | 182.0, 260.2 | 23 | (67.0, 400.0) |
| Lymphocytes (1000 per μL) | 1 | 0.7, 1.3 | 23 | (0.3, 2.9) |
| Monocytes (1000 per μL) | 0.8 | 0.6, 0.9 | 23 | (0.1, 1.7) |
| Basophils (1000 per μL) | 0 | 0.0, 0.1 | 23 | (0.0, 0.2) |
| Neutrophils (1000 per μL) | 10.3 | 7.0, 13.5 | 23 | (1.8, 29.2) |
| Eosinophils (1000 per μL) | 0.1 | 0.0, 0.1 | 23 | (0.0, 0.4) |
| Gender, female | – | – | 12 (40%) | – |
| Diabetes type II | – | – | 9 (30%) | – |
| PLT-Inhibition medication | – | – | 5 (16%) | – |
| Infection (detected) | – | – | 23 (77%) | – |
| Cause of fever | – | – | Bacterial: 13, Viral: 9, control: 5, autoimmune: 3 | |
Quantification and statistical analysis
The data obtained by our IFC is stored as raw image files (.RIF). The files may be accessed by using the Ideas 6.3. software. The statistical data is based on the gating strategy supplied [Figure 1]. A minimum of 1000 leukocytes and 15,000 platelets per patient were analyzed [Figure 3H]. Intensities are reported as median intensities, and percentages are reported as the mean. Statistical analyses were conducted using the R statistical software (version 4.1.2; R Foundation for Statistical Computing, Vienna, Austria). To evaluate the differences among groups, the Kruskal-Wallis test with Dunn’s Post Hoc Test was employed as data was not normally distributed.
Graphical representations of the data were generated using ggplot2.20
Correlation analyses were performed using the Spearman Rank Sum test in R.
Determination of the best anticoagulant tube for studying blood cell aggregates
As depicted in [Figure 4], citrate 3.2% was selected as the anticoagulant of choice based on its ability to maintain low CD62P expression on platelets within the first-hour post-collection, with less than 5% of platelets showing activation in healthy individuals. Furthermore, the incidence of PLT-WBC (Figure 4A) and PLT-PLT aggregates (Figure 4B) was minimal in the first hour after blood draw in citrate-treated samples, suggesting a closer representation of the natural physiological state. Citrated blood only displayed a small increase in cell activation markers of leukocytes compared to EDTA anticoagulated samples (Figure 4C). Heparin induced platelet activation (Figure 4D) and increased PLT-PLT aggregates at baseline. Although EDTA features the highest PLT concentrations (Figure 4E) and least PLT-WBC aggregates after 6 h we don’t recommend EDTA as our anticoagulant of choice as it is known to induce in-vitro platelet aggregation, pseudothrombocytopenia21 and morphological changes.22 To assess the impact of time delays in measurement on the samples, we conducted repeated measurements of five samples at hourly intervals for up to 6 h. The data underscores the importance of performing sample preparation within the first hour after blood collection to ensure optimal data quality. Beyond this time window, a strong increase in platelet and leukocyte activation (Figure 4C) was observed across all anticoagulants used. Additionally, CD31 (PECAM-1) on the platelet surface is being downregulated over time (Figure 4F), which might impact platelet binding behavior.
Figure 4.
Determination of optimal anticoagulant selection and measurement timing
The anticoagulants citrate 3.2%, natrium-heparin (17 IU/mL blood), and EDTA (1.8 mg/ml blood) were compared at basal conditions (without stimulation) over 6 h for healthy controls (n = 5). The time from the blood draw to the start of the experimental protocol was recorded. The whiskers express the 1st and 4th IQR, lines connect the mean. Percentages of the selected population in relation to the total population or the median marker intensities are displayed.
(A and B) The ratio of cell-aggregates of their parent population are compared.
(C) The combined intensities of CD69 and HLADR on the surface of WBCs (gate B-I-3) are compared over time.
(D) Platelet activation was evaluated by the ratio of PLTs with surface expression CD62P.
(E) The PLT concentration (based on gate B-II-4) was compared over time.
(F) The intensity of CD31 on PLTs was compared over time.
A WB protocol induces less platelet activation than a RBC lysis protocol
Next, we directly compared an RBC lysis protocol without fixation to our featured WB protocol for detecting cell aggregates and platelet activation. [Figure 5] Notably, the unstimulated WB sample exhibited significantly less CD62P expression on platelets compared to the lysis sample [Figures 5A and 5B].
The lysis sample demonstrated a marked increase in leukocyte-leukocyte and PLT-PLT aggregates, relative to WB [Figures 5C and 5D]. This observation was further corroborated by relatively and absolutely larger PLT-WBC aggregates detected [Figures 5E and 5F] in the lysis sample, suggesting that lysis induces cell activation and augments aggregate formation compared to WB.
Moreover, lysis increased the concentration of PLT-PLT and PLT-WBC aggregates [Figures 5G and 5H]. Such morphological alterations deviate from the physiological norm, rendering them undesirable for our study. If a WB analysis approach is not desired our analysis highlights the crucial role of using a combined Fix-Lyse solution in minimizing platelet activation induced by ADP,23 released by lysed RBCs. The fixative component of the solution halts any further activation of platelets by ADP and stabilizes platelet aggregates for up to 24 h.18
Limitations
The protocol described above is well-tested and usually provides robust results. Still some challenges remain. This protocol has been developed to immediately evaluate fresh, unfixed whole blood samples.
IFCs can currently handle about 2000 events per second. As most of these are RBCs in WB, measuring times of about 10 min are necessary to capture enough cells for the quantification of leukocyte-cell aggregates. This makes the detection of rare leukocyte subtypes challenging.
Additionally, incorporating a wide array of sample conditions could potentially lead to temporal batch effects through sample degradation causing misleading conclusions if not properly controlled or accounted for. It is strongly recommended to initiate sample processing within 60–80 min.
Sequential processing of multiple samples in a row may lead to clogging due to platelet deposition. To mitigate this, it is advisable to cleanse the cytometer with a 10% bleach solution followed by ddH2O after every fifth assay.
The incorporation of a washing step after staining results in the loss of a fraction of the initial platelet population. This step is necessary to prevent a possible clogging of the cytometers microfluidics and increases fluorescent signal quality by getting rid of unbound antibodies. Additionally, prompt and meticulous handling of the sample is important to avert unintended platelet activation and aggregate disruption or induction, which consequently restricts the scope of feasible experimental methodologies.
Starting with IDEAS 6.3 software, stored imaging data is encrypted using a proprietary algorithm. This hinders any open-source approaches to data analysis using the featured imaging cytometer.
Troubleshooting
Problem 1
Cells appear to be out of focus, smeared or blurred in the preview section of the IFC. Relevant for the acquisition section of the protocol.
Potential solution
To address issues related to autofocusing in the IFC, which can be caused by slight deviations in the microfluidics system, it is recommended to periodically flush the machine with a solution of 10% bleach followed by ddH2O. Additionally check if cell concentrations are within optimal range for your IFC and dilute the sample if concentrations are too high.
Restart the machine if the issues can’t be resolved that way.
Alternatively make use of the optional fixation step (sample preparation) to limit platelet aggregation.
Problem 2
No cells are visible at all, relevant for the acquisition section of the protocol.
Potential solution
Check if you are detecting events in the “All” gate of the INSPIRE acquisition software. If there are events detected and no cells are visible in bright field, try to manually recenter the image stream using the manual controls.
If manual recentering of the image stream did not improve the situation there might be an obstruction in the microfluidics. Flushing the IFC with 10% bleach followed by ddH2O might resolve the issue.
Additionally, issues with sample acquisition can lead to the formation of larger cell aggregates, which can adversely affect the fluidics system of the cytometer. To safeguard against this, it is possible to filter the sample using a 100 μm mesh before analysis.
Air bubbles in the sample disrupt the microfluidics. Make sure to eliminate any bubbles from the sample and purge fluidic lines if bubbles are detected by the IFC.
Finally, it is important to dilute the sample appropriately before measurement, in accordance with the concentration ranges required for your specific IFC.
Problem 3
It takes a long time to open a .RIF file, the raw image file format, for data analysis. See the data analysis section of the protocol.
Potential solution
Don’t record events that are not part of your target population. This is especially important for RBCs as these make up the majority of a WB sample.
Problem 4
Cells (mostly RBCs) are visible in the bright-field channel, but not cells of interest in the other relevant detector channels. (Whole blood staining (and stimulation)).
Potential solution
Adjust the brightness histogram of the channel of interest manually. If this does not resolve the issue, make sure that the individual antibodies are providing a strong signal. It is recommended to titrate antibodies according to current practice. There might be an issue with the antibody staining. In this case we recommend optimizing staining for the selected cell population on isolated cells.
Problem 5
Fluorescence specific to one channel is detected in a neighboring channel. (Data and statistical analysis).
Potential solution:
Check your cell populations visually in the stimulated sample to catch large PLT-PLT-aggregates which cause fluorescence spillover in all channels, as shown in Figure 6.
Figure 6.
Catching large PLT-PLT aggregates in the PLT-WBC gate
A sample stimulated with TRAP6 as described includes large PLT-PLT aggregates which can be confused with PLT-WBC aggregates. Abnormally high CD62P intensities give these aggregates away.
Correct your compensation matrix. If this does not resolve the issue, reduce the antibody concentration of the relevant channel to limit signal bleeding into the wrong detector at high signal intensities. If this is not helping neither, reduce laser power in the offending channel.
Problem 6
The samples cannot be processed within 2 h because of high sample counts, experimental design or geographical limitations. See sample collection.
Potential solution
Fix the WB samples within 1 h, stain the samples within 6 h. We recommend to not compare fixed to unfixed samples. Investigate cell populations of interest for possible morphological alterations and issues with antibody binding caused by fixation with PFA.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Oliver Hayden (oliver.hayden@tum.de).
Technical contact
Questions about the technical specifics of performing the protocol should be directed to the technical contact, Komal Javarappa (kompaddy@yahoo.com).
Materials availability
All materials used in the protocol are available commercially.
Data and code availability
Original data for figures in the paper is available below:
Krell, Johannes (2024), “Dataset for Protocol for rapid assessment and visualization of cell microaggregate formation in whole blood by Imaging Flow Cytometry,” Mendeley Data, V1, https://doi.org/10.17632/nxm7bnns98.1.
Contact oliver.hayden@tum.de for raw data requests.
Acknowledgments
This study was supported by grants from INTRA-CREATE THEMATIC GRANT “INTERSECTION OF ENGINEERING AND HEALTH” (NRF2019-THE002-0008) and DZIF (TTU 03.818_01) (to C.P.d.C.).
The authors are very grateful to Kerem Delikoyun, Qinayu Chen, and Kay Khine Maw for their help with sample logistics and Si Ko Myo for his support in administrative work in clinical study organization.
Author contributions
J.K. designed the experiments and performed the data analysis; J.K., K.K.J., and A.W. performed the experiments; J.K. and M.S. wrote and edited the manuscript. A.W. assisted in laboratory experiments; O.H., P.K., L.R., G.S., and M.S. supervised the study and edited the manuscript. A.L.F. and C.P.d.C. edited the manuscript. J.K. performed the revision of the manuscript.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xpro.2025.103598.
Contributor Information
Johannes Krell, Email: johannes.krell@mri.tum.de.
Oliver Hayden, Email: oliver.hayden@tum.de.
Supplemental information
References
- 1.Pluta K., Porębska K., Urbanowicz T., Gąsecka A., Olasińska-Wiśniewska A., Targoński R., Krasińska A., Filipiak K.J., Jemielity M., Krasiński Z. Platelet–Leucocyte Aggregates as Novel Biomarkers in Cardiovascular Diseases. Biology. 2022;11:224. doi: 10.3390/biology11020224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Totani L., Evangelista V. Platelet–Leukocyte Interactions in Cardiovascular Disease and Beyond. Arterioscler. Thromb. Vasc. Biol. 2010;30:2357–2361. doi: 10.1161/ATVBAHA.110.207480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Klenk C., Erber J., Fresacher D., Röhrl S., Lengl M., Heim D., Irl H., Schlegel M., Haller B., Lahmer T., et al. Platelet aggregates detected using quantitative phase imaging associate with COVID-19 severity. Commun. Med. 2023;3:161. doi: 10.1038/s43856-023-00395-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hottz E.D., Azevedo-Quintanilha I.G., Palhinha L., Teixeira L., Barreto E.A., Pão C.R.R., Righy C., Franco S., Souza T.M.L., Kurtz P., et al. Platelet activation and platelet-monocyte aggregate formation trigger tissue factor expression in patients with severe COVID-19. Blood. 2020;136:1330–1341. doi: 10.1182/blood.2020007252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hirsch J., Uzun G., Zlamal J., Singh A., Bakchoul T. Platelet-neutrophil interaction in COVID-19 and vaccine-induced thrombotic thrombocytopenia. Front. Immunol. 2023;14 doi: 10.3389/fimmu.2023.1186000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Assinger A., Schrottmaier W.C., Salzmann M., Rayes J. Platelets in Sepsis: An Update on Experimental Models and Clinical Data. Front. Immunol. 2019;10:1687. doi: 10.3389/fimmu.2019.01687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marquardt L., Anders C., Buggle F., Palm F., Hellstern P., Grau A.J. Leukocyte-platelet aggregates in acute and subacute ischemic stroke. Cerebrovasc. Dis. 2009;28:276–282. doi: 10.1159/000228710. [DOI] [PubMed] [Google Scholar]
- 8.Gawaz M.P., Mujais S.K., Schmidt B., Gurland H.J. Platelet-leukocyte aggregation during hemodialysis. Kidney Int. 1994;46:489–495. doi: 10.1038/ki.1994.299. [DOI] [PubMed] [Google Scholar]
- 9.Elalamy I., Chakroun T., Gerotziafas G.T., Petropoulou A., Robert F., Karroum A., Elgrably F., Samama M.-M., Hatmi M. Circulating platelet–leukocyte aggregates: A marker of microvascular injury in diabetic patients. Thromb. Res. 2008;121:843–848. doi: 10.1016/j.thromres.2007.07.016. [DOI] [PubMed] [Google Scholar]
- 10.Blair T.A., Frelinger A.L., 3rd Platelet surface marker analysis by mass cytometry. Platelets. 2020;31:633–640. doi: 10.1080/09537104.2019.1668549. [DOI] [PubMed] [Google Scholar]
- 11.Welch E.L., Crooks M.G., Hart S.P. Agreement between blood draw techniques for assessing platelet activation by flow cytometry. Platelets. 2019;30:530–534. doi: 10.1080/09537104.2018.1535705. [DOI] [PubMed] [Google Scholar]
- 12.Murphy S., Gardner F.H. Platelet preservation. Effect of storage temperature on maintenance of platelet viability--deleterious effect of refrigerated storage. N. Engl. J. Med. 1969;280:1094–1098. doi: 10.1056/NEJM196905152802004. [DOI] [PubMed] [Google Scholar]
- 13.Söderström A.C., Nybo M., Nielsen C., Vinholt P.J. The effect of centrifugation speed and time on pre-analytical platelet activation. Clin. Chem. Lab. Med. 2016;54:1913–1920. doi: 10.1515/cclm-2016-0079. [DOI] [PubMed] [Google Scholar]
- 14.Perez A.G.M., Lana J.F.S.D., Rodrigues A.A., Luzo A.C.M., Belangero W.D., Santana M.H.A. Relevant Aspects of Centrifugation Step in the Preparation of Platelet-Rich Plasma. ISRN Hematol. 2014;2014 doi: 10.1155/2014/176060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Petriz J., Bradford J.A., Ward M.D. No lyse no wash flow cytometry for maximizing minimal sample preparation. Methods. 2018;134–135:149–163. doi: 10.1016/j.ymeth.2017.12.012. [DOI] [PubMed] [Google Scholar]
- 16.Markey K.A., Gartlan K.H., Kuns R.D., MacDonald K.P.A., Hill G.R. Imaging the immunological synapse between dendritic cells and T cells. J. Immunol. Methods. 2015;423:40–44. doi: 10.1016/j.jim.2015.04.029. [DOI] [PubMed] [Google Scholar]
- 17.Wong K., Li X., Ma Y. Paraformaldehyde induces elevation of intracellular calcium and phosphatidylserine externalization in platelets. Thromb. Res. 2006;117:537–542. doi: 10.1016/j.thromres.2005.04.030. [DOI] [PubMed] [Google Scholar]
- 18.Finsterbusch M., Schrottmaier W.C., Kral-Pointner J.B., Salzmann M., Assinger A. Measuring and interpreting platelet-leukocyte aggregates. Platelets. 2018;29:677–685. doi: 10.1080/09537104.2018.1430358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rolling C.C., Barrett T.J., Berger J.S. Platelet-monocyte aggregates: molecular mediators of thromboinflammation. Front. Cardiovasc. Med. 2023;10 doi: 10.3389/fcvm.2023.960398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wickham H. Springer International Publishing; 2016. ggplot2: Elegant Graphics for Data Analysis. [Google Scholar]
- 21.Markewitz R., Sharifzadeh S.R., Junker R., Wandinger K.-P. EDTA-associated pseudothrombocytopenia: definition and real-world occurrence. J. Lab. Med. 2023;47:105–114. doi: 10.1515/labmed-2023-0032. [DOI] [Google Scholar]
- 22.White J.G., Escolar G. EDTA-induced changes in platelet structure and function: adhesion and spreading. Platelets. 2000;11:56–61. [PubMed] [Google Scholar]
- 23.Helms C.C., Marvel M., Zhao W., Stahle M., Vest R., Kato G.J., Lee J.S., Christ G., Gladwin M.T., Hantgan R.R., Kim-Shapiro D.B. Mechanisms of hemolysis-associated platelet activation. J. Thromb. Haemost. 2013;11:2148–2154. doi: 10.1111/jth.12422. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Original data for figures in the paper is available below:
Krell, Johannes (2024), “Dataset for Protocol for rapid assessment and visualization of cell microaggregate formation in whole blood by Imaging Flow Cytometry,” Mendeley Data, V1, https://doi.org/10.17632/nxm7bnns98.1.
Contact oliver.hayden@tum.de for raw data requests.